
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 07 October 2022
Sec. Pharmacogenetics and Pharmacogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.889473
Aim: In this study, we investigated the association between ABCC2 polymorphism and clopidogrel response as well as the associated hypothetical mechanism.
Methods: Chinese patients (213) with coronary artery disease (CAD) who underwent percutaneous coronary intervention (PCI) and received clopidogrel were recruited. Thereafter, their ADP-induced platelet inhibition rates (PAIR%) were determined via thromboelastometry. Further, the single-nucleotide polymorphisms (SNPs) of ABCC2 were genotyped using high-resolution melting curve (HRM)-PCR, while CYP2C19*2 and *3 polymorphisms were genotyped via real-time PCR.
Results: The allele frequencies of ABCC2 rs717620 were 74.88 and 25.12% for the C and T alleles, respectively. Further, ABCC2 rs717620 TT carriers exhibited significantly higher PAIR% values (72.60 ± 27.69) than both CT (61.44 ± 23.65) and CC carriers (52.72 ± 21.99) (p = 0.047 and p = 0.001, respectively), and ABCC2 rs717620 CT carriers showed significantly higher mean PAIR% values than ABCC2 rs717620 CC carriers (p = 0.011). However, the PAIR% values corresponding to ABCC2 rs2273697 and ABCC2 rs3740066 carriers were not different. Additionally, CYP2C19*2 AA carriers presented significantly lower PAIR% values than CYP2C19*2 GA (p = 0.015) and GG (p = 0.003) carriers, and CYP2C19*3 GA carriers also presented significantly lower PAIR% values than CYP2C19*3 GG carriers (p = 0.041). In patients with CYP2C19 extensive metabolizers (EM), ABCC2 rs717620 TT carriers showed significantly higher PAIR% values (89.77 ± 9.73) than CT (76.76 ± 26.00) and CC carriers (74.09 ± 25.29) (p = 0.040 and p = 0.009, respectively). In patients with CYP2C19 poor metabolizers (PM), ABCC2 rs717620 CC carriers showed significantly lower PAIR% values (51.72 ± 25.78) than CT carriers (75.37 ± 23.57) (p = 0.043). Furthermore, after adjusting for confounding factors, ABCC2 rs717620 was identified as a strong predictor of clopidogrel hyperreactivity.
Conclusion: We proposed a new target, ABCC2 rs717620, in the efflux pathway that affects individual responses to clopidogrel. The TT allele of ABCC2 rs717620 was also identified as an independent risk factor for clopidogrel hyperreactivity, and CYP2C19*2 and *3 showed association with an increased risk for clopidogrel resistance. Additionally, ABCC2 rs717620 may affect individual responses to clopidogrel via post-transcriptional regulation and interaction with CYP2C19. These findings provide new insights that may guide the accurate use of clopidogrel.
Clopidogrel is an antiplatelet agent that is widely used in patients with coronary artery disease (CAD) undergoing percutaneous coronary intervention (PCI) (Watanabe et al., 2019). However, pharmacodynamic responses to clopidogrel differ significantly between individuals (Akkaif et al., 2021). Further, CYP2C19 plays a vital role in clopidogrel transformation; thus, the Food and Drug Administration (FDA) recommends that tests be conducted for its identification prior to the commencement of clopidogrel treatment (Holmes et al., 2010). Specifically, clopidogrel shows high activity in patients with fast CYP2C19 metabolism (EM), while those with poor CYP2C19 metabolism (PM) often show clopidogrel resistance (Bolcato et al., 2021). Paradoxically, some studies have revealed that some patients with EM-type CYP2C19 show clopidogrel resistance rates as high as 34.6% ± 0.6% (Zhang et al., 2018), while 15.8% of patients with PM-type CYP2C19 experience bleeding events caused by clopidogrel hyperreactivity (Nagashima et al., 2013). Further, it has been observed that only 15% of clopidogrel is metabolized, while up to 85% is excreted, but CYP2C19 is only responsible for the metabolism of this drug (Simon et al., 2009). Thus, when CYP2C19 is the only factor considered to guide clopidogrel treatment, there remains a risk of bleeding or thrombosis. Furthermore, it is still unclear whether targets in other pathways are involved in individual responses to clopidogrel. Interestingly, the efflux pathway for clopidogrel has rarely been reported. ABCC2, which is an ATP binding cassette efflux transporter that belongs to the ATP-binding cassette (ABC) superfamily of transmembrane proteins, is primarily expressed in polarized cells, such as hepatocytes, intestinal epithelia, and renal proximal tubule cells (Zhu et al., 2022). Specifically, ABCC2 plays an important role in the use of ATP hydrolysis energy to transfer its binding substrate out of the plasma membrane and mediate the transmembrane transport of several exogenous and endogenous drug metabolites. Thus, altered ABCC2 functioning owing to ABCC2 single nucleotide polymorphisms (SNPs) can change the clearance, organ distribution, and absorption of several clinically important drugs, including antibiotics (e.g., ceftriaxone, rifampicin, and ampicillin), toxins, anti-hyperlipidemia inhibitors, and several cancer chemotherapy drugs (e.g., MTX, vinblastine, and irinotecan) (Jemnitz et al., 2010). ABCC2 rs717620, rs2273697, and rs3740066, are the three common SNP genotypes of ABCC2 in East-Asian (Han Chinese) ethnic population according to the SNP database of 1000 Genomes Project Database (www.1000genomes.org). These three SNPs are related to several clinical functions. For example, ABCC2 rs717620 is related to the low effect of simvastatin treatment on HDL-C level in Chinese Han population (Liu et al., 2018). Further, in the Asia Pacific epilepsy cohort, ABCC2 rs2273697 and rs3740066 polymorphisms were associated with antiepileptic drug resistance (Sha’Ari et al., 2014). It has also been observed that ABCC2 rs3740066 is significantly associated with the vomiting of docetaxel in patients with breast cancer (Jabir et al., 2018). Therefore, in this study, these three SNPs were selected for further research.
Interestingly, the relationships between ABCC2 gene polymorphisms, clopidogrel reactivity, and individual responses to clopidogrel have not yet been reported. Therefore, we investigated the effect of ABCC2 gene polymorphisms on the variability of responses to clopidogrel and the possible associated mechanisms. The results thus obtained will provide insights into the effect of ABCC2 polymorphisms on clopidogrel precision medicine, and the clarification of the possible associated mechanisms will also provide new ideas for the efflux of other ABCC2-dependent drugs in other diseases.
Blood samples were collected from 213 hospitalized patients with CAD aged 20–75 years, who underwent PCI between January 2020 and December 2021. Following their first PCI for CAD at the Department of Cardiology of the China-Japan Friendship Hospital, the patients received a daily maintenance clopidogrel dose of 75 mg for 7 days. Finally, samples were collected 6 h after the administration of the treatment on Day 7. Patients with liver, kidney, or lung failure, malignant tumors or hemorrhagic diseases were excluded. The study protocol was reviewed and approved by the Clinical Research Ethics Committee of the China-Japan Friendship Hospital (No. 2019-96-K64). All the study procedures were performed in accordance with the principles set forth in the Declaration of Helsinki.
The clopidogrel reactivity levels corresponding to the subjects were assessed using the TEG 5000 Thrombelastograph® Hemostasis Analyzer System (Haemoscope; Haemonetics Corporation, Braintree, MA, United States) and expressed as ADP-induced platelet inhibition rates (PAIR%). Further, on Day 7 after the PCI, maximal amplitudes (MA)-ADP, R values (coagulation reaction time), K angle (coagulation time), and coagulation indexes (CIs) were determined using the TEG 5000 system, and ADP% values were calculated using a software based on the following expression: PAIR (%) =
Genomic DNA was extracted from ethylenediamine tetra acetic acid (EDTA)-anticoagulated peripheral blood (4 ml) using the Thermo Kingfisher Flex system (BOKUN BIOTECH, Changchun, China) according to the manufacturer’s instructions and stored at −40°C until further analysis.
(1) Detection of ABCC2 gene polymorphism and genotyping
A high-resolution melting curve (HRM)-PCR ABCC2 genotyping assay was developed to detect ABCC2 gene polymorphisms. The detection assay is detailed in the Chinese patent application: “A primer design method and kit for the detection of human ABCC2 gene polymorphism” (CN Patent number ZL 201910110953.6). The HRM-PCR was performed with our ABCC2 genotyping assay using a LightCycler® 480 instrument (Roche, Basel, Switzerland) to detect the three common SNP genotypes of ABCC2 (rs717620, rs2273697, and rs3740066) in East-Asian ethnic population, as described in the introduction section. The genotypes of these three SNPs can be clearly and rapidly distinguished using this method, which is consistent with the sequence typing results.
(2) Determination of CYP2C19 gene polymorphisms and genotyping
Genetic testing for CYP2C19 revealed the alleles CYP2C19*1, CYP2C19*2 (variants in exons 2 and 5) and CYP2C19*3 (variants in exon 4) (Khaliq et al., 2000). Using the procedure of de De Morais et al. (1994) genomic DNA was extracted, amplified, and analyzed by restriction fragment length. In brief, the patients were divided into three metabolic groups based on CYP2C19 genotypes (CYP2C19 *1 [wild type], CYP2C19*2 rs4244285 [c.681G > A], and CYP2C19*3 rs4986893 [c.636G > A]), as follows: extensive metabolizers (EM; CYP2C19*1/*1), intermediate metabolizers (IM; CYP2C19*1/*2 or *1/*3), and poor metabolizers (PM; CYP2C19*2/*2 or *3/*3).
Statistical analyses were performed using SPSS software version 20.0 (IBM Corp, Armonk, NY, United States). Categorical data were summarized as counts (percentages), while continuous variables were expressed as the mean ± standard deviation (SD). The power to calculate sample size was estimated by the PS-Power Sample Size software (version 3.1.2). Two-group comparisons were performed by conducting the Wilcoxon rank-sum test or χ2 test, when appropriate. Deviation from Hardy-Weinberg equilibrium (HWE) for ABCC2 and CYP2C19 SNPs was assessed using the χ2 tests. Further, a stepwise multiple linear regression analysis was performed to assess the most appropriate SNP parameter combination for estimating PAIR%. A multivariate logistic regression model was also constructed to adjust for important confounders and to assess clopidogrel resistance/hyperreactivity. Effect size estimates were provided as odds ratios (ORs) and 95% CIs. All the statistical tests were two-tailed, and p values <0.05 were considered statistically significant.
We have used the RNA structure software (http://rna.urmc.rochester.edu/RNAstructureWeb/) to predict the effect of the rs717620 mutant allele on the RNA secondary structure of ABCC2. Using the ABCC2 rs717620 5′-UTR mRNA sequence as the input, the predicted secondary structure was verified to ensure that it has a negative minimum folding free energy (MFE) and a high minimum folding free energy index (MFEI), and the optimal mRNA 5′-UTR stem-loop secondary structures for wild-type and mutant ABCC2 rs717620 served as output.
Overall, 213 patients with CAD who underwent PCI were recruited, and their baseline characteristics are shown in Table 1. Basically, 211 (99.06%) of the patients were Han Chinese. Further, there were 23 (10.8%) patients in the HPR group and 190 (89.2%) in the LPR group, and the LPR group showed significantly higher high-density lipoprotein-cholesterol (HDL-C) levels than the HPR group (p = 0.029). Furthermore, there were no significant differences between the two groups with respect to sex, age, ethnicity (Han Chinese), platelet count (PLT), mean platelet volume (MPV), homocysteine (HCY) levels, total cholesterol (CHO) levels, triglyceride (TG) levels, or low-density lipoprotein cholesterol (LDL-C) levels.
The allele frequencies of all the five SNPs (ABCC2 rs717620, rs2273697, rs3740066, CYP2C19*2, and CYP2C19*3) in the patients were in accordance with the Hardy-Weinberg equilibrium (HWE). In ABCC2 rs717620, the allele frequencies were 74.88 and 25.12% for C- and T-alleles, respectively. In CYP2C19*2, they were 74.41 and 25.59% for G- and A-alleles, respectively, and in CYP2C19*3, they were 93.90 and 6.10% for G- and A-alleles, respectively. Frequency comparison with East-Asian (Han Chinese) reference subpopulations was shown in Supplement Table S1, indicated no significant difference between our study and reference subpopulations. The p-values of all the five SNPs (ABCC2 rs717620, rs2273697, rs3740066, CYP2C19*2, and CYP2C19*3) were 0.248, 0.123, 0.929, 0.117, 0.753, respectively, indicating that all the allele frequencies of ABCC2 rs717620, rs2273697, rs3740066, CYP2C19*2 (rs4244285), and CYP2C19 *3 (rs4986893) were consistent with the known East-Asian (Han Chinese) ethnic frequency reported in the 1000 Genomes Project Database (www.1000genomes.org).
Correlation analyses were performed to determine the correlation between the three ABCC2 genetic polymorphisms and the inhibitory effect of clopidogrel on platelet reactivity. Thus, it was observed that ABCC2 rs717620 TT carriers exhibited significantly higher PAIR% values (72.60 ± 27.69) than both the CT (61.44 ± 23.65) and CC carriers (52.72 ± 21.99) (p = 0.047 and p = 0.001, respectively). The ABCC2 rs717620 CT group also presented significantly higher mean PAIR% values (61.44 ± 23.65) than the ABCC2 rs717620 CC homozygotes (52.72 ± 21.99) (p = 0.011) (Figure 1A). However, neither ABCC2 rs2273697 nor ABCC2 rs3740066 polymorphisms affected PAIR% values (Figures 1B,C). Additionally, CYP2C19*2 AA carriers presented significantly lower mean PAIR% values (59.60 ± 26.07) than CYP2C19*2 GA carriers (74.84 ± 25.09) (p = 0.015). Our results also indicated that CYP2C19*2 AA carriers presented significantly lower mean PAIR% values (59.60 ± 26.07) than CYP2C19*2 GG carriers (77.63 ± 23.69) (p = 0.003) (Figure 1D). It was also evident that CYP2C19*3 GA carriers presented significantly lower mean PAIR% values (66.39 ± 25.68) than CYP2C19*3 GG carriers (78.91 ± 21.42) (p = 0.041). However, neither CYP2C19*3 AA and GA nor CYP2C19*3 AA and GG carriers presented any significant differences in PAIR% values (Figure 1E). Thus, it was observed that patients with CYP2C19 PM presented significantly lower PAIR% values (60.51 ± 27.37) than patients with IM (74.92 ± 25.94) or EM (77.18 ± 24.01) (p = 0.007 and p = 0.002, respectively) (Figure 1F). Based on the CYP2C19 metabolic phenotype, stratification analysis for the determination of the influence of ABCC2 rs717620 on PAIR% was further conducted. Conversely, the ABCC2 rs717620 polymorphism exerted an influence on the PAIR% values corresponding to patients with EM and PM type CYP2C19, but not on the values corresponding to patients with IM type CYP2C19. Further, in patients with EM type CYP2C19, the PAIR% values corresponding to ABCC2 rs717620 TT carriers were significantly higher (89.77 ± 9.73) than those corresponding to the CT (76.76 ± 26.00) and CC (74.09 ± 25.29) groups (p = 0.040 and p = 0.009, respectively). Notably, the average PAIR% value of the CYP2C19 EM group was 77.18, and relatively, when CYP2C19 EM was combined with ABCC2 rs717620 TT, the average PAIR% value was 89.77, indicative of a 12.59 increase. When the CYP2C19 EM group was also combined with ABCC2 rs717620 CT, the average PAIR% value was 76.76, i.e., it basically remained unchanged. Further, when the CYP2C19 EM group was combined with ABCC2 rs717620 CC, the average PAIR% value was 74.09, indicative of a 3.09 decrease. Furthermore, in patients with CYP2C19 PM, the PAIR% values corresponding to ABCC2 rs717620 CC carriers (51.72 ± 25.78) were significantly lower than those corresponding to CT carriers (75.37 ± 23.57) (p = 0.043). Additionally, the average PAIR% value of the CYP2C19 PM group was 60.51, and relatively, when the CYP2C19 PM group was combined with ABCC2 rs717620 CC, the average PAIR% value was 51.72, indicative of a decrease of 8.79. However, the PAIR% values corresponding to the ABCC2 rs717620 TT, CT, and CC groups were not significantly different among patients with IM type CYP2C19 (Figure 1G).
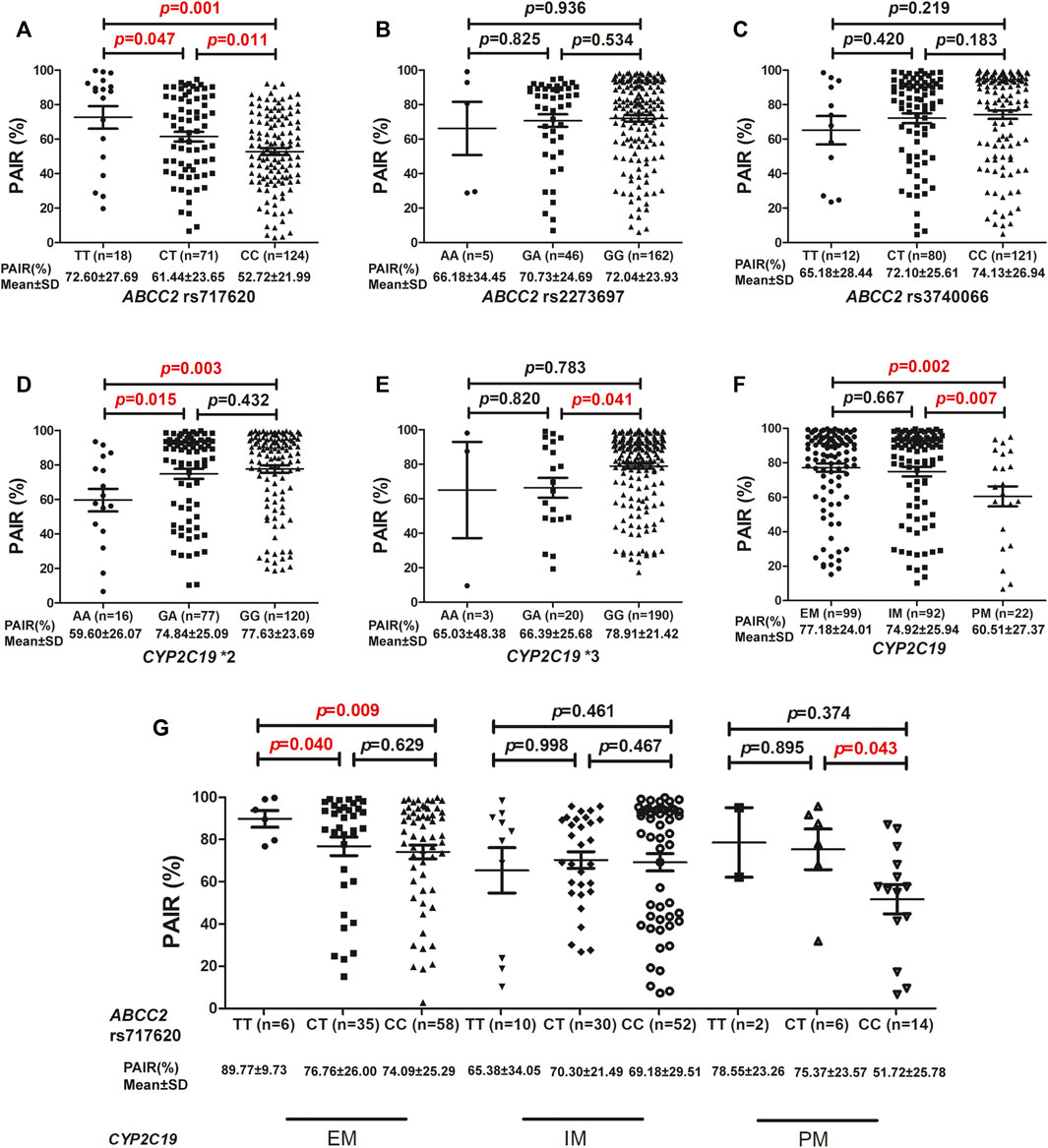
FIGURE 1. Association between ABCC2 and CYP2C19 based on PAIR% values. Association between PAIR% values and (A) ABCC2 rs717620, (B) ABCC2 rs2273697, (C) ABCC2 rs3740066, (D) CYP2C19*2, (E) CYP2C19*3, and (F) CYP2C19 metabolic phenotype distributions. (G) Influence of ABCC2 rs717620 polymorphism on the PAIR% values of different CYP2C19 metabolic phenotypes. PAIR(%), ADP-induced platelet aggregation inhibition rate.
Table 2 shows the strength of the relationships between PAIR% values and clinical variables based on stepwise linear regression analysis. The factors that remained significantly correlated with the PAIR% values were ABCC2 rs717620 (p < 0.001), CYP2C19 *2 (p < 0.001), CYP2C19 *3 (p < 0.001), and CYP2C19 metabolic phenotypes (p = 0.022) (Table 2). The removed factors, including ABCC2 rs2273697, ABCC2 rs3740066, sex, age, ethnicity (Han Chinese), PLT, MPV, HCY, CHO, TG, HDL-C, and LDL-C showed no significant correlation with the PAIR% values in the stepwise regression analysis. Further, the variance inflation factor (VIF) showed no multicollinearity between the factors (VIF <5), and the PAIR% variability (R2) that could be explained by the ABCC2 rs717620, CYP2C19 *2, CYP2C19 *3, and CYP2C19 metabolic phenotypes was 0.696, 0.506, 0.623, and 0.710, respectively, and 0.852 collectively.
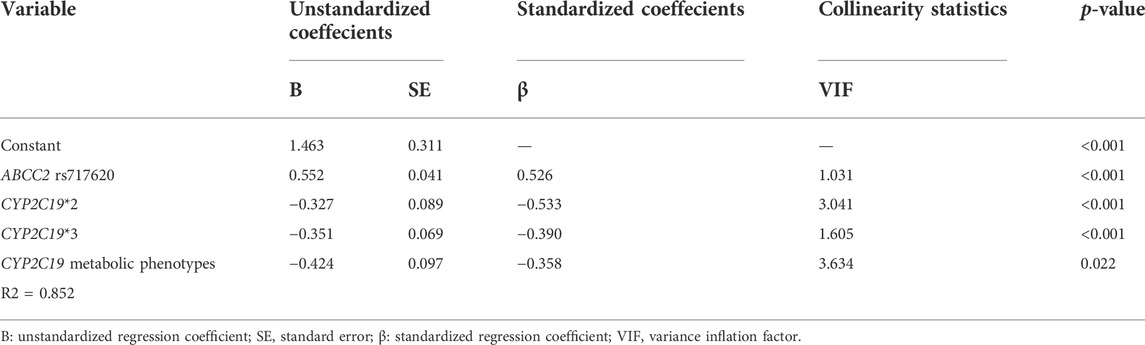
TABLE 2. Relationship between PAIR% values and clinical variables based on stepwise linear regression analysis.
To investigate whether the ABCC2 rs717620 gene polymorphism is a risk factor for clopidogrel resistance or hyperreactivity, we performed a multivariate logistic regression analysis after adjusting for the covariates that could potentially affect the PAIR% values, including ABCC2 rs2273697, ABCC2 rs3740066, sex, age, ethnicity (Han Chinese), PLT, MPV, HCY, CHO, TG, HDL-C, and LDL-C. For ABCC2 rs717620, three genetic models (additive, dominant, and recessive) were used to analyze the risk of clopidogrel resistance and hyperreactivity. The results showed that, in the three genetic models, ABCC2 rs717620 remained an independent risk factor for LPR (clopidogrel hyperreactivity) (CC vs. CT: OR, 5.132; 95% CI, 1.855–14.196; p = 0.002; CC vs. TT: OR, 9.962; 95% CI, 1.065–93.172; p = 0.044; CC vs. CT + TT: OR, 5.076; 95% CI, 1.953–13.193; p = 0.001; CC + CT vs. TT: OR, 1.655; 95% CI, 0.173–15.869; p = 0.002). Additionally, for CYP2C19*2 and CYP2C19*3, the additive model, out of three genetic model, was found to be adequate based on its metabolism function. CYP2C19*2 also appeared to be a protective factor for LPR (GG vs. GA: OR, 0.044; 95% CI, 0.002–0.970; p = 0.048; GG vs. AA: OR, 0.002, 95% CI, 0.001–0.343; p = 0.019), and CYP2C19*3 was identified as a major protective factor for LPR (GG vs. GA: OR, 0.041; 95% CI, 0.002–0.717; p = 0.029; GG vs. AA: OR, 0.010; 95% CI, 0.005–0.081; p = 0.002) (Table 3).
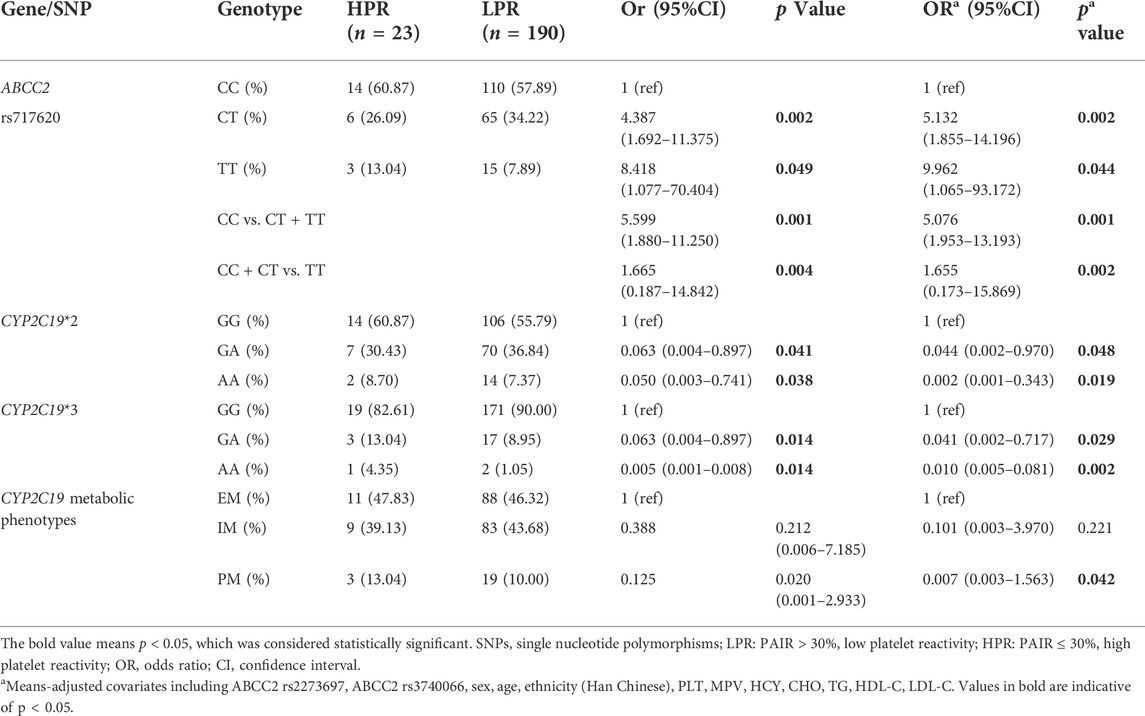
TABLE 3. Association between ABCC2 and CYP2C19 SNPs and clopidogrel hyperreactivity in 213 patients based on multivariate logistic regression analysis.
The ABCC2 transporter belongs to the family of ATP-binding cassette (ABC) transporters, in the liver, ABCC2 is localized on the bile canalicular membrane of hepatocytes (Toyoda et al., 2016), which transport substrates through cell membranes by binding and hydrolyzing ATP (Higgins, 2007) (Figure 2).
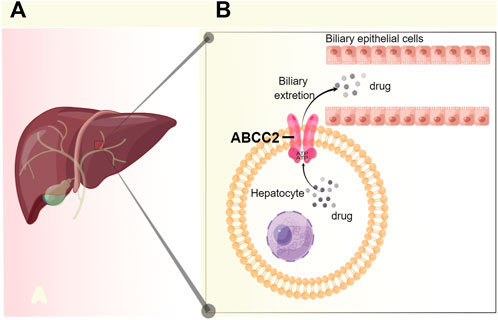
FIGURE 2. Schematic diagram of the anatomical, cell biological placement, and function of ABCC2 (A) In the liver, ABCC2 is localized on the bile canalicular membrane of hepatocytes. (B) ABCC2 transport drug by an ATP-dependent manner, and a part of drug is excreted from hepatocyte into bile by ABCC2. Figure adapted from ref. (Higgins, 2007; Toyoda et al., 2016). The figure was drawn using figdraw (https://www.figdraw.com/).
The mechanism underlying the effect of ABCC2 rs717620 on the differential regulation of clopidogrel was investigated. Thus, the cDNA sequence of the 5′UTR part-length of human ABCC2 rs717620 wild-type and mutant are shown in Figures 3A,B, respectively. Secondary structure modeling by our team revealed a change in the mRNA 5′-UTR secondary structure of ABCC2, i.e., the pre-experimental modeling showed that, in rs717620 allele C, the AUG site of the Kozak sequence recognized by the pre-translation initiation complex formed a single chain “ring” structure (Figure 3C). Further, in rs717620 allele T (U in mRNA), U at the -24 site was complementary to the Kozak sequence, forming a hairpin structure; thus, AUG of the Kozak sequence was on the “stem” structure (Figure 3D).
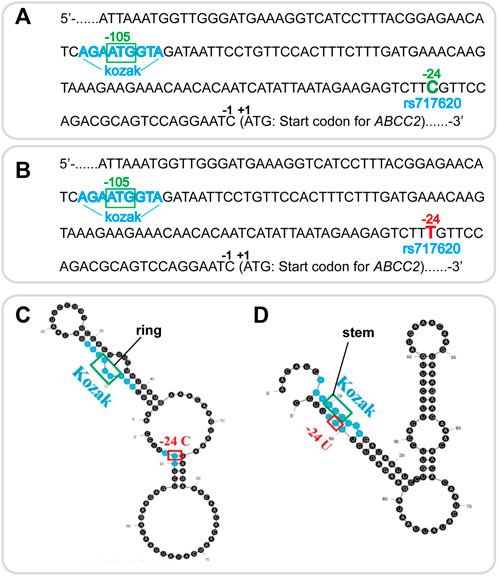
FIGURE 3. Wildtype and mutation in the mRNA 5′-UTR secondary structure of ABCC2 rs717620 based on secondary structure modeling.(A) cDNA sequence of the 5′UTR part-length of human ABCC2. The Kozak sequence is colored blue, and the upstream open reading frame beginning at −105 ATG is shown in the green box. The rs717620 C site at -24 is colored green. (B) rs717620 T site at −24 (colored red). (C) mRNA sequence of the 5′UTR region of human wildtype ABCC2 rs717620. In the C allele, the Kozak AUG sequence showed a “ring” structure, enabling Kozak to bind easily to the pre-translation initiation complex. (D) Complementary combination of the T allele and the Kozak sequence with AUG on the “stem”, showing the possibility of missing the opportunity for a complementary combination with the AUG of the Kozak sequence, resulting in missed scanning.
Based on the function of ABCC2, its anatomical and cell biological placement (Figure 2), and modeling of the mRNA secondary structure (Figure 3), and the association showed in our study (Figure 1; Tables 2,3), we proposed a hypothetical mechanism as follows: ABCC2 rs717620 possibly mediates individual differences in clopidogrel responses via post-transcriptional regulation (Figure 4). The specific steps of the hypothetical mechanism are as follows: ABCC2 rs717620 is located at position 24 in the 5′-UTR region of the ABCC2 mRNA, and when ABCC2 rs717620 T is transcribed to ABCC2 rs717620 U, the U and Kozak sequences of ABCC2 complement each other, making it difficult for the transcription initiation complex to complement and combine with AUG, resulting in “leaky scanning”. This leaky scanning of Kozak then significantly reduces the efficiency of ribosomal translation and ABCC2 protein expression. It also reduces the number of ABCC2 efflux transporters at the hepatocyte membrane. Further, a decrease in ABCC2 efflux transporters results in a decrease in the efflux of intracellular clopidogrel out of the plasma membrane, resulting in the increased accumulation and metabolic utilization of the drug. Furthermore, under the action of P450 enzymes, mainly CYP2C19, the amount of clopidogrel converted from inactive clopidogrel to 2-oxo-clopidogrel and then to active clopidogrel increases. This step is also regulated by the CYP2C19 gene polymorphism.
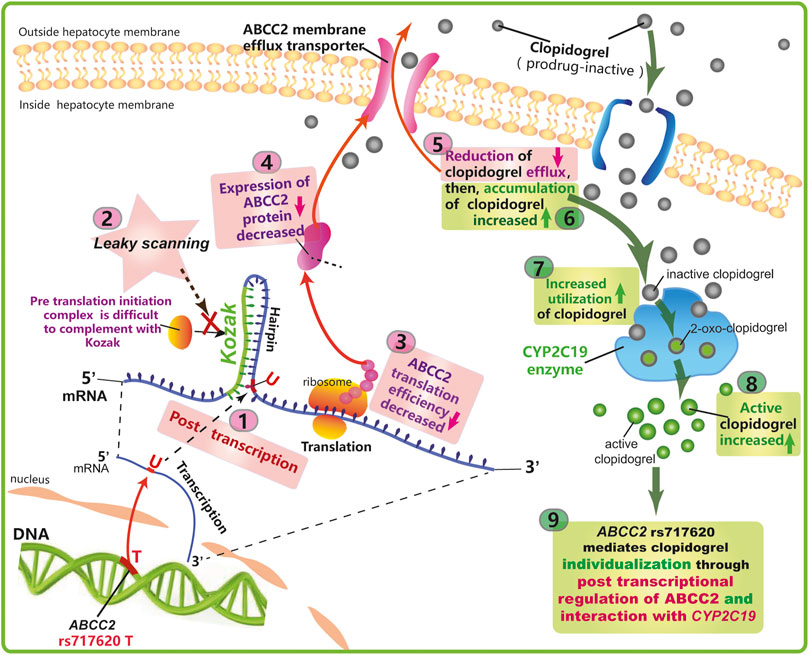
FIGURE 4. Proposed mechanism underlying the effect of ABCC2 rs717620 on the differential regulation of clopidogrel.
Taken together, this study showed that only a single CYP2C19 target cannot guide the use of clopidogrel. CYP2C19 needs to be detected together with the new target, ABCC2 rs717620 (and perhaps other unknown targets) to guide medical professionals in clopidogrel precision medicine. Our research group will explore this hypothesis in future mechanistic studies (Figure 5).
ABCC2 is an ATP binding cassette transporter, which is expressed, translocated, and inserted into the apical plasma membrane. Notably, ABCC2 can transfer its substrate, including glucuronide, glutathione, and the sulfate conjugates of several endogenous and exogenous substances, across membranes. The global variability of ABCC2 includes 27,843 SNPs, such as rs717620, rs2273697, rs3740066, rs927344, rs27843, etc. The global allele frequencies are 13.50, 18.65, 28.81, 0.54, 3.35, respectively in the SNP database of the 1000 Genomes Project Database (www.1000genomes.org).
The altered functioning of ABCC2 SNPs, especially ABCC2 rs717620, rs2273697, and rs3740066 can alter the clearance, distribution, and absorption of several clinically important drugs. For example, the ABCC2 rs717620 T variant is associated with an increased risk of hyperbilirubinemia and mortality in patients with drug-induced liver injury (Huang et al., 2021). It has also been observed that in patients with epilepsy, ABCC2 rs2273697 and rs3740066 polymorphisms increase blood carbamazepine concentrations (Sha’Ari et al., 2014). Further, the ABCC2 rs717620 genotype is significantly associated with an increased risk of a decrease in simvastatin dose use (Becker et al., 2013). However, there is no study on the relationship between ABCC2 and individual differences in clopidogrel response. It is well known that CYP2C19 plays a critical role in the biological activity of clopidogrel. Further, in patients with loss of function (LOF) alleles (CYP2C19*2 and CYP2C19*3), metabolic transformation slows down, resulting in higher platelet reactivity. Thus, the LOF variation of CYP2C19 affects the response and clinical outcome of clopidogrel. Further, mutations in the CYP2C19 gene have also been identified as strong predictors of platelet aggregation.
In this study, 213 Chinese patients with coronary heart disease were included and the influence of ABCC2 gene polymorphisms (rs717620, rs2273697, and rs3740066), CYP2C19*2, and CYP2C19*3 on clopidogrel response were analyzed. Consistent with most previous studies, the five SNP allele frequencies observed were similar to those reported for East-Asian (Han Chinese) subpopulations in the SNP database of the 1000 Genomes Project Database (www.1000genomes.org).
Further, the ABCC2 rs717620 efflux pathway was found to be closely related to the differential responses to clopidogrel. We also observed that ABCC2 rs717620 TT carriers exhibited significantly higher PAIR% values and increased risk of clopidogrel hyperreactivity. Thus, to the best of our knowledge, this is the first study to report that ABCC2 rs717620 gene polymorphism is significantly correlated with individual responses to clopidogrel. Further, carriers of CYP2C19*2 and CYP2C19*3 showed increased risk of clopidogrel resistance. This conclusion is consistent with that of a previous study (Li et al., 2018), and in addition to CYP2C19, N6AMT1 rs2254638 polymorphism also had an effect on clopidogrel resistance in Chinese patients with coronary heart disease. Thus, could be used as an independent biomarker for predicting clopidogrel resistance.
Additionally, ABCC2 rs717620 brought about significant differences in various metabolic CYP2C19 types. The difference in PAIR% among the ABCC2 rs717620 genotypes was observed in both CYP2C19 EM and PM genotype patients, suggesting that there may be interactions between ABCC2 rs717620 and CYP2C19 in the efflux and metabolic utilization of clopidogrel in vivo, implying that the TT genotype of ABCC2 rs717620 and the CYP2C19 EM metabolic type may jointly lead to low platelet reactivity and clopidogrel hyperreactivity; and that possibly, ABCC2 rs717620 CC and CYP2C19 PM work together to bring about high platelet reactivity and clopidogrel resistance. These findings suggests that not only CYP2C19, but the ABCC2 rs717620 of different genotypes also play a role in these patients in determining response to clopidogrel. The specific interaction needs to be studied further.
Stepwise multiple linear regression model showed that there were positive correlations between the ABCC2 rs717620 with the PAIR%, indicating that an increase in ABCC2 rs717620 T-allele would be associated with a higher PAIR% and clopidogrel hyperreactivity. In contrast, the negative correlations between the CYP2C19 *2, CYP2C19 *3, and CYP2C19 metabolic phenotypes and PAIR% values indicated that an increase in CYP2C19 A-allele would be associated with a lower PAIR% value and clopidogrel resistance.
Based on the multivariable-adjusted logistic regression analysis of additive, dominant, and recessive genetic models, we observed that the ABCC2 rs717620 T allele was an independent risk factor for clopidogrel hyperreactivity. Therefore, in addition to the metabolic pathway for CYP2C19 utilization, in this study, a new target, ABCC2 rs717620, of the efflux pathway, which was shown to exert a significant effect on individual responses to clopidogrel, was identified for the first time. Further, there were significant differences between CYP2C19*2 and CYP2C19*3, indicating that CYP2C19*2 and *3 were independent risk factors for clopidogrel resistance. Zhifu et al. (Wang et al., 2019) reported that Han Chinese patients with allele A in CYP2C19*2 and *3 are susceptible to high platelet reactivity and clopidogrel resistance after clopidogrel administration. Consistent with the results of several previous studies, our finding further supported the predictive role of CYP2C19 polymorphism in clopidogrel efficacy in East-Asian (Han Chinese) ethnic population.
ABCC2 rs717620 is located on the -24 site of the ABCC2 5ʹ-UTR mRNA (Zhang et al., 2010). Further, the role of the Kozak sequence is to combine with the pre-translation initiation complex to improve translation efficiency and protein expression (Li et al., 2018). Therefore, by modeling the ABCC2 5ʹ-UTR mRNA secondary structure, we proposed a hypothetical mechanism as follows: ABCC2 rs717620 may mediate individual differences in response to clopidogrel via post-transcriptional regulation. This implies that a decrease in clopidogrel efflux from inside hepatocyte membranes to outside hepatocyte membranes results in its increased accumulation as well as CYP2C19 metabolic utilization, consequently enhancing the reactivity of clopidogrel. (Kozak, 2005).Further, the hypothesis that ABCC2 gene polymorphisms affect individual differences in clopidogrel reactivity via changes in mRNA secondary structure, which needs to be investigated further, provides a new direction for future research. Several studies have shown that ABCC2 SNPs mediate the efflux of irinotecan, cisplatin, and methotrexate from intracellular to extracellular spaces in colorectal cancer, ovarian cancer, and acute lymphoblastic leukemia (Corpechot et al., 2020; Huang et al., 2021). Therefore, this hypothetical mechanism may also provide novel ideas regarding ABCC2 that will facilitate studies on the mechanism of other drugs in other diseases.
This study had some limitations. First, the rapid allele CYP2C19*17 was not genotyped (Sim et al., 2006), which would mis-classify the normal phenotype. This would be genotyped in our future research. Second, clinical information on comorbidity and concomitant medication were not all collected. This would be considerd in our future studies. Third, the clinical outcomes of the patients included in this study was not considered. This would also be supplemented in our future research. Fourth, the exact mechanisms were not comprehensively investigated. Thus, further studies are still needed in this regard.
Based on the results of this study, we proposed that the ABCC2 efflux pathway affects individual responses to clopidogrel, and also suggested a novel potential target, ABCC2 rs717620, i.e., our results confirmed that ABCC2 rs717620 as well as CYP2C19*2 and *3 are related to individual responses to clopidogrel, and there may be interactions between CYP2C19 and ABCC2 rs717620 metabolic types. Moreover, the TT of ABCC2 rs717620 was found to be an independent risk factor for clopidogrel hyperreactivity, while CYP2C19*2 and *3 were found to be independent risk factors for clopidogrel resistance. Further, preliminary mechanistic studies suggested that ABCC2 rs717620 alters the secondary structure of mRNA and causes leaky Kozak scanning. This post-transcriptional regulation may be the reason for the individual differences in clopidogrel repsonse; however, further indepth studies are still needed in this regard. We hope that the results of this study provide insights into new targets to facilitate personalized decision making regarding clopidogrel therapy. Moreover, further research on the hypothetical mechanism by which ABCC2 affects individualized difference in clopidogrel response may also provide new ideas regarding the mechanism of individualized differences in responses to other drugs dependent on ABCC2.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Clinical Research Ethics Committee of China-Japan Friendship Hospital (No. 2019-96-K64). The patients/participants provided their written informed consent to participate in this study.
LC designed the study, conducted experiments, and wrote the manuscript. CZ and MH recruited the subjects and genotyped them. MZ and PG analyzed the data. YC and LM edited the manuscript. All authors have contributed to the manuscript and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Grant No. 82074221), Key Clinical Specialty Project of Beijing (Grant No. 2020), National High Level Hospital Clinical Research Funding, Elite Medical Professionals Project of the China-Japan Friendship Hospital [No. ZRJY2021 (Grant no. ZRJY2021-GG03)], and the Scientific Research Fund of the China-Japan Friendship Hospital (Grant No. 2019-1-QN-50).
We thank Professor Cheng Xiao and Zongping Chang from the scientific research center of the China-Japan Friendship Hospital for their valuable suggestion regaring experimental design, and Wenquan Niu and Xiangling Deng from the China-Japan Friendship Hospital for their valuable guidance on data analysis. Thank you to my husband, child, and mother for enlightening my life.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.889473/full#supplementary-material
Akkaif, M. A., Daud, N., Sha'aban, A., Ng, M. L., Abdul Kader, M. A. S., Noor, D. A. M., et al. (2021). The role of genetic polymorphism and other factors on clopidogrel resistance (CR) in an asian population with coronary heart disease (CHD). Molecules 26 (7), 1987. doi:10.3390/molecules26071987
Becker, M. L., Elens, L. L., Visser, L. E., HofmAn, A., Uitterlinden, A. G., van Schaik, R. H. N., et al. (2013). Genetic variation in the ABCC2 gene is associated with dose decreases or switches to other cholesterol-lowering drugs during simvastatin and atorvastatin therapy. Pharmacogenomics J. 13 (3), 251–256. doi:10.1038/tpj.2011.59
Bolcato, L., Khouri, C., Veringa, A., Alffenaar, J. W. C., Yamada, T., Naito, T., et al. (2021). Combined impact of inflammation and pharmacogenomic variants on voriconazole trough concentrations: A meta-analysis of individual data. J. Clin. Med. 10 (10), 2089. doi:10.3390/jcm10102089
Corpechot, C., Barbu, V., Chazouilleres, O., Broue, P., Girard, M., Roquelaure, B., et al. (2020). Genetic contribution of ABCC2 to Dubin-Johnson syndrome and inherited cholestatic disorders. Liver Int. 40 (1), 163–174. doi:10.1111/liv.14260
De Morais, S. M., Wilkinson, G. R., Blaisdell, J., Meyer, U. A., NaKamura, K., and Goldstein, J. A. (1994). Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol. Pharmacol. 46 (4), 594–598.
Higgins, C. F. (2007). Multiple molecular mechanisms for multidrug resistance transporters. Nature 446 (7137), 749–757. doi:10.1038/nature05630
Holmes, D. J., Dehmer, G. J., Kaul, S., Leifer, D., O'Gara, P. T., and Stein, C. M. (2010). ACCF/AHA clopidogrel clinical alert: Approaches to the FDA "boxed warning": A report of the American College of Cardiology foundation task force on clinical expert consensus documents and the American heart association endorsed by the society for cardiovascular angiography and interventions and the society of thoracic surgeons. J. Am. Coll. Cardiol. 56 (4), 321–341. doi:10.1016/j.jacc.2010.05.013
Huang, Y. S., Chang, T. E., Perng, C. L., and Huang, Y-H. (2021). The association of transporter ABCC2 (MRP2) genetic variation and drug-induced hyperbilirubinemia. J. Chin. Med. Assoc. 84 (2), 129–135. doi:10.1097/JCMA.0000000000000470
Jabir, R. S., Ho, G. F., Annuar, M., and Stanslas, J. (2018). Association of allelic interaction of single nucleotide polymorphisms of influx and efflux transporters genes with nonhematologic adverse events of docetaxel in breast cancer patients. Clin. Breast Cancer 18 (5), e1173–e1179. doi:10.1016/j.clbc.2018.04.018
Jemnitz, K., Heredi-Szabo, K., Janossy, J., Ioja, E., Vereczkey, L., and Krajcsi, P. (2010). ABCC2/Abcc2: A multispecific transporter with dominant excretory functions. Drug Metab. Rev. 42 (3), 402–436. doi:10.3109/03602530903491741
Khaliq, Y., Gallicano, K., SeguIn, I., FyKe, K., CariGnan, G., Bulman, D., et al. (2000). Single and multiple dose pharmacokinetics of nelfinavir and CYP2C19 activity in human immunodeficiency virus-infected patients with chronic liver disease. Br. J. Clin. Pharmacol. 50 (2), 108–115. doi:10.1046/j.1365-2125.2000.00238.x
Kozak, M. (2005). Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37. doi:10.1016/j.gene.2005.06.037
Li, H., Zhang, Y. J., Li, M. P., Hu, X. L., Song, P. Y., Peng, L. M., et al. (2018). Association of N6AMT1 rs2254638 polymorphism with clopidogrel response in Chinese patients with coronary artery disease. Front. Pharmacol. 9, 1039. doi:10.3389/fphar.2018.01039
Liu, N., Yang, G., Hu, M., Cai, Y., Hu, Z., Jia, C., et al. (2018). Association of ABCC2 polymorphism and gender with high-density lipoprotein cholesterol response to simvastatin. Pharmacogenomics 19 (14), 1125–1132. doi:10.2217/pgs-2018-0084
Nagashima, Z., Tsukahara, K., Morita, S., Endo, T., Sugano, T., Hibi, K., et al. (2013). Platelet reactivity in the early and late phases of acute coronary syndromes according to cytochrome P450 2C19 phenotypes. J. Cardiol. 62 (3), 158–164. doi:10.1016/j.jjcc.2013.03.006
Sha'Ari, H. M., Haerian, B. S., Baum, L., Saruwatari, J., Tan, H. J., Rafia, M. H., et al. (2014). ABCC2 rs2273697 and rs3740066 polymorphisms and resistance to antiepileptic drugs in Asia Pacific epilepsy cohorts. Pharmacogenomics 15 (4), 459–466. doi:10.2217/pgs.13.239
Sim, S. C., Risinger, C., Dahl, M. L., Aklillu, E., Christensen, M., Bertilsson, L., et al. (2006). A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin. Pharmacol. Ther. 79 (1), 103–113. doi:10.1016/j.clpt.2005.10.002
Simon, T., Verstuyft, C., Mary-Krause, M., Quteineh, L., Drouet, E., Meneveau, N., et al. (2009). Genetic determinants of response to clopidogrel and cardiovascular events. N. Engl. J. Med. 360 (4), 363–375. doi:10.1056/NEJMoa0808227
Toyoda, Y., Takada, T., and Suzuki, H. (2016). Halogenated hydrocarbon solvent-related cholangiocarcinoma risk: Biliary excretion of glutathione conjugates of 1, 2-dichloropropane evidenced by untargeted metabolomics analysis. Sci. Rep. 6, 24586. doi:10.1038/srep24586
Wang, Z., Liu, Z., Wang, W., Fu, Y., Chen, W., Li, W., et al. (2019). Two common mutations within CYP2C19 affected platelet aggregation in Chinese patients undergoing PCI: A one-year follow-up study. Pharmacogenomics J. 19 (2), 157–163. doi:10.1038/s41397-018-0036-2
Watanabe, H., Domei, T., Morimoto, T., Natsuaki, M., Shiomi, H., Toyota, T., et al. (2019). Effect of 1-month dual antiplatelet therapy followed by clopidogrel vs 12-month dual antiplatelet therapy on cardiovascular and bleeding events in patients receiving PCI: The STOPDAPT-2 randomized clinical trial. JAMA 321 (24), 2414–2427. doi:10.1001/jama.2019.8145
Zhang, Y., Zhao, T., Li, W., and Vore, M. (2010). The 5'-untranslated region of multidrug resistance associated protein 2 (MRP2; ABCC2) regulates downstream open reading frame expression through translational regulation. Mol. Pharmacol. 77 (2), 237–246. doi:10.1124/mol.109.058982
Zhang, Q., Zhong, Z., Li, B., Liao, Z., Zhao, P., Ye, Z., et al. (2018). Effects of different CYP2C19 genotypes on prognosis of patients complicated with atrial fibrillation taking clopidogrel after PCI. Exp. Ther. Med. 16 (4), 3492–3496. doi:10.3892/etm.2018.6650
Zhu, Q., Hu, X., Liu, Y., Xie, Y., Xu, C., Lin, M., et al. (2022). Identification of single domain antibodies with insect cytotoxicity using phage-display antibody library screening and Plutella xylostella ATP-binding cassette transporter subfamily C member 2 (ABCC2) -based insect cell expression system. Int. J. Biol. Macromol. 209, 586–596. doi:10.1016/j.ijbiomac.2022.03.143
Keywords: ABCC2 polymorphism, clopidogrel response, percutaneous coronary intervention, individualized difference, precise medication
Citation: Chen L, Zheng C, Hao M, Gao P, Zhao M, Cao Y and Ma L (2022) Association of ABCC2 polymorphism with clopidogrel response in Chinese patients undergoing percutaneous coronary intervention. Front. Pharmacol. 13:889473. doi: 10.3389/fphar.2022.889473
Received: 07 March 2022; Accepted: 20 September 2022;
Published: 07 October 2022.
Edited by:
Daniel Frank Carr, University of Liverpool, United KingdomReviewed by:
Sojeong Yi, United States Food and Drug Administration, United StatesCopyright © 2022 Chen, Zheng, Hao, Gao, Zhao, Cao and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Ma, bGlhbmdtYTMyMUAxNjMuY29t; Yongtong Cao, Y2FveW9uZ3Rvbmc5MkBzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.