- 1Department of Pathology, College of Korean Medicine, Kyung Hee University, Seoul, South Korea
- 2Korean Medicine-Based Drug Repositioning Cancer Research Center, College of Korean Medicine, Kyung Hee University, Seoul, South Korea
- 3Department of Biotechnology and Genetic Engineering, Global Biotechnology and Biomedical Research Network (GBBRN), Faculty of Biological Sciences, Islamic University, Kushtia, Bangladesh
- 4Department of Biotechnology and Genetic Engineering, Faculty of Biological Sciences, Islamic University, Kushtia, Bangladesh
- 5ABEx Bio-Research Center, East Azampur, Bangladesh
Gastric cancer (GC), second most leading cause of cancer-associated mortality globally, is the cancer of gastrointestinal tract in which malignant cells form in lining of the stomach, resulting in indigestion, pain, and stomach discomfort. Autophagy is an intracellular system in which misfolded, aggregated, and damaged proteins, as well as organelles, are degraded by the lysosomal pathway, and avoiding abnormal accumulation of huge quantities of harmful cellular constituents. However, the exact molecular mechanism of autophagy-mediated GC management has not been clearly elucidated. Here, we emphasized the role of autophagy in the modulation and development of GC transformation in addition to underlying the molecular mechanisms of autophagy-mediated regulation of GC. Accumulating evidences have revealed that targeting autophagy by small molecule activators or inhibitors has become one of the greatest auspicious approaches for GC managements. Particularly, it has been verified that phytochemicals play an important role in treatment as well as prevention of GC. However, use of combination therapies of autophagy modulators in order to overcome the drug resistance through GC treatment will provide novel opportunities to develop promising GC therapeutic approaches. In addition, investigations of the pathophysiological mechanism of GC with potential challenges are urgently needed, as well as limitations of the modulation of autophagy-mediated therapeutic strategies. Therefore, in this review, we would like to deliver an existing standard molecular treatment strategy focusing on the relationship between chemotherapeutic drugs and autophagy, which will help to improve the current treatments of GC patients.
1 Introduction
Gastric cancer (GC) is one of the most common frequently gastrointestinal and deadly cancers with more than one million new cases diagnosed yearly, which is the largest reason among the cancer fatalities worldwide (Hoang and Vu 2021). In the initial phages of GC, surgery is the most suitable option (Lin et al., 2021b). Because of its peculiar as well as cunning characteristics of initial diagnostic signs and symptoms, GC may result; even a minority proportion of cases are being accurately recognized, although more than 60% of patients had local or distant metastasis just at the moment of testing (Sun et al., 2021). Therefore, chemotherapy-mediated treatment seems to be preferred for the largest percentage of patients who belong to middle and late stage of GC (Xiao et al., 2021), although numerous patients are cured, but their rates of survival are very low. Principle of pathogenesis and progress of GC cancer is also mostly unclear (Tang et al., 2021). Because of drug resistance, patients with GC often have no sensitivity to chemotherapy, which is the primary reason of chemotherapeutic failure and poor success probability (Riquelme et al., 2016).
Autophagy, engulfing dysregulated organelles and cellular macromolecules, is an evolutionarily preserved catabolic active process concerning the formation of autophagosomes, leading to breakdown of cellular constituents after fusion with lysosomes (Rahman and Rhim 2017). In addition, cellular degradation procedures mostly fall into two classes, macroautophagy, which is commonly known as autophagy, and ubiquitin–proteasome system (Rahman et al., 2020c; Uddin et al., 2020). Furthermore, autophagy regulates in the activation of several cancer-related genes, which inhibits tumor promotion and suppression (Botti et al., 2006; Maiuri et al., 2009). Recent studies have widely explained the involvement of autophagy in GC growth, metastasis, and forecasting (Xu et al., 2020a; Spirina et al., 2020; Xiu et al., 2021). Moreover, microRNAs (miRNAs), short (∼22 nucleotides in length) noncoding RNA molecules, can control gene expression at a posttranscriptional level, which has an important association in autophagy-mediated GC regulation. There are abundant convincing studies that showed inseparable association between miRNAs and GC (Shao et al., 2020). miRNAs affected GC, which includes oncogenesis, diagnosis, development, treatment, and prognosis, although many miRNAs have been linked to GC, and few could be useful to clinical practice (Ouyang et al., 2021), even though numerous miRNAs have been related to GC and few can be applied to clinical practice as well. Furthermore, diet-related natural ingredients may control autophagy in the GC cell that promote cancer cell chemosensitivity (Xu et al., 2020a). In the present study, we would like to represent an overview of the literature in association with autophagy modulation in GC treatment as well as the role of autophagy boosting and suppressing, which control GC growth and invasion as a potential treatment strategy and managements.
2 Molecular Mechanism of Autophagy Regulation and Cancer Progression
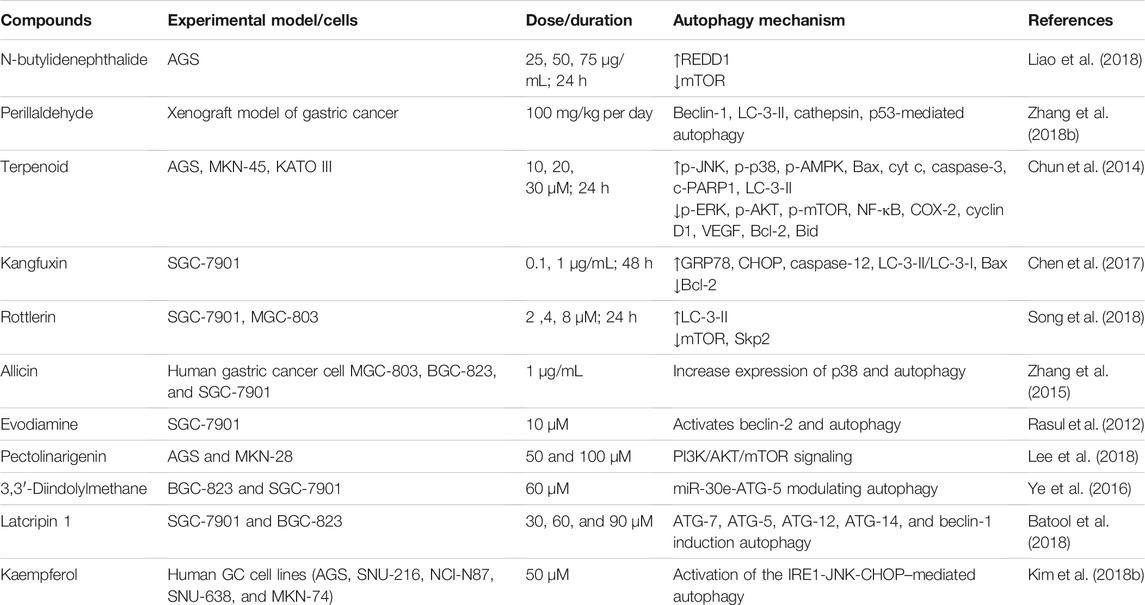
Autophagy is a self-digestion process that assists in maintaining cellular homeostasis through recycling unwanted or damaged toxic cellular organelles into the cells (Rahman and Rhim 2017; Rahman et al., 2021b). Autophagy modulation has been implicated to regulate several cancers and neurodegeneration (Rahman et al., 2020b; Rahman et al., 2020d). Generally, autophagy process might be introduced via the accumulation of preautophagosome structures formation, which is known as phagophore assembly sites (PASs) (Hurley and Young 2017; Rahman and Rhim 2017). Phosphatidylinositol 3-phosphate PI3K associated with endoplasmic reticulum (ER) has an essential role to imitate PAS formation (Kotani et al., 2018). AMPK, AMP-activated protein kinase, mTOR, mammalian target of rapamycin, and ULK1, unc-51 like autophagy activating kinase-1, have been facilitated to initiate phagophore formation in the process of autophagy induction (Alers et al., 2012). However, beclin-1, Vps34, and Vps15/p150 help to recruit formation of phagophore (Velazquez and Jackson 2018). After that, phagophore nucleation has been followed to elongate membrane to form autophagosome formation (Rubinsztein et al., 2012). Mature autophagosome binds to lysosome, which results to autolysosome formation (Kardideh et al., 2019). Finally, autolysosomes that contain inner cargos have been degraded by acid hydrolases, as well as produce nutrients and other recycling metabolites, resulting to maintenance of intracellular homeostasis inside the cells (Figure 1).
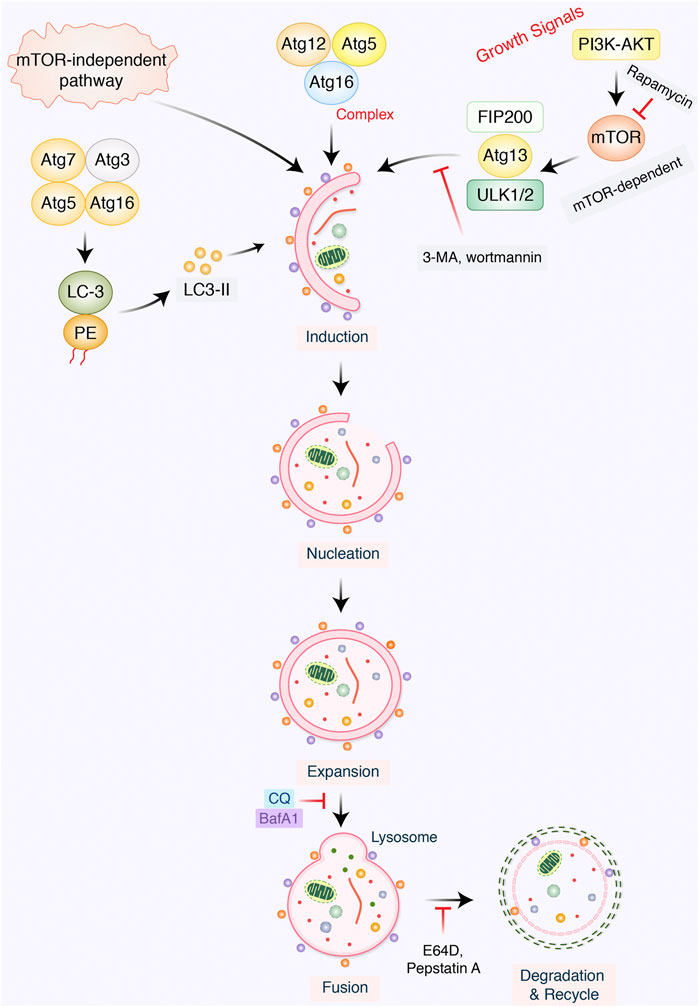
FIGURE 1. Mechanism of the autophagic pathway. Autophagy initiates via the formation of a macromolecular assembly structure. PI3K-AKT and mTOR contribute to the formation of the phagophore assembly site (PAS). ULK1/2, ATG-13, Vps34, and beclin-1 complex activate phagophore formation, which creates nucleation elongation as a result of autophagosome formation. Mature autophagosome and lysosome bind to form autolysosome formation. Eventually, autolysosomes are degraded via acid hydrolases, thereby releasing nutrients as well as recycling metabolites.
A direct connection has been found between autophagy and cancer (Liang et al., 1999). Recently, a huge number of researches have indicated that ATGs as well as associated pathways may crosstalk between oncogenes and tumor suppressors (Ariosa et al., 2021; Khaleel 2021). Certainly, collected data have supported that the role of autophagy in cancer is complicated, which may have opposite values in a context- as well as cell type–dependent manner (Li et al., 2021b). Autophagy determines whether the cancer is inhibited or activated under certain conditions. It has been mentioned that mTOR plays a central role in activating or protecting oncogenic cells via induction of autophagy (Xu et al., 2020b). In addition, inhibition of autophagy pathway may regulate cancer progression, as well as the influence of autophagy develops into either a death function or cellular survival function (Jung et al., 2020). The metabolism of cancer cells is strongly changed to retain their survival and proliferation under adverse microenvironmental situations. It was found that autophagy acts as an essential function in maintaining metabolic variations in cancer cells (Goldsmith et al., 2014), although autophagy is familiar to sustain neoplastic cell metabolism during stress, and the mutual relation between cancer cell metabolism and autophagy remains unknown. AMPK and mTOR have been recognized as the enteral signaling mechanisms which regulate autophagy through the regulation of amino acid as well as glucose levels (Alers et al., 2012). However, the specific metabolites, oxygen concentration, growth factors, ROS, ATP-to-ADP ratio, specific amino acid levels, palmitate, and oncogenes regulate autophagy initiation in addition to autophagosome formation. In addition, they regulate the balance via assimilating the autophagy-related signals in cancer (Singh and Cuervo 2011; Panda et al., 2015). Conspicuously, autophagy has been commonly recognized to play a “double role” as it can either delay or activate cancer initiation as well as progression (Rahman et al., 2020b; Patra et al., 2020).
The dual role of autophagy in cancer has been emphasized in autophagy regulation as well as cancer cell activation, which metabolism controls tumor growth and progression. Basically, the actual role of autophagy in GC prognosis, metastasis, and progression has not been widely deliberated yet. However, tumor metastasis sign has predicted advanced progression as well as poor prognosis of GC (Jin et al., 2014: ; Cai et al., 2020). Tumor metastasis process is complex, involving a series of pathological actions, for example, breakdown of epithelial-to-mesenchymal transition (EMT), extracellular matrix (ECM), tumor microenvironment formation, and tumor angiogenesis (Winkler et al., 2020). In addition, in tumor metastasis, the role of autophagy is supposed to be both prometastatic and antimetastatic effects (Poole and Macleod, 2021). Concerning GC, while autophagic cell death may prevent metastasis, most of the current results support the notion that autophagy facilitates tumor metastasis via affecting several aspects (Figure 2). In the present study, we would like to focus on the relationship with chemotherapeutic treatment of GC through modulation of autophagy pathway.
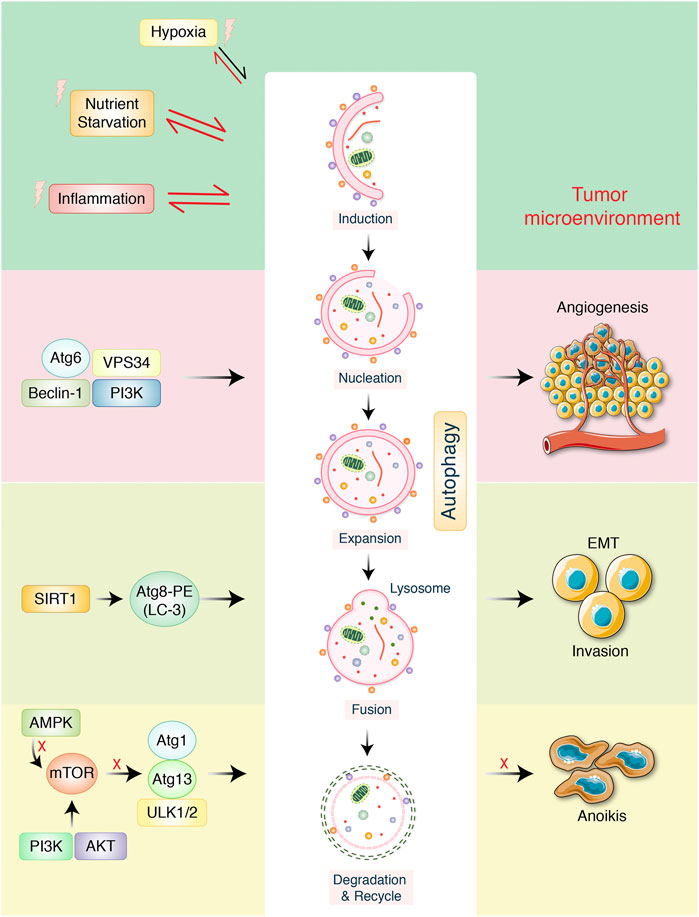
FIGURE 2. Autophagy-mediated metastasis formation in gastric cancer. The autophagy-related proteins are involved in regulation in cancer. Starvation, hypoxia, and inflammation might be stimulated during autophagic process which create tumor microenvironment. VPS34, ATG6, beclin-1, and PI3K increase tumor angiogenesis. Transcription factor SIRT1 activates autophagy improvement via inducing ATG8-LC-3-PE conjugation, which later encourages epithelial-to-mesenchymal transition (EMT), as well as tumor invasion. mTOR negatively regulates autophagy via inhibiting ATG13-ATG1-ULK1/2 protein complex. AMPK/PI3K regulates autophagy, which detached tumor cells to overcome anoikis.
3 Role and Regulation of Autophagy in GC Management
During gastric carcinogenesis, the complicated autophagy-mediated regulated pathways become more complicated, and it is necessary to properly investigate. Primarily, AMPK regulatory system interacts with PI3K-AKT signaling, although a multitude of transcription components regulate cell biological activities such as multiplication, development, apoptosis, and autophagy in anticancer activities. Clinical and pathological studies have shown that tumor tissues have lower levels of AMPK expression than normal tissues, and this is thought to be a contributing factor to emergence and progression of melanomas (Faubert et al., 2015; Scazzone et al., 2021). Perillaldehyde activates AMPK by phosphorylating LKB1 at the S307 and S428 regions, and AMPK induces GC cell autophagy by phosphorylating and activating ULK1, which restricts GC cell proliferation (Zhang et al., 2019). MTDH also performs a vital role in drug resistance, specifically in the resistance to 5-fluorouracil (5-FU), doxorubicin, CDDP, and etoposide, but also paclitaxel. MTDH governs ATG-5 expression through triggering AMPK phosphorylation, recommending that MTDH may invoke autophagy via the AMPK/ATG-5 signal transduction pathway and encourage drug tolerance in GC cells (Pei et al., 2018). Cancer cells with an abnormally active PI3K/AKT/mTOR pathway have a higher propensity to proliferate aggressively, extending their survival period and becoming resistant to chemotherapy (Zheng et al., 2021). A study has demonstrated that activating AKT makes GC cells more resistant to chemotherapy treatments such 5-FU, doxorubicin, mitomycin C, and cisplatin (5-FU) (Munshi et al., 2001). Autophagy is inhibited by the mTOR route, which negatively regulates; therefore, blocking the mTOR system is essential to triggering autophagy (Yap et al., 2008). Flavonoids can act as an anticancer element by inhibiting the PI3K/AKT/mTOR system, resulting in arresting G2/M cell cycle and autophagy, which induce death of GC cells (Raha et al., 2015). Furthermore, it has been shown that suppressing YWHAZ in BGC-823 cells inhibits the PI3K/AKT/mTOR regulatory mechanism, resulting in cell death and autophagy (Guo et al., 2018a). lncRNAs-HAGLROS associates mTORC1 that stimulate the mTORC1 regulatory system, boosting GC cell proliferation and sustaining its malignant state (Chen et al., 2018a). Furthermore, elevated production of HAGLROS correlates with the formation and lousy prediction of GC, according to new research findings (Chen et al., 2018a). A number of investigations suggested that long noncoding RNAs (lncRNAs) influence chemoresistance in a variety of cancers (Fan et al., 2014; Qu et al., 2016; Özeş et al., 2016). Moreover, ARHGAP5-AS1 is a novel drug-resistant lncRNA that increases in GC-resistant cells and can revert chemoresistance and afterward turned down (Zhu et al., 2019).
Autophagy is thought to be a beneficial process for inhibiting tumor development at numerous phases, conserving genetic consistency, removing intracellular supplies of reactive oxygen species (ROS), as well as sustaining bioenergetic activities (Wang et al., 2018; Weng et al., 2018). Autophagy, as both a cell’s stress feedback system to inside and outside stimuli as well as promote cellular damage also the life expectancy of cancer cells subjected to chemotherapeutics. (Scherz-Shouval et al., 2010). Autophagy separates cellular components for example mitochondria inside of cells, that also helps stop the propagation of pro-apoptotic elements inside of cells and thus aids the tumor type of cells avoiding apoptosis. Autophagy suppression in cancerous cells could also increase the toxic effects of anti-tumor prescription medications and backward drug resistance (Kumar et al., 2015; Belounis et al., 2016). Autophagy and apoptosis can be induced by apatinib and astragalus polysaccharides, however this polysaccharide inhibit metastasis of GC and invasion of GC. Whilst autophagy blockers could even significantly boost AGS apoptotic cell death, it appears that such increased autophagy caused by apatinib safe guards the cells from apoptosis. Because increased autophagy could have negative impacts on chemo, inhibiting autophagy can stimulate protective GC cells and improve the anti-tumor influence of chemotherapeutics (Wu et al., 2018b). Autophagy suppression appears to have a pro-apoptotic impact on peoples GC cells. Cinobufagin can cause the generation of ROS which causes apoptotic cell death as well as autophagic cell death through stimulating the ROS/JNK/p38 alignment. Steadily increasing proapoptotic protein expression, abnormal mitochondrial membrane potential, and elevated ROS manufacturing are observed while autophagy is interrupted, implying that autophagy suppression improves cinobufagin-induced cell death, which might take place in part via the mitochondrial-coded cell death passageway (Xiong et al., 2019).
Autophagy could indeed preserve cell equilibrium in the preliminary phases of tumorigenesis, inhibiting the incidence as well as advancement of GC. Autophagic cell death is distinct from apoptotic cell death. Autophagy and apoptosis coexist in GC cells, and their interplay governs cell death autonomously. Apoptosis occurs downstream of autophagy, as well as apoptotic cell death that occurs by autophagy (Liu et al., 2021a). For instance, the PI3K/AKT/mTOR sensing process, which can synchronously govern the destiny of GC cells, can control autophagy and apoptosis, respectively (Hu et al., 2021). Caffeine and theophylline, which are derivatives from methylxanthine, have been shown to suppress the PI3K/AKT/mTOR paths through activating PTEN. As a result, GC cells undergo apoptosis and autophagy, and their expansion is inhibited (Liu et al., 2019). ER stress and its unfolded protein response can also be linked to GC cell ability to survive, advancement, and medication resistance via biological processes such as autophagy. Melatonin induces cell autophagy via ER stress, helping to promote GC apoptotic cell death and preventing their growth, expansion, and invasion (Liu et al., 2019; Peng et al., 2019). Mitochondrial apoptosis can be induced by tetrandrine and also inhibit the AKT/mTOR pathway in HGC-27 cells causing autophagy and apoptosis, resulting in antitumor action and death of cells in gastric tumors. During the tetrandrine-induced antitumor procedure, autophagy and apoptosis work together to improve tumor cell death (Bai et al., 2018). Beclin-1 also serves as an important role enhancing autophagy-induced apoptosis resistance, and the expressions of beclin-1, Bcl-xl, and Bcl-2 are positively correlated with autophagy (Menon and Dhamija 2018). Beclin-1 can also stimulate the expression of Bcl-2 and Bcl-xl; an alternative is to suppress Bak and Bax protein levels while increasing levels of cleaved caspase in GC cells (Kang et al., 2011). This will prevent GC cells from undergoing apoptosis while boosting autophagy in GC cells (Maiuri et al., 2007; Du and Ji 2014).
4 Autophagy Markers Expressed in GC Progression
Because of the differences in biological and clinical characteristics, carcinoembryonic antigen and carbohydrate antigen 19-9 have been found to be the most common GC markers measured before and after surgery (Lin et al., 2020), although which preoperative or postoperative combined tumor markers have a more prognostic value has not been clear yet, as well as whether change of the preoperative and postoperative systemic inflammatory response (SIR) levels affects the prognosis of GC. Perioperative SIR variations were described as changes in the neutrophil–lymphocyte ratio, lymphocyte–monocyte ratio, systemic immune–inflammation index, and platelet–lymphocyte ratio (Lin et al., 2021c). Autophagy-mediated tumor metastasis in GC is described in Figure 2, which is modified from Qian and Yang in 2016 (Qian and Yang 2016). Inhibition of mTOR by cellular energy sensor AMPK activates autophagy, which acts a prosurvival role in cancer cells during ECM detachment (Mukhopadhyay et al., 2021). Improved autophagic process might prevent ECM detached cells from anoikis, in addition to contributing to luminal filling probably through providing sustained ATP sources (Santagostino et al., 2021). Therefore, autophagy has been considered as an adaptive strategy for separate cancer cells to overcome anoikis in the early stage of cancer progression (Rahman et al., 2020d; Ariosa et al., 2021). Besides, a class III histone deacetylase, silent mating type information regulation 1 (SIRT1), is augmented in tumor tissues, in addition to correlating with metastasis of GC in advanced lymph node (Mody et al., 2021). However, the regulatory effects of SIRT1 in EMT, as well as invasion ability of GC, were further confirmed by an in vitro study (Xu et al., 2021a). Significantly, it has been found that SIRT1 is a well-categorized autophagy mediator that is derived to initiate autophagy through deacetylation of LC-3 ATGs (Chen et al., 2021a). Collecting this observation, autophagic process is modulated via SIRT1 or other mediators that might play an essential role in tumor progression via modulating EMT as well as tumor cell invasion (Zia et al., 2021). Recently, it has been reported that autophagy encouraged the survival as well as invasive capability of SGC-7901 cells via facilitating the development of vasculogenic mimicry (Ahmadpour et al., 2021). Particularly, autophagy inhibition by beclin-1 silencing expression decreased cancer cell invasion and survival (Ding et al., 2014). Therefore, beclin-1–mediated autophagy might be considered as a tumor angiogenesis factor for GC. Pharmacologically, autophagy inhibition along with antiangiogenic therapy may combine as a promising way to overcome tumor angiogenesis. Moreover, concerning the tumor microenvironment, autophagy might be strongly encouraged via nutrient depletion, hypoxia, and inflammation (Poillet-Perez et al., 2021). However, it is also mentioned that activated autophagy shapes the tumor microenvironment through activating tumor angiogenesis, regulating inflammatory responses, and providing nutrient supply (Mukhopadhyay et al., 2021).
Based on The Cancer Genome Atlas database, gene expression data for GC patients undergoing numerous other molecular markers have been recognized to possess prognostic value and their expression patterns of autophagy-related genes (ATGs) in GC cells, which served as a hallmark of autophagy regulation (Liu et al., 2021b). Autophagy and GC are controlled by crucial kinases such as mTOR, PI3K/AKT, AMPK, and MAPK, as well as epidermal growth factor receptor, cell cycle mediators, vascular endothelial growth factor, cytokines, apoptosis-associated regulators, and miRNAs (Xiu et al., 2021). ATG5 has been found to participate in autophagosome elongation as a vital regulator and is crucial for autophagy, which is associated with chemotherapy resistance of maximum cancer cells (Wang et al., 2019). Moreover, single-strand conformation polymorphism analysis has been revealed that the frameshift mutations in ATG genes with mononucleotide repeats, including ATG-2B (mammalian ATG-2 homolog), ATG-9B (mammalian ATG-9 homolog), and ATG-12 have in common GC with high microsatellite instability subtypes (Kang et al., 2009). ABCC1 encodes the multidrug-resistant protein 1 (MRP1), which promotes the MDR phenotype in GC. However, MRP1 and ATG5 expression were found to be positively associated and ATG5 expression can sometimes lead to more forceful and malignant trait of GC, which could provide important data for effectively evaluating chemotherapy impact in GC patients (Xu et al., 2020a). ATG5 and MRP1 expressions are being worked as self-governing prognostic markers for predicting overall survival and disease-free survival in GC patients (Ge et al., 2014). A rate-limiting enzyme arginine synthesis pathway, arginine succinate synthase 1 (ASS1), is highly expressed in GC tissues (Silberman et al., 2019). Studies have found that ASS1 may be a useful prognostic marker for predicting survival and metastasis in patients with GC (Tsai et al., 2018; Jangra et al., 2020). The gene SP1, part of the SP1 multigene family, which is critical in the emergence and progression of malignancies, was found to be highly overexpressed in GC tissues, and this overexpression was closely associated with patient survival (Xu et al., 2018). SP1 has an adverse effect on autophagy regulation because it binds directly to the p62 promoter and raises p62’s expression level. As the SP1-p62 axis may contribute to the development of GC, it could serve as a prognostic indicator for the detection of GC (Xu et al., 2018). Moreover, newly synthesized LC-3’s C-terminus has been hydrolyzed via a cysteine protease known as ATG-4B exposing Gly-120 called LC-3-I (Agrotis and Ketteler 2020). LC-3-I has been processed via a series of ubiquitin-like reactions by the help of enzymes ATG3, ATG7, and ATG12–ATG5–ATG16, which become adjacent to the head group of the lipid phosphatidylethanolamine (PE), a class of phospholipids found in biological membranes (Rahman et al., 2021a). However, the lipid modified form of LC-3, which is known as LC-3-II, has been believed to be intricated in autophagosome membrane elongation as well as fusion events during autophagy process (Schaefer and Dikic 2021). However, the exact function and role of LC-3 in autophagy pathway are still investigated. In addition to the PB1 domain and the TB domain, p62, also known as sequestosome 1 (SQSTM1) autophagosome cargo protein, which targets other proteins that bind to it for selective autophagy, has several other domains such as the KIR, a ubiquitin-related area, and so on. A number of investigations have revealed that p62 levels are inversely associated with levels of GC autophagy (Weng et al., 2018). When autophagy occurs, the expression of beclin-1 increases because it is a yeast ATG6 homolog and is an essential activator of autophagy (Hu et al., 2015), and a reduction in beclin-1 expression in GC indicates a drop in autophagy (Zheng et al., 2019).
5 Role of miRNAs Through Regulation of Autophagy in GC
Alongside miRNAs, lncRNAs are essential to regulate autophagy, which can aid in the development of more effective treatment strategies and the identification of novel medicinal aims for the research of processes of resistance in GC (Roy 2021). In GC tissues, lncRNA MALAT1 activates autophagy via downregulating the tumor suppressor miR-204. miR-204 overexpression in GC cell lines CTC105 and CTC141 reduces transient receptor potential melastatin 3 (TRPM3), an activator of oncogenic autophagy-miR-204, which can be used as a target in GC therapy. Antiautophagy tumor suppressor, miR-30a, upsurges sensitivity to imatinib (IM) in gastrointestinal stromal tumor (GIST) cells alone with mouse xenograft models with specific target of miR-30a in association with beclin-1–driven autophagy in IM-resistant cells such as GIST-882 than the sensitive GIST-1 cell line (Shao et al., 2020), but counterintuitive that miR-183 could contribute to GC suppression (Li et al., 2019). Furthermore, miR-20a, an oncogene, is significantly expressed in GC, and it is anticipated that it will serve as an indicator for clinical identification (Xin et al., 2019). It has been found that miR-21 is overexpressed, and its anomalous expression has an important role in GC growth via modulating tumor suppressors PTEN and PDCD4 expression, which regulates migration, cell growth, invasion, and apoptosis (Li et al., 2014). According to the findings, the level of miR-1265 in GC samples was shown to be lower than in samples from nearby healthy cells. The calcium-binding protein (CAB39) gene is miR-1265’s target. CAB39 is an important element of the LKB1-STRAD-CAB39 combination (Brajenovic et al., 2004; Treiber et al., 2019), autophagy produced by the CAB39-LKB1-AMPK pathway is cancerous in GC cells and enhanced the phosphorylated of AMPK more than 100-fold (Hawley et al., 2003) at the Thr172 junction of LKB1 with STRAD and CAB39. As a result, miR-1265 slows GC development and autophagy by reducing the expression of CAB39 and controlling the AMPK-mTOR regulatory pathway (Xu et al., 2019). On the other hand, targeting miR-25-3p activated growth inhibition, invasion, and migration of GC cells in vitro, and in vivo delivery of miR-25-3p inhibitors significantly encouraged SCID mice–bearing human GC xenografts antitumor activity (Ning et al., 2020). An miRNA, miR-495-3p, has been linked to the development of malignant phenotypes in patients with GC. It is thought to be responsible for reversing MDR by inhibiting autophagy through modulation of the mTOR signaling pathway. MiRNAs-181a endorses EMT in esophageal squamous cell carcinoma through the transforming growth factor-β/Smad pathway (Xu et al., 2021b). It is also believed that miR-495-3p is responsible for reversing MDR by limiting autophagy and also that miR-495-3p is related to the malignant phenotype of GC patients (Chen et al., 2018b). In addition, certain miRNAs, such as miR-375 (Yuan et al., 2018), miR-21 (Gu et al., 2020), and miR-361-5p (Tian et al., 2018), which operate as autophagy blockers, control autophagy and limit GC activity via modulating the mTOR pathway (Figure 3 and Figure 4). Furthermore, the expression of miRNAs, such as miR-181a (Zhao et al., 2016), miR-30a (Du et al., 2018), miR-let-7a (Fan et al., 2018), miR-133a-3p (Zhang et al., 2018a), miR-532-3p (Guo et al., 2018b), and others, is connected with such a reduction in the capacity of GC cells to proliferate. It has been found that SLC7A11 is identified as a target of miR-375, which diminished the stemness of GC cells via activating SLC7A11-dependent ferroptosis (Ni et al., 2021). MALAT1, a competing endogenous RNA of miR-23b-3p, was shown to reduce the suppressive activities of miR-23b-3p on ATG-12, while simultaneously increasing the development of ATG-12, resulting in chemoinduced GC cell autophagy and drug resistance (YiRen et al., 2017). In addition, AKT and mTOR have been reported to be targeted via miR-495 overexpression of miR-495, which could prevent the growth in addition to induce the apoptosis of GC cells through blocking of PI3K/AKT/mTOR, which altered Bax, caspase-3/-9, and cyclin D1 expression (Ouyang et al., 2021). miR-153-3p facilitates ATG-7–mediated autophagy induction in fluorouracil resistance via the adenosine monophosphate (AMP)-activated protein kinase (AMPK)/ATG3 pathway in GC (Hou et al., 2020). Gastric carcinogenesis is complicated by the participation of miRNAs (Song and Meltzer 2012), and these autophagy-mediated processes must be further investigated.
FIGURE 3. MicroRNAs regulates autophagy-mediated cell proliferation and migration in gastric cancer. Overexpression of miR-375 inhibited the proliferation and migration of gastric cancer in vitro and xenograft nude mouse model. miRNA blocks autophagy via AKT/mTOR signaling pathway and regulating invasion as well as migration in epithelial-to-mesenchymal transition. In addition, most usually effective miRNAs control the transcriptional expression of upstream activators and inhibitors of autophagy in gastric cancer.
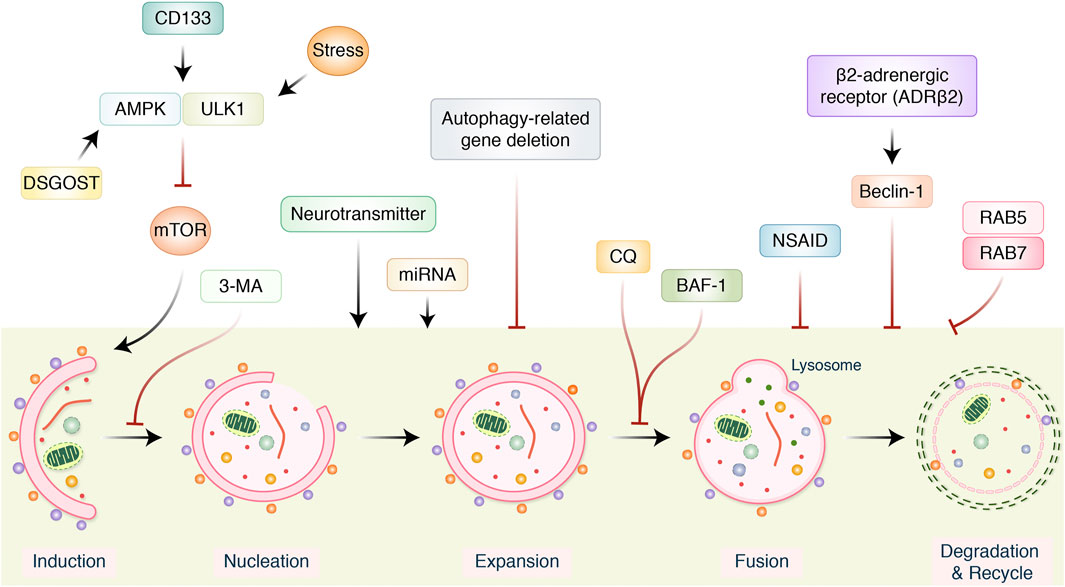
FIGURE 4. Several signaling pathways modulating autophagy in gastric cancer treatment. AMPK/ULK1 inhibits mTOR pathway, which positively activates autophagy induction. Neurotransmitter and miRNA regulate autophagy induction. However, autophagy-related gene (ATG), 3-metheylalanine (3-MA), chloroquine (CQ), bafilomycin A1 (BAF-1), and nonsteroidal anti-inflammatory drugs (NSAIDs) inhibited and modulated entire autophagy process. β2-Adrenergic receptor activates beclin-1 and inhibits autophagy.
6 Therapeutic Target and Treatment Strategy of Autophagy Modulation in GC
Therapeutic targeting of autophagy in GCs might be proposed to be an auspicious novel therapeutic approach. Meanwhile, both autophagy inhibitors and autophagy inducers may lead to inhibit cancer cell death, but at present, they are only in the clinical development stage for the treatment of GCs. Despite the fact that the association between autophagy and cancer still is debated, the participation of shared regulatory mechanisms (Table 1) makes autophagy an attractive therapeutic focus for the treatment of cancer (Khaleel 2021). Autophagy can be used to sensitize tumor cells to chemotherapy or radiation, therapy blocking its cytoprotective action, which leads to the destruction of antiapoptotic cells via autophagy (Gupta et al., 2021). Through the use of their mechanisms, autophagy demonstrated resistance to chemotherapy and radiotherapy, and it also hinders the effectiveness of anticancer medicine (Zhang et al., 2021). Inhibiting autophagy in cancerous cells can have adverse repercussions on the body, including such mitochondrial dysfunction, redox imbalance, nucleotide consumption, and reduction of energy supply (Poillet-Perez et al., 2021). As a result, inhibiting autophagy could be recruited to increase chemosensitivity as a therapeutic strategy. AMP-activated protein kinase α (AMPKα) has been shown to promote autophagy by triggering ULK1 and blocking mTOR/p70S6K process (Xu et al., 2020b). Stress-inducing neurotransmitter norepinephrine increases autophagic flux by AMPK-ULK1 pathway, as a result activating the tumor-promoting autophagy pathways and speeding up GC development (Zhi et al., 2019). Anticancer therapy efficacy may be improved using AMPK inhibitors; 3-methyladenine (3-MA), a class III PI3K inhibitor, suppresses autophagy by inhibiting autophagy from initial stage and preventing autophagosomes formation. Antimalarial medication chloroquine (CQ) is used to treat malaria, which also impedes autophagy (Lin et al., 2021a). There is no direct impact on organelle acidity (Park et al., 2021), but it suppresses autophagy by preventing the autophagosome and lysosome from joining (Honma et al., 2021). The effectiveness of antitumor therapy could be improved by using this supplement; CQ and 5-FU together have the potential to further reduce the number of GC stem cells (Xiao et al., 2017). As a result of blocking lysosome–autophagy fusion, BAF-1 shows lower LC-3-II levels while simultaneously elevating p62, which is one way that BAF-1 inhibits autophagy (Bai et al., 2018). However, there is no information on the connection between BAF-1 and the therapy of GC. In addition, chemokine CXCL12 encouraged mTOR activation and played an important role in GC cell peritoneal metastasis control (Mele et al., 2020).
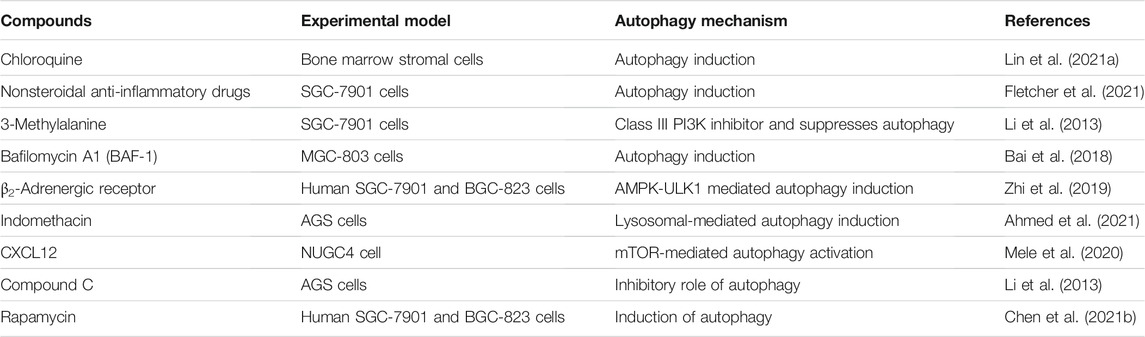
TABLE 1. Several molecular target and therapeutic role of different drugs in autophagy modulation in gastric cancer cells.
Recently, it has been found that inhibition of autophagy via silencing of beclin-1 protein expression decreased GC cell survival as well as invasion (Shafabakhsh et al., 2021). The deletion of ATGs then suppresses autophagic activity via siRNA- or miRNA-mediated silencing methods, which have been gaining attention (Schaefer and Dikic 2021). The use of sequence-specific DNA or RNA analogs can be used to build customized compounds with anticancer properties at a low cost that prevent the production of specific gene sequences with high specificity. In addition, several essential controllers of autophagy pathways, such as ATG-3, ATG-4B, ATG-4C, ATG-5, ATG-6, beclin-1, ATG-10, and ATG-12, can be targeted in this way in order to combat GC (Mandhair et al., 2021). It has already been discovered that enhanced ATG-5 activity was observed to be associated with a better patient survival and disorder survival in GC patients and that ATG-5 was abundantly expressed in drug-resistant GC cell lines. It has also been discovered that suppressing ATG-5 (siRNA-ATG-5-695) can restore sensitivity to chemotherapeutics in resistant cells (Xu et al., 2020a). GC cells treated with cinobufagin undergo apoptosis because of the inhibition of ATG-5 synthesis caused by siRNA, which can also increase ROS formation and activate a cell death pathway in mitochondria. The stimulation of autophagy in GC cells by norepinephrine is a critical component for the growth of GC (Krizanova et al., 2016; Le et al., 2016; Zhi et al., 2019). The β2-adrenergic receptor (ADRB2) is a variant of the adrenergic receptor that is responsible for catecholamine production in the body. Beclin-1 production can also reduce by deletion of the ADRB2 gene that may also inactivate the AMPK-ULK1 system; as a result, autophagy is reduced (Xiu et al., 2021). When it comes to the autophagy function, Rab5a, a part of the Rab group, is also engaged in intracellular material transport and protein classification, but it also plays a role in the process (He et al., 2021). In GC cells, activation of mTOR by Rab5a, an upstream regulator of mTOR, can suppress autophagy and increase pharmacological resistant through activation of mTOR (Li et al., 2021b). Because of this, a Rab5a mutation can reduce mTOR activity in SGC7901 cells while simultaneously increasing autophagy and reversing pharmaceutical tolerance (Li et al., 2021a). Nonsteroidal anti-inflammatory drugs (NSAIDs) target the epithelium of the gastrointestinal system (Fletcher et al., 2021). If these medications are used to treat inflammation and pain, they have a detrimental effect on the digestive epithelia, which is their primary negative impact (Fratter et al., 2021). However, because NSAIDs decrease carcinogenesis in gastrointestinal tissues, they are considered an adjuvant to chemotherapy (Wang et al., 2021). It has been demonstrated that indomethacin-treated AGS cells exhibit decreased lysosomal acid content and increased membrane permeability, impairing lysosomal role and cathepsin action; thus, inadequate deterioration of autophagic materials impairs autophagic flux, raising the susceptibility of GC cells to cytotoxic agents (Ahmed et al., 2021). Rapamycin increases intracellular ROS generation, as well as displays selective synergistic antitumor activity with EF24 in human GC cell lines SGC-7901 and BGC-823 (Chen et al., 2021b). Therefore, perturbation of autophagic modulation may be a possible approach for controlling and treating GC.
7 Phytochemicals for the Prevention and Treatment of GC via Autophagy
Phytochemicals have been demonstrated to be promising for regulating and controlling GC (Mitra and Dash 2018), which makes it possible for cellular components to degrade and be recycled in a controlled manner (Yao et al., 2021). Numerous phytochemicals and their autophagic activities are summarized in Table 2. Rottlerin, extracted from Mallotus philippensis Muell (Euphorbiaceae), induced autophagy and caspase-independent apoptosis against SGC-7901 and MGC-803 cells (Song et al., 2018). Evodiamine activates autophagy through beclin-2 expression in SGC-7901 GC cells (Rasul et al., 2012). Morus alba root extract, containing oxyresveratrol, has been found to accumulate ROS production and initiate autophagic and apoptotic cell death via FOXO-caspase-3 pathway (Kwon et al., 2015; Rahman et al., 2017). Cytotoxic activity on AGS, MKN-45, and KATO-III human GC cells via induction of caspase activation and autophagy via AKT/NF-κB pathway in AGS cells (Kang et al., 2021) have been demonstrated. 3,3′-Diindolylmethane modulates autophagy activation by miR-30e-ATG-5 in BGC-823 and SGC-7901 cells (Ye et al., 2016). Pectolinarigenin, a natural flavonoid present in Cirsium chanroenicum, downregulated PI3K/AKT/mTOR pathway via G2/M phase cell cycle arrest, apoptotic, and autophagic cell death in human GC cells (Lee et al., 2018). Flux analysis of autophagy and increase in the level of LC-3-II revealed induction of autophagy by the tuber of Amorphophallus konjac. G0/G1 phase cell cycle arrest has been detected by flow cytometry. Chen et al. determined apoptosis- and autophagy-inducing effects of kangfuxin, an organic extract of Periplaneta americana Linnaeus. (Blattidae), against SGC-7901 cell line (Chen et al., 2017). Proteins that mediate ER stress–mediated apoptosis including glucose-regulated protein 78 (GRP78), C/EBP-homologous protein (CHOP), and caspase-12 have been greatly upregulated in the group treated with kangfuxin. In addition, the LC-3-I/LC-3-II ratio and expression levels of beclin-1 were also higher in the kangfuxin group. Also, 3,3′-diindolylmethane, derived from cruciferous vegetables, increased the ATG-5 expression and LC-3 in GC cells in addition to decrease miRNAs-30e level (Ye et al., 2016). Besides, perillaldehyde, isolated from Perilla frutescens, increased AMPK phosphorylation, leading to autophagy via beclin-1, LC-3-II, cathepsin, p53, and caspase-3 in tumor xenograft model of GC in MFC mouse as well as GC9811-P human GC cells (Zhang et al., 2018b). Danggui-Sayuk-Ga-Osuyu-Saenggang-Tang (DSGOST), a traditional Chinese medicine, stimulates autophagy by triggering AMPK/ULK1 signaling, consequently activating EMT and exosomes to increase chemoresistance, and therefore, one of the mechanisms of GC resistance to DSGOST is survival, promoting autophagy. Using compound C, a well-known suppressor of AMPK, to impede DSGOST-mediated autophagy and diminish drug resistance, promotes killing of GC cells and decreases their drug tolerance (Kim et al., 2018a). In addition, natural plant components of Saussurea lappa Clarke, Dioscorea nipponica Makino, and Melandrium firmum have been found to induce antiproliferative and apoptotic functions (Rahman et al., 2013; Rahman et al., 2014; Rahman et al., 2015). Latcripin 1 (LP1) was found to arrest S-phase cell cycle as well as decrease matrix metalloproteinase 2 (MMP-2) and MMP-9 expression with induction formation of autophagosomes by AKT/mTOR pathway against GC cell lines SGC-7901 and BGC-823 (Batool et al., 2018). Allicin has been found to increase autophagy in human GC cell MGC-803, BGC-823, and SGC-7901 cells via regulation of p38 signaling (Zhang et al., 2015). Kaempferol, a flavonoid isolated from fruits and vegetables, induces autophagic cell death through IRE1-JNK-CHOP activation in response to ER stress in human GC cell lines (Kim et al., 2018b). Numerous phytochemicals and their response in GC by autophagy signaling are presented in Figure 5. Therefore, autophagy induction by phytochemical might possibly be targeted as a potential therapeutic approach to control GC.
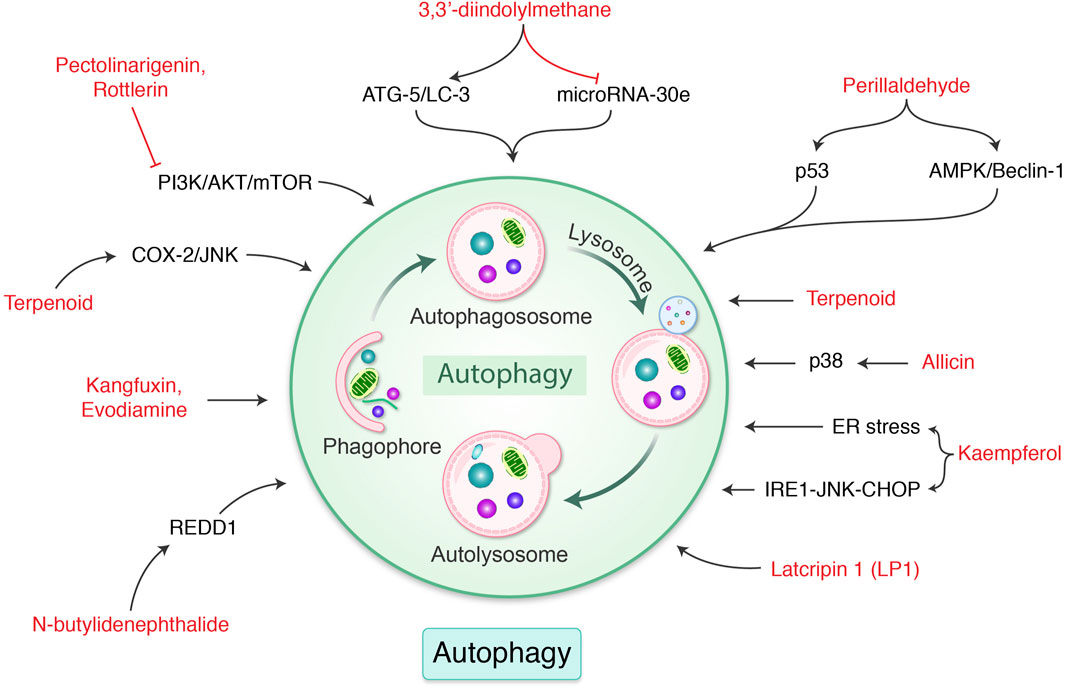
FIGURE 5. Phytochemicals modulate autophagy-mediated cell death in gastric cancer. Different naturally occurring molecules are regulated ER stress, p38, p53, ATG gene, COX-2, and mTOR pathway, which modulate autophagy in gastric cancer. Each compound is well-described in the text.
8 Role of Autophagy Activators and Inhibitors on GC
There are no clear evidences whether autophagy has a cancer-promoter or a cancer-suppressor function in GC. It has been found that autophagy plays an important role in the human cancer patient who suffers chemoresistance, and development of molecule that acts as activator or inhibitor for autophagy may be an open novel resource to treat GC (Mele et al., 2020; Mendes et al., 2021). CQ and hydroxychloroquine (HCQ) are approved by the Food and Drug Administration for clinical application, which can be blocking degradation and the fusion step of autophagosome as a result of regulating autophagy to illustrate the dual role of autophagy in cancer. At present, multiple types of tumors are treated by CQ and HCQ separately or mixed with chemotherapy (Lin et al., 2017). Moreover, it has been found that abnormal expression of autophagy genes may cause some cancer-related pathology. Kim et al. (2020) discovered that genipin, derived from Gardenia jasminoides, can boost p53, a tumor-suppressor protein p53 and DRAM, trigger apoptotic cell death and autophagy, and improve the susceptibility of AGS and MKN45 GC cell lines to OXA by increasing p53 and DRAM (Kim et al., 2020). According to Xu et al. (2018), tanshinone IIA can diminishes the expression of MRP1 and impede adriamycin efflux (ADM) (Mirzaei et al., 2021). The mixture of ADM and tanshinone IIA may also increase the sensitivity of GC cells to ADM by inducing autophagy and boosting the death of the cells (Xu et al., 2020a). TCM licorice contains a compound known as liqueritin that promotes beclin-1 expression and reduces p62 expression, which activates autophagy (Zhao et al., 2021). DSGOST, a traditional Korean herbal remedy induced by several chemotherapy medications, is often utilized to activate the AMPK/ULK1 pathway, increasing autophagy flux, inducing autophagy and apoptosis, and increasing the sensitivity of the GC cell lines AGS and SNU-638 (Kim et al., 2018a). Furthermore, excessive or aberrant activity of autophagic may activate cytotoxicity in addition to contributing to intracellular components’ improper degradation, which is essential for maintaining cancer cell survival in GC cells (Qian and Yang 2016). Researchers have discovered that the herbal remedies cucurbitacin B and phloretin have anticancer properties and reverse chemotherapy resistance in recent years (Choi 2019; Fabiani 2020). Several investigations on their mechanisms of action have revealed that cucurbitacin B inhibits CIP2A (cancerous inhibitor of protein phosphatase 2A), which then reactivates PP2A (protein phosphatase 2A), hence increasing PP2A-dependent mTORC1 inactivation and decreasing PP2A-independent mTORC1 activation (Liu et al., 2017). Phloretin suppresses ERK1/2 and MAPK p38 phosphorylation and enhances LC-3B II and beclin-1 expression, thereby initiating autophagy and increasing the susceptibility of GC cells to ADM in vitro (You et al., 2020). The presence of irregular glycosylation has long been considered a warning sign of cancer, and this has been linked to tumor growth, progression, metastasis, and resistance to chemotherapy (Wang et al., 2020). In GC cells, 5-FU was found to induce cell proliferation arrest, as well as autophagic cell death via beclin-1 upregulation, which significantly enhanced autophagy and reduced cancer cell growth. Therefore, inducing autophagy may efficiently control GC (Yang and Pan 2016).
Numerous therapeutic can promote protective autophagy in cancer cells, thereby preventing cancer cells from undergoing drug-induced apoptosis. One study demonstrated that oxaliplatin can partially antagonize apoptotic cell death in GC MGC803 cells that protect autophagy-induced cell death (Xu et al., 2011). Tunicamycin was first discovered as a natural antibacterial and anticancer chemical because it is an effective glycosylation inhibitor (Wu et al., 2018a). Inhibiting N-glycosylation with tunicamycin boosts ER stress and autophagy while increasing the susceptibility of GC cells to ADM and VCR (Wu et al., 2018a). In contrast, OGT inhibitor–mediated autophagy was significantly attenuated by 3-MA, a blocker of autophagosome formation, however, when pretreated with CQ (Rahman et al., 2019; Rahman et al., 2020a). Indomethacin, a well-known NSAID, has been used successfully as a coadjutant in the development of anticancer medicines (López-Contreras et al., 2020). In AGS cells, indomethacin increases OXA-induced cell death, increases p62 and NBR1 accumulation, impairs lysosomal activity, and inhibits autophagic destruction (Vallecillo-Hernández et al., 2018). DDP reduces MALAT1 expression, whereas propofol increases the inhibitory effects of miR-30e on ATG5 and autophagy, making GC highly susceptible to DDP both in vitro and in vivo (Ashrafizadeh et al., 2020; Tabnak et al., 2021).
9 Conclusion and Prospects
In tumor progression, autophagy plays a complicated task and diverse consequences, depending on what type of tumor and its phases. The role of autophagy has theoretical as well as clinical significance to maintain cellular homeostasis. In the initial stages of malignant transformation or cancer formation, autophagy appears to restrict tumor growth, but in the late stages, autophagy seems to enhance tumor survival as well as tolerance to chemotherapy. Several factors may control the intracellular autophagy level, which determines the effectiveness of antitumor therapies based on autophagy modulation in GC. While the PI3K and mTOR regulatory systems have been proven as major signaling routes governing autophagy, alternative autophagy-related mechanisms (p53, MAPK, or PTEN) must be investigated in further studies. The importance of miRNA and Helicobacter pylori in the control of autophagy in GC regulation is rapidly being recognized, while autophagy stimulators and blockers have obtained considerable clinical testing results in the treatment of GC, which has potential therapeutic approaches as well. Nevertheless, the dual role of autophagy therapy in GC should also be addressed. Many investigations, from the other side, have shown that autophagy and apoptosis can coexist or happen in consecutive order, and they can combine to modulate cancer regulation and management. Because of the extreme variation and unclear source of antitumor drug resistance, which causes inefficient medication and poor diagnosis in patients with progressive of GC, current research has summarized the complicated interaction involving autophagy and chemotherapy protest in GC. Although rapid recognition and monitoring of GC are critical stages in the therapeutic process, as a result, associated proteins and ATGs are accused of being engaged in autophagy methods and are predicted to get more new targets and diagnostic markers for GC treatments. Even more research into autophagy indicators may offer a new option for prognostic indicators and medicinal objectives for GC. Furthermore, more data suggest that too many organic ingredients can cause chemotherapy tolerance by controlling signal transduction. As a result, natural substances, either alone or in conjunction with autophagy modulators and/or chemotherapeutic medicines, might have a beneficial impact on drug-resistant malignancy in GC. However, additional research is required to discover molecular mechanisms and particular objectives, as well as to validate the efficacy and safeness of these treatments in medically significant in GC cancer models. It should be confirmed how autophagy functioning is controlled variably in GC, or which variables induce tissue-specific suppression and/or stimulation of autophagy.
Author Contributions
Idea and conceptualization by MAR. Figures are drawing by MHR. Writing and original draft preparation by KRA and MNP. Visualization and supervision by BK. All authors have read and agreed to the published version of the manuscripts.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1I1A2066868), the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1A5A2019413), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number : HF20C0116), and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HF20C0038).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agrotis, A., and Ketteler, R. (2020). On ATG4B as Drug Target for Treatment of Solid Tumours-The Knowns and the Unknowns. Cells 9, 53. doi:10.3390/cells9010053
Ahmadpour, S., Khodadust, F., Hormati, A., and Eivaziatashbeik, K. (2021). Pivotal Role of Peptides in Gastric Carcinoma: Diagnosis and Therapy. Int. J. Pept. Res. Ther. 27, 503–525. doi:10.1007/s10989-020-10104-9
Ahmed, M. A. E., Mohanad, M., Ahmed, A. A. E., Aboulhoda, B. E., and El-Awdan, S. A. (2021). Mechanistic Insights into the Protective Effects of Chlorogenic Acid against Indomethacin-Induced Gastric Ulcer in Rats: Modulation of the Cross Talk between Autophagy and Apoptosis Signaling. Life Sci. 275, 119370. doi:10.1016/j.lfs.2021.119370
Alers, S., Löffler, A. S., Wesselborg, S., and Stork, B. (2012). Role of AMPK-mTOR-Ulk1/2 in the Regulation of Autophagy: Cross Talk, Shortcuts, and Feedbacks. Mol. Cel Biol 32, 2–11. doi:10.1128/MCB.06159-11
Ariosa, A. R., Lahiri, V., Lei, Y., Yang, Y., Yin, Z., Zhang, Z., et al. (2021). A Perspective on the Role of Autophagy in Cancer. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166262. doi:10.1016/j.bbadis.2021.166262
Ashrafizadeh, M., Zarrabi, A., Hushmandi, K., Kalantari, M., Mohammadinejad, R., Javaheri, T., et al. (2020). Association of the Epithelial-Mesenchymal Transition (EMT) with Cisplatin Resistance. Int. J. Mol. Sci. 21, 4002. doi:10.3390/ijms21114002
Bai, X. Y., Liu, Y. G., Song, W., Li, Y. Y., Hou, D. S., Luo, H. M., et al. (2018). Anticancer Activity of Tetrandrine by Inducing Pro-death Apoptosis and Autophagy in Human Gastric Cancer Cells. J. Pharm. Pharmacol. 70, 1048–1058. doi:10.1111/jphp.12935
Batool, S., Joseph, T. P., Hussain, M., Vuai, M. S., Khinsar, K. H., Din, S. R. U., et al. (2018). LP1 from Lentinula Edodes C91-3 Induces Autophagy, Apoptosis and Reduces Metastasis in Human Gastric Cancer Cell Line SGC-7901. Int. J. Mol. Sci. 19, 2986. doi:10.3390/ijms19102986
Belounis, A., Nyalendo, C., Le Gall, R., Imbriglio, T. V., Mahma, M., Teira, P., et al. (2016). Autophagy Is Associated with Chemoresistance in Neuroblastoma. BMC cancer 16, 1–14. doi:10.1186/s12885-016-2906-9
Botti, J., Djavaheri-Mergny, M., Pilatte, Y., and Codogno, P. (2006). Autophagy Signaling and the Cogwheels of Cancer. Autophagy 2, 67–73. doi:10.4161/auto.2.2.2458
Brajenovic, M., Joberty, G., Küster, B., Bouwmeester, T., and Drewes, G. (2004). Comprehensive Proteomic Analysis of Human Par Protein Complexes Reveals an Interconnected Protein Network. J. Biol. Chem. 279, 12804–12811. doi:10.1074/jbc.M312171200
Cai, W. Y., Dong, Z. N., Fu, X. T., Lin, L. Y., Wang, L., Ye, G. D., et al. (2020). Identification of a Tumor Microenvironment-Relevant Gene Set-Based Prognostic Signature and Related Therapy Targets in Gastric Cancer. Theranostics 10, 8633–8647. doi:10.7150/thno.47938
Chen, J-F., Wu, P., Xia, R., Yang, J., Huo, X-Y., Gu, D-Y., et al. (2018). STAT3-induced lncRNA HAGLROS Overexpression Contributes to the Malignant Progression of Gastric Cancer Cells via mTOR Signal-Mediated Inhibition of Autophagy. Mol. Cancer 17, 1–16. doi:10.1186/s12943-017-0756-y
Chen, S., Wu, J., Jiao, K., Wu, Q., Ma, J., Chen, D., et al. (2018). MicroRNA-495-3p Inhibits Multidrug Resistance by Modulating Autophagy through GRP78/mTOR axis in Gastric Cancer. Cel Death Dis. 9, 1–12. doi:10.1038/s41419-018-0950-x
Chen, L., He, M., Zhang, M., Sun, Q., Zeng, S., Zhao, H., et al. (2021). The Role of Non-coding RNAs in Colorectal Cancer, with a Focus on its Autophagy. Pharmacol. Ther. 226, 107868. doi:10.1016/j.pharmthera.2021.107868
Chen, W., Zou, P., Zhao, Z., Chen, X., Fan, X., Vinothkumar, R., et al. (2021). Corrigendum to"Synergistic Antitumor Activity of Rapamycin and EF24 via Increasing ROS for the Treatment of Gastric Cancer. Redox Biol. 10, 78–89. doi:10.1016/j.redox.2016.09.006
Chen, P. P., Ma, X. Y., Lin, Q., Xu, H. L., Shi, H. X., Zhang, H. Y., et al. (2017). Kangfuxin Promotes Apoptosis of Gastric Cancer Cells through Activating ERstress and Autophagy. Mol. Med. Rep. 16, 9043–9050. doi:10.3892/mmr.2017.7716
Choi, B. Y. (2019). Biochemical Basis of Anti-cancer-effects of Phloretin—A Natural Dihydrochalcone. Molecules 24, 278. doi:10.3390/molecules24020278
Chun, J., Kang, M., and Kim, Y. S. (2014). A Triterpenoid Saponin from Adenophora Triphylla Var. Japonica Suppresses the Growth of Human Gastric Cancer Cells via Regulation of Apoptosis and Autophagy. Tumour Biol. 35, 12021–12030. doi:10.1007/s13277-014-2501-0
Ding, Y. P., Yang, X. D., Wu, Y., and Xing, C. G. (2014). Autophagy Promotes the Survival and Development of Tumors by Participating in the Formation of Vasculogenic Mimicry. Oncol. Rep. 31, 2321–2327. doi:10.3892/or.2014.3087
Du, X., Liu, B., Luan, X., Cui, Q., and Li, L. (2018). miR-30 Decreases Multidrug Resistance in Human Gastric Cancer Cells by Modulating Cell Autophagy. Exp. Ther. Med. 15, 599–605. doi:10.3892/etm.2017.5354
Du, Y., and Ji, X. (2014). Bcl‐2 Down‐regulation by Small Interfering RNA Induces Beclin1‐dependent Autophagy in Human SGC‐7901 Cells. Cel Biol. Int. 38, 1155–1162. doi:10.1002/cbin.10333
Fan, H., Jiang, M., Li, B., He, Y., Huang, C., Luo, D., et al. (2018). MicroRNA-let-7a Regulates Cell Autophagy by Targeting Rictor in Gastric Cancer Cell Lines MGC-803 and SGC-7901. Oncol. Rep. 39, 1207–1214. doi:10.3892/or.2018.6194
Fan, Y., Shen, B., Tan, M., Mu, X., Qin, Y., Zhang, F., et al. (2014). Long Non‐coding RNA UCA 1 Increases Chemoresistance of Bladder Cancer Cells by Regulating Wnt Signaling. FEBS J. 281, 1750–1758. doi:10.1111/febs.12737
Faubert, B., Vincent, E. E., Poffenberger, M. C., and Jones, R. G. (2015). The AMP-Activated Protein Kinase (AMPK) and Cancer: many Faces of a Metabolic Regulator. Cancer Lett. 356, 165–170. doi:10.1016/j.canlet.2014.01.018
Fletcher, R., Tong, J., Risnik, D., Leibowitz, B. J., Wang, Y-J., Concha-Benavente, F., et al. (2021). Non-steroidal Anti-inflammatory Drugs Induce Immunogenic Cell Death in Suppressing Colorectal Tumorigenesis. Oncogene 40, 2035–2050. doi:10.1038/s41388-021-01687-8
Fratter, A., Biagi, D., Giacomini, I., Montopoli, M., and Cocetta, V. (2021). Novel Adenosine Triphosphate-Based Nutraceutical Formulation to Prevent Non-steroidal Anti-inflammatory Drug Enteric Cell Toxicity: Preliminary In Vitro Evidence. J. Med. Food 24 (12), 1293–1303. doi:10.1089/jmf.2021.0019
Ge, J., Chen, Z., Huang, J., Chen, J., Yuan, W., Deng, Z., et al. (2014). Upregulation of Autophagy-Related Gene-5 (ATG-5) Is Associated with Chemoresistance in Human Gastric Cancer. PloS one 9, e110293. doi:10.1371/journal.pone.0110293
Goldsmith, J., Levine, B., and Debnath, J. (2014). Autophagy and Cancer Metabolism. Method Enzymol. 542, 25–57. doi:10.1016/B978-0-12-416618-9.00002-9
Gu, Y., Fei, Z., and Zhu, R. (2020). miR-21 Modulates Cisplatin Resistance of Gastric Cancer Cells by Inhibiting Autophagy via the PI3K/Akt/mTOR Pathway. Anti-cancer drugs 31, 385–393. doi:10.1097/CAD.0000000000000886
Guo, F., Jiao, D., Sui, G., Sun, L., Gao, Y., Fu, Q., et al. (2018). Anticancer Effect of YWHAZ Silencing via Inducing Apoptosis and Autophagy in Gastric Cancer Cells. Neoplasma 65, 693–700. doi:10.4149/neo_2018_170922N603
Guo, W., Chen, Z., Chen, Z., Yu, J., Liu, H., Li, T., et al. (2018). Promotion of Cell Proliferation through Inhibition of Cell Autophagy Signalling Pathway by Rab3IP Is Restrained by microRNA-532-3p in Gastric Cancer. J. Cancer 9, 4363. doi:10.7150/jca.27533
Gupta, P., Kumar, N., and Garg, M. (2021). Emerging Roles of Autophagy in the Development and Treatment of Urothelial Carcinoma of the Bladder. Expert Opin. Ther. Targets 25, 787–797. doi:10.1080/14728222.2021.1992384
Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Mäkelä, T. P., et al. (2003). Complexes between the LKB1 Tumor Suppressor, STRAD Alpha/beta and MO25 Alpha/beta Are Upstream Kinases in the AMP-Activated Protein Kinase cascade. J. Biol. 2, 28. doi:10.1186/1475-4924-2-28
He, H., Huang, J., Wu, S., Jiang, S., Liang, L., Liu, Y., et al. (2021). The Roles of GTPase-Activating Proteins in Regulated Cell Death and Tumor Immunity. J. Hematol. Oncol. 14, 1–15. doi:10.1186/s13045-021-01184-1
Hoang, H. T., and Vu, T. Q. (2021). P2-2 A Study of EOX as Adjuvant Chemotherapy post D2 Resection in Vietnamese Patients with Locally Advanced Gastric Cancer. Ann. Oncol. 32, S330. doi:10.1016/j.annonc.2021.05.671
Honma, Y., Miyagawa, K., Hara, Y., Hayashi, T., Kusanaga, M., Ogino, N., et al. (2021). Correlation of Hepatitis C Virus-Mediated Endoplasmic Reticulum Stress with Autophagic Flux Impairment and Hepatocarcinogenesis. Med. Mol. Morphol. 54, 108–121. doi:10.1007/s00795-020-00271-5
Hou, S. W., Guo, M. F., Xi, H. Y., Zhang, L. Q., Zhao, A. L., Hou, H., et al. (2020). MicroRNA-153-3p Sensitizes Melanoma Cells to Dacarbazine by Suppressing ATG5-Mediated Autophagy and Apoptosis. Transl Cancer Res. 9, 5626–5636. doi:10.21037/tcr-20-2660
Hu, S., Yin, J., Yan, S., Hu, P., Huang, J., Zhang, G., et al. (2021). Chaetocochin J, an Epipolythiodioxopiperazine Alkaloid, Induces Apoptosis and Autophagy in Colorectal Cancer via AMPK and PI3K/AKT/mTOR Pathways. Bioorg. Chem. 109, 104693. doi:10.1016/j.bioorg.2021.104693
Hu, Y-F., Lei, X., Zhang, H-Y., Ma, J-w., Yang, W-w., Chen, M-l., et al. (2015). Expressions and Clinical Significance of Autophagy-Related Markers Beclin1, LC3, and EGFR in Human Cervical Squamous Cell Carcinoma. OncoTargets Ther. 8, 2243. doi:10.2147/OTT.S86844
Hurley, J. H., and Young, L. N. (2017). Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 86, 225–244. doi:10.1146/annurev-biochem-061516-044820
Jangra, A., Arora, M. K., Kisku, A., and Sharma, S. (2020). The Multifaceted Role of Mangiferin in Health and Diseases: a Review. Adv. Tradit Med. 21, 619–643. doi:10.1007/s13596-020-00471-5
Jin, X., Zhu, Z., and Shi, Y. (2014). Metastasis Mechanism and Gene/protein Expression in Gastric Cancer with Distant Organs Metastasis. Bull. Cancer 101, 1–12. doi:10.1684/bdc.2013.1882
Jung, S., Jeong, H., and Yu, S. W. (2020). Autophagy as a Decisive Process for Cell Death. Exp. Mol. Med. 52, 921–930. doi:10.1038/s12276-020-0455-4
Kang, M. R., Kim, M. S., Oh, J. E., Kim, Y. R., Song, S. Y., Kim, S. S., et al. (2009). Frameshift Mutations of Autophagy-Related Genes ATG2B, ATG5, ATG9B and ATG12 in Gastric and Colorectal Cancers with Microsatellite Instability. J. Pathol. 217, 702–706. doi:10.1002/path.2509
Kang, R., Zeh, H. J., Lotze, M. T., and Tang, D. (2011). The Beclin 1 Network Regulates Autophagy and Apoptosis. Cell Death Differ 18, 571–580. doi:10.1038/cdd.2010.191
Kang, S. Y., Hwang, D., Shin, S., Park, J., Kim, M., Rahman, M. D. H., et al. (2021). Potential of Bioactive Food Components against Gastric Cancer: Insights into Molecular Mechanism and Therapeutic Targets. Cancers (Basel) 13, 4502. doi:10.3390/cancers13184502
Kardideh, B., Samimi, Z., Norooznezhad, F., Kiani, S., and Mansouri, K. (2019). Autophagy, Cancer and Angiogenesis: where Is the Link. Cell Biosci 9, 65. doi:10.1186/s13578-019-0327-6
Khaleel, R. A. (2021). Autophagy as a Molecular Target for Cancer Treatment. Eur. J. Pharm. Sci. 8, 1946–1962. doi:10.1016/j.ejps.2019.04.011
Kim, B. R., Jeong, Y. A., Kim, D. Y., Kim, J. L., Jeong, S., Na, Y. J., et al. (2020). Genipin Increases Oxaliplatin-Induced Cell Death through Autophagy in Gastric Cancer. J. Cancer 11, 460. doi:10.7150/jca.34773
Kim, T. W., Lee, S. Y., Kim, M., Cheon, C., Jang, B-H., Shin, Y. C., et al. (2018a). DSGOST Regulates Resistance via Activation of Autophagy in Gastric Cancer. Cel Death Dis. 9, 1–13. doi:10.1038/s41419-018-0658-y
Kim, T. W., Lee, S. Y., Kim, M., Cheon, C., and Ko, S. G. (2018b). Kaempferol Induces Autophagic Cell Death via IRE1-JNK-CHOP Pathway and Inhibition of G9a in Gastric Cancer Cells. Cell Death Dis 9, 875. doi:10.1038/s41419-018-0930-1
Kotani, T., Kirisako, H., Koizumi, M., Ohsumi, Y., and Nakatogawa, H. (2018). The Atg2-Atg18 Complex Tethers Pre-autophagosomal Membranes to the Endoplasmic Reticulum for Autophagosome Formation. P Natl. Acad. Sci. USA 115, 10363–10368. doi:10.1073/pnas.1806727115
Krizanova, O., Babula, P., and Pacak, K. (2016). Stress, Catecholaminergic System and Cancer. Stress 19, 419–428. doi:10.1080/10253890.2016.1203415
Kumar, A., Singh, U. K., and Chaudhary, A. (2015). Targeting Autophagy to Overcome Drug Resistance in Cancer Therapy. Future Med. Chem. 7, 1535–1542. doi:10.4155/fmc.15.88
Kwon, Y. H., Bishayee, K., Rahman, M. A., Hong, J. S., Lim, S. S., and Huh, S. O. (2015). Morus alba Accumulates Reactive Oxygen Species to Initiate Apoptosis via FOXO-Caspase 3-Dependent Pathway in Neuroblastoma Cells. Mol. Cell 38, 630–637. doi:10.14348/molcells.2015.0030
Le, C. P., Nowell, C. J., Kim-Fuchs, C., Botteri, E., Hiller, J. G., Ismail, H., et al. (2016). Chronic Stress in Mice Remodels Lymph Vasculature to Promote Tumour Cell Dissemination. Nat. Commun. 7, 1–14. doi:10.1038/ncomms10634
Lee, H. J., Venkatarame Gowda Saralamma, V., Kim, S. M., Ha, S. E., Raha, S., Lee, W. S., et al. (2018). Pectolinarigenin Induced Cell Cycle Arrest, Autophagy, and Apoptosis in Gastric Cancer Cell via PI3K/AKT/mTOR Signaling Pathway. Nutrients 10–1043. doi:10.3390/nu10081043
Li, H., He, C., Wang, X., Wang, H., Nan, G., and Fang, L. (2019). MicroRNA-183 Affects the Development of Gastric Cancer by Regulating Autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR Signals. Artif. Cell nanomedicine, Biotechnol. 47, 3163–3171. doi:10.1080/21691401.2019.1642903
Li, L., Zhou, L., Li, Y., Lin, S., and Tomuleasa, C. (2014). MicroRNA-21 Stimulates Gastric Cancer Growth and Invasion by Inhibiting the Tumor Suppressor Effects of Programmed Cell Death Protein 4 and Phosphatase and Tensin Homolog. J. BUON 19, 228–236.
Li, Q., Wang, J., Meng, X., Chen, W., Feng, J., and Mao, J. (2021). Identification of Autophagy‐related Gene and lncRNA Signatures in the Prognosis of HNSCC. Oral Dis. doi:10.1111/odi.13889
Li, Y., Gao, S., Du, X., Ji, J., Xi, Y., and Zhai, G. (2021). Advances in Autophagy as a Target in the Treatment of Tumours. J. Drug Target., 1–22. doi:10.1080/1061186x.2021.1961792
Li, Y., Zhang, J., Ma, H., Chen, X., Liu, T., Jiao, Z., et al. (2013). Protective Role of Autophagy in Matrineinduced Gastric Cancer Cell Death. Int. J. Oncol. 42, 1417–1426. doi:10.3892/ijo.2013.1817
Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., et al. (1999). Induction of Autophagy and Inhibition of Tumorigenesis by Beclin 1. Nature 402, 672–676. doi:10.1038/45257
Liao, K. F., Chiu, T. L., Huang, S. Y., Hsieh, T. F., Chang, S. F., Ruan, J. W., et al. (2018). Anti-Cancer Effects of Radix Angelica Sinensis (Danggui) and N-Butylidenephthalide on Gastric Cancer: Implications for REDD1 Activation and mTOR Inhibition. Cell Physiol Biochem 48, 2231–2246. doi:10.1159/000492641
Lin, C., Blessing, A. M., Pulliam, T. L., Shi, Y., Wilkenfeld, S. R., Han, J. J., et al. (2021). Inhibition of CAMKK2 Impairs Autophagy and Castration-Resistant Prostate Cancer via Suppression of AMPK-ULK1 Signaling. Oncogene 40, 1690–1705. doi:10.1038/s41388-021-01658-z
Lin, J-X., Xu, Y-C., Lin, W., Xue, F-Q., Ye, J-X., Zang, W-D., et al. (2021). Effectiveness and Safety of Apatinib Plus Chemotherapy as Neoadjuvant Treatment for Locally Advanced Gastric Cancer: A Nonrandomized Controlled Trial. JAMA Netw. Open 4, e2116240. doi:10.1001/jamanetworkopen.2021.16240
Lin, J. P., Lin, J. X., Ma, Y. B., Xie, J. W., Yan, S., Wang, J. B., et al. (2020). Prognostic Significance of Pre- and post-operative Tumour Markers for Patients with Gastric Cancer. Br. J. Cancer 123, 418–425. doi:10.1038/s41416-020-0901-z
Lin, J. X., Wang, Z. K., Huang, Y. Q., Xie, J. W., Wang, J. B., Lu, J., et al. (2021). Dynamic Changes in Pre- and Postoperative Levels of Inflammatory Markers and Their Effects on the Prognosis of Patients with Gastric Cancer. J. Gastrointest. Surg. 25, 387–396. doi:10.1007/s11605-020-04523-8
Lin, Y-C., Lin, J-F., Wen, S-I., Yang, S-C., Tsai, T-F., Chen, H-E., et al. (2017). Chloroquine and Hydroxychloroquine Inhibit Bladder Cancer Cell Growth by Targeting Basal Autophagy and Enhancing Apoptosis. Kaohsiung J. Med. Sci. 33, 215–223. doi:10.1016/j.kjms.2017.01.004
Liu, H., Song, J., Zhou, Y., Cao, L., Gong, Y., Wei, Y., et al. (2019). Methylxanthine Derivatives Promote Autophagy in Gastric Cancer Cells Targeting PTEN. Anti-cancer drugs 30, 347–355. doi:10.1097/CAD.0000000000000724
Liu, X., Duan, C., Ji, J., Zhang, T., Yuan, X., Zhang, Y., et al. (2017). Cucurbitacin B Induces Autophagy and Apoptosis by Suppressing CIP2A/PP2A/mTORC1 Signaling axis in Human Cisplatin Resistant Gastric Cancer Cells. Oncol. Rep. 38, 271–278. doi:10.3892/or.2017.5648
Liu, X., Zheng, Q., Yu, Q., Hu, Y., Cheng, Y., Shao, Z., et al. (2021). Apatinib Regulates the Growth of Gastric Cancer Cells by Modulating Apoptosis and Autophagy. Naunyn-Schmiedeberg's Arch. Pharmacol. 394, 1009–1018. doi:10.1007/s00210-020-02018-6
Liu, X. L., Ma, B., Chen, M. L., Zhang, Y. Q., Ma, Z., and Chen, H. (2021). Prognostic Autophagy-Related Genes of Gastric Cancer Patients on Chemotherapy. Front. Genet. 12. doi:10.3389/fgene.2021.720849
López-Contreras, F., Muñoz-Uribe, M., Pérez-Laines, J., Ascencio-Leal, L., Rivera-Dictter, A., Martin-Martin, A., et al. (2020). Searching for Drug Synergy against Cancer through Polyamine Metabolism Impairment: Insight into the Metabolic Effect of Indomethacin on Lung Cancer Cells. Front. Pharmacol. 10, 1670. doi:10.3389/fphar.2019.01670
Maiuri, M., Tasdemir, E., Criollo, A., Morselli, E., Vicencio, J., Carnuccio, R., et al. (2009). Control of Autophagy by Oncogenes and Tumor Suppressor Genes. Cel Death Differ. 16, 87–93. doi:10.1038/cdd.2008.131
Maiuri, M., Zalckvar, E., Kimchi, A., and Kroemer, G. (2007). Self-eating and Self-Killing: Crosstalk between Autophagy and Apoptosis. Nat. Rev. Mol. Cel Biol 8, 741–752. doi:10.1038/nrm2239
Mandhair, H. K., Novak, U., and Radpour, R. (2021). Epigenetic Regulation of Autophagy: A Key Modification in Cancer Cells and Cancer Stem Cells. World J. Stem Cell 13, 542–567. doi:10.4252/wjsc.v13.i6.542
Mele, L., Del Vecchio, V., Liccardo, D., Prisco, C., Schwerdtfeger, M., Robinson, N., et al. (2020). The Role of Autophagy in Resistance to Targeted Therapies. Cancer Treat. Rev. 88, 102043. doi:10.1016/j.ctrv.2020.102043
Mendes, F., Pereira, E., Martins, D., Tavares-Silva, E., Pires, A. S., Abrantes, A. M., et al. (2021). Oxidative Stress in Bladder Cancer: an Ally or an Enemy. Mol. Biol. Rep. 48, 1–12. doi:10.1007/s11033-021-06266-4
Menon, M. B., and Dhamija, S. (2018). Beclin 1 Phosphorylation - at the Center of Autophagy Regulation. Front Cel Dev Biol 6. doi:10.3389/fcell.2018.00137
Mirzaei, S., Zarrabi, A., Hashemi, F., Zabolian, A., Saleki, H., Azami, N., et al. (2021). Nrf2 Signaling Pathway in Chemoprotection and Doxorubicin Resistance: Potential Application in Drug Discovery. Antioxidants 10, 349. doi:10.3390/antiox10030349
Mitra, S., and Dash, R. (2018). Natural Products for the Management and Prevention of Breast Cancer. Evid. Based Complement. Alternat Med. 2018, 8324696. doi:10.1155/2018/8324696
Mody, D., Bouckaert, J., Savvides, S. N., and Gupta, V. (2021). Rational Design and Development of HDAC Inhibitors for Breast Cancer Treatment. Curr. Pharm. Des. 27, 4610–4629. doi:10.2174/1381612827666210917143953
Mukhopadhyay, S., Mahapatra, K. K., Praharaj, P. P., Patil, S., and Bhutia, S. K. (2021). Recent Progress of Autophagy Signaling in Tumor Microenvironment and its Targeting for Possible Cancer Therapeutics. Semin. Cancer Biol. S1044-579X (21), 00227–233. doi:10.1016/j.semcancer.2021.09.003
Munshi, R., Kandl, K. A., Carr-Schmid, A., Whitacre, J. L., Adams, A. E., and Kinzy, T. (2001). Overexpression of Translation Elongation Factor 1A Affects the Organization and Function of the Actin Cytoskeleton in Yeast. Genetics 157, 1425–1436. doi:10.1093/genetics/157.4.1425
Ni, H. W., Qin, H., Sun, C., Liu, Y. C., Ruan, G. J., Guo, Q. Q., et al. (2021). MiR-375 Reduces the Stemness of Gastric Cancer Cells through Triggering Ferroptosis. Stem Cel Res Ther 12, 325. doi:10.1186/s13287-021-02394-7
Ning, L., Zhang, M. S., Zhu, Q. L., Hao, F. Y., Shen, W. L., and Chen, D. (2020). miR-25-3p Inhibition Impairs Tumorigenesis and Invasion in Gastric Cancer Cells In Vitro and In Vivo. Bioengineered 11, 81–90. doi:10.1080/21655979.2019.1710924
Ouyang, J., Xie, Z. Z., Lei, X. Y., Tang, G. T., Gan, R. L., and Yang, X. Y. (2021). Clinical Crosstalk between microRNAs and Gastric Cancer (Review). Int. J. Oncol. 58, 7. doi:10.3892/ijo.2021.5187
Özeş, A. R., Miller, D. F., Özeş, O. N., Fang, F., Liu, Y., Matei, D., et al. (2016). NF-κB-HOTAIR axis Links DNA Damage Response, Chemoresistance and Cellular Senescence in Ovarian Cancer. Oncogene 35, 5350–5361. doi:10.1038/onc.2016.75
Panda, P. K., Mukhopadhyay, S., Das, D. N., Sinha, N., Naik, P. P., and Bhutia, S. K. (2015). Mechanism of Autophagic Regulation in Carcinogenesis and Cancer Therapeutics. Semin. Cel Dev Biol 39, 43–55. doi:10.1016/j.semcdb.2015.02.013
Park, N. Y., Jo, D. S., Kim, Y. H., Bae, J-E., Kim, J. B., Park, H. J., et al. (2021). Triamterene Induces Autophagic Degradation of Lysosome by Exacerbating Lysosomal Integrity. Arch. Pharm. Res. 44, 621–631. doi:10.1007/s12272-021-01335-5
Patra, S., Mishra, S. R., Behera, B. P., Mahapatra, K. K., Panigrahi, D. P., Bhol, C. S., et al. (2020). Autophagy-modulating Phytochemicals in Cancer Therapeutics: Current Evidences and Future Perspectives. Semin. Cancer Biol. S1044-579X (20), 30104–30108. doi:10.1016/j.semcancer.2020.05.008
Pei, G., Luo, M., Ni, X., Wu, J., Wang, S., Ma, Y., et al. (2018). Autophagy Facilitates Metadherin-Induced Chemotherapy Resistance through the AMPK/ATG5 Pathway in Gastric Cancer. Cell Physiol. Biochem. 46, 847–859. doi:10.1159/000488742
Peng, X., Tu, Y., San Fu, Y. X., Ma, C., Yang, Y., Wu, H., et al. (2019). 14-Deoxycoleon U-Induced Endoplasmic Reticulum Stress-Mediated Apoptosis, Autophagy, and Cell Cycle Arrest in Lung Adenocarcinoma. OncoTargets Ther. 12, 5955. doi:10.2147/OTT.S211933
Poillet-Perez, L., Sarry, J. E., and Joffre, C. (2021). Autophagy Is a Major Metabolic Regulator Involved in Cancer Therapy Resistance. Cell Rep 36, 109528. doi:10.1016/j.celrep.2021.109528
Poole, L. P., and Macleod, K. F. (2021). Mitophagy in Tumorigenesis and Metastasis. Cell Mol Life Sci 78, 3817–3851. doi:10.1007/s00018-021-03774-1
Qian, H. R., and Yang, Y. (2016). Functional Role of Autophagy in Gastric Cancer. Oncotarget 7, 17641–17651. doi:10.18632/oncotarget.7508
Qu, L., Ding, J., Chen, C., Wu, Z. J., Liu, B., Gao, Yi., et al. (2016). Exosome-transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 29, 653–668. doi:10.1016/j.ccell.2016.03.004
Raha, S., Yumnam, S., Hong, G. E., Lee, H. J., Saralamma, V. V. G., Park, H-S., et al. (2015). Naringin Induces Autophagy-Mediated Growth Inhibition by Downregulating the PI3K/Akt/mTOR cascade via Activation of MAPK Pathways in AGS Cancer Cells. Int. J. Oncol. 47, 1061–1069. doi:10.3892/ijo.2015.3095
Rahman, M. A., Bishayee, K., Sadra, A., and Huh, S. O. (2017). Oxyresveratrol Activates Parallel Apoptotic and Autophagic Cell Death Pathways in Neuroblastoma Cells. Biochim. Biophys. Acta Gen. Subj 1861, 23–36. doi:10.1016/j.bbagen.2016.10.025
Rahman, M. A., Cho, Y., Hwang, H., and Rhim, H. (2020a). Pharmacological Inhibition of O-GlcNAc Transferase Promotes mTOR-dependent Autophagy in Rat Cortical Neurons. Brain Sci. 10, 958. doi:10.3390/brainsci10120958
Rahman, M. A., Cho, Y., Nam, G., and Rhim, H. (2021a). Antioxidant Compound, Oxyresveratrol, Inhibits APP Production through the AMPK/ULK1/mTOR-Mediated Autophagy Pathway in Mouse Cortical Astrocytes. Antioxidants-Basel 10, 408. doi:10.3390/antiox10030408
Rahman, M. A., Hong, J. S., and Huh, S. O. (2015). Antiproliferative Properties of Saussurea Lappa Clarke Root Extract in SH-Sy5y Neuroblastoma Cells via Intrinsic Apoptotic Pathway. Anim. Cell Syst 19, 119–126. doi:10.1080/19768354.2015.1008041
Rahman, M. A., Hwang, H., Cho, Y., and Rhim, H. (2019). Modulation of O-GlcNAcylation Regulates Autophagy in Cortical Astrocytes. Oxid Med. Cel Longev 2019, 6279313. doi:10.1155/2019/6279313
Rahman, M. A., Kim, N. H., and Huh, S. O. (2013). Cytotoxic Effect of Gambogic Acid on SH-Sy5y Neuroblastoma Cells Is Mediated by Intrinsic Caspase-dependent Signaling Pathway. Mol. Cel Biochem 377, 187–196. doi:10.1007/s11010-013-1584-z
Rahman, M. A., Rahman, M. H., Hossain, M. S., Biswas, P., Islam, R., Uddin, M. J., et al. (2020b). Molecular Insights into the Multifunctional Role of Natural Compounds: Autophagy Modulation and Cancer Prevention. Biomedicines 8, 517. doi:10.3390/biomedicines8110517
Rahman, M. A., Rahman, M. R., Zaman, T., Uddin, M. S., Islam, R., Abdel-Daim, M. M., et al. (2020c). Emerging Potential of Naturally Occurring Autophagy Modulators against Neurodegeneration. Curr. Pharm. Des. 26, 772–779. doi:10.2174/1381612826666200107142541
Rahman, M. A., Rahman, M. S., Rahman, M. H., Rasheduzzaman, M., Mamun-Or-Rashid, A. N. M., Uddin, M. J., et al. (2021b). Modulatory Effects of Autophagy on APP Processing as a Potential Treatment Target for Alzheimer's Disease. Biomedicines 9, 5. doi:10.3390/biomedicines9010005
Rahman, M. A., and Rhim, H. (2017). Therapeutic Implication of Autophagy in Neurodegenerative Diseases. BMB Rep. 50, 345–354. doi:10.5483/bmbrep.2017.50.7.069
Rahman, M. A., Saha, S. K., Rahman, M. S., Uddin, M. J., Uddin, M. S., Pang, M. G., et al. (2020d). Molecular Insights into Therapeutic Potential of Autophagy Modulation by Natural Products for Cancer Stem Cells. Front. Cel Dev Biol 8. doi:10.3389/fcell.2020.00283
Rahman, M. A., Yang, H., Kim, N. H., and Huh, S. O. (2014). Induction of Apoptosis by Dioscorea Nipponica Makino Extracts in Human SH-Sy5y Neuroblastoma Cells via Mitochondria-Mediated Pathway. Anim. Cell Syst 18, 41–51. doi:10.1080/19768354.2014.880372
Rasul, A., Yu, B., Zhong, L., Khan, M., Yang, H., and Ma, T. (2012). Cytotoxic Effect of Evodiamine in SGC-7901 Human Gastric Adenocarcinoma Cells via Simultaneous Induction of Apoptosis and Autophagy. Oncol. Rep. 27, 1481–1487. doi:10.3892/or.2012.1694
Riquelme, I., Letelier, P., Riffo-Campos, A. L., Brebi, P., and Roa, J. C. (2016). Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int. J. Mol. Sci. 17, 424. doi:10.3390/ijms17030424
Roy, S. G. (2021). Regulation of Autophagy by miRNAs in Human Diseases. Nucleus Calcutta 64, 317–329. doi:10.1007/s13237-021-00378-9
Rubinsztein, D. C., Shpilka, T., and Elazar, Z. (2012). Mechanisms of Autophagosome Biogenesis. Curr. Biol. 22, R29–R34. doi:10.1016/j.cub.2011.11.034
Santagostino, S. F., Assenmacher, C. A., Tarrant, J. C., Adedeji, A. O., and Radaelli, E. (2021). Mechanisms of Regulated Cell Death: Current Perspectives. Vet. Pathol. 58, 596–623. doi:10.1177/03009858211005537
Scazzone, C., Agnello, L., Bivona, G., Lo Sasso, B., and Ciaccio, M. (2021). Vitamin D and Genetic Susceptibility to Multiple Sclerosis. Biochem. Genet. 59, 1–30. doi:10.1007/s10528-020-10010-1
Schaefer, L., and Dikic, I. (2021). Autophagy: Instructions from the Extracellular Matrix. Matrix Biol. 100, 1–8. doi:10.1016/j.matbio.2021.06.002
Scherz-Shouval, R., Weidberg, H., Gonen, C., Wilder, S., Elazar, Z., and Oren, M. (2010). p53-dependent Regulation of Autophagy Protein LC3 Supports Cancer Cell Survival under Prolonged Starvation. Proc. Natl. Acad. Sci. 107, 18511–18516. doi:10.1073/pnas.1006124107
Shafabakhsh, R., Arianfar, F., Vosough, M., Mirzaei, H. R., Mahjoubin-Tehran, M., Khanbabaei, H., et al. (2021). Autophagy and Gastrointestinal Cancers: the behind the Scenes Role of Long Non-coding RNAs in Initiation, Progression, and Treatment Resistance. Cancer Gene Ther. 28, 1229–1255. doi:10.1038/s41417-020-00272-7
Shao, G. Y., Zhao, Z. G., Zhao, W., Hu, G., Zhang, L. Y., Li, W., et al. (2020). Long Non-coding RNA MALAT1 Activates Autophagy and Promotes Cell Proliferation by Downregulating microRNA-204 Expression in Gastric Cancer. Oncol. Lett. 19, 805–812. doi:10.3892/ol.2019.11184
Silberman, A., Goldman, O., Assayag, O. B., Jacob, A., Rabinovich, S., Adler, L., et al. (2019). Acid-induced Downregulation of ASS1 Contributes to the Maintenance of Intracellular pH in Cancer. Cancer Res. 79, 518–533. doi:10.1158/0008-5472.CAN-18-1062
Singh, R., and Cuervo, A. M. (2011). Autophagy in the Cellular Energetic Balance. Cell Metab 13, 495–504. doi:10.1016/j.cmet.2011.04.004
Song, J., Zhou, Y., Gong, Y., Liu, H., and Tang, L. (2018). Rottlerin Promotes Autophagy and Apoptosis in Gastric Cancer Cell Lines. Mol. Med. Rep. 18, 2905–2913. doi:10.3892/mmr.2018.9293
Song, J. H., and Meltzer, S. J. (2012). MicroRNAs in Pathogenesis, Diagnosis, and Treatment of Gastroesophageal Cancers. Gastroenterology 143, 35–47. e32. doi:10.1053/j.gastro.2012.05.003
Spirina, L. V., Avgustinovich, A. V., Afanas' ev, S. G., Cheremisina, O. V., Volkov, M. Y., Choynzonov, E. L., et al. (2020). Molecular Mechanism of Resistance to Chemotherapy in Gastric Cancers, the Role of Autophagy. Curr. Drug Targets 21, 713–721. doi:10.2174/1389450120666191127113854
Sun, D., Hang, X., Ren, J., Sun, Q., Liu, Z., Wang, Y., et al. (2021) Identification of Five m6A-Relevant mRNAs Signature and Risk Score for the Prognostication of Gastric Cancer. doi:10.21203/rs.3.rs-489055/v1
Tabnak, P., Masrouri, S., Geraylow, K. R., Zarei, M., and Esmailpoor, Z. H. (2021). Targeting miRNAs with Anesthetics in Cancer: Current Understanding and Future Perspectives. Biomed. Pharmacother. 144, 112309. doi:10.1016/j.biopha.2021.112309
Tang, J., Zhang, C., Lin, J., Duan, P., Long, J., and Zhu, H. (2021). ALOX5‐5‐HETE Promotes Gastric Cancer Growth and Alleviates Chemotherapy Toxicity via MEK/ERK Activation. Cancer Med. 10, 5246–5255. doi:10.1002/cam4.4066
Tian, L., Zhao, Z., Xie, L., and Zhu, J. (2018). MiR-361-5p Suppresses Chemoresistance of Gastric Cancer Cells by Targeting FOXM1 via the PI3K/Akt/mTOR Pathway. Oncotarget 9, 4886. doi:10.18632/oncotarget.23513
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA Biogenesis and its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cel. Biol. 20, 5–20. doi:10.1038/s41580-018-0059-1
Tsai, C. Y., Chi, H. C., Chi, L. M., Yang, H. Y., Tsai, M. M., Lee, K. F., et al. (2018). Argininosuccinate Synthetase 1 Contributes to Gastric Cancer Invasion and Progression by Modulating Autophagy. FASEB J. 32, 2601–2614. doi:10.1096/fj.201700094R
Uddin, M. S., Al Mamun, A., Rahman, M. A., Behl, T., Perveen, A., Hafeez, A., et al. (2020). Emerging Proof of Protein Misfolding and Interaction in Multifactorial Alzheimer's Disease. Curr. Top. Med. Chem. 20, 2380–2390. doi:10.2174/1568026620666200601161703
Vallecillo-Hernández, J., Barrachina, M. D., Ortiz-Masiá, D., Coll, S., Esplugues, J. V., Calatayud, S., et al. (2018). Indomethacin Disrupts Autophagic Flux by Inducing Lysosomal Dysfunction in Gastric Cancer Cells and Increases Their Sensitivity to Cytotoxic Drugs. Scientific Rep. 8, 3593. doi:10.1038/s41598-018-21455-1
Velazquez, A. F. C., and Jackson, W. T. (2018). So Many Roads: the Multifaceted Regulation of Autophagy Induction. Mol. Cel Biol 38, e00303–18. doi:10.1128/MCB.00303-18
Wang, D., Cabalag, C. S., Clemons, N. J., and DuBois, R. N. (2021). Cyclooxygenases and Prostaglandins in Tumor Immunology and Microenvironment of Gastrointestinal Cancer. Gastroenterology 161, 1813–1829. doi:10.1053/j.gastro.2021.09.059
Wang, J-P., Chi, R-F., Liu, J., Deng, Y-Z., Han, X-B., Qin, F-Z., et al. (2018). The Role of Endogenous Reactive Oxygen Species in Cardiac Myocyte Autophagy. Physiol. Res. 67, 31–40. doi:10.33549/physiolres.933653
Wang, S., You, L., Dai, M., and Zhao, Y. (2020). Mucins in Pancreatic Cancer: A Well‐established but Promising Family for Diagnosis, Prognosis and Therapy. J. Cell Mol. Med. 24, 10279–10289. doi:10.1111/jcmm.15684
Wang, Z-C., Huang, F-Z., Xu, H-B., Sun, J-C., and Wang, C-F. (2019). MicroRNA-137 Inhibits Autophagy and Chemosensitizes Pancreatic Cancer Cells by Targeting ATG5. Int. J. Biochem. Cel. Biol. 111, 63–71. doi:10.1016/j.biocel.2019.01.020
Weng, J., Xiao, J., Mi, Y., Fang, X., Sun, Y., Li, S., et al. (2018). PCDHGA9 Acts as a Tumor Suppressor to Induce Tumor Cell Apoptosis and Autophagy and Inhibit the EMT Process in Human Gastric Cancer. Cel Death Dis. 9, 1–21. doi:10.1038/s41419-017-0189-y
Winkler, J., Abisoye-Ogunniyan, A., Metcalf, K. J., and Werb, Z. (2020). Concepts of Extracellular Matrix Remodelling in Tumour Progression and Metastasis. Nat. Commun. 11, 5120. doi:10.1038/s41467-020-18794-x
Wu, J., Chen, S., Liu, H., Zhang, Z., Ni, Z., Chen, J., et al. (2018a). Tunicamycin Specifically Aggravates ER Stress and Overcomes Chemoresistance in Multidrug-Resistant Gastric Cancer Cells by Inhibiting N-Glycosylation. J. Exp. Clin. Cancer Res. 37, 1–12. doi:10.1186/s13046-018-0935-8
Wu, J., Yu, J., Wang, J., Zhang, C., Shang, K., Yao, X., et al. (2018b). Astragalus Polysaccharide Enhanced Antitumor Effects of Apatinib in Gastric Cancer AGS Cells by Inhibiting AKT Signalling Pathway. Biomed. Pharmacother. 100, 176–183. doi:10.1016/j.biopha.2018.01.140
Xiao, H., Xiao, Y., Chen, P., Quan, H., Luo, J., and Huang, G. (2021). Association Among Blood Transfusion, Postoperative Infectious Complications, and Cancer-specific Survival in Patients with Stage II/III Gastric Cancer after Radical Gastrectomy: Emphasizing Benefit from Adjuvant Chemotherapy. Ann. Surg. Oncol. 28, 2394–2404. doi:10.1245/s10434-020-09102-4
Xiao, H., Xiong, L., Song, X., Jin, P., Chen, L., Chen, X., et al. (2017). Angelica Sinensis Polysaccharides Ameliorate Stress-Induced Premature Senescence of Hematopoietic Cell via Protecting Bone Marrow Stromal Cells from Oxidative Injuries Caused by 5-fluorouracil. Int. J. Mol. Sci. 18, 2265. doi:10.3390/ijms18112265
Xin, L., Zhou, L-Q., Liu, L., Yuan, Y-W., Zhang, H-T., and Zeng, F. (2019). METase Promotes Cell Autophagy via Promoting SNHG5 and Suppressing miR-20a in Gastric Cancer. Int. J. Biol. macromolecules 122, 1046–1052. doi:10.1016/j.ijbiomac.2018.09.051
Xiong, X., Lu, B., Tian, Q., Zhang, H., Wu, M., Guo, H., et al. (2019). Inhibition of Autophagy Enhances Cinobufagin-Induced Apoptosis in Gastric Cancer. Oncol. Rep. 41, 492–500. doi:10.3892/or.2018.6837
Xiu, T., Guo, Q., and Jing, F-B. (2021). Facing Cell Autophagy in Gastric Cancer–What Do We Know So Far. Int. J. Gen. Med. 14, 1647. doi:10.2147/IJGM.S298705
Xu, J-L., Yuan, L., Tang, Y-C., Xu, Z-Y., Xu, H-D., Cheng, X-D., et al. (2020). The Role of Autophagy in Gastric Cancer Chemoresistance: Friend or Foe. Front. Cel Develop. Biol. 8, 1484. doi:10.3389/fcell.2020.621428
Xu, J. J., Wang, X. Q., Wang, W. J., Zhang, L. H., and Huang, P. L. (2021). Candidate Oncogene circularNOP10 Mediates Gastric Cancer Progression by Regulating miR-204/SIRT1 Pathway. J. Gastrointest. Oncol. 12, 1428–1443. doi:10.21037/jgo-21-422
Xu, L., Qu, X. J., Liu, Y. P., Xu, Y. Y., Liu, J., Hou, K. Z., et al. (2011). Protective Autophagy Antagonizes Oxaliplatin-Induced Apoptosis in Gastric Cancer Cells. Chin. J. Cancer 30, 490–496. doi:10.5732/cjc.010.10518
Xu, R., Zhou, X. M., Li, Y. S., Ren, L., and He, X. R. (2021). MicroRNA181a Promotes Epithelialmesenchymal Transition in Esophageal Squamous Cell Carcinoma via the TGFbeta/Smad Pathway. Mol. Med. Rep. 23, 316. doi:10.3892/mmr.2021.11955
Xu, T., Sun, D., Chen, Y., and Ouyang, L. (2020). Targeting mTOR for Fighting Diseases: A Revisited Review of mTOR Inhibitors. Eur. J. Med. Chem. 199, 112391. doi:10.1016/j.ejmech.2020.112391
Xu, X. W., Pan, C. W., Yang, X. M., Zhou, L., Zheng, Z. Q., and Li, D. C. (2018). SP1 Reduces Autophagic Flux through Activating P62 in Gastric Cancer Cells. Mol. Med. Rep. 17, 4633–4638. doi:10.3892/mmr.2018.8400
Xu, Z., Li, Z., Wang, W., Xia, Y., He, Z., Li, B., et al. (2019). MIR-1265 Regulates Cellular Proliferation and Apoptosis by Targeting Calcium Binding Protein 39 in Gastric Cancer and, Thereby, Impairing Oncogenic Autophagy. Cancer Lett. 449, 226–236. doi:10.1016/j.canlet.2019.02.026
Yang, C., and Pan, Y. (2016). Fluorouracil Induces Autophagy-Related Gastric Carcinoma Cell Death through Beclin-1 Upregulation by miR-30 Suppression. Tumor Biol. 37, 15489–15494. doi:10.1007/s13277-015-3775-6
Yao, R-Q., Ren, C., Xia, Z-F., and Yao, Y-M. (2021). Organelle-specific Autophagy in Inflammatory Diseases: a Potential Therapeutic Target Underlying the Quality Control of Multiple Organelles. Autophagy 17, 385–401. doi:10.1080/15548627.2020.1725377
Yap, T. A., Garrett, M. D., Walton, M. I., Raynaud, F., de Bono, J. S., and Workman, P. (2008). Targeting the PI3K–AKT–mTOR Pathway: Progress, Pitfalls, and Promises. Curr. Opin. Pharmacol. 8, 393–412. doi:10.1016/j.coph.2008.08.004
Ye, Y., Fang, Y., Xu, W., Wang, Q., Zhou, J., and Lu, R. (2016). 3,3'-Diindolylmethane Induces Anti-human Gastric Cancer Cells by the miR-30e-ATG5 Modulating Autophagy. Biochem. Pharmacol. 115, 77–84. doi:10.1016/j.bcp.2016.06.018
YiRen, H., YingCong, Y., Sunwu, Y., Keqin, L., Xiaochun, T., Senrui, C., et al. (2017). Long Noncoding RNA MALAT1 Regulates Autophagy Associated Chemoresistance via miR-23b-3p Sequestration in Gastric Cancer. Mol. Cancer 16, 1–12. doi:10.1186/s12943-017-0743-3
You, Q., Xu, J., Zhu, Z., Hu, Z., and Cai, Q. (2020). Phloretin Flavonoid Exhibits Selective Antiproliferative Activity in Doxorubicin-Resistant Gastric Cancer Cells by Inducing Autophagy, Inhibiting Cell Migration and Invasion, Cell Cycle Arrest and Targeting ERK1/2 MAP Pathway. J. BUON 25, 308–313.
Yuan, K-T., Li, B-X., Yuan, Y-J., Tan, M., Tan, J-F., Dai, W-G., et al. (2018). Deregulation of microRNA-375 Inhibits Proliferation and Migration in Gastric Cancer in Association with Autophagy-Mediated AKT/mTOR Signaling Pathways. Techn. Cancer Res. Treat. 17, 1533033818806499. doi:10.1177/1533033818806499
Zhang, X., Li, Z., Xuan, Z., Xu, P., Wang, W., Chen, Z., et al. (2018). Novel Role of miR-133a-3p in Repressing Gastric Cancer Growth and Metastasis via Blocking Autophagy-Mediated Glutaminolysis. J. Exp. Clin. Cancer Res. 37, 1–22. doi:10.1186/s13046-018-0993-y
Zhang, X., Zhu, Y., Duan, W., Feng, C., and He, X. (2015). Allicin Induces Apoptosis of the MGC-803 Human Gastric Carcinoma Cell Line through the P38 Mitogen-Activated Protein Kinase/caspase-3 Signaling Pathway. Mol. Med. Rep. 11, 2755–2760. doi:10.3892/mmr.2014.3109
Zhang, Y., Liu, S., Feng, Q., Huang, X., Wang, X., Peng, Y., et al. (2018). Perilaldehyde Activates AMP-Activated Protein Kinase to Suppress the Growth of Gastric Cancer via Induction of Autophagy. J. Cel Biochem. doi:10.1002/jcb.27491
Zhang, Y., Liu, S., Feng, Q., Huang, X., Wang, X., Peng, Y., et al. (2019). Perilaldehyde Activates AMP‐activated Protein Kinase to Suppress the Growth of Gastric Cancer via Induction of Autophagy. J. Cell. Biochem. 120, 1716–1725. doi:10.1002/jcb.27491
Zhang, Z., Shi, J., Nice, E. C., Huang, C., and Shi, Z. (2021). The Multifaceted Role of Flavonoids in Cancer Therapy: Leveraging Autophagy with a Double-Edged Sword. Antioxidants 10, 1138. doi:10.3390/antiox10071138
Zhao, J., Lan, W., Peng, J., Guan, B., Liu, J., Zhang, M., et al. (2021). Babao Dan Reverses Multiple-Drug Resistance in Gastric Cancer Cells via Triggering Apoptosis and Autophagy and Inhibiting PI3K/AKT/mTOR Signaling. Evid. Based Complement. Alternat Med. 2021, 5631942. doi:10.1155/2021/5631942
Zhao, J., Nie, Y., Wang, H., and Lin, Y. (2016). MiR-181a Suppresses Autophagy and Sensitizes Gastric Cancer Cells to Cisplatin. Gene 576, 828–833. doi:10.1016/j.gene.2015.11.013
Zheng, W., Wu, C., Wu, X., Cai, Y., Liu, B., and Wang, C. (2021). Genetic Variants of Autophagy-Related Genes in the PI3K/Akt/mTOR Pathway and Risk of Gastric Cancer in the Chinese Population. Gene 769, 145190. doi:10.1016/j.gene.2020.145190
Zheng, Y., Tu, J., Wang, X., Yu, Y., Li, J., Jin, Y., et al. (2019). The Therapeutic Effect of Melatonin on GC by Inducing Cell Apoptosis and Autophagy Induced by Endoplasmic Reticulum Stress. OncoTargets Ther. 12, 10187. doi:10.2147/OTT.S226140
Zhi, X., Li, B., Li, Z., Zhang, J., Yu, J., Zhang, L., et al. (2019). Adrenergic Modulation of AMPK-dependent Autophagy by Chronic Stress Enhances Cell Proliferation and Survival in Gastric Cancer. Int. J. Oncol. 54, 1625–1638. doi:10.3892/ijo.2019.4753
Zhu, L., Zhu, Y., Han, S., Chen, M., Song, P., Dai, D., et al. (2019). Impaired Autophagic Degradation of lncRNA ARHGAP5-AS1 Promotes Chemoresistance in Gastric Cancer. Cel Death Dis. 10, 1–16. doi:10.1038/s41419-019-1585-2
Keywords: gastric cancer, autophagy, phytochemical, tumorigenesis, autophagy-related genes, autophagy modulator
Citation: Rahman MA, Ahmed KR, Rahman MH, Park MN and Kim B (2022) Potential Therapeutic Action of Autophagy in Gastric Cancer Managements: Novel Treatment Strategies and Pharmacological Interventions. Front. Pharmacol. 12:813703. doi: 10.3389/fphar.2021.813703
Received: 12 November 2021; Accepted: 13 December 2021;
Published: 28 January 2022.
Edited by:
Aditi Banerjee, University of Maryland, Baltimore, United StatesReviewed by:
Fen Pei, University of Pittsburgh, United StatesNur M. Kocaturk, MRC Protein Phosphorylation and Ubiquitylation Unit (MRC), United Kingdom
Copyright © 2022 Rahman, Ahmed, Rahman, Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Md. Ataur Rahman, YXRhdXIxOTgxcmFobWFuQGhvdG1haWwuY29t; Bonglee Kim, Ym9uZ2xlZWtpbUBraHUuYWMua3I=
†These authors have contributed equally to this work
 Md. Ataur Rahman
Md. Ataur Rahman Kazi Rejvee Ahmed4†
Kazi Rejvee Ahmed4† MD. Hasanur Rahman
MD. Hasanur Rahman Moon Nyeo Park
Moon Nyeo Park Bonglee Kim
Bonglee Kim