- 1Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Pre-Clinical Research Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia
- 3Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
Vascular dementia (VaD) is a general term used to describe difficulties in memory, reasoning, judgment, and planning caused by a reduced blood flow to the brain and consequent brain damage, in which microRNAs (miRNAs) are involved. Dracocephalum moldavica L. (D. moldavica) is traditionally used in the treatment of cardiovascular diseases as well as VaD, but the biomolecular mechanisms underlying its therapeutic effect are obscure. In the present study, the molecular mechanisms involved in the treatment of VaD by the total flavonoids from Dracocephalum moldavica L. (TFDM) were explored by the identification of miRNA profiling using bioinformatics analysis and experimental verification. A total of 2,562 differentially expressed miRNAs (DEMs) and 3,522 differentially expressed genes (DEGs) were obtained from the GSE120584 and GSE122063 datasets, in which the gene functional enrichment and protein-protein interaction network of 93 core targets, originated from the intersection of the top DEM target genes and DEGs, were established for VaD gene profiling. One hundred and eighty-five targets interacting with 42 flavonoids in the TFDM were included in a compound-target network, subsequently found that they overlapped with potential targets for VaD. These 43 targets could be considered in the treatment of VaD by TFDM, and included CaMKII, MAPK, MAPT, PI3K, and KDR, closely associated with the vascular protective effect of TFDM, as well as anti-oxidative, anti-inflammatory, and anti-apoptotic properties. The subsequent analysis of the compound-target gene-miRNA network indicated that eight miRNAs that mediated 43 targets had a close interaction with TFDM, suggesting that the neuroprotective effects were principally due to kaempferol, apigenin, luteolin, and quercetin, which were mostly associated with the miR-3184-3p/ESR1, miR-6762-3p/CDK1, miR-6777-3p/ESRRA, and other related axes. Furthermore, the in vitro oxygen-glucose deprivation (OGD) model demonstrated that the dysregulation of miR-3184-3p and miR-6875-5p found by qRT-PCR was consistent with the changes in the bioinformatics analysis. TFDM and its active compounds involving tilianin, luteolin, and apigenin showed significant effects on the upregulation of miR-3184-3p and downregulation of miR-6875-5p in OGD-injured cells, in line with the improved cell viability. In conclusion, our findings revealed the underlying miRNA-target gene network and potential targets of TFDM in the treatment of VaD.
Introduction
Vascular dementia (VaD) represents a group of syndromes characterized by cognitive impairment resulting from the death of the hypoxic brain tissue caused by the reduced perfusion from diseased brain blood vessels (Iadecola, 2013), causing approximately 15–20% of all dementia cases (O'Brien and Thomas, 2015). The pathogenesis of VaD is complex and is the result of a variety of cardiovascular or genetic risk factors associated with Apolipoprotein E (APOE), angiotensin I converting enzyme (ACE), paraoxonase 1 (PON1), or presenilin 1 (PSEN1) genes, or due to stroke (O’Brien and Thomas, 2015). Thus, there is no authorized treatment for VaD in clinical practice. The current therapies for VaD are mostly performed using cholinesterase inhibitors or memantine due to the lack of a reliable biomarker or an established molecular mechanism, and these compounds are moderately effective. However, among all forms of dementia, VaD is considered unique because it can be prevented if the intervention is provided early enough (Thal et al., 2012). Therefore, an exploration of specific biomarkers or target genes for VaD might be of help in the development of mechanism-targeted prevention and therapy.
Although the pathologic mechanism of VaD is not yet fully elucidated, it has been reported that non-coding RNAs such as microRNAs (miRNAs), long non-coding RNAs, and circular RNAs are involved in the pathophysiology of VaD, including cognitive impairment, vascular dysfunction, neuroinflammatory response, blood-brain barrier disruption, and synaptic loss (Toyama et al., 2018; Liu et al., 2019b; Huang et al., 2020; Vijayan and Reddy, 2020; Wang et al., 2020). miRNAs are enriched in the brain and responsible for the regulation of post-transcription of a wide range of proteins through the degradation or inhibition of their target mRNAs. In a rat model of VaD, a reduction in miR-126 expression in endothelial cells revealed the pathophysiological process mediating the cognitive impairment in the rats accompanied by a decreased cerebral blood flow and vessel patency, causing the activation of astrocytes and microglia, and a reduction in synaptic plasticity (Yu et al., 2019). It has been reported that the miR-93-mediated Toll-like receptor signaling pathway plays a functional role when cognition defects in VaD is treated by acupuncture (Wang et al., 2020). In addition, the involvement of miR-124 (Liu et al., 2019a), miR-210-5p (Ren et al., 2018), miR-132 (Yang et al., 2020a), and miR-7-5p (Xu et al., 2019) has been demonstrated in the development of VaD. Although it has been proposed that miRNAs represent therapeutic targets and diagnostic biomarkers for VaD, the varying expression profiles of miRNAs and disease characteristics of VaD are not fully understood.
Almost one hundred traditional Chinese medicines are used to treat VaD, including Ginkgo biloba L. (Wang et al., 2013), Salvia miltiorrhiza Bunge (Kim et al., 2015), Panax ginseng C.A.Mey. (Zhu et al., 2018), and Dracocephalum moldavica L. (D. moldavica) (Jia et al., 2017; Jiang et al., 2021). Many of these herbal medicines have roles and working mechanisms in the regulation of aberrant miRNAs when used to treat vascular cognitive decline. For instance, EGb761, the standardized extract from Ginkgo biloba L., protects against ischemia/reperfusion-induced injury in brain microvascular endothelial cells through the lncRNA Rmst/miR-150 axis (Qiao et al., 2020). Ginsenoside Rg1 and Rg2 extracted from Panax ginseng C.A.Mey. have inhibitory activity on miR-155-5p (Wang et al., 2018), miR-144 (Chu et al., 2019), and miR-216a (Chen et al., 2021), thus being potential therapeutic targets in the treatment of cerebral ischemia.
D. moldavica is a traditional Uygur medicine in China, widely used to treat coronary heart disease, angina, and atherosclerosis as well as for calming the nerves (Dastmalchi et al., 2007; Maimaitiyiming et al., 2014; Miernisha et al., 2016). Modern neuropharmacological studies confirmed that D. moldavica has beneficial effects on the central nervous systems, as demonstrated by the promotion of prolonged pentobarbital-induced sedation in mice (Martinez-Vazquez et al., 2012), neuroprotection against cerebral ischemia/reperfusion injury in rats (Jia et al., 2017), and cognitive improvement against scopolamine-induced deficits in mice (Deepa et al., 2020). The total flavonoids from D. moldavica (TFDM) represent a standardized extract from D. moldavica and function as a novel treatment for VaD as established by our research team, obtaining the support of a number of major national projects in China and patent authorization. Our ongoing study demonstrated that TFDM provides neuroprotection and improvement of learning and memory capabilities against beta-amyloid-induced toxicity in vitro and in vivo (Liu et al., 2018). The principal flavonoids in TFDM, such as tilianin, luteolin, apigenin, and quercetin improve the cognitive function in various types of cognitive decline (Jiang et al., 2018 and, 2021; Liu et al., 2009; Liu et al., 2013; Zhao et al., 2013). Furthermore, TFDM and its principal compounds reduce the cognitive deficit acting on different mechanisms, including anti-oxidative, anti-amyloidogenic, neurotrophic, anti-inflammatory, and anti-apoptotic pathways as a result of the regulation of the expression of a number of genes, such as amyloid-beta precursor protein (APP), beta-secretase 1 (BACE1), cAMP-responsive element-binding protein 1 (CREB), calcium/calmodulin-dependent protein kinase II gamma (CaMKII), mitogen-activated protein kinase (MAPK), and B-cell leukemia/lymphoma-2 (Bcl-2) (Liu et al., 2009; Zhao et al., 2013; Jiang et al., 2018; Jiang et al., 2020; Liu et al., 2018). However, the effect of TFDM in the improvement of VaD through the regulation of multiple genes remains unclear. Thus, the discovery of the mechanism of action could allow its potential use in the treatment of VaD.
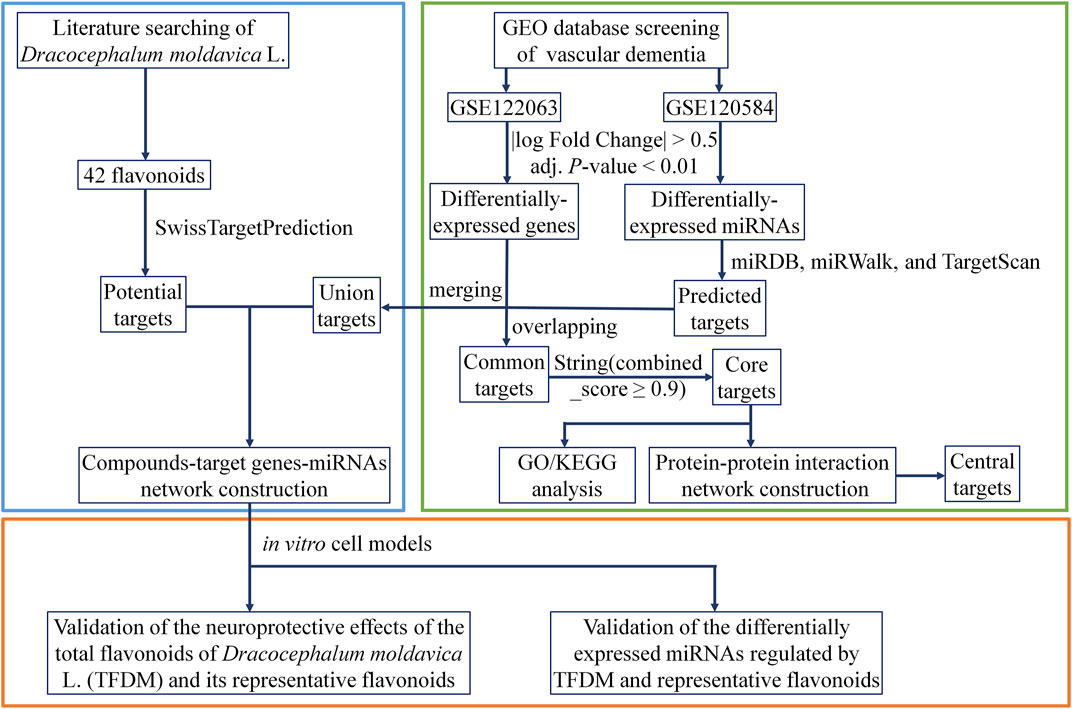
In the current study, bioinformatics approaches with biological verification were used to determine the miRNAs involved in the treatment of VaD by TFDM. Eight significant miRNAs that mediated 43 targets with a close interaction with TFDM were identified using the transcriptional profile network of VaD established from the Gene Expression Omnibus (GEO) database. The qRT-PCR assay revealed that the upregulation of miR-3184-3p and downregulation of miR-6875-5p might play critical roles in the treatment of VaD by TFDM. The workflow is displayed in Figure 1.
Materials and Methods
Dataset Acquisition
Two microarray datasets, GSE122063 (McKay et al., 2019) and GSE120584 (Shigemizu et al., 2019a), were selected from the GEO (https://www.ncbi.nlm.nih.gov/gds/) using “VaD” as the search term, with the species limited to “Homo sapiens”. The GSE122063 dataset includes gene expression profiles of the frontal and temporal cortex of patients with VaD, Alzheimer’s disease (AD), and non-demented controls (Control), and McKay et al. focused on the special changes in gene expression in samples of the frontal cortex of VaD patients, identifying the oxytocin receptors (OXTR) as significantly upregulated (McKay et al., 2019). In the present study, 11 samples from non-demented controls and 8 VaD samples were selected and subjected to analysis to obtain all the DEGs. Shigemizu et al. constructed risk prediction models for three subtypes of dementia, including AD, VaD, and dementia with Lewy bodies (DLB), using potential biomarkers from serum samples, and further analyzed the functional genes associated with DLB pathology (Shigemizu et al., 2019b). The samples of 91 VaD patients and normal subjects from the GSE120584 dataset were selected for miRNA analysis.
Data Processing and Differential Expression Analysis
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is an online analysis tool in the GEO database. It was used to screen differentially expressed genes (DEGs) between control and VaD tissue samples in the GSE122063 dataset, with a cut-off of |log Fold Change| > 0.5 and adjusted (adj.) p-value < 0.01. Volcano plots were generated to describe the distribution and expression of the DEGs that were identified. Differentially expressed miRNAs (DEMs) in VaD patients and normal subjects in the GSE120584 dataset were also analyzed using the GEO2R webtool. The six most upregulated and downregulated miRNAs were selected in ascending order by p-value.
Target Prediction of miRNAs
The target genes of the top DEMs were predicted using miRDB (http://mirdb.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), and TargetScan (http://www.targetscan.org/vert_72/) online analysis tools. Overlapping target genes were identified using the Venn diagrams and then used in additional analysis. TargetScan can predict the biological target of miRNA by searching for conserved sites that match the seed region of miRNA, which are ranked by cumulative weighted context++ scores of the sites and their probability of conserved targeting. In contrast, miRDB is designed to screen target genes with a target score ≥50 by analyzing thousands of miRNA target interactions from high-throughput sequencing experiments. The miRWalk database is based on the current miRBase, using miRNA names to search for miRNA-related target gene prediction data. In the above three databases, the “human” species was selected before the prediction of the target genes.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
GO analysis was performed to gain insight into the molecular mechanisms of VaD by molecular function (MF), cellular component (CC), and biological process (BP) domains of the core targets derived from the overlap of target genes of the most differentially expressed miRNAs and DEGs. KEGG pathway enrichment analysis was performed to define the signaling pathways associated with these core targets. The analysis used a threshold of adj. p-values < 0.01.
Construction of a Protein-Protein Interaction Network
The STRING 11.0 database (https://string-db.org/cgi/input) was used to assess the PPI network information of the core targets to discover the network of the interacting genes. A combined score of more than 0.9 was the comprehensive protein interaction score selected as the cut-off criterion. Cytoscape 3.7.0 software was then used to visualize the resulting PPI network. The four algorithms Maximal Clique Centrality (MCC), Density of Maximum Neighborhood Component (DMNC), Maximum Neighborhood Component (MNC), and DEGREE in the cytohubba plug-in were used to identify standard and central targets with high connectivity in the gene expression network.
Construction of Compound-Target Gene-miRNA Network for TFDM
Identification of Components and Target Screening
The flavonoids in D. moldavica were searched in the reported literature (Yang et al., 2013; Liu et al., 2019c; Wu et al., 2020). Then, query Canonical SMILES of flavonoids in Pubchem was used to predict potential target genes. The potential targets of these compounds were predicted with probability ≥0.05 using the online prediction tool SwissTargetPrediction (http://www.swisstargetprediction.ch/). This portal allows the estimation of the most likely macromolecular targets for small molecules considered biologically active. The prediction was based on the similarity of the 2D and 3D compounds.
Network Construction
Compound-target (C-T) and compound-target gene-miRNA (C-T-M) networks were constructed using Cytoscape. The eigenvector centrality parameter of the node topology was calculated using Gephi 0.9.2 software by the analysis of the importance of each node, which depends on the number of nodes of its neighbours and their importance. Therefore, it is considered an appropriate predictor in interactive networks. In the network, the nodes represent the compounds, the targets, and the miRNAs, while the edges represent the relationship between compounds and targets or between targets and miRNAs.
Drug Materials
Plant Source
D. moldavica from the Lamiaceae Martinov family belongs to the genus of Dracocephalum L. The samples used in the present work were produced in Jimusaer (Xinjiang province, China) in July 2017 (batch number: 20170713). The D. moldavica specimen (D170713) was deposited in the herbarium of the Xinjiang Institute of Materia Medica (Ürümqi, China) and identified by Prof. Jiang He, Xinjiang Institute of Materia Medica (Ürümqi, China).
Extraction of TFDM and Isolation of Tilianin
After confirmation of the herb, the powder of the aboveground parts (90 kg) was airdried at room temperature (RT), then refluxed three times with 40% EtOH at 100°C. Next, 3.2 kg crude extract was collected after evaporation of the filtered ethanol solution under reduced pressure. The crude extract was partitioned by column chromatography with a HPD600 resin and eluted with water, 50% EtOH, and 70% EtOH. The eluent was concentrated and dried to obtain TFDM, which was evaluated by high-performance liquid chromatography (HPLC).
The HPLC analysis was performed on a Sthermos U3000 HPLC system (Thermo Fisher Scientific, PA, USA). All separations were performed on a Welch Ultimater XB-C18 column (250 × 4.6 mm, 5 μm), with an injection volume of 20 μL. The mobile phase was composed of acetonitrile A and 0.5% formic acid aqueous solution B using a specific gradient elution (0–60 min, 20% A; 60–69 min, 50% A; 69–70 min, 80% A; 70–80 min, 20% A). The flow rate of the mobile phase was 1.0 ml min−1, and the temperature was maintained at 35°C. The components were quantified by the peak areas at 330 nm UV wavelength.
The purified product was collected by filtering the 70% EtOH eluate on a silica gel column (100–200 mesh, chloroform:methanol, 95:5–90:10–80:20) to remove the impurities. The structure of the compound was subsequently determined by the physicochemical and spectroscopic data, and the purity of the compound was 99% as revealed by HPLC analysis, yielding a total of 280 mg of tilianin, which was the same batch in our previous research (Liu et al., 2018).
Preparation of the Pharmaceutical Solution
Apigenin (4,5,7-trihydroxyflavone, 97%) and luteolin (3′,4′,5,7-tetrahydroxyflavone, 98%) used in this work were purchased from Acmec Biochemical (Shanghai, China). TFDM and other three active compounds were dissolved in dimethyl sulfoxide (DMSO, Sigma-Adrich, St. Louis, MO, USA) to a stock concentration of 100 mM and kept at a temperature of −20°C in the dark. The final dilution was freshly prepared at the time of the treatment by diluting the concentrated solution directly into the culture medium.
Cell Culture and Treatment
SH-SY5Y cells (ATCC; Manassas, VA, USA) were routinely cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA). Cells were seeded in 96-well plates at a density of 1 × 104 cells/well in sugar-free DMEM and subjected to the hypoxic condition of 93.7%N2/1.3%O2/5%CO2 for 24 h at 37°C in a tri-gas incubator (Wiggens, Straubenhardt, Germany) for the establishment of an in vitro OGD model. Cells were treated with different concentrations of TFDM (25–100 μg/ml), tilianin (8–32 μM), luteolin (2.5–10 μM), and apigenin (2.5–10 μM) in the standard medium in a normoxic condition for 12 h prior to OGD injury. Next, the medium was replaced with the same sugar-free one with the same compounds at the same concentration during the OGD process.
Cell Viability Assay
Cell survival was assessed using the cell counting kit-8 (CCK8; Vazyme Biotech, Nanjing, China) according to the manufacturer’s instructions and measured at 450 nm using a Spark 20M multimode microplate reader (Tecan Group Ltd., Mannedorf, Switzerland).
Expression of DEMs by Quantitative Reverse Transcription-Polymerase Chain Reaction
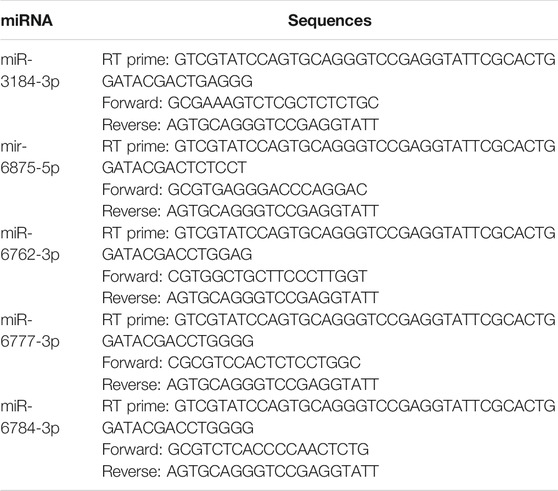
The qRT-PCR was used to confirm the changes of the predicted DEMs in the OGD-injured SH-SY5Y cells treated with TFDM and principal compounds. Briefly, total RNA was extracted using Trizol reagent (Invitrogen). miRNA was reverse-transcribed using a miRNA 1st Strand cDNA Synthesis Kit (by stem-loop) (Vazyme Biotech). PCR was performed using miRNA Universal SYBR qPCR Master Mix (Vazyme Biotech). Relative miRNA expression was calculated using the 2−ΔΔCT method. Results were normalized using small nuclear U6 RNA as the internal control. Primers are listed in Table 1.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism Version 7.0 software (GraphPad Prism Software; La Jolla, CA, USA). Data were analyzed using one-way ANOVA, followed by a Tukey’s multiple comparison test, or Student’s t-test. The results were expressed as mean ± standard deviation (SD). p-values less than 0.05 were considered statistically significant.
Results
Identification of DEGs in VaD
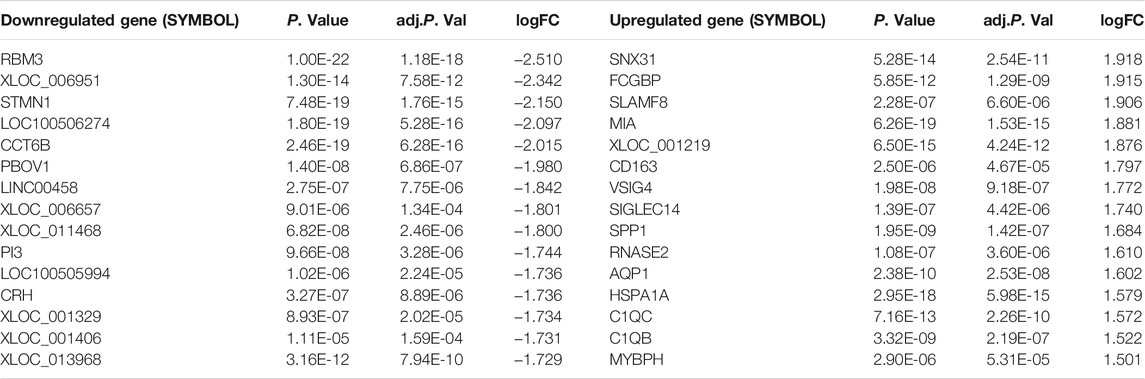
DEGs in VaD were analyzed in the GSE122063 dataset. Using the cut-off criteria |log Fold Change| > 0.5 and adj. p-value < 0.01, 1,478 significantly upregulated DEGs were identified and 2,044 were downregulated in the brains of patients with VaD compared with normal control brains, as presented in Figure 2A. The 15 most downregulated and upregulated genes whose expression changed significantly are listed in Table 2. The genes with a significantly increased expression included Nexin 31 (SNX31), Fc Fragment of IgG Binding Protein (FCGBP), SLAM Family Member 8 (SLAMF8), Maternal immune activation (MIA), and XLOC_001219, and those with a significantly decreased expression were RNA Binding Motif Protein 3 (RBM3), XLOC_006951, stathmin 1 (STMN1), LOC100506274, and chaperonin Containing TCP1 Subunit 6B (CCT6B).
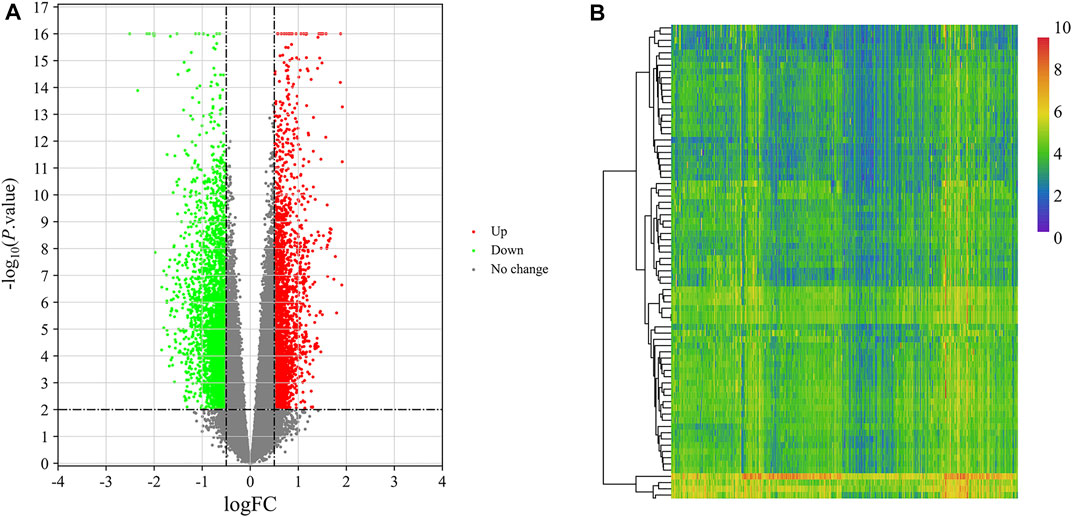
FIGURE 2. Identification of DEGs and DEMs in VaD. (A) Volcano plot of GSE122063 for the analysis of DEGs. (B) Heat map of the identified DEMs from the GSE120584 dataset.
Identification of DEMs in VaD
The miRNA dataset GSE120584 was screened for DEMs in VaD using the GEO2R web tool to compare VaD patients with the normal group. A total of 2,562 DEMs were identified from this microarray dataset, including 1,167 upregulated miRNAs and 1,395 downregulated miRNAs, as shown in a heat map (Figure 2B). The six most upregulated miRNAs were hsa-miR-3184-5p, hsa-miR-4667-5p, hsa-miR-6875-5p, hsa-miR-4746-3p, hsa-miR-4467, and hsa-miR-1268a, while the six most downregulated miRNAs were hsa-miR-6777-3p, hsa-miR-6784-3p, hsa-miR-3184-3p, hsa-miR-6762-3p, hsa-miR-4747-3p, and hsa-miR-3675-3p, as listed in Table 3.
Target Prediction, Core Target Analysis, and Functional Analysis
The target genes of the 12 most dysregulated DEMs were predicted using the online analysis tools miRDB, miRWalk, and TargetScan. A total of 2,495 targets were identified. The miRNA-gene regulatory network was mapped (Figure 3A) using Cytoscape software. Next, the screening was performed using the parameters |log Fold Change| > 0.5 and adj. p-value < 0.01, and 3,522 DEGs were identified. A total of 311 common targets were identified by analyzing the association between DEGs and the target genes of miRNAs (Figure 3B), considered core targets closely related to the principal pathophysiological process of VaD. STRING was then used to analyze the core targets, resulting in a total of 94 core targets with a combined_score ≥0.9, which were selected for further analysis. They were imported into Cytoscape 3.7.0 and the key associations were studied. The complex PPI network contained 94 nodes and 228 edges (Figure 3C).
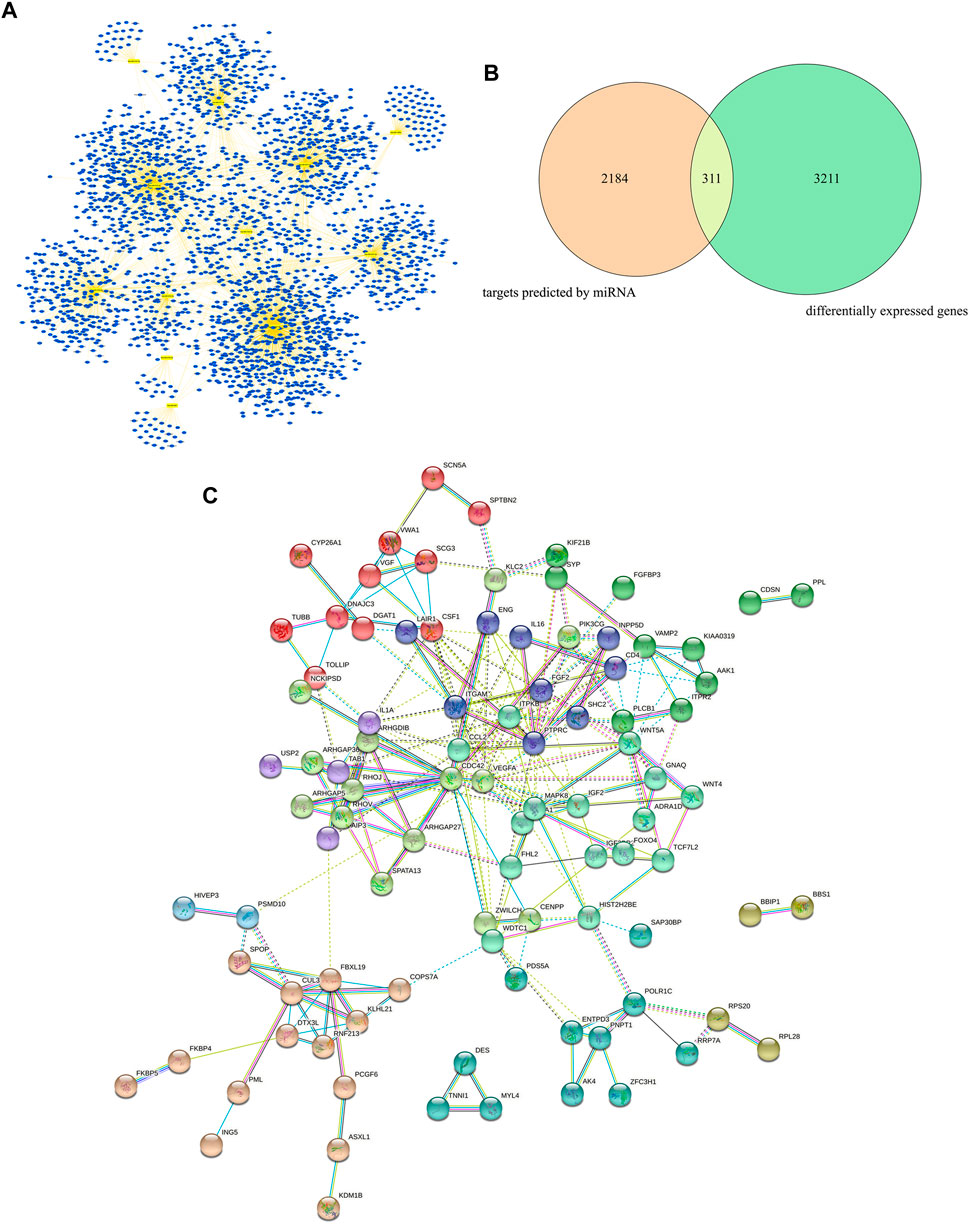
FIGURE 3. Target prediction, core target analysis, and functional analysis of DEGs and DEMs in VaD. (A) miRNA-gene regulatory network of the 12 most dysregulated DEMs and their predicted target genes. (B) Venn diagram of the intersection between miRNA-predicted targets and DEGs. (C) Protein-protein interaction analysis of 94 core targets using Cytoscape.
GO and KEGG pathway analyses were used to determine the enrichment of biological function and the pathways of these 94 core target genes. The GO annotation revealed that most significantly enriched pathways in BP were mostly enriched in post-translational protein modification, positive regulation of hydrolase activity, positive regulation of the MAPK cascade, and blood vessel development. The most significantly enriched pathways in MF were cytokine activity, receptor-ligand activity, ubiquitin-like protein ligase binding, and purine ribonucleoside binding. The CC terms were mainly represented by cytoplasmic vesicle membrane, secretory granule membrane, and cullin-RING ubiquitin ligase complex (Figure 4A).
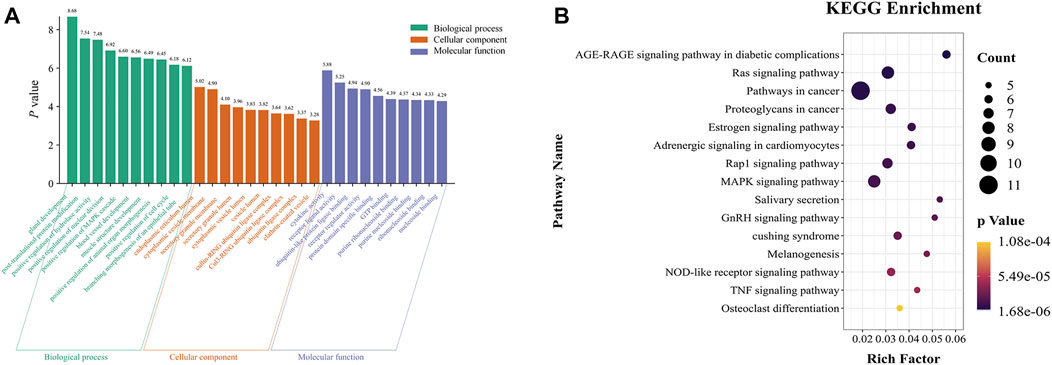
FIGURE 4. GO and KEGG pathway analysis of the 94 core targets. (A) Top 10 significantly enriched GO terms. (B) Top 15 significant KEGG pathway terms.
The signaling pathways enriched in KEGG analysis were ranked in ascending order according to the p-value. The results revealed that the selected core targets were significantly enriched in the advanced glycation end products (AGE) - receptor for AGE (RAGE) signaling pathway in diabetic complications, Ras signaling pathway, estrogen signaling pathway, Rap1 signaling pathway, MAPK signaling pathway, tumor necrosis factor (TNF) signaling pathway, and other signaling pathways (Figure 4B).
Construction of a PPI Network of Central DEGs
A number of central targets based on the MCC (Figure 5A), DMNC (Figure 5B), MNC (Figure 5C), and DEGREE (Figure 5D) algorithms in Cytoscape software were identified from the extraction of 94 core target gene-related proteins in VaD. Thirteen central targets with a high degree of connectivity were finally identified, such as Cullin 3 (CUL3), kelch like family member 21 (KLHL21), F-box and leucine-rich repeat protein 19 (FBXL19), Wnt family member 5A (WNT5A), DnaJ homolog subfamily C member 3 (DNAJC3), cluster of differentiation 4 (CD4), vesicle-associated membrane protein 2 (VAMP2), AP2-associated protein kinase 1 (AAK1), Dyslexia-associated protein KIAA0319 (KIAA0319), colony-stimulating factor 1 (CSF1), Von Willebrand factor A domain containing 1 (VWA1), Secretogranin III (SCG3), and COP9 Signalosome Subunit 7A (COPS7A) (Figure 5E).
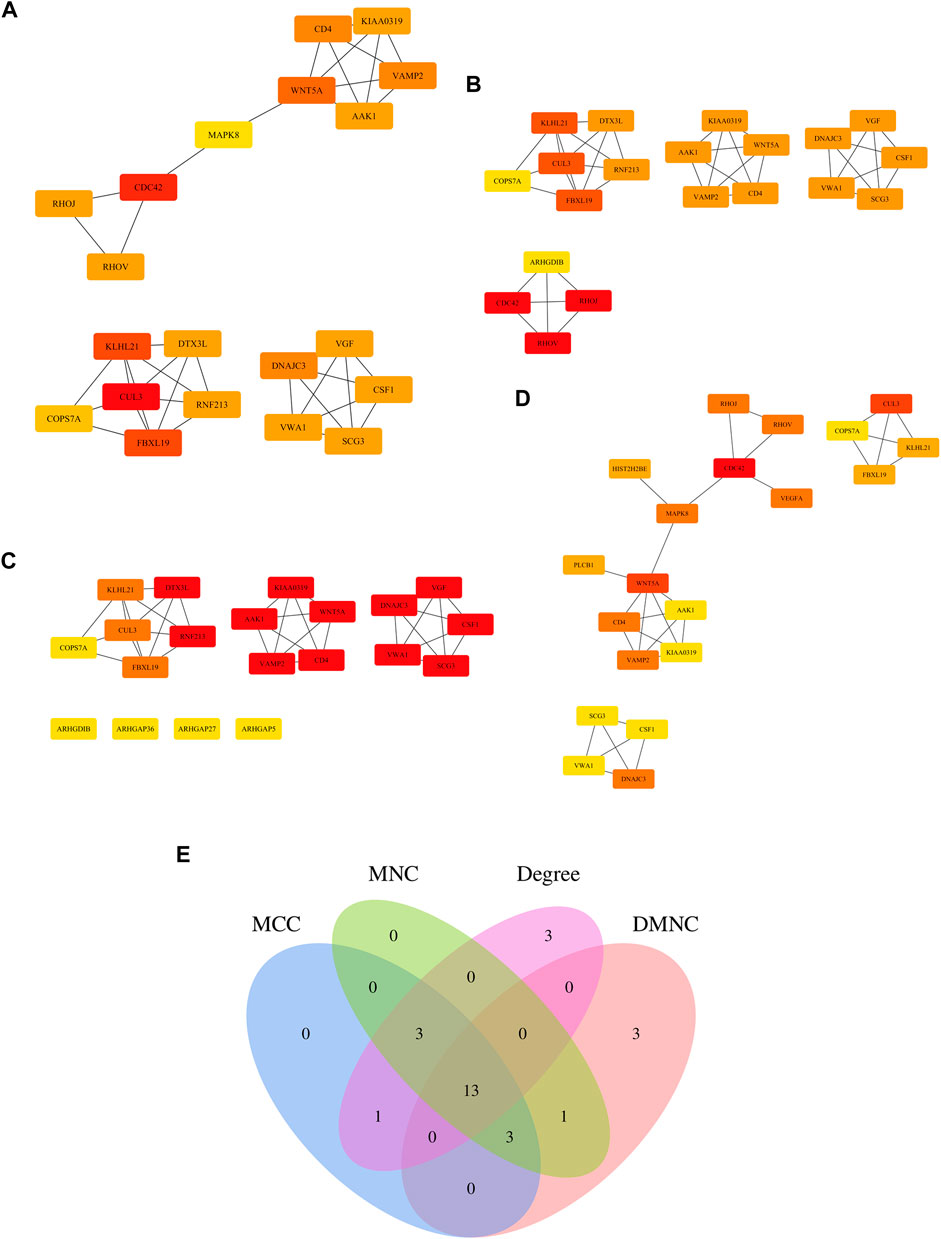
FIGURE 5. Determination of the central genes of the 94 core targets. (A–D) Four different metrics were used, such as Maximal Clique Centrality (MCC) (A), Density of Maximum Neighborhood Component (DMNC) (B), Maximum Neighborhood Component (MNC) (C), and DEGREE (D) to analyze central genes of the 94 core targets. (E) Venn diagram showing 13 overlapping central genes from these four metrics.
Construction of a C-T-M Network for TFDM
D. moldavica contains 42 flavonoids, including kaempferol, chrysoeriol, diosmetin, 8-hydroxy-salvigenin, apigenin, luteolin, salvigenin, 3-hydroxyflavone, isorhamnetin, quercetin, acacetin, gardenin A/B, and thymonin, as shown in Table 4. A total of 185 targets for TFDM were screened by target prediction using SwissTargetPrediction (Figure 6). Prior to the prediction of potential VaD-related targets for TFDM, a combination of 2,495 targets predicted by the 12 most dysregulated miRNAs and 3,522 differentially expressed target genes resulted in 5,706 combined targets that were considered potential targets and comprised a library related to the pathophysiology of VaD. Subsequently, genes common to both the potential VaD target library and targets predicted for TFDM were analyzed and selected (Figure 7A), yielding 43 common targets, classified as key targets for TFDM (Figures 7A,B), and considered those effective for the treatment of VaD. Furthermore, the corresponding miRNAs that bind to the common targets were identified and analyzed, and the results indicated that 8 miRNAs mediated 43 targets that potentially contributed to the neuroprotective effects of TFDM (consisting of 42 flavonoid compounds) on VaD (Figure 8). The 15 most active compounds, 15 vital mRNAs, and 8 miRNAs were found from the list of eigenvector centrality in descending order, as listed in Table 5.
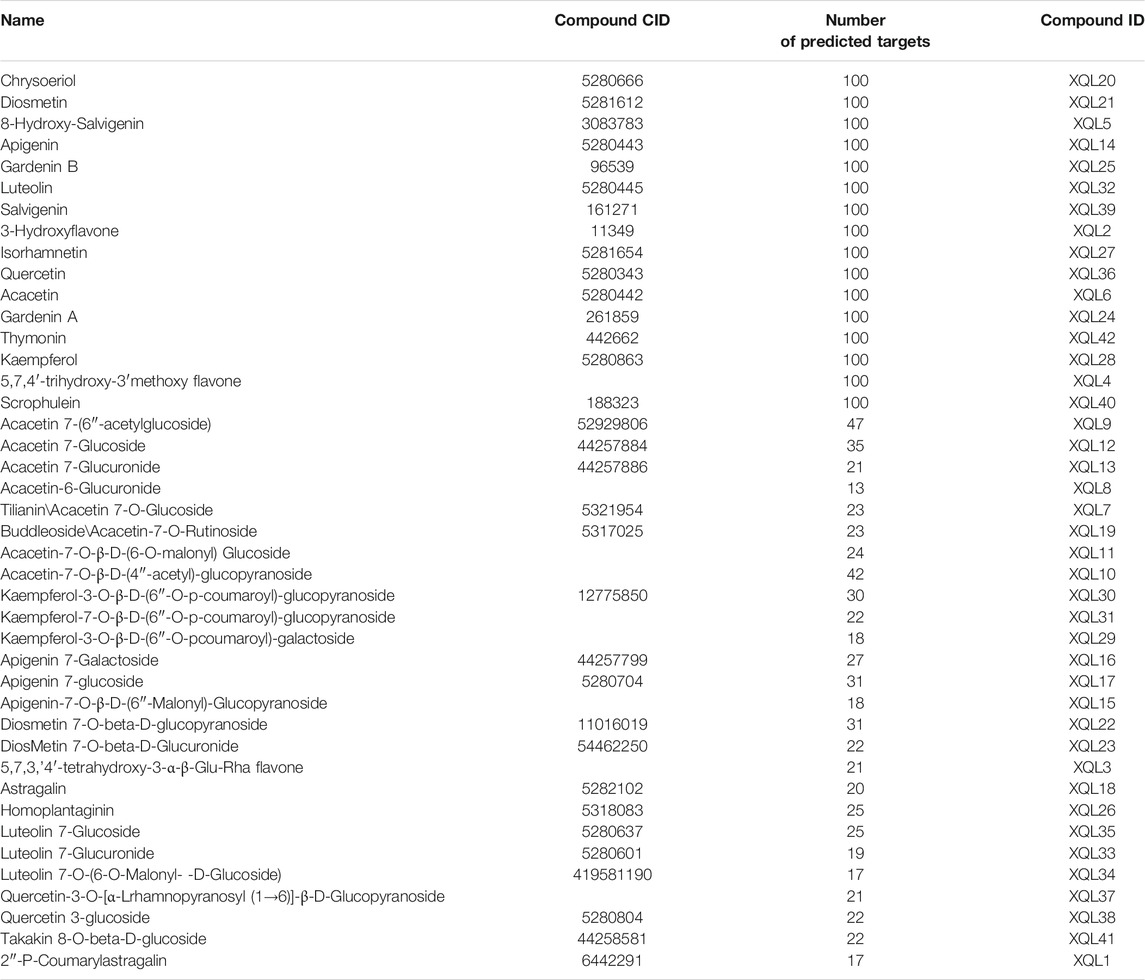
TABLE 4. Information including name, compound CID, number of predicted targets, and compound ID used in the present study on the 42 flavonoids in Dracocephalum moldavica L.
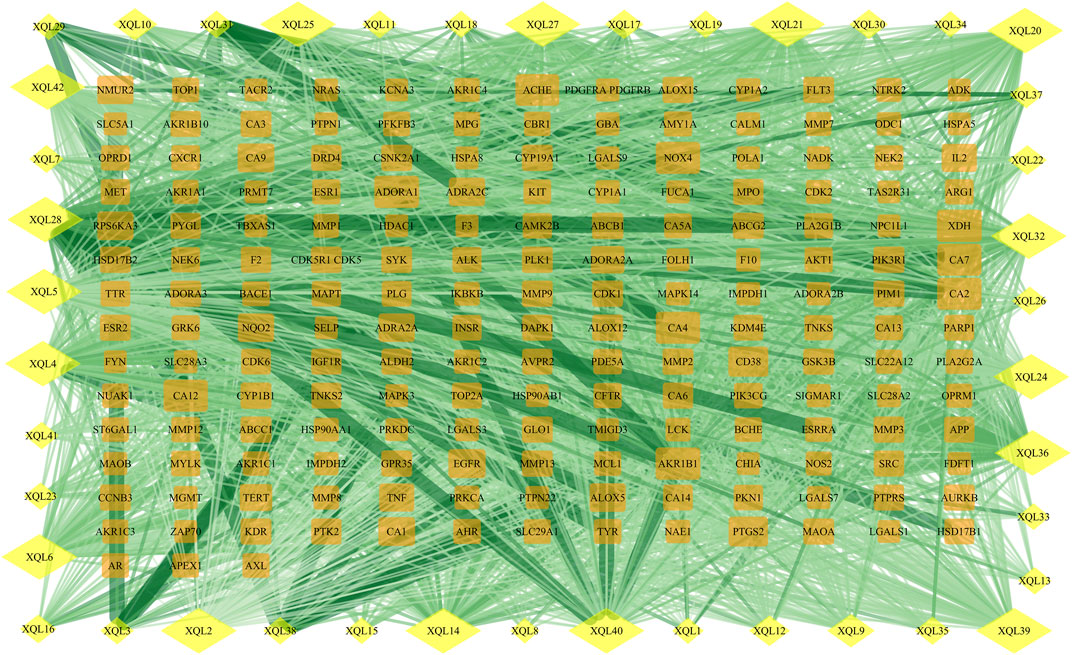
FIGURE 6. Protein network of TFDM-related targets. Yellow diamonds represent flavonoids, and orange rectangles represent potential targets of the compounds.
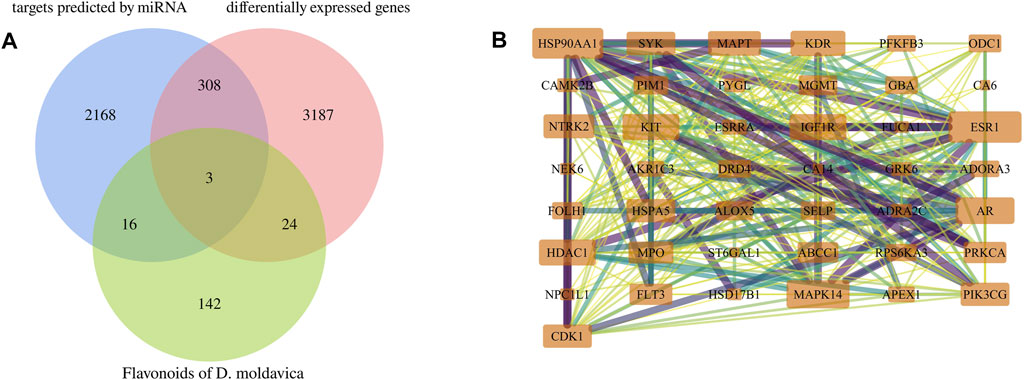
FIGURE 7. Identification of key targets for TFDM. (A) Intersection Venn diagram of potential targets of VaD and targets of TFDM. (B) Forty-three effective targets of TFDM for the treatment of VaD overlapped in the established potential VaD target library.
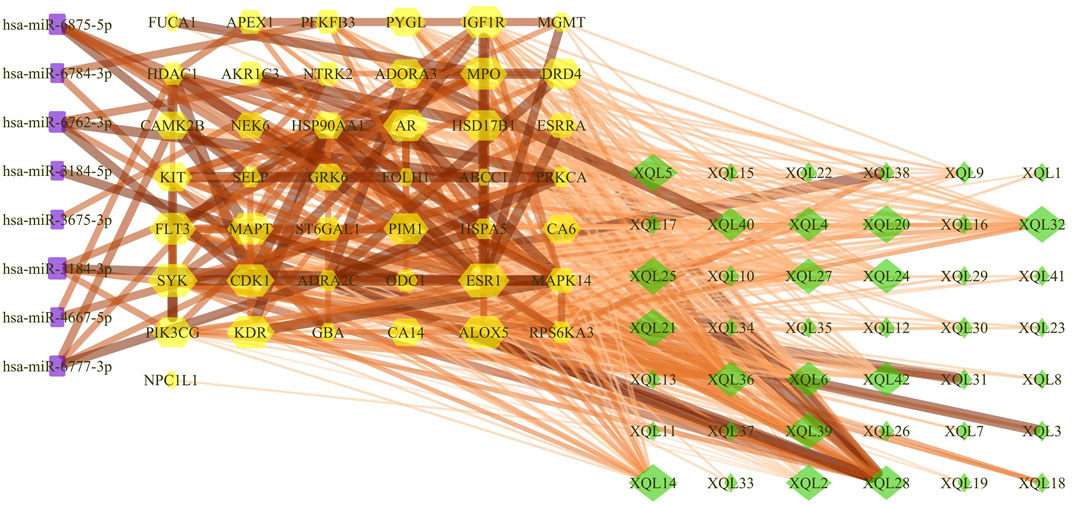
FIGURE 8. Compound-target gene-miRNA (C-T-M) network. The purple rectangle represents the miRNAs, the yellow hexagon represents the targets, and the green diamond represents the flavonoids of TFDM.
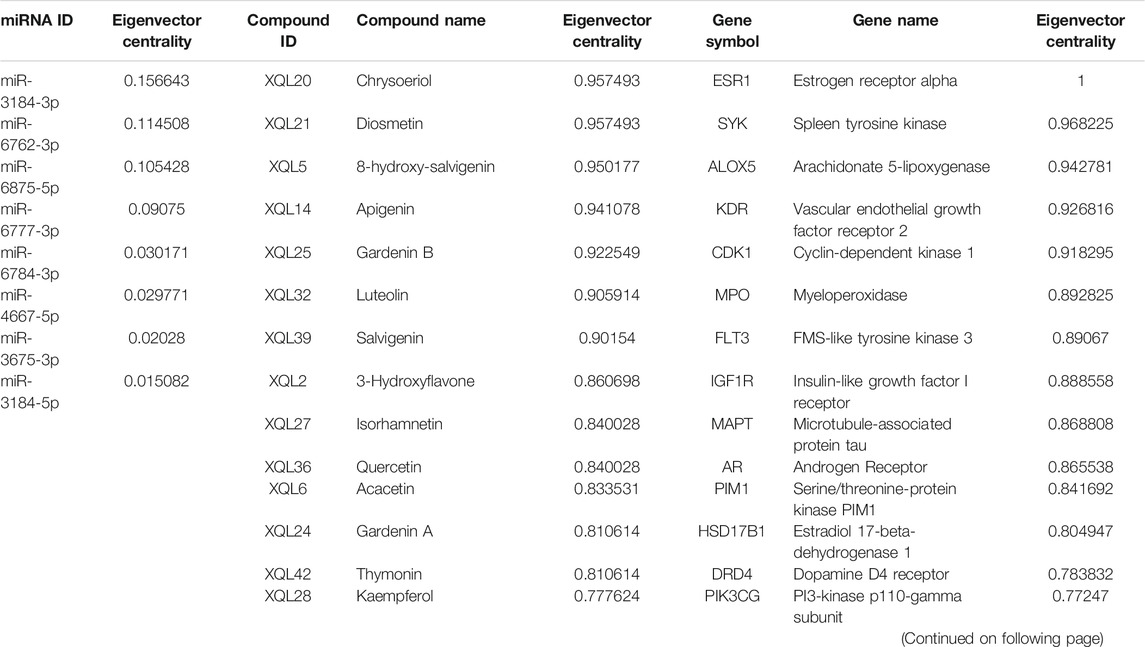
TABLE 5. Correspondence of top 15 compounds, target genes, and miRNAs based on eigenvector centrality within the predicted compound-mRNA-miRNA network of total flavonoids from Dracocephalum moldavica L. (TFDM).
Neuroprotection and Verification of DEMs Regulated by TFDM
Considering our previous research on TFDM, this compound and its representative flavonoids, including tilianin, luteolin, and apigenin, were selected to investigate the effects on the expression of predicted miRNAs. The fingerprint (approximately 71 peaks) of TFDM and the structure of tilianin, luteolin, and apigenin are shown in Figures 9A–D.
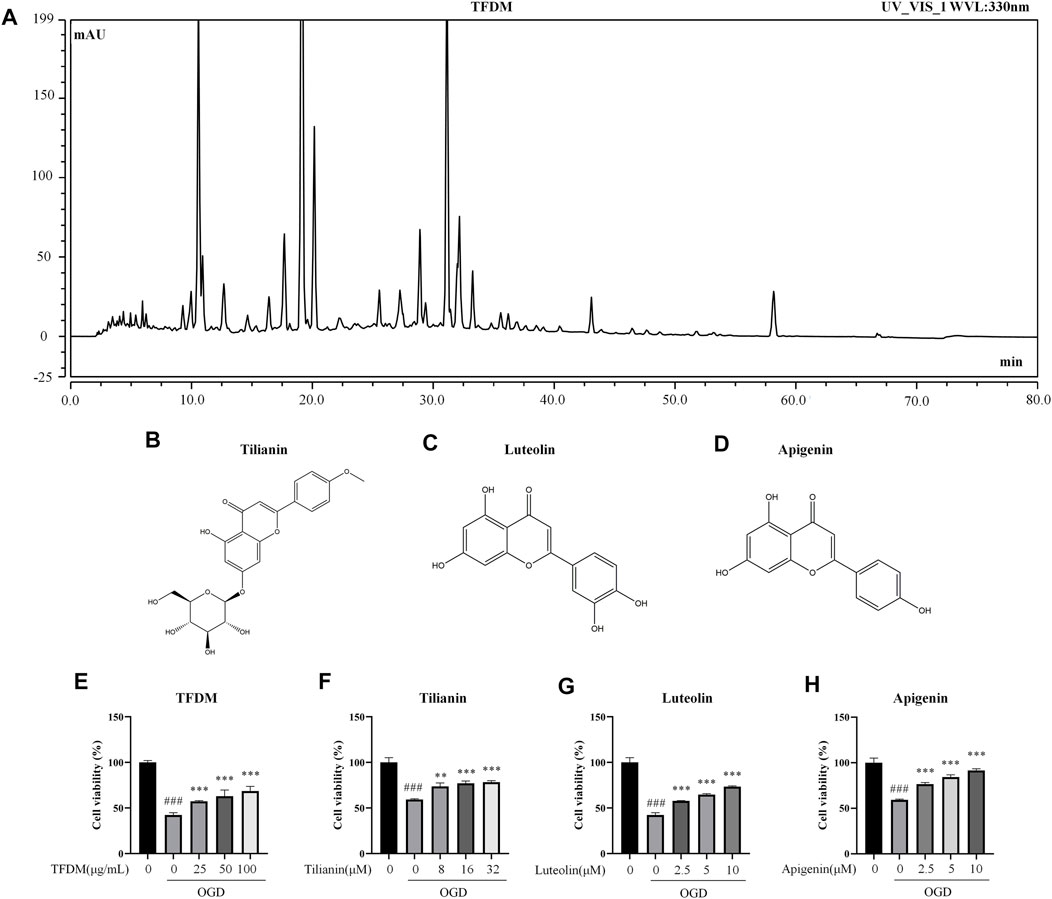
FIGURE 9. Fingerprint of TFDM, compound structures, and neuroprotective effects of TFDM as well as active compounds on neuronal cells against OGD injury. (A) HPLC chromatogram at 330 nm for TFDM. (B–D) Structure of tilianin (B), luteolin (C), and apigenin (D). (E) Improved cell viability of SH-SY5Y cells by TFDM after the exposure to OGD. (F–H) Improved cell viability of SH-SY5Y cells by tilianin (F), luteolin (G), and apigenin (H) after the exposure to OGD. Statistical analysis was performed using one-way ANOVA followed by a Tukey’s multiple comparison test. n = 3. ###p < 0.001 vs. control; **p < 0.01, ***p < 0.001 vs. OGD. Results are expressed as mean ± SD.
Before the verification of the predicted miRNAs regulated by TFDM, the neuroprotective effects of TFDM and its representative flavonoids on OGD-injured SH-SY5Y cells were measured. TFDM significantly improved the cell viability of OGD-injured SH-SY5Y cells in a dose-dependent manner at 25 μg/ml, 50 μg/ml, and 100 μg/ml (all p < 0.001), as shown in Figure 9E. The three active compounds from TFDM, such as tilianin, luteolin, and apigenin, also increased cell viability at the selected concentrations (p < 0.01–0.001, Figures 9F–H). These results suggested that TFDM and its three active compounds had neuroprotective effects against OGD-induced injury.
The relationship of the C-T-M network revealed that miR-3184-3p was a potential miRNA targeted by each flavonoid among the eight miRNAs. In addition, miR-6875-5p, miR-6777-3p, miR-6762-3p, and miR-6784-3p were targeted by 37, 17, 16, and 12 flavonoids from TFDM, respectively, suggesting that these five miRNAs might play a role in the treatment of VaD by TFDM. Therefore, miR-3184-3p, miR-6875-5p, miR-6777-3p, miR-6762-3p, and miR-6784-3p were selected to be confirmed in OGD-injured SH-SY5Y cells treated by TFDM.
According to the qRT-PCR results, the expression of miR-3184-3p was decreased, and miR-6875-5p was increased in response to the OGD injury (both p < 0.001, Figure 10A), consistent with the analysis of the C-T-M network. The exposure to OGD did not significantly changed the expression of miR-6784-3p compared with the control group (Figure 10A), while the expression of miR-6777-3p and miR-6762-3p was increased compared with the control group (p < 0.05 and 0.001), which was an opposite result compared to the C-T-M network. Thus, these results indicated that miR-3184-3p and miR-6875-5p were confirmed as having a significant aberrant change in the process of cerebral ischemia, which was consistent with previous bioinformatics analysis.
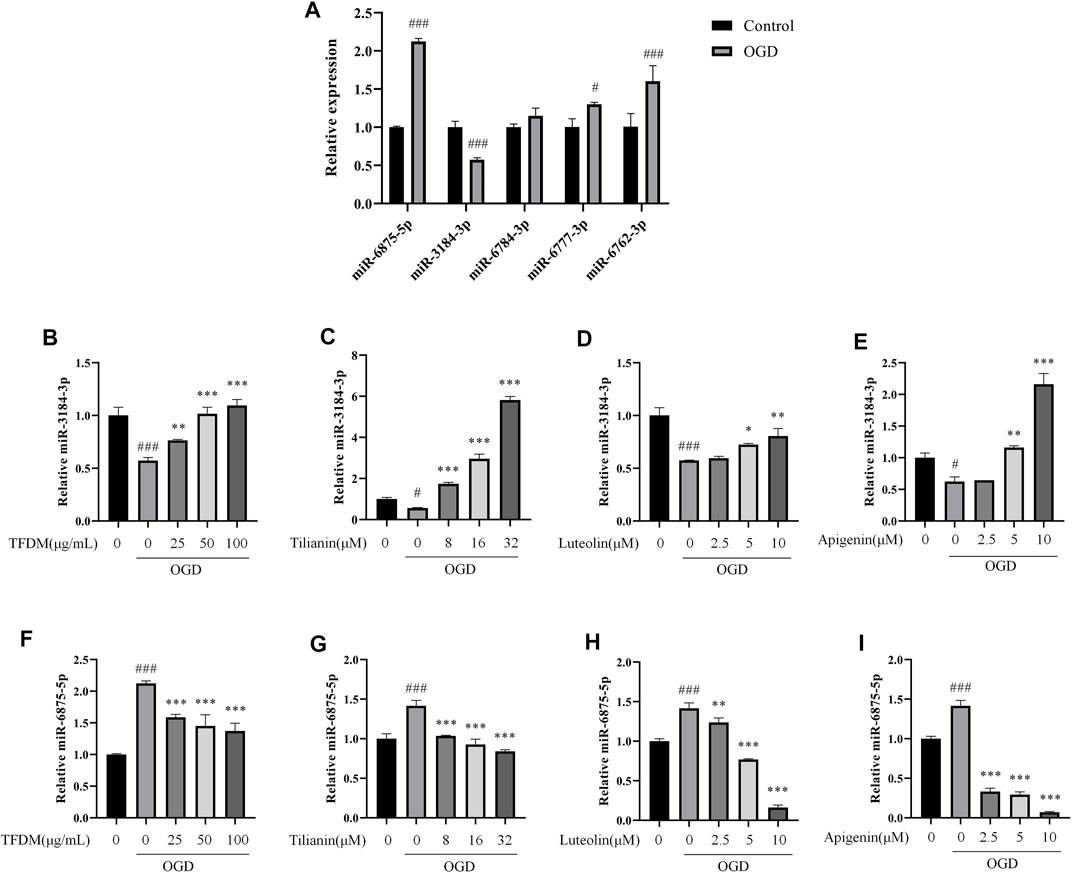
FIGURE 10. Regulation of miR-3184-3p and miR-6875-5p by TFDM and active compounds in OGD-injured SH-SY5Y cells. (A) Changes in the expression of miRNAs in OGD-injured SH-SY5Y cells compared with control cells. (B–E) Upregulation of miR-3184-3p in OGD-injured SH-SY5Y cells after the treatment with TFDM (B) and the active compounds tilianin (C), luteolin (D), and apigenin (E). (F–I) Downregulation of miR-6875-5p in OGD-injured SH-SY5Y cells after the treatment with TFDM (F) and the active compounds tilianin (G), luteolin (H), and apigenin (I). Statistical analysis was performed using Student’s t-test or one-way ANOVA followed by a Tukey’s multiple comparison test. n = 3. ###p < 0.001 vs. control; *p < 0.05, **p < 0.01, ***p < 0.001 vs. OGD. Results were expressed as mean ± SD.
Furthermore, miR-3184-3p was upregulated and miR-6875-5p was downregulated by TFDM treatment in a dose-dependent manner at 25 μg/ml, 50 μg/ml, and 100 μg/ml in OGD-injured SH-SY5Y cells compared with control cells (p < 0.01–0.001, Figures 10B,F), accompanied by the increase of cell viability. In the same way, tilianin, luteolin, and apigenin increased the expression of miR-3184-3p and decreased the expression of miR-6875-5p in OGD-injured SH-SY5Y cells compared with control cells (p < 0.05–0.001, Figures 10C–E and G-I). Therefore, these results suggested that miR-3184-3p and miR-6875-5p might play a role in the treatment of VaD by TFDM.
Discussion
To our knowledge, the present study is the first constructing a C-T-M network indicating the potential molecular mechanism of TFDM on the treatment of VaD. Subsequent confirmation provided evidence that miR-3184-3p and miR-6875-5p might be promising targets used by TFDM to exert therapeutic effects and synergistic action in the treatment of VaD.
In this study, 3,522 aberrantly expressed mRNAs were identified based on VaD microarray analysis, including 1,478 upregulated and 2,044 downregulated mRNAs. Among these genes with the most significant changes in expression, corticotropin-releasing hormone (CRH) and CD163 have a previously confirmed correlation with the occurrence and pathology of VaD (Heilig et al., 1995; Guo et al., 2018; Landis et al., 2018; Short et al., 2019). Several significantly upregulated genes in VaD were identified as first, including Heat shock 70 kDa protein 1A (HSPA1A), V-set and immunoglobulin domain containing 4 (VSIG4), and complement C1q B chain (C1QB), all defined as vascular disease risks (Dulin et al., 2010; Bos et al., 2017; Lyu et al., 2020).
A total of 2,562 miRNAs were significantly dysregulated, divided into 1,167 upregulated and 1,395 downregulated miRNAs. Based on a comprehensive study of data mining and bioinformatics analysis, the 12 most abnormally expressed miRNAs were acquired. Among these miRNAs, miR-6784-3p, miR-6762-3p, miR-4747-3p, and miR-4746-3p were reported for the first time. Other significantly expressed miRNAs, such as miR-6777-3p, miR-3184-3p, miR-3184-5p, miR-4667-5p, and miR-6875-5p, closely implicated with the occurrence and development of a variety of tumors (Miyamoto et al., 2015; Akazawa et al., 2019; Hindle et al., 2019; Zhou et al., 2020; Liu et al., 2021), were identified for the first time in VaD, suggesting a novel potential regulatory role in the pathology of this disease. miR-3675-3p is a unique biomarker representing a target in the inhibition of cell apoptosis through the MAPK signaling pathway (Zhang et al., 2020). As a novel miRNA identified in our analysis, the downregulation of miR-3675-3p might participate in the induction of neuronal apoptosis in VaD. miR-4467 and miR-1268a showed an increased expression in VaD in the present study. The former has previously been described as a promising and robust candidate biomarker in the diagnosis of neurodegenerative diseases by its analysis in body fluids (Denk et al., 2015), while the latter is significantly associated with cerebrovascular disorders (Sonoda et al., 2019) and hypertension (Li et al., 2020), predicting the risk of cerebrovascular diseases prior to the onset of stroke.
An overlap between DEGs in these miRNA-predicted targets and the large number of DEGs in VaD was constructed to gain insight into the molecular functions of the most abnormally expressed miRNAs in VaD, then the related biological function and pathways were analyzed. GO analysis indicated that the dysregulated mRNAs performed a crucial role in transcription, post-transcription, and protein modification. The primary biological processes in which the target genes of these selected aberrant miRNAs were involved included a variety of binding processes, including the binding of nucleotides and chromatin, cell differentiation, vascular events involving blood vessel development, muscle structure development, branching morphogenesis, and extensive regulation of receptor activity. The pathway enrichment analysis indicated the involvement of RAGE, Ras/MAPK, TNF, gonadotropin-releasing hormone (GnRH), and estrogen pathways, which are known as implicated in the pathogenesis of VaD (Emanuele et al., 2005; Belkhelfa et al., 2018; Wang et al., 2019; Yang et al., 2020b). Among these enriched pathways, the RAGE signaling pathway plays a role in inflammatory reactions through the proinflammatory cytokine overproduction mediated by the nuclear factor-κB (NF-κB), such as TNF-α (Vicente Miranda and Outeiro, 2010), both in the blood-brain barrier (BBB) and in the glial responses. MAPK is also a crucial signaling pathway in the pathological process of VaD, which destroys the BBB, activates inflammatory mediators, and induces programmed cell death (Mohamed et al., 2015). Thus, these predicted biological processes and signaling pathways indicate the correlation of pathological processes within miRNA-target genes, including neuroinflammation, vascular dysfunction, altered barrier permeability, and neuronal damage.
A total of 13 central genes were obtained through the topological analysis of the PPI network. Among them, WNT5A, a Wingless-type (Wnt) family member, induces different aspects of synaptic differentiation and plasticity in the hippocampal neurons (Varela-Nallar et al., 2010). WNT5A was a predicted target of miR-4667-5p in the present work, and the protein Wnt5a that it encodes is known to induce the rapid activation of the CaMKII (Farias et al., 2009), thereby reinforcing the Wnt/Ca2+ signaling pathway in the hippocampal neurons. Wnt5a also evokes cortical axon outgrowth and repression by calcium signaling pathways. It is required in the Wnt5a/CaMKII signaling pathway and is essential for the binding of tau that is phosphorylated at Ser262 (Li et al., 2014). In addition, dysfunction of the Wnt signaling is linked to an increased Aβ formation, neuroinflammation, and neurotoxicity (Li et al., 2011; Tapia-Rojas et al., 2016). Therefore, a change in WNT5A expression represents a considerable potential as an indicator of VaD pathology.
Other central genes may additionally be involved in different pathological processes of neurodegeneration. For instance, AAK1, a predicted gene regulated by miR-4667-5p, encodes the adaptor-associated kinase 1 and is a modulatory enzyme of the adaptor protein complex 2 (AP-2) responsible for the regulation of clathrin-mediated endocytosis (Smythe and Ayscough, 2003). In a murine model of AD periodic variation in AAK1 expression, it is closely correlated with cognitive decline (Fu et al., 2018). AAK1 inhibitors have been further developed to provide valuable treatments for disorders such as neuropathic pain, AD, and other neurodegenerative diseases (Abdel-Magid, 2017), indicating that AAK1 serves as a suitable biomarker of cognitive disorders, including VaD. By combining the analysis of various functionalities, the selected miRNAs and their central target genes were screened as biomarkers or potential targets for direct or indirect involvement in VaD.
Notably, the present study indicates that TFDM has multiple targets and influences numerous pathways that prevent and treat VaD. On the one hand, TFDM contains 42 flavonoids, reflecting the multi-component action. On the other hand, a single flavonoid component had multiple targets. Thus, a molecular network was established based on the target prediction of each flavonoid contained in TFDM using Cytoscape, which revealed 43 key genes related by TFDM against the pathophysiology of VaD. Among the flavonoids of TFDM, common neuroprotective compounds such as kaempferol, chrysoeriol, apigenin, luteolin, quercetin, and salvigenin exhibited a favorable target docking score performance. Furthermore, eigenvector centrality was used to express the importance of the nodes; those present in a free state (aglycones) exhibited much stronger potential activity toward the targets than the sugar-bound flavone glycosides.
Kaempferol, one of the active compounds in TFDM, is also an essential ingredient in the extracts of Ginkgo biloba leaves, having antioxidant properties by acting as a free radical scavenger (Ahlemeyer and Krieglstein, 2003). Kaempferol plays a role in the regulation of inflammation and apoptosis through the targeting of the PI3-kinase p110-gamma subunit (PIK3CG) (Shen et al., 2020). Luteolin, another active ingredient of TFDM, displays neuroprotective effects on the BBB by inhibiting fibrillary Aβ1–40-induced inflammation through the inhibition of the p38 MAPK signaling pathways, as reported by our previous work (Zhang et al., 2017). Quercetin, also reported by our research group, provides neurovascular protection by reducing oxidative damage, inactivating RAGE-mediated pathways, and maintaining cholinergic neuron function, thereby providing an alternative medication for neuroprotection (Liu et al., 2013). Tilianin, an indicative reference of TFDM, ameliorates the cognitive dysfunction and neuronal damage in VaD by modulating p-CaMKII-mediated long-term memory, oxidation and inflammation associated with the oxidized-CaMKII (Jiang et al., 2021). Taken together, these active compounds might be considered ingredients effective in the treatment of VaD. Furthermore, the underlying mechanisms and potential targets of these active ingredients in TFDM were located into the purpose of the current bioinformatics analysis. This might support the TFDM-mediated neuroprotection on the attenuation of the Aβ-induced cognitive decline and cardiocerebrovascular deficit by restoring the anti-amyloidogenic, neurotrophic, and MAPK pathways (Liu et al., 2018; Zeng et al., 2018).
The interactions between the targets and potential binding of miRNAs were analyzed to further explore the potential effective molecules and investigate the underlying mechanism used by these active compounds in TFDM. As demonstrated in the present study, miR-3184-3p might be a potential target gene of all the compounds in TFDM against VaD. miR-3184-3p targeting the PIK3CG was regulated by apigenin, luteolin, and quercetin against VaD. Another most enriched target miRNA was miR-6875-5p that could be regulated by 37 active components of TFDM. miR-6875-5p/G protein-coupled receptor kinase 6 (GRK6) and miR-6875-5p/RPS6KA3 might be responsible for the beneficial action of apigenin, luteolin, and tilianin. According to the results of this study, highly compound-enriched miR-6777-3p was targeted by apigenin, luteolin, quercetin, and kaempferol in the protection of blood vessels by regulating the expression of the steroid hormone receptor ERR1 (ESRRA) and CAMK2. In addition, other axes, including miR-3184-5p/ornithine decarboxylase (ODC1), miR-3675-3p/BDNF/NT-3 growth factors receptor (NTRK2), miR-4667-5p/Protein kinase C alpha type (PRKCA), miR-6784-3p/Lysosomal acid glucosylceramidase (GBA), and miR-6875-5p/MAPK14, are the most likely key mediators in the pathogenesis of VaD. Therefore, the discovery of novel miRNAs that mediate specific signaling pathways provides new insight into the function and molecular mechanisms of TFDM, which acts as a candidate for a multiple targeting therapy.
The expression of 5 miRNAs that were most closely related to TFDM in the treatment of ischemic injury was measured by qRT-PCR. The expression of miR-3184-3p and miR-6875-5p in OGD-injured neuronal cells matched the bioinformatics results, suggesting that both miRNAs might play essential roles in the VaD process. In line with the neuroprotective effects of TFDM with respect to OGD injury, the abnormal expression of miR-3184-3p and miR-6875-5p was restored by TFDM, as well as by the active compounds tilianin, luteolin, and apigenin.
Although the present study described putative new biomarkers for VaD, identifying a number of promising DEMs, the results require a greater depth of experimental confirmation, such as the function and interaction assays of key genes in vitro and in vivo. Based on bioinformatic analysis and experimental verification, miRNA-mediated RAGE signaling and CaMKII-induced multi-target pathways should be confirmed, as well as the underlying molecular basis of key genes and miRNA molecules in turn to confirm the therapeutic effects of TFDM on VaD.
Conclusion
In conclusion, differentially expressed miRNAs, mRNAs, and their functional properties during the pathogenesis of VaD were identified as contributors through bioinformatics analysis, thus revealing the therapeutic efficacy and molecular mechanisms of TFDM. The qRT-PCR validation suggested that miR-3184-3p and miR-6875-5p might be potential targets of TFDM, which might mediate multiple signaling pathways involved in the pathology of VaD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ML conducted the miRNA profiling analysis, performed the qRT-PCR experiments, and drafted the manuscript. GS completed the sample preparation, identified compounds, and revised the manuscript. HJ, LZ, and KZ performed miRNA-mRNA network analysis. YL performed the HPLC profiling of TFDM. GA participated in the mRNA and miRNA profiling analysis and revised the manuscript. RL and ZL designed the study, interpreted the results, and revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (U1803281) and CAMS Innovation Fund for Medical Science (2021-1-I2M-030). The Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia has funded this project, under grant no. (FP-176-43).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Prof Jianguo Xing, Key Laboratory of Uighur Medicine of Xinjiang Uygur Autonomous Region, Xinjiang Institute of Materia Medica for the provision of D. moldavica.
References
Abdel-Magid, A. F. (2017). Inhibitors of Adaptor-Associated Kinase 1 (AAK1) May Treat Neuropathic Pain, Schizophrenia, Parkinson's Disease, and Other Disorders. ACS Med. Chem. Lett. 8, 595–597. doi:10.1021/acsmedchemlett.7b00208
Ahlemeyer, B., and Krieglstein, J. (2003). Pharmacological Studies Supporting the Therapeutic Use of Ginkgo Biloba Extract for Alzheimer's Disease. Pharmacopsychiatry 36 (Suppl. 1), S8S14. doi:10.1055/s-2003-40454
Akazawa, Y., Mizuno, S., Fujinami, N., Suzuki, T., Yoshioka, Y., Ochiya, T., et al. (2019). Usefulness of Serum microRNA as a Predictive Marker of Recurrence and Prognosis in Biliary Tract Cancer after Radical Surgery. Sci. Rep. 9, 5925. doi:10.1038/s41598-019-42392-7
Belkhelfa, M., Beder, N., Mouhoub, D., Amri, M., Hayet, R., Tighilt, N., et al. (2018). The Involvement of Neuroinflammation and Necroptosis in the hippocampus during Vascular Dementia. J. Neuroimmunol 320, 48–57. doi:10.1016/j.jneuroim.2018.04.004
Bos, S., Phillips, M., Watts, G. F., Verhoeven, A. J. M., Sijbrands, E. J. G., and Ward, N. C. (2017). Novel Protein Biomarkers Associated with Coronary Artery Disease in Statin-Treated Patients with Familial Hypercholesterolemia. J. Clin. Lipidol. 11, 682–693. doi:10.1016/j.jacl.2017.03.014
Chen, Y., Wang, S., Yang, S., Li, R., Yang, Y., Chen, Y., et al. (2021). Inhibitory Role of Ginsenoside Rb2 in Endothelial Senescence and Inflammation Mediated by microRNA-216a. Mol. Med. Rep. 23, 415. doi:10.3892/mmr.2021.12054
Chu, S. F., Zhang, Z., Zhou, X., He, W. B., Chen, C., Luo, P., et al. (2019). Ginsenoside Rg1 Protects against Ischemic/reperfusion-Induced Neuronal Injury through miR-144/Nrf2/ARE Pathway. Acta Pharmacol. Sin. 40, 13–25. doi:10.1038/s41401-018-0154-z
Dastmalchi, K., Damien Dorman, H. J., Koşar, M., and Hiltunen, R. (2007). Chemical Composition and In Vitro Antioxidant Evaluation of a Water-Soluble Moldavian Balm (Dracocephalum Moldavica L.) Extract. LWT - Food Sci. Tech. 40, 239–248. doi:10.1016/j.lwt.2005.09.019
Deepa, P., Bae, H. J., Park, H. B., Kim, S. Y., Choi, J. W., Kim, D. H., et al. (2020). Dracocephalum Moldavica Attenuates Scopolamine-Induced Cognitive Impairment through Activation of Hippocampal ERK-CREB Signaling in Mice. J. Ethnopharmacol. 253, 112651. doi:10.1016/j.jep.2020.112651
Denk, J., Boelmans, K., Siegismund, C., Lassner, D., Arlt, S., and Jahn, H. (2015). MicroRNA Profiling of CSF Reveals Potential Biomarkers to Detect Alzheimer`s Disease. PLoS One 10, e0126423. doi:10.1371/journal.pone.0126423
Dulin, E., García-Barreno, P., and Guisasola, M. C. (2010). Extracellular Heat Shock Protein 70 (HSPA1A) and Classical Vascular Risk Factors in a General Population. Cell Stress Chaperones 15, 929–937. doi:10.1007/s12192-010-0201-2
Emanuele, E., D'Angelo, A., Tomaino, C., Binetti, G., Ghidoni, R., Politi, P., et al. (2005). Circulating Levels of Soluble Receptor for Advanced Glycation End Products in Alzheimer Disease and Vascular Dementia. Arch. Neurol. 62, 1734–1736. doi:10.1001/archneur.62.11.1734
Farías, G. G., Alfaro, I. E., Cerpa, W., Grabowski, C. P., Godoy, J. A., Bonansco, C., et al. (2009). Wnt-5a/JNK Signaling Promotes the Clustering of PSD-95 in Hippocampal Neurons. J. Biol. Chem. 284, 15857–15866. doi:10.1074/jbc.M808986200
Fu, X., Ke, M., Yu, W., Wang, X., Xiao, Q., Gu, M., et al. (2018). Periodic Variation of AAK1 in an Aβ1-42-Induced Mouse Model of Alzheimer's Disease. J. Mol. Neurosci. 65, 179–189. doi:10.1007/s12031-018-1085-3
Guo, L., Akahori, H., Harari, E., Smith, S. L., Polavarapu, R., Karmali, V., et al. (2018). CD163+ Macrophages Promote Angiogenesis and Vascular Permeability Accompanied by Inflammation in Atherosclerosis. J. Clin. Invest. 128, 1106–1124. doi:10.1172/JCI93025
Heilig, M., Sjögren, M., Blennow, K., Ekman, R., and Wallin, A. (1995). Cerebrospinal Fluid Neuropeptides in Alzheimer's Disease and Vascular Dementia. Biol. Psychiatry 38, 210–216. doi:10.1016/0006-3223(94)00239-Y
Hindle, A. G., Thoonen, R., Jasien, J. V., Grange, R. M. H., Amin, K., Wise, J., et al. (2019). Identification of Candidate miRNA Biomarkers for Glaucoma. Invest. Ophthalmol. Vis. Sci. 60, 134–146. doi:10.1167/iovs.18-24878
Huang, Y., Liao, X., Luo, J., Liu, H., Zhong, S., and Chen, J. (2020). Expression of Circular RNAs in the Vascular Dementia Rats. Neurosci. Lett. 735, 135087. doi:10.1016/j.neulet.2020.135087
Iadecola, C. (2013). The Pathobiology of Vascular Dementia. Neuron 80, 844–866. doi:10.1016/j.neuron.2013.10.008
Jia, J. X., Zhang, Y., Wang, Z. L., Yan, X. S., Jin, M., Huo, D. S., et al. (2017). The Inhibitory Effects of Dracocephalum Moldavica L. (DML) on Rat Cerebral Ischemia Reperfusion Injury. J. Toxicol. Environ. Health A. 80, 1206–1211. doi:10.1080/1528739410.1080/15287394.2017.1367139
Jiang, H., Ashraf, G. M., Liu, M., Zhao, K., Wang, Y., Wang, L., et al. (2021). Tilianin Ameliorates Cognitive Dysfunction and Neuronal Damage in Rats with Vascular Dementia via p-CaMKII/ERK/CREB and Ox-CaMKII-dependent MAPK/NF-κB Pathways. Oxid. Med. Cel. Longev. 2021, 6673967. doi:10.1155/2021/6673967
Jiang, H., Fang, J., Xing, J., Wang, L., Wang, Q., Wang, Y., et al. (2018). Tilianin Mediates Neuroprotection against Ischemic Injury by Attenuating CaMKII-dependent Mitochondrion-Mediated Apoptosis and MAPK/NF-κB Signaling. Life Sci. 216, 233–245. doi:10.1016/j.lfs.2018.11.035
Jiang, H., Xing, J., Fang, J., Wang, L., Wang, Y., Zeng, L., et al. (2020). Tilianin Protects against Ischemia/Reperfusion-Induced Myocardial Injury through the Inhibition of the Ca2+/Calmodulin-dependent Protein Kinase II-dependent Apoptotic and Inflammatory Signaling Pathways. Biomed. Res. Int. 2020, 5939715. doi:10.1155/2020/5939715
Kim, M. S., Bang, J. H., Lee, J., Kim, H. W., Sung, S. H., Han, J. S., et al. (2015). Salvia Miltiorrhiza Extract Protects white Matter and the hippocampus from Damage Induced by Chronic Cerebral Hypoperfusion in Rats. BMC Complement. Altern. Med. 15, 415. doi:10.1186/s12906-015-0943-6
Landis, R. C., Quimby, K. R., and Greenidge, A. R. (2018). M1/M2 Macrophages in Diabetic Nephropathy: Nrf2/HO-1 as Therapeutic Targets. Curr. Pharm. Des. 24, 2241–2249. doi:10.2174/1381612824666180716163845
Li, B., Zhong, L., Yang, X., Andersson, T., Huang, M., and Tang, S. J. (2011). WNT5A Signaling Contributes to Aβ-Induced Neuroinflammation and Neurotoxicity. PLoS One 6, e22920. doi:10.1371/journal.pone.0022920
Li, L., Fothergill, T., Hutchins, B. I., Dent, E. W., and Kalil, K. (2014). Wnt5a Evokes Cortical Axon Outgrowth and Repulsive Guidance by Tau Mediated Reorganization of Dynamic Microtubules. Dev. Neurobiol. 74, 797–817. doi:10.1002/dneu.22102
Li, Z., Chyr, J., Jia, Z., Wang, L., Hu, X., Wu, X., et al. (2020). Identification of Hub Genes Associated with Hypertension and Their Interaction with miRNA Based on Weighted Gene Coexpression Network Analysis (WGCNA) Analysis. Med. Sci. Monit. 26, e923514. doi:10.12659/MSM.923514
Liu, H., Li, Y., Zhang, J., Wu, N., Liu, F., Wang, L., et al. (2021). Erb‐B2 Receptor Tyrosine Kinase 2 is negatively regulated by the p53-responsive microRNA‐3184‐5p in cervical cancer cells. Oncol. Rep. 45, 95–106. doi:10.3892/or.2020.7862
Liu, Q. S., Jiang, H. L., Wang, Y., Wang, L. L., Zhang, J. X., He, C. H., et al. (2018). Total Flavonoid Extract from Dracoephalum Moldavica L. Attenuates β-amyloid-induced Toxicity through Anti-amyloidogenesic and Neurotrophic Pathways. Life Sci. 193, 214–225. doi:10.1016/j.lfs.2017.10.041
Liu, R., Gao, M., Qiang, G. F., Zhang, T. T., Lan, X., Ying, J., et al. (2009). The Anti-amnesic Effects of Luteolin against Amyloid Beta(25-35) Peptide-Induced Toxicity in Mice Involve the protection of Neurovascular Unit. Neuroscience 162, 1232–1243. doi:10.1016/j.neuroscience.2009.05.009
Liu, R., Zhang, T. T., Zhou, D., Bai, X. Y., Zhou, W. L., Huang, C., et al. (2013). Quercetin Protects against the Aβ(25-35)-Induced Amnesic Injury: Involvement of Inactivation of Rage-Mediated Pathway and Conservation of the NVU. Neuropharmacology 67, 419–431. doi:10.1016/j.neuropharm.2012.11.018
Liu, X., Feng, Z., Du, L., Huang, Y., Ge, J., Deng, Y., et al. (2019a). The Potential Role of MicroRNA-124 in Cerebral Ischemia Injury. Int. J. Mol. Sci. 21, 120. doi:10.3390/ijms21010120
Liu, X., Zhang, R., Wu, Z., Si, W., Ren, Z., Zhang, S., et al. (2019b). miR‐134‐5p/Foxp2/Syn1 is involved in cognitive impairment in an early vascular dementia rat model. Int. J. Mol. Med. 44, 1729–1740. doi:10.3892/ijmm.2019.4331
Liu, Y., Jin, M., and Wang, Z. (2019c). Advances in the Study of Flavonoids Content and Pharmacological Action of Dracocephalum Moldavica. Chin. J. Ethnomed. Ethnopharm. 28, 68–71.
Lyu, Q., Pang, X., Zhang, Z., Wei, Y., Hong, J., and Chen, H. (2020). Microglial V-Set and Immunoglobulin Domain-Containing 4 Protects against Ischemic Stroke in Mice by Suppressing TLR4-Regulated Inflammatory Response. Biochem. Biophys. Res. Commun. 522, 560–567. doi:10.1016/j.bbrc.2019.11.077
Maimaitiyiming, D., Hu, G., Aikemu, A., Hui, S. W., and Zhang, X. (2014). The Treatment of Uygur Medicine Dracocephalum Moldavica L on Chronic Mountain Sickness Rat Model. Pharmacogn. Mag. 10, 477–482. doi:10.4103/0973-1296.141817
Martínez-Vázquez, M., Estrada-Reyes, R., Martínez-Laurrabaquio, A., López-Rubalcava, C., and Heinze, G. (2012). Neuropharmacological Study of Dracocephalum Moldavica L. (Lamiaceae) in Mice: Sedative Effect and Chemical Analysis of an Aqueous Extract. J. Ethnopharmacol. 141, 908–917. doi:10.1016/j.jep.2012.03.028
McKay, E. C., Beck, J. S., Khoo, S. K., Dykema, K. J., Cottingham, S. L., Winn, M. E., et al. (2019). Peri-Infarct Upregulation of the Oxytocin Receptor in Vascular Dementia. J. Neuropathol. Exp. Neurol. 78, 436–452. doi:10.1093/jnen/nlz023
Miernisha, A., Bi, C. W., Cheng, L. K., Xing, J. G., Liu, J., Maiwulanjiang, M., et al. (2016). Badiranji Buya Keli, a Traditional Uyghur Medicine, Induces Vasodilation in Rat Artery: Signaling Mediated by Nitric Oxide Production in Endothelial Cells. Phytother. Res. 30, 16–24. doi:10.1002/ptr.5494
Miyamoto, S., Usami, S., Kuwabara, Y., Horie, T., Baba, O., Hakuno, D., et al. (2015). Expression Patterns of miRNA-423-5p in the Serum and Pericardial Fluid in Patients Undergoing Cardiac Surgery. PLoS One 10, e0142904. doi:10.1371/journal.pone.0142904
Mohamed, I. N., Ishrat, T., Fagan, S. C., and El-Remessy, A. B. (2015). Role of Inflammasome Activation in the Pathophysiology of Vascular Diseases of the Neurovascular Unit. Antioxid. Redox Signal. 22, 1188–1206. doi:10.1089/ars.2014.6126
O'Brien, J. T., and Thomas, A. (2015). Vascular Dementia. Lancet 386, 1698–1706. doi:10.1016/S0140-6736(15)00463-8
Qiao, P., Yan, H., and Wang, J. (2020). EGb761 Protects Brain Microvascular Endothelial Cells against Oxygen-Glucose Deprivation-Induced Injury through lncRNA Rmst/miR-150 Axis. Neurochem. Res. 45, 2398–2408. doi:10.1007/s11064-020-03099-8
Ren, Z., Yu, J., Wu, Z., Si, W., Li, X., Liu, Y., et al. (2018). MicroRNA-210-5p Contributes to Cognitive Impairment in Early Vascular Dementia Rat Model through Targeting Snap25. Front. Mol. Neurosci. 11, 388. doi:10.3389/fnmol.2018.00388
Shen, X., Yang, R., An, J., and Zhong, X. (2020). Analysis of the Molecular Mechanisms of the Effects of Prunella Vulgaris against Subacute Thyroiditis Based on Network Pharmacology. Evid. Based Complement. Alternat. Med. 2020, 9810709. doi:10.1155/2020/9810709
Shigemizu, D., Akiyama, S., Asanomi, Y., Boroevich, K. A., Sharma, A., Tsunoda, T., et al. (2019a). Risk Prediction Models for Dementia Constructed by Supervised Principal Component Analysis Using miRNA Expression Data. Commun. Biol. 2, 77. doi:10.1038/s42003-019-0324-7
Shigemizu, D., Akiyama, S., Asanomi, Y., Boroevich, K. A., Sharma, A., Tsunoda, T., et al. (2019b). A Comparison of Machine Learning Classifiers for Dementia with Lewy Bodies Using miRNA Expression Data. BMC Med. Genomics 12, 150. doi:10.1186/s12920-019-0607-3
Short, A. K., Maras, P. M., Pham, A. L., Ivy, A. S., and Baram, T. Z. (2019). Blocking CRH Receptors in Adults Mitigates Age-Related Memory Impairments Provoked by Early-Life Adversity. Neuropsychopharmacology 45, 515–523. doi:10.1038/s41386-019-0562-x
Smythe, E., and Ayscough, K. R. (2003). The Ark1/Prk1 Family of Protein Kinases. Regulators of Endocytosis and the Actin Skeleton. EMBO Rep. 4, 246–251. doi:10.1038/sj.embor.embor776
Sonoda, T., Matsuzaki, J., Yamamoto, Y., Sakurai, T., Aoki, Y., Takizawa, S., et al. (2019). Serum MicroRNA-Based Risk Prediction for Stroke. Stroke 50, 1510–1518. doi:10.1161/STROKEAHA.118.023648
Tapia-Rojas, C., Burgos, P. V., and Inestrosa, N. C. (2016). Inhibition of Wnt Signaling Induces Amyloidogenic Processing of Amyloid Precursor Protein and the Production and Aggregation of Amyloid-β (Aβ)42 Peptides. J. Neurochem. 139, 1175–1191. doi:10.1111/jnc.13873
Thal, D. R., Grinberg, L. T., and Attems, J. (2012). Vascular Dementia: Different Forms of Vessel Disorders Contribute to the Development of Dementia in the Elderly Brain. Exp. Gerontol. 47, 816–824. doi:10.1016/j.exger.2012.05.023
Toyama, K., Spin, J. M., Deng, A. C., Huang, T. T., Wei, K., Wagenhäuser, M. U., et al. (2018). MicroRNA-Mediated Therapy Modulating Blood-Brain Barrier Disruption Improves Vascular Cognitive Impairment. Arterioscler. Thromb. Vasc. Biol. 38, 1392–1406. doi:10.1161/ATVBAHA.118.310822
Varela-Nallar, L., Alfaro, I. E., Serrano, F. G., Parodi, J., and Inestrosa, N. C. (2010). Wingless-type Family Member 5A (Wnt-5a) Stimulates Synaptic Differentiation and Function of Glutamatergic Synapses. Proc. Natl. Acad. Sci. U S A. 107, 21164–21169. doi:10.1073/pnas.1010011107
Vicente Miranda, H., and Outeiro, T. F. (2010). The Sour Side of Neurodegenerative Disorders: the Effects of Protein Glycation. J. Pathol. 221, 13–25. doi:10.1002/path.2682
Vijayan, M., and Reddy, P. H. (2020). Non-Coding RNAs Based Molecular Links in Type 2 Diabetes, Ischemic Stroke, and Vascular Dementia. J. Alzheimers Dis. 75, 353–383. doi:10.3233/JAD-200070
Wang, J., Chen, W., and Wang, Y. (2013). A Ginkgo Biloba Extract Promotes Proliferation of Endogenous Neural Stem Cells in Vascular Dementia Rats. Neural Regen. Res. 8, 1655–1662. doi:10.3969/j.issn.1673-5374.2013.18.003
Wang, J., Li, D., Hou, J., and Lei, H. (2018). Protective Effects of Geniposide and Ginsenoside Rg1 Combination Treatment on Rats Following Cerebral Ischemia Are Mediated via Microglial microRNA-155-5p I-nhibition. Mol. Med. Rep. 17, 3186–3193. doi:10.3892/mmr.2017.8221
Wang, L., Yang, J. W., Lin, L. T., Huang, J., Wang, X. R., Su, X. T., et al. (2020). Acupuncture Attenuates Inflammation in Microglia of Vascular Dementia Rats by Inhibiting miR-93-Mediated TLR4/MyD88/NF-Κb Signaling Pathway. Oxid. Med. Cel. Longev. 2020, 8253904. doi:10.1155/2020/8253904
Wang, X., Wang, Y., Zhu, Y., Yan, L., and Zhao, L. (2019). Neuroprotective Effect of S-Trans, Trans-farnesylthiosalicylic Acid via Inhibition of RAS/ERK Pathway for the Treatment of Alzheimer's Disease. Drug Des. Devel. Ther. 13, 4053–4063. doi:10.2147/DDDT.S233283
Wu, C., Liu, H., Rong, X., Liu, J., Ding, W., Cheng, X., et al. (2020). Phytochemical Composition Profile and Space-Time Accumulation of Secondary Metabolites for Dracocephalum Moldavica Linn. Via UPLC-Q/TOF-MS and HPLC-DAD Method. Biomed. Chromatogr. 34, e4865. doi:10.1002/bmc.4865
Xu, H., Nie, B., Liu, L., Zhang, C., Zhang, Z., Xu, M., et al. (2019). Curcumin Prevents Brain Damage and Cognitive Dysfunction during Ischemic-Reperfusion through the Regulation of miR-7-5p. Curr. Neurovasc. Res. 16, 441–454. doi:10.2174/1567202616666191029113633
Yang, F. W., Wang, H., Wang, C., and Chi, G. N. (2020a). Upregulation of Acetylcholinesterase Caused by Downregulation of microRNA-132 Is Responsible for the Development of Dementia after Ischemic Stroke. J. Cel. Biochem. 121, 135–141. doi:10.1002/jcb.28985
Yang, L., Xing, J., He, C., and Yang, P. (2013). Chemical Constituents of Dracocephalum Moldavica L. And Their Pharmacological Activities. World Clin. Drugs 34, 226–231.
Yang, Y., Zhao, L., Li, N., Dai, C., Yin, N., Chu, Z., et al. (2020b). Estrogen Exerts Neuroprotective Effects in Vascular Dementia Rats by Suppressing Autophagy and Activating the Wnt/β-Catenin Signaling Pathway. Neurochem. Res. 45, 2100–2112. doi:10.1007/s11064-020-03072-5
Yu, P., Venkat, P., Chopp, M., Zacharek, A., Shen, Y., Ning, R., et al. (2019). Role of microRNA-126 in Vascular Cognitive Impairment in Mice. J. Cereb. Blood Flow. Metab. 39, 2497–2511. doi:10.1177/0271678X18800593
Zeng, C., Jiang, W., Yang, X., He, C., Wang, W., and Xing, J. (2018). Pretreatment with Total Flavonoid Extract from Dracocephalum Moldavica L. Attenuates Ischemia Reperfusion-Induced Apoptosis. Sci. Rep. 8, 17491. doi:10.1038/s41598-018-35726-4
Zhang, H., Zhang, M., Meng, L., Guo, M., Piao, M., Huang, Z., et al. (2020). Investigation of Key miRNAs and Their Target Genes Involved in Cell Apoptosis during Intervertebral Disc Degeneration Development Using Bioinformatics Methods. J. Neurosurg. Sci. doi:10.23736/S0390-5616.20.04773-6
Zhang, J. X., Xing, J. G., Wang, L. L., Jiang, H. L., Guo, S. L., and Liu, R. (2017). Luteolin Inhibits Fibrillary β-Amyloid1-40-Induced Inflammation in a Human Blood-Brain Barrier Model by Suppressing the P38 MAPK-Mediated NF-Κb Signaling Pathways. Molecules 22, 334. doi:10.3390/molecules22030334
Zhao, L., Wang, J. L., Liu, R., Li, X. X., Li, J. F., and Zhang, L. (2013). Neuroprotective, Anti-amyloidogenic and Neurotrophic Effects of Apigenin in an Alzheimer's Disease Mouse Model. Molecules 18, 9949–9965. doi:10.3390/molecules18089949
Zhou, F., Lei, Y., Xu, X., Zhou, H., Liu, H., Jiang, J., et al. (2020). LINC00355:8 Promotes Cell Proliferation and Migration with Invasion via the MiR-6777-3p/Wnt10b axis in Hepatocellular Carcinoma. J. Cancer 11, 5641–5655. doi:10.7150/jca.43831
Keywords: bioinformatics, Dracocephalum moldavica L., flavonoids, microRNA, network regulation, vascular dementia
Citation: Liu M, Shan G, Jiang H, Zeng L, Zhao K, Li Y, Ashraf GM, Li Z and Liu R (2021) Identification of miRNA and Their Regulatory Effects Induced by Total Flavonoids From Dracocephalum moldavica in the Treatment of Vascular Dementia. Front. Pharmacol. 12:796628. doi: 10.3389/fphar.2021.796628
Received: 17 October 2021; Accepted: 17 November 2021;
Published: 06 December 2021.
Edited by:
Yimin Cui, Peking University First Hospital, ChinaCopyright © 2021 Liu, Shan, Jiang, Zeng, Zhao, Li, Ashraf, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuorong Li, bGl6aHVvcm9uZ0BpbWIucHVtYy5lZHUuY24=; Rui Liu, bGl1cnVpQGltYi5wdW1jLmVkdS5jbg==
†These authors have contributed equally to this work
 Mimin Liu
Mimin Liu Guangzhi Shan
Guangzhi Shan Hailun Jiang
Hailun Jiang Li Zeng
Li Zeng Kaiyue Zhao
Kaiyue Zhao Yiran Li
Yiran Li Ghulam Md Ashraf
Ghulam Md Ashraf Zhuorong Li
Zhuorong Li Rui Liu
Rui Liu