- 1School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing, China
- 2National Institute of TCM Constitution and Preventive Medicine, Beijing University of Chinese Medicine, Beijing, China
- 3Tang Center for Herbal Medicine Research, The University of Chicago, Chicago, IL, United States
- 4Department of Anesthesia and Critical Care, The University of Chicago, Chicago, IL, United States
Accumulating knowledge has been achieved on DNA methylation participating in numerous cellular processes and multiple human diseases; however, few studies have addressed the pleiotropic role of DNA methylation in Chinese herbal medicine (CHM). CHM has been used worldwide for the prevention and treatment of multiple diseases. Newly developed epigenetic techniques have brought great opportunities for the development of CHM. In this review, we summarize the DNA methylation studies and portray the pleiotropic role of DNA methylation in CHM. DNA methylation serves as a mediator participating in plant responses to environmental factors, and thus affecting CHM medicinal plants growth and bioactive compound biosynthesis which are vital for therapeutic effects. Furthermore, DNA methylation helps to uncover the pharmaceutical mechanisms of CHM formulae, herbs, and herbal-derived compounds. It also provides scientific validation for constitution theory and other essential issues of CHM. This newly developed field of DNA methylation is up-and-coming to address many complicated scientific questions of CHM; it thus not only promotes disease treatment but also facilitates health maintenance.
Introduction
Chinese herbal medicine (CHM), originated in ancient China, has been used worldwide to prevent and treat different medical conditions (Ang-Lee et al., 2001; Pang et al., 2017). With multi-target and holistic approaches, CHMs have shown advantages in managing multi-factorial, systemic, and complex diseases, such as cancer, metabolic syndrome, and autoimmune diseases (Zhong et al., 2016). Newly developed biomedical technologies, such as “-omics” analysis, have triggered the transformation of the scientific paradigm and shifted from reductionism to holism (Kellogg et al., 2018; Lippolis et al., 2018). These new approaches have echoed the philosophy and therapeutics of CHM (Fardet and Rock, 2014; Delker and Mann, 2017; Hu and Su, 2017).
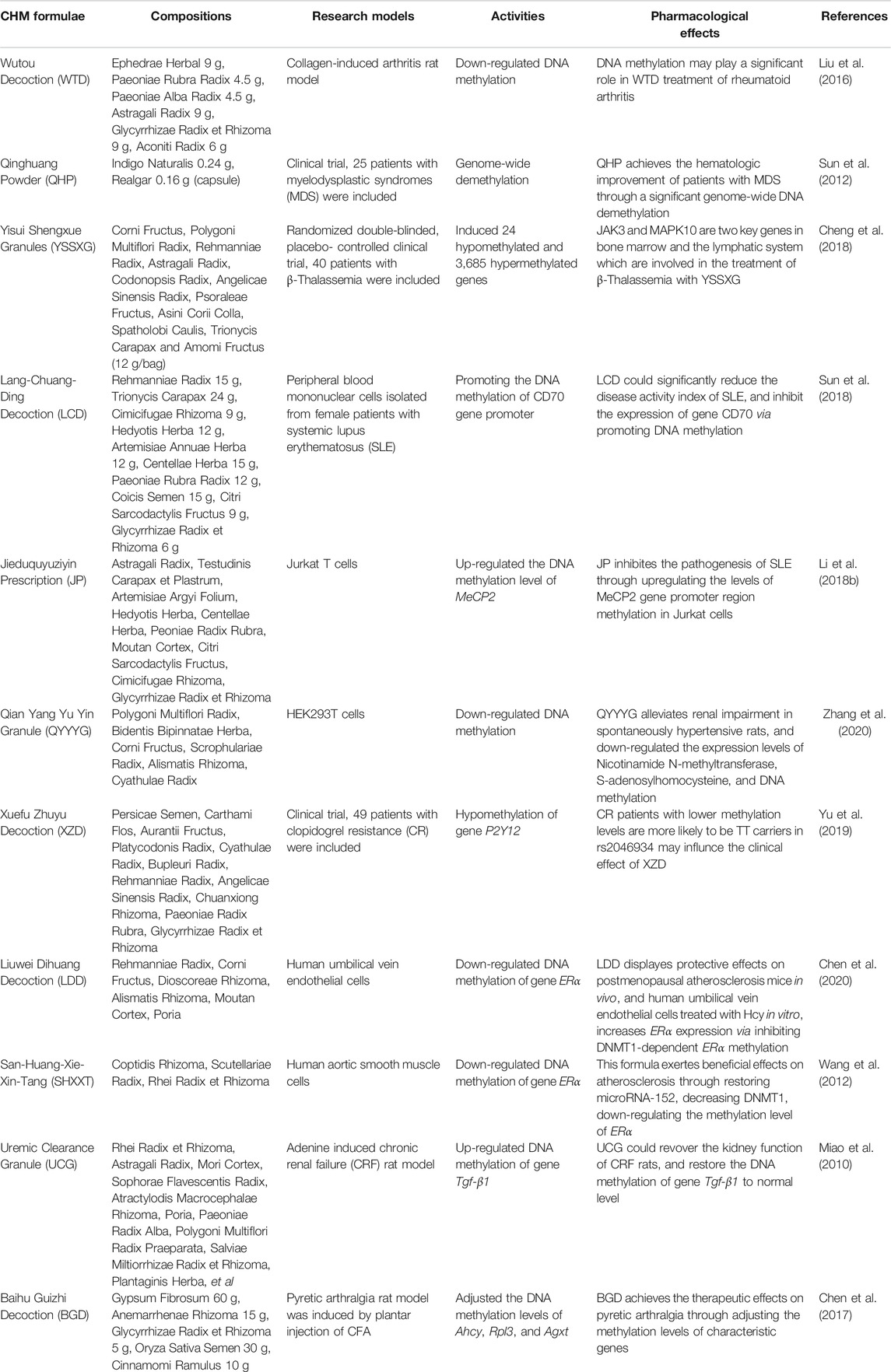
The neologism “-omics,” including genomics, proteomics, and metabolomics, refers to the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism (Kedaigle and Fraenkel, 2018). The current “-omics” studies provide new opportunities for the potential future development of the CHM (Yang et al., 2015). In the last decade, significant developments have been achieved in epigenomics studies, focusing on the complete set of epigenetic modifications. Unlike the classic Mendelian inheritance, which focuses on mutations of the DNA sequence, the epigenetic modifications are reversible and heritable; they can modify the gene expression without altering the DNA sequence (Elhamamsy, 2017; Feinberg, 2018) (Figure 1). Thus, epigenetic modifications play an essential role in regulating gene expression and are crucial for various cellular processes (Wilkinson and Gozani, 2017).
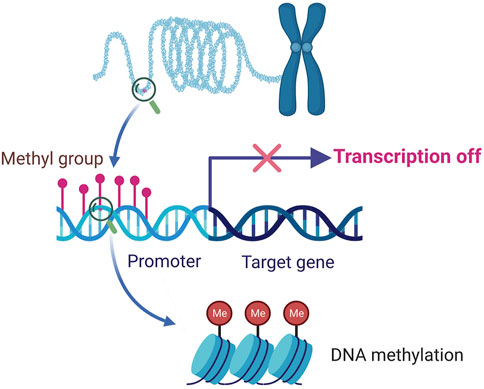
FIGURE 1. The primary function of DNA methylation. As one of the most important epigenetic mechanisms, DNA methylation refers to the process of methyl groups being added to the DNA molecule. When DNA methylation occurs in promoter regions, the gene transcription may be repressed without changing the DNA sequence.
DNA methylation is one of the most broadly studied and well-characterized epigenetic modifications (Nowacka-Zawisza and Wisnik, 2017; Draht et al., 2018; Islam et al., 2018). DNA methylation involves adding a methyl group to the 5-carbon position of the cytosine ring of DNA by DNA methyltransferases (Schubeler, 2015) (Figure 2). DNA methylation plays a crucial role in gene expression and regulation, and faulty methylation could induce devastating consequences, including various fatal human diseases, such as cancer, lupus, and cardiovascular diseases (Liu et al., 2017; Fernandez-Sanles et al., 2018; Imgenberg-Kreuz et al., 2018).
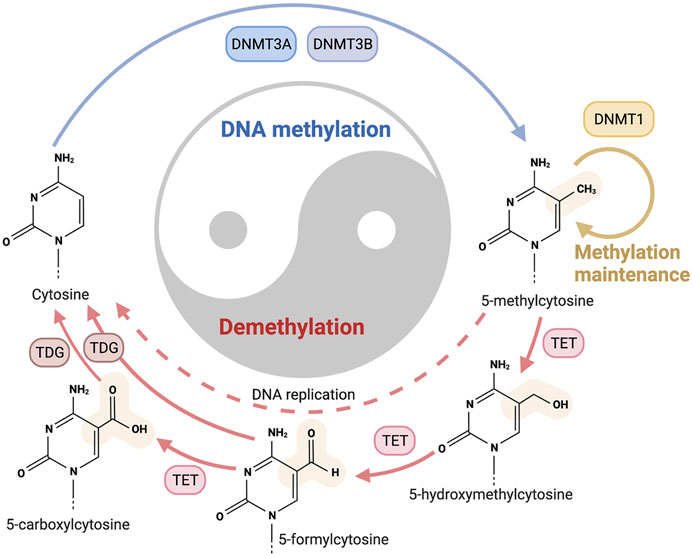
FIGURE 2. Processing of DNA methylation and demethylation, these two mechanisms may achieve a changeable balance. The formation and maintenance of DNA methylation need the catalysis of DNA methyltransferases (DNMTs) to produce 5-methyl-cytosine from cytosine (indicated with blue line). The demethylation process contains two pathways. The passive pathway refers to the dilution of the modified cytosines during DNA replications (indicated with red dashed lines). The active pathway of demethylation to reverse 5-methyl-cytosine to cytosine, enzymes of ten-eleven translocation (TET), and thymine DNA glycosylase (TDG) participate in this process (indicated with red lines).
Multiple technologies have been applied in the research of CHM, and fruitful outcomes have been achieved (Wang et al., 2013; Gu and Pei, 2017). The recent development of DNA methylation technique sheds new light on the CHM research, and many related studies have also been conducted, which helped to provide a better understanding of some puzzling issues (Hsieh et al., 2011; Liu et al., 2016; Yao et al., 2018b) (Figure 3). In this review, the application of DNA methylation in CHM research will be presented, and specific attention will be placed on its clinical utility; attempts will be made to elucidate the research trends in this field to benefit future studies.
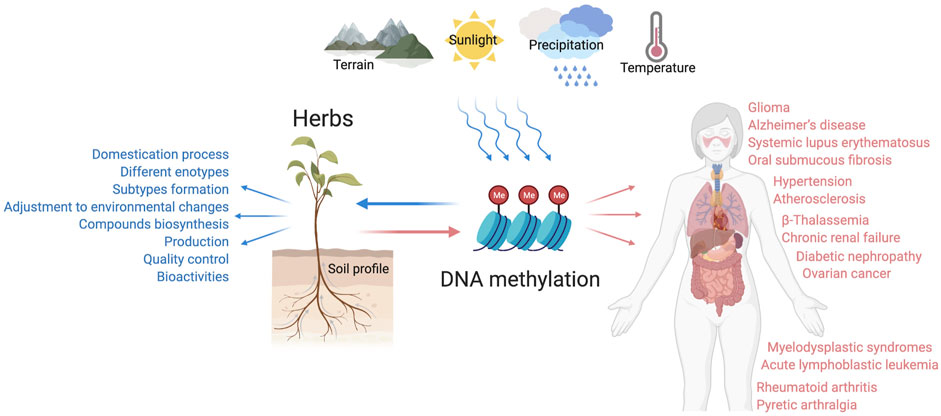
FIGURE 3. The pleiotropic role of DNA methylation in the growth and pharmacological activities of CHM botanical drugs. DNA methylation serves as a mediator mechanism for the medicinal plants to respond to the ever-changing environmental factors, which affect the domestication process, subtypes formation, compounds biosynthesis, and many other bioprocesses. DNA methylation is also indispensable for herbs to exert therapeutic functions related to multisystem diseases, such as atherosclerosis, hypertension, Alzheimer’s disease, ovarian cancer, rheumatoid arthritis, etc.
Concept and Biofunctions of DNA Methylation
DNA methylation is a dynamic state which reflects the changeable balance achieved by DNA methyltransferase-induced methylation and DNA demethylase-induced demethylation (Figure 2). DNA methylation exerts comprehensive biofunctions in plants and mammals, and displays great significance in the medical field.
Patterns of DNA Methylation and Demethylation
DNA methylation is a very common biological phenomenon that exists in most eukaryotic organisms including both animals and plants, although they have different manifestations. In mammals, DNA methylation mainly occurs in the context of CG to form a cytosine-guanine dinucleotide (CpG) site; while, in plants, there are more types of DNA methylation, it may occur in all cytosine sequence contexts of CG, CHG, and CHH (H = A, C, or T) (He et al., 2011).
In mammals, DNA methylation is catalyzed by DNA methyltransferases (DNMTs), i.e., DNMT1, DNMT3A, and DNMT3B. The de novo DNA methylation is mainly induced by DNMT3A and DNMT3B, and DNMT1 is crucial for the maintenance of DNA methylation (Figure 2). DNA methylation can be reversed by demethylation which contains passive and active pathways (Greenberg and Bourc’his, 2019). During DNA replication, DNA methylation may be diluted, and this process is the passive pathway of DNA demethylation. The active pathway of demethylation is catalyzed by enzymes of ten-eleven translocation (TET), and thymine DNA glycosylase (TDG) (Kohli and Zhang, 2013).
In plants, the de novo DNA methylation is mediated by three types of DNMTs: MET1, CMT3, and DRM2. The maintenance of CG methylation mainly relies on MET1 which is a homolog of DNMT1 in mammals, while the maintenance of CHG DNA methylation is catalyzed by CMT3. DRM2 induces de novo DNA methylation at all sequence contexts in plants through the RNA-directed DNA methylation (RdDM) pathway (He et al., 2011; Zhang et al., 2018). As for DNA demethylation in plants, the passive pathway is similar to mammals, whereas the active DNA demethylation pathway is distinct. Plants can directly remove the 5-methylcytosine (5-mC) base by 5-mC DNA glycosylases which is different from the way mammals initiate active DNA demethylation by the oxidation and deamination of 5-mC (Zhang et al., 2018).
Biofunctions of DNA Methylation
DNA methylation plays essential roles in diverse biological processes involving the regulation of gene expression, transposon silencing, chromosome interactions, and trait inheritance (Luo et al., 2018). The most noticeable molecular function of DNA methylation is gene regulation. When DNA methylation occurs in the promoter region it usually inhibits gene transcription, for DNA methylation can impede the binding of transcription factors to their motifs, thus the transcription process can be hindered (Yin et al., 2017) (Figure 1). DNA methylation also participates in the process of heterochromatin formation via recruiting the chromatin remodelers and modifiers by DNMT proteins to chromatin (Greenberg and Bourc’his, 2019). Except for genes, the main targets of DNA methylation are transposable elements, and transposons may harm genome stability by relocating DNA transposons or inserting retrotransposons. CpG-rich promoters control the expression of active retrotransposons. DNA methylation can induce the silence of transposons and promote the irreversible genetic inactivation of transposable elements through mutagenic deamination, and thus support genome stability (Barau et al., 2016). DNA methylation can also reprogram the development of both mammals and plants, exert regulatory effects in the bioprocesses of early embryogenesis, X chromosome inactivation, genomic imprinting, stem cell differentiation (He et al., 2011; Schubeler, 2015; Zhang et al., 2018). Aberrant DNA methylation is shown to be involved in the occurrence and development of multiple diseases, a host of promising diagnostic and therapeutic approaches based on DNA methylation has been explored and partially applied in the clinical practice (Dor and Cedar, 2018; Papanicolau-Sengos and Aldape, 2021) (Figure 3). DNA methylation research also sheds new light on the field of CHM, providing a perspective to unveil the obscure mechanisms behind the profound CHM clinical experience.
Dna Methylation in the Growth and Development of Medicinal Plants in Chinese Herbal Medicine
Planting and growth are the key basis for the curative effects of CHM herbs. DNA methylation is an effective mechanism that participates in multiple biological processes of the growth and development of medicinal plants, such as subtypes domestication, development, and the adjustment to dynamic environments (Table 1). Investigations of several selected CHM medicinal plants are presented below.
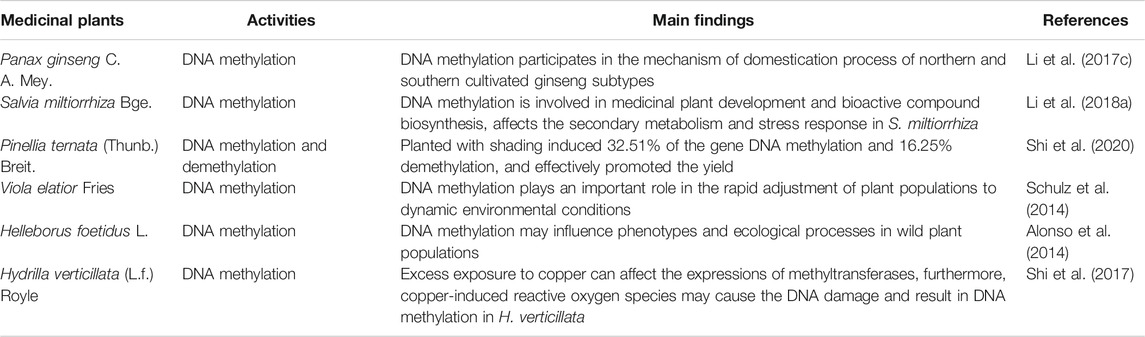
TABLE 1. DNA methylation in the growth and adaptation to ecological changes of CHM medicinal plants.
Panax ginseng C. A. Mey. (Araliaceae; Ginseng Radix et Rhizoma) originates in China and has been one of the most widely used herbal medicines globally, with a history of over a thousand years (Dai et al., 2017; Yao et al., 2018a). DNA methylation has been reported to be related to the domestication process and quality control of P. ginseng. There are several different subtypes of P. ginseng, and distinct phenotypic and genetic attributes could be identified among landraces. Through genome sequencing and phylogenetic analysis of wild and cultivated P. ginseng accessions, the northern cultivated P. ginseng and southern cultivated P. ginseng have undergone different selection pressures during the domestication process. Functional analyses revealed that DNA methylation is related to the different genes, which suggested that DNA methylation may participate in the domestication process of P. ginseng subtypes (Li MR. et al., 2017). The accumulation of ginsenosides is crucial for the quality control of Panax quinquefolium L. (Araliaceae; Panacis Quinquefolii Radix); the cold environment plays a decisive role in this process. It has been revealed that sufficient cold exposure duration in winter may induce sufficient DNA demethylation in tender leaves in early spring; this is closely correlated with the increased ginsenosides accumulation in the roots (Hao et al., 2020).
Salvia miltiorrhiza Bge. (Lamiaceae; Salviae Miltiorrhizae Radix et Rhizoma) is another frequently-used Chinese herbal medication (Yu et al., 2015; Zhang Y. et al., 2017). In plants, the epigenetic modification of cytosine DNA methylation was established and maintained by cytosine-5 DNA methyltransferases (C5-MTases). Through genome-wide analyses, eight putative SmC5-MTase genes were identified from the genome of S. miltiorrhiza. Moreover, the further transcript abundance analysis suggested the functional importance of SmC5-MTases in the secondary metabolism and stress response in S. miltiorrhiza. This research provided helpful information to reveal the role of DNA methylation in medicinal plant development and bioactive compound biosynthesis (Li J. et al., 2018).
Pinellia ternata (Thunb.) Breit. (Araceae; Pinelliae Rhizoma) is a typical Chinese herbal medicine with great demand in the Chinese market. It was found in planting that shade can effectively increase its yield. To explore its reasons, P. ternata grew under natural light, and 90% shading was selected as the control and experimental groups for genomic DNA methylation analysis using methylate sensitive amplification polymorphism (MSAP). The results showed that compared to the natural light group, shading induced 32.51% of the gene DNA methylation and 16.25% demethylation, which revealed that DNA methylation variations might participate in the mechanism of increased production of P. ternata under shading conditions (Shi et al., 2020).
Epigenetic modifications work as compensations for the relatively slow response of genetic adaptations. Through surveying the populations of Viola elatior Fries (Violaceae) from two adjacent habitat types, investigated by amplified fragment length polymorphisms and methylation-sensitive amplification polymorphisms analyses, researchers found that DNA methylation may have been playing an essential role in the rapid adjustment of plant populations to dynamic environmental conditions (Schulz et al., 2014). Furthermore, DNA methylation may also influence phenotypes and ecological processes in wild plant populations, and epigenetic variation can also predict the regional and local intraspecific functional diversity. Studies investigating the natural variation in global DNA cytosine methylation and its phenotypic correlations in the perennial herb Helleborus foetidus L. (Ranunculaceae) have confirmed this result (Alonso et al., 2014; Herrera et al., 2014; Medrano et al., 2014).
The environment is a significant factor that influences DNA methylation. Excess exposure to copper can induce DNA methylation changes in Hydrilla verticillata (L.f.) Royle (Hydrocharitaceae) in two different ways. On the one hand, the excess copper stress can affect the expressions of methyltransferases and then regulate the DNA methylation patterns; on the other hand, the copper-induced reactive oxygen species may cause DNA damage and then result in DNA methylation (Shi et al., 2017).
DNA methylation in botanical studies has revealed that this mechanism can explain how different environmental factors affect plants’ growth and adaptation to ecological changes. The botanical growth at different environmental conditions will likely affect the quality control of CHMs and subsequently affect their clinical therapeutic outcome.
DNA Methylation in the Pharmacological Mechanisms of Chinese Herbal Medicine
Due to the complexity of ingredients and the uncertainty of the pharmacological mechanisms, the term “black box” is usually to be applied to depict this dilemma of CHM. However, with the rapid advance of scientific research, the condition has been greatly improved, DNA methylation and other epigenetic approaches contribute markedly to revealing the pharmacological mechanisms of CHM formulae, botanical drugs, and herbal-derived compounds.
Chinese Herbal Medicine Formulae
Many classical CHM formulae with a long history have been widely used in various clinical departments, to uncover their pharmacological mechanisms through DNA methylation studies may also contribute the modern clinical practice. Estrogen receptor α (ERα) can protect against atherosclerosis (AS), the hypermethylation can reduce the expression of the gene ERα. Classical CHM formula Liuwei Dihuang Decoction (LDD) has been reported to enhance ERα expression to prevent AS. An in vitro study has shown that LDD protects human umbilical vein endothelial cells (HUVECs) from apoptosis induced by Hcy. Treatment with LDD significantly increased the expression of ERα and reduced the rate of ERα gene methylation by inhibiting the expression of DNMT1. The level of primary methyl donor SAM and the SAM/SAH ratio were both reduced by LDD. ApoE−/- mice were ovariectomized to establish a model of postmenopausal AS. The results displayed that LDD effectively prevented plaque formation and reduced the concentration of Hcy; furthermore, LDD upregulated ERα expression and inhibition of DNMT1 expression in atherosclerotic mice, validating the similar epigenetic regulation effects in HUVECs experiments (Chen et al., 2020).
Another CHM formula also displayed beneficial effects on AS via an epigenetic modification to regulate the expression of ERα. San-Huang-Xie-Xin-Tang (SHXXT) showed better effects compared with Statin to restore microRNA-152, decrease DNMT1, down-regulate the methylation level of ERα, and finally increase ERα expression in both cellular and animal studies (Wang et al., 2012).
Wutou Decoction (WTD) is a classical formula of CHM; it has been wildly applied in treating rheumatoid arthritis for many years (Zhang et al., 2015; Guo et al., 2017). Collagen-induced arthritis (CIA) rat model was established to explore the effects of WTD on the epigenetic modifications of rheumatoid arthritis. The data showed that the DNA methylation level was significantly down-regulated in group WTD than in group CIA, and H3 acetylation of peripheral blood mononuclear cells (PBMCs) was overexpressed in WTD compared with CIA. This result indicated that DNA methylation might play a significant role in WTD treatment of rheumatoid arthritis. WTD modulates DNA methylation and histone modifications, functioning as an anti-inflammatory potential (Liu et al., 2016).
Baihu Guizhi Decoction (BGD) has specific curative effects on pyretic arthralgia (PA), according to CHM practice. To investigate the characteristic methylation genes of the PA model showed that the whole-genome CpG island in the PA group was kept in a lower methylation state than the standard group. The intervention of BGD significantly ameliorated the foot swelling and pathological injury in PA models, decreased the levels of IL-1β, TNF-α, EGF, IL-17, and other inflammatory factors. It also inhibited the mRNA expression levels of down-regulated methylation genes Ahcy and Rpl3 and promoted the mRNA expression levels of upregulated methylation gene Agxt. The results indicated that BGD achieved the therapeutic effects on PA by adjusting characteristic genes’ methylation levels (Chen et al., 2017).
Xuefu Zhuyu Decoction (XZD) is a widely used CHM formula in treating multiple circulation system diseases. A group of 49 patients diagnosed with clopidogrel resistance (CR) were randomly divided into control and treatment groups, and the DNA methylation level of gene P2Y12 was inspected with a bisulfite pyrosequencing assay. Results indicated that XZD improved CR, and the P2Y12 polymorphisms and their methylation level influenced the drug effect. Patients with lower methylation levels of CpGI were more likely to be TT genotype in rs2046934 and received substantial benefits from XZD (Yu et al., 2019).
Besides the classical CHM formulae, DNA methylation research has also been conducted in pharmacological investigations into the modern composed formulae. Qian Yang Yu Yin Granule (QYYYG) is a Chinese herbal formulation that has been used to treat hypertensive nephropathy for decades in China. The proliferation of HEK293T cells induced by Angiotensin II was chosen to observe the epigenetic mechanism of QYYYG on renal damage. It was found that QYYYG inhibited HEK293T cells’ proliferation, down-regulated the expression of Nicotinamide N-Methyltransferase (NNMT), S-adenosylhomocysteine (SAH), acetyl-cortactin, and DNA methylation. The results revealed that QYYYG alleviated renal impairment in spontaneously hypertensive rats and inhibited the proliferation of HEK293T cells through the pathways linked to DNA methylation and other epigenetic mechanisms (Zhang et al., 2020).
Refractory cytopenia with multilineage dysplasia (RCMD) is one of the most common types of myelodysplastic syndromes (MDS) (Iwafuchi and Ito, 2018). Qinghuang Powder (QHP) has been applied to treat MDS for over 30 years and has proven effective in the CHM practice. To elucidate the mechanism of QHP in treating RCMD, clinical trials were conducted. Patients with MDS-RCMD were treated with QHP, and then bone marrow samples were obtained for further DNA methylation analysis. It was revealed that, through significant genome-wide DNA demethylation, QHP might achieve the hematologic improvement of MDS patients, suggesting this CHM formula may provide another choice for MDS treatment (Sun et al., 2012; Zhou et al., 2018).
β-Thalassemia, common in many parts of the world, is a hereditary disease resulting from red blood pigment hemoglobin (Sirisena et al., 2018). Yisui Shengxue Granules (YSSXG), a Chinese herbal medication, has been used as an alternative therapy for this disease. A double-blind, randomized, placebo-controlled trial was designed to examine the therapeutic effects of YSSXG and its influence on global DNA methylation. Apparent increases in hemoglobin, red blood cells, and fetal hemoglobin counts were observed in the treatment group. Moreover, DNA methylation examination screened 24 hypomethylated and 3,685 hypermethylated genes after the intervention. Further gene functional analysis suggested that JAK3 and MAPK10 were critical genes in the bone marrow and the lymphatic system; JAK3 was likely to be related to hematopoietic cytokines in the process of early hematopoiesis (Cheng et al., 2018).
Systemic lupus erythematosus (SLE) is a refractory autoimmune disease in which the body’s immune system mistakenly attacks healthy tissue in many parts of the body. It shows a high incidence rate in women of child-bearing age (De Risi-Pugliese et al., 2018; Di Battista et al., 2018). Clinical observation showed that Lang-Chuang-Ding Decoction (LCD), a CHM formula, could significantly reduce SLE disease activity index (SLEDAI); a further study aimed to investigate its effect on the DNA methylation of CD70 gene of patients with SLE. PBMCs isolated from female patients with SLE or healthy donors were cultured and treated with LCD medicated serum or normal serum. The mRNA and DNA methylation expressions of the CD70 gene were detected. The result showed that, by promoting the DNA methylation of CD70 gene promoter, LCD could inhibit the expression of gene CD70, which unveiled the potential mechanism of LCD to treat SLE (Sun et al., 2018). Furthermore, the Jieduquyuziyin Prescription (JP) is also an effective CHM formula for SLE. In vitro experiments with Jurkat T cells demonstrated that JP upregulated the levels of DNA methylation, MeCP2 mRNA, and protein as effectively as prednisone acetate, and thus may activate the MeCP2 gene by increasing the methylation level, thereby inhibiting the pathogenesis of SLE (Li R. et al., 2018).
Uremic Clearance Granule (UCG) has been used to treat chronic renal failure (CRF) in clinical practice for many years. Adenine-induced CRF was established in Wister rats to explore the mechanism of the therapeutic effects. After treatment with UCG, the kidney function of CRF rats was recovered. Tgf-β1 promoter was demethylated at some loci in the pathological control group. UCG could also recover the methylation level of gene Tgf-β1 and then regulate the transcription and translation of the gene, indicating that the DNA methylation may participate in the molecular mechanism of the therapeutic effect (Miao et al., 2010).
Chinese Herbal Medicine Botanical Drugs and Compounds
With multiple pharmacological effects, Ginseng Radix et Rhizoma is one of the world’s most commonly used natural products (Wan et al., 2017; Wang et al., 2018). Ginsenoside Rg3, a bioactive constituent isolated from Ginseng Radix et Rhizoma, has shown antitumorigenic activities (Wu et al., 2018). Moreover, a further study indicated that DNA methylation was involved in this bioprocess (Teng et al., 2017). It was demonstrated that ginsenoside Rg3 could reduce global genomic DNA methylation and alter cytosine methylation of the promoter regions of some related genes. This epigenetic modification was associated with increased cell growth inhibition in the HepG2 human hepatocarcinoma cell line (Teng et al., 2017). Ginsenoside Rg3 has also been shown to increase the expression of tumor suppressor von Hippel-Lindau (VHL) gene in ovarian cancer cells via suppressing DNMT3A expression and thus downregulating the DNA methylation level of the VHL promoter (Wang et al., 2021). The Warburg effect is associated with increased glycolysis and rapid energy production; it has been remarked as one of the metabolic hallmarks of cancer. Ginsenoside Rg3 could down-regulate DNA methyltransferase DNMT3A and decrease the methylation in the promoter region of miR-519a-5p precursor gene, thus upregulating miR-519a-5p to inhibit the HIF-1α-stimulated Warburg effect in ovarian cancer (Lu et al., 2020). On MCF-7 breast cancer cells, ginsenoside ginsenoside Rh2 intervention down-regulated the methylation level of LINE1, which is a global methylation marker; ginsenoside Rh2 also induced genome-wide DNA methylation changes mapped to genes associated with immune response and tumorigenesis, thereby exerting anti-tumor effects (Lee et al., 2018).
Salviae Miltiorrhizae Radix et Rhizoma has been widely used in CHM practice for centuries (Rongrong et al., 2016; Zhang Y. et al., 2017). Tanshinones, isolated from S. miltiorrhiza, is a class of abietane diterpene compounds that contain three major types, those are cryptotanshinone (CT), tanshinone IIA (T2A), and tanshinone I (T1). Oral submucous fibrosis (OSF) induced by chewing the areca nut has been considered a precancerous lesion. Tanshinone intervention could suppress arecoline-induced epithelial-mesenchymal transition in oral submucous fibrosis and reverse the hypermethylation of TP53 promoter induced by arecoline. Through down-regulating the DNA methylation level of the TP53 promoter, tanshinone recovered the expression of p53 and reactivated the p53 pathway. Thus, tanshinone played an inhibitory role in the progression of OSF (Zheng et al., 2018).
Tanshinones, especially the T1, showed a growth inhibition effect on breast cancer cells in a dose-dependent manner. T1 also significantly reduced acetylation levels of histone H3 associated with the Aurora-A gene, implying tanshinones could inhibit the growth of breast cancer cells through the epigenetic modification of Aurora-A expression (Gong et al., 2012). T2A could induce demethylation of Nrf2 and activate the Nrf2 antioxidative stress signaling pathway, potentially contributing to the attenuation of JB6 cellular transformation and anticancer effects (Wang et al., 2014). T2A also demonstrated a potential preventive effect on the high glucose-induced diabetic nephropathy cellular model, and DNA methylation may play a role in this process (Li W. et al., 2019).
Schisandrin B, derived from Schisandrae Chinensis Fructus, is an active agent who can exert various pharmacological activities. It has been proved to have significant protective effects against Alzheimer’s disease (AD) (Hu et al., 2018). Cell experiments revealed that schisandrin B could significantly inhibit the Aβ1-42 -induced damages in the SH-SY5Y neuronal cell line via the regulation of DNMT3A and DNMT1 mRNA expression then influence the DNA methylation level (Zhang M. et al., 2017). Further validation, such as in vivo experiments or clinical trials, is still needed to make this discovery go even more profound.
Curcumin is an active component of Curcumae Longae Rhizoma. Previous studies indicate curcumin possesses several biological activities beneficial to cancer treatment and prevention (Bano et al., 2018; Khorsandi et al., 2018; Nagaraju et al., 2018). Examined in human glioblastoma cells, DNA methylation may play a potential role in the process of curcumin to upregulate the RANK gene expression at both mRNA and protein levels. Curcumin induced a decreased methylation level of the RANK gene promoter, then reactivated gene RANK; thus, the alleviating effect to glioma could be achieved (Wu et al., 2013).
Triptolide is a component derived from the Chinese herb Tripterygium wilfordii Hook. f. and has potent immunosuppressive and anti-inflammatory effects. Triptolide and tripchlorolide exert beneficial effects on synapses in Alzheimer’s disease mice, and the underlying mechanism may be related to DNA methylation, for the compounds can inhibit the DNA methylation of Nlgn1 promoter in the hippocampus, and thus increase the expression of gene Nlgn1 (Lu et al., 2019). Evaluated in vitro, triptolide was shown to have the effect of inhibiting the growth and proliferation of acute lymphoblastic leukemia Jurkat cell line in dose-and time-dependent manners. Triptolide can also reverse the hypermethylation of antioncogene APC and upregulate the APC expression, which may provide a potential mechanism of triptolide’s protective effects (Wu et al., 2010).
As a monotherapy or adjunct therapy, many CHM formulae and bioactive compounds have displayed beneficial pharmacological activities in treating various modern diseases (Table 2, 3). Elucidating the mechanism of these activities is essential for the safety and effectiveness of CHM clinical application. However, the complexity of ingredients, the multi-targeting treatment strategy, and the guidance of the holistic concept all increase the ambiguity of CHM. The recent development of epigenetics, especially DNA methylation research, facilitates uncovering the hidden secrets of CHM. As an epigenetic modification, DNA methylation serves as an intermediate factor playing an essential role in the mechanism of CHM to exert therapeutic effects (Figure 3).
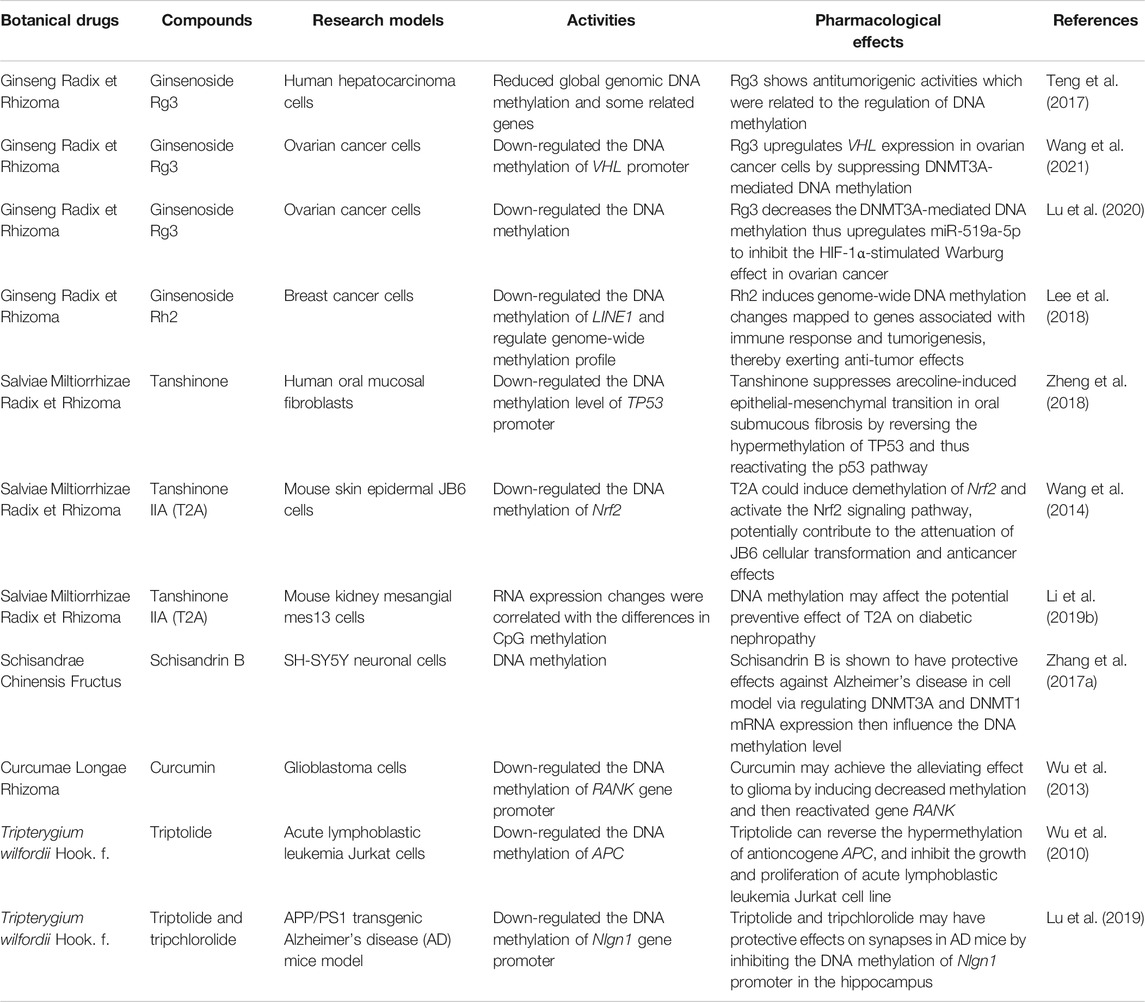
TABLE 3. DNA methylation participates in multiple pharmacological effects of botanical drugs and compounds.
DNA Methylation in Constitution Theory and Other Issues of Chinese Herbal Medicine
DNA methylation has not only been applied in pharmaceutical research but has also been demonstrated to be beneficial in elucidating basic theoretical research in CHM, for instance, the body constitution theory, syndrome differentiation, etc.
CHM uses a unique theoretical and practical system to diagnose and treat diseases. Syndrome differentiation is one of the primary approaches used in clinical practice and CHM higher education (Hua et al., 2017; Zhao, 2017). Based on the clinical information collected through the four diagnostic methods of observation, listening/smelling, inquiry, and palpation examination, the diagnosis of disease and syndrome can be drawn. However, if combined with constitution differentiation, the diagnosis could be more accurate. Constitution differentiation will also reveal more comprehensive information about the background of the disease (Xu et al., 2017), it enhances not only the disease treatment but also the disease prevention in CHM. Body constitution theory originates in the profound CHM clinical practice, which has been widely used in China and abroad and proved to be reliable and fruitful (Li L. et al., 2019). Constitution in CHM is the study of the intrinsic features of the human body, including congenital and acquired ones (Wang et al., 2011). There are nine basic constitution types in CHM, i.e., balanced constitution, qi-deficiency constitution, yang-deficiency constitution, yin-deficiency constitution, phlegm-dampness constitution, damp-heat constitution, blood stasis constitution, qi stagnation constitution, and inherited special constitution. Different constitution types have different characteristics and different predispositions. Therefore, based on the constitution identification, corresponding high-risk diseases can be predicted, creating the possibility of early prevention (Li M. et al., 2017; Ma et al., 2017; Liu et al., 2018).
Among the nine constitution types, the balanced constitution is thought to be the healthiest type. Phlegm-dampness Constitution (PC) is one of the most studied constitution types which predisposes individuals to complex metabolic disorders (Wang et al., 2013; Li L. et al., 2017). The theory of PC can effectively promote early prediction and prevention of metabolic diseases. It also provides a new perspective on the increasing prevalence of metabolic diseases and the related complications, which have already caused heavy medical and financial burdens in the global society (Wang et al., 2013). As a reversible epigenetic modification, DNA methylation can be used to explore the influence of postnatal factors on body constitution. Our group previously has applied the Infinium HumanMethylation450 BeadChip technique to explore the DNA methylation profiles of PC; through the DNA methylation research, the molecular mechanism of PC has been partially revealed (Yao et al., 2018b). DNA samples were extracted from peripheral blood mononuclear cells of PC and balanced constitution volunteers; the balanced constitution was set to be the control group. A genome-wide DNA methylation analysis was conducted, which indicated a genome-scale hyper-methylation pattern in PC and located 288 differentially methylated probes (DMPs). Based on these DMPs, 256 genes were mapped; some of these genes were metabolic-related and could be enriched in multiple metabolic gene pathways (Yao et al., 2018b). This research indicated PC has a metabolic disorder feature even in healthy individuals, thus, DNA methylation may play a potential role in this pathological mechanism. The study data should benefit the understanding of PC and explain why PC is prone to multiple metabolic diseases (Yao et al., 2018b; Tobi et al., 2018).
Syndrome differentiation is an essential theory in CHM diagnostics; however, the lack of biological basis limits its application and development in the context of modern science to a certain extent. The emerging DNA methylation research provides a new potential resolution to this issue. A study combined genome-wide DNA methylation and serum cytokine profiles was conducted to search the molecular bases of CHM syndrome classification in patients with chronic hepatitis B. As the results indicated that the hypomethylation of CpG loci in 5’ UTR of HLA-F may up-regulate HLA-F expression, furthermore, this parameter was significantly correlated with the cytokine levels of IL-12, MIP-1α, and MIP-1β, they may serve as a diagnostic marker for the syndrome differentiation of chronic hepatitis B (Hu et al., 2021). A similar approach was applied to explore the biomarker for syndrome differentiation in difficult-to-control asthma. Patients with hot syndrome showed hypermethylation at probe cg10791966 in gene ALDH3A1 with corresponding lower mRNA expression, implying that ALDH3A1 might be a potential biomarker for the syndrome differentiation of difficult-to-control asthma (Song et al., 2020). Besides the diagnostic application, aberrant DNA methylation can also serve as potential targets for CHM interventions, which may help to develop a novel clinical mode (Shen et al., 2021).
Discussion and Conclusion
Chinese medicine, including CHM and acupuncture, has been attracting more attention due to its profitable clinical performance. With the doctrine of holism as a core concept, CHM can address the limitations of western medicine’s “one target, one drug” strategy. With profound clinical experience, CHM provides effective treatment solutions for many modern refractory diseases. However, barriers still hinder further exploration of CHM, as many mechanisms and hidden pearls of wisdom still need to be unveiled. As outlined above, epigenetic research, especially DNA methylation, presents an opportunity to address these challenges.
DNA methylation research is intensely beneficial and meaningful to CHM. DNA methylation, a dynamic changing and reversible process, has the features of heritability and variability. It promotes examining how herb growth and development is affected by environmental factors which may affect the quality control and efficacy of CHM herbs. DNA methylation also illuminates the pharmaceutical mechanisms of CHM formulae and compounds, which is meaningful in facilitating the clinical application of CHM. DNA methylation provides scientific validation and proves the clinical feasibility of CHM body constitution theory, some specific DNA methylation loci can serve as potential biomarkers to realize the diagnostic of CHM syndromes and constitutions at a molecular level. In addition, DNA methylation is widely distributed in the human body and not limited to a particular organ or system. CHM is a holistic medical system, and herbal medications work in a systematic and multi-targeted way. From this point of view, methylation may help to disclose the molecular mechanism and locate the potential drug targetsof CHM, which would be extremely significant for clinical practice.
The newly developed field of DNA methylation is up-and-coming for addressing many complicated issues of CHM. It could not only promote clinical treatment but, more importantly, also facilitate disease prevention. At present, investigators are concentrating on the epigenetic research of the CHM constitution, which aims to elucidate the molecular basis of different body constitution types. The obtained results are expected to reveal why different constitution types make individuals susceptible to various medical conditions. Epigenetic research of body constitution will possibly lay the foundation for disease prediction and prevention, which is vital to alleviate the heavy health care burden in modern society.
Currently, the field of epigenetic research in CHM is in its infancy. Although some inspiring studies have been conducted, the number of related publications is still somewhat limited. By reviewing the documents that we retrieved, some publications were not well-founded enough to achieve a solid conclusion, so further validating experiments are needed. More innovative and specific scientific approaches, more systematic experimental design, and practical techniques are required to improve the quality of the research of this domain. Furthermore, more in-depth integration is needed in future studies between CHM and epigenetics to apply CHM theory and clinical practice better.
Author Contributions
All authors have made significant and substantive contributions to the study. C-SY and C-ZW conceived the idea of this review article. All authors have participated in the outline design, review of relevant literature, manuscript drafting, review, and final revisions.
Funding
This work was supported by funding from the National Natural Science Foundation of China (Grant Nos 81803970, 81973714); Young Talent Promotion Project of China Association of Chinese Medicine (CACM-2019-QNRC2-A01); Young Scientists Development Program of Beijing University of Chinese Medicine; and Fundamental Research Funds for the Central Universities (2020-JYB-XJSJJ-003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer (WHH) declared past co-authorships with several of the authors (CZW, JYW, HY, CSY) to the handling editor.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alonso, C., Pérez, R., Bazaga, P., Medrano, M., and Herrera, C. M. (2014). Individual Variation in Size and Fecundity Is Correlated with Differences in Global DNA Cytosine Methylation in the Perennial Herb Helleborus Foetidus (Ranunculaceae). Am. J. Bot. 101, 1309–1313. doi:10.3732/ajb.1400126
Ang-Lee, M. K., Moss, J., and Yuan, C. S. (2001). Herbal Medicines and Perioperative Care. JAMA 286, 208–216. doi:10.1001/jama.286.2.208
Bano, N., Yadav, M., and Das, B. C. (2018). Differential Inhibitory Effects of Curcumin between HPV+ve and HPV-Ve Oral Cancer Stem Cells. Front. Oncol. 8, 412. doi:10.3389/fonc.2018.00412
Barau, J., Teissandier, A., Zamudio, N., Roy, S., Nalesso, V., Hérault, Y., et al. (2016). The DNA Methyltransferase DNMT3C Protects Male Germ Cells from Transposon Activity. Science 354, 909–912. doi:10.1126/science.aah5143
Chen, H., Ju, S. H., Wei, J. P., Fu, W. J., Zheng, H., and Xu, S. J. (2017). Effect of Baihu Guizhi Decoction on Characteristic Methylation Genes Expression of Pyretic Arthralgia Rat Model. Zhongguo Zhong Yao Za Zhi 42, 332–340. doi:10.19540/j.cnki.cjcmm.20161222.013
Chen, Q., Zhang, Y., Meng, Q., Wang, S., Yu, X., Cai, D., et al. (2020). Liuwei Dihuang Prevents Postmenopausal Atherosclerosis and Endothelial Cell Apoptosis via Inhibiting DNMT1-Medicated ERα Methylation. J. Ethnopharmacol 252, 112531. doi:10.1016/j.jep.2019.112531
Cheng, Y. L., Zhang, X. H., Sun, Y. W., Wang, W. J., Huang, J., Chu, N. L., et al. (2018). Genomewide DNA Methylation Responses in Patients with Beta-Thalassemia Treated with Yisui Shengxue Granules. Chin. J. Integr. Med. 25 (7), 490–496. doi:10.1007/s11655-018-2777-9
Dai, D., Zhang, C. F., Williams, S., Yuan, C. S., and Wang, C. Z. (2017). Ginseng on Cancer: Potential Role in Modulating Inflammation-Mediated Angiogenesis. Am. J. Chin. Med. 45, 13–22. doi:10.1142/S0192415X17500021
De Risi-Pugliese, T., Cohen Aubart, F., Haroche, J., Moguelet, P., Grootenboer-Mignot, S., Mathian, A., et al. (2018). Clinical, Histological, Immunological Presentations and Outcomes of Bullous Systemic Lupus Erythematosus: 10 New Cases and a Literature Review of 118 Cases. Semin. Arthritis Rheum. 48, 83–89. doi:10.1016/j.semarthrit.2017.11.003
Delker, R. K., and Mann, R. S. (2017). From Reductionism to Holism: Toward a More Complete View of Development through Genome Engineering. Adv. Exp. Med. Biol. 1016, 45–74. doi:10.1007/978-3-319-63904-8_3
Di Battista, M., Marcucci, E., Elefante, E., Tripoli, A., Governato, G., Zucchi, D., et al. (2018). One Year in Review 2018: Systemic Lupus Erythematosus. Clin. Exp. Rheumatol. 36, 763–777. doi:10.1016/j.semarthrit.2017.11.003
Dor, Y., and Cedar, H. (2018). Principles of DNA Methylation and Their Implications for Biology and Medicine. Lancet 392, 777–786. doi:10.1016/S0140-6736(18)31268-6
Draht, M. X. G., Goudkade, D., Koch, A., Grabsch, H. I., Weijenberg, M. P., Van Engeland, M., et al. (2018). Prognostic DNA Methylation Markers for Sporadic Colorectal Cancer: a Systematic Review. Clin. Epigenetics 10, 35. doi:10.1186/s13148-018-0461-8
Elhamamsy, A. R. (2017). Role of DNA Methylation in Imprinting Disorders: an Updated Review. J. Assist. Reprod. Genet. 34, 549–562. doi:10.1007/s10815-017-0895-5
Fardet, A., and Rock, E. (2014). The Search for a New Paradigm to Study Micronutrient and Phytochemical Bioavailability: From Reductionism to Holism. Med. Hypotheses 82, 181–186. doi:10.1016/j.mehy.2013.11.035
Feinberg, A. P. (2018). The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N. Engl. J. Med. 378, 1323–1334. doi:10.1056/NEJMra1402513
Fernández-Sanlés, A., Sayols-Baixeras, S., Curcio, S., Subirana, I., Marrugat, J., and Elosua, R. (2018). DNA Methylation and Age-Independent Cardiovascular Risk, an Epigenome-Wide Approach: The REGICOR Study (REgistre GIroní del COR). Arterioscler Thromb. Vasc. Biol. 38, 645–652. doi:10.1161/ATVBAHA.117.310340
Gong, Y., Li, Y., Abdolmaleky, H. M., Li, L., and Zhou, J. R. (2012). Tanshinones Inhibit the Growth of Breast Cancer Cells through Epigenetic Modification of Aurora A Expression and Function. Plos One 7, e33656. doi:10.1371/journal.pone.0033656
Greenberg, M. V. C., and Bourc'his, D. (2019). The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat. Rev. Mol. Cel Biol 20, 590–607. doi:10.1038/s41580-019-0159-6
Gu, S., and Pei, J. F. (2017). Chinese Herbal Medicine Meets Biological Networks of Complex Diseases: A Computational Perspective. Evid. Based Compl Alt. 2017, 7198645. doi:10.1155/2017/7198645
Guo, Q., Zheng, K., Fan, D., Zhao, Y., Li, L., Bian, Y., et al. (2017). Wu-tou Decoction in Rheumatoid Arthritis: Integrating Network Pharmacology and In Vivo Pharmacological Evaluation. Front. Pharmacol. 8, 230. doi:10.3389/fphar.2017.00230
Hao, M., Zhou, Y., Zhou, J., Zhang, M., Yan, K., Jiang, S., et al. (2020). Cold-induced Ginsenosides Accumulation Is Associated with the Alteration in DNA Methylation and Relative Gene Expression in Perennial American Ginseng (Panax Quinquefolius L.) along with its Plant Growth and Development Process. J. Ginseng Res. 44, 747–755. doi:10.1016/j.jgr.2019.06.006
He, X. J., Chen, T., and Zhu, J. K. (2011). Regulation and Function of DNA Methylation in Plants and Animals. Cell Res 21, 442–465. doi:10.1038/cr.2011.23
Herrera, C. M., Medrano, M., and Bazaga, P. (2014). Variation in DNA Methylation Transmissibility, Genetic Heterogeneity and Fecundity-Related Traits in Natural Populations of the Perennial Herb Helleborus Foetidus. Mol. Ecol. 23, 1085–1095. doi:10.1111/mec.12679
Hsieh, H. Y., Chiu, P. H., and Wang, S. C. (2011). Epigenetics in Traditional Chinese Pharmacy: a Bioinformatic Study at Pharmacopoeia Scale. Evid. Based Complement. Alternat Med. 2011, 816714. doi:10.1093/ecam/neq050
Hu, X. L., Guo, C., Hou, J. Q., Feng, J. H., Zhang, X. Q., Xiong, F., et al. (2018). Stereoisomers of Schisandrin B Are Potent ATP Competitive GSK-3beta Inhibitors with Neuroprotective Effects against Alzheimer's Disease: Stereochemistry and Biological Activity. ACS Chem. Neurosci. 10 (2), 996–1007. doi:10.1021/acschemneuro.8b00252
Hu, X. Q., and Su, S. B. (2017). An Overview of Epigenetics in Chinese Medicine Researches. Chin. J. Integr. Med. 23, 714–720. doi:10.1007/s11655-016-2274-y
Hu, X. Q., Zhou, Y., Chen, J., Lu, Y. Y., Chen, Q. L., Hu, Y. Y., et al. (2021). DNA Methylation and Transcription of HLA-F and Serum Cytokines Relate to Chinese Medicine Syndrome Classification in Patients with Chronic Hepatitis B. Chin. J. Integr. Med, 1–8. doi:10.1007/s11655-021-3279-8
Hua, M., Fan, J., Dong, H., and Sherer, R. (2017). Integrating Traditional Chinese Medicine into Chinese Medical Education Reform: Issues and Challenges. Int. J. Med. Educ. 8, 126–127. doi:10.5116/ijme.58e3.c489
Imgenberg-Kreuz, J., Carlsson Almlöf, J., Leonard, D., Alexsson, A., Nordmark, G., Eloranta, M. L., et al. (2018). DNA Methylation Mapping Identifies Gene Regulatory Effects in Patients with Systemic Lupus Erythematosus. Ann. Rheum. Dis. 77, 736–743. doi:10.1136/annrheumdis-2017-212379
Islam, F., Tang, J. C., Gopalan, V., and Lam, A. K. (2018). Epigenetics: DNA Methylation Analysis in Esophageal Adenocarcinoma. Methods Mol. Biol. 1756, 247–256. doi:10.1007/978-1-4939-7734-5_21
Iwafuchi, H., and Ito, M. (2018). Differences in the Bone Marrow Histology between Childhood Myelodysplastic Syndrome with Multilineage Dysplasia and Refractory Cytopenia of Childhood without Multilineage Dysplasia. Histopathol. 74 (2), 239–247. doi:10.1111/his.13721
Kedaigle, A., and Fraenkel, E. (2018). Turning Omics Data into Therapeutic Insights. Curr. Opin. Pharmacol. 42, 95–101. doi:10.1016/j.coph.2018.08.006
Kellogg, R. A., Dunn, J., and Snyder, M. P. (2018). Personal Omics for Precision Health. Circ. Res. 122, 1169–1171. doi:10.1161/CIRCRESAHA.117.310909
Khorsandi, K., Hosseinzadeh, R., and Shahidi, F. K. (2018). Photodynamic Treatment with Anionic Nanoclays Containing Curcumin on Human Triple-Negative Breast Cancer Cells: Cellular and Biochemical Studies. J. Cel Biochem 120 (4), 4998–5009. doi:10.1002/jcb.27775
Kohli, R. M., and Zhang, Y. (2013). TET Enzymes, TDG and the Dynamics of DNA Demethylation. Nature 502, 472–479. doi:10.1038/nature12750
Lee, H., Lee, S., Jeong, D., and Kim, S. J. (2018). Ginsenoside Rh2 Epigenetically Regulates Cell-Mediated Immune Pathway to Inhibit Proliferation of MCF-7 Breast Cancer Cells. J. Ginseng Res. 42, 455–462. doi:10.1016/j.jgr.2017.05.003
Li, J., Li, C., and Lu, S. (2018a). Identification and Characterization of the Cytosine-5 DNA Methyltransferase Gene Family in Salvia Miltiorrhiza. PeerJ 6, e4461. doi:10.7717/peerj.4461
Li, R., Zhuang, A., Ma, J., Ji, L., Hou, X., Chen, H., et al. (2018b). Effect of Jieduquyuziyin Prescription-Treated Rat Serum on MeCP2 Gene Expression in Jurkat T Cells. In Vitro Cel Dev Biol Anim 54, 692–704. doi:10.1007/s11626-018-0295-x
Li, L., Feng, J., Yao, H., Xie, L., Chen, Y., Yang, L., et al. (2017a). Gene Expression Signatures for Phlegm-Dampness Constitution of Chinese Medicine. Sci. China Life Sci. 60, 105–107. doi:10.1007/s11427-016-0212-9
Li, M. H., Mo, S. M., Lv, Y. B., Tang, Z. H., and Dong, J. C. (2017b). A Study of Traditional Chinese Medicine Body Constitution Associated with Overweight, Obesity, and Underweight. Evid. Based Compl Alt. 2017, 7361896. doi:10.1155/2017/7361896
Li, M. R., Shi, F. X., Li, Y. L., Jiang, P., Jiao, L., Liu, B., et al. (2017c). Genome-Wide Variation Patterns Uncover the Origin and Selection in Cultivated Ginseng (Panax Ginseng Meyer). Genome Biol. Evol. 9, 2159–2169. doi:10.1093/gbe/evx160
Li, L., Yao, H., Wang, J., Li, Y., and Wang, Q. (2019a). The Role of Chinese Medicine in Health Maintenance and Disease Prevention: Application of Constitution Theory. Am. J. Chin. Med. 47, 495–506. doi:10.1142/S0192415X19500253
Li, W., Sargsyan, D., Wu, R., Li, S., Wang, L., Cheng, D., et al. (2019b). DNA Methylome and Transcriptome Alterations in High Glucose-Induced Diabetic Nephropathy Cellular Model and Identification of Novel Targets for Treatment by Tanshinone IIA. Chem. Res. Toxicol. 32, 1977–1988. doi:10.1021/acs.chemrestox.9b00117
Lippolis, J. D., Powell, E. J., Reinhardt, T. A., Thacker, T. C., and Casas, E. (2018). Symposium Review: Omics in Dairy and Animal Science-Promise, Potential, and Pitfalls. J. Dairy Sci. 101, 1–14. doi:10.3168/jds.2018-15267
Liu, G., Bin, P., Wang, T., Ren, W., Zhong, J., Liang, J., et al. (2017). DNA Methylation and the Potential Role of Methyl-Containing Nutrients in Cardiovascular Diseases. Oxid Med. Cel Longev 2017, 1670815. doi:10.1155/2017/1670815
Liu, J., Xu, F., Mohammadtursun, N., Lv, Y., Tang, Z., and Dong, J. (2018). The Analysis of Constitutions of Traditional Chinese Medicine in Relation to Cerebral Infarction in a Chinese Sample. J. Altern. Complement. Med. 24, 458–462. doi:10.1089/acm.2017.0027
Liu, Y. F., Wen, C. Y., Chen, Z., Wang, Y., Huang, Y., Hu, Y. H., et al. (2016). Effects of Wutou Decoction on DNA Methylation and Histone Modifications in Rats with Collagen-Induced Arthritis. Evid. Based Complement. Alternat Med. 2016, 5836879. doi:10.1155/2016/5836879
Lu, J., Chen, H., He, F., You, Y., Feng, Z., Chen, W., et al. (2020). Ginsenoside 20(S)-Rg3 Upregulates HIF-1α-Targeting miR-519a-5p to Inhibit the Warburg Effect in Ovarian Cancer Cells. Clin. Exp. Pharmacol. Physiol. 47, 1455–1463. doi:10.1111/1440-1681.13321
Lu, X., Yang, B., Yu, H., Hu, X., Nie, J., Wan, B., et al. (2019). Epigenetic Mechanisms Underlying the Effects of Triptolide and Tripchlorolide on the Expression of Neuroligin-1 in the hippocampus of APP/PS1 Transgenic Mice. Pharm. Biol. 57, 453–459. doi:10.1080/13880209.2019.1629463
Luo, C., Hajkova, P., and Ecker, J. R. (2018). Dynamic DNA Methylation: In the Right Place at the Right Time. Science 361, 1336–1340. doi:10.1126/science.aat6806
Ma, Y. L., Yao, H., Yang, W. J., Ren, X. X., Teng, L., and Yang, M. C. (2017). Correlation between Traditional Chinese Medicine Constitution and Dyslipidemia: A Systematic Review and Meta-Analysis. Evid. Based Complement. Alternat Med. 2017, 1896746. doi:10.1155/2017/1896746
Medrano, M., Herrera, C. M., and Bazaga, P. (2014). Epigenetic Variation Predicts Regional and Local Intraspecific Functional Diversity in a Perennial Herb. Mol. Ecol. 23, 4926–4938. doi:10.1111/mec.12911
Miao, X. H., Wang, C. G., Hu, B. Q., Li, A., Chen, C. B., and Song, W. Q. (2010). TGF-beta1 Immunohistochemistry and Promoter Methylation in Chronic Renal Failure Rats Treated with Uremic Clearance Granules. Folia Histochem. Cytobiol 48, 284–291. doi:10.2478/v10042-010-0001-7
Nagaraju, G. P., Benton, L., Bethi, S. R., Shoji, M., and El-Rayes, B. F. (2018). Curcumin Analogs: Their Roles in Pancreatic Cancer Growth and Metastasis. Int. J. Cancer 145 (1), 10–19. doi:10.1002/ijc.31867
Nowacka-Zawisza, M., and Wiśnik, E. (2017). DNA Methylation and Histone Modifications as Epigenetic Regulation in Prostate Cancer (Review). Oncol. Rep. 38, 2587–2596. doi:10.3892/or.2017.5972
Pang, B., Zhang, Y., Liu, J., He, L. S., Zheng, Y. J., Lian, F. M., et al. (2017). Prevention of Type 2 Diabetes with the Chinese Herbal Medicine Tianqi Capsule: A Systematic Review and Meta-Analysis. Diabetes Ther. 8, 1227–1242. doi:10.1007/s13300-017-0316-x
Papanicolau-Sengos, A., and Aldape, K. (2021). DNA Methylation Profiling: An Emerging Paradigm for Cancer Diagnosis. Annu. Rev. Pathol. doi:10.1146/annurev-pathol-042220-022304
Rongrong, W., Li, D., Guangqing, Z., Xiaoyin, C., and Kun, B. (2016). The Effects of Iontophoretic Injections of Salvia Miltiorrhiza on the Maturation of the Arteriovenous Fistula: A Randomized, Controlled Trial. Altern. Ther. Health Med. 22, 18–22.
Schübeler, D. (2015). Function and Information Content of DNA Methylation. Nature 517, 321–326. doi:10.1038/nature14192
Schulz, B., Eckstein, R. L., and Durka, W. (2014). Epigenetic Variation Reflects Dynamic Habitat Conditions in a Rare Floodplain Herb. Mol. Ecol. 23, 3523–3537. doi:10.1111/mec.12835
Shen, F. L., Zhao, Y. N., Yu, X. L., Wang, B. L., Wu, X. L., Lan, G. C., et al. (2021). Chinese Medicine Regulates DNA Methylation to Treat Haematological Malignancies: A New Paradigm of "State-Target Medicine. Chin. J. Integr. Med, 1–7. doi:10.1007/s11655-021-3316-7
Shi, D., Zhuang, K., Xia, Y., Zhu, C., Chen, C., Hu, Z., et al. (2017). Hydrilla Verticillata Employs Two Different Ways to Affect DNA Methylation under Excess Copper Stress. Aquat. Toxicol. 193, 97–104. doi:10.1016/j.aquatox.2017.10.007
Shi, J., Xiong, Y. J., Zhang, H., Meng, X., Zhang, Z. Y., Zhang, M. M., et al. (2020). Analysis of Shading on DNA Methylation by MSAP in Pinellia Ternata. Zhongguo Zhong Yao Za Zhi 45, 1311–1315. doi:10.19540/j.cnki.cjcmm.20200105.105
Sirisena, M., Birman, C. S., Mckibbin, A. J., and O'brien, K. J. (2018). Bilateral Auditory Ossicular Expansions in a Child with Beta-Thalassemia Major: Case Report and Literature Review. Int. J. Pediatr. Otorhinolaryngol. 112, 126–131. doi:10.1016/j.ijporl.2018.06.046
Song, W., Zheng, S., Li, M., Zhang, X., Cao, R., Ye, C., et al. (2020). Linking Endotypes to Omics Profiles in Difficult-To-Control Asthma Using the Diagnostic Chinese Medicine Syndrome Differentiation Algorithm. J. Asthma 57, 532–542. doi:10.1080/02770903.2019.1590589
Sun, J., Shao, T. J., Zhang, D. Y., Huang, X. Q., Xie, Z. J., and Wen, C. P. (2018). Effect of Lang-Chuang-Ding Decoction () on DNA Methylation of CD70 Gene Promoter in Peripheral Blood Mononuclear Cells of Female Patients with Systemic Lupus Erythematosus. Chin. J. Integr. Med. 24, 348–352. doi:10.1007/s11655-017-2804-2
Sun, S. Z., Ma, R., Hu, X. M., Yang, X. H., Xu, Y. G., Wang, H. Z., et al. (2012). Karyotype and DNA-Methylation Responses in Myelodysplastic Syndromes Following Treatment with Traditional Chinese Formula Containing Arsenic. Evid. Based Complement. Alternat Med. 2012, 969476. doi:10.1155/2012/969476
Teng, S., Wang, Y., Li, P., Liu, J., Wei, A., Wang, H., et al. (2017). Effects of R Type and S Type Ginsenoside Rg3 on DNA Methylation in Human Hepatocarcinoma Cells. Mol. Med. Rep. 15, 2029–2038. doi:10.3892/mmr.2017.6255
Tobi, E. W., Slieker, R. C., Luijk, R., Dekkers, K. F., Stein, A. D., Xu, K. M., et al. (2018). DNA Methylation as a Mediator of the Association between Prenatal Adversity and Risk Factors for Metabolic Disease in Adulthood. Sci. Adv. 4, eaao4364. doi:10.1126/sciadv.aao4364
Wan, J. Y., Wang, C. Z., Zhang, Q. H., Liu, Z., Musch, M. W., Bissonnette, M., et al. (2017). Significant Difference in Active Metabolite Levels of Ginseng in Humans Consuming Asian or Western Diet: The Link with Enteric Microbiota. Biomed. Chromatogr. 31, e3851. doi:10.1002/bmc.3851
Wang, C. Z., Yao, H., Zhang, C. F., Chen, L., Wan, J. Y., Huang, W. H., et al. (2018). American Ginseng Microbial Metabolites Attenuate DSS-Induced Colitis and Abdominal Pain. Int. Immunopharmacol 64, 246–251. doi:10.1016/j.intimp.2018.09.005
Wang, J., Li, Y., Ni, C., Zhang, H., Li, L., and Wang, Q. (2011). Cognition Research and Constitutional Classification in Chinese Medicine. Am. J. Chin. Med. 39, 651–660. doi:10.1142/S0192415X11009093
Wang, J., Wang, Q., Li, L., Li, Y., Zhang, H., Zheng, L., et al. (2013). Phlegm-dampness Constitution: Genomics, Susceptibility, Adjustment and Treatment with Traditional Chinese Medicine. Am. J. Chin. Med. 41, 253–262. doi:10.1142/S0192415X13500183
Wang, L., Han, X., Zheng, X., Zhou, Y., Hou, H., Chen, W., et al. (2021). Ginsenoside 20(S)-Rg3 Upregulates Tumor Suppressor VHL Gene Expression by Suppressing DNMT3A-Mediated Promoter Methylation in Ovarian Cancer Cells. Nan Fang Yi Ke Da Xue Xue Bao 41, 100–106. doi:10.12122/j.issn.1673-4254.2021.01.14
Wang, L., Zhang, C., Guo, Y., Su, Z. Y., Yang, Y., Shu, L., et al. (2014). Blocking of JB6 Cell Transformation by Tanshinone IIA: Epigenetic Reactivation of Nrf2 Antioxidative Stress Pathway. Aaps j 16, 1214–1225. doi:10.1208/s12248-014-9666-8
Wang, Y. S., Chou, W. W., Chen, K. C., Cheng, H. Y., Lin, R. T., and Juo, S. H. (2012). MicroRNA-152 Mediates DNMT1-Regulated DNA Methylation in the Estrogen Receptor α Gene. PLoS One 7, e30635. doi:10.1371/journal.pone.0030635
Wilkinson, A. W., and Gozani, O. (2017). Cancer Epigenetics: Reading the Future of Leukaemia. Nature 543, 186–188. doi:10.1038/nature21894
Wu, B., Yao, X., Nie, X., and Xu, R. (2013). Epigenetic Reactivation of RANK in Glioblastoma Cells by Curcumin: Involvement of STAT3 Inhibition. DNA Cel Biol 32, 292–297. doi:10.1089/dna.2013.2042
Wu, W., Zhou, Q., Zhao, W., Gong, Y., Su, A., Liu, F., et al. (2018). Ginsenoside Rg3 Inhibition of Thyroid Cancer Metastasis Is Associated with Alternation of Actin Skeleton. J. Med. Food 21, 849–857. doi:10.1089/jmf.2017.4144
Wu, X. M., Shen, J. Z., Shen, S. F., and Fan, L. P. (2010). Effect of Triptolide on Reversing Hypermethylation of Apc Gene in Jurkat Cells and its Possible Mechanisms. Zhongguo Shi Yan Xue Ye Xue Za Zhi 18, 866–872.
Xu, F., Zhang, Y., Cui, W., Yi, T., Tang, Z., and Dong, J. (2017). The Association between Metabolic Syndrome and Body Constitution in Traditional Chinese Medicine. Eur. J. Integr. Med. 14, 32–36. doi:10.1016/j.eujim.2017.08.008
Yang, Z., Wang, L., Zhang, F., and Li, Z. (2015). Evaluating the Antidiabetic Effects of Chinese Herbal Medicine: Xiao-Ke-An in 3T3-L1 Cells and KKAy Mice Using Both Conventional and Holistic Omics Approaches. BMC Complement. Altern. Med. 15, 272. doi:10.1186/s12906-015-0785-2
Yao, H., Wan, J. Y., Zeng, J., Huang, W. H., Sava-Segal, C., Li, L., et al. (2018a). Effects of Compound K, an Enteric Microbiome Metabolite of Ginseng, in the Treatment of Inflammation Associated colon Cancer. Oncol. Lett. 15, 8339–8348. doi:10.3892/ol.2018.8414
Yao, H., Mo, S., Wang, J., Li, Y., Wang, C. Z., Wan, J. Y., et al. (2018b). Genome-Wide DNA Methylation Profiles of Phlegm-Dampness Constitution. Cell Physiol Biochem 45, 1999–2008. doi:10.1159/000487976
Yin, Y., Morgunova, E., Jolma, A., Kaasinen, E., Sahu, B., Khund-Sayeed, S., et al. (2017). Impact of Cytosine Methylation on DNA Binding Specificities of Human Transcription Factors. Science 356, eaaj2239. doi:10.1126/science.aaj2239
Yu, L., Zhang, R., Li, P., Zheng, D., Zhou, J., Wang, J., et al. (2015). Traditional Chinese Medicine: Salvia Miltiorrhiza Enhances Survival Rate of Autologous Adipose Tissue Transplantation in Rabbit Model. Aesthet. Plast Surg 39, 985–991. doi:10.1007/s00266-015-0566-9
Yu, Q., Su, J., Zhu, K., Yang, S., Zhu, H., and Yu, J. (2019). The Effect of Xuefu Zhuyu Decoction on Clopidogrel Resistance and its Association with the P2Y12 Gene Polymorphisms and Promoter DNA Methylation. Pak J. Pharm. Sci. 32, 2565–2572.
Zhang, H., Lang, Z., and Zhu, J. K. (2018). Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cel Biol 19, 489–506. doi:10.1038/s41580-018-0016-z
Zhang, M., Zheng, H.-x., Gao, Y.-y., Zheng, B., Liu, J.-p., Wang, H., et al. (2017a). The Influence of Schisandrin B on a Model of Alzheimer's Disease Using β-amyloid Protein Aβ1-42-Mediated Damage in SH-Sy5y Neuronal Cell Line and Underlying Mechanisms. J. Toxicol. Environ. Health A 80, 1199–1205. doi:10.1080/15287394.2017.1367133
Zhang, Y., Xie, Y., Liao, X., Jia, Q., and Chai, Y. (2017b). A Chinese Patent Medicine Salvia Miltiorrhiza Depside Salts for Infusion Combined with Conventional Treatment for Patients with Angina Pectoris: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Phytomedicine 25, 100–117. doi:10.1016/j.phymed.2017.01.002
Zhang, S. F., Mao, X. J., Jiang, W. M., and Fang, Z. Y. (2020). Qian Yang Yu Yin Granule Protects against Hypertension-Induced Renal Injury by Epigenetic Mechanism Linked to Nicotinamide N-Methyltransferase (NNMT) Expression. J. Ethnopharmacol 255, 112738. doi:10.1016/j.jep.2020.112738
Zhang, Y., Bai, M., Zhang, B., Liu, C., Guo, Q., Sun, Y., et al. (2015). Uncovering Pharmacological Mechanisms of Wu-tou Decoction Acting on Rheumatoid Arthritis through Systems Approaches: Drug-Target Prediction, Network Analysis and Experimental Validation. Sci. Rep. 5, 9463. doi:10.1038/srep09463
Zhao, Z. Z. (2017). Retrospect and Prospect of Higher Education in Chinese Medicine in Hong Kong, China. Chin. J. Integr. Med. 23, 494–495. doi:10.1007/s11655-017-2814-0
Zheng, L., Guan, Z. J., Pan, W. T., Du, T. F., Zhai, Y. J., and Guo, J. (2018). Tanshinone Suppresses Arecoline-Induced Epithelial-Mesenchymal Transition in Oral Submucous Fibrosis by Epigenetically Reactivating the P53 Pathway. Oncol. Res. 26, 483–494. doi:10.3727/096504017X14941825760362
Zhong, X. X., He, Q. Y., Liao, J. Q., Yin, X. J., Zhao, G. F., and Li, M. (2016). The Compatibility Law of Chinese Patent Medicines for the Treatment of Coronary Heart Disease Angina Pectoris Based on Association Rules and Complex Network. Int. J. Clin. Exp. Med. 9, 9418–9424.
Zhou, Q. B., Yang, X. H., Wang, H. Z., Wang, D. X., Xu, Y. G., Hu, X. M., et al. (2018). Effect of Qinghuang Powder Combined with Bupi Yishen Decoction in Treating Patients with Refractory Cytopenia with Multilineage Dysplasia through Regulating DNA Methylation. Chin. J. Integr. Med. 25 (5), 354–359. doi:10.1007/s11655-018-2554-9
Keywords: epigenetics, DNA methylation, Chinese herbal medicine, constitution theory, ethnopharmacology
Citation: Guo W, Ma H, Wang C-Z, Wan J-Y, Yao H and Yuan C-S (2021) Epigenetic Studies of Chinese Herbal Medicine: Pleiotropic Role of DNA Methylation. Front. Pharmacol. 12:790321. doi: 10.3389/fphar.2021.790321
Received: 06 October 2021; Accepted: 22 November 2021;
Published: 07 December 2021.
Edited by:
George Qian Li, Western Sydney University, AustraliaReviewed by:
Qi Wang, Harbin Medical University, ChinaWei-Hua Huang, Central South University, China
Copyright © 2021 Guo, Ma, Wang, Wan, Yao and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Yi Wan, d2FuamlueWkxMTI4QDE2My5jb20=; Haiqiang Yao, aGFpcWlhbmd5YW9Ab3V0bG9vay5jb20=
†These authors have contributed equally to this work
 Wenqian Guo
Wenqian Guo Han Ma
Han Ma Chong-Zhi Wang
Chong-Zhi Wang Jin-Yi Wan
Jin-Yi Wan Haiqiang Yao
Haiqiang Yao Chun-Su Yuan3,4
Chun-Su Yuan3,4