
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 03 January 2022
Sec. Ethnopharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.766309
 Lulu Zhang1,2†
Lulu Zhang1,2† Weifeng Yang3†
Weifeng Yang3† Yajun Chu4†
Yajun Chu4† Bo Wen1
Bo Wen1 Yungchi Cheng5
Yungchi Cheng5 Tariq Mahmood6
Tariq Mahmood6 Mei Bao1,2
Mei Bao1,2 Feng Ge7
Feng Ge7 Li Li1
Li Li1 Jianfeng Yi2
Jianfeng Yi2 Chengqiang Du4*
Chengqiang Du4* Cheng Lu1*
Cheng Lu1* Yong Tan1*
Yong Tan1*Methicillin-resistant Staphylococcus aureus (MRSA) is a superbacterium, and when it forms biofilms, it is difficult to treat even with the first-line of antibiotic linezolid (LNZ). Reyanning mixture (RYN), a compound-based Chinese medicine formula, has been found to have inhibitory effects on biofilms. This study aims to explore the synergistic inhibitory effect and corresponding mechanisms of their (LNZ&RYN) combination on the planktonic as well as biofilm cells of MRSA. Broth microdilution and chessboard methods were employed for the determination of minimum inhibitory concentrations (MICs) and synergistic concentration of LNZ&RYN, respectively. The effect of the combined medication on biofilm and mature biofilm of MRSA were observed by biofilm morphology and permeability experiments, respectively. To unveil the molecular mechanism of action of the synergistic combination of LNZ and RYN, RT-PCR based biofilm-related gene expression analysis and ultra-high pressure liquid chromatography-time-of-flight mass spectrometry based endogenous metabonomic analysis were deployed. The results indicated that 1/16RYN as the best combined dose reduced LNZ (4 μg/ml) to 2 μg/ml. The combined treatment inhibited living MRSA before and after biofilm formation, removed the residual structure of dead bacteria in MRSA biofilms and affected the shape and size of bacteria, resulting in the improvement of biofilm permeability. The mechanism was that biofilm-related genes such as agrC, atlA, and sarA, as well as amino acid uptake associated with the metabolism of 3-dehydrocarnitine, kynurenine, L-leucine, L-lysine and sebacic acid were inhibited. This study provides evidence for the treatment of MRSA and its biofilms with LNZ combined with RYN.
Methicillin-resistant Staphylococcus aureus (MRSA) is commonly known as a “superbug”, and its infection leads to many refractory diseases such as pneumonia, bacteraemia, and skin and soft tissue infections (Patel, 2009). A clinical survey has shown that MRSA is highly prevalent in hospitals around the world, and the incidence of MRSA is more than 50% in Asia, North America and South America (Stefani et al., 2012). MRSA is difficult to treat, and when it forms a mature biofilm, its treatment will be more difficult. Biofilms are a structural group attached to living or inert surfaces formed by microbial cells that adhere to each other and are surrounded by their own extracellular polymeric substances (EPSs) to form a special membrane structure (Wu et al., 2015). As a natural barrier, the formation of biofilms is considered a way for microorganisms to adapt to harsh environments. Compared with planktonic bacteria, the resistance of mature biofilm bacteria to antibiotics and host immune defence is significantly improved and even antibiotics up to 100–1000 times the minimum inhibitory concentration (MIC) cannot kill bacteria in biofilms (David, 2003). Biofilms are one of the important reasons for MRSA resistance. More than 80% of bacterial infections are biofilm-mediated (Stella et al., 2021). How to destroy the structure of biofilms and effectively treat the related infections caused by MRSA and its biofilms has become a hotspot of scientific research.
Antibiotics are the primary and conventional drugs to treat MRSA infection. vancomycin is the first choice for the treatment of MRSA infection. With the long-term clinical use of Vancomycin, S. aureus with low sensitivity or even drug resistance to vancomycin has appeared (Gowrishankar et al., 2013). Linezolid (LNZ) is generally considered an alternative to vancomycin. It is an oxazolidinone antibiotic that inhibits protein synthesis by binding to 50 s ribosomal subunits (Wunderink et al., 2012). LNZ may be more effective in the treatment of hospital-acquired MRSA and ventilator-associated pneumonia than vancomycin (Wunderink et al., 2012). However, the treatment window of LNZ is narrow, and the probability of nephrotoxic adverse reactions is high. Moreover, MRSA has gradually become resistant to LNZ (Boudet et al., 2021).
Antibiotic resistance is currently a major challenge in antimicrobial therapy. There is a worldwide search for antibacterial products that can replace or compensate for the lack of antibiotics (Gowrishankar et al., 2015). Traditional plant medicine (TPM) has been used in the clinic for thousands of years. Many TPM extracts can regulate the quorum sensing (QS) system in vitro and reduce the adhesion ability of bacteria to inhibit the growth of bacteria and their biofilms (Karbasizade et al., 2017). However, the bacteriostatic ability of TPM is weaker than that of antibiotics. Through the complementary advantages of TPM and specific antibiotics, together they are expected to achieve better therapeutic effects. Studies have shown that when TPM is used in combination with antibiotics, TPM can inhibit and destroy drug-resistant biofilms; as a result, bacteria in biofilms can effectively contact antibiotics, and thus, the antibiotics kill bacteria to a greater extent (Dey et al., 2020). The Reyanning mixture (ReYanNingHeJi, RYN), a Chinese patent drug, is composed of Taraxacum, Polygonum cuspidatum, Sonchus brachyotus and Scutellaria barbata. Its active pharmaceutical ingredients contain emodin, rhubarb anthraquinone, polydatin and flavonoids. A study has shown that RYN has an inhibitory effect on S. aureus, among which taraxacum inhibits the biofilm of S. aureus (Kenny et al., 2015). Antibiotics combined with RYN in the clinic are usually used to treat bacterial infections such as pneumonia and suppurative tonsillitis in China. The therapeutic effect and mechanism of the combination of RYN and antibiotics in the treatment of MRSA and its biofilm infections are unclear.
In the post-genome era, life science research has entered a comprehensive, systematic and dynamic functional exploration stage. The combined application of a variety of high-throughput research strategies, such as genomics, transcriptomics, proteomics and metabonomics, is more conducive to the comprehensive exploration of cell life processes. The selection of appropriate methods and tools will provide important support for the reliability and credibility of the research. In this study, real-time fluorescence polymerase chain reaction (RT–PCR) was used to analyze the gene expression of MRSA and its biofilm after treatment with LNZ combined with RYN (Rishi et al., 2018). Metabonomics is a technique to explore the changes in all endogenous small molecular metabolites (molecular weight less than 1500 Da) in cells after changes in external conditions (Emara et al., 2011). This has improved our understanding of the changes in intracellular metabolites. By identifying the metabolites of MRSA after drug intervention, we can further understand the effects of drugs on the molecular metabolism of bacteria from the planktonic state to the formation of biofilms. These small molecular metabolites are also likely to become therapeutic targets. In biological signalling, genes, as signal sources, are responsible for regulating and controlling the production of phenotypic metabolites. Changes in gene expression could affect the type and content of metabolites. The combination of gene detection and metabonomics technology provides technical support for exploring the mechanism of RYN combined with antibiotics against MRSA. In the present study, the synergistic effects of RYN and LNZ on MRSA and its biofilms were confirmed through bacteriostatic experiments and morphological observations in vitro. Then, to explore the mechanisms of these synergistic effects, the expression of MRSA biofilm-related genes was examined using RT–PCR, and the changes of endogenous metabolites of MRSA in mature biofilms were analysed by cell metabonomics technique. This study aims to clarify the synergistic inhibitory effects and corresponding mechanisms of RYN combined with LNZ on MRSA and its biofilm to lay a foundation for the establishment and optimization of an anti-MRSA regimen of RYN combined with LNZ, and to provide guidance for more safer and more effective clinical use.
The Reyanning mixture (RYN, Batch No. 180411) was obtained from Tsinghua Deren Xi’an Happiness Pharmaceutical Co., Ltd., China. According to 2020 China Pharmacopoeia, 1000 ml of RYN is from 372 g Taraxacum (Compositae; Taraxacum mongolicum Hand. -Mazz.), 372 g Polygonum cuspidatum (Polygonaceae; Polygonum cuspidatum Siebold and Zucc.), 372 g Sonchus brachyotus (Compositae; Sonchus arvensis L.), and 186 g Scutellaria barbata (Lamiaceae; Scutellaria barbata D. Don.). These herbs were decocted twice in water. The temperature of decoction was maintained at 96–100°C. The duration of decoction was 2 h for the first time and 1 h for the second time. The ratio of the herbs to solvent was 1:8 for the first time and 1:6 for the second time. Twice decocted solutions were mixed, centrifuged and filtered. Then the filtered solution was concentrated at 60–80°C, 0.01–0.09 Mpa steam pressure, and 0.04–0.08 Mpa vacuum degree. The concentration was stopped when the relative density reached 1.03–1.08 (70°C). Then 1.5 g stevioside and 0.5 g ethyl hydroxybenzene were added to the concentrated solution, and all were boiled and mixed well.
The product quality testing standards stipulate that the active ingredient content of the drug shall not be less than 0.15 mg/ml for emodin and 0.35 mg/ml for polydatin. The quality of RYN was controlled by chromatography, and no traces of heavy metals, organic solvents or other contaminants were found. The results showed that the two main components were emodin (C15H10O5, 0.95 mg/ml) and polydatin (C20H22O8, 2.44 mg/ml) (Supplementary Figures S1, S2; Supplementary Tables S1, S2).
Linedzolid injection (LNZ, Batch No. 14K03U03) was from Pfizer, Norway. Tryptone and yeast extracts were obtained from Oxoid, United Kingdom. NaCl from Merck, Germany, and 2,3,5-triphenyltetrazolium chloride (TTC) was obtained from Amresco, USA. 2,3-Bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) and phenazine methosulfate (PMS) were purchased from Sigma–Aldrich, USA. LIVE/DEAD ®BacLightTM Bacterial Viability Kit (Batch No. Lmuri 7012), FilmTracer™ SYPRO Ruby biofilm matrix stain (Batch No. 953539), and glutaraldehyde (Batch No. G6527) were purchased from Sigma–Aldrich, United States. Ethanol (Batch No. 10009527) and tert-butanol (Batch No. 80023928) were purchased from the Sinopharmaceutical Group, China. LC–MS grade acetonitrile and HPLC grade methanol were purchased from the Merck Company (Dannstadt, Germany), and formic acid (spectroscopic grade) was purchased from the CNW Company. The experimental water was Watsons distilled water. All chemical standards were purchased from Sigma–Aldrich (MO, United States) unless otherwise specified.
MRSA was extracted and isolated from the sputum of inpatients in the Respiratory Department of Beijing Dongzhimen Hospital and maintained in cryogenic storage at −80°C on glass beads. The experimental strain was continuously cultured for 10 generations in the laboratory and repeatedly induced by related antibiotics for five generations, and there was no significant difference in MICs before and after the two, which proved that it had genetic stability. The working culture of the bacteria was maintained on an agar plate at 4°C and subcultured in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl/litre) before use. In short, a single MRSA colony was inoculated into LB culture medium and incubated with 280 rpm at 37°C for 24 h. The broth culture overnight was diluted with fresh medium to obtain an initial inoculation size of OD600 = 0.02.
MICs of RYN and LNZ were determined by a standard broth microdilution method in sterile 96-well microplates (Standards, N.C.F.C.L, 2000). Two-fold serial dilutions were made in LB broth over a range to give final concentrations of 1/2–1/1024 for RYN solution and 32–0.0625 μg/ml for LNZ. Then 100 μl of bacterial suspension (OD600 = 0.02) was added to each well.
The negative control was comprised of LB broth and the tested sample while the positive control was LB broth and bacterial suspension only. The final volume of each well was 200 μl. The MICs of the test samples were detected after 24 h of incubation at 37°C, followed by the addition of 30 μl of 2.5 mg/ml TTC and incubation for an additional 20 min at 37°C (Yang et al., 2018). Viable bacteria reduced the yellow dye to pink. The MIC was defined as the lowest sample concentration that prevented this change and exhibited complete inhibition of microbial growth (Fankam et al., 2011).
The combined effect of RYN with LNZ was evaluated by the checkerboard microdilution method, and the fractional inhibition concentration index (FICI) of the drug-drug interaction was obtained (Basri and Sandra, 2016). The concentration range of multiple dilutions was set according to the single use MIC of the drug (LNZ: 4–0.0625 μg/ml, RYN: 1/4–1/256RYN solution). Then, 100 μl of drug solution and 100 μl of bacterial suspension (OD600 = 0.02) were added to each well. After 24 h of incubation at 37°C, 40 μl of TTC was added to each well, and the plates were incubated again for 20 min. Wells containing the solution that turned pink comparable to that of the positive control were interpreted as positive for bacterial growth, while the wells containing colourless solutions were interpreted as negative for bacterial growth. The FICI evaluated the bacteriostatic effect of the combination of the two drugs: FICI= (A/MICA) + (B/MICB). A and B are the respective MICs of drugs in combined bacteriostasis. MICA and MICB drugs were used alone with the bacteriostatic MIC. The results were interpreted as synergy (FICI≤0.5), addition (0.5 < FICI≤1), indifference (1 < FICI≤2) or antagonism (FICI>2) (Yu et al., 2005). The experiments were performed in triplicate and the median FICI values were used in the analysis.
The XTT reduction assay is considered to be proportional to the metabolic activity of cells (Kannappan et al., 2017). The bacterial liquid was diluted to OD600 = 0.1 with LB medium containing 0.25% glucose (LB-G) and incubated at 37°C for 0 and 24 h. The corresponding concentrations of RYN and LNZ were prepared. The concentration of RYN is 1/4MIC (1/8RYN), 1/8MIC (1/16RYN), 1/16MIC (1/32RYN) and 1/32MIC (1/64RYN), while that of LNZ is 1/2MIC (2 μg/ml). The same amount of drug solution (100 μl) as the bacterial solution was added to the 96-well plate of the mature biofilm. After incubation at 37°C for 24 h, the planktonic cells were washed out. Forty microlitres of XTT-PMS solution was added to each well and incubated in the dark at 37°C for 20 min. The optical density was measured by an enzyme-labelled instrument at 450 and 655 nm.
Mature biofilms of MRSA were cultured according to our previous study (Yang et al., 2018) The structural parameters of individual cellular and extracellular components of biofilms formed by MRSA strains were evaluated by CLSM observations and digital image analysis (Cerca et al., 2012). Briefly, the biofilm was washed twice with LB-G after being exposed to RYN and LNZ for 24 h. The viability of cells in the biofilm was determined by SYTO9/PI staining, and the extracellular matrix of the biofilm was determined by Matrix dye. The quartile glass Petri dish was then incubated in the dark at 37°C for 20 min. After washing the dye with LB-G medium, 500–600 μl LB-G medium was added to each well to keep the well moist. CLSM image acquisition was performed using Olympus TM FluoView FV1000 (Olympus, Lisboa, Portugal) confocal scanning laser microscope. Three stacks of horizontal plane images (1024 × 1024 pixels) with a z-step of 5 µm were acquired for each well from three different randomly chosen areas (Gomes et al., 2011). For image analysis, original Olympus files (OIB format) were imported into the IMARIS 9.1 software package (Bitplane, Zurich, Switzerland). The individual components of biofilms were represented by fluorescence emitted by SYTO9 (from cells with live membranes), PI (from bacteria with dead membranes) and Matrix (from bacteria with extracellular matrix). Three independent experiments were performed for each MRSA strain.
MRSA mature biofilms were cultured on 5 × 5 mm diameter round basic slides (Agar Scientific, Stansted, United Kingdom) in the wells of a 24-well plate (Wang et al., 2011). The unbound suspension bacteria were discarded, and 2 μg/ml LNZ, 1/16RYN and 2 μg/ml LNZ+1/16RYN were added. Blank LB-G medium of the same volume was added to the control group. The plate was exposed to a box with constant temperature and humidity at 37°C for 24 h. The slides were then stabilized in 1 ml of 4% glutaraldehyde for 1 h, fixed with 4% osmium acid, and dehydrated in serial dilutions (50, 70, 90, 100% v/v) of ethanol for 20 min. Subsequently, the slides were immersed in tertbutyl alcohol for 1 h. The slides were air-dried for 2 days and then transferred to copper disks and dusted with gold (160 s, 40 mA). The samples were analysed under SEM (JEOL JSM-35CF, SEM, JEOL, Japan) under 20–25 kV voltage (Absalon et al., 2011; Krzysciak et al., 2017).
The effects of RYN and LNZ on the structure of biofilms were studied by simulating an experimental model of antibiotic permeation in vitro (Anderl et al., 2000). In a 6 cm Petri dish, 4 ml of bacterial liquid (OD600 = 0.1) and 4 ml of LB-G were added, the blue filter membrane was put into the bacterial solution and cultured in a constant temperature incubator at 37°C for 48 h, and a mature biofilm was formed on the filter membrane. The mature biofilm was supplemented with 2 μg/ml LNZ, 1/16RYN and 2 μg/ml LNZ+1/16RYN, and the control group was supplemented with the same volume of blank LB-G. Then the 6 cm Petri dish was placed in a constant temperature incubator and cultured at 37°C for 24 h. The MRSA biofilm was transferred from 6 cm plate to a plate containing vancomycin (500 μg/ml). The bacterial biofilm was covered with a filter membrane to form a bacterial biofilm sandwich, and 6 mm filter paper was placed on the sandwich structure. After 2, 4 and 6 h, the filter paper was removed and affixed to a plate coated with MRSA. The plate was placed in a constant temperature incubator and cultured at 37°C for 18 h, and the bacteriostatic zone was observed.
The preparation of total RNA from MRSA was performed using RNA protection reagent according to the manufacturer’s instructions (TIANGEN, China). Briefly, total RNA was prepared by lysostaphin extraction using OD600 = 0.1 of bacteria at 24 h, followed by further purification with an RNA prep Pure Cell/Bacteria Kit (TIANGEN, China) according to the manufacturer’s instructions. The quality and quantity of total RNA were confirmed by agarose electrophoresis and UV spectrophotometry, respectively.
Contaminating chromosomal DNA was removed by DNase treatment (TaKaRa, Japan). Purified S. aureus RNA was reverse transcribed into cDNA by the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Japan) and then subjected to RT–PCR analysis using an ABI 7500 thermocycler (Applied Biosystems, America). The relative quantification of MRSA strain scripts was determined by the ratio of expression of target transcripts relative to agrA, agrB, agrC, agrD, atlA, RNAⅢ, and sarA. The sequences of primers for RT–PCR experiments are provided in Table 1.
Intracellular metabolites from each biological replicate were extracted using a cold methanol extraction approach as previously reported (Zhu et al., 2014). MRSA bacteria were collected at 0 and 24 h after administration. To put it simply, 60% methanol water was placed in a refrigerator at −20°C for pre-cooling. The precultured bacterial liquid was loaded into a 15 ml centrifuge tube, and each tube was 7 ml. Methanol water was added to the centrifuge tube at a ratio of bacterial liquid: methanol water = 1:1, shaken by hand for 5 s, and then placed on the ice in a refrigerator. The bacterial solution was centrifugally quenched at 3000 rpm for 10 min at 4°C, and the supernatant was removed after centrifugation. The protein was precipitated by adding methanol to the sample, fully vortexed for 30 s and then centrifuged at 4°C 120000 rpm for 15 min, and the supernatant was extracted for detection.
Ultra-high-pressure liquid chromatography-time-of-flight mass spectrometry (UPLC-Q-TOF-MS) was described elsewhere and was used with minor modifications (Huang et al., 2013). Briefly, a 4 μl aliquot was chromatographed using an Agilent C18 analytical column (i.d. 100 × 2.1 mm, particle size 1.8 μm). Mobile phase A and mobile phase B were water/formic acid (99.9:0.1, v/v) and acetonitrile/formic acid (99.9:0.1, v/v), respectively, and the flow rate was 0.4 ml/min. The column eluent was directed to the mass spectrometer for analyses. When a Premier mass spectrometer operating in the positive ion electrospray mode was used, the instrumental parameters were set as follows: the capillary voltage was set to 3.0 kV; the sampling cone voltage was set to 35.0 V; the nitrogen drying gas was set at a constant flow rate of 600 L/h; the source temperature was 100°C; the desolvation temperature was 350°C; the cone gas flow was 50 L/h; and the extraction cone was 4 V. When a Premier mass spectrometer operating in the negative ion electrospray mode was used, the instrumental parameters were set as follows: the capillary voltage was set to 3.5 kV; the sampling cone voltage was set to 50 V; the nitrogen drying gas was set at a constant flow rate of 600 L/h; the source temperature was 100°C; the desolvation temperature was 350°C; the cone gas flow was 50 L/h; and the extraction cone was 4 V. MS/MS analysis was performed on the mass spectrometer set at different collision energies according to the stability of each metabolite. The time-of-flight analyser was used in the V mode and was tuned for maximum resolution (>10,000 resolving power, atm/z556.2771). The instrument was previously calibrated with sodium formate; the lock mass spray for precise mass determination was set by leucine enkephalin atm/z 556.2771 with a concentration of 0.5 ng/μL. Fifteen injections of QC samples were performed to equilibrate the UPLC–MS system before running the actual samples. QC samples were injected every six samples at regular intervals throughout the analytical run.
These data were preprocessed by Mass Profiler software (Agilent) and edited later in EXCEL2007 software. The final result is organized into a two-dimensional data matrix, including variables (retention time and mass-charge ratio), observation (sample) and peak intensity. The sample of this project obtained 6623 features in positive mode and 3581 features in the negative mode. All of the data are then normalized to the total signal integral. The edited data matrix was imported into SIMCA-P software (Umetrics AB, Umea, Sweden, version 13.0) for principal component analysis (PCA). The SAS statistical package (order no. 195557), version 9.1.3, was used for the statistical analysis. The attribute data were analysed using the χ-square test. The measurement data obtained indicated a normal distribution. Comparisons between multiple groups were analysed using analysis of variance. Metabolites with significant differences were screened by the VIP (VIP>1), p value (p < 0.01) and fold change value (FC < -3, FC > 3). Metabolic pathway analysis was performed using MetaboAnalyst software (version 3.0). The calculated p value was established on the basis of the pathway enrichment analysis whereas the pathway impact value was derived from the pathway topology analysis. Correlation analysis using was performed using the Pearson correlation coefficient with R (pheatmap package) software.
The MICs of LNZ and RYN to MRSA were 4 μg/ml (MICa) and 1/2RYN (MICb) solutions respectively, and the initial concentration of the chessboard method was determined accordingly (Table 2). When 2 μg/ml LNZ (1/2MICa)+1/32RYN (1/16MICb) was used as the MIC combination of MRSA (FICI = 0.625), the combination of LNZ and RYN showed an additive effect (Table 2). The experimental results show that the combination of RYN and LNZ can reduce the concentration of LNZ against MRSA (Supplementary Figure S3).
As shown in Figures 1A–C, when MRSA biofilms matured, except that 1/64RYN had no obvious inhibitory effect on the living bacteria in MRSA biofilms, 1/8RYN, 1/16RYN and 1/32RYN all had bacteriostatic effects in a dose-dependent manner. The r value of linear regression analysis is 0.959. LNZ (2 μg/ml) inhibited the living bacteria in MRSA biofilms, but the inhibitory effect was not as good as that of the 1/8RYN group and 1/16RYN group. Viable bacteria in MRSA biofilms were inhibited in the positive drug LNZ group, but the inhibitory effect was not as good as that the in 1/8RYN group and 1/16RYN group. It is suggested that both RYN and LNZ can destroy the structure of MRSA biofilm and infiltrate into the biofilm to kill the deep bacteria in the biofilm, but the effect of LNZ is weaker than that of high and middle dose of RYN.
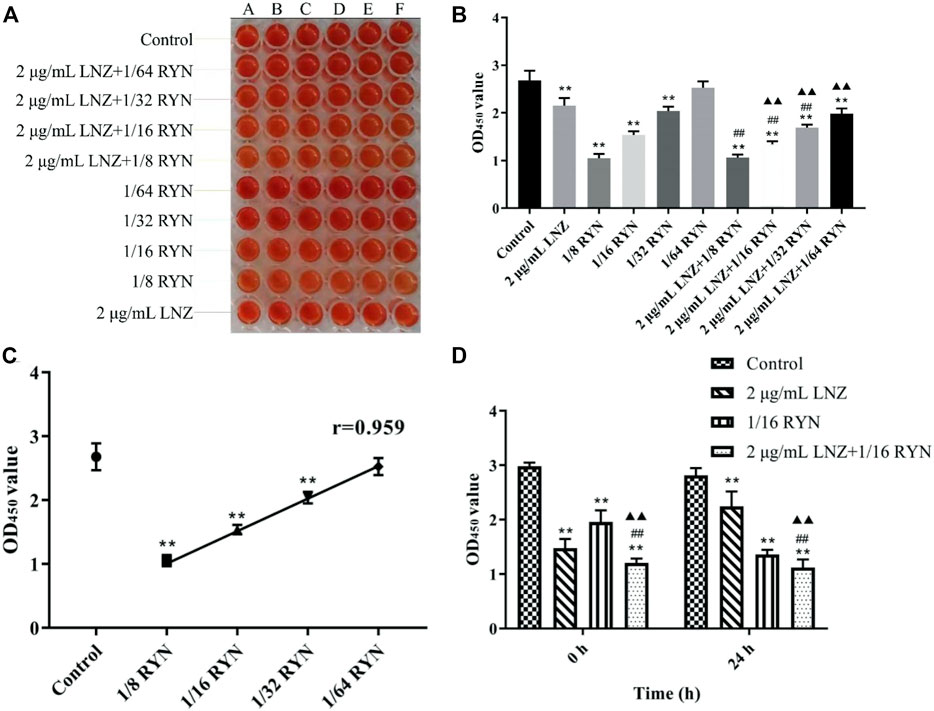
FIGURE 1. Effects of single and combined use of LNZ and RYN on living bacteria before and after MRSA biofilm production. (**p < 0.01 vs. control group; ##p < 0.01 vs. 2 μg/ml LNZ group; ▲▲p < 0.01 vs. 1/16RYN group.). (A) Results of XTT staining; (B) The effects of single and pairwise use of LNZ of 1/2 MIC and four doses of RYN on the number of living bacteria in MRSA biofilms; (C) The effect of RYN on the number of living bacteria in MRSA biofilm was dose-dependent; (D) Before and after the formation of biofilms, the effect of 2 μg/ml LNZ+1/16RYN was better than that of the two drugs.
As shown in Figure 1B, the effect of the combination of LNZ and RYN is better than respective those of the two drugs alone, and the combination significantly affected the change in living bacteria in biofilms. The combination effect is improved with increasing RYN concentration and dose. There were significant differences between 2 μg/ml LNZ+1/16RYN group and the 2 μg/ml LNZ group and the 1/16RYN group, and the effect was better than that of 2 μg/ml LNZ+1/32RYN group. This experiment shows that the best combined dose of RYN combined with LNZ is 2 μg/ml LNZ+1/16RYN.
As shown in Figure 1D, there was a significant difference in the amount of live MRSA bacteria in the effect of drugs before and after biofilm formation. Compared with control group, 2 μg/ml LNZ had a better inhibitory effect on MRSA living bacteria before biofilm formation than 1/16RYN at 0 h. In the case of the combination of the two drugs, the inhibitory effect of the 2 μg/ml LNZ+1/16RYN group on living MRSA before and after biofilm formation was greater than that of the two drugs alone. The results show that the combination of LNZ and RYN has a significant inhibitory effect on living MRSA before and after biofilm formation.
As shown in Figure 2A, the surface of the control group was rough, the fluorescence signal value of living bacteria was strong, the vitality of living bacteria in biofilms was strong, the distribution of bacteria was tight, and the fluorescence signal value of dead bacteria is weaker than that of living bacteria. Compared with the control group, the light spot of living bacteria in the 2 μg/ml LNZ group decreased slightly due to drug action, but the signal intensity was still larger, and the fluorescence signal values of dead bacteria in the two groups were similar. However, the fluorescence signal of living bacteria was significantly weakened and the fluorescence signal of dead bacteria in the 1/16RYN group was significantly stronger than those in the former two groups. The fluorescence signal values of living and dead bacteria in the combined group (2 μg/ml LNZ+1/16RYN) were significantly lower than those in the first three groups. Its surface is finer than that of other groups, the thickness of biofilm decreases obviously, and its distribution is sparse. The administration group did not have any inhibitory effect on the extracellular matrix of MRSA biofilms. These results suggested that RYN could destroy the protective effect of biofilms on the bacteria itself and that combination with LNZ could inhibit and kill bacteria in biofilms. It has not only inhibition and killing effects on living bacteria, but also scavenging effects on the residual structure of dead bacteria.
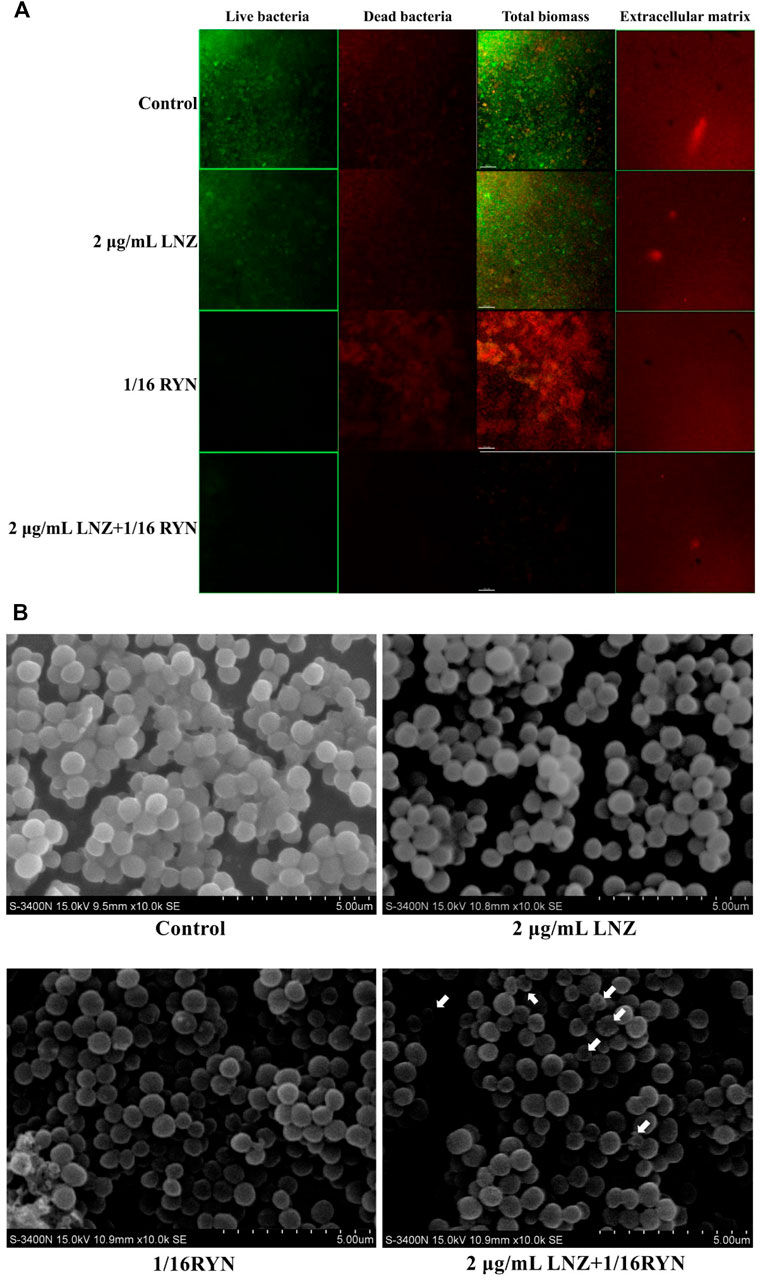
FIGURE 2. Imaging results of the effects of LNZ and RYN on MRSA biofilm live and dead bacteria and extracellular matrix. (A) CLSM. In the first three columns, green is the fluorescence signal of living bacteria, and red is the fluorescence signal of dead bacteria. The images show the inhibitory effect of LNZ and RYN on the maturity of MRSA biofilms, which is consistent with the results of the biofilm assay, and the combined group had a strong inhibitory effect on biofilms. There was no significant difference among the four groups in the extracellular matrix of the fourth column. The scale bar in images represents 150 µm. (B) SEM. In the mature stage of a MRSA biofilms without drug intervention (control group), MRSA grew in clumps and the structure of the cell arrangement was compact. In the mature stage of MRSA biofilms treated with 2 μg/ml LNZ and 1/16RYN, the structure of the cell arrangement was slightly loose. Combined 2 μg/ml LNZ+1/16RYN intervention in the mature stage of a MRSA biofilm resulted in a loose cell arrangement structure, a different cell size, and an empty shell. Original magnification: ×10000.
As shown in Figure 2B, the bacteria gathered on the MRSA biofilm surface of the control group were wrapped in a membranous structure, and the bacteria in the biofilm were uniform in size and normal in shape. There was a membrane structure in the MRSA of the positive drug group (2 μg/ml LNZ), and there was no significant difference in the size of bacteria between the 2 μg/ml LNZ group and the control group. In the 1/16RYN group, there was no obvious membrane structure in the outer layer of MRSA, the arrangement of bacteria was no longer compact, and the shape of bacteria was irregular and round. In the 2 μg/ml LNZ+1/16RYN group, there was no membrane structure in the outer layer of MRSA, the arrangement structure of bacteria was loose, the size of bacteria was different, and there were bacteria in the empty shell state (shown by white arrow). It is suggested that RYN has a destructive effect on MRSA biofilms, and the combination of the two drugs can increase the inhibitory effect on bacteria in the mature stage of MRSA biofilms.
As shown in Figure 3, there is no bacteriostatic zone in the control group, 2 μg/ml LNZ group and 1/16RYN group at 2, 4 and 6 h, while in the 2 μg/ml LNZ+1/16RYN group, there is a bacteriostatic zone at 4 and 6 h, and the size of bacteriostatic zone was 2 h (6 mm) < 4 h (7 mm) <6 h (10 mm). The results show that the combination of LNZ and RYN has an effect on the structure of the MRSA biofilm, and the permeability of the MRSA biofilm is improved compared with the single drug group.
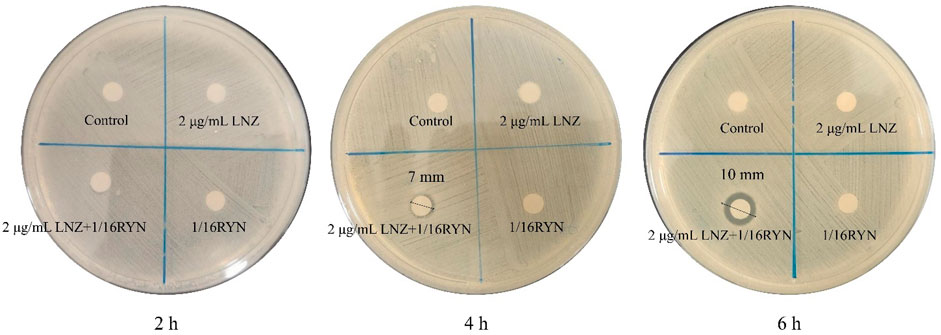
FIGURE 3. Effect of LNZ and RYN on the permeability of MRSA biofilms. There was no bacteriostatic zone in the control group, 2 μg/ml LNZ group and 1/16RYN group, but the bacteriostatic zone in the 2 μg/ml LNZ+1/16RYN group increased with the extension of action time [2 h (6 mm) < 4 h (7 mm) < 6 h (10 mm)].
The amplification curves of agrA, agrB, agrC, agrD, atlA, RNAⅢ and sarA were consistent, and the dissolution curves were all single peaks, indicating that the specificity of each gene primer was strong, and that the amplification product was single (Supplementary Figure S4).
The expression of the MRSA biofilm-related genes agrA, agrB, agrC, agrD, atlA, RNAⅢ and sarA was inhibited after drug intervention in the formation of biofilms (Figure 4). Compared with the control group, the expression of agrA, agrB, agrC, agrD, atlA, RNAⅢ and sarA was inhibited in the 2 μg/ml LNZ group and the 2 μg/ml LNZ+1/16RYN group. In the 1/16RYN group, the expression of six genes except for agrC was downregulated. In the comparison of RYN and LNZ alone, the inhibitory effect of agrD in the 1/16RYN group was higher than that in the 2 μg/ml LNZ group. In the comparison of drug use alone and in combination, the inhibitory effect of agrC, agrD, atlA and sarA in the 2 μg/ml LNZ+1/16RYN group is higher than that in the 2 μg/ml LNZ group, and the inhibitory effect of agrC, atlA and sarA is higher than that in the 1/16RYN group.
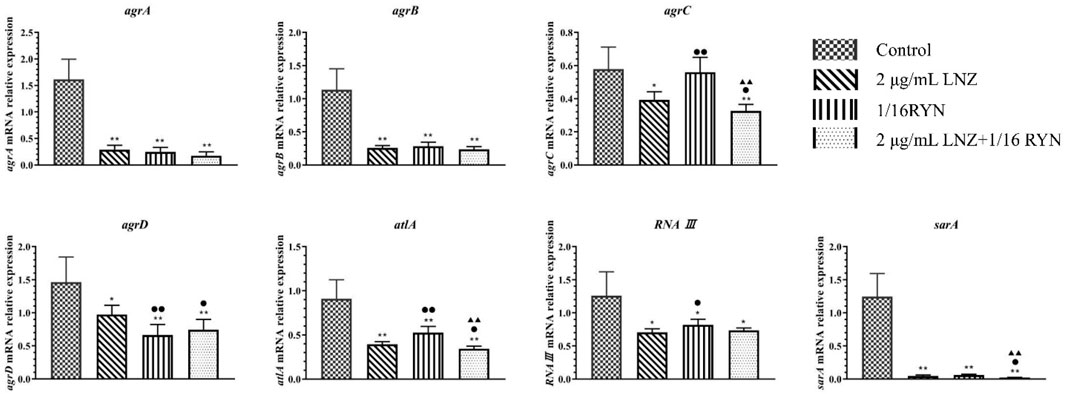
FIGURE 4. mRNA expression of biofilm-related genes after different drug treatments. (**p < 0.01, *p < 0.05 vs. control group; ▲▲p < 0.01, ▲p < 0.05 vs. 1/16RYN group; ●●p < 0.01, ●p < 0.05 vs. 2 μg/ml LNZ group.). Compared with the control group, the gene expression decreased after drug intervention. The expression of agrC, atlA and sarA decreased significantly compared with LNZ and RYN alone under the intervention of the combination of the two drugs.
UPLC-Q-TOF-MS analysis of the MRSA samples from the biofilm period indicated the presence of 6623 metabolite features in the positive mode and 3581 metabolite features in the negative mode, of which 2181 were putatively identified. The PCA scores of all metabolites in the six groups are shown in Figure 5A. The dataset was processed via an unsupervised statistical approach using PCA. PCA was used to investigate any subdata clustering, which was not evident. Furthermore, the dataset was assessed for outliers via a distance of observation (DModX) analysis, which indicated that no samples exceeded the threshold for rejecting a sample. However, some RYN and LNZ + RYN samples were intermixed with each other, although there was a tendency for the samples to be separated between the two groups. To further validate that the metabolic differences between MRSA were the result of metabolic consequences induced by LNZ, PLS-DA was used to compare the metabolic profiles of the three groups. Five groups of samples and QC could be separated in the PLS-DA score plot (Figure 5A).
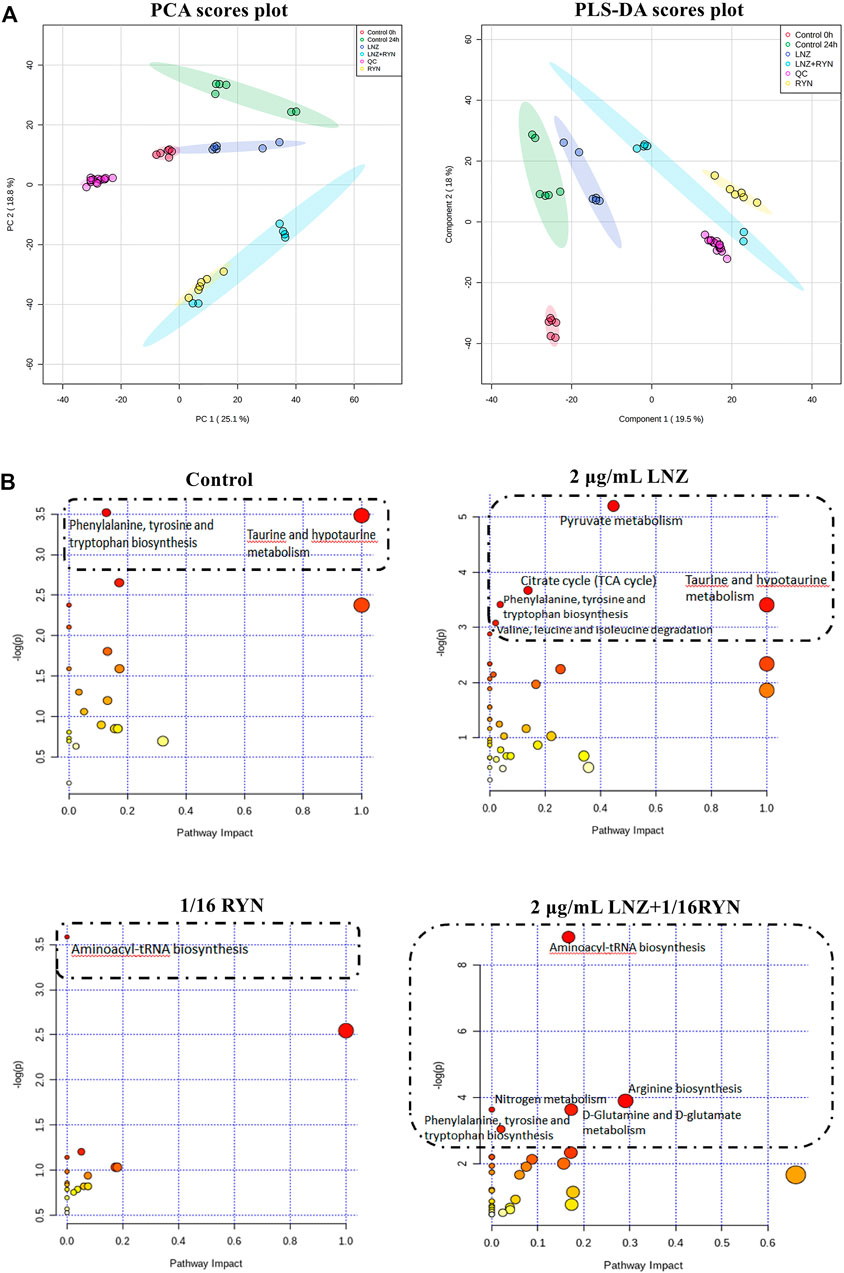
FIGURE 5. PCA and PLS-DA score plots and overview of pathway analysis metabolites. (A) PCA and PLS-DA score plots. PCA score plot of all samples including QC samples. PLS-DA score plot of all samples including QC samples. (B) overview of pathway analysis metabolites. The bubble chart of pathway enrichment analysis shows all matched pathways according to the path impact values (X-axis) of topological analysis and p value (Y-axis) of path enrichment analysis. Each node represents a biological pathway. Bubble area (radius) is proportional to the impact value of each pathway, with the colour denoting the significance from highest in red to lowest in white. The nodes in the dashed box represent the biological pathways that are significantly related to biofilm markers.
Twenty-seven differential metabolites were found in the comparison of the two control groups before and after biofilm formation (Table 3). These metabolites reflect the metabolic phenotype characteristics of MRSA biofilm formation, and are significantly related to taurine and hypotaurine metabolism and phenylalanine, tyrosine and tryptophan biosynthesis (Figure 5B, Supplementary Table S3).
Twenty-eight differential metabolites were found in the comparison of the LNZ group and the control group (Table 3). These metabolites reflect the pharmacodynamic characteristics of 2 μg/ml LNZ in MRSA biofilms and are significantly related to pyruvate metabolism, the citrate cycle (TCA cycle), phenylalanine, tyrosine and tryptophan biosynthesis, taurine and hypotaurine metabolism and valine, leucine and isoleucine degradation (Figure 5B, Supplementary Table S4).
Twenty-five differential metabolites were found in the comparison of the 1/16RYN group and the control group (Table 3). These metabolites reflect the pharmacodynamic characteristics of 1/16 RYN treating MRSA with biofilms and are significantly related to aminoacyl-tRNA biosynthesis (Figure 5B, Supplementary Table S5).
Thirty-two differential metabolites were found in the comparison of the 2 μg/ml LNZ+1/16RYN group and the control group (Table 3). These metabolites reflect the pharmacodynamic characteristics of 2 μg/ml LNZ+16RYN treated MRSA biofilms and are significantly related to aminoacyl-tRNA biosynthesis, arginine biosynthesis, nitrogen metabolism, D-glutamine and D-glutamate metabolism and phenylalanine, tyrosine and tryptophan biosynthesis (Figure 5B, Supplementary Table S6).
As shown in Table 3, there were 10 common metabolites in the control group, 2 μg/ml LNZ group, 1/16RYN group and 2 μg/ml LNZ+1/16RYN group. These metabolites are pharmacodynamic biomarkers of LNZ combined with RYN against MRSA. There were no metabolic pathways significantly related to these biomarkers. 3-Dehydrocarnitine decreases after biofilm formation in the control group. After intervention with 2 μg/ml LNZ and 1/16RYN alone, 3-dehydrocarnitine in cells increased. After the combined intervention of the two, 3-dehydrocarnitine increased significantly. The level of metabolites in the administration group was opposite to that in the control group, and the change trend in the combination group was consistent with that in the single group and larger than that in the single group (Figure 6A). Other metabolites similar to this change are kynurenine, L-leucine and L-lysine. Their levels increased after the biofilm matured but decreased after LNZ and RYN intervention, and the extent of the decrease increased after the combination of the two drugs. The intervention effects of LNZ and RYN on ADP-D-ribose, sebacic acid and sucrose were the opposite. The level of ADP-D-ribose decreased after biofilm formation, and LNZ intervention further promoted the reduction of ADP-D-ribose. The level of ADP-D-ribose increases after RYN intervention. The level of sebacic acid increases after the formation of biofilms, and LNZ promotes an increase in sebacic acid content. However, the level of sebacic acid decreased after RYN intervention, and increased after combined use. The level of sucrose decreases only in the LNZ group.
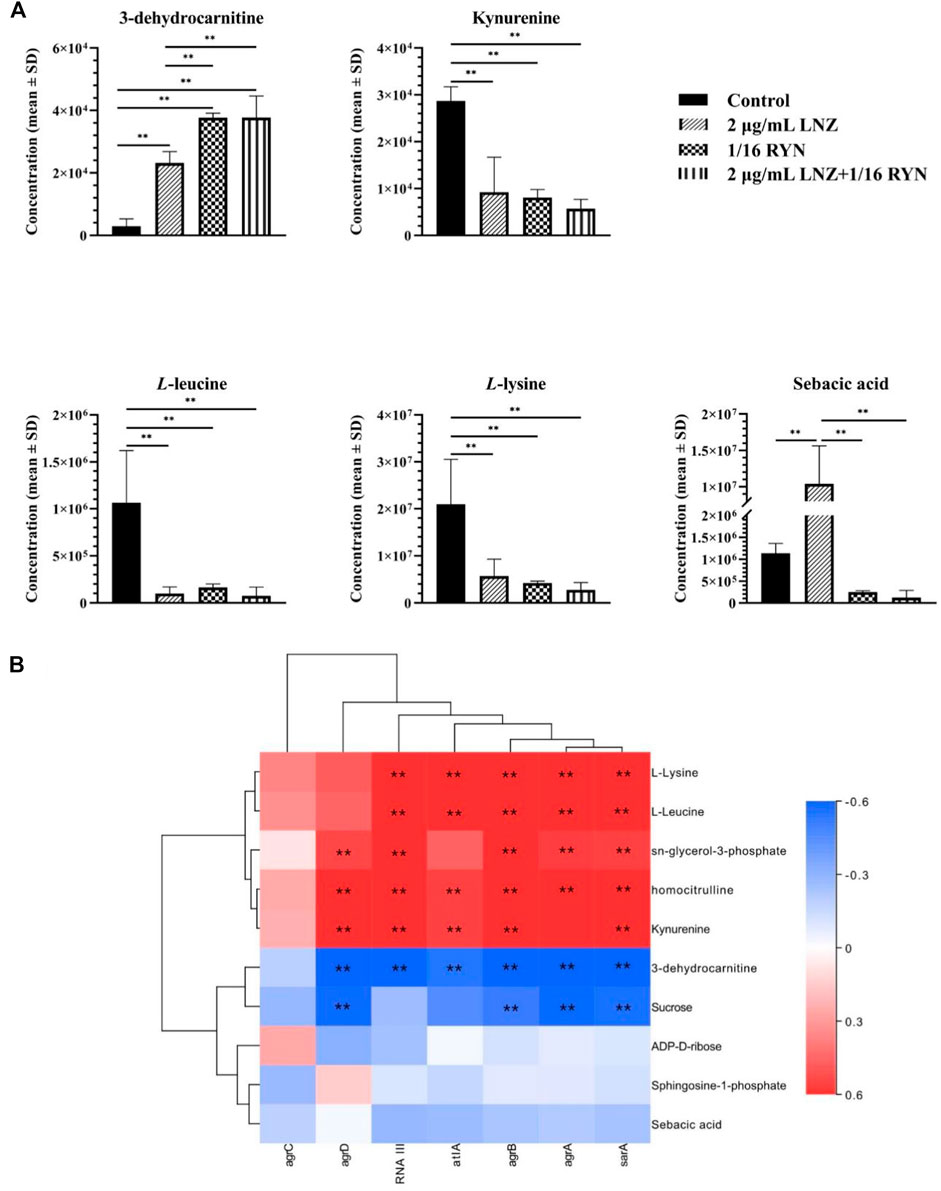
FIGURE 6. Peak intensities of pharmacodynamic biomarkers in different groups and its correlation with gene expression. (A) **p < 0.01. (B) Correlation analysis between pharmacodynamic biomarkers of RYN and LNZ and biofilm-associated genes. Significance levels are indicated as follows: **p < 0.01, red and blue indicate positive and negative correlations, respectively.
As shown in Figure 6B, unsupervised clustering yielded positive correlations between L-lysine, L-leucine, kynurenine, homocitrulline, sn-glycerol-3-phosphate and seven biofilm-associated genes. 3-Dehydrocarnitine, sucrose, ADP-D-ribose, sphingosine-1-phosphate, and sebacic acid were negatively correlated. Correlations between agrA, agrB, and sarA and the ten pharmacodynamic markers were similar, and correlations between agrC and the ten pharmacodynamic markers were the weakest.
Antibiotics are currently the first-choice drugs for the clinical treatment of bacterial infections. However, for some drug-resistant bacteria, the existing antibiotics are difficult to effectively treat them. MRSA, a highly pathogenic drug-resistant bacterium, seriously threatens human health. Biofilms are the cause of MRSA resistance, enhance the pathogenicity of MRSA and make the treatment of MRSA more difficult. The lowest inhibitory concentration of the antibiotic LNZ on MRSA biofilms is 32–64 times higher than that of LNZ on planktonic MRSA biofilms (Parra-Ruiz et al., 2012). RYN, a compound preparation of TPM, is often used for the treatment of MRSA and its biofilm infection in China. Therefore, in this study we explored whether the combination of RYN and LNZ has complementary advantages and revealed its mechanisms. The results showed that the combination of drugs could synergistically inhibit the growth of MRSA and its biofilm. The mechanisms of their synergistic antibacterial action lie in affecting physiological metabolic pathways such as MRSA amino acid, fat metabolism and cell membrane synthesis by reducing the transcription of the QS system and adhesion-related genes. Eventually the combined medication reduced the dose of LNZ and inhibited the formation of biofilms.
Bacteria in biofilms and bacteria in the suspended state have large differences in growth characteristics and nutrient absorption (El-Azizi et al., 2005). The ability of MRSA to form biofilms greatly affects its survival and virulence, and gives it great resistance to host immune system clearance and clinical drug therapy. It is possible that EPS, a constituent of biofilms, may facilitate the anti-drug properties of biofilms by preventing the bulk transport of antibiotics across the biofilm in a direct drug binding manner (Donlan, 2000). In our results, the disruption of biofilms by RYN greatly assisted LNZ in exerting a bacteriostatic effect on MRSA. As a TCM compound preparation, RYN which is rich in taraxacum, emodin and polydatin has good inhibitory properties on bacteria and their biofilms (Lau and Plotkin, 2013). Emodin, an anthraquinone derivative isolated from Polygonum cuspidatum and palm wood, has inhibitory effects on the biofilm formation of S. aureus, Pseudomonas aeruginosa and Streptococcus suis (Yan et al., 2017). Another study found that emodin extracted from the rhizome of Polygonum cuspidatum could destroy the integrity of the MRSA cell wall and cause the loss of intracellular components, thereby significantly inhibiting the growth of MRSA strains (Cao et al., 2015). Thus, emodin may be one of the key components by which RYN inhibits MRSA and its biofilm. Existing studies have shown that the coadministration of antibiotics and nonantibiotics could reduce the resistance of bacteria and enhance the antibacterial activity of drugs (Tyski, 2003). The bacteriostatic effect of the combination of RYN and LNZ was superior to the effect of each of the two drugs alone in the in vitro experimental results, and the dose of the combination of RYN with LNZ was reduced compared with the dose of LNZ alone. All of these results showed that RYN acts synergistically with antibiotic drugs as a non-antibiotic drug.
To further investigate the antimicrobial mechanism of RYN in combination with LNZ, we examined the QS system and adhesion genes associated with MRSA biofilm formation. Of the significantly changed genes we examined, sarA and agrC were associated with the QS system. SarA is an important transcriptional regulator in S. aureus that is closely related to biofilms, adhesion and haemolysis (Valle et al., 2003). Compared with planktonic bacteria, the transcription of sarA in, S. aureus was upregulated during the biofilm stage (Valle et al., 2003). SarA is a positive regulator of biofilm formation, in part because it inhibits the production of proteases and nucleases (Figure 7). The increase in bacterial extracellular protease can inhibit the activity of fibronectin binding protein and inhibit the adhesion of bacteria. Fibronectin binding protein is one of the surface adhesion factors of S. aureus, which can help bacteria adhere and colonize (Keane et al., 2007). The biofilm formation and fibronectin binding of SarA mutant strains are significantly lower than those of parent strains (Abdelhady et al., 2014). It is inferred that due to the deletion of sarA, the inhibition of protease by SarA disappears, the production of protease in the mutant increases, and the binding ability to fibronectin is significantly decreased, which will greatly affect the adhesion of MRSA and the production of biofilms (Abdelhady et al., 2014). In addition, the increase in nuclease production also affected the formation of biofilms due to the deletion of sarA (Beenken et al., 2010). Nuclease mutants could form abnormally thick biofilms, including the phenomenon of increased levels of matrix-related extracellular DNA (eDNA) (Figure 7) (Beenken et al., 2010). Bacteria release eDNA through autolysis during programmed cell death, which contributes to the formation of biofilms. The inhibition of nuclease can promote the formation of MRSA biofilms. Emodin has been reported to downregulate sarA and intervene in the release of eDNA (Yan et al., 2017). LNZ showed poor clearance from biofilms of S. aureus that were wild type for sarA. However, it was more sensitive to sarA mutant S. aureus, and the biofilm clearance rate was improved (Weiss et al., 2009). This suggests that SarA is an important factor in maintaining the stability of biofilms (Weiss et al., 2009). RYN caused the inhibition of SarA expression in MRSA leading to reduced MRSA biofilm production. This increases the permeability of LNZ to biofilms, thereby exerting an inhibitory effect on MRSA.
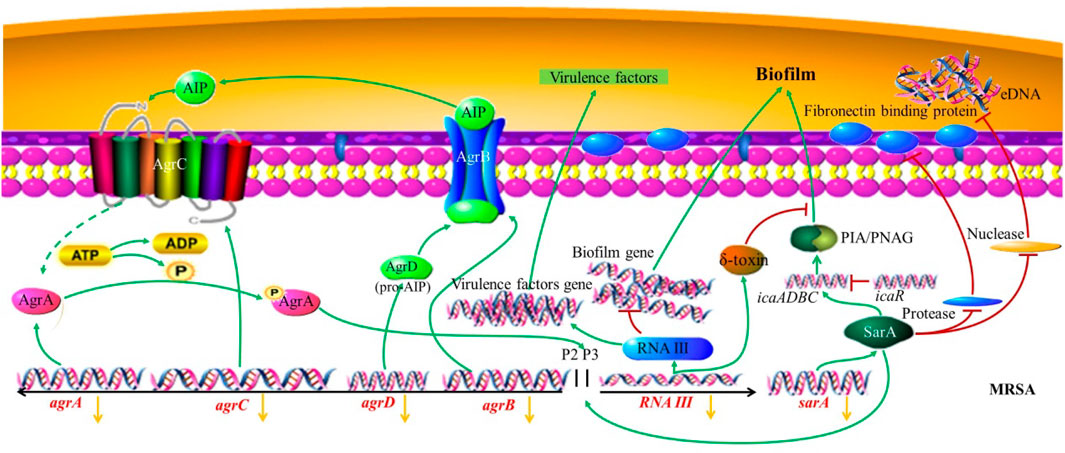
FIGURE 7. MRSA biofilm regulated by the QS system. The yellow arrow indicates that the combined action of RYN and LNZ leads to a decrease in the expression of the gene.
Both agr and sarA are the main regulators in biofilm formation, and the Agr system composed of agr is one of the main two-component signal transduction systems of S. aureus. The Agr-QS system showed downregulation of bacterial cell wall-associated adhesion factors, which could weaken the adhesion ability and indirectly inhibit the formation of initial biofilms (Boles and Horswill, 2008). It has been proven that inducing the expression of agr can lead to the separation of S. aureus cells from the established biofilms (Boles and Horswill, 2008). Compared with the QS system in other bacteria, agr, as an active QS system, hinders the development of biofilms by inhibiting the production of adhesion proteins and inducing the expression of matrix degradation enzymes (such as protease, nuclease and lipase) (Kong et al., 2006). It acts as a regulatory switch between staphylococcal planktonic cells and the biofilm state. The Agr system contains agrA, agrB, agrC and agrD 3 kb loci. When the bacteria reach the critical concentration, the signal molecule AIP precursor encoded by agrD is processed and secreted out of the cell by the AgrB transmembrane protein encoded by agrB (Zhang et al., 2002). AgrC, a homologous transmembrane receptor encoded by agrC, is the receptor of mature AIP (autoinducing peptide), and its combination can activate the autophosphorylation of transcriptional AgrA (Zhang et al., 2002). Agr can be induced by AIP, which is also necessary for mature biofilm dissemination (Boles and Horswill, 2008). AgrA can cooperate with SarA to participate in the activation of promoter 2 (P2) and promoter 3 (P3). P2 can regulate the agr system through a self-induction mechanism. P3 inhibits the formation of biofilms and turns on the expression of virulence factors by regulating the production of the agr effector molecule RNAⅢ (Zhang et al., 2002). As an inhibitor of protein synthesis, the concentration of LNZ used in clinical treatment has an important effect on the inhibition of agr activity, which can reduce the transcriptional level of RNAⅢ and inhibit the diseases mediated by invasive toxins caused by MRSA (Tsuji et al., 2012). This is in agreement with our experimental results. Although this inhibition of agr may not be conducive to the inhibition of biofilm growth, it has been reported that the effect of sarA on agr is epistatic during biofilm formation. The agr mutant has little effect on biofilm development in vitro, indicating that the biofilm is not dependent on the agr gene in vitro (Beenken et al., 2010). In addition, RNAⅢ of the P3 transcription unit of the Agr system can regulate virulence factors, which can help bacteria transition from the colonization stage in the early stage of infection to the stage of invasion and acquisition of host nutrients in the later stage of infection. Studies on animal infection have shown that the loss of Agr system function can significantly reduce the pathogenicity of S. aureus (Montgomery et al., 2010). This indicates that inhibition of the Agr system has an important role in reducing the pathogenicity of MRSA. Under the control of the QS system, the most important step to adjust bacteria from the planktonic state to the biofilm state is the adhesion between bacteria and the external environment so that bacteria can be colonized at a certain point and not easily be removed. Therefore, we examined the atlA gene related to MRSA adhesion. AtlA, encoded by the atlA gene, is the most important peptidoglycan hydrolase in staphylococci. AtlA, also known as a bifunctional autolysin precursor protein, can destroy the internal junction and cell wall structure of peptidoglycan, resulting in bacterial cell lysis and autolysis (Götz et al., 2014). In addition, the adhesion molecule-like function of AtlA is conducive to the initial adhesion between bacteria and substrate molecules, which plays an important role in the formation of biofilms and leads to the chronicity and recurrence of bacterial infection (Götz et al., 2014). Some studies have shown that the biofilm growth of atlA deletion mutants is inhibited (Biswas et al., 2006). In our results, the expression of atlA was further suppressed after the combination of the two drugs. This inhibition may lead to a decrease in the autolysis of bacteria, a decrease in the expression of AtlA protein, a decrease in the ability of bacteria to adhere to substrate molecules, and a decrease in the ability of biofilm formation. The combination of the two drugs can enhance the inhibition of the expression of agrC, atlA and sarA, to a certain extent, but the specific regulatory mechanism needs to be further studied.
Traditional antibiotics are designed to resist planktonic bacteria and treat metabolically exuberant bacteria, but the bacteria in biofilms are different from planktonic bacteria in metabolism (Davey and O’toole, 2000). Metabolic changes contribute to a higher tolerance of bacterial biofilms to therapeutic agents (Stewart, 2002). Our data suggest that the differential metabolites of planktonic MRSA and biofilm-producing MRSA may be related to the biosynthesis of phenylalanine, tyrosine and tryptophan (Figure 5B). This is consistent with previous reports that the uptake of amino acids by biofilms is an important factor that affects biofilm physiology (Ammons et al., 2014). On this basis, we concluded that there were 10 metabolites related to the combination of RYN and LNZ in the endogenous metabolites of MRSA. Among them, the levels of homocitrulline, kynurenine, L-leucine and L-lysine decreased significantly after the combination treatment (Table 3). S. aureus must adapt to various carbon and nitrogen sources when invading the host. Amino acids can be used by S. aureus catabolism as a secondary carbon source for survival and increment (Halsey et al., 2017). In biofilms, the consumption and secretion of bacterial metabolites increased significantly. Some studies have shown that biofilm cultures of S. aureus selectively absorb amino acids (Ammons et al., 2014). Exposure of S. aureus to subtle changes in temperature, pH and osmotic pressure will lead to significant changes in the composition of amino acids in the cytoplasm of S. aureus (Alreshidi et al., 2016). The decrease in amino acid uptake was accompanied by a decrease in amino acids in the cytoplasm, indicating that the metabolic activity of the bacteria decreased to adapt to changes in temperature, pH and osmotic pressure (Ammons et al., 2014). According to the existing literature, by observing the catabolism of amino acids during the growth of S. aureus, leucine in the culture medium showed a gradual consumption trend, indicating that it may be used in protein synthesis (Alreshidi et al., 2019). The uptake of leucine by S. aureus was very high in both the logarithmic growth phase and stable phase, and the uptake of leucine in the stable phase was 4 times that in the logarithmic growth phase (Alreshidi et al., 2019). This is consistent with our experimental results. Under ideal conditions, leucine, lysine and serine are the most commonly used amino acids in S. aureus culture medium. Studies have shown that leucine deficiency could lead to a 6–8 h growth lag of S. aureus (Kaiser et al., 2018). The stable period is a complex period in the growth of bacteria, which is physiologically equivalent to the biofilm period (Jananee and Preeti, 2017). In the pathway analysis, it was also found that biofilm formation and drug action were closely related to the nitrogen metabolism pathway, tryptophan biosynthetic pathway and proline biosynthetic pathway (Figure 5B). It is suggested that amino acids may play an important role in the growth of MRSA biofilms. Combined with our results, it can be concluded that the combination of the two drugs may have a synergistic and additive effect on inhibiting the uptake of amino acids, which may be an important factor affecting the inhibition of MRSA biofilm growth (Figure 8).
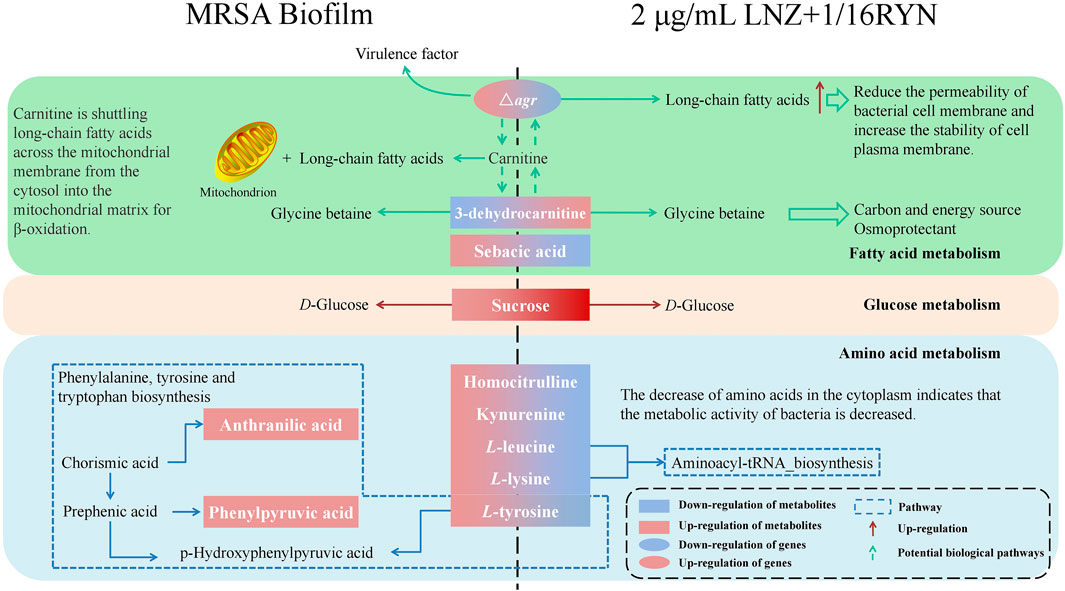
FIGURE 8. Effects of RYN combined with LNZ on MRSA metabolites. Boxes are differential metabolites and oval boxes are genes. Red boxes and red arrows indicate upregulation, and blue boxes indicate downregulation. Dashed boxes indicate pathways. Dashed arrows indicate possible potential biological pathways.
Our study involved four different states of MRSA biofilms, including no drug intervention, 2 μg/ml LNZ intervention alone, 1/16RYN intervention alone, and 2 μg/ml LNZ+1/16RYN combined intervention. 3-dehydrocarnitine in four different states of MRSA showed that the content of 3-dehydrocarnitine decreased with the formation of biofilms without drug intervention. The content of 3-dehydrocarnitine increased after drug intervention, and the effect of the combined drug was greater than that of the single drug. 3-Dehydrocarnitine is an intermediate in the decomposition of carnitine to glycine betaine (GB) (Wargo and Hogan, 2009). Carnitine can transport long-chain fatty acids to the mitochondria of animals, so that relatively large amounts of carnitine are often present in adipose metabolic tissue (Bremer, 1983). Most bacteria can decompose carnitine into betaine (GB, proline betaine, etc.) as osmotic protective agents. Osmotic protective agents can protect bacteria against an increase in external osmotic pressure, ensure the stability of osmotic pressure inside bacterial cells and maintain cell vitality, and cell metabolism will not produce negative interference.
Carnitine can also provide carbon and nitrogen sources for bacteria (Aurich and Lorenz, 1959). In our study, we found that the content of 3-dehydrocarnitine increased significantly under the combination of LNZ and RYN. This indicated that under drug intervention, MRSA significantly produced 3-dehydrocarnitine to adapt to changes in osmotic pressure in the culture medium after administration. In addition, glucose metabolism in which sucrose is involved as an important metabolite is also one of the important aspects influenced by the association of LNZ with RYN. Internal sugars are primarily used for glycolysis, biosynthesis of various components, such as cell walls and lipoteichoic acid, and intercellular polysaccharide biosynthesis. However, high-sugar environments also tend to inhibit the growth of staphylococci (Luo et al., 2020).
The metabolome is downstream of the gene regulatory network and is the embodiment of biological endpoint information. Functional changes in upstream macromolecules (genes, proteins) will ultimately be reflected at the metabolic level. Changes in biofilm-associated genes will affect the levels of biofilm-associated metabolic markers. agrA and agrB genes are significantly associated with markers of metabolic pathways of amino acid metabolism and fatty acid metabolism. Current studies have shown that the level of long-chain fatty acids in the plasma membrane of the agr mutant strain is higher than that of the wild type (Song et al., 2020). Long-chain fatty acids help reduce cell membrane permeability and stability, which may be related to the resistance of MRSA to antibiotics (Meadows and Wargo, 2015). Carnitine transports long-chain fatty acids from the cytoplasm to the mitochondrial matrix for β-oxidation. 3-dehydrocarnitine acts as an intermediate in the breakdown of carnitine to glycine betaine, and metabolite levels may be influenced by long-chain fatty acids in the plasma membrane. It is hypothesized that the increased accumulation of long-chain fatty acids following RYN and LNZ inhibition of agr may be related to the increased levels of 3-dehydrocarnitine, a metabolic marker bound by RYN and LNZ. This is subject to further validation. The correlation of sarA with coadministered pharmacodynamic markers is similar to that of agrA and agrB (Figure 6B), but no study has yet demonstrated an association.
The combination of LNZ and RYN has a synergistic inhibitory effect on MRSA and its biofilm. RYN helps LNZ break through the biofilm to come into contact with the dormant MRSA by destroying the biofilm structure and inhibiting the adhesion and aggregation of biofilms. The combined medication reduces the transcription of agrC, atlA and sarA and interferes with MRSA amino acid and fat metabolism to achieve the antibacterial effect of the synergistic inhibition of biofilm formation and killing of MRSA. This study lays the foundation and provides a theoretical basis for the combination of RYN and LNZ in the clinic. In further research, in vivo experiments will be carried out to verify the synergistic anti-MRSA and biofilm effects of RYN and LNZ to solve the problem of the clinical treatment of MRSA and its biofilm infections.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
LZ, WY, and YC contributed equally to this work and should be considered co-first authors. YT, CL, and WY conceived the experiments. LZ and MB conducted the experiments. YuC, YaC, and FG analyzed the results. BW, LL, and CD prepared the figure. LZ and WY wrote the main manuscript text. JY, CL, TM, YT, and WY wrote review and final editing. All the authors contributed to critical scrutiny of the literature, and its approval.
This work was supported by the National Science and Technology Major Project (2017YFC1703701); the Fundamental Research Funds for the Central Public Welfare Research Institutes (Z0735, Z0749); the Beijing TCM Science and Technology Development Fund Project (JJ2018-102); the Open Project of the State Key Laboratory of Innovative Natural Medicine and TCM Injections (QFSKL2018003); the CACMS Innovation Fund (CI2021A00704-2); the International Cooperation of China Academy of Chinese Medical Sciences (GH201909).
YaC and CD were employed by the company Tsing Hua De Ren Xi an Happiness Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.766309/full#supplementary-material
CLSM, Confocal laser scanning microscopy; eDNA, extracellular DNA; EPS,Extracellular polymeric substances; FC, Fold change; FICI, Fractional inhibition concentration index; GB, glycine betaine; LB, Luria-Bertani; LNZ, linezolid; MIC, Minimum inhibitory concentrations; MRSA, Methicillin-resistant Staphylococcus aureus; PCA, Principal component analysis; PMS, Phenazine methosulfate; QS, Quorum sensing; RT–PCR, Real-time fluorescence polymerase chain reaction; RYN, Reyanning mixture; SEM, Scanning electron microscope; TPM, Traditional plant medicine; TTC, 2,3,5-triphenyltetrazolium chloride; XTT, 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide.
Abdelhady, W., Bayer, A. S., Seidl, K., Moormeier, D. E., Bayles, K. W., Cheung, A., et al. (2014). Impact of Vancomycin on sarA-Mediated Biofilm Formation: Role in Persistent Endovascular Infections Due to Methicillin-Resistant Staphylococcus aureus. J. Infect. Dis. 209, 1231–1240. doi:10.1093/infdis/jiu007
Absalon, C., Van Dellen, K., Watnick, P. I., and Isberg, R. R. (2011). A Communal Bacterial Adhesin Anchors Biofilm and Bystander Cells to Surfaces. Plos Pathog. 7, e1002210. doi:10.1371/journal.ppat.1002210
Alreshidi, M. M., Dunstan, R. H., Macdonald, M. M., Gottfries, J., and Roberts, T. K. (2019). The Uptake and Release of Amino Acids by Staphylococcus aureus at Mid-exponential and Stationary Phases and Their Corresponding Responses to Changes in Temperature, pH and Osmolality. Front. Microbiol. 10, 3059. doi:10.3389/fmicb.2019.03059
Alreshidi, M. M., Hugh, D. R., Gottfries, J., Macdonald, M. M., Crompton, M. J., Ang, C.-S., et al. (2016). Changes in the Cytoplasmic Composition of Amino Acids and Proteins Observed in Staphylococcus aureus during Growth under Variable Growth Conditions Representative of the Human Wound Site. PLOS ONE 11, e0159662. doi:10.1371/journal.pone.0159662
Ammons, M. C. B., Tripet, B. P., Carlson, R. P., Kirker, K. R., Gross, M. A., Stanisich, J. J., et al. (2014). Quantitative NMR Metabolite Profiling of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Discriminates between Biofilm and Planktonic Phenotypes. J. proteome Res. 13, 2973–2985. doi:10.1021/pr500120c
Anderl, J. N., Franklin, M. J., and Stewart, P. S. (2000). Role of Antibiotic Penetration Limitation in Klebsiella pneumoniae Biofilm Resistance to Ampicillin and Ciprofloxacin. Antimicrob. Agents Chemother. 44, 1818–1824. doi:10.1128/aac.44.7.1818-1824.2000
Aurich, H., and Lorenz, I. (1959). On the Catabolism of Carnitine by Pseudomonas Pyocyanea. Acta Biol. Med. Ger. 3, 272–275.
Basri, D. F., and Sandra, V. (2016). Synergistic Interaction of Methanol Extract from Canarium Odontophyllum Miq. Leaf in Combination with Oxacillin against Methicillin-Resistant Staphylococcus aureus (MRSA) ATCC 33591. Int. J. Microbiol. 2016, 5249534. doi:10.1155/2016/5249534
Beenken, K. E., Mrak, L. N., Griffin, L. M., Zielinska, A., Shaw, L. N., Rice, K. C., et al. (2010). Epistatic Relationships between sarA and Agr in Staphylococcus aureus Biofilm Formation. PLOS ONE 5, e10790. doi:10.1371/journal.pone.0010790
Biswas, R., Voggu, L., Simon, U. K., Hentschel, P., Thumm, G., and Gotz, F. (2006). Activity of the Major Staphylococcal Autolysin Atl. Fems Microbiol. Lett. 259, 260–268. doi:10.1111/j.1574-6968.2006.00281.x
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated Dispersal of Staphylococcus aureus Biofilms. PLoS Pathog. 4, e1000052. doi:10.1371/journal.ppat.1000052
Boudet, A., Jay, A., Dunyach-Remy, C., Chiron, R., Lavigne, J. P., and Marchandin, H. (2021). In-Host Emergence of Linezolid Resistance in a Complex Pattern of Toxic Shock Syndrome Toxin-1-Positive Methicillin-Resistant Staphylococcus aureus Colonization in Siblings with Cystic Fibrosis. Toxins (Basel) 13 (5), 317. doi:10.3390/toxins13050317
Bremer, J. (1983). Carnitine – Metabolism and Functions. Physiol. Rev. 63, 1420–1480. doi:10.1152/physrev.1983.63.4.1420
Cao, F., Peng, W., Li, X., Liu, M., Li, B., Qin, R., et al. (2015). Emodin Is Identified as the Active Component of Ether Extracts from Rhizoma Polygoni Cuspidati, for Anti-MRSA Activity. Can. J. Physiol. Pharmacol. 93, 485–493. doi:10.1139/cjpp-2014-0465
Cerca, N., Gomes, F., Pereira, S., Teixeira, P., and Oliveira, R. (2012). Confocal Laser Scanning Microscopy Analysis of S. Epidermidis Biofilms Exposed to Farnesol, Vancomycin and Rifampicin. BMC Res. Notes 5, 244. doi:10.1186/1756-0500-5-244
Davey, M. E., and O'toole, G. A. (2000). Microbial Biofilms: from Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 64, 847. doi:10.1128/mmbr.64.4.847-867.2000
David, D. (2003). Understanding Biofilm Resistance to Antibacterial Agents. Nat. Rev. Drug Discov. 2, 114–122. doi:10.1038/nrd1008
Dey, P., Parai, D., Banerjee, M., Hossain, S. T., and Mukherjee, S. K. (2020). Naringin Sensitizes the Antibiofilm Effect of Ciprofloxacin and Tetracycline against Pseudomonas aeruginosa Biofilm. Int. J. Med. Microbiol. IJMM 3103, 151410. doi:10.1016/j.ijmm.2020.151410
Donlan, R. M. (2000). Role of Biofilms in Antimicrobial Resistance. Asaio J. 46, S47–52. doi:10.1097/00002480-200011000-00037
El-Azizi, M., Rao, S., Kanchanapoom, T., and Khardori, N. (2005). In Vitro activity of Vancomycin, Quinupristin/dalfopristin, and Linezolid against Intact and Disrupted Biofilms of Staphylococci. Ann. Clin. Microbiol. Antimicrobials 4, 2. doi:10.1186/1476-0711-4-2
Emara, S., Amer, S., Ali, A., Abouleila, Y., and Masujima, T. (2011). Single-Cell Metabolomics. Cham, Switzerland: Springer International Publishing.
Fankam, A. G., Kuete1, V., Voukeng, I. K., Kuiate, J. R., and Pages, J.-M. (2011). Antibacterial Activities of Selected Cameroonian Spices and Their Synergistic Effects with Antibiotics against Multidrug-Resistant Phenotypes. BMC Complement. Altern. Med. 11, 104. doi:10.1186/1472-6882-11-104
Gomes, F., Teixeira, P., Cerca, N., Azeredo, J., and Oliveira, R. (2011). Effect of Farnesol on Structure and Composition of Staphylococcus Epidermidis Biofilm Matrix. Curr. Microbiol. 63, 354–359. doi:10.1007/s00284-011-9984-3
Götz, F., Heilmann, C., and Stehle, T. (2014). Functional and Structural Analysis of the Major Amidase (Atl) in Staphylococcus. Int. J. Med. Microbiol. IJMM 304, 156–163. doi:10.1016/j.ijmm.2013.11.006
Gowrishankar, S., Arumugam, K., Ayyanar, K. S., and Balamurugan, K. (2015). Bacillus Amyloliquefaciens-Secreted Cyclic Dipeptide-Cyclo (L-Leucyl- L-Prolyl) Inhibits Biofilm and Virulence in Methicillin-Resistant Staphylococcus aureus. RSC Adv. 5 (116), 95788. doi:10.1039/C5RA11641D
Gowrishankar, S., Thenmozhi, R., Balaji, K., and Pandian, S. K. (2013). Emergence of Methicillin-Resistant, Vancomycin-Intermediate Staphylococcus aureus Among Patients Associated with Group A Streptococcal Pharyngitis Infection in Southern India. Infect. Genet. Evol. 14, 383–389. doi:10.1016/j.meegid.2013.01.002
Halsey, C. R., Lei, S., Wax J., K., Lehman, K. M., Nuxoll, S. A., Steinke, L., et al. (2017). Amino Acid Catabolism in Staphylococcus aureus and the Function of Carbon Catabolite Repression. mBio 8 (1), e01434–16. doi:10.1128/mBio.01434-16
Huang, H., Zhang, A., Cao, H., Lu, H., Wang, B., Xie, Q., et al. (2013). Metabolomic Analyses of Faeces Reveals Malabsorption in Cirrhotic Patients. Dig. Liver Dis. official J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 45, 677–682. doi:10.1016/j.dld.2013.01.001
Jananee, J., and Preeti, S. (2017). Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 8, 2000. doi:10.3389/fmicb.2017.02000
Kaiser, J. C., King, A. N., Grigg, J. C., Sheldon, J. R., Edgell, D. R., Murphy, M. E. P., et al. (2018). Repression of Branched-Chain Amino Acid Synthesis in Staphylococcus aureus Is Mediated by Isoleucine via CodY, and by a Leucine-Rich Attenuator Peptide. Plos Genet. 14, e1007159. doi:10.1371/journal.pgen.1007159
Kannappan, A., Sivaranjani, M., Srinivasan, R., Rathna, J., Pandian, S. K., and Ravi, A. V. (2017). Inhibitory Efficacy of Geraniol on Biofilm Formation and Development of Adaptive Resistance in Staphylococcus Epidermidis RP62A. J. Med. Microbiol. 66, 1506–1515. doi:10.1099/jmm.0.000570
Karbasizade, V., Dehghan, P., Sichani, M. M., Shahanipoor, K., Jafari, R., Yousefian, R., et al. (2017). Evaluation of Three Plant Extracts against Biofilm Formation and Expression of Quorum Sensing Regulated Virulence Factors in Pseudomonas aeruginosa. Pak J. Pharm. Sci. 30, 585–589.
Keane, F. M., Loughman, A., Valtulina, V., Brennan, M., Speziale, P., and Foster, T. J. (2007). Fibrinogen and Elastin Bind to the Same Region within the A Domain of Fibronectin Binding Protein A, an MSCRAMM of Staphylococcus aureus. Mol. Microbiol. 63, 711–723. doi:10.1111/j.1365-2958.2006.05552.x
Kenny, O., Brunton, N. P., Walsh, D., Hewage, C. M., Mcloughlin, P., and Smyth, T. J. (2015). Characterisation of Antimicrobial Extracts from Dandelion Root (Taraxacum officinale) Using LC-SPE-NMR. Phytotherapy Res. PTR 29, 526–532. doi:10.1002/ptr.5276
Kong, K.-F., Vuong, C., and Otto, M. (2006). Staphylococcus Quorum Sensing in Biofilm Formation and Infection. Int. J. Med. Microbiol. 296, 133–139. doi:10.1016/j.ijmm.2006.01.042
Krzysciak, W., Koscielniak, D., Papiez, M., Vyhouskaya, P., Zagorska-Swiezy, K., Kolodziej, I., et al. (2017). Effect of a Lactobacillus Salivarius Probiotic on a Double-Species Streptococcus Mutans and Candida Albicans Caries Biofilm. Nutrients 9, 1242. doi:10.3390/nu9111242
Lau, D., and Plotkin, B. (2013). Antimicrobial and Biofilm Effects of Herbs Used in Traditional Chinese Medicine. Nat. Product. Commun. 8, 1617–1620. doi:10.1177/1934578x1300801129
Luo, Z., Yue, S., Chen, T., She, P., Wu, Y., and Wu, Y. (2020). Staphylococcus aureusReduced Growth of under High Glucose Conditions Is Associated with Decreased Pentaglycine Expression. Front. Microbiol. 11, 537290. doi:10.3389/fmicb.2020.537290
Meadows, J. A., and Wargo, M. J. (2015). Carnitine in Bacterial Physiology and Metabolism. Microbiology (Reading, England) 161, 1161–1174. doi:10.1099/mic.0.000080
Montgomery, C. P., Boyle-Vavra, S., and Daum, R. S. (2010). Importance of the Global Regulators Agr and SaeRS in the Pathogenesis of CA-MRSA USA300 Infection. PLOS ONE 5, e15177. doi:10.1371/journal.pone.0015177
Parra-Ruiz, J., Bravo-Molina, A., Peña-Monje, A., and Herna´Ndez-Quero, J. (2012). Activity of Linezolid and High-Dose Daptomycin, Alone or in Combination, in an In Vitro Model of Staphylococcus aureus Biofilm. J. Antimicrob. Chemother. 67, 2682–2685. doi:10.1093/jac/dks272
Patel, M. (2009). Community-associated Meticillin-Resistant Staphylococcus aureus Infections: Epidemiology, Recognition and Management. Drugs 69, 693–716. doi:10.2165/00003495-200969060-00004
Rishi, E., Rishi, P., Therese, K. L., Ramasubban, G., Biswas, J., Sharma, T., et al. (2018). Culture and Reverse Transcriptase Polymerase Chain Reaction (RT–PCR) Proven Mycobacterium Tuberculosis Endophthalmitis: A Case Series. Ocul. Immunol. Inflamm. 26, 220–227. doi:10.1080/09273948.2016.1207786
Song, H. S., Choi, T. R., Han, Y. H., Park, Y. L., Park, J. Y., Yang, S. Y., et al. (2020). Increased Resistance of a Methicillin-Resistant Staphylococcus aureus Δagr Mutant with Modified Control in Fatty Acid Metabolism. AMB Express 10, 64. doi:10.1186/s13568-020-01000-y
Standards, N.C.F.C.L (2000). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically.
Stefani, S., Chung, D. R., Lindsay, J. A., Friedrich, A. W., Kearns, A. M., Westh, H., et al. (2012). Meticillin-resistant Staphylococcus aureus (MRSA): Global Epidemiology and Harmonisation of Typing Methods. Int. J. Antimicrob. Agents 39, 273–282. doi:10.1016/j.ijantimicag.2011.09.030
Stella, C., Danela, C., Barbara, P., Camilla, P., Elisa, G., Girolamo, C., et al. (2021). Therapeutic Strategies to Counteract Antibiotic Resistance in MRSA Biofilm-Associated Infections. ChemMedChem 16 (1), 65–80. doi:10.1002/cmdc.202000677
Stewart, P. S. (2002). Mechanisms of Antibiotic Resistance in Bacterial Biofilms. Int. J. Med. Microbiol. IJMM 292, 107. doi:10.1078/1438-4221-00196
Tsuji, B. T., Brown, T., Parasrampuria, R., Brazeau, D. A., Forrest, A., Kelchlin, P. A., et al. (2012). Front-Loaded Linezolid Regimens Result in Increased Killing and Suppression of the Accessory Gene Regulator System of Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 3712–3719. doi:10.1128/AAC.05453-11
Tyski, S. (2003). Non-antibiotics--drugs with Additional Antimicrobial Activity. Acta Pol. Pharm. 60, 401–404.
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J. M., Amorena, B., Penadés, J. R., et al. (2003). SarA and Not sigmaB Is Essential for Biofilm Development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. doi:10.1046/j.1365-2958.2003.03493.x
Wang, Y., Wang, T., Hu, J., Ren, C., Lei, H., Hou, Y., et al. (2011). Anti-biofilm Activity of TanReQing, a Traditional Chinese Medicine Used for the Treatment of Acute Pneumonia. J. Ethnopharmacol 134, 165. doi:10.1016/j.jep.2010.11.066
Wargo, M. J., and Hogan, D. A. (2009). Identification of Genes Required for Pseudomonas aeruginosa Carnitine Catabolism. Microbiology 155, 2411–2419. doi:10.1099/mic.0.028787-0
Weiss, E. C., Spencer, H. J., Daily, S. J., Weiss, B. D., and Smeltzer, M. S. (2009). Impact of sarA on Antibiotic Susceptibility of Staphylococcus aureus in a Catheter-Associated In Vitro Model of Biofilm Formation. Antimicrob. Agents Chemother. Open access 53, 2475–2482. doi:10.1128/aac.01432-08
Wu, H., Moser, C., Wang, H.-Z., Høiby, N., and Song, Z.-J. (2015). Strategies for Combating Bacterial Biofilm Infections. Int. J. Oral Sci. 7, 1–7. doi:10.1038/ijos.2014.65
Wunderink, R. G., Niederman, M. S., Kollef, M. H., Shorr, A. F., Kunkel, M. J., Baruch, A., et al. (2012). Linezolid in Methicillin-Resistant Staphylococcus aureus Nosocomial Pneumonia: A Randomized, Controlled Study. Clin. Infect. Dis. 5, 5. doi:10.1093/cid/cir895
Yan, X., Gu, S., Shi, Y., Cui, X., Wen, S., and Ge, J. (2017). The Effect of Emodin on Staphylococcus aureus Strains in Planktonic Form and Biofilm Formation In Vitro. Arch. Microbiol. 199, 1267–1275. doi:10.1007/s00203-017-1396-8
Yang, W., Liu, J., Blažeković, B., Sun, Y., Ma, S., Ren, C., et al. (2018). In Vitro antibacterial Effects of Tanreqing Injection Combined with Vancomycin or Linezolid against Methicillin-Resistant Staphylococcus aureus. BMC Complement. Altern. Med. 18, 169. doi:10.1186/s12906-018-2231-8
Yu, H.-H., Kim, K.-J., Cha, J.-D., Kim, H.-K., Lee, Y.-E., Choi, N.-Y., et al. (2005). Antimicrobial Activity of Berberine Alone and in Combination with Ampicillin or Oxacillin against Methicillin-Resistant Staphylococcus aureus. J. Med. Food 8, 454–461. doi:10.1089/jmf.2005.8.454
Zhang, L., Gray, L., Novick, R. P., and Ji, G. (2002). Transmembrane Topology of AgrB, the Protein Involved in the post-translational Modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277, 34736–34742. doi:10.1074/jbc.M205367200
Keywords: methicillin-resistant Staphylococcus aureus, biofilm, reyanning mixture, linezolid, synergistic inhibitory effect
Citation: Zhang L, Yang W, Chu Y, Wen B, Cheng Y, Mahmood T, Bao M, Ge F, Li L, Yi J, Du C, Lu C and Tan Y (2022) The Inhibition Effect of Linezolid With Reyanning Mixture on MRSA and its Biofilm is More Significant than That of Linezolid Alone. Front. Pharmacol. 12:766309. doi: 10.3389/fphar.2021.766309
Received: 28 August 2021; Accepted: 01 December 2021;
Published: 03 January 2022.
Edited by:
Dâmaris Silveira, University of Brasilia, BrazilReviewed by:
Shanmugaraj Gowrishankar, Alagappa University, IndiaCopyright © 2022 Zhang, Yang, Chu, Wen, Cheng, Mahmood, Bao, Ge, Li, Yi, Du, Lu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Tan, dGNtdGFueW9uZ0AxMjYuY29t; Cheng Lu, bHZfY2hlbmcwODE2QDE2My5jb20=; Chengqiang Du, ZGNxNTA2OEAxNjMuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.