
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 12 April 2021
Sec. Ethnopharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.594833
This article is part of the Research TopicExploration of Natural Product Leads for Multitarget-Based Treatment of Cancer - Computational to Experimental JourneyView all 15 articles
Hepatocellular carcinoma (HCC) is the fifth most common malignant tumor and the second leading cause of cancer-related death in the world. Plumbagin (PL) is a small molecule naphthoquinone compound isolated from Plumbago zeylanica L. that has important anticancer properties, but its mechanism requires further investigation. In this study, we used a comprehensive network pharmacology approach to study the mechanism of action of PL for the treatment of HCC. The method includes the construction of multiple networks; moreover, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to identify biological processes and signaling pathways. Subsequently, in vitro experiments were performed to verify the predicted molecular mechanisms obtained from the network pharmacology-based analysis. Network pharmacological analysis showed that PL may exert anti-HCC effects by enhancing reactive oxygen species (ROS) production to generate oxidative stress and by regulating the PI3K/Akt and MAPK signaling pathways. In vitro experiments confirmed that PL mainly mediates the production of ROS, regulates the PI3K/Akt and MAPK signaling pathways to promote apoptosis and autophagy, and shows significant therapeutic effects on HCC. In conclusion, our work proposes a comprehensive systems pharmacology approach to explore the potential mechanism of PL for the treatment of HCC.
Hepatocellular carcinoma (HCC) comprises a group of malignant tumors that seriously threaten human life. Surgical treatment is currently the accepted treatment of choice, but the high recurrence and high metastatic characteristics of HCC severely restrict the survival prognosis of HCC patients (Farazi and Depinho, 2006). In recent years, studies have found that traditional Chinese medicine can inhibit the growth and proliferation of HCC cells in various ways and has a prominent role in HCC treatment (Franco and Usatoff, 2001; Belghiti and Fuks, 2012). Therefore, it is a feasible research direction to search for potential liver cancer treatment drugs by screening and selecting biologically active ingredients that can effectively reduce liver cancer mortality (Su et al., 2019).
East Asian (China, Japan, and South Korea) and traditional Chinese medicine (TCM) have been widely used for disease prevention and treatment for more than two thousand years. Chinese herbal medicine is an important part of Chinese medicine, including plant medicine, animal medicine and mineral medicine. In recent years, Chinese medicine has received increasing attention (Lam et al., 2015). Plumbagin (PL) is a natural naphthoquinone compound isolated and purified from Plumbago zeylanica L., a traditional Chinese medicinal plant from China (Tilak et al., 2004; Sakamoto et al., 2011). A large number of studies have shown that PL has antitumor, anti-liver fibrosis, anti-hepatitis B virus, antiplatelet activity, anti-atherosclerosis and other effects (Panda and Kamble, 2016). Studies have shown that PL has significant inhibitory effects on leukemia, lung cancer, prostate cancer, melanoma, breast cancer and other malignant tumors (Subramaniya et al., 2011; Tian et al., 2011; Liu et al., 2017). Previous experimental studies have shown that PL can induce HCC SMMC-7721 cell apoptosis by inhibiting the epithelial-mesenchymal transition (EMT) and inhibit HCC cell angiogenesis and proliferation through the SDF-1-CXCR4/CXCR7 axis (Wei et al., 2019; Zhong et al., 2019).
However, more detailed therapeutic targets and signaling mechanisms of PL acting on HCC have not been revealed. Therefore, to further explore the relevant mechanisms of PL acting on HCC, this study aimed to identify therapeutic targets and signaling mechanisms through network pharmacology based on bioinformatics and verify them through relevant experiments in vitro. Network pharmacology is an emerging new method to explore the systemic mechanism of therapeutic compounds in diseases (Kohl et al., 2011; Szklarczyk et al., 2016a). The workflow is shown in Figure 1. Using network pharmacology to determine herbal targets and potential mechanisms is becoming increasingly important to save money, effort and time required for drug discovery and design (Győrffy et al., 2013; Zhang et al., 2013). Indeed, network pharmacology has successfully led to the construction and visualization of drug-disease-target networks, which are helpful for evaluating drug mechanisms from multiple perspectives (Gao et al., 2016; Fang et al., 2017). Therefore, network pharmacology can be used to determine the pharmacological target and mechanism of action of PL in HCC and provide guidance and evidence for future research on the use of PL to treat HCC.
The predicted genes were obtained by searching the keyword “plumbagin” in the STITCH database (http://stitch.embl.de) (Szklarczyk et al., 2016b). The chemical structural formula of PL was entered into the SuperPred database (http://prediction.charite.de) (Mathias et al., 2008) and the PhamMapper database (http://lilab.ecust.edu.cn/pharmmapper/help.php) (Li et al., 2017). Additionally, the UniProt database (https://www.UniProt.org/) was combined with the literature to collect PL-predictive genes (Rolf et al., 2004), and the Swiss Target Prediction database (http://www.swisstargetprediction.ch/) (Antoine et al., 2019) was used to supplement the gene prediction information. We predicted the target genes of the compound by selecting the compound with the highest “Tanimoto score” and set the “minimum required interaction score” to “high confidence (0.700)” in STITCH. The threshold of “gene probability” was set to >0.6, and the related genes of the predicted compounds in the “Swiss Target Forecast” were selected.
The significant prediction genes of HCC were retrieved from OncoDB.HCC (http://oncodb.hcc.ibms.sinica.edu.tw/) (Su et al., 2007) and the GeneCards database (https://www.genecards.org/) (Marilyn et al., 2010). OncoDB.HCC can effectively integrate data sets in public references, provide a multidimensional view of current HCC research, and is the first comprehensive HCC genome database. Potential genes associated with HCC were collected from GeneCards with the keyword “Hepatocellular carcinoma.”
Venny2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/) (Shade and Handelsman, 2012) was used to screen common targets related to PL and HCC. The Cytoscape v3.7.1 (Shannon et al., 2003) software was used to establish target disease and component-target network models, and the merge function was used to build component-target-disease network models. The results were analyzed to determine the relationships in the network model. The proteins were sorted according to their degree of binding.
Gene Ontology and KEGG pathway analyses can clarify the role of potential targets by gene function and signaling pathways. The Bioconductor package “org.Hs.eg.db” was installed and run in the R software (Stoll et al., 2005). The drug-disease common targets were converted into Entrez IDs, and then the “clusterProfiler” package was installed in the R software. According to the converted Entrez IDs, enrichment analysis of key target gene GO functions and analysis of KEGG signaling pathways were performed with p < 0.05. The results were output in the form of bar and bubble charts.
Plumbagin (PL) was purchased from Sigma-Aldrich (St. Louis, MO, United States) with purity ≥98%; the Acridine Orange (AO)/Ethidium bromide (EB) Double Stain Kit was from Solable Technology (Beijing, China); N-acetyl-l-cysteine, SB203580, and SB202190 were from Sigma-Aldrich (St. Louis, MO, United States); SC-79, MEK2206, 3-MA, and Z-VAD-FMK were from Selleck (Texas, United States); the BCA Protein Assay Kit, ROS Assay Kit, Annexin V-FITC Apoptosis Detection Kit and Cell lysis buffer for Western were all obtained from Beyotime Biotechnology (Shanghai, China); antibodies against Akt, phospho-Akt, mTOR, phospho-mTOR, p38 MAPK, phospho-p38 MAPK, PI3K, phospho-PI3K, LC3B, cleave-RP, and cleave-caspase 3 were from Cell Signaling Technology, Inc. (Boston, MA, United States); and β-p38 MAPK was purchased from Boster Technology (Wuhan, China).
The human HCC cell lines SMMC-7721 and BEL-7404 were purchased from the Shanghai Institute of Biological Sciences (Shanghai, China). The DMEM high-glucose medium required for human HCC SMMC-7721 cell culture and the RPMI-1640 medium required for human HCC BEL-7404 cells were from Thermo Fisher Scientific (MA, United States). The complete culture medium contained 10% fetal bovine serum (FBS), 100 μg/ml penicillin and 100 μg/ml streptomycin, which were obtained from Invitrogen (CA, United States). The cells were cultured in an incubator at 37°C with a humidified atmosphere of 5% CO2.
Cell operations were performed on an ultraclean workbench. After the cells were recovered, SMMC-7721 and BEL-7404 cells at 80% confluence were digested into seed plates and seeded in 96-well plates at a density of 5 × 103 cells/well. After 24 h, the cells were completely attached to the wall and then synchronized. Each group was equipped with four auxiliary holes. A negative control well was set up without seeding cells. After treatment with various concentrations of PL (0.5, 3, 6, and 10 μM) for 6, 12 or 24 h in a 5% CO2 incubator at 37°C, 20 μL of 5 mg/ml MTT solution was added to each well, the plates were wrapped in foil and protected from light, and then placed in an incubator to continue incubation for an additional 3–4 h. The absorbance value of each well was measured using a continuous spectrum scanning microplate reader (Thermo Fisher, MA, United States) at a wavelength of 560 nm.
SMMC-7721 and BEL-7404 cells in logarithmic growth phase were digested and plated. Three multi-wells were set up and inoculated into a 24-well culture plate according to the standard of 500 μL per well, after which a moderate concentration of PL was added to stimulate the cells for 24 h. The supernatant was discarded after the intervention, a final concentration of 1 μg/ml AO/EB was added, and the plate was incubated for 10 min in the dark. Observation by fluorescence microscopy (Olympus, Tokyo, Japan) revealed that AO produces green fluorescence in the cytosol and the nuclear compartment (emission peak between 530 and 550 nm). Fluorescence microscopy showed four cell morphologies. The nuclear chromatin of living cells was green and showed a normal structure; the nuclear chromatin of early apoptotic cells was green, though the shape was shrunken or rounded; the nuclear spot material of late apoptotic cells was orange-red, and the shape was solid or as round beads; and the nuclear chromatin of nonapoptotic dead cells was orange-red, with normal cell morphology.
ROS production was determined using a 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) assay (Beyotime, Shanghai, China). After treatment, SMMC-7721 and BEL-7404 cells were incubated with DCFH-DA for 30 min at 37°C in the dark. The cells were then harvested and suspended in PBS. The fluorescence intensity in each well was detected by an LSM 700 confocal microscope (Zeiss, Hamburg, Germany).
The instructions of the Annexin V-FITC Apoptosis Detection Kit were strictly followed to process each group of cells. The cells were digested with trypsin without EDTA and washed twice with prechilled PBS, and 10–50 × 104 cells were collected. The cells were then resuspended in 100 μL of 1×Binding Buffer, and then 5 μL each of Annexin V-FITC and PI staining solution were added. The lysates were then mixed gently and incubated at room temperature in the dark for 10 min. The cells were detected by flow cytometry within 1 h. Apoptosis rate = (number of apoptotic cells/total number of cells observed) × 100%.
The cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min. The cells were then blocked with 2% bovine serum albumin (BSA) at 37°C for 30 min, after which the cells were incubated with anti-LC3B (1:100, Cell Signaling Technology, Boston, United States) overnight at 4°C. The cells were then incubated with the corresponding secondary antibodies for 1 h at room temperature. The nuclei were stained with DAPI for 10 min and washed twice with PBS, and then images were captured using an LSM 700 confocal microscope (Zeiss, Hamburg, Germany).
SMMC-7721 and BEL-7404 cells treated with the preset intervention conditions were collected and proteins were extracted. After using BSA to measure the absorbance, a standard curve was prepared versus the standard, and the protein concentration of the sample was determined according to the standard curve. The samples were prepared according to the instructions for the BCA protein quantitation kit. Depending on the relative molecular weights of the proteins, 7.5–12.5% PAGE-SDS gel electrophoresis was performed, and the proteins were then transferred to PVDF membranes. The PVDF membranes were blocked with 5% skimmed milk powder for 1 h at room temperature. Then, the following appropriately diluted primary antibodies were added: anti-β-actin (1:200), PI3K (1:1000), p-PI3K (1:500), Akt (1:2000), p-Akt (1:500), mTOR (1:1000), p-mTOR (1:1000), p38 MAPK (1:1000), p-p38 MAPK (1:500), cleave-caspase 3 (1:1000), cleave-PARP (1:1000), and LC3B (1:1000). Next, the blots were incubated at 4°C overnight. After washing three times with PBS, the corresponding secondary antibody was added and incubated for 2 h at room temperature. After incubating with the luminescent solution from the ECL kit, the film was exposed in the cassette, the protein bands were developed, and Band Scan software was used to scan grayscale images.
Data are expressed as the means ± standard deviation (SD). Statistical analysis was performed using SPSS 22.0. One-way ANOVA was used for comparisons between groups, and the difference was statistically significant at p < 0.05.
The 3D chemical structure of PL was imported into the chemical substance interaction search tool STITCH and the SuperPred and PhamMapper databases for retrieval; the Swiss Target Prediction database was used to supplement the target information. Then, the corresponding targets were input into the UniProt database, and after removing the duplicates, 98 targets of PL were obtained; the list of targeted genes is shown in Supplementary Table S1. Potential genes related to HCC were collected from GeneCards with the keyword “hepatocellular carcinoma.” In addition, the potential targets of HCC were retrieved from the OncoDB.HCC database, and after removing the duplicates, 6192 targets related to HCC were obtained; the list of targeted genes is shown in Supplementary Table S2.
Among the 6192 HCC-related targets discovered, 84 targets were common targets corresponding to PL. Through Venny 2.1.0, AKT1, ALDH1A1, MTOR, NR0B2, FOXO3, FOXO1, MMP9, SOD1, JUND, RECQL, CXCR4, NQO1 and other common targets were obtained (Figure 2A and Table 1). The 84 drug-disease common targets of PL and HCC were input into the Cytoscape 3.7.1 software, and a network diagram of the “component-target-disease” interaction was drawn (Figure 2B). The above 84 common targets were entered into the STRING database, and the PPI network of protein interactions was analyzed (Figure 2C). The ranking of the top 30 targets is shown in Figure 2D.
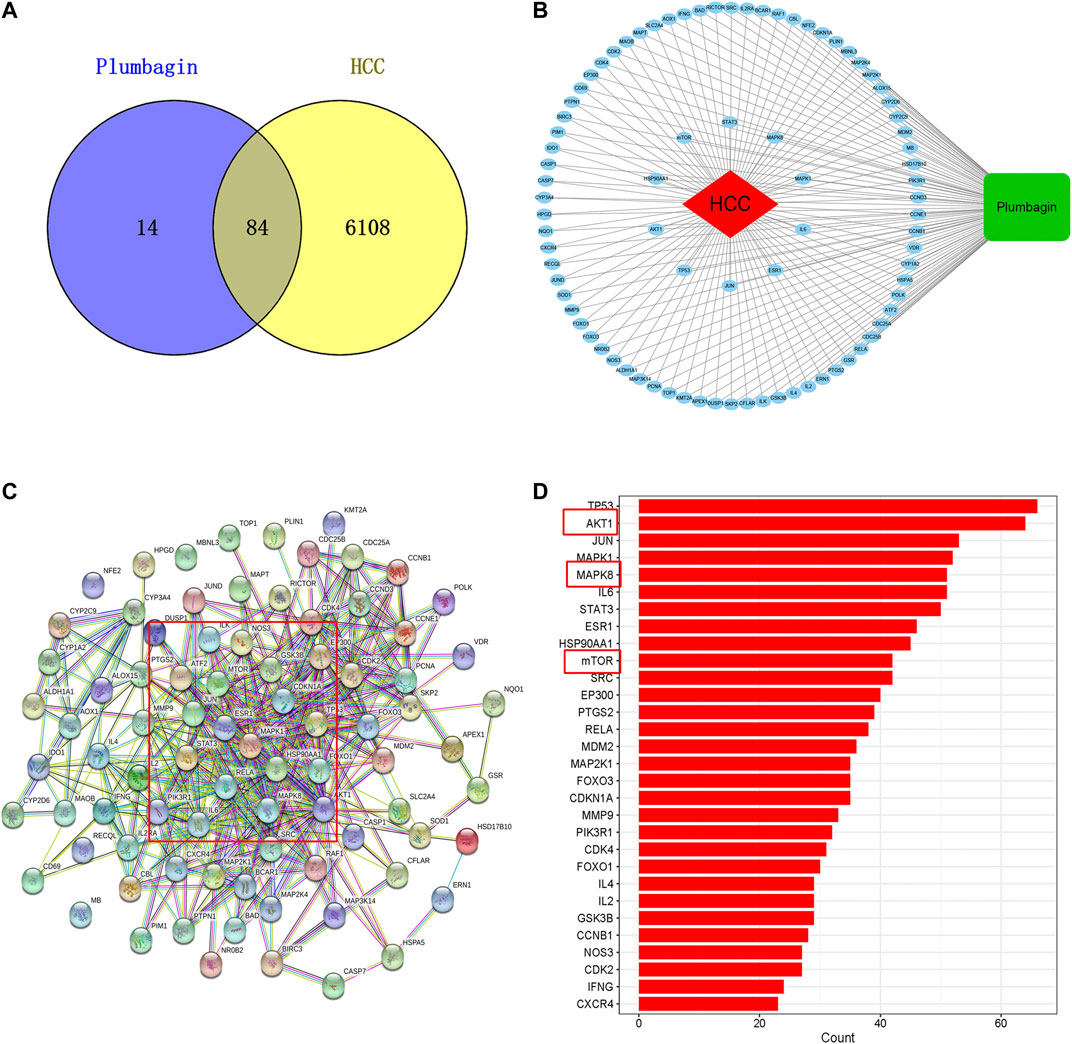
FIGURE 2. (A) Venn diagram of common targets of plumbagin (PL) in HCC. (B) Network of PL targets and HCC targets. The rectangle represents the compound PL, the ellipse represents 84 common targets, and the diamond represents HCC. The protein-protein interaction (PPI) network based on targets of PL in HCC. (C) Nodes represent different proteins. Edges represent protein-protein associations, and the line thickness indicates the strength of data support. (D) Bar plot of the protein-protein interaction (PPI) network. The X-axis represents the number of neighboring proteins of the target protein. The Y-axis represents the target proteins.
GO enrichment analysis was used to further study the biological processes, cellular components and molecular functions of the 84 common targets. As shown in Figure 3A, the intersection genes were enriched in 1,560 biological processes, and there were 164 biological processes with a number of enriched genes >10. The results indicated that the top 20 GO items with significant abundance (p < 0.001) are mainly related to biological processes such as oxidative stress, reactive oxygen synthesis, cell proliferation, and peptidyl-serine phosphorylation. Intersect gene enrichment analysis of the expression processes of the 27 cell components revealed that they are mainly related to transferase complex, transferring phosphorus-containing groups, transcription factor complex, and membrane raft (Figure 3B). The intersection genes were enriched in 100 processes related to molecular function, mainly related to protein serine/threonine kinase activity and phosphatase binding (Figure 3C). The above results indicate that PL correlates highly with anticancer activity, and PL may exert anti-HCC effects by participating in oxidative stress and reactive oxygen synthesis, regulating protein kinase phosphorylation.
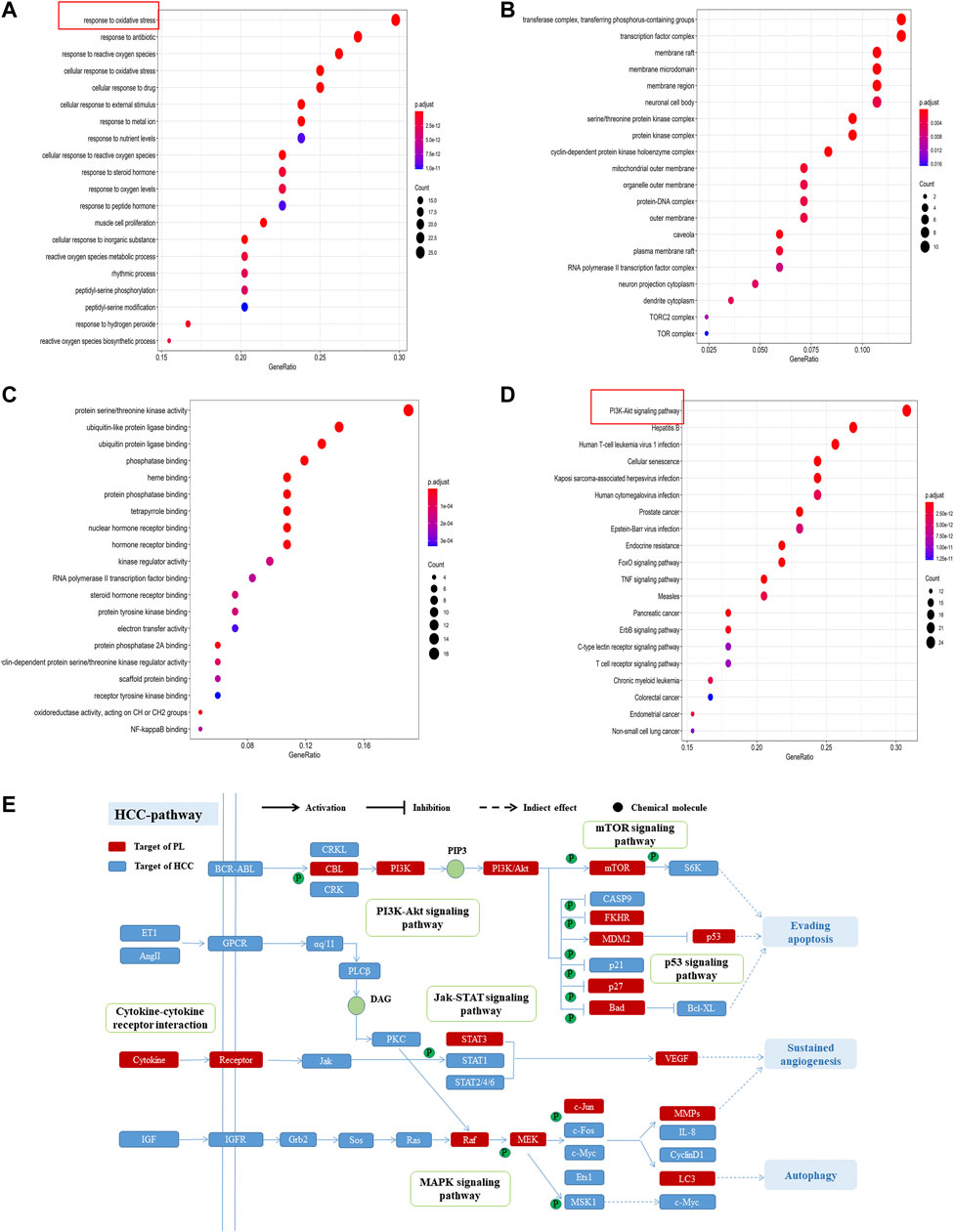
FIGURE 3. (A) GO enrichment analysis of biological processes of common targets of PL and HCC. The Y-axis represents significant GO biological processes, and the X-axis represents the counts of enriched targets. The gradient of color represents the different adjusted p values. (B) GO enrichment analysis of cell components of common targets of PL and HCC. The Y-axis represents significant cell components, and the X-axis represents the counts of enriched targets. The gradient of color represents the different adjusted p values. (C) GO enrichment analysis of the molecular functions of common targets of PL and HCC. The Y-axis represents significant GO molecular functions, and the X-axis represents the counts of enriched targets. The gradient of color represents the different adjusted p values. (D) KEGG analysis for common targets of PL and HCC. The Y-axis represents significant KEGG pathways, and the X-axis represents the ratio of enriched targets in a pathway to all common targets. The size of the nodes shows the count of targets, and the gradient of color represents the adjusted p value. (E) Distribution of target proteins of PL on the compressed “HCC pathway.” Five pathways form the tubular HCC pathway. Arrows show activation activity, T-arrows show inhibition activity, and segments represent indirect activation effects or inhibition effects.
Furthermore, of the 84 common targets, 78 genes participate in 154 KEGG pathways with adjusted p-values <0.01; after sorting the adjusted p-values, the top 20 pathways were analyzed (Figure 3D). The results show that the common targets of PL and HCC are mainly enriched in the PI3K-Akt, mTOR and MAPK signaling pathways. Then, considering the complex mechanism of PL to treat HCC, a complete “HCC pathway” was constructed by integrating the key pathways obtained through KEGG network analysis, as shown in Figure 3E. The results indicate that these integration pathways involve multiple pathophysiological modules, such as cell survival, proliferation, apoptosis, and angiogenesis. The PI3K/AKT pathway is responsible for regulating cell proliferation and apoptosis, and the mTOR/p38 MAPK pathway is responsible for regulating cell autophagy. Therefore, we performed in vitro experiments to validate that PL promotes apoptosis and autophagy through the PI3K-Akt/p38 MAPK signaling pathway, thereby explaining the potential therapeutic mechanism of PL in HCC treatment.
The MTT assay was found that PL inhibited BEL-7404 and SMMC-7721 cell proliferation (Lin et al., 2018). And the inhibitory effect of PL on BEL-7404 and SMMC-7721 cell proliferation was concentration- and time-dependent. After treatment with different concentrations of PL (0–10 μM) at three time points (6, 12 and 24 h), the cell proliferation rate decreased significantly (p < 0.05). Within the dose range of 3–10 μM, the cell growth inhibition rate reached greater than 50% within 24 h (Figures 4A,C). The half-inhibitory concentrations (IC50) of PL on BEL-7404 hepatocellular carcinoma cells were 4.689 μM at 6 h, 2.770 μM at 12 h, and 2.457 μM at 24 h (Figure 4B). The half-inhibitory concentrations (IC50) of PL on SMMC-7721 HCC cells were 36.05 μM at 6 h, 5.41 μM at 12 h, and 2.43 μM at 24 h (Figure 4D) (Lin et al., 2018).
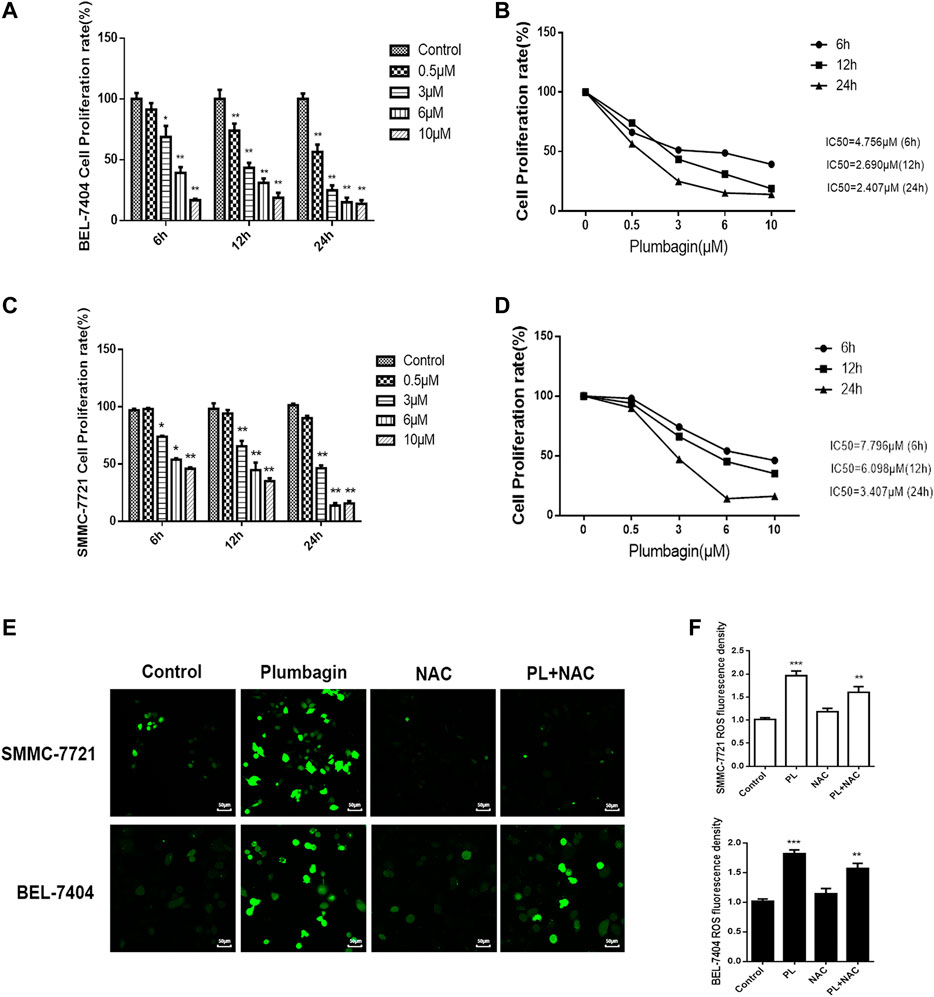
FIGURE 4. (A) Cell viability of BEL-7404 cells treated with various concentrations of PL (0.5, 3, 6, and 10 μM) for 6, 12 or 24 h *; p < 0.05, **; p < 0.01 ***; p < 0.001, compared with the 0 μM group. (B) The 50% inhibitory concentrations (IC50) of BEL-7404 cells were 4.756 μM at 6 h, 2.690 μM at 12 h, and 2.407 μM at 24 h. (C) Cell viability of SMMC-7721 cells treated with various concentrations of PL (0.5, 3, 6, and 10 μM) for 6, 12 or 24 h; p < 0.05, **; p < 0.01 ***; p < 0.001, compared with the 0 μM group. (D) The 50% inhibitory concentrations (IC50) of SMMC-7721 cells were 7.796 μM at 6 h, 6.098 μM at 12 h, and 3.407 μM at 24 h. (E) The production of ROS was measured by DCFH-DA to assess the level of oxidative stress. SMMC-7721 and BEL-7404 cells were pretreated with 10 mM NAC for 30 min and were then treated with 5 μM PL for 24 h, and fluorescence was detected by confocal fluorescence microscopy. Scale bar, 50 μm. (F) For statistical analysis, the mean DCFH-DA fluorescence intensity (representing cellular ROS level) was measured from 9 random fields for each culture. All values represent the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01 vs. normal control group, respectively.
Oxidative stress mediates the pathogenesis of liver cancer. The network pharmacology GO analysis results indicate that PL may mediate oxidative stress by activating the production of ROS and may have an anti-liver cancer effect. In our in vitro validation experiments, we treated BEL-7404 and SMMC-7721 cells with PL, the antioxidant NAC and PL+NAC and then detected levels of ROS in the cells using fluorescent probe technology. The results are shown in Figure 4E. After PL treatment, ROS expression levels in the two cell groups were significantly increased. Statistical analysis showed that after treatment with PL+NAC antioxidants, ROS expression levels were lower than in the PL group but higher than in the NAC group (Figure 4F). These findings indicate that PL may mediate oxidative stress through the production of ROS and may promote an anti-liver cancer effect.
SMMC-7721 and BEL-7404 cells were subjected to AO/EB staining (Lin et al., 2018). Observation under a fluorescence microscope showed that cells in the PL group emitted green fluorescence after AO staining. The nuclei presented one to several bead-shaped bodies of varying sizes, called apoptotic bodies, which are characterized by smooth and clear edges, and the fluorescence under the excitation of a fluorescence microscope is enhanced and uniform. These findings are consistent with the nuclear morphological features of apoptosis. In addition, the cell membrane may display a “budding” phenomenon. Late apoptotic cells with damaged cell membranes were simultaneously stained by EB and showed the same nuclear morphological characteristics, but the fluorescence showed orange-red, the green fluorescence of cells in the PL+NAC group decreased after AO staining, and the formation of apoptotic bodies was reduced, while the green fluorescence intensity was stronger than that of the NAC group alone (Figures 5A,B). These results indicate that PL may mediate apoptosis through ROS.
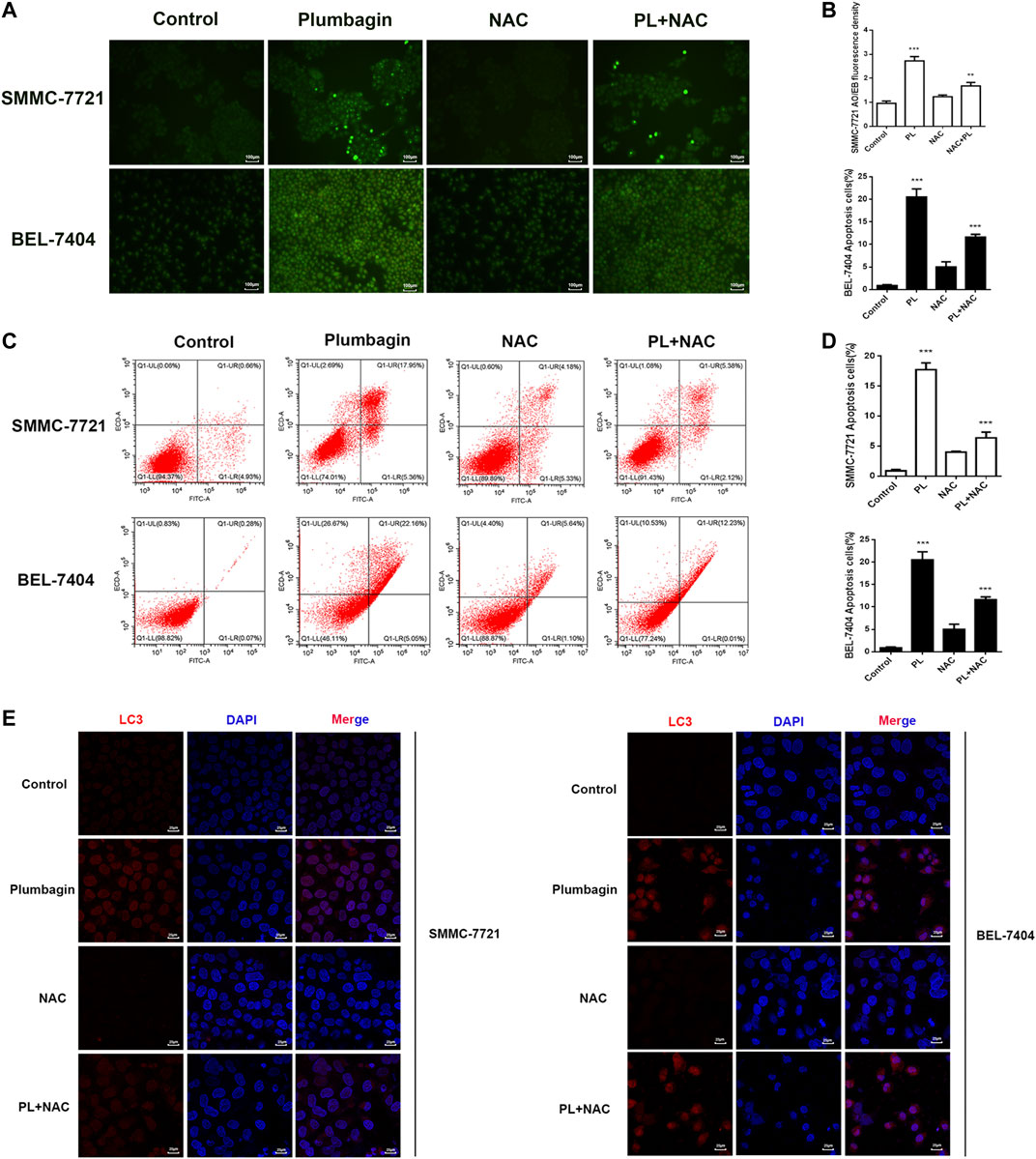
FIGURE 5. (A) AO/EB staining to detect the formation of PL-induced BEL-7404 cells and SMMC-7721 apoptotic bodies. Scale bar, 100 μm. All data are representative of at least three independent experiments. (B) Data were shown as the mean ± SD. *p < 0.05, **p < 0.01 vs. the normal control group. (C) SMMC-7721 and BEL-7404 cells were pretreated with 10 mM NAC for 30 min and were then treated with 5 μM PL for 24 h. Then, the ratio of apoptosis was measured and analyzed by Annexin V-FITC/PI staining and flow cytometry. (D) Data are shown as the mean ± SD. *p < 0.05, **p < 0.01 vs. the normal control group. (E) SMMC-7721 and BEL-7404 cells were pretreated with 10 mM NAC for 30 min and were then treated with 5 μM PL for 24 h. Immunofluorescence detection of LC3 expression. The expression of LC3 (red) was significantly higher. The morphology of SMMC-7721 and BEL-7404 cells had a fusiform shape rather than a rounded shape. Cell nuclei were defined by DAPI (blue). Scale bar, 25 μm.
Moreover, we used flow cytometry to further verify that PL may mediate apoptosis through ROS. As shown in Figures 5C,D, after PL treatment, the percentage of apoptosis increased significantly compared with the control group, mainly with regard to the percentage of late apoptotic cells. The level of apoptosis in the PL+NAC group was significantly reduced. Therefore, these findings further indicate that PL may mediate apoptosis through ROS.
The expression level of the autophagy marker LC3 in BEL-7404 and SMMC-7721 cells after PL treatment was assessed by immunofluorescence detection. As depicted in Figure 5E, the red fluorescence of the autophagy marker LC3 in the PL group showed a concentrated state, the fluorescence dot density increased, and the fluorescence intensity increased, indicating that the expression level of LC3 protein increased. In contrast, the fluorescence intensity decreased in the PL+NAC antioxidant group. These findings indicate that PL may induce autophagy by mediating the generation of ROS in HCC cells, thereby playing a role in the treatment of liver cancer.
Western blotting revealed no significant difference in expression of Akt and mTOR phosphorylated protein in the PL low concentration group compared with the control group in SMMC-7721 cells (p > 0.05). After intervention, the medium- and high-concentration groups of PL showed significantly reduced expression of PI3K Akt and mTOR phosphorylated proteins and upregulated expression of p38 MAPK phosphorylated proteins (Figure 6A). As shown in Figure 6B, when using PI3K, MAPK, Akt, and mTOR as internal reference total proteins for gray value analysis, the difference was statistically significant (p < 0.05). For BEL-7404 cells, there was no significant difference in expression levels of PI3K, MAPK, Akt and mTOR phosphorylated proteins in the PL low-concentration group compared with the control group (p > 0.05). Additionally, PI3K, Akt, and mTOR phosphorylated protein expression was significantly downregulated in the PL medium- and high-concentration groups, with increased expression of p38 MAPK phosphorylated protein (Figure 6C). As shown in Figure 6D, the difference was statistically significant when using PI3K, MAPK, Akt, and mTOR as internal reference total proteins for gray value analysis (p < 0.05).
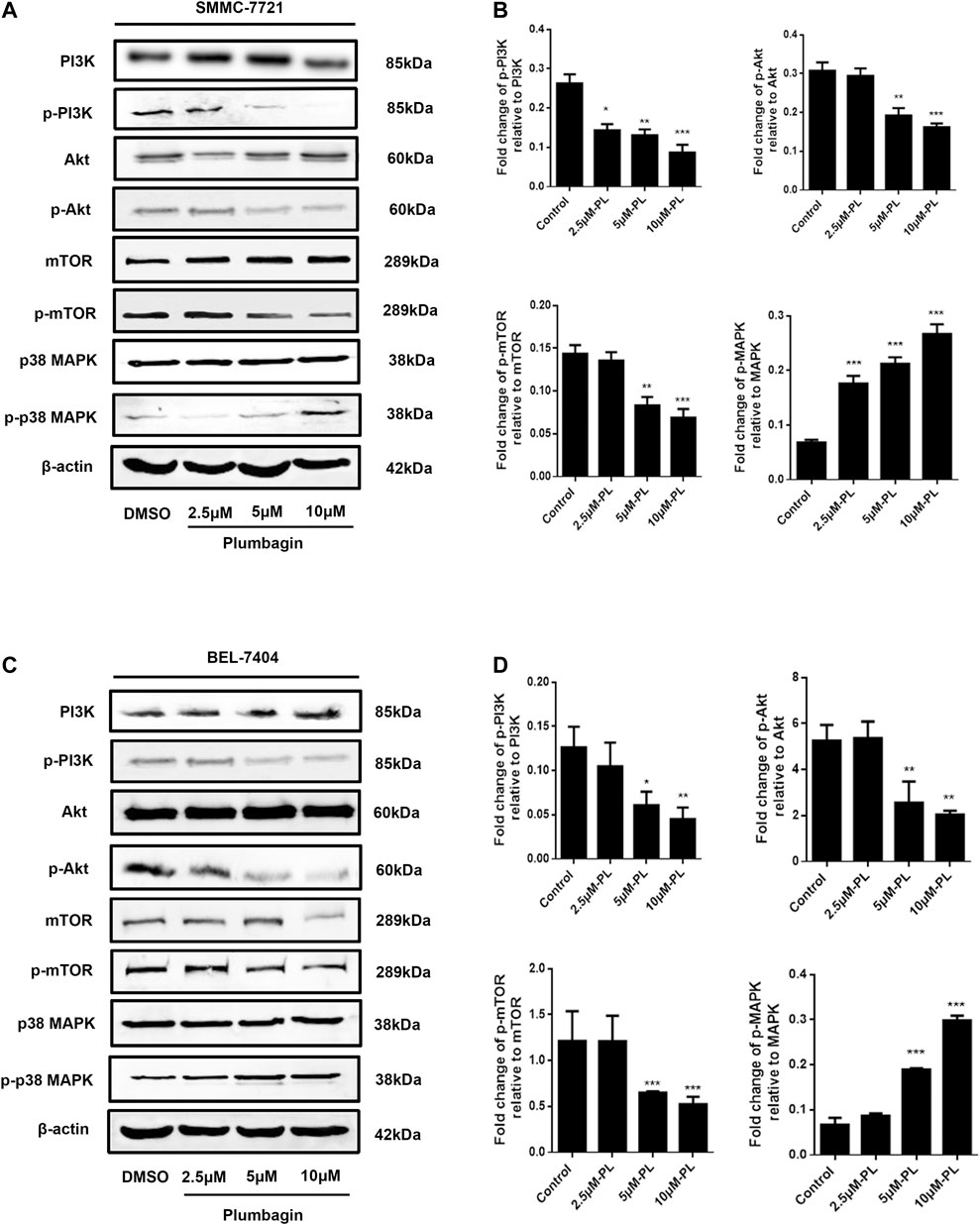
FIGURE 6. (A) Protein expression levels of PI3K, p-PI3K, Akt, p-Akt, mTOR, p-mTOR, MAPK, and p-MAPK in SMMC-7721 cells measured by Western blot after treatment with various concentrations of PL (0.5, 2.5, 5, and 10 μM). (B) PI3K, MAPK, Akt, and mTOR were used as internal reference total proteins for gray value analysis. Each bar represents the mean value ± standard deviation (SD) of the intensity of PL-induced cells across triplicate experiments. *p < 0.05, **p < 0.01 ***p < 0.001, compared with the control group. (C) Protein expression levels of PI3K, p-PI3K, AKT, p-AKT, mTOR, p-mTOR, MAPK, and p-MAPK in BEL-7404 cells measured by Western blot after treatment with various concentrations of PL (0.5, 2.5, 5, and 10 μM). (D) PI3K, MAPK, Akt, and mTOR were used as internal reference total proteins for gray value analysis. Each bar represents the mean value ± standard deviation (SD) of the intensity of PL-induced cells across triplicate experiments. *p < 0.05, **p < 0.01 ***p < 0.001, compared with the control group.
We employed Western blotting to verify that PL mediates apoptosis through regulation of the PI3K/Akt signaling pathway. The results showed that for the SMMC-7721 liver cancer cell line, expression of phosphorylated Akt was significantly downregulated in the PL group compared with the control group. At the same time, protein expression levels of cleave-caspase 3 and cleave-PARP were upregulated, but caspase 3 and PARP total proteins were decreased, as shown in Figures 7A,B. After pretreatment with the Akt agonist SC-79 and apoptosis-targeting Caspase inhibitor Z-VAD-FMK for 1 h followed by PL treatment for 24 h, p-Akt levels increased, and the levels of cleave-caspase 3 and cleave-PARP were reduced; however, caspase 3 and PARP total protein levels remained unchanged. Interestingly, after pretreatment with the Akt inhibitor MEK2206 for 1 h followed by PL incubation for 24 h, there was no significant difference in protein expression levels of p-Akt, cleave-caspase 3 and cleave-PARP compared with the PL treatment group alone.
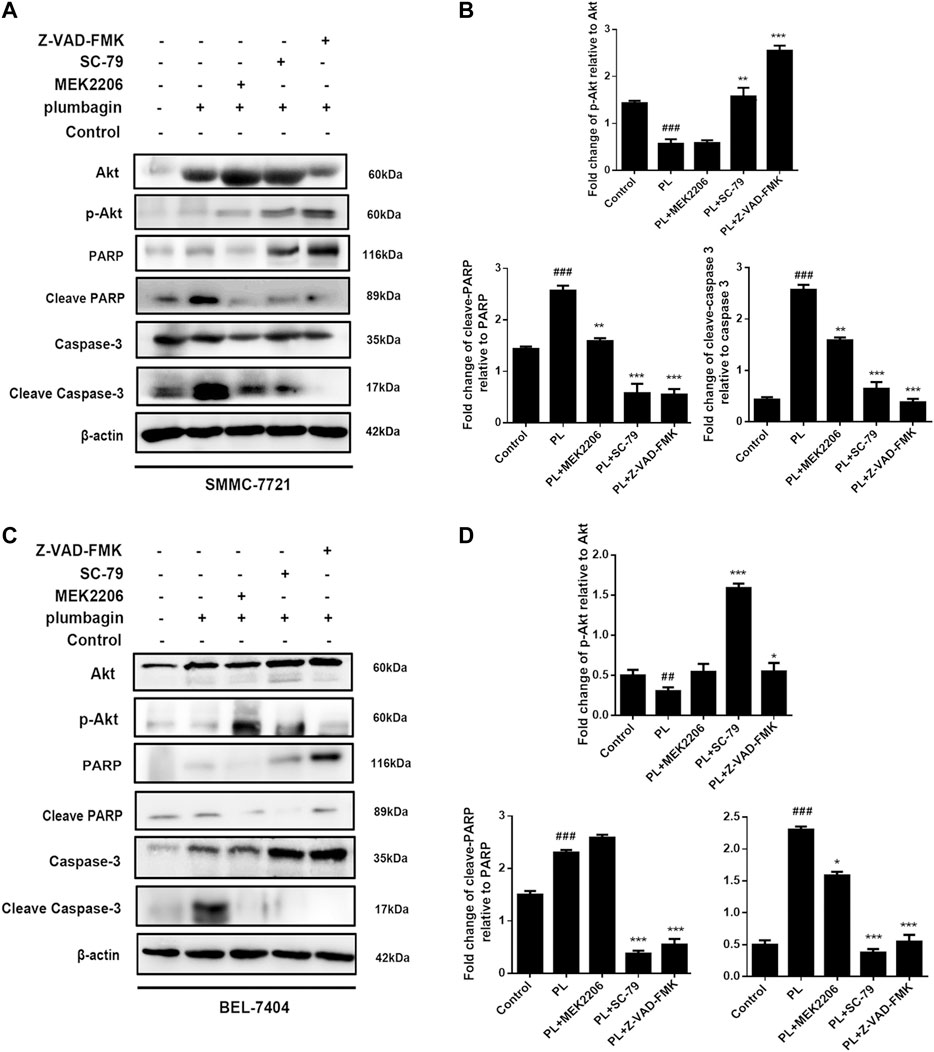
FIGURE 7. (A) Protein expression levels of p-Akt, Akt, PARP, cleaved PARP, caspase 3, and cleaved caspase 3 in SMMC-7721 cells measured by Western blot after treatment with 5 μM PL, 10 μM SC79 Akt agonist, 10 μM MEK2206 Akt inhibitor and 10 μM Z-VAD-FMK caspase inhibitor. (B) p-Akt, Akt, PARP, cleaved PARP, caspase 3, and cleaved caspase 3 were used as internal reference total proteins for gray value analysis. *p < 0.05, **p < 0.01 ***p < 0.001, compared with the PL group. #p < 0.05, ##p < 0.01 vs. the control group, respectively. (C) Protein expression levels of p-Akt, Akt, PARP, cleaved PARP, caspase 3, and cleaved caspase 3 in BEL-7404 cells measured by Western blot after treatment with 5 μM PL, 10 μM SC79 Akt agonist, 10 μM MEK2206 Akt inhibitor and 10 μM Z-VAD-FMK caspase inhibitor. (D) p-Akt, Akt, PARP, cleaved PARP, caspase 3, and cleaved caspase 3 were used as internal reference total proteins for gray value analysis. *p < 0.05, **p < 0.01 ***p < 0.001, compared with the PL group. #p < 0.05, ##p < 0.01 ###p < 0.001 vs. the control group.
In the BEL-7404 liver cancer cell line, pretreatment with the Akt agonist SC-79 and the apoptosis-targeting Caspase inhibitor Z-VAD-FMK for 1 h followed by PL treatment for 24 h upregulated p-Akt expression. Protein expression levels of cleave-caspase 3 and cleave-PARP were downregulated, which was consistent with the results for SMMC-7721 cells (Figures 7C,D). The above results indicate that PL mediates apoptosis by regulating the PI3K/Akt signaling pathway.
The experimental results showed that in SMMC-7721 cells, both PL and the autophagy agonist rapamycin upregulated the expression level of the autophagy marker protein LC3 and the phosphorylation level of p-p38 compared with the control group, and the difference was significant. In addition, the autophagy inhibitor 3-MA and the p38 MAPK inhibitors SB202190 and SB203580 jointly interfered in SMMC-7721 cells treated with PL. Compared with the PL treatment alone group, levels of LC3 were significantly downregulated in the p-p383-MA+PL group. These expression levels indicated suppression of PL-induced autophagy. In the SB202190+PL and SB203580+PL groups, LC3 expression was increased, whereas that of p-p38 was significantly decreased (Figures 8A,B).
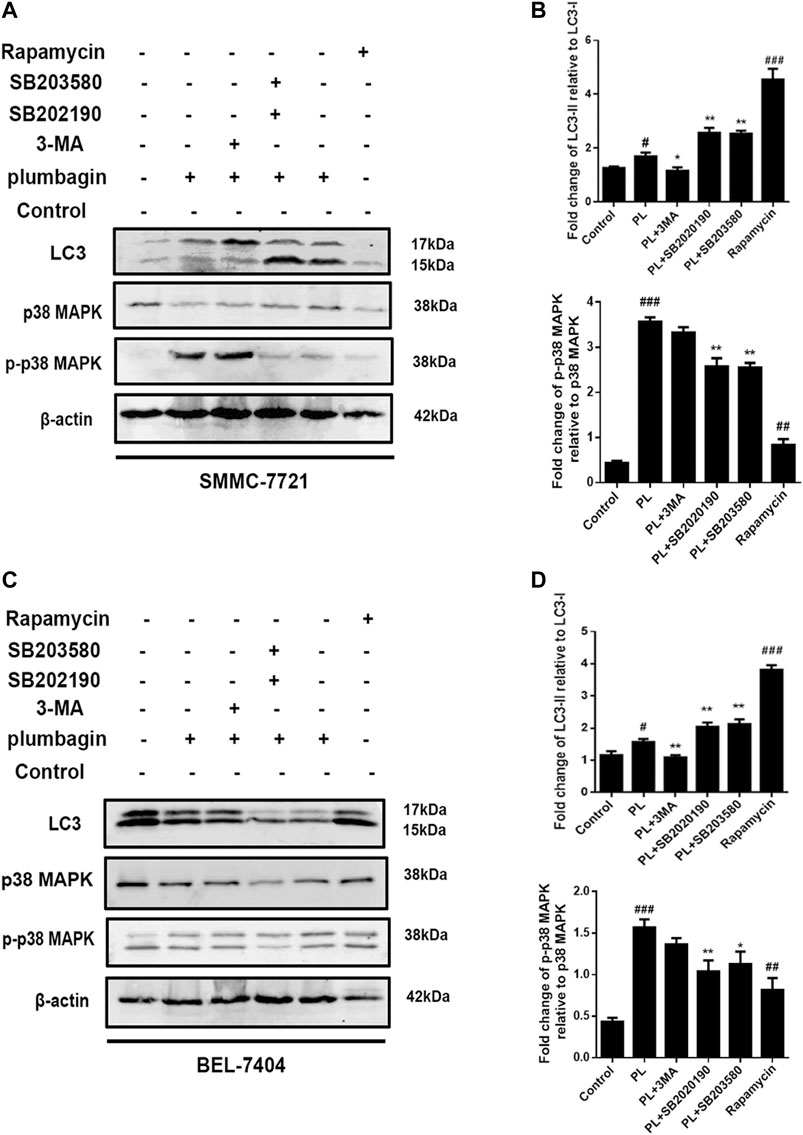
FIGURE 8. (A) Protein expression levels of LC3-I, LC3-II, p38 MAPK, and p-p38 MAPK in SMMC-7721 cells measured by Western blot after treatment with 5 μM PL, 10 μM 3-MA autophagy inhibitor, 10 μM rapamycin autophagy agonist, 10 μM SB202190, and 10 μM SB203580 p38 MAPK inhibitor. (B) LC3-I, LC3-II, p38 MAPK, and p-p38 MAPK were used as internal reference total proteins for gray value analysis. *p < 0.05, **p < 0.01 ***p < 0.001, compared with the PL group. #p < 0.05, ##p < 0.01 ###p < 0.001 vs. the control group. (C) Protein expression levels of LC3-I, LC3-II, p38 MAPK, and p-p38 MAPK in BEL-7404 cells measured by Western blot after treatment with 5 μM PL, 10 μM 3-MA autophagy inhibitor, 10 μM rapamycin autophagy agonist, 10 μM SB202190, and 10 μM SB203580 p38 MAPK inhibitor. (D) LC3-I, LC3-II, p38 MAPK, and p-p38 MAPK were used as internal reference total proteins for gray value analysis. *p < 0.05, **p < 0.01 ***p < 0.001, compared with the PL group. #p < 0.05, ##p < 0.01 ###p < 0.001 vs. the control group.
In BEL-7404 cells, expression levels of the autophagy marker protein LC3 in the PL and autophagy agonist rapamycin groups were significantly increased compared with the blank control group. In addition, the autophagy inhibitor 3-MA and the p38 MAPK inhibitors SB202190 and SB203580 acted in BEL-7404 cells treated with PL. Compared with the PL treatment group, the 3-MA+PL group showed significantly decreased levels of LC3 and p-p38. LC3 expression levels were increased in the SB202190+PL and SB203580+PL groups, while p38 phosphorylation expression levels were significantly reduced in the SB203580+PL group (Figures 8C,D). The above results indicate that PL may simultaneously upregulate the phosphorylation of the p38 MAPK protein and induce autophagy in HCC cells.
HCC is a malignant tumor with high morbidity and mortality in China and worldwide (Orcutt and Anaya, 2018 Jan-Mar). Due to its rapid proliferation and high metastasis and recurrence rates, it is not sensitive to the characteristics of conventional chemotherapy drugs and is prone to drug resistance; thus, treatment of HCC is facing a severe test. Although the current treatments for HCC include surgical resection, arterial intubation chemotherapy, radiotherapy and local treatment, the effects are not satisfactory. Moreover, substantial progress in the diagnosis and treatment of HCC has failed to greatly improve its efficacy. Therefore, international interest in finding new ways to treat HCC is also increasing. The development and application of new drugs is important for tumor treatment. The search for safe and effective anti-cancer drugs is a major research topic in the treatment of HCC. By exploring the regulation of multichannel signaling pathways, network pharmacology can improve the efficacy of drugs and the success rate of clinical trials, reducing the cost of drug discovery (Zhang et al., 2013). In this study, network pharmacology results from the network databases revealed core gene targets, and we explored the biological functions, pathways, and mechanisms of PL in HCC. The potential targets and signaling pathways of PL acting on HCC were further explored and verified through in vitro experiments. These findings indicate that PL is a promising resource with specific therapeutic effects on HCC.
In various human cancer cell lines, ROS is involved in autophagy, apoptosis and cell cycle arrest (Zou et al., 2017). Studies have shown that c-Jun induced by ROS is activated and regulated in HeLa cells, leading to apoptosis (Lo and Wang, 2013). In breast cancer MDA-MB-231 and MCF-7 cells, ROS-induced JNK activation induces apoptosis through mitochondrial membrane depolarization (Lebelo et al., 2020). In addition, antioxidant N-acetylcysteine (NAC) restored the depleted GSH contents of the new polyphenol conjugate DPP-23 in pancreatic cancer MIAPaCa-2 cells, further confirming that apoptosis and oxidation induced by DPP-23 stress are closely related. These findings also show that oxidative stress is an upstream event that induces apoptosis (Shin et al., 2014). Studies have shown that the anticancer effects of PL are mainly related to ROS generation, mitochondrial function, apoptosis and autophagy and related signaling pathways (Zhou et al., 2015). In our study, GO analysis results showed that the biological functions affected by the common targets of PL and HCC are mainly related to oxidative stress and ROS generation, and KEGG pathway analysis showed that the targets are mainly concentrated in the PI3K/Akt/and mTOR/MAPK signaling pathways. Our in vitro cell experiments confirmed that PL can mediate the generation of ROS and promote apoptosis. In addition, the phosphorylation levels of PI3K, Akt and mTOR proteins in the PI3K/Akt signaling pathway were downregulated. Our study also found that PL mainly mediates apoptosis through the mitochondrial apoptotic pathway. These results indicate that PL-induced apoptosis of human liver cancer cells is closely related to the increase in intracellular ROS, and ROS may be an upstream factor that regulates the PI3K/Akt/mTOR pathway. The interaction between PL-induced autophagy and apoptosis varies with different cell types, external stimuli and environments. Some studies have suggested that autophagy is a potential partner, antagonist or promoter of apoptosis. Our study found that the autophagy inhibitor 3-MA can attenuate PL-induced apoptosis of HCC cells. Therefore, we speculate that PL-induced autophagy can enhance apoptosis to a certain extent, and when autophagy is weakened by autophagy inhibitors, apoptosis will also be affected.
The p38 MAPK signaling pathway is thought to regulate autophagy. p38 mitogen-activated protein kinase (p38 MAPK) is a well-known kinase that is phosphorylated and participates in amino acid signal transduction in bacterial lipopolysaccharide, heat shock, osmotic stress and other environmental stress responses (Webber, 2010; Gupta et al., 2014). Recent studies have shown interaction between the p38α type of p38 MAPK and a new ATG9 binding partner, p38IP, to control the level of starvation-induced autophagy (Webber and Tooze, 2010). p38 MAPK has a dual role in autophagy: it is both a positive and a negative regulator. Importantly, phosphorylation of p38α leads to increased expression of the autophagy protein marker LC3 (Moruno-Manchón et al., 2013). Our study found that consistency between PL-induced autophagy in HCC cells and the positive effect of p38α in autophagy control. Overall, PL acts on SMMC-7721 and BEL-7404 cells to increase the phosphorylation level of p38α, thereby inducing autophagy.
Studies have also shown that the pyridinimidazole compound SB203580 is an inhibitor of p38 MAPK. It can inhibit the expression of p38 and can induce autophagy when acting on colorectal cancer cells. Prolonged inhibition can lead to cell death with autophagy characteristics (Lim et al., 2006). Our research also found that after co-intervention of the p38 MAPK inhibitors SB203580 and SB202190 with PL, the expression levels of LC3 increased, indicating that the level of autophagy increased. In addition, expression of p-p38/p38 was significantly reduced. These results indicate that using p38 MAPK inhibitors to reduce the phosphorylation levels of p38 can also induce autophagy, suggesting that this may be a potential mechanism that limits the degree of autophagy that cells can withstand and that p38α activity may determine the balance between cell survival and cell death during cell stress. As autophagy is becoming an important factor affecting many neurodegenerative diseases and cancers, p38 in the p38 MAPK signaling pathway may become a clinical target for autophagy control.
In conclusion, this study identified key targets of effects of PL against HCC through network pharmacology. In vitro experiments were conducted to verify the effective molecular mechanism by which PL targets HCC. This study shows that PL can promote apoptosis and induce autophagy through ROS-mediated PI3K/Akt and mTOR/MAPK signaling pathways and suggests that PL may exert anti-HCC effects through multiple targets and signaling pathways. This study also demonstrates that network pharmacology is of great significance for target screening and pathway prediction of drugs and diseases.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
DL, YF, YN, and WJ are the major contributors to this manuscript. YN conducted the analysis, wrote the first version of the manuscript, and finalized the manuscript. WJ downloaded the references and developed the graphs and tables in the manuscript. SS and LJ collected the data. DL (corresponding author) conceived and coordinated the study and critically evaluated the data. All authors read and approved the final manuscript.
The authors gratefully acknowledge financial support by the National Natural Science Foundation of China (No. 81760757), the Key projects of Guangxi Natural Science Foundation (2019JJD140032), and the Guangxi first-class discipline construction key project (2018XK020), The second batch of “Qihuang Project” high-level talent team cultivation project of Guangxi University of Traditional Chinese Medicine (2021001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All the authors thank all the people who helped and provided comments to complete this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.594833/full#supplementary-material.
Antoine, D., Olivier, M., and Vincent, Z. S. (2019). Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res.47, W357. doi:10.1093/nar/gkz382
Belghiti, J., and Fuks, D. (2012). Liver resection and transplantation in hepatocellular carcinoma. Liver Cancer 1, 71–82. doi:10.1159/000342403
Fang, J., Wang, L., Wu, T., Yang, C., Gao, L., Cai, H., et al. (2017). Network pharmacology-based study on the mechanism of action for herbal medicines in alzheimer treatment. J. Ethnopharmacol 196, 281–292. doi:10.1016/j.jep.2016.11.034
Farazi, P. A., and Depinho, R. A. (2006). Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687. doi:10.1038/nrc1934
Franco, D., and Usatoff, V. (2001). Resection of hepatocellular carcinoma. Hepatogastroenterology 48, 33–36. doi:10.1007/978-1-4615-0547-1_1
Gao, L., Wang, X., Niu, Y., Duan, D., Yang, X., Hao, J., et al. (2016). Molecular targets of Chinese herbs: a clinical study of hepatoma based on network pharmacology. Sci. Rep. 6, 24944. doi:10.1038/srep24944
Gupta, J., del Barco Barrantes, I., Igea, A., Sakellariou, S., Pateras, I. S., Gorgoulis, V. G., et al. (2014). Dual function of p38α MAPK in colon cancer: suppression of colitis-associated tumor initiation but requirement for cancer cell survival. Cancer cell 25, 484–500. doi:10.1016/j.ccr.2014.02.019
Győrffy, B., Surowiak, P., Budczies, J., and Lánczky, A. (2013). Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLOS ONE 8, e82241. doi:10.1371/journal.pone.0082241
Kohl, M., Wiese, S., and Warscheid, B. (2011). Cytoscape: software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303. doi:10.1007/978-1-60761-987-1_18
Lam, W., Jiang, Z., Guan, F., Huang, X., Hu, R., Wang, J., et al. (2015). Phy906(kd018), an adjuvant based on a 1800-year-old Chinese medicine, enhanced the anti-tumor activity of sorafenib by changing the tumor microenvironment. Sci. Rep. 5, 9384. doi:10.1038/srep09384
Lebelo, M. T., Joubert, A. M., and Visagie, M. H. (2020). Sulphamoylated estradiol analogue induces reactive oxygen species generation to exert its antiproliferative activity in breast cancer cell lines. Molecules 25, 4337. doi:10.3390/molecules25184337
Li, D., Ling, C., and Zhu, D. (2007). [Study on the quantitative evaluation on the degree of TCM basic syndromes often encountered in patients with primary liver cancer]. Zhongguo Zhong Xi Yi Jie He Za Zhi. 27, 602–5.
Li, Y., Chang, N., Han, Y., Zhou, M., Gao, J., Hou, Y., et al. (2017). Anti-inflammatory effects of Shufengjiedu capsule for upper respiratory infection via the ERK pathway. Biomed. Pharmacother. 94, 758–766. doi:10.1016/j.biopha.2017.07.118
Lim, S. J., Lee, Y. J., and Lee, E. (2006). P38mapk inhibitor sb203580 sensitizes human snu-c4 colon cancer cells to exisulind-induced apoptosis. Oncol. Rep. 16, 1131–1135.
Lin, Y., Chen, Y., Wang, S., Ma, J., Peng, Y., Yuan, X., et al. (2018). Plumbagin induces autophagy and apoptosis of smmc‐7721 cells in vitro and in vivo. J. Cell Biochem. 120, 9820-9830. doi:10.1002/jcb.28262
Liu, Y., Cai, Y., He, C., Chen, M., and Li, H. (2017). Anticancer properties and pharmaceutical applications of plumbagin: a review. Am. J. Chin. Med. 45, 423–441. doi:10.1142/S0192415X17500264
Lo, Y. L., and Wang, W. (2013). Formononetin potentiates epirubicin-induced apoptosis via ros production in hela cells in vitro. Chem. Biol. Interact 205, 188–197. doi:10.1016/j.cbi.2013.07.003
Marilyn, S., Irina, D., Justin, A., Naomi, R., Tsippi, I. S., Michael, S., et al. (2010). Genecards version 3: the human gene integrator. Database (Oxford). 2010, baq020. doi:10.1093/database/baq020
Mathias, D., Stefan, G., Jessica, A., Burghardt, W., and Robert, P. (2008). Superpred: Drug classification and target prediction. Nucleic Acids Res. 36 (suppl_2), W55-9. doi:10.1093/nar/gkn307
Moruno-Manchón, J. F., Pérez-Jiménez, E., and Knecht, E. (2013). Glucose induces autophagy under starvation conditions by a p38 mapk-dependent pathway. Biochem. J. 449, 497–506. doi:10.1042/bj20121122
Orcutt, S. T., and Anaya, D. A. (2018). Liver resection and surgical strategies for management of primary liver cancer. Cancer Control 25, 1073274817744621. doi:10.1177/1073274817744621
Panda, S., and Kamble, A. (2016). Genotypic and seasonal variation of plumbagin content from different populations of plumbago zeylanica L. In India. Proc. Natl. Acad. Sci., India, Sect. B. 86, 165–169. doi:10.1007/s40011-014-0432-6
Rolf, A., Amos, B., Wu, C. H., Barker, W. C., Brigitte, B., Serenella, F., et al. (2004). Uniprot: the universal protein knowledgebase. Nuclc Acids Res. 32, D115–D119. doi:10.1093/nar/gkh131
Sakamoto, S., Tanizaki, Y., Pongkitwitoon, B., Tanaka, H., and Morimoto, S. (2011). A chimera of green fluorescent protein with single chain variable fragment antibody against ginsenosides for fluorescence-linked immunosorbent assay. Protein Expr. Purif. 77, 124–130. doi:10.1016/j.pep.2011.01.010
Shade, A., and Handelsman, J. (2012). Beyond the venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi:10.1111/j.1462-2920.2011.02585.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shin, S. Y., Lee, J. M., Lee, M. S., Koh, D., Jung, H., Lim, Y., et al. (2014). Targeting cancer cells via the reactive oxygen species-mediated unfolded protein response with a novel synthetic polyphenol conjugate. Clin. Cancer Res. 20, 4302–4313. doi:10.1158/1078-0432.CCR-14-0424
Stoll, H., King, G., and Zeng, L. (2005). Whatif: R software for evaluating counterfactuals. J. Stat. Softw. 015. doi:10.18637/jss.v015.i04
Su, M., Guo, C., Liu, M., Liang, X., and Yang, B. (2019). Therapeutic targets of vitamin c on liver injury and associated biological mechanisms: a study of network pharmacology. Int. Immunopharmacol 66, 383–387. doi:10.1016/j.intimp.2018.11.048
Su, W., Chao, C., Yeh, S., Chen, D., Chen, P., and JouOncodb, Y. (2007). OncoDB.HCC: an integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 35, D727–D731. doi:10.1093/nar/gkl845
Subramaniya, B. R., Srinivasan, G., Sadullah, S. S., Davis, N., Subhadara, L. B., Halagowder, D., et al. (2011). Apoptosis inducing effect of plumbagin on colonic cancer cells depends on expression of cox-2. PLOS ONE. 6, e18695. doi:10.1371/journal.pone.0018695
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2016a). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi:10.1093/nar/gkw937
Szklarczyk, D., Santos, A., Von Mering, C., Jensen, L. J., Bork, P., and Kuhn, M. (2016b). Stitch 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–D384. doi:10.1093/nar/gkv1277
Tian, L., Yin, D., Ren, Y., Gong, C., Chen, A., and Guo, F. (2011). Plumbagin induces apoptosis via the p53 pathway and generation of reactive oxygen species in human osteosarcoma cells. Mol. Med. Rep. 5, 126–132. doi:10.3892/mmr.2011.624
Tilak, J. C., Adhikari, S., and Devasagayam, T. P. (2004). Antioxidant properties of plumbago zeylanica, an indian medicinal plant and its active ingredient, plumbagin. Redox Rep. 9, 219–227. doi:10.1179/135100004225005976
Webber, J. L. (2010). Regulation of autophagy by p38alpha MAPK. Autophagy 6, 292–293. doi:10.4161/auto.6.2.11128
Webber, J. L., and Tooze, S. A. (2010). Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 29, 27–40. doi:10.1038/emboj.2009.321
Wei, Y., Lv, B., Xie, J., Zhang, Y., Lin, Y., Wang, S., et al. (2019). Plumbagin promotes human hepatoma smmc-7721 cell apoptosis via caspase-3/vimentin signal-mediated Emt. Drug Des Devel Ther. Vol. 13, 2343–2355. doi:10.2147/dddt.s204787
Zhang, G. B., Li, Q. Y., Chen, Q. L., and Su, S. B. (2013). Network pharmacology: a new approach for Chinese herbal medicine research. Evid. Based Complement. Alternat Med. 2013, 621423–621429. doi:10.1155/2013/621423
Zhong, J., Li, J., Wei, J., Huang, D., Huo, L., Zhao, C., et al. (2019). Plumbagin restrains hepatocellular carcinoma angiogenesis by stromal cell-derived factor (sdf-1)/cxcr4-cxcr7 axis. Med. Sci. Monit. 25, 6110–6119. doi:10.12659/MSM.915782
Zhou, Z. W., Li, X. X., He, Z. X., Pan, S. T., Yang, Y., Zhang, X., et al. (2015). Induction of apoptosis and autophagy via sirtuin1- and PI3K/Akt/mTOR-mediated pathways by plumbagin in human prostate cancer cells. Drug Des. Devel Ther. 9, 1511. doi:10.2147/DDDT.S75976
Keywords: plumbagin, hepatocellular carcinoma, network pharmacology, ROS, PI3K/Akt pathway, MAPK pathway
Citation: Wei Y, Lin Y, Chen W, Liu S, Jin L and Huang D (2021) Computational and In Vitro Analysis of Plumbagin’s Molecular Mechanism for the Treatment of Hepatocellular Carcinoma. Front. Pharmacol. 12:594833. doi: 10.3389/fphar.2021.594833
Received: 14 August 2020; Accepted: 18 February 2021;
Published: 12 April 2021.
Edited by:
Marcus Scotti, Federal University of Paraíba, BrazilCopyright © 2021 Wei, Lin, Chen, Liu, Jin and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Delun Huang, aHVhbmdkZWx1bjA2MjFAZm94bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.