- 1Center for Global Health and Diseases, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 2Division of Clinical Pharmacology, Toxicology & Therapeutic Innovation, Children’s Mercy Kansas City, Kanas City, MO, United States
- 3Big Data Institute, Li Ka Shing Centre for Health Information and Discovery, University of Oxford, Oxford, United Kingdom
- 4Foundation for Innovative New Diagnostics, Geneva, Switzerland
- 5The National Malaria Control Program, Ministry of Health, Antananarivo, Madagascar
- 6University of Fianarantsoa, Fianarantsoa, Madagascar
Plasmodium vivax is one of the five human malaria parasite species, which has a wide geographical distribution and can cause severe disease and fatal outcomes. It has the ability to relapse from dormant liver stages (hypnozoites), weeks to months after clearance of the acute blood-stage infection. An 8-aminoquinoline drug primaquine (PQ) can clear the hypnozoites, and thus can be used as an anti-relapse therapeutic agent. Recently, a number of studies have found that its efficacy is compromised by polymorphisms in the cytochrome P450 2D6 (CYP2D6) gene; decreased or absence of CYP2D6 activity contributes to PQ therapeutic failure. The present study sought to characterize CYP2D6 genetic variation in Madagascar, where populations originated from admixture between Asian and African populations, vivax malaria is endemic, and PQ can be deployed soon to achieve national malaria elimination. In a total of 211 samples collected from two health districts, CYP2D6 decreased function alleles CYP2D6*10, *17, *29, *36+*10, and *41 were observed at frequencies of 3.55–17.06%. In addition, nonfunctional alleles were observed, the most common of which were CYP2D6*4 (2.13%), *5 (1.66%), and the *4x2 gene duplication (1.42%). Given these frequencies, 34.6% of the individuals were predicted to be intermediate metabolizers (IM) with an enzyme activity score (AS) ≤ 1.0; both the IM phenotype and AS ≤ 1.0 have been found to be associated with PQ therapeutic failure. Furthermore, the allele and genotype frequency distributions add to the archaeological and genomic evidence of Malagasy populations constituting a unique, Asian-African admixed origin. The results from this exploratory study provide fresh insights about genomic characteristics that could affect the metabolism of PQ into its active state, and may enable optimization of PQ treatment across human genetic diversity, which is critical for achieving P. vivax elimination.
Introduction
Five Plasmodium species are known to cause malaria in humans; these are, Plasmodium falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. Although P. vivax has often been regarded as causing a benign self-limiting infection, there is clear evidence that it can cause severe disease and fatality (Baird, 2009; Battle et al., 2012; Baird, 2013). It has a wide geographical distribution, with relatively high prevalence of infection in the South-East Asian and Western Pacific regions and in most of the Americas (Howes et al., 2016; Battle et al., 2019). Within Africa, countries around the Horn of Africa and Madagascar, which have a lower prevalence of the human Duffy-negative phenotype, once thought to provide complete protection against P. vivax (Miller et al., 1976), can sustain endemic transmission of this parasite (Menard et al., 2010; Howes et al., 2011; Howes et al., 2015; Twohig et al., 2019). There is now substantial evidence that P. vivax is endemic throughout Africa (Howes et al., 2015; Twohig et al., 2019), though local surveillance for this parasite remains rare (Zimmerman, 2017).
The challenges in controlling and eliminating vivax malaria are likely to be related to a number of unique aspects of P. vivax biology (Adams and Mueller, 2017; Bourgard et al., 2018), mainly its ability to relapse from long-lasting, dormant liver stages (the hypnozoites) (Imwong et al., 2007; White, 2011; White and Imwong, 2012; Battle et al., 2014). The hypnozoites persist in the liver, from weeks to months, and cause relapses after clearance of the acute blood-stage infection (White, 2011; Battle et al., 2014). As there are no diagnostic tests that can identify these dormant liver-stage infections, undetectable hypnozoite carriers are an important potential parasite reservoir for sustaining transmission and reintroduction, if not treated appropriately. From the 1950s until 2018, 8-aminoquinoline primaquine (PQ) was the only licensed drug available to clear the hypnozoites (John et al., 2012; Baird, 2019). In 2018, the United States Food and Drug Administration approved the use of a new hypnozoitocidal 8-aminoquinoline called tafenoquine (Baird, 2018; White, 2019). Although these two 8-aminoquinoline drugs are the only likely near-term anti-relapse therapies currently available, their safety and efficacy is severely limited by host genetic variation: patients with a deficiency in glucose-6-phosphate dehydrogenase (G6PD) enzyme activity levels can have acute hemolytic anemia (Baird et al., 2018; Baird, 2019; White, 2019). In addition, clinical and laboratory evidence suggested that the efficacy of PQ may depend on genetic variation in cytochrome P450 2D6 (CYP2D6) drug-metabolizing enzyme activity (Baird, 2018; Baird et al., 2018; Baird, 2019).
CYP2D6 is involved in the metabolism of up to 21% of drugs, many of which are commonly prescribed (Saravanakumar et al., 2019). Among these is PQ, which has been shown to be metabolized principally via CYP2D6 in animal models (Pybus et al., 2013; Potter et al., 2015), in vitro enzymatic assays (Fasinu et al., 2014; Fasinu et al., 2016a; Fasinu et al., 2016b; Saito et al., 2018; Camarda et al., 2019), and in human studies (Goncalves et al., 2017; Ariffin et al., 2019; Spring et al., 2019). CYP2D6 also contributes to the metabolism of tafenoquine (Marcsisin et al., 2014; Vuong et al., 2015). CYP2D6 is among the most extensively studied and characterized polymorphic drug-metabolizing enzymes (Taylor et al., 2020). Variation in the CYP2D6 gene includes single nucleotide variants, short insertions and deletions, as well as gene copy number variations (CNV). The latter encompass deletions of the entire gene, gene duplications and multiplications, and structural rearrangements with the highly similar CYP2D7 pseudogene (Nofziger et al., 2020). Currently, there are over 130 CYP2D6 “star” (*) alleles listed by the Pharmacogene Variation Consortium (PharmVar) (Nofziger et al., 2020). Many of the listed alleles contribute, in part or entirely, to altered rates of CYP2D6-mediated drug metabolism due to increased, decreased, or absent enzyme activity (Nofziger et al., 2020; Taylor et al., 2020). A system for translating CYP2D6 genotype to phenotype, and assigning individuals into four phenotype groups [ultrarapid metabolizers (UM), normal metabolizers (NM), intermediate metabolizers (IM), and poor metabolizers (PM)], has been developed by the Clinical Pharmacogenetic Implementation Consortium (CPIC) (Caudle et al., 2020).
In a clinical trial of a vaccine against P. vivax involving 25 healthy malaria-naïve adults, who were given a combination of chloroquine and PQ as treatment, two participants had multiple relapses of malaria (Bennett et al., 2013). One participant who experienced two relapses was genotyped as CYP2D6*4/*41 and classified as IM, whereas another participant who experienced three relapses was a PM with a CYP2D6*5/*6 genotype. In another study, an IM subject (genotyped as CYP2D6*5/*41) suffered multiple attacks of vivax malaria at approximately two-month intervals despite taking PQ as prescribed (Ingram et al., 2014). Since those findings were reported (Bennett et al., 2013; Ingram et al., 2014), there have been several other reports suggesting that decreased or absence of CYP2D6 activity contributes to PQ therapeutic failure (Baird et al., 2018; Dijanic et al., 2018; He et al., 2019; Martin Ramirez et al., 2020; Mat Salleh et al., 2020; Silvino et al., 2020). In contrast, in clinical trials using tafenoquine in combination with chloroquine, the IM phenotype was reported to have had no significant effect on the efficacy of tafenoquine in preventing P. vivax relapse; however, more data on PM persons are required (St Jean et al., 2016; Lacerda et al., 2019).
Madagascar is an island nation in the Indian Ocean, approximately 400 km (250 miles) off the East African coast. Archaeological and genomic data provide evidence of sequential waves of human migration over the last two millennia to Madagascar from Indonesia and from East Africa by Bantu populations, together with more limited later arrivals of Arab, European, Indian, and Chinese peoples. These events have resulted in a genetically cosmopolitan Malagasy society (Kusuma et al., 2015; Brucato et al., 2016; Kusuma et al., 2016; Pierron et al., 2017; Pierron et al., 2018). Plasmodium falciparum is the predominant malaria species in Madagascar, and P. vivax, P. malariae and P. ovale are also present. Plasmodium vivax is diagnosed at similar prevalence as P. falciparum by molecular methods in the western highlands fringe region (Howes et al., 2018). Irrespective of the parasite species involved, any malaria positive cases are treated with an age-adjusted dose of artesunate-amodiaquine. Plasmodium vivax-specific treatment of hypnozoites in the form of PQ for radical cure is not available, although vivax relapse has been reported (Andrianaranjaka et al., 2013). The Madagascar National Malaria Control (NMCP) Program 2018–2022 has declared its objective for achieving local elimination in low-burden highland districts while gradually pushing back residual transmission to coastal areas, en route to achieving national elimination (National Malaria Control Programme of Madagascar, 2017). Therefore, assessing PQ efficacy in the target P. vivax patient population is essential to support the malaria elimination goal. Although CYP2D6 genetic variation has been extensively studied in many populations, there are no published data for the Malagasy, a rather unique population with a rich admixture history between Asian and African ancestral populations and the potential for novel genotypic combinations.
To reduce this knowledge gap, the aim of the present study was to characterize CYP2D6 genetic variation in a Malagasy population exposed to P. vivax transmission. Given that CYP2D6 emerges as a major pathway for PQ bioactivation, and decreased or absence of enzyme activity may contribute to PQ therapeutic failure, CYP2D6 genetic variation and its functional implications must be understood to develop strategies and expectations for P. vivax elimination in Madagascar as well as other P. vivax endemic settings.
Methods
Subjects and Study Sites
This investigation is a part of our ongoing malaria epidemiological studies being conducted in the western highlands fringe region of Madagascar (Menard et al., 2010; Howes et al., 2017; Howes et al., 2018; Willie et al., 2018). The study was approved by the University Hospitals of Cleveland Institutional Review Board (#09-13-01), the Division of Microbiology and Infectious Diseases/NIAID/National Institutes of Health (NIH) (#13-0067), and the Madagascar Ministry of Health Ethics Committee (#099). Written informed consent was obtained from all subjects, or subject guardians, prior to enrollment.
The Malagasy population sampled included rural communities in the Mandoto district (n = 167) and a migrant population in the rural Ampasimpotsy community of Tsiroanomandidy district (n = 44) relocated from the capital city, Antananarivo. These communities are located in the highlands fringe region and self-identified as being predominantly of the Merina highland ethnic group (Howes et al., 2017). These populations were considered genetically representative of Malagasy populations targeted for PQ therapy.
Sample Collection and Processing
Samples were collected between March and August 2015 from apparently healthy individuals of all ages during a study screening for asymptomatic malaria infections and G6PD status. Most individuals were male (92%) due to the parallel objective of qualitative phenotypic G6PD characterization. Details of the communities surveyed have been previously documented (Howes et al., 2017). Two hundred μL of capillary blood was collected into K+-EDTA microtainers for DNA extraction and subsequent CYP2D6 genotyping. DNA was extracted from 50 to 100 μL of each sample using a QIAamp® DNA Micro Kit (QIAGEN, Germantown, MD). The remaining blood sample and extracted DNA were stored at −20°C.
CYP2D6 Genotype Analysis and Translation Into Phenotype
Samples were genotyped using commercially available TaqMan™ genotyping assays on a custom-designed OpenArray (Thermo Fisher Scientific, Waltham, MA), as recommended by the manufacturer on a QuantStudio™ 12K Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). Data were analyzed with the TaqMan™ Genotyper Software and manually inspected. A total of 29 SNPs were tested, which allowed the identification of the presence of the following allelic variants using star allele nomenclature per PharmVar: CYP2D6*2, *3, *4, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *29, *31, *35, *40, *41, *42, *44, *45, *49, *56, *59, *99, *100, and *101. CYP2D6*1 was assigned if no variants were identified. In addition, all samples were interrogated for the presence of CNVs including the CYP2D6*5 gene deletion, duplications, hybrid genes, and tandem arrangements utilizing a previously published quantitative multiplex method that interrogates four gene regions (Gaedigk et al., 2012). Long-range PCR reactions were performed to confirm and further characterize CNV results following published protocols (Gaedigk et al., 2007; Gaedigk et al., 2010; Gaedigk et al., 2019). Additional information is provided in Supplementary Table S1 using the PharmVar recommended template for reporting genotyping method details.
Star alleles were called using nomenclature detailed by PharmVar. Subsequently, activity scores (AS) were determined for each diplotype and translated into phenotype as recommended by CPIC (Caudle et al., 2020). Briefly, values of 0, 0.25, 0.5, or 1 were assigned to each allele and the sum of both values added to determine a diplotype’s AS. Alleles with gene duplications received a value double that of its single counterpart. Values assigned to each allele are provided in Table 1. AS groups were translated into phenotype as follows: UM, AS > 2.25; NM, AS = 1.25, 1.5, 2.0, and 2.25; IM, AS = 0.25, 0.5, 0.75, and 1.0; PM, AS = 0.
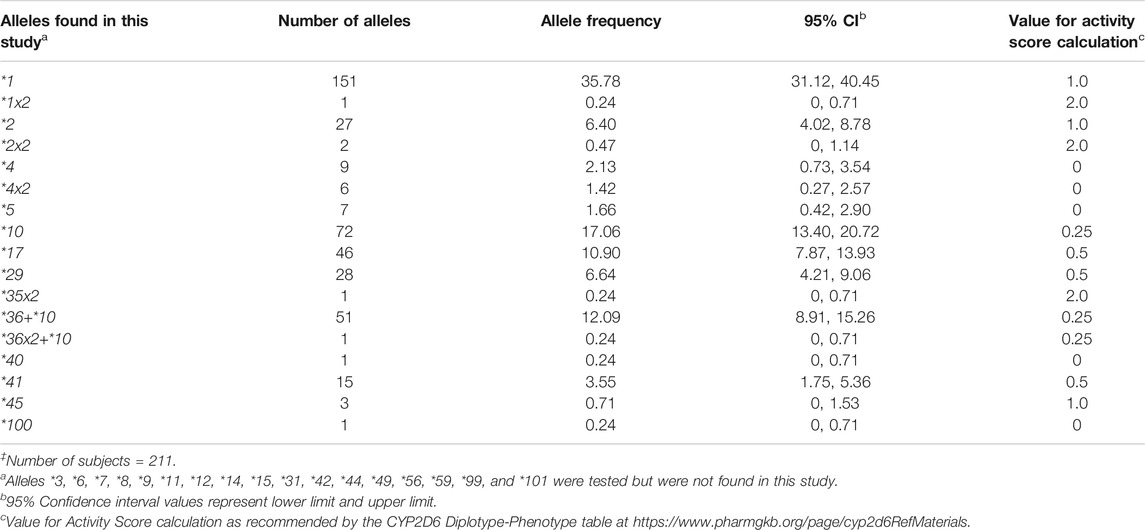
TABLE 1. CYP2D6 allele frequencies in the Madagascar study population‡.
Statistical Analysis
95% Confidence interval (95% CI) lower and upper limit values for the allele frequencies were calculated as described in http://awarnach.mathstat.dal.ca/∼joeb/biol3046/PDFs/PopGen1_HWE.pdf. Expected genotype numbers were calculated, and chi-square (χ2) deviation between the observed and expected numbers was calculated for each genotype. A χ2 value greater than 3.841 at one degree of freedom (d.f.) and α = 0.05 is considered significant (p < 0.05). The online calculator https://www.socscistatistics.com/pvalues/chidistribution.aspx was used to generate a p-value from the cumulative χ2 score.
Results
CYP2D6 Allele Frequencies
All alleles detected were counted and calculated for their respective frequencies (Table 1). Nine alleles were detected (CYP2D6*2, *4, *10, *17, *29, *40, *41, *45, and *100). In addition, we detected CYP2D6*5 gene deletions, gene duplications (CYP2D6*1x2, *2x2, *4x2, and *35x2), and alleles with tandem arrangements (CYP2D6*36+*10 and *36x2+*10). The most common nonfunctional allele was CYP2D6*4 (2.13%), followed by *5 (1.66%) and the *4x2 gene duplication (1.42%). Among the decreased function alleles, those with an AS of 0.25 (CYP2D6*10, 17.06%; *36+*10, 12.09%) were more common compared to those with an AS of 0.5 (CYP2D6*17, 10.90%; *29, 6.64%; *41, 3.55%). Alleles found in the Malagasy population and their frequencies are summarized in Table 1.
CYP2D6 Genotype Frequencies and Predicted Phenotypes
Forty-one genotypes were inferred, with frequencies ranging from 0.47% (1/211) to 14.22% (30/211) (Table 2). Six genotypes (CYP2D6*1/*1, *1/*2, *1/*10, *1/*17, *1/*29, and *1/*36+*10) were present at frequencies >5%, with CYP2D6*1/*10 being the most common (14.22%). Among the 41 observed genotypes, two (4.88%) were predicted to be UM (AS, 3.0), 18 (43.9%) NM (AS range, 1.25–2.25), and 21 (51.22%) IM (AS range, 0.25–1.0). No PM subjects were identified. Based on the low frequency of nonfunctional alleles observed in our population sample (24 of 422 alleles, or 5.67%), about 0.32% of the population would be expected to be PMs.
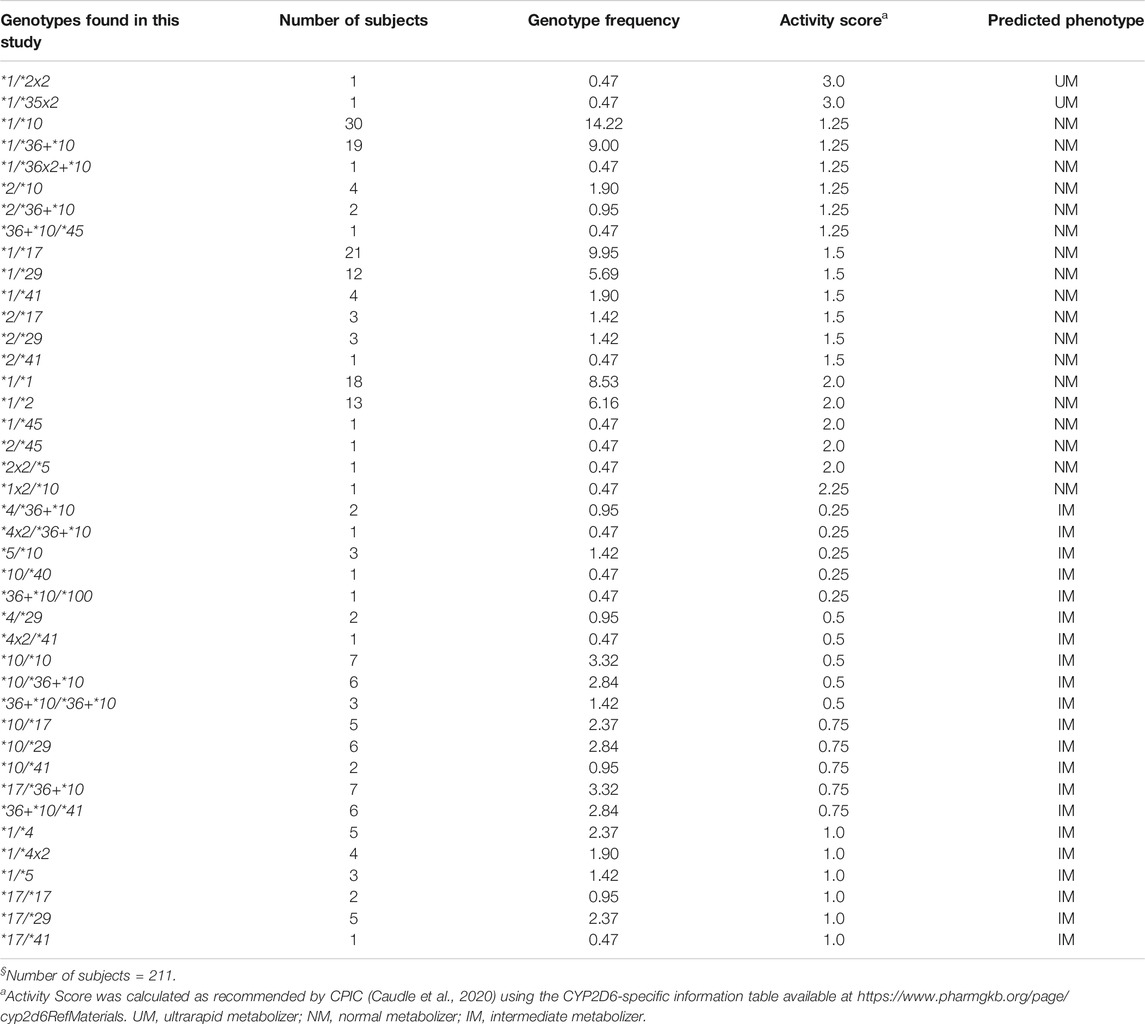
TABLE 2. CYP2D6 genotype frequencies and predicted phenotypes in the Madagascar study population§.
All six genotypes with frequencies >5% were predicted to be NM, and they were distributed equally among three AS groups [1.25, 1.5, and 2.0 (n = 2 each)]. Among the genotypes predicted to be NM, those with an AS of 1.25 were more common [six genotypes among 57 samples (27%)] compared to those with an AS of 1.5 [six genotypes among 44 samples (20.85%)] or 2.0 [five genotypes among 34 samples (16.11%)]. The distribution of the genotypes, predicted to be IM, according to their AS was as follows: AS 0.25, five genotypes among eight samples (3.79%); AS 0.5, five genotypes among 19 samples (9.01%); AS 0.75, five genotypes among 26 samples (12.32%); and AS 1.0, six genotypes among 20 samples (9.48%). Among those with an AS of 0.25 and 0.5, the nonfunctional CYP2D6*4, *4x2, and *5 alleles were present in a total of nine samples (33.33%). All genotypes, their frequencies, and translation via AS into phenotype are summarized in Table 2.
Chi-Square Statistic
A χ2 test-based comparison between the observed and expected genotype numbers for all 41 genotypes (Supplementary Table S2) showed that the total expected genotype number (n = 189) was significantly lower than the total observed genotype number (n = 211) [cumulative χ2 = 84.39, 23 d.f. (41−18; 1 d.f. for having a finite sample, and 17 d.f. for the total number of alleles), p < 0.00001]. The observed and expected genotype numbers did not differ from each other for 36 genotypes including the six genotypes with frequencies >5%. Five genotypes, four observed in one subject each, contributed to the overall deficit in the expected genotype number. These five genotypes are highlighted in Supplementary Table S2.
Discussion
CYP2D6 is one of the Tier 1 Very Important Pharmacogenes with extensive variation (https://www.pharmgkb.org/vip/PA166170264/overview). CYP2D6 allele frequencies differ considerably among ethnically distinct populations (Gaedigk et al., 2017). A comprehensive summary of allele frequencies across populations including source citations can be found on the PharmGKB website (https://www.pharmgkb.org/page/cyp2d6RefMaterials). Based on the latter, CYP2D6*4 has the highest frequency of all nonfunctional alleles in the European group (15–20%; elsewhere, 5–12%), whereas CYP2D6*5, observed at frequencies of 3–6% in most populations, occurs more frequently in South African populations. The CYP2D6*10 decreased function allele is the most common variant allele in the East Asian group (frequencies 9–64%; elsewhere, 2–8%), whereas CYP2D6*17 and *29 are more prominent in the Sub-Saharan African and African American/Afro-Caribbean groups, as compared to others (12–20% and 6–12%, respectively; elsewhere, 1–3% and 1–2%, respectively). CYP2D6*41 allele frequency is higher in Middle Eastern populations than in other ethnic groups (14–18%; elsewhere, 1–10%). Some alleles (e.g., CYP2D6*2, *4, or *5) are found at variable frequencies in almost every population studied, whereas others have only been found in a few populations to date (e.g., CYP2D6*100 and *101 in Trinidadians of Indian ancestry). Finally, populations including South African, Caribbean, and others with diverse founding populations and admixture often reveal unique allele frequency patterns (Nofziger et al., 2020).
In the Malagasy population, although some allele frequencies were comparable to those found in other populations, allele and genotype frequency distribution patterns were rather unique. This is particularly true for CYP2D6*10, *17, and *29. The CYP2D6*10 allele frequency (17.06%) is considerable higher compared to Europeans or Africans, although not quite as high as those observed across East Asia. In contrast, CYP2D6*17 and *29 allele frequencies (10.90 and 6.64%, respectively) are more similar to those in African than in other populations, albeit not as high as in some African populations. Of note was the high frequency (12.09%) of the CYP2D6*36+*10 tandem rearrangement, which is typically only seen in East Asians (16–30%). These predominantly East Asian or African alleles were observed to come together as CYP2D6*10/*17 (2.37%), CYP2D6*10/*29 (2.84%), and CYP2D6*17/*36+*10 (3.32%) genotypes. Finding these allelic variants at notable frequencies is in full agreement with the archaeological and other genomic evidence that Malagasy populations originate from Asian and African populations and constitute a unique, heavily admixed population. Furthermore, the discovery of a subject carrying the rare CYP2D6*100 allele underscores the need to thoroughly characterize populations with a high level of admixture.
Vivax malaria relapse following PQ treatment has predominantly been observed in patients carrying decreased function alleles such as CYP2D6*10, *36, and *41, and nonfunctional alleles such as CYP2D6*4 and *5 in various combinations giving rise to IM and PM phenotypes (Bennett et al., 2013; Ingram et al., 2014; Baird et al., 2018; Dijanic et al., 2018; He et al., 2019; Martin Ramirez et al., 2020; Mat Salleh et al., 2020; Silvino et al., 2020). In most cases where there was drug failure, the following genotypes were observed: CYP2D6*4/*41 (Bennett et al., 2013), *5/*41 (Ingram et al., 2014), *5/*10, *10/*10, and *10/*41 (Baird et al., 2018; Mat Salleh et al., 2020), *1/*4 (Martin Ramirez et al., 2020), and *2/*36 (He et al., 2019), all predicting IM status. In addition, there were two genotypes, CYP2D6*5/*6 (Bennett et al., 2013) and *4/*5 (Baird et al., 2018; Dijanic et al., 2018) that were reported to completely abolish enzyme activity and give rise to PM status. Interestingly, some studies have shown that relapses also occurred in patients with CYP2D6*1/*10 and *2/*10 (Baird et al., 2018) genotypes, predicting NM status. However, subjects in these studies (Bennett et al., 2013; Ingram et al., 2014; Baird et al., 2018; Dijanic et al., 2018; He et al., 2019; Martin Ramirez et al., 2020; Mat Salleh et al., 2020; Silvino et al., 2020) may harbor rare or novel alleles that may not have been detected by screening for the common allelic variants only. Similar results have been reported when the AS system (activity value >1.0, normal function; activity value ≤1.0, decreased function), instead of genotypes and/or predicted phenotypes, was considered to determine the odds or risk of relapse; AS ≤ 1.0 were significantly associated with higher odds or risk of relapse (Baird et al., 2018; Brasil et al., 2018; Silvino et al., 2020). In this current Malagasy sample set, the decreased function alleles were highly prevalent at 50.48%, whereas nonfunctional alleles were noticeably less common at 5.67%. Given these frequencies, 34.6% of the individuals are predicted to be IM with an AS ≤ 1.0, whereas PMs are likely rare at <1%. Therefore, the Malagasy population presents a unique opportunity to investigate PQ metabolism and therapeutic efficacy more systematically with respect to CYP2D6 genotypes. This is especially relevant considering that a new policy to start administration of PQ may soon be executed Madagascar NMCP (National Malaria Control Programme of Madagascar, 2017).
Given the importance of CYP2D6-mediated metabolism for PQ efficacy, the precise role that this enzyme plays in PQ metabolism and pharmacokinetics is now being studied in vivo (Tekwani et al., 2015; Avula et al., 2018). Although still limited, there is recent information on the consequences associated with differential CYP2D6 metabolism status of PQ (Goncalves et al., 2017; Ariffin et al., 2019; Spring et al., 2019). A pharmacokinetic study conducted in 14-year old African children showed that PM children (AS = 0.0) had higher levels of PQ, as compared to IM (AS = 0.5, 1.0), NM (AS = 1.5, 2.0), and UM (AS = 3.0) children, strongly suggesting compromised CYP2D6-mediated PQ metabolism (Goncalves et al., 2017). Another study investigated the effect of the 100C>T SNP on PQ metabolism in healthy volunteers from Malaysia and showed that individuals being heterozygous or homozygous for the T allele had less PQ biotransformed into an active metabolite (Ariffin et al., 2019). However, one limitation of this study was that only one SNP (100C>T) was tested as a key marker for CYP2D6*10. As this SNP is also present on a number of other alleles, including the nonfunctional CYP2D6*4 allele (reported to occur at ∼3% frequency in Malay populations, https://www.pharmgkb.org/page/cyp2d6RefMaterials), the CYP2D6*10 frequency is likely over-estimated, and the contribution of other allelic variants that have been reported in Malay populations (Teh et al., 2001; Chin et al., 2016; Goh et al., 2017) was not accounted for. A more comprehensive study assessed the impact of CYP2D6 metabolizer status on the plasma levels of suspected active phenolic metabolites of PQ (Spring et al., 2019). In this United States-based pharmacokinetic study, volunteers were characterized as genotypic NM (AS = 1.5, 2.0), IM (AS = 0.25, 0.5, 1.0), and PM (AS = 0.0). Consistent with the body of literature, data further corroborated that PQ metabolism was decreased in IM and PM individuals when compared to NM individuals (Spring et al., 2019). Collectively, these findings (Goncalves et al., 2017; Ariffin et al., 2019; Spring et al., 2019) provide an encouraging basis for performing studies in vivax-endemic areas, such as Madagascar, to further unravel the pathways of PQ metabolism and characterize the impact of variable CYP2D6 activity on P. vivax treatment outcomes. Furthermore, by comparing PQ metabolite profiles of subjects with different genotypes, one may be able to assess which CYP2D6 allelic variants impact the production of oxidative metabolites posited to be necessary for radical cure of P. vivax infection.
Being a unique population, Madagascar provides an opportunity to gain new insights into PQ metabolism and efficacy. First, in the present study, we tested for a discrete set of CYP2D6 alleles, selected out of over 130 alleles that have been defined to date (https://www.pharmgkb.org/page/cyp2d6RefMaterials). It is likely that some of the individuals in our study population may have undetected novel or rare alleles, including structural variants, which may or may not be functionally relevant. This limitation may influence the observed frequency of CYP2D6 alleles, especially the normal function CYP2D6*1 allele, which is assigned if no other SNPs are identified. Other alleles may default to CYP2D6*2, or other, however, depending whether they are on a CYP2D6*1-like or *2-like backbone. We refer to Nofziger et al. (Nofziger et al., 2020) for additional information and example of alleles defaulting to CYP2D6*10 assignment. Clearly, sequencing-based approaches will better identify novel or rare alleles than genotype panels. Second, variation in other drug-metabolizing enzymes, drug transporters and/or drug targets may contribute to PQ metabolism and response, and contribute overall to therapeutic efficacy (Li et al., 2003; Silvino et al., 2016; Sortica et al., 2017; Ariffin et al., 2019; Chamnanphon et al., 2020). An early in vitro study identified hepatic CYP1A2 and CYP2D6 as those responsible for the metabolism of PQ (Li et al., 2003). Other reports indicate that polymorphically expressed monoamine oxidase A (Ariffin et al., 2019), CYP2C8 (Silvino et al., 2016), CYP2C19 (Ariffin et al., 2019; Chamnanphon et al., 2020), UDP-glucuronosyltransferase 2B7 (Chamnanphon et al., 2020), ATP-binding cassette transporter G2 (Chamnanphon et al., 2020), and solute carrier organic anion transporters 1A2, 1B1, and 2B1 (Sortica et al., 2017) may also contribute to PQ metabolism, and thus play a role in variable treatment outcomes for vivax malaria. In addition, in a recent genome-wide association study, two signals (rs62103056 on chromosome 18 and a 30-kb intergenic region on chromosome 12) were significantly associated with tafenoquine efficacy in patients with P. vivax malaria (St Jean et al., 2020). Biological relevance and replication of these findings are, however, needed. It is therefore possible that additional variation in CYP2D6 and/or potential “modifiers” elsewhere in the genome may affect PQ metabolism. Untangling not only the complex routes of PQ metabolism, but also the contribution of genomic variation on metabolite formation, especially those that are active against P. vivax stages, are essential to eventually develop optimized PQ dosing strategies. Finally, another open question is substrate-specific allele function (Marcath et al., 2019). In other words, it is unknown whether, and if so to what extent, PQ is metabolized in a substrate-specific manner especially by alleles categorized as decreased function. This concern is highly relevant given the results of an in vitro study conducted by Saito et al. (Saito et al., 2018). In this study, PQ was used as a substrate and 5-hydroxylation was used as a measure of CYP2D6 activity. CYP2D6*17 retained 42% of activity compared to the CYP2D6*1 reference, confirming the decreased function status of this allelic variant (and value of 0.5 used to calculate AS). There was no 5-hydroxylation activity detected for CYP2D6*36 and several other nonfunctional alleles, confirming their no function status (and value of 0 used for AS calculation). Interestingly, no 5-hydroxylation activity was detected for CYP2D6*10 and CYP2D6*29. The absence of detectable in vitro activity for these alleles is especially concerning, given that both are relatively common in many populations plagued by malaria, including the Malagasy. Thus, future studies need to address whether the CPIC recommended method of translating AS into phenotype is appropriate to predict CYP2D6 activity for PQ treatment.
In conclusion, although allele and genotype frequencies need to be validated in a larger population sample, this snapshot study is highly informative as it revealed the presence of numerous decreased function alleles and predicts that a substantial proportion of this population has decreased CYP2D6 activity. In addition, this study provides new insight regarding the genetic admixture of CYP2D6 in a portion of the Malagasy population. Consistent with the historical peopling of Madagascar, the alleles associated with African and East Asian/Indonesian populations are among those most highly prevalent. Finally, current knowledge provides fresh insights about P. vivax elimination challenges for the global population at risk, and enables future studies to address those challenges with the goal to optimize PQ treatment across human genetic diversity. Such efforts would not only benefit successful P. vivax treatment, and eventual elimination, in Madagascar but all populations afflicted by this parasite.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the University Hospitals of Cleveland Institutional Review Board (#09–13-01), the Division of Microbiology and Infectious Diseases/NIAID/National Institutes of Health (NIH) (#13–0067), and the Madagascar Ministry of Health Ethics Committee (#099). Written informed consent was obtained from all subjects, or subject guardians, prior to enrollment. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
RM and PZ conceived the idea. RH, TR, and AR supervised the fieldwork in Madagascar. RM and AG performed the genotyping and analyzed the data, and wrote the manuscript. RH, TR, AR, and PZ edited the manuscript. All authors approved the final version.
Funding
This study was supported by grants from the National Institutes of Health, USA (R01AI097366 and 1R01AI148469–01 to P.A.Z.).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
RM dedicates this paper to Carolyn Myers, who just turned 80! Her life dedicated to Pediatric Pharmacology research, service to mankind, and spirituality is exemplary and infectious.
We thank all study participants, local health officials, field doctors, and project technicians for their participation and support. We also thank Erin Boone and Karim Pirani for technical assistance and analysis of genotype data, and Scott Williams for critically reading the final version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.654054/full#supplementary-material.
References
Adams, J. H., and Mueller, I. (2017). The biology of Plasmodium vivax. Cold Spring Harb Perspect. Med. 7 (9). doi:10.1101/cshperspect.a025585
Andrianaranjaka, V., Lin, J. T., Golden, C., Juliano, J. J., and Randrianarivelojosia, M. (2013). Activation of minority-variant Plasmodium vivax hypnozoites following artesunate + amodiaquine treatment in a 23-year old man with relapsing malaria in Antananarivo, Madagascar. Malar. J. 12, 177. doi:10.1186/1475-2875-12-177
Ariffin, N. M., Islahudin, F., Kumolosasi, E., and Makmor-Bakry, M. (2019). Effects of MAO-A and CYP450 on primaquine metabolism in healthy volunteers. Parasitol. Res. 118 (3), 1011–1018. doi:10.1007/s00436-019-06210-3
Avula, B., Tekwani, B. L., Chaurasiya, N. D., Fasinu, P., Dhammika Nanayakkara, N. P., Bhandara Herath, H. M. T., et al. (2018). Metabolism of primaquine in normal human volunteers: investigation of phase I and phase II metabolites from plasma and urine using ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Malar. J. 17 (1), 294. doi:10.1186/s12936-018-2433-z
Baird, J. K. (2019). 8-Aminoquinoline therapy for latent malaria. Clin. Microbiol. Rev. 32 (4). doi:10.1128/cmr.00011-19
Baird, J. K., Battle, K. E., and Howes, R. E. (2018). Primaquine ineligibility in anti-relapse therapy of Plasmodium vivax malaria: the problem of G6PD deficiency and cytochrome P-450 2D6 polymorphisms. Malar. J. 17 (1), 42. doi:10.1186/s12936-018-2190-z
Baird, J. K., Louisa, M., Noviyanti, R., Ekawati, L., Elyazar, I., Subekti, D., et al. (2018). Association of impaired cytochrome P450 2D6 activity genotype and phenotype with therapeutic efficacy of primaquine treatment for latent Plasmodium vivax malaria. JAMA Netw. Open 1 (4), e181449. doi:10.1001/jamanetworkopen.2018.1449
Baird, J. K. (2018). Tafenoquine for travelers' malaria: evidence, rationale and recommendations. J. Trav. Med 25 (1). doi:10.1093/jtm/tay110
Baird, J. K. (2013). Evidence and implications of mortality associated with acute Plasmodium vivax malaria. Clin. Microbiol. Rev. 26 (1), 36–57. doi:10.1128/cmr.00074-12
Baird, J. K. (2009). Resistance to therapies for infection by Plasmodium vivax. Cmr 22 (3), 508–534. doi:10.1128/cmr.00008-09
Battle, K. E., Karhunen, M. S., Bhatt, S., Gething, P. W., Howes, R. E., Golding, N., et al. (2014). Geographical variation in Plasmodium vivax relapse. Malar. J. 13, 144. doi:10.1186/1475-2875-13-144
Battle, K. E., Gething, P. W., Elyazar, I. R. F., Moyes, C. L., Sinka, M. E., Howes, R. E., et al. (2012). The global public health significance of Plasmodium vivax. Adv. Parasitol. 80, 1–111. doi:10.1016/b978-0-12-397900-1.00001-3
Battle, K. E., Lucas, T. C. D., Nguyen, M., Howes, R. E., Nandi, A. K., Twohig, K. A., et al. (2019). Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000-17: a spatial and temporal modelling study. The Lancet 394 (10195), 332–343. doi:10.1016/s0140-6736(19)31096-7
Bennett, J. W., Pybus, B. S., Yadava, A., Tosh, D., Sousa, J. C., McCarthy, W. F., et al. (2013). Primaquine failure and cytochrome P-450 2D6 in Plasmodium vivax malaria. N. Engl. J. Med. 369 (14), 1381–1382. doi:10.1056/nejmc1301936
Bourgard, C., Albrecht, L., Kayano, A., Sunnerhagen, P., and Costa, F. T. M. (2018). Plasmodium vivax biology: insights provided by genomics, transcriptomics and proteomics. Front Cel Infect Microbiol 8, 34. doi:10.3389/fcimb.2018.00034
Brasil, L. W., Rodrigues-Soares, F., Santoro, A. B., Almeida, A. C. G., Kuhn, A., Ramasawmy, R., et al. (2018). CYP2D6 activity and the risk of recurrence of Plasmodium vivax malaria in the Brazilian Amazon: a prospective cohort study. Malar. J. 17 (1), 57. doi:10.1186/s12936-017-2139-7
Brucato, N., Kusuma, P., Cox, M. P., Pierron, D., Purnomo, G. A., Adelaar, A., et al. (2016). Malagasy genetic ancestry comes from an historical Malay trading post in Southeast Borneo. Mol. Biol. Evol. 33 (9), 2396–2400. doi:10.1093/molbev/msw117
Camarda, G., Jirawatcharadech, P., Priestley, R. S., Saif, A., March, S., Wong, M. H. L., et al. (2019). Antimalarial activity of primaquine operates via a two-step biochemical relay. Nat. Commun. 10 (1), 3226. doi:10.1038/s41467-019-11239-0
Caudle, K. E., Sangkuhl, K., Whirl‐Carrillo, M., Swen, J. J., Haidar, C. E., Klein, T. E., et al. (2020). Standardizing CYP 2D6 genotype to phenotype translation: consensus recommendations from the clinical Pharmacogenetics implementation Consortium and Dutch Pharmacogenetics working group. Clin. Transl Sci. 13 (1), 116–124. doi:10.1111/cts.12692
Chamnanphon, M., Gaedigk, A., Puangpetch, A., Pasomsub, E., Chantratita, W., Longley, R. J., et al. (2020). Pharmacogene variation in Thai Plasmodium vivax relapse patients treated with a combination of primaquine and chloroquine. Pgpm Vol. 13, 1–12. doi:10.2147/pgpm.s201007
Chin, F. W., Chan, S. C., Abdul Rahman, S., Noor Akmal, S., and Rosli, R. (2016). CYP2D6 genetic polymorphisms and phenotypes in different ethnicities of Malaysian breast cancer patients. Breast J. 22 (1), 54–62. doi:10.1111/tbj.12518
Dijanic, C., Nickerson, J., Shakya, S., Dijanic, A., and Fabbri, M. (2018). Relapsing malaria: a case report of primaquine resistance. Case Rep. Infect. Dis. 2018, 9720823. doi:10.1155/2018/9720823
Fasinu, P. S., Avula, B., Tekwani, B. L., Nanayakkara, N. P., Wang, Y. H., Bandara Herath, H. M., et al. (2016a). Differential kinetic profiles and metabolism of primaquine enantiomers by human hepatocytes. Malar. J. 15, 224. doi:10.1186/s12936-016-1270-1
Fasinu, P. S., Tekwani, B. L., Avula, B., Chaurasiya, N. D., Nanayakkara, N. P., Wang, Y. H., et al. (2016b). Pathway-specific inhibition of primaquine metabolism by chloroquine/quinine. Malar. J. 15, 466. doi:10.1186/s12936-016-1509-x
Fasinu, P. S., Tekwani, B. L., Nanayakkara, N., Avula, B., Herath, H., Wang, Y.-H., et al. (2014). Enantioselective metabolism of primaquine by human CYP2D6. Malar. J. 13, 507. doi:10.1186/1475-2875-13-507
Gaedigk, A., Jaime, L. K., Bertino, J. S., Berard, A., Pratt, V. M., Bradfordand, L. D., et al. (2010). Identification of novel CYP2D7-2D6 hybrids: non-functional and functional variants. Front. Pharmacol. 1, 121. doi:10.3389/fphar.2010.00121
Gaedigk, A., Ndjountché, L., Divakaran, K., Dianne Bradford, L., Zineh, I., Oberlander, T. F., et al. (2007). Cytochrome P4502D6 (CYP2D6) gene locus heterogeneity: characterization of gene duplication events. Clin. Pharmacol. Ther. 81 (2), 242–251. doi:10.1038/sj.clpt.6100033
Gaedigk, A., Sangkuhl, K., Whirl-Carrillo, M., Klein, T., and Leeder, J. S. (2017). Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med. 19 (1), 69–76. doi:10.1038/gim.2016.80
Gaedigk, A., Turner, A., Everts, R. E., Scott, S. A., Aggarwal, P., Broeckel, U., et al. (2019). Characterization of reference materials for genetic testing of CYP2D6 alleles. J. Mol. Diagn. 21 (6), 1034–1052. doi:10.1016/j.jmoldx.2019.06.007
Gaedigk, A., Twist, G. P., and Leeder, J. S. (2012). CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics 13 (1), 91–111. doi:10.2217/pgs.11.135
Goh, L. L., Lim, C. W., Sim, W. C., Toh, L. X., and Leong, K. P. (2017). Analysis of genetic variation in CYP450 genes for clinical implementation. PLoS One 12 (1), e0169233. doi:10.1371/journal.pone.0169233
Goncalves, B. P., Pett, H., Tiono, A. B., Murry, D., Sirima, S. B., Niemi, M., et al. (2017). Age, weight, and CYP2D6 genotype are major determinants of primaquine pharmacokinetics in African children. Antimicrob. Agents Chemother. 61 (5). doi:10.1128/aac.02590-16
He, X., Pan, M., Zeng, W., Zou, C., Pi, L., Qin, Y., et al. (2019). Multiple relapses of Plasmodium vivax malaria acquired from West Africa and association with poor metabolizer CYP2D6 variant: a case report. BMC Infect. Dis. 19 (1), 704. doi:10.1186/s12879-019-4357-9
Howes, R. E., Chan, E. R., Rakotomanga, T. A., Schulte, S., Gibson, J., Zikursh, M., et al. (2017). Prevalence and genetic variants of G6PD deficiency among two Malagasy populations living in Plasmodium vivax-endemic areas. Malar. J. 16 (1), 139. doi:10.1186/s12936-017-1771-6
Howes, R. E., Patil, A. P., Piel, F. B., Nyangiri, O. A., Kabaria, C. W., Gething, P. W., et al. (2011). The global distribution of the Duffy blood group. Nat. Commun. 2, 266. doi:10.1038/ncomms1265
Howes, R. E., Reiner, R. C., Battle, K. E., Longbottom, J., Mappin, B., Ordanovich, D., et al. (2015). Plasmodium vivax transmission in Africa. Plos Negl. Trop. Dis. 9 (11), e0004222. doi:10.1371/journal.pntd.0004222
Howes, R. E., Battle, K. E., Mendis, K. N., Smith, D. L., Cibulskis, R. E., Baird, J. K., et al. (2016). Global epidemiology of Plasmodium vivax. Am. J. Trop. Med. Hyg. 95 (6 Suppl. l), 15–34. doi:10.4269/ajtmh.16-0141
Howes, R. E., Zikursh, M., Cramer, E. Y., Ramiranirina, B., Ratsimbasoa, A., Kang, S. Y., et al. (2018). Risk factors for malaria infection in Central Madagascar: insights from a cross-sectional population survey. Am. J. Trop. Med. Hyg. 99 (4), 995–1002. doi:10.4269/ajtmh.18-0417
Imwong, M., Snounou, G., Pukrittayakamee, S., Tanomsing, N., Kim, J. R., Nandy, A., et al. (2007). Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J. Infect. Dis. 195 (7), 927–933. doi:10.1086/512241
Ingram, R. J. H., Crenna-Darusallam, C., Soebianto, S., Noviyanti, R., and Baird, J. (2014). The clinical and public health problem of relapse despite primaquine therapy: case review of repeated relapses of Plasmodium vivax acquired in Papua New Guinea. Malar. J. 13, 488. doi:10.1186/1475-2875-13-488
John, G. K., Douglas, N. M., von Seidlein, L., Nosten, F., Baird, J., White, N. J., et al. (2012). Primaquine radical cure of Plasmodium vivax: a critical review of the literature. Malar. J. 11, 280. doi:10.1186/1475-2875-11-280
Kusuma, P., Brucato, N., Cox, M. P., Pierron, D., Razafindrazaka, H., Adelaar, A., et al. (2016). Contrasting linguistic and genetic origins of the Asian source populations of Malagasy. Sci. Rep. 6, 26066. doi:10.1038/srep26066
Kusuma, P., Cox, M. P., Pierron, D., Razafindrazaka, H., Brucato, N., Tonasso, L., et al. (2015). Mitochondrial DNA and the Y chromosome suggest the settlement of Madagascar by Indonesian sea nomad populations. BMC Genomics 16, 191. doi:10.1186/s12864-015-1394-7
Lacerda, M. V. G., Llanos-Cuentas, A., Krudsood, S., Lon, C., Saunders, D. L., Mohammed, R., et al. (2019). Single-dose tafenoquine to prevent relapse of Plasmodium vivax malaria. N. Engl. J. Med. 380 (3), 215–228. doi:10.1056/NEJMoa1710775
Li, X. Q., Bjorkman, A., Andersson, T. B., Gustafsson, L. L., and Masimirembwa, C. M. (2003). Identification of human cytochrome P(450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur. J. Clin. Pharmacol. 59 (5-6), 429–442. doi:10.1007/s00228-003-0636-9
Marcath, L. A., Pasternak, A. L., and Hertz, D. L. (2019). Challenges to assess substrate-dependent allelic effects in CYP450 enzymes and the potential clinical implications. Pharmacogenomics J. 19 (6), 501–515. doi:10.1038/s41397-019-0105-1
Marcsisin, S. R., Sousa, J. C., Reichard, G. A., Caridha, D., Zeng, Q., Roncal, N., et al. (2014). Tafenoquine and NPC-1161B require CYP 2D metabolism for anti-malarial activity: implications for the 8-aminoquinoline class of anti-malarial compounds. Malar. J. 13, 2. doi:10.1186/1475-2875-13-2
Martin Ramirez, A., Lombardia Gonzalez, C., Soler Maniega, T., Gutierrez Liarte, A., Domingo Garcia, D., Lanza Suarez, M., et al. (2020). Several Plasmodium vivax relapses after correct primaquine treatment in a patient with impaired cytochrome P450 2D6 function. Malar. J. 19 (1), 259. doi:10.1186/s12936-020-03326-1
Mat Salleh, N. H., Rahman, M. F. A., Samsusah, S., De Silva, J. R., Ng, D. C.-E., Ghozali, A. H., et al. (2020). Case report: recurrence of Plasmodium vivax malaria due to defective cytochrome P450 2D6 function in Pos Lenjang, Pahang, Malaysia. Trans. R. Soc. Trop. Med. Hyg. 114 (9), 700–703. doi:10.1093/trstmh/traa042
Menard, D., Barnadas, C., Bouchier, C., Henry-Halldin, C., Gray, L. R., Ratsimbasoa, A., et al. (2010). Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl. Acad. Sci. 107 (13), 5967–5971. doi:10.1073/pnas.0912496107
Miller, L. H., Mason, S. J., Clyde, D. F., and McGinniss, M. H. (1976). The resistance factor to Plasmodium vivax in blacks. N. Engl. J. Med. 295 (6), 302–304. doi:10.1056/nejm197608052950602
National Malaria Control Programme of Madagascar (2017). National strategic plan for malaria control in Madagascar 2018-2022. Progressive malaria elimination from Madagascar.
Nofziger, C., Turner, A. J., Sangkuhl, K., Whirl‐Carrillo, M., Agúndez, J. A. G., Black, J. L., et al. (2020). PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 107 (1), 154–170. doi:10.1002/cpt.1643
Pierron, D., Heiske, M., Razafindrazaka, H., Pereda-Loth, V., Sanchez, J., Alva, O., et al. (2018). Strong selection during the last millennium for African ancestry in the admixed population of Madagascar. Nat. Commun. 9 (1), 932. doi:10.1038/s41467-018-03342-5
Pierron, D., Heiske, M., Razafindrazaka, H., Rakoto, I., Rabetokotany, N., Ravololomanga, B., et al. (2017). Genomic landscape of human diversity across Madagascar. Proc. Natl. Acad. Sci. USA 114 (32), E6498–E6506. doi:10.1073/pnas.1704906114
Potter, B. M. J., Xie, L. H., Vuong, C., Zhang, J., Zhang, P., Duan, D., et al. (2015). Differential CYP 2D6 metabolism alters primaquine pharmacokinetics. Antimicrob. Agents Chemother. 59 (4), 2380–2387. doi:10.1128/aac.00015-15
Pybus, B. S., Marcsisin, S. R., Jin, X., Deye, G., Sousa, J. C., Li, Q., et al. (2013). The metabolism of primaquine to its active metabolite is dependent on CYP 2D6. Malar. J. 12, 212. doi:10.1186/1475-2875-12-212
Saito, T., Gutiérrez Rico, E. M., Kikuchi, A., Kaneko, A., Kumondai, M., Akai, F., et al. (2018). Functional characterization of 50 CYP2D6 allelic variants by assessing primaquine 5-hydroxylation. Drug Metab. Pharmacokinet. 33 (6), 250–257. doi:10.1016/j.dmpk.2018.08.004
Saravanakumar, A., Sadighi, A., Ryu, R., and Akhlaghi, F. (2019). Physicochemical properties, biotransformation, and transport pathways of established and newly approved medications: a systematic review of the top 200 most prescribed drugs vs. the FDA-approved drugs between 2005 and 2016. Clin. Pharmacokinet. 58 (10), 1281–1294. doi:10.1007/s40262-019-00750-8
Silvino, A. C., Costa, G. L., Araujo, F. C., Ascher, D. B., Pires, D. E., Fontes, C. J., et al. (2016). Variation in human cytochrome P-450 drug-metabolism genes: a gateway to the understanding of Plasmodium vivax relapses. PLoS One 11 (7), e0160172. doi:10.1371/journal.pone.0160172
Silvino, A. C. R., Kano, F. S., Costa, M. A., Fontes, C. J. F., Soares, I. S., de Brito, C. F. A., et al. (2020). Novel insights into Plasmodium vivax therapeutic failure: CYP2D6 activity and time of exposure to malaria modulate the risk of recurrence. Antimicrob. Agents Chemother. 64 (5). doi:10.1128/aac.02056-19
Sortica, V. A., Lindenau, J. D., Cunha, M. G., O Ohnishi, M. D., R Ventura, Â. K., Santos, S. E., et al. (2017). SLCO1A2, SLCO1B1 and SLCO2B1 polymorphisms influences chloroquine and primaquine treatment in Plasmodium vivax malaria. Pharmacogenomics 18 (15), 1393–1400. doi:10.2217/pgs-2017-0077
Spring, M. D., Sousa, J. C., Li, Q., Darko, C. A., Morrison, M. N., Marcsisin, S. R., et al. (2019). Determination of cytochrome P450 isoenzyme 2D6 (CYP2D6) genotypes and pharmacogenomic impact on primaquine metabolism in an active-duty US military population. J. Infect. Dis. 220 (11), 1761–1770. doi:10.1093/infdis/jiz386
St Jean, P. L., Xue, Z., Carter, N., Koh, G. C., Duparc, S., Taylor, M., et al. (2016). Tafenoquine treatment of Plasmodium vivax malaria: suggestive evidence that CYP2D6 reduced metabolism is not associated with relapse in the Phase 2b DETECTIVE trial. Malar. J. 15, 97. doi:10.1186/s12936-016-1145-5
St Jean, P. L., Koh, G. C. K. W., Breton, J. J., Espino, F. E. J., Hien, T. T., Krudsood, S., et al. (2020). Pharmacogenetic assessment of tafenoquine efficacy in patients with Plasmodium vivax malaria. Pharmacogenet Genomics 30 (7), 161–165. doi:10.1097/fpc.0000000000000407
Taylor, C., Crosby, I., Yip, V., Maguire, P., Pirmohamed, M., and Turner, R. M. (2020). A review of the important role of CYP2D6 in pharmacogenomics. Genes (Basel). 11 (11).1295 doi:10.3390/genes11111295
Teh, L. K., Ismail, R., Yusoff, R., Hussein, A., Isa, M. N., and Rahman, A. R. A. (2001). Heterogeneity of the CYP2D6 gene among Malays in Malaysia. J. Clin. Pharm. Ther. 26 (3), 205–211. doi:10.1046/j.1365-2710.2001.00347.x
Tekwani, B. L., Avula, B., Sahu, R., Chaurasiya, N. D., Khan, S. I., Jain, S., et al. (2015). Enantioselective pharmacokinetics of primaquine in healthy human volunteers. Drug Metab. Dispos 43 (4), 571–577. doi:10.1124/dmd.114.061127
Twohig, K. A., Pfeffer, D. A., Baird, J. K., Price, R. N., Zimmerman, P. A., Hay, S. I., et al. (2019). Growing evidence of Plasmodium vivax across malaria-endemic Africa. Plos Negl. Trop. Dis. 13 (1), e0007140. doi:10.1371/journal.pntd.0007140
Vuong, C., Xie, L. H., Potter, B. M. J., Zhang, J., Zhang, P., Duan, D., et al. (2015). Differential cytochrome P450 2D metabolism alters tafenoquine pharmacokinetics. Antimicrob. Agents Chemother. 59 (7), 3864–3869. doi:10.1128/aac.00343-15
White, N. J. (2011). Determinants of relapse periodicity in Plasmodium vivax malaria. Malar. J. 10, 297. doi:10.1186/1475-2875-10-297
White, N. J., and Imwong, M. (2012). Relapse. Adv. Parasitol. 80, 113–150. doi:10.1016/b978-0-12-397900-1.00002-5
White, N. J. (2019). Tafenoquine - a radical improvement?. N. Engl. J. Med. 380 (3), 285–286. doi:10.1056/nejme1816383
Willie, N., Mehlotra, R. K., Howes, R. E., Rakotomanga, T. A., Ramboarina, S., Ratsimbasoa, A. C., et al. (2018). Insights into the performance of SD Bioline Malaria Ag P.f/Pan rapid diagnostic test and Plasmodium falciparum histidine-rich protein 2 gene variation in Madagascar. Am. J. Trop. Med. Hyg. 98 (6), 1683–1691. doi:10.4269/ajtmh.17-0845
Keywords: CYP2D6, hypnozoite, Madagascar, malaria treatment, Plasmodium vivax, primaquine
Citation: Mehlotra RK, Gaedigk A, Howes RE, Rakotomanga TA, Ratsimbasoa AC and Zimmerman PA (2021) CYP2D6 Genetic Variation and Its Implication for Vivax Malaria Treatment in Madagascar. Front. Pharmacol. 12:654054. doi: 10.3389/fphar.2021.654054
Received: 15 January 2021; Accepted: 01 March 2021;
Published: 20 April 2021.
Edited by:
Amit V Pandey, University of Bern, SwitzerlandReviewed by:
Kailash C Pandey, National Institute of Malaria Research (ICMR), IndiaJulio Benitez, University of Extremadura, Spain
Copyright © 2021 Mehlotra, Gaedigk, Howes, Rakotomanga, Ratsimbasoa and Zimmerman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajeev K. Mehlotra, cmttQGNhc2UuZWR1; Peter A. Zimmerman, cGF6QGNhc2UuZWR1
†These authors have contributed equally to this work.
 Rajeev K. Mehlotra
Rajeev K. Mehlotra Andrea Gaedigk
Andrea Gaedigk Rosalind E. Howes
Rosalind E. Howes Tovonahary A. Rakotomanga5,6
Tovonahary A. Rakotomanga5,6 Arsene C. Ratsimbasoa
Arsene C. Ratsimbasoa Peter A. Zimmerman
Peter A. Zimmerman