- 1Department of Internal Medicine, Section of Gerontology and Geriatrics, Leiden University Medical Center, Leiden, Netherlands
- 2Department of Cardiology, Leiden University Medical Center, Leiden, Netherlands
- 3William Harvey Research Institute, Barts and The London School of Medicine, Queen Mary University of London, London, United Kingdom
- 4Barts NIHR Biomedical Research Unit, London, United Kingdom
- 5Department of Epidemiology, Erasmus MC - University Medical Center Rotterdam, Rotterdam, Netherlands
- 6Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, Netherlands
- 7Department of Pediatrics, University of California, San Francisco, San Francisco, CA, United States
- 8Institute for Translational Genomics and Population Sciences, Los Angeles BioMedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, United States
- 9Centre de Recherche de l’Institut Universitaire de Cardiologie et de Pneumologie de Québec, Québec City, QC, Canada
- 10Department of Medicine, Faculty of Medicine, Université Laval, Québec City, QC, Canada
- 11Division of Preventive Medicine, Brigham and Women’s Hospital, Boston, MA, United States
- 12Harvard Medical School, Boston, MA, United States
- 13Blizard institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom
- 14Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, United States
- 15Department of Epidemiology, University of Washington, Seattle, WA, United States
- 16Department of Health Services University of Washington, Seattle, WA, United States
- 17Department of Biostatistics, Boston University School of Public Health, Boston, MA, United States
- 18NHLBI Framingham Heart Study, Framingham, MA, United States
- 19Einthoven Laboratory for Experimental Vascular Medicine, Leiden University Medical Center, Leiden, Netherlands
- 20Netherlands Heart Institute, Utrecht, Netherlands
Background: The pharmacogenetic effect on cardiovascular disease reduction in response to statin treatment has only been assessed in small studies. In a pharmacogenetic genome wide association study (GWAS) analysis within the Genomic Investigation of Statin Therapy (GIST) consortium, we investigated whether genetic variation was associated with the response of statins on cardiovascular disease risk reduction.
Methods: The investigated endpoint was incident myocardial infarction (MI) defined as coronary heart disease death and definite and suspect non-fatal MI. For imputed single nucleotide polymorphisms (SNPs), regression analysis was performed on expected allelic dosage and meta-analysed with a fixed-effects model, inverse variance weighted meta-analysis. All SNPs with p-values <5.0 × 10−4 in stage 1 GWAS meta-analysis were selected for further investigation in stage-2. As a secondary analysis, we extracted SNPs from the Stage-1 GWAS meta-analysis results based on predefined hypotheses to possibly modifying the effect of statin therapy on MI.
Results: In stage-1 meta-analysis (eight studies, n = 10,769, 4,212 cases), we observed no genome-wide significant results (p < 5.0 × 10−8). A total of 144 genetic variants were followed-up in the second stage (three studies, n = 1,525, 180 cases). In the combined meta-analysis, no genome-wide significant hits were identified. Moreover, none of the look-ups of SNPs known to be associated with either CHD or with statin response to cholesterol levels reached Bonferroni level of significance within our stage-1 meta-analysis.
Conclusion: This GWAS analysis did not provide evidence that genetic variation affects statin response on cardiovascular risk reduction. It does not appear likely that genetic testing for predicting effects of statins on clinical events will become a useful tool in clinical practice.
Introduction
Therapy with statins, 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors, is widely used for the primary and secondary prevention of cardiovascular disease (Cholesterol Treatment Trialists et al., 2015). Statin therapy reduces LDL-cholesterol concentrations by 30–50% and is accompanied by a 20–30% risk reduction in cardiovascular events (Cholesterol Treatment Trialists et al., 2015). Behaviours (e.g. treatment adherence), medical conditions, and genetics all contribute to the inter-individual variation in lipid lowering response to statin therapy.
In addition to many smaller pharmacogenetic studies, a large genome-wide association study (GWAS), which investigated the genetic variation associated with the LDL cholesterol lowering response of statins identified four independent loci mapped to the APOE, LPA, SORT1, SCLO1B1 genes (Postmus et al., 2014a). While many pharmacogenetic studies have investigated the effect of statins on LDL-cholesterol lowering response, the pharmacogenetic effect on cardiovascular risk reduction in response to statin treatment has been studied in small candidate gene studies with limited success (Peters et al., 2008; Peters et al., 2010a; Peters et al., 2010b; Peters et al., 2011; Leusink et al., 2014; Li et al., 2015). The decrease in risk of cardiovascular events driven by statin therapy is largely due to the LDL lowering, but could also be in part be attributable by other ancillary mechanisms such as inflammation, thrombosis and anti-platelet mechanisms (Oesterle et al., 2017). Therefore, candidate gene studies into lipid lowering pathways might not capture all main pharmacogenetic effects responsible for the differential response to statin therapy with respect to cardiovascular events (Leusink et al., 2016).
Previously, three GWAS analyses have been performed to investigate the pharmacogenetics effect of statins on cardiovascular disease reduction, however two of these studies were relatively small and did not yield any genetic variants associated with differential cardiovascular event reduction by statins (Shiffman et al., 2012; Postmus et al., 2014b). The GWAS of Wei et al. demonstrated that the LPA gene could have a role in the differential response of statin on coronary events (Wei et al., 2018), but did not provide replication of this finding. In a pharmacogenetic GWAS study within the Genomic Investigation of Statin Therapy (GIST) consortium, we aimed to investigate whether genetic variation was associated with the response of statins on cardiovascular disease reduction, in particular myocardial infarction (MI).
Methods
Design and Contributing Studies
This study was conducted within the GIST consortium, which includes data from 11 large studies.
We conducted a two-stage design approach, in order to follow-up any potential discovery findings within independent data. The data in stage one comprised two randomized controlled clinical trials (RCTs) (ASCOT UK and PROSPER) and six observational cohort studies (ARIC, ASCOT UK OBS, CHS, FHS, HVH, RS) including n = 10,769 participants (4212 cases and 6557 controls): 7,215 in RCTs (658 cases and 6557 controls) and 3,554 cases from observational studies. The data in stage two comprised three studies: ASCOT SC OBS and RCT, and MESA, with n = 1,525 (180 cases and 1345 controls). The details for all participating studies are in Supplementary Table S1 and Supplementary Note S1.
Subjects
Only subjects of European descent were included. For RCTs all subjects using placebo and statin treatment were included in this analysis. For observational studies, incident MI cases after starting statin treatment were included as cases. Subjects with a previous MI event before statin treatment were eligible for inclusion. All participants gave written informed consent and the study was approved by all institutional ethics committees.
Outcome Definition
The investigated endpoint was incident MI (fatal and non-fatal) defined as coronary heart disease death and definite and suspect non-fatal MI (cases). Case definitions per cohort are described in Supplementary Note S1. For the RCTs the controls were defined as subjects who did not experience incident MI during the follow-up period. For observational studies, the control group consisted of subjects without an incident MI after initiating statin treatment.
Genotyping, GWAS Analysis and Statistical Models
Genotyping, quality control, data cleaning and HAPMAP imputation were performed independently in each study as outlined in Supplementary Table S2. All analyses were performed with the expected allelic dosages for the imputed single nucleotide polymorphisms (SNPs). Each study independently performed their GWAS on incident MI.
As both RCT and observational studies are included, we used two different statistical models to investigate the pharmacogenetic effect of statins on MI, both assuming the same underlying relation. For RCTs an additive genetic model was assumed and each SNP tested using a Cox-proportional hazards regression model with MI as the outcome, adjusted for statin use and including an interaction term of statin use and SNP. The main parameter of interest was the interaction term between statin use and the SNP allelic dosage.
For observational studies we used a case-only design, with incident MI cases included, where the outcome variable in the statistical model was a binary indicator variable for statin use according to whether or not the subject was taking any type of statin prior to the myocardial infarction event and with SNP dosage as the predictor. This was assessed with binary logistic regression.
Analyses were additionally adjusted for age-, sex- and study-specific covariates (for example, ancestry principal components or country).
Quality Control and Meta-Analysis
Centrally, within each study, SNPs with MAF <1% or imputation quality <0.3 were excluded from the analysis. QQ-plots were assessed for each study to check for between-study differences (Supplementary Figure S1). The software package METAL was used for performing a fixed effects, inverse variance weighted meta-analysis (Willer et al., 2010). The interaction betas from the RCTs and the genetic variants association betas from the case only studies were meta-analysed.
To correct for possible residual population stratification, genomic control was applied to each study within METAL by adjusting for the genomic inflation factor prior to meta-analysis.
Follow-up From Stage 1 to Stage 2
All SNPs with p-values <5.0 × 10−4 in the stage 1 GWAS meta-analysis were selected for further investigation in stage 2. A maximum of two SNPs per independent gene region were selected, based on the lowest p-value of statistical significance. A total of 144 SNPs, within 103 independent loci, were selected for follow-up in the second stage.
The studies in stage 2 provided the regression estimates for these follow-up SNPs, and these were meta-analysed together. Results from the stage 1 and stage 2 meta-analyses for the 144 follow-up SNPs were combined using a fixed-effects model, inverse variance weighted meta-analysis in METAL.
Significance Criteria
A SNP would be declared significant in stage 1 data alone if it reached the genome-wide statistical significance threshold of p < 5 × 10−8 in the meta-analysis. Next, a locus would also be considered significant if the top SNP at the locus reached genome-wide significance in the combined meta-analysis, with concordant direction of effect between stage 1 and stage 2.
Additional Analysis
As a secondary analysis, we performed a look-up within our Stage 1 GWAS meta-analysis results for SNPs of interest that would possibly be associated with a pharmacogenetic effect of statin therapy on myocardial infarction. SNPs significantly associated with coronary events and/or with LDL or HDL changes after statin treatment based on previous GWAS studies were considered SNPS of interest. First we performed a look-up of 23 SNPs which were known to have genome-wide significant association with coronary events from the CARDIOGRAM study (Schunkert et al., 2011; Nikpay et al., 2015) at the time of analysis. Secondly, we performed a look-up of five SNPs with known associations of pharmacogenetic effects of statin therapy on LDL lowering and HDL changes, based on the large GWAS studies in the GIST consortium (Postmus et al., 2014a; Postmus et al., 2016). For each look-up analysis, to control for multiple testing, we applied a Bonferroni correction, using a p-value threshold for statistical significance of 0.05/28 = p= 0.0018.
Results
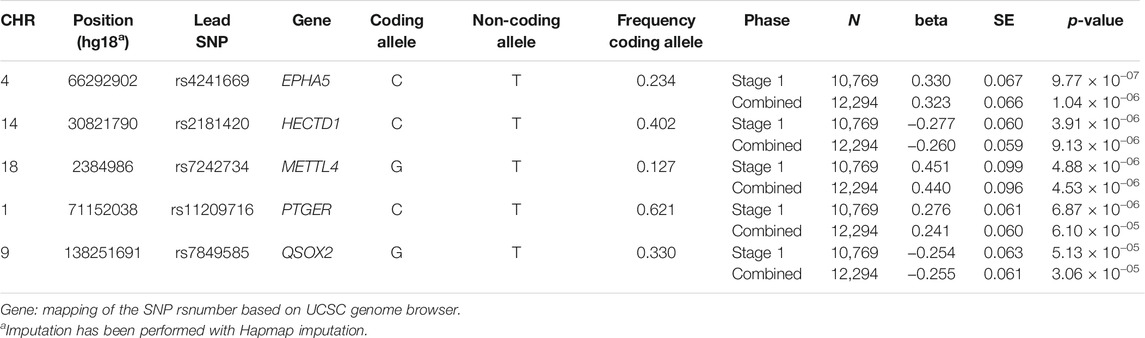
In the stage 1 meta-analysis (eight studies, 10,769 individuals, 4,212 cases), we observed no genome-wide significant results (p < 5.0 × 10−8) (Figure 1). The top signal was for rs4241669, mapping to EPHA5 (beta, se: 0.330, 0.067) with a p-value of 9.8 × 10−7. The beta can be interpreted as an estimate of the interaction between statin use and the SNP allelic dosage on incident MI risk. A total of 144 genetic variants with p-value <5.0 × 10−4 were followed-up in the second stage. In the combined meta-analysis of stage 1 and stage 2, again no genome wide significant results were observed (Table 1), hence our overall results do not show any genetic variants with evidence of association with differential risk reduction in MI in response to statins. In Table 1 we present results for the top five genetic loci with p < 5.0 × 10−6). None of the top loci are near to known genes involved in pathways related to lipids or cardiovascular mechanisms.
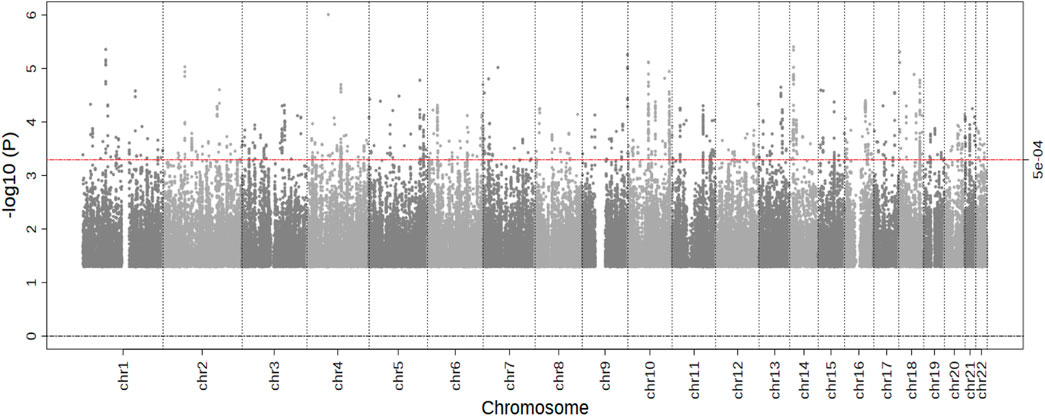
FIGURE 1. Results of the Stage 1 GWAS meta-analysis. Manhattan plot presenting the log10 p-values from the stage 1 meta-analysis (n = 10,769) on myocardial infarction risk after statin treatment. p values were generated using cox-proportional or logistic regression analysis. The blue line represents a p-value of 5.0 × 10−4. No SNPs reached the significance threshold of 5.0 × 10−8.
Based on the look-up of SNPs known to be associated with either coronary events or with LDL cholesterol response to statin treatment (Table 2), a coronary event associated SNP at the SORT 1 locus (rs599839) was most strongly associated with statin MI response, as was a SNP at this locus for the statin LDL response (rs646776). However none of the SNPs reached a Bonferroni level of significance within our stage 1 meta-analysis for statin response to MI risk (all p > 0.0018).
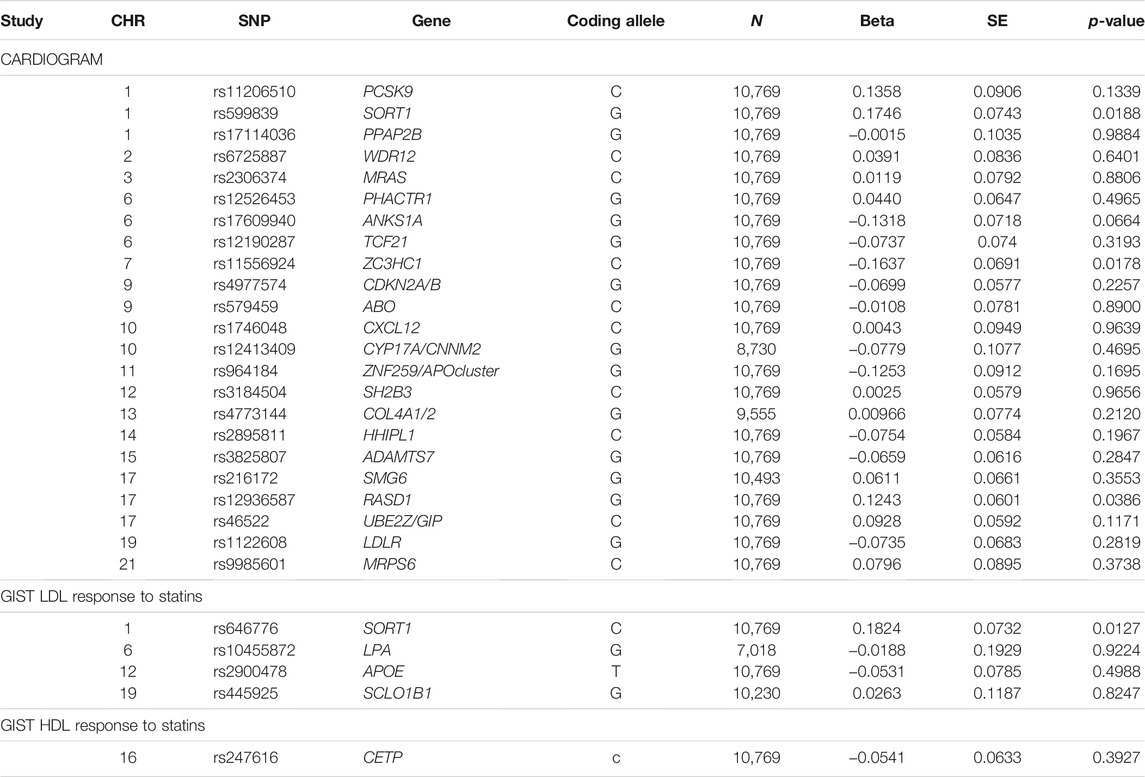
TABLE 2. Look-up of the most significant CARDIOGRAM on coronary events and previous top GIST loci with LDL and HDL response to statins within the Stage 1 meta-analysis results of this study.
Discussion
In this genome-wide association study we did not identify any genetic variants with significant evidence of association between inter-individual variation to statin therapy and differential risk reduction for MI. Furthermore, we did not identify significant genetic effects on MI risk with statin treatment for any known CHD-associated variants or any known variants associated with statin LDL response.
There have been three previous pharmacogenetic GWAS studies of the modification of clinical benefit by statins (Shiffman et al., 2012; Postmus et al., 2014b; Wei et al., 2018). The first study by Shiffman et al reported an association in the DNAJC5B gene, although this was not genome-wide significant (Shiffman et al., 2012), and the result has never been replicated. Like our study, the other GWAS analysis by Postmus et al. found no significant pharmacogenetic associations (Postmus et al., 2014b). Both studies, however, were not sufficiently large by current GWAS standards to detect genome-wide significant associations with small effects. The third study by Wei et al. found that variants within the LPA gene were significantly associated with residual cardiovascular risk in statin users, (Wei et al., 2018), but our GWAS study did not replicate that result (p = 0.9224).
Our study has some limitations. Although it is the largest pharmacogenetic GWAS analysis of clinical cardiovascular response to statins to date, the number of cases (n = 4,392) is still relatively small. Leusink et al. calculated that to reach genome wide significance with an interaction odds ratio of 1.1, 15,000 MI cases would be required (Leusink et al., 2016). Knowing that small effects in large samples would not be relevant for clinical practice, we hypothesized that our study may still have sufficient statistical power to detect any potential relevant variants with larger effects sizes, if they were to exist. Another limitation could be that we included subjects from both randomized controlled trials and observational studies, and this could have generated some noise and therefore less precision.
In conclusion, this meta-analysis of GWASs, the largest one performed up until now investigating the pharmacogenetics of statin therapy on clinical events, did not provide any evidence that genetic variation affects statin response on coronary outcomes. Taken together with results of previous studies it does not appear likely that genetic testing for predicting effects of statins on clinical events can be a useful tool in clinical practice.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/gap/, phs000930.v10.p1.
Ethics Statement
All participants gave written informed consent and the study was approved by all institutional ethics committees.
Author Contributions
ST, HW, DC, GH, PM, JR, BP, MC, RK, AC, and WJ designed and implemented the study. ST, HW, RN, ET, XL, BA, and DC provided per study study results and performed statistical analyses. ST, IP, HW, RN, RS, BP, RK, and WJ drafted the manuscript. All authors critically revised the manuscript for important intellectual content and approved the final paper.
Funding
The authors declare that this study received funding from Pfizer USA and Bristol-Myers Squibb USA. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. ASCOT. The Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT) trial was funded by an investigator-initiated grant from Pfizer USA. The study was investigator-led and was conducted, analyzed, and reported independently of the company. The Genome Wide Association Scan was funded by the National Institutes for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Unit at Barts and the NIHR Biomedical Research Centre at Imperial College, the International Centre for Circulatory Health Charity and the Medical Research Council through G952010. ARIC. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. CHS. This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, 75N92021D00006; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and U01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Framingham HS. The Framingham Heart Study work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine (Contract No. N01-HC-25195), its contract with Affymetrix, Inc. for genotyping services (Contract No. N02-HL-6-4278), and based upon analyses by Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Also supported by R01HL103612 (PI Psaty, subcontract PI, Vasan). HVH. This Heart and Vascular Health Study research was supported by NHLBI grants HL085251, HL073410, HL085251, and HL068986. MESA. The Multi-Ethnic Study of Atherosclerosis (MESA) and MESA SNP Health Association Resource (SHARe) are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support is provided by grants and contracts N01 HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and RR-024156. Additional funding was supported in part by the Clinical Translational Science Institute grant UL1RR033176 and is now at the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124. The authors thank the other investigators in the Pharmacogenetics Working Group, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. PROSPER. The Prospective Study of Pravastatin in the Elderly at Risk (PROSPER) trial was supported by an investigator-initiated grant from Bristol-Myers Squibb, USA. The study was conducted, analysed, and reported independently of the company. The GWAS project PHASE has received funding from the European Union’s Seventh Framework Programme (FP7/2007–2013) under grant agreement HEALTH-F2-2009-223004. A part of the genotyping was funded by The Netherlands Consortium for Healthy Ageing (NGI: 05060810). WJ is an established clinical investigator of The Netherlands Heart Foundation (2001 D 032). Rotterdam Study. The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission and Municipality of Rotterdam. This work was supported by the Netherlands Genomics Initiative (NGI) Netherlands Organization for Scientific Research (NOW; 050-060-810). TNT. The TNT study was funded by Pfizer, who also provided support for genotyping.
Conflict of Interest
The authors declare that this study received funding from Pfizer USA and Bristol-Myers Squibb USA.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.679857/full#supplementary-material
References
Cholesterol Treatment Trialists, C., Fulcher, J., O'Connell, R., Voysey, M., Emberson, J., Blackwell, L., et al. (2015). Efficacy and Safety of LDL-Lowering Therapy Among Men and Women: Meta-Analysis of Individual Data from 174,000 Participants in 27 Randomised Trials. Lancet 385 (9976), 1397–1405. doi:10.1016/S0140-6736(14)61368-4
Leusink, M., de Keyser, C. E., Onland-Moret, N. C., Hofman, A., Visser, L. E., Stricker, B. H., et al. (2014). No Association Between CYP3A4*22 and Statin Effectiveness in Reducing the Risk for Myocardial Infarction. Pharmacogenomics 15 (11), 1471–1477. doi:10.2217/pgs.14.90
Leusink, M., Onland-Moret, N. C., de Bakker, P. I., de Boer, A., and Maitland-van der Zee, A. H. (2016). Seventeen Years of Statin Pharmacogenetics: A Systematic Review. Pharmacogenomics 17 (2), 163–180. doi:10.2217/pgs.15.158
Li, J. H., Suchindran, S., Shah, S. H., Kraus, W. E., Ginsburg, G. S., and Voora, D. (2015). SLCO1B1 Genetic Variants, Long-Term Low-Density Lipoprotein Cholesterol Levels and Clinical Events in Patients Following Cardiac Catheterization. Pharmacogenomics 16 (5), 449–458. doi:10.2217/pgs.15.2
Nikpay, M., Goel, A., Won, H. H., Hall, L. M., Willenborg, C., Kanoni, S., et al. (2015). A Comprehensive 1,000 Genomes-Based Genome-wide Association Meta-Analysis of Coronary Artery Disease. Nat. Genet. 47 (10), 1121–1130. doi:10.1038/ng.3396
Oesterle, A., Laufs, U., and Liao, J. K. (2017). Pleiotropic Effects of Statins on the Cardiovascular System. Circ. Res. 120 (1), 229–243. doi:10.1161/CIRCRESAHA.116.308537
Peters, B. J., Maitland-van der Zee, A. H., Stricker, B. H., van Wieren-de Wijer, D. B., de Boer, A., Kroon, A. A., et al. (2008). Effectiveness of Statins in the Reduction of the Risk of Myocardial Infarction Is Modified by the GNB3 C825T Variant. Pharmacogenet Genomics 18 (7), 631–636. doi:10.1097/FPC.0b013e3283023fb2
Peters, B. J., Pett, H., Klungel, O. H., Stricker, B. H., Psaty, B. M., Glazer, N. L., et al. (2011). Genetic Variability Within the Cholesterol Lowering Pathway and the Effectiveness of Statins in Reducing the Risk of MI. Atherosclerosis 217 (2), 458–464. doi:10.1016/j.atherosclerosis.2011.06.023
Peters, B. J., Rodin, A. S., Klungel, O. H., Stricker, B. H., de Boer, A., and Maitland-van der Zee, A. H. (2010). Variants of ADAMTS1 Modify the Effectiveness of Statins in Reducing the Risk of Myocardial Infarction. Pharmacogenet Genomics 20 (12), 766–774. doi:10.1097/FPC.0b013e328340aded
Peters, B. J., Rodin, A. S., Klungel, O. H., van Duijn, C. M., Stricker, B. H., van't Slot, R., et al. (2010). Pharmacogenetic Interactions Between ABCB1 and SLCO1B1 Tagging SNPs and the Effectiveness of Statins in the Prevention of Myocardial Infarction. Pharmacogenomics 11 (8), 1065–1076. doi:10.2217/pgs.10.81
Postmus, I., Johnson, P. C., Trompet, S., de Craen, A. J., Slagboom, P. E., Devlin, J. J., et al. (2014). In Search for Genetic Determinants of Clinically Meaningful Differential Cardiovascular Event Reduction by Pravastatin in the PHArmacogenetic Study of Statins in the Elderly at Risk (PHASE)/PROSPER Study. Atherosclerosis 235 (1), 58–64. doi:10.1016/j.atherosclerosis.2014.04.009
Postmus, I., Trompet, S., Deshmukh, H. A., Barnes, M. R., Li, X., Warren, H. R., et al. (2014). Pharmacogenetic Meta-Analysis of Genome-wide Association Studies of LDL Cholesterol Response to Statins. Nat. Commun. 5, 5068. doi:10.1038/ncomms6068
Postmus, I., Warren, H. R., Trompet, S., Arsenault, B. J., Avery, C. L., Bis, J. C., et al. (2016). Meta-analysis of Genome-wide Association Studies of HDL Cholesterol Response to Statins. J. Med. Genet. 53 (12), 835–845. doi:10.1136/jmedgenet-2016-103966
Schunkert, H., König, I. R., Kathiresan, S., Reilly, M. P., Assimes, T. L., Holm, H., et al. (2011). Large-scale Association Analysis Identifies 13 New Susceptibility Loci for Coronary Artery Disease. Nat. Genet. 43 (4), 333–338. doi:10.1038/ng.784
Shiffman, D., Trompet, S., Louie, J. Z., Rowland, C. M., Catanese, J. J., Iakoubova, O. A., et al. (2012). Genome-wide Study of Gene Variants Associated with Differential Cardiovascular Event Reduction by Pravastatin Therapy. PLoS One 7 (5), e38240. doi:10.1371/journal.pone.0038240
Wei, W. Q., Li, X., Feng, Q., Kubo, M., Kullo, I. J., Peissig, P. L., et al. (2018). LPA Variants Are Associated with Residual Cardiovascular Risk in Patients Receiving Statins. Circulation 138 (17), 1839–1849. doi:10.1161/CIRCULATIONAHA.117.031356
Keywords: pharmacogenetics, statins, GWAS, cardiovascular disease, myocardial infarction
Citation: Trompet S, Postmus I, Warren HR, Noordam R, Smit RAJ, Theusch E, Li X, Arsenault B, Chasman DI, Hitman GA, Munroe PB, Rotter JI, Psaty BM, Caulfield MJ, Krauss RM, Cupples AL and Jukema WJ (2022) The Pharmacogenetics of Statin Therapy on Clinical Events: No Evidence that Genetic Variation Affects Statin Response on Myocardial Infarction. Front. Pharmacol. 12:679857. doi: 10.3389/fphar.2021.679857
Received: 01 July 2021; Accepted: 01 November 2021;
Published: 05 January 2022.
Edited by:
Vita Dolzan, University of Ljubljana, SloveniaReviewed by:
Theodora Katsila, National Hellenic Research Foundation, GreeceEvangelia Eirini Tsermpini, University of Patras, Greece
Copyright © 2022 Trompet, Postmus, Warren, Noordam, Smit, Theusch, Li, Arsenault, Chasman, Hitman, Munroe, Rotter, Psaty, Caulfield, Krauss, Cupples and Jukema. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stella Trompet, cy50cm9tcGV0QGx1bWMubmw=
 Stella Trompet
Stella Trompet Iris Postmus
Iris Postmus Helen R. Warren
Helen R. Warren Raymond Noordam
Raymond Noordam Roelof A. J. Smit
Roelof A. J. Smit Elizabeth Theusch7
Elizabeth Theusch7 Xiaohui Li
Xiaohui Li Benoit Arsenault
Benoit Arsenault Daniel I. Chasman
Daniel I. Chasman Graham A. Hitman
Graham A. Hitman Patricia B. Munroe
Patricia B. Munroe Jerome I. Rotter
Jerome I. Rotter Wouter J. Jukema
Wouter J. Jukema