- 1Department of Gastrointestinal and Anorectal Surgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing Medical University, Chongqing, China
- 2Department of Neuroscience, Legacy Research Institute, Portland, OR, United States
- 3Integrative Physiology and Neuroscience, Washington State University, Vancouver, WA, United States
- 4Legacy Tumor Bank, Legacy Research Institute, Portland, OR, United States
- 5Mid-Columbia Medical Center, The Dalles, OR, United States
Methylation is an important mechanism contributing to cancer pathology. Methylation of tumor suppressor genes and oncogenes has been closely associated with tumor occurrence and development. New insights regarding the potential role of the adenosine receptor-independent pathway in the epigenetic modulation of DNA methylation offer the possibility of new interventional strategies for cancer therapy. Targeting DNA methylation of cancer-related genes is a promising therapeutic strategy; drugs like 5-Aza-2′-deoxycytidine (5-AZA-CdR, decitabine) effectively reverse DNA methylation and cancer cell growth. However, current anti-methylation (or methylation modifiers) are associated with severe side effects; thus, there is an urgent need for safer and more specific inhibitors of DNA methylation (or DNA methylation modifiers). The adenosine signaling pathway is reported to be involved in cancer pathology and participates in the development of tumors by altering DNA methylation. Most recently, an adenosine metabolic clearance enzyme, adenosine kinase (ADK), has been shown to influence methylation on tumor suppressor genes and tumor development and progression. This review article focuses on recent updates on ADK and its two isoforms, and its actions in adenosine receptor-independent pathways, including methylation modification and epigenetic changes in cancer pathology.
1 Introduction
The relationship between cancer and DNA methylation was first described by Feinberg and Vogelstein, who revealed that changes in DNA methylation promote the development of invasive colorectal cancer (Feinberg and Vogelstein, 1983). This led to the hypothesis that epigenetic silencing of tumor suppressors promotes carcinogenesis, as well as the finding that reversing this silencing suppresses tumor growth and may prevent tumorigenesis (Feinberg and Vogelstein, 1983). Aberrant DNA methylation has been confirmed to influence the development of numerous human cancers (Nejman et al., 2014; Sun et al., 2014). DNA hypermethylation in cancer cells has been studied most extensively as targeting promoter regions, especially the tumor suppressor genes. The promoter region of tumor suppressor genes is structurally rich in CpG and focal hypermethylation often occurs in its promoter region (López-Moyado et al., 2019), which leads to gene silencing, genomic instability, cell apoptosis, altered DNA repair, and cell cycle control (Wu and Bekaii-Saab, 2012). Hypermethylation inactivates the transcription of tumor suppressor genes, but it does not change the sequence of the gene itself. The methylation process and status can potentially be reversed and regulated.
DNA methylation utilizes methyl from S-adenosylmethionine (SAM). DNA methyltransferase (DNMT) catalyzes DNA methylation by transferring the methyl group from SAM to a target adenine or cytosine at a specific DNA site (Zhao et al., 2015), SAM is thus irreversibly converted to S- adenosylhomocysteine (SAH). SAH is then converted into adenosine and homocysteine (Hcy) by S-adenosylhomocysteine hydrolase (SAHH). Studies showed that increased downstream adenosine product can reversely influent the SAH to Hcy and transmethylation. Blockade of an adenosine metabolic enzyme, adenosine kinase (ADK) results in reduced adenosine removal and causes adenosine accumulation, and also elevates SAH level (Boison et al., 2002); the increased SAH, as a potent inhibitor of all DNMT, allows reversal of aberrant DNA methylation and expression of antioncogene (James et al., 2002).
Of note, adenosine, as an essential biological molecule of life, plays an important role in various aspects of cancer pathology, such as tumor immunity, tissue ischemia, hypoxia, revascularization, and apoptosis (Fishman et al., 2009a; Antonioli et al., 2013). Adenosine can conduct its manipulatory effects via the G protein-coupled four subtypes of adenosine receptors, i.e., adenosine A1, A2A, A2B, and A3 receptors (A1R, A2AR, A2BR, and A3R) (Fredholm et al., 2005; Jacobson, 2009). The activation of adenosine receptors is primarily determined by the availability of extracellular levels of adenosine. In addition to the aforementioned receptor-dependent actions, adenosine also yields receptor-independent actions, which rely on metabolic and intracellular levels and the metabolism of adenosine (Boison and Yegutkin, 2019). ADK plays a crucial role in the regulation of both extracellular and intracellular adenosine levels (Jacobson and Reitman, 2020) and adenosine receptor-dependent and independent pathways, in coordination with other adenosine metabolizing enzymes (Boison and Yegutkin, 2019). We will briefly review adenosine metabolism with a focus on the relationship between receptor-independent pathways of adenosine and DNA methylation in cancer.
2 Deoxyribonucleic Acid Hypermethylation in Tumor-Suppressor Genes
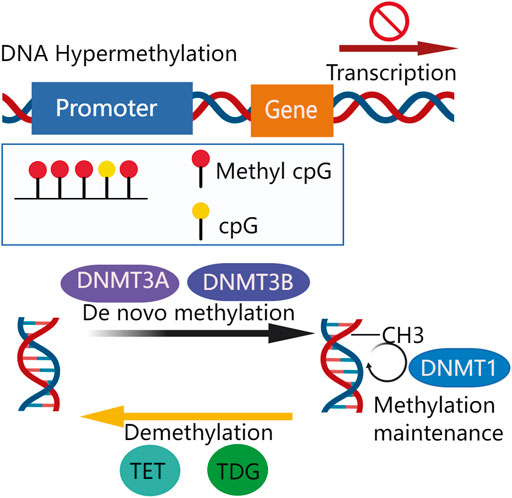
DNA methylation, one of the most abundant epigenetic modifications modulates gene expression and affects cellular processes of metabolism, survival, proliferation, and apoptosis, among others. (Weber et al., 2007; Baylin et al., 2001). Methylation occurs on cytosines within dinucleotide CpG islands (CGIs) which are rich in CpG and usually located at the promoter regions of genes (Oates et al., 2006). It is commonly associated with a transcriptionally repressed status. However, methylation-dependent transcriptional changes can result in both gain and loss of function depending on the gene region affected (Weber et al., 2007). DNA methylation consists of two functionally overlapped aspects: de novo and maintenance methylation. A new DNA methylation commonly yields 5-methylcytosine (5-mC), which is established by transferring the methyl group from S-adenosylmethionine (SAM) to cytosine at a CpG site by DNA methyltransferases DNMT3A and DNMT3B (Egger et al., 2006; Hung and Shen, 2003). DNMT3A and DNMT3B mediate de novo DNA methylation that does not require a DNA template with preexisting methylation (Okano et al., 1999) whereas DNMT1 contributes to maintaining methylation that involves replicating methylation patterns into a newly-synthesized DNA strand (Goyal et al., 2006). On the other hand, a demethylation system also exists, which includes ten-eleven translocation methylcytosine dioxygenases (TETs) and thymine DNA glycosylase (TDG)-base excision repair (BER) (Pan et al., 2017). The TETs catalyze the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine, and its downstream oxidation products: 5-formylcytosine and 5-carboxylcytosine, are removed by TDG of BER (Figure 1). Both methylation and demethylation systems contribute to the dynamically balanced methylation status of the genome (Weber et al., 2007).
De novo methylation is mediated by DNMT3A and DNMT3B to transfer methyl group (-CH3). Methylation is maintained by DNMT1. Demethylation of DNA is mediated by TET, TDG, and BER. A certain extent of promoter CpG island methylation impairs transcription, silencing gene expression.
Alternation in DNA methylation patterns is of importance in cancer pathology without affecting genome editing (Feinberg and Tycko, 2004) while DNA both hypermethylation and hypomethylation are seen in cancers (Das and Singal, 2004; Franco et al., 2008; Sinčić and Herceg, 2011). Cancer-associated methylome alterations are attributable to expressional changes of DNMTs (Morey et al., 2006; Gao et al., 2013; Micevic et al., 2017), which can result in increased genomic instability, expression of oncogenes, and/or decreased expression of tumor suppressor genes (Zhang et al., 2017; Valencia and Kadoch, 2019). Specifically, hypomethylation commonly occurs in oncogenes during cancer development and has been extensively reviewed (Mendizabal et al., 2017); in contrast, DNA hypermethylation is mostly found in tumor suppressor genes (Su et al., 2018). In the present review, we will focus on hypermethylation of tumor suppressor genes and possible adenosine regulations.
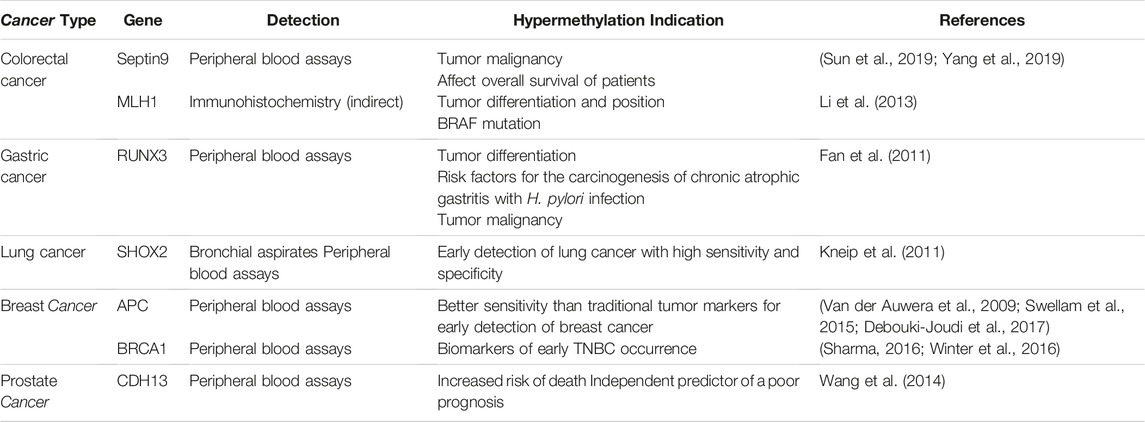
Hypermethylation resulting in epigenetic silencing was first demonstrated in the studies of retinoblastoma patients, in which hypermethylation was discovered in the promoter of the retinoblastoma tumor-suppressor (RB1) gene (Greger et al., 1989). Since then, a large number of tumor-suppressor genes have been identified as being silenced by DNA hypermethylation in tumorigenesis of different cancers. In colorectal cancers: 1) a cytokinesis-related gene Septin9 was identified highly correlated with the occurrence and development of colorectal cancer (Tanaka et al., 2002) and DNA methylation is the main mechanism regulating Septin9 gene expression (Sellin et al., 2011; Connolly et al., 2011), which mediates cytokinesis failure, leading to aneuploidy, centrosome amplification, and multipolar mitosis, eventually cause cell division and carcinogenesis (Sun et al., 2019; Cortez et al., 2016). In addition, the methylation level of the Septin9 gene is also considered to have clinical guiding significance due to the correlation with malignancy (Sun et al., 2019; Bae et al., 2017) and the overall survival of patients (Yang et al., 2019). Methylation of Septin9 in peripheral blood is the first blood DNA methylation marker approved by the US Food and Drug Administration (FDA) for CRC screening (Church et al., 2014), and is now widely used as a colorectal cancer biomarker (Xie et al., 2018). 2) MLH1, as the homolog of MutL, the main protein of the mismatch repair (MMR) system (Gelsomino et al., 2016), is silenced due to the hypermethylation of its promoter (Liu et al., 2017), resulting in deficient mismatch repair (dMMR) (Yamamoto and Imai, 2015). The replication errors of microsatellites (MS) cannot be corrected and accumulate continuously, resulting in microsatellite instability (MSI). Significance correlations were found in MLH1 promoter methylation and gender, tumor position, tumor differentiation, MSI, MLH1 protein expression, and v-RAF murine sarcoma viral oncogene homolog B1(BRAF) mutation in CRC patients (Li et al., 2013). In gastric cancer: runt-related transcription factor 3 (RUNX3) is an important downstream target of transforming growth factor-beta (TGFb) superfamily signaling, CpG silencing in the promoter region of regulated genes by hypermethylation is thought to be one of the mechanisms leading to loss of gene function (Fan et al., 2011). Through the detection of plasma samples, RUNX3 methylation level was considered to be a risk factor for gastric cancer metastasis and a potential indicator of gastric cancer progression (Fan et al., 2011). In breast cancer, the following genes are described: 1) ataxia-telangiectasia mutation (ATM) gene, a tumor suppressor plays a crucial role in maintaining genome integrity by activating cell cycle checkpoints and promoting the repair of DNA double-strand breaks (Wengner et al., 2020). Hypermethylation in ATM gene promoter downregulates ATM mRNA expression and positively correlates with increased tumor size and advanced disease stages III and IV (Begam et al., 2017; Cao et al., 2018). 2) a DNA repair gene, breast cancer 1 (BRCA1) - when a pathogenic mutation occurs, resulting in homologous recombination deficiency, the damaged DNA is difficult to repair, and it has been proved to easily lead to malignant tumors such as triple-negative breast cancer (TNBC) (Sharma, 2016). By comprehensively comparing the molecular biological characteristics of TNBC patients with BRCA1 hypermethylation and BRCA1 mutation, Dominik Glodzik et al. found the frequency of BRCA1 promoter hypermethylation correlates with clinicopathological variables, molecular subtypes, and patient outcomes in the early-stage of TNBC. This study indicated hypermethylation of the BRCA1 promoter region as a potential biomarker of early TNBC occurrence (Glodzik et al., 2020).
Together, the evidence indicates that DNA hypermethylation in the promoter region of tumor suppressors plays a crucial role in tumorigenesis, which is an epigenetic hallmark of various types of cancer. Table 1 lists representative tumor suppression genes with hypermethylation in their promoters. Indeed, the demethylation treatment strategy was proposed after discovering abnormal hypermethylation in tumors and researchers started the attempt to reverse hypermethylation (Issa, 2007). 5-Aza-2′-deoxycytidine (5-AZA-CdR, decitabine) (Karahoca and Momparler, 2013) was shown to have the ability to reverse DNA methylation, activate tumor suppressor genes, and promote apoptosis (Flohr and Breull, 1975), with possible mechanisms relied on the inhibition of DNMT1 (Chen et al., 2019). In a xenograft mouse model bearing the colon cancer line, HCT116, the 5-AZA-CdR was shown to demethylate the CDH13 gene, restoring its expression, resulting in a suppression of tumor growth (Ren and Huo, 2012). However, related experiments confirmed that gene re-expression in response to 5-AZA-CdR was transient and re-silenced upon drug removal (Bender et al., 1998; Egger et al., 2007). Besides, studies have also pointed out that 5-AZA-CdR treatment has always been interpreted with caution since the 5-AZA-CdR treatment can non-selectively affect the entire genome (Christman, 2002; Sigalotti et al., 2014). The non-selective demethylation yielded from 5-AZA-CdR may trigger serious adverse reactions, which limit its clinical use. Thus, methylation inhibitors with fewer side effects and higher selectivity on cancer cells are of interest for development.
3 Adenosine Regulations in Cancer
Adenosine is an endogenous purine nucleoside and an intermediary metabolite in DNA methylation. Adenosine accumulation has been observed in tumor tissues, which is associated with tumor growth, invasion, metastasis, and immune evasion in tumor pathology (Mastelic-Gavillet et al., 2019; Borodovsky et al., 2020; Wang et al., 2021). Adenosine has immunosuppressive effects on intratumoral immune populations (Stagg and Smyth, 2010). It can bind cell surface receptors and is secreted in a paracrine or autocrine manner or reverse regulate DNA methylation through substrate accumulation, thus exerting its biological effect. Major pathways regarding adenosine production, metabolic removal, and transportation across the cell membranes have been extensively reviewed otherwise (Boison and Yegutkin, 2019), we briefly summarize them as follows.
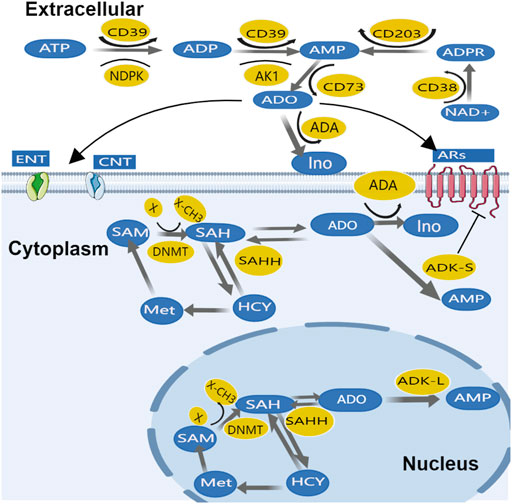
Adenosine production and transportation in cancer tissues are similar to physiological conditions; extracellular ATP and ADP can rapidly metabolize to adenosine monophosphate (AMP) majorly through two steps of dephosphorylation: 1) The first step, ATP and ADP are both converted to AMP by ecto-nucleoside triphosphate diphosphohydrolase-1 (CD39); then 2) AMP can generate adenosine by the final dephosphorylation reaction catalyzed by the enzyme ecto-5′-nucleotidase (CD73) (Fishman et al., 2009b) - this called CD39/CD73 pathway. Alternatively, cyclic ADP ribose hydrolase (CD38) can convert adenosine diphosphate ribose (ADPR) to AMP, this process can be regulated by ecto-nucleotide pyrophosphatase/phosphodiesterase 1, NPP1 (CD203a) (Gazzoli et al., 2002; Häusler et al., 2011). Afterward, CD73 converts AMP into adenosine–called CD38/CD203a pathway. In adenosine transportation across membranes, equilibrative nucleoside transporter (ENT) and concentrative nucleoside transporters (CNTs) play important roles (Song et al., 2017); Adenosine removal differs between intracellular and extracellular. Extracellular adenosine is converted to inosine by adenosine deaminase (ADA), which is widely expressed in the plasma as well as on the cell membrane. Inosine is then derivatized (removed from ribose) by purine nucleoside phosphorylase (PNP), which converts it to hypoxanthine. It is worth noting that ADA not only metabolizes adenosine, it also allosterically modulates ARs, resulting in a positive effect of amplifying downstream signals (Borea et al., 2018) including 1) enhanced AR1 sensitivity to adenosine (SU Xiaoyang, 2018); 2) interaction of ADA-CD26 complex in T cells with ADA-anchored protein in dendritic cells enhanced T cell proliferation (Pacheco et al., 2005), etc.
While the metabolism of intracellular adenosine is mainly dominated by ADK. The major adenosine removal enzyme ADK has two isoforms with distinguished subcellular expression patterns; while ADK short isoform (ADK-S) is expressed dominantly in cytosolic space, ADK long isoform (ADK-L) is solely located in the nuclei (Cui et al., 2009; Fedele et al., 2005). Intracellular adenosine is mainly removed by ADK-S, which converts adenosine to AMP (Boison and Yegutkin, 2019). Adenosine can also be directly inactivated on the cell surface by adenosine deaminase (ADA). In addition, adenosine metabolism also depends on adenosine phosphoribosyltransferase (APRT) to catalyze adenine reaction with ribose 1-phosphate to generate phosphate and adenosine in the nucleus. However, when energy consumption increases and/or energy supply is compromised, ATP is converted into AMP by adenylate kinase-1 (AK1) and nucleotide diphosphate kinase (NDPK), and then dephosphorylated into adenosine by 5-nucleotidase (Eltzschig et al., 2012). This process promotes extracellular ATP regeneration through a reversible phosphonate transfer reaction (Boison, 2013). The nucleoside transporters and adenosine removal enzymes maintain a dynamic balance between extracellular and intracellular adenosine (Figure 2). Due to mitochondria being the main source of ATP, mitochondrial bioenergy is related to adenosine homeostasis (Ashar et al., 2017).
Moreover, adenosine metabolism is a part of the transmethylation pathway, in which DNA can be methylated by DNMTs while SAM donates methyl group (-CH3) via a methyltransferase (MT) - catalyzed transmethylation reaction (Figure 2). Then, the SAM converted SAH is hydrolyzed to adenosine and Hcy by SAHH. Interestingly, the nuclear form of ADK-L drives methyl flux, enhancing DNA and histone methylation (Yegutkin, 2014).
Extracellular adenosine turnover is mediated by AR, ENT, and CNT. Factors that mediate adenosine production and removal include the enzymes CD39, CD73, ADK, and ADA. Additionally, intracellular adenosine metabolism depends on the cytoplasmic form of ADK-S and ADA. In the nucleus adenosine is part of the transmethylation pathway in which DNA is methylated by DNMT. ADK-L participates in driving the methyl groups through the transmethylation pathway affecting DNA and histone methylations. For the sake of clarity, only the most important enzymes are mentioned.
4 Adenosine Receptor-Dependent Pathway in Cancer
Substantial evidence indicates that adenosine mediates its physiological effects (Borea et al., 2018) as well as its pathophysiological actions in cancer (Fishman et al., 2009a; Franco et al., 2021) through the activation of four adenosine receptors (ARs), i.e., A1R, A2AR, A2BR, and A3R. Activation of ARs by specific ligands, agonists, or antagonists will regulate the occurrence and development of tumors through a series of signaling pathways (Borea et al., 2018; Franco et al., 2021). A1R has been studied mainly in glioblastoma (Synowitz et al., 2006; Fishman et al., 2009a), where A1R activation on microglia/macrophages in the tumor suppresses not only the production of cytokines such as interleukin-1β but also stromal metalloproteinase (MMP) (Tsutsui et al., 2004). Based on that, A1R is thought to have the effect of inhibiting tumor growth (Synowitz et al., 2006). Besides, what cannot be ignored is the important role of ARs in tumor immunity. In the tumor microenvironment, adenosine suppresses antitumor immunity, essentially through A2AR and A2BR (Buisseret et al., 2018). In particular, the A2AR, due to the high concentration of Ado in the tumor microenvironment, activates Gs-coupled A2AR and leads to an increase in cAMP, thereby inhibiting the activation of tumor lymphocytes (Fishman et al., 2009a; Merighi et al., 2019). Therefore, selective antagonism of A2AR can reduce cAMP levels, thereby enabling lymphocytes to effectively fight tumor cells (Franco et al., 2021). So far, a large number of clinical trials on A2AR/A2BR antagonists are also in progress (Franco et al., 2021). On the other hand, adenosine was observed to increase HIF1α protein accumulation under hypoxia situations through cell surface A3R interaction in various tumors (Merighi et al., 2005), and HIF1α plays an important role in tumor VEGF expression and angiogenesis (Merighi et al., 2005). Based on the relationship between tumor, hypoxia, and adenosine concentrations, A3R antagonists are considered to have a potential role in cancer therapy (Franco et al., 2021). Adenosine receptor-dependent pathway in cancer was already described in detail by Pier Andrea (Borea et al., 2018).
5 Adenosine Receptor-independent Pathway With Deoxyribonucleic Acid Methylation in Cancer
As an ATP metabolite, adenosine is released by all cell types and is shown to accumulate in tumor cells, which is associated with increased angiogenesis, high metabolism rate, and compromised hypoxia of the microenvironment (Losenkova et al., 2020). Accumulation of adenosine in the tumor microenvironment (TME) (de Lera Ruiz et al., 2014) has been proven to play an important role in tumor immunity, high concentrations of adenosine inhibit tumor immune effects (Ohta and Sitkovsky, 2001; Ohta et al., 2006; Ohta, 2016) and facilitate angiogenesis (van de Veen et al., 2020), which offers the possibility of targeting adenosine in cancer pathology and manipulation of adenosine actions represents a potential anti-cancer strategy. Meanwhile, solid tumors can maintain adenosine gradients - the adenosine levels in the tumor center are higher than in the peripheral area of the tumor (Ohta et al., 2006). High levels of adenosine are shown to hinder tumor growth and proliferation. For instance, peripheral tumor cells located in the parenchyma and stroma have been shown to have high proliferative and invasive abilities (Seetulsingh-Goorah, 2006) and their proliferation can be suppressed by adenosine (Seetulsingh-Goorah, 2006; Schiedel et al., 2013). Based on that, Sanna S. Virtanen et al. found adenosine with relatively high (10 μmol/L for the former and 50 μmol/L for the latter) concentrations showed the ability to inhibit tumor invasion and migration (Schiedel et al., 2013). Besides, incubation of human prostate carcinoma cell line PC-3 cells triggered a concentration-dependent increase in cAMP levels with increasing adenosine concentrations. However, in the presence of A2BR-selective antagonists, no changes in cAMP levels were observed (Schiedel et al., 2013). In addition, in a study on glioblastoma, Helena Marcelino et al. found that proliferation/viability of glioblastoma cells was significantly reduced after 30 μM doses of adenosine for three consecutive days. At the same time, the cocktail of adenosine receptor antagonists (Fredholm et al., 2001) was administered, but the tumor suppressor effect was not affected (Marcelino et al., 2021).
The above described discrepant effects of adenosine on pro-and anti-tumor cell growth suggest a possible involvement of multiple mechanisms. In other words, its inhibitory effect on proliferation is proposed beyond receptor-mediated adenosine activity (Virtanen et al., 2014), though the underlying mechanisms remain unclear. Possible metabolic contributors that determine high-adenosine level mediated inhibition may include extracellular adenosine deaminase activity, subsequent cellular uptake, interconversion of transported nucleosides, simultaneous inhibition of multiple protein kinases (Virtanen et al., 2014), as well as ADK actions. However, the potential involvement of multiple pathways in adenosine production, transportation, and metabolism, suggests the complexity of adenosine’s effect on tumor pathology.
Importantly, the metabolism of adenosine also affects the methylation process. When SAM/SAH is an important source of adenosine, it can reverse regulate DNA methylation through the substrate accumulation effect (Kloor and Osswald, 2004; Viré et al., 2006). Kai X et al., by observing the effects of different concentrations of adenosine (0, 1.5, 3.0, 4.5 mmol/L) and treatment time (24, 48, 72, 96 h) on the proliferation, apoptosis, and HMLH1 expression of human colorectal cancer cell SW480, found that after treating colorectal cancer cells with different concentrations of adenosine, the hypermethylation of tumor suppressor genes hMLH1 was reversed and inhibited the proliferation of tumor cells. This kind of positive effect increased with the addition of exogenous adenosine concentration and treatment time (Xie et al., 2014). Meanwhile, Li Q et al. found that after treating human colorectal cancer cells SW480 with adenosine (3.0 mmol/L) for 72 h, the activity of methyltransferase (DNMT1 and DNMT3A) in these cells was inhibited, and similar to the above finding the hypermethylation of tumor suppressor genes RECK was reversed (Li et al., 2015). Like the aforementioned, alternations in DNA methylation patterns impact the occurrence and development of tumors (Klutstein et al., 2016). Studies regarding adenosine and DNA methylation status have also been reported in non-tumor disorders such as epilepsy, showing that inhibition of DNA methyltransferase activity during adenosine release is associated with restoration of global DNA methylation levels (Williams-Karnesky et al., 2013), this suggests that adenosine manipulation is a potential strategy in cancer manipulation via DNA methylation.
However, side effects such as flushing, dyspnea, chest pain, hypotension, bradycardia, etc. make the usage of exogenous adenosine less feasible for cancer treatment (Pritchard et al., 2010; Galagudza et al., 2012; Gul et al., 2020). A further question is whether systemic adenosine leads to a reversal of global methylation status or affects the site that should have been hypomethylated. Another concern is adenosine receptor-mediated action showed a cancer-promoting effect. Conversely, accumulating evidence supports ADK as a therapeutic target in cancer (Boison and Yegutkin, 2019; Murugan et al., 2021). The expression of ADK was shown to be upregulated in specific cancer types, including colorectal cancer (Giglioni et al., 2008), and breast cancer (Wang and Yang, 2014; Shamloo et al., 2019). Most recently, it has been found that a significantly enhanced expression of ADK in specimens of patients with glioma, both the tumor center and peritumoral tissue (de Groot et al., 2012). The general increase of purine metabolizing enzymes including ADK may allow accelerated purine metabolism to support the growth of cancer (Vannoni et al., 2004; Giglioni et al., 2008).
6 Targeting Adenosine Kinase on Deoxyribonucleic Acid Methylation in Cancer
The above described receptor-independent pathway mechanisms of adenosine play important roles in various types of cells with diverse functions (Boison et al., 2002). As an essential adenosine removal enzyme, inhibition of ADK can be more effective to decrease the cellular reuptake of adenosine and thereby increase the ambient concentration of extracellular adenosine (Newby et al., 1983; Davies et al., 1984). ADK inhibition was hypothesized to function as a site- and event-specific modulator for adenosine levels (Yamamoto and Imai, 2015; Cortez et al., 2016). This also provides a new direction for the treatment of tumors–targeting overexpression of ADK to regulate onsite adenosine level and DNA methylation, thereby affecting the proliferation and apoptosis of tumor cells. ADK-based adenosine intervention can avoid the aforementioned side effects of systemic adenosine administration (Liu et al., 2019) and pharmacokinetics limitation of the very short half-life in circulation (Hwang et al., 2016). ADK inhibitors have been revealed to have anti-inflammatory, antinociceptive, and anticonvulsant features (McGaraughty et al., 2005), and is being considered for the treatment of various diseases, including diabetes (Annes et al., 2012) and diseases of the nervous system (Chen et al., 2016).
ADK inhibitor development was initially based on 5-iodotubercidin (5-ITU), and 5′-amino-5′-deoxyadenosine (Cottam et al., 1993; Wiesner et al., 1999; Chen et al., 2016). Since then, several types of ADK inhibitors have been developed, which are classified as nucleoside and non-nucleoside ADK inhibitors (Boison, 2013). Nucleoside ADK inhibitors are adenosine derivatives that have hydroxylated ribose or cyclopentane rings, and additional purines or pyrimidine heterocyclic bases (Gomtsyan and Lee, 2004). The 5-aza group of the purine ring is replaced by a carbon connected to iodine. These compounds bind to enzymes to competitively inhibit adenosine (McGaraughty et al., 2005). In contrast, non-nucleoside ADK inhibitors lack ribose or cyclopentane rings, while some of them are constructed on pyrimidine or pyridyl pyrimidine nuclei. The non-nucleoside ADK inhibitors have been shown to relieve pain and inflammation in animal models (McGaraughty et al., 2005). Some ADK inhibitors are based on 6-(het)aryl-7-deazapurine pro-nucleotides that can inhibit cell growth by strongly inhibiting ADK activity (Spácilová et al., 2010), however, the mechanism of this finding has not been further investigated. Helena Marcelino et al. tested the effect of two ADK inhibitors on tumor cells in experiments on glioblastoma, and the results suggested that both ITU (25 μM) and ABT702 (15 μM) affected cells proliferation/viability (Marcelino et al., 2021). Co-incubation of ITU (25 μM) and adenosine (30 μM) produced a strong and similar decrease in cell proliferation in both GBM cell lines compared to ITU alone, this suggests that only 25 mM ITU may be sufficient to generate the maximum accumulation of intracellular adenosine (Marcelino et al., 2021). Zhang LM et al. showed that 5-ITU with concentrations (1, 2, 4, 6, 8, 10 μmol/L) for 48 h could significantly inhibit proliferation and induced apoptosis in a colon cancer cell line HT-29 (Zhang and Xie, 2015). Compared to the inhibitory effect of each concentrations group on HT-29 cells, the 6 μmol/L group showed a better effect on HT-29 cells, and the tumor suppressor gene DLC-1 in HT-29 cells was up-regulated and its methylation level was decreased after being treated with 2, 4, and 6 μmol/L ITU, respectively, this effect increases with increasing concentration (Zhang and Xie, 2015). As discussed above, ADK may play a potential adenosine receptor-independent epigenetic function, however, current available ADK inhibitors have not yet been reported to have high selectivity to target ADK-L or ADK-S. To distinguish the role of ADK-S and ADK-L on the regulation of cytoplasmic or nuclear adenosine levels and their possible epigenetic functions, using genetic approaches may bring us the answer.
Targeted therapy is a new strategy for cancer treatment. The goal is to use gene therapy to suppress the endogenous expression of ADK, with or without selectively targeting its two isoforms, i.e., the nuclear ADK-L and cytosolic ADK-S (Chen, 2010). Previous studies identified two independent promoters driving the expression of ADK isoforms, suggesting that each of the two isoforms of ADK are independently regulated at the transcriptional level (Cui et al., 2011), and independent transcriptional regulation may in turn indicate distinct physiological functions of the two isoforms (Boison, 2013). Besides, distinguish expression locations of two isoforms indicate that ADK-L (vs. ADK-S) has a unique role in proliferation and differentiation - two main nuclear activities associated with cancer pathology (Cui et al., 2009; Kiese et al., 2016). In patients with grade II and III gliomas, both subtypes of ADK are increased in the tumor and peritumoral areas, in addition to the detection of tumor invasion in the peritumoral tissue suggesting that ADK is involved in glioma progression and ADK level elevations may be associated with epilepsy in glioma patients (Huang et al., 2015). Amir E et al. reported a high positive correlation between ADK-L expression and whole-genome methylation in HeLa cells, (Wahba et al., 2021). Most recently, Shen HY et al. revealed that the expression level of ADK-L in breast cancer tissue was elevated compared to adjacent tissues, while the ADK-S expression level had no significant change, by measuring the protein expression level (Shamloo et al., 2019). Selective knockout of ADK isoforms via CRISPR/Cas9-mediated approaches suppressed breast cancer cell migration and invasion, which with the elevation of a tumor-related enzyme, matrix metalloproteinases, and downregulation of cyclin D2 and THB1 (Shamloo et al., 2019). Williams Karnesky et al. transfected ADK deficient BHK-AK2 cells with ADK-L- or ADK-S-expressing plasmids (Williams-Karnesky et al., 2013). ADK-L receptors showed a 400% increase in overall DNA methylation compared to controls, while ADK-S receptors showed only a modest 50% increase in overall DNA methylation. While both isoforms of ADK are involved in the regulation of overall DNA methylation, the nuclear subtype is more effective in regulating DNA methylation (Williams-Karnesky et al., 2013). ADK-L affects epigenetic remodeling by regulating methyltransferase activity and is considered the preferred mechanism for adenosine clearance in the nuclei (Boison and Yegutkin, 2019). ADK-L is directly related to the S-adenosylmethionine-dependent transmethylation pathway, which drives DNA and histone methylation (Boison, 2013). ADK-S regulates extracellular adenosine concentration for the availability of ARs activation (Pignataro et al., 2007; Boison and Yegutkin, 2019).
These studies support the observed functional differences of ADK-L and ADK-S in cancer. While ADK-L and ADK-S control adenosine concentrations in the nucleus and cytoplasm/extracellular respectively, ADK-L may play a role in adenosine receptor-independent regulation of epigenetic functions, and ADK-S determines adenosine availability for activation of adenosine receptors (Pignataro et al., 2007; Williams-Karnesky et al., 2013). Additional experimental evidence is needed to evaluate this notion. Together, selective inhibition of ADK-L is indicated as a novel adenosine receptor-independent strategy to offer a new perspective on cancer therapy, which may achieve more precise cancer intervention than general ADK or ADK-S manipulation.
7 Prospect and Challenge
With the observations that ADK inhibitions with isoform- and site-selective manners enhance the beneficial effect of endogenous adenosine and avoid various side effects of systemic manipulation of adenosine and adenosine receptors, research on ADK has made considerable progress in recent years. The emergence of new molecular tools including genetic approaches has enabled deeper exploration of ADK function. Further characterization of the metabolism of adenosine in different subcellular contexts, including cytoplasm, nucleus, and extracellular space, is needed for potential targeted ADK therapy. Studies have shown that elevated adenosine levels are related to apoptosis in various cancers (Xie et al., 2014; Jafari et al., 2017), which may be attributed to nuclear ADK-L (vs. ADK-S). In addition, the ADK effects on epigenetics, especially DNA methylation, may also be through its direct interaction with other nuclear proteins (Wang et al., 2005; Mohannath et al., 2014) rather than its regulation on the adenosine level. We should always bear in mind the challenge that increased adenosine levels can: 1) inhibit immune and inflammatory responses; 2) stimulate angiogenesis: epigenetic regulation of pro-angiogenic genes by ADK, and is thought to be another mechanism by which ADK is involved in cancer (Murugan et al., 2021). Knockdown of ADK decreases the methylation level of the VEGFR2 promoter region, which elevates intracellular adenosine and promotes proliferation, migration, and angiogenesis of human endothelial cells (Xu et al., 2017)—all aspects that may promote tumor growth. Last but not least, the downregulation of ADK found in hepatocellular carcinoma cells (YH, 2017) suggests the diversity of ADK changes across cancers. In summary, additional studies are needed to fully understand the role of adenosine in cancer pathology and to reveal the anticancer potential of ADK inhibition.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The project was supported by grants from the Good Samaritan Foundation of Legacy Health: No. 750390799 (H-YS) and the General Project of Chongqing Natural Science Foundation: No. cstc2021jcyj-msxmX0112 (Y-XW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Annes, J. P., Ryu, J. H., Lam, K., Carolan, P. J., Utz, K., Hollister-Lock, J., et al. (2012). Adenosine Kinase Inhibition Selectively Promotes Rodent and Porcine Islet β-cell Replication. Proc. Natl. Acad. Sci. U. S. A. 109 (10), 3915–3920. doi:10.1073/pnas.1201149109
Antonioli, L., Blandizzi, C., Pacher, P., and Haskó, G. (2013). Immunity, Inflammation and Cancer: a Leading Role for Adenosine. Nat. Rev. Cancer 13 (12), 842–857. doi:10.1038/nrc3613
Ashar, F. N., Zhang, Y., Longchamps, R. J., Lane, J., Moes, A., Grove, M. L., et al. (2017). Association of Mitochondrial DNA Copy Number with Cardiovascular Disease. JAMA Cardiol. 2 (11), 1247–1255. doi:10.1001/jamacardio.2017.3683
Bae, J. M., Kim, J. H., Kwak, Y., Lee, D. W., Cha, Y., Wen, X., et al. (2017). Distinct Clinical Outcomes of Two CIMP-Positive Colorectal Cancer Subtypes Based on a Revised CIMP Classification System. Br. J. Cancer 116 (8), 1012–1020. doi:10.1038/bjc.2017.52
Baylin, S. B., Esteller, M., Rountree, M. R., Bachman, K. E., Schuebel, K., and Herman, J. G. (2001). Aberrant Patterns of DNA Methylation, Chromatin Formation and Gene Expression in Cancer. Hum. Mol. Genet. 10 (7), 687–692. doi:10.1093/hmg/10.7.687
Begam, N., Jamil, K., and Raju, S. G. (2017). Promoter Hypermethylation of the ATM Gene as a Novel Biomarker for Breast Cancer. Asian Pac J. Cancer Prev. 18 (11), 3003–3009. doi:10.22034/APJCP.2017.18.11.3003
Bender, C. M., Pao, M. M., and Jones, P. A. (1998). Inhibition of DNA Methylation by 5-Aza-2'-Deoxycytidine Suppresses the Growth of Human Tumor Cell Lines. Cancer Res. 58 (1), 95–101.
Boison, D. (2013). Adenosine Kinase: Exploitation for Therapeutic Gain. Pharmacol. Rev. 65 (3), 906–943. doi:10.1124/pr.112.006361
Boison, D., Scheurer, L., Zumsteg, V., Rülicke, T., Litynski, P., Fowler, B., et al. (2002). Neonatal Hepatic Steatosis by Disruption of the Adenosine Kinase Gene. Proc. Natl. Acad. Sci. U. S. A. 99 (10), 6985–6990. doi:10.1073/pnas.092642899
Boison, D., and Yegutkin, G. G. (2019). Adenosine Metabolism: Emerging Concepts for Cancer Therapy. Cancer Cell 36 (6), 582–596. doi:10.1016/j.ccell.2019.10.007
Borea, P. A., Gessi, S., Merighi, S., Vincenzi, F., and Varani, K. (2018). Pharmacology of Adenosine Receptors: The State of the Art. Physiol. Rev. 98 (3), 1591–1625. doi:10.1152/physrev.00049.2017
Borodovsky, A., Barbon, C. M., Wang, Y., Ye, M., Prickett, L., Chandra, D., et al. (2020). Small Molecule AZD4635 Inhibitor of A2AR Signaling Rescues Immune Cell Function Including CD103+ Dendritic Cells Enhancing Anti-tumor Immunity. J. Immunother. Cancer 8 (2). doi:10.1136/jitc-2019-000417
Buisseret, L., Pommey, S., Allard, B., Garaud, S., Bergeron, M., Cousineau, I., et al. (2018). Clinical Significance of CD73 in Triple-Negative Breast Cancer: Multiplex Analysis of a Phase III Clinical Trial. Ann. Oncol. 29 (4), 1056–1062. doi:10.1093/annonc/mdx730
Cao, X., Tang, Q., Holland-Letz, T., Gündert, M., Cuk, K., Schott, S., et al. (2018). Evaluation of Promoter Methylation of RASSF1A and ATM in Peripheral Blood of Breast Cancer Patients and Healthy Control Individuals. Int. J. Mol. Sci. 19 (3). doi:10.3390/ijms19030900
Chen, J., Wu, L., Xu, H., and Cheng, S. (2019). 5-Aza-CdR Regulates RASSF1A by Inhibiting DNMT1 to Affect Colon Cancer Cell Proliferation, Migration and Apoptosis. Cancer Manag. Res. 11, 9517–9528. doi:10.2147/CMAR.S229726
Chen, L. (2010). A Global Comparison between Nuclear and Cytosolic Transcriptomes Reveals Differential Compartmentalization of Alternative Transcript Isoforms. Nucleic Acids Res. 38 (4), 1086–1097. doi:10.1093/nar/gkp1136
Chen, Y. L., Zhang, Y. N., Wang, Z. Z., Xu, W. G., Li, R. P., and Zhang, J. D. (2016). Effects of Adenosine Metabolism in Astrocytes on Central Nervous System Oxygen Toxicity. Brain Res. 1635, 180–189. doi:10.1016/j.brainres.2016.01.026
Christman, J. K. (2002). 5-Azacytidine and 5-Aza-2'-Deoxycytidine as Inhibitors of DNA Methylation: Mechanistic Studies and Their Implications for Cancer Therapy. Oncogene 21 (35), 5483–5495. doi:10.1038/sj.onc.1205699
Church, T. R., Wandell, M., Lofton-Day, C., Mongin, S. J., Burger, M., Payne, S. R., et al. (2014). Prospective Evaluation of Methylated SEPT9 in Plasma for Detection of Asymptomatic Colorectal Cancer. Gut 63 (2), 317–325. doi:10.1136/gutjnl-2012-304149
Connolly, D., Yang, Z., Castaldi, M., Simmons, N., Oktay, M. H., Coniglio, S., et al. (2011). Septin 9 Isoform Expression, Localization and Epigenetic Changes during Human and Mouse Breast Cancer Progression. Breast Cancer Res. 13 (4), R76. doi:10.1186/bcr2924
Cortez, B. A., Rezende-Teixeira, P., Redick, S., Doxsey, S., and Machado-Santelli, G. M. (2016). Multipolar Mitosis and Aneuploidy after Chrysotile Treatment: a Consequence of Abscission Failure and Cytokinesis Regression. Oncotarget 7 (8), 8979–8992. doi:10.18632/oncotarget.6924
Cottam, H. B., Wasson, D. B., Shih, H. C., Raychaudhuri, A., Di Pasquale, G., and Carson, D. A. (1993). New Adenosine Kinase Inhibitors with Oral Antiinflammatory Activity: Synthesis and Biological Evaluation. J. Med. Chem. 36 (22), 3424–3430. doi:10.1021/jm00074a024
Cui, X. A., Agarwal, T., Singh, B., and Gupta, R. S. (2011). Molecular Characterization of Chinese Hamster Cells Mutants Affected in Adenosine Kinase and Showing Novel Genetic and Biochemical Characteristics. BMC Biochem. 12, 22. doi:10.1186/1471-2091-12-22
Cui, X. A., Singh, B., Park, J., and Gupta, R. S. (2009). Subcellular Localization of Adenosine Kinase in Mammalian Cells: The Long Isoform of AdK Is Localized in the Nucleus. Biochem. Biophys. Res. Commun. 388 (1), 46–50. doi:10.1016/j.bbrc.2009.07.106
Das, P. M., and Singal, R. (2004). DNA Methylation and Cancer. J. Clin. Oncol. 22 (22), 4632–4642. doi:10.1200/JCO.2004.07.151
Davies, L. P., Jamieson, D. D., Baird-Lambert, J. A., and Kazlauskas, R. (1984). Halogenated Pyrrolopyrimidine Analogues of Adenosine from Marine Organisms: Pharmacological Activities and Potent Inhibition of Adenosine Kinase. Biochem. Pharmacol. 33 (3), 347–355. doi:10.1016/0006-2952(84)90225-9
de Groot, M., Iyer, A., Zurolo, E., Anink, J., Heimans, J. J., Boison, D., et al. (2012). Overexpression of ADK in Human Astrocytic Tumors and Peritumoral Tissue Is Related to Tumor-Associated Epilepsy. Epilepsia 53 (1), 58–66. doi:10.1111/j.1528-1167.2011.03306.x
de Lera Ruiz, M., Lim, Y. H., and Zheng, J. (2014). Adenosine A2A Receptor as a Drug Discovery Target. J. Med. Chem. 57 (9), 3623–3650. doi:10.1021/jm4011669
Debouki-Joudi, S., Trifa, F., Khabir, A., Sellami-Boudawara, T., Frikha, M., Daoud, J., et al. (2017). CpG Methylation of APC Promoter 1A in Sporadic and Familial Breast Cancer Patients. Cancer Biomark. 18 (2), 133–141. doi:10.3233/CBM-160005
Egger, G., Aparicio, A. M., Escobar, S. G., and Jones, P. A. (2007). Inhibition of Histone Deacetylation Does Not Block Resilencing of P16 after 5-Aza-2'-Deoxycytidine Treatment. Cancer Res. 67 (1), 346–353. doi:10.1158/0008-5472.CAN-06-2845
Egger, G., Jeong, S., Escobar, S. G., Cortez, C. C., Li, T. W., Saito, Y., et al. (2006). Identification of DNMT1 (DNA Methyltransferase 1) Hypomorphs in Somatic Knockouts Suggests an Essential Role for DNMT1 in Cell Survival. Proc. Natl. Acad. Sci. U. S. A. 103 (38), 14080–14085. doi:10.1073/pnas.0604602103
Eltzschig, H. K., Sitkovsky, M. V., and Robson, S. C. (2012). Purinergic Signaling during Inflammation. N. Engl. J. Med. 367 (24), 2322–2333. doi:10.1056/NEJMra1205750
Fan, X. Y., Hu, X. L., Han, T. M., Wang, N. N., Zhu, Y. M., Hu, W., et al. (2011). Association between RUNX3 Promoter Methylation and Gastric Cancer: a Meta-Analysis. BMC Gastroenterol. 11, 92. doi:10.1186/1471-230X-11-92
Fedele, D. E., Gouder, N., Güttinger, M., Gabernet, L., Scheurer, L., Rülicke, T., et al. (2005). Astrogliosis in Epilepsy Leads to Overexpression of Adenosine Kinase, Resulting in Seizure Aggravation. Brain 128 (Pt 10), 2383–2395. doi:10.1093/brain/awh555
Feinberg, A. P., and Tycko, B. (2004). The History of Cancer Epigenetics. Nat. Rev. Cancer 4 (2), 143–153. doi:10.1038/nrc1279
Feinberg, A. P., and Vogelstein, B. (1983). Hypomethylation Distinguishes Genes of Some Human Cancers from Their Normal Counterparts. Nature 301 (5895), 89–92. doi:10.1038/301089a0
Fishman, P., Bar-Yehuda, S., Synowitz, M., Powell, J. D., Klotz, K. N., Gessi, S., et al. (2009). Adenosine Receptors and Cancer. Handb. Exp. Pharmacol. (193), 399–441. doi:10.1007/978-3-540-89615-9_14
Flohr, H., and Breull, W. (1975). Effect of Etafenone on Total and Regional Myocardial Blood Flow. Arzneimittelforschung 25 (9), 1400–1403.
Franco, R., Rivas-Santisteban, R., Navarro, G., and Reyes-Resina, I. (2021). Adenosine Receptor Antagonists to Combat Cancer and to Boost Anti-cancer Chemotherapy and Immunotherapy. Cells 10 (11). doi:10.3390/cells10112831
Franco, R., Schoneveld, O., Georgakilas, A. G., and Panayiotidis, M. I. (2008). Oxidative Stress, DNA Methylation and Carcinogenesis. Cancer Lett. 266 (1), 6–11. doi:10.1016/j.canlet.2008.02.026
Fredholm, B. B., Chen, J. F., Cunha, R. A., Svenningsson, P., and Vaugeois, J. M. (2005). Adenosine and Brain Function. Int. Rev. Neurobiol. 63, 191–270. doi:10.1016/S0074-7742(05)63007-3
Fredholm, B. B., IJzerman, A. P., Jacobson, K. A., Klotz, K. N., and Linden, J. (2001). International Union of Pharmacology. XXV. Nomenclature and Classification of Adenosine Receptors. Pharmacol. Rev. 53 (4), 527–552.
Galagudza, M., Korolev, D., Postnov, V., Naumisheva, E., Grigorova, Y., Uskov, I., et al. (2012). Passive Targeting of Ischemic-Reperfused Myocardium with Adenosine-Loaded Silica Nanoparticles. Int. J. Nanomedicine 7, 1671–1678. doi:10.2147/IJN.S29511
Gao, J., Wang, L., Xu, J., Zheng, J., Man, X., Wu, H., et al. (2013). Aberrant DNA Methyltransferase Expression in Pancreatic Ductal Adenocarcinoma Development and Progression. J. Exp. Clin. Cancer Res. 32, 86. doi:10.1186/1756-9966-32-86
Gazzoli, I., Loda, M., Garber, J., Syngal, S., and Kolodner, R. D. (2002). A Hereditary Nonpolyposis Colorectal Carcinoma Case Associated with Hypermethylation of the MLH1 Gene in Normal Tissue and Loss of Heterozygosity of the Unmethylated Allele in the Resulting Microsatellite Instability-High Tumor. Cancer Res. 62 (14), 3925–3928.
Gelsomino, F., Barbolini, M., Spallanzani, A., Pugliese, G., and Cascinu, S. (2016). The Evolving Role of Microsatellite Instability in Colorectal Cancer: A Review. Cancer Treat. Rev. 51, 19–26. doi:10.1016/j.ctrv.2016.10.005
Giglioni, S., Leoncini, R., Aceto, E., Chessa, A., Civitelli, S., Bernini, A., et al. (2008). Adenosine Kinase Gene Expression in Human Colorectal Cancer. Nucleosides Nucleotides Nucleic Acids 27 (6), 750–754. doi:10.1080/15257770802145629
Glodzik, D., Bosch, A., Hartman, J., Aine, M., Vallon-Christersson, J., Reuterswärd, C., et al. (2020). Comprehensive Molecular Comparison of BRCA1 Hypermethylated and BRCA1 Mutated Triple Negative Breast Cancers. Nat. Commun. 11 (1), 3747. doi:10.1038/s41467-020-17537-2
Gomtsyan, A., and Lee, C. H. (2004). Nonnucleoside Inhibitors of Adenosine Kinase. Curr. Pharm. Des. 10 (10), 1093–1103. doi:10.2174/1381612043452703
Goyal, R., Reinhardt, R., and Jeltsch, A. (2006). Accuracy of DNA Methylation Pattern Preservation by the Dnmt1 Methyltransferase. Nucleic Acids Res. 34 (4), 1182–1188. doi:10.1093/nar/gkl002
Greger, V., Passarge, E., Höpping, W., Messmer, E., and Horsthemke, B. (1989). Epigenetic Changes May Contribute to the Formation and Spontaneous Regression of Retinoblastoma. Hum. Genet. 83 (2), 155–158. doi:10.1007/BF00286709
Gul, M., Sheikh, M., Chaudhry, A., Gerges, L., Al Halabi, H., Feldman, E., et al. (2020). Role of Cardiac Nuclear Stress Perfusion Exam after Computed Tomographic Coronary Angiogram for Evaluation of Obstructive Coronary Artery Disease in Patients with Chest Pain. J. Thorac. Dis. 12 (9), 5067–5077. doi:10.21037/jtd-2019-pitd-12
Häusler, S. F., Montalbán del Barrio, I., Strohschein, J., Chandran, P. A., Engel, J. B., Hönig, A., et al. (2011). Ectonucleotidases CD39 and CD73 on OvCA Cells Are Potent Adenosine-Generating Enzymes Responsible for Adenosine Receptor 2A-dependent Suppression of T Cell Function and NK Cell Cytotoxicity. Cancer Immunol. Immunother. 60 (10), 1405–1418. doi:10.1007/s00262-011-1040-4
Huang, J., He, Y., Chen, M., Du, J., Li, G., Li, S., et al. (2015). Adenosine Deaminase and Adenosine Kinase Expression in Human Glioma and Their Correlation with Glioma-associated E-pilepsy. Mol. Med. Rep. 12 (5), 6509–6516. doi:10.3892/mmr.2015.4285
Hung, M. S., and Shen, C. K. (2003). Eukaryotic Methyl-CpG-Binding Domain Proteins and Chromatin Modification. Eukaryot. Cell 2 (5), 841–846. doi:10.1128/ec.2.5.841-846.2003
Hwang, D., Lee, J. M., and Koo, B. K. (2016). Physiologic Assessment of Coronary Artery Disease: Focus on Fractional Flow Reserve. Korean J. Radiol. 17 (3), 307–320. doi:10.3348/kjr.2016.17.3.307
Issa, J. P. (2007). DNA Methylation as a Therapeutic Target in Cancer. Clin. Cancer Res. 13 (6), 1634–1637. doi:10.1158/1078-0432.CCR-06-2076
Jacobson, K. A., and Reitman, M. L. (2020). Adenosine-Related Mechanisms in Non-adenosine Receptor Drugs. Cells 9 (4). doi:10.3390/cells9040956
Jacobson, K. A. (2009). Introduction to Adenosine Receptors as Therapeutic Targets. Handb. Exp. Pharmacol. 193.
Jafari, S. M., Joshaghani, H. R., Panjehpour, M., Aghaei, M., and Zargar Balajam, N. (2017). Apoptosis and Cell Cycle Regulatory Effects of Adenosine by Modulation of GLI-1 and ERK1/2 Pathways in CD44+ and CD24- Breast Cancer Stem Cells. Cell Prolif. 50 (4). doi:10.1111/cpr.12345
James, S. J., Melnyk, S., Pogribna, M., Pogribny, I. P., and Caudill, M. A. (2002). Elevation in S-Adenosylhomocysteine and DNA Hypomethylation: Potential Epigenetic Mechanism for Homocysteine-Related Pathology. J. Nutr. 132 (8 Suppl. l), 2361S–2366S. doi:10.1093/jn/132.8.2361S
Karahoca, M., and Momparler, R. L. (2013). Pharmacokinetic and Pharmacodynamic Analysis of 5-Aza-2'-Deoxycytidine (Decitabine) in the Design of its Dose-Schedule for Cancer Therapy. Clin. Epigenetics 5 (1), 3. doi:10.1186/1868-7083-5-3
Kiese, K., Jablonski, J., Boison, D., and Kobow, K. (2016). Dynamic Regulation of the Adenosine Kinase Gene during Early Postnatal Brain Development and Maturation. Front. Mol. Neurosci. 9, 99. doi:10.3389/fnmol.2016.00099
Kloor, D., and Osswald, H. (2004). S-adenosylhomocysteine Hydrolase as a Target for Intracellular Adenosine Action. Trends Pharmacol. Sci. 25 (6), 294–297. doi:10.1016/j.tips.2004.04.004
Klutstein, M., Nejman, D., Greenfield, R., and Cedar, H. (2016). DNA Methylation in Cancer and Aging. Cancer Res. 76 (12), 3446–3450. doi:10.1158/0008-5472.CAN-15-3278
Kneip, C., Schmidt, B., Seegebarth, A., Weickmann, S., Fleischhacker, M., Liebenberg, V., et al. (2011). SHOX2 DNA Methylation Is a Biomarker for the Diagnosis of Lung Cancer in Plasma. J. Thorac. Oncol. 6 (10), 1632–1638. doi:10.1097/JTO.0b013e318220ef9a
Li, Q., Wang, Y.-X., Shen, H.-Y., Xie, K., Zhang, L.-M., and Zeng, L.-H. (2015). Adenosine Interferes with Gene Methylation and its Mechanism RECK Human Colorectal Cancer SW480 Cells. J. Third Mil. Med. Univ. 37 (1), 46–50.
Li, X., Yao, X., Wang, Y., Hu, F., Wang, F., Jiang, L., et al. (2013). MLH1 Promoter Methylation Frequency in Colorectal Cancer Patients and Related Clinicopathological and Molecular Features. PloS One 8 (3), e59064. doi:10.1371/journal.pone.0059064
Liu, L., Nishihara, R., Qian, Z. R., Tabung, F. K., Nevo, D., Zhang, X., et al. (2017). Association between Inflammatory Diet Pattern and Risk of Colorectal Carcinoma Subtypes Classified by Immune Responses to Tumor. Gastroenterology 153 (6), 1517–e14. doi:10.1053/j.gastro.2017.08.045
Liu, Y. J., Chen, J., Li, X., Zhou, X., Hu, Y. M., Chu, S. F., et al. (2019). Research Progress on Adenosine in Central Nervous System Diseases. CNS Neurosci. Ther. 25 (9), 899–910. doi:10.1111/cns.13190
López-Moyado, I. F., Tsagaratou, A., Yuita, H., Seo, H., Delatte, B., Heinz, S., et al. (2019). Paradoxical Association of TET Loss of Function with Genome-wide DNA Hypomethylation. Proc. Natl. Acad. Sci. U. S. A. 116 (34), 16933–16942. doi:10.1073/pnas.1903059116
Losenkova, K., Zuccarini, M., Karikoski, M., Laurila, J., Boison, D., Jalkanen, S., et al. (2020). Compartmentalization of Adenosine Metabolism in Cancer Cells and its Modulation during Acute Hypoxia. J. Cell Sci. 133 (10). doi:10.1242/jcs.241463
Marcelino, H., Carvalho, T. M. A., Tomás, J., Teles, F. I., Honório, A. C., Rosa, C. B., et al. (2021). Adenosine Inhibits Cell Proliferation Differently in Human Astrocytes and in Glioblastoma Cell Lines. Neuroscience 467, 122–133. doi:10.1016/j.neuroscience.2021.05.019
Mastelic-Gavillet, B., Navarro Rodrigo, B., Décombaz, L., Wang, H., Ercolano, G., Ahmed, R., et al. (2019). Adenosine Mediates Functional and Metabolic Suppression of Peripheral and Tumor-Infiltrating CD8+ T Cells. J. Immunother. Cancer 7 (1), 257. doi:10.1186/s40425-019-0719-5
McGaraughty, S., Cowart, M., Jarvis, M. F., and Berman, R. F. (2005). Anticonvulsant and Antinociceptive Actions of Novel Adenosine Kinase Inhibitors. Curr. Top. Med. Chem. 5 (1), 43–58. doi:10.2174/1568026053386845
Mendizabal, I., Zeng, J., Keller, T. E., and Yi, S. V. (2017). Body-hypomethylated Human Genes Harbor Extensive Intragenic Transcriptional Activity and Are Prone to Cancer-Associated Dysregulation. Nucleic Acids Res. 45 (8), 4390–4400. doi:10.1093/nar/gkx020
Merighi, S., Battistello, E., Giacomelli, L., Varani, K., Vincenzi, F., Borea, P. A., et al. (2019). Targeting A3 and A2A Adenosine Receptors in the Fight against Cancer. Expert Opin. Ther. Targets 23 (8), 669–678. doi:10.1080/14728222.2019.1630380
Merighi, S., Benini, A., Mirandola, P., Gessi, S., Varani, K., Leung, E., et al. (2005). A3 Adenosine Receptors Modulate Hypoxia-Inducible Factor-1alpha Expression in Human A375 Melanoma Cells. Neoplasia 7 (10), 894–903. doi:10.1593/neo.05334
Micevic, G., Theodosakis, N., and Bosenberg, M. (2017). Aberrant DNA Methylation in Melanoma: Biomarker and Therapeutic Opportunities. Clin. Epigenetics 9, 34. doi:10.1186/s13148-017-0332-8
Mohannath, G., Jackel, J. N., Lee, Y. H., Buchmann, R. C., Wang, H., Patil, V., et al. (2014). A Complex Containing SNF1-Related Kinase (SnRK1) and Adenosine Kinase in Arabidopsis. PLoS One 9 (1), e87592. doi:10.1371/journal.pone.0087592
Morey, S. R., Smiraglia, D. J., James, S. R., Yu, J., Moser, M. T., Foster, B. A., et al. (2006). DNA Methylation Pathway Alterations in an Autochthonous Murine Model of Prostate Cancer. Cancer Res. 66 (24), 11659–11667. doi:10.1158/0008-5472.CAN-06-1937
Murugan, M., Fedele, D., Millner, D., Alharfoush, E., Vegunta, G., and Boison, D. (2021). Adenosine Kinase: An Epigenetic Modulator in Development and Disease. Neurochem. Int. 147, 105054. doi:10.1016/j.neuint.2021.105054
Nejman, D., Straussman, R., Steinfeld, I., Ruvolo, M., Roberts, D., Yakhini, Z., et al. (2014). Molecular Rules Governing De Novo Methylation in Cancer. Cancer Res. 74 (5), 1475–1483. doi:10.1158/0008-5472.CAN-13-3042
Newby, A. C., Holmquist, C. A., Illingworth, J., and Pearson, J. D. (1983). The Control of Adenosine Concentration in Polymorphonuclear Leucocytes, Cultured Heart Cells and Isolated Perfused Heart from the Rat. Biochem. J. 214 (2), 317–323. doi:10.1042/bj2140317
Oates, N. A., van Vliet, J., Duffy, D. L., Kroes, H. Y., Martin, N. G., Boomsma, D. I., et al. (2006). Increased DNA Methylation at the AXIN1 Gene in a Monozygotic Twin from a Pair Discordant for a Caudal Duplication Anomaly. Am. J. Hum. Genet. 79 (1), 155–162. doi:10.1086/505031
Ohta, A. (2016). A Metabolic Immune Checkpoint: Adenosine in Tumor Microenvironment. Front. Immunol. 7, 109. doi:10.3389/fimmu.2016.00109
Ohta, A., Gorelik, E., Prasad, S. J., Ronchese, F., Lukashev, D., Wong, M. K., et al. (2006). A2A Adenosine Receptor Protects Tumors from Antitumor T Cells. Proc. Natl. Acad. Sci. U. S. A. 103 (35), 13132–13137. doi:10.1073/pnas.0605251103
Ohta, A., and Sitkovsky, M. (2001). Role of G-Protein-Coupled Adenosine Receptors in Downregulation of Inflammation and Protection from Tissue Damage. Nature 414 (6866), 916–920. doi:10.1038/414916a
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 99 (3), 247–257. doi:10.1016/s0092-8674(00)81656-6
Pacheco, R., Martinez-Navio, J. M., Lejeune, M., Climent, N., Oliva, H., Gatell, J. M., et al. (2005). CD26, Adenosine Deaminase, and Adenosine Receptors Mediate Costimulatory Signals in the Immunological Synapse. Proc. Natl. Acad. Sci. U. S. A. 102 (27), 9583–9588. doi:10.1073/pnas.0501050102
Pan, F., Wingo, T. S., Zhao, Z., Gao, R., Makishima, H., Qu, G., et al. (2017). Tet2 Loss Leads to Hypermutagenicity in Haematopoietic Stem/progenitor Cells. Nat. Commun. 8, 15102. doi:10.1038/ncomms15102
Pignataro, G., Simon, R. P., and Boison, D. (2007). Transgenic Overexpression of Adenosine Kinase Aggravates Cell Death in Ischemia. J. Cereb. Blood Flow. Metab. 27 (1), 1–5. doi:10.1038/sj.jcbfm.9600334
Pritchard, E. M., Szybala, C., Boison, D., and Kaplan, D. L. (2010). Silk Fibroin Encapsulated Powder Reservoirs for Sustained Release of Adenosine. J. Control Release 144 (2), 159–167. doi:10.1016/j.jconrel.2010.01.035
Ren, J. Z., and Huo, J. R. (2012). 5-aza-2'-deoxycytidine-induced Inhibition of CDH13 Expression and its Inhibitory Effect on Methylation Status in Human Colon Cancer Cells In Vitro and on Growth of Xenograft in Nude Mice. Zhonghua Zhong Liu Za Zhi 34 (1), 6–10.
Schiedel, A. C., Lacher, S. K., Linnemann, C., Knolle, P. A., and Müller, C. E. (2013). Antiproliferative Effects of Selective Adenosine Receptor Agonists and Antagonists on Human Lymphocytes: Evidence for Receptor-independent Mechanisms. Purinergic Signal 9 (3), 351–365. doi:10.1007/s11302-013-9354-7
Seetulsingh-Goorah, S. P. (2006). Mechanisms of Adenosine-Induced Cytotoxicity and Their Clinical and Physiological Implications. Biofactors 27 (1-4), 213–230. doi:10.1002/biof.5520270119
Sellin, M. E., Holmfeldt, P., Stenmark, S., and Gullberg, M. (2011). Microtubules Support a Disk-like Septin Arrangement at the Plasma Membrane of Mammalian Cells. Mol. Biol. Cell 22 (23), 4588–4601. doi:10.1091/mbc.E11-09-0754
Shamloo, B., Kumar, N., Owen, R. H., Reemmer, J., Ost, J., Perkins, R. S., et al. (2019). Dysregulation of Adenosine Kinase Isoforms in Breast Cancer. Oncotarget 10 (68), 7238–7250. doi:10.18632/oncotarget.27364
Sharma, P. (2016). Biology and Management of Patients with Triple-Negative Breast Cancer. Oncologist 21 (9), 1050–1062. doi:10.1634/theoncologist.2016-0067
Sigalotti, L., Fratta, E., Coral, S., and Maio, M. (2014). Epigenetic Drugs as Immunomodulators for Combination Therapies in Solid Tumors. Pharmacol. Ther. 142 (3), 339–350. doi:10.1016/j.pharmthera.2013.12.015
Sinčić, N., and Herceg, Z. (2011). DNA Methylation and Cancer: Ghosts and Angels above the Genes. Curr. Opin. Oncol. 23 (1), 69–76. doi:10.1097/CCO.0b013e3283412eb4
Song, A., Zhang, Y., Han, L., Yegutkin, G. G., Liu, H., Sun, K., et al. (2017). Erythrocytes Retain Hypoxic Adenosine Response for Faster Acclimatization upon Re-ascent. Nat. Commun. 8, 14108. doi:10.1038/ncomms14108
Spácilová, P., Naus, P., Pohl, R., Votruba, I., Snásel, J., Zábranská, H., et al. (2010). CycloSal-phosphate Pronucleotides of Cytostatic 6-(Het)aryl-7-Deazapurine Ribonucleosides: Synthesis, Cytostatic Activity, and Inhibition of Adenosine Kinases. ChemMedChem 5 (8), 1386–1396. doi:10.1002/cmdc.201000192
Stagg, J., and Smyth, M. J. (2010). Extracellular Adenosine Triphosphate and Adenosine in Cancer. Oncogene 29 (39), 5346–5358. doi:10.1038/onc.2010.292
Su, J., Huang, Y. H., Cui, X., Wang, X., Zhang, X., Lei, Y., et al. (2018). Homeobox Oncogene Activation by Pan-Cancer DNA Hypermethylation. Genome Biol. 19 (1), 108. doi:10.1186/s13059-018-1492-3
Su Xiaoyang, L. S. (2018). YAN Yusheng. Res. Prog. metabolic Pathw. adenosine Regul. pro Inflamm. cytokines Cell Inj. J. Clin. Thorac. Cardiovasc. Surg. 25 (12).
Sun, D., Luo, M., Jeong, M., Rodriguez, B., Xia, Z., Hannah, R., et al. (2014). Epigenomic Profiling of Young and Aged HSCs Reveals Concerted Changes during Aging that Reinforce Self-Renewal. Cell stem Cell 14 (5), 673–688. doi:10.1016/j.stem.2014.03.002
Sun, J., Fei, F., Zhang, M., Li, Y., Zhang, X., Zhu, S., et al. (2019). The Role of mSEPT9 in Screening, Diagnosis, and Recurrence Monitoring of Colorectal Cancer. BMC Cancer 19 (1), 450. doi:10.1186/s12885-019-5663-8
Swellam, M., Abdelmaksoud, M. D., Sayed Mahmoud, M., Ramadan, A., Abdel-Moneem, W., and Hefny, M. M. (2015). Aberrant Methylation of APC and RARβ2 Genes in Breast Cancer Patients. IUBMB Life 67 (1), 61–68. doi:10.1002/iub.1346
Synowitz, M., Glass, R., Färber, K., Markovic, D., Kronenberg, G., Herrmann, K., et al. (2006). A1 Adenosine Receptors in Microglia Control Glioblastoma-Host Interaction. Cancer Res. 66 (17), 8550–8557. doi:10.1158/0008-5472.CAN-06-0365
Tanaka, M., Kijima, H., Itoh, J., Matsuda, T., and Tanaka, T. (2002). Impaired Expression of a Human Septin Family Gene Bradeion Inhibits the Growth and Tumorigenesis of Colorectal Cancer In Vitro and In Vivo. Cancer Gene Ther. 9 (6), 483–488. doi:10.1038/sj.cgt.7700460
Tsutsui, S., Schnermann, J., Noorbakhsh, F., Henry, S., Yong, V. W., Winston, B. W., et al. (2004). A1 Adenosine Receptor Upregulation and Activation Attenuates Neuroinflammation and Demyelination in a Model of Multiple Sclerosis. J. Neurosci. 24 (6), 1521–1529. doi:10.1523/JNEUROSCI.4271-03.2004
Valencia, A. M., and Kadoch, C. (2019). Chromatin Regulatory Mechanisms and Therapeutic Opportunities in Cancer. Nat. Cell Biol. 21 (2), 152–161. doi:10.1038/s41556-018-0258-1
van de Veen, W., Globinska, A., Jansen, K., Straumann, A., Kubo, T., Verschoor, D., et al. (2020). A Novel Proangiogenic B Cell Subset Is Increased in Cancer and Chronic Inflammation. Sci. Adv. 6 (20), eaaz3559. doi:10.1126/sciadv.aaz3559
Van der Auwera, I., Bovie, C., Svensson, C., Limame, R., Trinh, X. B., van Dam, P., et al. (2009). Quantitative Assessment of DNA Hypermethylation in the Inflammatory and Non-inflammatory Breast Cancer Phenotypes. Cancer Biol. Ther. 8 (23), 2252–2259. doi:10.4161/cbt.8.23.10133
Vannoni, D., Bernini, A., Carlucci, F., Civitelli, S., Di Pietro, M. C., Leoncini, R., et al. (2004). Enzyme Activities Controlling Adenosine Levels in Normal and Neoplastic Tissues. Med. Oncol. 21 (2), 187–195. doi:10.1385/MO:21:2:187
Viré, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C., et al. (2006). The Polycomb Group Protein EZH2 Directly Controls DNA Methylation. Nature 439 (7078), 871–874. doi:10.1038/nature04431
Virtanen, S. S., Kukkonen-Macchi, A., Vainio, M., Elima, K., Härkönen, P. L., Jalkanen, S., et al. (2014). Adenosine Inhibits Tumor Cell Invasion via Receptor-independent Mechanisms. Mol. Cancer Res. 12 (12), 1863–1874. doi:10.1158/1541-7786.MCR-14-0302-T
Wahba, A. E., Fedele, D., Gebril, H., AlHarfoush, E., Toti, K. S., Jacobson, K. A., et al. (2021). Adenosine Kinase Expression Determines DNA Methylation in Cancer Cell Lines. ACS Pharmacol. Transl. Sci. 4 (2), 680–686. doi:10.1021/acsptsci.1c00008
Wang, H., Buckley, K. J., Yang, X., Buchmann, R. C., and Bisaro, D. M. (2005). Adenosine Kinase Inhibition and Suppression of RNA Silencing by Geminivirus AL2 and L2 Proteins. J. Virol. 79 (12), 7410–7418. doi:10.1128/JVI.79.12.7410-7418.2005
Wang, J., Wang, Y., Chu, Y., Li, Z., Yu, X., Huang, Z., et al. (2021). Tumor-derived Adenosine Promotes Macrophage Proliferation in Human Hepatocellular Carcinoma. J. hepatology 74 (3), 627–637. doi:10.1016/j.jhep.2020.10.021
Wang, L., Lin, Y. L., Li, B., Wang, Y. Z., Li, W. P., and Ma, J. G. (2014). Aberrant Promoter Methylation of the Cadherin 13 Gene in Serum and its Relationship with Clinicopathological Features of Prostate Cancer. J. Int. Med. Res. 42 (5), 1085–1092. doi:10.1177/0300060514540631
Wang, L. F., and Yang, S. Y. (2014). Expression And Clinical Significance of Adenosine Kinase in Breast Cancer. 6, 475–477.
Weber, M., Hellmann, I., Stadler, M. B., Ramos, L., Pääbo, S., Rebhan, M., et al. (2007). Distribution, Silencing Potential and Evolutionary Impact of Promoter DNA Methylation in the Human Genome. Nat. Genet. 39 (4), 457–466. doi:10.1038/ng1990
Wengner, A. M., Scholz, A., and Haendler, B. (2020). Targeting DNA Damage Response in Prostate and Breast Cancer. Int. J. Mol. Sci. 21 (21). doi:10.3390/ijms21218273
Wiesner, J. B., Ugarkar, B. G., Castellino, A. J., Barankiewicz, J., Dumas, D. P., Gruber, H. E., et al. (1999). Adenosine Kinase Inhibitors as a Novel Approach to Anticonvulsant Therapy. J. Pharmacol. Exp. Ther. 289 (3), 1669–1677.
Williams-Karnesky, R. L., Sandau, U. S., Lusardi, T. A., Lytle, N. K., Farrell, J. M., Pritchard, E. M., et al. (2013). Epigenetic Changes Induced by Adenosine Augmentation Therapy Prevent Epileptogenesis. J. Clin. Invest. 123 (8), 3552–3563. doi:10.1172/JCI65636
Winter, C., Nilsson, M. P., Olsson, E., George, A. M., Chen, Y., Kvist, A., et al. (2016). Targeted Sequencing of BRCA1 and BRCA2 across a Large Unselected Breast Cancer Cohort Suggests that One-Third of Mutations Are Somatic. Ann. Oncol. 27 (8), 1532–1538. doi:10.1093/annonc/mdw209
Wu, C., and Bekaii-Saab, T. (2012). CpG Island Methylation, Microsatellite Instability, and BRAF Mutations and Their Clinical Application in the Treatment of Colon Cancer. Chemother. Res. Pract. 2012, 359041. doi:10.1155/2012/359041
Xie, K., Wang, Y.-X., Shen, H.-Y., Zhang, L.-M., Zeng, L.-H., and Shu, N.-B. (2014). Effect of Adenosine on hMLH1 Gene of Human Colorectal Cancer Cells. Nat. Med. J. China 2014 (94), 16. doi:10.3760/cma.j.issn.0376-2491.2014.16.016
Xie, L., Jiang, X., Li, Q., Sun, Z., Quan, W., Duan, Y., et al. (2018). Diagnostic Value of Methylated Septin9 for Colorectal Cancer Detection. Front. Oncol. 8, 247. doi:10.3389/fonc.2018.00247
Xu, Y., Wang, Y., Yan, S., Zhou, Y., Yang, Q., Pan, Y., et al. (2017). Intracellular Adenosine Regulates Epigenetic Programming in Endothelial Cells to Promote Angiogenesis. EMBO Mol. Med. 9 (9), 1263–1278. doi:10.15252/emmm.201607066
Yamamoto, H., and Imai, K. (2015). Microsatellite Instability: an Update. Arch. Toxicol. 89 (6), 899–921. doi:10.1007/s00204-015-1474-0
Yang, X., Xu, Z. J., Chen, X., Zeng, S. S., Qian, L., Wei, J., et al. (2019). Clinical Value of Preoperative Methylated Septin 9 in Chinese Colorectal Cancer Patients. World J. Gastroenterol. 25 (17), 2099–2109. doi:10.3748/wjg.v25.i17.2099
Yegutkin, G. G. (2014). Enzymes Involved in Metabolism of Extracellular Nucleotides and Nucleosides: Functional Implications and Measurement of Activities. Crit. Rev. Biochem. Mol. Biol. 49 (6), 473–497. doi:10.3109/10409238.2014.953627
Yh, L. (2017). Expression and Clinical Significance of Adenosine Kinase in Liver Cancer. J. Hubei Coll. Med. 36 (6), 504–507.
Zhang, L. M, Wang, Y.-X., Xie, K., Zeng, L.-H., and Li, Q. (2015). The Demethylation of DLC-1 Genes in HT-29 Cells by 5-iodonogenic Tuberculosis Journal of the Third Military. Medical University, 37(2), 106–110.
Zhang, Y., Zhang, L., Li, R., Chang, D. W., Ye, Y., Minna, J. D., et al. (2017). Genetic Variations in Cancer-Related Significantly Mutated Genes and Lung Cancer Susceptibility. Ann. Oncol. 28 (7), 1625–1630. doi:10.1093/annonc/mdx161
Keywords: DNA methylation, adenosine, receptor-independent pathway, adenosine kinase, ADK isoforms, ADK inhibitor, cancer therapy
Citation: Luo H-Y, Shen H-Y, Perkins RS and Wang Y-X (2022) Adenosine Kinase on Deoxyribonucleic Acid Methylation: Adenosine Receptor-Independent Pathway in Cancer Therapy. Front. Pharmacol. 13:908882. doi: 10.3389/fphar.2022.908882
Received: 31 March 2022; Accepted: 16 May 2022;
Published: 01 June 2022.
Edited by:
Ke-Wu Zeng, Peking University, ChinaReviewed by:
Mariachiara Zuccarini, University of Studies G. d’Annunzio Chieti and Pescara, ItalyElena De Marchi, University of Ferrara, Italy
Copyright © 2022 Luo, Shen, Perkins and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Xu Wang, MzAwODk3QGhvc3BpdGFsLmNxbXUuZWR1LmNu
†These authors have contributed equally to this work
 Hao-Yun Luo
Hao-Yun Luo Hai-Ying Shen
Hai-Ying Shen R. Serene Perkins4,5
R. Serene Perkins4,5