- 1Department of Pharmacy, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Hubei Province Clinical Research Center for Precision Medicine for Critical Illness, Wuhan, China
- 3St. Boniface Hospital Research Centre, Winnipeg, MB, Canada
Cancer has become the second greatest cause of death worldwide. Although there are several different classes of anticancer drugs that are available in clinic, some tough issues like side-effects and low efficacy still need to dissolve. Therefore, there remains an urgent need to discover and develop more effective anticancer drugs. MicroRNAs (miRNAs) are a class of small endogenous non-coding RNAs that regulate gene expression by inhibiting mRNA translation or reducing the stability of mRNA. An abnormal miRNA expression profile was found to exist widely in cancer cell, which induces limitless replicative potential and evading apoptosis. MiRNAs function as oncogenes (oncomiRs) or tumor suppressors during tumor development and progression. It was shown that regulation of specific miRNA alterations using miRNA mimics or antagomirs can normalize the gene regulatory network and signaling pathways, and reverse the phenotypes in cancer cells. The miRNA hence provides an attractive target for anticancer drug development. In this review, we will summarize the latest publications on the role of miRNA in anticancer therapeutics and briefly describe the relationship between abnormal miRNAs and tumorigenesis. The potential of miRNA-based therapeutics for anticancer treatment has been critically discussed. And the current strategies in designing miRNA targeting therapeutics are described in detail. Finally, the current challenges and future perspectives of miRNA-based therapy are conferred.
Introduction
When researchers first discovered miRNAs in 1993, they did not realize the importance of these miRNAs. Because the first miRNAs gene Lin-4 found from the C. elegans that controls the timing development in its life cycle is considered to be unique in the C. elegans. (Lee et al., 1993). However, hundreds of miRNAs were found in different species including mammals by traditional cloning and bioinformatics methods, which attracted the attention of scientists in various fields, especially for let-7 (Reinhart et al., 2000). Up to present, a total of 38,589 miRNAs have been recorded in miRBase (v222018, www.mirbase.org), an online miRNA database. MiRNA is a single chain non-coding endogenous RNA with a length of around 22 nucleotides, which is a post-transcription regulatory factor. It plays an important regulatory role mainly by inhibiting mRNA translation or reducing the stability of mRNA. More than 90% of miRNA is in the region of encoding protein genes or introns of the gene, and few of them are in the exon region of the gene. miRNAs located in intergenic regions have independent promoter elements, while those located in coding protein genes can share specific promoter elements with host genes in addition to their own independent promoters (Bartel, 2009).
The biosynthesis pathway of miRNA in animal cells is a complex process starting from nucleus to cytoplasm (Figure 1). For most of miRNAs, a primitive miRNA (pri-miRNA) is first formed from DNA sequences, and then processed into pre-miRNA with hairpin structure by Drosha and DGCR8 enzymes. Under the action of transporters composed of Ran-GTP and Exportin-5, pre-miRNA is transported to the cytoplasm and forms a double stranded miRNA under the processing of Dicer enzyme. A single stranded RNA that is cleaved from the double stranded miRNA is transported and assembled into a protein complex composed of Argonaute to form a RNA induced silencing complex (RISC), which can recognize the target gene and play an inhibitory role (Peng and Croce, 2016; Lou et al., 2018). A single miRNA could regulate the mRNA of more than one target gene, and each target gene mRNA could also be regulated by multiple miRNAs. The miRNA binding to the target mainly plays a regulatory role by post transcriptional inhibition of mRNA translation, or through a cleavage or degradation of mRNA (Wu, 2020a).
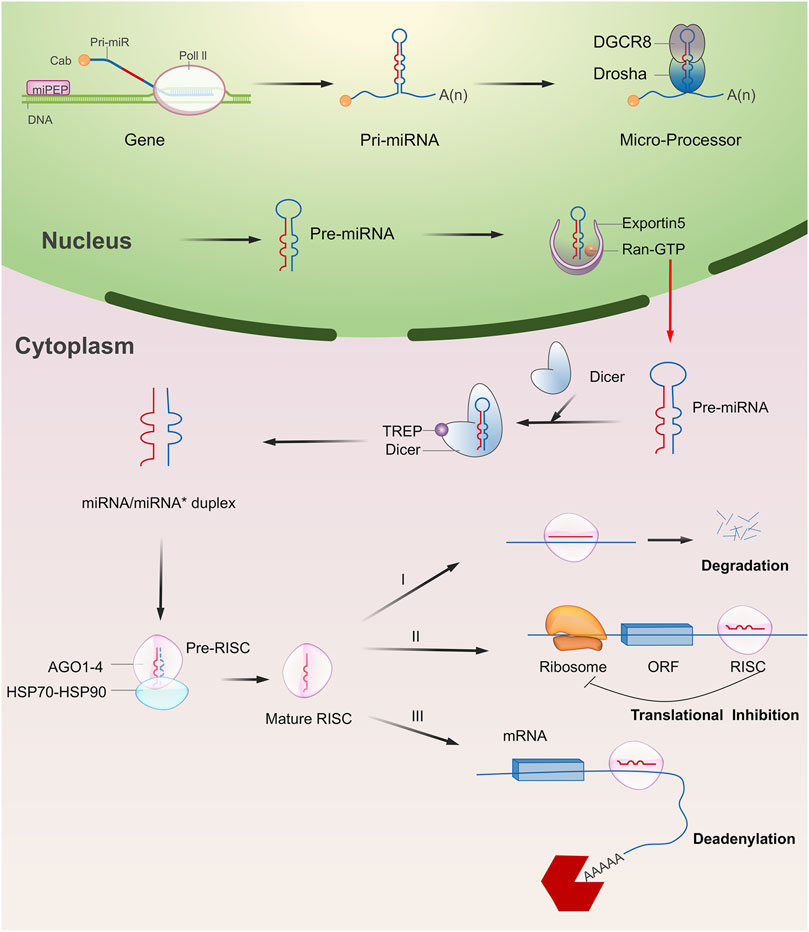
FIGURE 1. Overview of miRNA biogenesis and functions. Typically, miRNA begins with a transcription to generate the pri-miRNA. It was next processed into pre-miRNA with hairpin structure by Drosha and DGCR8 enzymes (microprocessor complex). The pre-miRNA is then exported to the cytoplasm and forms a double stranded miRNA under the processing of Dicer enzyme. A single stranded RNA that is cleaved from the double stranded miRNA is transported and assembled into a protein complex composed of Argonaute to form a RNA induced silencing complex (RISC). In most cases, RISC binds to target mRNAs to induce translational repression, deadenylation, and degradation.
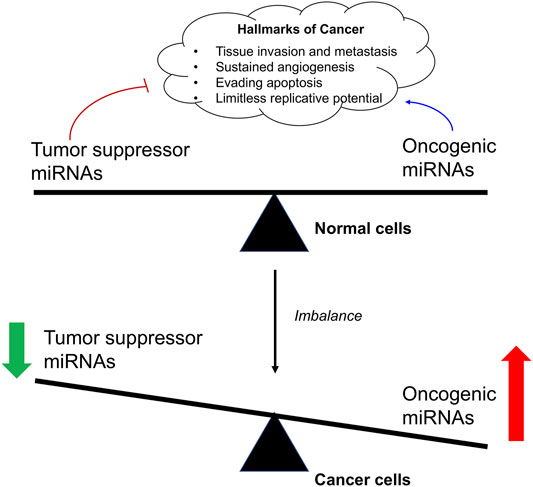
Although most of the miRNAs are located in the cytoplasm, some of them are located in the nucleus, directly regulating the transcription process at the DNA level (Pu et al., 2019). For example, miRNA-373 could up-regulate the expression level of E-cadherin after binding to the E-cadherin promoter (Wang et al., 2018a). MiRNA-mediated gene regulation is a basic post transcriptional regulatory pathway in human beings, which regulates 90% of protein-coding genes and participates in many cell biological processes (Makarova and Kramerov, 2007; Czimmerer et al., 2013). In cancer cells, mature miRNAs were found to play a crucial role in the cancer pathogenesis as an oncogenic or tumor suppressor agent because imbalance of miRNA regulation seem to be markedly associated to cancer cell proliferation, invasion, migration and metastasis, as well as apoptosis (He et al., 2020). The restoration of abnormal miRNA alterations in cancer cells using miRNA mimics or antagomirs could normalize the gene regulatory network and signaling pathways, and even reverse the phenotype (He et al., 2020; Otoukesh et al., 2020). Therefore, miRNA-based therapy provides a promising anti-cancer strategy for cancer therapy and miRNA could be also regarded as a good target for the development of anticancer drugs. In this review paper, we will summarize the latest research on the miRNA-based therapeutics for cancer and the development of anticancer drugs targeting miRNA.
MicroRNA and Cancers
Cancer is a genetic disease characterized by the uncontrollable cell proliferation and apoptosis with the tumor suppressor gene mutation (He et al., 2020). Since the emergence of recombinant DNA technology, to identify the underlying mutated genes that contributes to the development of a cancer has been the central goal of cancer research. For example, the transcription factors (myc genes), Src-family, epidermal growth factor receptor (EGFR), and Raf kinase have been performed extensive researches in cancer research (Elbadawy et al., 2019; Parkin et al., 2019). In recent years, emerging evidence has shown that miRNA is also closely related to the occurrence of various types of cancers (Song and Meltzer, 2012; Chen et al., 2015a; Takasaki, 2015; Gao et al., 2018). Studies have shown that a single miRNA may bind to up to 200 targets with different functions, including transcription factors, receptors and vectors. MiRNA may control about 30% of human mRNA expression involved in cell growth, differentiation and apoptosis (Figure 2). Moreover, the expression of some types of miRNAs is found to be significantly different between normal tissues and tumor tissues, suggesting its important role in tumor occurrence, development, invasion, and metastasis (Gaur et al., 2007; Lee et al., 2008).
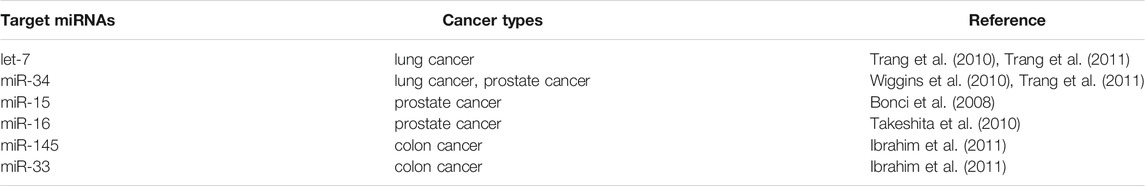
Based on their differential roles in the regulation of mRNA in cancer cells, miRNA are divided into oncogenic miRNAs, also known as oncomiRs, and tumor suppressor miRNAs (Table 1). For examples, the miR-15a and miR-16 were identified as the first tumor-suppressive miRNA, which negatively regulate bcl-2 (Cimmino et al., 2005). Bcl-2 is an anti-apoptotic gene, which is overexpressed in a variety of tumors, including leukemia and lymphoma (Warren et al., 2019). Therefore, the deletion or down-regulation of these two miRNAs leads to the increase of bcl-2 expression and promotes the occurrence of leukemia and lymphoma. Let-7, as one of the first identified miRNAs, is found to inhibit the expression of oncogene Ras (Johnson et al., 2005; Chirshev et al., 2019). About 15–30% of human tumors contain Ras mutation, it would cause cell transformation when the Ras mutation is activated with increased protein expression of bcl-2 (Kapoor et al., 2020). The analysis of 21 patients with different types of tumor showed that the expression of let-7 decreased significantly in twelve lung cancer patients, but only partially decreased in other tumor patients. In vitro tissue culture test also showed that the transient increase of let-7 in human lung cancer cells could inhibit the proliferation of cancer cells, which also indicated that let-7 might be a tumor suppressor gene in lung tissue (Takamizawa et al., 2004; Huang et al., 2020). It is also interesting to note that overall miRNA expression is lower in cancer cells than in normal cells. This suggests that a subgroup of miRNAs play a role in tumor inhibition, and the loss of their function may promote tumor occurrence. He et al. reported that three miRNAs encoded by miR-34 (miR-34 a-c) were found in human tumors by in vivo and in vitro tests, which were induced in p53 dependent cell cycle stress response or DNA damage (He et al., 2007). Among them, miR-34a is located on chromosome 1p36, and the deletion of this region could be observed in a variety of tumors (such as neurofibroma). The abnormal expression of these miRNAs down-regulates a variety of cell cycle regulatory genes, such as anti-apoptotic factor Bcl-2, resulting in cell cycle arrest and even apoptosis. Therefore, miR-34 plays an important role in the regulation of cell proliferation and apoptosis and may be used as a tumor suppressor gene.
Alternatively, oncogenic miRNAs that promote tumorigenesis with a highly expressed in tumor tissues have also been identified. The discovery of miR-17-92 provided the first functional evidence of such an oncomiR (Gruszka and Zakrzewska, 2018). In B-cell lymphoma and cell lines, the pri-miRNA and mature miRNAs of miR-17-92 were found to be significantly overexpressed (Mihailovich et al., 2015). In vivo examination confirmed that enhanced miR-17-92 expression would promote the formation of myc-induced B cell lymphoma and the incidence rate of lymphoma is faster and higher. Furthermore, the expression of miR-17-92 was found to be regulated by myc protein. Myc activates its expression by directly binding to miR-17-92 site on chromosome 13 to down-regulate the expression of transcription factor E2F1 protein, which could induce apoptosis. When a single member of miR-17-92 family was induced to co-express with c-myc, it did not promote the formation of tumor, indicating that the tumorigenic effect of miR-17 family may be caused by the interaction of all members of miR-17 family (Fuziwara and Kimura, 2015). All above suggest that miR-17-92 gene cluster is a potential human oncogene. Another oncomiR, miR-21, is found to be a significant up-regulation in several types of solid tumors, such as lung, breast, prostate, and malignant glioma, supporting its oncogenic role in cancer pathogenesis (Meq, 2013; Amani-Shalamzari et al., 2020; Shakeri et al., 2021). When miR-21 inhibitor was transfected into gastric cancer cell line HEK-293, the proliferation of cancer cells was inhibited, and the apoptosis of cancer cells was induced. It also increased the proportion of G1/S phase of cancer cells, which made the cells to be more sensitive to radiotherapy and chemotherapy (Zhang et al., 2011a). In animal models, when the nude mice were injected the MCF-7cells transfected with oligonucleotides complementary to miR-21, the tumor volume was significantly decreased as 50% compared with that of control groups, and the inhibition effect on tumor growth lasted for 2 weeks (Si et al., 2007). Some studies have also revealed that miR-483 family could significantly inhibit the expression of PDGFB and directly down regulate the proliferation, migration and other malignant behaviors of human umbilical vein endothelial like cells. It was also showed that miR-483 family was shown to down-regulate the phosphorylation of Akt protein in PI3K/Akt signaling pathway after negatively regulating PDGFB (Bertero et al., 2013).
From the above, the rise of miRNA research has made scientists gradually realize that miRNA plays an irreplaceable role in the complex molecular network of oncogenes and tumor suppressor genes. The expression of miRNAs in tumors could be reduced, deleted or increased, and its expression changes are related to gene deletion, translocation, amplification or virus infection. As miRNA is a regulatory molecule in the process of gene expression and protein translation, and it plays a pivotal role in the regulation of tumor occurrence. With the breakthrough of important theories on cancer pathogenesis and the solution of difficult problems in diagnosis and treatment, it will be possible to effectively control and treat cancer in extensive research in the future.
Potential of microRNA-Based Therapeutics for Anticancer Treatment
As the imbalance of miRNA expression level is associated with tumorigenesis, restoring miRNA function and inhibition of overexpressed miRNAs in cancer represent the two major approaches to develop miRNA-based cancer therapies (He et al., 2020) (Figure 3). Restoring miRNA function usually applied the miRNA mimics and some small molecules, which could enhance the function of endogenous miRNAs and restore the expression of tumor suppressive miRNAs, while inhibition of overexpressed miRNAs included the small molecule inhibitors, antagomiRs, and miRNA sponges, that specifically target oncomiRs which are overexpressed in cancer cells.
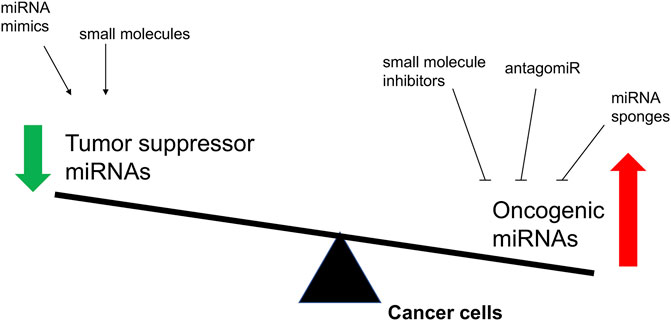
FIGURE 3. The overview of proposed strategies to regulate the biological activity of miRNAs involved in cancer. The objectives of these molecular interventions are the downregulation of oncomiRNAs or the restoring of tumor suppressor miRNAs.
Restoring miRNA Function in Cancer
Since those miRNAs play the role as tumor suppressors, restoration of reduced tumor suppressor miRNA function to normal level by miRNA mimics represents a promising cancer treatment strategy (To et al., 2020). Indeed, the gene therapy methods were applied to restore the gene function in cancer cells before the discovery of miRNAs. However, it achieved limited success because of the limitations of DNA plasmids and viral vectors (Roma-Rodrigues et al., 2020). In recent decades, the rapid development of miRNA technology has provided an alternative tools. The size of miRNA are considerably small than that of protein so that it could permeate into cell easily by some techniques. Esquela-Kerscher et al. demonstrated that a restoration of the miRNA let-7 could observably inhibit the tumor growth on mice models (Esquela-Kerscher et al., 2008). It was the first time for this study to confirm the tumor suppressor role of let-7 and its potential to use as the targets for cancer therapy. Since then, the therapy of restoring miRNA function by miRNA mimics has rapidly gained interest. The miRNA mimics used for restoring miRNA function are summarized in Table 2. Among them, the let-7 and miR-34 families were two members to be the most studied.
Studies have shown that let-7 family miRNAs inhibit well-known oncogenes, like NIRF, myc, HMGA, STAT3, and Ras, and the low expression of let-7 is found to be associated with poor prognosis in lung adenocarcinoma (Esquela-Kerscher et al., 2008). Hence, recovery of its activity may be a feasible strategy for lung cancer therapeutic. On the mouse lung cancer models, the effect of let-7 was examined using different delivery methods. The results showed that the let-7 mimics could reduce tumor growth, induce necrosis and de-repression of the direct let-7 targets Ras and CDK6 (Trang et al., 2010). As for miR-34 family, it was shown to be transcriptionally regulated by the tumor suppressor p53, and depletion or down-regulation of miR-34 has been found in several cancers (Hermeking, 2010). Through targeting CD44, miR-34a was shown to inhibit metastasis and reduce the growth of tumor (Wiggins et al., 2010). Similar with the supplement by let-7 mimics, when treated with the lipid-complexed miR-34a mimic on the mouse lung cancer models, the tumor volumes were significantly inhibited with a well-tolerated dose range (Wiggins et al., 2010). In the myc-driven liver cancer cells, the expression level of miR-26a was lower than in the normal health cells (Ji et al., 2009). With the replacement of miR-26a mimics to increase miR-26a levels, it would induce the cell cycle arrest through the inhibition of cyclin D2 and E2 (Kota et al., 2009). In vivo animals models, the tumor volumes were shown a sensitive response to the administration of miR-26a (Kota et al., 2009). All these above suggest that restoring miRNA function by miRNA mimics could provide a promising strategy for the cancer therapeutics.
Inhibition of Overexpressed miRNAs in Cancer
As for those overexpressed oncomiRs in cancer cells, the suppression of oncomiRs has been widely studied for the development of novel miRNA-based therapeutics (Table 3). The main inhibitors of miRNA included the small molecule inhibitors and the complementary oligonucleotides, such as anti-miRNA oligonucleotide (AMOS), locked nuclear acid (LNA)-AMOS, antagomirs and miRNA sponge. AMOS is the first miRNA inhibitor based on the principle of complementary with target miRNA sequence to neutralize the function of miRNA (Weiler et al., 2006). AMOS, in the form of a short DNA oligonucleotide strand, specifically combines with complementary endogenous miRNA or its precursor molecules to form stable DNA:RNA, which makes the endogenous miRNA occupied by AMO instead of binding to its target mRNA. miRNA is thus degraded by nuclease to achieve the effect of inhibiting miRNA (Garbett et al., 2010). A series of modified AMOS, such as 2′-O-methyl AMOS, 2′-O-methoxyethyl AMOS and LNA-AMOS, have emerged in the follow-up studies. LNA-AMOS are modified on the structure of AMOS. In detail, it forms a rigid ring through a connection of methylene at the positions of the 2′-oxygen and 4′-position of multiple nucleotides, which is embedded in the C3 position of sugar group. LNA-AMOS are more stable than AMOS, and it has higher selectivity and sensitivity (Weiler et al., 2006). Similar to antagomirs, miRNA sponges could be applied to inhibit miRNA functions by preventing stable binding to their targets. Instead of short oligonucleotide strands, these agents are longer nucleic acids, usually DNA plasmids or transcribed RNA, with several miRNA binding motives.
For example, miR-10b is associated with the metastatic properties with a overexpression in breast and esophagus tumors. Through the inhibition of miR-10b, treatment of mice with the cholesterol-conjugated antagomir-10b resulted in significantly reduced levels of tumor volumes compared with the vehicle group. It is well known that tumor angiogenesis significantly enhances the invasion and metastasis of tumor cells (De Ieso and Yool, 2018; Wang et al., 2018b). With the development of technology for miRNA research, the relationship between miRNA and tumor angiogenesis becomes more evidently. High expression of miRNA-132 in the endothelium of human tumors could promote pathological angiogenesis (Zhang et al., 2011b). By reducing the expression of miRNA-132, 2′-O-Me modified antimiR-132 was demonstrated a significant decrease in tumor burden and angiogenesis, indicating the potential for miR-132 as a target in anti-angiogenesis therapeutics. In glioma, miR-9 was found an up-regulation compared with normal cells and could promote the tumorigenesis and angiogenesis. It suggested that miR-9 is crucial for glioma pathogenesis and can be treated as a potential therapeutic target for glioma.
MiRNA as an Important Target for the Development of Anticancer Drugs
The investigation of drug targets has always been one of the most important contents in the research and development of new drugs. As a molecule widely involved in the regulation of gene expression, miRNA is undoubtedly an important object in the research of drug targets (Schmidt, 2014). Traditional drugs are mainly small chemical molecules targeting single protein in cancer which have certain limitations in clinical use. In contrast, miRNA has the natural characteristics of regulating multiple target genes, and is at the center of the multi-target regulatory network (Wang, 2011a). Moreover, the generation of miRNA is strictly regulated by signal pathway, which involves different important enzymes. MiRNA has such a complex and fine regulatory mechanism that the whole signal pathway has become a promising therapeutic drug or drug therapeutic targets in cancer (Vishnoi and Rani, 2017). Furthermore, miRNA-based drugs target molecules that cannot be target molecules that cannot be targeted by chemical drugs or antibody drugs, which is expected to make a breakthrough in diseases with poor efficacy of traditional drugs, especially cancer (Yu et al., 2020). Therefore, as drug targets, multi-target regulatory molecular miRNA has attracted more and more attention in the research and development of new drugs used in clinic.
Restoring miRNA by Small Molecules
The down-regulated expression of miRNA can be restored by some small molecular compounds, such as the hypomethylating agents (Galm et al., 2006). Decitabine or 5-azacytidine are two drugs for the treatment of myelodysplastic and they were found to increase the expression of several miRNAs (Lujambio et al., 2007). In addition, enoxacin was also demonstrated to promote the biosynthesis of several miRNAs. In the cell-cultured tests, an overall upregulation of miRNA expression was induced by the treatment of enoxacin (Melo et al., 2011). Moreover, enoxacin reduced the tumor growth by the upregulated expression of 24 mature miRNAs on the mice xenograft models (Melo et al., 2011). All these examples suggested the feasible role of restoring miRNA by small molecules for the anticancer therapeutics.
Restoring miRNA by Oligonucleotides
Another more specific approach for restoring miRNA is miRNA mimics. MiRNA mimics are chemically synthesized double stranded RNA molecules which regulate the function of miRNA by a simulation of endogenous miRNAs (Wang, 2011b). Because of the unstable status of miRNA mimics in the biological system, the core obstacle of the application is to develop an effective delivery system, like the nanoparticles, lipid emulsions, atelocollagen formulations, and adeno-associated virus. It was shown that the target delivery of miR-34a and let-7 mimics using the lipid emulsions could significantly inhibit the cancer progression on a colon xenograft mouse model (Trang et al., 2011). Using the adeno-associated virus as the carrier, the administration of miR-26a was demonstrated an inhibition of cancer cell proliferation and reducing tumor volume (Kota et al., 2009). Importantly, the strategy for restoring miRNA by the liposome-formulated miR-34 mimic (MRX34) has been developed to the clinical trials for the patients with liver cancer (Bader, 2012). The detailed information of MRX34 would be discussed in the subsequent sections.
Inhibiting miRNA by Oligonucleotides
Recent decades, the most frequently used approaches to block the function of miRNA are belongs to antisense oligonucleotides (ASO) and miRNA sponges (Roberts et al., 2020). The former includes the locked nucleic acids (LNAs) and antagomirs. LNA is a synthetic nucleic acid analogue containing bridged, bicyclic sugar moiety As a novel nucleotide derivative, it has attracted extensive attention in the field of pharmaceutical research and is expected to become a new breakthrough in the treatment of various diseases at the molecular level (Papargyri et al., 2020). It is a special bicyclic nucleotide derivative with one or more 2′-O, 4′-C-methylene in its structure-β-D-furan ribonucleic acid monomer, the 2′-O position and 4′-C position of ribose form oxygen methylene bridge, sulfur methylene bridge or amine methylene bridge through different shrinkage which are connected into a ring. This ring bridge locks the N configuration of furan sugar C3′- endotype and reduces the flexibility of ribose structure. Since LNA and DNA/RNA have the same phosphate skeleton in structure, it has good recognition ability and strong affinity for DNA and RNA. A higher expression of miR-21 was associated with the cancer initiation and progression of melanoma (Javanmard et al., 2020). In vitro studies using B16F10 cell line, a significant reduction was found in the number of transfected cells with LNA-anti-miR-21 and the transfected cells were shown an observable apoptosis. Moreover, the treatment of anti-miR-21 could inhibit the tumor growth in the xenograft mouse models (Javanmard et al., 2020).
Antagomir is a single stranded small RNA designed according to the mature sequence of microRNA and specially labeled and chemically modified (Scherr et al., 2007). It is an efficient blocker specially used to inhibit endogenous microRNA. Antagomirs often use thiophosphate instead of phosphate to covalently bind with cholesterol at the 3′-end of oligomer to prevent the complementary matching between miRNA and its target gene mRNA by competitive binding with mature miRNA in vivo, and inhibit the function of miRNA (Scherr et al., 2007). It has higher stability and inhibition effect in vivo and in vitro. Arefeh Kardani et al. reported that an inhibition of miR-155 in MCF-7 breast cancer cell lines was induced by the treatmen of gold nanoparticles functionalized with antagomir-155 (Kardani et al., 2020). In another study, the inhibition of miR-194 by antagomir-194 significantly reduced the proliferation of MCF-7 and MDA-MB-231 breast cancer cells (Chen et al., 2018).
As for miRNA sponge, it is another effective inhibitor of miRNA. It contains multiple miRNA binding sites (RBS) and can adsorb corresponding miRNA molecules like a sponge. After adsorption, miRNA cannot bind to its target molecules, which affects the function of miRNA (Kluiver et al., 2012). At present, it is found that the molecules that can act as miRNA sponge include long non coding RNA (lncrna) and circular RNA (circular RNA, circrna), these two RNAs can bind miRNA or compete for miRNA target molecules and play a negative regulatory role in miRNA. For example, Shu et al. developed a system to express circular inhibitors of miRNA targeting miR-223 and miR-21 as a sponge. It was shown a more potent suppression of miRNA functions than their linear counterparts for the inhibition of cancer cell growth (Shu et al., 2018).
Small Molecule Inhibitors of microRNA
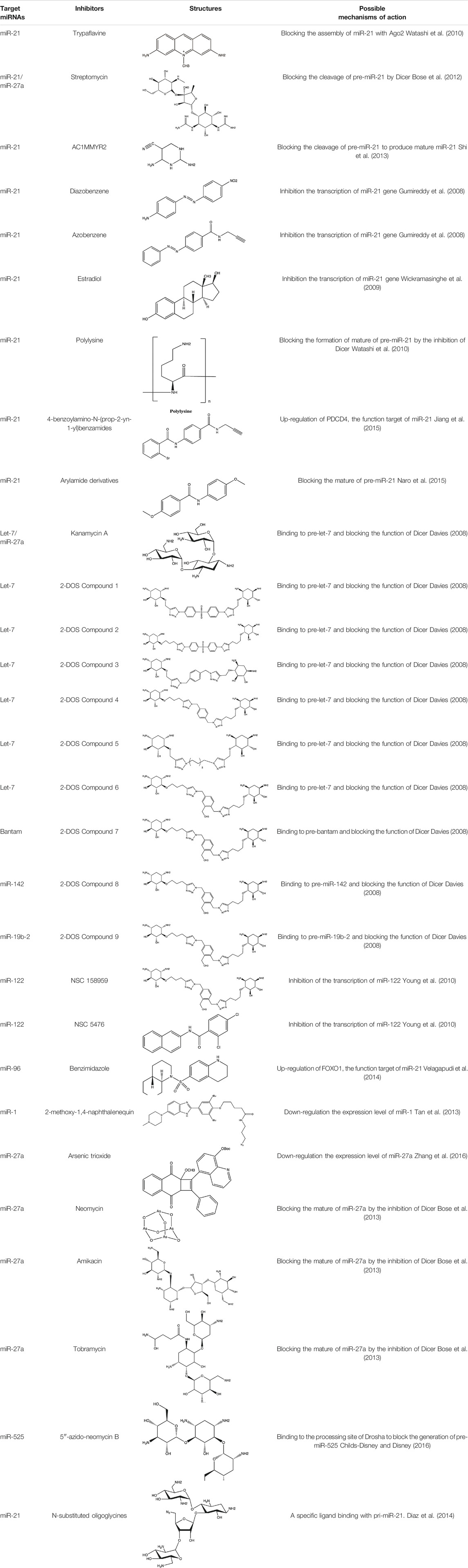
Since the biggest challenge for ASO applied in clinic is the poor cell-permeability for drug delivery, the recent trend is moving toward to the development of small-molecule drugs in the regulation of miRNA. Small molecules could cross the cell membrane by free diffusion, they modulate the function of miRNA like a microRNA mimic. Furthermore, small molecule inhibitors of microRNA are chemical compounds and thus traditional drug development can be applied (Wu, 2020b). At present, there are many kinds of miRNA specific chemical small molecule inhibitors, and their mechanisms are different. The target sites of inhibition and interference are throughout the whole process of miRNA gene expression, processing, maturation and function (Figure 4). The chemical structures of representative small molecule inhibitors were listed in Table 4.
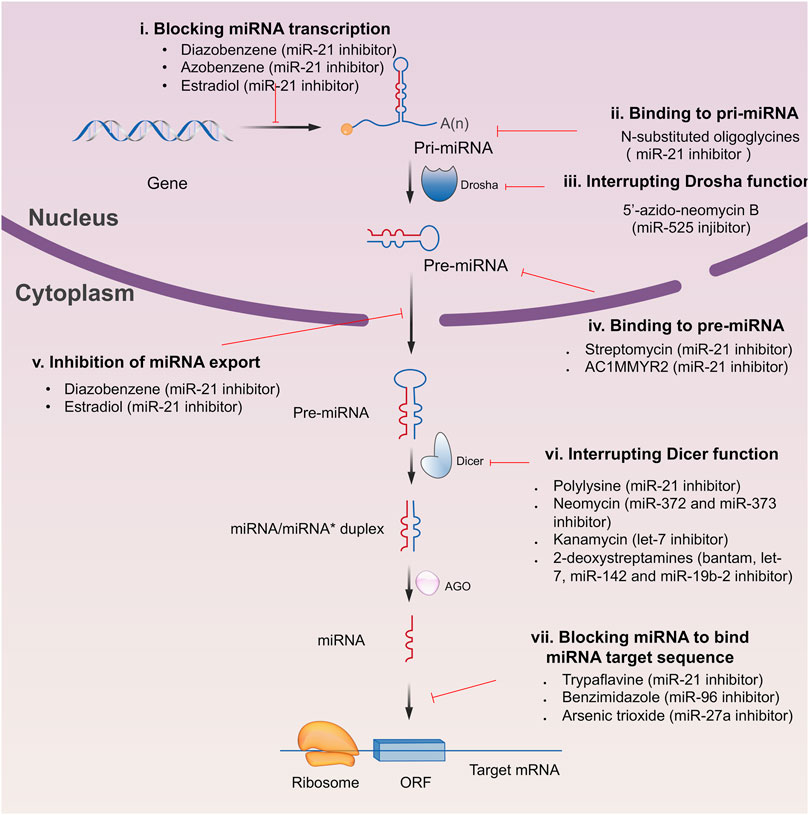
FIGURE 4. Schematic illustration of different inhibition mechanisms of miRNA specific small molecule inhibitors.
For example, miR-21 is one of the tumor-associated microRNAs (oncomiR) that was discovered earlier and recognized. It has been confirmed in a variety of tumor cells, including breast cancer, ovarian cancer, colon cancer, pancreatic cancer, thyroid cancer, etc. There is high expression in cancer which are closely related to the occurrence and development of tumor (Volinia et al., 2006; Tetzlaff et al., 2007; Asangani et al., 2008; Pan et al., 2010). At present, there are abundant literatures on the chemical small molecule inhibitors. Trypaflavine (TPF), a small molecule compound, was reported by Jiang Research Group in 2010, which significantly down-regulated the expression level of miR-21 (Watashi et al., 2010). Further experiments showed that TPF could inhibit the formation of RISC by blocking the assembly of miR-21 and Argonaute 2 (ago2) protein, leading to the down-regulation of the expression level of miR-21. Davies et al. found that kanamycin A could inhibit the expression of let-7 by binding to pre-let-7 and interfering with Dicer (Davies and Arenz, 2006). In addition, experiments showed that the inhibition rate reached 69 ± 3% after 2 h with the treatment of 100 μM of kanamycin A.
MIR-122 is a liver specific miRNA, which is highly expressed in the liver, accounting for about 72% of the total miRNA in adult liver. It is one of the earliest miRNAs with tissue-specific and high abundance expression (Girard et al., 2008). At present, it has been found that miR-122 plays an important role in regulating liver physiological functions such as the growth cycle of hepatocytes and fat metabolism (Esau et al., 2006). It also plays a key role in the occurrence and development of liver diseases such as acute and chronic liver injury, liver cirrhosis, alcoholic hepatitis, liver tumor and hepatitis C virus (HCV) infection (Jopling et al., 2005). Deiters’s group performed the research work on the discovery of small-molecule inhibitors of miR-122 and they successfully obtained two small-molecule inhibitors (NSC 158959 and NSC 5476) with specificity towards miR-122 (Young et al., 2010). Its target may be resulted from the transcription of miR-122 gene to pri-miR-122. As for miR-1, it is highly expressed in skeletal muscle cells, which has been proved to regulate the formation of skeletal muscle cells and the development of muscle and is closely related to the development of heart (Thum et al., 2008; Wystub et al., 2013). Using 4-naphthalenequinone as the basic skeleton, dozens of derivatives were obtained by photocyclization reaction (Tan et al., 2013). The 2-methoxy-1,4-naphthalenequin, which exerted specific inhibitory effect on miR-1, was screened out from these compounds. It was confirmed that 2-methoxy-1,4-naphthalenequin could significantly down-regulate the expression level of mature miR-1 in cells. However, the specific mechanism of this compound remains to be further studied.
Clinical Research Progress of miRNA-Related Drugs
In recent 20 years, the field of miRNA has made rapid progress and made great achievements in all directions. The three main discoverers, Victor Ambros, Gary Ruvkun and David Baulcombe won the Lasker Basic Medicine Award in 2008 and became hot candidates for the Nobel Prize in physiology and medicine for many years. At present, cooperated with academic laboratories, several pharmaceutical companies were launched miRNA clinical researches for anticancer therapeutics.
MRX34, a liposomal injection of miRNA-34a, developed by Mirnarx Therapeutics, Inc., entered the Phase I clinical trials in 2013 to evaluate the safety in patients with primary liver cancer or other selected solid tumors or hematologic malignancies (ClinicalTrials.gov, NCT number: 01829971) (Zhou et al., 2019). miRNA-34a is generally downregulated and acted as tumor suppressor in most of cancer cells by affecting more than 20 oncogenes to induce the cell apoptosis and cell cycle arrest (Welch et al., 2007). It has been shown that an increase of miRNA-34a in cancer cells would significantly inhibit the cell proliferation, suggesting a potential therapeutic strategy for cancer treatment (Yu et al., 2014). The AODNS is not permeable to enter cell by diffusion, a liposomal nanoparticle was thus employed the to carry the miRNA-34a for cancer treatment. The MRX34 is given intravenously for 5 days in a row and then 2 weeks off (total of 21 days). The trails was finally terminated as the occurrence of five immune related serious adverse events involving death of patients (Beg et al., 2017). However, the development of MRX34 indicated a feasible approach in miRNA drug discovery by using advanced formulations such as liposomal nanoparticles to increase the permeability of AODNS into the cell.
In 2014, the TargomiRs, a miR-16 based microRNA mimic, was developed by EnGeneIC Limited and underwent phase I clinical trial in patients as second or third line treatment for patients with recurrent malignant pleural mesothelioma and non-small cell lung cancer (ClinicalTrials.gov, NCT number: 02369198) (Reid et al., 2016). Several family members of miR-16 were found to be as tumor suppressors, showing a down-regulation in the broad variety of cancer cells. A restore of expression of miR-16 by a miR-16 based microRNA mimic could induce a significant inhibition of cell proliferation of some cancer cells in vitro models (Liu et al., 2008). As for the in vivo nude mice models, the miR-16 mimic need to be loaded by the nonliving bacterial minicells for the intravenous injection (Viteri and Rosell, 2018). Preliminary data revealed that the treatment of TargomiRs is controllable when the patients were exposed to five billion nanoparticles loaded with miR-16 once a week (van Zandwijk et al., 2017). The results from the phase I study were encouraging and no adverse effects were observed, therefore, TargomiR is expected to perform the phase II clinical trials in next.
MiR-155 plays a crucial role in promoting the growth and survival of the cancer cells because it is found to be highly expressed in certain lymphomas and leukemias (Poltronieri et al., 2013). In 2016, a phase I clinical trial of cobomarsen, an inhibitor of miR-155, was carried out to evaluate its safety, tolerability, pharmacokinetics and potential efficacy on the patients with cutaneous T-cell lymphoma (CTCL), chronic lymphocytic leukemia (CLL), diffuse large B-cell lymphoma (DLBCL), and adult T-cell leukemia/lymphoma (ATLL) (Anastasiadou et al., 2021). The preliminary data revealed that intratumoral injections of cobomarsen over a period of up to 15 days could improve the cutaneous lesions and without any observable adverse effects (Foss et al., 2018). Therefore, a phase II clinical trial was continued to study the efficacy and safety of cobomarsen for the treatment of CTCL and mycosis fungoides (MF) subtype in 2018 (ClinicalTrials.gov, NCT number: 03713320) (Anastasiadou et al., 2021). Although the study was terminated in December, 2020 because of some business reasons, it also encouraged to develop the microRNA as the cancer therapy.
A high expression level of miR-10b was found in glioblastoma patients by the analysis of the Cancer Genome Atlas data, indicating the oncogenic effects of miR-10b. Guessous et al confirmed that miR-10b is upregulated in human glioblastoma tissues, glioblastoma cell and stem cell lines while inhibition of miR-10b would reduce the cancer cell proliferation and inhibited invasion and migration, as well (Guessous et al., 2013). Hence, targeting miR-10b might be a good strategy for glioblastoma treatment. Furthermore, based on the critical function of anti-mir-10b in inhibition of glioblastoma growth, a clinical trial was performed for evaluating the expression Levels of microRNA-10b in patients with gliomas (clinicaltrial.gov, NCT number: 01849952). The estimated primary completion date would be May 2022. More studies are needed for confirming the effects of anti-glioblastoma in clinic.
Challenges and Perspectives of miRNA-Based Therapy
Since developing miRNA as new drug candidates is the ultimate aim in clinical settings, the most important issue is to ensure its safety and effectiveness. Although miRNA-based therapeutics has made some progresses, there are still some barriers that limit the further application of miRNA from bench to bedside. The first limitation is delivery efficiency. The miRNA delivery systems currently used are chemically synthesized with poor cellular uptake properties (Bader et al., 2011). To make use of their medicinal effects, the miRNA should be able to across the complicated circulatory systems and the cell membranes of different tissues (Cheng et al., 2015). Since it was difficult for the miRNAs to access target cells, the second obstacle is the specificity and off-target effects of miRNA. Different from siRNA, the property of “multi-targeting” of miRNA is double-edged sword. It could help to cure the diseases by affecting as many targets correlated with pathogenesis as possible. In the meantime, it caused the off-target effects. While targeting is sequence specific rather than gene specific, it is more challenging to specifically target since the off target effect only requires partial complementary binding of miRNA and target mRNA (Singh et al., 2011). The third concern is the miRNA-induced toxicity. Studies showed that some miRNAs could transcriptionally regulate the expression of drug metabolizing enzymes, such as cytochrome P450s (CYPs) and bile acid synthase CYP7A1 (Takagi et al., 2010). Deregulating the expression of CYPs by specific miRNA may weaken the metabolism of drug to induce drug accumulation, and eventually lead to toxicity. The fourth challenge is to overcome the issue of rapid clearance in blood system (Fortunato and Iorio, 2020). For the naked nucleotide based drugs, the biggest obstacle for their function in vivo are the quick degradation by nuclease and the drug escape from endosome during endocytosis.
Despite the mentioned limitations, miRNA still has broad application prospects in cancer treatment. Some miRNAs may be directly related to cancer by controlling cell proliferation, differentiation and apoptosis, while others may be indirectly related to cancer by targeting oncogenes and tumor suppressor genes. Research on the role of miRNA in the occurrence and development of malignant tumor has become a hot spot. With the development of molecular biology, the detection methods of miRNA have been improved, more and more miRNAs related to cancer has been identified, and the correlation between miRNA and target mRNA has been increasingly identified. The change of miRNA expression profile is an important factor in tumorigenesis and development. Detection of miRNA in tumor patients is advantageous for tumor diagnosis, treatment and prognosis, therefore miRNA may become a new biomarker of tumor. As miRNAs are involved in the main biological behaviors of tumors by regulating target genes, new research-based therapies based on miRNAs will bring positive application prospects in the future. The clinical application of miRNA is based on high efficiency, high sensitivity and low-cost detection methods. However, the current detection methods of miRNA have certain limitations, which hinder the wide use in clinic. After overcoming these problems, miRNA detection and treatment will be expected to smoothly enter the clinic and become a new target for cancer therapeutics.
Author Contributions
Conceptualization, KK-WA-Y and ZF; collected and reviewed the literature, ZF, LW, and FC; Data analysis and figure preparation, ZF and SL; writing-original draft preparation, ZF and KK-WA-Y; writing-review and final editing, KK-WA-Y and CS; funding acquisition, CS. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 82073402).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amani-Shalamzari, S., Agha Alinejad, H., Shahbazi, S., and Alizadeh, S. (2020). The Effect of Endurance Training on Expression of miR-21 and its Downstream in Breast Cancer Bearing Mice. J. Basic Res. Med. Sci. 7, 47–57.
Anastasiadou, E., Seto, A. G., Beatty, X., Hermreck, M., Gilles, M.-E., Stroopinsky, D., et al. (2021). Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 27, 1139–1149. doi:10.1158/1078-0432.ccr-20-3139
Aqeilan, R. I., Calin, G. A., and Croce, C. M. (2010). miR-15a and miR-16-1 in Cancer: Discovery, Function and Future Perspectives. Cell Death Differ 17, 215–220. doi:10.1038/cdd.2009.69
Asangani, I. A., Rasheed, S. A., Nikolova, D. A., Leupold, J. H., Colburn, N. H., Post, S., et al. (2008). MicroRNA-21 (miR-21) post-transcriptionally Downregulates Tumor Suppressor Pdcd4 and Stimulates Invasion, Intravasation and Metastasis in Colorectal Cancer. Oncogene 27, 2128–2136. doi:10.1038/sj.onc.1210856
Azizmohammadi, S., Safari, A., Azizmohammadi, S., Kaghazian, M., Sadrkhanlo, M., Yahaghi, E., et al. (2017). Molecular Identification of miR-145 and miR-9 Expression Level as Prognostic Biomarkers for Early-Stage Cervical Cancer Detection. QJM 110, 11–15. doi:10.1093/qjmed/hcw101
Bader, A. G., Brown, D., Stoudemire, J., and Lammers, P. (2011). Developing Therapeutic microRNAs for Cancer. Gene Ther. 18, 1121–1126. doi:10.1038/gt.2011.79
Bader, A. G. (2012). miR-34 - a microRNA Replacement Therapy Is Headed to the Clinic. Front. Genet. 3, 120. doi:10.3389/fgene.2012.00120
Bao, W., Wang, H. H., Tian, F. J., He, X. Y., Qiu, M. T., Wang, J. Y., et al. (2013). A TrkB-STAT3-miR-204-5p Regulatory Circuitry Controls Proliferation and Invasion of Endometrial Carcinoma Cells. Mol. Cancer 12, 1–19. doi:10.1186/1476-4598-12-155
Barrett-Lee, P. (2009). A Pleiotropically Acting microRNA, miR-31, Inhibits Breast Cancer Metastasis. Adv. Breast Cancer 6, 24–25.
Bartel, D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell 136, 215–233. doi:10.1016/j.cell.2009.01.002
Beg, M. S., Brenner, A. J., Sachdev, J., Borad, M., Kang, Y. K., Stoudemire, J., et al. (2017). Phase I Study of MRX34, a Liposomal miR-34a Mimic, Administered Twice Weekly in Patients with Advanced Solid Tumors. Invest. New Drugs 35, 180–188. doi:10.1007/s10637-016-0407-y
Bertero, T., Bourget-Ponzio, I., Puissant, A., Loubat, A., Mari, B., Meneguzzi, G., et al. (2013). Tumor Suppressor Function of miR-483-3p on Squamous Cell Carcinomas Due to its Pro-apoptotic Properties. Cell Cycle 12, 2183–2193. doi:10.4161/cc.25330
Bommer, G. T., Gerin, I., Feng, Y., Kaczorowski, A. J., Kuick, R., Love, R. E., et al. (2007). p53-mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr. Biol. 17, 1298–1307. doi:10.1016/j.cub.2007.06.068
Bonci, D., Coppola, V., Musumeci, M., Addario, A., Biffoni, M., Labbaye, C., et al. (2008). The miR-15a/miR-16-1 Cluster Controls Prostate Cancer Progression by Targeting Multiple Oncogenic Activities. Nat. Med. 14, 1271–1277. doi:10.1038/nm.1880
Bose, D., Jayaraj, G., Suryawanshi, H., Agarwala, P., Pore, S. K., Banerjee, R., et al. (2012). The Tuberculosis Drug Streptomycin as a Potential Cancer Therapeutic: Inhibition of miR-21 Function by Directly Targeting its Precursor. Angew. Chem. Int. Ed. Engl. 51, 1019–1023. doi:10.1002/anie.201106455
Bose, D., Jayaraj, G. G., Kumar, S., and Maiti, S. (2013). A molecular-beacon-based Screen for Small Molecule Inhibitors of miRNA Maturation. ACS Chem. Biol. 8, 930–938. doi:10.1021/cb300650y
Bou Kheir, T., Futoma-Kazmierczak, E., Jacobsen, A., Krogh, A., Bardram, L., Hother, C., et al. (2011). miR-449 Inhibits Cell Proliferation and Is Down-Regulated in Gastric Cancer. Mol. Cancer 10, 29–12. doi:10.1186/1476-4598-10-29
Chan, J. A., Krichevsky, A. M., and Kosik, K. S. (2005). MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res. 65, 6029–6033. doi:10.1158/0008-5472.CAN-05-0137
Chen, J., Sun, D., Chu, H., Gong, Z., Zhang, C., Gong, B., et al. (2015). Screening of Differential microRNA Expression in Gastric Signet Ring Cell Carcinoma and Gastric Adenocarcinoma and Target Gene Prediction. Oncol. Rep. 33, 2963–2971. doi:10.3892/or.2015.3935
Chen, P., Price, C., Li, Z., Li, Y., Cao, D., Wiley, A., et al. (2013). miR-9 Is an Essential Oncogenic microRNA Specifically Overexpressed in Mixed Lineage Leukemia-Rearranged Leukemia. Proc. Natl. Acad. Sci. U S A. 110, 11511–11516. doi:10.1073/pnas.1310144110
Chen, Q. Y., Jiao, D. M., Yan, L., Wu, Y. Q., Hu, H. Z., Song, J., et al. (2015). Comprehensive Gene and microRNA Expression Profiling Reveals miR-206 Inhibits MET in Lung Cancer Metastasis. Mol. Biosyst. 11, 2290–2302. doi:10.1039/c4mb00734d
Chen, W. X., Hu, Q., Qiu, M. T., Zhong, S. L., Xu, J. J., Tang, J. H., et al. (2013). miR-221/222: Promising Biomarkers for Breast Cancer. Tumour Biol. 34, 1361–1370. doi:10.1007/s13277-013-0750-y
Chen, Y., Wei, H., Liu, Y., and Zheng, S. (2018). Promotional Effect of microRNA-194 on Breast Cancer Cells via Targeting F-Box/WD Repeat-Containing Protein 7. Oncol. Lett. 15, 4439–4444. doi:10.3892/ol.2018.7842
Cheng, C. J., Bahal, R., Babar, I. A., Pincus, Z., Barrera, F., Liu, C., et al. (2015). MicroRNA Silencing for Cancer Therapy Targeted to the Tumour Microenvironment. Nature 518, 107–110. doi:10.1038/nature13905
Childs-Disney, J. L., and Disney, M. D. (2016). Small Molecule Targeting of a MicroRNA Associated with Hepatocellular Carcinoma. ACS Chem. Biol. 11, 375–380. doi:10.1021/acschembio.5b00615
Chirshev, E., Oberg, K. C., Ioffe, Y. J., and Unternaehrer, J. J. (2019). Let-7 as Biomarker, Prognostic Indicator, and Therapy for Precision Medicine in Cancer. Clin. Transl Med. 8, 24–14. doi:10.1186/s40169-019-0240-y
Cimmino, A., Calin, G. A., Fabbri, M., Iorio, M. V., Ferracin, M., Shimizu, M., et al. (2005). miR-15 and miR-16 Induce Apoptosis by Targeting BCL2. Proc. Natl. Acad. Sci. U S A. 102, 13944–13949. doi:10.1073/pnas.0506654102
Czimmerer, Z., Hulvely, J., Simandi, Z., Varallyay, E., Havelda, Z., Szabo, E., et al. (2013). A Versatile Method to Design Stem-Loop Primer-Based Quantitative PCR Assays for Detecting Small Regulatory RNA Molecules. PloS one 8, e55168. doi:10.1371/journal.pone.0055168
Davies, B. P., and Arenz, C. (2006). A Homogenous Assay for Micro RNA Maturation. Angew. Chem. Int. Ed. Engl. 45, 5550–5552. doi:10.1002/anie.200601332
De Ieso, M. L., and Yool, A. J. (2018). Mechanisms of Aquaporin-Facilitated Cancer Invasion and Metastasis. Front. Chem. 6, 135. doi:10.3389/fchem.2018.00135
Diaz, J. P., Chirayil, R., Chirayil, S., Tom, M., Head, K. J., and Luebke, K. J. (2014). Association of a Peptoid Ligand with the Apical Loop of Pri-miR-21 Inhibits Cleavage by Drosha. RNA 20, 528–539. doi:10.1261/rna.042911.113
Du, B., Wang, Z., Zhang, X., Feng, S., Wang, G., He, J., et al. (2014). MicroRNA-545 Suppresses Cell Proliferation by Targeting Cyclin D1 and CDK4 in Lung Cancer Cells. PloS one 9, e88022. doi:10.1371/journal.pone.0088022
Elbadawy, M., Usui, T., Yamawaki, H., and Sasaki, K. (2019). Emerging Roles of C-Myc in Cancer Stem Cell-Related Signaling and Resistance to Cancer Chemotherapy: a Potential Therapeutic Target against Colorectal Cancer. Int. J. Mol. Sci. 20, 2340. doi:10.3390/ijms20092340
Endo, H., Muramatsu, T., Furuta, M., Uzawa, N., Pimkhaokham, A., Amagasa, T., et al. (2013). Potential of Tumor-Suppressive miR-596 Targeting LGALS3BP as a Therapeutic Agent in Oral Cancer. Carcinogenesis 34, 560–569. doi:10.1093/carcin/bgs376
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M., et al. (2006). miR-122 Regulation of Lipid Metabolism Revealed by In Vivo Antisense Targeting. Cell Metab 3, 87–98. doi:10.1016/j.cmet.2006.01.005
Esquela-Kerscher, A., Trang, P., Wiggins, J. F., Patrawala, L., Cheng, A., Ford, L., et al. (2008). The Let-7 microRNA Reduces Tumor Growth in Mouse Models of Lung Cancer. Cell cycle 7, 759–764. doi:10.4161/cc.7.6.5834
Fletcher, C. E., Dart, D. A., Sita-Lumsden, A., Cheng, H., Rennie, P. S., and Bevan, C. L. (2012). Androgen-regulated Processing of the Oncomir miR-27a, Which Targets Prohibitin in Prostate Cancer. Hum. Mol. Genet. 21, 3112–3127. doi:10.1093/hmg/dds139
Fontana, L., Fiori, M. E., Albini, S., Cifaldi, L., Giovinazzi, S., Forloni, M., et al. (2008). Antagomir-17-5p Abolishes the Growth of Therapy-Resistant Neuroblastoma through P21 and BIM. PloS one 3, e2236. doi:10.1371/journal.pone.0002236
Fortunato, O., and Iorio, M. V. (2020). The Therapeutic Potential of MicroRNAs in Cancer: Illusion or Opportunity? Pharmaceuticals (Basel) 13, 438. doi:10.3390/ph13120438
Foss, F. M., Querfeld, C., Kim, Y. H., Pinter-Brown, L. C., William, B. M., Porcu, P., et al. (2018). Ph 1 Study of MRG-106, an Inhibitor of miR-155, in CTCL. Jco 36, 2511. doi:10.1200/jco.2018.36.15_suppl.2511
Fujita, Y., Kojima, K., Hamada, N., Ohhashi, R., Akao, Y., Nozawa, Y., et al. (2008). Effects of miR-34a on Cell Growth and Chemoresistance in Prostate Cancer PC3 Cells. Biochem. Biophys. Res. Commun. 377, 114–119. doi:10.1016/j.bbrc.2008.09.086
Fuziwara, C. S., and Kimura, E. T. (2015). Insights into Regulation of the miR-17-92 Cluster of miRNAs in Cancer. Front. Med. (Lausanne) 2, 64. doi:10.3389/fmed.2015.00064
Galm, O., Herman, J. G., and Baylin, S. B. (2006). The Fundamental Role of Epigenetics in Hematopoietic Malignancies. Blood Rev. 20, 1–13. doi:10.1016/j.blre.2005.01.006
Gambari, R., Brognara, E., Spandidos, D. A., and Fabbri, E. (2016). Targeting oncomiRNAs and Mimicking Tumor Suppressor miRNAs: Νew Trends in the Development of miRNA Therapeutic Strategies in Oncology (Review). Int. J. Oncol. 49, 5–32. doi:10.3892/ijo.2016.3503
Gandellini, P., Folini, M., Longoni, N., Pennati, M., Binda, M., Colecchia, M., et al. (2009). miR-205 Exerts Tumor-Suppressive Functions in Human Prostate through Down-Regulation of Protein Kinase Cepsilon. Cancer Res. 69, 2287–2295. doi:10.1158/0008-5472.CAN-08-2894
Gao, C., Zhou, C., Zhuang, J., Liu, L., Liu, C., Li, H., et al. (2018). MicroRNA Expression in Cervical Cancer: Novel Diagnostic and Prognostic Biomarkers. J. Cel Biochem 119, 7080–7090. doi:10.1002/jcb.27029
Gao, P., Xing, A. Y., Zhou, G. Y., Zhang, T. G., Zhang, J. P., Gao, C., et al. (2013). The Molecular Mechanism of microRNA-145 to Suppress Invasion-Metastasis cascade in Gastric Cancer. Oncogene 32, 491–501. doi:10.1038/onc.2012.61
Garbett, K. A., Horváth, S., Ebert, P. J., Schmidt, M. J., Lwin, K., Mitchell, A., et al. (2010). Novel Animal Models for Studying Complex Brain Disorders: BAC-Driven miRNA-Mediated In Vivo Silencing of Gene Expression. Mol. Psychiatry 15, 987–995. doi:10.1038/mp.2010.1
Garzon, R., Heaphy, C. E., Havelange, V., Fabbri, M., Volinia, S., Tsao, T., et al. (2009). MicroRNA 29b Functions in Acute Myeloid Leukemia. Blood 114, 5331–5341. doi:10.1182/blood-2009-03-211938
Gaur, A., Jewell, D. A., Liang, Y., Ridzon, D., Moore, J. H., Chen, C., et al. (2007). Characterization of microRNA Expression Levels and Their Biological Correlates in Human Cancer Cell Lines. Cancer Res. 67, 2456–2468. doi:10.1158/0008-5472.CAN-06-2698
Girard, M., Jacquemin, E., Munnich, A., Lyonnet, S., and Henrion-Caude, A. (2008). miR-122, a Paradigm for the Role of microRNAs in the Liver. J. Hepatol. 48, 648–656. doi:10.1016/j.jhep.2008.01.019
Gong, C., Yang, Z., Wu, F., Han, L., Liu, Y., and Gong, W. (2016). miR-17 Inhibits Ovarian Cancer Cell Peritoneal Metastasis by Targeting ITGA5 and ITGB1. Oncol. Rep. 36, 2177–2183. doi:10.3892/or.2016.4985
Gruszka, R., and Zakrzewska, M. (2018). The Oncogenic Relevance of miR-17-92 Cluster and its Paralogous miR-106b-25 and miR-106a-363 Clusters in Brain Tumors. Int. J. Mol. Sci. 19, 879. doi:10.3390/ijms19030879
Gu, Y., Cheng, Y., Song, Y., Zhang, Z., Deng, M., Wang, C., et al. (2014). MicroRNA-493 Suppresses Tumor Growth, Invasion and Metastasis of Lung Cancer by Regulating E2F1. PloS one 9, e102602. doi:10.1371/journal.pone.0102602
Guessous, F., Alvarado-Velez, M., Marcinkiewicz, L., Zhang, Y., Kim, J., Heister, S., et al. (2013). Oncogenic Effects of miR-10b in Glioblastoma Stem Cells. J. Neurooncol. 112, 153–163. doi:10.1007/s11060-013-1047-0
Gumireddy, K., Young, D. D., Xiong, X., Hogenesch, J. B., Huang, Q., and Deiters, A. (2008). Small-molecule Inhibitors of Microrna miR-21 Function. Angew. Chem. Int. Ed. Engl. 47, 7482–7484. doi:10.1002/anie.200801555
Han, C., Shen, J. K., Hornicek, F. J., Kan, Q., and Duan, Z. (2017). Regulation of microRNA-1 (miR-1) Expression in Human Cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 227–232. doi:10.1016/j.bbagrm.2016.12.004
He, B., Zhao, Z., Cai, Q., Zhang, Y., Zhang, P., Shi, S., et al. (2020). miRNA-Based Biomarkers, Therapies, and Resistance in Cancer. Int. J. Biol. Sci. 16, 2628–2647. doi:10.7150/ijbs.47203
He, L., He, X., Lim, L. P., De Stanchina, E., Xuan, Z., Liang, Y., et al. (2007). A microRNA Component of the P53 Tumour Suppressor Network. Nature 447, 1130–1134. doi:10.1038/nature05939
He, Z., Yu, L., Luo, S., Li, M., Li, J., Li, Q., et al. (2017). miR-296 Inhibits the Metastasis and Epithelial-Mesenchymal Transition of Colorectal Cancer by Targeting S100A4. BMC cancer 17, 1–10. doi:10.1186/s12885-017-3121-z
Hermeking, H. (2010). The miR-34 Family in Cancer and Apoptosis. Cel Death Differ 17, 193–199. doi:10.1038/cdd.2009.56
Hou, C., Sun, B., Jiang, Y., Zheng, J., Yang, N., Ji, C., et al. (2016). MicroRNA-31 Inhibits Lung Adenocarcinoma Stem-like Cells via Down-Regulation of MET-Pi3k-Akt Signaling Pathway. Anticancer Agents Med. Chem. 16, 501–518. doi:10.2174/1871520615666150824152353
Huang, L., Wang, L., Wang, L., Wu, Z., Jiang, F., Li, Q., et al. (2020). MicroRNA Let‐7b Inhibits Proliferation and Induces Apoptosis of Castration‐Resistant Prostate Cancer Cells by Blocking the Ras/Rho Signaling Pathway via NRAS. Clin. translational Sci.
Huang, T. C., Chang, H. Y., Chen, C. Y., Wu, P. Y., Lee, H., Liao, Y. F., et al. (2011). Silencing of miR-124 Induces Neuroblastoma SK-N-SH Cell Differentiation, Cell Cycle Arrest and Apoptosis through Promoting AHR. FEBS Lett. 585, 3582–3586. doi:10.1016/j.febslet.2011.10.025
Hudson, R. S., Yi, M., Esposito, D., Watkins, S. K., Hurwitz, A. A., Yfantis, H. G., et al. (2012). MicroRNA-1 Is a Candidate Tumor Suppressor and Prognostic Marker in Human Prostate Cancer. Nucleic Acids Res. 40, 3689–3703. doi:10.1093/nar/gkr1222
Humphreys, K. J., McKinnon, R. A., and Michael, M. Z. (2014). miR-18a Inhibits CDC42 and Plays a Tumour Suppressor Role in Colorectal Cancer Cells. PloS one 9, e112288. doi:10.1371/journal.pone.0112288
Ibrahim, A. F., Weirauch, U., Thomas, M., Grünweller, A., Hartmann, R. K., and Aigner, A. (2011). MicroRNA Replacement Therapy for miR-145 and miR-33a Is Efficacious in a Model of colon Carcinoma. Cancer Res. 71, 5214–5224. doi:10.1158/0008-5472.CAN-10-4645
Javanmard, S. H., Vaseghi, G., Ghasemi, A., Rafiee, L., Ferns, G. A., Esfahani, H. N., et al. (2020). Therapeutic Inhibition of microRNA-21 (miR-21) Using Locked-Nucleic Acid (LNA)-anti-miR and its Effects on the Biological Behaviors of Melanoma Cancer Cells in Preclinical Studies. Cancer Cel Int 20, 1–12. doi:10.1186/s12935-020-01394-6
Ji, J., Shi, J., Budhu, A., Yu, Z., Forgues, M., Roessler, S., et al. (2009). MicroRNA Expression, Survival, and Response to Interferon in Liver Cancer. N. Engl. J. Med. 361, 1437–1447. doi:10.1056/NEJMoa0901282
Jiang, C. S., Wang, X. M., Zhang, S. Q., Meng, L. S., Zhu, W. H., Xu, J., et al. (2015). Discovery of 4-Benzoylamino-N-(prop-2-Yn-1-Yl)benzamides as Novel microRNA-21 Inhibitors. Bioorg. Med. Chem. 23, 6510–6519. doi:10.1016/j.bmc.2015.08.007
Jiang, S., Zhang, H. W., Lu, M. H., He, X. H., Li, Y., Gu, H., et al. (2010). MicroRNA-155 Functions as an OncomiR in Breast Cancer by Targeting the Suppressor of Cytokine Signaling 1 Gene. Cancer Res. 70, 3119–3127. doi:10.1158/0008-5472.CAN-09-4250
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., et al. (2005). RAS Is Regulated by the Let-7 microRNA Family. Cell 120, 635–647. doi:10.1016/j.cell.2005.01.014
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M., and Sarnow, P. (2005). Modulation of Hepatitis C Virus RNA Abundance by a Liver-specific MicroRNA. Science 309, 1577–1581. doi:10.1126/science.1113329
Kang, W., Tong, J. H., Lung, R. W., Dong, Y., Zhao, J., Liang, Q., et al. (2015). Targeting of YAP1 by microRNA-15a and microRNA-16-1 Exerts Tumor Suppressor Function in Gastric Adenocarcinoma. Mol. Cancer 14, 52–10. doi:10.1186/s12943-015-0323-3
Kano, M., Seki, N., Kikkawa, N., Fujimura, L., Hoshino, I., Akutsu, Y., et al. (2010). miR-145, miR-133a and miR-133b: Tumor-Suppressive miRNAs Target FSCN1 in Esophageal Squamous Cell Carcinoma. Int. J. Cancer 127, 2804–2814. doi:10.1002/ijc.25284
Kapoor, I., Bodo, J., Hill, B. T., Hsi, E. D., and Almasan, A. (2020). Targeting BCL-2 in B-Cell Malignancies and Overcoming Therapeutic Resistance. Cell Death Dis 11, 1–11. doi:10.1038/s41419-020-03144-y
Kardani, A., Yaghoobi, H., Alibakhshi, A., and Khatami, M. (2020). Inhibition of miR-155 in MCF-7 Breast Cancer Cell Line by Gold Nanoparticles Functionalized with Antagomir and AS1411 Aptamer. J. Cel Physiol 235, 6887–6895. doi:10.1002/jcp.29584
Kato, T., Enomoto, A., Watanabe, T., Haga, H., Ishida, S., Kondo, Y., et al. (2014). TRIM27/MRTF-B-dependent Integrin β1 Expression Defines Leading Cells in Cancer Cell Collectives. Cell Rep 7, 1156–1167. doi:10.1016/j.celrep.2014.03.068
Kikkawa, N., Kinoshita, T., Nohata, N., Hanazawa, T., Yamamoto, N., Fukumoto, I., et al. (2014). microRNA-504 Inhibits Cancer Cell Proliferation via Targeting CDK6 in Hypopharyngeal Squamous Cell Carcinoma. Int. J. Oncol. 44, 2085–2092. doi:10.3892/ijo.2014.2349
Kluiver, J., Slezak-Prochazka, I., Smigielska-Czepiel, K., Halsema, N., Kroesen, B. J., and van den Berg, A. (2012). Generation of miRNA Sponge Constructs. Methods 58, 113–117. doi:10.1016/j.ymeth.2012.07.019
Kota, J., Chivukula, R. R., O'Donnell, K. A., Wentzel, E. A., Montgomery, C. L., Hwang, H. W., et al. (2009). Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell 137, 1005–1017. doi:10.1016/j.cell.2009.04.021
Lee, D. Y., Deng, Z., Wang, C. H., and Yang, B. B. (2007). MicroRNA-378 Promotes Cell Survival, Tumor Growth, and Angiogenesis by Targeting SuFu and Fus-1 Expression. Proc. Natl. Acad. Sci. U S A. 104, 20350–20355. doi:10.1073/pnas.0706901104
Lee, E. J., Baek, M., Gusev, Y., Brackett, D. J., Nuovo, G. J., and Schmittgen, T. D. (2008). Systematic Evaluation of microRNA Processing Patterns in Tissues, Cell Lines, and Tumors. RNA 14, 35–42. doi:10.1261/rna.804508
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75, 843–854. doi:10.1016/0092-8674(93)90529-y
Li, K. K., Pang, J. C., Lau, K. M., Zhou, L., Mao, Y., Wang, Y., et al. (2013). MiR-383 Is Downregulated in Medulloblastoma and Targets Peroxiredoxin 3 (PRDX3). Brain Pathol. 23, 413–425. doi:10.1111/bpa.12014
Li, L., and Wang, B. (2014). Overexpression of microRNA-30b Improves Adenovirus-Mediated P53 Cancer Gene Therapy for Laryngeal Carcinoma. Int. J. Mol. Sci. 15, 19729–19740. doi:10.3390/ijms151119729
Li, Y., Zu, L., Wang, Y., Wang, M., Chen, P., and Zhou, Q. (2015). miR-132 Inhibits Lung Cancer Cell Migration and Invasion by Targeting SOX4. J. Thorac. Dis. 7, 1563–1569. doi:10.3978/j.issn.2072-1439.2015.09.06
Liang, Z., Ahn, J., Guo, D., Votaw, J. R., and Shim, H. (2013). MicroRNA-302 Replacement Therapy Sensitizes Breast Cancer Cells to Ionizing Radiation. Pharm. Res. 30, 1008–1016. doi:10.1007/s11095-012-0936-9
Lichner, Z., Saleh, C., Subramaniam, V., Seivwright, A., Prud'homme, G. J., and Yousef, G. M. (2015). miR-17 Inhibition Enhances the Formation of Kidney Cancer Spheres with Stem Cell/Tumor Initiating Cell Properties. Oncotarget 6, 5567–5581. doi:10.18632/oncotarget.1901
Lin, Y., Liu, A. Y., Fan, C., Zheng, H., Li, Y., Zhang, C., et al. (2015). MicroRNA-33b Inhibits Breast Cancer Metastasis by Targeting HMGA2, SALL4 and Twist1. Sci. Rep. 5, 1–12. doi:10.1038/srep09995
Ling, N., Gu, J., Lei, Z., Li, M., Zhao, J., Zhang, H. T., et al. (2013). microRNA-155 Regulates Cell Proliferation and Invasion by Targeting FOXO3a in Glioma. Oncol. Rep. 30, 2111–2118. doi:10.3892/or.2013.2685
Liu, K., Li, X., Cao, Y., Ge, Y., Wang, J., and Shi, B. (2015). MiR-132 Inhibits Cell Proliferation, Invasion and Migration of Hepatocellular Carcinoma by Targeting PIK3R3. Int. J. Oncol. 47, 1585–1593. doi:10.3892/ijo.2015.3112
Liu, Q., Fu, H., Sun, F., Zhang, H., Tie, Y., Zhu, J., et al. (2008). miR-16 Family Induces Cell Cycle Arrest by Regulating Multiple Cell Cycle Genes. Nucleic Acids Res. 36, 5391–5404. doi:10.1093/nar/gkn522
Liu, T., Hu, K., Zhao, Z., Chen, G., Ou, X., Zhang, H., et al. (2015). MicroRNA-1 Down-Regulates Proliferation and Migration of Breast Cancer Stem Cells by Inhibiting the Wnt/β-Catenin Pathway. Oncotarget 6, 41638–41649. doi:10.18632/oncotarget.5873
Liu, W., Zabirnyk, O., Wang, H., Shiao, Y. H., Nickerson, M. L., Khalil, S., et al. (2010). miR-23b Targets Proline Oxidase, a Novel Tumor Suppressor Protein in Renal Cancer. Oncogene 29, 4914–4924. doi:10.1038/onc.2010.237
Lou, S., Sun, T., Li, H., and Hu, Z. (2018). Mechanisms of microRNA-Mediated Gene Regulation in Unicellular Model Alga Chlamydomonas Reinhardtii. Biotechnol. Biofuels 11, 1–12. doi:10.1186/s13068-018-1249-y
Lujambio, A., Ropero, S., Ballestar, E., Fraga, M. F., Cerrato, C., Setién, F., et al. (2007). Genetic Unmasking of an Epigenetically Silenced microRNA in Human Cancer Cells. Cancer Res. 67, 1424–1429. doi:10.1158/0008-5472.CAN-06-4218
Luo, W., Huang, B., Li, Z., Li, H., Sun, L., Zhang, Q., et al. (2013). MicroRNA-449a Is Downregulated in Non-small Cell Lung Cancer and Inhibits Migration and Invasion by Targeting C-Met. PloS one 8, e64759. doi:10.1371/journal.pone.0064759
Ma, J., Hong, L., Xu, G., Hao, J., Wang, R., Guo, H., et al. (2016). miR-483-3p Plays an Oncogenic Role in Esophageal Squamous Cell Carcinoma by Targeting Tumor Suppressor EI24. Cell Biol Int 40, 448–455. doi:10.1002/cbin.10585
Makarova, J. A., and Kramerov, D. A. (2007). Noncoding RNAs. Biochemistry (Mosc) 72, 1161–1178. doi:10.1134/s0006297907110016
Matsuzaki, J., and Suzuki, H. (2015). Role of MicroRNAs-221/222 in Digestive Systems. J. Clin. Med. 4, 1566–1577. doi:10.3390/jcm4081566
Melo, S., Villanueva, A., Moutinho, C., Davalos, V., Spizzo, R., Ivan, C., et al. (2011). Small Molecule Enoxacin Is a Cancer-specific Growth Inhibitor that Acts by Enhancing TAR RNA-Binding Protein 2-mediated microRNA Processing. Proc. Natl. Acad. Sci. U S A. 108, 4394–4399. doi:10.1073/pnas.1014720108
Mihailovich, M., Bremang, M., Spadotto, V., Musiani, D., Vitale, E., Varano, G., et al. (2015). miR-17-92 fine-tunes MYC Expression and Function to Ensure Optimal B Cell Lymphoma Growth. Nat. Commun. 6, 1–15. doi:10.1038/ncomms9725
Naro, Y., Thomas, M., Stephens, M. D., Connelly, C. M., and Deiters, A. (2015). Aryl Amide Small-Molecule Inhibitors of microRNA miR-21 Function. Bioorg. Med. Chem. Lett. 25, 4793–4796. doi:10.1016/j.bmcl.2015.07.016
Ng, W. L., Yan, D., Zhang, X., Mo, Y. Y., and Wang, Y. (2010). Over-expression of miR-100 Is Responsible for the Low-Expression of ATM in the Human Glioma Cell Line: M059J. DNA Repair (Amst) 9, 1170–1175. doi:10.1016/j.dnarep.2010.08.007
Okamoto, K., Ishiguro, T., Midorikawa, Y., Ohata, H., Izumiya, M., Tsuchiya, N., et al. (2012). miR-493 Induction during Carcinogenesis Blocks Metastatic Settlement of colon Cancer Cells in Liver. EMBO J. 31, 1752–1763. doi:10.1038/emboj.2012.25
Otoukesh, B., Abbasi, M., Gorgani, H. O., Farahini, H., Moghtadaei, M., Boddouhi, B., et al. (2020). MicroRNAs Signatures, Bioinformatics Analysis of miRNAs, miRNA Mimics and Antagonists, and miRNA Therapeutics in Osteosarcoma. Cancer Cel Int 20, 1–20. doi:10.1186/s12935-020-01342-4
Pan, X., Wang, Z. X., and Wang, R. (2010). MicroRNA-21: a Novel Therapeutic Target in Human Cancer. Cancer Biol. Ther. 10, 1224–1232. doi:10.4161/cbt.10.12.14252
Papargyri, N., Pontoppidan, M., Andersen, M. R., Koch, T., and Hagedorn, P. H. (2020). Chemical Diversity of Locked Nucleic Acid-Modified Antisense Oligonucleotides Allows Optimization of Pharmaceutical Properties. Mol. Ther. Nucleic Acids 19, 706–717. doi:10.1016/j.omtn.2019.12.011
Parkin, A., Man, J., Timpson, P., and Pajic, M. (2019). Targeting the Complexity of Src Signalling in the Tumour Microenvironment of Pancreatic Cancer: from Mechanism to Therapy. FEBS J. 286, 3510–3539. doi:10.1111/febs.15011
Pekarsky, Y., and Croce, C. M. (2015). Role of miR-15/16 in CLL. Cel Death Differ 22, 6–11. doi:10.1038/cdd.2014.87
Peng, Y., and Croce, C. M. (2016). The Role of MicroRNAs in Human Cancer. Signal. Transduct Target. Ther. 1, 15004–15009. doi:10.1038/sigtrans.2015.4
Poltronieri, P., D'Urso, P. I., Mezzolla, V., and D'Urso, O. F. (2013). Potential of Anti-cancer Therapy Based on Anti-miR-155 Oligonucleotides in Glioma and Brain Tumours. Chem. Biol. Drug Des. 81, 79–84. doi:10.1111/cbdd.12002
Pu, M., Chen, J., Tao, Z., Miao, L., Qi, X., Wang, Y., et al. (2019). Regulatory Network of miRNA on its Target: Coordination between Transcriptional and post-transcriptional Regulation of Gene Expression. Cell Mol Life Sci 76, 441–451. doi:10.1007/s00018-018-2940-7
Quintavalle, C., Garofalo, M., Zanca, C., Romano, G., Iaboni, M., del Basso De Caro, M., et al. (2012). miR-221/222 Overexpession in Human Glioblastoma Increases Invasiveness by Targeting the Protein Phosphate PTPμ. Oncogene 31, 858–868. doi:10.1038/onc.2011.280
Reid, G., Kao, S. C., Pavlakis, N., Brahmbhatt, H., MacDiarmid, J., Clarke, S., et al. (2016). Clinical Development of TargomiRs, a miRNA Mimic-Based Treatment for Patients with Recurrent Thoracic Cancer. Epigenomics 8, 1079–1085. doi:10.2217/epi-2016-0035
Reid, J. F., Sokolova, V., Zoni, E., Lampis, A., Pizzamiglio, S., Bertan, C., et al. (2012). miRNA Profiling in Colorectal Cancer Highlights miR-1 Involvement in MET-dependent Proliferation. Mol. Cancer Res. 10, 504–515. doi:10.1158/1541-7786.MCR-11-0342
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide Let-7 RNA Regulates Developmental Timing in Caenorhabditis elegans. Nature 403, 901–906. doi:10.1038/35002607
Roberts, T. C., Langer, R., and Wood, M. J. A. (2020). Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 19, 673–694. doi:10.1038/s41573-020-0075-7
Roma-Rodrigues, C., Rivas-García, L., Baptista, P. V., and Fernandes, A. R. (2020). Gene Therapy in Cancer Treatment: Why Go Nano? Pharmaceutics 12, 233. doi:10.3390/pharmaceutics12030233
Saito, Y., Nakaoka, T., and Saito, H. (2015). microRNA-34a as a Therapeutic Agent against Human Cancer. J. Clin. Med. 4, 1951–1959. doi:10.3390/jcm4111951
Savi, F., Forno, I., Faversani, A., Luciani, A., Caldiera, S., Gatti, S., et al. (2014). miR-296/Scribble axis Is Deregulated in Human Breast Cancer and miR-296 Restoration Reduces Tumour Growth In Vivo. Clin. Sci. (Lond) 127, 233–242. doi:10.1042/CS20130580
Scheibner, K. A., Teaboldt, B., Hauer, M. C., Chen, X., Cherukuri, S., Guo, Y., et al. (2012). MiR-27a Functions as a Tumor Suppressor in Acute Leukemia by Regulating 14-3-3θ. PloS one 7, e50895. doi:10.1371/journal.pone.0050895
Scherr, M., Venturini, L., Battmer, K., Schaller-Schoenitz, M., Schaefer, D., Dallmann, I., et al. (2007). Lentivirus-mediated Antagomir Expression for Specific Inhibition of miRNA Function. Nucleic Acids Res. 35, e149. doi:10.1093/nar/gkm971
Schmidt, M. F. (2014). Drug Target miRNAs: Chances and Challenges. Trends Biotechnol. 32, 578–585. doi:10.1016/j.tibtech.2014.09.002
Segura, M. F., Hanniford, D., Menendez, S., Reavie, L., Zou, X., Alvarez-Diaz, S., et al. (2009). Aberrant miR-182 Expression Promotes Melanoma Metastasis by Repressing FOXO3 and Microphthalmia-Associated Transcription Factor. Proc. Natl. Acad. Sci. U S A. 106, 1814–1819. doi:10.1073/pnas.0808263106
Selcuklu, S. D., Donoghue, M. T., Rehmet, K., de Souza Gomes, M., Fort, A., Kovvuru, P., et al. (2012). MicroRNA-9 Inhibition of Cell Proliferation and Identification of Novel miR-9 Targets by Transcriptome Profiling in Breast Cancer Cells. J. Biol. Chem. 287, 29516–29528. doi:10.1074/jbc.M111.335943
Sengupta, S., den Boon, J. A., Chen, I. H., Newton, M. A., Stanhope, S. A., Cheng, Y. J., et al. (2008). MicroRNA 29c Is Down-Regulated in Nasopharyngeal Carcinomas, Up-Regulating mRNAs Encoding Extracellular Matrix Proteins. Proc. Natl. Acad. Sci. U S A. 105, 5874–5878. doi:10.1073/pnas.0801130105
Shakeri, A., Ghanbari, M., Tasbandi, A., and Sahebkar, A. (2021). Regulation of microRNA‐21 Expression by Natural Products in Cancer. Phytotherapy Res.
Shao, J., Cao, J., Liu, Y., Mei, H., Zhang, Y., and Xu, W. (2015). MicroRNA-519a Promotes Proliferation and Inhibits Apoptosis of Hepatocellular Carcinoma Cells by Targeting FOXF2. FEBS Open Bio 5, 893–899. doi:10.1016/j.fob.2015.10.009
Shen, H., Wang, D., Li, L., Yang, S., Chen, X., Zhou, S., et al. (2017). MiR-222 Promotes Drug-Resistance of Breast Cancer Cells to Adriamycin via Modulation of PTEN/Akt/FOXO1 Pathway. Gene 596, 110–118. doi:10.1016/j.gene.2016.10.016
Shi, W., Gerster, K., Alajez, N. M., Tsang, J., Waldron, L., Pintilie, M., et al. (2011). MicroRNA-301 Mediates Proliferation and Invasion in Human Breast Cancer. Cancer Res. 71, 2926–2937. doi:10.1158/0008-5472.CAN-10-3369
Shi, Z., Zhang, J., Qian, X., Han, L., Zhang, K., Chen, L., et al. (2013). AC1MMYR2, an Inhibitor of Dicer-Mediated Biogenesis of Oncomir miR-21, Reverses Epithelial-Mesenchymal Transition and Suppresses Tumor Growth and Progression. Cancer Res. 73, 5519–5531. doi:10.1158/0008-5472.CAN-13-0280
Shi, Z. M., Wang, J., Yan, Z., You, Y. P., Li, C. Y., Qian, X., et al. (2012). MiR-128 Inhibits Tumor Growth and Angiogenesis by Targeting p70S6K1. PloS one 7, e32709. doi:10.1371/journal.pone.0032709
Shu, Y., Wu, K., Zeng, Z., Huang, S., Ji, X., Yuan, C., et al. (2018). A Simplified System to Express Circularized Inhibitors of miRNA for Stable and Potent Suppression of miRNA Functions. Mol. Ther. Nucleic Acids 13, 556–567. doi:10.1016/j.omtn.2018.09.025
Si, M. L., Zhu, S., Wu, H., Lu, Z., Wu, F., and Mo, Y. Y. (2007). miR-21-mediated Tumor Growth. Oncogene 26, 2799–2803. doi:10.1038/sj.onc.1210083
Singh, S., Narang, A. S., and Mahato, R. I. (2011). Subcellular Fate and Off-Target Effects of siRNA, shRNA, and miRNA. Pharm. Res. 28, 2996–3015. doi:10.1007/s11095-011-0608-1
Song, B., Ji, W., Guo, S., Liu, A., Jing, W., Shao, C., et al. (2014). miR-545 Inhibited Pancreatic Ductal Adenocarcinoma Growth by Targeting RIG-I. FEBS Lett. 588, 4375–4381. doi:10.1016/j.febslet.2014.10.004
Song, J. H., and Meltzer, S. J. (2012). MicroRNAs in Pathogenesis, Diagnosis, and Treatment of Gastroesophageal Cancers. Gastroenterology 143, 35–e2. doi:10.1053/j.gastro.2012.05.003
Song, J. J., and Li, W. (2019). MiR-10b Suppresses the Growth and Metastasis of Colorectal Cancer Cell by Targeting FGF13. Eur. Rev. Med. Pharmacol. Sci. 23, 576–587. doi:10.26355/eurrev_201901_16870
Sun, X. J., Liu, H., Zhang, P., Zhang, X. D., Jiang, Z. W., and Jiang, C. C. (2013). miR-10b Promotes Migration and Invasion in Nasopharyngeal Carcinoma Cells. Asian Pac. J. Cancer Prev. 14, 5533–5537. doi:10.7314/apjcp.2013.14.9.5533
Sun, Y., Bai, Y., Zhang, F., Wang, Y., Guo, Y., and Guo, L. (2010). miR-126 Inhibits Non-small Cell Lung Cancer Cells Proliferation by Targeting EGFL7. Biochem. Biophys. Res. Commun. 391, 1483–1489. doi:10.1016/j.bbrc.2009.12.098
Takagi, S., Nakajima, M., Kida, K., Yamaura, Y., Fukami, T., and Yokoi, T. (2010). MicroRNAs Regulate Human Hepatocyte Nuclear Factor 4alpha, Modulating the Expression of Metabolic Enzymes and Cell Cycle. J. Biol. Chem. 285, 4415–4422. doi:10.1074/jbc.M109.085431
Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., et al. (2004). Reduced Expression of the Let-7 microRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 64, 3753–3756. doi:10.1158/0008-5472.CAN-04-0637
Takasaki, S. (2015). Roles of microRNAs in Cancers and Development. Methods Mol. Biol. 1218, 375–413. doi:10.1007/978-1-4939-1538-5_24
Takeshita, F., Patrawala, L., Osaki, M., Takahashi, R. U., Yamamoto, Y., Kosaka, N., et al. (2010). Systemic Delivery of Synthetic microRNA-16 Inhibits the Growth of Metastatic Prostate Tumors via Downregulation of Multiple Cell-Cycle Genes. Mol. Ther. 18, 181–187. doi:10.1038/mt.2009.207
Tan, S., Li, R., Ding, K., Lobie, P. E., and Zhu, T. (2011). miR-198 Inhibits Migration and Invasion of Hepatocellular Carcinoma Cells by Targeting the HGF/c-MET Pathway. FEBS Lett. 585, 2229–2234. doi:10.1016/j.febslet.2011.05.042
Tan, S. B., Huang, C., Chen, X., Wu, Y., Zhou, M., Zhang, C., et al. (2013). Small Molecular Inhibitors of miR-1 Identified from Photocycloadducts of Acetylenes with 2-Methoxy-1,4-Naphthalenequinone. Bioorg. Med. Chem. 21, 6124–6131. doi:10.1016/j.bmc.2013.04.058
Tang, H., Yao, L., Tao, X., Yu, Y., Chen, M., Zhang, R., et al. (2013). miR-9 Functions as a Tumor Suppressor in Ovarian Serous Carcinoma by Targeting TLN1. Int. J. Mol. Med. 32, 381–388. doi:10.3892/ijmm.2013.1400
Taniguchi, K., Sugito, N., Kumazaki, M., Shinohara, H., Yamada, N., Nakagawa, Y., et al. (2015). MicroRNA-124 Inhibits Cancer Cell Growth through PTB1/PKM1/PKM2 Feedback cascade in Colorectal Cancer. Cancer Lett. 363, 17–27. doi:10.1016/j.canlet.2015.03.026
Tavazoie, S. F., Alarcón, C., Oskarsson, T., Padua, D., Wang, Q., Bos, P. D., et al. (2008). Endogenous Human microRNAs that Suppress Breast Cancer Metastasis. Nature 451, 147–152. doi:10.1038/nature06487
Tetzlaff, M. T., Liu, A., Xu, X., Master, S. R., Baldwin, D. A., Tobias, J. W., et al. (2007). Differential Expression of miRNAs in Papillary Thyroid Carcinoma Compared to Multinodular Goiter Using Formalin Fixed Paraffin Embedded Tissues. Endocr. Pathol. 18, 163–173. doi:10.1007/s12022-007-0023-7
Thomas, M., Lange-Grünweller, K., Weirauch, U., Gutsch, D., Aigner, A., Grünweller, A., et al. (2012). The Proto-Oncogene Pim-1 Is a Target of miR-33a. Oncogene 31, 918–928. doi:10.1038/onc.2011.278
Thum, T., Catalucci, D., and Bauersachs, J. (2008). MicroRNAs: Novel Regulators in Cardiac Development and Disease. Cardiovasc. Res. 79, 562–570. doi:10.1093/cvr/cvn137
Tian, Y., Luo, A., Cai, Y., Su, Q., Ding, F., Chen, H., et al. (2010). MicroRNA-10b Promotes Migration and Invasion through KLF4 in Human Esophageal Cancer Cell Lines. J. Biol. Chem. 285, 7986–7994. doi:10.1074/jbc.M109.062877
To, K. K. W., Fong, W., Tong, C. W. S., Wu, M., Yan, W., and Cho, W. C. S. (2020). Advances in the Discovery of microRNA-Based Anticancer Therapeutics: Latest Tools and Developments. Expert Opin. Drug Discov. 15, 63–83. doi:10.1080/17460441.2020.1690449
Trang, P., Medina, P. P., Wiggins, J. F., Ruffino, L., Kelnar, K., Omotola, M., et al. (2010). Regression of Murine Lung Tumors by the Let-7 microRNA. Oncogene 29, 1580–1587. doi:10.1038/onc.2009.445
Trang, P., Wiggins, J. F., Daige, C. L., Cho, C., Omotola, M., Brown, D., et al. (2011). Systemic Delivery of Tumor Suppressor microRNA Mimics Using a Neutral Lipid Emulsion Inhibits Lung Tumors in Mice. Mol. Ther. 19, 1116–1122. doi:10.1038/mt.2011.48
Tsang, W. P., Ng, E. K., Ng, S. S., Jin, H., Yu, J., Sung, J. J., et al. (2010). Oncofetal H19-Derived miR-675 Regulates Tumor Suppressor RB in Human Colorectal Cancer. Carcinogenesis 31, 350–358. doi:10.1093/carcin/bgp181
Tu, K., Liu, Z., Yao, B., Han, S., and Yang, W. (2016). MicroRNA-519a Promotes Tumor Growth by Targeting PTEN/PI3K/AKT Signaling in Hepatocellular Carcinoma. Int. J. Oncol. 48, 965–974. doi:10.3892/ijo.2015.3309
Ugalde, A. P., Ramsay, A. J., De La Rosa, J., Varela, I., Mariño, G., Cadiñanos, J., et al. (2011). Aging and Chronic DNA Damage Response Activate a Regulatory Pathway Involving miR-29 and P53. EMBO J. 30, 2219–2232. doi:10.1038/emboj.2011.124
van Zandwijk, N., Pavlakis, N., Kao, S. C., Linton, A., Boyer, M. J., Clarke, S., et al. (2017). Safety and Activity of microRNA-Loaded Minicells in Patients with Recurrent Malignant Pleural Mesothelioma: a First-In-Man, Phase 1, Open-Label, Dose-Escalation Study. Lancet Oncol. 18, 1386–1396. doi:10.1016/S1470-2045(17)30621-6
Velagapudi, S. P., Gallo, S. M., and Disney, M. D. (2014). Sequence-based Design of Bioactive Small Molecules that Target Precursor microRNAs. Nat. Chem. Biol. 10, 291–297. doi:10.1038/nchembio.1452
Vishnoi, A., and Rani, S. (2017). MiRNA Biogenesis and Regulation of Diseases: an Overview. Methods Mol. Biol. 1509, 1–10. doi:10.1007/978-1-4939-6524-3_1
Viteri, S., and Rosell, R. (2018). An Innovative Mesothelioma Treatment Based on miR-16 Mimic Loaded EGFR Targeted Minicells (TargomiRs). Transl Lung Cancer Res. 7, S1. doi:10.21037/tlcr.2017.12.01
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., et al. (2006). A microRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc. Natl. Acad. Sci. U S A. 103, 2257–2261. doi:10.1073/pnas.0510565103
Voorhoeve, P. M., Le Sage, C., Schrier, M., Gillis, A. J., Stoop, H., Nagel, R., et al. (2006). A Genetic Screen Implicates miRNA-372 and miRNA-373 as Oncogenes in Testicular Germ Cell Tumors. Cell 124, 1169–1181. doi:10.1016/j.cell.2006.02.037
Wang, C. Z., Yuan, P., and Li, Y. (2015). MiR-126 Regulated Breast Cancer Cell Invasion by Targeting ADAM9. Int. J. Clin. Exp. Pathol. 8, 6547–6553.
Wang, L., Qu, J., Zhou, L., Liao, F., and Wang, J. (2018). MicroRNA-373 Inhibits Cell Proliferation and Invasion via Targeting BRF2 in Human Non-small Cell Lung Cancer A549 Cell Line. Cancer Res. Treat. 50, 936–949. doi:10.4143/crt.2017.302
Wang, L., Yao, J., Shi, X., Hu, L., Li, Z., Song, T., et al. (2013). MicroRNA-302b Suppresses Cell Proliferation by Targeting EGFR in Human Hepatocellular Carcinoma SMMC-7721 Cells. BMC cancer 13, 448–449. doi:10.1186/1471-2407-13-448
Wang, S., Yan, Y., Cheng, Z., Hu, Y., and Liu, T. (2018). Sotetsuflavone Suppresses Invasion and Metastasis in Non-small-cell Lung Cancer A549 Cells by Reversing EMT via the TNF-Α/nf-Κb and PI3K/AKT Signaling Pathway. Cell Death Discov 4, 26–11. doi:10.1038/s41420-018-0026-9
Wang, Y. Y., Ye, Z. Y., Zhao, Z. S., Li, L., Wang, Y. X., Tao, H. Q., et al. (2013). Clinicopathologic Significance of miR-10b Expression in Gastric Carcinoma. Hum. Pathol. 44, 1278–1285. doi:10.1016/j.humpath.2012.10.014
Wang, Z. (2011). The Concept of Multiple-Target Anti-miRNA Antisense Oligonucleotide Technology, MicroRNA and Cancer. Springer, 51–57. doi:10.1007/978-1-60761-863-8_4
Wang, Z. (2011). The Guideline of the Design and Validation of MiRNA Mimics, MicroRNA and Cancer. Springer, 211–223. doi:10.1007/978-1-60761-863-8_15
Warren, C. F. A., Wong-Brown, M. W., and Bowden, N. A. (2019). BCL-2 Family Isoforms in Apoptosis and Cancer. Cel Death Dis 10, 1–12. doi:10.1038/s41419-019-1407-6
Watashi, K., Yeung, M. L., Starost, M. F., Hosmane, R. S., and Jeang, K. T. (2010). Identification of Small Molecules that Suppress microRNA Function and Reverse Tumorigenesis. J. Biol. Chem. 285, 24707–24716. doi:10.1074/jbc.M109.062976
Weiler, J., Hunziker, J., and Hall, J. (2006). Anti-miRNA Oligonucleotides (AMOs): Ammunition to Target miRNAs Implicated in Human Disease? Gene Ther. 13, 496–502. doi:10.1038/sj.gt.3302654
Welch, C., Chen, Y., and Stallings, R. L. (2007). MicroRNA-34a Functions as a Potential Tumor Suppressor by Inducing Apoptosis in Neuroblastoma Cells. Oncogene 26, 5017–5022. doi:10.1038/sj.onc.1210293
Wen, Q., Zhao, J., Bai, L., Wang, T., Zhang, H., and Ma, Q. (2015). miR-126 Inhibits Papillary Thyroid Carcinoma Growth by Targeting LRP6. Oncol. Rep. 34, 2202–2210. doi:10.3892/or.2015.4165
Wickramasinghe, N. S., Manavalan, T. T., Dougherty, S. M., Riggs, K. A., Li, Y., and Klinge, C. M. (2009). Estradiol Downregulates miR-21 Expression and Increases miR-21 Target Gene Expression in MCF-7 Breast Cancer Cells. Nucleic Acids Res. 37, 2584–2595. doi:10.1093/nar/gkp117
Wiggins, J. F., Ruffino, L., Kelnar, K., Omotola, M., Patrawala, L., Brown, D., et al. (2010). Development of a Lung Cancer Therapeutic Based on the Tumor Suppressor microRNA-34. Cancer Res. 70, 5923–5930. doi:10.1158/0008-5472.CAN-10-0655
Wu, K. J. (2020). The Role of miRNA Biogenesis and DDX17 in Tumorigenesis and Cancer Stemness. Biomed. J. 43, 107–114. doi:10.1016/j.bj.2020.03.001
Wu, P. (2020). Inhibition of RNA-Binding Proteins with Small Molecules. Nat. Rev. Chem. 4, 441–458. doi:10.1038/s41570-020-0201-4
Wuchty, S., Arjona, D., Li, A., Kotliarov, Y., Walling, J., Ahn, S., et al. (2011). Prediction of Associations between microRNAs and Gene Expression in Glioma Biology. PloS one 6, e14681. doi:10.1371/journal.pone.0014681
Würdinger, T., Tannous, B. A., Saydam, O., Skog, J., Grau, S., Soutschek, J., et al. (2008). miR-296 Regulates Growth Factor Receptor Overexpression in Angiogenic Endothelial Cells. Cancer cell 14, 382–393. doi:10.1016/j.ccr.2008.10.005
Wystub, K., Besser, J., Bachmann, A., Boettger, T., and Braun, T. (2013). miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development. Plos Genet. 9, e1003793. doi:10.1371/journal.pgen.1003793
Xia, J., Cao, T., Ma, C., Shi, Y., Sun, Y., Wang, Z. P., et al. (2018). miR-7 Suppresses Tumor Progression by Directly Targeting MAP3K9 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 13, 121–132. doi:10.1016/j.omtn.2018.08.012
Xia, Z., Liu, F., Zhang, J., and Liu, L. (2015). Decreased Expression of MiRNA-204-5p Contributes to Glioma Progression and Promotes Glioma Cell Growth, Migration and Invasion. PloS one 10, e0132399. doi:10.1371/journal.pone.0132399
Xiong, Y., Kotian, S., Zeiger, M. A., Zhang, L., and Kebebew, E. (2015). miR-126-3p Inhibits Thyroid Cancer Cell Growth and Metastasis, and Is Associated with Aggressive Thyroid Cancer. PloS one 10, e0130496. doi:10.1371/journal.pone.0130496
Xu, K., Chen, Z., Qin, C., and Song, X. (2014). miR-7 Inhibits Colorectal Cancer Cell Proliferation and Induces Apoptosis by Targeting XRCC2. Onco Targets Ther. 7, 325–332. doi:10.2147/OTT.S59364
Xu, Y., Jin, J., Liu, Y., Huang, Z., Deng, Y., You, T., et al. (2014). Snail-regulated MiR-375 Inhibits Migration and Invasion of Gastric Cancer Cells by Targeting JAK2. PloS one 9, e99516. doi:10.1371/journal.pone.0099516
Yamashita, R., Sato, M., Kakumu, T., Hase, T., Yogo, N., Maruyama, E., et al. (2015). Growth Inhibitory Effects of miR-221 and miR-222 in Non-small Cell Lung Cancer Cells. Cancer Med. 4, 551–564. doi:10.1002/cam4.412
Yang, H., Liu, P., Zhang, J., Peng, X., Lu, Z., Yu, S., et al. (2016). Long Noncoding RNA MIR31HG Exhibits Oncogenic Property in Pancreatic Ductal Adenocarcinoma and Is Negatively Regulated by miR-193b. Oncogene 35, 3647–3657. doi:10.1038/onc.2015.430
Yang, Z. G., Ma, X. D., He, Z. H., and Guo, Y. X. (2017). miR-483-5p Promotes Prostate Cancer Cell Proliferation and Invasion by Targeting RBM5. Int. Braz. J. Urol. 43, 1060–1067. doi:10.1590/S1677-5538.IBJU.2016.0595
Yoshino, H., Chiyomaru, T., Enokida, H., Kawakami, K., Tatarano, S., Nishiyama, K., et al. (2011). The Tumour-Suppressive Function of miR-1 and miR-133a Targeting TAGLN2 in Bladder Cancer. Br. J. Cancer 104, 808–818. doi:10.1038/bjc.2011.23
Young, D. D., Connelly, C. M., Grohmann, C., and Deiters, A. (2010). Small Molecule Modifiers of microRNA miR-122 Function for the Treatment of Hepatitis C Virus Infection and Hepatocellular Carcinoma. J. Am. Chem. Soc. 132, 7976–7981. doi:10.1021/ja910275u
Yu, A. M., Choi, Y. H., and Tu, M. J. (2020). RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 72, 862–898. doi:10.1124/pr.120.019554
Yu, G., Li, H., Wang, J., Gumireddy, K., Li, A., Yao, W., et al. (2014). miRNA-34a Suppresses Cell Proliferation and Metastasis by Targeting CD44 in Human Renal Carcinoma Cells. J. Urol. 192, 1229–1237. doi:10.1016/j.juro.2014.05.094
Zhang, B. G., Liu, B. Y., Yan, M., Chen, X. H., Yu, P. T., and Zhu, Z. G. (2011). Biological Behaviour and PTEN Expression Change after MicroRNA-21 Down-Regulation in Gastric Cancer Cells. J. Surg. Concepts Pract. 16, 562–567.
Zhang, S., Ma, C., Pang, H., Zeng, F., Cheng, L., Fang, B., et al. (2016). Arsenic Trioxide Suppresses Cell Growth and Migration via Inhibition of miR-27a in Breast Cancer Cells. Biochem. Biophys. Res. Commun. 469, 55–61. doi:10.1016/j.bbrc.2015.11.071
Zhang, Y., Chao, T., Li, R., Liu, W., Chen, Y., Yan, X., et al. (2009). MicroRNA-128 Inhibits Glioma Cells Proliferation by Targeting Transcription Factor E2F3a. J. Mol. Med. (Berl) 87, 43–51. doi:10.1007/s00109-008-0403-6
Zhang, Z., Liu, S., Shi, R., and Zhao, G. (2011). miR-27 Promotes Human Gastric Cancer Cell Metastasis by Inducing Epithelial-To-Mesenchymal Transition. Cancer Genet. 204, 486–491. doi:10.1016/j.cancergen.2011.07.004
Zhang, Z. G., Chen, W. X., Wu, Y. H., Liang, H. F., and Zhang, B. X. (2014). MiR-132 Prohibits Proliferation, Invasion, Migration, and Metastasis in Breast Cancer by Targeting HN1. Biochem. Biophys. Res. Commun. 454, 109–114. doi:10.1016/j.bbrc.2014.10.049
Zhou, L. Y., Qin, Z., Zhu, Y. H., He, Z. Y., and Xu, T. (2019). Current RNA-Based Therapeutics in Clinical Trials. Curr. Gene Ther. 19, 172–196. doi:10.2174/1566523219666190719100526
Zhu, S., Si, M. L., Wu, H., and Mo, Y. Y. (2007). MicroRNA-21 Targets the Tumor Suppressor Gene Tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328–14336. doi:10.1074/jbc.M611393200
Keywords: microRNA, anticancer therapeutics, drug target, MiRNA mimics, antagomirs, oncomirs, tumor suppressor miRNAs
Citation: Fu Z, Wang L, Li S, Chen F, Au-Yeung KK-W and Shi C (2021) MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 12:736323. doi: 10.3389/fphar.2021.736323
Received: 05 July 2021; Accepted: 10 August 2021;
Published: 25 August 2021.
Edited by:
Sanjun Shi, Chengdu University of Traditional Chinese Medicine, ChinaReviewed by:
Khalil Hajiasgharzadeh, Tabriz University of Medical Sciences, IranAlfredo Garcia Venzor, Instituto Nacional de Medicina Genómica (INMEGEN), Mexico
Ping Sun, University of Pittsburgh, United States
Saravanakumar Marimuthu, University of Nebraska Medical Center, United States
Ipek Erdogan, Izmir Institute of Technology, Turkey
Copyright © 2021 Fu, Wang, Li, Chen, Au-Yeung and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathy Ka-Wai Au-Yeung, S0F1WWV1bmdAc2JyYy5jYQ==; Chen Shi, MjE5MTM2OTA5QHFxLmNvbQ==
 Zhiwen Fu
Zhiwen Fu Liu Wang
Liu Wang Shijun Li
Shijun Li Fen Chen
Fen Chen Kathy Ka-Wai Au-Yeung3*
Kathy Ka-Wai Au-Yeung3* Chen Shi
Chen Shi