
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 15 April 2021
Sec. Pharmacology of Anti-Cancer Drugs
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.645864
This article is part of the Research Topic Chemo-Radiation-Resistance in Cancer Therapy View all 35 articles
Drug therapy is the primary treatment for patients with advanced cancer. The use of anticancer drugs will inevitably lead to drug resistance, which manifests as tumor recurrence. Overcoming chemoresistance may enable cancer patients to have better therapeutic effects. However, the mechanisms underlying drug resistance are poorly understood. E3 ubiquitin ligases (E3s) are a large class of proteins, and there are over 800 putative functional E3s. E3s play a crucial role in substrate recognition and catalyze the final step of ubiquitin transfer to specific substrate proteins. The diversity of the set of substrates contributes to the diverse functions of E3s, indicating that E3s could be desirable drug targets. The E3s MDM2, FBWX7, and SKP2 have been well studied and have shown a relationship with drug resistance. Strategies targeting E3s to combat drug resistance include interfering with their activators, degrading the E3s themselves and influencing the interaction between E3s and their substrates. Research on E3s has led to the discovery of possible therapeutic methods to overcome the challenging clinical situation imposed by drug resistance. In this article, we summarize the role of E3s in cancer drug resistance from the perspective of drug class.
Cancer is a multifactorial disease and is considered the most severe public health issue worldwide (Siegel et al., 2019). Drug therapy is the main treatment for patients with advanced cancer. The drugs currently used for tumor treatment include platinum drugs, antitumor antibiotics, alkylating agents, hormones, molecular targeting agents, and immunotherapy. The use of tumor drugs will inevitably lead to drug resistance, which manifests as tumor recurrence (Glickman and Sawyers, 2012; Vasan et al., 2019).
Several mechanisms have been found to underlie anticancer drug resistance, including the effects of cancer stem cells (CSCs), epithelial-mesenchymal transition (EMT), and DNA damage repair (DDR) (Gong et al., 2018). Identifying the key molecules in these processes can help us understand the occurrence of drug resistance, and these key molecules play an essential role in predicting and reversing resistance to anticancer drugs. However, the mechanisms have not yet been elucidated.
Protein ubiquitination-based modification can regulate various signal-mediated cell death responses and plays an essential role in the occurrence, development, and outcome of cancer (Nalepa et al., 2006). Ubiquitination refers to the process by which ubiquitin is covalently bound to target proteins under the catalysis of a series of enzymes. The ubiquitination process usually requires the cooperation of the E1 ubiquitin-activating enzymes (E1s), E2 ubiquitin-coupling enzymes (E2s), and E3 ubiquitin ligases (E3s) (Buetow and Huang, 2016). Mechanistically, ubiquitin is activated in an ATP-dependent manner, inducing a thioester bond with an E1. The moieties are then transferred to the active site cysteine of the E2. The E3 binds to both the E2∼Ub thioester and the substrate, catalyzing the transfer of ubiquitin from the active site cysteine of the E2 to the substrate lysine or N terminus (Tatham et al., 2013; Berndsen and Wolberger, 2014).
Ubiquitin chain topology determines the fate of ubiquitylated proteins (Ikeda and Dikic, 2008). Usually, ubiquitin-dependent proteolysis is associated with K48-linked and K11-linked ubiquitin chains. However, activation of signaling pathways is mainly dependent on K63‐linked or M1-linked ubiquitin chains. Ubiquitin-mediated proteolysis is essential for the maintenance of protein homeostasis because it removes misfolded or unwanted proteins. The non-proteasomal ubiquitin-mediated assembly of signaling complexes also plays a pivotal role in several cellular processes, such as autophagy, DNA repair, and endocytosis (Bennett and Harper, 2008; Chen and Sun, 2009; Cappadocia and Lima, 2018). Given these phenomena, it is understandable that the dysregulation of ubiquitination can lead to genetic and epigenetic alterations in cancer.
E3s are critical components in the ubiquitination reaction owing to their strict control of both substrate affinity and specificity (Zheng and Shabek, 2017). E3s are a large class of proteins, and there are over 800 putative functional E3s (Ciechanover, 2015). E3s have been classified into three subgroups: E3s containing really interesting new gene (RING) and UFD2 homology (U-box) domains, the 28-member homologous to E6AP carboxyl terminus (HECT) E3 family, and the 14‐member RING‐between‐RING (RBR) E3 family (Uchida and Kitagawa, 2016). The diversity of the E3 substrates contributes to the diverse functions of E3s (Li et al., 2014), and as a result, E3s are closely related to tumorigenesis because they regulate oncogenes and tumor suppressors (Senft et al., 2018). In addition, the substrate specificity of E3s suggests that they have promise as anticancer drug targets (Wang et al., 2017). Here, for the first time, we summarize the role of E3s in anticancer drug resistance from the perspective of drug class (Figure 1).
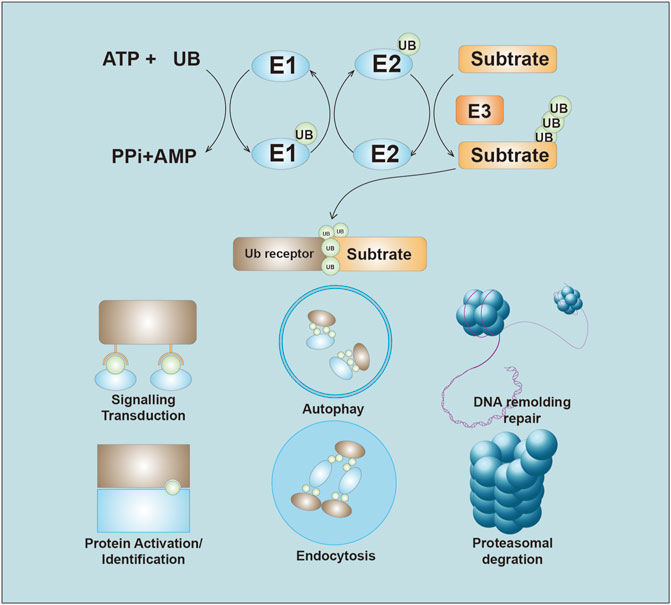
FIGURE 1. Overview of the Ubiquitin system. Ubiquitin is initiate through the thioester bond with E1 in the ATP-depend manner. The activated ubiquitin then be transferred to the E2. E3 ligases recognize and transfer the ubiquitin to substrate. The ubiquitin chains lead to several ending, including proteasome-mediated degradation, ubiquitination signals transduction, autophagy, DNA remolding and repair, Protein identification, and endocytosis.
Platinum drugs are widely used in the treatment of human cancers. The zinc-finger E3s MUL1 and UBR5 was found to be involved in platinum resistance. The E3 UBR5 was amplified and overexpressed in ovarian cancer (OC). Higher UBR5 expression was observed in platinum-resistant OC tumor tissue than in normal tissues. Overexpression of UBR5 induced cisplatin resistance in OC cells both in vivo and in vitro. Moreover, UBR5 knockdown via siRNA partly reversed platinum resistance in OC cells (O'Brien et al., 2008; Bradley et al., 2014). Mitochondrial E3 ubiquitin-protein ligase 1 (MUL1) is an E3 that interacts with and negatively regulates AKT. The degradation of AKT was found to lead to cisplatin sensitivity in OC cells (Lee et al., 2019). A decrease in EMT in cisplatin-resistant nasopharyngeal carcinoma (NPC) cells was observed after upregulation of NEDD4 in cells, suggesting that NEDD4 could be a novel therapeutic target for overcoming drug resistance in NPC (Feng et al., 2017).
Tripartite motif (TRIM) family proteins, most of which have E3 activities, control important cellular processes such as intracellular signaling, innate immunity, transcription, autophagy, and carcinogenesis (Hatakeyama, 2017). TRIM25 expression was identified to be significantly lower in the cisplatin-resistant non-small cell lung carcer (NSCLC) cell line A549 than in control cell lines (Qin et al., 2012). Overexpression of TRIM32 promoted degradation of Abi2, resulting in enhancement of cell growth, transforming activity, and cell motility. Moreover, TRIM32 suppressed the apoptosis induced by cisplatin in the hepatocellular carcinoma (HCC) cell line HEp2. Overexpression of TRIM32 in the HCC cell line also induced resistance to another platinum drug, oxaliplatin (Cui et al., 2016). Increased TRIM11 expression inhibits the apoptosis induced by cisplatin, and TRIM11 functions as an oncogene related to drug resistance both in vivo and in vitro. TRIM11 destabilized Daple in a p62-selective autophagic manner, further upregulating β-catenin expression to induce enhanced expression of ABCC9, which can transport chemotherapeutic drugs (Zhang et al., 2020). Autophagy can be a key mechanism of resistance to chemotherapy (Onorati et al., 2018). TRIM65 was found to be upregulated in NSCLC, and its overexpression promoted NSCLC cell resistance to cisplatin (Li et al., 2016). The inhibition of miR-138–5p attenuated the effects of TRIM65 knockdown on autophagy and cisplatin-induced apoptosis, suggesting that TRIM65 regulates cisplatin resistance in NSCLC by regulating miR-138–5p (Pan et al., 2019). TRIM59 was also found to be overexpressed in cisplatin-resistant A549 cells, and its overexpression in these cells resulted in increased cisplatin resistance. TRIM59 enhanced the ubiquitination of PTEN, a critical upstream regulator of HK2. The regulation of the PTEN/AKT/HK2 pathway induced by TRIM59 might provide insights into overcoming cancer resistance to cisplatin treatment (He and Liu, 2020).
Another type of RING-box-containing E3s, RING finger proteins (RNFs), play a role in platinum resistance. RNF38 was proposed as a biomarker of poor NSCLC prognosis, and its silencing increased the sensitivity of NSCLC cells to cisplatin (Wu et al., 2020). RNF138 was more highly expressed in cisplatin-resistant gastric cancer (GC) cell lines than in normal cell lines and modulated cisplatin resistance in these GC cells (Lu et al., 2018). Pellino family proteins (Pellino-1, 2, and 3) are E3s that contain C-terminal RING-like domains. Pellino-1 overexpression conferred NSCLC cells with resistance to the apoptosis induced by cisplatin or paclitaxel (Jeon et al., 2016).
The F-box-containing family member FBXW7 is one of the four subunits of the SKP1-cullin-F-box (SCF)-E3 complex, which functions in phosphorylation-dependent ubiquitination (Skaar et al., 2014). In NSCLC, FBXW7 upregulation significantly increased cisplatin chemosensitivity and abrogated the mesenchymal phenotype of NSCLC cell lines (Yu et al., 2013). Another report by Guodong X et al. indicated that FBXW7 could interact with Snai1 in NSCLC cell lines and directly degrade its expression, resulting in suppression of cisplatin and sorafenib resistance (Guodong et al., 2018). In colorectal carcinoma (CRC) cells, FBXW7 deficiency induced by mutation or loss can lead to the aberrant phosphorylation of p53 at serine 15 and further promote resistance to oxaliplatin. An understanding of the regulation of phospho-p53 (Ser15) by FBXW7 E3 activity could provide important clues for the clinical targetability of this axis (Perez-Losada et al., 2005; Li et al., 2015). In NPC, upregulation of FBXW7 significantly increased cisplatin-based chemosensitivity (Song et al., 2015). The CRL4 expression level was increased in cisplatin-resistant OC cells. CRL4 knockdown with shRNAs was able to reverse the cisplatin resistance of OC cells (Hu et al., 2019). In CRC, knockdown of CUL4A sensitized parental CRC cells to cisplatin (Englinger et al., 2017). CUL4B destabilized HP1α, a gene that suppresses the open confirmation of chromatin that is important for the DDR and DNA repair. The DDR and DNA repair are believed to be reasons for cisplatin resistance (Kim et al., 2017).
F-box-only proteins are the substrate-recognition component of the SCF-E3 complex. The relatively low level of FBXO22 in A549 cells contributes to an accumulation of CD147 and the cisplatin resistance of the cells (Wu et al., 2017). Additionally, FBXO21 was found to ubiquitinate and destabilized P-glycoprotein (P-gp), resulting in attenuation of multidrug resistance. However, a stem cell marker, CD44, was found to inhibit FBXO21-directed degradation of P-gp and promote multidrug resistance (Ravindranath et al., 2015). Using a microarray, another E3, FBXO32, was newly identified as a negative regulator of EMT in urothelial carcinoma (UC) tumors after they had acquired platinum resistance. FBXO32 dysregulation in platinum-resistant UC cells resulted in elevated expression of the EMT marker snail and decreased expression of E-cadherin (Tanaka et al., 2016).
SKP2, also known as F-box and leucine-rich repeat protein (FBXL1), is a member of the FBXL subfamily of F-box proteins and plays a pivotal role in cell cycle progression and proliferation. Evidence has shown that SKP2 can interact with Akt and facilitate its ubiquitination. K63-mediated AKT ubiquitination can be mediated by Skp2 and can regulate NPC to induce cisplatin resistance (Yu et al., 2019). Overexpression of SKP2 reduced the expression of p27Kip1, cyclin E, and p21Cip1, increased the proportion of S-phase cells, and increased resistance against cisplatin in NSCLC cell lines (Ishii et al., 2004).
E3s targeting P53 play a role in cisplatin resistance. MDM2, an important regulator of P53, controls cisplatin resistance in multiple cancers. MDM2 interacted and destabilized P53 and induced tumor cell resistance to cisplatin (Muscolini et al., 2011; Sheng et al., 2017). The genes encoding molecules that interfere with the interaction between MDM2 and p53 might also lead to drug resistance. Some genes, such as zinc-finger CCHC-type containing 10 (ZCCHC10) and NUMB, inhibit cisplatin resistance by interfering with P53 ubiquitination mediated by MDM2 (Colaluca et al., 2008; Ning et al., 2019). TRAF6, an E3 that controls p53 mitochondrial translocation, was found to be overexpressed in CRC tissues. TRAF6 overexpression negatively correlates with apoptosis and predicts poor response to cisplatin-based chemotherapy and radiotherapy (Zhang et al., 2016). Cisplatin can enhance the FLIP-p53-Itch interaction, inducing FLIP ubiquitination and degradation in a p53-and Itch-dependent manner. These results suggest that the modulation of FLIP content may be an effective strategy for overcoming chemoresistance in OC (Abedini et al., 2008; Abedini et al., 2010).
The U-box domain-containing E3 CHIP has been studied to help understand drug sensitivity. CHIP knockdown increases the proportion of cisplatin-sensitive cells. CHIP can act as an activator of Bcl-2 expression levels to suppress breast cancer (BC) malignant progression (Tsuchiya et al., 2015). In addition to CHIP’s role in BC, Dong‐E Tang et al. revealed that CHIP could ubiquitinate YAP1 at the K280 site by K48‐linked polyubiquitination and also generated a human GC cell line with resistance to cisplatin resistance (Tang et al., 2019). Microtubule-associated serine/threonine kinase 1 (MAST1) mediates cisplatin resistance in human cancers. Pan C et al. used a proteomics screen to identify CHIP-destabilized MAST1. The MAST1 destabilization resulted in hsp90B-induced sensitivity to cisplatin (Pan et al., 2019). ZFP91 knockdown reduced FOXA1 polyubiquitination, which decreased FOXA1 turnover and enhanced cellular sensitivity to cisplatin therapy (Tang et al., 2020). E3s and their corresponding substrates are summarized in Figure 2.
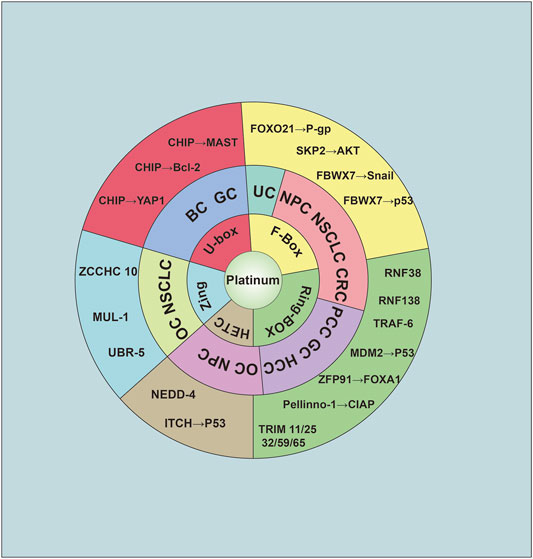
FIGURE 2. The relationship of E3 ligase and platinum resistance. The E3 was cataloged as five group: U-box, F-box, Ring-Box, HETC, and Zing finger. Abbreviation was as follow: BC: Breast cancer; GC: Gastric cancer; UC, urothelial carcinoma; NPC, nasopharyngeal carcinoma; NSCLC, non-small cell lung cancer; CRC, colorectal carcinoma; PCC, pancreatic carcinoma; HCC, hepatocellular carcinoma; OC, ovarian cancer.
Paclitaxel is an M-phase-specific plant drug initially derived from the Pacific yew and has become the first member of the taxane family to be used in cancer chemotherapy (Bernabeu et al., 2017). In a single-cell RNA sequencing analysis, protein ubiquitination was identified as the most differentially regulated pathway in docetaxel-resistant prostate cancer (PC) cells (Schnepp et al., 2020).
In NSCLC, BC and GC, silencing FBXW7 resulted in enhanced Taxol resistance (Yokobori et al., 2014; Gasca et al., 2016). However, in dormant BC cells, disruption of FBXW7 resulted in a shift of tumor cells from the quiescent state, rendering them susceptible to chemotherapy (Shimizu et al., 2019). miR-363 expression can promote chemoresistance by directly targeting FBXW7 (Zhang et al., 2016). Another Fox-containing E3, FBXW11, was also found to be involved in the development of Taxol resistance via associations with FOXO3a (Su et al., 2011). In PC, Skp2 silencing or using Skp2 inhibitors restored paclitaxel sensitivity in paclitaxel-resistant PC cells (Yang et al., 2016). EDD, an E3, promotes docetaxel resistance in hormone-refractory PC by regulating Wnt/β-Catenin signaling (Bian et al., 2020). In PC, SPOP knockout was found to confer resistance to cell death caused by docetaxel (Shi et al., 2019). Taxol decreased the expression of HDAC3 while increasing the expression of SIAH2 in melanoma cells. In addition, the E3 ligase SIAH2 can interact with HDAC3 and by so doing confers resistance to Taxol (Kim et al., 2015).
E3s targeting p53 also play roles in Taxol resistance. Pirh2, a RING finger-containing E3, can lead to polyubiquitination and proteasomal degradation of p53. The ectopic expression of Pirh2 enhanced cell proliferation, resistance to doxorubicin, and migration potential (Daks et al., 2016). The protease HAUSP is a critical component of the p53-Mdm2 pathway and acts as a specific deubiquitinase for p53 and Mdm2 and is thus essential for p53 regulation. HAUSP downregulation causes resistance to another plant-derived drug, camptothecin, and camptothecin-induced apoptosis (Becker et al., 2008). Parkin interferes with paclitaxel-induced microtubule assembly and stabilization by directly binding the microtubules on the outer cell surface. In addition, Parkin promotes the activity of paclitaxel to trigger multinucleation and apoptosis. Moreover, clinical data have revealed that the response of patients to preoperative paclitaxel therapy is correlated with Parkin expression (Wang et al., 2009).
Fludarabine is a DNA synthesis inhibitor (Lukenbill and Kalaycio, 2013). In vitro and in vivo experiments showed that COP1 overexpression reduced HG3 cell sensitivity to fludarabine treatment by promoting ubiquitin-dependent p53 degradation (Fu et al., 2018). This result indicates that E3s promoting P53 degradation can also be related to fludarabine resistance.
5-Fluorouracil (5-FU) inhibits thymidylate synthase from activating thymine-induced cell death and functions mainly as an S-phase antimetabolite (Longley et al., 2003). In BC, high preoperative expression of Skp2 was found to be associated with resistance to 5-FU therapy in 94% of patients (Davidovich et al., 2008). Cbl was decreased in 60% of human pancreatic ductal adenocarcinoma (PDAC) cases. Cbl knockdown increased PDAC resistance to gemcitabine and 5-FU (Kadera et al., 2015). TRIM47 is commonly overexpressed and related to poor prognosis in CRC patients. TRIM47 increases the ubiquitination and degradation of SMAD4. The overexpression of TRIM47 elevated CRC chemoresistance in response to 5-FU therapy (Liang et al., 2019). In CRC, high FBXW7 expression downregulated CRY2 through proteasomal degradation and increased CRC cell sensitivity to 5-FU (Fang et al., 2015). Cancer-associated fibroblasts (CAFs) play a pivotal role in creating the tumor microenvironment, which impacts adaptive resistance to chemotherapy (Kalluri, 2016). CRC cells cocultured with CAFs showed increased expression of RANBP2-type and C3HC4-type zinc-finger-containing 1 (RBCK1). Additionally, overexpression of RBCK1 was demonstrated in chemoresistant CRC tumors and CRC patients with poor prognosis. Exogenous expression of RBCK1 or RBCK1 inhibition was able to significantly influence 5-FU sensitivity in CRC cells (Liu et al., 2019).
Gemcitabine is another widely used S-phase antimetabolite drug (Mini et al., 2006). In pancreatic cancer (PCC) cells, FBW7 promoted gemcitabine sensitivity via upregulation of equilibrative nucleoside transporter 1 (ENT1) through lysosome inhibition (Hu et al., 2017). SMURF2 was downregulated in PCC tissues, and its expression was negatively associated with gemcitabine resistance. Upregulation of miR-15b was associated with degradation of SMURF2, and its expression was associated with EMT (Zhang et al., 2015). TRIM31 overexpression conferred gemcitabine resistance to PCC cells by promoting K63-linked polyubiquitination of tumor necrosis factor receptor-associated factor 2 (TRAF2) and sustained the activation of nuclear transcription factor kappa B (NF-κB) in PCC cells (Yu et al., 2018). Cul4A degraded TGFβ1, and its overexpression promoted resistance to gemcitabine in lung cancer. In vivo experiments, Cul4A-RNAi combined with gemcitabine chemotherapy inhibited lung cancer tumor growth, suggesting that this combination may provide a new approach for lung cancer treatment (Hung et al., 2015).
Asparaginase is a drug that selectively inhibits tumor cells by hydrolyzing asparagine (Asselin and Rizzari, 2015). FBXW7 overexpression can rescue Wnt-induced sensitization to asparaginase in FBXW7 mutant or wild-type leukaemias. In contrast, the FBXW7 R465C mutant, which has impaired binding to its canonical phosphodegron, abrogated this effect (Hinze et al., 2019).
Among various antitumour drugs, alkylating agents may be the most widely used category (Bhatt et al., 2017). Alkylating agents are cytotoxic drugs that combine with cell proteins and nucleic acids to kill tumor cells and have a direct toxic effect on cells (Lajous et al., 2019). Temozolomide (TMZ) is a DNA alkylating agent that can penetrate the blood-brain barrier. HERC3, an E3, promotes the ubiquitination-mediated degradation of SMAD7 and consequently activates the TGF-β pathway. Moreover, ectopic HERC3 expression was correlated with TMZ resistance in glioblastoma (GBM) cells (Li et al., 2019). Nucleolin (NCL) is overexpressed in GBM, and its overexpression was found to be positively relative to response to TMZ in GBM cells. The loss of MDM2-mediated NCL ubiquitination resulted in the inhibition of HDAC activity and sensitized GBM cells to TMZ (Ko et al., 2018).
Doxorubicin is a cytotoxic anthracycline antibiotic that is often used as a tumor chemotherapy agent. Using mass spectrometry analysis, Kamran M et al. found that AURKA restricted FBXL7-induced survivin ubiquitination and degradation in GC, resulting in the promotion of doxorubicin resistance (Kamran et al., 2017). Doxorubicin-resistant HCC cells showed decreased expression of FBXW7. HSF1 was found to play an essential role in transcriptional activation of MDR1 via FBXW7-mediated degradation (Mun et al., 2020). Expression of P-gp on cancer cell surfaces is a critical determinant of anticancer drug resistance (Lin and Yamazaki, 2003). In other words, the reversal of drug resistance can be achieved by modulating the ubiquitination of P-gp (Zhang et al., 2004). Via mass spectrometry analyses, FBXO15/Fbx15 was found to interact with P-gp (Katayama et al., 2013). The downregulation of P-gp expression by UBE2R1-and FBXO15-mediated ubiquitination boosted sensitivity to vincristine and doxorubicin (Katayama et al., 2016).
Zeb1, an influential EMT-related transcription factor, mediated cell resistance to doxorubicin treatment. In HCC doxorubicin-resistant cells, the downregulation of SIAH1 mediated the stability of Zeb1, aiding resensitization of cells to doxorubicin treatment (Long et al., 2019). RNF8 activated K63 ubiquitination of Twist, which induced its translocation to the nucleus for subsequent EMT and CSC functions, thereby conferring doxorubicin resistance (Lee et al., 2016). SMO stabilizes and activates TRAF6, suggesting that the SMO/TRAF6 axis can contribute to doxorubicin resistance in lymphoma (Qu et al., 2018).
Dysregulated cholesterol metabolism in cancer cells may lead to drug resistance. Lower expression of the E3 Trc8 produced a decreased ubiquitination rate of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCoAR), increased cholesterol synthesis, and increased cholesterol content in multidrug-resistant cells (Gelsomino et al., 2013). Overexpression of the E3 ubiquitin-protein ligase ZNRF2 improved cell survival in the presence of doxorubicin (Xiao et al., 2017). TRIM25 regulated p53 expression in NSCLC tissues and cell lines. Using TRIM25 RNAi increased the doxorubicin sensitivity of lung cancer cell lines (Qin et al., 2016). CUL2 knockdown enhanced cell sensitivity to doxorubicin treatment by regulating MAF1-mediated actin stress fiber integrity and apoptosis (Wang et al., 2019). MDM2, an E3 targeting p53 for degradation, can influence PC and BC cell sensitivity to doxorubicin (Lang et al., 2017; Cheteh et al., 2020). FKBP12 attenuated the cell toxicity of doxorubicin by binding to and degrading MDM2, disrupting the MDM2/MDM4 interaction, and inducing MDM2 self-ubiquitination (Liu et al., 2017). Cbl-b was found to be poorly expressed in multidrug-resistant GC and GC cells. In addition, Cbl was also found to induce cell resistance to adriamycin (Xu et al., 2017; X CHe et al., 2017; Zhang et al., 2015).
Endocrine therapy is an optional treatment for patients with hormone sensitivity, especially in PC and BC (Heinlein and Chang, 2004; Waks and Winer, 2019). Androgenic drugs such as abiraterone and enzalutamide can control the progression of PC.
Persistent androgen receptor (AR) activation leads to the loss of efficacy of anti-AR drugs in advanced PC. Reversal of this aberrant activation could be an ideal method for overcoming drug resistance. STUB1 disassociates AR/AR-V7 from HSP70, leading to AR/AR-V7 ubiquitination and degradation, which confers enzalutamide and abiraterone resistance (Liu et al., 2018). Inhibition of protein degradation by blocking Cullin-RING E3 complexes can interfere with the AR–ERG interaction, which is related to survival in PC (Rulina et al., 2016). AMFR can mediate the loss of 11b-hydroxysteroid dehydrogenase-2 (11b-HSD2), which inactivates cortisol, sustaining tumor cortisol concentrations to stimulate enzalutamide resistance. Reinstatement of 11b-HSD2 expression, or AMFR loss, reverses enzalutamide resistance in mouse xenograft tumors (Li et al., 2017).
Tamoxifen, a blocker of estrogen in breast cells, remains a cornerstone in the treatment of BC patients with estrogen receptor-positive tumors (Jordan, 2003). The RING finger protein TRIM2 is highly expressed in tamoxifen-resistant MCF-7 cells. TRIM2 was overexpressed in tamoxifen-resistant BC cells, which led to a reduction in Bim (Yin et al., 2017). The E3 HRD1 was downregulated in tamoxifen-resistant BC cells, and its knockdown significantly increased the survival of MCF7 cells treated with tamoxifen (Wang et al., 2017). The E3 RBCK1 regulated FKBPL stability at the posttranslational level via ubiquitination, and its downregulation increased sensitivity to tamoxifen treatment (Donley et al., 2014). The FBXW2-mediated downregulation of Sox2, a transcription factor conferring drug resistance, suppressed stem cell properties and overcame BC cell resistance to tamoxifen (Yin et al., 2019). The ubiquitin ligase c-Cbl was upregulated during tamoxifen-induced apoptosis of MCF-7 cells. Overexpression of c-Cbl significantly downregulated c-Src protein levels and tamoxifen-induced AKT activity (Yan et al., 2011). In addition, SIAH2 expression is significantly correlated with ER positivity in BC. SIAH2 sensitizes cells to tamoxifen through regulation of ER-a expression (Interiano et al., 2014).
The fusion of BCR (located on chromosome 22q11.2) and ABL1 (located on chromosome 9q34) leads to chronic myeloid leukemia (CML) (Baccarani et al., 2019). Imatinib is a tyrosine kinase inhibitor that can selectively inhibit BCR/ABL kinase activity and function as an effective therapy for CML. Smith PG et al. found that TGFβ played a key role in imatinib resistance by directly affecting c-Cbl-dependent Lyn ubiquitination and turnover, which resulted in bursts of Lyn kinase activity (Smith et al., 2012). TRAF6, an E3, facilitates the K63 ubiquitination of ULK1, resulting in reversal of imatinib resistance in CML cells (Han et al., 2019). LZTR1 acts as the regulator of RAS ubiquitination and MAPK pathway activation. Johannels W et al. reported that loss of LZTR1 expression could induce resistance to imatinib and rebastinib in CML cell lines (Bigenzahn et al., 2018).
Bortezomib is an effective proteasome inhibitor for cancer treatment that reversibly and selectively inhibits the 20S proteasome (Scott et al., 2016). Using an RNA microarray, researchers found that genes related to ubiquitination were differentially regulated in a bortezomib-resistant cell line (Park et al., 2014). NEDD4-1 ubiquitinates Akt and targets pAkt-Ser473 for proteasomal degradation. Low NEDD4-1 expression has been linked to poor prognosis in patients with multiple myeloma (MM), and NEDD4-1 knockdown results in bortezomib resistance in vitro and in vivo (Huang et al., 2020). Additionally, BAP1 depletion resulted in decreased gallbladder carcinoma (GBC) sensitivity to bortezomib (Hirosawa et al., 2018). Shen Y et al. demonstrated that silencing DTX3L improved the sensitivity to bortezomib in MM cell lines and increased the expression of apoptosis biomarkers (Shen et al., 2017). Malek E et al. observed increased CUL1 and SKP2 mRNA levels in patient CD138 + cells. Skp2 binds to Cullin-1 and Commd1 and synergistically enhances bortezomib-induced apoptosis (Malek et al., 2017).
Targeting HER2 with an inhibitor can be a treatment strategy for HER-2-positive BC or GC. c-Cbl and CHIP can interact and ubiquitinate HER2, which can be an effective strategy for combatting lapatinib resistance in HER2-positive cancer (Nunes et al., 2016; Huang et al., 2020). In addition, Skp2 silencing sensitized Her2-overexpressing tumors to trastuzumab treatment (Chan et al., 2012). MDM2 inhibition overcame lapatinib resistance in cells with either wild-type or mutant p53 and xenograft models, suggesting the potential of therapy directed against MDM2 for overcoming lapatinib resistance (Kurokawa et al., 2013). In GC, miR-223, which can regulate FBXW7, decreased GC cell sensitivity to trastuzumab (Eto et al., 2015). Jagged-1-mediated activation of Notch-1 can lead to trastuzumab resistance. The E3 Mindbomb-1 was required for Jagged-1 ubiquitination and subsequent Notch activation, which led to resistance to trastuzumab (Pandya et al., 2016).
Targeting aberrant EGFR expression in cancer cells is a promising treatment strategy for NSCLC. Activation of the Hippo-YAP pathway was correlated with EGFR inhibitor treatment (Kim and Myung, 2018). Wang H et al. identified tankyrase and its associated E3 RNF146 as positive YAP activity regulators by CRISPR screening. Tankyrase inhibition by RNF146 enhanced the growth inhibitory activity of EGFR inhibitors in NSCLC by inhibiting YAP signaling (Wang et al., 2016). FBXW7 regulated quiescence by targeting the c-MYC protein for ubiquitination. High levels of FBXW7 and low levels of c-MYC were observed in gefitinib-resistant cells with EGFR exon 19 deletion, suggesting that FBXW7 plays a pivotal role in the maintenance of gefitinib resistance in EGFR mutation-positive NSCLC (Hidayat et al., 2019). Cetuximab is a monoclonal antibody with a molecular target of EGFR. Specific silencing of Cbl-b expression increased the expression of EGFR and decreased the sensitivity of GC cells to cetuximab (Yu et al., 2016).
Regorafenib and sorafenib are multikinase inhibitors of RAS/RAF/MEK/ERK signaling that function to prevent tumors. CRC cells containing FBW7-inactivating mutations, including missense mutations in three arginine residues (R465, R479, and R505), were found to be insensitive to regorafenib and sorafenib (Tong et al., 2017). Nanog is a master transcriptional regulator of stemness in CSCs. The E3 FBXW8 ubiquitinates Nanog and suppresses Nanog expression, resulting in stemness enhancement and sorafenib resistance (Cao et al., 2019).
JQ1 and I-BET, two selective inhibitors of the bromodomain and extraterminal (BET) family, have shown promising early clinical trial outcomes. Xiangpeng Dai et al. found that cullin-3SPOP was responsible for promoting BET protein degradation. PC cell lines derived from individuals harboring SPOP mutations had increased resistance to BET inhibitor-induced cell growth arrest and apoptosis (Dai et al., 2017). Moreover, specific SPOP mutations could impair BET degradation (Janouskova et al., 2017).
BRAF inhibitors (BRAFi) and MEK inhibitors (MEKi) provide rapid disease control in patients with BRAF-mutant metastatic melanoma (Kuske et al., 2018). BRAFi/MEKi resistance triggers proteasomal degradation of AMPK-α1 and consequently drives autophagy and metabolic reprogramming in melanoma cells. Li YY et al. discovered that RING finger 44 (RNF44) could earmark AMPK-α1 for ubiquitination-mediated degradation in BRAFi-resistant melanoma cells (Li et al., 2017). PARP inhibitors (PARPi) are used clinically to treat BRCA-mutated breast tumors. FBXO5 assembles the active SCF ubiquitin ligase complex constitutively targeting RAD51 for degradation. This mechanism controls BC biology and sensitivity to PARPi (Marzio et al., 2019). The E3s involved in the response to non-platinum anticancer drugs are listed in Table 1.
The purpose of studying the mechanisms of resistance is to identify strategies to combat resistance. E3s control tumor drug resistance by specifically binding to drug resistance-related genes and controlling their expression. Therefore, targeting E3 ligases can serve as a potential and effective strategy for combatting drug resistance. In the current environment, studying the resistance mechanisms of all existing E3s is challenging and impractical. However, some E3s have shown their ability to combat resistance. Several kinds of E3s have been deemed “druggable”. First, E3s that target key pathway molecules, like Cbl, which targets EGFR, and NEDD4, which targets AKT, have potential (Zhao et al., 2020). Second, E3s targeting important oncogenes have also been proposed to be druggable. E3s like MDM2, which targets p53, FBXW2, which targets Sox2, and FBXW7, which targets MYC, have shown value in combating drug resistance.
Ongoing research mainly focuses on potential drugs that can be used in therapeutic applications targeting E3s. Such drugs must directly target the E3s to exert their effect; this requirement fits with several different tools, such as siRNAs, agonists, small molecule compounds that affect the binding of E3s to target proteins, and PROteolysis TArgeting Chimera (PROTAC) technology. Strategies targeting E3s include using small molecules or PROTACs that degrade E3s or interfere with the interaction between E3s and their substrates.
Small molecules can target different E3s by directly binding with them and inhibiting their enzymatic activity. As mentioned above, MDM2 is a selective E3 that binds to P53. Agents that inhibit MDM2 include Nutlin-3a, RG7112, NVP-CGM097, AMG-232 and MI319. The combination of traditional anticancer drugs with novel agents was better for drug efficacy than monotherapy (Azmi et al., 1990; Kojima et al., 2006; Vu et al., 2013; Reuther et al., 2018) (Figure 3). These data suggest that targeting relevant E3s with small molecules to overcome drug resistance could be effective. Agonists have also been shown to be effective in controlling drug resistance. The PPARγ agonist pioglitazone inhibited EGFR/MDM2 signaling-mediated PPARγ degradation and increased cancer cell sensitivity to chemotherapy drugs (Shi et al., 2016).
Another strategy for combating drug resistance is interfering with the interaction between E3s and their target substrates. Using chemical library screens, E Malek et al. identified a novel compound, DT204, that reduced Skp2 binding to Cullin-1 and Commd1 and synergistically enhanced bortezomib-induced apoptosis (Malek et al., 2017). Small molecules have also been used to interfere with the interaction between EGFR and its E3. The downregulation of EGFR ubiquitination inhibits the internalization of EGFR, which is an essential mechanism of EGFR activation and drug resistance (Yu et al., 2020).
Traditional drugs also facilitate the ubiquitination of E3s. Vitamin K3, an inhibitor of Siah2, promoted sensitivity of leukemia cells to imatinib (Huang et al., 2018). ATA is a molecular compound derived from Tanshinone IIA through chemical modification (Tian et al., 2010). A mechanistic study revealed that ATA promoted HER2 degradation by increasing c-Cbl and CHIP-mediated HER2 ubiquitination (Huang et al., 2020). Oridonin is a natural compound inducing oxidative stress that enhances CHIP targeting of BCR-ABL for ubiquitin-proteasome degradation, resulting in the enhancement of cancer cell death (Huang et al., 2017). Ginsenoside RD is another natural compound that increases the ubiquitination of multidrug resistance 1 (MDR1). Ginsenoside Rd treatment can reverse doxorubicin resistance in MCF-7/ADR cells (Pokharel et al., 2010).
PROTAC technology utilizes the ubiquitin‐protease system to target a specific protein for ubiquitination and degradation (Sakamoto et al., 2001). PROTAC technology targets proteins, including transcription factors, skeletal proteins, enzymes, and regulatory factors (Zhang et al., 2018). Targeting oncogene family proteins using PROTACs to overcome drug resistance has recently become a popular area of research.
PROTACs directly targeting oncogenes are a tool for overcoming drug resistance. Ibrutinib resistance can occur due to a cysteine to serine mutation (C481S) in the site normally covalently bound by ibrutinib. Alexandru D. Buhimschi et al. introduced MT-802, which is a PROTAC that induces ubiquitination-dependent degradation of wild-type and C481S-mutant BTK from PROTAC, which could be a novel tool for overcoming ibrutinib resistance (Buhimschi et al., 2018). CP5V (apcin-A-PEG5-VHL ligand 1), as an efficient Cdc20 PROTAC, can mediate degradation of the oncogene Cdc20 through the ubiquitous pathway and overcome resistance to taxane chemotherapy in BC by inhibiting mitotic slippage (Chi et al., 2019). The SH2-U-box targets both native and T315I-mutant BCR-ABL for ubiquitination and degradation and thus may serve as a tool for treating both imatinib-sensitive and imatinib-resistant CML (Ru et al., 2016).
In summary, E3s can play a role in tumor resistance by binding oncogenes or pathway proteins. Degrading E3s via different strategies or affecting their function can be used as strategies for anticancer treatment. However, E3s have some shortcomings in treating drug resistance. The first is the diversity of E3s, which prevents a complete understanding of all of the E3 functions and their corresponding target genes. The second is that E3s can bind to multiple different oncogenes. For example, TRIM family proteins perform different functions by binding to different oncogenes. The same E3 not only combines with one oncogene or tumor suppressor gene but also can interact with multiple oncogenes or tumor suppressor genes at the same time. Directly targeting E3s in anti-drug resistance research may lead to failure.
Drug resistance has been a prominent factor negatively affecting clinical treatment. Various E3s target oncogenes or tumor suppressors, affecting the sensitivity of tumor cells to different drug treatments. Strategies that target E3s to combat drug resistance include interfering with E3 activators, degrading E3s and affecting the interaction between E3s and their substrates. Some famous E3s, like MDM2, FBWX7, and SKP2, have been well studied and shown to have value for treating drug resistance. Research on E3s has led to the discovery of possible therapeutic methods to overcome the challenging clinical situation imposed by drug resistance.
YL was contributed to the acquisition, analysis, and interpretation of data. CD and CZ was contributed to the conception and design; revising the article for important intellectual content; final approval of the version to be published.
This work was supported by grants from National Natural Science Foundation of China (81372515), National Natural Science Youth Foundation of China (81602027,81702928), Natural Science Foundation General Program of Hunan Province (2018JJ3821), Natural Science Foundation Youth Program of Hunan Province (806288981033), Project of Scientific Research Plan of Hunan Provincial Health Commission (C2019185) and the Youth Foundation of Xiangya Hospital Central South University (2015Q01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abedini, M. R., Muller, E. J., Bergeron, R., Gray, D. A., and Tsang, B. K. (2010). Akt promotes chemoresistance in human ovarian cancer cells by modulating cisplatin-induced, p53-dependent ubiquitination of FLICE-like inhibitory protein. Oncogene 29 (1), 11–25. doi:10.1038/onc.2009.300
Abedini, M. R., Muller, E. J., Brun, J., Bergeron, R., Gray, D. A., and Tsang, B. K. (2008). Cisplatin induces p53-dependent FLICE-like inhibitory protein ubiquitination in ovarian cancer cells. Cancer Res. 68 (12), 4511–4517. doi:10.1158/0008-5472.Can-08-0673
Asselin, B., and Rizzari, C. (2015). Asparaginase pharmacokinetics and implications of therapeutic drug monitoring. Leuk. Lymphoma 56 (8), 2273–2280. doi:10.3109/10428194.2014.1003056
Azmi, A. S., Aboukameel, A., Banerjee, S., Wang, Z., Mohammad, M., Wu, J., et al. (1990). MDM2 inhibitor MI-319 in combination with cisplatin is an effective treatment for pancreatic cancer independent of p53 function. Eur. J. Cancer, 46 (6). Oxford, England, 1122–1131. doi:10.1016/j.ejca.2010.01.015
Baccarani, M., Rosti, G., and Soverini, S. (2019). Chronic myeloid leukemia: the concepts of resistance and persistence and the relationship with the BCR-ABL1 transcript type. Leukemia 33 (10), 2358–2364. doi:10.1038/s41375-019-0562-1
Becker, K., Marchenko, N. D., Palacios, G., and Moll, U. M. (2008). A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle (Georgetown, Tex) 7 (9), 1205–1213. doi:10.4161/cc.7.9.5756
Bennett, E. J., and Harper, J. W. (2008). DNA damage: ubiquitin marks the spot. Nat. Struct. Mol. Biol. 15 (1), 20–22. doi:10.1038/nsmb0108-20
Bernabeu, E., Cagel, M., Lagomarsino, E., Moretton, M., and Chiappetta, D. A. (2017). Paclitaxel: what has been done and the challenges remain ahead. Int. J. pharmaceutics 526 (1-2), 474–495. doi:10.1016/j.ijpharm.2017.05.016
Berndsen, C. E., and Wolberger, C. (2014). New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21 (4), 301–307. doi:10.1038/nsmb.2780
Bhatt, L., Sebastian, B., and Joshi, V. (2017). Mangiferin protects rat myocardial tissue against cyclophosphamide induced cardiotoxicity. J. Ayurveda Integr. Med. 8 (2), 62–67. doi:10.1016/j.jaim.2017.04.006
Bian, P., Dou, Z., Jia, Z., Li, W., and Pan, D. (2020). Activated Wnt/β-Catenin signaling contributes to E3 ubiquitin ligase EDD-conferred docetaxel resistance in prostate cancer. Life Sci. 254, 116816. doi:10.1016/j.lfs.2019.116816
Bigenzahn, J. W., Collu, G. M., Kartnig, F., Pieraks, M., Vladimer, G. I., Heinz, L. X., et al. (2018). LZTR1 is a regulator of RAS ubiquitination and signaling. Science (New York, NY) 362 (6419), 1171–1177. doi:10.1126/science.aap8210
Bradley, A., Zheng, H., Ziebarth, A., Sakati, W., Branham-O'Connor, M., Blumer, J. B., et al. (2014). EDD enhances cell survival and cisplatin resistance and is a therapeutic target for epithelial ovarian cancer. Carcinogenesis 35 (5), 1100–1109. doi:10.1093/carcin/bgt489
Buetow, L., and Huang, D. T. (2016). Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cel. Biol. 17 (10), 626–642. doi:10.1038/nrm.2016.91
Buhimschi, A. D., Armstrong, H. A., Toure, M., Jaime-Figueroa, S., and Chen, T. L. (2018). Targeting the C481S ibrutinib‐resistance mutation in bruton's tyrosine kinase using PROTAC‐mediated degradation. Biochemistry 57 (26), 3564–3575.
Cao, J., Zhao, M., Liu, J., Zhang, X., Pei, Y., Wang, J., et al. (2019). RACK1 promotes self-renewal and chemoresistance of cancer stem cells in human hepatocellular carcinoma through stabilizing Nanog. Theranostics 9 (3), 811–828. doi:10.7150/thno.29271
Cappadocia, L., and Lima, C. D. (2018). Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 118 (3), 889–918. doi:10.1021/acs.chemrev.6b00737
Chan, C. H., Li, C. F., Yang, W. L., Gao, Y., Lee, S. W., Feng, Z., et al. (2012). The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149 (5), 1098–1111. doi:10.1016/j.cell.2012.02.065
Chen, Z. J., and Sun, L. J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cel. 33 (3), 275–286. doi:10.1016/j.molcel.2009.01.014
Cheteh, E. H., Sarne, V., Ceder, S., Bianchi, J., Augsten, M., Rundqvist, H., et al. (2020). Interleukin-6 derived from cancer-associated fibroblasts attenuates the p53 response to doxorubicin in prostate cancer cells. Cell Death Discov 6, 42. doi:10.1038/s41420-020-0272-5
Chi, J. J., Li, H., Zhou, Z., Izquierdo-Ferrer, J., Xue, Y., Wavelet, C. M., et al. (2019). A novel strategy to block mitotic progression for targeted therapy. EBioMedicine 49, 40–54. doi:10.1016/j.ebiom.2019.10.013
Ciechanover, A. (2015). The unravelling of the ubiquitin system. Nat. Rev. Mol. Cel. Biol. 16 (5), 322–324. doi:10.1038/nrm3982
Colaluca, I. N., Tosoni, D., Nuciforo, P., Senic-Matuglia, F., Galimberti, V., Viale, G., et al. (2008). NUMB controls p53 tumour suppressor activity. Nature 451 (7174), 76–80. doi:10.1038/nature06412
Cui, X., Lin, Z., Chen, Y., Mao, X., Ni, W., Liu, J., et al. (2016). Upregulated TRIM32 correlates with enhanced cell proliferation and poor prognosis in hepatocellular carcinoma. Mol. Cell. Biochem. 421 (1-2), 127–137. doi:10.1007/s11010-016-2793-z
Dai, X., Gan, W., Li, X., Wang, S., Zhang, W., Huang, L., et al. (2017). Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 23 (9), 1063–1071. doi:10.1038/nm.4378
Daks, A., Petukhov, A., Fedorova, O., Shuvalov, O., Merkulov, V., Vasileva, E., et al. (2016). E3 ubiquitin ligase Pirh2 enhances tumorigenic properties of human non-small cell lung carcinoma cells. Genes & cancer 7 (11-12), 383–393. doi:10.18632/genesandcancer.123
Davidovich, S., Ben-Izhak, O., Shapira, M., Futerman, B., and Hershko, D. D. (2008). Over-expression of Skp2 is associated with resistance to preoperative doxorubicin-based chemotherapy in primary breast cancer. Breast cancer researchBCR 10 (4), R63. doi:10.1186/bcr2122
Donley, C., McClelland, K., McKeen, H. D., Nelson, L., Yakkundi, A., Jithesh, P. V., et al. (2014). Identification of RBCK1 as a novel regulator of FKBPL: implications for tumor growth and response to tamoxifen. Oncogene 33 (26), 3441–3450. doi:10.1038/onc.2013.306
Englinger, B., Mair, M., Miklos, W., Pirker, C., Mohr, T., van Schoonhoven, S., et al. (2017). Loss of CUL4A expression is underlying cisplatin hypersensitivity in colorectal carcinoma cells with acquired trabectedin resistance. Br. J. Cancer 116 (4), 489–500. doi:10.1038/bjc.2016.449
Eto, K., Iwatsuki, M., Watanabe, M., Ishimoto, T., Ida, S., Imamura, Y., et al. (2015). The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int. J. Cancer J. Int. du Cancer 136 (7), 1537–1545. doi:10.1002/ijc.29168
Fang, L., Yang, Z., Zhou, J., Tung, J. Y., Hsiao, C. D., Wang, L., et al. (2015). Circadian clock gene CRY2 degradation is involved in chemoresistance of colorectal cancer. Mol. Cancer Ther. 14 (6), 1476–1487. doi:10.1158/1535-7163.Mct-15-0030
Feng, S., Yang, G., Yang, H., Liang, Z., Zhang, R., Fan, Y., et al. (2017). NEDD4 is involved in acquisition of epithelial-mesenchymal transition in cisplatin-resistant nasopharyngeal carcinoma cells. Cel Cycle (Georgetown, Tex) 16 (9), 869–878. doi:10.1080/15384101.2017.1308617
Fu, C., Shi, X., Gong, Y., Wan, Y., Sun, Z., Shi, H., et al. (2018). Constitutively photomorphogenic 1 reduces the sensitivity of chronic lymphocytic leukemia cells to fludarabine through promotion of ubiquitin-mediated P53 degradation. Cell Physiol. Biochem. : Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 50 (6), 2314–2328. doi:10.1159/000495092
Gasca, J., Flores, M. L., Giráldez, S., Ruiz-Borrego, M., Tortolero, M., Romero, F., et al. (2016). Loss of FBXW7 and accumulation of MCL1 and PLK1 promote paclitaxel resistance in breast cancer. Oncotarget 7 (33), 52751–52765. doi:10.18632/oncotarget.10481
Gelsomino, G., Corsetto, P. A., Campia, I., Montorfano, G., Kopecka, J., Castella, B., et al. (2013). Omega 3 fatty acids chemosensitize multidrug resistant colon cancer cells by down-regulating cholesterol synthesis and altering detergent resistant membranes composition. Mol. Cancer 12, 137. doi:10.1186/1476-4598-12-137
Glickman, M. S., and Sawyers, C. L. (2012). Converting cancer therapies into cures: lessons from infectious diseases. Cell 148 (6), 1089–1098. doi:10.1016/j.cell.2012.02.015
Gong, J., Zhou, Y., Liu, D., and Huo, J. (2018). F-box proteins involved in cancer-associated drug resistance. Oncol. Lett. 15 (6), 8891–8900. doi:10.3892/ol.2018.8500
Guodong, X., Yuan, L., Meng, W., Xiang, L., Sida, Q., Xin, S., et al. (2018). FBXW7 suppresses epithelial-mesenchymal transition and chemo-resistance of non-small-cell lung cancer cells by targeting snai1 for ubiquitin-dependent degradation. Xi'an, China. Cell. Prolif. 51 (5). e12473. doi:10.1111/cpr.12473
Han, S. H., Korm, S., Han, Y. G., Choi, S. Y., Kim, S. H., Chung, H. J., et al. (2019). GCA links TRAF6-ULK1-dependent autophagy activation in resistant chronic myeloid leukemia. Autophagy, 15. 2076-2090. doi:10.1080/15548627.2019.1596492
Hatakeyama, S. (2017). TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochemical Sciences 42 (4), 297–311. doi:10.1016/j.tibs.2017.01.002
He, R., and Liu, H. (2020). TRIM59 knockdown blocks cisplatin resistance in A549/DDP cells through regulating PTEN/AKT/HK2. Gene 747, 144553. doi:10.1016/j.gene.2020.144553
Heinlein, C. A., and Chang, C. (2004). Androgen receptor in prostate cancer. Endocr. Rev. 25 (2), 276–308. doi:10.1210/er.2002-0032
Hidayat, M., Mitsuishi, Y., Takahashi, F., Tajima, K., Yae, T., Miyahara, K., et al. (2019). Role of FBXW7 in the quiescence of gefitinib-resistant lung cancer stem cells in EGFR-mutant non-small cell lung cancer. Bosn J. Basic Med. Sci. 19 (4), 355–367. doi:10.17305/bjbms.2019.4227
Hinze, L., Pfirrmann, M., Karim, S., Degar, J., McGuckin, C., Vinjamur, D., et al. (2019). Synthetic lethality of Wnt pathway activation and asparaginase in drug-resistant acute leukemias. Cancer cell 35 (4), 664–676. doi:10.1016/j.ccell.2019.03.004.e7
Hirosawa, T., Ishida, M., Ishii, K., Kanehara, K., Kudo, K., Ohnuma, S., et al. (2018). Loss of BAP1 expression is associated with genetic mutation and can predict outcomes in gallbladder cancer. PloS one 13 (11), e0206643. doi:10.1371/journal.pone.0206643
Hu, Q., Qin, Y., Zhang, B., Liang, C., Ji, S., Shi, S., et al. (2017). FBW7 increases the chemosensitivity of pancreatic cancer cells to gemcitabine through upregulation of ENT1. Oncol. Rep. 38 (4), 2069–2077. doi:10.3892/or.2017.5856
Hu, X., Meng, Y., Xu, L., Qiu, L., Wei, M., Su, D., et al. (2019). Cul4 E3 ubiquitin ligase regulates ovarian cancer drug resistance by targeting the antiapoptotic protein BIRC3. Cel Death Dis. 10 (2), 104. doi:10.1038/s41419-018-1200-y
Huang, B., Yip, W. K., Wei, N., and Luo, K. Q. (2020). Acetyltanshinone IIA is more potent than lapatinib in inhibiting cell growth and degrading HER2 protein in drug-resistant HER2-positive breast cancer cells. Cancer Lett. 490, 1–11. doi:10.1016/j.canlet.2020.06.010
Huang, H., Weng, H., Dong, B., Zhao, P., Zhou, H., and Qu, L. (2017). Oridonin triggers chaperon-mediated proteasomal degradation of BCR-ABL in leukemia. Scientific Rep. 7, 41525. doi:10.1038/srep41525
Huang, J., Lu, Z., Xiao, Y., He, B., Pan, C., Zhou, X., et al. (2018). Inhibition of Siah2 ubiquitin ligase by vitamin K3 attenuates chronic myeloid leukemia chemo-resistance in hypoxic microenvironment. Med. Sci. monitor : Int. Med. J. Exp. Clin. Res. 24, 727–735. doi:10.12659/msm.908553
Huang, X., Gu, H., Zhang, E., Chen, Q., Cao, W., Yan, H., et al. (2020). The NEDD4-1 E3 ubiquitin ligase: a potential molecular target for bortezomib sensitivity in multiple myeloma. Int. J. Cancer J. Int. du Cancer 146 (7), 1963–1978. doi:10.1002/ijc.32615
Hung, M. S., Chen, I. C., You, L., Jablons, D. M., Li, Y. C., Mao, J. H., et al. (2015). Knockdown of Cul4A increases chemosensitivity to gemcitabine through upregulation of TGFBI in lung cancer cells. Oncol. Rep. 34 (6), 3187–3195. doi:10.3892/or.2015.4324
Ikeda, F., and Dikic, I. (2008). Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: beyond the Usual Suspects' review series. EMBO Rep. 9 (6), 536–542. doi:10.1038/embor.2008.93
Interiano, R. B., Yang, J., Harris, A. L., and Davidoff, A. M. (2014). Seven in Absentia Homolog 2 (SIAH2) downregulation is associated with tamoxifen resistance in MCF-7 breast cancer cells. J. Surg. Res. 190 (1), 203–209. doi:10.1016/j.jss.2014.02.018
Ishii, T., Matsuse, T., Masuda, M., and Teramoto, S. (2004). The effects of S-phase kinase-associated protein 2 (SKP2) on cell cycle status, viability, and chemoresistance in A549 lung adenocarcinoma cells. Exp. Lung Res. 30 (8), 687–703. doi:10.1080/01902140490517818
Janouskova, H., El Tekle, G., Bellini, E., Udeshi, N. D., Rinaldi, A., Ulbricht, A., et al. (2017). Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat. Med. 23 (9), 1046–1054. doi:10.1038/nm.4372
Jeon, Y. K., Kim, C. K., Koh, J., Chung, D. H., and Ha, G. H. (2016). Pellino-1 confers chemoresistance in lung cancer cells by upregulating cIAP2 through Lys63-mediated polyubiquitination. Oncotarget 7 (27), 41811–41824. doi:10.18632/oncotarget.9619
Jordan, V. C. (2003). Tamoxifen: a most unlikely pioneering medicine. Nat. Rev. Drug Discov. 2 (3), 205–213. doi:10.1038/nrd1031
Kadera, B. E., Toste, P. A., Wu, N., Li, L., Nguyen, A. H., Dawson, D. W., et al. (2015). Low expression of the E3 ubiquitin ligase CBL confers chemoresistance in human pancreatic cancer and is targeted by epidermal growth factor receptor inhibition. Clin. Cancer Res. : official J. Am. Assoc. Cancer Res. 21 (1), 157–165. doi:10.1158/1078-0432.Ccr-14-0610
Kalluri, R. (2016). The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16 (9), 582–598. doi:10.1038/nrc.2016.73
Kamran, M., Long, Z. J., Xu, D., Lv, S. S., Liu, B., Wang, C. L., et al. (2017). Aurora kinase A regulates Survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis 6 (2), e298. doi:10.1038/oncsis.2016.80
Katayama, K., Fujiwara, C., Noguchi, K., and Sugimoto, Y. (2016). RSK1 protects P-glycoprotein/ABCB1 against ubiquitin-proteasomal degradation by downregulating the ubiquitin-conjugating enzyme E2 R1. Scientific Rep. 6, 36134. doi:10.1038/srep36134
Katayama, K., Noguchi, K., and Sugimoto, Y. (2013). FBXO15 regulates P-glycoprotein/ABCB1 expression through the ubiquitin--proteasome pathway in cancer cells. Cancer Sci. 104 (6), 694–702. doi:10.1111/cas.12145
Kim, H. B., and Myung, S. J. (2018). Clinical implications of the Hippo-YAP pathway in multiple cancer contexts. BMB Rep. 51 (3), 119–125. doi:10.5483/bmbrep.2018.51.3.018
Kim, J., Xu, S., Xiong, L., Yu, L., Fu, X., and Xu, Y. (2017). SALL4 promotes glycolysis and chromatin remodeling via modulating HP1α-Glut1 pathway. Oncogene 36 (46), 6472–6479. doi:10.1038/onc.2017.265
Kim, Y., Kim, H., Park, D., and Jeoung, D. (2015). miR-335 targets SIAH2 and confers sensitivity to anti-cancer drugs by increasing the expression of HDAC3. Mol. Cell 38 (6), 562–572. doi:10.14348/molcells.2015.0051
Ko, C. Y., Lin, C. H., Chuang, J. Y., Chang, W. C., and Hsu, T. I. (2018). MDM2 degrades deacetylated nucleolin through ubiquitination to promote glioma stem-like cell enrichment for chemotherapeutic resistance. Mol. Neurobiol. 55 (4), 3211–3223. doi:10.1007/s12035-017-0569-4
Kojima, K., Konopleva, M., McQueen, T., O'Brien, S., Plunkett, W., and Andreeff, M. (2006). Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood 108 (3), 993–1000. doi:10.1182/blood-2005-12-5148
Kurokawa, M., Kim, J., Geradts, J., Matsuura, K., Liu, L., Ran, X., et al. (2013). A network of substrates of the E3 ubiquitin ligases MDM2 and HUWE1 control apoptosis independently of p53. Sci. signaling 6 (274), ra32. doi:10.1126/scisignal.2003741
Kuske, M., Westphal, D., Wehner, R., Schmitz, M., Beissert, S., Praetorius, C., et al. (2018). Immunomodulatory effects of BRAF and MEK inhibitors: implications for Melanoma therapy. Pharmacol. Res. 136, 151–159. doi:10.1016/j.phrs.2018.08.019
Lajous, H., Lelièvre, B., Vauléon, E., Lecomte, P., and Garcion, E. (2019). Rethinking alkylating(-like) agents for solid tumor management. Trends Pharmacological Sciences 40 (5), 342–357. doi:10.1016/j.tips.2019.03.003
Lang, V., Aillet, F., Xolalpa, W., Serna, S., Ceccato, L., Lopez-Reyes, R. G., et al. (2017). Analysis of defective protein ubiquitylation associated to adriamycin resistant cells. Cel Cycle (Georgetown, Tex) 16 (24), 2337–2344. doi:10.1080/15384101.2017.1387694
Lee, H. J., Li, C. F., Ruan, D., Powers, S., Thompson, P. A., Frohman, M. A., et al. (2016). The DNA damage transducer RNF8 facilitates cancer chemoresistance and progression through twist activation. Mol. Cel. 63 (6), 1021–1033. doi:10.1016/j.molcel.2016.08.009
Lee, J., An, S., Jung, J. H., Kim, K., Kim, J. Y., An, I. S., et al. (2019). MUL1 E3 ligase regulates the antitumor effects of metformin in chemoresistant ovarian cancer cells via AKT degradation. Int. J. Oncol. 54 (5), 1833–1842. doi:10.3892/ijo.2019.4730
Li, G., Ci, W., Karmakar, S., Chen, K., Dhar, R., Fan, Z., et al. (2014). SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer cell 25 (4), 455–468. doi:10.1016/j.ccr.2014.02.007
Li, H., Li, J., Chen, L., Qi, S., Yu, S., Weng, Z., et al. (2019). HERC3-Mediated SMAD7 ubiquitination degradation promotes autophagy-induced EMT and chemoresistance in glioblastoma. Clin. Cancer Res. : official J. Am. Assoc. Cancer Res. 25 (12), 3602–3616. doi:10.1158/1078-0432.Ccr-18-3791
Li, J., Alyamani, M., Zhang, A., Chang, K. H., Berk, M., Li, Z., et al. (2017). Aberrant corticosteroid metabolism in tumor cells enables GR takeover in enzalutamide resistant prostate cancer.eLife 6. e20183. doi:10.7554/eLife.20183
Li, N., Lorenzi, F., Kalakouti, E., Normatova, M., Babaei-Jadidi, R., Tomlinson, I., et al. (2015). FBXW7-mutated colorectal cancer cells exhibit aberrant expression of phosphorylated-p53 at Serine-15. Oncotarget 6 (11), 9240–9256. doi:10.18632/oncotarget.3284
Li, Y., Ma, C., Zhou, T., Liu, Y., Sun, L., and Yu, Z. (2016). TRIM65 negatively regulates p53 through ubiquitination. Biochem. biophysical Res. Commun. 473 (1), 278–282. doi:10.1016/j.bbrc.2016.03.093
Li, Y. Y., Wu, C., Shah, S. S., Chen, S. M., Wangpaichitr, M., Kuo, M. T., et al. (2017). Degradation of AMPK-α1 sensitizes BRAF inhibitor-resistant melanoma cells to arginine deprivation. Mol. Oncol. 11 (12), 1806–1825. doi:10.1002/1878-0261.12151
Liang, Q., Tang, C., Tang, M., Zhang, Q., Gao, Y., and Ge, Z. (2019). TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J. Exp. Clin. Cancer Res. 38 (1), 159. doi:10.1186/s13046-019-1143-x
Lin, J. H., and Yamazaki, M. (2003). Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin. Pharmacokinet. 42 (1), 59–98. doi:10.2165/00003088-200342010-00003
Liu, C., Lou, W., Yang, J. C., Liu, L., and Gao, A. C. (2018). Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat. Commun. 9 (1). 4700. doi:10.1038/s41467-018-07178-x
Liu, M. L., Zang, F., and Zhang, S. J. (2019). RBCK1 contributes to chemoresistance and stemness in colorectal cancer (CRC). Biomed. Pharmacother. 118, 109250. doi:10.1016/j.biopha.2019.109250
Liu, T., Xiong, J., Yi, S., Zhang, H., Zhou, S., Gu, L., et al. (2017). FKBP12 enhances sensitivity to chemotherapy-induced cancer cell apoptosis by inhibiting MDM2. Oncogene 36 (12), 1678–1686. doi:10.1038/onc.2016.331
Long, L., Xiang, H., Liu, J., Zhang, Z., and Sun, L. (2019). ZEB1 mediates doxorubicin (Dox) resistance and mesenchymal characteristics of hepatocarcinoma cells. Exp. Mol. Pathol. 106, 116–122. doi:10.1016/j.yexmp.2019.01.001
Longley, D. B., Harkin, D. P., and Johnston, P. G. (2003). 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3 (5), 330–338. doi:10.1038/nrc1074
Lu, Y., Han, D., Liu, W., Huang, R., Ou, J., Chen, X., et al. (2018). RNF138 confers cisplatin resistance in gastric cancer cells via activating Chk1 signaling pathway. Cancer Biol. Ther. 19 (12), 1128–1138. doi:10.1080/15384047.2018.1480293
Lukenbill, J., and Kalaycio, M. (2013). Fludarabine: a review of the clear benefits and potential harms. Leuk. Res. 37 (9), 986–994. doi:10.1016/j.leukres.2013.05.004
Malek, E., Abdel-Malek, M. A., Jagannathan, S., Vad, N., Karns, R., Jegga, A. G., et al. (2017). Pharmacogenomics and chemical library screens reveal a novel SCF(SKP2) inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia 31 (3), 645–653. doi:10.1038/leu.2016.258
Marzio, A., Puccini, J., Kwon, Y., Maverakis, N. K., Arbini, A., Sung, P., et al. (2019). The F-box domain-dependent activity of EMI1 regulates PARPi sensitivity in triple-negative breast cancers. Mol. Cel. 73 (2), 224–237. doi:10.1016/j.molcel.2018.11.003.e6
Mini, E., Nobili, S., Caciagli, B., Landini, I., and Mazzei, T. (2006). Cellular pharmacology of gemcitabine. Ann. Oncol. 17 (Suppl. 5), v7–12. doi:10.1093/annonc/mdj941
Mun, G. I., Choi, E., Lee, Y., and Lee, Y. S. (2020). Decreased expression of FBXW7 by ERK1/2 activation in drug-resistant cancer cells confers transcriptional activation of MDR1 by suppression of ubiquitin degradation of HSF1. Cel Death Dis. 11 (5), 395. doi:10.1038/s41419-020-2600-3
Muscolini, M., Montagni, E., Palermo, V., Di Agostino, S., Gu, W., Abdelmoula-Souissi, S., et al. (2011). The cancer-associated K351N mutation affects the ubiquitination and the translocation to mitochondria of p53 protein. J. Biol. Chem. 286 (46), 39693–39702. doi:10.1074/jbc.M111.279539
Nalepa, G., Rolfe, M., and Harper, J. W. (2006). Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5 (7), 596–613. doi:10.1038/nrd2056
Ning, Y., Hui, N., Qing, B., Zhuo, Y., Sun, W., Du, Y., et al. (2019). ZCCHC10 suppresses lung cancer progression and cisplatin resistance by attenuating MDM2-mediated p53 ubiquitination and degradation. Cel Death Dis. 10 (6), 414. doi:10.1038/s41419-019-1635-9
Nunes, J., Zhang, H., Angelopoulos, N., Chhetri, J., Osipo, C., Grothey, A., et al. (2016). ATG9A loss confers resistance to trastuzumab via c-Cbl mediated Her2 degradation. Oncotarget 7 (19), 27599–27612. doi:10.18632/oncotarget.8504
O'Brien, P. M., Davies, M. J., Scurry, J. P., Smith, A. N., Barton, C. A., Henderson, M. J., et al. (2008). The E3 ubiquitin ligase EDD is an adverse prognostic factor for serous epithelial ovarian cancer and modulates cisplatin resistance in vitro. Br. J. Cancer 98 (6), 1085–1093. doi:10.1038/sj.bjc.6604281
Onorati, A. V., Dyczynski, M., Ojha, R., and Amaravadi, R. K. (2018). Targeting autophagy in cancer. Cancer 124 (16), 3307–3318. doi:10.1002/cncr.31335
Pan, C., Chun, J., Li, D., Boese, A. C., Li, J., Kang, J., et al. (2019). Hsp90B enhances MAST1-mediated cisplatin resistance by protecting MAST1 from proteosomal degradation. J. Clin. Invest. 129 (10), 4110–4123. doi:10.1172/jci125963
Pan, X., Chen, Y., Shen, Y., and Tantai, J. (2019). Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cel Death Dis. 10 (6), 429. doi:10.1038/s41419-019-1660-8
Pandya, K., Wyatt, D., Gallagher, B., Shah, D., Baker, A., Bloodworth, J., et al. (2016). PKCα attenuates jagged-1-mediated Notch signaling in ErbB-2-positive breast cancer to reverse trastuzumab resistance. Clin. Cancer Res. : official J. Am. Assoc. Cancer Res. 22 (1), 175–186. doi:10.1158/1078-0432.Ccr-15-0179
Park, J., Bae, E. K., Lee, C., Choi, J. H., Jung, W. J., Ahn, K. S., et al. (2014). Establishment and characterization of bortezomib-resistant U266 cell line: constitutive activation of NF-κB-mediated cell signals and/or alterations of ubiquitylation-related genes reduce bortezomib-induced apoptosis. BMB Rep. 47 (5), 274–279. doi:10.5483/bmbrep.2014.47.5.134
Perez-Losada, J., Mao, J. H., and Balmain, A. (2005). Control of genomic instability and epithelial tumor development by the p53-Fbxw7/Cdc4 pathway. Cancer Res. 65 (15), 6488–6492. doi:10.1158/0008-5472.Can-05-1294
Pokharel, Y. R., Kim, N. D., Han, H. K., Oh, W. K., and Kang, K. W. (2010). Increased ubiquitination of multidrug resistance 1 by ginsenoside Rd. Nutr. Cancer 62 (2), 252–259. doi:10.1080/01635580903407171
Qin, X., Chen, S., Qiu, Z., Zhang, Y., and Qiu, F. (2012). Proteomic analysis of ubiquitination-associated proteins in a cisplatin-resistant human lung adenocarcinoma cell line. Int. J. Mol. Med. 29 (5), 791–800. doi:10.3892/ijmm.2012.912
Qin, Y., Cui, H., and Zhang, H. (2016). Overexpression of TRIM25 in lung cancer regulates tumor cell progression. Techn. Cancer Res. Treat. 15 (5), 707–715. doi:10.1177/1533034615595903
Qu, C., Kunkalla, K., Vaghefi, A., Frederiksen, J. K., Liu, Y., Chapman, J. R., et al. (2018). Smoothened stabilizes and protects TRAF6 from degradation: a novel non-canonical role of smoothened with implications in lymphoma biology. Cancer Lett. 436, 149–158. doi:10.1016/j.canlet.2018.08.020
Ravindranath, A. K., Kaur, S., Wernyj, R. P., Kumaran, M. N., Miletti-Gonzalez, K. E., Chan, R., et al. (2015). CD44 promotes multi-drug resistance by protecting P-glycoprotein from FBXO21-mediated ubiquitination. Oncotarget 6 (28), 26308–26321. doi:10.18632/oncotarget.4763
Reuther, C., Heinzle, V., Nölting, S., Herterich, S., Hahner, S., Halilovic, E., et al. (2018). The HDM2 (MDM2) inhibitor NVP-CGM097 inhibits tumor cell proliferation and shows additive effects with 5-fluorouracil on the p53-p21-Rb-E2F1 cascade in the p53wild type neuroendocrine tumor cell line GOT1. Neuroendocrinology 106 (1), 1–19. doi:10.1159/000453369
Ru, Y., Wang, Q., Liu, X., Zhang, M., Zhong, D., Ye, M., et al. (2016). The chimeric ubiquitin ligase SH2-U-box inhibits the growth of imatinib-sensitive and resistant CML by targeting the native and T315I-mutant BCR-ABL. Scientific Rep. 6, 28352. doi:10.1038/srep28352
Rulina, A. V., Mittler, F., Obeid, P., Gerbaud, S., Guyon, L., Sulpice, E., et al. (2016). Distinct outcomes of CRL-Nedd8 pathway inhibition reveal cancer cell plasticity. Cel Death Dis. 7 (12), e2505. doi:10.1038/cddis.2016.395
Sakamoto, K. M., Kim, K. B., Kumagai, A., Mercurio, F., Crews, C. M., and Deshaies, R. J. (2001). Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. United States America 98 (15), 8554–8559. doi:10.1073/pnas.141230798
Schnepp, P. M., Shelley, G., Dai, J., Wakim, N., Jiang, H., Mizokami, A., et al. (2020). Single-cell transcriptomics analysis identifies nuclear protein 1 as a regulator of docetaxel resistance in prostate cancer cells. Mol. Cancer Res. : MCR 18 (9), 1290–1301. doi:10.1158/1541-7786.Mcr-20-0051
Scott, K., Hayden, P. J., Will, A., Wheatley, K., and Coyne, I. (2016). Bortezomib for the treatment of multiple myeloma. Cochrane Database Syst. Rev. 4, Cd010816. doi:10.1002/14651858.CD010816.pub2
Senft, D., Qi, J., and Ronai, Z. A. (2018). Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18 (2), 69–88. doi:10.1038/nrc.2017.105
Shen, Y., Sun, Y., Zhang, L., and Liu, H. (2017). Effects of DTX3L on the cell proliferation, adhesion, and drug resistance of multiple myeloma cells. Tumour Biol. : J. Int. Soc. Oncodevelopmental Biol. Med. 39 (6), 1010428317703941. doi:10.1177/1010428317703941
Sheng, W., Dong, M., Chen, C., Wang, Z., Li, Y., Wang, K., et al. (2017). Cooperation of Musashi-2, Numb, MDM2, and P53 in drug resistance and malignant biology of pancreatic cancer. FASEB J. : official Publ. Fed. Am. Societies Exp. Biol. 31 (6), 2429–2438. doi:10.1096/fj.201601240R
Shi, J., Zhang, W., You, M., Xu, Y., Hou, Y., and Jin, J. (2016). Pioglitazone inhibits EGFR/MDM2 signaling-mediated PPARγ degradation. Eur. J. Pharmacol. 791, 316–321. doi:10.1016/j.ejphar.2016.09.010
Shi, Q., Zhu, Y., Ma, J., Chang, K., Ding, D., Bai, Y., et al. (2019). Prostate Cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by upregulating Caprin1-dependent stress granule assembly. Mol. Cancer 18 (1), 170. doi:10.1186/s12943-019-1096-x
Shimizu, H., Takeishi, S., Nakatsumi, H., and Nakayama, K. I. (2019). Prevention of cancer dormancy by Fbxw7 ablation eradicates disseminated tumor cells. JCI Insight 4 (4). doi:10.1172/jci.insight.125138
Siegel, R. L., Miller, K. D., and Jemal, A. (2019). Cancer statistics. CA: a Cancer J. clinicians 69 (1), 7–34. doi:10.3322/caac.21551
Skaar, J. R., Pagan, J. K., and Pagano, M. (2014). SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 13 (12), 889–903. doi:10.1038/nrd4432
Smith, P. G., Tanaka, H., and Chantry, A. (2012). A novel co-operative mechanism linking TGFβ and Lyn kinase activation to imatinib resistance in chronic myeloid leukaemia cells. Oncotarget 3 (5), 518–524. doi:10.18632/oncotarget.500
Song, Y., Zhou, X., Bai, W., and Ma, X. (2015). FBW7 increases drug sensitivity to cisplatin in human nasopharyngeal carcinoma by downregulating the expression of multidrug resistance-associated protein. Tumour Biol. : J. Int. Soc. Oncodevelopmental Biol. Med. 36 (6), 4197–4202. doi:10.1007/s13277-015-3056-4
Su, J. L., Cheng, X., Yamaguchi, H., Chang, Y. W., Hou, C. F., Lee, D. F., et al. (2011). FOXO3a-Dependent mechanism of e1a-induced chemosensitization. Cancer Res. 71 (21), 6878–6887. doi:10.1158/0008-5472.Can-11-0295
Tanaka, N., Kosaka, T., Miyazaki, Y., Mikami, S., Niwa, N., Otsuka, Y., et al. (2016).Acquired platinum resistance involves epithelial to mesenchymal transition through ubiquitin ligase FBXO32 dysregulation. JCI Insight 1 (18), e83654. doi:10.1172/jci.insight.83654
Tang, D. E., Dai, Y., Lin, L. W., Xu, Y., Liu, D. Z., Hong, X. P., et al. (2019). STUB1 suppresseses tumorigenesis and chemoresistance through antagonizing YAP1 signaling. Cancer Sci. 110 (10), 3145–3156. doi:10.1111/cas.14166
Tang, D. E., Dai, Y., Xu, Y., Lin, L. W., Liu, D. Z., Hong, X. P., et al. (2020). The ubiquitinase ZFP91 promotes tumor cell survival and confers chemoresistance through FOXA1 destabilization. Carcinogenesis 41 (1), 56–66. doi:10.1093/carcin/bgz085
Tatham, M. H., Plechanovová, A., Jaffray, E. G., Salmen, H., and Hay, R. T. (2013). Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 453 (1), 137–145. doi:10.1042/bj20130244
Tian, H. L., Yu, T., Xu, N. N., Feng, C., Zhou, L. Y., Luo, H. W., et al. (2010). A novel compound modified from tanshinone inhibits tumor growth in vivo via activation of the intrinsic apoptotic pathway. Cancer Lett. 297 (1), 18–30. doi:10.1016/j.canlet.2010.04.020
Tong, J., Tan, S., Zou, F., Yu, J., and Zhang, L. (2017). FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene 36 (6), 787–796. doi:10.1038/onc.2016.247
Tsuchiya, M., Nakajima, Y., Waku, T., Hiyoshi, H., Morishita, T., Furumai, R., et al. (2015). CHIP buffers heterogeneous Bcl-2 expression levels to prevent augmentation of anticancer drug-resistant cell population. Oncogene 34 (35), 4656–4663. doi:10.1038/onc.2014.387
Uchida, C., and Kitagawa, M. (2016). RING-, HECT-, and RBR-type E3 ubiquitin ligases: involvement in human cancer. Curr. Cancer Drug Targets 16 (2), 157–174. doi:10.2174/1568009616666151112122801
Vasan, N., Baselga, J., and Hyman, D. M. (2019). A view on drug resistance in cancer. Nature 575 (7782), 299–309. doi:10.1038/s41586-019-1730-1
Vu, B., Wovkulich, P., Pizzolato, G., Lovey, A., Ding, Q., Jiang, N., et al. (2013). Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med. Chem. Lett. 4 (5), 466–469. doi:10.1021/ml4000657
Waks, A. G., and Winer, E. P. (2019). Breast cancer treatment: a review. Jama 321 (3), 288–300. doi:10.1001/jama.2018.19323
Wang, D., Ma, L., Wang, B., Liu, J., and Wei, W. (2017). E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 36 (4), 683–702. doi:10.1007/s10555-017-9703-z
Wang, H., Liu, B., Zhang, C., Peng, G., Liu, M., Li, D., et al. (2009). Parkin regulates paclitaxel sensitivity in breast cancer via a microtubule-dependent mechanism. J. Pathol. 218 (1), 76–85. doi:10.1002/path.2512
Wang, H., Lu, B., Castillo, J., Zhang, Y., Yang, Z., McAllister, G., et al. (2016). Tankyrase inhibitor sensitizes lung cancer cells to endothelial growth factor receptor (EGFR) inhibition via stabilizing angiomotins and inhibiting YAP signaling. J. Biol. Chem. 291 (29), 15256–15266. doi:10.1074/jbc.M116.722967
Wang, X., Rusin, A., Walkey, C. J., Lin, J. J., and Johnson, D. L. (2019). The RNA polymerase III repressor MAF1 is regulated by ubiquitin-dependent proteasome degradation and modulates drug resistance and apoptosis. J. Biol. Chem. 294. 19255-19268. 10.1074/jbc.RA119.008849
Wang, Y., Guo, A., Liang, X., Li, M., Shi, M., Li, Y., et al. (2017). HRD1 sensitizes breast cancer cells to Tamoxifen by promoting S100A8 degradation. Oncotarget 8 (14), 23564–23574. doi:10.18632/oncotarget.15797
Wu, B., Liu, Z. Y., Cui, J., Yang, X. M., Jing, L., Zhou, Y., et al. (2017). F-box protein FBXO22 mediates polyubiquitination and degradation of CD147 to reverse cisplatin resistance of tumor cells. Int. J. Mol. Sci. 18 (1). doi:10.3390/ijms18010212
Wu, C., Chen, L., Tao, H., Kong, L., and Hu, Y. (2020). Ring finger protein 38 induces the drug resistance of cisplatin in non-small cell lung cancer. Cel Biol. Int. doi:10.1002/cbin.11423
X Che, X., Zhang, Y., Qu, X., Guo, T., Ma, Y., Li, C., et al. (2017). The E3 ubiquitin ligase Cbl-b inhibits tumor growth in multidrug-resistant gastric and breast cancer cells. Neoplasma 64 (6), 887–892. doi:10.4149/neo_2017_610
Xiao, Q., Yang, Y., An, Q., and Qi, Y. (2017). MicroRNA-100 suppresses human osteosarcoma cell proliferation and chemo-resistance via ZNRF2. Oncotarget 8 (21), 34678–34686. doi:10.18632/oncotarget.16149
Xu, L., Zhang, Y., Qu, X., Che, X., Guo, T., Cai, Y., et al. (2017).E3 ubiquitin ligase cbl-b prevents tumor metastasis by maintaining the epithelial phenotype in multiple drug-resistant gastric and breast cancer cells. Neoplasia, 19 (4). New York, NY, 374–382. doi:10.1016/j.neo.2017.01.011
Yan, S. C., Liu, Y. P., Zhang, L. Y., Qu, J. L., Xu, L., Liu, J., et al. (2011). Ubiquitin ligase c-Cbl is involved in tamoxifen-induced apoptosis of MCF-7 cells by downregulating the survival signals. Acta Oncol. 50 (5), 693–699. doi:10.3109/0284186x.2010.543144
Yang, Y., Lu, Y., Wang, L., Mizokami, A., Keller, E. T., Zhang, J., et al. (2016). Skp2 is associated with paclitaxel resistance in prostate cancer cells. Oncol. Rep. 36 (1), 559–566. doi:10.3892/or.2016.4809
Yin, H., Zhu, Q., Liu, M., Tu, G., Li, Q., Yuan, J., et al. (2017). GPER promotes tamoxifen-resistance in ER+ breast cancer cells by reduced Bim proteins through MAPK/Erk-TRIM2 signaling axis. Int. J. Oncol. 51 (4), 1191–1198. doi:10.3892/ijo.2017.4117
Yin, Y., Xie, C. M., Li, H., Tan, M., and Sun, Y. (2019). The FBXW2–MSX2–SOX2 axis regulates stem cell property and drug resistance of cancer cells. Proc. Natl. Acad. ences 116 (41), 201905973.
Yokobori, T., Yokoyama, Y., Mogi, A., Endoh, H., Altan, B., Kosaka, T., et al. (2014). FBXW7 mediates chemotherapeutic sensitivity and prognosis in NSCLCs. Mol. Cancer Res. : MCR 12 (1), 32–37. doi:10.1158/1541-7786.Mcr-13-0341
Yu, C., Chen, S., Guo, Y., and Sun, C. (2018). Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics 8 (12), 3224–3236. doi:10.7150/thno.23259
Yu, H. G., Wei, W., Xia, L. H., Han, W. L., Zhao, P., Wu, S. J., et al. (2013). FBW7 upregulation enhances cisplatin cytotoxicity in non- small cell lung cancer cells. Asian Pac. J. Cancer Prev. : APJCP 14 (11), 6321–6326. doi:10.7314/apjcp.2013.14.11.6321
Yu, J. J., Zhou, D. D., Yang, X. X., Cui, B., Tan, F. W., Wang, J., et al. (2020). TRIB3-EGFR interaction promotes lung cancer progression and defines a therapeutic target. Nat. Commun. 11 (1), 3660. doi:10.1038/s41467-020-17385-0
Yu, P., Fan, Y., Qu, X., Zhang, J., Song, N., Liu, J., et al. (2016). Cbl-b regulates the sensitivity of cetuximab through ubiquitin-proteasome system in human gastric cancer cells. J. BUON : official J. Balkan Union Oncol. 21 (4), 867–873.
Yu, X., Wang, R., Zhang, Y., Zhou, L., Wang, W., Liu, H., et al. (2019). Skp2-mediated ubiquitination and mitochondrial localization of Akt drive tumor growth and chemoresistance to cisplatin. Oncogene 38 (50), 7457–7472. doi:10.1038/s41388-019-0955-7
Zhang, P. F., Sheng, L. L., Wang, G., Tian, M., Zhu, L. Y., Zhang, R., et al. (2016). miR-363 promotes proliferation and chemo-resistance of human gastric cancer via targeting of FBW7 ubiquitin ligase expression. Oncotarget 7 (23), 35284–35292. doi:10.18632/oncotarget.9169
Zhang, R., Li, S. W., Liu, L., Yang, J., Huang, G., and Sang, Y. (2020). TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis 9 (5), 45. doi:10.1038/s41389-020-0229-9
Zhang, W. L., Zhang, J. H., Wu, X. Z., Yan, T., and Lv, W. (2015). miR-15b promotes epithelial-mesenchymal transition by inhibiting SMURF2 in pancreatic cancer. Int. J. Oncol. 47 (3), 1043–1053. doi:10.3892/ijo.2015.3076
Zhang, X., Lee, H. C., Shirazi, F., Baladandayuthapani, V., Lin, H., Kuiatse, I., et al. (2018). Protein targeting chimeric molecules specific for bromodomain and extra-terminal motif family proteins are active against pre-clinical models of multiple myeloma. Leukemia 32 (10), 2224–2239. doi:10.1038/s41375-018-0044-x
Zhang, X., Li, C. F., Zhang, L., Wu, C. Y., Han, L., Jin, G., et al. (2016). TRAF6 restricts p53 mitochondrial translocation, apoptosis, and tumor suppression. Mol. Cel. 64 (4), 803–814. doi:10.1016/j.molcel.2016.10.002
Zhang, Y., Qu, X., Teng, Y., Li, Z., Xu, L., Liu, J., et al. (2015). Cbl-b inhibits P-gp transporter function by preventing its translocation into caveolae in multiple drug-resistant gastric and breast cancers. Oncotarget 6 (9), 6737–6748. doi:10.18632/oncotarget.3253
Zhang, Z., Wu, J. Y., Hait, W. N., and Yang, J. M. (2004). Regulation of the stability of P-glycoprotein by ubiquitination. Mol. Pharmacol. 66 (3), 395. doi:10.1124/mol.104.001966
Zhao, L., Qiu, T., Jiang, D., Xu, H., Zou, L., Yang, Q., et al. (2020). SGCE promotes breast cancer stem cells by stabilizing EGFR. Adv. Sci. (Weinh) 7 (14), 1903700. doi:10.1002/advs.201903700
Keywords: E3 ubiquitin ligase, cancer, cancer treatment, drugresistance, drug
Citation: Liu Y, Duan C and Zhang C (2021) E3 Ubiquitin Ligase in Anticancer Drugdsla Resistance: Recent Advances and Future Potential. Front. Pharmacol. 12:645864. doi: 10.3389/fphar.2021.645864
Received: 24 December 2020; Accepted: 24 February 2021;
Published: 15 April 2021.
Edited by:
Dexin Kong, Tianjin Medical University, ChinaReviewed by:
Pranav Gupta, Albert Einstein College of Medicine, United StatesCopyright © 2021 Liu, Duan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaojun Duan, ZHVhbmNqeHlAMTI2LmNvbQ==; Chunfang Zhang, emNmNjE2OUBvdXRsb29rLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.