- 1Department of Biological Repositories, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China
- 2Department of Pharmacy, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China
- 3Department of Pharmacy, Xiamen Maluan Bay Hospital, Xiamen, Fujian, China
- 4Animal Biosafety Level III Laboratory, Wuhan University School of Basic Medical Sciences, Wuhan, Hubei, China
- 5Department of Urology, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China
- 6Tumor Precision Diagnosis and Treatment Technology and Translational Medicine, Hubei Engineering Research Center, Wuhan, Hubei, China
- 7School of Pharmaceutical Sciences, Wuhan University, Wuhan, Hubei, China
Cordyceps is a genus of ascomycete fungi and used widely in fungal drugs. However, in-depth studies of the metabolites of wild Cordyceps species and their substituents are lacking. In this study, a liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based metabolomics analysis was carried out to comprehensively profile the metabolites in wild Chinese Cordyceps species (Ophiocordyceps sinensis (Berk.) G.H. Sung, J.M. Sung, Hywel-Jones and Spatafora 2007) from Naqu (NCs) and Yushu (YCs) and their substituents including artificially cultivated Cordyceps species (CCs) and mycelia. A total of 901 metabolites were identified in these samples, including lipids, amino acids, nucleosides, carbohydrates, organic acids, coenzymes, vitamins, alkaloids and their derivatives. Univariate and multivariate statistical analyses revealed remarkable differences and significantly different metabolites among them. Seventy amino acid-relevant metabolites were analyzed quantitatively in four samples for the first time. The four samples contained abundant L-glutamic acid and oxidized glutathione as well as multiple unique amino acid-relevant metabolites (e.g., 3-chloro-L-tyrosine, 6-aminocaproic acid, L-theanine, anserine, γ-glutamyl-cysteine). Collectively, our study provides rich metabolic information of wild Cordyceps species and their substituents, which could facilitate their quality control and optimal utilization.
Introduction
Cordyceps is a genus of ascomycete fungi. Cordyceps species are highly valued tonic foods and well-known fungal drugs. Studies have demonstrated the diverse pharmaceutical effects of Cordyceps species-based products (Kuo et al., 1996; Zhang et al., 2005; Hsu et al., 2008; Yang et al., 2011; Zhang et al., 2014; Mi et al., 2018; Cheng et al., 2020) and revealed various chemical constituents, such as nucleosides, lipids, saccharides, mannitol, and amino acids (Zhao et al., 2014; Liu et al., 2015).
However, Cordyceps species are rarely found in the wild, and their substituents are desired (Li et al., 2019). The fermented mycelia of wild Cordyceps species are popular, and have been made into medical and functional products in the forms of capsules, tablets, or granulations (Dong et al., 2015; Zhao et al., 2020). Besides, Sunshine Lake Pharma (Dongguan, China) has undertaken artificial cultivation of Chinese Cordyceps species. Ophiocordyceps sinensis (Berk.) G.H. Sung, J.M. Sung, Hywel-Jones & Spatafora 2007 (known previously as Cordyceps sinensis) is a fungus that grows on insects in the family Ophiocordycipitaceae (Li et al., 2019). O. sinensis is a form of wild Cordyceps species. The annual yield of cultured Cordyceps species (CCs) reaches tens of tons, accounting for ≥20% of the total natural resource (Li et al., 2019). However, whether wild Cordyceps species (e.g., O. sinensis) and such substituents have chemical homogeneity is controversial.
Metabolomics analysis has become a powerful approach for large-scale study of metabolites in biological samples (Patti et al., 2012). In general, two analytical methods are used to conduct metabolomics analysis: nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) (Perez et al., 2021). Compared with MS, NMR provides unique structural information. Nonetheless, it has limited sensitivity when measuring a large number of metabolites with a wide dynamic range (Lu et al., 2019; Perez et al., 2021). With regard to MS-based metabolic profiling, gas chromatography (GC) and liquid chromatography (LC) are hyphenated to MS for more accurate identification of metabolites. GC-MS often requires further derivatization of metabolites, and makes them more volatile and more thermally stable (Smart et al., 2010; Zhang et al., 2020). Nevertheless, LC-MS can detect thermally labile metabolites directly without derivatization, and is used more widely for metabolite profiling. LC-MS-based metabolomics analysis has been applied to analyze a certain class of metabolites (e.g., lipid, nucleoside, nucleobase, nucleotide, protein) (Fan et al., 2006; Mi et al., 2016; Cheng et al., 2017; Zhang et al., 2018; Jia et al., 2019; Lin et al., 2022) or dozens of metabolites (Wang et al., 2015; Zhang et al., 2015; Chen et al., 2018; He et al., 2019; Yao et al., 2019) in wild Cordyceps species and/or their substituents. However, few studies have profiled hundreds of metabolites in wild Cordyceps species or cultivated Cordyceps species and their mycelia for more comprehensive assessment of their chemical composition and alterations.
In this study, we leveraged liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) to comprehensively survey metabolites in wild Cordyceps species (O. sinensis) from Naqu (Tibet, China; NCs) and Yushu (Qinghai, China; YCs), CCs (O. sinensis), and mycelia (Hirsutella sinensis X.J. Liu, Y.L. Guo, Y.X. Yu & W. Zeng 1989) from Bailing capsules (BLs). Using univariate and multivariate statistical analyses, chemical differences among NCs, YCs, CCs, and BLs were evaluated. Furthermore, differential metabolites among four samples were screened out and enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was undertaken. Besides, 70 amino acid-relevant metabolites in these four samples were analyzed quantitatively. Taken together, this work provides a holistic insight into the metabolic differences in wild Cordyceps species and their substituents.
Materials and methods
Materials and reagents
Wild Cordyceps species (O. sinensis (Berk.) G.H. Sung, J.M. Sung, Hywel-Jones & Spatafora 2007), from Naqu (NCs) and Yushu (YCs) were acquired from Hubei Tianji Pharmaceuticals (Hubei, China) and Qinghai Yuanzu Biological Technology (Qinghai, China), respectively. Artificially cultivated Cordyceps species (O. sinensis; catalog number = 200111-I01) was purchased from Sunshine Lake Pharma (Guangdong, China). BLs containing H. sinensis X.J. Liu, Y.L. Guo, Y.X. Yu & W. Zeng 1989 (1911355) were obtained from Zhongmei East China Pharmaceuticals (Zhejiang, China). Methanol (I1069707004) and acetonitrile (JA095830) were sourced from Merck (Darmstadt, Germany). Formic acid (RH208519) was purchased from Aladdin (Shanghai, China). Standards for quantitative analysis of amino acid-relevant metabolites are shown in Supplementary Table S1. [2H2]L-threonine [IsoReag (Shanghai, China); 21J161-G1] was used as an internal standard and added to calibrate as-prepared Cordyceps samples, compensating for losses during sample preparation or variability during analytical determination.
Liquid chromatography-tandem mass spectrometry for metabolic profiling of wild and cultivated species of Cordyceps and mycelia
To extract metabolites, 50 mg of each sample was first weighed and homogenized with steel balls at 30 Hz for 3 min. Then, 1 ml of a methanol:water mixture (7:3) was added, and the mixture was vortex-mixed for 5 min followed by centrifugation (12,000 rpm, 10 min, 4°C). Subsequently, the supernatant (400 μl) was collected and stored at −20°C overnight. Finally, the supernatant was centrifuged again (12,000 rpm, 3 min, 4°C), and 200 μl of supernatant pipetted for LC-MS.
Chromatographic separation was undertaken on a Shim-pack CBM30A ultra-high-pressure liquid chromatography (UPLC) system (Shimadzu, Kyoto, Japan). An Acquity HSS T3 C18 column (2.1 mm × 100 mm, 1.8 μm; Waters, Milford, MA, United States) was used for LC and its temperature were maintained at 40°C. Mobile phase A was H2O with 0.1% formic acid. Mobile phase B was acetonitrile with 0.1% formic acid. Gradient elution was: 0–10 min, 5%–90% B; 10–11 min, 90% B; 11–11.1 min, 90%–5% B; 11.1–14 min, 5% B. The flow rate was 0.35 ml/min and the injection volume was 2 μl.
A Triple Quad™ 6500+ MS (Sciex, Framingham, MA, United States) instrument was operated in electrospray ionization in positive (ESI+) and negative (ESI−) modes with the following parameters: positive ion spray voltage, 5500 V; negative ion spray voltage, −4500 V; source temperature, 500°C; ion source gas 1, 50 psi; ion source gas 2, 50 psi; curtain gas, 25 psi; collision gas, high. A specific set of multiple reaction monitoring (MRM) transitions was recorded for each period according to the metabolites eluted within this period.
Liquid chromatography-tandem mass spectrometry for quantifying amino acid-relevant metabolites
Similarly, 50 mg of each sample was weighed and homogenized with steel balls at 30 Hz for 3 min. Then, 500 µl of a methanol:water (7:3) mixture containing [2H2]L-threonine was added. The mixture was vortex-mixed for 5 min followed by centrifugation (12,000 rpm, 10 min, 4°C). Subsequently, the supernatant (300 μl) was collected and stored at −20°C for 30 min. Finally, the supernatant was centrifuged again (12,000 rpm, 10 min, 4°C) and 200 μl of the supernatant was pipetted for LC-MS.
Chromatographic separation was achieved on an ExionLC AD UPLC system (Sciex) with an Acquity BEH Amide column (2.1 mm × 100 mm, 1.7 μm). The column temperature was maintained at 40°C. Mobile phase A was H2O with ammonium acetate (2 mM) and 0.04% formic acid. Mobile phase B was acetonitrile with ammonium acetate (2 mM) and 0.04% formic acid. Gradient elution was: 0–1.2 min, 90% B; 1.2–9 min, 90%–60% B; 9–10 min, 60%–40% B; 10–11 min, 40% B; 11–15 min, 40%–90% B. The flow rate was 0.35 ml/min, and the injection volume was 2 μl.
Acquisition of MS data was undertaken on a Triple Quad™ 6500+ MS system (Sciex) in MRM mode with the following parameters: positive ion spray voltage, 5500 V; negative ion spray voltage, −4500 V; source temperature, 550°C; ion source gas 1, 50 psi; ion source gas 2, 50 psi; curtain gas, 35 psi; collision gas, medium.
Data processing
Metabolites were identified by matching the retention time, precursor ions/product ions, and MS/MS spectra to the self-compiled MetWare database (MetWare Biological Science and Technology, Wuhan, China) and public databases [METLIN (https://massconsortium.com/), Human Metabolome Database (HMDB; https://hmdb.ca/), KEGG (www.genome.jp/)] using Analyst 1.6.3 (Sciex). The integration and calibration of chromatographic peaks were done with MultiQuant 3.0.3 (Sciex).
Statistical analyses
Principal component analysis (PCA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) were conducted in R (R Institute for Statistical Computing, Vienna, Austria) using the statistics function “prcomp” and “MetaboAnalystR” package, respectively. Hierarchical clustering analysis (HCA) and the Pearson correlation coefficient were carried out by the R package “ComplexHeatmap.”
Results and discussion
Liquid chromatography-tandem mass spectrometry-based wide-targeted metabolomics analysis for the characterization of metabolites in wild Cordyceps species and their substituents
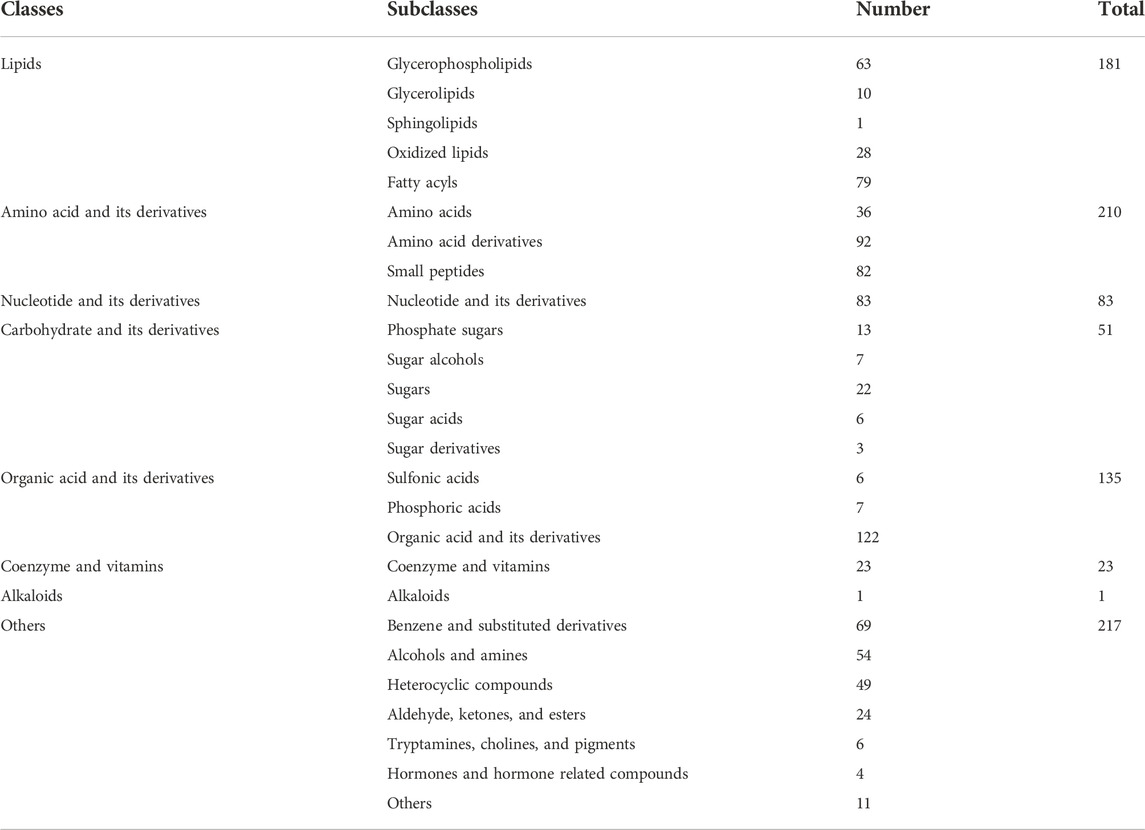
LC-MS-based strategies for metabolomics analysis can be targeted or untargeted (Patti et al., 2012). Untargeted metabolomics analysis is dependent upon high-resolution MS to measure metabolites without bias. Typically, targeted metabolomics analysis uses MRM on MS/MS to analyze a priori defined metabolites with high sensitivity and quantitative reproducibility. In this work, a LC-MS/MS-based wide-targeted metabolomics analysis was employed to comprehensively identify metabolites and assess their alterations in wild Chinese Cordyceps species (O. sinensis (Berk.) G.H. Sung, J.M. Sung, Hywel-Jones & Spatafora 2007; NCs and YCs) and their substituents, including artificially cultivated Cordyceps species (CCs) and fermented mycelia (BLs). Total ion chromatographs of sample metabolomic analysis in ESI+ and ESI− modes are shown in Supplementary Figure S1. A total of 901 metabolites were detected in pooled quality control (QC) samples comprising extracts of NCs, YCs, CCs, and BLs. Table 1 shows that the chemical constituents of samples included lipids, amino acids, nucleotides, carbohydrates, organic acids, vitamins, and their derivatives or other small molecules. Notably, more lipids, amino acids, organic acids, and their derivatives were detected.
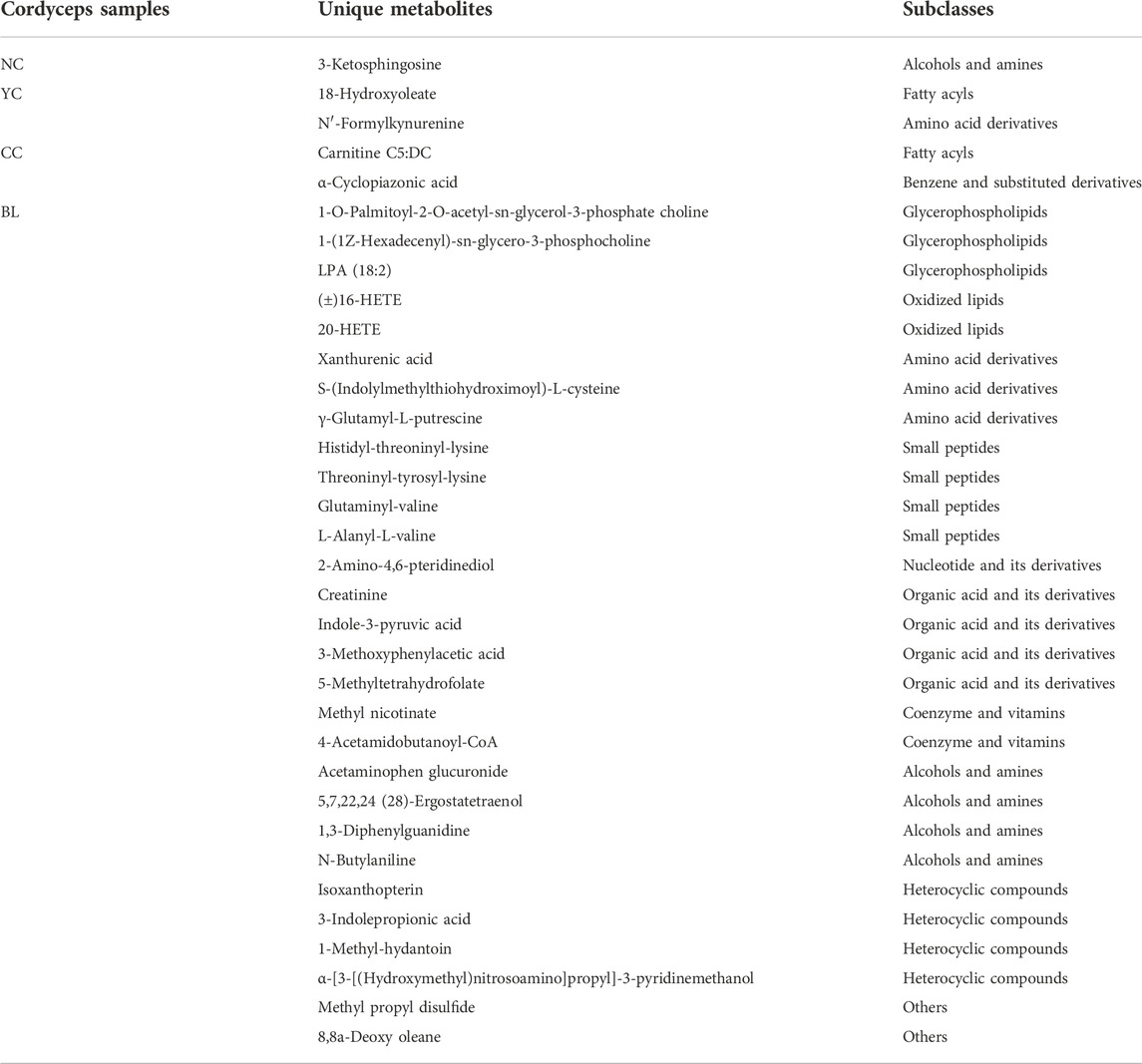
Specifically, in ESI + mode, 453, 466, 453, and 452 metabolites were detected in NCs, YCs, CCs, and BLs, respectively. In ESI− mode, 389, 385, 379, and 382 metabolites were detected in NCs, YCs, CCs, and BLs, respectively (Figure 1). Detailed information of metabolites detected in NCs, YCs, CCs, and BLs is displayed in Supplementary Table S2. A Venn diagram highlighted that 753 out of 901 metabolites were detected in these four samples (Figure 1B). Importantly, some widely used bioactive constituents or biomarkers were identified in all samples, such as nucleosides (e.g., cordycepin, adenosine), ergosterol, D-mannitol, and amino acids (e.g., tryptophan, glutamic acid) (Li et al., 2006; Yue et al., 2013; Zhao et al., 2014; Liu et al., 2015). Besides, 1, 2, 2, and 29 metabolites were only detected in NCs, YCs, CCs, and BLs, respectively (Figure 1B; Table 2). Collectively, we detected a wide variety of metabolites with large amounts in wild and cultivated Cordyceps species and mycelia, which facilitated comprehensive assessment of their differences.
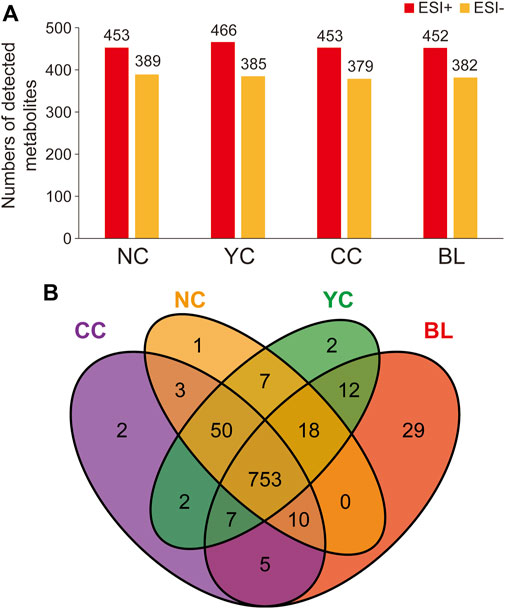
FIGURE 1. Metabolites identified in NCs, YCs, CCs, and BLs. (A) Numbers of metabolites detected from NCs, YCs, CCs, and BLs in ESI+ and ESI- modes, respectively. (B) Venn diagram of metabolites detected from NCs, YCs, CCs, and BLs.
Multivariate statistical analyses for assessment of differences between wild Cordyceps species and their substituents
To characterize differences between wild Cordyceps species and their substituents comprehensively, multivariate statistical analyses (PCA, OPLS-DA, clustering analysis, correlation analysis) were employed. Metabolites measured in QC samples with relative standard deviation <30% were used for subsequent analyses (Supplementary Table S2). Results of PCA (Figure 2A) and OPLS-DA (Figures 2B–F) clearly showed distinct sample clusters corresponding to NCs, YCs, CCs, and BLs, respectively. Besides, clustering analyses (Figure 2G) and correlation analyses (Figure 2H) indicated differential metabolites among NCs, YCs, CCs, and BLs. Notably, the differences between CCs and wild Cordyceps species (NCs and YCs) were less than those between BLs and wild Cordyceps species. The similarity between CCs and NCs was higher than that between CCs and YCs (Figure 2H). Taken together, these results suggested that metabolic differences were present among NCs, YCs, CCs, and BLs.
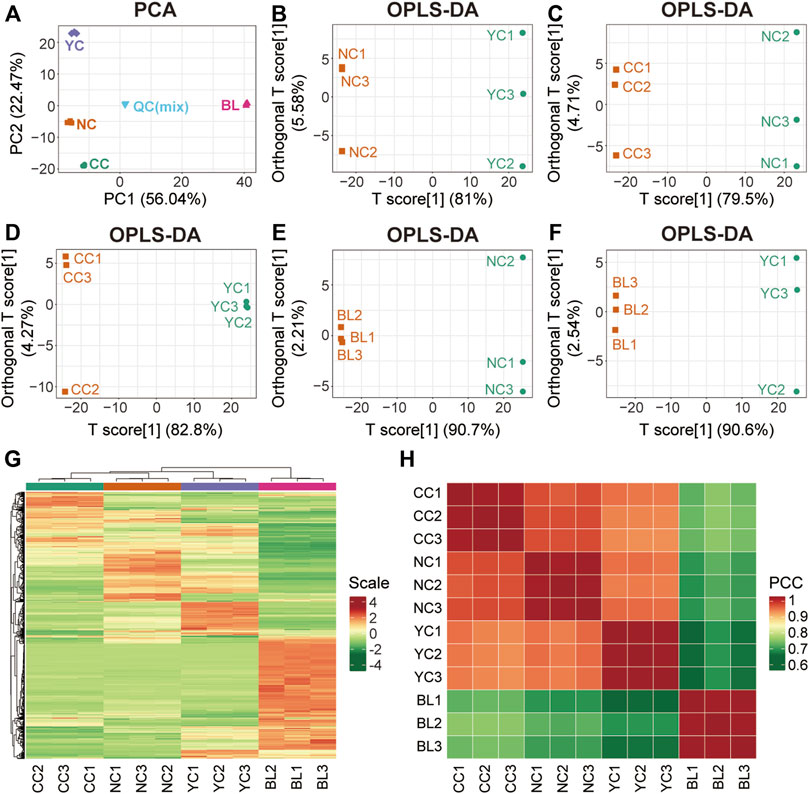
FIGURE 2. Multivariate statistical analyses of metabolites detected in NCs, YCs, CCs, and BLs. (A) PCA result. (B–F) Plots showing OPLS-DA scores. (G) Heatmap of hierarchical clustering analysis. (H) Heatmap of correlation analysis. PCC, Pearson’s correlation coefficient.
Differential metabolites between Cordyceps from Naqu and Yushu
Molecular features with variable importance in projection (VIP) ≥1 and fold change (FC) ≥2 or ≤0.5 were defined as significantly differential metabolites between two samples of Cordyceps species. A total of 365 differential metabolites (183 were up-regulated and 182 were down-regulated) between NCs and YCs were documented (Figure 3A). Furthermore, except for one alkaloid, all detected metabolite classes had an upward or downward trend in level (Supplementary Table S3). Furthermore, enrichment analysis using the KEGG database displayed that more differential metabolites between NCs and YCs were associated with biosynthesis of cofactors (Figure 3B). However, nine glycerolipids (MG (18:2 (9Z, 12Z)/0:0/0:0) [rac], glycidyl oleate, MAG (16:1) isomer, MAG (16:1), MG (20:5) isomer, MG (18:1 (9Z)/0:0/0:0) [rac], glycine linoleate, (±) 1,2-sebacyl glycerol (10:0), 2-palmitoyl-rac-glycerol) and one sphingolipid (Sphinganine 1-phosphate) were detected in both types of wild Cordyceps species. Besides, the contents of bioactive cordycepin (i.e., 3′-deoxyadenosine) and D-mannitol (i.e., cordycepic acid) were higher in NCs than YCs. Collectively, these data suggested that wild Cordyceps species from different habitats had some differential metabolites, indicating that local unique ecological factors may affect the chemical constituents of wild Cordyceps species.
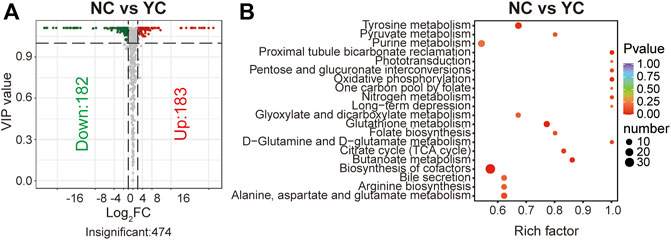
FIGURE 3. Differential analyses between NCs and YCs. (A) Volcano plot of differential metabolites between NCs and YCs. Differential metabolites were screened based on VIP ≥1 and FC ≥ 2 or ≤0.5. The fold change was the ratio of NCs to YCs. (B) Enrichment analysis of differential metabolites between NCs and YCs using the KEGG database. The top-20 enriched metabolic pathways (with p-values) are displayed.
Differential metabolites between wild and cultivated species of Cordyceps
Similarly, 356 significant differential metabolites (95 were up-regulated and 261 were down-regulated) were found between CCs and NCs, and 452 remarkably altered metabolites (147 were up-regulated and 305 were down-regulated) were found between CCs and YCs (VIP ≥1 and FC ≥ 2 or ≤0.5) (Figures 4A,B). Among them, there were 258 overlapped differential metabolites (including lipids, amino acids, nucleotides) between CCs and wild Cordyceps species (Figure 4C) and they are emboldened in Supplementary Tables S4, S5. Of note, the content of ergosterol was lower in CCs than in both types of wild Cordyceps species. Besides, six sugar acids (D-glucoronic acid, L-gulonolactone, D-glucarate, L-arabinonic acid-1,4-lactone, mucic acid, gluconic acid), and two sugar derivatives (N-acetyl-D-glucosamine, methyl beta-D-galactopyranoside) detected in wild and cultivated species of Cordyceps displayed no differences. Moreover, enrichment analyses using the KEGG database clearly showed that more significant differential metabolites between CCs and wild Cordyceps species were associated mainly with purine metabolism (Figures 4D,E). These results showed that CCs and wild Cordyceps species had different chemical constituents. On the one hand, this finding will provide promising indicators to discriminate artificially cultivated Cordyceps species from wild Cordyceps species (Zhang et al., 2015). On the other hand, this finding may encourage study of whether these differential metabolites between CCs and wild Cordyceps species are relevant to differences in their pharmacology.
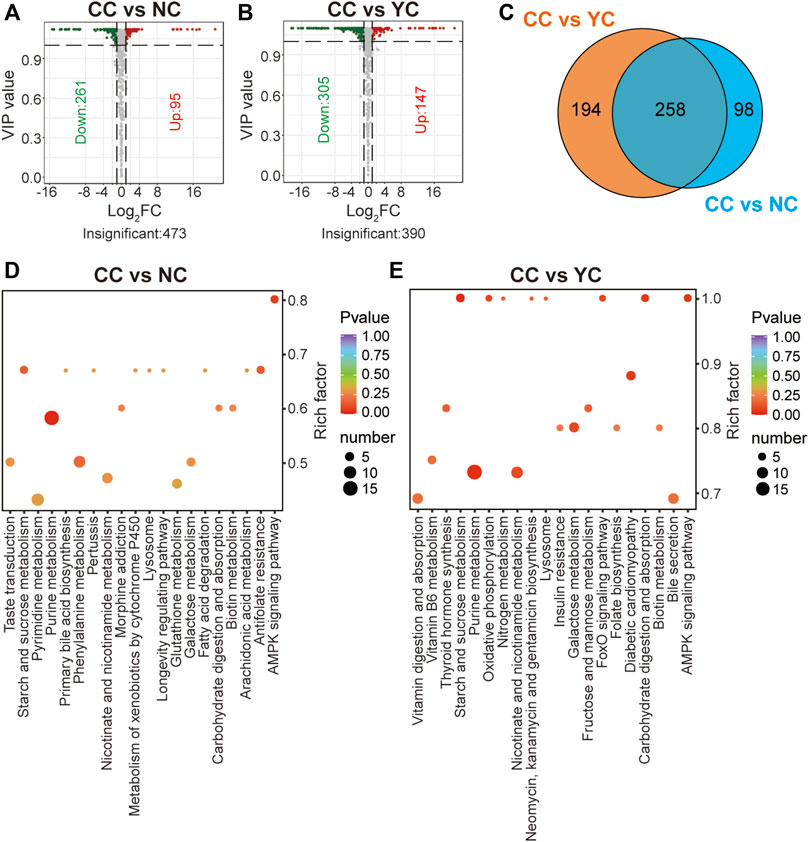
FIGURE 4. Differential analyses between CCs and wild Cordyceps species. (A) Volcano plot of differential metabolites between CCs and NCs. Differential metabolites were screened based on VIP ≥1 and FC ≥ 2 or ≤0.5. The fold change was the ratio of CCs to NCs. (B) Volcano plot of differential metabolites between CCs and YCs. Differential metabolites were screened based on VIP ≥1 and FC ≥ 2 or ≤0.5. The fold change was the ratio of CCs to YCs. (C) Venn diagram of differential metabolites between CCs and wild Cordyceps species. (D) Enrichment analysis of differential metabolites between CCs and NCs using the KEGG database. The top-20 KEGG enriched metabolic pathways (with p-values) are displayed. (E) Enrichment analysis of differential metabolites between CCs and YCs using the KEGG database. The top-20 KEGG enriched metabolic pathways (with p-values) are displayed.
Differential metabolites between bailing capsules and wild Cordyceps species
Compared with NCs, 625 metabolites were altered significantly in BLs (VIP ≥1; FC ≥ 2 or ≤0.5; 334 were up-regulated; 291 were down-regulated) (Figure 5A; Supplementary Table S6). Besides, 598 significantly different metabolites (324 were up-regulated and 274 were down-regulated) were found between BLs and YCs (Figure 5B; Supplementary Table S7). Figure 5C shows 522 overlapped differential metabolites between BLs and wild Cordyceps species, and they are emboldened in Supplementary Tables S6, S7. There were more differential metabolites (including lipids, amino acids, nucleotides) between BLs and wild Cordyceps species (Figures 5A,B) than those between CCs and wild Cordyceps species (Figures 4A,B). For the well-known bioactive constituents, the cordycepin content was higher in BLs than in wild Cordyceps species, whereas the ergosterol content was lower in BLs. Likewise, these differential metabolites between BLs and wild Cordyceps species could facilitate discrimination of adulteration in the processed products of wild Cordyceps species (Zhang et al., 2020). Besides, enrichment analyses using the KEGG database showed that differential metabolites between BLs and NCs and between BLs and YCs were located to different pathways (Figures 5D,E), which indicated that wild Cordyceps species from different habitats had metabolomic differences.
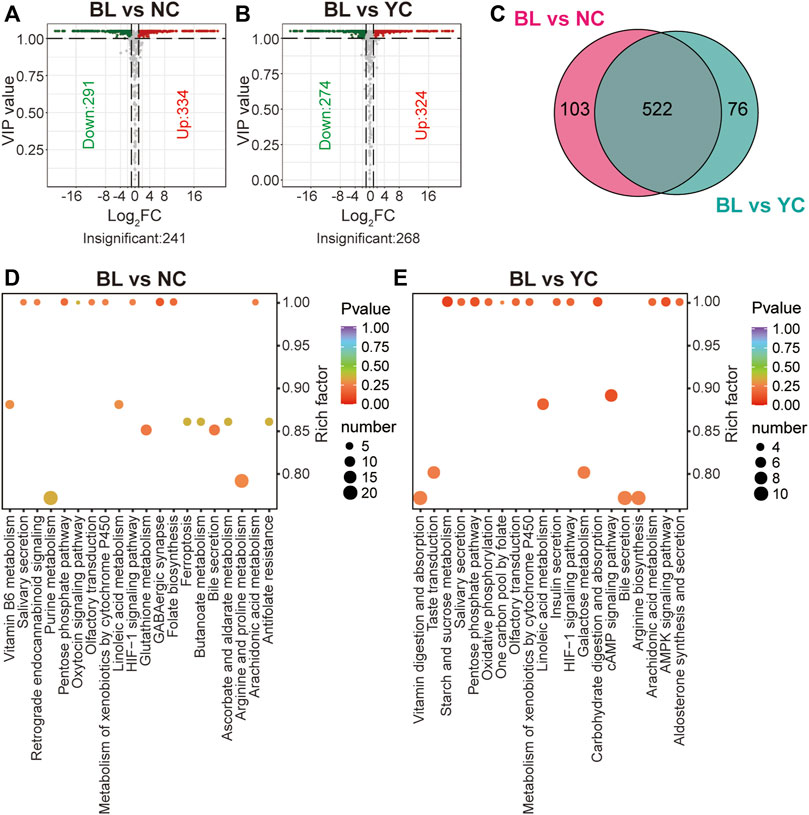
FIGURE 5. Differential analyses between BLs and wild Cordyceps species (A) Volcano plot of differential metabolites between BLs and NCs. Differential metabolites were screened based on VIP ≥1 and FC ≥ 2 or ≤0.5. The fold change was the ratio of BLs to NCs. (B) Volcano plot of differential metabolites between BLs and YCs. Differential metabolites were screened based on VIP ≥1 and FC ≥ 2 or ≤0.5. The fold change was the ratio of BLs to NCs. (C) Venn diagram of differential metabolites between BLs and wild Cordyceps species. (D) Enrichment analysis of differential metabolites between BLs and NCs using the KEGG database. The top-20 enriched pathways (with p-values) are displayed. (E) Enrichment analysis of differential metabolites between BLs and YCs using the KEGG database. The top-20 enriched pathways (with p-values) are displayed.
Quantitative analyses of amino acid-relevant metabolites in wild Cordyceps species and their substituents
Amino acids are important chemical and bioactive constituents in Cordyceps species (Zhang et al., 1991; Liu et al., 2015; Zhang et al., 2022) but have not been characterized comprehensively previously. Here, 70 amino acids and their relevant metabolites in four samples were analyzed quantitatively. Figures 6A,B show the quantitative results of some amino acids and their derivatives. Figure 6C displays the quantitative results of 11 small peptides. Compared with widely targeted metabolic analyses (Supplementary Figure S2), most of these amino acid-relevant metabolites detected by targeted quantitative analyses showed consistent trends in alteration across four samples (Figure 6). However, 11 amino acids and their derivatives were not detected by widely targeted metabolic analysis (nicotinuric acid, 5-hydroxy-tryptophan, homoserine, 3-iodo-L-tyrosine, 3-chloro-L-tyrosine, N-acetylaspartate, L-leucine, L-theanine, argininosuccinic acid, 2-aminobutyric acid, 4-acetamidobutyric acid) (Supplementary Figures S2A,B). Targeted quantitative analyses showed such undetected metabolites with relatively low abundance (Figures 6A,B).
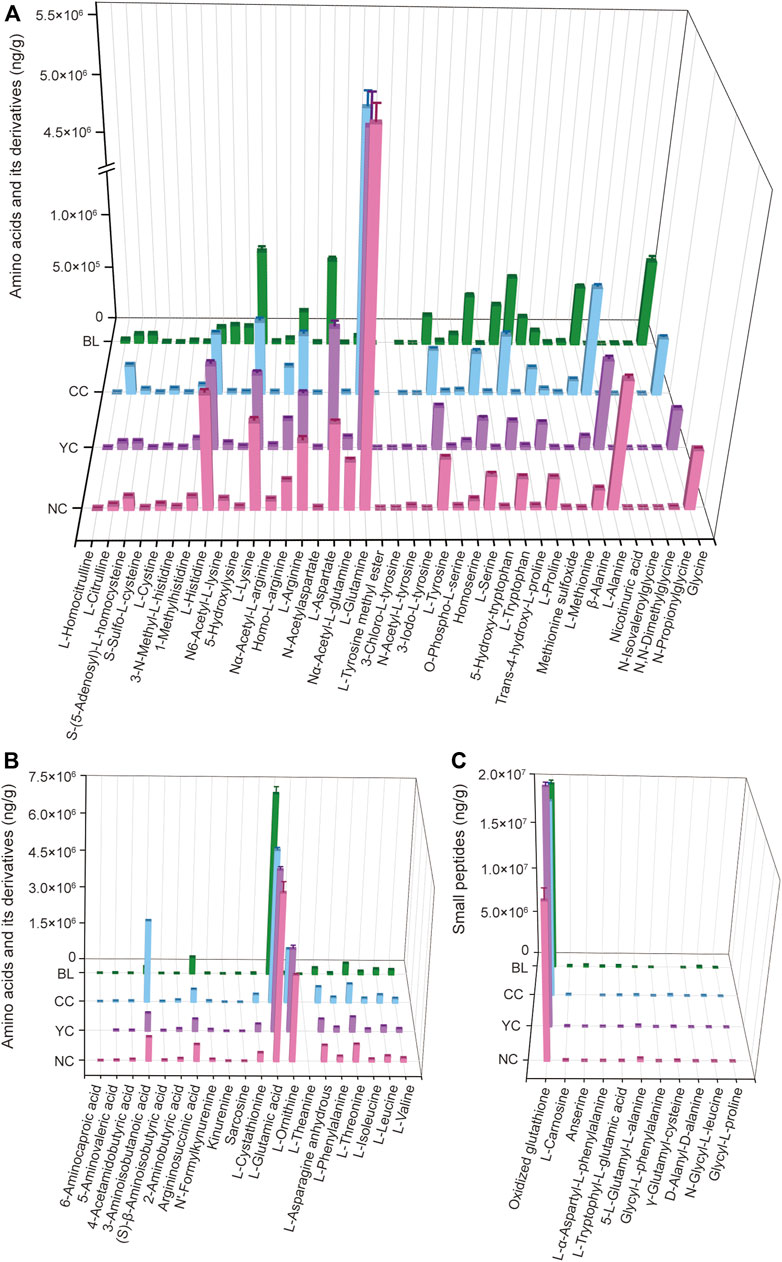
FIGURE 6. Absolute quantitative results of some amino acids and their derivatives (A,B) and small peptides (C) in wild and cultivated Cordyceps species and BLs.
Additionally, absolute quantitative results showed that L-glutamic acid and oxidized glutathione had a high level in wild and cultivated species of Cordyceps and mycelia (Figures 6B,C). Notably, the content of L-glutamine and L-ornithine was significantly higher in wild and cultivated species of Cordyceps than that in BLs (Figure 6A). Furthermore, some unique amino acid-relevant metabolites were noted: 1) 3-chloro-L-tyrosine was not detected in CCs and BLs, but was present in wild Cordyceps species (Figure 6A); 2) 6-aminocaproic acid was not detected in YCs but was present in NCs, CCs, and BLs (Figure 6B); 3) L-theanine was not detected in wild and cultivated species of Cordyceps but was present in BLs (Figure 6B); 4) anserine and γ-glutamyl-cysteine were not detected in CCs and BLs, respectively (Figure 6C).
Conclusion
In summary, this work performed a comparative metabolic profiling to comprehensively characterize metabolites and assess their alterations in wild Cordyceps species (O. sinensis (Berk.) G.H. Sung, J.M. Sung, Hywel-Jones & Spatafora 2007) and their substituents. LC-MS/MS-based widely targeted approach measured 901 metabolites in Cordyceps samples, including lipids, amino acids, nucleosides, carbohydrates, organic acids, coenzymes, vitamins, alkaloids and their derivatives. Univariate and multivariate statistical analyses revealed metabolic differences among wild Cordyceps species from different habitats, cultivated Cordyceps species, and mycelia, and covered all the detected metabolite classes. Enrichment analyses using the KEGG database clearly showed differential metabolic pathways among four samples. Importantly, some amino acid-relevant metabolites were found to be unique to wild Cordyceps species (e.g., 3-chloro-L-tyrosine) or their substituents (e.g., L-theanine). These differences revealed among wild and cultivated Cordyceps species and mycelia could facilitate rational utilization and better QC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
SG and ML designed the experiments, analyzed the data, and prepared the manuscript. DX, WZ, and MZ carried out the experiments and recorded the data. LZ optimized the language of the manuscript. SL and HH revised the manuscript. All authors agreed to submission of the final version of the manuscript.
Funding
This work was supported by the Traditional Chinese Medicine Research Project of Hubei Provincial Health Commission (ZY 2019M033), Science and Technology Innovation Cultivation Fund of Zhongnan Hospital of Wuhan University (znpy2019093), and Program of Excellent Doctoral (Postdoctoral) of Zhongnan Hospital of Wuhan University (ZNYB2019015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1036589/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Total ion chromatograms of LC-MS/MS-based metabolic analysis of Cordyceps samples in positive and negative ion modes.
SUPPLEMENTARY FIGURE S2 | Peak areas of amino acids and their derivatives (A,B) and small peptides (C) detected in NCs, YCs, CCs, and BLs.
References
Chen, L., Liu, Y., Guo, Q., Zheng, Q., and Zhang, W. (2018). Metabolomic comparison between wild Ophiocordyceps sinensis and artificial cultured Cordyceps militaris. Biomed. Chromatogr. 32, e4279. doi:10.1002/bmc.4279
Cheng, W., Zhang, X., Song, Q., Lu, W., Wu, T., Zhang, Q., et al. (2017). Determination and comparative analysis of 13 nucleosides and nucleobases in natural fruiting body of Ophiocordyceps sinensis and its substitutes. Mycology 8 (4), 318–326. doi:10.1080/21501203.2017.1385546
Cheng, J., Song, J., Wei, H., Wang, Y., Huang, X., Liu, Y., et al. (2020). Structural characterization and hypoglycemic activity of an intracellular polysaccharide from Sanghuangporus sanghuang mycelia. Int. J. Biol. Macromol. 164, 3305–3314. doi:10.1016/j.ijbiomac.2020.08.202
Dong, C., Guo, S., Wang, W., and Liu, X. (2015). Cordyceps industry in China. Mycology 6 (2), 121–129. doi:10.1080/21501203.2015.1043967
Fan, H., Li, S., Xiang, J., Lai, C., Yang, F., Gao, J., et al. (2006). Qualitative and quantitative determination of nucleosides, bases and their analogues in natural and cultured Cordyceps by pressurized liquid extraction and high performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC–ESI–MS/MS). Anal. Chim. Acta X. 567 (2), 218–228. doi:10.1016/j.aca.2006.03.032
He, Y., Zhang, W., Peng, F., Lu, R., Zhou, H., Bao, G., et al. (2019). Metabolomic variation in wild and cultured cordyceps and mycelia of Isaria cicadae. Biomed. Chromatogr. 33 (4), e4478. doi:10.1002/bmc.4478
Hsu, C. H., Sun, H. L., Sheu, J. N., Ku, M. S., Hu, C. M., Chan, Y., et al. (2008). Effects of the immunomodulatory agent Cordyceps militaris on airway inflammation in a mouse asthma model. Pediatr. Neonatol. 49 (5), 171–178. doi:10.1016/S1875-9572(09)60004-8
Jia, W., Shi, L., Zhang, F., Chang, J., and Chu, X. (2019). High-throughput screening of the nucleosides and nucleotides using characteristic structural fragments fusion. J. Pharm. Biomed. Anal. 175, 112787. doi:10.1016/j.jpba.2019.112787
Kuo, Y. C., Tsai, W. J., Shiao, M. S., Chen, C. F., and Lin, C. Y. (1996). Cordyceps sinensis as an immunomodulatory agent. Am. J. Chin. Med. 24 (2), 111–125. doi:10.1142/S0192415X96000165
Li, S. P., Yang, F. Q., and Tsim, K. W. K. (2006). Quality control of Cordyceps sinensis, a valued traditional Chinese medicine. J. Pharm. Biomed. Anal. 41 (5), 1571–1584. doi:10.1016/j.jpba.2006.01.046
Li, X., Liu, Q., Li, W., Li, Q., Qian, Z., Liu, X., et al. (2019). A breakthrough in the artificial cultivation of Chinese cordyceps on a large-scale and its impact on science, the economy, and industry. Crit. Rev. Biotechnol. 39 (2), 181–191. doi:10.1080/07388551.2018.1531820
Lin, M., Guo, S., Xie, D., Li, S., and Hu, H. (2022). Lipidomic profiling of wild cordyceps and its substituents by liquid chromatography-electrospray ionization-tandem mass spectrometry. Lwt-Food Sci. Technol. 163, 113497. doi:10.1016/j.lwt.2022.113497
Liu, Y., Wang, J., Wang, W., Zhang, H., Zhang, X., and Han, C. (2015). The chemical constituents and pharmacological actions of Cordyceps sinensis. Evid. Based. Complement. Altern. Med. 2015, 575063. doi:10.1155/2015/575063
Lu, Y., Zhi, Y., Miyakawa, T., and Tanokura, M. (2019). Metabolic profiling of natural and cultured Cordyceps by NMR spectroscopy. Sci. Rep. 9 (1), 7735. doi:10.1038/s41598-019-44154-x
Mi, J. N., Wang, J. R., and Jiang, Z. H. (2016). Quantitative profiling of sphingolipids in wild Cordyceps and its mycelia by using UHPLC-MS. Sci. Rep. 6, 20870. doi:10.1038/srep20870
Mi, J., Han, Y., Xu, Y., Kou, J., Li, W. J., Wang, J. R., et al. (2018). Deep profiling of immunosuppressive glycosphingolipids and sphingomyelins in wild cordyceps. J. Agric. Food Chem. 66 (34), 8991–8998. doi:10.1021/acs.jafc.8b02706
Patti, G. J., Yanes, O., and Siuzdak, G. (2012). Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13 (4), 263–269. doi:10.1038/nrm3314
Perez, D. S. L., Alseekh, S., Scossa, F., and Fernie, A. R. (2021). Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 18 (7), 733–746. doi:10.1038/s41592-021-01116-4
Smart, K. F., Aggio, R. B., Van Houtte, J. R., and Villas-Boas, S. G. (2010). Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 5 (10), 1709–1729. doi:10.1038/nprot.2010.108
Wang, J., Kan, L., Nie, S., Chen, H., Cui, S. W., Phillips, A. O., et al. (2015). A comparison of chemical composition, bioactive components and antioxidant activity of natural and cultured Cordyceps sinensis. LWT - Food Sci. Technol. 63 (1), 2–7. doi:10.1016/j.lwt.2015.03.109
Yang, M. L., Kuo, P. C., Hwang, T. L., and Wu, T. S. (2011). Anti-inflammatory principles from Cordyceps sinensis. J. Nat. Prod. 74 (9), 1996–2000. doi:10.1021/np100902f
Yao, C. L., Qian, Z. M., Tian, W. S., Xu, X. Q., Yan, Y., Shen, Y., et al. (2019). Profiling and identification of aqueous extract of Cordyceps sinensis by ultra-high performance liquid chromatography tandem quadrupole-orbitrap mass spectrometry. Chin. J. Nat. Med. 17 (8), 631–640. doi:10.1016/S1875-5364(19)30066-4
Yue, K., Ye, M., Zhou, Z., Sun, W., and Lin, X. (2013). The genus cordyceps: A chemical and pharmacological review. J. Pharm. Pharmacol. 65 (4), 474–493. doi:10.1111/j.2042-7158.2012.01601.x
Zhang, S. S., Zhang, D. S., Zhu, T. J., and Chen, X. Y. (1991). A pharmacological analysis of the amino acid components of Cordyceps sinensis Sacc. Yao Xue Xue Bao 26 (5), 326–330. doi:10.16438/j.0513-4870.1991.05.002
Zhang, W., Yang, J., Chen, J., Hou, Y., and Han, X. (2005). Immunomodulatory and antitumour effects of an exopolysaccharide fraction from cultivated Cordyceps sinensis (Chinese caterpillar fungus) on tumour-bearing mice. Biotechnol. Appl. Biochem. 42, 9–15. doi:10.1042/BA20040183
Zhang, H. W., Lin, Z. X., Tung, Y. S., Kwan, T. H., Mok, C. K., Leung, C., et al. (2014). Cordyceps sinensis (a traditional Chinese medicine) for treating chronic kidney disease. Cochrane Database Syst. Rev. 12, CD008353. doi:10.1002/14651858.CD008353.pub2
Zhang, J., Wang, P., Wei, X., Li, L., Cheng, H., Wu, Y., et al. (2015). A metabolomics approach for authentication of Ophiocordyceps sinensis by liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Food Res. Int. 76, 489–497. doi:10.1016/j.foodres.2015.07.025
Zhang, X., Liu, Q., Zhou, W., Li, P., Alolga, R. N., Qi, L. W., et al. (2018). A comparative proteomic characterization and nutritional assessment of naturally- and artificially-cultivated Cordyceps sinensis. J. Proteomics 181, 24–35. doi:10.1016/j.jprot.2018.03.029
Zhang, J., Yu, H., Li, S., Zhong, X., Wang, H., and Liu, X. (2020). Comparative metabolic profiling of Ophiocordyceps sinensis and its cultured mycelia using GC-MS. Food Res. Int. 134, 109241. doi:10.1016/j.foodres.2020.109241
Zhang, Y., Liu, J., Wang, Y., Sun, C., Li, W., Qiu, J., et al. (2022). Nucleosides and amino acids, isolated from Cordyceps sinensis, protected against cyclophosphamide-induced myelosuppression in mice. Nat. Prod. Res., 1–4. doi:10.1080/14786419.2022.2043307
Zhao, J., Xie, J., Wang, L. Y., and Li, S. P. (2014). Advanced development in chemical analysis of Cordyceps. J. Pharm. Biomed. Anal. 87, 271–289. doi:10.1016/j.jpba.2013.04.025
Keywords: wild Cordyceps species, cultivated Cordyceps species, mycelia, metabolic profiling, liquid chromatography-tandem mass spectrometry
Citation: Guo S, Lin M, Xie D, Zhang W, Zhang M, Zhou L, Li S and Hu H (2022) Comparative metabolic profiling of wild Cordyceps species and their substituents by liquid chromatography-tandem mass spectrometry. Front. Pharmacol. 13:1036589. doi: 10.3389/fphar.2022.1036589
Received: 04 September 2022; Accepted: 14 November 2022;
Published: 24 November 2022.
Edited by:
Kah Keng Wong, Universiti Sains Malaysia Health Campus, MalaysiaReviewed by:
Jianing Mi, Guangdong Provincial Hospital of Chinese Medicine, ChinaRichou Han, Guangdong Institute of Applied Biological Resources, China
Copyright © 2022 Guo, Lin, Xie, Zhang, Zhang, Zhou, Li and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhou, emhvdWxpX2plcnJ5QHdodS5lZHUuY24=; Sheng Li, bGlzaGVuZy16bnl5QHdodS5lZHUuY24=; Hankun Hu, aHVoYW5rdW5Ad2h1LmVkdS5jbg==
†These authors have contributed equally to this work
 Shan Guo1†
Shan Guo1† Wenqing Zhang
Wenqing Zhang Li Zhou
Li Zhou Sheng Li
Sheng Li Hankun Hu
Hankun Hu