- 1Hunan Normal University School of Medicine, Changsha, China
- 2Key Laboratory of Molecular Radiation Oncology Hunan Province, Changsha, China
- 3Xiangya Cancer Center, Xiangya Hospital, Central South University, Changsha, China
- 4Department of Radiology, Xiangya Hospital, Central South University, Changsha, China
Acetylation is considered as one of the most common types of epigenetic modifications, and aberrant histone acetylation modifications are associated with the pathological process of cancer through the regulation of oncogenes and tumor suppressors. Recent studies have shown that immune system function and tumor immunity can also be affected by acetylation modifications. A comprehensive understanding of the role of acetylation function in cancer is essential, which may help to develop new therapies to improve the prognosis of cancer patients. In this review, we mainly discussed the functions of acetylase and deacetylase in tumor, immune system and tumor immunity, and listed the information of drugs targeting these enzymes in tumor immunotherapy.
Introduction
The main driving force for tumor initiation and progression are not only the alterations in cancer cells but also the influences of immune system and tumor immune microenvironments (Bindea et al., 2013; Jones et al., 2019). As an important hallmark of cancer, tumor cells evade immune surveillance by suppressing the immune system and having low immunogenicity (Peng et al., 2019; Gao et al., 2020). For example, aberrant expression of immune checkpoints (ICs) components, such as the programmed cell death protein 1 (PD-1), cytotoxic T lymphocyte-associated protein 4 (CTLA-4), T-cell immunoglobulin and mucin domain-containing lymphocyte activation gene 3 (LAG-3), and LAG-3 with Ig and ITIM structural domains T-cell immune receptors (TIGIT), creates an immune destructive environment that promotes tumor cells escape immune destructions (Saleh et al., 2020). Cancer immunotherapies such as cancer vaccines, adoptive T-cell therapy (ACT), and immune checkpoint blockade (ICB), which kills cancer cells through employing the body’s own immune system, have achieved encouraging progress within the last decade (Wang S. et al., 2019). Despite these breakthroughs, only 10–30% of patients respond to and benefit from them, and the underlying reasons of these low benefits are due to the development of primary and acquired drug resistance, low response frequency of some cancers, and the heterogeneity of tumors (Sharma et al., 2017). Therefore, the urgent question is how to enhance patient responsivity and benefit from immunotherapy by targeting tumor cells or modulating tumor immune microenvironment.
Recently, there is growing evidence showing that epigenetic modification is essential to regulation of tumor immunity and immunotherapy, which provides a possible target for improving the outcome of immunotherapy. Epigenetic gene regulation, which alters gene expression and function without involving alterations in DNA sequences, is an important regulatory process in cellular biology. Several types of epigenetic modification were identified such as DNA methylation, histone modification, miRNA regulation, genomic imprinting, and chromosome remodeling, among which acetylation is considered one of the most common types of epigenetic modification (Lawrence et al., 2016). Histone acetylation is mediated by histone acetyltransferase (HAT), while deacetylation is mediated by histone deacetylase (HDAC). Acetylation of histones alters the secondary structure of the histone tail, leading to relaxation of the chromatin structure by increasing the distance between DNA and histones, opening up tracts of DNA and allowing for increased binding of transcription factor complexes to gene promoter sequences, thereby upregulating transcription (Kouzarides, 2007; Lawrence et al., 2016). In contrast, histone deacetylation usually promotes chromatin condensation and down-regulates the transcriptional level of related genes, and is usually accompanied by an increase in histone methylation of the same residue.
Besides, there is growing evidence that abnormal histone acetylation modification is related to the pathological process of cancer through regulating the expression of oncogenes and tumor suppressors (Audia and Campbell, 2016). For instance, previous studies have shown that changes in histone acetylation level, especially the loss of acetylated Lys16 of histone H4, is related to the development of many cancers and is a common feature in human tumor cells (Fraga et al., 2005). Due to the critical role of acetylation in tumorigenesis, several drugs targeting HDAC have been developed to treat cancer. Furthermore, recent studies have shown acetylation modification is essential for immune system function and tumor immunity.
Therefore, in view of the effects of acetylation modification on tumor cells and immune system, as well as the clinical use of the HATs and HDACs inhibitors, it is worth exploring whether HATs and HDACs could influence immunotherapy efficacy by altering the tumor immune microenvironment through acetylation regulation on tumor cells or immune cells, and whether targeting these enzymes may improve the efficacy of immunotherapy. In this review, we focus on how HATs and HDACs modulate tumor immunity and discuss the potential application of drugs targeting these enzymes to improve the outcome of immunotherapy.
HATs in Tumor Immunity Regulation
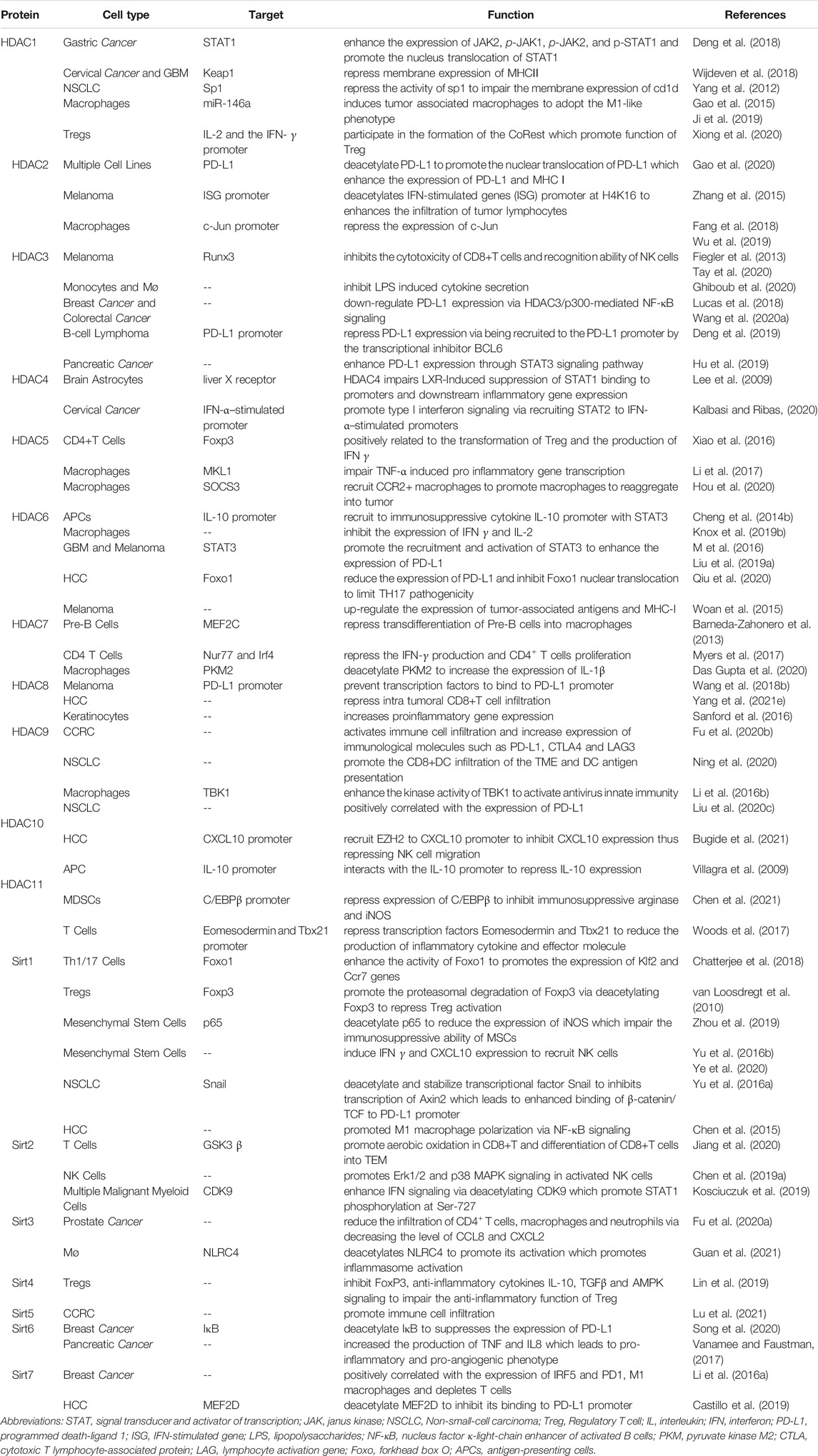
There are three major HAT subfamilies in human: the GCN5-related N-acetyltransferase (GNAT) subfamily including PCAF and GCN5; the MYST subfamily including TIP60, MOZ, MORF, MOF and HBO1; the p300 subfamily including p300, CBP. Besides these enzymes, HATs also include TAT1, ESCO1, ESCO2 and HAT1 (Narita et al., 2019). In addition to acetylated histones, HATs can also directly acetylate a range of transcription factors such as C/EBPα, FOXO1, and p53, resulting in the regulation of transcription factor activity and thereby regulation of cancer progression (Sheikh and Akhtar, 2019). At present, only a portion of HATs have been shown to be involved in tumor immunity regulation (Table 1).
P300/CBP-Associated Factor
P300/CBP-associated factor (PCAF), also named lysine acetyltransferase 2B (KAT2B) (Liu T. et al., 2019), is a HAT that mainly acetylates H3 histones, as well as a number of non-histone proteins that coordinate carcinogenic and tumor suppressive processes (Wang LT. et al., 2020). Previous studies have shown that the expression of PCAF is reduced as a tumor suppressor in esophageal, breast, ovarian, colorectal and pancreatic cancers, and that loss of PCAF expression is associated with poor prognosis in gastric cancer and may serve as a potential biomarker for invasive and aggressive tumors (Brasacchio et al., 2018). For example, it acts as a suppressor of HCC progression by promoting apoptosis through acetylation of glioma-associated oncogene homolog-1 (Gli1), histone H4 and the phosphatase and tensin homolog deleted on chromosome 10 (PTEN). On the contrary, PCAF was reported to be highly expressed in HCCs and to promote tumor progression via acetylation of phosphoglycerate kinase 1 (PGK1), pyruvate kinase M2 (PKM2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which subsequentially induces the Warburg effect and activates Akt signaling (Zheng et al., 2013; Wang T. et al., 2018).
With its modulatory effects on tumors, PCAF can also regulate immune system function. In a study of Foxp3+ Treg cells, PCAF was found to contribute to the inhibition of Treg cells apoptosis in response to TCR stimulation and to increase inducible Tregs (iTregs) production via IL-2 and TGF-β. In addition, PCAF acetylates Foxp3 to impair Treg cells function (Liu Y. et al., 2019). Another study has shown that overexpression of PCAF significantly suppressed the expression of pro-inflammatory genes TNF-α, IL-6 and CXCL10, suggesting that PCAF is a potential negative regulator of the inflammatory response of M1 macrophages (Wang et al., 2021).
There is limited evidence reporting the role of PCAF in tumor immunity regulation. A study reported that in lung adenocarcinoma tumor growth was compromised in PCAF−/− mice, with reduced infiltration of CD4+Foxp3+ Treg cells but increased infiltration of host CD8 T cells, indicating that targeting PCAF reduces tumor volume and improves anti-tumor immunity (Liu Y. et al., 2019). More research on the role of PACF in tumor immunity in other cancer types is needed in the future.
GCN5
General control non-depressible 5 (GCN5) mainly responsible for acetylation of H3K27, is the first histone acetyltransferase to be characterized in saccharomyces cerevisiae. GCN5 is highly expressed in a variety of human cancers and promotes cancer progression by participating in the acetylation of many non-histone proteins (Haque et al., 2021). An example is in prostate cancer, where upregulated GCN5 downregulates Egr-1 expression via the PI3K/PTEN/Akt signaling pathway, negatively affecting IL-6-induced prostate cancer cell metastasis and epithelial-mesenchymal transition (EMT) (Shao et al., 2018).
In the immune system, GCN5 was reported to regulate the development of Treg cells and the transcription of interferon (IFN)-stimulated gene (ISG) expression (Au-Yeung and Horvath, 2018). In Foxp3+ Treg cells, GCN5 deletion has no effect on Treg in vitro but inhibits Treg function in vivo, and impairs T-effector (Teff) cells function (Liu Y. et al., 2019). For invariant natural killer T (iNKT) cells, the deficiency of GCN5 blocks its development (Wang et al., 2017).
At present, studies on the role of GCN5 in tumor immunity are still relatively few. Overexpression of GCN5 and PCAF in solid tumors in vivo enhances immune surveillance and associated NKG2D-dependent tumor cell death (Hu et al., 2021). Knockdown of GCN5 and PCAF in osteosarcoma and lung cancer resulted impaired induction of the natural killer group 2D (NKG2D) ligand Rae-1 by IL-12 and the chemotherapeutic agent doxorubicin as inhibition of NKG2D ligand expression was associated with tumor cell death and accelerated tumor progression (Hu et al., 2017). On the contrary, in head and neck squamous cell carcinoma, GCN5 acetylates H3K27, which activates transcription of PD-L1 and galectin-9 to evade tumor immunity (Ma et al., 2020). More research is needed to explain this opposite trend and to provide more precise strategies for cancer therapy.
p300/CBP
p300 (E1A binding protein p300) and CBP[CREB] (cyclic-AMP response element binding protein) binding proteins are considered functional homologs, sharing 63% homology at the amino acid level and exhibiting high structural similarity and functional redundancy (Ogryzko et al., 1996; Iyer et al., 2004; He et al., 2021). p300/CBP as a vital transcriptional co-activator and HAT contributes to a variety of cellular activities and plays a role in immune-mediated diseases and cancers through chromatin remodeling and gene activation (Karamouzis et al., 2007; Dancy and Cole, 2015). Overexpression or mutations of p300/CBP are found in malignant tumor, such as prostate and breast cancers (Bouchal et al., 2011; Hickey et al., 2021).
Previous studies have found that p300 and CBP are essential for the development and function of Foxp3+ Treg cells. Inhibition of p300 and CBP impairs Foxp3+ Treg cells function and furthers antitumor immunity (Liu et al., 2013; Liu et al., 2014). p300/CBP promotes Treg differentiation by inducing T and B cells to secrete the pro-inflammatory cytokines IL2 and IL10 (Castillo et al., 2019). It was observed that Treg was reduced in follicular lymphoma in tissues carrying CBP/p300 loss-of-function mutations (Castillo et al., 2019). In breast and lung cancers, p300/CBP inhibition can restrict tumor progression by impairing the function of immunosuppressive cells such as regulatory T cells and MDSCs in tumor microenvironments (Liu et al., 2013; de Almeida Nagata et al., 2019).
Several studies reported p300 can induce the expression of PD-L1 in liver cancer to impair CD8+ T cell-mediated anti-tumor immunity (Xiang et al., 2020; Guo et al., 2021), probably via acetylation of myocyte enhancer factor 2D (MEF2D). Furthermore, targeting p300/CBP by small molecular inhibitors such as E7386 and A485 could remarkably enhance the efficacy of PD-L1 blockade therapy in prostate and breast cancers in preclinical mouse models (Liu J. et al., 2020; Yamada et al., 2021). Additionally, p300 has been reported to regulate tumor cell immunogenicity, p300 ablations prevent chemotherapy-induced processing and presentation of major histocompatibility class I (MHC-I) antigens and abrogating the rejection of low MHC-I-expressing tumors by reinvigorated CD8 cytotoxic T cells (CTLs) (Zhou et al., 2021).
Tip60
Tat-interactive protein 60-kDa (Tip60, also known as KAT5) is one of the MYST subfamily of HATs and was originally identified as a tat-interacting protein widely involved in DNA damage repair, cellular activity and carcinogenesis (Zhang et al., 2017; McGuire et al., 2019). The regulatory function of Tip60 on cancer is dependent on cancer types. As a tumor suppressor in most cancers such as breast cancer (Bassi et al., 2016), gastric cancer (Sakuraba et al., 2011) and advanced stage colorectal cancer (Mattera et al., 2009), downregulation of Tip60 leads to defective DNA repair inducing the accumulation of genetic mutations that can cause tumor progression (Bassi et al., 2016). On the contrary, Tip60 promotes prostate cancer progression via acetylation of androgen receptor (AR) to augment AR signaling (Halkidou et al., 2003; Shiota et al., 2010; Coffey et al., 2012).
Tip60 was found to be essential for the survival of thymic and peripheral Treg cells (Xiao et al., 2014). Overexpression of Tip60 promoted FOXP3-mediated transcriptional repression, suggesting that enhancing its function would alter T cell-mediated transcriptional inhibition (Li et al., 2007). For tumor immunity, studies have shown that Tip60 plays a critical role in fostering acetylation, dimerization and function in Treg cells, leading to tumor suppression. Accordingly, targeting ubiquitin-specific protease (Usp) 7, which controls Treg function primarily by stabilizing expression and promoting multimerization of Tip60 and Foxp3, limits tumor progression (Wang et al., 2016).
HAT1
Histone acetyltransferase 1 (HAT1), also known as KAT1, was the first identified HAT (Poziello et al., 2021), and is a type of B histone acetyltransferase responsible for acetylating newly synthesized histones (Yang G. et al., 2021). As an oncogene, HAT1 is overexpressed and related to poor prognosis in a variety of solid tumors, such as HCCs (Jin et al., 2017), nasopharyngeal carcinoma (Miao et al., 2018), and pancreatic cancer (Fan et al., 2019), and can be a therapeutic target (Carafa et al., 2018). This may be due to the fact that HAT1 is involved in chromatin assembly and DNA damage repair, and its alterations can induce tumor development, invasion and metastasis (Poziello et al., 2021). HAT1 is also a transcription factor that upregulates the expression of various genes such as Bcl2L12 and Fas, and promotes cancer cell proliferation (Fan et al., 2019).
Despite being the first HAT to be discovered, HAT1 is still one of the least studied enzymes of its class (Poziello et al., 2021). In pancreatic cancer, HAT1 was shown to function as an important regulator in cancer immunity through transcriptional upregulation of PD-L1 in tumor cells, indicating that HAT1 could be used as a novel diagnostic and prognostic marker in immune checkpoint blockade therapy (Fan et al., 2019).
Histone Deacetylases in Tumor Immunity Regulation
Based on the homologies between HDAC and yeast deacetylases, HDACs can be divided into four sub-classes: Class I HDACs (HDAC1, 2, 3 and 8), which are similar to yeast Rpd3, are widely expressed in vivo and mainly located in the nucleus. Class II HDACs (HDAC4, 5, 6, 7, 9 and 10) have been studied that expressed in the cytoplasm and nucleus and distributed specifically in tissues. Class III HDACs (Sirt1-7) are similar to yeast Sir2, and its enzyme activity requires NAD+. Class IV HDAC (HDAC11) is homologous to yeast Rpd3 and Hda1. (Li and Seto, 2016). Studies have reported that more than 75% of human cancer tissues possess highly expressed class I HDACs (Nakagawa et al., 2007), besides, high expression of HDACs and low acetylation of histones are common in cancer cells (Cacan et al., 2014; Cacan, 2016; Alqinyah et al., 2017). The regulatory role of HDACs on tumor immunity was summarized in Figure 1.
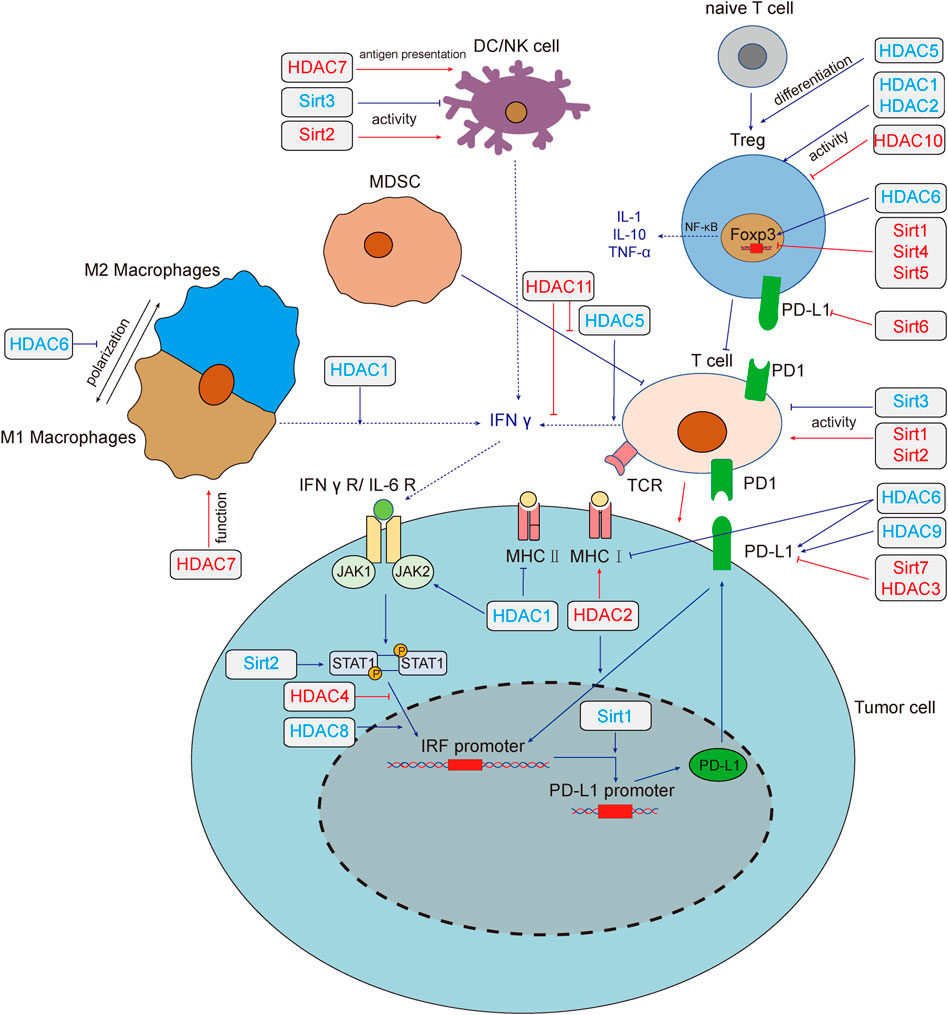
FIGURE 1. HADCs regulate tumor immunity and tumor immune microenvironment. Deacetylase regulates tumor immune response through multiple pathways. HDAC1,3,4,6,8,9 and Sirt1,2,6,7 regulate the expression of PD-L1 by regulating the IFN signaling pathway. HDAC1,2,6 regulates the expression of MHC, which affect the recognition of tumor cells by T cells. Sirt1, 2, 3 affect tumor immunity by changing the activity of T cells, HDAC1, 2, 10 could affect the function of Treg cells, and HDAC5 promotes the differentiation of naive T cells into Treg cells. Sirt2,3 could affect the activity of NK cells. HDAC7 both enhances the anti-tumor function of macrophages and the antigen presentation function of DC cells. HDAC1, 5, 6, 11 and Sirt1, 4, 5 modulate the tumor microenvironment by affecting the secretion of cytokines. Red lines and text colors represent the promoting effects of tumor immunity, while blue represents the tumor immunosuppression effects. Dotted lines indicate the regulation and function of cytokines. Abbreviations: T cell, T lymphocyte; NK, natural killer cell; DC, dendritic cells; MDSC, myeloid-derived suppressor cell; IFN-γ, interferon-γ; IFNγ R, interferon-γ receptor; IL, interleukin; JAK, Janus kinase; STAT, signal transducer and activator of transcription; TNF-α, tumor necrosis factor-α; Treg, regulatory T cell; IRF, interferon regulatory factor; PD-L1, programmed death-ligand 1; MHC, major histocompatibility complex; Sirt, sirtuins; HDAC, histone deacetylases.
Histone Deacetylase Family
The HDAC family is a histone deacetylase subfamily that contains HDAC1-11, whose activity is independent of NAD+. It has been observed in multiple cancers that HDACs promotes the proliferation of cancer cells by reducing the expression of the cyclin-dependent kinase inhibitor p21 or TGF-β, in addition, Class I HDACs can also promote cancer cell metastasis by inhibiting the expression of E-cadherin (Glozak and Seto, 2007).
HDAC family plays an important regulatory role in immune cells via its histone deacetylation activity. For instance, the effects of HDACs have been reflected in all aspects of T cells, including T cell development, peripheral immature T cell pool formation, T cell activation and differentiation into effector T cells, activation of regulatory T (Treg) cells, secretion of cytokines, and T cell immune function (Kumari et al., 2013; Cacan, 2017; Ellmeier and Seiser, 2018). Besides, ample evidences have indicated that HDACs regulate the macrophage development, differentiation, polarization, and activation through a variety of signal pathways (Mohammadi et al., 2018).
In addition, HDAC also participates in immunomodulatory networks in cancer cells such as STAT and NF-κB, which not only regulate the expression of molecules in the signal pathway but also controls the nucleus translocation and degradation of STAT and NF-κB signal molecules (Villagra et al., 2010). Target HDACs can increase the expression of antigen presentation molecules such as MHC I, MHC II, CD40 or promote their translocation (Maeda et al., 2000; Khan et al., 2008). HDAC inhibitors can also increase the expression of PD-L1 in tumor cells, and the combination of HDAC inhibitors and PD-1 blockers can delay tumor progression and improve survival rate (Woods et al., 2015).
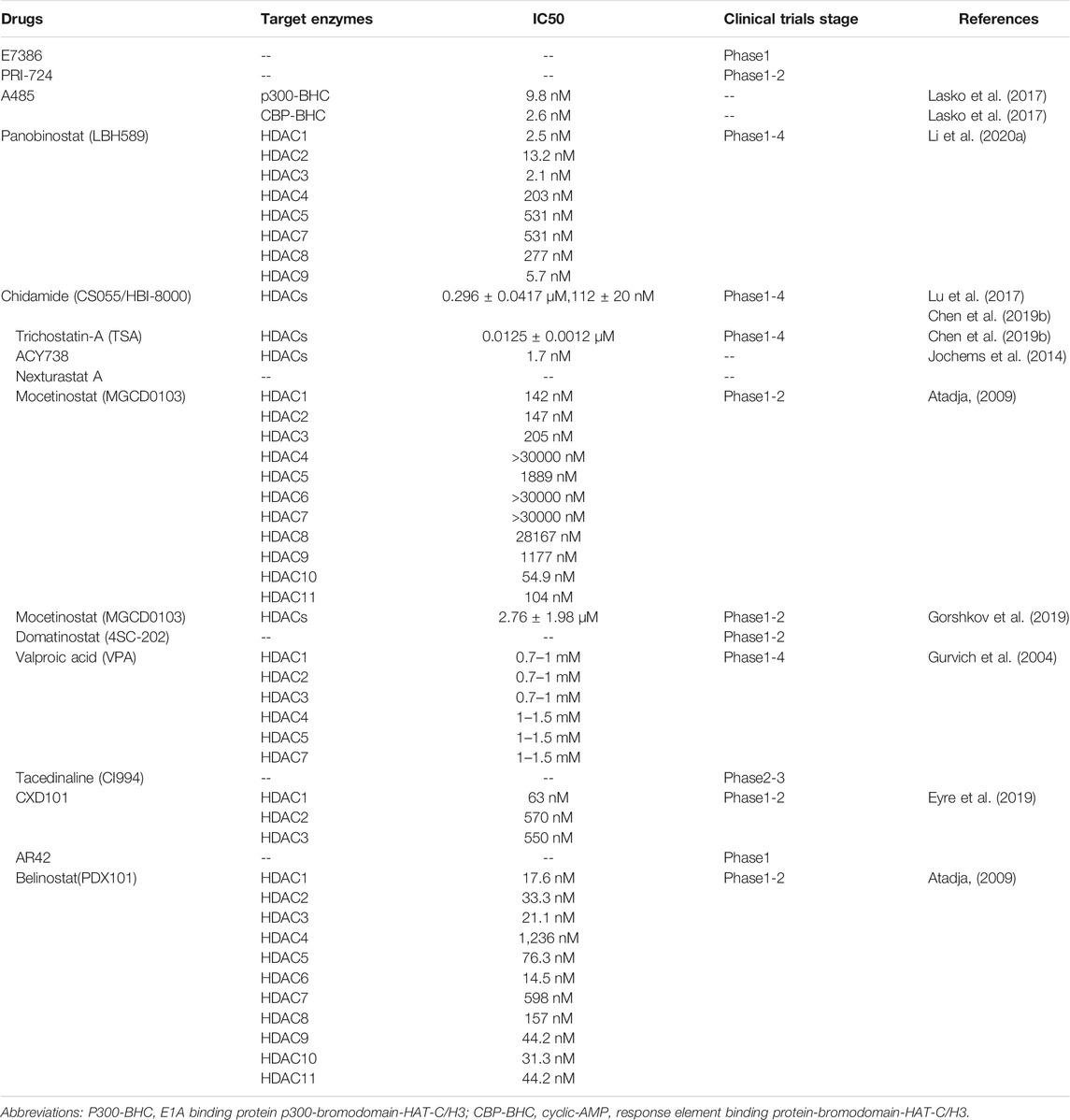
HDAC1
HDAC1, belonging to Class I, regulates N-terminal lysine residue deacetylation of histones to regulate gene transcription, cell cycle, growth, and apoptosis (Willis-Martinez et al., 2010). HDAC1 were significantly up-regulated in gastric cancer and could promote tumorigenesis and inhibit apoptosis (Yu et al., 2019). In addition, several recent studies have shown that HDAC1 inhibition is beneficial to the therapy of cancer, thus highlighting the cancer promoting function of HDAC1.
HDAC1/2 are required for normal T-cell development (Dovey et al., 2013), and HDAC1 regulates T cell-mediated autoimmunity by regulating CD4+T cells trafficking (Hamminger et al., 2021). Inhibition of HDAC1 promoted acetylation of histone H3/H4 in IFN-β1 promoter, and enhanced phosphorylation of interferon regulatory factor (IRF) 3 and its binding to the IFN-β promoter, which could lead to an anti-tumor immune response of macrophages microenvironment (Mounce et al., 2014; Meng et al., 2016). In addition, the role of HDAC1 in immune cells polarization has been highlighted. Several studies have shown that HDAC1 directly up-regulating the expression of miR-146a in macrophages, which induces tumor associated macrophages (TAMs) to adopt the M1-like phenotype. Besides, HDAC1 inhibition could promote the shift of microglia from M1 to M2 (Gao et al., 2015; Ji et al., 2019).
Mounting evidence shows that HDAC1 could regulate the expression of immune checkpoint molecules. HDAC1 inhibition could suppress the expression of PD-L1 induced by the IFN-γ signal pathway via down-regulating nucleus translocation of JAK2-mediated STAT1 in gastric cancer cells (Deng et al., 2018). In addition, the inhibition of HDAC1 could upregulate membrane expression of MHCⅡin cervical cancer and glioblastoma multiforme cells, and enhance the expression of CD1d in NSCLC cells (Yang et al., 2012; Wijdeven et al., 2018). HDAC1 also affects tumor immunity by regulating the secretion of cytokines. The CoREST complex, composed of HDAC1/2, LSD1, and scaffolding proteins Rcor1 and Rcor2, regulates a variety of immune and inflammatory responses. Targeting HDAC1/2 inhibits the binding of CoREST with IL-2 and the IFN-γ promoter, thereby promoting their expression, inhibiting the function of Treg, and enhancing anti-tumor immunity (Xiong et al., 2020).
HDAC2
Similar to HDAC1 in structure, HDAC2 has been shown vital in cardiac hypertrophy, Alzheimer’s disease, Parkinson’s and cancer (Kramer, 2009). HDAC2 inactivation can inhibit tumor cell growth and activate apoptosis by activating p53 and Bax (Jung et al., 2012). HDAC2 enhances proinflammatory cytokine production in LPS-stimulated macrophages by impairing the expression of c-Jun which are essential for the negative regulation of the inflammatory response (Wu et al., 2019) and inhibits the expression of plasminogen activator inhibitor 1 (PAI-1), TNF, and macrophage inflammatory protein-2 (MIP-2) in macrophage cells (Fang et al., 2018). Besides, studies have shown that HDAC2 enhances the infiltration of tumor lymphocytes and inhibits IL6 through its histone deacetylation function (Zhang et al., 2015; Xu et al., 2019).
P300-mediated PD-L1 acetylation at K263 inhibits the nucleus translocation of PD-L1, whereas HDAC2 deacetylate PD-L1 K263 and promote nucleus translocation of PD-L1. In the nucleus, PD-L1 interact with RelA and IRF proteins to form a positive feedback pathway to promote immune escape. Besides, treatment with HDAC2 inhibitors can also induce interferon type III related genes IL28A and IL28B, to activate STAT1 and increase the expression of MHC class I antigen presenting genes, thus achieving an improved immunotherapeutic effect (Gao et al., 2020).
HDAC3
HDAC3 plays an important role in apoptosis, cell progress, and DNA damage repair (Sarkar et al., 2020). In colorectal cancer and triple-negative breast cancer, the level of HDAC3 was upregulated. HDAC3 promotes the proliferation of colorectal cancer cells, HCC cells and glioma cells and inhibits the apoptosis of prostate cancer cells (Tong et al., 2020).
HDAC3 inhibition was reported to inhibit lipopolysaccharides (LPS) induced cytokine secretion in monocytes and M1 macrophages (Ghiboub et al., 2020). HDAC3 was reported to reduce the ratios of CD4+ and CD8+ T cells infiltration in colorectal carcinoma through upregulating B7x expression, which is related to a poor prognosis (Li et al., 2020b). HDAC3 inhibits the cytotoxicity of CD8+T cells and recognition ability of NK cells in melanoma cells by inhibiting genes encoding necessary cytotoxic proteins and key transcription factors (Fiegler et al., 2013; Tay et al., 2020).
Interestingly, depend on the cancer type, there are opposite reports on the regulation of PD-L1 by HDAC3. Studies have found that expression of PD-L1 is negatively correlated with HDAC3, suggesting that HDAC3 is a key inhibitor of PD-L1 transcription. Drugs targeting HDAC3 like resveratrol and pioglitazone upregulate PD-L1 expression in NSCLC, breast and colorectal cancer (Lucas et al., 2018; Wang H. et al., 2020). In B-cell lymphoma, HDAC3 is recruited to the PD-L1 promoter by the transcriptional inhibitor BCL6. In addition, HDAC3 inhibitors can also indirectly reduce the level of DNA methyltransferase 1 protein and activate PD-L1 transcription. HDAC3 inhibition combined with anti-PD-1/PD-L1 therapy can significantly improve the efficacy of B-cell lymphoma treatment (Deng et al., 2019). However, one study showed that the HDAC3/STAT3 pathway transcriptionally regulates PD-L1 expression in pancreatic ductal adenocarcinoma, and HDAC3 inhibitors reduce the protein and mRNA levels of PD-L1 in pancreatic cancer cells (Hu et al., 2019).
HDAC4
HDAC4, in class II, plays an important role in cell cycle progression and developmental events (Wang et al., 2014). HDAC4 was inhibited by miR-155 in human diffuse large B cell lymphoma (DLBCL) cells, resulting in up-regulation of downstream genes and induction of uncontrolled cell proliferation (Sandhu et al., 2012).
As an important signal pathway regulating tumor immunity, IFN signal promotes the phosphorylation of STAT and binds to the transcription factor interferon regulatory factor1 (IRF1) which promotes expression of PD-L1 (Kalbasi and Ribas, 2020). Although there is no direct report on the relationship between HDAC4 and tumor immunity, studies believe that HDAC4 can affect IFN signal. HDAC4 was reported to form a trimer with liver X receptor (LXR)α and pSTAT1, which block the binding of pSTAT1 to the promoter and inhibit the expression of d IRF1, TNF-α, and IL-6 in brain astrocytes (Lee et al., 2009). In cervical cancer cell line, HDAC4 was reported to promote type I interferon signaling via recruiting STAT2 to IFN-α–stimulated promoters (Lu et al., 2019).
HDAC5
HDAC5 is highly expressed in many tumors such as NSCLC, and melanoma but lowly expressed in gastric cancer (Yang J. et al., 2021). Several studies have shown that HDAC5 enhance the invasion and metastasis of neuroblastoma, pancreatic cancer and lung cancer. In addition, HDAC5 also inhibits the proliferation of tumor cells, which may be mediated by TGF-β (Yang J. et al., 2021).
HDAC5 has been reported to repress the production of proinflammatory cytokine in macrophages via TNF-α signaling (Li et al., 2017) and play an inhibitory role in regulating tumor microenvironments. HDAC5 deficient CD4+T cells lack the ability to transform into Tregs, while CD8+T cells impairs the ability to produce IFN- γ without HDAC5, which may offset the immune benefit resulting from decreased Treg function (Xiao et al., 2016). HDAC5-driven escape tumors exhibit a significant transition from neutrophils to macrophages. HDAC5 inhibits suppressor of cytokine signaling 3 (SOCS3), which leads to an increase of C-C motif chemokine ligand 2 (CCL2), recruits CCR2+ macrophages, promotes macrophages to reaggregate into tumor microenvironments, and promotes tumor recurrence (Cheng et al., 2014a; Hou et al., 2020).
HDAC6
HDAC6 protein is related to tumorigenesis, cell survival and metastasis of cancer cells (Aldana-Masangkay and Sakamoto, 2011). In breast cancer, HDAC6 promotes cell movement by acting on the nonhistone substrates, which enhance tumor cell movement, metastasis, and invasion. In addition, the expression of HDAC6 is closely related to endocytosis, and inhibits EGFR transport and degradation through α-tubulin deacetylation, thus activating cell proliferation through the downstream pathway of EGFR in NSCLC (Li et al., 2018).
HDAC6 and STAT3 form a complex and are recruited together to the immunosuppressive cytokine IL-10 promoter in antigen-presenting cells (APCs) to promote immune tolerance (Cheng et al., 2014b). In addition, NextA, a selective HDAC6 inhibitor, was found to increase the proportion of M1/M2 macrophages in tumor microenvironments, promoting IFN-γ and IL-2 levels and transformation of tumor microenvironments from “cold” to “hot”, thus enhancing the efficacy of immune checkpoint blocking therapy (Knox et al., 2019b).
HDAC6 has been also reported the two opposite regulation of PD-L1 in different tumor types. Several studies have reported that inhibition of HDAC6 decreases expression of PD-L1 in glioblastoma and melanoma. Selective inhibition of HDAC6 reduces the immunosuppressive activity of PD-L1 and leads to the recovery of host antitumor activity. A study suggested these effects may be mediated by recruitment and activation of STAT3 (M et al., 2016; Liu JR. et al., 2019). However, another study showed targeting HDAC6 can increase expression of PD-L1 and promote immunotherapy efficacy. Meanwhile, HDAC6 binds to cytoplasmic Foxo1 at K242 and deacetylates Foxo1 to weak the stability of Foxo1 and inhibit nucleus translocation, which limits IL-17–producing helper T (TH17) pathogenicity and antitumor effect in hepatocellular carcinoma (Qiu et al., 2020). Targeting HDAC6 in melanoma can up-regulate the expression of tumor-associated antigens and MHC-I molecules leading to enhance anti-tumor immunity (Woan et al., 2015).
HDAC7, 8, 9, 10, 11
Although the studies of HDAC7, 8, 9, 10 and 11 in tumorigenesis and progress have been widely reported, there is still a lack of research on their role in tumor immunity. Thereby, we summarize and describe the function of these enzymes in this part. HDAC7, 9, 10 belongs to the class Ⅱ HDACs, HDAC8 belongs to the class I HDACs, and HDAC11 is the only class IV HDAC, all of them affect the initial and progress of tumors in different ways. HDAC7 is considered a regulator of apoptosis in developing thymocytes (Dequiedt et al., 2003) and promoted cell proliferation through regulation of c-myc (Zhu et al., 2011). HDAC8 has been reported to promote the proliferation of hepatocellular carcinoma and inhibit apoptosis. On the contrary, targeting of HDAC8 inhibits the proliferation of lung cancer cell lines (Chakrabarti et al., 2015). HDAC9 is upregulated in various tumors such as glioblastoma, medulloblastoma (Yang C. et al., 2021). HDAC10 function as a tumor suppressor in stem-like lung adenocarcinoma and has been shown to interact with HDAC2 (Fischer et al., 2002; Li et al., 2020c). Notably, HDAC11 as the smallest HDAC isotype possesses very low deacetylase activity. Studies have shown that HDAC11 is highly expressed in prostate cancer, ovarian cancer, breast cancer, and NSCLC. Besides, targeting HDAC11 can enhance chemosensitivity (Liu SS. et al., 2020; Nunez-Alvarez and Suelves, 2021).
HDAC7, 8, 9, 10, 11 regulates immune function via affecting the polarization, antigen presentation, infiltration and activation of immune cells via various mechanisms. Previous studies have shown that HDAC7 and HDAC9 are involved in immune response mediated by immune effector cells. HDAC7 blocks the induction of genes that involved in macrophage immunity, phagocytosis, inflammation and cytokine production (Barneda-Zahonero et al., 2013). Besides, in uterine macrophages, HDAC9 deficiency promotes M2 macrophage polarization (Liu et al., 2021). HDAC9 also enhance immune response via enhancing dendritic cell antigen presentation and CD8+T cell TME infiltration (Ning et al., 2020). In addition, HDAC10 and HDAC11 promote the inhibition of immune response by immune regulatory cells. HDAC10 deletion Treg exhibited a stronger immune inhibitory effect and represses inflammation after intracerebral hemorrhage (Dahiya et al., 2020). MDSCs without HDAC11 exhibit stronger inhibitory activity against CD8+T cells via inducing high level of immunosuppressive enzymes expressed in CD8+T cells (Chen et al., 2021).
In addition, the secretion of cytokines is also an important apparent regulatory function of these enzymes. HADC7 represses the production of IFN-γ and promotes CD4+ T cells proliferation, as well as increasing the expression of the proinflammatory cytokine IL-1β in macrophage (Myers et al., 2017; Das Gupta et al., 2020). Similarly, in T cells, HDAC11 knockout increases the expression of transcription factors Eomesodermin and Tbx21, which reduce the production of inflammatory cytokines and increase the expression of IFN-γ (Woods et al., 2017). Besides, HDAC11 can bind to the promoter of IL-10 to inhibit IL-10 expression and induce inflammatory antigen-presenting cells to activate primordial antigen-specific CD4+T cells (Villagra et al., 2009).
In terms of tumor immunity, HDAC8, 9, and 10 have been reported to be related to the expression of immune checkpoint. HDAC8 can promote tumor immunity by inhibiting the expression of PD-L1. In melanoma cells, HDAC8 competitively inhibits the transcription factor to bind the PD-L1 promoter, thus inhibiting the expression of PD-L1 (Wang YF. et al., 2018). However, opposite studies have shown that HDAC8 inhibition increases the expression of NKG2D ligand in glioma cells which enhanced the recognition ability and cytotoxicity of NK cells, and activates immune cells in hepatocellular carcinoma resulting in an effective and lasting response to ICB (Yang W. et al., 2021; Mormino et al., 2021). On the contrary, HDAC9 and HDAC10 are opposite to HDAC8 in up-regulating PD-L1. One study has suggested that HDAC9 can significantly promote infiltration of immune cells and increase expression of immunological molecules such as PD-L1, CTLA4 and LAG3 in clear cell renal cell carcinoma (Fu Y. et al., 2020). In addition, the expression of HDAC10 in NSCLC was positively correlated with the expression of PD-L1, and the level of PD-L1 is significantly higher than paracancerous tissues, indicating a poor prognosis (Liu X. et al., 2020). Besides, HDAC10 inhibits NK cell-mediated antitumor immunity in hepatocellular carcinoma via recruiting EZH2 to block the CXCL10 promoter (Bugide et al., 2021).
Sirtuin Family
The mammalian Sirtuin protein family is a homologue of Sir2, which can regulate various processes in mammalian cells and play a crucial part in regulating aging, metabolism, gene transcription, and stress responses (such as neurodegeneration, diabetes, cardiovascular disease and many types of cancer) (Michan and Sinclair, 2007). Sirtuins plays an indispensable role in tumorigenesis and development through cellular effects to genomic instability (regulation of cell cycle, DNA repair, cell survival and apoptosis), such as modulating cancer-related metabolism and changing the tumor microenvironment. There are few studies available. (Palmirotta et al., 2016).
Sirt1
Sirt1 (Sirtuin 1) mainly locates in the nucleus, and can removes acetyl groups from proteins. Sirt1 can inhibit transcription by directly deacetylating histones H1 lys26, H3 lys9、lys14, and H4 lys16 and by recruiting other ribozymes to chromatin to promote histone and DNA methylation changes that regulate chromatin function (Vaquero et al., 2004).
At present, the research of Sirt1 in tumor immunity is gradually in-depth. In T cells, Sirt1 enhances the activity of Foxo1 by deacetylating Foxo1, and promotes the expression of Klf2 and Ccr7 genes, thereby improving the anti-tumor immune response of T cells (Chatterjee et al., 2018). Additionally, the deacetylation of FoxP3 induced by Sirt1 is critical to the immunosuppressive function of Treg, resulting in Foxp3 degradation and reduced Treg cell number and activity (van Loosdregt et al., 2010). Sirt1 could enhance the tumor killing ability of macrophages, and deacetylate K310 of the p65 subunit in NF-κB, thereby increasing the infiltration of CD8+T cells in tumors or increasing the expression of CXCL10, so as to recruit NK cells and macrophages into tumor microenvironments (Yu et al., 2016a; Yu et al., 2016b; Zhou et al., 2019; Ye et al., 2020). Sirt1 can enhance the activity of NF-κB by promoting the phosphorylation of p65 and IκB, thereby promoting the polarization of M1 in hepatocellular carcinoma (Zhou et al., 2019).
In tumor microenvironments, Sirt1 participates in the immune response by activating the pro-inflammatory pathway (Chen et al., 2015). Pharmacological activation of Sirt1 increases the stability of the transcription factor snail, enhances the binding of β-catenin/TCF to the PD-L1 promoter, and promotes the expression of PD-L1 in NSCLC (Yang M. et al., 2021). Further research is needed on the specific regulation details of Sirt1 in tumor microenvironment.
Sirt2
Sirt2 is the only Sirtuin protein found mainly in the cytoplasm, and it is also expressed in mitochondria and nucleus (Wang Y. et al., 2019). Sirt2 plays an important role in cell differentiation, senescence, infection, inflammation, oxidative stress, and autophagy by regulating the function of important oncogenes, such as Myc and KRAS (Chen et al., 2020). Although there is clear evidence of that Sirt2 is abnormally expressed in various tumors, its causal relationship with tumorigenesis is still unclear, and its effect on the development of different kinds of tumors and its molecular mechanism are still contested (Song et al., 2016; Wang Y. et al., 2019).
Studies have found that the level of Sirt2 is positively correlated with CD8+ effector memory T (TEM) cells in peripheral T lymphocytes from breast cancer patients. Sirt2 can inhibits GSK3 β acetylation in CD8+T cells by promoting aerobic oxidation, which promotes the differentiation of CD8+T cells into TEM (Jiang et al., 2020). In chemically induced hepatocellular carcinoma mice, it has been found that sirt2 was induced in the immune microenvironment of hepatoma cells to enhance the tumor-killing activity of NK cells by promoting mitogen-activated protein kinase (MAPK) in activated NK cells (Chen M. et al., 2019). In addition, Sirt2 can regulate IFN-driven gene transcription by regulating the phosphorylation of Ser-727 on IFN I-dependent STAT1 through deacetylation of CDK9 Lys48 (Kosciuczuk et al., 2019).
Sirt3
Sirt3 plays an important role in mitochondrial function, aging, and carcinogenesis by targeting a series of key regulatory factors and their related pathways in tumors to regulate cell death (Chen et al., 2014). A study has shown that Sirt3 deacetylates NLRC4 to promote its activation, thereby promoting the activation of inflammasomes and mediating the production of the pro-inflammatory cytokine IL-1β in macrophages (Guan et al., 2021).
Blocking Sirt3 and Sirt6 can inhibit the activation of RIPK3 and MLKL in prostate cancer cells, thus enhancing necrotizing apoptosis and promoting infiltration of CD4+T cells, macrophages and neutrophils, so as to inhibit the progression of cancer (Fu W. et al., 2020).
Sirt4
Sirt4 is located in mitochondria and plays an important role in cellular metabolism and tumor biology (Tomaselli et al., 2020). It is mostly regarded as a tumor suppressor gene, and its expression is low in breast, gastric, and colon cancers (Huang and Zhu, 2018). Sirt4 can inhibit FoxP3, anti-inflammatory cytokines IL-10, TGFβ and AMPK signal in Treg cells, which impairs their anti-inflammatory function (Lin et al., 2019).
Sirt5
Sirt5 promotes cancer cell proliferation by targeting a variety of metabolic enzymes including GLS, SHMT2, and PKM2 (Abril et al., 2021). It can maintain histone acetylation and methylation levels in melanoma, thereby promoting the corresponding expression of MITF and c-myc (Milling and Edgar, 2019). It is reported that Sirt5 affects the tumor immune microenvironment through its succinylation function, which is negatively correlated with Treg infiltration in clear cell renal cell carcinoma (Lu et al., 2021).
Sirt6
Sirt6 is a NAD + -dependent histone H3K9 deacetylase that prevents genomic instability, maintains telomere integrity, and regulates metabolic homeostasis and DNA repair (Desantis et al., 2018). It inhibits IGF-Akt and NF-κβ signal transduction by the deacetylation of H3K9 (Sundaresan et al., 2012). High Sirt6 expression levels are observed in the immune system. It may be a possibly a negative regulator of immune cell function and metabolism, which related to neutrophil inactivation and increased the polarization to M2 macrophages (Pillai and Gupta, 2021).
Induction of Sirt6 expression in 4T1 cells inhibits the activation of NF-κB and suppresses the transcription of PD-L1 (Song et al., 2020), which suppresses the proliferation and transcription of PD-L1 in Treg cells (Vanamee and Faustman, 2017). In addition, overexpression of Sirt6 in pancreatic cancer cells increases the production of TNF and IL8 which has nothing to do with the activation of NF-κB, thus leading to pro-inflammatory and pro-angiogenic phenotype (Bauer et al., 2012).
Sirt7
Sirt7 plays an important role in ribosomal biogenesis, cellular stress response, genomic stability, metabolic regulation, aging, and cancer (Ford et al., 2006; Blank and Grummt, 2017; Tang et al., 2021). Compared with other nucleus-localized Sirts, Sirt7 exhibits weaker deacetylase activity. The enzyme activity of Sirt7 targets acetylated H3K18 and succinylated H3K122 (Li L. et al., 2016).
Sirt7 is high expressed in breast cancer, which is an indicator of poor prognosis, and the expression of Sirt7 is positively correlated with the expression of IRF5 and PD1, which increases M1 macrophages, and depletes T cells in the immune environment of breast cancer (Huo et al., 2020). Sirt7 knockout hepatocellular carcinoma cells expressed higher levels of PD-L1 by inhibiting the deacetylation of MEDF2D and reduced T cell infiltration and activation. Moreover, Sirt7 inhibition combined with PD1 blocking therapy can significantly improve the efficacy of hepatocellular carcinoma (Xiang et al., 2020).
Targeting Acetylation in Tumor Immunotherapy
Immune checkpoint blockades such as anti-PD-1/PD-L1 and anti-CTLA4 have shown promising effects in cancer treatment. However, only few patients benefit from immunotherapy. The combinations of immunotherapy and acetylation-modified drugs (HAT inhibitors and HDAC inhibitors) have attracted more attention in tumor treatment due to the role of acetylases in the regulation of tumor and tumor immunity.
Currently available HAT inhibitors are primarily target CBP and p300, including E7386 (Yamada et al., 2021), PRI-724 (Osawa et al., 2015; Osawa et al., 2019), and A485 (Lasko et al., 2017; Liu J. et al., 2020), which have shown promising effects in improving anti-PD1 immunotherapy. Among them, E7386 (Yamada et al., 2021) and PRI-724 (Osawa et al., 2015; Osawa et al., 2019) inhibit the interaction between CBP and/or β-catenin, which can increase the infiltration of CD8+ T cells. A485 (Lasko et al., 2017; Liu J. et al., 2020) directly targets p300/CBP and inhibits the secretion of exosomal PD-L1.
Correspondingly, partial HDAC inhibitors can also modulate immunotherapies. Some drugs enhance tumor immunity by up regulating PD-L1 in tumor cells, such as Panobinostat(LBH589) (Atadja, 2009; Woods et al., 2015), Chidamide (CS055/HBI-8000) (Ning et al., 2012; Yan et al., 2020; Que et al., 2021), Trichostatin-A (TSA) (Li et al., 2021) and ACY738 (Regna et al., 2016; Maharaj et al., 2020). Other drugs can achieve antitumor immunity by increasing the infiltration of cytotoxic cells, including Nexturastat A (Knox et al., 2019a), Mocetinostat (Boumber et al., 2011; Briere et al., 2018), Domatinostat (4SC-202) (Bretz et al., 2019), Chidamide (CS055/HBI-8000) (Ning et al., 2012; Yan et al., 2020; Que et al., 2021), Valproic acid (VPA) (Xie et al., 2018; Adeshakin et al., 2020), Tacedinaline (CI994) (Berger et al., 1990; el-Beltagi et al., 1993; LoRusso et al., 1996; Burke et al., 2020), CXD10 (Eyre et al., 2019; Blaszczak et al., 2021), Nexturastat A (Knox et al., 2019a), AR42 (Booth et al., 2017) and Trichostatin-A (TSA) (Li et al., 2021), which can improve the infiltration of macrophages, NK cells, and neutrophils. In addition, Mocetinostat (Boumber et al., 2011; Briere et al., 2018), Chidamide (CS055/HBI-8000) (Ning et al., 2012; Yan et al., 2020; Que et al., 2021), Valproic acid (VPA) (Xie et al., 2018; Adeshakin et al., 2020), Trichostatin-A (Li et al., 2021) and Belinostat (PDX101) (Atadja, 2009; Llopiz et al., 2019) can also play an immune regulatory role by reducing the infiltration of immunosuppressive cells such as myeloid-derived suppressor cell (MDSC) and T-regulatory cells (Tregs). Finally, ACY738 (Regna et al., 2016; Maharaj et al., 2020) enhances MHC-restricted antigen presentation by upregulating tumor cell MHC-I expression.
Conclusion
Over the last few decades, we have recognized acetylation plays an important role in the regulation of protein function, chromatin structure and gene expression. Research into this field has covered metabolism, immunity, cell cycle, DNA damage repair, apoptosis and autophagy. New discoveries reveal the evidence that acetylation may also influence the tumor immunity through several pathways including regulating the expression and function of immune checkpoint molecules and antigens in tumor cells, as well as processes such as infiltration, secretion of cytokines and antigen presentation by immune cells. Emerging interest in acetylation research has been centered on the use of HAT or HDAC inhibitors that have shown great potential for improving immunotherapeutic outcomes. Further studies should focus on safety and the best way to combine these drugs with various immunotherapies to conquer cancer in the future.
Author Contributions
JL, XH and LZ designed and wrote the manuscript. WL and RZ reviewed and critically read the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by the Hunan Science and Technology Department Plan Project (2018XK2304), National Natural Science Foundations of China (82,071,895 to WL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abril, Y. L. N., Fernandez, I. R., Hong, J. Y., Chiang, Y. L., Kutateladze, D. A., Zhao, Q., et al. (2021). Pharmacological and Genetic Perturbation Establish SIRT5 as a Promising Target in Breast Cancer. Oncogene 40 (9), 1644–1658. doi:10.1038/s41388-020-01637-w
Adeshakin, A. O., Yan, D., Zhang, M., Wang, L., Adeshakin, F. O., Liu, W., et al. (2020). Blockade of Myeloid-Derived Suppressor Cell Function by Valproic Acid Enhanced Anti-PD-L1 Tumor Immunotherapy. Biochem. Biophys. Res. Commun. 522 (3), 604–611. doi:10.1016/j.bbrc.2019.11.155
Aldana-Masangkay, G. I., and Sakamoto, K. M. (2011). The Role of HDAC6 in Cancer. J. Biomed. Biotechnol. 2011, 875824. doi:10.1155/2011/875824
Alqinyah, M., Maganti, N., Ali, M. W., Yadav, R., Gao, M., Cacan, E., et al. (2017). Regulator of G Protein Signaling 10 (Rgs10) Expression Is Transcriptionally Silenced in Activated Microglia by Histone Deacetylase Activity. Mol. Pharmacol. 91 (3), 197–207. doi:10.1124/mol.116.106963
Atadja, P. (2009). Development of the Pan-DAC Inhibitor Panobinostat (LBH589): Successes and Challenges. Cancer Lett. 280 (2), 233–241. doi:10.1016/j.canlet.2009.02.019
Au-Yeung, N., and Horvath, C. M. (2018). Histone H2A.Z Suppression of Interferon-Stimulated Transcription and Antiviral Immunity Is Modulated by GCN5 and BRD2. iScience 6, 68–82. doi:10.1016/j.isci.2018.07.013
Audia, J. E., and Campbell, R. M. (2016). Histone Modifications and Cancer. Cold Spring Harb Perspect. Biol. 8 (4), a019521. doi:10.1101/cshperspect.a019521
Barneda-Zahonero, B., Román-González, L., Collazo, O., Rafati, H., Islam, A. B., Bussmann, L. H., et al. (2013). HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages. Plos Genet. 9 (5), e1003503. doi:10.1371/journal.pgen.1003503
Bassi, C., Li, Y. T., Khu, K., Mateo, F., Baniasadi, P. S., Elia, A., et al. (2016). The Acetyltransferase Tip60 Contributes to Mammary Tumorigenesis by Modulating DNA Repair. Cell Death Differ 23 (7), 1198–1208. doi:10.1038/cdd.2015.173
Bauer, I., Grozio, A., Lasigliè, D., Basile, G., Sturla, L., Magnone, M., et al. (2012). The NAD+-dependent Histone Deacetylase SIRT6 Promotes Cytokine Production and Migration in Pancreatic Cancer Cells by Regulating Ca2+ Responses. J. Biol. Chem. 287 (49), 40924–40937. doi:10.1074/jbc.M112.405837
Berger, M. R., Richter, H., Seelig, M. H., Eibl, H., and Schmähl, D. (1990). New Cytostatics-Mmore Activity and Less Toxicity. Cancer Treat Rev. 17 (2-3), 143–154. doi:10.1016/0305-7372(90)90039-i
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 39 (4), 782–795. doi:10.1016/j.immuni.2013.10.003
Blank, M. F., and Grummt, I. (2017). The Seven Faces of SIRT7. Transcription 8 (2), 67–74. doi:10.1080/21541264.2016.1276658
Blaszczak, W., Liu, G., Zhu, H., Barczak, W., Shrestha, A., Albayrak, G., et al. (2021). Immune Modulation Underpins the Anti-cancer Activity of HDAC Inhibitors. Mol. Oncol. 1, 1. doi:10.1002/1878-0261.12953
Booth, L., Roberts, J. L., Poklepovic, A., Kirkwood, J., and Dent, P. (2017). HDAC Inhibitors Enhance the Immunotherapy Response of Melanoma Cells. Oncotarget 8 (47), 83155–83170. doi:10.18632/oncotarget.17950
Bouchal, J., Santer, F. R., Höschele, P. P., Tomastikova, E., Neuwirt, H., and Culig, Z. (2011). Transcriptional Coactivators P300 and CBP Stimulate Estrogen Receptor-Beta Signaling and Regulate Cellular Events in Prostate Cancer. Prostate 71 (4), 431–437. doi:10.1002/pros.21257
Boumber, Y., Younes, A., and Garcia-Manero, G. (2011). Mocetinostat (MGCD0103): a Review of an Isotype-specific Histone Deacetylase Inhibitor. Expert Opin. Investig. Drugs 20 (6), 823–829. doi:10.1517/13543784.2011.577737
Brasacchio, D., Busuttil, R. A., Noori, T., Johnstone, R. W., Boussioutas, A., and Trapani, J. A. (2018). Down-regulation of a Pro-apoptotic Pathway Regulated by PCAF/ADA3 in Early Stage Gastric Cancer. Cell Death Dis 9 (5), 442. doi:10.1038/s41419-018-0470-8
Bretz, A. C., Parnitzke, U., Kronthaler, K., Dreker, T., Bartz, R., Hermann, F., et al. (2019). Domatinostat Favors the Immunotherapy Response by Modulating the Tumor Immune Microenvironment (TIME). J. Immunother. Cancer 7 (1), 294. doi:10.1186/s40425-019-0745-3
Briere, D., Sudhakar, N., Woods, D. M., Hallin, J., Engstrom, L. D., Aranda, R., et al. (2018). The Class I/IV HDAC Inhibitor Mocetinostat Increases Tumor Antigen Presentation, Decreases Immune Suppressive Cell Types and Augments Checkpoint Inhibitor Therapy. Cancer Immunol. Immunother. 67 (3), 381–392. doi:10.1007/s00262-017-2091-y
Bugide, S., Gupta, R., Green, M. R., and Wajapeyee, N. (2021). EZH2 Inhibits NK Cell-Mediated Antitumor Immunity by Suppressing CXCL10 Expression in an HDAC10-dependent Manner. Proc. Natl. Acad. Sci. U S A. 118 (30). doi:10.1073/pnas.2102718118
Burke, B., Eden, C., Perez, C., Belshoff, A., Hart, S., Plaza-Rojas, L., et al. (2020). Inhibition of Histone Deacetylase (HDAC) Enhances Checkpoint Blockade Efficacy by Rendering Bladder Cancer Cells Visible for T Cell-Mediated Destruction. Front Oncol. 10, 699. doi:10.3389/fonc.2020.00699
Cacan, E., Ali, M. W., Boyd, N. H., Hooks, S. B., and Greer, S. F. (2014). Inhibition of HDAC1 and DNMT1 Modulate RGS10 Expression and Decrease Ovarian Cancer Chemoresistance. PLoS One 9 (1), e87455. doi:10.1371/journal.pone.0087455
Cacan, E. (2017). Epigenetic-mediated Immune Suppression of Positive Co-stimulatory Molecules in Chemoresistant Ovarian Cancer Cells. Cell Biol Int 41 (3), 328–339. doi:10.1002/cbin.10729
Cacan, E. (2016). Histone Deacetylase-1-Mediated Suppression of FAS in Chemoresistant Ovarian Cancer Cells. Anticancer Res. 36 (6), 2819–2826.
Carafa, V., Nebbioso, A., Cuomo, F., Rotili, D., Cobellis, G., Bontempo, P., et al. (2018). RIP1-HAT1-SIRT Complex Identification and Targeting in Treatment and Prevention of Cancer. Clin. Cancer Res. 24 (12), 2886–2900. doi:10.1158/1078-0432.CCR-17-3081
Castillo, J., Wu, E., Lowe, C., Srinivasan, S., McCord, R., Wagle, M. C., et al. (2019). CBP/p300 Drives the Differentiation of Regulatory T Cells through Transcriptional and Non-transcriptional Mechanisms. Cancer Res. 79 (15), 3916–3927. doi:10.1158/0008-5472.CAN-18-3622
Chakrabarti, A., Oehme, I., Witt, O., Oliveira, G., Sippl, W., Romier, C., et al. (2015). HDAC8: a Multifaceted Target for Therapeutic Interventions. Trends Pharmacol. Sci. 36 (7), 481–492. doi:10.1016/j.tips.2015.04.013
Chatterjee, S., Daenthanasanmak, A., Chakraborty, P., Wyatt, M. W., Dhar, P., Selvam, S. P., et al. (2018). CD38-NAD+Axis Regulates Immunotherapeutic Anti-tumor T Cell Response. Cell Metab 27 (1), 85–e8. doi:10.1016/j.cmet.2017.10.006
Chen, G., Huang, P., and Hu, C. (2020). The Role of SIRT2 in Cancer: A Novel Therapeutic Target. Int. J. Cancer 147 (12), 3297–3304. doi:10.1002/ijc.33118
Chen, J., Cheng, F., Sahakian, E., Powers, J., Wang, Z., Tao, J., et al. (2021). HDAC11 Regulates Expression of C/EBPβ and Immunosuppressive Molecules in Myeloid-Derived Suppressor Cells. J. Leukoc. Biol. 109 (5), 891–900. doi:10.1002/JLB.1A1119-606RRR
Chen, M., Xu, M., Zhu, C., Wang, H., Zhao, Q., and Zhou, F. (2019a). Sirtuin2 Enhances the Tumoricidal Function of Liver Natural Killer Cells in a Mouse Hepatocellular Carcinoma Model. Cancer Immunol. Immunother. 68 (6), 961–971. doi:10.1007/s00262-019-02337-5
Chen, X., Lu, Y., Zhang, Z., Wang, J., Yang, H., and Liu, G. (2015). Intercellular Interplay between Sirt1 Signalling and Cell Metabolism in Immune Cell Biology. Immunology 145 (4), 455–467. doi:10.1111/imm.12473
Chen, X., Zhao, S., Li, H., Wang, X., Geng, A., Cui, H., et al. (2019b). Design, Synthesis and Biological Evaluation of Novel Isoindolinone Derivatives as Potent Histone Deacetylase Inhibitors. Eur. J. Med. Chem. 168, 110–122. doi:10.1016/j.ejmech.2019.02.032
Chen, Y., Fu, L. L., Wen, X., Wang, X. Y., Liu, J., Cheng, Y., et al. (2014). Sirtuin-3 (SIRT3), a Therapeutic Target with Oncogenic and Tumor-Suppressive Function in Cancer. Cell Death Dis 5, e1047. doi:10.1038/cddis.2014.14
Cheng, F., Lienlaf, M., Perez-Villarroel, P., Wang, H. W., Lee, C., Woan, K., et al. (2014a). Divergent Roles of Histone Deacetylase 6 (HDAC6) and Histone Deacetylase 11 (HDAC11) on the Transcriptional Regulation of IL10 in Antigen Presenting Cells. Mol. Immunol. 60 (1), 44–53. doi:10.1016/j.molimm.2014.02.019
Cheng, F., Lienlaf, M., Wang, H. W., Perez-Villarroel, P., Lee, C., Woan, K., et al. (2014b). A Novel Role for Histone Deacetylase 6 in the Regulation of the Tolerogenic STAT3/IL-10 Pathway in APCs. J. Immunol. 193 (6), 2850–2862. doi:10.4049/jimmunol.1302778
Coffey, K., Blackburn, T. J., Cook, S., Golding, B. T., Griffin, R. J., Hardcastle, I. R., et al. (2012). Characterisation of a Tip60 Specific Inhibitor, NU9056, in Prostate Cancer. PLoS One 7 (10), e45539. doi:10.1371/journal.pone.0045539
Dahiya, S., Beier, U. H., Wang, L., Han, R., Jiao, J., Akimova, T., et al. (2020). HDAC10 Deletion Promotes Foxp3+ T-Regulatory Cell Function. Sci. Rep. 10 (1), 424. doi:10.1038/s41598-019-57294-x
Dancy, B. M., and Cole, P. A. (2015). Protein Lysine Acetylation by P300/CBP. Chem. Rev. 115 (6), 2419–2452. doi:10.1021/cr500452k
Das Gupta, K., Shakespear, M. R., Curson, J. E. B., Murthy, A. M. V., Iyer, A., Hodson, M. P., et al. (2020). Class IIa Histone Deacetylases Drive Toll-like Receptor-Inducible Glycolysis and Macrophage Inflammatory Responses via Pyruvate Kinase M2. Cell Rep 30 (8), 2712–e8. e2718. doi:10.1016/j.celrep.2020.02.007
de Almeida Nagata, D. E., Chiang, E. Y., Jhunjhunwala, S., Caplazi, P., Arumugam, V., Modrusan, Z., et al. (2019). Regulation of Tumor-Associated Myeloid Cell Activity by CBP/EP300 Bromodomain Modulation of H3K27 Acetylation. Cell Rep 27 (1), 269–e4. e264. doi:10.1016/j.celrep.2019.03.008
Deng, R., Zhang, P., Liu, W., Zeng, X., Ma, X., Shi, L., et al. (2018). HDAC Is Indispensable for IFN-γ-Induced B7-H1 Expression in Gastric Cancer. Clin. Epigenetics 10 (1), 153. doi:10.1186/s13148-018-0589-6
Deng, S., Hu, Q., Zhang, H., Yang, F., Peng, C., and Huang, C. (2019). HDAC3 Inhibition Upregulates PD-L1 Expression in B-Cell Lymphomas and Augments the Efficacy of Anti-PD-L1 Therapy. Mol. Cancer Ther. 18 (5), 900–908. doi:10.1158/1535-7163.MCT-18-1068
Dequiedt, F., Kasler, H., Fischle, W., Kiermer, V., Weinstein, M., Herndier, B. G., et al. (2003). HDAC7, a Thymus-specific Class II Histone Deacetylase, Regulates Nur77 Transcription and TCR-Mediated Apoptosis. Immunity 18 (5), 687–698. doi:10.1016/s1074-7613(03)00109-2
Desantis, V., Lamanuzzi, A., and Vacca, A. (2018). The Role of SIRT6 in Tumors. Haematologica 103 (1), 1–4. doi:10.3324/haematol.2017.182675
Dovey, O. M., Foster, C. T., Conte, N., Edwards, S. A., Edwards, J. M., Singh, R., et al. (2013). Histone Deacetylase 1 and 2 Are Essential for normal T-Cell Development and Genomic Stability in Mice. Blood 121 (8), 1335–1344. doi:10.1182/blood-2012-07-441949
el-Beltagi, H. M., Martens, A. C., Lelieveld, P., Haroun, E. A., and Hagenbeek, A. (1993). Acetyldinaline: a New Oral Cytostatic Drug with Impressive Differential Activity against Leukemic Cells and normal Stem Cells-Ppreclinical Studies in a Relevant Rat Model for Human Acute Myelocytic Leukemia. Cancer Res. 53 (13), 3008–3014.
Ellmeier, W., and Seiser, C. (2018). Histone Deacetylase Function in CD4+ T Cells. Nat. Rev. Immunol. 18 (10), 617–634. doi:10.1038/s41577-018-0037-z
Eyre, T. A., Collins, G. P., Gupta, A., Coupe, N., Sheikh, S., Whittaker, J., et al. (2019). A Phase 1 Study to Assess the Safety, Tolerability, and Pharmacokinetics of CXD101 in Patients with Advanced Cancer. Cancer 125 (1), 99–108. doi:10.1002/cncr.31791
Fan, P., Zhao, J., Meng, Z., Wu, H., Wang, B., Wu, H., et al. (2019). Overexpressed Histone Acetyltransferase 1 Regulates Cancer Immunity by Increasing Programmed Death-Ligand 1 Expression in Pancreatic Cancer. J. Exp. Clin. Cancer Res. 38 (1), 47. doi:10.1186/s13046-019-1044-z
Fang, W. F., Chen, Y. M., Lin, C. Y., Huang, H. L., Yeh, H., Chang, Y. T., et al. (2018). Histone Deacetylase 2 (HDAC2) Attenuates Lipopolysaccharide (LPS)-induced Inflammation by Regulating PAI-1 Expression. J. Inflamm. (Lond) 15, 3. doi:10.1186/s12950-018-0179-6
Fiegler, N., Textor, S., Arnold, A., Rölle, A., Oehme, I., Breuhahn, K., et al. (2013). Downregulation of the Activating NKp30 Ligand B7-H6 by HDAC Inhibitors Impairs Tumor Cell Recognition by NK Cells. Blood 122 (5), 684–693. doi:10.1182/blood-2013-02-482513
Fischer, D. D., Cai, R., Bhatia, U., Asselbergs, F. A., Song, C., Terry, R., et al. (2002). Isolation and Characterization of a Novel Class II Histone Deacetylase, HDAC10. J. Biol. Chem. 277 (8), 6656–6666. doi:10.1074/jbc.M108055200
Ford, E., Voit, R., Liszt, G., Magin, C., Grummt, I., and Guarente, L. (2006). Mammalian Sir2 Homolog SIRT7 Is an Activator of RNA Polymerase I Transcription. Genes Dev. 20 (9), 1075–1080. doi:10.1101/gad.1399706
Fraga, M. F., Ballestar, E., Villar-Garea, A., Boix-Chornet, M., Espada, J., Schotta, G., et al. (2005). Loss of Acetylation at Lys16 and Trimethylation at Lys20 of Histone H4 Is a Common Hallmark of Human Cancer. Nat. Genet. 37 (4), 391–400. doi:10.1038/ng1531
Fu, W., Li, H., Fu, H., Zhao, S., Shi, W., Sun, M., et al. (2020a). The SIRT3 and SIRT6 Promote Prostate Cancer Progression by Inhibiting Necroptosis-Mediated Innate Immune Response. J. Immunol. Res. 2020, 8820355. doi:10.1155/2020/8820355
Fu, Y., Piao, C., Zhang, Z., Zhu, Y., Sun, S., Bi, J., et al. (2020b). Decreased Expression and Hypomethylation of HDAC9 lead to Poor Prognosis and Inhibit Immune Cell Infiltration in clear Cell Renal Cell Carcinoma. Urol. Oncol. 38 (9), 740–e9. e749. doi:10.1016/j.urolonc.2020.03.006
Gao, J., Wang, D., Liu, D., Liu, M., Ge, Y., Jiang, M., et al. (2015). Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand Induces the Expression of Proinflammatory Cytokines in Macrophages and Re-educates Tumor-Associated Macrophages to an Antitumor Phenotype. Mol. Biol. Cell 26 (18), 3178–3189. doi:10.1091/mbc.E15-04-0209
Gao, Y., Nihira, N. T., Bu, X., Chu, C., Zhang, J., Kolodziejczyk, A., et al. (2020). Acetylation-dependent Regulation of PD-L1 Nuclear Translocation Dictates the Efficacy of Anti-PD-1 Immunotherapy. Nat. Cell Biol 22 (9), 1064–1075. doi:10.1038/s41556-020-0562-4
Ghiboub, M., Zhao, J., Li Yim, A. Y. F., Schilderink, R., Verseijden, C., van Hamersveld, P. H. P., et al. (2020). HDAC3 Mediates the Inflammatory Response and LPS Tolerance in Human Monocytes and Macrophages. Front Immunol. 11, 550769. doi:10.3389/fimmu.2020.550769
Glozak, M. A., and Seto, E. (2007). Histone Deacetylases and Cancer. Oncogene 26 (37), 5420–5432. doi:10.1038/sj.onc.1210610
Gorshkov, K., Sima, N., Sun, W., Lu, B., Huang, W., Travers, J., et al. (2019). Quantitative Chemotherapeutic Profiling of Gynecologic Cancer Cell Lines Using Approved Drugs and Bioactive Compounds. Transl Oncol. 12 (3), 441–452. doi:10.1016/j.tranon.2018.11.016
Guan, C., Huang, X., Yue, J., Xiang, H., Shaheen, S., Jiang, Z., et al. (2021). SIRT3-mediated Deacetylation of NLRC4 Promotes Inflammasome Activation. Theranostics 11 (8), 3981–3995. doi:10.7150/thno.55573
Guo, L., Li, H., Fan, T., Ma, Y., and Wang, L. (2021). Synergistic Efficacy of Curcumin and Anti-programmed Cell Death-1 in Hepatocellular Carcinoma. Life Sci. 279, 119359. doi:10.1016/j.lfs.2021.119359
Gurvich, N., Tsygankova, O. M., Meinkoth, J. L., and Klein, P. S. (2004). Histone Deacetylase Is a Target of Valproic Acid-Mediated Cellular Differentiation. Cancer Res. 64 (3), 1079–1086. doi:10.1158/0008-5472.can-03-0799
Halkidou, K., Gnanapragasam, V. J., Mehta, P. B., Logan, I. R., Brady, M. E., Cook, S., et al. (2003). Expression of Tip60, an Androgen Receptor Coactivator, and its Role in Prostate Cancer Development. Oncogene 22 (16), 2466–2477. doi:10.1038/sj.onc.1206342
Hamminger, P., Marchetti, L., Preglej, T., Platzer, R., Zhu, C., Kamnev, A., et al. (2021). Histone Deacetylase 1 Controls CD4+ T Cell Trafficking in Autoinflammatory Diseases. J. Autoimmun. 119, 102610. doi:10.1016/j.jaut.2021.102610
Haque, M. E., Jakaria, M., Akther, M., Cho, D. Y., Kim, I. S., and Choi, D. K. (2021). The GCN5: its Biological Functions and Therapeutic Potentials. Clin. Sci. (Lond) 135 (1), 231–257. doi:10.1042/CS20200986
He, Z. X., Wei, B. F., Zhang, X., Gong, Y. P., Ma, L. Y., and Zhao, W. (2021). Current Development of CBP/p300 Inhibitors in the Last Decade. Eur. J. Med. Chem. 209, 112861. doi:10.1016/j.ejmech.2020.112861
Hickey, T. E., Selth, L. A., Chia, K. M., Laven-Law, G., Milioli, H. H., Roden, D., et al. (2021). The Androgen Receptor Is a Tumor Suppressor in Estrogen Receptor-Positive Breast Cancer. Nat. Med. 27 (2), 310–320. doi:10.1038/s41591-020-01168-7
Hou, P., Kapoor, A., Zhang, Q., Li, J., Wu, C. J., Li, J., et al. (2020). Tumor Microenvironment Remodeling Enables Bypass of Oncogenic KRAS Dependency in Pancreatic Cancer. Cancer Discov. 10 (7), 1058–1077. doi:10.1158/2159-8290.CD-19-0597
Hu, G., He, N., Cai, C., Cai, F., Fan, P., Zheng, Z., et al. (2019). HDAC3 Modulates Cancer Immunity via Increasing PD-L1 Expression in Pancreatic Cancer. Pancreatology 19 (2), 383–389. doi:10.1016/j.pan.2019.01.011
Hu, J., Bernatchez, C., Zhang, L., Xia, X., Kleinerman, E. S., Hung, M. C., et al. (2017). Induction of NKG2D Ligands on Solid Tumors Requires Tumor-specific CD8+ T Cells and Histone Acetyltransferases. Cancer Immunol. Res. 5 (4), 300–311. doi:10.1158/2326-6066.CIR-16-0234
Hu, J., Xia, X., Zhao, Q., and Li, S. (2021). Lysine Acetylation of NKG2D Ligand Rae-1 Stabilizes the Protein and Sensitizes Tumor Cells to NKG2D Immune Surveillance. Cancer Lett. 502, 143–153. doi:10.1016/j.canlet.2020.12.002
Huang, G., and Zhu, G. (2018). Sirtuin-4 (SIRT4), a Therapeutic Target with Oncogenic and Tumor-Suppressive Activity in Cancer. Onco Targets Ther. 11, 3395–3400. doi:10.2147/OTT.S157724
Huo, Q., Li, Z., Cheng, L., Yang, F., and Xie, N. (2020). SIRT7 Is a Prognostic Biomarker Associated with Immune Infiltration in Luminal Breast Cancer. Front Oncol. 10, 621. doi:10.3389/fonc.2020.00621
Iyer, N. G., Ozdag, H., and Caldas, C. (2004). p300/CBP and Cancer. Oncogene 23 (24), 4225–4231. doi:10.1038/sj.onc.1207118
Ji, J., Wang, J., Yang, J., Wang, X. P., Huang, J. J., Xue, T. F., et al. (2019). The Intra-nuclear SphK2-S1p Axis Facilitates M1-To-M2 Shift of Microglia via Suppressing HDAC1-Mediated KLF4 Deacetylation. Front Immunol. 10, 1241. doi:10.3389/fimmu.2019.01241
Jiang, C., Liu, J., Guo, M., Gao, X., Wu, X., Bai, N., et al. (2020). The NAD-dependent Deacetylase SIRT2 Regulates T Cell Differentiation Involved in Tumor Immune Response. Int. J. Biol. Sci. 16 (15), 3075–3084. doi:10.7150/ijbs.49735
Jin, X., Tian, S., and Li, P. (2017). Histone Acetyltransferase 1 Promotes Cell Proliferation and Induces Cisplatin Resistance in Hepatocellular Carcinoma. Oncol. Res. 25 (6), 939–946. doi:10.3727/096504016X14809827856524
Jochems, J., Boulden, J., Lee, B. G., Blendy, J. A., Jarpe, M., Mazitschek, R., et al. (2014). Antidepressant-like Properties of Novel HDAC6-Selective Inhibitors with Improved Brain Bioavailability. Neuropsychopharmacology 39 (2), 389–400. doi:10.1038/npp.2013.207
Jones, P. A., Ohtani, H., Chakravarthy, A., and De Carvalho, D. D. (2019). Epigenetic Therapy in Immune-Oncology. Nat. Rev. Cancer 19 (3), 151–161. doi:10.1038/s41568-019-0109-9
Jung, K. H., Noh, J. H., Kim, J. K., Eun, J. W., Bae, H. J., Xie, H. J., et al. (2012). HDAC2 Overexpression Confers Oncogenic Potential to Human Lung Cancer Cells by Deregulating Expression of Apoptosis and Cell Cycle Proteins. J. Cell Biochem 113 (6), 2167–2177. doi:10.1002/jcb.24090
Kalbasi, A., and Ribas, A. (2020). Tumour-intrinsic Resistance to Immune Checkpoint Blockade. Nat. Rev. Immunol. 20 (1), 25–39. doi:10.1038/s41577-019-0218-4
Karamouzis, M. V., Konstantinopoulos, P. A., and Papavassiliou, A. G. (2007). Roles of CREB-Binding Protein (CBP)/p300 in Respiratory Epithelium Tumorigenesis. Cell Res 17 (4), 324–332. doi:10.1038/cr.2007.10
Khan, A. N., Gregorie, C. J., and Tomasi, T. B. (2008). Histone Deacetylase Inhibitors Induce TAP, LMP, Tapasin Genes and MHC Class I Antigen Presentation by Melanoma Cells. Cancer Immunol. Immunother. 57 (5), 647–654. doi:10.1007/s00262-007-0402-4
Knox, T., Sahakian, E., Banik, D., Hadley, M., Palmer, E., Noonepalle, S., et al. (2019a). Author Correction: Selective HDAC6 Inhibitors Improve Anti-PD-1 Immune Checkpoint Blockade Therapy by Decreasing the Anti-inflammatory Phenotype of Macrophages and Down-Regulation of Immunosuppressive Proteins in Tumor Cells. Sci. Rep. 9 (1), 14824. doi:10.1038/s41598-019-51403-6
Knox, T., Sahakian, E., Banik, D., Hadley, M., Palmer, E., Noonepalle, S., et al. (2019b). Selective HDAC6 Inhibitors Improve Anti-PD-1 Immune Checkpoint Blockade Therapy by Decreasing the Anti-inflammatory Phenotype of Macrophages and Down-Regulation of Immunosuppressive Proteins in Tumor Cells. Sci. Rep. 9 (1), 6136. doi:10.1038/s41598-019-42237-3
Kosciuczuk, E. M., Mehrotra, S., Saleiro, D., Kroczynska, B., Majchrzak-Kita, B., Lisowski, P., et al. (2019). Sirtuin 2-mediated Deacetylation of Cyclin-dependent Kinase 9 Promotes STAT1 Signaling in Type I Interferon Responses. J. Biol. Chem. 294 (3), 827–837. doi:10.1074/jbc.RA118.005956
Kouzarides, T. (2007). Chromatin Modifications and Their Function. Cell 128 (4), 693–705. doi:10.1016/j.cell.2007.02.005
Krämer, O. H. (2009). HDAC2: a Critical Factor in Health and Disease. Trends Pharmacol. Sci. 30 (12), 647–655. doi:10.1016/j.tips.2009.09.007
Kumari, A., Cacan, E., Greer, S. F., and Garnett-Benson, C. (2013). Turning T Cells on: Epigenetically Enhanced Expression of Effector T-Cell Costimulatory Molecules on Irradiated Human Tumor Cells. J. Immunother. Cancer 1, 17. doi:10.1186/2051-1426-1-17
Lasko, L. M., Jakob, C. G., Edalji, R. P., Qiu, W., Montgomery, D., Digiammarino, E. L., et al. (2017). Discovery of a Selective Catalytic P300/CBP Inhibitor that Targets Lineage-specific Tumours. Nature 550 (7674), 128–132. doi:10.1038/nature24028
Lawrence, M., Daujat, S., and Schneider, R. (2016). Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 32 (1), 42–56. doi:10.1016/j.tig.2015.10.007
Lee, J. H., Park, S. M., Kim, O. S., Lee, C. S., Woo, J. H., Park, S. J., et al. (2009). Differential SUMOylation of LXRalpha and LXRbeta Mediates Transrepression of STAT1 Inflammatory Signaling in IFN-Gamma-Stimulated Brain Astrocytes. Mol. Cell 35 (6), 806–817. doi:10.1016/j.molcel.2009.07.021
Li, B., Samanta, A., Song, X., Iacono, K. T., Bembas, K., Tao, R., et al. (2007). FOXP3 Interactions with Histone Acetyltransferase and Class II Histone Deacetylases Are Required for Repression. Proc. Natl. Acad. Sci. U S A. 104 (11), 4571–4576. doi:10.1073/pnas.0700298104
Li, L., Shi, L., Yang, S., Yan, R., Zhang, D., Yang, J., et al. (2016a). SIRT7 Is a Histone Desuccinylase that Functionally Links to Chromatin Compaction and Genome Stability. Nat. Commun. 7, 12235. doi:10.1038/ncomms12235
Li, T., Zhang, C., Hassan, S., Liu, X., Song, F., Chen, K., et al. (2018). Histone Deacetylase 6 in Cancer. J. Hematol. Oncol. 11 (1), 111. doi:10.1186/s13045-018-0654-9
Li, X., Jiang, Y., Peterson, Y. K., Xu, T., Himes, R. A., Luo, X., et al. (2020a). Design of Hydrazide-Bearing HDACIs Based on Panobinostat and Their P53 and FLT3-ITD Dependency in Antileukemia Activity. J. Med. Chem. 63 (10), 5501–5525. doi:10.1021/acs.jmedchem.0c00442
Li, X., Su, X., Liu, R., Pan, Y., Fang, J., Cao, L., et al. (2021). HDAC Inhibition Potentiates Anti-tumor Activity of Macrophages and Enhances Anti-PD-L1-mediated Tumor Suppression. Oncogene 40 (10), 1836–1850. doi:10.1038/s41388-020-01636-x
Li, X., Zhang, Q., Ding, Y., Liu, Y., Zhao, D., Zhao, K., et al. (2016b). Methyltransferase Dnmt3a Upregulates HDAC9 to Deacetylate the Kinase TBK1 for Activation of Antiviral Innate Immunity. Nat. Immunol. 17 (7), 806–815. doi:10.1038/ni.3464
Li, Y., Liu, Y., Zhao, N., Yang, X., Li, Y., Zhai, F., et al. (2020b). Checkpoint Regulator B7x Is Epigenetically Regulated by HDAC3 and Mediates Resistance to HDAC Inhibitors by Reprogramming the Tumor Immune Environment in Colorectal Cancer. Cell Death Dis 11 (9), 753. doi:10.1038/s41419-020-02968-y
Li, Y., and Seto, E. (2016). HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect. Med. 6 (10). doi:10.1101/cshperspect.a026831
Li, Y., Zhang, X., Zhu, S., Dejene, E. A., Peng, W., Sepulveda, A., et al. (2020c). HDAC10 Regulates Cancer Stem-like Cell Properties in KRAS-Driven Lung Adenocarcinoma. Cancer Res. 80 (16), 3265–3278. doi:10.1158/0008-5472.CAN-19-3613
Li, Z., Qin, H., Li, J., Yu, L., Yang, Y., and Xu, Y. (2017). HADC5 Deacetylates MKL1 to Dampen TNF-α Induced Pro-inflammatory Gene Transcription in Macrophages. Oncotarget 8 (55), 94235–94246. doi:10.18632/oncotarget.21670
Lin, W., Chen, W., Liu, W., Xu, Z., and Zhang, L. (2019). Sirtuin4 Suppresses the Anti-neuroinflammatory Activity of Infiltrating Regulatory T Cells in the Traumatically Injured Spinal Cord. Immunology 158 (4), 362–374. doi:10.1111/imm.13123
Liu, J., He, D., Cheng, L., Huang, C., Zhang, Y., Rao, X., et al. (2020a). p300/CBP Inhibition Enhances the Efficacy of Programmed Death-Ligand 1 Blockade Treatment in Prostate Cancer. Oncogene 39 (19), 3939–3951. doi:10.1038/s41388-020-1270-z
Liu, J. R., Yu, C. W., Hung, P. Y., Hsin, L. W., and Chern, J. W. (2019a). High-selective HDAC6 Inhibitor Promotes HDAC6 Degradation Following Autophagy Modulation and Enhanced Antitumor Immunity in Glioblastoma. Biochem. Pharmacol. 163, 458–471. doi:10.1016/j.bcp.2019.03.023
Liu, S. S., Wu, F., Jin, Y. M., Chang, W. Q., and Xu, T. M. (2020b). HDAC11: a Rising star in Epigenetics. Biomed. Pharmacother. 131, 110607. doi:10.1016/j.biopha.2020.110607
Liu, T., Wang, X., Hu, W., Fang, Z., Jin, Y., Fang, X., et al. (2019b). Epigenetically Down-Regulated Acetyltransferase PCAF Increases the Resistance of Colorectal Cancer to 5-Fluorouracil. Neoplasia 21 (6), 557–570. doi:10.1016/j.neo.2019.03.011
Liu, X., Wang, Y., Zhang, R., Jin, T., Qu, L., Jin, Q., et al. (2020c). HDAC10 Is Positively Associated with PD-L1 Expression and Poor Prognosis in Patients with NSCLC. Front Oncol. 10, 485. doi:10.3389/fonc.2020.00485
Liu, Y., Bao, C., Wang, L., Han, R., Beier, U. H., Akimova, T., et al. (2019c). Complementary Roles of GCN5 and PCAF in Foxp3+ T-Regulatory Cells. Cancers (Basel) 11 (4). doi:10.3390/cancers11040554
Liu, Y., Du, M., and Lin, H. Y. (2021). Histone Deacetylase 9 Deficiency Exaggerates Uterine M2 Macrophage Polarization. J. Cell Mol Med 25 (16), 7690–7708. doi:10.1111/jcmm.16616
Liu, Y., Wang, L., Han, R., Beier, U. H., Akimova, T., Bhatti, T., et al. (2014). Two Histone/protein Acetyltransferases, CBP and P300, Are Indispensable for Foxp3+ T-Regulatory Cell Development and Function. Mol. Cell Biol 34 (21), 3993–4007. doi:10.1128/MCB.00919-14
Liu, Y., Wang, L., Predina, J., Han, R., Beier, U. H., Wang, L. C., et al. (2013). Inhibition of P300 Impairs Foxp3⁺ T Regulatory Cell Function and Promotes Antitumor Immunity. Nat. Med. 19 (9), 1173–1177. doi:10.1038/nm.3286
Llopiz, D., Ruiz, M., Villanueva, L., Iglesias, T., Silva, L., Egea, J., et al. (2019). Enhanced Anti-tumor Efficacy of Checkpoint Inhibitors in Combination with the Histone Deacetylase Inhibitor Belinostat in a Murine Hepatocellular Carcinoma Model. Cancer Immunol. Immunother. 68 (3), 379–393. doi:10.1007/s00262-018-2283-0
LoRusso, P. M., Demchik, L., Foster, B., Knight, J., Bissery, M. C., Polin, L. M., et al. (1996). Preclinical Antitumor Activity of CI-994. Invest New Drugs 14 (4), 349–356. doi:10.1007/BF00180810
Lu, D., Yan, J., Wang, L., Liu, H., Zeng, L., Zhang, M., et al. (2017). Design, Synthesis, and Biological Evaluation of the First C-Met/HDAC Inhibitors Based on Pyridazinone Derivatives. ACS Med. Chem. Lett. 8 (8), 830–834. doi:10.1021/acsmedchemlett.7b00172
Lu, W., Che, X., Qu, X., Zheng, C., Yang, X., Bao, B., et al. (2021). Succinylation Regulators Promote Clear Cell Renal Cell Carcinoma by Immune Regulation and RNA N6-Methyladenosine Methylation. Front Cell Dev Biol 9, 622198. doi:10.3389/fcell.2021.622198
Lu, Y., Stuart, J. H., Talbot-Cooper, C., Agrawal-Singh, S., Huntly, B., Smid, A. I., et al. (2019). Histone Deacetylase 4 Promotes Type I Interferon Signaling, Restricts DNA Viruses, and Is Degraded via Vaccinia Virus Protein C6. Proc. Natl. Acad. Sci. U S A. 116 (24), 11997–12006. doi:10.1073/pnas.1816399116
Lucas, J., Hsieh, T. C., Halicka, H. D., Darzynkiewicz, Z., and Wu, J. M. (2018). Upregulation of PD-L1 E-xpression by R-esveratrol and P-iceatannol in B-reast and C-olorectal C-ancer C-ells O-ccurs via HDAC3/p300-mediated NF-κB S-ignaling. Int. J. Oncol. 53 (4), 1469–1480. doi:10.3892/ijo.2018.4512
M, L., P, K., M, S., J, P., K V, W., C, L., et al. (2016). Essential Role of HDAC6 in the Regulation of PD-L1 in Melanoma. Mol. Oncol. 10 (5), 735–750. doi:10.1016/j.molonc.2015.12.012
Ma, H., Chang, H., Yang, W., Lu, Y., Hu, J., and Jin, S. (2020). A Novel IFNα-Induced Long Noncoding RNA Negatively Regulates Immunosuppression by Interrupting H3K27 Acetylation in Head and Neck Squamous Cell Carcinoma. Mol. Cancer 19 (1), 4. doi:10.1186/s12943-019-1123-y
Maeda, T., Towatari, M., Kosugi, H., and Saito, H. (2000). Up-regulation of Costimulatory/adhesion Molecules by Histone Deacetylase Inhibitors in Acute Myeloid Leukemia Cells. Blood 96 (12), 3847–3856. doi:10.1182/blood.v96.12.3847.h8003847_3847_3856
Maharaj, K., Powers, J. J., Mediavilla-Varela, M., Achille, A., Gamal, W., Quayle, S., et al. (2020). HDAC6 Inhibition Alleviates CLL-Induced T-Cell Dysfunction and Enhances Immune Checkpoint Blockade Efficacy in the Eμ-TCL1 Model. Front Immunol. 11, 590072. doi:10.3389/fimmu.2020.590072
Mattera, L., Escaffit, F., Pillaire, M. J., Selves, J., Tyteca, S., Hoffmann, J. S., et al. (2009). The p400/Tip60 Ratio Is Critical for Colorectal Cancer Cell Proliferation through DNA Damage Response Pathways. Oncogene 28 (12), 1506–1517. doi:10.1038/onc.2008.499
McGuire, A., Casey, M. C., Shalaby, A., Kalinina, O., Curran, C., Webber, M., et al. (2019). Quantifying Tip60 (Kat5) Stratifies Breast Cancer. Sci. Rep. 9 (1), 3819. doi:10.1038/s41598-019-40221-5
Meng, J., Liu, X., Zhang, P., Li, D., Xu, S., Zhou, Q., et al. (2016). Rb Selectively Inhibits Innate IFN-β Production by Enhancing Deacetylation of IFN-β Promoter through HDAC1 and HDAC8. J. Autoimmun. 73, 42–53. doi:10.1016/j.jaut.2016.05.012
Miao, B. P., Zhang, R. S., Yang, G., Sun, J. J., Tang, Y. Y., Liang, W. F., et al. (2018). Histone Acetyltransferase 1 up Regulates Bcl2L12 Expression in Nasopharyngeal Cancer Cells. Arch. Biochem. Biophys. 646, 72–79. doi:10.1016/j.abb.2018.03.040
Michan, S., and Sinclair, D. (2007). Sirtuins in Mammals: Insights into Their Biological Function. Biochem. J. 404 (1), 1–13. doi:10.1042/BJ20070140
Milling, S., and Edgar, J. M. (2019). How T'reg-ulate Healing of the Injured Spinal Cord? Immunology 158 (4), 253–254. doi:10.1111/imm.13148
Mohammadi, A., Sharifi, A., Pourpaknia, R., Mohammadian, S., and Sahebkar, A. (2018). Manipulating Macrophage Polarization and Function Using Classical HDAC Inhibitors: Implications for Autoimmunity and Inflammation. Crit. Rev. Oncol. Hematol. 128, 1–18. doi:10.1016/j.critrevonc.2018.05.009
Mormino, A., Cocozza, G., Fontemaggi, G., Valente, S., Esposito, V., Santoro, A., et al. (2021). Histone-deacetylase 8 Drives the Immune Response and the Growth of Glioma. Glia 69 (11), 2682–2698. doi:10.1002/glia.24065
Mounce, B. C., Mboko, W. P., Kanack, A. J., and Tarakanova, V. L. (2014). Primary Macrophages Rely on Histone Deacetylase 1 and 2 Expression to Induce Type I Interferon in Response to Gammaherpesvirus Infection. J. Virol. 88 (4), 2268–2278. doi:10.1128/JVI.03278-13
Myers, D. R., Lau, T., Markegard, E., Lim, H. W., Kasler, H., Zhu, M., et al. (2017). Tonic LAT-HDAC7 Signals Sustain Nur77 and Irf4 Expression to Tune Naive CD4 T Cells. Cell Rep 19 (8), 1558–1571. doi:10.1016/j.celrep.2017.04.076
Nakagawa, M., Oda, Y., Eguchi, T., Aishima, S., Yao, T., Hosoi, F., et al. (2007). Expression Profile of Class I Histone Deacetylases in Human Cancer Tissues. Oncol. Rep. 18 (4), 769–774. doi:10.3892/or.18.4.769
Narita, T., Weinert, B. T., and Choudhary, C. (2019). Functions and Mechanisms of Non-histone Protein Acetylation. Nat. Rev. Mol. Cell Biol 20 (3), 156–174. doi:10.1038/s41580-018-0081-3
Ning, Y., Ding, J., Sun, X., Xie, Y., Su, M., Ma, C., et al. (2020). HDAC9 Deficiency Promotes Tumor Progression by Decreasing the CD8+ Dendritic Cell Infiltration of the Tumor Microenvironment. J. Immunother. Cancer 8 (1). doi:10.1136/jitc-2020-000529
Ning, Z. Q., Li, Z. B., Newman, M. J., Shan, S., Wang, X. H., Pan, D. S., et al. (2012). Chidamide (CS055/HBI-8000): a New Histone Deacetylase Inhibitor of the Benzamide Class with Antitumor Activity and the Ability to Enhance Immune Cell-Mediated Tumor Cell Cytotoxicity. Cancer Chemother. Pharmacol. 69 (4), 901–909. doi:10.1007/s00280-011-1766-x
Núñez‐Álvarez, Y., and Suelves, M. (2021). HDAC11: a Multifaceted Histone Deacetylase with Proficient Fatty Deacylase Activity and its Roles in Physiological Processes. FEBS J. 23, 1. doi:10.1111/febs.15895
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996). The Transcriptional Coactivators P300 and CBP Are Histone Acetyltransferases. Cell 87 (5), 953–959. doi:10.1016/s0092-8674(00)82001-2
Osawa, Y., Kojika, E., Nishikawa, K., Kimura, M., Osakaya, S., Miyauchi, H., et al. (2019). Programmed Cell Death Ligand 1 (PD-L1) Blockade Attenuates Metastatic colon Cancer Growth in cAMP-Response Element-Binding Protein (CREB)-binding Protein (CBP)/β-catenin Inhibitor-Treated Livers. Oncotarget 10 (32), 3013–3026. doi:10.18632/oncotarget.26892
Osawa, Y., Oboki, K., Imamura, J., Kojika, E., Hayashi, Y., Hishima, T., et al. (2015). Inhibition of Cyclic Adenosine Monophosphate (cAMP)-Response Element-Binding Protein (CREB)-binding Protein (CBP)/β-Catenin Reduces Liver Fibrosis in Mice. EBioMedicine 2 (11), 1751–1758. doi:10.1016/j.ebiom.2015.10.010
Palmirotta, R., Cives, M., Della-Morte, D., Capuani, B., Lauro, D., Guadagni, F., et al. (2016). Sirtuins and Cancer: Role in the Epithelial-Mesenchymal Transition. Oxid Med. Cell Longev 2016, 3031459. doi:10.1155/2016/3031459
Peng, M., Mo, Y., Wang, Y., Wu, P., Zhang, Y., Xiong, F., et al. (2019). Neoantigen Vaccine: an Emerging Tumor Immunotherapy. Mol. Cancer 18 (1), 128. doi:10.1186/s12943-019-1055-6
Pillai, V. B., and Gupta, M. P. (2021). Is Nuclear Sirtuin SIRT6 a Master Regulator of Immune Function? Am. J. Physiol. Endocrinol. Metab. 320 (3), E399–E414. doi:10.1152/ajpendo.00483.2020
Poziello, A., Nebbioso, A., Stunnenberg, H. G., Martens, J. H. A., Carafa, V., and Altucci, L. (2021). Recent Insights into Histone Acetyltransferase-1: Biological Function and Involvement in Pathogenesis. Epigenetics 16 (8), 838–850. doi:10.1080/15592294.2020.1827723
Qiu, W., Wang, B., Gao, Y., Tian, Y., Tian, M., Chen, Y., et al. (2020). Targeting Histone Deacetylase 6 Reprograms Interleukin-17-Producing Helper T Cell Pathogenicity and Facilitates Immunotherapies for Hepatocellular Carcinoma. Hepatology 71 (6), 1967–1987. doi:10.1002/hep.30960
Que, Y., Zhang, X.-L., Liu, Z.-X., Zhao, J.-J., Pan, Q.-Z., Wen, X.-Z., et al. (2021). Frequent Amplification of HDAC Genes and Efficacy of HDAC Inhibitor Chidamide and PD-1 Blockade Combination in Soft Tissue Sarcoma. J. Immunother. Cancer 9 (2), e001696. doi:10.1136/jitc-2020-001696
Regna, N. L., Vieson, M. D., Luo, X. M., Chafin, C. B., Puthiyaveetil, A. G., Hammond, S. E., et al. (2016). Specific HDAC6 Inhibition by ACY-738 Reduces SLE Pathogenesis in NZB/W Mice. Clin. Immunol. 162, 58–73. doi:10.1016/j.clim.2015.11.007
Sakuraba, K., Yokomizo, K., Shirahata, A., Goto, T., Saito, M., Ishibashi, K., et al. (2011). TIP60 as a Potential Marker for the Malignancy of Gastric Cancer. Anticancer Res. 31 (1), 77–79.
Saleh, R., Toor, S. M., Sasidharan Nair, V., and Elkord, E. (2020). Role of Epigenetic Modifications in Inhibitory Immune Checkpoints in Cancer Development and Progression. Front Immunol. 11, 1469. doi:10.3389/fimmu.2020.01469
Sandhu, S. K., Volinia, S., Costinean, S., Galasso, M., Neinast, R., Santhanam, R., et al. (2012). miR-155 Targets Histone Deacetylase 4 (HDAC4) and Impairs Transcriptional Activity of B-Cell Lymphoma 6 (BCL6) in the Eμ-miR-155 Transgenic Mouse Model. Proc. Natl. Acad. Sci. U S A. 109 (49), 20047–20052. doi:10.1073/pnas.1213764109
Sanford, J. A., Zhang, L. J., Williams, M. R., Gangoiti, J. A., Huang, C. M., and Gallo, R. L. (2016). Inhibition of HDAC8 and HDAC9 by Microbial Short-Chain Fatty Acids Breaks Immune Tolerance of the Epidermis to TLR Ligands. Sci. Immunol. 1 (4), 1. doi:10.1126/sciimmunol.aah4609
Sarkar, R., Banerjee, S., Amin, S. A., Adhikari, N., and Jha, T. (2020). Histone Deacetylase 3 (HDAC3) Inhibitors as Anticancer Agents: A Review. Eur. J. Med. Chem. 192, 112171. doi:10.1016/j.ejmech.2020.112171
Shao, G., Liu, Y., Ma, T., Zhang, L., Yuan, M., and Zhao, S. (2018). GCN5 Inhibition Prevents IL-6-induced Prostate Cancer Metastases through PI3K/PTEN/Akt Signaling by Inactivating Egr-1. Biosci. Rep. 38 (6), 1. doi:10.1042/BSR20180816
Sharma, P., Hu-Lieskovan, S., Wargo, J. A., and Ribas, A. (2017). Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168 (4), 707–723. doi:10.1016/j.cell.2017.01.017
Sheikh, B. N., and Akhtar, A. (2019). The many Lives of KATs - Detectors, Integrators and Modulators of the Cellular Environment. Nat. Rev. Genet. 20 (1), 7–23. doi:10.1038/s41576-018-0072-4
Shiota, M., Yokomizo, A., Masubuchi, D., Tada, Y., Inokuchi, J., Eto, M., et al. (2010). Tip60 Promotes Prostate Cancer Cell Proliferation by Translocation of Androgen Receptor into the Nucleus. Prostate 70 (5), 540–554. doi:10.1002/pros.21088
Song, H. Y., Biancucci, M., Kang, H. J., O'Callaghan, C., Park, S. H., Principe, D. R., et al. (2016). SIRT2 Deletion Enhances KRAS-Induced Tumorigenesis In Vivo by Regulating K147 Acetylation Status. Oncotarget 7 (49), 80336–80349. doi:10.18632/oncotarget.12015
Song, L., Chen, X., Mi, L., Liu, C., Zhu, S., Yang, T., et al. (2020). Icariin-induced Inhibition of SIRT6/NF-Κb Triggers Redox Mediated Apoptosis and Enhances Anti-tumor Immunity in Triple-Negative Breast Cancer. Cancer Sci. 111 (11), 4242–4256. doi:10.1111/cas.14648
Sundaresan, N. R., Vasudevan, P., Zhong, L., Kim, G., Samant, S., Parekh, V., et al. (2012). The Sirtuin SIRT6 Blocks IGF-Akt Signaling and Development of Cardiac Hypertrophy by Targeting C-Jun. Nat. Med. 18 (11), 1643–1650. doi:10.1038/nm.2961
Tang, M., Tang, H., Tu, B., and Zhu, W. G. (2021). SIRT7: a sentinel of Genome Stability. Open Biol. 11 (6), 210047. doi:10.1098/rsob.210047
Tay, R. E., Olawoyin, O., Cejas, P., Xie, Y., Meyer, C. A., Weng, Q. Y., et al. (2020). Hdac3 Is an Epigenetic Inhibitor of the Cytotoxicity Program in CD8 T Cells. J. Exp. Med. 217 (7), 1. doi:10.1084/jem.20191453
Tomaselli, D., Steegborn, C., Mai, A., and Rotili, D. (2020). Sirt4: A Multifaceted Enzyme at the Crossroads of Mitochondrial Metabolism and Cancer. Front Oncol. 10, 474. doi:10.3389/fonc.2020.00474
Tong, L., Liang, H., Zhuang, H., Liu, C., and Zhang, Z. (2020). The Relationship between HDAC3 and Malignant Tumors: A Mini Review. Crit. Rev. Eukaryot. Gene Expr. 30 (3), 279–284. doi:10.1615/CritRevEukaryotGeneExpr.2020034380
van Loosdregt, J., Vercoulen, Y., Guichelaar, T., Gent, Y. Y., Beekman, J. M., van Beekum, O., et al. (2010). Regulation of Treg Functionality by Acetylation-Mediated Foxp3 Protein Stabilization. Blood 115 (5), 965–974. doi:10.1182/blood-2009-02-207118
Vanamee, É. S., and Faustman, D. L. (2017). TNFR2: A Novel Target for Cancer Immunotherapy. Trends Mol. Med. 23 (11), 1037–1046. doi:10.1016/j.molmed.2017.09.007
Vaquero, A., Scher, M., Lee, D., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2004). Human SirT1 Interacts with Histone H1 and Promotes Formation of Facultative Heterochromatin. Mol. Cell 16 (1), 93–105. doi:10.1016/j.molcel.2004.08.031
Villagra, A., Cheng, F., Wang, H. W., Suarez, I., Glozak, M., Maurin, M., et al. (2009). The Histone Deacetylase HDAC11 Regulates the Expression of Interleukin 10 and Immune Tolerance. Nat. Immunol. 10 (1), 92–100. doi:10.1038/ni.1673
Villagra, A., Sotomayor, E. M., and Seto, E. (2010). Histone Deacetylases and the Immunological Network: Implications in Cancer and Inflammation. Oncogene 29 (2), 157–173. doi:10.1038/onc.2009.334
Wang, H., Fu, C., Du, J., Wang, H., He, R., Yin, X., et al. (2020a). Enhanced Histone H3 Acetylation of the PD-L1 Promoter via the COP1/c-Jun/HDAC3 axis Is Required for PD-L1 Expression in Drug-Resistant Cancer Cells. J. Exp. Clin. Cancer Res. 39 (1), 29. doi:10.1186/s13046-020-1536-x
Wang, L., Kumar, S., Dahiya, S., Wang, F., Wu, J., Newick, K., et al. (2016). Ubiquitin-specific Protease-7 Inhibition Impairs Tip60-dependent Foxp3+ T-Regulatory Cell Function and Promotes Antitumor Immunity. EBioMedicine 13, 99–112. doi:10.1016/j.ebiom.2016.10.018
Wang, L. T., Liu, K. Y., Jeng, W. Y., Chiang, C. M., Chai, C. Y., Chiou, S. S., et al. (2020b). PCAF-mediated Acetylation of ISX Recruits BRD4 to Promote Epithelial-Mesenchymal Transition. EMBO Rep. 21 (2), e48795. doi:10.15252/embr.201948795
Wang, S., He, Z., Wang, X., Li, H., and Liu, X. S. (2019a). Antigen Presentation and Tumor Immunogenicity in Cancer Immunotherapy Response Prediction. Elife 8, 1. doi:10.7554/eLife.49020
Wang, T., Yao, W., Shao, Y., Zheng, R., and Huang, F. (2018a). PCAF fine-tunes Hepatic Metabolic Syndrome, Inflammatory Disease, and Cancer. J. Cell Mol Med 22 (12), 5787–5800. doi:10.1111/jcmm.13877
Wang, X., Li, H., Chen, S., He, J., Chen, W., Ding, Y., et al. (2021). P300/CBP-associated Factor (PCAF) Attenuated M1 Macrophage Inflammatory Responses Possibly through KLF2 and KLF4. Immunol. Cell Biol 99 (7), 724–736. doi:10.1111/imcb.12455
Wang, Y., Yang, J., Hong, T., Chen, X., and Cui, L. (2019b). SIRT2: Controversy and Multiple Roles in Disease and Physiology. Ageing Res. Rev. 55, 100961. doi:10.1016/j.arr.2019.100961
Wang, Y., Yun, C., Gao, B., Xu, Y., Zhang, Y., Wang, Y., et al. (2017). The Lysine Acetyltransferase GCN5 Is Required for iNKT Cell Development through EGR2 Acetylation. Cell Rep 20 (3), 600–612. doi:10.1016/j.celrep.2017.06.065
Wang, Y. F., Liu, F., Sherwin, S., Farrelly, M., Yan, X. G., Croft, A., et al. (2018b). Cooperativity of HOXA5 and STAT3 Is Critical for HDAC8 Inhibition-Mediated Transcriptional Activation of PD-L1 in Human Melanoma Cells. J. Invest Dermatol. 138 (4), 922–932. doi:10.1016/j.jid.2017.11.009
Wang, Z., Qin, G., and Zhao, T. C. (2014). HDAC4: Mechanism of Regulation and Biological Functions. Epigenomics 6 (1), 139–150. doi:10.2217/epi.13.73
Wijdeven, R. H., van Luijn, M. M., Wierenga-Wolf, A. F., Akkermans, J. J., van den Elsen, P. J., Hintzen, R. Q., et al. (2018). Chemical and Genetic Control of IFNγ-Induced MHCII Expression. EMBO Rep. 19 (9), 1. doi:10.15252/embr.201745553
Willis-Martinez, D., Richards, H. W., Timchenko, N. A., and Medrano, E. E. (2010). Role of HDAC1 in Senescence, Aging, and Cancer. Exp. Gerontol. 45 (4), 279–285. doi:10.1016/j.exger.2009.10.001
Woan, K. V., Lienlaf, M., Perez-Villaroel, P., Lee, C., Cheng, F., Knox, T., et al. (2015). Targeting Histone Deacetylase 6 Mediates a Dual Anti-melanoma Effect: Enhanced Antitumor Immunity and Impaired Cell Proliferation. Mol. Oncol. 9 (7), 1447–1457. doi:10.1016/j.molonc.2015.04.002
Woods, D. M., Sodré, A. L., Villagra, A., Sarnaik, A., Sotomayor, E. M., and Weber, J. (2015). HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol. Res. 3 (12), 1375–1385. doi:10.1158/2326-6066.CIR-15-0077-T
Woods, D. M., Woan, K. V., Cheng, F., Sodré, A. L., Wang, D., Wu, Y., et al. (2017). T Cells Lacking HDAC11 Have Increased Effector Functions and Mediate Enhanced Alloreactivity in a Murine Model. Blood 130 (2), 146–155. doi:10.1182/blood-2016-08-731505
Wu, C., Li, A., Hu, J., and Kang, J. (2019). Histone Deacetylase 2 Is Essential for LPS-Induced Inflammatory Responses in Macrophages. Immunol. Cell Biol 97 (1), 72–84. doi:10.1111/imcb.12203
Xiang, J., Zhang, N., Sun, H., Su, L., Zhang, C., Xu, H., et al. (2020). Disruption of SIRT7 Increases the Efficacy of Checkpoint Inhibitor via MEF2D Regulation of Programmed Cell Death 1 Ligand 1 in Hepatocellular Carcinoma Cells. Gastroenterology 158 (3), 664–e24. doi:10.1053/j.gastro.2019.10.025
Xiao, H., Jiao, J., Wang, L., O'Brien, S., Newick, K., Wang, L. C., et al. (2016). HDAC5 Controls the Functions of Foxp3(+) T-Regulatory and CD8(+) T Cells. Int. J. Cancer 138 (10), 2477–2486. doi:10.1002/ijc.29979
Xiao, Y., Nagai, Y., Deng, G., Ohtani, T., Zhu, Z., Zhou, Z., et al. (2014). Dynamic Interactions between TIP60 and P300 Regulate FOXP3 Function through a Structural Switch Defined by a Single Lysine on TIP60. Cell Rep 7 (5), 1471–1480. doi:10.1016/j.celrep.2014.04.021
Xie, Z., Ago, Y., Okada, N., and Tachibana, M. (2018). Valproic Acid Attenuates Immunosuppressive Function of Myeloid-Derived Suppressor Cells. J. Pharmacol. Sci. 137 (4), 359–365. doi:10.1016/j.jphs.2018.06.014
Xiong, Y., Wang, L., Di Giorgio, E., Akimova, T., Beier, U. H., Han, R., et al. (2020). Inhibiting the Coregulator CoREST Impairs Foxp3+ Treg Function and Promotes Antitumor Immunity. J. Clin. Invest 130 (4), 1830–1842. doi:10.1172/JCI131375
Xu, P., Ye, S., Li, K., Huang, M., Wang, Q., Zeng, S., et al. (2019). NOS1 Inhibits the Interferon Response of Cancer Cells by S-Nitrosylation of HDAC2. J. Exp. Clin. Cancer Res. 38 (1), 483. doi:10.1186/s13046-019-1448-9
Yamada, K., Hori, Y., Inoue, S., Yamamoto, Y., Iso, K., Kamiyama, H., et al. (2021). E7386, a Selective Inhibitor of the Interaction between β-Catenin and CBP, Exerts Antitumor Activity in Tumor Models with Activated Canonical Wnt Signaling. Cancer Res. 81 (4), 1052–1062. doi:10.1158/0008-5472.CAN-20-0782
Yan, Z., Yao, S., Liu, Y., Zhang, J., Li, P., Wang, H., et al. (2020). Durable Response to Sintilimab and Chidamide in a Patient with Pegaspargase- and Immunotherapy-Resistant NK/T-Cell Lymphoma: Case Report and Literature Review. Front Oncol. 10, 608304. doi:10.3389/fonc.2020.608304
Yang, C., Croteau, S., and Hardy, P. (2021a). Histone Deacetylase (HDAC) 9: Versatile Biological Functions and Emerging Roles in Human Cancer. Cell Oncol. 44, 997–1017. doi:10.1007/s13402-021-00626-9
Yang, G., Yuan, Y., Yuan, H., Wang, J., Yun, H., Geng, Y., et al. (2021b). Histone Acetyltransferase 1 Is a Succinyltransferase for Histones and Non-histones and Promotes Tumorigenesis. EMBO Rep. 22 (2), e50967. doi:10.15252/embr.202050967
Yang, J., Gong, C., Ke, Q., Fang, Z., Chen, X., Ye, M., et al. (2021c). Insights into the Function and Clinical Application of HDAC5 in Cancer Management. Front Oncol. 11, 661620. doi:10.3389/fonc.2021.661620
Yang, M., Li, Z., Tao, J., Hu, H., Li, Z., Zhang, Z., et al. (2021d). Resveratrol Induces PD-L1 Expression through Snail-Driven Activation of Wnt Pathway in Lung Cancer Cells. J. Cancer Res. Clin. Oncol. 147 (4), 1101–1113. doi:10.1007/s00432-021-03510-z
Yang, P. M., Lin, P. J., and Chen, C. C. (2012). CD1d Induction in Solid Tumor Cells by Histone Deacetylase Inhibitors through Inhibition of HDAC1/2 and Activation of Sp1. Epigenetics 7 (4), 390–399. doi:10.4161/epi.19373
Yang, W., Feng, Y., Zhou, J., Cheung, O. K., Cao, J., Wang, J., et al. (2021e). A Selective HDAC8 Inhibitor Potentiates Antitumor Immunity and Efficacy of Immune Checkpoint Blockade in Hepatocellular Carcinoma. Sci. Transl Med. 13 (588), 1. doi:10.1126/scitranslmed.aaz6804
Ye, F., Jiang, J., Zong, C., Yang, X., Gao, L., Meng, Y., et al. (2020). Sirt1-Overexpressing Mesenchymal Stem Cells Drive the Anti-tumor Effect through Their Pro-inflammatory Capacity. Mol. Ther. 28 (3), 874–888. doi:10.1016/j.ymthe.2020.01.018
Yu, Y., Liu, Y., Zong, C., Yu, Q., Yang, X., Liang, L., et al. (2016a). Mesenchymal Stem Cells with Sirt1 Overexpression Suppress Breast Tumor Growth via Chemokine-dependent Natural Killer Cells Recruitment. Sci. Rep. 6, 35998. doi:10.1038/srep35998
Yu, Y., Zhang, Q., Meng, Q., Zong, C., Liang, L., Yang, X., et al. (2016b). Mesenchymal Stem Cells Overexpressing Sirt1 Inhibit Prostate Cancer Growth by Recruiting Natural Killer Cells and Macrophages. Oncotarget 7 (44), 71112–71122. doi:10.18632/oncotarget.12737
Yu, Z., Zeng, J., Liu, H., Wang, T., Yu, Z., and Chen, J. (2019). Role of HDAC1 in the Progression of Gastric Cancer and the Correlation with lncRNAs. Oncol. Lett. 17 (3), 3296–3304. doi:10.3892/ol.2019.9962
Zhang, Q., Zhao, K., Shen, Q., Han, Y., Gu, Y., Li, X., et al. (2015). Tet2 Is Required to Resolve Inflammation by Recruiting Hdac2 to Specifically Repress IL-6. Nature 525 (7569), 389–393. doi:10.1038/nature15252
Zhang, X., Wu, J., and Luan, Y. (2017). Tip60: Main Functions and its Inhibitors. Mini Rev. Med. Chem. 17 (8), 675–682. doi:10.2174/1389557516666160923125031
Zheng, X., Gai, X., Ding, F., Lu, Z., Tu, K., Yao, Y., et al. (2013). Histone Acetyltransferase PCAF Up-Regulated Cell Apoptosis in Hepatocellular Carcinoma via Acetylating Histone H4 and Inactivating AKT Signaling. Mol. Cancer 12 (1), 96. doi:10.1186/1476-4598-12-96
Zhou, B., Yang, Y., and Li, C. (2019). SIRT1 Inhibits Hepatocellular Carcinoma Metastasis by Promoting M1 Macrophage Polarization via NF-Κb Pathway. Onco Targets Ther. 12, 2519–2529. doi:10.2147/OTT.S195234
Zhou, Y., Bastian, I. N., Long, M. D., Dow, M., Li, W., Liu, T., et al. (2021). Activation of NF-Κb and P300/CBP Potentiates Cancer Chemoimmunotherapy through Induction of MHC-I Antigen Presentation. Proc. Natl. Acad. Sci. USA 118 (8), e2025840118. doi:10.1073/pnas.2025840118
Zhu, C., Chen, Q., Xie, Z., Ai, J., Tong, L., Ding, J., et al. (2011). The Role of Histone Deacetylase 7 (HDAC7) in Cancer Cell Proliferation: Regulation on C-Myc. J. Mol. Med. (Berl) 89 (3), 279–289. doi:10.1007/s00109-010-0701-7
Glossary
HAT histone acetyltransferase
HDAC histone deacetylase
ICs immune checkpoints
PD-1 programmed cell death protein 1
PD-L1 programmed death-ligand 1
CTLA-4 cytotoxic T lymphocyte-associated protein 4
LAG lymphocyte activation gene
ACT adoptive T-cell therapy
ICB immune checkpoint blockade
MHC major histocompatibility class
CTLs cytotoxic T cells
IL interleukin
Tregs regulatory T cells
IRF interferon regulatory factor
IFN interferon
TNF tumor necrosis factor
NK natural killer cell
Sirt Sirtuin
NF- κ B nucleus factor κ-light-chain enhancer of activated B cells
CXCL C-X-C motif chemokine ligand
EMT epithelial-mesenchymal transition
MDSCs myeloid-derived suppressor cells
NSCLC Non-small-cell carcinoma
Lys lysine
Ser serine
JAK Janus kinase
STAT signal transducer and activator of transcription
TAM tumor associated macrophage
CoREST repressor element 1 silencing transcription factor corepressor
DLBCL diffuse large B cell lymphoma
Foxo forkhead box O
NCOR2 nucleus receptor co-repressor 2
EZH enhancer of zeste homolog 2
APC antigen-presenting cell
TEM effector memory T
RIPK receptor interacting protein kinase
MLKL mixed lineage kinase domain like pseudokinase
TGF transforming growth factor
AMPK AMP-activated protein kinase
GBM glioblastoma
Keywords: acetylated modifications, tumor immunity, acetylases, deacetylases, cancer immunotherapy
Citation: Lu J, He X, Zhang L, Zhang R and Li W (2021) Acetylation in Tumor Immune Evasion Regulation. Front. Pharmacol. 12:771588. doi: 10.3389/fphar.2021.771588
Received: 06 September 2021; Accepted: 05 November 2021;
Published: 22 November 2021.
Edited by:
Yi-Chao Zheng, Zhengzhou University, ChinaCopyright © 2021 Lu, He, Zhang, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzheng Li, d2VuemhlbjcyN0AxNjMuY29t; Ran Zhang, WlIxMzk3MUBodW5udS5lZHU=
†These authors have contributed equally to this work and share first authorship
 Jun Lu
Jun Lu Xiang He
Xiang He Lijuan Zhang
Lijuan Zhang Ran Zhang1*
Ran Zhang1* Wenzheng Li
Wenzheng Li