
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Pharmacol. , 26 September 2022
Sec. Pharmacogenetics and Pharmacogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1013527
This article is part of the Research Topic The Role of Pharmacogenomics in Addressing Health Disparities: The Path, The Promise, and The Barriers View all 7 articles
The emergence of Coronavirus disease 2019 (COVID-19) has changed the world in an irrevocable direction. The changes have affected all vital sectors, including finance, education, communications, research, and, undoubtedly, the medical practice. Every field related to human health explored how to escalate its performance and updated its research priorities to respond to the situation imposed by the devastating pandemic. Pharmacogenomics (PGx) is not an exception. Multiple researchers in the field investigated the available PGx information related to medications used in COVID-19 management, and various reviews have appeared since late 2020 till today (Takahashi et al., 2020; Badary, 2021; Fricke-Galindo and Falfán-Valencia, 2021; Biswas et al., 2022; Franczyk et al., 2022). Most of these reviews concluded that the evidence of the pharmacogenomic interactions for drugs used in the new Coronavirus-induced severe respiratory syndrome (SARS-CoV2) infection management is not strong enough to warrant testing. Herein, there are common findings among these reviews that deserve highlighting. Moreover, extrapolating these findings into populations underrepresented in PGx research that were challenged by the pandemic, equally to all other populations, can reflect on future practices. The following insights explore the probability of the situation imposed by COVID-19 being an opportunity, rather than a threat, for largescale PGx adoption.
The earliest publication of SARS-CoV2 treatment PGx was a literature review by Takahashi and colleagues (Takahashi et al., 2020), which was available online in August 2020. In their work, the authors listed the available therapeutic options for SARS-CoV2 infection used in the US healthcare system and in interventional clinical trials registered on ClinicalTrials.gov at the time of publication. The authors reviewed the pharmacogenetic biomarkers found to be associated with these drugs. None of the studied drugs had approved PGx-guided dosing guidelines, and few had moderate to low significance PGx associations. The illustrated candidate response and toxicity biomarkers were retrieved from previous research in diseases other than COVID-19. The authors highlighted that no PGx data is available from SARS-CoV2 patients yet.
Nevertheless, Takahashi and colleagues used the association between abacavir, the anti-HIV agent, and HLA-B*57:01 to exemplify the success of pharmacogenomics implementation in infectious diseases. At the same time, they underscored the time between identifying HIV and recognizing this PGx association and its implementation success (Takahashi et al., 2020). Compared to HIV-induced acquired immunodeficiency, the COVID-19-induced illness burden on global public health is unmatched, and the urgent need for better control measures is substantially different.
On the other hand, and due to limited PGx data for most of the antiviral drugs re-purposed for COVID-19, some reviews illustrated the available sporadic reports about PGx associations from the literature (Badary, 2021; Fricke-Galindo and Falfán-Valencia, 2021; Franczyk et al., 2022). The listed studies’ outcomes were primarily inconclusive and limited by sample size or conflicting results. Nevertheless, demonstrating them collectively displays the current deficiency in PGx research on drugs used in infectious diseases compared to PGx research on non-communicable conditions medications.
One common outcome of the COVID-19-PGx reviews is listing the candidate PGx effectors on drugs used for SARS-CoV2. For example, SLCO1B1*4 was recorded as a candidate effector on lopinavir clearance. The variations in this allele frequency among populations were briefly highlighted, and its frequency was reported as 14% in Europeans, 6% in Africans, and 0.3% in East Asians (Takahashi et al., 2020). However, in some Middle Eastern populations, this allele’s prevalence reaches 20%–45% (Pasanen et al., 2008; Al-Mahayri et al., 2020). The high prevalence of a deficient SLCO1B1 allele could be associated with increased clearance of lopinavir in these ethnicities. Although there has been a recommendation against using lopinavir in SARS-CoV2 in the early 2021 (COVID-19 Treat Guidelines, 2022), SLCO1B1 is active in the transportation of other antivirals, like atazanavir.
Furthermore, in a more recent SARS-CoV2 drugs PGx review (Biswas et al., 2022), the authors went beyond the PGx interactions of first-line COVID-19 used therapeutics to consider all medications used in supportive therapy, drug-drug interactions, drug-herbal interactions, besides the genetic biomarkers of disease severity. In their review, Biswas and colleagues listed the most critical PGx-biomarkers that interact with drugs used in COVID-19 main therapeutic agents (i.e., antivirals) and supportive therapies (i.e., corticosteroids, antiplatelets, anticoagulants, non-steroidal anti-inflammatory agents, etc.). Accordingly, they suggested a panel of genes of interest that can be used to assess the impact of PGx interactions on COVID-19 and to establish COVID-19 precision medicine. These genes include CYP3A4/5, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, ABCB1, and SLCO1B1, among others. These specific genes illustrate a wide range of inter-population and inter-ethnic variability, as reported in studies from Africa and Southeast Asia (da Rocha et al., 2021; Runcharoen et al., 2021). Table 1 lists some of these variants and demonstrates the difference in minor allele frequencies between Europeans, commonly represented in genetic data, and selected examples of rarely studied populations. Indeed, the lack of diversity in genomic data has an unforeseen effect on global health. For instance, although the African populations exhibit the most diverse genomes among human populations, the most minor data about COVID-19 host genomes came from Africa. Simultaneously, African populations suffer from the highest burden of infectious diseases (Zhang et al., 2022). However, whether the PGx variations in the actionable genes among populations can explain any differences in COVID-19 treatment outcomes was not evaluated, though not possible to exclude given the lack of such studies.
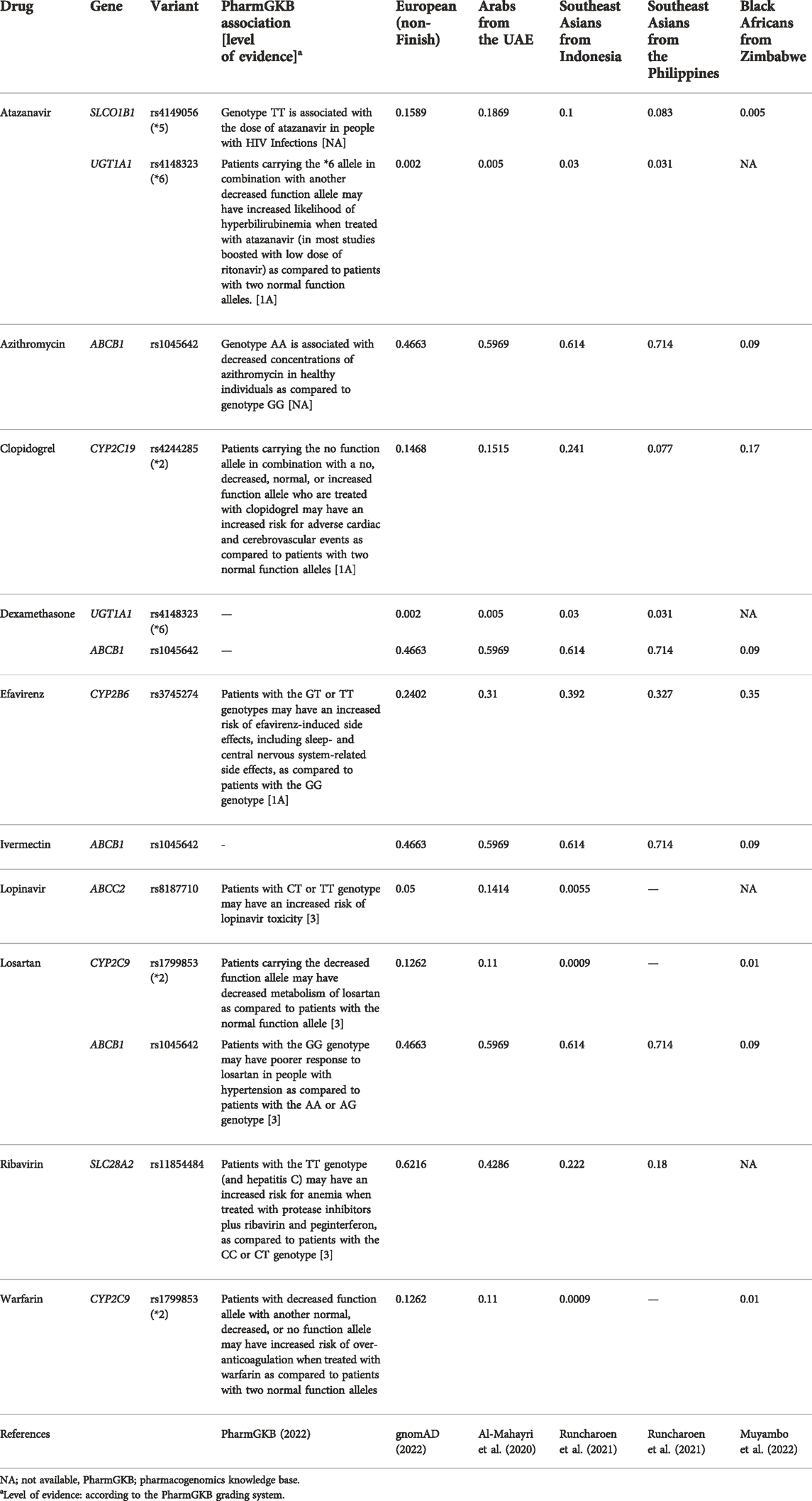
TABLE 1. Examples of the suggested gene-drug pairs related to the management of COVID-19 and the frequencies of their minor alleles in Europeans compared to selected rarely-studied populations.
According to the mentioned COVID-19 PGx reviews, a significant limitation hindering PGx data use for COVID-19 patients is the unavailability of the patient’s PGx profile when admitted for acute illness. Pre-emptive PGx testing would be one solution for this limitation, while providing a reactive point-of-care PGx testing is another. Stevenson and colleagues have shown that in a cohort of patients hospitalized for COVID-19, nine in 10 patients had at least one order of a drug with a PGx recommendation. Moreover, through a simulation analysis, the authors found that 17 treatment modifications per 100 patients would be feasible if pre-emptive PGx data were available in the EHRs of these patients at the time of admission. The authors also emphasized that the multigene-PGx results are projected to positively impact patients’ healthcare years after hospital discharge (Stevenson et al., 2021).
In a similar attempt to explore the PGx-data potential impact on COVID-19 outcomes, Sahana and coworkers analyzed the PGx variants from the “IndiGen” dataset, the dataset of 1000 + Indian whole genome sequences. The authors concluded that the population-specific PGx variation landscape, if utilized, could have contributed to designing population-specific clinical trials and expediting decision-making throughout the pandemic (Sahana et al., 2021).
Another critical point to consider, specifically during a pandemic when few therapeutic options are available, is that any PGx testing is limited by proving its cost-effectiveness (Takahashi et al., 2020). However, if preemptive PGx testing had been adopted and integrated into the health records, PGx data would have proved its cost-effectiveness, and such limitations would have been eliminated. It has been repeatedly shown that almost 96%–99% of healthy individuals carry an actionable PGx variant if tested through a multigene testing (Chanfreau-Coffinier et al., 2019; Tasa et al., 2019).
Nevertheless, even if the previous controversial points were resolved and the PGx data is available at the point of admission, it cannot be utilized unless clinical guidelines are available at the hands of the clinician. This fact brings us back to the difficulty in generating such guidelines without extensive prospective studies, which should prove cost-effectiveness besides positive or equivalent clinical outcomes. There is a severe need to break this vicious circle before a new pandemic strikes. Utilizing novel machine learning approaches (Gaziano et al., 2021) and integrating epidemiological methodologies like Mendelian randomization (Khasawneh et al., 2022) can offer much-needed supporting evidence. Machine learning promises to fill the gap between translational research and clinical practice, which can empower healthcare systems through analyzing real-life data and building prediction models (Santus et al., 2021). Nevertheless, suppose such unconventional approaches fail to gain the practitioners’ and regulators’ confidence or fail to harness ethical concerns. In that case, PGx implementation might be stuck in the same vicious circle for a long time.
Interestingly, COVID-19 patients’ DNA biorepositories have already been established (COVID-19 hg, 2022). While these were collected to fuel COVID-19 research, extracting PGx data and returning it to patients’ HER should be considered whenever the collected consents and the governing regulations allow such a practice (Stevenson et al., 2021). Indeed, utilizing clinical and genomic data gathered during the pandemic can unleash opportunities for precision medicine. For example, the retrospective analysis of clinical and genomic data of 4,125 patients hospitalized for COVID-19 showed that intermediate or poor CYP2C19 metabolizers treated with remdesivir experienced higher alanine aminotransferase (ALT) elevations compared to normal, rapid, or ultrarapid metabolizers. Due to its emergency approval for COVID-19, there was limited data about remdesivir safety or pharmacokinetics before this report (Tuteja et al., 2022). Accordingly, this recent finding about CYP2C19-remdesivir interaction exemplifies the potential gains of the availability of PGx information in patients’ health records.
Herein, the minimal COVID-19 PGx available data could be seen as an unprecedented motivation to accelerate the PGx implementation process. With the decrease in genetic testing costs, the increase in the number of available techniques for genotyping, and the evolution in data science that allows optimal data storage and making sense of genetic information, there is an immense need to consider PGx testing in the population scale and maintain this information in medical health records. Having this information at the point of admission due to an acute illness or in the mid of a pandemic is, undoubtedly, a rich source for evaluating the practice of precision medicine in real life. While developed countries with more robust healthcare systems can be considered the ultimate candidates for taking the lead in PGx implementation, developing countries and those with emerging economies inhabited by poorly studied communities should explore investing in population-PGx as a plausible approach to achieving cost-effective precision health care.
To conclude, before the next pandemic actively hits, there is a need to seriously consider preemptive PGx testing implementation in different populations. Postponing such initiatives due to doubts about cost-effectiveness or performance feasibility threatens time, money, and probably the lives of people who could have been spared from unnecessary treatments or adverse drug events.
Conceptualization and writing by ZA-M.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Mahayri, Z. N., Patrinos, G. P., Wattanapokayakit, S., Iemwimangsa, N., Fukunaga, K., Mushiroda, T., et al. (2020). Variation in 100 relevant pharmacogenes among emiratis with insights from understudied populations. Sci. Rep. 10, 21310. doi:10.1038/s41598-020-78231-3
Badary, O. A. (2021). Pharmacogenomics and COVID-19: Clinical implications of human genome interactions with repurposed drugs. Pharmacogenomics J. 21, 275–284. doi:10.1038/s41397-021-00209-9
Biswas, M., Sawajan, N., Rungrotmongkol, T., Sanachai, K., Ershadian, M., and Sukasem, C. (2022). Pharmacogenetics and precision medicine approaches for the improvement of COVID-19 therapies. Front. Pharmacol. 13, 835136. doi:10.3389/fphar.2022.835136
Chanfreau-Coffinier, C., Hull, L. E., Lynch, J. A., DuVall, S. L., Damrauer, S. M., Cunningham, F. E., et al. (2019). Projected prevalence of actionable pharmacogenetic variants and level A drugs prescribed among us veterans health administration pharmacy users. JAMA Netw. Open 2, e195345. doi:10.1001/jamanetworkopen.2019.5345
COVID-19 hg (2022). COVID-19 host genetics initiative. Available at: https://www.covid19hg.org/(Accessed September 4, 2022).
COVID-19 Treat Guidelines (2022). Lopinavir/ritonavir and other HIV Protease Inhibitors. COVID-19 Treat. Guidel. Available at: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/lopinavir-ritonavir-and-other-hiv-protease-inhibitors/(Accessed July 7, 2022).
da Rocha, J. E. B., Othman, H., Botha, G., Cottino, L., Twesigomwe, D., Ahmed, S., et al. (2021). The extent and impact of variation in ADME genes in sub-saharan african populations. Front. Pharmacol. 12, 634016. doi:10.3389/fphar.2021.634016
Franczyk, B., Rysz, J., Miłoński, J., Konecki, T., Rysz-Górzyńska, M., and Gluba-Brzózka, A. (2022). Will the use of pharmacogenetics improve treatment efficiency in COVID-19? Pharmaceuticals 15, 739. doi:10.3390/ph15060739
Fricke-Galindo, I., and Falfán-Valencia, R. (2021). Pharmacogenetics approach for the improvement of COVID-19 treatment. Viruses 13, 413. doi:10.3390/v13030413
Gaziano, L., Giambartolomei, C., Pereira, A. C., Gaulton, A., Posner, D. C., Swanson, S. A., et al. (2021). Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat. Med. 27, 668–676. doi:10.1038/s41591-021-01310-z
gnomAD (2022). gnomAD-genome aggregation database. Available at: https://gnomad.broadinstitute.org/(Accessed August 7, 2022).
Khasawneh, L. Q., Al-Mahayri, Z. N., and Ali, B. R. (2022). Mendelian randomization in pharmacogenomics: The unforeseen potentials. Biomed. Pharmacother. Biomedecine Pharmacother. 150, 112952. doi:10.1016/j.biopha.2022.112952
Muyambo, S., Ndadza, A., Soko, N. D., Kruger, B., Kadzirange, G., Chimusa, E., et al. (2022). Warfarin pharmacogenomics for precision medicine in real-life clinical practice in southern Africa: Harnessing 73 variants in 29 pharmacogenes. Omics J. Integr. Biol. 26, 35–50. doi:10.1089/omi.2021.0199
Pasanen, M. K., Neuvonen, P. J., and Niemi, M. (2008). Global analysis of genetic variation in SLCO1B1. Pharmacogenomics 9, 19–33. doi:10.2217/14622416.9.1.19
PharmGKB (2022). PharmGKB. Available at: https://www.pharmgkb.org/(Accessed August 7, 2022).
Runcharoen, C., Fukunaga, K., Sensorn, I., Iemwimangsa, N., Klumsathian, S., Tong, H., et al. (2021). Prevalence of pharmacogenomic variants in 100 pharmacogenes among Southeast asian populations under the collaboration of the Southeast asian pharmacogenomics research network (SEAPharm). Hum. Genome Var. 8, 7–6. doi:10.1038/s41439-021-00135-z
Sahana, S., Sivadas, A., Mangla, M., Jain, A., Bhoyar, R. C., Pandhare, K., et al. (2021). Pharmacogenomic landscape of COVID-19 therapies from Indian population genomes. Pharmacogenomics 22, 603–618. doi:10.2217/pgs-2021–0028
Santus, E., Marino, N., Cirillo, D., Chersoni, E., Montagud, A., Santuccione Chadha, A., et al. (2021). Artificial intelligence-aided precision medicine for COVID-19: Strategic areas of research and development. J. Med. Internet Res. 23 (3), e22453. doi:10.2196/22453
Stevenson, J. M., Alexander, G. C., Palamuttam, N., and Mehta, H. B. (2021). Projected utility of pharmacogenomic testing among individuals hospitalized with COVID-19: A retrospective multicenter study in the United States. Clin. Transl. Sci. 14, 153–162. doi:10.1111/cts.12919
Takahashi, T., Luzum, J. A., Nicol, M. R., and Jacobson, P. A. (2020). Pharmacogenomics of COVID-19 therapies. NPJ Genom. Med. 5, 35–37. doi:10.1038/s41525-020-00143-y
Tasa, T., Krebs, K., Kals, M., Mägi, R., Lauschke, V. M., Haller, T., et al. (2019). Genetic variation in the Estonian population: Pharmacogenomics study of adverse drug effects using electronic health records. Eur. J. Hum. Genet. 27, 442–454. doi:10.1038/s41431-018-0300-6
Tuteja, S., Yu, Z., Wilson, O., Chen, H., Wendt, F., Chung, C. P., et al. (2022). Pharmacogenetic variants and risk of remdesivir‐associated liver enzyme elevations in Million Veteran Program participants hospitalized with COVID-19. Clin. Transl. Sci. 15, 1880–1886. doi:10.1111/cts.13313
Zhang, C., Verma, A., Feng, Y., Melo, M. C. R., McQuillan, M., Hansen, M., et al. (2022). Impact of natural selection on global patterns of genetic variation and association with clinical phenotypes at genes involved in SARS-CoV-2 infection. Proc. Natl. Acad. Sci. U. S. A. 119, e2123000119. doi:10.1073/pnas.2123000119
Keywords: pharmacogenomics (PGx), COVID-19, precision medicine, cost-effective, understudied groups
Citation: Al-Mahayri ZN (2022) Pharmacogenomics at the post-pandemic: If not now, then when?. Front. Pharmacol. 13:1013527. doi: 10.3389/fphar.2022.1013527
Received: 07 August 2022; Accepted: 09 September 2022;
Published: 26 September 2022.
Edited by:
Youssef M. Roman, Virginia Commonwealth University, United StatesReviewed by:
Md. Siddiqul Islam, Southeast University, BangladeshCopyright © 2022 Al-Mahayri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeina N. Al-Mahayri, WmVpbmEubWFoYXlyaUBnbWFpbC5jb20=, WmVpbmEubWFoYWlyaUB1YWV1LmFjLmFl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.