- 1Department of Orthopaedics, Xiangya Hospital, Central South University, Changsha, China
- 2National Institution of Drug Clinical Trial, Xiangya Hospital, Central South University, Changsha, China
- 3Departments of Clinical Pharmacology, Xinagya Hospital, Central South University, Changsha, China
- 4Institute of Clinical Pharmacology and Hunan Key Laboratory of Pharmacogenetics, Central South University, Changsha, China
- 5Lung Cancer and Gastrointestinal Unit, Department of Medical Oncology, Hunan Cancer Hospital, Affiliated Cancer Hospital of Xiangya School of Medicine, Changsha, China
- 6Hunan Clinical Research Center in Gynecologic Cancer, Hunan Cancer Hospital, Affiliated Cancer Hospital of Xiangya School of Medicine, Changsha, China
- 7Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, China
- 8Department of Pharmacy, The Second People’s Hospital of Huaihua City, Huaihua, China
- 9Department of Pharmacy, Xinagya Hospital, Central South University, Changsha, China
- 10National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China
Objective: The purpose of this study was to investigate the associations of genetic variants in double-strand break (DSB) repair pathway genes with prognosis in patients with lung cancer treated with platinum-based chemotherapy.
Methods: Three hundred ninety-nine patients with lung cancer who received platinum-based chemotherapy for at least two cycles were included in this study. A total of 35 single nucleotide polymorphisms (SNPs) in DSB repair, base excision repair (BER), and nucleotide excision repair (NER) repair pathway genes were genotyped, and were used to evaluate the overall survival (OS) and the progression-free survival (PFS) of patients who received platinum-based chemotherapy using Cox proportional hazard models.
Results: The PFS of patients who carried the MAD2L2 rs746218 GG genotype was shorter than that in patients with the AG or AA genotypes (recessive model: p = 0.039, OR = 5.31, 95% CI = 1.09–25.93). Patients with the TT or GT genotypes of TNFRSF1A rs4149570 had shorter OS times than those with the GG genotype (dominant model: p = 0.030, OR = 0.57, 95% CI = 0.34–0.95). We also investigated the influence of age, gender, histology, smoking, stage, and metastasis in association between SNPs and OS or PFS in patients with lung cancer. DNA repair gene SNPs were significantly associated with PFS and OS in the subgroup analyses.
Conclusion: Our study showed that variants in MAD2L2 rs746218 and TNFRSF1A rs4149570 were associated with shorter PFS or OS in patients with lung cancer who received platinum-based chemotherapy. These variants may be novel biomarkers for the prediction of prognosis of patients with lung cancer who receive platinum-based chemotherapy.
Introduction
Lung cancer has one of the highest rates of cancer-related mortality (Parkin et al., 1999; Siegel et al., 2021). Approximately 2.2 million new lung cancer cases and 1.8 million deaths resulting from lung cancer were reported in 2020, which was double the number reported 30 years earlier (Parkin et al., 1999; Siegel et al., 2021). The overall 5-year survival rate for lung cancer is less than 18% due to rapid progression and late-stage diagnosis (Alam et al., 2020). Lung cancer mainly consists of non–small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), which occur in 85:15 ratio (Shi and Sun, 2015; Schwartz and Cote, 2016). Surgery, radiotherapy, chemotherapy, immunotherapy, and targeted therapy are the primary approaches for the treatment of lung cancer. Specific management is contingent on staging and pathohistological type (Kalemkerian et al., 2018; Ettinger et al., 2019). Development of targeted therapies and immunotherapy has resulted in substantial clinical benefits. However, the majority of patients do not have activating mutations and do not experience long-term stable remission (Hirsch et al., 2017; Arbour and Riely, 2019). Chemotherapy is the main treatment for lung cancer, with platinum-based chemotherapy the most widely-used approach.
Platinum-based chemotherapy has been widely used as a therapeutic regimen to treat cancer, including patients with lung cancer, since the first platinum agent, cisplatin, was approved over 40 years ago (Rottenberg et al., 2021). Cisplatin, carboplatin, and oxaliplatin are the three main platinum-based antineoplastic drugs (Low et al., 2021). Cisplatin is the standard treatment for NSCLC, and is the first choice to treat patients with advanced cancers without treatable gene mutations (Kryczka et al., 2021). The platinum-doublet chemotherapy, platinum combined with etoposide, is recommended as a first-line treatment for late-stage SCLC (Thai et al., 2021; Zugazagoitia and Paz-Ares, 2022). Although platinum-based chemotherapy can improve survival rate, patients treated with platinum-based drugs often suffered from drug resistance, resulting in poor prognosis and therapeutic failure. There are many prognostic factors such as genetic polymorphisms, age, gender, histology type, smoking, metastasis, and clinical stage that have been reported to be connected to platinum-based chemotherapy sensitivity (Cescon et al., 2015; Chen et al., 2016; Yin et al., 2016). Therapeutic efficacy is unsatisfactory and unpredictable. Therefore, the identification of novel biomarkers may help to identify therapeutic avenues that can improve survival time.
Investigation of mechanisms of drug resistance is of great importance for the improvement of the prognosis of lung cancer in response to platinum-based chemotherapy. Given that platinum-based drugs induce DNA fragmentation through crosslinking with DNA to form DNA adducts, alteration of DNA repair mechanisms can affect tumor sensitivity to platinum drugs (Rottenberg et al., 2021). Base excision repair (BER), nucleotide excision repair (NER), DNA mismatch repair (MMR), homologous recombination (HR), and non-homologous end joining (NHEJ) are the major DNA repair pathways, among which HR and NHEJ are responsible for repairing double-strand breaks (DSBs). Furthermore, NHEJ plays an important role in the DNA damage response system (Gupta et al., 2018; Xu and Xu, 2020; Jiang et al., 2021), which can directly link the ends of DSBs by DNA ligase to promote ligation of DNA ends. This process is characterized by impedance of homologous DNA sequences, while HR uses the intact sister chromatid as a template (Huang and Dynan, 2002; Andres et al., 2015; Xing et al., 2015; Almohaini et al., 2016; Kulkarni et al., 2016; Menon and Povirk, 2017; Reid et al., 2017). Previous studies have shown the importance of double-strand break repair (DSBR) pathways in platinum chemotherapy resistance (Kryczka et al., 2021). A numbers of DNA repair genes have been confirmed to be related to platinum-based chemotherapy resistance in patients with lung cancer, including the XRCC5 and HSPB1 genes. However, few studies have focused on polymorphisms in the DSBR pathway genes.
We investigated the associations between SNPs in the MAD2L2, XPC, XRCC3, BRCA2, RAD52, NFKB1, NFKBIA, TNFRSF1A, or FASN genes and prognosis in patients with lung cancer who received platinum-based chemotherapy.
Patients and Methods
Patients and Data Collection
The inclusion criteria for 399 patients with lung cancer were as follows: 1) all patients attended at the Xiangya Hospital of Central South University or Affiliated Cancer Hospital of Xiangya School of Medicine (Changsha, Hunan, China) from August 2009 to May 2013; 2) patients with lung cancer received platinum-based chemotherapy for at least two cycles; 3) patients with lung cancer had not undergone surgery, radiotherapy, targeted drug therapy, or other biological therapy before chemotherapy. The research proposal was approved by the Ethics Committee of Xiangya Hospital, Central South University. All patients provided written informed consent prior to participating in the study.
The deadline for patient enrollment was 15 July 2019. Standard follow-up clinical data included age, gender, smoking history, histology classification, TNM stage, and metastasis. The two main data processing approaches were PFS, which was defined as the time period from diagnosis until disease progression. Patients without OS and PFS data were removed from the study at the final follow-up. The overall survival was calculated as the time between lung cancer diagnosis and follow-up or death.
Single Nucleotide Polymorphism Selection, DNA Extraction, and Genotyping
The SNPs genotyped in our study were ERCC1 SNPs (rs2298881), ERCC2 SNPs (rs1052555, rs238406), ERCC4 SNP (rs1799801), ERCC6 SNPs (rs2228527, rs3793784), XPC SNPs (rs2228000, rs2228001), XRCC1 SNP (rs25489), XRCC3 SNPs (rs1799794, rs861539), BRCA1 SNP (rs799917), BRCA2 SNPs (rs543304,rs206118), RAD51 SNPs (rs12593359, rs1801320, rs1801321), RAD52 SNPs (rs1051669, rs7963551), POLH/POLR1C SNP (rs6941583), MAD2L2 SNPs (rs2233004, rs746218, rs2233006), NFKB1 SNPs (rs230529, rs1585215, rs4648068), NFKBIA SNP (rs2233406), TNF SNP (rs1800629), TNFRSF1A SNPs (rs4149570, rs2234649), TNFRSF1B SNP (rs1061622), and FASN SNPs (rs1140616, rs2228309, rs4246445, rs4485435). Haploview was used to choose pair-wise tagging SNPs with pair wise r2 threshold ≥0.8, and all SNPs had a minor allele frequency (MAF) greater than 0.05 (Table 1).
All blood samples were collected and stored in EDTA tubes. We used a genomic DNA Purification Kit (Promega) to extract genomic DNA. Genotyping of all SNPs was performed using the Sequenom MassARRAY Genotyping Platform (Sequenom, San Diego, CA, United States).
Statistical Analysis
Logistic regression was used to select covariates using the Cox proportional hazard model. The covariates included age, gender, histologic type, smoking status, clinical stage, and metastasis status. Three analysis models (additive model: compares major allele homozygotes versus heterozygotes versus minor allele homozygotes; dominant model: compares major allele homozygous versus combined heterozygotes and minor allele homozygous groups; recessive model: compares major allele-carrying genotypes with homozygous variant genotype) were used to calculate the associations between SNPs and prognosis. In the association analyses, we divided the patients into two or three groups by their genotypes of the SNPs. In additive model and dominant model, patients with wild type were used as a control group; and in recessive model, patients with wild type and heterozygote were used as a control group. The Cox proportional hazard regression analysis was used to analyze OS and PFS. All data were analyzed using SPSS 18.0 software (SPSS Inc., Chicago, IL, United States), PLINK (version 1.9, http://pngu.mgh.harvard.edu/purcell/plink/), and R 4.1.0. The associations between PFS or OS and SNPs were calculated as odds ratio (OR) and their 95% confidence intervals (95% CI) using unconditional logistic regression.
Results
Demographic Characteristics of Patient Characteristics
Three hundred ninety-nine patients with lung cancer were enrolled in this study. All included patients had received platinum-based chemotherapy as the first-line treatment. The patients were 21–75 years old, with a median age of 56 years old. In this study, 317 (79.4%) patients were male and 82 (20.6%) were female. Furthermore, 152 (38.1%) patients were non-smokers and 247 (61.9%) were smokers. In addition, 311 (77.9%) patients had NSCLC and 88 (22.1%) had SCLC. Finally, 351 (88.0%) patients were at advanced stages (stage Ⅲ/Ⅳ/ED), and the remaining 48 (12.0%) were at early stages (stage I/II/LD) (Table 2).
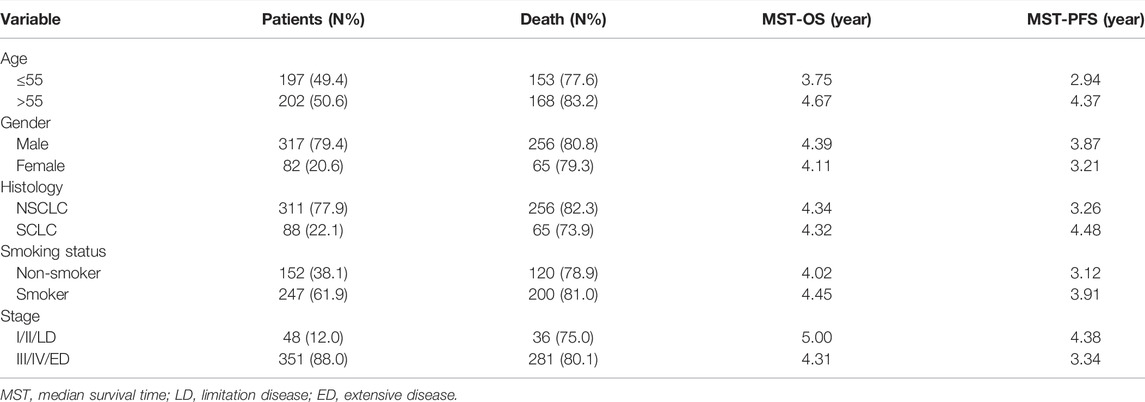
TABLE 2. Distribution of characteristics in patients with patients with lung cancer and prognosis analysis.
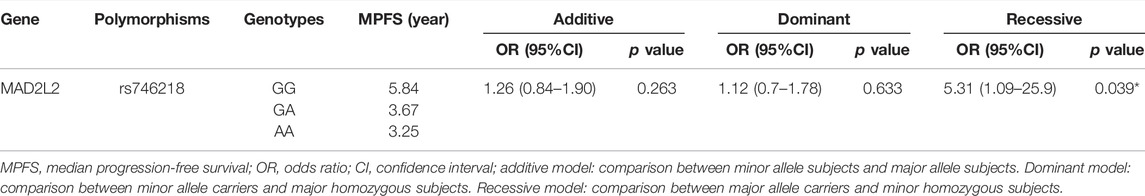
Association Between MAD2L2 rs746218 and PFS in Patients With Lung Cancer
Multivariate Cox regression adjusted for age, gender, histology type, smoking status, stage, and metastasis showed that the MAD2L2 rs746218 polymorphism was significantly related to PFS in patients with lung cancer in the recessive model (p = 0.039, OR = 5.31, and 95% CI = 1.09–25.93) (Table 3). Patients carrying the AG or AA genotype had a significantly longer PFS times than those carrying the GG genotype (Figure 1A). Compared with other SNPs, MAD2L2 rs746218 was significantly associated with PFS in the recessive model analysis in patients with lung cancer who received platinum-based chemotherapy.
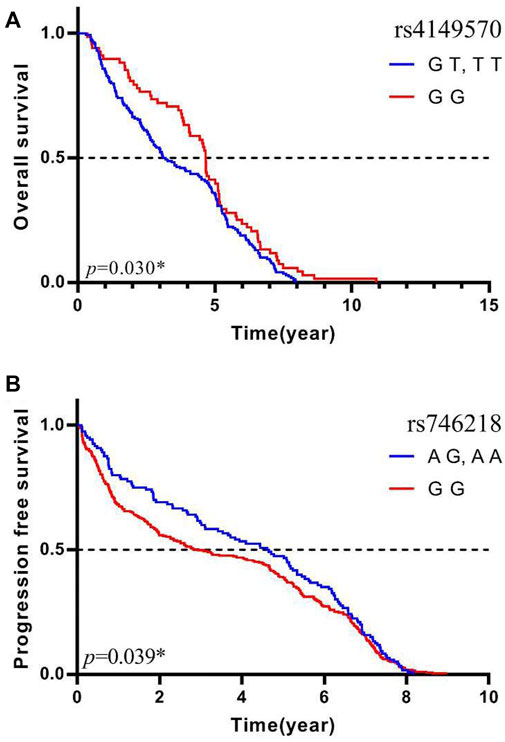
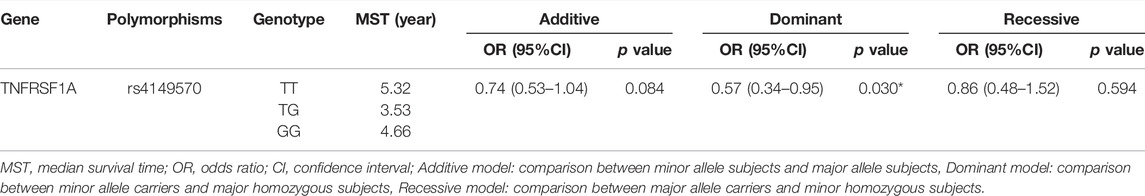
FIGURE 1. MAD2L2 rs746218 and TNFRSF1A rs4149570 were significantly associated with platinum-based chemotherapy prognosis in patients with lung cancer. (A) TNFRSF1A rs4149570 was significantly associated with OS. (B) MAD2L2 rs746218 was significantly associated with PFS.
Association Between TNFRSF1A rs4149570 and OS in Patients With Lung Cancer
Univariate Cox regression analysis was used to evaluate OS, and the results were adjusted for age, gender, histology type, smoking status, stage, and metastasis status. As shown in Table 4, TNFRSF1A rs4149570 was associated with OS in patients with lung cancer in the dominant model (p = 0.030, OR = 0.57, and 95% CI = 0.34–0.95). In the dominant model, the OS of patients who carried the rs4149570 GG genotype was significantly longer than that of patients carrying the TT or GT genotypes (Figure 1B). Compared with other SNPs, TNFRSF1A rs4149570 was most significantly associated with OS in the dominant model in patients with lung cancer who received platinum-based chemotherapy.
Stratification Analyses
In the stratification analyses, age (≤56, >56), smoking status (no, yes), gender (male, female), histological type (NSCLC, SCLC), metastasis (no, yes), and Stage (I/II/LD, III/IV/ED) were evaluated as covariates for associations between SNPs and PFS. The following SNPs were significantly associated with PFS: BRCA2 rs206118 in patients ≤56 years old (additive model: p = 0.039, OR = 0.56, and 95% CI = 0.33–0.97; dominant model: p = 0.041, OR = 0.52, and 95% CI = 0.27–0.97); XRCC3 rs1799794 in patients >56 years old (dominant model p = 0.036, OR = 2.03, and 95% CI = 1.05–3.92); NFKB1 rs230529 in patients >56 years old (recessive model p = 0.012, OR = 2.40, and 95% CI = 1.21–4.74) and NFKB1 rs1585215 in patients with SCLC (recessive model: p = 0.045, OR = 14.66, and 95% CI = 1.06–203.60); RAD52 rs7963551 in male patients (additive model: p = 0.046, OR = 1.49, and 95% CI = 1.01–2.22); NFKBIA rs2233406 (additive model: p = 0.029, OR = 6.73, and 95% CI = 1.22–37.16 dominant model: p = 0.029, OR = 6.73, 95% CI = 1.22–37.16); and TNFRSF1A rs4149570 in patients with lung cancer with stage III/IV cancer (dominant model: p = 0.050, OR = 0.26, and 95% CI = 0.07–1.00) (Table 5; Figure 2).
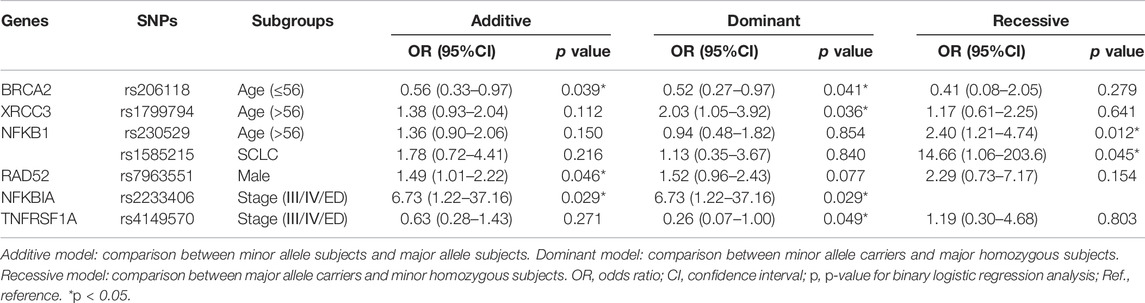
TABLE 5. Stratification analyses of Association between the seven polymorphisms and PFS in lung cancer patients.
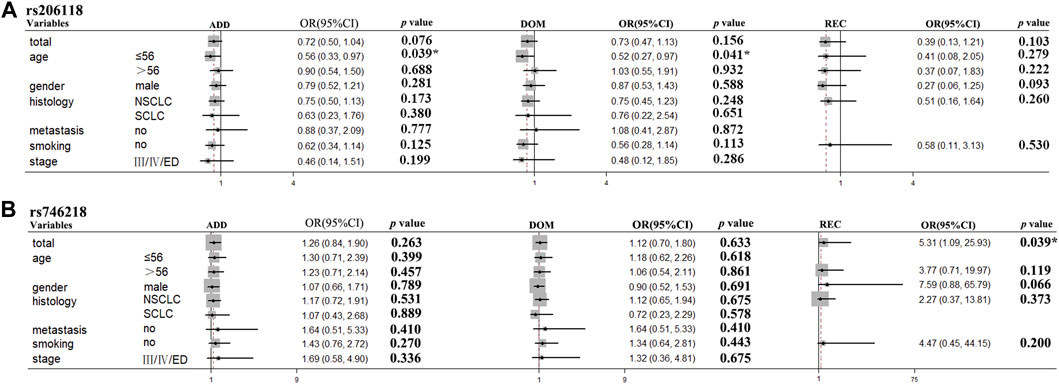
FIGURE 2. BRCA2 rs206118 and MAD2L2 rs746218 polymorphisms were significantly associated with survival time in the subgroups of patients with lung cancer treated with platinum-based chemotherapy. (A) BRCA2 rs206118 polymorphism was significantly associated with PFS time in patients less than 55 years of age. (B) MAD2L2 rs746218 polymorphism was significantly associated with PFS time in patients treated with platinum-based chemotherapy.
For OS stratification analyses, the results were as follows: TNFRSF1A rs4149570 in patients >56 years old (dominant model: p = 0.048, OR = 0.48, and 95% CI = 0.23–0.99); TNFRSF1A rs4149570 in patients with advanced stage cancer (additive model: p = 0.049, OR = 0.70, and 95% CI = 0.48–1.00; dominant model: p = 0.049, OR = 0.58, and 95% CI = 0.34–1.00); XRCC3 rs1799794 in patients with SCLC (dominant model: p = 0.048, OR = 2.27, and 95% CI = 1.01–5.13); XPC rs2228000 in non-smokers (dominant model: p = 0.023, OR = 2.53, and 95% CI = 1.13–5.64); and FASN rs4246445 in non-smokers (dominant model: p = 0.043, OR = 0.43, 95% CI = 0.19–0.97) (Table 6; Figure 3).
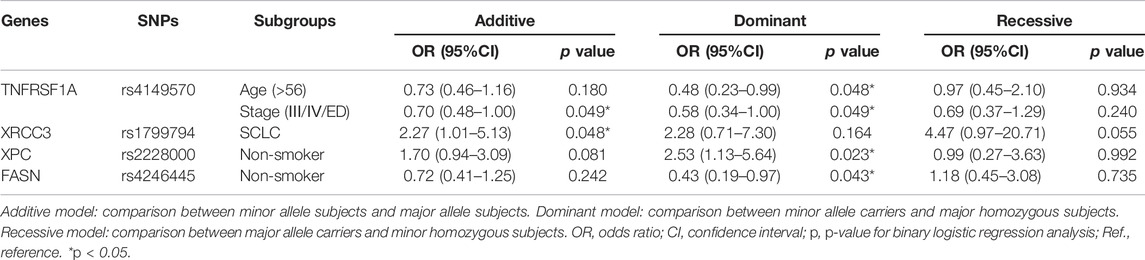
TABLE 6. Stratification analyses of association between the four polymorphisms and OS in lung cancer patients.
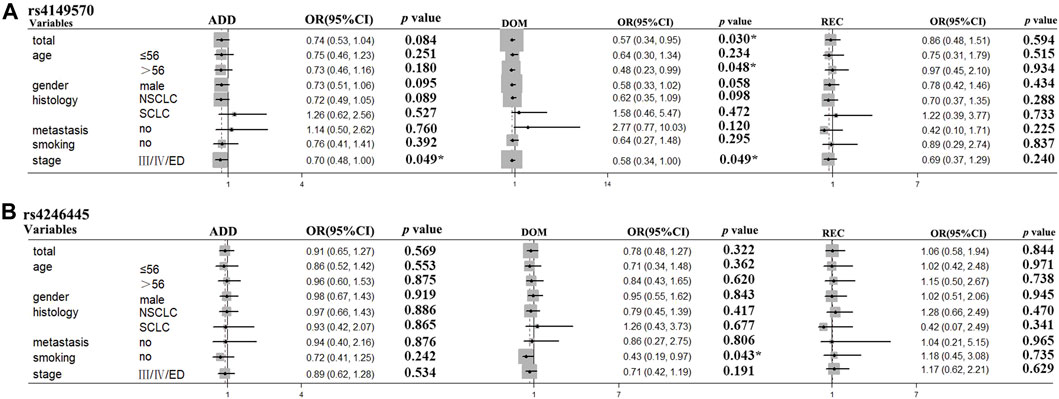
FIGURE 3. TNFRSF1A rs4149570 and FASN rs4246445 polymorphisms were significantly associated with survival times in the subgroups of patients with lung cancer treated with platinum-based chemotherapy. (A) TNFRSF1A rs4149570 polymorphism was significantly associated with OS time in patients less than 55 years of age. (B) FASN rs4246445 polymorphism was significantly associated with PFS time in patients who received platinum-based chemotherapy.
Our results showed that MAD2L2 rs746218 and TNFRSF1A rs4149570 were significantly associated with prognosis in patients with lung cancer who received platinum-based chemotherapy. The PFS time of patients with GG genotype (Median PFS: 3.45 (0.10–9.17) years) of MAD2L2 rs746218 was longer than that in patients with GA or AA genotypes (Median PFS: 2.56 (0.04–11.85) years). Furthermore, OS was longer in patients with the GG genotype of TNFRSF1A rs4149570 than that in patients with the AA or AG genotypes. In the subgroup analysis, BRCA2 rs206118, XRCC3 rs1799794, NFKB1 rs230529, RAD52 rs7963551, NFKB1 rs1585215, NFKBIA rs2233406, and TNFRSF1A rs4149570 were associated with the PFS time. Patients younger than 56 years old with the TT or TC genotype of rs206118 had longer PFS times than those with the CC genotype. For XRCC3 rs1799794, the AA and AG genotypes were associated with shorter PFS times in patients >56 years old. For NFKB1 rs230529, the AA and AG genotypes were associated with longer PFS times in patients greater than 56 years old). For NFKB1 rs1585215, patients with squamous cell carcinoma (SCC) patients carrying the GG genotype had significantly shorter PFS times. The TT and TG genotypes of RAD52 rs7963551 were associated with significantly shorter PFS times compared with the GG genotype in male patients. For NFKBIA rs2233406, the CC and CT genotypes were associated with shorter PFS times than the TT genotype in patients with stage III/IV cancer. For TNFRSF1A rs4149570 in patients with stage III/IV cancer, the TT and TG genotypes were associated with longer PFS times than the GG genotype. In the subgroup analyses, TNFRSF1A rs4149570, XRCC3 rs1799794, XPC rs2228000, and FASN rs4246445 were significantly associated with OS. In patients older than 56 years old, the TT and TG genotypes of TNFRSF1A rs4149570 were associated with longer OS than that associated with the GG genotype. For TNFRSF1A rs4149570, the TT and TG genotypes were associated with longer OS in patients with stage III/IV cancer. For XRCC3 rs1799794, the GG genotype was associated with longer OS in patients with SCLC. Non-smokers with the GG or GA genotypes of XPC rs2228000 had shorter OS than those with the AA genotype. Non-smokers with the AA or AG genotypes of FASN rs4246445 had longer OS than those with the GG genotype.
Discussion
Platinum chemotherapy is one of the most important approaches for treatment of lung cancer. Platinum agents are typically used in combination with other antitumor drugs, but efficacy is limited due to resistance (Garufi et al., 2020; Yu et al., 2020). The DNA repair system contributes to platinum resistance, which influences the curative effects of chemotherapy and negatively impacts the clinical outcomes (Simon et al., 2007; Sullivan et al., 2014; Jiang et al., 2019; Peng et al., 2020). Polymorphism research has shown that gene polymorphisms affect prognosis, the folate metabolism pathway, drug transporters, and metabolic enzymes. The DNA repair system is essential for maintaining genome integrity and preventing genome instability-associated diseases, such as lung cancer (Chen et al., 2014; Anoushirvani et al., 2019; Zhao et al., 2020). Polymorphisms in DNA repair genes play a significant role in the ability to repair DNA damage. The relationship between repair gene polymorphisms and platinum chemoresistance has received a great deal of attention with regard to sensitivity of lung cancer to chemotherapy (Longhese et al., 2010; Li et al., 2018; Makovec, 2019; Schmid et al., 2020). In this study, DNA repair gene polymorphisms were studied to identify significant biomarkers for the prediction of platinum-based chemotherapy response.
In this study, we also investigated the correlations between 35 polymorphisms in 9 DNA repair genes (XRCC3, BRCA2/ZAR1L, XPC, RAD52, MAD2L2, NFKB1, NFKBIA, TNFRSF1A, and FASN) with platinum-based chemotherapy prognosis in 399 patients with lung cancer. A previous study showed that XRCC3, BRCA2, and RAD52 were involved in the HR-mediated DBS repair. XRCC3 is a RAD51 paralog in the HR-mediated DBS repair pathway that assists RAD51 with HR initiation (Brenneman et al., 2002). BRCA2 is a tumor suppressor gene critical to multiple cellular processes including DNA repair, the cell cycle, and apoptosis (Cleary et al., 2020). Mutation of BRCA2 was shown to promote tumor sensitivity towards PARP inhibitors (Farmer et al., 2005). In addition, RAD52 has been shown to play a major role in facilitating restart of damaged replication forks (Mortensen et al., 2009). XPC is the main DNA damage sensor in NER, and MAD2L2 is a controller of NHEJ-mediated DBS repair (Van Cuijk et al., 2015; Vassel et al., 2020). NFKB1, NFKBIA, TNFRSF1A, and FASN were found to be related to DNA repair, which affects tumor sensitivity to DNA-damaging agents (Chui et al., 2010; Bredel et al., 2011; Park et al., 2012; Jones and Infante, 2015). Gene variations in these genes have been reported to correlate to onset and progression of several types of tumors.
Our results showed that MAD2L2 rs746218 and TNFRSF1A rs4149570 may be biomarkers for predicting the prognosis of patients with lung cancer in response to platinum-based chemotherapy. The MAD2L2 gene, which is essential for DNA repair, localizes to uncapped telomeres and promotes the non-homologous end-joining (NHEJ)-mediated fusion of deprotected chromosome ends and genomic instability. In addition, MAD2L2 can control DNA breaks by inhibiting 5’ end resection (Tomida et al., 2015; Dai et al., 2020). The TNFRSF1A gene plays a crucial role in non-small cell lung cancer growth, invasion, and metastasis (Lee et al., 2010; Fujikawa et al., 2014; Hu et al., 2019). The MAD2L2 containing new shield complex protein plays a critical role in the choice between homologous recombination (HR) and non-homologous end-joining (NHEJ)-mediated repair. Upregulation of MAD2L2 (also known as MAD2B or REV7) decreases DNA end resection, which increases NHEJ and chromosomal abnormalities, resulting mitotic catastrophe in PARP inhibitor treated HR-proficient cells. In addition, MAD2L2 can also inhibit end-resection in irradiation (IR)-induced DNA double-strand breaks (DSBs) (Boersma et al., 2015; Simonetta et al., 2018; De Krijger et al., 2021). MAD2L2 accelerates end-joining of DNA double-strand breaks in several settings, including immunoglobulin class switch recombination through ATM kinase activity (Xu et al., 2015; Batenburg et al., 2017; Noordermeer et al., 2018). Previous studies showed that MAD2L2 promoted DNA repair activity through 53BP1 and promotes NHEJ by inhibiting 5′ end resection downstream of RIF1 protein (Ghezraoui et al., 2018; Liang et al., 2020). Both MAD2L2 Rs746218 and TNFRSF1A rs4149570 are upstream transcript variants, and might affect gene expression by interacting with promoters to influence gene transcription. Therefore, MAD2L2 rs746218 could influence the choice between HR and NHEJ by affecting the expression of MAD2L2. In addition, the TNFRSF1A gene was shown to play a crucial role in NSCLS growth, invasion, and metastasis (Lee et al., 2010; Fujikawa et al., 2014; Hu et al., 2019). However, the mechanism by which the TNFRSF1A gene affects prognosis associated with platinum-based chemotherapy has not been characterized. Future studies should characterize the mechanism by which MAD2L2 rs746218 participates in the double-strand break repair pathway and the mechanism by which the TNFRSF1A gene contributes to platinum chemoresistance. Characterization of these mechanisms may allow for the development of new drug candidates and more effective use of combination therapies including platinum-based drugs and DNA repair regulators.
Our study was subject to the following limitations. Our study was a single-center study, which limits the generalizability of the results. In addition, the small sample size resulted in a broad confidence interval for MAD2L2 rs746218, and more samples are needed to confirm the results. Potential mechanisms by which MAD2L2 rs746218 and TNFRSF1A rs4149570 impacted prognosis in patients with lung cancer who received platinum-based chemotherapy were determined using TCGA data (https://portal.gdc.cancer.gov/). This analysis showed that low expression of TNFRSF1A in LUAD (lung adenocarcinoma) was associated with significantly longer PFS and OS (Figure 4). However, the mechanisms of these effects require further investigation.
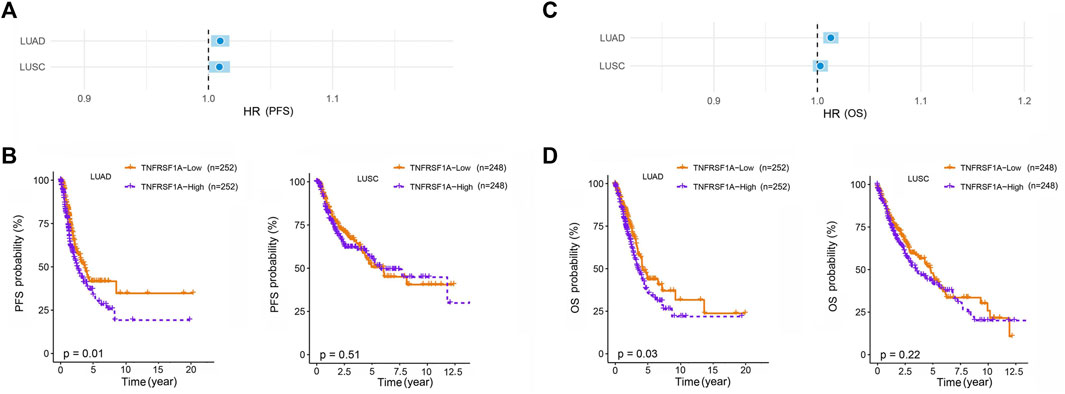
FIGURE 4. Association of the expression of TNFRSF1A with lung cancer prognosis in patients with LUAD (Lung adenocarcinoma) and LUSC (Lung squamous cell carcinoma). Low expression of TNFRSF1A in patients with LUAD was associated with significantly longer (A,B) progression-free survival (PFS) and overall survival (C,D) (OS).
In summary, our study showed that MAD2L2 rs746218 was significantly associated with platinum-based chemotherapy, and PFS and TNFRSF1A rs4149570 was significantly associated with OS time in patients with lung cancer treated with platinum-based chemotherapy. Polymorphisms of MAD2L2 rs746218 and TNFRSF1A rs4149570 polymorphisms may be biomarkers for predicting prognosis in patients with lung cancer treated with platinum-based chemotherapy.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Xiangya Hospital, Central South University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Study design was contributed by JC, J-YL, and L-MT. J-YL, TZ, JC, and ZW took the lead in data collection and data analysis, assisted by CL, H-XH, F-XD, M-RL, ML, and YW. Data interpretation was performed by J-YL, with assistance from the other authors. The manuscript was written primarily by J-YL and TZ, and revised by J-YY, Z-QL, L-MT, and JC. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Nature Science Foundation of China (81803640, 81603207) and Hunan Provincial Natural Science Foundation of China (2020JJ5885).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alam, H., Tang, M., Maitituoheti, M., Dhar, S. S., Kumar, M., Han, C. Y., et al. (2020). KMT2D Deficiency Impairs Super-enhancers to Confer a Glycolytic Vulnerability in Lung Cancer. Cancer Cell 37, 599. doi:10.1016/j.ccell.2020.03.005
Almohaini, M., Chalasani, S. L., Bafail, D., Akopiants, K., Zhou, T., Yannone, S. M., et al. (2016). Nonhomologous End Joining of Complex DNA Double-Strand Breaks with Proximal Thymine Glycol and Interplay with Base Excision Repair. DNA Repair (Amst) 41, 16–26. doi:10.1016/j.dnarep.2016.03.003
Andres, S. N., Schellenberg, M. J., Wallace, B. D., Tumbale, P., and Williams, R. S. (2015). Recognition and Repair of Chemically Heterogeneous Structures at DNA Ends. Environ. Mol. Mutagen 56, 1–21. doi:10.1002/em.21892
Anoushirvani, A. A., Aghabozorgi, R., Ahmadi, A., Arjomandzadegan, M., Khalili, S., Sahraei, M., et al. (2019). The Relationship between rs3212986C>A Polymorphism and Tumor Stage in Lung Cancer Patients. Cureus 11, e4423. doi:10.7759/cureus.4423
Arbour, K. C., and Riely, G. J. (2019). Systemic Therapy for Locally Advanced and Metastatic Non-small Cell Lung Cancer: A Review. JAMA 322, 764–774. doi:10.1001/jama.2019.11058
Batenburg, N. L., Walker, J. R., Noordermeer, S. M., Moatti, N., Durocher, D., and Zhu, X. D. (2017). ATM and CDK2 Control Chromatin Remodeler CSB to Inhibit RIF1 in DSB Repair Pathway Choice. Nat. Commun. 8, 1921. doi:10.1038/s41467-017-02114-x
Boersma, V., Moatti, N., Segura-Bayona, S., Peuscher, M. H., Van Der Torre, J., Wevers, B. A., et al. (2015). MAD2L2 Controls DNA Repair at Telomeres and DNA Breaks by Inhibiting 5' End Resection. Nature 521, 537–540. doi:10.1038/nature14216
Bredel, M., Scholtens, D. M., Yadav, A. K., Alvarez, A. A., Renfrow, J. J., Chandler, J. P., et al. (2011). NFKBIA Deletion in Glioblastomas. N. Engl. J. Med. 364, 627–637. doi:10.1056/NEJMoa1006312
Brenneman, M. A., Wagener, B. M., Miller, C. A., Allen, C., and Nickoloff, J. A. (2002). XRCC3 Controls the Fidelity of Homologous Recombination: Roles for XRCC3 in Late Stages of Recombination. Mol. Cell 10, 387–395. doi:10.1016/s1097-2765(02)00595-6
Cescon, D. W., She, D., Sakashita, S., Zhu, C. Q., Pintilie, M., Shepherd, F. A., et al. (2015). NRF2 Pathway Activation and Adjuvant Chemotherapy Benefit in Lung Squamous Cell Carcinoma. Clin. Cancer Res. 21, 2499–2505. doi:10.1158/1078-0432.CCR-14-2206
Chen, J., Wu, L., Wang, Y., Yin, J., Li, X., Wang, Z., et al. (2016). Effect of Transporter and DNA Repair Gene Polymorphisms to Lung Cancer Chemotherapy Toxicity. Tumour Biol. 37, 2275–2284. doi:10.1007/s13277-015-4048-0
Chen, J., Yin, J., Li, X., Wang, Y., Zheng, Y., Qian, C., et al. (2014). WISP1 Polymorphisms Contribute to Platinum-Based Chemotherapy Toxicity in Lung Cancer Patients. Int. J. Mol. Sci. 15, 21011–21027. doi:10.3390/ijms151121011
Chui, Y. L., Ching, A. K., Chen, S., Yip, F. P., Rowlands, D. K., James, A. E., et al. (2010). BRE Over-expression Promotes Growth of Hepatocellular Carcinoma. Biochem. Biophys. Res. Commun. 391, 1522–1525. doi:10.1016/j.bbrc.2009.12.111
Cleary, J. M., Aguirre, A. J., Shapiro, G. I., and D'andrea, A. D. (2020). Biomarker-Guided Development of DNA Repair Inhibitors. Mol. Cell 78, 1070–1085. doi:10.1016/j.molcel.2020.04.035
Dai, Y., Zhang, F., Wang, L., Shan, S., Gong, Z., and Zhou, Z. (2020). Structural Basis for Shieldin Complex Subunit 3-mediated Recruitment of the Checkpoint Protein REV7 during DNA Double-Strand Break Repair. J. Biol. Chem. 295, 250–262. doi:10.1074/jbc.RA119.011464
De Krijger, I., Föhr, B., Pérez, S. H., Vincendeau, E., Serrat, J., Thouin, A. M., et al. (2021). MAD2L2 Dimerization and TRIP13 Control Shieldin Activity in DNA Repair. Nat. Commun. 12, 5421. doi:10.1038/s41467-021-25724-y
Ettinger, D. S., Wood, D. E., Aggarwal, C., Aisner, D. L., Akerley, W., Bauman, J. R., et al. (2019). NCCN Guidelines Insights: Non-small Cell Lung Cancer, Version 1.2020. J. Natl. Compr. Canc Netw. 17, 1464–1472. doi:10.6004/jnccn.2019.0059
Farmer, H., Mccabe, N., Lord, C. J., Tutt, A. N., Johnson, D. A., Richardson, T. B., et al. (2005). Targeting the DNA Repair Defect in BRCA Mutant Cells as a Therapeutic Strategy. Nature 434, 917–921. doi:10.1038/nature03445
Fujikawa, K., Migita, K., Shigemitsu, Y., Umeda, M., Nonaka, F., Tamai, M., et al. (2014). MEFV Gene Polymorphisms and TNFRSF1A Mutation in Patients with Inflammatory Myopathy with Abundant Macrophages. Clin. Exp. Immunol. 178, 224–228. doi:10.1111/cei.12407
Garufi, G., Palazzo, A., Paris, I., Orlandi, A., Cassano, A., Tortora, G., et al. (2020). Neoadjuvant Therapy for Triple-Negative Breast Cancer: Potential Predictive Biomarkers of Activity and Efficacy of Platinum Chemotherapy, PARP- and Immune-Checkpoint-Inhibitors. Expert Opin. Pharmacother. 21, 687–699. doi:10.1080/14656566.2020.1724957
Ghezraoui, H., Oliveira, C., Becker, J. R., Bilham, K., Moralli, D., Anzilotti, C., et al. (2018). 53BP1 Cooperation with the REV7-Shieldin Complex Underpins DNA Structure-specific NHEJ. Nature 560, 122–127. doi:10.1038/s41586-018-0362-1
Gupta, R., Somyajit, K., Narita, T., Maskey, E., Stanlie, A., Kremer, M., et al. (2018). DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 173, 972. doi:10.1016/j.cell.2018.03.050
Hirsch, F. R., Scagliotti, G. V., Mulshine, J. L., Kwon, R., Curran, W. J., Wu, Y. L., et al. (2017). Lung Cancer: Current Therapies and New Targeted Treatments. Lancet 389, 299–311. doi:10.1016/S0140-6736(16)30958-8
Hu, Z., Li, H., Xie, R., Wang, S., Yin, Z., and Liu, Y. (2019). Genomic Variant in Porcine TNFRSF1A Gene and its Effects on TNF Signaling Pathway In Vitro. Gene 700, 105–109. doi:10.1016/j.gene.2019.03.046
Huang, J., and Dynan, W. S. (2002). Reconstitution of the Mammalian DNA Double-Strand Break End-Joining Reaction Reveals a Requirement for an Mre11/Rad50/NBS1-Containing Fraction. Nucleic Acids Res. 30, 667–674. doi:10.1093/nar/30.3.667
Jiang, M., Jia, K., Wang, L., Li, W., Chen, B., Liu, Y., et al. (2021). Alterations of DNA Damage Response Pathway: Biomarker and Therapeutic Strategy for Cancer Immunotherapy. Acta Pharm. Sin. B 11, 2983–2994. doi:10.1016/j.apsb.2021.01.003
Jiang, Y., Dai, H., Li, Y., Yin, J., Guo, S., Lin, S. Y., et al. (2019). PARP Inhibitors Synergize with Gemcitabine by Potentiating DNA Damage in Non-small-cell Lung Cancer. Int. J. Cancer 144, 1092–1103. doi:10.1002/ijc.31770
Jones, S. F., and Infante, J. R. (2015). Molecular Pathways: Fatty Acid Synthase. Clin. Cancer Res. 21, 5434–5438. doi:10.1158/1078-0432.CCR-15-0126
Kalemkerian, G. P., Loo, B. W., Akerley, W., Attia, A., Bassetti, M., Boumber, Y., et al. (2018). NCCN Guidelines Insights: Small Cell Lung Cancer, Version 2.2018. J. Natl. Compr. Canc Netw. 16, 1171–1182. doi:10.6004/jnccn.2018.0079
Kryczka, J., Kryczka, J., Czarnecka-Chrebelska, K. H., and Brzeziańska-Lasota, E. (2021). Molecular Mechanisms of Chemoresistance Induced by Cisplatin in NSCLC Cancer Therapy. Int. J. Mol. Sci. 22, 8885. doi:10.3390/ijms22168885
Kulkarni, S., Vella, E. T., Coakley, N., Cheng, S., Gregg, R., Ung, Y. C., et al. (2016). The Use of Systemic Treatment in the Maintenance of Patients with Non-small Cell Lung Cancer: A Systematic Review. J. Thorac. Oncol. 11, 989–1002. doi:10.1016/j.jtho.2016.03.007
Lee, E. B., Jeon, H. S., Yoo, S. S., Choi, Y. Y., Kang, H. G., Cho, S., et al. (2010). Polymorphisms in Apoptosis-Related Genes and Survival of Patients with Early-Stage Non-small-cell Lung Cancer. Ann. Surg. Oncol. 17, 2608–2618. doi:10.1245/s10434-010-1082-4
Li, Y. Q., Chen, J., Yin, J. Y., Liu, Z. Q., and Li, X. P. (2018). Gene Expression and Single Nucleotide Polymorphism of ATP7B Are Associated with Platinum-Based Chemotherapy Response in Non-small Cell Lung Cancer Patients. J. Cancer 9, 3532–3539. doi:10.7150/jca.26286
Liang, L., Feng, J., Zuo, P., Yang, J., Lu, Y., and Yin, Y. (2020). Molecular Basis for Assembly of the Shieldin Complex and its Implications for NHEJ. Nat. Commun. 11, 1972. doi:10.1038/s41467-020-15879-5
Longhese, M. P., Bonetti, D., Manfrini, N., and Clerici, M. (2010). Mechanisms and Regulation of DNA End Resection. EMBO J. 29, 2864–2874. doi:10.1038/emboj.2010.165
Low, H. B., Wong, Z. L., Wu, B., Kong, L. R., Png, C. W., Cho, Y. L., et al. (2021). DUSP16 Promotes Cancer Chemoresistance through Regulation of Mitochondria-Mediated Cell Death. Nat. Commun. 12, 2284. doi:10.1038/s41467-021-22638-7
Makovec, T. (2019). Cisplatin and beyond: Molecular Mechanisms of Action and Drug Resistance Development in Cancer Chemotherapy. Radiol. Oncol. 53, 148–158. doi:10.2478/raon-2019-0018
Menon, V., and Povirk, L. F. (2017). XLF/Cernunnos: An Important but Puzzling Participant in the Nonhomologous End Joining DNA Repair Pathway. DNA Repair (Amst) 58, 29–37. doi:10.1016/j.dnarep.2017.08.003
Mortensen, U. H., Lisby, M., and Rothstein, R. (2009). Rad52. Curr. Biol. 19, R676–R677. doi:10.1016/j.cub.2009.06.001
Noordermeer, S. M., Adam, S., Setiaputra, D., Barazas, M., Pettitt, S. J., Ling, A. K., et al. (2018). The Shieldin Complex Mediates 53BP1-dependent DNA Repair. Nature 560, 117–121. doi:10.1038/s41586-018-0340-7
Park, Y. Y., Jung, S. Y., Jennings, N. B., Rodriguez-Aguayo, C., Peng, G., Lee, S. R., et al. (2012). FOXM1 Mediates Dox Resistance in Breast Cancer by Enhancing DNA Repair. Carcinogenesis 33, 1843–1853. doi:10.1093/carcin/bgs167
Parkin, D. M., Pisani, P., and Ferlay, J. (1999). Global Cancer Statistics. CA Cancer J. Clin. 49, 3331. doi:10.3322/canjclin.49.1.33
Peng, S., Sun, Y., Luo, Y., Ma, S., Sun, W., Tang, G., et al. (2020). MFP-FePt-GO Nanocomposites Promote Radiosensitivity of Non-small Cell Lung Cancer via Activating Mitochondrial-Mediated Apoptosis and Impairing DNA Damage Repair. Int. J. Biol. Sci. 16, 2145–2158. doi:10.7150/ijbs.46194
Reid, D. A., Conlin, M. P., Yin, Y., Chang, H. H., Watanabe, G., Lieber, M. R., et al. (2017). Bridging of Double-Stranded Breaks by the Nonhomologous End-Joining Ligation Complex Is Modulated by DNA End Chemistry. Nucleic Acids Res. 45, 1872–1878. doi:10.1093/nar/gkw1221
Rottenberg, S., Disler, C., and Perego, P. (2021). The Rediscovery of Platinum-Based Cancer Therapy. Nat. Rev. Cancer 21, 37–50. doi:10.1038/s41568-020-00308-y
Schmid, S., Omlin, A., Higano, C., Sweeney, C., Martinez Chanza, N., Mehra, N., et al. (2020). Activity of Platinum-Based Chemotherapy in Patients with Advanced Prostate Cancer with and without DNA Repair Gene Aberrations. JAMA Netw. Open 3, e2021692. doi:10.1001/jamanetworkopen.2020.21692
Schwartz, A. G., and Cote, M. L. (2016). Epidemiology of Lung Cancer. Adv. Exp. Med. Biol. 893, 21–41. doi:10.1007/978-3-319-24223-1_2
Shi, Y., and Sun, Y. (2015). Medical Management of Lung Cancer: Experience in China. Thorac. Cancer 6, 10–16. doi:10.1111/1759-7714.12168
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer Statistics, 2021. CA A Cancer J. Clin. 71, 7–33. doi:10.3322/caac.21654
Simon, G. R., Ismail-Khan, R., and Bepler, G. (2007). Nuclear Excision Repair-Based Personalized Therapy for Non-small Cell Lung Cancer: from Hypothesis to Reality. Int. J. Biochem. Cell Biol. 39, 1318–1328. doi:10.1016/j.biocel.2007.05.006
Simonetta, M., De Krijger, I., Serrat, J., Moatti, N., Fortunato, D., Hoekman, L., et al. (2018). H4K20me2 Distinguishes Pre-replicative from Post-replicative Chromatin to Appropriately Direct DNA Repair Pathway Choice by 53BP1-RIF1-Mad2l2. Cell Cycle 17, 124–136. doi:10.1080/15384101.2017.1404210
Sullivan, I., Salazar, J., Majem, M., Pallarés, C., Del Río, E., Páez, D., et al. (2014). Pharmacogenetics of the DNA Repair Pathways in Advanced Non-small Cell Lung Cancer Patients Treated with Platinum-Based Chemotherapy. Cancer Lett. 353, 160–166. doi:10.1016/j.canlet.2014.07.023
Thai, A. A., Solomon, B. J., Sequist, L. V., Gainor, J. F., and Heist, R. S. (2021). Lung Cancer. Lancet 398, 535–554. doi:10.1016/S0140-6736(21)00312-3
Tomida, J., Takata, K., Lange, S. S., Schibler, A. C., Yousefzadeh, M. J., Bhetawal, S., et al. (2015). REV7 Is Essential for DNA Damage Tolerance via Two REV3L Binding Sites in Mammalian DNA Polymerase ζ. Nucleic Acids Res. 43, 1000–1011. doi:10.1093/nar/gku1385
Van Cuijk, L., Van Belle, G. J., Turkyilmaz, Y., Poulsen, S. L., Janssens, R. C., Theil, A. F., et al. (2015). SUMO and Ubiquitin-dependent XPC Exchange Drives Nucleotide Excision Repair. Nat. Commun. 6, 7499. doi:10.1038/ncomms8499
Vassel, F. M., Bian, K., Walker, G. C., and Hemann, M. T. (2020). Rev7 Loss Alters Cisplatin Response and Increases Drug Efficacy in Chemotherapy-Resistant Lung Cancer. Proc. Natl. Acad. Sci. U. S. A. 117, 28922–28924. doi:10.1073/pnas.2016067117
Xing, M., Yang, M., Huo, W., Feng, F., Wei, L., Jiang, W., et al. (2015). Interactome Analysis Identifies a New Paralogue of XRCC4 in Non-homologous End Joining DNA Repair Pathway. Nat. Commun. 6, 6233. doi:10.1038/ncomms7233
Xu, G., Chapman, J. R., Brandsma, I., Yuan, J., Mistrik, M., Bouwman, P., et al. (2015). REV7 Counteracts DNA Double-Strand Break Resection and Affects PARP Inhibition. Nature 521, 541–544. doi:10.1038/nature14328
Xu, Y., and Xu, D. (2020). Repair Pathway Choice for Double-Strand Breaks. Essays Biochem. 64, 765–777. doi:10.1042/EBC20200007
Yin, J. Y., Li, X., Zhou, H. H., and Liu, Z. Q. (2016). Pharmacogenomics of Platinum-Based Chemotherapy Sensitivity in NSCLC: toward Precision Medicine. Pharmacogenomics 17, 1365–1378. doi:10.2217/pgs-2016-0074
Yu, C., Wang, Z., Sun, Z., Zhang, L., Zhang, W., Xu, Y., et al. (2020). Platinum-Based Combination Therapy: Molecular Rationale, Current Clinical Uses, and Future Perspectives. J. Med. Chem. 63, 13397–13412. doi:10.1021/acs.jmedchem.0c00950
Zhao, L., Bao, C., Shang, Y., He, X., Ma, C., Lei, X., et al. (2020). The Determinant of DNA Repair Pathway Choices in Ionising Radiation-Induced DNA Double-Strand Breaks. Biomed. Res. Int. 2020, 4834965. doi:10.1155/2020/4834965
Keywords: lung cancer, platinum-based chemotherapy, prognosis, genetic polymorphisms, MAD2L2, TNFRSF1A
Citation: Liu J-Y, Zou T, Yin J-Y, Wang Z, Liu C, Huang H-X, Ding F-X, Lei M-R, Wang Y, Liu M, Liu Z-Q, Tan L-M and Chen J (2022) Genetic Variants in Double-Strand Break Repair Pathway Genes to Predict Platinum-Based Chemotherapy Prognosis in Patients With Lung Cancer. Front. Pharmacol. 13:915822. doi: 10.3389/fphar.2022.915822
Received: 08 April 2022; Accepted: 30 May 2022;
Published: 11 July 2022.
Edited by:
Zeming Liu, Huazhong University of Science and Technology, ChinaReviewed by:
Jun-Li Luo, The Scripps Research Institute, United StatesJessy Abraham, All India Institute of Medical Sciences Raipur, India
Copyright © 2022 Liu, Zou, Yin, Wang, Liu, Huang, Ding, Lei, Wang, Liu, Liu, Tan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Ming Tan, bGltaW5ndGFuQDEyNi5jb20=; Juan Chen, Y2oxMDI4QGNzdS5lZHUuY24=
 Jun-Yan Liu
Jun-Yan Liu Ting Zou2
Ting Zou2 Ji-Ye Yin
Ji-Ye Yin Zhan Wang
Zhan Wang Chong Liu
Chong Liu Ying Wang
Ying Wang Zhao-Qian Liu
Zhao-Qian Liu Juan Chen
Juan Chen