
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 10 February 2023
Sec. Experimental Pharmacology and Drug Discovery
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1086913
This article is part of the Research TopicSpotlight on Artificial Intelligence in Experimental Pharmacology and Drug DiscoveryView all 7 articles
Background: A steep increase in new drug applications has increased the overhead of writing technical documents such as medication guides. Natural language processing can contribute to reducing this burden.
Objective: To generate medication guides from texts that relate to prescription drug labeling information.
Materials and Methods: We collected official drug label information from the DailyMed website. We focused on drug labels containing medication guide sections to train and test our model. To construct our training dataset, we aligned “source” text from the document with similar “target” text from the medication guide using three families of alignment techniques: global, manual, and heuristic alignment. The resulting source-target pairs were provided as input to a Pointer Generator Network, an abstractive text summarization model.
Results: Global alignment produced the lowest ROUGE scores and relatively poor qualitative results, as running the model frequently resulted in mode collapse. Manual alignment also resulted in mode collapse, albeit higher ROUGE scores than global alignment. Within the family of heuristic alignment approaches, we compared different methods and found BM25-based alignments to produce significantly better summaries (at least 6.8 ROUGE points above the other techniques). This alignment surpassed both the global and manual alignments in terms of ROUGE and qualitative scoring.
Conclusion: The results of this study indicate that a heuristic approach to generating inputs for an abstractive summarization model increased ROUGE scores, compared to a global or manual approach when automatically generating biomedical text. Such methods hold the potential to significantly reduce the manual labor burden in medical writing and related disciplines.
The increased discovery and development rate of novel drugs has led to a steep increase in new drug applications submitted to health authorities. This has resulted in an increased volume of medical writing-related technical documents such as protocols, clinical study reports, summaries of drug efficacy and safety, clinical, and non-clinical summaries of pharmacology (ICH, 2022). These technical documents are the source of information for other types of documents such as plain-language summaries and medication guides written for patients, and prescription drug labeling, typically written for healthcare professionals.
Structured Product Labeling (SPL) is a Health Level Seven International standard which defines the content of human prescription drug labeling in an XML format. The “drug labeling” includes all published material accompanying a drug, such as the Prescribing Information, a technical document containing detailed information needed to use the drug safely and effectively. As of Release four of the SPL standard, 22,000 United States Food Drug Administration (FDA) informational product inserts have been encoded according to the standard (FDA, 2022).
SPL documents contain both the content of labeling (i.e., all text, tables and figures) for a product along with additional machine readable information (i.e., drug listing data elements). Drug listing data elements include information about the product (i.e., proprietary and non-proprietary names, ingredients, ingredient strengths, dosage forms, routes of administration, appearance, DEA schedule) and the packaging (i.e., package quantity and type). Large scale publicly accessible databases of SPL data are available, such as Dailymed (FDA, 2019).
Within SPL data, the medication guide is an electronic page or paper handout that accompanies the prescribing information (FDA, 2020). The medication guide does not contain any new information but instead represents a condensed summary of the most important information from all sections of the SPL. The medication guide is recommended by the FDA for certain products in order to provide information regarding known side effects of the product, as well as instructions for safe and effective storage and use. While there are exceptions, a typical medication guide presents information in sections, each headed by a guiding question. Six questions are used most frequently: “What is … ?“, “What is the most important information I should know about … ?“, “Who should not take … ?“, “How should I take … ?“, “What should I avoid while taking … ?“, “What are the possible side effects of … ?”
Medication guides are manually compiled by expert medical writers. Given the considerable length of drug labels, as well as the necessary quality checks, developing these documents is a time-consuming and costly process. An alternative to this approach is to use natural language processing techniques, such as text summarization, to automatically generate medication guides from SPL information.
We conducted a series of studies to test the hypothesis that text can be autogenerated from predefined source documents. Specifically, we present the application of a Pointer-Generator Network (PGN) summarization model to create medication guides from prescription drug labeling information. We compared the effectiveness of various input data selection and model training schemes. We choose to focus on autogenerating medication guides for two main reasons. First, there are thousands of medication guides publicly available. Second, the published medication guides represented an appropriate control for text generated by the algorithm.
The dataset was collected from the DailyMed website. This article used the September 2019 edition, providing the official drug labels in XML format. Out of the total 5,357 prescription drug labels 27% of files (1437) provided medication guides. For the remainder of this study we will focus only on these drugs with medication guides present. Medication guides varied in completeness and did not always address all six guiding questions. Table 1 lists the frequency of each question in the dataset alongside the average number of words dedicated to answering the respective question.
Text summarization can be divided into two broad categories: extractive (Nallapati et al., 2017; Zhou et al., 2018; Liu and Lapata, 2019) and abstractive (Rush et al., 2015; Nallapati et al., 2016; See et al., 2017). Given a source document, the goal is to produce a target summary that contains the relevant information in a semantically-condensed form, i.e., using fewer words. Extractive summarization methods produce summaries by selecting and concatenating subsequences of the initial document. For example, a word-level extractive summarization model might summarize the sentence “I am going to take Paracetamol tablets after dinner to reduce side effects of the COVID-19 vaccine, which are muscle aches and chills.” into “I am going to take Paracetamol tablets to reduce muscle aches and chills.” No novel n-grams are produced by the model, as it merely combines pieces of the original text. Similarly, a sentence-level extractive summarization model might summarize a document by selecting and concatenating the introductory sentences of each paragraph.
Abstractive summarization models, on the other hand, create summaries by producing novel n-grams that do not necessarily exist in the source text. The added flexibility can provide richer and more cohesive summaries, tying concepts together in ways that the source text does not. Using the previous word-level example, a strong abstractive summarization model could produce the summary “I will take Paracetamol tablets to reduce side effects of the COVID-19 vaccine”, even though “will” is not in the initial text.
Recurrent neural networks and attentional networks both provide strong frameworks to process sequential data via encoder-decoder architectures. Basic Seq2Seq models, which largely follow the encoder-decoder paradigm, have no built-in mechanism for ensuring that the generated information is factually or conceptually correct. For example, an abstractive summarization model might erroneously produce a summary that states “I am going to take two Paracetamol tablets”, when in fact that piece of information was never specified in the source text.
The PGN (illustrated in Figure 1) aims to solve this information accuracy problem by introducing a pointer mechanism. This allows the model to select and copy tokens from an input text, improving factuality (See et al., 2017).
Nevertheless, long document summarization is still an open challenge due to memory constraints. Particularly notable is the attention mechanism in the Transformer architecture, which requires O (n2) attention computations and parameters for an n-token document. In addition, the pointer mechanism requires another O (n2) computations and parameters (See et al., 2017).
We evaluated three training variants to learn to generate medication guides. This includes a one-shot approach using the entire document and entire medication guide, a manual alignment using domain knowledge, and heuristic alignment using a number of similarity approximations (see Figure 2). Table 2 visually illustrates these variants. All experiments were performed on 16GB Quadro RTX 5000 GPUs. Model hyperparameters and other reproducibility information are listed in Supplementary Material.
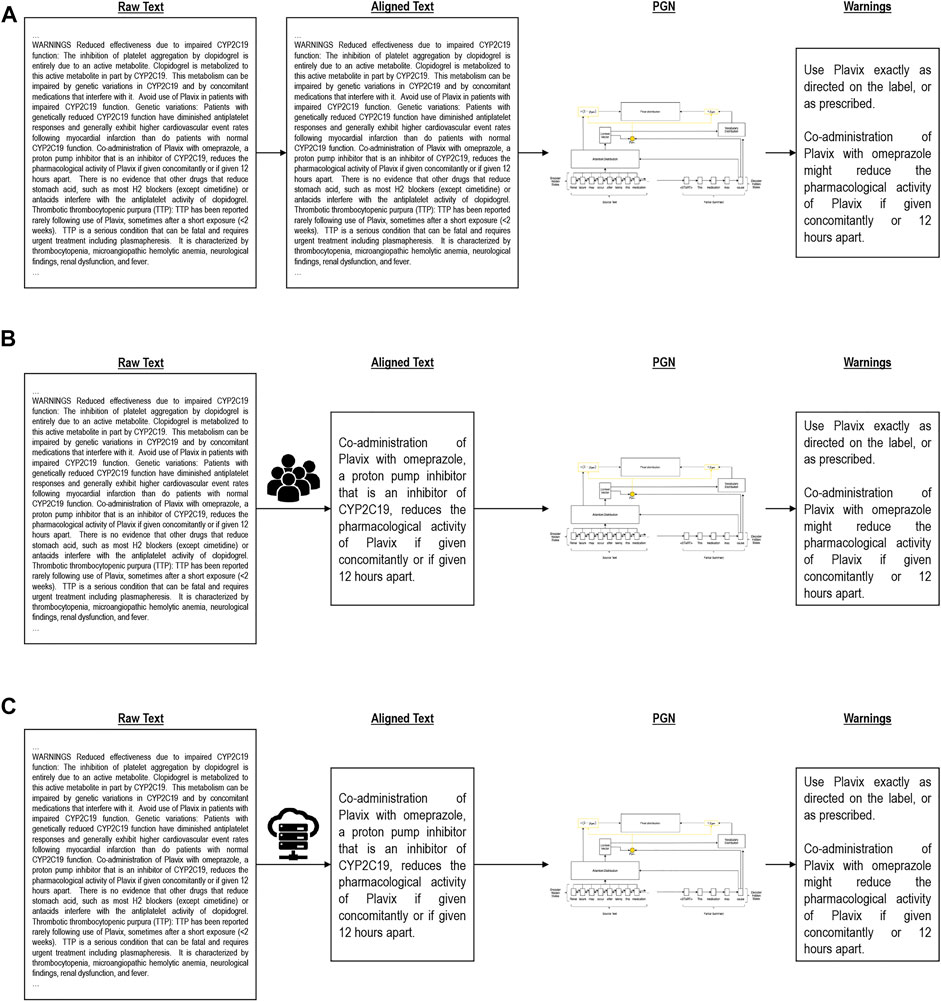
FIGURE 2. Alignment algorithms. (A) is Global alignment, (B) is Manual alignment, (C) is Heuristic alignment.
The global approach (Figure 2A) takes the entire drug label (without the medication guide) as input and attempts to generate the full medication guide in a single generation step. This baseline model, due to the O (n2) complexity of the attention mechanism, quickly reaches a memory bottleneck when scaled to fit longer documents such as drug labels. For relatively long documents, standard GPUs may become insufficient. In these cases, we cap the size of the input document to ensure no memory overflow occurs. Due to this loss of information, the quality of generated medication guides is expected to suffer.
Using domain knowledge, we manually selected sections of the input drug label corresponding to medication guide questions (Figure 2B). Instead of generating the entire medication guide at once, here we separately generate answers to each guiding question based on the manually identified drug label sections. This condition simulates an idealized setting and represents a headroom analysis of what an effective alignment can achieve.
As a more realistic divide and conquer condition, we split the input document into smaller subsections following the existing section dividers and again generate answers to each medication guide question separately (Figure 2C). Instead of relying on manual alignment, we explored a number of text similarity heuristics to model the similarity of drug label sections (input) to medication guide answers (output). The single subsection with the highest similarity is used for training, and the remainder of the label discarded. Like manual alignment, this allows us to generate smaller input documents that contain key information for summarization. The generalized heuristic alignment process is formalized in Appendix B. We explored five text similarity heuristics for automatic input/output alignment: (1) TF-IDF ‘best match’, (2) TF-IDF ‘average match’, (3) TF-IDF ‘median match’, (4) LSH + Jaccard Similarity, (5) BERT. The technical details of these similarity metrics are discussed in Appendix C.
The dataset was split into 60% training data, 20% testing, and 20% validation sets. The following results are from model runs on the test sets. Table 3 reports model performance in terms of F1 ROUGE-1, F1 ROUGE-2, and F1 ROUGE-L scores between generated and reference medication guides.
As expected, one-shot summarization of entire medication guides produced both quantitatively and qualitatively poor results. This is likely due to the model’s inability to attend over the entire input drug label, and thus only capturing a limited amount of the text to use for summarization. Furthermore, unlike other methods, there is no guarantee that the available (pre-cutoff) tokens contain salient information for producing summaries.
Qualitative inspection showed frequent mode collapse and often contained extremely repetitive or vague summaries. For example, the following summary or a highly similar version was produced for 21 input documents in a sample of 100:
“The risk of getting an ulcer or bleeding increases with: past history of stomach ulcers, or stomach or intestinal bleeding with use of NSAIDs taking medicines called corticosteroids, anticoagulants, SSRIs, or SNRIs increasing doses of NSAIDs longer use of NSAIDs smoking drinking alcohol older age poor health advanced liver disease bleeding problems past history of stomach ulcers, or stomach or intestinal bleeding with use of NSAIDs taking medicines called corticosteroids, anticoagulants, SSRIs, or SNRIs increasing doses of NSAIDs longer use of NSAIDs smoking drinking alcohol older age poor health advanced liver disease bleeding problems.”
This summary, while producing a decent ROUGE score for many documents, is rather unspecific to any drug and lacks the proper structure of a medication guide.
We compared manually selected input pairings from three different sections (“Contraindications”, “Interactions”, and “Warnings”). The manually aligned “Contraindications” section demonstrated the best performance overall, with higher ROUGE-1, ROUGE-2, and ROUGE-L scores than the global or any of the other manually aligned sections by at least six ROUGE points (Table 3). However, experiments for all three sections resulted in mode collapse, and qualitatively the generated summaries were consistently poor (Table 4). The model seemed to underperform for “Warnings” in particular, and we suspect this is because the source text tended to be significantly longer in this section.
Among automatic alignment techniques, the ‘median match’ TF-IDF approach demonstrated the best performance overall, with higher ROUGE-1, ROUGE-2, and ROUGE-L scores than any of the other heuristic alignment approaches by at least 6.8 ROUGE points (Table 3). The ‘median match’ TF-IDF heuristic also produced the most natural appearing medication guides. In some cases, it was able to generate the exact guide. The best automatic alignment technique even outperformed the manual alignment in terms of ROUGE score. We suspect that this is because the heuristic alignment’s use of text similarity metrics allowed for creating closer source/target matches as input for the PGN. Table 5 shows two examples of reference texts and their generated summaries created with this heuristic.
The main goal of this study was to test whether neural text generation is a viable approach to support the development of regulatory documents. In order to test this hypothesis we used medication guides as source documents because large datasets are publicly available, and they represent an appropriate control for automatically generated text. Our results indicate that the ROUGE scores increased when the model shifted from a global to a heuristic approach. These results are not entirely surprising as the global approach is more complex and, due to text length, prone to GPU memory issues.
These results are relevant for several reasons. First, to the best of our knowledge, this is the first time that an algorithm has been used to generate medication guides with robust accuracy.
Second, we believe that accurate generation of text can improve efficiency in medical writing. For example, medication guides are typically generated manually by medical communication specialists and medical writers who choose specific information from the labeling information mainly based on personal experience.
The results of this study also indicates that approximately only a third of sponsor-generated prescription information packages included medication guides. Furthermore, we found that there was a robust level of inconsistency in the medication guides with respect to addressing the six questions recommended by the FDA.
Overall, this study relied on medication guides to demonstrate feasibility of neural text generation. However, we believe that this use case study can be extended to different types of regulatory documents produced by medical writers. Neural text generation has the potential to greatly improve efficiency in medical writing and accelerate regulatory submissions. Potentially, this approach could also be used to create plain-language summaries and medication guides in different languages. Furthermore, more modern encoder-decoder transformers such as GPT-3 (Brown et al., 2020) may improve the performance of the approach presented here. Hovever, the strength of the PGN is that it allows both abstractive and extractive summarization as needed, so further studies comparing both approaches would be required.
Our study shows several limitations. First, the frequently inconsistent structure of medication guides hampers automated approaches. One such inconsistency is the frequency of the six guiding questions, displayed in Table 1. As shown in that table, each question appears a different number of times in the dataset. By having missing data (i.e., questions not asked and answered), the model has an uneven data distribution of sentences summarizing a certain section. Hence, sections that have more frequent similar questions discussed (i.e., contraindication section), are better summarized sections than those that do not have enough instances of uniformly framed questions (i.e. pediatric use section). As the size of datasets increase, this limitation may be at least partially remediated since question discussions will become more numerous and more data will lead to better document generation.
Second, ROUGE scores for summaries generated using the TF-IDF + Euclidean (“median”) aligned inputs are higher than any scores resulting from a manual alignment of inputs, and the generated summaries are similarly higher quality as determined by human evaluation. Manually aligning the “contributions” sections of the documents produces the next highest ROUGE scores, followed by the TF-IDF + Euclidean (“average”) aligned inputs. However, even with the best-performing median heuristic alignment, the generated summaries are not consistently comprehensive and informative. Though the BERT model does not initially show promising results, future work should include exploration of other semantic similarity metrics to improve input pairings, as the best inputs were aligned entirely by their lexical similarity. Another consideration is that the target summaries from the medication guide were often very short, ranging from as few as 12 tokens to around 200. The source text samples were comparatively lengthy, with as many as 2000 tokens, unlike (See et al., 2017), where the article was truncated to 400 tokens and the summary to 100 tokens for training. Achieving more consistent source and target lengths could possibly help the heuristic alignment and our model’s overall performance.
In this study, we generated medication guides by training a PGN using SPL from the DailyMed database and applying three different alignment techniques: global, manual, and heuristic. We found that heuristic alignment outperformed the other two methods, and showed potential to automatically generate medication guides to reduce the manual burden in medical writing.
Publicly available datasets were analyzed in this study. This data can be found here: https://dailymed.nlm.nih.gov/dailymed/spl-resources-all-drug-labels.cfm.
CE conceptualized and supervised the study. KP and CM preprocessed the dataset and created the inputs for the summarization model. DA ran and tested the models. AGA and RG contributed to the study design and manuscript preparation.
We acknowledge support by the Open Access Publishing Fund of the University of Tübingen.
Author CM was employed by the company Boston Lighthouse Innovations. Author RG was employed by the company Pfizer Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1086913/abstract#supplementary-material
Brown, T., Mann, B., Ryder, N., Subbiah, M., Kaplan, J. D., Dhariwal, P., et al. (2020). Language models are few-shot learners. Adv. neural Inf. Process. Syst. 33, 1877–1901.
FDA (2019). Dailymed spl resources: Download all drug labels. Available at: https://dailymed.nlm.nih.gov/dailymed/spl-resources-all-drug-labels.cfm (Accessed 10 30, 2020).
FDA (2020). Medication guides. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/medication-guide (Accessed 10 17, 2022).
FDA (2022). Structured product labeling resources. Available at: https://www.fda.gov/industry/fda-data-standards-advisory-board/structur ed-product-labeling-resources (Accessed 10 17, 2022).
ICH (2022). International council for harmonisation of technical requirements for pharmaceuticals for human use. Available at: https://https://ich.org/(Accessed 10 17, 2022).
Liu, Y., and Lapata, M. (2019). Text summarization with pretrained encoders. arXiv preprint arXiv:1908.08345.
Nallapati, R., dos santos, C. N., Zhou, B., Gulcehre, C., and Xiang, B. (2016). Abstractive text summarization using sequence-to-sequence rnns and beyond. arXiv preprint arXiv:1602.06023.
Nallapati, R., Zhai, F., and Zhou, B. (2017). “Summarunner: A recurrent neural network based sequence model for extractive summarization of documents,” in Thirty-first AAAI conference on artificial intelligence, San Francisco, California USA, 4 – 9 February 2017.
Rush, A. M., Chopra, S., and Weston, J. (2015). A neural attention model for abstractive sentence summarization. arXiv preprint arXiv:1509.00685.
See, A., Liu, P. J., and Manning, C. D. (2017). Get to the point: Summarization with pointer-generator networks. arXiv preprint arXiv:1704.04368.
Keywords: artificial intelligence-AI, medication guide, drug labeling, natural language generation, abstractive summarization, pointer generator network
Citation: Meyer C, Adkins D, Pal K, Galici R, Garcia-Agundez A and Eickhoff C (2023) Neural text generation in regulatory medical writing. Front. Pharmacol. 14:1086913. doi: 10.3389/fphar.2023.1086913
Received: 01 November 2022; Accepted: 25 January 2023;
Published: 10 February 2023.
Edited by:
Amit Khurana, University Hospital RWTH Aachen, GermanyReviewed by:
Anchi Pratibha, National Institute of Pharmaceutical Education and Research, IndiaCopyright © 2023 Meyer, Adkins, Pal, Galici, Garcia-Agundez and Eickhoff . This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carsten Eickhoff , Y2Fyc3Rlbi5laWNraG9mZkB1bmktdHVlYmluZ2VuLmRl
†Present Address: Ruggero Galici, Currently at Alexion, AstraZeneca Rare Disease, New Haven, CT, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.