
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 15 June 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.912303
Leukemia is a group of life-threatening hematological malignancies which is currently incurable and often accompanied by drug resistance or disease relapse. Understanding the pathogenesis of leukemia and finding specific therapeutic targets and biomarkers is of great importance to improve the clinical efficacy of leukemia. Exosome-derived ncRNAs have been demonstrated as critical components of intercellular communication and function as key facilitators in the leukemia biological process. This review outlines the current investigations of exosomal ncRNAs (including miRNA, circRNA, and lncRNA) as important mediators of leukemia and potential therapeutic targets and biomarkers for leukemia treatment. Moreover, we generally analyze the prospects and challenges for exosomal ncRNAs from the aspects of research and clinical application.
Leukemia is a group of hematological malignancies characterized by the abnormal proliferation of hematopoietic cells that infiltrate the bone marrow and invade the blood and other extramedullary tissues (Bhat et al., 2020). Over 250,000 people worldwide are diagnosed with leukemia each year, accounting for 2.5% of all cancers (Rodriguez-Abreu et al., 2007). The four major subtypes of leukemia are acute lymphocytic leukemia (ALL), chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), and chronic myeloid leukemia (CML), with different clinical features and prognoses (Rodriguez-Abreu et al., 2007) (Table 1). The fundamental form of treatment for leukemia is chemotherapy. But leukemia is incurable currently and patients often develop drug resistance or disease relapse (Mardani et al., 2019). In addition, the non-specific toxicity of traditional chemotherapeutic drugs to normal blood cells can cause serious hematological adverse reactions and affect the efficacy of the drugs. Therefore, demonstrating the pathogenesis of leukemia and finding specific therapeutic targets and biomarkers is of great importance to improve the clinical efficacy of leukemia patients.
Exosomes are intraluminal vesicles (ILV) of multivesicular bodies (MVB) and are typically 30–150 nm in diameter (Mashouri et al., 2019). A plethora of evidence indicated that exosomes could mediate tumor progression through participating in intercellular communication within the tumor microenvironment (TME) (Zhou et al., 2018; Mashouri et al., 2019). Exosomes can be secreted from cells by fusion of MVBs with the plasma membrane and internalized into recipient cells via endocytosis to release the encapsulated “cargo”, as well as regulated the biological processes (Figure 1). The composition of exosome cargo is heterogeneous and dynamic, which can reflect the unique profile of the constituents and more importantly, the physiological and pathological condition of their original cells or tissues. In addition to the proteins, nucleic acids, and lipids that have been detected in exosomes, non-coding RNAs (ncRNAs), a type of transcripts that are not translated into proteins, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), can also be encapsulated in exosomes and transmitted among cells. Among them, miRNAs are a type of small ncRNA of approximately 22 nucleotides in length with 5′-phosphate and 3′-hydroxyl ends (Slack and Chinnaiyan, 2019; Bhat et al., 2020). CircRNAs are single-stranded covalently closed circular transcripts without 5′ caps and 3′ tails (Qu et al., 2018). LncRNAs are linear transcripts with lengths exceeding 200 nucleotides (Bhat et al., 2020). It had been reported that ncRNAs encapsulated in exosomes were involved in various pathological processes of tumors, such as the cancerous transformation of stromal cells in the tumor microenvironment (Noh et al., 2020), drug resistance transmission among cells (Zhang Z. et al., 2020), proliferation, apoptosis, angiogenesis, and metastasis (Zheng et al., 2018; Xie et al., 2020) (Figure 2). These functions make exosomal ncRNAs become an important source for the discovery of new tumor therapeutic targets.
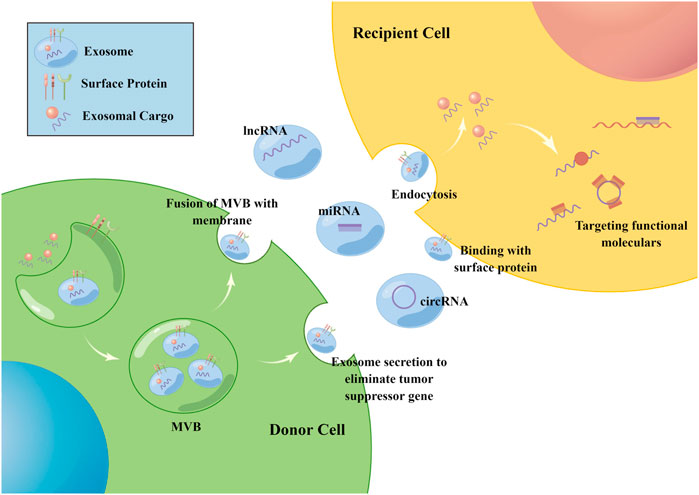
FIGURE 1. The biogenesis of exosomes. Exosomes are intraluminal vesicles of MVB and can be secreted from cells by fusion of MVBs with the plasma membrane, then internalized into recipient cells via endocytosis or the binding of surface proteins to release the encapsulated “cargo”. Non-coding RNAs including miRNA, circRNA and lncRNA can be encapsulated in exosomes and transmitted among cells. Once inside the recipient cells, ncRNAs can exert their function through targeting RNAs and proteins. The figure was drawn by Figdraw (www.figdraw.com).
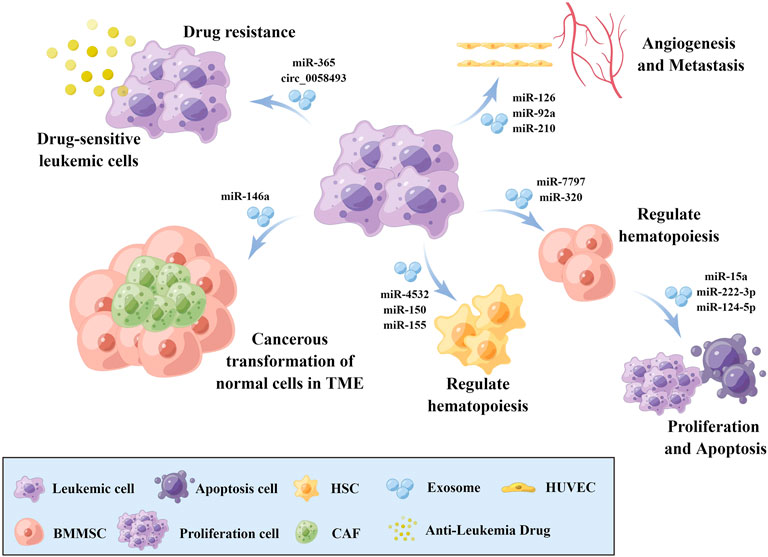
FIGURE 2. The role of exosomal ncRNAs in leukemia biology. Exosomal ncRNAs are involved in the cancerous transformation of stromal cells in the tumor microenvironment, drug resistance transmission, proliferation, apoptosis, angiogenesis, and metastasis via transmission among cells. BMMSCs: Bone marrow mesenchymal stem cell, HSCs: Hematopoietic stem cells, CAFs: Cancer-associated fibroblasts, HUVECs: Human umbilical vein endothelial cells, TME: Tumor microenvironment. The figure was drawn by Figdraw (www.figdraw.com).
Exosomes have been found in almost all the body fluids, such as plasma, serum, urine, semen, saliva, bronchial fluid, cerebral spinal fluid (CSF), breast milk, amniotic fluid, synovial fluid, tears, lymph, bile, and gastric acid (Guo et al., 2017; Doyle and Wang, 2019; Mashouri et al., 2019). Its detection in different body fluids is also evidence of its stability in a variety of adverse environments (Zhou et al., 2018). More importantly, exosomes can protect ncRNAs from ribonuclease degradation in blood circulation (Zhang et al., 2019). Furthermore, as exosomal ncRNAs can be stably detected in plasma or serum, they are regarded as a type of non-invasive liquid biopsy that may be more acceptable in clinical application. Besides, it has been confirmed in many types of cancer that the concentration of exosomes is much higher in the blood of cancer patients compared with healthy individuals, which can improve the detection sensitivity (Smolarz and Widlak, 2021). These characteristics highlight the potential of exosomal ncRNAs as both biomarkers and treatment vectors in tumor therapy.
The transfer of exosomal ncRNAs between cells and subsequent functional effects have been extensively demonstrated in numerous solid tumor studies, but their functions in leukemia have not been well explored. This review outlines the current understanding of exosomal ncRNAs expression patterns in different types of leukemia, the mechanisms that contribute to leukemia carcinogenesis, and their potential role in the clinical application (Table 2). Revealing the regulatory patterns of exosomal ncRNAs in leukemia can lead to the identification of novel therapeutic targets and biomarkers, ultimately opening new prospects for treatment, diagnosis, and prognostication of leukemia.
The most extensively studied class of ncRNA in exosomes to date is miRNA. MicroRNAs are short ncRNAs of approximately 22 nucleotides in length that regulate the expression of other RNAs, notably mRNAs, through binding between the 5’ end of the miRNA with complementary sequences in target RNAs (Slack and Chinnaiyan, 2019). Exosomal miRNAs have been implicated in a host of the biological process of cancer cells (Nicoloso et al., 2009) and have been reported to be involved in tumor cell proliferation, apoptosis, invasion, migration, angiogenesis, the transformation of the tumor microenvironment, and the occurrence and transfer of drug resistance. Exosomal miRNAs are expected to become a new type of therapeutic target and biomarker.
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, with a 5-year survival rate of 24% (Zhang F. et al., 2020). 30% of newly diagnosed patients cannot achieve complete remission, and about 50% of patients relapse after intensive chemotherapy (Thol et al., 2015). The main treatments for AML are allogeneic stem cell transplantation and intensive chemotherapy at present. However, the prognosis of AML patients, especially elderly patients, is poor (Dombret and Gardin, 2016; Shallis et al., 2019). Therefore, it is urgently needed to identify effective and reliable biomarkers and therapeutic targets for improving the prognosis of AML (Fang et al., 2020). Additionally, as the diagnosis of AML is often demonstrated by an increased number of myeloblasts in the bone marrow using marrow aspirate, the utilization of non-invasive tools for the diagnosis and management of AML will greatly reduce patients’ pain and the worldwide health burden (Fayyad-Kazan et al., 2013).
Bone marrow mesenchymal stem cell (BMMSC) plays a crucial role in regulating bone marrow hematopoietic function under physiological conditions and is considered to be a keystone of the hematopoietic stem cell niche (Zhang F. et al., 2020). Hematopoietic insufficiency is a hallmark of AML and MSCs from AML patients exhibit significant growth deficiency and impaired osteogenic differentiation capacity (Zhang F. et al., 2020). Yoshida et al. (2019) found that miR-7797 was significantly overexpressed in exosomes produced by AML cells, and could be transmitted into BMMSCs through exosomes, thereby regulating the Hippo-YAP signaling pathway, inhibiting the normal hematopoietic function of BMMSCs and promoting the development of AML (Horiguchi et al., 2016). Conversely, BMMSCs can also deliver ncRNAs through exosomes to regulate AML cell function. Zhang F. et al. (2020) found that miR-222-3p was significantly enriched in human BMMSCs exosomes which could be transmitted into AML cells, regulated the IRF2/INPP4B signaling pathway, inhibited the proliferation, and promoted the apoptosis of AML cells. Xu Y. C. et al. (2020) found that miR-124-5p mediated the inhibitory effect of BMMSCs exosomes on the cell cycle progression, AML cells proliferation, and apoptosis promotion. These findings indicate that exploring the BMMSCs-associated process may lead to the discovery of new therapeutic targets for AML.
AML is a genetically heterogeneous clonal disorder characterized by the accumulation of acquired genetic alterations in hematopoietic progenitor cells that alter normal mechanisms of self-renewal, proliferation, and differentiation, all of which are implicated in the process of hematopoiesis (Marcucci et al., 2011). Zhao et al. (2019) found that miR-4532 was significantly overexpressed in AML cell lines compared with CD34+ hematopoietic stem cells (HSCs) and enriched in AML cell exosomes. Exosomal miR-4532 could be transferred into HSCs and repress normal HSC hematopoiesis via activation of the LDOC1-dependent STAT3 signaling pathway. Hornick et al. (2016) found that exosomes derived from AML cells could destroy hematopoietic function through transferring miR-150 and miR-155 to hematopoietic stem cells and promote AML development. These findings indicate that AML cells can disturb normal hematopoietic function through exosome-derived miRNAs.
Emerging evidence has proved that AML originates from a small subpopulation of leukemic stem cells (LSCs) (Lapidot et al., 1994; Bonnet and Dick, 1997; Blair et al., 1998). LSCs are more resistant to conventional chemotherapy than ordinary leukemia cells and have the potential of self-renewal and initiating leukemia (Bonnet and Dick, 1997). Therefore, finding strategies to specifically eliminate LSCs may be necessary for solving AML recurrence or drug resistance (Peng et al., 2018). Hu et al. (2020) found that miR-34a was elevated in exosomes derived from LSCs. miR-34a could impede the secretion of exosomes which was promoted by RAB27B through inhibiting the expression of RAB27B. This process could increase the miR-34a level of LSCs in which miR-34a could inhibit HDAC2 expression and regulated the JAK1/STAT2/p53 pathway to promote the clearance of LSCs. However, LSCs took away the tumor-suppressive miR-34a by secreting exosomes, leading to the decrease of miR-34a in LSCs and the abnormal proliferation of LSCs (Hu et al., 2020). Similarly, miR-34c-5p has also been observed to promote LSCs clearance whereas RAB27B-mediated extracellular transfer of miR-34c-5p through exosomes partly promoted the miR-34c-5p decrease in LSCs, thus promoting the growth of LSCs (Peng et al., 2018). These studies suggest that inhibiting the extracellular transfer of tumor-suppressive miRNAs mediated by exosomes may be utilized to improve clinical response in future AML treatment.
Exosomal miRNAs also have a strong potential for clinical application as novel biomarkers for leukemia treatment. Upregulation of miR-125b in plasma exosomes was reported by Jiang and colleagues and was an independent prognostic factor for patients with intermediate-risk AML that indicated a higher risk of relapse and overall death (p < 0.001 for both) (Jiang et al., 2018). Lin et al. (2020) found that a high level of miR-532 in plasma exosomes of AML patients was positively correlated with overall survival (OS) (p < 0.05). Through screening and verification, Barrera-Ramirez et al. described upregulation of miR-101-3p and downregulation of miR-23b-5p, miR-339-3p, and miR-425-5p (p < 0.05) in MSCs exosomes of AML patients, compared to healthy donors (Barrera-Ramirez et al., 2017). The dysregulation of exosomal miRNAs and their association with AML prognosis demonstrate their potential as convenient, noninvasive biomarkers for future therapies.
Chronic myeloid leukemia (CML) is a clonal hemopoietic stem cell disorder characterized by a reciprocal translocation between the long arms of chromosomes 9 (ch9) and 22 (ch22) (Apperley, 2015). CML affects about 1-2/100,000 population per year, and accounts for 15% of all new cases of leukemia (Apperley, 2015). As the first-line drugs for CML treatment, tyrosine kinase inhibitors (TKIs) such as imatinib, dasatinib, and nilotinib have significantly improved the survival rate of CML patients. However, real-world research showed that within the first 3 years after diagnosis, 44% of CML patients on first-line treatment discontinued their first-line treatment, of which 21% were due to TKI intolerance and 19% were due to treatment failure (Geelen et al., 2017). Resistance to TKIs is one of the main causes of poor efficacy in CML patients, and the mechanism is still unclear.
Reciprocal interactions between leukemic cells and BMMSC remodel the normal niche into a malignant niche, leading to leukemia progression (Gao et al., 2019). The study of Gao and colleagues demonstrated that HNRNPA1 (a type of RNA-binding proteins) -mediated exosomal transfer of miR-320 from leukemia cells to BMMSC is an important process of leukemia progression and could be a potential therapeutic target for CML. As a tumor-suppressive miRNA, miR-320 was found to be upregulated in exosomes and secreted by leukemic cells, resulting in increased proliferation of the donor cells. Interestingly, the secreted exosomes were significantly endocytosed by adjacent BMMSC and inhibited osteogenesis partially via β-catenin inhibition, further promoting tumorigenesis. Besides, blast crisis CML (CML-BC) patients had a much higher miR-320 expression in plasma exosomes, compared to chronic phase CML (CML-CP) patients, indicating its clinical relevance (Gao et al., 2019). The study of Jiang et al. (2020) revealed the elevation of miR-711 in exosomes derived from K562 cells compared to parental cells exosomes. K562 cell-derived exosomal miR-711 could be transferred into BMMSCs and weaken the adhesive abilities (Jiang et al., 2020). miR-15a was shuttled by human BMMSC-exosomes which might be associated with the proliferation of CML cells (Zhang X. et al., 2020).
It has been reported that the interaction between leukemic cells and endothelial cells is associated with leukemia progression. Taverna et al. (2014) found that miR-126 was overexpressed in LAMA84 exosomes compared to parental cells and shuttled into human umbilical vein endothelial cells (HUVECs). miR-126 could change the bone marrow microenvironment via inhibiting the expression of chemokine CXCL12 and adhesion molecule VCAM1 in HUVECs, resulting in decreased motility and adhesion of LAMA84 cells, potentially facilitating disease progression (Taverna et al., 2014). The study of Umezu et al. (2013) showed that exosomes derived from K562 cells could transfer miR-92a into HUVECs and promote proliferation and angiogenesis. Tadokoro et al. (2013) found K562 exosomes could transfer miR-210 into HUVECs and promote angiogenesis. These data indicate the intercellular communication among exosomal miRNAs and endothelial cells demonstrates a possibility as a future therapeutic target due to its effect on the tumor microenvironment.
Exosomal miRNAs have also been implicated in the drug resistance of CML. The study of Min et al. (2018) demonstrated that miR-365 was significantly upregulated in exosomes derived from CML IM-resistant cells compared to those from sensitive cells. The subsequent results demonstrated that IM-resistant cells could transfer exosomal miR-365 into sensitive cells and inhibited the expression of BAX and Cleaved caspase-3, inducing drug resistance in sensitive cells (Min et al., 2018). This suggests CML cells may utilize exosome-derived miRNAs to improve chemoresistance.
The use of TKIs makes the life span of CML patients close to healthy people, but lifelong medication may cause unacceptable side effects such as bone marrow suppression or great economic burden. Therefore, TKIs discontinuation has become a research hotspot of CML in recent years. The study of Ohyashiki et al. (2016) demonstrated exosomal miR-215 was downregulated in CML patients who successfully discontinued IM, indicating the obvious potential of miR-215 in plasma exosomes as a prognostic marker for IM discontinuation.
In addition, cancer-associated cachexia (CAC) has a significant influence on the treatment tolerance and life quality, thus affecting the treatment response of CML patients. miR-92a-3p was found highly expressed in CML-derived exosomes which could be taken up by adipose tissue and suppressed the adipogenic ability of adipose-derived mesenchymal stem cells (ADSCs), resulting in reduced body fat and weight in mice (Wan et al., 2019).
Chronic lymphocytic leukemia (CLL) is a malignancy of CD5+ B cells and characterized by the accumulation of small, mature-appearing lymphocytes in the bone marrow, blood, and lymphoid tissues (Kipps et al., 2017). CLL is the most common type of leukemia in western countries with a highly variable clinical course and is currently incurable (Dubovsky et al., 2013; Hallek, 2019). It is urgent to clarify the pathogenesis of CLL and find effective therapeutic targets for CLL.
Cancer-associated fibroblasts (CAFs) are one of the most crucial components of the tumor microenvironment and may derive from fibroblasts, epithelial cells, and BMMSCs (Xing et al., 2010). The intercellular transmission of miRNA through exosomes is an important factor in CAFs formation. Paggetti and colleagues found that exosomes derived from CLL cells could transfer miR-150 and miR-146a into BMMSCs. They also found CLL cell exosomes could induce the transformation of BMMSCs into CAFs, but whether it was mediated by miR-150 and miR-146a was not clearly stated (Paggetti et al., 2015). Subsequently, the study of Yang et al. demonstrated CLL cells exosomes could transmit miR-146a into BMMSCs and promote the CAFs transformation by inhibiting the expression of tumor suppressor protein USP16 (Yang et al., 2020). Their involvement in CAFs transformation hints at the possibility of using exosomal miRNAs as future targets for CLL treatment.
The intercellular communication among nurturing cells and leukemia cells through exosome transmission is also an essential factor for the formation of the tumor microenvironment. Farahani et al. (2015) found that miR-202-3p was enriched in CLL exosomes and could be transmitted into human stromal cells (HS-5), altering the transcriptome in the recipient cells and enhancing their proliferation. More importantly, the expression of miR-202-3p in exosomes derived from CLL patients’ plasma showed a higher level compared to healthy donors, indicating the potential of plasma exosomal miR-202-3p as a diagnostic biomarker in CLL treatment (Farahani et al., 2015).
CLL is characterized by immune defects that prevent an efficient anti-tumor response. The increase of myeloid-derived suppressor cells (MDSCs) is an important factor of CLL immunodeficiency. The study of Bruns et al. (2017) revealed that the transmission of miR-155 through CLL exosomes could induce the increase of MDSCs and this immune regulatory interplay could be disrupted by vitamin D that negatively regulated miR-155 expression in CLL-cells. More importantly, the microRNA profiling of plasma-derived exosomes conducted earlier by Yeh et al. (2015) identified distinct exosome microRNA signatures, including miR-29a-c, miR-150, and miR-155 that were significantly higher in circulating CLL exosomes compared to exosomes from healthy donors (p < 0.001). Besides, a high level of miR-150 and miR-155 in exosomes were strongly associated with BCR activation, the pathway which was essential in CLL development (Yeh et al., 2015). These data demonstrate the strong potential and advantages of exosomal miRNAs as convenient, noninvasive biomarkers and therapeutic targets for future leukemia treatment due to their easily accessible in peripheral blood and the extensive effect on tumor biological process.
The estimated annual incidence of adult acute lymphocytic leukemia (ALL) is about one in 100,000. Most ALL occurs in children and adolescents younger than 20 years old, with cure rates approaching 90%. However, on the contrary, the therapeutic progress has been slow, with an average survival of 35% in adult patients aged 18–60 years (Bassan and Hoelzer, 2011; Man et al., 2017). Besides, relapsed leukemia is normally associated with high rates of treatment failure due to chemotherapy resistance (Haque and Vaiselbuh, 2020b). Exploring the relationships between ALL biological processes and exosome-associated non-coding RNAs can not only better clarify the pathogenesis of ALL but also provide novel therapeutic targets and biomarkers to improve ALL response. The study of Haque et al. revealed that exosomes derived from ALL cell lines promoted cellular proliferation not only in leukemia B cell lines but also in controlling human B cells. Furthermore, miR-181a was overexpressed in ALL cell exosomes and specific silencing of exosomal miR-181a reversed exosome-induced leukemia cell proliferation in vitro. These data suggested that exosomal miR-181a is a biologically active player with a functional role in leukemia cells and could be a novel target for growth suppression in ALL (Haque and Vaiselbuh, 2020a). Haque and colleagues further investigated the influence of vincristine and prednisone (the standard chemotherapeutic agents for ALL) on miR-181a expression in the first time diagnosed leukemia and relapsed leukemia. The consequences showed that vincristine and prednisone exposure did not change miR-181a expression either at the cellular or exosomal level in relapsed leukemia cell lines contrary to the first time diagnosed leukemia cells, where miR-181a expression was suppressed. More importantly, this non-suppressive nature of miR-181a made relapsed leukemia cells resistant to vincristine and prednisone. These data suggested that cellular and exosomal miR-181a played important roles in the chemoresistance of relapsed leukemia (Haque and Vaiselbuh, 2020b).
Circular RNAs (circRNAs) are a class of single-stranded closed RNA molecules without 5′ caps and 3′ tails that undergo a specific backsplicing from pre-mRNA. circRNAs are found to be widely expressed across eucaryons and expressed specifically in cells and tissues (Qu et al., 2018). CircRNAs can competitively inhibit the function of miRNAs, acting as an RNA sponge to block miRNAs from binding to their target mRNA, regulating the expression of the relevant protein, and playing essential roles in the different biological processes of numerous tumors (Zhong et al., 2018).
Formed by backsplicing events where a 5′ cap binds to a 3′ tail, circRNAs lack 5′ and 3′ terminal ends. As such, circRNAs are inherently resistant to the ribonuclease of degradation, which works by targeting the 5′ and 3′ termini. This results in the high stability of circRNAs in the cytoplasm, exosomes, and body fluid. In fact, plenty of circRNAs has been detected in the peripheral circulation of patients and tumor-derived exosomes (Zhou et al., 2018). Moreover, Tay et al. (2015) investigated the design and effect of miRNA sponge expression vectors and found that circularized miRNA sponges were resistant to miRNA-mediated RNA destabilization and displayed superior anti-tumor activities compared to the linear carriers in malignant melanoma cell lines. The unique stability of circRNA endows it with a more potent inhibitory effect on the oncogenic activity of miRNAs and the advantages as potential therapeutic targets and biomarkers for future clinical applications.
To date, the role of exosomal circRNAs in leukemia has not been explored to the same extent as miRNAs. Nevertheless, there have been several recent studies that have begun to investigate their function in leukemia. The research of Wang et al. (2021) revealed that circ_0009910 could be shuttled among AML cells via exosomes and regulated the expression of miR-5195-3p and GRB10, thus influencing the proliferation, apoptosis, and cell cycle progression of AML cells. Bi et al. (2021) found that oncogene circ_0004136 could promote AML development through regulating miR-570-3p/TSPAN3 axis while exosome-mediated circ_0004136 knockdown restrained AML cell malignant progression. Our recent study also demonstrated that upregulated circ_0058493 in exosomes derived from CML cells was associated with imatinib resistance (Zhong et al., 2021). Besides, Wu et al. (2020) identified a novel mitochondrial genome-derived circRNAs (Mc), mc-COX2. They further found that elevated mc-COX2 in exosomes from CLL plasma compared to normal controls was associated with the progression and prognosis of CLL.
Currently, the understanding of the role of exosomal circRNA in the molecular mechanisms and possible clinical applications in leukemia are very limited and need further study. It has been extensively verified in solid tumors that exosomal circRNAs participate in numerous cancer biological processes including tumorigenesis, proliferation, angiogenesis, metastasis, drug resistance, etc (Liu et al., 2020; Xie et al., 2020; Xu J. et al., 2020; Huang et al., 2021). Xie et al. (2020) reported that exosomal circSHKBP1 regulated the miR-582-3p/HUR/VEGF pathway, suppressed HSP90 degradation, and promoted GC progression. Besides, circSHKBP1 derived from serum exosomes was more abundant in GC patients than in healthy controls, making it a promising circulating biomarker of GC diagnosis and an exceptional candidate for further therapeutic exploration. Xu J. et al. (2020) found that exosomes were crucial for spreading circRNA-SORE-mediated sorafenib resistance among HCC tumor cells. In addition, HCC patients with relatively lower exosomal circRNA-SORE expression had a higher response rate towards sorafenib (80%) compared with patients with higher expression (25%), indicating exosomal circRNA-SORE was a promising prognostic biomarker or therapeutic target for HCC treatment (Xu J. et al., 2020). These studies suggest that exosomal circRNAs have a powerful role in the regulation of the tumor biological process and further investigation is urgently needed to elucidate the precise function of exosomal circRNAs in leukemia.
LncRNAs are defined as transcripts with lengths exceeding 200 nucleotides that are not translated into protein and most of them are markedly expressed in differentiated tissues or particular cancer types (Bhat et al., 2020). Compared to small ncRNAs, lncRNAs exhibit extensive mechanistic diversity to carry out their function. Briefly, lncRNAs can exert their function through binding to protein complexes, nucleic acids and acting as ceRNAs that inhibit the effect of miRNAs, similarly to circRNAs (Slack and Chinnaiyan, 2019).
LncRNAs are involved in many crucial biological processes such as chromatin modification, gene expression, and nuclear transport, and are functional regulators of carcinogenesis, apoptosis, tumor migration, and drug resistance. Its functional relevance in cancer indicates the possibility of using lncRNAs as future biomarkers and therapeutic targets for cancer treatments (Zhou et al., 2018).
The function of exosomal lncRNAs in leukemia progression has not been reported yet, but recent studies have demonstrated that exosomal lncRNAs play critical roles in other hematological tumors. Li et al. (2018) found that lncRNA RUNX2-AS1 in myeloma cells could be packed into exosomes and transmitted to MSCs. RUNX2-AS1 was able to form an RNA duplex with RUNX2 pre-mRNA and transcriptionally repressed RUNX2 expression, resulting in decreased osteogenic potential of MSCs. Therefore, exosomal lncRNA RUNX2-AS1 may serve as a potential therapeutic target for bone lesions in MM (Li et al., 2018). The study of Deng et al. revealed that LINC00461 was highly expressed in MSC-derived exosomes and further enhanced multiple myeloma (MM) cell proliferation, which might become a candidate for therapeutic applications (Deng et al., 2019). Further investigations of the regulatory function and the clinical relevance of exosomal lncRNAs in leukemia are needed.
The function of exosome-derived lncRNAs has been investigated in a wide range of solid tumors and the results showed that exosomal lncRNAs could affect tumor growth, metastasis, invasion, and prognosis by regulating the tumor microenvironment. By intercellular transfer through exosomes, lncRNAs can create a microenvironment suitable for the metastasis of tumor cells (Zhou et al., 2018). Wu et al. (2018) found that exosomal lnc-MMP2-2 was involved in increasing vascular endothelial cell permeability and enhancing the invasion of lung cancer cells by promoting MMP2 expression. Their findings suggested that exosomal lnc-MMP2-2 might represent a putative therapeutic target for lung cancer treatment. The study of Behera and colleagues demonstrated both in vitro and vivo that lnc-H19 in BMMSC-exosomes significantly promoted endothelial angiogenesis and osteogenesis through binding with miR-106 and activating Angpt1/Tie2-NO signaling pathway in mesenchymal and endothelial cells (Behera et al., 2021). These findings demonstrated the mediating ability of exosomal lncRNAs in the pathological process of tumors. The emerging field of exosomal lncRNAs in leukemia is worth further exploration.
Intercellular communication of cells via exosome-derived ncRNAs and subsequent functional effects have been demonstrated in numerous studies, including hematologic tumors and solid tumors, highlighting the potential of exosome-derived ncRNAs as functional mediators in tumors. On the one hand, exosomes derived from normal cells can transfer tumor suppressor genes into tumor cells, thus inhibiting tumor promotion. On the other hand, oncogenes can be shuttled from tumor cells into normal cells or sensitive cells, facilitating the formation of tumor microenvironment or drug resistance. Besides, tumor cells can reduce the cellular level of tumor suppressor genes through exosomes excretion, thus contributing to tumor promotion.
The regulatory function in the leukemia pathological process hints at the possibility of using exosomal ncRNAs as future therapeutic targets for leukemia treatments. According to how exosomal ncRNAs participate in tumor promotion, there are two research directions for future clinical application: 1) Increasing the level of tumor suppressor genes in a cancerous environment. a) Exosomes containing tumor suppressor genes with higher affinity for tumor tissues may be designed and synthesized through special membrane protein decoration. Then the functional exosomes are supposed to target the specific tumor microenvironment, increasing the level of tumor suppressor genes and inhibiting leukemia promotion. b) According to the research listed above (Table 2), functional ncRNAs are always enriched in exosomes and this phenomenon is controlled by certain loading and sorting mechanisms. Future studies should aim to regulate the loading and sorting process of exosomal ncRNAs and inhibit the secretion of tumor suppressor genes from tumor cells. 2) Decreasing the level of oncogenes in a cancerous environment. a) The protection of exosomes from enzymes in circulation makes it possible for the anti-RNAs loading in exosomes and being applied in gene therapy. In fact, Shtam et al. (2013) has transfected siRNA into exosomes and successfully silenced the expression of RAD51 using exosomes as vectors. b) As exosomes are critical components of cellular communication and the key facilitators in the exchange of information between cells (Zhou et al., 2018), it may be practical to inhibit the transmission of tumor-related exosomes in circulation or tumor microenvironment using target drugs or inhibitors.
To date, there have been abundant exosomal ncRNAs found to be dysregulated in leukemia and they are recognized as vital sources of diagnostic or prognostic biomarkers for future treatment. Exosomal ncRNAs are ideal biomarkers because of their: 1) Specificity: Specific types and concentration of ncRNAs in exosomes can represent the conditions of original cells or tissues; 2) Sensitivity: Emerging evidence have clarified that cancer patients have a larger number of circulating exosomes compared to healthy controls (Zhou et al., 2018). In addition, functional ncRNAs are always enriched in exosomes. These features will provide higher sensitivity for exosomal ncRNAs detection in clinical samples, compared to free ncRNAs in body fluids; 3) Stability: Exosomes can protect the contents from enzymes digesting during circulation. ncRNA levels in exosomes will not change significantly after exposure to a variety of stress conditions; 4) Accessibility: Most of the functional exosomal ncRNAs are distributed in blood and can be obtained in a non-invasive manner. Thus, the emerging field of exosomal ncRNAs as biomarkers in leukemia is worth our time and effort.
Despite the strong potential of exosomal ncRNAs as novel therapeutic targets and biomarkers, there are still lots of challenges in developing exosomal ncRNAs for clinical applications. From an investigative perspective, the functions of ncRNAs identified in current studies are usually based on the results obtained from exogenous overexpression of ncRNAs in vitro (such as transfecting miRNA mimic) or ncRNAs knockdown in overexpressed cell lines. However, it is hardly known whether the body produces a comparable amount of ncRNAs and promotes the same functions. Similarly, most reports mechanistically analyzing the role of exosomal ncRNAs are conducted in cell line systems or co-culture systems in vitro in which the purified exosomes preparations are incubated with intended recipient cells. It is not clear whether the amount of exosome or exosomal ncRNAs applied is within a physiological range. Exogenous supplementation of native and synthetic exosomes may exaggerate the function of exosomal ncRNAs (Sempere et al., 2017). Nevertheless, the critical role of exosomal ncRNAs in tumor development is beyond doubt.
As exosomal ncRNAs are recognized as vital sources of biomarkers for clinical use, it is important to identify the dysregulated ncRNAs in circulating exosomes. Next-generation sequencing (NGS) is a powerful and useful approach for comprehensive transcriptomic profiling and has become an increasingly popular application to analyze ncRNAs in exosomes (Buschmann et al., 2018). However, the circulating exosomes are derived from a variety of cell populations. Their heterogeneous origin will limit the accurate detection of disease-specific exosomes in peripheral blood samples. A vast number of exosomes released from other cell types will dilute the exosome population derived from tumor cells, which reduces the proportion of disease-associated ncRNAs in the sequencing libraries (Huang et al., 2013). Besides, there is no standardized methodology for exosome isolation. Although ultra-centrifugation (UC) is the commonly used method for exosome isolation, other approaches will gain preference when the sample volume is limited, which include filtration, precipitation, sedimentation to size-exclusion chromatography (SEC) and immunocapture (Buschmann et al., 2018). The details in exosome isolation protocols vary among studies and the purity is also difficult to maintain within a certain range, these factors may affect the accuracy and repeatability of the results of NGS or other exosome research (Buschmann et al., 2018). Additionally, compared to cell and tissue samples, the amount of total RNA extracted from exosomes will be far less, therefore a more sensitive exosomal RNA extraction kit is required to ensure the measurement accuracy. The kits from different brands with different protocols will affect the quality of secreted RNA, which may also influence the experiment consequences to some extent. Developing the standard and appropriate exosome isolation and detection methods is a critical step in all areas of exosome research.
From a therapeutic perspective, certain methods of exosome isolation may not be easily scalable to the higher throughput of a hospital laboratory. Although UC is a commonly used method of exosome isolation with high purity and yield, it may require a large volume of blood. The extraction efficiency is also limited by the number of wells in the ultracentrifuge machine, usually six, and the extremely long time the process costs (usually up to 18 h). There are various commercial exosome isolation kits based on filtration, precipitation, SEC and immunocapture which are time-saving and need less sample volume, but the difference of protocols will affect the accuracy and repeatability of detection results (Buschmann et al., 2018). There is a need to standardize methodology and develop technologies that make the isolation and detection of exosomes feasible and scalable to clinical use in hospitals (Desmond et al., 2019). In addition, the exosomes collected from serum or plasma may be contaminated by EVs released by irrelevant cells. How to detect and enrich exosomes secreted by specific tumor cells from complex environments, like heterogeneous cancer tissue or circulation, is also an obstacle for their clinical application. The specific proteins expressed on the membrane of exosomes reflect its origin to some extent. Antibodies designed to target the cell type-specific surface proteins on exosomes may be used to identify and isolate exosomes originating from specific tissues for more accurate clinical detection (Sempere et al., 2017). It has been reported that the epithelial cell adhesion molecule (EpCAM) could be used as a marker to isolate epithelial-derived EVs from the blood sample of colorectal cancer patients by immunoaffinity-capture (Ostenfeld et al., 2016). However, Rupp et al. (2011) demonstrated that EpCAM could be cleaved from exosomes via serum metalloproteinases, which may hamper the tumor exosome enrichment by immune-affinity isolation in breast cancer. Further investigations are required to find and develop specific and stable markers for exosome enrichment. Moreover, though the stability of exosome contents makes it possible to apply exosomes in gene therapy, research is still needed to ensure that exosomes containing therapeutic ncRNAs are able to safely escape from the immune system and accurately identify cancerous tissues.
Understanding the pathogenesis of leukemia and finding specific therapeutic targets and biomarkers is of great importance to improve the clinical efficacy of leukemia patients. Exosomal non-coding RNAs can mediate the communication between cells and the tumor microenvironment by exosome transmission, which is involved in regulating cancerous transformation of mesenchymal cells, fibroblasts, endothelial cells, and advancing tumor cell proliferation, angiogenesis, metastasis, and drug resistance. In addition, exosomal non-coding RNA has excellent characteristics such as tissue or cell specificity, circulation stability, making it a novel biomarker for leukemia identification, treatment monitoring, and efficacy prediction.
Despite the strong potential of exosomal ncRNAs as future therapeutic targets and biomarkers, there are still many challenges that need further research. First, standardization of exosome isolation and purification methodology may increase the accuracy and reliability of functional investigations and advance the clinical application of exosomal ncRNAs. Second, the biological functions of exosomal ncRNAs should be further confirmed in vivo and patient samples. The clinical relevance of exosomal ncRNAs in leukemia deserves more extensive exploration. Third, understanding the mechanisms by which donor cells manage to exquisitely load and sort specific ncRNAs cargo into exosomes and how these functional exosomes accurately identify distinct recipient cells without being attacked by the immune system will contribute to the clinical utility of exosomal ncRNAs.
Currently, the understanding of the regulatory function, molecular mechanisms, and clinical relevance of exosomal ncRNAs in leukemia is still limited, especially for circRNA and lncRNA. Further studies on exosomal ncRNAs in leukemia will not only profound impacts on clarifying the pathological process of leukemia but also lay the foundation for novel treatment methods that employ exosomal ncRNAs as biomarkers and therapeutic targets.
PX and LC proposed the subject of the review. B-JT, JX, and S-TH searched the literature. B-JT summarized the literature and drafted the manuscript. BS, PX, and LC revised the review. All authors approved the final version and participated in the submission.
This project was supported by grants from Scientific Research Project of Hunan Provincial Health Commission (202103040800), Natural Science Foundation of Hunan Province (2021JJ30969), Research Foundation of Wu Jieping Medical Foundation, Clinical Pharmacy Branch of Chinese Medical Association (No. 320.6750.19090-6), Research Project established by Chinese Pharmaceutical Association Hospital Pharmacy department (No. CPA-Z05-ZC-2021-002), the Fundamental Research Funds for the Central Universities of Central South University (2022ZZTS0953).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Apperley, J. F. (2015). Chronic Myeloid Leukaemia. Lancet 385 (9976), 1447–1459. doi:10.1016/s0140-6736(13)62120-0
Barrera-Ramirez, J., Lavoie, J. R., Maganti, H. B., Stanford, W. L., Ito, C., Sabloff, M., et al. (2017). Micro-RNA Profiling of Exosomes from Marrow-Derived Mesenchymal Stromal Cells in Patients with Acute Myeloid Leukemia: Implications in Leukemogenesis. Stem Cell Rev. Rep. 13 (6), 817–825. doi:10.1007/s12015-017-9762-0
Bassan, R., and Hoelzer, D. (2011). Modern Therapy of Acute Lymphoblastic Leukemia. J. Clin. Oncol. 29 (5), 532–543. doi:10.1200/jco.2010.30.1382
Behera, J., Kumar, A., Voor, M. J., and Tyagi, N. (2021). Exosomal lncRNA-H19 Promotes Osteogenesis and Angiogenesis through Mediating Angpt1/Tie2-NO Signaling in CBS-Heterozygous Mice. Theranostics 11 (16), 7715–7734. doi:10.7150/thno.58410
Bhat, A. A., Younes, S. N., Raza, S. S., Zarif, L., Nisar, S., Ahmed, I., et al. (2020). Role of Non-coding RNA Networks in Leukemia Progression, Metastasis and Drug Resistance. Mol. Cancer 19 (1), 57. doi:10.1186/s12943-020-01175-9
Bi, J., Pu, Y., and Yu, X. (2021). Exosomal Circ_0004136 Enhances the Progression of Pediatric Acute Myeloid Leukemia Depending on the Regulation of miR-570-3p/TSPAN3 axis. Anticancer Drugs 32, 802–811. doi:10.1097/cad.0000000000001068
Blair, A., Hogge, D. E., and Sutherland, H. J. (1998). Most Acute Myeloid Leukemia Progenitor Cells with Long-Term Proliferative Ability In Vitro and In Vivo Have the Phenotype CD34(+)/CD71(-)/HLA-DR-. Blood 92 (11), 4325–4335. doi:10.1182/blood.v92.11.4325.423k14_4325_4335
Bonnet, D., and Dick, J. E. (1997). Human Acute Myeloid Leukemia Is Organized as a Hierarchy that Originates from a Primitive Hematopoietic Cell. Nat. Med. 3 (7), 730–737. doi:10.1038/nm0797-730
Bruns, H., Böttcher, M., Qorraj, M., Fabri, M., Jitschin, S., Dindorf, J., et al. (2017). CLL-cell-mediated MDSC Induction by Exosomal miR-155 Transfer Is Disrupted by Vitamin D. Leukemia 31 (4), 985–988. doi:10.1038/leu.2016.378
Buschmann, D., Kirchner, B., Hermann, S., Märte, M., Wurmser, C., Brandes, F., et al. (2018). Evaluation of Serum Extracellular Vesicle Isolation Methods for Profiling miRNAs by Next-Generation Sequencing. J. Extracell. Vesicles 7 (1), 1481321. doi:10.1080/20013078.2018.1481321
Deng, M., Yuan, H., Liu, S., Hu, Z., and Xiao, H. (2019). Exosome-transmitted LINC00461 Promotes Multiple Myeloma Cell Proliferation and Suppresses Apoptosis by Modulating microRNA/BCL-2 Expression. Cytotherapy 21 (1), 96–106. doi:10.1016/j.jcyt.2018.10.006
Desmond, B. J., Dennett, E. R., and Danielson, K. M. (2019). Circulating Extracellular Vesicle MicroRNA as Diagnostic Biomarkers in Early Colorectal Cancer-A Review. Cancers (Basel) 12 (1). doi:10.3390/cancers12010052
Döhner, H., Weisdorf, D. J., and Bloomfield, C. D. (2015). Acute Myeloid Leukemia. N. Engl. J. Med. 373 (12), 1136–1152. doi:10.1056/NEJMra1406184
Dombret, H., and Gardin, C. (2016). An Update of Current Treatments for Adult Acute Myeloid Leukemia. Blood 127 (1), 53–61. doi:10.1182/blood-2015-08-604520
Doyle, L. M., and Wang, M. Z. (2019). Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 8 (7). doi:10.3390/cells8070727
Dubovsky, J. A., Chappell, D. L., Harrington, B. K., Agrawal, K., Andritsos, L. A., Flynn, J. M., et al. (2013). Lymphocyte Cytosolic Protein 1 Is a Chronic Lymphocytic Leukemia Membrane-Associated Antigen Critical to Niche Homing. Blood 122 (19), 3308–3316. doi:10.1182/blood-2013-05-504597
Fang, Z., Wang, X., Wu, J., Xiao, R., and Liu, J. (2020). High Serum Extracellular Vesicle miR-10b Expression Predicts Poor Prognosis in Patients with Acute Myeloid Leukemia. Cancer Biomark. 27 (1), 1–9. doi:10.3233/cbm-190211
Farahani, M., Rubbi, C., Liu, L., Slupsky, J. R., and Kalakonda, N. (2015). CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS One 10 (10), e0141429. doi:10.1371/journal.pone.0141429
Fayyad-Kazan, H., Bitar, N., Najar, M., Lewalle, P., Fayyad-Kazan, M., Badran, R., et al. (2013). Circulating miR-150 and miR-342 in Plasma Are Novel Potential Biomarkers for Acute Myeloid Leukemia. J. Transl. Med. 11, 31. doi:10.1186/1479-5876-11-31
Gao, X., Wan, Z., Wei, M., Dong, Y., Zhao, Y., Chen, X., et al. (2019). Chronic Myelogenous Leukemia Cells Remodel the Bone Marrow Niche via Exosome-Mediated Transfer of miR-320. Theranostics 9 (19), 5642–5656. doi:10.7150/thno.34813
Geelen, I. G. P., Thielen, N., Janssen, J. J. W. M., Hoogendoorn, M., Roosma, T. J. A., Willemsen, S. P., et al. (2017). Treatment Outcome in a Population-Based, 'real-World' Cohort of Patients with Chronic Myeloid Leukemia. Haematologica 102 (11), 1842–1849. doi:10.3324/haematol.2017.174953
Guo, W., Gao, Y., Li, N., Shao, F., Wang, C., Wang, P., et al. (2017). Exosomes: New Players in Cancer (Review). Oncol. Rep. 38 (2), 665–675. doi:10.3892/or.2017.5714
Hallek, M. (2019). Chronic Lymphocytic Leukemia: 2020 Update on Diagnosis, Risk Stratification and Treatment. Am. J. Hematol. 94 (11), 1266–1287. doi:10.1002/ajh.25595
Haque, S., and Vaiselbuh, S. R. (2020a). Silencing of Exosomal miR-181a Reverses Pediatric Acute Lymphocytic Leukemia Cell Proliferation. Pharm. (Basel) 13 (9), 241. doi:10.3390/ph13090241
Haque, S., and Vaiselbuh, S. R. (2020b). Vincristine and Prednisone Regulates Cellular and Exosomal miR-181a Expression Differently within the First Time Diagnosed and the Relapsed Leukemia B Cells. Leuk. Res. Rep. 14, 100221. doi:10.1016/j.lrr.2020.100221
Horiguchi, H., Kobune, M., Kikuchi, S., Yoshida, M., Murata, M., Murase, K., et al. (2016). Extracellular Vesicle miR-7977 Is Involved in Hematopoietic Dysfunction of Mesenchymal Stromal Cells via poly(rC) Binding Protein 1 Reduction in Myeloid Neoplasms. Haematologica 101 (4), 437–447. doi:10.3324/haematol.2015.134932
Hornick, N. I., Doron, B., Abdelhamed, S., Huan, J., Harrington, C. A., Shen, R., et al. (2016). AML Suppresses Hematopoiesis by Releasing Exosomes that Contain microRNAs Targeting C-MYB. Sci. Signal 9 (444), ra88. doi:10.1126/scisignal.aaf2797
Hu, Y., Ma, X., Wu, Z., Nong, Q., Liu, F., Wang, Y., et al. (2020). MicroRNA-34a-mediated Death of Acute Myeloid Leukemia Stem Cells through Apoptosis Induction and Exosome Shedding Inhibition via Histone Deacetylase 2 Targeting. Iubmb Life 72 (7), 1481–1490. doi:10.1002/iub.2273
Huang, X., Yuan, T., Tschannen, M., Sun, Z., Jacob, H., Du, M., et al. (2013). Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genomics 14, 319. doi:10.1186/1471-2164-14-319
Huang, Y., Liang, B., and Chen, X. (2021). Exosomal Circular RNA Circ_0074673 Regulates the Proliferation, Migration, and Angiogenesis of Human Umbilical Vein Endothelial Cells via the microRNA-1200/MEOX2 axis. Bioengineered 12 (1), 6782–6792. doi:10.1080/21655979.2021.1967077
Jiang, L., Deng, T., Wang, D., and Xiao, Y. (2018). Elevated Serum Exosomal miR-125b Level as a Potential Marker for Poor Prognosis in Intermediate-Risk Acute Myeloid Leukemia. Acta Haematol. 140 (3), 183–192. doi:10.1159/000491584
Jiang, Y. H., Liu, J., Lin, J., Li, S. Q., Xu, Y. M., Min, Q. H., et al. (2020). K562 Cell-Derived Exosomes Suppress the Adhesive Function of Bone Marrow Mesenchymal Stem Cells via Delivery of miR-711. Biochem. Biophys. Res. Commun. 521 (3), 584–589. doi:10.1016/j.bbrc.2019.10.096
Kipps, T. J., Stevenson, F. K., Wu, C. J., Croce, C. M., Packham, G., Wierda, W. G., et al. (2017). Chronic Lymphocytic Leukaemia. Nat. Rev. Dis. Prim. 3, 16096. doi:10.1038/nrdp.2016.96
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A Cell Initiating Human Acute Myeloid Leukaemia after Transplantation into SCID Mice. Nature 367 (6464), 645–648. doi:10.1038/367645a0
Li, B., Xu, H., Han, H., Song, S., Zhang, X., Ouyang, L., et al. (2018). Exosome-mediated Transfer of lncRUNX2-AS1 from Multiple Myeloma Cells to MSCs Contributes to Osteogenesis. Oncogene 37 (41), 5508–5519. doi:10.1038/s41388-018-0359-0
Lin, X., Ling, Q., Lv, Y., Ye, W., Huang, J., Li, X., et al. (2020). Plasma Exosome-Derived microRNA-532 as a Novel Predictor for Acute Myeloid Leukemia. Cancer Biomark. 28 (2), 151–158. doi:10.3233/cbm-191164
Liu, D., Kang, H., Gao, M., Jin, L., Zhang, F., Chen, D., et al. (2020). Exosome-transmitted circ_MMP2 Promotes Hepatocellular Carcinoma Metastasis by Upregulating MMP2. Mol. Oncol. 14 (6), 1365–1380. doi:10.1002/1878-0261.12637
Man, L. M., Morris, A. L., and Keng, M. (2017). New Therapeutic Strategies in Acute Lymphocytic Leukemia. Curr. Hematol. Malig. Rep. 12 (3), 197–206. doi:10.1007/s11899-017-0380-3
Marcucci, G., Haferlach, T., and Döhner, H. (2011). Molecular Genetics of Adult Acute Myeloid Leukemia: Prognostic and Therapeutic Implications. J. Clin. Oncol. 29 (5), 475–486. doi:10.1200/jco.2010.30.2554
Mardani, R., Jafari Najaf Abadi, M. H., Motieian, M., Taghizadeh-Boroujeni, S., Bayat, A., Farsinezhad, A., et al. (2019). MicroRNA in Leukemia: Tumor Suppressors and Oncogenes with Prognostic Potential. J. Cell Physiol. 234 (6), 8465–8486. doi:10.1002/jcp.27776
Mashouri, L., Yousefi, H., Aref, A. R., Ahadi, A. M., Molaei, F., and Alahari, S. K. (2019). Exosomes: Composition, Biogenesis, and Mechanisms in Cancer Metastasis and Drug Resistance. Mol. Cancer 18 (1), 75. doi:10.1186/s12943-019-0991-5
Min, Q. H., Wang, X. Z., Zhang, J., Chen, Q. G., Li, S. Q., Liu, X. Q., et al. (2018). Exosomes Derived from Imatinib-Resistant Chronic Myeloid Leukemia Cells Mediate a Horizontal Transfer of Drug-Resistant Trait by Delivering miR-365. Exp. Cell Res. 362 (2), 386–393. doi:10.1016/j.yexcr.2017.12.001
Nicoloso, M. S., Spizzo, R., Shimizu, M., Rossi, S., and Calin, G. A. (2009). MicroRNAs--the Micro Steering Wheel of Tumour Metastases. Nat. Rev. Cancer 9 (4), 293–302. doi:10.1038/nrc2619
Noh, G. T., Kwon, J., Kim, J., Park, M., Choi, D. W., Cho, K. A., et al. (2020). Verification of the Role of Exosomal microRNA in Colorectal Tumorigenesis Using Human Colorectal Cancer Cell Lines. PLoS One 15 (11), e0242057. doi:10.1371/journal.pone.0242057
Ohyashiki, K., Umezu, T., Katagiri, S., Kobayashi, C., Azuma, K., Tauchi, T., et al. (2016). Downregulation of Plasma miR-215 in Chronic Myeloid Leukemia Patients with Successful Discontinuation of Imatinib. Int. J. Mol. Sci. 17 (4), 570. doi:10.3390/ijms17040570
Ostenfeld, M. S., Jensen, S. G., Jeppesen, D. K., Christensen, L. L., Thorsen, S. B., Stenvang, J., et al. (2016). miRNA Profiling of Circulating EpCAM(+) Extracellular Vesicles: Promising Biomarkers of Colorectal Cancer. J. Extracell. Vesicles 5, 31488. doi:10.3402/jev.v5.31488
Paggetti, J., Haderk, F., Seiffert, M., Janji, B., Distler, U., Ammerlaan, W., et al. (2015). Exosomes Released by Chronic Lymphocytic Leukemia Cells Induce the Transition of Stromal Cells into Cancer-Associated Fibroblasts. Blood 126 (9), 1106–1117. doi:10.1182/blood-2014-12-618025
Peng, D., Wang, H., Li, L., Ma, X., Chen, Y., Zhou, H., et al. (2018). miR-34c-5p Promotes Eradication of Acute Myeloid Leukemia Stem Cells by Inducing Senescence through Selective RAB27B Targeting to Inhibit Exosome Shedding. Leukemia 32 (5), 1180–1188. doi:10.1038/s41375-018-0015-2
Qu, S., Liu, Z., Yang, X., Zhou, J., Yu, H., Zhang, R., et al. (2018). The Emerging Functions and Roles of Circular RNAs in Cancer. Cancer Lett. 414, 301–309. doi:10.1016/j.canlet.2017.11.022
Rodriguez-Abreu, D., Bordoni, A., and Zucca, E. (2007). Epidemiology of Hematological Malignancies. Ann. Oncol. 18 (Suppl. 1), i3–i8. doi:10.1093/annonc/mdl443
Rupp, A. K., Rupp, C., Keller, S., Brase, J. C., Ehehalt, R., Fogel, M., et al. (2011). Loss of EpCAM Expression in Breast Cancer Derived Serum Exosomes: Role of Proteolytic Cleavage. Gynecol. Oncol. 122 (2), 437–446. doi:10.1016/j.ygyno.2011.04.035
Sempere, L. F., Keto, J., and Fabbri, M. (2017). Exosomal MicroRNAs in Breast Cancer towards Diagnostic and Therapeutic Applications. Cancers (Basel) 9 (7). doi:10.3390/cancers9070071
Shallis, R. M., Wang, R., Davidoff, A., Ma, X., and Zeidan, A. M. (2019). Epidemiology of Acute Myeloid Leukemia: Recent Progress and Enduring Challenges. Blood Rev. 36, 70–87. doi:10.1016/j.blre.2019.04.005
Shtam, T. A., Kovalev, R. A., Varfolomeeva, E. Y., Makarov, E. M., Kil, Y. V., and Filatov, M. V. (2013). Exosomes Are Natural Carriers of Exogenous siRNA to Human Cells In Vitro. Cell Commun. Signal 11, 88. doi:10.1186/1478-811x-11-88
Slack, F. J., and Chinnaiyan, A. M. (2019). The Role of Non-coding RNAs in Oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Smolarz, M., and Widlak, P. (2021). Serum Exosomes and Their miRNA Load-A Potential Biomarker of Lung Cancer. Cancers 13 (6), 1373. doi:10.3390/cancers13061373
Tadokoro, H., Umezu, T., Ohyashiki, K., Hirano, T., and Ohyashiki, J. H. (2013). Exosomes Derived from Hypoxic Leukemia Cells Enhance Tube Formation in Endothelial Cells. J. Biol. Chem. 288 (48), 34343–34351. doi:10.1074/jbc.M113.480822
Taverna, S., Amodeo, V., Saieva, L., Russo, A., Giallombardo, M., De Leo, G., et al. (2014). Exosomal Shuttling of miR-126 in Endothelial Cells Modulates Adhesive and Migratory Abilities of Chronic Myelogenous Leukemia Cells. Mol. Cancer 13, 169. doi:10.1186/1476-4598-13-169
Taverna, S., Fontana, S., Monteleone, F., Pucci, M., Saieva, L., De Caro, V., et al. (2016). Curcumin Modulates Chronic Myelogenous Leukemia Exosomes Composition and Affects Angiogenic Phenotype via Exosomal miR-21. Oncotarget 7 (21), 30420–30439. doi:10.18632/oncotarget.8483
Taverna, S., Giallombardo, M., Pucci, M., Flugy, A., Manno, M., Raccosta, S., et al. (2015). Curcumin Inhibits In Vitro and In Vivo Chronic Myelogenous Leukemia Cells Growth: a Possible Role for Exosomal Disposal of miR-21. Oncotarget 6 (26), 21918–21933. doi:10.18632/oncotarget.4204
Tay, F. C., Lim, J. K., Zhu, H., Hin, L. C., and Wang, S. (2015). Using Artificial microRNA Sponges to Achieve microRNA Loss-Of-Function in Cancer Cells. Adv. Drug Deliv. Rev. 81, 117–127. doi:10.1016/j.addr.2014.05.010
Terwilliger, T., and Abdul-Hay, M. (2017). Acute Lymphoblastic Leukemia: a Comprehensive Review and 2017 Update. Blood Cancer J. 7 (6), e577. doi:10.1038/bcj.2017.53
Thol, F., Schlenk, R. F., Heuser, M., and Ganser, A. (2015). How I Treat Refractory and Early Relapsed Acute Myeloid Leukemia. Blood 126 (3), 319–327. doi:10.1182/blood-2014-10-551911
Umezu, T., Ohyashiki, K., Kuroda, M., and Ohyashiki, J. H. (2013). Leukemia Cell to Endothelial Cell Communication via Exosomal miRNAs. Oncogene 32 (22), 2747–2755. doi:10.1038/onc.2012.295
Wan, Z., Chen, X., Gao, X., Dong, Y., Zhao, Y., Wei, M., et al. (2019). Chronic Myeloid Leukemia-Derived Exosomes Attenuate Adipogenesis of Adipose Derived Mesenchymal Stem Cells via Transporting miR-92a-3p. J. Cell Physiol. 234 (11), 21274–21283. doi:10.1002/jcp.28732
Wang, D., Ming, X., Xu, J., and Xiao, Y. (2021). Circ_0009910 Shuttled by Exosomes Regulates Proliferation, Cell Cycle and Apoptosis of Acute Myeloid Leukemia Cells by Regulating miR‐5195‐3p/GRB10 axis. Hematol. Oncol. 39, 390–400. doi:10.1002/hon.2874
Wu, D. M., Deng, S. H., Liu, T., Han, R., Zhang, T., and Xu, Y. (2018). TGF-β-mediated Exosomal Lnc-MMP2-2 Regulates Migration and Invasion of Lung Cancer Cells to the Vasculature by Promoting MMP2 Expression. Cancer Med. 7 (10), 5118–5129. doi:10.1002/cam4.1758
Wu, Z., Sun, H., Wang, C., Liu, W., Liu, M., Zhu, Y., et al. (2020). Mitochondrial Genome-Derived circRNA Mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic Acids 20, 801–811. doi:10.1016/j.omtn.2020.04.017
Xie, M., Yu, T., Jing, X., Ma, L., Fan, Y., Yang, F., et al. (2020). Exosomal circSHKBP1 Promotes Gastric Cancer Progression via Regulating the miR-582-3p/HUR/VEGF axis and Suppressing HSP90 Degradation. Mol. Cancer 19 (1), 112. doi:10.1186/s12943-020-01208-3
Xing, F., Saidou, J., and Watabe, K. (2010). Cancer Associated Fibroblasts (CAFs) in Tumor Microenvironment. Front. Biosci. (Landmark Ed. 15, 166–179. doi:10.2741/3613
Xu, J., Ji, L., Liang, Y., Wan, Z., Zheng, W., Song, X., et al. (2020). CircRNA-SORE Mediates Sorafenib Resistance in Hepatocellular Carcinoma by Stabilizing YBX1. Signal Transduct. Target Ther. 5 (1), 298. doi:10.1038/s41392-020-00375-5
Xu, Y. C., Lin, Y. S., Zhang, L., Lu, Y., Sun, Y. L., Fang, Z. G., et al. (2020). MicroRNAs of Bone Marrow Mesenchymal Stem Cell-Derived Exosomes Regulate Acute Myeloid Leukemia Cell Proliferation and Apoptosis. Chin. Med. J. Engl. 133 (23), 2829–2839. doi:10.1097/cm9.0000000000001138
Yang, Y., Li, J., and Geng, Y. (2020). Exosomes Derived from Chronic Lymphocytic Leukaemia Cells Transfer miR-146a to Induce the Transition of Mesenchymal Stromal Cells into Cancer-Associated Fibroblasts. J. Biochem. 168, 491–498. doi:10.1093/jb/mvaa064
Yeh, Y. Y., Ozer, H. G., Lehman, A. M., Maddocks, K., Yu, L., Johnson, A. J., et al. (2015). Characterization of CLL Exosomes Reveals a Distinct microRNA Signature and Enhanced Secretion by Activation of BCR Signaling. Blood 125 (21), 3297–3305. doi:10.1182/blood-2014-12-618470
Yoshida, M., Horiguchi, H., Kikuchi, S., Iyama, S., Ikeda, H., Goto, A., et al. (2019). miR-7977 Inhibits the Hippo-YAP Signaling Pathway in Bone Marrow Mesenchymal Stromal Cells. PLoS One 14 (3), e0213220. doi:10.1371/journal.pone.0213220
Zhang, F., Lu, Y., Wang, M., Zhu, J., Li, J., Zhang, P., et al. (2020). Exosomes Derived from Human Bone Marrow Mesenchymal Stem Cells Transfer miR-222-3p to Suppress Acute Myeloid Leukemia Cell Proliferation by Targeting IRF2/INPP4B. Mol. Cell Probes 51, 101513. doi:10.1016/j.mcp.2020.101513
Zhang, X., Yang, Y., Yang, Y., Chen, H., Tu, H., and Li, J. (2020). Exosomes from Bone Marrow Microenvironment-Derived Mesenchymal Stem Cells Affect CML Cells Growth and Promote Drug Resistance to Tyrosine Kinase Inhibitors. Stem Cells Int. 2020, 8890201. doi:10.1155/2020/8890201
Zhang, Z., Zhang, L., Yu, G., Sun, Z., Wang, T., Tian, X., et al. (2020). Exosomal miR-1246 and miR-155 as Predictive and Prognostic Biomarkers for Trastuzumab-Based Therapy Resistance in HER2-Positive Breast Cancer. Cancer Chemother. Pharmacol. 86 (6), 761–772. doi:10.1007/s00280-020-04168-z
Zhang, Z. Y., Li, Y. C., Geng, C. Y., Zhou, H. X., Gao, W., and Chen, W. M. (2019). Serum Exosomal microRNAs as Novel Biomarkers for Multiple Myeloma. Hematol. Oncol. 37 (4), 409–417. doi:10.1002/hon.2639
Zhao, C., Du, F., Zhao, Y., Wang, S., and Qi, L. (2019). Acute Myeloid Leukemia Cells Secrete microRNA-4532-Containing Exosomes to Mediate Normal Hematopoiesis in Hematopoietic Stem Cells by Activating the LDOC1-dependent STAT3 Signaling Pathway. Stem Cell Res. Ther. 10 (1), 384. doi:10.1186/s13287-019-1475-7
Zheng, R., Du, M., Wang, X., Xu, W., Liang, J., Wang, W., et al. (2018). Exosome-transmitted Long Non-coding RNA PTENP1 Suppresses Bladder Cancer Progression. Mol. Cancer 17 (1), 143. doi:10.1186/s12943-018-0880-3
Zhong, A. N., Yin, Y., Tang, B. J., Chen, L., Shen, H. W., Tan, Z. P., et al. (2021). CircRNA Microarray Profiling Reveals Hsa_circ_0058493 as a Novel Biomarker for Imatinib-Resistant CML. Front. Pharmacol. 12, 728916. doi:10.3389/fphar.2021.728916
Zhong, Y., Du, Y., Yang, X., Mo, Y., Fan, C., Xiong, F., et al. (2018). Circular RNAs Function as ceRNAs to Regulate and Control Human Cancer Progression. Mol. Cancer 17 (1), 79. doi:10.1186/s12943-018-0827-8
Keywords: exosome, non-coding RNA, leukemia, tumor development, therapeutic target, biomarker
Citation: Tang B-J, Sun B, Chen L, Xiao J, Huang S-T and Xu P (2022) The Landscape of Exosome-Derived Non-Coding RNA in Leukemia. Front. Pharmacol. 13:912303. doi: 10.3389/fphar.2022.912303
Received: 04 April 2022; Accepted: 27 May 2022;
Published: 15 June 2022.
Edited by:
Marco Ragusa, University of Catania, ItalyReviewed by:
Paula Alexandra Da Costa Martins, Maastricht University, NetherlandsCopyright © 2022 Tang, Sun, Chen, Xiao, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Xu, eHVwaW5nMTEwOUBjc3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.