- 1Department of Rehabilitation Medicine, The Affiliated Wenling Hospital of Wenzhou Medical University, Wenling, China
- 2School of Pharmacy, Jiangsu Provincial Engineering Research Center of Traditional Chinese Medicine External Medication Development and Application, Nanjing University of Chinese Medicine, Nanjing, China
- 3School of Life Sciences, Jilin University, Changchun, China
Although cancer has seriously threatened people’s health, it is also identified by the World Health Organization as a controllable, treatable and even curable chronic disease. Traditional Chinese medicine (TCM) has been extensively used to treat cancer due to its multiple targets, minimum side effects and potent therapeutic effects, and thus plays an important role in all stages of tumor therapy. With the continuous progress in cancer treatment, the overall efficacy of cancer therapy has been significantly improved, and the survival time of patients has been dramatically prolonged. In recent years, a series of advanced technologies, including nanotechnology, gene editing technology, real-time cell-based assay (RTCA) technology, and flow cytometry analysis technology, have been developed and applied to study TCM for cancer therapy, which efficiently improve the medicinal value of TCM and accelerate the research progress of TCM in cancer therapy. Therefore, the applications of these advanced technologies in TCM for cancer therapy are summarized in this review. We hope this review will provide a good guidance for TCM in cancer therapy.
Introduction
In recent years, cancer has become the disease with the second highest mortality rate among all diseases in the world, second only to cardiovascular and cerebrovascular diseases (Bhatt et al., 2017). Numerous methods have been established to treat cancer, such as chemotherapy, radiotherapy, surgery, and immunotherapy, etc. (Baskar et al., 2012; Tang et al., 2020). However, all these treatments also suffer from certain limitations. For example, surgery is limited to the minority of cancers, while radiotherapy often causes severe side effects and induces depression and appetite lose in patients (Wu et al., 2013). Chemotherapy is the most common therapeutic method, however, the resistance of tumor cells to the chemotherapy is the main reason that hinders the application of chemotherapy drugs (Tang et al., 2020). Therefore, it is urgent to find alternative drugs to treat tumors.
Nowadays, traditional Chinese medicine (TCM), one of the mainstream forms of alternative drugs, has been widely used to treat cancer due to its multiple targets, minimum side effects and potent therapeutic effects (Cui et al., 2004; Chen et al., 2008; Xie et al., 2016). TCM presents lower toxic effects, higher quality of life, and longer survival rate in cancer therapy (Xiang et al., 2019). Meanwhile, many TCM ingredients extracted from natural plants such as curcumin, resveratrol, berberine, quercetin, tanshinone also show strong antitumor effects including inhibition of the development, proliferation, angiogenesis, and metastasis of human cancer, thus play an important role in all stages of cancer therapy (Hu et al., 2013; Yang et al., 2015; Xu et al., 2016).
Recently, many advanced technologies had been widely applied to investigate TCM for cancer therapy. For example, TCM can be developed into nano-drug delivery system (TCM-NDDS) through nanotechnology (Zhang L et al., 2018; Gao et al., 2019), and the drawbacks of TCM such as poor bioavailability, low solubility, short plasma half-life can be excellent solved, and this will greatly improve the therapeutic value of TCM to treat and prevent cancer (Geng et al., 2022). Meanwhile, gene editing technology also provides a new strategy in TCM for cancer therapy (Rhee et al., 2018). Through gene editing technology, the molecular mechanisms underlying TCM for cancer therapy can be studied in detail, animal models can be established by this novel method, and all these will significantly reduce the difficulties in tumor therapy research (Concha-Benavente and Ferris, 2017; Shen, et al., 2021). Moreover, a series of physiological states such as cell viability, migration, and growth changes can be reflected in a real time way by real-time cell-based assay (RTCA) technology with the advantages of high accuracy and repeatability (Wang T. X, et al., 2013; Yan et al., 2018). In addition, flow cytometry analysis technology, one of the qualitative and quantitative analysis of advanced cell technology, is applied to quickly determine the biological properties of cells or biological particle under a fast linear flow state in TCM study due to its high sensitivity, fast analysis, high accuracy, wide measurement parameters and high throughput (Adan et al., 2017; McKinnon, 2018; Feng and Zhao, 2021). These advanced technologies have been extensively applied in the researches of lymphoma, cervical cancer and other solid tumors, thus promoting the development of TCM (Concha-Benavente and Ferris, 2017; Shi M. et al., 2021).
As is known to all, advanced technology makes great contributions in TCM for cancer therapy, and thus promoting the rapid development of TCM, and the shortcomings of TCM in actual application are also maken up by interdisciplinary research (Zheng et al., 2022). Therefore, in order to efficiently enhance the therapeutic value of TCM and accelerate the research progress of TCM in cancer therapy, application of advanced technology (e.g., nanotechnology, gene editing technology, RTCA technology, flow cytometry analysis technology, etc.) in TCM for cancer therapy were summarized, and the main areas of this review is summarized in Figure 1.
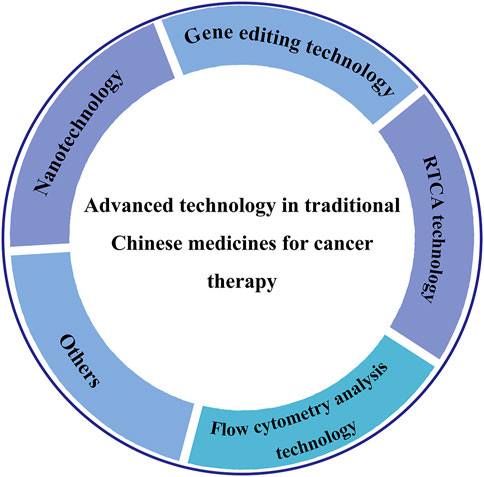
FIGURE 1. The main topics of this review. The main topics include the application of advanced technologies such as nanotechnology, gene editing technology, RTCA technology, flow cytometry analysis technology in TCM for cancer therapy.
The application of advanced technology
Many advanced technologies including nanotechnology, gene editing technology, RTCA technology, flow cytometry analysis technology had been extensively used to investigate the effect of TCM for cancer therapy, as shown in Table 1. In this section, we summarize the research development of these technologies in recent years.
Nanotechnology
As is well known, the therapeutic effects of TCM are mainly attributable to the active ingredients inside (Li et al., 2018). According to their differences in chemical structures, the ingredients in TCM can be classified into terpenoids, alkaloids, sesquiterpenes, polysaccharides, glycosides, saponins, etc. (Puglia et al., 2017). However, the physicochemical properties of some active ingredients such as poor solubility, low bioavailability, short plasma half-life may limit their application (Gao et al., 2019). In order to solve these drawbacks, nanotechnology was introduced into TCM in recently years (Zhang Q et al., 2018; Gao et al., 2019; Bayda et al., 2020). Nanotechnology is a science that involves detection, design, fabrication, and application of certain chemicals and materials on the level of nanoscale. TCM can be carefully developed into TCM-NDDS through nanotechnology, which will greatly enhance the therapeutic value of TCM to treat and even prevent cancer (Ma et al., 2019).
Various TCM-NDDS (e.g., liposomes, nanoparticles, micelles, polymer nanoparticles, inorganic nanomaterials) have been meticulously designed to improve the therapeutic value of TCM by nanotechnology, as summarized in Table 1. For example, Zhang et al. (Zhang L et al., 2018) designed and synthesized TCM-NDDS which used hollow nanocages MOF-5 as drug carrier to load oleanolic acid (MOF@OA). Its drug-loading capacity and cytotoxicity were evaluated, and results showed that MOF improved the drug-loading capacity of OA and the kinetic and therapeutic efficiency of OA. Similarly, Zhao et al. (Zhao et al., 2021) constructed a nanocarrier using the mesoporous silica nanoparticles (MSNs) which was further modified by the cancer cell plasma membrane. After modification, TCM isoimperatorin (ISOIM) was loaded inside the empty nanocarrierinside (CCM@MSNs-ISOIM). Their antitumor effects, anti-lymphoma mechanism, and the biosafety in vivo were characterized (the prepared scheme and targeted therapeutic mechanisms in lymphoma of CCM@MSNs-ISOIM were presented in Figure 2). This nano-platform presented high drug-loading capacity, good biocompatibility and excellent tumor targeting. Meanwhile, it also showed the characteristics of anti-phagocytosis and low pH sensitivity.
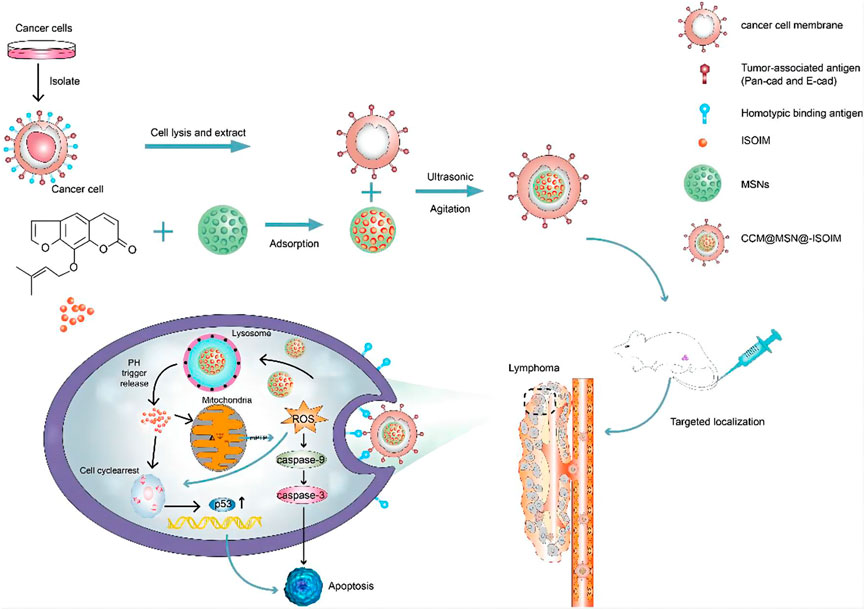
FIGURE 2. The scheme diagram of CCM@MSNs-ISOIM construction and its targeted therapeutic mechanisms in lymphoma (Zhao et al., 2021). In this work, a nano-platform based on MSNs which camouflaged by the cancer cell membrane was constructed. The nano-platform has the characteristics of anti-phagocytosis, high drug-loading capacity, low pH sensitivity, good biocompatibility and active targeting, promoting mitochondrial-mediated apoptosis.
Additionally, in order to overcome the critical drawbacks of celastrol (e.g., low water-solubility, short plasma half-life, and notable systemic toxicity) and improve its antitumor efficacy, Geng et al. (Geng et al., 2022) designed a novel polymer structure, poly (2-(N-oxide-N, N-dimethylamino)ethylmethacrylate)-block-poly (2-hydroxy- ethyl methacrylate) (OPDMA-HEMA), and used it for drug delivery (OPDMA-Cela). The OPDMA-Cela resulted in potent immunogenic cell death and decreased the PD-L1 protein level. Moreover, compared with free celastrol, OPDMA-Cela showed a higher anticancer efficacy, minimum systemic toxicity with enhanced solubility in water. Similarly, Gao et al. (Gao et al., 2019) prepared menthol-modified casein nanocarrier loaded with 10-hydroxycamptothecin (HCPT-M-CA-NPs) for glioma targeting therapy. The results showed that the drug accumulation in the tumor site was greatly improved by HCPT-M-CA-NPs, and the median survival time of intracranial glioma-bearing mice was significantly extended.
In summary, nanotechnology, as an emerging technology in recent years, has received extensive attention from researchers. TCM designed and developed into nano-drug delivery system through nanotechnology can efficiently solved the drawbacks of TCM including poor solubility, low bioavailability, short half-life, thus enhancing the therapeutic use of TCM to treat and prevent cancer.
Gene editing technology
As an emerging technology, gene editing technology is also extensively applied in cancer therapy in TCM. Gene editing technology is a technology that artificially inserts, deletes and replaces gene sites containing genetic information in the nucleotide sequence (Rhee et al., 2018). It is mainly composed of zinc finger nucleases (ZFNs) technology, transcription activator-like effector nucleases (TALENs) technology and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas) technology (Puchta et al., 1993; Rouet et al., 1994). At present, due to the complex experimental process and high off-target effect, ZFNs and TALENs technologies have been gradually replaced by CRISPR-Cas technology. Therefore, we will focus our discussion on CRISPR/Cas technology in this review (Adli, 2018).
CRISPR/Cas9 technology, as the major type of CRISPR-Cas technology, has been extensively applied in cancer research because of its simple design, high efficiency, low cost and simultaneous multi-point editing (Gesner et al., 2011; Sapranauskas et al., 2011; Sashital et al., 2011). CRISPR/Cas9 system is mainly composed of trans-activated RNA gene expression CRISPR RNA (trans-activating crRNA, tracrRNA), Cas9 protein-coding gene with endonuclease, and specific targeting CRISPR RNA (crRNA) (Jinek et al., 2012; Wen et al., 2015). Its principle is very simple, as shown in Figure 3 (El-Mounadi et al., 2020). According to the dimer structure of crRNA and tracrRNA, a new single-stranded chimeric sgRNA (single guide RNA) can be designed to specifically identify the target gene sequence, and then further combined with Cas9 protein to form a targeted cutting complex which specifically cut the target DNA and break the double strand of DNA (Bhaya et al., 2011; Jinek, Chylinski et al., 2012; Jackson et al., 2014; Qu et al., 2015). In theory, CRISPR/Cas9 technology can achieve editing of any genomic site by synthesizing sgRNA that targets to various genomic sites (Concha-Benavente and Ferris, 2017; Shi S. J. et al., 2021).
FIGURE 3. Schematic diagram of CRISPR/Cas9 technology. According to the dimer structure of crRNA and tracrRNA, a new single-stranded chimeric sgRNA (single guide RNA) can be designed to specifically identify the target gene sequence, and then further combined with Cas9 protein to form a targeted cutting complex which specifically cut the target DNA and break the double strand of DNA.
CRISPR/Cas9 technology provides a new strategy in TCM for cancer therapy, thus promoting the research progress of tumor therapy in recent years (Maus et al., 2014; Corrales et al., 2017; Ren and Zhao, 2017). It can significantly reduce the difficulties of tumor therapy research and has been extensively applied to the research of lymphoma, cervical cancer and other solid tumors (Concha-Benavente and Ferris, 2017; Shi M. et al., 2021). In order to investigate the roles of proteins in apoptosis and necroptosis, Hu et al. (Hu et al., 2013) knocked out the related gene by CRISPR/Cas9 technology. The results revealed that necrosomes, which is composed of receptor-interacting protein kinase 1, receptor-interacting protein 3 and mixed lineage kinase domain-like protein, underwent necroptosis when caspase-8 function was lost. They further investigated the cytotoxicity of bufalin (a component of TCM) in glioma cell with the gene knockout by CRISPR/Cas9. The results indicated that glioma cells with the knockout of caspase-8 were more sensitive to bufalin than those without caspase-8, and bufalin may be a potential drug for glioma therapy. Shi et al. (Shi S. J. et al., 2021) edited KV1.3 coding gene KCNA3 through this technology, and a KV1.3 knockout (KO) cell model was successfully generated. The results showed that Loureirin B, a component extracted from Resina Draconis, could inhibit Ca2+ influx and IL-2 secretion in Jurkat T cells through the inhibition of KV1.3 and STIM1/Orail targets. In addition, in order to explore new targeting, Wu et al. (Wu et al., 2018) knocked-down of dihydroorotate dehydrogenase by CRISPR/Cas9 technology, which resulted in apoptosis and normal differentiation of acute myeloid leukemia cells. This study indicated that dihydroorotate dehydrogenase may be a potential therapeutic target in acute myeloid leukemia. Moreover, they further verified that isobavachalcone, which is derived from psoralea corylifolia, could act as an advanced dihydroorotate dehydrogenase inhibitor to trigger apoptosis and differentiation of acute myeloid leukemia cells, and offered a useful therapeutic target for acute myeloid leukemia treatment. In addition, Zhong et al. (Zhong et al., 2020) used CRISPR/Cas9 technology to knockout ten-eleven translocation 1 (TET1) gene and investigated the effects of chrysin, a natural flavonoid commonly found in honey, with the protein expression of TET in gastric cancer (GC) cells. The results indicated that chrysin was effective against tumors by regulating TET1 expression in GC. Zhong et al. (Zhong et al., 2020) showed that knockdown of H19 and miR675 by CRISPR/Cas9 technology could induced p53 expression and ultimately promoted cell apoptosis in MHCC97H cells. Moreover, Galangin could also promote cell apoptosis by inducing H19 and miR675 expression in MHCC97H cells, presenting a crucial role in hepatocarcinogenesis. Zhou et al. (Zhou et al., 2020) found that knockout of the F-box protein S-phase kinase-associated protein 2 (Skp2) by CRISPR/Cas9 technology could inhibit Hexokinase 2 (HK2) and glycolysis, and affected colorectal cancer cell growth rate in vitro and in vivo. Dioscin, which is a natural steroid saponin derived from a variety of plants, could reduce the Skp2 protein expression by reducing the stability of Skp2, promoting Skp2 ubiquitination, inhibiting Skp2 expression and thus delaying tumor growth in vivo, and all these suggested that enhancement of ubiquitination-dependent Skp2 turnover had great potential for cancer therapy. In summary, all these samples suggest the great potential of CRISPR/Cas9 technology in the TCM study.
For CRISPR/Cas9 technology, it mainly relies on a sgRNA-Cas9 complex, thereby avoiding time-consuming and labor-intensive protein construction process (Ding et al., 2013; Shah et al., 2013; Gupta and Musunuuru, 2014). Meanwhile, it also presents the advantages of simple operation, high gene editing efficiency and good versatility (Cong et al., 2013; Chen et al., 2015). At present, CRISPR/Cas9 technology as an advanced technology was extensively applied in TCM for cancer therapy, promoting its research progress. Though this technology has the aforementioned advantages, CRISPR/Cas9 technology also has some limitations. For example, this technology has a high off-target efficiency, which may lead to the binding of sgRNA to other sites. Meanwhile, the off-target site is generally difficult to find, and thus the whole-genome sequence should be evaluated to determine the off-target site. In addition, the T7 promoter contained in sgRNA synthesized in vitro is limited to the transcribed target gene sequence, which limits the selection of target sites to a certain extent (Qiu, 2016; Lei et al., 2020).
Real-time cell-based assay technology
As a new cell detection technology, RTCA technology is a transient cell sensing continuous recording system based on the principle of resistance and impedance (Wang T. X et al., 2013). RTCA system integrates the microelectronic cell sensor chip into the bottom of the cell detection plate which is suitable for cell attachment and growth. And then, a real-time, dynamic and quantitative cell impedance detection sensing system that tracks changes in cell morphology, proliferation and differentiation is constructed (Stefanowicz-Hajduk et al., 2016). The biological state and changes of cells, and cell dynamics in real time without labeling and damage are monitored by RTCA (Turker sener et al., 2017).
RTCA is mainly composed of biosensor board, impedance measurement unit, impedance conversion unit, real-time analysis and data processing unit (Wang S. Y et al., 2013). The electrical impedance measured by the biosensor board can detect and evaluate the growth state, morphological changes, death number and adhesion degree of cells in real time (Burmakina et al., 2018). The electrode impedance measured by the impedance measurement unit is mainly determined by the ionic environment on the electrode surface and the bulk solution (Meindl et al., 2013), and thus, changes in impedance could reflect changes in cell biological states (e.g., quantity, mass, size, attachment state) (as shown in Figure 4). The impedance conversion unit is mainly composed of analog-to-digital converter (ADC). When the cell condition changes, ADC can record the analog electrical signal in real time manner and automatically convert it into digital signal, which can reflect a series of biological conditions such as cell viability, migration, and growth changes (Wang T. X et al., 2013).
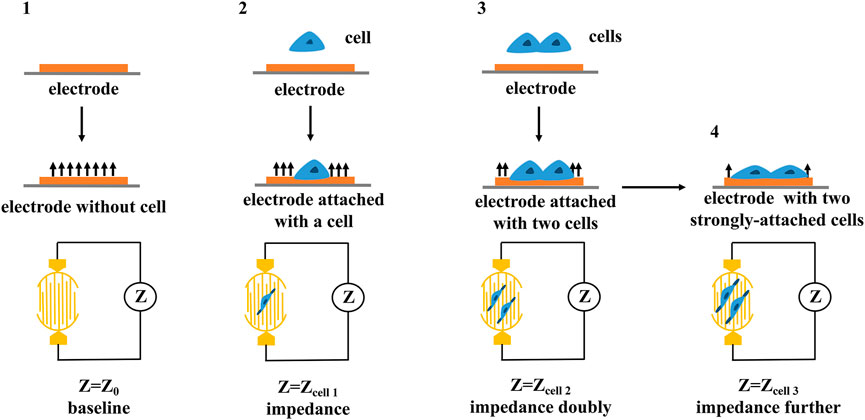
FIGURE 4. The principle of cellular impedance detection. The cell biological states including quantity, mass, size, attachment state can be reflected by the changes in impedance.
Compared with traditional methods, RTCA has many advantages such as less cell consumption, high throughput, rapid and simple operation (Meindl et al., 2013; Kho et al., 2015). It can be used to verify the function and growth rule of different types of cells and the mechanism of different drugs on tumor cells (Kho et al., 2015), thus RTCA was widely applied to various tumor therapy. Yang et al. (Yang et al., 2020) analyzed the inhibitory ability of different concentrations of chelidonine hydrochloride on cervical cancer cells by combining MTT method, xCELLigence RTCA S16 and colony formation assay. The results showed that the inhibitory effect of chelidonine hydrochloride on cervical cancer cells was dose-dependent, suggesting chelidonine hydrochloride had obvious antitumor effect. In addition, Liu et al. (Liu et al., 2020) applied RTCA technology to monitor the proliferation of nasopharyngeal carcinoma CNE2 cells in a real time, and found that different concentrations of coumarin compound isoimperatorin had significant inhibitory effect on nasopharyngeal carcinoma CNE2 cells. Calibasi-Kocal et al. (Calibasi-Kocal et al., 2019) found that curcumin had anti-proliferation activity against both HCT-116 and metastatic colorectal cancer cells by xCELLigence RTCA DP analysis technique. Lin et al. (Lin et al., 2020) evaluated the effect of curcumin micelles on GC cell growth by combing MTS cell proliferation assay, flow cytometry, RTCA and xenotransplantation. The results showed that curcumin micelles could significantly inhibit cell proliferation, colony formation and apoptosis of GC. Curcumin micelles could also significantly inhibit tumor growth in vivo, suggesting that curcumin could be used in the treatment of GC therapy. Li et al. (Li et al., 2020) investigated the anti-breast cancer effect (e.g., MCF10A, MCF7, SK-BR-3 and MDA-MB-231) of oleandrin derived from oleander leaves by RTCA technology. The results revealed that oleandrin exhibited no detectable effect on MCF10A, and had a certain inhibiting effect on other breast cancer cell. Similarly, Yang et al. (Yang et al., 2014) detected the inhibitory effect of salvia miltiorrhiza and ginseng on MCF-7 by RTCA technology. The salvia miltiorrhiza and ginseng presented a continuous inhibitory effect on MCF-7. Meanwhile, the combination of salvia miltiorrhiza and ginseng had a certain selectivity for breast cancer therapy.
RTCA technology overcomes the limitations of the traditional methods such as long time, large interference, and large damage to cells. Moreover, most cells in micropores can be detected in a short time and the detection data has higher accuracy and repeatability (Otero-Gonzalez et al., 2012; Yan et al., 2018). However, RTCA can only indirectly monitor the activity of adherent cells based on cellular impedance, suspended cells require some auxiliary methods (Martinez-Serra et al., 2014). In addition, special microplates are required when the RTCA instruments are used in different experiments, therefore the cost will be increased.
Flow cytometry analysis technology
Flow cytometry analysis technology, one of the qualitative and quantitative analysis of advanced cell technology, is a detection method that can quickly determine the biological properties of cells or biological particle under a fast linear flow state (Ormerod, 2002). It has the advantages of high sensitivity, fast analysis, high accuracy, wide measurement parameters and high throughput (Adan, et al., 2017; McKinnon, 2018; Feng and Zhao, 2021). The principle of flow cytometry analysis technology is relatively simple, and its schematic diagram is shown in Figure 5. Flow cytometry is mainly composed of flow chamber, liquid flow system, light source, optical system, signal collection, signal conversion system, computer and analysis system (Jani et al., 2001). After suspension samples (body fluid, blood, etc.) or single cell stained with fluorescence dyes are prepared into suspension samples, they are wrapped in sheath liquid and passed through the detection area in a single row. Fluorescence signals, which reflect the intensity of cell membrane surface antigen, are captured by fluorescence detector, and then, amplified and converted into an electrical signal by a photomultiplitizer. The continuous electrical signal is converted into a digital signal which can be recorded by a computer and processed into an image for in-depth analysis (Yang et al., 2017; Shi M. et al., 2021).
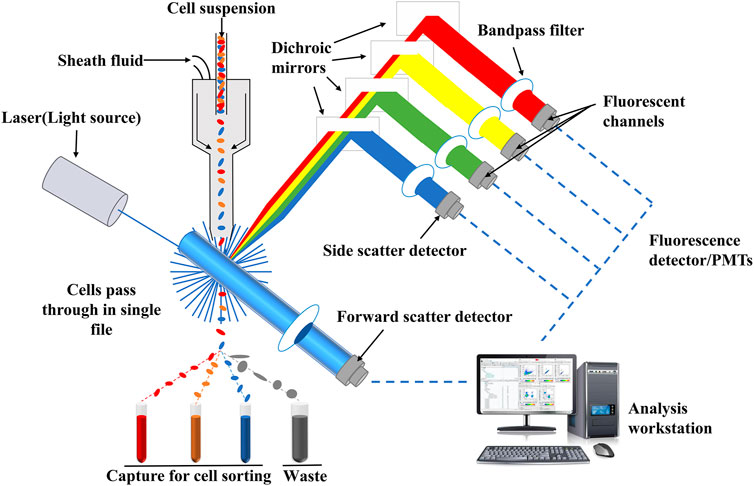
FIGURE 5. Schematic diagram of flow cytometry. Fluorescence signals which reflect the intensity of cell membrane surface antigen can be captured by fluorescence detector, and then, amplified and converted into an electrical signal by a photomultiplitizer. The continuous electrical signal is converted into a digital signal which can be recognized by a computer and processed into an image for in-depth analysis.
Recently, with the development of science and technology, the performance of flow cytometer has been constantly improved, and the operation has become more and more flexible. It has become an indispensable detection tool in medical research and is often used in cancer treatment research in TCM. Fan et al. (Fan et al., 2020) found that dihydroartemisinin (DHA) could inhibit GC cell proliferation by flow cytometry combined with other technologies. DHA inhibited the occurrence and invasion of GC by regulating the cyclin D1-CDK4-Rb signaling pathway, which provided a guiding strategy for GC therapy. Meanwhile, Gong et al. (Gong et al., 2021) found that curcumin at the doses of 5, 15 and 30 μM could induce the apoptosis of renal carcinoma cells in a dose-dependent manner by flow cytometry. Zhang et al. (Zhang et al., 2017) investigated the effects of different concentrations of puerarin on the survival and apoptosis of SMMC-7721cells by combining MTT method with flow cytometry. The results showed that puerarin inhibited the proliferation of SMMC-7721 cells in a dose- and time-dependent manner, suggesting that puerarin may be a potential anti-hepatocellular carcinoma substance. Sun et al. (Sun and Liu, 2021) investigated the apoptosis of human tongue squamous cell within 24 h by flow cytometry, and found that osthole induced cell apoptosis in a concentration-dependent manner, which was attributed to the promotion of apoptosis, inhibition of autophagic flow and autophagy pathway injury. Zhu et al. (Zhu et al., 2021) investigated the role of trichosanthin (TCS) in cervical cancer by CCK-8 method and flow cytometry. They found that TCS inhibited cell viability, induced cell apoptosis as well as cell migration in a time- and dose-dependent manner. Similarly, Huang et al. (Huang et al., 1999) used flow cytometry to investigate the effect of Dongxia Pill on 116 tumor cell lines including nasopharyngeal carcinoma cells in vitro. They found that Dongxia Pill had significant inhibitory effect on the growth of 6 human tumor cells, and the inhibited rate increased with the increase of drug concentration. Moreover, Dongxia Pill could increase the G2 phase of nasopharyngeal carcinoma cells and induce cell apoptosis. Moreover, Hsu et al. (Hsu et al., 2008) used flow cytometry to investigate the anticancer effects of Huang-lian-jie-du-tang on HepG2 and PLC/PRF/5 cell. Huang-lian-jie-du-tang significantly reduced the levels of cyclin A, cyclin B1, Cdc2 and Cdc25C, resulting in cell cycle arrest. Meanwhile, the expression of Bcl-XL in HepG2 and PLC/PRF/5 cells were all decreased, thus inhibiting cell survival signal. It is suggested that Huang-lian-jie-du-tang has potential anticancer effect on human liver cancer cells.
Flow cytometry analysis technology had been extensively applied in TCM study due to its advantages of fast detection, high precision and accuracy (McKinnon, 2018). The target phenotype for tumor cell subsets (including lineage and functional balance) can be determined through multiple parameter flow of flow cytometry. Moreover, the effects of drugs on cytokines, cell adhesion molecules expression, immune related cell subsets, intracellular enzyme expression can be also investigated by flow cytometry. However, flow cytometry analysis technology requires samples to be in single-cell suspension, otherwise it will affect the experimental results (Otto et al., 2016; Ozawa et al., 2016). This will limit some of its application in TCM study such as in vivo evaluation.
Other technologies
As mentioned above, nanotechnology, gene editing technology, RTCA technology, and flow cytometry analysis technology have all been extensively applied in TCM for cancer therapy, promoting their research process. Besides, the live cell dynamic imaging technology and cell energy metabolism technology have also been applied in TCM study.
The live cell dynamic imaging technology integrates the incubator and microscope system to keep cells viable, allowing long-term dynamic process experiments of cell migration, interaction, and response to environmental perturbations. At present, it is mainly used in the detection of dynamic processes of living cells such as cell proliferation/inhibition, cell migration/invasion, cell culture quality control/culture optimization, etc. (Li et al., 2021).
Energy metabolism has been recognized as a key factor in cellular function, from tumorigenesis to immune cell activation. Cell energy metabolism technology can simultaneously measure and analyze mitochondrial oxygen consumption rate (OCR) and extra cellular acidification rate (ECAR) in living cells in real-time, quantitatively and automatically without invading and destroying the sample (Yi et al., 2012; Pelletier et al., 2014). This technology is often used to study the dependence of cancer cells on energy-providing substrates (Patsoukis et al., 2015). The most common of which is the dependence of cancer cells on glutamine, which can reveal the weakness of cancer cells and thus provide a basis for finding drug targets (Hensley et al., 2013; Wang et al., 2015).
Summary and perspective
In recent decades, increasing numbers of patients have been attracted to TCM for cancer treatment because TCM can regulate oncogenes and tumor suppressor genes, and microenvironment. Some current clinical trials of TCM in cancer treatment, were summarized in this review, as shown in Table 2 (The data were obtained from https://clinicaltrials.gov/). In order to promote the development of TCM in cancer therapy, some advanced technologies including nanotechnology, gene editing technology, RTCA technology, flow cytometry analysis technology had been widely applied in recent years. In this review, we summarize the application of these advanced technology in TCM for cancer therapy. Owing to poor solubility, low bioavailability, short half-life of some ingredient in TCM, nanotechnology was introduced to develop into various TCM-NDDS. The shortcomings of TCM can be effectively solved, and the medicinal value of TCM to treat and prevent cancer is enhanced. In addition, CRISPR/Cas9 technology, as an emerging technology, has also been extensively used in cancer research due to its advantages of simple design, high efficiency, low cost and simultaneous multi-point editing. CRISPR/Cas9 technology can achieve editing of any genomic site by synthesizing sgRNA that targets to various genomic sites (Concha-Benavente and Ferris, 2017; Shi S. J. et al., 2021), reducing the difficulties of tumor therapy research. Moreover, the roles of proteins in apoptosis and necroptosis can also be investigated through CRISPR/Cas9 technology, which promote the mechanism research of cancer. Besides, as a new cell detection technology, RTCA technology can be used to monitor the biological state and changes of cells, and cell dynamics in real time without labeling and damaging. The function and growth rule of different cells can also be determined. And owing to high sensitivity, fast analysis, high accuracy and large throughput, flow cytometry analysis technology is used to qualitatively and quantitatively analyze of the biological properties of cells. These advanced technologies all promote the research process of TCM for cancer therapy. However, some challenges still exist, and many efforts still need to be performed. For instance, the encapsulation efficiency of some TCM-NDDS is low, and the targeting is lack. Therefore, the preparation methods of TCM-NDDS need to be further explored and their preparation parameters should be further optimized. In addition, CRISPR/Cas9 technology shows a high off-target efficiency, and the off-target site is also generally difficult to find. RTCA can only indirectly monitor the activity of adherent cells, and special microplates are required when the RTCA instruments was used in different experiments, therefore increasing its cost. In view of this situation, some advanced technologies still need to be developed and introduced to study TCM for the treatment of cancer. Meanwhile, a variety of technologies can be combined to investigate the mechanism of inhibiting tumor growth and inducing tumor cell apoptosis. Only integrating with modern science and technology, and making full use of RTCA, flow cytometry technology and other advanced technologies, can TCM accelerate the pace of modernization and internationalization.
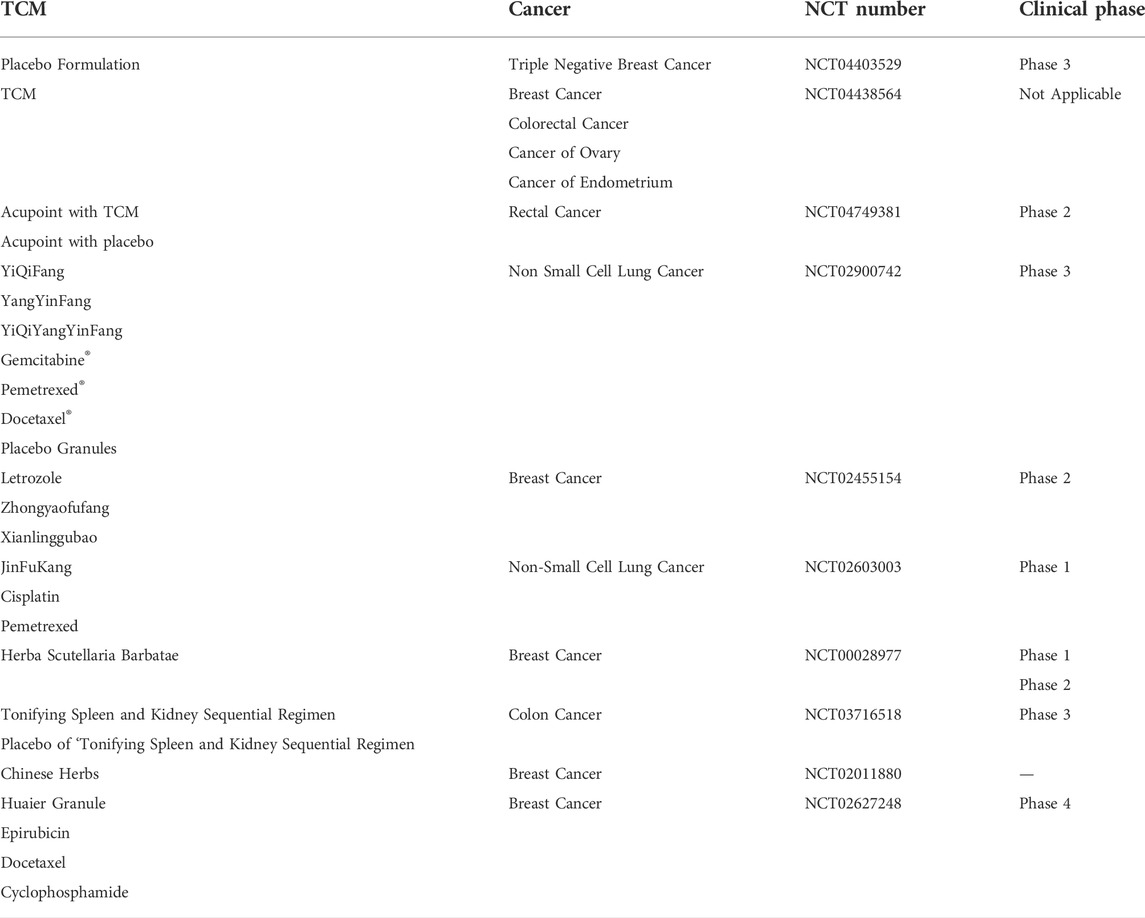
TABLE 2. The current clinical trials of TCM in cancer treatment. The date are obtained from https://clinicaltrials.gov/.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The project was supported by the National Natural Science Foundation of China (81773910, 82074004) and National Key R&D Program of China (No. 2019YFC1710603).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adan, A., Alizada, G., Kiraz, Y., Baran, Y., and Nalbant, A. (2017). Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 37, 163–176. doi:10.3109/07388551.2015.1128876
Adli, M. (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9, 1911. doi:10.1038/s41467-018-04252-2
Baskar, R., Lee, K. A., Yeo, R., and Yeoh, K. W. (2012). Cancer and radiation therapy: Current advances and future directions. Int. J. Med. Sci. 9, 193–199. doi:10.7150/ijms.3635
Bayda, S., Adeel, M., Tuccinardi, T., Cordani, M., and Rizzolio, F. (2020). The history of nanoscience and nanotechnology: From chemical-physical applications to nanomedicine. Molecules 25, E112. doi:10.3390/molecules25010112
Bhatt, A. P., Redinbo, M. R., and Bultman, S. J. (2017). The role of the microbiome in cancer development and therapy. Ca. Cancer J. Clin. 67, 326–344. doi:10.3322/caac.21398
Bhaya, D., Davison, M., and Barrangou, R. (2011). CRISPR-cas systems in bacteria and archaea: Versatile small rnas for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. doi:10.1146/annurev-genet-110410-132430
Burmakina, G., Bliznetsov, K., and Malogolovkin, A. (2018). Real-time analysis of the cytopathic effect of African swine fever virus. J. Virol. Methods 257, 58–61. doi:10.1016/j.jviromet.2018.04.003
Calibasi-Kocal, G., Pakdemirli, A., Bayrak, S., Ozupek, N. M., Sever, T., Basbinar, Y., et al. (2019). Curcumin effects on cell proliferation, angiogenesis and metastasis in colorectal cancer. J. Buon. 24, 1482–1487.
Chen, S. D., Sanjana, N. E., Zheng, K. J., Shalem, O., Lee, K., Shi, X., et al. (2015). Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260. doi:10.1016/j.cell.2015.02.038
Chen, Z., Gu, K., Zheng, Y., Zheng, W., Lu, W., and Shu, X. O. (2008). The use of complementary and alternative medicine among Chinese women with breast cancer. J. Altern. Complement. Med. 14, 1049–1055. doi:10.1089/acm.2008.0039
Concha-Benavente, F., and Ferris, R. L. (2017). Oncogenic growth factor signaling mediating tumor escape from cellular immunity. Curr. Opin. Immunol. 45, 52–59. doi:10.1016/j.coi.2017.01.004
Cong, L., Ran, F. A., Cox, D., Lin, S. L., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Corrales, L., Matson, V., Flood, B., Spranger, S., and Gajewski, T. F. (2017). Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 27, 96–108. doi:10.1038/cr.2016.149
Cui, Y., Shu, X. O., Gao, Y. T., Wen, W. Q., Ruan, Z. X., Jin, F., et al. (2004). Use of complementary and alternative medicine by Chinese women with breast cancer. Breast Cancer Res. Treat. 85, 263–270. doi:10.1023/B:BREA.0000025422.26148.8d
Ding, Q. R., Regan, S. N., Xia, Y. L., Oostrom, L. A., Cowan, C. A., and Musunuru, K. (2013). Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394. doi:10.1016/j.stem.2013.03.006
El-Mounadi, K., Morales-Floriano, M. L., and Garcia-Ruiz, H. (2020). Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 11, 56. doi:10.3389/fpls.2020.00056
Fan, H. N., Zhu, M. Y., Peng, S. Q., Zhu, J. S., Zhang, J., and Qu, G. Q. (2020). Dihydroartemisinin inhibits the growth and invasion of gastric cancer cells by regulating cyclin D1-CDK4-Rb signaling. Pathol. Res. Pract. 216, 152795. doi:10.1016/j.prp.2019.152795
Feng, R. T., and Zhao, J. Z. (2021). Influence of methodological factors on experimental results of immunophenotypic analysis of blood lymphocytes by flow cytometry. Clin. Res. Cardiol. 29, 14–15+31.
Gao, C. F., Liang, J. M., Zhu, Y., Ling, C. L., Cheng, Z. K., Li, R. X., et al. (2019). Menthol-modified casein nanoparticles loading 10-hydroxycamptothecin for glioma targeting therapy. Acta Pharm. Sin. B 9, 843–857. doi:10.1016/j.apsb.2019.01.006
Geng, Y., Xiang, J. J., Shao, S. Q., Tang, J. B., and Shen, Y. Q. (2022). Mitochondria-targeted polymer-celastrol conjugate with enhanced anticancer efficacy. J. Control. Release 342, 122–133. doi:10.1016/j.jconrel.2022.01.002
Gesner, E. M., Schellenberg, M. J., Garside, E. L., George, M. M., and MacMillan, A. M. (2011). Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 18, 688–692. doi:10.1038/nsmb.2042
Gong, X. L., Jiang, L., Li, W., Liang, Q. B., and Li, Z. (2021). Curcumin induces apoptosis and autophagy inhuman renal cell carcinoma cells via Akt/mTOR suppression. Bioengineered 12, 5017–5027. doi:10.1080/21655979.2021.1960765
Gupta, R. M., and Musunuru, K. (2014). Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-cas9. J. Clin. Invest. 124, 4154–4161. doi:10.1172/JCI72992
Hensley, C. T., Wasti, A. T., and DeBerardinis, R. J. (2013). Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Invest. 123, 3678–3684. doi:10.1172/JCI69600
Hsu, Y. L., Kuo, P. L., Tzeng, T. F., Sung, S. C., Yen, M. H., Lin, L. T., et al. (2008). Huang-lian-jie-du-tang, a traditional Chinese medicine prescription, induces cell-cycle arrest and apoptosis in human liver cancer cells in vitro and in vivo. J. Gastroenterol. Hepatol. 23, e290–e299. doi:10.1111/j.1440-1746.2008.05390.x
Hu, Y. Y., Wang, S. P., Wu, X., Zhang, J. M., Chen, R., Chen, M. W., et al. (2013). Chinese herbal medicine-derived compounds for cancer therapy: A focus on hepatocellular carcinoma. J. Ethnopharmacol. 149, 601–612. doi:10.1016/j.jep.2013.07.030
Huang, C. Y., Chen, T. K., Zhu, D. M., and Huang, Q. Q. (2020). Enhanced tumor targeting and radiotherapy by quercetin loaded biomimetic nanoparticles. Front. Chem. 8, 225. doi:10.3389/fchem.2020.00225
Huang, P. C., Deng, H. H., Zhao, M. L., Huang, T. Y., and Yu, L. R. (1999). Effect of Chinese medicine compound“dong xia wan”on 6 human tumor cell lines in vitro. Cancer Res. Preve. Tre. 05, 330–331. doi:10.3971/j.issn.1000-8578.1999.05.004
Jackson, R. N., Golden, S. M., van Erp, P. B., Carter, J., Westra, E. R., Brouns, S. J., et al. (2014). Structural biology crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345, 1473–1479. doi:10.1126/science.1256328
Jani, I. V., Janossy, G., Iqbal, A., Mhalu, F. S., Lyamuya, E. F., Biberfeld, G., et al. (2001). Affordable CD4(+) T cell counts by flow cytometry II. The use of fixed whole blood in resource-poor settings. J. Immunol. Methods 257, 145–154. doi:10.1016/s0022-1759(01)00458-6
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable Dual-RNA-Guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Kho, D., MacDonald, C., Johnson, R., Unsworth, C. P., O'Carroll, S. J., du Mez, E., et al. (2015). Application of xCELLigence RTCA biosensor technology for revealing the profile and window of drug responsiveness in real time. Biosens. (Basel) 5, 199–222. doi:10.3390/bios5020199
Lei, L. F., Cai, Y. G., Wang, H. B., Wu, X. S., and Ye, X. L. (2020). Research progress and application of genome editing techniques in zebrafish. Genomics Appl. Biol. 39, 591–598. doi:10.13417/j.gab.039.000591
Li, L., Feng, Y., Hong, Y. L., Lin, X., and Shen, L. (2018). Recent advances in drug delivery system for bioactive glycosides from traditional Chinese medicine. Am. J. Chin. Med. 46, 1791–1824. doi:10.1142/s0192415x18500908
Li, W. W., Ji, L. Y., Tian, J., Tang, W. Z., Shan, X. L., Zhao, P., et al. (2021). Ophiopogonin D alleviates diabetic myocardial injuries by regulating mitochondrial dynamics. J. Ethnopharmacol. 271, 113853. doi:10.1016/j.jep.2021.113853
Li, X. X., Wang, D. Q., Sui, C. G., Meng, F. D., Sun, S. L., Zheng, J., et al. (2020). Oleandrin induces apoptosis via activating endoplasmic reticulum stress in breast cancer cells. Biomed. Pharmacother. 124, 109852. doi:10.1016/j.biopha.2020.109852
Lin, X. K., Wang, L. H., Zhao, L. Q., Zhu, Z., Chen, T. Z., Chen, S. N., et al. (2020). Curcumin micelles suppress gastric tumor cell growth by upregulating ROS generation, disrupting redox equilibrium and affecting mitochondrial bioenergetics. Food Funct. 11, 4146–4159. doi:10.1039/d0fo00260g
Liu, J., He, L., Hu, J., Li, K. R., Zhou, F. L., Hu, M., et al. (2020). Isoimperatorin induces apoptosis of nasopharyngeal carcinoma cells via the MAPK/ERK1/2 signaling pathway. Evid. Based. Complement. Altern. Med. 2020, 2138186. doi:10.1155/2020/2138186
Ma, Z., Zhang, B., Fan, Y. Q., Wang, M., Kebebe, D., Li, J. W., et al. (2019). Traditional Chinese medicine combined with hepatic targeted drug delivery systems: A new strategy for the treatment of liver diseases. Biomed. Pharmacother. 117, 109128. doi:10.1016/j.biopha.2019.109128
Martinez-Serra, J., Gutierrez, A., Munoz-Capo, S., Navarro-Palou, M., Ros, T., Amat, J. C., et al. (2014). xCELLigence system for real-time label-free monitoring of growth and viability of cell lines from hematological malignancies. Onco. Targets. Ther. 7, 985–994. doi:10.2147/OTT.S62887
Maus, M. V., Grupp, S. A., Porter, D. L., and June, C. H. (2014). Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123, 2625–2635. doi:10.1182/blood-2013-11-492231
McKinnon, K. M. (2018). Flow cytometry: An overview. Curr. Protoc. Immunol. 120, 5 1 1–511. doi:10.1002/cpim.40.1
Meindl, C., Absenger, M., Roblegg, E., and Frohlich, E. (2013). Suitability of cell-based label-free detection for cytotoxicity screening of carbon nanotubes. Biomed. Res. Int. 2013, 564804. doi:10.1155/2013/564804
Ormerod, M. G. (2002). Investigating the relationship between the cell cycle and apoptosis using flow cytometry. J. Immunol. Methods 265, 73–80. doi:10.1016/s0022-1759(02)00071-6
Otero-Gonzalez, L., Sierra-Alvarez, R., Boitano, S., and Field, J. A. (2012). Application and validation of an impedance-based real time cell analyzer to measure the toxicity of nanoparticles impacting human bronchial epithelial cells. Environ. Sci. Technol. 46, 10271–10278. doi:10.1021/es301599f
Otto, G., Lamote, A., Deckers, E., Dumont, V., Delahaut, P., Scippo, M. L., et al. (2016). A flow-cytometry-based method for detecting simultaneously five allergens in a complex food matrix. J. Food Sci. Technol. 53, 4179–4186. doi:10.1007/s13197-016-2402-x
Ozawa, S., Okabe, S., and Ishii, S. (2016). Specific single-cell isolation of escherichia colio157 from environmental water samples by using flow cytometry and fluorescence-activated cell sorting. Foodborne Pathog. Dis. 13, 456–461. doi:10.1089/fpd.2016.2125
Patsoukis, N., Bardhan, K., Chatterjee, P., Sari, D., Liu, B., Bell, L. N., et al. (2015). PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692. doi:10.1038/ncomms7692
Pelletier, M., Billingham, L. K., Ramaswamy, M., and Siegel, R. M. (2014). Extracellular flux analysis to monitor glycolytic rates and mitochondrial oxygen consumption. Methods Enzymol. 542, 125–149. doi:10.1016/B978-0-12-416618-9.00007-8
Puchta, H., Dujon, B., and Hohn, B. (1993). Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21, 5034–5040. doi:10.1093/nar/21.22.5034
Puglia, C., Lauro, M. R., Tirendi, G. G., Fassari, G. E., Carbone, C., Bonina, F., et al. (2017). Modern drug delivery strategies applied to natural active compounds. Expert Opin. Drug Deliv. 14, 755–768. doi:10.1080/17425247.2017.1234452
Qiu, R. Z. (2016). Research and application of gene editing technologies:an ethical perspective. J. Med. Philos. 37, 1–7. doi:10.12014/J.ISSN.1002-0772.2016.07a.01
Qu, L., Li, H. S., Jiang, Y. H., and Dong, C. S. (2015). The molecular mechanism of CRISPR/Cas9 system and its application in gene therapy of human diseases. Hereditas 37, 974–982. doi:10.16288/j.yczz.15-109
Ren, J. T., and Zhao, Y. B. (2017). Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell 8, 634–643. doi:10.1007/s13238-017-0410-x
Rhee, J.-K., Jung, Y. C., Kim, K. R., Yoo, J., Kim, J., Lee, Y. J., et al. (2018). Impact of tumor purity on immune gene expression and clustering analyses across multiple cancer types. Cancer Immunol. Res. 6, 87–97. doi:10.1158/2326-6066.Cir-17-0201
Rouet, P., Smih, F., and Jasin, M. (1994). Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 91, 6064–6068. doi:10.1073/pnas.91.13.6064
Sapranauskas, R., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P., and Siksnys, V. (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. doi:10.1093/nar/gkr606
Sashital, D. G., Jinek, M., and Doudna, J. A. (2011). An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 18, 680–687. doi:10.1038/nsmb.2043
Shah, S. A., Erdmann, S., Mojica, F. J., and Garrett, R. A. (2013). Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biol. 10, 891–899. doi:10.4161/rna.23764
Shi, M., Shen, Z., Zhang, N., Wang, L., Yu, C., and Yang, Z. (2021). CRISPR/Cas9 technology in disease research and therapy: A review. Sheng Wu Gong Cheng Xue Bao 37, 1205–1228. doi:10.13345/j.cjb.200401
Shi, S. J., Zhao, Q. R., Ke, C. H., Long, S. R., Zhang, F., Zhang, X., et al. (2021). Loureirin B exerts its immunosuppressive effects by inhibiting STIM1/Orai1 and KV1.3 channels. Front. Pharmacol. 12, 685092. doi:10.3389/fphar.2021.685092
Stefanowicz-Hajduk, J., Adamska, A., Bartoszewski, R., and Ochocka, J. R. (2016). Reuse of E-plate cell sensor arrays in the xCELLigence Real-Time Cell Analyzer. Biotechniques 61, 117–122. doi:10.2144/000114450
Sun, X. Y., and Liu, X. (2021). Effects of osthole on apoptosis and autophagy of human tongue cancer tca8113 cells. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 43, 501–506. doi:10.3881/j.issn.1000-503X.13663
Tang, S. M., Deng, X. T., Zhou, J., Li, Q. P., Ge, X. X., and Miao, L. (2020). Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects. Biomed. Pharmacother. 121, 109604. doi:10.1016/j.biopha.2019.109604
Turker Sener, L., Albeniz, G., Dinc, B., and Albeniz, I. (2017). iCELLigence real-time cell analysis system for examining the cytotoxicity of drugs to cancer cell lines. Exp. Ther. Med. 14, 1866–1870. doi:10.3892/etm.2017.4781
Wang, Q., Hardie, R. A., Hoy, A. J., van Geldermalsen, M., Gao, D., Fazli, L., et al. (2015). Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 236, 278–289. doi:10.1002/path.4518
Wang, S. Y., Wang, X. J., and Ma, J. (2013). Progress in real time xCELLigence analysis system on drug cardiotoxicity screening. Chin. J. Pharmacol. Toxicol. 27, 908–912. doi:10.3867/j.issn.1000-3002.2013.05.026
Wang, T. X., Hu, N., Cao, J. Y., Wu, J. Y., Su, K. Q., and Wang, P. (2013). A cardiomyocyte-based biosensor for antiarrhythmic drug evaluation by simultaneously monitoring cell growth and beating. Biosens. Bioelectron. 49, 9–13. doi:10.1016/j.bios.2013.04.039
Wen, D., Danquah, M., Chaudhary, A. K., and Mahato, R. I. (2015). Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J. Control. Release 219, 237–247. doi:10.1016/j.jconrel.2015.08.011
Wu, D., Wang, W., Chen, W., Lian, F., Lang, L., Huang, Y., et al. (2018). Pharmacological inhibition of dihydroorotate dehydrogenase induces apoptosis and differentiation in acute myeloid leukemia cells. Haematologica 103, 1472–1483. doi:10.3324/haematol.2018.188185
Wu, H., Xie, J. S., Pan, Q. R., Wang, B. B., Hu, D. Q., and Hu, X. (2013). Anticancer agent shikonin is an incompetent inducer of cancer drug resistance. Plos One 8, e52706. doi:10.1371/journal.pone.0052706
Xiang, Y. N., Cuo, Z. M., Zhu, P. F., Chen, J., and Huang, Y. Y. (2019). Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med. 8, 1958–1975. doi:10.1002/cam4.2108
Xie, J., Yang, Z. G., Zhou, C. G., Zhu, J., Lee, R. J., and Teng, L. S. (2016). Nanotechnology for the delivery of phytochemicals in cancer therapy. Biotechnol. Adv. 34 (4), 343–353. doi:10.1016/j.biotechadv.2016.04.002
Xu, X. H., Li, T., Fong, C. M. V., Chen, X. P., Chen, X. J., Wang, Y. T., et al. (2016). Saponins from Chinese medicines as anticancer agents. Molecules 21, 1326. doi:10.3390/molecules21101326
Yan, G., Du, Q., Wei, X., Miozzi, J., Kang, C., Wang, J., et al. (2018). Application of real-time cell electronic analysis system in modern pharmaceutical evaluation and analysis. Molecules 23, 3280. doi:10.3390/molecules23123280
Yan, G. J., Sun, W., Pei, Y. F., Yang, Z. G., Wang, X. B., Sun, Y., et al. (2018). A novel release kinetics evaluation of Chinese compound medicine: Application of the xCELLigence RTCA system to determine the release characteristics of Sedum sarmentosum compound sustained-release pellets. Saudi Pharm. J. 26 (3), 445–451. doi:10.1016/j.jsps.2017.07.007
Yang, L. T., He, L., He, H. N., and Wu, Q. (2017). The application of flow cytometry in testing bacteria of raw milk. J. Guangxi Norm. Univ. 35, 112–116. doi:10.16088/j.issn.1001-6600.2017.02.017
Yang, T. F., Xu, R., Su, Q., Wang, H. Y., Liu, F., Dai, B. L., et al. (2020). Chelerythrine hydrochloride inhibits proliferation and induces mitochondrial apoptosis in cervical cancer cells via PI3K/BAD signaling pathway. Toxicol. Vitro 68, 104965. doi:10.1016/j.tiv.2020.104965
Yang, X. W., Yang, S., Chai, H. Y., Yang, Z. G., Lee, R. J., Liao, W. W., et al. (2015). A novel isoquinoline derivative anticancer agent and its targeted delivery to tumor cells using transferrin-conjugated liposomes. PLoS One 10 (8), e0136649. doi:10.1371/journal.pone.0136649
Yang, Y., Yan, X. J., Bi, L., Chen, S. S., Zhu, J. J., and Chen, W. P. (2014). Optimization of effective component formula from active ingredients of Salvia Miltiorrhiza and Panax Ginseng through orthogonal design method to resist breast cancer. Chin. Pharmacol. Bull. 30, 1605–1611. doi:10.3969/j.issn.1001-1978.2014.11.025
Yi, W., Clark, P. M., Mason, D. E., Keenan, M. C., Hill, C., Goddard, W. A., et al. (2012). Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980. doi:10.1126/science.1222278
Zhang, L., Chen, Y., Shi, R., Rang, T. G., Pang, G. S., Wang, B. R., et al. (2018). Synthesis of hollow nanocages MOF-5 as drug delivery vehicle to solve the load-bearing problem of insoluble antitumor drug oleanolic acid (OA). Inorg. Chem. Commun. 96, 20–23. doi:10.1016/j.inoche.2018.07.029
Zhang, Q., Huang, X., Pu, Y. Q., Yi, Y. X., Zhang, T., and Wang, B. (2018). pH-sensitive and biocompatible quercetin-loaded GO-PEA-HA carrier improved antitumour efficiency and specificity. Artif. Cells Nanomed. Biotechnol. 46, S28–S37. doi:10.1080/21691401.2018.1489261
Zhang, W. G., Yin, X. C., Liu, X. F., Meng, K. W., Tang, K., Huang, F. L., et al. (2017). Puerarin induces hepatocellular carcinoma cell apoptosis modulated by mapk signaling pathways in a dose-dependent manner. Anticancer Res. 37, 4425–4431. doi:10.21873/anticanres.11837
Zhao, Q. Q., Sun, X. Y., Wu, B., Shang, Y. H., Huang, X. Y., Dong, H., et al. (2021). Construction of homologous cancer cell membrane camouflage in a nano-drug delivery system for the treatment of lymphoma. J. Nanobiotechnology 19, 8. doi:10.1186/s12951-020-00738-8
Zheng, Y. H., Wang, Y., Xia, M. Y., Gao, Y., Zhang, L., Song, Y. N., et al. (2022). The combination of nanotechnology and traditional Chinese medicine (TCM) inspires the modernization of TCM: Review on nanotechnology in TCM-based drug delivery systems. Drug Deliv. Transl. Res. 12, 1306–1325. doi:10.1007/s13346-021-01029-x
Zhong, X., Huang, S., Liu, D., Jiang, Z., Jin, Q., Li, C., et al. (2020). Galangin promotes cell apoptosis through suppression of H19 expression in hepatocellular carcinoma cells. Cancer Med. 9, 5546–5557. doi:10.1002/cam4.3195
Zhong, X., Liu, D., Jiang, Z., Li, C., Chen, L., Xia, Y., et al. (2020). Chrysin induced cell apoptosis and inhibited invasion through regulation of tet1 expression in gastric cancer cells. Onco. Targets. Ther. 13, 3277–3287. doi:10.2147/OTT.S246031
Zhou, L., Yu, X., Li, M., Gong, G., Liu, W., Li, T., et al. (2020). Cdh1-mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine 51, 102570. doi:10.1016/j.ebiom.2019.11.031
Keywords: traditional Chinese medicine, nanotechnology, CRISPR/Cas9, real-time cell-based assay, flow cytometry
Citation: Ke G, Zhang J, Gao W, Chen J, Liu L, Wang S, Zhang H and Yan G (2022) Application of advanced technology in traditional Chinese medicine for cancer therapy. Front. Pharmacol. 13:1038063. doi: 10.3389/fphar.2022.1038063
Received: 06 September 2022; Accepted: 28 September 2022;
Published: 13 October 2022.
Edited by:
Xuefeng Li, Guangzhou Medical University, ChinaReviewed by:
Tongzheng Liu, Jinan University, ChinaJingjing Zhang, The Ohio State University, United States
Copyright © 2022 Ke, Zhang, Gao, Chen, Liu, Wang, Zhang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan Zhang, lct@jlu.edu.cn; Guojun Yan, yanguojun@njucm.edu.cn
 Gaofeng Ke1
Gaofeng Ke1