- 1Infectious Diseases Institute, Guangzhou Eighth People’s Hospital, Guangzhou Medical University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Translational Cancer Research of Chinese Medicines, Joint International Research Laboratory of Translational Cancer Research of Chinese Medicines, International Institute for Translational Chinese Medicine, School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
- 3Institute of Precision Medicine, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 4Institute of Molecular Rhythm and Metabolism, School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
Editorial on the Research Topic
Recent advances in novel therapeutic molecules and targets for inflammatory diseases
Inflammatory diseases are composed of numerous disorders and conditions that are characterized by inflammation, such as inflammatory bowel disease, hepatitis, and rheumatoid arthritis (Okin et al., 2012). In pathological conditions with inflammatory diseases, the immune system mistakenly attacks healthy cells or tissues, resulting in chronic pain, redness, swelling, stiffness, and damage to the body (Marchetti et al., 2005). Inflammatory diseases have been linked to several potential causes, including diet, stress, and sleep disorders. Anti-inflammatory drugs help to prevent or minimize disease progression. However, common medications used are frequently accompanied by serious adverse effects. There is an urgent need to develop new therapies for inflammatory diseases and elucidate the critical genes and inner mechanism.
Diagnostic biomarkers are useful in the therapeutics of diseases at multiple aspects along the patient’s diagnostic and treatment course. There are various inflammatory biomarkers including cytokines/chemokines, acute phase proteins, immune-related effectors, reactive oxygen and nitrogen species, prostaglandins and cyclooxygenase-related factors, transcription factors, and growth factors (Brenner et al., 2014). Lin et al. demonstrated that lncRNA DLEU2 in the intestinal mucosa was dysregulated upon gut inflammation and could act as a diagnostic biomarker for ulcerative colitis (Lin et al.). They identified DLEU2 as an anti-inflammatory lncRNA which inhibits gut inflammation by negatively regulating the NF-κB signaling pathway (Lin et al.). Huang et al. reported that MHR (monocyte to high-density lipoprotein ratio) and MAR (monocyte to apolipoprotein A1 ratio) were ideal pro-inflammatory markers to reflect bone homeostasis imbalance caused by chronic inflammation in the bone microenvironment of postmenopausal women with type 2 diabetes mellitus (Huang et al.). These researchers expanded the Research Topic of biomarkers to inflammatory diseases.
Finding the right therapeutic targets is the most important approach in anti-inflammatory drug discovery. Many targets are responsible for the anti-inflammatory actions, such as inhibition of cytokine signaling, decreasing leucocyte activation, chemotaxis, and recruitment. Researchers have characterized several targets in this Research Topic. K-Ras is a well-studied oncogene. Qi et al. reported that inhibition of K-RasG13D mutation promoted cancer stemness and inflammation via RAS/ERK pathway (Qi et al.). This finding may be important for understanding the effects of K-RasG13D mutation on promoting cancer stemness and inflammation when employing targeted therapies to K-RasG13D mutations in clinical practice. Promyelocytic leukemia zinc finger protein (PLZF) is a transcription factor that acts in regulating a variety of biological processes, such as spermatogenesis, stem cell maintenance, immune regulation, and invariant natural killer T cell (iNKT) development. Hu et al. demonstrated that PLZF, upregulated in mice with non-alcoholic fatty liver disease, was an essential regulator of hepatic lipid and glucose metabolism (Hu et al.). PLZF activates SREBP-1c gene transcription by binding directly to the promoter, inducing repressor-to-activator conversion.
Target-based small molecule drug discovery is an important research direction for inflammatory diseases. Schepetkin et al. designed, synthesized, and evaluated several novel analogs of O-substituted tryptanthrin oxime derivatives as c-Jun N-terminal kinase (JNK) inhibitors. Some of these compounds had a high affinity for JNK1-3 and potently inhibited LPS-induced nuclear NF-κB/AP-1 activation and IL-6 production in human monocytic cells (Schepetkin et al.). Ginsenoside Rc (Rc) is a major component of Panax ginseng. Tang et al. reported that Rc potentially ameliorated inflammatory response and barrier function to protect the gut from DSS-induced colitis (Tang et al.). Zhong et al. showed that Rc alleviated acetaminophen-induced hepatotoxicity by relieving inflammation and oxidative stress (Zhong et al.). Both studies indicated that Rc could be a ligand of FXR by binding with the protein domain. Thus, signaling inhibitors and ligands of nuclear receptors may represent novel anti-inflammatory therapies for the inhibition of multiple cytokines and inflammatory signaling pathways.
Overall, we identified a series of attractive diagnostic biomarkers, therapeutic targets, and compounds for the intervention of inflammatory diseases (Figure 1). These biomarkers and targets consist of lncRNA, oncogene, protein of signaling pathways, and transcriptional factor (e.g., nuclear receptor). Therapeutic agents include natural (e.g., Ginsenoside Rc) and synthetic compounds (e.g., O-substituted tryptanthrin oxime derivatives) (Figure 1). The researchers also delineated intricate gene-chemical relationships and molecular mechanisms in inflammatory disease development. All these findings may yield novel anti-inflammatory lead structures with proven efficacy and pharmacokinetics in vivo to manage inflammatory diseases in the future. Further studies would identify the patients who would benefit from a particular anti-inflammatory drug action, making treatment for disease state more personalized and effective.
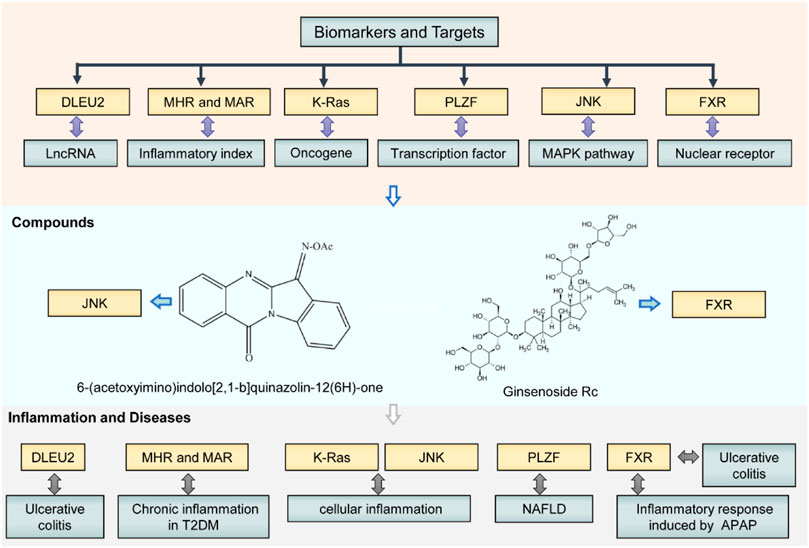
FIGURE 1. Biomarkers, targets, and compounds for the management of inflammatory diseases in this Research Topic. These biomarkers and targets consist of lncRNA, oncogene, key protein of signaling pathways, and transcriptional factor (e.g., nuclear receptor). Therapeutic agents include natural (e.g., Ginsenoside Rc) and synthetic compounds (e.g., O-substituted tryptanthrin oxime derivatives). NAFLD, non-alcoholic fatty liver disease; T2DM, type 2 diabetes mellitus; APAP, acetaminophen.
Author contributions
FL drafted the manuscript; SGW, XC and SW reviewed and edited the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82104238 to FL, 82000612 to SGW).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Brenner, D. R., Scherer, D., Muir, K., Schildkraut, J., Boffetta, P., Spitz, M. R., et al. (2014). A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol. Biomarkers Prev. 23 (9), 1729–1751. doi:10.1158/1055-9965.EPI-14-0064
Marchetti, B., and Abbracchio, M. P. (2005). To be or not to be (inflamed)--is that the question in anti-inflammatory drug therapy of neurodegenerative disorders? Trends Pharmacol. Sci. 26 (10), 517–525. doi:10.1016/j.tips.2005.08.007
Keywords: inflammatory diseases, biomarkers, targets, inflammation, compounds
Citation: Li F, Wang S, Chao X and Wang S (2023) Editorial: Recent advances in novel therapeutic molecules and targets for inflammatory diseases. Front. Pharmacol. 13:1121821. doi: 10.3389/fphar.2022.1121821
Received: 12 December 2022; Accepted: 19 December 2022;
Published: 04 January 2023.
Edited and reviewed by:
Dieter Steinhilber, Goethe University Frankfurt, GermanyCopyright © 2023 Li, Wang, Chao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Li, bGlfZmVuZ2ZlbmdAMTYzLmNvbQ==; Shaogui Wang, d2FuZ3NoYW9ndWlAZ3p1Y20uZWR1LmNu; Xiaojuan Chao, Y2h4ajIwMThAMTYzLmNvbQ==; Shuai Wang, d2FuZ3M5MUAxNjMuY29t
 Feng Li1*
Feng Li1* Shaogui Wang
Shaogui Wang Xiaojuan Chao
Xiaojuan Chao Shuai Wang
Shuai Wang