
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 19 July 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.955218
This article is part of the Research Topic New Insights into the Histone Lysine Specific Demethylase (LSD1) Inhibitors for Cancer Treatment View all 4 articles
Epigenetics has emerged as a prime focus area in the field of cancer research. Lysine-specific demethylase 1A (LSD1), the first discovered histone demethylase, is mainly responsible for catalysing demethylation of histone 3 lysine 4 (H3K4) and H3K9 to activate or inhibit gene transcription. LSD1 is abnormally expressed in various cancers and participates in cancer proliferation, apoptosis, metastasis, invasion, drug resistance and other processes by interacting with regulatory factors. Therefore, it may serve as a potential therapeutic target for cancer. This review summarises the major oncogenic mechanisms mediated by LSD1 and provides a reference for developing novel and efficient anticancer strategies targeting LSD1.
Cancer is a life-threatening disease that seriously threatens human health and life (Liu et al., 2020). The number of new cancer cases exceeded 19.29 million in 2021, with approximately 9.96 million deaths reported worldwide (Siegel et al., 2021). Mechanisms underlying tumorigenesis are usually driven by genetic mutations, especially mutations that occur in cancer suppressor genes and (or) proto-oncogenes (Wang et al., 2018; Fang et al., 2020). Several recent studies have shown that abnormal epigenetic modifications, such as DNA and histone modifications, play essential roles in regulating tumorigenesis and the proliferation and differentiation of cancer stem cells (Chen J. et al., 2020; Ghasemi et al., 2021; Zhao and Peng, 2022). In addition, epigenetic reprogramming and unlocking phenotypic plasticity have been reported as the hallmarks of cancer (Gupta et al., 2019; Hanahan, 2022; Saltarella et al., 2022).
Histone octamers are composed of two H2A, H2B, H3, and H4 subunits, and each core histone has a folding region and an amino-terminal domain. Various covalent modifications such as acetylation, phosphorylation, methylation, ubiquitination, and glycosylation can occur at the amino terminus of histones, which affect the chromatin structure and activation or inhibition of transcription (Yang et al., 2021a; Yang et al., 2021b). Shi and Tsukada (2013) discovered the first histone demethylase in 2004, a lysine-specific histone demethylase 1 (LSD1). The discovery of LSD1 indicates that histone methylation modification is a dynamic process that can regulate not only the methylation of histones but also the interaction of histones with other functional proteins. Numerous studies have shown that LSD1 is responsible for regulating the transcriptional activation and repression of specific genes, X chromosome inactivation and viral pathogenesis and plays an essential role in embryonic and cancer development (Miller et al., 2021). This review summarises the research progress of LSD1 in various cancers and highlights the mechanism of action of LSD1 underlying its anticancer activity (Figure 1).
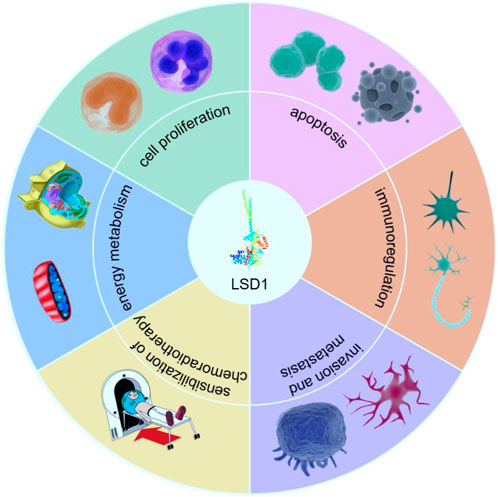
FIGURE 1. The role of LSD1 in tumorigenesis and development. LSD1 participates in cancer progression by regulating multiple critical physiological processes, such as the proliferation, invasion, metastasis, energy metabolism, immune regulation, and drug resistance of cells (drawn using tools from PNGBAG; https://www.pngbag.com/, copyright ©2020 PNGBAG.COM).
LSD1, also known as KDM1A, AOF2, BHC110, KIAA0601, NPAO, and p110b, is a flavin adenine dinucleotide (FAD)-dependent monoamine oxidase located in the nucleus (Anand and Marmorstein, 2007). It mainly consists of three domains, namely, the N-terminal SWIRM (Swi3p/Rsc8p/Moira), C-terminal amine oxidase (AOL) and central Tower domains (Figure 2) (Karakaidos et al., 2019; Majello et al., 2019). The SWIRM domain is highly conserved and can recognise and bind to histones, whereas the AOL domain mainly binds to histone substrates (Chen et al., 2006; Castelli et al., 2018). The Tower domain extends two parallel alpha helices outward from the AOL domain to facilitate interactions with other co-regulators/cofactors, including FAD. Typically, the SWIRM and AOL domains are connected and overlap to form a spherical shape (Chen et al., 2006; Kaniskan et al., 2018). The Tower domain is formed by two helices extending outward from the C-terminal AOL domain, thereby dividing the AOL domain into the following two parts: the FAD-binding domain and a catalytically active centre. The main difference between LSD1 and LSD2 is that the N-terminus of LSD2 has a zinc finger domain (Zn-CW), which is necessary for binding to its methylated substrate (Zhang et al., 2013). LSD1+8a, a subtype generated by alternative splicing of LSD1, is involved in the differentiation of neuronal cells via demethylation of H3K9me2/1 (Jotatsu et al., 2017).
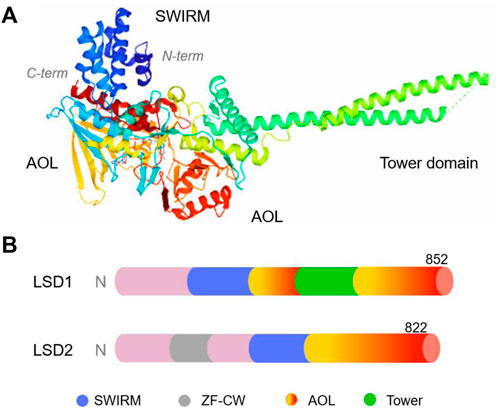
FIGURE 2. The structure of human lysine-specific demethylase (LSD) protein. (A) Three-dimensional structure of human LSD1 protein (PDB No.: 2Z5U), Copyright © 2007 Elsevier Inc. (B) Schematic illustration of the structure of LSD1 and LSD2.
The primary biological function of LSD1 is the demethylation of histone or non-histone proteins (Gu et al., 2020). Some standard arginine and lysine methylation sites on histones or non-histones proteins include histone 3 lysine 4 (H3K4), H3K9, H3K27, H3K36, H3K79, and histone 4 lysine 20 (H4K20). Methylation at H3K4, H3K36, and H3K79 sites is involved in the repression of gene transcription (Hyun et al., 2017). LSD1 can remove the monomethyl (Me1) or dimethyl (Me2) group from lysine residues and can specifically demethylate the H3K4 site to inhibit its transcriptional activity (Wang and Cole, 2020). In addition, LSD1 can interact with REST/CoREST/co-regulatory factors to exert a regulatory effect (non-methylation function) (Gu et al., 2020; Perillo et al., 2020).
Abnormal expression of LSD1 in various cancers promotes cancer progression and is closely related to the survival and prognosis of patients (Figure 3) (Ramírez-Ramírez et al., 2020; Zhang et al., 2020). Given that LSD1 is a potential therapeutic target for cancer, many targeted inhibitors of LSD1 with excellent anticancer activity have been reported, and some of these inhibitors have entered clinical trials (Maes et al., 2018; Wimalasena et al., 2020; Fang et al., 2021; Yang et al., 2022).
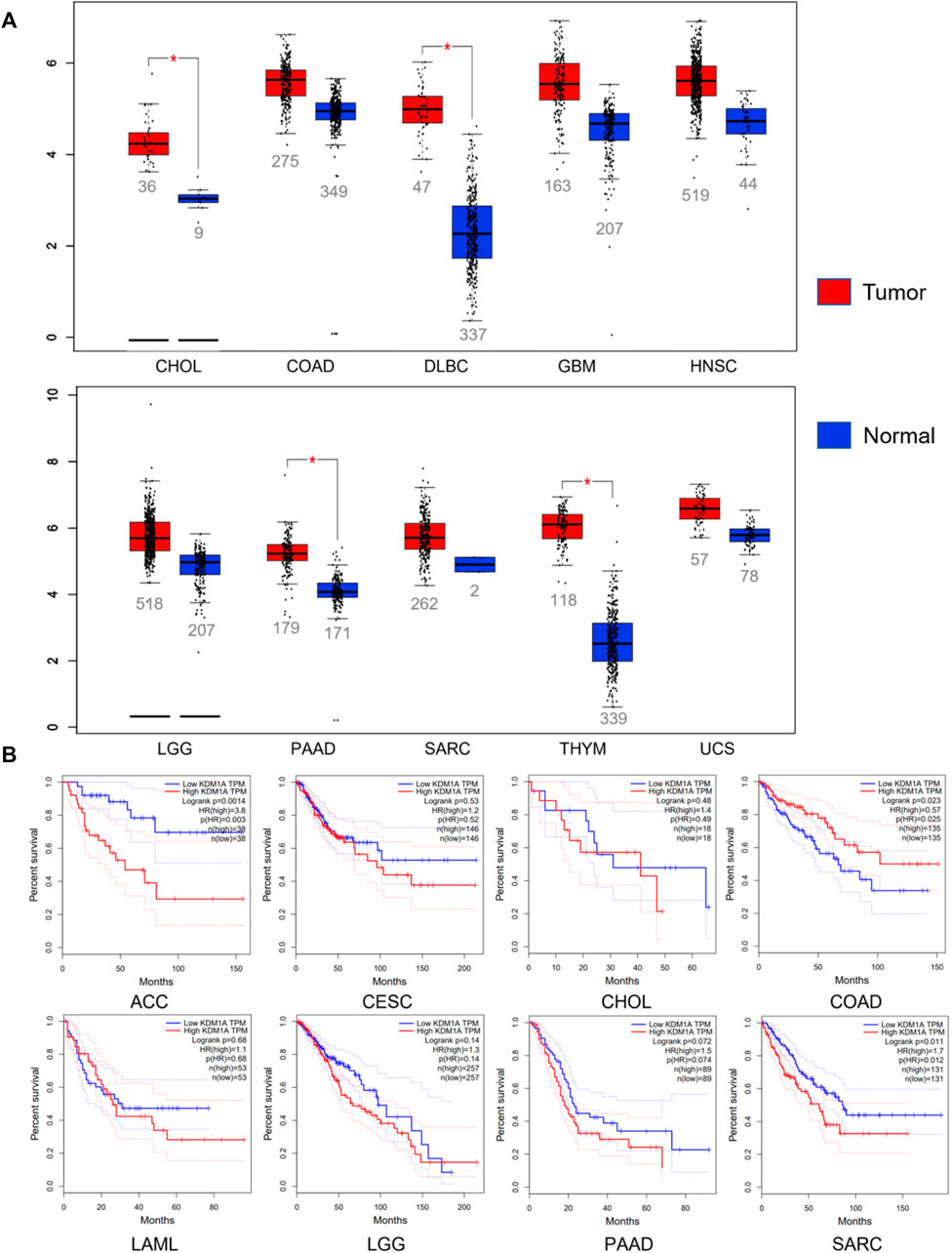
FIGURE 3. Relationship between LSD1 and cancers. (A) LSD1 is aberrantly expressed in various cancers. (B) Analysis of the correlation between LSD1 expression and overall survival (OS). Data from the GEPIA database, http://gepia.cancer-pku.cn/index.html.
p53 is a well-known cancer suppressor and transcriptional activator regulated by many post-translational modifications, including lysine methylation. LSD1 promotes the interaction of p53 with the coactivator 53BP1 (p53-binding protein 1) by removing the monomethyl (K370me1) and dimethyl (K370me2) groups at K370 (Chen L. et al., 2020). However, the direct interaction of LSD1 with p53 inhibits p53-mediated transcriptional activation and apoptosis, resulting in altering the chromatin structure and inhibiting the development of the cancer marker alpha-fetoprotein (AFP) (Wen and Wang, 2022). Therefore, downregulating the expression of LSD1 and (or) inhibiting its activity can promote cancer cell apoptosis. The intrinsically disordered C-terminal domain of p53 inhibits LSD1 activity, and direct interaction between the two proteins may contribute to their functional crosstalk (Speranzini et al., 2017).
Overexpression of LSD1 can strongly inhibit p53 in prostate cancer (PCa) and promote androgen-independent (AI) transformation of PCa and LNCaP cells in an androgen-deficient setting (Li X. et al., 2016). Low doses of the LSD1 inhibitor HCI-2509 can significantly alter the cell cycle and expression of p53, MYCN and hypoxia pathway-related genes in neuroblastoma (Gupta et al., 2018).
Terminal deoxynucleotidyl transferase (TdT)-interacting factor 1 (TdIF1) is a ubiquitously expressed protein that binds to TdT polymerase (Zhang et al., 2018). It is abundantly expressed in lung cancer, is associated with a poor prognosis and is a potential cancer-promoting factor and therapeutic target (Liu et al., 2021). Silencing or inhibiting TdIF1 can inhibit the migration and invasion of cancer cells and cancer growth. LSD1 binds to TdIF1 and is recruited to the E-cadherin promoter region, where it activates transcription to induce epithelial–mesenchymal transdifferentiation (EMT) and promote the invasion and migration of cancer cells (Zhang et al., 2019; Liu et al., 2021). Therefore, simultaneous inhibition or silencing of LSD1 and TdIF1 exerts synergistic effects on anticancer proliferation, migration and invasion.
Growth factor independence 1 (GFI1) and the closely related protein GFI1B are major regulators of both early haematopoiesis and haematopoietic stem cells, and their aberrant activation has been implicated in human medulloblastoma and haematological malignancies (Beauchemin and Möröy, 2020; Ravasio et al., 2020). GFI1 and GFI1B are major proteins that interact with LSD1, recruit histone-modifying enzymes to the promoters and enhancers of target genes through the N-terminal SNAG domain to regulate the expression of target genes, such as HDAC and LSD1, and play an essential role in cell proliferation and differentiation (Maiques-Diaz et al., 2018; Beauchemin and Möröy, 2020; Tatsumi et al., 2020). In addition, they are involved in the recruitment of CoREST complexes to chromatin in myeloid cells (Van Bergen and Van Der Reijden, 2019). An irreversible inhibitor of LSD1, T-3775440, selectively suppressed the proliferation of SCLC cells overexpressing GFI1B (Takagi et al., 2017).
Polo-like kinase 1 (PLK1) is a serine/threonine kinase that is a key regulator of eukaryotic cell division. Because PLK1 is highly expressed in cancer cells and is associated with the poor prognosis of many cancers, it has emerged as a new target for developing many anticancer drugs (Li Z. et al., 2020; Yu et al., 2021).
LSD1 can recognise and bind to the promoter region of PLK1 to regulate its expression, which regulates the expression of cell division-related genes. However, the interaction between p53 and PLK1 is negatively regulated (Li Y. et al., 2020; Jung et al., 2021). p53 inhibits transcription at the PLK1 promoter, whereas PLK1 inhibits the function of p53 by directly binding to it or inactivating it by promoting its degradation (Dufies et al., 2021; Zhang C. et al., 2022; Hirschler-Laszkiewicz et al., 2022). Therefore, LSD1 may regulate the cell cycle and proliferation through the p53/PLK1 signalling axis.
Hypoxia-inducible factor 1-alpha (HIF-1α) is a key protein that regulates the expression and synthesis of cytokines and growth mediators in cells during hypoxia (Yang et al., 2021c). It plays an essential role in the formation of tumor blood vessels and the proliferation, metastasis, invasion, apoptosis, energy metabolism, and drug resistance of tumor cells (Li G. et al., 2021). The expression of HIF-1α is positively correlated with the malignancy and poor prognosis of cancers. Therefore, inhibiting the expression of HIF-1α and blocking hypoxia signal transduction mediated by it may help develop novel therapeutic strategies for cancers.
The stability of HIF-1α is regulated by lysine methylation. SET7/9 methyltransferase adds a methyl group to HIF-1α, which subsequently triggers protein degradation via the ubiquitin–proteasome pathway. However, LSD1 demethylates HIF1α at K391, thus protecting HIF-1α from ubiquitin-mediated protein degradation, and directly inhibits PHD2-mediated hydroxylation of HIF-1α (Baek and Kim, 2016; Lee et al., 2017; Yang et al., 2017). In addition, LSD1 is highly expressed in cancers, potently stabilises HIF1α and enhances the transcriptional activity of downstream target genes, such as VEGF, which can induce cancer angiogenesis. Cellular senescence is a state of permanent cell cycle arrest, which strongly affects the development, invasion and prognosis of cancers. In glioblastoma, silencing/inhibiting LSD1 downregulates HIF-1α, which inhibits the growth and migration of cancer cells and induces cellular senescence (Saccà et al., 2019).
Snail is an important regulator of EMT-associated zinc finger structures. It is involved in the immune escape, immune regulation, drug resistance of cancer cells, and maintenance of cancer cell stemness. And it also recruits LSD1 to suppress the expression of breast cancer susceptibility gene 1 (BRCA1) (Li H. M. et al., 2020; Pan et al., 2021). The interaction of LSD1 with the SNAG domain at the N-terminus of Snail inhibits the expression of E-cadherin.
An inhibitor of LSD1, parnate, blocks Snail-dependent inhibition of the E-cadherin promoter and inhibits the migration and invasion of cancer cells without affecting their proliferation (Ferrari-Amorotti et al., 2013; Ferrari-Amorotti et al., 2014). It has been reported that parnate treatment inhibited bone marrow homing/transplantation of Snail2-expressing K562 cells. Furthermore, the knockdown of LSD1 in triple-negative breast cancer (TNBC) can significantly inhibit the proliferation and invasion of cancer cells and metastasis (Bai et al., 2017). LSD1 interacts with Snail1 to mediate the ectopic expression of Snail1, thus increasing the risk of AML in mice (Carmichael et al., 2020).
Zinc finger proteins (ZNFs) are the largest family of transcription factors in the human genome with extensive and important molecular biological functions. In addition to regulating the transcription of downstream target genes by interacting with different functional domains and trans-regulatory elements, ZNFs can recruit other chromosome modifiers and interact with different partner proteins to inhibit or promote gene transcription. ZFPs are abnormally expressed in cancer cells and participate in the occurrence and development of cancers by regulating gene transcription and translation.
ZNF217, a C2H2 zinc finger transcription factor, acts as a key effector in stimulating embryonic immortalisation and oncogenicity in various cancer-related processes. Downregulation of ZNF217 can inhibit the proliferation, invasion and EMT of HCC cells (Si et al., 2019). Mechanistic studies have shown that ZNF217 interacts with CoREST, LSD1, HDAC, and C-terminal binding protein (CtBP) to inhibit the transcriptional activity of CDH1 (Li Y. et al., 2021). SP-2509 inhibits the viability of cancer cells by blocking the binding of LSD1 to ZNF217 by inhibiting LSD1-independent function in castration-resistant PCa (Sehrawat et al., 2018).
Immune checkpoints are inhibitory signals in the immune system, which mainly maintain normal immune function by regulating immune activation (Sun et al., 2020; Zhang and Sun, 2020). The binding of programmed death-ligand 1/2 (PD-L1/PD-L2) expressed on cancer cells with programmed cell death protein 1 (PD-1) expressed on cytotoxic T cells triggers inhibitory signalling, leading to immune escape owing to T cell depletion (Dyck and Mills, 2017).
Knockdown/inhibition of LSD1 in cancer cells upregulates the expression of repetitive elements, including ERV, induces dsRNA stress and activates type 1 interferons, which stimulate anticancer T cell immunity and suppress cancer growth (Sheng et al., 2018). The expression of LSD1 in TNBC is inversely correlated with immunoregulatory factors such as cytotoxic T cell-attracting chemokine (C-C motif) ligand 5 (CCL5). Inhibition of LSD1 increases the expression of effector T cell-attracting chemokine factors and PD-L1 and promotes the trafficking of CD8+ T lymphocytes in the microenvironment of TNBC (Qin et al., 2019). Knockout of LSD1 or inhibition of LSD1 activity can also enhance the immune response of T cells in various cancers, such as cervical, ovarian, gastric, and oral cancers (Soldi et al., 2020; Xu et al., 2021; Alhousami et al., 2022; Shen et al., 2022). Therefore, LSD1 inhibition may be an effective adjuvant for immunotherapy in patients with poorly immunogenic cancers.
Silent information regulator 1 (SIRT1) is a class III histone deacetylase that is dependent on nicotinamide adenine dinucleotide and is involved in the regulation of glucose and lipid metabolism, insulin secretion, oxidative stress, organ metabolism and tumorigenesis (Soni et al., 2021; Jalgaonkar et al., 2022). The highly conserved SWIRM domain of LSD1 interacts with SIRT to form a functional complex, which mediates the activity of SIRT1. The interaction between SIRT1 and LSD1 plays a conserved and cooperative role in H3K4 demethylation to inhibit Notch signalling-mediated gene transcription (Cao et al., 2021). SIRT1 and LSD1 are aberrantly expressed in cancers and play an antagonistic role in DNA repair and mutation acquisition (Boila and Sengupta, 2020; Wang et al., 2020). Moreover, the expression of LSD1 and SIRT1 is negatively correlated with the prognosis and survival of patients.
Mammalian target of rapamycin complex 1 (mTORC1) is an evolutionarily conserved serine/threonine protein kinase responsible for the regulation of cell proliferation and metabolism (Battaglioni et al., 2022; Huang et al., 2022). The activity of mTORC1 is regulated or feedback-regulated by a variety of intracellular signaling factors, such as growth factors, protein levels, energy levels, and intracellular oxygen partial pressure (Wang et al., 2021; Li et al., 2022). mTORC1 can participate in cancer progression by phosphorylating its substrates to regulate intracellular protein synthesis, material metabolism (lipid, nucleotide, and glucose) and autophagy.
mTORC1 is a key regulator of autophagy, and its activity is directly and positively correlated with the level of LSD1 (Li et al., 2019). Activated LSD1 alters the levels of phosphorylated mTOR1 and AKT through the mTOR signaling pathway, thereby reducing autophagy in tumour cells (Feng S. et al., 2016; Ma et al., 2022). Inhibition of LSD1 in drug-resistant leukaemia helps detect aberrantly activated mTORC1; therefore, activation of mTORC1 may be a pro-survival mechanism for cancer cells (Abdel-Aziz et al., 2020). Simultaneous knockdown or pharmacological inhibition of mTORC1 and LSD1 significantly enhanced primary cell differentiation and reduced the proportion of primary human AML cells (Deb et al., 2020).
F-box and WD-40 domain protein 7 (FBXW7) is an important recognition factor in the ubiquitin–proteasome degradation pathway. It is also a typical cancer suppressor, which mediates the ubiquitination and degradation of various oncoproteins and regulates the occurrence and development of malignant cancers.
LSD1 recognises and binds to FBXW7 and inhibits its dimerisation, resulting in autoubiquitination/degradation of FBXW7 (Lan et al., 2019). Inhibiting the activity/expression of LSD1 exerts anticancer effects by stabilising/upregulating the accumulation of FBXW7. Knockdown/silencing of LSD1 or overexpression of FBXW7 in PCa cells can significantly reduce the activity of oncoproteins such as c-MYC and NOTCH-1. The oncogenic activity of LSD1 depends on the interaction of LSD1 with FBXW7, independent of its demethylase activity (Lan et al., 2019), which is supported by functional validation of the allosteric inhibitors SP-2509 and GSK-2879552 (Qin et al., 2020).
Long noncoding RNAs (lncRNAs) are non-protein-coding RNA molecules with a length of >200 nucleotides (Ghafouri-Fard and Taheri, 2020). They are involved in the growth, proliferation, metastasis and drug resistance of cancers and other related processes and are abnormally expressed in various cancers, such as gastric, lung, breast, liver, and endometrial cancers (Chen et al., 2016; Kim et al., 2018; Raeisi et al., 2019). LSD1 can bind to lncRNAs to exert oncogenic effects.
Enhancers of LSD1 interact with lncRNAs and are recruited to the promoter regions of cancer suppressors, such as KLF2, LATS2, and P21, to inhibit their transcriptional activity (Li W. et al., 2016; Zang et al., 2016). LSD1 can activate p62-mediated antioxidative pathways by upregulating certain lncRNAs (Ma et al., 2021). In addition, FOXP4-AS1 and DUXAP8 can recognise and bind to LSD1 and upregulate the expression of LSD1 to accelerate cancer progression (Chen et al., 2019; Gong et al., 2019).
The energy metabolism of cancer cells is significantly different from that of normal cells; that is, despite the presence of sufficient oxygen, cancer cells use glycolysis as the main energy production method, which is called the Warburg effect (Bose et al., 2021; Cook et al., 2021). Overexpressed LSD1 in cancer cells promotes glucose uptake and glycolytic activity and upregulates the expression of GLUT1 and glycolytic enzymes; however, it strongly downregulates the expression of mitochondrial metabolism-related genes (Luo et al., 2021). Knockdown or pharmacological inhibition of LSD1 can activate the transcriptional activity of the gluconeogenesis genes FBP1 and G6Pase, resulting in increased de novo glucose synthesis and decreased intracellular glycogen content (Wang D. et al., 2022). Inhibition of LSD1 activity in oesophageal cancer cells can significantly reduce the extracellular acidification rate (ECAR) and increase the oxygen consumption rate (OCR) and OCR/ECAR ratio (Kosumi et al., 2016). LSD1 may contribute to the malignant behaviour of oesophageal cancer by regulating metabolism, glycolytic pathway, and mitochondrial respiration. In addition, LSD1 plays an important role in regulating adaptive thermogenesis and lipid metabolism and may be a novel target for treating obesity (Wang D. et al., 2022).
The highly conserved Notch signalling pathway regulates essential life processes such as cell differentiation, proliferation and angiogenesis via interaction among adjacent cells or paracrine/endocrine effects (Trindade and Duarte, 2020; Misiorek et al., 2021). Cancer cells use the Notch signalling pathway to form a microenvironment of cancer-promoting factors by promoting the secretion of cancer-related inflammatory cytokines, such as IFN-γ, which regulates immune cell function; and IL-1β and CCL2, which mediate immune infiltration, the release of senescence-related cytokines and immunosuppression, thereby suppressing the immune response (Cortesi et al., 2021; Wang L. et al., 2022; Zhou et al., 2022).
The interaction between LSD1 and NOTCH1 inhibits the expression and downstream signalling of NOTCH1. ORY-1001, a selective inhibitor of LSD1, can activate the Notch pathway and strongly inhibit cancer growth in chemotherapy-resistant PDX models (Augert et al., 2019). However, the expression of LSD1 and NOTCH3 in clinical specimens of HCC is strongly negatively correlated with the survival of patients. This negative correlation may result from the increased expression of LSD1 in cancer-associated fibroblasts (CAFs), which regulate driving NOTCH3-mediated self-renewal of cancer stem cells (Liu et al., 2018).
Phosphatidylinositol-3-kinase/protein kinase B (PI3K/Akt) is a well-known growth and development signalling pathway in organisms, which plays a crucial role in cell proliferation, apoptosis, cell cycle, DNA repair, and protein synthesis (Hoxhaj and Manning, 2020). PI3K/AKT not only mediates life activities such as the proliferation, migration, metastasis, and self-renewal of cancer cells but also regulates the self-renewal ability of cancer stem cells (Yang et al., 2020).
Silencing/knockout of LSD1 inhibited the invasion and metastasis of gastric cancer cells owing to the significant downregulation of VEGF-C, p-PI3K, PI3K, p-AKT, AKT, VEGFR-3, MMP-2, and MMP-9 (Pan et al., 2019). LSD1 can enhance AR signalling and activate the PI3K/AKT pathway in PCa by increasing p85 gene expression in the absence of AR (Wang et al., 2019). Similarly, TCP, an inhibitor of LSD1, can block the binding of LSD1 to the promoter regions of NOTCH3, Hes1, and CR2, thereby reducing Notch and PI3K/Akt/mTOR signalling (Hou et al., 2019). Moreover, high expression of LSD1 was detected in patients with colorectal cancer with mutations in the catalytic subunit of PI3K, and PIK3CA-mutated colorectal cancer cells were found to be dependent on LSD1 for growth (Miller et al., 2020).
The Wnt/β-Catenin signalling pathway plays a key role in regulating cell pluripotency and carcinogenesis and is closely related to important processes such as cancer proliferation, metastasis and chemotherapy resistance (Zhang and Wang, 2020; Liu et al., 2022). LSD1 can activate the Wnt/β-Catenin signalling pathway by downregulating DKK1 in colorectal cancer. After pharmacological inhibition or silencing of LSD1, the translocation of β-Catenin to the nucleus is significantly reduced, and the transcription of the target gene c-Myc is downregulated. Combination therapy with LSD1 inhibitors and 5-FU strongly inhibited Wnt/β-Catenin signalling and DNA synthesis, significantly inhibiting the proliferation, migration, and growth of cancer cells in a tumor xenograft-bearing model (Peng et al., 2020). LSD1 is necessary to emerge cancer stem cells after long-term sorafenib treatment. LSD1 inhibitors downregulate the expression of multiple regulators of the Wnt/β-Catenin signalling pathway and increase the sensitivity of cancer cells to sorafenib (Huang et al., 2017). LSD1 can downregulate the Wnt pathway antagonists APC2 and DKK1 through demethylation in thyroid cancer, thus inhibiting APC2 transcription and activating the HIF-1α/DKK1 axis to regulate cancer progression (Zhang W. et al., 2022).
As the first discovered histone demethylase, LSD1 has promoted the research progress of epigenetics. LSD1 is a key regulator of the proliferation, apoptosis, differentiation, invasion, metastasis, drug resistance, and cancer stemness and hence is a promising therapeutic target for cancers (Feng Z. et al., 2016; Wang et al., 2016; Alsaqer et al., 2017; Cusan et al., 2018; Verigos et al., 2019; Tayari et al., 2021).
This review summarises the cancer-promoting regulatory mechanisms of LSD1 in lung, breast, gastric, colorectal, liver, bladder, leukaemia, and other cancers. LSD1 regulates different signalling pathways, such as the Wnt/β-Catenin, PI3K/AKT signalling, EMT-related, Notch signalling and ubiquitin–proteasome pathway, and target proteins, such as the classic cancer suppressor star protein p53, HIFs, and immune checkpoints, in different types of cancer to mediate cancer progression. However, because the occurrence and development of cancers are very complex biological processes, all oncogenic factors or pathways mediated by LSD1 could not be described in this review. Some examples include SOX2, which maintains cancer cell stemness; sex hormone receptors (AR and ER); the transcriptional repressor ZNF516; retinoic acid-related orphan receptor alpha (RORα); aryl hydrocarbon receptors; ubiquitin-specific protease 28 and miRNAs (Park et al., 2016; Bai et al., 2017; Khanal et al., 2017; Kim et al., 2017; Li et al., 2017; Macheleidt et al., 2018; Cuyàs et al., 2020; Yu et al., 2020; Zhang et al., 2021). However, the detailed mechanisms of LSD1 underlying carcinogenesis remain unclear and warrant further investigation. For example, does or how LSD1 is involved in ferroptosis and pyroptosis, and whether LSD1 interact with SITR6?
To date, many LSD1 inhibitors have been reported, with some having excellent anticancer activity. Among these inhibitors, ORY-1001 and GSK2879552 have entered clinical trials, thus validating the clinical application of LSD1 as a therapeutic target for cancers and incredibly encouraging the development of strategies targeting LSD1 for treating cancers. Furthermore, inhibiting LSD1 protein synthesis or promoting its degradation may be a potential anticancer strategy (Qi et al., 2020). Tumorigenesis patterns may be induced by point-to-net or multi-point-to-net cascades involving numerous regulatory factors/signalling pathways. In the future, a cure for cancer may be developed if LSD1-like master switches involved in the regulation of cascade reactions related to tumorigenesis and cancer development or target proteins with relatively more regulatory functions can be identified or if a multitarget regulation strategy can be developed. Therefore, refining the oncogenic mechanism of LSD1 will help develop promising tumor therapeutic drugs or strategies.
CY and DL conceived and designed the whole project. CY and DL drafted the manuscript. SZ collected and organized relevant literature. LZ, YZ, and ZZ revised the manuscript. All authors reviewed and approved the final manuscript.
The work was supported by the National Natural Science Foundation of China (82104477, U19A2010, and 81891012), special support from China Postdoctoral Science Foundation (2019M663456 and 2019TQ0044), Xinglin Scholar Research Promotion Project of Chengdu University of TCM (BSH2019008), National Interdisciplinary Innovation Team of Traditional Chinese Medicine (ZYYCXTD-D-202209), the Macao Science and Technology Development Fund (FDCT 007/2020/ALC), the Shenzhen-Hong Kong-Macau S&T Program (Category C) (SGDX2020110309420200), and the Research Fund of University of Macau (CPG2022-00005-ICMS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Aziz, A. K., Pallavicini, I., Ceccacci, E., Meroni, G., Saadeldin, M. K., Varasi, M., et al. (2020). Tuning mTORC1 activity dictates the response of acute myeloid leukemia to LSD1 inhibition. Haematologica 105, 2105–2117. doi:10.3324/haematol.2019.224501
Alhousami, T., Diny, M., Ali, F., Shin, J., Kumar, G., Kumar, V., et al. (2022). Inhibition of LSD1 attenuates oral cancer development and promotes therapeutic efficacy of immune checkpoint blockade and YAP/TAZ inhibition. Mol. Cancer Res. 20, 712–721. doi:10.1158/1541-7786.Mcr-21-0310
Alsaqer, S. F., Tashkandi, M. M., Kartha, V. K., Yang, Y. T., Alkheriji, Y., Salama, A., et al. (2017). Inhibition of LSD1 epigenetically attenuates oral cancer growth and metastasis. Oncotarget 8, 73372–73386. doi:10.18632/oncotarget.19637
Anand, R., and Marmorstein, R. (2007). Structure and mechanism of lysine-specific demethylase enzymes. J. Biol. Chem. 282, 35425–35429. doi:10.1074/jbc.R700027200
Augert, A., Eastwood, E., Ibrahim, A. H., Wu, N., Grunblatt, E., Basom, R., et al. (2019). Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci. Signal. 12, eaau2922. doi:10.1126/scisignal.aau2922
Baek, S. H., and Kim, K. I. (2016). Regulation of HIF-1α stability by lysine methylation. BMB Rep. 49, 245–246. doi:10.5483/bmbrep.2016.49.5.053
Bai, J. W., Chen, M. N., Wei, X. L., Li, Y. C., Lin, H. Y., Chen, M., et al. (2017). The zinc-finger transcriptional factor Slug transcriptionally downregulates ERα by recruiting lysine-specific demethylase 1 in human breast cancer. Oncogenesis 6, e330. doi:10.1038/oncsis.2017.38
Battaglioni, S., Benjamin, D., Wälchli, M., Maier, T., and Hall, M. N. (2022). mTOR substrate phosphorylation in growth control. Cell 185, 1814–1836. doi:10.1016/j.cell.2022.04.013
Beauchemin, H., and Möröy, T. (2020). Multifaceted actions of GFI1 and GFI1B in hematopoietic stem cell self-renewal and lineage commitment. Front. Genet. 11, 591099. doi:10.3389/fgene.2020.591099
Boila, L. D., and Sengupta, A. (2020). Evolving insights on histone methylome regulation in human acute myeloid leukemia pathogenesis and targeted therapy. Exp. Hematol. 92, 19–31. doi:10.1016/j.exphem.2020.09.189
Bose, S., Zhang, C., and Le, A. (2021). Glucose metabolism in cancer: The Warburg effect and beyond. Adv. Exp. Med. Biol. 1311, 3–15. doi:10.1007/978-3-030-65768-0_1
Cao, Y., Tang, L., Du, K., Paraiso, K., Sun, Q., Liu, Z., et al. (2021). Anterograde regulation of mitochondrial genes and FGF21 signaling by hepatic LSD1. JCI Insight 6, 147692. doi:10.1172/jci.insight.147692
Carmichael, C. L., Wang, J., Nguyen, T., Kolawole, O., Benyoucef, A., De Mazière, C., et al. (2020). The EMT modulator SNAI1 contributes to AML pathogenesis via its interaction with LSD1. Blood 136, 957–973. doi:10.1182/blood.2019002548
Castelli, G., Pelosi, E., and Testa, U. (2018). Targeting histone methyltransferase and demethylase in acute myeloid leukemia therapy. Onco. Targets. Ther. 11, 131–155. doi:10.2147/ott.S145971
Chen, J., Zhao, J., Ding, J., Wang, Z., Du, J., Wu, C., et al. (2020a). Knocking down LSD1 inhibits the stemness features of colorectal cancer stem cells. Braz J. Med. Biol. Res. 53, e9230. doi:10.1590/1414-431x20209230
Chen, L., Liu, S., and Tao, Y. (2020b). Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 5, 90. doi:10.1038/s41392-020-0196-9
Chen, R. Y., Ju, Q., Feng, L. M., Yuan, Q., and Zhang, L. (2019). The carcinogenic complex lncRNA FOXP4-AS1/EZH2/LSD1 accelerates proliferation, migration and invasion of gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 23, 8371–8376. doi:10.26355/eurrev_201910_19148
Chen, Y., Yang, Y., Wang, F., Wan, K., Yamane, K., Zhang, Y., et al. (2006). Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc. Natl. Acad. Sci. U. S. A. 103, 13956–13961. doi:10.1073/pnas.0606381103
Chen, Z., Li, J. L., Lin, S., Cao, C., Gimbrone, N. T., Yang, R., et al. (2016). cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J. Clin. Invest. 126, 2267–2279. doi:10.1172/jci85250
Cook, K. M., Shen, H., Mckelvey, K. J., Gee, H. E., and Hau, E. (2021). Targeting glucose metabolism of cancer cells with dichloroacetate to radiosensitize high-grade gliomas. Int. J. Mol. Sci. 22, 7265. doi:10.3390/ijms22147265
Cortesi, M., Zanoni, M., Pirini, F., Tumedei, M. M., Ravaioli, S., Rapposelli, I. G., et al. (2021). Pancreatic cancer and cellular senescence: Tumor microenvironment under the spotlight. Int. J. Mol. Sci. 23, 254. doi:10.3390/ijms23010254
Cusan, M., Cai, S. F., Mohammad, H. P., Krivtsov, A., Chramiec, A., Loizou, E., et al. (2018). LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPα-dependent enhancers in AML. Blood 131, 1730–1742. doi:10.1182/blood-2017-09-807024
Cuyàs, E., Gumuzio, J., Verdura, S., Brunet, J., Bosch-Barrera, J., Martin-Castillo, B., et al. (2020). The LSD1 inhibitor iadademstat (ORY-1001) targets SOX2-driven breast cancer stem cells: A potential epigenetic therapy in luminal-B and HER2-positive breast cancer subtypes. Aging (Albany NY) 12, 4794–4814. doi:10.18632/aging.102887
Deb, G., Wingelhofer, B., Amaral, F. M. R., Maiques-Diaz, A., Chadwick, J. A., Spencer, G. J., et al. (2020). Pre-clinical activity of combined LSD1 and mTORC1 inhibition in MLL-translocated acute myeloid leukaemia. Leukemia 34, 1266–1277. doi:10.1038/s41375-019-0659-6
Dufies, M., Verbiest, A., Cooley, L. S., Ndiaye, P. D., He, X., Nottet, N., et al. (2021). Plk1, upregulated by HIF-2, mediates metastasis and drug resistance of clear cell renal cell carcinoma. Commun. Biol. 4, 166. doi:10.1038/s42003-021-01653-w
Dyck, L., and Mills, K. H. G. (2017). Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 47, 765–779. doi:10.1002/eji.201646875
Fang, Y., Yang, C., Teng, D., Su, S., Luo, X., Liu, Z., et al. (2021). Discovery of higenamine as a potent, selective and cellular active natural LSD1 inhibitor for MLL-rearranged leukemia therapy. Bioorg. Chem. 109, 104723. doi:10.1016/j.bioorg.2021.104723
Fang, Y., Yang, C., Yu, Z., Li, X., Mu, Q., Liao, G., et al. (2020). Natural products as LSD1 inhibitors for cancer therapy. Acta Pharm. Sin. B 11, 621–631. doi:10.1016/j.apsb.2020.06.007
Feng, S., Jin, Y., Cui, M., and Zheng, J. (2016a). Lysine-specific demethylase 1 (LSD1) inhibitor S2101 induces autophagy via the AKT/mTOR pathway in SKOV3 ovarian cancer cells. Med. Sci. Monit. 22, 4742–4748. doi:10.12659/msm.898825
Feng, Z., Yao, Y., Zhou, C., Chen, F., Wu, F., Wei, L., et al. (2016b). Pharmacological inhibition of LSD1 for the treatment of MLL-rearranged leukemia. J. Hematol. Oncol. 9, 24. doi:10.1186/s13045-016-0252-7
Ferrari-Amorotti, G., Chiodoni, C., Shen, F., Cattelani, S., Soliera, A. R., Manzotti, G., et al. (2014). Suppression of invasion and metastasis of triple-negative breast cancer lines by pharmacological or genetic inhibition of slug activity. Neoplasia 16, 1047–1058. doi:10.1016/j.neo.2014.10.006
Ferrari-Amorotti, G., Fragliasso, V., Esteki, R., Prudente, Z., Soliera, A. R., Cattelani, S., et al. (2013). Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res. 73, 235–245. doi:10.1158/0008-5472.Can-12-1739
Ghafouri-Fard, S., and Taheri, M. (2020). Long non-coding RNA signature in gastric cancer. Exp. Mol. Pathol. 113, 104365. doi:10.1016/j.yexmp.2019.104365
Ghasemi, S., Xu, S., Nabavi, S. M., Amirkhani, M. A., Sureda, A., Tejada, S., et al. (2021). Epigenetic targeting of cancer stem cells by polyphenols (cancer stem cells targeting). Phytother. Res. 35, 3649–3664. doi:10.1002/ptr.7059
Gong, A., Huang, Z., Ge, H., Cai, Y., and Yang, C. (2019). The carcinogenic complex lncRNA DUXAP8/EZH2/LSD1 accelerates the proliferation, migration and invasion of colorectal cancer. J. buon 24, 1830–1836.
Gu, F., Lin, Y., Wang, Z., Wu, X., Ye, Z., Wang, Y., et al. (2020). Biological roles of LSD1 beyond its demethylase activity. Cell. Mol. Life Sci. 77, 3341–3350. doi:10.1007/s00018-020-03489-9
Gupta, P. B., Pastushenko, I., Skibinski, A., Blanpain, C., and Kuperwasser, C. (2019). Phenotypic plasticity: Driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 24, 65–78. doi:10.1016/j.stem.2018.11.011
Gupta, S., Doyle, K., Mosbruger, T. L., Butterfield, A., Weston, A., Ast, A., et al. (2018). Reversible LSD1 inhibition with HCI-2509 induces the p53 gene expression signature and disrupts the MYCN signature in high-risk neuroblastoma cells. Oncotarget 9, 9907–9924. doi:10.18632/oncotarget.24035
Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46. doi:10.1158/2159-8290.CD-21-1059
Hirschler-Laszkiewicz, I., Festa, F., Huang, S., Moldovan, G. L., Nicolae, C., Dhoonmoon, A., et al. (2022). The human ion channel TRPM2 modulates cell survival in neuroblastoma through E2F1 and FOXM1. Sci. Rep. 12, 6311. doi:10.1038/s41598-022-10385-8
Hou, G., Zhao, Q., Zhang, M., Wang, P., Ye, H., Wang, Y., et al. (2019). LSD1 regulates Notch and PI3K/Akt/mTOR pathways through binding the promoter regions of Notch target genes in esophageal squamous cell carcinoma. Onco. Targets. Ther. 12, 5215–5225. doi:10.2147/ott.S207238
Hoxhaj, G., and Manning, B. D. (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88. doi:10.1038/s41568-019-0216-7
Huang, M., Chen, C., Geng, J., Han, D., Wang, T., Xie, T., et al. (2017). Targeting KDM1A attenuates Wnt/β-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett. 398, 12–21. doi:10.1016/j.canlet.2017.03.038
Huang, W. Q., Zou, Y., Tian, Y., Ma, X. F., Zhou, Q. Y., Li, Z. Y., et al. (2022). Mammalian target of rapamycin as the therapeutic target of vascular proliferative diseases: Past, present, and future. J. Cardiovasc. Pharmacol. 79, 444–455. doi:10.1097/fjc.0000000000001208
Hyun, K., Jeon, J., Park, K., and Kim, J. (2017). Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324. doi:10.1038/emm.2017.11
Jalgaonkar, M. P., Parmar, U. M., Kulkarni, Y. A., and Oza, M. J. (2022). SIRT1-FOXOs activity regulates diabetic complications. Pharmacol. Res. 175, 106014. doi:10.1016/j.phrs.2021.106014
Jotatsu, T., Yagishita, S., Tajima, K., Takahashi, F., Mogushi, K., Hidayat, M., et al. (2017). LSD1/KDM1 isoform LSD1+8a contributes to neural differentiation in small cell lung cancer. Biochem. Biophys. Rep. 9, 86–94. doi:10.1016/j.bbrep.2016.11.015
Jung, Y., Kraikivski, P., Shafiekhani, S., Terhune, S. S., and Dash, R. K. (2021). Crosstalk between Plk1, p53, cell cycle, and G2/M DNA damage checkpoint regulation in cancer: Computational modeling and analysis. NPJ Syst. Biol. Appl. 7, 46. doi:10.1038/s41540-021-00203-8
Kaniskan, H., Martini, M. L., and Jin, J. (2018). Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 118, 989–1068. doi:10.1021/acs.chemrev.6b00801
Karakaidos, P., Verigos, J., and Magklara, A. (2019). LSD1/KDM1A, a gate-keeper of cancer stemness and a promising therapeutic target. Cancers (Basel) 11, E1821. doi:10.3390/cancers11121821
Khanal, T., Choi, K., Leung, Y. K., Wang, J., Kim, D., Janakiram, V., et al. (2017). Loss of NR2E3 represses AHR by LSD1 reprogramming, is associated with poor prognosis in liver cancer. Sci. Rep. 7, 10662. doi:10.1038/s41598-017-11106-2
Kim, J., Piao, H. L., Kim, B. J., Yao, F., Han, Z., Wang, Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715. doi:10.1038/s41588-018-0252-3
Kim, K., Lee, J. M., Yu, Y. S., Kim, H., Nam, H. J., Moon, H. G., et al. (2017). RORα2 requires LSD1 to enhance tumor progression in breast cancer. Sci. Rep. 7, 11994. doi:10.1038/s41598-017-12344-0
Kosumi, K., Baba, Y., Sakamoto, A., Ishimoto, T., Harada, K., Nakamura, K., et al. (2016). Lysine-specific demethylase-1 contributes to malignant behavior by regulation of invasive activity and metabolic shift in esophageal cancer. Int. J. Cancer 138, 428–439. doi:10.1002/ijc.29714
Lan, H., Tan, M., Zhang, Q., Yang, F., Wang, S., Li, H., et al. (2019). LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc. Natl. Acad. Sci. U. S. A. 116, 12311–12320. doi:10.1073/pnas.1902012116
Lee, J. Y., Park, J. H., Choi, H. J., Won, H. Y., Joo, H. S., Shin, D. H., et al. (2017). LSD1 demethylates HIF1α to inhibit hydroxylation and ubiquitin-mediated degradation in tumor angiogenesis. Oncogene 36, 5512–5521. doi:10.1038/onc.2017.158
Li, G., Ko, C. N., Li, D., Yang, C., Wang, W., Yang, G. J., et al. (2021a). A small molecule HIF-1α stabilizer that accelerates diabetic wound healing. Nat. Commun. 12, 3363. doi:10.1038/s41467-021-23448-7
Li, H. M., Bi, Y. R., Li, Y., Fu, R., Lv, W. C., Jiang, N., et al. (2020a). A potent CBP/p300-Snail interaction inhibitor suppresses tumor growth and metastasis in wild-type p53-expressing cancer. Sci. Adv. 6, eaaw8500. eaaw8500. doi:10.1126/sciadv.aaw8500
Li, L., Liu, X., He, L., Yang, J., Pei, F., Li, W., et al. (2017). ZNF516 suppresses EGFR by targeting the CtBP/LSD1/CoREST complex to chromatin. Nat. Commun. 8, 691. doi:10.1038/s41467-017-00702-5
Li, W., Sun, M., Zang, C., Ma, P., He, J., Zhang, M., et al. (2016a). Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 7, e2225. doi:10.1038/cddis.2016.126
Li, X., Li, T., Chen, D., Zhang, P., Song, Y., Zhu, H., et al. (2016b). Overexpression of lysine-specific demethylase 1 promotes androgen-independent transition of human prostate cancer LNCaP cells through activation of the AR signaling pathway and suppression of the p53 signaling pathway. Oncol. Rep. 35, 584–592. doi:10.3892/or.2015.4362
Li, X., Tian, X., Liu, T., Li, M., Wang, W., Wang, P., et al. (2022). Hydroxytyrosol alleviated hypoxia-mediated PC12 cell damage through activating PI3K/AKT/mTOR-HIF-1α signaling. Oxid. Med. Cell. Longev. 2022, 8673728. doi:10.1155/2022/8673728
Li, Y., Tao, L., Zuo, Z., Zhou, Y., Qian, X., Lin, Y., et al. (2019). ZY0511, a novel, potent and selective LSD1 inhibitor, exhibits anticancer activity against solid tumors via the DDIT4/mTOR pathway. Cancer Lett. 454, 179–190. doi:10.1016/j.canlet.2019.03.052
Li, Y., Wu, H., Wang, Q., and Xu, S. (2021b). ZNF217: The cerberus who fails to guard the gateway to lethal malignancy. Am. J. Cancer Res. 11, 3378–3405.
Li, Y., Zhao, Z. G., Luo, Y., Cui, H., Wang, H. Y., Jia, Y. F., et al. (2020b). Dual targeting of Polo-like kinase 1 and baculoviral inhibitor of apoptosis repeat-containing 5 in TP53-mutated hepatocellular carcinoma. World J. Gastroenterol. 26, 4786–4801. doi:10.3748/wjg.v26.i32.4786
Li, Z., Yang, C., Li, X., Du, X., Tao, Y., Ren, J., et al. (2020c). The dual role of BI 2536, a small-molecule inhibitor that targets PLK1, in induction of apoptosis and attenuation of autophagy in neuroblastoma cells. J. Cancer 11, 3274–3287. doi:10.7150/jca.33110
Liu, C., Liu, L., Chen, X., Cheng, J., Zhang, H., Zhang, C., et al. (2018). LSD1 stimulates cancer-associated fibroblasts to drive notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 78, 938–949. doi:10.1158/0008-5472.Can-17-1236
Liu, J., Xiao, Q., Xiao, J., Niu, C., Li, Y., Zhang, X., et al. (2022). Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 7, 3. doi:10.1038/s41392-021-00762-6
Liu, Q., Xiong, J., Xu, D., Hao, N., Zhang, Y., Sang, Y., et al. (2021). TdIF1-LSD1 Axis regulates epithelial-mesenchymal transition and metastasis via histone demethylation of E-cadherin promoter in lung cancer. Int. J. Mol. Sci. 23, 250. doi:10.3390/ijms23010250
Liu, X., Zhou, M., Wang, F., Mubarik, S., Wang, Y., Meng, R., et al. (2020). Secular trend of cancer death and incidence in 29 cancer groups in China, 1990-2017: A joinpoint and age-period-cohort analysis. Cancer Manag. Res. 12, 6221–6238. doi:10.2147/cmar.S247648
Luo, J., Wang, H., Wang, L., Wang, G., Yao, Y., Xie, K., et al. (2021). lncRNA GAS6-AS1 inhibits progression and glucose metabolism reprogramming in LUAD via repressing E2F1-mediated transcription of GLUT1. Mol. Ther. Nucleic Acids 25, 11–24. doi:10.1016/j.omtn.2021.04.022
Ma, L., Xu, A., Kang, L., Cong, R., Fan, Z., Zhu, X., et al. (2021). LSD1-Demethylated LINC01134 confers oxaliplatin resistance through SP1-induced p62 transcription in HCC. Hepatology 74, 3213–3234. doi:10.1002/hep.32079
Ma, T., Li, A., Guo, Y., Li, S., Li, M., Feng, S., et al. (2022). KDM1A/LSD1 as a promising target in various diseases treatment by regulating autophagy network. Biomed. Pharmacother. 148, 112762. doi:10.1016/j.biopha.2022.112762
Macheleidt, I. F., Dalvi, P. S., Lim, S. Y., Meemboor, S., Meder, L., Käsgen, O., et al. (2018). Preclinical studies reveal that LSD1 inhibition results in tumor growth arrest in lung adenocarcinoma independently of driver mutations. Mol. Oncol. 12, 1965–1979. doi:10.1002/1878-0261.12382
Maes, T., Mascaró, C., Tirapu, I., Estiarte, A., Ciceri, F., Lunardi, S., et al. (2018). ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell 33, 495–511. e412. doi:10.1016/j.ccell.2018.02.002
Maiques-Diaz, A., Spencer, G. J., Lynch, J. T., Ciceri, F., Williams, E. L., Amaral, F. M. R., et al. (2018). Enhancer activation by pharmacologic displacement of LSD1 from GFI1 induces differentiation in acute myeloid leukemia. Cell Rep. 22, 3641–3659. doi:10.1016/j.celrep.2018.03.012
Majello, B., Gorini, F., Saccà, C. D., and Amente, S. (2019). Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers (Basel) 11, E324. doi:10.3390/cancers11030324
Miller, S. A., Policastro, R. A., Savant, S. S., Sriramkumar, S., Ding, N., Lu, X., et al. (2020). Lysine-specific demethylase 1 mediates AKT activity and promotes epithelial-to-mesenchymal transition in PIK3CA-mutant colorectal cancer. Mol. Cancer Res. 18, 264–277. doi:10.1158/1541-7786.Mcr-19-0748
Miller, S. A., Policastro, R. A., Sriramkumar, S., Lai, T., Huntington, T. D., Ladaika, C. A., et al. (2021). LSD1 and aberrant DNA methylation mediate persistence of enteroendocrine progenitors that support BRAF-mutant colorectal cancer. Cancer Res. 81, 3791–3805. doi:10.1158/0008-5472.Can-20-3562
Misiorek, J. O., Przybyszewska-Podstawka, A., Kałafut, J., Paziewska, B., Rolle, K., Rivero-Müller, A., et al. (2021). Context matters: NOTCH signatures and pathway in cancer progression and metastasis. Cells 10, E94. Cells. doi:10.3390/cells10010094
Pan, H. M., Lang, W. Y., Yao, L. J., Wang, Y., and Li, X. L. (2019). shRNA-interfering LSD1 inhibits proliferation and invasion of gastric cancer cells via VEGF-C/PI3K/AKT signaling pathway. World J. Gastrointest. Oncol. 11, 622–633. doi:10.4251/wjgo.v11.i8.622
Pan, T., Yu, Z., Jin, Z., Wu, X., Wu, A., Hou, J., et al. (2021). Tumor suppressor lnc-CTSLP4 inhibits EMT and metastasis of gastric cancer by attenuating HNRNPAB-dependent Snail transcription. Mol. Ther. Nucleic Acids 23, 1288–1303. doi:10.1016/j.omtn.2021.02.003
Park, U. H., Kang, M. R., Kim, E. J., Kwon, Y. S., Hur, W., Yoon, S. K., et al. (2016). ASXL2 promotes proliferation of breast cancer cells by linking ERα to histone methylation. Oncogene 35, 3742–3752. doi:10.1038/onc.2015.443
Peng, W., Zhang, H., Tan, S., Li, Y., Zhou, Y., Wang, L., et al. (2020). Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer. Ther. Adv. Med. Oncol. 12, 1758835920937428. doi:10.1177/1758835920937428
Perillo, B., Tramontano, A., Pezone, A., and Migliaccio, A. (2020). LSD1: More than demethylation of histone lysine residues. Exp. Mol. Med. 52, 1936–1947. doi:10.1038/s12276-020-00542-2
Qi, W., Yang, Z., Feng, Y., Li, H., Che, N., Liu, L., et al. (2020). Gli1 regulates stemness characteristics in gastric adenocarcinoma. Diagn. Pathol. 15, 60. doi:10.1186/s13000-020-00949-5
Qin, X. K., Du, Y., Liu, X. H., and Wang, L. (2020). LSD1 promotes prostate cancer cell survival by destabilizing FBXW7 at post-translational level. Front. Oncol. 10, 616185. doi:10.3389/fonc.2020.616185
Qin, Y., Vasilatos, S. N., Chen, L., Wu, H., Cao, Z., Fu, Y., et al. (2019). Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 38, 390–405. doi:10.1038/s41388-018-0451-5
Raeisi, F., Abolfathi, M., Ahmadi-Naji, R., Iranparast, S., Noshadi, E., Akbari, A., et al. (2019). ARA lncRNA, is upregulated in liver and breast tumor tissues. Mol. Biol. Rep. 46, 77–82. doi:10.1007/s11033-018-4447-6
Ramírez-Ramírez, R., Gutiérrez-Angulo, M., Peregrina-Sandoval, J., Moreno-Ortiz, J. M., Franco-Topete, R. A., Cerda-Camacho, F. J., et al. (2020). Somatic deletion of KDM1A/LSD1 gene is associated to advanced colorectal cancer stages. J. Clin. Pathol. 73, 107–111. doi:10.1136/jclinpath-2019-206128
Ravasio, R., Ceccacci, E., Nicosia, L., Hosseini, A., Rossi, P. L., Barozzi, I., et al. (2020). Targeting the scaffolding role of LSD1 (KDM1A) poises acute myeloid leukemia cells for retinoic acid-induced differentiation. Sci. Adv. 6, eaax2746. doi:10.1126/sciadv.aax2746
Saccà, C. D., Gorini, F., Ambrosio, S., Amente, S., Faicchia, D., Matarese, G., et al. (2019). Inhibition of lysine-specific demethylase LSD1 induces senescence in Glioblastoma cells through a HIF-1α-dependent pathway. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 535–546. doi:10.1016/j.bbagrm.2019.03.004
Saltarella, I., Apollonio, B., Lamanuzzi, A., Desantis, V., Mariggiò, M. A., Desaphy, J.-F., et al. (2022). The landscape of lncRNAs in multiple myeloma: Implications in the “hallmarks of cancer”. Clin. Perspect. Ther. Oppor. 14, 1963. doi:10.3390/cancers14081963
Sehrawat, A., Gao, L., Wang, Y., Bankhead, A., Mcweeney, S. K., King, C. J., et al. (2018). LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc. Natl. Acad. Sci. U. S. A. 115, E4179-E4188. doi:10.1073/pnas.1719168115
Shen, D. D., Pang, J. R., Bi, Y. P., Zhao, L. F., Li, Y. R., Zhao, L. J., et al. (2022). LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in gastric cancer. Mol. Cancer 21, 75. doi:10.1186/s12943-022-01557-1
Sheng, W., Lafleur, M. W., Nguyen, T. H., Chen, S., Chakravarthy, A., Conway, J. R., et al. (2018). LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563. e519. doi:10.1016/j.cell.2018.05.052
Shi, Y. G., and Tsukada, Y. (2013). The discovery of histone demethylases. Cold Spring Harb. Perspect. Biol. 5, a017947. doi:10.1101/cshperspect.a017947
Si, W., Zhao, Y., Zhou, J., Zhang, Q., and Zhang, Y. (2019). The coordination between ZNF217 and LSD1 contributes to hepatocellular carcinoma progress and is negatively regulated by miR-101. Exp. Cell Res. 379, 1–10. doi:10.1016/j.yexcr.2019.03.017
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. Ca. Cancer J. Clin. 71, 7–33. doi:10.3322/caac.21654
Soldi, R., Ghosh Halder, T., Weston, A., Thode, T., Drenner, K., Lewis, R., et al. (2020). The novel reversible LSD1 inhibitor SP-2577 promotes anti-tumor immunity in SWItch/Sucrose-NonFermentable (SWI/SNF) complex mutated ovarian cancer. PLoS One 15, e0235705. doi:10.1371/journal.pone.0235705
Soni, S. K., Basu, P., Singaravel, M., Sharma, R., Pandi-Perumal, S. R., Cardinali, D. P., et al. (2021). Sirtuins and the circadian clock interplay in cardioprotection: Focus on sirtuin 1. Cell. Mol. Life Sci. 78, 2503–2515. doi:10.1007/s00018-020-03713-6
Speranzini, V., Ciossani, G., Marabelli, C., and Mattevi, A. (2017). Probing the interaction of the p53 C-terminal domain to the histone demethylase LSD1. Arch. Biochem. Biophys. 632, 202–208. doi:10.1016/j.abb.2017.07.021
Sun, L., Wang, X., Saredy, J., Yuan, Z., Yang, X., Wang, H., et al. (2020). Innate-adaptive immunity interplay and redox regulation in immune response. Redox Biol. 37, 101759. doi:10.1016/j.redox.2020.101759
Takagi, S., Ishikawa, Y., Mizutani, A., Iwasaki, S., Matsumoto, S., Kamada, Y., et al. (2017). LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res. 77, 4652–4662. doi:10.1158/0008-5472.Can-16-3502
Tatsumi, G., Kawahara, M., Yamamoto, R., Hishizawa, M., Kito, K., Suzuki, T., et al. (2020). LSD1-mediated repression of GFI1 super-enhancer plays an essential role in erythroleukemia. Leukemia 34, 746–758. doi:10.1038/s41375-019-0614-6
Tayari, M. M., Santos, H. G. D., Kwon, D., Bradley, T. J., Thomassen, A., Chen, C., et al. (2021). Clinical responsiveness to all-trans retinoic acid is potentiated by LSD1 inhibition and associated with a quiescent transcriptome in myeloid malignancies. Clin. Cancer Res. 27, 1893–1903. doi:10.1158/1078-0432.Ccr-20-4054
Trindade, A., and Duarte, A. (2020). Notch signaling function in the angiocrine regulation of tumor development. Cells 9, E2467. Cells. doi:10.3390/cells9112467
Van Bergen, M., and Van Der Reijden, B. A. (2019). Targeting the GFI1/1B-CoREST complex in acute myeloid leukemia. Front. Oncol. 9, 1027. doi:10.3389/fonc.2019.01027
Verigos, J., Karakaidos, P., Kordias, D., Papoudou-Bai, A., Evangelou, Z., Harissis, H. V., et al. (2019). The histone demethylase LSD1/?dm1a mediates chemoresistance in breast cancer via regulation of a stem cell Program. Cancers (Basel) 11, E1585. doi:10.3390/cancers11101585
Wang, D., Kuang, Y., Zhang, G., Xiao, K., and Liu, Y. (2022a). Lysine-specific demethylase 1 in energy metabolism: A novel target for obesity. J. Nutr., nxac080. doi:10.1093/jn/nxac080
Wang, L. H., Wu, C. F., Rajasekaran, N., and Shin, Y. K. (2018). Loss of tumor suppressor gene function in human cancer: An overview. Cell. Physiol. biochem. 51, 2647–2693. doi:10.1159/000495956
Wang, L., Lankhorst, L., and Bernards, R. (2022b). Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer 22, 340–355. doi:10.1038/s41568-022-00450-9
Wang, L., Tang, J., Wang, L., Tan, F., Song, H., Zhou, J., et al. (2021). Oxidative stress in oocyte aging and female reproduction. J. Cell. Physiol. 236, 7966–7983. doi:10.1002/jcp.30468
Wang, P., Wang, Z., and Liu, J. (2020). Role of HDACs in normal and malignant hematopoiesis. Mol. Cancer 19, 5. doi:10.1186/s12943-019-1127-7
Wang, Y., Zhu, Y., Wang, Q., Hu, H., Li, Z., Wang, D., et al. (2016). The histone demethylase LSD1 is a novel oncogene and therapeutic target in oral cancer. Cancer Lett. 374, 12–21. doi:10.1016/j.canlet.2016.02.004
Wang, Z. A., and Cole, P. A. (2020). The chemical biology of reversible lysine post-translational modifications. Cell Chem. Biol. 27, 953–969. doi:10.1016/j.chembiol.2020.07.002
Wang, Z., Gao, S., Han, D., Han, W., Li, M., Cai, C., et al. (2019). LSD1 activates PI3K/AKT signaling through regulating p85 expression in prostate cancer cells. Front. Oncol. 9, 721. doi:10.3389/fonc.2019.00721
Wen, J., and Wang, D. (2022). Deciphering the PTM codes of the tumor suppressor p53. J. Mol. Cell Biol. 13, 774–785. doi:10.1093/jmcb/mjab047
Wimalasena, V. K., Wang, T., Sigua, L. H., Durbin, A. D., and Qi, J. (2020). Using chemical epigenetics to target cancer. Mol. Cell 78, 1086–1095. doi:10.1016/j.molcel.2020.04.023
Xu, S., Wang, X., Yang, Y., Li, Y., and Wu, S. (2021). LSD1 silencing contributes to enhanced efficacy of anti-CD47/PD-L1 immunotherapy in cervical cancer. Cell Death Dis. 12, 282. doi:10.1038/s41419-021-03556-4
Yang, C., Fang, Y., Luo, X., Teng, D., Liu, Z., Zhou, Y., et al. (2022). Discovery of natural product-like spirooxindole derivatives as highly potent and selective LSD1/KDM1A inhibitors for AML treatment. Bioorg. Chem. 120, 105596. doi:10.1016/j.bioorg.2022.105596
Yang, C., Wang, K., Liang, Q., Tian, T. T., and Zhong, Z. (2021a). Role of NSD1 as potential therapeutic target in tumor. Pharmacol. Res. 173, 105888. doi:10.1016/j.phrs.2021.105888
Yang, C., Wang, K., Zhou, Y., and Zhang, S. L. (2021b). Histone lysine methyltransferase SET8 is a novel therapeutic target for cancer treatment. Drug Discov. Today 26, 2423–2430. doi:10.1016/j.drudis.2021.05.004
Yang, C., Zhong, Z. F., Wang, S. P., Vong, C. T., Yu, B., Wang, Y. T., et al. (2021c). HIF-1: Structure, biology and natural modulators. Chin. J. Nat. Med. 19, 521–527. doi:10.1016/s1875-5364(21)60051-1
Yang, L., Shi, P., Zhao, G., Xu, J., Peng, W., Zhang, J., et al. (2020). Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 5, 8. doi:10.1038/s41392-020-0110-5
Yang, S. J., Park, Y. S., Cho, J. H., Moon, B., An, H. J., Lee, J. Y., et al. (2017). Regulation of hypoxia responses by flavin adenine dinucleotide-dependent modulation of HIF-1α protein stability. Embo J. 36, 1011–1028. doi:10.15252/embj.201694408
Yu, Z., Deng, P., Chen, Y., Liu, S., Chen, J., Yang, Z., et al. (2021). Inhibition of the PLK1-coupled cell cycle machinery overcomes resistance to oxaliplatin in colorectal cancer. Adv. Sci. 8, e2100759. doi:10.1002/advs.202100759
Yu, Z., Liu, H., Fan, J., Chen, F., and Liu, W. (2020). MicroRNA-155 participates in the expression of LSD1 and proinflammatory cytokines in rheumatoid synovial cells. Mediat. Inflamm. 2020, 4092762. doi:10.1155/2020/4092762
Zang, C., Nie, F. Q., Wang, Q., Sun, M., Li, W., He, J., et al. (2016). Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget 7, 11696–11707. doi:10.18632/oncotarget.7077
Zhang, C., Xu, H., Sui, X., Chen, L., Chen, B., Lv, H., et al. (2022a). Icaritin inhibits PLK1 to activate DNA damage response in NK/T cell lymphoma and increases sensitivity to GELOX regime. Mol. Ther. Oncolytics 25, 288–304. doi:10.1016/j.omto.2022.04.012
Zhang, J., Zhao, D., Li, Q., Du, X., Liu, Y., Dai, X., et al. (2019). Upregulation of LSD1 promotes migration and invasion in gastric cancer through facilitating EMT. Cancer Manag. Res. 11, 4481–4491. doi:10.2147/cmar.S186649
Zhang, J., Zhu, J., Zheng, G., Wang, Q., Li, X., Feng, Y., et al. (2021). Co-expression of miR155 or LSD1 shRNA increases the anti-tumor functions of CD19 CAR-T cells. Front. Immunol. 12, 811364. doi:10.3389/fimmu.2021.811364
Zhang, Q., Qi, S., Xu, M., Yu, L., Tao, Y., Deng, Z., et al. (2013). Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Res. 23, 225–241. doi:10.1038/cr.2012.177
Zhang, W., Ruan, X., Li, Y., Zhi, J., Hu, L., Hou, X., et al. (2022b). KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/β-catenin signaling pathway. Theranostics 12, 1500–1517. doi:10.7150/thno.66142
Zhang, Y. H., and Sun, H. X. (2020). Immune checkpoint molecules in pregnancy: Focus on regulatory T cells. Eur. J. Immunol. 50, 160–169. doi:10.1002/eji.201948382
Zhang, Y., and Wang, X. (2020). Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 13, 165. doi:10.1186/s13045-020-00990-3
Zhang, Y., Wang, Z., Huang, Y., Ying, M., Wang, Y., Xiong, J., et al. (2018). TdIF1: A putative oncogene in NSCLC tumor progression. Signal Transduct. Target. Ther. 3, 28. doi:10.1038/s41392-018-0030-9
Zhang, Y., Wu, T., Wang, Y., Zhao, X., Zhao, B., Zhao, X., et al. (2020). The R251Q mutation of LSD1 promotes invasion and migration of luminal breast cancer cells. Int. J. Biol. Macromol. 164, 4000–4009. doi:10.1016/j.ijbiomac.2020.08.221
Zhao, Y., and Peng, H. (2022). The role of N6-methyladenosine (m6A) methylation modifications in hematological malignancies. Cancers 14, 332. doi:10.3390/cancers14020332
Keywords: anticancer activity, demethylation, epigenetics, histone modifications, LSD1, lysine methylation, signaling pathway, targets
Citation: Yang C, Li D, Zang S, Zhang L, Zhong Z and Zhou Y (2022) Mechanisms of carcinogenic activity triggered by lysine-specific demethylase 1A. Front. Pharmacol. 13:955218. doi: 10.3389/fphar.2022.955218
Received: 28 May 2022; Accepted: 29 June 2022;
Published: 19 July 2022.
Edited by:
Chao Han, China Pharmaceutical University, ChinaReviewed by:
Zhongrui Li, Nanjing Medical University, ChinaCopyright © 2022 Yang, Li, Zang, Zhang, Zhong and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhangfeng Zhong, emhhbmdmZW5nemhvbmdAdW0uZWR1Lm1v; Yingtang Zhou, emhvdXlpbmd0YW5nQHpqb3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.