
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Pharmacol. , 25 April 2022
Sec. Pharmacogenetics and Pharmacogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.886377
This article is part of the Research Topic Translation and Implementation of Pharmacogenomic Testing in Daily Clinical Practice: Considering Current Challenges and Future Needs View all 6 articles
Adverse drug reactions (ADR) remain the major problems in healthcare. Most severe ADR are unpredictable, dose-independent and termed as type B idiosyncratic reactions. Recent pharmacogenomic studies have demonstrated the strong associations between severe ADR and genetic markers, including specific HLA alleles (e.g., HLA-B*15:02/HLA-B*57:01/HLA-A*31:01 for carbamazepine-induced severe cutaneous adverse drug reactions [SCAR], HLA-B*58:01 for allopurinol-SCAR, HLA-B*57:01 for abacavir-hypersensitivity, HLA-B*13:01 for dapsone/co-trimoxazole-induced SCAR, and HLA-A*33:01 for terbinafine-induced liver injury), drug metabolism enzymes (such as CYP2C9*3 for phenytoin-induced SCAR and missense variant of TPMT/NUDT15 for thiopurine-induced leukopenia), drug transporters (e.g., SLCO1B1 polymorphism for statin-induced myopathy), and T cell receptors (Sulfanilamide binding into the CDR3/Vα of the TCR 1.3). This mini review article aims to summarize the current knowledge of pharmacogenomics of severe ADR, and the potentially clinical use of these genetic markers for avoidance of ADR.
Adverse drug reaction (ADR) remains one of the leading causes of death around the world (Shoshi et al., 2015). More than 100,000 people have been reported to die by ADR every year (Alomar, 2014), and most severe ADR belongs to type B unpredictable reactions, which are rare, no connection to the dosage, and occur in individuals with an underlying genetic predisposition (Pirmohamed et al., 2004; Uetrecht, 2007). Type B ADR can be presented as skin injury and liver injury. Skin injury is classified from mild maculopapular exanthema (MPE) to life-threatening severe cutaneous adverse drug reactions (SCAR), including drug reactions with eosinophilia and systemic symptoms (DRESS), Stevens–Johnson syndrome (SJS), and toxic epidermal necrolysis (TEN). Although SCAR are rare, they affect approximately 2% of all hospitalized patients (Valeyrie-Allanore et al., 2007), with an incidence between 2 and 7 cases of SJS/TEN cases/million/per year (Mockenhaupt et al., 2008; Levi et al., 2009; Sassolas et al., 2010; Sekula et al., 2013) and 1/1,000 to 1/10,000 cases of DRESS (Amante et al., 2009). The mortality of DRESS, SJS, and TEN are approximately 2%, 1∼10%, and > 30%, respectively (Roujeau and Stern, 1994; Kardaun et al., 2013; Chung et al., 2016a; Mockenhaupt, 2017; Wang et al., 2018; Tsai et al., 2019). Furthermore, ADR also identified to induce hepatic toxicity, called as drug-induced liver injury (DILI). Approximately 10% of DILI patients may progress to acute liver failure (Yip et al., 2015), and the mortality of DILI is up to 7% (Björnsson and Björnsson, 2017). The incidence of DILI is estimated to be 1 to 10 per 100,000 new users (Yip et al., 2015). Since severe ADR can abe easily confused with other aetiologies of liver damage or renal impairment, the diagnosis of “drug-induced” and culprit drug are sometime difficult to determine. DILI can be further categorized into two classes, allergic and non-allergic. Allergic DILI is often related to HLA genetic factor and results in abnormal immune response; non-allergic DILI, on the other hand, is mostly the result of accumulation of related reagents within liver (Kuna et al., 2018).
In this review, we summarize the currently identified genetic biomarkers of severe ADR, especially focusing on genetic variants of human leukocyte antigens (HLA), T cell receptor (TCR), drug-metabolizing enzymes, and drug-transporters (Figure 1). Up to present, the U.S. Food and Drug Administration (FDA) has labeled more than 180 approved drugs with genetic factors (Administration, 2021).
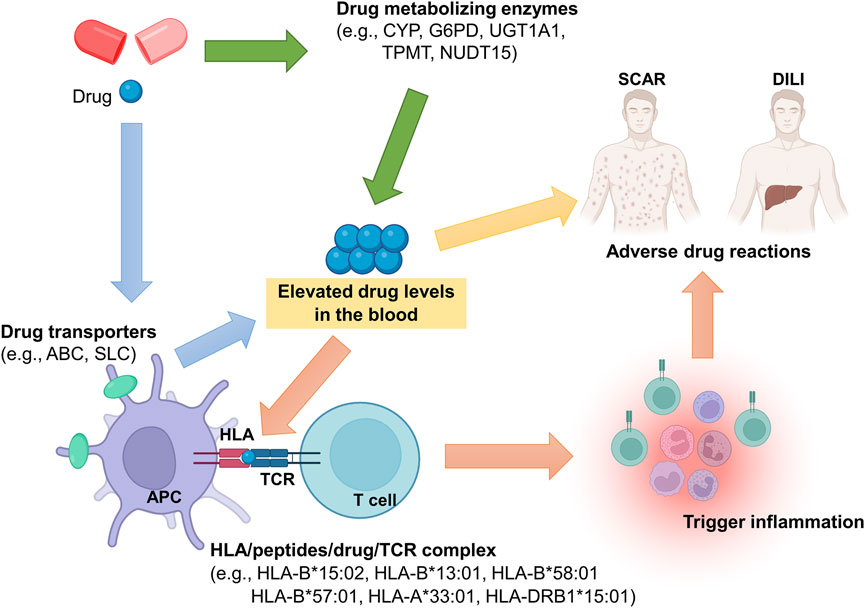
FIGURE 1. Potential genetic determinants involved in pathogenesis of severe ADR. Genetic polymorphisms in drug metabolizing enzymes or drug transporters may alter their function, and then elevated drug levels in the blood, resulting in ADR occurrence. Also, the drug may trigger immune responses through HLA/drug/TCR complex. In the HLA/drug/TCR model, HLA is considered as the key molecular for induction of ADR. Taken together, genetic polymorphisms of HLA, drug metabolizing enzyme, drug transporter, and TCR play important roles in ADR pathogenesis.
Type B idiosyncratic reactions is thought to be elicited by the excessive activation of CD4+ and CD8+ T-lymphocytes (Lerch and Pichler, 2004). Drugs or their reactive metabolites considered as foreign antigens that bind to receptors, activating the immune reactions. HLA are the primary immune anchors for presenting foreign antigens and responsible for pathogenesis of SCAR and DILI (Phillips et al., 2011; Chung et al., 2016a; Stephens et al., 2021). The highly polymorphic properties of HLA molecules among individuals provide diverse opportunities for interactions with various drugs. A specific type of HLA protein may have a higher affinity toward drug/metabolite antigens, presenting the antigen to TCRs, resulting in the activation of T lymphocytes, clonal expansion, skin inflammation, organ damage, and epidermal detachment.
The increasing data have been found a link between HLA alleles and severe ADR (Table 1) in the last two decades. Carbamazepine (CBZ), belongs to aromatic and antiepileptic drug, is one of the common culprit drug(s) of SJS/TEN in different ethnic groups (Roujeau et al., 1995). HLA-B*15:02 is firstly reported to be strongly associated to carbamazepine (CBZ)-induced SJS/TEN in Chinese population (odds ratio [OR] = 2504) (Chung et al., 2004), and the association is latterly validated in different populations, such as Thai, Malaysian, Chinese, and Indian patients (Hung et al., 2006; Locharernkul et al., 2008; Mehta et al., 2009; Tassaneeyakul et al., 2010; Cheung et al., 2013; Tangamornsuksan et al., 2013; Chung et al., 2016b). Furthermore, it’s been proven that HLA-A*31:01 is associated with CBZ-induced hypersensitivity (Kim et al., 2011; McCormack et al., 2011; Ozeki et al., 2011), especially for DRESS patients (OR = 13.2) (Genin et al., 2014). Recently, HLA-B*57:01 is also identified to be associated with CBZ-induced SJS/TEN in Europeans (OR = 9.0) (Mockenhaupt et al., 2019). The phenotype-specific and ethnicity-specific are found in CBZ-induced SCAR patients. Oxcarbazepine (OXC) is another aromatic and antiepileptic drug that has a similar structure of carbamazepine, and HLA-B*15:02 allele is also found to be associated with OXC-induced SJS/TEN (OR = 27.9) (Chen et al., 2017). Furthermore, Asian patients carry the alleles of HLA-B*15:02, HLA-B*13:01, and HLA-B*51:01, have found a higher risk to induce phenytoin-induced SCAR (Chung et al., 2014; Su et al., 2019).
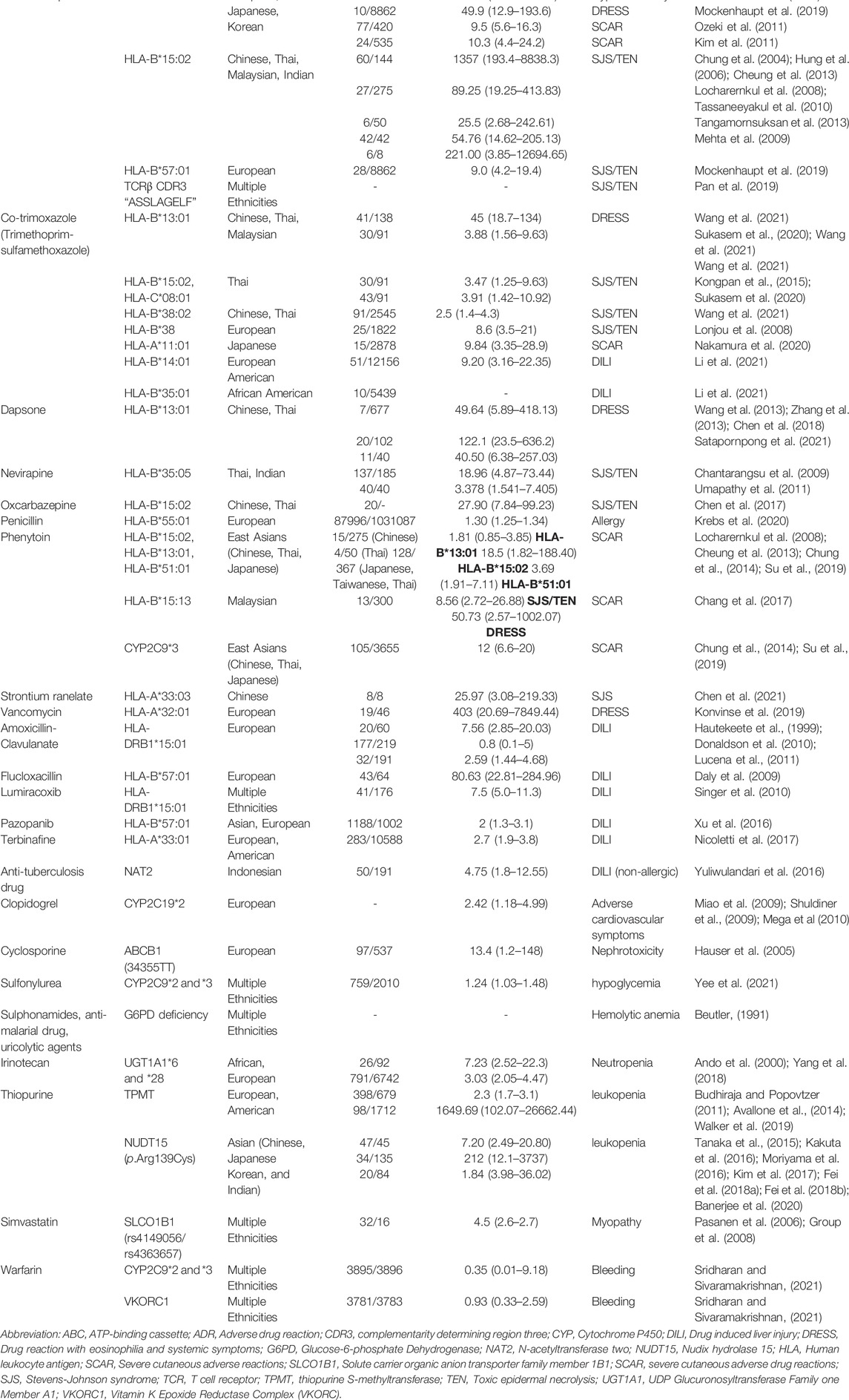
TABLE 1. Genetic associations with severe ADR in HLA, TCR, drug metabolism enzymes, and drug transporters.
Allopurinol is classified as a xanthine oxidase inhibitor and used to treat gout; however, it is known as one of the most common causes of SJS/TEN (Wang et al., 2019). Hung et al. have firstly identified that HLA-B*58:01 is strongly associated with allopurinol-induced SCAR in Chinese population (OR = 580.3) (Hung et al., 2005). This association was then verified in Japanese, South Korean, Thai, Hong Kong, European, Australia, and Portugal patients (Chung et al., 2007; Kaniwa et al., 2008; Lonjou et al., 2008; Tassaneeyakul et al., 2009; Kang et al., 2011; Lee et al., 2012; Ng et al., 2016).
Abacavir is effectively for treatment with HIV infection, and it has been reported that hypersensitivity reactions induced by abacavir is strongly associated with HLA-B*57:01 in Australia’s, U.S. and European populations (Hetherington et al., 2002; Mallal et al., 2002; Sousa-Pinto et al., 2015). In addition, HLA-A*02:06 is strongly associated with acetaminophen-related SJS/TEN with severe ocular complications in Japan population (Ueta et al., 2019).
HLA-B*13:01 has been recently reported to be associated with DRESS induced by sulfonamide, including dapsone (Wang et al., 2013; Zhang et al., 2013; Chen et al., 2018; Liu et al., 2019; Satapornpong et al., 2021), salazosulfapyridine (Yang et al., 2014), and co-trimoxazole (sulfamethoxazole-trimethoprim) (Wang et al., 2021) in Chinese or Thai populations, while HLA-A*11:01 is found to be associated with sulfonamide-related SCAR in Japanese population (Nakamura et al., 2020). The phenotype-specific is also observed in sulfonamide-induced ADR; for example, HLA-B*38:02 and HLA-B*15:02 was found to be associated with co-trimoxazole-induced SJS/TEN (Lonjou et al., 2008; Wang et al., 2021), but not with co-trimoxazole-induced DRESS.
Recently, Konvinse, et al. reported that HLA-A*32:01 is strongly associated with vancomycin-induced DRESS in a population of European ancestry (Konvinse et al., 2019), and the genome-wide association study (GWAS) conducted by Krebs et al. shows that HLA-B*55:01 is a genetic marker for penicillin allergy in United States, United Kingdom, and Estonian populations (OR = 1.4) (Krebs et al., 2020). Chen et al. further revealed that HLA-A*33:03 is associated with strontium ranelate-SJS (OR = 25.9) (Chen et al., 2021).
In addition to SCAR, several studies have identified the correlations between allergic DILI and specific HLA alleles. Amoxicillin-clavulanate (AC) is an antibiotic medication used to treat a variety of bacterial infections, but it is also considered as one of the most common culprit drugs of DILI (holding up to 10 ∼ 13% of DILI patients) (Andrade et al., 2005). The AC-induced DILI has been proved to be highly associated with HLA-DRB1*15:01 (Hautekeete et al., 1999). A GWAS study conducted by Lucena et al. has confirmed the HLA-DRB1*15:01 association and two novel HLA alleles associated with AC-induced DILI are further identified: HLA-A*02:01 in White European patients and HLA-B*18:01 in Spanish patients (Lucena et al., 2011). Both HLA class I and II alleles influence susceptibility to AC-induced DILI. Another common DILI inducing drug, lumiracoxib, is a COX-2 selective inhibitor nonsteroidal anti-inflammatory drug, like AC-induced DILI, has been identified that HLA-DRB1*15:01 is correlated with lumiracoxib-induced DILI (OR = 5.0) (Singer et al., 2010).
Flucloxacillin, belongs a narrow-spectrum beta-lactam antibiotic and used widely to treat patients with staphylococcal infections, is also a common cause of DILI. Daly et al. previously identified HLA-B*57:01 is strongly associated with flucloxacillin-induced DILI (OR = 80.6) (Daly et al., 2009). The same allele as HLA-B*57:01 is associated with pazopanib-induced DILI in Europeans (Xu et al., 2016). In fact, HLA-B*57:01 is also found to be strongly associated with abacavir hypersensitivity and CBZ-induced SJS/TEN in European descendants. These results suggest that HLA-B*57:01 is regarded as the most common risk allele for severe ADR, including SCAR and DILI, in European descendants.
Currently, Li et al. identified that HLA-B*14:01 allele is the highest associated HLA with co-trimoxazole (sulfamethoxazole-trimethoprim)-related DILI in European Americans (OR = 9.2), while HLA-B*35:01 is the most associated allele in African Americans (Li et al., 2021). In the recent research using the GWAS study, Nicoletti et al. discovered that HLA-A*33:01 is associated with DILI, especially with terbinafine-induced liver injury (OR = 40.5) (Nicoletti et al., 2017).
In addition to HLA alleles, several studies have shown that specific TCRs play important roles in the pathogenesis of severe ADR (Pirmohamed and Park, 2003; Pan et al., 2019). Pan et al. identified a public TCR composed of a TCRα complementarity determining region 3 (CDR3) “VFDNTDKLI” paired with a TCRβ CDR3 “ASSLAGELF” in clonotypes derived from patients of Asian and European descent with CBZ-induced SJS/TEN (Abel et al., 2008), which may explain how patients with different HLA alleles associated with different ethnicities can develop similar hypersensitivity reactions. This drug-specific TCR shows phenotype-specificity in an HLA-B*15:02-favored manner. In addition, Zhao et al. reported a promiscuous immune response associated with HLA Class-II‒-restricted T cells in patients with dapsone-induced DRESS (Zhao et al., 2021), but the detailed interactions and mechanisms that underlie HLA-B*13:01/dapsone–restricted CD8+ T cell responses remain poorly understood. The recent discovery of HLA genetic predispositions and oligoclonal and clonotype-specific TCR usages (Ko et al., 2011; Chung et al., 2015a) support the concept that an immune synapse involving an HLA–drug–TCR interaction is essential for inducing type B idiosyncratic ADR.
The gene polymorphism in drug metabolizing enzymes have also been attributed to ADR. Although previous studies shows that it have mainly been involved in dose-dependent mild ADR, a number of researches revealed that genetic defects of drug metabolizing enzymes also be responsible for the development of type B ADR (Pirmohamed and Park, 2003). The divergences in individual metabolism and drug clearance may contribute to occurrence and prognosis of ADR.
Cytochrome P450 (CYP) belongs to a superfamily of heme-containing enzymes responsible for oxidative biotransformation of a broad list of molecules (Kalgutkar et al., 2007). Modifications of its activity can be brought by the genetic polymorphisms, which may result in three phenotypes, such as poor, extensive, and ultra-rapid metabolizers (Sikka et al., 2005). There are at least 57 human genes known to code for CYP enzymes. CYP2D6, CYP2C9 and CYP2C19 genes were found to be responsible in 40% of biotransformation of drug, however, they were also regarded as one of the major susceptibility factors for ADR (Nebert and Russell, 2002; Zhou et al., 2009).
CYP2D6 accounts for the metabolism of 25% of drugs, and its polymorphism is highly relevant in altered enzymatic activity and ADR (Zhou, 2009). CYP2D6*3, *4, *5 and *17 are associated with poor metabolizers, and gene duplication of more than two normally-functioning alleles with ultra-rapid metabolizers (Zhou et al., 2009). Its substrates are mostly lipophilic and include antiarrhythmics, antipsychotics, antidepressants, opioids and some beta-blockers (Gardiner and Begg, 2006). One meta-analysis recommended reducing 50% of tricyclic antidepressant dose in patients who are CYP2D6 poor metabolizers (CYP2D6*4/*4 carriers) (Kirchheiner et al., 2004). Likewise, ultra-rapid metabolizers taking codeine may increase its active metabolite, morphine, resulting in life-threatening toxicity in patients taking the standard dose (Crews et al., 2012). Recently, a case report study identified two patients with CYP2D6*4 variant may be involved in severe ADR induced by quetiapine (Stäuble et al., 2021).
CYP2C9 contributes to 15% of metabolizing activity to drugs (Daly et al., 2017). Its substrates include anticoagulants, sulfonylureas, and some nonsteroidal anti-inflammatory drugs (Gardiner and Begg, 2006). CYP2C9 genotype is an important predictor of warfarin-induced bleeding. In a meta-analysis study, patients with CYP2C9*2 and CYP2C9*3 alleles are poor metabolizers who are at a greater risk of bleeding, requiring lower doses of warfarin (Sanderson et al., 2005). Further studies showed that the shorter time to achieve therapeutic international normalized ratio (INR) for warfarin is observed in patients with both CYP2C9*2 and *3 and vitamin K epoxide reductase complex (VKORC1C1173T) genes (Sridharan and Sivaramakrishnan, 2021). CYP2C9 was also responsible for metabolism of phenytoin. CYP2C9*3 can reduce the clearance of phenytoin and has been found to be associated with development of phenytoin-induced SCAR (Chung et al., 2014). In addition, CYP2C9*2 and *3 alleles are found to enhance hypoglycemic effect in patients treated with sulfonylureas (Yee et al., 2021).
CYP2C19 metabolizes anti-depressants and proton pump inhibitors. Clopidogrel was metabolized into its active substance by CYP2C19. Loss of function in CYP2C19*2 and *3 alleles was associated with decrease in efficacy leading to increased ischemic complications (Miao et al., 2009; Shuldiner et al., 2009; Mega et al., 2010; Paré et al., 2010). Furthermore, a meta-analysis study demonstrated that poor metabolizers with CYP2C19 polymorphisms (CYP2C19*1, *2, and *17) are associated with increased risks in neurological, sexual and gastrointestinal side effects in patients taking citalopram/escitalopram (Fabbri et al., 2018).
Glucose-6-phosphate dehydrogenase (G6PD) is an important enzyme involved in red blood cell (RBC) oxidation through pentose phosphate pathway. Patients with G6PD deficiency are at a risk of hemolytic anemia after treatment with sulphonamides, anti-malarial drugs and uricolytic agents (Beutler, 1991). G6PD deficiency has also been reported to involve in primaquine- and dapsone-induced acute hemolytic anemia (Luzzatto and Seneca, 2014).
The genetic polymorphism of uridine diphospho glucuronosyltransferase 1A1 (UGT1A1*28) has been reported to reduce the UGT1A1 enzymatic activity and result in irinotecan-induced neutropenia (Ando et al., 2000). Further analysis study shows that Asians with the higher presence of UGT1A1*28 are more at a risk in developing irinotecan-induced toxicity compared to Western populations. Also, patients carried UGT1A1*6 are likely to develop irinotecan-induced toxicity (Yang et al., 2018).
N-acetyl transferase 2 (NAT2) is an acetylator enzyme found in the liver and gastrointestinal tract that reacts with drugs like dapsone, isoniazid, hydralazine, and sulfonamindes (Sim et al., 2014). Studies regarding its polymorphisms are responsible for its slow acetylator phenotype. It has been reported that patients with slow phenotype of NAT2 are associated with anti-tuberculosis nonallergic drug-induced liver injury (Yuliwulandari et al., 2016).
Thiopurine-induced leukopenia has been found to be associated with polymorphisms in thiopurine S-methyltransferase (TPMT) and Nudix Hydrolase 15 (NUDT15) genes, which encode TPMT and nudix hydrolase enzyme, respectively. Both enzymes are involved in thiopurine-containing drug metabolism such as azathioprine (Eichelbaum et al., 2006; Yang et al., 2015a). In meta-analysis studies, TPMT*3C variant is known to be associated with an increased risk in thiopurine-induced leukopenia in European descendants (Budhiraja and Popovtzer, 2011; Avallone et al., 2014; Walker et al., 2019). On the other hand, NUDT15 R139C (rs116855232, NUDT15*3) variant carriers are strongly associated with thiopurine-induced leukopenia in Asian populations, including Chinese, Japanese, Korean, and Indian populations (Tanaka et al., 2015; Kakuta et al., 2016; Moriyama et al., 2016; Kim et al., 2017; Fei et al., 2018a; Fei et al., 2018b; Banerjee et al., 2020).
Drug transporters, responsible for influx and efflux of drugs, are categorized into two superfamilies: ATP-binding cassette (ABC) family, and solute carrier (SLC) family (International Transporter et al., 2010). Studies of correlation between drug transporter genes and ADR have increased noticeably. Associations of polymorphisms in ABCB1 gene with cyclosporine-induced nephrotoxicity have been identified (Hauser et al., 2005). ABCB1 also involved in ADR of osmotic-release oral system methylphenidate in adolescents (Kim et al., 2013). Furthermore, a meta-analysis study shows that patients carried ABCC2 3972T > T and ABCG2 34G > A genes are at a higher risk of irinotecan-induced neutropenia and diarrhea, respectively (Zaïr and Singer, 2016).
On the other hand, SLC drug transporter family has a well-known association with statin-related ADR (Niemi et al., 2006; Pasanen et al., 2006). Evidence revealed that the presence of C allele of rs4149056 and homozygous CC of rs4363657 of SLCO1B1 show an increased risk to develop statin-induced myopathy (König et al., 2006; Group et al., 2008). Further study reported a significant association between patients carried SLCO1B1 T521C and myopathy induced by statins, including simvastatin, rosuvastatin and ceruvastatin (Xiang et al., 2018; Carr et al., 2019; Turner et al., 2020). It has also been reported that SLC6A3 rs28363170 is associated with haloperidol-related ADR (Zastrozhin et al., 2017), SLC22A2 rs316019 is associated with cisplatin-induced ototoxicity in cancer patients (Langer et al., 2020), and S allele of SLC6A4 is involved in serotonin inhibitors-induced mania and gastrointestinal ADR (Zhu et al., 2017).
Patients with chronic kidney disease (CKD) and renal impairment may significantly delay drug clearance and metabolism, resulting in an increased risk of allopurinol-SCAR development and poor prognosis (Chung et al., 2015b), Furthermore, increased risks of allopurinol hypersensitivity have been significantly associated with female sex, CKD, cardiovascular disease (CVD) (Carnovale et al., 2014), allopurinol use starting after 60 years of age, and an initial dosage >100 mg/day. Allopurinol-associated mortality has found to be higher in patients with CKD, CVD, and older age (Yang et al., 2015b). Allopurinol prescribed for patients with asymptomatic hyperuricemia with underlying CKD or CVD also show an increased risk of hypersensitivity reactions and mortality (Yang et al., 2015b).
Genetic HLA patterns associated with SCAR and DILI development have been identified for many drugs, and several pharmacogenetic markers have been successfully applied in clinical practice. Cost-effectiveness studies have examined the application of genetic testing before drug treatment to prevent SCAR development (Hughes et al., 2004; Ke et al., 2017; Plumpton et al., 2017), indicating that genetic screening is an important severe ADR prevention strategy. In fact, there are four prospective clinical trials have been conducted worldwide to demonstrate the clinical utility of HLA tests (including HLA-A*31:01, B*15:02, B*57:01, and B*58:01 genetic screening) (Mallal et al., 2008; Chen et al., 2011; Amstutz et al., 2014; University, 2017; Ke et al., 2019).
So far, a preventive genetic test for HLA-B*15:02 among potential new users of CBZ is supported by the national health insurance programs in Taiwan, Singapore, Hong Kong, Thailand, and mainland China (Chen et al., 2011; Tiamkao et al., 2013; Chen et al., 2014). The U.S. FDA further recommend genetic HLA-A*31:01 screening prior to the use of CBZ, and genetic HLA-B*15:02 screening before oxcarbazepine treatment, especially with ethnicities with high probability of HLA-B*15:02, such as Chinese and Thai. Recently, a trial is ongoing involving screening HLA to reduce ADR. (Identifier: NCT03184597).
Genetic HLA-B*57:01 testing prior to abacavir treatment for HIV treatment is widely used in clinical practice (Mallal et al., 2008) and is recommended by the U.S. FDA, European Medicines Agency, and Canada Health. However, HLA-B*57:01 genetic screening did not present a good result for new users before flucloxacillin treatment due to its low positive predictive value with 0.12% (17, 67). And, another HLA allele, HLA-B*57:03, is also found to be associated with DILI induced by flucloxacillin (141).
HLA-B*58:01 screening is commonly employed to protect patients from the risk of allopurinol-induced SCAR (Khanna et al., 2012). The American College of Rheumatology guidelines for the management of gout has recommended genetic HLA-B*58:01 testing prior to allopurinol use since 2012 (Khanna et al., 2012). Several medical centers in Hong Kong, Thailand, Korea, Taiwan, and mainland China provide such pre-screening (Ke et al., 2019). Furthermore, HLA-B*13:01 testing is recommended for new patients with leprosy being initiated on dapsone therapy in China (Liu et al., 2019); an ongoing clinical trial is examining the efficacy of CYP2C9*3 and HLA-B alleles screening to prevention of phenytoin-induced SCAR in China population (Chang et al., 2020).
The U.S. FDA has recommended genetic testing of TPMT and NUDT15 polymorphisms prior to the use of thiopurine, especially for azathioprine. The British Society of Rheumatology guidelines have recommended that TPMT testing prior to prescribing azathioprine in Europeans (Chakravarty et al., 2008). As genetic NUDT15 has shown to be strongly associated with thiopurine-related leukopenia in Asian populations, the preventive test of NUDT15 for azathioprine has recently discussed to support by the national health insurance in China and Taiwan, but it still not approved.
With the current available literature, there is an expanding number of published papers regarding genetic polymorphisms associated with severe ADR. Recently, the high-throughput technologies, such as whole genome sequencing (WGS) and whole exome sequencing (WES), have provided a rapid method to screen the genetic variants for patient and transformed the landscape of genetic biomarkers research. The use of pharmacogenetic testing, both reactively and preemptively, have been successful in terms of response to treatment. Studies have showed that reactive testing could explain or predict the treatment outcome during drug administration, while preemptive testing can prevent severe ADR that may occur. A number of studies have supported the use of pharmacogenetic testing in terms of cost-effectiveness. These studies have shown that testing lessens the cost compared to the addressing the life-threatening severe ADR developed. To achieve success of its use, standard implementation process of pharmacogenetic testing should be taken in place. The knowledge and expertise of the people involved, strong financial support, integrated data systems and holistic team approach will be deemed necessary. It is more necessary to promote the education of genetic testing for physicians in district hospital and community clinics. Pharmacogenetic testing will become a cornerstone to the concept of personalized or precision medicine.
C-WW contributed to the conception. C-WW, IP, and W-HL writing of the manuscript. W-HC reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 110-2320-B-182A-014-MY3, 110-2326-B-182A-003-), and Chang Gung Memorial Hospital (CORPG3J0321-3 and CORPG1J0011-3). Furthermore, the Figure 1 is created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abel, S., Paturel, L., and Cabié, A. (2008). Abacavir Hypersensitivity. N. Engl. J. Med. 358 (23), 2515–2516.
Administration, U. S. F. a. D. (2021). Table of Pharmacogenomic Biomarkers in Drug Labeling. Available from: https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling.
Alomar, M. J. (2014). Factors Affecting the Development of Adverse Drug Reactions (Review Article). Saudi Pharm. J. 22 (2), 83–94. doi:10.1016/j.jsps.2013.02.003
Amante, M. F., Filippini, A. V., Cejas, N., Lendoire, J., Imventarza, O., and Parisi, C. (2009). Dress Syndrome and Fulminant Hepatic Failure Induced by Lamotrigine. Ann. Hepatol. 8 (1), 75–77. doi:10.1016/s1665-2681(19)31817-4
Amstutz, U., Shear, N. H., Rieder, M. J., Hwang, S., Fung, V., Nakamura, H., et al. (2014). Recommendations for HLA-B*15:02 and HLA-A*31:01 Genetic Testing to Reduce the Risk of Carbamazepine-Induced Hypersensitivity Reactions. Epilepsia 55 (4), 496–506. doi:10.1111/epi.12564
Ando, Y., Saka, H., Ando, M., Sawa, T., Muro, K., Ueoka, H., et al. (2000). Polymorphisms of UDP-Glucuronosyltransferase Gene and Irinotecan Toxicity: a Pharmacogenetic Analysis. Cancer Res. 60 (24), 6921.
Andrade, R. J., Lucena, M. I., Fernández, M. C., Pelaez, G., Pachkoria, K., García-Ruiz, E., et al. (2005). Drug-induced Liver Injury: an Analysis of 461 Incidences Submitted to the Spanish Registry over a 10-year Period. Gastroenterology 129 (2), 512–521. doi:10.1016/j.gastro.2005.05.006
Avallone, E. V., Pica, R., Cassieri, C., Zippi, M., Paoluzi, P., and Vernia, P. (2014). Azathioprine Treatment in Inflammatory Bowel Disease Patients: Type and Time of Onset of Side Effects. Eur. Rev. Med. Pharmacol. Sci. 18 (2), 165–170.
Banerjee, R., Ravikanth, V. V., Pal, P., Bale, G., Avanthi, U. S., Goren, I., et al. (2020). NUDT15 C415T Variant Compared with TPMT Genotyping in Predicting Azathioprine-Induced Leucopenia: Prospective Analysis of 1014 Inflammatory Bowel Disease Patients in India. Aliment. Pharmacol. Ther. 52 (11-12), 1683–1694. doi:10.1111/apt.16137
Beutler, E. (1991). Glucose-6-phosphate Dehydrogenase Deficiency. N. Engl. J. Med. 324 (3), 169–174. doi:10.1056/NEJM199101173240306
Björnsson, E. S., and Björnsson, H. K. (2017). Mortality Associated with Drug-Induced Liver Injury (DILI). Transl Gastroenterol. Hepatol. 2, 114. doi:10.21037/tgh.2017.11.16
Budhiraja, P., and Popovtzer, M. (2011). Azathioprine-related Myelosuppression in a Patient Homozygous for TPMT*3A. Nat. Rev. Nephrol. 7 (8), 478–484. doi:10.1038/nrneph.2011.74
Carnovale, C., Venegoni, M., and Clementi, E. (2014). Allopurinol Overuse in Asymptomatic Hyperuricemia: a Teachable Moment. JAMA Intern. Med. 174 (7), 1031–1032. doi:10.1001/jamainternmed.2014.1427
Carr, D. F., Francis, B., Jorgensen, A. L., Zhang, E., Chinoy, H., Heckbert, S. R., et al. (2019). Genomewide Association Study of Statin-Induced Myopathy in Patients Recruited Using the UK Clinical Practice Research Datalink. Clin. Pharmacol. Ther. 106 (6), 1353–1361. doi:10.1002/cpt.1557
Chakravarty, K., McDonald, H., Pullar, T., Taggart, A., Chalmers, R., Oliver, S., et al. (2008). BSR/BHPR Guideline for Disease-Modifying Anti-rheumatic Drug (DMARD) Therapy in Consultation with the British Association of Dermatologists. Rheumatology (Oxford) 47 (6), 924–925. doi:10.1093/rheumatology/kel216a
Chang, C. C., Ng, C. C., Too, C. L., Choon, S. E., Lee, C. K., Chung, W. H., et al. (2017). Association of HLA-B*15:13 and HLA-B*15:02 with Phenytoin-Induced Severe Cutaneous Adverse Reactions in a Malay Population. Pharmacogenomics J. 17 (2), 170–173. doi:10.1038/tpj.2016.10
Chang, C. J., Chen, C. B., Hung, S. I., Ji, C., and Chung, W. H. (2020). Pharmacogenetic Testing for Prevention of Severe Cutaneous Adverse Drug Reactions. Front. Pharmacol. 11, 969. doi:10.3389/fphar.2020.00969
Chantarangsu, S., Mushiroda, T., Mahasirimongkol, S., Kiertiburanakul, S., Sungkanuparph, S., Manosuthi, W., et al. (2009). HLA-B*3505 Allele Is a strong Predictor for Nevirapine-Induced Skin Adverse Drug Reactions in HIV-Infected Thai Patients. Pharmacogenet Genomics 19 (2), 139–146. doi:10.1097/FPC.0b013e32831d0faf
Chen, C. B., Chen, Y. E., Chu, M. T., Wang, C. W., Hui, R. C., Lu, C. W., et al. (2021). The Risk of Anti-osteoporotic Agent-Induced Severe Cutaneous Adverse Drug Reactions and Their Association with HLA. J. Eur. Acad. Dermatol. Venereol. 35 (3), 712–720. doi:10.1111/jdv.16924
Chen, C. B., Hsiao, Y. H., Wu, T., Hsih, M. S., Tassaneeyakul, W., Jorns, T. P., et al. (2017). Risk and Association of HLA with Oxcarbazepine-Induced Cutaneous Adverse Reactions in Asians. Neurology 88 (1), 78–86. doi:10.1212/WNL.0000000000003453
Chen, P., Lin, J. J., Lu, C. S., Ong, C. T., Hsieh, P. F., Yang, C. C., et al. (2011). Carbamazepine-induced Toxic Effects and HLA-B*1502 Screening in Taiwan. N. Engl. J. Med. 364 (12), 1126–1133. doi:10.1056/NEJMoa1009717
Chen, W.-T., Wang, C.-W., Lu, C.-W., Chen, C.-B., Lee, H.-E., Hung, S.-L., et al. (2018). The Function of HLA-B*13:01 Involved in the Pathomechanism of Dapsone-Induced Severe Cutaneous Adverse Reactions. J. Invest. Dermatol. 138 (7), 1546–1554. doi:10.1016/j.jid.2018.02.004
Chen, Z., Liew, D., and Kwan, P. (2014). Effects of a HLA-B*15:02 Screening Policy on Antiepileptic Drug Use and Severe Skin Reactions. Neurology 83 (22), 2077–2084. doi:10.1212/WNL.0000000000001034
Cheung, Y. K., Cheng, S. H., Chan, E. J., Lo, S. V., Ng, M. H., and Kwan, P. (2013). HLA-B Alleles Associated with Severe Cutaneous Reactions to Antiepileptic Drugs in Han Chinese. Epilepsia 54 (7), 1307–1314. doi:10.1111/epi.12217
Chung, W. H., Chang, W. C., Lee, Y. S., Wu, Y. Y., Yang, C. H., Ho, H. C., et al. (2014). Genetic Variants Associated with Phenytoin-Related Severe Cutaneous Adverse Reactions. JAMA 312 (5), 525–534. doi:10.1001/jama.2014.7859
Chung, W. H., Chang, W. C., Stocker, S. L., Juo, C. G., Graham, G. G., Lee, M. H., et al. (2015). Insights into the Poor Prognosis of Allopurinol-Induced Severe Cutaneous Adverse Reactions: the Impact of Renal Insufficiency, High Plasma Levels of Oxypurinol and Granulysin. Ann. Rheum. Dis. 74 (12), 2157–2164. doi:10.1136/annrheumdis-2014-205577
Chung, W. H., Hung, S. I., and Chen, Y. T. (2007). Human Leukocyte Antigens and Drug Hypersensitivity. Curr. Opin. Allergy Clin. Immunol. 7 (4), 317–323. doi:10.1097/ACI.0b013e3282370c5f
Chung, W. H., Hung, S. I., Hong, H. S., Hsih, M. S., Yang, L. C., Ho, H. C., et al. (2004). Medical Genetics: a Marker for Stevens-Johnson Syndrome. Nature 428 (6982), 486. doi:10.1038/428486a
Chung, W. H., Pan, R. Y., Chu, M. T., Chin, S. W., Huang, Y. L., Wang, W. C., et al. (2015). Oxypurinol-Specific T Cells Possess Preferential TCR Clonotypes and Express Granulysin in Allopurinol-Induced Severe Cutaneous Adverse Reactions. J. Invest. Dermatol. 135 (9), 2237–2248. doi:10.1038/jid.2015.165
Chung, W. H., Wang, C. W., and Dao, R. L. (2016). Severe Cutaneous Adverse Drug Reactions. J. Dermatol. 43 (7), 758–766. doi:10.1111/1346-8138.13430
Chung, W. H., Wang, C. W., and Dao, R. L. (2016). Severe Cutaneous Adverse Drug Reactions. J. Dermatol. 43 (7), 758–766. doi:10.1111/1346-8138.13430
Crews, K. R., Gaedigk, A., Dunnenberger, H. M., Klein, T. E., Shen, D. D., Callaghan, J. T., et al. (2012). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for Codeine Therapy in the Context of Cytochrome P450 2D6 (CYP2D6) Genotype. Clin. Pharmacol. Ther. 91 (2), 321–326. doi:10.1038/clpt.2011.287
Daly, A. K., Donaldson, P. T., Bhatnagar, P., Shen, Y., Pe'er, I., Floratos, A., et al. (2009). HLA-B*5701 Genotype Is a Major Determinant of Drug-Induced Liver Injury Due to Flucloxacillin. Nat. Genet. 41 (7), 816–819. doi:10.1038/ng.379
Daly, A. K., Rettie, A. E., Fowler, D. M., and Miners, J. O. (2017). Pharmacogenomics of CYP2C9: Functional and Clinical Considerations. J. Pers Med. 8 (1). doi:10.3390/jpm8010001
Donaldson, P. T., Daly, A. K., Henderson, J., Graham, J., Pirmohamed, M., Bernal, W., et al. (2010). Human Leucocyte Antigen Class II Genotype in Susceptibility and Resistance to Co-amoxiclav-induced Liver Injury. J. Hepatol. 53 (6), 1049–1053. doi:10.1016/j.jhep.2010.05.033
Eichelbaum, M., Ingelman-Sundberg, M., and Evans, W. E. (2006). Pharmacogenomics and Individualized Drug Therapy. Annu. Rev. Med. 57, 119–137. doi:10.1146/annurev.med.56.082103.104724
Fabbri, C., Tansey, K. E., Perlis, R. H., Hauser, J., Henigsberg, N., Maier, W., et al. (2018). Effect of Cytochrome CYP2C19 Metabolizing Activity on Antidepressant Response and Side Effects: Meta-Analysis of Data from Genome-wide Association Studies. Eur. Neuropsychopharmacol. 28 (8), 945–954. doi:10.1016/j.euroneuro.2018.05.009
Fei, X., Shu, Q., Hua, B. Z., Wang, S. Y., Chen, Z. Y., Ge, W. H., et al. (2018). NUDT15 R139C Variation Increases the Risk of Azathioprine-Induced Toxicity in Chinese Subjects: Case Report and Literature Review. Medicine (Baltimore) 97 (17), e0301. doi:10.1097/MD.0000000000010301
Fei, X., Shu, Q., Zhu, H., Hua, B., Wang, S., Guo, L., et al. (2018). NUDT15 R139C Variants Increase the Risk of Azathioprine-Induced Leukopenia in Chinese Autoimmune Patients. Front. Pharmacol. 9, 460. doi:10.3389/fphar.2018.00460
Gardiner, S. J., and Begg, E. J. (2006). Pharmacogenetics, Drug-Metabolizing Enzymes, and Clinical Practice. Pharmacol. Rev. 58 (3), 521–590. doi:10.1124/pr.58.3.6
Genin, E., Chen, D. P., Hung, S. I., Sekula, P., Schumacher, M., Chang, P. Y., et al. (2014). HLA-A*31:01 and Different Types of Carbamazepine-Induced Severe Cutaneous Adverse Reactions: an International Study and Meta-Analysis. Pharmacogenomics J. 14 (3), 281–288. doi:10.1038/tpj.2013.40
Group, S. C., Link, E., Parish, S., Armitage, J., Bowman, L., Heath, S., et al. (2008). SLCO1B1 Variants and Statin-Induced Myopathy-Aa Genomewide Study. N. Engl. J. Med. 359 (8), 789–799. doi:10.1056/NEJMoa0801936
Hauser, I. A., Schaeffeler, E., Gauer, S., Scheuermann, E. H., Wegner, B., Gossmann, J., et al. (2005). ABCB1 Genotype of the Donor but Not of the Recipient Is a Major Risk Factor for Cyclosporine-Related Nephrotoxicity after Renal Transplantation. J. Am. Soc. Nephrol. 16 (5), 1501–1511. doi:10.1681/ASN.2004100882
Hautekeete, M. L., Horsmans, Y., Van Waeyenberge, C., Demanet, C., Henrion, J., Verbist, L., et al. (1999). HLA Association of Amoxicillin-Clavulanate-Iinduced Hepatitis. Gastroenterology 117 (5), 1181–1186. doi:10.1016/s0016-5085(99)70404-x
Hetherington, S., Hughes, A. R., Mosteller, M., Shortino, D., Baker, K. L., Spreen, W., et al. (2002). Genetic Variations in HLA-B Region and Hypersensitivity Reactions to Abacavir. Lancet 359 (9312), 1121–1122. doi:10.1016/S0140-6736(02)08158-8
Hughes, D. A., Vilar, F. J., Ward, C. C., Alfirevic, A., Park, B. K., and Pirmohamed, M. (2004). Cost-effectiveness Analysis of HLA B*5701 Genotyping in Preventing Abacavir Hypersensitivity. Pharmacogenetics 14 (6), 335–342. doi:10.1097/00008571-200406000-00002
Hung, S. I., Chung, W. H., Jee, S. H., Chen, W. C., Chang, Y. T., Lee, W. R., et al. (2006). Genetic Susceptibility to Carbamazepine-Induced Cutaneous Adverse Drug Reactions. Pharmacogenet Genomics 16 (4), 297–306. doi:10.1097/01.fpc.0000199500.46842.4a
Hung, S. I., Chung, W. H., Liou, L. B., Chu, C. C., Lin, M., Huang, H. P., et al. (2005). HLA-B*5801 Allele as a Genetic Marker for Severe Cutaneous Adverse Reactions Caused by Allopurinol. Proc. Natl. Acad. Sci. U S A. 102 (11), 4134–4139. doi:10.1073/pnas.0409500102
International Transporter, Giacomini, K. M., Huang, S.-M., Tweedie, D. J., Benet, L. Z., Brouwer, K. L. R., Chu, X., et al. (2010). Membrane Transporters in Drug Development. Nat. Rev. Drug Discov. 9 (3), 215.. doi:10.1038/nrd3028
Kakuta, Y., Naito, T., Onodera, M., Kuroha, M., Kimura, T., Shiga, H., et al. (2016). NUDT15 R139C Causes Thiopurine-Induced Early Severe Hair Loss and Leukopenia in Japanese Patients with IBD. Pharmacogenomics J. 16 (3), 280–285. doi:10.1038/tpj.2015.43
Kalgutkar, A. S., Obach, R. S., and Maurer, T. S. (2007). Mechanism-based Inactivation of Cytochrome P450 Enzymes: Chemical Mechanisms, Structure-Activity Relationships and Relationship to Clinical Drug-Drug Interactions and Idiosyncratic Adverse Drug Reactions. Curr. Drug Metab. 8 (5), 407–447. doi:10.2174/138920007780866807
Kang, H. R., Jee, Y. K., Kim, Y. S., Lee, C. H., Jung, J. W., Kim, S. H., et al. (2011). Positive and Negative Associations of HLA Class I Alleles with Allopurinol-Induced SCARs in Koreans. Pharmacogenet Genomics 21 (5), 303–307. doi:10.1097/FPC.0b013e32834282b8
Kaniwa, N., Saito, Y., Aihara, M., Matsunaga, K., Tohkin, M., Kurose, K., et al. (2008). HLA-B Locus in Japanese Patients with Anti-epileptics and Allopurinol-Related Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis. Pharmacogenomics 9 (11), 1617–1622. doi:10.2217/14622416.9.11.1617
Kardaun, S. H., Sekula, P., Valeyrie-Allanore, L., Liss, Y., Chu, C. Y., Creamer, D., et al. (2013). Drug Reaction with Eosinophilia and Systemic Symptoms (DRESS): an Original Multisystem Adverse Drug Reaction. Results from the Prospective RegiSCAR Study. Br. J. Dermatol. 169 (5), 1071–1080. doi:10.1111/bjd.12501
Ke, C. H., Chung, W. H., Tain, Y. L., Huang, Y. B., Wen, Y. H., Chuang, H. Y., et al. (2019). Utility of Human Leukocyte Antigen-B*58: 01 Genotyping and Patient Outcomes. Pharmacogenet Genomics 29 (1), 1–8. doi:10.1097/FPC.0000000000000359
Ke, C. H., Chung, W. H., Wen, Y. H., Huang, Y. B., Chuang, H. Y., Tain, Y. L., et al. (2017). Cost-effectiveness Analysis for Genotyping before Allopurinol Treatment to Prevent Severe Cutaneous Adverse Drug Reactions. J. Rheumatol. 44 (6), 835–843. doi:10.3899/jrheum.151476
Khanna, D., Fitzgerald, J. D., Khanna, P. P., Bae, S., Singh, M. K., Neogi, T., et al. (2012). 2012 American College of Rheumatology Guidelines for Management of Gout. Part 1: Systematic Nonpharmacologic and Pharmacologic Therapeutic Approaches to Hyperuricemia. Arthritis Care Res. (Hoboken) 64 (10), 1431–1446. doi:10.1002/acr.21772
Kim, S. H., Lee, K. W., Song, W. J., Kim, S. H., Jee, Y. K., Lee, S. M., et al. (2011). Carbamazepine-induced Severe Cutaneous Adverse Reactions and HLA Genotypes in Koreans. Epilepsy Res. 97 (1-2), 190–197. doi:10.1016/j.eplepsyres.2011.08.010
Kim, S. W., Lee, J. H., Lee, S. H., Hong, H. J., Lee, M. G., and Yook, K. H. (2013). ABCB1 c.2677G>T Variation Is Associated with Adverse Reactions of OROS-Methylphenidate in Children and Adolescents with ADHD. J. Clin. Psychopharmacol. 33 (4), 491–498. doi:10.1097/JCP.0b013e3182905a8d
Kim, S. Y., Shin, J. H., Park, J. S., Kang, S. Y., Nam, T. S., Kim, J. K., et al. (2017). NUDT15 p.R139C Variant Is Common and Strongly Associated with Azathioprine-Induced Early Leukopenia and Severe Alopecia in Korean Patients with Various Neurological Diseases. J. Neurol. Sci. 378, 64–68. doi:10.1016/j.jns.2017.04.041
Kirchheiner, J., Nickchen, K., Bauer, M., Wong, M. L., Licinio, J., Roots, I., et al. (2004). Pharmacogenetics of Antidepressants and Antipsychotics: the Contribution of Allelic Variations to the Phenotype of Drug Response. Mol. Psychiatry 9 (5), 442–473. doi:10.1038/sj.mp.4001494
Ko, T. M., Chung, W. H., Wei, C. Y., Shih, H. Y., Chen, J. K., Lin, C. H., et al. (2011). Shared and Restricted T-Cell Receptor Use Is Crucial for Carbamazepine-Induced Stevens-Johnson Syndrome. J. Allergy Clin. Immunol. 128 (6), 1266–e11. doi:10.1016/j.jaci.2011.08.013
Kongpan, T., Mahasirimongkol, S., Konyoung, P., Kanjanawart, S., Chumworathayi, P., Wichukchinda, N., et al. (2015). Candidate HLA Genes for Prediction of Co-trimoxazole-induced Severe Cutaneous Reactions. Pharmacogenet Genomics 25 (8), 402–411. doi:10.1097/FPC.0000000000000153
König, J., Seithel, A., Gradhand, U., and Fromm, M. F. (2006). Pharmacogenomics of Human OATP Transporters. Naunyn Schmiedebergs Arch. Pharmacol. 372 (6), 432–443. doi:10.1007/s00210-006-0040-y
Konvinse, K. C., Trubiano, J. A., Pavlos, R., James, I., Shaffer, C. M., Bejan, C. A., et al. (2019). HLA-A*32:01 Is Strongly Associated with Vancomycin-Induced Drug Reaction with Eosinophilia and Systemic Symptoms. J. Allergy Clin. Immunol. 144 (1), 183–192. doi:10.1016/j.jaci.2019.01.045
Krebs, K., Bovijn, J., Zheng, N., Lepamets, M., Censin, J. C., Jürgenson, T., et al. (2020). Genome-wide Study Identifies Association between HLA-B∗55:01 and Self-Reported Penicillin Allergy. Am. J. Hum. Genet. 107 (4), 612–621. doi:10.1016/j.ajhg.2020.08.008
Kuna, L., Bozic, I., Kizivat, T., Bojanic, K., Mrso, M., Kralj, E., et al. (2018). Models of Drug Induced Liver Injury (DILI) - Current Issues and Future Perspectives. Curr. Drug Metab. 19 (10), 830–838. doi:10.2174/1389200219666180523095355
Langer, T., Clemens, E., Broer, L., Maier, L., Uitterlinden, A. G., de Vries, A. C. H., et al. (2020). Usefulness of Current Candidate Genetic Markers to Identify Childhood Cancer Patients at Risk for Platinum-Induced Ototoxicity: Results of the European PanCareLIFE Cohort Study. Eur. J. Cancer 138, 212–224. doi:10.1016/j.ejca.2020.07.019
Lee, M. H., Stocker, S. L., Anderson, J., Phillips, E. J., Nolan, D., Williams, K. M., et al. (2012). Initiating Allopurinol Therapy: Do We Need to Know the Patient's Human Leucocyte Antigen Status? Intern. Med. J. 42 (4), 411–416. doi:10.1111/j.1445-5994.2011.02567.x
Lerch, M., and Pichler, W. J. (2004). The Immunological and Clinical Spectrum of Delayed Drug-Induced Exanthems. Curr. Opin. Allergy Clin. Immunol. 4 (5), 411–419. doi:10.1097/00130832-200410000-00013
Levi, N., Bastuji-Garin, S., Mockenhaupt, M., Roujeau, J. C., Flahault, A., Kelly, J. P., et al. (2009). Medications as Risk Factors of Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in Children: a Pooled Analysis. Pediatrics 123 (2), e297–304. doi:10.1542/peds.2008-1923
Li, Y. J., Phillips, E. J., Dellinger, A., Nicoletti, P., Schutte, R., Li, D., et al. (2021). Human Leukocyte Antigen B*14:01 and B*35:01 Are Associated with Trimethoprim-Sulfamethoxazole Induced Liver Injury. Hepatology 73 (1), 268–281. doi:10.1002/hep.31258
Liu, H., Wang, Z., Bao, F., Wang, C., Sun, L., Zhang, H., et al. (2019). Evaluation of Prospective HLA-B*13:01 Screening to Prevent Dapsone Hypersensitivity Syndrome in Patients with Leprosy. JAMA Dermatol. 155 (6), 666–672. doi:10.1001/jamadermatol.2018.5360
Locharernkul, C., Loplumlert, J., Limotai, C., Korkij, W., Desudchit, T., Tongkobpetch, S., et al. (2008). Carbamazepine and Phenytoin Induced Stevens-Johnson Syndrome Is Associated with HLA-B*1502 Allele in Thai Population. Epilepsia 49 (12), 2087–2091. doi:10.1111/j.1528-1167.2008.01719.x
Lonjou, C., Borot, N., Sekula, P., Ledger, N., Thomas, L., Halevy, S., et al. (2008). A European Study of HLA-B in Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis Related to Five High-Risk Drugs. Pharmacogenet Genomics 18 (2), 99–107. doi:10.1097/FPC.0b013e3282f3ef9c
Lucena, M. I., Molokhia, M., Shen, Y., Urban, T. J., Aithal, G. P., Andrade, R. J., et al. (2011). Susceptibility to Amoxicillin-Clavulanate-Induced Liver Injury Is Influenced by Multiple HLA Class I and II Alleles. Gastroenterology 141 (1), 338–347. doi:10.1053/j.gastro.2011.04.001
Luzzatto, L., and Seneca, E. (2014). G6PD Deficiency: a Classic Example of Pharmacogenetics with On-Going Clinical Implications. Br. J. Haematol. 164 (4), 469–480. doi:10.1111/bjh.12665
Mallal, S., Nolan, D., Witt, C., Masel, G., Martin, A. M., Moore, C., et al. (2002). Association between Presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and Hypersensitivity to HIV-1 Reverse-Transcriptase Inhibitor Abacavir. Lancet 359 (9308), 727–732. doi:10.1016/s0140-6736(02)07873-x
Mallal, S., Phillips, E., Carosi, G., Molina, J. M., Workman, C., Tomazic, J., et al. (2008). HLA-B*5701 Screening for Hypersensitivity to Abacavir. N. Engl. J. Med. 358 (6), 568–579. doi:10.1056/NEJMoa0706135
McCormack, M., Alfirevic, A., Bourgeois, S., Farrell, J. J., Kasperavičiūtė, D., Carrington, M., et al. (2011). HLA-A*3101 and Carbamazepine-Induced Hypersensitivity Reactions in Europeans. N. Engl. J. Med. 364 (12), 1134–1143. doi:10.1056/NEJMoa1013297
Mega, J. L., Simon, T., Collet, J. P., Anderson, J. L., Antman, E. M., Bliden, K., et al. (2010). Reduced-function CYP2C19 Genotype and Risk of Adverse Clinical Outcomes Among Patients Treated with Clopidogrel Predominantly for PCI: a Meta-Analysis. JAMA 304 (16), 1821–1830. doi:10.1001/jama.2010.1543
Mehta, T. Y., Prajapati, L. M., Mittal, B., Joshi, C. G., Sheth, J. J., Patel, D. B., et al. (2009). Association of HLA-B*1502 Allele and Carbamazepine-Induced Stevens-Johnson Syndrome Among Indians. Indian J. Dermatol. Venereol. Leprol. 75 (6), 579–582. doi:10.4103/0378-6323.57718
Miao, J., Liu, R., and Li, Z. (2009). Cytochrome P-450 Polymorphisms and Response to Clopidogrel. N. Engl. J. Med. 3 (21), 2250.. doi:10.4330/wjc.v3.i5.153
Mockenhaupt, M. (2017). Epidemiology of Cutaneous Adverse Drug Reactions. Allergol. Select 1 (1), 96–108. doi:10.5414/ALX01508E
Mockenhaupt, M., Viboud, C., Dunant, A., Naldi, L., Halevy, S., Bouwes Bavinck, J. N., et al. (2008). Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis: Assessment of Medication Risks with Emphasis on Recently Marketed Drugs. The EuroSCAR-Study. J. Invest. Dermatol. 128 (1), 35–44. doi:10.1038/sj.jid.5701033
Mockenhaupt, M., Wang, C. W., Hung, S. I., Sekula, P., Schmidt, A. H., Pan, R. Y., et al. (2019). HLA-B*57:01 Confers Genetic Susceptibility to Carbamazepine-Induced SJS/TEN in Europeans. Allergy 74 (11), 2227–2230. doi:10.1111/all.13821
Moriyama, T., Nishii, R., Perez-Andreu, V., Yang, W., Klussmann, F. A., Zhao, X., et al. (2016). NUDT15 Polymorphisms Alter Thiopurine Metabolism and Hematopoietic Toxicity. Nat. Genet. 48 (4), 367–373. doi:10.1038/ng.3508
Nakamura, R., Ozeki, T., Hirayama, N., Sekine, A., Yamashita, T., Mashimo, Y., et al. (2020). Association of HLA-A*11:01 with Sulfonamide-Related Severe Cutaneous Adverse Reactions in Japanese Patients. J. Invest. Dermatol. 140 (8), 1659–e6. doi:10.1016/j.jid.2019.12.025
Nebert, D. W., and Russell, D. W. (2002). Clinical Importance of the Cytochromes P450. Lancet 360 (9340), 1155–1162. doi:10.1016/S0140-6736(02)11203-7
Ng, C. Y., Yeh, Y. T., Wang, C. W., Hung, S. I., Yang, C. H., Chang, Y. C., et al. (2016). Impact of the HLA-B(*)58:01 Allele and Renal Impairment on Allopurinol-Induced Cutaneous Adverse Reactions. J. Invest. Dermatol. 136 (7), 1373–1381. doi:10.1016/j.jid.2016.02.808
Nicoletti, P., Aithal, G. P., Bjornsson, E. S., Andrade, R. J., Sawle, A., Arrese, M., et al. (2017). Association of Liver Injury from Specific Drugs, or Groups of Drugs, with Polymorphisms in HLA and Other Genes in a Genome-wide Association Study. Gastroenterology 152 (5), 1078–1089. doi:10.1053/j.gastro.2016.12.016
Niemi, M., Pasanen, M. K., and Neuvonen, P. J. (2006). SLCO1B1 Polymorphism and Sex Affect the Pharmacokinetics of Pravastatin but Not Fluvastatin. Clin. Pharmacol. Ther. 80 (4), 356–366. doi:10.1016/j.clpt.2006.06.010
Ozeki, T., Mushiroda, T., Yowang, A., Takahashi, A., Kubo, M., Shirakata, Y., et al. (2011). Genome-wide Association Study Identifies HLA-A*3101 Allele as a Genetic Risk Factor for Carbamazepine-Induced Cutaneous Adverse Drug Reactions in Japanese Population. Hum. Mol. Genet. 20 (5), 1034–1041. doi:10.1093/hmg/ddq537
Pan, R. Y., Chu, M. T., Wang, C. W., Lee, Y. S., Lemonnier, F., Michels, A. W., et al. (2019). Identification of Drug-specific Public TCR Driving Severe Cutaneous Adverse Reactions. Nat. Commun. 10 (1), 3569. doi:10.1038/s41467-019-11396-2
Paré, G., Mehta, S. R., Yusuf, S., Anand, S. S., Connolly, S. J., Hirsh, J., et al. (2010). Effects of CYP2C19 Genotype on Outcomes of Clopidogrel Treatment. N. Engl. J. Med. 363 (18), 1704–1714. doi:10.1056/NEJMoa1008410
Pasanen, M. K., Neuvonen, M., Neuvonen, P. J., and Niemi, M. (2006). SLCO1B1 Polymorphism Markedly Affects the Pharmacokinetics of Simvastatin Acid. Pharmacogenet Genomics 16 (12), 873–879. doi:10.1097/01.fpc.0000230416.82349.90
Phillips, E. J., Chung, W. H., Mockenhaupt, M., Roujeau, J. C., and Mallal, S. A. (2011). Drug Hypersensitivity: Pharmacogenetics and Clinical Syndromes. J. Allergy Clin. Immunol. 127 (3 Suppl. l), S60–S66. doi:10.1016/j.jaci.2010.11.046
Pirmohamed, M., James, S., Meakin, S., Green, C., Scott, A. K., Walley, T. J., et al. (2004). Adverse Drug Reactions as Cause of Admission to Hospital: Prospective Analysis of 18 820 Patients. BMJ 329 (7456), 15–19. doi:10.1136/bmj.329.7456.15
Pirmohamed, M., and Park, B. K. (2003). Cytochrome P450 Enzyme Polymorphisms and Adverse Drug Reactions. Toxicology 192 (1), 23–32. doi:10.1016/s0300-483x(03)00247-6
Plumpton, C. O., Alfirevic, A., Pirmohamed, M., and Hughes, D. A. (2017). Cost Effectiveness Analysis of HLA-B*58:01 Genotyping Prior to Initiation of Allopurinol for Gout. Rheumatology (Oxford) 56 (10), 1729–1739. doi:10.1093/rheumatology/kex253
Roujeau, J. C., Kelly, J. P., Naldi, L., Rzany, B., Stern, R. S., Anderson, T., et al. (1995). Medication Use and the Risk of Stevens-Johnson Syndrome or Toxic Epidermal Necrolysis. N. Engl. J. Med. 333 (24), 1600–1607. doi:10.1056/NEJM199512143332404
Roujeau, J. C., and Stern, R. S. (1994). Severe Adverse Cutaneous Reactions to Drugs. N. Engl. J. Med. 331 (19), 1272–1285. doi:10.1056/NEJM199411103311906
Sanderson, S., Emery, J., and Higgins, J. (2005). CYP2C9 Gene Variants, Drug Dose, and Bleeding Risk in Warfarin-Treated Patients: a HuGEnet Systematic Review and Meta-Analysis. Genet. Med. 7 (2), 97–104. doi:10.1097/01.gim.0000153664.65759.cf
Sassolas, B., Haddad, C., Mockenhaupt, M., Dunant, A., Liss, Y., Bork, K., et al. (2010). ALDEN, an Algorithm for Assessment of Drug Causality in Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis: Comparison with Case-Control Analysis. Clin. Pharmacol. Ther. 88 (1), 60–68. doi:10.1038/clpt.2009.252
Satapornpong, P., Pratoomwun, J., Rerknimitr, P., Klaewsongkram, J., Nakkam, N., Rungrotmongkol, T., et al. (2021). HLA-B*13 :01 Is a Predictive Marker of Dapsone-Induced Severe Cutaneous Adverse Reactions in Thai Patients. Front. Immunol. 12, 661135. doi:10.3389/fimmu.2021.661135
Sekula, P., Dunant, A., Mockenhaupt, M., Naldi, L., Bouwes Bavinck, J. N., Halevy, S., et al. (2013). Comprehensive Survival Analysis of a Cohort of Patients with Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis. J. Invest. Dermatol. 133 (5), 1197–1204. doi:10.1038/jid.2012.510
Shoshi, A., Hoppe, T., Kormeier, B., Ogultarhan, V., and Hofestädt, R. (2015). GraphSAW: a Web-Based System for Graphical Analysis of Drug Interactions and Side Effects Using Pharmaceutical and Molecular Data. BMC Med. Inform. Decis. Mak 15, 15. doi:10.1186/s12911-015-0139-5
Shuldiner, A. R., O'Connell, J. R., Bliden, K. P., Gandhi, A., Ryan, K., Horenstein, R. B., et al. (2009). Association of Cytochrome P450 2C19 Genotype with the Antiplatelet Effect and Clinical Efficacy of Clopidogrel Therapy. JAMA 302 (8), 849–857. doi:10.1001/jama.2009.1232
Sikka, R., Magauran, B., Ulrich, A., and Shannon, M. (2005). Bench to Bedside: Pharmacogenomics, Adverse Drug Interactions, and the Cytochrome P450 System. Acad. Emerg. Med. 12 (12), 1227–1235. doi:10.1197/j.aem.2005.06.027
Sim, E., Abuhammad, A., and Ryan, A. (2014). Arylamine N-Acetyltransferases: from Drug Metabolism and Pharmacogenetics to Drug Discovery. Br. J. Pharmacol. 171 (11), 2705–2725. doi:10.1111/bph.12598
Singer, J. B., Lewitzky, S., Leroy, E., Yang, F., Zhao, X., Klickstein, L., et al. (2010). A Genome-wide Study Identifies HLA Alleles Associated with Lumiracoxib-Related Liver Injury. Nat. Genet. 42 (8), 711–714. doi:10.1038/ng.632
Sousa-Pinto, B., Pinto-Ramos, J., Correia, C., Gonçalves-Costa, G., Gomes, L., Gil-Mata, S., et al. (2015). Pharmacogenetics of Abacavir Hypersensitivity: A Systematic Review and Meta-Analysis of the Association with HLA-B*57:01. J. Allergy Clin. Immunol. 136 (4), 1092–e3. doi:10.1016/j.jaci.2015.03.019
Sridharan, K., and Sivaramakrishnan, G. (2021). A Network Meta-Analysis of CYP2C9, CYP2C9 with VKORC1 and CYP2C9 with VKORC1 and CYP4F2 Genotype-Based Warfarin Dosing Strategies Compared to Traditional. J. Clin. Pharm. Ther. 46 (3), 640–648. doi:10.1111/jcpt.13334
Stäuble, C. K., Lampert, M. L., Mikoteit, T., Hatzinger, M., Hersberger, K. E., and Meyer Zu Schwabedissen, H. E. (2021). Severe Adverse Drug Reactions to Quetiapine in Two Patients Carrying CYP2D6*4 Variants: A Case Report. Int. J. Mol. Sci. 22 (12). doi:10.3390/ijms22126480
Stephens, C., Lucena, M. I., and Andrade, R. J. (2021). Genetic Risk Factors in the Development of Idiosyncratic Drug-Induced Liver Injury. Expert Opin. Drug Metab. Toxicol. 17 (2), 153–169. doi:10.1080/17425255.2021.1854726
Su, S. C., Chen, C. B., Chang, W. C., Wang, C. W., Fan, W. L., Lu, L. Y., et al. (2019). HLA Alleles and CYP2C9*3 as Predictors of Phenytoin Hypersensitivity in East Asians. Clin. Pharmacol. Ther. 105 (2), 476–485. doi:10.1002/cpt.1190
Sukasem, C., Pratoomwun, J., Satapornpong, P., Klaewsongkram, J., Rerkpattanapipat, T., Rerknimitr, P., et al. (2020). Genetic Association of Co-trimoxazole-induced Severe Cutaneous Adverse Reactions Is Phenotype-specific: HLA Class I Genotypes and Haplotypes. Clin. Pharmacol. Ther. 108 (5), 1078–1089. doi:10.1002/cpt.1915
Tanaka, Y., Kato, M., Hasegawa, D., Urayama, K. Y., Nakadate, H., Kondoh, K., et al. (2015). Susceptibility to 6-MP Toxicity Conferred by a NUDT15 Variant in Japanese Children with Acute Lymphoblastic Leukaemia. Br. J. Haematol. 171 (1), 109–115. doi:10.1111/bjh.13518
Tangamornsuksan, W., Chaiyakunapruk, N., Somkrua, R., Lohitnavy, M., and Tassaneeyakul, W. (2013). Relationship between the HLA-B*1502 Allele and Carbamazepine-Induced Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis: a Systematic Review and Meta-Analysis. JAMA Dermatol. 149 (9), 1025–1032. doi:10.1001/jamadermatol.2013.4114
Tassaneeyakul, W., Jantararoungtong, T., Chen, P., Lin, P. Y., Tiamkao, S., Khunarkornsiri, U., et al. (2009). Strong Association between HLA-B*5801 and Allopurinol-Induced Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in a Thai Population. Pharmacogenet Genomics 19 (9), 704–709. doi:10.1097/FPC.0b013e328330a3b8
Tassaneeyakul, W., Tiamkao, S., Jantararoungtong, T., Chen, P., Lin, S. Y., Chen, W. H., et al. (2010). Association between HLA-B*1502 and Carbamazepine-Induced Severe Cutaneous Adverse Drug Reactions in a Thai Population. Epilepsia 51 (5), 926–930. doi:10.1111/j.1528-1167.2010.02533.x
Tiamkao, S., Jitpimolmard, J., Sawanyawisuth, K., and Jitpimolmard, S. (2013). Cost Minimization of HLA-B*1502 Screening before Prescribing Carbamazepine in Thailand. Int. J. Clin. Pharm. 35 (4), 608–612. doi:10.1007/s11096-013-9777-9
Tsai, Y. G., Liou, J. H., Hung, S. I., Chen, C. B., Chiu, T. M., Wang, C. W., et al. (2019). Increased Type 2 Innate Lymphoid Cells in Patients with Drug Reaction with Eosinophilia and Systemic Symptoms Syndrome. J. Invest. Dermatol. 139 (8), 1722–1731. doi:10.1016/j.jid.2018.10.048
Turner, R. M., Radman, I., Bozina, N., and Alfirevic, A. (2020). Pharmacogenetics and Statin-Related Myopathy: what Do We Know? Pharmacogenomics 21 (12), 821–825. doi:10.2217/pgs-2020-0041
Ueta, M., Nakamura, R., Saito, Y., Tokunaga, K., Sotozono, C., Yabe, T., et al. (2019). Association of HLA Class I and II Gene Polymorphisms with Acetaminophen-Related Stevens-Johnson Syndrome with Severe Ocular Complications in Japanese Individuals. Hum. Genome 6, 50. doi:10.1038/s41439-019-0082-6
Uetrecht, J. (2007). Idiosyncratic Drug Reactions: Current Understanding. Annu. Rev. Pharmacol. Toxicol. 47, 513–539. doi:10.1146/annurev.pharmtox.47.120505.105150
Umapathy, S., Pawar, A., Bajpai, S., Pazare, A. R., and Ghosh, K. (2011). HLA Involvement in Nevirapine-Induced Dermatological Reaction in Antiretroviral-Treated HIV-1 Patients. J. Pharmacol. Pharmacother. 2 (2), 114–115. doi:10.4103/0976-500X.81905
University, S. A. H. o. G. M. (2017). HLA Screening in Reducing the Risk of Antiepileptic Drug-Induced Cutaneous Adverse Reactions. HLA Associated Important Dis. 88, 2018. Available from: https://clinicaltrials.gov/ct2/show/NCT03184597?term=NCT03184597&draw=2&rank=1. doi:10.1212/WNL.0000000000004008
Valeyrie-Allanore, L., Sassolas, B., and Roujeau, J. C. (2007). Drug-induced Skin, Nail and Hair Disorders. Drug Saf. 30 (11), 1011–1030. doi:10.2165/00002018-200730110-00003
Walker, G. J., Harrison, J. W., Heap, G. A., Voskuil, M. D., Andersen, V., Anderson, C. A., et al. (2019). Association of Genetic Variants in NUDT15 with Thiopurine-Induced Myelosuppression in Patients with Inflammatory Bowel Disease. JAMA 321 (8), 773–785. doi:10.1001/jama.2019.0709
Wang, C. W., Tassaneeyakul, W., Chen, C. B., Chen, W. T., Teng, Y. C., Huang, C. Y., et al. (2021). Whole Genome Sequencing Identifies Genetic Variants Associated with Co-trimoxazole Hypersensitivity in Asians. J. Allergy Clin. Immunol. 147 (4), 1402–1412. doi:10.1016/j.jaci.2020.08.003
Wang, C. W., Yang, L. Y., Chen, C. B., Ho, H. C., Hung, S. I., Yang, C. H., et al. (2018). Randomized, Controlled Trial of TNF-α Antagonist in CTL-Mediated Severe Cutaneous Adverse Reactions. J. Clin. Invest. 128 (3), 985–996. doi:10.1172/JCI93349
Wang, H., Yan, L., Zhang, G., Chen, X., Yang, J., Li, M., et al. (2013). Association between HLA-B*1301 and Dapsone-Induced Hypersensitivity Reactions Among Leprosy Patients in China. J. Invest. Dermatol. 133 (11), 2642–2644. doi:10.1038/jid.2013.192
Wang, Y. H., Chen, C. B., Tassaneeyakul, W., Saito, Y., Aihara, M., Choon, S. E., et al. (2019). The Medication Risk of Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in Asians: The Major Drug Causality and Comparison with the US FDA Label. Clin. Pharmacol. Ther. 105 (1), 112–120. doi:10.1002/cpt.1071
Xiang, Q., Chen, S. Q., Ma, L. Y., Hu, K., Zhang, Z., Mu, G. Y., et al. (2018). Association between SLCO1B1 T521C Polymorphism and Risk of Statin-Induced Myopathy: a Meta-Analysis. Pharmacogenomics J. 18 (6), 721–729. doi:10.1038/s41397-018-0054-0
Xu, C. F., Johnson, T., Wang, X., Carpenter, C., Graves, A. P., Warren, L., et al. (2016). HLA-B*57:01 Confers Susceptibility to Pazopanib-Associated Liver Injury in Patients with Cancer. Clin. Cancer Res. 22 (6), 1371–1377. doi:10.1158/1078-0432.CCR-15-2044
Yang, C. Y., Chen, C. H., Deng, S. T., Huang, C. S., Lin, Y. J., Chen, Y. J., et al. (2015). Allopurinol Use and Risk of Fatal Hypersensitivity Reactions: A Nationwide Population-Based Study in Taiwan. JAMA Intern. Med. 175 (9), 1550–1557. doi:10.1001/jamainternmed.2015.3536
Yang, F., Gu, B., Zhang, L., Xuan, J., Luo, H., Zhou, P., et al. (2014). HLA-B*13:01 Is Associated with Salazosulfapyridine-Induced Drug Rash with Eosinophilia and Systemic Symptoms in Chinese Han Population. Pharmacogenomics 15 (11), 1461–1469. doi:10.2217/pgs.14.69
Yang, J. J., Landier, W., Yang, W., Liu, C., Hageman, L., Cheng, C., et al. (2015). Inherited NUDT15 Variant Is a Genetic Determinant of Mercaptopurine Intolerance in Children with Acute Lymphoblastic Leukemia. J. Clin. Oncol. 33 (11), 1235–1242. doi:10.1200/JCO.2014.59.4671
Yang, Y., Zhou, M., Hu, M., Cui, Y., Zhong, Q., Liang, L., et al. (2018). UGT1A1*6 and UGT1A1*28 Polymorphisms Are Correlated with Irinotecan-Induced Toxicity: A Meta-Analysis. Asia Pac. J. Clin. Oncol. 14 (5), e479–e489. doi:10.1111/ajco.13028
Yee, J., Heo, Y., Kim, H., Yoon, H. Y., Song, G., and Gwak, H. S. (2021). Association between the CYP2C9 Genotype and Hypoglycemia Among Patients with Type 2 Diabetes Receiving Sulfonylurea Treatment: A Meta-Analysis. Clin. Ther. 43 (5), 836–e4. doi:10.1016/j.clinthera.2021.03.008
Yip, V. L., Alfirevic, A., and Pirmohamed, M. (2015). Genetics of Immune-Mediated Adverse Drug Reactions: a Comprehensive and Clinical Review. Clin. Rev. Allergy Immunol. 48 (2-3), 165–175. doi:10.1007/s12016-014-8418-y
Yuliwulandari, R., Susilowati, R. W., Wicaksono, B. D., Viyati, K., Prayuni, K., Razari, I., et al. (2016). NAT2 Variants Are Associated with Drug-Induced Liver Injury Caused by Anti-tuberculosis Drugs in Indonesian Patients with Tuberculosis. J. Hum. Genet. 61 (6), 533–537. doi:10.1038/jhg.2016.10
Zaïr, Z. M., and Singer, D. R. (2016). Efflux Transporter Variants as Predictors of Drug Toxicity in Lung Cancer Patients: Systematic Review and Meta-Analysis. Pharmacogenomics 17 (9), 1089–1112. doi:10.2217/pgs-2015-0006
Zastrozhin, M. S., Brodyansky, V. M., Skryabin, V. Y., Grishina, E. A., Ivashchenko, D. V., Ryzhikova, K. A., et al. (2017). Pharmacodynamic Genetic Polymorphisms Affect Adverse Drug Reactions of Haloperidol in Patients with Alcohol-Use Disorder. Pharmgenomics Pers Med. 10, 209–215. doi:10.2147/PGPM.S140700
Zhang, F. R., Liu, H., Irwanto, A., Fu, X. A., Li, Y., Yu, G. Q., et al. (2013). HLA-B*13:01 and the Dapsone Hypersensitivity Syndrome. N. Engl. J. Med. 369 (17), 1620–1628. doi:10.1056/NEJMoa1213096
Zhao, Q., Almutairi, M., Tailor, A., Lister, A., Harper, N., Line, J., et al. (2021). HLA Class-II‒Restricted CD8+ T Cells Contribute to the Promiscuous Immune Response in Dapsone-Hypersensitive Patients. J. Invest. Dermatol. 141 (10), 2412–e2. doi:10.1016/j.jid.2021.03.014
Zhou, S. F., Liu, J. P., and Chowbay, B. (2009). Polymorphism of Human Cytochrome P450 Enzymes and its Clinical Impact. Drug Metab. Rev. 41 (2), 89–295. doi:10.1080/03602530902843483
Zhou, S. F. (2009). Polymorphism of Human Cytochrome P450 2D6 and its Clinical Significance: Part I. Clin. Pharmacokinet. 48 (11), 689–723. doi:10.2165/11318030-000000000-00000
Keywords: adverse drug reactions, drug-induced liver injury, CYP, human leukocyte antigens, drug transporter, stevens-johnson syndrome, toxic epidermal necrolysis
Citation: Wang C-W, Preclaro IAC, Lin W-H and Chung W-H (2022) An Updated Review of Genetic Associations With Severe Adverse Drug Reactions: Translation and Implementation of Pharmacogenomic Testing in Clinical Practice. Front. Pharmacol. 13:886377. doi: 10.3389/fphar.2022.886377
Received: 28 February 2022; Accepted: 08 April 2022;
Published: 25 April 2022.
Edited by:
Wojciech Miltyk, Medical University of Bialystok, PolandReviewed by:
Taisei Mushiroda, RIKEN Center for Integrative Medical Sciences (IMS), JapanCopyright © 2022 Wang, Preclaro, Lin and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Hung Chung, d2VuaHVuZ2NodW5nQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.