Abstract
Cutaneous malignancies, including basal cell carcinoma, cutaneous squamous cell carcinoma, and cutaneous melanoma, are common human tumors. The incidence of cutaneous malignancies is increasing worldwide, and the leading cause of death is malignant invasion and metastasis. The molecular biology of oncogenes has drawn researchers’ attention because of the potential for targeted therapies. Noncoding RNAs, including microRNAs, long noncoding RNAs, and circular RNAs, have been studied extensively in recent years. This review summarizes the aspects of noncoding RNAs related to the metastasis mechanism of skin malignancies. Continuous research may facilitate the identification of new therapeutic targets and help elucidate the mechanism of tumor metastasis, thus providing new opportunities to improve the survival rate of patients with skin malignancies.
1 Introduction
Skin cancer is a common malignancy, and the most common types are basal cell carcinoma (BCC), cutaneous squamous cell carcinoma (CSCC), and cutaneous malignant melanoma (CMM) (Sato et al., 2021) (Figure 1). BCC and CSCC are more prevalent than melanoma, and most patients receive prompt treatment leading to a better long-term prognosis (Bednarski et al., 2021; Reddy et al., 2021). BCC and CSCC originate from epidermal keratin-forming cells and have a lower mortality than melanoma; the lesion is confined to the site of origin, and the treatment is thus straightforward (Cives et al., 2020; Krasowska et al., 2021). CMM is a difficult-to-treat metastatic malignancy that originates from epidermal melanocytes and is associated with a high mortality (Fujimura and Aiba 2020). When detected early, melanoma is treatable by surgical excision; however, rapid invasion and metastasis are the main reasons for the lower survival time after advanced treatment (Allegra et al., 2020). Genetic changes can induce the transformation of normal cells into cancer cells, and cancer cells can become malignant after cell division.
FIGURE 1
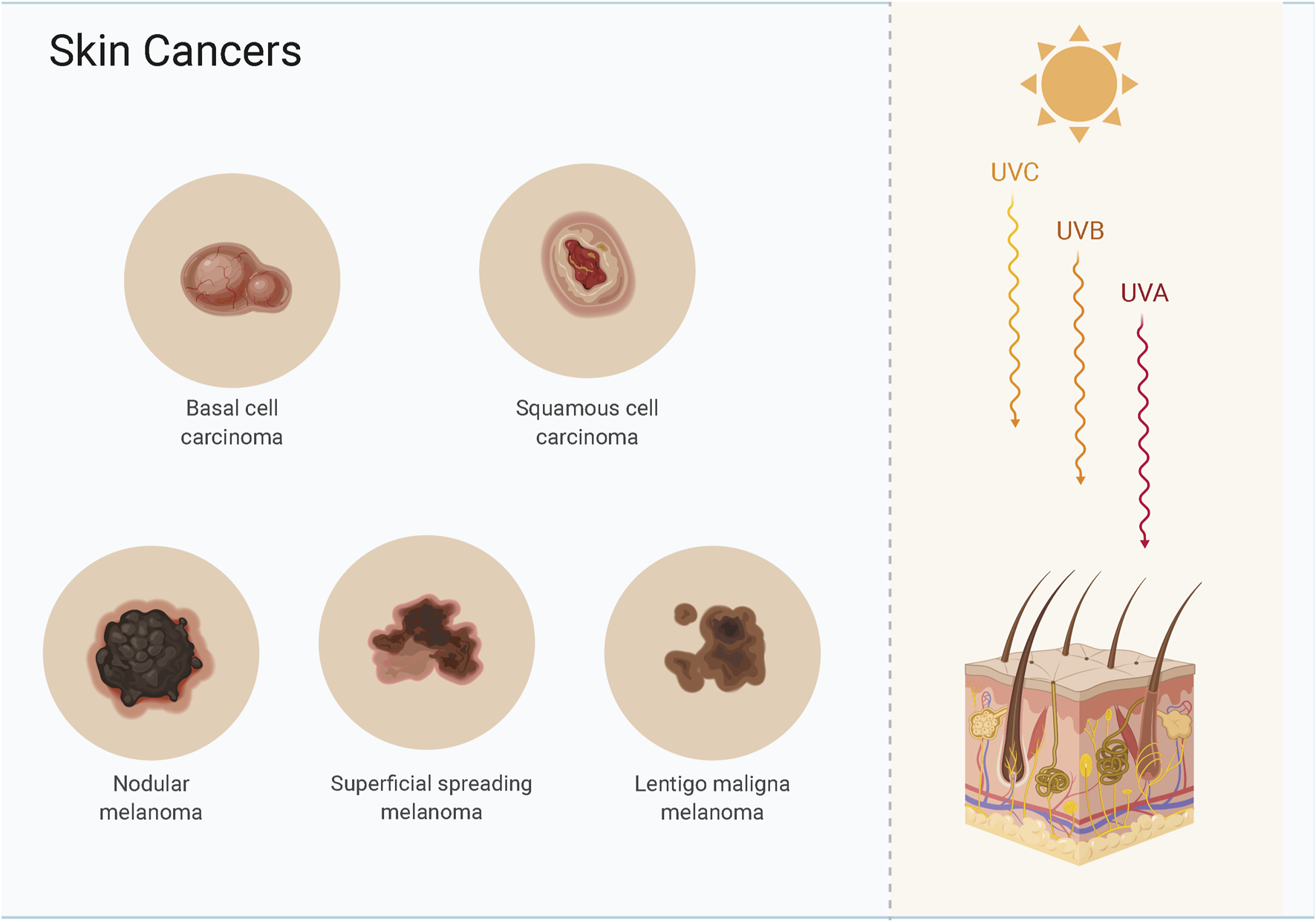
Skin cancers include basal cell carcinoma, cutaneous squamous cell carcinoma and melanoma.
Noncoding RNAs (ncRNAs) include microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) (He et al., 2021; Mercer et al., 2022). NcRNAs perform their respective biological functions at the RNA level (Xiaoqing Li et al., 2021). NcRNAs bind to various molecular targets to initiate specific cellular responses and regulate gene expression, intracellular signaling, epigenetic modifications, and other functions. The ncRNA is vital in the occurrence and development of tumors including skin cancer (Ding et al., 2021; Winkle et al., 2021). Accumulating evidence highlights the importance of ncRNAs in skin cancer. For example, circRNA_0016418 cross-talks with miR-625 and suppresses the progression of human skin melanoma (Zou et al., 2019). LncRNA MEG3 acts as a sponge for miR-21 and promotes melanoma growth and metastasis (Wu et al., 2020). The function of ncRNAs in cutaneous cancers was described previously (Zhou et al., 2022). Here, we provide an update of recent findings on the role of ncRNAs in skin cancer.
2 Noncoding RNAs in cancer
2.1 MiRNAs
MiRNAs are essential ncRNAs that regulate protein biosynthesis by modulating transcription (Kakumani 2022; Singh et al., 2022). The synthesis of miRNAs is catalyzed by RNA polymerase II, which produces primary miRNAs that enter the nucleus. Drosha, a family of ribonucleases, further catalyzes the production of precursor miRNAs with a hairpin structure (Asakiya et al., 2022; Quirico et al., 2022). This double-stranded product consists of a mature miRNA guide strand and a miRNA guest strand. Both miRNAs can be loaded into the RNA-induced silencing complex to degrade or inhibit translation of mRNA, thereby affecting protein expression (Shunhao Zhang et al., 2021; Das et al., 2022; Diener et al., 2022). A single miRNA can target hundreds of mRNAs, affecting the expression and interactions of many genes and participating in the regulation of numerous physiological and pathological processes. The biogenesis and biological function of miRNAs is described in Figure 2.
FIGURE 2
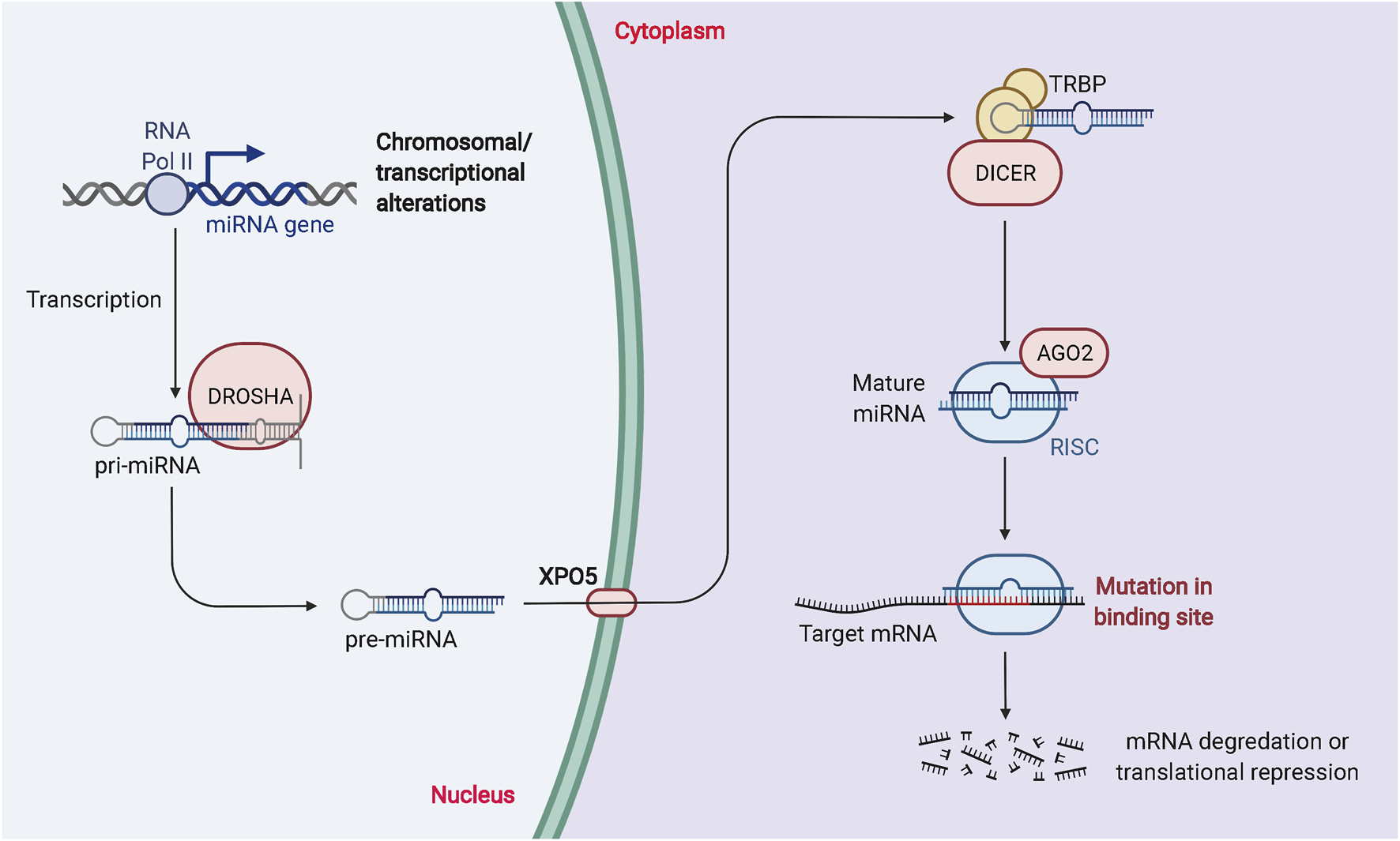
The biogenesis and biological function of miRNAs. RNA Pol II or III can regulate the transcription of pri-miRNAs, and DROSHA and DGCR8 can process the nuclear processing of pri-miRNAs into pre-miRNAs. Exportin 5 is involved in the processing of pre-miRNAs for nuclear export. Dicer and TRBP modulation regulate the cytoplasmic processing of pre-miRNAs into mature miRNA duplexes. miRNA duplexes include passenger strands and mature miRNAs. The passenger strand is degraded, and the mature miRNA strand is integrated into RISC to mediate translational repression or mRNA degradation depending on the degree of complementarity to the mRNA target.
2.2 LncRNAs
LncRNAs are defined as non-protein coding RNAs with transcripts longer than 200 bp (Huang et al., 2022; Yang et al., 2022), and they are considered byproducts of RNA polymerase II transcription with no biological function (Fernandes et al., 2021; Kong et al., 2021; Nojima and Proudfoot 2022). At present, there is no unified classification standard for lncRNAs (Nojima and Proudfoot 2022). According to the localization, lncRNAs are divided into cytoplasmic lncRNAs and nuclear lncRNAs, some of which are located in both the nucleus and the cytoplasm (Jianfei Tang et al., 2021). LncRNAs play different regulatory functions according to cellular localization (Cui et al., 2021). In the cytoplasm, lncRNAs act as competing endogenous RNAs (ceRNAs) to compete with miRNAs for binding and contribute to the release of target mRNAs. In tumors, abnormally expressed lncRNAs may break this balance, resulting in the abnormal expression of tumor-promoting genes or tumor-suppressor genes, and promoting the malignant progression of tumors. The ceRNA mechanism of lncRNAs is displayed in Figure 3. LncRNAs can be classified into five types according to the location in the genome relative to the protein-coding genes. The righteous lncRNAs overlap with exon regions of code-capable genes. The transcription of antisense lncRNAs begins with the reverse transcription of protein-coding genes. Bidirectional lncRNA expression start sites are very close to those of neighboring coding genes on the antisense strands. Basal lncRNAs originate from intronic regions, and intergenic lncRNAs are located in the interval between two genes on the chromosome (Cui et al., 2021; Jin et al., 2021). Based on their molecular function, lncRNAs are classified into decoy, guide, and backbone molecules. Decoy molecules are sufficient to induce transcription factors and inhibit the transcription of downstream genes. Guide molecules can bind to DNA or proteins at the same site and guide them to the site of action, enhancing the transcriptional activity of genes. The backbone molecules act as scaffolds for protein complexes, forming nucleic acid-protein complexes with target proteins and transferring the enzyme molecules involved in epigenetics to the protein complexes (Liu et al., 2022; Mercer et al., 2022). LncRNAs play critical regulatory roles in life activities and biological processes such as development, gene expression, stem cell differentiation, cell proliferation, and metastasis, indicating the close correlation to the occurrence and development of human diseases (DeSouza et al., 2021; Di Lu et al., 2021).
FIGURE 3
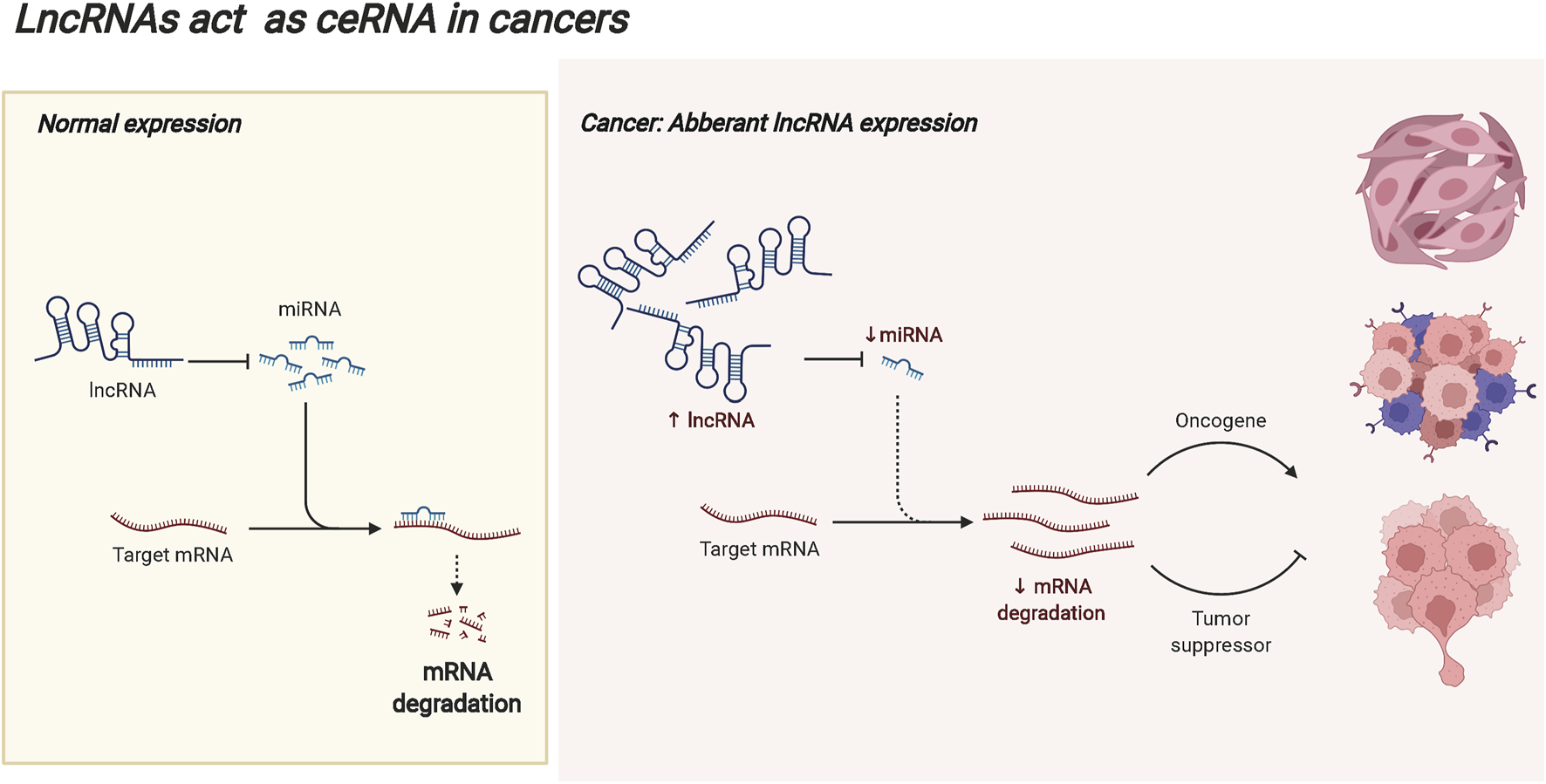
The ceRNA mechanism of lncRNAs. In the cytoplasm, lncRNAs can act as competing endogenous RNAs (ceRNAs) to compete with miRNAs for binding and contribute to the release of target mRNAs. In the occurrence and progression of tumors, abnormally expressed lncRNAs may break this balance, resulting in the abnormal expression of tumor-promoting genes or tumor suppressor genes, and promoting the malignant progression of tumors.
2.3 CircRNAs
CircRNAs are closed-loop molecules with unique structures that are widely distributed in animal and plant cells and regulate gene expression. They exist independently of proteins without being affected by exonucleases (Yue Zhang et al., 2022). Because of their low abundance and the limitation of detection technology, circRNAs were initially ignored as abnormal products of RNA splicing (Wen et al., 2022). The development of high-throughput sequencing technology has led to the identification of an increasing number of circRNAs (Kai Wang et al., 2022). Most circRNAs originate from exons in the coding regions of genes, whereas others originate from the 3′-UTR, 5′-UTR, introns, intergenic regions and antisense RNAs (Caba et al., 2021). CircRNAs are classified into four types, namely, exonic circRNAs (ecircRNAs), circular intronic RNAs (ciRNAs), exonic-intronic circRNAs (eiciRNAs), and tRNA intronic circRNAs (tricRNAs) (Wenqing Zhang et al., 2021). The most abundant and well studied circRNAs are exonucleotide circRNAs, which account for more than 80% of the total and are mainly localized in the cytoplasm. CiRNAs and eiciRNAs are abundant in the nucleus, and most circRNAs produced by different species are relatively evolutionarily conserved. The expression of the same circRNA varies greatly between diseased and non-diseased tissues and between different tissues or periods because of trans-shear (Tian Tian et al., 2021). In addition, circRNAs with covalent closed-loop structures are more stable than related linear mRNAs in vivo because they do not have 5′ caps or 3′ Poly A tails, which makes them highly resistant to the nucleic acid exonuclease (RNAse) (Zeng et al., 2021). Currently, there are three hypothetical models to explain the possible mechanisms of exon circRNA production, RNA binding protein (RBP)-mediated cyclization, intron pairing-driven cyclization, and lasso-driven cyclization (Kim et al., 2021; Sinha et al., 2021). The RBP connects two non-adjacent introns at both ends of the sequence, promoting the formation of a loop by bringing the two ends close together and then forming an exonic loop RNA by splicing. This model is defined as intron pairing-driven cyclization. The lasso-driven cyclization suggests that the pre-mRNA is in a half-folded state. During the transcription process, the non-adjacent exons approach each other and are driven by transacting factors to create a lasso. The intron sequence is removed by splicing in the lasso structure to form the circRNA. The biogenesis and biological function of circRNAs are shown in Figure 4.
FIGURE 4
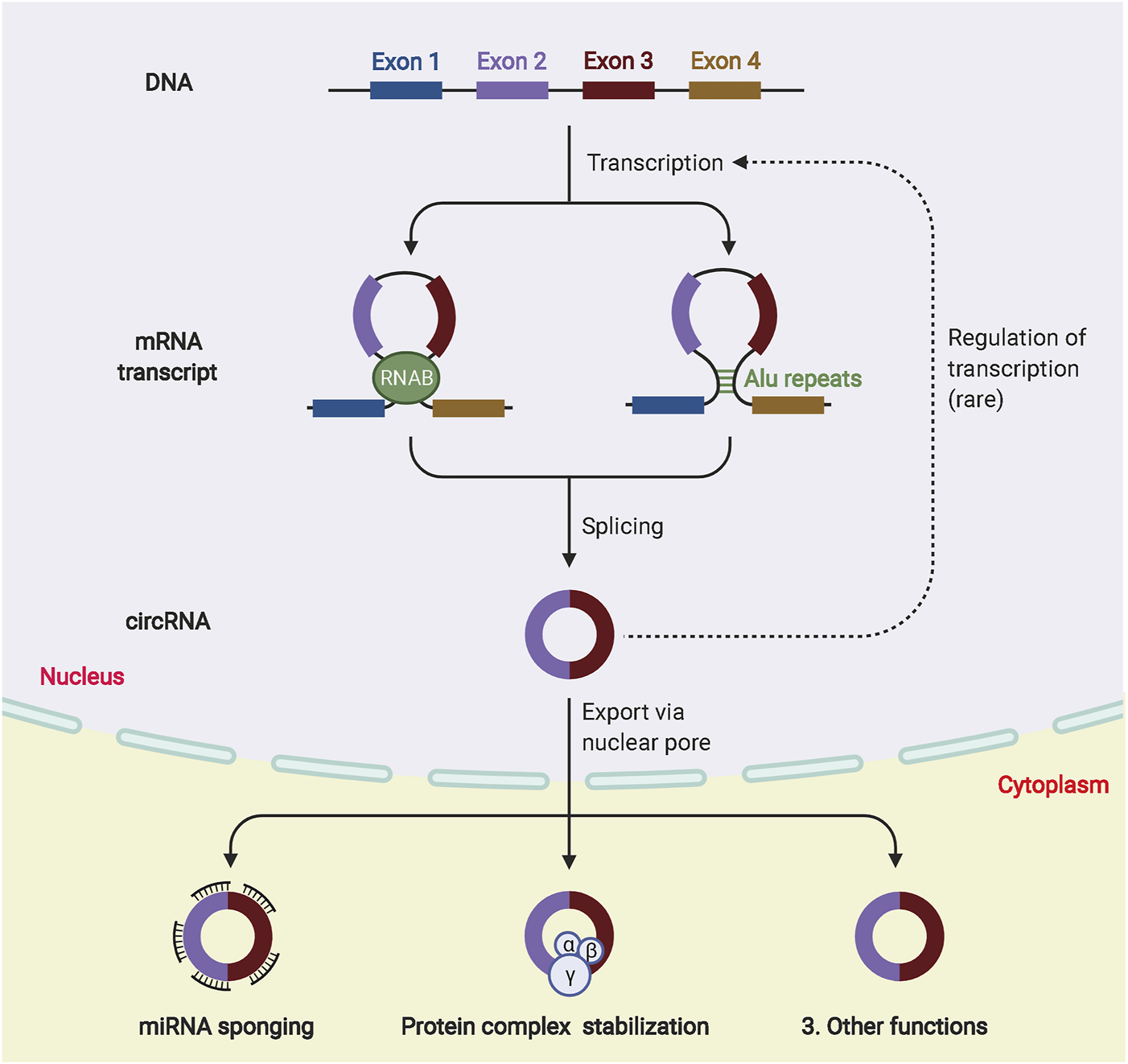
The biogenesis and biological function of circRNAs. Different circRNAs can be generated from pre-mRNA by back-splicing. According to the synthesis mechanism, circRNA can be divided into ecircRNA, ciRNA and eiciRNA. In the nucleus, circRNAs can participate in transcriptional regulation. In the cytoplasm, circRNAs are involved in many biological processes, such as miRNA sponge formation and binding to RBP.
3 Noncoding RNAs in skin cancer
NcRNAs play an essential role in the occurrence and development of skin cancers. This review summarizes the noncoding RNA aspects related to the metastasis mechanism of skin malignancies. The information may help identify new therapeutic targets related to the mechanism of tumor metastasis and provide new opportunities to improve the survival rate and quality of life of patients with skin malignancies.
3.1 MiRNAs in skin cancer
Sand et al. (Sand et al., 2012) identified 16 miRNAs that are highly expressed in BCC tissues and 10 miRNAs significantly downregulated in BCC tissues compared with adjacent skin by microarray miRNA analysis. MiR-203 inhibits BCC cell proliferation, differentiation, and tumor growth by targeting c-JUN as an oncogenic factor in BCC (Sonkoly et al., 2012). MiR-451a inhibits BCC cell growth and colony formation by targeting TBX1, which may serve as an effective therapeutic target for BCC (Sun and Jiang 2018). MiR-18a expression is significantly increased in BCC tissues and cells and promotes BCC malignant progression by targeting the Akt/mTOR/Beclin 1/LC3 axis (Mi et al., 2020). MiR-17-92 cluster members and miR-143–145 cluster members are involved in BCC progression as pro-oncogenes and oncogenes, respectively (Sand et al., 2017). The miRNAs involved in BCC are listed in Table 1.
TABLE 1
| miRNAs | Expression | Function role | Related genes | References |
|---|---|---|---|---|
| miR-203 | Down | Inhibit cell proliferation and differentiation and tumor growth | c-JUN | Sonkoly et al. (2012) |
| miR-451a | Down | Inhibit cell growth and colony formation | TBX1 | Sun and Jiang, (2018) |
| miR-18a | Up | Promote tumor malignant progression | Akt/mTOR/Beclin 1/LC3 | Mi et al. (2020) |
| miR-17-92 | Up | Promote tumor malignant progression | / | Sand et al. (2017) |
| miR-143-145 | Down | Inhibit tumor malignant progression | / | Sand et al. (2017) |
Functional characterization of miRNAs in basal cell carcinoma.
Various miRNAs are upregulated in CSCC and function as oncogenes to promote the malignant behavior of CSCC (Table 2 and Table 3). MiR-365 promotes the malignant progression of CSCC by targeting nuclear factor I B (NFIB), which may serve as a potential therapeutic target (Meijuan Zhou et al., 2014). MiR-31 expression promotes the malignant progression of tumors by increasing migration, invasion, and colony formation (Shao Hua Wang et al., 2014). MiR-365b and miR-135b target leucine zipper tumor suppressor 1 in CSCC tissues to promote migration and invasiveness (Olasz et al., 2015). Increased miR-365 expression promotes the malignant progression of tumors by targeting cyclin-dependent kinase 6 (Zhou et al., 2015). MiR-346 inhibits SRC kinase signaling inhibitor 1 expression and promotes CSCC cell proliferation and migration (Chen et al., 2016). Cadherin 1 and matrix metaloproteases (MMPs) are the downstream targets of miR-199a-5p and miR-217 in CSCC tissues, and their upregulation promotes malignant progression (Wang et al., 2016). Multiple miRNAs are upregulated in CSCC and regulate various downstream signaling targets to promote tumor cell proliferation, migration, and invasion. MiR-27 regulates PTRF (Bai et al., 2017), miR-31 regulates rhobtb1 (Lin et al., 2017), miR-506 regulates laminin subunit gamma 1 (Jian Zhou et al., 2019), miR-125b regulates MMP 13 (Xu et al., 2012), miR-124/214 regulates ERK1/2 (Yamane et al., 2013), and miR-196a promotes malignant behavior in the same cellular signaling pattern (Wei Zhang et al., 2022). In addition to promoting proliferation and migration, numerous miRNAs inhibit cancer cell apoptosis to enhance the malignant behavior. MiR-365 expression is upregulated in CSCC and inhibits apoptosis by targeting BAX (Zhou et al., 2017). MiR-142-5p targets PTEN (Bai et al., 2018), miR-186 targets reticulophagy regulator 1 (Xinde Hu et al., 2019), and miR-320a regulates autophagy related 2B (Bi et al., 2021) to inhibit cancer cell apoptosis. Programmed cell death 5 is the dominant downstream target of miR-766, the upregulation of which inhibits the apoptosis of CSCC cells (Liu et al., 2020). Increased expression of miR-22 decreases CSCC chemosensitivity by targeting the Wnt/β-catenin signaling axis, thus promoting malignant progression (Yuan et al., 2021). MiR-486-3p targets flotillin 2 (FLOT2) to promote CSCC cell proliferation, migration, and tumorigenicity (Li et al., 2022). Angiogenesis is a common antineoplastic target for drugs and ncRNAs. MiR-361-5p inhibits SCC cell proliferation and angiogenesis by targeting VEGFA (Kanitz et al., 2012).
TABLE 2
| miRNAs | Expression | Function role | Related genes | References |
|---|---|---|---|---|
| miR-365 | Up | Promote tumor malignant progression | NFIB/CDK6/BAX | (Meijuan Zhou et al., 2014; Zhou et al., 2015) |
| miR-31 | Up | Promote cell migration, invasion, and colony formation | RhoBTB1 | (Aoxue Wang et al., 2014; Lin et al., 2017) |
| miR-135b | Up | Increase the motility and aggressiveness of cancer cells | LZTS1 | Olasz et al. (2015) |
| miR-346 | Up | Promote cell proliferation and migration | SRCIN1 | Chen et al. (2016) |
| miR-199a-5p | Up | Promote tumor malignant progression | CDH1/MMPs | Wang et al. (2016) |
| miR-217 | Up | Promote cell growth, cell cycle and invasion | PTRF | Bai et al. (2017) |
| miR-142-5p | Up | Promote cell growth and inhibit cell apoptosis | PTEN | Bai et al. (2018) |
| miR-506 | Up | Promote cell viability, migration, and invasion | LAMC1 | Wu Zhou et al. (2019) |
| miR-186 | Up | Promote cell growth and inhibit cell apoptosis | RETREG1 | Yan Hu et al. (2019) |
| miR-766 | Up | Promote cell proliferation, migration, invasion and inhibit cell apoptosis | PDCD5/MMPs | Liu et al. (2020) |
| miR-320a | Up | Promote cell growth and inhibit cell apoptosis | ATG2B | Bi et al. (2021) |
| miR-22 | Up | Promote chemosensitivity | Wnt/β-catenin | Yuan et al. (2021) |
| miR-486-3p | Up | Promote cell proliferation, migration and tumorigenicity | FLOT2 | Li et al. (2022) |
| miR-196a | Up | Promote cell proliferation, migration, invasion | — | Wei Zhang et al. (2022) |
Functional characterization of upregulated miRNAs in cutaneous squamous cell carcinoma.
TABLE 3
| miRNAs | Expression | Function role | Related genes | References |
|---|---|---|---|---|
| miR-125b | Down | Inhibit proliferation, colony formation, migration and invasion | MMP13 | Xu et al. (2012) |
| miR-124/-214 | Down | Inhibit cell proliferation | ERK1/2 | Yamane et al. (2013) |
| miR-361-5p | Down | Inhibit cell invasion and angiogenesis | VEGFA | Kanitz et al. (2012) |
| miR-20a | Down | Suppress cell proliferation and metastasis | LIMK1 | Jianda Zhou et al. (2014) |
| miR-199a | Down | Inhibit tumor growth and metastasis | CD44 | Shao Hua Wang et al. (2014) |
| miR-199a-5p | Down | Inhibit cell invasion | BCAM, FZD6 and DDR1 | Kim et al. (2015) |
| miR-148a | Down | Inhibit cell proliferation and metastasis | MAP3K4 and MAP3K9 | Bai et al. (2018) |
| miR-204 | Down | Inhibit tumor malignant progression | PTPN11 | Toll et al. (2016) |
| miR-497 | Down | Inhibit tumor malignant progression | SERPINE-1 | Mizrahi et al. (2018) |
| miR-497 | Down | Inhibit cell proliferation | FAM114A2 | Wei et al. (2018) |
| miR-3619-5p | Down | Inhibit cell proliferation and cisplatin resistance | KPNA4 | Zhang et al. (2019) |
| miR-30a-5p | Down | Inhibit tumor malignant progression | FOXG1 | Zhang et al. (2019) |
| miR-199a-5p | Down | Inhibit tumor malignant progression | Sirt1/CD44ICD | Lu et al. (2020) |
| miR-27a | Down | Inhibit tumor malignant progression | EGFR | Yinghui Wang et al. (2019) |
| miR-125b | Down | Inhibit cell proliferation and promote cell apoptosis | STAT3 | Ke Tian et al. (2020) |
| miR-216b | Down | Inhibit tumor malignant progression | TPX2 | Feng et al. (2020) |
| miR-10a | Down | Inhibit tumor malignant progression | SDC-1 | Xiong et al. (2020) |
| miR-214 | Down | Inhibit tumor malignant progression | VEGFA and Bcl-2 | Ma et al. (2020) |
| miR-451a | Down | Inhibit tumor malignant progression | PDPK1 | Fu et al. (2021) |
| miR-130a | Down | Inhibit tumor malignant progression | ACVR1 | Lohcharoenkal et al. (2021) |
| miR-573 | Down | Inhibit cell proliferation, migration and invasion | PICSAR | Kai Wang et al. (2022) |
| miR-30c | Down | Inhibit cell proliferation and cisplatin resistance | SIRT1 | Liu et al. (2022) |
Functional characterization of downregulated miRNAs in cutaneous squamous cell carcinoma.
Decreased expression of various miRNAs results in CSCC suppression. MiR-20a is downregulated in CSCC tissues and inhibits cell proliferation and metastasis by targeting LIM domain kinase 1 (Jianda Zhou et al., 2014). Decreased expression of miR-199a affects the interaction between CD44 and Ezrin and inhibits CSCC proliferation and metastasis (Kanitz et al., 2012). MiR-199a-5p targets BCAM, FZD6, and DDR1 (Kim et al., 2015), and miR-148a targets MAP3K4 and MAP3K9 to inhibit the proliferation, invasiveness, and metastasis of CSCC cells (Luo et al., 2015). MiR-204 is downregulated in CSCC tissues and regulates protein tyrosine phosphatase non-receptor type 11 (PTPN11), modulating the signal transducer and activator of transcription 3 (STAT3) and MAPK signaling pathways to inhibit the malignant progression of CSCC (Toll et al., 2016). MiR-497 expression is decreased in CSCC tissues and inhibits the malignant progression of CSCC by targeting PTPN11. Decreased expression of miR-497 inhibits the malignant progression of CSCC by targeting PTPN11 and SERPINE-1 in the AKT/mTOR and EMT processes (Mizrahi et al., 2018) and the FAM114A2 signaling pathway (Wei et al., 2018). MiR-3619-5p is downregulated in CSCC and modulates karyopherin subunit alpha four to inhibit proliferation and cisplatin resistance (Zhang et al., 2019). MiR-30a-5p (Shao et al., 2019) and miR-199a-5p are downregulated in CSCC tissues and modulate forkhead box G1 (FOXG1) to inhibit CSCC cell migration, invasion, and colony formation. MiR-199a-5p also inhibits the migration and tumorigenicity of CSCC by targeting Sirt1/CD44ICD (Lu et al., 2020). MiR-27a regulates EGFR (Yinghui Wang et al., 2019), miR-125b regulates STAT3 (Ke Tian et al., 2020), miR-216b regulates TPX2 (Feng et al., 2020), and miR-10a regulates SDC-1 (Xiong et al., 2020) to inhibit CSCC cell proliferation, migration, and invasion. MiR-214 is downregulated in CSCC tissues and inhibits migration and invasion by regulating VEGFA, Bcl-2, and the Wnt/β-catenin pathway (Feng et al., 2020). Downregulation of miR-451a and miR-130a inhibits CSCC cell proliferation, migration, invasion, and EMT by regulating the PI3K/AKT signaling pathway through 3-phosphoinositide dependent protein kinase 1 (Fu et al., 2021). Decreased miR-130a expression inhibits CSCC proliferation, cell motility, and invasion by regulating the ACVR1 and BMP/SMAD pathways (Lohcharoenkal et al., 2021). MiR-573 and miR-30c are downregulated in CSCC tissues and inhibit proliferation, migration, and invasion by modulating PICSAR (Lipeng Wang et al., 2022) and SIRT1 (Liu et al., 2022).
In summary, miRNAs play an essential role in the development and progression of CMM, promoting or inhibiting malignant progression by regulating cell proliferation, invasion, metastasis, drug resistance, the immune microenvironment, cell cycle progression, and apoptosis. Detailed information is provided in Table 4, Table 5, Table 6.
TABLE 4
| Cellular process | miRNAs | Related genes | References |
|---|---|---|---|
| Cell proliferation | miR-377 | E2F3 and MAP3K7 | Zehavi et al. (2015) |
| miR-664 | PLP2 | Ding et al. (2015) | |
| miR-21 | PTEN | Saldanha et al. (2016) | |
| miR-330-5p | TYR and PDIA3 | Su et al. (2016) | |
| miR-4262 | KLF6 | Zhang et al. (2016) | |
| miR-135b | LATS2 | Xinde Hu et al. (2019) | |
| miR-181 | STAT3-AKT | He et al. (2020a) | |
| miR-140-3p | ABHD2 | He et al. (2020b) | |
| miR-34a | ZEB1 | Xu et al. (2021) | |
| Cell cycle | miR-193b | CCND1 | Chen et al. (2010) |
| miR-186 | — | Su et al. (2018) | |
| miR-21-5p | CDKN2C | Yang et al. (2020) | |
| Cell apoptosis | miR-125b | — | (Glud et al., 2011; Nyholm et al., 2014) |
| miR-92a-3p | MYCBP2 | Venza et al. (2016) | |
| miR-365 | BCL2 | Zhu et al. (2018) | |
| miR-128 | CCL18 | Song et al. (2018) |
Functional characterization of miRNAs in cell proliferation, cell cycle and apoptosis of melanoma.
TABLE 5
| Cellular process | miRNAs | Related genes | References |
|---|---|---|---|
| Migration | miR-532-5p | RUNX3 | Kitago et al. (2009) |
| MiR-125b | / | Glud et al. (2010) | |
| miR-200 and miR-203 | E-cadherin | van Kempen et al. (2012) | |
| miR-21 | TIMP3 | Martin del Campo et al. (2015) | |
| miR-21 | PDCD4 | Jiao et al. (2015) | |
| Metastasis | miR-23a | ATG12 | Guo et al. (2017) |
| miR-367 | PTEN | Long et al. (2018) | |
| miR-509-3p | GPC6 | Li et al. (2019) | |
| miR-152-5p | TXNIP | Li et al. (2020) |
Functional characterization of miRNAs in cell migration and metastasis of melanoma.
TABLE 6
| Cellular process | miRNAs | Related genes | References |
|---|---|---|---|
| Drug resistance | miR-514a | NF1 | Stark et al. (2015) |
| miR-579-3p | Verofini | Wang et al. (2018) | |
| miR-26a | HMGB1- dabrafenib | Yan Yu et al. (2019) | |
| miR-205 | INPPL1 | Sánchez-Sendra et al. (2020) | |
| miR-92a-3p | Dasatinib | Yuxiong Jiang et al. (2021) | |
| Tumor microenvironment | miR-125b-5p | LIPA | Gerloff et al. (2020) |
Functional characterization of miRNAs in drug resistance and the tumor microenvironment of melanoma.
3.2 LncRNAs in skin cancer
Sand et al. (2016a) identified 1851 upregulated lncRNAs and 2165 downregulated lncRNAs in microarrays of BCC compared with non-lesioned skin. Further exploration demonstrated the biological roles and molecular mechanisms of these differentially expressed lncRNAs. Lnc-PICSAR is upregulated in DDP-resistant CSCC cells and regulates the miR-485-5p/REV3-like DNA directed polymerase zeta catalytic subunit (REV3L) signaling axis (Wang et al., 2020). LINC00963 is increased in CSCC tissues and promotes malignant progression by upregulating SOX4 expression through the uptake of miR-1193 (Jingwen Wang et al., 2019). LINC00641 is downregulated in CSCC cell lines and inhibits CSCC growth and metastasis by downregulating miR-424 expression (Quan Liu et al., 2021). LINC00162 is specifically expressed in CSCC but not in keratin-forming cells in normal skin. Overexpression of LINC00162 promotes CSCC tumorigenesis in vitro and in vivo (Piipponen et al., 2016). LINC00319 is increased in CSCC and inhibits apoptosis, promoting cell proliferation, cell cycle progression, cell migration, and invasion. Mechanistic studies indicate that LINC00319 may exert an oncogenic function in CSCC by binding to miR-1207-5p and promoting the expression of cyclin-dependent kinase 3 (Li et al., 2018). LINC00520 regulates EGFR expression and inactivates the PI3K/Akt pathway, thereby inhibiting the development of CSCC (Mei and Zhong, 2019). USF1 activates LINC01048 to promote CSCC proliferation and inhibit apoptosis by interacting with TATA-box binding protein associated factor 15 (TAF15) to upregulate YAP1 (Chen et al., 2019). ALA-PDT promotes TINCR expression through the ERK1/2-SP3 pathway to induce apoptosis and autophagy, leading to the malignant progression of CSCC (Xiaobo Zhou et al., 2019). HOX transcript antisense RNA (HOTAIR) is elevated in CSCC cells and promotes migration, proliferation, and EMT, possibly by binding to miR-326 and promoting PRA1 domain family member 2 (PRAF2) expression (Guo Jun Yu et al., 2019). HOTAIR promotes CSCC stemness and progression by upregulating Sp1 and modulating miR-199a (Chen et al., 2022). LINC00346 is highly expressed in CSCC tissues to promote CSCC malignant progression by activating STAT3 and MMP expression (Piipponen et al., 2020). Small Cajal body-specific RNA 2 is upregulated in CSCC tissues and promotes proliferation and invasion by suppressing miR-342-3p expression (Zhang et al., 2020a). PICSAR functions as an oncogene by regulating the miR-125b/YAP1 signaling axis, and EZR-AS1 regulates the PI3K/AKT signaling pathway to promote CSCC cell proliferation and invasion and inhibit apoptosis (Qirong Lu et al., 2021). Overexpression of H19 promotes malignant behavior and induces apoptosis. H19 promotes the expression of the EMT-related marker miR-675 and inhibits p53 expression (Shunhao Zhang et al., 2021). Hcp5 can upregulate EZH2 expression by competitively binding to miR-138-5p, promoting autophagy and suppressing apoptosis by regulating the STAT3/VEGFR2 pathway, leading to malignant progression of CSCC (Zou et al., 2021). Neat1 promotes autophagy and decreases apoptosis by binding to miR-361-5p, promoting CSCC proliferation and invasion by activating the Wnt pathway (Shiqiu Jiang et al., 2021). NEAT1 promotes CSCC proliferation, invasion, and metastasis by regulating the expression of metalloplasmic proteins (Gong et al., 2022).
FOXD2-AS1 expression is increased in cutaneous melanoma tissue specimens and cell lines (Ren et al., 2019), inhibiting proliferation, migration, and invasion of cutaneous melanoma cells by regulating phospho-Akt expression (Wu Zhou et al., 2019). CPS1-IT1 inhibits melanoma metastasis by competitively binding to BRG1 and impairing CYR61 expression. Linc00961 is decreased in cutaneous melanoma tissues and functions as a ceRNA to regulate the miR-367/PTEN axis and inhibit cell proliferation and invasion (Mu et al., 2019). LncRNA FOXD3-AS1 promotes the proliferation, invasion, and migration of CMM by binding to miR-325 and promoting MAP3K2 expression as a potential cause of cutaneous melanoma (Wei et al., 2019). FENDRR suppresses MMP2 and MMP9 and antagonizes the JNK/c-Jun pathway to promote proliferation, migration, and invasion of melanoma (Shuang Chen et al., 2020). LncRNA-TTN-AS1 promotes TTN transcription and increases the stability of TTN mRNA thus inducing tumor progression. LINC00518 is upregulated in CMM and induces radioresistance of CMM by regulating glycolysis through a miR-33a-3p/HIF-1α negative feedback loop (Yan Liu et al., 2021). LINC01116 promotes cell proliferation, migration, invasion and EMT to sponge miR-3612 by regulating GDF11 and SDC3, thereby promoting melanoma progression (Yanhua Wang et al., 2022). LncRNA Tincr is downregulated in CMM, which promotes CTGF, CCN1, and AXL expression, leading to cell proliferation, invasion, and apoptosis inhibition in CMM cell lines. p53 upregulates PURPL in melanoma to promote cell proliferation, colony formation, migration, and invasion, and inhibiting apoptosis (Shuo Han et al., 2021). Mechanistic studies show that PURPL promotes mTOR-mediated phosphorylation of ULK1 at Ser757 by physically interacting with mTOR and ULK1, limiting the autophagic response to suppress apoptosis. TEX41 is activated by IRF4 and binds to miR-103a-3p to upregulate C1QB, thereby promoting cell proliferation, migration, and invasion while inhibiting apoptosis, suggesting that TEX41 is a potential therapeutic target for melanoma (Zheng et al., 2021). Linc00518 is upregulated in CMM tissues and promotes malignant progression via the miR-526b-3p/EIF5A2 axis (Xu et al., 2022).
Taken together, these data indicate that lncRNAs function as tumor-promoters or tumor-suppressors in CMM by interacting with miRNAs and regulating protein expression.
3.3 CircRNAs in skin cancer
The detailed information of circRNAs in skin cancers is shown in Table 7.
TABLE 7
| Cancer | circRNAs | Expression | Function role | Related genes | References |
|---|---|---|---|---|---|
| Basal cell carcinoma | Circ_0005795 | Up | Promote cell viability, colony formation, and suppress cell apoptosis | miR-1231 and caspase-3 | Yating Li et al. (2021) |
| Circ_NCKAP1 | Up | Promote cell proliferation, inhibit cell apoptosis | miR-148b-5p/HSP90 axis | Fan et al. (2021) | |
| Malignant melanoma | circRNA_0084043 | Up | promote melanoma cell proliferation, invasion and migration | miR-153-3p/Snail axis | Luan et al. (2018) |
| hsa_circ_0025039 | Up | promote cell proliferation, colony formation ability, invasion and glucose metabolism in melanoma cells | miR-198/CDK4 | Bian et al. (2018) | |
| circMTUS1 | Up | promote cell proliferation | hsa-miR-622/hsa-miR-1208 | Shang et al. (2019) | |
| Circular RNA ITCH | Up | downregulate GLUT1 and suppresses glucose uptake in melanoma to inhibit cancer cell proliferation | GLUT1 | Lin et al. (2021) | |
| circMYC | Up | promote the proliferation of human melanoma cells and Mel-CV cells. repress Mel-CV cell glycolysis and LDHA activities | Mel-CV cells/miR-1236/c-MYC-SRSF1 axis | Jin et al. (2020) | |
| CDR1as | Down | inhibit malignant progression | miR-7/LINC00632/IGF2BP3 | Hanniford et al. (2020) | |
| circ_0084043 | Up | promote cell proliferation, migration and invasion, facilitate apoptosis in A375 and SK-MEL-28 cells | miRNA (miR)-429/TRIB2 | Zhibing Chen et al. (2020) | |
| circ-FOXM1 | Up | promote cell proliferation, invasion, and glycolysis and facilitated cell apoptosis | miR-143-3p/FLOT2/MTT assay | Ke Tian et al. (2020) | |
| circ_0002770 | Up | promote cell invasion, migration, and proliferation | miR-331-3p/DUSP5 and TGFBR1 | Qian et al. (2020) | |
| circ_0020710 | Up | promoted melanoma cell proliferation, migration and invasion | miR-370-3p/CXCL12 | Wei et al. (2020) | |
| circ 0001591 | Up | promoted cell growth and cell invasion and reduced apoptotic rate of melanoma | ROCK1/PI3K/AKT/ROCK1/miR-431-5p | Yin et al. (2021) | |
| circ_0079593 | Up | Promotes cell proliferation, cell cycle progression, migration, invasion, inhibits cell apoptosis, and promotes tumor growth | miR-573/ABHD2 | Zhao et al. (2021) | |
| CircRNA_0082835 | Up | Promotes proliferation, invasion and migration of melanoma cells and regulates cell cycle levels | EZH2/miR-429 | Sun et al. (2021) | |
| circZNF609 | Up | Promotes the invasion, migration and proliferation of melanoma cells and inhibits apoptosis | miR-138-5p/SIRT7 axis | Yan Liu et al. (2021) | |
| circVANGL1 | Up | Promotes proliferation, migration and invasion of melanoma cells | miR-150-5p/TGF-β | Zhou et al. (2021) | |
| Cutaneous squamous cell carcinoma | circ_0070934 | Up | associated with tumor aggressiveness | miR-1238/miR-1247–5p | An et al. (2019) |
| hsa_circ_0070934 | Up | Promotes the invasion and proliferation potential of CSCC cells and inhibits apoptosis | miR-1236-3p/HOXB7 | Zhang et al. (2020a) | |
| circPVT1 | Up | Promote CSCC cell proliferation and migration | / | Shuang Chen et al. (2020) | |
| circRNA_001937 | Up | Promote CSCC progression and inhibit apoptosis | miRNA-597-3p/FOSL2 | Gao et al. (2020) | |
| circSEC24A | Up | Promotes cell proliferation, migration, invasion and glycolysis, inhibits apoptosis | miR-1193/MAP3K9 axis | Xiaoyan Lu et al. (2021a) | |
| hsa_circ_0001360 | Down | Inhibits the proliferation, migration and invasion of SCL-1 cells and promotes apoptosis | / | Chen et al. (2021) | |
| Circ_0067772 | Up | Promotes proliferation, migration and invasion of CSCC cells | miR-1238-3p/FOXG1 axis | Ziwei Li et al. (2021) | |
| hsa_circ_0070934 | Up | Promote CSCC cell proliferation, cell cycle process, migration, invasion, and inhibit apoptosis | miR-136-5p/PRAF2 axis | Xiong et al. (2021) | |
| circ-CYP24A1 | Up | Promote cell proliferation, migration and invasion, inhibit apoptosis | CDS2、MAVS 和 SOGA | Zhang et al. (2021a) | |
| hsa_circ_0008234 | Up | Increased the cell viability and colony formation of cSCC cells | miR-127-5p/ADCY7 | Cai et al. (2021) | |
| circFADS2 | Down | Inhibits CSCC cell proliferation, metastasis and glycolysis | miR-766-3p/HOXA9 | Zhang et al. (2021b) | |
| circ-LARP1B | Up | Promotes cell viability, colony-forming ability, migration, invasion, cell cycle progression and glycolysis of CSCC cells, and inhibits apoptosis | miR-515-5p/TPX2 axis | Yan Wang et al. (2022) | |
| circ_0001821 | Up | Promotes cell viability, colony formation, cell cycle progression and metastasis, and inhibits apoptosis in vitro and promotes tumor growth in vivo | miR-148a-3p/EGFR axis, PI3K/Akt pathway | Yue Zhang et al. (2022) |
Functional characterization of circRNAs in skin cancers.
A circRNA microarray identified 23 upregulated and 48 downregulated circRNAs in BCC with a potential function in regulating the development of BCC. However, the underlying mechanism needs to be examined (Sand et al., 2016b). Circ_0005795 expression is increased in BCC tissues, and knockdown of circ_0005795 inhibits cell viability, colony formation, and anti-apoptotic protein levels while increasing caspase-3 activity (Yating Li et al., 2021). Circ_0005795 exerts its pro-tumor effects by binding to miR-1231. Circ_NCKAP1 promotes the malignant progression of cutaneous BCC by regulating the miR-148b-5p/HSP90 signaling axis (Fan et al., 2021).
Circ_0070934 is expressed aberrantly in CSCC. Overexpression of circ_0070934 promotes cell proliferation, invasion, and migration, and inhibits apoptosis (An et al., 2019). The hsa_circ_0070934 binds to miR-1236-3p and regulates homeobox B7 expression to promote the malignant progression of CSCC (Zhang et al., 2020b). GSPS downregulates the expression of hsa_circ_0070934 and inhibits CSCC cell proliferation, cell cycle progression, migration, and invasion, and promotes apoptosis. However, GSPS also plays an oncogene role in CSCC by regulating the hsa_circ_0070934/miR-136-5p/PRA1 domain family member 2 (PRAF2) axis (Xiong et al., 2021). CircRNA_001937 promotes the malignant progression of CSCC by regulating the miR-597-3p/FOSL2 pathway as determined through ex vivo assays (Gao et al., 2020). CircSEC24A and MAP3K9 are upregulated in CSCC tissues, whereas the expression of miR-1193 is decreased (Ziwei Lu et al., 2021). Inhibition of circSEC24A inhibits malignant behavior and glycolysis to induced apoptosis, whereas overexpression of circSEC24A promotes tumor growth. CircSEC24A may act as a molecular sponge for miR-1193 and thus regulate the expression of MAP3K9 (Chen et al., 2021). The hsa_circ_0001360 inhibits cell proliferation, migration, and invasion and promotes apoptosis, acting as an antioncogene in CSCC. Circ_0067772 is upregulated in CSCC tissues and cells, and overexpression of circ_0067772 regulates FOXG1 by binding to miR-1238-3p, thereby promoting cell proliferation, migration, and invasion. Inhibition of circ_0067772 suppresses tumor growth (Xiaoqing Li et al., 2021).
Zhang et al. (2021c) also screened differentially expressed circRNAs in CSCC exosomes by RNA-seq analysis and identified 25 upregulated and 76 downregulated exosomal circRNAs. Circ-CYP24A1 promotes CSCC malignant progression by inducing the expression of CDS2, MAVS, and SOGA. Hsa_circ_0008234 promotes CSCC malignant progression by targeting miR-127-5p to regulate adenylate cyclase 7 expression (Cai et al., 2021). Circfads2 is downregulated in CSCC tissues and inhibits CSCC cell proliferation, metastasis, and glycolysis (Wenqing Zhang et al., 2021). Mechanistic studies suggest that circfads2 regulates the miR-766-3p/HOXA9 axis to inhibit CSCC progression, and is thus a potential therapeutic target for CSCC. Circ-LARP1B/miR-515-5p/TPX2 microtubule nucleation factor regulatory axis is involved in the malignant progression of CSCC by promoting cell viability, colony formation, migration, invasion, cell cycle progression, and glycolysis and inhibiting apoptosis (Yan Wang et al., 2022). Circ_0001821 plays a pro-oncogenic role in CSCC development by regulating the miR-148a-3p/EGFR signaling axis and PI3K/Akt pathway (Yue Zhang et al., 2022).
Luan et al. (Luan et al., 2018) found that Circ_0084043 is upregulated in melanoma tissues and functions as a sponge for miR-153-3p to upregulate Snail expression and thus promote melanoma cell proliferation, invasion, and migration. The hsa_circ_0025039 promotes CDK4 expression by binding to miR-198, promoting cell proliferation, colony-forming ability, invasion, and glucose metabolism (Bian et al., 2018). Circ_0084043 expression is increased in melanoma tissues and promotes the malignant development of melanoma by regulating the miR-429/TRIB2 signaling axis and Wnt/β-catenin signaling pathway (Xu E Chen et al., 2020). Circ-FOXM1 upregulates FLOT2 by binding to miR-143-3p, promoting cell proliferation, invasion, and glycolysis, playing a tumorigenic role in vivo (Shan Tian et al., 2020). The circRNA circ_0002770 promotes dual specificity phosphatase 5 and transforming growth factor beta receptor 1 expression by binding to miR-331-3p, thereby promoting the proliferation, invasion, and migration of melanoma cells, leading to malignant tumor progression (Qian et al., 2020). Wei et al. (Wei et al., 2020) showed that circ_0020710 expression is increased in melanoma tissues and promotes proliferation, migration, and invasion. increased circ_0020710 upregulates CXCL12 expression by sponging miR-370-3p, which is associated with cytotoxic lymphocyte depletion. Circ 0001591 expression is increased in melanoma patient sera, which promotes cell growth and invasion and inhibits apoptosis (Yin et al., 2021). Circ 0001591 upregulates ROCK1 expression by binding to miR-431-5p and promotes malignant progression of melanoma by regulating the ROCK1/PI3K/AKT signaling pathway. Circ_0079593 regulates ABHD2 expression through miR-573 and promotes cell proliferation, migration, and invasion and inhibits apoptosis (Zhao et al., 2021). Circ_0082835 is upregulated in melanoma tissues and binds to miR-429 to promote the proliferation, invasion, and migration of melanoma cells, as well as regulating cell cycle progression (Sun et al., 2021). CircZNF609 inhibits DNA damage and promotes the malignant progression of melanoma by regulating the miR-138-5p/SIRT7 signaling axis (Quan Liu et al., 2021). The expression of circVANGL1 is increased in melanoma tissues and cell lines, and overexpression of circVANGL1 promotes the proliferation, migration, and invasion of melanoma cells (Zhou et al., 2021). Taken together, these findings indicate that lncRNAs function as tumor-promoters or tumor-suppressors involved in the malignancy of cutaneous melanoma through their ceRNA activity for miRNAs.
4 Future prospects and conclusion
Various miRNAs are involved in the regulation of skin cancer development and metastasis. The differential expression of miRNAs at different stages of skin cancer provides a new basis and novel direction for developing biomarkers and therapeutic targets. Further studies on the mechanism underlying the function of miRNAs will facilitate the development of new therapeutic strategies for the treatment of skin cancers, which will improve the outcomes of patients with advanced skin cancer. LncRNAs are critical epigenetic factors that regulate the development, progression, metastasis, and resistance to therapeutic agents in skin cancers. Although the majority of lncRNAs are expressed at low levels, the value of lncRNAs as early diagnostic, predictive, and therapeutic targets for skin cancer is becoming increasingly evident. The complexity of lncRNAs increases the difficulty of their biological investigation because the same lncRNAs function differently in distinct tumor types. Exploring the functions of lncRNAs and elucidating the molecular regulatory mechanisms involved in the development and progression of skin cancer is a new research direction.
CircRNAs have drawn increased attention in life science and medical research. CircRNAs competitively bind to miRNAs and regulate multiple tumor signaling pathways, suggesting the potential of circRNAs as biomarkers for detecting related diseases. Research on circRNAs in tumors is still in its infancy, with only a fraction of functional circRNAs available. The current circRNA database is incomplete, and there are no databases of circRNAs related to tumor prognosis for functional prediction after RNA sequencing. In addition, the mechanism of circRNAs in tumorigenesis remains to be fully elucidated. Their interaction with mRNA and its diagnostic applications show potential for clinical practice. CircRNAs show great potential in tumor diagnosis, prognosis, and treatment with high clinical value. We believe that continuing efforts and practical investigation will clarify the value of circRNAs for precision medicine in clinical diagnosis and treatment. The cross-talk of ncRNAs is complex and highly tissue specific. Therefore, exploring their functions in many cell types or tissues is essential to determine specificity and therapeutic effects. Despite the challenges associated with the application of ncRNAs, extensive research efforts have been devoted to extend investigation to ncRNA clinical trials.
This review summarized the mechanisms underlying the role of ncRNAs in the occurrence, metastasis, and drug resistance of skin cancers. Their potential application for the early diagnosis, prognosis assessment, and targeted therapy has been suggested. Several ncRNAs have undergone different phases of clinical trials. For example, a miR-122-related drug (miravirsen) and ncRNA-associated nanoparticles have been the subject of clinical trials (Gebert et al., 2014). The miRNA mimic MIRX34 was used in a phase I trial in patients with primary liver cancer (Bouchie 2013). Therefore, further exploration of clinical biosignatures and functional cell assays is needed. In addition, non-invasive methods should be considered for ncRNA harvesting.
Statements
Author contributions
Original draft preparation, allocation, supplementation and editing: SL and FL. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Liaoning Cancer Hospital and Institute (Shenyang) and China Medical University (Shenyang).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- BCC
basal cell carcinoma
- CSCC
cutaneous squamous cell carcinoma
- CM
cutaneous melanoma
- miRNAs
microRNAs
- lncRNAs
long non-coding RNAs
- circRNAs
circular RNAs
- BRAF
B-Raf proto-oncogeneserine/threonine kinase
- ncRNA
non-coding RNA
- mRNA
messenger ribonucleic acid
- ciRNA
circular intronic RNA
- eiciRNA
exon-intron circular RNA
- RBP
RNA binding protein
- TBX1
T-box transcription factor 1
- NFIB
nuclear factor I B
- LZTS1
leucine zipper tumor suppressor 1
- CDK6
cyclin dependent kinase 6
- SRCIN1
SRC kinase signaling inhibitor 1
- CDH1
cadherin 1
- LAMC1
laminin subunit gamma 1
- RETREG1
reticulophagy regulator 1
- PDCD5
programmed cell death 5
- ATG2B
autophagy related 2B
- FLOT2
flotillin 2
- LIMK1
LIM domain kinase 1
- STAT3
signal transducer and activator of transcription 3
- PTPN11
protein tyrosine phosphatase non-receptor type 11
- FAM114A2
family with sequence similarity 114 member A2
- KPNA4
karyopherin subunit alpha 4
- FOXG1
forkhead box G1
- TPX2TPX2
TPX2 microtubule nucleation factor
- PDPK1
3-phosphoinositide dependent protein kinase 1
- REV3L
REV3 like, DNA directed polymerase zeta catalytic subunit
- TAF15
TATA-box binding protein associated factor 15
- HOTAIR
HOX transcript antisense RNA
- PRAF2PRAF2
PRA1 domain family member 2
- SCARNA2
small Cajal body-specific RNA 2
- MAP3K2
mitogen-activated protein kinase kinase kinase 2
- PURPL
p53 level regulator
- HOXB7
homeobox B7
- PRAF2PRAF2
PRA1 domain family member 2
- TPX2TPX2
TPX2 microtubule nucleation factor
- DUSP5
dual specificity phosphatase 5
- TGFBR1
transforming growth factor beta receptor 1
References
1
Allegra A. Pioggia G. Tonacci A. Musolino C. Gangemi S. (2020). Oxidative stress and photodynamic therapy of skin cancers: Mechanisms, challenges and promising developments. Antioxidants (Basel)9 (5), E448. 10.3390/antiox9050448
2
An X. Liu X. Ma G. Li C. (2019). Upregulated circular RNA circ_0070934 facilitates cutaneous squamous cell carcinoma cell growth and invasion by sponging miR-1238 and miR-1247-5p. Biochem. Biophys. Res. Commun.513 (2), 380–385. 10.1016/j.bbrc.2019.04.017
3
Aoxue Wang A. Landén N. X. Meisgen F. Lohcharoenkal W. Ståhle M. Sonkoly E. et al (2014). MicroRNA-31 is overexpressed in cutaneous squamous cell carcinoma and regulates cell motility and colony formation ability of tumor cells. PLoS One9 (7), e103206. 10.1371/journal.pone.0103206
4
Asakiya C. Zhu L. Yuhan J. Zhu L. Huang K. Xu W. et al (2022). Current progress of miRNA-derivative nucleotide drugs: Modifications, delivery systems, applications. Expert Opin. Drug Deliv.19 (4), 435–450. 10.1080/17425247.2022.2063835
5
Bai M. Zhang M. Long F. Yu N. Zeng A. Wang X. et al (2017). MiR-217 promotes cutaneous squamous cell carcinoma progression by targeting PTRF. Am. J. Transl. Res.9 (2), 647–655.
6
Bai X. Zhou Y. Chen P. Yang M. Xu J. (2018). MicroRNA-142-5p induces cancer stem cell-like properties of cutaneous squamous cell carcinoma via inhibiting PTEN. J. Cell. Biochem.119 (2), 2179–2188. 10.1002/jcb.26379
7
Bednarski I. A. Ciążyńska M. Wódz K. Dróżdż I. Skibińska M. Narbutt J. et al (2021). Hippo signaling pathway as a new potential target in non-melanoma skin cancers: A narrative review. Life (Basel)11 (7), 680. 10.3390/life11070680
8
Bi X. Jiang Z. Luan Z. Qiu D. (2021). Crocin exerts anti-proliferative and apoptotic effects on cutaneous squamous cell carcinoma via miR-320a/ATG2B. Bioengineered12 (1), 4569–4580. 10.1080/21655979.2021.1955175
9
Bian D. Wu Y. Song G. (2018). Novel circular RNA, hsa_circ_0025039 promotes cell growth, invasion and glucose metabolism in malignant melanoma via the miR-198/CDK4 axis. Biomed. Pharmacother.108, 165–176. 10.1016/j.biopha.2018.08.152
10
Bouchie A. (2013). First microRNA mimic enters clinic. Nat. Biotechnol.31 (7), 577. 10.1038/nbt0713-577
11
Caba L. Florea L. Gug C. Dimitriu D. C. Gorduza E. V. (2021). Circular RNA-is the circle perfect?Biomolecules11 (12), 1755. 10.3390/biom11121755
12
Cai L. Wang Y. Wu J. Wu G. (2021). Hsa_circ_0008234 facilitates proliferation of cutaneous squamous cell carcinoma through targeting miR-127-5p to regulate ADCY7. Arch. Dermatol. Res.314, 541–551. 10.1007/s00403-021-02261-8
13
Chen J. Feilotter H. E. Paré G. C. Zhang X. Pemberton J. G. Garady C. et al (2010). MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am. J. Pathol.176 (5), 2520–2529. 10.2353/ajpath.2010.091061
14
Chen B. Pan W. Lin X. Hu Z. Jin Y. Chen H. et al (2016). MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumour Biol.37 (2), 2765–2771. 10.1007/s13277-015-4046-2
15
Chen L. Chen Q. Kuang S. Zhao C. Yang L. Zhang Y. et al (2019). USF1-induced upregulation of LINC01048 promotes cell proliferation and apoptosis in cutaneous squamous cell carcinoma by binding to TAF15 to transcriptionally activate YAP1. Cell Death Dis.10 (4), 296. 10.1038/s41419-019-1516-2
16
Chen P. Li C. Huang H. Liang L. Zhang J. Li Q. et al (2021). Circular RNA profiles and the potential involvement of down-expression of hsa_circ_0001360 in cutaneous squamous cell carcinogenesis. FEBS Open Bio11 (4), 1209–1222. 10.1002/2211-5463.13114
17
Chen J. Hou S. F. Tang F. J. Liu D. S. Chen Z. Z. Zhang H. L. et al (2022). HOTAIR/Sp1/miR-199a critically regulates cancer stemness and malignant progression of cutaneous squamous cell carcinoma. Oncogene41 (1), 99–111. 10.1038/s41388-021-02014-x
18
Cives M. Mannavola F. Lospalluti L. Sergi M. C. Cazzato G. Filoni E. et al (2020). Non-melanoma skin cancers: Biological and clinical features. Int. J. Mol. Sci.21 (15), E5394. 10.3390/ijms21155394
19
Cui X. Y. Zhan J. K. Liu Y. S. (2021). Roles and functions of antisense lncRNA in vascular aging. Ageing Res. Rev.72, 101480. 10.1016/j.arr.2021.101480
20
Das M. K. Haugen Ø P. Haugen T. B. (2022). Diverse roles and targets of miRNA in the pathogenesis of testicular germ cell tumour. Cancers (Basel)14 (5), 1190. 10.3390/cancers14051190
21
DeSouza P. A. Qu X. Chen H. Patel B. Maher C. A. Kim A. H. et al (2021). Long, noncoding RNA dysregulation in glioblastoma. Cancers (Basel)13 (7), 1604. 10.3390/cancers13071604
22
Di Lu D. Sun L. Li Z. Mu Z. (2021). lncRNA EZR-AS1 knockdown represses proliferation, migration and invasion of cSCC via the PI3K/AKT signaling pathway. Mol. Med. Rep.23 (1), 76. 10.3892/mmr.2020.11714
23
Diener C. Keller A. Meese E. (2022). Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet.38, 613–626. 10.1016/j.tig.2022.02.006
24
Ding Z. Jian S. Peng X. Liu Y. Wang J. Zheng L. et al (2015). Loss of MiR-664 expression enhances cutaneous malignant melanoma proliferation by upregulating PLP2. Med. Baltim.94 (33), e1327. 10.1097/md.0000000000001327
25
Ding L. Wang R. Shen D. Cheng S. Wang H. Lu Z. et al (2021). Role of noncoding RNA in drug resistance of prostate cancer. Cell Death Dis.12 (6), 590. 10.1038/s41419-021-03854-x
26
Fan Z. X. Xi W. Miao X. Y. Li L. Y. Miao G. Y. (2021). Circ_NCKAP1 promotes skin basal cell carcinoma progression by sponging the miR-148b-5p/HSP90 axis. Eur. Rev. Med. Pharmacol. Sci.25 (17), 5355–5364. 10.26355/eurrev_202109_26642
27
Feng C. Zhang H. L. Zeng A. Bai M. Wang X. J. (2020). Tumor-Suppressive MicroRNA-216b binds to TPX2, activating the p53 signaling in human cutaneous squamous cell carcinoma. Mol. Ther. Nucleic Acids20, 186–195. 10.1016/j.omtn.2020.01.022
28
Fernandes M. Marques H. Teixeira A. L. Medeiros R. (2021). miRNA- and lncRNA-based therapeutics for non-hodgkin's lymphoma: Moving towards an RNA-guided precision medicine. Cancers (Basel)13 (24), 6324. 10.3390/cancers13246324
29
Fu J. Zhao J. Zhang H. Fan X. Geng W. Qiao S. et al (2021). MicroRNA-451a prevents cutaneous squamous cell carcinoma progression via the 3-phosphoinositide-dependent protein kinase-1-mediated PI3K/AKT signaling pathway. Exp. Ther. Med.21 (2), 116. 10.3892/etm.2020.9548
30
Fujimura T. Aiba S. (2020). Significance of immunosuppressive cells as a target for immunotherapies in melanoma and non-melanoma skin cancers. Biomolecules10 (8), E1087. 10.3390/biom10081087
31
Gao L. Jin H. J. Zhang D. Lin Q. (2020). Silencing circRNA_001937 may inhibit cutaneous squamous cell carcinoma proliferation and induce apoptosis by preventing the sponging of the miRNA-597-3p/FOSL2 pathway. Int. J. Mol. Med.46 (5), 1653–1660. 10.3892/ijmm.2020.4723
32
Gebert L. F. Rebhan M. A. Crivelli S. E. Denzler R. Stoffel M. Hall J. et al (2014). Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res.42 (1), 609–621. 10.1093/nar/gkt852
33
Gerloff D. Lützkendorf J. Moritz R. K. C. Wersig T. Mäder K. Müller L. P. et al (2020). Melanoma-derived exosomal miR-125b-5p educates tumor associated macrophages (TAMs) by targeting lysosomal acid lipase A (lipa). Cancers (Basel)12 (2), E464. 10.3390/cancers12020464
34
Glud M. Rossing M. Hother C. Holst L. Hastrup N. Nielsen F. C. et al (2010). Downregulation of miR-125b in metastatic cutaneous malignant melanoma. Melanoma Res.20 (6), 479–484. 10.1097/CMR.0b013e32833e32a1
35
Glud M. Manfé V. Biskup E. Holst L. Dirksen A. M. Hastrup N. et al (2011). MicroRNA miR-125b induces senescence in human melanoma cells. Melanoma Res.21 (3), 253–256. 10.1097/CMR.0b013e328345333b
36
Gong Z. Shen G. Huang C. Zhang J. Ji J. (2022). Downregulation of lncRNA NEAT1 inhibits the proliferation of human cutaneous squamous cell carcinoma in vivo and in vitro. Ann. Transl. Med.10 (2), 79. 10.21037/atm-21-6916
37
Guo Jun Yu G. J. Sun Y. Zhang D. W. Zhang P. (2019). Long non-coding RNA HOTAIR functions as a competitive endogenous RNA to regulate PRAF2 expression by sponging miR-326 in cutaneous squamous cell carcinoma. Cancer Cell Int.19, 270. 10.1186/s12935-019-0992-x
38
Guo W. Wang H. Yang Y. Guo S. Zhang W. Liu Y. et al (2017). Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics7 (8), 2231–2249. 10.7150/thno.18835
39
Hanniford D. Ulloa-Morales A. Karz A. Berzoti-Coelho M. G. Moubarak R. S. Sánchez-Sendra B. et al (2020). Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell37 (1), 55–70. e15. 10.1016/j.ccell.2019.12.007
40
He Y. Yang Y. Liao Y. Xu J. Liu L. Li C. et al (2020a). miR-140-3p inhibits cutaneous melanoma progression by disrupting AKT/p70S6K and JNK pathways through ABHD2. Mol. Ther. Oncolytics17, 83–93. 10.1016/j.omto.2020.03.009
41
He Y. Yang Y. Xu J. Liao Y. Liu L. Deng L. et al (2020b). IL22 drives cutaneous melanoma cell proliferation, migration and invasion through activation of miR-181/STAT3/AKT axis. J. Cancer11 (9), 2679–2687. 10.7150/jca.40974
42
He C. Wang K. Gao Y. Wang C. Li L. Liao Y. et al (2021). Roles of noncoding RNA in reproduction. Front. Genet.12, 777510. 10.3389/fgene.2021.777510
43
Huang W. Li H. Yu Q. Xiao W. Wang D. O. (2022). LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res.41 (1), 100. 10.1186/s13046-022-02319-z
44
Jian Zhou J. Zhang Y. Han Z. Dong Z. Cao T. Wei A. et al (2019). miR-506 contributes to malignancy of cutaneous squamous cell carcinoma via targeting of P65 and LAMC1. Cell Cycle18 (3), 333–345. 10.1080/15384101.2019.1568747
45
Jianda Zhou J. Liu R. Luo C. Zhou X. Xia K. Chen X. et al (2014). MiR-20a inhibits cutaneous squamous cell carcinoma metastasis and proliferation by directly targeting LIMK1. Cancer Biol. Ther.15 (10), 1340–1349. 10.4161/cbt.29821
46
Jianfei Tang J. Fang X. Chen J. Zhang H. Tang Z. (2021). Long non-coding RNA (lncRNA) in oral squamous cell carcinoma: Biological function and clinical application. Cancers (Basel)13 (23), 5944. 10.3390/cancers13235944
47
Jiao J. Fan Y. Zhang Y. (2015). Expression and clinicopathological significance of microRNA-21 and programmed cell death 4 in malignant melanoma. J. Int. Med. Res.43 (5), 672–678. 10.1177/0300060515583707
48
Jin C. Dong D. Yang Z. Xia R. Tao S. Piao M. et al (2020). CircMYC regulates glycolysis and cell proliferation in melanoma. Cell biochem. Biophys.78 (1), 77–88. 10.1007/s12013-019-00895-0
49
Jin H. Du W. Huang W. Yan J. Tang Q. Chen Y. et al (2021). lncRNA and breast cancer: Progress from identifying mechanisms to challenges and opportunities of clinical treatment. Mol. Ther. Nucleic Acids25, 613–637. 10.1016/j.omtn.2021.08.005
50
Jingwen Wang J. Li C. Xu L. Yang C. Zhang X. (2019). MiR-1193 was sponged by LINC00963 and inhibited cutaneous squamous cell carcinoma progression by targeting SOX4. Pathol. Res. Pract.215 (10), 152600. 10.1016/j.prp.2019.152600
51
Kai Wang K. Li M. Zhang T. Xu C. Yu F. Duan H. et al (2022). LINC01116 facilitates melanoma 1 progression via sequestering miR-3612 and up-regulating GDF11 and SDC3. Arch. Med. Res.53 (1), 44–50. 10.1016/j.arcmed.2021.06.008
52
Kakumani P. K. (2022). AGO-RBP crosstalk on target mRNAs: Implications in miRNA-guided gene silencing and cancer. Transl. Oncol.21, 101434. 10.1016/j.tranon.2022.101434
53
Kanitz A. Imig J. Dziunycz P. J. Primorac A. Galgano A. Hofbauer G. F. et al (2012). The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PLoS One7 (11), e49568. 10.1371/journal.pone.0049568
54
Ke Tian K. Liu W. Zhang J. Fan X. Liu J. Zhao N. et al (2020). MicroRNA-125b exerts antitumor functions in cutaneous squamous cell carcinoma by targeting the STAT3 pathway. Cell. Mol. Biol. Lett.25, 12. 10.1186/s11658-020-00207-y
55
Kim B. K. Kim I. Yoon S. K. (2015). Identification of miR-199a-5p target genes in the skin keratinocyte and their expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci.79 (2), 137–147. 10.1016/j.jdermsci.2015.05.005
56
Kim E. Kim Y. K. Lee S. V. (2021). Emerging functions of circular RNA in aging. Trends Genet.37, 819–829. 10.1016/j.tig.2021.04.014
57
Kitago M. Martinez S. R. Nakamura T. Sim M. S. Hoon D. S. (2009). Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin. Cancer Res.15 (9), 2988–2994. 10.1158/1078-0432.Ccr-08-3172
58
Kong H. Sun M. L. Zhang X. A. Wang X. Q. (2021). Crosstalk among circRNA/lncRNA, miRNA, and mRNA in osteoarthritis. Front. Cell Dev. Biol.9, 774370. 10.3389/fcell.2021.774370
59
Krasowska D. Gerkowicz A. Mlak R. Leziak M. Małecka-Massalska T. Krasowska D. et al (2021). Risk of nonmelanoma skin cancers and Parkinson's disease-meta-analysis and systematic review. Cancers (Basel)13 (4), 587. 10.3390/cancers13040587
60
Li F. Liao J. Duan X. He Y. Liao Y. (2018). Upregulation of LINC00319 indicates a poor prognosis and promotes cell proliferation and invasion in cutaneous squamous cell carcinoma. J. Cell. Biochem.119 (12), 10393–10405. 10.1002/jcb.27388
61
Li Y. Li M. Shats I. Krahn J. M. Flake G. P. Umbach D. M. et al (2019). Glypican 6 is a putative biomarker for metastatic progression of cutaneous melanoma. PLoS One14 (6), e0218067. 10.1371/journal.pone.0218067
62
Li K. Tang M. Tong S. Wang C. Sun Q. Lv M. et al (2020). BRAFi induced demethylation of miR-152-5p regulates phenotype switching by targeting TXNIP in cutaneous melanoma. Apoptosis25 (3-4), 179–191. 10.1007/s10495-019-01586-0
63
Li X. Yuan Y. Wang Y. Xie K. Lu S. Chen F. et al (2022). MicroRNA-486-3p promotes the proliferation and metastasis of cutaneous squamous cell carcinoma by suppressing flotillin-2. J. Dermatol. Sci.105 (1), 18–26. 10.1016/j.jdermsci.2021.11.005
64
Lin N. Zhou Y. Lian X. Tu Y. (2017). MicroRNA-31 functions as an oncogenic microRNA in cutaneous squamous cell carcinoma cells by targeting RhoTBT1. Oncol. Lett.13 (3), 1078–1082. 10.3892/ol.2017.5554
65
Lin Q. Jiang H. Lin D. (2021). Circular RNA ITCH downregulates GLUT1 and suppresses glucose uptake in melanoma to inhibit cancer cell proliferation. J. Dermatol. Treat.32 (2), 231–235. 10.1080/09546634.2019.1654069
66
Lipeng Wang L. Hou S. Li J. Tian T. Hu R. Yu N. et al (2022). Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis. Bioengineered13 (1), 1209–1223. 10.1080/21655979.2021.2019172
67
Liu P. Shi L. Ding Y. Luan J. Shan X. Li Q. et al (2020). MicroRNA-766 promotes the proliferation, migration and invasion, and inhibits the apoptosis of cutaneous squamous cell carcinoma cells by targeting PDCD5. Onco. Targets. Ther.13, 4099–4110. 10.2147/ott.S222821
68
Liu T. Jiang F. Yu L. Y. Wu Y. Y. (2022). Lidocaine represses proliferation and cisplatin resistance in cutaneous squamous cell carcinoma via miR-30c/SIRT1 regulation. Bioengineered13 (3), 6359–6370. 10.1080/21655979.2022.2031419
69
Lohcharoenkal W. Li C. Das Mahapatra K. Lapins J. Homey B. Sonkoly E. et al (2021). MiR-130a acts as a tumor suppressor MicroRNA in cutaneous squamous cell carcinoma and regulates the activity of the BMP/SMAD pathway by suppressing ACVR1. J. Invest. Dermatol.141 (8), 1922–1931. 10.1016/j.jid.2021.01.028
70
Long J. Luo J. Yin X. (2018). miR-367 enhances the proliferation and invasion of cutaneous malignant melanoma by regulating phosphatase and tensin homolog expression. Mol. Med. Rep.17 (5), 6526–6532. 10.3892/mmr.2018.8663
71
Lu R. H. Xiao Z. Q. Zhou J. D. Yin C. Q. Chen Z. Z. Tang F. J. et al (2020). MiR-199a-5p represses the stemness of cutaneous squamous cell carcinoma stem cells by targeting Sirt1 and CD44ICD cleavage signaling. Cell Cycle19 (1), 1–14. 10.1080/15384101.2019.1689482
72
Luan W. Shi Y. Zhou Z. Xia Y. Wang J. (2018). circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem. Biophys. Res. Commun.502 (1), 22–29. 10.1016/j.bbrc.2018.05.114
73
Luo Q. Li W. Zhao T. Tian X. Liu Y. Zhang X. et al (2015). Role of miR-148a in cutaneous squamous cell carcinoma by repression of MAPK pathway. Arch. Biochem. Biophys.583, 47–54. 10.1016/j.abb.2015.07.022
74
Ma X. Wu D. Zhang X. Shao X. Hu G. (2020). microRNA-214 prevents traits of cutaneous squamous cell carcinoma via VEGFA and bcl-2. Technol. Cancer Res. Treat.19, 1533033820980098. 10.1177/1533033820980098
75
Martin del Campo S. E. Latchana N. Levine K. M. Grignol V. P. Fairchild E. T. Jaime-Ramirez A. C. et al (2015). MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: In vivo effects of MiR-21 inhibitor. PLoS One10 (1), e0115919. 10.1371/journal.pone.0115919
76
Mei X. L. Zhong S. (2019). Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin. Med. J.132 (4), 454–465. 10.1097/cm9.0000000000000070
77
Meijuan Zhou M. Zhou L. Zheng L. Guo L. Wang Y. Liu H. et al (2014). miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB). PLoS One9 (6), e100620. 10.1371/journal.pone.0100620
78
Mercer T. R. Munro T. Mattick J. S. (2022). The potential of long noncoding RNA therapies. Trends Pharmacol. Sci.43 (4), 269–280. 10.1016/j.tips.2022.01.008
79
Mi X. Lai K. Yan L. Xie S. Qiu X. Xiao S. et al (2020). miR-18a expression in basal cell carcinoma and regulatory mechanism on autophagy through mTOR pathway. Clin. Exp. Dermatol.45 (8), 1027–1034. 10.1111/ced.14322
80
Mizrahi A. Barzilai A. Gur-Wahnon D. Ben-Dov I. Z. Glassberg S. Meningher T. et al (2018). Alterations of microRNAs throughout the malignant evolution of cutaneous squamous cell carcinoma: The role of miR-497 in epithelial to mesenchymal transition of keratinocytes. Oncogene37 (2), 218–230. 10.1038/onc.2017.315
81
Mu X. Mou K. H. Ge R. Han D. Zhou Y. Wang L. J. et al (2019). Linc00961 inhibits the proliferation and invasion of skin melanoma by targeting the miR-367/PTEN axis. Int. J. Oncol.55 (3), 708–720. 10.3892/ijo.2019.4848
82
Nojima T. Proudfoot N. J. (2022). Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol.23, 389–406. 10.1038/s41580-021-00447-6
83
Nyholm A. M. Lerche C. M. Manfé V. Biskup E. Johansen P. Morling N. et al (2014). miR-125b induces cellular senescence in malignant melanoma. BMC Dermatol.14, 8. 10.1186/1471-5945-14-8
84
Olasz E. B. Seline L. N. Schock A. M. Duncan N. E. Lopez A. Lazar J. et al (2015). MicroRNA-135b regulates leucine zipper tumor suppressor 1 in cutaneous squamous cell carcinoma. PLoS One10 (5), e0125412. 10.1371/journal.pone.0125412
85
Piipponen M. Nissinen L. Farshchian M. Riihilä P. Kivisaari A. Kallajoki M. et al (2016). Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J. Invest. Dermatol.136 (8), 1701–1710. 10.1016/j.jid.2016.03.028
86
Piipponen M. Nissinen L. Riihilä P. Farshchian M. Kallajoki M. Peltonen J. et al (2020). p53-Regulated long noncoding RNA PRECSIT promotes progression of cutaneous squamous cell carcinoma via STAT3 signaling. Am. J. Pathol.190 (2), 503–517. 10.1016/j.ajpath.2019.10.019
87
Qian P. Linbo L. Xiaomei Z. Hui P. (2020). Circ_0002770, acting as a competitive endogenous RNA, promotes proliferation and invasion by targeting miR-331-3p in melanoma. Cell Death Dis.11 (4), 264. 10.1038/s41419-020-2444-x
88
Qirong Lu Q. Guo P. Liu A. Ares I. Martinez-Larranaga M. R. Wang X. et al (2021). The role of long noncoding RNA in lipid, cholesterol, and glucose metabolism and treatment of obesity syndrome. Med. Res. Rev.41 (3), 1751–1774. 10.1002/med.21775
89
Quan Liu Q. Cui W. Yang C. Du L. P. (2021). Circular RNA ZNF609 drives tumor progression by regulating the miR-138-5p/SIRT7 axis in melanoma. Aging (Albany NY)13 (15), 19822–19834. 10.18632/aging.203394
90
Quirico L. Orso F. Cucinelli S. Paradzik M. Natalini D. Centonze G. et al (2022). miRNA-guided reprogramming of glucose and glutamine metabolism and its impact on cell adhesion/migration during solid tumor progression. Cell. Mol. Life Sci.79 (4), 216. 10.1007/s00018-022-04228-y
91
Reddy P. Yao M. Patel M. (2021). Investigative landscape in advanced non-melanoma skin cancers. Curr. Treat. Options Oncol.22 (7), 56. 10.1007/s11864-021-00853-0
92
Ren W. Zhu Z. Wu L. (2019). FOXD2-AS1 correlates with the malignant status and regulates cell proliferation, migration, and invasion in cutaneous melanoma. J. Cell. Biochem.120 (4), 5417–5423. 10.1002/jcb.27820
93
Saldanha G. Potter L. Lee Y. S. Watson S. Shendge P. Pringle J. H. et al (2016). MicroRNA-21 expression and its pathogenetic significance in cutaneous melanoma. Melanoma Res.26 (1), 21–28. 10.1097/cmr.0000000000000216
94
Sánchez-Sendra B. Serna E. Navarro L. González-Muñoz J. F. Portero J. Ramos A. et al (2020). Transcriptomic identification of miR-205 target genes potentially involved in metastasis and survival of cutaneous malignant melanoma. Sci. Rep.10 (1), 4771. 10.1038/s41598-020-61637-4
95
Sand M. Skrygan M. Sand D. Georgas D. Hahn S. A. Gambichler T. et al (2012). Expression of microRNAs in basal cell carcinoma. Br. J. Dermatol.167 (4), 847–855. 10.1111/j.1365-2133.2012.11022.x
96
Sand M. Bechara F. G. Sand D. Gambichler T. Hahn S. A. Bromba M. et al (2016a). Circular RNA expression in basal cell carcinoma. Epigenomics8 (5), 619–632. 10.2217/epi-2015-0019
97
Sand M. Bechara F. G. Sand D. Gambichler T. Hahn S. A. Bromba M. et al (2016b). Long-noncoding RNAs in basal cell carcinoma. Tumour Biol.37 (8), 10595–10608. 10.1007/s13277-016-4927-z
98
Sand M. Hessam S. Amur S. Skrygan M. Bromba M. Stockfleth E. et al (2017). Expression of oncogenic miR-17-92 and tumor suppressive miR-143-145 clusters in basal cell carcinoma and cutaneous squamous cell carcinoma. J. Dermatol. Sci.86 (2), 142–148. 10.1016/j.jdermsci.2017.01.012
99
Sato S. Sawada Y. Nakamura M. (2021). STING signaling and skin cancers. Cancers (Basel)13 (22), 5603. 10.3390/cancers13225603
100
Shan Tian S. Han G. Lu L. Meng X. (2020). Circ-FOXM1 contributes to cell proliferation, invasion, and glycolysis and represses apoptosis in melanoma by regulating miR-143-3p/FLOT2 axis. World J. Surg. Oncol.18 (1), 56. 10.1186/s12957-020-01832-9
101
Shang Q. Li Y. Wang H. Ge S. Jia R. (2019). Altered expression profile of circular RNAs in conjunctival melanoma. Epigenomics11 (7), 787–804. 10.2217/epi-2019-0029
102
Shao Hua Wang S. H. Zhou J. D. He Q. Y. Yin Z. Q. Cao K. Luo C. Q. et al (2014). MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int. J. Clin. Exp. Pathol.7 (10), 7131–7141.
103
Shao J. Liang J. Zhong S. (2019). miR-30a-5p modulates traits of cutaneous squamous cell carcinoma (cSCC) via forkhead box protein G1 (FOXG1). Neoplasma66 (6), 908–917. 10.4149/neo_2018_181205N923
104
Shiqiu Jiang S. Liu H. Zhang J. Zhang F. Fan J. Liu Y. et al (2021). MMP1 regulated by NEAT1/miR-361-5p axis facilitates the proliferation and migration of cutaneous squamous cell carcinoma via the activation of Wnt pathway. Cancer Biol. Ther.22 (5-6), 381–391. 10.1080/15384047.2021.1941583
105
Shuang Chen S. Ding J. Wang Y. Lu T. Wang L. Gao X. et al (2020). RNA-seq profiling of circular RNAs and the oncogenic role of circPVT1 in cutaneous squamous cell carcinoma. Onco. Targets. Ther.13, 6777–6788. 10.2147/ott.S252233
106
Shunhao Zhang S. Sun J. Gu M. Wang G. Wang X. (2021). Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer. Cancer Med.10 (24), 8725–8740. 10.1002/cam4.4398
107
Shuo Han S. Li X. Wang K. Zhu D. Meng B. Liu J. et al (2021). PURPL represses autophagic cell death to promote cutaneous melanoma by modulating ULK1 phosphorylation. Cell Death Dis.12 (11), 1070. 10.1038/s41419-021-04362-8
108
Singh A. Jain D. Pandey J. Yadav M. Bansal K. C. Singh I. K. et al (2022). Deciphering the role of miRNA in reprogramming plant responses to drought stress. Crit. Rev. Biotechnol. [Epub ahead of print], 1–15. 10.1080/07388551.2022.2047880
109
Sinha T. Panigrahi C. Das D. Chandra P. A. (2021). Circular RNA translation, a path to hidden proteome. Wiley Interdiscip. Rev. RNA13, e1685. 10.1002/wrna.1685
110
Song H. Tao Y. Ni N. Zhou X. Xiong J. Zeng X. et al (2018). miR-128 targets the CC chemokine ligand 18 gene (CCL18) in cutaneous malignant melanoma progression. J. Dermatol. Sci.91 (3), 317–324. 10.1016/j.jdermsci.2018.06.011
111
Sonkoly E. Lovén J. Xu N. Meisgen F. Wei T. Brodin P. et al (2012). MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis1 (3), e3. 10.1038/oncsis.2012.3
112
Stark M. S. Bonazzi V. F. Boyle G. M. Palmer J. M. Symmons J. Lanagan C. M. et al (2015). miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget6 (19), 17753–17763. 10.18632/oncotarget.3924
113
Su B. B. Zhou S. W. Gan C. B. Zhang X. N. (2016). MiR-330-5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. J. Surg. Res.203 (2), 434–440. 10.1016/j.jss.2016.03.021
114
Su B. B. Zhou S. W. Gan C. B. Zhang X. N. (2018). MiR-186 inhibits cell proliferation and invasion in human cutaneous malignant melanoma. J. Cancer Res. Ther.14, S60–S64. 10.4103/0973-1482.157340
115
Sun H. Jiang P. (2018). MicroRNA-451a acts as tumor suppressor in cutaneous basal cell carcinoma. Mol. Genet. Genomic Med.6 (6), 1001–1009. 10.1002/mgg3.473
116
Sun Y. Hou Z. Luo B. Li C. Liu J. Liu J. et al (2021). Circular RNA circRNA_0082835 promotes progression and lymphatic metastasis of primary melanoma by sponging microRNA miRNA-429. Bioengineered12 (1), 4159–4173. 10.1080/21655979.2021.1953822
117
Tian Tian T. Zhao Y. Zheng J. Jin S. Liu Z. Wang T. et al (2021). Circular RNA: A potential diagnostic, prognostic, and therapeutic biomarker for human triple-negative breast cancer. Mol. Ther. Nucleic Acids26, 63–80. 10.1016/j.omtn.2021.06.017
118
Toll A. Salgado R. Espinet B. Díaz-Lagares A. Hernández-Ruiz E. Andrades E. et al (2016). MiR-204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression. Mol. Cancer15 (1), 53. 10.1186/s12943-016-0537-z
119
van Kempen L. C. van den Hurk K. Lazar V. Michiels S. Winnepenninckx V. Stas M. et al (2012). Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch.461 (4), 441–448. 10.1007/s00428-012-1309-9
120
Venza M. Visalli M. Beninati C. Benfatto S. Teti D. Venza I. et al (2016). miR-92a-3p and MYCBP2 are involved in MS-275-induced and c-myc-mediated TRAIL-sensitivity in melanoma cells. Int. Immunopharmacol.40, 235–243. 10.1016/j.intimp.2016.09.004
121
Wang S. Cao K. E. He Q. Yin Z. Zhou J. (2016). miR-199a-5p induces cell invasion by suppressing E-cadherin expression in cutaneous squamous cell carcinoma. Oncol. Lett.12 (1), 97–101. 10.3892/ol.2016.4602
122
Wang X. Qu H. Dong Y. Wang G. Zhen Y. Zhang L. et al (2018). Targeting signal-transducer-and-activator-of-transcription 3 sensitizes human cutaneous melanoma cells to BRAF inhibitor. Cancer Biomark.23 (1), 67–77. 10.3233/cbm-181365
123
Wang D. Zhou X. Yin J. Zhou Y. (2020). Lnc-PICSAR contributes to cisplatin resistance by miR-485-5p/REV3L axis in cutaneous squamous cell carcinoma. Open Life Sci.15 (1), 488–500. 10.1515/biol-2020-0049
124
Wei X. H. Gu X. L. Zhou X. T. Ma M. Lou C. X. (2018). miR-497 promotes the progression of cutaneous squamous cell carcinoma through FAM114A2. Eur. Rev. Med. Pharmacol. Sci.22 (21), 7348–7355. 10.26355/eurrev_201811_16272
125
Wei X. Gu X. Ma M. Lou C. (2019). Long noncoding RNA HCP5 suppresses skin cutaneous melanoma development by regulating RARRES3 gene expression via sponging miR-12. Onco. Targets. Ther.12, 6323–6335. 10.2147/ott.S195796
126
Wei C. Y. Zhu M. X. Lu N. H. Liu J. Q. Yang Y. W. Zhang Y. et al (2020). Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol. Cancer19 (1), 84. 10.1186/s12943-020-01191-9
127
Wei Zhang W. Yang X. Lv J. An S. (2022). Research on correlations of miR-196a expression with progression and prognosis of cutaneous squamous cell carcinoma. Clin. Cosmet. Investig. Dermatol.15, 97–105. 10.2147/ccid.S323414
128
Wen S. Y. Qadir J. Yang B. B. (2022). Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med.28 (5), 405–420. 10.1016/j.molmed.2022.03.003
129
Wenqing Zhang W. Zhou K. Zhang X. Wu C. Deng D. Yao Z. et al (2021). Roles of the H19/microRNA-675 axis in the proliferation and epithelial-mesenchymal transition of human cutaneous squamous cell carcinoma cells. Oncol. Rep.45 (4), 39. 10.3892/or.2021.7990
130
Winkle M. El-Daly S. M. Fabbri M. Calin G. A. (2021). Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov.20 (8), 629–651. 10.1038/s41573-021-00219-z
131
Wu L. Zhu L. Li Y. Zheng Z. Lin X. Yang C. et al (2020). LncRNA MEG3 promotes melanoma growth, metastasis and formation through modulating miR-21/E-cadherin axis. Cancer Cell Int.20, 12. 10.1186/s12935-019-1087-4
132
Wu Zhou W. Zhang S. Li J. Li Z. Wang Y. Li X. et al (2019). lncRNA TINCR participates in ALA-PDT-induced apoptosis and autophagy in cutaneous squamous cell carcinoma. J. Cell. Biochem.120 (8), 13893–13902. 10.1002/jcb.28662
133
Xiaobo Zhou X. Rao Y. Sun Q. Liu Y. Chen J. Bu W. et al (2019). Long noncoding RNA CPS1-IT1 suppresses melanoma cell metastasis through inhibiting Cyr61 via competitively binding to BRG1. J. Cell. Physiol.234 (12), 22017–22027. 10.1002/jcp.28764
134
Xiaoqing Li X. Kong Y. Li H. Xu M. Jiang M. Sun W. et al (2021). CircRNA circ_0067772 aggravates the malignant progression of cutaneous squamous cell carcinoma by regulating miR-1238-3p/FOXG1 axis. Genes Genomics43 (5), 491–501. 10.1007/s13258-021-01074-3
135
Xiaoyan Lu X. Gan Q. Gan C. (2021a). Circular RNA circSEC24A promotes cutaneous squamous cell carcinoma progression by regulating miR-1193/MAP3K9 Axis. Onco. Targets. Ther.14, 653–666. 10.2147/ott.S275691
136
Xinde Hu X. Liu Y. Ai P. He S. Liu L. Chen C. et al (2019). MicroRNA-186 promotes cell proliferation and inhibits cell apoptosis in cutaneous squamous cell carcinoma by targeting RETREG1. Exp. Ther. Med.17 (3), 1930–1938. 10.3892/etm.2019.7154
137
Xiong Z. Jiang B. Li G. (2020). Downregulation of miR-10a inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion by targeting Syndecan-1. Int. J. Clin. Exp. Pathol.13 (10), 2502–2512.
138
Xiong W. Wu L. Tang R. Zhang Q. Guo Q. Song S. (2021). Grape seed proanthocyanidins (GSPs) inhibit the development of cutaneous squamous cell carcinoma by regulating the hsa_circ_0070934/miR-136-5p/PRAF2 Axis. Cancer Manag. Res.13, 4359–4371. 10.2147/cmar.S302084
139
Xu E Chen X. E. Chen P. Chen S. Lu J. Ma T. Shi G. et al (2020). Long non-coding RNA FENDRR inhibits migration and invasion of cutaneous malignant melanoma cells. Biosci. Rep.40 (3), BSR20191194. 10.1042/bsr20191194
140
Xu N. Zhang L. Meisgen F. Harada M. Heilborn J. Homey B. et al (2012). MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem.287 (35), 29899–29908. 10.1074/jbc.M112.391243
141
Xu Y. Guo B. Liu X. Tao K. (2021). miR-34a inhibits melanoma growth by targeting ZEB1. Aging (Albany NY)13 (11), 15538–15547. 10.18632/aging.203114
142
Xu J. Zhang F. Lin M. Tong Y. (2022). LINC00518 affects the proliferation, invasion and migration of cutaneous malignant melanoma cells via miR-526b-3p/EIF5A2 axis. Acta Biochim. Pol.69 (1), 101–111. 10.18388/abp.2020_5746
143
Yamane K. Jinnin M. Etoh T. Kobayashi Y. Shimozono N. Fukushima S. et al (2013). Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J. Mol. Med.91 (1), 69–81. 10.1007/s00109-012-0935-7
144
Yan Hu Y. Wang Q. Zhu X. H. (2019). MiR-135b is a novel oncogenic factor in cutaneous melanoma by targeting LATS2. Melanoma Res.29 (2), 119–125. 10.1097/cmr.0000000000000524
145
Yan Liu Y. He D. Xiao M. Zhu Y. Zhou J. Cao K. et al (2021). Long noncoding RNA LINC00518 induces radioresistance by regulating glycolysis through an miR-33a-3p/HIF-1α negative feedback loop in melanoma. Cell Death Dis.12 (3), 245. 10.1038/s41419-021-03523-z
146
Yan Wang Y. Wu C. Du Y. Li Z. Li M. Hou P. et al (2022). Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer21 (1), 13. 10.1186/s12943-021-01484-7
147
Yan Yu Y. Xiang N. Lin M. Huang J. W. Zhang J. Cheng B. et al (2019). miR- 26a sensitizes melanoma cells to dabrafenib via targeting HMGB1-dependent autophagy pathways. Drug Des. devel. Ther.13, 3717–3726. 10.2147/dddt.S225671
148
Yang Z. Liao B. Xiang X. Ke S. (2020). miR-21-5p promotes cell proliferation and G1/S transition in melanoma by targeting CDKN2C. FEBS Open Bio10 (5), 752–760. 10.1002/2211-5463.12819
149
Yang L. Li L. P. Yi H. C. (2022). DeepWalk based method to predict lncRNA-miRNA associations via lncRNA-miRNA-disease-protein-drug graph. BMC Bioinforma.22 (12), 621. 10.1186/s12859-022-04579-0
150
Yanhua Wang Y. Tang S. Lv J. (2022). MicroRNA-573 inhibits cell proliferation, migration and invasion and is downregulated by PICSAR in cutaneous squamous cell carcinoma. Bosn. J. Basic Med. Sci.22 (2), 229–237. 10.17305/bjbms.2021.6301
151
Yating Li Y. Li Y. Li L. (2021). Circular RNA hsa_Circ_0005795 mediates cell proliferation of cutaneous basal cell carcinoma via sponging miR-1231. Arch. Dermatol. Res.313 (9), 773–782. 10.1007/s00403-020-02174-y
152
Yin D. Wei G. Yang F. Sun X. (2021). Circular RNA has circ 0001591 promoted cell proliferation and metastasis of human melanoma via ROCK1/PI3K/AKT by targeting miR-431-5p. Hum. Exp. Toxicol.40 (2), 310–324. 10.1177/0960327120950014
153
Yinghui Wang Y. Deng X. Dai Y. Niu X. Zhou M. (2019). miR-27a downregulation promotes cutaneous squamous cell carcinoma progression via targeting EGFR. Front. Oncol.9, 1565. 10.3389/fonc.2019.01565
154
Yuan S. Zhang P. Wen L. Jia S. Wu Y. Zhang Z. et al (2021). miR-22 promotes stem cell traits via activating Wnt/β-catenin signaling in cutaneous squamous cell carcinoma. Oncogene40 (39), 5799–5813. 10.1038/s41388-021-01973-5
155
Yue Zhang Y. Zhang X. Xu Y. Fang S. Ji Y. Lu L. et al (2022). Circular RNA and its roles in the occurrence, development, diagnosis of cancer. Front. Oncol.12, 845703. 10.3389/fonc.2022.845703
156
Yuxiong Jiang Y. Zhang C. Zhang J. Han D. Shi X. (2021). Comprehensive analysis of the prognosis and biological significance for IFIT family in skin cutaneous melanoma. Int. Immunopharmacol.101, 108344. 10.1016/j.intimp.2021.108344
157
Zehavi L. Schayek H. Jacob-Hirsch J. Sidi Y. Leibowitz-Amit R. Avni D. et al (2015). MiR-377 targets E2F3 and alters the NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol. Cancer14, 68. 10.1186/s12943-015-0338-9
158
Zeng X. Yuan X. Cai Q. Tang C. Gao J. (2021). Circular RNA as an epigenetic regulator in chronic liver diseases. Cells10 (8), 1945. 10.3390/cells10081945
159
Zhang D. Li Z. Zhang Y. Tu C. Huo J. Liu Y. et al (2016). miR-4262 promotes the proliferation of human cutaneous malignant melanoma cells through KLF6-mediated EGFR inactivation and p21 upregulation. Oncol. Rep.36 (6), 3657–3663. 10.3892/or.2016.5190
160
Zhang M. Luo H. Hui L. (2019). MiR-3619-5p hampers proliferation and cisplatin resistance in cutaneous squamous-cell carcinoma via KPNA4. Biochem. Biophys. Res. Commun.513 (2), 419–425. 10.1016/j.bbrc.2019.03.203
161
Zhang D. W. Wu H. Y. Zhu C. R. Wu D. D. (2020a). CircRNA hsa_circ_0070934 functions as a competitive endogenous RNA to regulate HOXB7 expression by sponging miR-1236-3p in cutaneous squamous cell carcinoma. Int. J. Oncol.57 (2), 478–487. 10.3892/ijo.2020.5066
162
Zhang Z. Jia M. Wen C. He A. Ma Z. (2020b). Long non-coding RNA SCARNA2 induces cutaneous squamous cell carcinoma progression via modulating miR-342-3p expression. J. Gene Med.22 (12), e3242. 10.1002/jgm.3242
163
Zhang Z. Guo H. Yang W. Li J. (2021a). Exosomal circular RNA RNA-seq profiling and the carcinogenic role of exosomal circ-cyp24a1 in cutaneous squamous cell carcinoma. Front. Med.8, 675842. 10.3389/fmed.2021.675842
164
Zhang Z. Huang Q. Yu L. Zhu D. Li Y. Xue Z. et al (2021b). The role of miRNA in tumor immune escape and miRNA-based therapeutic strategies. Front. Immunol.12, 807895. 10.3389/fimmu.2021.807895
165
Zhang Z. Sun H. Hou J. Li L. Wu L. (2021c). Circular RNA circFADS2 inhibits the progression of cutaneous squamous cell carcinoma by regulating miR-766-3p/HOXA9 axis. Histol. Histopathol.37 (4), 335–348. 10.14670/hh-18-409
166
Zhao F. Jia Z. Feng Y. Li Z. Feng J. (2021). Circular RNA circ_0079593 enhances malignant melanoma progression by the regulation of the miR-573/ABHD2 axis. J. Dermatol. Sci.102 (1), 7–15. 10.1016/j.jdermsci.2021.01.008
167
Zheng Y. Zhou W. Li M. Xu R. Zhang S. Liu Y. et al (2021). IRF4-activated TEX41 promotes the malignant behaviors of melanoma cells by targeting miR-103a-3p/C1QB axis. BMC Cancer21 (1), 1339. 10.1186/s12885-021-09039-1
168
Zhibing Chen Z. Chen J. Wa Q. He M. Wang X. Zhou J. et al (2020). Knockdown of circ_0084043 suppresses the development of human melanoma cells through miR-429/tribbles homolog 2 axis and Wnt/β-catenin pathway. Life Sci.243, 117323. 10.1016/j.lfs.2020.117323
169
Zhou L. Wang Y. Ou C. Lin Z. Wang J. Liu H. et al (2015). microRNA-365-targeted nuclear factor I/B transcriptionally represses cyclin-dependent kinase 6 and 4 to inhibit the progression of cutaneous squamous cell carcinoma. Int. J. Biochem. Cell Biol.65, 182–191. 10.1016/j.biocel.2015.06.009
170
Zhou L. Gao R. Wang Y. Zhou M. Ding Z. (2017). Loss of BAX by miR-365 promotes cutaneous squamous cell carcinoma progression by suppressing apoptosis. Int. J. Mol. Sci.18 (6), E1157. 10.3390/ijms18061157
171
Zhou H. Wu J. Leng S. Hou C. Mo L. Xie X. et al (2021). Knockdown of circular RNA VANGL1 inhibits TGF-β-induced epithelial-mesenchymal transition in melanoma cells by sponging miR-150-5p. J. Cell. Mol. Med.25 (23), 10837–10845. 10.1111/jcmm.16887
172
Zhou W. D. Shao L. Dong L. Zhang R. H. Li Y. F. Li H. Y. et al (2022). Circulating MicroRNAs as quantitative biomarkers for diagnosis and prognosis of uveal melanoma. Front. Oncol.12, 854253. 10.3389/fonc.2022.854253
173
Zhu Y. Wen X. Zhao P. (2018). MicroRNA-365 inhibits cell growth and promotes apoptosis in melanoma by targeting BCL2 and cyclin D1 (CCND1). Med. Sci. Monit.24, 3679–3692. 10.12659/msm.909633
174
Ziwei Li Z. Tian P. Huang T. Huang J. (2021). Noncoding-RNA-mediated regulation in response to macronutrient stress in plants. Int. J. Mol. Sci.22 (20), 11205. 10.3390/ijms222011205
175
Zou Y. Wang S. S. Wang J. Su H. L. Xu J. H. (2019). CircRNA_0016418 expedites the progression of human skin melanoma via miR-625/YY1 axis. Eur. Rev. Med. Pharmacol. Sci.23 (24), 10918–10930. 10.26355/eurrev_201912_19795
176
Zou S. Gao Y. Zhang S. (2021). lncRNA HCP5 acts as a ceRNA to regulate EZH2 by sponging miR-138-5p in cutaneous squamous cell carcinoma. Int. J. Oncol.59 (2), 56. 10.3892/ijo.2021.5236
Summary
Keywords
skin cancers, non-coding RNA, malignant progression, molecular biology, metastasis
Citation
Liu F and Li S (2022) Non-coding RNAs in skin cancers:Biological roles and molecular mechanisms. Front. Pharmacol. 13:934396. doi: 10.3389/fphar.2022.934396
Received
02 May 2022
Accepted
11 July 2022
Published
10 August 2022
Volume
13 - 2022
Edited by
Yue Hou, Northeastern University, China
Reviewed by
Deniz Bartsch, University Hospital of Cologne, Germany
Gabriel Tao, Merck (United States), United States
Updates
Copyright
© 2022 Liu and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenglong Li, lishenglong@cancerhosp-ln-cmu.com
This article was submitted to Pharmacology of Anti-Cancer Drugs, a section of the journal Frontiers in Pharmacology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.