- Department of Cancer Immunology, Genentech, Inc., South San Francisco, CA, United States
Autophagy is a catabolic process that targets its cargo for lysosomal degradation. In addition to its function in maintaining tissue homeostasis, autophagy is recognized to play a context-dependent role in cancer. Autophagy may inhibit tumor initiation under specific contexts; however, a growing body of evidence supports a pro-tumorigenic role of this pathway in established disease. In this setting, autophagy drives treatment resistance, metabolic changes, and immunosuppression both in a tumor-intrinsic and extrinsic manner. This observation has prompted renewed interest in targeting autophagy for cancer therapy. Novel genetic models have proven especially insightful, revealing unique and overlapping roles of individual autophagy-related genes in tumor progression. Despite identification of pharmacologically actionable nodes in the pathway, fundamental challenges still exist for successful therapeutic inhibition of autophagy. Here we summarize the current understanding of autophagy as a driver of resistance against targeted and immuno-therapies and highlight knowledge gaps that, if addressed, may provide meaningful advances in the treatment of cancer.
Introduction
Autophagy is an evolutionarily conserved lysosomal degradative pathway that digests diverse cellular cargo. These include cytosol itself, proteins, lipids, organelles and intracellular pathogens. Given its role in cellular quality control, autophagy is unsurprisingly involved in numerous pathophysiological conditions. Since Christian de Duve coined the term “autophagy,” our understanding of this pathway has evolved from a relatively simple non-selective catabolic process to a highly targeted mechanism by which specific cargo is identified for lysosomal turnover (Klionsky et al., 2003; Dikic and Elazar, 2018; Mizushima, 2018; Levine and Kroemer, 2019; Melia et al., 2020). Selective autophagy utilizes “autophagy receptor” proteins which bridge the cargo-of-interest to the autophagosome (reviewed in Kirkin, 2020). Each form of selective autophagy is named after its cargo (e.g., protein aggregates: aggrephagy, lipids: lipophagy, pathogens: xenophagy, organelles: mitophagy, pexophagy, ribophagy, ER-phagy, and nucleophagy). Autophagy receptors such as p62/SQSTM1, NDP52, OPTN and NBR1 consist of a ubiquitin-associated-domain and an LC3-interacting region. This allows them to bridge specific cargo (often ubiquitinated as a consequence of a specific cellular state/stress response) to autophagosomes by binding to members of the ATG8 family, such as LC3 (Zaffagnini and Martens, 2016; Kirkin and Rogov, 2019). Three types of autophagy have been characterized based on how cargo is delivered to the lysosome: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). Macroautophagy is the most extensively studied form of autophagy and the focus of this review. Molecular mechanisms underlying microautophagy and CMA are reviewed in detail elsewhere (Kaushik and Cuervo, 2018; Oku and Sakai, 2018).
Macroautophagy (herein autophagy) is regulated in a stepwise manner by multi-subunit complexes of autophagy-related proteins (Figure 1). The Unc51-like kinase (ULK) complex is a Ser/Thr protein kinase complex that phosphorylates multiple substrate proteins to initiate autophagy. ULK1/2 activity promotes stabilization of the Class lll phosphatidylinositol 3-kinase (PIK3C3) complex, where Vps34 phosphorylates phosphatidylinositol (PI) to generate PI-(3)-phosphate (Ptdins(3)P), a critical membrane targeting signal for the autophagosome elongation machinery. Importantly, Vps34 forms different subcomplexes with distinct functions. Complex l consists of Beclin 1, p150, ATG14L, and NRBF2 and regulates autophagy induction. Separately, complex II utilizes UVRAG instead of ATG14L and governs vesicular trafficking. Finally, complex III consists of RUBICON, a RUN-domain containing protein that binds UVRAG and negatively regulates autophagosome formation via regulation of the RAB7-GTPase (Matsunaga et al., 2009; Zhong et al., 2009; Ohashi et al., 2019; Bhargava et al., 2020). Thus, Vps34 functions as an upstream regulator of autophagosome formation, endosome maturation and membrane trafficking via its membership in distinct signaling complexes.
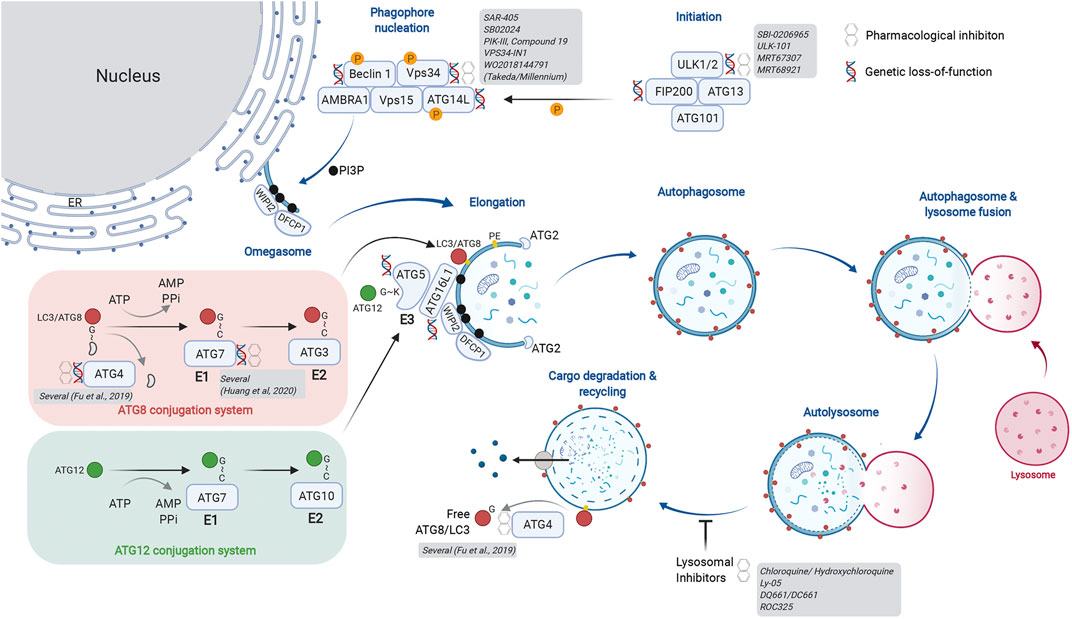
FIGURE 1. Actionable nodes of the autophagy pathway. The process of autophagy begins with generation of the initiation complex. Its substrates include factors critical for phagophore nucleation. This is also called the Vps34 complex and is involved in initiation of autophagy as well as endosome maturation. The lipid kinase activity of Vps34 generates PI3P on the target membrane, forming the omegasome upon which the autophagy nucleation complex forms. The ER is depicted as a membrane source for the omegasome, but other membranous compartments have also been described. The autophagy elongation machinery includes an E1-E2-E3-like process that ultimately results in lipidation of ATG8 family proteins, thereby identifying a mature autophagosome. Two conjugation systems are illustrated: the ATG8 conjugation system (red) transfers LC3/ATG8 to the E2-like protein ATG3. The ATG12 conjugation system (green) generates the E3-like complex by transferring ATG12 onto ATG5. Association of the ATG5-12 fusion with ATG16L1 forms the ATG16L1 complex; this acts in an E3-like manner to transfer ATG8/LC3 onto phosphatidyl ethanolamine (PE), thereby completing the lipidation process. Cysteine proteases of the ATG4 family cleave ATG8/LC3 from the cytosolic face of the autolysosome for re-use. ATG4 family proteins are critical in the initial activation of ATG8/LC3 by exposing the C-terminal glycine for conjugation. As depicted, several members of core autophagy machinery have been assessed in genetic and pharmacological models for their role in cancer biology. Additional details of genetic models are provided in Table 1. A more extensive list of pharmacological inhibitors is provided in Table 2. Created with BioRender.com.
Membrane PI3P generated by Vps34 are docking sites for FYVE domain containing proteins such as WIPI2, which in turn recruits the autophagosome elongation complex. Here, ubiquitin-like proteins of the ATG8 family (ATG8 in yeast; LC3A, B, C, GABARAPs, and GATE-16 proteins in mammals) are post-translationally modified by lipidation, first by exposure of a C-terminal glycine and subsequent conjugation to phosphatidylethanolamine (PE). There are parallels between the ubiquitin E1-E2-E3 conjugation machinery and the autophagosome elongation complex. ATG7 acts as the “E1” enzyme for its substrates ATG8 or ATG12. Following E1-mediated activation, ATG8 is transferred to the “E2-like” protein ATG3. The ATG12-ATG5-ATG16L1 complex acts as an “E3-like” enzyme to transfer ATG8 to PE on the growing autophagosome membrane. Importantly, generation of the ATG12-ATG5-ATG16L1 complex requires transfer of ATG12 from ATG7 using a separate conjugation machinery involving ATG10, another “E2-like” protein. Once autophagosomes enclose cargo, they undergo heterotypic fusion with lysosomes for substrate degradation by lysosomal hydrolases. Finally, ATG4 family proteins act as cysteine proteases to cleave and expose C-terminal glycine on ATG8 proteins (e.g., G120 of pro-LC3B) for participation in the elongation complex. They also cleave lipidated ATG8 proteins on the mature autophagosome to recycle them and maintain a cellular pool of non-lipidated ATG8. As depicted in Figure 1, our current understanding of the autophagic machinery provides multiple nodes that are genetically validated and attractive pharmacological targets.
This review summarizes our current understanding of how autophagy drives disease progression via altered metabolism and immunosuppression in tumor-intrinsic and extrinsic contexts. We catalogue the growing list of actionable targets in the pathway, and finally discuss current gaps in knowledge to successfully modulate autophagy for the treatment of cancer.
The Complicated Role of Autophagy in Cancer
Initial genetic studies using Becn1 (encoding Beclin 1) heterozygous mice demonstrated spontaneous malignancies and HBV-induced hepatocellular carcinogenesis in murine models (Qu et al., 2003; Yue et al., 2003). Autophagy activity in Becn1+/− mice was reduced compared to controls. Since outgrowing tumors did not show loss of the remaining allele, Becn1 was proposed as a haplo-insufficient tumor suppressor. Comparatively, deletion of downstream autophagy genes Atg5 or Atg7 permitted development of benign tumors that failed to progress to malignant cancers (Takamura et al., 2011; Guo et al., 2013; Strohecker et al., 2013; Rao et al., 2014; Yang et al., 2014). Loss of the autophagy-related gene Atg7 in intestinal epithelial cells was shown to attenuate tumor growth driven by loss of adenomatous polyposis coli (APC), a major tumor suppressor in colorectal cancer. However, simultaneous deletion of Atg7 and the tumor suppressor Tp53 initiated adenoma formation in the presence of wild type APC (Levy et al., 2015; Trentesaux et al., 2020). Thus, loss of autophagy-related genes can promote opposing outcomes for tumor growth depending on the driving oncogenic or tumor-suppressor. Defective autophagy can promote tumor initiation via multiple pathways. For instance, tumor cells derived from autophagy-deficient cells frequently exhibit accumulation of abnormal mitochondria due to lack of mitophagy, consequently suffering enhanced oxidative stress, DNA damage and potentially developing aneuploidy (Mathew et al., 2009). Autophagy also impacts cellular senescence in a context dependent manner. As a key feature of aging, senescence prevents malignant transformation by arresting cell division. While basal autophagy may counteract senescence by maintaining cellular fitness, autophagy is associated with oncogene-induced senescence (Dou et al., 2015; Kang et al., 2015). Autophagy-mediated turnover of p62/SQSTM1 (p62) can also contribute to tumor suppression. As an autophagy receptor, p62 bridges cargo to autophagosomes, but itself is a substrate for autophagic turnover. A known non-canonical function of p62 is regulation of the transcription factor Nrf2 (nuclear factor erythroid 2–related factor 2; discussed later) (Komatsu et al., 2010; Inami et al., 2011; Ichimura et al., 2013). A relatively limited number of studies suggest autophagy as a tumor suppressor in a cancer cell-extrinsic manner. p62 in the tumor microenvironment (TME), specifically in CAFs or adipocytes appears to suppress tumor growth (Valencia et al., 2014; Duran et al., 2016; Umemura et al., 2016). Autophagy has also shown to promote dendritic cell-mediated CD8+ T cell priming, thus enhancing tumor cell killing through immunogenic ATP release (Michaud et al., 2011).
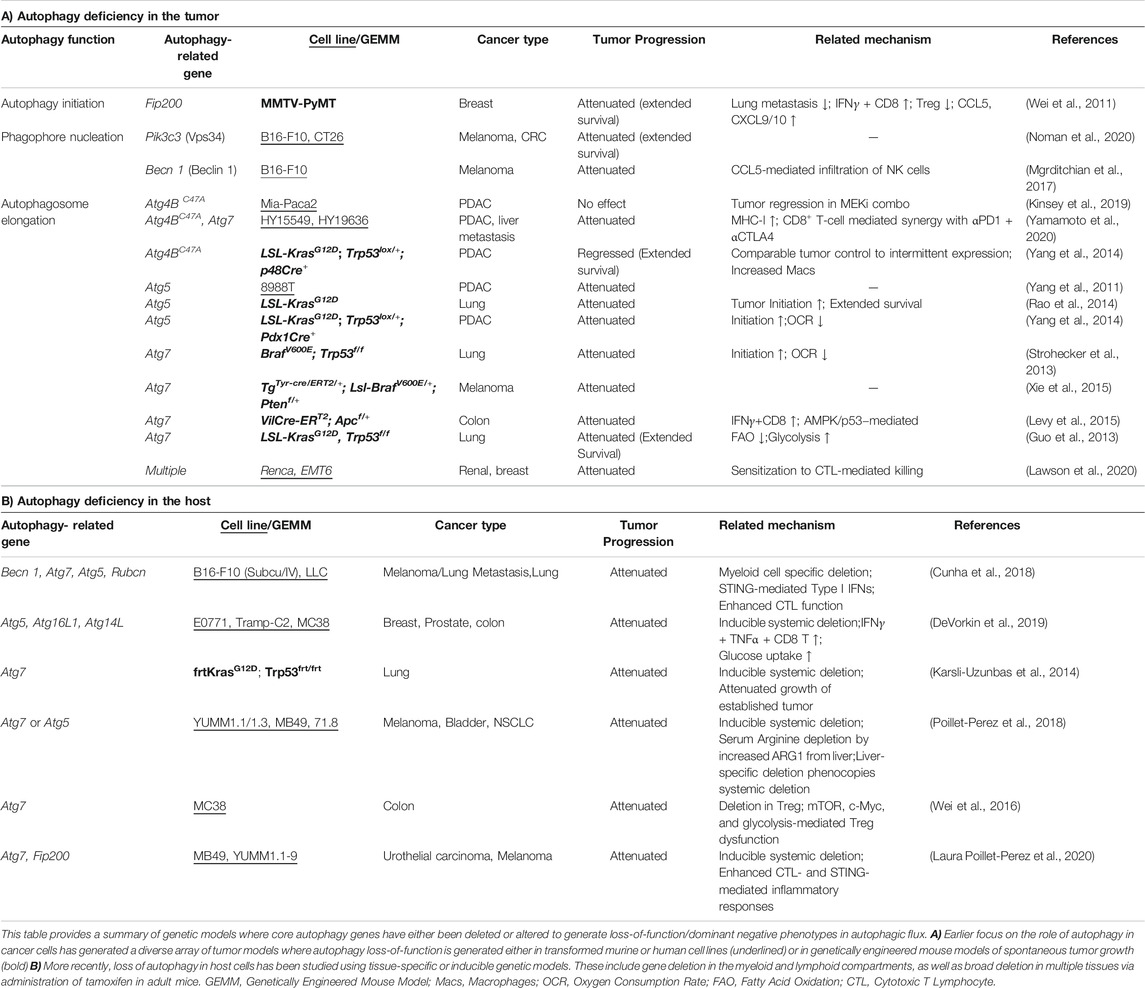
In contrast to its role as a tumor suppressor, an increasing number of studies propose a pro-tumorigenic role for autophagy in a tumor cell-intrinsic or extrinsic manner. While some tumor cells induce autophagy as a survival mechanism against diverse stress conditions including therapies, nutrient deprivation, or hypoxia, pancreatic cancer cells exhibit elevated autophagy activity even under basal conditions (Guo et al., 2011; Lock et al., 2011; Yang et al., 2011). Initial observations that autophagy was dispensable for basal growth of multiple cancer cell lines in 2D cell culture systems or xenograft models led to a pause in the pursuit of autophagy inhibitors (Eng et al., 2016). However, emerging data using more relevant model systems are proving that enhanced autophagy is indeed a critical driver of treatment resistance and tumor progression in vivo (Table 1). The use of nutrient-rich 2D culture systems and immunocompromised models may account for discrepancies observed between earlier studies and more recent work. Earlier studies of germline autophagy-related gene knockout mice showed autophagy to be a vital pathway for mammalian development, maintenance and expansion of immune cells and hematopoietic stem cells (reviewed in Kuma et al., 2017; Clarke and Simon, 2019); it was thus assumed that autophagy inhibition would compromise immune cell function in the context of cancer. However, recent work using elegant genetic models reveals that in fact, autophagy suppresses anti-tumor immunity. As listed in Table 1, tissue-specific deletion of autophagy-related genes in the myeloid, lymphoid and broader host cellular compartments have demonstrated that perturbing the pathway accelerates tumor clearance via multiple mechanisms (Table 1A – tumor intrinsic; Table 1B – tumor extrinsic). The following sections discuss our emerging understanding of how autophagy promotes therapeutic resistance and fosters tumor progression.
Tumor-Intrinsic Autophagy
Autophagy Supports Tumor Cell Metabolism
Autophagy supplies tumor cells with metabolic substrates by degrading glycogen, lipid droplets, damaged proteins or organelles (Guo et al., 2011; Yang et al., 2011; Guo et al., 2013; Strohecker et al., 2013; Rao et al., 2014; Yang et al., 2014). In addition, autophagy maintains the pool of functional mitochondria, critical for survival under nutrient or oxygen limiting conditions, and especially for fatty acid oxidation. Accumulation of lipid droplets and defective mitochondria by Atg7 deficiency attenuated Ras-driven tumor proliferation (Guo et al., 2013; Strohecker et al., 2013). Atg7 loss also attenuated BrafV600E-driven lung tumorigenesis by depleting glutamine, thereby impairing mitochondrial respiration. Inhibition of autophagy by knocking down Atg7 and Atg12 attenuated cancer cell glycolysis and cellular transformation by oncogenic Ras (Lock et al., 2011). Reduced glucose uptake was also observed upon Fip200 deletion in the MMTV-PyMT breast cancer model where tumorigenesis is driven by Polyoma virus-mediated Ras, Src, and PI3K activation (Wei et al., 2011). Inhibition of autophagy in tumor cells could therefore sensitize them to metabolic stresses and uncover additional dependencies for survival.
Autophagy Promotes Therapeutic Resistance
Responses to primary therapy include ER stress, hypoxia, and mTOR inhibition, all of which induce autophagy as a survival mechanism (Rebecca and Amaravadi, 2016; Amaravadi et al., 2019). Hence, autophagy inhibition may reverse resistance to targeted therapies but be comparatively less efficacious on its own. Specific driver mutations such as Ras, BRAF, LKB1 are known to sensitize cancer cells to autophagy inhibition (Amaravadi et al., 2019). In particular, Ras-driven pancreatic cancer is a relevant indication where enhanced autophagy is observed upon pharmacological MAP kinase pathway inhibition (Bryant et al., 2019; Kinsey et al., 2019; Lee et al., 2019). Similarly, BRAF-mutations also promote autophagy upon treatment with BRAF or MEK inhibitors (Levy et al., 2014; Ma et al., 2014; Mulcahy Levy et al., 2014; Xie et al., 2015; Mulcahy Levy et al., 2017). In both these settings, combining inhibitors of growth factor signaling along with autophagy provides a synergistic effect, revealing therapeutic efficacy in otherwise non-responsive disease. More recently, selective autophagy of MHC-I by pancreatic cancer cells was shown to promote resistance to immunotherapy, with genetic or pharmacological inhibition of autophagy re-sensitizing cancer cells to checkpoint blockade (Yamamoto et al., 2020). However, class I MHC as well as co-stimulatory or co-inhibitory proteins (e.g., CD40, CD80/86, PD-L1/2) are established interferon response genes. Autophagy is acknowledged as a potent inhibitor of type I and II interferon responses (Martin P. K. et al., 2018; Samie et al., 2018; Orvedahl et al., 2019; Wang et al., 2019; Wang Y. T. et al., 2020; Lawson et al., 2020). Thus, numerous autophagy-dependent mechanisms may govern cellular MHC-I levels. It will be important to confirm whether this observation is unique to pancreatic tumors or a broader phenomenon, given that loss of antigen presentation by tumor cells is a highly relevant mechanism for resistance to immunotherapy (Zaretsky et al., 2016; Patel et al., 2017; Burr et al., 2019).
Autophagy Protects Against Cell Death
Autophagy can provide tumor cells protection against cell death (Guo et al., 2011; Lock et al., 2011). Immunogenic death of tumor cells by pyroptosis (Wang Q. et al., 2020; Zhang et al., 2020) and necroptosis (Yatim et al., 2015; Snyder et al., 2019) is understood to enhance cellular immunity against cancer by multiple mechanisms including antigen release and adjuvant effects generated by dying cells. Intriguingly, a recent genome-wide assessment of cancer cell-intrinsic mechanisms of resistance against T cell mediated killing revealed autophagy to be a common cyto-protective pathway in murine models (Lawson et al., 2020). Autophagy inhibits necroptosis in intestinal epithelial and innate immune cells by regulating turnover of key components such as RIPK1 and RIPK3 (Matsuzawa-Ishimoto et al., 2017; Lim et al., 2019). In contrast, autophagosomes are shown to provide a platform for the necrosome in prostate epithelial and rhabdomyosarcoma cells (Basit et al., 2013; Goodall et al., 2016). The extent to which this occurs in additional cell types and its significance in vivo remains to be determined. Overall, there is consensus that autophagy is a relevant mechanism for cancer cell evasion of cell death (reviewed in Towers et al., 2020), but we have limited understanding of the clinical setting(s) where this dependency can be safely exploited.
Autophagy Impacts Immune Cell Infiltration of Tumors
Autophagy was reported to coordinate IL-6 secretion in RAS-driven invasion (Lock et al., 2014). Also, non-classical protein secretion was proposed as a mechanism connecting autophagy activity with increased secretion of cytokines known to modulate inflammation and tumorigenesis (Kraya et al., 2015). However, deletion of core autophagy-related genes in immune and cancer cells consistently enhances release of pro-inflammatory cytokines and chemokines (Saitoh et al., 2008; Wei et al., 2011; Murthy et al., 2014; Maycotte et al., 2015; Lee et al., 2016; Mgrditchian et al., 2017; Samie et al., 2018; DeVorkin et al., 2019; Lim et al., 2019; Cotzomi-Ortega et al., 2020; Noman et al., 2020). For example, Mgrditchian et al demonstrated that CCL5 was critical for NK cell infiltration and efficacy in B16F10 melanoma tumors. This is highly relevant for immunotherapy, since the above chemokines are critical for T and NK cell infiltration into solid tumors and strongly associated with a response to immune checkpoint inhibition (Dangaj et al., 2019; House et al., 2020). Thus, inhibition of tumor cell autophagy can reshape the TME by enhanced immune cell recruitment and function.
Tumor-Extrinsic Autophagy
Host Autophagy Feeds Tumors
The TME governs several aspects of disease progression (Sahai et al., 2020). Autophagy in the local TME as well as distal host tissues can promote tumor growth by providing critical nutrients. Studies in diverse model systems have shown that autophagy in the host TME epithelial cells (D. melanogaster tumor progression models), stromal cells (murine stellate cells, pancreatic ductal adenocarcinoma orthotopic grafts), or distal tissue (murine hepatocytes, subcutaneous melanoma grafts) provides amino acids such as alanine and arginine to support tumor cell survival (Karsli-Uzunbas et al., 2014; Sousa et al., 2016; Katheder et al., 2017; Poillet-Perez et al., 2018). These studies indicate that established tumors enhance autophagic flux in the host, thus shifting the metabolic set-point of numerous cell types including the immune system. This metabolic competition often favors the tumor, since co-opting host amino acids and glucose directly supports tumor growth while crippling the effector function of potentially tumoricidal lymphocytes such as cytotoxic T cell and NK cells (Chang et al., 2015; Mah et al., 2017; Konjar and Veldhoen, 2019; Terren et al., 2019). It is important to note that not all models exhibit sensitivity to autophagy inhibition in the host. Similarly, modulation of adaptive immunity may not play a role in autophagy-mediated tumor growth in certain settings (Poillet-Perez et al., 2018). Thus, it is critical to assess the impact of a specific autophagy-related gene across multiple models to determine its impact on tumor progression, metabolic reprogramming and immunosuppression.
Autophagy Reprograms Innate Immune Cells
Myeloid cells include macrophages, dendritic cells, monocytes and granulocytes and comprise the innate immune response. As professional phagocytes and antigen presenting cells, macrophages and dendritic cells are critical for shaping the tumor cytokine milieu as well as antigen-specific immunity. Myeloid cell-specific deletion of genes involved in autophagosome elongation and LC3-associated phagocytosis [LAP, reviewed in (Heckmann and Green, 2019)] enhanced anti-tumor immunity, attenuating tumor growth and metastasis in multiple syngeneic tumor models (Alissafi et al., 2018; Cunha et al., 2018). Here, defective autophagy was primarily generated in macrophages, monocytes and granulocytes, resulting in a preferential inflammatory or “M1” polarization of macrophages along with reduced suppressive capacity of myeloid derived suppressor cells. The impact of these changes included enhanced type I interferon responses along with an increase in polyfunctional cytotoxic T lymphocytes. Similar outcomes were observed upon pharmacological inhibition of lysosomal activity using Chloroquine [CQ; (Chen et al., 2018)]. In contrast, loss of Vps34 (encoded by Pik3c3) in dendritic cells as well as monocytes/macrophages promoted lung metastasis by B16F10 melanoma cells (Parekh et al., 2017). The broad regulatory role of Vps34 in membrane trafficking beyond autophagy may explain the divergent phenotypes generated by loss of Pik3c3 versus downstream autophagy-specific genes such as Atg5. These findings also reinforce the importance of comparing multiple autophagy pathway genes within a model system to determine if altered autophagy truly underlies the phenotypic outcome. Beyond anti-tumor immunity, deletion of autophagy-related genes in myeloid cells has resulted in enhanced type I/II interferon response, autoimmunity and anti-microbial immunity in numerous studies (Marchiando et al., 2013; Martin P. K. et al., 2018; Samie et al., 2018; Wang Y. T. et al., 2020). Altogether, accumulating evidence strongly supports inhibition of autophagy and related pathways as a mechanism to promote innate inflammation by myeloid cells.
Autophagy Regulates T Cell Function
Constitutive deletion of core autophagy-related genes in the T lymphocyte lineage has demonstrated its requirement for the development of thymocytes and peripheral T cells, along with maintenance of regulatory T cells (Tregs) (Pua et al., 2007; Kabat et al., 2016; Parekh et al., 2017). Thymocytes rapidly induce autophagy upon TCR engagement to meet bioenergetic needs (Stephenson et al., 2009; Hubbard et al., 2010). Additionally, loss of autophagy in T cells has shown to compromise their ability to generate antigen-specific memory following infection (Puleston et al., 2014; Xu et al., 2014). Intriguingly, defective autophagy in Tregs or in adult mice dramatically enhances anti-tumor immunity via loss of Tregs or generation of a potent effector memory T cell pool in pre-clinical models (Xu et al., 2014; DeVorkin et al., 2019). Recent studies have highlighted the need for sophisticated genetic models to delineate the role of autophagy in lymphocyte developmental versus function in adult tissues (DeVorkin et al., 2019; Laura Poillet-Perez, 2020). DeVorkin et al demonstrated that inducible deletion of Atg5 in adult mice enhanced T cell glycolytic metabolism while maintaining oxidative phosphorylation (OXPHOS). This shifted CD8+ T cells to an effector memory phenotype with increased IFNγ and TNFα production, consistent with the known requirement of glycolysis for optimal effector T cell function (Cham et al., 2008; Chang et al., 2013; Gubser et al., 2013). Notably, inhibition of T cell checkpoints such as PD-1 (encoded by Pdcd1) also promotes a shift toward glycolytic metabolism, and germline loss of Pdcd1 or its ligand PD-L1 impairs T cell memory while driving a terminally differentiated effector phenotype. In contrast, pharmacological PD-1 inhibition drives effector T lymphocyte functions while augmenting memory as the pharmacodynamic effect of PD-1 inhibition wanes (Odorizzi et al., 2015; Patsoukis et al., 2015; Ahn et al., 2018; Verma et al., 2019; Pauken et al., 2020). It appears that enhanced autophagy in the T cell compartment during tumor progression generates a competitive disadvantage for limiting metabolites, thereby curbing productive anti-tumor effector function. It must also be acknowledged that regulation of glycolytic metabolism promotes the maintenance of memory T cell pools (Sukumar et al., 2013; Zhang and Romero, 2018). Thus, timing and duration of autophagy inhibition should be key considerations to optimally induce an effector T cell response against cancer while retaining the ability to develop T cell memory against relevant tumor antigens.
Adaptation Against Autophagy Inhibition
While tumor cells can induce autophagy as a survival mechanism against therapies, compensatory responses to inhibition of autophagy are also being revealed. For instance, a reversible model of Atg5 deletion showed that while Atg5 deficient mice (ATG5i) exhibited tissue inflammation and degeneration, eventually succumbing to these phenotypes, there was no sign of overt tumor development (Cassidy et al., 2020). These mice also showed accelerated aging that was reverted by restoring Atg5 expression (R-Atg5i). Of note, restoring autophagy promoted spontaneous tumor development, suggesting that prolonged autophagy inhibition may select for additional stress-response pathways which accelerate disease progression when autophagy is restored. Recent studies demonstrate that overactivation of the oxidative stress response by the transcription factor Nrf2 is a dominant consequence of autophagy-related gene deletion in cancer cells (Kerins et al., 2019; Towers et al., 2019). The association between autophagy inhibition and Nrf2 was reported even earlier, predominantly due to accumulation of p62 (Komatsu et al., 2010; Inami et al., 2011; Ichimura et al., 2013). The Nrf2-Keap1 (kelch-like ECH-associated protein 1) pathway is a critical defense mechanism against oxidative stress. Keap1 tightly regulates Nrf2 activity by promoting its proteasomal degradation under basal conditions. Upon oxidative stress, post-translational cysteine modification of Keap1 unleashes Nrf2, where it induces an anti-oxidant transcriptional program. p62 binds Keap1 at the Nrf2-binding site, and over-abundant p62 outcompetes Nrf2, leading to non-canonical activation of Nrf2-mediated transcription (Komatsu et al., 2010). Nrf2-mediated induction of target genes is known to be associated with human cancers (Hayes and McMahon, 2009) and its regulation by p62 is responsible for development of human hepatocellular carcinoma (Inami et al., 2011; Takamura et al., 2011). Advances in phenotypic screening provide a novel opportunity to identify additional mechanisms of resistance to autophagy inhibition. Investments in this area of research will allow us to better predict how cancers may circumvent defective autophagy even when combined with current therapies.
Hurdles to Successful Pharmacological Inhibition of Autophagy
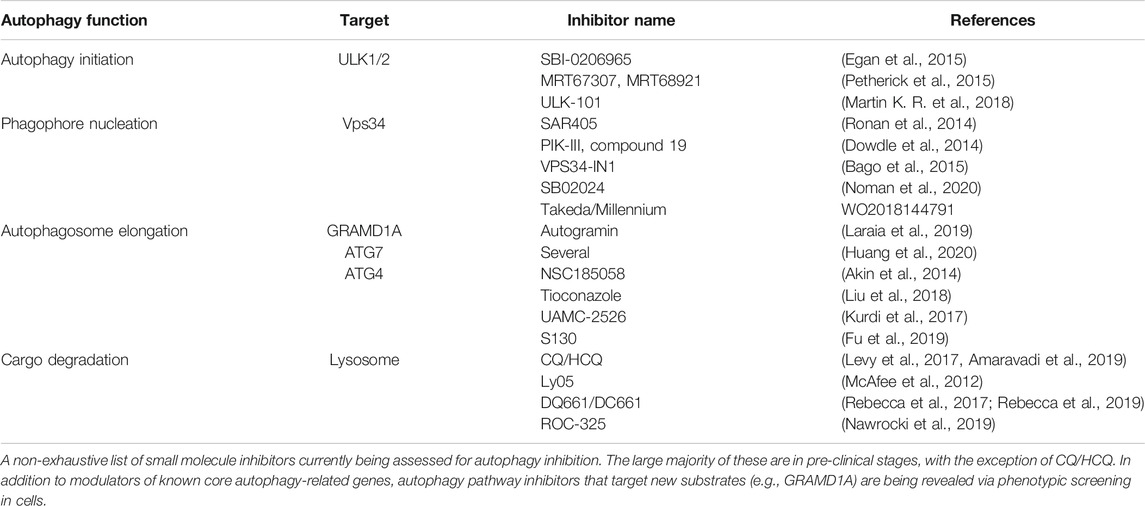
To date, lysomotropic agents such as CQ and hydroxychloroquine (HCQ) are the only candidates undergoing clinical assessment for inhibition of autophagy in cancer. CQ/HCQ have been used in malaria and rheumatologic disorders; they have been repurposed in combination with other agents for treatment of cancers. In numerous studies, CQ in combination with other agents showed beneficial outcomes (Levy et al., 2017; Amaravadi et al., 2019). Additionally, more potent lysosomal inhibitors have been developed, such as Lys05, DQ661, DC661, and ROC-325 (McAfee et al., 2012; Rebecca et al., 2017; Nawrocki et al., 2019; Rebecca et al., 2019). Although some studies with CQ/HCQ suggest clinical benefits, their potency and specificity toward autophagy pathway inhibition remain outstanding concerns (Maycotte et al., 2012). The quest to identify more specific autophagy modulators has driven efforts targeting earlier steps of the pathway (depicted in Figure 1). Most prominent are inhibitors of the Class III lipid kinase Vps34 (patent WO2018144791; Dowdle et al., 2014; Ronan et al., 2014; Bago et al., 2015; Honda et al., 2016; Noman et al., 2020) and the upstream kinases ULK1/2 (Egan et al., 2015; Petherick et al., 2015; Martin K. R. et al., 2018; Chaikuad et al., 2019). SB02024, developed by Sprint Bioscience together with SAR-405 by Sanofi Pharma revealed that inhibition of Vps34 attenuated tumor growth and extended survival in multiple pre-clinical models (Noman et al., 2020). Consistent with genetic observations, pharmacological Vps34 inhibition promoted tumor infiltration by NK and CD8+ T cells. It also demonstrated synergy with immune checkpoint inhibitors such as anti-PD-L1 or PD-1. Further down the pathway, inhibitors of the E3-like enzyme ATG7 and the ATG4 family of cysteine proteases present intriguing opportunities (Akin et al., 2014; Kurdi et al., 2017; Liu et al., 2018; Fu et al., 2019; Huang et al., 2020). Nonetheless, genetic models still comprise the large majority of evidence supporting more specific nodes of autophagy as targets. Beyond canonical members of the pathway, genetic phenotypic screens using engineered reporter cell lines have provided new insights for modulation of autophagic flux (DeJesus et al., 2016; Morita et al., 2018; Kerins et al., 2019; Shoemaker et al., 2019). In addition, pharmacological screens with the same cellular tools have revealed inhibitors with new mechanisms of action. For instance, autogramins were identified by an image-based phenotypic screen in EGFP-LC3 overexpressing MCF7 cells (Laraia et al., 2019). Autogramins selectively target GRAMD1A, which is required for autophagosome biogenesis by modulating cholesterol distribution around autophagosome initiation site. For all the above examples (including canonical autophagy genes), a number of unknowns still exist and require improved understanding in order to make meaningful progress for pharmacological modulation of autophagy. Below, we discuss some of these knowledge gaps.
Identifying Accurate Pharmacodynamic and Predictive Biomarkers of Autophagic Flux
As detailed above, inhibiting autophagy has revealed multiple molecular outcomes which cumulatively impact tumor progression. However, there is a paucity of accurate biomarkers to quantify therapeutic perturbation of autophagy. Classically, accumulation of autophagy receptors such as p62 is acknowledged as a direct pathway biomarker. More recently, immunomodulatory proteins such as MHC-I, TRIF, RIPK1, RIPK3, and STING have shown to be directly modulated by - and in turn impact - autophagy (Matsuzawa-Ishimoto et al., 2017; Samie et al., 2018; Gui et al., 2019; Lim et al., 2019; Liu et al., 2019; Yamamoto et al., 2020). Measuring turnover of autophagic cargo is a valuable cell-associated readout; however, we now appreciate that different cell types within a complex microenvironment exhibit varying kinetics and dynamics of autophagic flux. Additionally, the need for multiple tissue biopsies to measure cargo turnover over course of a treatment poses practical challenges in a therapeutic setting. Thus, identification of peripheral or biofluid-based surrogates would be highly valuable in understanding the magnitude and durability of autophagy inhibition generated by a therapeutic agent. For example, CCL5/RANTES was discussed as a peripheral biomarker for Vps34 inhibition (Mgrditchian et al., 2017; Noman et al., 2020). Although CCL5 is proposed as a direct target of autophagy, it is also a component of the interferon response that is consistently shown to be enhanced upon suppression of autophagy, thus complicating its interpretation as a bona fide autophagic substrate (Martin P. K. et al., 2018; Samie et al., 2018; Wang Y. T. et al., 2020). Additionally, metabolic profiling in pre-clinical models suggest non-essential amino acids Arginine and Alanine as viable circulating biomarkers (Sousa et al., 2016; Poillet-Perez et al., 2018). It will be valuable to determine whether these candidates are 1) meaningful PD biomarkers in a clinical setting, and 2) reflect the kinetics of pharmacological autophagy inhibition. Since the above studies generate chronic or genetic loss of autophagy, a return of autophagic flux is not measured. It is more likely that intermittent modulation of autophagy will be utilized via pulsatile dosing in patients, as systemic, long-term suppression of this pathway may not be desirable. Identification of facile biomarkers that faithfully report inhibition as well as normalization of autophagic flux will prove highly valuable in evaluating therapeutic options.
Accumulating evidence supports autophagy as a mechanism for resistance against targeted, radiation and chemotherapies (Santana-Codina et al., 2017). More recent demonstration of autophagy-mediated resistance to MAP-kinase pathway inhibition in pancreatic cancer have prompted clinical assessment of chloroquine in combination with trametinib (Bryant et al., 2019; Kinsey et al., 2019) [Clinicaltrials.gov NCT03979651]. Emerging pre-clinical studies demonstrate a critical role for autophagy pathway genes in immunosuppression and immune-evasion by cancer. Intriguingly, this is driven by autophagy in the tumor cell as well as components of the TME including myeloid cells and T lymphocytes (Wei et al., 2016; Cunha et al., 2018; DeVorkin et al., 2019; Yamamoto et al., 2020). To identify patients who would benefit most from autophagy inhibition, it will be critical to identify determinants of elevated autophagic flux and measure their correlation with treatment-associated disease progression.
Non-Canonical Roles of Autophagy Pathway Genes
A growing number of genes associated with autophagic flux also perform autophagy-independent functions. For instance, the Class III PI-3 Kinase Vps34 is well known to regulate endocytic sorting as well as autophagosome formation (Rostislavleva et al., 2015; Stjepanovic et al., 2017). The initiation complex kinases ULK1 and 2 are involved in autophagy-independent lysosomal targeting of ferritin, stress granule degradation, ER-Golgi trafficking of cargo and axon guidance (Joo et al., 2016; Goodwin et al., 2017; Wang et al., 2018; Wang et al., 2019). In phagocytes, components of the Vps34 complex III, autophagosome elongation and maturation machineries have also shown to drive LAP and endocytosis (LANDO) (Cunha et al., 2018; Heckmann et al., 2019). ATG5, ATG16L1, and ATG4 have been implicated in secretory pathways such as membrane exocytosis, leaderless cytokine secretion and exosome release (Murrow et al., 2015; Zhang et al., 2015; Guo et al., 2017; Keller et al., 2020). These non-canonical roles may well be consequential for immunomodulation by autophagy-related genes in the TME.
Comparing Genetic Models With Pharmacological Inhibition
Genetic mouse models clearly demonstrate a role for autophagy genes in mammalian development as well as tumor progression (reviewed in Kuma et al., 2017; Table 1). Broad functions of Vps34 and ULK1/2 are consistent with embryonic lethality conferred by their germline deficiency. In contrast, genes involved with the autophagosome elongation machinery consistently exhibit perinatal lethality. Thus, even though enhanced autophagy is associated with therapeutic resistance, immunosuppression and disease progression in cancer, the margin of safety should be a key consideration when assessing pharmacological inhibition of autophagy. As highlighted by the murine models discussed above, limitations of genetic loss-of-function prevent a complete assessment of therapeutic autophagy modulation. These include the inability to tune pathway inhibition, the constitutive deletion of targeted gene(s) and the inability to rescue or re-introduce autophagy following its inhibition. These may be particularly important features consider, since it is expected that complete, chronic inhibition of the pathway will likely be detrimental to a durable immune response against cancer. Indeed, recent evidence suggests that pharmacological inhibition of autophagy does not compromise adaptive immunity, consistent with the normal development of mice harboring hypomorphic loss-of-function in core genes such as Atg16L1 (Cadwell et al., 2008; Hubbard-Lucey et al., 2014; Starobinets et al., 2016; Noman et al., 2020). Moreover, the developmental roles of autophagy-related genes suggest that genetic loss-of-function models do not recapitulate the phenotypic outcomes of pharmacological inhibition, which is transient and incomplete. Thus, careful phenotypic assessment of cellular phenotypes upon pharmacological versus genetic inhibition of autophagy is necessary to delineate which outcomes are consequences of disrupted cell development vs. effector function.
Concluding Remarks
Novel insights into the immunomodulatory functions of autophagy have driven a resurgence of interest in its pharmacological modulation for numerous diseases. While augmenting autophagy is a relevant therapeutic avenue for autoimmunity, neuroinflammation and chronic inflammatory diseases, its role in cancer has remained pleiotropic. In the setting of established disease, sustained autophagy is acknowledged as a critical mechanism for treatment resistance and immune-evasion. Successful modulation of autophagy will depend on pharmacological approaches which safely diminish autophagic flux to promote meaningful immune responses against cancer, while at the same time allowing for the emergence of durable protection as determined by antigen-specific cellular immunity. The growing number of pharmacological and genetic approaches to modulate autophagy predicts a promising future for its therapeutic targeting to benefit patients battling cancer.
Author Contributions
All authors directly contributed to the content of the work and approved its publication.
Conflict of Interest
JL and AM are employees of Genentech, Inc. and shareholders in Roche.
References
Ahn, E., Araki, K., Hashimoto, M., Li, W., Riley, J. L., Cheung, J., et al. (2018). Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115 (18), 4749–4754. doi:10.1073/pnas.1718217115
Akin, D., Wang, S. K., Habibzadegah-Tari, P., Law, B., Ostrov, D., Li, M., et al. (2014). A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 10 (11), 2021–2035. doi:10.4161/auto.32229
Alissafi, T., Hatzioannou, A., Mintzas, K., Barouni, R. M., Banos, A., Sormendi, S., et al. (2018). Autophagy orchestrates the regulatory program of tumor-associated myeloid-derived suppressor cells. J. Clin. Invest. 128 (9), 3840–3852. doi:10.1172/JCI120888
Amaravadi, R. K., Kimmelman, A. C., and Debnath, J. (2019). Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9 (9), 1167–1181. doi:10.1158/2159-8290.CD-19-0292
Bago, R., Sommer, E., Malik, N., Munson, M. J., Prescott, A. R., Davies, P., et al. (2015). Characterization of VPS34-IN1, a specific inhibitor of Vps34 reveals that the phosphatidylinositol 3-phosphate binding SGK3 protein kinase is regulated by class III PI-3 kinase. Mol. Cancer Ther. 14 (7). doi:10.1158/1538-8514.Pi3k14-A04
Basit, F., Cristofanon, S., and Fulda, S. (2013). Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 20 (9), 1161–1173. doi:10.1038/cdd.2013.45
Bhargava, H. K., Tabata, K., Byck, J. M., Hamasaki, M., Farrell, D. P., Anishchenko, I., et al. (2020). Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex. Proc. Natl. Acad. Sci. U.S.A. 117, 17003–17010. doi:10.1073/pnas.2008030117
Bryant, K. L., Stalnecker, C. A., Zeitouni, D., Klomp, J. E., Peng, S., Tikunov, A. P., et al. (2019). Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 25 (4), 628–640. doi:10.1038/s41591-019-0368-8
Burr, M. L., Sparbier, C. E., Chan, K. L., Chan, Y. C., Kersbergen, A., Lam, E. Y. N., et al. (2019). An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell 36 (4), 385–401. doi:10.1016/j.ccell.2019.08.008
Cadwell, K., Liu, J. Y., Brown, S. L., Miyoshi, H., Loh, J., Lennerz, J. K., et al. (2008). A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal paneth cells. Nature 456 (7219), 259–263. doi:10.1038/nature07416
Cassidy, L. D., Young, A. R. J., Young, C. N. J., Soilleux, E. J., Fielder, E., Weigand, B. M., et al. (2020). Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat. Commun. 11 (1), 307. doi:10.1038/s41467-019-14187-x
Chaikuad, A., Koschade, S. E., Stolz, A., Zivkovic, K., Pohl, C., Shaid, S., et al. (2019). Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2). Biochem. J. 476 (5), 875–887. doi:10.1042/BCJ20190038
Cham, C. M., Driessens, G., O'Keefe, J. P., and Gajewski, T. F. (2008). Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 38 (9), 2438–2450. doi:10.1002/eji.200838289
Chang, C. H., Curtis, J. D., Maggi, L. B., Faubert, B., Villarino, A. V., O'Sullivan, D., et al. (2013). Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153 (6), 1239–1251. doi:10.1016/j.cell.2013.05.016
Chang, C. H., Qiu, J., O’Sullivan, D., Buck, M. D., Noguchi, T., Curtis, J. D., et al. (2015). Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162 (6), 1229–1241. doi:10.1016/j.cell.2015.08.016
Chen, D., Xie, J., Fiskesund, R., Dong, W., Liang, X., Lv, J., et al. (2018). Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 9 (1), 873. doi:10.1038/s41467-018-03225-9
Clarke, A. J., and Simon, A. K. (2019). Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 19 (3), 170–183. doi:10.1038/s41577-018-0095-2
Cotzomi-Ortega, I., Rosas-Cruz, A., Ramirez-Ramirez, D., Reyes-Leyva, J., Rodriguez-Sosa, M., Aguilar-Alonso, P., et al. (2020). Autophagy inhibition induces the secretion of macrophage migration inhibitory factor (MIF) with autocrine and paracrine effects on the promotion of malignancy in breast cancer. Biology (Basel) 9 (1), 20. doi:10.3390/biology9010020
Cunha, L. D., Yang, M., Carter, R., Guy, C., Harris, L., Crawford, J. C., et al. (2018). LC3-Associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell 175 (2), 429–441. doi:10.1016/j.cell.2018.08.061
Dangaj, D., Bruand, M., Grimm, A. J., Ronet, C., Barras, D., Duttagupta, P. A., et al. (2019). Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell 35 (6), 885–900.e10. doi:10.1016/j.ccell.2019.05.004
DeJesus, R., Moretti, F., McAllister, G., Wang, Z. C., Bergman, P., Liu, S. M., et al. (2016). Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. elife 5, e17290. doi:10.7554/eLife.17290
DeVorkin, L., Pavey, N., Carleton, G., Comber, A., Ho, C., Lim, J., et al. (2019). Autophagy regulation of metabolism is required for CD8(+) T cell anti-tumor immunity. Cell Rep. 27 (2), 502–513. doi:10.1016/j.celrep.2019.03.037
Dikic, I., and Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19 (6), 349–364. doi:10.1038/s41580-018-0003-4
Dou, Z., Xu, C., Donahue, G., Shimi, T., Pan, J. A., Zhu, J., et al. (2015). Autophagy mediates degradation of nuclear lamina. Nature 527 (7576), 105–109. doi:10.1038/nature15548
Dowdle, W. E., Nyfeler, B., Nagel, J., Elling, R. A., Liu, S., Triantafellow, E., et al. (2014). Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 16 (11), 1069–1079. doi:10.1038/ncb3053
Duran, A., Hernandez, E. D., Reina-Campos, M., Castilla, E. A., Subramaniam, S., Raghunandan, S., et al. (2016). p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell 30 (4), 595–609. doi:10.1016/j.ccell.2016.09.004
Egan, D. F., Chun, M. G., Vamos, M., Zou, H., Rong, J., Miller, C. J., et al. (2015). Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59 (2), 285–297. doi:10.1016/j.molcel.2015.05.031
Eng, C. H., Wang, Z., Tkach, D., Toral-Barza, L., Ugwonali, S., Liu, S., et al. (2016). Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc. Natl. Acad. Sci. U.S.A. 113 (1), 182–187. doi:10.1073/pnas.1515617113
Fu, Y. Y., Huang, Z. Y., Hong, L., Lu, J. H., Feng, D., Yin, X. M., et al. (2019). Targeting ATG4 in cancer therapy. Cancers (Basel) 11 (5), 649. doi:10.3390/cancers11050649
Goodall, M. L., Fitzwalter, B. E., Zahedi, S., Wu, M., Rodriguez, D., Mulcahy-Levy, J. M., et al. (2016). The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev. Cell 37 (4), 337–349. doi:10.1016/j.devcel.2016.04.018
Goodwin, J. M., Dowdle, W. E., DeJesus, R., Wang, Z., Bergman, P., Kobylarz, M., et al. (2017). Autophagy-independent lysosomal targeting regulated by ULK1/2-fip200 and ATG9. Cell Rep. 20 (10), 2341–2356. doi:10.1016/j.celrep.2017.08.034
Gubser, P. M., Bantug, G. R., Razik, L., Fischer, M., Dimeloe, S., Hoenger, G., et al. (2013). Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 14 (10), 1064–1072. doi:10.1038/ni.2687
Gui, X., Yang, H., Li, T., Tan, X., Shi, P., Li, M., et al. (2019). Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567 (7747), 262–266. doi:10.1038/s41586-019-1006-9
Guo, J. Y., Chen, H. Y., Mathew, R., Fan, J., Strohecker, A. M., Karsli-Uzunbas, G., et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25 (5), 460–470. doi:10.1101/gad.2016311
Guo, H., Chitiprolu, M., Roncevic, L., Javalet, C., Hemming, F. J., Trung, M. T., et al. (2017). Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev. Cell 43 (6), 716–730. doi:10.1016/j.devcel.2017.11.018
Guo, J. Y., Karsli-Uzunbas, G., Mathew, R., Aisner, S. C., Kamphorst, J. J., Strohecker, A. M., et al. (2013). Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 27 (13), 1447–1461. doi:10.1101/gad.219642.113
Hayes, J. D., and McMahon, M. (2009). NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 34 (4), 176–188. doi:10.1016/j.tibs.2008.12.008
Heckmann, B. L., and Green, D. R. (2019). LC3-associated phagocytosis at a glance. J Cell Sci 132 (5), jcs231472. doi:10.1242/jcs.222984
Heckmann, B. L., Teubner, B. J. W., Tummers, B., Boada-Romero, E., Harris, L., Yang, M., et al. (2019). LC3-Associated endocytosis facilitates beta-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell 178 (3), 536–551. doi:10.1016/j.cell.2019.05.056
Honda, A., Harrington, E., Cornella-Taracido, I., Furet, P., Knapp, M. S., Glick, M., et al. (2016). Potent, selective, and orally bioavailable inhibitors of VPS34 provide chemical tools to modulate autophagy in Vivo. ACS Med. Chem. Lett. 7 (1), 72–76. doi:10.1021/acsmedchemlett.5b00335
House, I. G., Savas, P., Lai, J. Y., Chen, A. X. Y., Oliver, A. J., Teo, Z. L., et al. (2020). Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 26 (2), 487–504. doi:10.1158/1078-0432.Ccr-19-1868
Huang, S. C., Adhikari, S., Brownell, J. E., Calderwood, E. F., Chouitar, J., D'Amore, N. R., et al. (2020). Discovery and optimization of pyrazolopyrimidine sulfamates as ATG7 inhibitors. Bioorg. Med. Chem. 28 (19), 115681. doi:10.1016/j.bmc.2020.115681
Hubbard, V. M., Valdor, R., Patel, B., Singh, R., Cuervo, A. M., and Macian, F. (2010). Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 185 (12), 7349–7357. doi:10.4049/jimmunol.1000576
Hubbard-Lucey, V. M., Shono, Y., Maurer, K., West, M. L., Singer, N. V., Ziegler, C. G., et al. (2014). Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity 41 (4), 579–591. doi:10.1016/j.immuni.2014.09.011
Ichimura, Y., Waguri, S., Sou, Y. S., Kageyama, S., Hasegawa, J., Ishimura, R., et al. (2013). Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 51 (5), 618–631. doi:10.1016/j.molcel.2013.08.003
Inami, Y., Waguri, S., Sakamoto, A., Kouno, T., Nakada, K., Hino, O., et al. (2011). Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 193 (2), 275–284. doi:10.1083/jcb.201102031
Joo, J. H., Wang, B., Frankel, E., Ge, L., Xu, L., Iyengar, R., et al. (2016). The noncanonical role of ULK/ATG1 in ER-to-golgi trafficking is essential for cellular homeostasis. Mol. Cell 62 (4), 491–506. doi:10.1016/j.molcel.2016.04.020
Kabat, A. M., Harrison, O. J., Riffelmacher, T., Moghaddam, A. E., Pearson, C. F., Laing, A., et al. (2016). The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. elife 5, e12444. doi:10.7554/eLife.12444
Kang, C., Xu, Q., Martin, T. D., Li, M. Z., Demaria, M., Aron, L., et al. (2015). The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349 (6255), aaa5612. doi:10.1126/science.aaa5612
Karsli-Uzunbas, G., Guo, J. Y., Price, S., Teng, X., Laddha, S. V., Khor, S., et al. (2014). Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4 (8), 914–927. doi:10.1158/2159-8290.CD-14-0363
Katheder, N. S., Khezri, R., O’Farrell, F., Schultz, S. W., Jain, A., Rahman, M. M., et al. (2017). Microenvironmental autophagy promotes tumour growth. Nature 541 (7637), 417–420. doi:10.1038/nature20815
Kaushik, S., and Cuervo, A. M. (2018). The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19 (6), 365–381. doi:10.1038/s41580-018-0001-6
Keller, M. D., Ching, K. L., Liang, F. X., Dhabaria, A., Tam, K., Ueberheide, B. M., et al. (2020). Decoy exosomes provide protection against bacterial toxins. Nature 579 (7798). doi:10.1038/s41586-020-2066-6
Kerins, M. J., Liu, P., Tian, W., Mannheim, W., Zhang, D. D., and Ooi, A. (2019). Genome-wide CRISPR screen reveals autophagy disruption as the convergence mechanism that regulates the NRF2 transcription factor. Mol. Cell Biol. 39 (13), e00037-19. doi:10.1128/MCB.00037-19
Kinsey, C. G., Camolotto, S. A., Boespflug, A. M., Guillen, K. P., Foth, M., Truong, A., et al. (2019). Protective autophagy elicited by RAF-->MEK-->ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25 (4), 620–627. doi:10.1038/s41591-019-0367-9
Kirkin, V. (2020). History of the selective autophagy research: how did it begin and where does it stand today? J. Mol. Biol. 432 (1), 3–27. doi:10.1016/j.jmb.2019.05.010
Kirkin, V., and Rogov, V. V. (2019). A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell 76 (2), 268–285. doi:10.1016/j.molcel.2019.09.005
Klionsky, D. J., Cregg, J. M., Dunn, W. A., Emr, S. D., Sakai, Y., Sandoval, I. V., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5 (4), 539–545. doi:10.1016/s1534-5807(03)00296-x
Komatsu, M., Kurokawa, H., Waguri, S., Taguchi, K., Kobayashi, A., Ichimura, Y., et al. (2010). The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12 (3), 213–223. doi:10.1038/ncb2021
Konjar, S., and Veldhoen, M. (2019). Dynamic metabolic state of tissue resident CD8 T cells. Front. Immunol. 10, 1683. doi:10.3389/fimmu.2019.01683
Kraya, A. A., Piao, S., Xu, X., Zhang, G., Herlyn, M., Gimotty, P., et al. (2015). Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy 11 (1), 60–74. doi:10.4161/15548627.2014.984273
Kuma, A., Komatsu, M., and Mizushima, N. (2017). Autophagy-monitoring and autophagy-deficient mice. Autophagy 13 (10), 1619–1628. doi:10.1080/15548627.2017.1343770
Kurdi, A., Cleenewerck, M., Vangestel, C., Lyssens, S., Declercq, W., Timmermans, J. P., et al. (2017). ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem. Pharmacol. 138, 150–162. doi:10.1016/j.bcp.2017.06.119
Laraia, L., Friese, A., Corkery, D. P., Konstantinidis, G., Erwin, N., Hofer, W., et al. (2019). The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis. Nat. Chem. Biol. 15 (7), 710–720. doi:10.1038/s41589-019-0307-5
Poillet-Perez, L., Sharp, D. S., Yang, Y., Laddha, S. V., Ibrahim, M., Bommareddy, K. P., et al. (2020). Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nature cancer 1, 923–934. doi:10.1038/s43018-020-00110-7
Lawson, K. A., Sousa, C. M., Zhang, X., Kim, E., Akthar, R., Caumanns, J. J., et al. (2020). Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. doi:10.1038/s41586-020-2746-2
Lee, C. S., Lee, L. C., Yuan, T. L., Chakka, S., Fellmann, C., Lowe, S. W., et al. (2019). MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl. Acad. Sci. U.S.A. 116 (10), 4508–4517. doi:10.1073/pnas.1817494116
Lee, J. P., Foote, A., Fan, H., Peral de Castro, C., Lang, T., Jones, S. A., et al. (2016). Loss of autophagy enhances MIF/macrophage migration inhibitory factor release by macrophages. Autophagy 12 (6), 907–916. doi:10.1080/15548627.2016.1164358
Levine, B., and Kroemer, G. (2019). Biological functions of autophagy genes: a disease perspective. Cell 176 (1–2), 11–42. doi:10.1016/j.cell.2018.09.048
Levy, J., Cacheux, W., Bara, M. A., L'Hermitte, A., Lepage, P., Fraudeau, M., et al. (2015). Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat. Cell Biol. 17 (8), 1062–U1440. doi:10.1038/ncb3206
Levy, J. M., Thompson, J. C., Griesinger, A. M., Amani, V., Donson, A. M., Birks, D. K., et al. (2014). Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 4 (7), 773–780. doi:10.1158/2159-8290.CD-14-0049
Levy, J. M. M., Towers, C. G., and Thorburn, A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17 (9), 528–542. doi:10.1038/nrc.2017.53
Lim, J., Park, H., Heisler, J., Maculins, T., Roose-Girma, M., Xu, M., et al. (2019). Autophagy regulates inflammatory programmed cell death via turnover of RHIM-domain proteins. elife 8, e44452. doi:10.7554/eLife.44452
Liu, D., Wu, H., Wang, C., Li, Y., Tian, H., Siraj, S., et al. (2019). STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 26 (9), 1735–1749. doi:10.1038/s41418-018-0251-z
Liu, P. F., Tsai, K. L., Hsu, C. J., Tsai, W. L., Cheng, J. S., Chang, H. W., et al. (2018). Drug repurposing screening identifies tioconazole as an ATG4 inhibitor that suppresses autophagy and sensitizes cancer cells to chemotherapy. Theranostics 8 (3), 830–845. doi:10.7150/thno.22012
Lock, R., Kenific, C. M., Leidal, A. M., Salas, E., and Debnath, J. (2014). Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 4 (4), 466–479. doi:10.1158/2159-8290.CD-13-0841
Lock, R., Roy, S., Kenific, C. M., Su, J. S., Salas, E., Ronen, S. M., et al. (2011). Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 22 (2), 165–178. doi:10.1091/mbc.E10-06-0500
Ma, X. H., Piao, S. F., Dey, S., McAfee, Q., Karakousis, G., Villanueva, J., et al. (2014). Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Invest. 124 (3), 1406–1417. doi:10.1172/JCI70454
Mah, A. Y., Rashidi, A., Keppel, M. P., Saucier, N., Moore, E. K., Alinger, J. B., et al. (2017). Glycolytic requirement for NK cell cytotoxicity and cytomegalovirus control. JCI Insight 2 (23), e95128. doi:10.1172/jci.insight.95128
Marchiando, A. M., Ramanan, D., Ding, Y., Gomez, L. E., Hubbard-Lucey, V. M., Maurer, K., et al. (2013). A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe 14 (2), 216–224. doi:10.1016/j.chom.2013.07.013
Martin, K. R., Celano, S. L., Solitro, A. R., Gunaydin, H., Scott, M., O'Hagan, R. C., et al. (2018). A potent and selective ULK1 inhibitor suppresses autophagy and sensitizes cancer cells to nutrient stress. iScience 8, 74–84. doi:10.1016/j.isci.2018.09.012
Martin, P. K., Marchiando, A., Xu, R., Rudensky, E., Yeung, F., Schuster, S. L., et al. (2018). Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat. Microbiol 3 (10), 1131–1141. doi:10.1038/s41564-018-0229-0
Mathew, R., Karp, C. M., Beaudoin, B., Vuong, N., Chen, G., Chen, H. Y., et al. (2009). Autophagy suppresses tumorigenesis through elimination of p62. Cell 137 (6), 1062–1075. doi:10.1016/j.cell.2009.03.048
Matsunaga, K., Saitoh, T., Tabata, K., Omori, H., Satoh, T., Kurotori, N., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11 (4), 385–396. doi:10.1038/ncb1846
Matsuzawa-Ishimoto, Y., Shono, Y., Gomez, L. E., Hubbard-Lucey, V. M., Cammer, M., Neil, J., et al. (2017). Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214 (12), 3687–3705. doi:10.1084/jem.20170558
Maycotte, P., Aryal, S., Cummings, C. T., Thorburn, J., Morgan, M. J., and Thorburn, A. (2012). Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8 (2), 200–212. doi:10.4161/auto.8.2.18554
Maycotte, P., Jones, K. L., Goodall, M. L., Thorburn, J., and Thorburn, A. (2015). Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol. Cancer Res. 13 (4), 651–658. doi:10.1158/1541-7786.MCR-14-0487
McAfee, Q., Zhang, Z., Samanta, A., Levi, S. M., Ma, X. H., Piao, S., et al. (2012). Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl. Acad. Sci. U.S.A. 109 (21), 8253–8258. doi:10.1073/pnas.1118193109
Melia, T. J., Lystad, A. H., and Simonsen, A. (2020). Autophagosome biogenesis: from membrane growth to closure. J. Cell Biol. 219 (6), e202002085. doi:10.1083/jcb.202002085
Mgrditchian, T., Arakelian, T., Paggetti, J., Noman, M. Z., Viry, E., Moussay, E., et al. (2017). Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 114 (44), E9271–E9279. doi:10.1073/pnas.1703921114
Michaud, M., Martins, I., Sukkurwala, A. Q., Adjemian, S., Ma, Y., Pellegatti, P., et al. (2011). Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334 (6062), 1573–1577. doi:10.1126/science.1208347
Mizushima, N. (2018). A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20 (5), 521–527. doi:10.1038/s41556-018-0092-5
Morita, K., Hama, Y., Izume, T., Tamura, N., Ueno, T., Yamashita, Y., et al. (2018). Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217 (11), 3817–3828. doi:10.1083/jcb.201804132
Mulcahy Levy, J. M., Foreman, N. K., and Thorburn, A. (2014). Using BRAF(V600E) as a marker of autophagy dependence in pediatric brain tumors. Autophagy 10 (11), 2077–2078. doi:10.4161/auto.36138
Mulcahy Levy, J. M., Zahedi, S., Griesinger, A. M., Morin, A., Davies, K. D., Aisner, D. L., et al. (2017). Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. elife 6, e19671. doi:10.7554/eLife.19671
Murrow, L., Malhotra, R., and Debnath, J. (2015). ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17 (3). 300–310. doi:10.1038/ncb3112
Murthy, A., Li, Y., Peng, I., Reichelt, M., Katakam, A. K., Noubade, R., et al. (2014). A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506 (7489), 456–462. doi:10.1038/nature13044
Nawrocki, S. T., Han, Y., Visconte, V., Przychodzen, B., Espitia, C. M., Phillips, J., et al. (2019). The novel autophagy inhibitor ROC-325 augments the antileukemic activity of azacitidine. Leukemia 33 (12), 2971–2974. doi:10.1038/s41375-019-0529-2
Noman, M. Z., Parpal, S., Van Moer, K., Xiao, M. L., Yu, Y., Viklund, J., et al. (2020). Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci. Adv. 6 (18), eaax7881. doi:10.1126/sciadv.aax7881
Odorizzi, P. M., Pauken, K. E., Paley, M. A., Sharpe, A., and Wherry, E. J. (2015). Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212 (7), 1125–1137. doi:10.1084/jem.20142237
Ohashi, Y., Tremel, S., and Williams, R. L. (2019). VPS34 complexes from a structural perspective. J. Lipid Res. 60 (2), 229–241. doi:10.1194/jlr.R089490
Oku, M., and Sakai, Y. (2018). Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays 40 (6), e1800008. doi:10.1002/bies.201800008
Orvedahl, A., McAllaster, M. R., Sansone, A., Dunlap, B. F., Desai, C., Wang, Y. T., et al. (2019). Autophagy genes in myeloid cells counteract IFNgamma-induced TNF-mediated cell death and fatal TNF-induced shock. Proc. Natl. Acad. Sci. U.S.A. 116 (33), 16497–16506. doi:10.1073/pnas.1822157116
Parekh, V. V., Pabbisetty, S. K., Wu, L., Sebzda, E., Martinez, J., Zhang, J., et al. (2017). Autophagy-related protein Vps34 controls the homeostasis and function of antigen cross-presenting CD8alpha(+) dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 114 (31), E6371–E6380. doi:10.1073/pnas.1706504114
Patel, S. J., Sanjana, N. E., Kishton, R. J., Eidizadeh, A., Vodnala, S. K., Cam, M., et al. (2017). Identification of essential genes for cancer immunotherapy. Nature 548 (7669), 537–542. doi:10.1038/nature23477.
Patsoukis, N., Bardhan, K., Chatterjee, P., Sari, D., Liu, B., Bell, L. N., et al. (2015). PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692. doi:10.1038/ncomms7692
Pauken, K. E., Godec, J., Odorizzi, P. M., Brown, K. E., Yates, K. B., Ngiow, S. F., et al. (2020). The PD-1 pathway regulates development and function of memory CD8(+) T cells following respiratory viral infection. Cell Rep. 31 (13), 107827. doi:10.1016/j.celrep.2020.107827
Petherick, K. J., Conway, O. J. L., Mpamhanga, C., Osborne, S. A., Kamal, A., Saxty, B., et al. (2015). Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290 (18), 11376–11383. doi:10.1074/jbc.C114.627778
Poillet-Perez, L., Xie, X., Zhan, L., Yang, Y., Sharp, D. W., Hu, Z. S., et al. (2018). Autophagy maintains tumour growth through circulating arginine. Nature 563 (7732), 569–573. doi:10.1038/s41586-018-0697-7
Pua, H. H., Dzhagalov, I., Chuck, M., Mizushima, N., and He, Y. W. (2007). A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204 (1), 25–31. doi:10.1084/jem.20061303
Puleston, D. J., Zhang, H., Powell, T. J., Lipina, E., Sims, S., Panse, I., et al. (2014). Autophagy is a critical regulator of memory CD8(+) T cell formation. elife 3, e03706. doi:10.7554/eLife.03706
Qu, X., Yu, J., Bhagat, G., Furuya, N., Hibshoosh, H., Troxel, A., et al. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112 (12), 1809–1820. doi:10.1172/JCI20039
Rao, S., Tortola, L., Perlot, T., Wirnsberger, G., Novatchkova, M., Nitsch, R., et al. (2014). A dual role for autophagy in a murine model of lung cancer. Nat. Commun. 5, 3056. doi:10.1038/ncomms4056
Rebecca, V. W., and Amaravadi, R. K. (2016). Emerging strategies to effectively target autophagy in cancer. Oncogene 35 (1), 1–11. doi:10.1038/onc.2015.99
Rebecca, V. W., Nicastri, M. C., Fennelly, C., Chude, C. I., Barber-Rotenberg, J. S., Ronghe, A., et al. (2019). PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov. 9 (2), 220–229. doi:10.1158/2159-8290.CD-18-0706
Rebecca, V. W., Nicastri, M. C., McLaughlin, N., Fennelly, C., McAfee, Q., Ronghe, A., et al. (2017). A unified approach to targeting the Lysosome's degradative and growth signaling roles. Cancer Discov. 7 (11), 1266–1283. doi:10.1158/2159-8290.CD-17-0741
Ronan, B., Flamand, O., Vescovi, L., Dureuil, C., Durand, L., Fassy, F., et al. (2014). A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10 (12), 1013–1019. doi:10.1038/nchembio.1681
Rostislavleva, K., Soler, N., Ohashi, Y., Zhang, L., Pardon, E., Burke, J. E., et al. (2015). Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350 (6257), aac7365. doi:10.1126/science.aac7365
Sahai, E., Astsaturov, I., Cukierman, E., DeNardo, D. G., Egeblad, M., Evans, R. M., et al. (2020). A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 20 (3), 174–186. doi:10.1038/s41568-019-0238-1
Saitoh, T., Fujita, N., Jang, M. H., Uematsu, S., Yang, B. G., Satoh, T., et al. (2008). Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456 (7219), 264–268. doi:10.1038/nature07383
Samie, M., Lim, J., Verschueren, E., Baughman, J. M., Peng, I., Wong, A., et al. (2018). Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat. Immunol. 19 (3), 246–254. doi:10.1038/s41590-017-0042-6
Santana-Codina, N., Mancias, J. D., and Kimmelman, A. C. (2017). The role of autophagy in cancer. Annu. Rev. Cell Biol. 1, 19–39. doi:10.1146/annurev-cancerbio-041816-122338
Shoemaker, C. J., Huang, T. Q., Weir, N. R., Polyakov, N. J., Schultz, S. W., and Denic, V. (2019). CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 17 (4), e2007044. doi:10.1371/journal.pbio.2007044
Snyder, A. G., Hubbard, N. W., Messmer, M. N., Kofman, S. B., Hagan, C. E., Orozco, S. L., et al. (2019). Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci Immunol 4 (36), eaaw2004. doi:10.1126/sciimmunol.aaw2004
Sousa, C. M., Biancur, D. E., Wang, X., Halbrook, C. J., Sherman, M. H., Zhang, L., et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536 (7617), 479–483. doi:10.1038/nature19084
Starobinets, H., Ye, J., Broz, M., Barry, K., Goldsmith, J., Marsh, T., et al. (2016). Antitumor adaptive immunity remains intact following inhibition of autophagy and antimalarial treatment. J. Clin. Invest. 126 (12), 4417–4429. doi:10.1172/JCI85705
Stephenson, L. M., Miller, B. C., Ng, A., Eisenberg, J., Zhao, Z., Cadwell, K., et al. (2009). Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy 5 (5), 625–635. doi:10.4161/auto.5.5.8133
Stjepanovic, G., Baskaran, S., Lin, M. G., and Hurley, J. H. (2017). Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol. Cell 67 (3), 528–534. doi:10.1016/j.molcel.2017.07.003
Strohecker, A. M., Guo, J. Y., Karsli-Uzunbas, G., Price, S. M., Chen, G. J., Mathew, R., et al. (2013). Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 3 (11), 1272–1285. doi:10.1158/2159-8290.CD-13-0397
Sukumar, M., Liu, J., Ji, Y., Subramanian, M., Crompton, J. G., Yu, Z., et al. (2013). Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123 (10), 4479–4488. doi:10.1172/JCI69589
Takamura, A., Komatsu, M., Hara, T., Sakamoto, A., Kishi, C., Waguri, S., et al. (2011). Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25 (8), 795–800. doi:10.1101/gad.2016211
Terren, I., Orrantia, A., Vitalle, J., Zenarruzabeitia, O., and Borrego, F. (2019). NK cell metabolism and tumor microenvironment. Front. Immunol. 10, 2278. doi:10.3389/fimmu.2019.02278
Towers, C. G., Fitzwalter, B. E., Regan, D., Goodspeed, A., Morgan, M. J., Liu, C. W., et al. (2019). Cancer cells upregulate NRF2 signaling to adapt to autophagy inhibition. Dev. Cell 50 (6), 690–703. doi:10.1016/j.devcel.2019.07.010
Towers, C. G., Wodetzki, D., and Thorburn, A. (2020). Autophagy and cancer: modulation of cell death pathways and cancer cell adaptations. J. Cell Biol. 219 (1), e201909033. doi:10.1083/jcb.201909033
Trentesaux, C., Fraudeau, M., Pitasi, C. L., Lemarchand, J., Jacques, S., Duche, A., et al. (2020). Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proc. Natl. Acad. Sci. U.S.A. 117 (20), 11136–11146. doi:10.1073/pnas.1917174117
Umemura, A., He, F., Taniguchi, K., Nakagawa, H., Yamachika, S., Font-Burgada, J., et al. (2016). p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell 29 (6), 935–948. doi:10.1016/j.ccell.2016.04.006
Valencia, T., Kim, J. Y., Abu-Baker, S., Moscat-Pardos, J., Ahn, C. S., Reina-Campos, M., et al. (2014). Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell 26 (1), 121–135. doi:10.1016/j.ccr.2014.05.004
Verma, V., Shrimali, R. K., Ahmad, S., Dai, W., Wang, H., Lu, S., et al. (2019). PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1(+)CD38(hi) cells and anti-PD-1 resistance. Nat. Immunol. 20 (9), 1231–1243. doi:10.1038/s41590-019-0441-y
Wang, B., Iyengar, R., Li-Harms, X., Joo, J. H., Wright, C., Lavado, A., et al. (2018). The autophagy-inducing kinases, ULK1 and ULK2, regulate axon guidance in the developing mouse forebrain via a noncanonical pathway. Autophagy 14 (5), 796–811. doi:10.1080/15548627.2017.1386820
Wang, B., Maxwell, B. A., Joo, J. H., Gwon, Y., Messing, J., Mishra, A., et al. (2019). ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol. Cell 74 (4), 742–757 e748. doi:10.1016/j.molcel.2019.03.027
Wang, Q., Wang, Y., Ding, J., Wang, C., Zhou, X., Gao, W., et al. (2020). A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature 579 (7799), 421–426. doi:10.1038/s41586-020-2079-1
Wang, Y. T., Zaitsev, K., Lu, Q., Li, S., Schaiff, W. T., Kim, K. W., et al. (2020). Select autophagy genes maintain quiescence of tissue-resident macrophages and increase susceptibility to Listeria monocytogenes. Nat Microbiol 5 (2), 272–281. doi:10.1038/s41564-019-0633-0
Wei, H., Wei, S., Gan, B., Peng, X., Zou, W., and Guan, J. L. (2011). Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 25 (14), 1510–1527. doi:10.1101/gad.2051011.
Wei, J., Long, L., Yang, K., Guy, C., Shrestha, S., Chen, Z., et al. (2016). Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 17 (3), 277–285. doi:10.1038/ni.3365
Xie, X., Koh, J. Y., Price, S., White, E., and Mehnert, J. M. (2015). Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov. 5 (4), 410–423. doi:10.1158/2159-8290.CD-14-1473
Xu, X., Araki, K., Li, S., Han, J. H., Ye, L., Tan, W. G., et al. (2014). Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 15 (12), 1152–1161. doi:10.1038/ni.3025
Yamamoto, K., Venida, A., Yano, J., Biancur, D. E., Kakiuchi, M., Gupta, S., et al. (2020). Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105. doi:10.1038/s41586-020-2229-5
Yang, A., Rajeshkumar, N. V., Wang, X., Yabuuchi, S., Alexander, B. M., Chu, G. C., et al. (2014). Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 4 (8), 905–913. doi:10.1158/2159-8290.CD-14-0362
Yang, S., Wang, X., Contino, G., Liesa, M., Sahin, E., Ying, H., et al. (2011). Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25 (7), 717–729. doi:10.1101/gad.2016111
Yatim, N., Jusforgues-Saklani, H., Orozco, S., Schulz, O., Barreira da Silva, R., Reis e Sousa, C., et al. (2015). RIPK1 and NF-kappaB signaling in dying cells determines cross-priming of CD8(+) T cells. Science 350 (6258), 328–334. doi:10.1126/science.aad0395
Yue, Z., Jin, S., Yang, C., Levine, A. J., and Heintz, N. (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100 (25), 15077–15082. doi:10.1073/pnas.2436255100
Zaffagnini, G., and Martens, S. (2016). Mechanisms of selective autophagy. J. Mol. Biol. 428 (9 Pt A), 1714–1724. doi:10.1016/j.jmb.2016.02.004
Zaretsky, J. M., Garcia-Diaz, A., Shin, D. S., Escuin-Ordinas, H., Hugo, W., Hu-Lieskovan, S., et al. (2016). Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375 (9), 819–829. doi:10.1056/NEJMoa1604958
Zhang, L., and Romero, P. (2018). Metabolic control of CD8(+) T cell fate decisions and antitumor immunity. Trends Mol. Med. 24 (1), 30–48. doi:10.1016/j.molmed.2017.11.005
Zhang, M., Kenny, S. J., Ge, L., Xu, K., and Schekman, R. (2015). Translocation of interleukin-1 beta into a vesicle intermediate in autophagy-mediated secretion. elife 4, e11205. doi:A10.7554/eLife.11205.
Zhang, Z., Zhang, Y., Xia, S., Kong, Q., Li, S., Liu, X., et al. (2020). Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature 579 (7799), 415–420. doi:10.1038/s41586-020-2071-9
Keywords: autoph, agy, immunotherapy, cancer, oncology, immunology
Citation: Lim J and Murthy A (2020) Targeting Autophagy to Treat Cancer: Challenges and Opportunities. Front. Pharmacol. 11:590344. doi: 10.3389/fphar.2020.590344
Received: 01 August 2020; Accepted: 09 October 2020;
Published: 23 November 2020.
Edited by:
Marco Tafani, Sapienza University of Rome, ItalyReviewed by:
Inmaculada Martinez-Reyes, Helmholtz Association of German Research Centers (HZ), GermanyPaola Maycotte, National Council of Science and Technology (CONACYT), Mexico
Copyright © 2020 Lim and Murthy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aditya Murthy, bXVydGh5LmFkaXR5YUBnZW5lLmNvbQ==
 Junghyun Lim
Junghyun Lim Aditya Murthy
Aditya Murthy