
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Pharmacol. , 09 December 2020
Sec. Pharmacogenetics and Pharmacogenomics
Volume 11 - 2020 | https://doi.org/10.3389/fphar.2020.587451
This article is part of the Research Topic Coronavirus Disease (COVID-19): Molecular Mechanisms, Translational Approaches and Therapeutics View all 118 articles
 Mohamed Abu-Farha1†
Mohamed Abu-Farha1† Salman Al-Sabah2
Salman Al-Sabah2 Maha M. Hammad1
Maha M. Hammad1 Prashantha Hebbar3
Prashantha Hebbar3 Arshad Mohamed Channanath3
Arshad Mohamed Channanath3 Sumi Elsa John3
Sumi Elsa John3 Ibrahim Taher4
Ibrahim Taher4 Abdulrahman Almaeen4
Abdulrahman Almaeen4 Amany Ghazy4,5
Amany Ghazy4,5 Anwar Mohammad1
Anwar Mohammad1 Jehad Abubaker1*
Jehad Abubaker1* Hossein Arefanian6
Hossein Arefanian6 Fahd Al-Mulla3*†
Fahd Al-Mulla3*† Thangavel Alphonse Thanaraj3*†
Thangavel Alphonse Thanaraj3*†COVID-19 is caused by Severe Acute Respiratory Syndrome Coronavirus-2, which has infected over thirty eight million individuals worldwide. Emerging evidence indicates that COVID-19 patients are at a high risk of developing coagulopathy and thrombosis, conditions that elevate levels of D-dimer. It is believed that homocysteine, an amino acid that plays a crucial role in coagulation, may also contribute to these conditions. At present, multiple genes are implicated in the development of these disorders. For example, single-nucleotide polymorphisms (SNPs) in FGG, FGA, and F5 mediate increases in D-dimer and SNPs in ABO, CBS, CPS1 and MTHFR mediate differences in homocysteine levels, and SNPs in TDAG8 associate with Heparin-induced Thrombocytopenia. In this study, we aimed to uncover the genetic basis of the above conditions by examining genome-wide associations and tissue-specific gene expression to build a molecular network. Based on gene ontology, we annotated various SNPs with five ancestral terms: pulmonary embolism, venous thromboembolism, vascular diseases, cerebrovascular disorders, and stroke. The gene-gene interaction network revealed three clusters that each contained hallmark genes for D-dimer/fibrinogen levels, homocysteine levels, and arterial/venous thromboembolism with F2 and F5 acting as connecting nodes. We propose that genotyping COVID-19 patients for SNPs examined in this study will help identify those at greatest risk of complications linked to thrombosis.
The COVID-19 outbreak, which began in China’s Hubei Province, was declared a pandemic by the World Health Organization on March 11, 2020. One of the most challenging features of COVID-19 is its outcome variability, as some afflicted are asymptomatic, while others experience mild-to-severe flu-like symptoms or even death (Bai et al., 2020; Day, 2020; Kimball et al., 2020; Mizumoto et al., 2020; Sakurai et al., 2020; Zhou et al., 2020). In consideration of the dramatic rise in the number of infected people and limited health resources worldwide, it would be especially advantageous to identify which patients will be most in need of critical care. Notably, the clinical status and presence of concurrent comorbidities in COVID-19 patients are good indicators of an individual’s prognosis. In addition, assessing blood levels of primarily the coagulation factors (D-dimer and fibrinogen) and homocysteine may be informative (Velavan and Meyer, 2020).
Initially, pneumonia and shortness of breath were believed to be the primary causes of death in COVID-19 patients, yet growing research reveals that thrombosis, a consequence of the deterioration of coagulation factors, may actually be the leading offender (Becker, 2020; Bikdeli et al., 2020; Levi et al., 2020; Tang et al., 2020; Yuki et al., 2020; Zhou et al., 2020). The incidence of thrombotic events in COVID-19 patients admitted to the intensive care unit can be as high as 31% (venous thromboembolic events are the most common) (Abou-Ismail et al., 2020; Klok et al., 2020). Numerous studies are presently investigating the pathophysiology of coagulopathy specific to COVID-19, so as to derive mechanistic insights that can direct interventions (Abou-Ismail et al., 2020). Elevated levels of D-dimer represent the initial manifestation of coagulopathy in critically ill patients, followed by abnormal prothrombin and partial thromboplastin times, and finally low platelet counts in later stages (Connors and Levy, 2020; Iba et al., 2020).
D-dimer is a fibrin degradation product (FDP), otherwise known as a protein fragment that circulates in the bloodstream after a clot is broken down by fibrinolysis. Clinically speaking, D-dimer levels are evaluated when monitoring a patient’s response to anticoagulants; a decrease is illustrative of the therapy’s efficacy. Interestingly, D-dimer levels will stay elevated in patients taking anticoagulants and suffering from conditions such as systemic lupus erythematosus, obesity or various infections (Greenberg, 2017) because they tend to trigger extravascular fibrin formation or prevent the anticoagulant from suppressing fibrin formation. Studies have found that D-dimer levels associate with a heightened risk of coronary heart, cardiovascular, and peripheral arterial diseases, as well as acute ischemic stroke, venous thrombosis, and atherosclerosis (Folsom et al., 2016; Soomro et al., 2016).
In this study, we focus on D-dimer levels as a biomarker for coagulopathy in COVID-19 patients. Second, we discuss homocysteine levels and thrombocytopenia, because the former correlates with D-dimer levels and the latter is induced by heparin when treating coagulopathy in COVID-19 patients. For all biomarkers, their molecular genetic basis and contribution to coagulopathy-related complications are examined. Further, we present differences in allele frequencies for associated genetic risk variants across continental populations.
We carried out a literature search using Medline (PubMed) for published studies using terms such as D-dimer, homocysteine, heparin and embolism. We chose search terms subjectively and iteratively. We also checked the reference lists of potentially relevant articles for additional citations and used the “related citations” search key in PubMed to identify similar papers.
Established genotype-trait association signals identified through global genome-wide association studies (GWAS) are available from the NHGRI-EBI GWAS Catalog (Buniello et al., 2019). Genetic variants and genes associated with traits and disorders discussed in this study were obtained by querying the GWAS Catalog available at https://www.ebi.ac.uk/gwas/. In order to identify genetic variants that regulate genes of interest in tissue-specific manner, we examined genotype-tissue expression data using GTEX v8 (https://www.gtexportal.org). GTEX is a comprehensive public resource to study tissue-specific gene expression and regulation. The GTEx Portal provides open access to data including gene expression, QTLs, and histology images.
As regards querying the GWAS Catalog, selection criteria was based on the P-values denoting the significance for association; associations at genome-wide significance (P< 5.0E-08) were given preference; the P-value threshold was relaxed when only few associations were seen at genome-wide significance. As regards querying the GTEX, selection criteria was based on significant P-values and Q-values.
Allele frequencies at genetic variants in continental populations were extracted from 1000 Genomes Project Phase 3 (Auton et al., 2015) data available through Ensembl genome browser (http://www.ensembl.org). As regards Middle East populations, genetic variants data published previously on Kuwaitis (Hebbar et al., 2020; John et al., 2018) (available from our in-house databases), Qataris (Fakhro et al., 2016) (as available from http://clingen.igib.res.in/almena/) and Iranians (Fattahi et al., 2019) (as available from http://www.iranome.ir/) were used.
The eXploring Genomic Relations (XGR) tool (Fang et al., 2016) was used to derive gene-gene interaction networks based on the list of GWAS-annotated study variants and other GWAS-annotated SNPs that are in Linkage Disequilibrium (LD) with the study variants. XGR uses databases on functional annotation such as STRINGs (Fang et al., 2016) presenting protein-protein interaction networks and Reactome or PathwayCommons (Cerami et al., 2011) presenting biological pathways to identify an optimal gene network that is likely to be modulated by input SNPs and their LD SNPs from a genome region of specified length around the study variants. To do this, XGR first defines and scores genes that are likely under the genetic influence of the study GWAS SNPs by way of performing the following three steps: (a) scoring the study SNPs (and their GWAS LD SNPs) by the p-values for association with the GWAS trait; (b) scoring the genes based on genomic proximity to quantify their genetic modulation by the study SNPs and their LD SNPs; and (c) scoring the networks to identify a maximum-scoring gene subnetwork. The resultant gene subnetwork of functional interactions with high confidence score >=700 from the study variants was considered in our study. The resources of GWAS Catalog and GTEX and the XGR tool was last accessed on 17th June 2020.
The trait names as used in GWAS Catalog are standardized using well-defined Experimental Factor Ontology (EFO) terms in the contexts of Disease, Phenotype and Biological Processes (Welter et al., 2014). SNPs associated with one or more related traits grouped together are annotated by such an EFO term. A SNP mapped to an EFO term can also be annotated by its parental ancestor term. The XGR tool (Fang et al., 2016) considers additional SNPs that are in strong LD (r2 > 0.8) with a given list of study variants and performs a hypergeometric test. It thereby outputs a list of the parental terms with FDR (false discovery rate) values to which the study variants are associated with.
Various studies have reported that elevated D-dimer levels (> 2 ug/mL) can increase the risk of fatality in COVID-19 adult patients (Levi et al., 2020; Zhou et al., 2020; Tang et al., 2020; Leonard-Lorant et al., 2020; Zhang et al., 2020; Cui et al., 2020; Chen et al., 2020; Gao et al., 2020; Wang et al., 2020; Chen et al., 2020; Huang et al., 2020) (Table 1). Additionally, high levels have been observed during advanced stages of other viral infections, such as HIV and Ebola (Lippi and Favaloro, 2020). Infection by SARS-CoV-2, in particular, results in a cytokine storm that injures endothelial cells and leads to a massive release of plasminogen activators. It is possible that this phenomenon is the reason why COVID-19 patients with severe outcomes display significantly elevated levels of D-dimer and FDPs. For children infected with SARS-CoV-2, a two to three-fold increase in D-dimer levels has been observed as the disease progressed from mild to moderate (Qiu et al., 2020), and those with obesity are presumed to be at highest risk of complications from this virus because obese children are reported to have higher D-dimer levels. Therefore, in addition to comorbidities such as diabetes, cardiovascular disease, hypertension and lipid disorders (Abu-Farha et al., 2020; Bosso et al., 2020), increased D-dimer levels are a risk factor for death in COVID-19 patients. As such, D-dimer levels should be carefully monitored throughout treatment.
Global genome-wide association studies (GWAS), as deposited in the GWAS Catalog (Buniello et al., 2019), report that approximately 16 genetic variants associate with D-dimer levels in healthy individuals. Of these, three non-coding single-nucleotide polymorphisms (SNPs) are linked to high levels of D-dimer in healthy individuals: rs13109457-A [an intergenic variant between fibrinogen gamma and alpha (FGG and FGA), effect size is 6.3%], rs12029080-G (an intronic variant from an lncRNA novel transcript, AC093117.1, effect size is 10.4%), and rs6687813-A (an intergenic variant between F5 and Z99572.1, effect size is 10.1%) (Supplementary Table S1A). Of note: the remaining 13 SNPs are not discussed here because they carry an effect size of < 0.1%.
Importantly, the three aforementioned SNPs are all common variants (i.e., their minor allele frequency is > 5%), but their frequency differs substantially across continental populations (Supplementary Table S1A and Supplementary Figure S1). For example, rs13109457-A occurs at 43% frequency in East Asia, but this value is halved across the Middle East; rs12029080-G occurs at 7% frequency in Africa, and this value is more than tripled for all other populations; rs6687813-A occurs at 0% frequency in East Asia, but is > 5% in all other populations; rs17031315_T, which downregulates expression of FGA and FGG, occurs at 12% frequency in Africans and ad-mixed Americans, but is quite rare in other populations. These variations may align with differences in COVID-19 severity across populations.
Furthermore, the genes that harbor the three top SNPs associate with disorders (some are often comorbid in COVID-19 patients) and traits relating to coagulation. An examination of the GWAS Catalog revealed that variants of FGG associate with thromboembolism and glycine and fibrinogen levels; variants of FGA associate with stroke, thromboembolism, and fibrinogen levels; variants of AC093117.1 associate with type 2 diabetes and end-stage coagulation; variants of F5 associate with ischemic stroke, thromboembolism, peripheral artery disease, and uric acid and cytokine network levels; variants of Z99572.1 associate with the coagulants, Factors II and V.
Upon examining Genotype-Tissue Expression (GTEx) data (https://www.gtexportal.org), we found that the above SNPs regulate genes involved in the production of proteins necessary for coagulation, or linked to disorders displaying elevations of D-dimer. In specific, rs13109457 (FGG, FGA) regulates PLRG1 (associated with fibrinogen) and TLR2 (associated with body fat); rs12029080 regulates F3 (a coagulant); rs6687813 (F5, Z99572.1) regulates SLC19A2 (associated with the heart’s QT interval), NME7 (associated with the QT interval, thromboembolism, blood pressure, and coronary artery disease), and METTL18 (associated with amnesia and hemorrhage).
Since FGA, FGG, and F5 are expressed in the liver, we also searched for expression quantitative trait loci (eQTLs) regulating expression of these genes in liver. GTEx identified rs17031315-A_T, an intronic variant of DCHS2, as downregulating FGG and FGA via an effect size of 1.7 units (Supplementary Table S1A). DCHS2 participates in homophilic cell adhesion via plasma membrane adhesion molecules and is related to cytokine, bilirubin, and fibrinogen counts, as well as disorders comorbid with COVID-19 (i.e., velopharyngeal dysfunction, venous thromboembolism, liver disease, jaundice, anemia, and thyroid disorders).
Another clinical parameter showing potential significance in COVID-19 patients is homocysteine levels (Ponti et al., 2020; Yang et al., 2020), as research reveals homocysteine levels associate positively with venous and pulmonary thromboembolism (r = 0.368 and r = 0.285, respectively) (Previtali et al., 2011). In an investigation by Todua et al. (Todua et al., 2017), analyses uncovered a correlation coefficient of 0.557 between the levels of D-dimer and homocysteine in patients suffering from conditions of pulmonary arterial thromboembolism.
The GWAS Catalog lists 48 signals involving 44 SNPs from 34 genetic loci (including genes for von Willebrand factor and NADPH oxidase 4) associated with homocysteine levels. The top six signals with an effect size > 0.15 units are from CBS, ABO, CPS1, MTHFR, (RPL24P4 and GNMT), and (ALDH1L1 and ALDH1L1-AS2) (Supplementary Table S1A). Recently, a GWAS on an Italian and Spanish cohort of COVID-19 patients showed that the ABO blood group locus, from the above list, is associated with respiratory failure (Ellinghaus et al., 2020; Ellinghaus et al., 2020). Moreover, a SNP in CPS1 appears to be linked to a heightened risk of persistent pulmonary hypertension in newborns (Pearson et al., 2001), and two missense variants [rs1801133_G > A (A222V) and rs1801131_T > G (E429A)] in MTFHR have been found to impact homocysteine pathways (Brustolin et al., 2010). These top association signals vary in allele frequencies across populations (Supplementary Table S1 and Supplementary Figure S1). As such, examining variants in these genes among COVID-19 patients will provide insight into their risk of clots, heart disease, stroke, and hypertension.
Thrombocytopenia, a condition characterized by low platelet counts, is a common complication in COVID-19 patients and associates with an increased risk of in‐hospital mortality (Lippi et al., 2020; Yang et al., 2020). There is currently limited evidence on how it arises following infection by SARS-CoV-2 (Xu et al., 2020), although one study found thrombocytopenia was induced by the medications, Pegylated interferon and Ribavirin (Tanaka et al., 2011).
COVID-19 is tied to an increased incidence of thrombotic and inflammatory events, which contribute to its severe morbidity and mortality. Heparin appears to be the preferred anticoagulant for COVID-19 treatment because in addition to its ability to inhibit the generation of thrombin, it is anti-inflammatory (Thachil, 2020). In a study involving 449 COVID-19 patients, no difference in 28-day mortality was found between the 99 that received heparin and those who did not (Tang et al., 2020). However, a difference in this outcome variable was observed between heparin users and non-users when researchers assessed patients with high D-dimer levels and sepsis-induced coagulopathy, which suggests that not all COVID-19 patients will benefit from anticoagulants. The study recommended that anticoagulants should only be given to COVID-19 patients with elevated D-dimer levels.
The GWAS Catalog lists ten variants from ZNF84 and AC106799.2 (an lncRNA) that associate with HIT (Supplementary Table S1A). Karnes et al. (Karnes et al., 2015) observed additional associations from SNPs near TDAG8, which governs production of the G-protein coupled receptor, TDAG8, to induce T-cell death (Choi et al., 1996) and attenuate immune-mediated inflammation (Onozawa et al., 2011; Onozawa et al., 2012). For the majority of these variants, allele frequencies show considerable variation across global populations (Supplementary Table S1 and Supplementary Figure S1).
Transaldolase 1 (TALDO1) is a rate-limiting metabolic enzyme involved in the nonoxidative pentose phosphate pathway, providing ribose-5-phosphate and NADPH for nucleic acid and lipid biosynthesis, respectively. It is believed that malfunctions in TALDO1 are due to mutations in TALDO1 such as R192C observed in patients from United Arab Emirates (Al-Shamsi et al., 2015). TALDO1-deficient patients present various clinical problems, including coagulopathy, thrombocytopenia, hepatosplenomegaly, anemia, hepatic fibrosis, and liver and renal tubular dysfunction (Loeffen et al., 2012; Banne et al., 2016). Progressive coagulopathy is a sensitive parameter indicating liver dysfunction in patients with TALDO deficiency (Lipiński et al., 2018).
From our research, we observed an interesting link between TALDO1 and interferon‐induced transmembrane 3 (IFITM3) in the COVID-19 pathway. IFITM3 produces a protein that blocks viruses from fusing with cellular membranes (Thevarajan et al., 2020). The rs12252-C variant of IFITM3 has been linked to severe influenza (Everitt et al., 2012; Wang et al., 2014) and has been observed in a patient with COVID-19 (Thevarajan et al., 2020). GTEx data indicates that rs77216754, an intergenic variant located between TALDO1 and PDDC1, downregulates IFITM3 and upregulates TALDO1. Collectively, this information points to a relationship between TALDO1 and COVID-19 in the context of coagulopathy. Of note, the minor allele frequency of rs77216754, an eQTL variant, is highest in African populations (32.5%) compared to the rest of the world (4 to 9%) (Supplementary Table S1A and Supplementary Figure S1).
Table 2 lists studies that examined the occurrence of thromboembolic complications in COVID-19 patients. Their cohorts differed by presence of pre-existing conditions, types and amounts of medications that were administered, severity of COVID-19, age, and sex. However, all experienced a high occurrence of VTE and pulmonary thrombosis.
The GWAS Catalog lists 13 signals that associate with PE and deep vein thrombosis. These signals correspond to SNPs in ABO, AL357518, COX7A2L, F2, F5, F11-AS1, FUNDC2, (GAPDHP50, ICE2P2), KNG1, (LRAT, FGG), PROCR, SLC44A2, and TSPAN15. Moreover, for signals that associate with VTE, the GWAS Catalog lists 14 signals that are of genome-wide significance and display an odds ratio of ≥1.75. In specific, these signals derive from ABO, AC022182, AC073115, (AP005230, AP005262), (CD, AL118508), F2, F5, LEMD3, LRP4, LY86, NME7, and (THBD, AL118508).
Like the cases of variants associated with the previously mentioned disorders, the top association signals for PE and VTE vary in allele frequency across populations (Supplementary Table S1A and Supplementary Figure S1). For example, rs60942712-T and rs17490626-G that associate with PE occur at > 10% frequency among Europeans, but are absent in East Asians. Additionally, rs73692310-T and rs28496996-G that associate with VTE occur at a frequency between 7 and 10% in African populations but are non-existent in other continental populations.
The eXploring Genomic Relations (XGR) tool (Fang et al., 2016) annotated the study variants with five ancestral terms: PE, VTE, vascular diseases, cerebrovascular disorders, and stroke (Supplementary Table S1B); the parental terms of PE, stroke and cerebrovascular disorders were associated with intergenic region of (AL357518.2, AL357518.1), FUNDC2, TSPAN15, F2, PROCR, F11-AS1, ABO, F5, intergenic region of (GAPDHP50 and ICE2P2), KNG1, COX7A2L, intergenic region of (LRAT, FGG), and SLC44A2; those of VTE and vascular diseases were associated with the above gene loci and additionally with LEMD3, AC022182.2, NME7, intergenic region of (CD93, AL118508.3), intergenic region of (THBD, AL118508.3), intergenic region of (AP005230.1,AP005262.1), LY86 and AC073115.2. Moreover, it identified a maximum-scoring gene regulatory network based on the linkage disequilibrium (LD) of the SNPs from a region of 0.05 MB (Supplementary Table S2 and Supplementary Figure S2). The network presents three clusters of genes, each containing hallmark genes involved in the regulation of D-dimer/fibrinogen, homocysteine levels and arterial/venous thromboembolism disorders. Particularly, genes that interact to influence D-dimer and fibrinogen levels include F5, FGA, FGB, and FGG; homocysteine levels are influenced by CBS and MTHFR; and arterial/venous thromboembolism disorders are influenced by F2, F3, F8, F11, CHRM4, KLKB1, CD93, THBD, and KNG1. Interestingly, F2 and F5 were discovered to be connecting nodes between these three clusters, although an obesity-related gene (IGFBP3) and a muscular dystrophy-related gene (ADCY1) were found to play prominent roles as well. As the region exploring LD becomes enlarged to 0.5 MB (Supplementary Table S3 and Supplementary Figure S3), the network brings additional genes into clusters that help govern D-dimer and fibrinogen counts, thrombosis, and obesity-related traits. Furthermore, genes relating to the following clinical markers for thrombosis also appeared: Partial thromboplastin time, Blood pressure/pulse pressure, Lipids, Eosinophil and blood cell counts, Cell adhesion measurements, Ubiquitination and degradation and Brain volume.
The data presented so far in this review enabled us to establish potential molecular link connecting some of the key genes in the coagulation process. This includes association of FGA, FGG and F5 with D-dimer levels, MTHFR with homocysteine levels and F2, F3, F8, F11, KLKB1 and KNG1 with thromboembolism. Figure 1 summarizes how these key genes could be linked to the coagulation process when a patient is infected with SARS-CoV-2. The coronavirus infection results in an injury to the endothelial cells leading to the activation of the coagulation cascade; both the intrinsic as well as the extrinsic pathway. Within the extrinsic pathway, tissue factor (encoded by F3) initiates the cascade to activate the common pathway of coagulation. Within the intrinsic pathway, which is also known as kallikrein/kinin system, pre-kallikrein is converted to plasma kallikrein (encoded by KLKB1 gene) via the action of kininogen (encoded by KNG1 gene) (Loof et al., 2014). This then results in the activation of Factor XI (encoded by F11) which initiates the activation of the common pathway of coagulation. The common pathway consists of several coagulation factors that are activated in parallel to eventually convert prothrombin (encoded by F2) to thrombin (Loof et al., 2014).
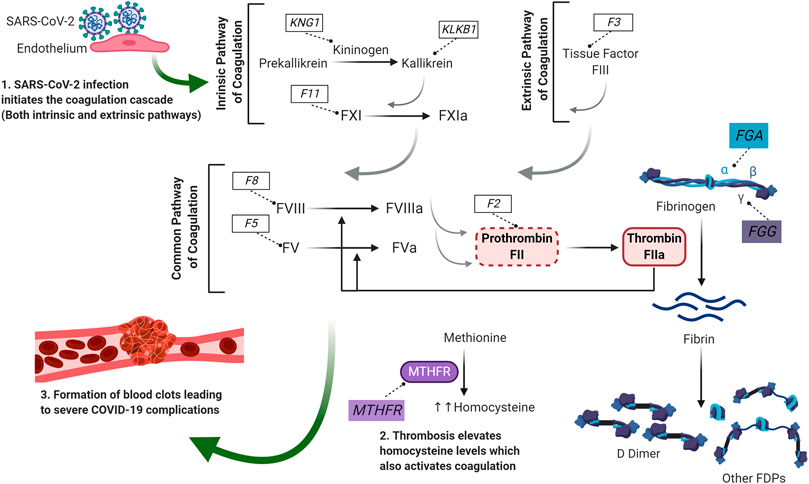
FIGURE 1. The molecular link between key genes in coagulation during COVID-19 infection. SARS-CoV-2 infection results in an injury in the endothelium which leads to the activation of the coagulation cascade; both the intrinsic and the extrinsic pathway. Within the extrinsic pathway, tissue factor (encoded by F3) initiates the cascade to activate the common pathway of coagulation. Within the intrinsic pathway, prekallikrein is converted to kallikrein via the action of kininogen and factor XI is activated. The activation of Factor XI initiates the common pathway of coagulation and leads to the activation of several coagulation factors including factor VIII and factor V. Thrombin converts Fibrinogen to Fibrin. Fibrin is then broken down to D-dimer and other FDPs. This SARS-CoV-2-induced thrombosis also leads to elevated levels of homocysteine which can eventually activate the coagulation cascade. Genes of interest are presented in boxes. For purpose of clarity, this illustration is only highlighting the genes of interest that were picked by XGR analysis to associate with thrombosis. Grey arrows indicate activation of other coagulation factors. F: Coagulation Factor, Fa: activated coagulation factor. FDPs: Fibrin degradation products, FGA: Fibrinogen alpha chain, FGG: Fibrinogen gamma chain, KLKB1: kallikrein, KNG1: Kininogen 1, MTHFR: methylenetetrahydrofolate reductase.
Thrombin plays an essential role in coagulation since it can activate the coagulation factors in the common pathway including FVIII (encoded by F8) and FV (encoded by F5) as well as convert fibrinogen to fibrin. Fibrinogen is a glycoprotein composed of three different polypeptide chains; alpha, beta and gamma (Mosesson et al., 2001; Undas and Ariëns, 2011). While FGA gene encodes the fibrinogen alpha chain (α chain), FGG gene encodes the fibrinogen gamma chain (γ chain). Fibrinolysis is afterwards initiated where fibrin is broken down to multiple fibrin degradation products (FDPs), one of which is D-dimer (Weisel and Litvinov, 2017). This thrombosis that is caused by the coronavirus infection can also lead to elevated levels of homocysteine which is synthesized from methionine by methylenetetrahydrofolate reductase (MTHFR). Elevated homocysteine levels could also activate the coagulation cascade resulting in D-dimer production. Eventually, all these elements collectively result in the formation of blood clots leading to severe complications (including death) that were documented in some COVID-19 patients.
There is emerging evidence that coagulopathy and thrombosis are a common finding besides pneumonia in patients suffering from COVID-19, especially in severe courses. Thus, any strategy carried out to predict the development of such conditions is relevant for improving the clinical approach to this viral infection. This study proposes a gene-gene interaction network with three clusters of hallmark genes which have an influence for D-dimer/fibrinogen levels, homocysteine levels, and arterial/venous thromboembolism. The study further hypothesizes that genotyping COVID-19 patients for SNPs in these genes helps not only to identify patients with greatest risk of thromboembolic complications but also the outcome of patients treated with heparin according to their genotypes.
Our knowledge about COVID-19 continues to expand as research groups dedicate tremendous efforts to unraveling the mysteries of this viral infection. At present, thrombosis is emerging as a common outcome that can turn fatal. Measuring D-dimer levels is routinely done to assess thrombosis, and elevated levels are frequently observed in severely afflicted COVID-19 patients. For this reason, they could be considered as an early biomarker and predictor of in-hospital mortality. The top three genetic risk variants (rs13109457-A, rs12029080-G, and rs6687813-A) that associate with significant increases in D-dimer levels are located in FGG, FGA, and F5. The data presented in this review and the cited studies from literature enabled us to establish the molecular connection between some of the key genes (FGA, FGG, F5, MTHFR, KLKB1, KNG1, F2, F8 and F11) and D-dimer and homocysteine levels during coagulation (Figure 1 and Supplementary Figure S2).
The intronic variant, rs17031315, from DCHS2 has been shown to downregulate FGG and FGA in the liver and GWA studies have tied DCHS2 to various disorders that are often comorbid with COVID-19. At least ten variants from ZNF84 and an lncRNA, as well as SNPs in TDAG8, associate with HIT. The eQTL variant, rs77216754, downregulates IFITM3 and upregulates TALDO1, and thereby connects with COVID-19 in the context of coagulopathy and thrombocytopenia. Levels of homocysteine correlate with those of D-dimer, influencing coagulation and thereby thrombosis. At least six variants that mediate > 0.15 unit changes in homocysteine levels have been identified in ABO, CBS, CPS1, MTHFR, (RPL24P4 and GNMT) and (ALDH1L1 and ALDH1L1-AS2). Of these, the ABO locus has recently been found to associate with respiratory failure in Italian and Spanish COVID-19 patients. We illustrate the differences in allele frequencies for key variants across continental populations.
SARS-CoV-2 infections are associated with cardiovascular diseases (CVDs) in multiple ways. Thrombosis is a common underlying pathology of the three major cardiovascular disorders: ischemic heart disease (acute coronary syndrome), stroke, and venous thromboembolism (VTE). The focus of the current review was to identify genetic variants that could predict COVID-19 patients at high risk of developing cardiovascular disorder of VTE. Apart from the markers of D-Dimer and homocysteine levels discussed in this study, other cardiovascular markers such as troponin levels appear to associate with the severity of COVID-19 and the development of different CVDs (Driggin et al., 2020; Lippi et al., 2020).
A recent state-of-art review by Bikdeli et al. (Bikdeli et al., 2020) emphasizes the need for high-quality multi-disciplinary data to understand how thrombotic disease and COVID-19 interact with one another. The authors rightly point out that elucidation of disease presentation/outcomes in COVID-19 patients and pre-existing or incident thromboembolic disorders will help in developing treatment strategies for thromboembolism in COVID-19 patients. Large-scale registries of patients with thromboembolic disorders, such as RIETE (Bikdeli et al., 2018) coordinated by S&H Medical Science Service in Spain, ACS COVID-19 Registry (that captures clinical data in COVID-19 patients) by American College of Surgeons (https://www.facs.org/quality-programs/covid19-registry) and AHA COVID-19 CVD registry (that captures cardiovascular data) by American Heart Association are being created. Similar large-scale efforts towards building registries collecting data on the genetic basis of interaction of thrombotic disease and COVID-19 with one another are in order. As a contribution towards this effort, we propose a set of SNPs for interested researchers and clinicians to genotype in COVID-19 patients to identify those with greatest risk of thromboembolic complications. The above-cited state-of-art review by Bikdeli et al. (Bikdeli et al., 2020) further discusses the importance of considering the drug-drug interactions between anti-coagulants and the investigational therapies newly developed for COVID-19 treatment and presents a potential algorithm to be used when making treatment decisions. In this context, the SNPs that we report to genotype in COVID-19 patients to identify the outcome of heparin-induced thrompocytopenia in patients treated with heparin according to their genotypes are meaningful.
Absence of access to genotype data for the variants reported in this review from COVID-19 patients is a limitation of the study. Thus, differences in allele frequencies of the risk variants could not be evaluated in COVID-19 patients versus healthy people nor between COVID-19 patients with thromboembolic complications versus less severe COVID-19 patients. However, the review presents SNPs, by way of proposing a gene-gene interaction network involving three clusters of hallmark genes which have influence for D-dimer/fibrinogen levels, homocysteine levels and arterial/venous thromboembolism, for genotyping in COVID-19 patients to identify those with greatest risk of thromboembolic complications.
In conclusion, it is likely that different populations will respond uniquely to SARS-CoV-2, due, at least partially, to genetic variations. In this analysis, we identified a list of SNPs and genes (Supplementary Tables S1, S3) that can be used as biomarkers to predict the development of coagulation. Furthermore, a gene-gene interaction network constructed using the study’s variants and their LD SNPs uncovered three clusters of genes involved in the regulation of D-dimer, fibrinogen, and homocysteine levels and arterial/venous thromboembolism disorders. Upon widening the considered genome region for LD SNPs to 0.5 MB, the optimal genetic network accrued genes that help contribute to obesity-related traits, partial thromboplastin time, blood pressure, lipid generation, white and red blood cell counts, cellular adhesion properties, ubiquitination, cellular degradation, and brain volume. We propose that genotyping COVID-19 patients for SNPs examined in this study will help identify those at greatest risk of complications linked to thrombosis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This research was funded by Kuwait Foundation for Advancement in Sciences, grant number KFAS PN20-93MM-02.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Kuwait Foundation for Advancement of Sciences for institutional funding.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.587451/full#supplementary-material.
Abou-Ismail, M. Y., Diamond, A., Kapoor, S., Arafah, Y., and Nayak, L. (2020). The hypercoagulable state in COVID-19: incidence, pathophysiology, and management. Thromb. Res. 194, 101–115. doi:10.1016/j.thromres.2020.06.029
Abu-Farha, M., Thanaraj, T. A., Qaddoumi, M. G., Hashem, A., Abubaker, J., and Al-Mulla, F. (2020). The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci. 21 (10), 3544. doi:10.3390/ijms21103544
Al-Shamsi, A. M., Ben-Salem, S., Hertecant, J., and Al-Jasmi, F. (2015). Transaldolase deficiency caused by the homozygous p.R192C mutation of the TALDO1 gene in four Emirati patients with considerable phenotypic variability. Eur. J. Pediatr. 174 (5), 661–668. doi:10.1007/s00431-014-2449-5
Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., Korbel, J. O., et al. (2015). A global reference for human genetic variation. Nature 526 (7571), 68–74. doi:10.1038/nature15393
Bai, Y., Yao, L., Wei, T., Tian, F., Jin, D. Y., Chen, L., et al. (2020). Presumed asymptomatic carrier transmission of COVID-19. Jama 323 (14), 1406–1407. doi:10.1001/jama.2020.2565
Banne, E., Meiner, V., Shaag, A., Katz-Brull, R., Gamliel, A., Korman, S., et al. (2016). Transaldolase deficiency: a new case expands the phenotypic spectrum. JIMD reports 26, 31–36. doi:10.1007/8904_2015_474
Becker, R. C. (2020). COVID-19 update: covid-19-associated coagulopathy. J. Thromb. Thrombolysis 50 (1), 54–67. doi:10.1007/s11239-020-02134-3
Beun, R., Kusadasi, N., Sikma, M., Westerink, J., and Huisman, A. (2020). Thromboembolic events and apparent heparin resistance in patients infected with SARS-CoV-2. Int. J. Lab. Hematol. 42 (Suppl. 1), 19–20. doi:10.1111/ijlh.13230
Bikdeli, B., Jimenez, D., Hawkins, M., Ortíz, S., Prandoni, P., Brenner, B., et al. (2018). Rationale, design and methodology of the computerized registry of patients with venous thromboembolism (RIETE). Thromb. Haemostasis 118 (1), 214–224. doi:10.1160/th17-07-0511
Bikdeli, B., Madhavan, M. V., Jimenez, D., Chuich, T., Dreyfus, I., Driggin, E., et al. (2020). COVID-19 and thrombotic or thromboembolic disease: implications for prevention, antithrombotic therapy, and follow-up: JACC state-of-the-art review. J. Am. Coll. Cardiol. 75 (23), 2950–2973. doi:10.1016/j.jacc.2020.04.031
Bompard, F., Monnier, H., Saab, I., Tordjman, M., Abdoul, H., Fournier, L., et al. (2020). Pulmonary embolism in patients with COVID-19 pneumonia. Eur. Respir. J. 56 (1), 2001365. doi:10.1183/13993003.01365-2020
Bosso, M., Thanaraj, T. A., Abu-Farha, M., Alanbaei, M., Abubaker, J., and Al-Mulla, F. (2020). The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol Ther Methods Clin Dev 18, 321–327. doi:10.1016/j.omtm.2020.06.017
Brustolin, S., Giugliani, R., and Félix, T. M. (2010). Genetics of homocysteine metabolism and associated disorders. Brazilian J. Med. Biol. Res. = Revista brasileira de pesquisas medicas e biologicas 43 (1), 1–7. doi:10.1590/s0100-879x2009007500021
Buniello, A., MacArthur, J. A. L., Cerezo, M., Harris, L. W., Hayhurst, J., Malangone, C., et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47 (D1), D1005–D1012. doi:10.1093/nar/gky1120
Cerami, E. G., Gross, B. E., Demir, E., Rodchenkov, I., Babur, O., Anwar, N., et al. (2011). Pathway Commons, a web resource for biological pathway data. Nucleic acids research 39, D685–D690. doi:10.1093/nar/gkq1039
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395 (10223), 507–513. doi:10.1016/s0140-6736(20)30211-7
Chen, T., Wu, D., Chen, H., Yan, W., Yang, D., Chen, G., et al. (2020). Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 368, m1091. doi:10.1136/bmj.m1091 Available at: www.icmje.org/coi_disclosure.pdf and declare: support from the Tongji Hospital for Pilot Scheme Project and the Chinese National Thirteenth Five Years Project in Science and Technology, National Commission of Health, People’s Republic of China, for the submitted work; no financial relationships with any organisation that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
Choi, J. W., Lee, S. Y., and Choi, Y. (1996). Identification of a putative G protein-coupled receptor induced during activation-induced apoptosis of T cells. Cell. Immunol. 168 (1), 78–84. doi:10.1006/cimm.1996.0051
Connors, J. M., and Levy, J. H. (2020). COVID-19 and its implications for thrombosis and anticoagulation. Blood 135 (23), 2033–2040. doi:10.1182/blood.2020006000 Squibb, Abbott, Portola, and Pfizer; and received research funding to the institution from CSL Behring. J.H.L. serves on research, data safety, or advisory committees for CSL Behring, Instrumentation Labs, Janssen, Merck, and Octapharma.
Cui, S., Chen, S., Li, X., Liu, S., and Wang, F. (2020). Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J. Thromb. Haemost. 18 (6), 1421–1424. doi:10.1111/jth.14830
Day, M. (2020). Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ 368, m1165. doi:10.1136/bmj.m1165
Driggin, E., Madhavan, M. V., Bikdeli, B., Chuich, T., Laracy, J., Biondi-Zoccai, G., et al. (2020). Cardiovascular considerations for patients, health care workers, and health systems during the COVID-19 pandemic. J. Am. Coll. Cardiol. 75 (18), 2352–2371. doi:10.1016/j.jacc.2020.03.031
Ellinghaus, D., Degenhardt, F., Bujanda, L., Buti, M., Albillos, A., Invernizzi, P., et al. (2020). Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 383, 1522–1534. doi:10.1056/NEJMoa2020283
Ellinghaus, D., Degenhardt, F., Bujanda, L., Buti, M., Albillos, A., Invernizzi, P., et al. (2020). The ABO blood group locus and a chromosome 3 gene cluster associate with SARS-CoV-2 respiratory failure in an Italian-Spanish genome-wide association analysis. medRxiv doi:10.1101/2020.05.31.20114991
Everitt, A. R., Clare, S., Pertel, T., John, S. P., Wash, R. S., Smith, S. E., et al. (2012). IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484 (7395), 519–523. doi:10.1038/nature10921
Fakhro, K. A., Staudt, M. R., Ramstetter, M. D., Robay, A., Malek, J. A., Badii, R., et al. (2016). The Qatar genome: a population-specific tool for precision medicine in the Middle East. Human Genome Var. 3, 16016. doi:10.1038/hgv.2016.16
Fang, H., Knezevic, B., Burnham, K. L., and Knight, J. C. (2016). XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med. 8 (1), 129. doi:10.1186/s13073-016-0384-y
Fattahi, Z., Beheshtian, M., Mohseni, M., Poustchi, H., Sellars, E., Nezhadi, S. H., et al. (2019). Iranome: a catalog of genomic variations in the Iranian population. Hum. Mutat. 40 (11), 1968–1984. doi:10.1002/humu.23880
Folsom, A. R., Gottesman, R. F., Appiah, D., Shahar, E., and Mosley, T. H. (2016). Plasma d-dimer and incident ischemic stroke and coronary heart disease: the atherosclerosis risk in communities study. Stroke 47 (1), 18–23. doi:10.1161/strokeaha.115.011035
Gao, Y., Li, T., Han, M., Li, X., Wu, D., Xu, Y., et al. (2020). Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 92 (7), 791–796. doi:10.1002/jmv.25770
Greenberg, C. S. (2017). The role of D-dimer testing in clinical hematology and oncology. Clin. Adv. Hematol. Oncol. 15 (8), 580–583.
Grillet, F., Behr, J., Calame, P., Aubry, S., and Delabrousse, E. (2020). Acute pulmonary embolism associated with COVID-19 pneumonia detected by pulmonary CT angiography. Radiology 296, E186–E188. doi:10.1148/radiol.2020201544
Hebbar, P., Abu-Farha, M., Alkayal, F., Nizam, R., Elkum, N., Melhem, M., et al. (2020). Genome-wide association study identifies novel risk variants from RPS6KA1, CADPS, VARS, and DHX58 for fasting plasma glucose in Arab population. Sci. Rep. 10 (1), 152. doi:10.1038/s41598-019-57072-9
Helms, J., Tacquard, C., Severac, F., Leonard-Lorant, I., Ohana, M., Delabranche, X., et al. (2020). High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Medicine 46 (6), 1089–1098. doi:10.1007/s00134-020-06062-x
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 395 (10223), 497–506. doi:10.1016/s0140-6736(20)30183-5
Iba, T., Levy, J. H., Levi, M., Connors, J. M., and Thachil, J. (2020). Coagulopathy of coronavirus disease 2019. Critical care medicine. doi:10.1097/ccm.0000000000004458 Behring, instrumentation labs, Janssen, Merck, and Octapharma. Dr. Connors’ institution received funding from CSL Behring; received funding from Abbott (consulting), Bristol-Myers Squibb (consulting), and Portola (scientific advisory board). The remaining authors have disclosed that they do not have any potential conflicts of interest.
John, S. E., Antony, D., Eaaswarkhanth, M., Hebbar, P., Channanath, A. M., Thomas, D., et al. (2018). Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci. Rep. 8 (1), 16583. doi:10.1038/s41598-018-34815-8
Karnes, J. H., Cronin, R. M., Rollin, J., Teumer, A., Pouplard, C., Shaffer, C. M., et al. (2015). A genome-wide association study of heparin-induced thrombocytopenia using an electronic medical record. Thromb. Haemostasis 113 (4), 772–781. doi:10.1160/th14-08-0670
Kimball, A., Hatfield, K. M., Arons, M., James, A., Taylor, J., Spicer, K., et al. (2020). Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility - king county, Washington, March 2020. MMWR Morb. Mortal. Wkly Rep. 69 (13), 377–381. doi:10.15585/mmwr.mm6913e1 Journal Editors form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
Klok, F. A., Kruip, M., van der Meer, N. J. M., Arbous, M. S., Gommers, D., Kant, K. M., et al. (2020). Confirmation of the high cumulative incidence of thrombotic complications in critically ill ICU patients with COVID-19: an updated analysis. Thromb. Res. 191, 148–150. doi:10.1016/j.thromres.2020.04.041
Klok, F. A., Kruip, M., van der Meer, N. J. M., Arbous, M. S., Gommers, D., Kant, K. M., et al. (2020). Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 191, 145–147. doi:10.1016/j.thromres.2020.04.013
Leonard-Lorant, I., Delabranche, X., Severac, F., Helms, J., Pauzet, C., Collange, O., et al. (2020). Acute pulmonary embolism in COVID-19 patients on CT angiography and relationship to D-dimer levels. Radiology 296, E189–E191. doi:10.1148/radiol.2020201561
Levi, M., Thachil, J., Iba, T., and Levy, J. H. (2020). Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 7 (6), e438–e440. doi:10.1016/s2352-3026(20)30145-9
Lipiński, P., Pawłowska, J., Stradomska, T., Ciara, E., Jankowska, I., Socha, P., et al. (2018). Long-term systematic monitoring of four polish transaldolase deficient patients. JIMD Rep. 42, 79–87. doi:10.1007/8904_2017_83
Lippi, G., and Favaloro, E. J. (2020). D-Dimer is associated with severity of coronavirus disease 2019: a pooled analysis. Thromb. Haemostasis 120 (5), 876–878. doi:10.1055/s-0040-1709650
Lippi, G., Lavie, C. J., and Sanchis-Gomar, F. (2020). Cardiac troponin I in patients with coronavirus disease 2019 (COVID-19): evidence from a meta-analysis. Prog. Cardiovasc. Dis. 63 (3), 390–391. doi:10.1016/j.pcad.2020.03.001
Lippi, G., Plebani, M., and Henry, B. M. (2020). Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clinica Chimica Acta 506, 145–148. doi:10.1016/j.cca.2020.03.022
Llitjos, J. F., Leclerc, M., Chochois, C., Monsallier, J. M., Ramakers, M., Auvray, M., et al. (2020). High incidence of venous thromboembolic events in anticoagulated severe COVID-19 patients. J. Thromb. Haemost. 18 (7), 1743–1746. doi:10.1111/jth.14869
Lodigiani, C., Iapichino, G., Carenzo, L., Cecconi, M., Ferrazzi, P., Sebastian, T., et al. (2020). Venous and arterial thromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb. Res. 191, 9–14. doi:10.1016/j.thromres.2020.04.024
Loeffen, Y. G., Biebuyck, N., Wamelink, M. M., Jakobs, C., Mulder, M. F., Tylki-Szymańska, A., et al. (2012). Nephrological abnormalities in patients with transaldolase deficiency. Nephrol. Dial. Transplant. 27 (8). , 3224–3227. doi:10.1093/ndt/gfs061
Longchamp, A., Longchamp, J., Manzocchi‐Besson, S., Whiting, L., Haller, C., Jeanneret, S., et al. (2020). Venous thromboembolism in critically ill patients with covid‐19: results of a screening study for deep vein thrombosis. Res. Pract. Thromb. Haemost. 4 (5), 842–847. doi:10.1002/rth2.12376
Loof, T. G., Deicke, C., and Medina, E. (2014). The role of coagulation/fibrinolysis during Streptococcus pyogenes infection. Front. Cell. Infect. Microbiol. 4, 128. doi:10.3389/fcimb.2014.00128
Middeldorp, S., Coppens, M., van Haaps, T. F., Foppen, M., Vlaar, A. P., Müller, M. C. A., et al. (2020). Incidence of venous thromboembolism in hospitalized patients with COVID-19. J. Thromb. Haemost. 18, 1995–2002. doi:10.1111/jth.14888
Mizumoto, K., Kagaya, K., Zarebski, A., and Chowell, G. (2020). Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan. Euro Surveill. 25 (10), 2000180. doi:10.2807/1560-7917.es.2020.25.10.2000180
Mosesson, M. W., Siebenlist, K. R., and Meh, D. A. (2001). The structure and biological features of fibrinogen and fibrin. Ann. N. Y. Acad. Sci. 936, 11–30. doi:10.1111/j.1749-6632.2001.tb03491.x
Onozawa, Y., Fujita, Y., Kuwabara, H., Nagasaki, M., Komai, T., and Oda, T. (2012). Activation of T cell death-associated gene 8 regulates the cytokine production of T cells and macrophages in vitro. Eur. J. Pharmacol. 683 (1–3), 325–331. doi:10.1016/j.ejphar.2012.03.007
Onozawa, Y., Komai, T., and Oda, T. (2011). Activation of T cell death-associated gene 8 attenuates inflammation by negatively regulating the function of inflammatory cells. Eur. J. Pharmacol. 654 (3), 315–319. doi:10.1016/j.ejphar.2011.01.005
Pearson, D. L., Dawling, S., Walsh, W. F., Haines, J. L., Christman, B. W., Bazyk, A., et al. (2001). Neonatal pulmonary hypertension--urea-cycle intermediates, nitric oxide production, and carbamoyl-phosphate synthetase function. N. Engl. J. Med. 344 (24), 1832–1838. doi:10.1056/nejm200106143442404
Poissy, J., Goutay, J., Caplan, M., Parmentier, E., Duburcq, T., Lassalle, F., et al. (2020). Pulmonary embolism in COVID-19 patients: awareness of an increased prevalence. Circulation 142 184–186. doi:10.1161/circulationaha.120.047430
Ponti, G., Ruini, C., and Tomasi, A. (2020). Homocysteine as a potential predictor of cardiovascular risk in patients with COVID-19. Med. Hypotheses 143, 109859. doi:10.1016/j.mehy.2020.109859
Previtali, E., Bucciarelli, P., Passamonti, S. M., and Martinelli, I. (2011). Risk factors for venous and arterial thrombosis. Blood Transfus. 9 (2), 120–138. doi:10.2450/2010.0066-10
Qiu, H., Wu, J., Hong, L., Luo, Y., Song, Q., and Chen, D. (2020). Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect. Dis. 20 (6), 689–696. doi:10.1016/s1473-3099(20)30198-5
Sakurai, A., Sasaki, T., Kato, S., Hayashi, M., Tsuzuki, S. I., Ishihara, T., et al. (2020). Natural history of asymptomatic SARS-CoV-2 infection. N. Engl. J. Med. 383, 885–886. doi:10.1056/NEJMc2013020
Soomro, A. Y., Guerchicoff, A., Nichols, D. J., Suleman, J., and Dangas, G. D. (2016). The current role and future prospects of D-dimer biomarker. Eur. Heart J. Cardiovasc. Pharmacother. 2 (3), 175–184. doi:10.1093/ehjcvp/pvv039
Tanaka, Y., Kurosaki, M., Nishida, N., Sugiyama, M., Matsuura, K., Sakamoto, N., et al. (2011). Genome-wide association study identified ITPA/DDRGK1 variants reflecting thrombocytopenia in pegylated interferon and ribavirin therapy for chronic Hepatitis C. Hum. Mol. Genet. 20 (17), 3507–3516. doi:10.1093/hmg/ddr249
Tang, N., Bai, H., Chen, X., Gong, J., Li, D., and Sun, Z. (2020). Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 18 (5), 1094–1099. doi:10.1111/jth.14817
Tang, N., Li, D., Wang, X., and Sun, Z. (2020). Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 18 (4), 844–847. doi:10.1111/jth.14768
Thachil, J. (2020). The versatile heparin in COVID-19. J. Thromb. Haemost. 18 (5), 1020–1022. doi:10.1111/jth.14821
Thevarajan, I., Nguyen, T. H. O., Koutsakos, M., Druce, J., Caly, L., van de Sandt, C. E., et al. (2020). Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 26, 453–455. doi:10.1038/s41591-020-0819-2 Sciences, Merck, Viiv Healthcare and Leidos; and honoraria for advisory boards and educational activities (Gilead Sciences, Merck, Viiv Healthcare and Abbvie).
Todua, F., Akhvlediani, M., Vorobiova, E., Tsivtsivadze, G., and Baramidze, A. (2017). Homocysteine and D-dimer levels and multilayer computed tomography for diagnosing pulmonary artery thromboembolism. Vessel Plus 1, 38–42. doi:10.20517/2574-1209.2016.06
Undas, A., and Ariëns, R. A. (2011). Fibrin clot structure and function: a role in the pathophysiology of arterial and venous thromboembolic diseases. Arterioscler. Thromb. Vasc. Biol. 31 (12), e88–e99. doi:10.1161/atvbaha.111.230631
Velavan, T. P., and Meyer, C. G. (2020). Mild versus severe COVID-19: laboratory markers. Int. J. Infect. Dis. 95, 304–307. doi:10.1016/j.ijid.2020.04.061
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., et al. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. Jama 323 (11), 1061–1069. doi:10.1001/jama.2020.1585
Wang, Z., Zhang, A., Wan, Y., Liu, X., Qiu, C., Xi, X., et al. (2014). Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U. S. A. 111 (2), 769–774. doi:10.1073/pnas.1321748111
Weisel, J. W., and Litvinov, R. I. (2017). Fibrin formation, structure and properties. Subcell. Biochem. 82, 405–456. doi:10.1007/978-3-319-49674-0_13
Welter, D., MacArthur, J., Morales, J., Burdett, T., Hall, P., Junkins, H., et al. (2014). The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42 (Database issue), D1001–D1006. doi:10.1093/nar/gkt1229
Wichmann, D., Sperhake, J. P., Lütgehetmann, M., Steurer, S., Edler, C., Heinemann, A., et al. (2020). Autopsy findings and venous thromboembolism in patients with COVID-19. Ann. Intern. Med. doi:10.7326/m20-2003
Xu, P., Zhou, Q., and Xu, J. (2020). Mechanism of thrombocytopenia in COVID-19 patients. Ann. Hematol. 99 (6), 1205–1208. doi:10.1007/s00277-020-04019-0
Yang, X., Yang, Q., Wang, Y., Wu, Y., Xu, J., Yu, Y., et al. (2020). Thrombocytopenia and its association with mortality in patients with COVID-19. J. Thromb. Haemost. 18 (6), 1469–1472. doi:10.1111/jth.14848
Yang, Z., Shi, J., He, Z., Lü, Y., Xu, Q., Ye, C., et al. (2020). Predictors for imaging progression on chest CT from coronavirus disease 2019 (COVID-19) patients. Aging 12 (7), 6037–6048. doi:10.18632/aging.102999
Yuki, K., Fujiogi, M., and Koutsogiannaki, S. (2020). COVID-19 pathophysiology: a review. Orlando, FL: Clinical immunology, 215, 108427.
Zhang, L., Yan, X., Fan, Q., Liu, H., Liu, X., Liu, Z., et al. (2020). D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J. Thromb. Haemost. 18 (6), 1324–1329. doi:10.1111/jth.14859
Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395 (10229), 1054–1062. doi:10.1016/s0140-6736(20)30566-3
Keywords: coagulopathy, COVID-19, heparin, d-dimer, homocysteine, pulmonary embolism, thrombocytopenia, venous thromboembolism
Citation: Abu-Farha M, Al-Sabah S, Hammad MM, Hebbar P, Channanath AM, John SE, Taher I, Almaeen A, Ghazy A, Mohammad A, Abubaker J, Arefanian H, Al-Mulla F and Thanaraj TA (2020) Prognostic Genetic Markers for Thrombosis in COVID-19 Patients: A Focused Analysis on D-Dimer, Homocysteine and Thromboembolism. Front. Pharmacol. 11:587451. doi: 10.3389/fphar.2020.587451
Received: 26 July 2020; Accepted: 28 October 2020;
Published: 09 December 2020.
Edited by:
Paola Patrignani, University of Studies G. d'Annunzio Chieti and Pescara, ItalyReviewed by:
Félix Javier Jiménez-Jiménez, Hospital Universitario del Sureste, SpainCopyright © 2020 Abu-Farha, Al-Sabah, Hammad, Hebbar, Channanath, John, Taher, Almaeen, Ghazy, Mohammad, Abubaker, Arefanian, Al-Mulla and Thanaraj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jehad Abubaker, amVoYWQuYWJ1YmFrZXJAZGFzbWFuaW5zdGl0dXRlLm9yZw==; Fahd Al-Mulla, ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=; Thangavel Alphonse Thanaraj, YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.