
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 28 January 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.826112
 David Speck1
David Speck1 Gunnar Kleinau1
Gunnar Kleinau1 Mark Meininghaus2
Mark Meininghaus2 Antje Erbe3,4
Antje Erbe3,4 Alexandra Einfeldt3,4
Alexandra Einfeldt3,4 Michal Szczepek1
Michal Szczepek1 Patrick Scheerer1,5*
Patrick Scheerer1,5* Vera Pütter3,4*
Vera Pütter3,4*G-protein coupled receptors (GPCR) transduce extracellular stimuli into the cell interior and are thus centrally involved in almost all physiological-neuronal processes. This essential function and association with many diseases or pathological conditions explain why GPCRs are one of the priority targets in medical and pharmacological research, including structure determination. Despite enormous experimental efforts over the last decade, both the expression and purification of these membrane proteins remain elusive. This is attributable to specificities of each GPCR subtype and the finding of necessary experimental in vitro conditions, such as expression in heterologous cell systems or with accessory proteins. One of these specific GPCRs is the leucine-rich repeat domain (LRRD) containing GPCR 7 (LGR7), also termed relaxin family peptide receptor 1 (RXFP1). This receptor is characterized by a large extracellular region of around 400 amino acids constituted by several domains, a rare feature among rhodopsin-like (class A) GPCRs. In the present study, we describe the expression and purification of RXFP1, including the design of various constructs suitable for functional/biophysical studies and structure determination. Based on available sequence information, homology models, and modern biochemical and genetic tools, several receptor variations with different purification tags and fusion proteins were prepared and expressed in Sf9 cells (small-scale), followed by an analytic fluorescence-detection size-exclusion chromatography (F-SEC) to evaluate the constructs. The most promising candidates were expressed and purified on a large-scale, accompanied by ligand binding studies using surface plasmon resonance spectroscopy (SPR) and by determination of signaling capacities. The results may support extended studies on RXFP1 receptor constructs serving as targets for small molecule ligand screening or structural elucidation by protein X-ray crystallography or cryo-electron microscopy.
In 1926, Frederick Hisaw set the starting point for relaxin research when he transferred blood plasma from a pregnant to a virgin guinea pig, thereby triggering pubic ligament relaxation in the recipient animal (Bylander et al., 1987). The relaxin sequence was determined in 1984 (Fields and Larkin, 1981; Yamamoto et al., 1981; Hudson et al., 1984) after successful isolation from the decidua and placenta. Following several years, the corresponding relaxin binding partner was identified (Hsu et al., 2000; Hsu et al., 2002). Based on sequence similarities with previously discovered leucine-rich repeat containing receptors (LGRs), this relaxin receptor was originally termed LGR7 but later renamed RXFP1 (Braun et al., 1991; Hsu et al., 1998; Bathgate et al., 2006). RXFP1 was traditionally purported to be involved in pregnancy and parturition processes; however, a sex-independent receptor-transcript distribution has been shown (e.g., kidney, brain, and cardiac cells) (Kohsaka et al., 1998; Hsu et al., 2000; Hsu et al., 2002; Samuel et al., 2004a; Luna et al., 2004), provoking the assumption of a variety of different physiological functions. Knock-out of the Lgr7 (Rxfp1) gene in mice, as observed for the relaxin knock-out, resulted in prolonged birth duration and increased infant mortality (Kamat et al., 2004; Krajnc-Franken et al., 2004). Receptor or ligand knock-out also led to an age-dependent increase in tissue fibrosis, whereby relaxin was studied extensively as an anti-fibrotic therapeutic (Samuel et al., 2003; Samuel et al., 2004a; Samuel et al., 2004b; McBride et al., 2017; Samuel et al., 2017).
RXFP1 activates intracellularly at least three different Gα variants, namely Gαs, Gαi, and Gαo (Hsu et al., 2000; Hsu et al., 2002; Halls et al., 2006; Halls et al., 2009a; Halls et al., 2009b), whereby the type of associated G-protein is also dependent on the cell type and receptor expressing tissue. Finally, RXFP1 and identified splicing variants are recognized as important key players in physiology and are also related to various disease states. Generally, RXFP1 or receptor variants are involved in cancer development, as well as skin, kidney, cardiac, liver, and lung fibrosis (reviewed in Chen et al. (2020).
RXFP1 is classified as a class A rhodopsin-like GPCR (Petrie et al., 2015). The superfamily of GPCR in humans comprises more than 800 members. They transduce a huge diversity of extracellular stimuli into the cell interior for cell-signaling regulation (Limbird, 2004). GPCRs are related to more than 100 human diseases, including cancer and endocrine pathologies (O'Hayre et al., 2013; Schöneberg et al., 2004; Vassart and Costagliola, 2011). This fact, combined with their omnipresence in nearly all physiological processes, reasons why these receptors are a strong focus of academic research or pharmacological investigations, including structure determination (Heyder et al., 2021) and drug development (Garland, 2013; Errey and Fiez-Vandal, 2020).
RXFP1 and RXFP2 form the LGR group C (Wilkinson et al., 2005). The LGR group A includes the homologous lutropin receptor (LHCGR) (Troppmann et al., 2013), the thyrotropin receptor (TSHR) (Kleinau et al., 2013; Kleinau et al., 2017), and the follitropin receptor (FSHR) (Ulloa-Aguirre and Zariñán, 2016) (Supplementary Figure S1). LGR4−6 constitute LGR group B (Park et al., 2005). LGRs are more complex in their extracellular architecture compared to other class A GPCRs. RXFP1 exposes a large extracellular part (∼400 amino acids) that is constituted by the Leucine-rich repeat domain (LRRD) and the low-density lipoprotein class A (LDLa) domain (Hsu et al., 2000), which are connected via a linker region (Supplementary Figure S1, Figure 1). Such module-like extracellular architecture is found mainly in other GPCR classes as the class B (secretin and adhesion) GPCRs (Nordström et al., 2009). Nonetheless, missing structural information for group C LGRs hampers a more complete functional understanding of this receptor. Moreover, only a few peptidic ligands were developed, with most relaxin derivatives (Sudo et al., 2003; Halls et al., 2005; Hossain et al., 2010; Hossain et al., 2011; Chan et al., 2012; Hossain et al., 2016), and only first non-peptidic ligands have been identified, such as the small molecule agonist ML290 (EC50 of 94 nM) (Xiao et al., 2010; Xiao et al., 2013; Wilson et al., 2018). Altogether, RXFP1 is a highly interesting GPCR from several aspects; however, considerable detailed information for the comprehensive understanding of this receptor or for modifying its signaling activity remains lacking. Our study aimed to express RXFP1 diverse receptor variants suitable for functional/biophysical studies and structure determination.
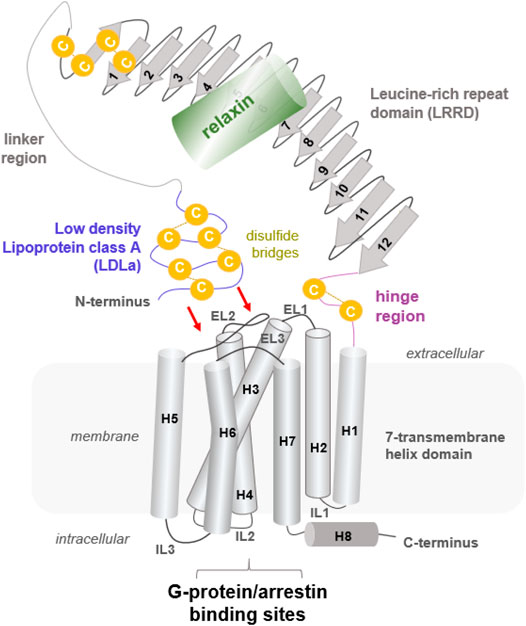
FIGURE 1. Putative structural arrangement of the different putative RXFP1 components. The RXFP1 receptor shows typical GPCR features as the seven transmembrane helices and exposes specific properties significant for LGRs in the class A GPCRs, such as a large extracellular part constituted by several interrelated domains. Their exact spatial arrangement to one another remains unknown, but it is assumed that the N-terminal LDLa domain interacts directly with extracellular loops (indicated by red arrows). The endogenous ligand relaxin, a peptide hormone, binds to the LRRD and simultaneously to the linker region.
The human RXFP1 is constituted by 757 amino acids, with half assigned to the N-terminal extracellular part (Kong et al., 2010). A homology model of the RXFP1 was created to predict and visualize putative structural properties and more rationally guide the design of different receptor variants for experimental studies. Moreover, we complexed the RXFP1 model with the ligand relaxin to recapitulate already reported (e.g., (Büllesbach and Schwabe, 2005)) amino acid residues involved in intermolecular interactions.
Template structures - The assumed structural architecture of RXFP1 is constituted by several interconnected domains (Kong et al., 2010; Sethi et al., 2016a) (Figure 1). To date, there is very little direct structural information published (Hopkins et al., 2007). For modeling a putative RXFP1 structure, the following templates of evolutionarily related class A GPCRs were used, accompanied by ab initio modeling of receptor regions without any template:
(1) For the N-terminal low-density lipoprotein class A (LDLa, amino acids Gly21-Gly63), structural information is available ((Berman et al., 2000), Protein database http://www.rcsb.org ID: 2JM4 (Hopkins et al., 2007)).
(2) The linker region (Asp64-Glu94, numbering with signal peptide) between the LDLa and the LRRD (Figure 1) was added to the LDLa domain without any structural template or structure predictions.
(3) The LRRD was constructed according to the FSHR LRRD structure (PDB ID: 4AY9 (Jiang et al., 2012), RXFP1 sequence Cys95-Ile370).
(4) The extracellular RXFP1 hinge region connecting the putative LRRD and the transmembrane domain (His371-Glu400) was inserted without any template structure.
(5) For modeling the RXFP1 seven transmembrane helix domains (TMD) in an active state conformation (As401-Gln701), the available structural complex of the β2-adrenoreceptor in complex with the Gs protein was utilized (PDB ID: 3SN6 (Rasmussen et al., 2011)).
In addition,
(6) The extracellular loop 2 (EL2) was constructed based on the EL2 of the opsin receptor (PDB ID: 3DQB (Scheerer et al., 2008)) because the amino acid constitution and length of the loop is more similar between opsin and RXFP1 compared to between RXFP1 and the β2-adrenoreceptor (detailed alignment not shown).
(7) The already determined relaxin-3 structure as an evidenced RXFP1 agonist (Sudo et al., 2003) was used as an agonistic peptide ligand (PDB ID: 2FHW, (Rosengren et al., 2006)).
(8) An inactive state RXFP1 model of the TMD structure for comparison with the active state conformation was designed by using the determined inactive state structure of the β2-adrenoreceptor (PDB ID: 2RH1) (Cherezov et al., 2007).
Template modifications prior to model composition - Minimal template modifications at the FSHR LRRD for the RXFP1 LRRD structure modeling were manual adjustments in repeat length by deletions or insertions of several amino acids. For modeling the RXFP1 TMD, the β2-adrenoreceptor template was modified by deleting the fused T4-lysozyme, the bound agonistic ligand, and Gs protein. Moreover, the EL1 and EL3 loop lengths were adjusted manually to respective RXFP1 loop lengths. Amino acid residues of the third intracellular loop (IL3) of the RXFP1 were added manually to the template because of the missing IL3 structure in the template. A similar procedure was used to prepare the inactive state conformation of the β2-adrenoreceptor; however, without deletion of the Gs protein.
Further details of modeling methods are provided in the Supplementary Material. Based on this information, we predicted the length and constitution of several designed and tested receptor constructs. These constructs can be constituted by partial receptor variants including specific or single domains, as reported for endogenously expressed splice variants (Chen et al., 2020).
Synthetic gene fragments encoding different truncated versions of human RXFP1 protein sequence were integrated into the baculovirus pVL1393 (hereafter abbreviated as pVL) transfer vector or pcDNA6.2 (abbreviated as pcDNA). All constructs (see Figure 4) carried an influenza hemagglutinin secretion tag at the N-terminus followed mostly by a FLAG peptide sequence-tag (FLAG-tag). The C-terminus carried a polyhistidine tag (His-tag) with a subsequent TEV cleavage site, an AVI-tag, and a 1D4-tag. An eGFP-fusion protein-tag was either located at the N-terminus between the FLAG-tag and the receptor (detachable using a PreScission™ protease cleavage site) or at the C-terminus between the TEV cleavage site and the AVI-tag.
All constructs were expressed in Sf9 (Spodoptera frugiperda) insect cells (Expression Systems) using the flashBAC baculovirus system (Oxford Expression Technologies) according to the manufacturer’s instructions. Sf9 cells were cultivated in suspension in ESF921 medium (Expression System).
In 2.5 ml small-scale test expressions, Sf9 cells were infected at a density of 4.0 x 106 cells/ml with high-titer viral stock at different multiplicities of infection (MOI). Cells were harvested by centrifugation 48 or 72 h after infection and stored at −20°C until purification.
For large-scale recombinant protein production, Sf9 cells at densities of 4.0 x 106 cells/ml were infected with high-titer viral stock at an MOI of 0.3. Cells were incubated for 48 h at 27°C in a cell wave bag at 22 rpm. Cells were harvested by centrifugation, flash-frozen, and stored at −80°C until use.
All purification steps were performed at 4°C or on ice. Biomass corresponding to 2 L of insect cell culture was thawed on ice and resuspended in lysis buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol and 0.1 mM TCEP) supplemented with cOmplete™ protease inhibitor Cocktail (Roche Applied Science). The receptor was solubilized by the addition of 1% (w/v) n-dodectyl-ß-D-maltoside (DDM, Anatrace), 0.1% (w/v) cholesteryl-hemi-succinate (CHS, Sigma), 10 strokes in a glass douncer, followed by 1 h incubation under gentle agitation at 4°C. The insoluble fraction was removed by centrifugation (150,000 rcf, 45 min). Detergent solubilized RXFP1 constructs were captured by gravity flow on 2 ml M2 resin (Anti-Flag M2-agarose, Sigma) pre-equilibrated with solubilization buffer supplemented with 1% DDM (w/v). M2 resin was washed with buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 0.1% DDM, 0.01% CHS, 10% glycerol, and 0.1 mM TCEP) and eluted with the same buffer supplemented with 0.4 mg/ml flag peptide. Immediately after elution, RXFP1 was subsequently concentrated with a 100 kDa Vivaspin Turbo concentrator (Sartorius) to a final volume of 500 µl. The sample was further purified by size-exclusion chromatography using a Superose 6 Increase 10/300 GL column (GE Healthcare) pre-equilibrated with gel filtration running buffer (50 mM HEPES pH7.5, 150 mM NaCl, 0.1% DDM, 0.01% CHS, 10% glycerol, and 0.1 mM TCEP). The monomeric receptor peak was pooled. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C. Binding activity was measured by SPR.
Forty microliters of Flag elution fractions were applied on an analytic Superose 6 increase 5/15 GL column (GE Healthcare). From each elution fraction, 5 µl were transferred into a 96-well plate supplemented with a 95 µl gel filtration buffer. The GFP signal of the different fractions was measured in a Tecan M1000 plate reader.
The nano Differential Scanning Fluorimetry method (nanoDSF) is based on the intrinsic tryptophan, tyrosine, and phenylalanine fluorescence. This method measures the fluorescence intensity ratio between the fluorescent amino acids in 10 μl capillaries. Once a molecule unfolds, the aromatic amino acid residues sites alter and trigger changes in the fluorescence. The Prometheus NT.48 nanoDSF (NanoTemper Technologies) was used to measure the ratio (350:330 nm) as a function of temperature. In a typical nanoDSF experiment, the intrinsic fluorescence intensity ratio (350:330 nm) is constantly measured and recorded. For analysis of the results, the intrinsic fluorescence intensity ratio (or the first derivative of the ratio) was plotted as a function of temperature, yielding a protein unfolding curve. In the experiment, we used standard glass capillaries from NanoTemper, with a sample volume of a maximum of 10 μl. For each experiment, at least three replicate experiments were performed.
nanoDSF data analysis was performed using the PR.ThermControl v2.0.4 software (NanoTemper Technologies). The thermal transition (unfolding) temperature (Tm) was obtained in the post-run data analysis. The Tm values can be used to assess the thermal stability of the analyzed protein.
All tested receptor constructs were captured at 20°C via a biotinylated Avi-tag on an SA-chip. Immobilization buffer contained 50 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% DDM, 0.01% CHS, and 0.1 mM TCEP. The obtained immobilization level was between 3000 and 5000 RU. A Biacore T200 was used for all experiments. Binding assays were performed at 20°C, with running buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 0.01% BSA, 0.1% DDM, and 0.01% CHS. Relaxin was injected at a concentration series of 0.41–100 nM. Relaxin H2 (human) from Bachem (#40399910.0200) and R&D Systems (#3596-RN/CF, >97% by SDS-PAGE, activity measured by its ability to induce cAMP accumulation in THP-1 human acute monocytic leukemia cells) was used in SPR and cellular assays, respectively.
HEK293 cells were cultured in Dulbecco’s modified eagle medium (DMEM) high glucose (Gibco), supplemented with 10% fetal bovine serum, 1.5% HEPES, 1% minimal essential medium non-essential amino acids (MEM NEAA), and 1% sodium pyruvate, in a humidified incubator at 37°C and 5% CO2. Twenty-four hours prior to transfection, HEK293 cells were seeded into six-well plates (Cellstar® 6-well Cell culture multiwell plate, Greiner Bio-One) with 5 x 105 cells per well in 5 ml cell culture medium, and incubated at 37°C and 5% CO2. Transfection was performed using GenJet (Signagen) according to the manufacturer’s protocol using 2 µg DNA in 100 µl cell culture medium per well without supplements and 6 µl GenJet in 100 µl cell culture medium per well without supplements. Cells were incubated with the transfection reagents for approximately 20 h at 37°C and 5% CO2. Subsequently, transfection reagents were removed from the cells and exchanged for cell culture medium with supplements. The cells were incubated at 37°C and 5% CO2 for 24 h before seeding for testing.
Twenty-four hours prior to performing the assay, cells were seeded into 384-well plates (Cellstar® 384 well black µClear, Greiner Bio-One) with 8000 cells per well in 30 µl cell culture medium with supplements. The assay was performed using HTRF Gs protein dynamic kit reagents (Cisbio). After aspiration of the old medium, the cells were washed with 1x Tyrode buffer supplemented with 2 mM calcium chloride, followed by stimulation for 1 h (37°C and 5% CO2) with relaxin or forskolin in Tyrode buffer supplemented with 2 mM CaCl2 and 0.05% BSA, before cAMP-d2 (10 µl) and anti-cAMP-Cryptate (10 µl) were added to the solution. HTRF ratio (620 nm /665 nm) was measured after 1 h incubation at RT using a BMG PheraStar Plus and plotted against the log ligand concentration.
So far known and postulated from the receptor sequence, RXFP1 consists of a TMD typical of GPCRs, comprising seven helices and six helix-interconnecting loops (Figure 1). However, the large extracellular part (Supplementary Figure S1, Figure 2) is atypical for class A GPCRs but is observed similarly in all evolutionary related LGR subtypes (Hsu et al., 2000). The extracellular part comprises an LDLa domain, a linker region between the LDLa and the LRRD, which is connected via a hinge region to TM1. The peptidic ligand relaxin is likely bound at two binding sites, the LRRD and the LDLa linker. This receptor part supposedly directly interacts with the ELs (Kong et al., 2013; Diepenhorst et al., 2014; Sethi et al., 2016b) in an as yet undeciphered mode of interaction. Yet, the N-terminal LDLa domain is mandatory for receptor signal regulation (Scott et al., 2006; Hopkins et al., 2007). The LDLa fold is stabilized by three cysteine bridges and an additional calcium ion binding site.
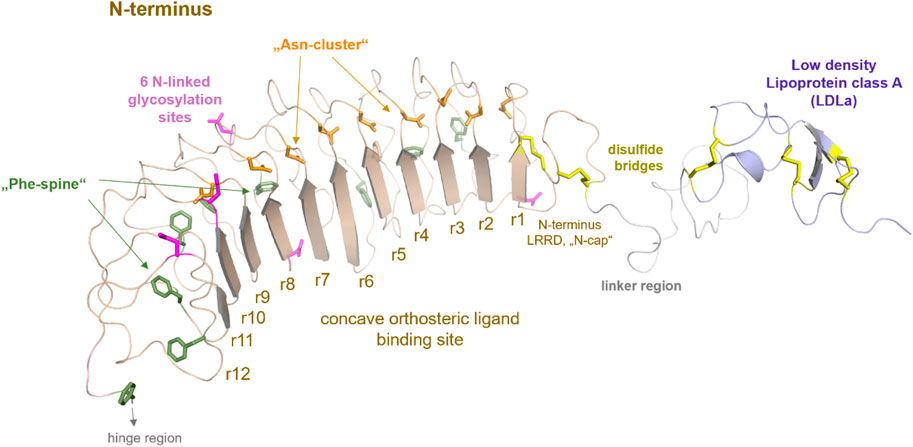
FIGURE 2. Putative structural details of extracellular RXFP1 modules. The extracellular RXFP1 N-terminus consists of four modules according to structural templates of homologous receptors and sequence properties: (i) The LDLa domain involved in signaling regulation (also complexed with a calcium ion (Hopkins et al., 2007)). (ii) The linker region between the LDLa and the LRRD. (iii) The LRRD with 12 repeating elements is each constituted by approximately 20−25 amino acids. The LRRD fold is stabilized by: 1. Two N-terminal cysteine bridges (N-cap); 2. A “Phe-spine” constituted by phenylalanines in the backbone of almost every repeat; 3. By an asparagine-enriched cluster (named Asn-cluster”) after the β-strand of each repeat, finally forming the concave β-sheet ligand binding site (Büllesbach and Schwabe, 2005). In the extracellular part, six potential glycosylation sites (Yan et al., 2008) are allocated (magenta sticks). (iv) The hinge region is short (∼30 amino acids) compared to other LGRs, such as glycoprotein hormone receptors (GPHR (Kleinau and Krause, 2009)), and contains one disulfide bridge.
The linker region between the LDLa and the LRRD is somewhat unfolded, though the length as a spacer is important for correct receptor function (Nordström et al., 2009). Although this linker has no specific folding in the ligand-unbound state, certain residues are assumed to contribute to ligand binding and interaction of the ECD with the extracellular loops (Sethi et al., 2016b).
Two cysteine bridges stabilize the N-terminal fold of the LRRD, as known from the FSHR or TSHR (Kleinau et al., 2013). Of note, the LRRD shows specific structural properties involved in stabilizing the domain fold (Figure 2) (Kajava, 1998; Kobe and Kajava, 2001; Enkhbayar et al., 2004; Bella et al., 2008).
A structural fold for the short hinge region connecting the LRRD to the TMD cannot yet be predicted. Nonetheless, several LGRs share one intrinsic activation mechanism, described as the release of a “tethered ligand” (Kleinau et al., 2013; Schöneberg et al., 2016; Schulze et al., 2020). For GPHRs, several studies have demonstrated that parts of the extracellular region act as an intramolecular “tethered” ligand that is activated by hormone binding (Zhang et al., 2000; Vlaeminck-Guillem et al., 2002; Kleinau et al., 2004; Kleinau et al., 2013; Brüser et al., 2016). Such an intrinsic mechanism is also proposed for the class C LGRs, whereby the tethered ligand is located in the LDLa domain (Sudo et al., 2003; Scott et al., 2006; Hopkins et al., 2007) and is likely silent in the ligand-unoccupied state. Therefore, the correct (functional) spatial distance of the LDLa domain in RXFP1 to the potentially interacting ELs (Hopkins et al., 2007; Diepenhorst et al., 2014) may be strikingly dependent on the length of the LDLa-LRRD linker region (Sethi et al., 2016b). It is noteworthy that for class A and C LGRs, involvement of the EL2 in the signal transduction process has been suggested (Ryu et al., 1998; Nishi et al., 2002; Sudo et al., 2003; Kleinau et al., 2007; Kleinau et al., 2008; Dupakuntla et al., 2012; Sethi et al., 2016b).
One of our goals was to identify functionally active RXFP1 variants that can be isolated in sufficient quantity for further in vitro studies. For small-scale expression tests in Spodoptera frugiperda (Sf9) cells, the human RXFP1 sequence was cloned into the pVL1393 vector. Based on the structural-functional receptor model described above, N- and C-terminal receptor truncations were combined in several receptor variants. The constructs were designed under the following aspects: 1) deletion of the N-terminal LDLa domain (Met82 in the linker region), 2) deletion of the entire extracellular part (Pro391 as start residue), and 3) deletion of the receptor C-terminus (deletion at Ser707) (Figure 3 and Figure 4). For signaling experiments, human RXFP1 variants were cloned into the pCDNA6.2 expression vector.
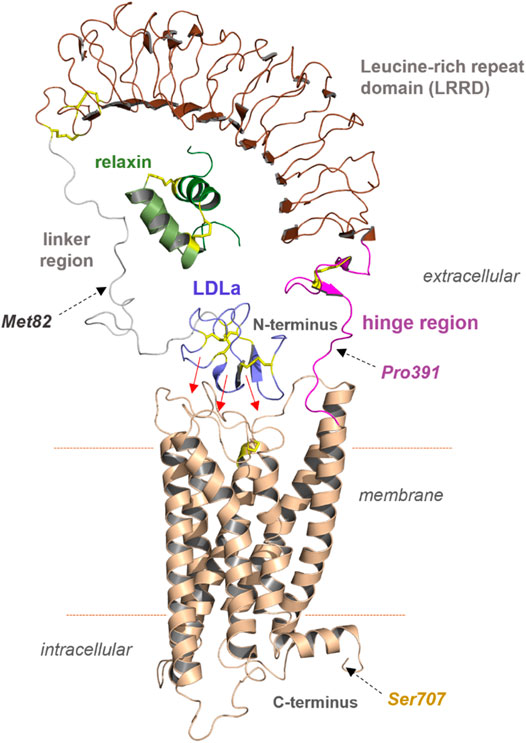
FIGURE 3. Predicted structural features of RXFP1. The assembled receptor parts and docked ligand relaxin visualizes principal features of the receptor and complex; however, it remains unknown how the diverse domains are arranged spatially to each other. Of note, the N-terminal LDLa domain is assumed to interact directly with extracellular loops or even parts of the TMD (indicated by red arrows). The endogenous ligand relaxin binds to both the LRRD and also potentially to the linker region simultaneously. Met82, Pro391, and Ser707 were used as breakpoints for the construction of diverse receptor variants.
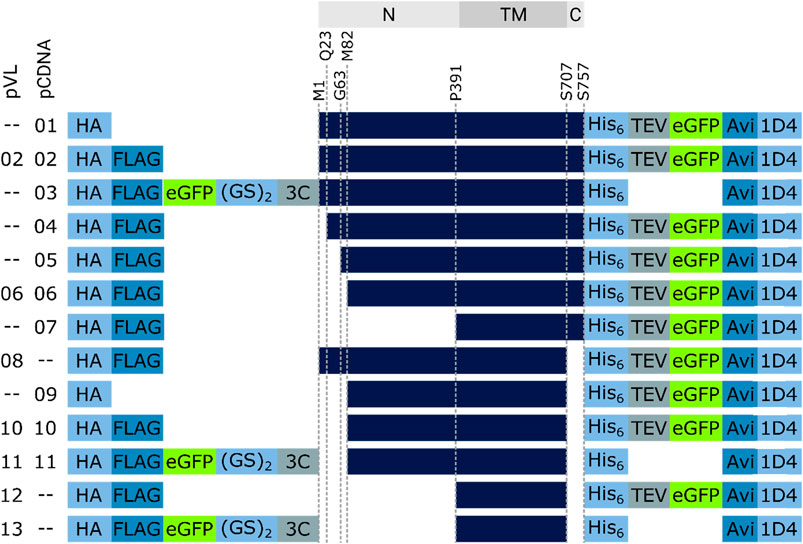
FIGURE 4. RXFP1 construct design. To assess the receptor region in which changes were made, the location is referenced above the constructs (gray bars, N = extracellular N-terminus, TM = transmembrane region, C = intracellular C-terminus). Several constructs do not differ in terms of GPCR sequence but in the position and number of tags used (HA = human influenza hemagglutinin signal sequence, GS = linker sequence, 3C & TEV = protease cleavage sequences, Avi = biotin-acceptor peptide, 1D4 = rhodopsin derived epitope tag).
To estimate the amount of soluble receptor, 5 g of cell paste was purified using the FLAG-tag present in all constructs. To assess the impact of the changes on protein yield, all RXFP1variants were compared with the wild-type RXFP1 (WT- RXFP1) (pVL-02) construct (67 µg/L, Figure 5). The C-terminal truncation of residues 708 to 757 and the N-terminal truncation of amino acids 1 to 82 (and, therefore, the deletion of the LDLa domain) had no drastic impact on final protein yield (numbers pVL-06 & pVL-08). Only the combination of N- (with and without LRRD) and C-terminal deletions resulted in a receptor yield of over 100 µg/L (numbers pVL-10/11 & pVL-12/13). The direct comparison with respect to the eGFP position showed a significantly better yield when located at the N-terminus. In summary, both the shortening of the receptor on both ends and the N-terminal fusion to an eGFP tripled the final protein yield compared to the WT construct (numbers pVL-02 vs. pVL-13).
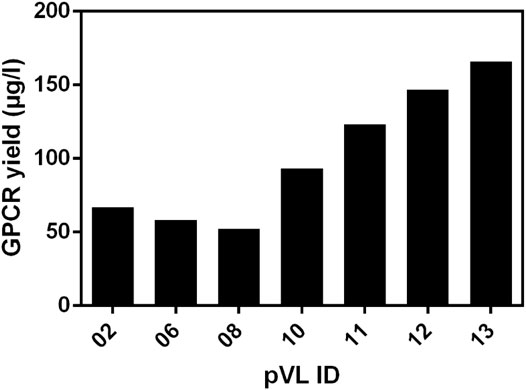
FIGURE 5. Comparison of the soluble receptor yield. Different RXFP1 variants (truncations and tag positions) were expressed in mid-scale and subsequently purified using the FLAG-tag for ANTI-FLAG® M2 affinity chromatography. The final yield was determined by nanodrop microvolume UV-Vis spectrophotometer measurement and normalized to µg/L expression volume.
Following the first purification step, clear effects of the different truncations on the yield of soluble RXFP1 were observed. Analytic F-SEC experiments of the elution fractions from the ANTI-FLAG® M2 affinity purification revealed strong differences in the overall yield and the distribution between large soluble aggregates and monodisperse protein for different constructs (Figure 6A).
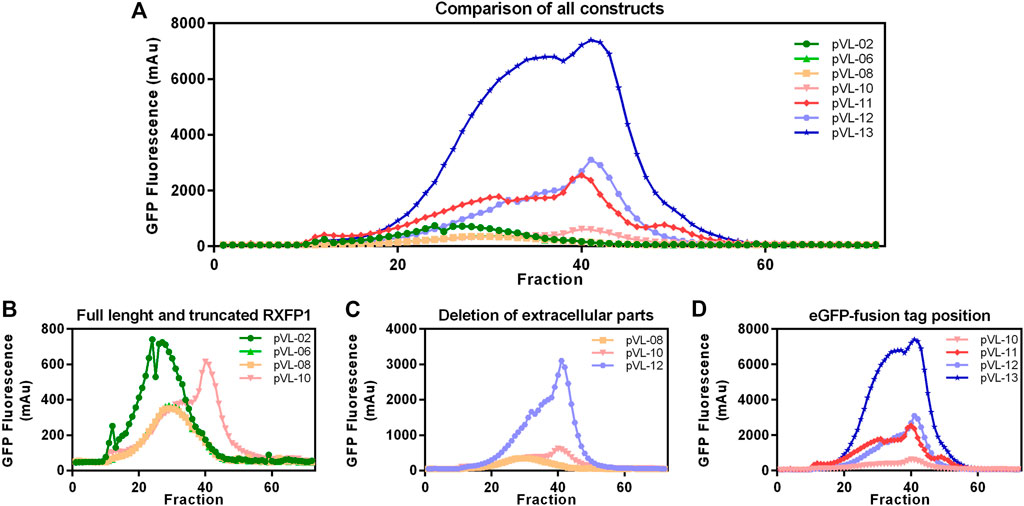
FIGURE 6. Construct characterization using F-SEC chromatography. Mid-scale ANTI-FLAG® M2 affinity purified RXFP1 variants were analyzed based on their eGFP fluorescence signal. (A) Overview of all chromatograms. (B) Comparison between WT-RXFP1and N- and C-terminal truncations. (C) Impact of the deletion of extracellular parts. (D) Impact of the eGFP-fusion tag position.
By comparing constructs with different N- and C- terminal truncations most constructs were shown to elute as soluble aggregates (pVL-02, pVL-06, pVL-08, Figure 6B). Only the combination of deleting the first 89 amino acids and the last 50 amino acids yielded monodisperse protein (pVL-10).
Since the removal of the LDLa domain had a positive effect on the monomeric GPCR RXFP1 yield, the LRRD was also removed in the next step (Figure 6C). Shortening the construct to the transmembrane region resulted in a significant increase in overall GFP fluorescence (600 mAu for pVL-10 vs. 3000 mAu for pVL-12) and a more prominent monomer protein peak. To investigate the effects of the eGFP position on the properties of the purified RXFP1 variant, the eGFP-fusion tag of constructs pVL-10 and pVL-12 was relocated to the N-terminus (pVL-11 and pVL-13). Direct comparison of the two truncated constructs shows an increase of the monomeric peak (fraction 40) when the eGFP-fusion tag was shifted from the C-terminus to the N-terminus (Figure 6D). Here, the best RXFP1 construct yield was obtained with the pVL-13 construct (FLA-eGFP-P391-S707-Avi) with a peak maximum of over 6000 mAu. If the focus had been placed only on the final protein yield, the construct pVL-13 with its high protein yield would have been a suitable candidate for all further experiments. However, since the ligand binding properties of RXFP1 should remain unaltered (e.g., for subsequent small-molecule screening), pVL-11 (FLA-eGFP-P391-S707-Avi) was used for further experimental work.
The cell pellet from a 2L Sf9 expression culture was thawed in solubilization buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, 2 mM CaCl2, 0.5 mM EDTA, cOmplete™ protease inhibitor cocktail tablet (Roche Applied Science), 0.1 mM TCEP, and 2 mM biotin) without detergent. DDM/CHS was added to a final concentration of 2%, and cells were further disrupted by a douncing procedure. Solubilization was performed at 4°C for 1 h with gentle agitation. To separate the cell debris from the solubilized fraction, centrifugation was performed at 25 000 rcf for 45 min, and the supernatant was bound to 3 ml ANTI-FLAG® M2 agarose resin for 2 h (at 4°C with overhead rotation). After separating the solubilizate from the ANTI-FLAG® M2 agarose beads, with a rinsing step comprising 13 column volumes (CV) wash buffer (HEPES pH 7.5, 300 mM NaCl, 10% glycerol, 2 mM CaCl2, 0.1% DDM, 0.01% CHS, 0.1 mM TCEP) was performed. The receptor was subsequently eluted (50 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% DDM, 0.01% CHS, 0.1 mM TCEP, 200 µg/ml Flag peptide, and 5 mM EDTA) and separated into multiple fractions, with total volume of 6 ml. The eGFP fluorescence of all fractions was measured and combined accordingly. Following concentration using 100 kDa cut-off concentrator, the sample was finally polished using a Superose 6 10/30 increase column. Size-exclusion chromatogram revealed two main peaks in the region of interest between 20 and 35 ml (Figure 7A). Both peaks contained RXFP1 (as shown in the SDS-PAGE Figure 7B); however, only the second peak includes the monomeric receptor. The protein behaves smaller in the SDS-PAGE than the actual size. MS analytics (in gel digest) proved the integrity of the protein.
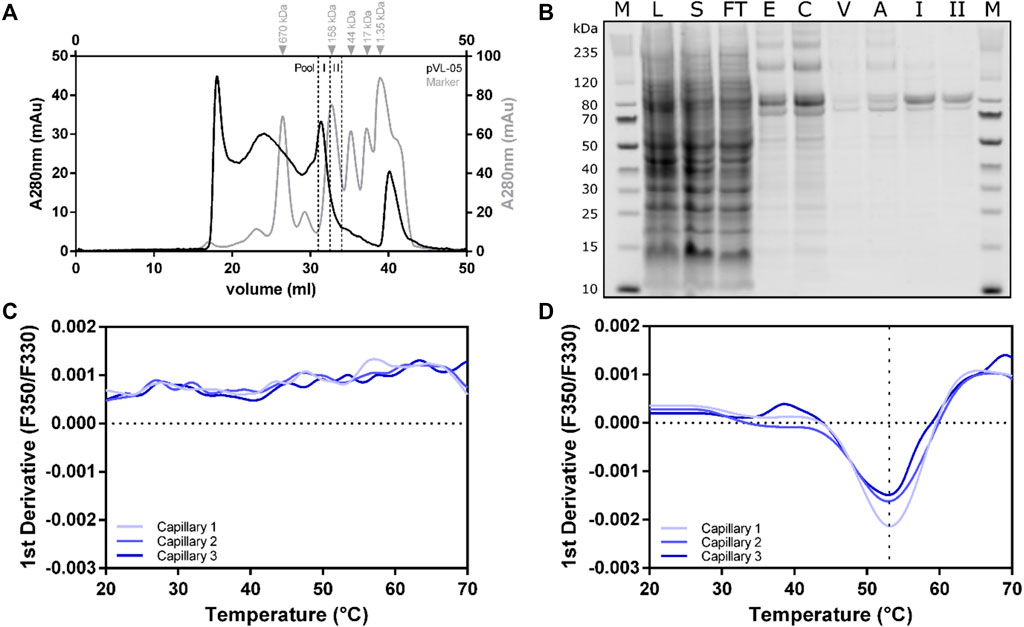
FIGURE 7. Large-scale purification of RXFP1 constructs. (A) SEC chromatogram of pVL-11 2L large-scale purification. (B) SDS-PAGE shows the course of pVL-11 purification, the expected size of the cut construct is 76 kDa (M = size standard, L = lysate, S = solubilizate, TF = flow through, E = M2 eluate, C= concentrated M2 eluate as SEC input, V = void volume peak, A = aggregate peak fraction, I = pool fraction I monomeric peak RXFP1, II = pool fraction II monomeric peak RXFP1). (C) nanoDSF melting curve of pVL-11 aggregate peak fraction. (D) nanoDSF melting curve of monomeric GPCR from the pool fraction I.
Thermal stability of the different F-SEC peak fraction pools (I and II) were analyzed with nano-differential scanning fluorimetry (nanoDSF) (Figures 7C,D). While the sample from the aggregated peak fraction had no melting curve, a melting temperature of approximately 53°C could be determined for the monomeric fraction I peak, which indicates a proper folded protein. Based on these results, the monomeric peak fraction I was used for subsequent ligand binding experiments using SPR.
Purified RXFP1 constructs pVL-11(FLAG-eGFP-M82-S707-Avi) and pVL-13 (FLAG-eGFP-P391-S7079-Avi) (purified using the same procedure) were used for SPR experiments. As both constructs carry a biotinylated Avi-tag at the C-terminus, they could be captured via the biotin on streptavidin sensor chip. The interaction with ligand relaxin was measured to prove that the purified protein is pharmacologically active, and the truncations did not lead to loss of relaxin binding capability. These SPR experiments clearly showed a concentration-dependent binding of relaxin to the purified pVL-11 construct with nanomolar affinity (Kd = 20 nM ±1,5) (Figure 8A), which is in accordance with previous reports (Chan et al., 2012; Halls et al., 2005). The RXFP1 construct without extracellular domain shows no dose-dependent ligand-response, confirming the importance of this domain for relaxin binding (Figure 8B), which is also in agreement with previous studies (Wu et al., 2016).
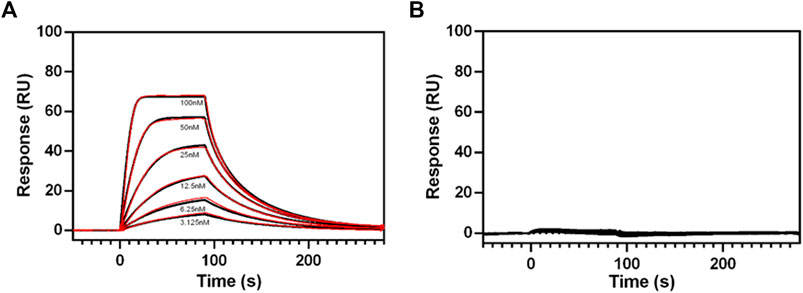
FIGURE 8. SPR sensograms of relaxin binding to different RXFP1 constructs. (A) Dose-dependent binding curves of relaxin (by adding with 3.125, 6.25, 12.5, 25, 50, and 100 nM relaxin concentration to the receptor fixed on the SPR chip) for RXFP1 pVL-11(FLAG-eGFP-M82-S707- AVI), and (B) for RXFP1 pVL-13(FLAG-eGFP-P391-S707-AVI). The red lines represent kinetic fits. Experiments were performed in triplicates.
To investigate the suitability of the different constructs in cellular assays, the cAMP signal was measured after stimulation with 2 nM relaxin (Figure 9). The activity of WT-RXFP1 (amino acids 1−757) without any tags was used as a reference for all other receptor variants.
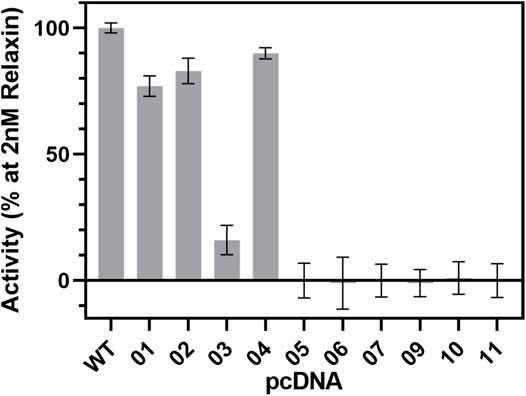
FIGURE 9. cAMP signaling of RXFP1 variants. All RXFP1 pCDNA constructs used for cAMP signaling assays are numbered according to Figure 4. Activity of all RXFP1 variants (WT and constructs 1−11) normalized to the cAMP signal of fully activated WT-RXFP1 using 2 nM relaxin and performed in quadruplicates.
N-terminal HA- or FLAG-tags combined with C-terminal His-eGFP-Avi-1D4 tags changed the functional activity of the construct slightly compared to WT-RXFP1, as shown by their ability to increase intracellular cAMP levels using 2 nM relaxin as an RXFP1 agonist. Removal of the first 23 amino acids did not dramatically affect cAMP accumulation. However, when the eGFP-fusion tag was shifted from the C-terminus to the N-terminus, the cAMP signal decreased to approximately 20% compared with the WT-RXFP1. Further shortening the N-terminus beyond the first 23 amino acids (with a resulting removal of the LDLa and LRR domains) resulted in a complete loss of signaling.
We here describe the design of various RXFP1 constructs with the aim of generating receptor variants suitable for functional and biophysical studies or structure determination. Several RXFP1 variants with different lengths and (technically required) tags were cloned and screened for sufficient protein expression. Most promising candidates were expressed in large-scale approaches accompanied by ligand binding studies and signaling assays. The expression of GPCRs is generally a difficult task, especially if they exert large extracellular domains in addition to the transmembrane helices. Indeed, the typical approach of shortening the N- and C-terminal regions of a GPCR resulted in increased expression in the case of RXFP1 as well as with enrichment in monomeric receptor fractions. Moving the eGFP-fusion tag to the N-terminus finally led to a sufficient yield for SPR analysis.
Moreover, we found that removing the LDLa domain leads to enhanced monomeric receptor expression without losing the ability to bind its natural ligand relaxin, even if the affinity decreases compared to the WT-RXFP1, which is consistent with other studies (Sethi et al., 2016b). However, this construct cannot be used for signaling experiments because cAMP accumulation no longer occurs after relaxin addition. Further truncation toward the transmembrane region of the receptor resulted in the highest possible receptor expression; however, neither relaxin binding nor a cAMP response to ligand stimulation was observable (Table 1).
RXFP1 variants pVL-11 (FLAG-eGFP-M82-S707-Avi) and pVL-13 (FLAG-eGFP-P391-S7079-Avi) are particularly suitable for SPR ligand screening and subsequent structural elucidation. In the case of pVL-11, ligands could be potentially found that, like relaxin, require the LRRD for high-affinity binding. In the case of construct pVL-13, it should be possible to identify compounds that bind in the transmembrane region. For the cAMP assay, the construct would then have to be extended by the LDLa module.
Despite these described efforts in our study, some limitations and future needs should be addressed. First, RXFP1 is known to be a receptor that can activate several signaling pathways besides cAMP (Gs), such as ERK1/2, p38MAPK, Gi3, or cGMP (Halls et al., 2007; Bathgate et al., 2013; Kocan et al., 2017). These signaling pathways are of utmost importance to capture the full pharmacological and physiological spectrum of this receptor. Therefore, in addition to the cAMP accumulation studied here, it will be interesting to test signaling-competent receptor variants for their ability to activate the extended set of signaling pathways. This is also particularly interesting because the cell systems used here (Sf9 insect cells for expression, HEK293 cells for functional characterization) differ from cell types of organs (such as ovary, testis, cervix, breast, endometrial epithelial cells) that naturally express this receptor (Kong et al., 2010) in terms of the amount of G-proteins or other intracellular factors and effectors. In addition, there are different glycosylation patterns for proteins in insect cells (e.g. (Marada et al., 2015), which is also relevant for RXFP1 with six putative glycosylation sites. These sites have effects on receptor expression and cAMP accumulation, but not on ligand binding (Yan et al., 2008). This, in turn, suggests that experimental approaches with “artificial” cell systems overexpressing the receptor may vary in terms of the nature of glycosylation or intracellular response to stimulation. These circumstances must generally be taken into account for in vitro data obtained, which also applies to our study. However, the cell systems used here are currently common experimental tools (e.g. (Shukla et al., 2006)) for the subsequent structural and biophysical methods, which at least allow comparability between different experimental studies. Finally, it should be mentioned that RXFP1 is capable of binding three different relaxin proteins (Patil et al., 2017) and that our relaxin H2-binding competent constructs designed here should be tested for the binding of other ligand-peptides to assess the binding specificities in detail.
Anyhow, our findings could pave the way for further structural elucidation by protein X-ray crystallography or cryo-electron microscopy methods of this unique GPCR, which shows diverse, interesting structural features merely by analyses of the sequence (Supplementary Figure S1) and obtained homology models. These specificities are related to the module-like architecture of the RXFP1, the interplay between LDLa and TMD, or specific amino acids in the transmembrane domain (Figure 10).
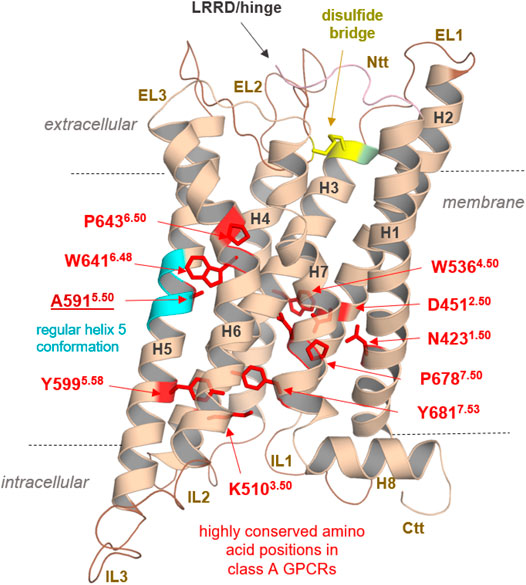
FIGURE 10. Specificities of RXFP1 and common class A GPCR properties. Typical highly conserved class A GPCR amino acids are highlighted as red sticks in the TMD model of the RXFP1. They are responsible for specific receptor functions as structural fold, expression, and signal transduction. In addition, they determine structural features in the helices (as the prolines, e.g., P6436.50), or they participate as intramolecular switches (as W6416.48) regulating the transition between active and inactive state conformations (D4512.50). Of note, the TMH5 conformation is likely regular compared to most other class A GPCRs with a proline-induced kink (cyan helix region) because RXFP1 has an alanine at this position. Superscripted numbers according to the unifying Ballesteros & Weinstein numbering for class A GPCRs (Ballesteros and Weinstein, 1995). An additional structural factor of importance shared between the LGRs is homo- or hetero-oligomerization, a widely recognized property of many GPCRs (e.g., (Smith and Milligan, 2010; Tena-Campos et al., 2014)), including receptors of relevance in endocrinology (Kleinau et al., 2016). Cooperative effects between receptor monomers arranged as dimers or oligomers (homomers) are long known to be obligatory and impact LGR groups A and C ligand binding properties (Chazenbalk et al., 1996; Urizar et al., 2005; Svendsen et al., 2009; Zoenen et al., 2012). Furthermore, interactions with non-LGR receptors (heteromerization), such as angiotensin receptors (AT1R and AT2R), have been identified in vivo, specifically for RXFP1 (Chow et al., 2014; Chow et al., 2019). This fact drastically widens the spectrum of the potential functional and physiological importance of this receptor. Finally, the RXFP1 is a highly interesting and pharmacologically relevant class A GPCR with unique structural and functional features, which needs further advanced experimental elucidation.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
DS conducted and evaluated functional studies of RXFP1; AE, DS, and MS performed cell culture, expression, and purification of RXFP1; VP, MS, GK, and PS designed the RXFP1 constructs; MM conducted and evaluated the cAMP signaling assays; GK designed and prepared the protein models and model calculations of RXFP1; AEi performed the SPR assays; VP supervised the SPR assay; DS and GK wrote the first manuscript draft and prepared the figures; DS, GK, MM, MS, PS, and VP analyzed the data and wrote the final manuscript; GK, MS, PS, and VP supervised the research; PS and VP designed and funded the project.
This work was supported by a Bayer AG research cooperation with the Charité—Universitätsmedizin Berlin (to PS and VP) and additional funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through CRC 1423, project number 421152132, subprojects A01/A05/Z03 (to PS), through CRC 1365, project number 394046635, subproject A03 (to PS); through CRC 1078, subproject B06 (to PS); through Germany’s Excellence Strategies—EXC2008/1 (UniSysCat)—390540038 (to PS). PS acknowledge the Einstein Center of Catalysis (EC2).
AE, AEi, MM, and VP were employees of Bayer AG and may have additional stock options. VP was also employed by NUVISAN ICB GmbH. The authors declare that this study received funding from Bayer AG. The funder had the following involvement in the study: study design, and collection, analysis, interpretation of data.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Sabine Dämmig, Tobias Ewering, Anja Koch, and Melina Kehl for expert technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.826112/full#supplementary-material
Ballesteros, J. A., and Weinstein, H. (1995). [19] Integrated Methods for the Construction of Three-Dimensional Models and Computational Probing of Structure-Function Relations in G Protein-Coupled Receptors. Methods Neurosci. 25, 366–428. doi:10.1016/s1043-9471(05)80049-7
Bathgate, R. A., Halls, M. L., van der Westhuizen, E. T., Callander, G. E., Kocan, M., and Summers, R. J. (2013). Relaxin Family Peptides and Their Receptors. Physiol. Rev. 93, 405–480. doi:10.1152/physrev.00001.2012
Bathgate, R. A., Ivell, R., Sanborn, B. M., Sherwood, O. D., and Summers, R. J. (2006). International Union of Pharmacology LVII: Recommendations for the Nomenclature of Receptors for Relaxin Family Peptides. Pharmacol. Rev. 58, 7–31. doi:10.1124/pr.58.1.9
Bella, J., Hindle, K. L., McEwan, P. A., and Lovell, S. C. (2008). The Leucine-Rich Repeat Structure. Cell Mol Life Sci 65, 2307–2333. doi:10.1007/s00018-008-8019-0
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The Protein Data Bank. Nucleic Acids Res. 28, 235–242. doi:10.1093/nar/28.1.235
Braun, T., Schofield, P. R., and Sprengel, R. (1991). Amino-terminal Leucine-Rich Repeats in Gonadotropin Receptors Determine Hormone Selectivity. EMBO J. 10, 1885–1890. doi:10.1002/j.1460-2075.1991.tb07714.x
Brüser, A., Schulz, A., Rothemund, S., Ricken, A., Calebiro, D., Kleinau, G., et al. (2016). The Activation Mechanism of Glycoprotein Hormone Receptors with Implications in the Cause and Therapy of Endocrine Diseases. J. Biol. Chem. 291, 508–520. doi:10.1074/jbc.M115.701102
Büllesbach, E. E., and Schwabe, C. (2005). The Trap-like Relaxin-Binding Site of the Leucine-Rich G-Protein-Coupled Receptor 7. J. Biol. Chem. 280, 14051–14056. doi:10.1074/jbc.M500030200
Bylander, J. E., Frieden, E. H., and Adams, W. C. (1987). Effects of Porcine Relaxins upon Uterine Hypertrophy and Protein Metabolism in Mice. Proc. Soc. Exp. Biol. Med. 185, 76–80. doi:10.3181/00379727-185-42519
Chan, L. J., Rosengren, K. J., Layfield, S. L., Bathgate, R. A., Separovic, F., Samuel, C. S., et al. (2012). Identification of Key Residues Essential for the Structural Fold and Receptor Selectivity within the A-Chain of Human Gene-2 (H2) Relaxin. J. Biol. Chem. 287, 41152–41164. doi:10.1074/jbc.M112.409284
Chazenbalk, G. D., Kakinuma, A., Jaume, J. C., McLachlan, S. M., and Rapoport, B. (1996). Evidence for Negative Cooperativity Among Human Thyrotropin Receptors Overexpressed in Mammalian Cells. Endocrinology 137, 4586–4591. doi:10.1210/endo.137.11.8895321
Chen, T. Y., Li, X., Hung, C. H., Bahudhanapati, H., Tan, J., Kass, D. J., et al. (2020). The Relaxin Family Peptide Receptor 1 (RXFP1): An Emerging Player in Human Health and Disease. Mol. Genet. Genomic Med. 8, e1194. doi:10.1002/mgg3.1194
Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., et al. (2007). High-resolution crystal Structure of an Engineered Human Beta2-Adrenergic G Protein-Coupled Receptor. Science 318, 1258–1265. doi:10.1126/science.1150577
Chow, B. S., Kocan, M., Bosnyak, S., Sarwar, M., Wigg, B., Jones, E. S., et al. (2014). Relaxin Requires the Angiotensin II Type 2 Receptor to Abrogate Renal Interstitial Fibrosis. Kidney Int. 86, 75–85. doi:10.1038/ki.2013.518
Chow, B. S. M., Kocan, M., Shen, M., Wang, Y., Han, L., Chew, J. Y., et al. (2019). AT1R-AT2R-RXFP1 Functional Crosstalk in Myofibroblasts: Impact on the Therapeutic Targeting of Renal and Cardiac Fibrosis. J. Am. Soc. Nephrol. 30, 2191–2207. doi:10.1681/ASN.2019060597
Diepenhorst, N. A., Petrie, E. J., Chen, C. Z., Wang, A., Hossain, M. A., Bathgate, R. A., et al. (2014). Investigation of Interactions at the Extracellular Loops of the Relaxin Family Peptide Receptor 1 (RXFP1). J. Biol. Chem. 289, 34938–34952. doi:10.1074/jbc.M114.600882
Dupakuntla, M., Pathak, B., Roy, B. S., and Mahale, S. D. (2012). Extracellular Loop 2 in the FSH Receptor Is Crucial for Ligand Mediated Receptor Activation. Mol. Cel Endocrinol 362, 60–68. doi:10.1016/j.mce.2012.05.008
Enkhbayar, P., Kamiya, M., Osaki, M., Matsumoto, T., and Matsushima, N. (2004). Structural Principles of Leucine-Rich Repeat (LRR) Proteins. Proteins 54, 394–403. doi:10.1002/prot.10605
Errey, J. C., and Fiez-Vandal, C. (2020). Production of Membrane Proteins in Industry: The Example of GPCRs. Protein Expr. Purif. 169, 105569. doi:10.1016/j.pep.2020.105569
Fields, P. A., and Larkin, L. H. (1981). Purification and Immunohistochemical Localization of Relaxin in the Human Term Placenta. J. Clin. Endocrinol. Metab. 52, 79–85. doi:10.1210/jcem-52-1-79
Garland, S. L. (2013). Are GPCRs Still a Source of New Targets? J. Biomol. Screen. 18, 947–966. doi:10.1177/1087057113498418
Halls, M. L., Bathgate, R. A., and Summers, R. J. (2006). Relaxin Family Peptide Receptors RXFP1 and RXFP2 Modulate cAMP Signaling by Distinct Mechanisms. Mol. Pharmacol. 70, 214–226. doi:10.1124/mol.105.021691
Halls, M. L., Bond, C. P., Sudo, S., Kumagai, J., Ferraro, T., Layfield, S., et al. (2005). Multiple Binding Sites Revealed by Interaction of Relaxin Family Peptides with Native and Chimeric Relaxin Family Peptide Receptors 1 and 2 (LGR7 and LGR8). J. Pharmacol. Exp. Ther. 313, 677–687. doi:10.1124/jpet.104.080655
Halls, M. L., Hewitson, T. D., Moore, X. L., Du, X. J., Bathgate, R. A., and Summers, R. J. (2009). Relaxin Activates Multiple cAMP Signaling Pathway Profiles in Different Target Cells. Ann. N. Y Acad. Sci. 1160, 108–111. doi:10.1111/j.1749-6632.2008.03814.x
Halls, M. L., van der Westhuizen, E. T., Bathgate, R. A., and Summers, R. J. (2007). Relaxin Family Peptide Receptors-Fformer Orphans Reunite with Their Parent Ligands to Activate Multiple Signalling Pathways. Br. J. Pharmacol. 150, 677–691. doi:10.1038/sj.bjp.0707140
Halls, M. L., van der Westhuizen, E. T., Wade, J. D., Evans, B. A., Bathgate, R. A., and Summers, R. J. (2009). Relaxin Family Peptide Receptor (RXFP1) Coupling to G(alpha)i3 Involves the C-Terminal Arg752 and Localization within Membrane Raft Microdomains. Mol. Pharmacol. 75, 415–428. doi:10.1124/mol.108.051227
Heyder, N. A., Kleinau, G., Speck, D., Schmidt, A., Paisdzior, S., Szczepek, M., et al. (2021). Structures of Active Melanocortin-4 Receptor-Gs-Protein Complexes with NDP-α-MSH and Setmelanotide. Cell Res 31, 1176–1189. doi:10.1038/s41422-021-00569-8
Hopkins, E. J., Layfield, S., Ferraro, T., Bathgate, R. A., and Gooley, P. R. (2007). The NMR Solution Structure of the Relaxin (RXFP1) Receptor Lipoprotein Receptor Class A Module and Identification of Key Residues in the N-Terminal Region of the Module that Mediate Receptor Activation. J. Biol. Chem. 282, 4172–4184. doi:10.1074/jbc.M609526200
Hossain, M. A., Kocan, M., Yao, S. T., Royce, S. G., Nair, V. B., Siwek, C., et al. (2016). A Single-Chain Derivative of the Relaxin Hormone Is a Functionally Selective Agonist of the G Protein-Coupled Receptor, RXFP1. Chem. Sci. 7, 3805–3819. doi:10.1039/c5sc04754d
Hossain, M. A., Rosengren, K. J., Samuel, C. S., Shabanpoor, F., Chan, L. J., Bathgate, R. A., et al. (2011). The Minimal Active Structure of Human Relaxin-2. J. Biol. Chem. 286, 37555–37565. doi:10.1074/jbc.M111.282194
Hossain, M. A., Samuel, C. S., Binder, C., Hewitson, T. D., Tregear, G. W., Wade, J. D., et al. (2010). The Chemically Synthesized Human Relaxin-2 Analog, B-R13/17k H2, Is an RXFP1 Antagonist. Amino Acids 39, 409–416. doi:10.1007/s00726-009-0454-1
Hsu, S. Y., Kudo, M., Chen, T., Nakabayashi, K., Bhalla, A., van der Spek, P. J., et al. (2000). The Three Subfamilies of Leucine-Rich Repeat-Containing G Protein-Coupled Receptors (LGR): Identification of LGR6 and LGR7 and the Signaling Mechanism for LGR7. Mol. Endocrinol. 14, 1257–1271. doi:10.1210/mend.14.8.0510
Hsu, S. Y., Liang, S. G., and Hsueh, A. J. (1998). Characterization of Two LGR Genes Homologous to Gonadotropin and Thyrotropin Receptors with Extracellular Leucine-Rich Repeats and a G Protein-Coupled, Seven-Transmembrane Region. Mol. Endocrinol. 12, 1830–1845. doi:10.1210/mend.12.12.0211
Hsu, S. Y., Nakabayashi, K., Nishi, S., Kumagai, J., Kudo, M., Sherwood, O. D., et al. (2002). Activation of Orphan Receptors by the Hormone Relaxin. Science 295, 671–674. doi:10.1126/science.1065654
Hudson, P., John, M., Crawford, R., Haralambidis, J., Scanlon, D., Gorman, J., et al. (1984). Relaxin Gene Expression in Human Ovaries and the Predicted Structure of a Human Preprorelaxin by Analysis of cDNA Clones. EMBO J. 3, 2333–2339. doi:10.1002/j.1460-2075.1984.tb02135.x
Jiang, X., Liu, H., Chen, X., Chen, P. H., Fischer, D., Sriraman, V., et al. (2012). Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor. Proc. Natl. Acad. Sci. U S A. 109, 12491–12496. doi:10.1073/pnas.1206643109
Kajava, A. V. (1998). Structural Diversity of Leucine-Rich Repeat Proteins. J. Mol. Biol. 277, 519–527. doi:10.1006/jmbi.1998.1643
Kamat, A. A., Feng, S., Bogatcheva, N. V., Truong, A., Bishop, C. E., and Agoulnik, A. I. (2004). Genetic Targeting of Relaxin and Insulin-like Factor 3 Receptors in Mice. Endocrinology 145, 4712–4720. doi:10.1210/en.2004-0515
Kleinau, G., Claus, M., Jaeschke, H., Mueller, S., Neumann, S., Paschke, R., et al. (2007). Contacts between Extracellular Loop Two and Transmembrane helix Six Determine Basal Activity of the Thyroid-Stimulating Hormone Receptor. J. Biol. Chem. 282, 518–525. doi:10.1074/jbc.M606176200
Kleinau, G., Jaeschke, H., Mueller, S., Raaka, B. M., Neumann, S., Paschke, R., et al. (2008). Evidence for Cooperative Signal Triggering at the Extracellular Loops of the TSH Receptor. FASEB J. 22, 2798–2808. doi:10.1096/fj.07-104711
Kleinau, G., Jäschke, H., Neumann, S., Lättig, J., Paschke, R., and Krause, G. (2004). Identification of a Novel Epitope in the Thyroid-Stimulating Hormone Receptor Ectodomain Acting as Intramolecular Signaling Interface. J. Biol. Chem. 279, 51590–51600. doi:10.1074/jbc.M404748200
Kleinau, G., and Krause, G. (2009). Thyrotropin and Homologous Glycoprotein Hormone Receptors: Structural and Functional Aspects of Extracellular Signaling Mechanisms. Endocr. Rev. 30, 133–151. doi:10.1210/er.2008-0044
Kleinau, G., Müller, A., and Biebermann, H. (2016). Oligomerization of GPCRs Involved in Endocrine Regulation. J. Mol. Endocrinol. 57, R59–R80. doi:10.1530/JME-16-0049
Kleinau, G., Neumann, S., Grüters, A., Krude, H., and Biebermann, H. (2013). Novel Insights on Thyroid-Stimulating Hormone Receptor Signal Transduction. Endocr. Rev. 34, 691–724. doi:10.1210/er.2012-1072
Kleinau, G., Worth, C. L., Kreuchwig, A., Biebermann, H., Marcinkowski, P., Scheerer, P., et al. (2017). Structural-Functional Features of the Thyrotropin Receptor: A Class A G-Protein-Coupled Receptor at Work. Front. Endocrinol. (Lausanne) 8, 86. doi:10.3389/fendo.2017.00086
Kobe, B., and Kajava, A. V. (2001). The Leucine-Rich Repeat as a Protein Recognition Motif. Curr. Opin. Struct. Biol. 11, 725–732. doi:10.1016/s0959-440x(01)00266-4
Kocan, M., Sarwar, M., Ang, S. Y., Xiao, J., Marugan, J. J., Hossain, M. A., et al. (2017). ML290 Is a Biased Allosteric Agonist at the Relaxin Receptor RXFP1. Sci. Rep. 7, 2968. doi:10.1038/s41598-017-02916-5
Kohsaka, T., Min, G., Lukas, G., Trupin, S., Campbell, E. T., and Sherwood, O. D. (1998). Identification of Specific Relaxin-Binding Cells in the Human Female. Biol. Reprod. 59, 991–999. doi:10.1095/biolreprod59.4.991
Kong, R. C., Petrie, E. J., Mohanty, B., Ling, J., Lee, J. C., Gooley, P. R., et al. (2013). The Relaxin Receptor (RXFP1) Utilizes Hydrophobic Moieties on a Signaling Surface of its N-Terminal Low Density Lipoprotein Class A Module to Mediate Receptor Activation. J. Biol. Chem. 288, 28138–28151. doi:10.1074/jbc.M113.499640
Kong, R. C., Shilling, P. J., Lobb, D. K., Gooley, P. R., and Bathgate, R. A. (2010). Membrane Receptors: Structure and Function of the Relaxin Family Peptide Receptors. Mol. Cel Endocrinol 320, 1–15. doi:10.1016/j.mce.2010.02.003
Krajnc-Franken, M. A., van Disseldorp, A. J., Koenders, J. E., Mosselman, S., van Duin, M., and Gossen, J. A. (2004). Impaired Nipple Development and Parturition in LGR7 Knockout Mice. Mol. Cel Biol 24, 687–696. doi:10.1128/mcb.24.2.687-696.2004
Limbird, L. E. (2004). The Receptor Concept: a Continuing Evolution. Mol. Interv. 4, 326–336. doi:10.1124/mi.4.6.6
Luna, J. J., Riesewijk, A., Horcajadas, J. A., Van Os Rd, Rd., Domínguez, F., Mosselman, S., et al. (2004). Gene Expression Pattern and Immunoreactive Protein Localization of LGR7 Receptor in Human Endometrium throughout the Menstrual Cycle. Mol. Hum. Reprod. 10, 85–90. doi:10.1093/molehr/gah019
Marada, S., Navarro, G., Truong, A., Stewart, D. P., Arensdorf, A. M., Nachtergaele, S., et al. (2015). Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling. Plos Genet. 11, e1005473. doi:10.1371/journal.pgen.1005473
McBride, A., Hoy, A. M., Bamford, M. J., Mossakowska, D. E., Ruediger, M. P., Griggs, J., et al. (2017). In Search of a Small Molecule Agonist of the Relaxin Receptor RXFP1 for the Treatment of Liver Fibrosis. Sci. Rep. 7, 10806. doi:10.1038/s41598-017-10521-9
Nishi, S., Nakabayashi, K., Kobilka, B., and Hsueh, A. J. (2002). The Ectodomain of the Luteinizing Hormone Receptor Interacts with Exoloop 2 to Constrain the Transmembrane Region: Studies Using Chimeric Human and Fly Receptors. J. Biol. Chem. 277, 3958–3964. doi:10.1074/jbc.M109617200
Nordström, K. J., Lagerström, M. C., Wallér, L. M., Fredriksson, R., and Schiöth, H. B. (2009). The Secretin GPCRs Descended from the Family of Adhesion GPCRs. Mol. Biol. Evol. 26, 71–84. doi:10.1093/molbev/msn228
O'Hayre, M., Vázquez-Prado, J., Kufareva, I., Stawiski, E. W., Handel, T. M., Seshagiri, S., et al. (2013). The Emerging Mutational Landscape of G Proteins and G-Protein-Coupled Receptors in Cancer. Nat. Rev. Cancer 13, 412–424. doi:10.1038/nrc3521
Park, J. I., Semyonov, J., Chang, C. L., and Hsu, S. Y. (2005). Conservation of the Heterodimeric Glycoprotein Hormone Subunit Family Proteins and the LGR Signaling System from Nematodes to Humans. Endocrine 26, 267–276. doi:10.1385/ENDO:26:3:267
Patil, N. A., Rosengren, K. J., Separovic, F., Wade, J. D., Bathgate, R. A. D., and Hossain, M. A. (2017). Relaxin Family Peptides: Structure-Activity Relationship Studies. Br. J. Pharmacol. 174, 950–961. doi:10.1111/bph.13684
Petrie, E. J., Lagaida, S., Sethi, A., Bathgate, R. A., and Gooley, P. R. (2015). In a Class of Their Own - RXFP1 and RXFP2 Are Unique Members of the LGR Family. Front. Endocrinol. (Lausanne) 6, 137. doi:10.3389/fendo.2015.00137
Rasmussen, S. G., DeVree, B. T., Zou, Y., Kruse, A. C., Chung, K. Y., Kobilka, T. S., et al. (2011). Crystal Structure of the β2 Adrenergic Receptor-Gs Protein Complex. Nature 477, 549–555. doi:10.1038/nature10361
Rosengren, K. J., Lin, F., Bathgate, R. A., Tregear, G. W., Daly, N. L., Wade, J. D., et al. (2006). Solution Structure and Novel Insights into the Determinants of the Receptor Specificity of Human Relaxin-3. J. Biol. Chem. 281, 5845–5851. doi:10.1074/jbc.M511210200
Ryu, K., Lee, H., Kim, S., Beauchamp, J., Tung, C. S., Isaacs, N. W., et al. (1998). Modulation of High Affinity Hormone Binding. Human Choriogonadotropin Binding to the Exodomain of the Receptor Is Influenced by Exoloop 2 of the Receptor. J. Biol. Chem. 273, 6285–6291. doi:10.1074/jbc.273.11.6285
Samuel, C. S., Royce, S. G., Hewitson, T. D., Denton, K. M., Cooney, T. E., and Bennett, R. G. (2017). Anti-fibrotic Actions of Relaxin. Br. J. Pharmacol. 174, 962–976. doi:10.1111/bph.13529
Samuel, C. S., Unemori, E. N., Mookerjee, I., Bathgate, R. A., Layfield, S. L., Mak, J., et al. (2004). Relaxin Modulates Cardiac Fibroblast Proliferation, Differentiation, and Collagen Production and Reverses Cardiac Fibrosis In Vivo. Endocrinology 145, 4125–4133. doi:10.1210/en.2004-0209
Samuel, C. S., Zhao, C., Bathgate, R. A., Bond, C. P., Burton, M. D., Parry, L. J., et al. (2003). Relaxin Deficiency in Mice Is Associated with an Age-Related Progression of Pulmonary Fibrosis. FASEB J. 17, 121–123. doi:10.1096/fj.02-0449fje
Samuel, C. S., Zhao, C., Bond, C. P., Hewitson, T. D., Amento, E. P., and Summers, R. J. (2004). Relaxin-1-deficient Mice Develop an Age-Related Progression of Renal Fibrosis. Kidney Int. 65, 2054–2064. doi:10.1111/j.1523-1755.2004.00628.x
Scheerer, P., Park, J. H., Hildebrand, P. W., Kim, Y. J., Krauss, N., Choe, H. W., et al. (2008). Crystal Structure of Opsin in its G-Protein-Interacting Conformation. Nature 455, 497–502. doi:10.1038/nature07330
Schöneberg, T., Kleinau, G., and Brüser, A. (2016). What Are They Waiting For?-Tethered Agonism in G Protein-Coupled Receptors. Pharmacol. Res. 108, 9–15. doi:10.1016/j.phrs.2016.03.027
Schöneberg, T., Schulz, A., Biebermann, H., Hermsdorf, T., Römpler, H., and Sangkuhl, K. (2004). Mutant G-Protein-Coupled Receptors as a Cause of Human Diseases. Pharmacol. Ther. 104, 173–206. doi:10.1016/j.pharmthera.2004.08.008
Schulze, A., Kleinau, G., Neumann, S., Scheerer, P., Schöneberg, T., and Brüser, A. (2020). The Intramolecular Agonist Is Obligate for Activation of Glycoprotein Hormone Receptors. FASEB J. 34, 11243–11256. doi:10.1096/fj.202000100R
Scott, D. J., Layfield, S., Yan, Y., Sudo, S., Hsueh, A. J., Tregear, G. W., et al. (2006). Characterization of Novel Splice Variants of LGR7 and LGR8 Reveals that Receptor Signaling Is Mediated by Their Unique Low Density Lipoprotein Class A Modules. J. Biol. Chem. 281, 34942–34954. doi:10.1074/jbc.M602728200
Sethi, A., Bruell, S., Patil, N., Hossain, M. A., Scott, D. J., Petrie, E. J., et al. (2016). The Complex Binding Mode of the Peptide Hormone H2 Relaxin to its Receptor RXFP1. Nat. Commun. 7, 11344. doi:10.1038/ncomms11344
Sethi, A., Bruell, S., Patil, N., Akhter Hossain, M., Scott, D. J., Petrie, E. J., et al. (2016). The Complex Binding Mode of the Peptide Hormone H2 Relaxin to its Receptor RXFP1. Nat. Commun. 7, 11344. doi:10.1038/ncomms11344
Shukla, A. K., Reinhart, C., and Michel, H. (2006). Comparative Analysis of the Human Angiotensin II Type 1a Receptor Heterologously Produced in Insect Cells and Mammalian Cells. Biochem. Biophys. Res. Commun. 349, 6–14. doi:10.1016/j.bbrc.2006.07.210
Smith, N. J., and Milligan, G. (2010). Allostery at G Protein-Coupled Receptor Homo- and Heteromers: Uncharted Pharmacological Landscapes. Pharmacol. Rev. 62, 701–725. doi:10.1124/pr.110.002667
Sudo, S., Kumagai, J., Nishi, S., Layfield, S., Ferraro, T., Bathgate, R. A., et al. (2003). H3 Relaxin Is a Specific Ligand for LGR7 and Activates the Receptor by Interacting with Both the Ectodomain and the Exoloop 2. J. Biol. Chem. 278, 7855–7862. doi:10.1074/jbc.M212457200
Svendsen, A. M., Vrecl, M., Knudsen, L., Heding, A., Wade, J. D., Bathgate, R. A., et al. (2009). Dimerization and Negative Cooperativity in the Relaxin Family Peptide Receptors. Ann. N. Y Acad. Sci. 1160, 54–59. doi:10.1111/j.1749-6632.2009.03835.x
Tena-Campos, M., Ramon, E., Rivera, D., Borroto-Escuela, D. O., Romero-Fernandez, W., Fuxe, K., et al. (2014). G-protein-coupled Receptors Oligomerization: Emerging Signaling Units and New Opportunities for Drug Design. Curr. Protein Pept. Sci. 15, 648–658. doi:10.2174/1389203715666140901094248
Troppmann, B., Kleinau, G., Krause, G., and Gromoll, J. (2013). Structural and Functional Plasticity of the Luteinizing Hormone/choriogonadotrophin Receptor. Hum. Reprod. Update 19, 583–602. doi:10.1093/humupd/dmt023
Ulloa-Aguirre, A., and Zariñán, T. (2016). The Follitropin Receptor: Matching Structure and Function. Mol. Pharmacol. 90, 596–608. doi:10.1124/mol.116.104398
Urizar, E., Montanelli, L., Loy, T., Bonomi, M., Swillens, S., Gales, C., et al. (2005). Glycoprotein Hormone Receptors: Link between Receptor Homodimerization and Negative Cooperativity. EMBO J. 24, 1954–1964. doi:10.1038/sj.emboj.7600686
Vassart, G., and Costagliola, S. (2011). G Protein-Coupled Receptors: Mutations and Endocrine Diseases. Nat. Rev. Endocrinol. 7, 362–372. doi:10.1038/nrendo.2011.20
Vlaeminck-Guillem, V., Ho, S. C., Rodien, P., Vassart, G., and Costagliola, S. (2002). Activation of the cAMP Pathway by the TSH Receptor Involves Switching of the Ectodomain from a Tethered Inverse Agonist to an Agonist. Mol. Endocrinol. 16, 736–746. doi:10.1210/mend.16.4.0816
Wilkinson, T. N., Speed, T. P., Tregear, G. W., and Bathgate, R. A. (2005). Coevolution of the Relaxin-like Peptides and Their Receptors. Ann. N. Y Acad. Sci. 1041, 534–539. doi:10.1196/annals.1282.080
Wilson, K. J., Xiao, J., Chen, C. Z., Huang, Z., Agoulnik, I. U., Ferrer, M., et al. (2018). Optimization of the First Small-Molecule Relaxin/insulin-like Family Peptide Receptor (RXFP1) Agonists: Activation Results in an Antifibrotic Gene Expression Profile. Eur. J. Med. Chem. 156, 79–92. doi:10.1016/j.ejmech.2018.06.008
Wu, Q. P., Zhang, L., Shao, X. X., Wang, J. H., Gao, Y., Xu, Z. G., et al. (2016). Application of the Novel Bioluminescent Ligand-Receptor Binding Assay to Relaxin-RXFP1 System for Interaction Studies. Amino Acids 48, 1099–1107. doi:10.1007/s00726-015-2146-3
Xiao, J., Huang, Z., Chen, C. Z., Agoulnik, I. U., Southall, N., Hu, X., et al. (2013). Identification and Optimization of Small-Molecule Agonists of the Human Relaxin Hormone Receptor RXFP1. Nat. Commun. 4, 1953. doi:10.1038/ncomms2953
Xiao, J., Chen, C. Z., Huang, Z., Agoulnik, I. U., Ferrer, M., Southall, N., et al. (2010). Discovery, Optimization, and Biological Activity of the First Potent and Selective Small-Molecule Agonist Series of Human Relaxin Receptor 1 (RXFP1). Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): National Center for Biotechnology Information (US).
Yamamoto, S., Kwok, S. C., Greenwood, F. C., and Bryant-Greenwood, G. D. (1981). Relaxin Purification from Human Placental Basal Plates. J. Clin. Endocrinol. Metab. 52, 601–604. doi:10.1210/jcem-52-4-601
Yan, Y., Scott, D. J., Wilkinson, T. N., Ji, J., Tregear, G. W., and Bathgate, R. A. (2008). Identification of the N-Linked Glycosylation Sites of the Human Relaxin Receptor and Effect of Glycosylation on Receptor Function. Biochemistry 47, 6953–6968. doi:10.1021/bi800535b
Zhang, M., Tong, K. P., Fremont, V., Chen, J., Narayan, P., Puett, D., et al. (2000). The Extracellular Domain Suppresses Constitutive Activity of the Transmembrane Domain of the Human TSH Receptor: Implications for Hormone-Receptor Interaction and Antagonist Design. Endocrinology 141, 3514–3517. doi:10.1210/endo.141.9.7790
Keywords: G-protein coupled receptors (GPCR), leucine-rich repeat containing receptor 7 (LGR7), relaxin family peptide receptor 1 (RXFP1), fluorescence-detection size-exclusion chromatography (FSEC), protein engineering, surface plasmon resonance spectroscopy (SPR)
Citation: Speck D, Kleinau G, Meininghaus M, Erbe A, Einfeldt A, Szczepek M, Scheerer P and Pütter V (2022) Expression and Characterization of Relaxin Family Peptide Receptor 1 Variants. Front. Pharmacol. 12:826112. doi: 10.3389/fphar.2021.826112
Received: 30 November 2021; Accepted: 31 December 2021;
Published: 28 January 2022.
Edited by:
Alexander S. Sobolev, Lomonosov Moscow State University, RussiaReviewed by:
Guillermo Romero, University of Pittsburgh, United StatesCopyright © 2022 Speck, Kleinau, Meininghaus, Erbe, Einfeldt, Szczepek, Scheerer and Pütter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Scheerer, cGF0cmljay5zY2hlZXJlckBjaGFyaXRlLmRl; Vera Pütter, dmVyYS5wdWV0dGVyQG51dmlzYW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.