
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 10 November 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1004632
This article is part of the Research Topic Antimycobacterial Drug Discovery: Molecular Therapeutics and Target Identification: Volume II View all 6 articles
 M. Korycka-Machała1
M. Korycka-Machała1 M. Kawka2
M. Kawka2 J. Lach3
J. Lach3 R. Płocińska1
R. Płocińska1 A. Bekier2
A. Bekier2 B. Dziadek2
B. Dziadek2 A. Brzostek1
A. Brzostek1 P. Płociński1,4
P. Płociński1,4 D. Strapagiel3
D. Strapagiel3 M. Szczesio5
M. Szczesio5 K. Gobis6
K. Gobis6 J. Dziadek1*
J. Dziadek1*It was recently reported that 4-substituted picolinohydrazonamides carrying hydrophilic cyclic amines, such as morpholine and pyrrolidine, at the end of their thiosemicarbazide chain have potent antimycobacterial activity in vitro at concentrations below 1 μg/ml. Here, two selected compounds, 2,4-disubstituted pyridine derivatives 11 and 15, revealed significant bactericidal activity against Mycobacterium tuberculosis localized intracellularly within human macrophages, as well as against biofilm-forming tubercle bacilli. Mutants were selected that were resistant to the investigated compounds at an efficiency similar to that identified in the presence of the first line antituberculosis drug rifampicin. The resistant mutants were viable in the presence of the tested compounds exclusively on solid media. Genome-wide sequencing of the mutants selected in the presence of compound 11 revealed the accumulation of nonsynonymous mutations in the mmpR5 gene encoding a transcriptional repressor of the MmpS5-MmpL5 efflux pump, whose upregulation has been associated with bedaquiline resistance. The depletion of MmpR5 in wild-type M. tuberculosis using CRISPR–Cas9 technology increased the resistance of this strain to compound 11. Mass spectrometry-based proteomics (LC–MS/MS) of wild-type tubercle bacilli growing in subinhibitory concentrations of compounds 11 or 15 revealed 15 overproduced proteins not detectable in the control cells, including virulence-related proteins.
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is one of the most serious bacterial pathogens, claiming 1.5 million lives each year. Tubercle bacilli are intracellular pathogens, and their life cycle includes long states of persistence. Therefore, M. tuberculosis is relatively hard to eradicate and poses a challenge for effective chemotherapy. Tuberculosis therapy lasts 6–24 months, depending on the drug susceptibility of the infecting bacterial strain and its metabolic state. In particular, the treatment of multidrug-resistant tuberculosis (MDR-TB) is very expensive, very difficult for patients to follow, carries a burden of side effects and is successful in only approximately 54% of cases (Jagielski et al., 2016). However, even tuberculosis caused by drug-sensitive strains requires 6 months and a cocktail of four drugs used daily to prevent the selection of drug-resistant M. tuberculosis mutants. The rise of drug resistance among M. tuberculosis strains in recent years and the phenomenon of HIV (human immunodeficiency virus)–M. tuberculosis coinfection are serious public health challenges worldwide. Therefore, the development of alternative medical strategies based on a new generation of drugs is desperately needed to effectively cure MDR-TB, reduce the duration of current therapies and minimize the toxicity and cost of the antituberculosis agents used (Zumla et al., 2015). A new hope for the improved treatment of MDR-TB is based on bedaquiline and delamanid, drugs recently approved by American and European drug agencies (FDA/EMA) that are under investigation in clinical trials (https://clinicaltrials.gov/ct2/show/NCT02754765). Delamanid is a promising nitroimidazole with the potential to decrease the duration of anti-TB treatment, thereby reducing the mortality rate of MDR-TB (Ryan and Lo, 2014). Bedaquiline is a diarylquinolone with activity against MDR-TB that is able to kill both actively replicating and dormant mycobacteria, thus shortening the duration of a treatment regime (Koul et al., 2008).
An interesting group of compounds with wide biological activity, including antibacterial, antiviral, cytotoxic, anticonvulsant, and analgesic activities, are thiosemicarbazides (Nevagi et al., 2014; Cihan-Ustundag et al., 2016). These compounds carry sulfur and nitrogen atoms and can easily form hydrogen bonds with proteins. Some derivatives of thiosemicarbazides, such as 2-butyl-4-chloroimidazole-based substituted piperazine-thiosemicarbazone hybrids, manganese complexes derived from 2-acetylpyridine-N (4)-R-thiosemicarbazones and 4-nitropyrrole-semicarbazide conjugates, have been reported due to their anti-M. tuberculosis activity (Jallapally et al., 2014; Oliveira et al., 2014; Rane et al., 2014). More recently, three of 30 investigated imidazole-thiosemicarbazide derivatives appeared to be active against M. tuberculosis in vitro, with only one found to be active against bacilli that had been engulfed by human macrophages (Bekier et al., 2021). The selected compounds also presented inhibitory activity against mycobacterial biofilms.
The potent FtsZ inhibitor (E20) carrying a 2,4-disubstituted-6- thiophenyl-pyrimidine and a chiral amino quinuclidine moiety was not effective against bacteria in the phenotypic screening (Chan et al., 2013). On the other hand, 2,4-disubstituted-6-thiophenyl-pyrimidine derivative (F20) showed antibacterial activity against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF). It was reported that F20 inhibits the activity of GTPase, disrupts FtsZ polymerization, and finally impairs bacterial cell division leading to cell death (Fang et al., 2019). Other compounds carrying pyrimidine ring linked to the benzimidazole orbenzothiazole moieties through an acrylonitrile bridge, were evaluated for their in vitro antibacterial activity against S. aureus, Bacillus subtilis, Escherichia coli, and Pseudomonas aeruginosa. The docking studies indicated that the compounds inhibit the β-lactamase enzyme through covalent bonding with Ser70 (AlNeyadi et al., 2017). A new class of azolyl pyrimidines linked by diamino sulfone moiety displayed antimicrobial (B. subtilis) and antifungal (Aspergillus niger) activity (Butta et al., 2016). The high-throughput screening allowed to select 190 substituted simple or fused pyrimidines with modest activity against bacterial growth, including M. tuberculosis, but many active compounds showed significant cytotoxicity. Substituted pyrimidines 2-(3,5-dimethyl-pyrazol-1-yl) group with various 4-phenylamino and 4-cycloakylamino and 5-carboxyethyl substituents showed higher activity/selectivity ratios than closely related pyrimidines substituted with 2-(2-pyridyl), 4- phenylthio, and 5-methoxy groups (Maddry et al., 2009).
Nineteen novel cycloalkyloaminothiosemicarbazide derivatives were synthesized recently and evaluated against M. tuberculosis. Compounds carrying a morpholine ring at the end of the thiosemicarbazide chain exhibited activity against tubercle bacilli growing in vitro. One compound presented high specificity against M. tuberculosis and very low cytotoxicity, as determined for human dermal fibroblasts and mouse melanoma cell lines (Krause et al., 2020).
Here, we selected two of the most promising cycloalkyloaminothiosemicarbazide derivatives to test their activity against tubercle bacilli localized inside human macrophages, as well as against biofilm-forming M. tuberculosis. The frequency of mutations resulting drug-resistant strains was also determined. Whole-genome sequencing analysis of resistant mutants allowed us to identify that mutations accumulated in the mmpR5 gene increased resistance to the tested compounds, as well as to the EMA/FDA approved drug bedaquiline.
The presented compounds, from a chemical point of view, belong to the group of 2,4-disubstituted pyridine derivatives (Krause et al., 2020). In the 2-position, they have a thiosemicarbazone group linked to a hydrazonamide moiety. Compounds were analyzed using the SwissADME service (Swiss Institute of Bioinformatics 2021) (Daina and Zoete, 2016; Daina et al., 2017) and ProTOX II service (Banerjee et al., 2018) to obtain computational pharmacokinetic and toxicological profiles. Both of the compounds are predicted to have a high absorption in the gastrointestinal tract, which may make them effective drugs (Table 1, Supplementary Figure S1).
M. tuberculosis H37Rv was grown at 37°C in Middlebrook 7H10 (Difco, Baltimore, MD, United States) medium supplemented with 10% OADC (oleic acid-albumin-dextrose-catalase). The liquid cultures were grown in Middlebrook 7H9 broth (Difco, Baltimore, MD, United States) supplemented with OADC and 0.05% Tween-80 (pH = 7).
The MIC90/50 for M. tuberculosis was determined on solid and/or liquid media supplemented with various concentrations of the tested chemical agents. The 2,4-disubstituted pyridine derivatives were dissolved in dimethyl sulfoxide (DMSO) and added directly to the growth medium. The final concentration of DMSO in the medium never exceeded 0.1% (vol/vol), and DMSO did not affect the growth of the bacilli.
The Microplate Alamar Blue Assay (MABA test) was applied to define the MIC value as described by Franzblau et al. (1998). The susceptibility of the tested strains was assessed based on the change in color from blue to pink based on visual inspection. Wells containing only bacteria, medium, or compound were used as controls in this experiment, and the MABA test was repeated independently three times.
The mutants resistant to the tested compounds were selected on 7H10/OADC solid media supplemented with the 11 (6 and 8 μg/ml) and 15 (9 and 15 μg/ml) compounds. The frequency of resistance to 11 and 15 was determined as described previously (Korycka-Machala et al., 2019). Briefly, approximately 300,000 cells of M. tuberculosis culture at an OD of 0.8–1.0, were used to inoculate 100 ml of Middlebrook 7H9 supplemented with 10% Middlebrook OADC, 0.05% tween 80, and 0.005% glycerol, giving a total cell count of 10,000 cells per 4 ml culture. This volume was divided to start 20 cultures of 4 ml each. Cultures were grown at 37°C until reaching an OD of 1.0. Then the cultures were spun at 4000 RPM for 10 min at 4°C and resuspended in 250 μl of 7H9/OADC/tween/glycerol and spotted onto 7H10/OADC/tween/glycerol plates supplemented with 0.5, 2.0 μg/ml rifampicin, 2.0 μg/ml streptomycin, 6.0, 8.0 μg/ml 11, and 9.0, 15.0 μg/ml 15.
The effect of compounds 11 and 15 on mature mycobacterial biofilms was determined as described previously (Bekier et al., 2021). Briefly, to develop the biofilm, M. tuberculosis was cultured to an OD600 = 1 in 7H9/OADC supplemented with 0.05% Tween 80. Next, the inoculum was added at a ratio of 1:100 v/v to Sauton’s medium, and then the obtained mixture was dispensed into each well of a 24-well plate (2.5 ml/well). The plate was covered with a lid, protected with parafilm, and incubated in humidity at 37°C for 5 weeks. Next, the medium under the mature biofilm was replaced with 2.5 ml of Sauton’s medium supplemented with 0.1% casitone and the compounds in various concentrations (compounds were not added to controls). The plates were then covered, sealed with parafilm, and incubated at 37°C for 48 h. The viability of the bacilli was determined by fluorescence measurements (excitation: 550 nm, emission: 590 nm) in the presence of resazurin (375 µl of 0.02% resazurin per well) after 90 min of incubation using the multimode microplate reader SpectraMax® i3 (Syngen). The no compound control was used to represent 100% viability. The results were expressed as the percent viability compared to untreated Mycobacterium biofilm. We also investigated the effect of compounds 11 and 15 on biofilm formation. In the analysis, the diluted inoculum of M. tuberculosis in Sauton’s medium was supplemented with 0.6 and 0.4 μg/ml of compounds 15 and 11, respectively, and dispersed into each well (2.5 ml). The plates were protected by parafilm and incubated in a humidified incubator at 37°C for 5 weeks. The biofilm was photographed and the medium was replaced with 2.5 ml of Sauton’s medium supplemented with 0.1% casein hydrolysate and 375 µl of a 0.02% resazurin solution, and the plate was incubated for 90 min. Further, 200 µl of each sample was transferred to a 96-well plate, and the amount of resorufin was detected using a SpectraMax® i3 multimode microplate reader (Molecular Devices, San Jose, CA, United States) at excitation 560 nm, emission 590 nm. No compound samples were used as a control of 100% viability.
Human monocytes were isolated from commercially available (Regional Blood Donation Station, Lodz, Poland) and freshly prepared buffy coats from healthy human blood donors (Korycka-Machala et al., 2019; Kawka et al., 2021). The cultures of differentiated human monocyte-derived macrophages (MDMs) were extensively washed to remove any nonadherent cells, left resting overnight, and incubated with culture medium supplemented with various concentrations of the tested compounds. The viability of macrophages was determined after 48 h of incubation with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma, St. Louis, MO, United States), as described previously (Korycka-Machala et al., 2017). Additionally, the MDMs were infected with tubercle bacilli at an MOI of 1:10 as described by Korycka-Machala et al. (2017). Two hours after infection, the extracellularly located bacteria were extensively washed out with culture medium, followed by an additional 1 h incubation in medium supplemented with 1 g/L gentamicin (Sigma, St. Louis, MO, United States); finally, the samples were washed three times with Iscove’s medium supplemented with 2% human AB serum (Sigma, St. Louis, MO, United States). Next, culture medium with or without (control) the test compounds at a concentration of 1x MIC90 was added to independent cultures of the infected macrophages, followed by incubation at 37°C for 48 h under a humidified atmosphere of 10% CO2–90% air. Finally, the macrophages were lysed with 0.1% SDS, and the number of CFUs (colony forming units) was determined as previously described (Korycka-Machala et al., 2020).
The sequencing libraries were prepared using the Nextera XT DNA sample preparation protocol (Illumina Inc., San Diego, CA, United States). A total of 1 ng of genomic DNA isolated from the wild-type and ten individual mutants was used for the preparation of paired-end libraries according to the manufacturer’s instructions. Whole-genome shotgun sequencing and in silico analysis were performed as described previously (Korycka-Machala et al., 2019; Korycka-Machala et al., 2020). Sequences of the seven mutants selected for compound 11 and the three mutants selected for compound 15 have been deposited in GenBank under accession no. PRJNA843930.
The depletion of the MmpR5 protein in M. tuberculosis was performed by applying the CRISPR/Cas9 strategy optimized by Rock et al. (2017). The sgRNA probe carrying 20 nucleotide target sequences followed by a strong (GGAAA) PAM site was introduced into the chromosomal DNA using the pLJR965 integration vector (Supplementary Figure S2). The silencing of mmpR5 transcripts in the presence of the inducer anhydrotetracycline (aTc, 100 ng/ml) was confirmed by qRT–PCR. RNA was isolated from induced and control cultures using TRIzol LS reagent (Invitrogen, Carlsbad, CA, United States) and mechanical disruption (FastPrep-24, MP Biomedicals Eschwege, Germany). qRT–PCR analysis was performed as described previously using sigA gene expression as the internal control (Korycka-Machala et al., 2020). The relative fold change was determined using the double delta Ct method (2−ΔΔCT). The reaction was carried out at 60°C. The 208 bp in length sigA product was obtained using RvsigA-F (CCTACGCTACGTGGYGGATTCG) forward and RvsigA-R (TGGATTTCCAGCACCTTCTCCG) reverse primers. The 136 bp in length mmpR5 product was obtained using RvmmpR5-F (TGGCTGACGTGGGGCTGAGG) forward and RvmmpR5-R (CCGGTTCGCTGGCTGTATCGC) reverse primers, the 78 bp in length mmpS5 was obtained using RvmmpS5-F (ATCACCTGCCGAATCACCGT) forward and RvmmpS5-R (GCAGTAGGTCAGGGCATCCA) reverse, the 181 bp in length mmpL5 using RvmmpL5-F (ACACCCTCGACGGAATCGAC) forward and RvmmpL5-R (TGATCCTGCAGCCCTTCCTG) reverse primers.
The mass spectrometry analyses of the whole-cell protein lysates obtained according to previously published methodologies (Plocinski et al., 2019) were performed as a service at the Institute of Biochemistry and Biophysics PAS on a Q Exactive high-performance mass spectrometer using an experimental pipeline reported elsewhere (Goralczyk-Binkowska et al., 2020). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD035807.
4-Substituted picolinohydrazoamides were reported as a new class of antitubercular agents active against tubercle bacilli at low concentrations and presenting limited cytotoxicity on human dermal fibroblasts and mouse melanoma cell lines (Krause et al., 2020). The two compounds, 11 and 15, differ only in the substituents at the 4-position of the pyridine ring. However, the nature of these substituents greatly influences the LogP values associated with bioavailability and the ease of penetration through biological barriers. The pyrrolidine-containing derivative 15 is less lipophilic (LogP 1.06). The nitrogen atom present in the pyrrolidine group shows basic properties, which translates into the ability to bind protons and thus better interaction with proton donating targets. The LogP value for derivative 11 (2.56) is within the range of 1.3–4.1 required for potential anti-tuberculosis chemotherapeutic agents (Santos et al., 2020), but the sulfur atom present in it does not show a tendency to be protonated. Hence, presumably, there is a difference in the antimycobacterial activity observed for the two compounds (Table 1).
Mycobacterium tuberculosis is an intracellular pathogen able to propagate inside host alveolar macrophages during infection, where it is protected from drugs unable to penetrate inside human phagocytes. Here, we evaluated the activity of the two most potent compounds, 11 and 15, against intracellularly located tubercle bacilli. The optical density (OD600) of M. tuberculosis in vitro culture and CFU analyses confirmed the strong in vitro antitubercular effect of both compounds, with MIC99 values as low as 0.8 and 1.5 μg/ml for 11 and 15, respectively (Supplementary Figure S3). MDMs were differentiated from peripheral blood monocytes that were isolated from buffy coats of healthy human blood donors and used to determine the cytotoxicity of the investigated compounds. Compounds 11 and 15 exhibited cytotoxic effects (IC50 values) against human phagocytes at concentrations that exceeded 1.2 and 1.5 μg/ml, respectively (Supplementary Table S1). To test the activity of the investigated compounds against M. tuberculosis engulfed by human macrophages, tubercle bacilli at an MOI of 1:10 were used for infection of the MDMs. After 2 h of phagocytosis, the extracellular bacteria were washed out, and the remaining cell membrane-attached bacilli were killed by gentamicin. The macrophages carrying M. tuberculosis were then exposed to the compounds at a 1x MIC concentration and incubated for 48 h. The number of viable, intracellularly located bacilli was determined by CFU enumeration. A significant (p ≤ 0.00064, p ≤ 0.0001) decrease in the number of viable bacilli was observed in the macrophages treated with compounds 11 and 15, respectively (Figure 1), compared to the control cells, which indirectly suggests an ability of the compounds to cross the target cell membrane. Efficient penetration inside macrophages was also previously reported for compounds such as thio-functionalized carbohydrate derivatives (Korycka-Machala et al., 2017), 1H-benzo [d]imidazole derivatives (Korycka-Machala et al., 2019), and imidazole-thiosemicarbazide derivatives (Bekier et al., 2021).
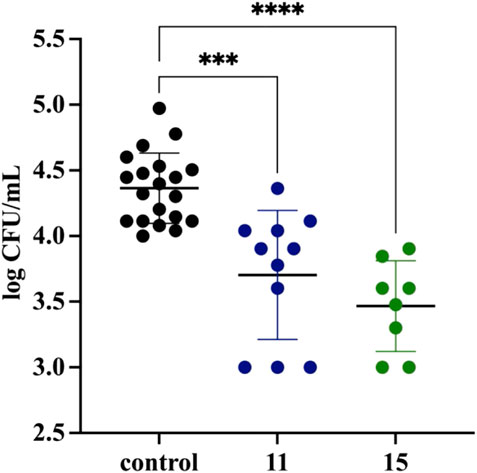
FIGURE 1. Inhibitory effect of compounds 11 and 15 on the intracellular growth of M. tuberculosis in human monocyte-derived macrophages. The intracellular growth of the pathogen in the experimental (11/15-treated) samples versus the control (untreated) sample is presented as a log CFU/mL, which represents the ratio of the CFU number determined for the 11 (blue) and 15 (green) treated samples to the CFU number estimated in the control sample. Error bars indicate standard deviations. Data were analyzed using the Mann–Whitney U test. The differences between control cells and compound-treated samples were statistically significant, with p = 0.0006 (***) and p < 0.0001 (****) for compounds 11 and 15, respectively. The statistical analysis was done using SigmaStat v. 4.0.
The specific composition of the cell wall allows mycobacteria to adhere to surfaces, as well as to form biofilms in the air–media interfaces (Esteban and Garcia-Coca, 2017; Dokic et al., 2021), making them prevalent in almost all environments. Antibiotics have difficulty penetrating biofilms, mainly because the bacteria in the biofilm are in a dormant form, but also because the biofilm creates anaerobic areas where some compounds are not active; these factors favor the development of drug resistance. The drug resistance of biofilm-forming microorganisms, including M. tuberculosis, may result in treatment failure with drugs that are normally active against the same bacteria in the planktonic state (Islam et al., 2012). It was reported that M. tuberculosis can develop a biofilm in vitro (Ojha et al., 2008); however, its role in the pathogenesis of tuberculosis has not yet been elucidated. Biofilms composed of tubercle bacilli in vivo are considered to be due to caseous necrosis and cavitation formation in the lungs. Therefore, potential new anti-TB drugs should present bactericidal effects against intra- or extracellularly located planktonic bacilli, as well as against biofilm-forming bacteria. To test whether compounds 11 and 15 are active against M. tuberculosis in a biofilm, we grew bacteria for 5 weeks in Sauthon’s medium as described in the Methods section. Next, the medium under the biofilm was replaced with a fresh medium supplemented or not with the investigated compounds, and the cultures were further incubated for 48 h. The resazurin-based determination of the viability of the tubercle bacilli in the tested and control biofilm cultures allowed us to identify the dose-dependent bactericidal effect of both compounds on the biofilm composed of M. tuberculosis. The presence of 0.2, 0.4, and 0.6 μg/ml of compound 11 decreased the viability of M. tuberculosis biofilms by approximately 13, 24, and 30%, respectively (p < 0.0027, p < 0.0001 for 0.2 and 0.4–0.6, respectively). Compound 15 affected the viability of biofilm-forming bacilli at concentrations of 0.3, 0.6, and 1.5 μg/ml by approximately 21, 26, and 40%, respectively (p < 0.0001) (Figure 2, Supplementary Table S2). Further, we investigated the effect of compounds 11 and 15 on biofilm development. The supplementation of media with 0.4 and 0.6 μg/ml of compounds 11 and 15, respectively inhibited the biofilm formation. The effect was more pronounced for compound 11 with about 80% of decrease in the viability of tubercle bacilli (Supplementary Figure S4). A similar analysis was made by Bekier and others for the imidazole-thiosemicarbazide derivative (ITD-13). The investigated compound at the MIC90 concentration was able to inhibit the formation of biofilms and decrease the tubercle bacilli viability by approximately 90%. However, mature biofilm treated with ITD-13 at a 1x MIC concentration decreased the viability of the bacilli by only approximately 15% (Bekier et al., 2021).
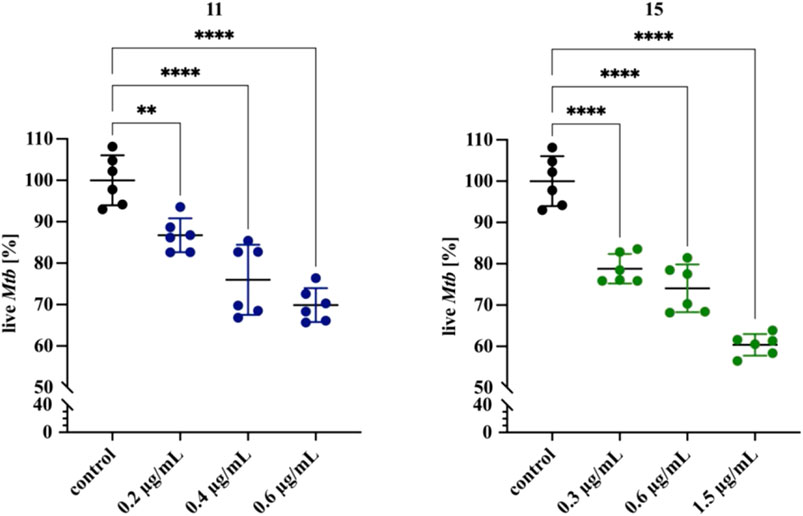
FIGURE 2. Effects of compounds 11 and 15 on mature biofilms composed of M. tuberculosis. Error bars indicate the standard error of the mean (SD). ** depicts the value with significant differences at p=0.0027, **** depicts the values with significant differences at p<0.0001. Data were compared using one-way ANOVA followed by Dunnett’s test. No compound control is represented by black dots. Blue and green dots represent compound 11 and 15, respectively. The concentrations of compounds are depicted at X-axis. The statistical analysis was done using SigmaStat v. 4.0. The graph was prepared using GraphPad Prism 9 version 9.3.1 (350).
The acquired resistance of M. tuberculosis to anti-TB drugs is typically due to the accumulation of mutations in a gene encoding a drug target, an enzyme activating a given prodrug within the bacilli, or a protein involved in the transport of a drug. The frequency of acquiring resistance to particular anti-TB drugs differs depending on the number of possible mutations that can result in drug resistance (David, 1970). The calculated mutation rate for both investigated compounds was similar to that identified for the most potent anti-TB drug rifampicin and much higher compared to streptomycin (Table 2). Moreover, the mutants selected on plates supplemented with compounds 11 or 15 at a concentration of 10x MIC were not viable when inoculated in broth carrying the same concentrations of compounds.
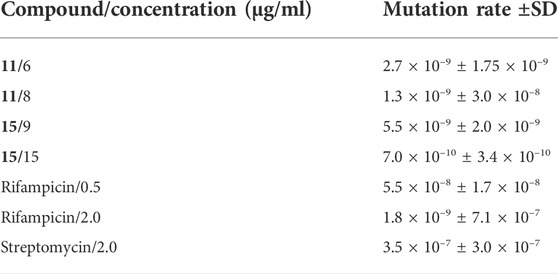
TABLE 2. The mutation rate of tubercle bacilli calculated for the investigated compounds and anti-TB drugs.
The Next Generation Sequencing (NGS) of whole DNA isolated from seven mutants grown on 7H10/OADC supplemented with 8 μg/ml of compound 11 and 3 mutants grown in the presence of 9 μg/ml of compound 15 revealed that all investigated mutants resistant to 11 carried mutations in various locations in the rv0678 gene coding for the MmpR5 protein (Table 3 and Supplementary Table S3). One of three mutants selected in the presence of compound 15 carried a nucleotide substitution in the gene encoding the MmpL5 protein, which is controlled by the MmpR5 regulator and was reported to be part of the MmpS5-MmpL5 efflux system related to azole resistance in M. tuberculosis (Milano et al., 2009). None of the selected mutants carried mutations in an essential gene or in a gene encoding an enzyme that could modify the compounds used. The very low frequency of acquired resistance to compounds 11 and 15, the viability of the mutants exclusively on solid media, and the accumulation of mutations in the putative efflux-pump system suggested that the resistance phenotype was due to the enhanced detoxification of the bacilli carrying mutations in either the mmpR5 or mmpL5 genes. The lack of mutations in genes encoding a target protein(s) could suggest a multitarget mode of action for both compounds used. In such a case, the selection of resistant mutants would require mutations of more than a single target and might happen with lower efficiency than a mutation enhancing the detoxification of bacilli by the efflux-pump system.
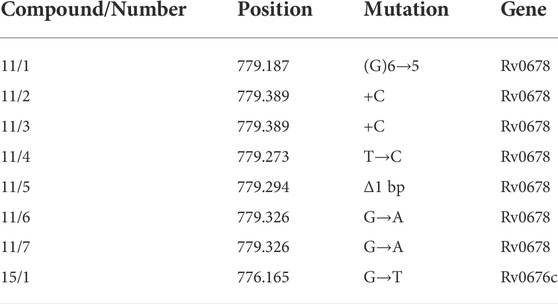
TABLE 3. The mutations identified in mmpR5 (rv0678) and mmpL5 (rv0676c) genes of mutants resistant to compounds 11 and 15.
The inactivation of MmpR5, which leads to the upregulation of the MmpS5-MmpL5 efflux pump, was related to low level resistance to various drugs, including clofazimine and bedaquiline (Hartkoorn et al., 2014). Therefore we decided to use the bedaquiline to confirm the phenotype of CRISPR–Cas9mmpR5 mutant. MmpS5-MmpL5 efflux system affects particular compounds acting on or affected by the electron transport chain (azoles, clofazimine, and bedaquiline). However, it was reported (Villellas et al., 2017) that MmpS5-MmpL5 mutations affect the resistance of Mtb also to telithromycin and partially to rifampicin (not statistically significant) but not to rifabutin, amikacin, ofloxacin, streptomycin, and isoniazid. We verified the role of the mutations in mmpR5 in the acquired resistance to the investigated compounds by constructing an M. tuberculosis CRISPR–Cas9 mutant with an inducible (anhydrotetracycline, aTc) depletion of the MmpR5 regulator. M. tuberculosis mutants carrying a 20-nucleotide mmpR5 target sequence followed by a strong PAM site were analyzed by qRT–PCR to determine the mmpR5, mmpL5 and mmpS5 mRNA levels in the presence and absence of the inducer (aTc). The supplementation of the M. tuberculosis CRISPR–Cas9mmpR5 culture with aTc depleted the mRNA for mmpR5 by approximately 50%. Moreover, we observed the 10x increased expression for transcripts of mmpL5 and mmpS5 in M. tuberculosis CRISPR–Cas9mmpR5 strain (Figure 3A). On the other hand, the control strain carrying an empty CRISPR–Cas9 vector was not affected in the presence of aTc. By applying the Alamar-Blue assay, we also observed an increased resistance of M. tuberculosis CRISPR–Cas9mmpR5 to bedaquiline in the presence of aTc, as expected. The MIC, determined to be 0.04 μg/ml for the wild-type strain, increased to 0.39 μg/ml for the CRISPR–Cas9mmpR5 mutant (Figure 3B). Furthermore, we used the CRISPR–Cas9mmpR5 mutant to determine the MIC value for compound 11 in the MmpR5 depletion background. The MIC was determined on 7H10/OADC plates supplemented with aTc and the investigated compound and was found to increase from 0.4 to 3 μg/ml in the presence of the inducer (Figure 3C).
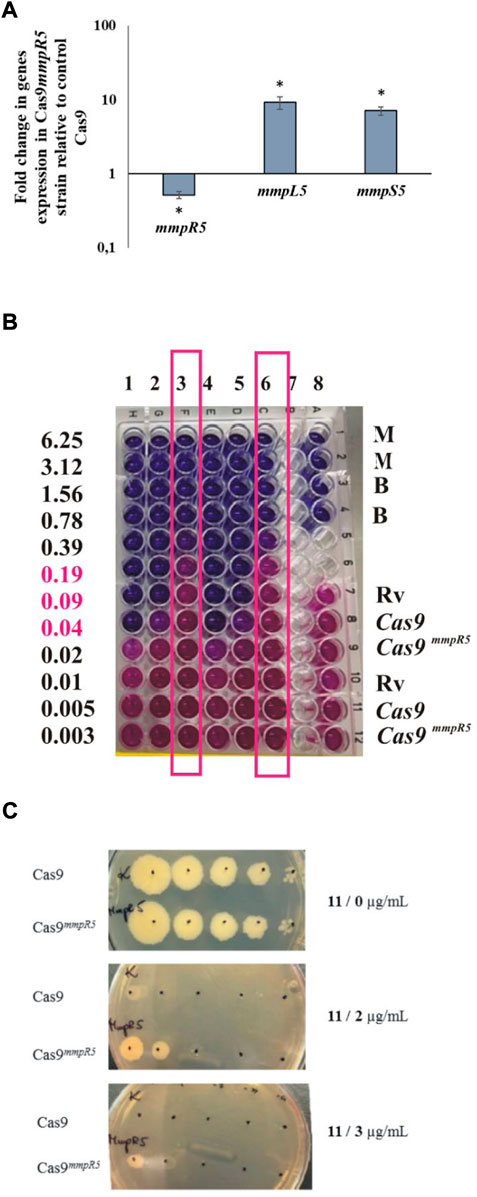
FIGURE 3. Depletion of MmpR5 affects the resistance of M. tuberculosis to compounds 11, 15, and bedaquiline. (A) Expression profile of mmpR5, mmpL5, mmpS5 genes in CRISPR Cas9mmpR5 M. tuberculosis mutant. The mutant M. tuberculosis strain CRISPR–Cas9mmpR5 (Cas9 mmpR5) and the control CRISPR–Cas9 strain expressing the “empty” CRISPRi/dCas9 vector (cas9) were grown in 7H9/OADC broth for 16 h at 37°C with the addition of anhydrotetracycline (100 ng/μl) to deplete MmpR5. Transcript levels were determined using qRT–PCR and SYBR green chemistry. The expression level of mmpR5, mmpL5, mmpS5 was normalized to the sigA housekeeping gene and compared to the control strain. The calibrator was the control strain carrying an empty vector, + aTc. Statistical significance was determined using Student’s t test (mmpR5 -p = 0.007663, mmpS5 -p = 0.0004, mmpL5 -p =0.0014 ) and Microsoft Excel/Office 365. (B) The depletion of MmpR5 affects the resistance of tubercle bacilli to bedaquiline (BDQ) as determined by the Alamar blue assay. Columns 1 and 4, M. tuberculosis H37Rv + BDQ; columns 2 and 5, M. tuberculosis CRISPR–Cas9 + BDQ; columns 3 and 6, M. tuberculosis CRISPR–Cas9mmpR5 + BDQ; column 8, controls: M (7H9 medium), B (BDQ µg/mL), Rv (M. tuberculosis H37Rv), Cas9 (M. tuberculosis CRISPR–Cas9), Cas9mmpR5 (M. tuberculosis CRISPR–Cas9mmpR5). Columns 3 and 6 representing M. tuberculosis CRISPR–Cas9mmpR5 + BDQ were framed. The growth of bacteria (pink color) detected at concentrations of BDQ 0.19, 0.09, and 0.04 (pink fonts) was detected for M. tuberculosis CRISPR–Cas9mmpR5 + BDQ exclusively. (C) Growth on solid medium in the presence of compound 11. Growth of M. tuberculosis CRISPR–Cas9 (Cas9) and M. tuberculosis CRISPR–Cas9mmpR5 (Cas9mmpR5) on the control 7H10/OADC plate (1) and on plates supplemented with 2 µg/mL (2) and 3 µg/mL (3) of compound 11.
Subinhibitory concentrations of antibiotics affect bacterial cells and lead to the global modification of their transcriptome and proteome in response to toxic substances. In the set of genes upregulated in the presence of a tested drug, one could identify those that specifically respond to the presence of the drug, not as a response to the growth inhibition per se. The monitoring of M. tuberculosis gene expression in the presence of isoniazid (INH), performed by DNA microarray, revealed several type II fatty acid synthase enzymes, including AcpM and KasA, directly involved in the FAS2 processes inhibited by INH (Wilson et al., 1999). Another group identified efflux, transport, and virulence genes presenting resistance-dependent differences in response to RMP (de Knegt et al., 2013). Waddell SJ et al. (2004) applied a DNA microarray to compare the transcriptional response of tubercle bacilli to six compounds, including INH, isoxyl, and tetrahydrolipstatin. The comparison of profiles allowed the authors to define a set of genes responding to each drug tested and drug-specific changes that may reflect the mode of action of a given drug. Liang Chen et al. (2018) evaluated the changes in DNA methylation and gene expression of bacilli treated with RMP and INH, identifying 68 genes upregulated and 63 genes downregulated by both drugs. To evaluate the global response of M. tuberculosis to 2,4-disubstituted pyridine derivatives, we applied LC–MS/MS technology to determine the compound-induced changes directly at the protein level. Tubercle bacilli were induced with subinhibitory concentrations (0.5x MIC) of compounds 11 and 15 for 24 h, and the total proteins were isolated and subjected to analysis as described previously (Brzostek et al., 2021). The comparison of compound-treated samples with the untreated control revealed compound-dependent changes in the proteome of M. tuberculosis. We focused our analysis on proteins below the detection level in uninduced culture and enriched significantly in the presence of the tested compounds in all three biological repeats of the experiment. Following treatment of bacilli with compounds 11 and 15, we identified 53 and 35 proteins, respectively, meeting such criteria (Supplementary Table S4). Fifteen proteins were significantly upregulated in the presence of both chemicals used (Figure 4). The analysis of potential pathways upregulated in the presence of subinhibitory concentrations of compounds 11 and 15 performed using ShinyGO v0.741 software identified PPE family proteins significantly enriched in the presence of compound 11 (Supplementary Table S5). Two of these proteins, Rv1773c and potentially essential Rv1990c (Griffin et al., 2011), were annotated as putative transcriptional regulators. The PPE family is represented by PPE31 (Rv1807); however, PPE32 and PPE33, which are encoded by genes of the same operon, were exclusively induced by compound 11. PPE31 was reported to be a virulence-associated factor that modulates innate immune responses to mycobacterial infection (Feng et al., 2021). This protein was also detected in the global transcriptional response of M. tuberculosis to vancomycin (Provvedi et al., 2009). Compounds 11 and 15 also induced the Rv3230c oxidoreductase; this protein was reported to be a biologically relevant electron transfer partner for membrane-bound stearoyl-CoA delta (9)-desaturase (DesA3), which produces oleic acid, a precursor of mycobacterial membrane phospholipids and triglycerides (Chang Y, 2006). Other proteins overproduced in the presence of both compounds were Rv3717, described as a zinc-dependent amidase able to hydrolyze peptidoglycan (Li et al., 2018), and Rv1076, annotated as esterase/lipase LipU presenting activity for the hydrolysis of short carbon chain substrates (Li et al., 2017). The overproduction in the presence of either compound was also detected for possible toxin VapC2 (Rv0301). VapC toxins are PIN domain endonucleases that in M. tuberculosis cleave RNAs essential for decoding at the ribosomal A-site contributing in the persistence process (Winther et al., 2016). The other overproduced proteins were 17-kDa CadI (Rv2641) reported as aq protein induced by cadmium in M. bovis and M. tuberculosis) (Hotter et al., 2001), and several conserved hypothetical proteins of unknown function (Rv2808, Rv3054c, Rv1906c, Rv0141c, Rv1312, and Rv1227c). Based on our results we are not able to say whether the above proteins are overproduced as a stress response or, some of them, are involved in the cellular processes affected directly by the subinhibitory concentrations of the tested compounds.
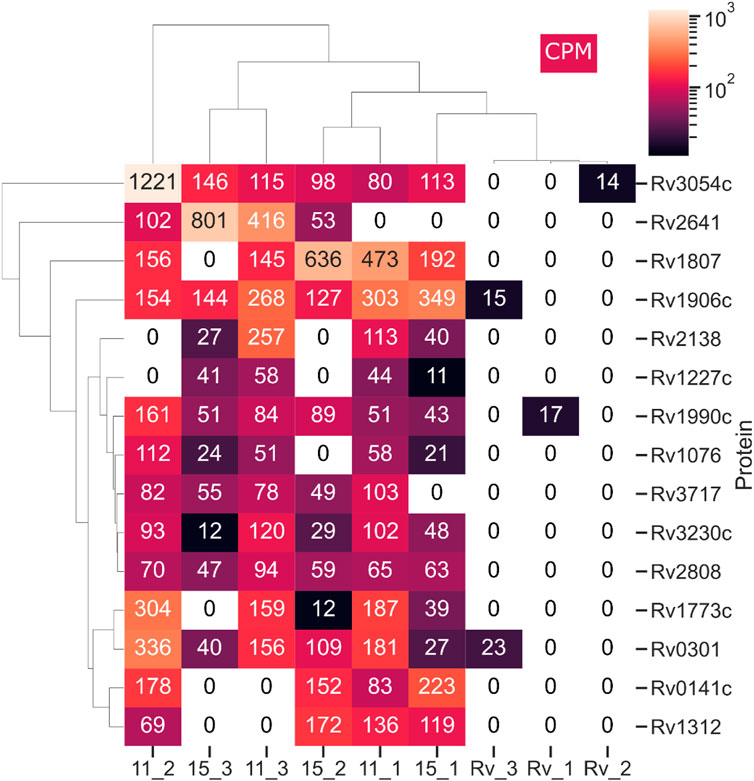
FIGURE 4. Proteins detectable by LC–MS/MS in M. tuberculosis treated with compounds 11 and 15 but not in the control, untreated cells. This heat map was prepared using the Seaborn package - Python programming. CPM represents counts per million. The total protein was isolated from 3 biological repeats of M. tuberculosis culture supplemented with subinhibitory (0.5 MIC) concentrations of compounds 11 (11_1-3), 15 (15_1-3) and untreated control (Rv_1-3). The isolated protein samples were analyzed on Q Exactive high-performance mass spectrometer.
Taking all of the above into account, we found that 2,4-disubstituted pyridine derivatives are effective against intracellularly deposited tubercle bacilli, as well as against M. tuberculosis biofilms. The acquired resistance to the investigated compounds occurs at a low efficiency, and selected resistant mutants are not viable in liquid media. The resistance mechanism is related to the upregulation of the efflux pump MmpS5-MmpL5. Subinhibitory concentrations of the investigated compounds induce the expression of various virulence-associated genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD035807. Genomics data is available at BioProject: PRJNA843930.
MK-M - performed the construction and selection of M. tuberculosis mutants and their growth analysis; MK – performed biofilm formation experiments; JL - sequenced and analyzed the genomes of selected mutants; RP – completed qPCR analysis; AB - performed biofilm formation experiments; BD - performed macrophage related experiments; AMB – BSL3 experiments; PP – proteomics, data analysis; DS - sequenced and analyzed the genomes of selected mutants; MS – performer computational pharmacokinetic and toxicological profiles; KG – compounds chemical synthesis and analysis; JD conceived of, designed and coordinated the study and drafted and finalized the manuscript.
The study was partially supported by grants of National Science Centre, Poland UMO-2017/25/B/NZ7/00124 (KG) and the Ministry of Science and Higher Education, POL-OPENSCREEN, DIR/WK/2018/06 (MK-M, JD).
We thank Jeremy Rock and Sarah Fortune for providing us with the pLJR965 vector and detailed instructions for the generation of Cas9-regulated strains in M. tuberculosis. The authors thank the mass spectrometry service at the Institute of Biochemistry and Biophysics PAS in Warsaw for MS analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1004632/full#supplementary-material
Alneyadi, S. S., Salem, A. A., Ghattas, M. A., Atatreh, N., and Abdou, I. M. (2017). Antibacterial activity and mechanism of action of the benzazole acrylonitrile-based compounds: In vitro, spectroscopic, and docking studies. Eur. J. Med. Chem. 136, 270–282. doi:10.1016/j.ejmech.2017.05.010
Banerjee, P., Eckert, A. O., Schrey, A. K., and Preissner, R. (2018). ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263. doi:10.1093/nar/gky318
Bekier, A., Kawka, M., Lach, J., Dziadek, J., Paneth, A., Gatkowska, J., et al. (2021). Imidazole-thiosemicarbazide derivatives as potent anti-Mycobacterium tuberculosis compounds with antibiofilm activity. Cells 10, 3476. doi:10.3390/cells10123476
Brzostek, A., Plocinski, P., Minias, A., Ciszewska, A., Gasior, F., Pawelczyk, J., et al. (2021). Dissecting the RecA-(in)dependent response to mitomycin C in Mycobacterium tuberculosis using transcriptional profiling and proteomics analyses. Cells 10, 1168. doi:10.3390/cells10051168
Butta, R. D. S., Adivireddyv, P., and Venkatapuram, P. (2016). Synthesis and antimicrobial activity of azolyl pyrimidines. J. Heterocycl. Chem. 54, 524–530.
Chan, F. Y., Sun, N., Neves, M. A., Lam, P. C., Chung, W. H., Wong, L. K., et al. (2013). Identification of a new class of FtsZ inhibitors by structure-based design and in vitro screening. J. Chem. Inf. Model. 53, 2131–2140. doi:10.1021/ci400203f
Chang Y, F. B., and Fox, B. G. (2006). Identification of Rv3230c as the NADPH oxidoreductase of a two-protein DesA3 acyl-CoA desaturase in Mycobacterium tuberculosis H37Rv. Biochemistry 45 (45), 13476–13486. doi:10.1021/bi0615285
Cihan-Ustundag, G., Gursoy, E., Naesens, L., Ulusoy-Guzeldemirci, N., and Capan, G. (2016). Synthesis and antiviral properties of novel indole-based thiosemicarbazides and 4-thiazolidinones. Bioorg. Med. Chem. 24, 240–246. doi:10.1016/j.bmc.2015.12.008
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. doi:10.1038/srep42717
Daina, A., and Zoete, V. (2016). A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 11, 1117–1121. doi:10.1002/cmdc.201600182
David, H. L. (1970). Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 20, 810–814. doi:10.1128/am.20.5.810-814.1970
De Knegt, G. J., Bruning, O., Ten Kate, M. T., De Jong, M., Van Belkum, A., Endtz, H. P., et al. (2013). Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberc. (Edinb) 93, 96–101. doi:10.1016/j.tube.2012.10.013
Dokic, A., Peterson, E., Arrieta-Ortiz, M. L., Pan, M., Di Maio, A., Baliga, N., et al. (2021). Mycobacterium abscessus biofilms produce an extracellular matrix and have a distinct mycolic acid profile. Cell Surf. 7, 100051. doi:10.1016/j.tcsw.2021.100051
Esteban, J., and Garcia-Coca, M. (2017). Mycobacterium biofilms. Front. Microbiol. 8, 2651. doi:10.3389/fmicb.2017.02651
Fang, Z., Li, Y., Zheng, Y., Li, X., Lu, Y. J., Yan, S. C., et al. (2019). Antibacterial activity and mechanism of action of a thiophenyl substituted pyrimidine derivative. RSC Adv. 9, 10739–10744. doi:10.1039/c9ra01001g
Feng, S., Hong, Z., Zhang, G., Li, J., Tian, G. B., Zhou, H., et al. (2021). Mycobacterium PPE31 contributes to host cell death. Front. Cell. Infect. Microbiol. 11, 629836. doi:10.3389/fcimb.2021.629836
Franzblau, S. G., Witzig, R. S., Mclaughlin, J. C., Torres, P., Madico, G., Hernandez, A., et al. (1998). Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36, 362–366. doi:10.1128/JCM.36.2.362-366.1998
Ghose, A. K., Viswanadhan, V. N., and Wendoloski, J. J. (1999). A knowledge based approach in designing combinatorial and medicinal chemistry libraries for drug discovery: 1. Qualitative and quantitative definitions of a drug like molecule. Abstr. Pap. Am. Chem. Soc. 217, U708.
Goralczyk-Binkowska, A., Jasinska, A., Dlugonski, A., Plocinski, P., and Dlugonski, J. (2020). Laccase activity of the ascomycete fungus Nectriella pironii and innovative strategies for its production on leaf litter of an urban park. PLoS One 15, e0231453. doi:10.1371/journal.pone.0231453
Griffin, J. E., Gawronski, J. D., Dejesus, M. A., Ioerger, T. R., Akerley, B. J., and Sassetti, C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251. doi:10.1371/journal.ppat.1002251
Hartkoorn, R. C., Uplekar, S., and Cole, S. T. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2979–2981. doi:10.1128/AAC.00037-14
Hotter, G. S., Wilson, T., and Collins, D. M. (2001). Identification of a cadmium-induced gene in Mycobacterium bovis and Mycobacterium tuberculosis. FEMS Microbiol. Lett. 200, 151–155. doi:10.1111/j.1574-6968.2001.tb10707.x
Islam, M. S., Richards, J. P., and Ojha, A. K. (2012). Targeting drug tolerance in mycobacteria: A perspective from mycobacterial biofilms. Expert Rev. anti. Infect. Ther. 10, 1055–1066. doi:10.1586/eri.12.88
Jagielski, T., Minias, A., Van Ingen, J., Rastogi, N., Brzostek, A., Zaczek, A., et al. (2016). Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin. Microbiol. Rev. 29, 239–290. doi:10.1128/CMR.00055-15
Jallapally, A., Addla, D., Yogeeswari, P., Sriram, D., and Kantevari, S. (2014). 2-Butyl-4-chloroimidazole based substituted piperazine-thiosemicarbazone hybrids as potent inhibitors of Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 24, 5520–5524. doi:10.1016/j.bmcl.2014.09.084
Kawka, M., Brzostek, A., Dzitko, K., Kryczka, J., Bednarek, R., Plocinska, R., et al. (2021). Mycobacterium tuberculosis binds human serum amyloid A, and the interaction modulates the colonization of human macrophages and the transcriptional response of the pathogen. Cells 10, 1264. doi:10.3390/cells10051264
Korycka-Machala, M., Brzostek, A., Dziadek, B., Kawka, M., Poplawski, T., Witczak, Z. J., et al. (2017). Evaluation of the mycobactericidal effect of thio-functionalized carbohydrate derivatives. Molecules 22, 812–826. doi:10.3390/molecules22050812
Korycka-Machala, M., Pawelczyk, J., Borowka, P., Dziadek, B., Brzostek, A., Kawka, M., et al. (2020). PPE51 is involved in the uptake of disaccharides by Mycobacterium tuberculosis. Cells 9, E603. doi:10.3390/cells9030603
Korycka-Machala, M., Viljoen, A., Pawelczyk, J., Borowka, P., Dziadek, B., Gobis, K., et al. (2019). 1H-Benzo[d]Imidazole derivatives affect MmpL3 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 63, e0044119. doi:10.1128/AAC.00441-19
Koul, A., Vranckx, L., Dendouga, N., Balemans, W., Van Den Wyngaert, I., Vergauwen, K., et al. (2008). Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 283, 25273–25280. doi:10.1074/jbc.M803899200
Krause, M., Foks, H., Ziembicka, D., Augustynowicz-Kopec, E., Glogowska, A., Korona-Glowniak, I., et al. (2020). 4-Substituted picolinohydrazonamides as a new class of potential antitubercular agents. Eur. J. Med. Chem. 190, 112106. doi:10.1016/j.ejmech.2020.112106
Li, C., Li, Q., Zhang, Y., Gong, Z., Ren, S., Li, P., et al. (2017). Characterization and function of Mycobacterium tuberculosis H37Rv lipase Rv1076 (LipU). Microbiol. Res. 196, 7–16. doi:10.1016/j.micres.2016.12.005
Li, X., He, J., Fu, W., Cao, P., Zhang, S., and Jiang, T. (2018). Effect of Mycobacterium tuberculosis Rv3717 on cell division and cell adhesion. Microb. Pathog. 117, 184–190. doi:10.1016/j.micpath.2018.02.034
Liang Chen, H. L., Chen, T., Yu, L., Guo, H., Chen, Y., Chen, M., et al. (2018). Genome-wide DNA methylation and transcriptome changes in Mycobacterium tuberculosis with rifampicin and isoniazid resistance. Int. J. Clin. Exp. Pathol. 11 (6), 3036–3045.
Lipinski, C. A., Lombardo, F., Dominy, B. W., and Feeney, P. J. (2012). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 64, 4–17. doi:10.1016/j.addr.2012.09.019
Maddry, J. A., Ananthan, S., Goldman, R. C., Hobrath, J. V., Kwong, C. D., Maddox, C., et al. (2009). Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis 89, 354–363. doi:10.1016/j.tube.2009.07.006
Milano, A., Pasca, M. R., Provvedi, R., Lucarelli, A. P., Manina, G., Ribeiro, A. L., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberc. (Edinb) 89, 84–90. doi:10.1016/j.tube.2008.08.003
Muegge, I., Heald, S. L., and Brittelli, D. (2001). Simple selection criteria for drug-like chemical matter. J. Med. Chem. 44, 1841–1846. doi:10.1021/jm015507e
Nevagi, R. J., Dhake, A. S., Narkhede, H. I., and Kaur, P. (2014). Design, synthesis and biological evaluation of novel thiosemicarbazide analogues as potent anticonvulsant agents. Bioorg. Chem. 54, 68–72. doi:10.1016/j.bioorg.2014.04.002
Ojha, A. K., Baughn, A. D., Sambandan, D., Hsu, T., Trivelli, X., Guerardel, Y., et al. (2008). Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69, 164–174. doi:10.1111/j.1365-2958.2008.06274.x
Oliveira, C. G., Da, S. M. P. I., Souza, P. C., Pavan, F. R., Leite, C. Q., Viana, R. B., et al. (2014). Manganese(II) complexes with thiosemicarbazones as potential anti-Mycobacterium tuberculosis agents. J. Inorg. Biochem. 132, 21–29. doi:10.1016/j.jinorgbio.2013.10.011
Perez-Riverol, Y., Bai, J., Bandla, C., Garcia-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi:10.1093/nar/gkab1038
Plocinski, P., Macios, M., Houghton, J., Niemiec, E., Plocinska, R., Brzostek, A., et al. (2019). Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 47, 5892–5905. doi:10.1093/nar/gkz251
Provvedi, R., Boldrin, F., Falciani, F., Palu, G., and Manganelli, R. (2009). Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiol. Read. 155, 1093–1102. doi:10.1099/mic.0.024802-0
Rane, R. A., Naphade, S. S., Bangalore, P. K., Palkar, M. B., Shaikh, M. S., and Karpoormath, R. (2014). Synthesis of novel 4-nitropyrrole-based semicarbazide and thiosemicarbazide hybrids with antimicrobial and anti-tubercular activity. Bioorg. Med. Chem. Lett. 24, 3079–3083. doi:10.1016/j.bmcl.2014.05.018
Rock, J. M., Hopkins, F. F., Chavez, A., Diallo, M., Chase, M. R., Gerrick, E. R., et al. (2017). Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2, 16274. doi:10.1038/nmicrobiol.2016.274
Ryan, N. J., and Lo, J. H. (2014). Delamanid: First global approval. Drugs 74, 1041–1045. doi:10.1007/s40265-014-0241-5
Santos, C. S., Reis, M., and Martins, F. (2020). Lipophilicity assessment of some isoniazid derivatives active against Mycobacterium tuberculosis. Colloids Surfaces A Physicochem. Eng. Aspects 559, 124820. doi:10.1016/j.colsurfa.2020.124820
Veber, D. F., Johnson, S. R., Cheng, H. Y., Smith, B. R., Ward, K. W., and Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623. doi:10.1021/jm020017n
Villellas, C., Coeck, N., Meehan, C. J., Lounis, N., De Jong, B., Rigouts, L., et al. (2017). Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J. Antimicrob. Chemother. 72, 684–690. doi:10.1093/jac/dkw502
Waddell Sj, S. R., Laing, K., Kremer, L., Reynolds, R. C., and Gs, B. (2004). The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tuberc. (Edinb) 84 (3-4), 263–274. doi:10.1016/j.tube.2003.12.005
Wilson, M., Derisi, J., Kristensen, H. H., Imboden, P., Rane, S., Brown, P. O., et al. (1999). Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. U. S. A. 96, 12833–12838. doi:10.1073/pnas.96.22.12833
Winther, K., Tree, J. J., Tollervey, D., and Gerdes, K. (2016). VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 44, 9860–9871. doi:10.1093/nar/gkw781
Keywords: pyridine derivatives, tuberculosis, drug resistance, Mtb inhibitors, MmpR5, biofilm
Citation: Korycka-Machała M, Kawka M, Lach J, Płocińska R, Bekier A, Dziadek B, Brzostek A, Płociński P, Strapagiel D, Szczesio M, Gobis K and Dziadek J (2022) 2,4-Disubstituted pyridine derivatives are effective against intracellular and biofilm-forming tubercle bacilli. Front. Pharmacol. 13:1004632. doi: 10.3389/fphar.2022.1004632
Received: 27 July 2022; Accepted: 26 October 2022;
Published: 10 November 2022.
Edited by:
Gert Kruger, University of KwaZulu-Natal, South AfricaReviewed by:
Martin Schepelmann, Medical University of Vienna, AustriaCopyright © 2022 Korycka-Machała, Kawka, Lach, Płocińska, Bekier, Dziadek, Brzostek, Płociński, Strapagiel, Szczesio, Gobis and Dziadek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Dziadek, amR6aWFkZWtAY2JtLnBhbi5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.