
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Pharmacol. , 09 March 2023
Sec. Obstetric and Pediatric Pharmacology
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1082066
This article is part of the Research Topic Emerging Researchers in Frontiers in Pharmacology: Obstetric and Pediatric Pharmacology 2022 View all 13 articles
Background: Epithelial ovarian cancer (EOC) is the most common type of ovarian cancer. About 90% of ovary tumors are epithelial. The current treatment for EOC involves surgical debulking of the tumors followed by a combination of chemotherapy. While most patients achieve complete remission, many EOCs will recur and develop chemoresistance. The cancer cells can adapt to several stress stimuli, becoming resistant. Therefore, new ways to fight resistant cells during the disease are being studied. Recently, exosomes, which reflect cell behavior in normal and pathological conditions such as epithelial ovarian cancer, are of academic interest as new biomarkers for diagnosis and therapy. Consequently, the current study aimed to investigate the research output of exosomes in EOC.
Method: A bibliometric method was used for analyzing publications on exosome and epithelial ovarian cancer from the beginning to 15 October 2022 by searching keywords in Scopus, PubMed and Google scholar. Annual scientific publications, authors, citations, journals, co-authorships, and keywords co-occurrence were analyzed and plotted using Microsoft Office Excel and VOS viewer. 39 original journal articles and 3 reviews have been published since 2015 up to 15 October 2022.
Results: The findings showed that China is the top country in research output, international collaborations, organization, author, and sponsorship. The top journals were the Journal of Ovarian Research, Oncotarget, and Tumor Biology, all in the United States. The top institution was Shanghai Jiao Tong University in China. The top author was Xipeng Wang. Co-occurrence analysis showed that academics’ interest is toward:1) 1) Exosomes as prognostic biomarkers of EOC as well as their role in the proliferation and migration of cells. 2) The role of exosomes in metastasis through different mechanisms; 3) The role of exosomes in epithelial-mesenchymal transition of ovarian cancer cells; 4) The diagnostic role of EVs in EOC; and 5) Conferring chemoresistance in EOC through the exosomal transfer of miRNAs.
Conclusion: Research on the exosome and EOC has an increasing trend, and China is much more involved than other countries in research, financial support, and international cooperation. These findings could aid researcher in understanding novel ideas and subjects interested by sponsors in this field.
Despite decades of efforts for improving the effectiveness of treatment approaches, epithelial ovarian cancer (EOC) is still the most lethal of all gynecological malignancies (Obermair et al., 2021). From a clinical point of view, the standard approach of EOC management is still debulking surgery and chemotherapy (Gadducci et al., 2022; Lodewijk et al., 2022). However, innovative surgical and medical developments were associated with only marginal survival improvements. The negative prognosis of patients suffering from OCE is normal because of late-stage diagnosis and the chemoresistance development during the illness period (Ma et al., 2014; Akter et al., 2022; Wanyama et al., 2022). Overdiagnosed, EOC remains an asymptomatic disease until the development of diffuse peritoneal carcinomatosis, including abdominal disease (van Baal et al., 2018). In addition, the transvaginal ultrasound combined with tumor markers carbohydrate antigen 125 (CA-125) serum level dosage as a promising screening method attracted attention, but yet had limitations in identifying EOC as an early-stage disease (Fishman et al., 2005; Olivier et al., 2006). Accordingly, 70% of patients diagnosed with EOC are frequently at the end stage of the disease with a 5-year survival rate of less than 40% (Meng et al., 2016a), so the necessity of developing non-invasive tools with highly sensitive for ensuring an early-stage diagnosis is one of the main challenges in biomarker investigate. Drug resistance development is another significant point in EOC investigations (Liu et al., 2018; Bukowski et al., 2020). In this regard, new advanced methods have been presented to date, precise approaches for detecting and treating different cancers (Bartosh et al., 2016; Baghban et al., 2022; Miri et al., 2022). One of the most newly known biomarkers of diseases is extracellular nano-vesicles, called exosomes (Hubert et al., 2015; Nawaz et al., 2016; Yousafzai et al., 2018; Ghafourian et al., 2022). Exosomes are produced by cells and carry several genetic materials and proteins playing key roles in the signaling and crosstalk of cells (Mashouri et al., 2019; Afshar et al., 2021; Bayat et al., 2021; Zhankina et al., 2021; Akbar et al., 2022). It has been reported that exosomes and their cargoes are an appropriate tool for maintaining the homeostasis of cancer tissues as they are able to mediate intercellular communication (Salido-Guadarrama et al., 2014; Caruso Bavisotto et al., 2019). Moreover, researchers propose that a panel of exosome-derived circulating miRNAs may aid to come for diagnosing early-onset of cancer and monitoring disease over cancer therapy (Wang and Chen, 2014). In this regard, increasing attention has been concentrated on the function of exosomes and their molecular cargo in EOC (Li and Wang, 2017).
In light of the above-mentioned, the present bibliometrics study focuses on analyzing and examining the scientific productivity developed specifically on exosomes in EOC up to 15 October 2022 to observe its evolution. In this context, the scientific productivity by year, countries, subject areas, organizations, sponsors, authors, citations, keyword co-occurrence, and co-authorships mapping related to exosomes and EOC were extracted from the titles and keywords of the included documents and analyzed. According to the best of our knowledge, there is no similar study on the subject of the exosome in EOC. The findings and statistics attained are of great value for academics working on this topic or beginning their initial steps.
The current research focuses on bibliometric analysis, which applies methods and software that allow identification, documentation and combination of diverse properties of the knowledge area (Donthu et al., 2021). Scientific mapping is performed according to the obtained data and different networks are resulted that define their relations between diverse factors. The proper expression of these networks will determine the upcoming lines of strengthening the scientific production of this field. Documents are tracked and selected according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Figure 1) (Moher et al., 2009). The data for this study, dating from the beginning to 2022 (15 October 2022), will be extracted using the PoP software (version 8) from Scopus, PubMed and Google scholar using the following search terms in title and keywords: “epithelial ovarian cancer” and “exosomes” or “exosome” or “exosomal” or “extracellular vesicles” or “extracellular vesicle”. Then, those articles investigating ovarian cancer in general (which also includes epithelial ones) or microvesicles and extracellular vesicles (which also contain exosomes) were excluded as this study aims to investigate the studies focusing on only EOC and only exosomes. All the information including metadata on citation information and abstract, among other information will be exported in CSV format to the Microsoft Office Excel software for data analysis after merging the obtained files. Additionally, the VOSviewer software bibliometric analysis software, which enabled to build diverse illustrations of scientific mapping, will be employed to generate the collaboration and word co-occurrence networks.
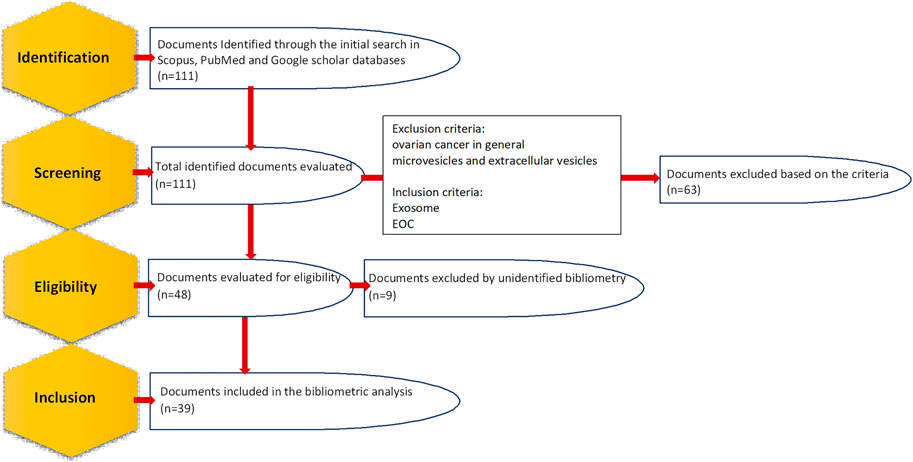
FIGURE 1. Steps of identifying and selecting documents according to preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines.
The primary research yielded 111 documents in Scopus, PubMed and Google scholar. Only articles published in English were included. After the exclusion of duplicate, irrelevant journal articles and non-journal articles, 39 articles on exosome and EOC were selected for bibliometric analysis; 36 of which were original articles (Zhang et al., 2016a; Meng et al., 2016b; Zhang et al., 2016b; Labani-Motlagh et al., 2016; Ying et al., 2016; Chen et al., 2017; Hu et al., 2017; Li et al., 2017; Wu et al., 2017; Chen et al., 2018; Qiu et al., 2018; Zhang et al., 2018; Zhou et al., 2018; Zhang et al., 2019a; Keserű et al., 2019; Tang et al., 2019; Zhu et al., 2019; Cheng et al., 2020; Li et al., 2020; Lu et al., 2020; Luo and Gui, 2020; Maeda et al., 2020; Masoumi-Dehghi et al., 2020; Alharbi et al., 2021; Xiong et al., 2021a; Cai et al., 2021; Gao et al., 2021; Li et al., 2021; Liu et al., 2021; Ma et al., 2021; Zhu et al., 2022a; Chen et al., 2022; Jeon et al., 2022; Lai et al., 2022; Yang et al., 2022), and the rest of them were review articles (Li and Wang, 2017; Lucidi et al., 2020; Shiao et al., 2021). The first English article is “Characterization of exosomes derived from ovarian cancer cells and normal ovarian epithelial cells by nanoparticle tracking analysis” in 2015 (Zhang et al., 2016a). The scientific production evolution has been illustrated in Figure 2. 2021 is the year with the greatest scientific production with 8 articles and a 4-fold increase in publication compared to the first year, 2015. In the case of 2022, as the year is not completed it is soon to judge about.
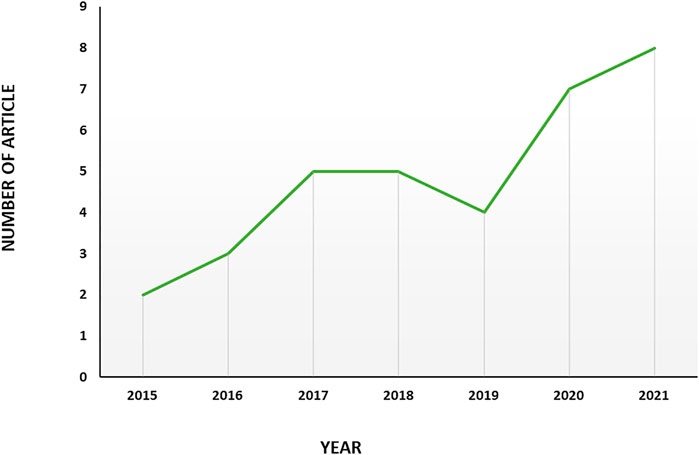
FIGURE 2. Trend of productivity by years in terms of number of publications in the field of exosome and EOC.
Selected articles on exosome and OEC has been published in the context of 11 areas based on area categories of the Dimension database. “Biomedical and clinical sciences” has the maximum publication share (37.36%), followed by “oncology and carcinogenesis” (34.06%), “biological sciences” (6.50%), “biochemistry and cell biology” (4.58%), “multidisciplinary” (3.30%) “multidisciplinary” (3.30%) “multidisciplinary” (7.69%), “biochemistry and cell biology” and “immunology” (5.49%), “chemical sciences”, “microbiology” and “medicinal and biomolecular chemistry” (2.20%) and “medical biotechnology”, “clinical sciences” and “reproductive medicine” (1.10%) (Figure 3).
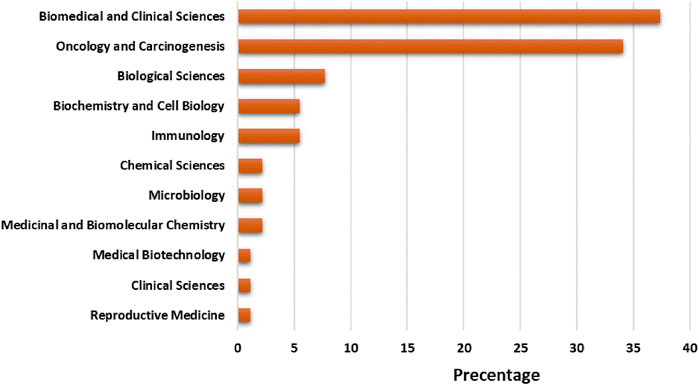
FIGURE 3. Trend of productivity by subject areas in terms of number of publications in the field of exosome and EOC.
A total of 14 countries including China, Germany, Sweden, Hungary, Japan, the United States, Italy, Iran, Thailand, South Korea, Taiwan, Australia, Saudi Arabia, and Chile had published at least one article related to the exosome and EOC from 2015 to 2022. As observed in Figure 4, China with 29 is the top country.
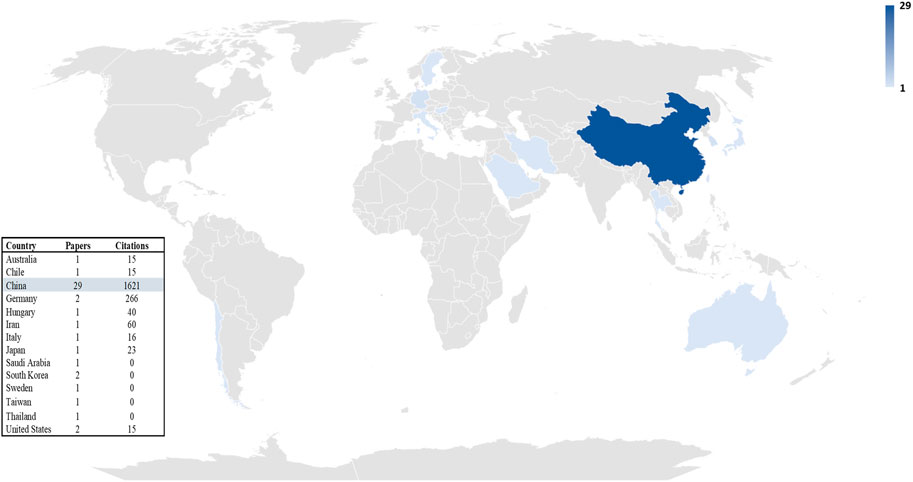
FIGURE 4. Trend of productivity by countries in terms of number of publications in the field of exosome and EOC.
In order to analyze country co-authorship or international collaboration, all 14 countries were included in the analysis. This analysis resulted in 9 clusters that 3 of which contains only two or more country. These three clusters contain 8 countries and 2 out of these 3 clusters are linked together. These results have been illustrated in Figure 5. It shows that authors in eight countries have international collaboration. Among these countries, China has the best international collaboration.
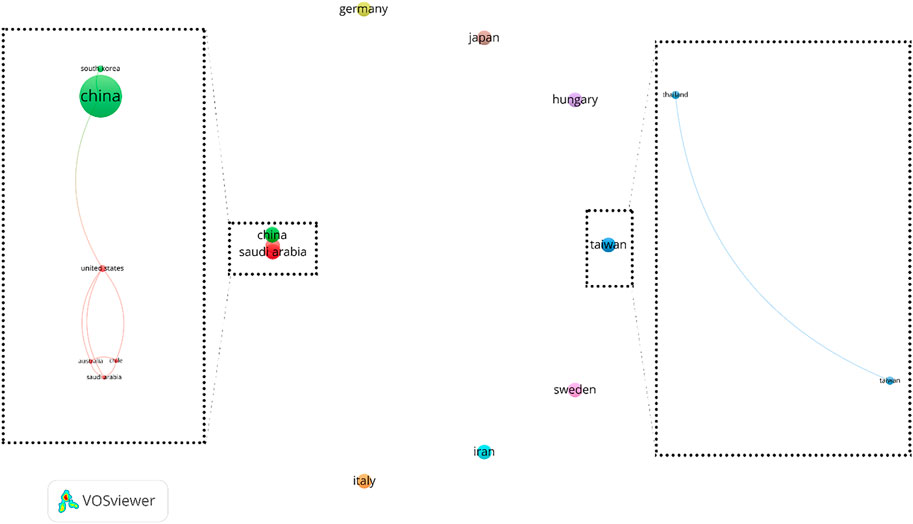
FIGURE 5. Country co-authorship networks (criteria: countries with at least 1 article in the field of exosome and OEC).
A total of 63 organizations including 25 hospitals and 38 universities and colleges published at least one journal article from 2015 up to 15 October 2022. Among these organizations, the Shanghai Jiao Tong University in China with 6 articles is ranked as the first producer university and the Tongji University in China with 5 articles is ranked as the second one. Table 1 shows the ranking of organizations with more than one journal article in this field between 2015 to 15 October 2022.
To analyze the organizations’ collaboration, the organizations’ co-authorship network was obtained using VOSviewer software (Figure 6). All 63 identified organizations were included in the primary analysis and it resulted in 26 clusters that 9 of them has no collaboration with other organizations. The largest cluster contains 8 organizations including, which has been shown in detail in Figure 6.
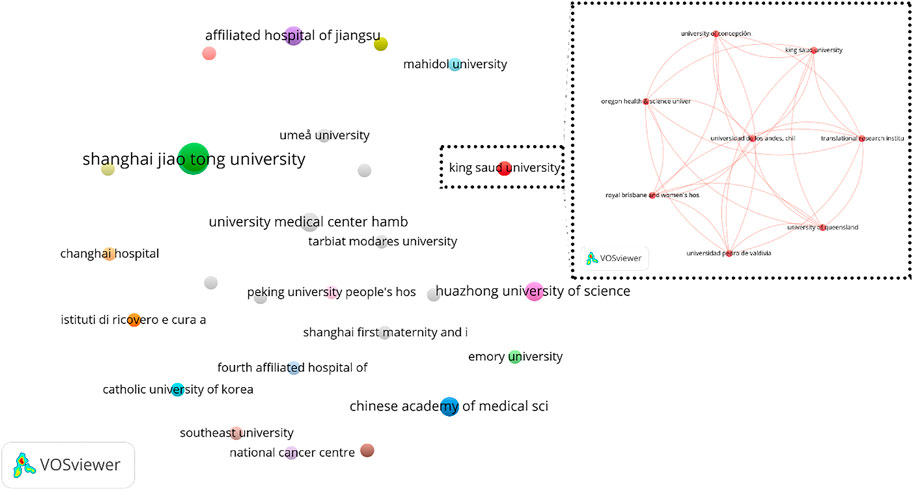
FIGURE 6. Organization co-authorship network (criteria: organizations with at least 1 article in the field of exosome and OEC).
245 authors are identified in 39 articles implying a productivity index of 0.16 articles per author. Authors with more than 2 articles in the field of exosome and EOC have been presented in Table 2. Xipeng Wang (Ying et al., 2016; Chen et al., 2017; Li and Wang, 2017; Wu et al., 2017; Chen et al., 2018; Zhou et al., 2018; Li et al., 2020), Xinjing Wang (Ying et al., 2016; Chen et al., 2017; Wu et al., 2017; Chen et al., 2018; Zhou et al., 2018; Li et al., 2020), and Qinyi Zhu (Ying et al., 2016; Chen et al., 2017; Wu et al., 2017; Zhou et al., 2018; Li et al., 2020) are the most productive author with 7, 6, and 5 articles in the field of exosome and EOC. Most citations are also related to the same authors with values of 887, 811, and 651, respectively. It is worth notable that these authors are in the same group.
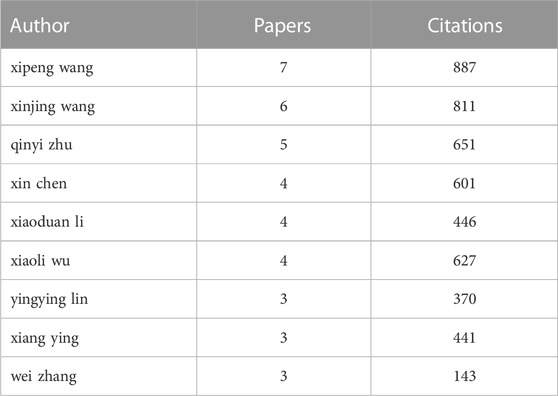
TABLE 2. Most productive authors in terms of number of publications in the field of exosome and EOC.
Regarding the author collaboration trend in producing journal articles, there is no single authorship document and all articles display a contribution of two or more authors that authors in 25 of which have different affiliations. The participation of several authors with diverse affiliations in one manuscript displays thematic maturity (Berelson, 1952; López López, 1996).
For authors’ co-authorship analysis, only the authors with at least 2 articles were included in this analysis; so, 25 of 246 authors were entered into the network analysis. As it was demonstrated in Figures 6, 7 groups were resulted by clustering the co-authorships among these 25 authors. Only 2 clusters link together and the rest of them have no link with each other. In this network, a total of 70 links were found between authors.
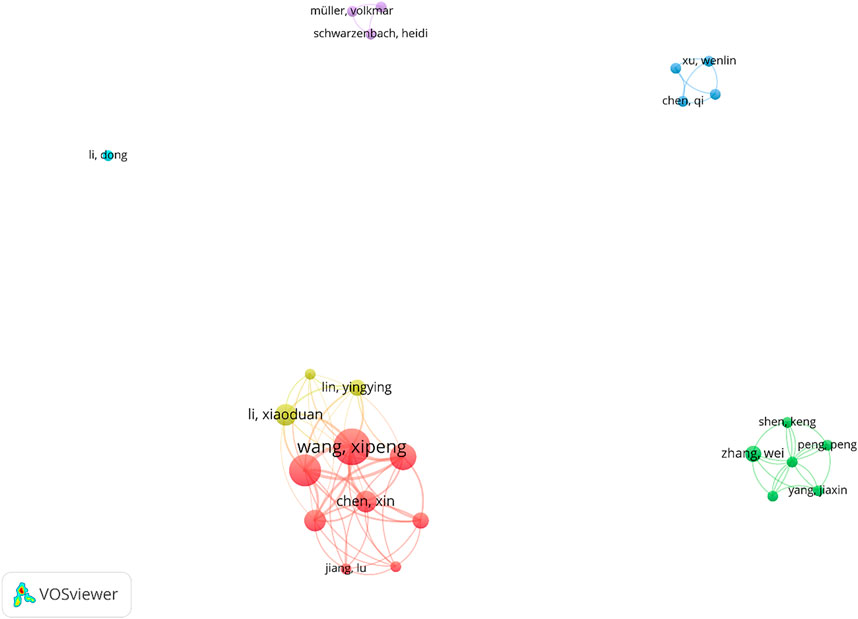
FIGURE 7. Author co-authorship networks (criteria: authors with at least 2 articles in the field of exosome and OEC).
A primary analysis of citations showed that the selected articles had been cited 2,101 times, and the mean citation to each article was 53.87 times. 2018 is the year with the maximum number of citations, with 549 citations (26.13%). For articles with the maximum citations, three original articles by Ying et al. (Ying et al., 2016) with 272 citations, Zhou et al. (Zhou et al., 2018) with 230 citations and Meng et al. (Meng et al., 2016b) with 220 citations were first, second, and third, respectively. Table 3 indicates ten articles with the maximum citations in detail. As observed 7 out of these 10 articles focused on the delivery of microRNAs (miRNAs) by exosomes.
On the other hand, the authors’ co-citation analysis (ACA) indicates how often authors of the former articles are co-cited by authors of subsequent articles. These groups cause clusters with central nodes whose size shows the co-citation trend reached by each author. Figure 8 demonstrates a scientific mapping by ACA. The structure contains 10,504 authors, 15 of which meet the threshold recognized in fifteen citations, resulting in 3 clusters. The authors with the maximum citations were: Xipeng Wang (33 co-citations), Klaus Pantel (28 co-citations), and Heidi Schwarzenbach (27 co-citations).
In fact, each cluster in Figure 8 forms a “school of thought” (Zupic and Čater, 2015), which allows us to observe the methods shared between the authors. Cluster 1, which is red, is composed of 7 academics including Xin Chen (15 co-citations), Ahmedin Jemal (24 co-citations), Rebecca l. Siegel (18 co-citations), Xinjing Wang (19 co-citations), Xipeng Wang (33 co-citations), Xiaoli Wu (17 co-citations), and Qinyi Zhu (17 co-citations). This school of thought focuses on different accepts of cancer research (Ying et al., 2016; Siegel et al., 2019). The green cluster 2 is made up of five authors, who focus on cancer biomarkers: Cicek Gercel-Taylor (17 co-citations), Klaus Pantel (28 co-citations), Volkmar Müller (16 co-citations), Heidi Schwarzenbach (27 co-citations), and Douglas D. Taylor (17 co-citations). Among the main lines of this school are tumor-derived exosomes as diagnostic biomarkers of ovarian cancer (Taylor and Gercel-Taylor, 2008) and exosomal microRNAs as EOC markers (Pan et al., 2018). Finally, cluster 3, which is blue, concentrates on 3 researchers: (17 co-citations), Anil K. Sood (17 co-citations), and Clotilde Théry (17 co-citations). These authors address the functions of extracellular vesicles in cancer therapy (Zhao et al., 2022a; Singh et al., 2022).
According to the bibliographic coupling of resources, a total of 30 journals with five clusters were identified (Figure 9). The most productive ones are the Journal of ovarian research, Oncotarget, and Tumor biology, with three articles each. The Oncotarget has the maximum number of citations received by accumulating 492 (Table 4). A total number of 25 journals only published one article in the field of exosome and OEC. 9 out of which (15 out of 39 documents) were published mostly in the United States. 19 out of 25 journals are Q1 suggesting the high quality of the research in this field.

FIGURE 9. Bibliographic coupling of journals (criteria: journals with at least one article in the field of exosome and OEC).
The main financial sponsors for “exosome and EOC” research was presented in Table 5. The obtained results showed that the National Natural Science Foundation of China was the top funding sponsor with sponsoring 20 projects on this topic.
Despite the current relevance of using keywords in some analyses, 3 articles without the keyword section were recognized after reviewing selected articles. In the rest of the articles, the co-occurrence analysis of author keywords is used. For this purpose, at least 2 author keyword co-occurrences were considered as the criteria. 20 out of 114 diverse keywords met the inclusion criteria. As revealed in Figure 10, the analysis of involved keywords resulted in 5 clusters. The keywords with the maximum number of co-occurrences in each group are as follows: Exosomes (14 co-citations), ovarian cancer (12 co-citations), epithelial ovarian cancer (10 co-citations), exosome (9 co-citations), biomarker (4 co-citations), prognosis (4 co-citations), EOC (3 co-citations), biomarkers (3 co-citations), extracellular vesicles (3 co-citations), hypoxia (3 co-citations), microRNAs (3 co-citations), metastasis (2 co-citations), macrophages (2 co-citations), epithelial-mesenchymal transition (2 co-citations), epithelial ovarian cancer (EOC) (2 co-citations), diagnosis (2 co-citations), chemoresistance (2 co-citations), migration (2 co-citations), proliferation (2 co-citations), proteomics (2 co-citations).
The present research mainly purposed to study the international research activities in the field of exosome and EOC from the start of data published in this field up to 15 October 2022. It is emphasized that this research included studies focused on EOC, not ovarian cancer in general and exosomes alone, not microvesicles and extracellular vesicles. There are several bibliometric reviews on different aspects of ovarian cancer (Wang et al., 2016; El Bairi et al., 2021; Gupta et al., 2021; Khedkar et al., 2021; Liu et al., 2022; Xu and Li, 2022) but there is no bibliometric review on EOC and applications of exosomes in EOC. The analysis of productivity by year showed that despite the growth of research interest in exosomes in different fields, research on exosomes and EOC has opened up in recent decades, and it is in the early stages. According to the findings of the current study, China is ranked the first publisher of journal articles in this field. This can be contributed to a large number of highly ranked institutions in publication and more funding sponsors in this region. However, Wang B et al., in 2019 and Wang Y et al., in 2017 stated that the United States ranked as the first producer in the field of exosomes and China ranked second at that time (Wang et al., 2017; Wang et al., 2019). Moreover, Yuanxia Liu et al. reported that the United States and UK are the first and second article producer from inception to 2021 (Liu et al., 2022). When it comes to funding sponsors of the research, Chinese sponsors especially the National Natural Science Foundation of China are impressive. This is since China efforts to impact high-tech technologies and promote their research and development sections and businesses.
Regarding the journal that publishes the articles, the Journal of Ovarian Research is the top publisher of journal articles in the field of exosome and EOC based on our analysis. Wang Y et al. and Wang B et al. stated that Plos One, Journal of Biological Chemistry, and Scientific Reports are the top publishers of articles in the exosome field (Wang et al., 2017; Wang et al., 2019). Furthermore, Bernardo Pereira Cabral et al. reported that Oncotarget led published most cancer-related articles between 2012–2017 (Cabral et al., 2018). All of these journals are among high-ranking journals showing the field of study is novel and high-tech.
Citation analysis showed that 7 out of these 10 most cited articles focused on the delivery of microRNAs (miRNAs) by exosomes. The maximum co-occurrence author keywords showed the research flashpoints in the studied area over the study period. According to this analysis, the academics are interested to work on: 1) Exosomes as prognostic biomarkers of EOC and their role in the proliferation and migration of cells. 2) The role of exosomes in metastasis through different mechanisms; 3) The role of exosomes in epithelial-mesenchymal transition of ovarian cancer cells; 4) The diagnostic role of EVs in EOC; and 5) Conferring chemoresistance in EOC through Exosomal transfer of miRNAs.
The sampling bias is one of the key limitations of this study. As mentioned, this study focuses on research investigating specifically EOC and exosome. Therefore, sampling criteria excluded the published reports on extracellular vesicles, microvesicles and ovarian cancer, while exosomes are a subclass of extracellular vesicles and microvesicles. Moreover, EOC is one type of ovarian cancer. Therefore, considering exosome and EOC alone cannot make a general conclusion for highly cited or most productive authors in this field of ovarian cancer.
To reduce this limitation, according to citation analysis results which showed most cited articles focused on the delivery of miRNAs by exosomes, an extra search was conducted using the PubMed database on studies in the field of ovarian cancer and exosomal miRNA. 20 other articles other than those found above were found in this field. These articles are related to different countries including the United States, China, Hungary, Australia, Korea, Japan, Germany and the UK. The most cited article in this area is related to the United States with 487 citations, which is considerable. The information related to these articles has been listed in Table 6.
The present research findings showed the increasing trend of research activities in the field of exosomes and EOC. The scientific production was made up of 39 journal articles found in the international databases of Scopus, PubMed and Google scholar. China is the main contributor to the publications, institutions, funding, and international collaborations on the studied topic. The United States ranked first in terms of journals published articles on the studied topic. Moreover, results show that the scientists working in this area are mostly interested into apply microRNA delivery using exosomes in EOC contexts. However, it showed be noted that these results only related to studies in the field of EOC, not ovarian cancer in general or nanoscale extracellular vesicles, not all scales extracellular vesicles. These findings could be helpful for academics working in the field of exosomes and EOC to develop their studies and find new pipelines along the way.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Conceptualization, NB; methodology, NB; software, NB; validation, NB; formal analysis, NB; investigation, NB; resources, NB, MU, and IN; data curation, NB; writing—original draft preparation, NB; writing—review and editing, MU and IN; visualization, NB and MU; supervision, NB; project administration, NB All authors have read and agreed to the published version of the manuscript.
MU was employed by the company Department of Cancer Immunology, Genentech Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afshar, A., Zare, M., Farrar, Z., Hashemi, A., Baghban, N., Khoradmehr, A., et al. (2021). Exosomes of mesenchymal stem cells as nano-cargos for anti-SARS-CoV-2 asRNAs. Mod. Med. Laboratory J. 4, 11–18.
Akbar, A., Malekian, F., Baghban, N., Kodam, S. P., and Ullah, M. (2022). Methodologies to isolate and purify clinical grade extracellular vesicles for medical applications. Cells 11, 186. doi:10.3390/cells11020186
Akter, S., Rahman, M. A., Hasan, M. N., Akhter, H., Noor, P., Islam, R., et al. (2022). Recent advances in ovarian cancer: Therapeutic strategies, potential biomarkers, and technological improvements. Cells 11, 650. doi:10.3390/cells11040650
Alharbi, M., Lai, A., Sharma, S., Kalita-de Croft, P., Godbole, N., Campos, A., et al. (2021). Extracellular vesicle transmission of chemoresistance to ovarian cancer cells is associated with hypoxia-induced expression of glycolytic pathway proteins, and prediction of epithelial ovarian cancer disease recurrence. Cancers 13, 3388. doi:10.3390/cancers13143388
Baghban, N., Khoradmehr, A., Nabipour, I., Tamadon, A., and Ullah, M. (2022). The potential of marine-based gold nanomaterials in cancer therapy: A mini-review. Gold Bull. 55, 53–63. doi:10.1007/s13404-021-00304-6
Bayat, F., Afshar, A., and Baghban, N. (2021). Algal cells-derived extracellular vesicles: A review with special emphasis on their antimicrobial effects. Front. Microbiol. 12, 785716. doi:10.3389/fmicb.2021.785716
Berelson, B. (1952). Content analysis in communication research. Washington, DC: American Psychological Association
Bukowski, K., Kciuk, M., and Kontek, R. (2020). Mechanisms of multidrug resistance in cancer chemotherapy. Int. J. Mol. Sci. 21, 3233. doi:10.3390/ijms21093233
Cabral, B. P., da Graça Derengowski Fonseca, M., and Mota, F. B. (2018). The recent landscape of cancer research worldwide: A bibliometric and network analysis. Oncotarget 9, 30474–30484. doi:10.18632/oncotarget.25730
Cai, J., Gong, L., Li, G., Guo, J., Yi, X., and Wang, Z. (2021). Exosomes in ovarian cancer ascites promote epithelial–mesenchymal transition of ovarian cancer cells by delivery of miR-6780b-5p. Cell death Dis. 12, 210–217. doi:10.1038/s41419-021-03490-5
Caruso Bavisotto, C., Scalia, F., Marino Gammazza, A., Carlisi, D., Bucchieri, F., Conway de Macario, E., et al. (2019). Extracellular vesicle-mediated cell–cell communication in the nervous system: Focus on neurological diseases. Int. J. Mol. Sci. 20, 434. doi:10.3390/ijms20020434
Chen, L., Wang, K., Li, L., Zheng, B., Zhang, Q., Zhang, F., et al. (2022). Plasma exosomal miR-1260a, miR-7977 and miR-192-5p as diagnostic biomarkers in epithelial ovarian cancer. Future Oncol. 18, 2919–2931. doi:10.2217/fon-2022-0321
Chen, X., Ying, X., Wang, X., Wu, X., Zhu, Q., and Wang, X. (2017). Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 38, 522–528. doi:10.3892/or.2017.5697
Chen, X., Zhou, J., Li, X., Wang, X., Lin, Y., and Wang, X. (2018). Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. 435, 80–91. doi:10.1016/j.canlet.2018.08.001
Cheng, L., Zhang, K., Qing, Y., Li, D., Cui, M., Jin, P., et al. (2020). Proteomic and lipidomic analysis of exosomes derived from ovarian cancer cells and ovarian surface epithelial cells. J. ovarian Res. 13, 9–13. doi:10.1186/s13048-020-0609-y
Chevillet, J. R., Kang, Q., Ruf, I. K., Briggs, H. A., Vojtech, L. N., Hughes, S. M., et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U. S. A. 111, 14888–14893. doi:10.1073/pnas.1408301111
Dai, Y., Cao, Y., Köhler, J., Lu, A., Xu, S., and Wang, H. (2021). Unbiased RNA-Seq-driven identification and validation of reference genes for quantitative RT-PCR analyses of pooled cancer exosomes. BMC genomics 22, 27. doi:10.1186/s12864-020-07318-y
Donthu, N., Kumar, S., Mukherjee, D., Pandey, N., and Lim, W. M. (2021). How to conduct a bibliometric analysis: An overview and guidelines. J. Bus. Res. 133, 285–296. doi:10.1016/j.jbusres.2021.04.070
El Bairi, K., Al Jarroudi, O., and Afqir, S. (2021). Tracing ovarian cancer research in Morocco: A bibliometric analysis. Gynecol. Oncol. Rep. 37, 100777. doi:10.1016/j.gore.2021.100777
Fishman, D. A., Cohen, L., Blank, S. V., Shulman, L., Singh, D., Bozorgi, K., et al. (2005). The role of ultrasound evaluation in the detection of early-stage epithelial ovarian cancer. Am. J. Obstetrics Gynecol. 192, 1214–1221. doi:10.1016/j.ajog.2005.01.041
Gadducci, A., Cosio, S., and Lippolis, P. V. (2022). Hyperthermic intraperitoneal chemotherapy in the management of primary epithelial ovarian cancer: A debated issue for gynecologic oncologists. Anticancer Res. 42, 4659–4665. doi:10.21873/anticanres.15970
Gao, L., Nie, X., Gou, R., Hu, Y., Dong, H., Li, X., et al. (2021). Exosomal ANXA2 derived from ovarian cancer cells regulates epithelial-mesenchymal plasticity of human peritoneal mesothelial cells. J. Cell. Mol. Med. 25, 10916–10929. doi:10.1111/jcmm.16983
Ghafourian, M., Mahdavi, R., Akbari Jonoush, Z., Sadeghi, M., Ghadiri, N., Farzaneh, M., et al. (2022). The implications of exosomes in pregnancy: Emerging as new diagnostic markers and therapeutics targets. Cell Commun. Signal. 20, 51–19. doi:10.1186/s12964-022-00853-z
Gupta, A., Agrawal, M. A., Jaggi, K., and Goswami, A. D. (2021). Bibliometric analysis of emerging technologies in the field of computer science helping in ovarian cancer research.
Horie, K., Nanashima, N., Yokoyama, Y., Yoshioka, H., and Watanabe, J. (2022). Exosomal MicroRNA as biomarkers for diagnosing or monitoring the progression of ovarian clear cell carcinoma: A pilot study. Molecules 27, 3953. doi:10.3390/molecules27123953
Hu, Y., Li, D., Wu, A., Qiu, X., Di, W., Huang, L., et al. (2017). TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. 393, 60–67. doi:10.1016/j.canlet.2017.02.009
Hubert, A., Subra, C., Jenabian, M.-A., Labrecque, P.-F. T., Tremblay, C., Laffont, B., et al. (2015). Elevated abundance, size, and MicroRNA content of plasma extracellular vesicles in viremic HIV-1+ patients: Correlations with known markers of disease progression. J. Acquir. immune Defic. syndromes 70, 219–227. doi:10.1097/QAI.0000000000000756
Jeon, H., Seo, S. M., Kim, T. W., Ryu, J., Kong, H., Jang, S. H., et al. (2022). Circulating exosomal miR-1290 for diagnosis of epithelial ovarian cancer. Curr. Issues Mol. Biol. 44, 288–300. doi:10.3390/cimb44010021
Kanlikilicer, P., Bayraktar, R., Denizli, M., Rashed, M. H., Ivan, C., Aslan, B., et al. (2018). Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine 38, 100–112. doi:10.1016/j.ebiom.2018.11.004
Kanlikilicer, P., Rashed, M. H., Bayraktar, R., Mitra, R., Ivan, C., Aslan, B., et al. (2016). Ubiquitous release of exosomal tumor suppressor miR-6126 from ovarian cancer cells. Cancer Res. 76, 7194–7207. doi:10.1158/0008-5472.CAN-16-0714
Keserű, J. S., Soltész, B., Lukács, J., Márton, É., Szilágyi-Bónizs, M., Penyige, A., et al. (2019). Detection of cell-free, exosomal and whole blood mitochondrial DNA copy number in plasma or whole blood of patients with serous epithelial ovarian cancer. J. Biotechnol. 298, 76–81. doi:10.1016/j.jbiotec.2019.04.015
Khedkar, V., Fernandes, C., Desai, D., Mansi, R., Chavan, G., Tidke, S. K., et al. (2021). Bibliometric analysis of named entity recognition for chemoinformatics and biomedical information extraction of ovarian cancer. Lincoln: Library Philosophy and Practice, University of Nebraska, 1–10.
Kim, S., Choi, M. C., Jeong, J. Y., Hwang, S., Jung, S. G., Joo, W. D., et al. (2019). Serum exosomal miRNA-145 and miRNA-200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas. J. Cancer 10, 1958–1967. doi:10.7150/jca.30231
Kobayashi, M., Salomon, C., Tapia, J., Illanes, S. E., Mitchell, M. D., and Rice, G. E. (2014). Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200. J. Transl. Med. 12, 4. doi:10.1186/1479-5876-12-4
Kobayashi, M., Sawada, K., Miyamoto, M., Shimizu, A., Yamamoto, M., Kinose, Y., et al. (2020). Exploring the potential of engineered exosomes as delivery systems for tumor-suppressor microRNA replacement therapy in ovarian cancer. Biochem. biophysical Res. Commun. 527, 153–161. doi:10.1016/j.bbrc.2020.04.076
Labani-Motlagh, A., Israelsson, P., Ottander, U., Lundin, E., Nagaev, I., Nagaeva, O., et al. (2016). Differential expression of ligands for NKG2D and DNAM-1 receptors by epithelial ovarian cancer-derived exosomes and its influence on NK cell cytotoxicity. Tumor Biol. 37, 5455–5466. doi:10.1007/s13277-015-4313-2
Lai, H., Guo, Y., Tian, L., Wu, L., Li, X., Yang, Z., et al. (2022). Protein panel of serum-derived small extracellular vesicles for the screening and diagnosis of epithelial ovarian cancer. Cancers 14, 3719. doi:10.3390/cancers14153719
Li, T., Lin, L., Liu, Q., Gao, W., Chen, L., Sha, C., et al. (2021). Exosomal transfer of miR-429 confers chemoresistance in epithelial ovarian cancer. Am. J. cancer Res. 11, 2124–2141.
Li, W., Zhang, X., Wang, J., Li, M., Cao, C., Tan, J., et al. (2017). TGFβ1 in fibroblasts-derived exosomes promotes epithelial-mesenchymal transition of ovarian cancer cells. Oncotarget 8, 96035–96047. doi:10.18632/oncotarget.21635
Li, X., Tang, M., Zhu, Q., Wang, X., Lin, Y., and Wang, X. (2020). The exosomal integrin α5β1/AEP complex derived from epithelial ovarian cancer cells promotes peritoneal metastasis through regulating mesothelial cell proliferation and migration. Cell. Oncol. 43, 263–277. doi:10.1007/s13402-019-00486-4
Li, X., and Wang, X. (2017). The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol. cancer 16, 92–10. doi:10.1186/s12943-017-0659-y
Liu, J., Yoo, J., Ho, J. Y., Jung, Y., Lee, S., Hur, S. Y., et al. (2021). Plasma-derived exosomal miR-4732-5p is a promising noninvasive diagnostic biomarker for epithelial ovarian cancer. J. ovarian Res. 14, 59–14. doi:10.1186/s13048-021-00814-z
Liu, Y., Liu, Q., and Jiang, X. (2022). Bibliometric analysis of hotspots and frontiers in cancer-related fatigue among ovarian cancer survivors. PloS one 17, e0274802. doi:10.1371/journal.pone.0274802
Liu, Y., Tang, J., Liu, D., Zhang, L., He, Y., Li, J., et al. (2018). Increased autophagy in EOC re-ascites cells can inhibit cell death and promote drug resistance. Cell Death Dis. 9, 419. doi:10.1038/s41419-018-0449-5
Lodewijk, I., Bernardini, A., Suárez-Cabrera, C., Bernal, E., Sánchez, R., Garcia, J., et al. (2022). Genomic landscape and immune-related gene expression profiling of epithelial ovarian cancer after neoadjuvant chemotherapy. NPJ Precis. Oncol. 6, 7–11. doi:10.1038/s41698-021-00247-3
López López, P. (1996). La investigación bibliométrica, Introducción a la bibliometría. Valencia, España: Promolibro, 43–63.
Lu, S., Liu, W., Shi, H., and Zhou, H. (2020). Exosomal miR-34b inhibits proliferation and the epithelial-mesenchymal transition by targeting Notch2 in ovarian cancer. Oncol. Lett. 20, 2721–2728. doi:10.3892/ol.2020.11837
Lucidi, A., Buca, D., Ronsini, C., Tinari, S., Bologna, G., Buca, D., et al. (2020). Role of extracellular vesicles in epithelial ovarian cancer: A systematic review. Int. J. Mol. Sci. 21, 8762. doi:10.3390/ijms21228762
Luo, Y., and Gui, R. (2020). Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J. Gynecol. Oncol. 31, e75. doi:10.3802/jgo.2020.31.e75
Ma, R., Ye, X., Cheng, H., Cui, H., and Chang, X. (2021). Tumor-derived exosomal circRNA051239 promotes proliferation and migration of epithelial ovarian cancer. Am. J. Transl. Res. 13, 1125–1139.
Ma, X., Wang, Y., Sheng, H., Tian, W., Qi, Z., Teng, F., et al. (2014). Prognostic significance of thrombocytosis, platelet parameters and aggregation rates in epithelial ovarian cancer. J. Obstetrics Gynaecol. Res. 40, 178–183. doi:10.1111/jog.12151
Maeda, K., Sasaki, H., Ueda, S., Miyamoto, S., Terada, S., Konishi, H., et al. (2020). Serum exosomal microRNA-34a as a potential biomarker in epithelial ovarian cancer. J. ovarian Res. 13, 47–49. doi:10.1186/s13048-020-00648-1
Mashouri, L., Yousefi, H., Aref, A. R., Molaei, F., and Alahari, S. K. (2019). Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. cancer 18, 75–14. doi:10.1186/s12943-019-0991-5
Masoumi-Dehghi, S., Babashah, S., and Sadeghizadeh, M. (2020). microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF-κB signaling pathways. J. Cell Commun. Signal. 14, 233–244. doi:10.1007/s12079-020-00548-5
Meng, X., Müller, V., Milde-Langosch, K., Trillsch, F., Pantel, K., and Schwarzenbach, H. (2016). “Circulating cell-free miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer,” in Circulating nucleic acids in serum and plasma–CNAPS IX (Springer), 3–8.
Meng, X., Müller, V., Milde-Langosch, K., Trillsch, F., Pantel, K., and Schwarzenbach, H. (2016). Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 7, 16923–16935. doi:10.18632/oncotarget.7850
Miri, M. R., Zare, A., Saberzadeh, J., Baghban, N., Nabipour, I., and Tamadon, A. (2022). Anti-lung cancer marine compounds: A review. Ther. Innovation Regul. Sci. 56, 191–205. doi:10.1007/s43441-022-00375-3
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., and Group, P. (2009). Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 151, 264–269. doi:10.7326/0003-4819-151-4-200908180-00135
Nawaz, M., Fatima, F., Nazarenko, I., Ekström, K., Murtaza, I., Anees, M., et al. (2016). Extracellular vesicles in ovarian cancer: Applications to tumor biology, immunotherapy and biomarker discovery. Expert Rev. proteomics 13, 395–409. doi:10.1586/14789450.2016.1165613
Nazarenko, I., Rupp, A. K., and Altevogt, P. (2013). Exosomes as a potential tool for a specific delivery of functional molecules. Methods Mol. Biol. Clift. N.J.) 1049, 495–511. doi:10.1007/978-1-62703-547-7_37
Obermair, A., Beale, P., Scott, C. L., Beshay, V., Kichenadasse, G., Simcock, B., et al. (2021). Insights into ovarian cancer care: Report from the ANZGOG ovarian cancer webinar series 2020. J. Gynecol. Oncol. 32, e95. doi:10.3802/jgo.2021.32.e95
Olivier, R. I., Lubsen-Brandsma, M. A., Verhoef, S., and van Beurden, M. (2006). CA125 and transvaginal ultrasound monitoring in high-risk women cannot prevent the diagnosis of advanced ovarian cancer. Gynecol. Oncol. 100, 20–26. doi:10.1016/j.ygyno.2005.08.038
Padda, J., Khalid, K., Khedr, A., Patel, V., Al-Ewaidat, O. A., Tasnim, F., et al. (2021). Exosome-derived microRNA: Efficacy in cancer. Cureus 13, e17441. doi:10.7759/cureus.17441
Pan, C., Stevic, I., Müller, V., Ni, Q., Oliveira-Ferrer, L., Pantel, K., et al. (2018). Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol. Oncol. 12, 1935–1948. doi:10.1002/1878-0261.12371
Pink, R. C., Samuel, P., Massa, D., Caley, D. P., Brooks, S. A., and Carter, D. R. (2015). The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol. Oncol. 137, 143–151. doi:10.1016/j.ygyno.2014.12.042
Qiu, J.-J., Lin, X.-J., Tang, X.-Y., Zheng, T.-T., Lin, Y.-Y., and Hua, K.-Q. (2018). Exosomal metastasis-associated lung adenocarcinoma transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int. J. Biol. Sci. 14, 1960–1973. doi:10.7150/ijbs.28048
Salido-Guadarrama, I., Romero-Cordoba, S., Peralta-Zaragoza, O., Hidalgo-Miranda, A., and Rodriguez-Dorantes, M. (2014). MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. OncoTargets Ther. 7, 1327–1338. doi:10.2147/OTT.S61562
Shiao, M.-S., Chang, J.-M., Lertkhachonsuk, A.-A., Rermluk, N., and Jinawath, N. (2021). Circulating exosomal miRNAs as biomarkers in epithelial ovarian cancer. Biomedicines 9, 1433. doi:10.3390/biomedicines9101433
Siegel, R. L., Miller, K. D., and Jemal, A. (2019). Cancer statistics. CA a cancer J. Clin. 69, 7–34. doi:10.3322/caac.21551
Singh, S., Goyal, D., Karthikeyan, R., Kumar, S., Malik, P. S., and Elangovan, R. (2022). RNA profile of immuno-magnetically enriched exosomes isolated from Non-Small Cell Lung Cancer.
Soltész, B., Lukács, J., Szilágyi, E., Márton, É., Szilágyi Bónizs, M., Penyige, A., et al. (2019). Expression of CD24 in plasma, exosome and ovarian tissue samples of serous ovarian cancer patients. J. Biotechnol. 298, 16–20. doi:10.1016/j.jbiotec.2019.03.018
Su, Y. Y., Sun, L., Guo, Z. R., Li, J. C., Bai, T. T., Cai, X. X., et al. (2019). Upregulated expression of serum exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J. Ovarian Res. 12, 6. doi:10.1186/s13048-018-0477-x
Tang, X., Liu, S., Liu, Y., Lin, X., Zheng, T., Liu, X., et al. (2019). Circulating serum exosomal aHIF is a novel prognostic predictor for epithelial ovarian cancer. OncoTargets Ther. 12, 7699–7711. doi:10.2147/OTT.S220533
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. doi:10.1016/j.ygyno.2008.04.033
Bartosh, T. J., Ullah, M., Zeitouni, S., Beaver, J., and Prockop, D. J. (2016). Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc. Natl. Acad. Sci. 113 E6447–E6456. doi:10.1073/pnas.1612290113
van Baal, J., van Noorden, C. J. F., Nieuwland, R., Van de Vijver, K. K., Sturk, A., van Driel, W. J., et al. (2018). Development of peritoneal carcinomatosis in epithelial ovarian cancer: A review. J. Histochem. Cytochem. official J. Histochem. Soc. 66, 67–83. doi:10.1369/0022155417742897
Wang, B., Xing, D., Zhu, Y., Dong, S., and Zhao, B. (2019). The state of exosomes research: A global visualized analysis. BioMed Res. Int. 2019, 1495130. doi:10.1155/2019/1495130
Wang, Q., Sun, L., Wang, Y., and Wang, S. (2016). A bibliometric study on review over 1992-2014 literature on life quality of ovarian cancer patients in China. Modern Clinical Nursing, 47–50.
Wang, W.-T., and Chen, Y.-Q. (2014). Circulating miRNAs in cancer: From detection to therapy. J. Hematol. Oncol. 7, 86–89. doi:10.1186/s13045-014-0086-0
Wang, Y., Wang, Q., Wei, X., Shao, J., Zhao, J., Zhang, Z., et al. (2017). Global scientific trends on exosome research during 2007–2016: A bibliometric analysis. Oncotarget 8, 48460–48470. doi:10.18632/oncotarget.17223
Wanyama, F. M., Tauber, R., Mokomba, A., Nyongesa, C., and Blanchard, V. (2022). The burden of hepatitis B, hepatitis C, and human immunodeficiency viruses in ovarian cancer patients in nairobi, Kenya. Infect. Dis. Rep. 14, 433–445. doi:10.3390/idr14030047
Wu, Q., Wu, X., Ying, X., Zhu, Q., Wang, X., Jiang, L., et al. (2017). Suppression of endothelial cell migration by tumor associated macrophage-derived exosomes is reversed by epithelial ovarian cancer exosomal lncRNA. Cancer Cell Int. 17, 62–13. doi:10.1186/s12935-017-0430-x
Xiao, F., Xiao, S., and Xue, M. (2019). miR-139 controls viability of ovarian cancer cells through apoptosis induction and exosome shedding inhibition by targeting ATP7A. Onco Targets Ther. 12, 10727–10737. doi:10.2147/OTT.S221236
Xiong, C., Sun, Z., Yu, J., and Lin, Y. (2021). Exosome component 4 promotes epithelial ovarian cancer cell proliferation, migration, and invasion via the wnt pathway. Front. Oncol. 11, 797968. doi:10.3389/fonc.2021.797968
Xiong, J., He, X., Xu, Y., Zhang, W., and Fu, F. (2021). MiR-200b is upregulated in plasma-derived exosomes and functions as an oncogene by promoting macrophage M2 polarization in ovarian cancer. J. Ovarian Res. 14, 74. doi:10.1186/s13048-021-00826-9
Xu, C., and Li, X. (2022). Research trends in the early diagnosis of ovarian cancer during 2001–2020: A bibliometric analysis. Eur. J. Gynaecol. Oncol. 43, 321–334.
Yang, S., Zhao, H., Xiao, W., Shao, L., Zhao, C., and Sun, P. (2022). Extracellular vesicle-packaged miR-181c-5p from epithelial ovarian cancer cells promotes M2 polarization of tumor-associated macrophages via the KAT2B/HOXA10 axis. J. Gene Med. 24, e3446. doi:10.1002/jgm.3446
Ying, X., Wu, Q., Wu, X., Zhu, Q., Wang, X., Jiang, L., et al. (2016). Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 7, 43076–43087. doi:10.18632/oncotarget.9246
Yousafzai, N. A., Wang, H., Wang, Z., Zhu, Y., Zhu, L., Jin, H., et al. (2018). Exosome mediated multidrug resistance in cancer. Am. J. cancer Res. 8, 2210–2226.
Zeh, N., Schneider, H., Mathias, S., Raab, N., Kleemann, M., Schmidt-Hertel, S., et al. (2019). Human CAP cells represent a novel source for functional, miRNA-loaded exosome production. PLoS One 14, e0221679. doi:10.1371/journal.pone.0221679
Zhang, H., and Lu, B. (2020). microRNAs as biomarkers of ovarian cancer. Expert Rev. anticancer Ther. 20, 373–385. doi:10.1080/14737140.2020.1760095
Zhang, H., Xu, S., and Liu, X. (2019). MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol. Lett. 17, 5601–5607. doi:10.3892/ol.2019.10220
Zhang, S., Zhang, X., Fu, X., Li, W., Xing, S., and Yang, Y. (2018). Identification of common differentially-expressed miRNAs in ovarian cancer cells and their exosomes compared with normal ovarian surface epithelial cell cells. Oncol. Lett. 16, 2391–2401. doi:10.3892/ol.2018.8954
Zhang, W., Ou, X., and Wu, X. (2019). Proteomics profiling of plasma exosomes in epithelial ovarian cancer: A potential role in the coagulation cascade, diagnosis and prognosis. Int. J. Oncol. 54, 1719–1733. doi:10.3892/ijo.2019.4742
Zhang, W., Peng, P., Kuang, Y., Yang, J., Cao, D., You, Y., et al. (2016). Characterization of exosomes derived from ovarian cancer cells and normal ovarian epithelial cells by nanoparticle tracking analysis. Tumor Biol. 37, 4213–4221. doi:10.1007/s13277-015-4105-8
Zhang, W., Yang, J., Cao, D., You, Y., Shen, K., and Peng, P. (2016). Regulation of exosomes released from normal ovarian epithelial cells and ovarian cancer cells. Tumor Biol. 37, 15763–15771. doi:10.1007/s13277-016-5394-2
Zhankina, R., Baghban, N., Askarov, M., Saipiyeva, D., Ibragimov, A., Kadirova, B., et al. (2021). Mesenchymal stromal/stem cells and their exosomes for restoration of spermatogenesis in non-obstructive azoospermia: A systemic review. Stem Cell Res. Ther. 12, 229–312. doi:10.1186/s13287-021-02295-9
Zhao, L., Corvigno, S., Ma, S., Celestino, J., Fleming, N. D., Hajek, R. A., et al. (2022). Molecular profiles of serum-derived extracellular vesicles in high-grade serous ovarian cancer. Cancers 14, 3589. doi:10.3390/cancers14153589
Zhao, Z., Shuang, T., Gao, Y., Lu, F., Zhang, J., He, W., et al. (2022). Targeted delivery of exosomal miR-484 reprograms tumor vasculature for chemotherapy sensitization. Cancer Lett. 530, 45–58. doi:10.1016/j.canlet.2022.01.011
Zhou, J., Li, X., Wu, X., Zhang, T., Zhu, Q., Wang, X., et al. (2018). Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol. Res. 6, 1578–1592. doi:10.1158/2326-6066.CIR-17-0479
Zhu, X.-L., Wang, H.-J., Wang, X.-R., Wu, D., Ji, X., Xu, L., et al. (2022). IL-6 secretion of CD4+ T cells stimulated by LC3-positive extracellular vesicles in human epithelial ovarian cancer. Clin. Transl. Oncol. 24, 2222–2230. doi:10.1007/s12094-022-02883-y
Zhu, X., Shen, H., Yin, X., Yang, M., Wei, H., Chen, Q., et al. (2019). Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 38, 81–14. doi:10.1186/s13046-019-1095-1
Zhu, Z., Chen, Z., Wang, M., Zhang, M., Chen, Y., Yang, X., et al. (2022). Detection of plasma exosomal miRNA-205 as a biomarker for early diagnosis and an adjuvant indicator of ovarian cancer staging. J. Ovarian Res. 15, 27. doi:10.1186/s13048-022-00961-x
Keywords: MicroRNAs, female, cancer, ovarian, extracellular vesicle
Citation: Baghban N, Ullah M and Nabipour I (2023) The current trend of exosome in epithelial ovarian cancer studies: A bibliometric review. Front. Pharmacol. 14:1082066. doi: 10.3389/fphar.2023.1082066
Received: 27 October 2022; Accepted: 20 February 2023;
Published: 09 March 2023.
Edited by:
Qiwei Yang, The University of Chicago, United StatesReviewed by:
Muhammad Nawaz, University of Gothenburg, SwedenCopyright © 2023 Baghban, Ullah and Nabipour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neda Baghban, bmVkYS5iYWdoYmFuQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.