- 1School of Mathematics and Statistics, South-Central University for Nationalities, Wuhan, China
- 2College of Informatics, Huazhong Agricultural University, Wuhan, China
- 3School of Computer Science, Wuhan University, Wuhan, China
MiRNAs can regulate genes encoding specific proteins which are related to the efficacy of drugs, and predicting miRNA-drug resistance associations is of great importance. In this work, we propose an attentive multimodal graph convolution network method (AMMGC) to predict miRNA-drug resistance associations. AMMGC learns the latent representations of drugs and miRNAs from four graph convolution sub-networks with distinctive combinations of features. Then, an attention neural network is employed to obtain attentive representations of drugs and miRNAs, and miRNA-drug resistance associations are predicted by the inner product of learned attentive representations. The computational experiments show that AMMGC outperforms other state-of-the-art methods and baseline methods, achieving the AUPR score of 0.2399 and the AUC score of 0.9467. The analysis demonstrates that leveraging multiple features of drugs and miRNAs can make a contribution to the miRNA-drug resistance association prediction. The usefulness of AMMGC is further validated by case studies.
1 Introduction
Drug development is an expensive and time-consuming process, and wet experiments are needed to select the drug targets and ensure the safety as well as the effectiveness of drugs. Although there are more than 25,000 protein-coding genes in the human genome, approved drugs can only target about 600 specific disease-related proteins (Hopkins and Groom, 2002; Overington et al., 2006). MicroRNAs (miRNAs) as one type of non-coding RNAs are identified as potential targets (Huang et al., 2019) due to their involvement in regulating the expression of related genes, and over-expressed/under-expressed expression of miRNAs could down-regulate/up-regulate genes with protein products necessary for drug efficacy/inhibiting drug function, and thus variations in miRNA profiling of patients is the cause of different therapeutics for individuals, especially the drug resistance. A thorough understanding of the impact of miRNA expression on drug resistance is important for drug discovery.
A few efforts have been made on predicting resistance associations between miRNAs and drugs. Dai et al. constructed the ncDR database (Dai et al., 2017), which contains comprehensive information about miRNA-drug resistance associations and laid a solid foundation for further computational analysis. By formulating known miRNA-drug resistance associations as a bipartite graph with miRNA and drug nodes, Huang et al. (Huang et al., 2019) proposed a graph convolution based miRNA-drug resistance association prediction model, named GCMDR, which uses miRNA expression profile and drug fingerprint as features. GCMDR produces satisfying results, but it still remains the space for improvement. A high-accuracy prediction model usually requires diverse information, which reflects different characteristics of drugs, miRNAs, and miRNA-drug resistance associations. Although miRNA expression profiles and drug substructures play important roles in the association prediction, more features about miRNAs and drugs should be taken into account to improve the performances. In recent years, the graph learning methods, especially graph neural networks (GNN), showed great success in biomedical association prediction (Mudiyanselage et al., 2020; Yu et al., 2020; Fu et al., 2021; Lei et al., 2021; Liu et al., 2021; Yang and Lei, 2021; Zhang et al., 2021a). Thus, it is necessary to develop GNN-based multimodal method to address above mentioned issues and improve the miRNA-drug resistance association prediction.
In this work, we propose a novel method, namely attentive multimodal graph convolutional network (AMMGC), to predict miRNA-drug resistance associations. We construct four graph convolution sub-networks that leverage distinctive combinations of features including drug fingerprints, drug label encoding, miRNA expression profiles, and miRNA GO-based similarities, and learn the latent representations of drugs and miRNAs from sub-networks independently. Then, an attention neural network is employed to obtain attentive representations of drugs and miRNAs from their respective latent representations. Finally, miRNA-drug resistance associations are predicted by the inner product of the attentive representations. The computational experiments show that AMMGC outperforms state-of-the-art and baseline methods. The experimental analyses reveal that leveraging multiple features of drugs and miRNAs can enhance the performance of the miRNA-drug resistance association prediction.
2 Materials
In this paper, we use a miRNA-drug resistance association dataset from the published work (Huang et al., 2019), which contains 3,338 miRNA-drug resistance associations between 754 miRNAs and 106 drugs. Moreover, we collect some side information of drugs and miRNAs to build our prediction model.
We consider two features of the drugs to give more insights into their characteristics. One is the PubChem substructure fingerprint (Wang et al., 2009), which represents drugs as 920-dimensional binary vectors. The other is the label/integer encoding of SMILES of drugs (Öztürk et al., 2018), in which each label of SMILES is represented by a integer (e.g., “C”:1, “O”:2, “N”:3, “ = ”:4, etc.). Considering the varied lengths of drug canonical SMILES, we set the dimensions of representations as 85, and then a drug is represented by an 85-dimensional vector. For the miRNAs, we consider two features to describe their biological information. One is the miRNA expression profile (Gillis et al., 2007), and the miRNA expression profile has 172 dimensions representing the expression levels of a single type of miRNAs in 172 different human tissues and cell lines. The other is the miRNA Gene Ontology (GO) functional similarity described in (Yang et al., 2018). The GO functional similarity has 2,587 dimensions, and each dimension denotes the similarity scores between the miRNA and other miRNAs concerning their gene regulation functions. Since the above features are not available for all miRNAs, we use the average of those miRNAs whose values are known to estimate the missing values of other miRNAs.
3 Methods
3.1 Problem Definition
Given p drugs, q miRNAs, and their resistance associations, our task is to predict novel miRNA-drug resistance associations by leveraging features of drugs and miRNAs as well as known associations. Formally, the associations between p drugs and q miRNAs can be formulated as an undirected graph, in which drugs and miRNAs are taken as nodes and their associations are taken as edges. The graph can be represented by a (p + q) × (p + q) adjacency matrix A, defined as
3.2 Graph Convolutional Networks
Graph convolutional network (Kipf and Welling, 2017) employs convolution operation over graphs to learn node embeddings by using local graph structure and node features. Formally, given an undirected graph G with the node feature matrix X and the adjacency matrix A, a graph convolutional network updates embeddings of graph nodes with the following rule:
where
3.3 Attentive Multimodal Graph Convolutional Network-Based Method
As shown in Figure 1, AMMGC constructs four graph convolution sub-networks with distinctive combinations of drug and miRNA features to obtain node embeddings from different views. After that, AMMGC applies an attention neural network to combine these node embeddings in different views for the miRNA-drug resistance association prediction.
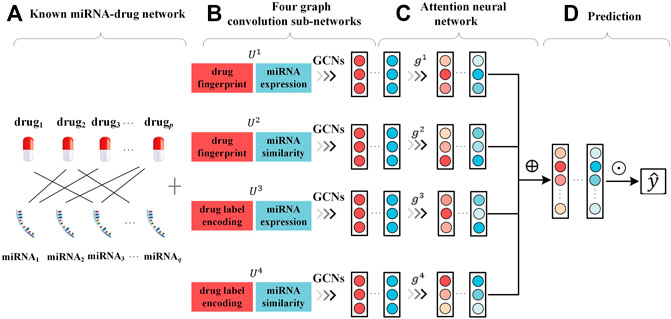
FIGURE 1. Framework of proposed method AMMGC (A). Known miRNA-drug resistance association network. (B). Learning node embeddings independently from four graph convolution sub-networks based on different feature combinations. (C). Applying an attention neural network to learn attentive node embeddings. ⊕ denotes the concatenation operation. (D). Using embeddings of drugs and miRNAs to produce the scores of miRNA-drug pairs. ⊙ denotes the inner product of drug embeddings and miRNA embeddings.
As discussed in section 2, we have two drug features and two miRNA features, which are drug Pubchem fingerprint (
where W0 and W1 are weight matrices of two GCN layers,
where
Graph convolution sub-networks contain different types of information, and the node embeddings in a specific modal can only reflect information from one aspect. It is necessary to fuse multiple node embeddings in different views. It is assumed that embeddings in different modals are not equally contributed to the associations, and we employ an attention neural network to learn the weight of multiple node embeddings. For each modal u, we first project the original embedding matrix to an unnormalized matrix gu = WuEu + bu. To reduce the computational complexity, we let gu to be a vector, and form the weighted embedding matrix by
The prediction score
where Zi and Zp+j denote the i-th attentive drug embedding the j-th attentive miRNA embedding.
3.4 Model Training
In the model training with two steps, the node embeddings are learned from sub-networks and then are served as the input of the attention neural network to make predictions.
The graph convolution sub-network for each modal u is trained independently, and we use the binary cross-entropy loss function formulated as:
where N denotes the number of samples (miRNA-drug pairs), ys is the true label of s-th sample and
For the attention neural network, we adopt the weighted binary cross-entropy classification loss function as:
where
We minimize the loss L1 and L2 using Adam optimizer (Abadi et al., 2016), and set the learning rate equal to 0.01. In sub-networks, we set the hidden units of two GCN layers to 128 and 256. More details of parameter settings can be seen in section 4.4.
4 Experiments
4.1 Experimental Setting
We adopt 5-fold cross-validation (5-CV) to evaluate the performance of AMMGC. The known miRNA-drug resistance associations are randomly equally divided into five subsets. In each fold, one subset of known associations is used for testing, and the remaining four subsets of associations are used for training. Specifically, we use four subsets of association pairs as positive instances and randomly select an equal number of samples from other pairs as negative instances to train the prediction model. We adopt the following evaluation metrics: the area under the precise-recall curve (AUPR), the area under the receiver-operating characteristic curve (AUC), F1-measure (F1), accuracy (ACC), and recall (REC). To avoid the bias of data split, we implement 10 runs of 5-fold cross-validation for each model, and the average metric scores and standard deviations are calculated.
4.2 Performance Comparison
We compare our method with four baselines and one state-of-the-art miRNA-drug resistance association prediction method:
• Collaborative filtering(CF): Collaborative filtering (Su and Khoshgoftaar, 2009) is a classical recommendation algorithm.
• Label propagation(LP): We consider the LP model mentioned in (Zhang et al., 2021b) as a baseline. The LP method propagates the existing miRNA-drug association information in the network to predict new associations.
• Graph factorization(GF): Graph factorization (Ahmed et al., 2013) is a factorization-based network embedding method.
• Structural Deep Network Embedding (SDNE): SDNE (Wang et al., 2016) is a deep learning-based network embedding method.
• GCMDR: GCMDR (Huang et al., 2019) makes use of graph convolution to build a latent factor model, which utilizes miRNA expression profile and drug fingerprint.
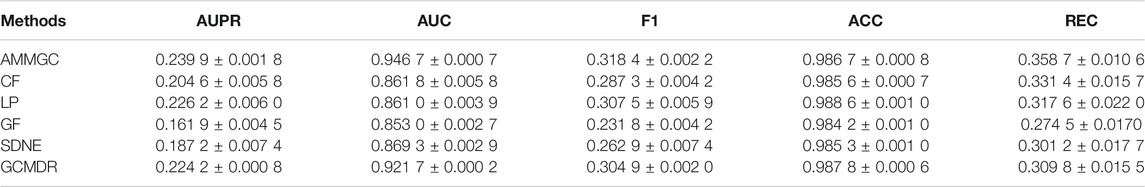
The 5-CV performances of all prediction models are shown in Table 1. In general, the proposed method AMMGC significantly outperforms four baselines in terms of most metrics. Specifically, AMMGC achieves an AUC of 0.946 7, which is almost 7.7% higher than that of the four baselines. This result shows that combining multiple features of drugs and miRNAs benefits the prediction of miRNA-drug pairs more than only using known miRNA-drug associations. Moreover, AMMGC also performs almost 4.89% higher than GCMDR on REC metric which is important for the study, verifying the efficacy and superiority of AMMGC. The substantial improvement of AMMGC over these compared methods is mainly attributed to two aspects: 1) AMMGC leverages the advantages of multiple features and applies them to build multimodal graph convolution sub-networks, while GCMDR only considers drug fingerprint and miRNA expression profile to build the prediction model; 2) AMMGC considers different contributions of embeddings learned from multimodal graph convolution, and assigns them different weights by applying the attention mechanism.
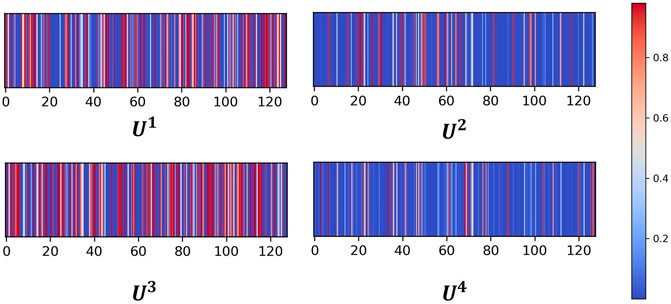
The attention neural network is one critical component of AMMGC, and it can measure and quantify the importance of each modal. Further, we visualize the attention coefficients for four modals in Figure 2. In general, four modals have different attention weights, and modal 1 and modal 3 have relatively higher weights than modal 2 and modal 4. This result shows AMMGC can well leverage different features to make predictions.
4.3 Ablation Analysis
In this section, we consider several variants of AMMGC to demonstrate the importance of the attention mechanism, multimodal and biological features. We provide a detailed analysis as follows.
• AMMGC without attention network (w/o AN): We assign the equal weights to all dimensions of multiple node embeddings from four graph convolution sub-networks.
• AMMGC without biochemical feature (w/o BF): Instead of using biological features of drugs and miRNAs, we simply employ one-hot encoding to build the graph convolution network.
• AMMGC without multimodal (w/o MM): Instead of using mutlimodal graph convolutional network, we directly concatenate two drug features and two miRNA features to build the prediction model.
• AMMGC with modal 1 (w U1): We train the AMMGC only with the modal 1.
• AMMGC with modal 2 (w U2): We train the AMMGC only with the modal 2.
• AMMGC with modal 3 (w U3): We train the AMMGC only with the modal 3.
• AMMGC with modal 4 (w U4): We train the AMMGC only with the modal 4.
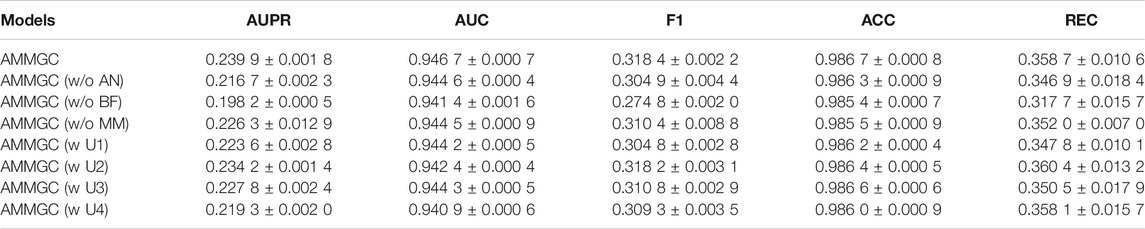
The ablation results are shown in Table 2. The performance of AMMGC is much better than that of AMMGC without attention network in terms of all evaluation metrics, revealing that the attention neural network can exploit different contributions of embeddings for the prediction task and further improve the prediction performance. To verify the effectiveness of the graph convolution sub-networks, we compare AMMGC with AMMGC without Biofeature. Since biological features of drugs and miRNAs play vital roles in miRNA-drug resistance associations, the AUPR value drops by 4.17% and the REC value drops by 4.1% without them. More importantly, AMMGC performs better than AMMGC without Multimodal and the single-modal graph convolution models (AMMGC w U1/U2/U3/U4), indicating that the multimodal model can leverage diverse information to achieve better performances.
4.4 Parameter Sensitivity Analysis
In this section, we investigate the sensitivity of parameters in AMMGC, including the number of graph convolution layers in sub-networks, the dimension of embeddings, and the negative sampling size.
For graph convolution sub-networks, we consider the number of layers from 1 to 6, and the results are shown in Figure 3A. The 2-layer GCN model achieves the best performance among all models, because 1-layer GCN may not learn sufficient information and more layers could lead to over-smoothing. In fact, related works have also shown that the two-layer GCN model can bring enough information and performs well (Li et al., 2019).
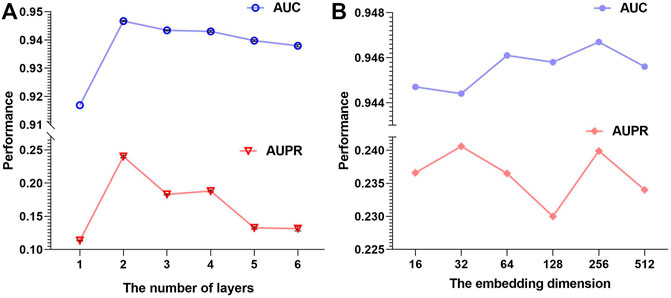
FIGURE 3. (A) AUPR and AUC of AMMGC with different GCN layers. (B) AUPR and AUC of AMMGC with different embedding dimensions.
Embedding dimension is an important factor for our proposed method, which could directly influence the model performance. We fix the hidden units of the first GCN layer to 128 for all four graph convolution sub-networks and change the hidden units of the second GCN layer in {16, 32, 64, 128, 256, 512}. Thus, the final embedding dimension is in a range of {64, 128, 256, 512, 1024, 2048} according to Equation 4. From Figure 3B, we can see that different embedding dimensions lead to different performances. In general, the AUPR value varies around 0.235 and the AUC value varies around 0.946, which proves that our model performs stably if the embedding dimension is selected in an appropriate range. We finally choose the embedding dimension is 256 since the model with this setting has superior AUC and AUPR performance.
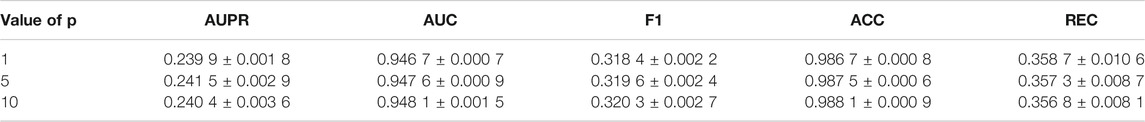
Since there are only positive samples in the dataset, negative samples are needed to conduct semi-supervised training on the prediction model. Thus, we randomly sample unlabeled miRNA-drug pairs to generate negative samples, and the size of negative sampling is fixed in each iteration. We conduct the experiments to evaluate the influence of the ratio p of the negative sampling size to that of the positive sampling size on the prediction performance. As shown in Table 3, AMMGC produces the greater scores on AUC, F1, and ACC with the increase of p, while producing the best AUPR and REC scores when p is set to 5. This result demonstrates the negative samples have an influence on the training of the AMMGC. Although the increase in the proportion of negative samples can bring slight performance improvements, the model takes more time to be convergent, so we choose to set p = 1 as the result of AMMGC.
4.5 Case Studies
In this section, we conduct case studies to test the capability of AMMGC in predicting novel miRNA-drug resistance associations. We train the AMMGC model with all known miRNA-drug resistance associations, then use the trained model to predict novel associations, which are further validated by public literature.
The results of case studies are shown in Table 4. According to Liao et al.’s research (Liao et al., 2015), hsa-mir-146a is found to influence the biologic features and prognosis of gastric cancer patients, and is associated with clinical characteristics in gastric cancer patients treated with adjuvant oxaliplatin and fluoropyrimidines. The association between gemcitabine and hsa-mir-145 is reported in Papadopoulos et al.’s work (Papadopoulos et al., 2015). In their study, hsa-mir-145 was significantly affected by gemcitabine treatment in T24 cells. Specifically, miR-145 levels were found to be dramatically upregulated (40.5-fold) during the first 36 h of treatment. In Hummel et al.’s research, they found Cisplatin can alter miRNA expression in esophageal cancer cells including hsa-mir-425 (Hummel et al., 2011). These studies show that AMMGC has the great potential of identifying novel miRNA-drug resistance associations.
5 Conclusion
In this work, we propose a deep learning-based method AMMGC to predict miRNA-drug resistance associations. AMMGC integrates multiple features of miRNAs and drugs to build graph convolution sub-networks, and learns node embeddings in different views. To obtain more comprehensive node embeddings, AMMGC employs an attention neural network to learn the contributions of different embeddings and assign them different weights for the final prediction. Experiment results demonstrate that integrating multiple features to build multimodal sub-networks is of vital importance for miRNA-drug resistance association prediction, and attention mechanism is also the key point to improve the performance.
There are several directions for our future study. On the one hand, AMMGC integrates side information about drugs and miRNAs under the graph convolutional network framework. More side information, such as interaction between drugs, miRNA-gene and gene-ontology relationship might be useful for predicting miRNA-drug resistance associations, and we hope to further investigate their usefulness. On the other hand, AMMGC is a general link prediction method, and it is promising to solve other related tasks.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://github.com/scz760904126/AMMGC.
Author Contributions
YN designed the study and drafted the manuscript. CS and YG implemented the algorithm and drafted the manuscript. WZ drafted the manuscript.
Funding
This work was supported by Fundamental Research Funds for the Central Universities. data collection, data analysis, data interpretation or writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abadi, M., Barham, P., Chen, J., Chen, Z., Davis, A., Dean, J., et al. (2016). “Tensorflow: A System for Large-Scale Machine Learning,” in 12th USENIX Symposium on Operating Systems Design and Implementation (Savannah, GA: OSDI 16), 265–283.
Ahmed, A., Shervashidze, N., Narayanamurthy, S., Josifovski, V., and Smola, A. J. (2013). “Distributed Large-Scale Natural Graph Factorization,” in Proceedings of the 22nd International Conference on World Wide Web, 37–48. doi:10.1145/2488388.2488393
Dai, E., Yang, F., Wang, J., Zhou, X., Song, Q., An, W., et al. (2017). ncDR: a Comprehensive Resource of Non-coding RNAs Involved in Drug Resistance. Bioinformatics 33, 4010–4011. doi:10.1093/bioinformatics/btx523
Fu, H., Huang, F., Liu, X., Qiu, Y., and Zhang, W. (2021). MVGCN: Data Integration through Multi-View Graph Convolutional Network for Predicting Links in Biomedical Bipartite Networks. Bioinformatics 9, btab651. doi:10.1093/bioinformatics/btab651
Gillis, A. J., Stoop, H. J., Hersmus, R., Oosterhuis, J. W., Sun, Y., Chen, C., et al. (2007). High-throughput Micrornaome Analysis in Human Germ Cell Tumours. J. Pathol. 213, 319–328. doi:10.1002/path.2230
Hopkins, A. L., and Groom, C. R. (2002). The Druggable Genome. Nat. Rev. Drug Discov. 1, 727–730. doi:10.1038/nrd892
Huang, Y. A., Hu, P., Chan, K. C. C., and You, Z. H. (2019). Graph Convolution for Predicting Associations between miRNA and Drug Resistance. Bioinformatics 36, 851–858. doi:10.1093/bioinformatics/btz621
Hummel, R., Wang, T., Watson, D. I., Michael, M. Z., Van der Hoek, M., Haier, J., et al. (2011). Chemotherapy-induced Modification of Microrna Expression in Esophageal Cancer. Oncol. Rep. 26, 1011–1017. doi:10.3892/or.2011.1381
Kipf, T. N., and Welling, M. (2017). “Semi-supervised Classification with Graph Convolutional Networks,” in 5th International Conference on Learning Representations (ICLR 2017).
Lei, X., Tie, J., and Pan, Y. (2021). Inferring Metabolite-Disease Association Using Graph Convolutional Networks. Ieee/acm Trans. Comput. Biol. Bioinf. 1, 1. doi:10.1109/TCBB.2021.3065562
Li, C., Liu, H., Hu, Q., Que, J., and Yao, J. (2019). A Novel Computational Model for Predicting microRNA-Disease Associations Based on Heterogeneous Graph Convolutional Networks. Cells 8, 977. doi:10.3390/cells8090977
Liao, Y. Q., Liao, Y. L., Li, J., Peng, L. X., Wan, Y. Y., and Zhong, R. (2015). Polymorphism in Mir-146a Associated with Clinical Characteristics and Outcomes in Gastric Cancer Patients Treated with Adjuvant Oxaliplatin and Fluoropyrimidines. Onco Targets Ther. 8, 2627–2633. doi:10.2147/OTT.S89635
Liu, X., Song, C., Huang, F., Fu, H., Xiao, W., and Zhang, W. (2021). GraphCDR: a Graph Neural Network Method with Contrastive Learning for Cancer Drug Response Prediction. Brief. Bioinform. 1, bbab457. doi:10.1093/bib/bbab457.Bbab457
Mudiyanselage, T. B., Lei, X., Senanayake, N., Zhang, Y., and Pan, Y. (2020). “Graph Convolution Networks Using Message Passing and Multi-Source Similarity Features for Predicting Circrna-Disease Association,” in 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 343–348. doi:10.1109/BIBM49941.2020.9313455
Overington, J. P., Al-Lazikani, B., and Hopkins, A. L. (2006). How many Drug Targets Are There. Nat. Rev. Drug Discov. 5, 993–996. doi:10.1038/nrd2199
Öztürk, H., Özgür, A., and Ozkirimli, E. (2018). DeepDTA: Deep Drug-Target Binding Affinity Prediction. Bioinformatics 34, i821–i829. doi:10.1093/bioinformatics/bty593
Papadopoulos, E. I., Yousef, G. M., and Scorilas, A. (2015). Gemcitabine Impacts Differentially on Bladder and Kidney Cancer Cells: Distinct Modulations in the Expression Patterns of Apoptosis-Related Micrornas and Bcl2 Family Genes. Tumour Biol. 36, 3197–3207. doi:10.1007/s13277-014-2190-8
Su, X., and Khoshgoftaar, T. M. (2009). A Survey of Collaborative Filtering Techniques. Adv. Artif. Intelligence 2009, 1–19. doi:10.1155/2009/421425
Wang, D., Cui, P., and Zhu, W. (2016). “Structural Deep Network Embedding,” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data mining, 1225–1234. doi:10.1145/2939672.2939753
Wang, Y., Xiao, J., Suzek, T. O., Zhang, J., Wang, J., and Bryant, S. H. (2009). PubChem: a Public Information System for Analyzing Bioactivities of Small Molecules. Nucleic Acids Res. 37, W623–W633. doi:10.1093/nar/gkp456
Yang, J., and Lei, X. (2021). Predicting Circrna-Disease Associations Based on Autoencoder and Graph Embedding. Inf. Sci. 571, 323–336. doi:10.1016/j.ins.2021.04.073
Yang, Y., Fu, X., Qu, W., Xiao, Y., and Shen, H. B. (2018). MiRGOFS: a GO-Based Functional Similarity Measurement for miRNAs, with Applications to the Prediction of miRNA Subcellular Localization and miRNA-disease Association. Bioinformatics 34, 3547–3556. doi:10.1093/bioinformatics/bty343
Yu, Z., Huang, F., Zhao, X., Xiao, W., and Zhang, W. (2020). Predicting Drug-Disease Associations through Layer Attention Graph Convolutional Network. Brief Bioinform 22. doi:10.1093/bib/bbaa243
Zhang, G., Li, M., Deng, H., Xu, X., Liu, X., and Zhang, W. (2021a). SGNNMD: Signed Graph Neural Network for Predicting Deregulation Types of miRNA-Disease Associations. Brief. Bioinform. 8, bbab464. doi:10.1093/bib/bbab464.Bbab464
Keywords: miRNA-drug resistance association, graph convolutional network, multimodal, deep learning, attention neural network
Citation: Niu Y, Song C, Gong Y and Zhang W (2022) MiRNA-Drug Resistance Association Prediction Through the Attentive Multimodal Graph Convolutional Network. Front. Pharmacol. 12:799108. doi: 10.3389/fphar.2021.799108
Received: 21 October 2021; Accepted: 20 December 2021;
Published: 12 January 2022.
Edited by:
Xiujuan Lei, Shaanxi Normal University, ChinaReviewed by:
Hao Chen, Novartis Institutes for BioMedical Research, United StatesTao Zeng, Guangzhou labratory, China
Copyright © 2022 Niu, Song, Gong and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanqing Niu, bml1eWFucWluZ0BtYWlsLnNjdWVjLmVkdS5jbg==
 Yanqing Niu1*
Yanqing Niu1* Congzhi Song
Congzhi Song Wen Zhang
Wen Zhang