
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 18 May 2021
Sec. Pharmacogenetics and Pharmacogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.672769
This article is part of the Research TopicPharmacogenomics of Adverse Drug Reactions (ADRs)View all 14 articles
 Zhao-Yang Chen1†
Zhao-Yang Chen1† Yang-Hui Zhu1†
Yang-Hui Zhu1† Ling-Yan Zhou1
Ling-Yan Zhou1 Wei-Qiao Shi1
Wei-Qiao Shi1 Zhou Qin1
Zhou Qin1 Bin Wu1
Bin Wu1 Yu Yan1
Yu Yan1 Yu-Wen Pei1
Yu-Wen Pei1 Ning-Ning Chao2
Ning-Ning Chao2 Rui Zhang3
Rui Zhang3 Mi-Ye Wang3
Mi-Ye Wang3 Ze-Hao Su4
Ze-Hao Su4 Xiao-Jun Lu5*
Xiao-Jun Lu5* Zhi-Yao He1,6*
Zhi-Yao He1,6* Ting Xu1,6
Ting Xu1,6The aim of this study was to investigate the correlation between genetic polymorphisms of azathioprine-metabolizing enzymes and adverse reactions of myelosuppression. To this end, a retrospective analysis was performed on 1,419 Chinese patients involving 40 different diseases and 3 genes: ITPA (94C>A), TPMT*3 (T>C), and NUDT15 (415C>T). Strict inclusion and exclusion criteria were established to collect the relative cases, and the correlation between azathioprine and myelosuppression was evaluated by adverse drug reaction criteria. The mutation rates of the three genes were 29.32, 3.73, and 21.92% and grades I to IV myelosuppression occurred in 54 (9.28%) of the 582 patients who took azathioprine. The highest proportion of myelosuppression was observed in 5 of the 6 (83.33%) patients carrying the NUDT15 (415C>T) TT genotype and 12 of the 102 (11.76%) patients carrying the NUDT15 (415C>T) CT genotype. Only the NUDT15 (415C>T) polymorphism was found to be associated with the adverse effects of azathioprine-induced myelosuppression (odds ratio [OR], 51.818; 95% CI, 5.280–508.556; p = 0.001), which suggested that the NUDT15 (415C>T) polymorphism could be an influencing factor of azathioprine-induced myelosuppression in the Chinese population. Epistatic interactions between ITPA (94C>A) and NUDT15 (415C>T) affect the occurrence of myelosuppression. Thus, it is recommended that the genotype of NUDT15 (415C>T) and ITPA (94C>A) be checked before administration, and azathioprine should be avoided in patients carrying a homozygous NUDT15 (415C>T) mutation. This study is the first to investigate the association between genetic polymorphisms of these three azathioprine-metabolizing enzymes and myelosuppression in a large number of cases with a diverse range of diseases.
Azathioprine (AZA) is a classic immunosuppressant that is widely used for post-transplant rejection, severe rheumatoid arthritis, systemic lupus erythematosus, pemphigus (Joly et al., 2020), inflammatory bowel disease (Ran et al., 2020), dermatomyositis, and other diseases (Mack et al., 2020). It is also recommended for the treatment of immune checkpoint inhibitor-related renal and musculoskeletal adverse events (Thompson et al., 2020). However, AZA can also cause drug adverse reactions (ADRs), including myelosuppression, hepatotoxicity, gastrointestinal reactions (nausea, vomiting, and diarrhea), and alopecia (Food and Drug Administration, 2018). Among them, myelosuppression is particularly harmful and could result in leukopenia, thrombocytopenia, pancytopenia, and even some life-threatening conditions (Panda et al., 2018).
AZA is metabolized into the active 6-thioguanine nucleotides (6-TNGs) by a series of enzymes in vivo (Yang et al., 2014; Moon and Loftus, 2016; Kishibe et al., 2018; Wang et al., 2018). AZA is first metabolized to 6-mercaptopurine (6-MP) by glutathione S-transferase (GST), and then converted into 6-thioinosine monophosphate (6-TIMP) with the help of hypoxanthine-guanine phosphoribosyl transferase (HGPRT). Subsequently, 6-TIMP is dehydrogenized into 6-thioxanthosine monophosphate (6-TXMP) by inosine monophosphate dehydrogenase (IMPDH), and then further metabolized to 6-TNGs by guanosine monophosphate synthetase (GMPS), which finally integrates into DNA and RNA molecules to exert cytotoxic and immunosuppressive effects. Moreover, 6-TGTP also binds to Rac1, and inactivates it by regulating the Vav-Rac1 signaling pathway in T lymphocytes; this results in the inhibition of Rac1 target genes, such as nuclear factor kappa beta (NF-κβ), finally leading to the increased apoptosis of activated T lymphocytes (Tiede et al., 2003; Poppe et al., 2006) (Supplementary Figure S1).
To reduce the risk of ADRs resulting from the use of AZA, researchers have attempted to establish AZA-metabolizing enzymes to predict the occurrence of myelosuppression and liver toxicity and adjust the dosage according to the genotype. The Clinical Pharmacogenetics Implementation Consortium (CPIC) first published guidelines for adjusting the dose of AZA based on the thiopurine S-methyltransferase (TPMT) polymorphism in 2011 (Relling et al., 2011), which were later updated in 2013 and 2018 (Relling et al., 2013; Relling et al., 2019). Currently, the guidelines recommend that patients with a normal TPMT metabolizer can use the standard recommended dose, those with intermediate metabolizers are recommended to use 30–80% of the normal dose, and those with poor metabolizers with nonmalignant conditions are not recommended to use AZA. Patients with poor TPMT metabolizers with malignancy are recommended to reduce the daily dose by 10-fold and to receive the dose thrice weekly instead of daily. Although some previous studies have investigated the association of AZA-induced ADRs with TPMT, inosine triphosphate pyrophosphatase (ITPA), nucleoside diphosphate-liked moiety X motif 15 (NUDT15), GST, multidrug resistance protein 4 (MRP4), HGPRT, IMPDH, and xanthine oxidase (XO), the results were varying due to ethnic differences in gene distribution (Krishnamurthy et al., 2008; Kudo et al., 2009; Yang et al., 2014; Burgis, 2016; Choi et al., 2019; Yang et al., 2019). Moreover, the dosage recommended in the CPIC guidelines is inaccurate in that it cannot be individualized among individuals of different races and regions. Among these genes, the most well-studied are ITPA, TPMT, and NUDT15. Some studies have reported that mutations in ITPA have no association with AZA-induced myelosuppression (Al-Judaibi et al., 2016; Steponaitiene et al., 2016). Other studies have indicated that the incidence of hepatotoxicity increases with a high TPMT enzyme activity, and that there is a high risk of myelosuppression with a low TPMT enzyme activity, due to its homozygous mutation. The Food and Drug Administration (FDA) recommends that the TPMT genotype of patients should be determined before using AZA (FDA). However, studies have shown that the frequency of TPMT gene mutations in the Asian population is only approximately 1.5–3%, thereby showing a high specificity but a low sensitivity. However, Asians have a low tolerance to AZA and a high incidence of leukopenia, which makes it necessary to explore predictive genes suitable for the Asian population specifically. In recent years, some studies have shown that NUDT15 might be highly correlated with AZA-induced myelosuppression in Asians (Chao et al., 2017; Wang et al., 2018; Banerjee et al., 2020; Kang et al., 2020), and the CPIP guideline also recommends that the NUDT15 genotype should be determined prior to the administration of AZA (2018) (Relling et al., 2019).
AZA is widely used in clinical settings, and genetic testing is essential for patients who need to take this drug for an extended duration. The abovementioned genes, ITPA, TPMT, and NUDT15, are currently being tested at the West China Hospital of Sichuan University, and the dose of AZA is being adjusted by doctors in accordance with the results of genetic tests to avoid adverse reactions. However, it has been found clinically that some patients with no mutations in these genes suffered myelosuppression, while others with homozygous mutations did not. To provide a reference for the analysis of genetic test results and accurate medication, this study was performed to explore the correlation between the polymorphism of these three genes and AZA-induced myelosuppression. As large-volume analytical studies, especially those involving diverse diseases, remain rare, this study is particularly important given that we examined a large number of cases with various diseases.
All included cases were collected from outpatient, emergency, and inpatient data of the West China Hospital of Sichuan University.
Related data of patients who underwent genetic testing of AZA-metabolizing enzyme genes from January 2016 to January 2019 in our hospital were extracted from the database. After the removal of duplicates, the patient information, including age, sex, clinical department, diagnosis, white blood cell count (WBC), and AZA daily dose, was compiled using the hospital information system. To determine myelosuppression, patients taking AZA who had complete routine blood examination results were included, while those who did not receive AZA or had incomplete WBC records were excluded.
According to the Common Terminology Criteria for Adverse Events (CTCAEs) version 5.0 published by the United States Department of Health and Human Services and the hospital leukocyte count index standard, myelosuppression was defined as a WBC count <3.5 × 109/l; a WBC count of 3–3.5 × 109/l was defined as grade I, 2–3 × 109/l as grade II, 1–2 × 109/l as grade III, and <1 × 109/l as grade IV. Adverse drug reaction correlation evaluation criteria of the National Medical Products Administration of China were used to evaluate AZA and myelosuppression correlation, and the Naranjo score was also used when the judgment results were controversial (National Health Commission PRC, 2011; Naranjo et al., 1981). The result “possible” was considered to be an adverse reaction of myelosuppression, and the results were judged by two clinical pharmacists after a double cross-check.
Microsoft Office Excel 2010 was used to input data, and SPSS 25.0 (IBM Corp., Armonk, NY, United States) was used for statistical analysis. Continuous variables are presented as mean ± SD. The independent t-test or Mann–Whitney U test was used to investigate the difference between two unrelated groups, and the one-way analysis of variance was used for comparison between multiple groups. Categorical variables were compared by the chi-square test or Fisher's test, and the Bonferroni correction was used for pairwise comparison between groups. The related factors of myelosuppression were analyzed by logistic regression analysis. Chi-square goodness-of-fit was used to confirm the agreement of the ITPA (94C>A), TPMT*3 (T>C), and NUDT15 (415C>T) genotype frequencies with the expected frequencies (Hardy–Weinberg equilibrium). The multifactor dimensionality reduction (MDR) method was used to examine gene–gene interactions, and MDR Permutation Testing software (version 1.0 Beta 2) was used for replacement testing. P-values < 0.05 were considered statistically significant.
A total of 1,419 available cases were covered in this study, including 742 (52.29%) inpatients and 677 (47.71%) outpatients/emergency patients (Figure 1). Among them, there were more female patients (65.19%), and the average age was 45.96 ± 14.41 years. Of the total cases, males comprised 494 (34.81%), with an average age of 42.74 ± 16.32 years. Nineteen departments were involved in the study, and the study population included 40 diseases, including pemphigus, inflammatory bowel disease, and autoimmune hepatitis (Supplementary Figure S2), among which, pemphigus was the most common (347, 24.45%).
Among the 1,419 patients, 1,279 (90.13%) had ITPA (94C>A) (rs1127354), TPMT*3 (T>C) (rs1142345), and NUDT15 (415C>T) (rs116855232), while 140 (9.87%) had TPMT*3 (T>C) and NUDT15 (415C>T). The genotypes of all patients are shown in Supplementary Table S1. The ITPA (94C>A), TPMT*3 (T>C), and NUDT15 (415C>T) genotype distributions were in Hardy–Weinberg equilibrium (p = 0.959, 0.811, and 0.406, respectively). The mutation rate of TPMT*3 (T>C) was the lowest (3.73%), similar to the previous reports in Asian populations (Kumagai et al., 2001; Chen et al., 2014; van et al., 2014). The respective proportions of wild type, heterozygous mutation, and homozygous mutation of the three genes were statistically significant (p = 1.374 × 10−68), and pairwise comparison between different genotypes by chi-square test also showed statistical significance. The data are shown in Table 1.
A total of 617 (43.48%) patients had AZA administration records, 582 (94.33%) of whom had complete routine blood examination results. Myelosuppression occurred in 54 (9.28%) patients, and the incidence of myelosuppression with grades from I to IV was 37.04% (20/54), 42.59% (23/54), 11.11% (6/54), and 9.26% (5/54), respectively.
A total of 516 patients carrying the ITPA (94C>A) genotype were included, among whom, 48 (9.30%) suffered from myelosuppression (grades I-IV). Patients carrying the ITPA (94C>A) AA genotype had the highest risk of myelosuppression, with an incidence of 25.00%, although this was not statistically significant. The mean daily dose (MDD) of AZA was significantly different among patients with different genotypes (p = 0.002). Multiple comparison results showed that only the AZA doses between the AC and CC genotype were significantly different (p = 0.323 × 10−3) (Supplementary Table S2).
A total of 582 cases with the TPMT*3 (T>C) genotype were included, among whom, 54 (9.28%) had ADRs of myelosuppression (grades I-IV). Similarly, the incidence of myelosuppression varied according to genotype; patients carrying the TPMT*3 (T>C) TC heterozygous mutation had the highest rate (20.00%), but there were no significant differences among the three groups. There were also no significant differences among the MDD of AZA among the different genotypes (Supplementary Table S2).
The number of patients carrying NUDT15 (415C>T) gene was 582, 54 (9.28%) of whom had ADRs of myelosuppression (grades I-IV). The incidence of myelosuppression varied by genotype; patients carrying the NUDT15 (415C>T) TT homozygote mutant had the highest rate (83.33%; 3 [60.00%] with grade IV), and the difference among the three genotypes was significant (p = 0.008 × 10−3). After the Bonferroni correction, there was a statistically significant difference in the myelosuppression rate between patients with the TT genotype and the other two genotypes (CT and CC) (TT, CC: p = 7.707 × 10−11 and TT, CT: p = 0.003 × 10−3), and there was no statistically significant difference between the CT and CC genotypes (p = 0.194). The WBC count was significantly different among the three genotypes (p = 0.002), but the results of multiple comparisons showed that only the TT and CC genotypes were significantly different (p = 0.002). The difference in the MDD of AZA among different genotypes was also significant (p = 0.010), but the results of multiple comparisons showed that it was only statistically significant between the CT and CC genotypes (p = 0.003) (Supplementary Table S2).
Our results show that all of the patients who carried the NUDT15 (415C>T) TT homozygote mutation, regardless the genotypes of TPMT*3 (T>C) and ITPA (94C>A), had a higher incidence of myelosuppression, and mostly at grade IV. Moreover, 20 of 313 (6.39%) patients carried the wild-type versions of these three genes, one of whom had grade IV myelosuppression. One patient carried a homozygote mutation of TPMT*3 (T>C), with wild-type NUDT15 (415C>T) and ITPA (94C>A), and did not suffer myelosuppression (Table 2).
Based on the occurrence of myelosuppression, all of the 582 patients who received AZA treatment and had routine blood examinations were divided into two groups: group A suffered myelosuppression and group B did not. The results showed that there were more females in both groups, but there was no significant difference in the ratio of males to females (p = 0.240), ages (p = 0.866), nor the MDD of AZA (p = 0.410) between the two groups of patients. The number of patients carrying different genotypes of ITPA (94C>A) and TPMT*3 (T>C) was also not significantly different between the two groups, although the number of patients carrying NUDT15 (415C>T) mutations was significantly different (p = 0.008 × 10−3) (Table 3).
Binary logistic regression analysis of myelosuppression-related factors showed that the risk of myelosuppression was significantly higher in patients with an NUDT15 (415C > T) TT genotype than in those with the wild type (odds ratio [OR], 51.818; 95% CI, 5.280–508.556; p = 0.001). Beyond this, no significant difference in the incidence of myelosuppression was found among the patients with other genotypes of these three genes compared with their corresponding wild type. Other factors, including age (p = 0.722), sex (p = 0.075), and dose (p = 0.490), had no significant association with the incidence of myelosuppression (Table 4).
The gene–gene interaction results are shown in Table 5. The NUDT15 (415C>T) locus had the highest testing balanced accuracy among the 3 SNPs. The optimal interaction models include NUDT15 (415C>T) and ITPA (94C>A) with a maximum cross-validation (CV) consistency of 10 out of 10 and a maximum testing balanced accuracy of 0.5921 (p < 0.05 on the basis of 1000-fold permutation testing).
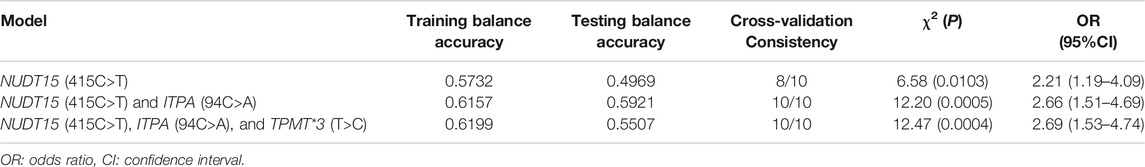
TABLE 5. Epistatic interactions between the variants of azathioprine metabolism influencing myelosuppression.
The retrospective case analysis was performed on a large number of relative cases involving various diseases. Current reports are mostly limited to a certain disease or a type [such as inflammatory bowel disease (Al-Judaibi et al., 2016; Wang et al., 2018; Walker et al., 2019; Kang et al., 2020), autoimmune disease (Fei et al., 2018; Fan et al., 2019), and acute lymphoblastic leukemia (Yang et al., 2015; Zhu et al., 2018)], and have generally included a small number of study subjects, with a focus on only one or two genes. In this study, 40 diseases were included, which covered all of the indications for AZA, and the correlation between genetic polymorphisms of AZA-metabolizing enzymes and AZA-induced myelosuppression were compared under different pathological states.
The most common single-nucleotide polymorphism (SNP) loci of ITPA in the population are 94C>A and IVS2 + 21A>C, and the mutation of ITPA (94C>A) leads to a high risk of ADRs (Arenas et al., 2007). Mutations at this locus affect protein expression by reducing the expression of the full length transcript, decreasing the catalytic activity and stability, and altering mRNA splicing events such as missplicing of exons 2 and 3. Finally, these mutations result in a poor expression of an unstable, catalytically compromised protein, and affect the activity of ITPA (Burgis, 2016). The reported frequency of the ITPA (94C>A) A allele is higher in Asians (11–19%) than in Caucasian, Hispanics, and Africans (1–7%) (Maeda et al., 2005; Hawwa et al., 2008; Okada et al., 2009). In this study, the mutation rate of ITPA (94C > A) was 29.32%, and the frequency of carrying the ITPA (94C>A) A allele was 16.02%, which was consistent with that reported in the current Asian population. Odahara et al. reported that the mutation rate of this gene was 39.6% in 48 Japanese inflammatory bowel disease (IBD) patients, and that the incidence of leukopenia in patients carrying this mutation was 36.8% (Odahara et al., 2015). However, the study indicated that leukopenia cannot be clearly attributed to the ITPA (94C>A) mutation as there may be other influencing factors. Moreover, Wroblova et al. reported a 13.8% mutation rate of the ITPA (94C > A) gene in 188 IBD patients, but no confirmed association was found between its polymorphism and myelosuppression toxicity (Wroblova et al., 2012). These studies also indicated that ITPA genetic polymorphisms may be associated with influenza-like symptoms, arthralgia, and pancreatitis (Zabala-Fernandez et al., 2011; Wroblova et al., 2012). However, the sample sizes of previous studies have been limited, and relatively new studies are lacking. The number of patients included in our study is large, and the result is representative of the Chinese population. In our study, the incidence of myelosuppression in patients with different ITPA (94C>A) genotypes, from high to low, were homozygous mutation, heterozygous mutation, and wild type. Nevertheless, there was no significant difference in the incidence of myelosuppression among these three genotypes. Moreover, a correlation factor analysis suggested that compared to the wild type, homozygous, and heterozygous mutations in patients carried a high risk of myelosuppression, although this was not significant. Therefore, there was no significant correlation between the ITPA (94C>A) gene polymorphism and myelosuppression.
Collie-Duguid et al. reported that the rate of TPMT gene mutation was 10.1% (20/199) in Caucasians, 2.0% (2/99) in Southwest Asians, and 4.7% (9/192) in Chinese (Collie-Duguid et al., 1999). Common mutation loci in TPMT include TPMT*2, TPMT*3A, TPMT*3B, and TPMT*3C. TPMT*3A is the dominant locus in Caucasians, and TPMT*3C is the most common locus in Southeast Asian, African, and African–American populations. TPMT has ten exons, eight of which encode the 28-kDa protein. Nucleotide transversion (G238C) at one locus of TPMT*2 leads to the substitution of a rigid proline for a more flexible alanine residue at codon 80 (Krynetski et al., 1995). This mutation causes changes in the tertiary structure of the TPMT protein, which reduces the stability and catalytic ability of the protein. TPMT*3A contains two single nucleotide transversions, G460A in exon 7 and A719G in exon 10, which leads to amino acid substitutions at codon 154 (Ala > Thr) and codon 240 (Tyr > Cys) (Sahasranaman et al., 2008). TPMT*3B and TPMT*3C both have only one mutation locus, G460A in exon 7 and A719G in exon 10, respectively (Zelinkova et al., 2006). These variants destabilize the TPMT protein, and reduce its binding affinity to 6-MP (Naushad et al., 2021). In our study, the mutation rate of TPMT*3 (T>C) was the lowest, at 3.73%, which was close to the previously reported mutation rate of 2.90% (15/522) in the Japanese population (Kumagai et al., 2001), and 3.17% (4/126) and 4.60% (4/87) in the Chinese population (Chen et al., 2014; Fei et al., 2018). Moreover, in these two studies (Chen et al., 2014; Fei et al., 2018), all of the mutant genotypes of TPMT were heterozygous and no homozygous mutation was found. The higher number of participants in our study could better reflect the mutation rate of this gene in the Chinese population (low). Chen et al. suggested that the TPMT gene polymorphism in Chinese SLE patients had a low sensitivity to predict leukopenia, resulting in a limited clinical value; therefore, they recommended that the AZA dose should be adjusted by monitoring the enzyme activity of TPMT (Chen et al., 2014). Two other studies on Chinese patients with autoimmune diseases demonstrated that the polymorphism of TPMT was not clearly associated with AZA-induced leukopenia (Fei et al., 2018; Fan et al., 2019). Although a meta-analysis of 14 published studies, involving 2276 patients with IBD, showed an association between the TPMT polymorphism and AZA-induced myelosuppression in Caucasians (p < 0.00001; pooled OR, 6.97; 95% CI, 3.89–12.47), no significant correlation was found in Asians (p = 0.12) (Liu et al., 2015). In our study, only two patients carried the TPMT*3 (T>C) CC genotype, one of whom had an AZA treatment history but no myelosuppression. The incidence of myelosuppression in patients with the TPMT*3 (T>C) TC genotype was significantly higher than that in patients with the wild genotype, but the difference was not statistically significant. The correlation factor analysis showed that patients with TPMT*3 (T>C) TC had a higher risk of myelosuppression (OR, 2.420; 95% CI, 0.655–8.946) those with the wild type, but the difference was not significant (p = 0.185). Therefore, there was no correlation between the polymorphism of TPMT*3 (T>C) and myelosuppression. Some studies in Western countries demonstrated a correlation between the TPMT gene polymorphism and the ADR of blood toxicity (Zabala-Fernandez et al., 2011; Al-Judaibi et al., 2016; Steponaitiene et al., 2016); however, for the Asian population, especially Chinese, there was no significant association between the TPMT gene polymorphism and myelosuppression.
There are four common mutation loci of NUDT15, including rs116855232, rs554405994, rs186364861, and rs147390019 (Moriyama et al., 2016), the most common of which is rs116855232 (415C>T, protein sequence p.Arg139Cys). Studies have reported that the NUDT15 (415C>T) mutation does not affect enzymatic activity but does adversely affect protein stability (Valerie et al., 2016). This may be due to the loss of supportive intramolecular bonds, leading to a rapid degradation of proteasomes in cells. Other reports noted that NUDT15 variants have no impact on the binding of “dGTP” to the NUDT protein. The NUDT15 (415C>T) mutation increases aggregation “hot spots” and induces unfavorable torsion in the protein (Naushad et al., 2021). The frequency of mutation for this locus (15–30%) is higher in East Asian populations, including Japanese (Kakuta et al., 2018; Tanaka et al., 2018), Chinese (Chao et al., 2017; Fei et al., 2018), Koreans (Kim et al., 2017; Yi et al., 2018), and Indians (Banerjee et al., 2020), while the mutation rate is low in Caucasians (Yang et al., 2015; Walker et al., 2019) (European: 0.5–0.8% and Hispanic: 7.7%). Some studies have investigated the mutation rate of NUDT15 (415C>T) in the Chinese population, but the majority have had small sample sizes. Fan et al. reported an NUDT15 (415C>T) mutation rate of 17.45% (26/149) in Chinese patients with autoimmune hepatitis, among whom, only 2 patients (1.34%) had homozygous mutations (Fan et al., 2019). Fei et al. studied 87 Chinese patients with autoimmune diseases, and found an NUDT15 (415C>T) mutation rate of 32.18%, with only one patient (1.15%) carrying a homozygous mutation (Fei et al., 2018). In the current study, the NUDT15 (415C>T) mutation rate was 21.92%, and 28 (1.97%) patients had an NUDT15 (415C>T) homozygous mutation; these results are higher than those reported by Fan et al. but lower than those of Fei et al. In addition, Kakuta and colleagues found a 25.27% mutation rate of this gene among 1,282 Japanese patients (Kakuta et al., 2018), which was close to the 21.92% observed in our study. Therefore, it is conceivable that our data truly reflect the mutation rate of NUDT15 in the Chinese population. Current studies suggest that polymorphism of NUDT15 is significantly associated with leukopenia or myelosuppression (Moriyama et al., 2016; Chao et al., 2017; Kim et al., 2017; Fei et al., 2018; Kakuta et al., 2018; Wang et al., 2018; Fan et al., 2019; Kang et al., 2020). Moreover, the risk of adverse reactions has been found to be much higher in people carrying homozygous mutations than in those with the wild genotype. Our results showed that the rate of AZA-induced myelosuppression in patients carrying the NUDT15 (415C>T) TT genotype was as high as 83.33%, and the incidence of grade IV myelosuppression was 60.00%, while patients with a heterozygous mutation and wild type had rates of 11.76 and 7.81%, respectively. Moreover, the incidence of myelosuppression was significantly different among patients with homozygous mutations, heterozygous mutations, and wild type (p = 0.008 × 10−3). Given that there was no significant difference in the MDD of AZA among these patient groups, the interference of dose difference on the incidence of myelosuppression could be eliminated. The correlation factor analysis showed that compared to the wild type, people carrying the homozygous mutant genotype had an extremely high risk of myelosuppression (OR, 51.818; 95% CI, 5.280–508.556; p = 0.001). The mutation frequency of NUDT15 (415C>T) was 21.92%, which was significantly higher than the 3.73% of TPMT*3 (T>C) (p = 1.868 × 10−46). Additionally, the analysis of factors associated with myelosuppression showed that the polymorphism of NUDT15 (415C>T) was significantly associated with myelosuppression; thus, the NUDT15 (415C>T) polymorphism is a promising predictor of AZA-induced myelosuppression in the Chinese population. According to the results, it is recommended to test the genotype of NUDT15 (415C>T) before taking AZA, and AZA should be avoided in patients with a homozygous mutant genotype.
In the current study, the overall incidence of myelosuppression was 9.28%, which was close to the 8.05% (12/149) and 8.7% (81/935) described previously in Chinese and Indian populations (Fan et al., 2019; Banerjee et al., 2020), but lower than 18.17–23.81% reported in the other studies mentioned above (Kim et al., 2017; Fei et al., 2018; Kakuta et al., 2018; Yang et al., 2019). This difference may be due to the inherent limitations of the retrospective study and incomplete information on medication and examination which may lead to the absence of myelosuppression records for some patients. In addition, within the 582 patients with medication records, 83 of patients received an adjusted dose of AZA according to the gene test results to the safe range. The analysis of myelosuppression-related factors showed that sex, age, the MDD of AZA, and polymorphisms of ITPA (94C>A) and TPMT*3 (T>C) had no significant association with myelosuppression, and that only the polymorphism of NUDT15 (415C>T) was an influencing factor. The analysis of different combinations of genotypes indicate that patients with the NUDT15 (415C>T) T allele were prone to suffer from myelosuppression and those who carried the NUDT15 (415C>T) TT genotype faced an even high risk. The results of gene–gene interactions showed that NUDT15 (415C>T) had the highest testing balanced accuracy, which also proved that this gene locus had a strong correlation with myelosuppression. There might be an interaction between ITPA (94C>A) and NUDT15 (415C>T) loci, which together affected the occurrence of myelosuppression induced by azathioprine. At present, the epistatic interactions among the above three gene loci had not been reported. This study was the first to analyze the gene–gene interactions among ITPA, NUDT15, and TPMT. We also found that in patients whose three genes were the wild type, 20 (6.39%) of them suffered from myelosuppression, and one case was grade IV. This finding suggests that these genes are not sufficient to predict myelosuppression in all patients, and there may be other relative metabolic enzyme genes that remain to be explored in future studies (Inman et al., 2018).
This study has some limitations. First, the retrospective nature of the study meant that the information was incomplete in some cases, and some medication records and test results were missing; in particular, one patient with the TPMT*3 (T>C) CC genotype had no medication records, which resulted in the exclusion of this population. In addition, as the genotype detection of ITPA (94C>A) was only initiated in our hospital in the last few years, there were 140 cases in whom only TPMT*3 (T>C) and NUDT15 (415C>T) were detected, with no information on the ITPA (94C>A) genotype. The degree of AZA-induced myelosuppression could only be evaluated by the information provided in the cases, which cannot be used to reconstruct the actual situation at the time, making it difficult to truly evaluate the severe grades of myelosuppression.
Our findings suggest that the polymorphism of NUDT15 (415C>T) is a significantly relative factor in the context of AZA-induced myelosuppression, and epistatic interactions between ITPA (94C>A) and NUDT15 (415C>T) affect the occurrence of myelosuppression. Therefore, it is recommended to test these two genes prior to administration of AZA. In people carrying a homozygous mutation of NUDT15 (415C>T), the risk of myelosuppression is very high, and therefore AZA should be avoided during their treatment. However, in our hospital, the cost of the detection of these three metabolic enzyme genes is 628 times that of one tablet of AZA (100 mg). Thus, the detection of ITPA (94C>A) and TPMT*3 (T>C) is not necessarily recommended for economic reasons but only to test the genotype of NUDT15 (415C>T) for patients who have difficulty in paying medical expenses. Moreover, there may be other AZA-metabolizing enzyme genes that could better predict the incidence of AZA-induced myelosuppression, and further investigations are needed.
All datasets presented in this study are included in the article/Supplementary Material.
This study was performed according to the recommendations of the ethical guidelines and approved by the Biomedical Ethics Committee of West China Hospital of Sichuan University (Nos. 2020973 and 2021402). All patients were exempt from providing informed consent.
Z-YC, Y-HZ, L-YZ, and Z-YH wrote the manuscript; X-JL and Z-YH designed the research; Z-YC, Y-HZ, ZQ, and Y-WP performed the research; Z-YC, Y-HZ, W-QS, and Z-YH analyzed the data; BW, YY, N-NC, RZ, M-YW, Z-HS, X-JL, and TX contributed new reagents/analytical tools.
This work was supported by the National Key Research and Development Program of China (2020YFC2008302), the Sichuan Science and Technology program (2019YFG0266), and the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC18028, 2021HXFH064).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.672769/full#supplementary-material
Al-Judaibi, B., Schwarz, U. I., Huda, N., Dresser, G. K., Gregor, J. C., Ponich, T., et al. (2016). Genetic Predictors of Azathioprine Toxicity and Clinical Response in Patients with Inflammatory Bowel Disease. J. Popul. Ther. Clin. Pharmacol. 23 (1), 26–36.
Arenas, M., Duley, J., Sumi, S., Sanderson, J., and Marinaki, A. (2007). The ITPA c.94C>A and g.IVS2+21A>C Sequence Variants Contribute to Missplicing of the ITPA Gene. Biochim. Biophys. Acta 1772 (1), 96–102. doi:10.1016/j.bbadis.2006.10.006
Banerjee, R., Ravikanth, V. V., Pal, P., Bale, G., Avanthi, U. S., Goren, I., et al. (2020). NUDT15 C415T Variant Compared with TPMT Genotyping in Predicting Azathioprine-Induced Leucopenia: Prospective Analysis of 1014 Inflammatory Bowel Disease Patients in India. Aliment. Pharmacol. Ther. 52 (11-12), 1683–1694. doi:10.1111/apt.16137
Burgis, N. E. (2016). A Disease Spectrum for ITPA Variation: Advances in Biochemical and Clinical Research. J. Biomed. Sci. 23 (1), 73. doi:10.1186/s12929-016-0291-y
Chao, K., Wang, X., Cao, Q., Qian, J., Wu, K., Zhu, X., et al. (2017). Combined Detection of NUDT15 Variants Could Highly Predict Thiopurine-Induced Leukopenia in Chinese Patients with Inflammatory Bowel Disease: A Multicenter Analysis. Inflamm. Bowel Dis. 23 (9), 1592–1599. doi:10.1097/mib.0000000000001148
Chen, D. Y., Lian, F., Yuan, S. W., Wang, Y. X., Zhan, Z. P., Ye, Y. J., et al. (2014). Association of Thiopurine Methyltransferase Status with Azathioprine Side Effects in Chinese Patients with Systemic Lupus Erythematosus. Clin. Rheumatol. 33 (4), 499–503. doi:10.1007/s10067-013-2441-x
Choi, R., Sohn, I., Kim, M. J., Woo, H. I., Lee, J. W., Ma, Y., et al. (2019). Pathway Genes and Metabolites in Thiopurine Therapy in Korean Children with Acute Lymphoblastic Leukaemia. Br. J. Clin. Pharmacol. 85 (7), 1585–1597. doi:10.1111/bcp.13943
Collie-Duguid, E. S., Pritchard, S. C., Powrie, R. H., Sludden, J., Collier, D. A., Li, T., et al. (1999). The Frequency and Distribution of Thiopurine Methyltransferase Alleles in Caucasian and Asian Populations. Pharmacogenetics 9 (1), 37–42. doi:10.1097/00008571-199902000-00006
Fan, X., Yin, D., Men, R., Xu, H., and Yang, L. (2019). NUDT15 Polymorphism Confer Increased Susceptibility to Thiopurine-Induced Leukopenia in Patients with Autoimmune Hepatitis and Related Cirrhosis. Front. Pharmacol. 10, 346. doi:10.3389/fphar.2019.00346
Fei, X., Shu, Q., Zhu, H., Hua, B., Wang, S., Guo, L., et al. (2018). NUDT15 R139C Variants Increase the Risk of Azathioprine-Induced Leukopenia in Chinese Autoimmune Patients. Front. Pharmacol. 9, 460. doi:10.3389/fphar.2018.00460
Food and Drug Administration (2018). IMURAN ® (azathioprine) Available: https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/016324s039lbl (Accessed 20 01, 2021).
Hawwa, A. F., Millership, J. S., Collier, P. S., Vandenbroeck, K., Mccarthy, A., Dempsey, S., et al. (2008). Pharmacogenomic Studies of the Anticancer and Immunosuppressive Thiopurines Mercaptopurine and Azathioprine. Br. J. Clin. Pharmacol. 66 (4), 517–528. doi:10.1111/j.1365-2125.2008.03248.x
Inman, G. J., Wang, J., Nagano, A., Alexandrov, L. B., Purdie, K. J., Taylor, R. G., et al. (2018). The Genomic Landscape of Cutaneous SCC Reveals Drivers and a Novel Azathioprine Associated Mutational Signature. Nat. Commun. 9 (1), 3667. doi:10.1038/s41467-018-06027-1
Joly, P., Horvath, B., Patsatsi, A., Uzun, S., Bech, R., Beissert, S., et al. (2020). Updated S2K Guidelines on the Management of Pemphigus Vulgaris and Foliaceus Initiated by the European Academy of Dermatology and Venereology (EADV). J. Eur. Acad. Dermatol. Venereol. 34 (9), 1900–1913. doi:10.1111/jdv.16752
Kakuta, Y., Kawai, Y., Okamoto, D., Takagawa, T., Ikeya, K., Sakuraba, H., et al. (2018). NUDT15 Codon 139 Is the Best Pharmacogenetic Marker for Predicting Thiopurine-Induced Severe Adverse Events in Japanese Patients with Inflammatory Bowel Disease: a Multicenter Study. J. Gastroenterol. 53 (9), 1065–1078. doi:10.1007/s00535-018-1486-7
Kang, B., Kim, T. J., Choi, J., Baek, S. Y., Ahn, S., Choi, R., et al. (2020). Adjustment of Azathioprine Dose Should Be Based on a Lower 6-TGN Target Level to Avoid Leucopenia in NUDT15 Intermediate Metabolisers. Aliment. Pharmacol. Ther. 52 (3), 459–470. doi:10.1111/apt.15810
Kim, S. Y., Shin, J. H., Park, J. S., Kang, S. Y., Nam, T. S., Kim, J. K., et al. (2017). NUDT15 p.R139C Variant Is Common and Strongly Associated with Azathioprine-Induced Early Leukopenia and Severe Alopecia in Korean Patients with Various Neurological Diseases. J. Neurol. Sci. 378, 64–68. doi:10.1016/j.jns.2017.04.041
Kishibe, M., Nozaki, H., Fujii, M., Iinuma, S., Ohtsubo, S., Igawa, S., et al. (2018). Severe Thiopurine-Induced Leukocytopenia and Hair Loss in Japanese Patients with Defective NUDT15 Variant: Retrospective Case-Control Study. J. Dermatol. 45 (10), 1160–1165. doi:10.1111/1346-8138.14588
Krishnamurthy, P., Schwab, M., Takenaka, K., Nachagari, D., Morgan, J., Leslie, M., et al. (2008). Transporter-mediated Protection against Thiopurine-Induced Hematopoietic Toxicity. Cancer Res. 68 (13), 4983–4989. doi:10.1158/0008-5472.Can-07-6790
Krynetski, E. Y., Schuetz, J. D., Galpin, A. J., Pui, C. H., Relling, M. V., and Evans, W. E. (1995). A Single Point Mutation Leading to Loss of Catalytic Activity in Human Thiopurine S-Methyltransferase. Proc. Natl. Acad. Sci. U S A. 92 (4), 949–953. doi:10.1073/pnas.92.4.949
Kudo, M., Saito, Y., Sasaki, T., Akasaki, H., Yamaguchi, Y., Uehara, M., et al. (2009). Genetic Variations in the HGPRT, ITPA, IMPDH1, IMPDH2, and GMPS Genes in Japanese Individuals. Drug Metab. Pharmacokinet. 24(6), 557–564. doi:10.2133/dmpk.24.557
Kumagai, K., Hiyama, K., Ishioka, S., Sato, H., Yamanishhi, Y., Mcleod, H. L., et al. (2001). Allelotype Frequency of the Thiopurine Methyltransferase (TPMT) Gene in Japanese. Pharmacogenetics 11(3), 275–278. doi:10.1097/00008571-200104000-00012
Liu, Y. P., Xu, H. Q., Li, M., Yang, X., Yu, S., Fu, W. L., et al. (2015). Association between Thiopurine S-Methyltransferase Polymorphisms and Azathioprine-Induced Adverse Drug Reactions in Patients with Autoimmune Diseases: A Meta-Analysis. Plos One 10 (12). e0144234. doi:10.1371/journal.pone.0144234
Mack, C. L., Adams, D., Assis, D. N., Kerkar, N., Manns, M. P., Mayo, M. J., et al. (2020). Diagnosis and Management of Autoimmune Hepatitis in Adults and Children: 2019 Practice Guidance and Guidelines from the American Association for the Study of Liver Diseases. Hepatology 72 (2), 671–722. doi:10.1002/hep.31065
Maeda, T., Sumi, S., Ueta, A., Ohkubo, Y., Ito, T., Marinaki, A. M., et al. (2005). Genetic Basis of Inosine Triphosphate Pyrophosphohydrolase Deficiency in the Japanese Population. Mol. Genet. Metab. 85 (4), 271–279. doi:10.1016/j.ymgme.2005.03.011
Moon, W., and Loftus, E. V. (2016). Review Article: Recent Advances in Pharmacogenetics and Pharmacokinetics for Safe and Effective Thiopurine Therapy in Inflammatory Bowel Disease. Aliment. Pharmacol. Ther. 43 (8), 863–883. doi:10.1111/apt.13559
Moriyama, T., Nishii, R., Perez-Andreu, V., Yang, W., Klussmann, F. A., Zhao, X., et al. (2016). NUDT15 Polymorphisms Alter Thiopurine Metabolism and Hematopoietic Toxicity. Nat. Genet. 48 (4), 367–373. doi:10.1038/ng.3508
Naranjo, C. A., Busto, U., Sellers, E. M., Sandor, P., Ruiz, I., Roberts, E. A., et al. (1981). A Method for Estimating the Probability of Adverse Drug Reactions. Clin. Pharmacol. Ther. 30 (2), 239–245. doi:10.1038/clpt.1981.154
National Health Commission, PRC (2011). Administrative Measures for Reporting and Monitoring of Adverse Drug Reactions. Available at: http://www.nhc.gov.cn/cmssearch/xxgk/getManuscriptXxgk.htm?id=b442a66fc52b4793a57160002ac2a1a9. [Accessed January 20, 2021]
Naushad, S. M., Janaki Ramaiah, M., Kutala, V. K., Hussain, T., and Alrokayan, S. A. (2021). Pharmacogenetic Determinants of Thiopurines in an Indian Cohort. Pharmacol. Rep. 73 (1), 278–287. doi:10.1007/s43440-020-00158-3
Odahara, S., Uchiyama, K., Kubota, T., Ito, Z., Takami, S., Kobayashi, H., et al. (2015). A Prospective Study Evaluating Metabolic Capacity of Thiopurine and Associated Adverse Reactions in Japanese Patients with Inflammatory Bowel Disease (IBD). Plos One 10 (9). e0137798. doi:10.1371/journal.pone.0137798
Okada, Y., Nakamura, K., Hiromura, K., Nojima, Y., Horiuchi, R., and Yamamoto, K. (2009). Pro32Thr Polymorphism of Inosine Triphosphate Pyrophosphatase Gene Predicts Efficacy of Low-Dose Azathioprine for Patients with Systemic Lupus Erythematosus. Clin. Pharmacol. Ther. 85 (5), 527–530. doi:10.1038/clpt.2008.261
Panda, B. K., Umarje, S., and Diwan, A. (2018). Azathioprine-Induced Pancytopenia and Septic Complications: A Probable Cause of Death. J. Pharm. Pract. 31 (5), 510–513. doi:10.1177/0897190017729521
Poppe, D., Tiede, I., Fritz, G., Becker, C., Bartsch, B., Wirtz, S., et al. (2006). Azathioprine Suppresses Ezrin-radixin-moesin-dependent T Cell-APC Conjugation through Inhibition of Vav Guanosine Exchange Activity on Rac Protein. J. Immunol. 176(1), 640–651. doi:10.4049/jimmunol.176.1.640
Ran, Z., Wu, K., Matsuoka, K., Jeen, Y. T., Wei, S. C., Ahuja, V., et al. (2020). Asian Organization for Crohn's and Colitis and Asia Pacific Association of Gastroenterology Practice Recommendations for Medical Management and Monitoring of Inflammatory Bowel Disease in Asia. J. Gastroenterol. Hepatol. 16 (1), 17-25. doi:10.1111/jgh.15185
Relling, M. V., Gardner, E. E., Sandborn, W. J., Schmiegelow, K., Pui, C. H., Yee, S. W., et al. (2011). Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing. Clin. Pharmacol. Ther. 89 (3), 387–391. doi:10.1038/clpt.2010.320
Relling, M. V., Gardner, E. E., Sandborn, W. J., Schmiegelow, K., Pui, C. H., Yee, S. W., et al. (2013). Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing: 2013 Update. Clin. Pharmacol. Ther. 93 (4), 324–325. doi:10.1038/clpt.2013.4
Relling, M. V., Schwab, M., Whirl-Carrillo, M., Suarez-Kurtz, G., Pui, C. H., Stein, C. M., et al. (2019). Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin. Pharmacol. Ther. 105 (5), 1095–1105. doi:10.1002/cpt.1304
Sahasranaman, S., Howard, D., and Roy, S. (2008). Clinical Pharmacology and Pharmacogenetics of Thiopurines. Eur. J. Clin. Pharmacol. 64 (8), 753–767. doi:10.1007/s00228-008-0478-6
Steponaitiene, R., Kupcinskas, J., Survilaite, S., Varkalaite, G., Jonaitis, L., Kiudelis, G., et al. (2016). TPMT and ITPA Genetic Variants in Lithuanian Inflammatory Bowel Disease Patients: Prevalence and Azathioprine-Related Side Effects. Adv. Med. Sci. 61 (1), 135–140. doi:10.1016/j.advms.2015.09.008
Tanaka, Y., Nakadate, H., Kondoh, K., Nakamura, K., Koh, K., and Manabe, A. (2018). Interaction between NUDT15 and ABCC4 Variants Enhances Intolerability of 6-mercaptopurine in Japanese Patients with Childhood Acute Lymphoblastic Leukemia. Pharmacogenomics J. 18 (2), 275–280. doi:10.1038/tpj.2017.12
Thompson, J. A., Schneider, B. J., Brahmer, J., Andrews, S., Armand, P., Bhatia, S., et al. (2020). NCCN Guidelines Insights: Management of Immunotherapy-Related Toxicities, Version 1.2020. J. Natl. Compr. Canc Netw. 18 (3), 230–241. doi:10.6004/jnccn.2020.0012
Tiede, I., Fritz, G., Strand, S., Poppe, D., Dvorsky, R., Strand, D., et al. (2003). CD28-dependent Rac1 Activation Is the Molecular Target of Azathioprine in Primary Human CD4+ T Lymphocytes. J. Clin. Invest. 111 (8), 1133–1145. doi:10.1172/jci16432
Valerie, N. C., Hagenkort, A., Page, B. D., Masuyer, G., Rehling, D., Carter, M., et al. (2016). NUDT15 Hydrolyzes 6-Thio-DeoxyGTP to Mediate the Anticancer Efficacy of 6-Thioguanine. Cancer Res. 76 (18), 5501–5511. doi:10.1158/0008-5472.CAN-16-0584
Van, G, T., Van Schaik, R. H., and Hesselink, D. A. (2014). Pharmacogenetics and Immunosuppressive Drugs in Solid Organ Transplantation. Nat. Rev. Nephrol. 10 (12), 725–731. doi:10.1038/nrneph.2014.172
Walker, G. J., Harrison, J. W., Heap, G. A., Voskuil, M. D., Andersen, V., Anderson, C. A., et al. (2019). Association of Genetic Variants in NUDT15 with Thiopurine-Induced Myelosuppression in Patients with Inflammatory Bowel Disease. Jama 321 (8), 773–785. doi:10.1001/jama.2019.0709
Wang, H. H., He, Y., Wang, H. X., Liao, C. L., Peng, Y., Tao, L. J., et al. (2018). Comparison of TPMT and NUDT15 Polymorphisms in Chinese Patients with Inflammatory Bowel Disease. World J. Gastroenterol. 24 (8), 941–948. doi:10.3748/wjg.v24.i8.941
Wroblova, K., Kolorz, M., Batovsky, M., Zboril, V., Suchankova, J., Bartos, M., et al. (2012). Gene Polymorphisms Involved in Manifestation of Leucopenia, Digestive Intolerance, and Pancreatitis in Azathioprine-Treated Patients. Dig. Dis. Sci. 57 (9), 2394–2401. doi:10.1007/s10620-012-2163-y
Yang, J. J., Landier, W., Yang, W., Liu, C., Hageman, L., Cheng, C., et al. (2015). Inherited NUDT15 Variant Is a Genetic Determinant of Mercaptopurine Intolerance in Children with Acute Lymphoblastic Leukemia. J. Clin. Oncol. 33 (11), 1235–1242. doi:10.1200/jco.2014.59.4671
Yang, J. J., Whirl-Carrillo, M., Scott, S. A., Turner, A. J., Schwab, M., Tanaka, Y., et al. (2019). Pharmacogene Variation Consortium Gene Introduction: NUDT15. Clin. Pharmacol. Ther. 105 (5), 1091–1094. doi:10.1002/cpt.1268
Yang, J., Wang, P. L., Qin, Z. F., Jia, M. M., Zhang, C. M., Tian, X. K., et al. (2019). NUDT15 and TPMT Genetic Polymorphisms Are Related to Azathioprine Intolerance in Chinese Patients with Rheumatic Diseases. Genet. Test. Mol. Biomarkers 23 (10), 751–757. doi:10.1089/gtmb.2018.0313
Yang, S. K., Hong, M., Baek, J., Choi, H., Zhao, W. T., Jung, Y. S., et al. (2014). A Common Missense Variant in NUDT15 Confers Susceptibility to Thiopurine-Induced Leukopenia. Nat. Genet. 46(9), 1017- doi:10.1038/ng.3060
Yi, E. S., Choi, Y. B., Choi, R., Lee, N. H., Lee, J. W., Yoo, K. H., et al. (2018). NUDT15 Variants Cause Hematopoietic Toxicity with Low 6-TGN Levels in Children with Acute Lymphoblastic Leukemia. Cancer Res. Treat. 50 (3), 872–882. doi:10.4143/crt.2017.283
Zabala-Fernandez, W., Barreiro-De Acosta, M., Echarri, A., Carpio, D., Lorenzo, A., Castro, J., et al. (2011). A Pharmacogenetics Study of TPMT and ITPA Genes Detects a Relationship with Side Effects and Clinical Response in Patients with Inflammatory Bowel Disease Receiving Azathioprine. J. Gastrointest. Liver Dis. 20 (3), 247–253.
Zelinkova, Z., Derijks, L. J., Stokkers, P. C., Vogels, E. W., Van Kampen, A. H., Curvers, W. L., et al. (2006). Inosine Triphosphate Pyrophosphatase and Thiopurine S-Methyltransferase Genotypes Relationship to Azathioprine-Induced Myelosuppression. Clin. Gastroenterol. Hepatol. 4 (1), 44–49. doi:10.1016/j.cgh.2005.10.019
Zhu, Y. P., Yin, D. D., Su, Y. L., Xia, X. Y., Moriyama, T., Nishii, R., et al. (2018). Combination of Common and Novel Rare NUDT15 Variants Improves Predictive Sensitivity of Thiopurine-Induced Leukopenia in Children with Acute Lymphoblastic Leukemia. Haematologica 103 (7), E293–E295. doi:10.3324/haematol.2018.187658
Keywords: azathioprine, ITPA, TPMT, NUDT15, myelosuppression, adverse drug reaction
Citation: Chen Z-Y, Zhu Y-H, Zhou L-Y, Shi W-Q, Qin Z, Wu B, Yan Y, Pei Y-W, Chao N-N, Zhang R, Wang M-Y, Su Z-H, Lu X-J, He Z-Y and Xu T (2021) Association Between Genetic Polymorphisms of Metabolic Enzymes and Azathioprine-Induced Myelosuppression in 1,419 Chinese Patients: A Retrospective Study. Front. Pharmacol. 12:672769. doi: 10.3389/fphar.2021.672769
Received: 26 February 2021; Accepted: 27 April 2021;
Published: 18 May 2021.
Edited by:
Moneeza Kalhan Siddiqui, University of Dundee, United KingdomReviewed by:
Francisco Abad-Santos, Princess University Hospital, SpainCopyright © 2021 Chen, Zhu, Zhou, Shi, Qin, Wu, Yan, Pei, Chao, Zhang, Wang, Su, Lu, He and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Jun Lu, bHV4aWFvanVuMTk3MkAxNjMuY29t; Zhi-Yao He, aGV5YW9kZUAxNjMuY29t, emhpeWFvaGVAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.