- 1The Eighth Affiliated Hospital, Sun Yat-sen University, Shenzhen, China
- 2School of Chemistry and Chemical Engineering, Henan Engineering Research Center of Chiral Hydroxyl Pharmaceutical, Henan Normal University, Xinxiang, China
- 3Institute of Agro-Food Science and Technology Shandong Academy of Agricultural Sciences, Key Laboratory of Agro-Products Processing Technology of Shandong Province, Key Laboratory of Novel Food Resources Processing Ministry of Agriculture, Jinan, China
- 4Department of Neurology, The First Affiliated Hospital of Henan University of Science and Technology, Luoyang, China
Chemotherapy is the most common clinical treatment for non-small cell lung cancer (NSCLC), but low efficiency and high toxicity of current chemotherapy drugs limit their clinical application. Therefore, it is urgent to develop hypotoxic and efficient chemotherapy drugs. Theophylline, a natural compound, is safe and easy to get, and it can be used as a modified scaffold structure and hold huge potential for developing safe and efficient antitumor drugs. Herein, we linked theophylline with different azide compounds to synthesize a new type of 1,2,3-triazole ring-containing theophylline derivatives. We found that some theophylline1,2,3-triazole compounds showed a good tumor-suppressive efficacy. Especially, derivative d17 showed strong antiproliferative activity against a variety of cancer cells in vitro, including H460, A549, A2780, LOVO, MB-231, MCF-7, OVCAR3, SW480, and PC-9. It is worth noting that the two NSCLC cell lines H460 H and A549 are sensitive to compound d17 particularly, with IC50 of 5.929 ± 0.97 μM and 6.76 ± 0.25 μM, respectively. Compound d17 can significantly induce cell apoptosis by increasing the ratio of apoptotic protein Bax/Bcl-2 by downregulating the expression of phosphorylated Akt protein, and it has little toxicity to normal hepatocyte cells LO2 at therapeutic concentrations. These data indicate that these theophylline acetic acid-1,2,3-triazole derivatives may be potential drug candidates for anti-NSCLC and are worthy of further study.
Introduction
It is reported that lung cancer is the deadliest cancer in men in developed countries (26.2%) and developing countries (22.3%) (Bray et al., 2018; Siegel et al., 2019). In 2020, there were 2.2 million new lung cancer cases worldwide, accounting for 11.4% of the total global new cases; the death toll from lung cancer was 1.782 million, accounting for 18.0% of the total global cancer deaths (Sung et al., 2021). Lung cancer falls into two categories, non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is the most common type of lung cancer, further divided into squamous cell carcinoma (SCC), large cell carcinoma (LCC), and adenocarcinoma (AC) (Goldstraw et al., 2011; Travis et al., 2011). AC (accounting for 50% of total NSCLC cases) and SCC (accounting for 30% of total NSCLC cases) are the most common types of NSCLC (Lee and Cheah, 2019). Chemotherapy is the most commonly used treatment of NSCLC, but both single-agent chemotherapy and combination chemotherapy will bring a series of serious side effects, such as hair loss, anemia, nausea, and vomiting (Miller et al., 2016). Therefore, it is extremely urgent to design a safe, efficient, and less side-effect chemotherapy drug.
It is estimated that methylxanthine-containing compounds, such as pentoxifylline (Figure 1A), can improve the efficacy of radiotherapy and chemotherapy and are used as chemotherapy sensitivity modifiers (Misirlioglu et al., 2007); caffeine (Figure 1B) and theophylline (Figure 1C) can enhance the toxicity of doxorubicin to tumor cells (Motegi et al., 2013; Yung-Lung Chang et al., 2017; David Osarieme et al., 2019; Liu et al., 2019). When theophylline is used in combination with gemcitabine or cisplatin, it has been found that theophylline can induce apoptosis in a variety of tumor cells (Hirsh et al., 2004). As a natural medicine, theophylline has a wide range of sources and low biological toxicity. Therefore, theophylline as a basic modified scaffold structure provides hope for developing safe and efficient antitumor drugs (Abou-Zied et al., 2019).
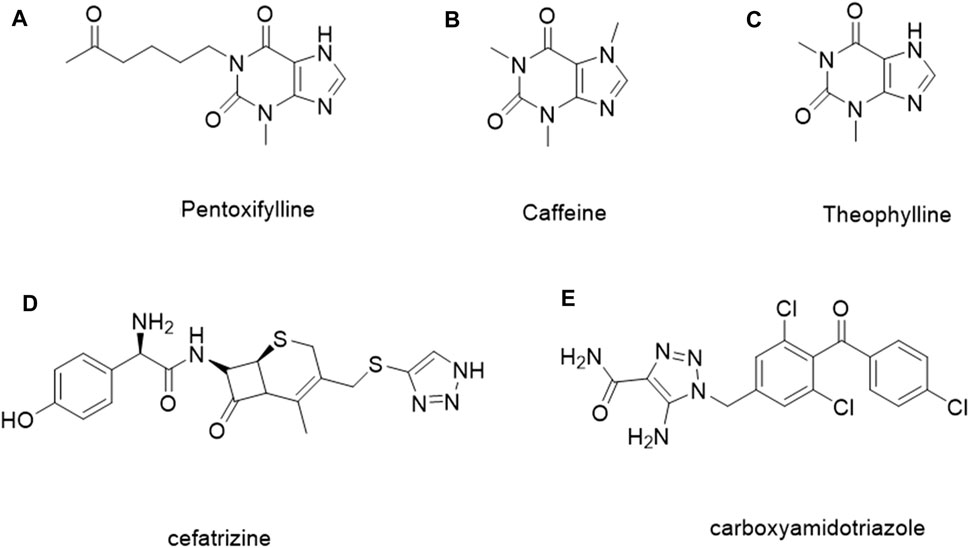
FIGURE 1. Examples of the methylxanthine-containing compounds and the reported 1, 2, 3-triazole derivatives for treating tumors.
1, 2, 3-Triazole, as an important nitrogen heterocyclic structure, plays an important role in compound design and synthesis (Majeed et al., 2013). Compounds with the 1, 2, 3-triazole ring generally show good inhibitory activity against cancer, inflammation, and microorganisms (Rohrig et al., 2012; Zhao et al., 2012; Chen et al., 2017; Al-Blewi et al., 2018; Sakly et al., 2018). In addition, the 1, 2, 3-triazole ring can be easily constructed by the copper-catalyzed azide and alkyne cycloaddition reaction, which reduces the difficulty of synthesis and further improves the application potential. In addition, some compounds containing 1, 2, 3-triazole, such as ceftriaxone (Figure 1D) and carboxamide triazole (Figure 1E), have been used in clinics or are undergoing clinical trials for cancer treatment (Xu et al., 2019; Vanaparthi et al., 2020). Tazobactam is also used as an antibacterial agent (Karlowsky et al., 2020; Lob et al., 2020; Los-Arcos et al., 2020). 1, 2, 3-Triazole can hybridize with other anticancer pharmacophores or act as a linker connecting two anticancer pharmacophores, which make it in the design and synthesis of antitumor compounds widely (Bozorov et al., 2019; Aouad et al., 2021; Liang et al., 2021).
Based on the above, we combined the advantages of theophylline and 1, 2, 3-triazole, hoping to develop a novel series of safe and efficient theophylline-containing 1, 2, 3-triazole ring derivatives for the treatment of NSCLC. We expect that this combination will improve the antitumor activity of such compounds and solve safety issues. For example, recent studies demonstrate that a novel series of benzimidazole derivatives have cell-cycle inhibition and apoptotic effects against a panel of selected human cancer cell lines (Atmaca et al., 2020; Atmaca et al., 2021). The structural modification of this series of compounds holds great potential that leads to the discovery of a series of novel antitumor chemical compounds which combine the advantages of the original molecule with the introduced additional functional groups.
Results and Discussion
Chemistry
The strategy for preparing target compound d is shown in Scheme 1. Compound 2 was obtained after reaction of theophylline acetic acid (compound 1) and 4-aminophenylacetylene. The target compounds d1–d29 were gained through click reaction of compound 2 with different azido compounds. The reaction conditions of these operations were gentle and easy to control. The structures of the key intermediates and all target compounds were confirmed by nuclear magnetic resonance (1H NMR and 13C NMR) and high-resolution mass spectrometry (HRMS) (in Supplementary Material).
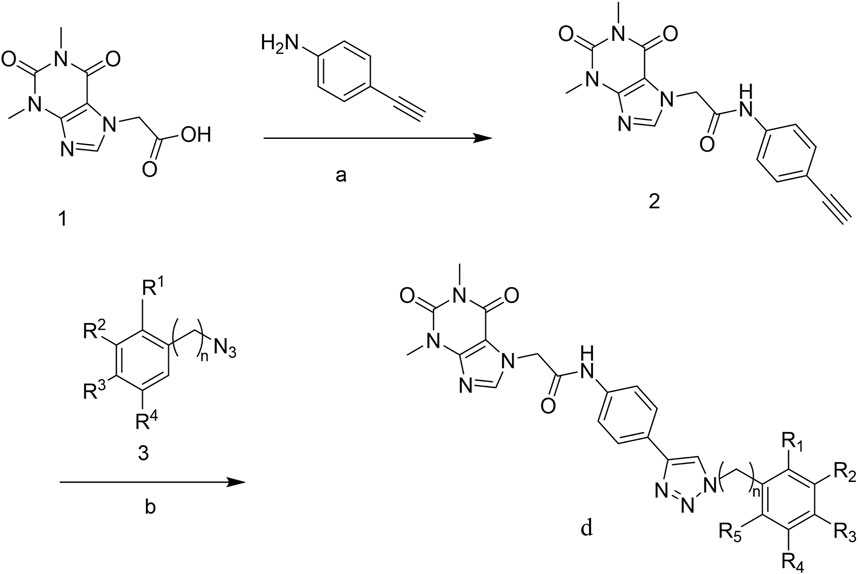
SCHEME 1. Reagents and conditions: (A) Theophylline acetic acid, 4-aminophenylacetylene, AHTU, and DIPEA were stirred in the DMF solvent 24 h at room temperature; (B) click reaction of copper sulfate water and sodium ascorbate in a solvent (tert-butanol: tetrahydrofuran: water = 1:1:1 at 85°C).
In Vitro Antitumor Activity Study
IC50 values were obtained from three independent experiments. These results are reported as the average ± SD.
Proliferative Activity of Nine Human Cancer Cell Lines Was Inhibited by Theophylline-1,2,3-Triazole Derivatives
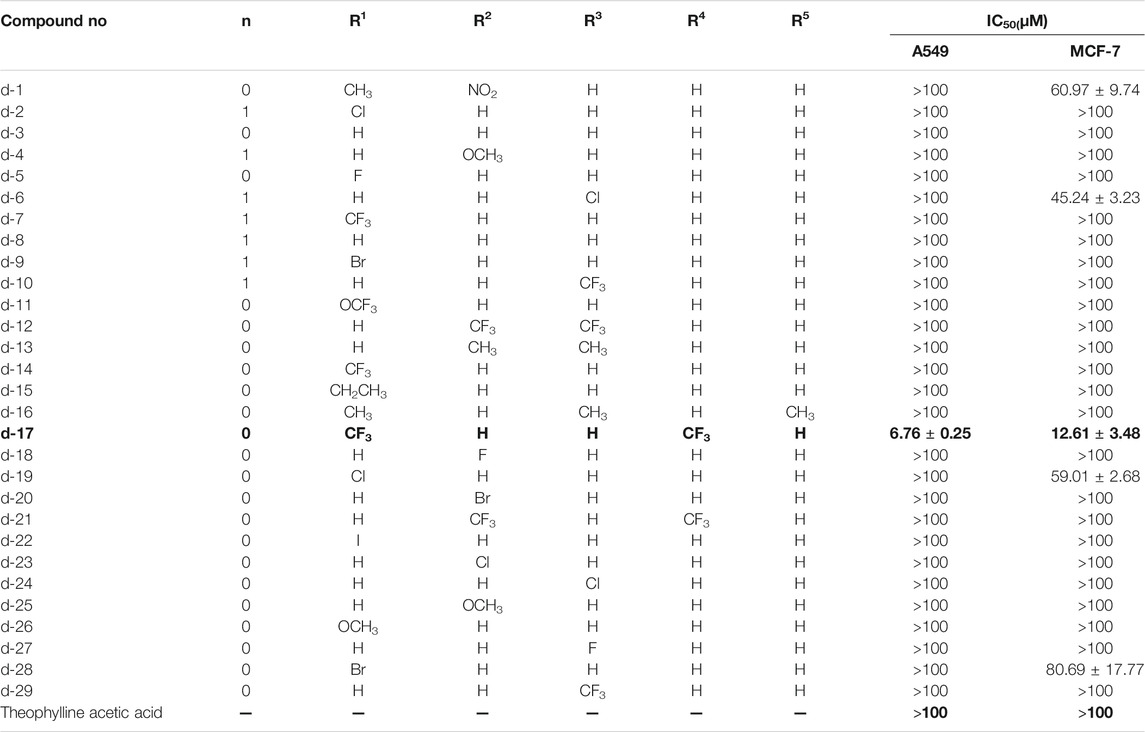
In order to screen out compounds with excellent antitumor activity from 31 theophylline acetic acid derivatives, we selected two tumor cells lines, A549 and MCF-7, as the treatment objects. The CCK8 assay was used to evaluate the effect of this series of theophylline acetic acid derivatives on A549 and MCF-7 proliferative activity. As shown in Table 1, both A549 and MCF-7 are not sensitive to theophylline acetic acid [half-maximal inhibitory concentration (IC50) >100 μM]. A549 is only sensitive to d17 (IC50 = 6.76 ± 0.25) but not sensitive to theophylline acetic acid and other theophylline-1, 2, 3-triazole derivatives. For MCF−7, d1 (IC50 = 60.97 ± 9.74), d6 (IC50 = 45.24 ± 3.23), d17 (IC50 = 12.61 ± 3.48), d19 (IC50 = 59.01 ± 2.68), and d28 (IC50 = 80.69 ± 17.77) are sensitive. Although the number of compounds sensitive to MCF-7 is more than A549, A549 has the best sensitivity to compound d17 (IC50 = 6.76 ± 0.25), and MCF−7 also shows moderate sensitivity to compound d17, so we chose compound d17 to carry out the study.
To confirm the antitumor activity of compound d17 and screen out the most sensitive cell line to compound d17, we added seven cell lines, H460, A2780, LOVO, MB-231, OVCAR3, SW480, and PC9, as treatment objects. As shown in Table 2, compound d17 showed strong antiproliferative and cytotoxicity to these nine cancer cell lines, H460 (IC50 = 5.93 ± 0.97 μM), A549 (IC50 = 6.76 ± 0.25 μM), A2780 (IC50 = 26.84 ± 6.96 μM), LOVO (IC50 = 37.42 ± 0.82 μM), MB-231 (IC50 = 18.78 ± 3.84 μM), MCF-7 (IC50 = 12.61 ± 1.76 μM), OVCAR3 (IC50 = 29.33 ± 6.20 μM), SW480 (IC50 = 15.66 ± 2.37 μM), and PC9 (IC50 = 18.20 ± 14.15 μM). Among these nine cell lines, H460 and A549 are the most sensitive cell lines to compound d17, with IC50 of 5.93 ± 0.97 μM Figure 2A and 8.926 μM (Figure 2B), respectively. In addition, we also measured the cytotoxicity of compound d17 to normal liver cells LO2 (Figure 2C), and the results showed that at an effective therapeutic concentration (8 μM), the cytotoxicity of d17 to normal liver cells was almost 0; when the compound concentration reached 16 μM, it had a little inhibitory effect on LO2.

TABLE 2. Antiproliferative activities of compounds d17 against nine human cancer cell lines and normal liver cell lines.
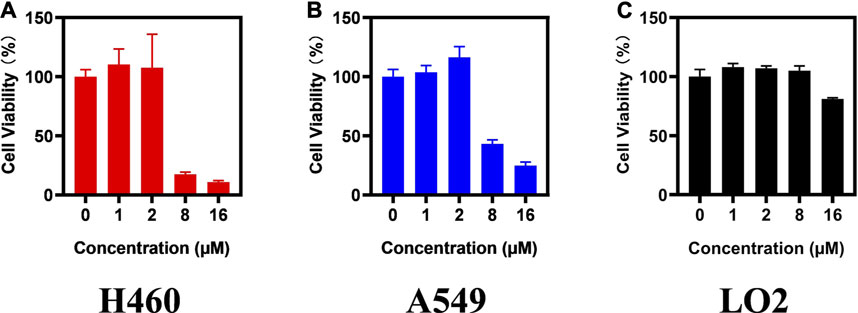
FIGURE 2. Compound d17 supresses H460 and A549 cancer cells. H460 (A), A549 (B), and LO2 (C) cells were exposed to compound d17 with indicated concentrations for 72 h, and cell viability was assessed by the CCK-8 assay, n = 3. *p-value < 0.05, **p-value < 0.01, and ***p-value < 0.001 (one-way ANOVA, followed by Tukey’s post-test).
To further evaluate the anti-NSCLC activity of compound d17, we used LIVE/DEAD staining. As shown in Figure 3, the number of dead cells increased as the concentration of compound d17 increased, which was consistent with the results of CCK8 determination. In short, these results indicate that compound d17 can effectively inhibit the proliferative activity of NSCLC and has little cytotoxicity to normal hepatocytes at effective therapeutic concentrations.
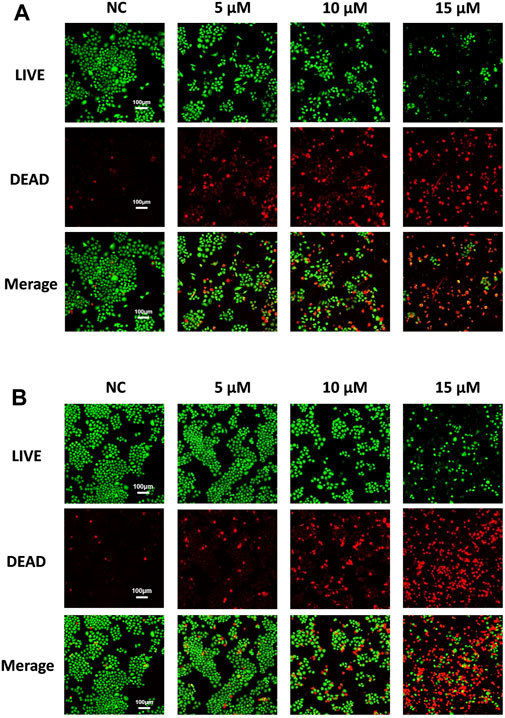
FIGURE 3. Compound d17 suppresses H460 and A549 cancer cells. Fluorescence images of (A) H460 and (B) A549 cells exposed to compound d17 with indicated concentrations for 48 h and then stained with the red/green kit; green indicates live cells, and red indicates dead cells.
Theophylline1, 2, 3-Triazole Derivatives Suppress NSCLC Cell Lines by Inducing Apoptosis
To clarify whether the antiproliferative effect is related to cell apoptosis, H460 and A549 cells were treated with different concentrations (5, 10, and 15 μM) of compound d17 for 48 h and then detected by flow cytometry. As shown in Figure 4, we observed significant apoptosis in H460 and A549 cells exposed to different concentrations of d17. The proportions of H460 apoptotic cells treated with compound d17 were 11.19% (5 μM), 24.89% (10 μM), and 40.09% (15 μM), while the proportions of A549 apoptotic cells treated with compound d17 were 8.55% (5 μM), 12.47% (10 μM), and 26.76% (15 μM). These results suggested that compound d17 considerably promoted the apoptosis of lung cancer cell lines H460 and A549 in a concentration-dependent manner.
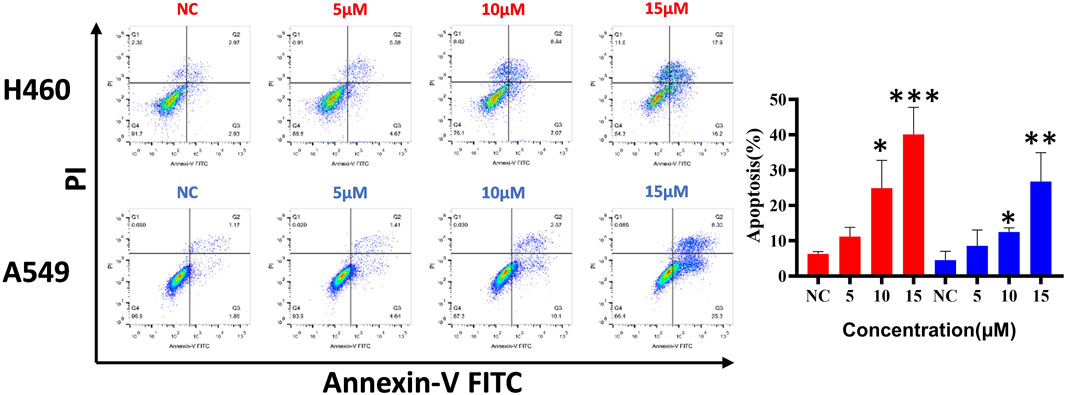
FIGURE 4. Compound d17 induced apoptosis of H460 and A549. Flow cytometry analysis data from three independent experiments were summarized and shown. NC, negative control. *p-value < 0.05, **p-value < 0.01, and ***p-value < 0.001 (one-way ANOVA, followed by Tukey’s post-test).
Theophylline1, 2, 3-Triazole Derivatives Trigger Apoptosis by Suppressing Phosphorylation of Akt Protein
In order to further explore the mechanism of d17-induced apoptosis in NSCLC, western blot was used to detect apoptosis-related markers Bax, Bcl-2 (Figure 5A), and Akt (Figure 5B). As shown in Figure 5, after H460 cells were treated with 0.1% DMSO as control or different concentrations of compound d17 for 24 h, total cell protein analysis showed that the p-Akt protein level in H460 cells was lower than that in the control group, and the ratio of p-Akt/Akt is also lower than that in the control group, and as the drug concentration increases, the ratio of p-Akt/Akt decreases. The levels of apoptosis inhibitor protein Bcl-2 and apoptosis marker protein Bax both decreased with the increase of drug concentration, but the ratio of Bax/Bcl-2 increased with the increase of drug concentration. Phosphorylated Akt protein can inhibit apoptosis by inhibiting the function of Bax protein, and various studies have reported that the overexpression of phosphorylated AKT (p-AKT) is a key defect in many types of solid tumors (Atmaca et al., 2017; Brown and Banerji, 2017; Shariati and Meric-Bernstam, 2019; Song et al., 2019; Iida et al., 2020). Compound d17 can inhibit the phosphorylation of Akt protein, which indicates that compound d17 can increase the ratio of apoptotic protein Bax/Bcl-2 and promotes NSCLC cell apoptosis by inhibiting the phosphorylation of Akt protein.
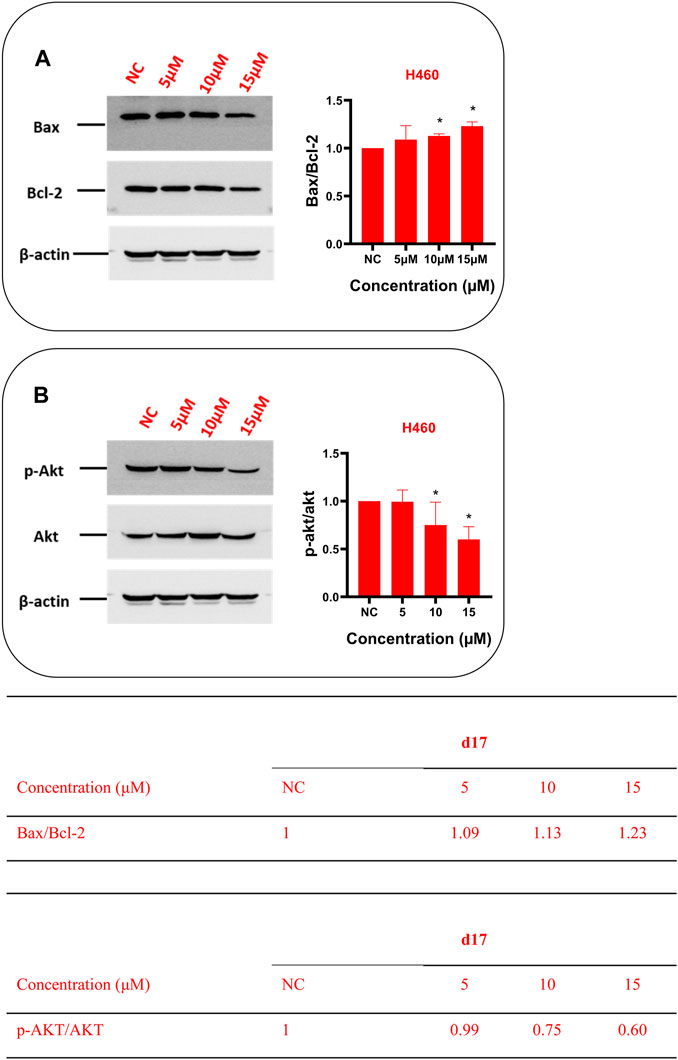
FIGURE 5. Compound d17 suppressed Akt phosphorylation and its transduction of downstream signaling Bax and Bcl-2 in NSCLC cells. Western blot was used to detect apoptosis-related markers Bax, Bcl-2 (A), and Akt (B). Protein bands (left images) and quantification (right images and tables below) are presented. NC, negative control. *p-value < 0.05, **p-value < 0.01, and ***p-value < 0.001 (one-way ANOVA, followed by Tukey’s post-test).
Conclusion
In a word, we designed and synthesized a series of theophylline derivatives containing the 1, 2, 3-triazole ring and evaluated their antiproliferative activity on nine kinds of cancer cells. Some of these compounds showed significant antitumor activity compared to theophylline acetic acid against one or more cancer cell lines used in this study. Among them, compound d17 showed strong antiproliferation and cytotoxicity to all nine kinds of cancer cells, and the two NSCLC, H460 and A549, show the most sensitivity to compound d17 particularly. We revealed the potential mechanism of d17-induce NSCLC cell death is that compound d17 through inhibiting Akt protein phosphorylation to induce mitochorylation appotosis. Current research shows that when appropriate substituents are introduced into the original molecule, the structural diversity of drugs can be expanded. Future research will focus on improving the anticancer activity and pharmacokinetic properties of these compounds.
Experimental Section
General Experimental Procedures
The theophylline acetic acid, 4-aminophenylacetylene, and azido compounds were purchased from Aladdin (CHINA). The RPMI-1640 medium, Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), trypsin, and phosphate-buffered saline (PBS) were purchased from Gibco (United States). The cell Counting Kit-8 (CCK-8) was purchased from Abmole (United States). An Annexin V/propidium iodide (PI) staining kit was purchased from BD Biosciences (United States). Akt, AKT1 (phospho S473), and the secondary antibodies of antirabbit and antimouse were purchased from Cell Signaling Technology, Inc. (United States). NSCLC cell lines PC-9, H460, and A549 and other cancer cell lines A2780, LOVO, MB-231, MCF-7, OVCAR3, and SW480 were obtained from ATCC.
Chemistry
The general procedures of preparation for erlotinib and compounds d1–d29 were described in the section of results. The structures of all target compounds were confirmed by nuclear magnetic resonance (1H NMR and 13C NMR) and high-resolution mass spectrometry (HRMS) as below.
Theophylline acetic acid [compound 1 (5 g, 0.02 mol)], 4-aminophenylacetylene (3.69 g, 0.0 3 mol), HATU (12.96 g, 0.03 mol), and DIPEA (8.13 g, 0.06 mol) were added together into a 500 ml reaction flask in DMF, stirring for 24 h at room temperature under nitrogen protection. The reaction process was monitored by thin-layer chromatography (TLC). After the reaction was completed, DMF was removed with an oil pump; dichloromethane was added and washed with saturated salt water; the organic phase was combined, dried with anhydrous sodium sulfate, and concentrated in vacuum to obtain solid compound 2.
Benzyl bromide and sodium azide were stirred in a solvent (acetone: water = 4:1) for 24 h at room temperature to produce benzyl azide 3 (n = 1). Aniline is added to the solvent (water:hydrochloric acid = 1:1) and stirred (below 5°), and then, sodium nitrite is dissolved in water, slowly dripping in the solvent (water:hydrochloric acid = 1:1). Finally, sodium azide is dissolved in water, slowly dripping in the solvent (water:hydrochloric acid = 1:1) too, reacting for 24 h to obtain phenyl azide 3 (n = 0).
The azide compound (1.2 mmol) and compound 2 (1.0 mmol) were added to 15 ml of a mixed solvent (tetrahydrofuran:water:tert-butanol = 1:1:1). Anhydrous copper sulfate (0.1 mmol) and sodium ascorbate (0.2 mmol) were added, and the mixture was stirred at 80°C for 8 h. Upon completion of the reaction (monitored by TLC), the mixture was extracted with dichloromethane (15 ml × 3). All the organic phases were continuously washed with water and brine, dried with anhydrous sodium sulfate, and concentrated in vacuum. The residue was purified by column chromatography (dichloromethane:methanol = 20∶1) to obtain the target compounds d1–d29 in the white powder form.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-methyl-3-nitrophenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d1). 1H NMR (400 MHz, DMSO-d6): δ 10.58 (s, 1H), 8.97 (s, 1H), 8.19 (d, J = 7.3, 1H), 8.09 (s, 1H), 7.94–7.89 (m, 3H), 7.74–7.69 (m, 3H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H), 2.24 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.50, 155.00, 151.49, 151.26, 148.44, 147.00, 144.26, 139.11, 138.17, 131.46, 128.74, 128.50, 126.51, 126.09, 125.80, 123.66, 119.89, 106.95, 49.25, 29.95, 27.94, 14.45. HR MS (ESI) m/z: calcd for C24H22 N9O5 [M + H]+ 516.1744, found 516.1741.
N-(4-(1-(2-chlorobenzyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d2). 1H NMR (400 MHz, DMSO) δ 10.53 (s, 1H), 8.53 (s, 1H), 8.08 (s, 1H), 7.82 (d, J = 8.3, 2H), 7.65 (d, J = 8.3, 2H), 7.54 (d, J = 7.6, 1H), 7.43–7.37 (m, 2H), 7.28 (d, J = 7.0, 1H), 5.75 (d, J = 7.5, 2H), 5.22 (s, 2H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.41, 154.98, 151.49, 148.43, 146.69, 144.25, 138.75, 133.64, 133.09, 130.99, 130.73, 130.10, 128.25, 126.36, 126.29, 121.85, 119.80, 106.93, 51.23, 49.22, 29.94, 27.92. HR MS (ESI) m/z: calcd for C24H22 ClN8O3 [M + H]+ 505.1503, found 505.1501.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-phenyl-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d3). 1H NMR (400 MHz, DMSO-d6) δ 10.58 (s, 1H), 9.23 (s, 1H), 8.09 (s, 1H), 7.93 (dd, J1 = 14.5, J2 = 8.2, 4H), 7.71 (d, J = 8.4, 2H), 7.64 (t, J = 7.7, 2H), 7.52 (t, J = 7.3, 1H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.50, 155.00, 151.50, 148.45, 147.54, 144.27, 139.05, 137.14, 129.16, 126.46, 126.01, 120.44, 119.90, 106.96, 49.26, 29.96, 27.95. HR MS (ESI) m/z: calcd for C23H21N8O3 [M + H]+ 457.1737, found 457.1737.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d4). 1H NMR (400 MHz, DMSO-d6) δ 10.53 (s, 1H), 8.55 (s, 1H), 8.08 (s, 1H), 7.81 (d, J = 8.3, 2H), 7.65 (d, J = 8.3, 2H), 7.30 (t, J = 7.8, 1H), 6.91 (dd, J1 = 16.6, J2 = 8.0, 3H), 5.60 (s, 2H), 5.22 (s, 2H), 3.75 (s, 3H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.40, 159.94, 154.98, 151.49, 148.43, 146.86, 144.25, 138.71, 137.89, 130.43, 126.46, 126.24, 121.51, 120.45, 119.81, 114.20, 113.95, 106.94, 55.59, 53.40, 29.94, 27.93. HR MS (ESI) m/z: calcd for C25H25N8O4 [M + H]+ 501.1999, found 501.2004.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-fluorophenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d5). 1H NMR (400 MHz, DMSO-d6) δ 10.59 (s, 1H), 9.01 (s, 1H), 8.10 (s, 1H), 7.92 (s, 3H), 7.67 (d, J = 31.5, 4H), 7.48 (s, 1H), 5.24 (s, 2H), 3.46 (d, J = 4.5, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 144.26, 139.10, 126.55, 126.46, 126.10, 119.88, 49.25, 40.40, 40.19, 29.95, 27.94. HR MS (ESI) m/z: calcd for C23H20FN8O3 [M + H]+ 475.1642, found 475.1651.
N-(4-(1-(4-chlorobenzyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d6). 1H NMR (400 MHz, DMSO-d6) δ 10.51 (s, 1H), 8.54 (s, 1H), 8.08 (s, 1H), 7.80 (d, J = 8.7, 2H), 7.64 (d, J = 8.7, 2H), 7.48–7.43 (m, 2H), 7.37 (d, J = 8.5, 2H), 5.64 (s, 2H), 5.22 (s, 2H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.40, 154.99, 151.50, 148.45, 146.92, 144.26, 138.74, 135.45, 133.34, 129.26, 126.26, 121.56, 119.84, 106.95, 52.67, 49.22, 29.94, 27.93. HR MS (ESI) m/z: calcd for C24H22ClN8O3 [M + H]+ 505.1503, found 505.1504.
2-(1,3-dimethyl-2-oxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-(trifluoromethyl)benzyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d7). 1H NMR (400 MHz, DMSO-d6) δ 10.55 (s, 1H), 8.55 (s, 1H), 8.08 (s, 1H), 7.82 (s, 3H), 7.65 (d, J = 44.0, 4H), 7.24 (d, J = 4.4, 1H), 5.83 (s, 2H), 5.23 (s, 2H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.42, 154.98, 148.43, 146.83, 144.26, 138.81, 133.73, 130.70, 129.39, 126.73, 126.67, 126.30, 122.09, 119.80, 106.94, 52.47, 50.16, 49.23, 39.99.29.95, 27.93, 7.64. HR MS (ESI) m/z: calcd for C25H22F3N8O3 [M + H]+ 539.1767, found 539.1766.
N-(4-(1-benzyl-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d8). 1H NMR (400 MHz, DMSO-d6) δ 10.53 (s, 1H), 8.56 (s, 1H), 8.09 (s, 1H), 7.80 (s, 2H), 7.68–7.62 (m, 2H), 7.42–7.31 (m, 5H), 5.64 (s, 2H), 5.22 (s, 2H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.41, 146.87, 144.25, 136.47, 129.26, 128.62, 128.36, 126.24, 121.52, 119.81, 53.48, 49.23, 40.16, 29.94, 27.93. HR MS (ESI) m/z: calcd for C24H23N8O3 [M + H]+ 471.1893, found 471.1903.
N-(4-(1-(2-bromobenzyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d9). 1H NMR (400 MHz, DMSO-d6) δ 10.51 (s, 1H), 8.51 (s, 1H), 8.08 (s, 1H), 7.82 (d, J = 8.6, 2H), 7.68 (dd, J1 = 21.2, J2 = 8.3, 3H), 7.45–7.40 (m, 1H), 7.33 (d, J = 16.8, 1H), 7.22 (d, J = 8.9, 1H), 5.72 (s, 2H), 5.22 (s, 2H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.40, 154.99, 151.50, 148.44, 146.70, 144.26, 138.76, 135.26, 133.38, 128.80, 126.39, 126.30, 123.34, 121.88, 119.84, 106.95, 53.57, 49.23, 29.94, 27.92. HR MS (ESI) m/z: calcd for C24H22BrN8O3 [M + H]+ 549.0998, found 549.1008.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(4-(trifluoromethyl)benzyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d10). 1H NMR (400 MHz, DMSO-d6) δ 10.53 (s, 1H), 8.60 (s, 1H), 8.08 (s, 1H), 7.79 (dd, J1 = 16.5, J1 = 8.4, 4H), 7.66 (d, J = 8.7, 2H), 7.54 (d, J = 8.1, 2H), 5.77 (s, 2H), 5.23 (s, 2H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.42, 154.99, 151.49, 148.44, 146.99, 144.26, 141.15, 138.79, 129.07, 126.35, 126.28, 126.22, 126.18, 121.81, 119.82, 106.94, 52.81, 49.23, 29.95, 27.93. HR MS (ESI) m/z: calcd for C25H22F3N8O3 [M + H]+ 539.1767, found 539.1776.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-(trifluoromethoxy)phenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide(d11). 1H NMR (400 MHz, DMSO-d6) δ 10.58 (s, 1H), 8.98 (s, 1H), 8.10 (s, 1H), 7.91 (t, J = 7.5, 3H), 7.76– 8 (m, 5H), 5.25 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.49, 155.00, 151.50, 148.45, 146.97, 144.26, 141.61, 139.11, 132.10, 130.20, 129.30, 128.02, 126.50, 125.76, 123.04, 119.94, 106.95, 49.25, 40.23, 29.94, 27.92. HR MS (ESI) m/z: calcd for C24H20F3N8O4 [M + H]+ 541.1560, found 541.1568.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(3-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d12). 1H NMR (400 MHz, DMSO-d6) δ 10.60 (s, 1H), 9.41 (s, 1H), 8.32 (s, 2H), 8.09 (s, 1H), 7.91 (d, J = 9.1, 4H), 7.73 (d, J = 8.4, 2H), 5.25 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.52, 155.00, 151.50, 148.45, 147.79, 144.26, 139.18, 137.58, 131.88, 126.48, 125.74, 124.28, 119.94, 116.98, 106.96, 49.26, 29.96, 27.94. HR MS (ESI) m/z: calcd for C24H20F3N8O3 [M + H]+ 525.1610, found 525.1623.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(m-tolyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d13). 1H NMR (400 MHz, DMSO-d6) δ 10.58 (s, 1H), 9.20 (s, 1H), 8.09 (s, 1H), 7.91 (d, J = 6.8, 2H), 7.79 (s, 1H), 7.72 (s, 3H), 7.50 (t, J = 6.5, 1H), 7.33 (s, 1H), 5.76 (s, 2H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.48, 154.99, 151.49, 148.44, 147.45, 144.25, 140.13, 139.01, 137.09, 130.18, 129.70, 126.41, 126.05, 120.82, 119.89, 117.49, 106.95, 55.37, 49.25, 29.94, 27.92, 21.42. HR MS (ESI) m/z: calcd for C24H23N8O3 [M + H]+ 471.1893, found 471.1906.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d14). 1H NMR (400 MHz, DMSO-d6) δ 10.59 (s, 1H), 9.28 (s, 1H), 8.09 (s, 1H), 7.87 (dd, J1 = 19.7, J2 = 7.2, 4H), 7.78–7.64 (m, 3H), 7.37 (s, 1H), 5.25 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 154.99, 151.50, 144.26, 132.41, 132.32, 126.47, 119.93, 119.72, 116.31, 55.36, 49.25, 40.21, 29.94, 27.92. HR MS (ESI) m/z: calcd for C24H20F3N8O3 [M + H]+ 525.1610, found 525.1619.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-ethylphenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d15). 1H NMR (400 MHz, DMSO-d6) δ 10.58 (s, 1H), 8.87 (s, 1H), 8.10 (s, 1H), 7.92 (d, J = 8.5, 2H), 7.71 (d, J = 8.6, 2H), 7.58 (s, 2H), 7.42 (s, 2H), 5.25 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H), 2.52 (s, 2H), 1.06 (t, J = 7.5, 3H). 13C NMR (100 MHz, DMSO-d6) δ 144.25, 130.69, 130.35, 127.45, 126.87, 126.40, 123.28, 119.83, 49.24, 29.94, 27.93, 24.27, 15.36. HR MS (ESI) m/z: calcd for C25H25N8O3 [M + H]+ 485.2050, found 485.2060.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-mesityl-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d16). 1H NMR (400 MHz, DMSO-d6) δ 10.57 (s, 1H), 8.72 (s, 1H), 8.09 (s, 1H), 7.90 (d, J = 8.5, 2H), 7.70 (d, J = 8.5, 2H), 7.12 (s, 2H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H), 2.34 (s, 3H), 1.94 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.45, 155.01, 151.50, 148.44, 144.28, 140.03, 138.88, 134.95, 126.37, 126.29, 123.33, 119.82, 49.23, 40.41, 29.95, 27.94, 17.36. HR MS (ESI) m/z: calcd for C26H27N8O3 [M + H]+ 499.2206, found 499.2216.
N-(4-(1-(2,5-bis(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d17). 1H NMR (400 MHz, DMSO-d6) δ 10.57 (s, 1H), 9.02 (s, 1H), 8.40 (s, 1H), 8.30 (q, J = 8.4, 2H), 8.10–8.07 (m, 1H), 7.91 (d, J = 7.4, 2H), 7.72 (d, J = 7.5, 2H), 5.24 (s, 2H), 3.49–3.46 (m, 3H), 3.23–3.20 (m, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.52, 155.01, 151.51, 148.45, 146.87, 144.27, 139.18, 129.79, 127.10, 126.52, 125.55, 119.93, 106.95, 49.25, 29.96, 27.94. HR MS (ESI) m/z: calcd for C25H19F6N8O3 [M + H]+ 593.1484, found 593.1491.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(3-fluorophenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d18). 1H NMR (400 MHz, DMSO-d6) δ 10.59 (s, 1H), 9.28 (s, 1H), 8.09 (s, 1H), 7.87 (d, J = 12.5, 4H), 7.72 (s, 3H), 7.37 (s, 1H), 5.25 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.52, 155.00, 151.50, 148.45, 147.65, 144.27, 139.15, 132.43, 132.34, 126.48, 125.78, 119.94, 119.74, 116.33, 115.75, 108.03, 107.76, 106.95, 49.25, 29.95, 27.93. HR MS (ESI) m/z: calcd for C23H20FN8O3 [M + H]+ 475.1642, found 475.1641.
N-(4-(1-(2-chlorophenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d19). 1H NMR (400 MHz, DMSO-d6) δ 10.58 (s, 1H), 8.97 (s, 1H), 8.09 (s, 1H), 7.91 (d, J = 8.4, 2H), 7.79 (dd, J = 15.4, 7.6, 2H), 7.71 (d, J = 8.4, 2H), 7.67–7.59 (m, 2H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 151.52, 144.27, 126.45, 122.85, 122.77, 119.94, 119.86, 117.37, 117.14, 49.25, 40.44, 29.95, 27.94. HR MS (ESI) m/z: calcd for C23H20ClN8O3 [M + H]+ 491.1347, found 491.1354.
N-(4-(1-(3-bromophenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d20). 1H NMR (400 MHz, DMSO-d6) δ 10.59 (s, 1H), 9.31 (s, 1H), 8.20 (s, 1H), 8.09 (d, J = 4.1, 1H), 8.00 (t, J = 5.5, 1H), 7.89 (t, J = 6.1, 2H), 7.72 (dd, J1 = 8.3, J2 = 3.8, 3H), 7.59 (s, 1H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.51, 154.99, 151.49, 148.44, 147.65, 144.25, 139.14, 138.23, 132.36, 131.83, 126.45, 125.79, 122.95, 119.93, 119.72, 119.34, 106.95, 49.25, 40.21, 29.95, 27.93. HR MS (ESI) m/z: calcd for C23H20BrN8O3 [M + H]+ 535.0842, found 535.0840.
N-(4-(1-(3,5-bis(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d21). 1H NMR (400 MHz, DMSO-d6) δ 10.60 (s, 1H), 9.55 (s, 1H), 8.67 (s, 2H), 8.28 (s, 1H), 8.09 (s, 1H), 7.90 (d, J = 7.4, 2H), 7.74 (d, J = 7.6, 2H), 5.25 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.54, 155.00, 151.50, 148.46, 147.99, 144.25, 139.31, 132.55, 132.21, 126.49, 125.48, 124.63, 120.95, 120.18, 119.99, 106.96, 55.34, 49.26, 29.93, 27.91. HR MS (ESI) m/z: calcd for C25H19F6N8O3 [M + H]+ 593.1484, found 593.1487.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-iodophenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d22). 1H NMR (400 MHz, DMSO-d6) δ 10.58 (s, 1H), 8.90 (s, 1H), 8.13–8.09 (m, 2H), 7.91 (d, J = 8.5, 2H), 7.71 (d, J = 8.6, 2H), 7.64 (d, J = 4.2, 2H), 7.39 (dt, J1 = 8.6, J2 = 4.5, 1H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.47, 155.00, 151.50, 148.45, 146.68, 144.27, 140.33, 140.23, 138.99, 132.46, 129.92, 128.50, 126.40, 126.03, 123.38, 119.88, 106.95, 96.39, 49.25, 49.07, 40.20, 29.96, 27.95. HR MS (ESI) m/z: calcd for C23H20IN8O3 [M + H]+ 583.0703, found 583.0704.
N-(4-(1-(3-chlorophenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d23). 1H NMR (400 MHz, DMSO-d6) δ 10.59 (s, 1H), 9.31 (s, 1H), 8.09 (d, J = 5.7, 2H), 7.97 (d, J = 8.2, 1H), 7.90 (d, J = 8.4, 2H), 7.69 (dd, J1 = 24.6, J2 = 8.2, 3H), 7.59 (d, J = 8.0, 1H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.51, 154.99, 151.50, 148.45, 147.66, 144.26, 139.15, 138.16, 134.71, 132.16, 128.93, 126.46, 125.78, 120.19, 119.93, 119.75, 118.98, 106.95, 49.25, 29.95, 27.94. HR MS (ESI) m/z: calcd for C23H20ClN8O3 [M + H]+ 491.1347, found 491.1348.
N-(4-(1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d24). 1H NMR (400 MHz, DMSO-d6) δ 10.56 (s, 1H), 9.25 (s, 1H), 8.09 (s, 1H), 7.98 (d, J = 8.8, 2H), 7.89 (d, J = 8.6, 2H), 7.71 (d, J = 8.8, 4H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.51, 155.00, 151.50, 148.45, 147.67, 144.27, 139.12, 135.92, 133.39, 130.40, 126.47, 125.84, 122.09, 119.91, 119.66, 106.95, 49.25, 29.96, 27.94. HR MS (ESI) m/z: calcd for C23H20ClN8O3 [M + H]+ 491.1347, found 491.1351.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(3-methoxyphenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d25). 1H NMR (400 MHz, DMSO-d6) δ 10.50 (s, 1H), 8.54 (s, 1H), 8.08 (s, 1H), 7.80 (d, J = 8.7, 2H), 7.64 (d, J = 8.7, 2H), 7.30 (t, J = 7.9, 1H), 6.91 (d, J = 23.8, 3H), 5.22 (s, 2H), 3.75 (s, 3H), 3.46 (s, 3H), 3.20 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.40, 154.99, 130.43, 126.25, 121.51, 120.46, 119.84, 114.22, 113.98, 55.60, 53.42, 40.24, 29.94. HR MS (ESI) m/z: calcd for C24H23N8O4 [M + H]+ 487.1842, found 487.1771.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d26). 1H NMR (400 MHz, DMSO-d6) δ 10.56 (s, 1H), 8.84 (s, 1H), 8.09 (s, 1H), 7.91 (d, J = 7.1, 2H), 7.74–7.64 (m, 3H), 7.55 (s, 1H), 7.34 (d, J = 7.8, 1H), 7.17 (s, 1H), 5.24 (s, 2H), 3.88 (s, 3H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 131.28, 126.41, 126.33, 126.23, 123.29, 121.33, 119.85, 113.46, 56.60, 49.24, 40.22, 29.93, 27.92. HR MS (ESI) m/z: calcd for C24H23N8O4 [M + H]+ 487.1842, found 487.1854.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(4-fluorophenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d27). 1H NMR (400 MHz, DMSO-d6) δ 10.56 (s, 1H), 9.20 (s, 1H), 8.09 (s, 1H), 7.99 (d, J = 12.4, 2H), 7.89 (d, J = 8.1, 2H), 7.71 (d, J = 8.2, 2H), 7.49 (t, J = 8.5, 2H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 151.52, 144.27, 126.45, 122.85, 122.77, 119.94, 119.86, 117.37, 117.14, 49.25, 40.44, 29.95, 27.94. HR MS (ESI) m/z: calcd for C23H20FN8O3 [M + H]+ 475.1642, found 475.1648.
N-(4-(1-(2-bromophenyl)-1H-1,2,3-triazol-4-yl)phenyl)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamide (d28). 1H NMR (400 MHz, DMSO-d6) δ 10.56 (s, 1H), 8.94 (s, 1H), 8.09 (s, 1H), 7.93 (d, J = 20.7, 3H), 7.77–7.56 (m, 5H), 5.24 (s, 2H), 3.47 (d, J = 3.0, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.47, 155.00, 151.50, 148.45, 146.66, 144.26, 139.02, 136.72, 134.13, 132.50, 129.46, 129.17, 126.44, 125.95, 123.52, 119.92, 119.38, 49.25, 40.23, 29.95, 27.93. HR MS (ESI) m/z: calcd for C23H20BrN8O3 [M + H]+ 535.0842, found 535.0848.
2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N-(4-(1-(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl)phenyl)acetamide (d29). 1H NMR (400 MHz, DMSO-d6) δ 10.57 (s, 1H), 9.38 (s, 1H), 8.21 (d, J = 7.7, 2H), 8.10–8.00 (m, 3H), 7.92 (d, J = 7.9, 2H), 7.72 (d, J = 7.8, 2H), 5.24 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 165.53, 155.00, 151.50, 148.45, 144.27, 139.89, 139.22, 127.79, 127.75, 126.53, 125.67,120.80, 119.93, 119.77, 106.96, 49.26, 29.96, 27.94. HR MS (ESI) m/z: calcd for C24H20F3N8O3 [M + H]+ 525.1610, found 25.1621.
Bioexperiment
Cell Culture and Treatment
Human non-small cell lung cancer cell lines PC-9, H460, and A549 were cultured with the RIPM-1640 complete medium containing 10% FBS and 1% penicillin–streptomycin at 37°C in a 5% CO2 humidification environment. Other tumor cell lines A2780, LOVO, MB-231, MCF-7, OVCAR-3, and SW480 were cultured with the DMEM complete medium containing 10% FBS and 1% penicillin–streptomycin at 37°C in 5% CO2 humidification environment too. All compounds were dissolved in DMSO to prepare 100 mM mother liquor and then used complete the medium to prepare different working concentrations.
Cell Counting Kit-8 (CCK-8) for Cell Proliferation and Cytotoxicity Assays
Cells in the logarithmic growth phase were seeded into 96-well plates (2000–4,000 cells/well). 24 h after cell implantation, the cells were treated with different concentrations of the compound (1, 2, 8, 16 μM) for 72 h, and 0.1% DMSO was used as a negative control. Finally, the CCK8 reagent was added and incubated for 1–4 h at 37°C. The absorbance of each well was detected at a 450 nm wavelength by a multifunctional microplate reader (Thermo Fisher Varioskan Luk). The cell survival rate of the negative control group was regarded as 100%, and the half-maximal inhibitory concentration (IC50) of the compounds was calculated by Graph Pad Prism 8.0 software.
Live/Dead Cell Imaging
LIVE/DEAD cell analysis was carried out using a laser confocal fluorescence microscope using the LIVE/DEAD kit. In brief, H460 and A549 (3 × 103–5 × 103 cells/well) cells were seeded in 96-well plates incubating for 24 h, and then, cells were treated with various concentrations of compound d17 (5, 10, 15 μM) for 48 h and 0.1% DMSO was used as a control. After various concentrations, compound d17 cells were stained with the LIVE/DEAD Cell Imaging Kit for 15–20 min and then observed and photographed using a fluorescence microscope (LSM880 with Fast Airyscan).
Flow Cytometry Detection for Cell Apoptosis
The cell apoptosis assay was carried out using the Annexin V/PI apoptosis kit and flow cytometry (BD LSRFortessaTM Flow Cytometer). Briefly, H460 and A549 cells in the logarithmic growth phase were seeded into 6-well plates (4.0×105∼6.0×105 cells/well). 24 h after cell implantation, the cells were treated with different concentrations of compound d17 (5, 10, 15 μM) for 48 h, and 0.1%DMSO was used as a negative control. All cells (including those in the supernatant) were collected after trypsin digestion and washed with PBS; then, the cells were gently resuspended with 100 μL Annexin V-FITC binding solution and then incubated with 2.5 μL Annexin V-FITC and 5 μL of propidium iodide (PI) staining solution in dark at room temperature for 20–30 min. Finally, cell apoptosis of each well was detected by flow cytometry. The percentage of apoptosis was analyzed by Flowjo software.
Western Blot Analysis
H460 and A549 cells in the logarithmic growth phase were seeded into 6-well plates (4.0 × 105∼6.0 × 105 cells/well). 24 h after cell implantation, the cells were treated with different concentrations of compound d17 (5, 10, 15 μM) for 24 h, and 0.1% DMSO was used as a negative control. The supernatant was discarded, and the cells were collected by trypsin digestion and washed once with PBS. Then, the cells were lysed on ice with 100 μL of RIPA lysis buffer containing protease and the phosphatase inhibitor for 30 min. Finally, the total protein extract was obtained by centrifugation at 12,000 RPM at 4 degrees for 10 min. The proteins were isolated by electrophoresis with 12.5% sodium dodecyl sulfate polyacrylamide gel. After electrophoresis, the proteins were transferred to the NC membrane and then sealed with 5% skim milk prepared by TBS-T [150 mM NaCl, 10 mM Tris (pH 7.4), and 0.1% Tween20] at room temperature for 1 h. After sealing, 1:1,000 diluted solution of anti-Bax (D2E11), anti-Bcl-2 (124), anti-Akt (PAN) (C67E7), anti-Akt1 (PhosphoS473) (EP2109Y), and the anti-β-actin (8H10D10) primary antibody was incubated overnight at 4°Cand then washed with TBS-T for 5 min (three times). Incubation was carried out with 1:2000 diluted solution of the antirabbit or antimouse secondary antibody for 1 h at room temperature, and finally, washing was carried out with TBS-T for 5 min (three times) to obtain protein strips through chemiluminescence. The protein expression level and proportion were quantitatively analyzed by ImageJ software.
Statistical Analysis
All values are presented as means ± SD. The significant differences are determined using GraphPad Prism 8 software. The significant differences between the two groups are confirmed using Student’s t-test. All experiments are considered to be statistically significant using one-way ANOVA, followed by Tukey’s post test (significant difference at p < 0.05).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author/s.
Author Contributions
MY, QL, and JxY conceived the study, designed the experiments, and supervised all research. LM synthesized all compounds. JhY, LM, LX, RZ, and YL carried out the experiments and analyzed the data.
Funding
This study was supported by the National Natural Science Foundation of China (NO. 81972488).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewersAny product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fphar.2021.753676/full#supplementary-material
References
Abou-Zied, H. A., Youssif, B. G. M., Mohamed, M. F. A., Hayallah, A. M., and Abdel-Aziz, M. (2019). EGFR inhibitors and apoptotic inducers: Design, synthesis, anticancer activity and docking studies of novel xanthine derivatives carrying chalcone moiety as hybrid molecules. Bioorg. Chem. 89, 102997. doi:10.1016/j.bioorg.2019.102997
Al-Blewi, F. F., Almehmadi, M. A., Aouad, M. R., Bardaweel, S. K., Sahu, P. K., Messali, M., et al. (2018). Design, synthesis, ADME prediction and pharmacological evaluation of novel benzimidazole-1,2,3-triazole-sulfonamide hybrids as antimicrobial and antiproliferative agents. Chem. Cent. J. 12 (1), 110. doi:10.1186/s13065-018-0479-1
Aouad, M. R., Khan, D. J. O., Said, M. A., Al-Kaff, N. S., Rezki, N., Ali, A. A., et al. (2021). Novel 1,2,3-Triazole Derivatives as Potential Inhibitors against Covid-19 Main Protease: Synthesis, Characterization, Molecular Docking and DFT Studies. Chemistry Select 6 (14), 3468–3486. doi:10.1002/slct.202100522
Atmaca, H., İlhan, S., Batır, M. B., Pulat, Ç. Ç., Güner, A., and Bektaş, H. (2020). Novel benzimidazole derivatives: Synthesis, in vitro cytotoxicity, apoptosis and cell cycle studies. Chem. Biol. Interact 327, 109163. doi:10.1016/j.cbi.2020.109163
Atmaca, H., Özkan, A. N., and Zora, M. (2017). Novel ferrocenyl pyrazoles inhibit breast cancer cell viability via induction of apoptosis and inhibition of PI3K/Akt and ERK1/2 signaling. Chem. Biol. Interact 263, 28–35. doi:10.1016/j.cbi.2016.12.010
Atmaca, H., Ilhan, S., Yilmaz, E. S., and Zora, M. (2021). 4‐Propargyl‐substituted 1 H ‐pyrroles induce apoptosis and autophagy via extracellular signal‐regulated signaling pathway in breast cancer. Arch. Pharm., e2100170. doi:10.1002/ardp.202100170
Bozorov, K., Zhao, J., and Aisa, H. A. (2019). 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview. Bioorg. Med. Chem. 27 (16), 3511–3531. doi:10.1016/j.bmc.2019.07.005
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492
Brown, J. S., and Banerji, U. (2017). Maximising the potential of AKT inhibitors as anti-cancer treatments. Pharmacol. Ther. 172, 101–115. doi:10.1016/j.pharmthera.2016.12.001
Chen, H.-J., Jiang, Y.-J., Zhang, Y.-Q., Jing, Q.-W., Liu, N., Wang, Y., et al. (2017). New triazole derivatives containing substituted 1,2,3-triazole side chains: Design, synthesis and antifungal activity. Chin. Chem. Lett. 28 (4), 913–918. doi:10.1016/j.cclet.2016.11.027
David Osarieme, E., Modupe, D. T., and Oluchukwu, O. P. (2019). The Anticancer Activity of Caffeine - A Review. Arch. Clin. Biomed. Res. 03, 05. doi:10.26502/acbr.50170077
Goldstraw, P., Ball, D., Jett, J. R., Le Chevalier, T., Lim, E., Nicholson, A. G., et al. (2011). Non-small-cell lung cancer. Lancet 378 (9804), 1727–1740. doi:10.1016/s0140-6736(10)62101-0
Hirsh, L., Dantes, A., Suh, B. S., Yoshida, Y., Hosokawa, K., Tajima, K., et al. (2004). Phosphodiesterase inhibitors as anti-cancer drugs. Biochem. Pharmacol. 68 (6), 981–988. doi:10.1016/j.bcp.2004.05.026
Iida, M., Harari, P. M., Wheeler, D. L., and Toulany, M. (2020). Targeting AKT/PKB to improve treatment outcomes for solid tumors. Mutat. Res. 819-820, 111690. doi:10.1016/j.mrfmmm.2020.111690
Karlowsky, J. A., Kazmierczak, K. M., Young, K., Motyl, M. R., and Sahm, D. F. (2020). In vitro activity of ceftolozane/tazobactam against phenotypically defined extended-spectrum β-lactamase (ESBL)-positive isolates of Escherichia coli and Klebsiella pneumoniae isolated from hospitalized patients (SMART 2016). Diagn. Microbiol. Infect. Dis. 96 (4), 114925. doi:10.1016/j.diagmicrobio.2019.114925
Lee, S. S., and Cheah, Y. K. (2019). The Interplay between MicroRNAs and Cellular Components of Tumour Microenvironment (TME) on Non-Small-cell Lung Cancer (NSCLC) Progression. J. Immunol. Res. 2019, 3046379. doi:10.1155/2019/3046379
Liang, T., Sun, X., Li, W., Hou, G., and Gao, F. (2021). 1,2,3-Triazole-Containing Compounds as Anti-Lung Cancer Agents: Current Developments, Mechanisms of Action, and Structure-Activity Relationship. Front. Pharmacol. 12, 661173. doi:10.3389/fphar.2021.661173
Liu, H., Song, J., Zhou, Y., Cao, L., Gong, Y., Wei, Y., et al. (2019). Methylxanthine derivatives promote autophagy in gastric cancer cells targeting PTEN. Anticancer Drugs 30 (4), 347–355. doi:10.1097/CAD.0000000000000724
Lob, S. H., Hoban, D. J., Young, K., Motyl, M. R., and Sahm, D. F. (2020). Activity of ceftolozane-tazobactam and comparators against Pseudomonas aeruginosa from patients in different risk strata - SMART United States 2016-2017. J. Glob. Antimicrob. Resist. 20, 209–213. doi:10.1016/j.jgar.2019.07.017
Los-Arcos, I., Burgos, J., Falcó, V., and Almirante, B. (2020). An overview of ceftolozane sulfate + tazobactam for treating hospital acquired pneumonia. Expert Opin. Pharmacother. 21 (9), 1005–1013. doi:10.1080/14656566.2020.1739269
Majeed, R., Sangwan, P. L., Chinthakindi, P. K., Khan, I., Dangroo, N. A., Thota, N., et al. (2013). Synthesis of 3-O-propargylated betulinic acid and its 1,2,3-triazoles as potential apoptotic agents. Eur. J. Med. Chem. 63, 782–792. doi:10.1016/j.ejmech.2013.03.028
Miller, K. D., Nogueira, L., Mariotto, A. B., Rowland, J. H., Yabroff, K. R., Alfano, C. M., et al. (2019). Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 69 (1), 363–385. doi:10.3322/caac.2155110.3322/caac.21565
Miller, K. D., Siegel, R. L., Lin, C. C., Mariotto, A. B., Kramer, J. L., Rowland, J. H., et al. (2016). Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 66 (4), 271–289. doi:10.3322/caac.21349
Misirlioglu, C. H., Demirkasimoglu, T., Kucukplakci, B., Sanri, E., and Altundag, K. (2007). Pentoxifylline and alpha-tocopherol in prevention of radiation-induced lung toxicity in patients with lung cancer. Med. Oncol. 24 (3), 308–311. doi:10.1007/s12032-007-0006-z
Motegi, T., Katayama, M., Uzuka, Y., and Okamura, Y. (2013). Evaluation of anticancer effects and enhanced doxorubicin cytotoxicity of xanthine derivatives using canine hemangiosarcoma cell lines. Res. Vet. Sci. 95 (2), 600–605. doi:10.1016/j.rvsc.2013.06.011
Röhrig, U. F., Majjigapu, S. R., Grosdidier, A., Bron, S., Stroobant, V., Pilotte, L., et al. (2012). Rational design of 4-aryl-1,2,3-triazoles for indoleamine 2,3-dioxygenase 1 inhibition. J. Med. Chem. 55 (11), 5270–5290. doi:10.1021/jm300260v
Sakly, R., Edziri, H., Askri, M., Knorr, M., Strohmann, C., and Mastouri, M. (2018). One-pot four-component domino strategy for the synthesis of novel spirooxindole-pyrrolidine/pyrrolizidine-linked 1,2,3-triazole conjugates via stereo- and regioselective [3+2] cycloaddition reactions: In vitro antibacterial and antifungal studies. Comptes Rendus Chim. 21 (1), 41–53. doi:10.1016/j.crci.2017.11.009
Shariati, M., and Meric-Bernstam, F. (2019). Targeting AKT for cancer therapy. Expert Opin. Investig. Drugs 28 (11), 977–988. doi:10.1080/13543784.2019.1676726
Song, M., Bode, A. M., Dong, Z., and Lee, M. H. (2019). AKT as a Therapeutic Target for Cancer. Cancer Res. 79 (6), 1019–1031. doi:10.1158/0008-5472.CAN-18-2738
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Travis, W. D., Brambilla, E., Noguchi, M., Nicholson, A. G., Geisinger, K. R., Yatabe, Y., et al. (2011). International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 6 (2), 244–285. doi:10.1097/JTO.0b013e318206a221
Vanaparthi, S., Bantu, R., Jain, N., Janardhan, S., and Nagarapu, L. (2020). Synthesis and anti-proliferative activity of a novel 1,2,3-triazole tethered chalcone acetamide derivatives. Bioorg. Med. Chem. Lett. 30 (16), 127304. doi:10.1016/j.bmcl.2020.127304
Xu, Z., Zhao, S. J., and Liu, Y. (2019). 1,2,3-Triazole-containing hybrids as potential anticancer agents: Current developments, action mechanisms and structure-activity relationships. Eur. J. Med. Chem. 183, 111700. doi:10.1016/j.ejmech.2019.111700
Yung-Lung Chang, Y-J. H., Ying, C., Wang3, Y., and Shih-Ming, H. (2017). Theophylline exhibits anti-cancer activity via suppressing SRSF3 in cervical and breast cancer cell lines. Oncotarget 8, 101461–101474. doi:10.18632/oncotarget.21464
Keywords: theophylline, 1,2,3-triazole, apoptosis, NSCLC, antitumor
Citation: Ye J, Mao L, Xie L, Zhang R, Liu Y, Peng L, Yang J, Li Q and Yuan M (2021) Discovery of a Series of Theophylline Derivatives Containing 1,2,3-Triazole for Treatment of Non-Small Cell Lung Cancer. Front. Pharmacol. 12:753676. doi: 10.3389/fphar.2021.753676
Received: 05 August 2021; Accepted: 10 September 2021;
Published: 26 October 2021.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyReviewed by:
Rana Jahanban-Esfahlan, Tabriz University of Medical Sciences, IranHarika Atmaca, Celal Bayar University, Turkey
Copyright © 2021 Ye, Mao, Xie, Zhang, Liu, Peng, Yang, Li and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miaomiao Yuan, eXVhbm1tMjAxOUAxNjMuY29t; Qingjiao Li, bGlxajIzQG1haWwuc3lzdS5lZHUuY24=; Jianxue Yang, RG9jeWp4MTk2OUAxMjYuY29t
†These authors have contributed equally to this work
 Jiahui Ye
Jiahui Ye Longfei Mao
Longfei Mao Luoyijun Xie1†
Luoyijun Xie1† Rongjun Zhang
Rongjun Zhang Qingjiao Li
Qingjiao Li Miaomiao Yuan
Miaomiao Yuan