
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 17 November 2022
Sec. Ethnopharmacology
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1032741
This article is part of the Research TopicA New Frontier for Traditional Medicine Research - Multi-Omics ApproachesView all 13 articles
 Yingying Miao1
Yingying Miao1 Qin Zhang1
Qin Zhang1 Zihang Yuan1
Zihang Yuan1 Jie Wang1
Jie Wang1 Yunxia Xu1
Yunxia Xu1 Yuanyuan Chai1
Yuanyuan Chai1 Min Du1
Min Du1 Qinwei Yu1
Qinwei Yu1 Luyong Zhang1,2*
Luyong Zhang1,2* Zhenzhou Jiang1,3*
Zhenzhou Jiang1,3*Tripterygium wilfordii multiglycoside (GTW), extracted and purified from the peeled roots of T. wilfordii Hook.f. (TwHF), is a well-known traditional Chinese medicine and applied to various autoimmune diseases clinically. However, it has been reported to cause severe liver injury. At present, the mechanism underlying GTW-induced hepatotoxicity remain poorly defined. Here, we evaluated the effects of GTW on mouse liver and elucidated the associated mechanisms via label-free proteomics combined with bioinformatics analysis. Male C57BL/6J mice were randomly divided into normal group, a low-dose GTW (70 mg/kg) group and a high-dose GTW (140 mg/kg) group. After 1-week administration, GTW dose-dependently induced hepatotoxicity. Further analysis showed that GTW could act on the intestinal immune network for IgA production pathway, which plays an important role in maintaining intestinal homeostasis and influences the crosstalk between gut and liver. Western blots confirmed that GTW could decrease pIgR protein expression in the liver and ileum, and, as a result, the secretion of IgA into gut lumen was reduced. Further validation showed that intestinal barrier integrity was impaired in GTW-treated mice, promoting bacteria transferring to the liver and triggering proinflammatory response. Our study demonstrated that gut-liver axis may play a vital part in the progression of GTW-induced hepatotoxicity, which provides guidance for basic research and clinical application of GTW.
Drug-induced hepatotoxicity is one of the major reasons for liver injury, the most serious of which is acute liver failure characterized by severe hepatocyte death (Andrea Iorga, 2017). Herbal drugs, with complex and diverse constituents, are effect triggers of liver injury leading to acute and chronic liver diseases (Stournaras and Tziomalos, 2015). Tripterygium wilfordii multiglycoside (GTW) is a well-known Chinese herbal medicine that is extracted and purified from the peeled roots of T. wilfordii Hook.f. (TwHF) and mostly used as tablets in clinical. The clinical dosage of GTW tablets is 1–1.5 mg/kg/day for adults (Gong et al., 2020). According to the standard of GTW tablets (WS3–B-33350-98-2011), stipulated by National Medical Products Administration (China), the contents of triptolide and wilforlide A in each GTW tablet (10 mg) shall not exceed and not less than10 μg, respectively (Dai et al., 2022). Because of its anti-inflammatory and immune-suppressive effects, GTW has been widely applied to various autoimmune diseases, for example, systemic lupus erythematosus (Zhou et al., 2018), nephrotic syndrome (Xu and Jiang, 2009) and rheumatoid arthritis (Tao et al., 2001; Marks, 2011). However, the adverse effects of GTW, especially liver injury, restrict its clinical application (Zhang et al., 2016; Liu et al., 2018). Since the liver is responsible for metabolism and detoxification, it is imperative to explore the mechanisms of GTW-induced liver injury for its safe clinical use.
Although studies have reported that GTW could cause liver damage when administered chronically or at very high doses (Zhang et al., 2012; Peng et al., 2021), generally, investigations on GTW-induced hepatotoxicity and associated molecular mechanisms are still few. Therefore, the effects of GTW on liver and the underlying mechanisms require further investigation.
Proteomics currently emerges as a comprehensive and powerful tool for detecting functional proteins and discovering molecular targets, therefore, it is more and more widely used to explore the pharmacological and toxicological mechanisms of herbal medicine (Bennett and Devarajan, 2018; Zhang et al., 2022). Numerous studies on drug-induced liver injury have been reported to identify differentially expressed proteins (DEPs) and explore associated mechanisms via label-free proteomics (Satoh et al., 2014; Dragoi et al., 2018). Although a previous study has used proteomics to detect DEPs in the livers of mice administered triptolide (TP), an essential bioactive but toxic component of TwHF (Li et al., 2017), at present, there are almost no investigations on liver proteome changes in GTW-induced hepatotoxic models.
In the present study, we evaluated the effects of GTW on mice livers and elucidated the associated mechanisms via label-free proteomics combined with bioinformatics analysis. By analyzing the identified DEPs and enriched functional pathways between the control and GTW groups, we found that GTW could act on the intestinal immune network for IgA production pathway, suggesting an important role of gut-liver axis in mediating GTW-induced hepatotoxicity. To our knowledge, our research was the first indication of the effects of GTW on mice liver proteome, which is expected to provide more insights into the mechanism of GTW-induced hepatotoxicity.
GTW (>98%, Batch No. 1507702) was obtained from Zhejiang DND Drug Factory (Zhejiang, China). The chemical profile of GTW was determined by HPLC. Standard chemicals including triptolide, triptonide, celastrol, and wilforlide A were purchased from National Institute for Food and Drug Control (Beijing, China), and wilforgine and wilforine were obtained from Chengdu Push Bio-technology Co., Ltd. (Sichuan, China). The conditions of HPLC were listed in Table 1.
Male C57BL/6J mice (6–8 weeks) were supplied by Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). Mice were housed in conditions with controlled light (12 h light/dark cycle), temperature (24 ± 2°C), and humidity (50%–60%) and had adequate food and tap water. All experiments on mice were performed under the guidelines of Ethical Committee of China Pharmaceutical University (Ethics approval No. 2022-05-003).
GTW in powder was suspended in 0.5% CMC-Na and administered to mice by gavage. Based on the resource equation method (Charan and Kantharia, 2013), one of the methods of sample size calculation in animal studies, 18 male C57BL/6J mice were randomly divided into the control group, GTW-L and GTW-H groups, with 6 mice in each group. The dosage and dosing time of GTW administration were chosen based on our previous studies (Zhang et al., 2012; Miao et al., 2019; Yuan et al., 2019). The doses selected for GTW in animal experiments were 70 mg/kg (GTW-L) and 140 mg/kg (GTW-H), which are approximately 5 times and 10 times mouse equivalent dosage (mouse equivalent dose was 9.1–13.65 mg/kg/day), respectively. After GTW administration for 1 week, mice were sacrificed.
Serum ALT, AST, and ALP detection kits were obtained from Whitman Biotech (Nanjing, China). All examinations were performed in accordance with the protocols.
Fragments from mice tissues were fixed in 4% paraformaldehyde overnight, embedded in paraffin, and then sliced for H&E and immunohistochemistry (IHC) staining to observe the pathological changes.
Mouse tissue sections were fixed in 4% paraformaldehyde for 15 min, washed in PBS and permeabilized with 0.1% Triton X-100 for 10 min. After blocked with 5% goat serum for 1 h at room temperature, they were incubated with primary antibodies at 4°C overnight. Then, samples were incubated with secondary antibodies and DAPI (1:1000) at room temperature for 1 h, and further imaged with a confocal laser scanning microscope (Olympus, Lake Success, LY). The following antibodies were used: mouse anti-occludin (1:200, #33-1500, Thermo Fisher), APC anti-mouse F4/80 antibody (1:50, #123115, BioLegend), Alexa Fluor 488-conjugated anti-mouse IgG (1:1000, ab150113, abcam).
Label-free proteomics analysis of liver samples from the control and GTW-H (140 mg/kg) groups was performed with the help of Novogene Co., Ltd. (Beijing, China). The experimental procedures and methods were listed in Supplementary Materials and Methods.
Gene Ontology (GO) annotation analysis of the identified DEPs was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (version 6.8) (Huang et al., 2009). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEPs was performed using KEGG Orthology-Based Annotation System (KOBAS) (version 3.0) (Bu et al., 2021), and the connection between pathways were visualized using Cytoscape with ClueGO and CluePedia applications (Bindea et al., 2009; Mlecnik and Bindea, 2013). p-value less than 0.05 was considered statistically significant.
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://www.string-db.org/) was used to establish protein-protein interaction (PPI) network of the DEPs (Szklarczyk et al., 2017). After visualization of the network by Cytoscape, the densely connected protein modules in the PPI network was detected using Molecular Complex Detection (MCODE) app with default parameter settings (Bader and Hogue, 2003). Furthermore, bioinformatics analysis for the top cluster, including biological process annotation and KEGG enrichment pathway analysis, was conducted according to the above-mentioned methods.
Mouse tissue samples were homogenized in RIPA buffer (Beyotime, China) supplemented with protease and phosphatase inhibitors (Bimake, China). After protein quantification assayed with a BCA method (Beyotime, China), the lysis was mixed with loading buffer (Bio-Rad, CA, United States) and denatured by heat. Then, protein was separated on SDS–PAGE and subsequently transferred onto PVDF membranes by electroblotting. Next, the membrane was blocked with 5% bovine serum albumin for 1 h at room temperature, followed by incubating with primary antibody at 4°C overnight. Finally, the membrane was incubated with HRP-conjugated polyclonal secondary antibody at room temperature for 1 h, and further visualized using an ECL detection kit (Tanon, Shanghai, China). The antibodies used in the study are as follows: rabbit anti-β-ACTIN (AC026, ABclonal), rabbit anti-pIgR (A6130, ABclonal), mouse anti-Occludin (#33-1500, Thermo Fisher).
Mouse tissue samples were homogenized in Trizol reagent for RNA extraction. Approximately 1 μg of RNA was converted to cDNA after quantification with Nanodrop 2000 (Thermo, DE, United States). Target genes were analyzed by real-time PCR (RT-PCR) using SYBR Green on Stepone Plus (Thermo, DE, United States) with specific primers. Primers used were listed on Table 2, and the β-actin gene was used for normalization.
Mouse fecal samples were suspended in PBS containing protease inhibitor, and centrifuged at 12,000 × g for 15 min. After centrifugation, the supernatant was collected and diluted 1/1000 for fecal IgA ELISA. Serum was diluted 1/2000 for IgA ELISA. Mouse IgA ELISA kit was obtained from Neobioscience Technology Co., Ltd. (Shenzhen, China).
Mouse serum TNFα concentration was measured using commercially available ELISA kits according to the manufacturer’s protocols. Mouse TNFα ELISA kit was obtained from Neobioscience Technology Co., Ltd. (Shenzhen, China).
Fluorescence in situ hybridization (FISH) analysis was conducted according to the protocol from GenePharma (Shanghai, China). Briefly, liver frozen sections were fixed in 4% paraformaldehyde for 15 min, washed in PBS, and incubated with proteinase K for 15 min. Then, slices were washed and incubated with 5 ng/μl of probes at 37°C for 12–16 h. After incubation, slices were washed in PBS and then stained with DAPI at room temperature for 20 min, and further imaged with a confocal laser scanning microscope (Olympus, Lake Success, LY). A mix of probes was synthesized by GenePharma, as shown in Table 3. The non-Eub probe was used as a negative control.
Data are presented as mean ± SEM. The differences between two groups were analyzed using Student’s t-test, and the differences among multiple groups were analyzed using one-way analysis of variance (ANOVA). Statistical analysis and graphing were performed using GraphPad Prism 9 software (GraphPad Software, Inc., San Diego, CA, United States). p-value less than 0.05 was considered statistically significant.
In this study, the fingerprint of GTW was determined by an HPLC method, and components including triptolide, triptonide, wilforgine, celastrol, wilforine, and wilforlide A were detected and quantified (Figure 1). According to Table 4, the contents of triptolide and wilforlide A meted the quality standards of GTW (WS3-B-3350-98-2011).
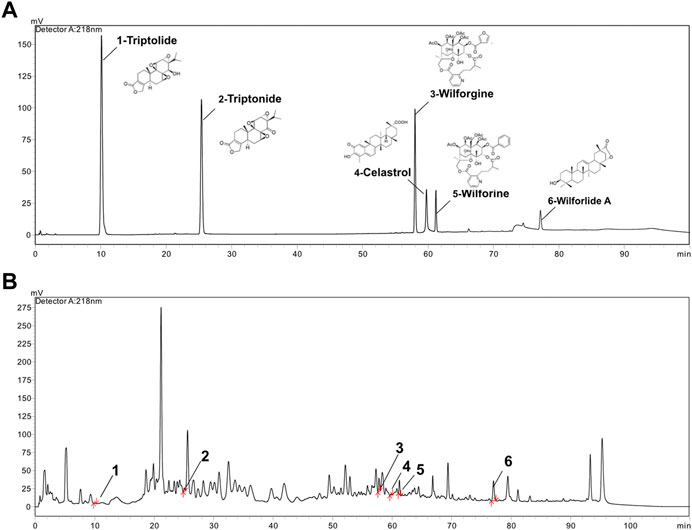
FIGURE 1. HPLC analysis of standard mixture and GTW. (A) The chromatograph of triptolide, triptonide, wilforgine, celastrol, wilforine, and wilforlide A standard mixture. (B) The HPLC chromatograph of GTW.
The body weights of experimental mice were recorded during 1-week administration of GTW. Compared with the control, GTW-H could prevent mice weight gain (Figure 2A). Meanwhile, liver tissue index of 140 mg/kg GTW-treated mice was slightly increased (Figure 2B). Serum aminotransferase levels were measured to evaluate whether GTW caused liver injury. As shown in Figures 2C–E, only 140 mg/kg GTW caused a significant increase in serum ALT and AST levels, suggesting hepatocellular injury. The effects of GTW on liver structure were examined by histopathological evaluation. Compared with the normal group, nuclear pyknosis was observed in the liver sections of GTW-H group (Figure 2F), suggesting apoptotic cell death. In addition, increased MPO immunopositive areas revealed recruitment and infiltration of neutrophils in GTW-H treated liver (Figure 2G). Taken together, these results demonstrate that GTW administration could induce hepatotoxicity in a dose-dependent manner.
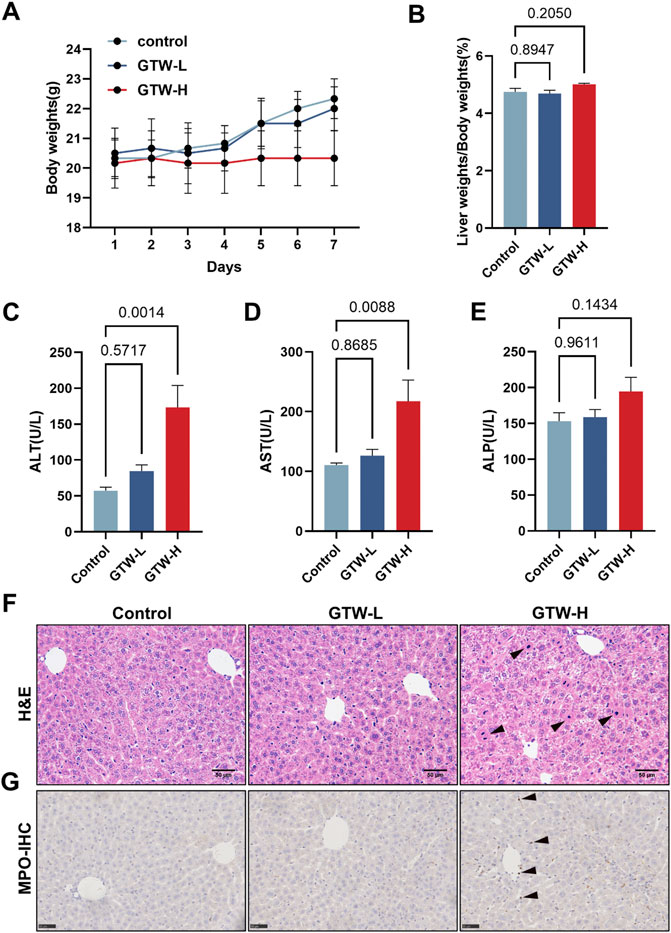
FIGURE 2. GTW administration caused liver injury in mice. (A) Changes in body weights (n = 6). (B) Liver tissue index calculated by tissue and body weights (n = 6). (C–E) Serum aminotransferase levels, including ALT, AST, and ALP (n = 6). (F) Representative H&E staining images of liver sections (scale bar = 50 μm), and nuclear pyknosis was pointed with black arrows. (G) Representative immunohistochemical measurements of liver in each group (scale bar = 50 μm). MPO positive areas were pointed with black arrows. Data are presented as mean ± SEM. p < 0.05 was considered statistically significant.
To obtain a comprehensive overview of the mechanism underlying GTW-induced hepatotoxicity, label-free quantitative proteomics was performed to identify the DEPs between the control group and GTW-H (hereinafter referred to as GTW) group. After three replicated biological analyses with high reliability, 3,670 proteins were recognized, of which 2,641 proteins were identified in both groups (Supplementary Table S1). Meanwhile, the proteomics profile of GTW group showed a clear separation from the control group (Figure 3A). Next, we screened DEPs by comparing protein abundance between the groups (fold change (FC) ≥ 2 and ≤ 0.5; p-value < 0.05) and identified 155 DEPs with 46 upregulated and 109 downregulated proteins (Figure 3B; Supplementary Table S2). The accuracy of the selected DEPs was measured by cluster analysis. As shown in Figure 3C, the identified DEPs were clearly distinguished between the two groups, suggesting that these DEPs may represent key changes in mice livers after GTW administration.
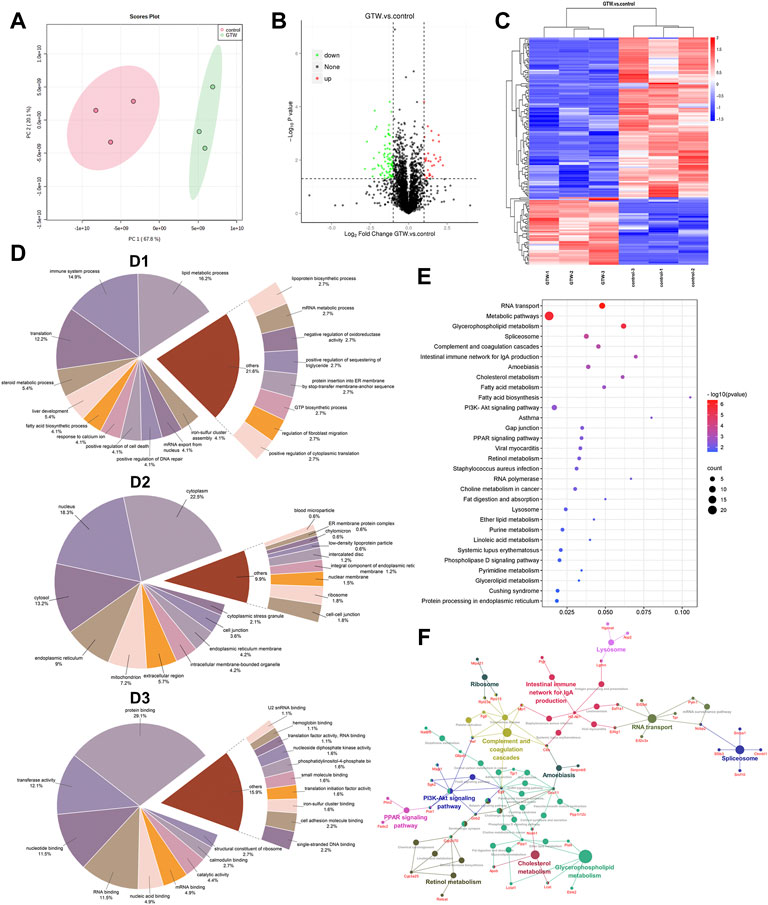
FIGURE 3. Results of proteomics combined with bioinformatics analysis of mice livers after GTW exposure. (A) PCA analysis of identified proteins. (B) Volcano plot. Proteins upregulated or downregulated were presented as red or green points, respectively, and black points represented proteins with no significant changes (|log2FC| ≥ 1 and p < 0.05). (C) Heatmaps of DEPs. Rows represented DEPs and columns represented samples with three biological replicates. The signal values of DEPs were shown as colored bars, that is, the high expressed DEPs were colored in red, and the low expressed proteins were colored in blue. (D) GO annotation classification analysis of DEPs. (D1: Biological process. D2: Cellular component. D3: Molecular function.) (E) KEGG enrichment analysis of DEPs (p < 0.05). The ordinate represented the pathway term, and the abscissa represented the ratio of DEP numbers involved in a KEGG pathway to all identified protein numbers annotated in this pathway. The number of DEPs was represented by dot size, and the p-value was indicated by dot color. (F) The connection between pathways.
The GO annotation analysis on DEPs was carried out and classified into three GO terms: biological process (BP), cellular component (CC), and molecular function (MF). According to BP results (Figure 3D1), lipid metabolic process (16.2%), immune system process (14.9%), and translation (12.2%) were the top three processes. In CC analysis (Figure 3D2), the largest proportions of DEPs were assigned to the cytoplasm (22.5%), nucleus (18.3%), and cytosol (13.2%). In MF analysis (Figure 3D3), protein binding (29.1%), transferase activity (12.1%), nucleotide binding (11.5%), and RNA binding (11.5%) accounted for the major proportions.
To better understand the functional categories, KEGG pathway analysis of DEPs was conducted. The top 30 pathways were shown in Figure 3E, including RNA transport, glycerophospholipid metabolism, spliceosome, complement and coagulation cascade, and intestinal immune network for IgA production. Figure 3F shows the connection between them.
To further determine the key pathways involved in GTW-induced hepatotoxicity, PPI analysis and significant cluster identification were performed. By using the STRING database, PPI network of the DEPs was conducted to explore their interactions. We observed that a total of 114 nodes and 432 edges showed interconnectivity (Figure 4A). The significant interconnected regions in this PPI network were analyzed, and the top three clusters were recognized (Figures 4B–D; Supplementary Table S3). Cluster 1 (Figure 4B) had the highest score, indicating that it would play an important role in regulating this PPI network, thus, BP annotation and KEGG pathway analysis were performed. According to Table 5, cluster 1 was closely related to immune system process, response to lipopolysaccharide, and response to bacterium, suggesting that abnormal immune response would be the main reason for GTW-induced hepatotoxicity. Further KEGG analysis showed that intestinal immune network for IgA production pathway would be involved in the liver toxicity.
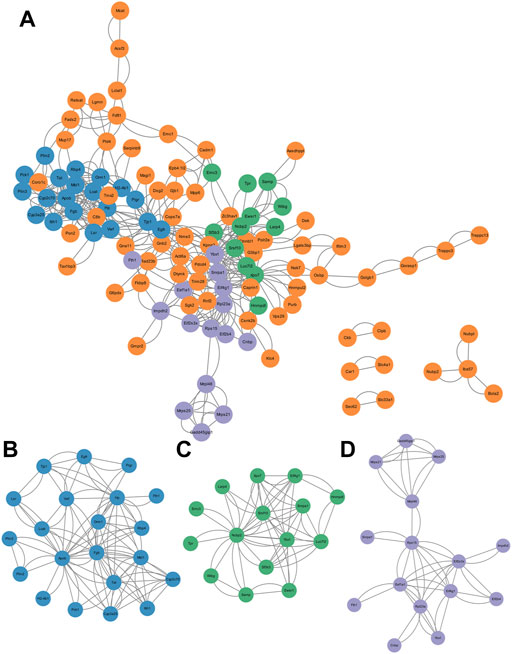
FIGURE 4. Construction and cluster analysis of PPI network. (A) PPI network of DEPs visualized by Cytoscape. (B–D) The top three clusters screened by MCODE, including cluster 1 (score = 5.4, nodes = 21, edges = 108), cluster 2 (score = 4.4, nodes = 15, edges = 62), and cluster 3 (score = 3.6 nodes = 15, edges = 50).
IgA, transported by polymeric immunoglobulin receptor (pIgR), is a major immunoglobulin isotype in the gut and plays a crucial role in maintaining gut barrier (Gutzeit et al., 2014). The interaction between gut and liver makes liver vulnerable to disrupted intestinal homeostasis (Seki and Schnabl, 2012). Intestinal barrier dysfunction could promote bacterial translocation to the liver and trigger proinflammatory response, contributing to the development of liver diseases (Wang et al., 2015; Tripathi et al., 2018). Thus, we assumed that GTW should affect the intestinal immune network for IgA production pathway and disturb intestinal homeostasis, contributing to abnormal immune response in livers of GTW-treated mice.
Next, we evaluate the effects of GTW on the intestinal immune network for IgA production pathway. Results from proteomics indicated that GTW could downregulate pIgR protein expression (Figure 5A). Immunoblots confirmed that pIgR protein in liver was decreased in GTW group (Figure 5B). In addition, we assessed the expression of pIgR in gut and found that pIgR protein expression was reduced in the ileum but not significantly changed in the colon after GTW exposure (Figure 5B). As a consequence, the reduction of pIgR resulted in a decreased secretion of IgA into the gut lumen (Figure 5C), and a buildup of serum IgA compared to the normal group (Figure 5D). These results suggest that GTW could inhibit intestinal immune network for IgA production pathway via disturbing pIgR/IgA system.
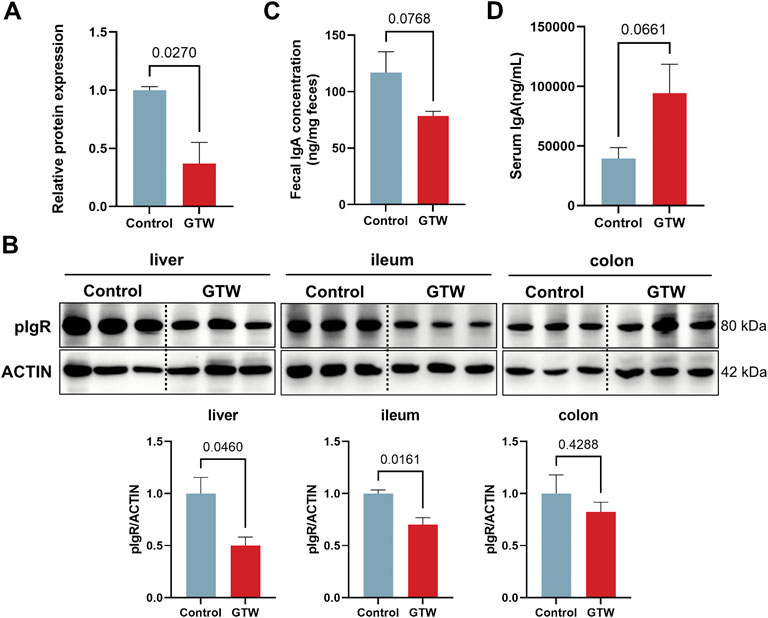
FIGURE 5. GTW affected the activation of intestinal immune network for IgA production pathway. (A) Relative pIgR protein expression according to proteomics results (n = 3). (B) The protein levels of pIgR in liver, ileum and colon were determined by western blot (n = 3). (C) IgA levels in feces (n = 5) and (D) serum (n = 5) were measured after GTW treatment. The band density was calculated using ImageJ software. Data are presented as mean ± SEM. p < 0.05 was considered statistically significant.
The intestinal immune network for IgA production pathway plays a crucial role in maintaining mucosal homeostasis and gut barrier (Brown and Kloppel, 1989; Mantis et al., 2011). Therefore, we assessed whether the intestinal barrier was altered in GTW-treated mice. Histopathological examination showed that in GTW-treated mice ileum, the villi were sparse and broken and the crypt depth was reduced (Figure 6A). In addition, GTW could damage the colon, manifested by extensive loss of epithelial cells, destruction of crypts, and inflammatory cell infiltration (Figure 6B). As a reliable sign of intestinal barrier disruption, we further analyzed the expression of tight junction protein occludin in the ileum and colon. As shown in Figure 6C, the ileal and colonic protein levels of occludin were decreased. Consistently, the fluorescence intensity of intestinal epithelial occludin was much lower in GTW-treated mice than in control mice (Figure 6D). These results point towards impaired intestinal barrier integrity and show consistency with disrupted pIgR/IgA system.
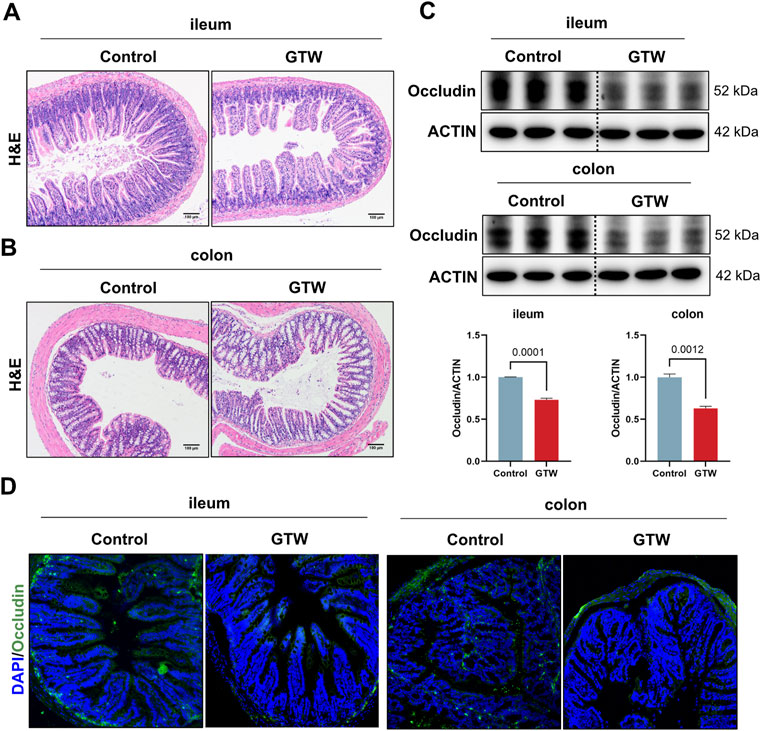
FIGURE 6. The effects of GTW on intestinal barrier integrity. (A,B) Representative H&E staining images of ileum and colon tissues (scale bar = 100 μm). (C) The protein levels of occludin in the ileum and colon were determined by western blot (n = 3). (D) Representative images of immunofluorescent staining of occludin (green) in ileum and colon sections (20x). The nuclei were visualized with DAPI (blue). The band density was calculated using ImageJ software. Data are presented as mean ± SEM. p < 0.05 was considered statistically significant.
The liver, a key immune organ, is positioned to receive gut-derived products via the portal vein, implying that it could be severely affected by a disrupted intestinal homeostasis (Schnabl, 2013; Wang et al., 2021). The disruption of gut barrier allows bacterial translocation (Albillos et al., 2020). We thus detected bacteria in the liver. The 16S rRNA gene expression in GTW-treated mice livers were significantly increased (Figure 7A), in addition, FISH analysis confirmed that bacterial invasion to the liver was promoted after GTW exposure (Figure 7B). In agreement with this result, clear signs of inflammatory response in GTW-treated mice livers were observed, with an increase of F4/80 labeled macrophages (Figure 7C) and of hepatic TLR2 and TLR4 mRNA levels (Figure 7D). In addition, increased mRNA levels of proinflammatory cytokine TNFα (Figure 7E) and increased serum TNFα concentration (Figure 7F) further confirmed liver inflammation. Thus, these findings suggest that GTW-caused intestinal barrier impairment could increase bacterial translocation to the liver and trigger hepatic inflammation, which may contribute to GTW-induced liver toxicity.
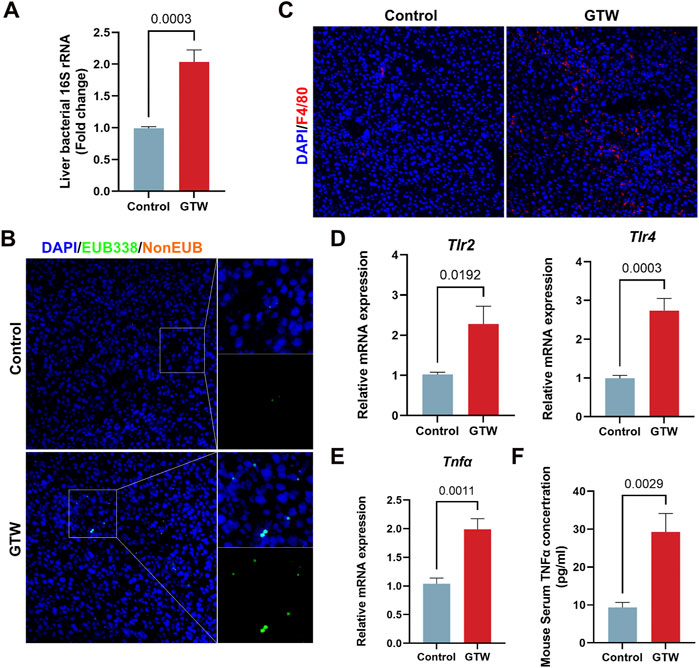
FIGURE 7. GTW administration promoted bacterial translocation to the liver and triggered hepatic inflammation. (A) Liver bacterial 16S rRNA expression measured by RT-PCR (n = 6). (B) Bacteria detected in the liver using FISH with probes EUB338 targeting eubacteria 16S rRNA region (20x). The nuclei were stained with DAPI (blue) and bacteria stained with EUB338 (green). (C) Representative images of immunofluorescent staining of F4/80 (red) in liver (20x). DAPI was used to visualize nuclei (blue). (D) Hepatic TLR2 and TLR4 mRNA expression levels were detected by RT-PCR (n = 6). (E) The expression levels of proinflammatory cytokine TNFα in liver were measured using RT-PCR analysis (n = 6). (F) Serum TNFα concentration in GTW-treated mice were detected by ELISA (n = 6). Data are presented as mean ± SEM. p < 0.05 was considered statistically significant.
GTW is a stable extract purified from the peeled roots of TwHF and extensively applied for autoimmune diseases in China. Three major ingredients, including diterpenoids (e.g., triptolide and triptonide), sesquiterpene alkaloids (e.g., wilforgine and wilforine), and triterpenoids (e.g., celastrol and wilforlide A), are thought to be contributed to the pharmacodynamics activities of GTW (Luo et al., 2016). In the study, we used an HPLC method to characterize the fingerprint of GTW, and calculated the contents of 6 representative components. It was found that among them, wilforlide A showed the highest content (the mean value was 4460.75 μg/g) while triptolide showed the lowest (the mean value was 5.25 μg/g). These results meted the quality standards of GTW (WS3-B-3350-98-2011) and showed consistency with previous studies (Luo et al., 2016).
The clinical application of GTW is restricted due to its adverse effects, especially liver injury. TP is concerned as an essential bioactive but toxic component of TwHF (Li et al., 2014b). Previous studies demonstrated that liver damage caused by TP was mainly associated with mitochondrial dysfunction (Fu et al., 2011), oxidative stress (Li et al., 2014a), abnormal lipid and glucose metabolism (Wang et al., 2013; Jiang et al., 2016), and inflammation (Wang et al., 2014). Compared to TP, studies about GTW-induced liver injury are few and associated signaling mechanisms remain poorly understood. In addition, to study the toxic effects of GTW, it is more appropriate to take GTW as a whole than to focus only on TP. In this study, we conducted experiments in male C57BL/6J mice to evaluate GTW-induced hepatotoxicity. ALT and AST are two reliable markers reflecting hepatocellular injury while ALP reflects the extent of cholestasis (Cockeram, 1998). After 1-week administration, GTW at high dose significantly increased serum ALT and AST levels but had minor effects on ALP, indicating hepatocellular damage. Further examinations confirmed that GTW could cause hepatocyte apoptosis and liver inflammation. Similar to our results, a recent study reported that GTW induced hepatotoxicity in zebrafish via increased inflammation and enhanced apoptosis (DUAN et al., 2021). In fact, we also investigated the hepatotoxic effects of GTW on female mice. By detecting serum aminotransferase levels, we found that whereas there was no significant gender difference, the hepatotoxic effects of GTW showed a better dose-dependent manner in male mice than in female (Supplementary Figure S1).
Based on label-free proteomics combined with bioinformatics analysis, intestinal immune network for IgA production pathway was predicted to be involved in GTW-induced liver toxicity. IgA is a major immunoglobulin isotype in the gut and plays a crucial role in maintaining intestinal homeostasis (Mantis et al., 2013). Transport of IgA from the lamina propria to the mucosal surface and the release of secretory IgA onto the gut lumen require pIgR, a protein expressed on mucosal epithelial cells (Kaetzel et al., 1991; Schneeman et al., 2005). Besides lamina propria-produced IgA, liver-derived IgA is also an important source of total gut IgA in mouse, which is secreted to bile by pIgR on hepatocytes and cholangiocytes (Brown and Kloppel, 1989; Turula and Wobus, 2018). Secretory IgA has been reported to favor the maintenance of gut barrier by regulating microbiota composition, preventing bacteria invasion, and downregulating immune responses in the intestinal mucosal (Strugnell and Wijburg, 2010; Mantis et al., 2011). Deficiency in secretory IgA would alter gut microbiome and increase intestinal permeability (Inamine and Schnabl, 2018). Accumulating evidence indicates that controlling gut microbial composition is critical for maintaining intestinal homeostasis (Albillos et al., 2020). Gut bacterial dysbiosis can lead to a direct interaction between the microbiota and epithelial cells, promoting inflammatory response and enhancing gut permeability, thereby increasing bacterial translocation and affecting the development of chronic liver diseases, including ALD, NAFLD and NASH(Mao et al., 2015; Cheng et al., 2018; Wang et al., 2020; Wang et al., 2021). Results from our study demonstrate that GTW could significantly reduce pIgR protein expression in the liver and ileum, as a result, a reduction in fecal IgA was observed. In agreement with reduced fecal IgA levels, serum IgA concentration was increased, which was consistent with previous study (Shimada et al., 1999). These findings suggest that GTW could act on intestinal immune network for IgA production pathway via inhibiting pIgR/IgA system, it raises the possibility that GTW would disrupt intestinal homeostasis, which might be associated with altered microbiota composition, and as a consequence, the increased intestinal inflammation, the impaired gut barrier integrity and the enhancement of intestinal permeability.
We thus assess whether GTW caused intestinal barrier dysfunction. Histopathological analysis showed that GTW administration caused damage to ileum and colon, in addition, obvious inflammation was observed in the colon of GTW-treated mice. Tight junctions, sealing adjacent intestinal epithelial cells together, play a central role in gut barrier maintenance (Chelakkot et al., 2018; Lee et al., 2018). Numerous studies have reported that damage to tight junctions contributes to intestinal barrier dysfunction (Lee, 2015; Chen et al., 2017). In our study, we found that the expression of tight junction protein occludin in the ileum and colon were decreased after GTW exposure, indicating impaired intestinal barrier integrity. The liver, the first organ for receiving gut-derived products through the portal vein, is vulnerable to disrupted intestinal homeostasis (Inamine and Schnabl, 2018). Gut barrier disruption enhances intestinal permeability and promotes bacteria and/or bacterial products (e.g., lipopolysaccharides) transferring to the liver, triggering a proinflammatory cascade, thereby inducing or exacerbating a range of hepatic diseases (Albillos et al., 2020; Ghosh et al., 2020). In our study, increased bacteria were found in the livers of GTW-treated mice while livers of control mice were harboring few bacteria. Toll-like receptors (TLR) have been shown to recognize microbial pattern recognition receptors and stimulate immune response (Kawasaki and Kawai, 2014), we thus assessed hepatic TLR2 and TLR4 expression. In agreement with increased bacterial translocation, TLR2 and TLR4 mRNA levels in GTW-treated mice livers were significantly elevated. In addition, F4/80 immunofluorescent staining revealed more macrophages in the livers of GTW-treated mice, and MPO immunohistochemical staining revealed increased recruitment and infiltration of neutrophils. The proinflammatory response was further confirmed by the increase of hepatic TNFα mRNA expression and of serum TNFα concentration. Taken together, our study suggests that GTW could disrupt intestinal barrier integrity, promoting the translocation of bacteria to the liver and increasing hepatic inflammation. Furthermore, our findings suggest a key role of gut-liver axis in GTW-induced liver injury. In fact, according to clinical reports, GTW has the highest incidence of gastrointestinal upset, ranging from diarrhea, nausea, vomiting, and even colitis (Zhang et al., 2016), it raises the possibility that GTW could disrupt gut barrier and thereby affect the progression of liver toxicity. However, the effects of GTW on gut microbiome and the role of intestinal microbiota dysbiosis in GTW-induced hepatotoxicity require validation in our further study, in addition, our study just did a preliminary exploration, more experiments need to be conducted to investigate the crosstalk between gut and liver in GTW-induced hepatotoxic model and to identify potential molecular targets.
Outcomes obtained from this study reveal novel insights into the mechanism of GTW-induced hepatotoxicity, but, inevitably, there are limitations. First, these findings were based on the analysis of livers in mice, which may have inconsistencies with clinical results. Second, the effects of GTW on gut microbiome and the crosstalk between gut and liver injury required further exploration. Third, in addition to the gut-liver axis, other functional pathways and targets related to GTW-induced hepatotoxicity need to be validated. Furthermore, potential ingredients and/or toxic core structures, contributing to hepatotoxicity, need to be explored to promote the clinical application of GTW.
Our findings demonstrate that GTW could induce liver injury. Based on label-free proteomics combined with bioinformatics analysis, we found that GTW may act on intestinal immune network for IgA production pathway and impair intestinal barrier integrity, therefore, the bacterial translocation to the liver and hepatic inflammation were increased. In summary, our study reveals a novel insight into the mechanism of GTW-induced hepatotoxicity that the crosstalk between gut and liver may play a crucial part in the progression of GTW-induced liver toxicity. However, more experimental data are needed to verify this.
The datasets of proteomics analysis presented in this study can be found in ProteomeXchange Consortium with the dataset identifier PXD038084.
The animal study was reviewed and approved by Ethical Committee of China Pharmaceutical University.
Conceptualization, YM and ZJ; methodology, YM and ZY; validation, QZ and MD; investigation, JW, YX, and YC; writing—original draft preparation, YM; writing—review and editing, ZJ; supervision, QY; project administration, LZ and ZJ; funding acquisition, LZ and ZJ.
This research was funded by the National Natural Science Foundation of China (82074114, 81973562, 81773995); the Innovation Team Projects in Universities of Guangdong Province (No. 2018KCXTD016); “Double First-Class” University project (CPU2018GY33); the Postgraduate Research Practice Innovation Program of Jiangsu Province (KYCX20_0669).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1032741/full#supplementary-material
Albillos, A., Gottardi, A. D., and Rescigno, M. (2020). The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 72, 558–577. doi:10.1016/j.jhep.2019.10.003
Andrea Iorga, L. D., and Kaplowitz, N. (2017). Drug-induced liver injury: Cascade of events leading to cell death, apoptosis or necrosis. Int. J. Mol. Sci. 18, E1018. doi:10.3390/ijms18051018
Bader, G. D., and Hogue, C. W. V. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 4, 2–27. doi:10.1186/1471-2105-4-2
Bennett, M. R., and Devarajan, P. (2018). The future role of proteomics in the understanding of acute kidney injury. Expert Rev. Proteomics 15, 191–192. doi:10.1080/14789450.2018.1443007
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO : A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi:10.1093/bioinformatics/btp101
Brown, W. R., and Kloppel, T. M. (1989). The liver and IgA: Immunological, cell biological and clinical implications. Hepatology 9, 763–784. doi:10.1002/hep.1840090518
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., and He, Z., (2021). KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi:10.1093/nar/gkab447
Charan, J., and Kantharia, N. (2013). How to calculate sample size in animal studies? J. Pharmacol. Pharmacother. 4, 303–306. doi:10.4103/0976-500X.119726
Chelakkot, C., Ghim, J., and Ryu, S. H. (2018). Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 50, 103. doi:10.1038/s12276-018-0126-x
Chen, W. Y., Wang, M., Zhang, J., Barve, S. S., McClain, C. J., and Joshi-Barve, S. (2017). Acrolein disrupts tight junction proteins and causes endoplasmic reticulum stress-mediated epithelial cell death leading to intestinal barrier dysfunction and permeability. Am. J. Pathol. 187, 2686–2697. doi:10.1016/j.ajpath.2017.08.015
Cheng, C., Tan, J., Qian, W., Zhang, L., and Hou, X. (2018). Gut inflammation exacerbates hepatic injury in the high-fat diet induced NAFLD mouse: Attention to the gut-vascular barrier dysfunction. Life Sci. 209, 157–166. doi:10.1016/j.lfs.2018.08.017
Cockeram, A. W. (1998). Canadian association of gastroenterology Practice guidelines: Evaluation of dysphagia. Can. J. Gastroenterol. 12, 409–413. doi:10.1155/1998/303549
Dai, M., Peng, W., Zhang, T., Zhao, Q., Ma, X., and Cheng, Y., (2022). Metabolomics reveals the role of PPARα in Tripterygium Wilfordii-induced liver injury. J. Ethnopharmacol. 289, 115090. doi:10.1016/j.jep.2022.115090
Dragoi, D., Benesic, A., Pichler, G., Kulak, N. A., Bartsch, H. S., and Gerbes, A. L. (2018). Proteomics analysis of monocyte-derived hepatocyte-like cells identifies integrin beta 3 as a specific biomarker for drug-induced liver injury by Diclofenac. Front. Pharmacol. 9, 699–710. doi:10.3389/fphar.2018.00699
Duan, X. Y., Ma, R. J., Hsiao, C. Der, Jiang, Z. Z., Zhang, L. Y., and Zhang, Y., (2021). Tripterygium wilfordii multiglycoside-induced hepatotoxicity via inflammation and apoptosis in zebrafish. Chin. J. Nat. Med. 19, 750–757. doi:10.1016/S1875-5364(21)60078-X
Fu, Q., Huang, X., Shu, B., Xue, M., Zhang, P., and Wang, T., (2011). Inhibition of mitochondrial respiratory chain is involved in triptolide-induced liver injury. Fitoterapia 82, 1241–1248. doi:10.1016/j.fitote.2011.08.019
Ghosh, S. S., Wang, J., Yannie, P. J., and Ghosh, S. (2020). Intestinal barrier function and metabolic/liver diseases. Liver Res. 4, 81–87. doi:10.1016/j.livres.2020.03.002
Gong, L., Wang, L., Cao, Y., and Li, C. (2020). Rapid induction of clinical remission in sapho syndrome using high-dose Tripterygium glycosides: A case report. Med. Baltim. 99, e21102. doi:10.1097/MD.0000000000021102
Gutzeit, C., Magri, G., and Cerutti, A. (2014). Intestinal IgA production and its role in host-microbe interaction. Immunol. Rev. 260, 76–85. doi:10.1111/imr.12189
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Inamine, T., and Schnabl, B. (2018). Immunoglobulin A and liver diseases. J. Gastroenterol. 53, 691–700. doi:10.1007/s00535-017-1400-8
Jiang, Z., Huang, X., Huang, S., Guo, H., Wang, L., and Li, X., (2016). Sex-related differences of lipid metabolism induced by triptolide: The possible role of the LXRα/SREBP-1 signaling pathway. Front. Pharmacol. 7, 87–10. doi:10.3389/fphar.2016.00087
Kaetzel, C. S., Robinson, J. K., Chintalacharuvu, K. R., Vaerman, J. P., and Lamm, M. E. (1991). The polymeric immunoglobulin receptor (secretory component) mediates transport of immune complexes across epithelial cells: A local defense function for IgA. Proc. Natl. Acad. Sci. U. S. A. 88, 8796–8800. doi:10.1073/pnas.88.19.8796
Kawasaki, T., and Kawai, T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5, 461–468. doi:10.3389/fimmu.2014.00461
Lee, B., Moon, K. M., and Kim, C. Y. (2018). Tight junction in the intestinal epithelium: Its association with diseases and regulation by phytochemicals. J. Immunol. Res. 2018, 2645465. doi:10.1155/2018/2645465
Lee, S. H. (2015). Intestinal permeability regulation by tight junction: Implication on inflammatory bowel diseases. Intest. Res. 13, 11–18. doi:10.5217/ir.2015.13.1.11
Li, J., Shen, F., Guan, C., Wang, W., Sun, X., and Fu, X., (2014). Activation of Nrf2 protects against triptolide-induced hepatotoxicity. PLoS One 9, e100685. doi:10.1371/journal.pone.0100685
Li, M., Hu, T., Tie, C., Qu, L., Zheng, H., and Zhang, J. (2017). Quantitative proteomics and targeted fatty acids analysis reveal the damage of triptolide in liver and kidney. Proteomics 17, 1700001–1700025. doi:10.1002/pmic.201700001
Li, X. J., Jiang, Z. Z., and Zhang, L. Y. (2014b). Triptolide: Progress on research in pharmacodynamics and toxicology. J. Ethnopharmacol. 155, 67–79. doi:10.1016/j.jep.2014.06.006
Liu, L., Zhao, H., Sun, X., Zheng, Q., Luo, Y., and Ru, Y., (2018). Efficacy and safety of Tripterygium wilfordii hook F for chronic urticaria: A systematic review and meta-analysis. BMC Complement. Altern. Med. 18, 243–312. doi:10.1186/s12906-018-2305-7
Luo, H., Wu, X., Huang, H., Chen, S., Yang, W., and Zhang, L., (2016). Simultaneous determination of triptolide, tripterifordin, celastrol and nine sesquiterpene alkaloids in Tripterygium preparations using high-performance liquid chromatography-triple quadrupole mass spectrometry. J. Pharm. Biomed. Anal. 117, 195–204. doi:10.1016/j.jpba.2015.08.043
Mantis, N. J., Rol, N., and Corthésy, B. (2013). Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 4, 603–611. doi:10.1038/mi.2011.41
Mantis, N. J., Rol, N., and Corthésy, B. (2011). Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 4, 603–611. doi:10.1038/mi.2011.41
Mao, J., Tang, H., Zhao, T., Tan, X., Bi, J., and Wang, B., (2015). Intestinal mucosal barrier dysfunction participates in the progress of nonalcoholic fatty liver disease. Int. J. Clin. Exp. Pathol. 8, 3648–3658.
Marks, W. H. (2011). Tripterygium wilfordii hook F. Versus sulfasalazine in the treatment of rheumatoid arthritis: A well-designed clinical trial of a botanical demonstrating effectiveness. Fitoterapia 82, 85–87. doi:10.1016/j.fitote.2010.11.024
Miao, Y. Y., Luo, L., Shu, T., Wang, H., Jiang, Z. Z., and Zhang, L. Y. (2019). Study on difference of liver toxicity and its molecular mechanisms caused by Tripterygium wilfordii multiglycoside and equivalent amount of triptolid in rats. Zhongguo Zhong Yao Za Zhi 44, 3468–3477. doi:10.19540/j.cnki.cjcmm.20190301.002
Mlecnik, B., and Bindea, G. (2013). Systems biology CluePedia Cytoscape plugin : Pathway insights using integrated experimental and in silico data. Bioinformatics, 29, 661–663. doi:10.1093/bioinformatics/btt019
Peng, W., Dai, M. Y., Bao, L. J., Zhu, W. F., and Li, F. (2021). FXR activation prevents liver injury induced by Tripterygium wilfordii preparations. Xenobiotica. 51, 716–727. doi:10.1080/00498254.2021.1900626
Satoh, S., Otsuka, Y., Ozeki, Y., Itoh, K., Hashiguchi, A., and Yamazaki, K., (2014). Label-free visualization of acetaminophen-induced liver injury by high-speed stimulated Raman scattering spectral microscopy and multivariate image analysis. Pathol. Int. 64, 518–526. doi:10.1111/pin.12206
Schnabl, B. (2013). Linking intestinal homeostasis and liver disease. Curr. Opin. Gastroenterol. 29, 264–270. doi:10.1097/MOG.0b013e32835ff948
Schneeman, T. A., Bruno, M. E. C., Schjerven, H., Johansen, F.-E., Chady, L., and Kaetzel, C. S. (2005). Regulation of the polymeric ig receptor by signaling through TLRs 3 and 4: Linking innate and adaptive immune responses. J. Immunol. 175, 376–384. doi:10.4049/jimmunol.175.1.376
Seki, E., and Schnabl, B. (2012). Role of innate immunity and the microbiota in liver fibrosis: Crosstalk between the liver and gut. J. Physiol. 590, 447–458. doi:10.1113/jphysiol.2011.219691
Shimada, S., Kawaguchi-Miyashita, M., Kushiro, A., Sato, T., Nanno, M., and Sako, T., (1999). Generation of polymeric immunoglobulin receptor-deficient mouse with marked reduction of secretory IgA. J. Immunol. 163, 5367–5373. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10553061.
Stournaras, E., and Tziomalos, K. (2015). Herbal medicine-related hepatotoxicity. World J. Hepatol. 7, 2189–2193. doi:10.4254/wjh.v7.i19.2189
Strugnell, R. A., and Wijburg, O. L. C. (2010). The role of secretory antibodies in infection immunity. Nat. Rev. Microbiol. 8, 656–667. doi:10.1038/nrmicro2384
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., and Simonovic, M., (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi:10.1093/nar/gkw937
Tao, X., Cush, J. J., Garret, M., and Lipsky, P. E. (2001). A phase I study of ethyl acetate extract of the Chinese antirheumatic herb Tripterygium wilfordii Hook F in rheumatoid arthritis. J. Rheumatol. 28, 2160–2167.
Tripathi, A., Debelius, J., Brenner, D. A., Karin, M., Loomba, R., and Schnabl, B., (2018). The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411. doi:10.1038/s41575-018-0011-z
Turula, H., and Wobus, C. E. (2018). The role of the polymeric immunoglobulin receptor and secretory immunoglobulins during mucosal infection and immunity. Viruses 10, 2377–E315. doi:10.3390/v10050237
Wang, F., Cui, Q., Zeng, Y., and Chen, P. (2020). Gut microbiota-an important contributor to liver diseases. Nan Fang. Yi Ke Da Xue Xue Bao 40, 595–600. doi:10.12122/j.issn.1673-4254.2020.04.23
Wang, J., Jiang, Z., Ji, J., Wang, X., Wang, T., and Zhang, Y., (2013). Gene expression profiling and pathway analysis of hepatotoxicity induced by triptolide in Wistar rats. Food Chem. Toxicol. 58, 495–505. doi:10.1016/j.fct.2013.04.039
Wang, L., Llorente, C., Hartmann, P., Yang, A. M., Chen, P., and Schnabl, B. (2015). Methods to determine intestinal permeability and bacterial translocation during liver disease. J. Immunol. Methods 421, 44–53. doi:10.1016/j.jim.2014.12.015
Wang, R., Tang, R., Li, B., Ma, X., Schnabl, B., and Tilg, H. (2021). Gut microbiome, liver immunology, and liver diseases. Cell. Mol. Immunol. 18, 4–17. doi:10.1038/s41423-020-00592-6
Wang, X., Jiang, Z., Cao, W., Yuan, Z., Sun, L., and Zhang, L. (2014). Th17/Treg imbalance in triptolide-induced liver injury. Fitoterapia 93, 245–251. doi:10.1016/j.fitote.2014.01.006
Xu, G., Jiang, D., and Xu, C. (2009). Tripterygium wilfordii hook F treatment for idiopathic refractory nephrotic syndrome in adults : A meta-analysis. Nephron. Clin. Pract. 111 (4), c223–c228. doi:10.1159/000208990
Yuan, Z., Zhang, H., Hasnat, M., Ding, J., Chen, X., and Liang, P., (2019). A new perspective of triptolide-associated hepatotoxicity: Liver hypersensitivity upon LPS stimulation. Toxicology 414, 45–56. doi:10.1016/j.tox.2019.01.005
Zhang, C., Sun, P. P., Guo, H. T., Liu, Y., Li, J., and He, X. J., (2016). Safety profiles of Tripterygium wilfordii hook F: A systematic review and meta-analysis. Front. Pharmacol. 7, 402–408. doi:10.3389/fphar.2016.00402
Zhang, M., Guo, F., Li, X., Xian, M., Wang, T., and Wu, H., (2022). Yi-Xin-Shu capsule ameliorates cardiac hypertrophy by regulating RB/HDAC1/GATA4 signaling pathway based on proteomic and mass spectrometry image analysis. Phytomedicine. 103, 154185. doi:10.1016/j.phymed.2022.154185
Zhang, Y., Jiang, Z., Xue, M., Zhang, S., Wang, Y., and Zhang, L. (2012). Toxicogenomic analysis of the gene expression changes in rat liver after a 28-day oral Tripterygium wilfordii multiglycoside exposure. J. Ethnopharmacol. 141, 170–177. doi:10.1016/j.jep.2012.02.015
Keywords: Tripterygium wilfordii multiglycoside, hepatotoxicity, label-free proteomics, intestinal immune network for IgA production, gut-liver axis
Citation: Miao Y, Zhang Q, Yuan Z, Wang J, Xu Y, Chai Y, Du M, Yu Q, Zhang L and Jiang Z (2022) Proteomics analysis reveals novel insights into the mechanism of hepatotoxicity induced by Tripterygium wilfordii multiglycoside in mice. Front. Pharmacol. 13:1032741. doi: 10.3389/fphar.2022.1032741
Received: 31 August 2022; Accepted: 07 November 2022;
Published: 17 November 2022.
Edited by:
Yiider Tseng, Shandong University of Traditional Chinese Medicine, ChinaReviewed by:
Yisheng He, The Chinese University of Hong Kong, Shenzhen, ChinaCopyright © 2022 Miao, Zhang, Yuan, Wang, Xu, Chai, Du, Yu, Zhang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luyong Zhang, bHl6aGFuZ0BjcHUuZWR1LmNu; Zhenzhou Jiang, YmVhZ2xlamlhbmdAY3B1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.