
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Pharmacol. , 30 June 2022
Sec. Pharmacology of Ion Channels and Channelopathies
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.929684
This article is part of the Research Topic Ion Channel Structure and Drug Discovery Accelerated by Cryo-EM View all 11 articles
Mitiglinide is a highly selective fast-acting anti-diabetic drug that induces insulin secretion by inhibiting pancreatic KATP channels. However, how mitiglinide binds KATP channels remains unknown. Here, we show the cryo-EM structure of the SUR1 subunit complexed with mitiglinide. The structure reveals that mitiglinide binds inside the common insulin secretagogue-binding site of SUR1, which is surrounded by TM7, TM8, TM16, and TM17. Mitiglinide locks SUR1 in the NBD-separated inward-facing conformation. The detailed structural analysis of the mitiglinide-binding site uncovers the molecular basis of its high selectivity.
More than 400 million people are living with diabetes worldwide, and type 2 diabetes (T2DM) accounts for nearly 90% of patients with diabetes (Chatterjee et al., 2017). Dysfunction of insulin secretion is one of the hallmarks of T2DM (Daryabor et al., 2020). Previous studies have established that the KATP channel plays an essential role in insulin secretion and is a drug target for diabetes (Gribble and Reimann, 2003).
The KATP channel is a hetero-octamer complex composed of four pore-forming Kir6 (kir6.1 or Kir6.2) subunits and four regulatory sulfonylureas receptor subunits (SUR1 or SUR2). KATP channels can sense intracellular ATP/ADP ratio using the inhibitory ATP binding site on Kir6 and the activating Mg–ADP binding site on SUR, coupling the metabolism status of the cell to its membrane potential (Ashcroft and Gribble, 1998). KATP channels are widely distributed in many tissues, including the brain (Ashford et al., 1988), muscles (Noma, 1983; Spruce et al., 1985), and endocrine cells (Cook and Hales, 1984), and perform important physiological functions. In pancreatic β-cells, KATP channels are mainly formed by Kir6.2 and SUR1, and play key roles in controlling insulin secretion (Ashcroft and Rorsman, 1989). When blood glucose level increases, the intracellular ATP/ADP ratio also increases accordingly, leading to the suppression of the KATP channel activity. The inhibited potassium efflux through the KATP channel induces depolarization of the β-cell membrane, resulting in the subsequent activation of voltage-gated calcium channel, calcium influx, and insulin secretion (Rorsman and Trube, 1985; Nichols, 2006). Mutations in genes encoding either Kir6.2 or SUR1 cause disorders in insulin secretion, such as congenital hyperinsulinemia and neonatal diabetes (Ashcroft, 2005). Small-molecule drugs that inhibit the pancreatic KATP channel are widely used to boost insulin secretion for the treatment of diabetes and are therefore named insulin secretagogues (ISs) (Gribble and Reimann, 2003).
Insulin secretagogues are chemically diverse small molecules, including sulphonylureas, such as glibenclamide (GBM), and glinides, such as repaglinide (RPG) and mitiglinide (MTG) (Wu et al., 2020). ISs bind to the SUR subunits to inhibit the KATP channels (Wu et al., 2020). MTG ((+)-monocalcium bis [(2S)-2-benzyl-3-(cis-hexahydro-2-isoindolinyl carbonyl)propionate]dihydrate), also named KAD-1229, is a glinide that was developed for the treatment of postprandial hyperglycemia (Pratley et al., 2001) and has been approved for the treatment of patients with T2DM in Japan (brand name, Glufast, approved in 2004). MTG has an immediate and short-lasting effect on hypoglycemic action in the postprandial glucose-load state in clinical trials (Phillippe and Wargo, 2013). In vitro experiments supported that MTG increases insulin release from a pancreatic β-cell line MIN6 cell (Mogami et al., 1994; Reimann et al., 2001). The insulin responses to chronic MTG treatment were comparable to chronic RPG or GBM treatment in MIN6 cells (Reimann et al., 2001). MTG can displace [3H]-GBM binding to HIT-15 cells with an IC50 of 13 nM (Ohnota et al., 1994), suggesting that they might share an overlapped binding site. Notably, MTG is highly selective toward SUR1 over SUR2 (Reimann et al., 2001; Sunaga et al., 2001), and GBM is moderately selective, while RPG is non-selective (Quast et al., 2004). Recent structural studies have uncovered the binding modes of GBM and RPG (Wu et al., 2020), but the exact binding mode of MTG on SUR1 remains unknown. Here, we present the cryo-EM structure of the SUR1 subunit complexed with MTG, allowing the direct visualization of how MTG binds SUR1.
FreeStyle 293-F (Thermo Fisher Scientific) suspension cells were cultured in SMM 293-TI (Sino Biological Inc.) supplemented with 1% fetal bovine serum (FBS) at 37°C, with 6% CO2 and 70% humidity. Sf9 insect (Thermo Fisher Scientific) suspension cells were cultured in Sf-900 III SFM medium (Thermo Fisher Scientific) at 27°C.
We made a truncated Mesocricetus auratus SUR1 construct maSUR1core (from 208 to C terminal) based on the previous work by Ding et al. (2019). We added an N-terminal GFP and MBP tag, a PreScission protease cleavage site, KNtp of mmKir6.2 (Ding et al., 2019), and a GS-rich linker before SUR1core. The construct was made in a modified BacMam vector (Li et al., 2017).
Wild-type SUR1 or its mutants together with CGFP-tagged Kir6.2 were transfected into FreeStyle 293F suspension cells using polyethyleneimine at the cell density of 1.0×106 cells/ml. Cells were cultured in FreeStyle expression medium supplemented with 1% FBS for 24 h and then seeded into 12-mm dishes for adhesion before recording. Macroscopic currents were recorded in the inside-out mode at +60 mV by Axon-patch 200B amplifier (Axon Instruments, United States). Patch electrodes were pulled by a horizontal microelectrode puller (P-1000, Sutter Instrument Co., United States) to 2.0–3.0 MΩ resistance. Both pipette and bath solution was based on KINT buffer, containing (mM): 140 KCl, 1 EGTA, and 10 HEPES (pH, 7.4, KOH). For mitiglinide (Targetmol) and RPG (Abcam) inhibition, both the stock solutions (10 mM mitiglinide and 100 mM RPG) were dissolved in DMSO, stored at −20°C, and diluted into KINT buffer to the final concentrations before recording. ATP (Sigma) and ADP (Sigma) stocks were prepared on ice, and stored at −20°C. ATP and ADP were dissolved in water and adjusted to pH = 7 by KOH (Sigma). The nucleotide concentration was determined by its extinction coefficient and absorption at A259 nm. Signals were acquired at 5 kHz and low-pass filtered at 1 kHz. Data were further analyzed by pclampfit 10.0 software.
SUR1core was expressed as described previously (Ding et al., 2019), and the purification process was carried out with minor modifications. For protein purification, membrane pellets were homogenized in TBS (20 mM Tris–HCl; pH, 7.5; 150 mM NaCl) and solubilized in TBS with 1% GDN (Anatrace), supplemented with protease inhibitors (1 mg/ml leupeptin, 1 mg/ml pepstatin, 1 mg/ml aprotinin, and 1 mM PMSF), 1 mM EDTA, and 1 mM EGTA for 30 min at 4°C. Unsolubilized materials were discarded after centrifugation at 100,000 g for 30 min and the supernatant was loaded onto a 5-ml StrepTactin 4FF (Smart Lifesciences) packed column. The Strep column was washed with TBS buffer supplemented with 0.01% GDN, protease inhibitors (1 mg/ml leupeptin, 1 mg/ml pepstatin, and 1 mg/ml aprotinin), 1 mM EDTA, and 1 mM EGTA. Then the column was washed using TBS supplemented with 0.01% GDN, 3mM MgCl2, and 1 mM ATP. The last washing step buffer was TBS supplemented with 0.01% GDN. The SUR1core was eluted with TBS (50 mM Tris–HCl; pH, 7.5; 150 mM NaCl) supplemented with 0.006% GDN and 5 mM desthiobiotin. The eluate was concentrated and supplemented with PreScission protease overnight. The next day, SUR1core was further purified using Superose 6 increase (GE Healthcare) column running with TBS supplemented with 0.006% GDN. Peak fractions were collected and concentrated to A280 = 10 (50 μM). The purified protein was used for cryo-EM sample preparation.
100 µM mitiglinide was added to the sample and incubated for 20 min before centrifugation at 40,000 rpm for 30 min using a TLA55 rotor (Beckman). Cryo-EM grids were prepared with Vitrobot Mark IV (FEI) and GIG R0.6/1 holey carbon grids, which were glow discharged for 120 s using air before adding the protein sample. A 2.5-µl sample was applied to the glow-discharged grid, and then the grid was blotted at blotting force at level 2 for 2 s at 100% humidity and 10°C, before it was plunge frozen in liquid ethane.
Cryo-grids were screened on a Talos Arctica microscope (Thermo Fisher Scientific) operated at 200 kV. Grids of good quality were further loaded onto a Titan Krios microscope (Thermo Fisher Scientific) operated at 300 kV for data collection. Images were collected using a K2 camera (Gatan) mounted post a Quantum energy filter with a 20-eV slit and operated under super-resolution mode with a pixel size of 1.045 Å at the object plane, controlled by software Serial EM. Defocus values were set from −1.5 to −1.8 μm for data collection. The dose rate on the detector was 8.078 e− pixel−1 s−1, and the total exposure was 54.3 e−A−2. Each 6.72 s movie was dose-fractioned into 32 frames.
A total of 1,181 movies were collected and the movies were exposure filtered, gain corrected, motion corrected, mag-distortion corrected, and binned with MotionCor2 (Zheng et al., 2017), producing dose-weighted and summed micrographs. CTF models of dose-weighted micrographs were determined using Gctf (Zhang, 2016). Gautomatch (developed by Kai Zhang, MRC-LMB) was used for auto-picking. Data processing was initially performed using Relion_3.0 (Zivanov et al., 2018). Particles were extracted from dose-weighted micrographs. After 2D classification (379K particles) and 3D classification with C1 symmetry, 130K particles with good transmembrane domain (TMD) densities were re-extracted and re-centered. In this stage, the mitiglinide density can be observed. The remaining particles were subjected to focused 3D classification with a TMD mask to select around 20K particles (seed particles) with good mitiglinide density. Then the 379K particles were subjected to seed-facilitated 3D classification (Wang et al., 2021) to produce 130K particles in CryoSPARC2 (Punjani et al., 2017). These particles were subjected to non-uniform refinement, CTF refinement, and local non-uniform refinement in cryoSPARC2 to generate a high-resolution map.
SUR1 ABC transporter domain in the previously reported KATP model (PDB ID: 6BJ1) was docked into the density map using UCSF Chimera (Pettersen et al., 2004). The model was manually rebuilt in Coot (Emsley et al., 2010) and refined against the density map using Phenix (Afonine et al., 2018). Figures were prepared using Pymol and UCSF Chimera X (Pettersen et al., 2020).
Global resolution estimations of cryo-EM density maps are based on the 0.143 Fourier Shell Correlation criterion (Rosenthal and Henderson, 2003). Electrophysiological data reported were analyzed with pClampfit 10.0 software. The number of biological replicates (N) and the relevant statistical parameters for each experiment (such as a mean or standard error) are described in figure legends. No statistical methods were used to pre-determine sample sizes.
Previous cryo-EM studies on KATP channels in complex with GBM (Martin et al., 2017; Wu et al., 2018) or RPG (Ding et al., 2019) have established that the ABC transporter region of the SUR subunit harbors the IS-binding site. Therefore, we used a truncated construct (SUR1core) that encompasses 208-1582 of SUR1, including TMD1, NBD1, TMD2, and NBD2 (Figure 1A and Supplementary Figure S1), for subsequent structure determination. Purified SUR1core protein was supplemented with ATP and MTG for cryo-EM sample preparation and data collection. Image processing yielded a reconstruction with an overall resolution of 3.27 Å (Figure 1B,C and Supplementary Figure S1,S2). High-quality local map allows the unambiguous identification of the MTG molecule (Figure 1D–G).
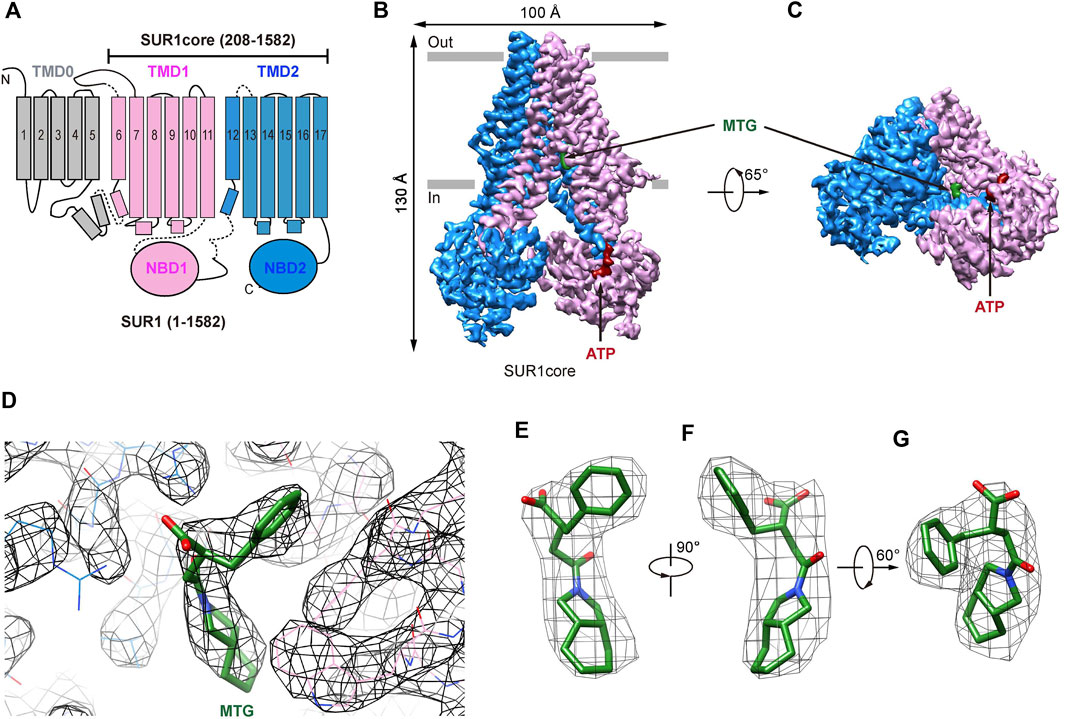
FIGURE 1. Structure of SUR1 complexed with MTG. (A) Topology of SUR1 subunits. TMD, transmembrane domain; NBD, nucleotide-binding domain. Transmembrane helices are shown as cylinders. TMD0 fragment, TMD1-NBD1, and TMD2-NBD2 are shown in gray, pink, and blue, respectively. (B) Side view of the cryo-EM density of the SUR1core. ATP and MTG are shown in red and green, respectively. (C) Bottom view of the cryo-EM density of the SUR1core. (D) Densities of MTG and its surrounding amino acids. (E–G) MTG densities viewed from different angles.
In the presence of ATP and MTG, SUR1core shows an inward-facing conformation with its central vestibule widely open to the cytosol (Figure 1B,C). ATP binds to the NBD1 and MTG binds inside a pocket in the central vestibule of SUR1 (Figure 1B–D). The MTG-binding pocket is formed by residues on TM7 and TM8 of TMD1 and TM16 and TM17 of TMD2 (Figure 2A,B). The interactions between MTG and SUR1 involve both polar interactions and hydrophobic interactions (Figure 2A,B). The central negatively charged carboxyl group of MTG makes electrostatic interactions with positively charged R1246 on TM16 and R1300 on TM17 (Figure 2A,B). The benzene ring of MTG stacks with the phenyl group of F433 (Figure 2A,B). The bulky hexahydro-2-isoindoline group binds in a hydrophobic pocket surrounded by L1241 and T1242 of TMD2 and I381, I385, F433, and W430 of TMD1 (Figure 2A,B). To understand the role of R1246 and R1300 on the inhibitory function of MTG, we mutated them into alanines individually. We found that both R1246A and R1300A mutations significantly decreased the inhibitory effect of MTG (Figure 2Dand Supplementary Figure S3), emphasizing their importance on MTG binding and inhibition.
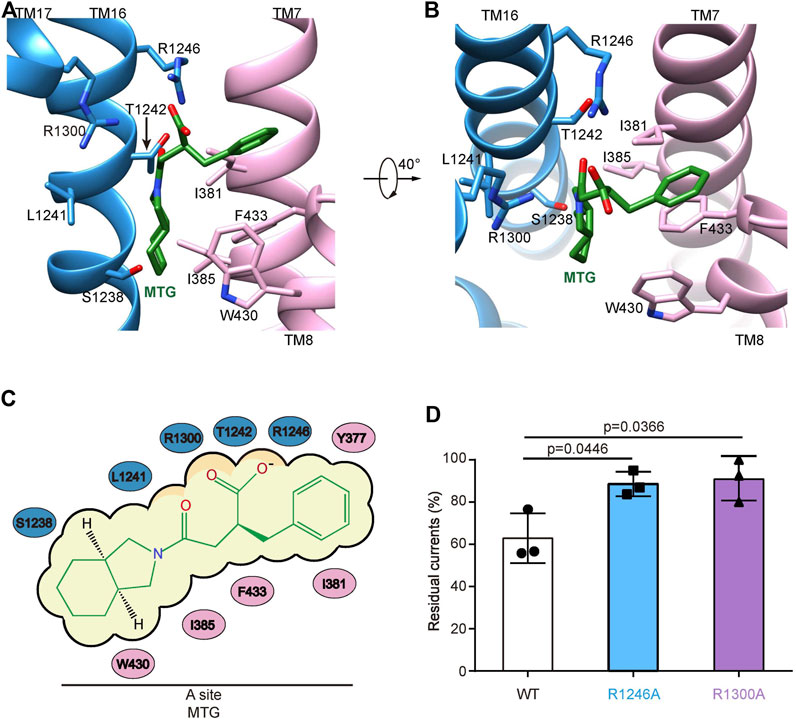
FIGURE 2. The MTG-binding pocket in SUR1. (A,B) Close-up views of the MTG-binding site. TMD1, TMD2, and MTG are shown in pink, blue, and green, respectively. (C) Cartoon representation of the interaction between MTG and SUR1. (D) Wild-type and mutated KATP channels inhibition by 10 μM MTG. Data are shown as mean ± SD and n = 3 independent patches. p was calculated by unpaired two-tailed t-test and indicated above.
Accumulated studies on the structure–activity relationship of IS indicate that there are two overlapping binding sites for IS on SUR, the A-site and the B-site (Yan et al., 2006) (Figure 3A). GBM binds in both A- and B-sites (Wu et al., 2018), while RPG binds in the B-site (Ding et al., 2019; Martin et al., 2019). Our structure shows that MTG mainly binds in the A-site (Figure 3A,B). MTG-interacting residues are almost identical between SUR1 and SUR2, except that T1242 is replaced by a serine in SUR2 and S1238 is replaced by tyrosine in SUR2 (Figure 3C). Notably, S1238 is near the A-site (Figure 2A,B), and previous studies found that S1238 is the key determinant for the selectivity of certain IS, such as glibenclamide, tolbutamide, and nateglinide (Ashfield et al., 1999; Hansen et al., 2002). In agreement with this, the hexahydro-2-isoindoline group of MTG is near the S1238, and the replacement of S1238 with a tyrosine would generate sterical clashes with MTG (Figure 3D,E), abolishing its binding.
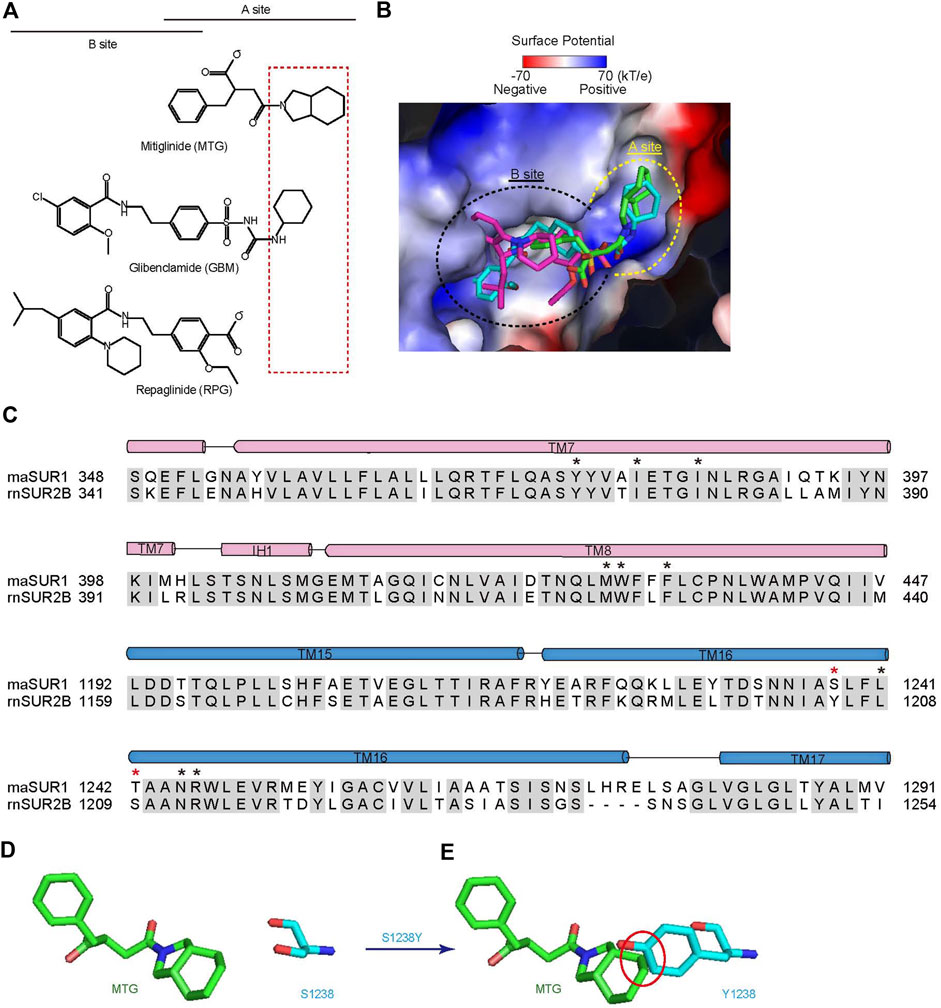
FIGURE 3. MTG binds in the A-site of SUR1. (A) Chemical structures of MTG (A-site ligand), RPG (B-site ligand), and GBM (A + B site ligand). The “SUR Isotype Selectivity Determinative Moiety” (SISDM) is boxed in red. (B) Structural superposition of MTG, RPG (PDB ID: 6BJ3), and GBM (PDB ID: 6BAA) in their binding pocket. Insulin secretagogues are shown as sticks. The electrostatic surface of SUR1 is calculated with Pymol. (C) Sequence alignment of MTG binding pocket between Mesocricetus auratus SUR1 (maSUR1) and Rattus norvegicus (rnSUR2). The identical and different MTG-interacting residues are highlighted by black and red asterisks above. (D) The relative position of MTG (green) and S1238 side chain (cyan). (E) The mutation of S1238 to Tyr (cyan) would generate sterical clashes with MTG (green). Clashes with inter-atom distance smaller than 2.2 Å non-bond distance are circled in red.
The structures of SUR1 in complex with IS are now available for RPG, GBM, and MTG. Among them, MTG has the highest selectivity toward SUR1 over SUR2 (Quast et al., 2004). Notably, SUR isotype-selectivity is highly clinically relevant because SUR1-containing KATP channels are mainly distributed in pancreatic endocrine cells, while SUR2-containing KATP channels have broad distribution in cardiac muscles and vascular smooth muscles, participating in several key physiological processes in the cardiovascular system, such as the preservation of cardio-protection under ischemic conditions. Moreover, patients with diabetes often have cardiovascular diseases, such as coronary heart disease. Therefore, it is important to consider the off-target side effect of IS treatment for diabetes. The high selectivity of MTG (1,000×) might mitigate the underlying side effects of inhibiting SUR2-containing KATP channels. In agreement with this theory, it was reported that MTG could preserve the cardio-protective effect of ischemic preconditioning while GBM could not (Ogawa et al., 2007), emphasizing the potential benefit of using highly selective IS for diabetes. In addition, the high selectivity of MTG is also explored to design 18F-labeled positron emission tomography tracers for the measurement of β-cell mass in vivo during the progression of diabetes, aiming to better understand the pathogenesis, to facilitate early diagnosis, and to develop novel therapeutics for diabetes (Kimura et al., 2014). Despite the early observation of high selectivity of MTG, how this is achieved was not well understood previously. Our current structure of SUR1 complexed with MTG shows that the hexahydro-2-isoindoline group is close to S1238, the key residue that is different between SUR1 and SUR2. Therefore, we designate this large hexahydro-2-isoindoline as the “SUR Isotype Selectivity Determinative Moiety” (SISDM). Furthermore, structural comparisons among RPG, GBM, and MTG reveal that the size of SISDM correlates well with their selectivity: larger SISDM confers better SUR-isotype selectivity. Therefore, our structure provides mechanistic insight into the high selectivity of MTG and paves the way for further development of next-generation IS with high selectivity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at http://www.wwpdb.org/, 7WIT. https://www.ebi.ac.uk/pdbe/emdb/, EMD-32535.
LC initiated the project. MW purified protein, prepared the cryo-EM sample, and performed electrophysiology experiments. MW and J-XW collected the data. MW processed the data, and built and refined the model with the help of LC. All authors contributed to the manuscript preparation.
The work is supported by grants from the National Natural Science Foundation of China (91957201, 31870833, and 31821091 LC) and the Center for Life Sciences (CLS). This work is also supported by Peking University Ge Li and Ning Zhao Life Science Research Fund for Young Scientists (LGZNQN202102 to LC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cryo-EM data collection was supported by the Electron Microscopy Laboratory and Cryo-EM platform of Peking University with the assistance of Xuemei Li, Zhenxi Guo, Bo Shao, Xia Pei, and Guopeng Wang. Part of the structural computation was also performed on the Computing Platform of the Center for Life Science and High-Performance Computing Platform of Peking University. We thank the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with negative stain EM.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.929684/full#supplementary-material
Afonine, P. V., Poon, B. K., Read, R. J., Sobolev, O. V., Terwilliger, T. C., Urzhumtsev, A., et al. (2018). Real-space Refinement in PHENIX for Cryo-EM and Crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544. doi:10.1107/S2059798318006551
Ashcroft, F. M. (2005). ATP-sensitive Potassium Channelopathies: Focus on Insulin Secretion. J. Clin. Invest. 115, 2047–2058. doi:10.1172/JCI25495
Ashcroft, F. M., and Gribble, F. M. (1998). Correlating Structure and Function in ATP-Sensitive K+ Channels. Trends Neurosci. 21, 288–294. doi:10.1016/s0166-2236(98)01225-9
Ashcroft, F. M., and Rorsman, P. (1989). Electrophysiology of the Pancreatic Beta-Cell. Prog. Biophys. Mol. Biol. 54, 87–143. doi:10.1016/0079-6107(89)90013-8
Ashfield, R., Gribble, F. M., Ashcroft, S. J., and Ashcroft, F. M. (1999). Identification of the High-Affinity Tolbutamide Site on the SUR1 Subunit of the K(ATP) Channel. Diabetes 48, 1341–1347. doi:10.2337/diabetes.48.6.1341
Ashford, M. L., Sturgess, N. C., Trout, N. J., Gardner, N. J., and Hales, C. N. (1988). Adenosine-5'-triphosphate-sensitive Ion Channels in Neonatal Rat Cultured Central Neurones. Pflugers Arch. 412, 297–304. doi:10.1007/BF00582512
Chatterjee, S., Khunti, K., and Davies, M. J. (2017). Type 2 Diabetes. Lancet 389, 2239–2251. doi:10.1016/S0140-6736(17)30058-2
Cook, D. L., and Hales, C. N. (1984). Intracellular ATP Directly Blocks K+ Channels in Pancreatic B-Cells. Nature 311, 271–273. doi:10.1038/311271a0
Daryabor, G., Atashzar, M. R., Kabelitz, D., Meri, S., and Kalantar, K. (2020). The Effects of Type 2 Diabetes Mellitus on Organ Metabolism and the Immune System. Front. Immunol. 11, 1582. doi:10.3389/fimmu.2020.01582
Ding, D., Wang, M., Wu, J. X., Kang, Y., and Chen, L. (2019). The Structural Basis for the Binding of Repaglinide to the Pancreatic KATP Channel. Cell Rep. 27, 1848–e1844. doi:10.1016/j.celrep.2019.04.050
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and Development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501. doi:10.1107/S0907444910007493
Gribble, F. M., and Reimann, F. (2003). Differential Selectivity of Insulin Secretagogues: Mechanisms, Clinical Implications, and Drug Interactions. J. Diabetes Complicat. 17, 11–15. doi:10.1016/s1056-8727(02)00272-6
Hansen, A. M., Christensen, I. T., Hansen, J. B., Carr, R. D., Ashcroft, F. M., and Wahl, P. (2002). Differential Interactions of Nateglinide and Repaglinide on the Human Beta-Cell Sulphonylurea Receptor 1. Diabetes 51, 2789–2795. doi:10.2337/diabetes.51.9.2789
Kimura, H., Matsuda, H., Fujimoto, H., Arimitsu, K., Toyoda, K., Mukai, E., et al. (2014). Synthesis and Evaluation of 18F-Labeled Mitiglinide Derivatives as Positron Emission Tomography Tracers for β-cell Imaging. Bioorg. Med. Chem. 22, 3270–3278. doi:10.1016/j.bmc.2014.04.059
Li, N., Wu, J. X., Ding, D., Cheng, J., Gao, N., and Chen, L. (2017). Structure of a Pancreatic ATP-Sensitive Potassium Channel. Cell 168, 101–e10. doi:10.1016/j.cell.2016.12.028
Martin, G. M., Kandasamy, B., DiMaio, F., Yoshioka, C., and Shyng, S. L. (2017). Anti-diabetic Drug Binding Site in a Mammalian KATP Channel Revealed by Cryo-EM. Elife 6, e31054. doi:10.7554/eLife.31054
Martin, G. M., Sung, M. W., Yang, Z., Innes, L. M., Kandasamy, B., David, L. L., et al. (2019). Mechanism of Pharmacochaperoning in a Mammalian KATP Channel Revealed by Cryo-EM. Elife 8, e46417. doi:10.7554/eLife.46417
Mogami, H., Shibata, H., Nobusawa, R., Ohnota, H., Satou, F., Miyazaki, J., et al. (1994). Inhibition of ATP-Sensitive K+ Channel by a Non-sulfonylurea Compound KAD-1229 in a Pancreatic Beta-Cell Line, MIN 6 Cell. Eur. J. Pharmacol. 269, 293–298. doi:10.1016/0922-4106(94)90036-1
Nichols, C. G. (2006). KATP Channels as Molecular Sensors of Cellular Metabolism. Nature 440, 470–476. doi:10.1038/nature04711
Noma, A. (1983). ATP-regulated K+ Channels in Cardiac Muscle. Nature 305, 147–148. doi:10.1038/305147a0
Ogawa, K., Ikewaki, K., Taniguchi, I., Takatsuka, H., Mori, C., Sasaki, H., et al. (2007). Mitiglinide, a Novel Oral Hypoglycemic Agent, Preserves the Cardioprotective Effect of Ischemic Preconditioning in Isolated Perfused Rat Hearts. Int. Heart. J. 48, 337–345. doi:10.1536/ihj.48.337
Ohnota, H., Koizumi, T., Tsutsumi, N., Kobayashi, M., Inoue, S., and Sato, F. (1994). Novel Rapid- and Short-Acting Hypoglycemic Agent, a Calcium(2s)-2-Benzyl-3-(cis-Hexahydro-2-Isoindolinylcarbonyl) Propionate (KAD-1229) that Acts on the Sulfonylurea Receptor: Comparison of Effects between KAD-1229 and Gliclazide. J. Pharmacol. Exp. Ther. 269, 489–495.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera-Aa Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Pettersen, E. F., Goddard, T. D., Huang, C. C., Meng, E. C., Couch, G. S., Croll, T. I., et al. (2020). UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 30 (1), 70–82. doi:10.1002/pro.3943
Phillippe, H. M., and Wargo, K. A. (2013). Mitiglinide for Type 2 Diabetes Treatment. Expert Opin. Pharmacother. 14, 2133–2144. doi:10.1517/14656566.2013.834048
Pratley, R. E., Foley, J. E., and Dunning, B. E. (2001). Rapid Acting Insulinotropic Agents: Restoration of Early Insulin Secretion as a Physiologic Approach to Improve Glucose Control. Curr. Pharm. Des. 7, 1375–1397. doi:10.2174/1381612013397348
Punjani, A., Rubinstein, J. L., Fleet, D. J., and Brubaker, M. A. (2017). cryoSPARC: Algorithms for Rapid Unsupervised Cryo-EM Structure Determination. Nat. Methods 14, 290–296. doi:10.1038/nmeth.4169
Quast, U., Stephan, D., Bieger, S., and Russ, U. (2004). The Impact of ATP-Sensitive K+ Channel Subtype Selectivity of Insulin Secretagogues for the Coronary Vasculature and the Myocardium. Diabetes 53 (Suppl. 3), S156–S164. doi:10.2337/diabetes.53.suppl_3.s156
Reimann, F., Proks, P., and Ashcroft, F. M. (2001). Effects of Mitiglinide (S 21403) on Kir6.2/SUR1, Kir6.2/SUR2A and Kir6.2/SUR2B Types of ATP-Sensitive Potassium Channel. Br. J. Pharmacol. 132, 1542–1548. doi:10.1038/sj.bjp.0703962
Rorsman, P., and Trube, G. (1985). Glucose Dependent K+-channels in Pancreatic Beta-Cells Are Regulated by Intracellular ATP. Pflugers Arch. 405, 305–309. doi:10.1007/BF00595682
Rosenthal, P. B., and Henderson, R. (2003). Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-Particle Electron Cryomicroscopy. J. Mol. Biol. 333, 721–745. doi:10.1016/j.jmb.2003.07.013
Spruce, A. E., Standen, N. B., and Stanfield, P. R. (1985). Voltage-dependent ATP-Sensitive Potassium Channels of Skeletal Muscle Membrane. Nature 316, 736–738. doi:10.1038/316736a0
Sunaga, Y., Gonoi, T., Shibasaki, T., Ichikawa, K., Kusama, H., Yano, H., et al. (2001). The Effects of Mitiglinide (KAD-1229), a New Anti-diabetic Drug, on ATP-Sensitive K+ Channels and Insulin Secretion: Comparison with the Sulfonylureas and Nateglinide. Eur. J. Pharmacol. 431, 119–125. doi:10.1016/s0014-2999(01)01412-1
Wang, N., Jiang, X., Zhang, S., Zhu, A., Yuan, Y., Xu, H., et al. (2021). Structural Basis of Human Monocarboxylate Transporter 1 Inhibition by Anti-cancer Drug Candidates. Cell 184, 370–e13. doi:10.1016/j.cell.2020.11.043
Wu, J. X., Ding, D., Wang, M., and Chen, L. (2020). Structural Insights into the Inhibitory Mechanism of Insulin Secretagogues on the Pancreatic ATP-Sensitive Potassium Channel. Biochemistry 59, 18–25. doi:10.1021/acs.biochem.9b00692
Wu, J. X., Ding, D., Wang, M., Kang, Y., Zeng, X., and Chen, L. (2018). Ligand Binding and Conformational Changes of SUR1 Subunit in Pancreatic ATP-Sensitive Potassium Channels. Protein Cell 9, 553–567. doi:10.1007/s13238-018-0530-y
Yan, F. F., Casey, J., and Shyng, S. L. (2006). Sulfonylureas Correct Trafficking Defects of Disease-Causing ATP-Sensitive Potassium Channels by Binding to the Channel Complex. J. Biol. Chem. 281, 33403–33413. doi:10.1074/jbc.M605195200
Zhang, K. (2016). Gctf: Real-Time CTF Determination and Correction. J. Struct. Biol. 193, 1–12. doi:10.1016/j.jsb.2015.11.003
Zheng, S. Q., Palovcak, E., Armache, J. P., Verba, K. A., Cheng, Y., and Agard, D. A. (2017). MotionCor2: Anisotropic Correction of Beam-Induced Motion for Improved Cryo-Electron Microscopy. Nat. Methods 14, 331–332. doi:10.1038/nmeth.4193
Keywords: KATP, SUR1, mitiglinide, insulin secretagogues, ABC transporter, diabetes
Citation: Wang M, Wu J-X and Chen L (2022) Structural Insights Into the High Selectivity of the Anti-Diabetic Drug Mitiglinide. Front. Pharmacol. 13:929684. doi: 10.3389/fphar.2022.929684
Received: 27 April 2022; Accepted: 25 May 2022;
Published: 30 June 2022.
Edited by:
Tian-Le Xu, Shanghai Jiao Tong University, ChinaReviewed by:
Huaiyu Yang, East China Normal University, ChinaCopyright © 2022 Wang, Wu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Chen, Y2hlbmxlaTIwMTZAcGt1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.