- 1Medical Biology Research Center, Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
- 2Molecular Virology Lab, Department of Microbiology, School of Biology, College of Science, University of Tehran, Tehran, Iran
- 3Department of Hematology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
- 4Medical Technology Research Center, Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
- 5Institute of Biodiversity and Ecosystem Research, Bulgarian Academy of Sciences, Sofia, Bulgaria
- 6Departamento de Ciencias del Ambiente, Facultad de Química y Biología, Universidad de Santiago de Chile, Santiago, Chile
The SARS-CoV-2 virus was reported for the first time in Wuhan, Hubei Province, China, and causes respiratory infection. This pandemic pneumonia killed about 1,437,835 people out of 61,308,161cases up to November 27, 2020. The disease’s main clinical complications include fever, recurrent coughing, shortness of breath, acute respiratory syndrome, and failure of vital organs that could lead to death. It has been shown that natural compounds with antioxidant, anticancer, and antiviral activities and RNA interference agents could play an essential role in preventing or treating coronavirus infection by inhibiting the expression of crucial virus genes. This study aims to introduce a summary of coronavirus’s genetic and morphological structure and determine the role of miRNAs, siRNAs, chemical drugs, and natural compounds in stimulating the immune system or inhibiting the virus’s structural and non-structural genes that are essential for replication and infection of SARS-CoV-2.
Introduction
Over the past 50 years, a wide range of human and animal diseases have been caused by coronaviruses (CoVs). Since the emergence of Severe Acute Respiratory Syndrome (SARS-CoV) Coronavirus in 2003, a considerable number of new Human coronaviruses (H-CoVs) have been identified (Ding et al., 2003). The appearance of Middle East Respiratory Syndrome (MERS-CoV) coronavirus in 2012 and the continuous occurrence of human cases further intensified attention to the importance of studying these viruses from different aspects (Centers for Disease Control and Prevention, 2012). Tolerance of new mutations and recombination has led to the evolution of this group of viruses, giving them the ability to transmit between species. In most cases, CoV infections are self-limiting, and after completing a reasonable period, the body recovers from the illness (Enjuanes et al., 2000). However, some strains of this family cause severe infections and have been the cause of widely spread epidemics during the last two decades (Mahase, 2020).
The SARS-CoV-2 virus was reported for the first time in Wuhan, Hubei Province, China, and causes respiratory infection. According to WHO statistics, on November 27, 2020, Coronavirus disease 2019 (COVID-19) killed about 1,437,835 people out of 61,308,161cases, and the most significant number of deaths and infected people were in the Americas (Chen et al., 2020b). In recent years, many treatment strategies such as prescribing antibiotics, the application of antiviral drugs (including Human Immunodeficiency Virus 1 (HIV-1) protease inhibitors, oseltamivir, and ribavirin), different corticosteroids, interferons, and natural human immunoglobulin have been used for treating patients suffering from H-COVs. Recently, most treatment strategies for CoVs such as MERS-CoV, SARS-CoV, and Severe Acute Respiratory Syndrome coronavirus-2 (SARS-CoV-2) have been based on inhibiting viral agents involved in the replication, infection, or induction of host immune system agents to combat viral infection (Li and De Clercq, 2020; Prajapat et al., 2020). Numerous studies are being conducted today to prove the role of various biomolecules, including RNA interference (RNAi), fusion inhibitors, neutralizing antibodies, plant, or microbial metabolites, as antiviral compounds (Prajapat et al., 2020).
This review article tries to present some information about the biology, replication, infusion, and pathogenesis of coronavirus. Also, it has decided to explain the existing treatment strategies based on chemical drugs, natural compounds, small interfering RNA (siRNAs), and microRNA (miRNAs) that can be used to fight against coronavirus infection, especially SARS-CoV-2. For this purpose, keywords including natural product, flavonoid, polyphenols, phytochemicals, microRNA, siRNA, Coronavirus, COVID-19 up to August 2020 were searched and evaluated using Scopus, PubMed, WOS, and Google Scholar databases.
Coronavirus
Coronavirus Classification and Taxonomy
Genome organization, genome homology, reproduction strategies in a host, and the virion’s structural traits are criteria for CoVs classification by the International Committee for Taxonomy of Viruses (ICTV) (Casals et al., 1980). In terms of phylogenetics, CoVs belong to Coronaviridae, as the class of Nidovirals. Coronaviridae family includes two sub-families: Orthocoronavirinae and Torovirinae. The sub-family of Orthocoronavirinae consists of four genera: Alpha, Beta, Gamma, and Delta coronaviruses that are responsible for infection on a vast range of hosts, from mammalians to birds (King et al., 2012). Human infection with CoVs was first reported in 1965 (Hamre and Procknow, 1966). H-CoV 229E and CoV-NL63 are human pathogens of the genus Alpha-CoV, causing common cold (Mcintosh et al., 1970; Monto, 1974). Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-CoV-1), MERS-CoV, and SARS-CoV-2 are phylogenetically classified in the Beta-CoVs genus and have caused a high percentage of mortality in the human population during the last two decades (Zhou et al., 2020). The most common CoVs genes used for phylogenetic studies are nsp12 (RNA dependent RNA polymerase), nsp5 (chymotrypsin-like protease), nsp13 (helicase), nucleocapsid (N), and spike protein (S) (ICTV, 2009).
Genomic Structure
CoVs are positive single-strand RNA with 32–46% G + C content. They have the biggest genome size among all known RNA viruses (about 26.4–31.7 kb) (Lai and Cavanagh, 1997; Enjuanes et al., 2000). Depending on the strain, the genome in CoVs contains a different number of open reading frames (ORF). Nevertheless, ORF1a, ORFb1, envelope (E), protein S, protein N, and membrane protein (M) are present in all Human-CoV strains (Cavanagh, 1995; Enjuanes et al., 2000). The SARS-CoV-2 genome size varies from 29.8 to 29.9 kb and encodes 9,860 amino acids. The ORF1ab in SARS-CoV-2 is over 21 KB in length and covers two-thirds of the entire genome (Cascella et al., 2020). According to recent reports, the ORF portion of the SARS-CoV-2 genome has a lower CG dinucleotide percentage than other coronaviruses, so ORF RNA translation is highly efficient (Wang Y. et al., 2020).
Coronavirus Replication Process
The replication cycle of CoVs occurs in several stages (Figure 1). In the initial stage, the virus binding proteins are attached to appropriate receptors on the host cell’s surface. Then membrane fusion occurs, which finally leads to virus genome insertion in the host cell. In the second step, the RNA polymerase-dependent RNA gene is activated to translate the virus genome into structural proteins (Ziebuhr, 2005). During these steps, in addition to viral factors, some host cell factors can also enhance or inhibit this process.
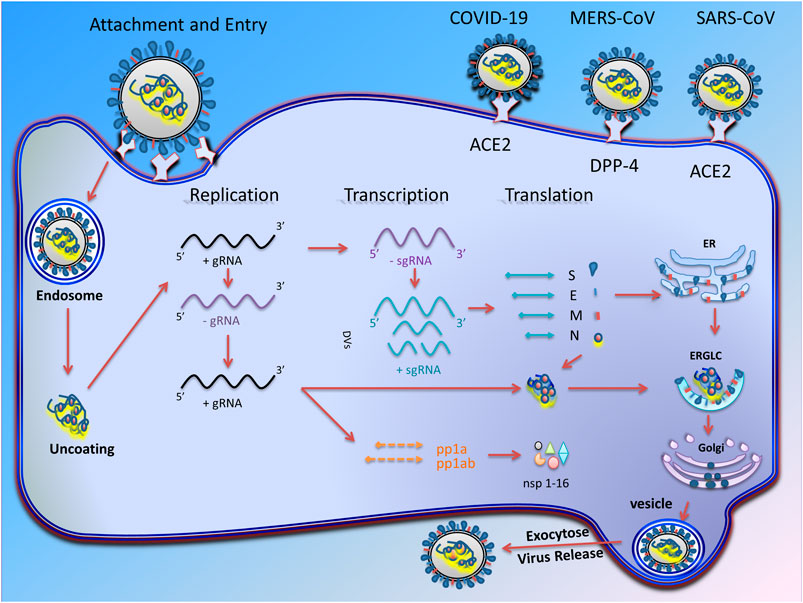
FIGURE 1. Transcription and replication of coronavirus. Schematic picture showing the coronavirus infection starts with attachment of virus S proteins to receptors on the host cells. After successful fusion, ribonucleocapsid enters the cell cytoplasm and loses its coat to mRNA is released. RNA-dependent RNA synthesis in CoVs includes two different genome replication processes to achieve multiple copies of genomic RNAs (gRNAs) and transcription of sub-genomic RNAs (sg mRNAs) coding structural and accessory proteins. The gRNA eventually producing 16 non-structural proteins (nsp-16) that are involved in virus replication. After assembly and budding, the full virus particles are transferred to the cytoplasmic membrane and finally released through the exocytosis process.
Attachment; Entry and Cellular Factors Involvement
The first stage of virus entry starts with the attachment of S proteins to host ligands. Most HCoVs use endocytosis pathways for their entrance, while others can use direct membrane fusion (Gallagher and Buchmeier, 2001). The ectodomain S protein consists of a receptor-binding subunit (S1) and a membrane subunit (S2). After binding to the host cell surface receptors, the S1 subunit leads to a structural change in S2. The S2 proteolytic domain then fuses the virus membrane into the host membrane, leading to the viral genome’s entry into the host cell. A partial mutation in the S protein that changes the amino acid sequence can significantly affect the virus’s pathogenesis and tissue or cell susceptibility to infection (Li, 2016).
HCoV-NL63, SARS-CoV, and SARS-CoV-2, use Angiotensin-Converting Enzyme 2 (ACE2) to contaminate cells (Moore et al., 2004; Zhang et al., 2020a). Both types of alveolar cells (I and II types) and the endothelial cells express ACE2. It has been shown that MERS-CoV uses Dipeptidyl peptidase 4 (DPP4), a physiological ligand for adenosine deaminase, which is extensively expressed by the endothelial cells in various tissues (Raj et al., 2013). However, many host factors can limit the entry of the virus. According to cell culture mechanisms, a family of Interferon-Inducible Transmembrane proteins (IFITM) could limit S protein-dependent access in HCoV-229E and HCoV-NL63, which would also lead to intense reduction of the infection in SARS-CoV and MERS-CoV (Huang et al., 2011; Wrensch et al., 2014). It seems that IFITM inhibits the membrane fusion by preventing the virus fusion to the cell membrane, endosomal membranes, or through membrane fluidity. Moreover, it was shown that human coronavirus HCoV-OC43 utilizes IFITM2 and IFITM3 as receptors to facilitate cell entry (Zhao et al., 2014).
Transcription and Replication
In CoVs, RNA-dependent RNA polymerase amplifies the virus genome in two ways and eventually produces several copies of genomic RNAs (gRNAs) and subgenomic RNAs (sg mRNAs) (Ziebuhr, 2005). Generally, CoVs replication is initiated with the translation of ORF1a to produce polyprotein1a (pp1a) with 4,382 amino acids and polyprotein1ab (pp1ab) with 7,073 amino acids. Then, the ORF1b is translated through the ribosomal frameshifting mechanism (Lai, 1990; Ziebuhr, 2005). Eventually, each of these polyproteins is cleaved to produce 16 non-structural proteins (nsp-16), which are involved in the virus replication cycle (Mcintosh and Peiris, 2009). Some of the non-structural proteins (Table 1) that have critical roles in the virus’s life cycle include helicase (nsp13), RNA-dependent RNA polymerase (RdRP) (nsp12 polymerase), 3-chymotripsin-like protease (3CLpro) (nsp5 protease), and papain-like protease (PLpro) (nsp3 protease) (Lai, 1990; Sawicki et al., 2005).
Virus Assembly and Egress
The structural proteins (Table 2) and some membrane accessory proteins are translated inside ER-bound ribosomes, while N protein is translated via free ribosomes in the cytoplasm (Nal et al., 2005). One of the distinct characteristics of CoVs is virion aggregation inside the ER and budding toward the Golgi apparatus (Cawood et al., 2007). Moreover, E, S, and N proteins interpolation to virions is modulated by M protein heterotypic interaction in the budding locus (Opstelten et al., 1995). After assembly and budding, the full virus particles are transferred to the cytoplasmic membrane and finally are released through the exocytosis process (Figure 1).
Coronaviruses Pathogenesis
Clinically, the pathogenesis of coronaviruses is divided into three stages. In the viremia phase, the virus enters the peripheral blood after the lungs are infected. Thus, the virus can reach its target tissues, such as the heart, kidneys, and gastrointestinal tract, through the bloodstream. The second phase is the pneumonia phase. Then the patient enters the recovery phase if the immune system can overcome the virus’s attack at this stage. However, in patients with immunodeficiency, high blood pressure, diabetes, and the elderly, the immune system cannot effectively manage the infection, and a critical stage of the disease occurs (Peiris et al., 2003; Guan et al., 2020).
Treatment
Vaccination is the best way to control the COVID-19 pandemic, and many efforts are being made to produce it. Nevertheless, its production is time-consuming because it must first be proven to be immunogenic and effective. Therefore, prevention is currently the best treatment. There are two essential strategies for treatment this disease. The first step is a general treatment that reduces the symptoms and clinical complications of the patients, and the second is drug treatment.
General Treatment
The available treatment options for these patients include measuring and monitoring of vital signs such as heart function, kidney, liver, respiratory rate, and oxygen therapy if necessary. Moreover, patients must be getting enough water, sufficient calories, balance for electrolytes, and use of fever medicines such as acetaminophen and ibuprofen (Wang J. et al., 2020; Zimmermann and Curtis, 2020). Although the use of corticosteroids such as methylprednisolone in the SARS-CoV epidemic has been shown to improve some clinical symptoms, it is not common in COVID-19. Nevertheless, it could be used only temporarily in patients with severe illness such as dyspnea and Acute Respiratory Distress Syndrome (ARDS) (Huang et al., 2020; Russell et al., 2020; Zhang et al., 2020b). Herbal medicines have the property to be used as a dietary supplement for relieving and reduce respiratory symptoms and other symptoms of COVID-19. They can improve the general condition of patients with mild disease severity. Althaea officinalis L. (Malvaceae), Commiphora myrrha (T.Nees) Engl. (Burseraceae), Glycyrrhiza glabra L. (Fabaceae), Hedera helix L. (Araliaceae), and Sambucus nigra L. (Viburnaceae) are some of the herbal medicines can be used as adjunctive therapy for mild COVID-19 (Silveira et al., 2020).
Coronavirus Drug Treatment
SARS-CoV-2 is a new virus from the Coronaviruses family, with 79% genomic similarity to SARS-CoV and 51.8% to MERS-CoV. Therefore, since the risk of SARS-CoV-2 vertical infection is the same as these two types of viruses, their treatment can be similar. Consequently, drugs that were previously effective for the treatment of SARS-CoV and MERS-CoV may also have therapeutic potential for the treatment of COVID-19 (Ren et al., 2020). Although a definitive cure for this new virus has not yet been discovered, previous studies suggest that drugs such as western medicines and natural products may have potential efficiency against COVID-19 and could be used to reduce the severity of the disease.
Interferon
Interferon type I is a member of the innate immune system produced and secreted in response to viral infections, including alpha and beta-interferon. Interferons exert their antiviral activity in two ways 1) cytotoxic T lymphocytes and macrophages stimulate the immune system to kill the virus by increasing or stimulating natural killer cells, 2) or inhibit virus replication in the host cell (Samuel, 2001; Sadler and Williams, 2008; Fensterl and Sen, 2009). Previous studies have shown that interferon-α, particularly its recombinant form (INF- α2b), can alleviate symptoms and shorten the disease course of respiratory tract viral infections in the early phase (caused by influenza, SARS-CoV, and MERS-CoV) through inhibiting virus replication and decreasing the virus load. Therefore, these molecules may be useful in the treatment of COVID-19 (Zheng et al., 2004; Danesh et al., 2011; Falzarano et al., 2013; Khalid et al., 2015). However, according to the latest update (3.2020) of the COVID-19 Treatment Guidelines Panel, there is no comment about its therapeutic effect on COVID-19 (COVID-19 Treatment Guidelines Panel).
Lopinavir/Ritonavir
Lopinavir/ritonavir is a protease inhibitor that has previously been used against HIV due to its antiviral activity. Nevertheless, later in the SARS-CoV (an epidemic that occurred in 2003), it was discovered that lopinavir/ritonavir by 3CLpro-inhibitor action could be useful for coronavirus treatment (Chu et al., 2004; Su et al., 2019). Due to the structural similarity of 3CLpro in SARS-CoV2 and SARS-CoV, previous studies have suggested that this drug may be useful for treating SARS-CoV2 (Uzunova et al., 2020). Nevertheless, later it was found that these drugs do not have the desired effect in treating this infectious disease, and the coronavirus disease 2019 treatment guidelines (last updated: November 3, 2020) announced that its use is not recommended, except in a clinical trial (COVID-19 Treatment Guidelines Panel, 2019).
Ribavirin
Ribavirin is a nucleoside analog that has antiviral activity. Co-administration of lopinavir/ritonavir has a significant therapeutic effect against SARS-CoV and may reduce ARDS risk (Chu et al., 2004). However, according to the latest update (November 3, 2020) of the COVID-19 Treatment Guidelines Panel, there is no comment about its therapeutic effect on COVID-19 (COVID-19 Treatment Guidelines Panel).
Chloroquine
Chloroquine is an antiparasitic drug used against malaria, but recently it has been shown to have antiviral activity by blocking virus infection and could suppress SARS-CoV-2 infection in vitro (Wang M. et al., 2020; Colson et al., 2020). Although many physicians and specialists initially considered this drug to treat COVID-19, its use is no longer recommended today (COVID-19 Treatment Guidelines Panel).
Favipiravir
One of the promising drugs in the treatment of COVID-19 is favipiravir. This drug has an inhibitory effect on RNA polymerase, and its early clinical trial showed that not only is the antiviral activity of this drug more significant than lopinavir/ritonavir, but it has fewer side effects (Cai et al., 2020). However, according to the latest update (November 3, 2020) of the COVID-19 Treatment Guidelines Panel, there is no comment about its therapeutic effect on COVID-19 (COVID-19 Treatment Guidelines Panel).
Remdesivir
Remdesivir is one of the drugs that can be used against RNA viruses such as SARS-CoV/MERS-CoV. In vivo and in vitro studies have shown that this adenosine analog by inhibiting RdRP has a therapeutic effect against COVID-19 and could be a choice for treating this new pandemic infectious disease (Holshue et al., 2020; Wang M. et al., 2020). This drug is currently recommended to treat COVID-19 according to the latest update (November 3, 2020) of the COVID-19 Treatment Guidelines Panel (COVID-19 Treatment Guidelines Panel).
Arbidol
Arbidol is an antiviral compound for prophylaxis and treatment of influenza. An in vitro study has proved that arbidol has a direct antiviral activity on SARS-CoV replication (Wang X. et al., 2020). A clinical trial of this drug on SARS-CoV-2 has also shown that it reduces high flow nasal catheter (HFNC) oxygen therapy, enhances the process of viral clearance and improves focal absorption on radiologic images (Xu et al., 2020a). Arbidol could inhibit host cell adhesion and SARS-CoV-2 spike glycoprotein trimerization. Furthermore, the simultaneous use of arbidol by Lopinavir/ritonavir has a more significant therapeutic effect than the Lopinavir/ritonavir group (Deng et al., 2020). However, according to the latest update (November 3, 2020) of the COVID-19 Treatment Guidelines Panel, there is no comment about its therapeutic effect on COVID-19 (COVID-19 Treatment Guidelines Panel).
Antithrombotic Therapy
Anticoagulants, like heparins, play a vital role in preventing arterial thromboembolism in patients with heart arrhythmias. One of the leading causes of death in patients with COVID-19 is myocardial and stroke infarction for unknown reasons. Therefore, if hospitalized patients with COVID-19 due to hemophilia or similar diseases do not have anticoagulant contraindications, administration of a prophylactic dose plays a vital role in reducing the complications COVID-19 in patients with symptoms of blood coagulation (such as low levels of antithrombin, increased fibrinogen and di-dimer) (Godino et al., 2020).
Natural Products as Antiviral Agents
Natural products are biochemical mixtures produced by living organisms in nature The effectiveness of natural compounds in ancient medicine was introduced thousands of years ago (Ji et al., 2009). These compounds, with various pharmacological traits, have antioxidant, anticancer, anti-inflammatory, and antiviral activities. Today, many of the chemical drugs used in medicine are derived from natural compounds (Frabasile et al., 2017). The therapeutic strategy for natural products and chemical drugs is to target the protein molecules needed for each stage of the virus’s life cycle (Dias et al., 2012). This section discusses the natural compounds’ role in the inhibition of coronavirus infection (Table 3; Figure 2; Figure 3; and Figure 4). It is important to emphasize that within the compounds listed in Table 3, only epigallocatechin-3-gallate (NCT04446065) and resveratrol and zinc picolinate combination therapy (NCT04542993) has been investigated in clinical phase-2 trials against COVID-19 infection.
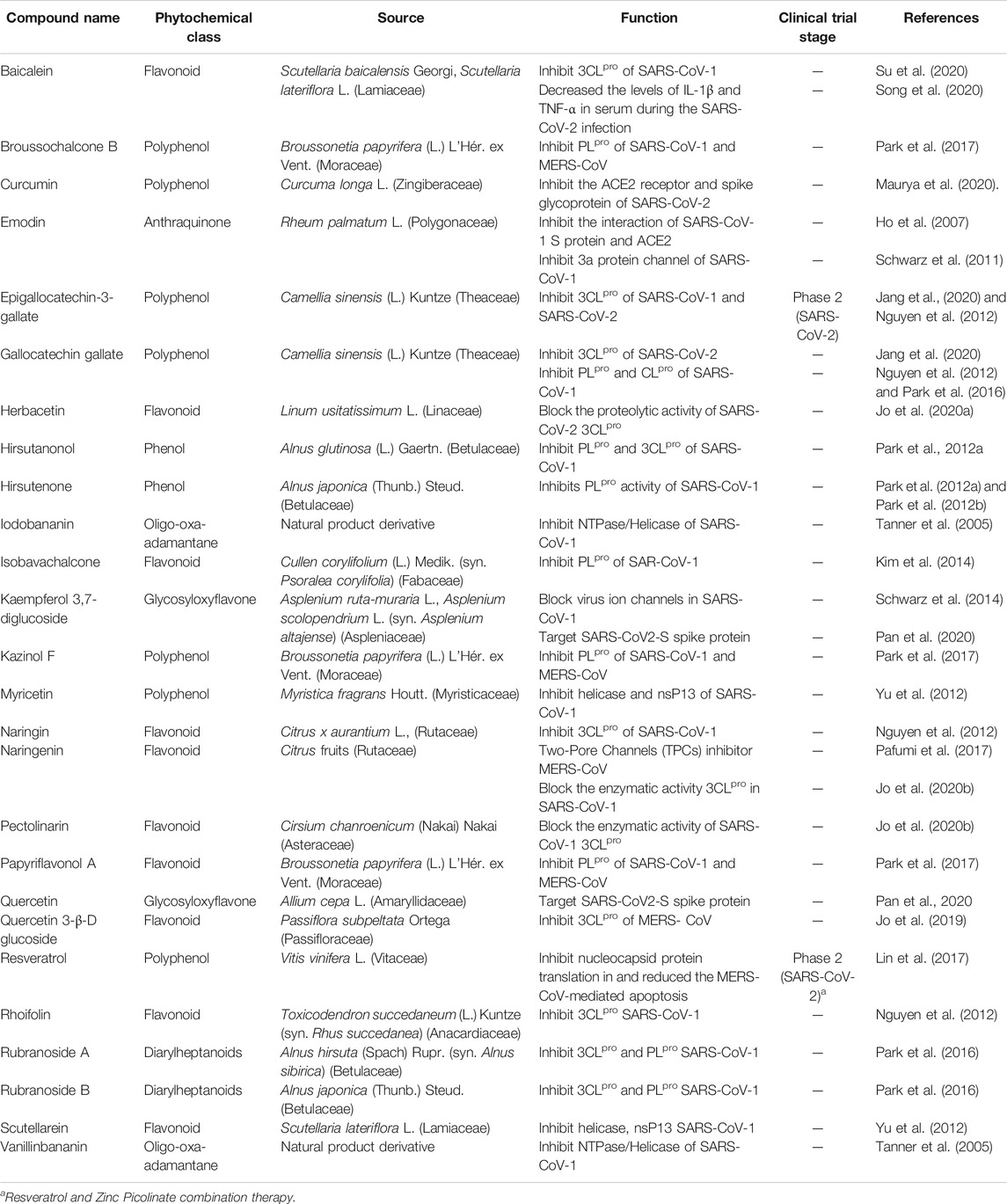
TABLE 3. Different types of phytochemicals by their sources and function in the coronavirus infection.
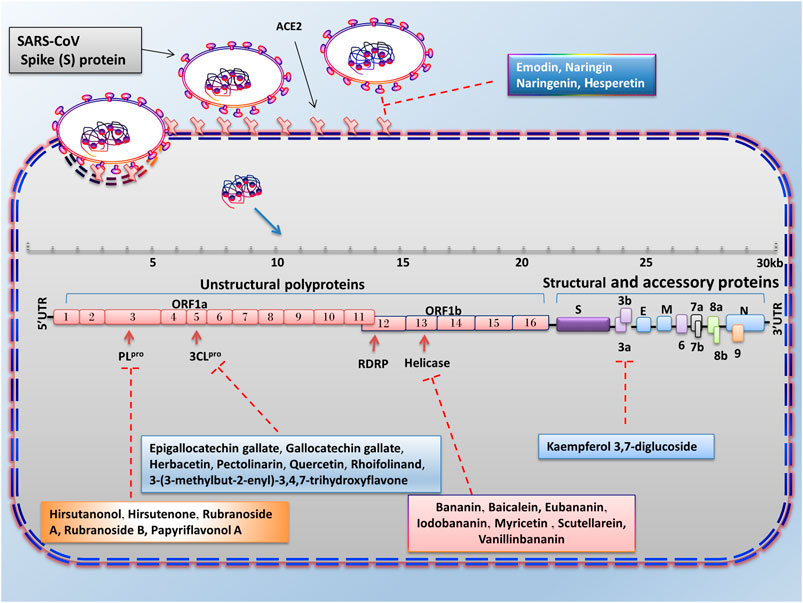
FIGURE 3. Advantages of employing natural compounds against infection and replication of SARS-CoV. Some of the natural compounds by reducing the expression of 3CLpro, PLpro, helicase, and 3a genes can play a therapeutic role in inhibiting the SARS-CoV infection.
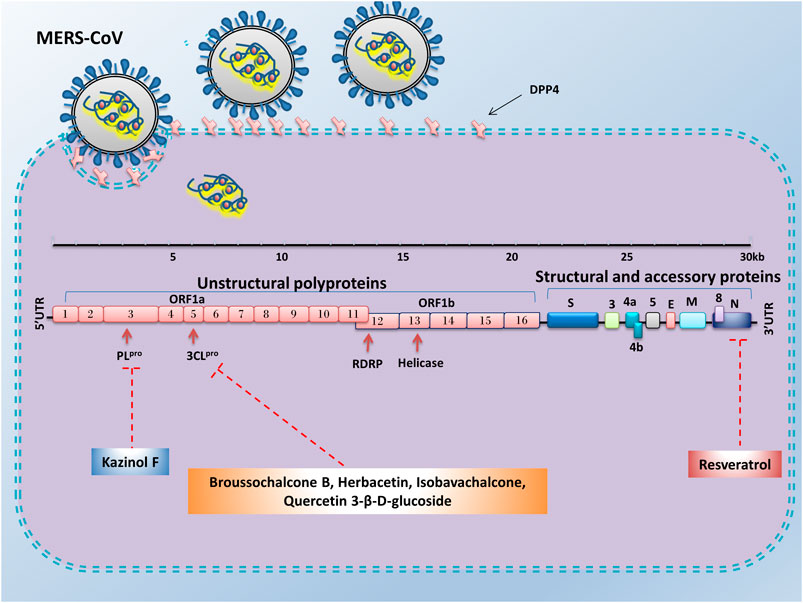
FIGURE 4. Advantages of employing natural compounds against infection and replication of MERS-CoV. Some of the natural compounds by reducing the expression of nucleocapsid, 3CLpro, and PLpro genes could play a therapeutic role in inhibiting the MERS-CoV infection.
Targeting S Protein, ACE2, and TMPRSS2 to Inhibit Cell Attachment and Entry
When a coronavirus enters the host cell, it must first, through its S protein, bind to the specific receptors at the surface of host cells. The particular receptor for SARS-CoV and SARS-CoV-2 is the ACE2 protein. To reduce the viral entry strength, the expression of the virus S protein can be inhibited or the expression of ACE2 in host cells can be reduced by natural products. Studies have shown that the amino acid sequence of SARS-CoV Spike protein is approximately 76.47%, similar to SARS-CoV-2 (Xu et al., 2020b). Since virus S protein is highly glycosylated, the use of plant-derived lectins, which tend to bind to glycosylated proteins, can prevent the virus from binding to receptors on the cell surface. Previous studies have shown that herbal compounds could be used as inhibitors for the human immune deficiency virus and SARS-CoV feline (Pyrc et al., 2007). Emodin belongs to the anthraquinones compounds (a group of natural compounds), which have anti-inflammatory, anticancer, and antioxidant activities (Izhaki, 2002). In a dose-dependent manner, emodin could significantly inhibit the ACE2 and S protein interaction in the biotinylated Enzyme-Linked Immunosorbent Assay (ELISA) method and Vero E6 cell line. Therefore, we can consider this compound as an antiviral therapeutic agent in the treatment of SARS-CoV (Ho et al., 2007). Some drugs and substances could reduce the expression of the ACE2 receptor. As seen, interleukin-4 and interferon-γ decreased this receptor in Vero E6 cells (De Lang et al., 2006). Molecular docking and structure-based drug design study reveal that berberine, thebaine, mangiferin, piperine, nimbin, and curcumin have an inhibitory effect against ACE2 receptor and spike glycoprotein of SARS-CoV-2. Among these, curcumin and nimbin have shown the highest interaction with ACE2 receptors and spike glycoprotein than other natural compounds and chemical drugs such as hydroxychloroquine, nafamostat, and captopril (Maurya et al., 2020).
The citrus peel is rich in flavonoid and alkaloid compounds (such as naringin, naringenin, hesperetin, and hesperidin) that play an essential role in maintaining gastrointestinal health and improving the immune system. Naringin is an anti-inflammatory substance that decreases the inflammatory effect in LPS-induced RAW 264.7 macrophages and LPS-treated rats. Therefore, it can inhibit the increase of pro-inflammatory cytokines such as IL-6, IL-1β, iNOS, and COX-2 induced by LPS treatment. Molecular docking investigation showed that naringin, naringenin, and hesperetin could bind to ACE2, such as chloroquine. Furthermore, these compounds can bind to ACE2 at lower binding energy levels than chloroquine (docking energy of 5.7 kcal/mol). However, more in vitro and in vivo experimentation is needed to determine whether these compounds are more effective than chloroquine (da Silva Antonio et al., 2020). However, it should be noted that inhibition of ACE2 due to its important physiological activities (such as homeostasis of blood pressure, protection against pulmonary cell destruction, electrolyte retention, and water) may have detrimental effects on patients’ life. In addition to ACE2, Transmembrane Protein Serine 2 (TMPRSS2) is another essential protein for SARS-CoV-1 and SARS-CoV-2 infection. TMPRSS2 induces viral fusion with the host cell membrane through irreversible structural changes in viral S protein (Hoffmann et al., 2020). As a result, the inhibition of TMPRSS2 in the animal model’s airways decreases the severity of lung damage after infection by SARS-CoV and MERS-CoV (Chikhale et al., 2020). Therefore, one of the strategies that could block the entry and restrict the pathogenesis of SARS-CoV-2 is the inhibition of TMPRSS2. The results of two independent studies using computational biology and molecular docking showed that some natural compounds such as neohesperidin, myricitrin, quercitrin, naringin, icariin, citicoline, bianthraquinone, isogemichalcone B, and (-)-epigallocatechin-(3-O-methyl) gallate could be used as a TMPRSS inhibitor (Chikhale et al., 2020; Rahman et al., 2020).
Targeting Helicase and Inhibiting Virus Replication
SARS-CoV-2 Helicase protein is encoded by the nsp 13, a gene located downstream of the RdRP. The inhibition of this protein could reduce virus replication (Ivanov et al., 2004). Helicases could separate the double-stranded Nucleic Acid (NA) by using free energy that is obtained from the hydrolysis of Nucleoside Triphosphate (NTP). Therefore, this protein is a potential target for antiviral drug development. Previous studies revealed that various natural products could suppress helicase (unwinding or ATPase) activity. In 2005, Tanner et al. investigated the effect of adamantane-derived bananins on SARS-CoV helicase.
Their results showed that iodobananin, vanillinbananin, bananin, and eubananin had an inhibitory effect on the helicase protein’s ATPase activity with IC50 rates in the range 0.5–2.8 µM. Furthermore, using Fluorescence Resonance Energy Transfer (FRET) it was found that these four compounds could also inhibit unwinding helicase activity. The authors also observed that these compounds were ineffective against E. coli helicase but only affected SARS helicase, so they did not have a general helicase inhibitor activity. Finally, the results of cytopathic effects in fetal rhesus kidney-4 cells (FRhK-4) and RT-PCR demonstrated that these compounds could reduce virus replication without being toxic to the cell (Tanner et al., 2005). In 2012, the results of a study by Yu et al. showed that of the sixty-four natural compounds (flavonoids), only myricetin and scutellarein could reduce SARS-CoV helicase activity. The events showed that these two compounds could reduce Helicase ATPase activity by up to 90% at a dose of 10 µM without any toxicity effects on the healthy breast cell (MCF10A). Moreover, further analysis has shown that they do not change the helicase unwinding activity (Yu et al., 2012). Baicalein is another natural product that has an inhibitory effect on SARS-CoV (nsp13) helical activity. Studies have shown that this compound has no inhibitory effect on the dsDNA-unwinding activity of nsP13; however, it can decrease helicase ATPase activity up to 60% with an IC50 value 0.47 μM (Keum et al., 2013).
Targeting 3CLPRO, PLPRO Protease and Inhibiting Virus Protein Processing
Another important antiviral strategy is the use of specific inhibitors against viral proteases such as 3CLpro and PLpro, which play an essential role in the processing and maturation of proteins and virus replication. The SARS-CoV-2 genome (positive single-stranded RNA), after translation, is capable of producing two polypeptides, pp1a and pp1ab. Finally, these two polypeptides, cleaved by 3CLpro or PLpro activity, produce sixteen non-structural proteins (Thiel et al., 2003; Muramatsu et al., 2016). Flavonoids are a group of phenolic compounds found in plants with anti-inflammatory, antiviral, antioxidant, and anticancer activities. There have been many reports of reduced coronavirus infection with these compounds (Tapas et al., 2008). In vitro studies in Pichia pastoris GS115 showed that epigallocatechin-3-gallate, quercetin, and gallocatechin gallate could reduce the expression of SARS-CoV 3CLpro. Molecular docking experiments and kinetic enzyme studies also revealed that these three compounds with IC50 rates in the range 47–73 µM could reduce protease activity up to 80%. In the meantime, gallocatechin gallate’s effect was more significant than the other compounds (Nguyen et al., 2012). Rhoifolinand, herbacetin, and pectolinarin are other flavonoid compounds that have an adverse influence on 3CLpro. Studies with FRET protease assays and absorption spectroscopic studies have shown that these compounds can bind to 3CLpro and significantly inhibit its protease activity (drug concentration less than 40 µM in IC50) (Jo et al., 2020b). In another study, Jo et al. examined the inhibitory effects of several flavonoids compounds against MERS-CoV 3CLpro. Their results reveal the isobavachalcone, quercetin 3-β-D-glucoside, and herbacetin had noticeable inhibitory actions with IC50 values of 35.85, 37.03, 40.59 µM, respectively (Jo et al., 2019). The 3CLpro of SARS-CoV-2 is highly conserved among all CoVs and has approximately 96% similarity with SARS-CoV-1. Due to its essential role in viral replication, it is a potential therapeutic target for COVID-19 (Xu et al., 2020c). Since natural products derived from microbial sources have a unique chemical diversity compared to plant products, more than 50% of FDA-approved natural compound-based medicines are derived from microbial compounds. The virtual screening and molecular binding by Sayed et al. showed that citriquinochroman, holyrine B, proximicin C, and several other microbial compounds could inhibit 3CLpro of SARS-CoV-2 (Sayed et al., 2020). In addition to microbial compounds, molecular docking and MD simulation studies have shown that some marine natural products including hydroxypentafuhalol, pentaphlorethol B, and luteolin-7-rutinoside (ΔG about −14.6 to −10.7 kcal/mol) can also inhibit 3CLpro of SARS-CoV-2 (Gentile et al., 2020).
The Papain-like protease (PLpro), is another protease that controls the proliferation of SARS-CoV-2 and is known as a potential target for treating this virus. Previous studies have shown that Alnus japonica (Thunb.) Steud. (Betulaceae) has anticancer, anti-inflammatory, and anti-influenza properties. The study of Kim et al. showed that the natural phenolic compounds (diarylheptanoids) prepared from this plant could change the proteolytic activity of PLpro. The fluorometric assay results showed that among the nine extracted substances, hirsutenone, hirsutanonol, rubranoside B, and rubranoside A had a dose-dependent inhibitory effect against PLpro. Among these compounds, hirsutenone had a remarkable inhibitory effect on SARS-CoV PLpro (IC50 = 4.1 µM) and 3CLpro (IC50 = 36.2 µM) enzyme activity. Further study showed that this substance, containing an α, β-unsaturated carbonyl group with a catechol moiety in the backbone, and the presence of this structure played an essential role in its inhibitory effects (Park et al., 2012a). In addition, polyphenols derived from the root of Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) have been shown to have good inhibitory potential against SARS-CoV and MERS-CoV proteases. Park et al. investigated the inhibitory effect of ten natural compounds derived from this plant on SARS-CoV and MERS-CoV proteases. The effect of these compounds on SARS-CoV proteases showed that the papyriflavonol A (Broussonol E) was the most effective inhibitor of PLpro (IC50 value 3.7 µM) and 3-(3-methylbut-2-enyl)-3,4,7-trihydroxyflavone was the most useful inhibitor of 3CLpro (IC50 value of 30.2 µM). Additionally, broussochalcone B (Bavachalcone), with a concentration of 27.9 µM (IC50) and kazinol F with a concentration of 39.5 µM (IC50), were able to reduce the activity of MERS-CoV 3CLpro and MERS-CoV PLpro, respectively (Park et al., 2017).
Targeting Nucleocapsid (N) Protein to Inhibit Virus Infection and Replication
Resveratrol is a natural compound with anti-inflammatory, antioxidant, and anticancer properties (Yeung et al., 2019; Filardo et al., 2020). The previous study demonstrated that this compound has antiviral activity and can inhibit viral infections caused by Herpes Simplex Virus (HSV), Respiratory Syncytial Virus (RSV), and Epstein-Barr Virus (EBV) (Faith et al., 2006; Zang et al., 2011; De Leo et al., 2012). Also, resveratrol by repressing the expression of MERS-CoV N protein in the Vero E6 cell line, could reduce RNA expression, viral yield, and replication of MERS-CoV. Additionally, it significantly decreases the virus’s infection and enhances infected cells’ survival by repressing Caspase 3 cleavage (Lin et al., 2017). Other natural compounds that can reduce coronavirus infection risk include the alkaloids fangchinoline, cepharanthine, and tetrandrine. The results of a study by Kim et al. showed that these compounds could inhibit the expression of pro-inflammatory cytokines (IFN-α1, IL-6, IFN-β1, IL-8, and IL-1) caused by human coronavirus OC43 infection in the MRC-5 cell line. These three natural compounds can also decrease the OC43 replication by inhibiting N protein expression and reducing the cytotoxic effect of this virus in the MRC-5 cell line, and increasing the proliferation and survival of MRC-5 human lung cells (Kim et al., 2019).
Targeting 3A Protein and Inhibiting Virus Release
The production and release of the virus require some ion channels in the host cell membrane. Therefore, inhibition of these ion channels played a significant role in inhibiting viral infections, so one antiviral strategy is to use compounds that can restrain these channels (Liang and Li, 2010). One of these ion channels is the cation-selective channel (3a protein) generated by the ORF3a of the SARS-CoV genome. Kaempferol glycoside is a natural flavonol found in a variety of plants. Previous studies have shown that this compound has antiviral properties (IC50 value of 2.3 μM) and can inhibit the expression of 3a protein SARS CoV (an ion channel) in Xenopus oocyte as a model system (Schwarz et al., 2014).
RNA Interference
RNA interference (RNAi) are RNA molecules found in many eukaryotes that inhibit gene expression by targeted 3UTR of mRNA molecules. Today, siRNA and miRNA are the most common type of RNAi used for gene silencing. RNAi origin can be endogenous (originating in the cell) and exogenous (coming from a virus or laboratory tools). Exogenous RNAi can be transmitted to cells using electroporation, viral vectors, liposomes, and calcium phosphate (Sohrab et al., 2018). When inserting into the cell, synthetic siRNAs are cleaved by Dicer in the cytoplasm. After placement in the RNA-Induced Silencing Complex (RISC) and Ago2, RNAi molecules (Figure 5) become single-stranded RNA and can destroy the target mRNA or inhibit its translation (Ding et al., 2018).
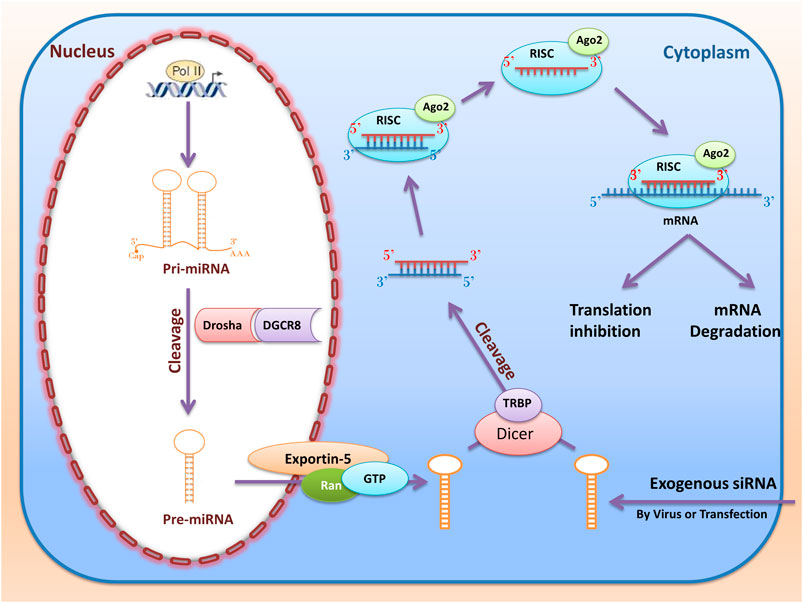
FIGURE 5. RNA interference (miRNAs and siRNAs) biogenesis and function. The miRNAs first transcribed from the nucleus genome as pri-miRNA. Then pri-miRNA cleavage with Drosha and DGCR and converted to pre-miRNA. Then, RanGTP and exportin 5 cause the pre miRNA to be transported from the nucleus to the cytoplasm and cleavage by Dicer and TRBP. Ultimately, after entering the RISC complex, mature miRNA can be attached to the target mRNA and perform their function by destroying mRNA or inhibiting the translation. When inserting into the cell, synthetic siRNAs are cleaved by Dicer in the cytoplasm. This dsRNA enters RISC, and if cold finds the target mRNA, the mRNA is cleaved by the RISC and Ago2.
The Therapeutic Effect of siRNA on Coronaviruses
Synthetic siRNAs have about 21–23 bp length and perform their role by inhibiting gene expression at the post-transcription level. Unlike miRNAs, each siRNA is designed against a specific gene, so it can only impede that gene expression (Taxman et al., 2006). The siRNAs are first inserted into the cell as long double-stranded RNAs and then are cleaved by RNase III (Dicer) in the cytoplasm to become small dsRNAs of approximately twenty-one base pairs. This dsRNA later enters the RISC and converts to single-stranded RNA (ssRNA). If RISC and siRNAs complex could find the specific target site on mRNA, it could cleave the mRNA, and cellular exonucleases could invade to destroy the target mRNA (Wu and Chan, 2006). Studies have shown that this type of RNAi has the potential to act as antiviral agent to reduce replication and infection of many viruses such as HIV, Flock House Virus (FHV), Hepatitis C Virus (HCV), and Hepatitis B Virus (HBV) (Hamasaki et al., 2003; Liu et al., 2017; Shahid et al., 2017; Taning et al., 2018). Previous studies have shown that siRNA could target different parts of the virus genome to reduce the replication and infection of SARS-CoV (Figure 6A). For example, targeting the nsp1 gene (from nucleotides 250–786) by siRNA is one of the best ways to control SARS-CoV. Because targeting this area of the genome inhibits virus propagation, pathogenesis, and replication in Vero E6 cells (Ni et al., 2005). Another strategy that can help to suppress infection and replication of SARS-CoV is inhibiting the virus S protein or its specific receptor ACE2.
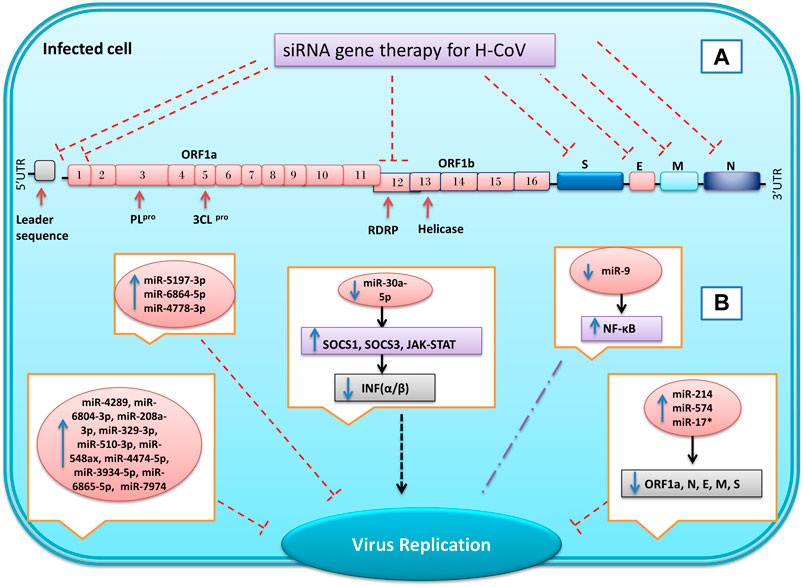
FIGURE 6. Inhibition of genes expression of SARS coronavirus using miRNAs and synthetic small interfering RNAs (siRNA). By designing specific siRNAs, it is possible to inhibit the expression of the virus’s structural and non-structural genes for reduce the replication and infusion of the virus (A). In coronavirus infection, the microRNAs expression of host cells changes in response to infection. Some of these changes are the cell’s response to the infection, and others are caused by the virus, which can eventually lead to a reduction or increase in virus replication and infection (B) Inhibition  , Induction
, Induction  , not define
, not define  , Up-regulation
, Up-regulation  , Down-regulation
, Down-regulation  .
.
The SARS-CoV Spike protein is an essential viral surface glycoprotein for identifying target cells and interacting with ACE2 host cell receptors (Gallagher and Buchmeier, 2001). A study by Zhang et al. reveals that inhibiting expression of S protein using specific siRNAs could reduce the viral titers, infection, and replication of SARS-CoV in Vero E6 and 293T cells (Zhang et al., 2004). RNAi technology is also a useful instrument to suppress ACE2 expression at the host cells’ surface to counteract SARS-CoV infection. In addition to the lungs, ACE2 is expressed on the surface of the bronchial, renal, duodenum, colon, gastrointestinal, and cardiovascular cells (Donoghue et al., 2000). Inhibition of the ACE2 protein in Vero E6 cell lines using siRNA could reduce replication, copies number, and infection of severe acute respiratory syndrome-associated coronavirus (Lu et al., 2008). Li et al. investigated the effect of siSC5 (nsp12 region) and siSC2 (spike protein) against SARS-CoV on in vitro and in vivo models. Their results showed that these two siRNAs by inhibited virus replication in FRHK-4 cells could reduce the virus infection’s effects and symptoms without having any toxicity effect on Rhesus macaque (dosages of 10–40 mg/kg) (Li et al., 2005a).
The spike protein structure determines the type of host cells which could be infected by a virus. However, variations of S protein are seen among the various strains of coronavirus. Therefore, one of the essential strategies for controlling viral infections targets the conserved genome areas between various coronaviruses (Kuo et al., 2000). Hence, Wang et al. tried to use two specific siRNAs against the conserved sequence of the SARS-CoV. The study results showed that by targeting two regions (14,450–14,468 and 15,877–15,895), encoding RNA polymerase could inhibit expression of RNA polymerase, N protein, 3CLpro and also reduce replication and cytopathic effect of the virus (Wang et al., 2004). Among the two COVID-19 proteases, the nsp3 sequence encoding papain-like protease is less conserved, while the nsp5 sequence encoding 3CLpro is highly conserved and can be selected as a potential target of siRNA for COVID-19 treatment (Liu et al., 2020).
The RdRP gene encodes a key enzyme for the replication of the virus. In SARS-CoV-2, this gene is located in ORF1b by 645 bp long and has been reported to be highly conserved, so it has the potential to target several siRNAs. Also, investigations of the RdRP sequence have shown that this gene has no genetic similarity to human genes and other coronaviruses (Wu et al., 2020). Lu et al.’s investigation shows using specific siRNA can reduce SARS-CoV RdRP expression by more than 90% in HeLa and 239T cell lines, leading to inhibition of plaque formation in Vero E6 cells. Therefore, its suppression could be considered a suitable therapeutic target in COVID-19 patients (Lu et al., 2004). The siRNAs could target other essential coronavirus genes. For example, by designing three different siRNAs, Shi et al. were able to reduce the expression of N, E, and M genes of SARS-CoV in Vero E6 cells (Yi et al., 2005). Furthermore, the replication of SARS-CoV could be inhibited by targeting the leader sequence using specific siRNA in Vero E6 cells (Li et al., 2005b).
Although previous studies have shown that RNAi has antiviral potential, it appears that viruses can use these molecules to their advantage. MERS-CoV could infect both human and bat cell lines. Nevertheless, its pathogenic power and replication are varied between the bat (Eptesicus fuscus) cells and human (A549, MRC5, and Huh7) cell lines. In human cells, MERS-CoV shut-down interferon antiviral responses in the innate immunity system, unlike in bat cells by inhibition of IRF3 (a critical activator for INF β expression) (Banerjee et al., 2019).
Complemented palindromic small RNAs (cpsRNAs) are a group of small RNAs produced by mammalian and invertebrate viruses. There are sequences in the SARS-CoV (ORF3b) genome that have the origin of a cpsRNA called SARS-CoV-cpsR-19. The apoptosis assay events indicate that SARS-CoV-cpsR-19 could induce apoptosis in HeLa cells by increasing the caspase 3 and BAX/BCL2 ratio and may play an essential role in SARS-CoV pathogenesis (Liu et al., 2018). It has been shown that some viruses (such as the Ebola virus and influenza A) could preserve themselves from RNAi-based immune systems and facilitate their replication by using Viral Suppressors of RNA silencing (VSR). Studies of Cui et al. have shown a short hairpin RNA (a novel VSR) in the SARS-CoV nucleocapsid protein sequence that defeats RNAi-triggered suppression (Cui et al., 2015). Moreover, N protein overexpression in Neuro-2a cells efficiently inhibits Dicer-mediated dsRNA cleavage and could increase replication and titration of MHV-A59 (a close relative to SARS-CoV in Coronaviridae family) viruses. The SARS-CoV-2 (N) nucleocapsid protein can also act as an escape agent from the immune system and contribute to its pathogenicity. N proteins have high homology (94%) of the amino acid sequences among coronaviruses. A recent study has shown that the N protein of SARS-CoV-2 has VSR activity that can antagonize RNAi in both effectors (recognition and cleavage of viral dsRNA by Dicer) and initiation (siRNA biogenesis) steps (Mu et al., 2020).
MicroRNAs
The miRNAs are a group of small non-coding RNAs with twenty-two nucleotides of length. These molecules are first transcribed from the nucleus genome (exons, introns, and intergenic regions) as pri-miRNA with a length of two hundred nucleotides to several thousand nucleotides (Booton and Lindsay, 2014). It is noteworthy that each pri-miRNA can be a precursor to several mature miRNAs. Then, first processing of pri-miRNA starts with RNase III (Drosha) and its cofactor (DGCR) to form pre-miRNA, a hairpin with a length of about sixty nucleotides. In the next step, RanGTP and exportin 5 cause the pre-miRNA to be transported from the nucleus to the cytoplasm. Dicer and TRBP performed the second processing in the cytoplasm to create a double-stranded RNA molecule with a length of about twenty-two nucleotides (Bartel, 2004). Ultimately, after entering the RISC complex, one of the strings (passenger strand) is destroyed, and mature miRNA (guide strand) can be attached to the target mRNA. MiRNAs perform their function by destroying mRNA or inhibiting the translation (Vazquez, 2006). Previous studies have shown that a miRNA alone can regulate the expression of several different genes by its function. Meanwhile, various miRNAs can simultaneously control the expression of one mRNA. However, about 60% of human genes can be regulated by these molecules (Kalhori et al., 2020). For the first time, Lecellier et al. (2005) reported that cellular miR-32 could reduce virus replication of the primate foamy virus (PFV-1) by targeting the viral RNA genome (Lecellier et al., 2005).
MicroRNA Regulates the Innate Immune System, Virus Replication, and Pathogenesis of Coronavirus
The innate immune system is the body’s first defense against viruses and bacteria. This system’s principal cells include macrophages, dendritic cells, natural killer cells, monocytes, and granulocytes (Leon-Icaza et al., 2019). Viruses, to increase replication, suppress the host’s innate immune system via reducing INF α/β production. For example, Japanese Encephalitis Virus (JEV), Dengue Virus (DENV), and Enterovirus 71 (EV71) are able to inhibit the overexpression of INF α/β in response to viral infection by enhancing the expression of miR-146a in infected cells (Wu et al., 2013; Ho et al., 2014; Sharma et al., 2015). Viruses can also decrease the innate immune system by inhibiting the expression or function of some miRNAs. For instance, in oligodendroglioma cells, the Borna Disease Virus (BDV) can inhibit the expression of miR-155 by its specific phosphoprotein, thus inhibiting INF α/β overexpression in response to viral infection and reducing the innate immune system (Zhai et al., 2013). Therefore, one of the main antiviral components of the intrinsic immune system is type 1 interferons (INF α/β).
Coronaviruses can prevent the induction of the immune system in response to viral infection with different strategies. In vivo and in vitro studies of Ma et al. on the Transmissible Gastroenteritis Virus (TGEV), a member of the alpha-coronavirus family, showed that this virus could downregulate miR-30a-5p expression. Furthermore, they found that virus replication was facilitated by reducing IFN-I signaling cascades via removing the inhibitory effect of miR-30a-5p on INF negative regulators (such as SOCS1, SOCS3, and JAK-STAT) (Ma et al., 2018). Therefore, the overexpression of miRNAs in infected cells could increase the innate immune system and may be considered as a therapeutic approach for treatment.
Although it has previously been reported that miRNAs play a significant role in regulating the eukaryotic gene, subsequent studies showed that these nano molecules can also alter the virus’s replication to increase or decrease its infection (Figure 6B). For example, miR-122 plays an essential role in the pathogenesis and replication of the Hepatitis C virus. The miR-122, to increase the virus’s stability and replication bind to the 5′ non-translated regions (NTRs) of the virus and repress RNA degradation via exonucleases. Therefore, the knockout of this miRNA in Huh-7 cells could reduce HCV replication (Jopling et al., 2005). In contrast, miR-32 could negatively alter the replication of the PFV-1 by targeting viral genes in the human HEK-293T cell line (Lecellier et al., 2005).
Some viruses have sequences (hairpin) in their genomes similar to miRNAs and can regulate the gene expression of the host cell or virus (Grundhoff and Sullivan, 2011; Kincaid and Sullivan, 2012). A computational approach study by Hassan et al. showed there are several hairpins in the genome of the MERS that could act as a precursor for thirteen miRNAs, which were significantly similar to human miRNAs. Their study showed ten miRNAs (miR-4289, miR-6804-3p, miR-208a-3p, miR-329-3p, miR-510-3p, miR-548ax, miR-4474-5p, miR-3934-5p, miR-6865-5p, and miR-7974) of these, miRNAs do not have any known specific biological function in humans or animals at all. Nevertheless, miR-18a, miR-628, and miR-342-3p had a biological role in humans related to Basal Cell Carcinoma (BCC) of the skin, malignant glioblastoma, and late-stage prion disease, respectively (Hasan et al., 2014). Numerous studies and reports suggest that some miRNAs have antiviral activity and can be used against influenza, HIV, HBV, and poliovirus (PV) (Sanghvi and Steel, 2012; Zhang et al., 2013; Shim et al., 2016; Hamada-Tsutsumi et al., 2019). On the other hand, viruses could alter the gene and miRNA expression profile in the host cell. For example, miR-146a and miR-130b upregulated by the human T Cell Leukemia Virus (HTLV-1) in PBMC cells (Bouzar and Willems, 2008).
Nucleocapsid (N) protein is a structural protein that has the same function in all coronaviruses. The human coronavirus CoV-OC43 could inhibit miR-9 function by its N protein and increase NF-κB expression in 253T cells. However, it is unclear whether upregulation NF-κB is a suitable response for virus replication or a secondary inhibitor for virus replication (Lai et al., 2014). Infection of bronchoalveolar stem cells (BASICs) by SARS-CoV reveals that this virus could upregulate the expression of miR-574-5p, miR-214, and miRNAs-17* 2–4 fold. Moreover, overexpression of these miRNAs could repress SARS-CoV replication by targeting the four viral structure proteins (E, S, M, N), and orf1a (Mallick et al., 2009). Unfortunately, to date, not many in vivo and in vitro studies have been performed on RNAi’s role in inhibiting the COVID-19, and most studies have been performed base on bioinformatics and in silico studies. Qingfei Paidu decoction (QFPD) contains twenty-one traditional Chinese medicines that have been used to treat COVID-19 since February 7, 2020. Chen et al.’s molecular docking study revealed that QFPD can bind to structural and non-structural proteins of COVID-19. They also found that miR-183 and miR-130A/B/301 predict targets of QFPD, and QFPD by these microRNAs may exert anti-SARS-CoV-2 activity (Chen et al., 2020a). In a bioinformatics approach study performed by Khan et al., it was found that several miRNAs can have antiviral properties in infections caused by SARS-CoV-1 and SARS-CoV-2. For example, evidence has shown miR-323a-5p, miR-622, miR-198, and miR-654-5p for SARS-CoV-1 and miR-323a-5p, miR-20b-5p, miR-17-5p for SARS-CoV-2 have antiviral roles by targeting the ORF1ab and the S region (Khan et al., 2020).
Therefore, it is likely miRNA with low side effects can be used as a therapeutic agent for the COVID-19 treatment. Differences in miRNA expression profiles in individuals are probably one reason why COVID-19 causes death in some people and causes only brief symptoms in others. As a result, microRNAs have recently emerged as a critical factor in increasing or inhibiting the potential of viral infection. We hope that clinical and preclinical research can use them in gene therapy as antiviral agents soon.
Conclusion
Today, the whole world is suffering from a pandemic disease called COVID-19, which has caused deaths in many developed and developing countries. Despite all the advances in human medicine, we have not yet been able to find a suitable treatment for this viral disease. The use of molecular or pharmacological methods to control infection or virus replication requires identifying essential genes involved in infection and replication of the virus. Two strategies are suggested to treat this disease. The first step is to reduce the virus’s infection by preventing the virus from attaching to its specific receptor. The next step is to reduce the virus’s replication by inhibiting the virus’s structural and non-structural genes. Medicinal plants and natural products are a good option for preventing and treating viral infections, especially COVID-19, due to their lower cost, lower side effects, and natural origin compared with chemical drugs. These compounds could increase efficiency and strengthen the host immune system against many infections and diseases due to their inherent properties. In the treatment of COVID-19, these compounds can reduce virus infection or replication by repressing the virus’s coupling to the host cell’s receptors or by inhibiting the expression of structural and non-structural genes. Moreover, RNAi (siRNA and miRNA) could inhibit viral infections, especially COVID-19, by inhibiting essential virus genes or inducing a host immune system. Therefore, the simultaneous use of natural compounds and RNAi can play a critical role in the treatment of SARS-CoV-2 and restraining this pandemic pneumonia.
Author Contributions
The study was conceptualized by MS and EA. The methodology was given by EA and MF. Writing and original draft preparation was done by MK and FS. Writing, review, and editing were done by MF, IA, and JE. Funding acquisition was provided by MK and JE. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Kermanshah University of Medical Sciences (grant numbers 990512). JE gratefully acknowledges funding from CONICYT (PAI/ACADEMIA No 79160109).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahn, D.-G., Choi, J.-K., Taylor, D. R., and Oh, J.-W. (2012). Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 57, 2095–2104. doi:10.1007/s00705-012-6751404-x10.1007/s00705-012-1404-x
An, S., Chen, C.-J., Yu, X., Leibowitz, J. L., and Makino, S. (1999). Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 73, 7853–7859. doi:10.1128/JVI.73.9.7853-7859.1999
Angelini, M. M., Akhlaghpour, M., Neuman, B. W., and Buchmeier, M. J. (2013). Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 4, e00524–e00513. doi:10.1128/mBio.00524-13
Angelini, M. M., Neuman, B. W., and Buchmeier, M. J. (2014). Untangling membrane rearrangement in the nidovirales. DNA Cell Biol. 33, 122–127. doi:10.1089/dna.2013.2304
Banerjee, A., Falzarano, D., Rapin, N., Lew, J., and Misra, V. (2019). Interferon regulatory factor 3-mediated signaling limits Middle-East respiratory syndrome (MERS) coronavirus propagation in cells from an insectivorous bat. Viruses 11, 152. doi:10.3390/v11020152
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi:10.1016/s0092-8674(04)00045-5
Booton, R., and Lindsay, M. A. (2014). Emerging role of MicroRNAs and long noncoding RNAs in respiratory disease. Chest 146, 193–204. doi:10.1378/chest.13-2736
Bosch, B. J., Van Der Zee, R., De Haan, C. A., and Rottier, P. J. (2003). The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811. doi:10.1128/jvi.77.16.8801-8811.2003
Bouvet, M., Debarnot, C., Imbert, I., Selisko, B., Snijder, E. J., Canard, B., et al. (2010). In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 6 (4), e1000863. doi:10.1371/journal.ppat.1000863
Bouvet, M., Imbert, I., Subissi, L., Gluais, L., Canard, B., and Decroly, E. (2012). RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci USA 109, 9372–9377. doi:10.1073/pnas.1201130109
Bouzar, A. B., and Willems, L. (2008). How HTLV-1 may subvert miRNAs for persistence and transformation. Retrovirology 5, 101. doi:10.1186/1742-4690-5-101
Cai, Q., Yang, M., Liu, D., Chen, J., Shu, D., Xia, J., et al. (2020). Experimental treatment with Favipiravir for COVID-19: an open-label control study. Engineering 6 (10), 1192–1198. doi:10.1016/j.eng.2020.03.007
Casals, J., Tignor, G. H., Lack, E. E., Jenson, B., Smith, H. G., Healy, G. B., et al. (1980). International committee for Taxonomy of viruses. Intervirology 14, 228. doi:10.1159/000149188
Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. C., and Di Napoli, R. (2020). “Features, evaluation and treatment coronavirus (COVID-19),” in Statpearls (Treasure Island, FL: StatPearls Publishing).
Cavanagh, D. (1995). “The coronavirus surface glycoprotein,” in The coronaviridae. Boston, MA: Springer, 73–113. doi:10.1007/978-1-4899-1531-3_5
Cawood, R., Harrison, S. M., Dove, B. K., Reed, M. L., and Hiscox, J. A. (2007). Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle 6, 863–867. doi:10.4161/cc.6.7.4032
Centers for Disease Control and Prevention (2012). Severe respiratory illness associated with a novel coronavirus--Saudi Arabia and Qatar, 2012. MMWR Morb. Mortal. Wkly. Rep. 61 (40), 820.
Chen, J., Wang, Y. K., Gao, Y., Hu, L. S., Yang, J. W., Wang, J. R., et al. (2020a). Protection against COVID-19 injury by qingfei paidu decoction via anti-viral, anti-inflammatory activity and metabolic programming. Biomed. Pharmacother. 129, 110281. doi:10.1016/j.biopha.2020.110281
Chen, Z. M., Fu, J. F., Shu, Q., Chen, Y. H., Hua, C. Z., Li, F. B., et al. (2020b). Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J. Pediatr. 16, 240–246. doi:10.1007/s12519-020-00345-5
Chikhale, R. V., Gupta, V. K., Eldesoky, G. E., Wabaidur, S. M., Patil, S. A., and Islam, M. A. (2020). Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 1–16, 1–16. doi:10.1080/07391102.2020.1798813
Chu, C., Cheng, V., Hung, I., Wong, M., Chan, K., Chan, K., et al. (2004). Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59, 252–256. doi:10.1136/thorax.2003.012658
Colson, P., Rolain, J.-M., and Raoult, D. (2020). Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents 55 (3), 105923. doi:10.1016/j.ijantimicag.2020.105923
Cornillez-Ty, C. T., Liao, L., Yates, J. R., Kuhn, P., and Buchmeier, M. J. (2009). Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 83, 10314–10318. doi:10.1128/JVI.00842-09
COVID-19 Treatment Guidelines Panel (2019). Coronavirus disease 2019 (COVID-19) treatment guidelines. Bethesda, MD: National Institutes of HealthAvailable at: https://www.covid19treatmentguidelines.nih.gov/
Cui, L., Wang, H., Ji, Y., Yang, J., Xu, S., Huang, X., et al. (2015). The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. virol. 89, 9029–9043. doi:10.1128/jvi.01331-15
Danesh, A., Cameron, C. M., León, A. J., Ran, L., Xu, L., Fang, Y., et al. (2011). Early gene expression events in ferrets in response to SARS coronavirus infection versus direct interferon-alpha2b stimulation. Virology 409, 102–112. doi:10.1016/j.virol.2010.10.002
da Silva Antonio, A., Wiedemann, L. S. M., and Veiga-Junior, V. F. (2020). Natural products' role against COVID-19. RSC Adv. 10, 23379–23393. doi:10.1039/D0RA03774E
De Lang, A., Osterhaus, A. D., and Haagmans, B. L. (2006). Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology 353, 474–481. doi:10.1016/j.virol.2006.06.011
De Leo, A., Arena, G., Lacanna, E., Oliviero, G., Colavita, F., and Mattia, E. (2012). Resveratrol inhibits Epstein Barr Virus lytic cycle in Burkitt’s lymphoma cells by affecting multiple molecular targets. Antiviral Res. 96, 196–202. doi:10.1016/j.antiviral.2012.09.003
Deng, L., Li, C., Zeng, Q., Liu, X., Li, X., Zhang, H., et al. (2020). Arbidol combined with LPV/r versus LPV/r alone against corona virus disease 2019: a retrospective cohort study. J. Infect. 81 (1), e1–e5. doi:10.1016/j.jinf.2020.03.002
Denison, M. R., Graham, R. L., Donaldson, E. F., Eckerle, L. D., and Baric, R. S. (2011). Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 8, 270–279. doi:10.4161/rna.8.2.15013
Dias, D. A., Urban, S., and Roessner, U. (2012). A historical overview of natural products in drug discovery. Metabolites 2, 303–336. doi:10.3390/metabo2020303
Ding, S. W., Han, Q., Wang, J., and Li, W. X. (2018). Antiviral RNA interference in mammals. Curr Opin Immunol 54, 109–114. doi:10.1016/j.coi.2018.06.010
Ding, Y., Wang, H., Shen, H., Li, Z., Geng, J., Han, H., et al. (2003). The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200, 282–289. doi:10.1002/path.1440
Donoghue, M., Hsieh, F., Baronas, E., Godbout, K., Gosselin, M., Stagliano, N., et al. (2000). A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res 87, e1–e9. doi:10.1161/01.res.87.5.e1
Enjuanes, L., Brian, D., Cavanagh, D., Holmes, K., Lai, M., Laude, H., et al. (2000). Coronaviridae. Virus Taxonomy, Classification and Nomenclature of viruses. Editors M. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelliet al. (New York, NY: Academic Press), 835–849.
Faith, S. A., Sweet, T. J., Bailey, E., Booth, T., and Docherty, J. J. (2006). Resveratrol suppresses nuclear factor-kappaB in herpes simplex virus infected cells. Antiviral Res. 72, 242–251. doi:10.1016/j.antiviral.2006.06.011
Falzarano, D., De Wit, E., Martellaro, C., Callison, J., Munster, V. J., and Feldmann, H. (2013). Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci. Rep. 3, 1686. doi:10.1038/srep01686
Fensterl, V., and Sen, G. C. (2009). Interferons and viral infections. BioFactors 35, 14–20. doi:10.1002/biof.6
Filardo, S., Di Pietro, M., Mastromarino, P., and Sessa, R. (2020). Therapeutic potential of resveratrol against emerging respiratory viral infections. Pharmacol Ther. 214, 107613. doi:10.1016/j.pharmthera.2020.107613
Frabasile, S., Koishi, A. C., Kuczera, D., Silveira, G. F., Verri, W. A., Dos Santos, C. N. D., et al. (2017). The citrus flavanone naringenin impairs dengue virus replication in human cells. Sci. Rep. 7, 41864. doi:10.1038/srep41864
Gallagher, T. M., and Buchmeier, M. J. (2001). Coronavirus spike proteins in viral entry and pathogenesis. Virology 279, 371–374. doi:10.1006/viro.2000.0757
Gentile, D., Patamia, V., Scala, A., Sciortino, M. T., Piperno, A., and Rescifina, A. (2020). Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar. Drugs 18 (4), 225. doi:10.3390/md18040225
Godino, C., Scotti, A., Maugeri, N., Mancini, N., Fominskiy, E., Margonato, A., et al. (2020). Antithrombotic therapy in patients with COVID-19? -rationale and evidence. Int. J. Cardiol. 35 (9), 2698–2706. doi:10.1016/j.ijcard.2020.09.064
Grundhoff, A., and Sullivan, C. S. (2011). Virus-encoded microRNAs. Virology 411, 325–343. doi:10.1016/j.virol.2011.01.002
Guan, W.-J., Ni, Z.-Y., Hu, Y., Liang, W.-H., Ou, C.-Q., He, J.-X., et al. (2020). Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382 (18), 1708–1720. doi:10.1056/NEJMoa2002032
Hamada‐Tsutsumi, S., Naito, Y., Sato, S., Takaoka, A., Kawashima, K., Isogawa, M., et al. (2019). The antiviral effects of human microRNA miR-302c-3p against hepatitis B virus infection. Aliment Pharmacol. Ther. 49, 1060–1070. doi:10.1111/apt.15197
Hamasaki, K., Nakao, K., Matsumoto, K., Ichikawa, T., Ishikawa, H., and Eguchi, K. (2003). Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett 543, 51–54. doi:10.1016/S0014-5793(03)00400-9
Hamre, D., and Procknow, J. J. (1966). A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121, 190–193. doi:10.3181/00379727-121-30734
Hasan, M. M., Akter, R., Ullah, M., Abedin, M., Ullah, G., and Hossain, M. (2014). A computational approach for predicting role of human microRNAs in MERS-CoV genome. Adv. bioinformatics 2014, 967946. doi:10.1155/2014/967946
Ho, B. C., Yu, I. S., Lu, L. F., Rudensky, A., Chen, H. Y., Tsai, C. W., et al. (2014). Inhibition of miR-146a prevents enterovirus-induced death by restoring the production of type I interferon. Nat. Commun. 5, 3344. doi:10.1038/ncomms4344
Ho, T.-Y., Wu, S.-L., Chen, J.-C., Li, C.-C., and Hsiang, C.-Y. (2007). Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 74, 92–101. doi:10.1016/j.antiviral.2006.04.014
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280. doi:10.1016/j.cell.2020.02.052
Holshue, M. L., Debolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936. doi:10.1056/NEJMoa2001191
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi:10.1016/S0140-6736(20)30183-5
Huang, I.-C., Bailey, C. C., Weyer, J. L., Radoshitzky, S. R., Becker, M. M., Chiang, J. J., et al. (2011). Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7 (1), e1001258. doi:10.1371/journal.ppat.1001258
ICTV (2009). Virus taxonomy: 2009 release. Available at: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/209/orthomyxoviridae (Accessed November 30, 2020).
Ivanov, K. A., Thiel, V., Dobbe, J. C., Van Der Meer, Y., Snijder, E. J., and Ziebuhr, J. (2004). Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78, 5619–5632. doi:10.1128/JVI.78.11.5619-5632.2004
Izhaki, I. (2002). Emodin–a secondary metabolite with multiple ecological functions in higher plants. New Phytol. 155, 205–217. doi:10.1046/j.1469-8137.2002.00459.x
Jang, M., Park, Y.-I., Cha, Y.-E., Park, R., Namkoong, S., Lee, J. I., et al. (2020). Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid. Based Complement. Alternat. Med. 2020, 5630838. doi:10.1155/2020/5630838
Ji, H. F., Li, X. J., and Zhang, H. Y. (2009). Natural products and drug discovery. Can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? EMBO Rep. 10, 194–200. doi:10.1038/embor.2009.12
Jo, S., Kim, H., Kim, S., Shin, D. H., and Kim, M. S. (2019). Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 94, 2023–2030. doi:10.1111/cbdd.13604
Jo, S., Kim, S., Kim, D. Y., Kim, M.-S., and Shin, D. H. (2020a). Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhib. Med. Chem. 35, 1539–1544. doi:10.1080/14756366.2019.169048010.1080/14756366.2020.1801672
Jo, S., Kim, S., Shin, D. H., and Kim, M.-S. (2020b). Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 35, 145–151. doi:10.1080/14756366.2019.1690480
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M., and Sarnow, P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581. doi:10.1126/science.1113329
Kalhori, M. R., Arefian, E., Atanaki, F. F., Kavousi, K., and Soleimani, M. (2020). miR-548x and miR-4698 controlled cell proliferation by affecting the PI3K/AKT signaling pathway in glioblastoma cell lines. Sci. Rep. 10, 1–12. doi:10.1038/s41598-020-57588-5
Keum, Y.-S., Lee, J. M., Yu, M.-S., Chin, Y.-W., and Jeong, Y.-J. (2013). Inhibition of SARS coronavirus helicase by baicalein. Bull Korean Chem. Soc. 34, 3187–3188. doi:10.5012/bkcs.2013.34.11.3187
Khalid, M., Al Rabiah, F., Khan, B., Al Mobeireek, A., Butt, T. S., and Al Mutairy, E. (2015). Ribavirin and interferon-α2b as primary and preventive treatment for Middle East respiratory syndrome coronavirus: a preliminary report of two cases. Antivir. Ther. (Lond) 20, 87–91. doi:10.3851/IMP2792
Khan, M. A., Sany, M. R. U., Islam, M. S., and Islam, A. (2020). Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 11, 765. doi:10.3389/fgene.2020.00765
Kim, D. E., Min, J. S., Jang, M. S., Lee, J. Y., Shin, Y. S., Park, C. M., et al. (2019). Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 9, 696. doi:10.3390/biom9110696
Kim, D. W., Seo, K. H., Curtis-Long, M. J., Oh, K. Y., Oh, J.-W., et al. (2014). Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzyme Inhib. Med. Chem. 29, 59–63. doi:10.1371/journal.ppat.100301810.3109/14756366.2012.753591
Kincaid, R. P., and Sullivan, C. S. (2012). Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 8, e1003018. doi:10.1371/journal.ppat.1003018
King, A. M., Adams, M. J., Carstens, E. B., and Lefkowitz, E. J. (2012). “Virus taxonomy,” in Ninth report of the international committee on Taxonomy of viruses (London, United Kingdom: Elsevier Academic Press), 486–487.
Kuo, L., Godeke, G.-J., Raamsman, M. J., Masters, P. S., and Rottier, P. J. (2000). Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74, 1393–1406. doi:10.1128/JVI.74.3.1393-1406.2000
Lai, F. W., Stephenson, K. B., Mahony, J., and Lichty, B. D. (2014). Human coronavirus OC43 nucleocapsid protein binds microRNA 9 and potentiates NF-κB activation. J. Virol. 88, 54–65. doi:10.1128/JVI.02678-13
Lai, M. M., and Cavanagh, D. (1997). The molecular biology of coronaviruses. Adv. Virus Res. 48, 1–100. doi:10.1016/S0065-3527(08)60286-9
Lai, M. M. (1990). Coronavirus: organization, replication and expression of genome. Annu. Rev. Microbiol. 44, 303. doi:10.1146/annurev.mi.44.100190.001511
Lecellier, C. H., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., et al. (2005). A cellular microRNA mediates antiviral defense in human cells. Science 308, 557–560. doi:10.1126/science.1108784
Lei, J., Kusov, Y., and Hilgenfeld, R. (2018). Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 149, 58–74. doi:10.1016/j.antiviral.2017.11.001
Leon-Icaza, S. A., Zeng, M., and Rosas-Taraco, A. G. (2019). MicroRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA 1 (1), 1. doi:10.1186/s41544-018-0004-7
Li, B.-J., Tang, Q., Cheng, D., Qin, C., Xie, F. Y., Wei, Q., et al. (2005a). Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 11, 944–951. doi:10.1038/nm1280
Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261. doi:10.1146/annurev-virology-110615-042301
Li, G., and De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV. Nat. Rev. Drug. Discov. 19, 149–150. doi:10.1038/d41573-020-00016-0
Li, T., Zhang, Y., Fu, L., Yu, C., Li, X., Li, Y., et al. (2005b). SiRNA targeting the leader sequence of SARS-CoV inhibits virus replication. Gene Ther. 12, 751–761. doi:10.1038/sj.gt.3302479
Liang, X., and Li, Z.-Y. (2010). Ion channels as antivirus targets. Virol. Sin. 25, 267–280. doi:10.1007/s12250-010-3136-y
Lin, C.-W., Tsai, F.-J., Wan, L., Lai, C.-C., Lin, K.-H., Hsieh, T.-H., et al. (2005). Binding interaction of SARS coronavirus 3CL(pro) protease with vacuolar-H+ ATPase G1 subunit. FEBS Lett. 579, 6089–6094. doi:10.1016/j.febslet.2005.09.075
Lin, S.-C., Ho, C.-T., Chuo, W.-H., Li, S., Wang, T. T., and Lin, C.-C. (2017). Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 17, 144. doi:10.1186/s12879-017-2253-8
Liu, C., Chen, Z., Hu, Y., Ji, H., Yu, D., Shen, W., et al. (2018). Complemented palindromic small RNAs first discovered from SARS coronavirus. Genes 9, 442. doi:10.3390/genes9090442
Liu, C., Liang, Z., and Kong, X. (2017). Efficacy analysis of combinatorial siRNAs against HIV derived from one double hairpin RNA precursor. Front. Microbiol. 8, 1651. doi:10.3389/fmicb.2017.01651
Liu, C., Zhou, Q., Li, Y., Garner, L. V., Watkins, S. P., Carter, L. J., et al. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 6, 315–331. doi:10.1021/acscentsci.0c00272
Liu, D., Yuan, Q., and Liao, Y. (2007). Coronavirus envelope protein: a small membrane protein with multiple functions. Cell Mol. Life Sci. 64, 2043–2048. doi:10.1007/s00018-007-7103-1
Lu, A., Zhang, H., Zhang, X., Wang, H., Hu, Q., Shen, L., et al. (2004). Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology 324, 84–89. doi:10.1016/j.virol.2004.03.031
Lu, C.-Y., Huang, H.-Y., Yang, T.-H., Chang, L.-Y., Lee, C.-Y., and Huang, L.-M. (2008). siRNA silencing of angiotensin-converting enzyme 2 reduced severe acute respiratory syndrome-associated coronavirus replications in Vero E6 cells. Eur. J. Clin. Microbiol. Infect. Dis. 27, 709–715. doi:10.1007/s10096-008-0495-5
Ma, Y., Wang, C., Xue, M., Fu, F., Zhang, X., Li, L., et al. (2018). The coronavirus transmissible gastroenteritis virus evades the type I interferon response through IRE1α-mediated manipulation of the microRNA miR-30a-5p/SOCS1/3 axis. J. Virol. 92, e00728–e00718. doi:10.1128/JVI.00728-18
Mahase, E. (2020). Coronavirus covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ 368, m641. doi:10.1136/bmj.m641
Mallick, B., Ghosh, Z., and Chakrabarti, J. (2009). MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS One 4 (11), e7837. doi:10.1371/journal.pone.0007837
Maurya, V. K., Kumar, S., Prasad, A. K., Bhatt, M. L. B., and Saxena, S. K. (2020). Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virusdisease 31, 179–193. doi:10.1007/s13337-020-00598-8
Mcintosh, K., Kapikian, A. Z., Turner, H. C., Hartley, J. W., Parrott, R. H., and Chanock, R. M. (1970). Seroepidemiologic studies of coronavirus infection in adults and children. Am. J. Epidemiol. 91, 585–592. doi:10.1093/oxfordjournals.aje.a121171
Mcintosh, K., and Peiris, J. (2009). “Coronaviruses,” in Clinical virology. 3rd Edn. (Washington, DC: American Society of Microbiology Press), 1155–1171. doi:10.1128/9781555815981.ch51
Miknis, Z. J., Donaldson, E. F., Umland, T. C., Rimmer, R. A., Baric, R. S., and Schultz, L. W. (2009). Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 83, 3007–3018. doi:10.1128/JVI.01505-08
Moore, M. J., Dorfman, T., Li, W., Wong, S. K., Li, Y., Kuhn, J. H., et al. (2004). Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78, 10628–10635. doi:10.1128/JVI.78.19.10628-10635.2004
Mu, J., Xu, J., Zhang, L., Shu, T., Wu, D., Huang, M., et al. (2020). SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 63, 1–4. doi:10.1007/s11427-020-1692-1
Muramatsu, T., Takemoto, C., Kim, Y.-T., Wang, H., Nishii, W., Terada, T., et al. (2016). SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U.S.A 113, 12997–13002. doi:10.1073/pnas.1601327113
Nal, B., Chan, C., Kien, F., Siu, L., Tse, J., Chu, K., et al. (2005). Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 86, 1423–1434. doi:10.1099/vir.0.80671-0
Nguyen, T. T. H., Woo, H.-J., Kang, H.-K., Kim, Y.-M., Kim, D.-W., Ahn, S.-A., et al. (2012). Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 34, 831–838. doi:10.1007/s10529-011-0845-8
Ni, B., Shi, X., Li, Y., Gao, W., Wang, X., and Wu, Y. (2005). Inhibition of replication and infection of severe acute respiratory syndrome-associated coronavirus with plasmid-mediated interference RNA. Antivir. Ther. (Lond) 10, 527–533.
Opstelten, D. J., Raamsman, M., Wolfs, K., Horzinek, M. C., and Rottier, P. (1995). Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131, 339–349. doi:10.1083/jcb.131.2.339
Pafumi, I., Festa, M., Papacci, F., Lagostena, L., Giunta, C., Gutla, V., et al. (2017). Naringenin impairs two-pore channel 2 activity and inhibits VEGF-induced angiogenesis. Sci. Rep. 7, 1–11. doi:10.1038/s41598-017-04974-1
Pan, B., Fang, S., Zhang, J., Pan, Y., Liu, H., Wang, Y., et al. (2020). Chinese herbal compounds against SARS-CoV-2: puerarin and quercetin impair the binding of viral S-protein to ACE2 receptor. Comput. Struct. Biotechnol. J. 18, 3518–3527. doi:10.1016/j.csbj.2020.11.010
Park, J.-Y., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, S.-J., Kim, D., et al. (2012a). Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 35, 2036. doi:10.1248/bpb.b12-00623
Park, J.-Y., Kim, J. H., Kim, Y. M., Jeong, H. J., Kim, D. W., Park, K. H., et al. (2012b). Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 20 (19), 5928–5935. doi:10.1016/j.bmc.2012.07.038
Park, J.-Y., Ko, J.-A., Kim, D. W., Kim, Y. M., Kwon, H.-J., Jeong, H. J., et al. (2016). Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 31 (1), 23–30. doi:10.3109/14756366.2014.1003215
Park, J.-Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., et al. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 32, 504–512. doi:10.1080/14756366.2016.1265519
Peiris, J. S. M., Chu, C.-M., Cheng, V. C.-C., Chan, K., Hung, I., Poon, L. L., et al. (2003). Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361, 1767–1772. doi:10.1016/s0140-6736(03)13412-5
Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., et al. (2020). Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 52 (1), 56–65. doi:10.4103/ijp.IJP_115_20
Pyrc, K., Berkhout, B., and Van Der Hoek, L. (2007). Antiviral strategies against human coronaviruses. Infect. Disord. Drug. Targets 7, 59–66. doi:10.2174/187152607780090757
Rahman, N., Basharat, Z., Yousuf, M., Castaldo, G., Rastrelli, L., and Khan, H. (2020). Virtual screening of natural products against type II Transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2). Molecules 25 (10), 2271. doi:10.3390/molecules25102271
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254. doi:10.1038/nature12005
Ren, L.-L., Wang, Y.-M., Wu, Z.-Q., Xiang, Z.-C., Guo, L., Xu, T., et al. (2020). Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. (Engl) 133 (9), 1015–1024. doi:10.1097/CM9.0000000000000722
Ricagno, S., Egloff, M.-P., Ulferts, R., Coutard, B., Nurizzo, D., Campanacci, V., et al. (2006). Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U.S.A. 103, 11892–11897. doi:10.1073/pnas.0601708103
Russell, C. D., Millar, J. E., and Baillie, J. K. (2020). Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 395, 473–475. doi:10.1016/S0140-6736(20)30317-2
Sadler, A. J., and Williams, B. R. (2008). Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568. doi:10.1038/nri2314
Samuel, C. E. (2001). Antiviral actions of interferons. Clin Microbiol Rev 14, 778–809. doi:10.1128/CMR.14.4.778-809.2001
Sanghvi, V. R., and Steel, L. F. (2012). RNA silencing as a cellular defense against HIV-1 infection: progress and issues. FASEB J. 26, 3937–3945. doi:10.1096/fj.12-210765
Sawicki, S. G., Sawicki, D. L., Younker, D., Meyer, Y., Thiel, V., Stokes, H., et al. (2005). Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1 (4), e39. doi:10.1371/journal.ppat.0010039
Sayed, A. M., Alhadrami, H. A., El-Gendy, A. O., Shamikh, Y. I., Belbahri, L., Hassan, H. M., et al. (2020). Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (M(pro)). Microorganisms 8 (7), 970. doi:10.3390/microorganisms8070970
Schwarz, S., Sauter, D., Wang, K., Zhang, R., Sun, B., Karioti, A., et al. (2014). Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 80, 177–182. doi:10.1055/s-0033-1360277
Schwarz, S., Wang, K., Yu, W., Sun, B., and Schwarz, W. (2011). Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Res. 90, 64–69. doi:10.1016/j.antiviral.2011.02.008
Shahid, I., Almalki, W. H., Alrabia, M. W., Mukhtar, M. H., Almalki, S. S. R., Alkahtani, S. A., et al. (2017). In vitro inhibitory analysis of consensus siRNAs against NS3 gene of hepatitis C virus 1a genotype. Asian Pac. J. Trop. Med. 10, 701–709. doi:10.1016/j.apjtm.2017.07.011
Sharma, N., Verma, R., Kumawat, K. L., Basu, A., and Singh, S. K. (2015). miR-146a suppresses cellular immune response during Japanese encephalitis virus JaOArS982 strain infection in human microglial cells. J. Neuroinflammation 12, 30. doi:10.1186/s12974-015-0249-0
Shim, B.-S., Wu, W., Kyriakis, C. S., Bakre, A., Jorquera, P. A., Perwitasari, O., et al. (2016). MicroRNA-555 has potent antiviral properties against poliovirus. J. Gen. Virol. 97, 659–668. doi:10.1099/jgv.0.000372
Silveira, D., Prieto-Garcia, J. M., Boylan, F., Estrada, O., Fonseca-Bazzo, Y. M., Jamal, C. M., et al. (2020). COVID-19: is there evidence for the use of herbal medicines as adjuvant symptomatic therapy? Front. Pharmacol. 11, 581840. doi:10.3389/fphar.2020.581840
Siu, K.-L., Kok, K.-H., Ng, M.-H. J., Poon, V. K., Yuen, K.-Y., Zheng, B.-J., et al. (2009). Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 284, 16202–16209. doi:10.1074/jbc.M109.008227
Sohrab, S. S., El-Kafrawy, S. A., Mirza, Z., Kamal, M. A., and Azhar, E. I. (2018). Design and delivery of therapeutic siRNAs: application to MERS-coronavirus. Curr. Pharm. Des. 24, 62–77. doi:10.2174/1381612823666171109112307
Song, J., Zhang, L., Xu, Y., Yang, D., Yang, S., Zhang, W., et al. (2020). The comprehensive study on the therapeutic effects of baicalein for the treatment of COVID-19 in vivo and in vitro. Biochem. Pharmacol. 183, 114302. doi:10.1016/j.bcp.2020.114302
Su, B., Wang, Y., Zhou, R., Jiang, T., Zhang, H., Li, Z., et al. (2019). Efficacy and tolerability of lopinavir/ritonavir- and efavirenz-based initial antiretroviral therapy in HIV-1-Infected patients in a tertiary care hospital in beijing, China. Front. Pharmacol. 10, 1472. doi:10.3389/fphar.2019.01472
Su, H.-X., Yao, S., Zhao, W.-F., Li, M.-J., Liu, J., Shang, W.-J., et al. (2020). Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta. Pharmacol. Sin. 41, 1167–1177. doi:10.1038/s41401-020-0483-6
Taning, C. N., Christiaens, O., Li, X., Swevers, L., Casteels, H., Maes, M., et al. (2018). Engineered flock house virus for targeted gene suppression through RNAi in fruit flies (Drosophila melanogaster) in vitro and in vivo. Front. Physiol. 9, 805. doi:10.3389/fphys.2018.00805
Tanner, J. A., Watt, R. M., Chai, Y.-B., Lu, L.-Y., Lin, M. C., Peiris, J. M., et al. (2003). The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 278, 39578–39582. doi:10.1074/jbc.C300328200
Tanner, J. A., Zheng, B.-J., Zhou, J., Watt, R. M., Jiang, J.-Q., Wong, K.-L., et al. (2005). The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 12, 303–311. doi:10.1016/j.chembiol.2005.01.006
Tapas, A. R., Sakarkar, D., and Kakde, R. (2008). Flavonoids as nutraceuticals: a review. Trop J. Pharm. Res. 7, 1089–1099. doi:10.4314/tjpr.v7i3.14693
Taxman, D. J., Livingstone, L. R., Zhang, J., Conti, B. J., Iocca, H. A., Williams, K. L., et al. (2006). Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 6, 7. doi:10.1186/1472-6750-6-7
Te Velthuis, A. J., Van Den Worm, S. H., and Snijder, E. J. (2012). The SARS-coronavirus nsp7+ nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res 40, 1737–1747. doi:10.1093/nar/gkr893
Thiel, V., Ivanov, K. A., Putics, A., Hertzig, T., Schelle, B., Bayer, S., et al. (2003). Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84, 2305–2315. doi:10.1099/vir.0.19424-0
Uzunova, K., Filipova, E., Pavlova, V., and Vekov, T. (2020). Insights into antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine affecting the new SARS-CoV-2. Biomed. Pharmacother. 131, 110668. doi:10.1016/j.biopha.2020.110668
Vazquez, F. (2006). Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 11, 460–468. doi:10.1016/j.tplants.2006.07.006
Wang, J., Qi, H., Bao, L., Li, F., and Shi, Y. (2020). A contingency plan for the management of the 2019 novel coronavirus outbreak in neonatal intensive care units. Lancet Child Adolesc. Health 4 (4), 258–259. doi:10.1016/S2352-4642(20)30040-7
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. doi:10.1038/s41422-020-0282-0
Wang, X., Cao, R., Zhang, H., Liu, J., Xu, M., Hu, H., et al. (2020). The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 6, 28. doi:10.1038/s41421-020-0169-8
Wang, Y., Mao, J., Wang, G., Qiu, Z., Yao, Q., and Chen, K. (2020). Human SARS-CoV-2 has evolved to reduce CG dinucleotide in its open reading frames. Res. Squ. 10, 12331. doi:10.1038/s41598-020-69342-y
Wang, Z., Ren, L., Zhao, X., Hung, T., Meng, A., Wang, J., et al. (2004). Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J. Virol. 78, 7523–7527. doi:10.1128/JVI.78.14.7523-7527.2004
Woo, P. C., Huang, Y., Lau, S. K., and Yuen, K.-Y. (2010). Coronavirus genomics and bioinformatics analysis. viruses 2, 1804–1820. doi:10.3390/v2081803
Wrensch, F., Winkler, M., and Pöhlmann, S. (2014). IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses 6, 3683–3698. doi:10.3390/v6093683
Wu, C.-H., Yeh, S.-H., Tsay, Y.-G., Shieh, Y.-H., Kao, C.-L., Chen, Y.-S., et al. (2009). Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 284, 5229–5239. doi:10.1074/jbc.M805747200
Wu, C.-J., and Chan, Y.-L. (2006). Antiviral applications of RNAi for coronavirus. Expert Opin. Investig. Drugs 15, 89–97. doi:10.1517/13543784.15.2.89
Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. doi:10.1038/s41586-020-2008-3
Wu, S., He, L., Li, Y., Wang, T., Feng, L., Jiang, L., et al. (2013). MiR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J. Infect 67, 329–341. doi:10.1016/j.jinf.2013.05.003
Xu, K., Chen, Y., Yuan, J., Yi, P., Ding, C., Wu, W., et al. (2020a). Clinical efficacy of arbidol in patients with 2019 novel coronavirus-infected pneumonia: a retrospective cohort study. SSRN Elect. J. 2020, 3542148. doi:10.2139/ssrn.3542148
Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., et al. (2020b). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 63, 457–460. doi:10.1007/s11427-020-1637-5
Xu, Z., Peng, C., Shi, Y., Zhu, Z., Mu, K., Wang, X., et al. (2020c). Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv 2020, 921627. doi:10.1101/2020.01.27.921627
Yeung, A. W. K., Aggarwal, B. B., Balacheva, A., Barreca, D., Battino, M., Belwal, T., et al. (2019). Resveratrol, a popular dietary supplement for human and animal health: quantitative research literature analysis. Animal Sci. Papers Reports 36 (4), 345–358.
Yi, S., De Hua, Y., Xiong, J., Jie, J., Huang, B., and Jin, Y. X. (2005). Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res. 15, 193–200. doi:10.1038/sj.cr.7290286
Yu, M.-S., Lee, J., Lee, J. M., Kim, Y., Chin, Y.-W., Jee, J.-G., et al. (2012). Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 22, 4049–4054. doi:10.1016/j.bmcl.2012.04.081
Zang, N., Xie, X., Deng, Y., Wu, S., Wang, L., Peng, C., et al. (2011). Resveratrol-mediated gamma interferon reduction prevents airway inflammation and airway hyperresponsiveness in respiratory syncytial virus-infected immunocompromised mice. J. Virol. 85, 13061–13068. doi:10.1128/JVI.05869-11
Zhai, A., Qian, J., Kao, W., Li, A., Li, Y., He, J., et al. (2013). Borna disease virus encoded phosphoprotein inhibits host innate immunity by regulating miR-155. Antiviral Res. 98, 66–75. doi:10.1016/j.antiviral.2013.02.009
Zhang, H., Li, Z., Li, Y., Liu, Y., Liu, J., Li, X., et al. (2013). A computational method for predicting regulation of human microRNAs on the influenza virus genome. BMC Syst. Biol. 7 (S2), S3. doi:10.1186/1752-0509-7-S2-S3
Zhang, H., Penninger, J. M., Li, Y., Zhong, N., and Slutsky, A. S. (2020a). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46 (4), 586–590. doi:10.1007/s00134-020-05985-9
Zhang, J., Zhou, L., Yang, Y., Peng, W., Wang, W., and Chen, X. (2020b). Therapeutic and triage strategies for 2019 novel coronavirus disease in fever clinics. Lancet Respir. Med. 8 (3), e11–e12. doi:10.1016/S2213-2600(20)30071-0
Zhang, Y., Li, T., Fu, L., Yu, C., Li, Y., Xu, X., et al. (2004). Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett. 560, 141–146. doi:10.1016/S0014-5793(04)00087-0
Zhao, G., Shi, S.-Q., Yang, Y., and Peng, J.-P. (2006). M and N proteins of SARS coronavirus induce apoptosis in HPF cells. Cell Biol. Toxicol. 22, 313–322. doi:10.1007/s10565-006-0077-1
Zhao, X., Guo, F., Liu, F., Cuconati, A., Chang, J., Block, T. M., et al. (2014). Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. U.S.A 111, 6756–6761. doi:10.1073/pnas.1320856111
Zheng, B., He, M.-L., Wong, K.-L., Lum, C. T., Poon, L. L., Peng, Y., et al. (2004). Potent inhibition of SARS-associated coronavirus (SCOV) infection and replication by type I interferons (IFN-α/β) but not by type II interferon (IFN-γ). J. Interferon Cytokine Res. 24, 388–390. doi:10.1089/1079990041535610
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv 2020, 914952. doi:10.1101/2020.01.22.914952
Ziebuhr, J. (2005). The coronavirus replicase. Coronavirus Rep. Rev. Gen. 287, 57–94. doi:10.1007/3-540-26765-4_3
Keywords: coronavirus, miRNA, siRNA, natural products, phytochemicals, SARS-CoV-2
Citation: Kalhori MR, Saadatpour F, Arefian E, Soleimani M, Farzaei MH, Aneva IY and Echeverría J (2021) The Potential Therapeutic Effect of RNA Interference and Natural Products on COVID-19: A Review of the Coronaviruses Infection. Front. Pharmacol. 12:616993. doi: 10.3389/fphar.2021.616993
Received: 13 October 2020; Accepted: 14 January 2021;
Published: 23 February 2021.
Edited by:
Per-Johan Jakobsson, Karolinska Institutet (KI), SwedenReviewed by:
Priyia Pusparajah, Monash University Malaysia, MalaysiaChun Yang, The First Affiliated Hospital of Nanjing Medical University, China
Copyright © 2021 Kalhori, Saadatpour, Arefian, Soleimani, Farzaei, Aneva and Echeverría. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Hosein Farzaei, bWguZmFyemFlaUBnbWFpbC5jb20=; Javier Echeverría, amF2aWVyLmVjaGV2ZXJyaWFtQHVzYWNoLmNs
 Mohammad Reza Kalhori
Mohammad Reza Kalhori Fatemeh Saadatpour
Fatemeh Saadatpour Ehsan Arefian2
Ehsan Arefian2 Masoud Soleimani
Masoud Soleimani Mohammad Hosien Farzaei
Mohammad Hosien Farzaei Ina Yosifova Aneva
Ina Yosifova Aneva Javier Echeverría
Javier Echeverría