- 1Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2College of Public Health, Hebei Medical University, Shijiazhuang, China
ZeXie Decoction (ZXD) is a traditional Chinese medicine composed of Alisma orientalis (Sam.) Juzep. and Atractylodes macrocephala Koidz. ZXD has been widely used to treat non-alcoholic fatty liver disease (NAFLD). The mechanistic basis for the pharmacological activity of ZXD, however, remains poorly understood. In this study, we used a network pharmacology approach and investigated the association between ZXD and NAFLD. We identified the active ingredients of ZXD and screened the potential targets of these ingredients, after which a database of relevant NAFLD-related targets were constructed and several enrichment analyses were performed. Furthermore, the ethanol and aqueous extracts of ZXD were prepared and experimental pharmacology validation was conducted using RT-qPCR of the non-alcoholic fatty liver disease (NAFLD) model in Sprague-Dawley (SD) rats. As a result, a herb-compound-target-pathway network model was developed, and HMGCR, SREBP-2, MAPK1, and NF-κBp65 targets were validated. The gene expression results of these four targets were consistent with those of the network pharmacology prediction. Using an integration strategy, we revealed that ZXD could treat NAFLD by targeting HMGCR, SREBP-2, MAPK1, and NF-κBp65.
Introduction
Rising obesity rate has partially been linked to the increasing prevalence of non-alcoholic fatty liver disease (NAFLD), characterized by hepatic steatosis unrelated to alcohol consumption (Than and Newsome, 2015). More than 25% of people suffer from NAFLD worldwide. A significant cause of human morbidity, NAFLD, can progress to severe conditions, including fibrosis, cirrhosis, and hepatoma (Tandon, 2013; Angulo et al., 2015). Treatment options for NAFLD are limited and are centered mainly around bariatric surgery and lifestyle changes.
Traditional Chinese medicine formulations (TCMFs) are traditional medicines that are used clinically for many years. ZeXie Decoction (ZXD) is one of the TCMFs composed of Alismatis Rhizoma (ZX, the rhizome of Alisma orientale (Sam.) Juzep.) and Atractylodis Macrocephalae Rhizoma (BZ, the rhizome of Atractylodes macrocephala Koidz.) in the ratio of 5:2 on a dry weight basis. ZX contains bioactive triterpenoid, including alisol A, alisol B, alisol A 23-acetate, alisol A 24-acetate, alisol B 23-acetate and so forth. BZ contains bioactive triterpenoid, including atractylenolide I, atractylenolide II, atractylenolide III and so forth (Hoang et al., 2016; Liu et al., 2019). Several mechanistic studies have found that ZXD, ZX, BZ, and other active derivatives could treat NAFLD through the modulation of insulin, and PI3K/AKT/NF-κB and PPAR pathways, thereby suppressing inflammation and lowering lipid levels in treated individuals (Dan et al., 2011; Song et al., 2014; Li et al., 2016; Bi et al., 2017; Tang et al., 2017; Wu et al., 2018). ZXD is a promising complementary therapy for the treatment of NAFLD.
TCMFs are typically designed as multi-component multi-target pharmaceutical agents, in contrast to modern drugs typically designed to target a single protein (Zhang et al., 2019). Presently, the molecular mechanisms underlying the efficacy of many TCMFs remain uncertain. In the past, experimental model systems were leveraged for pharmacological studies of TCMFs. However, developments in systems biology have led to more widespread use of network pharmacology-based analyses of traditional medicine preparations (Lee et al., 2018; Zuo et al., 2018; Huang et al., 2020). Network pharmacology tools offer an ideal opportunity to understand the complex mechanisms through which TCMFs function in vivo.
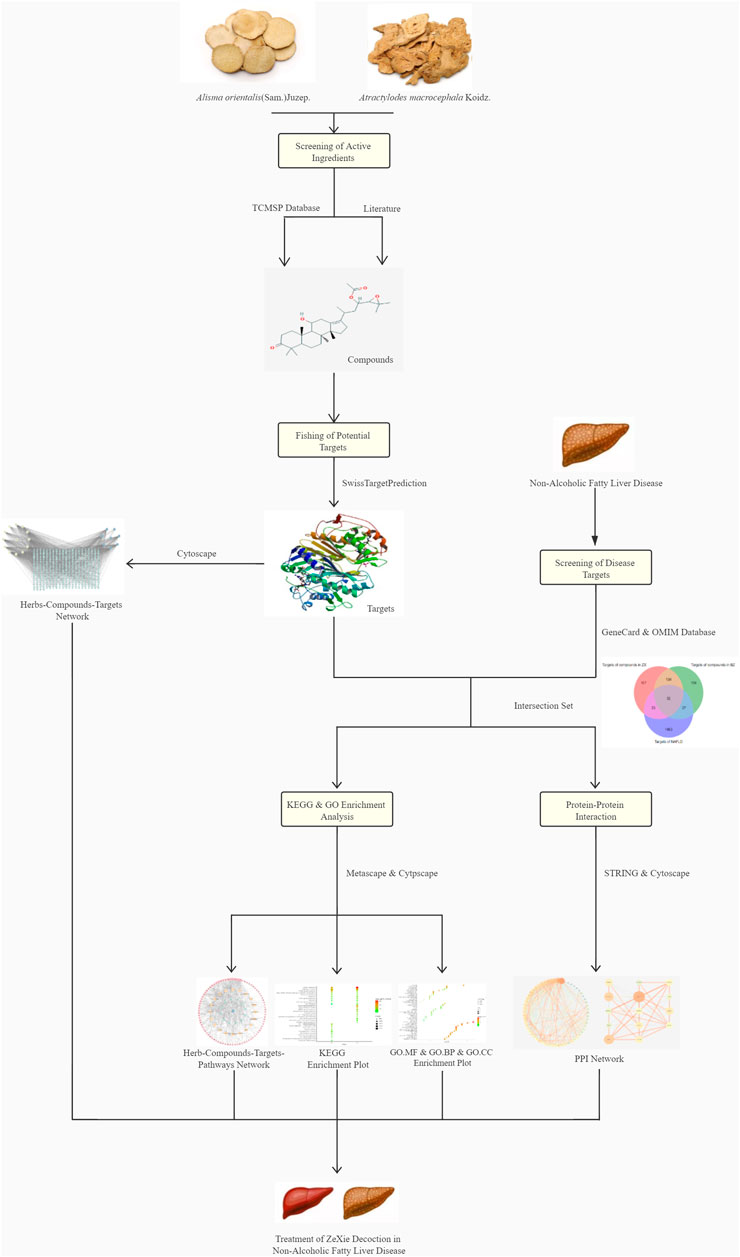
In our previous work (Chang et al., 2021), we studied ethanol and aqueous extracts of ZXD using ultra-performance liquid chromatography coupled with mass spectrometry to identify the major components of ZXD. Based on this work, published literature, and database sources, we developed a herb-compound-target-pathway network model and performed enrichment analysis to investigate the relationship between ZXD and NAFLD treatment. The network pharmacology framework is depicted in Figure 1. Basing on the non-alcoholic fatty liver disease (NAFLD) model in Sprague-Dawley (SD) rats, the prediction of the mechanism was validated by RT-qPCR analysis in vivo.
Materials and Methods
Experimental Animals
A total of 72 male SD rats weighting 200 ± 20 g (Beijing Vital River Laboratory Animal Technology Co. Ltd.) were housed in a room at a constant temperature of 23 ± 3°C, a constant humidity of 50 ± 10%, under a 12 h light/dark cycle. The rats were fed UV-disinfected fodder and drinking water. The animal experiment was approved by the Laboratory Animal Center, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College. The rats were adaptively bred for 1 week in the facility before the experiments.
Reagents
The dried rhizomes of ZX (Batch number: DD6081) and BZ (Batch number: DD8061) were purchased from Beijing Huamiao Pharmaceutical Co., Ltd. (Beijing, China). Their qualities were in accordance with the Chinese Pharmacopoeia. The specimens of ZX and BZ have been preserved in the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College.
PrimeScript RT reagent kit, TRIzol reagent, and DNA marker were purchased from TaKaRa Bio (Beijing, China). The total cholesterol (TC), triglycerides (TG), aspartate aminotransferase (AST), alanine aminotransferase (ALT) assay kit were purchased from Beijing Solarbio Science and Technology Co. Ltd (Beijing, China). Urethane, pioglitazone hydrochloride (PH) and paraformaldehyde were purchased from Macklin Reagent Company (Shanghai, China).
Sample Preparation
The samples of ZXD were prepared by combining ZX and BZ in a 5:2 ratio (w/w). The samples were crushed and refluxed thrice with 10-times the volume of 95% ethanol for 2 h. The filtrates were then pooled and dried using a rotary evaporator under vacuum, yielding 14.29% ethanol extract.
Qualitative Analysis
The comparative study of the ethanol and aqueous extracts of ZXD using UPLC-DAD-Q-TOF-MS was performed in our previous study to simultaneously identify the major chemical components in these two extracts (Chang et al., 2021). This analysis was conducted using an Waters ACQUITY UPLC system with a binary solvent delivery system and an autosampler. An Waters Acquity UPLC HSS T3 column (1.7 μm, 2.1 mm × 100 mm; Waters, MA, United States) was used to conduct this analysis at 30°C. Acetonitrile (A) and formic acid in water (0.1%, v/v) (B) were used as the mobile phase in this analysis, with 0.2 ml/min being selected as the flow rate. A linear gradient elution approach was used with the following settings: 0–10 min, 0–1% A; 10–15 min, 1–3% A; 15–20 min, 3–10% A; 20–25 min, 10–15% A; 25–30 min, 15–20% A; 30–35 min, 20–30% A; 35–40 min, 30–60% A; 40–45 min, 60–80% A; 45–50 min, 80–90% A; 50–55 min, 90–100% A. A 2 µL injection volume was used for all analysis, with 15°C as the injection temperature.
A Q-TOF analyzer in the SYNAPT G2 HDMS system (Waters, Manchester, United Kingdom) was used for tandem MS analyses. Positive ionization mode was used to acquire ESI spectra in the m/z 50–1,200 mass range with the following settings: a 120°C source temperature; 3.0 kV as the chosen capillary voltage; a desolvation gas temperature of 450°C and a gas flow rate of 800 L/h; a 40.0 V sampling cone voltage and a 50 L/h cone gas rate; 10–40 eV as the collision energy. The Masslynx 4.1 software package was used to acquire all data.
The Study of Network Pharmacology
Bioactive Ingredient Identification
The widely used Traditional Chinese Medicine System Pharmacology database and Analysis Platform (Ru et al., 2014) (TCMSP, https://tcmspw.com/tcmsp.php) was utilized to screen the active ingredients within ZX, BZ, and ZXD. In total, 101 herbal ingredients were identified using the keywords “Alisma Orientale (Sam.) Juzep.” and “Atractylodes Macrocephala Koidz.”. The potential active ingredients were then selected using an oral bioavailability (OB) threshold of ≥30% and a drug-likeness (DL) threshold of ≥0.18 (Leeson and SPRINGTHORPE, 2007; Xu et al., 2012). In addition, any compounds in ZXD, ZX, or BZ that have previously been reported to be highly pharmacologically active were also included in this analysis (Wu et al., 2021). Two-dimensional (2D) structures of all compounds were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and used for target prediction efforts.
Target Prediction
TheSwissTargetPrediction platform (David et al., 2013) (http://www.swisstargetprediction.ch/index.php) was utilized to identify the putative targets of the compounds retrieved from the PubChem database. The Swiss library contains over 3,000 proteins and 370,000 active ingredients, making it ideal for such efforts. Files of the identified bioactive ingredients with the suffix “SDF” were used as inputs for the SwissTargetPrediction platform, and probable macromolecular targets of these compounds were identified based on structural similarity. Only the SwissTargetPrediction platform was used for this predictive analysis to ensure the clarity of results. The species was set to Homo sapiens for this analysis and all subsequent analyses.
NAFLD-Related Target Identification
Annotated and predicted NAFLD-related targets were identified using the Online Mendelian Inheritance in Man (OMIM, https://omim.org/) (Hamosh et al., 2002) and GeneCards (https://www.genecards.org/) (Stelzer et al., 2016) databases. We used “Non-alcoholic fatty liver” and “Non-alcoholic fatty liver disease” as search terms and integrated the search results to develop a NAFLD-specific disease target database. NAFLD-related targets that were also identified as putative targets of ZXD bioactive ingredients were retained for further analysis.
Network Construction and Enrichment Analyses
Cytoscape 3.7.2 (Shannon et al., 2003) (https://cytoscape.org/) was used to construct a herb-compound-target network, with nodes corresponding to herbs, compounds, and targets and edges representing between-node interactions. Node sizes were scaled based on degree values determined using Cytoscape Network Analyzer such that smaller nodes were associated with smaller degree values.
The potential functional roles of targets overlapping between ZXD and NAFLD target lists were evaluated via Metascape (Zhou et al., 2019) (https://metascape.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, and Gene Ontology (GO) enrichment analyses. All genes were used as the enrichment background for KEGG analyses, with significantly enriched terms meeting the following criteria: count ≥3, p < 0.01, enrichment factor >1.5. These terms were grouped and clustered based on the membership similarity. For GO analyses, the molecular functions (MFs), biological processes (BPs), and cellular components (CCs) for which these targets were enriched were identified and visualized using enrichment plots.
The ZX and BZ herb-compound-target-pathway networks were then plotted based on the results of the above enrichment analyses.
Protein-Protein Interaction Analysis
Protein-protein interaction (PPI) analysis was conducted to identify which ZXD targets are most crucial in the context of disease pathology. STRING 11.0 (Szklarczyk et al., 2017) (https://string-db.org/) database was used to conduct PPI analysis of overlapping ZXD and NAFLD targets and KEGG pathway analysis. For the PPI analysis, active interaction sources were text mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The analysis range was only set to input proteins, and output obtained in the form of files with the suffix “TSV” was visualized using Cytoscape 3.7.2; NetworkAnalyzer was used to assess the degree and combined scores.
All macromolecular targets were retrieved from the Uniprot database (Alex, 2019) (https://www.uniprot.org/) to confirm their gene symbols. The plots described above were designed based on these symbols to ensure statistical normalization.
Experimental Pharmacology Validation
Experimental Design
Seventy-four male SD rats were randomly divided into two groups according to their diets, including blank (n = 13) and model group (n = 61). The blank group rats were fed with normal chew diets (NCD, Beijing Hfk Bioscience Co., Ltd.), and the model group rats were fed with high-fat diets containing 41% fat (HFD, Beijing Hfk Bioscience Co., Ltd.) for 12 weeks to induce NAFLD. After 12 weeks of modeling, one rat liver tissue from each group was used for histopathological evaluation to validate the NAFLD model. Afterwards, a 4-weeks intervention period with ZXD and positive control drug started. Rats from model group were randomly and equally assigned to five subgroups: vehicle, positive control, low-dose, middle-dose and high-dose groups (n = 12 each). The vehicle group and positive control group rats were orally administered distilled water and 10 mg/kg/d PH, respectively. The low-dose, middle-dose and high-dose group rats were orally administered 100, 200 and 400 mg/kg/d ZXD, respectively. The doses of ZXD and PH were selected based on the previous studies (Hong et al., 2010; Gong et al., 2018; Yang et al., 2019). During the 4-week intervention period, the blank group rats were fed with NCD and the remaining five group rats were fed with HFD. Animals were given free access to feed and drink. The animal weights and food consumption were monitored daily. At the end of intervention period, animals were fasted for 16 h and then anesthetized with an intraperitoneal injection of urethane (20%, 1 g/kg). The biological samples (blood, liver and urine) were collected and stored at -80°C to conduct the histopathological evaluation and serum biochemical assay.
Histopathological Evaluation
Hematoxylin and Eosin (HE) Staining
Fresh liver tissues were fixed with 4% paraformaldehyde for 24 h. The fixed tissues were then embedded in paraffin, cut into thin slices and stained with HE. Light microscopy was used to determine the histopathological changes.
Oil Red O (ORO) Staining
Frozen liver tissues were cut into thin sections and then mounted on glass slides. The slides were fixed in pre-cold 4% paraformaldehyde and rinsed with distilled water. The rinsed slides were placed in isopropanol solution and then incubated in ORO solution for 10 min. Afterwards, the ORO solution was removed and the slides were rinsed with running tap water for 5 min. Light microscopy was used to determine the histopathological changes.
Serum Biochemical Assays
The blood samples were collected and left standing for 30 min. The samples were centrifuged for 10 min at 3,000 g to separate the serum for biochemical assays. The total cholesterol (TC), triglycerides (TG), aspartate aminotransferase (AST), alanine aminotransferase (ALT) levels were measured according to the kit instructions.
RT-qPCR
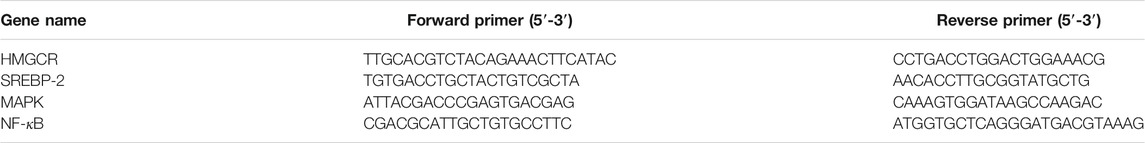
RT-qPCR was then performed to measure the hepatic expression of the predicted NAFLD-related genes. Total RNA was collected with the RNA isolation kit according to the instructions. The purity of the collected RNA was detected by the NanoDrop ND-2000 spectrophotometer, and the 260/280 ratios were calculated. PrimeScript™ RT reagent kit with gDNA eraser was used to carry out the reverse transcription reactions. The Realtime-PCR was performed on the ABI7500 fast system. The calculation of relative gene expression was based on the 2−ΔΔCT method. The primers used in the experiments are listed in Table 1.
Statistical Analysis
Statistical analysis was performed using SPSS 23.0 software, and the results are presented as mean ± SD. One-way analysis of variance was used for comparison, and p-value<0.05 was considered statistically significant. The data graphs were generated using GraphPad Prism 8.
Results
Active Ingredients of ZXD
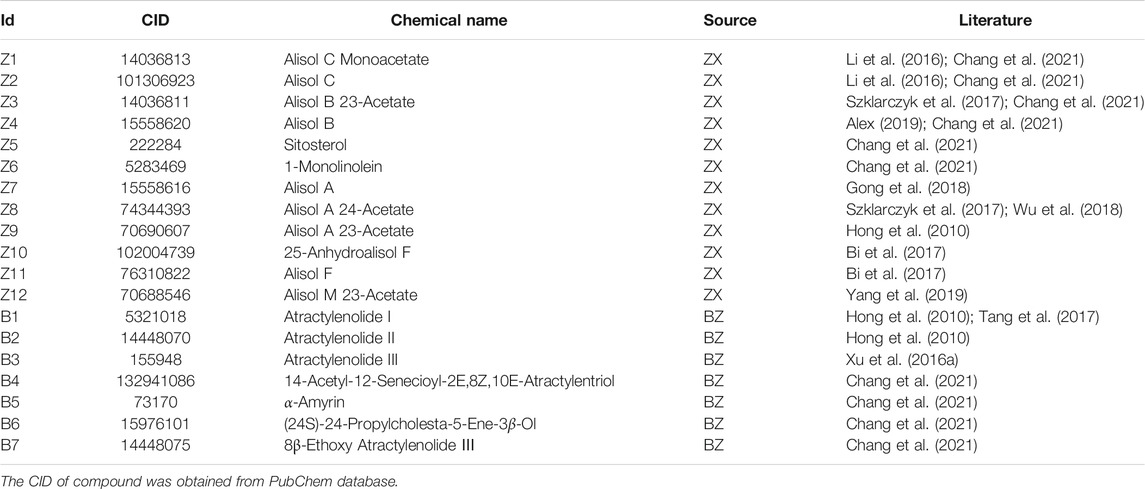
In our initial search of the TCMSP database, we identified 101 active ingredients of potential relevance. Of these, we selected six ingredients in ZX (Z1 to Z6) and four ingredients in BZ (B4 to B7) based upon their OB and DL values. In addition, we selected nine ingredients (Z7 to Z12 and B1 to B3) that have previously been reported to exhibit a high degree of pharmacological activity in ZXD preparations (Lin, 2012; Xu et al., 2016a; Chao et al., 2016; Song et al., 2017; Xu et al., 2018; Ho et al., 2019; Wu et al., 2021). These ingredients were numbered Z1-Z12 and B1-B7, and compound CID was confirmed with the Pubchem database (Table 2).
Identification of ZXD and NAFLD Targets
These 19 active compounds were next introduced into the Swiss platform to identify putative macromolecular targets. We identified 325 ZX targets and 327 BZ targets, and 186 overlapping targets (Supplementary Table 1). In addition, the OMIM and GeneCards databases were used to identify 1575 NAFLD-related targets, of which 122 targets overlapped with our list of ZXD targets (Supplementary Table 2) (Figure 2). These overlapping targets were retained, and corresponding Uniprot IDs were determined and used to identify corresponding gene symbols for each target.
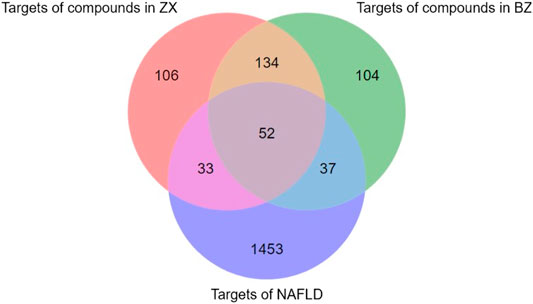
FIGURE 2. The venn diagram of targets in ZX, BZ and NAFLD. Totally 1575 NAFLD-related targets were identified, of which 122 targets overlapped with our list of ZXD targets.
These 19 active ingredients and 122 targets were incorporated into a herbs-compounds-targets network wherein node size was scaled according to the degree values (Figure 3). The targets with the highest degree values included HMGCR, MAPK1, and SREBP-2, suggesting that these targets may play critical roles in the context of NAFLD treatment.
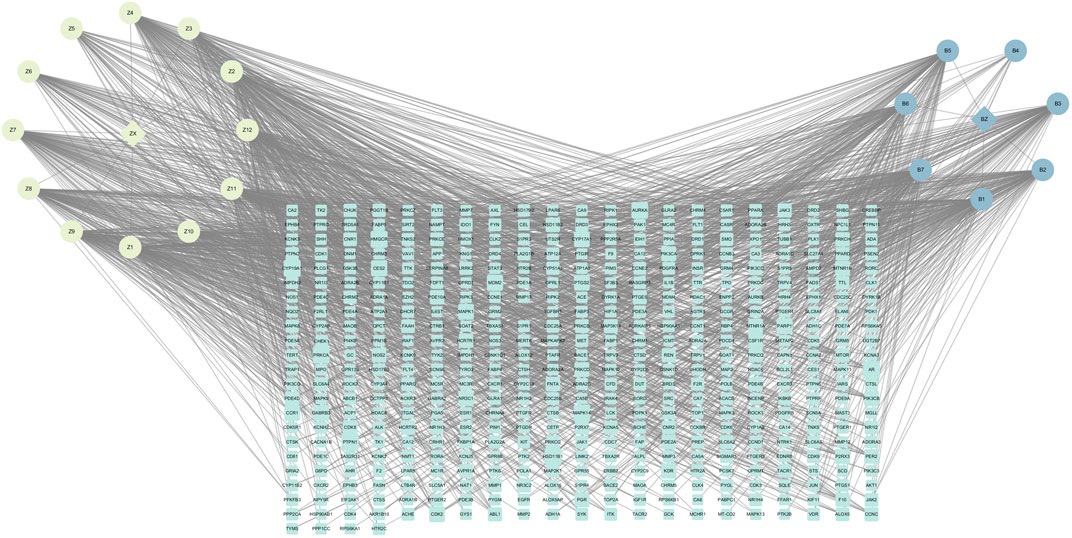
FIGURE 3. The herbs-compounds-targets network of 143 nodes. These 19 active ingredients and 122 targets were incorporated into a herbs-compounds-targets network wherein node size was scaled according to the degree values. The light-yellow nodes represent ZX and its compounds, the light blue nodes represent BZ and its compounds and the light green nodes represent the gene symbols. The size of node is proportional to the degree value and the edges represent interactions between the nodes.
Functional Enrichment and PPI Network Analyses
Metascape platform was used to carry out KEGG pathway and GO enrichment analyses of the 122 overlapping ZXD and NAFLD targets. Results were ranked based on ascending q-values, corresponding to the degree of correlation with these targets, and the top 20 of these enriched pathways were plotted (Supplementary Table 3) (Figures 4–6). These targets were enriched for the lipid metabolism GO BP, suggesting that this pathway may be linked to the pharmacological activity of ZXD. Other identified targets associated with this pathway were also retained for further analysis (Supplementary Table 4).
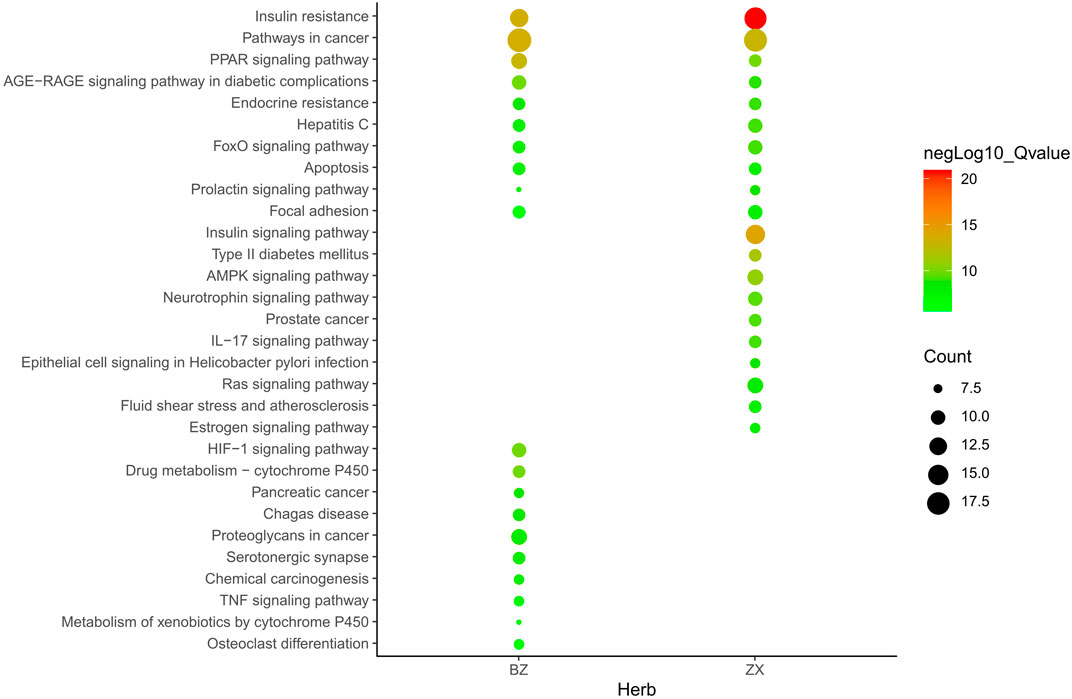
FIGURE 4. The Enrichment plot of KEGG pathway analysis. The size and color of nodes represent the count and–log10(q) values of pathways, respectively.
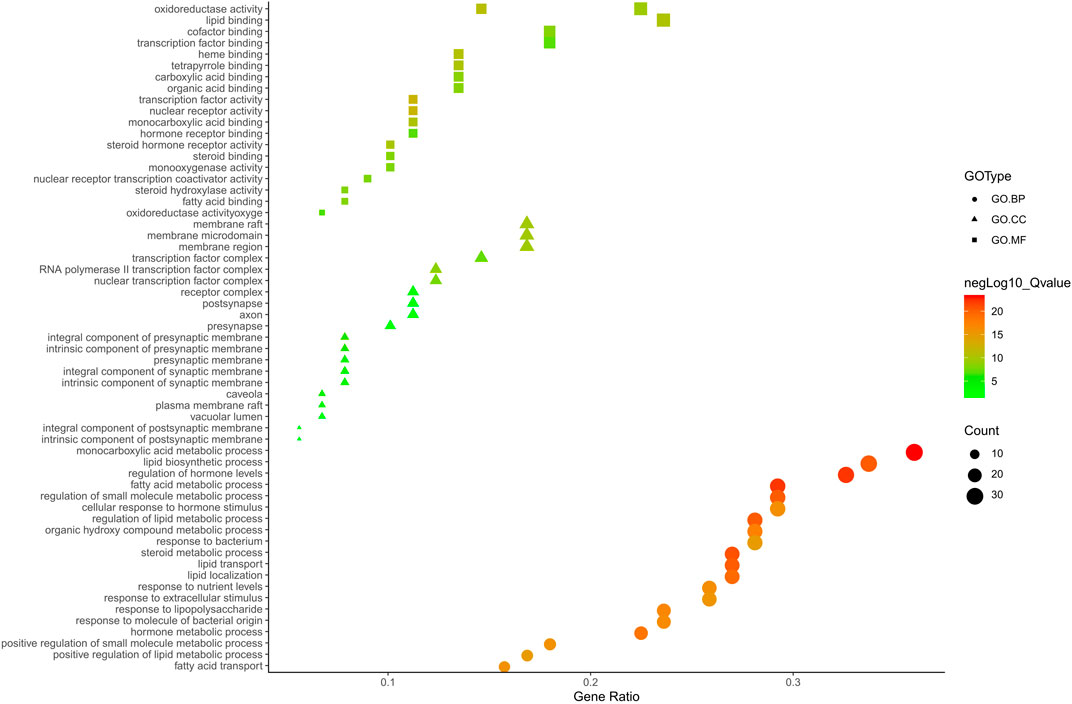
FIGURE 5. The GO. MF, GO. BP and GO. CC enrichment analysis plot of ZX. The size and color of nodes represent the count and–log10(q) values of molecular functions, biological processes and cellular components items, respectively.
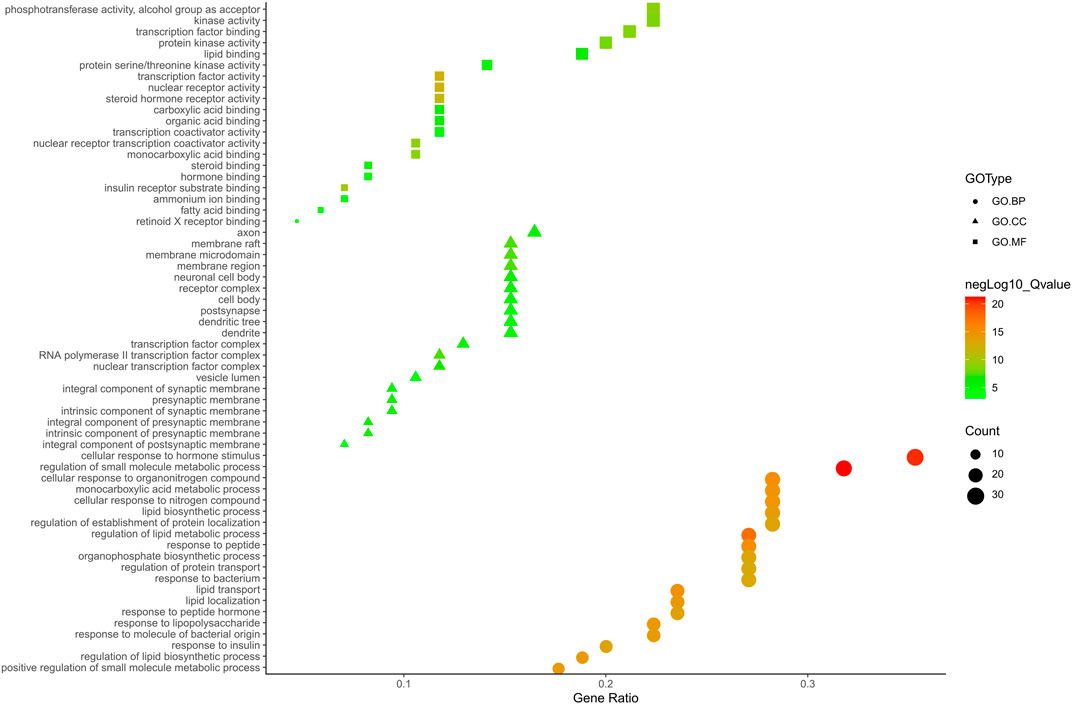
FIGURE 6. The GO. MF, GO. BP and GO. CC enrichment analysis plot of BZ. The size and color of nodes represent the count and–log10(q) values of molecular functions, biological processes and cellular components items, respectively.
KEGG analyses revealed that insulin-related signaling and PI3K/AKT/NF-κB pathways are closely associated with ZXD-mediated NAFLD treatment. Using these results, we constructed a herb-compound-target-pathway network (Figures 7, 8).
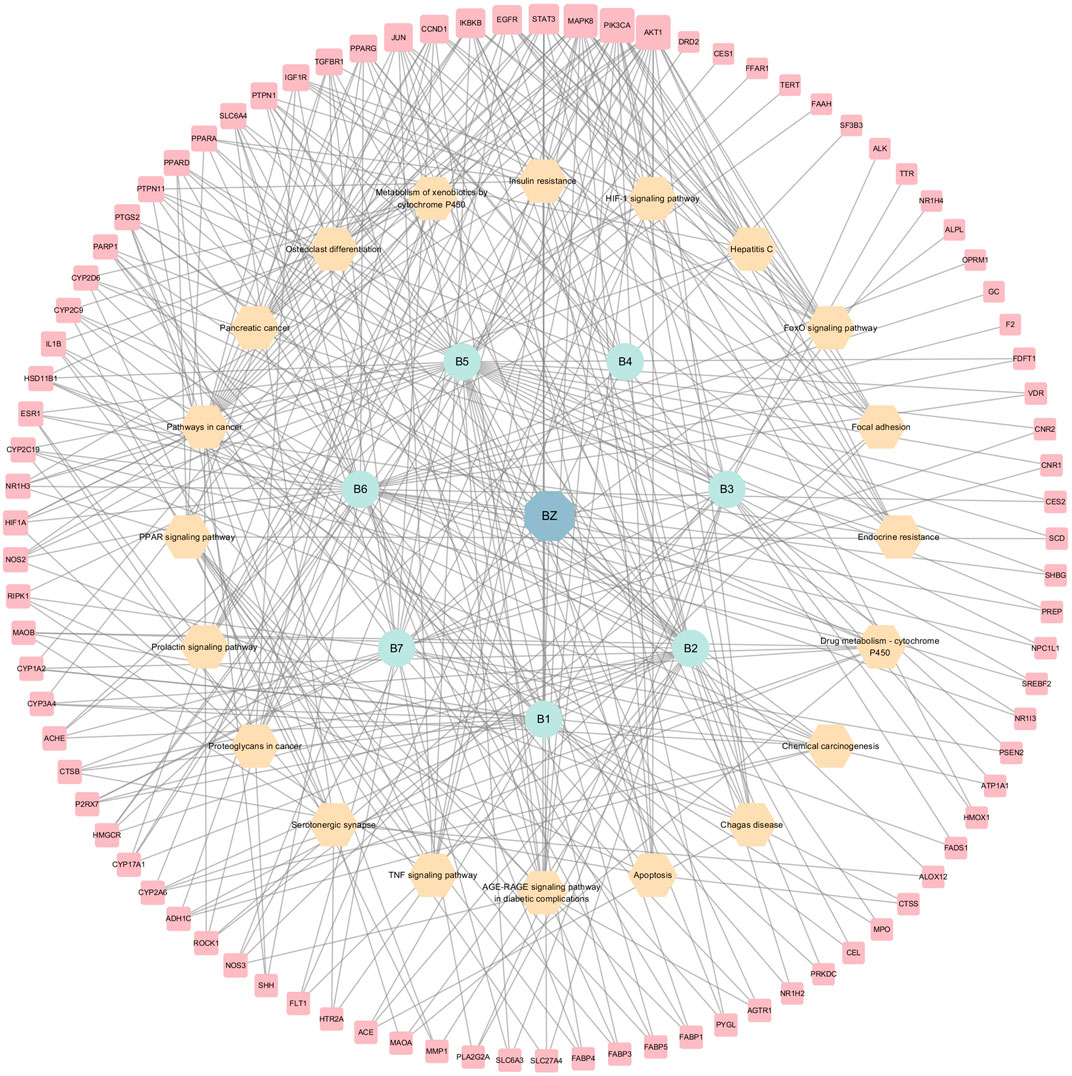
FIGURE 7. The herb-compounds-targets-pathways network of BZ. The light red nodes represent gene symbols, the light-yellow nodes represent pathways, the light green nodes represent compounds and the light blue nodes represent BZ. The size of node of gene symbol is proportional to the degree value and the edges represent interactions between the nodes.
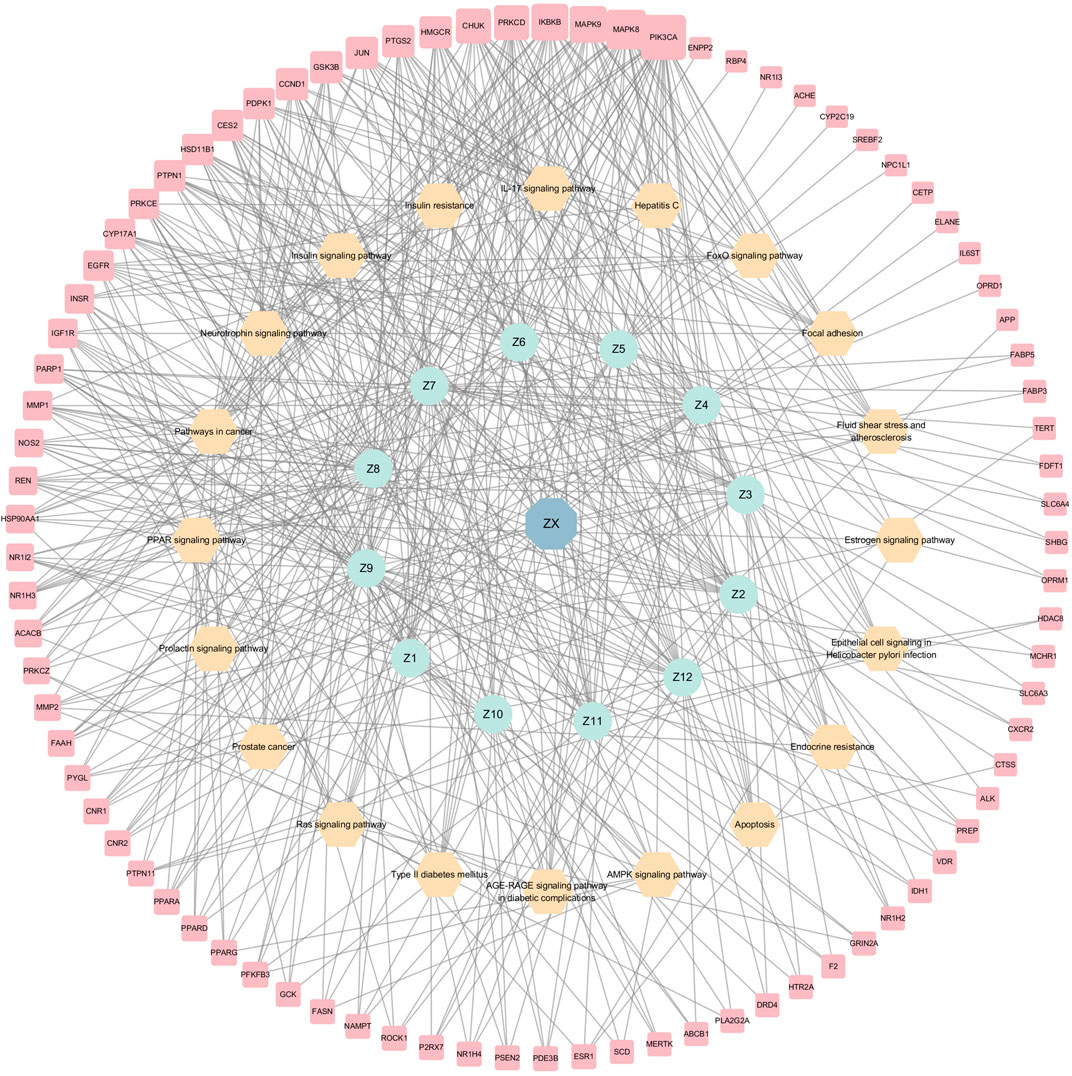
FIGURE 8. The herb-compounds-targets-pathways network of ZX. The light red nodes represent gene symbols, the light-yellow nodes represent pathways, the light green nodes represent compounds and the light blue nodes represent ZX. The size of node of gene symbol is proportional to the degree value and the edges represent interactions between the nodes.
To explore the interactions among the above targets in-depth, we constructed a PPI network using the STRING database with a medium confidence interaction threshold (0.4). Targets were then scored based on these predicted interactions and ranked in ascending order, retaining the targets that exhibited a combined score of ≥0.90 for further analysis (Supplementary Table 5). The degree values, centrality, and other parameters associated with these targets were then calculated using the Cytoscape Network Analyzer tool (Supplementary Table 6; Figures 9, 10). The data revealed precise, relevant interactions between these targets and highlighted the PI3K/AKT/NF-κB pathway as an essential target worthy of further investigation in the context of NAFLD treatment using ZXD. When all 122 overlapping targets were plotted, the resultant data also suggested that mitogen-activated protein kinases (MAPKs), fatty acid-binding proteins (FABPs), and matrix metalloproteinases (MMPs) may warrant future investigation in a therapeutic context.

FIGURE 9. The PPI network of all matched targets. The size of nodes represents their degree values and the width of edges represents interactions between the nodes. The color represents their degree values and combined scores, which changes from blue to orange represent values from low to high.
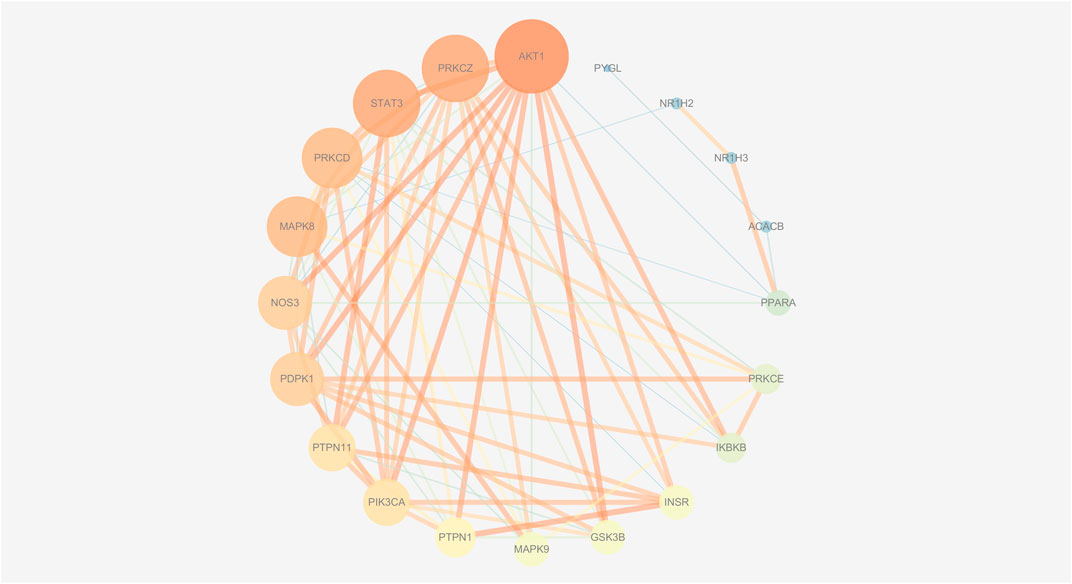
FIGURE 10. The PPI network of insulin-related targets. The size of nodes represents their degree values and the width of edges represents interactions between the nodes. The color represents their degree values and combined scores, which changes from blue to orange represent values from low to high.
Experimental Validation of Network Pharmacology
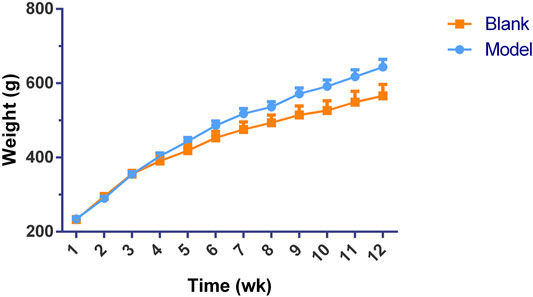
Significant differences (p < 0.01) in body weights were observed between the model and blank group rats that were fed HFD and NCD for 12 weeks, respectively (Figure 11). Furthermore, the HE-stained liver tissues of the model group rats revealed significant differences compared with the tissues of the blank group rats (Figure 12 A-B; Figures 13A,B). Specifically, liver tissue sections from the blank group rats had intact, clear boundaries; The hepatocytes had a cord-like arrangement and were radially distributed around the central vein, and the cell nuclei was clearly visible. In contrast, swelling of hepatocyte, deposition of big lipid drops and hepatic ballooning degeneration were observed in liver tissue sections from the model group rats. The differences implied that the NAFLD modeling was successful. Afterwards, we treated NAFLD rats with the ethanol extract of ZXD across a range of concentrations (100, 200 and 400 mg/kg/d, respectively) for 4 weeks, and PH (10 mg/kg/d) was used as a positive control. We observed significant differences in the histopathological evaluation (Figure 12 C-F; Figures 13C–F) and serum biochemical assay (Figure 14) data of different groups. The curative effect of ZXD on NAFLD was found to be dose-dependent in rats at concentrations 100, 200 and 400 mg/kg/d.
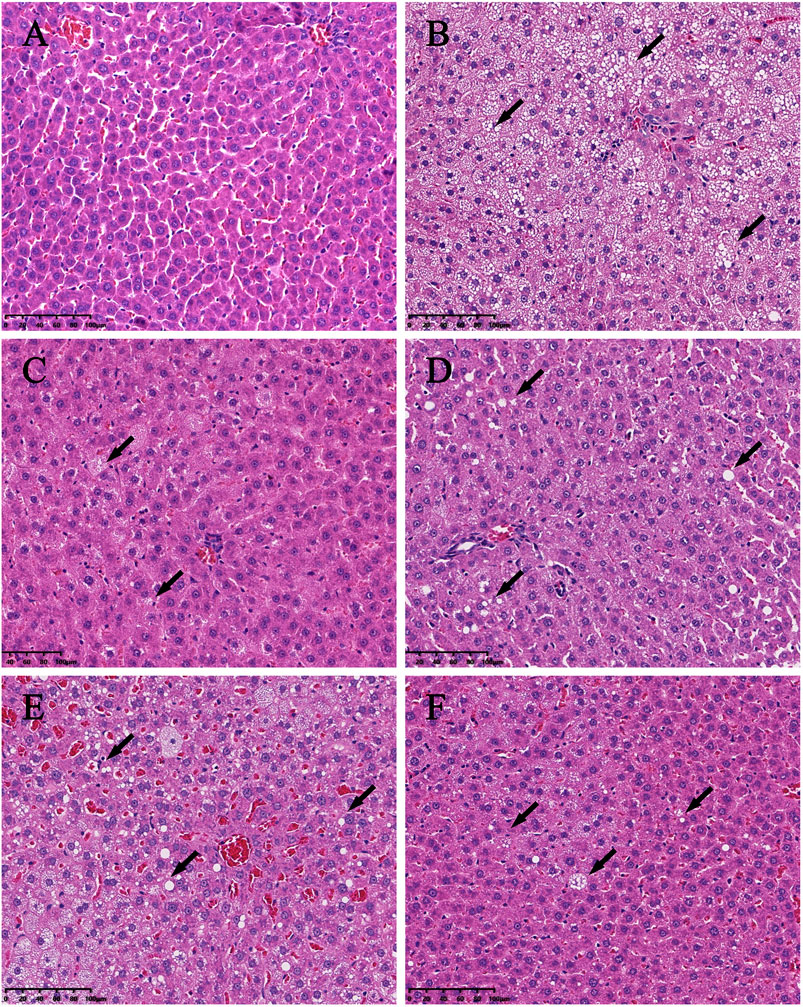
FIGURE 12. Histopathologic evaluation of liver tissues by HE staining (200×). (A) blank; (B) model; (C) positive control; (D) high-dose; (E) middle-dose; (F) low-dose. The lipid drops are indicated by arrows.
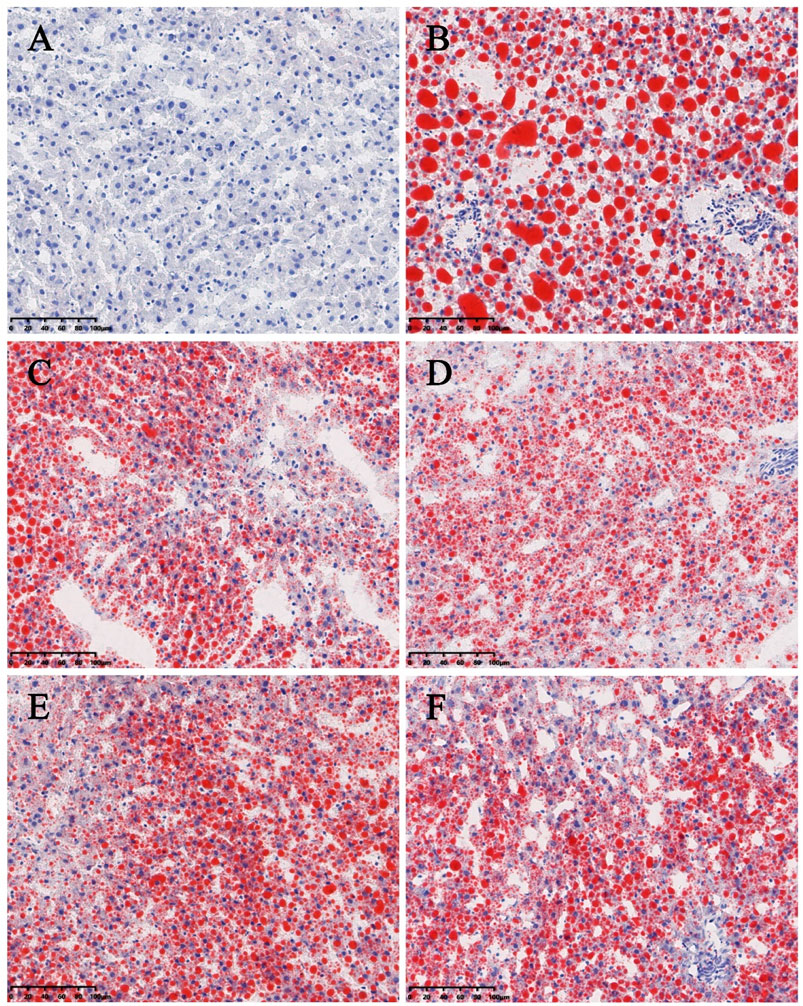
FIGURE 13. Histopathologic evaluation of liver tissues by ORO staining (200×). (A) blank; (B) model; (C) positive control; (D) high-dose; (E) middle-dose; (F) low-dose. Compared with the model group rats, the treated group rats reduced accumulation of lipid droplet remarkably.
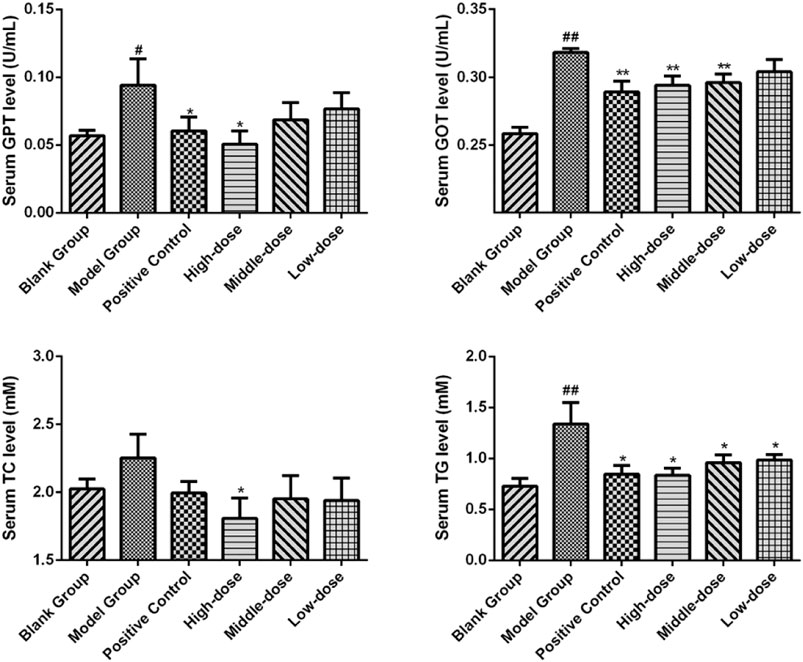
FIGURE 14. The serum biochemical parameter of rats after the 4-weeks intervention period. *p < 0.05 and **p < 0.01 vs. model group rats; #p < 0.05 and ##p < 0.01 vs. blank group rats.
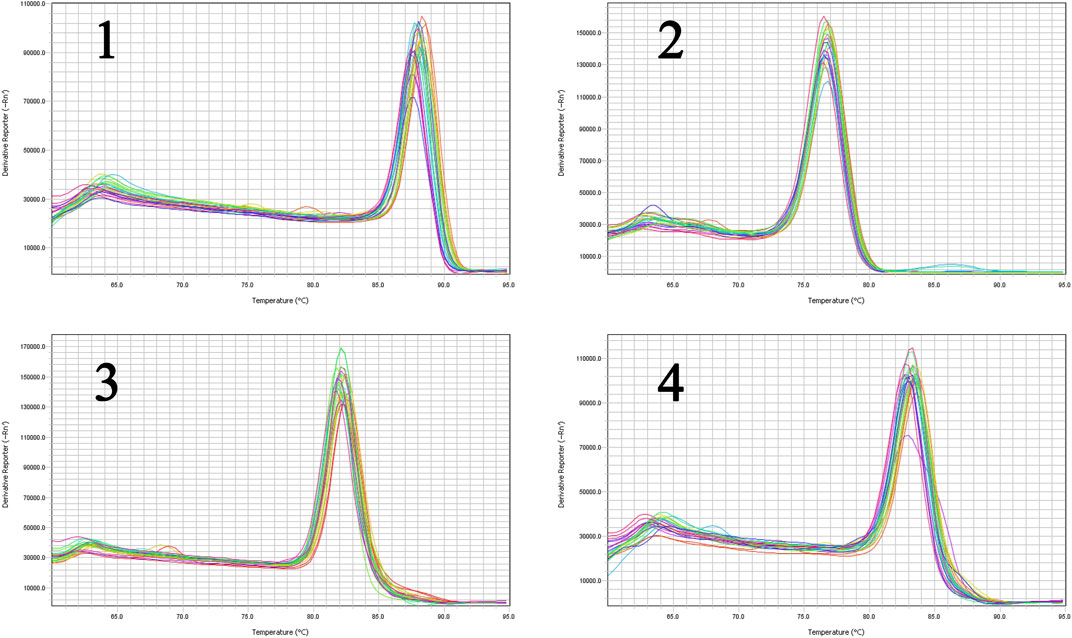
The liver tissues of blank, model and high-dose group rats were further conducted RT-qPCR. The results indicated that the gene expression of SREBP-2 and HMGCR of high-dose group rats decreased significantly (p < 0.05) in comparison with the blank group. The expression of MAPK1 and NF-κBp65 demonstrated an attenuation trend, which was however not significant (Figure 15). Figures 16, 17 show the amplification and melting curves of these four genes (Figures 16, 17).
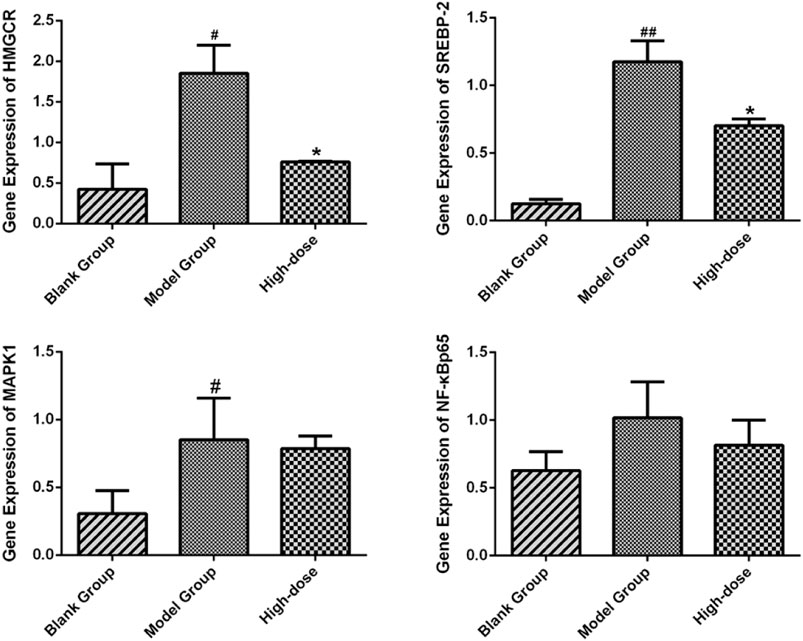
FIGURE 15. Expression of SREBP-2, HMGCR, NF-κB and MAPK1 in SD rats. *p < 0.05 and **p < 0.01 vs. model group rats; #p < 0.05 and ##p < 0.01 vs. blank group rats.
FIGURE 16. The amplification curve of the four major genes (1, SREBP-2; 2, HMGCR; 3, MAPK1; 4, NF-κB).
Discussion
Identification of NAFLD-Related Targets of ZXD Treatment
ZXD is one of the TCMFs that exhibits anti-inflammatory and lipid-lowering properties. ZXD has been used to treat NAFLD because of hyperlipidemia- and inflammation-related etiology of this disease. However, the multi-component multi-target nature of ZXD and other TCMFs has made it difficult to firmly establish the molecular mechanisms underlying their pharmacological activity. In our previous study, we used LC-MS to conduct ZXD quality control analysis (Chang et al., 2021). This analysis, together with a review of relevant pharmacologically active compounds documented in the literature, revealed that alisol A (Ho et al., 2019), alisol A 24-acetate (Li et al., 2016), alisol A 23-acetate (Lin, 2012), 25-anhydroalisol F (Bi et al., 2017), alisol F (Bi et al., 2017), and alisol M 23-acetate (Xu et al., 2018) in ZX and atractylenolide I (Chao et al., 2016; Tang et al., 2017), atractylenolide II (Chao et al., 2016), and atractylenolide III (Song et al., 2017) in BZ are highly potent despite not meeting the predictive thresholds of the TCMSP platform (OB ≥ 30% and DL ≥ 0.18). As such, OB and DL alone are not sufficient to predict the active compounds within TCMFs.
The Swiss Target Platform was then used to identify putative NAFLD-related target genes. Further, we constructed a herb-compound-target network that identified HMGCR, MAPK1, and SREBP-2 as targets with high degree values that may be relevant in the context of ZXD-mediated NAFLD treatment.
Targets Selected for Validation
HMGCR is the rate-limiting enzyme that regulates cholesterol synthesis. Patients with NAFLD are reported to have high serum levels of total cholesterol (TC) (Puri et al., 2007; Caballero et al., 2009). The gene expression of HMGCR, however, showed an abnormally elevated serum level of TC. The expression of HMGCR is regulated by SREBP, especially by SREBP-2, a transcription factor primarily involved in the synthesis of cholesterol and fatty acid (Brown and GOLDSTEIN, 1997; Wu et al., 2013). Therefore, we hypothesized that HMGCR and SREBP-2 induce the onset and development of NAFLD by affecting the serum level of TC. There are reports that ZXD suppresses the expression of HMGCR and SREBP-2 (Zheng and WU, 2010; Dan et al., 2011). Further, the major pharmacological components of ZXD - alisol B 23-acetate and alisol A 24-acetate - could bind to HMGCR competitively (Xu et al., 2016b).
MAPK signaling plays a central role in many physiological and pathological processes associated with inflammation and NAFLD (Cao et al., 2017; Jiang et al., 2019). MAPK1, MAPK8, and MAPK9 were high-scoring targets in our analysis and may thus be associated with NAFLD treatment. The pharmacologically active components of BZ are reported to modulate MAPK signaling (Huang et al., 2014). For instance, atractylenolide I inhibits MAPK by suppressing TLR4/MyD88 pathway activity, while atractylenolide II and III suppress inflammation by affecting MAPK phosphorylation.
PI3K/AKT/NF-κB pathways were found to be closely associated with ZXD-mediated NAFLD treatment in PPI network analyses. Since many studies (Wang et al., 2014; Fan et al., 2017) have confirmed the relationship between NAFLD and the PI3K/AKT/NF-κB pathway, NF-κBp65, the critical target of the PI3K/AKT/NF-κB signaling pathway, was selected to validate our findings. Furthermore, HMGCR, MAPK1, SREBP-2, and NF-κBp65 were selected for further validation through RT-qPCR.
RT-qPCR Experimental Validation Using in vivo NAFLD Model in SD Rats
The NAFLD model in rats was established using HFD and the success of modeling was verified by comparing changes in body weight and liver tissue section. The curative effect of ZXD was evaluated by histopathological evaluation and serum biochemical assays, and ZXD ameliorated lipid accumulation and treated NAFLD in a dose-dependent manner.
For validation purposes, RT-qPCR was performed to test the expression of HMGCR, MAPK1, SREBP-2, and NF-κBp65. ZXD significantly attenuated the expression of HMGCR and SREBP-2 (p < 0.05). The expression of MAPK1 and NF-κBp65 demonstrated an attenuation trend, which was however not significant (p > 0.05). The results indicated that our network pharmacology-based prediction model of molecular mechanism is reliable to some extent.
In future studies, we plan to perform a more in-depth investigation of the molecular mechanisms of ZXD based on the gut-liver axis to study the relationship between gut microbiota and NAFLD, using many novel databases of gut microbiota.
Conclusion
In conclusion, we constructed a herb-compound-target-pathway network. Using biological enrichment analyses, we determined that ZXD can treat NAFLD in part by modulating the key targets - HMGCR, SREBP-2, MAPK1, and NF-κBp65. The experimental validation was performed through NAFLD model in SD rats. RT-qPCR analyses revealed that ZXD could significantly attenuated the expression of the four genes. The validation results indicated that our network pharmacology-based prediction model of molecular mechanism is reliable, which can be utilized for pilot mechanism investigation of TCMFs.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
YS provided the way of thinking. JW and FZ completed the design of framework of this manuscript. JW conducted the network pharmacology study. JW, FZ and HR completed the animal experiments. XC, JW and ZL investigated the background of the present review. XC, YS and WJ were responsible for supervision, review and editing. JW wrote the manuscript and FZ was responsible for visualization.
Funding
This work was financially supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) (grant No. 2016-I2M-1-012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.722016/full#supplementary-material
References
Alex, B. (2019). UniProt: a Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 47 (D1), D506–D515. doi:10.1093/nar/gky1049
Angulo, P., Kleiner, D. E., Dam-Larsen, S., Adams, L. A., Bjornsson, E. S., Charatcharoenwitthaya, P., et al. (2015). Liver Fibrosis, but No Other Histologic Features, Is Associated with Long-Term Outcomes of Patients with Nonalcoholic Fatty Liver Disease[J]. Gastroenterology 149 (2), 278–281. doi:10.1053/j.gastro.2015.04.043
Bi, X., Wang, P., Ma, Q., Han, L., Wang, X., Mu, Y., et al. (2017). Anti-Inflammatory Activities and Liver Protection of Alisol F and 25-Anhydroalisol F through the Inhibition of MAPK, STAT3, and NF-κB Activation In Vitro and In Vivo. Molecules 22 (6), 951. doi:10.3390/molecules22060951
Brown, M. S., and Goldstein, J. L. (1997). The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor. Cell 89 (3), 331–340. doi:10.1016/s0092-8674(00)80213-5
Caballero, F., Fernández, A., De Lacy, A. M., Fernández-Checa, J. C., Caballería, J., and García-Ruiz, C. (2009). Enhanced Free Cholesterol, SREBP-2 and StAR Expression in Human NASH. J. Hepatol. 50 (4), 789–796. doi:10.1016/j.jhep.2008.12.016
Cao, Q., Guo, J., Qian, D., Ma, H., Peng, Y., Shang, E., et al. (2017). Liposome Encapsulation Attenuated Venenum Bufonis Induced Vascular Irritation in Rabbit Ear Vein via Regulating TLR/MAPK/NF-κB Pathway. RSC Adv. 7 (44), 27431–27440. doi:10.1039/c7ra01820g
Chang, X. Y., Li, Z. Z., Wu, J. S., Jin, W. Y., Shi, Y., et al. (2021). Identification of the Triterpenoids in the Extracts of ZeXie Decoction and Investigation of Their Effects on Nonalcoholic Fatty Liver[J]. Prog. Mod. Biomed. 21 (1), 1–12.
Chao, C. L., Huang, H. C., Lin, H. C., Chang, T. C., and Chang, W. L. (2016). Sesquiterpenes from Baizhu Stimulate Glucose Uptake by Activating AMPK and PI3K. Am. J. Chin. Med. 44 (5), 963–979. doi:10.1142/S0192415X16500531
Dan, H., Wu, J., Peng, M., Hu, X., Song, C., Zhou, Z., et al. (2011). Hypolipidemic Effects of Alismatis Rhizome on Lipid Profile in Mice Fed High-Fat Diet. Saudi Med. J. 32 (7), 701–707.
David, G., Olivier, M., and Vincent, Z. (2013). Shaping the Interaction Landscape of Bioactive Molecules[J]. Bioinformatics 29 (23), 3073–3079.
Fan, H., Ma, X., Lin, P., Kang, Q., Zhao, Z., Wang, L., et al. (2017). Scutellarin Prevents Nonalcoholic Fatty Liver Disease (NAFLD) and Hyperlipidemia via PI3K/AKT-dependent Activation of Nuclear Factor (Erythroid-Derived 2)-Like 2 (Nrf2) in Rats. Med. Sci. Monit. 23, 5599–5612. doi:10.12659/msm.907530
Gong, J., Ding, Y., Gan, Z. Y., Wang, H. W., Zhu, S. M., Li, H. Q., et al. (2018). Therapeutic Effects of Alisma Orientale Extract on Nonalcoholic Fatty Liver Disease in Rats and its Mechanism[J]. Chin. J. Comp. Med. 7 (28), 68–76.
Hamosh, A., Scott, A. F., Amberger, J., Bocchini, C., Valle, D., and Mukusick, V. A. (2002). Online Mendelian Inheritance in Man (OMIM), a Knowledgebase of Human Genes and Genetic Disorders[J]. Nucleic Acids Res. 30 (1), 52–55. doi:10.1093/nar/30.1.52
Ho, C., Gao, Y., Zheng, D., Liu, Y., Shan, S., Fang, B., et al. (2019). Alisol A Attenuates High-Fat-Diet-Induced Obesity and Metabolic Disorders via the AMPK/ACC/SREBP-1c Pathway. J. Cell Mol Med 23 (9945), 5108–5118. doi:10.1111/jcmm.14380
Hoang, L. S., Tran, M. H., Lee, J. S., Ngo, Q. M., Woo, M. H., and Min, B. S. (2016). Inflammatory Inhibitory Activity of Sesquiterpenoids from Atractylodes Macrocephala Rhizomes. Chem. Pharm. Bull. (Tokyo) 64 (5), 507–511. doi:10.1248/cpb.c15-00805
Hong, X., Tang, H., Wu, L., and Li, L. (2010). Protective Effects of the Alisma Orientalis Extract on the Experimental Nonalcoholic Fatty Liver Disease. J. Pharm. Pharmacol. 58 (10), 1391–1398. doi:10.1211/jpp.57.10.0013
Huang, J., Guo, W., Cheung, F., Tan, H. Y., Wang, N., and Feng, Y. (2020). Integrating Network Pharmacology and Experimental Models to Investigate the Efficacy of Coptidis and Scutellaria Containing Huanglian Jiedu Decoction on Hepatocellular Carcinoma. Am. J. Chin. Med. 48 (1), 161–182. doi:10.1142/S0192415X20500093
Huang, J. M., Zhang, G. N., Shi, Y., Zha, X., Zhu, Y., Wang, M. M., et al. (2014). Atractylenolide-I Sensitizes Human Ovarian Cancer Cells to Paclitaxel by Blocking Activation of TLR4/MyD88-dependent Pathway. Sci. Rep. 4 (3840), 3840–3847. doi:10.1038/srep03840
Jiang, J., Yan, L., Shi, Z., Wang, L., Shan, L., and Efferth, T. (2019). Hepatoprotective and Anti-inflammatory Effects of Total Flavonoids of Qu Zhi Ke (Peel of Citrus Changshan-Huyou) on Non-alcoholic Fatty Liver Disease in Rats via Modulation of NF-κB and MAPKs. Phytomedicine 64, 153082. doi:10.1016/j.phymed.2019.153082
Lee, A. Y., Park, W., Kang, T. W., Cha, M. H., and Chun, J. M. (2018). Network Pharmacology-Based Prediction of Active Compounds and Molecular Targets in Yijin-Tang Acting on Hyperlipidaemia and Atherosclerosis. J. Ethnopharmacol 221, 151–159. doi:10.1016/j.jep.2018.04.027
Leeson, P. D., and Springthorpe, B. (2007). The Influence of Drug-like Concepts on Decision-Making in Medicinal Chemistry. Nat. Rev. Drug Discov. 6 (11), 881–890. doi:10.1038/nrd2445
Li, X. Y., Wang, X. H., Huang, X. Q., Wu, T. T., Xu, W., and Wu, S. S. (2016). Development of a Cell-Based Peroxisome Proliferator-Activated Receptors (PPARs) Screening Model and its Application for Evaluation of Triterpenoids Isolate from Alismatis Rhizoma. Zhongguo Zhong Yao Za Zhi 41 (21), 4015–4022. doi:10.4268/cjcmm20162121
Lin, H. R. (2012). Triterpenes from Alisma Orientalis Act as Farnesoid X Receptor Agonists. Bioorg. Med. Chem. Lett. 22 (14), 4787–4792. doi:10.1016/j.bmcl.2012.05.057
Liu, S. S., Sheng, W. L., Li, Y., Zhang, S. S., Zhu, J. J., Gao, H. M., et al. (2019). Chemical Constituents from Alismatis Rhizoma and Their Anti-inflammatory Activities In Vitro and In Vivo. Bioorg. Chem. 92, 103226. doi:10.1016/j.bioorg.2019.103226
Puri, P., Baillie, R. A., Wiest, M. M., Mirshahi, F., Choudhury, J., Cheung, O., et al. (2007). A Lipidomic Analysis of Nonalcoholic Fatty Liver Disease. Hepatology 46 (4), 1081–1090. doi:10.1002/hep.21763
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a Database of Systems Pharmacology for Drug Discovery from Herbal Medicines. J. Cheminform 6 (1), 13. doi:10.1186/1758-2946-6-13
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Song, C., Huang, X., Lu, K., Peng, M., Yu, S., and Fang, N. (2014). The Rationality of the Hypolipidemic Effect of Alismatis Rhizoma Decoction, a Classical Chinese Medicine Formula in High-Fat Diet-Induced Hyperlipidemic Mice. Iran J. Pharm. Res. 13 (2), 641–649.
Song, M. Y., Jung, H. W., Kang, S. Y., and Park, Y. K. (2017). Atractylenolide III Enhances Energy Metabolism by Increasing the SIRT-1 and PGC1α Expression with AMPK Phosphorylation in C2C12 Mouse Skeletal Muscle Cells. Biol. Pharm. Bull. 40 (3), 339–344. doi:10.1248/bpb.b16-00853
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinformatics 54 (1), 1–33. doi:10.1002/cpbi.5
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 45 (Database issue), D362–D368. doi:10.1093/nar/gkw937
Tandon, R. K. (2013). Emergence of Non-alcoholic Fatty Liver Disease (NAFLD). J. Assoc. Physicians India 61 (7), 445–446.
Tang, X. M., Liao, Z. K., Huang, Y. W., Lin, X., and Wu, L. C. (2017). Atractylenolide Ⅰ Protects against Lipopolysaccharide-Induced Disseminated Intravascular Coagulation by Anti-inflammatory and Anticoagulation Effect. Asian Pac. J. Trop. Med. 10 (6), 582–587. doi:10.1016/j.apjtm.2017.06.007
Than, N. N., and Newsome, P. N. (2015). A Concise Review of Non-alcoholic Fatty Liver Disease[J]. Atherosclerosis 239 (1), 192–202. doi:10.1016/j.atherosclerosis.2015.01.001
Wang, C., Chi, Y., Li, J., Miao, Y., Li, S., Su, W., et al. (2014). FAM3A Activates PI3K p110α/Akt Signaling to Ameliorate Hepatic Gluconeogenesis and Lipogenesis. Hepatology 59 (5), 1779–1790. doi:10.1002/hep.26945
Wu, C., Jing, M., Yang, L., Jin, L., Ding, Y., Lu, J., et al. (2018). Alisol A 24-acetate Ameliorates Nonalcoholic Steatohepatitis by Inhibiting Oxidative Stress and Stimulating Autophagy through the AMPK/mTOR Pathway. Chem. Biol. Interact 291, 111–119. doi:10.1016/j.cbi.2018.06.005
Wu, J. S., Chang, X. Y., Li, Z. Z., Jin, W. Y., Shi, Y., et al. (2021). Research Progress on Molecular Mechanism of Lipid-Lowering and Anti-inflammatory Effect of Zexietang[J]. Chin. J. Exp. Traditional Med. Formulae 27 (2), 224–232.
Wu, N., Sarna, L. K., Hwang, S. Y., Zhu, Q., Wang, P., Siow, Y. L., et al. (2013). Activation of 3-Hydroxy-3-Methylglutaryl Coenzyme A (HMG-CoA) Reductase during High Fat Diet Feeding. Biochim. Biophys. Acta 1832 (10), 1560–1568. doi:10.1016/j.bbadis.2013.04.024
Xu, F., Lu, C., Wu, Q., Gu, W., Chen, J., Fang, F., et al. (2018). Studies on the Lipid-Regulating Mechanism of Alisol-Based Compounds on Lipoprotein Lipase. Bioorg. Chem. 80 (2018), 347–360. doi:10.1016/j.bioorg.2018.07.001
Xu, F., Yu, H., Lu, C., Chen, J., and Gu, W. (2016). The Cholesterol-Lowering Effect of Alisol Acetates Based on HMG-CoA Reductase and its Molecular Mechanism[J]. Evid.-based Complement. Altern. Med. 2016 (6), 1–11.
Xu, F., Yu, H., Lu, C., Chen, J., and Gu, W. (2016). The Cholesterol-Lowering Effect of Alisol Acetates Based on HMG-CoA Reductase and its Molecular Mechanism[J]. Evidence-Based Complement. Altern. Med. 2016, 1–11.
Xu, X., Zhang, W., Huang, C., Li, Y., Yu, H., Wang, Y., et al. (2012). A Novel Chemometric Method for the Prediction of Human Oral Bioavailability. Int. J. Mol. Sci. 13 (6), 6964–6982. doi:10.3390/ijms13066964
Yang, J. M., Sun, Y., Wang, M., Zhang, X. L., Zhang, S. J., Gao, Y. S., et al. (2019). Regulatory Effect of a Chinese Herbal Medicine Formula on Non-alcoholic Fatty Liver Disease. World J. Gastroenterol. 25 (34), 5105–5119. doi:10.3748/wjg.v25.i34.5105
Zhang, R., Zhu, X., Bai, H., and Ning, K. (2019). Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 10, 123. doi:10.3389/fphar.2019.00123
Zheng, C., and Wu, S. (2010). Screening Lipid-Reducing Active Components from Rhizoma Alismatis by Computer Simulation[J]. Rehabil. Med. 20 (3), 34–36.
Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A. H., Tanaseichuk, O., et al. (2019). Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 10 (1), 1523. doi:10.1038/s41467-019-09234-6
Keywords: ZeXie decoction, network pharmacology, RT-qPCR analysis, non-alcoholic fatty liver disease, integrating strategy
Citation: Wu J, Zhang F, Ruan H, Chang X, Wang J, Li Z, Jin W and Shi Y (2021) Integrating Network Pharmacology and RT-qPCR Analysis to Investigate the Mechanisms Underlying ZeXie Decoction-Mediated Treatment of Non-alcoholic Fatty Liver Disease. Front. Pharmacol. 12:722016. doi: 10.3389/fphar.2021.722016
Received: 08 June 2021; Accepted: 26 August 2021;
Published: 09 September 2021.
Edited by:
Luca Rastrelli, University of Salerno, ItalyReviewed by:
Ina Yosifova Aneva, Bulgarian Academy of Sciences, BulgariaWan-Wan Lin, National Taiwan University, Taiwan
Copyright © 2021 Wu, Zhang, Ruan, Chang, Wang, Li, Jin and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Shi, c2hpeXVlMTAyOUAxMjYuY29t
 Jiashuo Wu
Jiashuo Wu Fangqing Zhang1
Fangqing Zhang1