
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 26 July 2021
Sec. Pharmacogenetics and Pharmacogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.713178
This article is part of the Research Topic Pharmacogenomics: From Bench to Bedside and Back Again View all 8 articles
A correction has been applied to this article in:
Corrigendum: Validation of single nucleotide variant assays for human leukocyte antigen haplotypes HLA-B*15:02 and HLA-A*31:01 across diverse ancestral backgrounds
The human leukocyte antigen haplotypes HLA-B*15:02 and HLA-A*31:01 have been linked to life-threatening adverse drug reactions to the anticonvulsants carbamazepine and oxcarbazepine. Identification of these haplotypes via pharmacogenetic techniques facilitates implementation of precision medicine to prevent such reactions. Using reference samples from diverse ancestral origins, we investigated the test analytical validity (i.e., ability to detect whether or not the haplotypes were present or absent) of TaqMan assays for single nucleotide variants previously identified as potentially being able to “tag” these haplotypes. A TaqMan custom assay for rs10484555 and an inventoried assay for rs17179220 and were able to identify with 100% sensitivity and 100% specificity HLA-B*15:02 and HLA-A*31:01 respectively. A custom assay for rs144012689 that takes into account a neighboring single nucleotide variant with manual calling was also able to identify HLA-B*15:02 with 100% sensitivity and 100% specificity. A custom assay for rs1061235 identified HLA-A*31:01 with 100% sensitivity and 95% specificity. The slight reduction in specificity for the latter was owing to another haplotype (HLA-A*33:03) also being detected. While any positive call using the rs1061235 assay could therefore be further investigated, as the presence of the HLA-A*31:01 haplotype confers adverse drug reaction risk, the absence of false negatives (indexed by sensitivity) is more important than false positives. In summary, we present validated TaqMan assay methodology for efficient detection of HLA haplotypes HLA-B*15:02 and HLA-A*31:01. Our data are relevant for other genotyping technologies that identify, or have the potential to identify, these haplotypes using single nucleotide variants.
Adverse drug reactions (ADRs) pose a significant burden to the healthcare system, comprising a significant portion of hospital admissions, mortality, and overall healthcare costs (Baker et al., 2004; Pirmohamed et al., 2004). Globally, the cost of drug-related morbidity and mortality in 2000 was estimated at $177.4 billion, a figure that nearly doubled the previous 1995 estimate (Ernst and Grizzle, 2001). In addition to the strictly economical cost, the distribution of ADRs across the population highlights disparities within the healthcare system. In a United States study, it was found that older individuals, men, Black persons, and individuals residing in extremely rural areas (counties with less than 10,000 residents) experienced higher mortality associated with ADRs (Shepherd et al., 2012). This indicates the need for action to prevent ADRs to ensure that subsets of our population are not facing disproportionate challenges in their access to and quality of care.
Pharmacogenomics provides the opportunity to prevent ADRs. HLA-A and HLA-B are genes encoding human leukocyte antigens (HLAs), part of the class I major histocompatibility complex (MHC) (Wieczorek et al., 2017). This complex is a group of cell surface proteins involved in the presentation of antigenic peptides to T cells to induce an immune response (Wieczorek et al., 2017). The region that encodes the HLA complex consists of ∼260 genes in a ∼4-Mb span on chromosomal region 6p21.3, base pair positions 29,640,000–33,120,000 from the Genome Reference Consortium Human Build 37, hg1 (Trowsdale and Knight, 2013). The most polymorphic HLA class I and class II proteins are each expressed from three gene regions (MHC class I: HLA-A, -B, -C; MHC class II: (HLA-DR, -DP, -DQ), which are all highly polymorphic (Trowsdale and Knight, 2013). The HLA complex is the most polymorphic human genetic system, with over 8000 functional class I variants (Choo, 2007; Illing et al., 2017). Variations in the HLA genes play an important role in determining susceptibility to infection and to autoimmune disease, and can contribute to the recognition of drugs and their metabolites as foreign antigens by T-cell receptors with a consequent immunological reaction (Fan et al., 2017).
Specifically, the HLA haplotypes HLA-B*15:02 and HLA-A*31:01 are predictive of severe cutaneous reactions to the anticonvulsant drug carbamazepine, also prescribed as a mood stabilizer in psychiatry (Fan et al., 2017; Gui et al., 2018; Carvalho Henriques et al., 2020c). Cutaneous ADRs are one of the most common types of ADRs, typically occur in the first two to three months of drug use, and can range from a mild skin rash to a life-threatening reaction such as Stevens-Johnson syndrome and the more severe form of this, known as toxic epidermal necrolysis (SJS/TEN). SJS/TEN is characterized by rapid blistering, cutaneous and muscle detachment, and has a mortality rate of 10–30% (Fan et al., 2017; Mullan et al., 2019). Performing testing for these haplotypes can therefore be potentially life saving, with a Hong Kong study finding that testing for HLA-B*15:02 reduced the incidence of SJS/TEN induced by carbamazepine from 0.24 to 0% (Chen, Liew, and Kwan, 2014). The HLA-A*31:01 haplotype has a frequency of 2–5% in Northern European populations, with its presence increasing the risk of carbamazepine-induced SJS/TEN from 5 to 26% (Mccormack et al., 2011). As a result, before prescribing carbamazepine, it is advised that a genetic test be performed to ensure the patient does not carry either of these risk haplotypes (Amstutz et al., 2014).
Although genetic testing can greatly improve patient care and reduce the potential for ADRs, the uptake of screening has been limited because of the technical and economic barriers associated with conventional HLA haplotyping methods. Due to the highly polymorphic nature of the complex, the “gold-standard” for HLA haplotyping is sequencing-based methods and other specialized techniques, such as polymerase chain reaction-based sequence-specific priming and microbead hybridization (Dunckley, 2012; Lazaro et al., 2013; Edgerly and Weimer, 2018). However, these strategies are time consuming and expensive; the mean cost for testing in North America is $363.65 USD (Verbelen, Weale, and Lewis, 2017). This may limit the availability of such testing in patient groups in which it should be offered. Current United States Food and Drug Administration (FDA) recommendations for carbamazepine state that “prior to initiating carbamazepine therapy, testing for HLA-B*1502 should be performed in patients with ancestry in populations in which HLA-B*1502 may be present” and that “HLA-B*1502 is found almost exclusively in patients with ancestry across broad areas of Asia” (US National Library of Medicine, 2021). As a result, in many jurisdictions, HLA-B*15:02 testing for hypersensitivity to carbamazepine is offered only to Asians (Fang et al., 2019; Alberta Precision Laboratories, 2021; US National Library of Medicine, 2021).
SNV based methods of genotyping permit high throughput economical typing (Erlichster, Goudey, Skafidas, and Kwan, 2019) and therefore using such to “tag” the HLA haplotypes offers a potential route of making the identification of the at risk haplotypes more widely available (Petersdorf and O’hUigin, 2019). While previous studies had identified potential haplotype tagging SNVs for HLA-B*15:02 and HLA-A*31:01, there is no consensus regarding which SNVs can be used across ancestral groups. In fact, some of the literature is contradictory. For example rs2844682 and rs3909184 have been previously suggested as tag SNVs for HLA-B*15:02 (de Bakker et al., 2006; He et al., 2015), while another study concluded that these SNVs had no clinical value for identifying HLA-B*15:02 carriers (Zhu et al., 2015). In addition, many studies have restricted their populations to individuals from a particular ethnic group (de Bakker et al., 2006; Liu et al., 2015; Maekawa et al., 2015; Zhou et al., 2016; Gui et al., 2018). Further, methodologies vary greatly between studies, with some estimating validity based on databases (Erlichster et al., 2019) and others using genotyping (He et al., 2015; Fang et al., 2019).
A haplotype-tagging SNV method of screening US patients for HLA-B*15:02 without ethnicity-based preselection was able identify more than twice the number of carriers at risk of a carbamazepine-related ADRs than screening patients of Asian ancestry alone (Fang et al., 2019). This supports a prior suggestion that the FDA recommendations be revised to capture the broader range of patients who may be at risk for ADRs in response to carbamazepine (Payne, 2014). The aim of this study was therefore to generate validated SNV assays for the identification of HLA-B*15:02 and HLA-A*31:01 across a range of ancestral groups.
A qualitative literature review was conducted to select four potential tagging SNVs for investigation across ancestral groups (Tables 1, 2). Criteria for candidate tag SNV selection included: recency of the publication, population size, ancestry, method of tag selection, and validation method. No studies were excluded. Two SNVs were selected for each haplotype so that the two could be tested in parallel to determine if one had greater sensitivity and specificity than the other for their respective haplotypes - as a method of confirming prior literature on each SNV whilst allowing for possible novel findings. SNVs rs10484555 and rs144012689 were chosen to tag HLA-B*15:02 and SNVs rs17179220 and rs1061235 were chosen to tag HLA-A*31:01.
The TaqMan method of SNV genotyping (Shen, Abdullah, and Wang, 2009) has been used in other studies on HLA haplotype-tagging SNVs (He et al., 2015; Fang et al., 2019) and was used in this study on a ViiA7 Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer’s protocol, with an increase in the number of PCR amplification cycles from 40 to 50 where necessary. One of the selected SNVs, rs17179220, had an available TaqMan assay (Thermo Fisher Scientific, 2021): assay ID C__33415939_10. Custom assays for the other three were designed (Table 3). For rs1061235 and rs10484555, sequence was obtained from the National Center for Biotechnology Information (NCBI) website and submitted via Thermo Fisher’s custom TaqMan SNP genotyping assay design tool. For rs144012689, design was more complex, owing to the other SNV two base pairs away (rs2596496), which has the potential to interfere with assay performance, as identified by Fang et al. (2019). We requested two assays for the rs144012689, one additionally matching the nucleotide base C at the nearby rs2596496, and one to cover the nucleotide base G at rs2596496.
Samples were genotyped in duplicate, with an in-house automated method of conducting quality control on the data arising from the two technical replicates, and repeats being conducted as necessary. Data were analyzed using QuantStudio real-time PCR software, with manual adjustment of the genotyping result was conducted if required, based on the amplification data, in a manner that we have previously used (Carvalho Henriques et al., 2020a; Carvalho Henriques et al., 2020b). The software is unable to accurately assign calls when the number of clusters of samples in the allelic discrimination plots is greater than three or if there are too few numbers in specific clusters, so manual calls can ensure usable data can still be extracted.
Twenty-nine samples with haplotypes identified by the 1000 Genomes Project (The 1000 Genomes Project Consortium et al., 2015) or the GeT-RM Collaborative Project were purchased from the Coriell Institute for Medical Research. The 1000 Genomes Project determined haplotypes via whole genome sequencing, targeted exome sequencing, and high-density single nucleotide polymorphism (SNP) microarrays. The GeT-RM Collaborative project determined haplotypes via next generation sequencing and compared results to those from sequence-specific oligonucleotide and sequence-specific priming technologies. A further nine samples had haplotypes catalogued in the UCLA International HLA DNA Exchange. This resource determined haplotypes via cross-reference between multiple labs globally, which employed various techniques including sequence-specific oligonucleotide technology and next generation sequencing. All samples had either known ancestral background, race, or region of sample acquisition.
Rs10484555 was identified as a tag SNV for HLA-B*15:02 by two recent studies, one of them being Erlichster et al., described above. They found rs10484555 to have 100% sensitivity and 98.7% specificity for tagging HLA-B*15:02. Liu et al. (2015) identified tag SNVs by sequencing in 880 Han Chinese samples, confirmed the results in 500 additional samples, and reported rs10484555 to have 100% sensitivity and 99.3% specificity for tagging HLA-B*15:02 in this ancestral group.
Rs144012689 was identified as a tag SNV for HLA-B*15:02 by two recent studies. Gui et al. (2018) used sequencing data from 184 Han Chinese samples and confirmed their findings in a reference panel of 10,986 of the same ancestry. They found rs144012689 to have 96.7% sensitivity and 99.3% specificity for tagging HLA-B*15:02 in Han Chinese. Fang et al. (2019) identified rs144012689 as a potential tag for HLA-B*15:02, and tested association in 28,897 individuals in the United States, reporting 100% sensitivity and 99.97% specificity. The specificity was not 100% owing to the presence of rs144012689 in the HLA-B*15:13 haplotype, which is found at approximately 1/10 the frequency of HLA-B*15:02 (only eight identified in the sample of 28,897). This study also designed a TaqMan custom SNP Genotyping Assay for rs144012689, which they found to be 100% concordant with their sequencing results.
Rs31451122, rs3909184, and rs2844682 were also identified from literature as possible tag SNVs for HLA-B*15:02, but were not selected for further analysis. Rs31451122 was identified by a large-scale sequencing study (Zhou, et al., 2016), though samples were only from the Han Chinese population. Rs3909184 was identified by two studies, though one found this SNV to only have a sensitivity of 31.8% and a specificity of 80.4% for the tagging of HLA-B*15:02 (He et al., 2015). Rs2844682 was identified solely in the Han Chinese population, and specific linkage disequilibrium results or tagging ability was not provided (de Bakker et al., 2006).
Rs17179220 was identified by two recent studies as a tag SNV for HLA-A*31:01 (Erlichster et al., 2019; Zhou, et al., 2016). The study by Erlichster et al. used five large HLA reference panels, comprising 16,749 total samples, for tag SNV discovery and then validated all identified SNVs on a set of 955 ancestrally diverse samples from the 1000 Genomes dataset. Their aim was to identify tags that could function across multiple ethnicities; their results of rs17179220 having 100% sensitivity, and 99.6% specificity for tagging HLA-A*31:01 take into account African, admixed American, East Asian, and European populations. The study by Zhou et al. sequenced the entire HLA region in 20,635 individuals of Han Chinese ancestry and found a linkage disequilibrium r2 value of 0.972 between rs17179220 and the HLA-A*31:01 haplotype.
Rs1061235 was identified as a tag SNV for HLA-A*31:01 by three studies. de Bakker et al. (2006) used 361 samples of diverse ancestry from the International HapMap project for tag SNV discovery via PCR sequence-specific oligonucleotide priming (PCR-SSOP), and tested transferability on 996 other samples. Across all ancestral groups in which rs1061235 was identified, the r2 value was 1.000. Thorstensen et al. (2014) used a polymerase chain reaction-restriction fragment length polymorphism assay to investigate rs1061235 as a tag for HLA-A*31:01 in Norwegians. In a set of 204 samples, their assay had 100% sensitivity and 99.5% specificity for tagging HLA-A*31:01. The specificity was not 100% since rs1061235 may also be found on HLA-A*33:01 and HLA-A*33:03. Given the lower frequency of the HLA-A*33 haplotypes as compared to HLA*31:01 in Norwegians, the authors did not view this a major concern. However, these variants are more frequent in North America and the authors suggested that more work would have to be done before adopting rs1061235 as a tag SNV for HLA-A*31:01 in a multi-ethnic population such as Canada. The most recent study (He et al., 2015) tested 160 samples of diverse ancestry using a custom-designed rs1061235 TaqMan SNP Genotyping assay. They found the sensitivity to be 100% and the specificity to be 84% for tagging HLA-A*31:01.
Rs2571375, rs1150738, rs3869066, rs259945, and rs41541222 were also identified from literature as possible tag SNVs for HLA-A*31:01, but were not selected for further analysis. This was primarily due to a lack of data on these SNVs outside of select Asian ancestral groups. Rs2571375 was identified by only one study that was conducted in a Han Chinese population (Gui et al., 2018). Rs1150738, rs3869066, and rs259945 were all identified in one study in a Japanese population (Maekawa et al., 2015). Rs41541222 was identified by one study in an Asian population in 2012, but has not since been reported in any more recent literature (Aoki et al., 2012).
Table 4 summarizes the results for each assay. The assay targeting rs10484555, ANPRV67, identified HLA-B*15:02 with 100% sensitivity and 100% specificity (Figure 1). There were no false positives or false negatives, but sample 755 failed to amplify on multiple runs, so no result could be obtained. If included, the sensitivity and specificity of the assay would both be reduced to 97%. The sample worked well on other assays, so it is likely that nearby SNVs, insertions, or deletions interfered with the ability of the probe to bind to that sample. Sample NA17019 required manual calling owing to weak amplification of the FAM dye (Figure 2).
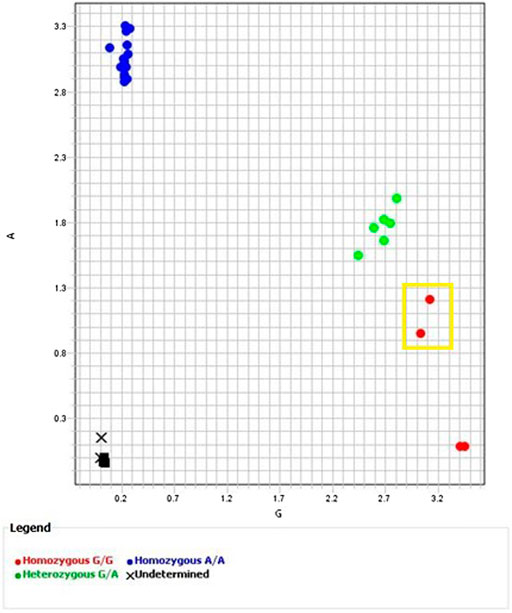
FIGURE 1. Allelic Discrimination Plot for ANPRV67 (rs10484555) Black “X”s represent sample 755, which failed to amplify. Axes indicate relative fluorescence of VIC (x-axis) and FAM (y-axis) dyes. Sample NA17019 surrounded by the yellow box. Though software assigned a homozygous G/G call, the position on the allelic discrimination plot and multicomponent plot (Figure 2 (A)) clearly indicate a heterozygous G/A call.
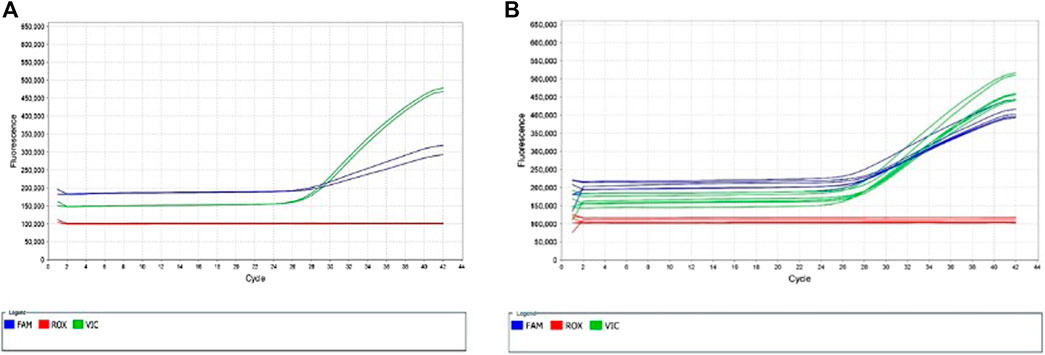
FIGURE 2. Multicomponent Plots for ANPRV67 (rs10484555). (A) Fluorescence of both VIC and FAM dyes indicate a heterozygous call for sample NA17019. A homozygous G/G sample would show solely fluorescence of the VIC dye. (B) Fluorescence of both VIC and FAM dyes from remainder of heterozygous samples (green circles in Figure 1, samples 738, HG00463, NA23090).
The assay targeting rs144012689 with the nucleotide base C at rs2596496, AN2XMRN, showed poor amplification for the majority of samples (Figure 3), with the exception of one group of samples showing strong amplification of the VIC dye (NA17235, NA17256, NA18545, 689, 691, 755). The assay targeting rs144012689 with the nucleotide base G at rs2596496, AN33GCK, showed strong amplification of most samples (Figure 4), with the exception of the same group as above. As these samples amplified well with assay AN2XMRN, we inferred that they have the C nucleotide at rs2596496. Manual calling of the group of six samples with assay AN33GCK (Figure 4) resulted in 100% sensitivity and specificity for the identification of HLA-B*15:02.
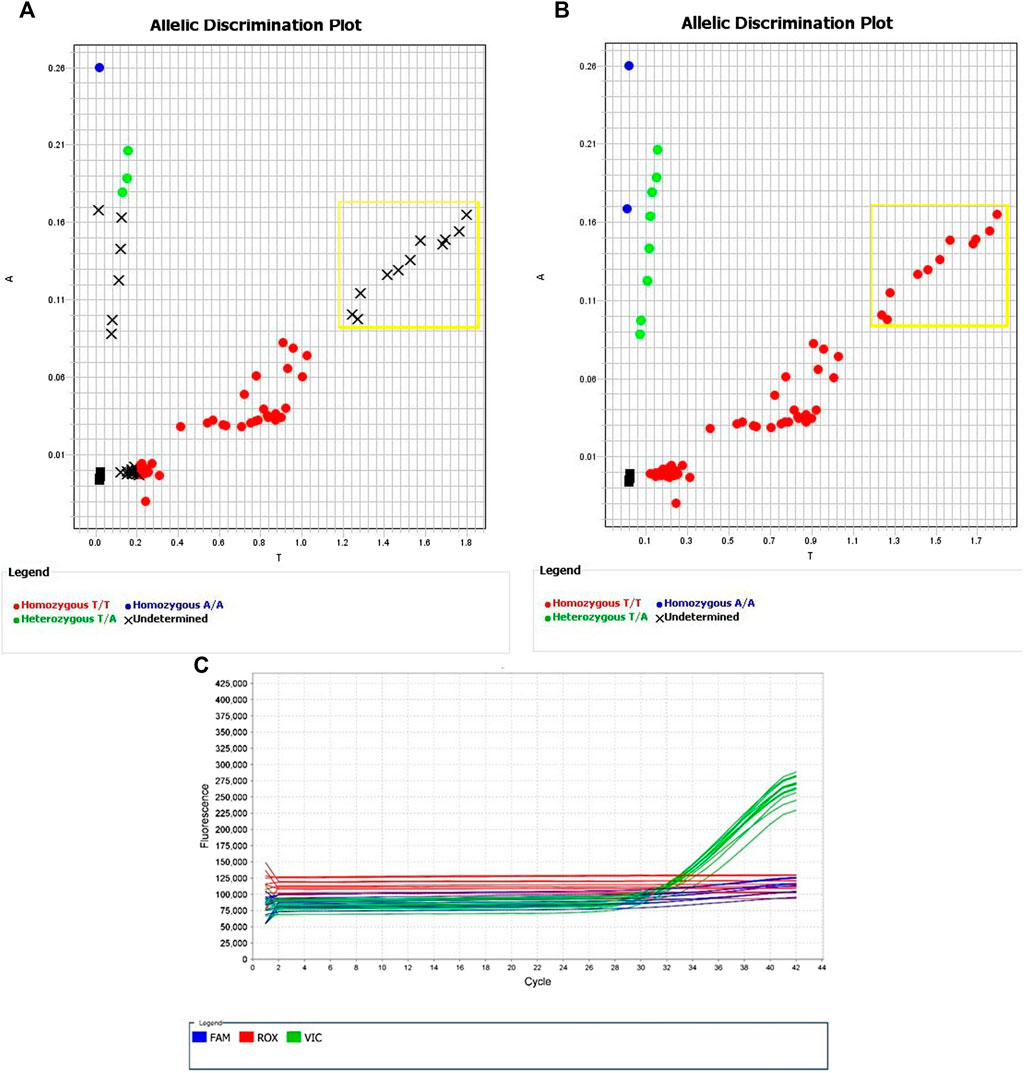
FIGURE 3. Allelic Discrimination Plots and Multicomponent Plot for AN2XMRN (rs144012689, rs2596496C)Samples in the yellow box showed most significant amplification (NA17235, NA17256, NA18545, 689, 691, 755), in comparison to the rest of the samples. This group likely has the nucleotide base C at rs2596496. (A) shows the software-generated calls, and (B) shows manually assigned calls based on relative fluorescence. (C) Fluorescence of VIC dye on multicomponent plot for NA17235, NA17256, NA18545, 689, 691, 755 (samples in yellow box) indicates homozygous T/T call.
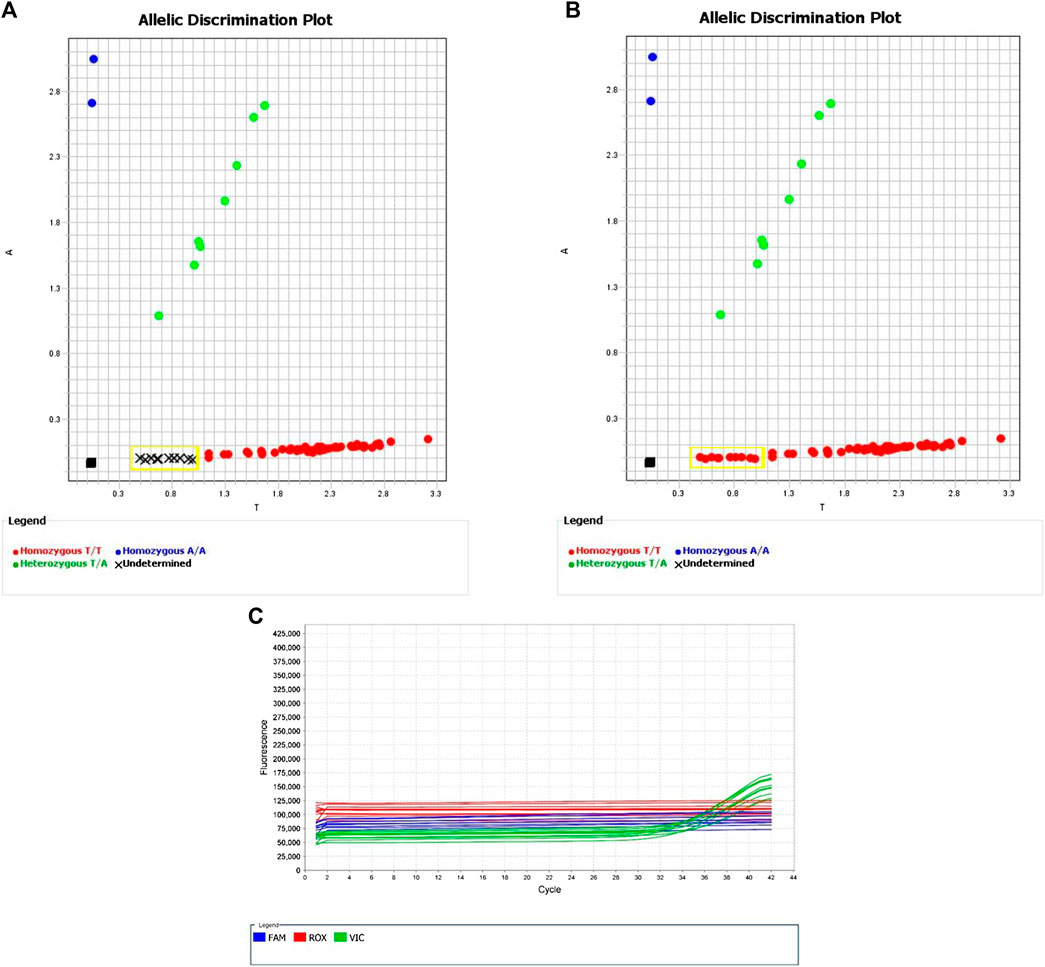
FIGURE 4. Allelic Discrimination Plots and Multicomponent Plot for AN33GCK (rs144012689, rs2596496G)Black “X”s indicate samples for which the automatically generated calls failed. Samples in yellow box are the ones that showed the most significant amplification on assay AN2XMRN (NA17235, NA17256, NA18545, 689, 691, 755). (A) shows the software-generated calls, and (B) shows manually assigned calls based on relative fluorescence. (C) Fluorescence of VIC dye on multicomponent plot for NA17235, NA17256, NA18545, 689, 691, 755 (samples in yellow box) indicates homozygous T/T call.
The assay targeting rs17179220, C__33415939_10, identified HLA-A*31:01 with 100% sensitivity and 100% specificity. Using the default genotyping settings in the Quant Studio software, amplification occurred late in the reaction and sample discrimination was challenging; however, increasing the number of PCR amplification cycles from 40 to 50 made the results much clearer.
The assay targeting rs1061235, ANNK49J, identified HLA-A*31:01 with 100% sensitivity and 95% specificity. All three false positives were due to the presence of rs1061235 on the haplotype HLA-A*33:03.
Our data are consistent with rs10484555 and rs17179220 being the best tagging SNVs for HLA-B*15:02 and HLA-A*31:01 respectively, across diverse ancestral backgrounds.
Assay ANPRV67 for rs10484555 had 100% sensitivity and specificity for HLA-B*15:02 (excluding one sample, 755, that failed to amplify on multiple attempts). We suggest further investigation, such as sequencing, of sample 755 and one sample that required manual calling (NA17019) is warranted. Our data are consistent with the database prediction of Erlichster et al. (2019) of 100% sensitivity and 98.7% specificity of rs10484555 for tagging HLA-B*15:02 in a diverse population, and with the 100% sensitivity and 99.3% specificity for tagging HLA-B*15:02 in the Han Chinese population reported by Liu et al. (2015).
Assay AN33GCK for rs144012689 with the nucleotide base G at rs2596496, in combination with manual calling, resulted in 100% sensitivity and 100% specificity for the identification of HLA-B*15:02. However, if another nucleotide other than C or G is present at rs2596496, which is theoretically possible (National Center for Biotechnology Information, 2020), the assay may not work. Even though there are no frequency data available for the other possible nucleotide variants, we suggest further custom assays for the other nucleotide possibilities at rs2596496 be developed to rule out this possibility. The presence of the C or G nucleotide within our sample set is not population-specific, as the aforementioned group of samples we hypothesize to have the C nucleotide at rs2596496 are from European, Han Chinese, and Hispanic populations. As well, there is the potential for false positives in individuals with the HLA-B*15:13 haplotype. This haplotype has a global frequency of 0.08%, in comparison to a frequency of 0.91% for HLA-B*15:02 (Fang et al., 2019), which is small but not negligible. Overall, for an HLA-B*15:02 haplotyping strategy, we suggest using assay ANPRV67 for rs10484555 first, and if any output such as that seen with samples 755 and NA17019 results, assay AN33GCK for rs144012689 may be additionally used.
Assay C__33415939_10 for rs17179220 had 100% sensitivity and specificity for HLA-A*31:01. This is comparable to the database prediction of Erlichster et al. (2019) of 100% sensitivity and 99.6% specificity of rs17179220 for tagging HLA-A*31:01. The assay performs best when the number of PCR amplification cycles is increased to 50 (increasing total run time from 90 to 110 min). Assay ANNK49J for rs1061235 also identified HLA-A*31:01 with 100% sensitivity, with its specificity impacted by this SNV also being present in the HLA-A*33:01 and HLA-A*33:03 haplotypes (Thorstensen et al., 2014). A study in an ancestrally-diverse group of Canadian children (Amstutz et al., 2013) found that out of 20 children carrying the minor allele of rs1061235, 12 had a haplotype of HLA-A*31:01, five had a haplotype of HLA-A*33:03, and three had a haplotype of HLA-A*33:01. As a result, they concluded that rs1061235 is not an optimal tag for HLA-A*31:01 in ancestrally diverse populations. However, they did find that the rs1061235 minor allele was overrepresented among carbamazepine hypersensitivity cases in patients not carrying the HLA-A*31:01 haplotype, suggesting a possible association of the HLA-A*33 haplotypes with such hypersensitivity. Further research regarding this is required.
The selection of positive controls, especially for minor alleles that are less represented within a sample set, has the potential to improve clustering and the ability of the QuantStudio software to make accurate automatic calls. For this purpose, we recommend NA23093 as a homozygous HLA-B*15:02 control and HG00463 or NA23090 as a heterozygous HLA-B*15:02 control. Numerous Coriell samples would be suitable for heterozygous HLA-A*31:01 controls (NA17039, NA17229, NA17235, NA17277, NA17281). A limitation of our study is that owing to the limited availability of reference samples with relevant haplotypes, our analysis was limited to only 38 samples, primarily of European ancestry, with Hispanic, Han Chinese, Filipino, and Black American representation. It has been previously noted that population sets that include larger numbers of those of different ancestry may see higher rates of false positives (Liu et al., 2015). Nonetheless, as the ADRs associated with these haplotypes on prescribing carbamazepine or oxcarbazepine may be fatal, individuals may not know the full details of their ancestry, and certainly testing HLA-B*15:02 is indicated in individuals had physician-reported ethnicity other than Asian (Fang et al., 2019) we recommend using these SNVs to identify these haplotypes in individuals of any ancestral group. Another potential limitation in the translation of this work to the clinical laboratory setting is the need for occasional manual calls. This is in fact not unusual with this and other techniques in this setting: data would need to be reviewed before exporting files or reporting to patients. This process is not particularly lengthy or challenging but does require some knowledge of the TaqMan assays and use of the QuantStudio software. Different population background and sample quality may be relevant to the amount of data that requires manual manipulation. As mentioned, the proximity of nearby SNVs to the SNVs of interest can interfere with assay performance, and certain SNVs may be more prevalent in certain ancestral groups. Poor sample quality or low concentration may also increase the prevalence of data that requires manual calls.
In summary, we have validated a method of detection of HLA-B*15:02 and HLA-A*31:01 that at <$1USD per sample plus labour of less than 2 h is orders of magnitude lower than the cost of haplotyping by the current “gold-standard” methods. A recent study that aimed to assess the cost-effectiveness of pre-emptive pharmacogenomic testing for HLA haplotypes, based on their relative frequencies worldwide, concluded that for carbamazepine, pre-emptive genotyping of HLA-B*15:02 would be cost-effective across most of East and South Asia, whereas HLA-A*31:01 testing would be likely to be cost-effective globally (Zhou et al., 2021). However, their calculations assumed the cost per sample to be $40, which is much greater than our estimated cost of TaqMan SNP genotyping.
Our findings are relevant for other technologies that rely on SNV detection for the identification of HLA haplotypes. For example, the Ion AmpliSeq Pharmacogenomics Panel uses rs1061235 to tag HLA-A*31:01. Although we and other investigators have found that the specificity of this SNV as a haplotype tagging marker is not 100%, as carbamazepine hypersensitivity has been found in patients with the rs1061235 minor allele but not carrying the HLA-A*31:01 haplotype (Amstutz et al., 2013), the use of this SNV to screen for carbamazepine hypersensitivity should be adequate. Further, the sensitivity is 100% in our data and that of others, and given the severity of the potential ADRs, the absence of false negatives is more important than the absence of false positives. Agena Biosciences use markers including rs10484555 for HLA-B*15:02 and rs1061235 for HLA-A*31:01, which should be fine. Pharmacoscan includes rs1061235 as a screen for HLA-A*31:01 as well. Cross-validation of our method could be undertaken versus the multiple technologies above described in a manner that we have previously described (Carvalho Henriques, et al., 2020a; 2020c).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/articles/dataset/EDS_files_raw_data_from_QuantStudio_for_Validation_of_Single_Nucleotide_Variant_Assays_for_Human_Leukocyte_Antigen_Haplotypes_HLA-B_15_02_and_HLA-A_31_01_Across_Diverse_Ancestral_Backgrounds/14977929.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
KA and AB were responsible for study design; AB and KA selected markers for validation; AB conducted the assay analytical validation with some assistance from XH; data analysis was conducted by AB with some assistance from KA; AB and KA were responsible for manuscript drafting, revisions, and finalization for submission. All authors approved the final version of the manuscript for submission.
The work reported herein was funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) Undergraduate Student Research Award to AB, an Alberta Innovates Strategic Research Project (SRP51_PRIME - Pharmacogenomics for the Prevention of Adverse Drug Reactions in mental health; G2018000868), a Canada Foundation for Innovation (CFI), John R. Evans Leaders Fund (JELF) grant (32147 - Pharmacogenetic translational biomarker discovery) with matching funding from the Alberta Innovation and Advanced Education Small Equipment Grants Program, and the Faculty of Medicine and Dentistry at the University of Alberta.
KA is a member of the Pharmacogene Variation Consortium (PharmVar), a member of the Genetic Testing Committee of the International Society of Psychiatric Genetics, and of the Clinical Pharmacogenetics Implementation Consortium.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Anne Halpin from the Histocompatibility Laboratory of Alberta Precision Laboratories in Edmonton, Alberta, for her assistance with reference samples.
Alberta Precision Laboratories (2021). Alberta Precision Laboratories Test Directory. Available at: https://www.albertahealthservices.ca/webapps/labservices/indexAPL.asp?zoneid=1&dslurchText=HLA&submit=Submit&upperTest=-1&lowerTest=-1javascript:alert(document.%20lastModified (Accessed March 20, 2021).
Amstutz, U., Ross, C. J. D., Castro-Pastrana, L. I., Rieder, M. J., Shear, N. H., Hayden, M. R., et al. (2013). HLA-A 31:01 and HLA-B 15:02 as Genetic Markers for Carbamazepine Hypersensitivity in Children. Clin. Pharmacol. Ther. 94, 1. doi:10.1038/clpt.2013.55
Amstutz, U., Shear, N. H., Rieder, M. J., Hwang, S., Fung, V., Nakamura, H., et al. (2014). Recommendations for HLA-B*15:02 and HLA-A*31:01 Genetic Testing to Reduce the Risk of Carbamazepine-Induced Hypersensitivity Reactions. Epilepsia 55, 4. doi:10.1111/epi.12564
Aoki, M., Hosono, N., Takata, S., Nakamura, Y., Kamatani, N., and Kubo, M. (2012). New Pharmacogenetic Test for Detecting an HLA-A*31: 01 Allele Using the InvaderPlus Assay. Pharmacogenet. Genomics. 22, 6. doi:10.1097/FPC.0b013e3283527c40
Baker, G. R., Norton, P. G., Flintoft, V., Blais, R., Brown, A., Cox, J., et al. (2004). The Canadian Adverse Events Study: the Incidence of Adverse Events Among Hospital Patients in Canada. CMAJ 170, 1678–1686. doi:10.1503/cmaj.1040498
Bettinotti, M. P., Mitsuishi, Y., Bibee, K., Lau, M., and Terasaki, P. I. (1997). Comprehensive Method for the Typing of HLA-A, B, and C Alleles by Direct Sequencing of PCR Products Obtained from Genomic DNA. J. Immunother. 20, 6. doi:10.1097/00002371-199711000-00001
Carvalho Henriques, B., Buchner, A., Hu, X., Yavorskyy, V., Wang, Y., Martens, K., et al. (2020a). Cross-validation of Technologies for Genotyping CYP2D6 and CYP2C19. BioRxiv. doi:10.1101/2019.12.24.870295
Carvalho Henriques, B., Buchner, A., Hu, X., Yavorskyy, V., Wang, Y., Martens, K., et al. (2020b). Cross-validation of Technologies for Genotyping CYP2D6 and CYP2C19. Transl Psychiatry Under Rev. doi:10.1101/2019.12.24.870295
Carvalho Henriques, B., Yang, E. H., Lapetina, D., Carr, M. S., Yavorskyy, V., Hague, J., et al. (2020c). How Can Drug Metabolism and Transporter Genetics Inform Psychotropic Prescribing?. Front. Genet. 11. doi:10.3389/fgene.2020.491895
Chen, Z., Liew, D., and Kwan, P. (2014). Effects of a HLA-B*15:02 Screening Policy on Antiepileptic Drug Use and Severe Skin Reactions. Neurology 83, 22. doi:10.1212/wnl.0000000000001034
Choo, S. Y. (2007). The HLA System: Genetics, Immunology, Clinical Testing, and Clinical Implications. Yonsei Med. J. 48, 1. doi:10.3349/ymj.2007.48.1.11
de Bakker, P., McVean, G., Sabeti, P., Mirette, M. M., Green, T., Marchini, J., et al. (2006). A High-Resolution HLA and SNP Haplotype Map for Disease Association Studies in the Extended Human MHC. Nat. Genet. 38, 10. doi:10.1038/ng1885
Dunckley, H. (2012). “HLA Typing by SSO and SSP Methods,” in Immunogenetics. Methods Mol. Biol. (Methods Protocols) 882, 9–25. doi:10.1007/978-1-61779-842-9_2
Edgerly, C. H., and Weimer, E. T. (2018). The Past, Present, and Future of HLA Typing in transplantation.” in HLA Typing. Methods Mol. Biol. 1802, 1–10. doi:10.1007/978-1-4939-8546-3_1
Erlichster, M., Goudey, B., Skafidas, E., and Kwan, P. (2019). Cross-ethnicity Tagging SNPs for HLA Alleles Associated with Adverse Drug Reaction. Pharmacogenomics J. 19, 3. doi:10.1038/s41397-018-0039-z
Ernst, F. R., and Grizzle, A. J. (2001). Drug-related Morbidity and Mortality: Updating the Cost-Of-Illness Model. J. Am. Pharm. Assoc. 41, 2. doi:10.1016/s1086-5802(16)31229-3
Fan, W., Shiao, M., Hui, R. C., Su, S., Wang, C., Chang, Y., et al. (2017). HLA Association with Drug-Induced Adverse Reactions. J. Immunol. Res. 2017, 3186328. doi:10.1155/2017/3186328
Fang, H., Xu, X., Kaur, K., Dedek, M., Zhu, G., Riley, B. J., et al. (2019). A Screening Test for HLA-B*15:02 in a Large United States Patient Cohort Identifies Broader Risk of Carbamazepine-Induced Adverse Events. Front. Pharmacol. 10, 149. doi:10.3389/fphar.2019.00149
Gui, H., Kwok, M., Baum, L., Sham, P. C., Kwan, P., and Cherny, S. S. (2018). SNP-based HLA Allele Tagging, Imputation and Association with Antiepileptic Drug-Induced Cutaneous Reactions in Hong Kong Han Chinese. Pharmacogenomics J. 18, 2. doi:10.1038/tpj.2017.11
He, Y., Hoskins, J. M., Clark, S., Campbell, N. H., Wagner, K., Motsinger-Reif, A. A., et al. (2015). Accuracy of SNPs to Predict Risk of HLA Alleles Associated with Drug-Induced Hypersensitivity Events across Racial Groups. Pharmacogenomics 16, 8. doi:10.2217/pgs.15.41
Illing, P. T., Purcell, A. W., and McCluskey, J. (2017). The Role of HLA Genes in Pharmacogenomics: Unravelling HLA Associated Adverse Drug Reactions. Immunogenetics 69, 617. doi:10.1007/s00251-017-1007-5
Lazaro, A., Tu, B., Yang, R., Xiao, Y., Kariyawasam, K., Ng, J., et al. (2013). “Human Leukocyte Antigen (HLA) Typing by DNA Sequencing,” in Transplantation Immunology. Methods Mol. Biol. (Methods Protocols) 1034, 161–195. doi:10.1007/978-1-62703-493-7_9
Liu, X., Sun, J., Yu, H., Chen, H., Wang, J., Zou, H., et al. (2015). Tag SNPs for HLA-B Alleles that Are Associated with Drug Response and Disease Risk in the Chinese Han Population. Pharmacogenomics J. 15, 5. doi:10.1038/tpj.2015.7
Maekawa, K., Nakamura, R., Kaniwa, N., Mizusawa, S., Kitamoto, A., Kitamoto, T., et al. (2015). Japan Pharmacogenomics Data Science ConsortiumDevelopment of a Simple Genotyping Method for the HLA-A*31:01-tagging SNP in Japanese. Pharmacogenomics 16, 15. doi:10.2217/pgs.15.115
Mccormack, M., Alfirevic, A., Bourgeois, S., Farrell, J. J., Kasperavičiūtė, D., Carrington, M., et al. (2011). HLA-A*3101 and Carbamazepine-Induced Hypersensitivity Reactions in Europeans. N. Engl. J. Med. 364, 12. doi:10.1056/NEJMoa1013297
Mullan, K. A., Anderson, A., Illing, P. T., Kwan, P., Purcell, A. W., and Mifsud, N. A. (2019). HLA‐associated Antiepileptic Drug‐induced Cutaneous Adverse Reactions. HLA 93, 6. doi:10.1111/tan.13530
National Center for Biotechnology Information (2020). Rs2596496 Reference SNP (Rs) Report. Available at: https://www.ncbi.nlm.nih.gov/snp/rs2596496 (Accessed March 20, 2021).
Payne, P. W. (2014). Ancestry-based Pharmacogenomics, Adverse Reactions and Carbamazepine: Is the FDA Warning Correct?. Pharmacogenomics J. 14, 5. doi:10.1038/tpj.2014.14
Petersdorf, E. W., and O'hUigin, C. (2019). The MHC in the Era of Next-Generation Sequencing: Implications for Bridging Structure with Function. Hum. Immunol. 80, 1. doi:10.1016/j.humimm.2018.10.002
Pirmohamed, M., James, S., Meakin, S., Green, C., Scott, A. K., Walley, T. J., et al. (2004). Adverse Drug Reactions as Cause of Admission to Hospital: Prospective Analysis of 18 820 Patients. BMJ 329, 7456. doi:10.1136/bmj.329.7456.15
Shen, G. Q., Abdullah, K. G., and Wang, Q. K. (2009). “The TaqMan Method for SNP Genotyping,” in Singe Nucleotide Polymorphisms. Methods Mol. Biol. (Methods Protocols) 578, 273–306. doi:10.1007/978-1-60327-411-1_19
Shepherd, G., Mohorn, P., Yacoub, K., and May, D. W. (2012). Adverse Drug Reaction Deaths Reported in United States Vital Statistics, 1999-2006. Ann. Pharmacother. 46, 2. doi:10.1345/aph.1p592
The Genomes Project Consortium (2015). A Global Reference for Human Genetic Variation. Nature 526, 7571. doi:10.1038/nature15393
Thermo Fisher Scientific (2021). Predesigned TaqMan SNP Genotyping Assays. Available at: https://www.thermofisher.com/ca/en/home/life-science/pcr/real-time-pcr/real-time-pcr-assays/snp-genotyping-taqman-assays/single-tube-snp-genotyping.html (Accessed March 20, 2021).
Thorstensen, K., Kvitland, M., Shirzadi, M., Helde, G., Moen, T., and Brodtkorb, E. (2014). Carbamazepine-induced Cutaneous Reactions: A Simple Assay to Identify Patients Carrying the HLA-A31:01 Allele. Scand. J. Clin. Lab. Invest. 74, 7. doi:10.3109/00365513.2014.921835
Trowsdale, J., and Knight, J. C. (2013). Major Histocompatibility Complex Genomics and Human Disease. Annu. Rev. Genomics Hum. Genet. 14, 301. doi:10.1146/annurev-genom-091212-153455
US National Library of Medicine (2021). CARBAMAZEPINE- Carbamazepine Capsule. extended release [package insert]Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=7a1e523a-b377-43dc-b231-7591c4c888ea (Accessed March 20, 2021).
Verbelen, M., Weale, M. E., and Lewis, C. M. (2017). Cost-effectiveness of Pharmacogenetic-Guided Treatment: Are We There yet?. Pharmacogenomics J. 17, 5. doi:10.1038/tpj.2017.21
Wieczorek, M., Abualrous, E. T., Sticht, J., Alvaro-Benito, M., Stolzenberg, S., Noe, F., et al. (2017). Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 8, 292. doi:10.3389/fimmu.2017.00292
Zhou, F., Cao, H., Zuo, X., Zhang, T., Zhang, X., Liu, X., et al. (2016). Deep Sequencing of the MHC Region in the Chinese Population Contributes to Studies of Complex Disease. Nat. Genet. 48, 7. doi:10.1038/ng.3576
Zhou, Y., Krebs, K., Milani, L., and Lauschke, V. M. (2021). Global Frequencies of Clinically Important HLA Alleles and Their Implications for the Cost‐effectiveness of Preemptive Pharmacogenetic Testing. Clin. Pharmacol. Ther. 109, 1. doi:10.1002/cpt.1944
Keywords: HLA antigens, adverse drug reactions, carbamazepine, psychiatry, pharmacogenetics, precision medicine, single nucleotide variants, oxcarbazepine
Citation: Buchner A, Hu X and Aitchison KJ (2021) Validation of Single Nucleotide Variant Assays for Human Leukocyte Antigen Haplotypes HLA-B*15:02 and HLA-A*31:01 Across Diverse Ancestral Backgrounds. Front. Pharmacol. 12:713178. doi: 10.3389/fphar.2021.713178
Received: 22 May 2021; Accepted: 15 July 2021;
Published: 26 July 2021.
Edited by:
Christina Mitropoulou, The Golden Helix Foundation, United KingdomReviewed by:
Seik-Soon Khor, National Center For Global Health and Medicine, JapanCopyright © 2021 Buchner, Hu and Aitchison. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherine J. Aitchison, a2FpdGNoaXNAdWFsYmVydGEuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.