
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 05 September 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.985084
 Xu Cao1†
Xu Cao1† Hening Chen1†
Hening Chen1† Zhiguo Li2†
Zhiguo Li2† Xiaoke Li1,3
Xiaoke Li1,3 Xianzhao Yang1,3
Xianzhao Yang1,3 Qiushuo Jin1,4
Qiushuo Jin1,4 Yijun Liang1
Yijun Liang1 Jiaxin Zhang1,3
Jiaxin Zhang1,3 Meiyue Zhou1
Meiyue Zhou1 Ningyi Zhang1
Ningyi Zhang1 Guang Chen1,3*
Guang Chen1,3* Hongbo Du1,3*
Hongbo Du1,3* Xiaobin Zao1,4*
Xiaobin Zao1,4* Yong’an Ye1,3*
Yong’an Ye1,3*The Chinese traditional medicine KangXianYiAi formula (KXYA) is used to treat hepatic disease in the clinic. Here we aim to confirm the therapeutic effects and explore the pharmacological mechanisms of KXYA on hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). We first collected and analyzed clinical data of 40 chronic hepatitis B (CHB) patients with precancerous liver lesions under KXYA treatment. Then, the cell viability, migration, cell cycle, and apoptosis of HepAD38 cells with KXYA treatment were examined. Next, we performed network pharmacological analysis based on database mining to obtain the key target pathways and genes of KXYA treatment on HBV-related HCC. We finally analyzed the expression of the key genes between normal and HBV-related HCC tissues in databases and measured the mRNA expression of the key genes in HepAD38 cells after KXYA treatment. The KXYA treatment could reduce the liver nodule size of CHB patients, suppress the proliferation and migration capabilities, and promote apoptosis of HepAD38 cells. The key pathways of KXYA on HBV-related HCC were Cancer, Hepatitis B, Viral carcinogenesis, Focal adhesion, and PI3K-Akt signaling, and KXYA treatment could regulate the expression of the key genes including HNF4A, MAPK8, NR3C1, PTEN, EGFR, and HDAC1. The KXYA exhibited a curative effect via inhibiting proliferation, migration, and promoting apoptosis of HBV-related HCC and the pharmacological mechanism was related to the regulation of the expression of HNF4A, MAPK8, NR3C1, PTEN, EGFR, and HDAC1.
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the third leading cause of death worldwide (Sung et al., 2021). Moreover, more than 80% of HCC occurrences were related to hepatitis B virus (HBV) infection (Zhu et al., 2016), which makes the pathogenesis and treatment of HBV-related HCC still research hotspots (Couri and Pillai, 2019). During the tumorigenesis process of HBV-related HCC, HBV had oncogenic effects through its direct action and interaction with the host. Previous studies have found that the HBV gene fragments could be integrated into human chromosomes and lead to abnormal expression and function of tumor-related genes in hepatocytes (Li et al., 2014), and HBV proteins are also carcinogenic, such as HBx protein, which could promote the occurrence of HCC by stabilizing protein of Cyclin D1 (Chen et al., 2015). Moreover, HBV infection could cause chronic inflammation of the liver, which leads to repeated necrosis and regeneration of hepatocytes, increasing the accumulation of mutant genes and malignant transformation (Neuveut et al., 2010). Based on high-throughput sequencing data, the mutation rates of certain tumor-associated genes were different in HBV-related HCC and other HCC (Amaddeo et al., 2015). Therefore, exploring the molecular mechanism of HBV-related HCC will provide a more precise direction for the diagnosis and treatment of HCC.
To treat HBV-related HCC, there are two aspects needed to consider, chronic HBV infection and liver malignancy. However, the current treatment for HBV-related HCC is less than satisfactory. On the one hand, present available antiviral therapies were not yet able to achieve a complete cure for chronic hepatitis B (CHB) patients on account of the persistent existence of covalently closed circular DNA (cccDNA) of HBV (Lee and Banini, 2019). On the other hand, the existing treatments of HCC could not significantly reduce the mortality rate (GBD 2019 Diseases and Injuries Collaborators, 2020), including targeted drugs, interventional, surgical, and others. Hence, it is necessary and urgent to develop new drugs and treatments for HBV-related HCC.
Traditional Chinese medicine (TCM) has shown certain advantages for HBV-related HCC with its multi-targets and multi-pathways mode of function (Xi and Minuk, 2018). Our previous study found that a TCM formula based on Chaihu and Huangqi could improve the curative effect of CHB in a clinical trial (Li F et al., 2020). Thus, we take the KangXianYiAi (KXYA) formula mainly composed of Chaihu, Huangqi, Shanyao, and Baijiezi herbs to treat HBV-related HCC. In the current study, we first observed the therapeutic effect via analyzing the liver nodules change in CHB patients of a clinical case series with KXYA treatment for at least half a year. We next validated the effects of KXYA in HBV-related hepatoma cell line, HepAD38, on proliferation, migration, and apoptosis. To further explore the mechanism of KXYA on HBV-related HCC, we used the weighted gene co-expression network analysis (WGCNA) and network pharmacology, which have emerged as powerful tools to explore the connection between drugs and disease and obtain the key targets and pathways (Wang et al., 2020; Cao et al., 2021).
Our results showed that the KXYA treatment could inhibit liver nodules development and reduce serum virology indicators of CHB patients. And KXYA exerted inhibiting effects on the proliferation and migration abilities of HepAD38 cells and promoted cell apoptosis. Furthermore, we constructed an interaction network of active compound targets, disease target genes, and pathological stage-related genes for KXYA treatment on HBV-related HCC. Through the interaction network, we screened and obtained 12 key target genes. Finally, we analyzed the expression of the key target genes in normal and HBV-related HCC tissues by public data mining, and in HepAD38 cells with KXYA treatment by real-time quantitative PCR. The detailed research strategy of the study has shown in Supplementary Figure S1. To sum up, this study presented a deeper insight into the tumorigenesis mechanism of HBV-related HCC and offered more evidence and targets for the TCM treatment of HBV-related HCC.
The granule of the KXYA formula is composed of Radix Bupleuri (Chaihu in Chinese; 8 g), Astragalus membranaceus (Huangqi in Chinese; 16 g), Rhizoma Dioscoreae (Shanyao in Chinese; 15 g), and Sinapis Semen (Baijiezi; 3.6 g). The daily dose of KXYA granules for human adults is 0.609 g/kg, and the standard weight of adults is 70 kg. The herbs of the KXYA formula were obtained and authenticated by the Nanning Peili Pharmaceutical Co., Ltd., under the guidance of the Chinese Pharmacopeia 2015 edition (batch number: 170,407), and the specific drug granule preparation process was shown in the Supplementary Table S1. The production process of the dry paste powder of KXYA is as follows. Take the prescription amount of medicinal herbs, add 23 times of water, decoct for 2 h, filter, concentrate the filtrate into a clear paste, dry it at 70°C vacuum (−0.040–0.090 MPa) for 60 h, get the dry paste, crush it, pass through 50 mesh, mix well and pack separately. Water was purified using a Milli-Q system (Millipore, Billerica, MA, Uthe SA). In the cell experiment, the dried paste powder of KXYA formula was dissolved in dimethylsulfoxide (DMSO) to 500 mg/ml and stored at −20°C, and the KXYA solution was added to cell culture with different finaconcentrationson and use 0.1% DMSO as control. The granule of KXYA is used in human clinical case series, and the granule is brewed orally in hot water at 40°C–50°C.
We collected data from 40 patients with precancerous liver lesions in Dongzhimen Hospital from January 2008 to January 2020 retrospectively. Diagnostic criteria of HBV-related liver precancerous lesions: the liver cirrhosis caused by HBV infection; the size of liver nodules ≥1.0 cm under B-scan ultrasonography; the liver cancer was excluded by dynamic-enhanced MRI, dynamic-enhanced CT or contrast-enhanced ultrasound. And meeting the above three conditions at the same time can be diagnosed as HBV-related liver precancerous lesions. Reference guide for the definition of liver precancerous lesions (International Working Party, 1995; Cong et al., 2016). All the patients were treated with Entecavir (ETV) combined with d the KXYA formula for more than 6 months, and the liver nodule size was recorded by ultrasound or MRI. The basic clinic information of the patients was in Supplementary Table S2. The ethical batch number of this study is DZMEC-KY-2019-131, and the informed consent of the study has been obtained from the patients.
Hepatoma cell lines including HepG2, Huh7, SK-Hep1, and HepAD38 were purchased from the American Type Culture Collection (Manassas, VA, United States) and maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin and 5% CO2. For HepAD38, which was stably integrated with the HBV genome (Li X et al., 2020), the media were additionally supplemented with 400 μg/ml G418 sulfate (InvivoGen) (Ladner et al., 1997).
5 × 103 HepAD38 cells were seeded in 96-well plates with six duplications, after being incubated for 24 h, treated cells with 50, 100, 250, and 500 μg/ml of the gradient-concentration of KXYA. After KXYA treatment for 24, 48, and 72 h, a CCK-8 assay kit (Solarbio, CN) was carried out to assess the ability of cell growth by measuring the absorbance at the wavelength of 450 nm by the TECAN infinite M200 Multimode microplate reader (Tecan, Mechelen, Belgium). For hepatoma cell lines, 5 × 103 Huh7, HepG2, and SK-Hep1 cells were seeded in 96-well plates with six duplications, after being incubated for 24 h, the cells were treated with KXYA (500 μg/ml) for 48 h. The detection method was the same as before, and the data normalized whereas taking the average number of the control groups as 1.
5 × 105 HepAD38 cells were seeded in 6-well plates. After being incubated for 24 h, the cells were treated with KXYA (500 μg/ml) for 48 h. HepAD38 cells were stained with 0.1% DAPI (GlpBio, United States) for 10 min, washed with PBS, and photographed to observe the morphological changes of the cells, and we counted and calculated the ratio of apoptotic cells to total cells in three visual fields. To detect apoptosis, the phosphatidylserine level on the cell surface was estimated with Annexin V-FITC and PI apoptosis detection kit (Solarbio, CN). Apoptosis and necrosis of cells were analyzed in a flow cytometer (Beckman, United States). Results were expressed as the percentage of apoptotic or necrotic cells from total cells. The experiments were performed in triplicate and repeated 3 times.
To detect the cell cycle, 5 × 105 HepAD38 cells were seeded in 6-well plates. After being incubated for 24 h, the cells were treated with KXYA (500 μg/ml) for 48 h. Cells were collected and fixed with 70% ethanol at 4°C overnight. After fixation, the cells were stained with propidium iodide (PI, 50 μg/ml, Solarbio, CN) for 30 min in dark. Then the samples were analyzed with a flow cytometer (Beckman, United States). The experiments were performed in triplicate and repeated 3 times.
5 × 105 HepAD38 cells were seeded in the 12-well plate. 12 h later, the cell monolayer was scratched with a sterile 10-μl pipette tip to generate a line-shaped wound. Then the cells were cultured in DMEM without FBS and treated with KXYA (500 μg/ml). After 48 h, images of the scratches were acquired with a digital camera. The scratch areas were quantified using Imagepro-plus 6.0 software (Media Cybernetics Inc., United States). The experiments were performed in triplicate and repeated 3 times, and the data normalized whereas taking the average number of the control groups as 1.
We searched the components of herbs in KXYA from CNKI (https://cnki.net/) (Hu and Xiao, 2018) and TCMSP (http://tcmspw.com/tcmsp.php) (Ru et al., 2014) databases, and the conditions for determining the active ingredient are oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18. The corresponding compound chemical structure was acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov) (Kim et al., 2016). Then the putative targets of active compounds were predicted with SwissTargetPrediction (http://swisstargetprediction.ch/) (Daina et al., 2019) and STITCH (http://stitch.embl.de/) (Szklarczyk et al., 2016) databases respectively, with the probability value ≥0.1 in SwissTargetPrediction database and confidence score ≥0.15 in the STITCH database.
The HBV-related HCC disease targets were gathered from GeneCards (https://genecards.org/) (Stelzer et al., 2016), NCBI Gene (https://ncbi.nlm.nih.gov/gene/) (Brown et al., 2015), OMIM (https://omim.org) (Amberger et al., 2015), and DisGeNET (https://disgenet.org/) (Pinero et al., 2020) databases. The search condition was using the keywords “HBV hepatocellular carcinoma or hepatitis B Virus hepatocellular carcinoma or Hepatitis B Virus-Related Hepatocellular Carcinoma” and selecting the organisms “Homo sapiens”. We set the search terms to {[“HBV” (All Fields) AND “hepatocellular carcinoma” (All Fields)] AND “H. sapiens” (porgn)} AND “Expression profiling by array” (Filter), and found the proper microarray dataset GSE62232 in GEO database (https://ncbi.nlm.nih.gov/gds), which contained 10 HBV-related HCC and 10 normal liver samples (Schulze et al., 2015).
WGCNA is a systems biology method that is used to correlate the modules to clinical traits through a soft-threshold algorithm, and Edmonson’s pathological grade was suitable clinical information for analysis (Qian et al., 2020). Using the R package of WGCNA in the SangerBox database (http://sangerbox.com/tool) (Langfelder and Horvath, 2008), we identified significant gene modules with the expression data in GSE62232. A weighted adjacency matrix was constructed according to a power function amn = |cmn|β (cmn = Pearson’s correlation between gene m and gene n; amn = adjacency between gene m and gene n). A scale-free co-expression network is constructed by the appropriate β value to increase the similarity matrix, and the adjacent matrix was transformed into a topological overlap matrix. The dynamic tree cut algorithm was used to detect gene modules. To identify hub modules, we set soft-thresholding power as 14 (scale-free R2 = 0.85), and minimal module size as 20. The gene significance and module membership were respectively defined by the correlation coefficient of each module gene and each trait. In general, the higher the Pearson’s correlation coefficient relevant to the module the more important clinical significance.
The KXYA compound-putative targets and the hub module genes were mapped with the HBV-related HCC targets to obtain the overlapped genes (OGEs). Inputting the OGEs into the STRING database (https://string-db.org/) (Szklarczyk et al., 2019) with confidence scores ≥0.4 and the species limited to “H. sapiens”, then exported protein-protein interaction (PPI) data. The PPI data was analyzed by Cytoscape 3.8.0 (https://cytoscape.org/) (Kohl et al., 2011) and the non-connection genes were removed. We used the Analyzer plugin to analyze the PPI network and get the Degree, by taking over a double median of Degree to get the preliminary hub network (Zhang et al., 2014). Then, using CytoNCA plugin analyzes the preliminary hub network to get network topological parameters: Betweenness Connectivity (BC), Closeness Connectivity (CNC), and Degree Connectivity (DC) (Yu et al., 2019). We took the excess median of BC, CC, and DC to obtain a hub network. The MCODE application calculated the hub network to get the key module. The genes in key module from MCO are called key genes (Saito et al., 2012).
We performed respectively enrichment analyses of KXYA compound-putative targets and key genes by DAVID (https://david.ncifcrf.gov/) (Dennis et al., 2003). The functional enrichment analysis included the Gene Ontology (GO) and the Kyoto Encyclopedia of Gene and Genome (KEGG), and the GO analysis consists of Biological Process (BP), Molecular Function (MF), and Cellular Component (CC). The filtering of retrieval results is with a threshold value of p < 0.05 and counts in descending order. The main results have been shown as bubble plots. To evaluate the clinical prognostic significance of key genes in hepatitis-related HCC and HCC, we analyzed the correlation between the mRNA expression of key genes and the survival of patients in Kaplan Meier (http://kmplot.com/) (Menyhárt et al., 2018). The patients were divided into high expression group and low expression group, and log-rank p-values were used to evaluate the overall survival (OS) of the two groups.
To explore the expression of key genes in HBV-related HCC, we verified the mRNA expression of key genes in the HBV-related HCC group versus the normal liver tissue group in the GSE62232 dataset, and outliers within the group were removed using a Box plot in SPSS 25.0 software (SPSS Inc., United States). Then we observed the expression of mRNA of key genes between HBV-related HCC tissues group and normal liver tissues group by The Cancer Genome Atlas (TCGA). To further explore the expression trends of key genes, the mRNA expression of key genes was verified in liver cancer data from TCGA and Oncomine (https://oncomine.org) (Rhodes et al., 2004) databases, and the protein situation of key genes was explored in HCC data from the HPA database (https://proteinatlas.org) (Uhlén et al., 2015).
The 5 × 105 HepAD38 cells were seeded in 6-well plates. After being incubated for 24 h, the cells were treated with KXYA (500 μg/ml) for 48 h. Total RNAs were extracted using the RaPure Total RNA Mini Kit (Magen, CN) according to the manufacturer’s instructions. The reverse transcription of total RNA to cDNA was performed with a qPCR RT Master Mix kit (TOYOBO, JAN). The quantitative Real-time PCR (qRT-PCR) was performed using the Real-time PCR Detection System (Agilent Technologies, United States) with the SYBR Green Real-time PCR Master Mix (TOYOBO, JAN). The primers used in this study are provided in Supplementary Table S3, using GAPDH as an internal control gene. The experiments were performed in triplicate and repeated 3 times.
Data for graphing was processed with GraphPad Prism 8.0 software (GraphPad Software Inc., United States). Statistical analyses were performed using the SPSS 25.0 statistical software package. Data were expressed as the mean ± SD, two groups using student’s t-test and more than two groups using one-way ANOVA. Differences between groups are considered to be statistically significant if values of p < 0.05.
A total of 40 HBV-related liver precancerous patients whose serum HBV DNA levels were lower than the detection limit (less than 1 × 102 IU/ml) and who had ultrasound or MRI exams before and after KXYA combined with ETV treatment were studied. And the enhanced MRI images of the typical case before and after KXYA treatment have shown in Figure 1A. Through the ultrasound test, the sizes of liver precancerous nodules before and after KXYA treatment were calculated. The result showed that after KXYA treatment, there were twenty-nine patients (72.5%) whose liver nodules became smaller or unaltered, eight (20%) patients’ liver nodules became larger, and there were three (7.5%) patients who had no response to treatment and processed to HCC. Except for patients who developed HCC, the liver precancerous nodules of the other patients after treatment with KXYA were significantly decreased (p < 0.05, Figure 1B). Meanwhile, we also analyzed the HBV serum indicators of the patients. The results showed that after treatment, the serum HBsAg (p < 0.0001, Figure 1C) and HBeAg (p < 0.01, Figure 1D) of these patients were significantly decreased, but there were not significant changes of serum HBsAb (Figure 1E), HBeAb (Figure 1F), and HBcAb (Figure 1G). The detailed information on HBV serum virology indicators were shown in Supplementary Table S4. These results indicated that KXYA could effectively inhibit the process of HBV-related HCC.
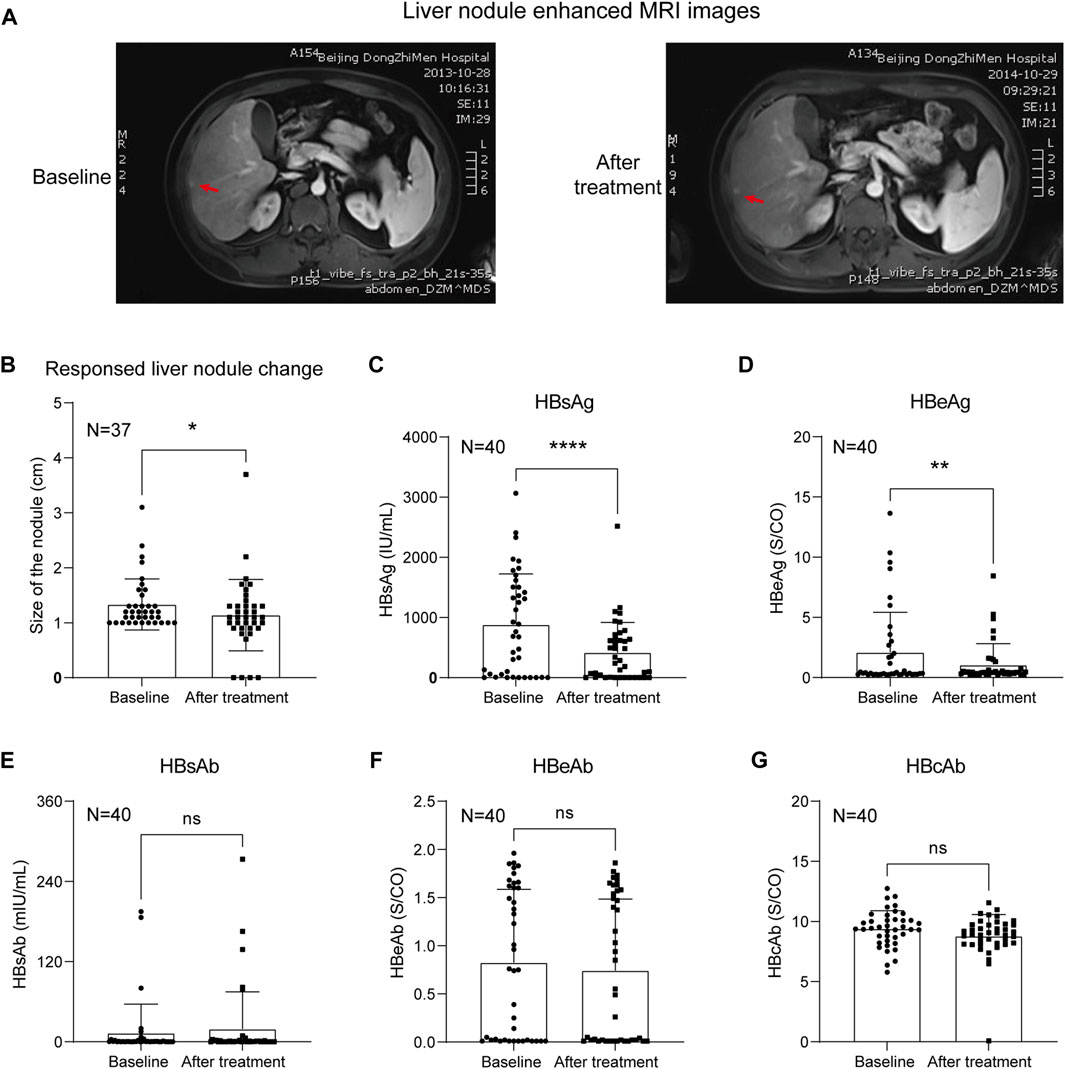
FIGURE 1. KXYA combined ETV treatment decreased the liver nodules of CHB patients. (A) The typical enhanced MRI image of the liver nodule in treatment case, red arrows pointed to the nodules. (B) Analysis of the liver nodules’ change in patients based on ultrasound. (C) Patients’ serological index HBsAg. (D) Patients’ serological index HBeAg. (E) Patients’ serological index HBsAb. (F) Patients’ serological index HBeAb. (G) Patients’ serological index HBcAb. Data are presented as the mean ± SD, p-value was calculated by performing paired Student’s t-test, *p < 0.05, **p < 0.01, ****p < 0.0001.
To confirm the inhibition effect of KXYA on HBV-related HCC, we treated HepAD38 cells with KXYA and detected the cell viability and migration activity. The CCK-8 result showed that, after treatment with different concentrations of the KXYA, the cell viability was significantly decreased in dose-dependent (Figure 2A), and the half-maximal inhibitory concentration (IC50) of KXYA with 48 h of treatment was 60 μg/ml (Figure 2B). Accordingly, for the no-HBV hepatoma cells including HepG2, Huh7, and SK-Hep1, KXYA also exhibited an inhibiting effect (Figure 2C). Meanwhile, the result of the cell wound scratch assay showed HepAD38 had a slower healing rate after KXYA treatment (Figure 2D). These results further supported the clinic curative effect of KXYA on HBV-related HCC via inhibiting cell proliferation and migration abilities, which might act by targeting both virus and cells.
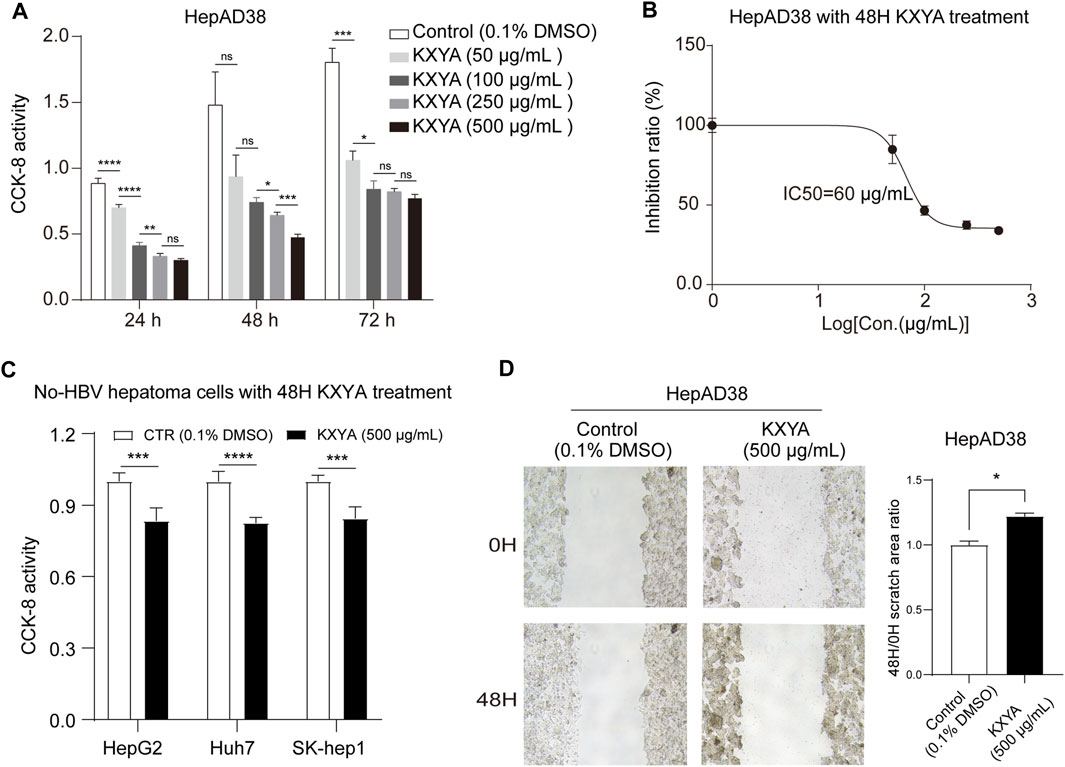
FIGURE 2. KXYA suppressed proliferation and migration of HepAD38 cells. (A) HepAD38 cells was treated with time- and dose-increased KXYA, and the cell viability was detected with CCK-8. (B) HepAD38 cells was treated with dose-increased concentration of KXYA for 48 h and the inhibitory concentration 50 (IC50) was calculated. (C) HepG2, Huh7, and SK-Hep1 cells were treated with KXYA (500 μg/ml) for 48 h, and the cell viability was detected with CCK-8. (D) The representative images and statistical analysis of migration assay for HepAD38 cells at 0 and 48 h with KXYA treatment. Data are presented as the mean ± SD, p-value was calculated by performing un-pair Student’s t-test, *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****p < 0.0001; ns, not significant.
The cell proliferation decrease is usually caused by cell cycle arrest or cell death (Vermeulen et al., 2003). We found that KXYA treatment could result in a solidified nuclear morphology for cellular morphology and increased the cell apoptosis count ratio (Figure 3A). To confirm how KXYA functioned, we performed cell apoptosis and cycle tests. The flow cytometry test showed that the ratio of apoptotic and necrotic cells were both increased after KXYA treatment (Figure 3B). Furthermore, the cell cycle analysis result showed that the G0/G1 phase was decreased, the S phase was unaltered, and the G2/M phase was increased after KXYA treatment (Figure 3C). The G2/M cell cycle arrest always causes cell apoptosis. These results indicated that KXYA could lead to G2/M cell cycle arrest and cause HepAD38 cells apoptosis.
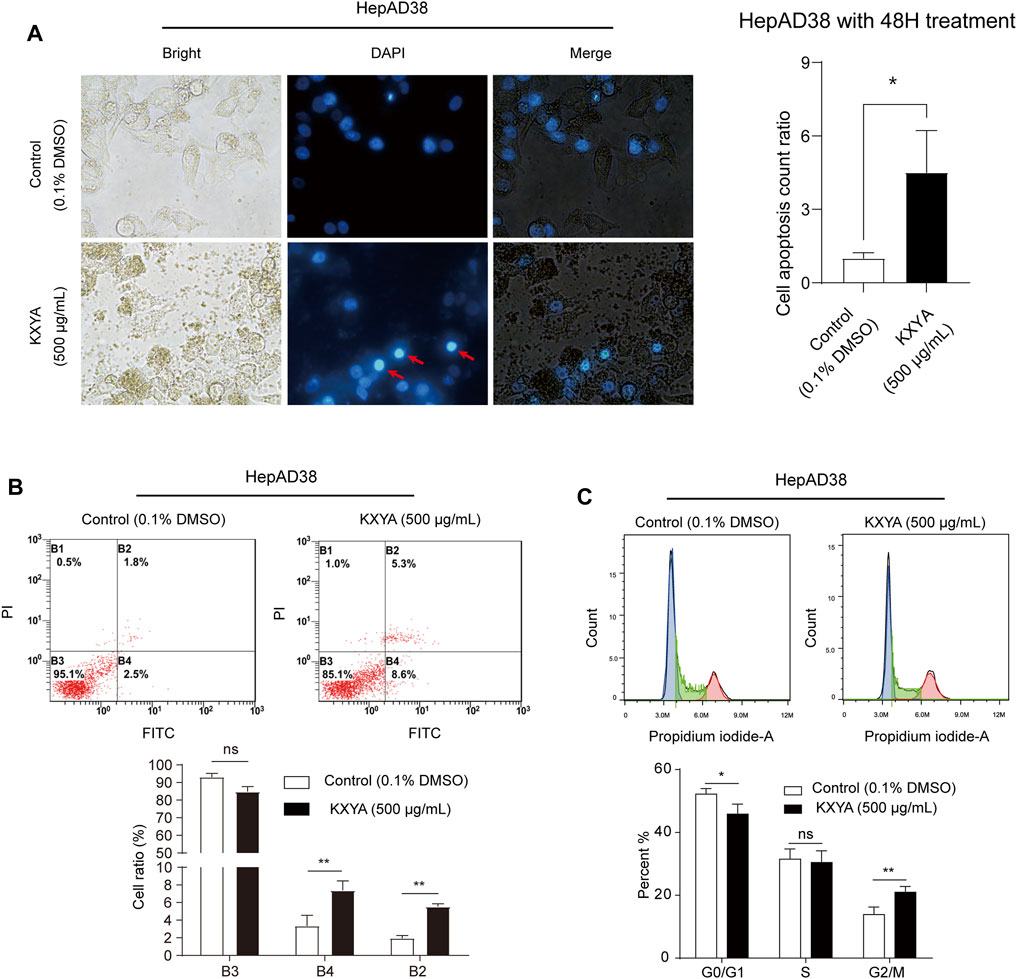
FIGURE 3. KXYA promoted apoptosis of HepAD38 cells. (A) HepAD38 cells was treated with KXYA (500 μg/ml) for 48 h, the representative images were DAPI fluorescence, bright and merge images, and statistical analysis of cell apoptosis count ratio. (B) HepAD38 cells was treated with KXYA (500 μg/ml) for 48 h, then performed PI/Annexin V-FITC fluorescent staining and flow cytometry. (C) HepAD38 cells was treated with KXYA (500 μg/ml) for 48 h, then performed PI fluorescent staining and flow cytometry. Data are presented as the mean ± SD, p-value was calculated by performing un-pair Student’s t-test, *, p < 0.05; **, p < 0.01; ns, not significant.
We firstly analyzed the four herbs in KXYA by CNKI and TCMSP databases, and the keywords were Chaihu, Huangqi, Shanyao, and Jiezi. The detailed information on all components in KXYA was in Supplementary Table S5. Next, according to the active components, we identified a total of 1,610 putative targets in KXYA by SwissTargetPrediction and STITCH analysis (Supplementary Table S6). We next analyzed HBV-related HCC disease targets, and subsequently obtained 5,819 disease targets of HBV-related HCC (Supplementary Table S7). To get the pathological grade-related genes of HBV-related HCC, we next used the GSE62232 dataset to perform a WGCNA analysis. There were 20 samplers in the sample clustering (Figure 4A), and we set a soft threshold power of β = 14 (scale-free R2 = 0.85) to ensure a scale-free network (Figure 4B). After merging similar clusters, we identified modules that contained similar gene patterns (Figure 4C). From the top-5 heatmap of module-trait relationships (Figure 4D), we found that the 2 modules, Coral and Antiquewhite, were the most correlated with Edmonson’s pathological grade (Supplementary Table S8). Also, the gene significances for Edmonson in Coral and Antiquewhite modules were plotted, and the minuscule p-values indicated that they were highly correlated to the disease (Figure 4E). The Coral module positively correlated with Edmonson contained 745 genes, and the Antiquewhite module negatively correlated with Edmonson contained 6,034 genes, which were in Supplementary Table S9.
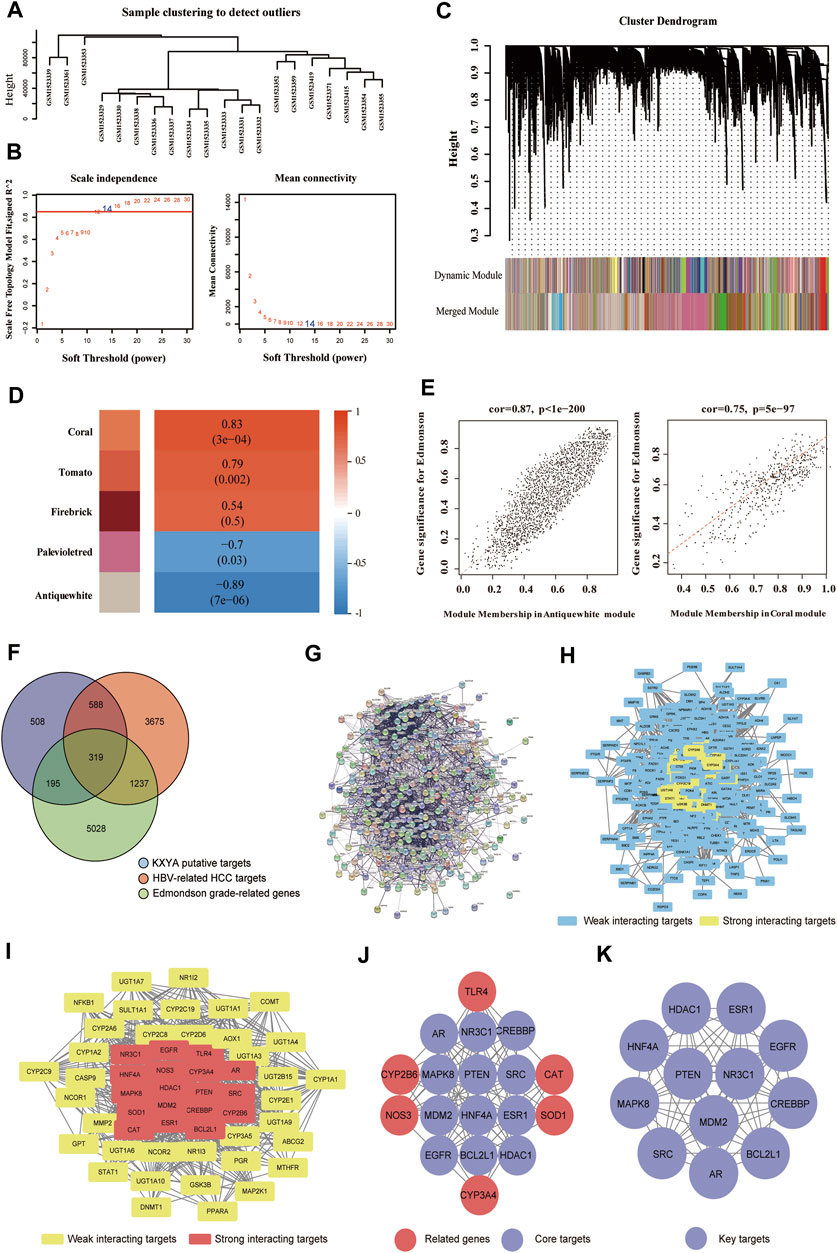
FIGURE 4. Obtained the key targets of KXYA on HBV-related HCC by network pharmacology analysis. (A) The sample clustering of the samples used in GSE62232 database. (B) The scale-free network analysis for samples in GSE62232 database. (C) The clustering dendrograms and modules for samples in GSE62232 database. (D) The heatmap of the top-5 modules, ranked by correlation between module genes and clinical traits of pathological grade. (E) The scatter plot of genes in the Coral and Antiquewhite modules. (F) The overlapped genes (OGEs) of KXYA treatment on HBV-related HCC, the blue circle represents KXYA putative targets, the red circle represents HBV-related HCC targets, and the green circle represents Edmonson grade-related genes. (G) The protein-protein interaction network of OGEs in STRING. (H) The protein-protein interaction network of OGEs in Cytoscape, the blue rectangles represent weak interacting target genes, the yellow rectangles represent strong interacting target genes. (I) The preliminary hub network of OGEs, the yellow rectangles represent weak interacting target genes, and the red rectangles represent strong interacting target genes. (J) The hub network of OGEs, the red circles represent related target genes, and the purple circles represent core target genes. (K) The Key module of OGEs, the purple circles represent key target genes. Pearson’s correlation test was used to calculate a correlation between two variable.
After taking the intersection of putative targets, we obtained 319 OGEs (Figure 4F) and constructed a PPI network of OGEs through the STRING database (Figure 4G). While, in the PPI network, there were 316 proteins imported into the Cytoscape to further obtain interaction networks (Figure 4H). Filtering with the network topological parameters, as Degree >30, we got a preliminary hub network of 54 interacting nodes (Figure 4I). With setting BC > 0.011, CC > 0.458, and DC > 39.5, we identified 18 highly connected nodes as the hub network (Figure 4J). We further analyzed the hub network with the MCODE plugin and finally obtained the key module with 12 genes, which were AR, BCL2L1, CREBBP, EGFR, ESR1, HDAC1, HNF4A, MAPK8, MDM2, NR3C1, PTEN, and SRC (Figure 4K).
The GO and KEGG analyses were performed for the putative targets and 12 key targets by the David database, and the specific information was in Supplementary Table S10. The GO enrichment analysis revealed that the GO-BP of KXYA putative target genes concentrated primarily on the oxidation-reduction process, signal transduction, and positive regulation of transcription from RNA polymerase II promoter (Figure 5A). And the KEGG enrichment analysis suggested that KXYA putative target genes were primarily associated with the metabolic, pathways in cancer and neuroactive ligand-receptor interaction (Figure 5B). To further explore the biological function, the key target genes were significantly related to multiple GO-BP, including negative regulation of the apoptotic process, positive regulation of transcription from RNA polymerase II promoter, and positive regulation of cell proliferation (Figure 5C). And the pathway enrichment results suggested that the key targets were mostly involved in pathways of cancer, prostate cancer, and thyroid hormone signaling pathway (Figure 5D). Based on the aforementioned results, both the putative targets and key targets of KXYA are enriched in pathways in cancer, hepatitis B, viral carcinogenesis, focal adhesion, microRNAs in cancer, proteoglycans in cancer, and PI3K-Akt signaling.
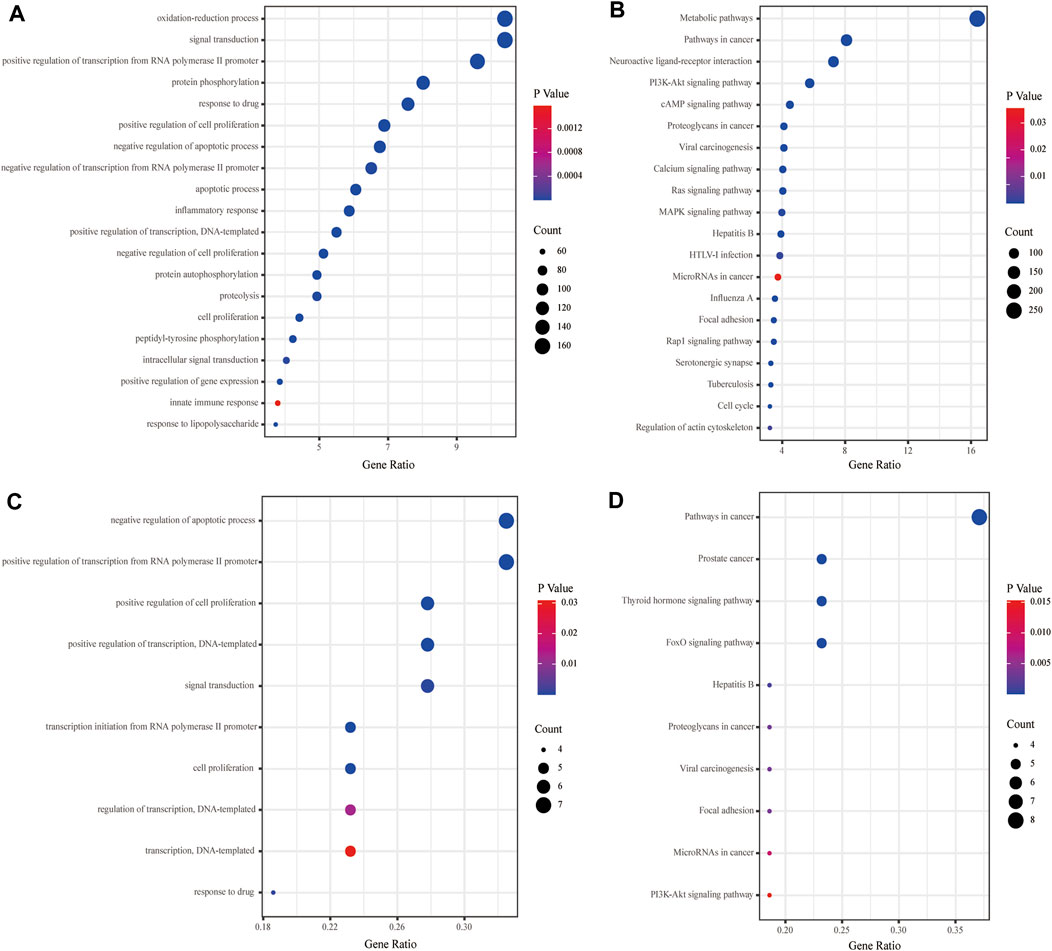
FIGURE 5. The functional enrichment analyses of the KXYA putative-target genes and the key genes. (A) Enriched biological processes of the KXYA putative-target genes. (B) Enriched KEGG pathways of the KXYA putative-target genes. (C) Enriched biological processes of the key genes. (D) Enriched KEGG pathways of the key genes.
To further explore the correlations between the key genes and HBV-related HCC, we observed the relationship between key genes and prognosis in TCGA data. There were 364 patients with liver cancer in TCGA database, among which 111 patients had the risk factors of hepatitis virus and no alcohol consumption. When analyzing the overall survival of the two groups of patients, the basic information and risk factors of patients and the pathology of liver cancer were consistent. The survival analysis for the HCC patients with hepatitis risk factor were performed and the results showed that patients with higher expression of ESR1, MAPK8, HNF4A, BCL2L1, PTEN, and EGFR had a better 5-year overall survival (Figures 6A–F), while those with lower expression of HDAC1 genes had a better prognosis (Figure 6L). Meanwhile, the prognosis of key genes in all TCGA HCC samples was also analyzed, and different from the previous analysis, the 5-year overall survival prognosis of HCC patients was better in the higher expression group of AR, CREBBP, EGFR, ESR1, HNF4A, MDM2, NR3C1, and PTEN (Supplementary Figures S2A–H), while the prognosis was better in the lower expression group of HDAC1 and SRC (Supplementary Figures S2K,L). Overall, these results indicated that the expression of the key genes was related to HBV-related HCC patients’ survival and played an important role during hepatocarcinogenesis.

FIGURE 6. The survival prognostic analysis of the key genes in hepatitis-related HCC. (A) The overall survival analysis of ESR1. (B) The overall survival analysis of MAPK8. (C) The overall survival analysis of HNF4A. (D) The overall survival analysis of BCL2L1. (E) The overall survival analysis of PTEN. (F) The overall survival analysis of EGFR. (G) The overall survival analysis of NR3C1. (H) The overall survival analysis of AR. (I) The overall survival analysis of CREBBP. (J) The overall survival analysis of MDM2. (K) The overall survival analysis of SRC. (L) The overall survival analysis of HDAC1.The association of the key genes’ expression in tumor tissues and the prognosis of hepatitis-related HCC patients (n = 111) in TCGA database. Survival was calculated using Kaplan-Meier’s method and compared using log-rank test.
We further analyzed the expression of the key genes in the public databases. In the GSE62232 dataset, the results showed that compared to normal liver tissues, BCL2L1, CREBBP, HDAC1, MDM2, and NR3C1 genes were significantly upregulated in HBV-related HCC tissues, while AR, EGFR, ESR1, HNF4A, MAPK8, and PTEN genes were significantly downregulated (Figure 7A). In the TCGA database, compared to normal liver tissues, HDAC1 and SRC were significantly upregulated in HBV-related HCC tissues, and AR, CREBBP, EGFR, ESR1, MAPK8, MDM2, NR3C1, and PTEN genes’ expression were significantly downregulated (Figure 7B), and the same results were observed between para-HCC and HCC tissues in all TCGA samples (Figure 7C). In the Oncomine database, we analyzed the expression of the key genes between normal liver tissues and HCC tissues, the results showed that among eight datasets, the expressions of BCL2L1, HNF4A, MDM2, NR3C1, and SRC were elevated in HCC tissues, while AR, CREBBP, EGFR, ESR1, MAPK8, and PTEN expressions were decreased (Figure 7D). The above analyses were performed at the mRNA level. At the protein level, we analyzed the expression of the key targets in the HPA database. The results of immunohistochemistry indicated that the protein expression of AR, BCL2L1, CREBBP, EGFR, MAPK8, and PTEN was downregulated (Supplementary Figures S3A–F), while NR3C1, HNF4A, and HDAC1 were upregulated in the HCC sample compared to normal liver tissues (Supplementary Figures S3J–L).
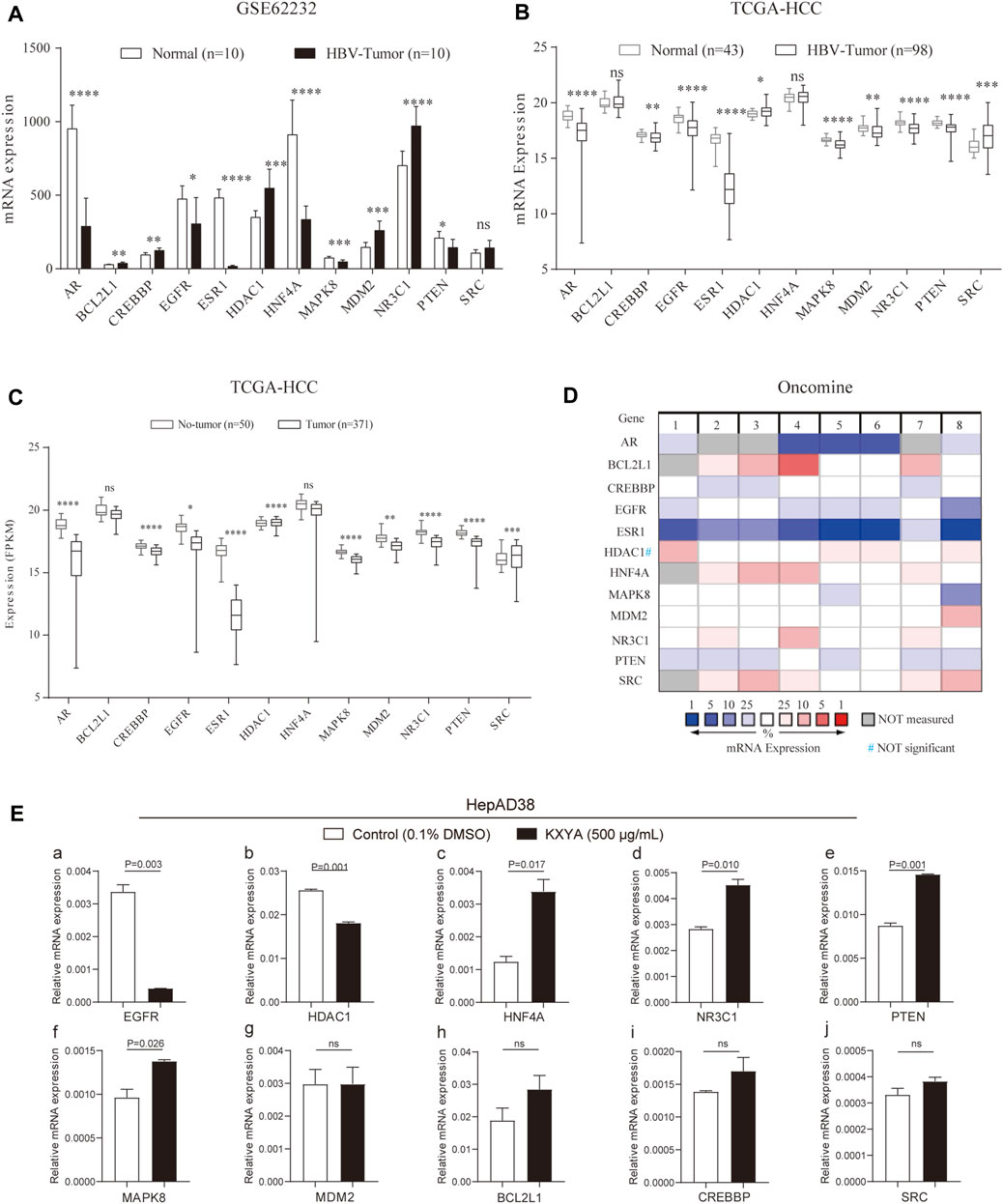
FIGURE 7. The mRNA expression of the key genes in public databases and HepAD38cells with KXYA treatment. (A) The mRNA expression of the key genes in GSE62232. (B) The mRNA expression of the key genes in HBV-related HCC of TCGA. (C) The mRNA expression of the key genes in HCC of TCGA. (D) The mRNA expression of the key genes in HCC of Oncomine. (E) HepAD38 cells was treated with KXYA (500 μg/ml) for 48 h and the key genes’ mRNA expression were detected by qRT-PCR, GAPDH was used as internal reference. Data are presented as the mean ± SD, p-value was calculated by performing un-pair Student’s t-test, ns, not significant.
Finally, to confirm the regulating effect of KXYA on target genes, we treated HepAD38 cells with KXYA (500 μg/ml) for 48 h and detected the target genes’ expression via qRT-PCR. And the results showed that among the target genes, EGFR and HDAC1 were downregulated after KXYA treatment (Figures 7A,B,E), while HNF4A, NR3C1, PTEN, and MAPK8 were upregulated (Figures 7C–F). These results indicated that the key targets’ expression alteration might play an important role during the HCC process. And during the treatment of KXYA on HBV-related HCC, expression of EGFR, HDAC1, HNF4A, MAPK8, NR3C1, and PTEN could be regulated, which were considered as further research targets.
HBV infection is one of the main reasons for hepatocarcinogenesis (Deng et al., 2016), and abnormalities of multiple genes are involved in the occurrence and progression of HBV-related HCC (Gómez-Moreno and Garaigorta, 2017). Currently, the treatment for HBV-related HCC is mainly the standardized treatment of HCC combined with anti-HBV therapy. However, due to the presence of virus reactivation, tumor recurrence, and other conditions, the clinic’s curative effect is not good enough (Li and Wang, 2016). In particular, for these low-level viremia CHB patients, even with ETV monotherapy, the incidence of HCC was still high, which suggested the importance of other more effective therapies (Kim et al., 2017; Yin et al., 2021). TCM is composed of a variety of compounds, with the characteristics of multi-target and holistic therapeutic effect (Zhou et al., 2014), which may be conducive to the treatment of HBV-related HCC. Here, we explored the function and mechanism of TCM formula KXYA on HBV-related HCC.
For hepatocarcinogenesis, there were several retrospective studies on liver nodules based on imaging have shown that the canceration rate of intrahepatic nodules without intervention was about 15%–35% after a median follow-up of 1 year (Motosugi et al., 2011; Kim et al., 2012; Hyodo et al., 2013). In our study, the canceration rate after the intervention of KXYA combined with ETV was 7.5% and there were 72.5% of patients had stable or smaller liver nodules. Furthermore, we found that KXYA could significantly inhibit the proliferation and migration abilities, and promote apoptosis of HepAD38 cells, and could also inhibit the cell viability of no-HBV hepatoma cells in vitro experiments. These results indicated that KXYA had a broad-spectrum effect on anti-HCC, and its inhibition effect on HBV-related HCC might be related to the regulation of the viability and apoptosis of HBV-infected or cancerous hepatocytes. Furthermore, the patients in our study have been treated with ETV monotherapy for a long time previously, which made the serum HBV DNA levels of patients before and after KXYA treatment all less than 100 IU/ml to detect. While other HBV serum indicators could be detected. And we found that HBsAg and HBeAg of patients were both decreased after KXYA treatment, while there was no significant change of HBsAb and HBeAb. The above results suggested KXYA might exert the function of accelerating HBV clearance by promoting the death of pathological cells infected with HBV. However, it was not clear whether KXYA has direct anti-HBV effects or functions on host immune activation, which needed further randomized controlled clinical trials, in vitro and in vivo experiments.
Meanwhile, the KXYA formula had a wide range of pharmacological targets, which made it is difficult to further investigate its efficacy mechanism in depth. Thus, we then used methods of network pharmacology and WGCNA to explore the mechanism of KXYA treatment on HBV-related HCC. We first constructed an interaction network between KXYA and HBV-related HCC, and after taking intersections with the targets of the disease and drugs, 12 key genes of KXYA treatment on HBV-related HCC were obtained, including AR, BCL2L1, CREBBP, EGFR, ESR1, HDAC1, HNF4A, MAPK8, MDM2, NR3C1, PTEN, and SRC. To further explore the expression and function of these key genes in HBV-related HCC, we performed GO and KEGG analysis and the results showed that the biological functions of potential targets and key genes of KXYA were mainly associated with pathways in cancer, hepatitis B, viral carcinogenesis, focal adhesion, microRNAs in cancer, proteoglycans in cancer, and PI3K-Akt signaling, which indicated the closed correlation between the key genes to HBV-related HCC. We next analyzed the expression of key genes in GSE62232 and TCGA databases, and the results showed that compared with normal liver tissues, the expression of AR, EGFR, ESR1, MAPK8, and PTEN were downregulated in HBV-related HCC tissues, and HDAC1 expression was upregulated. Meanwhile, we observed that in hepatitis-related HCC, patients with higher expression of BCL2L1, EGFR, ESR1, HNF4A, MAPK8, and PTEN, and lower expression of HDAC1 had a better clinical prognosis. These results indicated that the key genes we obtained might play an important role in HBV-related HCC.
Additionally, according to previous reports, most of the key genes we obtained exert certain anti- or promoting HCC functions. In HBV-related HCC, the decreased expression of HNF4A enhances potential oncogenic GALNT10 protein activity (Wu et al., 2015). In the early stages of HCC, HNF4A could inhibit cell proliferation of HCC through the miR-122-adam17 pathway (Yang et al., 2020), and downregulation of HNF4A could lead to loss of hepatocyte characteristics (Ramesh and Ganesan, 2016). The higher expression of MAPK8 is correlated to the lower tumor recurrence for HCC patients (Gao et al., 2018), and upregulation of MAPK8 could inhibit the proliferation, migration, and invasion abilities of mouse HCC cell lines (Zhang et al., 2016). NR3C1 encodes glucocorticoid receptors and is considered to be a tumor suppressor gene, which controls cell proliferation, differentiation, and apoptosis (Matthews et al., 2015). PTEN plays a critical role in antiviral innate immunity and the development of HCC, and HBV infection could downregulate the expression of PTEN via increasing N6 methyladenosine modification of PTEN RNA, which leads to its degradation (Kim et al., 2020). The HBx promotes cell proliferation by disturbing the cross-talk between PTEN and miR-181a in HBV-related hepato-carcinogenesis (Tian et al., 2017). EGFR (Crouchet et al., 2022) and HDAC1 (Rivas et al., 2021) have a role in promoting hepatocarcinogenesis. The upregulation of EGFR promotes the progression of HCC and leads to sorafenib resistance (Pang et al., 2019). HDAC1 expression is correlated with downregulation of PTEN, and poor prognosis of HCC patients (Wang et al., 2017).
PTEN, HNF4A, HDAC1, EGFR, and MAPK8 were involved in proliferation, migration, and apoptosis in HCC. And by qRT-PCR assay, we found that KXYA treatment could upregulate the expression of HNF4A, MAPK8, NR3C1, and PTEN, and downregulated the expression of EGFR and HDAC1 in HepAD38 cells. Through previous research and our experimental results, KXYA could induce cell apoptosis by increasing the mRNA expression of PTEN and HNF4A, thereby inhibiting the proliferation of HBV related hepatoma cell lines. To conclude, these key genes may be potential targets for HBV-related HCC pathogenesis and therapy of KXYA. However, we need to further explore the expression and function of the key genes with KXYA treatment in vivo with clinic samples and animal models.
In summary, our results demonstrated that the KXYA formula could exert the treatment effect on HBV-related HCC by regulating hepatocyte proliferation, migration, and apoptosis. During the treatment, the expression of HNF4A, MAPK8, NR3C1, PTEN, EGFR, and HDAC1 was regulated by KXYA, indicating their important roles during the tumorigenesis and progression of HBV-related HCC and might be the targets for the clinical diagnosis and therapy.
The datasets analyzed during the current study are available from the corresponding author on reasonable request. All the relevant data is provided within the paper and its supporting information files.
This study was approved by medical ethics regulations of the Medical Ethical Committee of Dongzhimen Hospital, Beijing University of Chinese Medicine (Beijing, China), and the ethical batch number of this study is DZMEC-KY-2019-131.
Conceptualization: YY, HD, XZ, and GC. Data curation: HC, ZL, NZ. Experiments: XC, XZ, HC, QJ, and YL. Formal analysis: JZ, HC, XL, and XY. Methodology: GC, XC, and MZ. Writing-original draft and Manuscript revision: HD, XZ, XC, and HC
This work was supported by Fundamental Research Funds for the Central Universities, Beijing University of Chinese Medicine, (Grant Number 2021-JYB-XJSJJ055); and (Grant Number 2021-JYB-XJSJJ056); and the China National Science and Technology major projects, 13th 5-year plan (Grant Number 2018ZX10725505).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.985084/full#supplementary-material
Amaddeo, G., Cao, Q., Ladeiro, Y., Imbeaud, S., Nault, J., Jaoui, D., et al. (2015). Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut 64 (5), 820–829. doi:10.1136/gutjnl-2013-306228
Amberger, J., Bocchini, C., Schiettecatte, F., Scott, A., and Hamosh, A. (2015). OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43, D789–D798. doi:10.1093/nar/gku1205
Brown, G. R., Hem, V., Katz, K. S., Ovetsky, M., Wallin, C., Ermolaeva, O., et al. (2015). Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 43, D36–D42. Database issue). doi:10.1093/nar/gku1055
Cao, X., Zao, X., Xue, B., Chen, H., Zhang, J., Li, S., et al. (2021). The mechanism of TiaoGanYiPi formula for treating chronic Hepatitis B by network pharmacology and molecular docking verification. Sci. Rep. 11 (1), 8402. doi:10.1038/s41598-021-87812-9
Chen, X., Zhang, L., Zheng, S., Zhang, T., Li, M., Zhang, X., et al. (2015). Hepatitis B virus X protein stabilizes Cyclin D1 and increases Cyclin D1 nuclear accumulation through ERK-mediated inactivation of GSK-3β. Cancer Prev. Res. 8 (5), 455–463. (Philadelphia, Pa.). doi:10.1158/1940-6207.capr-14-0384
Cong, W., Bu, H., Chen, J., Dong, H., Zhu, Y., Feng, L., et al. (2016). Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J. Gastroenterol. 22 (42), 9279–9287. doi:10.3748/wjg.v22.i42.9279
Couri, T., and Pillai, A. (2019). Goals and targets for personalized therapy for HCC. Hepatol. Int. 13 (2), 125–137. doi:10.1007/s12072-018-9919-1
Crouchet, E., Li, S., Sojoodi, M., Bandiera, S., Fujiwara, N., El Saghire, H., et al. (2022). Hepatocellular carcinoma chemoprevention by targeting the angiotensin-converting enzyme and EGFR transactivation. JCI insight 7 (13), e159254. doi:10.1172/jci.insight.159254
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–W364. doi:10.1093/nar/gkz382
Deng, M., Hou, J., Hu, J., Wang, S., Chen, M., Chen, L., et al. (2016). Hepatitis B virus mRNAs functionally sequester let-7a and enhance hepatocellular carcinoma. Cancer Lett. 383 (1), 62–72. doi:10.1016/j.canlet.2016.09.028
Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4 (5), R60. doi:10.1186/gb-2003-4-9-r60
Gao, X., Shan, W., Liu, X., Zhang, J., Zheng, J., and Yao, H. (2018). JNK1/2 and ERK1/2 provides vital clues about tumor recurrence and survival in hepatocellular carcinoma patients. Future Oncol. 14 (24), 2471–2481. doi:10.2217/fon-2018-0171
GBD 2019 Diseases and Injuries Collaborators (2020). Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the global burden of disease study 2019. Lancet (London, Engl. 396 (10258), 1204–1222. doi:10.1016/s0140-6736(20)30925-9
Gómez-Moreno, A., and Garaigorta, U. (2017). Hepatitis B virus and DNA damage response: Interactions and consequences for the infection. Viruses 9 (10), E304. doi:10.3390/v9100304
Hu, L., and Xiao, H. (2018). Development and research advance of pharmacognosy field based on CNKI. Zhongguo Zhong yao za zhi = China J. Chin. materia medica 43 (4), 689–695. doi:10.19540/j.cnki.cjcmm.20171208.004
Hyodo, T., Murakami, T., Imai, Y., Okada, M., Hori, M., Kagawa, Y., et al. (2013). Hypovascular nodules in patients with chronic liver disease: risk factors for development of hypervascular hepatocellular carcinoma. Radiology 266 (2), 480–490. doi:10.1148/radiol.12112677
International Working Party (1995). Terminology of nodular hepatocellular lesions. Hepatology 22 (3), 983–993. doi:10.1016/0270-9139(95)90324-0
Kim, G., Imam, H., Khan, M., Mir, S., Kim, S., Yoon, S., et al. (2020). HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73, 533. doi:10.1002/hep.31313
Kim, J. H., Sinn, D. H., Kang, W., Gwak, G. Y., Paik, Y. H., Choi, M. S., et al. (2017). Low-level viremia and the increased risk of hepatocellular carcinoma in patients receiving entecavir treatment. Hepatology 66 (2), 335–343. doi:10.1002/hep.28916
Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., et al. (2016). PubChem substance and compound databases. Nucleic Acids Res. 44 (D1), D1202–D1213. doi:10.1093/nar/gkv951
Kim, Y., Lee, W., Park, M., Kim, S., Rhim, H., and Choi, D. (2012). Hypovascular hypointense nodules on hepatobiliary phase gadoxetic acid-enhanced MR images in patients with cirrhosis: Potential of DW imaging in predicting progression to hypervascular HCC. Radiology 265 (1), 104–114. doi:10.1148/radiol.12112649
Kohl, M., Wiese, S., and Warscheid, B. (2011). Cytoscape: software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303. doi:10.1007/978-1-60761-987-1_18
Ladner, S. K., Otto, M. J., Barker, C. S., Zaifert, K., Wang, G. H., Guo, J. T., et al. (1997). Inducible expression of human Hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41 (8), 1715–1720. doi:10.1128/AAC.41.8.1715
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559. doi:10.1186/1471-2105-9-559
Lee, H. M., and Banini, B. A. (2019). Updates on chronic HBV: Current challenges and future goals. Curr. Treat. Options Gastroenterol. 17 (2), 271–291. doi:10.1007/s11938-019-00236-3
Li, F., Wang, Z., Hu, F., and Su, L. (2020). Cell culture models and animal models for HBV study. Adv. Exp. Med. Biol. 1179, 109–135. doi:10.1007/978-981-13-9151-4_5
Li, L., and Wang, H. (2016). Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 379 (2), 191–197. doi:10.1016/j.canlet.2015.07.018
Li, X., Zhang, J., Yang, Z., Kang, J., Jiang, S., Zhang, T., et al. (2014). The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. J. Hepatol. 60 (5), 975–984. doi:10.1016/j.jhep.2013.12.014
Li, X., Zhou, D., Chi, X., Li, Q., Wang, L., Lu, B., et al. (2020). Entecavir combining Chinese herbal medicine for HBeAg-positive chronic Hepatitis B patients: a randomized, controlled trial. Hepatol. Int. 14 (6), 985–996. doi:10.1007/s12072-020-10097-z
Matthews, L., Berry, A., Morgan, D., Poolman, T., Bauer, K., Kramer, F., et al. (2015). Glucocorticoid receptor regulates accurate chromosome segregation and is associated with malignancy. Proc. Natl. Acad. Sci. U. S. A. 112 (17), 5479–5484. doi:10.1073/pnas.1411356112
Menyhárt, O., Nagy, Á., and Győrffy, B. (2018). Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 5 (12), 181006. doi:10.1098/rsos.181006
Motosugi, U., Ichikawa, T., Sano, K., Sou, H., Onohara, K., Muhi, A., et al. (2011). Outcome of hypovascular hepatic nodules revealing no gadoxetic acid uptake in patients with chronic liver disease. J. Magn. Reson. Imaging 34 (1), 88–94. doi:10.1002/jmri.22630
Neuveut, C., Wei, Y., and Buendia, M. A. (2010). Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 52 (4), 594–604. doi:10.1016/j.jhep.2009.10.033
Pang, L., Xu, L., Yuan, C., Li, X., Zhang, X., Wang, W., et al. (2019). Activation of EGFR-KLF4 positive feedback loop results in acquired resistance to sorafenib in hepatocellular carcinoma. Mol. Carcinog. 58 (11), 2118–2126. doi:10.1002/mc.23102
Pinero, J., Ramirez-Anguita, J. M., Sauch-Pitarch, J., Ronzano, F., Centeno, E., Sanz, F., et al. (2020). The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48 (D1), D845–D855. doi:10.1093/nar/gkz1021
Qian, X., Chen, Z., Chen, S., Liu, L., and Zhang, A. (2020). Integrated analyses identify immune-related signature associated with qingyihuaji formula for treatment of pancreatic ductal adenocarcinoma using network pharmacology and weighted gene Co-expression network. J. Immunol. Res. 2020, 7503605. doi:10.1155/2020/7503605
Ramesh, V., and Ganesan, K. (2016). Integrative functional genomic delineation of the cascades of transcriptional changes involved in hepatocellular carcinoma progression. Int. J. Cancer 139 (7), 1586–1597. doi:10.1002/ijc.30195
Rhodes, D., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, N.Y.) 6 (1), 1–6. doi:10.1016/s1476-5586(04)80047-2
Rivas, M., Johnston, M., Gulati, R., Kumbaji, M., Margues Aguiar, T., Timchenko, L., et al. (2021). HDAC1-Dependent repression of markers of hepatocytes and P21 is involved in development of pediatric liver cancer. Cell. Mol. Gastroenterol. Hepatol. 12 (5), 1669–1682. doi:10.1016/j.jcmgh.2021.06.026
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13. doi:10.1186/1758-2946-6-13
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nat. Methods 9 (11), 1069–1076. doi:10.1038/nmeth.2212
Schulze, K., Imbeaud, S., Letouzé, E., Alexandrov, L., Calderaro, J., Rebouissou, S., et al. (2015). Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47 (5), 505–511. doi:10.1038/ng.3252
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 54, 1–3131 30 33. doi:10.1002/cpbi.5 30
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613. doi:10.1093/nar/gky1131
Szklarczyk, D., Santos, A., von Mering, C., Jensen, L. J., Bork, P., and Kuhn, M. (2016). STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44 (D1), D380–D384. doi:10.1093/nar/gkv1277
Tian, Y., Xiao, X., Gong, X., Peng, F., Xu, Y., Jiang, Y., et al. (2017). HBx promotes cell proliferation by disturbing the cross-talk between miR-181a and PTEN. Sci. Rep. 7, 40089. doi:10.1038/srep40089
Uhlén, M., Fagerberg, L., Hallström, B., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Sci. (New York, N.Y.) 347 (6220), 1260419. doi:10.1126/science.1260419
Vermeulen, K., Berneman, Z. N., and Van Bockstaele, D. R. (2003). Cell cycle and apoptosis. Cell Prolif. 36 (3), 165–175. doi:10.1046/j.1365-2184.2003.00267.x
Wang, H., Kohashi, K., Yoshizumi, T., Okumura, Y., Tanaka, Y., Shimokawa, M., et al. (2017). Coexpression of SALL4 with HDAC1 and/or HDAC2 is associated with underexpression of PTEN and poor prognosis in patients with hepatocellular carcinoma. Hum. Pathol. 64, 69–75. doi:10.1016/j.humpath.2017.03.007
Wang, L., Tan, N., Wang, H., Hu, J., Diwu, W., and Wang, X. (2020). A systematic analysis of natural α-glucosidase inhibitors from flavonoids of Radix scutellariae using ultrafiltration UPLC-TripleTOF-MS/MS and network pharmacology. BMC Complement. Med. Ther. 20 (1), 72. doi:10.1186/s12906-020-2871-3
Wu, Q., Liu, H., Liu, Y., Liu, W., Pan, D., Zhang, W., et al. (2015). Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in Hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 290 (2), 1170–1185. doi:10.1074/jbc.M114.601203
Xi, S. Y., and Minuk, G. Y. (2018). Role of traditional Chinese medicine in the management of patients with hepatocellular carcinoma. World J. Hepatol. 10 (11), 799–806. doi:10.4254/wjh.v10.i11.799
Yang, G., Zhang, M., Zhao, Y., Pan, Y., Kan, M., Li, J., et al. (2020). HNF-4α inhibits hepatocellular carcinoma cell proliferation through mir-122-adam17 pathway. PLoS One 15 (3), e0230450. doi:10.1371/journal.pone.0230450
Yin, G. Q., Li, J., Zhong, B., Yang, Y. F., and Wang, M. R. (2021). New therapeutic options for persistent low-level viremia in patients with chronic Hepatitis B virus infection: Increase of entecavir dosage. World J. Gastroenterol. 27 (8), 666–676. doi:10.3748/wjg.v27.i8.666
Yu, G., Luo, Z., Zhou, Y., Zhang, L., Wu, Y., Ding, L., et al. (2019). Uncovering the pharmacological mechanism of Carthamus tinctorius L. on cardiovascular disease by a systems pharmacology approach. Biomed. Pharmacother. = Biomedecine Pharmacother. 117, 109094. doi:10.1016/j.biopha.2019.109094
Zhang, Y., Guo, X., Wang, D., Li, R., Li, X., Xu, Y., et al. (2014). A systems biology-based investigation into the therapeutic effects of Gansui Banxia Tang on reversing the imbalanced network of hepatocellular carcinoma. Sci. Rep. 4, 4154. doi:10.1038/srep04154
Zhang, Y., Wu, J., Wang, Z., Xia, D., Li, G., You, Y., et al. (2016). Inhibitory effects of shRNA on expression of JNK1 and migration and invasion in mouse hepatocellular carcinoma cell lines mediated by ultrasound-targeted microbubble destruction. Biomed. Pharmacother. = Biomedecine Pharmacother. 78, 1–7. doi:10.1016/j.biopha.2015.12.026
Zhou, X., Menche, J., Barabási, A., and Sharma, A. (2014). Human symptoms-disease network. Nat. Commun. 5, 4212. doi:10.1038/ncomms5212
Zhu, R., Seto, W., Lai, C., and Yuen, M. (2016). Epidemiology of hepatocellular carcinoma in the asia-pacific region. Gut Liver 10 (3), 332–339. doi:10.5009/gnl15257
HCC Hepatocellular carcinoma
HBV hepatitis B virus
CHB chronic hepatitis B
cccDNA covalently closed circular DNA
TCM traditional Chinese medicine
KXYA KangXianYiAi
WGCNA weighted gene co-expression network analysis
ETV Entecavir
DMEM dulbecco’s modified eagle medium
FBS fetal bovine serum
DMSO dimethylsulfoxide
PI propidium iodide
OB oral bioavailability
DL drug-likeness
OGEs overlapped genes
PPI protein-protein interaction
BC Betweenness Connectivity
CNC Closeness Connectivity
DC Degree Connectivity
GO Gene Ontology
KEGG Kyoto Encyclopedia of Gene and Genome
BP Biological Process
MF Molecular Function
CC Cellular Component
OS overall survival
TCGA The Cancer Genome Atlas
qRT-PCR Quantitative Real-time PCR
AR Androgen receptor
BCL2L1 BCL2 like 1
CREBBP CREB binding protein
EGFR Epidermal growth factor receptor
ESR1 Estrogen receptor 1
HDAC1 Histone deacetylase 1
HNF4A Hepatocyte nuclear factor 4 alpha
MAPK8 Mitogen-activated protein kinase 8
MDM2 MDM2 proto-oncogene
NR3C1 Nuclear receptor subfamily 3 group C member 1
PTEN phosphatase and tensin homolog
SRC SRC proto-oncogene, non-receptor tyrosine kinase
Keywords: traditional Chinese medicine, HBV-related hepatocellular carcinoma, network pharmacological, bioinformatics, KangXianYiAi formula
Citation: Cao X, Chen H, Li Z, Li X, Yang X, Jin Q, Liang Y, Zhang J, Zhou M, Zhang N, Chen G, Du H, Zao X and Ye Y (2022) Network pharmacology and in vitro experiments-based strategy to investigate the mechanisms of KangXianYiAi formula for hepatitis B virus-related hepatocellular carcinoma. Front. Pharmacol. 13:985084. doi: 10.3389/fphar.2022.985084
Received: 03 July 2022; Accepted: 01 August 2022;
Published: 05 September 2022.
Edited by:
Dongwen Lyu (Lv), The University of Texas Health Science Center at San Antonio, United StatesReviewed by:
Danmeng Li, University of Florida, United StatesCopyright © 2022 Cao, Chen, Li, Li, Yang, Jin, Liang, Zhang, Zhou, Zhang, Chen, Du, Zao and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang Chen, Z2NoZW5AaG1zLmhhcnZhcmQuZWR1; Hongbo Du, ZHVob25nYm90Y21AMTI2LmNvbQ==; Xiaobin Zao, QTM0MTdAYnVjbS5lZHUuY24=; Yong’an Ye, eWV5b25nYW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.