- 1Department of Nephrology, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Hunan Key Laboratory of Kidney Disease and Blood Purification, Changsha, China
The kidney is an energy-consuming organ, and cellular metabolism plays an indispensable role in kidney-related diseases. Caveolin-1 (Cav-1), a multifunctional membrane protein, is the main component of caveolae on the plasma membrane. Caveolae are represented by tiny invaginations that are abundant on the plasma membrane and that serve as a platform to regulate cellular endocytosis, stress responses, and signal transduction. However, caveolae have received increasing attention as a metabolic platform that mediates the endocytosis of albumin, cholesterol, and glucose, participates in cellular metabolic reprogramming and is involved in the progression of kidney disease. It is worth noting that caveolae mainly depend on Cav-1 to perform the abovementioned cellular functions. Furthermore, the mechanism by which Cav-1 regulates cellular metabolism and participates in the pathophysiology of kidney diseases has not been completely elucidated. In this review, we introduce the structure and function of Cav-1 and its functions in regulating cellular metabolism, autophagy, and oxidative stress, focusing on the relationship between Cav-1 in cellular metabolism and kidney disease; in addition, Cav-1 that serves as a potential therapeutic target for treatment of kidney disease is also described.
1 Introduction
Kidney disease is currently a challenging public health problem worldwide and is receiving increasing attention. The kidney is a highly metabolically active organ, and cellular energy metabolism is very important for maintaining kidney homeostasis (Gewin, 2021). In addition to inflammation, metabolic disorders are another form of diseased kidney dysfunction and play a key role in renal fibrosis (Kang et al., 2015). The scaffold protein caveolin-1 (Cav-1) on the cell membrane is a key protein to maintain energy homeostasis by regulating energy metabolism and mediating the signal transduction of glucose and lipid metabolism (Baudrand et al., 2016), which is closely associated with metabolic-related diseases such as diabetes (Bonds et al., 2019; Haddad et al., 2020), obesity (Chang et al., 2017), cardiovascular disease (Mayurasakorn et al., 2018), and cancer (Sotgia et al., 2012; Zhang Z. et al., 2020). With increased research on Cav-1, it has been demonstrated that Cav-1 plays an important role in the process of renal fibrosis (Shihata et al., 2017). However, the mechanism by which Cav-1 regulates cellular metabolism and kidney disease is not clearly understood. This review comprehensively describes the structure, expression, and regulation as well as the associated signaling pathway of Cav-1, which may be a potential drug target for metabolic-related kidney disease. Specifically, we describe the role of Cav-1 in regulating cellular glucose and lipid metabolism, cellular oxidative stress, and autophagy.
2 Caveolin Family
The caveolin family consists of three members, namely, caveolin-1 (Cav-1), caveolin-2 (Cav-2), and caveolin-3 (Cav-3). Cav-1 was identified in 1989 by Glenney et al. as the first family member of caveolin proteins (Glenney, 1989). They isolated a 22 kDa cytoskeleton protein from Rous sarcoma virus–transformed chicken fibroblasts, which was a substrate for tyrosine phosphorylation (Glenney, 1989). Subsequently, Rothberg et al. found that the 22 kDa membrane protein was part of the plasma membrane of the cell and named it caveolin (Rothberg et al., 1992). In addition, it has been demonstrated that the VIP21 protein (21 kDa) from canine renal epithelial cells (MDCK) has only eight–amino acid (aa) sequences that differ from human Cav-1. VIP21 is equivalent to caveolin in canines; thus, Cav-1 is also known as VIP21 (Glenney, 1992; Kurzchalia et al., 1992). Furthermore, caveolin was officially renamed Cav-1 in 1996 by Scherer et al. (Scherer et al., 1996). After they identified a vesicular protein–related protein, Cav-2, of about 20 kDa by microsequencing the vesicular protein–enriched membrane of adipocytes (Scherer et al., 1996). Subsequently, Tang et al. (Tang et al., 1996) found the same DNA sequence as CAV1 in rat hearts using a gene probe, namely, CAV3 (about 17 KDa), which was 65% identical and 85% similar to CAV1, whereas CAV1 and CAV2 had 38% identical and 58% similar DNA sequences.
CAV1 and CAV2 are both located on human chromosome 7q31.1, whereas CAV3 is located on 3p25 (Engelman et al., 1998; Williams and Lisanti, 2004). Cav-1 exists in two isoforms (α and β), and each monomer has a hairpin conformation capable of dimerization (Fujimoto et al., 2000; Kogo and Fujimoto, 2000; Williams and Lisanti, 2004). Cav-1α contains 1–178 (24 kDa) residues, whereas Cav-1β contains 32–178 (21 kDa) residues (Scherer et al., 1995). In addition, the protein translation start site of Cav-1α starts from the first aa, whereas Cav-1β starts from the 32nd aa residue; thus, their N-terminal sequences are different (Scherer et al., 1995). Previous studies have suggested that these two monomers are encoded by different mRNAs (Kogo and Fujimoto, 2000). Furthermore, it is worth noting that only Cav-1α has tyrosine-14 phosphorylation sites but is not seen in Cav-1β due to the absence of a specific sequence at the N-terminus (Scherer et al., 1995; Vainonen et al., 2004). Cav-1 and Cav-2 are coexpressed in a variety of mammalian cells [such as endothelial cells (ECs), vascular smooth muscle cells, type I pneumocytes, and liver and kidney cells], whereas Cav-3 is mainly expressed not only in muscle cell but also in other cell types, such as astrocytes (Ikezu et al., 1998; Williams and Lisanti, 2004). Interestingly, it has been reported that Cav-1 is mainly in the glomeruli of the kidney (Breton et al., 1998; Sörensson et al., 2002; Moriyama et al., 2011). Furthermore, the subcellular localization of caveolins has been shown to be located in Golgi apparatus, endoplasmic reticulum (ER), mitochondria, nucleus, peroxisomes, lipid droplets (LDs) (Fridolfsson et al., 2014), and mitochondria-associated membranes (MAMs) (Sala-Vila et al., 2016). The Cav-1 protein sequence consists of four domains: an N-terminal domain (1–81 aa), an oligomerization domain (61–101 aa) including the scaffolding domain (82–101 aa), an intramembrane domain (102–134 aa), and a C-terminal domain (135–178 aa) (Filippini and D'Alessio, 2020; Root et al., 2015; Wong et al., 2020). Both the N-terminus and C-terminus of Cav-1 are exposed to the cytoplasm, and the C-terminal domain contains ubiquitination sites, whereas its N-terminal domain contains two phosphorylation sites: tyrosine-14 (Li et al., 1996b) and serine-80 (Schlegel et al., 2001). Tyrosine-14 (Y14) can be phosphorylated by the Src tyrosine kinase family (such as Src, Abl, and Fyn) (Wong et al., 2020) to regulate signaling proteins, whereas serine-80 (S80) phosphorylated Cav-1 is mainly targeted to the ER and participates in the secretion of Cav-1 (Schlegel et al., 2001). The C-terminal domain contains three palmitoylation sites (Cys133/143/156), and the palmitoylation site and the intramembrane domain (in the form of a U-shaped conformation) are embedded in the cell lipid bilayer (Hoop et al., 2012; Root et al., 2015; Park et al., 2019). In addition, the scaffold domain of Cav-1 carrying a cholesterol recognition aa consensus can bind cholesterol (Yang et al., 2014; Krishna and Sengupta, 2019) and other subtypes of caveolin, which is necessary for the formation of caveolin homo or hetero oligomers (Epand et al., 2005; Hoop et al., 2012).
Emerging evidence has shown that the scaffold domain of Cav-1 (CSD) plays a key role in tumor progression and cellular metabolic reprogramming (Bernatchez, 2020; Gopu et al., 2020). The CSD is a highly hydrophobic region composed of a 20-aa stretch of caveolin residues (Li et al., 1996a; Wong et al., 2021). It can bind proteins (such as src family kinases and G protein subunits) that contain a caveolin binding motif (CBM) consensus (Li et al., 1996a; Collins et al., 2012). The CBM includes three motifs: ΩXΩXXXXΩXXΩ, ΩXXXΩXXΩ, and the combination sequence ΩXΩXXXXΩXΩ (Ω is a Phe, Tyr, or Trp residue, X can be any aa) (Collins et al., 2012). Couet et al. found that the epidermal growth factor receptor (EGFR) kinase interacts directly with the caveolin scaffolding domain via its conserved caveolin binding motif, which may mediate caveolae signaling (Couet et al., 1997). However, it remains controversial whether CSD/CBM binding effectively mediates caveolae-related signaling. For instance, Collins et al. (Collins et al., 2012) stated that the CBM in most signal proteins is buried and difficult to access, and CBM sequences in caveolae-associated proteins are not abundant; thus, the interaction of CBM/CSD-mediated caveolae signaling needs to be reassessed (Collins et al., 2012). Nevertheless, Zakrzewicz et al. determined that the glycolytic enzyme enolase 1 with a CBM corecognition sequence can be transported to the cell membrane and combined with the CSD of Cav-1, which may affect cell migration and cell invasion (Zakrzewicz et al., 2014). It is worth noting that the consensus CBM of enolase 1 is not buried in the protein core but exposed on the surface of the protein, which makes it accessible for interactions with the CSD (Zakrzewicz et al., 2014). Furthermore, Okada et al. confirmed that the CSD domain of Cav-1 plays a key role in the cell cycle, migration, and proliferation of cancer cells and may provide a platform for specific signal transduction (Okada et al., 2019). In summary, caveolin may regulate other protein activities through CSD binding to the CBM, and its activity depends on binding to the CBM of other proteins.
3 Regulation of Caveolin-1 Expression
The expression of Cav-1 is regulated by three ways: genomic epigenetic modification, transcription, and posttranscriptional regulation mechanisms. The genomic epigenetic modification of CAV1 can be divided into DNA methylation modification and histone modification (such as methylation, acetylation, phosphorylation, and ubiquitination) (Yan et al., 2020). Zschocke et al. (Zschocke et al., 2002) found that ectopic expression of estrogen receptor caused DNA methylation or histone deacetylation changes of CAV1 in nerve cells, thereby regulating its mRNA expression. Subsequently, Sanders et al. (Sanders et al., 2017) found that after treatment of normal fibroblasts with transforming growth factor–β1 (TGF-β1), the mRNA expression of CAV1 was downregulated through histone modification (H3 lysine 4 trimethylation) instead of DNA methylation, which may be associated with idiopathic pulmonary fibrosis. In addition, Yan et al. reviewed the possible mechanism of CAV1 DNA methylation in chronic lung disease and regarded it as a possible target for early diagnosis and treatment of the chronic lung disease (Yan et al., 2020). However, whether its application as a biomarker requires further research.
The promoter sequence of CAV1 includes three G+C-rich potential sterol regulatory elements (SREs), a CAAT sequence, a Sp1 consensus sequence (Bist et al., 1997; Dasari et al., 2006; Chen et al., 2011), and a functional peroxisome proliferator response element (Llaverias et al., 2004). Therefore, the increase in intracellular free cholesterol can regulate the mRNA expression of CAV1 by stimulating the binding of SRE-binding protein 1 (SPEBP-1) to the cholesterol regulatory element in the CAV1 promoter (Bist et al., 1997). Other transcription factors, including P53 (Bist et al., 2000), c-myc (Xie et al., 2003), GAT-binding factor 6 (GAT-6) (Boopathi et al., 2011), NF-κB (Thangavel et al., 2019), forkhead box O (FoxO) (van den Heuvel et al., 2005), and FoxM1 (Huang et al., 2012), can also bind to the related E-BOX of the promoter of the CAV1 and regulate its transcription. These findings suggest that the Cav-1 may have a potential regulatory role in cellular metabolism, inflammation, and fibrosis.
Many factors contribute to the posttranscriptional regulation of CAV1, such as microRNA (miRNA), long noncoding RNAs (lncRNA), and circular RNA (circRNA), as well as other proteins. Among them, miRNAs, including miR-199a-5p (Zhong et al., 2020), miR-96 (Chen et al., 2020), miR-124 (Torrejón et al., 2017), miR-103/107 (Zhang et al., 2015), miR-204 (Huang et al., 2019), miR-130a (Wang et al., 2016), and miR-103-3p (Wang et al., 2021), have been demonstrated that to recognize homologous CAV1 mRNA and cause the degradation of CAV1 mRNA or inhibit its translation. In addition, lncRNAs, including lncRNA ANRIL (Zhong et al., 2020), lnc-BMP1-1 (Ling et al., 2020), and lncRNA IMFLNC1 (Wang J. et al., 2020), can regulate the protein expression and function of Cav-1. A recent study has shown that circRNA though some miRNAs, playing key role in posttranscriptional regulation of CAV1 (Luo Z. et al., 2020; Zhu et al., 2020). Zhao et al. (Li et al., 2020) found that the circRNA TADA2A, contains rich binding sites of miR526b, which plays a competitive inhibitory effect, thereby releasing the inhibitory effect of miRNA526b on CAV1 and then increasing the expression level of Cav-1. Furthermore, some proteins, such as Cavin-1 (Hansen and Nichols, 2010; Liu et al., 2014), flotillin-1 (Vassilieva et al., 2009), and RNA-binding protein HuR (Cao et al., 2021) can also affect the protein expression and stability of Cav-1. E3 ubiquitin ligase ZNRF1 and catalase induce Cav-1 ubiquitinated (Rungtabnapa et al., 2011; Burana et al., 2016; Lee et al., 2017). In addition, the Src kinase can bind to the Tyr14 site of Cav-1 and then induces its phosphorylation, leading to instability and degradation of Cav-1 (Yoon et al., 2019). Furthermore, it has been reported that Cav-1 can be degraded by palmitic acid–induced autophagy to promote astrocyte apoptosis and inflammation (Chen et al., 2018), which indicates that the expression of Cav-1 is not only regulated by noncoding RNA but that its stability and activity are also regulated by other proteins and enzymes as well as autophagy, which, in turn, affects the function of the Cav-1 protein.
4 Caveolin-1 and Cellular Metabolism
4.1 The Role of Caveolin-1 in the Formation of Caveolae on the Cell Membrane
An important physiological function of Cav-1 is to act as the core component of caveolae and participate in its biogenesis (Figure 1). Caveolae consist of a special membrane invagination on lipid rafts and are abundant in the plasma membrane of many mammalian cells (Parton et al., 2018). They are mainly composed of integral membrane proteins (caveolin proteins), peripheral membrane proteins (cavin proteins), and lipids (including cholesterol, sphingolipids, phosphatidylserine, glycosphingolipids, and sphingomyelin) assembled on the plasma membrane (Hirama et al., 2017), shaped like a bulb or flask, with a diameter about 50–100 nm (Cheng and Nichols, 2016). The caveolin family is the main scaffold protein involved in the formation of caveolae, which mainly refers to Cav-1; whereas in the muscle cells, it mainly refers to Cav-3 (Rothberg et al., 1992). The cavin family (including cavin-1, cavin-2, cavin-3, and cavin-4) assists in the formation of caveolae (Hill et al., 2008; Kovtun et al., 2015). Cavin-1, also known as polymerase I and transcript release factor, can bind to caveolin in a lipid-dependent manner, thereby stabilizing the curvature of caveolae (Hill et al., 2008). Cavin-2 (serum deprivation protein response) can directly bind to cavin-1 and target cavin-1 to participate in the biogenesis of caveolae by regulating the size of caveolae and inducing the expansion of caveolae-derived membrane-like tubules (Hansen et al., 2009). Cavin-3 (sdr-related gene product that binds to c-kinase) mainly regulates the membrane targeting function of caveolae (McMahon et al., 2009; Parton et al., 2018). Cavin-4, also known as muscle-restricted coiled-coil protein (MURC), is mainly restricted to expression in muscle cells and may not be important for the formation of caveolae (Parton et al., 2018). In addition, other components, such as Eps-15 homologous domain composition 2 (EHD2), PACSIN2/Syndapin II (PACSIN2), and receptor tyrosine kinase-like orphan receptor 1 (ROR1) (Yamaguchi et al., 2016) can also play an important role in modulating the function of caveolae (Parton, 2018).
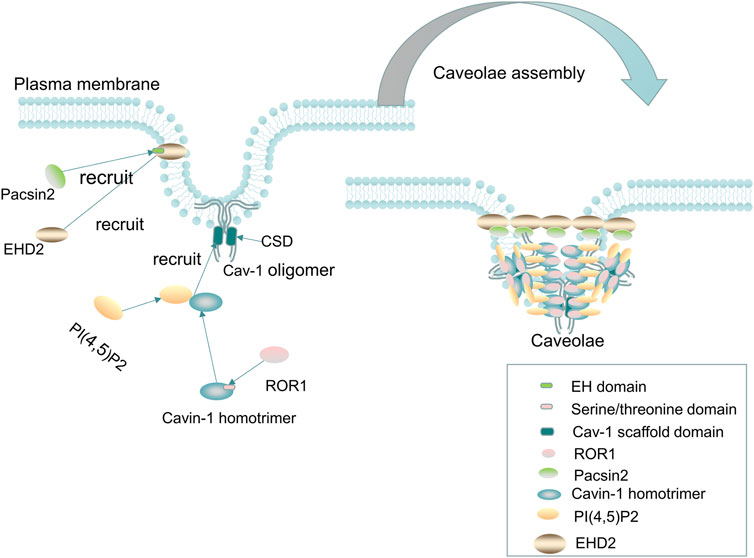
FIGURE 1. Cav-1 and caveolae. Cav-1 is essential for the formation of caveolae. The cholesterol-containing scaffold protein domain (CSD) of Cav-1 binds to cholesterol on the plasma membrane, and then, the Cavin-1 complex, EHD2, PACSIN2, and ROR1 are recruited to form mature caveolae. The recruited cavin-1 first binds to lipids [such as phosphatidylserine, and PI(4,5)P2] and then binds to the cholesterol-containing scaffold protein domain of Cav-1 to stabilize the curvature of the membrane. Two-thirds of the kinase domain of ROR1, as the scaffold protein of cavin-1 and Cav-1, binds to cavin-1, thereby promoting the binding of Cav-1 and preventing autophagy degradation of Cav-1. In addition, EHD2 is recruited to the neck of caveolae to form a loop through oligomerization, and then, Pacsin2 binds to the EH domain of EHD2 to maintain the stability of Caveolae and the distribution of Cav-1 on the plasma membrane. Without EHD2, the stability of caveolae and the distribution of Cav-1 on the plasma membrane will be difficult to maintain, resulting in dissociation of the caveolae from the plasma membrane.
The formation of caveolae mainly involves the following steps: first, caveolins, as integral membrane proteins, are synthesized in the ER, and then, Cav-1 and Cav-2 monomers are cotranslationally inserted into the membrane of the ER and form oligomerized 8S-Cav oligomers, which contain 7–14 caveolins (Han et al., 2020). The complex of oligomerized 8S-Cav oligomers is transported to the Golgi complex in the form of coat protein II (COPII) vesicle-dependent transport machinery, where Cav-1 assembles into a cholesterol-rich complex (a 70s-Cav complex composed of 18–25 8S-Cav subunits), and the 70s-Cav complex is subsequently transported and partially inserted into the plasma membrane and binds to cholesterol on the plasma membrane through its CSD domain, recruiting the cavin-1 complex, EHD2, PACSIN2, and ROR1 to form mature caveolae (Parton and del Pozo, 2013; Busija et al., 2017; Parton et al., 2018; Parton et al., 2020b).
The recruited peripheral protein cavin-1 first binds to lipids [such as phosphatidylserine and PI(4,5)P2] and then binds to the cholesterol-containing scaffold protein domain of Cav-1 to stabilize the curvature of the membrane and produce classic spherical caveolae (Hill et al., 2008). Plasma membrane insertion of EHD2 requires the binding of ATP and is oligomerized to form a ring in the neck of caveolae (Morén et al., 2012; Hoernke et al., 2017), and then, Pacsin2 binds to the EH domain of EHD2 to maintain the stability of caveolae and the distribution of Cav-1 on the plasma membrane (Hubert et al., 2020a). In addition, excessive lipid accumulation in the caveolae can cause caveolae to scission from the plasma membrane, especially in the absence of EHD2 restriction (Hubert et al., 2020b). Furthermore, EHD2 was found to regulate lipid metabolism, and loss of EHD2 caused caveolae to dissociate from the plasma membrane, while increasing fatty acid intake and promoting lipid deposition and the size of LDs (Matthaeus et al., 2020). ROR1, as a scaffold protein of cavin-1 and Cav-1, can interact with cavin-1 through its richest C-terminal serine/threonine domain, and two-thirds of the kinase domain binds to cavin-1, thus promoting the binding of Cav-1 and cavin-1 on the plasma membrane and maintaining Cav-1 expression by inhibiting lysosomal-dependent degradation and corresponding vesicle formation (Yamaguchi et al., 2016). When caveolae lack the abovementioned caveolae-related proteins to maintain their stability or subjected to mechanical stimuli (such as membrane tension or stress stimuli), Cav-1 and cavin can be released into the cytoplasm, and non-caveolae Cav-1 and cavin can recovered and reorganized into caveolae on the plasma membrane or be degraded by lysosomes (Parton et al., 2020b). In addition, non-caveola Cav-1 may also have an important physiological role independent of being a constituent of caveolae.
4.2 Caveolin-1 Mediates Protein Membrane Targeting, Endocytosis, and Signal Transduction
Cav-1 regulates various enzyme activities or receptor expression and targets them to the cell membrane. It has been found that the increased expression of Cav-1 can recruit glycolytic enzymes [phosphofructokinase (Vallejo and Hardin, 2005), aldolase (Raikar et al., 2006), and fatty acid translocase (CD36) (Ring et al., 2006)] and nervous system–related receptors [such as neurotrophin receptor, P2Y2 nucleotide receptor (Martinez et al., 2016), and heme oxygenase-1 (HO-1)] displaces to the cell membrane, where they directly binds the CSD of Cav-1 and then mediates downstream signal molecule transduction or fatty acid uptake (Abumrad et al., 2021), eventually affecting cellular metabolism.
Cav-1 not only affects membrane targeting of corresponding proteins but also mediates the endocytosis of some viruses, enzymes, macromolecular substances, and receptors. For example, Cav-1 participates in the endocytosis of BK virus (Moriyama et al., 2007), simian virus 40 (Damm et al., 2005), hepatitis B virus (Macovei et al., 2010), and human immunodeficiency virus (Huang et al., 2007; Mergia, 2017). However, unlike the abovementioned viruses, the SARS coronavirus entry into host cells through a novel clathrin-independent and caveolae-independent endocytic pathway (Filippini and D'Alessio, 2020; Wang et al., 2008). In addition, Cav-1 might mediate the endocytosis of receptor activin-like kinase 1 (ALK-1), which involves the bone formation protein-9 (BMP-9)/Cav-1/ALK-1 signaling pathway (Tao et al., 2020). The low expression of BMP-9 in lung ECs of mice with a knockout the CAV1 blocks the endocytosis of ALK-1, thereby reducing the endocytosis of ALK-1–mediated low-density lipoprotein (LDL) (Tao et al., 2020). Furthermore, previous studies have found that Cav-1 can mediate the endocytosis of macromolecular protein substances, such as insulin-like growth factor (IGF)–binding protein 5 (Yamaguchi et al., 2011), albumin (Chatterjee et al., 2017), glucose transporter 4 (Glut-4) (Yuan et al., 2007), LDL, gap junction protein connexin 36 (Cx36) (Kotova et al., 2020), receptors including GM-CSF receptor β (Zsiros et al., 2019), TGF-β1 receptor (Siegert et al., 2018), and glucagon receptors (Krilov et al., 2011). Interestingly, Han et al. (Han et al., 2019) found that, in hepatocytes, Cav-1 can regulate the expression of metabolic genes induced by TGF-β. In addition, the above proteins were coexpressed with Cav-1 and could be transendocytosed, which is essential for inflammation, fibrosis, and insulin-related signal transduction, eventually affecting the cellular metabolism (Campos et al., 2019), growth, and senescence (Volonte and Galbiati, 2020). Moreover, Cav-1 also regulates T cell antigen receptor (TCR) and B cell antigen receptor (BCR) signal transduction and regulates the innate inflammatory immune response (Fiala and Minguet, 2018).
Cav-1 is also involved in the regulation of various signal transduction pathways by recruiting multiple receptors, such as receptor tyrosine kinase, G protein–coupled receptors, G proteins, protein kinases, and phosphatases (Boscher and Nabi, 2012), binds to the sequence of the CSD of Cav-1, and then positively or negatively regulates downstream signal transduction. For example, Eph receptor tyrosine kinases (EphB1 and EphA2 receptor) are positively regulated by Cav-1 and activate downstream signaling molecules, such as extracellular regulatory protein kinase (ERK) and protein kinase B (PKB or AKT) signal transduction (Vihanto et al., 2006). In addition, the insulin receptor (IR) (Chen et al., 2008) can be activated by phosphorylated Cav-1 and binds to LDL receptor–related protein 6, thereby mediating the Akt-mTORC1 signaling pathway and regulating aerobic glycolysis in cancer cells (Tahir et al., 2013).
Cav-1 negatively regulates the membrane receptors of the tyrosine kinase family [such as EGFR (Janković et al., 2017; Yang et al., 2018) and TGF-β (Lee et al., 2007; Oliveira et al., 2017)] and then modulates cell proliferation and metastasis. Similarly, G protein–coupled receptors [such as G protein–coupled receptor kinase 2 (GRK2) (Liu et al., 2017), angiotensin receptor (Ang-II) (Ishizaka et al., 1998), and P2Y2 receptor (Martínez et al., 2019)] are also regulated by Cav-1. Liu et al. (Liu et al., 2017) established a rat model of liver injury and found that the expression of phosphorylated Cav-1 increased and that Cav-1 interacted with GRK2, thereby inhibiting eNOS activity. In addition, Cav-1 promotes Ang-II–mediated Akt and EGFR signaling to cause glomerular mesangial cell hypertrophy (Umesalma et al., 2016), whereas Ang-II–mediated calcium influx is reduced (Adebiyi et al., 2014). In addition, Cav-1 can also regulates the P2Y2 receptor to increase the activity of pERK1/2 and Akt, promoting the survival of astrocytoma cells and interfering with the process of brain injury (Martínez et al., 2019). Recently, the TCR and BCR signaling pathways were found to be regulated by the abovementioned Cav-1 (Tomassian et al., 2011; Fiala and Minguet, 2018), which not only positively regulates Toll-like receptor-9 (TLR-9) to promote MyD88-mediated TRAF3 and IRF3 signal transduction (Yang et al., 2019) but also negatively regulates TLR-9 and TLR-4 to intensify the downstream inflammation cascade and promote the progression of diabetes (Zhu et al., 2017). These data indicate that Cav-1 has a huge signal transduction network, which plays an important role in cellular biological activities, including cell metabolism and growth.
4.3 Caveolin-1 and Lipid Metabolism
4.3.1 Caveolin-1 and Biogenesis of Lipid Droplets
LDs are organelles with special structures that are ubiquitous in most eukaryotic cells and are involved in regulating energy metabolism and maintaining cellular homeostasis (Walther and Farese, 2012; Olzmann and Carvalho, 2019; Henne et al., 2020). The hydrophobic core of LDs is composed of triacylglycerols (TGs) and sterol esters (SEs) as neutral lipids and is surrounded by an ER-derived phospholipid monolayer, which is decorated with integral and peripheral proteins (Bersuker and Olzmann, 2017; Gao et al., 2019). The synthesis of neutral lipids is the first step in the formation of LDs. In mammalian cells, it is mainly catalyzed by ER diacylglycerol acyltransferase (DGAT) enzyme (DGAT1/2) and cholesterol acyltransferase (ACAT) enzyme (ACAT1/2) synthesis; among them, DGAT catalyzes the synthesis of TGs, whereas ACAT catalyzes the synthesis of SEs (Walther and Farese, 2012; Walther et al., 2017). The newly synthesized neutral lipid forms a lens-like structure between the leaflets of the ER bilayer and then can gradually fuse into larger and more stable lenses and buds from the ER to form nascent LDs (Renne et al., 2020). During this process, a continuous supply of phospholipids from the ER is needed to assist in the further expansion of LDs (Renne et al., 2020). In addition, some specific proteins are transported to the surface of LDs to promote their growth and expansion, such as glycerol-3-phosphate acyltransferase 4, DGAT2, adipose triglyceride lipase (ATGL), and other proteins, reaching the surface of LDs through ER-LD membrane bridges (Wilfling et al., 2014; Walther et al., 2017). Cytoplasmic proteins such as Perilipin family proteins and CCT1 are targeted to LDs through their hydrophobic domains (Walther et al., 2017). The perilipin family proteins PLINs1-5 are the main LD-related proteins and mainly refer to PLIN1 (perilipin A), PLIN2 (adipophilin), PLIN3 (TIP47), PLIN4 (S3-12), and PLIN5 (MLDP) (Itabe et al., 2017), which may regulate intracellular lipolysis. PLIN1 is highly expressed in mature adipocytes and forms a complex with α/β-hydrolase D5 (ABHD5) (CGI-58) on the surface of LDs under basic conditions (Sztalryd and Brasaemle, 2017). When PLIN1 is phosphorylated by cAMP, phosphorylated CGI-58 dissociates from it and binds to phosphorylated ATGL to activate triglyceride hydrolysis (Sztalryd and Brasaemle, 2017). In addition, cytoplasmic hormone-sensitive lipase (HSL) is phosphorylated by PKA and can be transported to the surface of LDs to participate in lipolysis (Frühbeck et al., 2014; Itabe et al., 2017). Furthermore, herein, we describe an important integral membrane protein, Cav-1, related to LD biogenesis and metabolism.
Cav-1 is one of the resident proteins of LDs and is involved in LD biogenesis (Pezeshkian et al., 2018). Roy et al. proposed that caveolin plays a cholesterol-trafficking role, and dominant-negative caveolin (CavDGV) can inhibit the signal transduced by H-Ras and change the distribution of cholesterol (Roy et al., 1999). Pol et al. showed that mutant caveolin protein (Cav-3DGV) accumulates in the ER and is targeted to the limiting membrane of LDs (Pol et al., 2001). However, Blouin et al. found that the expression level of Cav-1 affects the size of LDs, and in adipocytes lacking CAV1, the species of phospholipids on the LD surface are reduced, and only small LDs can be formed (Blouin et al., 2010). The accumulation of Cav-1 in the ER is targeted to LDs, which is related to its hydrophobic domain, especially its COOH-terminal domain sequence (Ostermeyer et al., 2004). Robenek et al. proposed a LD biogenesis model and hypothesized that the process of LD formation involves the synthesize lipid on the ER membrane, accumulation in the center of the bilayer to form a disc, and separation of Cav-1 from the ER membrane into LDs (Robenek et al., 2004). Furthermore, they stated that Cav-1 is not limited to the outer membrane monolayer around the LD but also present in the core of LDs (Robenek et al., 2004). Conversely, Cav-1 tends to be distributed in the lipid bilayer at the edge of the triolein lens and does not affect the curvature of the lipid lens but affects the distribution of surface neutral lipids and phospholipids on the LD surface (Pezeshkian et al., 2018). Moreover, CAV1 null adipocytes/fat pads increase protein kinase A (PKA) activity, which leads to phosphorylation of HSL and perilipin followed by the activation of lipolysis (Cohen et al., 2004). In addition, CAV1 gene knockout in the ECs increase the autocrine activity of prostaglandin I2 (PGI2), which acts as a stimulus to activate the cAMP/PKA pathway to promote the phosphorylation of HSL to increase lipolysis and reduce the formation of LDs, but it does not reduced triglyceride synthesis or fatty acid uptake (Kuo et al., 2018), suggesting that Cav-1 may play an important role in LD accumulation (Figure 2). Interestingly, previous studies have shown that exogenous addition of fatty acids may act as a signal to promote caveolin targeting LDs in nonadipocyte types (Liu et al., 2004; Pol et al., 2004). Similarly, Le Lay et al. found that, in adipocytes, an increase in exogenous cholesterol promotes the activation of Src, which triggers dynamin-dependent caveolae budding and trafficking of Cav-1 from the plasma membrane to LDs (Le Lay et al., 2006). Recently, the role of LD organelles (such as the ER, mitochondria, peroxisomes, lysosomes, and nucleus) in regulating lipid metabolism has attracted more attention (Barbosa et al., 2015; Barbosa and Siniossoglou, 2017; Suzuki, 2017; Geltinger et al., 2020). Yokomori et al. proposed that the transport of Cav-1 from the ER to LDs may be related to liver cirrhosis, and the connection between the ER and LDs may be a potential mechanism (Yokomori et al., 2019). Because Cav-1 is an important component of the membrane of each suborganelle, the role of Cav-1 in the communication of organelles and lipid metabolism may need to be addressed in the future.
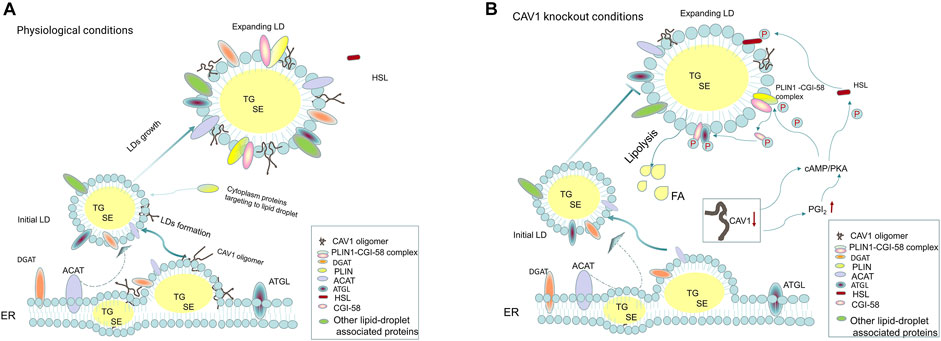
FIGURE 2. Cav-1 and lipid droplet biogenesis. (A) Under physiological conditions, the initial step of lipid droplets (LDs) formation is the synthesis of neutral lipids (TGs and SEs) by DGAT and ACAT on the ER, which serve as the core of LDs. As the LDs gradually expand, LD-associated proteins and ER-derived monolayer phospholipids are distributed around neutral lipids, promoting the initial lipid droplets to become enlarged lipid droplets. Cytoplasmic proteins, such as PLIN family proteins, are involved in regulating lipid droplet metabolism and can target the lipid droplet surface, among which PLIN1 binds to CGI-58 protein to form the PLIN1-CGI-58 complex. The newly synthesized caveolin-1 is inserted into the bilayer of the ER and fused and transferred to the LDs. Caveolin-1 is involved in LD biosynthesis; on the one hand, it may affect the distribution of phospholipids on the surface of LDs, and on the other hand, it is essential for the expansion of LDs. (B) When CAV1 is knocked out in cells (such as adipocytes and endothelial cells), the following processes may be affected: 1. growth and expansion of new lipid droplets are hindered; 2. the reduction in caveolin-1 expression can increase the phosphorylation of HSL and PLIN1 in a cAMP-dependent manner to enhance lipolysis. The latter is regulated by the PGI2/cAMP/PKA pathway, leading to phosphorylation of PLIN1 and CGI-58. Phosphorylated CGI-58 dissociates from PLIN1 and activates ATGL to co-activate lipolysis.
4.3.2 Caveolin-1 Regulates Cholesterol Homeostasis
Cav-1 has a high affinity for cholesterol and participates in the regulation of cellular cholesterol homeostasis, including cholesterol transport, cholesterol signal transduction, and cholesterol metabolism (Fielding and Fielding, 1995; Smart et al., 1996; Ikonen and Parton, 2000). Cholesterol metabolism includes endogenous synthesis, exogenous uptake, esterification, and efflux (Chang et al., 2006). The endogenous synthesis of cholesterol is regulated by SREBPs, which have three subtypes, namely, SREBP1a, SREBP1c, and SREBP2 (DeBose-Boyd and Ye, 2018). Among them, SREBP2 is mainly involved in cholesterol metabolism, SREBP1c participates in fatty acid synthesis, and SREBP1a regulates both cholesterol and fatty acid synthesis (Luo J. et al., 2020). SREBP2 is considered to be a key mediator of cholesterol biosynthesis, is synthesized in the ER, and then interacts with insulin-induced gene-1 protein (INSG-1) and SREBP cleavage activation protein (SCAP). When cholesterol in the ER is exhausted, INSG-1 can be degraded by lysosomes, and the SREBP2 and SCAP complexes are sorted into COPII-coated vesicles and escorted to the Golgi apparatus, where SREBP2 is processed by site 1 protease (S1P) and site 2 protease (S2P) cleavage, exposing the active N-terminal domain, and enters the nucleus to combine with the promoter regulatory element (SRE), thereby activating the transcription of cholesterol synthesis genes (Afonso et al., 2018; Brown et al., 2018; Yan et al., 2021). Interestingly, a previous study has shown that Cav-1 plays an important role in the transport of intracellular cholesterol, transporting newly synthesized cholesterol from the ER to caveolae and participating in the composition of the cell membrane (Smart et al., 1996). In skin fibroblasts, the expression of Cav-1 is also affected by cellular cholesterol levels, SREBP1 can inhibit the transcription of CAV1, and low levels of Cav-1 can further affect cholesterol homeostasis (Bist et al., 1997). As described in the review by Bosch et al., the loss of CAV1 in cells (such as fibroblasts) leads to the accumulation of cholesterol on the ER, which may be transported to mitochondria via MAMs, and the accumulation of cholesterol on mitochondria leads to mitochondrial dysfunction and increased reactive oxygen species (ROS) production, eventually affecting cellular metabolism (Bosch et al., 2011). Pol et al. found that, in hamster kidney (BHK) cells transfected with mutant CAV3 (Cav-3DGV), a dominant negative mutant, intracellular cholesterol transport to the cell membrane was blocked, resulting in the redistribution of cholesterol, which was characterized by reduced plasma membrane cholesterol, accumulation of free cholesterol in the lysosomes, and reduced cholesterol efflux and synthesis (Pol et al., 2001). Furthermore, Frank et al. demonstrated that the loss of CAV1 in mouse embryonic fibroblasts (MEFs) and mouse peritoneal macrophages leads to an increased accumulation of cholesterol in the ER, and the accumulation of cholesterol can increase the activity of acyl-CoA-cholesterol acyltransferase (ACAT) and reduce the synthesis of free cholesterol (Frank et al., 2006). ACAT (including ACAT1/2) is an enzyme that is involved in the esterification of cholesterol, and its role is to esterify free cholesterol and prevent free cholesterol from accumulating freely in cells and then causing lipotoxicity (Luo J. et al., 2020). Recently, Xu et al. reported that overexpression of ACAT1/2 in 3T3-L1 adipocytes promoted the colocalization of Cav-1 and free cholesterol on the surface of LDs, impairing the function of adipocytes and cholesterol homeostasis (Xu et al., 2019). This information indicated that Cav-1 is closely related to cholesterol homeostasis and that Cav-1 may play a key intermediate mediator role in cellular cholesterol metabolism.
To maintain cholesterol homeostasis, cells not only esterify free cholesterol but also need to remove excess cholesterol through efflux. Current knowledge indicates that the scavenger receptor-B1 (SR-BI) and ATP-binding cassette (ABC) transporter family proteins ABCA1 and ABCG1 can escort cholesterol in macrophages to various extracellular receptors (such as apoA-I, HDL, and LDL) and then transport them to the peripheral blood, thereby reducing the burden of cholesterol in the cell (Sharma and Agnihotri, 2019; Plummer et al., 2021). Furthermore, it has been found that Cav-1 and SR-BI are simultaneously upregulated in differentiated THP-1 macrophages, and their coexpression promotes the efflux of cholesteryl esters to HDL (Matveev et al., 1999). However, another study found that SR-BI–mediated selective cholesteryl ester uptake in human embryonic kidney 293 or Fischer rat thyroid (FRT) cells was not affected by Cav-1 expression (Wang et al., 2003). In addition, deficiency CAV1 gene in macrophages had a slight effect on ABCA1-mediated cellular cholesterol efflux to apoA-I but did not affect SR-BI and ABCG1-mediated cholesterol efflux to the HDL receptor (Frank et al., 2006), which indicates that the Cav-1 can regulate intracellular cholesterol efflux in different cell lines.
4.4 Caveolin-1 and Glucose Metabolism
It is currently known that Cav-1 is involved in regulating glucose metabolism (Nwosu et al., 2016; Gopu et al., 2020). The loss of CAV1 causes impairs glucose homeostasis and dyslipidemia, which is reversed by downstream aldosterone (MR) inhibition (Baudrand et al., 2016). Significantly increased CAV1 mRNA in the peripheral blood of patients with metabolic syndrome has been observed (de Souza et al., 2020). In contrast, Luo et al. found that mRNA expression of CAV1 is decreased in patients with type 2 diabetes, which may occur through direct binding of miR-103 to CAV1 (Luo M. et al., 2020). Furthermore, Fachim et al. demonstrated that changes in lifestyle (such as exercise or diet) can cause DNA methylation in the CAV1 transcript region and affect its expression in adipose tissue and peripheral blood cells in patients with type 2 diabetes (Fachim et al., 2020). They observed that decreased expression of Cav-1 in adipose tissue but increased expression in peripheral blood (Fachim et al., 2020). However, whether the changes of Cav-1 in other organs of patients with type 2 diabetes will affect other signaling proteins to regulate the progression of diabetes is worthy of further investigation. Furthermore, current evidence from various studies has shown that Cav-1 affects glucose metabolism by regulating a variety of glucose uptake transporters (Yuan et al., 2007; Elvira et al., 2013; Varela-Guruceaga et al., 2018). Lee et al. found that the expression of Cav-1 affects SGLT1 receptor–mediated glucose uptake on renal tubular epithelial cells by affecting the cAMP/Epac/PKA signaling pathway (Lee et al., 2012). In addition, the enhanced expression of ERK, p38MAPK, and NF-κB can increase the activity of SGLT1 and promote its binding to Cav-1 in renal tubular epithelial cells to increase glucose uptake (Lee et al., 2012). In addition, Cav-1 can upregulate the expression of SGLT1 (Elvira et al., 2013), indicating that Cav-1 may play a key role in glucose uptake by renal tubules, but the role of blood glucose in diabetes may need to be studied in the future.
Moreover, Cav-1 can regulate GLUT4 to mediate glucose uptake, which is related to insulin-related signaling pathways (Yuan et al., 2007). Long-term high glucose stimulates adipocytes and reduces the sensitivity of Cav-1 to insulin and inhibits IR and PKB (AKT-2) phosphorylation, which affect the GLUT4 in the intracellular glucose storage vesicles to transport to the plasma membrane caveolae and bind to Cav-1 and subsequently mediate glucose uptake (Palacios-Ortega et al., 2016). In contrast, in a high-glucose environment, the caveolae-related coiled-coil protein (NECC2) is highly expressed on adipocytes, which is closely linked to insulin-related glucose uptake by triggering insulin induction of NECC2 transport to the cell surface and binding to Cav-1 (Trávez et al., 2018). At this point, the IR binds to the scaffold domain of Cav-1 and then activates the PI3K/Akt signaling pathway (Trávez et al., 2018). However, Cav-1 regulates glucose metabolism in tumor cells mainly by regulating glucose transporter 3 (GLUT3/SLC2A3) uptake of glucose, which leads to increased aerobic glycolysis and increases intracellular ATP production, thus maintaining the growth and survival of tumor cells (Ali et al., 2019). Further investigations have shown that the abnormally increased expression of Cav-1 in tumor cells is related to the methylation of CpG sites, and the upregulation of Cav-1 expression then increases the activity of HMGA1 and promotes the translocation of HMGA1 into the nucleus, where HMGA1 binds to the promoter of GLUT3 and promotes its transcription (Ha et al., 2012). In addition, Cav-1 also affects mitochondrial function, increasing ATP production and inhibiting phosphorylation of AMPK at the Thr172 residue and modulating autophagy by the AMPK-TP53/p53 pathway in cancer cells (Ha and Chi, 2012). Further research demonstrated that the hypoxia response elements (HREs) of hypoxia inducible factor-1α (HIF-1α) bind to the promoter of CAV1 under hypoxic conditions and inhibit the transcript level of CAV1, reducing the translocation of GLUT4 to the plasma membrane and resulting in decreased glucose uptake (Varela-Guruceaga et al., 2018). In addition, glucose uptake mediated by GLUT4 membrane translocation is related to AMP-activated protein kinase (AMPK), which is a key sensor of glucose metabolism (Hu et al., 2017). Therefore, it is worth noting that Cav-1 plays an indispensable role in regulating cellular metabolism and may be a novel or important target in metabolic-related diseases.
4.5 Caveolin-1 and Autophagy
Autophagy plays an important role in cellular metabolism, and a recent study reported that autophagy are closely related to cellular metabolism and cell survival (Tan and Miyamoto, 2015). Participation in the regulation of autophagy is one of the functions of Cav-1. Cav-1 can bind to the autophagy-related protein 5 (ATG-5), ATG12, and ATG5-ATG12 complex in lung epithelial cells and inhibit the formation and function of autophagosomes (Chen et al., 2014). When the cells are under stress (such as starvation), knockout of CAV1 gene in MEFs can promote the formation of late autophagy lysosomes, enhance autophagic flux and promote cell survival, and it is involved in tumor progression (Shi et al., 2015). Deficiency of CAV1 gene in human aortic ECs causes connexin-43 and ATG5 from the plasma membrane to combine with several ATGs (such as ATG12, ATG16, and IP3R), which are involved in the initial steps of autophagosome formation to promote the formation of autophagosomes, thus inhibiting vascular inflammation and arterial wall macrophage infiltration (Zhang X. et al., 2020). However, in cisplatin-treated lung cancer cells, silencing CAV1 with siRNA inhibits the activation of Rho-related coiled-coil kinase 1 (ROCK1), thereby affecting Parkin-related mitochondrial autophagy and protecting against mitochondrial apoptosis and functional damage (Liu et al., 2020). In addition, under oxidative stress, Cav-1 phosphorylates at tyrosine-14, binds to beclin-1, and promotes its translocation to mitochondria, promoting mitochondrial autophagy (Nah et al., 2017). Recent studies have shown that the HIF-1α–Cav-1 signaling axis mediates autophagy to regulate cellular metabolism and promote the survival and metastasis of breast cancer cells (Wang N. et al., 2020). In addition, the close relationship between Cav-1 and autophagy plays an important role in regulating lipid metabolism. Bai et al. found that, in human umbilical vein ECs treated with high glucose, autophagy degradation of Cav-1 mediated by the AMPK/mTOR/PIK3C3 pathway is inhibited, thereby increasing the expression of Cav-1 and cavin, and then, LC3 is recruited and bound, subsequently inhibiting Cav-1-CAVIN1-LC3B-mediated autophagy degradation of Cav-1. This process eventually manifests as the increased expression of Cav-1 to promote the formation of caveolae and mediates the increase in LDL endocytosis (Bai et al., 2020). Interestingly, Xue et al. found that overexpression of CAV1 in L02 cells treated with alcohol and oleic acid can inhibit the Akt/mTOR pathway, thereby activating autophagy and alleviating cellular lipid deposition (Xue et al., 2020). The above information indicates that Cav-1 has different roles in the regulation of autophagy and cellular metabolism in different cell lines.
4.6 Caveolin-1 and Oxidative Stress
Oxidative stress is the key factor in cellular metabolism (Liu et al., 2019). It has been demonstrated that Cav-1 regulates oxidative stress and participates in the process of cellular life. Under oxidative stress, the expression of Cav-1 is increased in nucleus pulposus cells and related to premature senescence, whereas silencing the expression of CAV1 can reduce the protein expression of p53 and p21 to protect cells from senescence (Ding et al., 2017). In contrast, under oxidative stress, Cav-1 is abundantly expressed in chondrocytes and transferred from the plasma membrane to the nucleus to participate in the repair of DNA damage (Goutas et al., 2020). In addition, overexpression of CAV1 in rhabdomyosarcoma cells, the cell cycle is blocked in the G2/M phase, which is accompanied by the reduced expression of p21, p16, and cleaved caspase-3, whereas the production of catalase is increased; therefore, Cav-1 enhances DNA repair and protects against cellular senescence and apoptosis from oxidative stress (Codenotti et al., 2021). However, interestingly, Goutas et al. did not observe a displacement of Cav-1 and the DNA damage repair in patients with osteoarthritis (Goutas et al., 2020). Thus, the role of Cav-1 in DNA repair in cells requires further confirmation.
There is a closed relationship between oxidative stress and inflammation. Studies have shown that the Cav-1 regulates oxidative stress and affects inflammation. Wang et al. found that high-fat diet-fed ApoE−/− mice with atherosclerosis show increased activity of JNK-related signals, oxidative stress, and inflammation, whereas these changes were rescued in mice with a double knockout of ApoE and CAV1 (Wang et al., 2018). In contrast, in mice with liver injury induced by carbon tetrachloride (CCl4), CAV1 gene knockout can aggravate oxidative stress and activate the TGF-β signaling pathway and the production of proinflammatory factors, such as IL-1β and IL-6, leading to liver fibrosis (Ji et al., 2018). Furthermore, in E11 murine kidney podocytes, Cav-1 promotes the production of antioxidant enzymes and inhibits the oxidative stress response induced by H2O2, alleviating the inflammatory damage of podocytes (Chen et al., 2017). Conversely, Cav-1 also affects the energy conversion of cells (Fernández-Rojo et al., 2012). Recently, Shao et al. found that knockdown of CAV1 with shRNA in pancreatic stellate cells promotes the production of ROS, and the production of ROS further reduces the expression of Cav-1 (Shao et al., 2020). This Cav-1-ROS positive feedback induces the conversion of cell energy metabolism to glycolysis, and the products of glycolysis promote cell energy production through mitochondrial oxidative phosphorylation, which further promotes the proliferation of pancreatic cancer cells (Shao et al., 2020). This information indicates that Cav-1 plays an important role in regulating oxidative stress and inflammation by affecting cellular metabolism.
4.7 Other Functions of Caveolin-1: Mechanical Sensing and Vesicle Transport
Cav-1, as the main component protein of caveolae, is involved not only in regulating metabolism but also in mechanical transduction and vesicle transport. Cav-1 phosphorylation is essential for caveolae to perform signal transduction, endocytosis (Nabi and Le, 2003) and lipid transport (Pilch and Liu, 2011); enzymes (Coelho-Santos et al., 2016), viruses (Xing et al., 2020), and LDL (Gerbod-Giannone et al., 2019) enter the cell through caveolae-dependent endocytosis, which is related to phosphorylation of Cav-1. Caveolae act as a cell membrane sensor (Parton and del Pozo, 2013) and mechanical sensor (Sinha et al., 2011), which can quickly adapt to sudden and acute mechanical stress stimulation (Sinha et al., 2011). When the cell responds to a variety of stimuli, such as osmotic/stretch, shear, ultraviolet, and oxidative stimuli, the tension of the cell membrane increases, which promotes caveolae disassembly (Parton et al., 2020a). The above stimuli cause caveolae flattening and may eventually induce cavin dissociation, changes in lipid distribution on the cell membrane, and Cav-1 tyrosine phosphorylation (Parton et al., 2020a). Phosphorylated Cav-1 can regulate actin to detach caveolae from the plasma membrane and enter the cytoplasm (Zimnicka et al., 2016) and then traffic to LDs, which may be involved in cell lipid metabolism (Matthaeus and Taraska, 2020). It is particularly noteworthy that the relationship between Cav-1 and lipid metabolism has aroused widespread research interest. Recent studies have found that Cav-1 regulates lipid metabolism and participates in kidney-related diseases (Chen et al., 2016; Mitrofanova et al., 2019). Therefore, we focused on the relationship between Cav-1 regulation of cellular metabolism and the kidney.
5 Caveolin-1 and Kidney Disease
5.1 Acute Kidney Disease
Cav-1 is involved in the pathophysiology of acute kidney injury (AKI). Zager et al. found that the expression of Cav-1 was increased in ischemia ± reperfusion–induced AKI mice and verified that the destruction of the caveolae in the damage of renal tubular epithelial cells, which causes cholesterol and Cav-1 are separated from the plasma membrane, leading to free cholesterol deposits in the lumen of the renal tubules and an increased level of urine Cav-1 (Zager et al., 2002). Accordingly, Cav-1 was considered a possible biomarker of AKI (Zager et al., 2002). In addition, proximal renal tubular injury leads to increased destruction of caveolae, and Cav-1 translocates into the cytoplasm; activates the expression of PDGFR-β, EGFR, and Rho guanosine triphosphatase (GTPase) signaling proteins; and participates in the process of renal tubular cell regeneration (Mahmoudi et al., 2003; Fujigaki et al., 2007). Likely, in ischemic reperfusion AKI mice treated with EPO, a significant increase in the expression of Cav-1 in blood and kidney tissue is observed (Kongkham et al., 2016). In addition, Cav-1 has also been found to be highly expressed in apoptotic tubular cells, although it remains controversial whether Cav-1 plays a role in promoting repair or apoptosis in AKI (Mahmoudi et al., 2003). Notably, Moore et al. reported that Cav-1 is a tissue fibrosis inhibitor, and its genetic polymorphisms are associated with renal transplantation fibrosis and allogeneic transplantation failure (Moore et al., 2010). Furthermore, another recent study has demonstrated increased expression of Cav-1 in the serum of patients with kidney transplant, which associated with a decreased incidence of tubulointerstitial rejection (Emmerich et al., 2021). From the above information, it can be seen that Cav-1 has different roles in AKI, and its detailed mechanism still lacks experimental verification.
5.2 Glomerulus Nephritis
Although glomerulonephritis is an immune-mediated disease, recent studies have shown that consistent changes in the kidney transcriptome are consistent with the metabolic reprogramming of different forms of glomerulonephritis (Grayson et al., 2018), which may indicate that abnormal cellular metabolism also plays an important role in this disease. Tamai et al. showed, for the first time, that caveolae are present in mesangial cells and the Cav-1 is located on caveolae, as detection by electron microscope (Tamai et al., 2001). Furthermore, they demonstrated that Cav-1 can bind to PDGF receptors to mediate the PDGF pathway and modulate mesangial proliferative glomerulonephritis (Tamai et al., 2001). In addition, Ostalska-Nowicka et al. verified that the expression of Cav-1 in parietal epithelial cells is significantly lower in children diagnosed with focal segmental glomerulosclerosis and lupus glomerulonephritis than in those with minimal change disease, Schönlein-Henoch glomerulopathy, or in controls (Ostalska-Nowicka et al., 2007). Furthermore, the high expression level of Cav-1 in glomerular ECs is positively correlated with proteinuria, and it is suggested that Cav-1 mediates EC endocytosis of albumin and participates in the progression of glomerular-related diseases (Moriyama et al., 2011; Moriyama et al., 2017). Although current studies suggest that Cav-1 may regulate the progression of glomerular associated diseases by endocytosing of macromolecules (cholesterol or albumin), the mechanism by which Cav-1 modulates the development of glomerular diseases requires further study.
5.3 Diabetic Kidney Disease
Previous studies have shown that Cav-1 has antifibrotic properties by regulating cell proliferation, migration and adhesion, as well as inhibiting the TGF-β signaling pathway in diabetic kidney disease (DKD) (Gvaramia et al., 2013; Shihata et al., 2017; Van Krieken and Krepinsky, 2017). In addition, Cav-1 might also regulate glucose uptake and mediate the endocytosis of urinary albumin; therefore, it is considered a potential therapeutic target for DKD (Van Krieken and Krepinsky, 2017). Arya et al. found that, in rats with diabetic nephropathy, the highly expressed Cav-1 can bind to nitric oxide synthase (eNOS) and inhibit its signal transduction, thereby reducing the production of NO and eventually leading to increased level of serum urea nitrogen, blood creatinine and urine protein, whereas the above changes can be reversed by Cav-1 inhibitors (Arya et al., 2011). Conversely, in renal mesangial cells treated with high glucose, Cav-1 phosphorylated by Src kinase can activate RhoA, which may be related to the development of glomerular matrix accumulation in DKD (Wu et al., 2014). Xie et al. showed that, in mesangial cells treated with high glucose, RhoA/Rock promotes the translocation of NF-kB into the nucleus to increase the transcription level of inflammatory factors such as ICAM-1, TGF-β1, and FN and ultimately leads to the production of mesangial cell matrix (Xie et al., 2013). Furthermore, high glucose has been shown to induce an increase in the production of mitochondrial ROS (mtROS) and vascular endothelial growth factor (VEGF), and mtROS in glomerular ECs, which activates Src kinase to phosphorylate Cav-1, leading to increased albumin endocytosis and massive proteinuria (Xie et al., 2013). Furthermore, under high-glucose conditions, phosphorylation of Cav-1 upregulates the expression of TLR-4 and promotes the secretion of pro-inflammatory factors, such as TNF-α, IL-6, IL-1β, and MCP-1 in podocytes and accelerates the process of DKD (Sun et al., 2014b). More importantly, diabetic mice with deficiency of CAV1 gene in mesangial cells showed a significant inhibitory effect on the PKCβ1/ROS/RhoA/Rho-kinase signaling pathway; the expression of TGFβ1, FN, and collagen I is reduced, whereas the expression of AMPK is upregulated (Zhang et al., 2012; Guan et al., 2013). However, how Cav-1 regulates AMPK expression affects the development of DKD is unknown. Another study found that CAV1 knockout in mesangial cells of diabetic mice upregulates the protein expression of follistatin, which neutralizes and inhibits activin, ultimately decreasing proteinuria, glomerular sclerosis, and extracellular matrix accumulation (Zhang et al., 2019). SMPDL3b is an enzyme related to lipid metabolism, which can downregulate the expression of ceramide-1-phosphate and affect the phosphorylation of AKT, leading to podocyte damage (Mitrofanova et al., 2019). In mice carrying a specific knockout of the SMPDL3b in podocytes, the Cav-1 combines with IRB to transduce insulin signals and phosphorylate Akt, which reduces podocyte damage and delays the progression of DKD (Mitrofanova et al., 2019). These data suggest that Cav-1, a cell metabolism related molecule, plays a critical role in kidney injury in DKD.
5.4 Clear Cell Renal Cell Carcinoma
Metabolic reprogramming in clear cell renal cell carcinoma (RCC) has been recognized (Wettersten et al., 2017). It has been reported that Cav-1 inhibits breast cancer stem cells through metabolic reprogramming (Wang S. et al., 2020). Recent evidence has shown that Cav-1 is considered one of the possible prognostic biomarkers of clear cell RCC (Waalkes et al., 2011). Cav-1, which is increased significantly in RCC kidney tissue, can regulate the growth and metastasis of cancer cells (Joo et al., 2004; Steffens et al., 2011). In addition, the expression of Cav-1 and phosphorylated ERK-1/2 in local RCC tissues is also considered to be a predictor of metastasis of RCC (Campbell et al., 2013). The Cav-1/AKT/mTOR axis has been shown to promote the proliferation of cancer cells and vascular metastasis (Campbell et al., 2008). Recently, Zhang et al. found that Cav-1 binds to oxidized low-density apolipoprotein receptor 1 (LOX-1) to induce lipid deposition and promote tumor cell proliferation, but this effect can be reversed by celastrol (Zhang et al., 2021). Although Cav-1 participates in the metabolism of cancer cells and regulates the survival of cancer cells, the relationship between Cav-1 and RCC merits in-depth study.
6 Therapeutic Promising
Because Cav-1 plays an important role in cellular metabolism and activities related to cellular life, especially the development of various kidney diseases, recent studies targeting Cav-1 for the treatment of various diseases, especially kidney diseases, have become a research hotspot (Table 1). Here, we focus on some chemical compounds, drugs, and various extracts from traditional Chinese medicine, which target Cav-1 or its associated signaling pathway and summarize their application in kidney diseases.
6.1 Caveolin-1 Inhibitor
6.1.1 Chemical Compound
Methyl-β-cyclodextrin (MβCD), filipin, and daidzein are the more common inhibitors of Cav-1 (Woodman et al., 2004; Peng et al., 2007; Gao et al., 2014). Recent studies have shown that the effects of these drugs on the functions of Cav-1 may be secondary to the disruption of caveolae. MβCD and filipin can destroy the structure of caveolae and reduce Cav-1 phosphorylation and downstream signaling pathways, such as the Cav-1/RhoA and Scr/Cav-1/EGFR/Akt signaling pathways, resulting in the prevention of fibronectin and collagen I production and alleviation of glomerulosclerosis (Peng et al., 2007; Zhang et al., 2007). In addition, MβCD can affect the signal transduction of ANG-II receptors and reduce glomerular mesangial hyperplasia (Adebiyi et al., 2014). Previous studies have shown that Cav-1 located on caveolae of glomerular ECs (HRGECs) mediates albumin entry into glomerular ECs and transcytosis, leading to proteinuria in glomerular diseases, whereas MβCD can reverse the abovementioned changes (Moriyama et al., 2015; Moriyama et al., 2017). In addition, Daidzein, another inhibitor of Cav-1 (Sharma et al., 2012; Gao et al., 2014), has been found to have anti-inflammatory, antioxidative stress, and renoprotective effects in the mice with AKI induced by cisplatin (Meng et al., 2017; Tomar et al., 2020). Furthermore, Daidzein can also reverse the pathological changes in diabetic rats and reduces urine protein, blood creatinine, and urea nitrogen by inhibiting the Cav-1/eNOS/NO pathway (Arya et al., 2011). In addition, Daidzein has a potential effect on diabetes, and its complications are mediate by regulating cellular metabolism, including glucose and lipid metabolism, as well as oxidative stress (Das et al., 2018). Daidzein improves hyperglycemia, insulin resistance, dyslipidemia, obesity, and inflammation (Das et al., 2018). These data suggest that Cav-1 inhibitors can affect the progression of kidney disease by regulating cellular metabolism, but the precise mechanism is still unclear.
6.1.2 Traditional Chinese Medicine Extracts
Curcumin, as an extract from traditional herbal medicine, has multiple functions, such as antioxidation, anti-inflammatory, and antifibrosis activities (Santos-Parker et al., 2017; Tsuda, 2018; Kong et al., 2020). It can regulate the fibrosis process of kidney disease through Cav-1–related signaling pathways (Sun et al., 2017). Under high-glucose conditions, the Cav-1/TLR-4 signaling pathway mediates pro-inflammatory factors, such as TNF-α, IL-6, IL-1β, and MCP-1, to induce the reversal of podocyte damage in response to curcumin (Sun et al., 2014b). In addition, Curcumin can also inhibit the phosphorylation of Cav-1 under high-glucose conditions and alleviate the oxidative stress and apoptosis of podocytes, as well as the epithelial-mesenchymal transition (EMT) and proteinuria of podocytes, eventually delaying the progression of diabetic mice (Sun et al., 2014a; Sun et al., 2016).
Salidroside (SAL) is an active ingredient isolated from the traditional Chinese medicine rhodiola that has a protective effect on kidney diseases. Wu et al. found that SAL upregulates AMPK and downregulates Src kinase under high-glucose conditions, thereby inhibiting Cav-1 phosphorylation, inhibiting glomerular ECs (GECs) albumin endocytosis, and reducing albuminuria in diabetic mice (Wu et al., 2016). Xue et al. found that SAL can activate the Sirt1/PGC-1α axis to promote mitochondrial biogenesis and alleviate pathological changes in diabetic mice, as shown by decrease in urine albumin, blood urea nitrogen and serum creatinine (Xue et al., 2019). Furthermore, SAL inhibits the TLR4/NF-κB and MAPK pathways to prevent inflammation and fibrosis, protecting kidney function (Li et al., 2019). Previous studies have shown that Cav-1 regulates the TLR4 signaling pathway and participates in the inflammatory response of podocytes (Sun et al., 2014b). Whether SAL regulates the Cav-1/TLR4 pathway to reduce renal inflammatory damage requires follow-up research.
Catalpol was previously found to have neuroprotective effects in diabetic mice by increasing the expression of Cav-1 and PKC (Zhou et al., 2014). The review by Bai et al. summarized that the ability of catalpol to reduce kidney damage by improving lipid metabolism and IGF-1 signal transduction (Bai et al., 2019). On the basis of the function of Cav-1 in regulating lipid metabolism, catalpol may play a key role in kidney disease by regulating the expression of Cav-1.
6.1.3 Rock Inhibitor: Fasudil
It has been reported that Cav-1 plays a key role in regulating the RhoA/Rock pathway, which is involved in inflammation and apoptosis in kidney disease (Peng et al., 2007; Wu et al., 2014; Nozaki et al., 2015; Zhao et al., 2015). Fasudil, a Rock inhibitor, blocks the VEGF/Src/Cav-1/signaling pathway to alleviate renal inflammation, glomerulosclerosis, and proteinuria in diabetic mice (Xie et al., 2013; Jin et al., 2015). In addition, Rock inhibitor also plays an important role in cellular metabolism. In high-fat–fed mice, fasudil can activate AMPK, thereby promoting lipid metabolism (Noda et al., 2014; Noda et al., 2015). Furthermore, in diabetic mice, the RhoA/ROCK/NF-κB signaling pathway is inhibited when pancreatic islets are transplanted into mice, and the production of podocyte inflammatory factors such as IL-6 and MCP-1, is reduced, reversing podocyte damage (Huang et al., 2021). Recently, the role of Rho family GTPases in regulating cell glucose metabolism and maintaining glucose homeostasis has also received increasing attention (Møller et al., 2019; Machin et al., 2021). Therefore, whether Rock inhibitors indirectly affect Cav-1–related signaling pathways to regulate kidney energy metabolism still needs to be addressed.
6.1.4 Noncoding RNA
Many studies have shown that noncoding RNAs regulate the expression of Cav-1 and affect its downstream events. It has been found that miR-204 (Hall et al., 2014) and circAKT1 (Zhu et al., 2020) affect the progression of kidney disease by regulating Cav-1. Hall et al. found that, in VHL (−) RCC cells, miR-204 indirectly affects the transient receptor potential melastatin 3 (TRPM3)–induced autophagy by inhibiting the expression of Cav-1, which, in turn, affects the development of RCC (Hall et al., 2014). In addition, Zhu et al. found that circAKT1, which is highly expressed in clear cell RCC, promotes the proliferation and progression of renal cancer cells by upregulating the expression of Cav-1 by sponging miR-338-3p (Zhu et al., 2020). However, Mehta et al. recently found that CAV1 deficiency in glomerular mesangial cells can inhibit miR299a-5p, which may posttranscriptionally regulate the expression of follostatin, thereby exerting an anti-renal fibrosis effect (Mehta et al., 2021). The above studies may indicate that noncoding RNAs are involved in the progression of Cav-1–mediated kidney disease.
7 Conclusion and Perspectives
Cav-1 is a metabolism-related membrane protein in a variety of cell types, which is involved in the pathophysiology of a variety of diseases by a large signaling network system. Despite recent studies showing robust evidence for the critical role of Cav-1 in metabolic disorders, oxidative stress, and autophagy, most of the studies indicated that Cav-1 is a potential therapeutic target in cancer and cardiovascular diseases, whereas few studies have been conducted in kidney disease. Because cellular metabolic homeostasis is critical in kidney disease, here, we propose that Cav-1 is closely linked to kidney disease through the regulation of cellular metabolism. When cells are subjected to stress and other stimuli, the expression of Cav-1 is increased, and it is then translocated to the cell membrane. At the membrane, it recruits specific receptors and molecules such as IGF-IR, glucose transporters, and LOX-1 into the caveolae, mediating downstream signal transduction and affecting cellular metabolism including glucose and lipid metabolism. Conversely, in vascular ECs treated with high glucose, autophagy induced by the AMPK-MTOR-PIK3C3 pathway is blocked to reduce the degradation of Cav-1 by autophagy, whereas the increased expression of Cav-1 inhibits the formation of downstream autophagosomes by recruiting and binding to LC3B, thus further inhibiting the autophagic degradation of Cav-1 and leading to an increase in caveolae formation. This process mediates the increase in endocytosis of low-density apolipoprotein and affects cellular metabolism (Bai et al., 2020). As mentioned above, Cav-1 may have a potential role in kidney disease by regulating cellular metabolism (Figure 3). In addition, we also describe Cav-1 and its related signaling molecules as potential therapeutic targets based on the use of related inhibitors and extract from traditional medicines in various kidney diseases. This review may open up new horizons for future investigations of the role of Cav-1 in kidney and other disease.
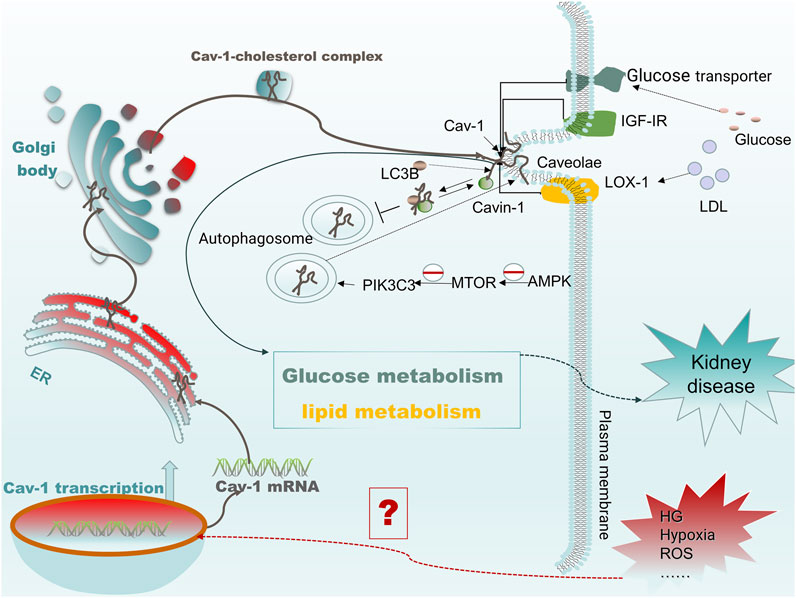
FIGURE 3. Cav-1 mediates protein membrane targeting and regulates cellular metabolism. When cells are subjected to harmful external stimuli (such as high glucose and oxidative stress), the transcription levels of CAV1 increase, which promotes the formation of caveolae on the cell membrane. The expressed Cav-1 can then recruit receptors such as IGF-IR, glucose transporter, and LOX-1 to caveolae and bind there to Cav-1 and subsequently regulate glucose or lipid metabolism by mediating specific receptor signals. In addition, under high glucose conditions, blocking autophagy induced by the AMPK-MTOR-PIK3C3 pathway reduces the degradation of Cav-1 by autophagy, resulting in increased expression of cavin and Cav-1. The high expression of Cav-1 recruits LC3B and binds together to inhibit the formation of autophagosomes, further inhibiting autophagic degradation of Cav-1 and leading to an increase in the formation of caveolae, which eventually mediates increased endocytosis of low-density apolipoprotein.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (81730018) and the National Key R&D Program of China (2018YFC1314002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abumrad, N. A., Cabodevilla, A. G., Samovski, D., Pietka, T., Basu, D., and Goldberg, I. J. (2021). Endothelial Cell Receptors in Tissue Lipid Uptake and Metabolism. Circ. Res. 128 (3), 433–450. doi:10.1161/circresaha.120.318003
Adebiyi, A., Soni, H., John, T. A., and Yang, F. (2014). Lipid Rafts Are Required for Signal Transduction by Angiotensin II Receptor Type 1 in Neonatal Glomerular Mesangial Cells. Exp. Cel Res 324 (1), 92–104. doi:10.1016/j.yexcr.2014.03.011
Afonso, M. S., Machado, R. M., Lavrador, M. S., Quintao, E. C. R., Moore, K. J., and Lottenberg, A. M. (2018). Molecular Pathways Underlying Cholesterol Homeostasis. Nutrients 10 (6), 760. doi:10.3390/nu10060760
Ali, A., Levantini, E., Fhu, C. W., Teo, J. T., Clohessy, J. G., Goggi, J. L., et al. (2019). CAV1 - GLUT3 Signaling Is Important for Cellular Energy and Can Be Targeted by Atorvastatin in Non-small Cell Lung Cancer. Theranostics 9 (21), 6157–6174. doi:10.7150/thno.35805
Arya, A., Yadav, H. N., and Sharma, P. L. (2011). Involvement of Vascular Endothelial Nitric Oxide Synthase in Development of Experimental Diabetic Nephropathy in Rats. Mol. Cel Biochem 354 (1-2), 57–66. doi:10.1007/s11010-011-0805-6
Bai, X., Yang, X., Jia, X., Rong, Y., Chen, L., Zeng, T., et al. (2020). CAV1-CAVIN1-LC3B-mediated Autophagy Regulates High Glucose-Stimulated LDL Transcytosis. Autophagy 16 (6), 1111–1129. doi:10.1080/15548627.2019.1659613
Bai, Y., Zhu, R., Tian, Y., Li, R., Chen, B., Zhang, H., et al. (2019). Catalpol in Diabetes and its Complications: A Review of Pharmacology, Pharmacokinetics, and Safety. Molecules 24 (18), 3302. doi:10.3390/molecules24183302
Barbosa, A. D., Savage, D. B., and Siniossoglou, S. (2015). Lipid Droplet-Organelle Interactions: Emerging Roles in Lipid Metabolism. Curr. Opin. Cel Biol 35, 91–97. doi:10.1016/j.ceb.2015.04.017
Barbosa, A. D., and Siniossoglou, S. (2017). Function of Lipid Droplet-Organelle Interactions in Lipid Homeostasis. Biochim. Biophys. Acta Mol. Cel Res 1864 (9), 1459–1468. doi:10.1016/j.bbamcr.2017.04.001
Baudrand, R., Gupta, N., Garza, A. E., Vaidya, A., Leopold, J. A., Hopkins, P. N., et al. (2016). Caveolin 1 Modulates Aldosterone-Mediated Pathways of Glucose and Lipid Homeostasis. J. Am. Heart Assoc. 5 (10), e003845. doi:10.1161/jaha.116.003845
Bernatchez, P. (2020). Endothelial Caveolin and its Scaffolding Domain in Cancer. Cancer Metastasis Rev. 39 (2), 471–483. doi:10.1007/s10555-020-09895-6
Bersuker, K., and Olzmann, J. A. (2017). Establishing the Lipid Droplet Proteome: Mechanisms of Lipid Droplet Protein Targeting and Degradation. Biochim. Biophys. Acta Mol. Cel Biol Lipids 1862 (10 Pt B), 1166–1177. doi:10.1016/j.bbalip.2017.06.006
Bist, A., Fielding, C. J., and Fielding, P. E. (2000). p53 Regulates Caveolin Gene Transcription, Cell Cholesterol, and Growth by a Novel Mechanism. Biochemistry 39 (8), 1966–1972. doi:10.1021/bi991721h
Bist, A., Fielding, P. E., and Fielding, C. J. (1997). Two Sterol Regulatory Element-like Sequences Mediate Up-Regulation of Caveolin Gene Transcription in Response to Low Density Lipoprotein Free Cholesterol. Proc. Natl. Acad. Sci. U S A. 94 (20), 10693–10698. doi:10.1073/pnas.94.20.10693
Blouin, C. M., Le Lay, S., Eberl, A., Köfeler, H. C., Guerrera, I. C., Klein, C., et al. (2010). Lipid Droplet Analysis in Caveolin-Deficient Adipocytes: Alterations in Surface Phospholipid Composition and Maturation Defects. J. Lipid Res. 51 (5), 945–956. doi:10.1194/jlr.M001016
Bonds, J. A., Shetti, A., Bheri, A., Chen, Z., Disouky, A., Tai, L., et al. (2019). Depletion of Caveolin-1 in Type 2 Diabetes Model Induces Alzheimer's Disease Pathology Precursors. J. Neurosci. 39 (43), 8576–8583. doi:10.1523/jneurosci.0730-19.2019
Boopathi, E., Gomes, C. M., Goldfarb, R., John, M., Srinivasan, V. G., Alanzi, J., et al. (2011). Transcriptional Repression of Caveolin-1 (CAV1) Gene Expression by GATA-6 in Bladder Smooth Muscle Hypertrophy in Mice and Human Beings. Am. J. Pathol. 178 (5), 2236–2251. doi:10.1016/j.ajpath.2011.01.038
Bosch, M., Marí, M., Gross, S. P., Fernández-Checa, J. C., and Pol, A. (2011). Mitochondrial Cholesterol: a Connection between Caveolin, Metabolism, and Disease. Traffic 12 (11), 1483–1489. doi:10.1111/j.1600-0854.2011.01259.x
Boscher, C., and Nabi, I. R. (2012). Caveolin-1: Role in Cell Signaling. Adv. Exp. Med. Biol. 729, 29–50. doi:10.1007/978-1-4614-1222-9_3
Breton, S., Lisanti, M. P., Tyszkowski, R., McLaughlin, M., and Brown, D. (1998). Basolateral Distribution of Caveolin-1 in the Kidney. Absence from H+-atpase-coated Endocytic Vesicles in Intercalated Cells. J. Histochem. Cytochem. 46 (2), 205–214. doi:10.1177/002215549804600209
Brown, M. S., Radhakrishnan, A., and Goldstein, J. L. (2018). Retrospective on Cholesterol Homeostasis: The Central Role of Scap. Annu. Rev. Biochem. 87, 783–807. doi:10.1146/annurev-biochem-062917-011852
Burana, D., Yoshihara, H., Tanno, H., Yamamoto, A., Saeki, Y., Tanaka, K., et al. (2016). The Ankrd13 Family of Ubiquitin-Interacting Motif-Bearing Proteins Regulates Valosin-Containing Protein/p97 Protein-Mediated Lysosomal Trafficking of Caveolin 1. J. Biol. Chem. 291 (12), 6218–6231. doi:10.1074/jbc.M115.710707
Busija, A. R., Patel, H. H., and Insel, P. A. (2017). Caveolins and Cavins in the Trafficking, Maturation, and Degradation of Caveolae: Implications for Cell Physiology. Am. J. Physiol. Cel Physiol 312 (4), C459–c477. doi:10.1152/ajpcell.00355.2016
Campbell, L., Al-Jayyoussi, G., Gutteridge, R., Gumbleton, N., Griffiths, R., Gumbleton, S., et al. (2013). Caveolin-1 in Renal Cell Carcinoma Promotes Tumour Cell Invasion, and in Co-operation with pERK Predicts Metastases in Patients with Clinically Confined Disease. J. Transl Med. 11, 255. doi:10.1186/1479-5876-11-255
Campbell, L., Jasani, B., Edwards, K., Gumbleton, M., and Griffiths, D. F. (2008). Combined Expression of Caveolin-1 and an Activated AKT/mTOR Pathway Predicts Reduced Disease-free Survival in Clinically Confined Renal Cell Carcinoma. Br. J. Cancer 98 (5), 931–940. doi:10.1038/sj.bjc.6604243
Campos, A., Burgos-Ravanal, R., González, M. F., Huilcaman, R., Lobos González, L., and Quest, A. F. G. (2019). Cell Intrinsic and Extrinsic Mechanisms of Caveolin-1-Enhanced Metastasis. Biomolecules 9 (8), 314. doi:10.3390/biom9080314
Cao, S., Xiao, L., Wang, J., Chen, G., and Liu, Y. (2021). The RNA-Binding Protein HuR Regulates Intestinal Epithelial Restitution by Modulating Caveolin-1 Gene Expression. Biochem. J. 478 (1), 247–260. doi:10.1042/bcj20200372
Chang, C. C., Chen, C. Y., Wen, H. C., Huang, C. Y., Hung, M. S., Lu, H. C., et al. (2017). Caveolin-1 Secreted from Adipose Tissues and Adipocytes Functions as an Adipogenesis Enhancer. Obesity (Silver Spring) 25 (11), 1932–1940. doi:10.1002/oby.21970
Chang, T. Y., Chang, C. C., Ohgami, N., and Yamauchi, Y. (2006). Cholesterol Sensing, Trafficking, and Esterification. Annu. Rev. Cel Dev Biol 22, 129–157. doi:10.1146/annurev.cellbio.22.010305.104656
Chatterjee, M., Ben-Josef, E., Robb, R., Vedaie, M., Seum, S., Thirumoorthy, K., et al. (2017). Caveolae-Mediated Endocytosis Is Critical for Albumin Cellular Uptake and Response to Albumin-Bound Chemotherapy. Cancer Res. 77 (21), 5925–5937. doi:10.1158/0008-5472.Can-17-0604
Chen, J., Capozza, F., Wu, A., Deangelis, T., Sun, H., Lisanti, M., et al. (2008). Regulation of Insulin Receptor Substrate-1 Expression Levels by Caveolin-1. J. Cel Physiol 217 (1), 281–289. doi:10.1002/jcp.21498
Chen, W., Chen, Y., Qin, L., Li, A., Zhao, X., Wang, X., et al. (2011). Transcription Factor Sp1 Is Essential for the Regulation of the Porcine Caveolin-1 Gene. DNA Cel Biol 30 (7), 491–497. doi:10.1089/dna.2010.1202
Chen, Y., Liu, C., Xie, B., Chen, S., Zhuang, Y., and Zhang, S. (2020). miR-96 Exerts an Oncogenic Role in the Progression of Cervical Cancer by Targeting CAV-1. Mol. Med. Rep. 22 (1), 543–550. doi:10.3892/mmr.2020.11101
Chen, Y. H., Lin, W. W., Liu, C. S., Hsu, L. S., Lin, Y. M., and Su, S. L. (2016). Caveolin-1 Expression Ameliorates Nephrotic Damage in a Rabbit Model of Cholesterol-Induced Hypercholesterolemia. PLoS One 11 (4), e0154210. doi:10.1371/journal.pone.0154210
Chen, Y. H., Lin, W. W., Liu, C. S., and Su, S. L. (2017). H2O2 Induces Caveolin-1 D-egradation and I-mpaired M-itochondrial F-unction in E11 P-odocytes. Mol. Med. Rep. 16 (5), 7841–7847. doi:10.3892/mmr.2017.7497
Chen, Z., Nie, S. D., Qu, M. L., Zhou, D., Wu, L. Y., Shi, X. J., et al. (2018). The Autophagic Degradation of Cav-1 Contributes to PA-induced Apoptosis and Inflammation of Astrocytes. Cell Death Dis 9 (7), 771. doi:10.1038/s41419-018-0795-3
Chen, Z. H., Cao, J. F., Zhou, J. S., Liu, H., Che, L. Q., Mizumura, K., et al. (2014). Interaction of Caveolin-1 with ATG12-ATG5 System Suppresses Autophagy in Lung Epithelial Cells. Am. J. Physiol. Lung Cel Mol Physiol 306 (11), L1016–L1025. doi:10.1152/ajplung.00268.2013
Cheng, J. P. X., and Nichols, B. J. (2016). Caveolae: One Function or Many. Trends Cel Biol 26 (3), 177–189. doi:10.1016/j.tcb.2015.10.010
Codenotti, S., Marampon, F., Triggiani, L., Bonù, M. L., Magrini, S. M., Ceccaroli, P., et al. (2021). Caveolin-1 Promotes Radioresistance in Rhabdomyosarcoma through Increased Oxidative Stress protection and DNA Repair. Cancer Lett. 505, 1–12. doi:10.1016/j.canlet.2021.02.005
Coelho-Santos, V., Socodato, R., Portugal, C., Leitão, R. A., Rito, M., Barbosa, M., et al. (2016). Methylphenidate-triggered ROS Generation Promotes Caveolae-Mediated Transcytosis via Rac1 Signaling and C-src-dependent Caveolin-1 Phosphorylation in Human Brain Endothelial Cells. Cell Mol Life Sci 73 (24), 4701–4716. doi:10.1007/s00018-016-2301-3
Cohen, A. W., Razani, B., Schubert, W., Williams, T. M., Wang, X. B., Iyengar, P., et al. (2004). Role of Caveolin-1 in the Modulation of Lipolysis and Lipid Droplet Formation. Diabetes 53 (5), 1261–1270. doi:10.2337/diabetes.53.5.1261
Collins, B. M., Davis, M. J., Hancock, J. F., and Parton, R. G. (2012). Structure-based Reassessment of the Caveolin Signaling Model: Do Caveolae Regulate Signaling through Caveolin-Protein Interactions. Dev. Cel 23 (1), 11–20. doi:10.1016/j.devcel.2012.06.012
Couet, J., Sargiacomo, M., and Lisanti, M. P. (1997). Interaction of a Receptor Tyrosine Kinase, EGF-R, with Caveolins. Caveolin Binding Negatively Regulates Tyrosine and Serine/threonine Kinase Activities. J. Biol. Chem. 272 (48), 30429–30438. doi:10.1074/jbc.272.48.30429
Damm, E. M., Pelkmans, L., Kartenbeck, J., Mezzacasa, A., Kurzchalia, T., and Helenius, A. (2005). Clathrin- and Caveolin-1-independent Endocytosis: Entry of Simian Virus 40 into Cells Devoid of Caveolae. J. Cel Biol 168 (3), 477–488. doi:10.1083/jcb.200407113
Das, D., Sarkar, S., Bordoloi, J., Wann, S. B., Kalita, J., and Manna, P. (2018). Daidzein, its Effects on Impaired Glucose and Lipid Metabolism and Vascular Inflammation Associated with Type 2 Diabetes. Biofactors 44 (5), 407–417. doi:10.1002/biof.1439
Dasari, A., Bartholomew, J. N., Volonte, D., and Galbiati, F. (2006). Oxidative Stress Induces Premature Senescence by Stimulating Caveolin-1 Gene Transcription through P38 Mitogen-Activated Protein kinase/Sp1-Mediated Activation of Two GC-Rich Promoter Elements. Cancer Res. 66 (22), 10805–10814. doi:10.1158/0008-5472.Can-06-1236
de Souza, G. M., de Albuquerque Borborema, M. E., de Lucena, T. M. C., da Silva Santos, A. F., de Lima, B. R., de Oliveira, D. C., et al. (2020). Caveolin-1 (CAV-1) up Regulation in Metabolic Syndrome: All Roads Leading to the Same End. Mol. Biol. Rep. 47 (11), 9245–9250. doi:10.1007/s11033-020-05945-y
DeBose-Boyd, R. A., and Ye, J. (2018). SREBPs in Lipid Metabolism, Insulin Signaling, and beyond. Trends Biochem. Sci. 43 (5), 358–368. doi:10.1016/j.tibs.2018.01.005
Ding, L., Zeng, Q., Wu, J., Li, D., Wang, H., Lu, W., et al. (2017). Caveolin-1 R-egulates O-xidative S-tress-induced S-enescence in N-ucleus P-ulposus C-ells P-rimarily via the P-53/p21 S-ignaling P-athway I-n vitro. Mol. Med. Rep. 16 (6), 9521–9527. doi:10.3892/mmr.2017.7789
Elvira, B., Honisch, S., Almilaji, A., Pakladok, T., Liu, G., Shumilina, E., et al. (2013). Up-regulation of Na(+)-Coupled Glucose Transporter SGLT1 by Caveolin-1. Biochim. Biophys. Acta 1828 (11), 2394–2398. doi:10.1016/j.bbamem.2013.06.007
Emmerich, F., Zschiedrich, S., Reichenbach-Braun, C., Süsal, C., Minguet, S., Pauly, M. C., et al. (2021). Low Pre-transplant Caveolin-1 Serum Concentrations Are Associated with Acute Cellular Tubulointerstitial Rejection in Kidney Transplantation. Molecules 26 (9), 2648. doi:10.3390/molecules26092648
Engelman, J. A., Zhang, X. L., Galbiati, F., and Lisanti, M. P. (1998). Chromosomal Localization, Genomic Organization, and Developmental Expression of the Murine Caveolin Gene Family (Cav-1, -2, and -3). Cav-1 and Cav-2 Genes Map to a Known Tumor Suppressor Locus (6-A2/7q31). FEBS Lett. 429 (3), 330–336. doi:10.1016/s0014-5793(98)00619-x
Epand, R. M., Sayer, B. G., and Epand, R. F. (2005). Caveolin Scaffolding Region and Cholesterol-Rich Domains in Membranes. J. Mol. Biol. 345 (2), 339–350. doi:10.1016/j.jmb.2004.10.064
Fachim, H. A., Siddals, K., Malipatil, N., Donn, R. P., Moreno, G. Y., Dalton, C. F., et al. (2020). Lifestyle Intervention in Individuals with Impaired Glucose Regulation Affects Caveolin-1 Expression and DNA Methylation. Adipocyte 9 (1), 96–107. doi:10.1080/21623945.2020.1732513
Fernández-Rojo, M. A., Restall, C., Ferguson, C., Martel, N., Martin, S., Bosch, M., et al. (2012). Caveolin-1 Orchestrates the Balance between Glucose and Lipid-dependent Energy Metabolism: Implications for Liver Regeneration. Hepatology 55 (5), 1574–1584. doi:10.1002/hep.24810
Fiala, G. J., and Minguet, S. (2018). Caveolin-1: The Unnoticed Player in TCR and BCR Signaling. Adv. Immunol. 137, 83–133. doi:10.1016/bs.ai.2017.12.002
Fielding, P. E., and Fielding, C. J. (1995). Plasma Membrane Caveolae Mediate the Efflux of Cellular Free Cholesterol. Biochemistry 34 (44), 14288–14292. doi:10.1021/bi00044a004
Filippini, A., and D'Alessio, A. (2020). Caveolae and Lipid Rafts in Endothelium: Valuable Organelles for Multiple Functions. Biomolecules 10 (9), 1218. doi:10.3390/biom10091218
Frank, P. G., Cheung, M. W., Pavlides, S., Llaverias, G., Park, D. S., and Lisanti, M. P. (2006). Caveolin-1 and Regulation of Cellular Cholesterol Homeostasis. Am. J. Physiol. Heart Circ. Physiol. 291 (2), H677–H686. doi:10.1152/ajpheart.01092.2005
Fridolfsson, H. N., Roth, D. M., Insel, P. A., and Patel, H. H. (2014). Regulation of Intracellular Signaling and Function by Caveolin. Faseb j 28 (9), 3823–3831. doi:10.1096/fj.14-252320
Frühbeck, G., Méndez-Giménez, L., Fernández-Formoso, J.-A., Fernández, S., and Rodríguez, A. (2014). Regulation of Adipocyte Lipolysis. Nutr. Res. Rev. 27 (1), 63–93. doi:10.1017/s095442241400002x
Fujigaki, Y., Sakakima, M., Sun, Y., Goto, T., Ohashi, N., Fukasawa, H., et al. (2007). Immunohistochemical Study on Caveolin-1alpha in Regenerating Process of Tubular Cells in Gentamicin-Induced Acute Tubular Injury in Rats. Virchows Arch. 450 (6), 671–681. doi:10.1007/s00428-007-0417-4
Fujimoto, T., Kogo, H., Nomura, R., and Une, T. (2000). Isoforms of Caveolin-1 and Caveolar Structure. J. Cel Sci 113 Pt 19, 3509–3517. doi:10.1242/jcs.113.19.3509
Gao, M., Huang, X., Song, B. L., and Yang, H. (2019). The Biogenesis of Lipid Droplets: Lipids Take center Stage. Prog. Lipid Res. 75, 100989. doi:10.1016/j.plipres.2019.100989
Gao, Y., Zhao, Y., Pan, J., Yang, L., Huang, T., Feng, X., et al. (2014). Treadmill Exercise Promotes Angiogenesis in the Ischemic Penumbra of Rat Brains through Caveolin-1/VEGF Signaling Pathways. Brain Res. 1585, 83–90. doi:10.1016/j.brainres.2014.08.032
Geltinger, F., Schartel, L., Wiederstein, M., Tevini, J., Aigner, E., Felder, T. K., et al. (2020). Friend or Foe: Lipid Droplets as Organelles for Protein and Lipid Storage in Cellular Stress Response, Aging and Disease. Molecules 25 (21), 5053. doi:10.3390/molecules25215053
Gerbod-Giannone, M. C., Dallet, L., Naudin, G., Sahin, A., Decossas, M., Poussard, S., et al. (2019). Involvement of Caveolin-1 and CD36 in Native LDL Endocytosis by Endothelial Cells. Biochim. Biophys. Acta Gen. Subj 1863 (5), 830–838. doi:10.1016/j.bbagen.2019.01.005
Gewin, L. S. (2021). Sugar or Fat? Renal Tubular Metabolism Reviewed in Health and Disease. Nutrients 13 (5), 1580. doi:10.3390/nu13051580
Glenney, J. R. (1992). The Sequence of Human Caveolin Reveals Identity with VIP21, a Component of Transport Vesicles. FEBS Lett. 314 (1), 45–48. doi:10.1016/0014-5793(92)81458-x
Glenney, J. R. (1989). Tyrosine Phosphorylation of a 22-kDa Protein Is Correlated with Transformation by Rous Sarcoma Virus. J. Biol. Chem. 264 (34), 20163–20166. doi:10.1016/s0021-9258(19)47038-5
Gopu, V., Fan, L., Shetty, R. S., Nagaraja, M. R., and Shetty, S. (2020). Caveolin-1 Scaffolding Domain Peptide Regulates Glucose Metabolism in Lung Fibrosis. JCI Insight 5 (19), e137969. doi:10.1172/jci.insight.137969
Goutas, A., Papathanasiou, I., Mourmoura, E., Tsesmelis, K., Tsezou, A., and Trachana, V. (2020). Oxidative Stress Response Is Mediated by Overexpression and Spatiotemporal Regulation of Caveolin-1. Antioxidants (Basel) 9 (8), 766. doi:10.3390/antiox9080766
Grayson, P. C., Eddy, S., Taroni, J. N., Lightfoot, Y. L., Mariani, L., Parikh, H., et al. (2018). Metabolic Pathways and Immunometabolism in Rare Kidney Diseases. Ann. Rheum. Dis. 77 (8), 1226–1233. doi:10.1136/annrheumdis-2017-212935
Guan, T. H., Chen, G., Gao, B., Janssen, M. R., Uttarwar, L., Ingram, A. J., et al. (2013). Caveolin-1 Deficiency Protects against Mesangial Matrix Expansion in a Mouse Model of Type 1 Diabetic Nephropathy. Diabetologia 56 (9), 2068–2077. doi:10.1007/s00125-013-2968-z
Gvaramia, D., Blaauboer, M. E., Hanemaaijer, R., and Everts, V. (2013). Role of Caveolin-1 in Fibrotic Diseases. Matrix Biol. 32 (6), 307–315. doi:10.1016/j.matbio.2013.03.005
Ha, T. K., and Chi, S. G. (2012). CAV1/caveolin 1 Enhances Aerobic Glycolysis in colon Cancer Cells via Activation of SLC2A3/GLUT3 Transcription. Autophagy 8 (11), 1684–1685. doi:10.4161/auto.21487
Ha, T. K., Her, N. G., Lee, M. G., Ryu, B. K., Lee, J. H., Han, J., et al. (2012). Caveolin-1 Increases Aerobic Glycolysis in Colorectal Cancers by Stimulating HMGA1-Mediated GLUT3 Transcription. Cancer Res. 72 (16), 4097–4109. doi:10.1158/0008-5472.Can-12-0448
Haddad, D., Al Madhoun, A., Nizam, R., and Al-Mulla, F. (2020). Role of Caveolin-1 in Diabetes and its Complications. Oxid Med. Cel Longev 2020, 9761539. doi:10.1155/2020/9761539
Hall, D. P., Cost, N. G., Hegde, S., Kellner, E., Mikhaylova, O., Stratton, Y., et al. (2014). TRPM3 and miR-204 Establish a Regulatory Circuit that Controls Oncogenic Autophagy in clear Cell Renal Cell Carcinoma. Cancer Cell 26 (5), 738–753. doi:10.1016/j.ccell.2014.09.015
Han, B., Porta, J. C., Hanks, J. L., Peskova, Y., Binshtein, E., Dryden, K., et al. (2020). Structure and Assembly of CAV1 8S Complexes Revealed by Single Particle Electron Microscopy. Sci. Adv. 6 (49), eabc6185. doi:10.1126/sciadv.abc6185
Han, M., Nwosu, Z. C., Piorońska, W., Ebert, M. P., Dooley, S., and Meyer, C. (2019). Caveolin-1 Impacts on TGF-β Regulation of Metabolic Gene Signatures in Hepatocytes. Front. Physiol. 10, 1606. doi:10.3389/fphys.2019.01606
Hansen, C. G., Bright, N. A., Howard, G., and Nichols, B. J. (2009). SDPR Induces Membrane Curvature and Functions in the Formation of Caveolae. Nat. Cel Biol 11 (7), 807–814. doi:10.1038/ncb1887
Hansen, C. G., and Nichols, B. J. (2010). Exploring the Caves: Cavins, Caveolins and Caveolae. Trends Cel Biol 20 (4), 177–186. doi:10.1016/j.tcb.2010.01.005
Henne, M., Goodman, J. M., and Hariri, H. (2020). Spatial Compartmentalization of Lipid Droplet Biogenesis. Biochim. Biophys. Acta Mol. Cel Biol Lipids 1865 (1), 158499. doi:10.1016/j.bbalip.2019.07.008
Hill, M. M., Bastiani, M., Luetterforst, R., Kirkham, M., Kirkham, A., Nixon, S. J., et al. (2008). PTRF-cavin, a Conserved Cytoplasmic Protein Required for Caveola Formation and Function. Cell 132 (1), 113–124. doi:10.1016/j.cell.2007.11.042
Hirama, T., Das, R., Yang, Y., Ferguson, C., Won, A., Yip, C. M., et al. (2017). Phosphatidylserine Dictates the Assembly and Dynamics of Caveolae in the Plasma Membrane. J. Biol. Chem. 292 (34), 14292–14307. doi:10.1074/jbc.M117.791400
Hoernke, M., Mohan, J., Larsson, E., Blomberg, J., Kahra, D., Westenhoff, S., et al. (2017). EHD2 Restrains Dynamics of Caveolae by an ATP-dependent, Membrane-Bound, Open Conformation. Proc. Natl. Acad. Sci. U S A. 114 (22), E4360–e4369. doi:10.1073/pnas.1614066114
Hoop, C. L., Sivanandam, V. N., Kodali, R., Srnec, M. N., and van der Wel, P. C. (2012). Structural Characterization of the Caveolin Scaffolding Domain in Association with Cholesterol-Rich Membranes. Biochemistry 51 (1), 90–99. doi:10.1021/bi201356v
Hu, R., Yan, H., Fei, X., Liu, H., and Wu, J. (2017). Modulation of Glucose Metabolism by a Natural Compound from Chloranthus Japonicus via Activation of AMP-Activated Protein Kinase. Sci. Rep. 7 (1), 778. doi:10.1038/s41598-017-00925-y
Huang, C., Qiu, Z., Wang, L., Peng, Z., Jia, Z., Logsdon, C. D., et al. (2012). A Novel FoxM1-Caveolin Signaling Pathway Promotes Pancreatic Cancer Invasion and Metastasis. Cancer Res. 72 (3), 655–665. doi:10.1158/0008-5472.Can-11-3102
Huang, C., Zhou, Y., Huang, H., Zheng, Y., Kong, L., Zhang, H., et al. (2021). Islet Transplantation Reverses Podocyte Injury in Diabetic Nephropathy or Induced by High Glucose via Inhibiting RhoA/ROCK/NF-κB Signaling Pathway. J. Diabetes Res. 2021, 9570405. doi:10.1155/2021/9570405
Huang, G., Lou, T., Pan, J., Ye, Z., Yin, Z., Li, L., et al. (2019). MiR-204 Reduces Cisplatin Resistance in Non-small Cell Lung Cancer through Suppression of the caveolin-1/AKT/Bad Pathway. Aging (Albany NY) 11 (7), 2138–2150. doi:10.18632/aging.101907
Huang, J. H., Lu, L., Lu, H., Chen, X., Jiang, S., and Chen, Y. H. (2007). Identification of the HIV-1 Gp41 Core-Binding Motif in the Scaffolding Domain of Caveolin-1. J. Biol. Chem. 282 (9), 6143–6152. doi:10.1074/jbc.M607701200
Hubert, M., Larsson, E., and Lundmark, R. (2020a). Keeping in Touch with the Membrane; Protein- and Lipid-Mediated Confinement of Caveolae to the Cell Surface. Biochem. Soc. Trans. 48 (1), 155–163. doi:10.1042/bst20190386
Hubert, M., Larsson, E., Vegesna, N. V. G., Ahnlund, M., Johansson, A. I., Moodie, L. W., et al. (2020b). Lipid Accumulation Controls the Balance between Surface Connection and Scission of Caveolae. Elife 9, e55038. doi:10.7554/eLife.55038
Ikezu, T., Ueda, H., Trapp, B. D., Nishiyama, K., Sha, J. F., Volonte, D., et al. (1998). Affinity-purification and Characterization of Caveolins from the Brain: Differential Expression of Caveolin-1, -2, and -3 in Brain Endothelial and Astroglial Cell Types. Brain Res. 804 (2), 177–192. doi:10.1016/s0006-8993(98)00498-3
Ikonen, E., and Parton, R. G. (2000). Caveolins and Cellular Cholesterol Balance. Traffic 1 (3), 212–217. doi:10.1034/j.1600-0854.2000.010303.x
Ishizaka, N., Griendling, K. K., Lassègue, B., and Alexander, R. W. (1998). Angiotensin II Type 1 Receptor: Relationship with Caveolae and Caveolin after Initial Agonist Stimulation. Hypertension 32 (3), 459–466. doi:10.1161/01.hyp.32.3.459
Itabe, H., Yamaguchi, T., Nimura, S., and Sasabe, N. (2017). Perilipins: a Diversity of Intracellular Lipid Droplet Proteins. Lipids Health Dis. 16 (1), 83. doi:10.1186/s12944-017-0473-y
Janković, J., Tatić, S., Božić, V., Živaljević, V., Cvejić, D., and Paskaš, S. (2017). Inverse Expression of Caveolin-1 and EGFR in Thyroid Cancer Patients. Hum. Pathol. 61, 164–172. doi:10.1016/j.humpath.2016.10.020
Ji, D. G., Zhang, Y., Yao, S. M., Zhai, X. J., Zhang, L. R., Zhang, Y. Z., et al. (2018). Cav-1 Deficiency Promotes Liver Fibrosis in Carbon Tetrachloride (CCl4)-Induced Mice by Regulation of Oxidative Stress and Inflammation Responses. Biomed. Pharmacother. 102, 26–33. doi:10.1016/j.biopha.2018.03.016
Jin, J., Peng, C., Wu, S. Z., Chen, H. M., and Zhang, B. F. (2015). Blocking VEGF/Caveolin-1 Signaling Contributes to Renal protection of Fasudil in Streptozotocin-Induced Diabetic Rats. Acta Pharmacol. Sin 36 (7), 831–840. doi:10.1038/aps.2015.23
Joo, H. J., Oh, D. K., Kim, Y. S., Lee, K. B., and Kim, S. J. (2004). Increased Expression of Caveolin-1 and Microvessel Density Correlates with Metastasis and Poor Prognosis in clear Cell Renal Cell Carcinoma. BJU Int. 93 (3), 291–296. doi:10.1111/j.1464-410x.2004.04604.x
Kang, H. M., Ahn, S. H., Choi, P., Ko, Y. A., Han, S. H., Chinga, F., et al. (2015). Defective Fatty Acid Oxidation in Renal Tubular Epithelial Cells Has a Key Role in Kidney Fibrosis Development. Nat. Med. 21 (1), 37–46. doi:10.1038/nm.3762
Kogo, H., and Fujimoto, T. (2000). Caveolin-1 Isoforms Are Encoded by Distinct mRNAs. Identification of Mouse Caveolin-1 mRNA Variants Caused by Alternative Transcription Initiation and Splicing. FEBS Lett. 465 (2-3), 119–123. doi:10.1016/s0014-5793(99)01730-5
Kong, D., Zhang, Z., Chen, L., Huang, W., Zhang, F., Wang, L., et al. (2020). Curcumin Blunts Epithelial-Mesenchymal Transition of Hepatocytes to Alleviate Hepatic Fibrosis through Regulating Oxidative Stress and Autophagy. Redox Biol. 36, 101600. doi:10.1016/j.redox.2020.101600
Kongkham, S., Sriwong, S., and Tasanarong, A. (2016). Erythropoietin Administration Promotes Expression of VEGF in Renal Ischemic–Reperfusion Injury in Rat Model. J. Med. Assoc. Thai 99 (Suppl. 4), S246–S255.
Kotova, A., Timonina, K., and Zoidl, G. R. (2020). Endocytosis of Connexin 36 Is Mediated by Interaction with Caveolin-1. Int. J. Mol. Sci. 21 (15). doi:10.3390/ijms21155401
Kovtun, O., Tillu, V. A., Ariotti, N., Parton, R. G., and Collins, B. M. (2015). Cavin Family Proteins and the Assembly of Caveolae. J. Cel Sci 128 (7), 1269–1278. doi:10.1242/jcs.167866
Krilov, L., Nguyen, A., Miyazaki, T., Unson, C. G., Williams, R., Lee, N. H., et al. (2011). Dual Mode of Glucagon Receptor Internalization: Role of PKCα, GRKs and β-arrestins. Exp. Cel Res 317 (20), 2981–2994. doi:10.1016/j.yexcr.2011.10.001
Krishna, A., and Sengupta, D. (2019). Interplay between Membrane Curvature and Cholesterol: Role of Palmitoylated Caveolin-1. Biophys. J. 116 (1), 69–78. doi:10.1016/j.bpj.2018.11.3127
Kuo, A., Lee, M. Y., Yang, K., Gross, R. W., and Sessa, W. C. (2018). Caveolin-1 Regulates Lipid Droplet Metabolism in Endothelial Cells via Autocrine Prostacyclin-Stimulated, cAMP-Mediated Lipolysis. J. Biol. Chem. 293 (3), 973–983. doi:10.1074/jbc.RA117.000980
Kurzchalia, T. V., Dupree, P., Parton, R. G., Kellner, R., Virta, H., Lehnert, M., et al. (1992). VIP21, a 21-kD Membrane Protein Is an Integral Component of Trans-golgi-network-derived Transport Vesicles. J. Cel Biol 118 (5), 1003–1014. doi:10.1083/jcb.118.5.1003
Le Lay, S., Hajduch, E., Lindsay, M. R., Le Lièpvre, X., Thiele, C., Ferré, P., et al. (2006). Cholesterol-induced Caveolin Targeting to Lipid Droplets in Adipocytes: a Role for Caveolar Endocytosis. Traffic 7 (5), 549–561. doi:10.1111/j.1600-0854.2006.00406.x
Lee, C. Y., Lai, T. Y., Tsai, M. K., Chang, Y. C., Ho, Y. H., Yu, I. S., et al. (2017). The Ubiquitin Ligase ZNRF1 Promotes Caveolin-1 Ubiquitination and Degradation to Modulate Inflammation. Nat. Commun. 8, 15502. doi:10.1038/ncomms15502
Lee, E. K., Lee, Y. S., Han, I. O., and Park, S. H. (2007). Expression of Caveolin-1 Reduces Cellular Responses to TGF-Beta1 through Down-Regulating the Expression of TGF-Beta Type II Receptor Gene in NIH3T3 Fibroblast Cells. Biochem. Biophys. Res. Commun. 359 (2), 385–390. doi:10.1016/j.bbrc.2007.05.121
Lee, Y. J., Kim, M. O., Ryu, J. M., and Han, H. J. (2012). Regulation of SGLT Expression and Localization through Epac/PKA-dependent Caveolin-1 and F-Actin Activation in Renal Proximal Tubule Cells. Biochim. Biophys. Acta 1823 (4), 971–982. doi:10.1016/j.bbamcr.2011.12.011
Li, J., Li, P., Zhang, G., Qin, P., Zhang, D., and Zhao, W. (2020). CircRNA TADA2A Relieves Idiopathic Pulmonary Fibrosis by Inhibiting Proliferation and Activation of Fibroblasts. Cel Death Dis 11 (7), 553. doi:10.1038/s41419-020-02747-9
Li, R., Guo, Y., Zhang, Y., Zhang, X., Zhu, L., and Yan, T. (2019). Salidroside Ameliorates Renal Interstitial Fibrosis by Inhibiting the TLR4/NF-Κb and MAPK Signaling Pathways. Int. J. Mol. Sci. 20 (5), 1103. doi:10.3390/ijms20051103
Li, S., Couet, J., and Lisanti, M. P. (1996a). Src Tyrosine Kinases, Galpha Subunits, and H-Ras Share a Common Membrane-Anchored Scaffolding Protein, Caveolin. Caveolin Binding Negatively Regulates the Auto-Activation of Src Tyrosine Kinases. J. Biol. Chem. 271 (46), 29182–29190. doi:10.1074/jbc.271.46.29182
Li, S., Seitz, R., and Lisanti, M. P. (1996b). Phosphorylation of Caveolin by Src Tyrosine Kinases. The Alpha-Isoform of Caveolin Is Selectively Phosphorylated by V-Src In Vivo. J. Biol. Chem. 271 (7), 3863–3868. doi:10.1074/jbc.271.7.3863
Ling, X., Li, Y., Qiu, F., Lu, X., Yang, L., Chen, J., et al. (2020). Down Expression of Lnc-BMP1-1 Decreases that of Caveolin-1 Is Associated with the Lung Cancer Susceptibility and Cigarette Smoking History. Aging (Albany NY) 12 (1), 462–480. doi:10.18632/aging.102633
Liu, A., Wu, Q., Guo, J., Ares, I., Rodríguez, J. L., Martínez-Larrañaga, M. R., et al. (2019). Statins: Adverse Reactions, Oxidative Stress and Metabolic Interactions. Pharmacol. Ther. 195, 54–84. doi:10.1016/j.pharmthera.2018.10.004
Liu, L., Xu, H. X., Wang, W. Q., Wu, C. T., Chen, T., Qin, Y., et al. (2014). Cavin-1 Is Essential for the Tumor-Promoting Effect of Caveolin-1 and Enhances its Prognostic Potency in Pancreatic Cancer. Oncogene 33 (21), 2728–2736. doi:10.1038/onc.2013.223
Liu, P., Ying, Y., Zhao, Y., Mundy, D. I., Zhu, M., and Anderson, R. G. (2004). Chinese Hamster Ovary K2 Cell Lipid Droplets Appear to Be Metabolic Organelles Involved in Membrane Traffic. J. Biol. Chem. 279 (5), 3787–3792. doi:10.1074/jbc.M311945200
Liu, S., Premont, R. T., Singh, S., and Rockey, D. C. (2017). Caveolin 1 and G-Protein-Coupled Receptor Kinase-2 Coregulate Endothelial Nitric Oxide Synthase Activity in Sinusoidal Endothelial Cells. Am. J. Pathol. 187 (4), 896–907. doi:10.1016/j.ajpath.2016.11.017
Liu, Y., Fu, Y., Hu, X., Chen, S., Miao, J., Wang, Y., et al. (2020). Caveolin-1 Knockdown Increases the Therapeutic Sensitivity of Lung Cancer to Cisplatin-Induced Apoptosis by Repressing Parkin-Related Mitophagy and Activating the ROCK1 Pathway. J. Cel Physiol 235 (2), 1197–1208. doi:10.1002/jcp.29033
Llaverias, G., Vázquez-Carrera, M., Sánchez, R. M., Noé, V., Ciudad, C. J., Laguna, J. C., et al. (2004). Rosiglitazone Upregulates Caveolin-1 Expression in THP-1 Cells through a PPAR-dependent Mechanism. J. Lipid Res. 45 (11), 2015–2024. doi:10.1194/jlr.M400049-JLR200
Luo, J., Yang, H., and Song, B. L. (2020a). Mechanisms and Regulation of Cholesterol Homeostasis. Nat. Rev. Mol. Cel Biol 21 (4), 225–245. doi:10.1038/s41580-019-0190-7
Luo, M., Xu, C., Luo, Y., Wang, G., Wu, J., and Wan, Q. (2020b). Circulating miR-103 Family as Potential Biomarkers for Type 2 Diabetes through Targeting CAV-1 and SFRP4. Acta Diabetol. 57 (3), 309–322. doi:10.1007/s00592-019-01430-6
Luo, Z., Rong, Z., Zhang, J., Zhu, Z., Yu, Z., Li, T., et al. (2020c). Circular RNA circCCDC9 Acts as a miR-6792-3p Sponge to Suppress the Progression of Gastric Cancer through Regulating CAV1 Expression. Mol. Cancer 19 (1), 86. doi:10.1186/s12943-020-01203-8
Machin, P. A., Tsonou, E., Hornigold, D. C., and Welch, H. C. E. (2021). Rho Family GTPases and Rho GEFs in Glucose Homeostasis. Cells 10 (4), 915. doi:10.3390/cells10040915
Macovei, A., Radulescu, C., Lazar, C., Petrescu, S., Durantel, D., Dwek, R. A., et al. (2010). Hepatitis B Virus Requires Intact Caveolin-1 Function for Productive Infection in HepaRG Cells. J. Virol. 84 (1), 243–253. doi:10.1128/jvi.01207-09
Mahmoudi, M., Willgoss, D., Cuttle, L., Yang, T., Pat, B., Winterford, C., et al. (2003). In Vivo and In Vitro Models Demonstrate a Role for Caveolin-1 in the Pathogenesis of Ischaemic Acute Renal Failure. J. Pathol. 200 (3), 396–405. doi:10.1002/path.1368
Martínez, M., Martínez, N. A., Miranda, J. D., Maldonado, H. M., and Silva Ortiz, W. I. (2019). Caveolin-1 Regulates P2Y2 Receptor Signaling during Mechanical Injury in Human 1321N1 Astrocytoma. Biomolecules 9 (10), 622. doi:10.3390/biom9100622
Martinez, N. A., Ayala, A. M., Martinez, M., Martinez-Rivera, F. J., Miranda, J. D., and Silva, W. I. (2016). Caveolin-1 Regulates the P2Y2 Receptor Signaling in Human 1321N1 Astrocytoma Cells. J. Biol. Chem. 291 (23), 12208–12222. doi:10.1074/jbc.M116.730226
Matthaeus, C., Lahmann, I., Kunz, S., Jonas, W., Melo, A. A., Lehmann, M., et al. (2020). EHD2-mediated Restriction of Caveolar Dynamics Regulates Cellular Fatty Acid Uptake. Proc. Natl. Acad. Sci. U S A. 117 (13), 7471–7481. doi:10.1073/pnas.1918415117
Matthaeus, C., and Taraska, J. W. (2020). Energy and Dynamics of Caveolae Trafficking. Front Cel Dev Biol 8, 614472. doi:10.3389/fcell.2020.614472
Matveev, S., van der Westhuyzen, D. R., and Smart, E. J. (1999). Co-expression of Scavenger Receptor-BI and Caveolin-1 Is Associated with Enhanced Selective Cholesteryl Ester Uptake in THP-1 Macrophages. J. Lipid Res. 40 (9), 1647–1654. doi:10.1016/s0022-2275(20)33410-6
Mayurasakorn, K., Hasanah, N., Homma, T., Homma, M., Rangel, I. K., Garza, A. E., et al. (2018). Caloric Restriction Improves Glucose Homeostasis, yet Increases Cardiometabolic Risk in Caveolin-1-Deficient Mice. Metabolism 83, 92–101. doi:10.1016/j.metabol.2018.01.012
McMahon, K. A., Zajicek, H., Li, W. P., Peyton, M. J., Minna, J. D., Hernandez, V. J., et al. (2009). SRBC/cavin-3 Is a Caveolin Adapter Protein that Regulates Caveolae Function. Embo j 28 (8), 1001–1015. doi:10.1038/emboj.2009.46
Mehta, N., Li, R., Zhang, D., Soomro, A., He, J., Zhang, I., et al. (2021). miR299a-5p Promotes Renal Fibrosis by Suppressing the Antifibrotic Actions of Follistatin. Sci. Rep. 11 (1), 88. doi:10.1038/s41598-020-80199-z
Meng, H., Fu, G., Shen, J., Shen, K., Xu, Z., Wang, Y., et al. (2017). Ameliorative Effect of Daidzein on Cisplatin-Induced Nephrotoxicity in Mice via Modulation of Inflammation, Oxidative Stress, and Cell Death. Oxid Med. Cel Longev 2017, 3140680. doi:10.1155/2017/3140680
Mergia, A. (2017). The Role of Caveolin 1 in HIV Infection and Pathogenesis. Viruses 9 (6), 129. doi:10.3390/v9060129
Mitrofanova, A., Mallela, S. K., Ducasa, G. M., Yoo, T. H., Rosenfeld-Gur, E., Zelnik, I. D., et al. (2019). SMPDL3b Modulates Insulin Receptor Signaling in Diabetic Kidney Disease. Nat. Commun. 10 (1), 2692. doi:10.1038/s41467-019-10584-4
Møller, L. L. V., Klip, A., and Sylow, L. (2019). Rho GTPases-Emerging Regulators of Glucose Homeostasis and Metabolic Health. Cells 8 (5), 434. doi:10.3390/cells8050434
Moore, J., McKnight, A. J., Simmonds, M. J., Courtney, A. E., Hanvesakul, R., Brand, O. J., et al. (2010). Association of Caveolin-1 Gene Polymorphism with Kidney Transplant Fibrosis and Allograft Failure. Jama 303 (13), 1282–1287. doi:10.1001/jama.2010.356
Morén, B., Shah, C., Howes, M. T., Schieber, N. L., McMahon, H. T., Parton, R. G., et al. (2012). EHD2 Regulates Caveolar Dynamics via ATP-Driven Targeting and Oligomerization. Mol. Biol. Cel 23 (7), 1316–1329. doi:10.1091/mbc.E11-09-0787
Moriyama, T., Marquez, J. P., Wakatsuki, T., and Sorokin, A. (2007). Caveolar Endocytosis Is Critical for BK Virus Infection of Human Renal Proximal Tubular Epithelial Cells. J. Virol. 81 (16), 8552–8562. doi:10.1128/jvi.00924-07
Moriyama, T., Sasaki, K., Karasawa, K., Uchida, K., and Nitta, K. (2017). Intracellular Transcytosis of Albumin in Glomerular Endothelial Cells after Endocytosis through Caveolae. J. Cel Physiol 232 (12), 3565–3573. doi:10.1002/jcp.25817
Moriyama, T., Takei, T., Itabashi, M., Uchida, K., Tsuchiya, K., and Nitta, K. (2015). Caveolae May Enable Albumin to Enter Human Renal Glomerular Endothelial Cells. J. Cel Biochem 116 (6), 1060–1069. doi:10.1002/jcb.25061
Moriyama, T., Tsuruta, Y., Shimizu, A., Itabashi, M., Takei, T., Horita, S., et al. (2011). The Significance of Caveolae in the Glomeruli in Glomerular Disease. J. Clin. Pathol. 64 (6), 504–509. doi:10.1136/jcp.2010.087023
Nabi, I. R., and Le, P. U. (2003). Caveolae/raft-dependent Endocytosis. J. Cel Biol 161 (4), 673–677. doi:10.1083/jcb.200302028
Nah, J., Yoo, S. M., Jung, S., Jeong, E. I., Park, M., Kaang, B. K., et al. (2017). Phosphorylated CAV1 Activates Autophagy through an Interaction with BECN1 under Oxidative Stress. Cel Death Dis 8 (5), e2822. doi:10.1038/cddis.2017.71
Noda, K., Godo, S., Saito, H., Tsutsui, M., and Shimokawa, H. (2015). Opposing Roles of Nitric Oxide and Rho-Kinase in Lipid Metabolism in Mice. Tohoku J. Exp. Med. 235 (3), 171–183. doi:10.1620/tjem.235.171
Noda, K., Nakajima, S., Godo, S., Saito, H., Ikeda, S., Shimizu, T., et al. (2014). Rho-kinase Inhibition Ameliorates Metabolic Disorders through Activation of AMPK Pathway in Mice. PLoS One 9 (11), e110446. doi:10.1371/journal.pone.0110446
Nozaki, Y., Kinoshita, K., Hino, S., Yano, T., Niki, K., Hirooka, Y., et al. (2015). Signaling Rho-Kinase Mediates Inflammation and Apoptosis in T Cells and Renal Tubules in Cisplatin Nephrotoxicity. Am. J. Physiol. Ren. Physiol 308 (8), F899–F909. doi:10.1152/ajprenal.00362.2014
Nwosu, Z. C., Ebert, M. P., Dooley, S., and Meyer, C. (2016). Caveolin-1 in the Regulation of Cell Metabolism: a Cancer Perspective. Mol. Cancer 15 (1), 71. doi:10.1186/s12943-016-0558-7
Okada, S., Raja, S. A., Okerblom, J., Boddu, A., Horikawa, Y., Ray, S., et al. (2019). Deletion of Caveolin Scaffolding Domain Alters Cancer Cell Migration. Cell Cycle 18 (11), 1268–1280. doi:10.1080/15384101.2019.1618118
Oliveira, S. D. S., Castellon, M., Chen, J., Bonini, M. G., Gu, X., Elliott, M. H., et al. (2017). Inflammation-induced Caveolin-1 and BMPRII Depletion Promotes Endothelial Dysfunction and TGF-β-Driven Pulmonary Vascular Remodeling. Am. J. Physiol. Lung Cel Mol Physiol 312 (5), L760–l771. doi:10.1152/ajplung.00484.2016
Olzmann, J. A., and Carvalho, P. (2019). Dynamics and Functions of Lipid Droplets. Nat. Rev. Mol. Cel Biol 20 (3), 137–155. doi:10.1038/s41580-018-0085-z
Ostalska-Nowicka, D., Nowicki, M., Zachwieja, J., Kasper, M., and Witt, M. (2007). The Significance of Caveolin-1 Expression in Parietal Epithelial Cells of Bowman's Capsule. Histopathology 51 (5), 611–621. doi:10.1111/j.1365-2559.2007.02844.x
Ostermeyer, A. G., Ramcharan, L. T., Zeng, Y., Lublin, D. M., and Brown, D. A. (2004). Role of the Hydrophobic Domain in Targeting Caveolin-1 to Lipid Droplets. J. Cel Biol 164 (1), 69–78. doi:10.1083/jcb.200303037
Palacios-Ortega, S., Varela-Guruceaga, M., Martínez, J. A., de Miguel, C., and Milagro, F. I. (2016). Effects of High Glucose on Caveolin-1 and Insulin Signaling in 3T3-L1 Adipocytes. Adipocyte 5 (1), 65–80. doi:10.1080/21623945.2015.1122856
Park, S., Glover, K. J., and Im, W. (2019). U-shaped Caveolin-1 Conformations Are Tightly Regulated by Hydrogen Bonds with Lipids. J. Comput. Chem. 40 (16), 1570–1577. doi:10.1002/jcc.25807
Parton, R. G. (2018). Caveolae: Structure, Function, and Relationship to Disease. Annu. Rev. Cel Dev Biol 34, 111–136. doi:10.1146/annurev-cellbio-100617-062737
Parton, R. G., and del Pozo, M. A. (2013). Caveolae as Plasma Membrane Sensors, Protectors and Organizers. Nat. Rev. Mol. Cel Biol 14 (2), 98–112. doi:10.1038/nrm3512
Parton, R. G., Kozlov, M. M., and Ariotti, N. (2020a). Caveolae and Lipid Sorting: Shaping the Cellular Response to Stress. J. Cel Biol 219 (4), e201905071. doi:10.1083/jcb.201905071
Parton, R. G., McMahon, K. A., and Wu, Y. (2020b). Caveolae: Formation, Dynamics, and Function. Curr. Opin. Cel Biol 65, 8–16. doi:10.1016/j.ceb.2020.02.001
Parton, R. G., Tillu, V. A., and Collins, B. M. (2018). Caveolae. Curr. Biol. 28 (8), R402–r405. doi:10.1016/j.cub.2017.11.075
Peng, F., Wu, D., Ingram, A. J., Zhang, B., Gao, B., and Krepinsky, J. C. (2007). RhoA Activation in Mesangial Cells by Mechanical Strain Depends on Caveolae and Caveolin-1 Interaction. J. Am. Soc. Nephrol. 18 (1), 189–198. doi:10.1681/asn.2006050498
Pezeshkian, W., Chevrot, G., and Khandelia, H. (2018). The Role of Caveolin-1 in Lipid Droplets and Their Biogenesis. Chem. Phys. Lipids 211, 93–99. doi:10.1016/j.chemphyslip.2017.11.010
Pilch, P. F., and Liu, L. (2011). Fat Caves: Caveolae, Lipid Trafficking and Lipid Metabolism in Adipocytes. Trends Endocrinol. Metab. 22 (8), 318–324. doi:10.1016/j.tem.2011.04.001
Plummer, A. M., Culbertson, A. T., and Liao, M. (2021). The ABCs of Sterol Transport. Annu. Rev. Physiol. 83, 153–181. doi:10.1146/annurev-physiol-031620-094944
Pol, A., Luetterforst, R., Lindsay, M., Heino, S., Ikonen, E., and Parton, R. G. (2001). A Caveolin Dominant Negative Mutant Associates with Lipid Bodies and Induces Intracellular Cholesterol Imbalance. J. Cel Biol 152 (5), 1057–1070. doi:10.1083/jcb.152.5.1057
Pol, A., Martin, S., Fernandez, M. A., Ferguson, C., Carozzi, A., Luetterforst, R., et al. (2004). Dynamic and Regulated Association of Caveolin with Lipid Bodies: Modulation of Lipid Body Motility and Function by a Dominant Negative Mutant. Mol. Biol. Cel 15 (1), 99–110. doi:10.1091/mbc.e03-06-0368
Raikar, L. S., Vallejo, J., Lloyd, P. G., and Hardin, C. D. (2006). Overexpression of Caveolin-1 Results in Increased Plasma Membrane Targeting of Glycolytic Enzymes: the Structural Basis for a Membrane Associated Metabolic Compartment. J. Cel Biochem 98 (4), 861–871. doi:10.1002/jcb.20732
Renne, M. F., Klug, Y. A., and Carvalho, P. (2020). Lipid Droplet Biogenesis: A Mystery "unmixing". Semin. Cel Dev Biol 108, 14–23. doi:10.1016/j.semcdb.2020.03.001
Ring, A., Le Lay, S., Pohl, J., Verkade, P., and Stremmel, W. (2006). Caveolin-1 Is Required for Fatty Acid Translocase (FAT/CD36) Localization and Function at the Plasma Membrane of Mouse Embryonic Fibroblasts. Biochim. Biophys. Acta 1761 (4), 416–423. doi:10.1016/j.bbalip.2006.03.016
Robenek, M. J., Severs, N. J., Schlattmann, K., Plenz, G., Zimmer, K. P., Troyer, D., et al. (2004). Lipids Partition Caveolin-1 from ER Membranes into Lipid Droplets: Updating the Model of Lipid Droplet Biogenesis. Faseb j 18 (7), 866–868. doi:10.1096/fj.03-0782fje
Root, K. T., Plucinsky, S. M., and Glover, K. J. (2015). Recent Progress in the Topology, Structure, and Oligomerization of Caveolin: a Building Block of Caveolae. Curr. Top. Membr. 75, 305–336. doi:10.1016/bs.ctm.2015.03.007
Rothberg, K. G., Heuser, J. E., Donzell, W. C., Ying, Y. S., Glenney, J. R., and Anderson, R. G. (1992). Caveolin, a Protein Component of Caveolae Membrane coats. Cell 68 (4), 673–682. doi:10.1016/0092-8674(92)90143-z
Roy, S., Luetterforst, R., Harding, A., Apolloni, A., Etheridge, M., Stang, E., et al. (1999). Dominant-negative Caveolin Inhibits H-Ras Function by Disrupting Cholesterol-Rich Plasma Membrane Domains. Nat. Cel Biol 1 (2), 98–105. doi:10.1038/10067
Rungtabnapa, P., Nimmannit, U., Halim, H., Rojanasakul, Y., and Chanvorachote, P. (2011). Hydrogen Peroxide Inhibits Non-small Cell Lung Cancer Cell Anoikis through the Inhibition of Caveolin-1 Degradation. Am. J. Physiol. Cel Physiol 300 (2), C235–C245. doi:10.1152/ajpcell.00249.2010
Sala-Vila, A., Navarro-Lérida, I., Sánchez-Alvarez, M., Bosch, M., Calvo, C., López, J. A., et al. (2016). Interplay between Hepatic Mitochondria-Associated Membranes, Lipid Metabolism and Caveolin-1 in Mice. Sci. Rep. 6, 27351. doi:10.1038/srep27351
Sanders, Y. Y., Liu, H., Scruggs, A. M., Duncan, S. R., Huang, S. K., and Thannickal, V. J. (2017). Epigenetic Regulation of Caveolin-1 Gene Expression in Lung Fibroblasts. Am. J. Respir. Cel Mol Biol 56 (1), 50–61. doi:10.1165/rcmb.2016-0034OC
Santos-Parker, J. R., Strahler, T. R., Bassett, C. J., Bispham, N. Z., Chonchol, M. B., and Seals, D. R. (2017). Curcumin Supplementation Improves Vascular Endothelial Function in Healthy Middle-Aged and Older Adults by Increasing Nitric Oxide Bioavailability and Reducing Oxidative Stress. Aging (Albany NY) 9 (1), 187–208. doi:10.18632/aging.101149
Scherer, P. E., Okamoto, T., Chun, M., Nishimoto, I., Lodish, H. F., and Lisanti, M. P. (1996). Identification, Sequence, and Expression of Caveolin-2 Defines a Caveolin Gene Family. Proc. Natl. Acad. Sci. U S A. 93 (1), 131–135. doi:10.1073/pnas.93.1.131
Scherer, P. E., Tang, Z., Chun, M., Sargiacomo, M., Lodish, H. F., and Lisanti, M. P. (1995). Caveolin Isoforms Differ in Their N-Terminal Protein Sequence and Subcellular Distribution. Identification and Epitope Mapping of an Isoform-specific Monoclonal Antibody Probe. J. Biol. Chem. 270 (27), 16395–16401. doi:10.1074/jbc.270.27.16395
Schlegel, A., Arvan, P., and Lisanti, M. P. (2001). Caveolin-1 Binding to Endoplasmic Reticulum Membranes and Entry into the Regulated Secretory Pathway Are Regulated by Serine Phosphorylation. Protein Sorting at the Level of the Endoplasmic Reticulum. J. Biol. Chem. 276 (6), 4398–4408. doi:10.1074/jbc.M005448200
Shao, S., Qin, T., Qian, W., Yue, Y., Xiao, Y., Li, X., et al. (2020). Positive Feedback in Cav-1-ROS Signalling in PSCs Mediates Metabolic Coupling between PSCs and Tumour Cells. J. Cel Mol Med 24 (16), 9397–9408. doi:10.1111/jcmm.15596
Sharma, B., and Agnihotri, N. (2019). Role of Cholesterol Homeostasis and its Efflux Pathways in Cancer Progression. J. Steroid Biochem. Mol. Biol. 191, 105377. doi:10.1016/j.jsbmb.2019.105377
Sharma, S., Singh, M., and Sharma, P. L. (2012). Ameliorative Effect of Daidzein: a Caveolin-1 Inhibitor in Vascular Endothelium Dysfunction Induced by Ovariectomy. Indian J. Exp. Biol. 50 (1), 28–34.
Shi, Y., Tan, S. H., Ng, S., Zhou, J., Yang, N. D., Koo, G. B., et al. (2015). Critical Role of CAV1/caveolin-1 in Cell Stress Responses in Human Breast Cancer Cells via Modulation of Lysosomal Function and Autophagy. Autophagy 11 (5), 769–784. doi:10.1080/15548627.2015.1034411
Shihata, W. A., Putra, M. R. A., and Chin-Dusting, J. P. F. (2017). Is There a Potential Therapeutic Role for Caveolin-1 in Fibrosis. Front. Pharmacol. 8, 567. doi:10.3389/fphar.2017.00567
Siegert, A. M., Serra-Peinado, C., Gutiérrez-Martínez, E., Rodríguez-Pascual, F., Fabregat, I., and Egea, G. (2018). Altered TGF-β Endocytic Trafficking Contributes to the Increased Signaling in Marfan Syndrome. Biochim. Biophys. Acta Mol. Basis Dis. 1864 (2), 554–562. doi:10.1016/j.bbadis.2017.11.015
Sinha, B., Köster, D., Ruez, R., Gonnord, P., Bastiani, M., Abankwa, D., et al. (2011). Cells Respond to Mechanical Stress by Rapid Disassembly of Caveolae. Cell 144 (3), 402–413. doi:10.1016/j.cell.2010.12.031
Smart, E. J., Ying, Ys., Donzell, W. C., and Anderson, R. G. (1996). A Role for Caveolin in Transport of Cholesterol from Endoplasmic Reticulum to Plasma Membrane. J. Biol. Chem. 271 (46), 29427–29435. doi:10.1074/jbc.271.46.29427
Sörensson, J., Fierlbeck, W., Heider, T., Schwarz, K., Park, D. S., Mundel, P., et al. (2002). Glomerular Endothelial Fenestrae In Vivo Are Not Formed from Caveolae. J. Am. Soc. Nephrol. 13 (11), 2639–2647. doi:10.1097/01.asn.0000033277.32822.23
Sotgia, F., Martinez-Outschoorn, U. E., Howell, A., Pestell, R. G., Pavlides, S., and Lisanti, M. P. (2012). Caveolin-1 and Cancer Metabolism in the Tumor Microenvironment: Markers, Models, and Mechanisms. Annu. Rev. Pathol. 7, 423–467. doi:10.1146/annurev-pathol-011811-120856
Steffens, S., Schrader, A. J., Blasig, H., Vetter, G., Eggers, H., Tränkenschuh, W., et al. (2011). Caveolin 1 Protein Expression in Renal Cell Carcinoma Predicts Survival. BMC Urol. 11, 25. doi:10.1186/1471-2490-11-25
Sun, L. N., Chen, Z. X., Liu, X. C., Liu, H. Y., Guan, G. J., and Liu, G. (2014a). Curcumin Ameliorates Epithelial-To-Mesenchymal Transition of Podocytes In Vivo and In Vitro via Regulating Caveolin-1. Biomed. Pharmacother. 68 (8), 1079–1088. doi:10.1016/j.biopha.2014.10.005
Sun, L. N., Liu, X. C., Chen, X. J., Guan, G. J., and Liu, G. (2016). Curcumin Attenuates High Glucose-Induced Podocyte Apoptosis by Regulating Functional Connections between Caveolin-1 Phosphorylation and ROS. Acta Pharmacol. Sin 37 (5), 645–655. doi:10.1038/aps.2015.159
Sun, L. N., Yang, Z. Y., Lv, S. S., Liu, X. C., Guan, G. J., and Liu, G. (2014b). Curcumin Prevents Diabetic Nephropathy against Inflammatory Response via Reversing Caveolin-1 Tyr14 Phosphorylation Influenced TLR4 Activation. Int. Immunopharmacol 23 (1), 236–246. doi:10.1016/j.intimp.2014.08.023
Sun, X., Liu, Y., Li, C., Wang, X., Zhu, R., Liu, C., et al. (2017). Recent Advances of Curcumin in the Prevention and Treatment of Renal Fibrosis. Biomed. Res. Int. 2017, 2418671. doi:10.1155/2017/2418671
Suzuki, M. (2017). Regulation of Lipid Metabolism via a Connection between the Endoplasmic Reticulum and Lipid Droplets. Anat. Sci. Int. 92 (1), 50–54. doi:10.1007/s12565-016-0378-2
Sztalryd, C., and Brasaemle, D. L. (2017). The Perilipin Family of Lipid Droplet Proteins: Gatekeepers of Intracellular Lipolysis. Biochim. Biophys. Acta Mol. Cel Biol Lipids 1862 (10 Pt B), 1221–1232. doi:10.1016/j.bbalip.2017.07.009
Tahir, S. A., Yang, G., Goltsov, A., Song, K. D., Ren, C., Wang, J., et al. (2013). Caveolin-1-LRP6 Signaling Module Stimulates Aerobic Glycolysis in Prostate Cancer. Cancer Res. 73 (6), 1900–1911. doi:10.1158/0008-5472.Can-12-3040
Tamai, O., Oka, N., Kikuchi, T., Koda, Y., Soejima, M., Wada, Y., et al. (2001). Caveolae in Mesangial Cells and Caveolin Expression in Mesangial Proliferative Glomerulonephritis. Kidney Int. 59 (2), 471–480. doi:10.1046/j.1523-1755.2001.059002471.x
Tan, V. P., and Miyamoto, S. (2015). HK2/hexokinase-II Integrates Glycolysis and Autophagy to Confer Cellular protection. Autophagy 11 (6), 963–964. doi:10.1080/15548627.2015.1042195
Tang, Z., Scherer, P. E., Okamoto, T., Song, K., Chu, C., Kohtz, D. S., et al. (1996). Molecular Cloning of Caveolin-3, a Novel Member of the Caveolin Gene Family Expressed Predominantly in Muscle. J. Biol. Chem. 271 (4), 2255–2261. doi:10.1074/jbc.271.4.2255
Tao, B., Kraehling, J. R., Ghaffari, S., Ramirez, C. M., Lee, S., Fowler, J. W., et al. (2020). BMP-9 and LDL Crosstalk Regulates ALK-1 Endocytosis and LDL Transcytosis in Endothelial Cells. J. Biol. Chem. 295 (52), 18179–18188. doi:10.1074/jbc.RA120.015680
Thangavel, C., Gomes, C. M., Zderic, S. A., Javed, E., Addya, S., Singh, J., et al. (2019). NF-κB and GATA-Binding Factor 6 Repress Transcription of Caveolins in Bladder Smooth Muscle Hypertrophy. Am. J. Pathol. 189 (4), 847–867. doi:10.1016/j.ajpath.2018.12.013
Tomar, A., Kaushik, S., Khan, S. I., Bisht, K., Nag, T. C., Arya, D. S., et al. (2020). The Dietary Isoflavone Daidzein Mitigates Oxidative Stress, Apoptosis, and Inflammation in CDDP-Induced Kidney Injury in Rats: Impact of the MAPK Signaling Pathway. J. Biochem. Mol. Toxicol. 34 (2), e22431. doi:10.1002/jbt.22431
Tomassian, T., Humphries, L. A., Liu, S. D., Silva, O., Brooks, D. G., and Miceli, M. C. (2011). Caveolin-1 Orchestrates TCR Synaptic Polarity, Signal Specificity, and Function in CD8 T Cells. J. Immunol. 187 (6), 2993–3002. doi:10.4049/jimmunol.1101447
Torrejón, B., Cristóbal, I., Rojo, F., and García-Foncillas, J. (2017). Caveolin-1 Is Markedly Downregulated in Patients with Early-Stage Colorectal Cancer. World J. Surg. 41 (10), 2625–2630. doi:10.1007/s00268-017-4065-9
Trávez, A., Rabanal-Ruiz, Y., López-Alcalá, J., Molero-Murillo, L., Díaz-Ruiz, A., Guzmán-Ruiz, R., et al. (2018). The Caveolae-Associated Coiled-Coil Protein, NECC2, Regulates Insulin Signalling in Adipocytes. J. Cel Mol Med 22 (11), 5648–5661. doi:10.1111/jcmm.13840
Tsuda, T. (2018). Curcumin as a Functional Food-Derived Factor: Degradation Products, Metabolites, Bioactivity, and Future Perspectives. Food Funct. 9 (2), 705–714. doi:10.1039/c7fo01242j
Umesalma, S., Houwen, F. K., Baumbach, G. L., and Chan, S. L. (2016). Roles of Caveolin-1 in Angiotensin II-Induced Hypertrophy and Inward Remodeling of Cerebral Pial Arterioles. Hypertension 67 (3), 623–629. doi:10.1161/hypertensionaha.115.06565
Vainonen, J. P., Aboulaich, N., Turkina, M. V., Strålfors, P., and Vener, A. V. (2004). N-terminal Processing and Modifications of Caveolin-1 in Caveolae from Human Adipocytes. Biochem. Biophys. Res. Commun. 320 (2), 480–486. doi:10.1016/j.bbrc.2004.05.196
Vallejo, J., and Hardin, C. D. (2005). Expression of Caveolin-1 in Lymphocytes Induces Caveolae Formation and Recruitment of Phosphofructokinase to the Plasma Membrane. Faseb j 19 (6), 586–587. doi:10.1096/fj.04-2380fje
van den Heuvel, A. P., Schulze, A., and Burgering, B. M. (2005). Direct Control of Caveolin-1 Expression by FOXO Transcription Factors. Biochem. J. 385 (Pt 3), 795–802. doi:10.1042/bj20041449
Van Krieken, R., and Krepinsky, J. C. (2017). Caveolin-1 in the Pathogenesis of Diabetic Nephropathy: Potential Therapeutic Target. Curr. Diab Rep. 17 (3), 19. doi:10.1007/s11892-017-0844-9
Varela-Guruceaga, M., Milagro, F. I., Martínez, J. A., and de Miguel, C. (2018). Effect of Hypoxia on Caveolae-Related Protein Expression and Insulin Signaling in Adipocytes. Mol. Cel Endocrinol 473, 257–267. doi:10.1016/j.mce.2018.01.026
Vassilieva, E. V., Ivanov, A. I., and Nusrat, A. (2009). Flotillin-1 Stabilizes Caveolin-1 in Intestinal Epithelial Cells. Biochem. Biophys. Res. Commun. 379 (2), 460–465. doi:10.1016/j.bbrc.2008.12.118
Vihanto, M. M., Vindis, C., Djonov, V., Cerretti, D. P., and Huynh-Do, U. (2006). Caveolin-1 Is Required for Signaling and Membrane Targeting of EphB1 Receptor Tyrosine Kinase. J. Cel Sci 119 (Pt 11), 2299–2309. doi:10.1242/jcs.02946
Volonte, D., and Galbiati, F. (2020). Caveolin-1, a Master Regulator of Cellular Senescence. Cancer Metastasis Rev. 39 (2), 397–414. doi:10.1007/s10555-020-09875-w
Waalkes, S., Eggers, H., Blasig, H., Atschekzei, F., Kramer, M. W., Hennenlotter, J., et al. (2011). Caveolin 1 mRNA Is Overexpressed in Malignant Renal Tissue and Might Serve as a Novel Diagnostic Marker for Renal Cancer. Biomark Med. 5 (2), 219–225. doi:10.2217/bmm.11.12
Walther, T. C., Chung, J., and Farese, R. V. (2017). Lipid Droplet Biogenesis. Annu. Rev. Cel Dev Biol 33, 491–510. doi:10.1146/annurev-cellbio-100616-060608
Walther, T. C., and Farese, R. V. (2012). Lipid Droplets and Cellular Lipid Metabolism. Annu. Rev. Biochem. 81, 687–714. doi:10.1146/annurev-biochem-061009-102430
Wang, D. X., Pan, Y. Q., Liu, B., and Dai, L. (2018). Cav-1 Promotes Atherosclerosis by Activating JNK-Associated Signaling. Biochem. Biophys. Res. Commun. 503 (2), 513–520. doi:10.1016/j.bbrc.2018.05.036
Wang, H., Yang, P., Liu, K., Guo, F., Zhang, Y., Zhang, G., et al. (2008). SARS Coronavirus Entry into Host Cells through a Novel Clathrin- and Caveolae-independent Endocytic Pathway. Cell Res 18 (2), 290–301. doi:10.1038/cr.2008.15
Wang, J., Chen, M. Y., Chen, J. F., Ren, Q. L., Zhang, J. Q., Cao, H., et al. (2020a). LncRNA IMFlnc1 Promotes Porcine Intramuscular Adipocyte Adipogenesis by Sponging miR-199a-5p to Up-Regulate CAV-1. BMC Mol. Cel Biol 21 (1), 77. doi:10.1186/s12860-020-00324-8
Wang, L., Connelly, M. A., Ostermeyer, A. G., Chen, H. H., Williams, D. L., and Brown, D. A. (2003). Caveolin-1 Does Not Affect SR-BI-Mediated Cholesterol Efflux or Selective Uptake of Cholesteryl Ester in Two Cell Lines. J. Lipid Res. 44 (4), 807–815. doi:10.1194/jlr.M200449-JLR200
Wang, L., Zhao, Y., Gang, S., Geng, T., Li, M., Xu, L., et al. (2021). Inhibition of miR-103-3p Preserves Neurovascular Integrity through Caveolin-1 in Experimental Subarachnoid Hemorrhage. Neuroscience 461, 91–101. doi:10.1016/j.neuroscience.2021.03.007
Wang, M. D., Wang, Y., Xia, Y. P., Dai, J. W., Gao, L., Wang, S. Q., et al. (2016). High Serum MiR-130a Levels Are Associated with Severe Perihematomal Edema and Predict Adverse Outcome in Acute ICH. Mol. Neurobiol. 53 (2), 1310–1321. doi:10.1007/s12035-015-9099-0
Wang, N., Muhetaer, G., Zhang, X., Yang, B., Wang, C., Zhang, Y., et al. (2020b). Sanguisorba Officinalis L. Suppresses Triple-Negative Breast Cancer Metastasis by Inhibiting Late-phase Autophagy via Hif-1α/Caveolin-1 Signaling. Front. Pharmacol. 11, 591400. doi:10.3389/fphar.2020.591400
Wang, S., Wang, N., Zheng, Y., Yang, B., Liu, P., Zhang, F., et al. (2020c). Caveolin-1 Inhibits Breast Cancer Stem Cells via C-Myc-Mediated Metabolic Reprogramming. Cel Death Dis 11 (6), 450. doi:10.1038/s41419-020-2667-x
Wettersten, H. I., Aboud, O. A., Lara, P. N., and Weiss, R. H. (2017). Metabolic Reprogramming in clear Cell Renal Cell Carcinoma. Nat. Rev. Nephrol. 13 (7), 410–419. doi:10.1038/nrneph.2017.59
Wilfling, F., Haas, J. T., Walther, T. C., and Farese, R. V. (2014). Lipid Droplet Biogenesis. Curr. Opin. Cel Biol 29, 39–45. doi:10.1016/j.ceb.2014.03.008
Williams, T. M., and Lisanti, M. P. (2004). The Caveolin Proteins. Genome Biol. 5 (3), 214. doi:10.1186/gb-2004-5-3-214
Wong, T. H., Dickson, F. H., Timmins, L. R., and Nabi, I. R. (2020). Tyrosine Phosphorylation of Tumor Cell Caveolin-1: Impact on Cancer Progression. Cancer Metastasis Rev. 39 (2), 455–469. doi:10.1007/s10555-020-09892-9
Wong, T. H., Khater, I. M., Joshi, B., Shahsavari, M., Hamarneh, G., and Nabi, I. R. (2021). Single Molecule Network Analysis Identifies Structural Changes to Caveolae and Scaffolds Due to Mutation of the Caveolin-1 Scaffolding Domain. Sci. Rep. 11 (1), 7810. doi:10.1038/s41598-021-86770-6
Woodman, O. L., Missen, M. A., and Boujaoude, M. (2004). Daidzein and 17 Beta-Estradiol Enhance Nitric Oxide Synthase Activity Associated with an Increase in Calmodulin and a Decrease in Caveolin-1. J. Cardiovasc. Pharmacol. 44 (2), 155–163. doi:10.1097/00005344-200408000-00003
Wu, D., Yang, X., Zheng, T., Xing, S., Wang, J., Chi, J., et al. (2016). A Novel Mechanism of Action for Salidroside to Alleviate Diabetic Albuminuria: Effects on Albumin Transcytosis across Glomerular Endothelial Cells. Am. J. Physiol. Endocrinol. Metab. 310 (3), E225–E237. doi:10.1152/ajpendo.00391.2015
Wu, S. Z., Peng, F. F., Li, J. L., Ye, F., Lei, S. Q., and Zhang, B. F. (2014). Akt and RhoA Activation in Response to High Glucose Require Caveolin-1 Phosphorylation in Mesangial Cells. Am. J. Physiol. Ren. Physiol 306 (11), F1308–F1317. doi:10.1152/ajprenal.00447.2013
Xie, X., Peng, J., Chang, X., Huang, K., Huang, J., Wang, S., et al. (2013). Activation of RhoA/ROCK Regulates NF-Κb Signaling Pathway in Experimental Diabetic Nephropathy. Mol. Cel Endocrinol 369 (1-2), 86–97. doi:10.1016/j.mce.2013.01.007
Xie, Z., Zeng, X., Waldman, T., and Glazer, R. I. (2003). Transformation of Mammary Epithelial Cells by 3-phosphoinositide- Dependent Protein Kinase-1 Activates Beta-Catenin and C-Myc, and Down-Regulates Caveolin-1. Cancer Res. 63 (17), 5370–5375. Available at: https://cancerres.aacrjournals.org/content/canres/63/17/5370.full.pdf.
Xing, Y., Wen, Z., Gao, W., Lin, Z., Zhong, J., and Jiu, Y. (2020). Multifaceted Functions of Host Cell Caveolae/Caveolin-1 in Virus Infections. Viruses 12 (5), 487. doi:10.3390/v12050487
Xu, Y., Du, X., Turner, N., Brown, A. J., and Yang, H. (2019). Enhanced Acyl-CoA:cholesterol Acyltransferase Activity Increases Cholesterol Levels on the Lipid Droplet Surface and Impairs Adipocyte Function. J. Biol. Chem. 294 (50), 19306–19321. doi:10.1074/jbc.RA119.011160
Xue, H., Li, P., Luo, Y., Wu, C., Liu, Y., Qin, X., et al. (2019). Salidroside Stimulates the Sirt1/PGC-1α axis and Ameliorates Diabetic Nephropathy in Mice. Phytomedicine 54, 240–247. doi:10.1016/j.phymed.2018.10.031
Xue, W., Wang, J., Jiang, W., Shi, C., Wang, X., Huang, Y., et al. (2020). Caveolin-1 Alleviates Lipid Accumulation in NAFLD Associated with Promoting Autophagy by Inhibiting the Akt/mTOR Pathway. Eur. J. Pharmacol. 871, 172910. doi:10.1016/j.ejphar.2020.172910
Yamaguchi, T., Lu, C., Ida, L., Yanagisawa, K., Usukura, J., Cheng, J., et al. (2016). ROR1 Sustains Caveolae and Survival Signalling as a Scaffold of Cavin-1 and Caveolin-1. Nat. Commun. 7, 10060. doi:10.1038/ncomms10060
Yamaguchi, Y., Yasuoka, H., Stolz, D. B., and Feghali-Bostwick, C. A. (2011). Decreased Caveolin-1 Levels Contribute to Fibrosis and Deposition of Extracellular IGFBP-5. J. Cel Mol Med 15 (4), 957–969. doi:10.1111/j.1582-4934.2010.01063.x
Yan, F., Su, L., Chen, X., Wang, X., Gao, H., and Zeng, Y. (2020). Molecular Regulation and Clinical Significance of Caveolin-1 Methylation in Chronic Lung Diseases. Clin. Transl Med. 10 (1), 151–160. doi:10.1002/ctm2.2
Yan, R., Cao, P., Song, W., Qian, H., Du, X., Coates, H. W., et al. (2021). A Structure of Human Scap Bound to Insig-2 Suggests How Their Interaction Is Regulated by Sterols. Science 371 (6533), eabb2224. doi:10.1126/science.abb2224
Yang, G., Xu, H., Li, Z., and Li, F. (2014). Interactions of Caveolin-1 Scaffolding and Intramembrane Regions Containing a CRAC Motif with Cholesterol in Lipid Bilayers. Biochim. Biophys. Acta 1838 (10), 2588–2599. doi:10.1016/j.bbamem.2014.06.018
Yang, J., Zhu, T., Zhao, R., Gao, D., Cui, Y., Wang, K., et al. (2018). Caveolin-1 Inhibits Proliferation, Migration, and Invasion of Human Colorectal Cancer Cells by Suppressing Phosphorylation of Epidermal Growth Factor Receptor. Med. Sci. Monit. 24, 332–341. doi:10.12659/msm.907782
Yang, Z., Wang, L., Yu, H., Wang, R., Gou, Y., Zhang, M., et al. (2019). Membrane TLR9 Positive Neutrophil Mediated MPLA Protects against Fatal Bacterial Sepsis. Theranostics 9 (21), 6269–6283. doi:10.7150/thno.37139
Yokomori, H., Ando, W., and Oda, M. (2019). Caveolin-1 Is Related to Lipid Droplet Formation in Hepatic Stellate Cells in Human Liver. Acta Histochem. 121 (2), 113–118. doi:10.1016/j.acthis.2018.10.008
Yoon, H. J., Kim, D. H., Kim, S. J., Jang, J. H., and Surh, Y. J. (2019). Src-mediated Phosphorylation, Ubiquitination and Degradation of Caveolin-1 Promotes Breast Cancer Cell Stemness. Cancer Lett. 449, 8–19. doi:10.1016/j.canlet.2019.01.021
Yuan, T., Hong, S., Yao, Y., and Liao, K. (2007). Glut-4 Is Translocated to Both Caveolae and Non-caveolar Lipid Rafts, but Is Partially Internalized through Caveolae in Insulin-Stimulated Adipocytes. Cel Res 17 (9), 772–782. doi:10.1038/cr.2007.73
Zager, R. A., Johnson, A., Hanson, S., and dela Rosa, V. (2002). Altered Cholesterol Localization and Caveolin Expression during the Evolution of Acute Renal Failure. Kidney Int. 61 (5), 1674–1683. doi:10.1046/j.1523-1755.2002.00316.x
Zakrzewicz, D., Didiasova, M., Zakrzewicz, A., Hocke, A. C., Uhle, F., Markart, P., et al. (2014). The Interaction of Enolase-1 with Caveolae-Associated Proteins Regulates its Subcellular Localization. Biochem. J. 460 (2), 295–307. doi:10.1042/bj20130945
Zhang, B., Peng, F., Wu, D., Ingram, A. J., Gao, B., and Krepinsky, J. C. (2007). Caveolin-1 Phosphorylation Is Required for Stretch-Induced EGFR and Akt Activation in Mesangial Cells. Cell Signal 19 (8), 1690–1700. doi:10.1016/j.cellsig.2007.03.005
Zhang, C. J., Zhu, N., Wang, Y. X., Liu, L. P., Zhao, T. J., Wu, H. T., et al. (2021). Celastrol Attenuates Lipid Accumulation and Stemness of Clear Cell Renal Cell Carcinoma via CAV-1/LOX-1 Pathway. Front. Pharmacol. 12, 658092. doi:10.3389/fphar.2021.658092
Zhang, D., Gava, A. L., Van Krieken, R., Mehta, N., Li, R., Gao, B., et al. (2019). The Caveolin-1 Regulated Protein Follistatin Protects against Diabetic Kidney Disease. Kidney Int. 96 (5), 1134–1149. doi:10.1016/j.kint.2019.05.032
Zhang, X., Ramírez, C. M., Aryal, B., Madrigal-Matute, J., Liu, X., Diaz, A., et al. (2020a). Cav-1 (Caveolin-1) Deficiency Increases Autophagy in the Endothelium and Attenuates Vascular Inflammation and Atherosclerosis. Arterioscler Thromb. Vasc. Biol. 40 (6), 1510–1522. doi:10.1161/atvbaha.120.314291
Zhang, Y., Peng, F., Gao, B., Ingram, A. J., and Krepinsky, J. C. (2012). High Glucose-Induced RhoA Activation Requires Caveolae and PKCβ1-Mediated ROS Generation. Am. J. Physiol. Ren. Physiol 302 (1), F159–F172. doi:10.1152/ajprenal.00749.2010
Zhang, Y., Qu, X., Li, C., Fan, Y., Che, X., Wang, X., et al. (2015). miR-103/107 Modulates Multidrug Resistance in Human Gastric Carcinoma by Downregulating Cav-1. Tumour Biolcell Mol. Life Sci. 36 (4), 2277–2285. doi:10.1007/s13277-014-2835-7
Zhang, Z., Gao, Z., Rajthala, S., Sapkota, D., Dongre, H., Parajuli, H., et al. (2020b). Metabolic Reprogramming of normal Oral Fibroblasts Correlated with Increased Glycolytic Metabolism of Oral Squamous Cell Carcinoma and Precedes Their Activation into Carcinoma Associated Fibroblasts. 77 (46), 1115–1133. doi:10.1007/s00018-019-03209-y
Zhao, R., Liu, K., Huang, Z., Wang, J., Pan, Y., Huang, Y., et al. (2015). Genetic Variants in Caveolin-1 and RhoA/ROCK1 Are Associated with Clear Cell Renal Cell Carcinoma Risk in a Chinese Population. PLoS One 10 (6), e0128771. doi:10.1371/journal.pone.0128771
Zhong, W., Li, Y. C., Huang, Q. Y., and Tang, X. Q. (2020). lncRNA ANRIL Ameliorates Oxygen and Glucose Deprivation (OGD) Induced Injury in Neuron Cells via miR-199a-5p/CAV-1 Axis. Neurochem. Res. 45 (4), 772–782. doi:10.1007/s11064-019-02951-w
Zhou, H., Liu, J., Ren, L., Liu, W., Xing, Q., Men, L., et al. (2014). Relationship between [corrected] Spatial Memory in Diabetic Rats and Protein Kinase Cγ, Caveolin-1 in the hippocampus and Neuroprotective Effect of Catalpol. Chin. Med. J. (Engl) 127 (5), 916–923. doi:10.3760/cma.j.issn.0366-6999.20132137
Zhu, Q., Zhan, D., Zhu, P., Chong, Y., and Yang, Y. (2020). CircAKT1 Acts as a Sponge of miR-338-3p to Facilitate clear Cell Renal Cell Carcinoma Progression by Up-Regulating CAV1. Biochem. Biophys. Res. Commun. 532 (4), 584–590. doi:10.1016/j.bbrc.2020.08.081
Zhu, T., Meng, Q., Ji, J., Zhang, L., and Lou, X. (2017). TLR4 and Caveolin-1 in Monocytes Are Associated with Inflammatory Conditions in Diabetic Neuropathy. Clin. Transl Sci. 10 (3), 178–184. doi:10.1111/cts.12434
Zimnicka, A. M., Husain, Y. S., Shajahan, A. N., Sverdlov, M., Chaga, O., Chen, Z., et al. (2016). Src-dependent Phosphorylation of Caveolin-1 Tyr-14 Promotes Swelling and Release of Caveolae. Mol. Biol. Cel 27 (13), 2090–2106. doi:10.1091/mbc.E15-11-0756
Zschocke, J., Manthey, D., Bayatti, N., van der Burg, B., Goodenough, S., and Behl, C. (2002). Estrogen Receptor Alpha-Mediated Silencing of Caveolin Gene Expression in Neuronal Cells. J. Biol. Chem. 277 (41), 38772–38780. doi:10.1074/jbc.M205664200
Keywords: caveolin-1 (Cav-1), cellular metabolism, kidney disease, oxidative stress, autophagy
Citation: Luo S, Yang M, Zhao H, Han Y, Jiang N, Yang J, Chen W, Li C, Liu Y, Zhao C and Sun L (2021) Caveolin-1 Regulates Cellular Metabolism: A Potential Therapeutic Target in Kidney Disease. Front. Pharmacol. 12:768100. doi: 10.3389/fphar.2021.768100
Received: 31 August 2021; Accepted: 08 November 2021;
Published: 10 December 2021.
Edited by:
Dan-Qian Chen, Northwest University, ChinaReviewed by:
Yaming Jiu, Institut Pasteur of Shanghai (CAS), ChinaJoan Krepinsky, McMaster University, Canada
Cecilia Jacques G. de Almeida, Oswaldo Cruz Foundation, Brazil
Copyright © 2021 Luo, Yang, Zhao, Han, Jiang, Yang, Chen, Li, Liu, Zhao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Sun, c3VubGluQGNzdS5lZHUuY24=
 Shilu Luo
Shilu Luo Ming Yang
Ming Yang Hao Zhao
Hao Zhao Yachun Han1,2
Yachun Han1,2 Chenrui Li
Chenrui Li Lin Sun
Lin Sun