
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 13 October 2022
Sec. Inflammation Pharmacology
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1002741
This article is part of the Research Topic Inflammation and Immune Diseases: Molecular Targets for Pathogenesis and Treatment View all 7 articles
The pathogenesis of systemic lupus erythematosus (SLE) is related to immune homeostasis imbalance. Epigenetic mechanisms have played a significant role in breaking immune tolerance. Enhancer of zeste homolog 2 (EZH2), the specific methylation transferase of lysine at position 27 of histone 3, is currently found to participate in the pathogenesis of SLE through affecting multiple components of the immune system. This review mainly expounds the mechanisms underlying EZH2-mediated disruption of immune homeostasis in SLE patients, hoping to provide new ideas in the pathogenesis of SLE and new targets for future treatment.
Systemic lupus erythematosus (SLE), is a highly heterogeneous autoimmune disease, characterized by damage of multiple systems and extensive presence of autoantibodies (Rapoport and Bloch, 2012). The occurrence of SLE is related to the excessive immune response to self-antigens and the lack of immune suppression, which in turn leads to a breakdown of immune tolerance. Nonetheless, the direct etiology of SLE is still not clear. Many factors are involved in the pathogenesis, including genetics, epigenetics, and environmental factors (Weeding and Sawalha, 2018; Xu et al., 2019).
Nowadays, epigenetic mechanisms have gradually been recognized as crucial factors in SLE pathogenesis, and epigenetics has become a focus of research (Teruel and Sawalha, 2017; Hedrich, 2018; Navarro quiroz et al., 2019; Wardowska, 2020). DNA hypomethylation in CD4+ T cells and B cells is a major factor contributing to SLE pathogenesis and the degree of hypomethylation is correlated with the disease activity (Javierre et al., 2010; Garaud et al., 2011; Long et al., 2016a; Wu et al., 2016a; Karagianni and Tzioufas, 2019). There are a lot of demethylated target genes identified in SLE, such as ITGAL, TET3, PRF1, MMP14, CSF3R, IFNGR2, etc (Javierre and Richardson, 2011; Yang et al., 2013; Zhang et al., 2013; Liu et al., 2014; Zhao et al., 2014; Wu et al., 2016b; Liao et al., 2017; Zhao et al., 2017). Various non-coding RNAs also play an important role in SLE pathogenesis through their functions in humoral and cellular immune responses (Yan et al., 2014; Honarpisheh et al., 2018). In addition, many other reports demonstrated changes in methylation or acetylation of Histone3(H3) and Histone4(H4) in SLE patients. There are significant increases of H3K4me3 in peripheral blood mononuclear cells (PBMCs) of SLE identified by chromatin immunoprecipitation linked to the microarray (ChIP-chip) approach and increased H3K4me3 is associated with the pathogenesis of the SLE (Dai et al., 2010). Increased H4 acetylation (ac) in monocytes of SLE is related to tumor-necrosis factor (TNF)αand type I interferon pathway, leading to the overproduction of proinflammatory cytokines (Sullivan et al., 2007; Zhang et al., 2010).
As a histone methylation transferase, enhancer of zeste homologue 2 (EZH2) inhibits the expression of target genes by catalyzing the formation of H3K27me3 during the development and differentiation of immune cells (Kuzmichev et al., 2002). Recent studies have identified that the expression of EZH2 is altered in SLE patients, and targeted therapy against EZH2 ameliorates the clinical performance of lupus mice (Rohraff et al., 2019; Schrezenmeier et al., 2019). Therefore, this review mainly reveals how EZH2 participates in the pathogenesis of SLE through immune equilibrium dysregulation, hoping to provide novel ideas for the treatment of SLE.
EZH2, a histone methylation transferase containing the SET domain, is a catalytic subunit of polycomb repressive complex 2 (PRC2) (Czermin et al., 2002; Kuzmichev et al., 2002). The C-terminal SET domain of EZH2 can exert the biological activity of methyltransferase (Vire et al., 2006; Poepsel et al., 2018). Structurally, PRC2 is composed of four core components (Figure 1): EZH2 or its close homolog EZH1, embryonic ectoderm development (EED), suppressor of zeste 12 (SUZ12), and retinoblastoma associated protein 46/48 (RbAp46/48, also known as RBBP7/4), as well as three free proteins: AEBP2, PCLs and JARID2 (Margueron and Reinberg, 2011; Cyrus et al., 2019; Hsu et al., 2019). EZH2 alone cannot exert its biological activity of methyltransferase, but it can play an important catalytic function when it interacts with other non-catalytic subunits such as SUZ12 and EED (Cyrus et al., 2019; Hsu et al., 2019; Zhu et al., 2019).
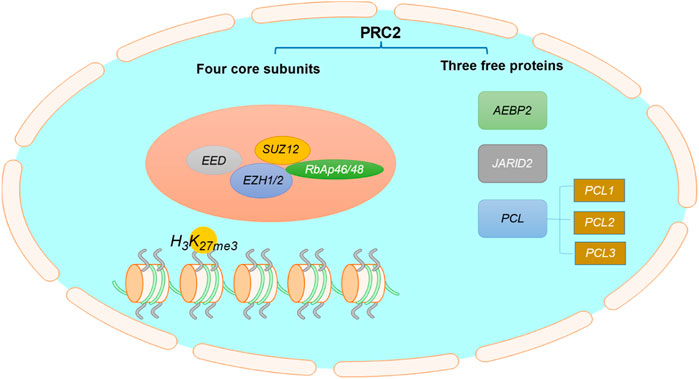
FIGURE 1. The structure of PRC2. PRC2 is composed of four core subunits: EZH1/2, EED, SUZ12, RbAp46/48, and three free proteins: AEBP2, PCLs and JARID2.
EZH2 exists in both the nucleus and the cytoplasm. Its most critical function is to suppress gene expression by promoting histone methylation and DNA methylation in the nucleus. EZH2 is also able to catalyze non-histones and plays a role in cytoplasmic methylation. In most cases, EZH2 exerts its biological functions through the formation of the PRC2 complex. In addition, EZH2 exerts some non-classical functions independent of the PRC2 complex under certain conditions.
The canonical biological function of EZH2 is to catalyze the lysine trimethylation of histone 3 at position 27(H3K27me3) (Vire et al., 2006). H3K27me3 is an inhibitory histone marker. The formation of H3K27me3 makes the chromosome structure denser, thereby inhibiting the expression of target genes (Czermin et al., 2002). As a histone methyltransferase, EZH2 is related to DNA replication, embryonic development, biosynthesis, cell cycle, invasiveness, and lymphocyte activation (Neo et al., 2014; Yan et al., 2016).
EZH2 is overactive in various cancer cells through functionally acquired mutations and overexpression. Previous studies have confirmed that multiple EZH2 targets in cancers (Table 1) belong to the cell cycle protein dependent kinase inhibitor (CKI) family, which negatively regulates the cell cycle (Kim and Roberts, 2016; Sha et al., 2016; Zhao et al., 2016; Tabbal et al., 2019; Wang et al., 2019). Clinical trials for EZH2 inhibitors are already underway in the field of cancer. At present, the clinical trials targeting EZH2 in tumors have indeed made great progress. There is a total of fifty-seven clinical trials regarding targeting EZH2 in cancers available on ClinicalTrials (https://clinicaltrials.gov/), nine of which have been completed (Table 2). Most new drugs targeting EZH2 are still in the preclinical phase or phase 1/2 of clinical trials, but tazemetostat was approved for the treatment of epithelioid sarcoma (ES) and preliminarily showed promising effects on follicular lymphoma (FL) (Straining and Eighmy, 2022). This is a milestone of the EZH2-targeted therapy.
DNA methylation is one of the most important epigenetic modifications, which can cause changes in chromatin structure, DNA conformation, DNA stability, and how DNA interacts with proteins, thereby regulating gene expression. EZH2 can directly control DNA methylation via recruiting DNA methyltransferases (DNMTs) (Vire et al., 2006), facilitating the binding of DNMTs to EZH2 target genes. EZH2 is also required for DNA methylation of EZH2 target promoters, supported by EZH2 controlled CpG methylation through direct physical contact with DNMTs. EZH2 serves as a recruitment platform for DNMTs, indicating that histone modification and DNA methylation jointly regulate the epigenetic state (Cedar and Bergman, 2009).
In addition to the canonical role of EZH2 as a histone methyltransferase in nucleus, it is clear that EZH2 can also methylate non-histones in the cytoplasm and cell nucleus. The non-histone methylation function of EZH2 also depends on the formation of PRC2 complex. EZH2 can regulate cell migration, invasion and adhesion by mediating the methylation of cytoplasmic proteins. Cytoplasmic EZH2 can regulate actin polymerization and cell mobility in various cell types by methylating Vav1 protein (Su et al., 2005). EZH2 can regulate the migration and adhesion dynamically in neutrophils and dendritic cells (DCs) by direct methylation of cytosolic talin (Loh et al., 2018). EZH2 disrupts the link between actin and talin by polymerizing the actin cytoskeleton (F-actin) and blocking the binding motif of talin, thereby aggravating the migration and invasion of cells. Moreover, cytoplasmic PRC2 can promote the proliferation of CD4+T cells and CD8+T cells by methylating Vav1 and Vav1 associated protein Nck (Dobenecker et al., 2018). In the nucleus, EZH2 can also methylate transcription factors, such as GATA4 and RORα, leading to functional repression (He et al., 2012) and proteasomal degradation (Lee et al., 2012). Furthermore, EZH2 can interact with STAT3 and directly methylate STAT3, resulting in enhanced nuclear localization and chromatin of STAT3, thereby exacerbating breast cancer (Yang et al., 2021; Zhao et al., 2021).
Apart from being designated as a methyltransferase, EZH2 can also play an important role independent of PRC2 in certain situations. EZH2 directly methylates the transcription factor PLZF of the natural killer T (NKT) cell lineage, leading to its ubiquitination and subsequent degradation, which impacts the development of the immune system (Vasanthakumar et al., 2017). EZH2 can also be converted into a transcriptional activator, as reported in a natural killer T cell lymphoma (NKTL) disease model (Yan et al., 2013; Yan et al., 2016). EZH2 can directly promote the transcription of cyclin D1 independent of its methyltransferase activity, inducing the growth of NK tumor cells in NKTL (Yan et al., 2013). The phosphorylation of EZH2 by janus kinase 3 (JAK3) promotes the PRC2 complex’s dissociation, switching EZH2 to a transcriptional activator, associated with a set of upregulated genes involved with DNA replication, cell cycle, biosynthesis, stemness and invasiveness (Yan et al., 2016). JAK3 inhibitors can significantly reduce the growth of NKTL cells in an EZH2 phosphorylation-dependent manner, while drugs that inhibit EZH2 methyltransferase activity have no such effect, which provides a new option for the treatment. EZH2 enhances breast cancer initiation by directly binding to the NOTCH1 promoter and activates NOTCH1 signaling independent of its catalytic H3K27me3 activity (Gonzalez et al., 2014). EZH2 exerts distinct non-histone methyltransferase roles in ER-negative and positive breast cancers. In ER-negative breast cancer, EZH2 physically interacts with RelA/RelB to promote the expression of NF-κB targets. Interestingly, EZH2 acts inversely in ER-positive luminal breast cancer cells and suppresses the expression of NF-κB target genes by interacting with ER and directing repressive histone methylation to its promoters (Lee et al., 2011). Consequently, EZH2 functions in breast cancer as a double-sided molecule that, acts as a transcriptional activator or repressor of NF-κB targets depending on the cellular context. What’s more, EZH2 interacts directly with estrogen receptor and β-catenin through transactivation in its two N-terminal domains, thereby linking estrogen and Wnt signaling pathways to promote cell cycle progression and ER-positive breast cancer progression (Shi et al., 2007). In addition, EZH2 acts as a transcriptional activator to activate androgen receptor (AR) gene transcription by directly occupying its promoter to promote prostate carcinogenesis (Kim et al., 2018). Another non-canonical function of EZH2 is to phosphorylate H2BY37 and induce transcriptional activation of ATG in colon cancer cells (Liu et al., 2020).
Collectively, EZH2 has multiple biological functions and plays a different role in different cells and diseases (Figure 2). With the in-depth study of EZH2, it will help us to better understand the function of EZH2 and provide new ideas for targeting EZH2 in tumor therapy.
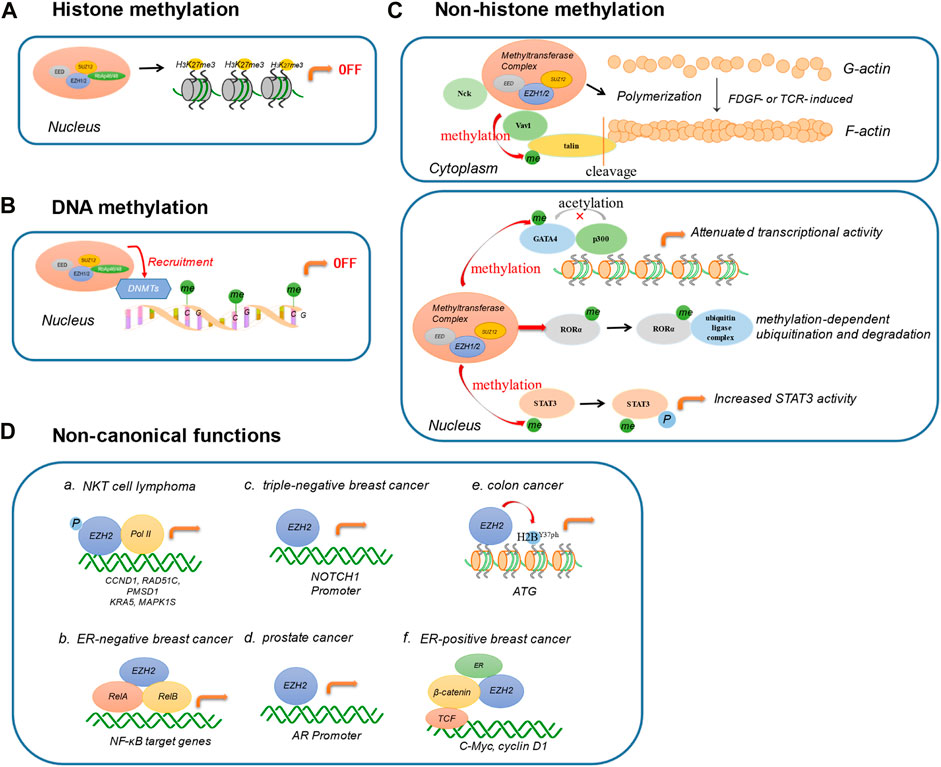
FIGURE 2. Biological functions of EZH2. (A) Histone methylation. EZH2 catalyzes the lysine trimethylation of histone 3 at position 27. (B) DNA methylation. EZH2 directly controls DNA methylation via recruiting DNA methyltransferases (DNMTs). (C) Non-histone methylation. EZH2 regulates cell migration, invasion, and adhesion by mediating the methylation of cytoplasmic proteins, including Vav1, Nck, and talin. In the nucleus, EZH2 can also methylate transcription factors for diverse functions, such as GATA4, RORα, and STAT3. (D) Non-canonical functions. EZH2 exerts non-canonical functions in a PRC2-independent complex. a. In NKT cell lymphoma, the phosphorylation of EZH2 directly binds with Pol II involved in DNA replication, cell cycle, biosynthesis, stemness, and invasiveness. b. In ER-negative breast cancer, EZH2 interacts with RelA/RelB to promote the expression of NF-κB targets. c. In triple-negative breast cancer, EZH2 activates NOTCH1 signaling by directly binding to NOTCH1 promoter. d. EZH2 combines with AR promoter in prostate cancer. e. EZH2 regulates H2B phosphorylation and then induces the transcription activation of ATG in colon cancer. f. EZH2 interacts directly with estrogen receptor and β-catenin, promoting ER-positive breast cancer progression. (DNMTs: DNA methyltransferases; ER: estrogen receptor; AR: androgen receptor).
The immune system is a dynamic and fine-tuned buffering system, containing both effector and regulatory components. Effector T cells, activated B cells and M1 macrophages induce immune response and inflammation by generating proinflammatory cytokines, chemokines and antibodies. In contrast, regulatory T cells, regulatory B cells and M2 macrophages suppress the immune response and mediate immune tolerance through producing suppressive mediators (O’neill et al., 2016). Numerous cells and molecules function cooperatively in both innate and adaptive immunity. The homeostasis of immune system is the cornerstone of the body’s fight against infections and tumors, and an imbalance between the effector and regulatory components of the immune system can lead to autoimmune diseases. EZH2 affects the homeostasis of the immune system through multiple aspects (Huntington and Gray, 2018; Horwitz et al., 2019). For example, EZH2 mediates the development, differentiation, activation, proliferation and cytokine secretion of immune cells (Nutt et al., 2020). In the following part, we will describe the effects of EZH2 on innate immunity and adaptive immunity.
Adaptive immunity, mediated by T cells and B cells, is induced by certain antigens and is antigen specific. Compared with innate immunity, adaptive immunity has many characteristics, such as self-limitation, self-tolerance and immune memory. The development and differentiation of adaptive lymphoid cells require EZH2. The following section will cover detailed mechanisms underlying the EZH2-mediated-regulation of T and B cells in the adaptive immune response.
T cells occupy a central position in the adaptive immune response by mediating cellular immune responses and humoral immune responses against thymus-dependent antigens. The differentiation and function of different T cell subsets are accompanied by EZH2-mediated transcriptome remodeling.
Helper T cells assist T and B cell immune responses. Stimulated by different antigens and cytokines, naive CD4+T cells can differentiate into different effector cells, such as Th1, Th2, Th17, Th22, and follicular helper T cells (Tfh). EZH2 plays a vital role in the differentiation and maturation of different subpopulations of CD4+T cells, but its role is still controversial and the mechanism is not completely clear (Koyanagi et al., 2005; Jacob et al., 2008; Wei et al., 2009; Kanno et al., 2012; Tumes et al., 2013; Tong et al., 2014; Zhang et al., 2014; Dupage et al., 2015; Yang et al., 2015; Karantanos et al., 2016; Li et al., 2018; Chen et al., 2020; Zhu et al., 2020).
Th1 cells express interferon (IFN)-γ/transcription factor T-bet and participate in the immune response towards intracellular pathogens. An increment in Th1 can cause autoimmune diseases. Th2 cells express interleukin (IL)-4 and Gata3 and limit worm infection, involved in allergies and parasitic infections. EZH2 binds directly to Tbx21 (encoding T-bet) and Gata3 (encoding Gata3), inhibiting the transcription activity of two TFs, and ultimately suppressing the differentiation of naive CD4+T cells into Th1 and Th2 cells (Koyanagi et al., 2005; Tumes et al., 2013; Zhang et al., 2014). Besides, EZH2 inhibits the differentiation of Th1 and Th2 cells by direct epigenetic labeling and silencing of genes that encode lineage-specific cytokines, such as Ifng and II13 (Koyanagi et al., 2005; Tumes et al., 2013; Zhang et al., 2014). Moreover, the lack of EZH2 accelerates the death of effector Th cells through death receptor-mediated external and internal apoptotic pathways (Zhang et al., 2014). LncRNA GAS5 can inhibit Th1 differentiation and promote Th2 differentiation through EZH2 and T-bet in allergic rhinitis (Zhu et al., 2020). Paradoxically, EZH2 can also induce Th1 cell differentiation by increasing T-bet stability and inducing the secreting of Th1 cytokines (Jacob et al., 2008; Tong et al., 2014).
Tfh cells produce IL-21 and help B cells participate in humoral immunity. The increase in Tfh causes antibody-related autoimmune diseases. Tfh differentiation is a multi-stage process involving many transcription factors that drive the specification and functional maturity of the Tfh lineage (Liu et al., 2013; Crotty, 2014). In early committed Tfh cells, the expression of EZH2 and H3K27me3 modification is upregulated. EZH2 specifically stabilizes the chromatin plasticity of a set of genes critical for Tfh fate commitment, notably B-cell lymphoma 6 (Bcl6), to direct Tfh cell commitment in a mouse model of acute lymphocytic choriomeningitis virus (LCMV) infection (Chen et al., 2020). Contrary to the well-known function as a chromatin repressor, EZH2 indirectly maintains the chromatin accessibility and reinforces the expression of Bcl6 (Baumjohann et al., 2011; Choi et al., 2013). EZH2 can inhibit the expression of p19Arf in the Cdkn2a locus, thereby promoting Tfh lineage stereotypes and functional maturity by shielding Tfh cells from Bcl6 inhibition, and promoting Tfh cell survival by protecting them from p53 induction (Li et al., 2018). These findings determine that EZH2 is an integrator of epigenetic and transcriptional regulatory mechanisms that can program the fate, survival and functional maturity of Tfh.
Th17 cells produce IL-17, express Rorc, and defend against extracellular bacteria and fungi. Th17 cells play an important role in autoinflammation and autoimmunity. Th22 cells produce IL-22, participating in body defense functions. Excessive Th22 causes inflammatory diseases and autoimmune diseases (Kanno et al., 2012; Jia and Wu, 2014). It is generally believed that Th17 and Th22 are closely related to the occurrence of autoimmune diseases, but there is little research about the role of H3K27me3 in Th17/22. EZH2 suppresses Th17 differentiation (Yang et al., 2015). Inhibition of the H3K27me3 demethylase KDM6A/B leads to an overall increase in the inhibitory H3K27me3 histone markers, resulting in the inhibition of the key transcription factor RORγt during Th17 differentiation, which in turn leads to metabolic reprogramming (Cribbs et al., 2020). The Th22 cell-derived IL-22 activates STAT3 and targets PRC2 to induce colon cancer cell proliferation via H3K27me3 (Sun et al., 2016).
Tregs negatively regulate the immune response by inhibiting the activation and proliferation of CD4+and CD8+T cells to maintain immune homeostasis. However, Tregs exhibit plastic differentiation and have specific functions of Th cells, such as the secretion of Th-related cytokines and the expression of specific transcription factors in Th cells in autoimmune diseases (Qiu et al., 2020). Foxp3 is specifically expressed in Tregs, which is essential for the differentiation and function of Tregs (Ziegler, 2006; Josefowicz et al., 2012). The interaction between Foxp3 and EZH2 only occurs in activated Treg cells (Arvey et al., 2014). Foxp3 can bind to EZH2 to inhibit transcription, thereby regulating the Treg cell phenotype (Kwon et al., 2017). In rheumatoid arthritis patients, downregulation of EZH2 in CD4+ T cells suppresses Foxp3 transcription by down-regulating RUNX1 and up-regulating SMAD7, finally leading to inhibition of Treg differentiation (Xiao et al., 2020). EZH2 mediates Tregs activation via IFN-γ and IL-4 downregulation, along with the consequent inducing of Foxp3 (Karantanos et al., 2016). Also, the lack of EZH2 can lead to a reduction in the number of Tregs and impaired function (Yang et al., 2015). Follicular regulatory T (Tfr) cells are a subset of regulatory T cells that control the production of antibodies by inhibiting the help of B cells mediated by Tfh. Foxp3 and EZH2 are required for the maintenance of Tfr cellular transcriptional programs, and the absence of Foxp3 or EZH2 may lead to a weakened inhibitory effect of Tfr cells on B cell responses (Hou et al., 2019).
The CD8+ subset of T cells proliferates and differentiates into cytotoxic T lymphocytes (CTLs), which express cytotoxic granules and kill infected cells. EZH2 can regulate the proliferation and apoptosis of naive CD8+T cells (Karantanos et al., 2016). MiR-26a is a negative regulator of CTL function in the tumor microenvironment (TME). EZH2 is negatively correlated to miR-26a in CTL, and inhibition of EZH2 in CTL impairs the function of secreting granzyme B/IFN-γin TME (Long et al., 2016b). The absence of EZH2 results in increased expression of cyclin-dependent kinase inhibitors Cdkn2a (p16 and Arf) and Cdkn1c (p57) in activated naive CD8+T cells (Chen et al., 2018), thereby inhibiting cell proliferation.
The main functions of B cells are to produce antibody mediating humoral immune response, present antigens and produce cytokines to participate in immune regulation. Multiple stages of B cell development in bone marrow and B cell immune response outside the bone marrow have been affected by EZH2. In bone marrow, EZH2 is necessary for the normal maturation from pro-B to pre-B stage through different mechanisms. In the early development of B cells, the lack of EZH2 causes B cell development to stay in the stage of differentiation from pro-B cells to pre-B cells (Su et al., 2003). The major mechanism is that EZH2 can regulate the heavy chain VDJ rearrangement and promote the formation of pre-BCR, so that pro-B cells can develop to the pre-B cell stage. At the same time, the effect of EZH2 on the expression of Igκ chain also directly affects the early differentiation of B cells in bone marrow (Mandal et al., 2011). However, EZH2 does not affect the survival of mature B cells in the spleen, and the ratio of marginal zone B cells to follicular B cells, which indicates that EZH2 does not affect the maintenance of B cell homeostasis in surrounding lymphoid tissues (Guo et al., 2018). Outside the bone marrow, EZH2 mediates both T-cell-dependent and T-cell-independent immune responses. The expression of EZH2 in naïve B cells in the resting state is extremely low, and the expression is rapidly increased in activated B cells and germinal center (GC) B cells (Su et al., 2003; Guo et al., 2018). EZH2 promotes the generation of GC while maintaining GC function through the following mechanisms (Velichutina et al., 2010; Beguelin et al., 2013; Caganova et al., 2013; Beguelin et al., 2016; Beguelin et al., 2017). Firstly, EZH2 can promote the rapid division and proliferation of GC B cells by inhibiting CKI to promote the formation of GC. Secondly, EZH2 can cooperate with the anti-apoptotic protein BCL2 to inhibit cell apoptosis, and then maintain the GC morphology. Lastly, in the GC B cells, EZH2 can inhibit the expression of terminal differentiation-related genes, sustain the germinal center B cell phenotype, and then maintain the germinal center reaction. B cell–specific deletion of EZH2 leads to a loss of GC formation, thereby leading to defects in the formation of plasma cells (PCs). Moreover, EZH2 promotes the final differentiation of peripheral mature B cells into plasma cells via extra-follicular immune response or T cell-independent immune response (Guo et al., 2018). The expression of EZH2 in PCs is further increased than in activated B cells. EZH2 controls transcriptional changes during plasma cells differentiation controlling several B and plasma cell genes through PRC2 and H3K27me3 dependent mechanisms (Scharer et al., 2018; Herviou et al., 2019).
EZH2 affects not only the quantity but also the quality of PCs. EZH2-defficient PCs display an impaired function of antibody secretion. The main mechanism is that EZH2 affects the unfolded protein response in PCs and affects the normal metabolic functions of PCs, including oxidative phosphorylation and glycolysis (Guo et al., 2018).
Innate immunity is the initial response to microbes that prevents, controls, or eliminates the host’s infection. Innate lymphoid cells (ILCs), include natural killer (NK) cells, DCs, mast cells, and ILC groups 1–3. Relatively, little research has been done on the role of EZH2 in NK cells, DCs.
NK cells can directly kill infected cells by releasing particles containing perforin and granzyme, or indirectly activate phagocytic microbes to phagocytic macrophages through IFN-γ release in the early stages of infection, thereby inhibiting virus replication (Vivier et al., 2008; Vivier et al., 2011). EZH2 regulates NK cells development and functions through its histone methyltransferase activity. In hematopoietic stem and progenitor cells (HSPC), EZH2-inhibitors gave rise to increased NK precursors, leading to up-regulation of IL-15R (CD122) and NKG2D activated receptors, which were associated with enhanced NK cell expansion and cytotoxicity against tumor cells (Yin et al., 2015a). And EZH2-inhibitors treatment enhances NK cell eradication of hepatocellular carcinoma cells in an NKG2D ligand-dependent manner (Bugide et al., 2018). These findings may provide insights into the contribution of epigenetic regulation to the origin of NK cells, and reveal that EZH2 inhibitors can directly or indirectly inhibit tumor growth by mobilizing NK cells.
DCs are the most powerful antigen-presenting cells (APCs) in the body at the center of initiating, regulating, and maintaining the immune response, which can efficiently ingest, process and present antigens (Macri et al., 2018). The abnormal regulation of their homeostasis and function is involved in the occurrence of autoimmune diseases (Kaewraemruaen et al., 2019). EZH2 is necessary to activate DCs, mediating epigenetic modification in allergen immunotherapy (Li et al., 2019). EZH2 controls cell adhesion and migration in DCs through methylation of the cytoplasmic regulatory protein talin (Gunawan et al., 2015). And EZH2 controls skin tolerance by regulating different subsets of skin DCs through methylation of talin1 (Loh et al., 2018).
Inheritance, epigenetics, sex hormones, infection, environment and other factors are all involved in lupus pathogenesis. Immune factors and non-immune factors are all closely related to the occurrence of lupus. Cell populations of both innate immune system (monocytes, macrophages and dendritic cells, natural killer cells, and innate lymphoid cells) and adaptive immune system (T cells and B cells) display hyper-responsiveness or hypo-responsiveness through epigenetic reprogramming in SLE. Abnormal proliferation and activation of B cells in SLE patients lead to peripheral immune tolerance deficiency and terminal differentiation of autoreactive plasma cells, which in turn produce autoantibodies (Barnas et al., 2019). T cells promote autoimmune responses and amplify systemic inflammation in SLE by producing inflammatory cytokines and assisting B cells to secret antibodies (Chen and Tsokos, 2021). EZH2 impacts the delicate balance of immune homeostasis in SLE patients by regulating T, B, and other immune cells (Rohraff et al., 2019). Compared with controls, SLE patients present an increased expression of EZH2 in PBMCs (Wu et al., 2021), neutrophils, monocytes, B cells, and CD4+T cells (Rohraff et al., 2019), which may involve the activation of type I interferon signaling pathway and Notch signaling pathway (Amsen et al., 2004; Ciofani and Zuniga-Pflucker, 2005; Cruickshank and Ulgiati, 2010; Zhao et al., 2016; Huang et al., 2017; Arima et al., 2018; Wu et al., 2021). MRL/lpr mice treated with EZH2 inhibitors exhibit lowered anti-dsDNA antibodies, decreased inflammatory cytokines, improved symptoms, and reduced mortality, indicating that EZH2 is a promising therapeutic target for lupus.
The dysfunction of CD4+T cells is critical in the pathogenesis of lupus (Rapoport and Bloch, 2012; Yin et al., 2015b; Dolff et al., 2019). Cytokine production, complement activation, and clonal B cell activation secondary to CD4+ T cellular immune disorders in lupus lead to the overproduction of autoantibodies and tissue damage (Comte et al., 2015).
EZH2 and H3K27me3 levels are elevated in CD4+ T cells of SLE. Over-activated rapamycin complex 1 (mTORC1) signaling and elevated glycolysis down-regulate miR-26a and miR-101 in patients with SLE, resulting in increased expression of their target EZH2 (Barnas et al., 2019; Chen and Tsokos, 2021). EZH2 upregulation in CD4+ T increases DNA methylation of the F11R gene encoding junctional adhesion molecule A (JAM-A), resulting in an imbalance of T cells and increased adhesion to endothelial cells in SLE patients. (Tsou et al., 2018). Inhibition of EZH2 and mTOR or glycolysis may have a particular therapeutic effect on SLE.
Increased BCL-6 on the miR-142 promoter up-regulates H3K27me3 (repressive mark) and down-regulates H3K9/14ac (activating mark), resulting in a decrease in miR-142-3p/5p expression, which leads to CD4+T cell over-activity in SLE (Ding et al., 2012; Ding et al., 2019). Another study (Sawalha, 2019) suggests that BCL-6 and EZH2 cooperate to inhibit miR-142-3p/5p expression in SLE CD4+T cells through epigenetic modification. MiR-142-3p/5p is anti-inflammatory, and when suppressed, CD4+T cells overexpress inflammatory molecules, including CD40L, ICOS, and IL-21, leading to an immune imbalance.
CD8+T cells exhibit decreased effector function and cytolytic activity in SLE, leading to an increased infection rate (Kis-toth et al., 2016; Comte et al., 2017). However, the involved mechanisms are not understood. CD38 is elevated in CD8+ T cells in SLE patients with increased rates of infections. The degranulation, secretion of granzyme and perforin of CD8+CD38highT cells are all decreased. CD38 leads to increased acetylation of EZH2 by inhibiting the acetylase Sirtuin1. Acetylated EZH2 leads to a decrease in the expression of RUNX3, a transcription factor associated with CD8+ T cells, which impairs the function of CD8+ T cells (Katsuyama et al., 2020). And inhibiting EZH2 can restore the cytolytic function of CD8+CD38high T cells, which may provide promise to overcome the incidence of infections in SLE patients (Chakraborty and Mehrotra, 2020). But inhibiting EZH2 can also impair the suppressive functions of Tregs, which can lead to spontaneous autoimmunity (Dupage et al., 2015). Although the inhibition of EZH2damages the function of Tregs, animal experiments confirm that the application of EZH2 inhibitors in lupus mice can inhibit autoimmunity and improve lupus-related symptoms. Therefore, from a global perspective, it can be considered that EZH2 mediates lupus autoimmunity and EZH2 inhibitors can improve the destruction of immune tolerance. However, there is no research exploring the changes in EZH2 expression in Treg and the impact of EZH2 on the Treg function in SLE. This will be the direction of future research on the pathogenesis of lupus.
Abnormal B cells also play a vital role in the pathogenesis of SLE. Dysregulation of B cell transcription factors, cytokines and B cell-T cell interaction can result in aberrant B cell maturation and autoantibody production (Yap and Chan, 2019). The peripheral immune deficiency of B cells may be related to the activation of toll-like receptors (TLR) pathway in SLE (Tsokos et al., 2016). EZH2 is increased in B cells and positively correlates with disease activity and autoantibody production. EZH2 is almost exclusively increased in plasmablasts, but not in naive and memory B cells in lupus (Schrezenmeier et al., 2019). mTORC1 is activated in B cells and regulates plasma cell differentiation in SLE patients. In the presence of methionine, Syk, and mTORC1 activation can co-induce the expression of EZH2 (Zhang et al., 2020). EZH2 induces H3K27me3 at the BTB and CNC homology 2 (BACH2) locus and inhibits BACH2 expression, thereby causing the expression of B lymphocyte induced maturation protein 1(BLIMP1) and X-box binding protein 1 (XBP1) and differentiation of plasmablasts. These results indicate that methionine activates signaling by controlling B cell immune metabolism and plays an important role in the differentiation of B cells into plasmablasts through epigenomic modification. And EZH2-inhibitor suppresses autoantibody production and GC formation in bm12 induced lupus-like chronic graft-versus-host disease (cGVHD) (Zheng et al., 2020). Therefore, we speculate that EZH2 may promote B cell proliferation and inhibit B cell apoptosis by regulating cyclin-dependent kinase inhibitors in SLE, thereby abnormally proliferating plasma cells, leading to the production of autoantibodies.
The pathogenesis of lupus also involves a variety of ILCs. The number of NK cells in SLE patients is reduced and their function is impaired. (Hervier et al., 2011). EZH2 inhibits the development of NK cells, as described in the third part. However, there is no research investigating whether NK cell reduction in SLE patients is associated with elevated EZH2 levels. This may be worth further research to provide new ideas for uncovering the pathogenesis of lupus. And more notably, EZH2 may participate in the pathogenesis of SLE by regulating differentiation and functions of ILCs.
In general, EZH2 mediates the development of SLE through the disruption of immune homeostasis in a variety of ways (Figure 3; Table 3). The role of EZH2 in CD4+ T cells and B cells has been widely reported, and it is likely that EZH2 in these cells has a greater impact on the pathogenesis of SLE than other immune cells. EZH2 expression is elevated in CD4+ T cells in lupus, leading to increased cytokine secretion and increased T cell adhesion. EZH2 is associated with decreased CD8 cytotoxic function in lupus-infected patients. More importantly, EZH2 can inhibit the expression of BACH2 and promote the differentiation of plasma cells, which is closely related to autoantibody production. The application of EZH2 inhibitors improved lupus-like symptoms in MRL/lpr mice, lowered the production of type I interferon in NZW/NZB F1 mice and reduced GC formation and autoantibody secretion in bm12 induced lupus-like cGVHD. These studies verified that EZH2 is essential in the pathogenesis of SLE by governing the function of multiple immune cells. Although multiple medications have been approved for the treatment of SLE, there is still a proportion of patients with poor prognosis. Therefore, the development of new drugs for lupus is of great significance. Clinical studies on EZH2 inhibitors in the field of oncology have confirmed the efficacy and safety of EZH2 inhibition therapy. In addition, a variety of medications treating SLE such as rituximab, cyclophosphamide, and methotrexate were originally used in hematological malignancy and other cancers field. Based on the research of EZH2 in lupus patients and mouse models, EZH2 inhibition may become a promising treatment for SLE.
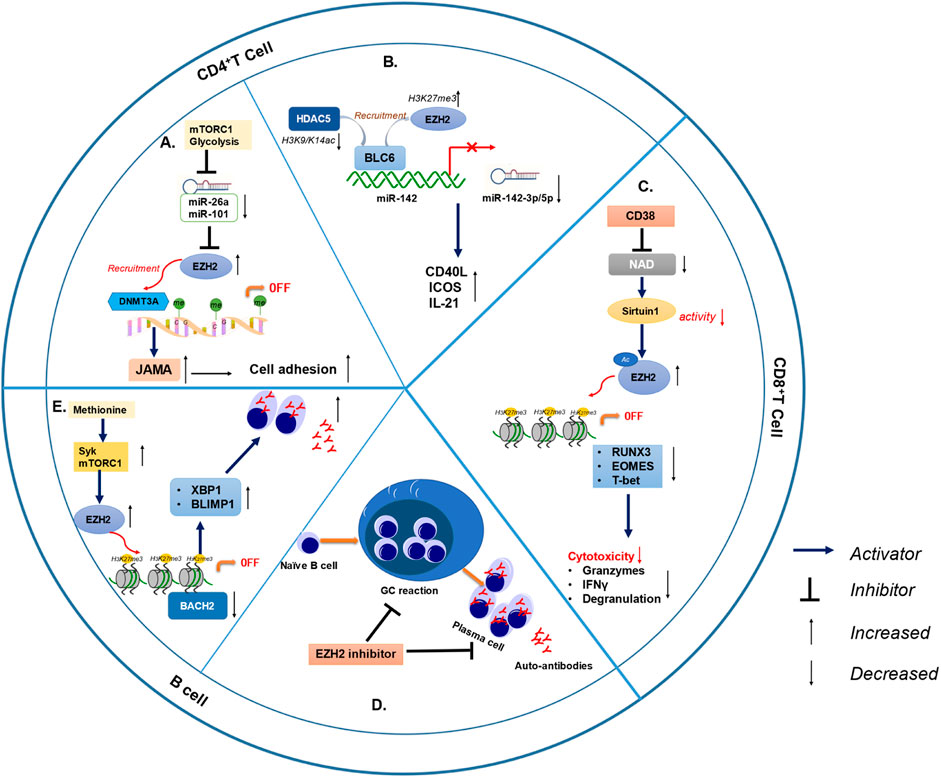
FIGURE 3. EZH2 is involved in lupus pathogenesis through different immune cells. (A) MiR-26 and miR-101 was inhibited by higher glycolysis and mTOR1, limiting post-transcriptional suppression of EZH2 and leading to increased EZH2 levels in SLE CD4+ T cells. Elevated EZH2 result in increased JAM-A expression and CD4+ T cell adhesion of lupus. (B) BCL-6 recruited EZH2 and HDAC5, resulting in increased H3K27me3 (repressive mark) and decreased H3K9/K14ac (activating marks), which lead to epigenetic repression of miR-142 and reduced miR-142-3p/5p expression in lupus CD4+T cells. In addition, CD4+T cells secret more inflammatory molecules when miR-142-3p/5p was decreased, including CD40L, ICOS and IL-21. (C) The CD38/NAD/Sirtuin1/EZH2 axis increased the methylation of RUNX3, EOMES, and T-bet, in turn, reducing cytolytic CD8+ T cell functions in SLE patients. (D) EZH2 inhibitor reduced autoantibody production and GC formation in bm12 induced lupus-like cGVHD. (E) Syk and mTORC1 activation synergistically induced EZH2 expression in the presence of methionine. EZH2 induced H3K27me3 at the BACH2 locus and repressed BACH2 expression, thereby inducing increased BLIMP1 and XBP1 expression and plasmablast differentiation. (mTOR1: mechanistic target of rapamycin complex 1; GC: germinal center; cGVHD: chronic graft versus host disease).
As an epigenetic regulator, EZH2 has various biological functions, the most important of which is to play the role of histone methyltransferase. EZH2 promotes the initiation and maintenance of immune and inflammatory responses by involving in the development, activation and differentiation of various immune cells. EZH2 is abnormally elevated in lupus and is associated with immune homeostasis imbalance and disruption of autoimmune tolerance in SLE patients. Therefore, EZH2 may serve as an important biological marker for the diagnosis and target for the treatment of SLE.
YY drafted this manuscript, as the first author. KL, ML, MG, and HZ revised the manuscript. All authors have read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81901664, 82070018), Natural Science Foundation of Hunan Province (2020JJ5894), and the Fundamental Research Funds of Central South University for postgraduate students (2021zzts0317).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amsen, D., Blander, J. M., Lee, G. R., Tanigaki, K., Honjo, T., and Flavell, R. A. (2004). Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117 (4), 515–526. doi:10.1016/s0092-8674(04)00451-9
Arima, H., Nishikori, M., Otsuka, Y., Kishimoto, W., Izumi, K., Yasuda, K., et al. (2018). B cells with aberrant activation of Notch1 signaling promote Treg and Th2 cell-dominant T-cell responses via IL-33. Blood Adv. 2 (18), 2282–2295. doi:10.1182/bloodadvances.2018019919
Arvey, A., VAN DER Veeken, J., Samstein, R. M., Feng, Y., Stamatoyannopoulos, J. A., and Rudensky, A. Y. (2014). Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 15 (6), 580–587. doi:10.1038/ni.2868
Barnas, J. L., Looney, R. J., and Anolik, J. H. (2019). B cell targeted therapies in autoimmune disease. Curr. Opin. Immunol. 61, 92–99. doi:10.1016/j.coi.2019.09.004
Baumjohann, D., Okada, T., and Ansel, K. M. (2011). Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187 (5), 2089–2092. doi:10.4049/jimmunol.1101393
Beguelin, W., Popovic, R., Teater, M., Jiang, Y., Bunting, K. L., Rosen, M., et al. (2013). EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23 (5), 677–692. doi:10.1016/j.ccr.2013.04.011
Beguelin, W., Rivas, M. A., Calvo Fernandez, M. T., Teater, M., Purwada, A., Redmond, D., et al. (2017). EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat. Commun. 8 (1), 877. doi:10.1038/s41467-017-01029-x
Beguelin, W., Teater, M., Gearhart, M. D., Calvo Fernandez, M. T., Goldstein, R. L., Cardenas, M. G., et al. (2016). EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell 30 (2), 197–213. doi:10.1016/j.ccell.2016.07.006
Bugide, S., Green, M. R., and Wajapeyee, N. (2018). Inhibition of Enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 115 (15), E3509–E18. doi:10.1073/pnas.1802691115
Caganova, M., Carrisi, C., Varano, G., Mainoldi, F., Zanardi, F., Germain, P. L., et al. (2013). Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Invest. 123 (12), 5009–5022. doi:10.1172/JCI70626
Cedar, H., and Bergman, Y. (2009). Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 10 (5), 295–304. doi:10.1038/nrg2540
Chakraborty, P., and Mehrotra, S. (2020). CD38: Modulating histone methyltransferase EZH2 activity in SLE. Trends Immunol. 41 (3), 187–189. doi:10.1016/j.it.2020.01.008
Chen, G., Subedi, K., Chakraborty, S., Sharov, A., Lu, J., Kim, J., et al. (2018). Ezh2 regulates activation-induced CD8+ T cell cycle progression via repressing Cdkn2a and Cdkn1c expression. Front. Immunol. 9, 549. doi:10.3389/fimmu.2018.00549
Chen, P. M., and Tsokos, G. C. (2021). T cell abnormalities in the pathogenesis of systemic lupus erythematosus: An update. Curr. Rheumatol. Rep. 23 (2), 12. doi:10.1007/s11926-020-00978-5
Chen, X., Cao, G., Wu, J., Wang, X., Pan, Z., Gao, J., et al. (2020). The histone methyltransferase EZH2 primes the early differentiation of follicular helper T cells during acute viral infection. Cell. Mol. Immunol. 17 (3), 247–260. doi:10.1038/s41423-019-0219-z
Choi, Y. S., Yang, J. A., Yusuf, I., Johnston, R. J., Greenbaum, J., Peters, B., et al. (2013). Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 190 (8), 4014–4026. doi:10.4049/jimmunol.1202963
Ciofani, M., and Zuniga-Pflucker, J. C. (2005). Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6 (9), 881–888. doi:10.1038/ni1234
Coit, P., Dozmorov, M. G., and Merrill, J. T. (2016). Epigenetic reprog ramming in naive CD4+ T cells favoring T cell activation and Non-Th1 Eff ector T Cell immune response as an early event in lupus flares [J]. Arthritis Rheumatol. 68 (9), 2200–2209.
Comte, D., Karampetsou, M. P., and Tsokos, G. C. (2015). T cells as a therapeutic target in SLE. Lupus 24 (4-5), 351–363. doi:10.1177/0961203314556139
Comte, D., Karampetsou, M. P., Yoshida, N., Kis-Toth, K., Kyttaris, V. C., and Tsokos, G. C. (2017). Signaling lymphocytic activation molecule family member 7 engagement restores defective effector CD8+ T cell function in systemic lupus erythematosus. Arthritis Rheumatol. 69 (5), 1035–1044. doi:10.1002/art.40038
Cribbs, A. P., Terlecki-Zaniewicz, S., Philpott, M., Baardman, J., Ahern, D., Lindow, M., et al. (2020). Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proc. Natl. Acad. Sci. U. S. A. 117 (11), 6056–6066. doi:10.1073/pnas.1919893117
Crotty, S. (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41 (4), 529–542. doi:10.1016/j.immuni.2014.10.004
Cruickshank, M. N., and Ulgiati, D. (2010). The role of notch signaling in the development of a normal B-cell repertoire. Immunol. Cell Biol. 88 (2), 117–124. doi:10.1038/icb.2009.73
Cyrus, S., Burkardt, D., and Weaver, D. D. (2019). PRC2-complex related dysfunction in overgrowth syndromes: A review of EZH2, EED, and SUZ12 and their syndromic phenotypes. Am. J. Med. Genet. C Semin. Med. Genet.
Czermin, B., Melfi, R., Mccabe, D., Seitz, V., Imhof, A., and Pirrotta, V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 (2), 185–196. doi:10.1016/s0092-8674(02)00975-3
Dai, Y., Zhang, L., and Hu, C. (2010). Genome-wide analysis of histone H3 lysine 4 trimethylation by ChIP-chip in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Clin. Exp. Rheumatol. 28 (2), 158–168.
Ding, S., Liang, Y., Zhao, M., Liang, G., Long, H., Zhao, S., et al. (2012). Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 64 (9), 2953–2963. doi:10.1002/art.34505
Ding, S., Zhang, Q., and Luo, S. (2019). BCL-6 suppresses miR-142-3p/5p expression in SLE CD4(+) T cells by modulating histone methylation and acetylation of the miR-142 promoter [J]. Cell Mol. Immunol.
Dobenecker, M. W., Park, J. S., Marcello, J., McCabe, M. T., Gregory, R., Knight, S. D., et al. (2018). Signaling function of PRC2 is essential for TCR-driven T cell responses. J. Exp. Med. 215 (4), 1101–1113. doi:10.1084/jem.20170084
Dolff, S., Scharpenberg, C., Specker, C., Kribben, A., Witzke, O., and Wilde, B. (2019). IL-22 production of effector CD4+ T-cells is altered in SLE patients. Eur. J. Med. Res. 24 (1), 24. doi:10.1186/s40001-019-0385-6
Dupage, M., Chopra, G., Quiros, J., Rosenthal, W. L., Morar, M. M., Holohan, D., et al. (2015). The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 42 (2), 227–238. doi:10.1016/j.immuni.2015.01.007
Garaud, S., Youinou, P., and Renaudineau, Y. (2011). DNA methylation and B-cell autoreactivity. Adv. Exp. Med. Biol. 711, 50–60. doi:10.1007/978-1-4419-8216-2_5
Gonzalez, M. E., Moore, H. M., Li, X., Toy, K. A., Huang, W., Sabel, M. S., et al. (2014). EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc. Natl. Acad. Sci. U. S. A. 111 (8), 3098–3103. doi:10.1073/pnas.1308953111
Gunawan, M., Venkatesan, N., Loh, J. T., Wong, J. F., Berger, H., Neo, W. H., et al. (2015). The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat. Immunol. 16 (5), 505–516. doi:10.1038/ni.3125
Guo, M., Price, M. J., Patterson, D. G., Barwick, B. G., Haines, R. R., Kania, A. K., et al. (2018). EZH2 represses the B cell transcriptional program and regulates antibody-secreting cell metabolism and antibody production. J. Immunol. 200 (3), 1039–1052. doi:10.4049/jimmunol.1701470
He, A., Shen, X., Ma, Q., Cao, J., von Gise, A., Zhou, P., et al. (2012). PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 26 (1), 37–42. doi:10.1101/gad.173930.111
Hedrich, C. M. (2018). Mechanistic aspects of epigenetic dysregulation in SLE. Clin. Immunol. 196, 3–11. doi:10.1016/j.clim.2018.02.002
Hervier, B., Beziat, V., Haroche, J., Mathian, A., Lebon, P., Ghillani-Dalbin, P., et al. (2011). Phenotype and function of natural killer cells in systemic lupus erythematosus: Excess interferon-γ production in patients with active disease. Arthritis Rheum. 63 (6), 1698–1706. doi:10.1002/art.30313
Herviou, L., Jourdan, M., Martinez, A. M., Cavalli, G., and Moreaux, J. (2019). EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation. Leukemia 33 (8), 2047–2060. doi:10.1038/s41375-019-0392-1
Honarpisheh, M., Kohler, P., VON Rauchhaupt, E., and Lech, M. (2018). The involvement of MicroRNAs in modulation of innate and adaptive immunity in systemic lupus erythematosus and lupus nephritis. J. Immunol. Res. 2018, 4126106. doi:10.1155/2018/4126106
Horwitz, D. A., Fahmy, T. M., Piccirillo, C. A., and La Cava, A. (2019). Rebalancing immune homeostasis to treat autoimmune diseases. Trends Immunol. 40 (10), 888–908. doi:10.1016/j.it.2019.08.003
Hou, S., Clement, R. L., Diallo, A., Blazar, B. R., Rudensky, A. Y., Sharpe, A. H., et al. (2019). FoxP3 and Ezh2 regulate Tfr cell suppressive function and transcriptional program. J. Exp. Med. 216 (3), 605–620. doi:10.1084/jem.20181134
Hsu, J. H., Rasmusson, T., and Robinson, J. (2019). EED-targeted PROTACs degrade EED, EZH2, and SUZ12 in the PRC2 complex. Cell Chem. Biol.
Hu, N., Qiu, X., and Luo, Y. (2008). Abnormal histone modification patterns in lupus CD4+ T cells [J]. J. Rheumatol. 35 (5), 804–810.
Huang, F., Zhao, J. L., Wang, L., Gao, C. C., Liang, S. Q., An, D. J., et al. (2017). miR-148a-3p mediates notch signaling to promote the differentiation and M1 activation of macrophages. Front. Immunol. 8, 1327. doi:10.3389/fimmu.2017.01327
Huntington, N. D., and Gray, D. H. (2018). Immune homeostasis in health and disease. Immunol. Cell Biol. 96 (5), 451–452. doi:10.1111/imcb.12043
Jacob, E., Hod-Dvorai, R., Schif-Zuck, S., and Avni, O. (2008). Unconventional association of the polycomb group proteins with cytokine genes in differentiated T helper cells. J. Biol. Chem. 283 (19), 13471–13481. doi:10.1074/jbc.M709886200
Javierre, B. M., Fernandez, A. F., Richter, J., Al-Shahrour, F., Martin-Subero, J. I., Rodriguez-Ubreva, J., et al. (2010). Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20 (2), 170–179. doi:10.1101/gr.100289.109
Javierre, B. M., and Richardson, B. (2011). A new epigenetic challenge: Systemic lupus erythematosus. Adv. Exp. Med. Biol. 711, 117–136. doi:10.1007/978-1-4419-8216-2_9
Jia, L., and Wu, C. (2014). The biology and functions of Th22 cells. Adv. Exp. Med. Biol. 841, 209–230. doi:10.1007/978-94-017-9487-9_8
Josefowicz, S. Z., Lu, L. F., and Rudensky, A. Y. (2012). Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564. doi:10.1146/annurev.immunol.25.022106.141623
Kaewraemruaen, C., Ritprajak, P., and Hirankarn, N. (2019). Dendritic cells as key players in systemic lupus erythematosus. Asian Pac J. Allergy Immunol.
Kanno, Y., Vahedi, G., Hirahara, K., Singleton, K., and O'Shea, J. J. (2012). Transcriptional and epigenetic control of T helper cell specification: Molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 30, 707–731. doi:10.1146/annurev-immunol-020711-075058
Karagianni, P., and Tzioufas, A. G. (2019). Epigenetic perspectives on systemic autoimmune disease. J. Autoimmun. 104, 102315. doi:10.1016/j.jaut.2019.102315
Karantanos, T., Chistofides, A., Barhdan, K., Li, L., and Boussiotis, V. A. (2016). Regulation of T Cell differentiation and function by EZH2. Front. Immunol. 7, 172. doi:10.3389/fimmu.2016.00172
Katsuyama, E., Suarez-Fueyo, A., Bradley, S. J., Mizui, M., Marin, A. V., Mulki, L., et al. (2020). The CD38/NAD/SIRTUIN1/EZH2 Axis mitigates cytotoxic CD8 T cell function and identifies patients with SLE prone to infections. Cell Rep. 30 (1), 112–123. doi:10.1016/j.celrep.2019.12.014
Kim, J., Lee, Y., Lu, X. D., Song, B., Fong, K. W., Cao, Q., et al. (2018). Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 25 (10), 2808–2820. doi:10.1016/j.celrep.2018.11.035
Kim, K. H., and Roberts, C. W. (2016). Targeting EZH2 in cancer. Nat. Med. 22 (2), 128–134. doi:10.1038/nm.4036
Kis-toth, K., Comte, D., Karampetsou, M. P., Kyttaris, V. C., Kannan, L., Terhorst, C., et al. (2016). Selective loss of signaling lymphocytic activation molecule family member 4-positive CD8+ T cells contributes to the decreased cytotoxic cell activity in systemic lupus erythematosus. Arthritis Rheumatol. 68 (1), 164–173. doi:10.1002/art.39410
Koyanagi, M., Baguet, A., Martens, J., Margueron, R., Jenuwein, T., and Bix, M. (2005). EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 280 (36), 31470–31477. doi:10.1074/jbc.M504766200
Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2002). Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16 (22), 2893–2905. doi:10.1101/gad.1035902
Kwon, H. K., Chen, H. M., Mathis, D., and Benoist, C. (2017). Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 18 (11), 1238–1248. doi:10.1038/ni.3835
Lee, J. M., Lee, J. S., Kim, H., Kim, K., Park, H., Kim, J.-Y., et al. (2012). EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 48 (4), 572–586. doi:10.1016/j.molcel.2012.09.004
Lee, S. T., Li, Z., Wu, Z., Aau, M., Guan, P., Karuturi, R. K. M., et al. (2011). Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 43 (5), 798–810. doi:10.1016/j.molcel.2011.08.011
Li, F., Zeng, Z., Xing, S., Gullicksrud, J. A., Shan, Q., Choi, J., et al. (2018). Ezh2 programs TFH differentiation by integrating phosphorylation-dependent activation of Bcl6 and polycomb-dependent repression of p19Arf. Nat. Commun. 9 (1), 5452. doi:10.1038/s41467-018-07853-z
Li, H., Wen, Y. H., Wu, S. L., Chen, D., Luo, X., Xu, R., et al. (2019). Epigenetic modification of enhancer of zeste homolog 2 modulates the activation of dendritic cells in allergen immunotherapy. Int. Arch. Allergy Immunol. 180 (2), 120–127. doi:10.1159/000500882
Liao, W., Li, M., Wu, H., Jia, S., Zhang, N., Dai, Y., et al. (2017). Down-regulation of MBD4 contributes to hypomethylation and overexpression of CD70 in CD4+ T cells in systemic lupus erythematosus. Clin. Epigenetics 9, 104. doi:10.1186/s13148-017-0405-8
Liu, H. W., Lin, H. L., Yen, J. H., Tsai, W. C., Chiou, S. S., Chang, J. G., et al. (2014). Demethylation within the proximal promoter region of human estrogen receptor alpha gene correlates with its enhanced expression: Implications for female bias in lupus. Mol. Immunol. 61 (1), 28–37. doi:10.1016/j.molimm.2014.05.002
Liu, X., Nurieva, R. I., and Dong, C. (2013). Transcriptional regulation of follicular T-helper (Tfh) cells. Immunol. Rev. 252 (1), 139–145. doi:10.1111/imr.12040
Liu, Z., Yang, L., Zhong, C., and Zhou, L. (2020). EZH2 regulates H2B phosphorylation and elevates colon cancer cell autophagy. J. Cell. Physiol. 235 (2), 1494–1503. doi:10.1002/jcp.29069
Loh, J. T., Lim, T. J. F., Ikumi, K., Matoba, T., Janela, B., Gunawan, M., et al. (2018). Ezh2 controls skin tolerance through distinct mechanisms in different subsets of skin dendritic cells. Iscience 10, 23–39. doi:10.1016/j.isci.2018.11.019
Long, H., Xiang, T., Luo, J., Li, F., Lin, R., Liu, S., et al. (2016). The tumor microenvironment disarms CD8+ T lymphocyte function via a miR-26a-EZH2 axis. Oncoimmunology 5 (12), e1245267. doi:10.1080/2162402X.2016.1245267
Long, H., Yin, H., Wang, L., Gershwin, M. E., and Lu, Q. (2016). The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J. Autoimmun. 74, 118–138. doi:10.1016/j.jaut.2016.06.020
Macri, C., Pang, E. S., Patton, T., and O'Keeffe, M. (2018). Dendritic cell subsets. Semin. Cell Dev. Biol. 84, 11–21. doi:10.1016/j.semcdb.2017.12.009
Mandal, M., Powers, S. E., Maienschein-Cline, M., Bartom, E. T., Hamel, K. M., Kee, B. L., et al. (2011). Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12 (12), 1212–1220. doi:10.1038/ni.2136
Margueron, R., and Reinberg, D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469 (7330), 343–349. doi:10.1038/nature09784
Navarro quiroz, E., Chavez-Estrada, V., Macias-Ochoa, K., Ayala-Navarro, M. F., Flores-Aguilar, A. S., Morales-Navarrete, F., et al. (2019). Epigenetic mechanisms and posttranslational modifications in systemic lupus erythematosus. Int. J. Mol. Sci. 20 (22), E5679. doi:10.3390/ijms20225679
Neo, W. H., Lim, J. F., Grumont, R., Gerondakis, S., and Su, I. H. (2014). c-Rel regulates Ezh2 expression in activated lymphocytes and malignant lymphoid cells. J. Biol. Chem. 289 (46), 31693–31707. doi:10.1074/jbc.M114.574517
Nutt, S. L., Keenan, C., and Chopin, M. (2020). EZH2 function in immune cell development. Biol. Chem.
O'neill, L. A., Kishton, R. J., and Rathmell, J. (2016). A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16 (9), 553–565. doi:10.1038/nri.2016.70
Poepsel, S., Kasinath, V., and Nogales, E. (2018). Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25 (2), 154–162. doi:10.1038/s41594-018-0023-y
Qiu, R., Zhou, L., Ma, Y., Zhou, L., Liang, T., Shi, L., et al. (2020). Regulatory T cell plasticity and stability and autoimmune diseases. Clin. Rev. Allergy Immunol. 58 (1), 52–70. doi:10.1007/s12016-018-8721-0
Rapoport, M., and Bloch, O. (2012). Systemic lupus erythematosus. N. Engl. J. Med. 366 (6), 574. doi:10.1056/NEJMc1115196
Rohraff, D. M., He, Y., Farkash, E. A., Schonfeld, M., Tsou, P. S., and Sawalha, A. H. (2019). Inhibition of EZH2 ameliorates lupus-like disease in MRL/lpr mice. Arthritis Rheumatol. 71 (10), 1681–1690. doi:10.1002/art.40931
Sawalha, A. H. (2019). BCL-6 and EZH2 cooperate to epigenetically repress anti-inflammatory miR-142-3p/5p in lupus CD4+T cells. Cell Mol. Immunol. 18. doi:10.1038/s41423-019-0288-z
Scharer, C. D., Barwick, B. G., Guo, M. Y., Bally, A. P. R., and Boss, J. M. (2018). Plasma cell differentiation is controlled by multiple cell division-coupled epigenetic programs. Nat. Commun. 9, 1698. doi:10.1038/s41467-018-04125-8
Schrezenmeier, E., Lino, A. C., and Alexander, T. (2019). EZH2 in B cell subsets. Arthritis Rheumatol.
Sha, M.-Q., Zhao, X.-L., Li, L., Li, L.-H., Li, Y., Dong, T.-G., et al. (2016). EZH2 mediates lidamycin-induced cellular senescence through regulating p21 expression in human colon cancer cells. Cell Death Dis. 7 (11), e2486. doi:10.1038/cddis.2016.383
Shi, B., Liang, J., Yang, X., Wang, Y., Zhao, Y., Wu, H., et al. (2007). Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 27 (14), 5105–5119. doi:10.1128/MCB.00162-07
Straining, R., and Eighmy, W. (2022). Tazemetostat: EZH2 inhibitor. J. Adv. Pract. Oncol. 13 (2), 158–163. doi:10.6004/jadpro.2022.13.2.7
Su, I. H., Basavaraj, A., Krutchinsky, A. N., Hobert, O., Ullrich, A., Chait, B. T., et al. (2003). Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4 (2), 124–131. doi:10.1038/ni876
Su, I. H., Dobenecker, M. W., Dickinson, E., Oser, M., Basavaraj, A., Marqueron, R., et al. (2005). Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 121 (3), 425–436. doi:10.1016/j.cell.2005.02.029
Sullivan, K. E., Suriano, A., Dietzmann, K., Lin, J., Goldman, D., and Petri, M. A. (2007). The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin. Immunol. 123 (1), 74–81. doi:10.1016/j.clim.2006.12.008
Sun, D., Lin, Y., Hong, J., Chen, H., Nagarsheth, N., Peng, D., et al. (2016). Th22 cells control colon tumorigenesis through STAT3 and Polycomb Repression complex 2 signaling. Oncoimmunology 5 (8), e1082704. doi:10.1080/2162402X.2015.1082704
Tabbal, H., Septier, A., Mathieu, M., Drelon, C., Rodriguez, S., Djari, C., et al. (2019). EZH2 cooperates with E2F1 to stimulate expression of genes involved in adrenocortical carcinoma aggressiveness. Br. J. Cancer 121 (5), 384–394. doi:10.1038/s41416-019-0538-y
Teruel, M., and Sawalha, A. H. (2017). Epigenetic variability in systemic lupus erythematosus: What we learned from genome-wide DNA methylation studies. Curr. Rheumatol. Rep. 19 (6), 32. doi:10.1007/s11926-017-0657-5
Tong, Q., He, S., Xie, F., Mochizuki, K., Liu, Y., Mochizuki, I., et al. (2014). Ezh2 regulates transcriptional and posttranslational expression of T-bet and promotes Th1 cell responses mediating aplastic anemia in mice. J. Immunol. 192 (11), 5012–5022. doi:10.4049/jimmunol.1302943
Tsokos, G. C., Lo, M. S., Reis, P. C., and Sullivan, K. E. (2016). New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12 (12), 716–730. doi:10.1038/nrrheum.2016.186
Tsou, P. S., Coit, P., Kilian, N. C., and Sawalha, A. H. (2018). EZH2 modulates the DNA methylome and controls T cell adhesion through junctional adhesion molecule A in lupus patients. Arthritis Rheumatol. 70 (1), 98–108. doi:10.1002/art.40338
Tumes, D. J., Onodera, A., Suzuki, A., Shinoda, K., Endo, Y., Iwamura, C., et al. (2013). The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity 39 (5), 819–832. doi:10.1016/j.immuni.2013.09.012
Vasanthakumar, A., Xu, D., Lun, A. T., Kueh, A. J., van Gisbergen, K. P., Iannarella, N., et al. (2017). A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 18 (4), 619–631. doi:10.15252/embr.201643237
Velichutina, I., Shaknovich, R., Geng, H., Johnson, N. A., Gascoyne, R. D., Melnick, A. M., et al. (2010). EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 116 (24), 5247–5255. doi:10.1182/blood-2010-04-280149
Vire, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C., et al. (2006). The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439 (7078), 871–874. doi:10.1038/nature04431
Vivier, E., Raulet, D. H., Moretta, A., Caligiuri, M. A., Zitvogel, L., Lanier, L. L., et al. (2011). Innate or adaptive immunity? The example of natural killer cells [J]. Science 331 (6013), 44–49. doi:10.1126/science.1198687
Vivier, E., Tomasello, E., Baratin, M., Walzer, T., and Ugolini, S. (2008). Functions of natural killer cells. Nat. Immunol. 9 (5), 503–510. doi:10.1038/ni1582
Wang, J., He, C., and Gao, P. (2019). HNF1B-mediated repression of SLUG is suppressed by EZH2 in aggressive prostate cancer. Oncogene.
Wardowska, A. (2020). The epigenetic face of lupus: Focus on antigen-presenting cells. Int. Immunopharmacol. 81, 106262. doi:10.1016/j.intimp.2020.106262
Weeding, E., and Sawalha, A. H. (2018). Deoxyribonucleic acid methylation in systemic lupus erythematosus: Implications for future clinical practice. Front. Immunol. 9, 875. doi:10.3389/fimmu.2018.00875
Wei, G., Wei, L., Zhu, J., Zang, C., Hu-Li, J., Yao, Z., et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30 (1), 155–167. doi:10.1016/j.immuni.2008.12.009
Wu, H., Huang, X., Qiu, H., Zhao, M., Liao, W., Yuan, S., et al. (2016). High salt promotes autoimmunity by TET2-induced DNA demethylation and driving the differentiation of Tfh cells. Sci. Rep. 6, 28065. doi:10.1038/srep28065
Wu, H., Zhao, M., Tan, L., and Lu, Q. (2016). The key culprit in the pathogenesis of systemic lupus erythematosus: Aberrant DNA methylation. Autoimmun. Rev. 15 (7), 684–689. doi:10.1016/j.autrev.2016.03.002
Wu, L., Jiang, X., Qi, C., Zhang, C., Qu, B., and Shen, N. (2021). EZH2 inhibition interferes with the activation of type I interferon signaling pathway and ameliorates lupus nephritis in NZB/NZW F1 mice. Front. Immunol. 12, 653989. doi:10.3389/fimmu.2021.653989
Xiao, X. Y., Li, Y. T., Jiang, X., Ji, X., Lu, X., Yang, B., et al. (2020). EZH2 deficiency attenuates Treg differentiation in rheumatoid arthritis. J. Autoimmun. 108, 102404. doi:10.1016/j.jaut.2020.102404
Xu, W. D., Fu, L., Liu, X. Y., Wang, J. M., Yuan, Z. C., Su, L. C., et al. (2019). Association between TL1A gene polymorphisms and systemic lupus erythematosus in a Chinese Han population. J. Cell. Physiol. 234 (12), 22543–22553. doi:10.1002/jcp.28818
Yan, J., Li, B., Lin, B., Lee, P. T., Chung, T. H., Tan, J., et al. (2016). EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood 128 (7), 948–958. doi:10.1182/blood-2016-01-690701
Yan, J., Ng, S. B., Tay, J. L., Lin, B., Koh, T. L., Tan, J., et al. (2013). EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 121 (22), 4512–4520. doi:10.1182/blood-2012-08-450494
Yan, S., Yim, L. Y., Lu, L., Lau, C. S., and Chan, V. S. F. (2014). MicroRNA regulation in systemic lupus erythematosus pathogenesis. Immune Netw. 14 (3), 138–148. doi:10.4110/in.2014.14.3.138
Yang, Q., Zhao, S., Shi, Z., Cao, L., Liu, J., Pan, T., et al. (2021). Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 40 (1), 120. doi:10.1186/s13046-021-01901-1
Yang, W., Tang, H., Zhang, Y., Tang, X., Zhang, J., Sun, L., et al. (2013). Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet. 92 (1), 41–51. doi:10.1016/j.ajhg.2012.11.018
Yang, X. P., Jiang, K., Hirahara, K., Vahedi, G., Afzali, B., Sciume, G., et al. (2015). EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci. Rep. 5, 10643. doi:10.1038/srep10643
Yap, D. Y. H., and Chan, T. M. (2019). B cell abnormalities in systemic lupus erythematosus and lupus nephritis-role in pathogenesis and effect of immunosuppressive treatments. Int. J. Mol. Sci. 20 (24), E6231. doi:10.3390/ijms20246231
Yin, J., Leavenworth, J. W., Li, Y., Luo, Q., Xie, H., Liu, X., et al. (2015). Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc. Natl. Acad. Sci. U. S. A. 112 (52), 15988–15993. doi:10.1073/pnas.1521740112
Yin, Y., Choi, S. C., Xu, Z., Perry, D. J., Seay, H., Croker, B. P., et al. (2015). Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 7 (274), 274ra18. doi:10.1126/scitranslmed.aaa0835
Zhang, M., Iwata, S., and Hajime, M. (2020). Methionine commits cells to differentiate into plasmablasts through epigenetic regulation of BTB and CNC homolog 2 by the methyltransferase enhancer of zeste homolog 2 [J]. Arthritis Rheumatol. 72. doi:10.1002/art.41208
Zhang, Y., Kinkel, S., Maksimovic, J., Bandala-Sanchez, E., Tanzer, M. C., Naselli, G., et al. (2014). The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood 124 (5), 737–749. doi:10.1182/blood-2013-12-544106
Zhang, Y., Zhao, M., Sawalha, A. H., Richardson, B., and Lu, Q. (2013). Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J. Autoimmun. 41, 92–99. doi:10.1016/j.jaut.2013.01.005
Zhang, Z., Song, L., Maurer, K., Petri, M. A., and Sullivan, K. E. (2010). Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 11 (2), 124–133. doi:10.1038/gene.2009.66
Zhao, E., Maj, T., Kryczek, I., Li, W., Wu, K., Zhao, L., et al. (2016). Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 17 (1), 95–103. doi:10.1038/ni.3313
Zhao, H., Wang, L., Luo, H., Li, Q. Z., and Zuo, X. (2017). TNFAIP3 downregulation mediated by histone modification contributes to T-cell dysfunction in systemic lupus erythematosus. Rheumatol. Oxf. 56 (5), 835–843. doi:10.1093/rheumatology/kew508
Zhao, M., Liu, S., Luo, S., Wu, H., Tang, M., Cheng, W., et al. (2014). DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J. Autoimmun. 54, 127–136. doi:10.1016/j.jaut.2014.07.002
Zhao, Y., Hu, Z., Li, J., and Hu, T. (2021). EZH2 exacerbates breast cancer by methylating and activating STAT3 directly. J. Cancer 12 (17), 5220–5230. doi:10.7150/jca.50675
Zhen, Y., Smith, R. D., and Finkelman, F. D. (2020). Ezh2-mediated epig enetic modification is required for allogeneic T cell-induced lupus disease [J]. Arthritis Res. Ther. 22 (1), 133.
Zheng, X., Tsou, P. S., and Sawalha, A. H. (2020). Increased expression of EZH2 is mediated by higher glycolysis and mTORC1 activation in lupus CD4+ T cells. Immunometabolism 2 (2), e200013. doi:10.20900/immunometab20200013
Zhu, K., DU, D., and Yang, R. (2019). Identification and assessments of novel and potent small-molecule inhibitors of EED-EZH2 interaction of polycomb repressive complex 2 by computational methods and biological evaluations. Chem. Pharm. Bull.
Zhu, X., Wang, X., Wang, Y., and Zhao, Y. (2020). Exosomal long non-coding RNA GAS5 suppresses Th1 differentiation and promotes Th2 differentiation via downregulating EZH2 and T-bet in allergic rhinitis. Mol. Immunol. 118, 30–39. doi:10.1016/j.molimm.2019.11.009
Keywords: EZH2, systemic lupus erythematosus, T cells, B cells, immune homeostasis
Citation: Yang Y, Liu K, Liu M, Zhang H and Guo M (2022) EZH2: Its regulation and roles in immune disturbance of SLE. Front. Pharmacol. 13:1002741. doi: 10.3389/fphar.2022.1002741
Received: 25 July 2022; Accepted: 23 September 2022;
Published: 13 October 2022.
Edited by:
Feng Lai Yuan, The Affiliated Hospital of Jiangnan University, ChinaReviewed by:
Jianchun Wu, University of Illinois at Chicago, United StatesCopyright © 2022 Yang, Liu, Liu, Zhang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huali Zhang, emhhbmdodWFsaUBjc3UuZWR1LmNu; Muyao Guo, Z211eWFvQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.