
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 09 September 2021
Sec. Pharmacology of Anti-Cancer Drugs
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.709324
This article is part of the Research Topic Non-coding RNA and Wnt/β-catenin Signaling Pathway in Human Cancer View all 8 articles
Cisplatin is a common chemotherapeutic drug that has been used to treat of numerous tumors, including testicular, lung, bladder, ovarian, liver and head and neck cancers. Although clinical chemotherapy based on cisplatin has shown a remarkable therapeutic effect, the resistance to cisplatin becomes increasingly obvious as a patient uses it for a prolonged period. It not only affects the prognosis of these tumors, but also causes the recurrence of cancer and decreases the overall survival rate. The development of cisplatin resistance involves several mechanisms, including DNA damage repair, ATP-binding cassette (ABC) transporter, autophagy, cancer stem cells (CSCs), epithelial–mesenchymal transition (EMT), and other related signaling pathways. Interestingly, these mechanisms have been found to be influenced by circular RNAs (circRNAs) to regulate tumor proliferation, invasion, chemosensitivity, and other biological behaviors in the tumor microenvironment (TME). In recent years, circRNAs in cisplatin resistance in tumors, especially lung cancer and gastric cancer, have gradually drawn peoples’ attention. This review summarizes recent studies on the functions and mechanisms of circRNAs in cisplatin resistance. We emphasize that circRNA can be used as a promising target gene to improve drug resistance and therapeutic efficacy.
Dr. Rosenborg accidentally discovered cisplatin in 1965 and predicted that it could inhibit cancer cell division (Rosenberg et al., 1965). About 50 years ago, cisplatin was approved by the US Food and Drug Administration (FDA) for the treatment of testicular cancer (Kartalou and Essigmann, 2001). Thereafter, cisplatin has become a common chemotherapeutic drug for numerous tumors, including testicular, lung, bladder, ovarian, liver, and head and neck cancers (Dasari and Tchounwou, 2014). It has numerous anticancer mechanisms. It is generally believed to interact with the nucleophilic N7 locus of purine base on DNA; as a result, DNA is damaged, and related signaling pathways are activated, thereby causing tumor cell apoptosis (Dasari and Tchounwou, 2014; Rocha et al., 2018; Ghosh, 2019) (Figure 1). With the continuous application of cisplatin in the chemotherapy of various tumors, its clinical efficacy has been found to be unsatisfactory. Cisplatin often develops resistance as patients use it for a prolonged period (Cocetta et al., 2019). Therefore, the cisplatin resistance mechanism of tumors has been widely studied to improve the survival rate and long-term prognosis of patients. For example, the accumulation of cisplatin and DNA compounds should be reduced by increasing the efflux or inhibiting the influx, lowering the toxicity of cisplatin with antioxidants (e.g., glutathione), enhancing DNA repair and DNA methylation, changing membrane protein transport, and activating or inactivating epithelial–mesenchymal transition (EMT) and other related pathways (Shen et al., 2012; Amable, 2016). Cisplatin resistance is also associated with cancer stem cells (CSCs). In addition, J.A.Ferreira et al. emphasized the functions of protein glycosylation in drug resistance (Ferreira et al., 2016). Besides, autophagy can promote the apoptosis of multidrug-resistant (MDR) cells, improve drug sensitivity, and protect cancer cells from the toxicity of chemotherapy drugs (Li et al., 2017). In recent years, increasing evidence has shown that the exocrine vesicles secreted from tumor cells can change the tumor microenvironment consequently, tumor growth is facilitated, and the resistance of chemotherapeutic drugs is promoted (Chiarugi and Cirri, 2016). Multiple mechanisms play roles in cisplatin resistance. These mechanisms are elaborated later.
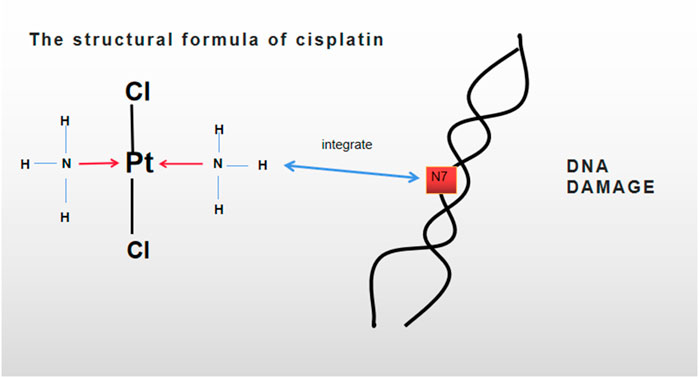
FIGURE 1. Molecular formula of cisplatin is Cl2H4N2Pt. DNA damage was caused by integrating cisplatin with the nucleophilic N7 locus of a purine base in a double-helix DNA.
Noncoding RNAs (NcRNAs) can be divided into microRNAs (miRNAs), lncRNAs, and circRNAs (Iorio and Croce, 2017). CircRNAs were first discovered from a plant virus 40 years ago (Sanger et al., 1976). They have become a new hotspot in RNA research. CircRNAs are covalently closed-loop RNA molecules, which can splice the pre-RNA encoded by the coding gene through the interaction between reverse splicing and canonical splicing (Chen and Yang, 2015; Belousova et al., 2018). CircRNAs can be divided into three types, namely, exonic circRNAs, circular intron circRNAs, and exon–intron circRNAs (Meng et al., 2017) (Figure 2). They have no polyadenylation tail with 5′–3′ polarity (Chen and Yang, 2015). In mammals, circRNAs act as a sponge of miRNAs or combine with RNA-related proteins to form complexes to regulate gene expression and act as a gene regulator (Han et al., 2018). Some circRNAs can modulate gene transcription through RNA polymerase II in the gene promoter region. In addition, CircRNAs can also negatively regulate classical splicing through the interaction of the splicing factor muscleblind (MBL) (Su et al., 2019a). Recent studies have shown that circRNAs play an important role in the treatment of nervous system diseases, vascular diseases, and tumors (Qu et al., 2015). CircRNAs can be used as a biomarker of various cancers. They regulate tumor proliferation, invasion, chemosensitivity, and other biological behaviors by sponging miRNA or directly targeting a gene. Similar to lncRNAs, circRNAs and miRNAs also form a competing endogenous RNA (ceRNA) network (Su et al., 2019a; Verduci et al., 2019). The expression of circRNAs in tumor cells may be upregulated or downregulated compared with that in the surrounding normal tissues. Its effect may trigger tumor cells to respond positively or negatively to chemotherapeutic drugs.
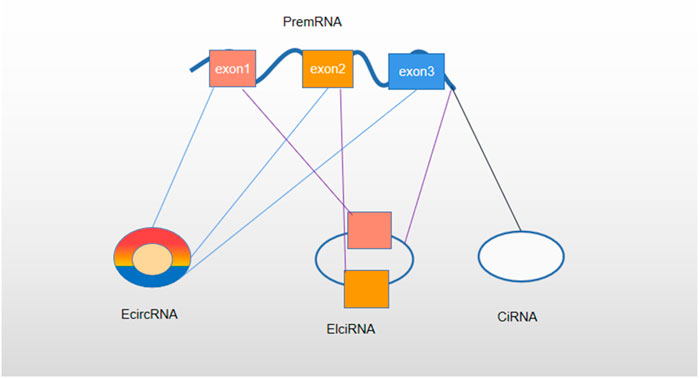
FIGURE 2. Classification of circRNA. Based on the composition, circRNAs are classified as exonic circRNAs (ecircRNAs), circular intron circRNAs (ciRNAs), and exon–intron circRNAs (EIciRNAs).
Studies have gradually explored the regulatory effect of circRNAs on cisplatin resistance. This review summarizes recent studies on the functions and mechanisms of circRNAs in cisplatin resistance.
CeRNA is an abbreviation of competing endogenous RNAs (Qi et al., 2015). CeRNAs comprise lncRNAs, circRNAs, protein-coding RNAs, tRNAs, rRNAs, and pseudogene RNAs (Qu et al., 2015). Theoretically, most ceRNAs have miRNA reaction elements (MREs). CeRNAs with the same MREs in single cells can regulate the transcription and translation of parental RNA by targeting the same or similar miRNA (An et al., 2017). The formation of a ceRNA network needs three basic conditions. First, a ceRNA should have a high expression level so that it is not affected by and interfered with the inhibition of downstream miRNAs on target genes. Second, only when the number of MREs is sufficient, circRNA can perform its biological function. The distribution and concentration of ceRNAs and miRNAs will also affect the ceRNA network. Third, different MREs have various effects on ceRNAs and miRNAs, which play a major role in ceRNA NET. Different circRNAs can bind to the same miRNA, but the nucleotide compositions of MREs may be partially different (Zhang et al., 2019) (Figure 3). In recent years, the studies on the mechanism of circRNA in cisplatin resistance are mostly based on the ceRNA network.
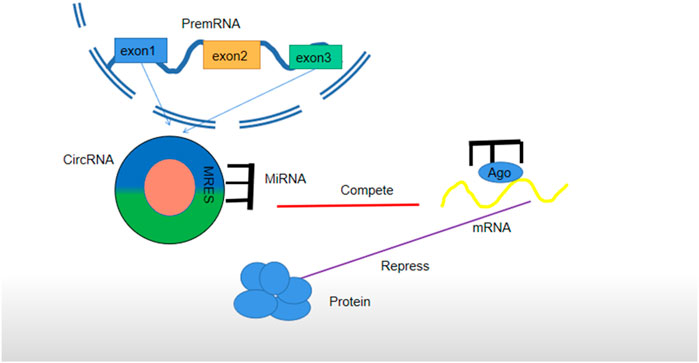
FIGURE 3. Regulatory mechanism of circRNA as ceRNA. CircRNA is cut off from a precursor RNA to sponge miRNA, block the expression of an miRNA-targeted gene (mRNA) in cells, and form a circRNA–miRNA–mRNA network. It has the same role in exosomes secreted by cells. Each miRNA can form a silent complex with an Argonaute (AGO) protein. An miRNA pairs with a target transcript, and the AGO protein promotes either target instability or transcriptional suppression.
Autophagy is a process through which starvation-induced lysosomes capture and degrade intracellular proteins and organelles and recycle intracellular components to maintain survival and metabolism (Amaravadi et al., 2016). Autophagy can be divided into macroautophagy, microautophagy, and concomitant factor-mediated autophagy (Guo et al., 2018). Macroautophagy is the most studied and classic form; in this process, autophagosomes transfer degraded substrates to lysosomes in the cytosol (Feng et al., 2014) (Figure 4). About 30 genes have been identified as autophagy-related genes (ATGs) in genetic analogues of many mammals and yeasts (Klionsky, 2012). For example, the deletion of Beclin-1, a mammalian gene of yeast ATG6, can promote tumorigenesis and support autophagy to inhibit tumor development (Qu et al., 2003). LC3 is an ubiquitin-like protein that combines with phosphatidylethanolamine (PE) to form a compound. This compound can interact with an ATG12-ATG5-ATG16L1 complex to confer the LC3–PE compound with the ability to produce an E3-like activity and participate in autophagy substrate degradation (Ichimiya et al., 2020). Autophagy disorders can lead to many diseases, such as Alzheimer’s disease, microbial infections, cardiomyopathy, diabetes, and even cancers (Levine and Kroemer, 2008; Sridhar et al., 2012).At early stages, autophagy is considered to inhibit cancer occurrence. However, at late stages, autophagy is the mechanism that promotes tumor cell survival. Autophagy plays a two-way regulation in tumor cells; thus, it has attracted considerable attention in tumor research (Sridhar et al., 2012). Many studies have also investigated the mechanism of autophagy in tumor chemoresistance. These studies have found that autophagy also plays a two-way role in the regulation of antitumor drugs. In the case of MDR, autophagy may be activated as a protective mechanism to protect tumor cells from the toxicity of chemotherapeutic drugs. However, autophagy can also induce tumor cells to die. Therefore, in this respect, autophagy can promote the sensitivity of chemotherapeutic drugs (Li et al., 2017). Most research results have shown that autophagy can promote cisplatin resistance in tumor cells. Studies have yet to determine whether autophagy can promote tumor cell apoptosis and the chemotherapeutic sensitivity of tumor cells remains unknown.
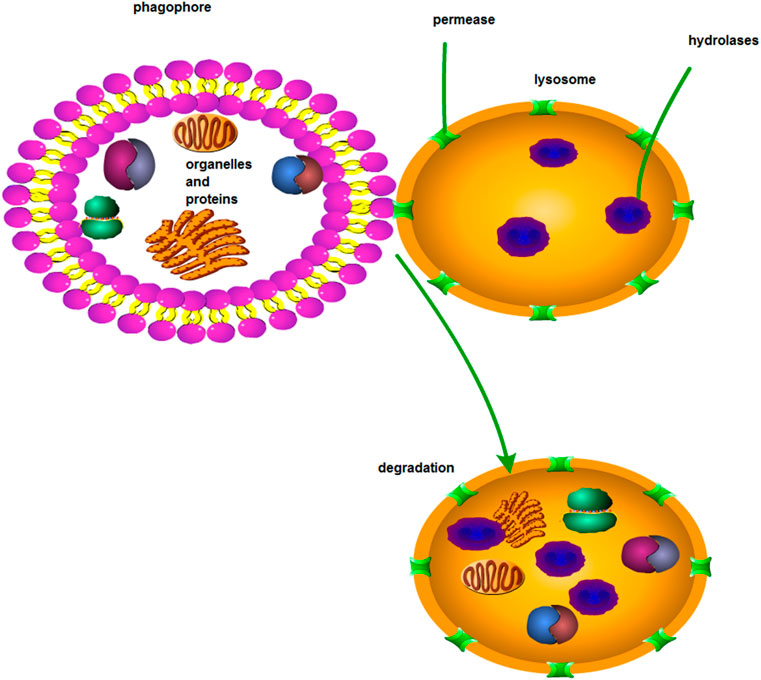
FIGURE 4. Macroautophagy. Through macroautophagy, substances in solute are transported to lysosomes via the autophagosome of a bilayer lipid.
ATP-binding cassette (ABC) transporters were discovered in the 1970s. They are energy-dependent transport systems of substrate-binding proteins (SBPs), which are activated by ATP hydrolysis (Theodoulou and Kerr, 2015). They can transport the solutes from the inside to the outside of cells (Figure 5). To date, 48 members of the ABC transporter protein family have been identified according to the sequence and structure of the ABC domain, and they are divided into seven A–G families. The ABC transporters can be mainly divided into three categories, namely, importers (prokaryotes), exporters (eukaryotes and prokaryotes), and ABCs repaired and translated by DNA. In short, importers import all kinds of molecules, whereas exporters export them (Paolini et al., 2015). Therefore, they influence the pharmacokinetics of chemotherapy (Paolini et al., 2015). Therefore, they influence the pharmacokinetics of chemotherapy. An ABC transporter identifies a series of antitumor drugs without structural correlation in cancer cells and transfers them outside the cell by using the energy of ATP hydrolysis (Stefan, 2019). Multidrug resistance protein 1 (MDR1) is also called P-glycoprotein. Its gene is known as ABCB1. The gene of breast cancer resistance protein (BCRP) is called ABCG2. The gene of multidrug resistance-related protein 1 (MRP1) is ABCC1. These three proteins are the most important ABC transporters in the mechanism of chemotherapeutic drug resistance in tumors (Hellsberg et al., 2015). CircRNA induces cisplatin resistance by regulating the expression of ABC transporter-related genes.
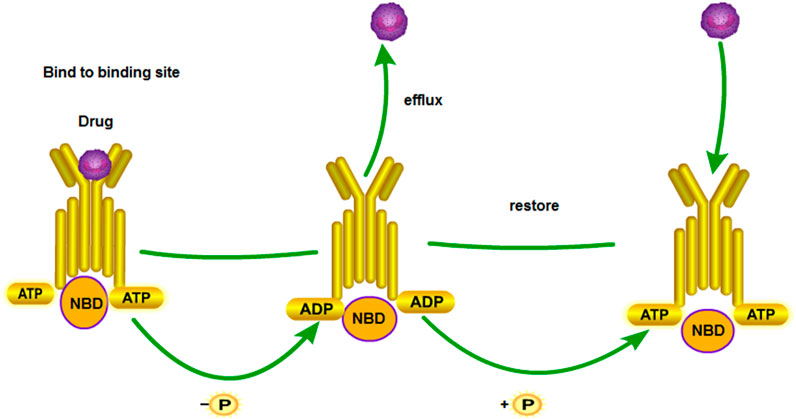
FIGURE 5. MDR1 drug transport mechanism. NBDs are nucleotide-binding domains. The substrate enters inside the protein. ATP binds to the binding site of NBD. ATP releases energy to change the conformation of transmembrane proteins and releases drugs out of the cell membrane. Subsequently, the next ATP is hydrolyzed to restore protein conformation. The above process is repeated.
Cancer stem cells (CSCs) are pluripotent tumor cells derived from normal stem cells, which have a self-renewal ability and a strong differentiation potential (Ajani et al., 2015; Lathia and Liu, 2017). The concept of CSCs has been widely recognized in the last decade, and CSCs are related to the growth and development of most malignant tumors (Chen et al., 2017). However, CSCs are generally insensitive to chemotherapeutic drugs. Although tumor cell chemoresistance occurs frequently, CSCs seem to be particularly resistant to chemotherapy. This resistance is due to the slower cell cycle of CSCs than that of many cancer cells targeted by chemotherapeutic drugs; their division speed is also not as fast as that of normal cells (Chang, 2016). CSCs are located in a special environment composed of fibroblasts and endothelial cells, mesenchymal cells, and immune cells. This environment is called a niche. Adjacent cells maintain CSCs through related signaling pathways and promote the endogenous drug resistance of CSCs (Prieto-Vila et al., 2017). Experiments have demonstrated that chemotherapy and radiotherapy can increase the quantity of CSCs and make tumor cells transform into CSCs (Chang, 2016). Moreover, several mechanisms, including EMT and ABC transporter, and signaling pathways including the Wnt pathway, participate in the drug resistance of CSCs. They are introduced in the following sections (Prieto-Vila et al., 2017). Therefore, to explore the role of CSCs in cisplatin resistance in tumors, pathways including fibroblast growth factor receptor (FGFR) or transforming growth factor-β (TGF-β) should be investigated to explore the role of CSCs in cisplatin resistance in tumors (Robey et al., 2018) (Figure 6). In discussing the role of circRNAs in cisplatin resistance, studies on the interaction of multiple mechanisms, including CSCs, are presented to discuss the role of circRNAs in cisplatin resistance.
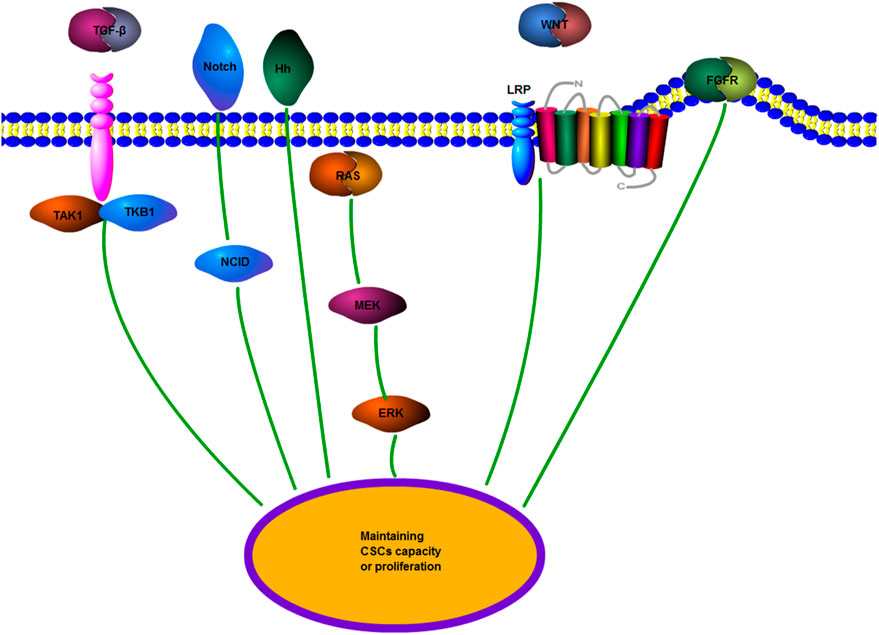
FIGURE 6. CSCs regulate drug resistance-related signaling pathways. TGF-β, transforming growth factor-β; Wnt/β-catenin; Hh, Hedgehog: Notch; FGFR, fibroblast growth factor receptor; and MEK, mitogen-activated protein kinase. Multiple signaling pathways promote the transformation of tumor cells to CSCs to maintain the features of CSCs or promote the growth and proliferation of CSCs.
Exosomes are nanosized bilayer vesicles composed of extracellular lipids, which contain some biomolecules, such as lipids, proteins, and nucleic acids. Exosomes were first believed to exclude useless cellular components; later, they were found to be involved in many physiological and pathological processes, even cancers (He et al., 2018). In addition, primary tumor biomarkers are transferred by exosomes to distant organs to achieve tumor metastasis (García-Olmo et al., 1999). These molecular markers include miRNAs, lncRNAs, and circRNAs. Thus, the effect of exosomes on tumors should be further studied. The drug resistance of exosomes in tumors has been extensively studied. Exosomes can promote tumor drug resistance through the following ways: firstly, exocrine molecular substances can compete with some anticancer chemotherapeutic drugs and bind to oncogenic targets to produce drug resistance; Secondly, exosomes transfer the drug resistance of drug-resistant cells to sensitive cells; Thirdly, tumor cells secrete chemotherapeutic drugs outside cells through exosomes (Zhang et al., 2018a). This principle applies to cisplatin resistance in tumor cells. CircRNAs secreted by exosomes promote related tumor cells to produce resistance to chemotherapeutic drugs. Similarly, circRNAs secreted by exosomes can regulate other targeted genes or other pathways, such as CSCs and Wnt, to regulate the biological characteristics of tumor cells.
Chemotherapeutic drugs, such as cisplatin, can damage the DNA. Accordingly, a mechanism called DNA damage repair (DDR) maintains the stability of DNA (including DNA in cancer cells) in our body. These damages are repaired through many methods, including nucleotide excision repair (NER), mismatch repair, base excision repair, and homology-directed repair (Gavande et al., 2016). The key genes encoding DNA damage response and DNA repair include BRCA1 and BRCA2 germline mutation, which leads to cancer susceptibility syndrome. The exposure of these tissues to carcinogens makes them more likely to become cancerous (Brown et al., 2017). In terms of drug resistance, excision repair cross-complementing (ERCC1) forms an ERCC1–XPF enzyme complex, which can repair the DNA damage caused by chemotherapeutic drugs via NER. ERCC1 overexpression is negatively correlated with the clinical outcome of platinum therapy (Zheng, 2017). Therefore, ERCC1 can be used as an indicator of cisplatin resistance, indicating that it plays a role in DNA repair (Huang et al., 2019a). DNA polymerase ζ plays a role in the microhomology-mediated repair of break-induced replication. REV3L is the largest catalytic subunit in DNA polymerase ζ, indicating that it also participates in DNA repair (Martin and Wood, 2019). Therefore, REV3L overexpression regulated by circRNAs can promote cisplatin resistance (Pang et al., 2020).
The lineage transition between epithelial and mesenchymal cells shows that polarized epithelial cells lose their adhesion properties and obtain a mesenchymal phenotype; this process is called EMT (Chen et al., 2017). The role of EMT in tumor drug resistance has been gradually recognized and explored. In the mouse model of Fischer et al., cyclophosphamide treatment significantly reduces the amount of primary epithelial tumors, but the amount of tumor cells with a positive EMT is not significantly reduced. This finding indicates that EMT may be involved in the formation of drug resistance, including TGF and other factors in an EMT-mediated signaling pathway (Du and Shim, 2016). In colon cancer cells, TGF-β upregulation can promote EMT and Adriamycin resistance (Li et al., 2015). As mentioned before, CSCs can interact with EMT to regulate the biological characteristics of tumors. They are related to multiple pathways. Similarly, EMT also includes TGF‐β, Notch, Wnt, KRAS, and phosphatidylinositol 3-kinase (PI3K) pathways (Singh et al., 2018) (Figure 7).
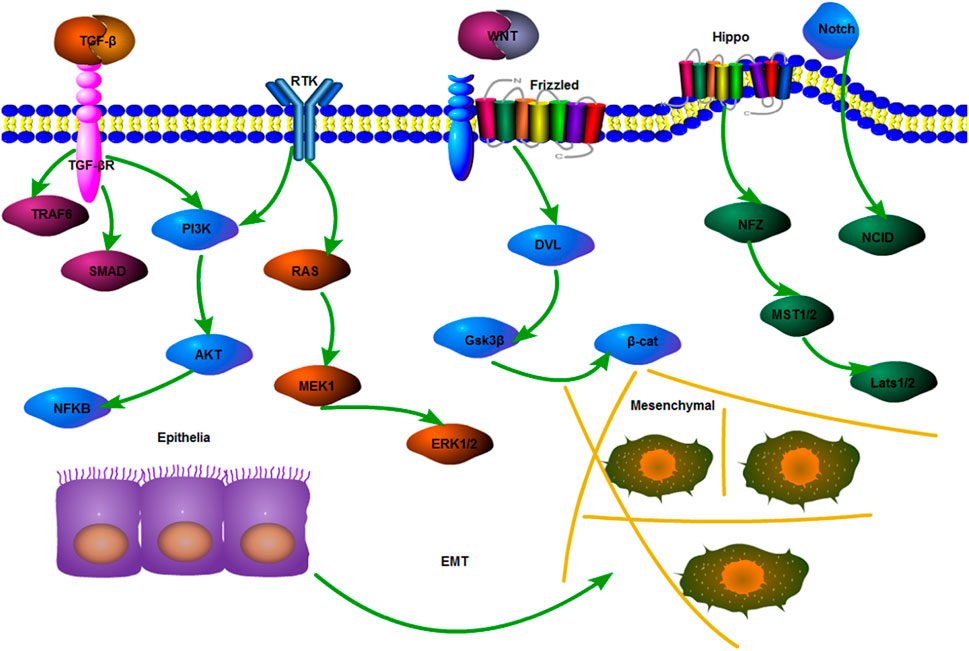
FIGURE 7. Mechanisms of EMT-related pathways. Mechanisms of EMT-related pathways include the most classical pathway, that is, TGF-β regulates the expression of a downstream cell attachment gene to lower by activating the Smad complex. Other pathways that can maintain and promote EMT include PI3K/Akt, Wnt, Hippo, and Notch.
The Wnt signaling pathway is involved in the embryonic development and homeostasis of normal adults and many normal processes. The human Wnt protein family contains 19 glycoproteins. This signaling pathway is not only involved in the proliferation, renewal, and survival of normal human cells but also implicated in diabetes, Parkinson’s disease, and cancers (Duchartre et al., 2016). The Wnt signaling pathway can be roughly divided into two categories, namely, β-catenin-dependent classical type and nonclassical type. The β-catenin-dependent classical pathway is the most frequently studied pathway. APC mutations in CRC have been discovered for many years. A study on human CRC specimens and mouse models has found that different types of APC mutations can activate different Wnt/β-catenin classical pathways to promote tumor formation and proliferation (Zhan et al., 2017). The Wnt signaling pathway is necessary for most stem cells, including normal tissue stem cells or tumor stem cells (Nusse and Clevers, 2017). It also plays a role in tumor invasion and metastasis and related chemoresistance.
The KRAS pathway is involved in the formation of cisplatin resistance. KRAS is the most common mutation of the RAS gene, which plays a key role in the occurrence of malignant tumors (Chen et al., 2019a). KRAS is a signaling pathway that regulates cell proliferation, apoptosis, and other biological activities through linear sensing signal molecules from the cell membrane to the nucleus (Liu et al., 2019). It also participates in cell cycle. When working against cancers, it can promote tumor cells to go into the S phase from the G1 phase and prevent cell apoptosis induced by remaining at the G0 phase (Nussinov et al., 2017). Therefore, KRAS activation can prevent cell apoptosis induced by cisplatin and other chemotherapeutic drugs.
PI3K regulates the signal transduction of human insulin and the proliferation and survival of cells. In addition, the mutated PI3K pathway controls the growth and proliferation of cancer cells (Lien et al., 2017). Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that can phosphorylate downstream signaling molecules. EGFR activation can trigger multiple downstream signal transduction pathways. The PI3K/Akt/mTOR pathway is one of the classical pathways. It regulates cell division, migration, proliferation, and differentiation (Li et al., 2016). CC-chemokine ligand 2 (CCL2) secreted from tumor-associated macrophages promotes tamoxifen resistance by activating the PI3K pathway (Li et al., 2020a). Therefore, the mechanism of the PI3K pathway in chemotherapy resistance in tumors has attracted increasing attention.
Many mechanisms regulate the cisplatin resistance of circRNA in several cancers (Figure 8). We discuss these mechanisms in several kinds of cancer in the following sections.
Lung cancer is the leading cause of death in men and the second leading cause of death in women worldwide (Torre et al., 2016). Many studies have investigated the efficacy of chemotherapy after lung cancer surgery, and some of them have proven the benefits of cisplatin-based neoadjuvant chemotherapy (Nagasaka and Gadgeel, 2018). (Figure 9).
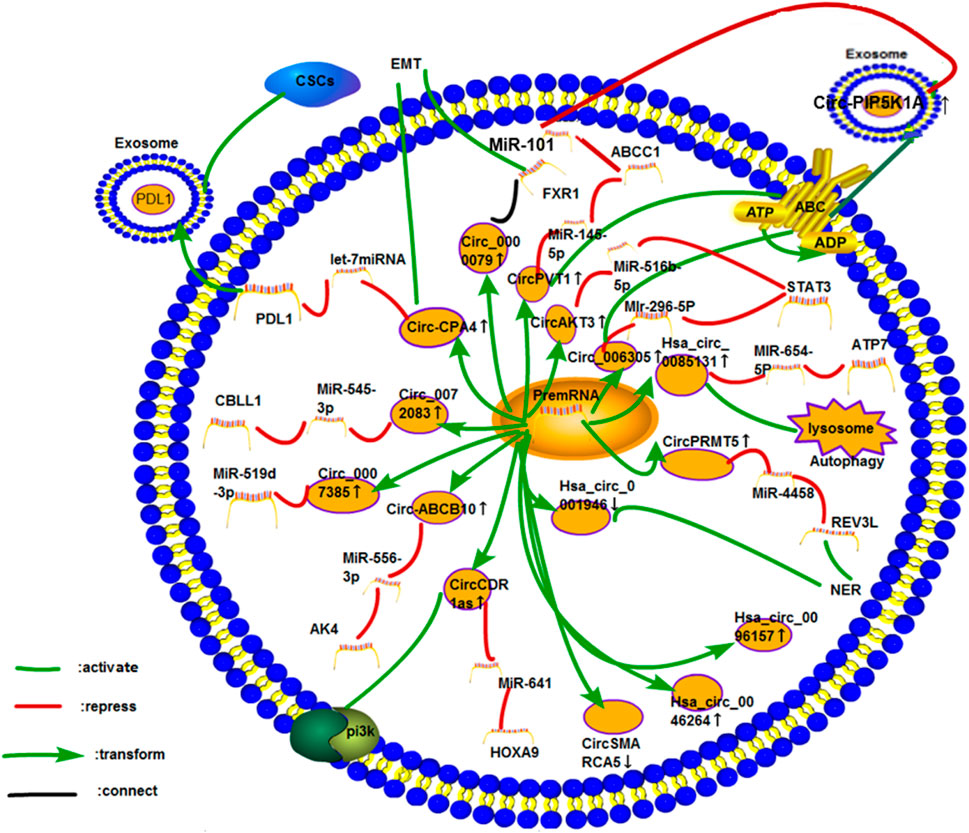
FIGURE 9. Cisplatin resistance mechanism of circRNA in lung cancer. So far, 16 circRNAs have been studied in lung cancer. The mechanisms of cisplatin resistance in these articles include the most common ceRNA NET, exosomes, PI3K pathway, autophagy, ABC transporter, DDR (NER), and CSCs. Three circRNAs, namely, hsa_circ_0096157, hsa_circ_0046264, and CircAMARCA5, play roles in cisplatin resistance in lung cancer. However, their specific mechanisms have not been explored.
CeRNA NET has been extensively studied in the drug resistance of lung cancer cells. Circ_0072083 could enhance the resistance of non-small cell lung cancer (NSCLC) to cisplatin via the miR-545-3p/CBLL1 axis. MiR-545-3p could significantly inhibit the proliferation, cell cycle, and invasion of NSCLC cells and promote cisplatin-induced apoptosis (Li et al., 2020b). The expression of circ_0007385 in NSCLC tissues and cell lines was higher than that in adjacent normal tissues, and this finding was associated with low overall survival (OS) rate. Circ_0007385 could regulate the high expression of HMGB1 in lung cancer tissues by sponging miR-519d-3p, so as to promote the proliferation, invasion, and cisplatin resistance of tumors (Ye et al., 2020).
As early as 2017, circ-ABCB10 has been reported to promote the proliferation and invasion of breast cancer cells by sponging miR-1271 (Liang et al., 2017). The knockdown of circ-ABCB10 expression could upregulate the sensitivity of miR-556-3p to cisplatin in lung cancer cells, whereas upregulating AK4 could reverse this phenomenon (Wu et al., 2020). CircAKT3 was revealed to inhibit the homeostasis of glycolysis through miR-516-5P/STAT3 to increase the cisplatin resistance of lung cancer cells (Xu et al., 2020a). Yu Dong et al. had reported that the expression of circ_0076305 was upregulated in drug-resistant NSCLC. Circ_0076305 could enhance the STAT3 expression by sponging miR-296-5P and the resistance of NSCLC to cisplatin (Dong et al., 2019). Western blot analysis showed that upregulating circ_0076305 could improve the protein expression of P-GP (MDR1) and MRP1. As mentioned before, MDR1 and MRP1 are ABC transporters. The ABC transporter has also been reported to be involved in lung cancer. CircPVT1, a ceRNA of miR-145-5p, could regulate the expression of ABCC1 and lower the sensitivity of pulmonary adenocarcinoma to pemetrexed and cisplatin (Zheng and Xu, 2020). Huasong Lu et al. detected the specimens of patients with NSCLC through PCR and found that the expression of circPVT1in the drug-resistant group was higher than that in the drug-sensitivity group when patients were treated with a combination of cisplatin and gemcitabine. The high expression of circPVT1 was also associated with the survival rate of patients (Lu et al., 2020a).
Few studies have focused on exosomes in lung cancer. Circ-CPA4 may regulate PD-L1 by activating the miRNA of let-7 to promote the proliferation, invasion, EMT, and cisplatin resistance of lung cancer cells. Circ-CPA4 knockdown could lower the expression of cyclin D1 and Bcl-2 and reduce the tumorigenesis in mice during heterotransplantation. Some studies had shown that exosomes of NSCLC containing PD-L1 could form immune escape by increasing the mRNA levels of stem cell-related signal and inactivating CD8 T cells in order to enhance cisplatin resistance (Hong et al., 2020). Anti-PD-1/PD-L1 has been widely used to treat solid tumors, but its curative effect is unsatisfactory (Lei et al., 2020). Therefore, PD-1/PD-L1 blocking reagent can be used in combination with cisplatin and other chemotherapeutic drugs. Na Shao et al. demonstrated that exosomal circ_PIP5K1A could promote cisplatin resistance by targeting miR-101 and consequently modulating ABCC1 expression in NSCLC (Shao et al., 2021).
Some studies have investigated the role of EMT-related genes in tumor drug resistance. Chen et al. revealed that the level of circ_0000079 (CiR79) in patients with NSCLC, especially those who were resistant to cisplatin, was significantly decreased. Low circ_0000079 levels were correlated with a low OS rate. Circ_0000079 could block the formation of the FXR1/PRCKI complex by combining with FXR1 to inhibit cell invasion and improve the chemosensitivity of NSCLC. FXR1/PRKCI-mediated glycogen synthesizes kinase 3β and activator protein-1 phosphorylation to inhibit a snail protein level (Chen et al., 2020a). Except NSCLC, FXR1 is related to poor prognosis in some cancers, including ovarian cancer, breast cancer, and head and neck squamous carcinoma. It is related to the PKC expression and the iota and epithelial transition (Raheja and Gandhi, 2016). The snail gene plays an important role in EMT in tumors and the proliferation of tumor cells (Chen et al., 2020a).
In the previous description of circ-CPA4, stem cells are mentioned. The role of CSCs in the cisplatin resistance of lung cancer cannot be ignored. The circRNA CDR1as/miR-641/HOXA9 axis could regulate the apoptosis of stem cells and enhance cisplatin resistance in NSCLC. In cisplatin-resistant NSCLC cells, the overexpression of circRNA CDR1as had been reported to increase the mRNA levels of stem cell signals (SOX2, OCT4, and Nanog). This phenomenon could be reversed by upregulating miR-641 and downregulating HOXA9 (Zhao et al., 2020).
Autophagy helps tumor cells respond to intracellular and environmental stresses, such as hypoxia, malnutrition, and cancer chemotherapy. Autophagy inhibition can improve the therapeutic effect of patients with advanced cancers (Amaravadi et al., 2019). The role of the ceRNA network in regulating tumor autophagy in chemoresistance has been confirmed. Hsa_circ_0085131 could be used as a ceRNA of miR-654-5P to release ATP7, which could enhance the autophagy and cisplatin resistance of lung cancer cells (Kong, 2020).
DDR can be completed by NER. Hsa_circ_0001946 in NSCLC cells was downregulated, and it could inhibit cell proliferation, invasion, and migration. The downregulation of hsa_circ_0001946 could activate the NER signaling pathway and reduce the sensitivity of cisplatin (Huang et al., 2019b). In addition, the expression of circRNA PRMT5 in cisplatin-resistant NSCLC had been revealed to lower its sensitivity to cisplatin. Moreover, the inhibition on the sensitivity of NSCLC to cisplatin was recovered through miR-4458 inhibition or REV3L upregulation by silencing circRNA PRMT5 (Pang et al., 2020). Therefore, studying the REV3L upstream genes in cisplatin resistance is of great significance.
Yuqiang Mao et al. found that circRNA CDR1as could enhance the pemetrexed and cisplatin resistance of lung adenocarcinoma in neoadjuvant chemotherapy through the EGFR/PI3K signaling pathway. Western blot analysis shows that the expression of EGFR and PI3K proteins in drug-resistant cell lines increases. In addition, the silencing of circRNA CDR1as could lead to the sensitivity of lung cancer cells to chemotherapeutic drugs. This effect can be blocked by upregulating EGFR (Mao and Xu, 2020). Huasong Lu et al. also reported that hsa_circ_0096157 could regulate the cisplatin resistance of NSCLC by regulating cell proliferation, apoptosis, and cell cycle (Lu et al., 2020b). The expression of circ-SMARC5 was found to reduce the proliferation of NSCLC and improve its chemosensitivity to cisplatin and gemcitabine (Tong, 2020). Liu et al. found that the expression of hsa_circ_0046264 in lung cancer was significantly upregulated, and this upregulation was associated with tumor proliferation, invasion, and stage. The expression of hsa-circ_0046264 was high in cisplatin-resistant cell lines (Liu et al., 2020a). The cisplatin resistance mechanism of these circRNAs has not been discussed and should be further studied.
GC is the fifth-most common cancer in the world. Generally, most patients are diagnosed with GC at an advanced stage. Systemic chemotherapy is still the main treatment for patients with advanced GC (Wagner et al., 2017). Cisplatin is commonly used in clinical chemotherapy. Some chemotherapy regimens, including docetaxel, cisplatin, and 5-fluorouracil, have been proven to be effective in the treatment of GC (Li et al., 2019a). (Figure 10).
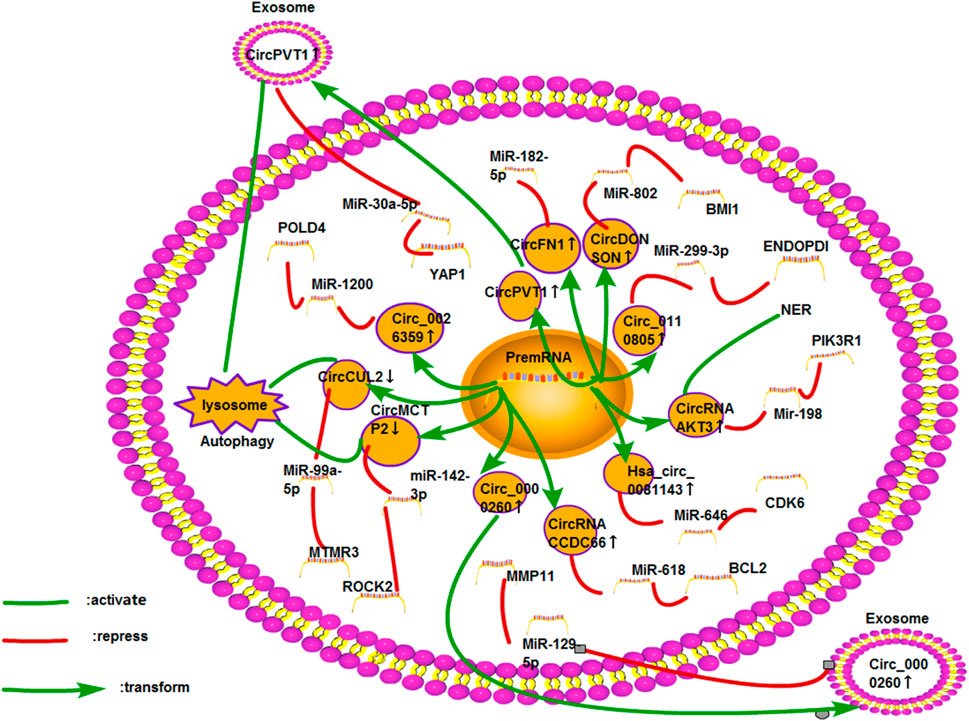
FIGURE 10. Cisplatin resistance mechanism of circRNA in GC. At present, 11 circRNAs are found to be related to cisplatin resistance in GC. The mechanisms involved include ceRNA NET, exosomes, autophagy, and DNA damage and repair.
Many studies have investigated the effect of circRNAs on the cisplatin resistance of GC, and most of them are based on the ceRNA network. Minghui Xue et al. reported that the expression of hsa_circ_0081143 in GC cells was upregulated, which was related to the proliferation, invasion, and metastasis of tumors. Hsa_circ_0081143 can also enhance the cisplatin resistance of GC cells via the miR-646/CDK6 pathway (Xue et al., 2019a). Circ_0110805 was overexpressed in cisplatin-resistant GC tissues or cells. Cisplatin resistance could be enhanced through the miR-299-3P/ENDOPDI axis (Yang et al., 2020a). The ceRNA network for circRNA /miR-646/ miR-299-3P should be further studied.
Lower OS rate, recurrence-free survival, and cisplatin resistance rate were associated with high circ_0026359 levels. Circ_0026359 could enhance the cisplatin resistance of GC via the miR-1200/POLD4 pathway (Zhang et al., 2020a). CircRNA 0001785 was shown to promote the proliferation of osteosarcoma and inhibit its apoptosis through the miR-1200/HOXB2 axis (Li et al., 2019b). Therefore, we should explore whether circRNA 0001785 also has a chemotherapy mechanism, including cisplatin resistance, in osteosarcoma or GC. Xiao Xu Huang et al. first found that circFN1 could improve the survival rate and cisplatin resistance of GC cells by sponging miR-182-5p; thus, it could be used as a new therapeutic target for cisplatin-resistant patients with GC (Huang et al., 2020). Cheng Yang et al. had found that circFN1 could promote the resistance of liver cancer cells to sorafenib (Yang et al., 2020b). Quantitative reverse transcription PCR (qRT-PCR) showed that the expression level of circRNA CCD66 in GC tissues was higher than that in normal tissues, especially in cisplatin-resistant cell lines. In terms of the mechanism, circRNA CCD66 could inhibit apoptosis by targeting the miR-618/BCL2 axis (Zhang et al., 2020b). Recently, circRNA CCD66 had been found to promote the resistance of CRC to oxaliplatin by regulating autophagy (Lin et al., 2020). CircDONSON was upregulated in GC cells. The transfection of si-circDONSON could lower the IC50 value of DDP, indicating that circDONSON could promote cisplatin resistance of GC. Studies had shown that circDONSON inhibited apoptosis and cisplatin resistance by sponging miR-802 (Liu et al., 2020b).
Studies have also investigated autophagy in GC. QRT-PCR showed that the level of circCUL2 in GC tissues significantly lowered than that in normal tissues. In GC cells overexpressing circCUL2, the levels of autophagosomes were low. Western blot analysis showed that BCL-2 expression is similar to the trend of autophagosomes. CircCUL2 was revealed to regulate the sensitivity of cisplatin via the miR-142-3P/ROCK2 axis (Peng et al., 2020). In comparison with chemosensitive cell lines, downregulateing the expression of circMCTP2 could activate autophagy in cisplatin-resistant GC cells. The heterotransplantation model showed the same trend. The overexpression of circMCTP2 could inhibit autophagy and enhance cisplatin sensitivity via the miR-99a-5p/MTMR3 axis (Sun et al., 2020a). Therefore, autophagy can enhance cisplatin resistance. In terms of the exosomes of GC cells, autophagy is also involved. In GC, circPVT1 from exosomes could regulate autophagy through the miR-30A-5P/YAP1 axis and enhance cisplatin resistance (Yao, 2020). The abnormal expression of YAP1 in HCC, GC, CRC, and lung cancer is regarded as a sign of poor prognosis (Shibata et al., 2018). Therefore, the regulation of the drug resistance of tumor cells through the ceRNA network should be studied by targeting the YAP1 gene. In addition, the expression of circ_0000260 in cisplatin-resistant gastric adenocarcinoma tissues and exosomes from the serum was higher than that in sensitive tumor tissues. A dual-luciferase experiment had demonstrated that miR-129-5p was the downstream target gene of circ_0000260, and MMP11 was the direct target gene of miR-129-5p (Liu et al., 2020c).
The expression of circAKT3 was upregulated in cisplatin-resistant GC, which had a strong invasive capacity. Through detecting the levels of caspase-3 and BRCA1, circAKT3 could inhibit apoptosis and promote DDR. On the basis of miRanda, RNAhybrid, GeneChip, and other databases, Huang Xiaoxu et al. revealed that circAKT3 could regulate the expression of PI3KR1 by sponging miR-198 (Huang et al., 2019a). As mentioned above, circAKT3 may enhance the cisplatin resistance of lung cancer cells (Xu et al., 2020a). This phenomenon indicates that circAKT3 may be a valuable gene for studying other cancers. However, the expression of circAKT3 was downregulated in renal clear cell cancer, thereby reducing its metastatic ability through the miR-296-3P/E-cadherin signaling pathway (Xue et al., 2019b). However, the study does not explore the next study on its antitumor effect.
Bladder cancer is the ninth-most common cancer in the world (Gong et al., 2020a). Cisplatin-based chemotherapy has been used to treat muscle-invasive bladder cancer. However, its curative effect remains unsatisfactory (Schardt et al., 2019). (Figure 11).
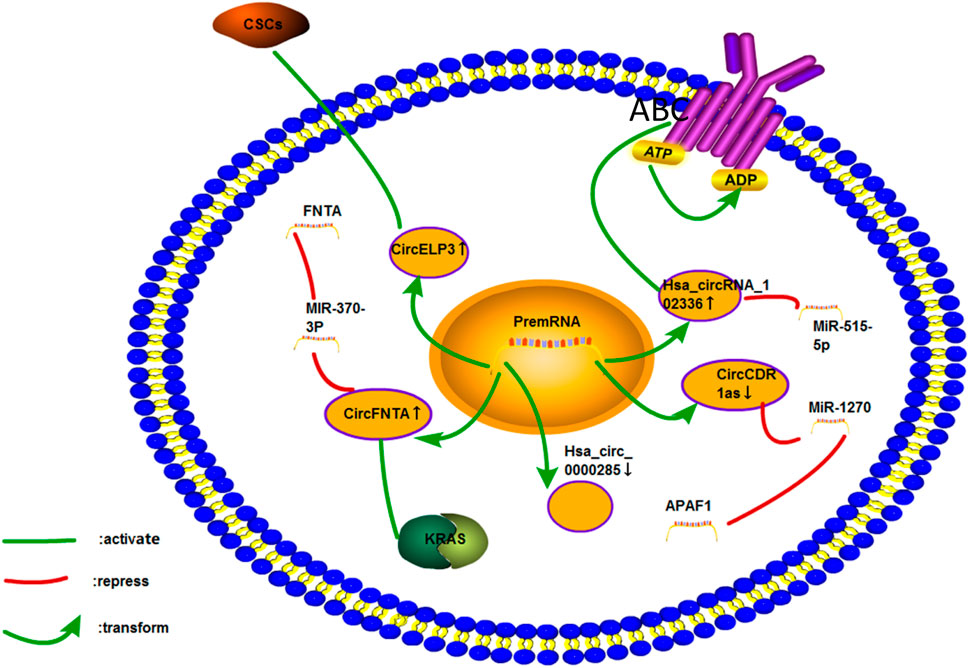
FIGURE 11. Cisplatin resistance mechanism of circRNA in bladder cancer. Only five circRNAs associated with the cisplatin resistance of bladder cancer have been studied. The mechanisms involved include ceRNA NET, CSCs, ABC transporter, and KRAS pathways.
The circRNA CDR1as could promote the resistance of lung cancer to chemotherapeutic drugs (Mao and Xu, 2020; Zhao et al., 2020). It could also promote the apoptosis and sensitivity of bladder cancer cells to cisplatin by sponging miR-1270 and regulating APAF1 (Yuan et al., 2019). Thus, this circRNA seems to have different roles in different cancers. Therefore, its role in the pathogenesis and chemoresistance of different types of cancers should be investigated. The ABC transporter plays an essential role in the cisplatin resistance of bladder cancer. On the basis of the GSE92675 database, Pengfeng Gong et al. found that the upregulation of circ_102336 expression in bladder cancer tissues and cells was associated with tumor proliferation and low survival rate. Circ_102336 could also promote the cisplatin resistance of bladder cancer cells and regulate apoptosis and ABC transport route by sponging miR-515-5p (Gong et al., 2020a). CircRNAs could also promote oxaliplatin resistance in GC by targeting miR-515-5p (Zhong et al., 2020). The androgen acceptor (AR) represses ADAR2 to enhance the expression of circFNTA. CircFNTA could enhance cisplatin resistance of bladder cancer cells through the miR-370-3P/FNTA pathway and activating the KRAS pathway (Chen et al., 2020b).
Yinjie Su et al. found that hypoxia could increase the level of circELP3. They also found that the high expression of circELP3 could inhibit the apoptosis of cancer cells and promote cisplatin resistance by targeting tumor stem-like cells (Su et al., 2019b). Hypoxia could promote the expression of some ncRNAs, so as to enhance the proliferation of bladder cancer cells. Under hypoxic condition, the expression of hsa_circRNA_403658 in bladder cancer was increased, and hsa_circRNA_403658 activated IDHA-mediated aerobic glycolysis to promote the proliferation of bladder cancer cells (Wei et al., 2019). Hypoxia could also induce circNRIP1 to increase its expression and improve the resistance of GC to 5-fluorouracil (Xu et al., 2020b). A previous study involving qPCR showed that hsa_circ_0000285 could improve the sensitivity of patients with bladder cancer to cisplatin, which may be related to the stage, differentiation, and metastasis of tumors (Chi et al., 2019). However, the study did not explore the specific mechanism of drug resistance. Further studies on the mechanisms involved in some articles should be performed.
Liver cancer is the second leading cause of cancer death worldwide (Pratama et al., 2019). Cisplatin is the standard drug for transcatheter arterial chemoembolization in the treatment of liver cancer in Japan (Ikeda, 2019). However, cisplatin resistance often occurs during chemotherapy and consequently lowers the survival rate of patients (Chen et al., 2019b). (Figure 12).
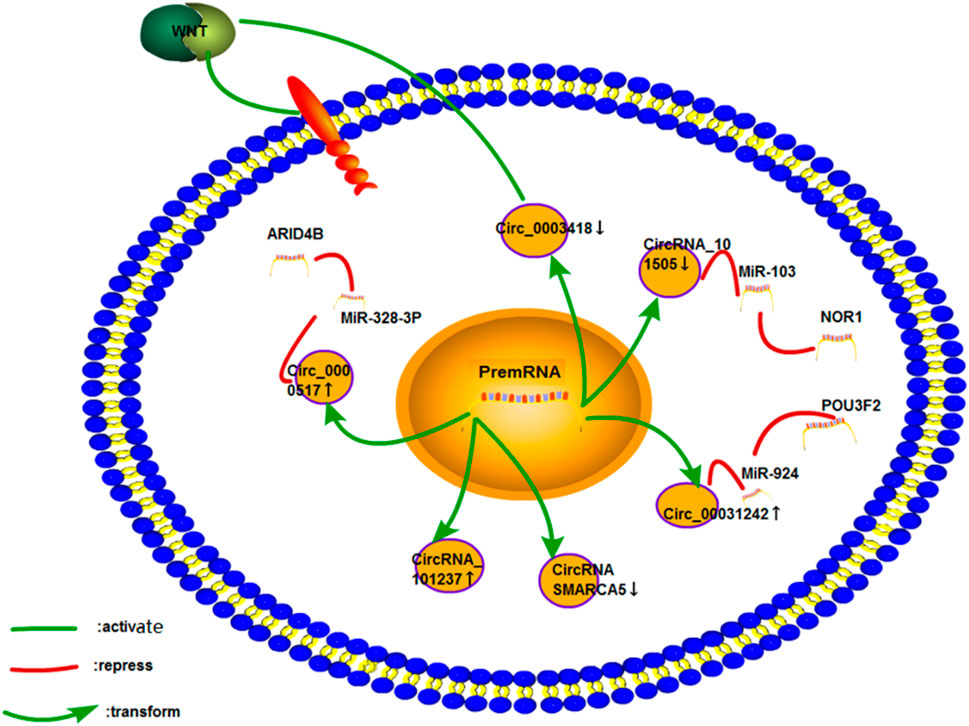
FIGURE 12. Cisplatin resistance mechanism of circRNA in liver cancer. Only six studies on liver cancer have been performed, and few studies on the mechanism of drug resistance have been conducted. The mechanism of the cisplatin resistance of CircRNA_101237 and CircRNA SMARCA5 has not been studied.
Hepatobiliary carcinoma (HCC) is the most common among all liver cancers (Chen et al., 2019b). Yanwei Luo et al. found a new circRNA (circRNA_101505) that was downregulated in the cisplatin resistance of HCC. Bioinformatic prediction and luciferase assay revealed that miR-103 was a direct downstream target of circRNA_101505. CircRNA_101505 could enhance the sensitivity of cisplatin to liver cancer cell lines through miR-103/NOR1 (Luo et al., 2019). Wei Fan et al. used qRT-PCR and Western blot analysis to examine the gene and protein expression levels of circ_0031242, miR-924, and POU3F2. They found that the miR-924/POU3F2 axis could be regulated to promote the cisplatin resistance, proliferation, and invasion of hepatoma cells by circ_0031242. The same results were observed in xenografts (Fan et al., 2021). The upregulation of the circ_0000517 expression in cisplatin-resistant hepatocellular carcinoma cells could promote cell cycle arrest and apoptosis through the miR-328-3P/ARID4B axis. The effect of circ_0000517 on glycolysis in liver cancer cells was analyzed through an ECAR assay. Circ_0000517 could reduce cell glycolysis (Zhao et al., 2021).
Circ_0003418 was found to lower cisplatin resistance in HCC. Silencing circ_0003418 could enhance cisplatin resistance in HCC via the Wnt/catenin signaling pathway (Chen et al., 2019b). Shuping Zhou et al. showed that circRNA_101237 was not only related to the proliferation, invasion, metastasis, and stage of HCC; it also enhanced the resistance of HCC to cisplatin (Zhou et al., 2020). However, the exact drug resistance mechanism of circRNA_101237 remains unclear.
Intrahepatic cholangiocarcinoma is the second-most common type of primary cancer of the liver. Qi Lu et al. conducted a subgroup analysis on 92 patients with intrahepatic cholangiocarcinoma. They found that the expression of circ-SMARCA5 in cancer tissues decreased, and this was negatively correlated with ECOG grading and TNM stage. The overexpression of circ-SMARCA5 could increase the sensitivity of ICC cells to cisplatin and gemcitabine (Lu and Fang, 2020).
Osteosarcoma is one of the most common malignant bone tumors in young individuals (Zhang et al., 2018b; Gong et al., 2020b; Hu et al., 2020). The treatment regime covers a few weeks of chemotherapy before and after surgery. The main treatment method includes methotrexate, adriamycin, and cisplatin (Tang et al., 2019). (Figure 13).
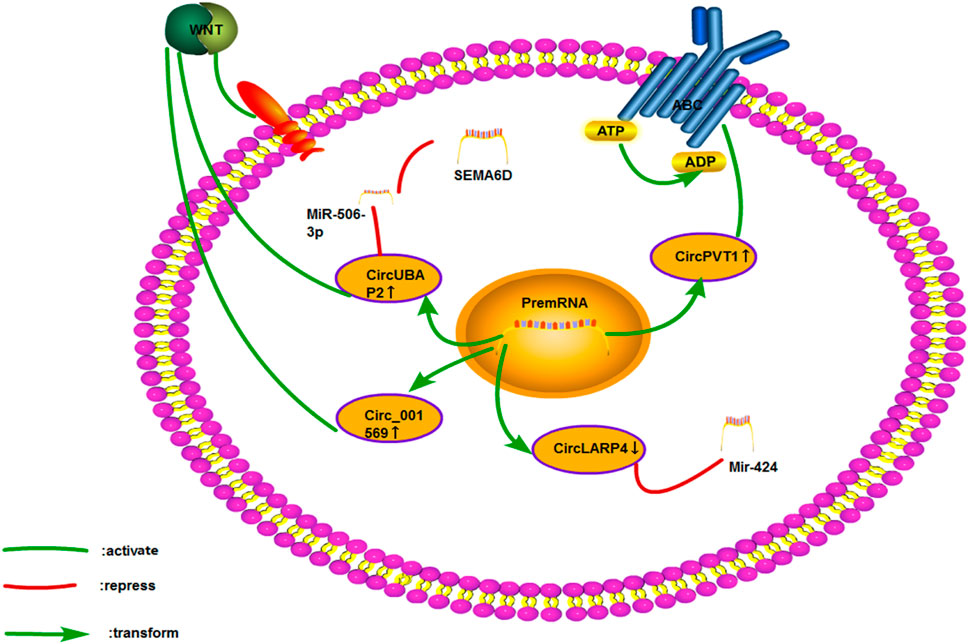
FIGURE 13. Cisplatin resistance mechanism of circRNA in osteosarcoma. Only four papers have been published on the cisplatin resistance of circRNA in osteosarcoma, which involves ceRNA NET, Wnt signaling pathway, and ABC transporter.
Yuhang Hu et al. evaluated 72 patients with osteosarcoma. They found that circRNA LARP4 was negatively correlated with the Enneking staging of osteosarcoma. In osteosarcoma cell lines with overexpressed circRNA LARP4, the IC50 value related to cisplatin and adriamycin was significantly lowered. Similarly, the circRNA LARP4 could increase its sensitivity to chemotherapy by sponging miR-424 (Hu et al., 2020). Recent studies had shown that the circRNA LARP4 could promote the sensitivity of breast cancer to adriamycin (Zhang et al., 2020c). Through the miR-506-3P/SEMA6D axis, circUBAP2 was reported to activate the Wnt/catenin signaling pathway and promote the proliferation, invasion, and cisplatin resistance of osteosarcoma. The expression of SEMA6D was upregulated in GC and esophageal cancer. However, few studies have investigated its drug resistance mechanism (Gong et al., 2020b).
Circ_001569 was also associated with the Wnt signaling pathway in osteosarcoma. Upregulating the expression of circ_001569 in osteosarcoma was correlated with distant metastasis and TNM staging, and the resistance of osteosarcoma cells to cisplatin was enhanced by activating the Wnt/catenin signaling pathway (Zhang et al., 2018b). As mentioned earlier, circPVT1 could regulate the expression of ABCC1 and promote cisplatin resistance in lung cancer (Zheng and Xu, 2020). Zhu Kun-Peng et al. found that circPVT1 could promote the resistance of osteosarcoma to cisplatin and adriamycin. Silencing circPVT1 was found to lower the expression of the drug resistance-related gene ABCB1 (Kun-Peng et al., 2018).
Thyroid cancer is a common malignant tumor (Liu et al., 2018a). It can be divided into five pathological types: papillary thyroid carcinoma (PTC), follicular thyroid cancer, poorly differentiated thyroid cancer, medullary thyroid carcinoma, and undifferentiated thyroid cancer (Liu et al., 2018a; Liu et al., 2018b). The most aggressive type is anaplastic thyroid cancer (ATC) (Zhang et al., 2014). The treatment of thyroid cancer includes surgery, radioactive iodine therapy, and chemotherapy (Carling and Udelsman, 2014). Cisplatin is the foundation of chemotherapy, but its effect is unsatisfactory, especially for ATC (Zhang et al., 2014).
Liu et al. confirmed that circEIF6 (HSA) could enhance the cisplatin resistance of cells in PTC and ATC. CircEIF6 regulates the TGF-β expression by sponging miR-144-3P, promotes the autophagy of thyroid cancer cells, and enhances cisplatin resistance (Liu et al., 2018b). TGF-β is an important gene in EMT mechanism. EMT may participate in the above process. However, the author did not discuss it.
Cervical carcinoma is the second-most common malignant tumor in women, and ovarian carcinoma is the most common cause of death in females (Vaughan et al., 2011; Peng et al., 2016). Cisplatin is still the main chemotherapy regimen in both cancers (Vargas-Hernández et al., 2014; Feng et al., 2017). However, cisplatin resistance has affected the curative effect and survival rate of patients (Waldmann et al., 2013; Peng et al., 2016).
The differential expression of circRNA CDR1as in bladder cancer and NSCLC had been reported. circRNA CDR1as was highly expressed in NSCLC cells, whereas it is weakly expressed in bladder cancer cells (Yuan et al., 2019; Zhao et al., 2020). Zhao et al. recently found that circRNA CDR1as was also poorly expressed in ovarian carcinoma and could enhance the sensitivity of cisplatin through the miR-1270/SCAI axis (Zhao et al., 2019). Therefore, circRNA CDR1as has a huge potential in the research on cisplatin resistance. In terms of exosomes, studies have explored ovarian carcinoma. Luo Yanwei Luo et al. Found that high levels of the exosomal circFOXP1 in cisplatin-resistant ovarian carcinoma could regulate the expression of CEBPG and FMNL3 through miR-22 and miR-150-3P to improve the cisplatin resistance of ovarian carcinoma (Luo and Gui, 2020).
Few studies have been performed on cervical cancer. In one study, Hsa_circ_0023404 could promote the invasion, metastasis, and cisplatin resistance of cervical cancer through the miR-5047/VEGFA axis. In addition, using the autophagy inhibitor 3-MA could recover the inhibitory effect of hsa_circ_0023404 on autophagy-induced apoptosis and improve the sensitivity of chemotherapy (Guo et al., 2019).
Nasopharyngeal carcinoma is a common head and neck cancer worldwide (He et al., 2015). Cisplatin is still the main chemotherapeutic drug in the standard chemotherapy regimen for patients with advanced nasopharyngeal carcinoma (Tang et al., 2018). However, numerous patients have a poor response to chemotherapy, resulting in cisplatin resistance (Liu et al., 2018c).
Qiongna Dong et al. found that the overexpression of hsa_circ_002807 was related to the staging, invasion, and metastasis of nasopharyngeal carcinoma, and hsa_circ_002807 could enhance the resistance to cisplatin and paclitaxel (Qiongna et al., 2020). However, its mechanism remains to be elucidated. The effect of circRNAs on cisplatin resistance in nasopharyngeal carcinoma in terms of autophagy should be further investigated.
OSCC is a highly invasive malignant tumor and the most common malignant tumor in the head and neck (Bray et al., 2018; Pai et al., 2019). Surgery, postoperative radiotherapy, and chemotherapy are the main treatment methods. The main drug in chemotherapy is cisplatin (Pai et al., 2019). However, the emergence of chemotherapy resistance decreases the curative effect of chemotherapeutic drugs (Pérez-Sayáns et al., 2010).
Circ_0001971 had recently been found to increase significantly in OSCC. Circ_0001971 could promote the proliferation, invasion, migration, and cisplatin resistance of tumors by sponging miR-194 and miR-204. Western blot analysis showed that the EMT-related proteins cyclin D1 and N-cadherin and the expression of E-cadherin increased significantly after silencing circ_0001971 (Tan et al., 2020). Therefore, EMT plays an important role in it. ABC transporters participate in the cisplatin resistance of OSCC. The expression of circ_0109291 in cisplatin-resistant OSCC cell lines was upregulated, thereby inhibiting apoptosis. Circ_0109291 could promote cisplatin resistance via the miR-188-3P/ABCB1 axis (Gao et al., 2020).
Laryngocarcinoma is one of the most common cancers in the respiratory system, head, and neck (Liu et al., 2016; Steuer et al., 2017). It is more common in men than in women (Steuer et al., 2017). Its main treatment strategy is chemotherapy, especially patients at an advanced stage (Liu et al., 2016). However, drug resistance occurs with the long-term use of chemotherapeutic drugs (Liang et al., 2014). One of the most common chemotherapeutic drugs for laryngocarcinoma is cisplatin. Its initial effect is good, but cisplatin resistance occurs over time (Tian et al., 2017).
Xuehan Yi et al. found that circ_0004507 could promote the proliferation, invasion, migration, and cisplatin resistance of laryngocarcinoma cells by sponging miR-873. Circ_0004507 was revealed to enhance the expression and translation of MDR1 (ABCB1) and ABCC1, so as to regulate cisplatin resistance (Yi et al., 2020). Recently, circPGAM1 had been reported to inhibit apoptosis of laryngocarcinoma cells and enhance cisplatin resistance. MiR-376A was a direct downstream target of circPGAM1, which could inhibit the expression of ATG2A to improve the sensitivity of cisplatin (Feng et al., 2021).
In recent years, the mortality of patients with esophageal cancer has increased significantly. Patients with esophageal cancer are always diagnosed at an advanced stage in addition to the high incidence rate, so chemotherapy becomes important (Huang and Yu, 2018). The basic drug used in chemotherapy is cisplatin (Su et al., 2019c). Cisplatin remains effective at early stages, but the emergence of cisplatin resistance gradually decreases the survival of patients with esophageal cancer (Hou et al., 2017; Zou et al., 2020).
CircRNA_001275 was significantly increased in cisplatin-resistant esophageal cancer tissues and cells. Through the miR-370-3P/WNT7A axis, circRNA_001275 could inhibit apoptosis and promote the proliferation and drug resistance of esophageal cancer cells (Zou et al., 2020). The mechanism of cisplatin resistance in esophageal cancer remains unknown, but it should be further explored in terms of autophagy and signaling pathways.
Colon cancer is the third-most common cancer in men and the second-most common cancer in women, accounting for about 10 and 9.2%, respectively (Pan et al., 2017). The combination of surgery, chemotherapy, and radiation has been the most common treatment for rectal cancer. Chemotherapy is used before, during, and after surgery and has a different role (Karpisheh et al., 2019). Patients with advanced colon cancer are still treated with cisplatin-based combination chemotherapy and radiotherapy to improve the quality of life and prolong the survival time (Scott et al., 2009).
The luciferase experiment showed a targeting relationship between miR-487a-3P and circ_0020095. The in vitro and in vivo silencing of circ_0020095 could inhibit colon cancer proliferation and invasion and enhance cisplatin sensitivity via the miR-487A-3P/SOX9 axis (Sun et al., 2020b). This finding also provides further insights into targeted treatments for colon cancer and even rectal cancer.
Research on circRNA in cisplatin resistance has attracted increasing attention in recent years, and most studies are based on the ceRNA network. The ceRNA network is essential for studying the mechanism of cisplatin resistance. The ceRNA network gives us a good framework in studies. In this large framework, one or several mechanisms involve the combined action in other mechanisms to explore tumor proliferation, invasion, and drug resistance. For example, studies on exosomes should consider whether CSCs participate in this process and detect CSC-related pathways, such as FGFR and MEK, or the mRNA level of related genes (SOX2, OCT4, and Nanog). In addition, the expression of RAS gene, ABCG2 (ABC transporter) and REV3L (DNA repair) may be a future research direction. Autophagy can promote and inhibit tumor proliferation and growth, which are a two-way mechanism. However, recent research has shown that circRNA can promote drug resistance in cisplatin resistance by activating autophagy. In the future, studies should detect the expression of more ATGs, such as P62 and LC3, and explore whether they can inhibit drug resistance. The exosome PD-L1 influences the cisplatin resistance of lung cancer by affecting the immune system. This finding suggests that immunotherapy combined with chemotherapy may improve the postoperative survival rate of patients with cancer. N6-methyladenosine (m6A), a common RNA including ncRNAs (circRNA) methylation modification, has gradually entered the field of vision in the proliferation of tumor cells. However, the role of circRNA m6A in the regulation of chemotherapeutic drugs such as cisplatin still need to be explored further. The cisplatin resistance mechanism of circRNA in different cancers is summarized in this review to provide a basis for future studies on circRNAs. Therefore, this review may provide a new idea for the clinical improvement of cisplatin resistance Table 1.
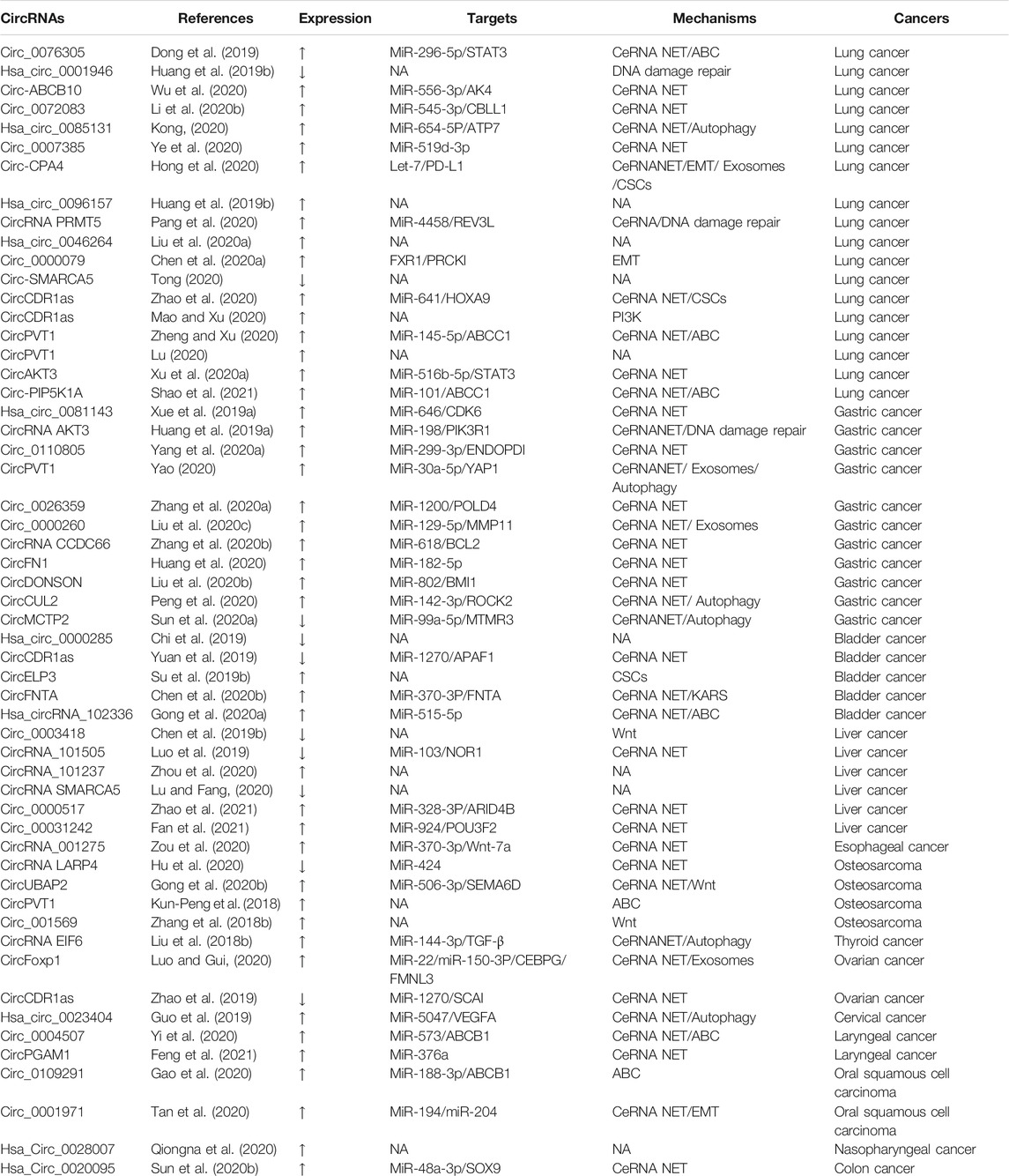
TABLE 1. Collection of papers on circRNA in cisplatin resistance. Expression refers to the circRNA expression in cancers. Targets correspond to targeted genes in circRNA downstream. Mechanism denotes the drug resistance mechanism of circRNA in various tumors. NA: not applicable.
QM and YL collected the related reports, drafted and wrote the manuscript. CL and XL helped revise the manuscript. CH, YX, and LT provided direction and guidance throughout the preparation of this manuscript. All authors read and approved the final manuscript.
This work was supported by China Postdoctoral Science Foundation (2020TQ0253, 2020M682927).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ajani, J. A., Song, S., Hochster, H. S., and Steinberg, I. B. (2015). Cancer Stem Cells: the Promise and the Potential. Semin. Oncol. 42 (Suppl. 1), S3–S17. doi:10.1053/j.seminoncol.2015.01.001
Amable, L. (2016). Cisplatin Resistance and Opportunities for Precision Medicine. Pharmacol. Res. 106, 27–36. doi:10.1016/j.phrs.2016.01.001
Amaravadi, R., Kimmelman, A. C., and White, E. (2016). Recent Insights into the Function of Autophagy in Cancer. Genes Dev. 30 (17), 1913–1930. doi:10.1101/gad.287524.116
Amaravadi, R. K., Kimmelman, A. C., and Debnath, J. (2019). Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 9 (9), 1167–1181. doi:10.1158/2159-8290.CD-19-0292
An, Y., Furber, K. L., and Ji, S. (2017). Pseudogenes Regulate Parental Gene Expression via ceRNA Network. J. Cel Mol Med 21 (1), 185–192. doi:10.1111/jcmm.12952
Belousova, E. A., Filipenko, M. L., and Kushlinskii, N. E. (2018). Circular RNA: New Regulatory Molecules. Bull. Exp. Biol. Med. 164 (6), 803–815. doi:10.1007/s10517-018-4084-z
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492
Brown, J. S., O'Carrigan, B., Jackson, S. P., and Yap, T. A. (2017). Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 7 (1), 20–37. doi:10.1158/2159-8290.CD-16-0860
Carling, T., and Udelsman, R. (2014). Thyroid Cancer. Annu. Rev. Med. 65, 125–137. doi:10.1146/annurev-med-061512-105739
Chang, J. C. (2016). Cancer Stem Cells: Role in Tumor Growth, Recurrence, Metastasis, and Treatment Resistance. Medicine (Baltimore) 95 (Suppl. 1), S20–S25. doi:10.1097/MD.0000000000004766
Chen, C., Zhang, M., and Zhang, Y. (2020). Circ_0000079 Decoys the RNA-Binding Protein FXR1 to Interrupt Formation of the FXR1/PRCKI Complex and Decline Their Mediated Cell Invasion and Drug Resistance in NSCLC. Cel Transpl. 29, 963689720961070. doi:10.1177/0963689720961070
Chen, H., Liu, S., Li, M., Huang, P., and Li, X. (2019). circ_0003418 Inhibits Tumorigenesis and Cisplatin Chemoresistance through Wnt/β-Catenin Pathway in Hepatocellular Carcinoma. Onco Targets Ther. 12, 9539–9549. doi:10.2147/OTT.S229507
Chen, J., Sun, Y., Ou, Z., Yeh, S., Huang, C. P., You, B., et al. (2020). Androgen Receptor-Regulated circFNTA Activates KRAS Signaling to Promote Bladder Cancer Invasion. EMBO Rep. 21 (4), e48467. doi:10.15252/embr.201948467
Chen, L. L., and Yang, L. (2015). Regulation of circRNA Biogenesis. RNA Biol. 12 (4), 381–388. doi:10.1080/15476286.2015.1020271
Chen, S., Li, F., Xu, D., Hou, K., Fang, W., and Li, Y. (2019). The Function of RAS Mutation in Cancer and Advances in its Drug Research. Curr. Pharm. Des. 25 (10), 1105–1114. doi:10.2174/1381612825666190506122228
Chen, T., You, Y., Jiang, H., and Wang, Z. Z. (2017). Epithelial-mesenchymal Transition (EMT): A Biological Process in the Development, Stem Cell Differentiation, and Tumorigenesis. J. Cel Physiol 232 (12), 3261–3272. doi:10.1002/jcp.25797
Chi, B. J., Zhao, D. M., Liu, L., Yin, X. Z., Wang, F. F., Bi, S., et al. (2019). Downregulation of Hsa_circ_0000285 Serves as a Prognostic Biomarker for Bladder Cancer and Is Involved in Cisplatin Resistance. Neoplasma 66 (2), 197–202. doi:10.4149/neo_2018_180318N185
Chiarugi, P., and Cirri, P. (2016). Metabolic Exchanges within Tumor Microenvironment. Cancer Lett. 380 (1), 272–280. doi:10.1016/j.canlet.2015.10.027
Cocetta, V., Ragazzi, E., and Montopoli, M. (2019). Mitochondrial Involvement in Cisplatin Resistance. Int. J. Mol. Sci. 20 (14). doi:10.3390/ijms20143384
Dasari, S., and Tchounwou, P. B. (2014). Cisplatin in Cancer Therapy: Molecular Mechanisms of Action. Eur. J. Pharmacol. 740, 364–378. doi:10.1016/j.ejphar.2014.07.025
Dong, Y., Xu, T., Zhong, S., Wang, B., Zhang, H., Wang, X., et al. (2019). Circ_0076305 Regulates Cisplatin Resistance of Non-small Cell Lung Cancer via Positively Modulating STAT3 by Sponging miR-296-5p. Life Sci. 239, 116984. doi:10.1016/j.lfs.2019.116984
Du, B., and Shim, J. S. (2016). Targeting Epithelial-Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 21 (7). doi:10.3390/molecules21070965
Duchartre, Y., Kim, Y. M., and Kahn, M. (2016). The Wnt Signaling Pathway in Cancer. Crit. Rev. Oncol. Hematol. 99, 141–149. doi:10.1016/j.critrevonc.2015.12.005
Fan, W., Chen, L., Wu, X., and Zhang, T. (2021). Circ_0031242 Silencing Mitigates the Progression and Drug Resistance in DDP-Resistant Hepatoma Cells by the miR-924/POU3F2 Axis. Cancer Manag. Res. 13, 743–755. doi:10.2147/CMAR.S272851
Feng, B., Chen, K., Zhang, W., Zheng, Q., and He, Y. (2021). circPGAM1 Enhances Autophagy Signaling during Laryngocarcinoma Drug Resistance by Regulating miR-376a. Biochem. Biophys. Res. Commun. 534, 966–972. doi:10.1016/j.bbrc.2020.10.063
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The Machinery of Macroautophagy. Cell Res 24 (1), 24–41. doi:10.1038/cr.2013.168
Feng, Y., Zou, W., Hu, C., Li, G., Zhou, S., He, Y., et al. (2017). Modulation of CASC2/miR-21/PTEN Pathway Sensitizes Cervical Cancer to Cisplatin. Arch. Biochem. Biophys. 623-624, 20–30. doi:10.1016/j.abb.2017.05.001
Ferreira, J. A., Peixoto, A., Neves, M., Gaiteiro, C., Reis, C. A., Assaraf, Y. G., et al. (2016). Mechanisms of Cisplatin Resistance and Targeting of Cancer Stem Cells: Adding Glycosylation to the Equation. Drug Resist. Updat 24, 34–54. doi:10.1016/j.drup.2015.11.003
Gao, F., Han, J., Wang, Y., Jia, L., Luo, W., Zeng, Y., et al. (2020). Circ_0109291 Promotes the Cisplatin Resistance of Oral Squamous Cell Carcinoma by Sponging miR-188-3p to Increase ABCB1 Expression. Cancer Biother. Radiopharm. 4. doi:10.1089/cbr.2020.3928
García-Olmo, D., García-Olmo, D. C., Ontañón, J., Martinez, E., and Vallejo, M. (1999). Tumor DNA Circulating in the Plasma Might Play a Role in Metastasis. The Hypothesis of the Genometastasis. Histol. Histopathol 14 (4), 1159–1164. doi:10.14670/HH-14.1159
Gavande, N. S., VanderVere-Carozza, P. S., Hinshaw, H. D., Jalal, S. I., Sears, C. R., Pawelczak, K. S., et al. (2016). DNA Repair Targeted Therapy: The Past or Future of Cancer Treatment? Pharmacol. Ther. 160, 65–83. doi:10.1016/j.pharmthera.2016.02.003
Ghosh, S. (2019). Cisplatin: The First Metal Based Anticancer Drug. Bioorg. Chem. 88, 102925. doi:10.1016/j.bioorg.2019.102925
Gong, P., Xu, R., Zhuang, Q., and He, X. (2020). A Novel Circular RNA (hsa_circRNA_102336), a Plausible Biomarker, Promotes the Tumorigenesis by Sponging miR-515-5p in Human Bladder Cancer. Biomed. Pharmacother. 126, 110059. doi:10.1016/j.biopha.2020.110059
Gong, X., Li, W., Dong, L., and Qu, F. (2020). Correction to: CircUBAP2 Promotes SEMA6D Expression to Enhance the Cisplatin Resistance in Osteosarcoma through Sponging miR-506-3p by Activating Wnt/β-Catenin Signaling Pathway. J. Mol. Histol. 51 (4), 471–340. doi:10.1007/s10735-020-09894-5
Guo, F., Liu, X., Cai, H., and Le, W. (2018). Autophagy in Neurodegenerative Diseases: Pathogenesis and Therapy. Brain Pathol. 28 (1), 3–13. doi:10.1111/bpa.12545
Guo, J., Chen, M., Ai, G., Mao, W., Li, H., and Zhou, J. (2019). Hsa_circ_0023404 Enhances Cervical Cancer Metastasis and Chemoresistance through VEGFA and Autophagy Signaling by Sponging miR-5047. Biomed. Pharmacother. 115, 108957. doi:10.1016/j.biopha.2019.108957
Han, B., Chao, J., and Yao, H. (2018). Circular RNA and its Mechanisms in Disease: From the Bench to the Clinic. Pharmacol. Ther. 187, 31–44. doi:10.1016/j.pharmthera.2018.01.010
He, C., Zheng, S., Luo, Y., and Wang, B. (2018). Exosome Theranostics: Biology and Translational Medicine. Theranostics 8 (1), 237–255. doi:10.7150/thno.21945
He, Y. Q., Xue, W. Q., Shen, G. P., Tang, L. L., Zeng, Y. X., and Jia, W. H. (2015). Household Inhalants Exposure and Nasopharyngeal Carcinoma Risk: a Large-Scale Case-Control Study in Guangdong, China. BMC Cancer 15, 1022. doi:10.1186/s12885-015-2035-x
Hellsberg, E., Montanari, F., and Ecker, G. F. (2015). The ABC of Phytohormone Translocation. Planta Med. 81 (6), 474–487. doi:10.1055/s-0035-1545880
Hong, W., Xue, M., Jiang, J., Zhang, Y., and Gao, X. (2020). Circular RNA Circ-CPA4/ Let-7 miRNA/PD-L1 axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in Non-small Cell Lung Cancer (NSCLC). J. Exp. Clin. Cancer Res. 39 (1), 149. doi:10.1186/s13046-020-01648-1
Hou, X. F., Xu, L. P., Song, H. Y., Li, S., Wu, C., and Wang, J. F. (2017). ECRG2 Enhances the Anti-cancer Effects of Cisplatin in Cisplatin-Resistant Esophageal Cancer Cells via Upregulation of P53 and Downregulation of PCNA. World J. Gastroenterol. 23 (10), 1796–1803. doi:10.3748/wjg.v23.i10.1796
Hu, Y., Gu, J., Shen, H., Shao, T., Li, S., Wang, W., et al. (2020). Circular RNA LARP4 Correlates with Decreased Enneking Stage, Better Histological Response, and Prolonged Survival Profiles, and it Elevates Chemosensitivity to Cisplatin and Doxorubicin via Sponging microRNA-424 in Osteosarcoma. J. Clin. Lab. Anal. 34 (2), e23045. doi:10.1002/jcla.23045
Huang, F. L., and Yu, S. J. (2018). Esophageal Cancer: Risk Factors, Genetic Association, and Treatment. Asian J. Surg. 41 (3), 210–215. doi:10.1016/j.asjsur.2016.10.005
Huang, M. S., Liu, J. Y., Xia, X. B., Liu, Y. Z., Li, X., Yin, J. Y., et al. (2019). Hsa_circ_0001946 Inhibits Lung Cancer Progression and Mediates Cisplatin Sensitivity in Non-small Cell Lung Cancer via the Nucleotide Excision Repair Signaling Pathway. Front. Oncol. 9, 508. doi:10.3389/fonc.2019.00508
Huang, X., Li, Z., Zhang, Q., Wang, W., Li, B., Wang, L., et al. (2019). Circular RNA AKT3 Upregulates PIK3R1 to Enhance Cisplatin Resistance in Gastric Cancer via miR-198 Suppression. Mol. Cancer 18 (1), 71. doi:10.1186/s12943-019-0969-3
Huang, X. X., Zhang, Q., Hu, H., Jin, Y., Zeng, A. L., Xia, Y. B., et al. (2020). A Novel Circular RNA circFN1 Enhances Cisplatin Resistance in Gastric Cancer via Sponging miR-182-5p. J. Cel Biochem. doi:10.1002/jcb.29641
Ichimiya, T., Yamakawa, T., Hirano, T., Yokoyama, Y., Hayashi, Y., Hirayama, D., et al. (2020). Autophagy and Autophagy-Related Diseases: A Review. Int. J. Mol. Sci. 21 (23). doi:10.3390/ijms21238974
Ikeda, K. (2019). Recent Advances in Medical Management of Hepatocellular Carcinoma. Hepatol. Res. 49 (1), 14–32. doi:10.1111/hepr.13259
Iorio, M. V., and Croce, C. M. (2017). MicroRNA Dysregulation in Cancer: Diagnostics, Monitoring and Therapeutics. A Comprehensive Review. EMBO Mol. Med. 9 (6), 852. doi:10.15252/emmm.201707779
Karpisheh, V., Nikkhoo, A., Hojjat-Farsangi, M., Namdar, A., Azizi, G., Ghalamfarsa, G., et al. (2019). Prostaglandin E2 as a Potent Therapeutic Target for Treatment of colon Cancer. Prostaglandins Other Lipid Mediat 144, 106338. doi:10.1016/j.prostaglandins.2019.106338
Kartalou, M., and Essigmann, J. M. (2001). Mechanisms of Resistance to Cisplatin. Mutat. Res. 478 (1-2), 23–43. doi:10.1016/s0027-5107(01)00141-5
Klionsky, D. J. (2012). Look People, "Atg" Is an Abbreviation for "autophagy-related." That's it. Autophagy 8 (9), 1281–1282. doi:10.4161/auto.21812
Kong, R. (2020). Circular RNA Hsa_circ_0085131 Is Involved in Cisplatin-Resistance of Non-small-cell Lung Cancer Cells by Regulating Autophagy. Cell Biol Int 44 (9), 1945–1956. doi:10.1002/cbin.11401
Kun-Peng, Z., Xiao-Long, M., and Chun-Lin, Z. (2018). Overexpressed circPVT1, a Potential New Circular RNA Biomarker, Contributes to Doxorubicin and Cisplatin Resistance of Osteosarcoma Cells by Regulating ABCB1. Int. J. Biol. Sci. 14 (3), 321–330. doi:10.7150/ijbs.24360
Lathia, J. D., and Liu, H. (2017). Overview of Cancer Stem Cells and Stemness for Community Oncologists. Target. Oncol. 12 (4), 387–399. doi:10.1007/s11523-017-0508-3
Lei, Q., Wang, D., Sun, K., Wang, L., and Zhang, Y. (2020). Resistance Mechanisms of Anti-pd1/pdl1 Therapy in Solid Tumors. Front Cel Dev Biol 8, 672. doi:10.3389/fcell.2020.00672
Levine, B., and Kroemer, G. (2008). Autophagy in the Pathogenesis of Disease. Cell 132 (1), 27–42. doi:10.1016/j.cell.2007.12.018
Li, B., Chen, L., Luo, H. L., Yi, F. M., Wei, Y. P., and Zhang, W. X. (2019). Docetaxel, Cisplatin, and 5-fluorouracil Compared with Epirubicin, Cisplatin, and 5-fluorouracil Regimen for Advanced Gastric Cancer: A Systematic Review and Meta-Analysis. World J. Clin. Cases 7 (5), 600–615. doi:10.12998/wjcc.v7.i5.600
Li, D., Ji, H., Niu, X., Yin, L., Wang, Y., Gu, Y., et al. (2020). Tumor-associated Macrophages Secrete CC-Chemokine Ligand 2 and Induce Tamoxifen Resistance by Activating PI3K/Akt/mTOR in Breast Cancer. Cancer Sci. 111 (1), 47–58. doi:10.1111/cas.14230
Li, H., Liu, F., and Qin, W. (2020). Circ_0072083 Interference Enhances Growth-Inhibiting Effects of Cisplatin in Non-small-cell Lung Cancer Cells via miR-545-3p/CBLL1 axis. Cancer Cel Int 20, 78. doi:10.1186/s12935-020-1162-x
Li, J., Liu, H., Yu, J., and Yu, H. (2015). Chemoresistance to Doxorubicin Induces Epithelial-Mesenchymal Transition via Upregulation of Transforming Growth Factor β Signaling in HCT116 colon Cancer Cells. Mol. Med. Rep. 12 (1), 192–198. doi:10.3892/mmr.2015.3356
Li, S., Pei, Y., Wang, W., Liu, F., Zheng, K., and Zhang, X. (2019). Circular RNA 0001785 Regulates the Pathogenesis of Osteosarcoma as a ceRNA by Sponging miR-1200 to Upregulate HOXB2. Cell Cycle 18 (11), 1281–1291. doi:10.1080/15384101.2019.1618127
Li, X., Wu, C., Chen, N., Gu, H., Yen, A., Cao, L., et al. (2016). PI3K/Akt/mTOR Signaling Pathway and Targeted Therapy for Glioblastoma. Oncotarget 7 (22), 33440–33450. doi:10.18632/oncotarget.7961
Li, Y. J., Lei, Y. H., Yao, N., Wang, C. R., Hu, N., Ye, W. C., et al. (2017). Autophagy and Multidrug Resistance in Cancer. Chin. J. Cancer 36 (1), 52. doi:10.1186/s40880-017-0219-2
Liang, D. P., Huang, T. Q., Li, S. J., and Chen, Z. J. (2014). Knockdown of S100A4 Chemosensitizes Human Laryngeal Carcinoma Cells In Vitro through Inhibition of Slug. Eur. Rev. Med. Pharmacol. Sci. 18 (22), 3484–3490.
Liang, H. F., Zhang, X. Z., Liu, B. G., Jia, G. T., and Li, W. L. (2017). Circular RNA Circ-ABCB10 Promotes Breast Cancer Proliferation and Progression through Sponging miR-1271. Am. J. Cancer Res. 7 (7), 1566–1576.
Lien, E. C., Dibble, C. C., and Toker, A. (2017). PI3K Signaling in Cancer: beyond AKT. Curr. Opin. Cel Biol 45, 62–71. doi:10.1016/j.ceb.2017.02.007
Lin, Y. C., Yu, Y. S., Lin, H. H., and Hsiao, K. Y. (2020). Oxaliplatin-Induced DHX9 Phosphorylation Promotes Oncogenic Circular RNA CCDC66 Expression and Development of Chemoresistance. Cancers (Basel) 12 (3). doi:10.3390/cancers12030697
Liu, F., Tai, Y., and Ma, J. (2018). LncRNA NEAT1/let-7a-5p axis Regulates the Cisplatin Resistance in Nasopharyngeal Carcinoma by Targeting Rsf-1 and Modulating the Ras-MAPK Pathway. Cancer Biol. Ther. 19 (6), 534–542. doi:10.1080/15384047.2018.1450119
Liu, F., Zhang, J., Qin, L., Yang, Z., Xiong, J., Zhang, Y., et al. (2018). Circular RNA EIF6 (Hsa_circ_0060060) Sponges miR-144-3p to Promote the Cisplatin-Resistance of Human Thyroid Carcinoma Cells by Autophagy Regulation. Aging (Albany NY) 10 (12), 3806–3820. doi:10.18632/aging.101674
Liu, J., Feng, L., Zhang, H., Zhang, J., Zhang, Y., Li, S., et al. (2018). Effects of miR-144 on the Sensitivity of Human Anaplastic Thyroid Carcinoma Cells to Cisplatin by Autophagy Regulation. Cancer Biol. Ther. 19 (6), 484–496. doi:10.1080/15384047.2018.1433502
Liu, J., Tang, Q., Li, S., and Yang, X. (2016). Inhibition of HAX-1 by miR-125a Reverses Cisplatin Resistance in Laryngeal Cancer Stem Cells. Oncotarget 7 (52), 86446–86456. doi:10.18632/oncotarget.13424
Liu, P., Wang, Y., and Li, X. (2019). Targeting the Untargetable KRAS in Cancer Therapy. Acta Pharm. Sin B 9 (5), 871–879. doi:10.1016/j.apsb.2019.03.002
Liu, S., Wu, M., and Peng, M. (2020). Circ_0000260 Regulates the Development and Deterioration of Gastric Adenocarcinoma with Cisplatin Resistance by Upregulating MMP11 via Targeting MiR-129-5p. Cancer Manag. Res. 12, 10505–10519. doi:10.2147/CMAR.S272324
Liu, Y., Xu, J., Jiang, M., Ni, L., and Ling, Y. (2020). CircRNA DONSON Contributes to Cisplatin Resistance in Gastric Cancer Cells by Regulating miR-802/BMI1 axis. Cancer Cel Int 20, 261. doi:10.1186/s12935-020-01358-w
Liu, Z. H., Yang, S. Z., Chen, X. T., Shao, M. R., Dong, S. Y., Zhou, S. Y., et al. (2020). Correlations of Hsa_circ_0046264 Expression with Onset, Pathological Stage and Chemotherapy Resistance of Lung Cancer. Eur. Rev. Med. Pharmacol. Sci. 24 (18), 9511–9521. doi:10.26355/eurrev_202009_23036
Lu, H., Xie, X., Chen, Q., Cai, S., Liu, S., and Bao, C., (2020a). Clinical Significance of circPVT1 in Patients with Non-small Cell Lung Cancer Who Received Cisplatin Combined with Gemcitabine Chemotherapy. Tumori 107, 204–208. doi:10.1177/0300891620941940
Lu, H., Xie, X., Wang, K., Chen, Q., Cai, S., Liu, D., et al. (2020b). Circular RNA Hsa_circ_0096157 Contributes to Cisplatin Resistance by Proliferation, Cell Cycle Progression, and Suppressing Apoptosis of Non-small-cell Lung Carcinoma Cells. Mol. Cel Biochem 475 (1–2), 63–77. doi:10.1007/s11010-020-03860-1
Lu, Q., and Fang, T. (2020). Circular RNA SMARCA5 Correlates with Favorable Clinical Tumor Features and Prognosis, and Increases Chemotherapy Sensitivity in Intrahepatic Cholangiocarcinoma. J. Clin. Lab. Anal. 34 (4), e23138. doi:10.1002/jcla.23138
Luo, Y., Fu, Y., Huang, R., Gao, M., Liu, F., Gui, R., et al. (2019). CircRNA_101505 Sensitizes Hepatocellular Carcinoma Cells to Cisplatin by Sponging miR-103 and Promotes Oxidored-nitro Domain-Containing Protein 1 Expression. Cell Death Discov 5, 121. doi:10.1038/s41420-019-0202-6
Luo, Y., and Gui, R. (2020). Circulating Exosomal circFoxp1 Confers Cisplatin Resistance in Epithelial Ovarian Cancer Cells. J. Gynecol. Oncol. 31 (5), e75. doi:10.3802/jgo.2020.31.e75
Mao, Y., and Xu, R. (2020). Circular RNA CDR1-AS Contributes to Pemetrexed and Cisplatin Chemoresistance through EGFR/PI3K Signaling Pathway in Lung Adenocarcinoma. Biomed. Pharmacother. 123, 109771. doi:10.1016/j.biopha.2019.109771
Martin, S. K., and Wood, R. D. (2019). DNA Polymerase ζ in DNA Replication and Repair. Nucleic Acids Res. 47 (16), 8348–8361. doi:10.1093/nar/gkz705
Meng, X., Li, X., Zhang, P., Wang, J., Zhou, Y., and Chen, M. (2017). Circular RNA: an Emerging Key Player in RNA World. Brief Bioinform 18 (4), 547–557. doi:10.1093/bib/bbw045
Nagasaka, M., and Gadgeel, S. M. (2018). Role of Chemotherapy and Targeted Therapy in Early-Stage Non-small Cell Lung Cancer. Expert Rev. Anticancer Ther. 18 (1), 63–70. doi:10.1080/14737140.2018.1409624
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169 (6), 985–999. doi:10.1016/j.cell.2017.05.016
Nussinov, R., Tsai, C. J., and Jang, H. (2017). A New View of Pathway-Driven Drug Resistance in Tumor Proliferation. Trends Pharmacol. Sci. 38 (5), 427–437. doi:10.1016/j.tips.2017.02.001
Pai, S., Bamodu, O. A., Lin, Y. K., Lin, C. S., Chu, P. Y., Chien, M. H., et al. (2019). CD47-SIRPα Signaling Induces Epithelial-Mesenchymal Transition and Cancer Stemness and Links to a Poor Prognosis in Patients with Oral Squamous Cell Carcinoma. Cells 8 (12). doi:10.3390/cells8121658
Pan, P., Skaer, C., Yu, J., Zhao, H., Ren, H., Oshima, K., et al. (2017). Berries and Other Natural Products in the Pancreatic Cancer Chemoprevention in Human Clinical Trials. J. Berry Res. 7 (3), 147–161. doi:10.3233/JBR-170159
Pang, J., Ye, L., Zhao, D., Zhao, D., and Chen, Q. (2020). Circular RNA PRMT5 Confers Cisplatin-Resistance via miR-4458/REV3L axis in Non-small-cell Lung Cancer. Cel Biol Int 44 (12), 2416–2426. doi:10.1002/cbin.11449
Paolini, A., Baldassarre, A., Del Gaudio, I., and Masotti, A. (2015). Structural Features of the ATP-Binding Cassette (ABC) Transporter ABCA3. Int. J. Mol. Sci. 16 (8), 19631–19644. doi:10.3390/ijms160819631
Peng, L., Sang, H., Wei, S., Li, Y., Jin, D., Zhu, X., et al. (2020). circCUL2 Regulates Gastric Cancer Malignant Transformation and Cisplatin Resistance by Modulating Autophagy Activation via miR-142-3p/ROCK2. Mol. Cancer 19 (1), 156. doi:10.1186/s12943-020-01270-x
Peng, L., Yuan, X., Jiang, B., Tang, Z., and Li, G. C. (2016). LncRNAs: Key Players and Novel Insights into Cervical Cancer. Tumour Biol. 37 (3), 2779–2788. doi:10.1007/s13277-015-4663-9
Pérez-Sayáns, M., Somoza-Martín, J. M., Barros-Angueira, F., Diz, P. G., Rey, J. M., and García-García, A. (2010). Multidrug Resistance in Oral Squamous Cell Carcinoma: The Role of Vacuolar ATPases. Cancer Lett. 295 (2), 135–143. doi:10.1016/j.canlet.2010.03.019
Pratama, M. Y., Pascut, D., Massi, M. N., and Tiribelli, C. (2019). The Role of microRNA in the Resistance to Treatment of Hepatocellular Carcinoma. Ann. Transl Med. 7 (20), 577. doi:10.21037/atm.2019.09.142
Prieto-Vila, M., Takahashi, R. U., Usuba, W., Kohama, I., and Ochiya, T. (2017). Drug Resistance Driven by Cancer Stem Cells and Their Niche. Int. J. Mol. Sci. 18 (12). doi:10.3390/ijms18122574
Qi, X., Zhang, D. H., Wu, N., Xiao, J. H., Wang, X., and Ma, W. (2015). ceRNA in Cancer: Possible Functions and Clinical Implications. J. Med. Genet. 52 (10), 710–718. doi:10.1136/jmedgenet-2015-103334
Qiongna, D., Jiafeng, Z., Yalin, H., Ping, H., Chuan, Z., Xiaojie, J., et al. (2020). Implication of Hsa_circ_0028007 in Reinforcing Migration, Invasion, and Chemo-Tolerance of Nasopharyngeal Carcinoma Cells. J. Clin. Lab. Anal. 34 (9), e23409. doi:10.1002/jcla.23409
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: A New star of Noncoding RNAs. Cancer Lett. 365 (2), 141–148. doi:10.1016/j.canlet.2015.06.003
Qu, X., Yu, J., Bhagat, G., Furuya, N., Hibshoosh, H., Troxel, A., et al. (2003). Promotion of Tumorigenesis by Heterozygous Disruption of the Beclin 1 Autophagy Gene. J. Clin. Invest. 112 (12), 1809–1820. doi:10.1172/JCI20039
Raheja, R., and Gandhi, R. (2016). FXR1: Linking Cellular Quiescence, Immune Genes and Cancer. Cell Cycle 15 (20), 2695–2696. doi:10.1080/15384101.2016.1215692
Robey, R. W., Pluchino, K. M., Hall, M. D., Fojo, A. T., Bates, S. E., and Gottesman, M. M. (2018). Revisiting the Role of ABC Transporters in Multidrug-Resistant Cancer. Nat. Rev. Cancer 18 (7), 452–464. doi:10.1038/s41568-018-0005-8
Rocha, C. R. R., Silva, M. M., Quinet, A., Cabral-Neto, J. B., and Menck, C. F. M. (2018). DNA Repair Pathways and Cisplatin Resistance: an Intimate Relationship. Clinics (Sao Paulo) 73 (Suppl. 1), e478s. doi:10.6061/clinics/2018/e478s
Rosenberg, B., Vancamp, L., and Krigas, T. (1965). INHIBITION OF CELL DIVISION IN ESCHERICHIA COLI BY ELECTROLYSIS PRODUCTS FROM A PLATINUM ELECTRODE. Nature 205, 698–699. doi:10.1038/205698a0
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. U S A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Schardt, J., Roth, B., and Seiler, R. (2019). Forty Years of Cisplatin-Based Chemotherapy in Muscle-Invasive Bladder Cancer: Are We Understanding How, Who and when? World J. Urol. 37 (9), 1759–1765. doi:10.1007/s00345-018-2544-8
Scott, N. A., Susnerwala, S., Gollins, S., Myint, A. S., and Levine, E. (2009). Preoperative Neo-Adjuvant Therapy for Curable Rectal Cancer-Rreaching a Consensus 2008. Colorectal Dis. 11 (3), 245–248. doi:10.1111/j.1463-1318.2008.01636.x
Shao, N., Song, L., and Sun, X. (2021). Exosomal circ_PIP5K1A Regulates the Progression of Non-small Cell Lung Cancer and Cisplatin Sensitivity by miR-101/ABCC1 axis. Mol. Cel Biochem. doi:10.1007/s11010-021-04083-8
Shen, D. W., Pouliot, L. M., Hall, M. D., and Gottesman, M. M. (2012). Cisplatin Resistance: a Cellular Self-Defense Mechanism Resulting from Multiple Epigenetic and Genetic Changes. Pharmacol. Rev. 64 (3), 706–721. doi:10.1124/pr.111.005637
Shibata, M., Ham, K., and Hoque, M. O. (2018). A Time for YAP1: Tumorigenesis, Immunosuppression and Targeted Therapy. Int. J. Cancer 143 (9), 2133–2144. doi:10.1002/ijc.31561
Singh, M., Yelle, N., Venugopal, C., and Singh, S. K. (2018). EMT: Mechanisms and Therapeutic Implications. Pharmacol. Ther. 182, 80–94. doi:10.1016/j.pharmthera.2017.08.009
Sridhar, S., Botbol, Y., Macian, F., and Cuervo, A. M. (2012). Autophagy and Disease: Always Two Sides to a Problem. J. Pathol. 226 (2), 255–273. doi:10.1002/path.3025
Stefan, S. M. (2019). Multi-target ABC Transporter Modulators: what Next and where to Go? Future Med. Chem. 11 (18), 2353–2358. doi:10.4155/fmc-2019-0185
Steuer, C. E., El-Deiry, M., Parks, J. R., Higgins, K. A., and Saba, N. F. (2017). An Update on Larynx Cancer. CA Cancer J. Clin. 67 (1), 31–50. doi:10.3322/caac.21386
Su, L. L., Chang, X. J., Zhou, H. D., Hou, L. B., and Xue, X. Y. (2019). Exosomes in Esophageal Cancer: A Review on Tumorigenesis, Diagnosis and Therapeutic Potential. World J. Clin. Cases 7 (8), 908–916. doi:10.12998/wjcc.v7.i8.908
Su, M., Xiao, Y., Ma, J., Tang, Y., Tian, B., Zhang, Y., et al. (2019). Circular RNAs in Cancer: Emerging Functions in Hallmarks, Stemness, Resistance and Roles as Potential Biomarkers. Mol. Cancer 18 (1), 90. doi:10.1186/s12943-019-1002-6
Su, Y., Yang, W., Jiang, N., Shi, J., Chen, L., Zhong, G., et al. (2019). Hypoxia-elevated circELP3 Contributes to Bladder Cancer Progression and Cisplatin Resistance. Int. J. Biol. Sci. 15 (2), 441–452. doi:10.7150/ijbs.26826
Sun, G., Li, Z., He, Z., Wang, W., Wang, S., Zhang, X., et al. (2020). Circular RNA MCTP2 Inhibits Cisplatin Resistance in Gastric Cancer by miR-99a-5p-Mediated Induction of MTMR3 Expression. J. Exp. Clin. Cancer Res. 39 (1), 246. doi:10.1186/s13046-020-01758-w
Sun, Y., Cao, Z., Shan, J., Gao, Y., Liu, X., Ma, D., et al. (2020). Hsa_circ_0020095 Promotes Oncogenesis and Cisplatin Resistance in Colon Cancer by Sponging miR-487a-3p and Modulating SOX9. Front. Cel Dev Biol 8, 604869. doi:10.3389/fcell.2020.604869
Tan, X., Zhou, C., Liang, Y., Lai, Y. F., and Liang, Y. (2020). Circ_0001971 Regulates Oral Squamous Cell Carcinoma Progression and Chemosensitivity by Targeting miR-194/miR-204 In Vitro and In Vivo. Eur. Rev. Med. Pharmacol. Sci. 24 (5), 2470–2481. doi:10.26355/eurrev_202003_20515
Tang, L. Q., Chen, D. P., Guo, L., Mo, H. Y., Huang, Y., Guo, S. S., et al. (2018). Concurrent Chemoradiotherapy with Nedaplatin versus Cisplatin in Stage II-IVB Nasopharyngeal Carcinoma: an Open-Label, Non-inferiority, Randomised Phase 3 Trial. Lancet Oncol. 19 (4), 461–473. doi:10.1016/S1470-2045(18)30104-9
Tang, Q. X., Wang, L. C., Wang, Y., Gao, H. D., and Hou, Z. L. (2019). Efficacy of Methotrexate, Doxorubicin, and Cisplatin for Osteosarcoma: Study Protocol for a Systematic Review of Randomized Controlled Trial. Medicine (Baltimore) 98 (6), e14442. doi:10.1097/MD.0000000000014442
Theodoulou, F. L., and Kerr, I. D. (2015). ABC Transporter Research: Going strong 40 Years on. Biochem. Soc. Trans. 43 (5), 1033–1040. doi:10.1042/BST20150139
Tian, L., Zhang, J., Ren, X., Liu, X., Gao, W., Zhang, C., et al. (2017). Overexpression of miR-26b Decreases the Cisplatin-Resistance in Laryngeal Cancer by Targeting ATF2. Oncotarget 8 (45), 79023–79033. doi:10.18632/oncotarget.20784
Tong, S. (2020). Circular RNA SMARCA5 May Serve as a Tumor Suppressor in Non-small Cell Lung Cancer. J. Clin. Lab. Anal. 34 (5), e23195. doi:10.1002/jcla.23195
Torre, L. A., Siegel, R. L., and Jemal, A. (2016). Lung Cancer Statistics. Adv. Exp. Med. Biol. 893, 1–19. doi:10.1007/978-3-319-24223-1_1
Vargas-Hernández, V. M., Moreno-Eutimio, M. A., Acosta-Altamirano, G., and Vargas-Aguilar, V. M. (2014). Management of Recurrent Epithelial Ovarian Cancer. Gland Surg. 3 (3), 198–202. doi:10.3978/j.issn.2227-684X.2013.10.01
Vaughan, S., Coward, J. I., Bast, R. C., Berchuck, A., Berek, J. S., Brenton, J. D., et al. (2011). Rethinking Ovarian Cancer: Recommendations for Improving Outcomes. Nat. Rev. Cancer 11 (10), 719–725. doi:10.1038/nrc3144
Verduci, L., Strano, S., Yarden, Y., and Blandino, G. (2019). The circRNA-microRNA Code: Emerging Implications for Cancer Diagnosis and Treatment. Mol. Oncol. 13 (4), 669–680. doi:10.1002/1878-0261.12468
Wagner, A. D., Syn, N. L., Moehler, M., Grothe, W., Yong, W. P., Tai, B. C., et al. (2017). Chemotherapy for Advanced Gastric Cancer. Cochrane Database Syst. Rev. 8 (8), Cd004064. doi:10.1002/14651858.CD004064.pub4
Waldmann, A., Eisemann, N., and Katalinic, A. (2013). Epidemiology of Malignant Cervical, Corpus Uteri and Ovarian Tumours - Current Data and Epidemiological Trends. Geburtshilfe Frauenheilkd 73 (2), 123–129. doi:10.1055/s-0032-1328266
Wei, Y., Zhang, Y., Meng, Q., Cui, L., and Xu, C. (2019). Hypoxia-induced Circular RNA has_circRNA_403658 Promotes Bladder Cancer Cell Growth through Activation of LDHA. Am. J. Transl Res. 11 (11), 6838–6849.
Wu, Z., Gong, Q., Yu, Y., Zhu, J., and Li, W. (2020). Knockdown of Circ-ABCB10 Promotes Sensitivity of Lung Cancer Cells to Cisplatin via miR-556-3p/AK4 axis. BMC Pulm. Med. 20 (1), 10. doi:10.1186/s12890-019-1035-z
Xu, G., Li, M., Wu, J., Qin, C., Tao, Y., and He, H. (2020). Circular RNA circNRIP1 Sponges microRNA-138-5p to Maintain Hypoxia-Induced Resistance to 5-Fluorouracil through HIF-1α-dependent Glucose Metabolism in Gastric Carcinoma. Cancer Manag. Res. 12, 2789–2802. doi:10.2147/CMAR.S246272
Xu, Y., Jiang, T., Wu, C., and Zhang, Y. (2020). CircAKT3 Inhibits Glycolysis Balance in Lung Cancer Cells by Regulating miR-516b-5p/STAT3 to Inhibit Cisplatin Sensitivity. Biotechnol. Lett. 42 (7), 1123–1135. doi:10.1007/s10529-020-02846-9
Xue, D., Wang, H., Chen, Y., Shen, D., Lu, J., Wang, M., et al. (2019). Circ-AKT3 Inhibits clear Cell Renal Cell Carcinoma Metastasis via Altering miR-296-3p/E-Cadherin Signals. Mol. Cancer 18 (1), 151. doi:10.1186/s12943-019-1072-5
Xue, M., Li, G., Fang, X., Wang, L., Jin, Y., and Zhou, Q. (2019). hsa_circ_0081143 Promotes Cisplatin Resistance in Gastric Cancer by Targeting miR-646/CDK6 Pathway. Cancer Cel Int 19, 25. doi:10.1186/s12935-019-0737-x
Yang, C., Dong, Z., Hong, H., Dai, B., Song, F., Geng, L., et al. (2020). circFN1 Mediates Sorafenib Resistance of Hepatocellular Carcinoma Cells by Sponging miR-1205 and Regulating E2F1 Expression. Mol. Ther. Nucleic Acids 22, 421–433. doi:10.1016/j.omtn.2020.08.039
Yang, X., Zhang, Q., and Guan, B. (2020). Circ_0110805 Knockdown Enhances Cisplatin Sensitivity and Inhibits Gastric Cancer Progression by miR-299-3p/ENDOPDI Axis. Onco Targets Ther. 13, 11445–11457. doi:10.2147/OTT.S279563
Yao, W., Guo, P., Mu, Q., and Wang, Y. (2020). Exosome-Derived Circ-PVT1 Contributes to Cisplatin Resistance by Regulating Autophagy, Invasion, and Apoptosis via miR-30a-5p/YAP1 Axis in Gastric Cancer Cells. Cancer Biother. Radiopharm. 36, 347–359. doi:10.1089/cbr.2020.3578
Ye, Y., Zhao, L., Li, Q., Xi, C., Li, Y., and Li, Z. (2020). circ_0007385 Served as Competing Endogenous RNA for miR-519d-3p to Suppress Malignant Behaviors and Cisplatin Resistance of Non-small Cell Lung Cancer Cells. Thorac. Cancer 11 (8), 2196–2208. doi:10.1111/1759-7714.13527
Yi, X., Chen, W., Li, C., Chen, X., Lin, Q., Lin, S., et al. (2020). Circular RNA Circ_0004507 Contributes to Laryngeal Cancer Progression and Cisplatin Resistance by Sponging miR-873 to Upregulate Multidrug Resistance 1 and Multidrug Resistance Protein 1. Head Neck 43, 928–941. doi:10.1002/hed.26549
Yuan, W., Zhou, R., Wang, J., Han, J., Yang, X., Yu, H., et al. (2019). Circular RNA Cdr1as Sensitizes Bladder Cancer to Cisplatin by Upregulating APAF1 Expression through miR-1270 Inhibition. Mol. Oncol. 13 (7), 1559–1576. doi:10.1002/1878-0261.12523
Zhan, T., Rindtorff, N., and Boutros, M. (2017). Wnt Signaling in Cancer. Oncogene 36 (11), 1461–1473. doi:10.1038/onc.2016.304
Zhang, C., Ji, Q., Yang, Y., Li, Q., and Wang, Z. (2018). Exosome: Function and Role in Cancer Metastasis and Drug Resistance. Technol. Cancer Res. Treat. 17, 1533033818763450. doi:10.1177/1533033818763450
Zhang, H., Yan, J., Lang, X., and Zhuang, Y. (2018). Expression of Circ_001569 Is Upregulated in Osteosarcoma and Promotes Cell Proliferation and Cisplatin Resistance by Activating the Wnt/β-Catenin Signaling Pathway. Oncol. Lett. 16 (5), 5856–5862. doi:10.3892/ol.2018.9410
Zhang, P., Wu, W., Chen, Q., and Chen, M. (2019). Non-Coding RNAs and Their Integrated Networks. J. Integr. Bioinform 16 (3). doi:10.1515/jib-2019-0027
Zhang, Q., Miao, Y., Fu, Q., Hu, H., Chen, H., Zeng, A., et al. (2020). CircRNACCDC66 Regulates Cisplatin Resistance in Gastric Cancer via the miR-618/BCL2 axis. Biochem. Biophys. Res. Commun. 526 (3), 713–720. doi:10.1016/j.bbrc.2020.03.156
Zhang, X., Su, X., Guo, Z., Jiang, X., and Li, X. (2020). Circular RNA La-Related RNA-Binding Protein 4 Correlates with Reduced Tumor Stage, as Well as Better Prognosis, and Promotes Chemosensitivity to Doxorubicin in Breast Cancer. J. Clin. Lab. Anal. 34 (7), e23272. doi:10.1002/jcla.23272
Zhang, Y., Yang, W. Q., Zhu, H., Qian, Y. Y., Zhou, L., Ren, Y. J., et al. (2014). Regulation of Autophagy by miR-30d Impacts Sensitivity of Anaplastic Thyroid Carcinoma to Cisplatin. Biochem. Pharmacol. 87 (4), 562–570. doi:10.1016/j.bcp.2013.12.004
Zhang, Z., Yu, X., Zhou, B., Zhang, J., and Chang, J. (2020). Circular RNA Circ_0026359 Enhances Cisplatin Resistance in Gastric Cancer via Targeting miR-1200/POLD4 Pathway. Biomed. Res. Int. 2020, 5103272. doi:10.1155/2020/5103272
Zhao, Y., Sun, Y., Yang, J., Zhu, Z., and Jia, X. (2021). WITHDRAWN: Circ_0000517 Contributes to Hepatocellular Carcinoma Progression by Upregulating ARID4B via Sponging miR-328-3p. Cell Signal, 109950. doi:10.1016/j.cellsig.2021.109950
Zhao, Y., Zheng, R., Chen, J., and Ning, D. (2020). CircRNA CDR1as/miR-641/HOXA9 Pathway Regulated Stemness Contributes to Cisplatin Resistance in Non-small Cell Lung Cancer (NSCLC). Cancer Cel Int 20, 289. doi:10.1186/s12935-020-01390-w
Zhao, Z., Ji, M., Wang, Q., He, N., and Li, Y. (2019). Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol. Ther. Nucleic Acids 18, 24–33. doi:10.1016/j.omtn.2019.07.012
Zheng, F., and Xu, R. (2020). CircPVT1 Contributes to Chemotherapy Resistance of Lung Adenocarcinoma through miR-145-5p/ABCC1 axis. Biomed. Pharmacother. 124, 109828. doi:10.1016/j.biopha.2020.109828
Zheng, H. C. (2017). The Molecular Mechanisms of Chemoresistance in Cancers. Oncotarget 8 (35), 59950–59964. doi:10.18632/oncotarget.19048
Zhong, Y., Wang, D., Ding, Y., Tian, G., and Jiang, B. (2020). Circular RNA Circ_0032821 Contributes to Oxaliplatin (OXA) Resistance of Gastric Cancer Cells by Regulating SOX9 via miR-515-5p. Biotechnol. Lett. 43, 339–351. doi:10.1007/s10529-020-03036-3
Zhou, S., Wei, J., Wang, Y., and Liu, X. (2020). Cisplatin Resistance-Associated circRNA_101237 Serves as a Prognostic Biomarker in Hepatocellular Carcinoma. Exp. Ther. Med. 19 (4), 2733–2740. doi:10.3892/etm.2020.8526
Zou, F. W., Yang, S. Z., Li, W. Y., Liu, C. Y., Liu, X. H., Hu, C. H., et al. (2020). circRNA_001275 Upregulates Wnt7a Expression by Competitively Sponging miR‐370‐3p to Promote Cisplatin Resistance in Esophageal Cancer. Int. J. Oncol. 57 (1), 151–160.
ABC ATP-binding cassette
AR androgen receptor
ATC anaplastic thyroid cancer
BCRP breast cancer resistance protein
CCL2 CC-chemokine ligand 2
CeRNA NET competing endogenous RNA network
CircRNA circular RNA
CRC colorectal cancer
CSCs cancer stem cells
DDR DNA damage repair
EGFR epidermal growth factor receptor
EMT epithelial–mesenchymal transition
ERCC1 excision repair cross-complementing 1
FGFR fibroblast growth factor receptor
FDA Food and Drug Administration
GC gastric cancer
HCC hepatocellular carcinoma
Hh Hedgehog
LncRNA long noncoding
m6A N6-methyladenosine
MBL muscle blind
MDR multidrug resistance
MDR1 multidrug resistance protein 1
MEK mitogen-activated protein kinase
MiRNA/MiR microRNA
MREs miRNA reaction elements
MRP1 multidrug resistance-related protein 1
NcRNA noncoding RNA
NER nucleotide excision repair
OS overall survival
OSCC oral squamous cell carcinoma
PD1 programmed cell death protein-1
PD-L1 programmed cell death-ligand 1
PE phosphatidylethanolamine
PI3K phosphatidylinositol 3-kinase
PTC papillary thyroid carcinoma
QRT-PCR quantitative reverse transcription PCR
SBP substrate-binding proteins
TGF-β transforming growth factor-β
TME tumor microenvironment
Keywords: circular RNA, cisplatin resistance, cancer, non-coding RNA, gene
Citation: Mu Q, Lv Y, Luo C, Liu X, Huang C, Xiu Y and Tang L (2021) Research Progress on the Functions and Mechanism of circRNA in Cisplatin Resistance in Tumors. Front. Pharmacol. 12:709324. doi: 10.3389/fphar.2021.709324
Received: 13 May 2021; Accepted: 30 August 2021;
Published: 09 September 2021.
Edited by:
Zhong Liu, Jinan University, ChinaReviewed by:
Zhaofeng Liang, Jiangsu University, ChinaCopyright © 2021 Mu, Lv, Luo, Liu, Huang, Xiu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunming Huang, SHVhbmdjaHVubWluZzU4ODhAMTYzLmNvbQ==; Youcheng Xiu, eW91Y2hlbmd4aXUyMDAwQDE2My5jb20=; Longguang Tang, bGd0YW5nMjAyMEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.