
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 13 December 2022
Sec. Gastrointestinal and Hepatic Pharmacology
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1080226
This article is part of the Research Topic Gut Microbiota-Mediated Natural Products Metabolism and Its Role in the Pharmacokinetics and Pharmacodynamics View all 3 articles
 Bo Shen1†
Bo Shen1† Cui Zhou1†
Cui Zhou1† Tianyi Gu1†
Tianyi Gu1† Zhenyang Shen1
Zhenyang Shen1 Yuecheng Guo1
Yuecheng Guo1 Weiming Dai1
Weiming Dai1 Yang Liu2
Yang Liu2 Jie Zhang2
Jie Zhang2 Lungen Lu1*
Lungen Lu1* Hui Dong1*
Hui Dong1*Background: Liver fibrosis is a common outcome of the pathological progression of chronic liver disease; however, no specific and effective therapeutic agent has been approved for its treatment. We investigated the effects of Kuhuang on liver fibrosis and the underlying mechanisms of action.
Materials and methods: To induce hepatic fibrosis, either 3,5-diethoxycarbonyl-1,4-dihydro-collidine (DDC) diet was administered, or bile duct ligation (BDL) surgery was performed on C57BL/6 mice. Kuhuang was orally administered to mice for 7 days before and after bile duct ligation or 4 weeks with a DDC diet. Hematoxylin and eosin, Sirius red staining, and immunohistochemical analyses were performed to evaluate hepatic pathology. Hepatic interferon-β (IFN-β) levels were measured using an enzyme-linked immunosorbent assay. RNA sequencing was performed to examine the gene expression profiles of liver tissues. The mRNA expression of inflammatory, profibrotic, and bile acid (BA)-related genes was further validated by qRT-PCR. A targeted metabolomics assay revealed the alteration of the hepatic bile acid (BA) composition. The composition of the gut microbiota was determined via 16S rRNA sequencing.
Results: Treatment with Kuhuang attenuated liver fibrosis and reduced the inflammatory response in bile duct ligation and DDC mouse models. In addition, the hepatic IFN signaling pathway was activated following Kuhuang treatment. Kuhuang treatment also significantly decreased hepatic levels of both primary and secondary BAs. In addition, Kuhuang treatment altered gut microbiota composition, with an increased abundance of interferon-inducing Akkermansia and decreased abundance of bile salt hydrolase-producing Lactobacillus, Clostridium, and Bifidobacterium. Furthermore, the abundance of Akkermansia was positively correlated with the hepatic mRNA expression levels of Ifna4, Ifnb, and Isg15, whereas that of Lactobacillus, Clostridium-sensu-stricto-1, and Bifidobacterium was positively correlated with levels of bile acid synthesis-related genes.
Conclusion: Our results suggest that Kuhuang plays a protective role during the progression of liver fibrosis, potentially by altering the composition of the gut microbiota, which consequently activates interferon signaling and inhibits bile acid synthesis in the liver.
Liver fibrosis, characterized by excessive extracellular matrix deposition, is a common pathological process in various chronic liver diseases, including viral hepatitis, alcohol-related liver disease, non-alcoholic fatty liver disease, autoimmune hepatitis, cholestatic liver disease, drug-related hepatitis, and genetic diseases (Guo et al., 2022b). Persisting and iterating liver fibrosis tends to develop into cirrhosis, a severe health burden globally (Ginès et al., 2021). However, no specific and effective therapeutic agents have been approved to treat liver cirrhosis. Fortunately, in rare cases, fibrosis may be reversible (Lee et al., 2015). Therefore, discovering drugs for liver fibrosis therapy is urgent to meet this clinical need, considering the high morbidity and mortality associated with chronic liver disease.
Kuhuang, a traditional Chinese herbal compound—composed of Yin Chin, Da Huang, Ku Shen, Da Qingye, and Chai Hu—has been proven clinically effective in treating cholestasis for many years (Xu et al., 2009). Previous studies have provided convincing evidence for the protective role of Kushen, an important component of Kuhuang, in liver fibrosis and carcinoma. In a model of liver fibrosis, the compound Kushen injection (CKI) repressed hepatic stellate cell (HSC) activation and protected against hepatocellular carcinoma by regulating TGFβ/Smad7 signaling (Yang et al., 2021). In addition, Yang et al. (2020) revealed that CKI could alleviate immunosuppression in the tumor-associated microenvironment by activating proinflammatory responses, suggesting that Kushen might also participate in regulating inflammatory responses. However, as Kuhuang is composed of multiple components besides Kushen, other mechanisms must be involved in its ability to alleviate liver fibrosis. To date, the mechanisms of action of Kuhuang in liver disease have not been elucidated.
Bile acids (BAs), the main constituents of bile, not only function as important digestive fluids but also as essential signaling molecules. When exposed to excessive BAs, bile ducts and hepatocytes are prone to injury, accompanied by mitochondrial dysfunction and recruitment and infiltration of inflammatory cells (Li et al., 2017; Trauner et al., 2017; Zhang et al., 2019a). BAs play an essential role in HSC proliferation and activation (Svegliati-Baroni et al., 2005; Liu et al., 2018). Constant ductular injury, enduring inflammation, and HSC activation eventually lead to liver fibrosis (Liu et al., 2018). In the liver, the classic pathway of BA synthesis is regulated by the BA-activated farnesol X receptor (Fxr), which subsequently facilitates slight heterodimer partner-dependent inhibition of cholesterol 7α-hydroxylase (encoded by CYP7A1) (Lu et al., 2000). In addition, activated Fxr-FGF15/19 signaling in the ileum suppresses BA synthesis through CYP7A1 (Katafuchi and Makishima, 2022). The gut microbiota is another crucial modulator in controlling BA homeostasis by linking the circulation of BAs between the liver and gut (Jiang et al., 2022). Bile salt hydrolase (BSH) activity is essential for the deconjugation of primary BAs by gut microbiota (Mori et al., 2022) and is mostly facilitated by Gram-positive bacteria, such as Lactobacillus (Jayashree et al., 2014), Bifidobacterium (Kim et al., 2005), Enterococcus (Wijaya et al., 2004), and Clostridium spp. (Rossocha et al., 2005) as well as in commensal Gram-negative Bacteroides spp. (Long et al., 2017). In turn, BAs are crucial players in modulating the composition of gut microbiota (Xu et al., 2022). Targeting the gut microbiota–BA axis may be a promising therapeutic strategy for liver fibrosis.
Although Kuhuang has been proven to be effective in alleviating cholestasis, the underlying mechanism remains unclear. To explore the mechanism of Kuhuang, we performed hepatic transcriptomics and targeted metabolomics analyses, as well as 16S rRNA sequencing of fecal samples in a mouse model of cholestasis. We found that Kuhuang could alleviate hepatic inflammatory response, BA accumulation, and liver fibrosis. The potential mechanism of Kuhuang may involve the activation of interferon signaling and inhibition of BA synthesis by altering gut microbiota composition.
A total of 25 male C57BL/6J mice (8 weeks old) were divided randomly into five groups (n = 5 per group): Chow group fed a chow diet; 3,5-diethoxycarbonyl-1,4-dihydro-collidine (DDC)+Kuhuang (KH) group fed a DDC diet (Trophic Animal Feed High-tech Company, Nantong, China) plus oral KH administration; DDC + H2O group fed a DDC diet plus oral H2O administration; bile duct ligation (BDL)+KH group treated with BDL under anesthetic condition plus oral KH administration, and BDL + H2O group treated with BDL plus oral H2O administration. For the BDL model (n = 5 per group), Kuhuang dissolved in ddH2O (20 mg/g body weight, BDL + KH group) (Zhang et al., 2019) or ddH2O (BDL + H2O group) was administered orally every day for 7 days before and after BDL, respectively. For the DDC model (n = 5 per group), Kuhuang (DDC + KH group) or ddH2O (DDC + H2O group) were administered orally every day for 4 weeks along with a DDC diet. The chow group (n = 5) received no intervention. Kuhuang granules were provided by Suzhou Leiyunshang Pharmacology Group (Suzhou, China). Animals were maintained in a specific pathogen-free environment with unrestrained access to food and water. The Animal Ethics Committee of the Shanghai General Hospital, Shanghai Jiaotong University School of Medicine, approved all animal-related experiments. Mice were treated with 3% phenobarbital sodium before sacrifice. Blood, liver, and fecal samples were collected. Liver tissues were either stored at −80°C for protein and RNA extraction or fixed with formalin, followed by paraffin embedding for histopathology analysis.
Total RNA was isolated from cells and tissues using the AG RNAex Pro Reagent (AG21102, Accurate Biotechnology, Co., Ltd, Hunan, China) according to the standard protocol. Subsequently, the isolated RNA was reverse-transcribed into cDNA using a reverse-transcriptase kit (Xinbeibio, Shanghai, China). qPCR was conducted with SYBR Green Master mix (Yeason, Shanghai, China) on a ViiATM Real-Time PCR instrument. The relative expression of the target genes was quantified by normalization to GAPDH. Primer sequences were as follows: Gapdh Forward primer: TCTCCTGCGACTTCAACAReverse primer: TGTAGCCGTATTCATTGTCA Il1β Forward primer: CCACAGACCTTCCAGGAGAATGReverse primer: GTGCAGTTCAGTGATCGTACAGG Il6 Forward primer: ACAACCTGAACCTTCCAAAGATGReverse primer: TATACCTCAAACTCCAAAAGACCAG Tnfα Forward primer: CACTTCGAAACCTGGGATTCAGReverse primer: GGTCTCCAGATTCCAGATGTCAG Col1a1 Forward primer: TCAGAGGCGAAGGCAACAGT Reverse primer: CCCCAAGTTCCGGTGTGA Atca2Forward primer: GCTGAAGTATCCGATAGAACACG Reverse primer: GGTCTCAAACATAATCTGGGTCA Il1raForward primer: TCAGATCTGCACTCAATGCC Reverse primer: CTGGTGTTTGACCTGGGAGT Ifn4 Forward primer: AGGATTTTGGATTCCCCTTGReverse primer: TATGTCCTCACAGCCAGCAG Ifnb Forward primer: AGCTCCAAGAAAGGACGAACATReverse primer: GCCCTGTAGGTGAGGGTTGATCT Isg15Forward primer: CAGGACGGTCTTACCCTTTCC Reverse primer: AGGCTCGCTGCAGTTCTGTAC Cyp7a1 Forward primer: TGGAATAAGGAGAAGGAAAGTAReverse primer: TGTGTCCAAATGCCTTCGCAGA Cyp8b1 Forward primer: CCTCTGGACAAGGGTTTTGTG Reverse primer: GCACCGTGAAGACATCCCC Cyp7b1Forward primer: GGAGCCACGACCCTAGATG Reverse primer: GCCATGCCAAGATAAGGAAGC Fxr Forward primer: TGTGAGGGCTGCAAAGGTTTReverse primer: ACATCCCCATCTCTCTGCAC Fgf15 Forward primer: GAGGACCAAAACGAACGAAATTReverse primer: ACGTCCTTGATGGCAATCG.
Liver tissues were immediately fixed in 4% paraformaldehyde once resected and then embedded in paraffin after 24–48 h. The embedded tissues were sliced into 4 µm thick sections for histological experiments. The inflammatory response and ballooning were assessed by H&E staining. In addition, collagen accumulation in the liver was evaluated using Sirius red staining. All images were captured using a light microscope, and the positive areas were quantified using ImageJ software (NIH, Bethesda, MD, United States).
After deparaffinization, formalin-fixed paraffin-embedded tissues were processed for antigen retrieval using an EDTA antigen retrieval solution (Sangon, Shanghai, China). Immunostaining blocking buffer (Sangon, Shanghai, China) was used to block non-specific antibody binding. The sections were incubated with primary antibodies against α-SMA (1:500, Abcam, 32,575) or F4/80 (1:500, Cell Signaling Technology, 30325S) overnight at 4°C. The slices were then washed and incubated with a horseradish peroxidase-labeled secondary antibody (1:200, Beyotime Biotechnology, Shanghai, China) for 1 h at room temperature. Positive areas were visualized using 3,3-diaminobenzidine tetrahydrochloride and counterstained with hematoxylin. Images were acquired using a fluorescence microscope (Leica). The positive fields were quantified using ImageJ (National Institutes of Health, Bethesda, MD, United States).
The levels of interferon-β (IFN-β) in the liver tissues were detected using enzyme-linked immunosorbent assay (ELISA) assay kits (MULTISCIENCES, Hangzhou, China). All procedures followed the manufacturer’s instructions.
The expression profiles of liver tissues were detected using RNA-seq. Bulk liver RNA was isolated using the AG RNAex Pro Reagent (AG21102, Accurate Biotechnology, Co., Ltd., Hunan, China) according to the manufacturer’s protocol. RNA was sequenced by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The data were analyzed using the online Majorbio Cloud Platform. DESeq was used to analyze differential gene expression. Differentially expressed genes (DEGs) were clustered using hierarchical cluster analysis. Gene ontology (GO) enrichment analysis of DEGs was performed using cluster Profiler.
Stool samples were collected after the mice were sacrificed. The total DNA was extracted using A QIAamp PowerFecal DNA Kit (QIAGEN, Germany). The 16S rRNA V3-V4 region amplified products were subjected to deep sequencing using the Illumina® MiSeq™ II platform. Mothur software was used to remove ambiguous, chimeric, contaminated, and short sequences (length < 350 bp). Operational taxonomic units (OTUs) of high-quality sequences were identified by mapping to sequences in the SILVA database with 97% similarity. Statistical differences in OTU abundance were determined at a threshold of linear discriminant analysis score >3. Alpha and beta diversity analyses were performed using Mothur and Bray–Curtis method, respectively.
Liver samples were subjected to protein precipitation using acidified and ice-cold MeOH. The supernatant was acquired after vortexing for 60 s, grinding at 55 Hz for 1 min, ultrasonication at 25°C for 10 min, storage at −20°C for 30 min, and centrifugation at 12,000 rpm at 4°C for 10 min. Then, 300 μl of the supernatant was reconstituted in 600 μl ddH2O, followed by filtration through a 0.22 μm membrane. Finally, 25% of the 200 μl mixture in 30% MeOH solvent was injected into the LC-MS system for detection. The chromatographic and mass spectrometry conditions for the ultra-performance liquid chromatography system analysis were used as previously described (Yang et al., 2017; Hu et al., 2020).
Statistical analysis was performed using the SPSS software (version 18.0; SPSS Inc., Chicago, IL, United States). Student’s t-test was applied for two groups, whereas a one-way analysis of variance was applied for more than two groups. Statistical significance was set at p < 0.05.
Mice that underwent BDL surgery to induce cholestatic liver fibrosis were divided into two groups: Kuhuang and H2O (control group). H&E staining showed less inflammatory cell infiltration and centrilobular necrosis in the liver of the Kuhuang group than in control (Figure 1A). Fewer macrophages were observed, and hepatic mRNA expression of Il6, Il1β, and Tnfα was downregulated in the liver of Kuhuang mice, indicating that Kuhuang treatment attenuated the liver inflammatory response (Figures 1B, C). In addition, Sirius red staining and IHC for α-SMA (Figures 1A, D) revealed that the hepatic profibrotic area was significantly reduced in the Kuhuang group. The mRNA levels of profibrogenic genes (Acta2 and Col1a1) were downregulated upon Kuhuang treatment (Figure 1E). These results indicate that Kuhuang treatment ameliorated hepatic inflammation and liver fibrosis in BDL mice.
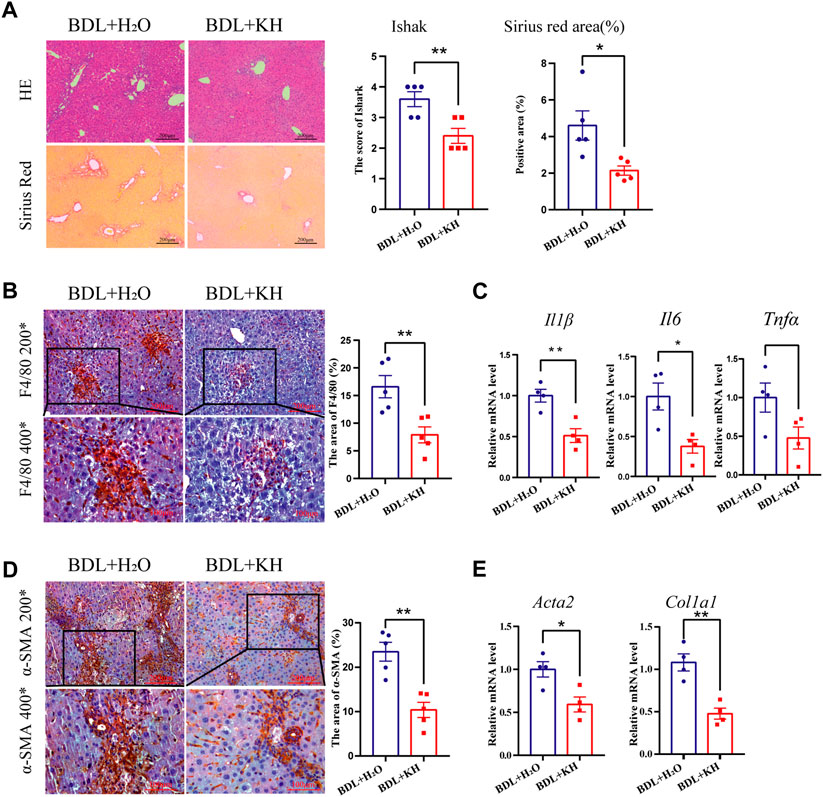
FIGURE 1. Kuhuang treatment ameliorates hepatic inflammation and fibrosis in BDL mice. (A) Representative images of H&E and sirius red staining of liver sections (left panel) of BDL mice. Quantification of Ishak score and Sirius red-positive areas are shown in the right panel. (B) Representative images (left panel) and quantification (right panel) of F4/80-positive areas. (C) QRT-PCR analyses inflammation-related genes (Il6, Il1β, and Tnfα) in liver tissue. (D) Representative images (left panel) and quantification (right panel) of α-SMA positive fibrosis areas. (E) QRT-PCR analysis of fibrosis-related genes (Col1a1 and Acta2) in liver tissue. The data are presented as mean ± SEM. N = 4–5 per group. The data were analyzed with Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations, KH, Kuhuang; BDL, bile duct ligation.
Cholestatic liver injury was also induced in mice by a 4-week DDC diet and oral administration of either Kuhuang or H2O. H&E staining showed less inflammatory cell infiltration and centrilobular necrosis in the liver of the Kuhuang group than in control (Figure 2A). Fewer macrophages were observed, and hepatic mRNA expression of Il6, Il1β, and Tnfα was downregulated in the liver of Kuhuang mice, indicating that Kuhuang treatment attenuated the liver inflammatory response (Figures 2B, C). In addition, Sirius red staining and IHC for α-SMA revealed that the hepatic profibrotic area was significantly reduced in the Kuhuang group (Figures 2A, D). The mRNA levels of profibrogenic genes (Acta2 and Col1a1) were downregulated upon Kuhuang treatment (Figure 2E). These results indicate that Kuhuang treatment ameliorates hepatic inflammation and liver fibrosis in DDC mice.
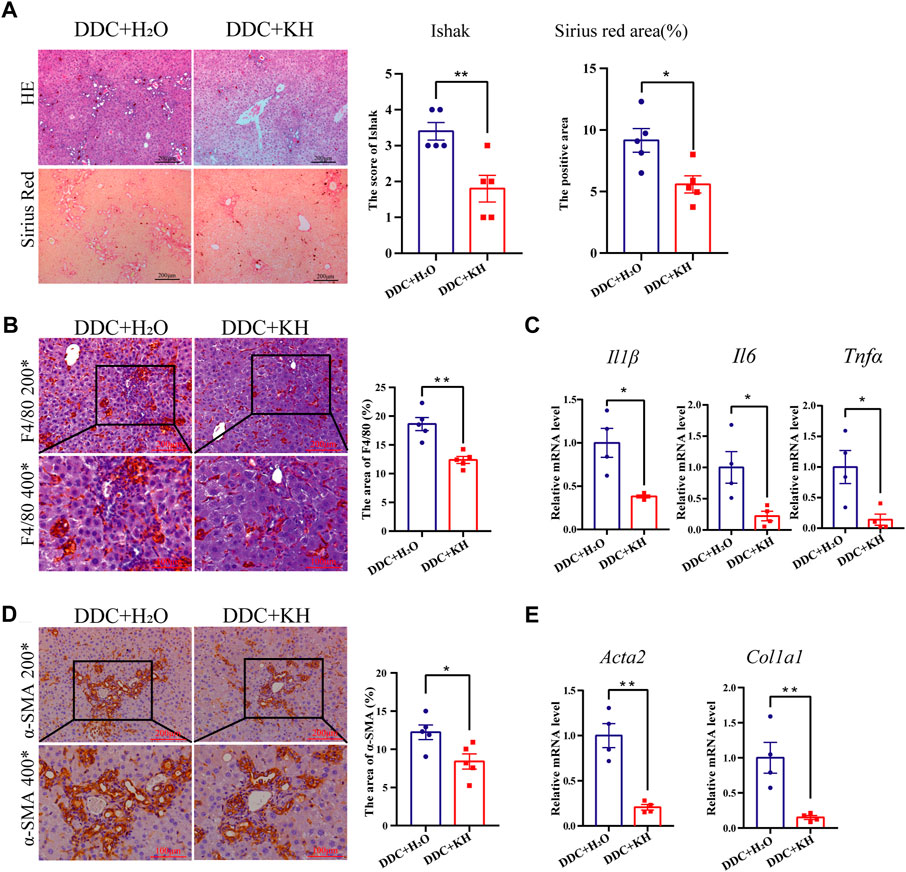
FIGURE 2. Kuhuang treatment ameliorates hepatic inflammation and fibrosis in DDC mice. (A) Representative images of H&E and Sirius red staining of liver sections (left panel) of DDC mice. Quantification of Ishak score and Sirius red-positive areas are shown in the right panel. (B) Representative images (left panel) and quantification (right panel) of F4/80-positive areas. (C) QRT-PCR analysis of inflammation-related genes (Il6, Il1β, and Tnfα) in liver tissue. (D) Representative images (left panel) and quantification (right panel) of α-SMA positive fibrosis areas. (E) QRT-PCR analysis of fibrosis-related genes (Col1a1 and Acta2) in liver tissue. The data are presented as mean ± SEM. N = 4–5 per group. The data were analyzed with Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: KH, Kuhuang; DDC, 3,5-diethoxycarbonyl-1,4-dihydro-collidine.
We performed RNA-seq analysis of the liver tissues from Kuhuang and control groups in DDC mice to explore the potential mechanisms involved in alleviating hepatic inflammation and liver fibrosis by Kuhuang treatment. A total of 982 genes were differentially expressed, of which 612 genes were upregulated and 370 were downregulated in Kuhuang-treated mice compared to the control (Figure 3A). Principal component analysis (PCA) identified a distinct gene expression pattern between the two groups, with PC1 explaining 32.68% of the variation and PC2 explaining 19.35% of the variation (Figure 3B). Gene Ontology (GO) enrichment analysis revealed that DEGs were enriched in pathways associated with interferon regulation, such as response to interferon-α, response to interferon-β, and cellular response to interferon-α (Figure 3C). As shown in Figure 3D, genes involved in interferon-related pathways, such as Igtp, Iif44, Iif206, Ifit1, Ifit3, and Isg15, were significantly upregulated after Kuhuang treatment (Figure 3D). These data indicate that Kuhuang treatment altered the hepatic gene expression pattern, especially related to the interferon pathway.
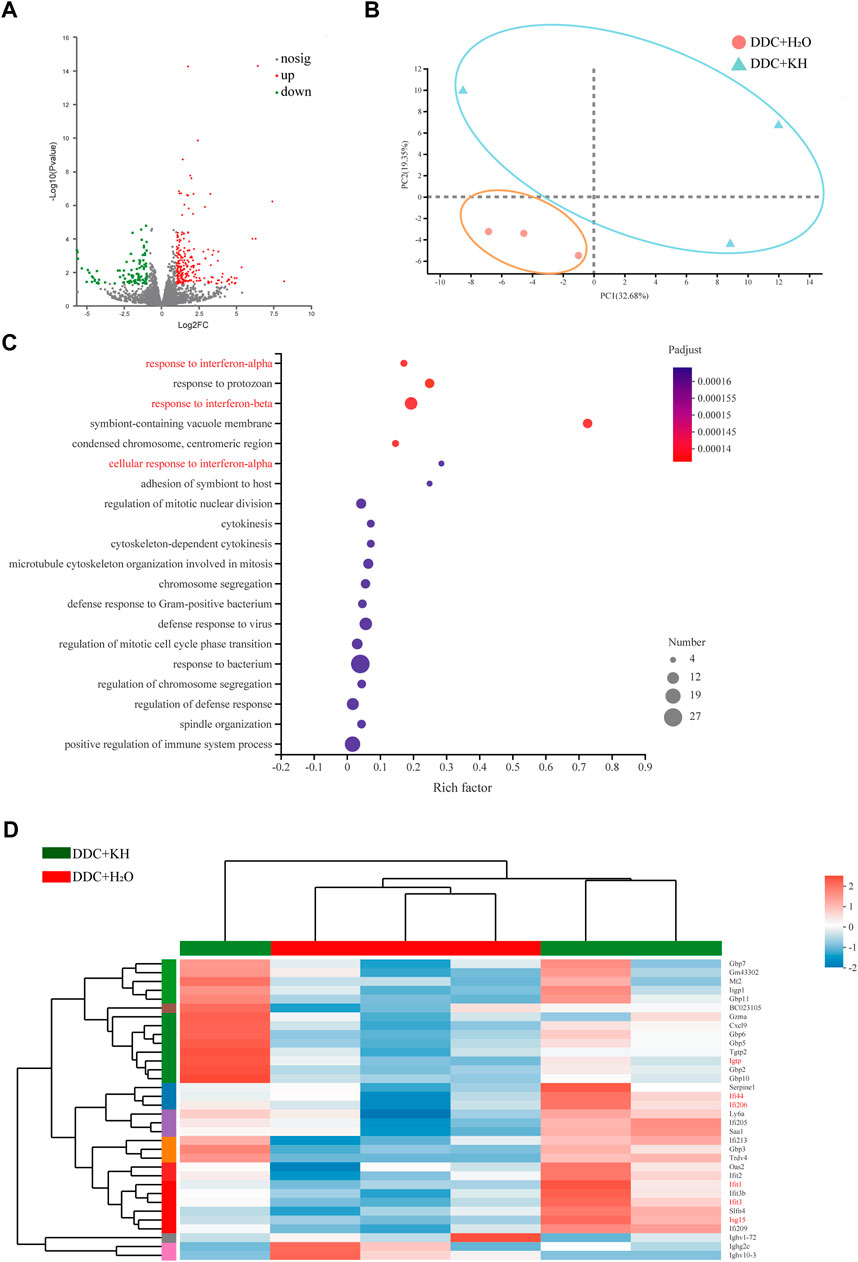
FIGURE 3. Kuhuang treatment upregulates interferon-related pathways in DDC mice. (A) Volcano plot of differentially expressed genes (612 upregulated and 370 downregulated) between DDC + KH and DDC + H2O group. (B) PCA of gene expression profiles of DDC + KH and DDC + H2O group. (C) GO enrichment of genes differentially expressed between DDC + KH and DDC + H2O group. (D) Heatmap of genes differentially expressed between DDC + KH and DDC + H2O group. Abbreviations: KH, Kuhuang; DDC, 3,5-diethoxycarbonyl-1,4-dihydro-collidine.
Next, we explored the influence of Kuhuang treatment on the composition of gut microbiota through 16S rRNA sequencing of fecal samples from DDC mice treated with Kuhuang or H2O, as well as from mice fed a chow diet. Principal coordinate analysis (PCOA) showed a distinct deviation along PC1 (explaining 34.78% of variation) and PC2 (explaining 23.66% of variation) among groups, suggesting that either the DDC diet or Kuhuang treatment would lead to alterations in gut microbiota (Figure 4A). Notably, Kuhuang treatment increased the diversity of gut microbiota (as indicated by sobs), which was significantly decreased in DDC mice compared with mice fed a chow diet, implying that Kuhuang might be able to rescue gut microbiota dysbiosis induced by the DDC diet (Figure 4B). Further analysis showed that, compared with DDC mice treated with H2O, the abundance of Verrucomicrobiota was remarkably higher at the phylum level in mice treated with Kuhuang (Figure 4C). At the genus level, the abundance of Akkermansia, Staphylococcus, and Corynebacterium was significantly higher, whereas that of Clostridium_sensu_stricto_1, Turicibacter, and Bifidobacterium was significantly lower in mice treated with Kuhuang (Figure 4D). LEfSe analysis revealed differential gut microbiota between mice treated with Kuhuang and with H2O. As shown in Figure 4E, Akkermansia, Staphylococcus, Corynebacterium, and Lachnospiraceae FCS020 group were more abundant, while Clostridium-sensu-stricto-1, Turicibacter, Bifidobacterium, and Lactobacillus were less abundant upon Kuhuang treatment. These results indicate that Kuhuang treatment altered the composition of the gut microbiota and rescued dysbiosis induced by the DDC diet.
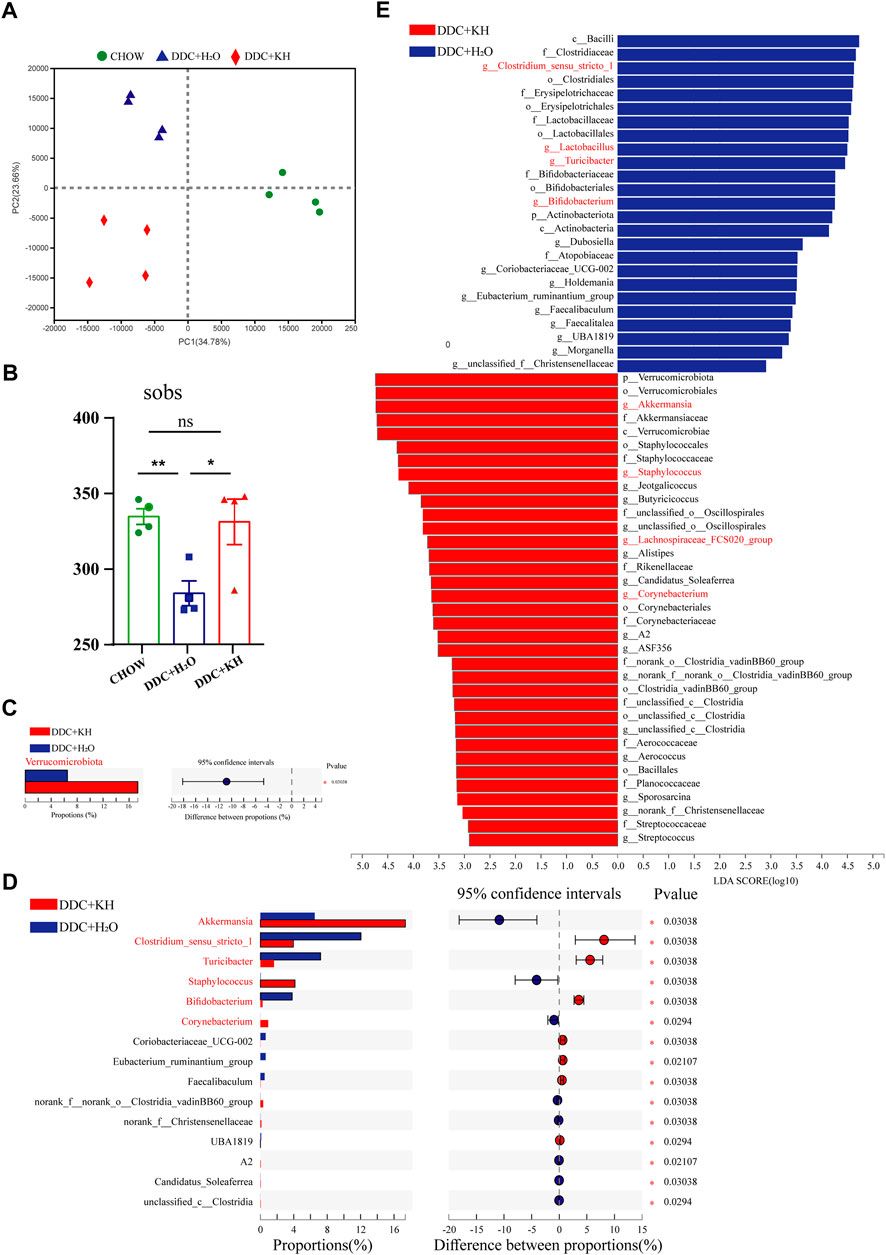
FIGURE 4. Kuhuang treatment altered gut microbiota composition in DDC mice. (A) Bray–Curtis distance-based PCOA of gut microbiota diversity of Chow, DDC + KH, and DDC + H2O groups. (B) Sobs in Chow, DDC + KH, and DDC + H2O groups. The data are presented as mean ± SEM. N = 4–5 per group. The data were analyzed with Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. (C) Gut microbiota with statistically differential abundance at phylum level between DDC + KH and DDC + H2O groups. (D) Gut microbiota with statistically differential abundance at genus level between DDC + KH and DDC + H2O groups. (E) LEfSe analysis of differentially abundant microbes between DDC + KH and DDC + H2O groups. Abbreviations: KH, Kuhuang; DDC, 3,5-diethoxycarbonyl-1,4-dihydro-collidine.
Furthermore, the composition of hepatic bile acids was examined using a targeted metabolomic assay in mice fed a DDC diet. PCA analysis showed distinct clustering along PC1 (explaining 33.3% of the variation) and PC2 (explaining 22.4% of the variation) between mice treated with Kuhuang and H2O (Figure 5A). Remarkably, Kuhuang treatment significantly decreased the levels of both primary bile acids (CA, CDCA, and GCDCA) and secondary bile acids (DCA, GCA, CDCA-G, and THCA), suggesting that Kuhuang treatment may reduce BAs in the liver (Figures 5B, C).
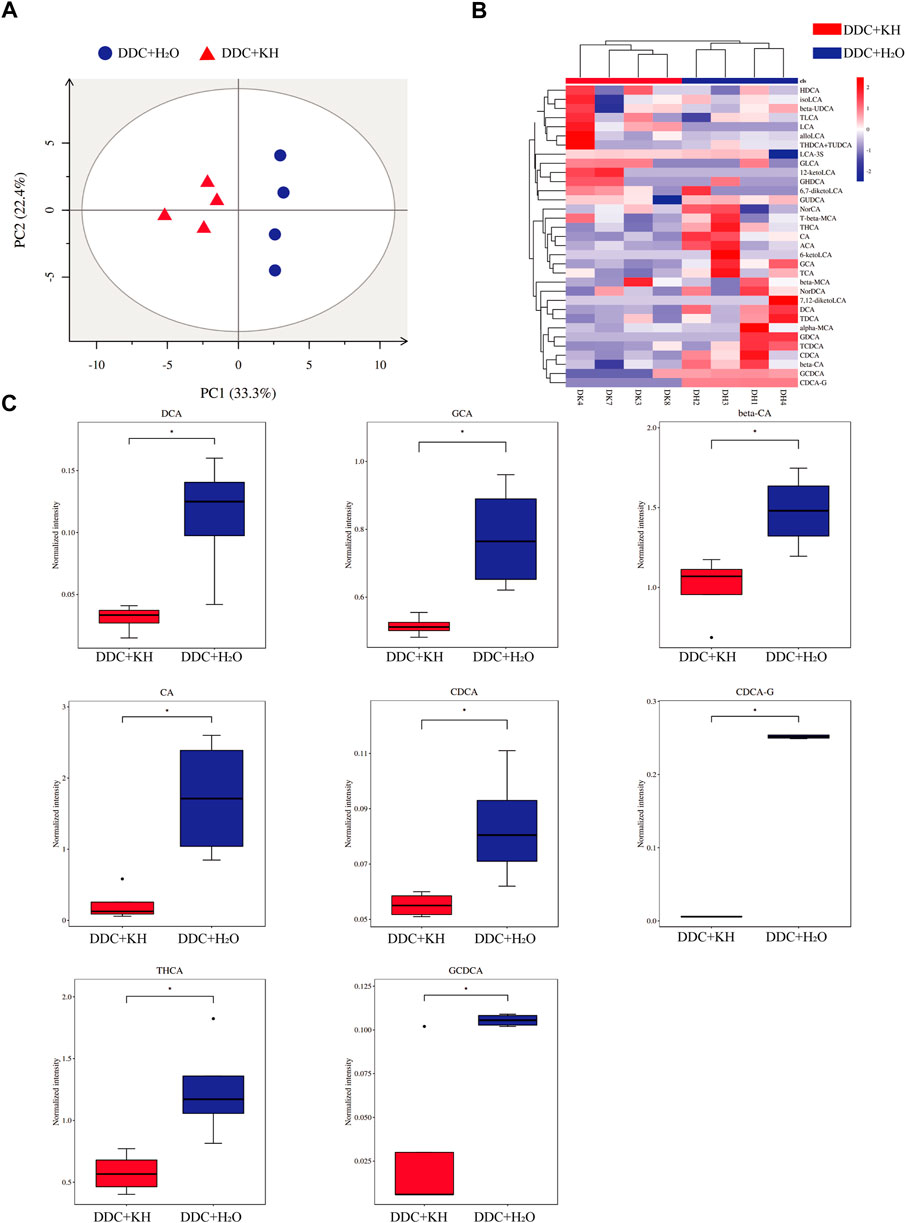
FIGURE 5. Kuhuang treatment altered hepatic BA levels in DDC mice. (A) PCA of hepatic BA composition of DDC + KH and DDC + H2O groups. (B) Heatmap of hepatic BAs with statistically different abundance between DDC + KH and DDC + H2O groups. (C) Histogram of normalized intensity of DCA, GCA, beta-CA, CA, CDCA, CDCA-G, THCA, and GCDCA in DDC + KH and DDC + H2O groups. The data are presented as mean ± SEM. N = 4–5 per group. The data were analyzed with Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: KH, Kuhuang; DDC, 3,5-diethoxycarbonyl-1,4-dihydro-collidine.
Based on our findings from RNA-seq analysis, we further validated the hepatic mRNA expression levels of interferon-related genes (Ifna4, Ifnb, Isg15, and Il1ra) by quantitative RT-PCR. Kuhuang treatment significantly increased the hepatic mRNA expression levels of Ifna4, Ifnb, and Isg15 and decreased that of Il1ra (Figure 6A). In addition, significant upregulation of IFNβ levels in the liver tissue of mice treated with Kuhuang was confirmed by ELISA (Figure 6B). Correlation analysis showed that the abundance of gut Akkermansia, Corynebacterium, and Staphylococcus was positively correlated with the hepatic mRNA expression levels of Ifna4, Ifnb, and Isg15, and that of Corynebacterium was negatively correlated with Il1ra expression level. The abundance of Lactobacillus, Clostridium-sensu-stricto-1, Bifidobacterium, and Turicibacter was negatively correlated with Ifna4, Ifnb, and Isg15 expression levels, and that of Lactobacillus was positively correlated with Il1ra expression level (Figure 6C). These results implied that the gut microbiota might participate in the regulation of interferon-related genes, which is consistent with previous investigations demonstrating that microbiota-derived stimulators could prompt type I interferon production by targeting interferon-related genes (Lam et al., 2021).
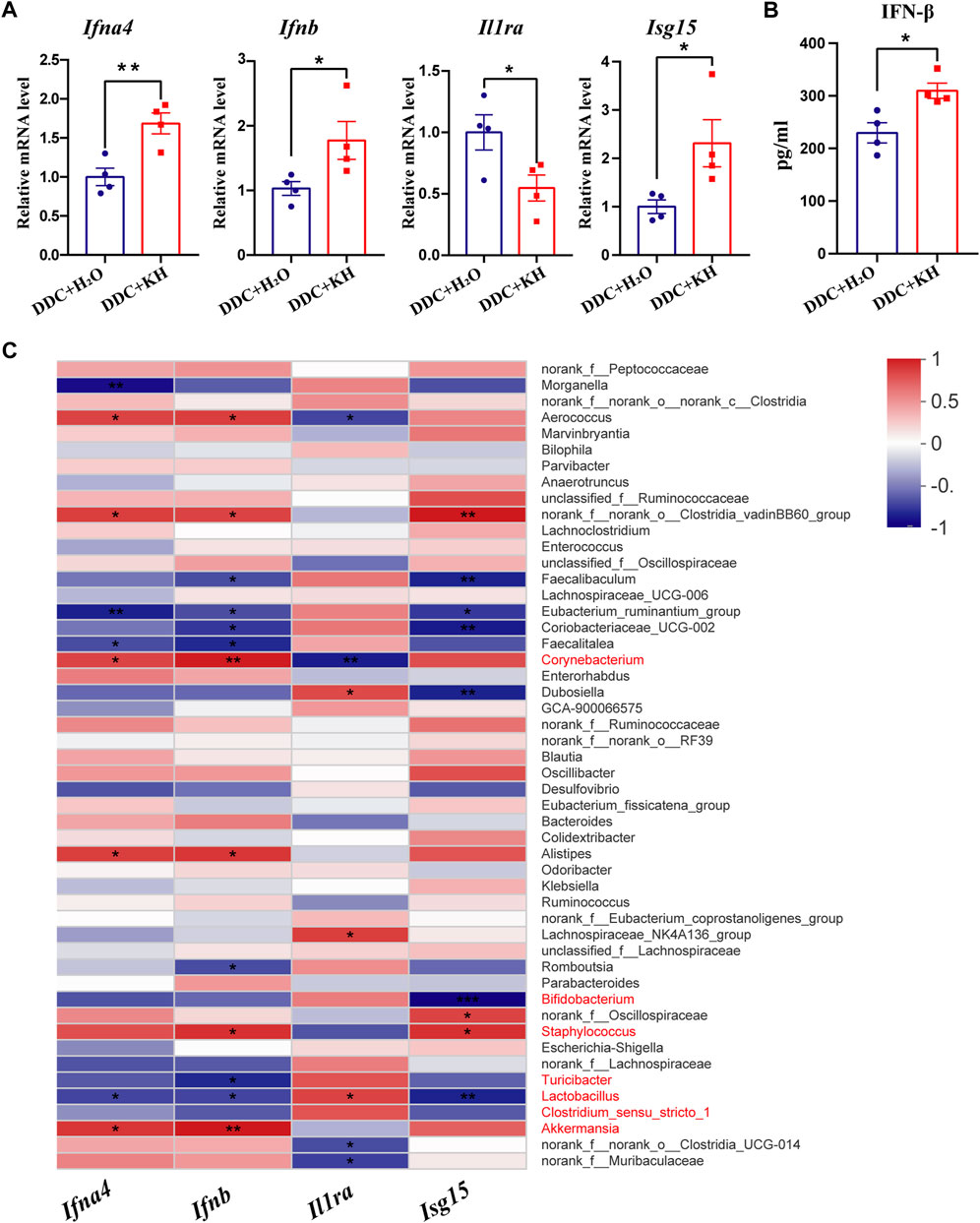
FIGURE 6. Correlation of gut microbiota abundance and hepatic IFN-related genes in DDC mice. (A) QRT-PCR analysis of IFN-related genes (Ifna4, Ifnb, Il1ra, and Isg15) in liver tissue. (B) ELISA analysis of hepatic IFN-β level. The data are presented as mean ± SEM. N = 4–5 per group. The data were analyzed with Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. (C) Correlation analysis between the expression levels of IFN-related genes and gut microbiota abundance. Abbreviations: KH, Kuhuang; DDC, 3,5-diethoxycarbonyl-1,4-dihydro-collidine.
By analyzing the results of 16S rRNA sequencing, we found that the abundance of bile salt hydrolase (BSH)-producing gut microbiota, such as Lactobacillus, Clostridium, and Bifidobacterium, were less abundant in DDC mice treated with Kuhuang than in those treated with H2O (Figure 7A). We also found that compared with DDC mice treated with H2O, the mRNA expression of Fxr and Fgf15 was significantly upregulated in the ileum of DDC mice treated with Kuhuang (Figure 7B). Furthermore, hepatic mRNA expression of BA synthesis-related genes (Cyp7a1, Cyp8b1, and Cyp7b1) was downregulated, whereas that of Fxr was upregulated significantly in DDC mice treated with Kuhuang (Figure 7C). Correlation analysis showed that the abundance of gut Bifidobacterium, Clostridium-sensu-stricto-1, Turicibacter, and Lactobacillus was positively correlated with BA synthesis-related genes (Cyp7a1, Cyp8b1, and Cyp7b1), and Turicibacter and Lactobacillus were negatively correlated with Fxr. In addition, the abundances of gut Corynebacterium, Staphylococcus, and Akkermansia were negatively correlated with BA synthesis-related genes (Cyp7a1, Cyp8b1, and Cyp7b1) and positively correlated with Fxr (Figure 7D). These data suggest that Kuhuang-induced gut microbiota alteration may also regulate hepatic genes related to BA synthesis.
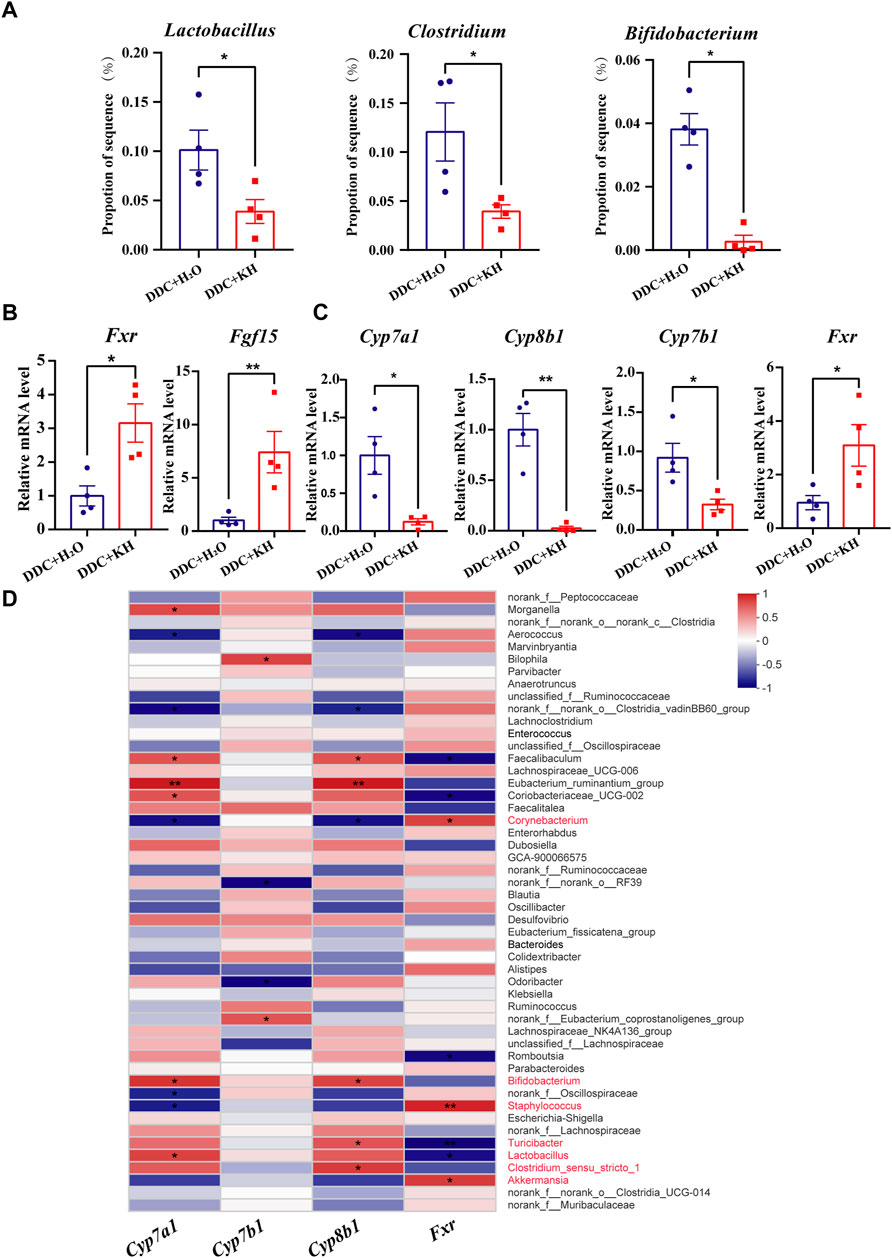
FIGURE 7. Correlation of gut microbiota abundance and hepatic BA synthesis-related genes in DDC mice. (A) Histogram of Lactobacillus, Clostridium, and Bifidobacterium proportions in gut microbiota in DDC + KH and DDC + H2O groups. (B) QRT-PCR analysis of Fxr and Fgf15 in ileum tissue. (C) QRT-PCR analysis of BA synthesis-related genes (Cyp7a1, Cyp7b1, Cyp8b1and Fxr) in liver tissue. The data are presented as mean ± SEM. N = 4–5 per group. The data were analyzed with Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. (D) Correlation analysis between the expression levels of BA synthesis-related genes and gut microbiota abundance. Abbreviations: KH, Kuhuang; DDC, 3,5-diethoxycarbonyl-1,4-dihydro-collidine.
Kuhuang has been reported to relieve jaundice symptoms and reduce elevated liver enzyme levels in clinical practice (Zhao et al., 2017). In this study, we found that Kuhuang treatment significantly alleviates the progression of liver fibrosis in both the DDC and BDL mouse models. In addition, Kuhuang treatment mitigated liver inflammation. These results suggest that Kuhuang is a potential drug for treating liver fibrosis. The abundance of gut Firmicutes and Bacteroidetes was decreased, and that of Proteobacteria and Fusobacteria is increased in patients with liver cirrhosis, suggesting that the gut microbiota played an important role in the development of liver fibrosis (Preveden et al., 2017). Our study showed that Kuhuang treatment significantly altered gut microbiota composition with an increased abundance of Akkermansia, Staphylococcus, Corynebacterium, and Lachnospiraceae FCS020 group and decreased abundance of Turicibacter, Clostridium-sensu-stricto-1, Bifidobacterium, and Lactobacillus in a mouse model of liver fibrosis. Clostridium_sensu_stricto_1 was known to be enriched with increased severity of liver fibrosis (Xiang et al., 2022). Furthermore, decreased abundance of gut Clostridium_sensu_stricto-1 ameliorated liver inflammation in mice with non-alcoholic steatohepatitis (Rom et al., 2020) or alcoholic liver disease (Guo et al., 2022a). Thus, we hypothesize that Kuhuang treatment attenuates liver fibrosis through Clostridium-mediated alleviation of inflammation. The Kuhuang treatment also increased the abundance of Lachnospiraceae FCS020 group. Lachnospiraceae participated in the synthesis of short-chain fatty acids (SCFAs) (Louis and Flint, 2017), which was critical in ensuring the integrity of the intestinal mucosal barrier (Parada Venegas et al., 2019). Our previous studies showed that disruption of the intestinal barrier was associated with the severity of liver fibrosis (Shen et al., 2021). Therefore, Kuhuang treatment may improve liver fibrosis through gut Lachnospiraceae-induced SCFAs to enhance the integrity of the intestinal mucosal barrier. In addition, Akkermansia played a similar role in protecting the intestinal barrier, reducing plasma endotoxin levels, and inhibiting intestinal inflammation and metabolic disorders (Depommier et al., 2019). Thus, Kuhuang may alleviate liver inflammation and fibrosis by increasing the abundance of Akkermansia, which inhibits intestinal inflammation and decreases endotoxin levels. In short, altered gut composition upon Kuhuang treatment may improve liver fibrosis by preventing intestinal endotoxins from entering the liver.
In addition to the gut microbiota, Kuhuang treatment altered the gene expression pattern in the liver. The interferon signaling-related genes (Ifna4, Ifnb, and Isg15) were significantly upregulated. Type I interferon protected against liver injury and inhibits inflammatory responses (Petrasek et al., 2011). In addition, α-interferon could effectively alleviate the progression of liver fibrosis in patients with chronic hepatitis C (Serejo et al., 2001). Furthermore, correlation analysis indicated that the abundance of Akkermansia, Corynebacterium, and Staphylococcus increased upon Kuhuang treatment and was positively correlated with the hepatic expression of Ifna4, Ifnb, and Isg15. Interestingly, the abundance of Lactobacillus, Clostridium-sensu-stricto-1, Bifidobacterium, and Turicibacter decreased upon Kuhuang treatment and was negatively correlated with Ifna4, Ifnb, and Isg15 expression. The results suggest that Kuhuang treatment may inhibit liver inflammation and fibrosis by regulating the liver’s interferon signaling (activating type I interferon), mediated by modulating microbiota composition. Lam et al. (2021) showed that Akkermansia produced c-di-AMP and induced the production of type I interferon. Type I interferon might alleviate liver fibrosis by inhibiting HSC activation and reducing immunological reactions (Tanabe et al., 2007; Shimozono et al., 2015). Our study found that Kuhuang treatment reduced the hepatic expression levels of profibrotic genes (Acta2 and Col1a1) and inflammatory cytokines (Il6, Il1β, and Tnfα) while upregulating interferon signaling-related genes (Ifna4, Ifnb, and Isg15). Thus, we hypothesize that Kuhuang may alleviate liver inflammation and fibrosis by increasing the abundance of Akkermansia, which leads to activating the type I interferon pathway in the liver.
Furthermore, our results showed that Kuhuang treatment significantly decreased the conjugated and non-conjugated BAs in the profibrotic liver. It significantly decreased the hepatic mRNA expression levels of Cyp7a1 and Cyp8b1, cytochrome P450 enzymes that are crucial for BA synthesis in the classical pathway. Notably, the classical pathway accounts for approximately 75% of BA synthesis (Thomas et al., 2008). In addition, Kuhuang treatment significantly decreased the mRNA expression level of Cyp7b1 in the liver, the key enzyme that regulates BA synthesis via an alternative pathway (Russell, 2003). Kuhuang treatment also significantly increased hepatic mRNA expression level of Fxr. In the liver, Fxr substantially inhibits BA synthesis (Lu et al., 2000). Thus, Kuhuang treatment may suppress BA retention by inhibiting Fxr-mediated BA synthesis in the liver. Cholestasis in the liver is an important factor contributing to the development of liver fibrosis (Ghonem et al., 2015). Cholestasis and destruction of the bile duct damage the bile duct cells and hepatocytes, further promoting the progression of liver fibrosis. Therefore, Kuhuang treatment may attenuate liver inflammation and fibrosis by inhibiting BA synthesis. In addition, the abundance of gut Turicibacter and Lactobacillus, which decreased upon Kuhuang treatment, was negatively correlated with the hepatic expression of Fxr. Meanwhile, the abundance of Corynebacterium, Staphylococcus, and Akkermansia, which increased upon Kuhuang treatment, was positively correlated with Fxr expression in the liver, suggesting that gut microbiota alteration may eventually result in the inhibition of hepatic Fxr-mediated BA synthesis. In addition, we found that Kuhuang treatment significantly reduced the abundance of BSH-producing Lactobacillus, Clostridium, and Bifidobacterium, which might result in increased levels of intestinal conjugated BAs, such as LCA and TCA, activators of Fxr (Wahlström et al., 2016; Xiang et al., 2021; Münzker et al., 2022). We observed that the expression levels of Fxr and Fgf15 in the ileum significantly increased after Kuhuang treatment. Fxr-FGF15 activation in the ileum can further inhibit the hepatic expression of BA synthesis-related genes and BA synthesis in the liver (Stofan and Guo, 2020; Katafuchi and Makishima, 2022). In addition, correlation analysis showed that the abundance of BSH-producing Lactobacillus, Clostridium, and Bifidobacterium was positively correlated with the expression levels of genes related to BA synthesis (Cyp7a1, Cyp7b1 or Cyp8b1). Thus, we hypothesize that Kuhuang treatment may inhibit hepatic BA synthesis by activating the Fxr-FGF15 in the ileum mediated by altering the abundance of BSH-producing gut microbiota.
In summary, our study indicates that Kuhuang attenuates inflammation and BA accumulation and eventually improves fibrosis in a cholestatic mouse model. Kuhuang treatment alters the expression of interferon signaling- and BA synthesis-related genes and decreases hepatic BA levels. Furthermore, alterations in the gut microbiota induced by Kuhuang, especially the increased abundance of interferon-inducing Akkermansia and decreased abundance of BSH-producing Lactobacillus, Clostridium, and Bifidobacterium, play potential roles in alleviating liver inflammation and BA accumulation by inhibiting intestinal endotoxin, activating hepatic interferon signaling, and suppressing hepatic BA synthesis (Figure 8). Nevertheless, more experimental and clinical evidence is needed before Kuhuang can be applied to treat liver fibrosis.
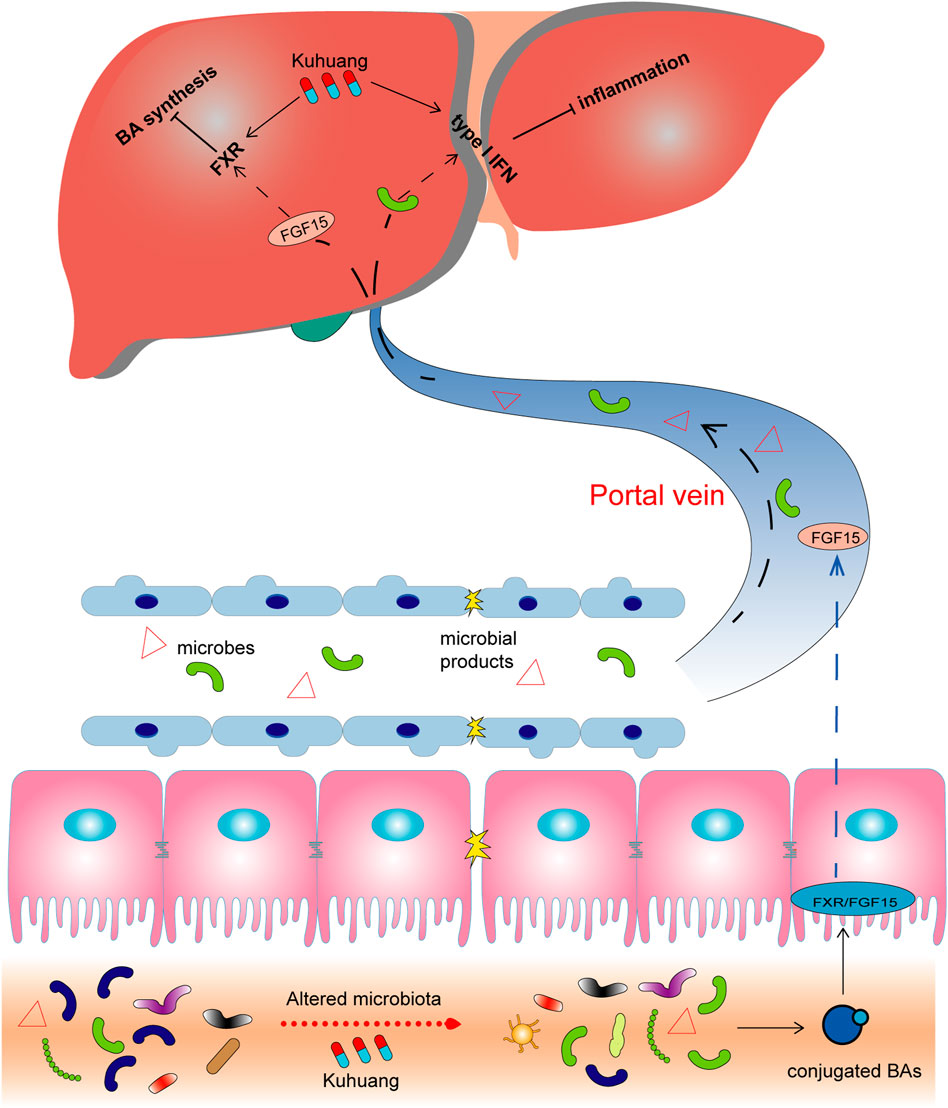
FIGURE 8. Schematic illustration of potential mechanisms of Kuhuang. Kuhuang may inhibit BA synthesis and inflammation responses in the liver by upregulating interferon signaling-related genes while downregulating genes associated with BA synthesis. Kuhuang also alters the composition of gut microbiota, consequently benefiting the maintenance of the intestinal barrier and increasing the level of conjugated BAs. Conjugated BAs activate intestinal Fxr/FGF15 signaling, followed by hepatic Fxr activation, which further inhibits BA synthesis in the liver. In addition, gut microbes and microbial products that translocate into the liver can also inhibit hepatic inflammation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject; PRJNA893596.
The animal study was reviewed and approved by the Shanghai General hospital, Shanghai Jiaotong University School of Medicine.
LL and HD designed the study. BS, CZ, TG, ZS, WD, and YG conducted the experiments and analyzed the data. BS, CZ, and TG drafted the manuscript. LL and HD revised the manuscript. All authors contributed to the manuscript and approved the final version.
This study was supported by the National Natural Science Foundation of China (81970528), the Shanghai Municipal Science and Technology Commission Research Project (22ZR1449700), and the Shanghai General Hospital Start-up Fund (02.06.01.20.01).
Authors YL and JZ were employed by Suzhou Leiyunshang Pharmacology Group.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Depommier, C., Everard, A., Druart, C., Plovier, H., Van Hul, M., Vieira-Silva, S., et al. (2019). Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 25 (7), 1096–1103. doi:10.1038/s41591-019-0495-2
Ghonem, N. S., Assis, D. N., and Boyer, J. L. (2015). Fibrates and cholestasis. Hepatology 62 (2), 635–643. doi:10.1002/hep.27744
Ginès, P., Krag, A., Abraldes, J. G., Solà, E., Fabrellas, N., and Kamath, P. S. (2021). Liver cirrhosis. Lancet 398 (10308), 1359–1376. doi:10.1016/s0140-6736(21)01374-x
Guo, W. L., Cao, Y. J., You, S. Z., Wu, Q., Zhang, F., Han, J. Z., et al. (2022a). Ganoderic acids-rich ethanol extract from Ganoderma lucidum protects against alcoholic liver injury and modulates intestinal microbiota in mice with excessive alcohol intake. Curr. Res. Food Sci. 5, 515–530. doi:10.1016/j.crfs.2022.02.013
Guo, X., Li, Y., Wang, W., Wang, L., Hu, S., Xiao, X., et al. (2022b). The construction of preclinical evidence for the treatment of liver fibrosis with quercetin: A systematic review and meta-analysis. Phytother. Res. 36 (10), 3774–3791. doi:10.1002/ptr.7569
Hu, T., An, Z., Shi, C., Li, P., and Liu, L. (2020). A sensitive and efficient method for simultaneous profiling of bile acids and fatty acids by UPLC-MS/MS. J. Pharm. Biomed. Anal. 178, 112815. doi:10.1016/j.jpba.2019.112815
Jayashree, S., Pooja, S., Pushpanathan, M., Rajendhran, J., and Gunasekaran, P. (2014). Identification and characterization of bile salt hydrolase genes from the genome of Lactobacillus fermentum MTCC 8711. Appl. Biochem. Biotechnol. 174 (2), 855–866. doi:10.1007/s12010-014-1118-5
Jiang, B., Yuan, G., Wu, J., Wu, Q., Li, L., and Jiang, P. (2022). Prevotella copri ameliorates cholestasis and liver fibrosis in primary sclerosing cholangitis by enhancing the FXR signalling pathway. Biochim. Biophys. Acta. Mol. Basis Dis. 1868 (3), 166320. doi:10.1016/j.bbadis.2021.166320
Katafuchi, T., and Makishima, M. (2022). Molecular basis of bile acid-FXR-FGF15/19 signaling Axis. Int. J. Mol. Sci. 23 (11), 6046. doi:10.3390/ijms23116046
Kim, G. B., Brochet, M., and Lee, B. H. (2005). Cloning and characterization of a bile salt hydrolase (bsh) from Bifidobacterium adolescentis. Biotechnol. Lett. 27 (12), 817–822. doi:10.1007/s10529-005-6717-3
Lam, K. C., Araya, R. E., Huang, A., Chen, Q., Di Modica, M., Rodrigues, R. R., et al. (2021). Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 184 (21), 5338–5356.e21. doi:10.1016/j.cell.2021.09.019
Lee, Y. A., Wallace, M. C., and Friedman, S. L. (2015). Pathobiology of liver fibrosis: A translational success story. Gut 64 (5), 830–841. doi:10.1136/gutjnl-2014-306842
Li, Y., Tang, R., Leung, P. S. C., Gershwin, M. E., and Ma, X. (2017). Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun. Rev. 16 (9), 885–896. doi:10.1016/j.autrev.2017.07.002
Liu, Z., Zhang, Z., Huang, M., Sun, X., Liu, B., Guo, Q., et al. (2018). Taurocholic acid is an active promoting factor, not just a biomarker of progression of liver cirrhosis: Evidence from a human metabolomic study and in vitro experiments. BMC Gastroenterol. 18 (1), 112. doi:10.1186/s12876-018-0842-7
Long, S. L., Gahan, C. G. M., and Joyce, S. A. (2017). Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 56, 54–65. doi:10.1016/j.mam.2017.06.002
Louis, P., and Flint, H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19 (1), 29–41. doi:10.1111/1462-2920.13589
Lu, T. T., Makishima, M., Repa, J. J., Schoonjans, K., Kerr, T. A., Auwerx, J., et al. (2000). Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6 (3), 507–515. doi:10.1016/s1097-2765(00)00050-2
Mori, H., Svegliati Baroni, G., Marzioni, M., Di Nicola, F., Santori, P., Maroni, L., et al. (2022). Farnesoid X receptor, bile acid metabolism, and gut microbiota. Metabolites 12 (7), 647. doi:10.3390/metabo12070647
Münzker, J., Haase, N., Till, A., Sucher, R., Haange, S. B., Nemetschke, L., et al. (2022). Functional changes of the gastric bypass microbiota reactivate thermogenic adipose tissue and systemic glucose control via intestinal FXR-TGR5 crosstalk in diet-induced obesity. Microbiome 10 (1), 96. doi:10.1186/s40168-022-01264-5
Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10, 277. doi:10.3389/fimmu.2019.00277
Petrasek, J., Dolganiuc, A., Csak, T., Kurt-Jones, E. A., and Szabo, G. (2011). Type I interferons protect from Toll-like receptor 9-associated liver injury and regulate IL-1 receptor antagonist in mice. Gastroenterology 140 (2), 697–708. doi:10.1053/j.gastro.2010.08.020
Preveden, T., Scarpellini, E., Milić, N., Luzza, F., and Abenavoli, L. (2017). Gut microbiota changes and chronic hepatitis C virus infection. Expert Rev. Gastroenterol. Hepatol. 11 (9), 813–819. doi:10.1080/17474124.2017.1343663
Rom, O., Liu, Y., Liu, Z., Zhao, Y., Wu, J., Ghrayeb, A., et al. (2020). Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci. Transl. Med. 12 (572), eaaz2841. doi:10.1126/scitranslmed.aaz2841
Rossocha, M., Schultz-Heienbrok, R., von Moeller, H., Coleman, J. P., and Saenger, W. (2005). Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 44 (15), 5739–5748. doi:10.1021/bi0473206
Russell, D. W. (2003). The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72, 137–174. doi:10.1146/annurev.biochem.72.121801.161712
Serejo, F., Costa, A., Oliveira, A. G., Ramalho, F., Batista, A., De Moura, M. C., et al. (2001). Alpha-interferon improves liver fibrosis in chronic hepatitis C: Clinical significance of the serum N-terminal propeptide of procollagen type III. Dig. Dis. Sci. 46 (8), 1684–1689. doi:10.1023/a:1010649403659
Shen, B., Wang, J., Guo, Y., Gu, T., Shen, Z., Zhou, C., et al. (2021). Dextran sulfate sodium salt-induced colitis aggravates gut microbiota dysbiosis and liver injury in mice with non-alcoholic steatohepatitis. Front. Microbiol. 12, 756299. doi:10.3389/fmicb.2021.756299
Shimozono, R., Nishimura, K., Akiyama, H., Funamoto, S., Izawa, A., Sai, T., et al. (2015). Interferon-β mediates signaling pathways uniquely regulated in hepatic stellate cells and attenuates the progression of hepatic fibrosis in a dietary mouse model. J. Interferon Cytokine Res. 35 (6), 464–473. doi:10.1089/jir.2014.0096
Stofan, M., and Guo, G. L. (2020). Bile acids and FXR: Novel targets for liver diseases. Front. Med. 7, 544. doi:10.3389/fmed.2020.00544
Svegliati-Baroni, G., Ridolfi, F., Hannivoort, R., Saccomanno, S., Homan, M., De Minicis, S., et al. (2005). Bile acids induce hepatic stellate cell proliferation via activation of the epidermal growth factor receptor. Gastroenterology 128 (4), 1042–1055. doi:10.1053/j.gastro.2005.01.007
Tanabe, J., Izawa, A., Takemi, N., Miyauchi, Y., Torii, Y., Tsuchiyama, H., et al. (2007). Interferon-beta reduces the mouse liver fibrosis induced by repeated administration of concanavalin A via the direct and indirect effects. Immunology 122 (4), 562–570. doi:10.1111/j.1365-2567.2007.02672.x
Thomas, C., Pellicciari, R., Pruzanski, M., Auwerx, J., and Schoonjans, K. (2008). Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7 (8), 678–693. doi:10.1038/nrd2619
Trauner, M., Fuchs, C. D., Halilbasic, E., and Paumgartner, G. (2017). New therapeutic concepts in bile acid transport and signaling for management of cholestasis. Hepatology 65 (4), 1393–1404. doi:10.1002/hep.28991
Wahlström, A., Sayin, S. I., Marschall, H. U., and Bäckhed, F. (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell. Metab. 24 (1), 41–50. doi:10.1016/j.cmet.2016.05.005
Wijaya, A., Hermann, A., Abriouel, H., Specht, I., Yousif, N. M., Holzapfel, W. H., et al. (2004). Cloning of the bile salt hydrolase (bsh) gene from Enterococcus faecium FAIR-E 345 and chromosomal location of bsh genes in food enterococci. J. Food Prot. 67 (12), 2772–2778. doi:10.4315/0362-028x-67.12.2772
Xiang, H., Liu, Z., Xiang, H., Xiang, D., Xiao, S., Xiao, J., et al. (2022). Dynamics of the gut-liver axis in rats with varying fibrosis severity. Int. J. Biol. Sci. 18 (8), 3390–3404. doi:10.7150/ijbs.69833
Xiang, J., Zhang, Z., Xie, H., Zhang, C., Bai, Y., Cao, H., et al. (2021). Effect of different bile acids on the intestine through enterohepatic circulation based on FXR. Gut Microbes 13 (1), 1949095. doi:10.1080/19490976.2021.1949095
Xu, J., Xie, S., Chi, S., Zhang, S., Cao, J., and Tan, B. (2022). Protective effects of taurocholic acid on excessive hepatic lipid accumulation via regulation of bile acid metabolism in grouper. Food Funct. 13 (5), 3050–3062. doi:10.1039/d1fo04085e
Xu, X., Ling, Q., Gao, F., He, Z. L., Xie, H. Y., and Zheng, S. S. (2009). Hepatoprotective effects of marine and kuhuang in liver transplant recipients. Am. J. Chin. Med. 37 (1), 27–34. doi:10.1142/s0192415x09006643
Yang, T., Shu, T., Liu, G., Mei, H., Zhu, X., Huang, X., et al. (2017). Quantitative profiling of 19 bile acids in rat plasma, liver, bile and different intestinal section contents to investigate bile acid homeostasis and the application of temporal variation of endogenous bile acids. J. Steroid Biochem. Mol. Biol. 172, 69–78. doi:10.1016/j.jsbmb.2017.05.015
Yang, Y., Sun, M., Li, W., Liu, C., Jiang, Z., Gu, P., et al. (2021). Rebalancing TGF-β/Smad7 signaling via Compound kushen injection in hepatic stellate cells protects against liver fibrosis and hepatocarcinogenesis. Clin. Transl. Med. 11 (7), e410. doi:10.1002/ctm2.410
Yang, Y., Sun, M., Yao, W., Wang, F., Li, X., Wang, W., et al. (2020). Compound kushen injection relieves tumor-associated macrophage-mediated immunosuppression through TNFR1 and sensitizes hepatocellular carcinoma to sorafenib. J. Immunother. Cancer 8 (1), e000317. doi:10.1136/jitc-2019-000317
Zhang, M., Tan, C., Tang, Q., Fan, H., Liu, X. X., and Wang, W. Z. (2019b). Effect of compound Kuhuang decoction on expression of Th17/Tregs differentiation related transcription factors in colonic tissues of experimental colitis mice. Chin. Archives Traditional Chin. Med. 37 (4), 807–809. doi:10.13193/j.issn.1673-7717.2019.04.008
Zhang, Y., Lu, Y., Ji, H., and Li, Y. (2019a). Anti-inflammatory, anti-oxidative stress and novel therapeutic targets for cholestatic liver injury. Biosci. Trends 13 (1), 23–31. doi:10.5582/bst.2018.01247
Zhao, J., Liao, X., Zhao, H., Yang, J., Zou, W., Wang, L., et al. (2017). Efficacy and safety of kuhuang injection in treating viral hepatitis: Systematic review and meta-analysis of randomized controlled trials. Zhongguo Zhong Yao Za Zhi 42 (20), 4007–4026. doi:10.19540/j.cnki.cjcmm.20170901.007
Acta2 actin alpha 2 smooth muscle
BA bile acid
BDL bile duct ligation
BSH bile salt hydrolase
CKI compound kushen injection
Col1a1 collagen type 1 α
CYP7a1 cytochrome P450 family 7 subfamily A member 1
CYP7b1 cytochrome P450 family 7 subfamily B member 1
CYP8b1 cytochrome P450 family 8 subfamily B member 1
DDC 3,5-diethoxycarbonyl-1,4-dihydro-collidine
DEG differentially expressed gene
ELISA enzyme-linked immunosorbent assay
FGF 15 fibroblast growth factor 15
Fxr farnesol X receptor
GAPDH glyceraldehyde-3-phosphate dehydrogenase
GO gene ontology
H&E hematoxylin and eosin
HSC hepatic stellate cells
Ifna4 interferon alpha 4
IFN-β interferon-β
IL1 β interleukin 1β
Il1ra interleukin 1 receptor antagonist
IL6 interleukin 6
Isg15 ISG15 ubiquitin-like modifier
OTUs operational taxonomic units
PCA principal component analysis
QRT-PCR quantitative real-time polymerase chain reaction
RNA-seq RNA sequencing
SCFA short-chain fatty acid
TNF α tumor necrosis factor α
Keywords: Kuhuang, liver fibrosis, bile acids, interferon, gut microbiota
Citation: Shen B, Zhou C, Gu T, Shen Z, Guo Y, Dai W, Liu Y, Zhang J, Lu L and Dong H (2022) Kuhuang alleviates liver fibrosis by modulating gut microbiota-mediated hepatic IFN signaling and bile acid synthesis. Front. Pharmacol. 13:1080226. doi: 10.3389/fphar.2022.1080226
Received: 26 October 2022; Accepted: 02 December 2022;
Published: 13 December 2022.
Edited by:
Xuan Zeng, Sun Yat-sen University, ChinaReviewed by:
Ze-Hua Zhao, Qilu Hospital, Shandong University, ChinaCopyright © 2022 Shen, Zhou, Gu, Shen, Guo, Dai, Liu, Zhang, Lu and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lungen Lu, bHVuZ2VuLmx1QHNoZ2guY24=; Hui Dong, a2Vyd2luYzIwMDJAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.