- Synergia Life Sciences Pvt. Ltd., Mumbai, India
Neuropathic pain is a chronic pain condition seen in patients with diabetic neuropathy, cancer chemotherapy-induced neuropathy, idiopathic neuropathy as well as other diseases affecting the nervous system. Only a small percentage of people with neuropathic pain benefit from current medications. The complexity of the disease, poor identification/lack of diagnostic and prognostic markers limit current strategies for the management of neuropathic pain. Multiple genes and pathways involved in human diseases can be regulated by microRNA (miRNA) which are small non-coding RNA. Several miRNAs are found to be dysregulated in neuropathic pain. These miRNAs regulate expression of various genes associated with neuroinflammation and pain, thus, regulating neuropathic pain. Some of these key players include adenylate cyclase (Ac9), toll-like receptor 8 (Tlr8), suppressor of cytokine signaling 3 (Socs3), signal transducer and activator of transcription 3 (Stat3) and RAS p21 protein activator 1 (Rasa1). With advancements in high-throughput technology and better computational power available for research in present-day pharmacology, biomarker discovery has entered a very exciting phase. We dissect the architecture of miRNA biological networks encompassing both human and rodent microRNAs involved in the development of neuropathic pain. We delineate various microRNAs, and their targets, that may likely serve as potential biomarkers for diagnosis, prognosis, and therapeutic intervention in neuropathic pain. miRNAs mediate their effects in neuropathic pain by signal transduction through IRAK/TRAF6, TLR4/NF-κB, TXIP/NLRP3 inflammasome, MAP Kinase, TGFβ and TLR5 signaling pathways. Taken together, the elucidation of the landscape of signature miRNA regulatory networks in neuropathic pain will facilitate the discovery of novel miRNA/target biomarkers for more effective management of neuropathic pain.
Introduction
Neuropathic pain is defined as “pain induced by a lesion or disease of the somatosensory nervous system” by International Association for the Study of Pain (Bouhassira and Attal, 2019). The overall prevalence of neuropathy-derived pain in the general populace is 7–10%. Neuropathic pain is more frequently diagnosed in women (8%) as compared to men (5.7%) (Murphy et al., 2020). The prevalence of neuropathic pain in the United Kingdom and in the United States is 1 and 2% respectively (Smith and Torrance, 2012). Neuropathic pain affects 20–26.4% of diabetic patients (Bouhassira et al., 2013) and 20% of patients with herpes zoster in the United States (Johnson and Rice, 2014; Saguil et al., 2017). 48–74% patients with low back-related leg pain (Harrisson et al., 2020) and 40% of people after surgery suffer from neuropathic pain (Johansen et al., 2012). Studies have found that about 8.1–17.9% of the Canadian population is affected by neuropathic pain (VanDenKerkhof et al., 2016). Another study showed that the East Asian population has a low incidence of neuropathic pain (3.2%) (Inoue et al., 2017).
Peripheral neuropathic pain and central neuropathic pain are two different types of neuropathic pain. Postherpetic neuralgia, diabetic neuropathy, and causalgia are examples of peripheral neuropathic pain, caused by an injury or dysfunction in the PNS (peripheral nervous system) (McCarberg et al., 2017). Central neuropathic pain, such as thalamic pain, post-stroke pain, and post-spinal cord injury pain, is caused by an injury or dysfunction in the CNS (central nervous system) (Colloca et al., 2017). Neuropathic pain can also be divided into two types: stimulus-evoked and stimulus-independent. Mechanical, thermal, or chemical stimulation causes hyperalgesia and allodynia, which are indications of stimulus-evoked pain and stimulus-independent pain is typically categorized as shooting, stabbing, or burning (Kerstman et al., 2013).
Peripheral tissue injury in peripheral neuropathic pain results in release of inflammatory cytokines/mediators/chemokines [e.g., IL-1β, TGF-β, and chemokine (C-C motif) ligand 2 (CCL2)] as well as neurotrophic factors like nerve growth factor that sensitize nociceptors (Sun et al., 2021). This leads to dysregulated expression of ion channels in sensory neurons causing reduction of thermal and mechanical threshold of nociceptors which is known as peripheral sensitization. The abnormal excitation of peripheral neurons results in increased levels of neurotransmitters like substance P and glutamate in the spinal cord dorsal horn. This results in activation of neurokinin receptor and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)/N-Methyl-D-aspartic acid (NMDA) receptors causing long-lasting elevated excitability of dorsal horn neurons which is known as central sensitization (Woolf, 2011). Persistent stimulation of proinflammatory proteins and constant peripheral sensitization might induce central sensitization of spinal cord dorsal horn in diabetic neuropathic pain (Zhu D. et al., 2019).
Nociceptors are a type of sensory neurons which are triggered by noxious stimuli like heat, cold, mechanical force, or chemical stimulation. Based on the molecular mechanisms of nociception, nociceptors are classified as – thermal nociceptors, mechanical nociceptors, and chemical nociceptors (Tracey, 2017). Thermal nociceptors belong to transient receptor potential cation channel (TRP) receptor family of which the vanilloid variant (TRPV) is found in thermal nociceptive receptors that is responsible for thermal stimuli transduction. The TRPV family consists of TRPV1, TRPV2, TRPV3 and TRPV4 which are stimulated at various thermal temperatures or thermal stimuli (Frias and Merighi, 2016). Mechanical receptors or mechanoreceptors are stimulated by noxious mechanical force. While not much is known about mechanoreceptors, some inhibitory mechanoreceptors like (TWIK-related potassium) TREK and potassium voltage-gated channel subfamily A member 1 (Kv1.1) are known to be involved in nocifensive (defense against injury) behavior by maintaining the noxious mechanical force threshold significantly high to prevent hyperactivity of the mechanoreceptors (Armstrong and Herr, 2021). Chemical nociceptors are expressed by nociceptive neurons in response to harmful, noxious, or irritating chemicals. Apart from thermal stimuli, TRP receptor family can also detect noxious chemicals. While TRPV1 binds to prostaglandins, capsaicin, bradykinin, and other important pro-inflammatory molecules; TRPA1 subfamily is known to detect a wide variety of pungent and oily isothiocyanate compounds like cinnamaldehyde, mustard oils, and formaldehyde (Armstrong and Herr, 2021). Another example of noxious chemical receptors are the acid-sensing ion channels. They are stimulated by protons resulting in opening of their cation channels. They are found throughout the central as well as peripheral nervous system (St. John Smith, 2018).
In some individuals, there may be a possibility of a genetic basis for developing neuropathic pain. A study (Armero et al., 2012) conducted on the Caucasian population concluded that females with polymorphism in the TRPV1 gene are more susceptible to develop neuropathic pain. Met315Ile TRPV1 genotype only in females diagnosed with neuropathic pain, together with other physiological factors such as sex, might influence susceptibility to neuropathic pain (Armero et al., 2012). Kalfon et al. (2019) studied the association of single nucleotide polymorphism in transient receptor potential cation channel subfamily V member 1 (TRPV1) and nerve growth factor (NGF) and localized provoked vulvodynia and observed a significant relation between rs222747 of TRPV1 (c.945G>C, p.Ile315Met) and localized provoked vulvodynia in affected women. Genotyping analyses showed a critically high prevalence of polymorphism c.945G>C (rs222747) of TRPV1 and a single nucleotide polymorphism in the promoter region of NGF (rs11102930) in localized provoked vulvodynia women compared with controls. Substitution of the amino acid modifies the channel’s functional properties leading to increased TRPV1 protein expression because of an elevated copy number. The study further suggests that rs222747 “C” allele of TRPV1 to be a common genetic predisposition for other pain syndromes. Diabetic patients with a polymorphism in OPRM1 are susceptible to diabetic neuropathic pain (Zorina-Lichtenwalter et al., 2018). Black South Africans with polymorphism in KCNS1 are known to have HIV-associated sensory neuropathy (Zorina-Lichtenwalter et al., 2018).
One of the causes of neuropathic pain is the demyelination of peripheral nerve fibers. Demyelination destroys the molecular and structural features of the nerve fibers, which develops neuropathic pain (Wei et al., 2019). Vitamin K2 is believed to play a role in myelin repair and synthesis in the peripheral nervous system. Vitamin K2 activates and carboxylates Gla residues on GAS6 protein that is structurally related to anticoagulation factor protein S (vitamin K-dependent protein). GAS6, and protein S bind and activate the receptor tyrosine kinases of TAM (Tyro3, Axl, and Mer) family which increase myelin production as well as repair after myelin injury (Mehta, 2017). We have previously (Mehta et al., 2018) carried out an open-label observational study to determine the role of vitamin K2-7 in peripheral neuropathy associated with type-2 diabetic patients. In the 1st week, the visual analog score (VAS) score was 8–10 for T2DM patients. The VAS score had reduced to 1–2 in T2DM patients after treatment with vitamin K2-7 by the 12th week. The symptoms of PN had reduced persistently, which showed the effectiveness of vitamin K2-7 in management of peripheral neuropathy caused due to T2DM (Mehta et al., 2018). Proinflammatory cytokines such as TNFα and IL-1β are involved in neuropathic pain development through neuroinflammatory mechanisms (Li QY. et al., 2017). We have also demonstrated that vitamin K2-7 was able to inhibit TNFα and IL-1β gene expression in human monocyte-derived macrophages in a dose-dependent manner (Pan et al., 2016). In bones, primary activation of the RANKL/RANK (receptor activator for nuclear factor kappa B) system activates osteoclasts, which triggers the damage of bones and subsequently damages the peripheral sensory nerves around bones due to bone fracture. Damage to peripheral nerves can lead to neuropathic pain development (Zajączkowska et al., 2019). Activation of the RANKL/RANK system activates the nuclear factor kappa beta (NF-κB), an important regulator of inflammation, which leads to the activation of osteoclasts. Vitamin K2-7 prevents activation of the RANKL/RANK system by upregulating osteoprotegerin, a decoy receptor for RANKL. Vitamin K2-7 prevents the binding of RANKL to the RANK receptor which prevents activation of NF-κB and prevents activation of osteoclasts (Badmaev et al., 2011). Considering these properties of vitamin K2-7, it may likely serve as a potential therapeutic agent in the management of neuropathic pain.
MicroRNAs (miRNAs) are small single-stranded non-coding RNA molecules containing 19–25 nucleotides. miRNAs regulate almost every cellular process by regulating post-transcriptional gene expression and mRNA silencing (Bartel, 2018). miRNAs are found to be dysregulated in several diseases and regulate expression of various genes that are associated with different diseases. We have recently elucidated the non-coding RNA interactome including miRNA networks in cancer chemoprevention (Shah et al., 2021). We have also previously delineated the architecture of miRNA networks in mesothelioma (Gandhi and Nair, 2020), prostate adenocarcinoma (Nair et al., 2014; Nair and Kong, 2015a), cancer chemoprevention (Neelakandan et al., 2012; Nair and Kong, 2015b), as well as miRNA-lncRNA interactions (Nair, 2016). The term miRNA interactome includes cellular biomolecules, e.g., nucleic acids and proteins that interact with miRNA. In this review, we dissect the architecture of miRNA biological networks encompassing both human and rodent microRNAs involved in the development of neuropathic pain. We delineate various microRNAs, and their targets, that may likely serve as potential biomarkers for diagnosis, prognosis, and therapeutic intervention in neuropathic pain.
Upregulated microRNAs in Neuropathic Pain
Human microRNAs Upregulated in Neuropathic Pain
Leinders et al. (2016) investigated the role of hsa-miR-132-3p in white blood cells and sural nerve biopsies of patients with neuropathic pain. hsa-miR-132-3p was elevated in white blood cells as well as sural nerve biopsies of patients. In another study, Leinders et al. (2017) identified that hsa-miR-146a and hsa-miR-21 were upregulated in white blood cells of patients suffering from neuropathic pain. It was found that miR-21 was upregulated by 2.2-fold and hsa-miR-146a was upregulated by 10-fold. Li et al. (2017b) studied the expression of hsa-miR-199a-3p in patients with diabetic neuropathy (DN). hsa-miR-199a-3p was upregulated by ∼2.5-fold in patients with DN. Upregulation of hsa-miR-199a-3p inhibited extracellular serine protease inhibitor E2 (SerpinE2) expression. The downregulation of SerpinE2 by hsa-miR-199a-3p was thought to cause DN by boosting blood coagulation in the skin peripheral circulation. Tramullas et al. (2018) studied hsa-miR-30c-5p expression in individuals with neuropathic pain associated with leg ischemia. It was found that hsa-miR-30c-5p expression was increased in plasma and cerebrospinal fluid of neuropathic pain patients. Von Schack et al. (2011) used TaqMan Low Density Array (TLDA) and reported upregulation of hsa-miR-133b (10.2 fold). Heyn et al. (2016) conducted a study with patients suffering from neuropathic pain to determine the expression of miRNAs in neuropathic pain. Blood samples from neuropathic pain patients showed that hsa-miR-124a and hsa-miR-155 were upregulated by 2-fold. hsa-miR-124a and hsa-miR-155 reduced the expression of Sirtuin 1 (SIRT1) mRNA in patients, which led to the development of neuropathic pain. Thus, these miRNAs can be investigated as therapeutic targets in neuropathic pain.
Xu et al. (2014) conducted a study to determine the expressions of miRNAs in neuropathic pain. Using miRCURY LNA array, hsa-miR-22, hsa-miR-31-5p, and hsa-miR-133b were found to be upregulated by more than 2-fold. Hori et al. (2013) found that hsa-miR-28-3p and hsa-miR-223 were upregulated by more than 2 fold after miRNA expression profiling. Genda et al. (2013) studied the change in hsa-miR-124 expression neuropathic pain. hsa-miR-124 was upregulated by 2.26-fold. It was suggested that change in miRNA expression played a role in the maintenance and development of and therapy for neuropathic pain. Asahchop et al. (2018) conducted a study on HIV/AIDS patients diagnosed with symptomatic distal sensory polyneuropathy (sDSP). hsa-miR-455-3p was upregulated by 12-fold in HIV patients with sDSP as compared to non-sDSP HIV patients. Thus, hsa-miR-455-3p can be a potential biomarker for HIV-related sDSP. Chatterjee et al. (2018) conducted a study on individuals suffering from arsenic-induced peripheral neuropathy. It was found that hsa-miR-29a was upregulated by 3.63-fold in arsenic-exposed individuals with peripheral neuropathy. According to the finding, arsenic-induced peripheral neuropathy could be caused by the mir-29a/beta-catenin/PMP22 axis. Cao et al. (2019b) used skins of patients suffering from Postherpetic Neuralgia (PHN) to study the expression of miRNAs in PHN. hsa-miR-4491, hsa-miR-502-5p, hsa-miR-4528, hsa-miR-4721, hsa-miR-760, hsa-miR-495-3p, hsa-miR-382-5p, hsa-miR-4506, hsa-miR-1258, and hsa-miR-330-5p were upregulated by more than 5-fold in skin of PHN patients as compared to control. These miRNAs can be potential targets to treat PHN.
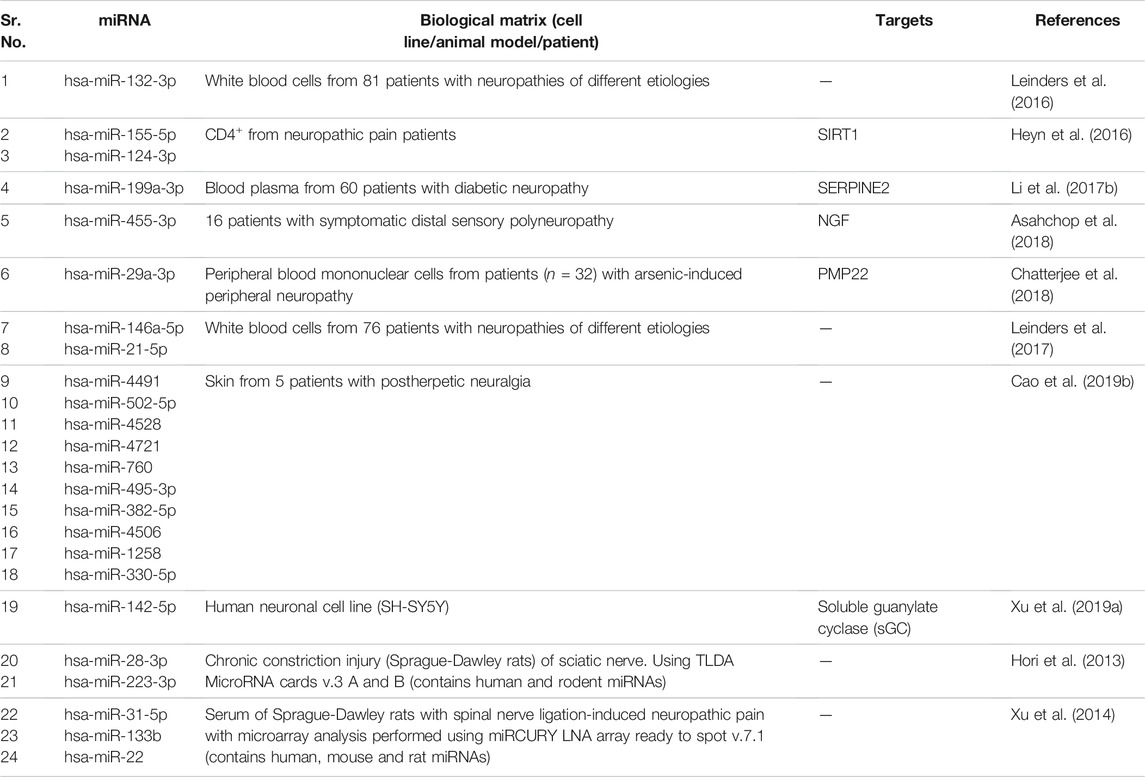
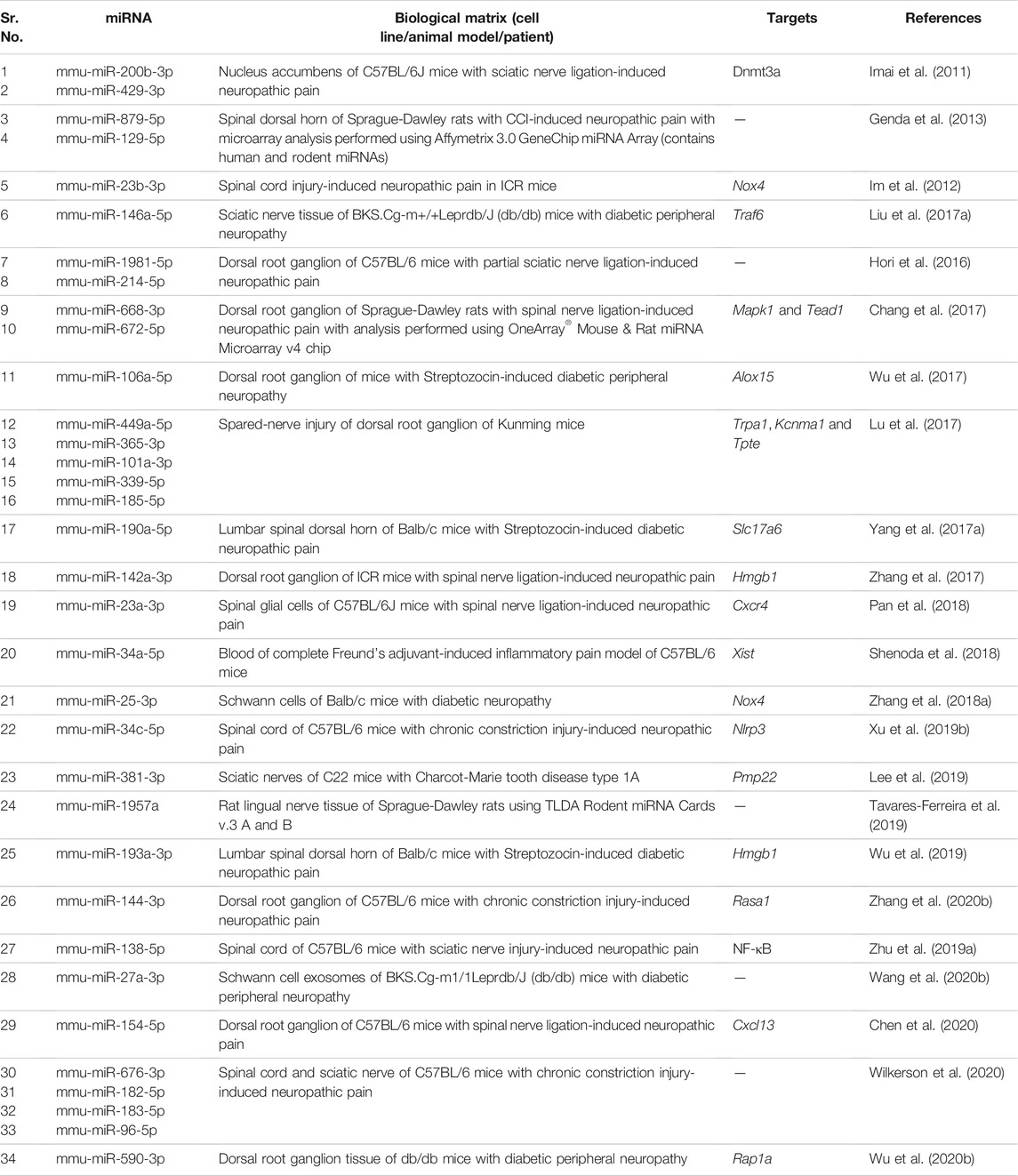
We summarize upregulated human microRNAs in neuropathic pain in Table 1. An in silico method was used to construct miRNA-miRNA and miRNA-target networks of upregulated human miRNAs in neuropathic pain as shown in Figures 1A,B respectively.
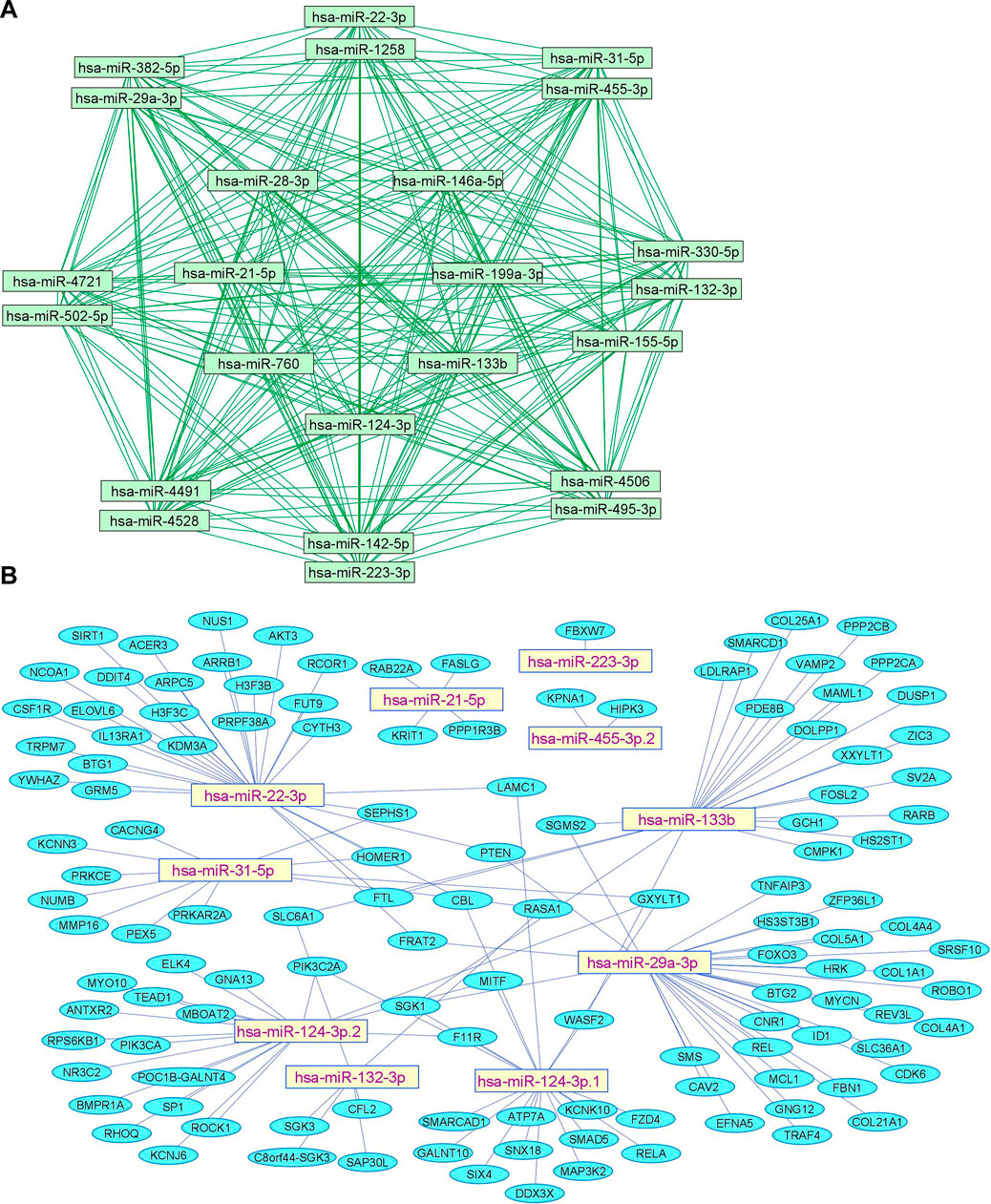
FIGURE 1. Upregulated human microRNA networks in neuropathic pain: (A) Human microRNA – microRNA network: Human miRNA-miRNA interaction network with 24 nodes and 276 edges. The networks were constructed using Cytoscape 3.8.2. (B) Human microRNA – gene target network: Architecture of networks of upregulated miRNAs in humans implicated in neuropathic pain showing interaction network of functionally enriched miRNAs with their targets with 137 nodes and 145 edges. The networks were constructed using Cytoscape 3.8.2. Using Mienturnet, the miRNA-target interactions were identified by TargetScan and the functionally enriched miRNAs were generated using the KEGG database.
Mouse microRNAs Upregulated in Neuropathic Pain
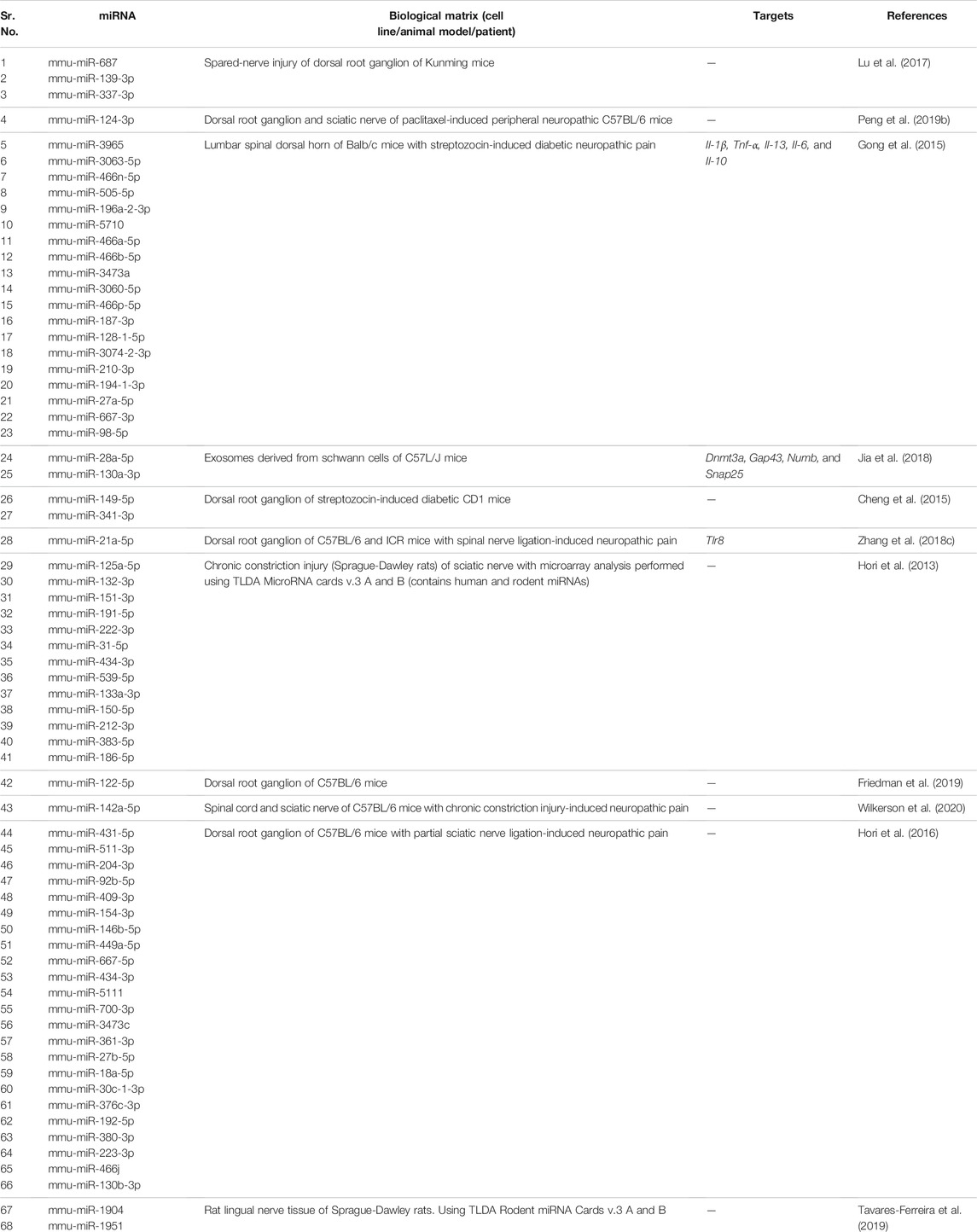
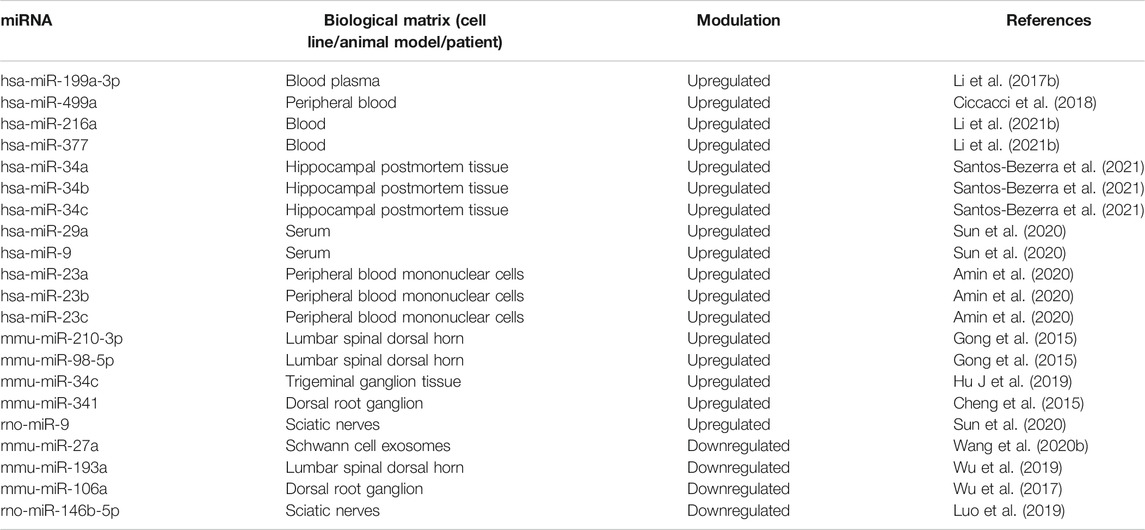
Gong et al. (2015) found that mmu-miR-98-5p and mmu-miR-210-3p were overexpressed in the diabetic neuropathic pain model of mice. Microarray studies showed that mmu-miR-98-5p regulated the Interleukin-6 (Il-6) gene. Thus, changes in miRNA level may affect inflammatory network homeostasis leading to the development of diabetic neuropathic pain. Jia et al. (2018) found out that overexpression of exosomal miRNA-28, -31a, and -130a in high glucose-stimulated Schwann cells of hyperglycemic mice led to the development of diabetic peripheral neuropathy (DPN). An increase in levels of exosomal miRNAs decreased the levels of proteins Dnmt3a, Numb, Snap25, and Gap43, which in turn led to the development of DPN. Exosomal miRNAs have been investigated as gene therapy for DPN. Thus, exosomes derived from mesenchymal stromal cells loaded with miR-146a (exo-146a) suppressed endothelial cell activation as well as peripheral blood inflammatory monocytes through inhibition of toll-like receptor (TLR)-4/NF-kappaB signaling pathway (Fan et al., 2021). Further, Numb is an endocytic protein that complexes with non-SUMOylated collapsin response mediator protein 2 (CRMP2) as well as E3 ubiquitin ligase Nedd4-2 and epidermal growth factor receptor pathway substrate 15 (Eps15). The complex then promotes clathrin-mediated endocytosis of voltage-gated sodium channels (NaV1.7) that plays a key role in neuronal excitability and neuropathic pain. It was observed that prevention of CRMP2 SUMOylation signaling pathway in CRMP2K374A/K374A female mice having neuropathic pain reversed mechanical allodynia (Gomez et al., 2021) likely through Numb/CRMP2/Nedd4-2/Eps15 axis. Hu et al. (2019) observed that mmu-miR-34c was significantly upregulated in trigeminal ganglion tissue of type 1 diabetic mice. Upregulation of mmu-miR-34c decreased the expression of proteins Microtubule-associated protein 1A/1B-light chain 3-II (LC3-II) and Autophagy Related 4B Cysteine Peptidase (Atg4B), which affected corneal nerve regeneration in diabetes leading to diabetic corneal neuropathy. Peng et al. (2019b) studied that mmu-miR-124-3p was overexpressed in the chemotherapy-induced peripheral neuropathy (CiPN) mice model. The increase in mmu-miR-124-3p corresponded with cold allodynia and degeneration of axon in dorsal root ganglion and sciatic nerve, which contributed to the development of CiPN. Zhang et al. (2018C) showed that overexpression of mmu-miR-21 caused neuropathic pain in mice. Overexpression of mmu-miR-21 led to overexpression of Toll Like Receptor 8 (Tlr8). An increase in TLR8 expression mediated ERK activation, production of inflammatory mediators, and neuronal hyperexcitability leading to neuropathic pain. Cheng et al. (2015) identified that mmu-miR-341 was upregulated in mice with diabetic polyneuropathy. Antagonizing the elevated levels of mmu-miR-341 improved electrophysiological, structural, and behavioral abnormalities of sensory neurons caused due to neuropathy. These findings showed that mmu-miR-341 caused abnormalities in sensory neurons of mice which led to diabetic polyneuropathy. Hori et al. (2013) reported that mmu-miR-125a-5p, -132, -191, -222, -212 and -133a were upregulated by more than 2 fold. Wilkerson et al. (2020) found that mmu-miR-142-5p was upregulated (∼20 fold) in sciatic nerves of CCI mice. Hori et al. (2016) determined expression profiles of miRNAs in neuropathic pain. mmu-miR-449a, -18a, -130b, and -223 were upregulated by more than 2-fold in sciatic nerves of mice.
We summarize upregulated mouse microRNAs in neuropathic pain in Table 2. An in silico method was used to construct miRNA-miRNA and miRNA-target networks of upregulated mouse miRNAs in neuropathic pain as shown in Figures 2A,B respectively.
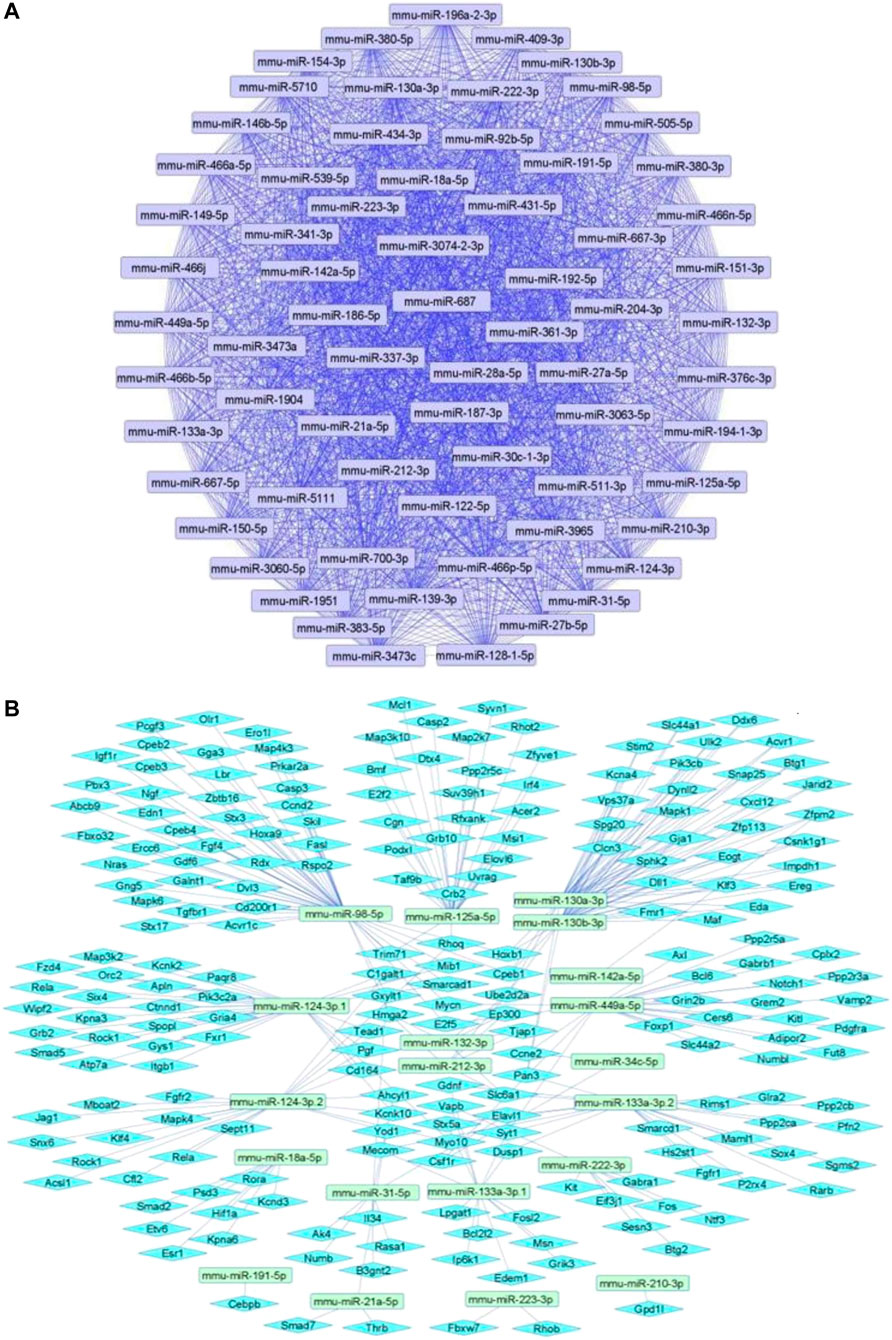
FIGURE 2. Upregulated mouse microRNA networks in neuropathic pain: (A) Mouse microRNA – microRNA network: Mouse miRNA-miRNA interaction network with 68 nodes and 2,278 edges. The networks were constructed using Cytoscape 3.8.2. (B) Mouse microRNA – gene target network: Architecture of networks of upregulated miRNAs in mouse implicated in neuropathic pain showing interaction network of functionally enriched miRNAs with their targets with 237 nodes and 287 edges. The networks were constructed using Cytoscape 3.8.2. Using Mienturnet, the miRNA-target interactions were identified by TargetScan and the functionally enriched miRNAs were generated using the KEGG database.
Rat microRNAs Upregulated in Neuropathic Pain
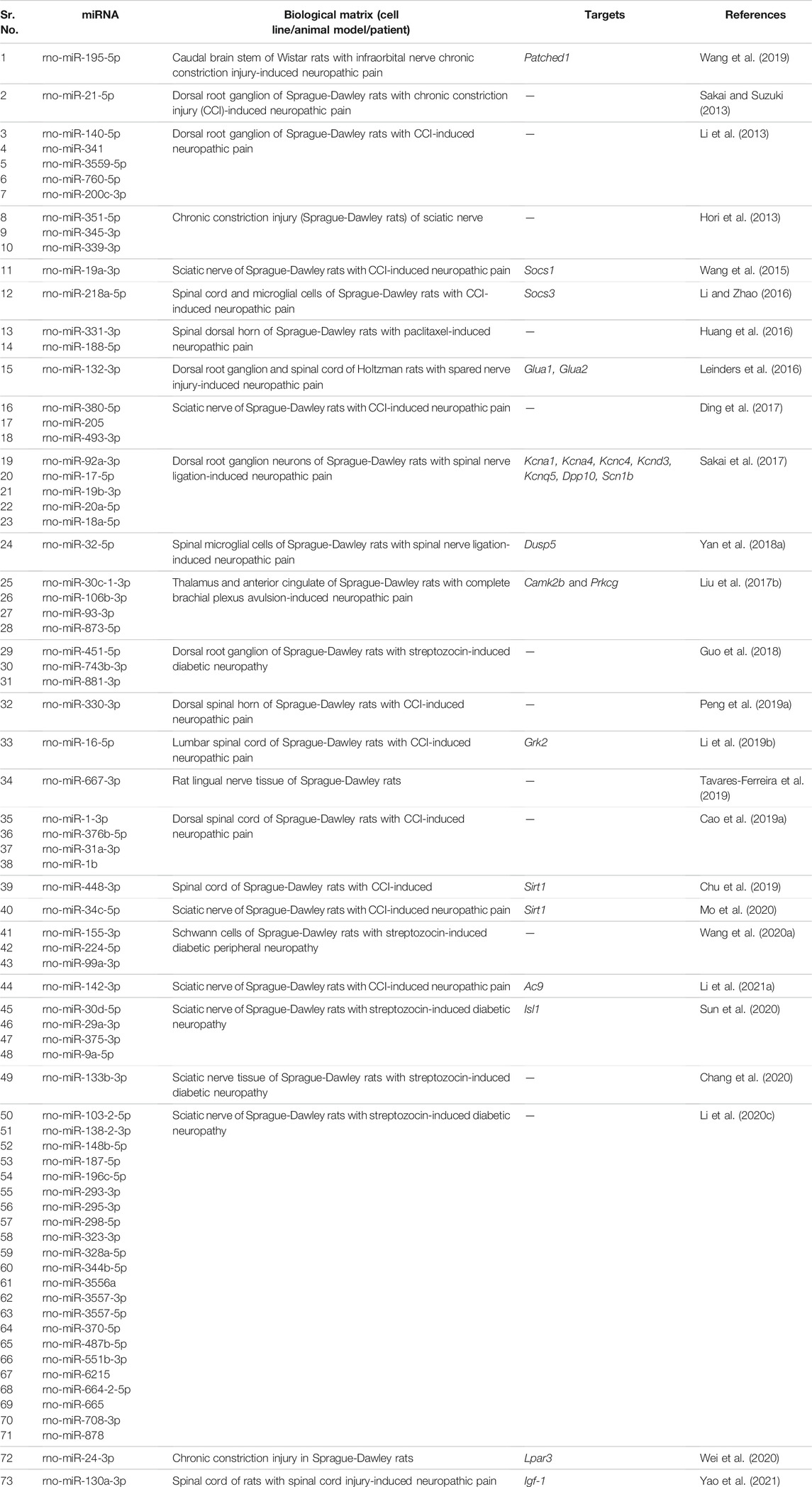
Li et al. (2021a) found that overexpression of rno-miR-142-3p developed neuropathic pain in chronic constriction injury (CCI) rat model. Adenylate cyclase 9 (Ac9) and cAMP levels were significantly reduced in CCI rats. A decrease in Ac9 level led to an increase in expression of inflammatory factors via reduced expression of cAMP/AMPK pathway-related proteins, thereby, leading to the development of neuropathic pain. Sakai and Suzuki (2013) demonstrated that upregulation of rno-miR-21 contributed to neuropathic pain in rats. An increase in rno-miR-21 in dorsal root ganglion was responsible for pain development. In addition, intrathecal Il-1β elevated rno-miR-21 expression in the dorsal root ganglion, resulting in neuropathic pain. Sun et al. (2020) suggested that rno-miR-9 was overexpressed in sciatic nerves of rats with diabetes. Increased expression of ISL LIM homeobox 1 (Isl1) led to activation of sonic hedgehog (SHH) signaling pathway. rno-miR-9 contributed to diabetic peripheral neuropathy through the SHH signaling pathway by binding to ISL1. Wang et al. (2019) observed that rno-miR-195 expression was decreased in the infraorbital nerve CCI rat model. On the contrary, expression of Patched1 decreased notably. mmu-miR-195 aggravated neuropathic pain by activating the SHH signaling pathway by binding Patched1. Wei et al. (2020) found that rno-miR-24-3p expression was increased in CCI rats. rno-miR-24-3p activated Wnt5a/β-Catenin signaling by decreasing the expression of Lpar3 to promote neuropathic pain development.
Li et al. (2019b) showed that rno-miR-15a and rno-miR-16 expression was remarkably increased in the CCI rat spinal cord. Thermal hyperalgesia and mechanical allodynia in CCI rats were reduced significantly when rno-miR-15a and rno-miR-16 were downregulated. G Protein-Coupled Receptor Kinase 2 (Grk2) was found to be a potential target of rno-miR-15a and rno-miR-16. Inhibition of rno-miR-15a and rno-miR-16 remarkably increased the expression of Grk2 in CCI rats. Notably, the silencing of Grk2 significantly reversed the effects of rno-miR-15a/16 downregulation. In conclusion, increased expression of rno-miR-15a and rno-miR-16 downregulated the expression of Grk2 leading to the development of neuropathic pain. Yan et al. (2018a) studied that rno-miR-32-5p was upregulated in spinal microglia of rats after spinal nerve ligation. Dual Specificity Phosphatase 5 (Dusp5) was found to be a target of rno-miR-32-5p. rno-miR-32-5p promoted neuropathic pain development by downregulating the expression of Dusp5. Li and Zhao (2016) studied that expression of rno-miR-218 was upregulated in rats after CCI. Suppressor Of Cytokine Signaling 3 (Socs3), a critical inflammatory mediator, was a direct target of rno-miR-218. rno-miR-218 downregulated the expression of Socs3, thus, activating JAK/STAT3 inflammatory signaling which led to the development of neuropathic pain.
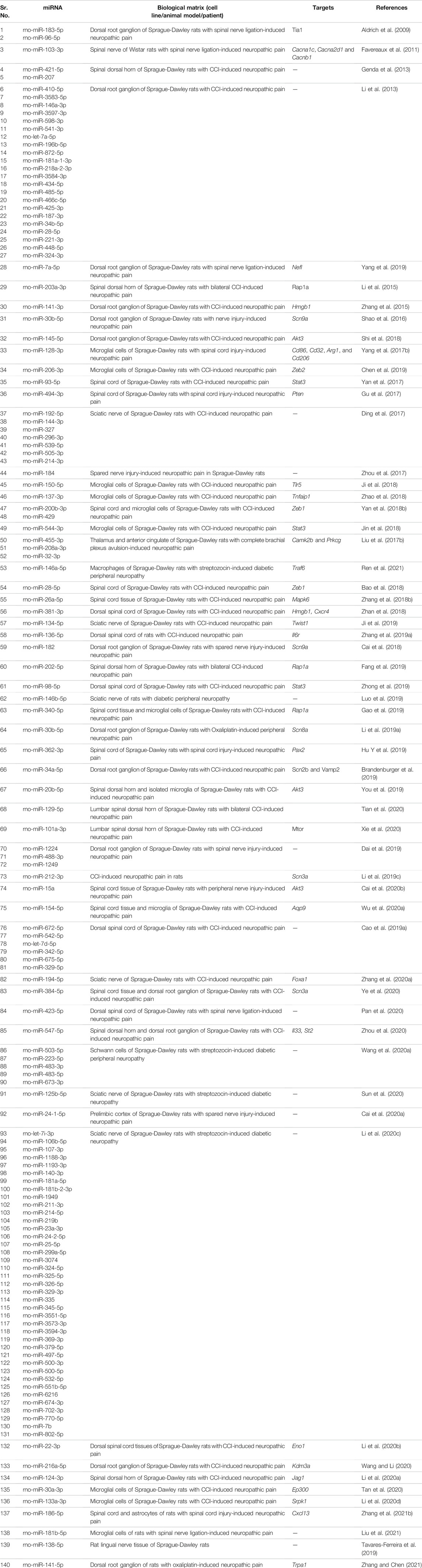
We summarize upregulated rat microRNAs in neuropathic pain in Table 3. An in silico method was used to construct miRNA-miRNA and miRNA-target networks of upregulated rat miRNAs in neuropathic pain as shown in Figures 3A,B respectively.
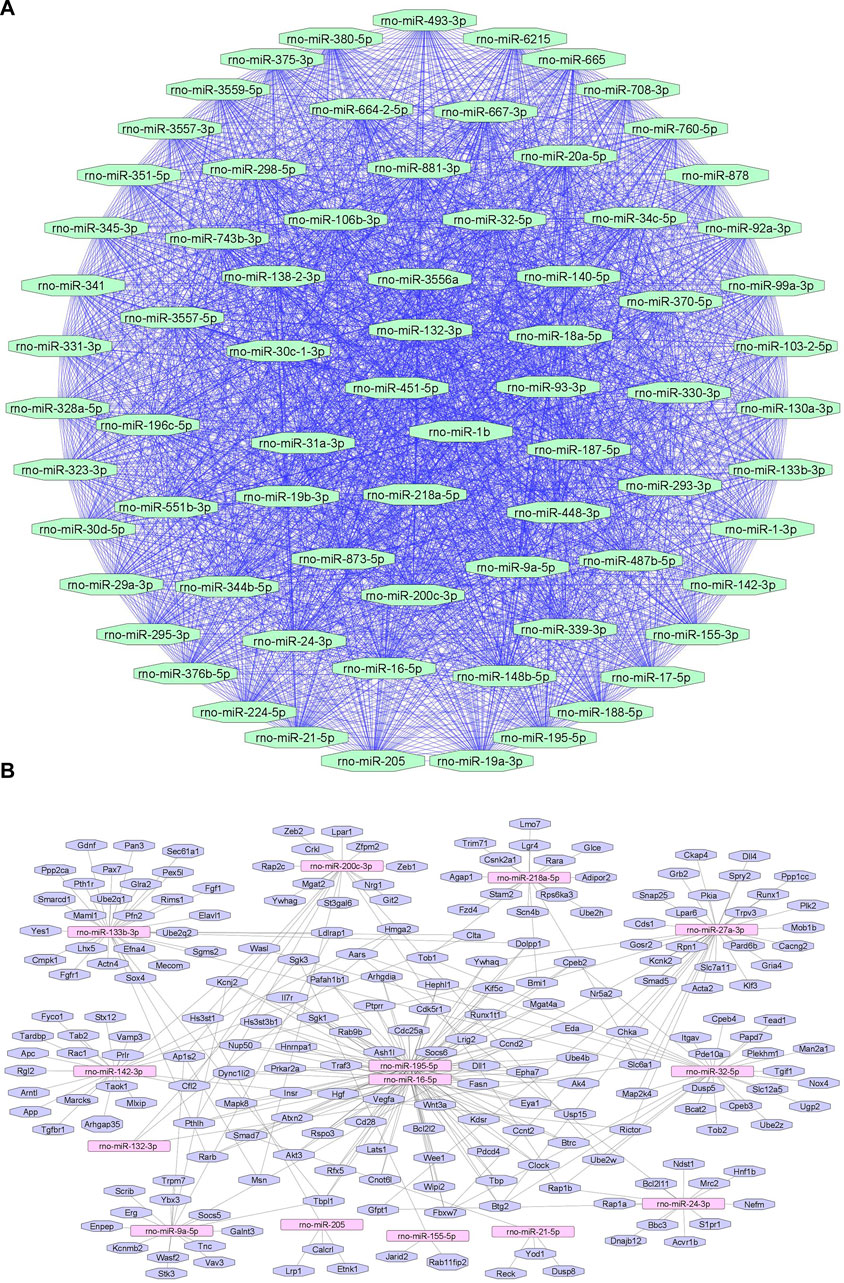
FIGURE 3. Upregulated rat microRNA networks in neuropathic pain: (A) Rat microRNA – microRNA network: Rat miRNA-miRNA interaction network with 73 nodes and 2,628 edges. The networks were constructed using Cytoscape 3.8.2. (B) Rat microRNA – gene target network: Architecture of networks of upregulated miRNAs in rat implicated in neuropathic pain showing interaction network of functionally enriched miRNAs with their targets with 205 nodes and 280 edges. The networks were constructed using Cytoscape 3.8.2. Using Mienturnet, the miRNA-target interactions were identified by TargetScan, and the functionally enriched miRNAs were generated using the KEGG database.
Downregulated microRNAs in Neuropathic Pain
Human microRNAs Downregulated in Neuropathic Pain
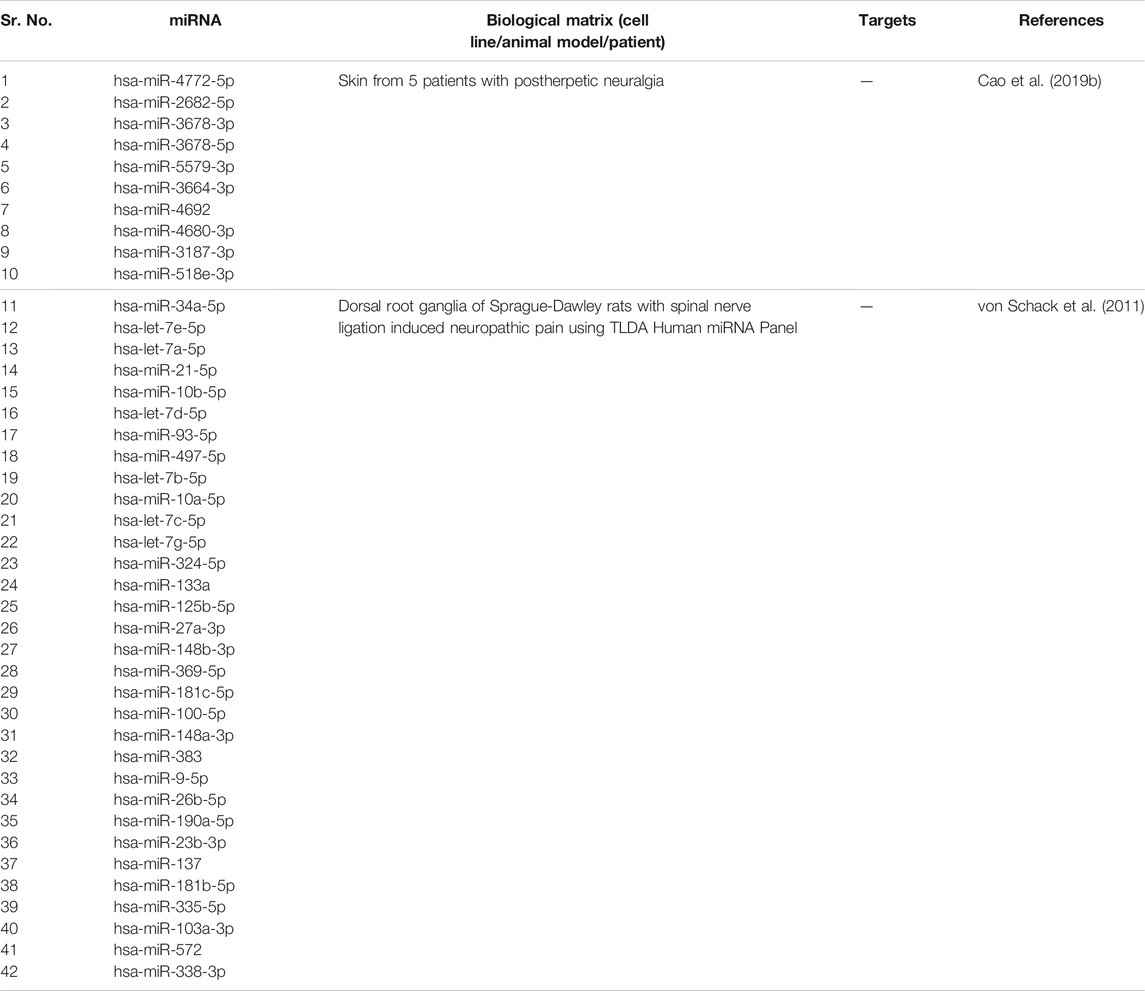
Liu et al. (2019) found that hsa-miR-101 expression was found to be decreased in plasma and sural nerve biopsies from patients with neuropathic pain. Reduction in hsa-miR-101 led to Nuclear Factor Kappa B (NF-κB) activation which contributed to the development of neuropathic pain. Von Schack et al. (2011) studied the expressions of miRNAs using TLDA miRNA panel. hsa-miR-103, hsa-miR-181b, hsa-miR-137, hsa-miR-23b, hsa-miR-26b, hsa-miR-148a, hsa-miR-181c, hsa-miR-148b, hsa-miR-125b, hsa-miR-133a, hsa-let-7a, hsa-let-7b, hsa-let-7c, hsa-let-7d, hsa-let-7e, hsa-let-7g, hsa-miR-10a, hsa-miR-497, hsa-miR-93, hsa-miR-10b, hsa-miR-21 and hsa-miR-34a were downregulated more than 2 fold. Cao et al. (2019b) collected skin samples of patients suffering from Postherpetic Neuralgia (PHN) to study the miRNA expression profile. More than 10-fold downregulation of following miRNAs was observed: hsa-miR-4772-5p, hsa-miR-2682-5p, hsa-miR-3678-3p, hsa-miR-3678-5p, hsa-miR-5579-3p, hsa-miR-3664-3p, hsa-miR-4692, hsa-miR-4680-3p, hsa-miR-3187-3p, and hsa-miR-518e-3p. These miRNAs could be potential targets for treating PHN.
We summarize downregulated human microRNAs in neuropathic pain in Table 4. An in silico method was used to construct miRNA-miRNA and miRNA-target networks of downregulated human miRNAs in neuropathic pain as shown in Figures 4A,B respectively.
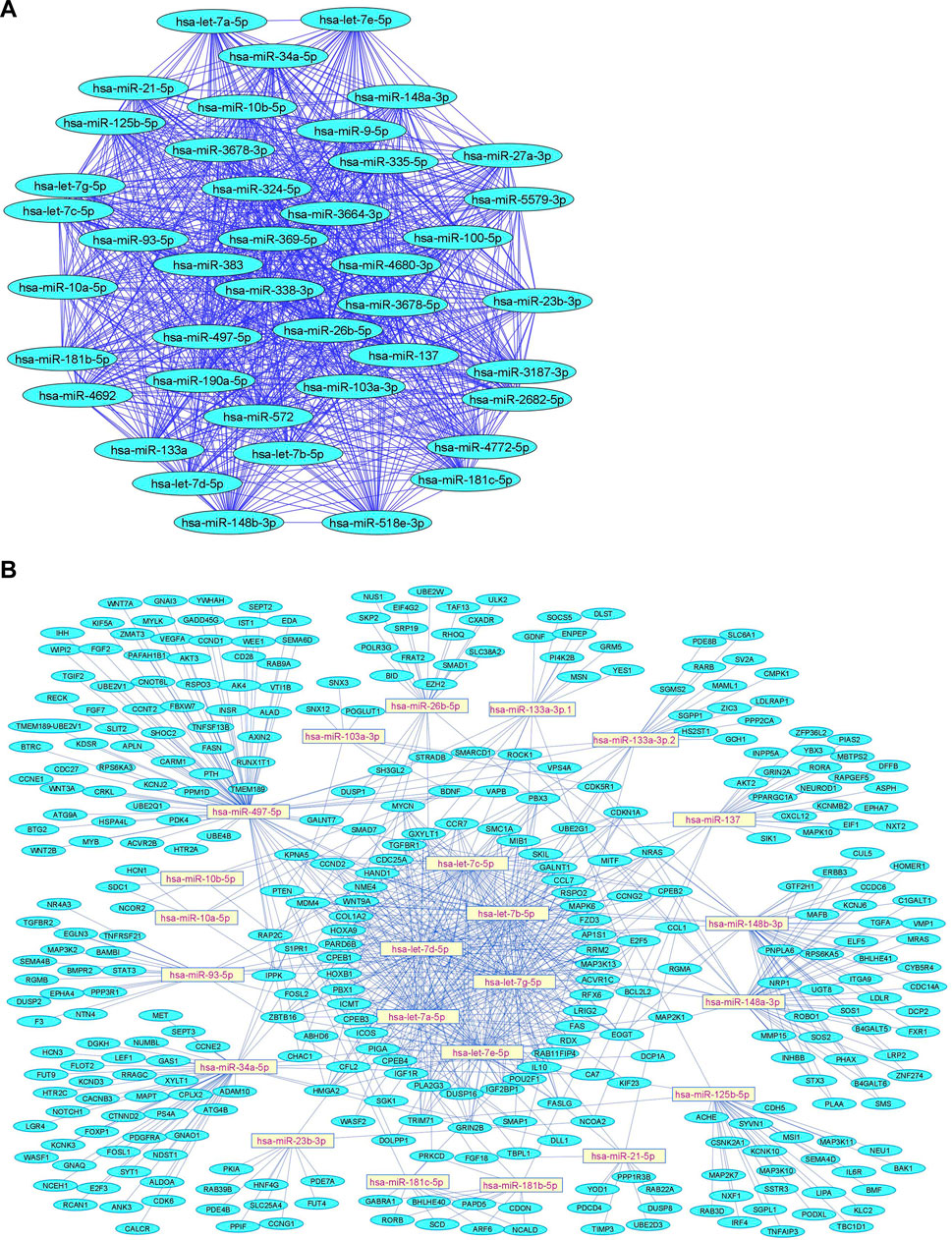
FIGURE 4. Downregulated human microRNA networks in neuropathic pain: (A) Human microRNA – microRNA network: Human miRNA-miRNA interaction network with 42 nodes and 861 edges. The networks were constructed using Cytoscape 3.8.2. (B) Human microRNA – gene target network: Architecture of networks of upregulated miRNAs in humans implicated in neuropathic pain showing interaction network of functionally enriched miRNAs with their targets with 380 nodes and 759 edges. The networks were constructed using Cytoscape 3.8.2. Using Mienturnet, the miRNA-target interactions were identified by TargetScan and the functionally enriched miRNAs were generated using the KEGG database.
Mouse microRNAs Downregulated in Neuropathic Pain
Zhu et al. (2019a) observed that mmu-miR-138 was decreased in mice after sciatic nerve injury. Decreased levels of mmu-miR-138 activated NF-κB pathway and inflammatory responses during nerve injury. Nerve injury resulted in neuropathic pain and subsequent pain hypersensitivity. Using the CCI model of neuropathic pain, Zhang et al. (2020b) found that mmu-miR-144 was downregulated after inducing CCI. Proinflammatory mediators like Il-6, Il-1β and Tnfα were significantly elevated after CCI induction. RAS P21 Protein Activator 1 (Rasa1), the target gene of mmu-miR-144, increased the levels of proinflammatory mediators after CCI. These results showed successful establishment of neuropathic pain in CCI-induced mice. Zhang et al. (2018a) observed that mmu-miR-25 was decreased in mice with diabetic peripheral neuropathy. Inhibition of mmu-miR-25 increased the levels of reactive oxygen species and overexpression of NADPH Oxidase 4 (Nox4). This imbalance was sufficient to aggravate the Schwann cells damage in sciatic nerves of diabetic mice, leading to diabetic peripheral neuropathy. Wang et al. (2020b) reported that a decrease in mmu-miR-27a in the exosomes derived from Schwann cells led to peripheral neuropathy in diabetic mice. A decrease in mmu-miR-27a led to the dysfunction of interaction between the axons and blood vessels that regulate peripheral nerve function, thus, contributing to diabetic peripheral neuropathy development. Wu et al. (2019) observed that expression of mmu-miR-193a was decreased in diabetic neuropathic pain. Downregulation of mmu-miR-193a increased the activity of inflammatory mediator protein High Mobility Group Box 1 (Hmgb1) which led to the progression of diabetic neuropathic pain.
Pan et al. (2018) observed that mmu-miR-23a was decreased in the pSNL model of neuropathic pain in mice. CXC chemokine receptor type 4 (Cxcr4) activity was increased and it targeted TXNIP/NLRP3 inflammasome axis to induce neuropathic pain in mice. Imai et al. (2011) observed that mmu-miR-200b and mmu-miR-429 expressions were decreased in nucleus accumbens neurons after sciatic nerve ligation in mice. Since Dnmt3a is a target of mmu-miR-200b and mmu-miR-429, the expression of DNA methyltransferase 3 alpha (Dnmt3a) was increased in nucleus accumbens. Double-immunolabeling with antibodies specific to Dnmt3a showed that increased Dnmt3a proteins were dominantly expressed in postsynaptic neurons in the nucleus accumbens area under a neuropathic pain-like state. Xu et al. (2019b) studied that mmu-miR-34c was downregulated in the CCI model of neuropathic pain. This led to an increase in inflammatory mediator Nlrp3 expression, which led to the development of neuropathic pain. Wilkerson et al. (2020) conducted a study on CNS tissues of a mouse model of neuropathic pain to elucidate microRNAs expressed in neuropathic pain. mmu-miR-182-5p, mmu-miR-96-5p and mmu-miR-183-5p were downregulated in CCI mice with mechanical allodynia. It was found that this microRNA cluster regulates more than 80% of genes related to neuropathic pain. Wu et al. (2017) determined that expression of mmu-miR-106a was decreased in mice with diabetic peripheral neuropathy. Increased 12/15-LOX expression induced mechanical allodynia and thermal hyperalgesia in mice. 12/15-LOX was observed to be a target of mmu-miR-106a. Hence, downregulation of mmu-miR-106a led to the progression of neuropathic pain. Lu et al. (2017) studied miRNA expression profile using microarray analysis and found that mmu-miR-101a and mmu-miR-365-3p were downregulated in mice.
We summarize downregulated mouse microRNAs in neuropathic pain in Table 5. An in silico method was used to construct miRNA-miRNA and miRNA-target networks of downregulated mouse miRNAs in neuropathic pain as shown in Figures 5A,B respectively.
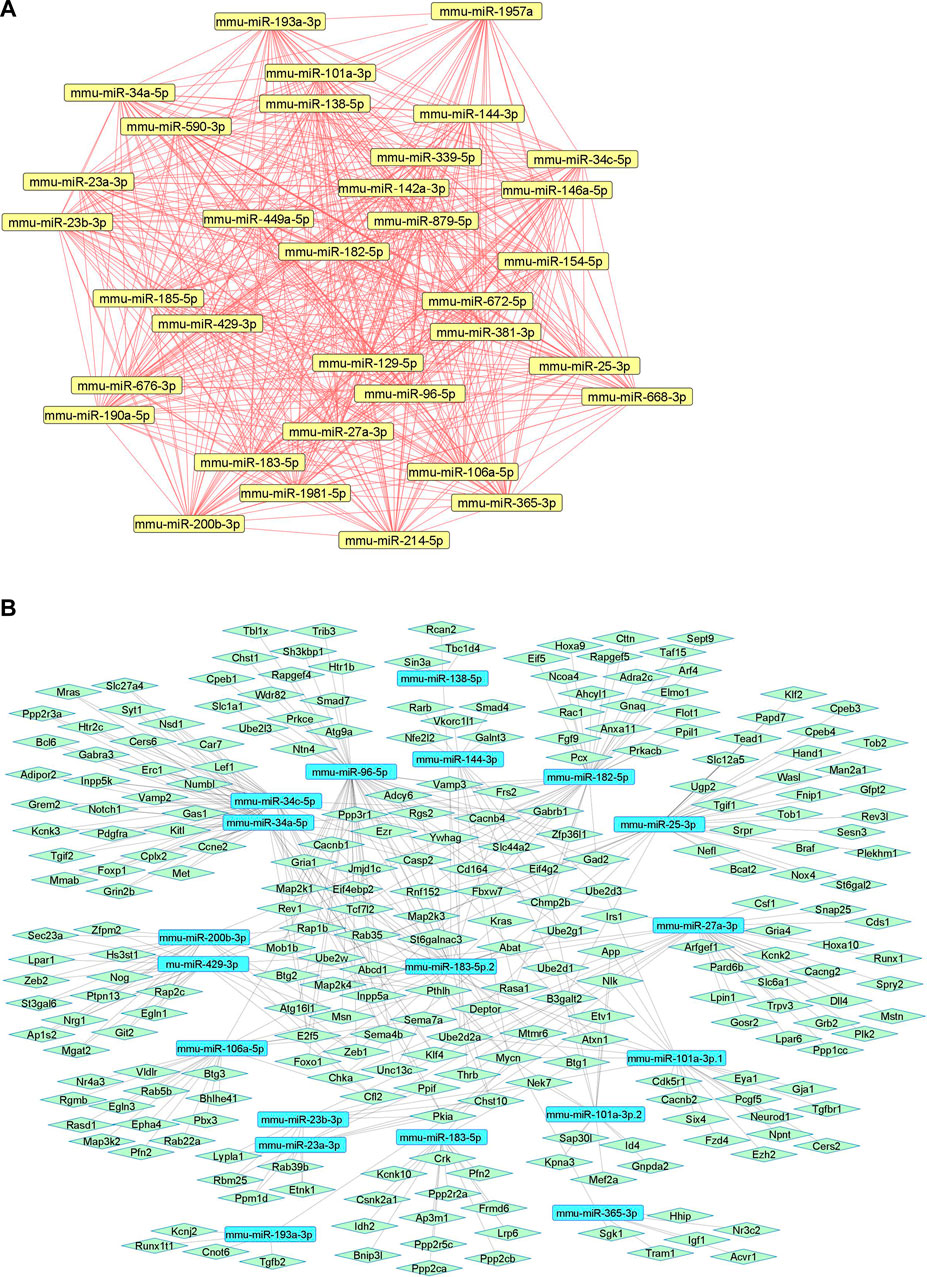
FIGURE 5. Downregulated mouse microRNA networks in neuropathic pain: (A) Mouse microRNA – microRNA network: Mouse miRNA-miRNA interaction network with 34 nodes and 561 edges. The networks were constructed using Cytoscape 3.8.2. (B) Mouse microRNA – gene target network: Architecture of networks of upregulated miRNAs in mouse implicated in neuropathic pain showing interaction network of functionally enriched miRNAs with their targets with 275 nodes and 413 edges. The networks were constructed using Cytoscape 3.8.2. Using Mienturnet, the miRNA-target interactions were identified by TargetScan and the functionally enriched miRNAs were generated using the KEGG database.
Rat microRNAs Downregulated in Neuropathic Pain
Zhong et al. (2019) found out that rno-miR-98 was downregulated in the CCI rat model. The study indicated that Signal Transducer and Activator of Transcription 3 (Stat3) was overexpressed, and it was found to be a probable target of rno-miR-98. Overexpression of Stat3 led to an increase in Il-6, Il-1β, and Tnfα expression, which contributed to development of neuropathic pain. Zhang et al. (2019b) observed that rno-miR-124-3p was remarkably downregulated in rats after CCI. The expression of Il-6, Il-1β, and Tnfα proteins increased greatly after CCI, which contributed to neuroinflammation. Downregulation of rno-miR-124-3p led to an increase in expression of Enhancer of zeste homolog 2 (Ezh2), a direct target of rno-miR-124-3p. Overexpression of Ezh2 and other inflammatory mediators led to the development of neuropathic pain. Li et al. (2020b) studied the effect of rno-miR-22-3p downregulation on the development of neuropathic pain. Downregulation of rno-miR-22-3p promoted the progression of neuropathic pain by regulating inflammation factors expression by directly targeting Enolase 1 (Eno1). Xie et al. (2020) found that rno-miR-101 was downregulated in lumbar spinal dorsal horns after CCI. mTOR (mRNA) was upregulated after CCI and was found to be a direct target of rno-miR-101. Activation of the mTOR signaling pathway was responsible for the onset, progression, and maintenance of neuropathic pain. Yan et al. (2017) observed that rno-miR-93 was downregulated in the spinal cord of CCI rats. Stat3 expression was upregulated. It was found that Stat3 was a direct target of rno-miR-93. Overexpression of rno-miR-93 remarkably reduced the expression of Stat3 in vitro and in vivo. Further, overexpression of Stat3 markedly reversed the rno-miR-93 overexpression-induced repressive effects on neuropathic pain development and neuroinflammation. In conclusion, the downregulation of rno-miR-93 and upregulation of Stat3 led to the development of neuropathic pain.
Zhang et al. (2018b) observed a significant decrease of rno-miR-26a-5p expression in the spinal cord tissues from CCI rats. Mitogen-Activated Protein Kinase 6 (Mapk6) was upregulated in CCI rats and was found to be a downstream target of rno-miR-26a-5p. Mapk6 upregulation led to the progression of neuropathic pain. Zhang et al. (2020c) found that rno-miR-128-3p was significantly downregulated in the spinal cord of CCI rats. Zinc Finger E-Box Binding Homeobox 1 (Zeb1), an inflammation mediator, was upregulated in CCI rats and was found to be a target of rno-miR-128-3p. Upregulated Zeb1 contributed to the development of neuropathic pain by promoting neuroinflammation. Miao et al. (2020) observed that rno-miR-183 was downregulated in the spinal dorsal horn of the CCI rat. Hdac2 reduced the expression of rno-miR-183 by deacetylating histone H4. By upregulating Hdac2 and activating the TXNIP-NLRP3 inflammasome axis, NF-κB p65 suppressed rno-miR-183 expression and generated an inflammatory response in rats, worsening neuropathic pain. Zhang et al. (2015) observed that rno-miR-141 expression was markedly decreased in CCI rats. Downregulation of rno-miR-141 led to an increase in the expression of Hmgb1. In CCI rats, Hmgb1 overexpression exacerbated mechanical allodynia and thermal hyperalgesia, as well as increased proinflammatory cytokines. This led to the development of neuropathic pain in rats. Yan et al. (2018b) found out that rno-miR-200b and rno-miR-429 were notably downregulated in CCI rat spinal cords. Zeb1 was predicted as the target of rno-miR-200b and rno-miR-429. Zeb1 expression was significantly increased in CCI rats, and overexpression of rno-miR-200b and rno-miR-429 significantly inhibited Zeb1 mRNA expression in rats. Knockdown of Zeb1 reduced neuropathic pain development. The findings suggested that rno-miR-200b/rno-miR-429, through targeting Zeb1, could be an essential regulator of neuropathic pain development. Sakai et al. (2013) observed that rno-miR-7a expression decreased in rats with neuropathic pain. β2 subunit of the voltage-gated sodium channel was found to be a target of rno-miR-7a. β2 subunit protein expression was increased in the dorsal root ganglion of rats, which led to the development of neuropathic pain.
We summarize downregulated rat microRNAs in neuropathic pain in Table 6. An in silico method was used to construct miRNA-miRNA and miRNA-target networks of downregulated rat miRNAs in neuropathic pain as shown in Figures 6A,B respectively.
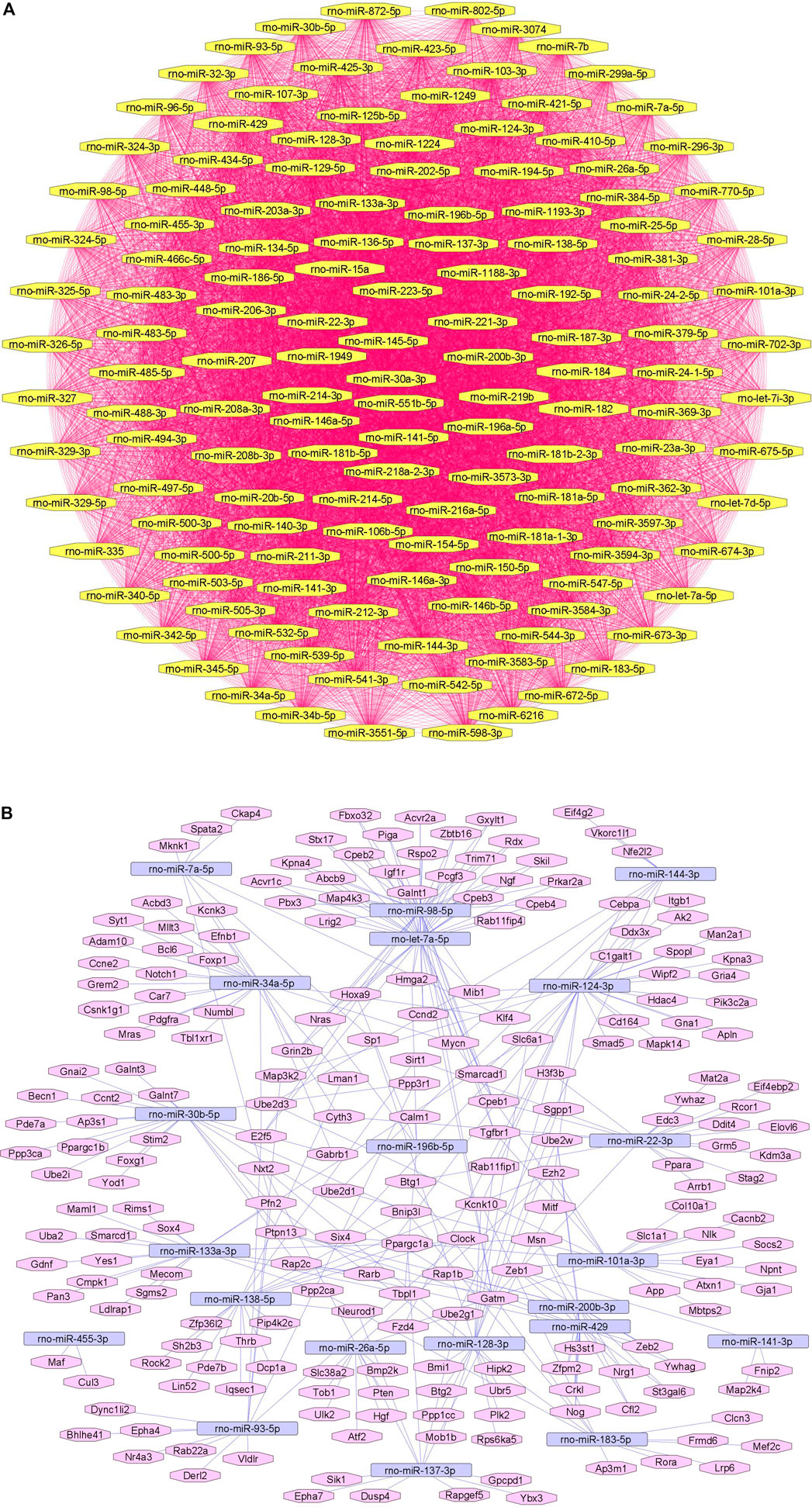
FIGURE 6. Downregulated rat microRNA networks in neuropathic pain: (A) Rat microRNA – microRNA network: Rat miRNA-miRNA interaction network with 140 nodes and 9,730 edges. The networks were constructed using Cytoscape 3.8.2. (B) Rat microRNA – gene target network: Architecture of networks of upregulated miRNAs in rats implicated in neuropathic pain showing interaction network of functionally enriched miRNAs with their targets with 239 nodes and 330 edges. The networks were constructed using Cytoscape 3.8.2. Using Mienturnet, the miRNA-target interactions were identified by TargetScan and the functionally enriched miRNAs were generated using the KEGG database.
microRNAs as Diagnostic and Prognostic Markers in Neuropathic Pain
Huang et al. (2017) compared the levels of miRNA in the blood of postherpetic neuralgia (PHN) and acute herpes zoster (AHZ) patients. 157 serum miRNAs were differentially expressed in PHN patients than in AHZ patients. In comparison to AHZ patients, 17 serum miRNAs from PHN patients were overexpressed and 139 were underexpressed. According to the results of qRT-PCR, the levels of miR-892b, miR-127-5p, miR-107, miR-486-3p, and miR-34c-5p were all considerably greater in PHN patients than in AHZ patients. These miRNAs can be used as diagnostic markers to detect the progression of postherpetic neuralgia. Dayer et al. (2019) evaluated the expression changes of 184 circulating miRNAs in plasma samples from individuals with various origins of persistent pain. Following statistical analysis, 7 circulating miRNAs were discovered that were differentially expressed depending on whether the pain was nociceptive or neuropathic. Two circulating miRNA signatures (hsa-miR-320a and hsa-miR-98-5p) accurately classified the pain type of 70% of patients in the validation set. To summarize, circulating miRNAs are promising biomarkers for identifying and characterizing chronic pain types, as well as for improving the treatment of chronic pain. Peng et al. (2019b) explored miR-124, miR-183, and miR-338 as diagnostic biomarkers in a CiPN mice model. Among the three miRNAs that were analyzed, only miR-124 was statistically significantly increased. Cold allodynia and axonal degeneration were caused by high levels of circulating miR-124 in both the DRG and the sciatic nerve. Hence, plasma levels of miR-124 may be a good diagnostic biomarker for CiPN. Chu et al. (2019) investigated the role of miR-448 as a prognostic biomarker in neuropathic pain. miR-448 was consistently increased in CCI rats, while miR-448 downregulation reduced thermal hyperalgesia and mechanical allodynia in CCI rats. In CCI rats, the expression levels of IL1, IL6, and TNF were substantially higher, but these effects were reversed after treatment with a miR-448 inhibitor. miR-448 increased neuropathic pain in CCI rats via controlling neuroinflammation. Hence, upregulated miR-448 could be used for the prognosis of neuropathic pain.
Regulatory Effects of miRNAs in Inflammation- and Diabetes-Associated Neuropathic Pain
Regulatory Effects of miRNAs in Inflammation-Associated Neuropathic Pain
We discuss herein a few examples of specific miRNAs that play key regulatory roles in inflammation-associated neuropathic pain.
hsa-miR-101
Liu et al. (2019) studied the expression of miRNAs in plasma samples of patients with neuropathic pain and reported a significant downregulation of miR-101. KPNB1, an important regulator for NF-κB/p65 nuclear importing, was identified as a direct target of miR-101 in human embryonic kidney HEK293T cells. Thus, miR-101 inhibits NF-κB signaling via targeting KPNB1 resulting in downregulation of inflammatory cytokines IL-1β and TNF-α.
mmu-miR-128
Yang et al. (2017b) reported that miR-128 was downregulated in murine microglial BV2 cells (treated with spinal cord segment-derived conditioned medium of male Sprague-Dawley rats following spinal cord injury) and that overexpression of miR-128 altered the M1/M2 microglial gene expression. M1 phenotypic markers like CD32 and CD86 were significantly downregulated while M2 phenotypic markers like CD206 and Arg1 were upregulated. Moreover, inflammatory cytokines like IL-6, TNFβ and TNFα were markedly suppressed following upregulation of miR-128. Further, it was reported that phosphorylated p38 (phospho-p38) was downregulated after overexpression of miR-128 suggesting a key role for miR-28 in the pathogenesis of neuropathic pain.
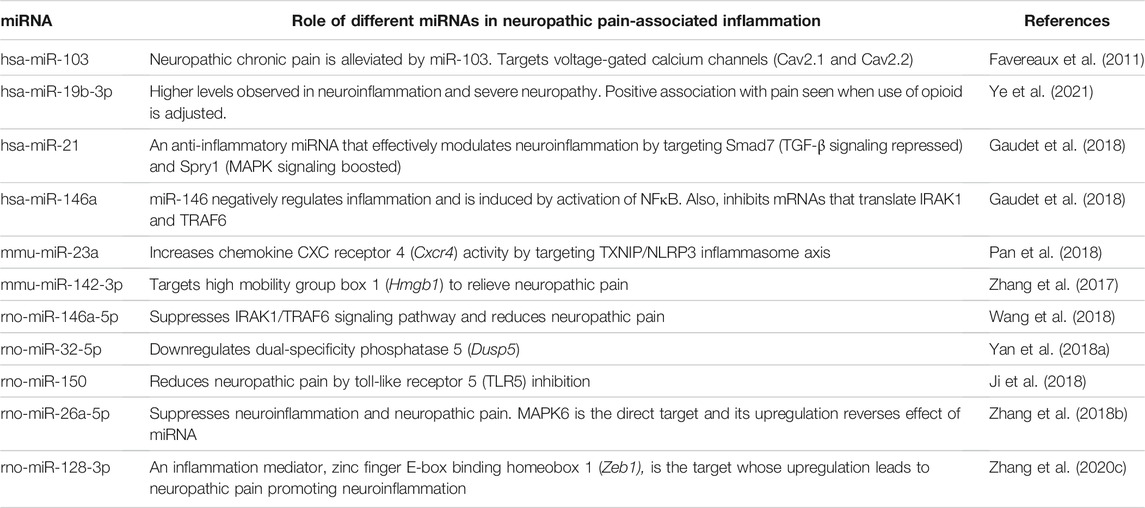
mmu-miR-23a
In pathogen-free adult male C57BL/6J wild-type mice, Pan et al. (2018) studied the functional regulatory role of miR-23a in pain and its association with chemokine CXC receptor 4 (CXCR4) which has been implicated in neuropathic pain. It was observed that expression of CXCR4 was increased in murine spinal glial cells induced with neuropathic pain via partial sciatic nerve ligation (pSNL). miR-23a was observed to bind directly to CXCR4-3′UTR resulting in downregulation of spinal CXCR4. Finally, downregulation of miR-23a increased thioredoxin-interacting protein (TXNIP) which is linked with induction of NOD-like receptor protein 3 (NLRP3) inflammasome resulting in elevated pain behavior.
rno-miR-146a-5p
miR-146a-5p plays an important role in downregulation of IL-1 receptor (toll/interleukin-1 receptor; TIR) and toll-like receptor (TLR4) signaling pathways. TLR4 is activated in neuropathic pain as a key innate immune receptor (Li et al., 2014). TLR4 activation leads to production of proinflammatory cytokines like TNF-α and IL-6 due to nuclear translocation of NF-κB via activation of TRAF6 and IRAK1 (Allette et al., 2017). In a chronic constriction injury model using Sprague-Dawley rats, Wang et al. (2018) demonstrated that NF-κB-dependent miR-146a-5p suppressed IRAK1/TRAF6 which plays a major role in TIR signaling pathway. Further, miR-146a-5p alleviated neuropathic pain by suppressing IRAK1 and TRAF6 via inhibition of TLR4/NF-κB signaling pathway.
rno-miR-34c-5p
miR-34c-5p was found to be involved in neuropathic pain via SIRT1 and STAT3 signaling pathway. In a chronic constriction injury model using male Sprague-Dawley rats, it was reported (Mo et al., 2020) that SIRT1 is suppressed by miR-34c-5p resulting in activation of STAT3 signaling pathway. This promoted the release of inflammatory factors like TNF-α, IL-6 and IL-1β eventually inducing neuropathic pain.
We summarize in Table 7 the role of different miRNAs in neuropathic pain-associated inflammation.
Regulatory Effects of miRNAs in Diabetes-Associated Neuropathic Pain
We discuss herein a few examples of specific miRNAs that play key regulatory roles in diabetes-associated neuropathic pain.
hsa-miR-199a-3p
In a study conducted by Li et al. (2017b), 2), miR-199a-3p was reported to be downregulated in plasma samples of diabetic patients as compared to healthy controls. miR-199a targeted SERPINE2 by binding to the 3′UTR of SERPINE2 and promoted coagulation resulting in the development of diabetic neuropathy. miR-199a-3p was also found to suppress tissue plasminogen activator (tPA) pathway via regulation of SERPINE2 expression which lies upstream of the tPA pathway.
mmu-miR-193a
Wu et al. (2019) reported downregulation of miR-193a to alleviate neuropathic pain in male Balb/c mice induced with diabetes by streptozotocin. miR-193a targeted high mobility group box protein 1 (HMGB1) by binding to the HMGB1 3′-UTR region. HMGB1 proteins are key proinflammatory mediators resulting in abnormal inflammation response. Upregulation of miR-193a showed a downregulation of inflammatory cytokines like IL-6, IL-1β, and TNF-α in the lumbar spinal dorsal horn of diabetic mice.
rno-miR-9
miR-9 was reported (Sun et al., 2020) to be highly upregulated in STZ-induced Sprague-Dawley rats. Insulin gene enhancer binding protein-1 (ISL1) was identified as a target of miR-9 which bound to 3′UTR of ISL1. ISL1 plays a key role in activation of insulin gene transcription of pancreatic beta-cells. Further, ISL1 modulated the sonic hedgehog (SHH) signaling pathway to improve diabetic peripheral neuropathy. miR-9 inhibited the expression of ISL1 as well as SHH signaling pathway resulting in development of diabetic peripheral neuropathy.
Since diabetes plays an important role in the etiopathogenesis of neuropathic pain, we summarize key microRNA changes in neuropathic pain in Table 8 including miRNAs of human, mouse and rat origin. Further, we summarize various signaling pathways modulated by miRNAs in neuropathic pain in Figure 7 for the benefit of the reader.
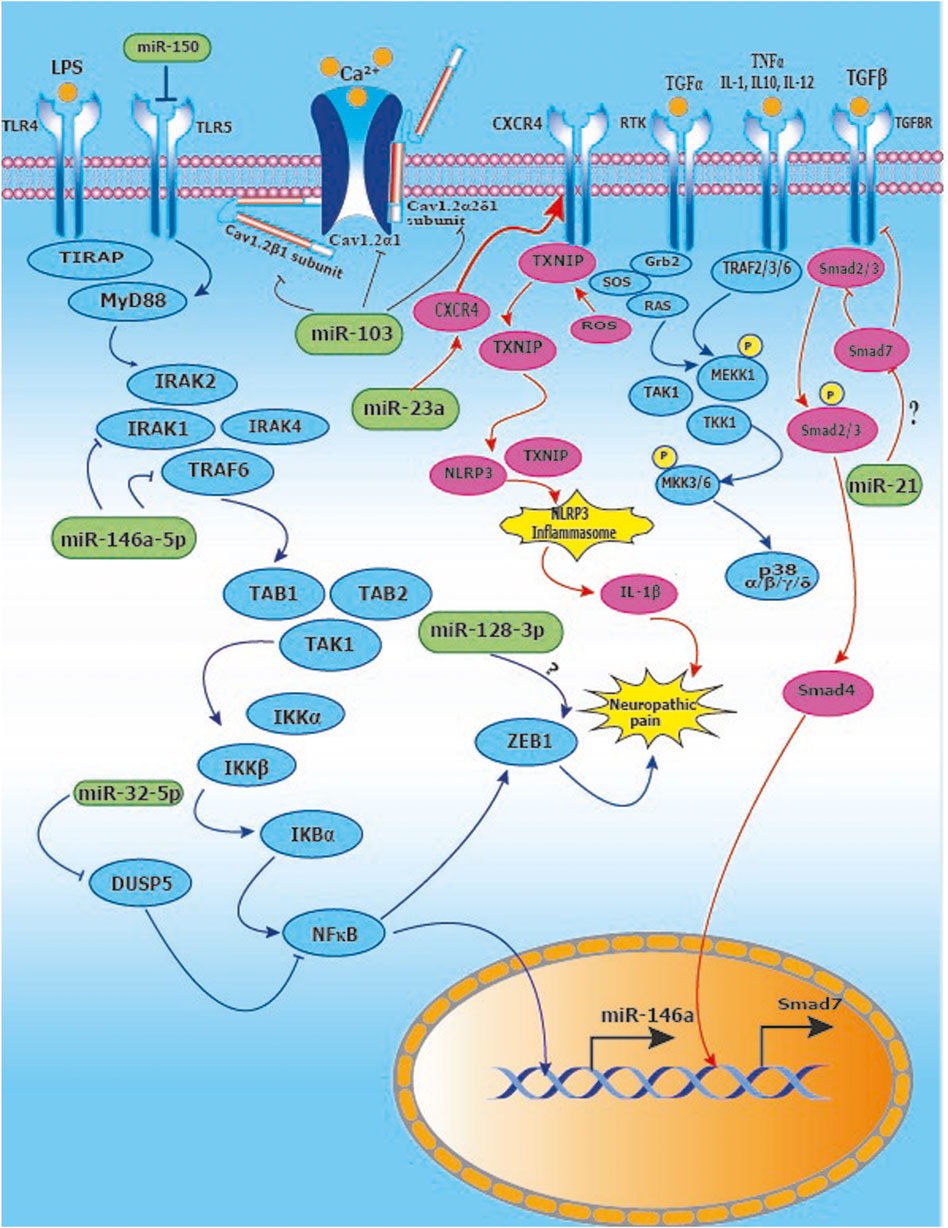
FIGURE 7. Signaling pathways modulated by miRNAs in neuropathic pain: (A) Activation of TLR4 and downstream IRAK/TRAF6 signaling leads to upregulation of NFĸB and ZEB1 leading to neuropathic pain. miR-146a-5p inhibits IRAK1 and TRAF6 which are upstream of NFĸB. Further, miR-32-5p inhibits DUSP5 resulting in suppression of NFkB signaling. miR-128-3p interacts with ZEB1, however, the nature of the interaction remains to be explored; (B) Inducing miR-103 suppresses the expression of subunits Cav1.2-α1, Cav1.2-α2δ1 and Cav1.2-β1 of Cav1.2-comprising L-type calcium channel thus relieving neuropathic pain; (C) Suppression of miR-23a results in upregulation of CXCR4 which mediates neuropathic pain via the TXNIP/NLRP3 inflammasome axis; (D) The p38 MAP Kinase pathway is implicated in neuropathic pain and miR-26a-5p downregulates MKK3/6 upstream of p38, whereas miR-21 inhibits Spry1 leading to RAS inhibition in the MAP Kinase signaling pathway.
miRNAs and Epigenetic Mechanisms of Neuropathic Pain
Epigenetic events such as covalent histone modifications and DNA methylation regulate gene expression (Liang et al., 2015). Enzymes involved in these processes are histone acetyltransferase (HAT) and histone deacetylases (HDACs) for histone modification; and DNA methyltransferases (DNMTs) and demethylation enzymes (translocation dioxygenases) for DNA methylation. A study reported that CpG islands present in promoter region of miR-129 were hypermethylated by complete Freund’s adjuvant (CFA). This process of methylation modulated chronic inflammatory pain by targeting Ca2+/calmodulin-dependent protein kinase γ (CaMKIIγ) (Liang et al., 2015) and (Pan et al., 2014). Given the epigenetic mechanisms involved in neuropathic pain, there is potential for therapeutic intervention by targeting miRNAs and/or their targets. The epigenetic alterations mediated by miRNAs result in either degradation of target mRNAs or translational repression. Epigenetic mutations are not reversed when they are persistent for a long period without any intervention, this is termed as “metabolic memory” of the target cell wherein the epigenetic alterations occurred (Yamunadevi et al., 2021). Further, Liu et al. (2020) reported an isoform of DNA methyltransferase (DNMT3a) that hypermethylates the promoter region of miR-214-3p. This led to the inhibition of expression of miR-214-3p in rats with spinal nerve ligation. Zebularine, a DNMT inhibitor, abrogated the suppression of miR-214-3p expression, resulting in the reduction of cytosine methylation in the promoter region; therefore, decrease in colony-stimulating factor-1 (CSF1) was observed as miR-214-3p negatively regulated expression of CSF1. As CSF1 decreased, neuropathic pain decreased subsequently. Thus, neuroinflammation and neuropathic pain were induced by increased production of CSF1 as miR-214-3p was epigenetically suppressed by DNMT3a (Liu et al., 2020). Furthermore, Tan et al. (2020) studied the role of miR-30a-3p in sciatic nerve CCI Sprague Dawley rats. rno-miR-30a-3p targeted E-cadherin transcriptional activator (EP300) which further upregulated brain-derived neurotrophic factor (BDNF); this resulted in increased neuropathic pain as EP300 and BDNF both were directly involved in neuropathic pain (Tan et al., 2020).
Kcna2, a voltage-dependent potassium channel mRNA, is inhibited by a conserved lncRNA, Kcna2 antisense RNA. This results in the decreased expression of Kcna2 channel. The decrease in voltage-dependent potassium channel resulted in alleviation of neuropathic pain and increase in excitability (Zhao et al., 2013). Further, methyl-CpG-binding domain protein (MBD1) is an epigenetic repressor that modulates gene transcription. Using MBD1-deficient (Mbd1−/−) mice with spinal nerve ligation, it was demonstrated that MBD1 recruited DNMT into the gene promoters of Kcna2 and Oprm1 gene and repressed their expression. Hence, regulation of DNMT-controlled expression of Kcna2 gene in dorsal root ganglion neurons led to neuropathic pain (Mo et al., 2018). Interestingly, Zhang et al. (2021a) reported the key role of voltage-gated potassium channels (Kv) in regulation of neuropathic pain induced by nerve injury. In dorsal root ganglion and spinal cord of naïve and CCI Sprague Dawley rats, neuron excitability and Kv currents were examined which showed that the downregulation of Kv1.2 induced hypersensitivity in naïve rats. As Kv1.2 was downregulated, the expression of miR-137 was increased which targeted Kcna2 and regulated it. Hence, by inhibiting miR-137, the Kv1.2 expression was upregulated restoring excitability and abnormal currents. As a result, when voltage-gated potassium channels (Kv1.2) was restored, it contributed to alleviation of neuropathic pain proving to be a novel therapeutic target (Zhang J. et al., 2021).
Conclusion and Future Perspectives
Complexity, progressive nature, and improper identification of neuropathic pain make it difficult to manage. Several clinical studies have been carried out in recent years in order to alleviate the poor quality of life associated with neuropathic pain using tricyclic antidepressants, opioid analgesics, physical and psychological therapies. However, these treatments are not sufficient for the management of neuropathic pain. Given the improper diagnosis of neuropathic pain, efforts have been made to identify miRNAs that can serve as biomarkers of neuropathic pain. Indeed, several miRNAs are modulated in the etiopathogenesis of neuropathic pain. It is, hence, important to study miRNA-miRNA and miRNA-gene target networks and evaluate the miRNA interactome in the preventative or therapeutic management of neuropathic pain. Moreover, it is clear that neuropathic pain is driven by neuroinflammation and nerve damage. Various miRNAs regulated in neuroinflammation and nerve damage may be especially useful as biomarkers for diagnosis and as therapeutic targets for the management of neuropathic pain. Taken together, experimental research aimed at deepening our knowledge of the miRNA interactome will be necessary in the near future to evaluate these exciting candidate biomarkers in the management of neuropathic pain.
Author Contributions
SN conceived of the manuscript. DM and SN edited the manuscript. YG wrote the manuscript and prepared the tables and figures under the supervision of SN. AP and NJ revised the manuscript with additional details and SA prepared the signaling figure under the supervision of SN. All authors read and approved the manuscript.
Funding
This work was funded by Synergia Life Sciences Pvt. Ltd., Mumbai, India.
Conflict of Interest
The authors are employees of Synergia Life Sciences Pvt. Ltd., Mumbai, India.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
Gla, gamma-carboxyglutamate; GAS6, growth arrest-specific 6; DN, diabetic neuropathy; PN, peripheral neuropathy; DPN, diabetic peripheral neuropathy; T2DM, type 2 diabetes mellitus; RANKL/RANK, receptor-activator for nuclear factor kappa B ligand; TLDA, TaqMan low density array; CCI, chronic constriction injury; DRG, dorsal root ganglion; PHN, post-herpetic neuralgia; AHZ, acute herpes zoster; hsa, Homo sapiens; mmu, Mus musculus; rno, Rattus norvegicus.
References
Akyuz, G., and Kenis, O. (2014). Physical Therapy Modalities and Rehabilitation Techniques in the Management of Neuropathic Pain. Am. J. Phys. Med. Rehabil. 93, 253–259. doi:10.1097/PHM.0000000000000037
Aldrich, B. T., Frakes, E. P., Kasuya, J., Hammond, D. L., and Kitamoto, T. (2009). Changes in Expression of Sensory Organ-specific microRNAs in Rat Dorsal Root Ganglia in Association with Mechanical Hypersensitivity Induced by Spinal Nerve Ligation. Neuroscience 164, 711–723. doi:10.1016/j.neuroscience.2009.08.033
Allette, Y. M., Kim, Y., Randolph, A. L., Smith, J. A., Ripsch, M. S., and White, F. A. (2017). Decoy Peptide Targeted to Toll-IL-1R Domain Inhibits LPS and TLR4-Active Metabolite Morphine-3 Glucuronide Sensitization of Sensory Neurons. Sci. Rep. 7, 3741. doi:10.1038/s41598-017-03447-9
Amin, K. N., Umapathy, D., Anandharaj, A., Ravichandran, J., Sasikumar, C. S., Chandra, S. K. R., et al. (2020). miR-23c Regulates Wound Healing by Targeting Stromal Cell-Derived Factor-1α (SDF-1α/CXCL12) Among Patients with Diabetic Foot Ulcer. Microvasc. Res. 127, 103924. doi:10.1016/j.mvr.2019.103924
Armero, P., Muriel, C., López, M., Santos, J., and González-Sarmiento, R. (2012). Análisis de polimorfismos del gen TRPV1 en pacientes españoles con dolor neuropático. Medicina Clínica 139, 1–4. doi:10.1016/j.medcli.2011.10.028
Armstrong, S. A., and Herr, M. J. (2021). Physiology, Nociception. StatPearls Publishing. Available at: https://www.ncbi.nlm.nih.gov/books/NBK551562/ (Accessed November 19, 2021).
Asahchop, E. L., Branton, W. G., Krishnan, A., Chen, P. A., Yang, D., Kong, L., et al. (2018). HIV-associated Sensory Polyneuropathy and Neuronal Injury Are Associated with miRNA-455-3p Induction. JCI Insight 3, e122450. doi:10.1172/jci.insight.122450
Badmaev, V., Mehta, D., Jonas, R., Rosenbush, S., and Hulse, S. (2011). Evolving story of Bone Health and the Nutritional Support. NutraCos 2011, 2–5.
Bao, Y., Wang, S., Xie, Y., Jin, K., Bai, Y., and Shan, S. (2018). MiR-28-5p Relieves Neuropathic Pain by Targeting Zeb1 in CCI Rat Models. J. Cel. Biochem. 119, 8555–8563. doi:10.1002/jcb.27096
Baron, R. (2009). “Neuropathic Pain: A Clinical Perspective,” in Sensory Nerves Handbook of Experimental Pharmacology. Editors B. J. Canning, and D. Spina (Berlin, Heidelberg: Springer Berlin Heidelberg), 3–30. doi:10.1007/978-3-540-79090-7_1
Bouhassira, D., and Attal, N. (2019). The Multiple Challenges of Neuropathic Pain. Neurosci. Lett. 702, 6–10. doi:10.1016/j.neulet.2018.11.054
Bouhassira, D., Letanoux, M., and Hartemann, A. (2013). Chronic Pain with Neuropathic Characteristics in Diabetic Patients: A French Cross-Sectional Study. PLoS One 8, e74195. doi:10.1371/journal.pone.0074195
Brandenburger, T., Johannsen, L., Prassek, V., Kuebart, A., Raile, J., Wohlfromm, S., et al. (2019). MiR-34a Is Differentially Expressed in Dorsal Root Ganglia in a Rat Model of Chronic Neuropathic Pain. Neurosci. Lett. 708, 134365. doi:10.1016/j.neulet.2019.134365
Cai, G., Zhu, Y., Zhao, Y., Chen, J., Guo, C., Wu, F., et al. (2020a). Network Analysis of miRNA and mRNA Changes in the Prelimbic Cortex of Rats with Chronic Neuropathic Pain: Pointing to Inflammation. Front. Genet. 11, 612. doi:10.3389/fgene.2020.00612
Cai, L., Liu, X., Guo, Q., huang, Q., Zhang, Q., and Cao, Z. (2020b). MiR-15a Attenuates Peripheral Nerve Injury-Induced Neuropathic Pain by Targeting AKT3 to Regulate Autophagy. Genes Genomics 42, 77–85. doi:10.1007/s13258-019-00881-z
Cai, W., Zhao, Q., Shao, J., Zhang, J., Li, L., Ren, X., et al. (2018). MicroRNA-182 Alleviates Neuropathic Pain by Regulating Nav1.7 Following Spared Nerve Injury in Rats. Sci. Rep. 8, 16750. doi:10.1038/s41598-018-34755-3
Cao, S., Yuan, J., Zhang, D., Wen, S., Wang, J., Li, Y., et al. (2019a). Transcriptome Changes in Dorsal Spinal Cord of Rats with Neuropathic Pain. J. Pain Res. 12, 3013–3023. doi:10.2147/JPR.S219084
Cao, S., Zhang, D., Yuan, J., Liu, C., Zhou, W., Zhang, L., et al. (2019b). MicroRNA and Circular RNA Expression in Affected Skin of Patients with Postherpetic Neuralgia. J. Pain Res. 12, 2905–2913. doi:10.2147/JPR.S221615
Cavalli, E., Mammana, S., Nicoletti, F., Bramanti, P., and Mazzon, E. (2019). The Neuropathic Pain: An Overview of the Current Treatment and Future Therapeutic Approaches. Int. J. Immunopathol Pharmacol. 33, 2058738419838383. doi:10.1177/2058738419838383
Chang, H. L., Wang, H. C., Chunag, Y. T., Chou, C. W., Lin, I. L., Lai, C. S., et al. (2017). miRNA Expression Change in Dorsal Root Ganglia after Peripheral Nerve Injury. J. Mol. Neurosci. 61, 169–177. doi:10.1007/s12031-016-0876-7
Chang, L. L., Wang, H. C., Tseng, K. Y., Su, M. P., Wang, J. Y., Chuang, Y. T., et al. (2020). Upregulation of miR-133a-3p in the Sciatic Nerve Contributes to Neuropathic Pain Development. Mol. Neurobiol. 57, 3931–3942. doi:10.1007/s12035-020-01999-y
Chatterjee, D., Bandyopadhyay, A., Sarma, N., Basu, S., Roychowdhury, T., Roy, S. S., et al. (2018). Role of microRNAs in Senescence and its Contribution to Peripheral Neuropathy in the Arsenic Exposed Population of West Bengal, India. Environ. Pollut. 233, 596–603. doi:10.1016/j.envpol.2017.09.063
Chen, M., Yang, Y., Zhang, W., Li, X., Wu, J., Zou, X., et al. (2020). Long Noncoding RNA SNHG5 Knockdown Alleviates Neuropathic Pain by Targeting the miR-154-5p/CXCL13 Axis. Neurochem. Res. 45, 1566–1575. doi:10.1007/s11064-020-03021-2
Chen, Z. L., Liu, J. Y., Wang, F., and Jing, X. (2019). Suppression of MALAT1 Ameliorates Chronic Constriction Injury-Induced Neuropathic Pain in Rats via Modulating miR-206 and ZEB2. J. Cel Physiol 234, 15647–15653. doi:10.1002/jcp.28213
Cheng, C., Kobayashi, M., Martinez, J. A., Ng, H., Moser, J. J., Wang, X., et al. (2015). Evidence for Epigenetic Regulation of Gene Expression and Function in Chronic Experimental Diabetic Neuropathy. J. Neuropathol. Exp. Neurol. 74, 804–817. doi:10.1097/NEN.0000000000000219
Chu, Y., Ge, W., and Wang, X. (2019). MicroRNA-448 Modulates the Progression of Neuropathic Pain by Targeting Sirtuin 1. Exp. Ther. Med. 18, 4665–4672. doi:10.3892/etm.2019.8165
Ciccacci, C., Latini, A., Greco, C., Politi, C., D'Amato, C., Lauro, D., et al. (2018). Association between a MIR499A Polymorphism and Diabetic Neuropathy in Type 2 Diabetes. J. Diabetes Complications 32, 11–17. doi:10.1016/j.jdiacomp.2017.10.011
Colloca, L., Ludman, T., Bouhassira, D., Baron, R., Dickenson, A. H., Yarnitsky, D., et al. (2017). Neuropathic Pain. Nat. Rev. Dis. Primers 3, 17002. doi:10.1038/nrdp.2017.2
Dai, D., Wang, J., Jiang, Y., Yuan, L., Lu, Y., Zhang, A., et al. (2019). Small RNA Sequencing Reveals microRNAs Related to Neuropathic Pain in Rats. Braz. J. Med. Biol. Res. 52, e8380. doi:10.1590/1414-431X20198380
Davies, M., Brophy, S., Williams, R., and Taylor, A. (2006). The Prevalence, Severity, and Impact of Painful Diabetic Peripheral Neuropathy in Type 2 Diabetes. Diabetes Care 29, 1518–1522. doi:10.2337/dc05-2228
Dayer, C. F., Luthi, F., Le Carré, J., Vuistiner, P., Terrier, P., Benaim, C., et al. (2019). Differences in the miRNA Signatures of Chronic Musculoskeletal Pain Patients from Neuropathic or Nociceptive Origins. PLoS ONE 14, e0219311. doi:10.1371/journal.pone.0219311
Ding, M., Shen, W., and Hu, Y. (2017). The Role of miR-539 in the Anterior Cingulate Cortex in Chronic Neuropathic Pain. Pain Med. 18, 2433–2442. doi:10.1093/pm/pnx004
Dound, Y., Mehta, D., Jadhav, S., Bhave, A., Devale, M., and Vaidya, A. B. (2018). A Novel Potential Role of Vitamin K2-7 in Relieving Peripheral Neuropathy. J. Pharmacol. Pharmacother. 9, 180. doi:10.4103/jpp.JPP_72_18
Fan, B., Chopp, M., Zhang, Z. G., and Liu, X. S. (2021). Treatment of Diabetic Peripheral Neuropathy with Engineered Mesenchymal Stromal Cell-Derived Exosomes Enriched with microRNA-146a Provide Amplified Therapeutic Efficacy. Exp. Neurol. 341, 113694. doi:10.1016/j.expneurol.2021.113694
Fang, B., Wei, L., Dong, K., Niu, X., Sui, X., and Zhang, H. (2019). miR-202 Modulates the Progression of Neuropathic Pain through Targeting RAP1A. J. Cel. Biochem. 120, 2973–2982. doi:10.1002/jcb.27025
Favereaux, A., Thoumine, O., Bouali-Benazzouz, R., Roques, V., Papon, M. A., Salam, S. A., et al. (2011). Bidirectional Integrative Regulation of Cav1.2 Calcium Channel by microRNA miR-103: Role in Pain. EMBO J. 30, 3830–3841. doi:10.1038/emboj.2011.249
Freynhagen, R., and Baron, R. (2009). The Evaluation of Neuropathic Components in Low Back Pain. Curr. Pain Headache Rep. 13, 185–190. doi:10.1007/s11916-009-0032-y
Frias, B., and Merighi, A. (2016). Capsaicin, Nociception and Pain. Molecules 21, 797. doi:10.3390/molecules21060797
Friedman, T. N., Yousuf, M. S., Catuneanu, A., Desai, M., Juźwik, C. A., Fournier, A. E., et al. (2019). Profiling the microRNA Signature of the Peripheral Sensory Ganglia in Experimental Autoimmune Encephalomyelitis (EAE). J. Neuroinflammation 16, 223. doi:10.1186/s12974-019-1600-7
Gandhi, M., and Nair, S. (2020). New Vistas in Malignant Mesothelioma: MicroRNA Architecture and NRF2/MAPK Signal Transduction. Life Sci. 257, 118123. doi:10.1016/j.lfs.2020.118123
Gao, L., Pu, X., Huang, Y., and Huang, J. (2019). MicroRNA-340-5p Relieved Chronic Constriction Injury-Induced Neuropathic Pain by Targeting Rap1A in Rat Model. Genes Genomics 41, 713–721. doi:10.1007/s13258-019-00802-0
Gaudet, A. D., Fonken, L. K., Watkins, L. R., Nelson, R. J., and Popovich, P. G. (2018). MicroRNAs: Roles in Regulating Neuroinflammation. Neuroscientist 24, 221–245. doi:10.1177/1073858417721150
Gauthier, A., Breuer, J., Carrington, D., Martin, M., and Rémy, V. (2009). Epidemiology and Cost of Herpes Zoster and post-herpetic Neuralgia in the United Kingdom. Epidemiol. Infect. 137, 38–47. doi:10.1017/S0950268808000678
Genda, Y., Arai, M., Ishikawa, M., Tanaka, S., Okabe, T., and Sakamoto, A. (2013). microRNA Changes in the Dorsal Horn of the Spinal Cord of Rats with Chronic Constriction Injury: A TaqMan® Low Density Array Study. Int. J. Mol. Med. 31, 129–137. doi:10.3892/ijmm.2012.1163
Gomez, K., Ran, D., Madura, C. L., Moutal, A., and Khanna, R. (2021). Non-SUMOylated CRMP2 Decreases NaV1.7 Currents via the Endocytic Proteins Numb, Nedd4-2 and Eps15. Mol. Brain 14, 20. doi:10.1186/s13041-020-00714-1
Gong, Q., Lu, Z., Huang, Q., Ruan, L., Chen, J., Liang, Y., et al. (2015). Altered microRNAs Expression Profiling in Mice with Diabetic Neuropathic Pain. Biochem. Biophys. Res. Commun. 456, 615–620. doi:10.1016/j.bbrc.2014.12.004
Gu, S., Xie, R., Liu, X., Shou, J., Gu, W., and Che, X. (2017). Long Coding RNA XIST Contributes to Neuronal Apoptosis through the Downregulation of AKT Phosphorylation and Is Negatively Regulated by miR-494 in Rat Spinal Cord Injury. Int. J. Mol. Sci. 18, 732. doi:10.3390/ijms18040732
Guo, G., Liu, Y., Ren, S., Kang, Y., Duscher, D., Machens, H. G., et al. (2018). Comprehensive Analysis of Differentially Expressed microRNAs and mRNAs in Dorsal Root Ganglia from Streptozotocin-Induced Diabetic Rats. PLoS ONE 13, e0202696. doi:10.1371/journal.pone.0202696
Harrisson, S. A., Ogollah, R., Dunn, K. M., Foster, N. E., and Konstantinou, K. (2020). Prevalence, Characteristics, and Clinical Course of Neuropathic Pain in Primary Care Patients Consulting with Low Back-Related Leg Pain. Clin. J. Pain 36, 813–824. doi:10.1097/AJP.0000000000000879
Hayakawa, K., Okazaki, R., Ishii, K., Ueno, T., Izawa, N., Tanaka, Y., et al. (2012). Phosphorylated Neurofilament Subunit NF-H as a Biomarker for Evaluating the Severity of Spinal Cord Injury Patients, a Pilot Study. Spinal Cord 50, 493–496. doi:10.1038/sc.2011.184
Heyn, J., Luchting, B., Hinske, L. C., Hübner, M., Azad, S. C., and Kreth, S. (2016). miR-124a and miR-155 Enhance Differentiation of Regulatory T Cells in Patients with Neuropathic Pain. J. Neuroinflammation 13, 248. doi:10.1186/s12974-016-0712-6
Hori, N., Narita, M., Yamashita, A., Horiuchi, H., Hamada, Y., Kondo, T., et al. (2016). Changes in the Expression of IL-6-Mediated MicroRNAs in the Dorsal Root Ganglion under Neuropathic Pain in Mice. Synapse 70, 317–324. doi:10.1002/syn.21902
Hori, Y., Goto, G., Arai-Iwasaki, M., Ishikawa, M., and Sakamoto, A. (2013). Differential Expression of Rat Hippocampal microRNAs in Two Rat Models of Chronic Pain. Int. J. Mol. Med. 32, 1287–1292. doi:10.3892/ijmm.2013.1504
Hu, J., Hu, X., and Kan, T. (2019). MiR-34c Participates in Diabetic Corneal Neuropathy via Regulation of Autophagy. Invest. Ophthalmol. Vis. Sci. 60, 16–25. doi:10.1167/iovs.18-24968
Hu, Y., Liu, Q., Zhang, M., Yan, Y., Yu, H., and Ge, L. (2019). MicroRNA-362-3p Attenuates Motor Deficit Following Spinal Cord Injury via Targeting Paired Box Gene 2. J. Integr. Neurosci. 18, 57–64. doi:10.31083/j.jin.2019.01.12
Huang, Y., Li, X., Tao, G., Zhu, T., and Lin, J. (2017). Comparing Serum microRNA Levels of Acute Herpes Zoster Patients with Those of Postherpetic Neuralgia Patients. Medicine (Baltimore) 96, e5997. doi:10.1097/MD.0000000000005997
Huang, Z. Z., Wei, J. Y., Ou-Yang, H. D., Li, D., Xu, T., Wu, S. L., et al. (2016). mir-500-Mediated GAD67 Downregulation Contributes to Neuropathic Pain. J. Neurosci. 36, 6321–6331. doi:10.1523/JNEUROSCI.0646-16.2016
Im, Y. B., Jee, M. K., Choi, J. I., Cho, H. T., Kwon, O. H., and Kang, S. K. (2012). Molecular Targeting of NOX4 for Neuropathic Pain after Traumatic Injury of the Spinal Cord. Cell Death Dis. 3, e426. doi:10.1038/cddis.2012.168
Imai, S., Saeki, M., Yanase, M., Horiuchi, H., Abe, M., Narita, M., et al. (2011). Change in MicroRNAs Associated with Neuronal Adaptive Responses in the Nucleus Accumbens under Neuropathic Pain. J. Neurosci. 31, 15294–15299. doi:10.1523/JNEUROSCI.0921-11.2011
Inoue, S., Taguchi, T., Yamashita, T., Nakamura, M., and Ushida, T. (2017). The Prevalence and Impact of Chronic Neuropathic Pain on Daily and Social Life: A Nationwide Study in a Japanese Population. Eur. J. Pain 21, 727–737. doi:10.1002/ejp.977
Ji, L. J., Shi, J., Lu, J. M., and Huang, Q. M. (2018). MiR-150 Alleviates Neuropathic Pain via Inhibiting Toll-like Receptor 5. J. Cel. Biochem. 119, 1017–1026. doi:10.1002/jcb.26269
Ji, L. J., Su, J., Xu, A. L., Pang, B., and Huang, Q. M. (2019). MiR‐134‐5p Attenuates Neuropathic Pain Progression through Targeting Twist1. J. Cel. Biochem. 120, 1694–1701. doi:10.1002/jcb.27486
Jia, L., Chopp, M., Wang, L., Lu, X., Szalad, A., and Zhang, Z. G. (2018). Exosomes Derived from High-Glucose-Stimulated Schwann Cells Promote Development of Diabetic Peripheral Neuropathy. FASEB J 32, fj201800597R–6922. doi:10.1096/fj.201800597R
Jin, H., Du, X. J., Zhao, Y., and Xia, D. L. (2018). XIST/miR-544 axis Induces Neuropathic Pain by Activating STAT3 in a Rat Model. J. Cel. Physiol. 233, 5847–5855. doi:10.1002/jcp.26376
Johansen, A., Romundstad, L., Nielsen, C. S., Schirmer, H., and Stubhaug, A. (2012). Persistent Postsurgical Pain in a General Population: Prevalence and Predictors in the Tromsø Study. Pain 153, 1390–1396. doi:10.1016/j.pain.2012.02.018
Johnson, R. W., and Rice, A. S. (2014). Clinical Practice. Postherpetic Neuralgia. N. Engl. J. Med. 371, 1526–1533. doi:10.1056/NEJMcp1403062
Kalfon, L., Azran, A., Farajun, Y., Golan-Hamu, O., Toben, A., Abramov, L., et al. (2019). Localized Provoked Vulvodynia: Association with Nerve Growth Factor and Transient Receptor Potential Vanilloid Type 1 Genes Polymorphisms. J. Low Genit Tract Dis. 23, 58–64. doi:10.1097/LGT.0000000000000445
Kerstman, E., Ahn, S., Battu, S., Tariq, S., and Grabois, M. (2013). “Neuropathic Pain,” in Handbook of Clinical Neurology (Amsterdam, Netherlands: Elsevier), 175–187. doi:10.1016/B978-0-444-52901-5.00015-0
Lee, J. S., Kwak, G., Kim, H. J., Park, H. T., Choi, B. O., and Hong, Y. B. (2019). miR-381 Attenuates Peripheral Neuropathic Phenotype Caused by Overexpression of PMP22. Exp. Neurobiol. 28, 279–288. doi:10.5607/en.2019.28.2.279
Leinders, M., Üçeyler, N., Pritchard, R. A., Sommer, C., and Sorkin, L. S. (2016). Increased miR-132-3p Expression Is Associated with Chronic Neuropathic Pain. Exp. Neurol. 283, 276–286. doi:10.1016/j.expneurol.2016.06.025
Leinders, M., Üçeyler, N., Thomann, A., and Sommer, C. (2017). Aberrant microRNA Expression in Patients with Painful Peripheral Neuropathies. J. Neurol. Sci. 380, 242–249. doi:10.1016/j.jns.2017.07.041
Li, H., Fan, L., Zhang, Y., Cao, Y., and Liu, X. (2020a). SNHG16 Aggravates Chronic Constriction Injury-Induced Neuropathic Pain in Rats via Binding with miR-124-3p and miR-141-3p to Upregulate JAG1. Brain Res. Bull. 165, 228–237. doi:10.1016/j.brainresbull.2020.09.025
Li, H., Huang, Y., Ma, C., Yu, X., Zhang, Z., and Shen, L. (2015). MiR-203 Involves in Neuropathic Pain Development and Represses Rap1a Expression in Nerve Growth Factor Differentiated Neuronal PC12 Cells. Clin. J. Pain 31, 36–43. doi:10.1097/AJP.0000000000000070
Li, H., Shen, L., Ma, C., and Huang, Y. (2013). Differential Expression of miRNAs in the Nervous System of a Rat Model of Bilateral Sciatic Nerve Chronic Constriction Injury. Int. J. Mol. Med. 32, 219–226. doi:10.3892/ijmm.2013.1381
Li, L., Luo, Y., Zhang, Y., Wei, M., Zhang, M., Liu, H., et al. (2020b). CircZNF609 Aggravates Neuropathic Pain via miR-22-3p/ENO1 axis in CCI Rat Models. Gene 763, 145069. doi:10.1016/j.gene.2020.145069
Li, L., Shao, J., Wang, J., Liu, Y., Zhang, Y., Zhang, M., et al. (2019a). MiR-30b-5p Attenuates Oxaliplatin-Induced Peripheral Neuropathic Pain through the Voltage-Gated Sodium Channel Nav1.6 in Rats. Neuropharmacology 153, 111–120. doi:10.1016/j.neuropharm.2019.04.024
Li, L., and Zhao, G. (2016). Downregulation of microRNA-218 Relieves Neuropathic Pain by Regulating Suppressor of Cytokine Signaling 3. Int. J. Mol. Med. 37, 851–858. doi:10.3892/ijmm.2016.2455
Li, Q. Y., Xu, H. Y., and Yang, H. J. (2017a). Effect of Proinflammatory Factors TNF-α,IL-1β, IL-6 on Neuropathic Pain. Zhongguo Zhong Yao Za Zhi 42, 3709–3712. doi:10.19540/j.cnki.cjcmm.20170907.004
Li, T., Wan, Y., Sun, L., Tao, S., Chen, P., Liu, C., et al. (2019b). Inhibition of MicroRNA-15a/16 Expression Alleviates Neuropathic Pain Development through Upregulation of G Protein-Coupled Receptor Kinase 2. Biomol. Ther. (Seoul) 27, 414–422. doi:10.4062/biomolther.2018.073
Li, X., Wang, S., Yang, X., and Chu, H. (2021a). miR-142-3p Targets AC9 to Regulate Sciatic Nerve Injury-induced N-europathic P-ain by R-egulating the cAMP/AMPK S-ignalling P-athway. Int. J. Mol. Med. 47, 561–572. doi:10.3892/ijmm.2020.4824
Li, Y., Gao, Y., Gong, Y., Guo, Y., Wang, L., Liu, Q., et al. (2020c). Treatment with Tang-Luo-ning Altered the microRNA Expression Profile in Rats with Diabetic Peripheral Neuropathy. Bioengineered 11, 841–851. doi:10.1080/21655979.2020.1797282
Li, Y., Zhang, H., Zhang, H., Kosturakis, A. K., Jawad, A. B., and Dougherty, P. M. (2014). Toll-like Receptor 4 Signaling Contributes to Paclitaxel-Induced Peripheral Neuropathy. J. Pain 15, 712–725. doi:10.1016/j.jpain.2014.04.001
Li, Y., Zhang, X., Fu, Z., and Zhou, Q. (2019c). MicroRNA-212-3p Attenuates Neuropathic Pain via Targeting Sodium Voltage-Gated Channel Alpha Subunit 3 (NaV 1.3). Curr. Neurovasc. Res. 16, 465–472. doi:10.2174/1567202616666191111104145
Li, Y. B., Wu, Q., Liu, J., Fan, Y. Z., Yu, K. F., and Cai, Y. (2017b). miR-199a-3p Is Involved in the Pathogenesis and Progression of Diabetic Neuropathy through Downregulation of SerpinE2. Mol. Med. Rep. 16, 2417–2424. doi:10.3892/mmr.2017.6874
Li, Y., Ma, W.-G., and Li, X.-C. (2021b). Identification of Blood miR-216a, miR-377 and Their Target Genes ANGPTL4, GAP-43 and Serum of PPARG as Biomarkers for Diabetic Peripheral Neuropathy of Type 2 Diabetes. Clin. Lab. 67. doi:10.7754/Clin.Lab.2020.191220
Li, Z., Li, A., Yan, L., Yang, T., Xu, W., and Fan, P. (2020d). Downregulation of Long Noncoding RNA DLEU1 Attenuates Hypersensitivity in Chronic Constriction Injury-Induced Neuropathic Pain in Rats by Targeting miR-133a-3p/SRPK1 axis. Mol. Med. 26, 104. doi:10.1186/s10020-020-00235-6
Liang, L., Lutz, B. M., Bekker, A., and Tao, Y. X. (2015). Epigenetic Regulation of Chronic Pain. Epigenomics 7, 235–245. doi:10.2217/epi.14.75
Liu, J. C., Xue, D. F., Wang, X. Q., Ai, D. B., and Qin, P. J. (2019). MiR-101 Relates to Chronic Peripheral Neuropathic Pain through Targeting KPNB1 and Regulating NF-Κb Signaling. Kaohsiung J. Med. Sci. 35, 139–145. doi:10.1002/kjm2.12025
Liu, L., Xu, D., Wang, T., Zhang, Y., Yang, X., Wang, X., et al. (2020). Epigenetic Reduction of miR-214-3p Upregulates Astrocytic colony-stimulating Factor-1 and Contributes to Neuropathic Pain Induced by Nerve Injury. Pain 161, 96–108. doi:10.1097/j.pain.0000000000001681
Liu, X. S., Fan, B., Szalad, A., Jia, L., Wang, L., Wang, X., et al. (2017a). MicroRNA-146a Mimics Reduce the Peripheral Neuropathy in Type 2 Diabetic Mice. Diabetes 66, 3111–3121. doi:10.2337/db16-1182
Liu, Y., Sun, H., and Sun, Y. (2021). LncRNA P21, Downregulating miR-181b, Aggravates Neuropathic Pain by Upregulating Tnfaip1 and Inhibit the AKT/CREB axis. Brain Res. Bull. 171, 150–161. doi:10.1016/j.brainresbull.2021.03.005
Liu, Y., Wang, L., Lao, J., and Zhao, X. (2017b). Changes in microRNA Expression in the Brachial Plexus Avulsion Model of Neuropathic Pain. Int. J. Mol. Med. 41, 1509. doi:10.3892/ijmm.2017.3333
Lu, S., Ma, S., Wang, Y., Huang, T., Zhu, Z., and Zhao, G. (2017). Mus Musculus-microRNA-449a Ameliorates Neuropathic Pain by Decreasing the Level of KCNMA1 and TRPA1, and Increasing the Level of TPTE. Mol. Med. Rep. 16, 353–360. doi:10.3892/mmr.2017.6559
Luo, Q., Feng, Y., Xie, Y., Shao, Y., Wu, M., Deng, X., et al. (2019). Nanoparticle-microRNA-146a-5p Polyplexes Ameliorate Diabetic Peripheral Neuropathy by Modulating Inflammation and Apoptosis. Nanomedicine 17, 188–197. doi:10.1016/j.nano.2019.01.007
McCarberg, B., D'Arcy, Y., Parsons, B., Sadosky, A., Thorpe, A., and Behar, R. (2017). Neuropathic Pain: a Narrative Review of Etiology, Assessment, Diagnosis, and Treatment for Primary Care Providers. Curr. Med. Res. Opin. 33, 1361–1369. doi:10.1080/03007995.2017.1321532
Mehta, D. (2017). The Potential Protective Role of Vitamin K in Diabetic Neuropathy. NutraCos 2017, 20–21.
Miao, J., Zhou, X., Ji, T., and Chen, G. (2020). NF-κB P65-dependent Transcriptional Regulation of Histone Deacetylase 2 Contributes to the Chronic Constriction Injury-Induced Neuropathic Pain via the microRNA-183/TXNIP/NLRP3 axis. J. Neuroinflammation 17, 225. doi:10.1186/s12974-020-01901-6
Mo, K., Wu, S., Gu, X., Xiong, M., Cai, W., Atianjoh, F. E., et al. (2018). MBD1 Contributes to the Genesis of Acute Pain and Neuropathic Pain by Epigenetic Silencing of Oprm1 and Kcna2 Genes in Primary Sensory Neurons. J. Neurosci. 38, 9883–9899. doi:10.1523/JNEUROSCI.0880-18.2018
Mo, Y., Liu, B., Qiu, S., Wang, X., Zhong, L., Han, X., et al. (2020). Down-regulation of microRNA-34c-5p Alleviates Neuropathic Pain via the SIRT1/STAT3 Signaling Pathway in Rat Models of Chronic Constriction Injury of Sciatic Nerve. J. Neurochem. 154, 301–315. doi:10.1111/jnc.14998
Moulin, D., Boulanger, A., Clark, A. J., Clarke, H., Dao, T., Finley, G. A., et al. (2014). Pharmacological Management of Chronic Neuropathic Pain: Revised Consensus Statement from the Canadian Pain Society. Pain Res. Manag. 19, 328–335. doi:10.1155/2014/754693
Murphy, D., Lester, D., Clay Smither, F., and Balakhanlou, E. (2020). Peripheral Neuropathic Pain. NeuroRehabilitation 47, 265–283. doi:10.3233/NRE-208002
Nair, S. (2016). Current Insights into the Molecular Systems Pharmacology of lncRNA-miRNA Regulatory Interactions and Implications in Cancer Translational Medicine. AIMS Mol. Sci. 3, 104–124. doi:10.3934/molsci.2016.2.104
Nair, S., and Kong, A.-N. T. (2015b). Architecture of Signature miRNA Regulatory Networks in Cancer Chemoprevention. Curr. Pharmacol. Rep. 1, 89–101. doi:10.1007/s40495-014-0014-6
Nair, S., Liew, C., Oo Khor, T., Cai, L., and Kong, A.-N. (2014). Differential Signaling Regulatory Networks Governing Hormone Refractory Prostate Cancers. J. Chin. Pharm. Sci. 23, 511–524. doi:10.5246/jcps.2014.08.067
Nair, S., and Tony Kong, A.-N. (2015a). Elucidation of Regulatory Interaction Networks Underlying Human Prostate Adenocarcinoma. J. Chin. Pharm. Sci. 24, 12. doi:10.5246/jcps.2015.01.002
Neelakandan, K., Babu, P., and Nair, S. (2012). Emerging Roles for Modulation of microRNA Signatures in Cancer Chemoprevention. Curr. Cancer Drug Targets 12, 716–740. doi:10.2174/156800912801784875
Pan, M. H., Maresz, K., Lee, P. S., Wu, J. C., Ho, C. T., Popko, J., et al. (2016). Inhibition of TNF-α, IL-1α, and IL-1β by Pretreatment of Human Monocyte-Derived Macrophages with Menaquinone-7 and Cell Activation with TLR Agonists In Vitro. J. Med. Food 19, 663–669. doi:10.1089/jmf.2016.0030
Pan, X., Shen, C., Huang, Y., Wang, L., Xia, Z., and Rossi, S. (2020). Loss of SNHG4 Attenuated Spinal Nerve Ligation-Triggered Neuropathic Pain through Sponging miR-423-5p. Mediators Inflamm. 2020, 2094948. doi:10.1155/2020/2094948
Pan, Z., Shan, Q., Gu, P., Wang, X. M., Tai, L. W., Sun, M., et al. (2018). miRNA-23a/CXCR4 Regulates Neuropathic Pain via Directly Targeting TXNIP/NLRP3 Inflammasome Axis. J. Neuroinflammation 15, 29. doi:10.1186/s12974-018-1073-0
Pan, Z., Zhu, L. J., Li, Y. Q., Hao, L. Y., Yin, C., Yang, J. X., et al. (2014). Epigenetic Modification of Spinal miR-219 Expression Regulates Chronic Inflammation Pain by Targeting CaMKIIγ. J. Neurosci. 34, 9476–9483. doi:10.1523/JNEUROSCI.5346-13.2014
Park, S. B., Goldstein, D., Krishnan, A. V., Lin, C. S., Friedlander, M. L., Cassidy, J., et al. (2013). Chemotherapy-induced Peripheral Neurotoxicity: a Critical Analysis. CA Cancer J. Clin. 63, 419–437. doi:10.3322/caac.21204
Peng, C., Zhang, C., Su, Z., and Lin, D. (2019a). DGCR5 Attenuates Neuropathic Pain through Sponging miR-330-3p and Regulating PDCD4 in CCI Rat Models. J. Cel. Physiol. 234, 7292–7300. doi:10.1002/jcp.27487
Peng, Q., Mechanic, J., Shoieb, A., Pardo, I. D., Schaevitz, L., Fenyk-Melody, J., et al. (2019b). Circulating microRNA and Automated Motion Analysis as Novel Methods of Assessing Chemotherapy-Induced Peripheral Neuropathy in Mice. PLoS ONE 14, e0210995. doi:10.1371/journal.pone.0210995
Ren, W., Xi, G., Li, X., Zhao, L., Yang, K., Fan, X., et al. (2021). Long Non-coding RNA HCG18 Promotes M1 Macrophage Polarization through Regulating the miR-146a/TRAF6 axis, Facilitating the Progression of Diabetic Peripheral Neuropathy. Mol. Cel. Biochem. 476, 471–482. doi:10.1007/s11010-020-03923-3
Saguil, A., Kane, S., and Mercado, M. (2017). Herpes Zoster and Postherpetic Neuralgia: Prevention and Management. Am. Fam. Physician 96, 656.
Sakai, A., Saitow, F., Maruyama, M., Miyake, N., Miyake, K., Shimada, T., et al. (2017). MicroRNA Cluster miR-17-92 Regulates Multiple Functionally Related Voltage-Gated Potassium Channels in Chronic Neuropathic Pain. Nat. Commun. 8, 16079. doi:10.1038/ncomms16079
Sakai, A., Saitow, F., Miyake, N., Miyake, K., Shimada, T., and Suzuki, H. (2013). miR-7a Alleviates the Maintenance of Neuropathic Pain through Regulation of Neuronal Excitability. Brain 136, 2738–2750. doi:10.1093/brain/awt191
Sakai, A., and Suzuki, H. (2013). Nerve Injury-Induced Upregulation of miR-21 in the Primary Sensory Neurons Contributes to Neuropathic Pain in Rats. Biochem. Biophys. Res. Commun. 435, 176–181. doi:10.1016/j.bbrc.2013.04.089
Santos-Bezerra, D. P., Cavaleiro, A. M., Santos, A. S., Suemoto, C. K., Pasqualucci, C. A., Jacob-Filho, W., et al. (2021). Alcohol Use Disorder Is Associated with Upregulation of MicroRNA-34a and MicroRNA-34c in Hippocampal Postmortem Tissue. Alcohol. Clin. Exp. Res. 45, 64–68. doi:10.1111/acer.14505
Shah, D., Gandhi, M., Kumar, A., Cruz-Martins, N., Sharma, R., and Nair, S. (2021). Current Insights into Epigenetics, Noncoding RNA Interactome and Clinical Pharmacokinetics of Dietary Polyphenols in Cancer Chemoprevention. Crit. Rev. Food Sci. Nutr. 2021, 1–37. doi:10.1080/10408398.2021.1968786
Shao, J., Cao, J., Wang, J., Ren, X., Su, S., Li, M., et al. (2016). MicroRNA-30b Regulates Expression of the Sodium Channel Nav1.7 in Nerve Injury-Induced Neuropathic Pain in the Rat. Mol. Pain 12, 174480691667152. doi:10.1177/1744806916671523
Shenoda, B. B., Tian, Y., Alexander, G. M., Aradillas-Lopez, E., Schwartzman, R. J., and Ajit, S. K. (2018). miR-34a-mediated Regulation of XIST in Female Cells under Inflammation. J. Pain Res. 11, 935–945. doi:10.2147/JPR.S159458
Shi, J., Jiang, K., and Li, Z. (2018). MiR-145 Ameliorates Neuropathic Pain via Inhibiting Inflammatory Responses and mTOR Signaling Pathway by Targeting Akt3 in a Rat Model. Neurosci. Res. 134, 10–17. doi:10.1016/j.neures.2017.11.006
Sisignano, M., Lötsch, J., Parnham, M. J., and Geisslinger, G. (2019). Potential Biomarkers for Persistent and Neuropathic Pain Therapy. Pharmacol. Ther. 199, 16–29. doi:10.1016/j.pharmthera.2019.02.004
Smith, B. H., and Torrance, N. (2012). Epidemiology of Neuropathic Pain and its Impact on Quality of Life. Curr. Pain Headache Rep. 16, 191–198. doi:10.1007/s11916-012-0256-0
St. John Smith, E. (2018). Advances in Understanding Nociception and Neuropathic Pain. J. Neurol. 265, 231–238. doi:10.1007/s00415-017-8641-6
Sun, L., Peng, C., Joosten, E., Cheung, C. W., Tan, F., Jiang, W., et al. (2021). Spinal Cord Stimulation and Treatment of Peripheral or Central Neuropathic Pain: Mechanisms and Clinical Application. Neural Plast. 2021, 5607898. doi:10.1155/2021/5607898
Sun, Q., Zeng, J., Liu, Y., Chen, J., Zeng, Q. C., Chen, Y. Q., et al. (2020). microRNA-9 and -29a Regulate the Progression of Diabetic Peripheral Neuropathy via ISL1-Mediated Sonic Hedgehog Signaling Pathway. Aging (Albany NY) 12, 11446–11465. doi:10.18632/aging.103230
Szok, D., Tajti, J., Nyári, A., Vécsei, L., and Trojano, L. (2019). Therapeutic Approaches for Peripheral and Central Neuropathic Pain. Behav. Neurol. 2019, 8685954. doi:10.1155/2019/8685954
Tan, M., Shen, L., and Hou, Y. (2020). Epigenetic Modification of BDNF Mediates Neuropathic Pain via miR-30a-3p/EP300 axis in CCI Rats. Biosci. Rep. 40, BSR20194442. doi:10.1042/BSR20194442
Tavares-Ferreira, D., Lawless, N., Bird, E. V., Atkins, S., Collier, D., Sher, E., et al. (2019). Correlation of miRNA Expression with Intensity of Neuropathic Pain in Man. Mol. Pain 15, 1744806919860323. doi:10.1177/1744806919860323
Tian, J., Song, T., Wang, W., Wang, H., and Zhang, Z. (2020). miR-129-5p Alleviates Neuropathic Pain through Regulating HMGB1 Expression in CCI Rat Models. J. Mol. Neurosci. 70, 84–93. doi:10.1007/s12031-019-01403-y
Tramullas, M., Francés, R., de la Fuente, R., Velategui, S., Carcelén, M., García, R., et al. (2018). MicroRNA-30c-5p Modulates Neuropathic Pain in Rodents. Sci. Transl. Med. 10, eaao6299. doi:10.1126/scitranslmed.aao6299
VanDenKerkhof, E. G., Mann, E. G., Torrance, N., Smith, B. H., Johnson, A., and Gilron, I. (2016). An Epidemiological Study of Neuropathic Pain Symptoms in Canadian Adults. Pain Res. Manag. 2016, 9815750. doi:10.1155/2016/9815750
von Schack, D., Agostino, M. J., Murray, B. S., Li, Y., Reddy, P. S., Chen, J., et al. (2011). Dynamic Changes in the microRNA Expression Profile Reveal Multiple Regulatory Mechanisms in the Spinal Nerve Ligation Model of Neuropathic Pain. PLoS One 6, e17670. doi:10.1371/journal.pone.0017670
Wang, C., Jiang, Q., Wang, M., and Li, D. (2015). MiR-19a Targets Suppressor of Cytokine Signaling 1 to Modulate the Progression of Neuropathic Pain. Int. J. Clin. Exp. Pathol. 8, 10901–10907.
Wang, C., Xu, X., Chen, J., Kang, Y., Guo, J., Duscher, D., et al. (2020a). The Construction and Analysis of lncRNA-miRNA-mRNA Competing Endogenous RNA Network of Schwann Cells in Diabetic Peripheral Neuropathy. Front. Bioeng. Biotechnol. 8, 490. doi:10.3389/fbioe.2020.00490
Wang, L., Chopp, M., Szalad, A., Lu, X., Zhang, Y., Wang, X., et al. (2020b). Exosomes Derived from Schwann Cells Ameliorate Peripheral Neuropathy in Type 2 Diabetic Mice. Diabetes 69, 749–759. doi:10.2337/db19-0432
Wang, W., and Li, R. (2021). MiR-216a-5p Alleviates Chronic Constriction Injury-Induced Neuropathic Pain in Rats by Targeting KDM3A and Inactivating Wnt/β-Catenin Signaling Pathway. Neurosci. Res. 170, 255–264. doi:10.1016/j.neures.2020.08.001
Wang, X., Wang, H., Zhang, T., He, M., Liang, H., Wang, H., et al. (2019). Inhibition of MicroRNA-195 Alleviates Neuropathic Pain by Targeting Patched1 and Inhibiting SHH Signaling Pathway Activation. Neurochem. Res. 44, 1690–1702. doi:10.1007/s11064-019-02797-2
Wang, Z., Liu, F., Wei, M., Qiu, Y., Ma, C., Shen, L., et al. (2018). Chronic Constriction Injury-Induced microRNA-146a-5p Alleviates Neuropathic Pain through Suppression of IRAK1/TRAF6 Signaling Pathway. J. Neuroinflammation 15, 179. doi:10.1186/s12974-018-1215-4
Wei, M., Li, L., Zhang, Y., Zhang, M., and Su, Z. (2020). Downregulated Circular RNA zRANB1 Mediates Wnt5a/β-Catenin Signaling to Promote Neuropathic Pain via miR-24-3p/LPAR3 axis in CCI Rat Models. Gene 761, 145038. doi:10.1016/j.gene.2020.145038
Wei, Z., Fei, Y., Su, W., and Chen, G. (2019). Emerging Role of Schwann Cells in Neuropathic Pain: Receptors, Glial Mediators and Myelination. Front. Cel. Neurosci. 13, 116. doi:10.3389/fncel.2019.00116
Wilkerson, J. L., Jiang, J., Felix, J. S., Bray, J. K., da Silva, L., Gharaibeh, R. Z., et al. (2020). Alterations in Mouse Spinal Cord and Sciatic Nerve microRNAs after the Chronic Constriction Injury (CCI) Model of Neuropathic Pain. Neurosci. Lett. 731, 135029. doi:10.1016/j.neulet.2020.135029
Williams, A. C., Eccleston, C., and Morley, S. (20122012). Psychological Therapies for the Management of Chronic Pain (Excluding Headache) in Adults. Cochrane Database Syst. Rev. 11, CD007407. doi:10.1002/14651858.CD007407.pub3
Woolf, C. J. (2011). Central Sensitization: Implications for the Diagnosis and Treatment of Pain. Pain 152, S2–S15. doi:10.1016/j.pain.2010.09.030
Wu, B., Guo, Y., Chen, Q., Xiong, Q., and Min, S. (2019). MicroRNA-193a Downregulates HMGB1 to Alleviate Diabetic Neuropathic Pain in a Mouse Model. Neuroimmunomodulation 26, 250–257. doi:10.1159/000503325
Wu, J., Wang, C., and Ding, H. (2020a). LncRNA MALAT1 Promotes Neuropathic Pain Progression through the miR-154-5p/AQP9 axis in CCI R-at M-odels. Mol. Med. Rep. 21, 291–303. doi:10.3892/mmr.2019.10829
Wu, Y., Gu, Y., and Shi, B. (2020b). miR-590-3p Alleviates Diabetic Peripheral Neuropathic Pain by Targeting RAP1A and Suppressing Infiltration by the T Cells. Acta Biochim. Pol. 67, 587–593. doi:10.18388/abp.2020_5451
Wu, Y., Xu, D., Zhu, X., Yang, G., and Ren, M. (2017). MiR-106a Associated with Diabetic Peripheral Neuropathy through the Regulation of 12/15-LOX-Meidiated Oxidative/Nitrative Stress. Curr. Neurovasc. Res. 14, 117–124. doi:10.2174/1567202614666170404115912
Xie, T., Zhang, J., Kang, Z., Liu, F., and Lin, Z. (2020). miR-101 Down-Regulates mTOR Expression and Attenuates Neuropathic Pain in Chronic Constriction Injury Rat Models. Neurosci. Res. 158, 30–36. doi:10.1016/j.neures.2019.09.002
Xu, H., Yue, C., and Chen, L. (2019a). Post-transcriptional Regulation of Soluble Guanylate Cyclase that Governs Neuropathic Pain in Alzheimer's Disease. J. Alzheimers Dis. 71, 1331–1338. doi:10.3233/JAD-190743
Xu, J., E, X., Liu, H., Li, F., Cao, Y., Tian, J., et al. (2015). Tumor Necrosis Factor-Alpha Is a Potential Diagnostic Biomarker for Chronic Neuropathic Pain after Spinal Cord Injury. Neurosci. Lett. 595, 30–34. doi:10.1016/j.neulet.2015.04.004
Xu, L., Wang, Q., Jiang, W., Yu, S., and Zhang, S. (2019b). MiR-34c Ameliorates Neuropathic Pain by Targeting NLRP3 in a Mouse Model of Chronic Constriction Injury. Neuroscience 399, 125–134. doi:10.1016/j.neuroscience.2018.12.030
Xu, Y., Zhang, X., Pu, S., Wu, J., Lv, Y., and Du, D. (2014). Circulating microRNA Expression Profile: a Novel Potential Predictor for Chronic Nervous Lesions. Acta Biochim. Biophys. Sin (Shanghai) 46, 942–949. doi:10.1093/abbs/gmu090
Yamunadevi, A., Pratibha, R., Rajmohan, M., Mahendraperumal, S., and Ganapathy, N. (2021). Basics of Epigenetics and Role of Epigenetics in Diabetic Complications. J. Pharm. Bioallied Sci. 13, S336–S343. doi:10.4103/jpbs.JPBS_771_20
Yan, T., Zhang, F., Sun, C., Sun, J., Wang, Y., Xu, X., et al. (2018a). miR-32-5p-mediated Dusp5 Downregulation Contributes to Neuropathic Pain. Biochem. Biophys. Res. Commun. 495, 506–511. doi:10.1016/j.bbrc.2017.11.013
Yan, X. T., Ji, L. J., Wang, Z., Wu, X., Wang, Q., Sun, S., et al. (2017). MicroRNA-93 Alleviates Neuropathic Pain through Targeting Signal Transducer and Activator of Transcription 3. Int. Immunopharmacol. 46, 156–162. doi:10.1016/j.intimp.2017.01.027
Yan, X. T., Zhao, Y., Cheng, X. L., He, X. H., Wang, Y., Zheng, W. Z., et al. (2018b). Inhibition of miR-200b/miR-429 Contributes to Neuropathic Pain Development through Targeting Zinc finger E Box Binding Protein-1. J. Cel. Physiol. 233, 4815–4824. doi:10.1002/jcp.26284
Yang, D., Yang, Q., Wei, X., Liu, Y., Ma, D., Li, J., et al. (2017a). The Role of miR-190a-5p Contributes to Diabetic Neuropathic Pain via Targeting SLC17A6. J. Pain Res. 10, 2395–2403. doi:10.2147/JPR.S133755
Yang, F. R., Chen, J., Yi, H., Peng, L. Y., Hu, X. L., and Guo, Q. L. (2019). MicroRNA-7a Ameliorates Neuropathic Pain in a Rat Model of Spinal Nerve Ligation via the Neurofilament Light Polypeptide-dependent Signal Transducer and Activator of Transcription Signaling Pathway. Mol. Pain 15, 1744806919842464. doi:10.1177/1744806919842464
Yang, Z., Xu, J., Zhu, R., and Liu, L. (2017b). Down-Regulation of miRNA-128 Contributes to Neuropathic Pain Following Spinal Cord Injury via Activation of P38. Med. Sci. Monit. 23, 405–411. doi:10.12659/MSM.898788
Yao, L., Guo, Y., Wang, L., Li, G., Qian, X., Zhang, J., et al. (2021). Knockdown of miR-130a-3p Alleviates Spinal Cord Injury Induced Neuropathic Pain by Activating IGF-1/IGF-1R Pathway. J. Neuroimmunol. 351, 577458. doi:10.1016/j.jneuroim.2020.577458
Ye, G., Zhang, Y., Zhao, J., Chen, Y., Kong, L., Sheng, C., et al. (2020). miR-384-5p Ameliorates Neuropathic Pain by Targeting SCN3A in a Rat Model of Chronic Constriction Injury. Neurol. Res. 42, 299–307. doi:10.1080/01616412.2020.1723313
Ye, L., Morse, L. R., Falci, S. P., Olson, J. K., Shrivastava, M., Nguyen, N., et al. (2021). hsa-MiR-19a-3p and Hsa-MiR-19b-3p Are Associated with Spinal Cord Injury-Induced Neuropathic Pain: Findings from a Genome-wide MicroRNA Expression Profiling Screen. Neurotrauma Rep. 2, 424–439. doi:10.1089/neur.2021.0011
You, H., Zhang, L., Chen, Z., Liu, W., Wang, H., and He, H. (2019). MiR-20b-5p Relieves Neuropathic Pain by Targeting Akt3 in a Chronic Constriction Injury Rat Model. Synapse 73, e22125. doi:10.1002/syn.22125
Zajączkowska, R., Kocot-Kępska, M., Leppert, W., and Wordliczek, J. (2019). Bone Pain in Cancer Patients: Mechanisms and Current Treatment. Int. J. Mol. Sci. 20, 6047. doi:10.3390/ijms20236047
Zhan, L. Y., Lei, S. Q., Zhang, B. H., Li, W. L., Wang, H. X., Zhao, B., et al. (2018). Overexpression of miR-381 Relieves Neuropathic Pain Development via Targeting HMGB1 and CXCR4. Biomed. Pharmacother. 107, 818–823. doi:10.1016/j.biopha.2018.08.053
Zhang, D., Mou, J. Y., Wang, F., Liu, J., and Hu, X. (2019a). CRNDE Enhances Neuropathic Pain via Modulating miR-136/IL6R axis in CCI Rat Models. J. Cel. Physiol. 234, 22234–22241. doi:10.1002/jcp.28790
Zhang, H., and Chen, H. (2021). TRPA1 Involved in miR-141-5p-Alleviated Neuropathic Pain Induced by Oxaliplatin. Neuroreport 32, 284–290. doi:10.1097/WNR.0000000000001589
Zhang, J., Rong, L., Shao, J., Zhang, Y., Liu, Y., Zhao, S., et al. (2021a). Epigenetic Restoration of Voltage-Gated Potassium Channel Kv1.2 Alleviates Nerve Injury-Induced Neuropathic Pain. J. Neurochem. 156, 367–378. doi:10.1111/jnc.15117
Zhang, J., Zhang, H., and Zi, T. (2015). Overexpression of microRNA-141 Relieves Chronic Constriction Injury-Induced Neuropathic Pain via Targeting High-Mobility Group Box 1. Int. J. Mol. Med. 36, 1433–1439. doi:10.3892/ijmm.2015.2342
Zhang, P., Sun, H., and Ji, Z. (2021b). Downregulating lncRNA PVT1 Relieves Astrocyte Overactivation Induced Neuropathic Pain through Targeting miR-186-5p/CXCL13/CXCR5 Axis. Neurochem. Res. 46, 1457–1469. doi:10.1007/s11064-021-03287-0
Zhang, X., Chen, Q., Shen, J., Wang, L., Cai, Y., and Zhu, K. R. (2020a). miR-194 Relieve Neuropathic Pain and Prevent Neuroinflammation via Targeting FOXA1. J. Cel. Biochem. 121, 3278–3285. doi:10.1002/jcb.29598
Zhang, X., Guo, H., Xie, A., Liao, O., Ju, F., and Zhou, Y. (2020b). MicroRNA-144 Relieves Chronic Constriction Injury-Induced Neuropathic Pain via Targeting RASA1. Biotechnol. Appl. Biochem. 67, 294–302. doi:10.1002/bab.1854
Zhang, X., Zhang, Y., Cai, W., Liu, Y., Liu, H., Zhang, Z., et al. (2020c). MicroRNA-128-3p Alleviates Neuropathic Pain through Targeting ZEB1. Neurosci. Lett. 729, 134946. doi:10.1016/j.neulet.2020.134946
Zhang, Y., Liu, H. L., An, L. J., Li, L., Wei, M., Ge, D. J., et al. (2019b). miR-124-3p Attenuates Neuropathic Pain Induced by Chronic Sciatic Nerve Injury in Rats via Targeting EZH2. J. Cel. Biochem. 120, 5747–5755. doi:10.1002/jcb.27861
Zhang, Y., Song, C., Liu, J., Bi, Y., and Li, H. (2018a). Inhibition of miR-25 Aggravates Diabetic Peripheral Neuropathy. NeuroReport 29, 945–953. doi:10.1097/WNR.0000000000001058
Zhang, Y., Su, Z., Liu, H. L., Li, L., Wei, M., Ge, D. J., et al. (2018b). Effects of miR-26a-5p on Neuropathic Pain Development by Targeting MAPK6 in in CCI Rat Models. Biomed. Pharmacother. 107, 644–649. doi:10.1016/j.biopha.2018.08.005
Zhang, Y., Mou, J., Cao, L., Zhen, S., Huang, H., and Bao, H. (2017). MicroRNA-142-3p Relieves Neuropathic Pain by Targeting High Mobility Group Box 1. Int. J. Mol. Med. 41, 501. doi:10.3892/ijmm.2017.3222
Zhang, Z. J., Guo, J. S., Li, S. S., Wu, X. B., Cao, D. L., Jiang, B. C., et al. (2018c). TLR8 and its Endogenous Ligand miR-21 Contribute to Neuropathic Pain in Murine DRG. J. Exp. Med. 215, 3019–3037. doi:10.1084/jem.20180800
Zhao, X., Tang, Z., Zhang, H., Atianjoh, F. E., Zhao, J. Y., Liang, L., et al. (2013). A Long Noncoding RNA Contributes to Neuropathic Pain by Silencing Kcna2 in Primary Afferent Neurons. Nat. Neurosci. 16, 1024–1031. doi:10.1038/nn.3438
Zhao, Y., Li, S., Xia, N., Shi, Y., and Zhao, C. M. (2018). Effects of XIST/miR-137 axis on Neuropathic Pain by Targeting TNFAIP1 in a Rat Model. J. Cel. Physiol. 233, 4307–4316. doi:10.1002/jcp.26254
Zhong, L., Fu, K., Xiao, W., Wang, F., and Shen, L. L. (2019). Overexpression of miR‐98 Attenuates Neuropathic Pain Development via Targeting STAT3 in CCI Rat Models. J. Cel. Biochem. 120, 7989–7997. doi:10.1002/jcb.28076
Zhou, J., Xiong, Q., Chen, H., Yang, C., and Fan, Y. (2017). Identification of the Spinal Expression Profile of Non-coding RNAs Involved in Neuropathic Pain Following Spared Nerve Injury by Sequence Analysis. Front. Mol. Neurosci. 10, 91. doi:10.3389/fnmol.2017.00091
Zhou, J., Zhuang, T., Ma, P., Shan, L., Sun, X. D., Gong, S., et al. (2020). MicroRNA-547-5p-mediated Interleukin-33/suppressor of Tumorigenicity 2 Signaling Underlies the Genesis and Maintenance of Neuropathic Pain and Is Targeted by the Therapy with Bone Marrow Stromal Cells. Mol. Pain 16, 1744806920931737. doi:10.1177/1744806920931737
Zhu, B., Gao, J., Ouyang, Y., Hu, Z., and Chen, X. (2019a). Overexpression of miR138 Ameliorates Spared Sciatic Nerve Injury-Induced Neuropathic Pain through the Anti-inflammatory Response in Mice. J. Pain Res. 12, 3135–3145. doi:10.2147/JPR.S219462
Zhu, D., Fan, T., Huo, X., Cui, J., Cheung, C. W., and Xia, Z. (2019b). Progressive Increase of Inflammatory CXCR4 and TNF-Alpha in the Dorsal Root Ganglia and Spinal Cord Maintains Peripheral and Central Sensitization to Diabetic Neuropathic Pain in Rats. Mediators Inflamm. 2019, 4856156. doi:10.1155/2019/4856156
Keywords: microRNA, neuropathic pain, biomarker, neuropathy, network, target, noncoding RNA
Citation: Gada Y, Pandey A, Jadhav N, Ajgaonkar S, Mehta D and Nair S (2022) New Vistas in microRNA Regulatory Interactome in Neuropathic Pain. Front. Pharmacol. 12:778014. doi: 10.3389/fphar.2021.778014
Received: 16 September 2021; Accepted: 10 December 2021;
Published: 25 February 2022.
Edited by:
John Timothy Skamarauskas, University of Hertfordshire, United KingdomReviewed by:
Yibo Ying, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, ChinaSachchida Nand Rai, University of Allahabad, India
Vinod Tiwari, Indian Institute of Technology (BHU), India
Copyright © 2022 Gada, Pandey, Jadhav, Ajgaonkar, Mehta and Nair. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sujit Nair, c3VqaXQxMDhAZ21haWwuY29t
 Yash Gada
Yash Gada Amitkumar Pandey
Amitkumar Pandey Nikita Jadhav
Nikita Jadhav Saiprasad Ajgaonkar
Saiprasad Ajgaonkar Sujit Nair
Sujit Nair