
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Pharmacol. , 29 October 2020
Sec. Experimental Pharmacology and Drug Discovery
Volume 11 - 2020 | https://doi.org/10.3389/fphar.2020.595888
This article is part of the Research Topic Drug Repurposing for COVID-19 Therapy View all 50 articles
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is an enveloped, single-stranded RNA virus. Humans infected with SARS-CoV-2 develop a disease known as coronavirus disease 2019 (COVID-19) with symptoms and consequences including acute respiratory distress syndrome (ARDS), cardiovascular disorders, and death. SARS-CoV-2 appears to infect cells by first binding viral spike proteins with host protein angiotensin-converting enzyme 2 (ACE2) receptors; the virus is endocytosed following priming by transmembrane protease serine 2 (TMPRSS2). The process of virus entry into endosomes and its release from endolysosomes are key features of enveloped viruses. Thus, it is important to focus attention on the role of endolysosomes in SARS-CoV-2 infection. Indeed, coronaviruses are now known to hijack endocytic machinery to enter cells such that they can deliver their genome at replication sites without initiating host detection and immunological responses. Hence, endolysosomes might be good targets for developing therapeutic strategies against coronaviruses. Here, we focus attention on the involvement of endolysosomes in SARS-CoV-2 infection and COVID-19 pathogenesis. Further, we explore endolysosome-based therapeutic strategies to restrict SARS-CoV-2 infection and COVID-19 pathogenesis.
Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) that causes the pandemic disease known as coronavirus disease 2019 (COVID-19) (Contini et al., 2020; Gudbjartsson et al., 2020) is an enveloped virus that contains a large single-stranded RNA genome (Chan et al., 2020; Huang et al., 2020; Lu et al., 2020; Ren et al., 2020). SARS-CoV-2 belongs to the same beta-coronavirus family as does SARS-CoV that caused the SARS outbreak in China in 2002 (Cherry, 2004) and Middle East respiratory syndrome coronavirus (MERS-CoV) that caused the MERS outbreak in Saudi Arabia in 2012 (Zaki et al., 2012; Li and Du, 2019). Similar to other enveloped coronaviruses, SARS-CoV-2 enters host cells by endocytosis and uses host cell machinery for replication.
Spiked glycoproteins on the outer surface of coronaviruses are recognized by and bind to cell surface receptors such as angiotensin-converting enzyme 2 (ACE2) (Huang et al., 2006; Hoffmann et al., 2020b; Shang et al., 2020b) as well as possibly other co-receptors (Raj et al., 2013). Following binding, receptor-bound virus is endocytosed whereupon the viral genome is delivered into the cytoplasm; endocytosis mechanisms are pH-dependent and -independent (Dimitrov, 2004; White and Whittaker, 2016). Viruses that co-opt pH-independent mechanisms, an example of which is HIV-1, fuse with cell surface membranes and use endocytic pathways to achieve infection (White and Whittaker, 2016). Viruses that enter cells by pH-dependent mechanisms fuse with endosome membranes and use host factors associated with endosomes to enable viral entry into cells (Yang et al., 2004; White and Whittaker, 2016).
Coronaviruses use endolysosome-associated cathepsin B and L proteases under acidic conditions and are considered to be late penetrating viruses (late-entry kinetic mechanism) (Follis et al., 2006; Bosch et al., 2008; Millet and Whittaker, 2014; Coutard et al., 2020; Hoffmann et al., 2020a; Hoffmann et al., 2020b; Pranesh et al., 2020). Following entry, coronaviruses are released into the cytosol from endolysosomes or are targeted for degradation in lysosomes. In addition, some coronaviruses including SARS-CoV-2 can escape endolysosomes and replicate in autophagosome-like structures in the cytosol (Maier and Britton, 2012; Chen et al., 2014; Gassen et al., 2019; Gassen et al., 2020). Accordingly, it is important to focus attention on the role of endolysosomes in early stages of interactions between the virus and host cells as well as COVID-19 pathogenesis.
Endosomes are formed from plasma membrane invaginations; a process known as endocytosis. These acidic organelles are categorized further as early, late and recycling endosomes; all with different compositions and hydrogen ion (H+) content (Luzio et al., 2007; Huotari and Helenius, 2011; Gautreau et al., 2014). Rab4 and Rab5 are important components of early endosomes and function optimally at a pH range of 5.5–6.0. Early endosomes participate in signaling between the extracellular and intracellular environments (Pálfy et al., 2012; Villaseñor et al., 2016); they can recycle to plasma membranes thereby returning endocytosed constituents back to the cell surface (McCaffrey et al., 2001; Grant and Donaldson, 2009; Hsu and Prekeris, 2010). Alternatively, early endosomes can mature and transform into late endosomes (Bright et al., 2005; Luzio et al., 2007); these are differentiated from early endosomes by the expression of Rab7 and have an optimal pH range of 5.0–5.5 (Vanlandingham and Ceresa, 2009; Guerra and Bucci, 2016). Late endosomes can also recycle to plasma membranes (Guerra and Bucci, 2016), can produce multi-vesicular bodies from which extracellular vesicles (exosomes) originate, or can fuse with lysosomes (Piper and Luzio, 2001; Traub, 2010). The fusion of late endosomes with lysosomes generates endolysosomes under more acidic conditions ranging from pH 4.5–5.0 (Figure 1) (Mullock et al., 1998; Luzio et al., 2007; Luzio et al., 2010). The tight range of H+ concentrations in these organelles controls enzymatic activities as well as fusions between autophagolysosomes and lysosomes, and lysosomes and endosomes; pH also affects autophagy and other important cellular processes (Luzio et al., 2007; Luzio et al., 2010; Nakamura and Yoshimori, 2017). Vacuolar-ATPase (v-ATPase) activity largely regulates the acidic nature of endolysosomes and does so by controlling the flux of cations and anions via hydrolysis of free ATP that drives protons against their electrochemical gradient into the lumen of endolysosomes (Mindell, 2012; Halcrow et al., 2019a; Khan et al., 2019a).
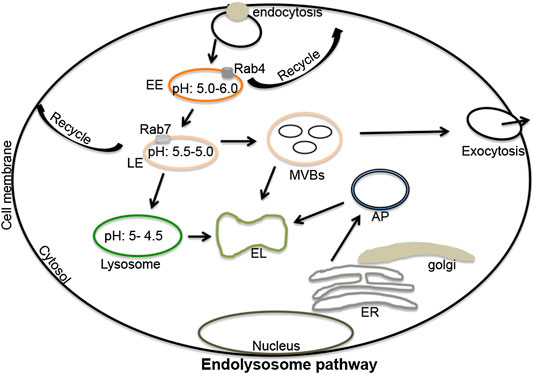
FIGURE 1. The endolysosome pathway: Extracellular signaling molecules upon binding to cell surface receptors can be engulfed by endocytosis. These endocytosed vesicles can mature and differentiate into early endosomes (pH 5.5–6.0), late endosomes (pH 5.5–5.0), lysosomes (pH 5.0–4.5), and endolysosomes (a fusion process of lysosomes and late endosomes). Various marker substances can differentiate early from late endosomes including Rab4 (early endosomes), and Rab5 and Rab7 (late endosomes). Both early and late endosomes regulate recycling processes that return constituent molecules back to plasma membranes. Late endosomes can produce multi-vesicular bodies, which can fuse with lysosomes or can be released from cells in the form of extracellular vesicles (exosomes). Lysosomes regulate the degradation of extracellular materials in endolysosomes produced by fusions with late endosomes. Lysosomes can also fuse with autophagosomes to form autolysosomes; sites where extracellular and intracellular components are degraded. EL, endolysosomes; ER, endoplasmic reticulum; EE, early endosomes; LE, late-endosomes; MVBs, multi-vesicular bodies; AP, autophagosomes; Rab, ras-related protein 4, 5 and 7).
Endolysosomes are involved in a wide range of cellular processes including membrane trafficking, catabolism of extracellular and intracellular components, immune responses and antigen presentation, cell secretions, and cell life and death (Eskelinen and Saftig, 2009; Munz, 2012; Repnik et al., 2013; Bright et al., 2016; Truschel et al., 2018; Khan et al., 2019a; Afghah et al., 2020). These acidic organelles have also been implicated in various pathological conditions; structural and functional changes have been reported in various neurodegenerative disorders as well as in cancer (Repnik et al., 2013; Bright et al., 2016; Davis, 2018; Halcrow et al., 2019a; Khan et al., 2019a). Because endolysosome pH regulates structural and functional features of endolysosomes, the involvement of v-ATPase in disease pathogenesis has received much attention and the v-ATPase complex has been targeted for therapeutic reasons. Indeed, inhibitors of v-ATPase and other strategies to keep endolysosomes from de-acidifying has shown benefit against diverse pathological conditions including different types of cancer (Whitton et al., 2018; Halcrow et al., 2019a; Halcrow et al., 2019b), neurological complications (Colacurcio and Nixon, 2016), and infectious diseases (Luzio et al., 2007).
Coronaviruses once endocytosed can avoid immune surveilence detection and degradation; thus enhancing infection (Hofmann and Pöhlmann, 2004; Belouzard et al., 2012; Shang et al., 2020a; Letko et al., 2020; Stower, 2020). SARS-CoV and MERS-CoV bind principally to dipeptidyl peptidase 4 while SARS-CoV-2 appears to bind mainly to ACE2; regardless, coronavirus spike proteins are activated by the host proteases TMPRSS2 or cathepsin B/L (Bosch et al., 2008; Shirato et al., 2013; Zhou et al., 2015; Hoffmann et al., 2020b; Pranesh et al., 2020). In addition, SARS-CoV-2 and MERS-CoV are activated by furin and this enhances viral entry especially in cells with lower expression levels of lysosomal cathepsin (Follis et al., 2006; Millet and Whittaker, 2014; Coutard et al., 2020; Hoffmann et al., 2020a).
Coronaviruses enter host cells by pH-dependent endocytosis (Yang et al., 2004; Burkard et al., 2014; Hoffmann et al., 2020b) and the acidic environment of endolysosomes is regulated not only by v-ATPase (Mindell, 2012), but also by Na+/K+-ATPase (Cain et al., 1989), mucolipin (TRPML1) channels (Li M. et al., 2017), big potassium channels (BK and MaxiK) (Khan et al., 2019b), Niemann-Pick type C (NPC1) (Wheeler et al., 2019a; Wheeler et al., 2019b; Höglinger et al., 2019; Lim et al., 2019), and two-pore channels (TPCs) (Marchant and Patel, 2015; Grimm et al., 2017; Khan et al., 2020). To date, TPCs and NPC1 have both been implicated in coronavirus infectivity.
TPCs are present in two forms; TPC1 and TPC2. TPC1s are mainly localized on early endosomes while TPC2s are mainly found on late endosomes/lysosomes (Brailoiu et al., 2009; Pitt et al., 2010; Zakon, 2012). Both subtypes of TPCs can help orchestrate interactions between endolysosomes and such viruses as Ebola (Sakurai et al., 2015), MERS-CoV (Gunaratne et al., 2018), and SARS-CoV-2 (Ou et al., 2020); TPCs regulate the trafficking of virus to late-endosomes/lysosomes following entry into cells. Not surprisingly then, TPC inhibitors can block entry of SARS-CoV-2 into cells and restrict the release of viral RNA into the cytosol (Figure 2) (Ou et al., 2020). TPCs are also involved in chloroquine-mediated endolysosome leakage and facilitated the release of HIV-1 Tat protein from endolysosomes thus enabling activation of HIV-1 LTR transactivation in the nucleus (Khan et al., 2020). Therefore, TPCs appear to promote virus entry and facilitate the release and transport of viral RNA to replication sites by inducing endolysosome permeability and depolarization.
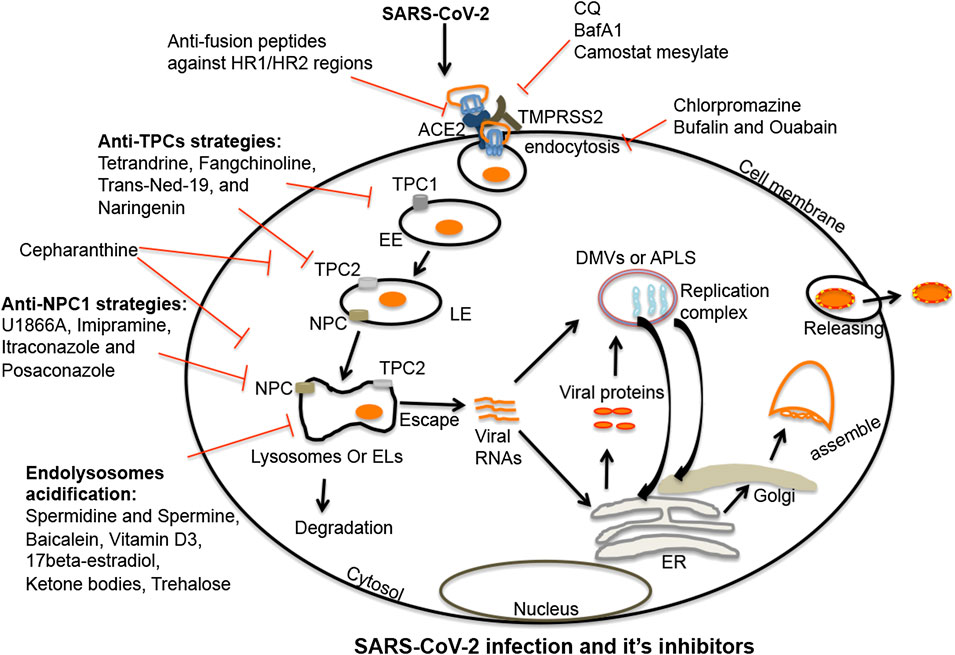
FIGURE 2. Endolysosome-mediated therapeutic strategies against SARS-CoV-2: SARS-CoV-2 enters cells following interactions between viral spike proteins and cell surface ACE2 receptors. Once endocytosed, spike proteins in endosomes are primed in late endosomes/lysosomes by cathepsin enzymes (B/L); this enhances virus entry. Post-fusion, virus is either released from or degraded in endolysosomes. SARS-CoV-2 once released from endolysosomes, enters the cytosol where it produces a replication complex to generate viral genomic and sub-genomic RNA. Following replication, viral structural proteins get inserted into the ER and move to the ERGIS (endoplasmic reticulum–Golgi intermediate compartment) secretory pathway for virus assembly. Following assembly, virions are transported to vesicles and released from cells by exocytosis. Thus, various stages are targetable for intervention. The first target might be fusion between spike proteins and host ACE2 receptors. A second target might be de-acidification of endolysosomes and blocking the priming of spike proteins by deactivating serine proteases. A third target might be clathrin-mediated endocytosis. Fourth, TPC and NPC1 inhibitors could effectively inhibit the virus infection by de-acidifying endolysosomes and blocking the trafficking of cholesterol. Endolysosome acidification may also be a therapeutic target because of its capacity to block the escape of viral RNA to the cytosol and enhance the degradation of the virus in lysosomes. Shown in the figure are multiple compounds and drugs capable of targeting each of these important steps in the virus cycle. SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; TMPRSS2, transmembrane protease serine 2; ACE2, angiotensin-converting enzyme 2; EL, endolysosome; EE, early endosome; LE, late-endosome; TPC1/2, two-pore channel 1 and 2; NPC1, Niemann-Pick disease type C1; viral RNAs, viral ribonucleic acids; DMVs, double-membrane vesicles; APLS, autophagosome-like structures; ER, endoplasmic reticulum.
NPC1 appears to also play a role in virus entry and infectivity. SARS-CoV enters into early endosomes, traffics to NPC1-positive late endosomes and lysosomes, and accesses highly active cathepsin L protease that triggers fusion mechanisms (Figure 2) (Shah et al., 2010; Zheng et al., 2018). MERS-CoV, Ebola, and SARS-CoV-2 use similar mechanisms to enter into host cells (Mingo et al., 2015; Zhou et al., 2016; Ballout et al., 2020).
Autophagy is a process by which extracellular and intracellular macromolecules are engulfed in and degraded by autophagolysosomes; structures formed by fusion of lysosomes with autophagosomes (Eskelinen and Saftig, 2009; Kenney and Benarroch, 2015; Yim and Mizushima, 2020). Autophagy is regulated by diverse proteins including autophagy-related-genes (ATGs), Beclin, ubiquitin-binding protein (p62), 5′-adenosine monophosphate-activated protein kinase, serine/threonine kinase 1 (Akt), and S-phase kinase-associated protein 2 (Skp2) (He and Klionsky, 2009; Badadani, 2012).
The process of autophagy degrades invading viruses, enhances antigen processing and presentation, and induces adaptive immune responses (Lee and Kim, 2007; Delgado et al., 2009; Richetta and Faure, 2013; Choi et al., 2018a). For example, toll-like receptors are pattern recognition receptors that sense viral RNA and DNA in endolysosomes, induce type I-interferon responses, and following induction of autophagy antiviral immune responses are decreased and invading viruses are degraded (Lee and Kim, 2007; Dalpke and Helm, 2012; Choi et al., 2018a). Autophagy has antiviral effects independent of the degradation process; interferon-γ can suppress replication of norovirus (Hwang et al., 2012; Baldridge et al., 2016; Biering et al., 2017). Additionally, viruses can modulate, escape, and inhibit autophagy at multiple steps to survive and replicate in host cells (Pattingre et al., 2005; Kyei et al., 2009; Chaumorcel et al., 2012).
Autophagy plays a role in viral infections including those caused by coronaviruses (Prentice et al., 2004a; Killian, 2012; Maier and Britton, 2012). Mouse hepatitis virus (MHV) has been used as a model for coronavirus infections (Prentice et al., 2004a); the replication complex of MHV generates double-membrane vesicles (DMVs) resembling autophagosomes (Snijder et al., 2006; Clementz et al., 2008; Gadlage et al., 2010) within which the autophagy markers LC3 and ATG12 colocalize (Prentice et al., 2004a). MHV replication is impaired when the autophagy marker ATG5 is knocked down (Prentice et al., 2004a). Replication proteins of SARS-CoV colocalize with LC3 and autophagy appears to play an important role in SARS-CoV replication (Prentice et al., 2004b). In contrast, SARS-CoV and MHV replication was not impaired when ATG5 and ATG7 were knocked down (Zhao et al., 2007; Reggiori et al., 2010; Schneider et al., 2012). The MERS-CoV and SARS-CoV associated protein, membrane-associated papain-like proteases, suppressed autophagy flux by blocking the fusion of lysosomes and autophagosomes (Chen X. et al., 2014; Gassen et al., 2019). Similarly, SARS-CoV-2 suppresses autophagy by modulating multiple autophagy regulatory factors (Gassen et al., 2020), by blocking the degradation of viral factors, and by increasing the formation of DMVs to promote virus replication. Induction of autophagy reduced the replication and infectivity of MERS-CoV (Gassen et al., 2019; Carmona-Gutierrez et al., 2020) and SARS-CoV-2 (Maier and Britton, 2012; Gassen et al., 2020).
Because endolysosomes influence coronavirus infections, these organelles might be targeted against SARS-CoV-2 infection and COVID-19 pathogenesis. Given the urgency of need and the tremendous costs involved in developing new drugs, a good approach to therapeutic drug development is the repurposing of drugs known to accumulate in and affect the function of endolysosomes. The diprotic weak base drugs chloroquine (CQ) and hydroxychloroquine (HCQ), that de-acidify endolysosomes, have shown effectiveness in controlling SARS-CoV-2 infection in in vitro studies, however the effectiveness of CQ/HCQ against COVID-19 has not been established for COVID-19 patients (Liu et al., 2020; Wang et al., 2020; Yao et al., 2020). Endolysosome de-acidification can restrict replication of SARS-CoV-2 because acidic conditions are necessary for SARS-CoV-2 to enter into and be released from host cells. In the context of SARS-CoV-2 infection, CQ and HCQ have been used in combination with azithromycin (Andreani et al., 2020; Carlucci et al., 2020); a weak base antibiotic known to accumulate in endolysosomes (Kong et al., 2017; Choi et al., 2018b; Andreani et al., 2020). Of course, CQ and HCQ have other pharmacological actions, but the involvement of endolysosome de-acidification in SARS-CoV-2 infection is supported by findings that other endolysosome de-acidification drugs; ammonium chloride, bafilomycin A1 and monensin all block coronavirus infections at the entry-level (Hoffmann et al., 2020b; Pranesh et al., 2020; Yang and Shen, 2020).
However, de-acidification may have other unintended consequences that might result in increased viral levels. Acidic conditions in endolysosomes are necessary for TLR-induced type-I interferon-mediated antiviral immune responses and antigen presentation (Dalpke and Helm, 2012; Munz, 2012; Choi et al., 2018a; Viret et al., 2020). Acidic endolysosomes are also important for autophagy, which is important for initiating innate immune responses and the degradation of viruses (Dalpke and Helm, 2012; Choi et al., 2018a). Accordingly, de-acidification of endolysosomes might hamper autophagy-mediated antiviral responses (Kužnik et al., 2011) by deactivating RNA sensors (Belizaire and Unanue, 2009; Kužnik et al., 2011; Kazi et al., 2013; Hussman, 2020; Offerhaus et al., 2020; Schrezenmeier and Dörner, 2020). Therefore, improving innate immune responses using synthetic RNAs, oligonucleotides, or small agonists of TLRs as well as type-I interferon treatment might improve clinical responses to CQ and HCQ (Dalpke and Helm, 2012; Freund et al., 2019; Hussman, 2020; Lee and Shin, 2020).
The spike protein of SARS-CoV-2 is necessary for viral entry into cells governed by receptor-mediated endocytosis (Hofmann and Pöhlmann, 2004; De Clercq, 2006; Burkard et al., 2014; Burkard et al., 2014; Zheng et al., 2018; Jiang et al., 2020; Ou et al., 2020; Shang et al., 2020a; Tay et al., 2020). SARS-CoV-2 spike is a trimer with three receptor-binding domains (RBDs) of S1 heads on top of a trimeric S2 stalk (Gui et al., 2017; Shang et al., 2020a; Walls et al., 2020). Following proteolytic cleavage, the RBD of S1 conformationally switches from a laid-down position to a standing-up position in order to facilitate fusion with cell membranes (Hofmann and Pöhlmann, 2004; Gui et al., 2017; Yuan et al., 2017); the laid-down position has a significantly higher binding capacity (Walls et al., 2020; Wrapp et al., 2020) and escapes host immune surveillance. These features of the spike protein might make development of vaccines and antibody-based therapies more challenging (Figure 2) (Rossmann, 1989; Sui et al., 2014; VanBlargan et al., 2016; Gui et al., 2017; Chu et al., 2020; Xia et al., 2020). Never-the-less, huge efforts are on-going to develop vaccines and antibody-based therapies based on the structural and binding properties of RBDs (Jiang et al., 2005; Du et al., 2014; Shang et al., 2020b; Tai et al., 2020). Additional sites for intervention against viral infection include the spike S2 stalk that contains HR1 and HR2 hydrophobic regions; stable six-helix-bundle (6-HB) structures that fuse the virus with the host cell membrane (Figure 2) (Bosch et al., 2003; Bosch et al., 2004; Aydin et al., 2014; Wang et al., 2019). These mechanisms might represent sites for intervention against viral replication because targeting these hydrophobic regions has been shown to restrict infection of HIV-1 (Kong et al., 2016; Yuan et al., 2019), SARS-CoV-2, and other coronaviruses (Bosch et al., 2004; Xia et al., 2019a; Xia et al., 2019b; Wang et al., 2019; Xia et al., 2020).
Post-fusion with plasma membranes, many viruses enter cells by endocytosis and clathrin-mediated endocytosis (Inoue et al., 2007; Wang et al., 2008). Therefore it is not surprising that the anti-schizophrenia drug chlorpromazine (Ban, 2007) that inhibits clathrin-mediated endocytosis inhibits infection by the coronaviruses MHV (Pu and Zhang, 2008), MERS-CoV (Burkard et al., 2014), and SARS-CoV (Inoue et al., 2007; Wang et al., 2008). Similarly, Na+/K+-ATPase pump-based inhibitors bufalin and ouabain restricted MERS-CoV infection (Burkard et al., 2014; Burkard et al., 2015; Amarelle and Lecuona, 2018) by inhibiting clathrin-mediated endocytosis (Ko et al., 2020). An additional FDA approved drug that might find use against COVID-19 is camostat mesylate that is used for the treatment of pancreatitis (Ramsey et al., 2019); it inhibited serine proteases and restricted MERS-CoV, SARS-CoV, and SARS-CoV-2 infections by inhibiting TMPRSS2 activity (Figure 2) (Shirato et al., 2013; Bojkova et al., 2020; Hoffmann et al., 2020b). Also, the cathepsin L inhibitors Z-FY (t-Bu)-DMK, K11777, and teicoplanin blocked the entry of SARS-CoV and MERS-CoV (Huang et al., 2006; Adedeji et al., 2013; Zhou et al., 2016; Baron et al., 2020). Accordingly, the aboved named agents might find use against SARS-CoV-2 infection (Figure 2) and the pathogenesis of COVID-19.
The coronavirus spike protein is activated under acidic conditions by the endolysosome proteases TMPRSS2 and cathepsins B, L; conditions that promote fusion with host cell membranes and entrance into cells (Hoffmann et al., 2020b). Consistent with this, de-acidification of endolysosomes by CQ, bafilomycinA1, and ammonium chloride have all been shown to deactivate TMPRSS2 and cathepsin B, L as well as suppress coronavirus infection (Figure 2) (Simmons et al., 2004; Vincent et al., 2005; Wang et al., 2008; Shirato et al., 2013; Al-Bari, 2017; Gao et al., 2020; Hoffmann et al., 2020b). Although mentioned earlier, it is important to consider more specifically the involvement of endolysosome-resident ion channels and proteins that regulate endolysosome pH including TPCs, NPC1, and v-ATPase.
TPCs are calcium- and sodium-permeable channels that regulate cell membrane trafficking and endolysosome pH (Wang et al., 2012; Lagostena et al., 2017; Khan et al., 2020). Because of the involvement of TPCs in the regulation of endolysosome pH it is not surprising that TPC activation increased the entry and trafficking of SARS-CoV-2 (Ou et al., 2020), MERS-CoV (Gunaratne et al., 2018) and Ebola (Sakurai et al., 2015) while the TPC inhibitors tetrandrine and Ned-19 significantly inhibited the entry and trafficking of viruses in host cells (Figure 2) (Ou et al., 2020). Moreover, apilimod and vaculin-1 restricted SARS-CoV-2 infection by reducing PIKfyve enzyme activity (Kang et al., 2020; Ou et al., 2020); PIKfyve is a regulator of PI(3,5)P2, an endogenous activator of TPCs (Dove et al., 2009; Kirsch et al., 2018). Further, the natural flavonoid naringenin inhibited TPCs (Tsai and Tsai, 2012; Pafumi et al., 2017; Benkerrou et al., 2019; Bai et al., 2020) and has antiviral activity against hepatitis C (HCV) (Nahmias et al., 2008), influenza A (Dong et al., 2014), Zika (Cataneo et al., 2019), and Dengue (Frabasile et al., 2017). Additionally, naringenin suppressed acute inflammation by inducing lysosome-mediated degradation of inflammatory cytokines (Jin et al., 2017) and ameliorated radiation-induced lung fibrosis (Zeng et al., 2018; Zhang et al., 2018). Thus, naringenin and other drugs targeting TPCs might be considered as possible therapeutic strategies against COVID-19 (Figure 2).
Niemann-Pick disease type C1 (NPC1) is an endolysosome-resident protein (Higgins et al., 1999) that regulates trafficking of late endosomes and lysosomes (Ko et al., 2001; Zhang et al., 2001; Ganley and Pfeffer, 2006; Sztolsztener et al., 2012), membrane trafficking of essential cellular factors such as cholesterol and sphingolipids (Chen et al., 2005; Infante et al., 2008; Kwon et al., 2009; Lange et al., 2012; Höglinger et al., 2019), and regulation of endolysosome pH and calcium (Elrick et al., 2012; Liu and Lieberman, 2019; Wheeler et al., 2019a). Impaired NPC1 is an underlying cause of Niemann-Pick disease; a lysosome storage disease (Lloyd-Evans et al., 2008; Schuchman and Desnick, 2017). NPC1 has been implicated in the infectivity of Ebola virus, MERS-CoV and SARS-CoV following late entry kinetics and access to cathepsin L in late endosomes and lysosomes (Shah et al., 2010; Mingo et al., 2015; Zhou et al., 2016; Zheng et al., 2018; Ballout et al., 2020; ). Because SARS-CoV-2 also uses similar cell entry and cleavage mechanisms, NPC1 might become a target against SARS-CoV-2 infection; the desired effect being endolysosome de-acidification and accumulation of lipids in endolysosomes (Zheng et al., 2018; Wheeler et al., 2019a; Ballout et al., 2020; Pranesh et al., 2020; Sturley et al., 2020). Indeed, increased levels of 25-hydroxycholesterol (25-HC) restricted viral infection of Filoviruses (Liu et al., 2013), Coronaviridae (ZhangY. et al., 2019b), and Flaviviridae (Chen Y. et al., 2014; Li et al., 2017). Elevated levels of 25-HC and 7-ketocholesterol (7-KC) (Willard et al., 2018) in NPC compromised cells may restrict infection by SARS-CoV-2. Moreover, available NPC1 inhibitors U1866A and imipramine inhibited several enveloped viruses including MERS-CoV and SARS-CoV (Wrensch et al., 2014), Ebola (Herbert et al., 2015; Lu et al., 2015), HIV-1 (Tang et al., 2009), HCV (Elgner et al., 2016), influenza A (Eckert et al., 2014), and chikungunya (Wichit et al., 2017) by de-acidifying endolysosomes and increasing lipid accumulation (Lange et al., 2012) (Figure 2). Also, the anti-fungal drugs itraconazole and posaconazole are not only inhibitors of NPC but also have antiviral activity (Strating et al., 2015; Trinh et al., 2017; Meutiawati et al., 2018; Rhoden et al., 2018; Schloer et al., 2019; Takano et al., 2019a; Takano et al., 2019b). In addition, cepharanthine, an inhibitor of TPC2 and NPC1, has antiviral activity (Figure 2) (Zhang et al., 2005; Matsuda et al., 2014; Lyu et al., 2017; Bailly, 2019; Kim et al., 2019; Fan et al., 2020; Rogosnitzky et al., 2020). Thus, TPCs and NPC1 might both attract attention as possible targets to block SARS-CoV-2 infection and suppress COVID-19.
The v-ATPase pump is an ion channel that is crucial for regulating endolysosome pH (Mindell, 2012); higher or lower activity levels of v-ATPase significantly affects endolysosome functions (Colacurcio and Nixon, 2016; Halcrow et al., 2019a). CQ, Baf A1 and ammonium chloride all cause de-acidification and deactivation of proteases in endolysosomes as well as inhibit coronavirus infections (Vincent et al., 2005; Gao et al., 2020; Hoffmann et al., 2020b). The SARS-CoV 3CLpro protease de-acidifies endolysosomes by direct interaction with the G1 subunit of v-ATPase (Lin et al., 2005) and blocks degradation of viral factors thereby enhancing virus replication. Endolysosome acidification may also restrict coronavirus infections by blocking the escape of viral RNA to the cytosol and promoting viral degradation in lysosomes (Carmona-Gutierrez et al., 2020; Gassen et al., 2020; Yang and Shen, 2020). A number of natural compounds acidify endolysosomes and might be tested for their ability to enhance coronavirus degradation; these include spermidine and spermine (Gassen et al., 2020), baicalein (Zhu et al., 2020), vitamin D3 (Hu et al., 2019; Daneshkhah et al., 2020), 17-beta-estradiol (Lipovka and Konhilas, 2014; Xiang et al., 2019; Khan, 2020; Suba, 2020), ketone bodies (Hui et al., 2012; Camberos-Luna et al., 2016), trehalose (Sharma et al., 2020), wogonin (Li et al., 2016), apigenin (Zhang X. et al., 2019), and butein (Ansari et al., 2018) (Figure 2).
The high fatality rate of COVID-19 especially among people with pre-existing co-morbities and rapidly increasing case numbers of SARS-CoV-2 infections has created a huge global need for effective therapeutic interventions against COVID-19. Because of the urgent need for therapeutics, re-purposing already approved pharmaceuticals might be the quickest available strategy. SARS-CoV-2 enters into endolysosomes where it can escape detection by immune surveillance and from there can traffic to the cytosol where it can propagate. Endolysosomes generally and endolysosome pH more specifically may represent important targets against SARS-CoV-2 replication and COVID-19 pathogenicity, and several compounds and drugs are available that may be repurposed for immediate testing. Reviewed above were several potential targets to block SARS-CoV-2 infection including endocytosis following binding of the spike protein with its receptor (ACE2), RNA replication and transcription, translation and proteolytic processing of viral proteins, virion assembly, and release from infected cells (Guy et al., 2020; Poduri et al., 2020); all targets involving the endolysosome system. In considering approaches against SARS-CoV-2 infection and COVID-19 pathogenesis, the involvement of endolysosomes should be considered.
All datasets presented in this study are included in the article/supplementary material.
All authors contributed equally to the manuscript.
P30GM100329, U54GM115458, R01MH100972, R01MH105329, R01MH119000, 2R01NS065957, and 2R01DA032444.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adedeji, A. O., Severson, W., Jonsson, C., Singh, K., Weiss, S. R., and Sarafianos, S. G. (2013). Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 87, 8017–8028. doi: 10.1128/jvi.00998-13
Afghah, Z., Chen, X., and Geiger, J. D. (2020). Role of endolysosomes and inter-organellar signaling in brain disease. Neurobiol. Dis. 134, 104670. doi: 10.1016/j.nbd.2019.104670
Al-Bari, M. A. A. (2017). Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 5, e00293. doi: 10.1002/prp2.293
Amarelle, L., and Lecuona, E. (2018). The antiviral effects of Na,K-ATPase inhibition: a minireview. Int. J. Mol. Sci. 19, 2154. doi: 10.3390/ijms19082154
Andreani, J., Le Bideau, M., Duflot, I., Jardot, P., Rolland, C., Boxberger, M., et al. (2020). In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb. Pathog. 145, 104228. doi: 10.1016/j.micpath.2020.104228
Ansari, M. Y., Ahmad, N., and Haqqi, T. M. (2018). Butein activates autophagy through AMPK/TSC2/ULK1/mTOR pathway to inhibit IL-6 expression in IL-1β stimulated human chondrocytes. Cell. Physiol. Biochem. 49, 932–946. doi: 10.1159/000493225
Aydin, H., Al-Khooly, D., and Lee, J. E. (2014). Influence of hydrophobic and electrostatic residues on SARS-coronavirus S2 protein stability: insights into mechanisms of general viral fusion and inhibitor design. Protein Sci. 23, 603–617. doi: 10.1002/pro.2442
Badadani, M. (2012). Autophagy mechanism, regulation, functions, and disorders. ISRN Cell Biol. 2012, 927064. doi: 10.5402/2012/927064
Bai, Y., Peng, W., Yang, C., Zou, W., Liu, M., Wu, H., et al. (2020). Pharmacokinetics and metabolism of naringin and active metabolite naringenin in rats, dogs, humans, and the differences between species. Front. Pharmacol. 11, 364. doi: 10.3389/fphar.2020.00364
Bailly, C. (2019). Cepharanthine: an update of its mode of action, pharmacological properties and medical applications. Phytomedicine 62, 152956. doi: 10.1016/j.phymed.2019.152956
Baldridge, M. T., Turula, H., and Wobus, C. E. (2016). Norovirus regulation by host and microbe. Trends Mol. Med. 22, 1047–1059. doi: 10.1016/j.molmed.2016.10.003
Ballout, R. A., Sviridov, D., Bukrinsky, M. I., and Remaley, A. T. (2020). The lysosome: a potential juncture between SARS-CoV-2 infectivity and Niemann-Pick disease type C, with therapeutic implications. FASEB. J. 34, 7253–7264. doi: 10.1096/fj.202000654R
Ban, T. A. (2007). Fifty years chlorpromazine: a historical perspective. Neuropsychiatr. Dis. Treat. 3, 495–500. PMID: 19300578; PMCID: PMC2655089
Baron, S. A., Devaux, C., Colson, P., Raoult, D., and Rolain, J.-M. (2020). Teicoplanin: an alternative drug for the treatment of COVID-19? Int. J. Antimicrob. Agents 55, 105944. doi: 10.1016/j.ijantimicag.2020.105944
Belizaire, R., and Unanue, E. R. (2009). Targeting proteins to distinct subcellular compartments reveals unique requirements for MHC class I and II presentation. Proc. Natl. Acad. Sci. U.S.A. 106, 17463–17468. doi: 10.1073/pnas.0908583106
Belouzard, S., Millet, J. K., Licitra, B. N., and Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4, 1011–1033. doi: 10.3390/v4061011
Benkerrou, D., Minicozzi, V., Gradogna, A., Milenkovic, S., Bodrenko, I. V., Festa, M., et al. (2019). A perspective on the modulation of plant and animal two pore channels (TPCs) by the flavonoid naringenin. Biophys. Chem. 25, 106246. doi: 10.1016/j.bpc.2019.106246
Biering, S. B., Choi, J., Halstrom, R. A., Brown, H. M., Beatty, W. L., Lee, S., et al. (2017). Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe 22, 74–85. e77. doi: 10.1016/j.chom.2017.06.005
Bojkova, D., McGreig, J. E., McLaughlin, K.-M., Masterson, S. G., Widera, M., Krähling, V., et al. (2020). SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles. bioRxiv doi: 10.1101/2020.04.03.024257
Bosch, B. J., Bartelink, W., and Rottier, P. J. M. (2008). Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82, 8887–8890. doi: 10.1128/jvi.00415-08
Bosch, B. J., Martina, B. E. E., van der Zee, R., Lepault, J., Haijema, B. J., Versluis, C., et al. (2004). Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 101, 8455–8460. doi: 10.1073/pnas.0400576101
Bosch, B. J., van der Zee, R., de Haan, C. A. M., and Rottier, P. J. M. (2003). The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003
Brailoiu, E., Churamani, D., Cai, X., Schrlau, M. G., Brailoiu, G. C., Gao, X., et al. (2009). Essential requirement for two-pore channel 1 in NAADP-mediated calcium signaling. J. Cell Biol. 186, 201–209. doi: 10.1083/jcb.200904073
Bright, N. A., Davis, L. J., and Luzio, J. P. (2016). Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr. Biol. 26, 2233–2245. doi: 10.1016/j.cub.2016.06.046
Bright, N. A., Gratian, M. J., and Luzio, J. P. (2005). Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15, 360–365. doi: 10.1016/j.cub.2005.01.049
Burkard, C., Verheije, M. H., Haagmans, B. L., van Kuppeveld, F. J., Rottier, P. J. M., Bosch, B.-J., et al. (2015). ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J. Virol. 89, 4434–4448. doi: 10.1128/jvi.03274-14
Burkard, C., Verheije, M. H., Wicht, O., van Kasteren, S. I., van Kuppeveld, F. J., Haagmans, B. L., et al. (2014). Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 10, e1004502. doi: 10.1371/journal.ppat.1004502
Cain, C. C., Sipe, D. M., and Murphy, R. F. (1989). Regulation of endocytic pH by the Na+,K+-ATPase in living cells. Proc. Natl. Acad. Sci. U.S.A. 86, 544–548. doi: 10.1073/pnas.86.2.544
Camberos-Luna, L., Gerónimo-Olvera, C., Montiel, T., Rincon-Heredia, R., and Massieu, L. (2016). The ketone body, β-hydroxybutyrate stimulates the autophagic flux and prevents neuronal death induced by glucose deprivation in cortical cultured neurons. Neurochem. Res. 41, 600–609. doi: 10.1007/s11064-015-1700-4
Carlucci, P., Ahuja, T., Petrilli, C. M., Rajagopalan, H., Jones, S., and Rahimian, J. (2020). Hydroxychloroquine and azithromycin plus zinc vs hydroxychloroquine and azithromycin alone: outcomes in hospitalized COVID-19 patients. medRxiv doi: 10.1099/jmm.0.001250
Carmona-Gutierrez, D., Bauer, M. A., Zimmermann, A., Kainz, K., Hofer, S. J., Kroemer, G., et al. (2020). Digesting the crisis: autophagy and coronaviruses. Model. Identif. Control 7, 119–128. doi: 10.15698/mic2020.05.715
Cataneo, A. H. D., Kuczera, D., Koishi, A. C., Zanluca, C., Silveira, G. F., Arruda, T. B. D., et al. (2019). The citrus flavonoid naringenin impairs the in vitro infection of human cells by Zika virus. Sci. Rep. 9, 16348. doi: 10.1038/s41598-019-52626-3
Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., et al. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 9, 221–236. doi: 10.1080/22221751.2020.1719902
Chaumorcel, M., Lussignol, M., Mouna, L., Cavignac, Y., Fahie, K., Cotte-Laffitte, J., et al. (2012). The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J. Virol. 86, 2571–2584. doi: 10.1128/jvi.05746-11
Chen, F. W., Gordon, R. E., and Ioannou, Y. A. (2005). NPC1 late endosomes contain elevated levels of non-esterified ('free') fatty acids and an abnormally glycosylated form of the NPC2 protein. Biochem. J. 390, 549–561. doi: 10.1042/bj20050236
Chen, X., Wang, K., Xing, Y., Tu, J., Yang, X., Zhao, Q., et al. (2014). Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell 5, 912–927. doi: 10.1007/s13238-014-0104-6
Chen, Y., Wang, S., Yi, Z., Tian, H., Aliyari, R., and Li, Y. (2014). Interferon-inducible cholesterol-25-hydroxylase inhibits hepatitis C virus replication via distinct mechanisms. Sci. Rep. 4, 7242. doi: 10.1038/srep07242
Cherry, J. D. (2004). The chronology of the 2002–2003 SARS mini pandemic. Paediatr. Respir. Rev. 5, 262–269. doi: 10.1016/j.prrv.2004.07.009
Choi, Y., Bowman, J. W., and Jung, J. U. (2018a). Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 16, 341–354. doi: 10.1038/s41579-018-0003-6
Choi, Y., Lim, H-S., Chung, D., Choi, J-g., and Yoon, D. (2018b). Risk evaluation of azithromycin-induced QT prolongation in real-world practice. BioMed Res. Int. 2018, 1574806. doi: 10.1155/2018/1574806
Chu, H., Chan, J. F.-W., Wang, Y., Yuen, T. T.-T., Chai, Y., and Hou, Y. (2020). Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 71, 1400–1409. doi: 10.1016/j.jcmgh.2020.09.017
Clementz, M. A., Kanjanahaluethai, A., O’Brien, T. E., and Baker, S. C. (2008). Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375, 118–129. doi: 10.1016/j.virol.2008.01.018
Colacurcio, D. J., and Nixon, R. A. (2016). Disorders of lysosomal acidification-the emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88. doi: 10.1016/j.arr.2016.05.004
Contini, C., Di Nuzzo, M., Barp, N., Bonazza, A., De Giorgio, R., Tognon, M., et al. (2020). The novel zoonotic COVID-19 pandemic: an expected global health concern. J. Infect. Dev. Ctries 14, 254–264. doi: 10.3855/jidc.12671
Coutard, B., Valle, C., de Lamballerie, X., Canard, B., Seidah, N. G., and Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 176, 104742. doi: 10.1016/j.antiviral.2020.104742
Dalpke, A. H., and Helm, M. (2012). RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol. 9, 828–842. doi: 10.4161/rna.20206
Daneshkhah, A., Agrawal, V., Eshein, A., Subramanian, H., Roy, H. K., and Backman, V. (2020). The possible role of vitamin D in suppressing cytokine storm and associated mortality in COVID-19 patients. medRxiv doi: 10.1101/2020.04.08.20058578.
Davis, L. (2018). The dynamics and function of the endolysosomal/lysosomal system. Apollo, Paris, Fance: OECD.
De Clercq, E. (2006). Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev. Anti-Infect. Ther. 4, 291–302. doi: 10.1586/14787210.4.2.291
Delgado, M., Singh, S., De Haro, S., Master, S., Ponpuak, M., Dinkins, C., et al. (2009). Autophagy and pattern recognition receptors in innate immunity. Immunol. Rev. 227, 189–202. doi: 10.1111/j.1600-065x.2008.00725.x
Dimitrov, D. S. (2004). Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2, 109–122. doi: 10.1038/nrmicro817
Dong, W., Wei, X., Zhang, F., Hao, J., Huang, F., Zhang, C., et al. (2014). A dual character of flavonoids in influenza A virus replication and spread through modulating cell-autonomous immunity by MAPK signaling pathways. Sci. Rep. 4, 7237. doi: 10.1038/srep07237
Dove, S. K., Michell, R. H., Kobayashi, T., Williams Fay, K., and Michell Robert, H. (2009). Inositol lipid-dependent functions in Saccharomyces cerevisiae: analysis of phosphatidylinositol phosphates. Biochem. J. 419, 1–16. doi: 10.1042/bj20081950
Du, L., Zhao, G., Yang, Y., Qiu, H., Wang, L., Kou, Z., et al. (2014). A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 88, 7045–7053. doi: 10.1128/jvi.00433-14
Eckert, N., Wrensch, F., Gärtner, S., Palanisamy, N., Goedecke, U., Jäger, N., et al. (2014). Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One 9, e97695. doi: 10.1371/journal.pone.0097695
Elgner, F., Ren, H., Medvedev, R., Ploen, D., Himmelsbach, K., Boller, K., et al. (2016). The intracellular cholesterol transport inhibitor U18666A inhibits the exosome-dependent release of mature hepatitis C virus. J. Virol. 90, 11181–11196. doi: 10.1128/jvi.01053-16
Elrick, M. J., Yu, T., Chung, C., and Lieberman, A. P. (2012). Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum. Mol. Genet. 21, 4876–4887. doi: 10.1093/hmg/dds324
Eskelinen, E.-L., and Saftig, P. (2009). Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 1793, 664–673. doi: 10.1016/j.bbamcr.2008.07.014
Fan, H.-H., Wang, L.-Q., Liu, W.-L., An, X.-P., Liu, Z.-D., He, X.-Q., et al. (2020). Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin. Med. J. 133, 1051–1056. doi: 10.1097/cm9.0000000000000797
Follis, K. E., York, J., and Nunberg, J. H. (2006). Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 350, 358–369. doi: 10.1016/j.virol.2006.02.003
Frabasile, S., Koishi, A. C., Kuczera, D., Silveira, G. F., Verri, W. A., Duarte Dos Santos, C. N, et al. (2017). The citrus flavanone naringenin impairs dengue virus replication in human cells. Sci. Rep. 7, 41864. doi: 10.1038/srep43976
Freund, I., Eigenbrod, T., Helm, M., and Dalpke, A. (2019). RNA modifications modulate activation of innate toll-like receptors. Genes 10, 92. doi: 10.3390/genes10020092
Gadlage, M. J., Sparks, J. S., Beachboard, D. C., Cox, R. G., Doyle, J. D., Stobart, C. C., et al. (2010). Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 84, 280–290. doi: 10.1128/jvi.01772-09
Ganley, I. G., and Pfeffer, S. R. (2006). Cholesterol accumulation sequesters Rab9 and disrupts late endosome function in NPC1-deficient cells. J. Biol. Chem. 281, 17890–17899. doi: 10.1074/jbc.m601679200
Gao, J., Tian, Z., and Yang, X. (2020). Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 14, 72–73. doi: 10.5582/bst.2020.01047
Gassen, N. C., Niemeyer, D., Muth, D., Corman, V. M., Martinelli, S., and Gassen, A. (2019). SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat. Commun. 10, 5770. doi: 10.1038/s41467-019-13659-4
Gassen, N. C., Papies, J., Bajaj, T., Dethloff, F., Emanuel, J., and Weckmann, K. (2020). Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. bioRxiv doi: 10.1101/2020.04.15.997254
Gautreau, A., Oguievetskaia, K., and Ungermann, C. (2014). Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb Perspect. Biol. 6, a016832. doi: 10.1101/cshperspect.a016832
Grant, B. D., and Donaldson, J. G. (2009). Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10, 597–608. doi: 10.1038/nrm2755
Grimm, C., Chen, C. C., Wahl-Schott, C., and Biel, M. (2017). Two-pore channels: catalyzers of endolysosomal transport and function. Front. Pharmacol. 8, 45. doi: 10.3389/fphar.2017.00045
Gudbjartsson, D. F., Helgason, A., Jonsson, H., Magnusson, O. T., Melsted, P., Norddahl, G. L., et al. (2020). Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 382, 2302–2315. doi: 10.1056/nejmoa2026116
Guerra, F., and Bucci, C. (2016). Multiple roles of the small GTPase Rab7. Cells 5, 34. doi: 10.3390/cells5030034
Gui, M., Song, W., Zhou, H., Xu, J., Chen, S., Xiang, Y., et al. (2017). Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 27, 119–129. doi: 10.1038/cr.2016.152
Gunaratne, G., Yang, Y., Li, F., Walseth, T., and Marchant, J. (2018). NAADP-dependent Ca 2+ signaling regulates Middle East respiratory syndrome-coronavirus pseudovirus translocation through the endolysosomal system. Cell Calcium 75, 30–34. doi: 10.1016/j.ceca.2018.08.003
Guy, R. K., DiPaola, R. S., Romanelli, F., and Dutch, R. E. (2020). Rapid repurposing of drugs for COVID-19. Science 368, 829–830. doi: 10.1126/science.abb9332
Infante, R. E., Wang, M. L., Radhakrishnan, A., Kwon, H. J., Brown, M. S., and Goldstein, J. L. (2008). NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad Sci. U. S.A. 105, 15287–15292. doi: 10.1073/pnas.0807328105
Höglinger, D., Burgoyne, T., Sanchez-Heras, E., Hartwig, P., Colaco, A., Newton, J., et al. (2019). NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10, 4276. doi: 10.1038/s41467-019-12152-2
Halcrow, P., Datta, G., Ohm, J. E., Soliman, M. L., Chen, X., and Geiger, J. D. (2019a) Role of endolysosomes and pH in the pathogenesis and treatment of glioblastoma. Cancer Rep. 2, doi: 10.1002/cnr1002.1177
Halcrow, P., Khan, N., Datta, G., Ohm, J. E., Chen, X., and Geiger, J. D. (2019b) Importance of measuring endolysosome, cytosolic, and extracellular pH in understanding the pathogenesis of and possible treatments for glioblastoma multiforme. Cancer Rep. 2. doi: 10.1002/cnr2.1193
He, C., and Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. doi: 10.1146/annurev-genet-102808-114910
Herbert, A. S., Davidson, C., Kuehne, A. I., Bakken, R., Braigen, S. Z., Gunn, K. E., et al. (2015). Niemann-Pick C1 is essential for ebolavirus replication and pathogenesis in vivo. mBio 6, e00565. doi: 10.1128/mbio.00565-15
Higgins, M. E., Davies, J. P., Chen, F. W., and Ioannou, Y. A. (1999). Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol. Genet. Metabol. 68, 1–13. doi: 10.1006/mgme.1999.2882
Hofmann, H., and Pöhlmann, S. (2004). Cellular entry of the SARS coronavirus. Trends Microbiol. 12, 466–472. doi: 10.1016/j.tim.2004.08.008
Hoffmann, M., Kleine-Weber, H., and Pöhlmann, S. (2020a) A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e775. doi: 10.1016/j.molcel.2020.04.022
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020b) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e278. doi: 10.1016/j.cell.2020.02.052
Hsu, V. W., and Prekeris, R. (2010). Transport at the recycling endosome. Curr. Opin. Cell Biol. 22, 528–534. doi: 10.1016/j.ceb.2010.05.008
Hu, W., Zhang, L., Li, M. X., Shen, J., Liu, X. D., Xiao, Z. G., et al. (2019). Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy 15, 707–725. doi: 10.1080/15548627.2018.1557835
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.1016/s0140-6736(20)30183-5
Huang, I.-C., Bosch, B. J., Li, F., Li, W., Lee, K. H., Ghiran, S., et al. (2006). SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281, 3198–3203. doi: 10.1074/jbc.m508381200
Hui, L., Chen, X., Bhatt, D., Geiger, N., Rosenberger, T., Haughey, N., et al. (2012). Ketone bodies protection against HIV-1 Tat-induced neurotoxicity. J. Neurochem. 122, 382–391. doi: 10.1111/j.1471-4159.2012.07764.x
Huotari, J., and Helenius, A. (2011). Endosome maturation. EMBO J. 30, 3481–3500. doi: 10.1038/emboj.2011.286
Hussman, J. P. (2020). Cellular and molecular pathways of COVID-19 and potential points of therapeutic intervention. Front. Pharmacol. 11, 1169. doi: 10.3389/fphar.2020.01169
Hwang, S., Maloney, N. S., Bruinsma, M. W., Goel, G., Duan, E., Zhang, L., et al. (2012). Nondegradative role of atg5-atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409. doi: 10.1016/j.chom.2012.03.002
Inoue, Y., Tanaka, N., Tanaka, Y., Inoue, S., Morita, K., Zhuang, M., et al. (2007). Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 81, 8722–8729. doi: 10.1128/jvi.00253-07
Jiang, S., He, Y., and Liu, S. (2005). SARS vaccine development. Emerg. Infect. Dis. 11, 1016–1020. doi: 10.3201/1107.050219
Jiang, S., Hillyer, C., and Du, L. (2020). Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 41, 355–359. doi: 10.1016/j.it.2020.03.007
Jin, L., Zeng, W., Zhang, F., Zhang, C., and Liang, W. (2017). Naringenin ameliorates acute inflammation by regulating intracellular cytokine degradation. J. Immunol. 199, 3466–3477. doi: 10.4049/jimmunol.1602016
Kang, Y.-L., Chou, Y.-Y., Rothlauf, P. W., Liu, Z., Piccinotti, S., Soh, T. K., et al. (2020). Inhibition of PIKfyve kinase prevents infection by EBOV and SARS-CoV-2. bioRxiv doi: 10.1101/2020.04.21.053058
Kazi, M. S., Saurabh, K., Rishi, P., and Rishi, E. (2013). Delayed onset chloroquine retinopathy presenting 10 years after long-term usage of chloroquine. Middle East Afr. J. Ophthalmol. 20, 89–91. doi: 10.4103/0974-9233.106404
Kenney, D. L., and Benarroch, E. E. (2015). The autophagy-lysosomal pathway. Neurology 85, 634–645. doi: 10.1212/wnl.0000000000001860
Khan, N. (2020). Possible protective role of 17β-estradiol against COVID-19. J. Allergy Infect. Dis. 1, 38–48. doi: 10.46439/allergy.1.010
Khan, N., Halcrow, P. W., Lakpa, K. L., Afghah, Z., Miller, N. M., Dowdy, S. F., et al. (2020). Two‐pore channels regulate Tat endolysosome escape and Tat‐mediated HIV‐1 LTR transactivation. FASEB J. 34, 4147–4162. doi: 10.1096/fj.201902534r
Khan, N., Haughey, N. J., Nath, A., and Geiger, J. D. (2019a) Involvement of organelles and inter-organellar signaling in the pathogenesis of HIV-1 associated neurocognitive disorder and Alzheimer’s disease. Brain Res. 1722, 146389. doi: 10.1016/j.brainres.2019.146389
Khan, N., Lakpa, K. L., Halcrow, P. W., Afghah, Z., Miller, N. M., Geiger, J. D, et al. (2019b) BK channels regulate extracellular Tat-mediated HIV-1 LTR transactivation. Sci. Rep. 9, 12285. doi: 10.1038/s41598-019-48777-y
Killian, M. (2012). Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res. Ther. 9, 16. doi: 10.1186/1742-6405-9-16
Kim, D., Min, J., Jang, M., Lee, J., Shin, Y., Park, C., et al. (2019). Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 9, 696. doi: 10.3390/biom9110696
Ko, D. C., Gordon, M. D., Jin, J. Y., and Scott, M. P. (2001). Dynamic movements of organelles containing niemann-pick C1 protein: NPC1 involvement in late endocytic events. MBoC 12, 601–614. doi: 10.1091/mbc.12.3.601
Ko, M., Chang, S. Y., Byun, S. Y., Choi, I., d’Alexandry d’Orengiani, A-L. P. H., Shum, D., et al. (2020). Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. bioRxiv doi: 10.1101/2020.02.25.965582
Kong, F. Y., Rupasinghe, T. W., Simpson, J. A., Vodstrcil, L. A., Fairley, C. K., McConville, M. J, et al. (2017). Pharmacokinetics of a single 1g dose of azithromycin in rectal tissue in men. PLoS One 12, e0174372. doi: 10.1371/journal.pone.0174372
Kong, R., Xu, K., Zhou, T., Acharya, P., Lemmin, T., Liu, K., et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. doi: 10.1126/science.aae0474
Kužnik, A., Benčina, M., Švajger, U., Jeras, M., Rozman, B., and Jerala, R. (2011). Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J. Immunol. 186, 4794–4804. doi: 10.4049/jimmunol.1000702
Kwon, H. J., Abi-Mosleh, L., Wang, M. L., Deisenhofer, J., Goldstein, J. L., Brown, M. S., et al. (2009). Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224. doi: 10.1016/j.cell.2009.03.049
Kwon, S. A., Kugemann, A., Carpaneto, A., Böckmann, R. A., and Dietrich, P. (2018). Phosphatidylinositol-3,5-bisphosphate lipid-binding-induced activation of the human two-pore channel 2. Cell. Mol. Life Sci. 75, 3803–3815. doi: 10.1007/s00018-018-2829-5
Kyei, G. B., Dinkins, C., Davis, A. S., Roberts, E., Singh, S. B., Dong, C., et al. (2009). Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. JJ. Cell Biol. 186, 255–268. doi: 10.1083/jcb.200903070
Lagostena, L., Festa, M., Pusch, M., and Carpaneto, A. (2017). The human two-pore channel 1 is modulated by cytosolic and luminal calcium. Sci. Rep. 7, 43900. doi: 10.1038/srep43900
Lange, Y., Ye, J., and Steck, T. L. (2012). Activation mobilizes the cholesterol in the late endosomes-lysosomes of niemann pick type C cells. PLoS One 7, e30051. doi: 10.1371/journal.pone.0030051
Lee, J. S., and Shin, E.-C. (2020). The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 20, 585–586. doi: 10.1038/s41577-020-00429-3
Lee, M. S., and Kim, Y.-J. (2007). Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 76, 447–480. doi: 10.1146/annurev.biochem.76.060605.122847
Letko, M., Marzi, A., and Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5, 562–569. doi: 10.1038/s41564-020-0688-y
Li, C., Deng, Y.-Q., Wang, S., Ma, F., Aliyari, R., Huang, X.-Y., et al. (2017) 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity 46, 446–456. doi: 10.1016/j.immuni.2017.02.012
Li, F., and Du, L. (2019). MERS coronavirus: an emerging zoonotic virus. Viruses 11, 663. doi: 10.3390/v11070663
Li, M., Zhang, W. K., Benvin, N. M., Zhou, X., Su, D., Li, H., et al. (2017) Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat. Struct. Mol. Biol. 24, 205–213. doi: 10.1038/nsmb.3362
Li, S.-J., Sun, S.-J., Gao, J., and Sun, F.-B. (2016). Wogonin induces Beclin-1/PI3K and reactive oxygen species-mediated autophagy in human pancreatic cancer cells. Oncol. Lett. 12, 5059–5067. doi: 10.3892/ol.2016.5367
Lim, C.-Y., Davis, O. B., Shin, H. R., Zhang, J., Berdan, C. A., Jiang, X., et al. (2019). ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 21, 1206–1218. doi: 10.1038/s41556-019-0391-5
Lin, C.-W., Tsai, F.-J., Wan, L., Lai, C.-C., Lin, K.-H., Hsieh, T.-H., et al. (2005). Binding interaction of SARS coronavirus 3CLproprotease with vacuolar-H+ATPase G1 subunit. FEBS Lett. 579, 6089–6094. doi: 10.1016/j.febslet.2005.09.075
Lipovka, Y., and Konhilas, J. P. (2014). Estradiol activates AMPK through interaction with extrogen receptor beta (15.4). FASEB J. 28, 15–14. doi: 10.1096/fasebj.28.1_supplement.15.4
Liu, E. A., and Lieberman, A. P. (2019). The intersection of lysosomal and endoplasmic reticulum calcium with autophagy defects in lysosomal diseases. Neurosci. Lett. 697, 10–16. doi: 10.1016/j.neulet.2018.04.049
Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Hu, H., et al. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Dis. 6, 1–4. doi: 10.1038/s41421-020-0156-0
Liu, S.-Y., Aliyari, R., Chikere, K., Li, G., Marsden, M. D., Smith, J. K., et al. (2013). Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38, 92–105. doi: 10.1016/j.immuni.2012.11.005
Lloyd-Evans, E., Morgan, A. J., He, X., Smith, D. A., Elliot-Smith, E., Sillence, D. J., et al. (2008). Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14, 1247–1255. doi: 10.1038/nm.1876
Lu, F., Liang, Q., Abi-Mosleh, L., Das, A., De Brabander, J. K., Goldstein, J. L, et al. (2015). Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife 4, e12177. doi: 10.7554/elife.12177
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/s0140-6736(20)30251-8
Luzio, J. P., Gray, S. R., and Bright, N. A. (2010). Endosome-lysosome fusion. Biochem. Soc. Trans. 38, 1413–1416. doi: 10.1042/bst0381413
Luzio, J. P., Pryor, P. R., and Bright, N. A. (2007). Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632. doi: 10.1038/nrm2217
Lyu, J., Yang, E. J., Head, S. A., Ai, N., Zhang, B., Wu, C., et al. (2017). Pharmacological blockade of cholesterol trafficking by cepharanthine in endothelial cells suppresses angiogenesis and tumor growth. Canc. Lett. 409, 91–103. doi: 10.1016/j.canlet.2017.09.009
Maier, H., and Britton, P. (2012). Involvement of autophagy in coronavirus replication. Viruses 4, 3440–3451. doi: 10.3390/v4123440
Marchant, J. S., and Patel, S. (2015). Two-pore channels at the intersection of endolysosomal membrane traffic. Biochem. Soc. Trans. 43, 434–441. doi: 10.1042/bst20140303
Matsuda, K., Hattori, S., Komizu, Y., Kariya, R., Ueoka, R., and Okada, S. (2014). Cepharanthine inhibited HIV-1 cell-cell transmission and cell-free infection via modification of cell membrane fluidity. Bioorg. Med. Chem. Lett 24, 2115–2117. doi: 10.1016/j.bmcl.2014.03.041
McCaffrey, M. W., Bielli, A., Cantalupo, G., Mora, S., Roberti, V., Santillo, M., et al. (2001). Rab4 affects both recycling and degradative endosomal trafficking. FEBS Lett. 495, 21–30. doi: 10.1016/s0014-5793(01)02359-6
Meutiawati, F., Bezemer, B., Strating, J. R. P. M., Overheul, G. J., Žusinaite, E., van Kuppeveld, F. J. M., et al. (2018). Posaconazole inhibits dengue virus replication by targeting oxysterol-binding protein. Antivir. Res. 157, 68–79. doi: 10.1016/j.antiviral.2018.06.017
Millet, J. K., and Whittaker, G. R. (2014). Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219. doi: 10.1073/pnas.1407087111
Mindell, J. A. (2012). Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69–86. doi: 10.1146/annurev-physiol-012110-142317
Mingo, R. M., Simmons, J. A., Shoemaker, C. J., Nelson, E. A., Schornberg, K. L., D'Souza, R. S., et al. (2015). Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+Endolysosomes is a rate-defining step. J. Virol. 89, 2931–2943. doi: 10.1128/jvi.03398-14
Mullock, B. M., Bright, N. A., Fearon, C. W., Gray, S. R., and Luzio, J. (1998). Fusion of lysosomes with late endosomes produces a hybrid organelle of intermediate density and is NSF dependent. J. Cell Biol. 140, 591–601. doi: 10.1083/jcb.140.3.591
Munz, C. (2012). Antigen processing for MHC class II presentation via autophagy. Front. Immunol. 3. doi: 10.3389/fimmu.2012.00009
Nahmias, Y., Goldwasser, J., Casali, M., van Poll, D., Wakita, T., Chung, R. T., et al. (2008). Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47, 1437–1445. doi: 10.1002/hep.22197
Nakamura, S., and Yoshimori, T. (2017). New insights into autophagosome-lysosome fusion. J. Cell Sci. 130, 1209–1216. doi: 10.1242/jcs.196352
Offerhaus, J. A., Wilde, A. A. M., and Remme, C. A. (2020). Prophylactic (hydroxy)chloroquine in COVID-19: potential relevance for cardiac arrhythmia risk. Heart Rhythm 17, 1480–1486. doi: 10.1016/j.hrthm.2020.07.001
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L, et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620. doi: 10.1038/s41467-020-15562-9
Pálfy, M., Reményi, A., and Korcsmáros, T. (2012). Endosomal crosstalk: meeting points for signaling pathways. Trends Cell Biol. 22, 447–456. doi: 10.1016/j.tcb.2012.06.004
Pafumi, I., Festa, M., Papacci, F., Lagostena, L., Giunta, C., Gutla, V., et al. (2017). Naringenin impairs two-pore channel 2 activity and inhibits VEGF-induced angiogenesis. Sci. Rep. 7, 5121. doi: 10.1038/s41598-017-04974-1
Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., et al. (2005). Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939. doi: 10.1016/j.cell.2005.07.002
Piper, R. C., and Luzio, J. P. (2001). Late endosomes: sorting and partitioning in multivesicular bodies. Traffic 2, 612–621. doi: 10.1034/j.1600-0854.2001.20904.x
Pitt, S. J., Funnell, T. M., Sitsapesan, M., Venturi, E., Rietdorf, K., Ruas, M., et al. (2010). TPC2 is a novel NAADP-sensitive Ca2+Release channel, operating as a dual sensor of luminal pH and Ca2+. J. Biol. Chem. 285, 35039–35046. doi: 10.1074/jbc.m110.156927
Poduri, R., Joshi, G., and Jagadeesh, G. (2020). Drugs targeting various stages of the SARS-CoV-2 life cycle: exploring promising drugs for the treatment of Covid-19. Cell. Signal. 74, doi: 10.1016/j.cellsig.2020.109721
Pranesh, P., Rajat, D., and Narendra, D. (2020). Targeting TMPRSS2 and cathepsin B/L together may Be synergistic against SARS-CoV-2 infection. doi: 10.26434/chemrxiv.12213125.v1
Prentice, E., Jerome, W. G., Yoshimori, T., Mizushima, N., and Denison, M. R. (2004a) Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279, 10136–10141. doi: 10.1074/jbc.m306124200
Prentice, E., McAuliffe, J., Lu, X., Subbarao, K., and Denison, M. R. (2004b) Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78, 9977–9986. doi: 10.1128/jvi.78.18.9977-9986.2004
Pu, Y., and Zhang, X. (2008). Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J. Virol. 82, 8112–8123. doi: 10.1128/jvi.00837-08
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H. W., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254. doi: 10.1038/nature12005
Ramsey, M. L., Nuttall, J., and Hart, P. A., (2019). A phase 1/2 trial to evaluate the pharmacokinetics, safety, and efficacy of NI-03 in patients with chronic pancreatitis: study protocol for a randomized controlled trial on the assessment of camostat treatment in chronic pancreatitis (TACTIC). Trials 20, 501. doi: 10.1186/s13063-019-3606-y
Reggiori, F., Monastyrska, I., Verheije, M. H., Calì, T., Ulasli, M., Bianchi, S., et al. (2010). Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508. doi: 10.1016/j.chom.2010.05.013
Ren, L.-L., Wang, Y.-M., Wu, Z.-Q., Xiang, Z.-C., Guo, L., Xu, T., et al. (2020). Identification of a novel coronavirus causing severe pneumonia in human. Chinese Med J 133, 1015–1024. doi: 10.1097/cm9.0000000000000722
Repnik, U., Česen, M. H., and Turk, B. (2013). The endolysosomal system in cell death and survival. Cold Spring Harb Perspect. Biol. 5, a008755. doi: 10.1101/cshperspect.a008755
Rhoden, E., Nix, W. A., Weldon, W. C., and Selvarangan, R. (2018). Antifungal azoles itraconazole and posaconazole exhibit potent in vitro antiviral activity against clinical isolates of parechovirus A3 (Picornaviridae). Antivir. Res. 149, 75–77. doi: 10.1016/j.antiviral.2017.11.011
Richetta, C., and Faure, M. (2013). Autophagy in antiviral innate immunity. Cell Microbiol. 15, 368–376. doi: 10.1111/cmi.12043
Rogosnitzky, M., Okediji, P., and Koman, I. (2020). Cepharanthine: a review of the antiviral potential of a Japanese-approved alopecia drug in COVID-19. Pharmacol. Rep. 72, 1–8. doi: 10.1007/s43440-020-00132-z
Rossmann, M. G. (1989). The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J. Biol. Chem. 264, 14587–14590. PMID: 2670920
Sakurai, Y., Kolokoltsov, A. A., Chen, C.-C., Tidwell, M. W., Bauta, W. E., Klugbauer, N., et al. (2015). Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 347, 995–998. doi: 10.1126/science.1258758
Schloer, S., Goretzko, J., Kühnl, A., Brunotte, L., Ludwig, S., and Rescher, U. (2019). The clinically licensed antifungal drug itraconazole inhibits influenza virus in vitro and in vivo. Emerg. Microb. Infect. 8, 80–93. doi: 10.1080/22221751.2018.1559709
Schneider, M., Ackermann, K., Stuart, M., Wex, C., Protzer, U., Schätzl, H. M., and Gilch, S. (2012). Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. J. Virol. 86, 10112–10122. doi: 10.1128/jvi.01001-12
Schrezenmeier, E., and Dörner, T. (2020). Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16, 155–166. doi: 10.1038/s41584-020-0372-x
Schuchman, E. H., and Desnick, R. J. (2017). Types A and B niemann-pick disease. Mol. Genet. Metabol. 120, 27–33. doi: 10.1016/j.ymgme.2016.12.008
Shah, P. P., Wang, T., Kaletsky, R. L., Myers, M. C., Purvis, J. E., Jing, H., et al. (2010). A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 78, 319–324. doi: 10.1124/mol.110.064261
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., and Li, F. (2020a) Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 11727–11734. doi: 10.1073/pnas.2003138117
Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., et al. (2020b) Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224. doi: 10.1038/s41586-020-2179-y
Sharma, V., Makhdoomi, M., Singh, L., Kumar, P., Khan, N., Singh, S., et al. (2020). Trehalose limits opportunistic mycobacterial survival during HIV co-infection by reversing HIV-mediated autophagy block. Autophagy 16, 1–20. doi: 10.1080/15548627.2020.1725374
Shirato, K., Kawase, M., and Matsuyama, S. (2013). Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561. doi: 10.1128/jvi.01890-13
Simmons, G., Reeves, J. D., Rennekamp, A. J., Amberg, S. M., Piefer, A. J., and Bates, P. (2004). Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 101, 4240–4245. doi: 10.1073/pnas.0306446101
Snijder, E. J., van der Meer, Y., Zevenhoven-Dobbe, J., Onderwater, J. J. M., van der Meulen, J., Koerten, H. K., et al. (2006). Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80, 5927–5940. doi: 10.1128/jvi.02501-05
Stower, H. (2020). Lopinavir-ritonavir in severe COVID-19. Nat. Med. 26, 465. doi: 10.1038/s41591-020-0849-9
Strating, J. R. P. M., van der Linden, L., Albulescu, L., Bigay, J., Arita, M., Delang, L., et al. (2015). Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 10, 600–615. doi: 10.1016/j.celrep.2014.12.054
Sturley, S., Rajakumar, T., Hammond, N., Higaki, K., Márka, Z., Márka, S, et al. (2020). Potential COVID-19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J. Lipid Res. 61, 972–982. doi: 10.1194/jlr.R120000851
Suba, Z. (2020). Prevention and therapy of COVID-19 via exogenous estrogen treatment for both male and female patients: prevention and therapy of COVID-19. J. Pharm. Pharm. Sci. 23, 75–85. doi: 10.18433/jpps31069
Sui, J., Deming, M., Rockx, B., Liddington, R. C., Zhu, Q. K., Baric, R. S., et al. (2014). Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J. Virol. 88, 13769–13780. doi: 10.1128/jvi.02232-14
Sztolsztener, M. E., Dobrzyn, A., Pikula, S., Tylki-Szymanska, A., and Bandorowicz-Pikula, J. (2012). Impaired dynamics of the late endosome/lysosome compartment in human Niemann-Pick type C skin fibroblasts carrying mutation in NPC1 gene. Mol. Biosyst. 8, 1197–1205. doi: 10.1039/c2mb05447g
Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., et al. (2020). Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 17, 613–620. doi: 10.1038/s41423-020-0400-4
Takano, T., Akiyama, M., Doki, T., and Hohdatsu, T. (2019a) Antiviral activity of itraconazole against type I feline coronavirus infection. Vet. Res. 50, 5. doi: 10.1186/s13567-019-0625-3
Takano, T., Wakayama, Y., and Doki, T. (2019b) Endocytic pathway of feline coronavirus for cell entry: differences in serotype-dependent viral entry pathway. Pathogens 8, 300. doi: 10.3390/pathogens8040300
Tang, Y., Leao, I. C., Coleman, E. M., Broughton, R. S., and Hildreth, J. E. K. (2009). Deficiency of niemann-pick type C-1 protein impairs release of human immunodeficiency virus type 1 and results in Gag accumulation in late endosomal/lysosomal compartments. J. Virol. 83, 7982–7995. doi: 10.1128/jvi.00259-09
Tay, M. Z., Poh, C. M., Rénia, L., MacAry, P. A., and Ng, L. F. P. (2020). The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374. doi: 10.1038/s41577-020-0311-8
Traub, L. M. (2010). The reverse logic of multivesicular endosomes. EMBO Rep. 11, 79–81. doi: 10.1038/embor.2009.281
Trinh, M. N., Lu, F., Li, X., Das, A., Liang, Q., De Brabander, J. K., et al. (2017). Triazoles inhibit cholesterol export from lysosomes by binding to NPC1. Proc. Natl. Acad. Sci. U.S.A. 114, 89–94. doi: 10.1073/pnas.1619571114
Truschel, S. T., Clayton, D. R., Beckel, J. M., Yabes, J. G., Yao, Y., Wolf-Johnston, A., et al. (2018). Age-related endolysosome dysfunction in the rat urothelium. PLoS One 13, e0198817. doi: 10.1371/journal.pone.0198817
Tsai, Y.-J., and Tsai, T.-H. (2012). Mesenteric lymphatic absorption and the pharmacokinetics of naringin and naringenin in the rat. J. Agric. Food Chem. 60, 12435–12442. doi: 10.1021/jf301962g
VanBlargan, L. A., Goo, L., and Pierson, T. C. (2016). Deconstructing the antiviral neutralizing-antibody response: implications for vaccine development and immunity. Microbiol. Mol. Biol. Rev. 80, 989–1010. doi: 10.1128/mmbr.00024-15
Vanlandingham, P. A., and Ceresa, B. P. (2009). Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284, 12110–12124. doi: 10.1074/jbc.m809277200
Villaseñor, R., Kalaidzidis, Y., and Zerial, M. (2016). Signal processing by the endosomal system. Curr. Opin. Cell Biol. 39, 53–60. doi: 10.1016/j.ceb.2016.02.002
Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., et al. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2, 69. doi: 10.1186/1743-422x-2-69
Viret, C., Rozières, A., Duclaux-Loras, R., Boschetti, G., Nancey, S., and Faure, M. (2020). Regulation of anti-microbial autophagy by factors of the complement system. Microb. Cell 7, 93–105. doi: 10.15698/mic2020.04.712
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., and Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6. doi: 10.2210/pdb6vxx/pdb
Wang, H., Yang, P., Liu, K., Guo, F., Zhang, Y., Zhang, G., and Jiang, C. (2008). SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 18, 290–301. doi: 10.1038/cr.2008.15
Wang, L., Xu, J., Kong, Y., Liang, R., Li, W., Li, J., et al. (2019). Engineering a novel antibody-peptide bispecific fusion protein against MERS-CoV. Antibodies 8, 53. doi: 10.3390/antib8040053
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. doi: 10.1038/s41422-020-0282-0
Wang, X., Zhang, X., Dong, X.-p., Samie, M., Li, X., Cheng, X., et al. (2012). TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383. doi: 10.1016/j.cell.2012.08.036
Wheeler, S., Haberkant, P., Bhardwaj, M., Tongue, P., Ferraz, M. J., Halter, D., et al. (2019a) Cytosolic glucosylceramide regulates endolysosomal function in Niemann-Pick type C disease. Neurobiol. Dis. 127, 242–252. doi: 10.1016/j.nbd.2019.03.005
Wheeler, S., Schmid, R., and Sillence, D. J. (2019b) Lipid-protein interactions in niemann-pick type C disease: insights from molecular modeling. Int. J. Mol. Sci. 20, 717. doi: 10.3390/ijms20030717
White, J. M., and Whittaker, G. R. (2016). Fusion of enveloped viruses in endosomes. Traffic 17, 593–614. doi: 10.1111/tra.12389
Whitton, B., Okamoto, H., Packham, G., and Crabb, S. J. (2018). Vacuolar ATPase as a potential therapeutic target and mediator of treatment resistance in cancer. Cancer Med. 7, 3800–3811. doi: 10.1002/cam4.1594
Wichit, S., Hamel, R., Bernard, E., Talignani, L., Diop, F., Ferraris, P., et al. (2017). Imipramine inhibits chikungunya virus replication in human skin fibroblasts through interference with intracellular cholesterol trafficking. Sci. Rep. 7, 3145. doi: 10.1038/s41598-017-03316-5
Willard, K., Elling, C., Stice, S., and Brindley, M. (2018). The oxysterol 7-ketocholesterol reduces Zika virus titers in vero cells and human neurons. Viruses 11, 20. doi: 10.3390/v11010020
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi: 10.1126/science.abb2507
Wrensch, F., Winkler, M., and Pöhlmann, S. (2014). IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses 6, 3683–3698. doi: 10.3390/v6093683
Xia, S., Lan, Q., Pu, J., Wang, C., Liu, Z., Xu, W., et al. (2019a) Potent MERS-CoV fusion inhibitory peptides identified from HR2 domain in spike protein of bat coronavirus HKU4. Viruses 11, 56. doi: 10.3390/v11010056
Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., et al. (2020). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30:343–355. doi: 10.1038/s41422-020-0305-x
Xia, S., Yan, L., Xu, W., Agrawal, A. S., Algaissi, A., Tseng, C.-T. K., et al. (2019b) A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 5, eaav4580. doi: 10.1126/sciadv.aav4580
Xiang, J., Liu, X., Ren, J., Chen, K., Wang, H.-l., Miao, Y.-y., et al. (2019). How does estrogen work on autophagy? Autophagy 15, 197–211. doi: 1 0.1080/15548627.2018.1520549
Yang, N., and Shen, H.-M. (2020). Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 16, 1724–1731. doi: 10.7150/ijbs.45498
Yang, Z.-Y., Huang, Y., Ganesh, L., Leung, K., Kong, W.-P., Schwartz, O., et al. (2004). pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 78, 5642–5650. doi: 10.1128/jvi.78.11.5642-5650.2004
Yao, X., Ye, F., Zhang, M., Cui, C., Huang, B., Niu, P., et al. (2020). In Vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 71, 732–739. doi: 10.1093/cid/ciaa237
Yim, W. W-Y., and Mizushima, N. (2020). Lysosome biology in autophagy. Cell Dis. 6, 6. doi: 10.1038/s41421-020-0141-7
Yuan, M., Cottrell, C. A., Ozorowski, G., van Gils, M. J., Kumar, S., Wu, N. C., et al. (2019). Conformational plasticity in the HIV-1 fusion peptide facilitates recognition by broadly neutralizing antibodies. Cell Host Microbe 25, 873–883.e875. doi: 10.1016/j.chom.2019.04.011
Yuan, Y., Cao, D., Zhang, Y., Ma, J., Qi, J., Wang, Q, et al. (2017). Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8, 15092. doi: 10.1038/ncomms15092
Zaki, A. M., Van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. M. E., and Fouchier, R. A. M. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. doi: 10.1056/nejmoa1211721
Zakon, H. H. (2012). Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc. Natl. Acad. Sci. U.S.A. 109, 10619–10625. doi: 10.1073/pnas.1201884109
Zeng, W., Jin, L., Zhang, F., Zhang, C., and Liang, W. (2018). Naringenin as a potential immunomodulator in therapeutics. Pharmacol. Res. 135, 122–126. doi: 10.1016/j.phrs.2018.08.002
Zhang, C., Zeng, W., Yao, Y., Xu, B., Wei, X., Wang, L., et al. (2018). Naringenin ameliorates radiation-induced lung injury by lowering IL-1β level. J. Pharmacol. Exp. Therapeut. 366, 341–348. doi: 10.1124/jpet.118.248807
Zhang, C. H., Wang, Y. F., Liu, X. J., Lu, J. H., Qian, C. W., Wan, Z. Y, et al. (2005). Antiviral activity of cepharanthine against severe acute respiratory syndrome coronavirus in vitro. Chin. Med. J. (Engl) 118, 493–496. PMID: 32818817; PMCID: PMC7418743
Zhang, M., Dwyer, N. K., Love, D. C., Cooney, A., Comly, M., Neufeld, E., et al. (2001). Cessation of rapid late endosomal tubulovesicular trafficking in Niemann-Pick type C1 disease. Proc. Natl. Acad. Sci. U.S.A. 98, 4466–4471. doi: 10.1073/pnas.081070898
Zhang, X., Bu, H., Jiang, Y., Sun, G., Jiang, R., Huang, X., et al. (2019) The antidepressant effects of apigenin are associated with the promotion of autophagy via the mTOR/AMPK/ULK1 pathway. Mol. Med. Rep. 20, 2867–2874. doi: 10.3892/mmr.2019.10491
Zhang, Y., Song, Z., Wang, M., Lan, M., Zhang, K., Jiang, P., et al. (2019) Cholesterol 25-hydroxylase negatively regulates porcine intestinal coronavirus replication by the production of 25-hydroxycholesterol. Vet. Microbiol. 231, 129–138. doi: 10.1016/j.vetmic.2019.03.004
Zhao, Z., Thackray, L. B., Miller, B. C., Lynn, T. M., Becker, M. M., Ward, E., et al. (2007). Coronavirus replication does not require the autophagy GeneATG5. Autophagy 3, 581–585. doi: 10.4161/auto.4782
Zheng, Y., Shang, J., Yang, Y., Liu, C., Wan, Y., Geng, Q., et al. (2018). Lysosomal proteases are a determinant of coronavirus tropism. J. Virol. 92, e01504–e01518. doi: 1 0.1128/jvi.01504-18
Zhou, N., Pan, T., Zhang, J., Li, Q., Zhang, X., Bai, C., et al. (2016). Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biol. Chem. 291, 9218–9232. doi: 10.1074/jbc.m116.716100
Zhou, Y., Vedantham, P., Lu, K., Agudelo, J., Carrion, R., Nunneley, J. W., et al. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 116, 76–84. doi: 10.1016/j.antiviral.2015.01.011
Keywords: endolysosome, endocytosis, two pore channel
Citation: Khan N, Chen X and Geiger JD (2020) Role of Endolysosomes in Severe Acute Respiratory Syndrome Coronavirus-2 Infection and Coronavirus Disease 2019 Pathogenesis: Implications for Potential Treatments. Front. Pharmacol. 11:595888. doi: 10.3389/fphar.2020.595888
Received: 17 August 2020; Accepted: 05 October 2020;
Published: 29 October 2020.
Edited by:
Rafael Maldonado, Pompeu Fabra University, SpainReviewed by:
Copyright © 2020 Khan, Chen and Geiger. This is an open-access article distributed under the terms of the . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan D. Geiger, am9uYXRoYW4uZ2VpZ2VyQHVuZC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.