- 1Department of Physiology, University of Kentucky, Lexington, KY, United States
- 2Department of Internal Medicine, Division of Pulmonary, Critical Care and Sleep Medicine, University of Kentucky, Lexington, KY, United States
Circadian rhythms are approximate 24-h biological cycles that optimize molecular and physiological functions to predictable daily environmental changes in order to maintain internal and organismal homeostasis. Environmental stimuli (light, feeding, activity) capable of altering the phase of molecular rhythms are important tools employed by circadian biologists to increase understanding of the synchronization of circadian rhythms to the environment and to each other within multicellular systems. The central circadian clock, located in the suprachiasmatic nucleus (SCN) of the hypothalamus is largely responsive to light and is thought to entrain the phase of peripheral clocks via neurohumoral signals. Mice are nocturnal and consume most of their food during the dark cycle. Early studies demonstrated that altered metabolic cues in the form of time restricted feeding, specifically, feeding mice during the light cycle, resulted in an uncoupling of molecular clocks in peripheral tissues with those from the SCN. These studies showed as much as a 12-h shift in gene expression in some peripheral tissues but not others. The shifts occurred without corresponding changes in the central clock in the brain. More recent studies have demonstrated that changes in cardiac physiology (heart rate, MAP) in response to time of food intake occur independent of the cardiac molecular clock. Understanding differences in the physiology/function and gene expression in other organs both independently and in relation to the heart in response to altered feeding will be important in dissecting the roles of the various clocks throughout the body, as well as, understanding their links to cardiovascular pathology.
Introduction
Homeostasis is classically defined as the maintenance of internal stability by the action and interaction of complex physiologic mechanisms. Central to the concept of homeostasis are negative feedback mechanisms that continually compensate for challenges to homeostasis. Negative feedback is responsible for maintaining homeostatically regulated variables within a relatively constant range (Richardson et al., 1998). Organisms across the biological spectra, from single cell bacteria to multicellular/multiorgan plants and animals, have evolved reflexive or reactive homeostatic mechanisms that work to respond to sudden behavioral or environmental homeostatic challenges. Organisms have also evolved anticipatory homeostatic mechanisms that alter the regulation of negative feedback mechanisms to prepare the organism for homeostatic challenges caused by predictable changes in the environment and behavior (Moore-Ede, 1986). Circadian rhythms are an example of an anticipatory homeostatic mechanism that enhance internal/organismal homeostatic capacities to predictable homeostatic challenges caused by 24-h rhythms in the environment and behavior. In this review, we use the framework of circadian homeostasis to understand how daily rhythms in behavior and the environment impact circadian mechanisms of homeostasis from the cellular/tissue to the whole organism level.
Circadian biologists work to understand how circadian homeostatic mechanisms promote a physiological advantage. Human lifestyles, which include high intensity light at night, shift work, travel across time zones, night time eating, etc. disrupt predictive behavioral and environmental entrainment cues that are critical for normal circadian homeostatic mechanisms (Bray et al., 2013). The disruption between behavior and environmental cycles has been linked to cancers, obesity, metabolic disease, and cardiovascular disease (West et al., 2017; Kelly et al., 2018; Maury, 2019; Resuehr et al., 2019; Shi et al., 2019; Delisle et al., 2021). Understanding the underlying mechanisms that disrupt circadian homeostatic mechanisms at the cellular and whole organism level will provide insight into how human lifestyles impact health and disease. Basic science studies in rats and mice show that feeding behavior in reference to the light cycle is central to the alignment of circadian homeostatic mechanisms at the cellular/tissue and whole organism level (Damiola et al., 2000; Bray et al., 2013; Schroder et al., 2014; Xin et al., 2021). These animal studies are exciting because they suggest that modifying feeding behavior might represent a powerful therapeutic tool to improve homeostasis and mitigate disease risks in people. In this review, we present the latest findings demonstrating how feeding behavior drives the timing of circadian homeostatic mechanisms with a focus on the heart. Importantly, we discuss how inverting normal feeding behavior leads to significant misalignment between canonical cellular circadian mechanisms in several tissues and physiological circadian rhythms central to homeostasis.
Time of Day Restricted Feeding and Molecular Rhythms
Circadian clocks serve a ubiquitous cellular timekeeping function that regulate the timing of physiological rhythms important for homeostasis. The canonical circadian clock mechanism expressed in most cells is a transcription translation feedback loop that cycles with a periodicity of about 24 h. In mammals, it is formed by four families of core clock genes [Bmal1, Clock, Per (Period), and Cry (Cryptochrome)] (Shearman et al., 2000; Mohawk et al., 2012; Buhr and Takahashi, 2013). BMAL1 and CLOCK are basic helix-loop-helix transcription factors that dimerize and initiate the transcription of the Per and Cry genes. Translated PER and CRY proteins repress BMAL1:CLOCK transactivation, and in so doing, decrease their own expression (Figure 1). As PER and CRY proteins are degraded, the cycle repeats. Components of the circadian clock also regulate the tissue-specific expression of genes outside the cellular timekeeping function and these genes, termed clock-controlled genes, are important for normal physiology (Panda et al., 2002a; Storch et al., 2002; Kornmann et al., 2007; Jeyaraj et al., 2012; Schroder et al., 2013). The importance of this molecular time keeping mechanism cannot be understated, because it is postulated to improve homeostasis by priming effector mechanisms prior to predictable behavioral and environmental challenges.
The location of the central clock in the suprachiasmatic nucleus (SCN) of the hypothalamus was first identified following surgical ablation of the SCN which resulted in arrhythmic behavior patterns (Stephan and Zucker, 1972). Light is the best characterized environmental time cue (zeitgeber), and light entrains the canonical circadian clock in the SCN. Light is transmitted to the SCN via the retinohypothalamic tract from the retina (Gooley et al., 2001; Panda et al., 2002b). The light entrained SCN synchronizes the clocks in peripheral tissues via neurohumoral cues, which in turn align the timing of the circadian clocks in the peripheral tissues to the light cycle. Mice are active and consume ∼80–90% of their calories during the dark cycle (Bray et al., 2013). It is now known that, in addition to light, the timing of feeding behavior can function to align or misalign canonical circadian clocks in the SCN and peripheral tissues (Damiola et al., 2000; Vollmers et al., 2009; Bray et al., 2013; Izumo et al., 2014; Schroder et al., 2014; Schroder et al., 2021; Xin et al., 2021; Zhang et al., 2021).
Time of day restricted feeding studies that restrict feeding behavior to the light (LRF) or dark (DRF) cycle demonstrate that restricting feeding behavior is a powerful technique to interrogate the interconnections between cellular circadian clocks, physiological circadian rhythms, metabolism, and the time of food (nutrient) intake. Restricted feeding studies that limit food availability to a certain time of day, for a certain period of time, have demonstrated that peripheral clocks respond to time of feeding with changes at both the molecular and physiological level to preserve homeostasis. DRF improved the phase alignment in rhythmic mRNA expression for Bmal1, Per2, and Dbp in the liver, kidney, and heart (Xin et al., 2021). (Figure 2) LRF, feeding mice when they are normally inactive, did not alter the phase of circadian clock signaling in the SCN, but LRF resulted in the misalignment and disruption in the rhythmic expression of clock genes in some (but not all) peripheral tissues (Bray et al., 2013; Xin et al., 2021). Compared to DRF, the phase in the rhythmic mRNA expression for Bmal1, Per2, and Dbp was only modestly impacted by LRF in heart or kidneys, but the phase of the rhythmic expression of Bmal1, Per2, and Dbp mRNA transcripts in the liver shifted by 10–12 h. Xin et al. is one of the few studies to examining sex as a variable in tissue specific gene expression. They found that although the liver clock and adipose clock maintained similar responses to inverted feeding between male and female mice, the kidney and heart clocks were less robust under LRF in male mice (Xin et al., 2021). So how does LRF cause a loss in the alignment or disruption in expression of peripheral clock genes?
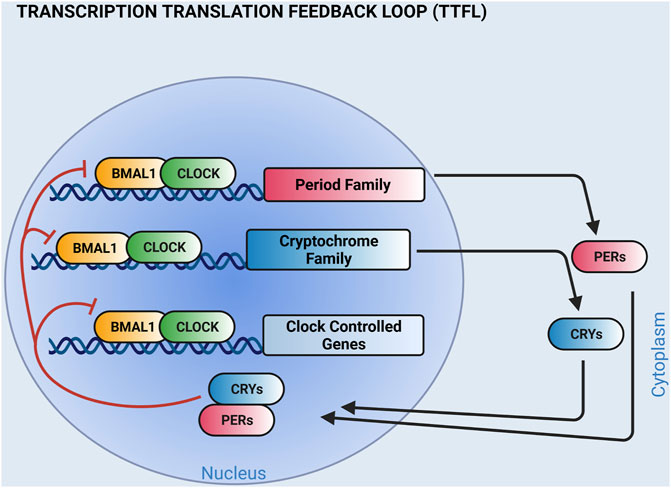
FIGURE 1. The molecular clock is a transcription translation feedback loop. Shown is a simplified cartoon of the circadian clock mechanism. BMAL1 and CLOCK transcription factor heterodimers bind E-boxes on the promoters of Period (PER 1,2, 3) and Cryptochrome (CRY 1, 2) family members to activate transcription. PER and CRY proteins dimerize and negatively feedback on BMAL1 and CLOCK. The circadian clock mechanism also regulates the tissue-specific expression of clock-controlled genes. Additional feedback loops that contribute to the circadian clock mechanism are not shown. (Created with BioRender.com).
The answer to how LRF results in phase misalignment or a disruption of peripheral clocks genes remains incomplete, but several clues can be found in earlier studies. One of which, employed Per1 deficient mouse embryonic fibroblasts (MEFs), which have a short period length in culture (assessed by Per2 gene expression), to examine entrainment in peripheral tissues (Pando et al., 2002). Collagen matrix embedded, Per1 deficient MEFs were implanted under the skin on the back of mice. After a few days, tissue (kidney, skeletal muscle, and MEF containing disks) was harvested from mice over a time course at 4-h intervals and Per2 mRNA in the implanted MEFs was shown to match the phase and period length of the host mouse isolated tissues. In addition, it was demonstrated that expression was food entrainable, most strikingly similar to that observed in the liver of the host mouse. More recent studies, employing long-term bioluminescence studies [RT-Biolumicorder (Saini et al., 2013)] in freely moving PER2:luciferase reporter mice, which express luciferase downstream of the Per2 promoter (Yoo et al., 2004), showed that the SCN was required for maintaining phase alignment between organs under constant conditions (ad libitum feeding and constant darkness). DRF and LRF in this study synchronized the phase of peripheral organs in whole body bioluminescence and activity rhythms of PER2:luciferase reporter mice, in both control and SCN ablated mice, with the phase of these rhythms shifting faster in the SCN ablated mice in response to LRF (Guan et al., 2020; Sinturel et al., 2021). Taken together, the aforementioned studies employing MEF implantation and whole animal bioluminescence suggest a complex regulation of feeding induced phase entrainment both systemically and within individual organs. The impact of feeding on phase likely involves regulatory mechanisms emanating from the SCN to synchronize gene expression between organs, as demonstrated by the bioluminescence studies, as well as, circulating and neurohumoral factors or metabolites, as demonstrated by the MEF implantation studies.
Identifying entrainment and food responsive re-entrainment factors for peripheral tissue gene expression has proven elusive. SCN regulated pathways including neurohumoral signaling (autonomic, glucocorticoids, insulin signaling, etc.) and daily changes in core body temperature have been shown to contribute (Balsalobre et al., 2000; Terazono et al., 2003; Buhr et al., 2010; Fougeray et al., 2022). One possible entrainment factor is glucocorticoid signaling. Glucocorticoid receptors are present in most cells in the body but not the SCN. Balsalobre et al. (2000) demonstrated that intraperitoneal injection of mice with dexamethasone, a glucocorticoid receptor agonist, resulted in phase shifts of Per1, Dbp and Reverb gene expression in peripheral tissues (Balsalobre et al., 2000). Of note, dexamethasone treatment is frequently used in circadian bioluminescence cell culture experiments to align molecular clocks (Izumo et al., 2006). Around the same time, Le Minh et al., showed that glucocorticoids slow clock gene phase shifts in the liver resulting from LRF, with little impact on the rate of clock gene phase resetting with a return to ad libitum conditions utilizing adrenalectomized and sham operated mice (Le Minh et al., 2001). These data suggested that glucocorticoid receptor signaling may serve to buffer the peripheral clock against phase shifts in response to transient alterations in feeding patterns and that differential expression/sensitivity of these receptors in tissues throughout the body may mediate changes in gene expression (Quatrini and Ugolini, 2021). Glucocorticoid receptor signaling may play a role in the apparent slow phase shifts observed in the circadian gene expression of peripheral tissues in response to LRF (7–10 days, liver) (Damiola et al., 2000). Another study demonstrated that circadian gene expression in the liver requires a functional insulin receptor to entrain the circadian clock mechanism to LRF (Fougeray et al., 2022). These data suggest an important role for insulin signaling/hepatocyte insulin sensitivity in regulating the liver circadian clock with important implications for maintaining blood glucose homeostasis, as well as how circadian disruption could effect metabolic homeostasis and contribute to metabolic disorders (e.g., metabolic disease, obesity and diabetes) (Fougeray et al., 2022). Future studies may identify tissue specific signaling mechanisms that regulate the circadian clock to alterations in the timing of feeding.
Time of Day Restricted Feeding and Circadian Rhythms: Canonical Circadian Clocks Not Required
Mouse models that are currently available to dissect the role of the molecular clock at the systems level and within individual tissues, suggest a direct link between the canonical circadian clock mechanism in the SCN, circadian clocks throughout the body, and circadian homeostasis of several homeostatically regulated physiological variables including mean arterial pressure (MAP). Studies utilizing SCN ablation, germline Bmal1 knock out mice (Bmal1−/−), or conditional systemic Bmal1−/− knock out mice show that these animals lose the circadian rhythm in MAP and HR (heart rate) (Yang et al., 2016).
MAP is normally homeostatically maintained within a narrowly defined range so organs and tissues can be perfused with blood to meet their metabolic demands (De Hert, 2012). The maintenance of MAP within constant range occurs by numerous nested negative feedback mechanisms that regulate cardiac output, total peripheral resistance, and blood volume (Nishida et al., 2012; Raven and Chapleau, 2014; Dampney, 2016). Central to the homeostatic regulation of MAP is heart rate and cardiac output (Richardson et al., 1998; De Hert, 2012). The importance of the heart as an effector in the homeostatic regulation of MAP is underscored by the fact that the loss of heart function causes sudden cardiac death, the worldwide leading cause of death (Srinivasan and Schilling, 2018). The autonomic nervous system dynamically regulates heart function on a beat-to-beat basis. Environmental and behavioral homeostatic challenges cause efferent sympathetic and parasympathetic-induced signaling to change cardiac output primarily through the modulation of HR. Like many other critically important homeostatic effector mechanisms, the autonomic nervous system not only changes HR in response to homeostatic challenges, but also in anticipation of daily environmental and behavioral cycles (Joyner and Limberg, 2014; Raven and Chapleau, 2014). It has been proposed that circadian homeostatic mechanisms prime effectors to improve their responsivity to predictable environmental and behavioral challenges. Normally basal HR increases in anticipation of the active cycle and decreases prior to the rest cycle. In the next section we discuss surprising new data that suggest the canonical circadian clocks in the SCN (i.e. the central clock) or heart do not appear to be directly responsible for priming the heart to generate these anticipatory rhythms in HR. Studies using LRF are now beginning to point to a role of another oscillator, perhaps the food entertainable oscillator (FEO), as being a driver in generating the circadian rhythm in HR in mice (Zhang et al., 2021).
Eat Your Heart Out Canonical Clocks
Lesioning the SCN (central clock) in the brain under ad libitum conditions results in a loss of circadian rhythm in HR in rats and mice (Sano et al., 1995; Scheer et al., 2001; Tong et al., 2013). A similar loss in the 24-h rhythm in HR is seen in mice undergoing both beta-adrenergic and muscarinic receptor blockade. These studies, along with the identification of neuronal pathways linking the SCN to autonomic nuclei, suggested that the circadian clock mechanism in the SCN modifies autonomic regulation to generate the circadian rhythm in HR. If true, then germline deletion of Bmal1 should also result in the loss of the circadian rhythm in the HR. Indeed, Bmal1−/− mice show a loss in the 24-h rhythms in sympathoadrenal function and HR (Lefta et al., 2012). The loss in the 24-h rhythm in HR was also seen in the inducible systemic deletion of Bmal1 in adult mice (Yang et al., 2016). To better understand how the canonical circadian clock in the heart may impact 24-h rhythms in HR, mouse models that disrupt the circadian clock mechanism by knocking out Bmal1 in a cardiac specific manner were developed (Schroder et al., 2013; Young et al., 2014; Yang et al., 2016; Zhang et al., 2021). Mouse models in which Bmal1 is deleted from the heart from birth or induced in adult cardiomyocytes showed a circadian and 24-h rhythm in HR. Taken together the data suggest the SCN and the canonical circadian clock mechanism outside the heart regulate autonomic activity to generate the daily rhythm in HR.
The model that the canonical clock in the SCN modifies autonomic signaling to generate the 24-h rhythm in HR was challenged when it was demonstrated that LRF shifted the phase in the 24-h rhythm of HR by ∼15 h but not the circadian clock mechanism in the SCN (Schroder et al., 2014). The shift in the 24-h rhythm in HR did not appear to be secondary to changes in the circadian clock in the heart, because LRF only caused a modest shift in the phase of the canonical circadian clock in the heart (Figure 2). Additional studies confirmed that the impact the LRF had on the phase of the 24-h rhythm in HR was not dependent on the canonical circadian clock mechanism in the heart, because mice in which the deletion of Bmal1 was induced in adult cardiomyocytes undergoing LRF showed a large phase shift in the 24-h rhythm of the HR similar to control mice (Schroder et al., 2021). Perhaps even more surprising were studies utilizing germline Bmal1−/− mice, which showed that the loss in the 24-h rhythm in HR was restored in mice undergoing LRF (Zhang et al., 2021). Together, the data now suggest a model, whereby, the canonical circadian clock mechanism is not needed to generate a 24-h rhythm in HR. The loss of the 24-h rhythm in HR after the ablation of the SCN or in the germline or inducible Bmal1 knock out mice may be attributed to arrhythmic feeding behaviors. The data suggest that the loss in the time of day feeding behavior in these mouse models might underlie the loss or damping of circadian rhythms in HR. There are several possibilities that might explain the dissociation between the phase in the 24-h rhythms of the canonical circadian clocks in the SCN, the heart, and other peripheral tissues and HR. One candidate is FEO. Consistent with the FEO contributing to the circadian rhythm in HR during LRF is that the circadian rhythm in HR aligns with time of feeding even in mice housed in constant darkness (free running conditions) (Zhang et al., 2021).
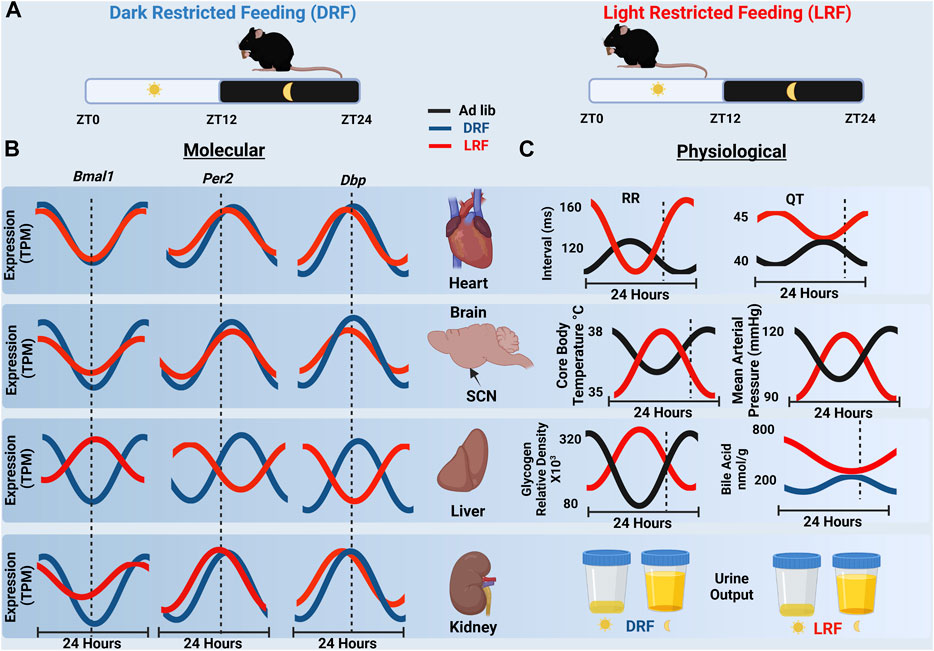
FIGURE 2. (A) Schematic representation of nighttime restricted feeding (DRF), left, and daytime restricted feeding (LRF), right. (B) A cartoon representation of RNA-seq expression data of Bmal1, Per2 and Dbp over 24 h in the heart, SCN, liver and kidney from mice undergoing DRF and LRF. The dashed line represents ZT12 which is the onset of dark cycle. Notice the ∼12-h shift in gene expression in the liver which is not present in the heart, SCN, or kidney. (TPM, transcripts per million) (Xin et al., 2021) (C) Organ specific physiological rhythms in response to ad libitum feeding, LRF or DRF are shown. RR and QT intervals rapidly shift in response to LRF (Schroder et al., 2014). Similar shifts were observed with core body temperature (10) and MAP (27) which are regulated by the SCN under ad libitum conditions. Liver rhythms in liver glycogen content (Pérez-Mendoza et al., 2014) and bile acid production (Ma et al., 2009) also shifted in response to LRF. With the exception of urine output (Zhang et al., 2021) all physiological rhythms shown shift by ∼12 h in response to LRF. (Created with BioRender.com).
Importantly, the LRF-induced dissociation between canonical circadian clock mechanisms in tissues and homeostatically regulated effector mechanisms and variables is not limited to the HR (Figure 2). There are several examples in mice undergoing LRF where phase of the 24-h rhythms in canonical circadian clocks in peripheral tissues stays closely aligned to the light cycle, but physiological rhythms align to feeding behavior. Examples include the MAP, core body temperature, liver glycogen content, and bile acid levels (but not urine output) (Figure 2). Future studies are needed to determine the impact that the FEO as well as the dissociation between canonical circadian clock signaling and these physiological rhythms have on overall homeostasis and health.
Discussion
Many studies have demonstrated the presence of the FEO, but the anatomical location and molecular underpinnings of the FEO are not yet known and understanding the role of FEO beyond food anticipatory activity is limited (Mendoza, 2007). Any discussion of HR being under the control of FEO is absent from the literature. In addition, genes responsible for the canonical molecular clock do not seem to impact FEO, as it remains intact in most circadian clock gene knockout models (Pendergast and Yamazaki, 2018).
Rodent models have demonstrated that feeding behavior in reference to the light cycle is central to the alignment of circadian homeostatic mechanisms and that LRF may cause misalignment between peripheral tissue molecular clocks and physiological rhythms important for systemic homeostasis. Components of this misalignment are likely present in a recent study examining a model of shift work in rats. They demonstrated that a restoration of circadian gene expression, with a lower amplitude, in the heart when food was restricted to non “work” hours was not sufficient to prevent significant cardiac fibrosis (Trott et al., 2022). Although a complete understanding of the impacts of feeding on homeostatic mechanisms at the molecular, cellular, tissue, organ and organismal level is currently unavailable, new scientific tools and animal models are beginning to offer some clarity.
Why don’t molecular rhythms phase shift to the same degree in all tissues in response to LRF? And why do phase shifts occur on a longer time scale (days to weeks), for instance genes in the heart and kidney do not shift very much while those in the liver shift ∼12 h after a week of LRF (Xin et al., 2021)? While the mechanisms responsible for these changes in gene expression may not yet be known perhaps this may reflect tissue specific roles of the molecular clock. For example, the phase of the circadian clock in the liver in mice shifts to align with feeding behavior in LRF mice. The shift in the liver circadian clock relies on the presence of hepatic insulin receptors (Fougeray et al., 2022). Tissues that have a limited phase shift like the heart and kidney are continuously functioning and gene expression may not shift as much as a result while the liver is highly metabolic and responsive to the presence of food, functioning to homeostatically regulate blood glucose, and may shift gene expression for appropriate food utilization. Peripheral tissues are subject to distinct homeostatic mechanisms and respond to different circulating metabolites and hormones secreted in response to feeding. For instance, the liver clock may shift in response to LRF because the liver is an effector for blood glucose, which is homeostatically regulated, to alter storage and release. This shift may initially be a reactive homeostatic response to the new time of feeding and then progress to an anticipatory homeostatic mechanism with prolonged restricted feeding.
In this review we focused primarily on HR because it is critical for cardiac output and loss of cardiac output results in sudden cardiac death. Since the 24-h rhythm in HR strongly depends on autonomic signaling, it might be that the FEO regulates the HR via the autonomic signaling. This could explain why LRF causes similar shifts in the phase of other physiological rhythms known to be impacted by the autonomic nervous system (e.g., MAP and body temperature). However, because MAP and core temperature are homeostatically regulated variables, their regulation is much more complex than HR. For example, MAP depends on the diameter of the arteries and veins, which impacts total peripheral resistance and blood distribution between the arteries and veins, as well as blood volume, which reflects the balance between fluid intake and fluid loss (passively or by the kidney).
Although the idea that FEO might drive the 24-h rhythm in HR is exciting, there is evidence that this might not be the case in all animal models. For example, recent LRF studies in rat show a very different type of dissociation between the canonical circadian clock in the heart and HR. Unlike mice, the phase in the canonical circadian clock in the heart realigns with feeding behavior (similar to the liver), but LRF for 5-days has less of an effect on the phase of the 24-h rhythm of HR than mice (Wu et al., 2010). Regardless, both mouse and rat animal models show LRF can cause dissociation between the phases of canonical circadian clocks in peripheral tissues and physiological rhythms. Understanding the mechanisms for this in different model systems will be important for future studies on reactive and circadian homeostasis.
The data strongly suggest that canonical circadian clock rhythms and physiological rhythms are regulated by distinct oscillator mechanisms. Lifestyles and disease that disrupt and misalign circadian and physiological rhythms have been shown to increase cardiovascular risk in humans (Scheer et al., 1999; Scheer et al., 2009; Chellappa et al., 2021). Understanding the impact that time of feeding has on cardiovascular homeostasis in animal models and humans will enable the development of novel targeted approaches to mitigate cardiovascular disease risk.
Author Contributions
ES and BD contributed and developed the concepts discussed. ES and BD wrote, edited, and prepared the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Heart Lung and Blood Institute grants R01HL153042 and R01HL141343.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Balsalobre, A., Brown, S. A., Marcacci, L., Tronche, F., Kellendonk, C., Reichardt, H. M., et al. (2000). Resetting of Circadian Time in Peripheral Tissues by Glucocorticoid Signaling. Science 289 (5488), 2344–2347. doi:10.1126/science.289.5488.2344
Bray, M. S., Ratcliffe, W. F., Grenett, M. H., Brewer, R. A., Gamble, K. L., and Young, M. E. (2013). Quantitative Analysis of Light-phase Restricted Feeding Reveals Metabolic Dyssynchrony in Mice. Int. J. Obes. (Lond) 37 (6), 843–852. doi:10.1038/ijo.2012.137
Buhr, E. D., and Takahashi, J. S. (2013). Molecular Components of the Mammalian Circadian Clock. Handb. Exp. Pharmacol. 217, 3–27. doi:10.1007/978-3-642-25950-0_1
Buhr, E. D., Yoo, S. H., and Takahashi, J. S. (2010). Temperature as a Universal Resetting Cue for Mammalian Circadian Oscillators. Science 330 (6002), 379–385. doi:10.1126/science.1195262
Chellappa, S. L., Qian, J., Vujovic, N., Morris, C. J., Nedeltcheva, A., Nguyen, H., et al. (2021). Daytime Eating Prevents Internal Circadian Misalignment and Glucose Intolerance in Night Work. Sci. Adv. 7 (49), eabg9910. doi:10.1126/sciadv.abg9910
Damiola, F., Le Minh, N., Preitner, N., Kornmann, B., Fleury-Olela, F., and Schibler, U. (2000). Restricted Feeding Uncouples Circadian Oscillators in Peripheral Tissues from the Central Pacemaker in the Suprachiasmatic Nucleus. Genes Dev. 14 (23), 2950–2961. doi:10.1101/gad.183500
Dampney, R. A. (2016). Central Neural Control of the Cardiovascular System: Current Perspectives. Adv. Physiol. Educ. 40 (3), 283–296. doi:10.1152/advan.00027.2016
De Hert, S. (2012). Physiology of Hemodynamic Homeostasis. Best. Pract. Res. Clin. Anaesthesiol. 26 (4), 409–419. doi:10.1016/j.bpa.2012.10.004
Delisle, B. P., George, A. L., Nerbonne, J. M., Bass, J. T., Ripplinger, C. M., Jain, M. K., et al. (2021). Understanding Circadian Mechanisms of Sudden Cardiac Death: A Report from the National Heart, Lung, and Blood Institute Workshop, Part 1: Basic and Translational Aspects. Circ Arrhythmia Electrophysiol. 14 (11), e010181. doi:10.1161/circep.121.010181
Fougeray, T., Polizzi, A., Régnier, M., Fougerat, A., Ellero-Simatos, S., Lippi, Y., et al. (2022). The Hepatocyte Insulin Receptor Is Required to Program the Liver Clock and Rhythmic Gene Expression. Cell Rep. 39 (2), 110674. doi:10.1016/j.celrep.2022.110674
Gooley, J. J., Lu, J., Chou, T. C., Scammell, T. E., and Saper, C. B. (2001). Melanopsin in Cells of Origin of the Retinohypothalamic Tract. Nat. Neurosci. 4 (12), 1165. doi:10.1038/nn768
Guan, D., Xiong, Y., Trinh, T. M., Xiao, Y., Hu, W., Jiang, C., et al. (2020). The Hepatocyte Clock and Feeding Control Chronophysiology of Multiple Liver Cell Types. Science 369 (6509), 1388–1394. doi:10.1126/science.aba8984
Izumo, M., Pejchal, M., Schook, A. C., Lange, R. P., Walisser, J. A., Sato, T. R., et al. (2014). Differential Effects of Light and Feeding on Circadian Organization of Peripheral Clocks in a Forebrain Bmal1 Mutant. Elife 3, 3. doi:10.7554/eLife.04617
Izumo, M., Sato, T. R., Straume, M., and Johnson, C. H. (2006). Quantitative Analyses of Circadian Gene Expression in Mammalian Cell Cultures. PLoS Comput. Biol. 2 (10), e136. doi:10.1371/journal.pcbi.0020136
Jeyaraj, D., Haldar, S. M., Wan, X., McCauley, M. D., Ripperger, J. A., Hu, K., et al. (2012). Circadian Rhythms Govern Cardiac Repolarization and Arrhythmogenesis. Nature 483 (7387), 96–99. doi:10.1038/nature10852
Joyner, M. J., and Limberg, J. K. (2014). Blood Pressure Regulation: Every Adaptation Is an Integration? Eur. J. Appl. Physiol. 114 (3), 445–450. doi:10.1007/s00421-013-2636-5
Kelly, R. M., Healy, U., Sreenan, S., McDermott, J. H., and Coogan, A. N. (2018). Clocks in the Clinic: Circadian Rhythms in Health and Disease. Postgrad. Med. J. 94 (1117), 653–658. doi:10.1136/postgradmedj-2018-135719
Kornmann, B., Schaad, O., Reinke, H., Saini, C., and Schibler, U. (2007). Regulation of Circadian Gene Expression in Liver by Systemic Signals and Hepatocyte Oscillators. Cold Spring Harb. Symp. Quant. Biol. 72, 319–330. doi:10.1101/sqb.2007.72.041
Le Minh, N., Damiola, F., Tronche, F., Schütz, G., and Schibler, U. (2001). Glucocorticoid Hormones Inhibit Food-Induced Phase-Shifting of Peripheral Circadian Oscillators. Embo J. 20 (24), 7128–7136. doi:10.1093/emboj/20.24.7128
Lefta, M., Campbell, K. S., Feng, H. Z., Jin, J. P., and Esser, K. A. (2012). Development of Dilated Cardiomyopathy in Bmal1-Deficient Mice. Am. J. Physiol. Heart Circ. Physiol. 303 (4), H475–H485. doi:10.1152/ajpheart.00238.2012
Ma, K., Xiao, R., Tseng, H. T., Shan, L., Fu, L., and Moore, D. D. (2009). Circadian Dysregulation Disrupts Bile Acid Homeostasis. PLoS One 4 (8), e6843. doi:10.1371/journal.pone.0006843
Maury, E. (2019). Off the Clock: From Circadian Disruption to Metabolic Disease. Int. J. Mol. Sci. 20 (7), 1597. doi:10.3390/ijms20071597
Mendoza, J. (2007). Circadian Clocks: Setting Time by Food. J. Neuroendocrinol. 19 (2), 127–137. doi:10.1111/j.1365-2826.2006.01510.x
Mohawk, J. A., Green, C. B., and Takahashi, J. S. (2012). Central and Peripheral Circadian Clocks in Mammals. Annu. Rev. Neurosci. 35, 445–462. doi:10.1146/annurev-neuro-060909-153128
Moore-Ede, M. C. (1986). Physiology of the Circadian Timing System: Predictive versus Reactive Homeostasis. Am. J. Physiol. 250 (5 Pt 2), R737–R752. doi:10.1152/ajpregu.1986.250.5.R737
Nishida, Y., Tandai-Hiruma, M., Kemuriyama, T., and Hagisawa, K. (2012). Long-term Blood Pressure Control: Is There a Set-point in the Brain? J. Physiol. Sci. 62 (3), 147–161. doi:10.1007/s12576-012-0192-0
Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., et al. (2002). Coordinated Transcription of Key Pathways in the Mouse by the Circadian Clock. Cell 109 (3), 307–320. doi:10.1016/s0092-8674(02)00722-5
Panda, S., Sato, T. K., Castrucci, A. M., Rollag, M. D., DeGrip, W. J., Hogenesch, J. B., et al. (2002). Melanopsin (Opn4) Requirement for Normal Light-Induced Circadian Phase Shifting. Science 298 (5601), 2213–2216. doi:10.1126/science.1076848
Pando, M. P., Morse, D., Cermakian, N., and Sassone-Corsi, P. (2002). Phenotypic Rescue of a Peripheral Clock Genetic Defect via SCN Hierarchical Dominance. Cell 110 (1), 107–117. doi:10.1016/s0092-8674(02)00803-6
Pendergast, J. S., and Yamazaki, S. (2018). The Mysterious Food-Entrainable Oscillator: Insights from Mutant and Engineered Mouse Models. J. Biol. Rhythms 33 (5), 458–474. doi:10.1177/0748730418789043
Pérez-Mendoza, M., Rivera-Zavala, J. B., and Díaz-Muñoz, M. (2014). Daytime Restricted Feeding Modifies the Daily Variations of Liver Gluconeogenesis: Adaptations in Biochemical and Endocrine Regulators. Chronobiology Int. 31 (7), 815–828. doi:10.3109/07420528.2014.908898
Quatrini, L., and Ugolini, S. (2021). New Insights into the Cell- and Tissue-Specificity of Glucocorticoid Actions. Cell Mol. Immunol. 18 (2), 269–278. doi:10.1038/s41423-020-00526-2
Raven, P. B., and Chapleau, M. W. (2014). Blood Pressure Regulation XI: Overview and Future Research Directions. Eur. J. Appl. Physiol. 114 (3), 579–586. doi:10.1007/s00421-014-2823-z
Resuehr, D., Wu, G., Johnson, R. L., Young, M. E., Hogenesch, J. B., and Gamble, K. L. (2019). Shift Work Disrupts Circadian Regulation of the Transcriptome in Hospital Nurses. J. Biol. Rhythms 34 (2), 167–177. doi:10.1177/0748730419826694
Richardson, D. C., Randall, D. C., and Speck, D. F. (1998). Cardiopulmonary System. Baltimore, MD: Fence Creek Publishing, LLC.
Saini, C., Liani, A., Curie, T., Gos, P., Kreppel, F., Emmenegger, Y., et al. (2013). Real-time Recording of Circadian Liver Gene Expression in Freely Moving Mice Reveals the Phase-Setting Behavior of Hepatocyte Clocks. Genes Dev. 27 (13), 1526–1536. doi:10.1101/gad.221374.113
Sano, H., Hayashi, H., Makino, M., Takezawa, H., Hirai, M., Saito, H., et al. (1995). Effects of Suprachiasmatic Lesions on Circadian Rhythms of Blood Pressure, Heart Rate and Locomotor Activity in the Rat. Jpn. Circ. J. 59 (8), 565–573. doi:10.1253/jcj.59.565
Scheer, F. A., Hilton, M. F., Mantzoros, C. S., and Shea, S. A. (2009). Adverse Metabolic and Cardiovascular Consequences of Circadian Misalignment. Proc. Natl. Acad. Sci. U. S. A. 106 (11), 4453–4458. doi:10.1073/pnas.0808180106
Scheer, F. A., Ter Horst, G. J., van Der Vliet, J., and Buijs, R. M. (2001). Physiological and Anatomic Evidence for Regulation of the Heart by Suprachiasmatic Nucleus in Rats. Am. J. Physiol. Heart Circ. Physiol. 280 (3), H1391–H1399. doi:10.1152/ajpheart.2001.280.3.H1391
Scheer, F. A., van Doornen, L. J., and Buijs, R. M. (1999). Light and Diurnal Cycle Affect Human Heart Rate: Possible Role for the Circadian Pacemaker. J. Biol. Rhythms 14 (3), 202–212. doi:10.1177/074873099129000614
Schroder, E. A., Burgess, D. E., Manning, C. L., Zhao, Y., Moss, A. J., Patwardhan, A., et al. (2014). Light Phase-Restricted Feeding Slows Basal Heart Rate to Exaggerate the Type-3 Long QT Syndrome Phenotype in Mice. Am. J. Physiol. Heart Circ. Physiol. 307 (12), H1777–H1785. doi:10.1152/ajpheart.00341.2014
Schroder, E. A., Lefta, M., Zhang, X., Bartos, D. C., Feng, H. Z., Zhao, Y., et al. (2013). The Cardiomyocyte Molecular Clock, Regulation of Scn5a, and Arrhythmia Susceptibility. Am. J. Physiol. Cell Physiol. 304 (10), C954–C965. doi:10.1152/ajpcell.00383.2012
Schroder, E. A., Burgess, D. E., Johnson, S. R., Ono, M., Seward, T., Elayi, C. S., et al. (2021). Timing of Food Intake in Mice Unmasks a Role for the Cardiomyocyte Circadian Clock Mechanism in Limiting QT-Interval Prolongation. Chronobiology Int. 39, 525–534. doi:10.1080/07420528.2021.2011307
Shearman, L. P., Sriram, S., Weaver, D. R., Maywood, E. S., Chaves, I., Zheng, B., et al. (2000). Interacting Molecular Loops in the Mammalian Circadian Clock. Science 288 (5468), 1013–1019. doi:10.1126/science.288.5468.1013
Shi, D., Chen, J., Wang, J., Yao, J., Huang, Y., Zhang, G., et al. (2019). Circadian Clock Genes in the Metabolism of Non-alcoholic Fatty Liver Disease. Front. Physiol. 10, 423. doi:10.3389/fphys.2019.00423
Sinturel, F., Gos, P., Petrenko, V., Hagedorn, C., Kreppel, F., Storch, K.-F., et al. (2021). Circadian Hepatocyte Clocks Keep Synchrony in the Absence of a Master Pacemaker in the Suprachiasmatic Nucleus or Other Extrahepatic Clocks. Genes Dev. 35 (5-6), 329–334. doi:10.1101/gad.346460.120
Srinivasan, N. T., and Schilling, R. J. (2018). Sudden Cardiac Death and Arrhythmias. Arrhythm. Electrophysiol. Rev. 7 (2), 111–117. doi:10.15420/aer.2018:15:2
Stephan, F. K., and Zucker, I. (1972). Circadian Rhythms in Drinking Behavior and Locomotor Activity of Rats Are Eliminated by Hypothalamic Lesions. Proc. Natl. Acad. Sci. U. S. A. 69 (6), 1583–1586. doi:10.1073/pnas.69.6.1583
Storch, K. F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F. C., Wong, W. H., et al. (2002). Extensive and Divergent Circadian Gene Expression in Liver and Heart. Nature 417 (6884), 78–83. doi:10.1038/nature744
Terazono, H., Mutoh, T., Yamaguchi, S., Kobayashi, M., Akiyama, M., Udo, R., et al. (2003). Adrenergic Regulation of Clock Gene Expression in Mouse Liver. Proc. Natl. Acad. Sci. U. S. A. 100 (11), 6795–6800. doi:10.1073/pnas.0936797100
Tong, M., Watanabe, E., Yamamoto, N., Nagahata-Ishiguro, M., Maemura, K., Takeda, N., et al. (2013). Circadian Expressions of Cardiac Ion Channel Genes in Mouse Might Be Associated with the Central Clock in the SCN but Not the Peripheral Clock in the Heart. Biol. Rhythm Res. 44 (4), 519–530. doi:10.1080/09291016.2012.704801
Trott, A. J., Greenwell, B. J., Karhadkar, T. R., Guerrero-Vargas, N. N., Escobar, C., Buijs, R. M., et al. (2022). Lack of Food Intake during Shift Work Alters the Heart Transcriptome and Leads to Cardiac Tissue Fibrosis and Inflammation in Rats. BMC Biol. 20 (1), 58. doi:10.1186/s12915-022-01256-9
Vollmers, C., Gill, S., DiTacchio, L., Pulivarthy, S. R., Le, H. D., and Panda, S. (2009). Time of Feeding and the Intrinsic Circadian Clock Drive Rhythms in Hepatic Gene Expression. Proc. Natl. Acad. Sci. U. S. A. 106 (50), 21453–21458. doi:10.1073/pnas.0909591106
West, A. C., Smith, L., Ray, D. W., Loudon, A. S. I., Brown, T. M., and Bechtold, D. A. (2017). Misalignment with the External Light Environment Drives Metabolic and Cardiac Dysfunction. Nat. Commun. 8 (1), 417. doi:10.1038/s41467-017-00462-2
Wu, T., Ni, Y., Zhuge, F., and Fu, Z. (2010). Resetting Process of Peripheral Circadian Gene Expression after the Combined Reversal of Feeding Schedule and Light/dark Cycle via a 24-h Light Period Transition in Rats. Physiol. Res. 59 (4), 581–590. doi:10.33549/physiolres.931818
Xin, H., Deng, F., Zhou, M., Huang, R., Ma, X., Tian, H., et al. (2021). A Multi-Tissue Multi-Omics Analysis Reveals Distinct Kineztics in Entrainment of Diurnal Transcriptomes by Inverted Feeding. iScience 24 (4), 102335. doi:10.1016/j.isci.2021.102335
Yang, G., Chen, L., Grant, G. R., Paschos, G., Song, W. L., Musiek, E. S., et al. (2016). Timing of Expression of the Core Clock Gene Bmal1 Influences its Effects on Aging and Survival. Sci. Transl. Med. 8 (324), 324ra16. doi:10.1126/scitranslmed.aad3305
Yoo, S. H., Yamazaki, S., Lowrey, P. L., Shimomura, K., Ko, C. H., Buhr, E. D., et al. (2004). PERIOD2::LUCIFERASE Real-Time Reporting of Circadian Dynamics Reveals Persistent Circadian Oscillations in Mouse Peripheral Tissues. Proc. Natl. Acad. Sci. U. S. A. 101 (15), 5339–5346. doi:10.1073/pnas.0308709101
Young, M. E., Brewer, R. A., Peliciari-Garcia, R. A., Collins, H. E., He, L., Birky, T. L., et al. (2014). Cardiomyocyte-specific BMAL1 Plays Critical Roles in Metabolism, Signaling, and Maintenance of Contractile Function of the Heart. J. Biol. Rhythms 29 (4), 257–276. doi:10.1177/0748730414543141
Keywords: circadian rhythm, heart, homeostasis, feeding, light cycle, heart rate
Citation: Schroder EA and Delisle BP (2022) Time Restricted Feeding to the Light Cycle Dissociates Canonical Circadian Clocks and Physiological Rhythms in Heart Rate. Front. Pharmacol. 13:910195. doi: 10.3389/fphar.2022.910195
Received: 01 April 2022; Accepted: 28 April 2022;
Published: 12 May 2022.
Edited by:
Nelson Chong, Nottingham Trent University, United KingdomReviewed by:
Michelle L. Gumz, University of Florida, United StatesCarolina Escobar, National Autonomous University of Mexico, Mexico
Copyright © 2022 Schroder and Delisle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth A. Schroder, eschr0@uky.edu; Brian P. Delisle, brian.delisle@uky.edu
 Elizabeth A. Schroder
Elizabeth A. Schroder