
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 12 April 2021
Sec. Ethnopharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.637273
This article is part of the Research Topic Network Pharmacology and Traditional Medicine: Setting the New Standards by Combining In silico and Experimental Work View all 40 articles
 Yunteng Xu1,2‡
Yunteng Xu1,2‡ Hui Li3,4‡
Hui Li3,4‡ Xiaojuan He3,4
Xiaojuan He3,4 Yanfeng Huang2,3
Yanfeng Huang2,3 Shengjie Wang3,4
Shengjie Wang3,4 Lili Wang2,3
Lili Wang2,3 Changlong Fu2,3
Changlong Fu2,3 Hongzhi Ye2,3
Hongzhi Ye2,3 Xihai Li1*
Xihai Li1* Tetsuya Asakawa5,6,7*†
Tetsuya Asakawa5,6,7*†This study aimed to identify whether the NF-κB signaling pathway plays a key role in the treatment of osteoarthritis (OA) with Bushen Zhuangjin Decoction (BZD) based on a typical network pharmacology approach (NPA). Four sequential experiments were performed: 1) conventional high-performance liquid chromatography (HPLC), 2) preliminary observation of the therapeutic effects of BZD, 3) NPA using three OA-related gene expression profiles, and 4) verification of the key pathway identified by NPA. Only one HPLC-verified compound (paeoniflorin) was identified from the candidate compounds discovered by NPA. The genes verified in the preliminary observation were also identified by NPA. NPA identified a key role for the NF-κB signaling pathway in the treatment of OA with BZD, which was confirmed by conventional western blot analysis. This study identified and verified NF-κB signaling pathway as the most important inflammatory signaling pathway involved in the mechanisms of BZD for treating OA by comparing the NPA results with conventional methods. Our findings also indicate that NPA is a powerful tool for exploring the molecular targets of complex herbal formulations, such as BZD.
Osteoarthritis (OA) is common in the aging population. It has been regarded as a severe public health concern since it remarkably reduces quality of life (Ashraf et al., 2018; Kwon et al., 2018). The mechanisms of OA are complicated and not fully understood. Many previous studies have documented the involvement of molecules and inflammatory signaling pathways in the development and progression of OA by regulation of the oxidative stress, cellular apoptosis, inflammatory response and microRNAs (Miyaki and Asahara, 2012; Mobasheri et al., 2015; Bouderlique et al., 2016; Saito and Tanaka, 2017). Of those, NF-κB signaling pathway plays a distinctive role in OA pathogenesis. Rogoglou and Papavassiliou discussed the role of NF-κB signaling pathway in OA and the potential pharmacological effects of suppression of the NF-κB signaling pathway (Rigoglou and Papavassiliou, 2013). Later, Lepetsos reported that NF-κB signaling pathway influences remodeling of the cartilage matrix, apoptosis of the chondrocytes, synovial inflammation, and the terminal chondrocyte differentiation (Lepetsos et al., 2019). Jimi et al. documented that NF-κB signaling pathway plays a key role in the regulation of the normal development and pathological destruction of cartilage (Jimi et al., 2019). Choi et al. therefore summarized that a comprehensive understanding of the roles of NF-κB signaling pathway is useful in the development of novel therapies against OA (Choi et al., 2019).
However, by far OA lacks effective therapy. Current mainstream treatments for OA, such as nonsteroidal anti-inflammatory drugs and opioids, are far from satisfactory since they can provide only temporary amelioration from symptoms (Yazici et al., 2017). It is urgent and indispensable to develop a novel and effective therapy for treating OA. In this context, many traditional Chinese medicine (TCM) treatments have been used to treat OA in China. Our laboratory carried out serials of studies to verified the efficacy of TCM medical plants for treating OA. Currently, we verified the therapeutic effects of Tougu Xiaotong capsule on tunicamycin-treated chondrocytes (Liang et al., 2019). More currently, we verified the therapeutic effects of Tougu Xiaotong capsule to treat OA in vivo and in vitro, in which the roles of p38 MAPK pathway were identidied (Li et al., 2020a; Li et al., 2020b). In addition to Tougu Xiaotong capsule, Bushen Zhuangjin Decoction (BZD) is another commonly used complex herbal formulation for treating OA in China. BZD is composed of 10 Chinese medical plants, including Radix Rehmanniae Preparata [Rehmannia glutinosa (Gaertn.) DC.)], Radix Angelicae Sinensis [Angelica sinensis (Oliv.) Diels], Radix Dipsaci [Dipsacus asperoides C.Y.Cheng and T.M.Ai], Radix Achyranthis Bidentatae [Radix Achyranthis Bidentatae], Poria [Smilax glabra Roxb], Pericarpium Citri Reticulatae Viride [Citrus × aurantium L], Fructus Corni [Cornus officinalis Siebold and Zucc], Cortex Eucommiae [Eucommia ulmoides Oliv], Radix Paeoniae Alba [Paeonia lactiflora Pall] and Cortex Acanthopanax Radicis [Eleutherococcus nodiflorus (Dunn) S.Y.Hu] (Supplementary Table S1). BZD is widely used to treat OA in China (Huang et al., 2017). Mi observed the efficacy of BZD for treating OA in 60 patients. He found that BZD contributed to relief the OA symptoms and the total response rate was 93.3% (Mi, 2013). Guan and Zhong compared the clinical outcomes in 80 patients with OA (BZD + rehabilitation vs. rehabilitation). They found that patients treated with BZD achieved better Lysholm knee scores (Guan and Zhong, 2017). Lin et al. observed the clinical outcomes of 220 patients with OA, of those 110 cases were treated with BZD (treatment group), and 110 cases were treated with glucosamine hydrochloride (control group). They found that the BZD group had better efficacy in alleviation of pain and amelioration of the Index of Severity for OA in comparison with the control group after 12-week treatments (Lin et al., 2017). A number of Chinese literatures reported the mechanisms of BZD are associated with suppressing the apoptosis of chondrocytes (Zhou et al., 2012), promoting the proliferation of the chondrocytes (Li Y. et al., 2020), protecting the injury of chondrocytes via upregulation of Sox 9 protein (Wu et al., 2020), downregulating the expression of ROCK, Cofilin,Phospho-Cofilin,LIMK1, and Phospho-LIMK1 proteins, which were relevant to damage of the cartilage in OA (Liang et al., 2015), decreasing the serum levels of tumor necrosis factor alpha (TNF-α) and Interleukin (IL)-6 in OA patients (Zhang, 2006) and animal models (Yang et al., 2020), and increasing the activity of the superoxidase dismutase in OA rabbits (Yao et al., 2005). Our laboratory also found that BZD presented the effects of suppressing the expression of the IL-1β, TNF-α and MMP-3 in synovial fluid in rat OA models (Li et al., 2014). However, literatures in English are limited. Our previous studies have found that BZD promotes the proliferation of chondrocytes by stimulating the cell cycle (Li et al., 2015) and suppresses endoplasmic reticulum stress-mediated apoptosis in an OA cell model (Lin et al., 2015). Evidence obtained from these literatures strongly implies that anti-inflammatory mechanism may play a role in the mechanisms of BZD. Due to the key roles of NF-κB signaling pathway in the inflammatory mechanisms of OA, we therefore speculated that NF-κB signaling pathway also plays a role in the mechanisms of BZD in treating OA. Nevertheless, by far, no study elucidates the role of NF-κB signaling pathway in BZD, which is desired to be further investigated.
A novel approach, known as the network pharmacology approach (NPA) has recently attracted interest. NPA combines systematic bioactive analysis and pharmacology. An NPA simultaneously searches for molecular targets from the ingredients of a complex herbal formulation and the target disease and identifies the therapeutic targets by determining the intersections shared by the formulation and disease. This approach is believed to be useful for elucidating the synergistic effects and interactions among compounds. It also identifies potential mechanisms of multi-component and multi-target drugs using the compound–compound, compound–target, and target–disease networks (Liu et al., 2016). NPA’s usefulness for exploring putative molecular targets and mechanisms of complex herbal formulation has been widely proved.
In the present study, we used a standard NPA to identify the underlying mechanisms of BZD’s effectiveness in treating OA. In terms of the key role of NF-κB signaling pathway in OA, we hypothesized that the NF-κB signaling pathway also plays a key role in the mechanisms of BZD for treating OA. To test this hypothesis, four sequential experiments were designed: 1) detection of the chemical components, 2) preliminary verification of BZD’s effects in lipopolysaccharide (LPS)-induced OA cell model, 3) NPA for BZD, and 4) validation of the pathway(s) identified by NPA. We attempted to identify the role of NF-κB signaling pathway in the treatment of OA with BZD by comparing the NPA results with conventional methods. Moreover, the findings of this study will prove that NPA is a robust tool to identify the molecular targets of a commonly used complex herbal formulation.
BZD medical plants, obtained from the Third People’s Hospital, affiliated with Fujian University of TCM (Fuzhou, China), were crushed and passed through a 20–40 mesh sieve. To establish the correct BZD ratio (Supplementary Table S1), we filtered 105 g of herbal powder using 840 ml of 67% ethanol and extracted by reflux. The filtrate was evaporated using a rotary evaporator (RE-2000; Shanghai Yarong Biochemistry Instrument Factory, Shanghai, China) and then dried to a constant weight in a vacuum drying oven (DZF-300; Shanghai Yiheng Scientific Instrument Co., Shanghai, China). BZD was dissolved in phosphate-buffered saline (PBS, HyClone Laboratories, Inc., Logan, UT, United States) to a stock concentration of 40 mg/ml and stored at −80°C. The working concentration of BZD was prepared by diluting the stock solution in PBS, filtering through a 0.22 µm filter, and storing at 4°C.
BZD extracts were analyzed by HPLC using an Agilent 1200 HPLC system (Agilent, Santa Clara, CA, United States) with an Agilent 5 TC-C18 (250*4.6 mm) column (Supplementary Figure S1). The analytical conditions included acetonitrile (A) and 0.2% phosphoric acid in water (B) as a mobile phase, a detection wavelength of 230 nm for morroniside (purity 98%, (Supplementary Figure S1E) and paeoniflorin (purity 98%, (Supplementary Figure S1F), a detection wavelength of 212 nm for asperosaponin VI (purity 98%, (Supplementary Figure S1G) (China Institute of Food and Drug test, Beijing, China), a flow rate of 0.8 ml/min, and a column temperature of 30°C.
Four-week-old male Sprague Dawley rats (BW: 90–120 g, n = 24) were purchased from the Shanghai Slack Laboratory Animal Co. (Shanghai, China). Rats were housed in the animal center at 60% humidity, 23°C room temperature, 12-h light/dark cycle (8:00 AM–8:00 PM), with freely available food and water. Animals were treated following the National Institute of Health Guidelines for the Care and Use of Laboratory Animals. All experiments were approved and supervised by the Animal Care and Use Committee of the Fujian University of TCM (Approval number: 2020015).
Chondrocytes were obtained and used to generate an LPS-induced cell model, as described previously (Liang et al., 2019; Li et al., 2020a; Li et al., 2020b). Briefly, chondrocytes were obtained from the knee joints of four rats at a time (six times in total). Cells were identified by Collagen II immunohistochemistry. Chondrocytes were exposed to 10 ng/ml LPS (Sigma-Aldrich, United States) for 8 h to establish the cell model (Li et al., 2020b).
Chondrocytes were treated with 12.5 μg/ml, 25 μg/ml, 50 μg/ml, 100 μg/ml, 200 μg/ml BZD and 10 ng/ml LPS for 8 h. MMP-9 and IL-6 levels in the cell supernatants were measured using a standard Enzyme-Linked Immunosorbent Assay as per the manufacturer’s instructions (R&D Systems, United States). Samples (50 µL) of the cell supernatants were analyzed using an enzyme labeling instrument (model ELx800; BioTek, Winooski, VT, United States) at a wavelength of 450 nm.
Chondrocytes were divided into a control group, a model group (LPS 10 ng/ml), and a BZD group (BZD 50 µg/ml + LPS 10 ng/ml) and incubated for 8 h. Total proteins were collected immediately using lysis buffer (Beyotime Institute of Biotechnology, Haimen, China), stored for 30 min on ice, and quantified using the bicinchoninic acid assay. A total of 20 µg protein was separated on 10% SDS-PAGE gels and transferred onto polyvinylidene fluoride membranes (Sigma-Aldrich, United States). The membranes were blocked with 5% non-fat milk and incubated with primary antibodies against tumor necrosis factor (TNF)-α (ab11564, abcam, United States), IL-1β (ab205924, abcam, United States), MyD88 (ab2064, abcam, United States), matrix metallopeptidase (MMP)-3 (ab52915, abcam, United States), TLR4 (ab217274, abcam, United States), NF-κB p65 (ab16502, abcam, United States), and GAPDH (5174s, Cell Signaling Technology, United States) overnight at 4°C. Goat anti-rabbit horseradish peroxidase-conjugated secondary antibody IgG (bs-0295G-HRP, Bioss, China) or goat anti-mouse horseradish peroxidase-conjugated secondary antibody IgG (bs-0296G-HRP, Bioss, China) was added to the membranes at room temperature. The immunocomplexes were visualized by the enhanced chemiluminescence method. The bands were quantified by scanning densitometry (Molecular Imager ChemiDoc X-Ray Spectroscopy System, cat. no. 170-8070; Bio-Rad). The blots were analyzed using Image Lab software with GAPDH as a control.
All BZD components were searched using the TCM systems pharmacology database and analysis platform (TCMSP, https://tcmspw.com/tcmsp.php) (Ru et al., 2014) as well as the SymMap (https://www.symmap.org/) (Wu Y. et al., 2019). Oral bioavailability (OB ≥ 30%) and drug-like (DL ≥ 0.18), which are commonly used screening methods for the chemical composition of TCM, were chosen as screening parameters. The OB value refers to the relative amount absorbed into the systemic blood circulation after the drug is administered by an extravascular route. DL refers to the similarity of a compound with a known drug, and the class of compounds having the potential to become drugs. One hundred and thirteen eligible compounds were obtained, two for RRP, two for RAS, eight for RD, 20 for RAB, 15 for P, five for PCRV, 20 for FC, 28 for CE, and 13 for RPA.
Differentially expressed genes identified for OA patients were obtained from the Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.Gov/geo/) (Clough and Barrett, 2016). The series GSE46750 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46750) (Lin et al., 2018) was selected to compare the gene expression profiles of inflammatory I and normal/reactive (N/R) synoviocytes from the synovium of the same OA patient. Differential expression patterns were identified in two regions of the synovium in 12 patients undergoing total knee arthroplasty. The series GSE51588 (https://www.ncbi.nlm.nih.gov/geo/quer y/acc.cgi?acc=GSE51588) (Gu et al., 2019) was also selected, which represents total RNA isolated from human OA (n = 20) and non-OA (n = 5) lateral and medial tibial plateaus of the knee. This profile was obtained by performing whole-genome microarray profiling of human osteoarthritic subchondral bone. The series GSE29746 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE29746) (Li et al., 2016) was selected to compare the gene expression profiles of OA synovial tissues with normal synovial fibroblasts from healthy individuals. The samples were derived from 11 healthy adult donors and 11 sex- and age-matched patients with OA. The original file was processed by a robust multiarray average algorithm with normalization of matrix data, and the relevant data were filtered using the Limma package to analyze the chip data twice, combining the p-value, and the difference multiple. The screening conditions for significantly differentially expressed genes were p < 0.05 with a |log 2 (fold change)| > 0.05.
The compound-target network for BZD was constructed and visualized using Cytoscape 3.7.2 software. Protein–protein interaction (PPI) data were obtained from the Database of Interacting Proteins (DIP™) (Xenarios et al., 2002), Biological General Repository for Interaction Datasets (Stark et al., 2006), Human Protein Reference Database (Goel et al., 2012), IntAct Molecular Interaction Database (IntAct) (Kerrien et al., 2012), Molecular INTeraction database (MINT) (Licata et al., 2012), and the biomolecular interaction network database (Alfarano et al., 2005) using the plugin Bisogenet of Cytoscape 3.7.2 software. The PPI networks for BZD putative targets and OA-related targets were visualized with Cytoscape software.
The protein–protein interaction networks for BZD and OA targets were drawn with the Biogenet plugin, and the intersection of the two networks was obtained by Cytoscape, which is the direct or indirect target regulatory network for BZD in OA. Using the network topology analysis plugin CytoNCA and filtering with Degree Centrality (DC), Betweenness Centrality (BC), Closeness Centrality (CCT), Eigenvector Centrality (EC), Local average connectivity-based method (LAC), and Network Centrality (NC), key genes were identified in the PPI network, and the core target was determined for BZD activity against OA.
The Gene Ontology database (GO, http://geneontology.org/) (Pomaznoy et al., 2018), which contains a molecular function (MF), biological process (BP), and cellular component (CC) data, was used to identify biological mechanisms from high-throughput genomic or transcriptome data. The functional categories were enriched within genes (false discovery rate [FDR] < 0.05), and the top 20 GO functional categories were selected. The KEGG, https://www.kegg.jp/) database (Kanehisa, 2002) was used to identify the function and biological correlation of candidate target genes. Cluster Profiler R package was used to visualize the GO and KEGG pathway data. The pathways which exhibited significant changes with an FDR < 0.05 were selected for further analysis. The genes that represented significantly regulated pathways were selected for gene-pathway network analysis. The gene-pathway network was constructed to screen key target genes involved in BZD activity.
A Venn diagram analysis was performed to identify the key target genes by finding intersections among the GSE46750, GSE51588 and GSE297 profiles. The results of the target gene identification for BZD and interactive PPI networks topological analysis were used to construct a Venn diagram. KEGG was then performed to further analyze the genes located at the intersections of the BZD target genes and the PPI networks’ topological analyses. Finally, the key pathways associated with BZD activity against OA were identified.
Based on the NPA results, western blot analysis was used to verify the targeted pathway’s involvement, which was determined to be NF-κB signaling (see the Results and Discussion section). Pyrrolidine dithiocarbamate (PDTC), an inhibitor of the NF-κB signaling pathway, was selected as a positive control. Chondrocytes were divided into a control group, a model group (LPS 10 ng/ml), a BZD group (BZD 50 µg/ml + LPS 10 ng/ml), and a PDTC group (PDTC10 µM + LPS 10 ng/ml), and treated for 8 h. The western blot procedure was done as described above. The primary antibodies used were NF-κB p65 (ab16502, Abcam, United States), IKK-α (ab32041, Abcam, United States), IKK-β (ab124957, Abcam, United States), MMP-3 (ab52915, Abcam, United States), Collagen II (ab34712, Abcam, United States), and GAPDH (5174s, Cell Signaling Technology, United States).
SPSS 20.0 software (SPSS Inc., Chicago, IL, United States) was used for statistical analyses. The results are presented as the mean ± standard deviation (SD). Data were analyzed by a two-way analysis of variance followed by Bonferroni’s posthoc correction for multiple comparisons. All the experiments were repeated independently three times. p < 0.05 was considered to be significantly different.
Three main components were identified in the BZD extract using HPLC: monoglucoside (peak 1), paeoniflorin (peak 2), and asperosaponin VI (peak 3) (Supplementary Figure S1). Thus, the concentrations of monoglucoside, paeoniflorin, and asperosaponin VI were used as quality control markers.
In this experiment, changes of MMP-9, IL-6 and NF-κB p65 levels were used as preliminary indicators of the activation (suppression) of the NF-κB signaling pathway.
LPS exposure significantly upregulated MMP-9 (Figure 1A) and IL-6 (Figure 1B) levels. When BZD was administrated for 8 h, the expression of both proteins decreased, whereas treatment with 50 µg/ml resulted in a significant reduction (p < 0.01 for MMP-9 and p < 0.05 for IL-6 vs. LPS-exposed chondrocytes, Figure 1). Therefore, we selected an 8 h exposure of 50 µg/ml BZD for subsequent experiments.
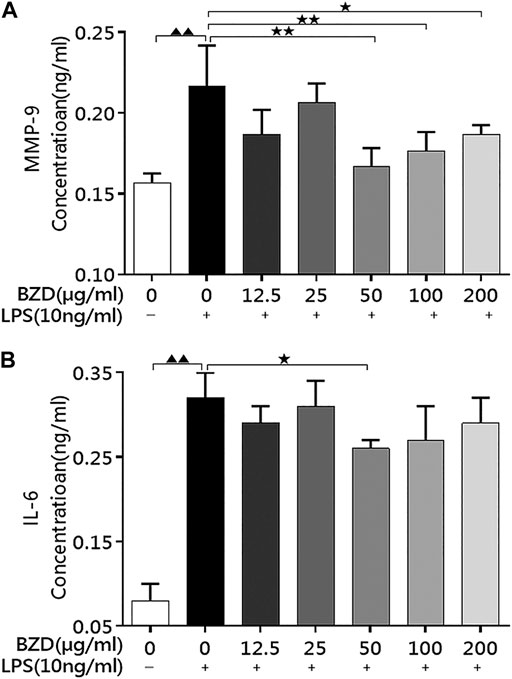
FIGURE 1. BZD administration decreased MMP-9 and IL-6 levels in LPS-exposed chondrocytes. (A) The MMP-9 levels in the culture medium after treatment with different concentrations of BZD for 8 h. (B) The IL-6 levels in the culture medium after treatment with different concentrations of BZD for 8 h ▲▲ means p < 0.01, vs. intact control; * means p < 0.05 and ** means p < 0.01, vs. LPS-exposed chondrocytes.
The results of western blot analysis revealed that a number of inflammatory-related genes with different mechanisms, including TNF-α, IL-1β, Myd88, MMP-3, TLR4, and NF-κB p65, exhibited similar changes. LPS exposure significantly upregulated protein expression, whereas BZD treatment significantly downregulated expression (Figure 2). The proteins involved in different mechanisms exhibited the same pattern, indicating that the mechanisms of BZD are complicated and multidimensional.
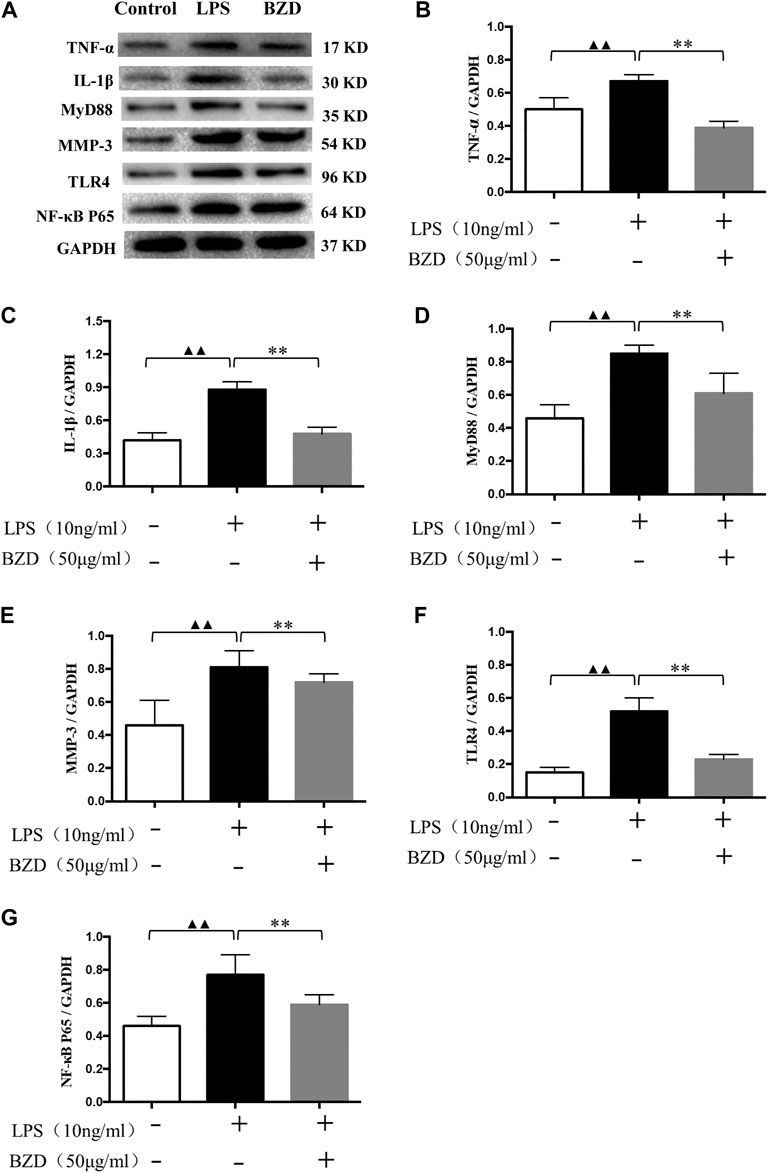
FIGURE 2. The effects of LPS and BZD exposure on protein expression. (A) Representative images of western blot analysis. (B–G) Relative protein expression of inflammatory-related genes. TNF-α (B), IL-1β (C), MyD88 (D), MMP-3 (E), TLR4 (F), and NF-κB p65 (G) exhibited a similar pattern. Protein expression was significantly upregulated by LPS exposure and significantly downregulated by BZD treatment. ▲▲ means p < 0.01, vs. intact control; ** means p < 0.01, vs. LPS-exposed chondrocytes.
Here, expression of MMP-9, IL-6 and NF-κB p65 significantly upregulated by the LPS exposure, whereas significantly downregulated by BZD treatment, strongly implied that activation (suppression) of the NF-κB signaling pathway plays a role in the effects of LPS exposure (BZD treatment). Therefore, we used NPA to explore relevant targets and pathways.
A total of 779, 4,531, and 610 OA-related targets were identified from the GEO database for GSE46750, GSE51588, and GSE29746. The volcano plot and heat maps are shown in Figures 3A–C. A total of 20 OA-related targets were located at the intersection of GSE46750, GSE51588, and GSE29746 (Figure 3D). After the duplications were removed, the remaining 98 compounds were considered to be candidate compounds from the TCM systems pharmacology database (Table 1).
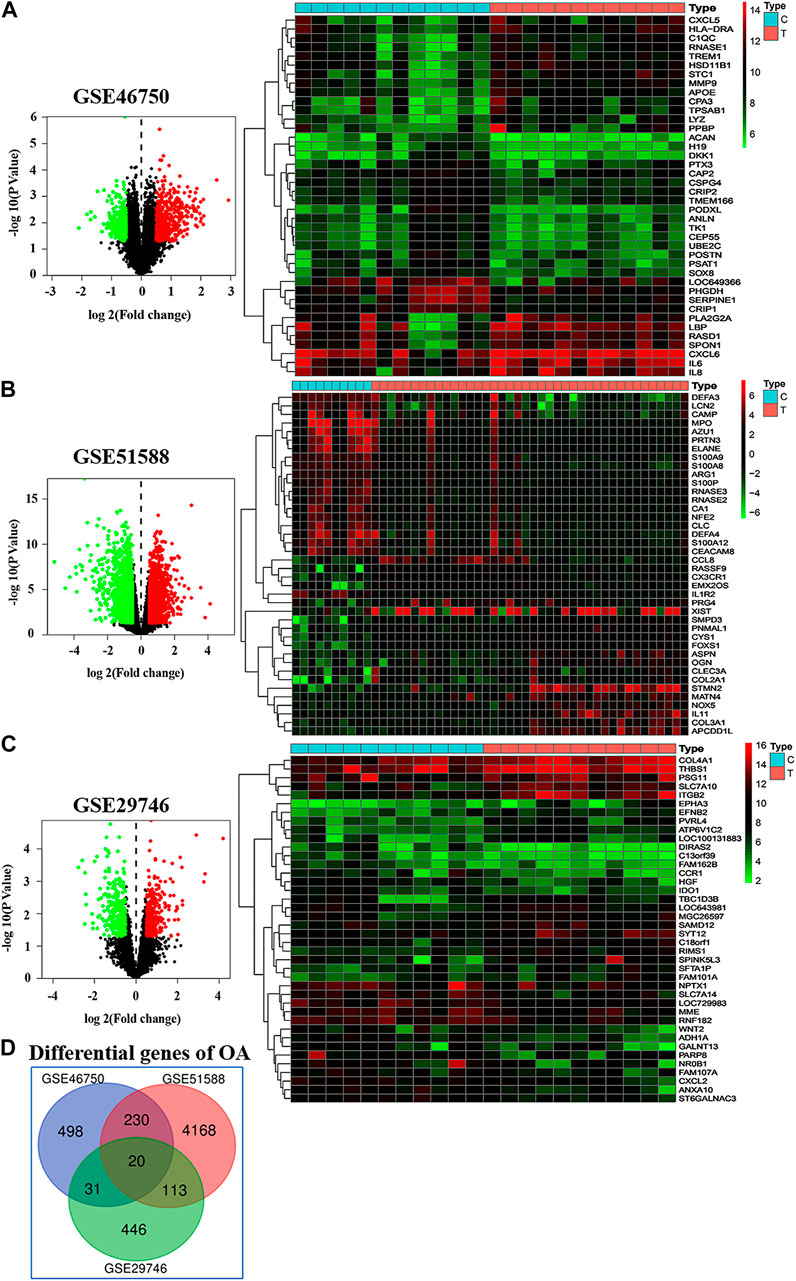
FIGURE 3. Differentially gene expression in GSE46750, GSE51588, and GSE29746. (A–C) In the volcano plot (left column), the abscissa represents the fold changes in gene expression, and the ordinate represents the statistical significance of the variations in gene expression. For GSE46750, there are 469 red dots representing significantly upregulated genes, and 310 green dots representing significantly downregulated genes (A). For GSE51588, there are 2,230 red dots and 2,301 green dots (B). For GSE29746, there are 291 red dots and 319 green dots (C). In the heat map (right column), the red pixels represent significantly upregulated genes, and the green represents significantly downregulated genes. (D) A total 20 OA associated targets are located at the intersection of GSE46750, GSE51588, and GSE29746 These include CCNA2, CKS2, CCNF, KIF11, ETS2, FRMD6, CENPN, POLQ, CCR1, CAP2, MATN2, TPX2, CENPE, PDE4B, DLGAP5, CD33, TNFSF10, ANGPTL2, LTC4S, and PMEPA1.
The compound–target network for BZD was constructed using the screened compounds and their targets (Figure 4). For GSE46750, the network contained 75 nodes (31 compounds for BZD and 44 compound targets) and 119 edges representing compound–target interactions (Figure 4A). For GSE51588, the network included 106 nodes (35 compounds for BZD and 71 compound targets) and 156 edges (Figure 4B). For GSE29746, the network included 38 nodes (23 compounds in BZD and 15 compound targets) and 47 edges (Figure 4C). The largest degrees for the three series were quercetin, which yielded a value of 46.43%.
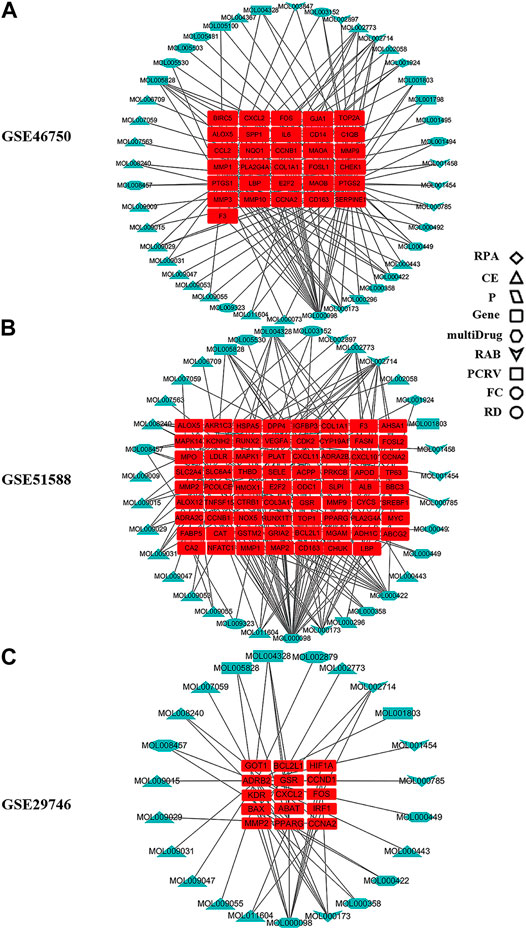
FIGURE 4. Compound–target network for BZD. The round red rectangles represent targets; the diamond, triangle, parallelogram, hexagon, quadrilateral arrow, rectangle, octagon, and ellipse represent the compounds from RPA, CE, P, multidrug, RAB, PCRV, FC, and RD, respectively. (A) GSE46750, (B) GSE51588, (C) GSE29746.
To explore the underlying mechanisms of BZD activity in OA, we merged the PPI networks of the putative BZD targets and OA-related targets to identify a set of candidate targets. For GSE46750, the network consisted of 1,054 nodes and 14,977 edges. The median degree of all nodes was 17, and nodes with more than 61° were identified as significant targets according to a previous study (Zeng et al., 2019). A secondary network consisting of significant (DC > 61) targets for BZD anti-OA activity was then constructed, which contained 139 nodes and 2,932 edges. The median values for BC and CCT were 75.38397 and 0.589744, respectively. The candidate targets were further screened, and 60 targets with BC > 75.38397 and CCT > 0.589744 were identified to generate the final network (Figure 5A). Thus, 60 target genes were eventually identified for BZD anti-OA activity in GSE46750. Similarly, for GSE51588, the network consisted of 3,265 nodes and 80,834 edges. Likewise, the median degree of all nodes was 30, and the nodes with more than 61° were identified as significant targets. The secondary network was constructed by the selection of targets with DC > 61 and contained 837 nodes and 35,888 edges. The median values for BC, CCT, EC, LAC, and NC were 338.4249, 0.517647, 0.023029, 17.30864, and 19.03273, respectively. The candidate targets were further screened and 262 targets with BC > 338.4249, CCT > 0.517647, EC > 0.023029, LAC > 17.30864 and NC > 19.03273 were identified to generate the final network (Figure 5B). Two hundred and sixty-two target genes were eventually identified in GSE46750. For GSE29746, the network consisted of 1,237 nodes and 19,308 edges. The median degree of all nodes was 19, and nodes with more than 61° were considered significant targets. The secondary network with DC > 61 contained 165 nodes and 3,693 edges. The median values for BC, CCT, EC, and LAC were 89.22116, 0.577465, 0.074042, and 15.8, respectively. The candidate targets were further screened and 33 targets with BC > 89.22116, CCT > 0.577465, EC > 0.074042, and LAC > 15.8 were identified to generate the final network (Figure 5C). Thirty-three target genes were eventually identified for GSE29746.
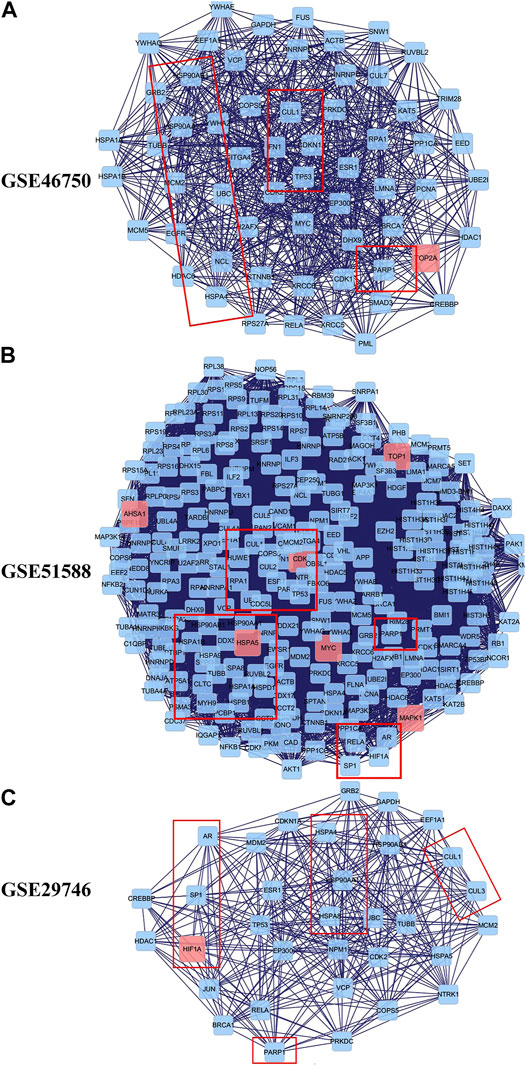
FIGURE 5. Identification of candidate targets for BZD activity against OA. The interactive PPI network topology analysis of BZD putative targets and OA-related targets were constructed for GSE46750 (A), GSE51588 (B), and GSE29746 (C).
We analyzed the GO and KEGG pathway using the Cluster Profiler R package to examine the 31 compound targets in GSE46750, 71 compound targets in GSE51588, and 15 compound targets in the GSE29746 group, as presented in Figure 4. For GSE46750, a total of 281 GO terms were significantly enriched (FDR < 0.05), with 252 associated with the BP, 6 with the CC, and 23 with MF categories. The highly enriched GO terms for the BP, CC, and MF included a response to lipopolysaccharide, response to a molecule of bacterial origin, collagen-containing extracellular matrix, condensed chromosome, endopeptidase activity, and metalloendopeptidase activity. For GSE51588, a total of 551 GO terms were significantly enriched (FDR < 0.05), with 501 associated with the BP, 23 with the CC, and 27 with MF categories. The highly enriched GO terms for the BP, CC, and MF included response to oxidative stress, response to nutrient levels, secretory granule lumen, cytoplasmic vesicle lumen, serine-type endopeptidase activity, and serine-type peptidase activity. For GSE29746, a total of 416 GO terms were significantly enriched (FDR < 0.05), with 378 associated with BP, 7 with the CC, and 31 with MF categories. The highly enriched GO terms for the BP, CC, and MF included regulation of membrane potential, response to alcohol, RNA polymerase II transcription factor complex, nuclear transcription factor complex, heat shock protein binding, and coenzyme binding. The top 20 terms are shown in Supplementary Figures S2–S4.
Figure 6 shows the pathways significantly influenced by BZD and OA, as identified by KEGG pathway analysis. For GSE46750, 41 considerably enriched pathways (FDR < 0.05) were identified, including the IL-17 signaling pathway, TNF signaling pathway, rheumatoid arthritis, serotonergic synapse, and cellular senescence. For GSE51588, 75 significantly enriched pathways (FDR < 0.05) were identified, including the AGE-RAGE signaling pathway associated with diabetic complications, hepatitis B, fluid shear stress, and atherosclerosis, transcriptional misregulation in cancer, and cellular senescence. For GSE29746, 44 significantly enriched pathways (FDR < 0.05) were identified, including Kaposi sarcoma-associated herpesvirus infection, human T-cell leukemia virus 1 infection, endocrine resistance, measles, and proteoglycans in cancer. Pathways common to the three series included IL-17, TNF, and NF-κB signaling.
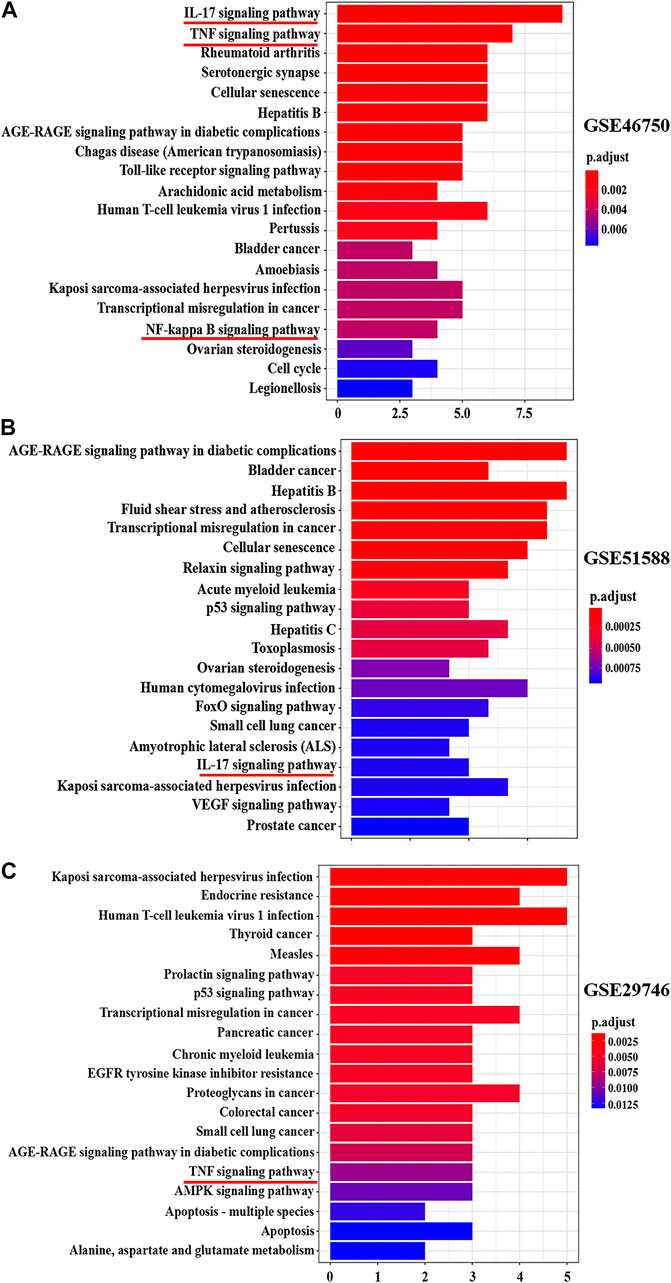
FIGURE 6. Top 20 KEGG pathway enrichment candidate targets for BZD activity against OA for each expression profile. Pathways with significant changes (FDR < 0.05) were identified. The vertical coordinates represent the KEGG pathway with significant enrichment, and the horizontal coordinates represent the number of differentially expressed genes in each pathway. The color of the bar graph indicates the significance of the enriched KEGG pathway, and the color gradient represents the size of the p-value. Inflammatory signaling pathways are underlined with a red bar. (A) GSE46750, (B) GSE51588, (C) GSE29746.
Figure 7 shows the gene-pathway network constructed based on significantly enriched pathways and genes. For GSE46750, the topological analysis involving 20 pathways and 26 genes was performed based on degree. The network diagram indicated that IL-6 had the highest degree and was, therefore, the core target gene. Several other genes also exhibited a large degree, including FOS, CXCL2, PTGS2, CD14, and E2F2 (Figure 7A). For GSE51588, the topological analysis involving 20 pathways and 39 genes was performed with a degree. The network diagram indicated that MAPK1 had the highest degree and was, therefore, the core target gene. Several other genes also exhibited a large degree, including MAPK14, CHUK, MYC, E2F2, and CYCS (Figure 7B). For GSE29746, the topological analysis involving 20 pathways and 13 genes was performed with a degree. The network diagram indicated that BAX had the highest degree and was, therefore, the core target gene. The other genes that also exhibited a high degree were CCND1, BCL2L1, and FOS (Figure 7C).
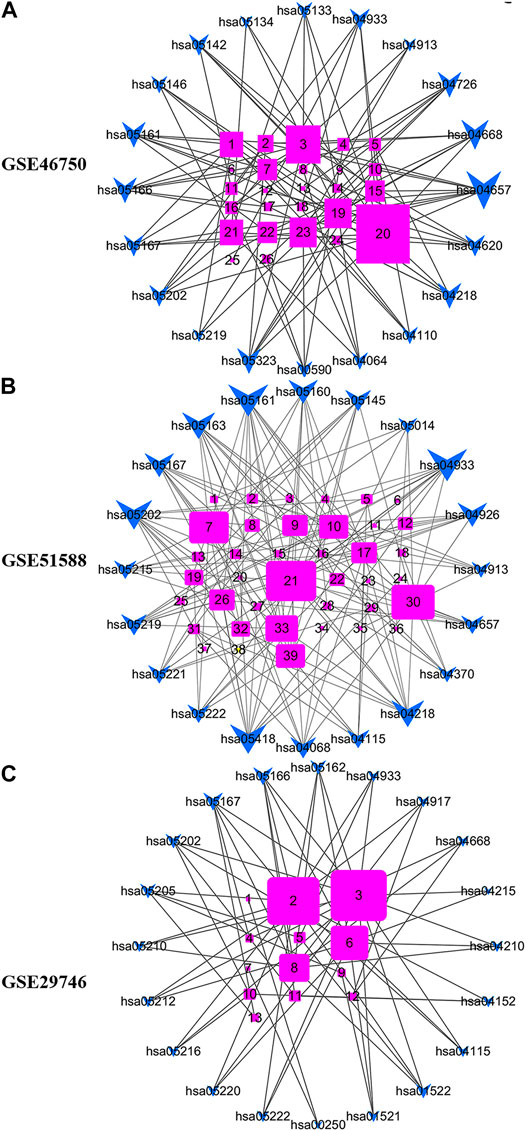
FIGURE 7. Gene-pathway network for BZD activity against OA. (A) For GSE46750, the topological analysis of 20 pathways and 26 genes was performed with degree. 1 = E2F2, 2 = MMP-3, 3 = FOS, 4 = ALOX5, 5 = CHEK1, 6 = BIRC5, 7 = CCNA2, 8 = LBP, 9 = MAOA, 10 = MMP-1, 11 = SERPINE1, 12 = SPP1, 13 = MAOB, 14 = C1QB, 15 = CCL2, 16 = PLA2G4A, 17 = FOSL1, 18 = COL1A1, 19 = CXCL2, 20 = IL6, 21 = CD14, 22 = MMP-9, 23 = PTGS2, 24 = CCNB1, 25 = F3, 26 = PTGS1. (B) For GSE51588, the topological analysis of 20 pathways and 39 genes was performed with degree. 1 = CAT, 2 = LDLR, 3 = MPO, 4 = THBD, 5 = IGFBP3, 6 = F3, 7 = CHUK, 8 = CCNA2, 9 = VEGFA, 10 = E2F2, 11 = AKR1C3, 12 = MMP-2, 13 = PLAT, 14 = MMP-1, 15 = COL1A1, 16 = ALOX5, 17 = CDK2, 18 = PLA2G4A, 19 = BCL2L1, 20 = GSTM2, 21 = MAPK1, 22 = PRKCB, 23 = BBC3, 24 = HMOX1, 25 = COL3A1, 26 = MMP-9, 27 = SELE, 28 = CXCL10, 29 = RUNX1T1, 30 = MAPK14, 31 = CCNB1, 32 = NFATC1, 33 = MYC, 34 = SLC2A4, 35 = CYP19A1, 36 = GRIA2, 37 = PPARG, 38 = RUNX2, 39 = CYCS. (C) For GSE29746, the topological analysis of 20 pathways and 13 genes was carried out with degree. 1 = GOT1, 2 = CCND1, 3 = BAX, 4 = CXCL2, 5 = MMP-2, 6 = BCL2L1, 7 = ABAT, 8 = FOS, 9 = KDR, 10 = PPARG, 11 = CCNA2, 12 = IRF1, 13 = HIF1A. The fuchsia squares represent target genes, and the blue quadrilateral arrow represents pathways. A large size represents a higher degree.
Figure 8 shows the results of the GO and KEGG analyses based on the Venn diagram analysis. Concerning the target gene analysis results, we found only one gene at the intersection of GSE46750, GSE51588, and GSE29746, namely CCNA2. Therefore, we enlarged the range of analysis to include the genes in the intersections of two profiles (showed as a red triangle). We associated 17 genes with the GO and KEGG analyses (Supplementary Figure S5), whereas three inflammatory-immune-related pathways were identified: IL-17, NF-κB, and TNF signaling pathways (Figure 8A). As for the results of the PPI network topology analysis action points, a total of 23 genes were located at the intersection of the three series and were subjected to GO and KEGG analysis (Supplementary Figures S6A–D). Three inflammatory-immune-related molecular functions were identified: NF-κB binding, MHC class II protein complex binding and MHC class I protein complex binding (Figure 8B). Twelve pathways converged at the intersection of GSE46750, GSE51588, and GSE29746. KEGG enrichment analysis revealed that they represented IL-17, NF-κB, and TNF signaling pathways (Supplementary Figure S6D). Based on the above results, we determined that the NF-κB signaling pathway would be the key target for further investigation.
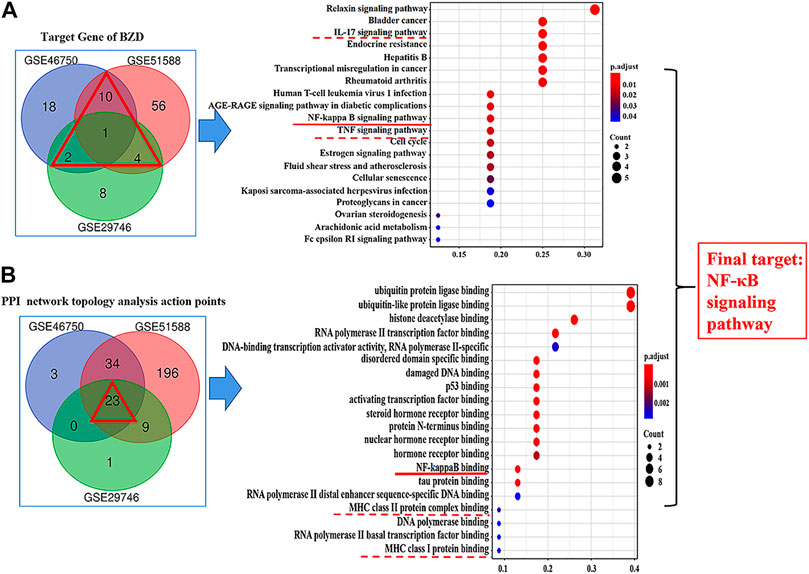
FIGURE 8. Results of the Gene ontology (GO) and KEGG analysis. (A) Results for the target genes of BZD. The intersection is shown by a red triangle, including 17 genes that were submitted to the GO and KEGG analysis. (B) Results for the PPI network topology analysis action points. The intersection is shown by a red triangle, including 23 genes that were submitted to the GO and KEGG analysis. The left column shows the results of the Venn diagram analysis. The right column shows the results of the KEGG pathway enrichment of candidate targets based on the Venn diagram analysis. The red under bars represent inflammatory-immune-related pathways. The vertical coordinates represent the KEGG pathway with significant enrichment, and the horizontal coordinates represent the number of differentially expressed genes in each pathway. The color of the bar graph indicates the significance of the enriched KEGG pathway, and the color gradient represents the size of the p-value. The size of each dot represents the number of genes.
The western blot analysis results revealed the expression of biomarkers affected by LPS, BZD, and PDTC, an inhibitor of the NF-κB signaling pathway. NF-κB P65, IKK-α, IKK-β, and MMP-3 exhibited similar expression patterns. They were upregulated by LPS exposure and downregulated by treatment with BZD and PDTC. The BZD group exhibited significant differences in all genes, whereas PDTC exhibited significant differences in NF-κB p65, IKK-β, and MMP-3. By contrast, Collagen II was downregulated by LPS exposure but upregulated by BZD and PDTC. Only the BZD group exhibited a significant difference. Therefore, the involvement of the NF-κB signaling pathway was confirmed (Figure 9).
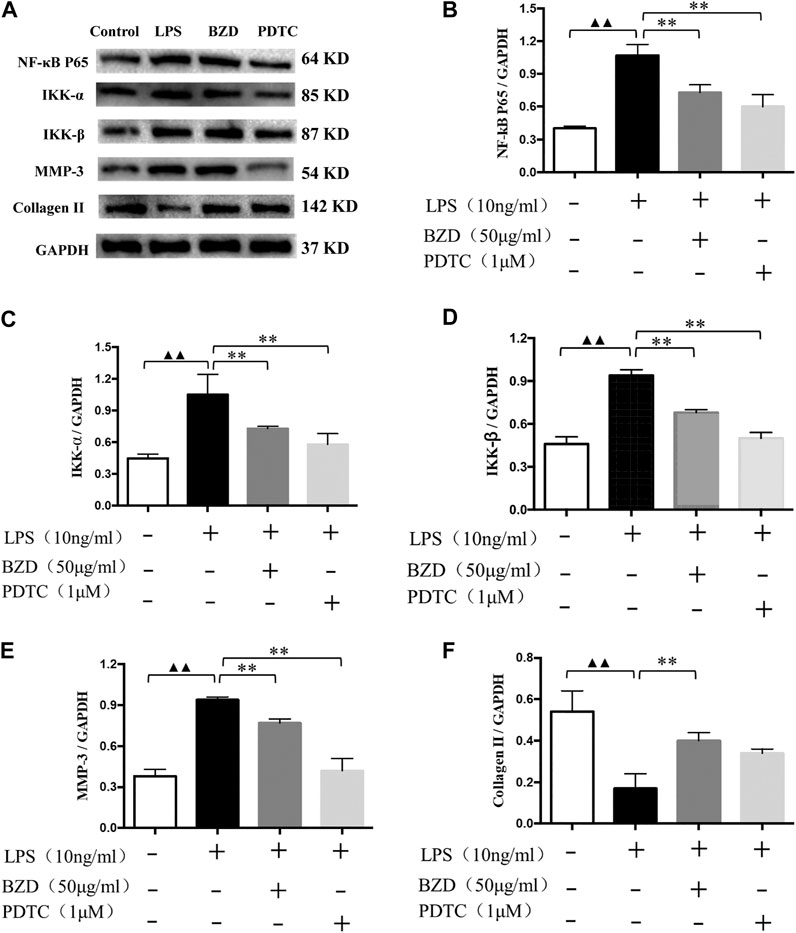
FIGURE 9. Verification of the targeted NF-κB signaling pathway. (A) Representative images of the western blot analysis. (B–F) Protein expression affected by LPS, BZD, and PDTC. LPS significantly upregulated the expression of NF-κB p65 (B), IKK-α (C), IKK-β (D), MMP-3 (E). These proteins were significantly downregulated by the administration of BZD. Treatment of the positive control with PDTC downregulated these genes and exhibited significant effects on NF-κB p65, IKK-β, and MMP-3. LPS significantly downregulated the expression of Collagen II (F). It was upregulated considerably by BZD treatment. Treatment with PDTC exhibited the same tendency but did not reach a significant difference. ▲▲ means p < 0.01, vs. intact control; ** means p < 0.01, vs. LPS-exposed chondrocytes.
In this study, we tested our hypothesis, namely NF-κB signaling pathway plays a key role in the mechanisms of BZD for treating OA by comparing the NPA results with conventional methods. In the first experiment, we identified the components of BZD using HPLC. In the second experiment, we performed a preliminary observation regarding the therapeutic effects of BZD in an OA cellular model created by LPS exposure. We identified several inflammatory-related genes that are activated by LPS exposure and suppressed by BZD. These genes are associated with different mechanisms; thus, the preliminary observation indicated that the complicated and multifold mechanisms involved in the effects of BZD on OA might be associated with multi-components, multi-targets, and multi-pathways. In the third experiment, we performed a standard NPA for BZD and identified 98 candidate compounds that were associated with OA. KEGG and GO enrichment analyses identified three inflammatory signaling pathways, IL-17, TNF, and NF-κB that were associated with BZD activity. We then performed a Venn diagram analysis consisting of the target genes and the PPI network topology analysis action point. The results indicated that the NF-κB signaling pathway plays a key role in the effects of BZD. These results were verified in the fourth experiment by indirectly verifying NF-κB signaling activity by western blot analysis. We found that BZD indeed contributed to reduced expression of NF-κB related genes, which were, conversely, upregulated by LPS exposure. Thus, the role of the NF-κB signaling pathway was confirmed. To our knowledge, this is the first study to utilize conventional experiments in combination with NPA to identify the key role of NF-κB signaling pathway in the trerapeutic effects of BZD against OA. These findings contribute to better understanding the pharmacological mechanisms of BZD for treating OA. Meanwhile, Our results indicate that NPA is a powerful tool to identify molecular targets incomplex herbal formulation commonly used in TCM.
It is known that expression of cytokines like MMP-9 and IL-6 is under the control of NF-κB signaling pathway (Epanchintsev et al., 2015). Our previous study using LPS-treated chondrocytes also found that MMP-9 and IL-6 were significantly upregulated by LPS exposure (Li et al., 2020b). We therefore believe that upregulation (downregulation) of MMP-9 and IL-6 can be used as an “indicator” of activation (suppression) of the NF-κB signaling pathway. We therefore first observed the changes of MMP-9 and IL-6. Our preliminary results revealed that MMP-9 and IL-6 levels were significantly increased by LPS exposure and decreased by BZD (Figure 1). MMP-9 and IL-6 are downregulated factors of NF-κB signaling pathway, perpetuating in OA by modulating inflammatory and catabolic mediators. MMP-9 degrades the collagenase fragments of Collagen II (Alves et al., 2014) by activation of the NF-κB signaling pathway. IL-6, as a pro-inflammatory cytokines, is also relevant to activation of the NF-κB signaling pathway (Pistolic et al., 2009; Liu et al., 2015). The changes of MMP-9 and IL-6 were in agreement with our previous studies in rat (Li et al., 2020b) and in rabbit (Wu G. et al., 2019), which contribute to exacerbating progression of OA. In addition, TNF-α, IL-1β, MyD88, MMP-3, TLR4, and NF-κB p65 were significantly upregulated by LPS, but suppressed by BZD treatment (Figure 2). It is known that IL-6, IL-1β, and TNF-α can induce the expression of MMPs, particularly MMP-3, MMP-9, and MMP-13, and inhibits the synthesis of Collagen II. These proteins play pivotal roles in cartilage matrix degeneration in OA. Pro-inflammatory cytokines such as IL-1β and TNF-α may mediate chondrocyte degeneration, which is associated with OA (Kapoor et al., 2011; Fei et al., 2019). MMP-3 directly degrades the extracellular matrix and indirectly affects the degeneration of the extracellular matrix by activating other latent MMPs (Honsawek et al., 2013). TLR4, MyD88, and NF-κB p65 represent another group of inflammatory-related genes that are associated with the TLR4/MyD88/NF-κB signaling pathway. Activation of TLR4 stimulates the MyD88 adaptor protein, resulting in IκB kinase activation and phosphorylation. This causes the release of cytosolic sequestered NF-κB p65 subunits and their translocation to the nucleus. NF-κB p65 activates gene transcription and protein synthesis of various inflammatory factors to regulate the inflammatory response. Changes in the expression of these genes by BZD exposure indicate that its effects and mechanisms of action are complicated and pleiotropic. However, the changes of MMP-9, IL-6, and the related genes stongly implied the key role of the NF-κB signaling pathway. We therefore further investigated the relevant genes and pathways using NPA.
Our analysis of the Gene-pathway network suggests that IL-6, MAPK1, and BAX exhibited the maximum degree, and may represent core targets for the GSE46750, GSE51588 and GSE29746 profiles. The other top three genes for GSE46750 (FOS, CXCL2, and PTGS2), GSE51588 (MAPK14, CHUK, and MYC), and GSE29746 (CCND1, BCL2L1, and FOS) were selected as key target genes (Figure 7). Comparing the results of experiment 2 with NPA, we confirmed increased levels of MMP-3, MMP-9, and IL-6 in LPS-exposed chondrocytes, whereas BZD decreased all levels of the proteins. MMP-3 was excluded from the top genes of GSE46750, and MMP-9 was excluded from GSE46750 and GSE51588. Genes belonging to the MMP family were identified as key targets of BZD in all three profiles. Moreover, further GO analysis revealed that these genes were associated with metalloendopeptidase and metallopeptidase activity, as shown in Supplementary Figure S5C. The other genes should be verified in future studies.
From the preliminary results of experiment 2, we supposed the key role of the NF-κB signaling pathway in the effects of BZD against OA, thus we performed the KEGG pathway enrichment analysis. The results identified 41 pathways in GSE46750, 75 pathways in GSE51588, and 44 pathways in the GSE29746 profiles. Inflammatory-related signaling pathways, including IL-17, TNF, and NF-κB signaling pathways, were identified from the intersection of the three profiles (Supplementary Figure S6D). The IL-17, TNF, and NF-κB signaling pathways were included in the top 20 signaling pathways of GSE46750, the IL-17 signaling pathway was included in the top 20 signaling pathways of GSE51588, and the TNF signaling pathway was included in the top 20 signaling pathways of GSE29746 (Figure 6). Thus, the IL-17, TNF, and NF-κB signaling pathways were identified as the top inflammatory-related signaling pathways associated with OA. With respect to the interactions of these pathways, activation of IL-17 triggers the activation of NF-κB (Schwandner et al., 2000). Similarly, activation of NF-κB is mediated by the activation of TNF (Jackson-Bernitsas et al., 2007), which subsequently induces activation of NF-κB (Mukhopadhyay et al., 2002; Jaco et al., 2017). Therefore, we hypothesized that the NF-κB signaling pathway is downstream of the IL-17 and TNF signaling pathways. The results of the Venn diagram analysis between the target gene and PPI network topology analysis action point also found that only the NF-κB signaling pathway was common to both analyses (Figure 8). Based on these results, we confirmed that the NF-κB signaling pathway contributes to the observed effects of LPS, BZD, and PDTC by measuring the expression of three proteins associated with the NF-κB signaling pathway. A final verification was performed using a traditional western blot analysis (Experiment 4). The expression of NF-κB p65, IKK-β, and MMP-3 was attenuated by BZD and PDTC treatment, thus confirming a key role for the NF-κB signaling pathway in the effects of BZD in OA (Figure 9).
We first used three OA-related expression profiles for NPA: GSE46750, GSE51588, and GSE29746. The compound–target networks for BZD were constructed using 44, 35, and 23 compounds, and 31, 71, and 15 compound targets for GSE46570, GSE51588, and GSE29746. Quercetin, wogonin, baicalein, and nobiletin acted on 21, 6, 6, and 6 targets in GSE4750, respectively. Quercetin, baicalein, kaempferol, and wogonin acted on 40, 13, 12, and 9 targets in GSE51588, respectively. Lastly, quercetin, wogonin, naringenin, and baicalein acted on 10, 5, 4, and 3 targets in GSE29746, respectively. Therefore, quercetin, wogonin, and baicalein were considered to be crucial pleiotropically active compounds associated with BZD activity. However, these compounds were not detected in BZD by HPLC analysis.
The PPI networks established for the BZD putative targets and OA-related targets were structured and merged to obtain candidate targets for BZD activity in OA. To achieve more accurate targets, 3, 6, and 5 parameters, including DC and BC, were set to screen nodes, and then to structure them into a new network. Finally, 60, 262, and 33 targets were identified from the GSE46750, GSE51588, and GSE29746 profiles, respectively (Figure 5). There were 23 common action points in the three working networks, including CUL, TP, HSP, and RARP types (Figure 8B).
Biological information on putative BZD targets was analyzed. The targets of BZD anti-OA activity included genes associated with the BP, CC, and MF by GO enrichment analysis. Our data also revealed that BZD regulates some BPs. For example, an effect on reducing cartilage degeneration may be associated with the suppression of the inflammatory response (Figure 6; Supplementary Figures S2–S4). A gene-pathway network was then constructed to investigate the core and key target genes for BZD anti-OA activity (Figure 7). Finally, a Venn diagram analysis was performed to identify the intersections shared among the three OA-related profiles.
Monoglucoside, paeoniflorin, and asperosaponin VI were selected as quality control markers for the BZD extract because they have been verified as quality control markers for FC, RPA, and RD in the pharmacopoeia of the People’s Republic of China (2015 version). Our HPLC results identified monoglucoside, paeoniflorin, and asperosaponin VI (Supplementary Figure S1), confirming these three compounds as quality control markers for the BZD extract. NPA identified 98 candidate compounds for the GSE46750, GSE51588, and GSE29746 profiles (Table 1) that were associated with OA. Interestingly, we found that paeoniflorin was included from the list of these candidate compounds, despite representing a core role in the effects of BZD on OA. The OB and DL values of paeoniflorin were 53.87 and 0.79, respectively.
Monoglucoside and asperosaponin VI were not included in the list of these candidate compounds. This result indicates that conventional methods, such as simply selecting certain compounds listed in the pharmacopoeia as pharmacologic quality control markers, may have certain limitations. Our findings suggest that the following processes may be a more reasonable method to identify the pharmacologic quality of control markers, particularly for complex herbal formulations with many ingredients. First, use NPA to identify the core bioactive compounds. Then, compare these compounds with the classical pharmacopoeia. Finally, identify the pharmacologic quality control markers. These processes require further verification in future studies using more complex herbal formulations.
Overall, the results of NPA, along with the conventional experiments, suggest that the mechanisms of BZD are complex and include multiple compounds and pathways, with NF-κB signaling playing a key role. Suppression of the NF-κB signaling pathway might be a key mechanism related to chondrocyte apoptosis, which is associated with cartilage degeneration.
In the present study, we used a set of four sequential experiments to identify and verify the key role of the NF-κB signaling pathway in the BZD efficacy against OA. Our results also indicate that NPA is a powerful toolfor exploring the molecular targets of complex herbal formulations and is useful to guide future studies of “target” compounds, genes, and pathways.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Animal Care and Use Committee of the Fujian University of TCM (Approval number: 2020015).
XL and TA designed the study. YX, HL, XH, YH, SW, LW, CF, HY, XL, and TA carried out the experiments and performed data analysis. YX, HL, XL, and TA wrote the manuscript. TA revised the manuscript. All of the authors have read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (Grant No. 81873319), the Project Funded by China Postdoctoral Science Foundation (Grant Nos. 2016M600625 and 2017T100591). This study is also supported by grants from the Japan Society for the Promotion of Science (Nos. 20791025, 24592157, 15k10358, and 18K08991).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Enago (www.enago.jp) for the English language review.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.637273/full#supplementary-material.
Alfarano, C., Andrade, C. E., Anthony, K., Bahroos, N., Bajec, M., Bantoft, K., et al. (2005). The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 33, D418–D424. doi:10.1093/nar/gki051
Alves, A. C., Albertini, R., dos Santos, S. A., Leal-Junior, E. C., Santana, E., Serra, A. J., et al. (2014). Effect of low-level laser therapy on metalloproteinase MMP-2 and MMP-9 production and percentage of collagen types I and III in a papain cartilage injury model. Lasers Med. Sci. 29 (3), 911–919. doi:10.1007/s10103-013-1427-x
Ashraf, S., Mapp, P. I., Shahtaheri, S. M., and Walsh, D. A. (2018). Effects of carrageenan induced synovitis on joint damage and pain in a rat model of knee osteoarthritis. Osteoarthr. Cartil. 26 (10), 1369–1378. doi:10.1016/j.joca.2018.07.001
Bouderlique, T., Vuppalapati, K. K., Newton, P. T., Li, L., Barenius, B., and Chagin, A. S. (2016). Targeted deletion of Atg5 in chondrocytes promotes age-related osteoarthritis. Ann. Rheum. Dis. 75 (3), 627–631. doi:10.1136/annrheumdis-2015-207742
Choi, M. C., Jo, J., Park, J., Kang, H. K., and Park, Y. (2019). NF-κB signaling pathways in osteoarthritic cartilage destruction. Cells 8 (7), 734. doi:10.3390/cells8070734
Clough, E., and Barrett, T. (2016). The gene expression omnibus database. Methods Mol. Biol. 1418, 93–110. doi:10.1007/978-1-4939-3578-9_5
Epanchintsev, A., Shyamsunder, P., Verma, R. S., and Lyakhovich, A. (2015). IL-6, IL-8, MMP-2, MMP-9 are overexpressed in Fanconi anemia cells through a NF-κB/TNF-α dependent mechanism. Mol. Carcinog. 54 (12), 1686–1699. doi:10.1002/mc.22240
Fei, J., Liang, B., Jiang, C., Ni, H., and Wang, L. (2019). Luteolin inhibits IL-1?-induced infammation in rat chondrocytes and attenuates osteoarthritis progression in a rat model. Biomed. Pharmacother. 109, 1586–1592. doi:10.1016/j.biopha.2018.09.161
Goel, R., Harsha, H. C., Pandey, A., and Prasad, T. S. (2012). Human protein reference database and human proteinpedia as resources for phosphoproteome analysis. Mol. Biosyst. 8 (2), 453–463. doi:10.1039/c1mb05340j
Gu, H. Y., Yang, M., Guo, J., Zhang, C., Lin, L. L., Liu, Y., et al. (2019). Identification of the biomarkers and pathological process of osteoarthritis: weighted gene co-expression network analysis. Front. Physiol. 10, 275. doi:10.3389/fphys.2019.00275
Guan, Y., and Zhong, Y. (2017). Clinical outcome of patients with osteoarthritis treated by the Bushen Zhuangjin Decoction. Doctor 7, 60–61. [in Chinese, with English summary]. doi:10.19604/j.cnki.dys.2017.07.033
Honsawek, S., Malila, S., Yuktanandana, P., Tanavalee, A., Deepaisarnsakul, B., and Parvizi, J. (2013). Association of MMP-3 (-1612 5A/6A) polymorphism with knee osteoarthritis in Thai population. Rheumatol. Int. 33 (2), 435–439. doi:10.1007/s00296-012-2371-y
Huang, S., Lin, J., and Li, X. (2017). Systematic review on treating knee osteoarthritis with Bushen Zhuangjin decoction. Rehabil. Med. 27 (3), 56 [in Chinese, with English summary]. doi:10.3724/SP.J.1329.2017.03056
Jackson-Bernitsas, D. G., Ichikawa, H., Takada, Y., Myers, J. N., Lin, X. L., Darnay, B. G., et al. (2007). Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-kappaB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene 26 (10), 1385–1397. doi:10.1038/sj.onc.1209945
Jaco, I., Annibaldi, A., Lalaoui, N., Wilson, R., Tenev, T., Laurien, L., et al. (2017). MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol. Cell 66 (5), 698–710. doi:10.1016/j.molcel.2017.05.003
Jimi, E., Fei, H., and Nakatomi, C. (2019). NF-κB signaling regulates physiological and pathological chondrogenesis. Int. J. Mol. Sci. 20 (24), 6275. doi:10.3390/ijms20246275
Kanehisa, M. (2002). “The KEGG database,” in Novartis Found Symp. Chichester, New York: John Wiley, 1999, Vol. 247, 91–52.
Kapoor, M., Martel-Pelletier, J., Lajeunesse, D., Pelletier, J. P., and Fahmi, H. (2011). Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7 (1), 33–42. doi:10.1038/nrrheum.2010.196
Kerrien, S., Aranda, B., Breuza, L., Bridge, A., Broackes-Carter, F., Chen, C., et al. (2012). The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40, D841–D846. doi:10.1093/nar/gkr1088
Kwon, J. Y., Lee, S. H., Na, H. S., Jung, K., Choi, J., Cho, K. H., et al. (2018). Kartogenin inhibits pain behavior, chondrocyte inflammation, and attenuates osteoarthritis progression in mice through induction of IL-10. Sci. Rep. 8 (1), 13832. doi:10.1038/s41598-018-32206-7
Lepetsos, P., Papavassiliou, K. A., and Papavassiliou, A. G. (2019). Redox and NF-κB signaling in osteoarthritis. Free Radical Biol. Med. 132, 90–100. doi:10.1016/j.freeradbiomed.2018.09.025
Li, H., Hao, Z., Zhao, L., Liu, W., Han, Y., Bai, Y., et al. (2016). Comparison of molecular mechanisms of rheumatoid arthritis and osteoarthritis using gene microarrays. Mol. Med. Rep. 13 (6), 4599–4605. doi:10.3892/mmr.2016.5144
Li, X., Chen, J., Liang, W., Li, H., Liu, F., Weng, X., et al. (2015). Bushen Zhuangjin decoction promotes chondrocyte proliferation by stimulating cell cycle progression. Exp. Ther. Med. 9 (3), 839–844. doi:10.3892/etm.2015.2214
Li, X., Liang, W., Dang, C., Ye, H., Zheng, C., Wu, G., et al. (2014). Empirical study on Bushen Zhuangjin decoction inhibiting inflammatory cytokine expression experiments to delay the degeneration of articular cartilage. Rheum. Arthritis 3 (5), 20–25. [in Chinese, with English summary]. doi:10.3969/j.issn.2095-4174.2014.05.005
Li, X., Zhang, Z., Liang, W., Zeng, J., Shao, X., Xu, L., et al. (2020a). Data on Tougu Xiaotong capsules may inhibit p38 MAPK pathway-mediated inflammation in vitro. Data Brief 28, 105023. doi:10.1016/j.dib.2019.105023
Li, X., Zhang, Z., Liang, W., Zeng, J., Shao, X., Xu, L., et al. (2020b). Tougu Xiaotong capsules may inhibit p38 MAPK pathway-mediated inflammation: in vivo and in vitro verification. J. Ethnopharmacol. 249, 112390. doi:10.1016/j.jep.2019.112390
Li, Y., Wang, W., and Qiu, Y. (2020). Effect of the reinforcing kidney and strengthening tendon decoction combined with glucosamine hydrochloride on the proliferation and apoptosis of knee chondrocytes in rats. Chin. J. Osteoporos. 26 (1), 25–30. doi:10.3969/j.issn.1006-7108.2020.01.006
Liang, J., Liu, W., Han, P., Qiu, Y., and Luo, Q. (2015). Influence of Bushen Zhuangjin decoction on cartilage morphology and skeleton proteins of ROCK, cofilin, phospho-cofilin, LIMK1 and phospho-LIMK1 in rabbits with knee osteoarthritis. J. Tradit. Chin. Med. Pharma. 10, 3732–3735. [in Chinese, with English summary].
Liang, W., Li, X., Hu, L., Ding, S., Kang, J., Shen, J., et al. (2019). An in vitro validation of the therapeutic effects of Tougu Xiaotong capsule on tunicamycin-treated chondrocytes. J. Ethnopharmacol. 229, 215–221. doi:10.1016/j.jep.2018.10.022
Licata, L., Briganti, L., Peluso, D., Perfetto, L., Iannuccelli, M., Galeota, E., et al. (2012). MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40 (D1), D857–D861. doi:10.1093/nar/gkr930
Lin, J., Wu, G., Zhao, Z., Huang, Y., Chen, J., Fu, C., et al. (2018). Bioinformatics analysis to identify key genes and pathways influencing synovial inflammation in osteoarthritis. Mol. Med. Rep. 18 (6), 5594–5602. doi:10.3892/mmr.2018.9575
Lin, M., Zhang, L., Wu, W., Wang, W., Zhu, D., Huang, S., et al. (2017). Clinical study on the treatment of liver-kidney deifciency type knee osteoarthritis with Bushen Zhuangjin tang. Rheum. Arthritis 6 (2), 15–17. [in Chinese, with English summary]. doi:10.3969/j.issn.2095-4174.2017.02.003
Lin, P., Weng, X., Liu, F., Ma, Y., Chen, H., Shao, X., et al. (2015). Bushen Zhuangjin decoction inhibits TM-induced chondrocyte apoptosis mediated by endoplasmic reticulum stress. Int. J. Mol. Med. 36 (6), 1519–1528. doi:10.3892/ijmm.2015.2387
Liu, H., Zeng, L., Yang, K., and Zhang, G. (2016). A network pharmacology approach to explore the pharmacological mechanism of xiaoyao powder on anovulatory infertility. Evid. Based Complement. Altern. Med. 2016, 2960372. doi:10.1155/2016/2960372
Liu, X., Ye, F., Xiong, H., Hu, D. N., Limb, G. A., Xie, T., et al. (2015). IL-1β induces IL-6 production in retinal Müller cells predominantly through the activation of p38 MAPK/NF-κB signaling pathway. Exp. Cell Res. 331 (1), 223–231. doi:10.1016/j.yexcr.2014.08.040
Mi, J. (2013). Clinical observation of efficacy of Bushen Zhuangjin Decoction in treating patients with osteoarthritis. Changsha, China: Hunan University of Chinese Medicine.
Miyaki, S., and Asahara, H. (2012). Macro view of microRNA function in osteoarthritis. Nat. Rev. Rheumatol. 8 (9), 543–552. doi:10.1038/nrrheum.2012.128
Mobasheri, A., Matta, C., Zákány, R., and Musumeci, G. (2015). Chondrosenescence: definition, hallmarks and potential role in the pathogenesis of osteoarthritis. Maturitas 80 (3), 237–244. doi:10.1016/j.maturitas.2014.12.003
Mukhopadhyay, A., Shishodia, S., Suttles, J., Brittingham, K., Lamothe, B., Nimmanapalli, R., et al. (2002). Ectopic expression of protein-tyrosine kinase Bcr-Abl suppresses tumor necrosis factor (TNF)-induced NF-kappa B activation and IkappaBalpha phosphorylation. Relationship with down-regulation of TNF receptors. J. Biol. Chem. 277 (34), 30622–30628. doi:10.1074/jbc.M204748200
Pistolic, J., Cosseau, C., Li, Y., Yu, J. J., Filewod, N. C., Gellatly, S., et al. (2009). Host defence peptide LL-37 induces IL-6 expression in human bronchial epithelial cells by activation of the NF-κB signaling pathway. J. Innate Immun. 1 (3), 254–267. doi:10.1159/000171533
Pomaznoy, M., Ha, B., and Peters, B. (2018). GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinform. 19 (1), 1–8. doi:10.1186/s12859-018-2533-3
Rigoglou, S., and Papavassiliou, A. G. (2013). The NF-κB signalling pathway in osteoarthritis. Int. J. Biochem. Cell Biol. 45 (11), 2580–2584. doi:10.1016/j.biocel.2013.08.018
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 6, 1–6. doi:10.1186/1758-2946-6-13
Saito, T., and Tanaka, S. (2017). Molecular mechanisms underlying osteoarthritis development: notch and NF-κB. Arthritis Res. Ther. 19 (1), 1–7. doi:10.1186/s13075-017-1296-y
Schwandner, R., Yamaguchi, K., and Cao, Z. (2000). Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J. Exp. Med. 191 (7), 1233–1240. doi:10.1084/jem.191.7.1233
Stark, C., Breitkreutz, B. J., Reguly, T., Boucher, L., Breitkreutz, A., and Tyers, M. (2006). BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539. doi:10.1093/nar/gkj109
Wu, F., Gao, H., Wang, G., Li, J., Huang, S., Jiang, X., et al. (2020). Protective mechanism of Bushen Zhuangjin decoction containing serum on IL-1β-induced chondrocyte injury in rats by overexpression of Sox9. J. Med. Res. 49 (7), 96–99. [in Chinese, with English summary]. doi:10.11969/j.issn.1673-548X.2020.07.021
Wu, G., Zhang, J., Chen, W., Chen, S., Huang, Y., Lin, R., et al. (2019). Tougu Xiaotong capsule exerts a therapeutic effect on knee osteoarthritis by regulating subchondral bone remodeling. Mol. Med. Rep. 19 (3), 1858–1866. doi:10.3892/mmr.2018.9778
Wu, Y., Zhang, F., Yang, K., Fang, S., Bu, D., Li, H., et al. (2019). SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 47 (D1), D1110–D1117. doi:10.1093/nar/gky1021
Xenarios, I., Salwinski, L., Duan, X. J., Higney, P., Kim, S. M., and Eisenberg, D. (2002). DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30 (1), 303–305. doi:10.1093/nar/30.1.303
Yang, B., Tian, C., Zhang, Z., and Chai, X. (2020). Regulatory effect of Zhuanggu Huoxue decoction on serum TNF-alpha and IL-6 in rats with knee osteoarthritis. Contemp. Med. Forum 18 (10), 203–206. [in Chinese, with English summary].
Yao, X., Li, H., Song, Y., Liu, Y., and Cai, Y. (2005). [An influence of modified “bushen zhuangjin tang” on the oxygen—free—radical metabolism in knee osteoarthritis: an experimental study]. J. Tradit. Chin. Orthop. Traumatol. 17 (9), 5–6. [in Chinese, with English summary]. doi:10.3969/j.issn.1001-6015.2005.09.002
Yazici, Y., McAlindon, T. E., Fleischmann, R., Gibofsky, A., Lane, N. E., Kivitz, A. J., et al. (2017). A novel Wnt pathway inhibitor, SM04690, for the treatment of moderate to severe osteoarthritis of the knee: results of a 24-week, randomized, controlled, phase 1 study. Osteoarthr. Cartil. 25 (10), 1598–1606. doi:10.1016/j.joca.2017.07.006
Zeng, Q., Li, L., Siu, W., Jin, Y., Cao, M., Li, W., et al. (2019). A combined molecular biology and network pharmacology approach to investigate the multi-target mechanisms of chaihu shugan san on Alzheimer’s disease. Biomed. Pharmacother. 120, 109370. doi:10.1016/j.biopha.2019.109370
Zhang, B. (2006). Effects of Bushen Zhuangjin decoction on serum levels of TNF-α and IL-6 in patients with osteoarthritis. Chin. Tradit. Herbal Drugs 9, 1389–1390. [in Chinese, with English summary]. doi:10.7501/j.issn.0253-2670.2006.9.611
Keywords: network pharmacology approach, complex herbal formulations, molecular targets, osteoarthritis, Bushen Zhuangjin decoction, NF-κB signaling pathway
Citation: Xu Y, Li H, He X, Huang Y, Wang S, Wang L, Fu C, Ye H, Li X and Asakawa T (2021) Identification of the Key Role of NF-κB Signaling Pathway in the Treatment of Osteoarthritis With Bushen Zhuangjin Decoction, a Verification Based on Network Pharmacology Approach. Front. Pharmacol. 12:637273. doi: 10.3389/fphar.2021.637273
Received: 03 December 2020; Accepted: 11 February 2021;
Published: 12 April 2021.
Edited by:
Yuanjia Hu, University of Macau, ChinaReviewed by:
Jinfan Tian, Capital Medical University, ChinaCopyright © 2021 Xu, Li, He, Huang, Wang, Wang, Fu, Ye, Li and Asakawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xihai Li, bGl4aWhhaWZ6QDE2My5jb20=; Tetsuya Asakawa, YXNha2F3YXQxOTcxQGdtYWlsLmNvbQ==
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.