- 1Shaanxi Institute of Microbiology, Xi’an, China
- 2Engineering Center of QinLing Mountains Natural Products, Shaanxi Academy of Sciences, Xi’an, China
- 3Xi’an Botanical Garden of Shaanxi Province, Institute of Botany of Shaanxi Province, Xi’an, China
To obtain novel fungi with potent β-glucosidase for minor ginsenoside production, Panax bipinnatifidus var. bipinnatifidus, which is a traditional medicinal plant containing various ginsenosides, was first employed to isolate endophytic fungi in this study. A total of 93 representative morphotype strains were isolated and identified according to ITS rDNA sequence analyses, and they were grouped into three phyla (Ascomycota, Basidiomycota, and Mucoromycota), five classes (Dothideomycetes, Sordariomycetes, Eurotiomycetes, Agaricomycetes, and Mucoromycetes), and 24 genera. Plectosphaerella (RA, 19.35%) was the most abundant genus, followed by Paraphoma (RA, 11.83%) and Fusarium (RA, 9.70%). The species richness index (S, 34) and the Shannon–Wiener index (H’, 3.004) indicated that P. bipinnatifidus harbored abundant fungal resources. A total of 26 endophytic fungal ethyl acetate extracts exhibited inhibitory activities against at least one pathogenic bacterium or fungus. In total, 11 strains showed strong β-glucosidase activities and also presented with the ability of ginsenoside biotransformation with varied glycoside-hydrolyzing pathways. Excitingly, three genera, namely, Ilyonectria, Sarocladium, and Lecanicillium, and all 11 taxa were first found to have the ability to transform ginsenosides in our study. The results indicated that P. bipinnatifidus could be a new fungi resource with potential novel natural compounds with antimicrobial activity and potent β-glucosidase for varied minor ginsenoside production.
Introduction
Ginseng, the root of Panax ginseng C. A. Meyer (Araliaceae), is called the king of all herbs in Asian countries for its various medicinal functions such as anticancer, blood vessel softening, antidiabetic, immunomodulatory, and antiaging effects (Ratan et al., 2021). It is well known that ginsenosides are the most valued and major active components in ginseng. Ginsenosides were consumed worldwide with over two billion USD worth in 2018, and future market demand is expected to be up to trillion USD by 2050 in pharmaceutical, cosmetic, and food industries (Li and Fan, 2020; Yao et al., 2021).
Until now, over 200 ginsenosides have been identified including major and minor ginsenosides. Major ginsenosides, including Rb1, Rb2, Rc, Rd, Re, and Rg1, account for more than 80% of the total ginsenosides in wild ginseng (Kim et al., 2022). However, deglycosylated minor ginsenosides, which are present in very low amounts in most natural ginseng plants, are generally more pharmaceutically active than major glycosylated ginsenosides because of higher bioavailability (Cui et al., 2016; Yang et al., 2021). Therefore, converting major ginsenosides to minor ginsenosides attracted lots of attention (Li and Fan, 2020; Siddiqi et al., 2021; Kim et al., 2022), and several preparations of minor ginsenosides were successfully commercialized with high price due to inefficient production (Li and Fan, 2020). The microbial fermentation and application of the isolated enzymes are the most feasible methods to solve the bottleneck problems because biocatalytic reactions have high substrate specificity, take place under mild conditions, produce just a few byproducts, and require simpler post-processing procedures (Li and Fan, 2020). Therefore, a reservoir of efficient enzymes or microorganisms for different minor ginsenoside production is an urgent requirement.
Endophytes generally inhabit healthy plant tissues without causing disease or injury to the host (Jia et al., 2016). Due to their special living environment and long term coexistence with their hosts, endophytes could produce diverse novel metabolites and are able to incorporate genetic information on bioactive substance biosynthesis from their host plants (Vasundhara et al., 2016). Therefore, endophytes have been widely considered valued resources of natural bioactive compounds and enzymes (Jia et al., 2016; Deshmukh et al., 2018; Gupta et al., 2020). However, considering the immense potential of endophytes for biotransformation of region- and stereo-selective production has remained largely unexplored, although the application of endophytes to biotransformation has been studied for several natural compound conversions such as favans (Agusta et al., 2005), alkaloids (Shibuya et al., 2003), and saponin (Cui et al., 2016; Cheng et al., 2021).
To obtain novel fungi with potent β-glucosidase for minor ginsenoside production, Panax bipinnatifidus var. bipinnatifidus, which is a traditional medicinal plant containing various ginsenosides and thus was supposed to harbor rich ginsenoside glucosidase resource, was first employed to isolate endophytic fungi in this study. Ginseng total saponins were used as substrates to examine the ginsenoside biotransformation potential of the isolated fungi. Also, antimicrobial activities of extracts of the fungal fermentation broth were determined. To the best of our knowledge, this is the first report on the diversity, antimicrobial, and ginsenoside biotransformation activities of cultivable endophytic fungi associated with P. bipinnatifidus.
Materials and Methods
Materials
Ginsenoside extracts from the roots of Panax ginseng C. A. Meyer. (saponin contents > 67%) and standard ginsenosides Rb1, Rb2, Rc, Rd, Re, F2, Rg3, Rh2, and Compound K (CK) were purchased from Chengdu DeSiTe Biological Technology Co., Ltd. Methanol and acetonitrile (HPLC grade) were obtained from Merck (Darmstadt, Germany). All other reagents and chemicals were at least of analytical grade.
Plant Collection and Isolation of Fungal Endophytes
In August 2020, wild and healthy P. bipinnatifidus samples were randomly collected from Yingpan town, Shaanxi province of China (33°48′21″ N, 108°56′23″ E, elevation, 1165 m), and were identified by associate researcher XinWei Shi at the Xi’an Botanical Garden of Shaanxi Province. These plants were carefully uprooted from their native habitats and placed in a sampling bag, which were immediately transported to the laboratory and stored in a refrigerator (4°C), as described previously (Tan et al., 2014). The endophytic fungal isolation procedure was carried out within 24 h of sample collection. The plant tissues were pretreated with the protocol mentioned by Qin et al. (2019). In brief, fifteen root tissues from healthy plants were washed sequentially in running tap water, ultrasonic bath (200 W, 10 min), and dried naturally at room temperature. Subsequently, the plant tissues were surface-sterilized with the method described by Tan et al. (2014) with minor modifications. In brief, the roots tissues were disinfected by sequential immersion in 70% ethanol for 1 min, 2.5% NaClO2 for 3 min, and 70% ethanol for 1 min. A total of 150 fragments (approximately 0.3 cm long and 0.3 cm wide) were excised from the pretreatment plant tissues. Tissue segments were plated on a PDA medium supplemented with amikacin sulfate (100 U/mL). The plates (90 mm) placed with seven segments were incubated at 28°C for 1 week. Emergent fungal colonies were re-cultured and photographed in a PDA medium. Finally, the pure isolates were stored at −80°C in a 20% glycerol solution in the Engineering Center of QinLing Mountains Natural Products, Shaanxi Provincial Institute of Microbiology.
Molecular Identification and Phylogenetic Analyses
The mycelia of endophytic fungi, which were purified and cultured on a PDA medium plate at 28°C for 7 days, were pulverized in liquid nitrogen with a mortar and pestle. The pretreated samples were used for the extraction of genomic DNA using the TaKaRa MiniBEST Bacterial Genomic DNA Extraction Kit (Dalian, China). Internal transcribed spacer (ITS) sequence amplification was conducted using the universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), according to the description by White et al. (1990) with minor modifications. Each 50 μl final reaction mixture contained 5.0 μL 10×Taq buffers, 4.0 μL 200 mmol/L dNTPs, 2.0 μL each primer (10 μM), 0.5 μL Ex Taq enzyme (TaKaRa, Dalian), 5.0 μL genomic DNA, and 31.5 μL ddH2O. The PCR assay was performed in a TProfessional Standard 96 Gradient thermal cycler (Biometra, Jena, Germany) programmed at 95°C for 5 min, 35 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, and 72°C for 10 min. 5 μL of each PCR product was analyzed electrophoretically in 1% (w/v) agarose gels stained with GelRed (Shanghai Generay Biotech Co., Ltd., China). All positive PCR products were subsequently purified and sequenced by BGI Biotechnology (Shenzheng, China). The aligned and edited sequences with MEGA 5.05 (Arizona State University, Tempe, United States) were used to sequence analysis in the National Center for Biotechnology Information (NCBI) GenBank database (http://www.ncbi.nlm.nih.gov/) and match similar sequences. The sequence with similarity over 97% is considered to be in the same genus. The evolutionary history was inferred, as described by Wei et al. (2018) and Felsenstein (1985). All sequences, which are aligned by MEGA 5.05 using Clustal W (Arizona State University, Tempe, United States) and deleted all positions containing gaps, were used to construct the maximum likelihood phylogenetic trees by MEGA software 5.05 (Stamatakis, 2006).
Crude Extract Preparation of Fungal Fermentation Broth
Five agar plugs (9 mm diameter) of each fungus, which were cultured on the PDA for 7 days, were inoculated in 250 ml flasks containing 50 ml of potato dextrose broth (PDB) (200 potatoes (peeled potatoes, cut into 1 cm pieces, boiled for 20 mins, and filtered to obtain filtrate), 20 dextrose, pH 6.0) and placed in a rotary shaker at 28°C and 230 rpm for 14 days. The fermentation broth was centrifuged at 8000 rpm for 8 min. 50 mL of the culture filtrate was extracted three times with an equal volume ethyl acetate, and the total organic phase was separated using a rotary evaporator in a 50°C water bath, as described by Xing et al. (2011) with minor modifications. The residue was weighed and diluted in pure methanol (10 mg/ml final concentration) and sterilized by filtration using an organic filter (0.22 μm, Shanyu Co., Ltd., China).
Antimicrobial Activity
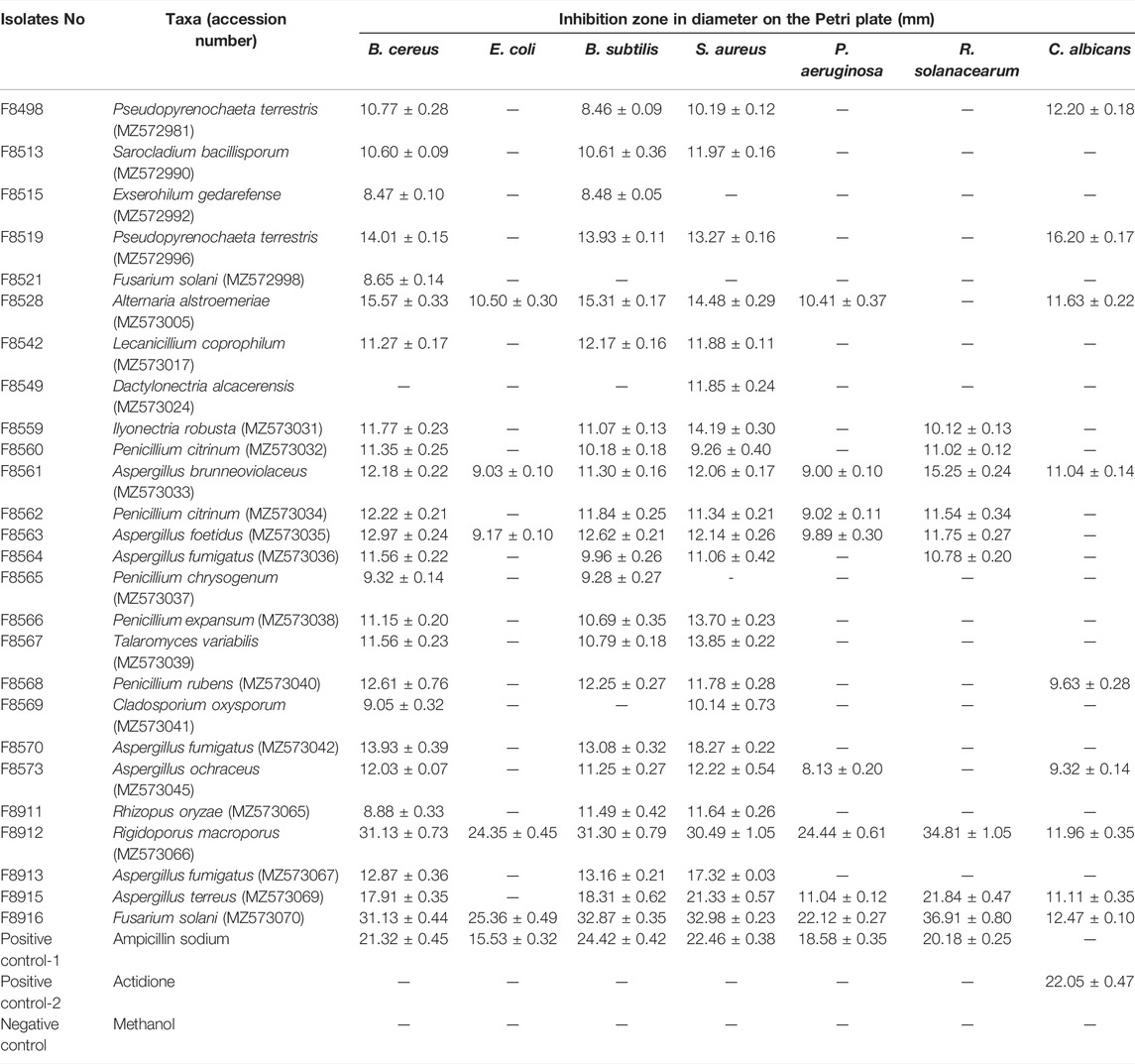
Seven pathogens such as Bacillus cereus, Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Pseudomonas aeruginosa, Xanthomonas oryzae pv. oryzae, and Candida albicans were used as test organisms to evaluate the antimicrobial activities of the ethyl acetate crude extracts from culture filtrates of 93 fungal strains using the agar diffusion protocol mentioned by Wang et al. (2019a). Precisely, the sterilized medium mixed with pre-cultured pathogens was poured into a Petri dish (90 mm) and allowed for solidification, and the agar blocks were removed with a sterilized toothpick. C. albicans was grown in Sabouraud agar medium at 28°C, and the other bacteria were grown in the Luria Bertani agar (LB) medium at 37°C. Concentrations of 10 mg/mLfor EtOAc extracts were added to the hole of the test plate and incubated at 37°C for 24 h for pathogenic bacteria or for 48 h for C. albicans. The aliquot of ampicillin sodium and actidione was dissolved in distilled water to a concentration of 1 mg/ml and used as a positive antimicrobial control. In addition, pure methanol was used as a negative control. Antimicrobial activities were evaluated by measuring the diameter of the inhibition zones. All experiments were replicated three times.
Screening of β-Glucosidase Producing Endophytic Fungi
The β-glucosidase activities were screened with the method mentioned by Cui et al. (2016). The isolates, which have grown on a PDA plate for 5 days, were inoculated on Esculin-R2A agar. The endophytic fungi with β-glucosidase activities can hydrolyze esculin and appear as colonies surrounded by a reddish-brown to dark brown zone. Esculin-R2A agar contains (g/L) Esculin 1, ferric citrate 0.5 with 15.2 R2A agar medium, and autoclaved at 121°C for 20 min.
Transformation of Ginseng Total Saponins
The isolates with β-glucosidase activities were inoculated into 250 ml flasks containing 30 ml of fermentation culture described by Ye et al. (2010) with minor modifications (20 g glucose, 10 g yeast powder, 0.5 g NaNO3, 1 g KH2PO4, 2 g Na2HPO4, 0.2 g FeSO4, 0.1 g ZnSO4, 0.2 g CuSO4, 1 g CaCl2, and 0.5 g MgSO4. 7H2O in 1 L of distilled water, initial pH of 6.5), and incubated at 28°C on a rotary shaker at 180 rpm. The ginsenoside extracts were first dissolved in methanol at a final concentration of 2 g/L and then supplied in equal volumes into the culture broth, which were incubated with the endophytic fungi and cultivated on a rotary shaker at 180 rpm for 72 h. The fungi were continually cultured for 48 h under these conditions; 2 ml of fungal suspension was aseptically removed from a shake flask culture and centrifuged to remove the precipitate. The residue was dissolved in methanol, and analysis of the biotransformation ability of ginsenosides was carried out by HPLC.
HPLC Analysis
Nine standard ginsenosides were first prepared in 1 g/L methanol solution, and the mixed standard samples containing nine standard ginsenosides were diluted to 100 mg/L with the methanol solution. Using the HPLC method described by Park et al. (2017) with minor modifications, all samples were quantitatively analyzed by HPLC (Waters, alliance separation module 2695, 2998 detector; Waters, Milford, MA, United States) using a C18 column (YMC-Pack ODS-AQ, 250 mm × 4.6 mm, 5 μm, YMC, Japan) with a column temperature of 30°C. The mobile phase was A (water) and B (acetonitrile). Gradient elution started with 77% solvent A (water) and 23% solvent B (acetonitrile) and was then changed to A from 77% to 48%, 0–12 min; A from 48% to 25%, 12–35 min; A from 48%, 35–36 min; and A from 48% to 77%, 36–42 min. All samples were detected by absorption at 203 nm, with an inject volume of 10 μL.
Diversity Analyses of the Endophytic Fungi
The isolation frequency (IF) was defined as the frequency of the occurrence of the specific endophytic fungi in total isolates based on the number of isolates (N). The relative abundance (RA) was calculated based on the number of all isolate numbers (N). The diversity of fungal species from P. bipinnatifidus was evaluated using the species richness index (S) and Shannon–Weiner index (H′), with the protocol mentioned by Fedor and Spellerberg (2013). The species richness index (S) was the number of endophytic fungal species obtained from the corresponding plant tissues. A total of 93 isolates were evaluated for the antimicrobial activities in this study.
Statistical Analyses
All results were expressed as the mean ± SEM. Graphs were prepared by Excel 2010 (Microsoft, United States).
Results
Isolation, Sequencing, Identification, and Diversity Analyses of the Endophytic Fungi from P. bipinnatifidus
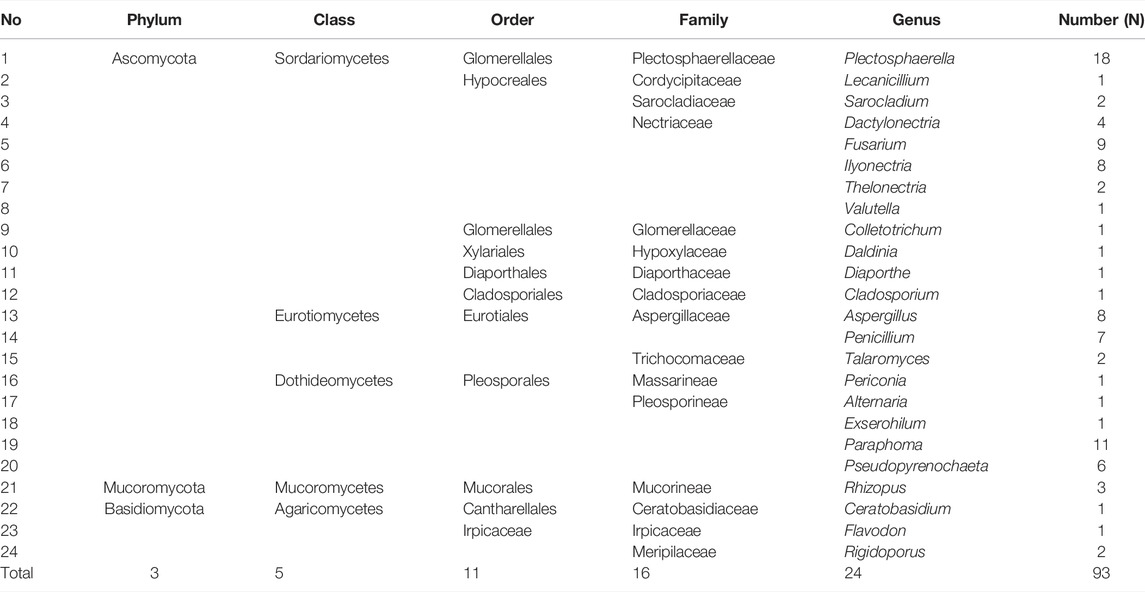
In this study, a total of 93 fungal colonies were successfully isolated from 150 tissue segments of P. bipinnatifidus with a potato dextrose agar (PDA) medium. As shown in Table 1, all isolates were grouped into 34 taxa by blasting their ITS rDNA regions using the BLAST in the NCBI GenBank database (Supplementary File S1) and constructing the phylogenetic trees using the maximum likelihood method (Supplementary File S2). The species richness index (S) and Shannon–Wiener index (H'), which are two important parameters for diversity analysis, were 34 and 3.004 for P. bipinnatifidus, respectively. As presented in Table 2, these isolates have been further grouped into three phyla (Ascomycota, Basidiomycota, and Mucoromycota), five classes (Sordariomycetes, Eurotiomycetes, Dothideomycetes, Mucoromycetes, and Agaricomycetes), 11 orders, 16 families, and 24 fungal genera.
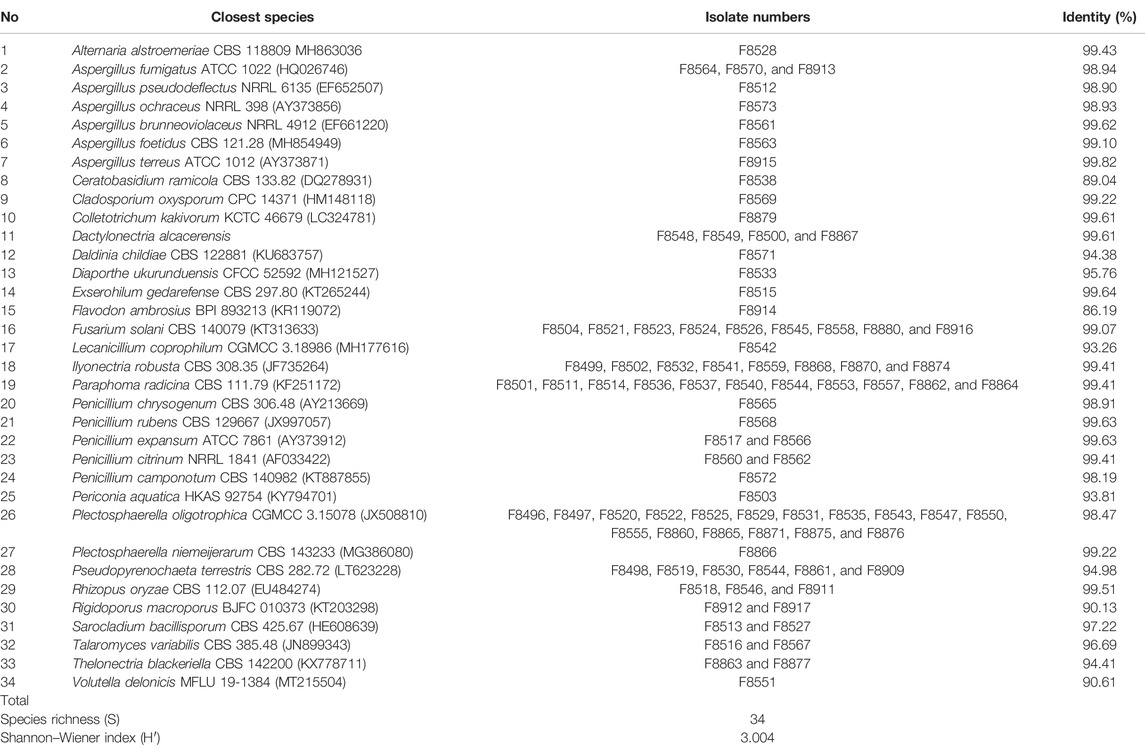
TABLE 1. Identification of endophytic fungi from P. bipinnatifidus by the Basic Local Alignment Search Tool (BLAST) in the GenBank database.
Relative Abundance Analyses of Endophytic Fungi From P. bipinnatifidus
As shown in Table 1, Plectosphaerella oligotrophica, Paraphoma radicina, and Fusarium solani were dominant species, and their IF were 18.28%, 11.83%, and 9.68%, respectively. The relative abundance (RA) of these isolates at genus, family, order, class, and phylum levels are shown in Figure 1(A–E), respectively. At the genus level, Plectosphaerella (RA, 19.35%) was the most abundant, followed by Paraphoma (RA, 11.83%) and Fusarium (RA, 9.70%). At the family level, Nectriaceae (RA, 25.81%), Pleosporineae (RA, 20.43%), and Plectosphaerellaceae (RA, 19.35%) were the three most abundant groups in this study. At the phylum level, the majority of fungi isolated from P. bipinnatifidus were identified as Ascomycota (RA, 92.47%), which represented three classes (Sordariomycetes (RA, 52.69%), Eurotiomycetes (RA, 21.51%), and Dothideomycetes (RA, 18.28%)). In addition, one class (Agaricomycetes, RA, 4.31%) belonged to Basidiomycota (RA, 4.31%), and one class (Mucoromycetes, RA, 3.23%) belonged to Mucoromycota (RA, 3.23%).
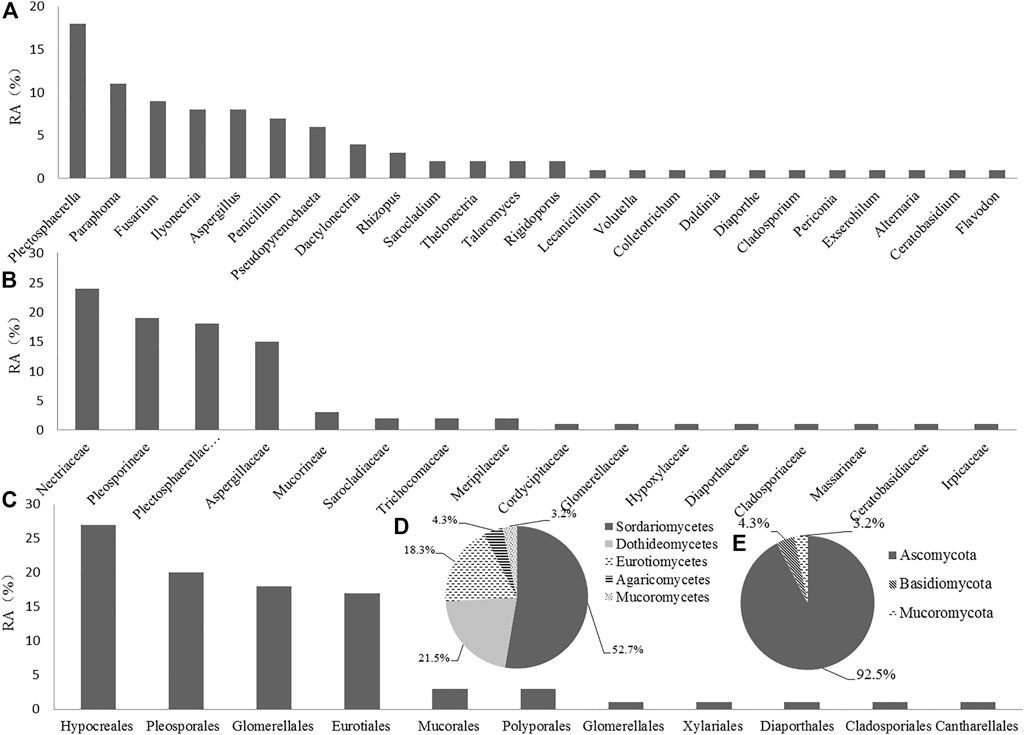
FIGURE 1. Relative abundance (RA, %) of endophytic fungi at the level of genus (A), family (B), order (C), class (D), and phylum (E).
Antimicrobial Activity Screening of the Ethyl Acetate Extracts From Endophytic Fungal Culture Filtrates
As shown in Table 3, 26 out of 93 endophytic fungal ethyl acetate extracts exhibited inhibitory activity against at least one pathogenic bacterium or fungus (Supplementary File S3). The other 67 extracts did not show inhibitory activities. These 26 strains belonged to genera of Alternaria, Aspergillus, Cladosporium, Dactylonectria, Exserohilum, Fusarium, Ilyonectria, Lecanicillium, Penicillium, Pseudopyrenochaeta, Rhizopus, Rigidoporus, Sarocladium, and Talaromyces.
Screening β-Glucosidase Activity Producing Endophytic Fungi
β-glucosidase activities were preliminarily screened for 93 endophytic fungi on plates using Esculin-R2A agar medium. Among these endophytes, 11 endophytic fungi, which were identified and named F8499 [Ilyonectria robusta (MZ572982)], F8504 [Fusarium solani (MZ572987)], F8512 [Aspergillus pseudodeflectus (MZ572989)], F8515 [Sarocladium bacillisporum (MZ572990)], F8542 [Lecanicillium coprophilum (MZ573017)], F8560 [Penicillium citrinum (MZ573032)], F8566 [Penicillium expansum (MZ573038)], F8567 [Talaromyces variabilis (MZ573039)], F8568 [Penicillium rubens (MZ573040)], F8569 [Cladosporium oxysporum (MZ573041)], and F8573 [Aspergillus ochraceus (MZ573045)], showed the strongest capacity of β-glucosidase-producing activity (Figure 2).
Biconversion of Major Ginsenosides From Ginseng Extracts
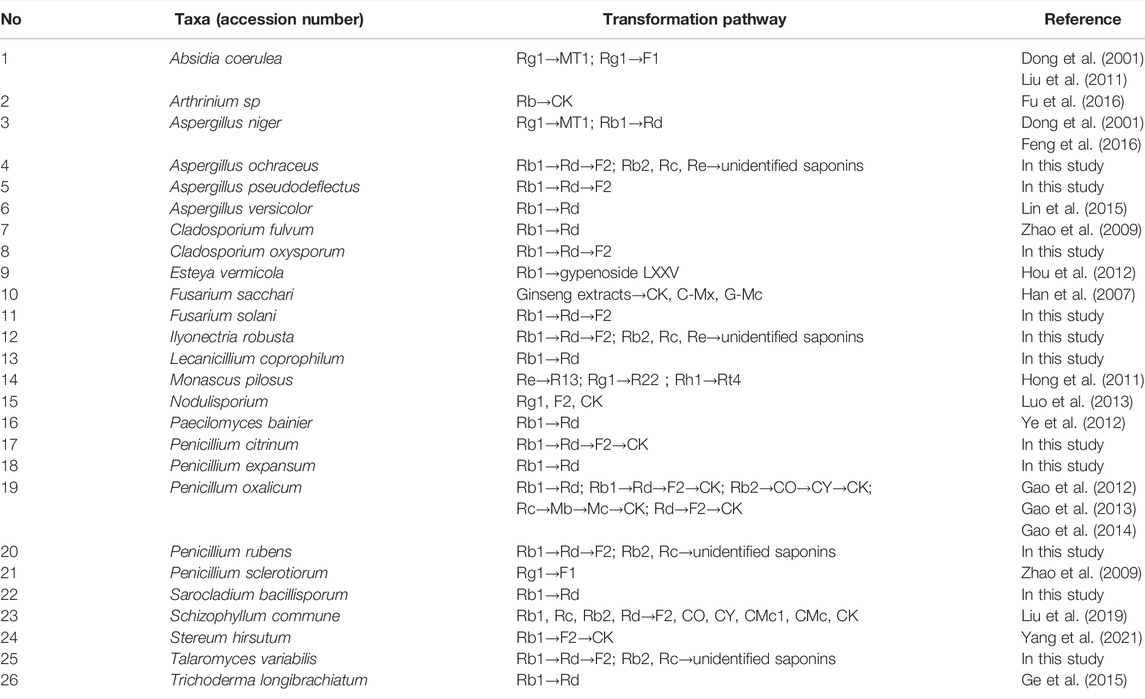
To evaluate the ginsenoside biotransformation ability of these endophytic fungi with β-glucosidase activity, we tested the composition of ginsenoside from fermentation broth before and after cultivating at the shaker level by HPLC methods (Figure 3). The results showed that 1) total saponins from ginseng extracts mainly contained major glycosylated ginsenosides such as Rb1, Rb2, Rc, Re, and Rd; 2) 11 endophytic fungi with β-glucosidase activity showed the capacity to biotransform major glycosylated ginsenosides to Rd, F2, CK, and the other unidentified saponins; and 3) all isolates could convert Rb1 to other ginsenosides, while Rb2 and Rc were transformed by four strains, and Re transformation was obtained by two strains. According to the literature reports (Geraldi, 2020), proposed transformation pathways for the isolates are summarized in Figure 4. Seven strains (F8499, F8504, F8512, F8567, F8568, F8569, and F8573) were able to transform ginsenoside Rb1 to F2 by the transformation pathway as follows: Rb1→Rd→F2; 3 strains (F8513, F8542, and F8566) were only able to transform ginsenoside Rb1 to Rd; F8560 was able to transform major ginsensides Rb1 to minor ginsensides CK directly as the following pathways: Rb1→Rd→F2→CK. Rb2, and Rc can be also transformed by endophytic fungi (F8499, F8567, F8568, F8573), but the transformation products were unidentified due to lack of adequate ginsenoside standards. Ginsenoside Re, which belonged to protopanaxatriol-type ginsenoside, can also be transformed by endophytic fungi (F8499 and F8573).
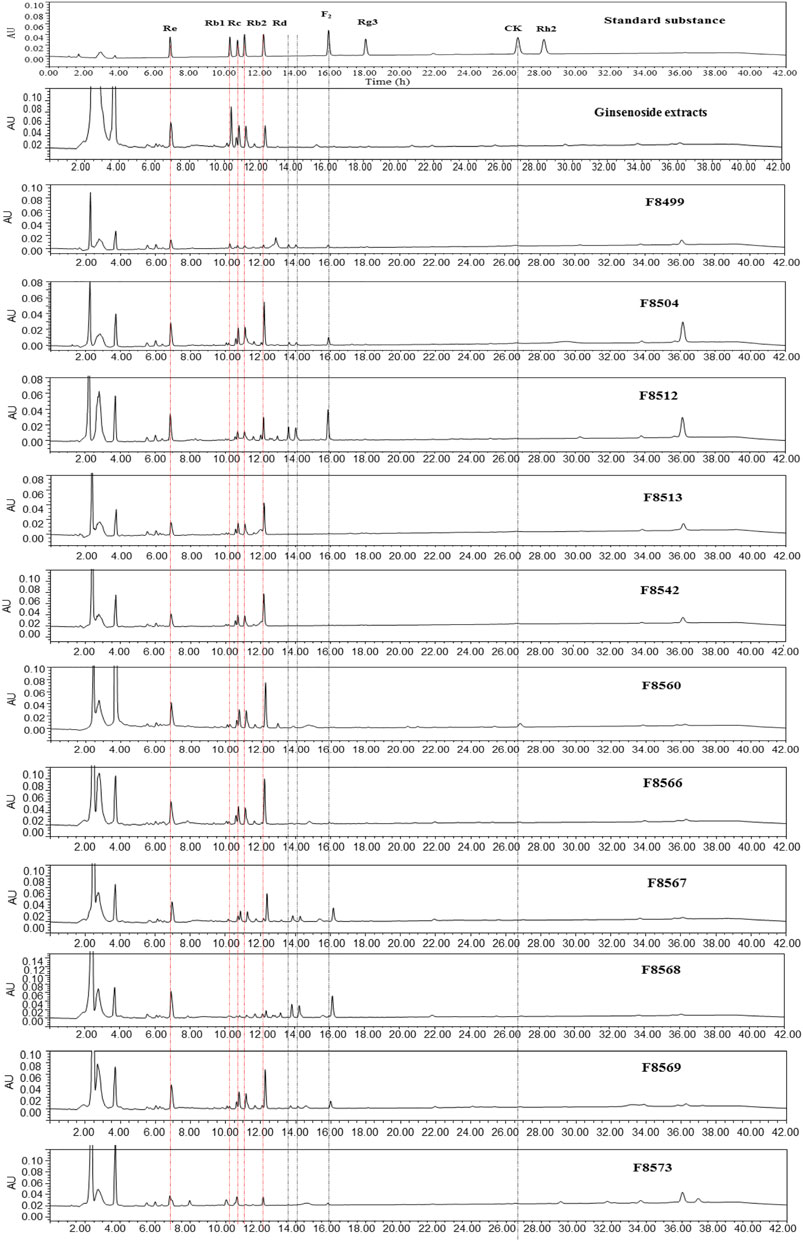
FIGURE 3. HPLC spectra of the ginsenoside standards and ginsenoside biotransformation products by 11 strains with β-glucosidase activity.
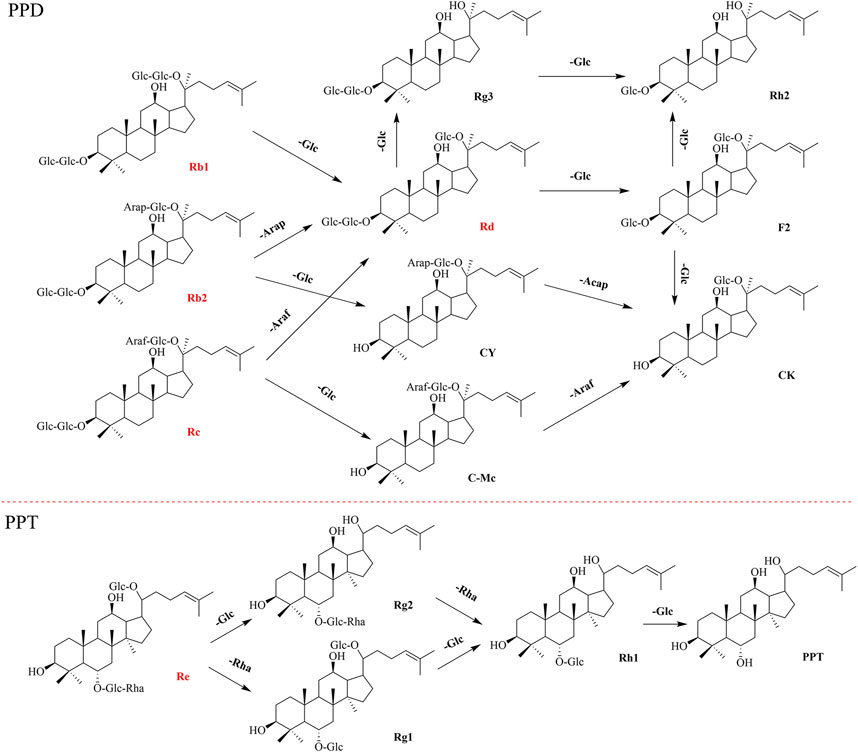
FIGURE 4. Schematic diagram of directed biotransformation of major ginsenosides (red section) of the extracts from P. ginseng roots. PPD (Protopanaxadiol-type ginsenoside) and PPT (Protopanaxatriol-type ginsenoside).
Discussion
Panax bipinnatifidus var. bipinnatifidus (also known as Panax japonicas var. major (Burkill) C.Y.Wu and Feng bipinnatifidus) is a perennial herb mainly distributed in China (Li et al., 2019). The root of P. bipinnatifidus, known as Zhuzi (means bead due to its appearance) ginseng locally, has been used as traditional medicine for thousands of years in Qinling Mountains, China. Modern pharmacological studies showed that the oleanane-type triterpenoid saponins (also known as ginsenosides) are the main chemical constituents in P. bipinnatifidus (Zhu et al., 2004; Tung et al., 2011). Based on the assumption that the endophytes incorporate genetic information on bioactive substance biosynthesis from their host plants, we supposed that endophytes from P. bipinnatifidus harbored rich enzyme resource for ginsenoside transformation. However, endophytic fungi resource associated with P. bipinnatifidus had remained unexplored.
In this study, 93 representative morphotype strains were isolated and identified from P. bipinnatifidus. var. bipinnatifidus in Qinling Mountains, China. Further diversity analysis showed that abundant fungal endophyte resources were harbored in this plant with the species richness index (S) of 34 and Shannon–Wiener index H′ of 3.004. The isolated fungi taxa also differed from those isolated from the different plants in the same region. In our previous study, it was found that there was some similarity between the endophytic fungi composition of C. japonicus Sieb (dominant genera: Colletotrichum, Aspergillus, and Diaporthe) and that of T. chinensis Baker (dominant genera: Collectotrichum, Fusarium, Aspergillus) from Yingpan town (Qinling Mountains region), Shaanxi province of China. However, the results obtained in this study (dominant genera: Plectosphaerella, Paraphoma, and Fusarium) showed significant difference from those of the aforementioned studies (An et al., 2020a; An et al., 2020b). Also, rare genera such as Lecanicillium, Sarocladium, Thelonectria, Valutella, Exserohilum, Flavodon, and Rigidoporus obtained in this study were quite different from those in our previous studies, including Biscogniauxia, Leptostroma, Stachybotrys, Didymella, Peniophora, Peniophora, Rhodotorula, Setophaeosphaeria, Clitopilus, Muyocopron, Thanatephorus, and Ceriporia. The result was in accordance with the previous studies that the endophytic fungal community composition was significantly influenced by the host geographic area, habitat environments, and growth cycle (Wang et al., 2019b). It was also suggested that highly abundant endophytic fungi resources remained unexplored in the Qinling Mountain area, which is the most important natural climatic boundary between the subtropical and warm temperate zones of China, and support an astonishingly high biodiversity (Ma et al., 2018) and are rich in medicinal plant resources.
Generally speaking, endophytes represent a wide source of unexplored and uncharacterized microorganisms capable of producing novel metabolites (Deshmukh et al., 2018), although they have been widely recognized as an important source of structurally diverse and pharmacologically active natural products (Katoch and Pull, 2017). Therefore, the isolates from P. bipinnatifidus were evaluated for their abilities to produce natural products with antimicrobial activity. The results showed that 26 out of 93 isolates exhibited inhibitory activity against at least one pathogenic bacterium or fungus. Among these, 26 strains (belonged to 14 genera) and seven genera, including Alternaria, Aspergillus, Cladosporium, Dactylonectria, Fusarium, Penicillium, and Talaromyces, have been widely reported to produce antimicrobial activities using natural products (Deshmukh et al., 2014; Deshmukh et al., 2018). Nonetheless, new antibiotics were still isolated from these genera of endophytes (Luyen et al., 2019; Qi et al., 2019; Zhang et al., 2021). In addition, the other seven genera such as Exserohilum, Lecanicillium, Ilyonectria, Rhizopus, Rigidoporus, Sarocladium, and Pseudopyrenochaeta with antimicrobial activity have rarely been reported to produce antimicrobial substances. Anyway, the results suggested these isolates have a potential for novel metabolite production.
Recent studies revealed that varied ginsenosides showed pharmaceutical potential with different health benefits because of the structural differences, such as the location of glycosides and the sugar type (Kim et al., 2018). Most ginsenosides contain a dammarane skeleton combined with various glycosidic residues, such as glucose, rhamnose, xylose, and arabinose. Among different carbohydrate hydrolases, β-glucosidase was considered the key enzyme for deglucosylated minor ginsenoside production (Song et al., 2017; Kim et al., 2022). Therefore, all 93 isolates from P. bipinnatifidus were preliminary screened for β-glucosidase activities by the agar selecting method, and 11 strains (belonging to 11 taxa, 8 genera) showed strong β-glucosidase activities. It is quite interesting that endophytic fungi isolated from P. bipinnatifidus represented a much higher percentage of strong β-glucosidase activities than those from the other two plants (Epimedium sp. and Herminium monorchis, in which saponins are not the main chemical constituents) in the same region (data not shown).
The biotransformation potential of the 11 isolates was examined with ginseng total saponins, and all strains presented activity of ginsenoside biotransformation with varied glycoside-hydrolyzing pathways. In view of literature reports, ginsenoside bioconversion by fungi with different origin is summarized in Table 4. It was found that transformation of protopanaxadiol-type ginsenoside Rb1/Rb2 (Ma et al., 2020; Kim et al., 2021) was mostly reported while transformation of protopanaxatriol-type ginsenoside Re by fungi was rare (Hong et al., 2011), probably due to the lack of microorganism resource with Re-specific glucosidase. Excitingly, two strains showed Re transformation activity in this study. In addition, the strain F8560, which was identified as Penicillium citrinum, was first found to have the ability to convert major ginsenoside Rb1 to minor ginsenoside CK. Further studies focusing on high efficiency production of CK with the enzyme from F8560 should be conducted since CK is one of the most valued minor ginsenosides, while the preparation of CK still remained costly (Ma et al., 2020; Kim et al., 2021). In addition, it is worth noting that three genera of Ilyonectria, Sarocladium, and Lecanicillium and all 11 taxa were first found to have the ability to transform ginsenosides in our study, which indicated that P. bipinnatifidus could be a new fungi resource with potent β-glucosidase for varied minor ginsenoside production.
Conclusion
In this study, the diversity, antimicrobial, and ginsenoside biotransformation potential of the endophytic fungi from P. bipinnatifidus were investigated for the first time. Our results illustrated that P. bipinnatifidus harbored abundant fungal endophytes resources, and a total of 93 representative morphotype strains were isolated and identified. Among 93 isolates, 26 strains exhibited inhibitory activities against at least one pathogenic bacterium or fungus, and 11 strains showed strong β-glucosidase activities and also presented with the ability of ginsenoside biotransformation with varied glycoside-hydrolyzing pathways. Excitingly, three genera, namely, Ilyonectria, Sarocladium, and Lecanicillium, and all 11 taxa were first found to have the ability to transform ginsenosides in our study. Overall, it was indicated that P. bipinnatifidus could be a new fungi resource with potential novel natural compounds with antimicrobial activity and potent β-glucosidase for varied minor ginsenoside production.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CA performed the experiments, wrote, and edited the manuscript. XS collected plant materials for the experiment. WX designed and supervised the project. All experiments were also performed by SM, CL, and HD.
Funding
This study was simultaneously supported by grant (2021YFC2101500) from the National Key R&D Program of China, Govt. of China, Beijing; grant (Nos 2018nk-01 and 2022k-01) from the Science and Technology Research Project of Shaanxi Province Academy of Sciences, Govt. of Shaanxi, Xi’an; and grant (Nos. 2017NY-192, 2018NY-152, 2019NY-209, and 2020JM-708) from Shaanxi Science and Technology Project, Govt. of Shaanxi, Xi’an.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the National Natural Science Foundation of China for financial support. We also acknowledge the Shaanxi Province Academy of Sciences for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.762862/full#supplementary-material
References
Agusta, A., Maehara, S., Ohashi, K., Simanjuntak, P., and Shibuya, H. (2005). Stereoselective Oxidation at C-4 of Flavans by the Endophytic Fungus Diaporthe Sp. Isolated from a Tea Plant. Chem. Pharm. Bull. (Tokyo) 53 (12), 1565–1569. doi:10.1248/cpb.53.1565
An, C., Ma, S., Shi, X., Xue, W., Liu, C., and Ding, H. (2020a). Diversity and Antimicrobial Activity of Endophytic Fungi Isolated from Chloranthus Japonicus Sieb in Qinling Mountains, China. Int. J. Mol. Sci. 21 (17), 5958. doi:10.3390/ijms21175958
An, C., Ma, S., Shi, X., Xue, W., Liu, C., and Ding, H. (2020b). Isolation, Diversity, and Antimicrobial Activity of Fungal Endophytes from Rohdea Chinensis (Baker) N.Tanaka (Synonym Tupistra Chinensis Baker) of Qinling Mountains, China. PeerJ 8, e9342. doi:10.7717/peerj.9342
Cheng, L., Zhang, H., Cui, H., Davari, M. D., Wei, B., Wang, W., et al. (2021). Efficient Enzyme-Catalyzed Production of Diosgenin: Inspired by the Biotransformation Mechanisms of Steroid Saponins in Talaromyces stollii CLY-6. Green. Chem. 23 (16), 5896–5910. doi:10.1039/d0gc04152a
Cui, L., Wu, S. Q., Zhao, C. A., and Yin, C. R. (2016). Microbial Conversion of Major Ginsenosides in Ginseng Total Saponins by Platycodon Grandiflorum Endophytes. J. Ginseng. Res. 40 (4), 366–374. doi:10.1016/j.jgr.2015.11.004
Deshmukh, S. K., Gupta, M. K., Prakash, V., and Saxena, S. (2018). Endophytic Fungi: A Source of Potential Antifungal Compounds. J. Fungi (Basel) 4 (3), 77. doi:10.3390/jof4030077
Deshmukh, S. K., Verekar, S. A., and Bhave, S. V. (2014). Endophytic Fungi: A Reservoir of Antibacterials. Front. Microbiol. 5, 715. doi:10.3389/fmicb.2014.00715
Dong, A., Cui, Y., Guo, H., Zheng, J., and Guo, D. (2001). Microbiological Transformation of Ginsenoside Rg1. J. Chine. Pharmac. Sci. 10 (3), 115–118. doi:10.1038/s41598-017-00262-0
Fedor, P. J., and Spellerberg, I. F. (2013). Shannon-wiener Index. Oikos Adavncing Ecol. 109, 203–207. doi:10.1016/b978-0-12-409548-9.00602-3
Felsenstein, J. (1985). Confidence Limits on Phylogenies: An Approach Using the Bootstrap. evolution 39 (4), 783–791. doi:10.1111/j.1558-5646.1985.tb00420.x
Feng, L., Xu, C., Li, Z., Li, J., Dai, Y., Han, H., et al. (2016). Microbial Conversion of Ginsenoside Rd from Rb1 by the Fungus Mutant Aspergillus niger Strain TH-10a. Prep. Biochem. Biotechnol. 46 (4), 336–341. doi:10.1080/10826068.2015.1031391
Fu, Y., Yin, Z. H., Wu, L. P., and Yin, C. R. (2016). Biotransformation of Ginsenoside Rb1 to Ginsenoside C-K by Endophytic Fungus Arthrinium Sp. GE 17-18 Isolated from Panax Ginseng. Lett. Appl. Microbiol. 63 (3), 196–201. doi:10.1111/lam.12606
Gao, J., Hu, Y., Ji, L., Wang, N., Wang, J., Tai, G., et al. (2013). A Novel Ginsenoside-Hydrolyzing Enzyme fromPenicillium Oxalicumand its Application in Ginsenoside Rd Production. Biocatal. Biotransformation 31 (6), 305–312. doi:10.3109/10242422.2013.857316
Gao, J., Wang, J., Cui, J., Wang, N., Bai, Y., Yuan, Y., et al. (2014). Purification and Characterization of Two Novel β-glucosidases fromPenicillium Oxalicumand Their Application in Bioactive Ginsenoside Production. Biocatal. Biotransformation 32 (4), 199–207. doi:10.3109/10242422.2014.934365
Gao, J., Xu, W., Fang, Q., Liang, F., Jin, R., Wu, D., et al. (2012). Efficient Biotransformation for Preparation of Pharmaceutically Active Ginsenoside Compound K by Penicillium oxalicum Sp. 68. Ann. Microbiol. 63 (1), 139–149. doi:10.1007/s13213-012-0454-3
Ge, F., Huang, Z., Yu, H., Wang, Y., and Liu, D. (2015). Transformation ofPanax Notoginsengsaponins by Steaming andTrichoderma Longibrachiatum. Biotechnol. Biotechnological Equipment 30 (1), 165–172. doi:10.1080/13102818.2015.1102611
Geraldi, A. (2020). Advances in the Production of Minor Ginsenosides Using Microorganisms and Their Enzymes. BIO. Integration. 1, 15–24. doi:10.15212/bioi-2020-0007
Gupta, S., Chaturvedi, P., Kulkarni, M. G., and Van Staden, J. (2020). A Critical Review on Exploiting the Pharmaceutical Potential of Plant Endophytic Fungi. Biotechnol. Adv. 39, 107462. doi:10.1016/j.biotechadv.2019.107462
Han, Y., Sun, B., Hu, X., Zhang, H., Jiang, B., Spranger, M. I., et al. (2007). Transformation of Bioactive Compounds by Fusarium Sacchari Fungus Isolated from the Soil-Cultivated Ginseng. J. Agric. Food Chem. 55 (23), 9373–9379. doi:10.1021/jf070354a
Hong, S. Y., Oh, J. H., and Lee, I. (2011). Simultaneous Enrichment of Deglycosylated Ginsenosides and Monacolin K in Red Ginseng by Fermentation with Monascus Pilosus. Biosci. Biotechnol. Biochem. 75 (8), 1490–1495. doi:10.1271/bbb.110195
Hou, J. G., Xue, J. J., Sun, M. Q., Wang, C. Y., Liu, L., Zhang, D. L., et al. (2012). Highly Selective Microbial Transformation of Major Ginsenoside Rb1 to Gypenoside Lxxv by Esteya Vermicola Cnu120806. J. Appl. Microbiol. 113 (4), 807–814. doi:10.1111/j.1365-2672.2012.05400.x
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., Han, T., et al. (2016). A Friendly Relationship between Endophytic Fungi and Medicinal Plants: A Systematic Review. Front. Microbiol. 7, 906. doi:10.3389/fmicb.2016.00906
Katoch, M., and Pull, S. (2017). Endophytic Fungi Associated with Monarda Citriodora, an Aromatic and Medicinal Plant and Their Biocontrol Potential. Pharm. Biol. 55 (1), 1528–1535. doi:10.1080/13880209.2017.1309054
Kim, M. J., Upadhyaya, J., Yoon, M. S., Ryu, N. S., Song, Y. E., Park, H. W., et al. (2018). Highly Regioselective Biotransformation of Ginsenoside Rb2 into Compound Y and Compound K by β-glycosidase Purified from Armillaria Mellea Mycelia. J. Ginseng. Res. 42 (4), 504–511. doi:10.1016/j.jgr.2017.07.001
Kim, S. A., Jeong, E. B., and Oh, D. K. (2021). Complete Bioconversion of Protopanaxadiol-type Ginsenosides to Compound K by Extracellular Enzymes from the Isolated Strain Aspergillus tubingensis. J. Agric. Food Chem. 69 (1), 315–324. doi:10.1021/acs.jafc.0c07424
Kim, S. Y., Lee, H. N., Hong, S. J., Kang, H. J., Cho, J. Y., Kim, D., et al. (2022). Enhanced Biotransformation of the Minor Ginsenosides in Red Ginseng Extract by Penicillium decumbens β-glucosidase. Enzyme Microb. Technol. 153, 109941. doi:10.1016/j.enzmictec.2021.109941
Li, J., Bai, D.-P., and Zhang, X.-F. (2019). SNP Discovery and Functional Annotation in the Panax Japonicus Var. Major Transcriptome. RSC Adv. 9 (37), 21513–21517. doi:10.1039/c8ra09495k
Li, W. N., and Fan, D. D. (2020). Biocatalytic Strategies for the Production of Ginsenosides Using Glycosidase: Current State and Perspectives. Appl. Microbiol. Biotechnol. 104 (9), 3807–3823. doi:10.1007/s00253-020-10455-9
Lin, F., Guo, X., and Lu, W. (2015). Efficient Biotransformation of Ginsenoside Rb1 to Rd by Isolated Aspergillus versicolor, Excreting β-glucosidase in the Spore Production Phase of Solid Culture. Antonie. Van. Leeuwenhoek. 108, 1117–1127. doi:10.1007/s10482-015-0565-5
Liu, X., Qiao, L., Xie, D., Zhang, Y., Zou, J., Chen, X., et al. (2011). Microbial Transformation of Ginsenoside-Rg₁ by Absidia Coerulea and the Reversal Activity of the Metabolites towards Multi-Drug Resistant Tumor Cells. Fitoterapia 82 (8), 1313–1317. doi:10.1016/j.fitote.2011.09.001
Liu, Z., Li, J. X., Wang, C. Z., Zhang, D. L., Wen, X., Ruan, C. C., et al. (2019). Microbial Conversion of Protopanaxadiol-type Ginsenosides by the Edible and Medicinal Mushroom Schizophyllum Commune: A Green Biotransformation Strategy. ACS. Omega. 4 (8), 13114–13123. doi:10.1021/acsomega.9b01001
Luo, S. L., Dang, L. Z., Li, J. F., Zou, C. G., Zhang, K. Q., and Li, G. H. (2013). Biotransformation of Saponins by Endophytes Isolated from Panax Notoginseng. Chem. Biodivers. 10 (11), 2021–2031. doi:10.1002/cbdv.201300005
Luyen, N. D., Huong, L. M., Thi Hong Ha, T., Cuong, L. H., Thi Hai Yen, D., Nhiem, N. X., et al. (2019). Aspermicrones A-C, Novel Dibenzospiroketals from the Seaweed-Derived Endophytic Fungus Aspergillus micronesiensis. J. Antibiot. (Tokyo) 72 (11), 843–847. doi:10.1038/s41429-019-0214-8
Ma, T., Hu, Y., Russo, I. M., Nie, Y., Yang, T., Xiong, L., et al. (2018). Walking in a Heterogeneous Landscape: Dispersal, Gene Flow and Conservation Implications for the Giant Panda in the Qinling Mountains. Evol. Appl. 11 (10), 1859–1872. doi:10.1111/eva.12686
Ma, Z., Mi, Y., Han, X., Li, H., Tian, M., Duan, Z., et al. (2020). Transformation of Ginsenoside via Deep Eutectic Solvents Based on Choline Chloride as an Enzymatic Reaction Medium. Bioproc. Biosyst Eng 43 (7), 1195–1208. doi:10.1007/s00449-020-02314-8
Park, B., Hwang, H., Lee, J., Sohn, S. O., Lee, S. H., Jung, M. Y., et al. (2017). Evaluation of Ginsenoside Bioconversion of Lactic Acid Bacteria Isolated from Kimchi. J. Ginseng. Res. 41 (4), 524–530. doi:10.1016/j.jgr.2016.10.003
Qi, X., Li, X., Zhao, J., He, N., Li, Y., Zhang, T., et al. (2019). Gkk1032c, a New Alkaloid Compound from the Endophytic Fungus Penicillium Sp. CPCC 400817 with Activity against Methicillin-Resistant S. aureus. J. Antibiot. (Tokyo) 72 (4), 237–240. doi:10.1038/s41429-019-0144-5
Ratan, Z. A., Haidere, M. F., Hong, Y. H., Park, S. H., Lee, J. O., Lee, J., et al. (2021). Pharmacological Potential of Ginseng and its Major Component Ginsenosides. J. Ginseng. Res. 45 (2), 199–210. doi:10.1016/j.jgr.2020.02.004
Shibuya, H., Kitamura, C., Maehara, S., Nagahata, M., Winarno, H., Simanjuntak, P., et al. (2003). Transformation of Cinchona Alkaloids into 1-N-Oxide Derivatives by Endophytic Xylaria Sp Isolated from Cinchona Pubescens. Chem. Pharm. Bull. (Tokyo) 51 (1), 71–74. doi:10.1248/cpb.51.71
Siddiqi, M. Z., Medjebouri, S., Liu, Q., Park, H. Y., Kim, G.-R., and Im, W.-T. (2021). Efficient Production of Various Minor Ginsenosides from PPD- and PPT-type Major Ginsenosides Using a Single Recombinant Bglfc Isolated from Flavobacterium Chilense. Biotechnol. Bioproc. E 26 (2), 232–246. doi:10.1007/s12257-020-0099-1
Song, X., Wu, H., Piao, X., Yin, Z., and Yin, C. (2017). Microbial Transformation of Ginsenosides Extracted from Panax Ginseng Adventitious Roots in an Airlift Bioreactor. Electron. J. Biotechnol. 26, 20–26. doi:10.1016/j.ejbt.2016.12.005
Stamatakis, A. (2006). Raxml-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 22 (21), 2688–2690. doi:10.1093/bioinformatics/btl446
Tan, X. M., Wang, C. L., Chen, X. M., Zhou, Y. Q., Wang, Y. Q., Luo, A. X., et al. (2014). In Vitro seed Germination and Seedling Growth of an Endangered Epiphytic Orchid, Dendrobium Officinale, Endemic to China Using Mycorrhizal Fungi (Tulasnella sp.). Scientia Horticulturae 165, 62–68. doi:10.1016/j.scienta.2013.10.031
Tung, N. H., Quang, T. H., Ngan, N. T. T., Minh, C. V., Anh, B. K., Long, P. Q., et al. (2011). Oleanolic Triterpene Saponins from the Roots of Panax Bipinnatifidus. Chem. Pharm. Bull. 59 (11), 1417–1420. doi:10.1248/cpb.59.1417
Vasundhara, M., Kumar, A., and Reddy, M. S. (2016). Molecular Approaches to Screen Bioactive Compounds from Endophytic Fungi. Front. Microbiol. 7, 1774. doi:10.3389/fmicb.2016.01774
Wang, S. S., Liu, J. M., Sun, J., Sun, Y. F., Liu, J. N., Jia, N., et al. (2019a). Diversity of Culture-independent Bacteria and Antimicrobial Activity of Culturable Endophytic Bacteria Isolated from Different Dendrobium Stems. Sci. Rep. 9, 10389. doi:10.1038/s41598-019-46863-9
Wang, Y. L., Gao, C., Chen, L., Ji, N. N., Wu, B. W., Lü, P. P., et al. (2019b). Community Assembly of Endophytic Fungi in Ectomycorrhizae of Betulaceae Plants at a Regional Scale. Front. Microbio.l 10, 3105. doi:10.3389/fmicb.2019.03105
Wei, W., Zhou, Y., Chen, F., Yan, X., Lai, Y., Wei, C., et al. (2018). Isolation, Diversity, and Antimicrobial and Immunomodulatory Activities of Endophytic Actinobacteria from Tea Cultivars Zijuan and Yunkang-10 (Camellia Sinensis Var. Assamica). Front. Microbiol. 9, 1304. doi:10.3389/fmicb.2018.01304
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” In PCR Protocols: A Guide to Methods And Applications. Editors M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (Berkeley, USA: Academic Press), 315–322.
Xing, Y. M., Chen, J., Cui, J. L., Chen, X. M., and Guo, S. X. (2011). Antimicrobial Activity and Biodiversity of Endophytic Fungi in Dendrobium Devonianum and Dendrobium Thyrsiflorum from Vietnam. Curr. Microbiol. 62 (4), 1218–1224. doi:10.1007/s00284-010-9848-2
Yang, W., Zhou, J., Harindintwali, J. D., and Yu, X. (2021). Production of Minor Ginsenosides by Combining Stereum Hirsutum and Cellulase. Plos. One. 16 (8), e0255899. doi:10.1371/journal.pone.0255899
Yao, L., Wang, J., He, J., Huang, L., and Gao, W. (2021). Endophytes, Biotransforming Microorganisms, and Engineering Microbial Factories for Triterpenoid Saponins Production. Crit. Rev. Biotechnol. 41 (2), 249–272. doi:10.1080/07388551.2020.1869691
Ye, L., Zhang, C., Li, J., Shi, X., and Feng, M. (2012). Effects of External Calcium on the Biotransformation of Ginsenoside Rb1 to Ginsenoside Rd by Paecilomyces Bainier 229-7. World J. Microbiol. Biotechnol. 28 (3), 857–863. doi:10.1007/s11274-011-0882-4
Ye, L., Zhou, C. Q., Zhou, W., Zhou, P., Chen, D. F., Liu, X. H., et al. (2010). Biotransformation of Ginsenoside Rb1 to Ginsenoside Rd by Highly Substrate-Tolerant Paecilomyces Bainier 229-7. Bioresour. Technol. 101 (20), 7872–7876. doi:10.1016/j.biortech.2010.04.102
Zhang, R., He, W., Wang, Y., Zhao, J., Zhou, R., Li, L., et al. (2021). New Butyrolactone Derivatives from the Endophytic Fungus Talaromyces Sp. CPCC 400783 of Reynoutria Japonica Houtt. J. Antibiot. 74 (4), 225–232. doi:10.1038/s41429-020-00388-w
Zhao, X., Wang, J., Li, J., Fu, L., Gao, J., Du, X., et al. (2009). Highly Selective Biotransformation of Ginsenoside Rb1 to Rd by the Phytopathogenic Fungus Cladosporium Fulvum (Syn. Fulvia Fulva). J. Ind. Microbiol. Biotechnol. 36 (5), 721–726. doi:10.1007/s10295-009-0542-y
Keywords: P. bipinnatifidus, endophytic fungi, diversity, ginsenoside, biotransformation
Citation: An C, Ma S, Shi X, Liu C, Ding H and Xue W (2022) Diversity and Ginsenoside Biotransformation Potential of Cultivable Endophytic Fungi Associated With Panax bipinnatifidus var. bipinnatifidus in Qinling Mountains, China. Front. Pharmacol. 13:762862. doi: 10.3389/fphar.2022.762862
Received: 23 August 2021; Accepted: 01 March 2022;
Published: 04 April 2022.
Edited by:
Ramsay Soup Teoua Kamdem, University of Bremen, GermanyReviewed by:
Pradeep Singh, Debre Tabor University, EthiopiaHemraj Chhipa, Agriculture University (Kota), India
Mukesh Kumar Awasthi, Northwest A&F University, China
Copyright © 2022 An, Ma, Shi, Liu, Ding and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjiao Xue, eC13ZW5qaWFvQDE2My5jb20=
 Chao An
Chao An Saijian Ma
Saijian Ma Xinwei Shi
Xinwei Shi Chen Liu
Chen Liu Hao Ding
Hao Ding Wenjiao Xue
Wenjiao Xue