
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 21 December 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1020759
This article is part of the Research TopicArtificial Intelligence and Machine Learning for Drug Discovery, Design and Repurposing: Methods and ApplicationsView all 8 articles
Background: Biomedical named entity recognition is one of the important tasks of biomedical literature mining. With the development of natural language processing technology, many deep learning models are used to extract valuable information from the biomedical literature, which promotes the development of effective BioNER models. However, for specialized domains with diverse and complex contexts and a richer set of semantically related entity types (e.g., drug molecules, targets, pathways, etc., in the biomedical domain), whether the dependencies of these drugs, diseases, and targets can be helpful still needs to be explored.
Method: Providing additional dependency information beyond context, a method based on the graph attention network and BERT pre-training model named MKGAT is proposed to improve BioNER performance in the biomedical domain. To enhance BioNER by using external dependency knowledge, we integrate BERT-processed text embeddings and entity dependencies to construct better entity embedding representations for biomedical named entity recognition.
Results: The proposed method obtains competitive accuracy and higher efficiency than the state-of-the-art method on three datasets, namely, NCBI-disease corpus, BC2GM, and BC5CDR-chem, with a precision of 90.71%, 88.19%, and 95.71%, recall of 92.52%, 88.05%, and 95.62%, and F1-scores of 91.61%, 88.12%, and 95.66%, respectively, which performs better than existing methods.
Conclusion: Drug, disease, and protein dependencies can allow entities to be better represented in neural networks, thereby improving the performance of BioNER.
The number of biomedical literature is increasing rapidly. On average, more than 3,000 new articles are published in peer-reviewed journals every day, excluding technical reports such as preprints and clinical trial reports in various archives. So far, PubMed (Roberts, 2001) has 33 million citations and abstracts in the biomedical literature. Reports containing valuable information about new discoveries and insights have been added to a large number of literature reports; meanwhile, the ever-increasing volume of biomedical literature has caused great challenges in extracting relevant and high-quality information. Therefore, there is an increasing demand for accurate biomedical text mining to extract information from the literature.
Named entity recognition (NER) is a fundamental task of natural language processing, which aims to identify named entities (NEs) in text, and is the basis for tasks such as network clustering (Su et al., 2021), disease module identification (Tian et al., 2020), relation extraction, knowledge graph construction, and link prediction (Su et al., 2020; Lei et al., 2021; Wang et al., 2021a; Wu et al., 2021; Cai et al., 2022). Benefiting from the use of neural network models (Ma and Hovy, 2016; Yang and Zhang, 2018) and pre-trained language models (LMs) (Akbik et al., 2018; Devlin et al., 2019), NER in natural language processing (NLP) has been extensively studied in general terms of NE types, such as person names, organizations, and departments, achieving human-level performance. The most representative model for NER is BiLSTM-CRF (Ma and Hovy, 2016; Tang et al., 2019), which utilizes a bidirectional long short-term memory (BiLSTM) network to encode biomedical texts and CRF to decode named entity labels. Ma and Hovy (2016) extended LSTM-CNNsCRF by utilizing a CNN and CRF. Zhang et al. (2018) used long short-term memory (LSTM) as a baseline model, combining multi-task learning and multi-step training to improve the performance of the clinical NER datasets. However, LSTM takes a long time to process long text; considering the context, it generally adopts a bidirectional structure (Kocaman and Talby, 2021), while BERT uses the attention mechanism, and the weight of each position of text relative to another position can be calculated in parallel, which is much faster than LSTM on the premise of sufficient computing resources. Other works replace the BiLSTM encoder with pre-trained language models, such as ELMo and BERT (Devlin et al., 2019), which consider deeper semantic features and farther contextual semantics, and those works obtain better experimental results. In addition, domain-specific pre-trained language models also bring improvement in clinical NER. In the past few years, the application of the recurrent neural network (RNN) (Li and Jiang, 2017; Ju et al., 2018; Hemati and Mehler, 20192019), convolutional neural network (CNN) (Korvigo et al., 2018; Zhu et al., 2018; Nie et al., 2021), and conditional random field (CRF) (Wang et al., 2018) in biomedical named entity recognition has promoted the development of biomedical text mining models. In recent years, in order to improve the performance of NER machine learning methods, external knowledge has been used as a complement to traditional contextual information, such as base information (Akbik et al., 2018), n-gram information (Li and Jiang, 2017), and part-of-speech (POS) tagging information (Devlin et al., 2019).
Biomedical named entity recognition (BioNER), which can help drug discovery (Wang et al., 2021b; Cai et al., 2022; Wang et al., 2022), is more challenging than it is in general fields. Researchers try to use various methods to improve the performance of BioNER, some introducing pre-trained models. Pre-trained transformer language models such as BERT (Devlin et al., 2019) and its variants such as RoBERTa (Liu et al., 2019) have brought significant performance gains on a variety of language tasks. BERT has been adopted in the biomedical domain. Lee et al. (2020) trained BERT on PubMed abstracts (PubMed) and PubMed central full-text articles (PMC) and proposed BioBERT for domain-specific language representation. Fang and Wang (2021) used a cased WordPiece vocabulary trained from a biomedical corpus, which also included all PubMed abstracts and 1 million PMC full-text articles, to promote Bioformer, which is a lighter model than BioBERT. BioNEs has more diverse context relations and richer semantic-related entities, such as drug molecules, targets, proteins, channels, and pathways. For example, “Alzheimer’s disease (AD) is a neurodegenerative disease characterized by progressive memory loss and dementia.” Here, there are four different descriptions of Alzheimer’s in this sentence which are “(AD),” “neurodegenerative disease,” “progressive memory loss,” and “dementia”—some of them are diseases, and some are symptoms. By observation, we find that if one of the words can be identified, then the other entities can be identified through dependencies. For instance, the disease entity “Alzheimer’s disease” can be identified through dependency “characterized by” and characteristic entity “progressive memory loss”. In addition, knowledge is also widely used in other text mining, while the one that contains the most dependencies is the knowledge graph. Some researchers have found that incorporating external knowledge can improve the performance of NER. The integration of external knowledge is used in deep learning models to improve their performances for NER. Wang et al. (2018) used distant supervision methods based on knowledge to improve the performances of clinical NER systems via injecting the representations of concepts I n KG into the representations of tokens. Wang et al. (2019) incorporated medical dictionaries into BiLSTM-CRF and achieved SOTA performance on the CCKS2017 dataset. Nie et al. (2021) used entities from the WikiData knowledge to promote NER models via concatenating concept embeddings and token embeddings. Xiong et al. (2022) integrated the boundary information on lexicon words from multiple knowledge graphs or knowledge graph(s) and lexicon(s). Chen et al. (2021) adopted both global co-reference relations and local dependency relations for building better entity mention representations for BioNER. However, most of them treated tokens in external knowledge equally also without considering the relationship between entities, which may be helpful for BioNER. Based on this consideration, we introduce a knowledge description by mining textual entity dependencies and expect to improve the NER performance. For example, the two entities in the knowledge graph, the diseases “Alzheimer’s disease” and “Senile Dementia,” belong to the “same as” entity dependence. If we can recognize that “Alzheimer’s disease” is a disease entity, we can more accurately infer whether the other word, “Senile Dementia,” is a disease entity according to the dependence.
In this study, we first perform word-level embeddings on biomedical domain text and knowledge graph entities using a BERT preprocessing model. Then, text word-level embeddings and entity dependencies are integrated using a graph neural network (GNN), specifically using graph attention (GAT) networks (Velickovic et al., 2018). GAT has shown good performance in many tasks, such as Chinese NER and short text classification (Hu et al., 2019; Sui et al., 2019). The experimental results show that the method proposed in this paper can obtain better word representation and further improve the recognition performance of biomedical named entities.
MKGAT mainly consists of five layers, including an embedding layer, a graph neural network layer, a knowledge fusion layer, and a decoding layer (Figure 1). Sentences and nodes in the knowledge graph are fed into the pre-trained LM embedding model Bioformer to obtain semantic representation vectors. Then, the node representation incorporates local dependencies by considering adjacent node information and updates the node representation vectors by feeding them into a GAT network. The semantic feature vector and the updated node feature vector are fed into a multi-dimensional encoder, which generates a more coupled vector through feature blending. The decoder is then applied to this vector, and finally, the label indicates the entity category is the output.
To leverage dependency knowledge, we use a knowledge graph named the natural brain knowledge graph (NBKG) constructed by ourselves (Figure 2). We mine external text entity dependencies through the NBKG, hoping to improve the BioNER performance. The NBKG is developed based on a large number of medical, botany, and pharmacy encyclopedias, including PubMed (Roberts, 2001), BIOFACQUIM (Pilón-Jiménez et al., 2019), CMAUP (Zeng et al., 2018), and DrugBank (Wishart et al., 2018) databases, which contain 30,926 drug compounds, 21,771 targets, and over 100 kinds of diseases in three categories: “Brain Disease,” “Drug,” and “Target.” The types of edges in the NBKG include the same entity type dependency, such as “same as” and “belong to” and different entity type dependencies, such as “caused by” and “act on.” For example, the disease entity “Alzheimer’s disease” has the same entity type dependency “same as” with the disease entity “senile dementia,” and also, “Alzheimer’s disease” has a different entity type dependency “caused by” with the protein entity “Mfn2.” We match text words with the NBKG entity, and in this way, the external entity dependency for words in a sentence is imported to improve the representation accuracy of the whole-text entity space.
To build a domain knowledge graph, ontology needs to be used to define concepts and express relationships between concepts, to identify named entities and extract relationships from the unstructured literature, such as PubMed, to integrate knowledge with structured knowledge, such as DrugBank, and to disambiguate semantics. Finally, knowledge will be sorted into the form of a knowledge graph according to the concepts, attributes, and relationships defined by ontology; the general process is shown in Figure 2.
Since the domain knowledge map is built, compared with extracting ontology structures from different databases, NBKG ontology is built manually, and the conceptual relationship displayed in the ontology is shown in Figure 3A. For unstructured knowledge, such as the PubMed literature, the finely tuned BioBERT is used to extract named entities and relationships. For entity normalization, we used the International Classification of Diseases, Tenth Edition (ICD-10), and other standards. The structure example of the final NBKG is shown in Figure 3B.
To obtain the context representation of each token of input sentence
where
where m is the number of nodes in the knowledge graph,
The external entity dependency introduced by the knowledge graph is considered to enhance the context representation of the token. The computational process of GAT (Velickovic et al., 2018) can be summarized as follows, given a graph
The graph attention mechanism is used to aggregate the information on neighbor nodes and their corresponding standardized attention scores, and the attention score
where
where
Similar to the method used by Chen et al. (2021), the sentence word embeddings
where
A conditional random field (CRF) is used for named entity label prediction. Given the input sentence
where
We tested the performance of the model on three general datasets in BioNER, NCBI-disease corpus (Dogan et al., 2014), BC2GM, and BC5CDR-chem (Li et al., 2016) datasets. The NCBI-disease corpus contains 793 PubMed abstracts, 6,892 disease mentions, and 790 unique disease concepts. BC2GM contains 20,703 labeled entities, and BC5CDR corpus consists of 1,500 PubMed articles with 4,409 annotated chemicals, which are used for the experiment. The NCBI-disease corpus is fully annotated at the mention and concept level to serve as a research resource for the biomedical natural language processing community. BC2GM is from a gene mention tagging task, as part of the BioCreative II challenge, which is concerned with the named entity extraction of gene and gene product mentions in the text. BC5CDR is introduced in BioCreative V CDR task corpus, which is a resource for chemical disease relation extraction.
The number of common disease concepts existing both in NBKG and NCBI-disease is 92, and the number of common gene/protein concepts existing both in NBKG and BC2GM is 1,468, which is displayed in the “common” column in the table, while the number of common gene/protein concepts existing both in NBKG and BC5CDR-chem is 257 (Table 1).
We use the “BIO” (B—begin, I—inside, and O—outside) labels to represent the boundaries of entity mentions. We set the maximum length of sentences to 175 for NCBI-disease corpus, 300 for BC2GM, and 200 for BC5CDR-chem, which covers over 99% of sentences. The hidden size of the BERT is set to 512. The dropout rate is set to 0.2 in BERT and the GNN updating layer. The Adam optimization algorithm is used as the optimizer with a learning rate of 1e-5. We use a layer GAT to model external knowledge with 50 hidden nodes. The number of epochs we set is 10, and the batch size is set to 35 at each epoch. Bioformer, pre-trained from the scratch on the same corpus as the vocabulary, which included 33 million PubMed abstracts and 1 million PMC full-text articles, is used to embed the sentence. We train the entities embedding on the NBKG using Bioformer. All hyper-parameters in Bioformer for entities embedding training are set to their default values except the following hyperparameters: the number of epochs is set to 10, and the batch size is set to 35. All experiments are conducted on RTX 3090.
We use several metrics to evaluate the model performance, including precision, recall, and F1 score. Precision is the ratio of true positives in the identified positive samples, which reflects the classification accuracy of the model for BioNEs tokens:
where TP indicates the number of positive classes predicted as positive classes, and FP indicates the number of negative classes predicted as positive classes. Recall represents the proportion of all positive samples in the test set that are correctly identified as positive samples, which reflects the ability of the model to distinguish BioNEs:
where FN indicates the number of positive classes predicted as negative classes. The F1 score is one of the most commonly used metrics in classification and information retrieval, reflecting the average performance of model precision and recall:
Using 3,924 disease BioNEs, 20,703 gene/protein BioNEs and 21,899 knowledge graph tokens, we make a comparison of our model with existing methods on the two datasets (Table 1).
Layered-BiLSTM-CRF without considering external knowledge was used for the test first. Then we compare our method with the SOTA method using BERT as the embedded layer, including BERT base, BioBERT v1.0, BioBERT v1.1, and MKRGCN considering external knowledge.
Layered-BiLSTM-CRF is a neural layered model for nested NER.
MTM-CW is a multi-task learning model which shares character-level embedding parameters, word-level embedding parameters, and character and word layer embedding parameters for NER.
BERT-base (Devlin et al., 2019) is a contextualized word representation model that is based on a masked language model and pre-trained using bidirectional transformers. We use the embeddings from it with BiLSTM-CRF architecture as a baseline.
BioBERT v1.0 (Lee et al., 2020) is a BERT-base pre-trained with 200 thousand PubMed abstracts and 270 thousand PMC full-text articles. We use the NER fine-tuning version of it with BiLSTM-CRF architecture as a baseline.
BioBERT v1.1 (Lee et al., 2020) is the same as BioBERT v1.0 but pre-trained with 1 million PubMed abstracts.
MKRGCN (Xiong et al., 2022) leverages lexicon of words in Chinese and domain knowledge graph concepts to consider the boundaries of NEs.
EnRel-G (Chen et al., 2021b) incorporates entity mention relations based on both global co-reference relations and local dependency relations by graph neural networks.
We run our model five times with different seeds, and report the precision, recall, and F1 score.
We have achieved the best results, as highlighted. MKGAT outperforms layered-BiLSTM-CRF, MTM-CW, BERT-base (Wiki + Books), BioBERT v1.0 (+PubMed + PMC), BioBERT v1.1 (+PubMed), and MKRGCN on the NCBI-disease corpus, BC2GM and BC5CDR-Chem datasets (Table 2). MKGAT achieves an F1 score of 90.78% on the NCBI-disease dataset, an F1 score of 88.12% on the BC2GM dataset, and an F1 score of 95.66% on the BC5CDR-chem dataset, higher than the existing best performances reported by Xiong et al. (2022) by 0.98% in F1 score on the NCBI-disease dataset and 0.31% in F1 score on the BC2GM dataset, respectively. Compared with Bioformer, MKGAT achieves higher F1 score by 1.42%, 2.91%, and 2.21% on the NCBI-disease corpus, BC2GM, and BC5CDR-chem datasets, respectively. The improvements from external knowledge encoding and the fusion layer are significant. In the case of the methods that can utilize KG concepts and local dependency, using them together may be more effective. For example, MKGAT performs better than MKRGCN on the NCBI-disease corpus but sometimes worse on the BC2GM (MKRGCN-max is higher than MKGAT-min). This result indicates that leveraging local dependency from external knowledge for NER is not simple. The reason may be that the representation space of article words is different from that of KG concepts. We can see that MKGAT performs better on NCBI-disease corpus than BC2GM as same as other models (Figure 4). This may be caused by the scale of the dataset and the length of the entities in them. MKGAT provides an effective attention mechanism to leverage local dependency for NER and also shows potential to be applied to other tasks.
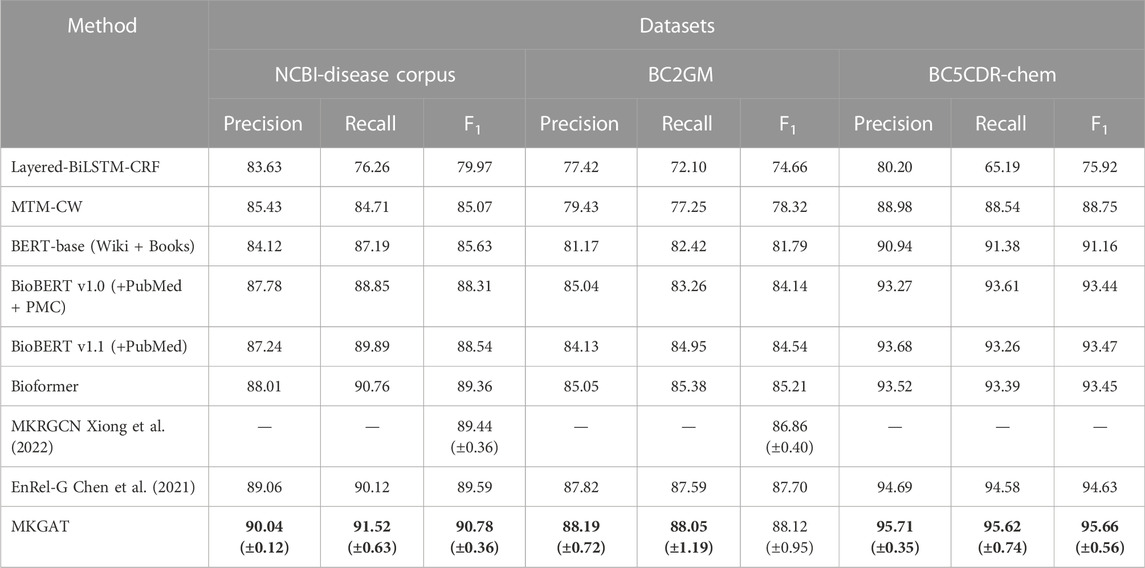
TABLE 2. Comparison of different methods on the NCBI-disease corpus, BC2GM and BC5CDR-Chem datasets.
We conduct an ablation study on the three BioNER datasets to evaluate the performance of the embedding mode and knowledge from KG on the MKGAT. The models with “concept” are the models using BERT to embed knowledge graph concepts without relation, and the models with “relation” are the models using the GNN to model knowledge graph concepts and their relations (Table 3). It is worth mentioning that the effect of the structure of BiLSTEM + CRF is compared. The results show that replacing the BiLSTM module with a full connection can still achieve better results, which may be related to our concern about the relationship from additional knowledge rather than context. Comparing the results of Bioformer with those of the Bioformer concept, we can see both the embedding mode and source of knowledge can bring improvements. The performance that Bioformer + GAT relation gets better scores on precision, recall, and F1 shows that considering relations of concepts can help find out a better representation space for all words in articles so as to identify entities more accurately.
We conduct a fine-grained study on the two NER datasets to identify which kind of entity can be recognized effectively in MKGAT.
It is shown that MKGAT achieves an F1 score of 90.33%, 91.81%, and 91.67% for labels “B,” “I,” and “O,” respectively, on the NCBI-disease corpus (Figure 5A), an F1 score of 88.05%, 88.32%, and 88.10% for labels “B,” “I”, and “O,” respectively, on the BC2GM (Figure 5B), and an F1 score of 95.69%, 95.32%, and 95.34% for labels “B,” “I,” and “O,” respectively, on the BC5CDR-chem dataset (Figure 5C). Compared with “B” and “O,” MKGAT achieved a higher score on label “I”. The reason may be that words with label “I” can be better characterized because of their greater quantity.
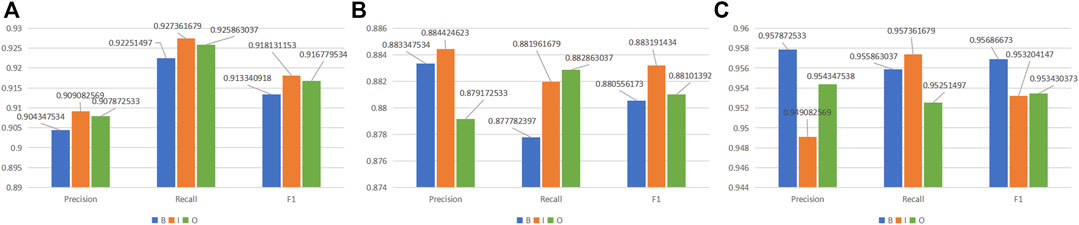
FIGURE 5. MKGAT performance for label “BIO” on (A) NCBI-disease, (B) BC2GM, and (C) BC5CDR-chem datasets.
From a pharmacological point of view, medical entities such as drugs, diseases, targets, and pathways are inseparable, and any of them will have an impact on the action of the drug. Then, for related research work in biomedicine, it is necessary to consider the dependencies between them, including BioNER. With the advent of the Internet era, especially with the outbreak of COVID-19, a wealth of information about diseases, drugs, and receptors has accumulated. However, most of the information is stored in the form of the literature. In addition to unstructured data, such as text data, many related semi-structured and unstructured data in different forms have also emerged, such as knowledge graphs. Therefore, it seems to us most natural to carry out biomedical named entity identification in the context of incorporating external knowledge. In this paper, we use a simple knowledge graph constructed by ourselves from PubMed (Roberts, 2001), BIOFACQUIM (Pilón-Jiménez et al., 2019), CMAUP (Zeng et al., 2018), and DrugBank (Wishart et al., 2018) databases. In order to take advantage of the dependencies in the knowledge graph, we match the entities in the graph with the words in the training sentences. To explore the effects of drug, disease, and target dependencies on BioNER, we constructed a model which first performs word-level embeddings on biomedical domain text and knowledge graph entities using a BERT preprocessing model. Then, text word-level embeddings and entity dependencies are integrated by using a graph neural network (GNN), specifically using graph attention (GAT) networks (Velickovic et al., 2018). GAT has shown good performance in many tasks, such as Chinese NER and short text classification (Hu et al., 2019; Sui et al., 2019).
To test the effect of the model, we conduct experiments using real-world datasets and compare them with seven currently more advanced methods (Table 2). MKGAT achieves an F1 score of 90.78% on the NCBI-disease dataset and an F1 score of 88.12% on the BC2GM dataset, higher than the existing best performances reported by Xiong et al. (2022) (Fang and Wang, 2021) by 0.98% in F1 score on the NCBI-disease corpus dataset and 0.31% in F1 score on the BC2GM dataset, respectively. Compared with Bioformer, MKGAT achieves higher F1 scores by 1.42% and 2.91% on the NCBI-disease dataset and the BC2GM dataset, respectively. The improvements from external knowledge encoding and the fusion layer are significant. Compared with MKRGCN (Xiong et al., 2022) of “integrating entity-related words with external knowledge,” our model is also very effective, indicating that “integrating entity dependencies” can also improve the accuracy of biomedical entity recognition. Compared with EnRel-G (Chen et al. (2021)), which incorporates entity-mentioned relations based on both global co-reference relations and local dependency relations, our model is more accurate, which means external knowledge brings more effective information. In addition, there are LSTM-based methods that have achieved good results in recent years, such as the SparkNLP’s BiLSTM-CNN structure model (Kocaman and Talby, 2021). However, because LSTM cannot perform parallel computing, this method is less efficient than the model proposed in this paper, so no further discussion will be made.
We also observed the performance of MKGAT using different modules on the three datasets (Table 3). It is found that when we do not use the pre-training model to fine-tune and only use the RNN to focus on the semantics within sentences, the performance degradation is serious, which confirms that compared with ordinary NEs, the semantic relationship of BioNEs is much more complex. The recognition accuracy is also unsatisfactory when we do not consider the data in the knowledge graph. This result confirms the effectiveness of our choice to incorporate external dependency knowledge in the process of identifying entities.
So is it really a dependency that affects the accuracy of the identified entity? We thought of this question and explored it. First, we only use the “entities in the knowledge graph corresponding to the words in the training set” to test. Then, we compare it with the test results using the “relational entities corresponding to the entities in the knowledge graph,” and we find that the results of integrating the relationship are better. However, it is worth noting that the improvement brought by dependencies is not much from the results alone. Why is the dependency relationship not improved so much? Later, we thought about whether the semantic relationship of BioNEs in texts is complicated, which leads to the confusion of recognition after incorporating the external dependency relationship. Therefore, we take the dependency entity as an attribute of BioNEs through GAT (Velickovic et al., 2018) and try to make the model focus on BioNEs themselves. The results show that it is helpful. Due to the complexity of the structure of BioNEs, such as “ (), numbers, etc.”, in order to explore the completeness of the model’s recognition, we also compared the boundary and internal recognition of BioNEs on three datasets (Figure 5). We found that although MKGAT can accurately identify NEs, the identification accuracy of boundaries is not as high as that of the interior, which also explains the direction of future research for us.
Although the model presented in this paper provides valuable insights into the application of pharmacological dependencies on BioNER, it is also important to consider the limitations to this study. As mentioned previously, it can be seen from the results that MKGAT is not accurate enough to identify the boundary of the entity; from the perspective of external knowledge, the knowledge graph constructed by us cannot cover all possible entities involved, resulting in the inaccurate recognition of individual BioNEs, and from the way of external knowledge integration, all kinds of local dependencies of entities in the knowledge graph are represented as an adjacent matrix, which may lose some information. It is important to remember, therefore, that the model presented here simply provides an idea for BioNER to leverage external knowledge.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
PH conceived the study. PH, XL, XW, and SW contributed to the study design. Data analysis was curated by PH, CG, and WC. The first draft of the manuscript was written by PH. XW, SW, CG, and WC critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Key Research and Development Project of China (2021YFA1000102 and 2021YFA1000103), the Natural Science Foundation of China (Grant nos. 61873280 and 61972416), Taishan Scholarship (tsqn201812029), the Foundation of Science and Technology Development of Jinan (201907116), the Shandong Provincial Natural Science Foundation (ZR2021QF023), the Fundamental Research Funds for the Central Universities (21CX06018A), the Spanish project PID 2019-106960GB-I00, and Juan de la Cierva IJC2018-038539-I.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akbik, A., Duncan, B., and Roland, V.. Contextual string embeddings for sequence labeling. In Proceedings of the 27th International Conference on Computational Linguistics, August 2018, Santa Fe, New Mexico, USA, pages 1638–1649.
Cai, L., Lu, C., Xu, J., Meng, Y., Wang, P., Fu, X., et al. (2022). Drug repositioning based on the heterogeneous information fusion graph convolutional network. Brief. Bioinform. 22 (6), bbab319. doi:10.1093/bib/bbab319
Chen, P., Ding, H., Araki, J., and Huang, R. (2021) Explicitly capturing relations between entity mentions via graph neural networks for domain-specific named entity recognition[C], Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing, August 2021, Volume 2.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K. (2019). Bert: Pre-training of deep bidirectional transformers for language understandingProceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language TechnologiesJuna 2019Minneapolis, Minnesota 1, Minneapolis, Minnesota: Association for Computational Linguistics, 4171–4186.
Dogan, R. I., Leaman, R., and Lu, Z. (2014). NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inf. 47, 1–10. doi:10.1016/j.jbi.2013.12.006
Fang, L., and Wang, K.. (2021) Team bioformer at BioCreative VII LitCovid track: Multic-label topic classification for COVID-19 literature with a compact BERT model[C], Proceedings of the seventh BioCreative challenge evaluation workshop, November 2021.
Hemati, W., and Mehler, A. (2019). LSTMVoter: Chemical named entity recognition using a conglomerate of sequence labeling tools. J. Cheminform. 11 (1), 3–7. doi:10.1186/s13321-018-0327-2
Hu, L., Yang, T., Shi, C., Ji, H., and Li, X. (2019) Heterogeneous graph attention networks for semi-supervised short text classification. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), November 2019, Hong Kong, China, pages 4823–4832.
Ju, M., Miwa, M., and Ananiadou, S.. (2018) A neural layered model for nested named entity recognition. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, June 2018, New Orleans, Louisiana, Volume 1, 1446–1459.
Kocaman, V., and Talby, D.. Biomedical named entity recognition at scale[C], Proceedings of the International Conference on Pattern Recognition, Milan, Italy. Springer, 2021: 635–646.
Korvigo, I., Holmatov, M., Zaikovskii, A., and Skoblov, M. (2018). Putting hands to rest: Efficient deep CNN-RNN architecture for chemical named entity recognition with no hand-crafted rules. J. Cheminform. 10 (1), 28–10. doi:10.1186/s13321-018-0280-0
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., et al. (2020). BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36 (4), 1234–1240. doi:10.1093/bioinformatics/btz682
Lei, L., Gao, Z., Wang, Y.-T., Zhang, M. W., Ni, J. C., Zheng, C. H., et al. (2021). Scmfmda: Predicting microRNA-disease associations based on similarity constrained matrix factorization. PLoS Comput. Biol. 17 (7), e1009165. doi:10.1371/journal.pcbi.1009165
Li, J., Sun, Y., Johnson, R. J., D., S., Chinh, H., R., L., et al. BioCreative V CDR task corpus: A resource for chemical disease relation extraction[J]. Database, 3455, 2016.
Li, L., and Jiang, Y., Biomedical named entity recognition based on the two channels and sentence-level reading control conditioned lstm-crf, Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Nov 2017. IEEE, 380.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen, D., et al. (2019) Roberta: A robustly optimized bert pretraining approach[J]. http://arxiv.org/abs/1907.11692.
Ma, X., and Hovy, E. (2016).End-to-end sequence labeling via bi-directional LSTM-CNNsCRF, Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, August 2016, Berlin, Germany, 1, Berlin, Germany: Association for Computational Linguistics, 1064–1074.
Nie, B., Ding, R., Xie, P., Huang, F., Qian, C., and Si, L. (2021) Knowledge-aware named entity recognition with alleviating heterogeneity. in Proceedings of the Thirty-Fifth AAAI Conference on Artificial Intelligence, AAAI 2021, February 2021, 13595–13603.
Pilón-Jiménez, B. A., Saldívar-González, F. I., Díaz-Eufracio, B. I., and Medina-Franco, J. L. (2019). Biofacquim: A Mexican compound database of natural products. Biomolecules 9 (1), 31. doi:10.3390/biom9010031
Roberts, R. J. (2001). PubMed central: The GenBank of the published literature. Natl. Acad. Sci. 26, 544.
Su, Y., Liu, C., Niu, Y., Cheng, F., and Zhang, X. (2021). A community structure enhancement based community detection algorithm for complex networks. IEEE Trans. Syst. Man. Cybern. Syst. 51 (5), 2833–2846. doi:10.1109/tsmc.2019.2917215
Su, Y., Sen, L., Zheng, C., and Zhang, X. (2020). A heuristic algorithm for identifying molecular signatures in cancer. IEEE Trans. Nanobioscience 19 (1), 132–141. doi:10.1109/TNB.2019.2930647
Sui, D., Chen, Y., Liu, K., Zhao, J., and Liu, S. (2019) Leverage lexical knowledge for Chinese named entity recognition via collaborative graph network. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLPIJCNLP), November 2019, Hong Kong, China, pages 3821–3831.
Tang, B., Jiang, D., Chen, Q., Wang, X., Yan, J., and Shen, Y. (2019) De-identification of clinical text via Bi-LSTM-CRF with neural Language Models. in Proceedings of the AMIA 2019, American Medical Informatics Association Annual Symposium, Washington, DC, USA, November 2019.
Tian, Y., Su, X., Su, Y., and Zhang, X. (2020). EMODMI:A multi-objective optimization based method to identify disease modules. IEEE Trans. Emerg. Top. Comput. Intell. 5 (4), 570–582. doi:10.1109/tetci.2020.3014923
Velickovic, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., and Bengio, Y. (2018).Graph attention networks, Proceedings of the 6th International Conference on Learning Representations, Vancouver, BC, Canada, April, 2018, Vancouver, BC, Canada: OpenReview.net.
Wang, H., Zhao, J., Su, Y., Zheng, C. H., et al. (2021a). scCDG: A Method based on DAE and GCN for scRNA-seq data Analysis. IEEE/ACM Trans. Comput. Biol. Bioinform., 3126641. doi:10.1109/tcbb.2021.3126641
Wang, Q. i., Zhou, Y., Gao, D., Xia, Y., and He, P. (2019). Incorporating dictionaries into deep neural networks for the Chinese clinical named entity recognition. J. Biomed. Inf. 92, 103133. doi:10.1016/j.jbi.2019.103133
Wang, S., Jiang, M., Zhang, S., Wang, X., Yuan, Q., Wei, Z., et al. (2021b). MCN-CPI: Multiscale convolutional network for compound-protein interaction prediction. Biomolecules 11 (8), 1119. doi:10.3390/biom11081119
Wang, S., Song, T., Zhang, S., Jiang, M., Wei, Z., and Li, Z. (2022). Molecular substructure tree generative model for de novo drug design. Brief. Bioinform. 23 (2), bbab592. doi:10.1093/bib/bbab592
Wang, Z., Qu, Y., Chen, L., Shen, J., Zhang, W., and Zhang, S. (2018). Label-aware double transfer learning for cross-specialty medical named entity recognition. Available at: https://arxiv.org/abs/1804.09021.
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46 (D1), D1074–D1082. doi:10.1093/nar/gkx1037
Wu, Q., Rui-Fen, C., Xia, J., Ni, J. C., Zheng, C. H., and Su, Y. (2021). Extra trees method for predicting LncRNA-disease association based on multi-layer graph embedding aggregation. IEEE/ACM Trans. Comput. Biol. Bioinform., 1. doi:10.1109/tcbb.2021.3113122
Xiong, Y., Peng, H., Xiang, Y., Wong, K. C., Chen, Q., Yan, J., et al. (2022). Leveraging Multi-source knowledge for Chinese clinical named entity recognition via relational graph convolutional network. J. Biomed. Inf. 128, 104035. doi:10.1016/j.jbi.2022.104035
Yang, J., and Zhang, Yue. Ncrf++: An opensource neural sequence labeling toolkit. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, July 2018, Melbourne, Australia.
Zeng, X., Zhang, P., Wang, Y., Qin, C., Chen, S., He, W., et al. (2018) Cmaup: A database of collective molecular activities of useful plants[J]. Nuclc Acids Res., 344, 66.
Zhang, Q., Li, Z., Feng, D., Li, D., Huang, Z., and Peng, Y. (2018) Multitask learning for Chinese named entity recognition. in Proceedings of the Advances in Multimedia Information Processing - PCM 2018 - 19th Pacific-Rim Conference on Multimedia, Hefei, China, September 2018, vol. 11165, 653–662.
Keywords: biomedical named entity recognition, pre-training model, graph attention network, external knowledge, entity dependencies
Citation: Han P, Li X, Wang X, Wang S, Gao C and Chen W (2022) Exploring the effects of drug, disease, and protein dependencies on biomedical named entity recognition: A comparative analysis. Front. Pharmacol. 13:1020759. doi: 10.3389/fphar.2022.1020759
Received: 16 August 2022; Accepted: 02 December 2022;
Published: 21 December 2022.
Edited by:
Joram Posma, Imperial College London, United KingdomReviewed by:
Tao Song, Polytechnic University of Madrid, SpainCopyright © 2022 Han, Li, Wang, Wang, Gao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xun Wang, d2FuZ3N5dW5AdXBjLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.