
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 22 June 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.918219
This article is part of the Research Topic Therapeutic Drug Monitoring and Clinical Toxicology of Anti-Cancer Drugs, volume II View all 13 articles
Background: Anlotinib is a small molecular multi-targeting tyrosine kinase inhibitor. Growing evidence indicates that treatment efficacy, and toxicity varies considerably between individuals. Therefore, this study aimed to investigate the relationship between cytochrome P450 (CYP450) gene polymorphisms, drug concentrations, and their adverse reactions in anlotinib-treated patients with lung cancer.
Methods: We enrolled 139 patients with lung cancer, treated with anlotinib. Twenty loci in the following five genes of the CYP450 family were genotyped: CYP450 family 3 subfamily A member 5 (CYP3A5), 3 subfamily A member 4 (CYP3A4), 2 subfamily C member 9 (CYP2C9), 2 subfamily C member 19 (CYP2C19), and 1 subfamily A member 2 (CYP1A2). Data on adverse reactions were collected from patients, and plasma anlotinib concentrations were measured.
Results: There were significant variances in plasma trough concentration (3.95–52.88 ng/ml) and peak plasma concentration (11.53–42.8 ng/ml) following administration of 8 mg anlotinib. Additionally, there were significant differences in the plasma trough concentration (5.65–81.89 ng/ml) and peak plasma concentration (18.01–107.18 ng/ml) following administration of 12 mg anlotinib. Furthermore, for CYP2C19-rs3814637, the peak plasma concentrations of mutant allele T carriers (TT+CT) were significantly higher than those of wildtypes (CC). For CYP2C19-rs11568732, the peak plasma concentrations of the mutant allele G carriers (GT+GG) were significantly higher than those of the wild-type (TT). More importantly, the incidence rates of hypertension and hemoptysis (peripheral lung cancer) with TT+CT in rs3814637 and GT+GG in rs11568732 were significantly higher than those with CC and TT.
Conclusions: The plasma trough and peak concentrations varied significantly for both 8 and 12 mg of anlotinib. Single-nucleotide polymorphisms in CYP2C19 are significantly associated with hypertension, hemoptysis, and anlotinib peak concentrations. Polymorphisms in CYP450 may explain inter-individual differences in anlotinib-related adverse reactions.
Lung cancer is one of the most common cancers, killing approximately 1.8 million people worldwide each year (Sung et al., 2021). It is also one of the most common causes of cancer-related deaths in China and worldwide (Cao et al., 2021). In addition to traditional drugs, new cytotoxic drugs, molecular-targeted therapies, and immune checkpoint inhibitors have provided new approaches for cancer treatment (Wu et al., 2021). Among them, molecular-targeted therapies have high specificity for tumor cells and low toxicity, and are considered promising cancer treatments (Herbst et al., 2018). Tyrosine kinase (TK) is an important component of the intracellular signal transduction system, transmitting conditional information from the extracellular domain or cytoplasm to the nucleus. TK is dysregulated in many tumor cells. Therefore, tyrosine kinase inhibitors (TKIs) have emerged as an effective approach for molecular-targeted therapies (Krause and Van Etten, 2005; Ferguson and Gray, 2018).
Anlotinib, a small molecular multi-targeted TKI, can inhibit tumor angiogenesis and proliferation (Syed, 2018). In vitro studies have shown that anlotinib inhibits tumor cell growth by inhibiting platelet-derived growth factor receptor β (PDGFRβ), vascular endothelial growth factor receptor 2/3 (VEGFR2/3), and stem cell factor receptor (c-Kit) (Chi et al., 2018; Lin et al., 2018; Liu et al., 2018). In vivo, anlotinib displayed broad activity in human tumor xenograft models (Sun et al., 2016). Anlotinib has shown promise in several cancer clinical trials. Consequently, anlotinib has been approved in China as a third-line treatment for patients with advanced non-small cell lung cancer (NSCLC), and small cell lung cancer. Despite the numerous benefits of anlotinib treatment, there is an increasing number of cases of treatment failure due to drug resistance and toxicity. Hypertension, elevated thyroid-stimulating hormone (TSH) levels, hand-foot syndrome (HFS), hepatotoxicity, and hemoptysis are the most prevalent adverse reactions to anlotinib (Han et al., 2018a; Han et al., 2018b). With the widespread use of anlotinib in clinical practice, adverse reactions have become increasingly concerning.
Currently, research on advanced lung cancer focuses on identifying the biological predictors of efficacy and adverse events, which lead to the individualization of treatment. Systemic exposure or intracellular drug concentrations can influence drug efficacy and adverse reactions, resulting in interpatient variation (Decosterd et al., 2015). Furthermore, the level of drug exposure is significantly associated with the pharmacokinetic properties (absorption, distribution, and metabolism) of the drug. Therefore, inter-individual variations in pharmacokinetics may influence the efficacy and adverse reactions of anlotinib.
Growing evidence suggests that genetic polymorphisms in cytochrome P450 (CYP450) may significantly influence inter-individual differences in drug responses and disposition (Evans and McLeod, 2003). The activities of these drug-metabolizing enzymes determine how quickly the drug is metabolized and can therefore influence the occurrence of adverse reactions. CYP450 oxidizes approximately half of the widely prescribed medications as well as most oral small-molecule anticancer medicines, such as TKIs (Parra-Guillen et al., 2017). It has been shown that the adverse reactions of TKI correlate with drug concentrations and metabolic enzyme gene polymorphisms (Liao et al., 2020), such as CYP3A5*3 for sorafenib-related severe hepatic and renal damage (Guo et al., 2018), and CYP3A4-rs4646437 and CYP3A5-rs776746 for sunitinib-related hypertension (Garcia-Donas et al., 2011; Diekstra et al., 2015; Diekstra et al., 2017). Studies have shown that P450-mediated oxidation is a key factor that affects the oral bioavailability, exposure, half-life, and interspecific differences in anlotinib. A variety of CYP450 enzymes are involved in anlotinib metabolism in vitro, with CYP3A4 and CYP3A5 being the most readily metabolized enzymes (Zhong et al., 2018). Wei et al. investigated the effects of anlotinib on CYP1A2, CYP2C6, CYP2D1, CYP2D2, and CYP3A1/2 in animals and discovered that anlotinib strongly induces CYP2D1 and CYP3A1/2 (Sun et al., 2017). Although in vitro and in vivo experiments have confirmed the effect of CYP450 enzymes on anlotinib metabolism, the relationship between CYP450 gene polymorphisms, plasma concentrations and clinical adverse reactions of anlotinib in patients with lung cancer remains unclear.
Accordingly, for the first time, we propose that individual differences in plasma drug concentrations, and adverse reactions in patients with lung cancer treated with anlotinib may be correlated with CYP450 polymorphisms. To validate this hypothesis, we examined the plasma concentrations of anlotinib in subjects, analyzing single-nucleotide polymorphisms (SNPs) at high-frequency mutation sites in CYP450. In addition, we examined the correlations between polymorphisms of these genes, plasma concentrations, and adverse reactions.
This single-center retrospective study was conducted in accordance with the Helsinki Declaration of the First Affiliated Hospital of Anhui Medical University, Hefei, China, between January 2020, and August 2021. The study protocol was reviewed and approved by the institutional ethics committee (No. Quick- PJ2019-14-15). Written informed consent was obtained from all patients. The inclusion criteria were as follows: 1) patients aged 18–80 years 2) gender was not limited; 3) patients taking more than two courses of anlotinib, with trackable complete information about adverse reactions during treatment; and 4) patients with clear pathological and imaging diagnoses of lung cancer, including non-small cell lung cancer and small cell lung cancer. The exclusion criteria were as follows: 1) moderate-to-severe hepatic and renal insufficiency, 2) allergy to anlotinib, 3) missing basic or adverse reaction information, 4) pregnant or lactating women, and 5) poor compliance.
Patients received a 2-week on/1-week off (2/1) schedule and a 12 or 8 mg once-daily dose for our study. As second-line treatment, anlotinib was combined with immune checkpoint inhibitors (carryilizumab and pabrolizumab). In addition, anlotinib alone was used as third-line treatment. None of the enrolled patients was taking drugs that affected the plasma levels of anlotinib (e.g., rifampicin, ketoconazole, and itraconazole). Peak concentration blood samples were collected at 8 a.m. on the day after the end of the course of treatment (day 15). Blood samples were collected 30 min before the start of the new course (day 22). The collected blood samples were centrifuged at 3,000 rpm for 5 min. The supernatant in the centrifuge tubes was collected and stored at −80°C until analysis, while the remaining samples were used for gene polymorphism analysis. An ACQUITY ultra-performance liquid chromatograph (UPLC, Waters, United States) combined with a Xevo TQ-S triple quadrupole mass spectrometer (Waters, United States) was used to determine the plasma concentrations of anlotinib. Gradient separation was performed on a PACQUITY UPLC BEH C18 column (1.7 µm 2.1 × 50 mm) (Waters, United States) at a temperature of 40°C and a flow rate of 0.5 ml/min. Data processing was performed using MassLynxV4.1 a data workstation (Waters, United States). A standard curve was established according to an internal reference to calculate the plasma concentrations of anlotinib. The results of methodological validation are presented in Supplementary Material S3.
DNA was extracted from the blood samples of patients using a blood genomic DNA extraction kit (Tiangen Biotechnology, Beijing, China) and stored at −80°C until analysis. The quality of the extracted DNA was strictly controlled using Nanodrop 2000 (Thermo Scientific, United States) and capillary electrophoresis (Qiagen, Switzerland). In this study, 20 loci in five different CYP450 genes were selected for polymorphism detection, including rs2242480, rs35599367, rs4646437, rs3735451, and rs4646460 in CYP3A4; rs1419745, rs4646450, rs15524, and rs3800959 in CYP3A5; rs11568732, rs12248560, rs12769205, rs3814637, rs4244285, and rs4986893 in CYP2C19; rs2069526, rs2470890, rs4646425, and rs4646427 in CYP1A2; and rs9332113 in CYP2C9. Primers for all gene loci in patients were designed using Sequenom (Assay Design Suite V2.0) online software. MassARRAY Analyzer Compac mass spectrometry was used to detect gene locus information. TYPER software was used to analyze the results and obtain the genotyping data.
All patients were followed-up in special clinics or wards between January 2020, and August 2021 to monitor anlotinib-induced adverse reactions. Adverse reactions were graded using the Common Terminology Criteria for Adverse Events version 5.0 (CTCAE 5.0). The results were recorded during an adverse reaction assessment.
All statistical analyses were performed using Statistical Products and Services Solutions, version 26 (SPSS 26.0). Descriptive statistics were used to summarize the demographic data and baseline characteristics of the patients receiving anlotinib. The mean and standard deviation were calculated for normally distributed data. Comparisons between groups for continuous variables were performed using an independent-sample t-test or one-way ANOVA. If the variance was not uniform, the Mann-Whitney U test was used. Chi-square and Fisher’s exact tests were used to compare differences in the Hardy-Weinberg balance test and adverse reactions among different genotypes. For all statistical analyses, a p-value < 0.05 was considered statistically significant.
In the current study, 139 patients were enrolled in the follow-up trials. The mean age of the study population was 62.97 ± 10.9 years. Of the patients, 87 (62.6%) were male and 52 (37.4%) were female. Only four patients (2.9%) had a family history of tumors. Twenty-four (17.3%) patients had a history of smoking. The detailed demographic and clinical characteristics of the patients are shown in Table 1.
A total of 73 of 139 patients with lung cancer received 8 mg anlotinib per day, and 66 patients received 12 mg anlotinib per day. The plasma concentrations of anlotinib are shown in Figure 1. The trough and peak concentrations of anlotinib (8 mg) were 17.59 ± 12.55 and 27.01 ± 7.97 ng/ml, ranging from 3.95 to 52.88 ng/ml and 11.53–42.8 ng/ml, respectively. In contrast, the trough and peak concentrations after 12 mg of anlotinib dosing were 23.51 ± 14.83 and 41.22 ± 27.15 ng/ml, ranging from 5.65 to 81.89 ng/ml and 18.01–107.18 ng/ml, respectively. There were significant differences between the trough and peak concentrations at a dosage of 8 versus 12 mg of anlotinib per day (p < 0.05).
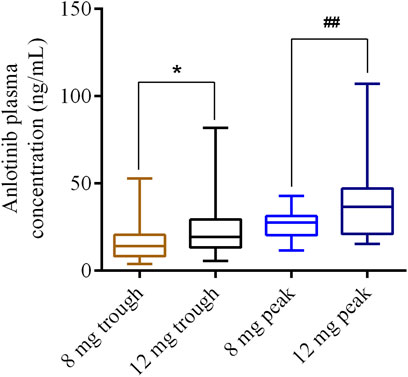
FIGURE 1. Distribution of anlotinib trough or peak concentrations for 8 and 12 mg dosages, respectively. *p < 0.05; ##p < 0.01.
The genotype status of 20 CYP450 loci was determined in 139 patients. For the CYP3A5-rs15524 polymorphism, the frequencies of the A and G alleles were 69.42 and 30.58%, respectively. The frequencies of the G and A alleles for rs4646450 were 71.22 and 28.78%, respectively. The frequencies of the C and T alleles for rs1419745 were 28.42 and 71.58%, and the frequencies of the A and G alleles for rs3800959 were 82.37 and 17.63%, respectively. For the CYP3A4-rs2242480 polymorphism, the frequencies of the T and C alleles were 25.54 and 74.46%, respectively. The frequencies of the C and T alleles of rs43735451 were 31.29 and 68.71%, respectively. The frequencies of the A and G alleles for rs4646437 were 11.87 and 88.13%, respectively, and those of the A and G alleles for rs4646440 were 24.10 and 75.90%, respectively. For the CYP2C9-rs9332113 polymorphism, the frequencies of the G and C alleles were 65.83 and 34.17%, respectively. For the CYP2C19-rs12769205 polymorphism, the frequencies of the G and A alleles were 33.81 and 66.19%, respectively. The frequencies of the T and C alleles of rs12248560 were 1.44 and 98.56%, respectively. The frequencies of the T and C alleles of rs3814637 were 8.27 and 91.73%, respectively. The frequencies of the A and G alleles of rs4986893 were 5.76 and 94.24%, respectively. The frequencies of the G and T alleles for rs11568732 were 8.27 and 91.73%, and those of the A and G alleles for rs4244285 were 33.45 and 66.55%, respectively. For the CYP1A2-rs4646427 polymorphism, the frequencies of the C and T alleles were 6.47 and 93.53% respectively. The frequencies of the G and T alleles were 6.47 and 93.53% for rs2069526, respectively. The frequencies of the T and C alleles were 13.67 and 86.33% for rs2470890 and 93.53 and 6.47% for rs4646425, respectively. All alleles followed–y the Weinberg equilibrium, except for CYP340-rs35599367. Genotype and allele frequencies of the study population are shown in Table 2.
Correlation analysis between different genotypes and the peak plasma concentrations of anlotinib showed that CYP2C19-rs3814637 and -rs11568732 were significantly associated with peak plasma concentrations. For CYP2C19-rs3814637, the peak plasma concentrations of the mutant allele T carriers (TT+CT) were significantly higher than those of the wildtypes (CC) (p = 0.035). For CYP2C19-rs11568732, the peak plasma concentrations of mutant allele G carriers (GT+GG) were significantly higher than those of the wild-type (TT) (p = 0.035), as shown in Table 3. The differences between the plasma concentrations of the other genotypes and that of anlotinib were not statistically significant.
After analyzing the correlation between different genotypes and adverse reactions of anlotinib, for all 20 of the SNPs, only the mutations of CYP2C19-rs3814637 and -rs11568732 genotypes were significantly associated with the occurrence of hypertension and hemoptysis (peripheral lung cancer) in the study population.
The incidence rate of hypertension in mutant allele T carriers (CT + TT) of rs3814637 was significantly higher than that in the wildtype (CC) (p = 0.034, odds ratio (OR) = 0.316, 95% confidence interval (CI): 0.12–0.835). The incidence rate of hypertension in mutated allele G carriers (GT + GG) of rs11568732 was significantly higher than that in the wild type (TT) (p = 0.034, OR = 0.316, 95% CI: 0.12–0.835).
The incidence rate of hemoptysis (peripheral lung cancer) in mutated allele G carriers (GT + GG) of rs11568732 was significantly higher than that in the wild type (TT) (p = 0.043, OR = 0.13, 95% CI: 0.02–0.845). The incidence rate of hemoptysis (peripheral lung cancer) in mutant allele T carriers (CT + TT) of rs3814637 was significantly higher than that in the wild type (CC) (p = 0.043, OR = 0.13, 95% CI: 0.02–0.845). There was no significant difference between rs11568732 and rs3814637 genotypes in the incidence of hemoptysis between central lung cancer (p > 0.05). Table 4 presents the results.
There were no significant differences in the incidence of proteinuria, hepatotoxicity, or other adverse reactions among the 20 SNP genotypes (p > 0.05).
Anlotinib is a multitarget drug that has been developed and marketed independently in China. A phase I clinical trial confirmed that anlotinib showed controlled toxicity at a dose of 12 mg once daily in a 2-week on/1-week off schedule (2/1) (Sun et al., 2016). However, adverse reactions to anlotinib in clinical practice often leads to dose reductions, or treatment discontinuation. In the present study, the most common adverse reactions associated with anlotinib were hypertension, hemoptysis, elevated TSH levels, hepatotoxicity, hypertriglyceridemia, proteinuria, dyspepsia, and HFS. Notably, hypertension was the most common adverse reaction during anlotinib treatment, which is consistent with the results of a phase II trial (Han et al., 2018b). In this study, plasma anlotinib concentrations showed large individual variations. In addition, 11 patients developed a grade 3 or higher incidence of hypertension during treatment, resulting in dose reductions in five patients and drug changes in six patients. This dramatic change can be explained not only by drug interactions and acquired factors but also by genetic factors, such as CYP450 gene polymorphisms. As CYP450 is regulated by genes, once the gene is mutated, CYP450 is regulated and synthesized, and drug metabolism via the enzyme is subsequently altered. Therefore, polymorphisms in CYP450 are the basis of race and individual differences in drug metabolism. In this study, we used a candidate gene approach to investigate the correlation between polymorphisms in genes encoding drug-metabolizing enzymes and anlotinib toxicity.
In humans, CYP3A4/5 has the largest impact on drug biotransformation, followed by CYP2D6, CYP2C6, CYP1A2, and other enzymes (Zanger and Schwab, 2013). Similarly, TKIs are metabolized by CYP450 enzymes, which affects drug concentrations. Imatinib is a cytochrome of CYP3A4 and CYP2C8 substrates that markedly increases plasma CYP3A4 substrate concentrations and reduces hepatic CYP3A4 activity in humans (Filppula et al., 2012). According to previous in vitro investigations, erlotinib is largely metabolized by CYP3A4, whereas CYP1A2 makes a minor contribution (Endo-Tsukude et al., 2018). The major metabolic enzymes of anlotinib are similar to those of imatinib and erlotinib. CYP3A4 and CYP3A5 play important roles in the metabolism of anlotinib.
It is known to all that the genetic mutations of metabolic enzymes do not necessarily lead to corresponding changes in blood exposure to substrate drugs. Currently, there is no evidence of the influence of CYP3A4/5 gene mutations on blood exposure to anlotinib. Anlotinib metabolism is regulated by multiple enzymes. In addition, as mentioned in the previous paragraph, even if CYP3A4/5 gene mutation alters its enzyme activity, this single degree of enzyme activity change does not necessarily affect the pharmacokinetic characteristics of anlotinib (Zhong et al., 2018). This process involves many complex mechanisms. For example, as the largest part (82%) of the intestinal CYP450 family, intestinal CYP3A is inhibited by anlotinib during intestinal absorption (the first site of drug exposure in the body’s metabolic system) and probably results in increased systemic exposure to anlotinib (Kaminsky and Zhang, 2003; Paine et al., 2006). There are many other factors.
This study provides evidence that anlotinib blood exposure is clinically associated not with CYP3A4 and CYP3A5 mutations but with CYP2C19 mutations. This is an interesting phenomenon, and further mechanistic evidence is required.
Previous in vivo studies have demonstrated that efficacy and adverse reactions are closely linked to genetic factors. However, few drug-metabolizing enzyme gene polymorphisms have been used as predictive factors of TKI toxicity (Scott, 2011; Patel, 2016). In this study, we found that the CYP2C19-rs3814637 and -rs11568732 genotypes were significantly associated with the occurrence of hypertension and hemoptysis. Studies have shown that CYP2C19 genotype can guide antiplatelet therapy (Martin et al., 2020). In addition, polymorphisms in the CYP2C19 gene have been reported to be positively associated with cardiovascular diseases, such as coronary artery disease and atherosclerosis (Ercan et al., 2008; Yang et al., 2010). Further studies showed that CYP2C19-rs11568732 was significantly associated with bleeding in patients with ST-segment elevation myocardial infarction who underwent primary percutaneous coronary intervention and treatment with clopidogrel (Novkovic et al., 2018). This finding is consistent with the results of our study, which showed that CYP2C19-rs11568732 was significantly associated with the occurrence of hemoptysis. Additionally, studies have shown that the CYP2C19*3 defective allele may contribute to a reduced risk of developing essential hypertension (Shin et al., 2012). However, few studies have investigated the correlation between CYP2C19-rs3814637 and -rs11568732 polymorphisms and hypertension. In this study, no association was observed between the different genotypes of other CYP450 loci and adverse reactions to anlotinib.
The epidermal growth factor receptor (EGFR) is a ligand receptor for epidermal growth factor (EGF). When EGF binds to EGFR, the signaling pathway is activated, leading to cell proliferation and differentiation. EGFR overexpression due to mutations or structural changes promotes carcinogenesis, invasion, and metastasis. Studies have shown that common mutations are associated with sensitivity to EGFR tyrosine kinase inhibitors (TKI) (Lynch et al., 2004; Paez et al., 2004). Anlotinib is a small-molecule, multi-target TKI. Currently, there is no evidence for an association between anlotinib and EGFR mutations. In this study, 17 patients were found to have EGFR mutations; however, no correlation was found between EGFR mutations and anlotinib-induced adverse reactions.
Based on this study, it is reasonable to speculate that inter-individual differences in anlotinib-related adverse reactions may be explained by CYP450 gene polymorphisms or different exposures, caused by CYP450 gene polymorphisms. Thus, the clinical application of anlotinib should be based on the genotyping of CYP450 in lung cancer, particularly for rs3814637 and rs11568732 of CYP2C19. This strategy may reduce the incidence of anlotinib-related adverse reactions. However, the underlying mechanisms by which CYP450 gene polymorphisms influence the variation in anlotinib-related adverse reactions and toxicity between individuals remain unclear, and could be a direction for further research.
Nevertheless, our study has a few limitations. First, it was a single-center prospective study with limited patient enrolment. Second, owing to the short follow-up time of the patients, there was no statistical information about progression-free survival and overall survival. Third, the anlotinib concentration may be influenced by other factors (e.g., coffee and drugs), which was not studied separately.
In summary, some CYP450 genotypes are significantly associated with adverse reactions to anlotinib in clinical practice, including hypertension and hemoptysis. Our results show that these genetic variants in CYP450 can explain inter-individual differences in anlotinib adverse reactions. Therefore, identifying CYP450 gene polymorphisms, particularly CYP2C19-rs3814637 and -rs11568732, before anlotinib treatment might be a potential personalized treatment approach.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by The Ethics Committee of the First Affiliated Hospital of Anhui Medical University approved this study. The patients/participants provided their written informed consent to participate in this study.
QX conceptualized and supervised the project; TT and GH carried out the study and interpreted the results; ZC and JJ drafted the initial version of the manuscript; LZ and ZX searched the literature and collected data; QX and XW modified the revised manuscript; All authors read and approved the manuscript.
This study was supported by the Clinical Research Fund of Wu Jieping Medical Foundation (320.6750.2020-03-6) and the Program of National Natural Science Foundation of China (82174011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to take the opportunity to thank the patients, their families, and all of the research members in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.918219/full#supplementary-material
Cao, W., Chen, H. D., Yu, Y. W., Li, N., and Chen, W. Q. (2021). Changing Profiles of Cancer Burden Worldwide and in China: a Secondary Analysis of the Global Cancer Statistics 2020. Chin. Med. J. Engl. 134 (7), 783–791. doi:10.1097/CM9.0000000000001474
Chi, Y., Fang, Z., Hong, X., Yao, Y., Sun, P., Wang, G., et al. (2018). Safety and Efficacy of Anlotinib, a Multikinase Angiogenesis Inhibitor, in Patients with Refractory Metastatic Soft-Tissue Sarcoma. Clin. Cancer Res. 24 (21), 5233–5238. doi:10.1158/1078-0432.CCR-17-3766
Decosterd, L. A., Widmer, N., Zaman, K., Cardoso, E., Buclin, T., and Csajka, C. (2015). Therapeutic Drug Monitoring of Targeted Anticancer Therapy. Biomark. Med. 9 (9), 887–893. doi:10.2217/bmm.15.78
Diekstra, M. H., Belaustegui, A., Swen, J. J., Boven, E., Castellano, D., Gelderblom, H., et al. (2017). Sunitinib-induced Hypertension in CYP3A4 Rs4646437 A-Allele Carriers with Metastatic Renal Cell Carcinoma. Pharmacogenomics J. 17 (1), 42–46. doi:10.1038/tpj.2015.100
Diekstra, M. H., Swen, J. J., Boven, E., Castellano, D., Gelderblom, H., Mathijssen, R. H., et al. (2015). CYP3A5 and ABCB1 Polymorphisms as Predictors for Sunitinib Outcome in Metastatic Renal Cell Carcinoma. Eur. Urol. 68 (4), 621–629. doi:10.1016/j.eururo.2015.04.018
Endo-Tsukude, C., Sasaki, J. I., Saeki, S., Iwamoto, N., Inaba, M., Ushijima, S., et al. (2018). Population Pharmacokinetics and Adverse Events of Erlotinib in Japanese Patients with Non-small-cell Lung Cancer: Impact of Genetic Polymorphisms in Metabolizing Enzymes and Transporters. Biol. Pharm. Bull. 41 (1), 47–56. doi:10.1248/bpb.b17-00521
Ercan, B., Ayaz, L., Ciçek, D., and Tamer, L. (2008). Role of CYP2C9 and CYP2C19 Polymorphisms in Patients with Atherosclerosis. Cell Biochem. Funct. 26 (3), 309–313. doi:10.1002/cbf.1437
Evans, W. E., and McLeod, H. L. (2003). Pharmacogenomics--drug Disposition, Drug Targets, and Side Effects. N. Engl. J. Med. 348 (6), 538–549. doi:10.1056/NEJMra020526
Ferguson, F. M., and Gray, N. S. (2018). Kinase Inhibitors: the Road Ahead. Nat. Rev. Drug Discov. 17 (5), 353–377. doi:10.1038/nrd.2018.21
Filppula, A. M., Laitila, J., Neuvonen, P. J., and Backman, J. T. (2012). Potent Mechanism-Based Inhibition of CYP3A4 by Imatinib Explains its Liability to Interact with CYP3A4 Substrates. Br. J. Pharmacol. 165 (8), 2787–2798. doi:10.1111/j.1476-5381.2011.01732.x
Garcia-Donas, J., Esteban, E., Leandro-García, L. J., Castellano, D. E., González del Alba, A., Climent, M. A., et al. (2011). Single Nucleotide Polymorphism Associations with Response and Toxic Effects in Patients with Advanced Renal-Cell Carcinoma Treated with First-Line Sunitinib: a Multicentre, Observational, Prospective Study. Lancet Oncol. 12 (12), 1143–1150. doi:10.1016/S1470-2045(11)70266-2
Guo, X. G., Wang, Z. H., Dong, W., He, X. D., Liu, F. C., and Liu, H. (2018). Specific CYP450 Genotypes in the Chinese Population Affect Sorafenib Toxicity in HBV/HCV-associated Hepatocellular Carcinoma Patients. Biomed. Environ. Sci. 31 (8), 586–595. doi:10.3967/bes2018.080
Han, B., Li, K., Wang, Q., Zhang, L., Shi, J., Wang, Z., et al. (2018a). Effect of Anlotinib as a Third-Line or Further Treatment on Overall Survival of Patients with Advanced Non-small Cell Lung Cancer: The ALTER 0303 Phase 3 Randomized Clinical Trial. JAMA Oncol. 4 (11), 1569–1575. doi:10.1001/jamaoncol.2018.3039
Han, B., Li, K., Zhao, Y., Li, B., Cheng, Y., Zhou, J., et al. (2018b). Anlotinib as a Third-Line Therapy in Patients with Refractory Advanced Non-small-cell Lung Cancer: a Multicentre, Randomised Phase II Trial (ALTER0302). Br. J. Cancer 118 (5), 654–661. doi:10.1038/bjc.2017.478
Herbst, R. S., Morgensztern, D., and Boshoff, C. (2018). The Biology and Management of Non-small Cell Lung Cancer. Nature 553 (7689), 446–454. doi:10.1038/nature25183
Kaminsky, L. S., and Zhang, Q. Y. (2003). The Small Intestine as a Xenobiotic-Metabolizing Organ. Drug Metab. Dispos. 31 (12), 1520–1525. doi:10.1124/dmd.31.12.1520
Krause, D. S., and Van Etten, R. A. (2005). Tyrosine Kinases as Targets for Cancer Therapy. N. Engl. J. Med. 353 (2), 172–187. doi:10.1056/NEJMra044389
Liao, D., Liu, Z., Zhang, Y., Liu, N., Yao, D., Cao, L., et al. (2020). Polymorphisms of Drug-Metabolizing Enzymes and Transporters Contribute to the Individual Variations of Erlotinib Steady State Trough Concentration, Treatment Outcomes, and Adverse Reactions in Epidermal Growth Factor Receptor-Mutated Non-small Cell Lung Cancer Patients. Front. Pharmacol. 11, 664. doi:10.3389/fphar.2020.00664
Lin, B., Song, X., Yang, D., Bai, D., Yao, Y., and Lu, N. (2018). Anlotinib Inhibits Angiogenesis via Suppressing the Activation of VEGFR2, PDGFRβ and FGFR1. Gene 654, 77–86. doi:10.1016/j.gene.2018.02.026
Liu, Z., Wang, J., Meng, Z., Wang, X., Zhang, C., Qin, T., et al. (2018). CD31-labeled Circulating Endothelial Cells as Predictor in Anlotinib-Treated Non-small-cell Lung Cancer: Analysis on ALTER-0303 Study. Cancer Med. 7, 3011–3021. doi:10.1002/cam4.1584
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., et al. (2004). Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-small-cell Lung Cancer to Gefitinib. N. Engl. J. Med. 350 (21), 2129–2139. doi:10.1056/NEJMoa040938
Martin, J., Williams, A. K., Klein, M. D., Sriramoju, V. B., Madan, S., Rossi, J. S., et al. (2020). Frequency and Clinical Outcomes of CYP2C19 Genotype-Guided Escalation and De-escalation of Antiplatelet Therapy in a Real-World Clinical Setting. Genet. Med. 22 (1), 160–169. doi:10.1038/s41436-019-0611-1
Novkovic, M., Matic, D., Kusic-Tisma, J., Antonijevic, N., Radojkovic, D., and Rakicevic, L. (2018). Analysis of the CYP2C19 Genotype Associated with Bleeding in Serbian STEMI Patients Who Have Undergone Primary PCI and Treatment with Clopidogrel. Eur. J. Clin. Pharmacol. 74 (4), 443–451. doi:10.1007/s00228-017-2401-5
Paez, J. G., Jänne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., et al. (2004). EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 304 (5676), 1497–1500. doi:10.1126/science.1099314
Paine, M. F., Hart, H. L., Ludington, S. S., Haining, R. L., Rettie, A. E., and Zeldin, D. C. (2006). The Human Intestinal Cytochrome P450 "pie". Drug Metab. Dispos. 34 (5), 880–886. doi:10.1124/dmd.105.008672
Parra-Guillen, Z. P., Berger, P. B., Haschke, M., Donzelli, M., Winogradova, D., Pfister, B., et al. (2017). Role of Cytochrome P450 3A4 and 1A2 Phenotyping in Patients with Advanced Non-small-cell Lung Cancer Receiving Erlotinib Treatment. Basic Clin. Pharmacol. Toxicol. 121 (4), 309–315. doi:10.1111/bcpt.12801
Patel, J. N. (2016). Cancer Pharmacogenomics, Challenges in Implementation, and Patient-Focused Perspectives. Pharmgenomics Pers. Med. 9, 65–77. doi:10.2147/PGPM.S62918
Scott, S. A. (2011). Personalizing Medicine with Clinical Pharmacogenetics. Genet. Med. 13 (12), 987–995. doi:10.1097/GIM.0b013e318238b38c
Shin, D. J., Kwon, J., Park, A. R., Bae, Y., Shin, E. S., Park, S., et al. (2012). Association of CYP2C19*2 and *3 Genetic Variants with Essential Hypertension in Koreans. Yonsei Med. J. 53 (6), 1113–1119. doi:10.3349/ymj.2012.53.6.1113
Sun, W., Wang, Z., Chen, R., Huang, C., Sun, R., Hu, X., et al. (2017). Influences of Anlotinib on Cytochrome P450 Enzymes in Rats Using a Cocktail Method. Biomed. Res. Int. 2017, 3619723. doi:10.1155/2017/3619723
Sun, Y., Niu, W., Du, F., Du, C., Li, S., Wang, J., et al. (2016). Safety, Pharmacokinetics, and Antitumor Properties of Anlotinib, an Oral Multi-Target Tyrosine Kinase Inhibitor, in Patients with Advanced Refractory Solid Tumors. J. Hematol. Oncol. 9 (1), 105. doi:10.1186/s13045-016-0332-8
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Syed, Y. Y. (2018). Anlotinib: First Global Approval. Drugs 78 (10), 1057–1062. doi:10.1007/s40265-018-0939-x
Wu, F., Wang, L., and Zhou, C. (2021). Lung Cancer in China: Current and Prospect. Curr. Opin. Oncol. 33 (1), 40–46. doi:10.1097/CCO.0000000000000703
Yang, Y. N., Wang, X. L., Ma, Y. T., Xie, X., Fu, Z. Y., Li, X. M., et al. (2010). Association of Interaction between Smoking and CYP 2C19*3 Polymorphism with Coronary Artery Disease in a Uighur Population. Clin. Appl. Thromb. Hemost. 16 (5), 579–583. doi:10.1177/1076029610364522
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 138 (1), 103–141. doi:10.1016/j.pharmthera.2012.12.007
Keywords: anlotinib, lung cancer, CYP450 gene polymorphisms, plasma concentration, adverse reactions
Citation: Tan T, Han G, Cheng Z, Jiang J, Zhang L, Xia Z, Wang X and Xia Q (2022) Genetic Polymorphisms in CYP2C19 Cause Changes in Plasma Levels and Adverse Reactions to Anlotinib in Chinese Patients With Lung Cancer. Front. Pharmacol. 13:918219. doi: 10.3389/fphar.2022.918219
Received: 12 April 2022; Accepted: 31 May 2022;
Published: 22 June 2022.
Edited by:
Miao Yan, Central South University, ChinaReviewed by:
Wei-Wei Jia, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Tan, Han, Cheng, Jiang, Zhang, Xia, Wang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Xia, eGlhcXVhbjIwMTBAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.