
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 15 July 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.950122
This article is part of the Research TopicPharmacological Mechanisms of Drugs Affecting Bone Formation and Bone ResorptionView all 16 articles
 Yin Cheng1†
Yin Cheng1† Haixia Liu1†
Haixia Liu1† Jing Li2
Jing Li2 Yujie Ma1
Yujie Ma1 Changheng Song1
Changheng Song1 Yuhan Wang1
Yuhan Wang1 Pei Li1
Pei Li1 Yanjing Chen1*
Yanjing Chen1* Zhiguo Zhang1*
Zhiguo Zhang1*Osteoclasts (OCs) are multinucleated cells that play a major role in osteolytic diseases such as osteoporosis. Monascin (Ms) is one of the active substances in the traditional Chinese medicine red yeast rice. Studies have found that red yeast rice can maintain bone health. In this study, the anti-osteoclastogenesis effects of Ms on RANKL-induced RAW264.7 cells were assessed, and the underlying mechanism was investigated. Ms exhibited inhibitory effects on OC differentiation and formation in a dose-dependent manner and suppressed the bone-resorbing activity of mature OCs. Ms blocked OCs-typical genes (c-Fos, NFATc1, CSTK, MMP-9, TRAP, ITG-β3, OSCAR and DC-STAMP). Furthermore, Ms treatment considerably inhibited the activation of MAPKs, JNK and p38. Taken together, Ms suppresses RANKL-induced osteoclastogenesis of RAW264.7 cells by restraining MAPKs signaling pathways and is a potential therapeutic option as a novel OC inhibitor to mitigate bone erosion.
The bones of human body provide surprisingly sophisticated support for the entire body as they protect the internal organs and are involved in metabolism (Karsenty and Ferron, 2012; Wang et al., 2021). As a mineralized connective tissue, bones are dynamically remodeled throughout life, consisting of three consecutive and cyclic phases: resorption, reversal and formation (Siddiqui and Partridge, 2016; Chen et al., 2018). Several bone diseases result from the disruption of the balance between osteoclasts (OCs) and osteoblasts. Excessive OC bone resorption results in osteoporosis and rheumatoid arthritis, characterized by low bone density, whereas the contrary may contribute to osteopetrosis, marked by high bone density (Jacome-Galarza et al., 2019; Blangy et al., 2020). Inhibiting OC formation and function is a viable strategy for treating low bone density diseases.
OCs are terminally differentiated macrophage multinucleated cells tightly attached to the bone surface, derived from myeloid mononuclear cells. Their differentiation depends on multiple factors (Asagiri and Takayanagi, 2007). Previous research reported the essential roles of the macrophage colony-stimulating factor (M-CSF) and the receptor activator of nuclear factor kappa-B ligand (RANKL) in OC differentiation, formation and function (Song et al., 2019; Zhao et al., 2019; Kitaura et al., 2020). M-CSF not only stimulates the proliferation and survival of the OC precursor cells but also upregulates RANK expression (Asagiri and Takayanagi, 2007). The RANKL-induced osteoclastogenesis is a sequential process comprising three stages: integration to its receptor RANK to recruit the adaptor molecule TNF receptor-associated factor 6 (TRAF6), resulting in mitogen-activated protein kinases (MAPKs) and nuclear factor-kappa B (NF-κB) activation, and promoting the expression of OCs-typical genes (Sun et al., 2021).
MAPKs signal pathways are indispensable in OC differentiation and bone resorption, including extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 signaling (Lee et al., 2018). ERKs regulate the transcription factors c-Fos and nuclear factor of activated T-cells, cytoplasmic (NFATc) 1, which are involved in the differentiation, migration and function of OCs(He et al., 2011). JNK signaling is activated by RANKL through inflammatory cytokines such as TRAF6, mainly affecting the fusion and tartrate-resistant acid phosphatase (TRAP) activity of OC precursors (Chang et al., 2008; Ikeda et al., 2008). RANKL-RANK signaling induces p38 phosphorylation and NF-κB activation (Matsumoto et al., 2000), which directly influences the fusion factors such as dendrocyte-expressed seven-transmembrane protein (DC-STAMP) and ITG-β3, as well as function factors, including TRAP, cathepsin K(CTSK) and matrix metalloproteinase 9 (MMP-9) (Mansky et al., 2002; Herbert et al., 2015).
Red yeast rice (RYR) has been used in East Asian countries for thousands of years as a food supplement or herbal medicine and is produced by the fermentation of rice with Monascus purpureus (Nguyen et al., 2017). According to numerous previous researches, RYR has been reported to maintain bone health (Wong and Rabie, 2008; Wang et al., 2015; Wu et al., 2020) and be effective against a variety of diseases such as hyperlipidemia, osteoporosis and cardiovascular disease (Wang et al., 2015; Liu et al., 2017; Xiong et al., 2017; Wang et al., 2019; Wu et al., 2020). Notably, Monascus purpureus has been shown to produce many functional monascus pigments, applied as a food colorant, functional foods or treatment for some diseases (Patakova, 2013). Yellow pigments monascin (Ms, Figure 1A) has a potential role in alleviating metabolic syndrome (Lin et al., 2017). It has anti-inflammatory, antioxidant and other properties (Lin et al., 2017; Zhang et al., 2020). The effect of Ms on OCs has not been investigated yet.
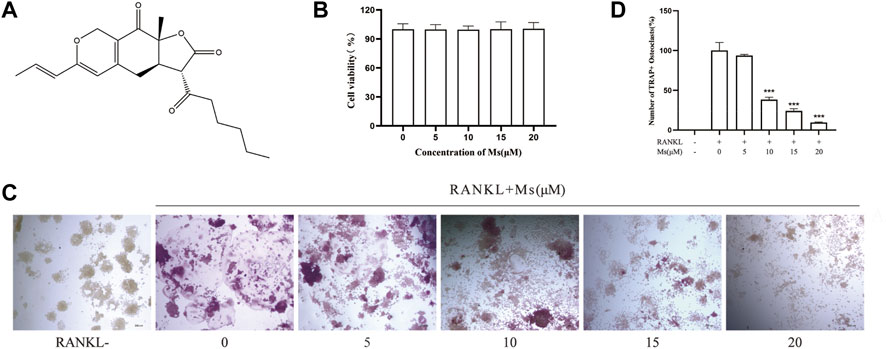
FIGURE 1. Inhibitory effect of Ms on RANKL-induced osteoclast formation in vitro. (A) Chemical structure of Ms. (B) Viability of RAW264.7 cells. RAW264.7 cells exposed to 5, 10, 15 and 20 μM Ms were assessed by CCK-8 kits after 72 h. (C) The osteoclast morphology after incubation with RANKL and Ms at 5, 10, 15 and 20 μM for 5 days. TRAP-positive cells (TRAP (+)) (nuclei≥3) were photographed by a light microscope, original magnification ×200 (Scale bar = 200 μm). (D) Quantitative assessment of TRAP-positive cells. The results represent the mean ± SD of three independent samples. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Control (RANKL+, Ms-).
This study explores the role of Ms in RANKL-induced OC differentiation and bone resorption in vitro and investigates its potential mechanism. Ms may provide a treatment strategy for osteoporosis.
RAW264.7 cells were purchased from the Institute of Basic Medicine of Peking Union Medical College (Beijing, China). High glucose Dulbecco minimum essential medium (DMEM) and fetal bovine serum (FBS) were sourced from Gibco Company (Rockford, IL, United States). Recombinant Mouse RANKL (462-TEC-010/CF) was obtained from R&D Systems (R&D, Minneapolis, United States). Leukocyte Acid Phosphatase (TRAP) Kits (G1492), bovine bone slices (IDS, Jelling, Denmark), 1% toluidine blue O solution in sodium borate (G3663) were obtained from the Solarbio company (Beijing, China). Monascin (Ms, CAS: 21,516-68-7, purity>98% by HPLC) was purchased from Chengdu Desite Bio-Technology Company (Chengdu, China). All cell culture plasticware was supplied by Corning (NY, United States). Primary antibodies of p-p38(Thr180/Tyr182), p-ERK (Thr202/Tyr204), p-JNK(Thr183/Tyr185) and NFATc1were products of Cell Signaling Technology (Beverly, MA, United States). Primary antibodies of c-Fos and secondary antibodies were purchased from the Proteintech group (Rocky Hill, United States).
RAW264.7 cells were cultured in DMEM supplemented with 10% FBS (complete DMEM) at 37 °C in a 5% CO2 incubator with humidified air.
To determine the effect of Ms on the viability of RAW264.7 cells, Cell Counting Kit-8 (CCK-8) assays were used. The cells at a density of 8 × 103 cells per well were seeded into a sterile 96-well plate, and incubated for 24 h to allow the cells to attach to the plate. After incubation with Ms at concentrations of 0–20 μM for 72 h, 100 μL 10% CCK-8 solution was added to replace the original medium, followed by 4 h of incubation at 37°C, and the absorbance at 450 nm was measured using a microplate reader.
TRAP staining was conducted to identify the morphology of mature OCs. RAW264.7 cells were cultured overnight in a 24-well plate at a density of 1×104 cells per well for adherence and then stimulated with 0, 5, 10, 15 and 20 μM Ms respectively in combination with RANKL 50 ng/ml. After 5 days, the cells were fixed with 4% paraformaldehyde for 5 min at 4°C and then stained with the TRAP staining kit according to the manufacturer’s instructions. Cells with more than three nuclei were identified as TRAP-positive multinucleated cells (TRAP (+)), which were counted and photographed using a light microscope.
RAW264.7 cells (8 × 103 cells per well) were seeded in a 96-well plate containing a sterile bovine bone slice, using the same methods of stimulation and cultivation as described previously. After a 7-days incubation period, all slices were washed in an ultrasonic instrument with 0.1% aqueous ammonia to remove the cells adhered to the surface. Subsequently, the bone slices were stained with 1% toluidine blue solution as described in the previous experiment described. Resorption pits were photographed using a microscope, and the percentage resorption was analyzed using Image-Pro Plus.
RAW264.7 cells at a density of 1 × 104 cells per well were induced in 24-well plates in a complete medium with RANKL (50 ng/ml) and Ms (0, 5, 10, 15 and 20 μM) for 5 days. The cells were fixed in 4% paraformaldehyde for 20 min, permeabilized with 0.1% TritonX-100 for 10 min, and stained with rhodamine-conjugated phalloidin for 40 min at 4 C to visualize F-actin cytoskeleton. Finally, the actin rings of mature OCs were captured using a fluorescence microscope. The fluorescence images were processed by Image-Pro Plus software.
RAW264.7 cells (4×104 cells per well) were seeded in 12-well plates using the same methods. Total RNA was extracted from cells by TRIzol reagent. After isolation, 3 μg total RNA from each sample was reverse transcribed into cDNA by Oligo (dT) and M-MLV reverse transcriptase. Thereafter, using SYBR Green detection and specific primers (listed in Table 1), the RT-generated DNA (0.5 μL) was amplified by a Bio-rad real-time PCR detection system. On completion of the reaction, gene expression values were calculated following the relative quantification method (2−ΔΔCT; means ± SD of triplicate determination).
The cells were washed three times with ice-cold PBS, and lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with Protease Inhibitor Cocktail and phenylmethylsulfonyl fluoride (PMSF) to obtain total protein. Proteins were quantitated using a bicinchoninic acid protein assay kit, separated on a 10% polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane. Subsequently, the membranes were blocked with 5% skimmed milk for 1 h at room temperature, and then incubated with the indicated primary antibodies including c-Fos (1:500), NFATc1 (1:1,000), p-p38 (1:1,000), p-JNK (1:1,000), p-ERK (1:1,000) and GAPDH (1:1,000) at 4 °C overnight. Then, the membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies for 2 h, visualized with the ECL system, and analyzed by ImageJ software.
Statistical analysis was performed with SPSS 25.0 software. The data are expressed as mean ± standard deviation (SD). Two-group comparisons were conducted using the Students’ t-test, multiple-group comparisons were conducted using one-way ANOVA, and any two-group comparisons were conducted by SNK-q test. Differences were considered to be statistically significant if p < 0.05.
Ms treatment at concentrations of 0–20 μM for 72 h did not affect the viability of OC precursors (Figure 1B). TRAP is considered a histochemical marker of OCs(Lv et al., 2015). A TRAP staining assay was used to explore whether Ms suppressed RANKL-induced OC formation. As shown in Figures 1C,D, the number of TRAP-positive multinuclear cells increased substantially in response to RANKL stimulation. Still, Ms treatment suppressed the purple giant multinucleated cells induced by RANKL in a dose-dependent manner. Collectively, these results indicate that Ms inhibits the OC formation and differentiation of RANKL-stimulated RAW264.7 cells.
Mature OCs play an essential role in polarization and adherence in bone resorption due to their unique morphological characteristics, consisting of an actin ring structure and a ruffled border membrane (Garbe et al., 2012; Georgess et al., 2014). In this study, F-actin rings were visualized by phalloidin staining and bone resorption pits were identified by toluidine blue staining. As illustrated in Figures 2A–D, numerous F-actin rings and bone resorption pits were generated in RANKL-induced RAW264.7-OCs, which were significantly reduced following Ms treatment in a dose-dependent manner. These results indicate that Ms inhibits the OC bone resorption.
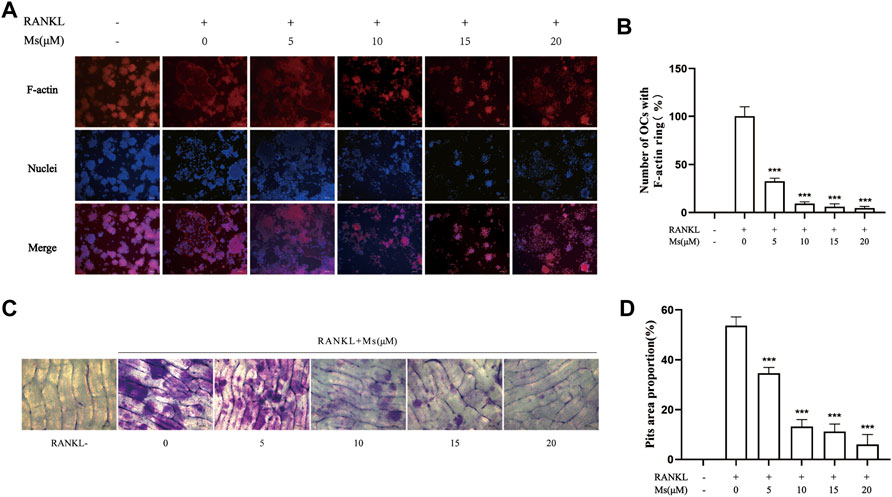
FIGURE 2. Ms inhibits formation of the F-actin ring and osteoclast resorptive function induced by RANKL in vitro. RAW264.7 macrophages were cultured in 24-well plates with or without Ms (5, 10, 15 and 20 μM) in the presence of 50 ng/ml RANKL for 5 days. (A) Representative images of F-actin rings; F-actin: red, nuclei: blue, original magnification ×200 (Scale bar = 200 μm) (B) Quantitative analyses of osteoclast numbers with intact F-actin ring. RAW264.7 macrophages were cultured on a bone slice in 96-well plates with or without Ms (5, 10, 15 and 20 μM) in the presence of 50 ng/ml RANKL for 7 days. (C) Resorption pits of osteoclasts were captured by a light microscope. Original magnification ×200 (Scale bar = 200 μm) (D) The resorbed areas were calculated using Image (J). The results represent the mean ± SD of three independent samples. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Control (RANKL+, Ms-).
Western blot analysis was carried out to investigate whether Ms inhibited c-Fos and NFATc1. As shown in Figures 3A,C, Ms significantly inhibited c-Fos and NFATc1 protein levels. Furthermore, RT-qPCR was performed to analyze expression levels of OC-specific genes in the absence or presence of Ms. As shown in Figures 3B,D, Ms had a strong inhibitory effect on c-Fos and NFATc1 genes, essential transcription factors and master regulators of osteoclastogenesis. In addition, we found that several NFATc1-regulated genes, including MMP-9, CTSK, TRAP, ITG-β3, OSCAR and DC-STAMP, were upregulated in RANKL-induced RAW264.7-OCs, but downregulated by Ms in a dose-dependent manner (Figure 4). These results suggest that Ms inhibits osteoclastogenesis via suppressing the expression of these genes.
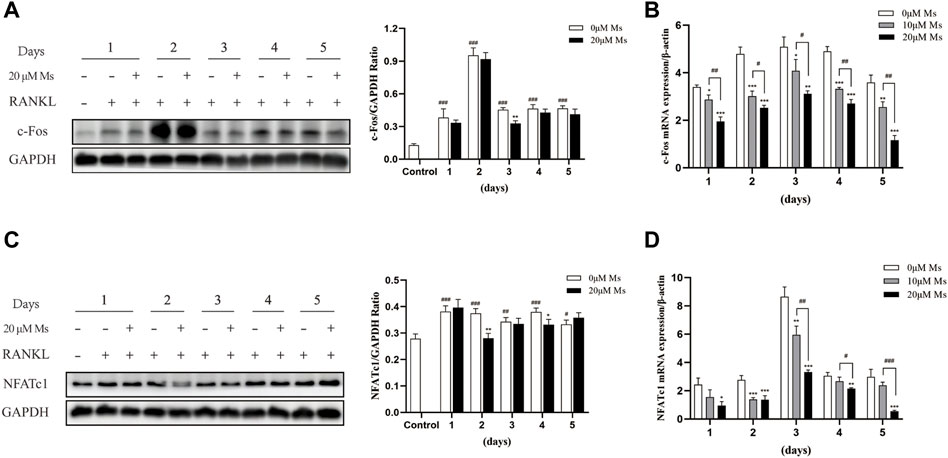
FIGURE 3. Effect of Ms on NFATc1 and c-Fos expression in RANKL-induced RAW264.7 cells. RAW264.7 cells were treated with varying concentrations of Ms with RANKL (50 ng/ml) for 5 days. The proteins were extracted to analyze c-Fos and NFATc1 expression by Western blot per day (A and C). The mRNA expression levels of c-Fos and NFATc1 genes were analyzed using RT-qPCR per day (B and D). Each point represents the mean ± SD. The experiments were repeated three times. #p < 0.05, ##p < 0.01 and ###p < 0.001 vs. Control group (RANKL-, Ms-); *p < 0.05, **p < 0.01 vs. ***p < 0.001 vs. corresponding time 0 μM Ms group (RANKL+, Ms-).
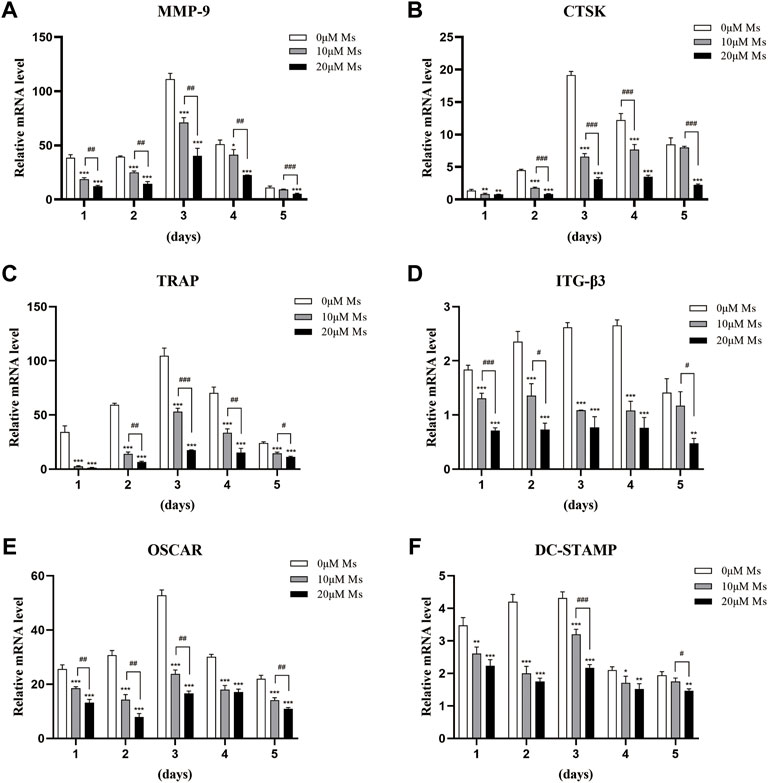
FIGURE 4. Ms inhibits the RANKL-induced expression of osteoclastic marker genes. RAW264.7 cells were treated with or without Ms (10 and 20 μM) in the presence of RANKL (50 ng/ml) after a 1–5 days culture period. The mRNA expression levels of osteoclast-specific genes, such as MMP-9 (A), CTSK (B), TRAP (C), ITG-β3 (D), OSCAR (E), and DC-STAMP (F), were analyzed using RT-qPCR. *p < 0.05, **p < 0.01 vs. ***p < 0.001 vs. corresponding time 0 μM Ms group (RANKL+, Ms-); #p < 0.05, ##p < 0.01 and ###p < 0.001 vs. corresponding time 10 μM Ms group (RANKL+, Ms+).
RANKL bind to its receptor RANK to activate a series of intracellular signaling pathways involved in OC differentiation, proliferation and survival (Leibbrandt and Penninger, 2009), including MAPKs pathways, which activate various transcription factors through phosphorylation (Ono and Nakashima, 2018). Firstly, western blot analysis was carried out to investigate whether Ms inhibited the activation and phosphorylation of MAPKs. Treatment with RANKL significantly increased the phosphorylation of JNK and p38 within 60 min, except ERK (Figure 5). Treatment with Ms for 15 min attenuated the phosphorylation of p38, JNK and ERK (Figure 5). These findings suggest that Ms inhibits RANKL induced osteoclastogenesis and bone resorption by suppressing MAPKs signal pathways in OCs.
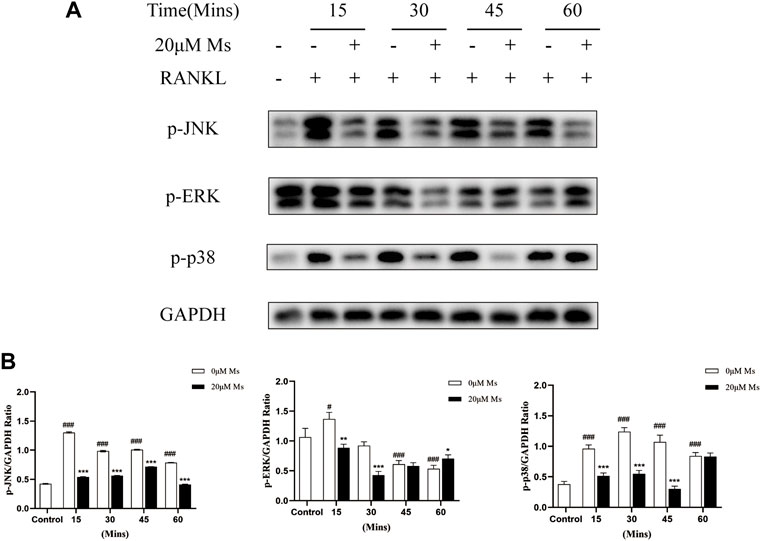
FIGURE 5. Ms inhibits RANKL-induced osteoclast differentiation by suppressing MAPKs signaling pathways. RAW264.7 cells were treated with or without Ms (20 μM) in the presence of RANKL. Protein levels of MAPKs activation, including JNK, ERK and p38, were detected by western blot at 0, 15, 30, 45 and 60 min. The results were expressed as means ± SD from three independent experiments. #p < 0.05, ##p < 0.01 and ###p < 0.001 vs. Control group (RANKL-, Ms-); *p < 0.05, **p < 0.01 vs. ***p < 0.001 vs. corresponding time 0 μM Ms group (RANKL+, Ms-).
Osteoclasts are essential in the complex process of bone remodeling and are related to lytic bone disorders, including osteoporosis and bone tumor metastasis, according to previous studies (Novack and Teitelbaum, 2008). Inhibiting OC formation is a promising strategy for these diseases. However, synthetic drugs usually cause severe side effects relative to natural components from herbal medicine. In this study, we performed a TRAP staining assay and observed that Ms inhibited RANKL-induced osteoclastogenesis. In addition, bone resorption was suppressed by Ms in vitro. Ms also suppressed OCs-typical genes in a dose-dependent manner. Furthermore, Ms inhibited the RANKL-induced phosphorylation of p38, JNK and ERK, and activation of c-Fos and NFATc1 in RANKL-induced RAW264.7-OCs.
The RAW264.7 cell linage is well-accepted and vastly used as a cellular model in osteoclastogenic research, supporting the exploration of the biological mechanisms of osteoclastogenesis (Collin-Osdoby and Osdoby, 2012). Mature OCs are giant, multinuclear, and polarized cells characterized by rearrangements of the actin cytoskeleton and a sealed compartment between the bone surface and basal membrane. Mature OCs are responsible for the digestion of the bone matrix (Boyle et al., 2003). In the first stage of this study, we defined TRAP-positive OCs and bone resorption area as indicators. The results revealed that Ms treatment significantly decreased the number of TRAP (+) cells, bone resorption area and F-actin rings compared to the RANKL-induced RAW264.7-OCs group. The findings suggest that Ms inhibits RANKL-mediated osteoclastogenesis and bone resorption. The underlying mechanisms of the inhibitory properties of Ms were investigated in subsequent experiments.
NFATc1 is a common target gene of both NF-κB and AP-1 in the early phase of RANKL-stimulated osteoclastogenesis. The former is recruited by the NFATc1 promoter, and the latter, containing c-Fos, assists NFATc1 (Ishida et al., 2002; Asagiri and Takayanagi, 2007). Cumulative evidence also indicates that c-Fos activation is a prerequisite for NFATc1 because RANKL-induced NFATc1 is completely abrogated in c-Fos-deficient cells (Takayanagi et al., 2002; Matsuo et al., 2004). In the second stage of our study, the essential transcription factors of c-Fos and NFATc1 were measured. Subsequently, we detected the OC-specific gene expression regulated by NFATc1, including TRAP, CTSK, MMP-9, OSCAR, DC-STAMP, and ITG-β3. Ms suppressed the activation of c-Fos and NFATc1 and reduced the expression of OC formation/function-related genes in a dose-dependent manner.
The stimuli of extracellular signaling transmission to coordinated intracellular molecules by MAPKs activation has been demonstrated to be significant for osteoclastogenesis (Lee et al., 2018; Qu et al., 2021). Both M-CSF and RANKL act through MAPKs, including ERK, JNK, and p38 signaling as cultured bone marrow/RAW264.7 cells differentiate into OCs (Ross, 2006; Lee et al., 2016). In the third stage of our study, we explored the inhibitory effect of Ms on the MAPKs signaling pathway. A series of experiments indicated that Ms attenuated the phosphorylation of p38, JNK and ERK. These results suggest that Ms inhibited RANKL-induced RAW264.7-OCs via blocking the MAPKs signaling pathways.
Taken together, this study was shown that Ms has potential anti-osteoclastogenic effects by inhibiting the MAPKs signaling pathways activation (Figure 6). Furthermore, Ms inhibits the transcription factors NFATc1 and c-Fos, resulting in decreased expression of OC-specific genes such as TRAP, CTSK, MMP-9, OSCAR, DC-STAMP, and ITG-β3. This study provides fundamental knowledge about the effects of Ms on osteoclastogenesis and insight into the therapeutic potential for diseases such as osteoporosis. In the future, animal studies will be performed to determine whether Ms retains its anti-osteoclastogenic effects and its impact on the MAPKs pathways in vivo.
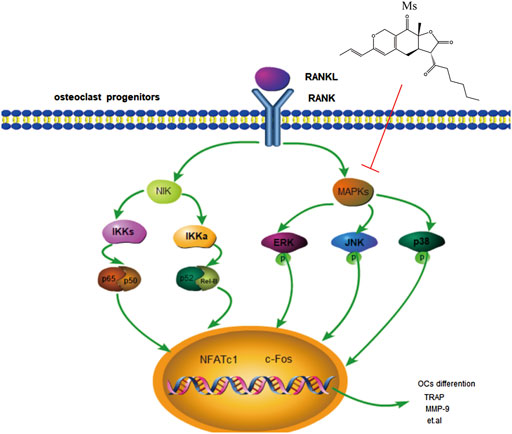
FIGURE 6. Schematic representation of the mechanism by which Ms inhibits osteoclastogenesis in RAW264.7 cells. RANKL induces MAPKs activation, including activation of JNK, p38 and ERK. Subsequently, the expression of osteoclast-typical genes, MMP-9, CTSK, TRAP, ITG-β3, OSCAR, and DC-STAMP are upregulated. Our results show that Ms inhibits osteoclastogenesis by inhibiting MAPKs activation. The sign (↓) means promoting. The sign (┴) means inhibiting.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
ZZ, YC and HL conceived the idea, drafted the manuscript and revised the manuscript. HL and JL conducted experimental guidance. YC performed the experiments and organized the manuscript. YM, CS, YW and PL helped to perform experiments.
This work was supported by the CACMS Innovation Fund (grant number CI2021A00107), the National Natural Science Foundation of China (grant number 82074297) and the Fundamental Research Funds for the Central Public Welfare Research Institutes (grant numbers YZ-202147, YZ-202108).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Asagiri, M., and Takayanagi, H. (2007). The molecular understanding of osteoclast differentiation. Bone 40 (2), 251–264. doi:10.1016/j.bone.2006.09.023
Blangy, A., Bompard, G., Guerit, D., Marie, P., Maurin, J., Morel, A., et al. (2020). The osteoclast cytoskeleton - Current understanding and therapeutic perspectives for osteoporosis. J. Cell Sci. 133 (13), jcs244798. doi:10.1242/jcs.244798
Boyle, W. J., Simonet, W. S., and Lacey, D. L. (2003). Osteoclast differentiation and activation. Nature 423 (6937), 337–342. doi:10.1038/nature01658
Chang, E. J., Ha, J., Huang, H., Kim, H. J., Woo, J. H., Lee, Y., et al. (2008). The JNK-dependent CaMK pathway restrains the reversion of committed cells during osteoclast differentiation. J. Cell Sci. 121 (15), 2555–2564. doi:10.1242/jcs.028217
Chen, X., Wang, Z., Duan, N., Zhu, G., Schwarz, E. M., Xie, C., et al. (2018). Osteoblast-osteoclast interactions. Connect. Tissue Res. 59 (2), 99–107. doi:10.1080/03008207.2017.1290085
Collin-Osdoby, P., and Osdoby, P. (2012). RANKL-mediated osteoclast formation from murine RAW 264.7 cells. Methods Mol. Biol. 816, 187–202. doi:10.1007/978-1-61779-415-5_13
Garbe, A. I., Roscher, A., Schüler, C., Lutter, A. H., Glösmann, M., Bernhardt, R., et al. (2012). Regulation of bone mass and osteoclast function depend on the F-actin modulator SWAP-70. J. Bone Min. Res. 27 (10), 2085–2096. doi:10.1002/jbmr.1670
Georgess, D., Machuca-Gayet, I., Blangy, A., and Jurdic, P. (2014). Podosome organization drives osteoclast-mediated bone resorption. Cell adh. Migr. 8 (3), 191–204. doi:10.4161/cam.27840
He, Y., Staser, K., Rhodes, S. D., Liu, Y., Wu, X., Park, S. J., et al. (2011). Erk1 positively regulates osteoclast differentiation and bone resorptive activity. PLoS One 6 (9), e24780. doi:10.1371/journal.pone.0024780
Herbert, B. A., Valerio, M. S., Gaestel, M., and Kirkwood, K. L. (2015). Sexual dimorphism in MAPK-activated protein kinase-2 (MK2) regulation of RANKL-induced osteoclastogenesis in osteoclast progenitor subpopulations. PLoS One 10 (5), e0125387. doi:10.1371/journal.pone.0125387
Ikeda, F., Matsubara, T., Tsurukai, T., Hata, K., Nishimura, R., Yoneda, T., et al. (2008). JNK/c-Jun signaling mediates an anti-apoptotic effect of RANKL in osteoclasts. J. Bone Min. Res. 23 (6), 907–914. doi:10.1359/jbmr.080211
Ishida, N., Hayashi, K., Hoshijima, M., Ogawa, T., Koga, S., Miyatake, Y., et al. (2002). Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 277 (43), 41147–41156. doi:10.1074/jbc.M205063200
Jacome-Galarza, C. E., Percin, G. I., Muller, J. T., Mass, E., Lazarov, T., Eitler, J., et al. (2019). Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature 568 (7753), 541–545. doi:10.1038/s41586-019-1105-7
Karsenty, G., and Ferron, M. (2012). The contribution of bone to whole-organism physiology. Nature 481 (7381), 314–320. doi:10.1038/nature10763
Kitaura, H., Marahleh, A., Ohori, F., Noguchi, T., Shen, W. R., Qi, J., et al. (2020). Osteocyte-related cytokines regulate osteoclast formation and bone resorption. Int. J. Mol. Sci. 21 (14), E5169. doi:10.3390/ijms21145169
Lee, K., Chung, Y. H., Ahn, H., Kim, H., Rho, J., Jeong, D., et al. (2016). Selective regulation of MAPK signaling mediates RANKL-dependent osteoclast differentiation. Int. J. Biol. Sci. 12 (2), 235–245. doi:10.7150/ijbs.13814
Lee, K., Seo, I., Choi, M. H., and Jeong, D. (2018). Roles of mitogen-activated protein kinases in osteoclast biology. Int. J. Mol. Sci. 19 (10), E3004. doi:10.3390/ijms19103004
Leibbrandt, A., and Penninger, J. M. (2009). RANKL/RANK as key factors for osteoclast development and bone loss in arthropathies. Adv. Exp. Med. Biol. 649, 100–113. doi:10.1007/978-1-4419-0298-6_7
Lin, C. H., Lin, T. H., and Pan, T. M. (2017). Alleviation of metabolic syndrome by monascin and ankaflavin: The perspective of monascus functional foods. Food Funct. 8 (6), 2102–2109. doi:10.1039/c7fo00406k
Liu, J. T., Chen, H. Y., Chen, W. C., Man, K. M., and Chen, Y. H. (2017). Red yeast rice protects circulating bone marrow-derived proangiogenic cells against high-glucose-induced senescence and oxidative stress: The role of heme oxygenase-1. Oxid. Med. Cell. Longev. 2017, 3831750. doi:10.1155/2017/3831750
Lv, Y. D., Wang, G. H., Xu, W. H., Tao, P. H., Lv, X. L., Wang, Y. Z., et al. (2015). Tartrate-resistant acid phosphatase 5b is a marker of osteoclast number and volume in RAW 264.7 cells treated with receptor-activated nuclear κB ligand. Exp. Ther. Med. 9 (1), 143–146. doi:10.3892/etm.2014.2071
Mansky, K. C., Sankar, U., Han, J. H., and Ostrowski, M. C. (2002). Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J. Biol. Chem. 277 (13), 11077–11083. doi:10.1074/jbc.M111696200
Matsumoto, M., Sudo, T., Saito, T., Osada, H., and Tsujimoto, M. (2000). Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-kappa B ligand (RANKL). J. Biol. Chem. 275 (40), 31155–31161. doi:10.1074/jbc.M001229200
Matsuo, K., Galson, D. L., Zhao, C., Peng, L., Laplace, C., Wang, K. Z., et al. (2004). Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 279 (25), 26475–26480. doi:10.1074/jbc.M313973200
Nguyen, T., Karl, M., and Santini, A. (2017). Red yeast rice.. Foods 6 (3), E19. doi:10.3390/foods6030019
Novack, D. V., and Teitelbaum, S. L. (2008). The osteoclast: Friend or foe? Annu. Rev. Pathol. 3, 457–484. doi:10.1146/annurev.pathmechdis.3.121806.151431
Ono, T., and Nakashima, T. (2018). Recent advances in osteoclast biology. Histochem. Cell Biol. 149 (4), 325–341. doi:10.1007/s00418-018-1636-2
Patakova, P. (2013). Monascus secondary metabolites: Production and biological activity. J. Ind. Microbiol. Biotechnol. 40 (2), 169–181. doi:10.1007/s10295-012-1216-8
Qu, Y. Y., Liu, X., Zong, S., Sun, H. X., Liu, S., Zhao, Y. R., et al. (2021). Protocatechualdehyde inhibits the osteoclast differentiation of RAW264.7 and BMM cells by regulating NF-κB and MAPK activity. Biomed Res. Int. 2021, 1–11. doi:10.1155/2021/6108999
Ross, F. P. (2006). M-CSF, c-Fms, and signaling in osteoclasts and their precursors. Ann. N. Y. Acad. Sci. 1068, 110–116. doi:10.1196/annals.1346.014
Siddiqui, J. A., and Partridge, N. C. (2016). Physiological bone remodeling: Systemic regulation and growth factor involvement. Physiol. (Bethesda) 31 (3), 233–245. doi:10.1152/physiol.00061.2014
Song, C., Yang, X., Lei, Y., Zhang, Z., Smith, W., Yan, J., et al. (2019). Evaluation of efficacy on RANKL induced osteoclast from RAW264.7 cells. J. Cell. Physiol. 234 (7), 11969–11975. doi:10.1002/jcp.27852
Sun, Y., Li, J., Xie, X., Gu, F., Sui, Z., Zhang, K., et al. (2021). Recent advances in osteoclast biological behavior. Front. Cell Dev. Biol. 9, 788680. doi:10.3389/fcell.2021.788680
Takayanagi, H., Kim, S., Koga, T., Nishina, H., Isshiki, M., Yoshida, H., et al. (2002). Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3 (6), 889–901. doi:10.1016/s1534-5807(02)00369-6
Wang, Y. F., Liu, W. T., Chen, C. Y., Ke, H. P., Jiang, H. L., Chen, X. L., et al. (2015). Anti-osteoporosis activity of red yeast rice extract on ovariectomy-induced bone loss in rats. Genet. Mol. Res. 14 (3), 8137–8146. doi:10.4238/2015.July.27.2
Wang, T. J., Lien, A. S., Chen, J. L., Lin, C. H., Yang, Y. S., Yang, S. H., et al. (2019). A randomized clinical efficacy trial of red yeast rice (monascus pilosus) against hyperlipidemia. Am. J. Chin. Med. 47 (2), 323–335. doi:10.1142/S0192415X19500150
Wang, H., Zheng, X., Zhang, Y., Huang, J., Zhou, W., Li, X., et al. (2021). The endocrine role of bone: Novel functions of bone-derived cytokines. Biochem. Pharmacol. 183, 114308. doi:10.1016/j.bcp.2020.114308
Wong, R. W., and Rabie, B. (2008). Chinese red yeast rice (Monascus purpureus-fermented rice) promotes bone formation. Chin. Med. 3, 4. doi:10.1186/1749-8546-3-4
Wu, B., Huang, J. F., He, B. J., Huang, C. W., and Lu, J. H. (2020). Promotion of bone formation by red yeast rice in experimental animals: A systematic review and meta-analysis. Biomed. Res. Int. 2020, 7231827. doi:10.1155/2020/7231827
Xiong, X., Wang, P., Li, X., Zhang, Y., and Li, S. (2017). The effects of red yeast rice dietary supplement on blood pressure, lipid profile, and C-reactive protein in hypertension: A systematic review. Crit. Rev. Food Sci. Nutr. 57 (9), 1831–1851. doi:10.1080/10408398.2015.1018987
Zhang, X., Liu, C., Tian, W., Zhang, H., Li, P., Wang, J., et al. (2020). Theoretical and experimental investigation of the antioxidative activity of monascin. Food Funct. 11 (7), 5915–5923. doi:10.1039/c9fo02410g
Keywords: monascin, osteoclast, RAW264.7, bone resorption, mapks, osteoporosis
Citation: Cheng Y, Liu H, Li J, Ma Y, Song C, Wang Y, Li P, Chen Y and Zhang Z (2022) Monascin abrogates RANKL-mediated osteoclastogenesis in RAW264.7 cells via regulating MAPKs signaling pathways. Front. Pharmacol. 13:950122. doi: 10.3389/fphar.2022.950122
Received: 22 May 2022; Accepted: 28 June 2022;
Published: 15 July 2022.
Edited by:
Dongwei Zhang, Beijing University of Chinese Medicine, ChinaReviewed by:
Cheng Xiao, China-Japan Friendship Hospital, ChinaCopyright © 2022 Cheng, Liu, Li, Ma, Song, Wang, Li, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjing Chen, Y2hlbnlqMDEwQDE2My5jb20=; Zhiguo Zhang, enpndGNtQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.