
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 21 August 2020
Sec. Ethnopharmacology
Volume 11 - 2020 | https://doi.org/10.3389/fphar.2020.01301
This article is part of the Research Topic Integrative Pharmacology-based Research on Traditional Medicine: Methodologies, Medical and Pharmacological Applications View all 73 articles
 Ajay Kumar1
Ajay Kumar1 Varinder Kaur2
Varinder Kaur2 Kritika Pandit1
Kritika Pandit1 Hardeep Singh Tuli3
Hardeep Singh Tuli3 Katrin Sak4
Katrin Sak4 Subheet Kumar Jain5
Subheet Kumar Jain5 Satwinderjeet Kaur1*
Satwinderjeet Kaur1*Onosma bracteata Wall. (Boraginaceae) is a highly valuable medicinal herb that is used for the treatment of fever, bronchitis, asthma, rheumatism, stomach irritation, and other inflammatory disorders. The present study aims to explore the hepatoprotective potential of ethanolic extract (Obeth) from O. bracteata aerial parts against carbon tetrachloride (CCl4) which causes hepatic damage in the male Wistar rats. Obeth showed effective radical quenching activity with an EC50 of 115.14 and 199.33 µg/mL in superoxide radical scavenging and lipid peroxidation analyses respectively along with plasmid DNA protective potential in plasmid nicking assay. The Obeth modulated mutagenicity of 2 Aminofluorine (2AF) in the pre-incubation mode of investigation (EC50 10.48 µg/0.1 mL/plate) in TA100 strain of Salmonella typhimurium. In in vivo studies, pretreatment of Obeth (50, 100, and 200 mg/kg) had the potential to normalize the biochemical markers aggravated by CCl4 (1mL/kg b.wt.) including liver antioxidative enzymes. Histopathological analysis also revealed the restoration of CCl4-induced liver histopathological alterations. Immunohistochemical studies showed that the treatment of Obeth downregulated the expression levels of p53 and cyclin D in hepatocytes. and downregulation in the Western blotting analysis revealed the downregulation of p-NF-kB, COX-2, and p53. HPLC data analysis showed the supremacy of major compounds namely, catechin, kaempferol, epicatechin, and Onosmin A in Obeth. The present investigation establishes the hepatoprotective and chemopreventive potential of O. bracteata against CCl4-induced hepatotoxicity via antioxidant defense system and modulation of the expression of proteins associated with the process of carcinogenesis in hepatic cells.
Global increase in the demand of industrial solvents in pharmaceuticals, paint and coating industry can raise the global solvent market to an estimated USD 12.31 billion by year 2026 as per Global Market Outlook report (2018). Recently, the use of industrial solvents has brought higher risks to both humans and animals. Industrial solvents cause environmental pollution and their acute and chronic exposure can cause cellular injuries and progressive genetic alterations (mutations) via activation of reactive oxygen species (ROS) with significant changes that may transform healthy cells to cancerous cells (Pizzino et al., 2017). The mutations caused alter the DNA which along with oxidative stress triggers destruction of tumor-suppressor genes and/or the stimulation of proto-oncogenes which contribute to the initiation of liver, breast, colon and prostate cancer (Maron and Ames, 1983; Canli et al., 2017).
The liver is the main organ essential for survival, which plays a crucial function in the body’s metabolism and detoxification process. The liver is a primary site in the body that is mostly involved in modulation and biotransformation of xenobiotic compounds mediated by cytochrome P450 (CYP) family of enzymes (Gu and Manautou, 2012). The chemicals like carbon tetrachloride (CCl4), acetylaminofluorene, thioacetamide, and polycyclic aromatic hydrocarbons are biotransformed into the transitional molecules and generate reactive oxygen species (ROS), including oxygen free radicals that induce hepatic damage (Karakus et al., 2011; Kaur et al., 2017; Kaur S. et al., 2019). Various hepatotoxic compounds and transitional radicals of oxidative reactions affect many proteins and encourage negative effects via oxidative stress (OS) or DNA damage (Rahal et al., 2014). The homeostatic balance of cellular redox is the main feature for combating the lethal insults of ROS, which are internally maintained by antioxidative defense systems. Any aberrant functioning to curb stress can lead to malfuctioning like cancer (Snezhkina et al., 2019). Carbon tetrachloride (CCl4) is a typical solvent which is used in the research for hepatic ailments. Firstly, liver can metabolize CCl4 by cytochrome P450 enzyme complex to produce toxic metabolites such as trichloromethyl free radicals (CCl3•) and trichloromethyl peroxyl radicals (CCl3OO•). These free radicals have capacity of eliminating hydrogen ions from fatty acids to induce lipid peroxidation. Hence, mutually CCl3• and CCl3OO• cause injury in the cell membrane, alteration in enzyme action and lastly cause hepatic damage or necrosis (Chao et al., 2019). The treatment of hepatic cancer still is one of the important tasks despite the newest developments in recognizing its biological basis. The deregulation of p53, COX-2, and NF-κB signaling is one of the central factors for the beginning of hepatic cancer (Alqahtani et al., 2019). p53 (a tumor suppressor gene) associated mutations are found in most of the hepatocellular carcinoma (HCC) (Aravalli et al., 2008). From the past few decades, the natural phytoconstituents from plants are recognized for restoring the health of humans (Che and Zhang, 2019). So, the diet rich in natural compounds has been shown to exert various healing effects via activating redox-signaling, phase-II detoxification enzymes, antioxidants, anti-inflammatory, and anti-carcinogenic properties (Wu et al., 2017).
Onosma bracteata Wall. family Boraginaceae widely knowns as ‘gaozaban’ is mainly distributed in India and Nepal in high altitude regions, spread in Jammu & Kashmir, Himachal Pradesh and Uttar Pradesh in north-western Himalayas (Ved et al., 2016). It is used as the main component for the preparation of drug in Unani and Ayurvedic medicinal system. It is usually used in Indian traditional medication due to its medicinal benefits viz. demulcent, diuretic, anti-aging, antioxidant, antibacterial, wound healing, antileprotic, spasmolytic, and tonic nature (Fareed et al., 2012). The entire parts of O. bracteata have pharmacological properties like anthelmintic, anti-inflammatory, antimicrobial, antiperoxidative, and radical scavenging (Albaqami et al., 2018; Imran et al., 2018; Farooq et al., 2019). It is reported that Onosma genus is a rich source of flavonoids, benzoquinones, naphthoquinones, shikonins, and onosmins (Kumar et al., 2013). However, to date, there are no records about its hepatoprotective activity.
Thus, we planned to explore the protective effects of Obeth from O. bracteata by using in vitro assays viz. antioxidant, antimutagenic and DNA protective potential. Further, in vivo studies were carried out to explore the hepatoprotective effects of Obeth via evaluation of serum marker enzymes, antioxidative enzymes and histopathological parameters. Modulatory effects on the expression level of p53, COX-2, NF-κB, and Cyclin D were studied using immunohistochemical (IHC) and Western blotting.
1-chloro-2, 4- dinitrobenzene (CDNB), 2- thiobarbituric acid (TBA), 5, 5-dithiobisnitrobenzoic acid (DTNB), Bovine Serum Albumin (BSA), Hydrogen peroxide (H2O2), malondialdehyde, nitro blue tetrazolium (NBT), and Silymarin were procured from Sigma (St. Louis, MO, USA). Carbon tetrachloride (CCl4), ethidium bromide (EtBr), low melting point agarose (LMPA), malondialdehyde (MDA), nicotinamide adenine dinucleotide (NADH), normal melting point agarose (NMPA), hydroxylamine hydrochloride, oxidized glutathione (GSSG), phenazine methosulfate (PMS), reduced glutathione (GSH), sodium pyruvate, and xylenol orange were obtained from HiMedia Pvt. Ltd., (Mumbai, India). The pBR322 (plasmid) DNA was purchased from Genei Pvt. Ltd. Bangalore, India. Cyclin-D (cyclin-dependent), p53 (tumor suppressor p53), COX-2, and Nf-κB antibodies were purchased from Cell signaling technology (Danvers, MA, USA). (PVDF) membrane (MDI, Ambala), ECL kit was procured from (Biorad, USA). Kits for analysis of serum parameters were purchased from Erba Mannheim (London, UK). The other chemicals used in the experiments were of Analytical grade.
The aerial parts including stems and leaves of Onosma bracteata Wall. were collected from Herbal Health Research Consortium (HHRC) Pvt. Ltd. Amritsar, Punjab (India), a Government of India approved Drug analysis laboratory. Mr. Viney (Research Officer), at the HHRC Pvt, Ltd. identified and authenticated the plant material by analyzing organoleptic, microscopic and pharmacognostic characteristics. The specimen with accession no: GAZ-03 was submitted in the herbarium of HHRC Pvt. Ltd. Amritsar, Punjab (India).
The plant material was washed with running tap water and dried at 40°C. The material was crushed to powder form (2 Kg) and soaked with 80% ethanol by using the maceration method. The filtered material was evaporated using Rotavapor (Buchi Rotavapor R-210, Switzerland) and was dried to yield 120 g (6%) of the ethanolic extract (Obeth).
The radical scavenging activity of Obeth was conducted according to the method suggested by Nishikimi et al. (1972), with little modifications. Initially, 0.06 M NBT and 0.156 M NADH were mixed to the different Obeth concentrations (25–400 μg/mL), along with the addition of 0.468 M PMS. In the control samples, Obeth was substituted by methanol in all of the above solutions. The mixtures were kept at room temperature for 20 min. In the incubation phase, the yellow-colored NBT was reduced to blue colour and the absorbance was recorded at 560 nm.
The percentage decrease of NBT was represented as:
where ODC = Absorbance of control solution.
ODS = Absorbance of the sample solution.
For this experiment, the procedure was adopted as proposed by Halliwell and Gutteridge (1989), with a little amendment. The lipid origin (10% homogenous egg), 150 mM KCl, and Obeth (25–400 mg/mL) were combined in this assay. In place of Obeth, methanol was used as a control. To continue lipo-oxidation to the above reaction mixture, 10 mM FeCl3 was added followed by incubation at 37°C for 30 min. Subsequently, HCl solution (comprising TCA, TBA and BHT) was added to the mixture and the resultant mixture was heat treated for 1 h at 90°C accompanied by cooling and centrifugation at 2,500 rpm for 15 min. Lastly, the absorbance of pink coloured supernatant formed was recorded at 532 nm.
The estimation of the percent inhibition (anti-lipoperoxidation activity) was based on the formula below:
where ODC = Absorbance of control solution.
ODS = Absorbance of the sample solution.
To determine the DNA protective ability of Obeth or capacity to scavenge the hydroxyl radical (•OH), the plasmid nicking assay was conducted as per the procedure given by Lee et al. (2002) with few amendments recommended by Robin et al. (2015). Different concentrations (25–100 µg/mL) of Obeth from O. bracteata were mixed with freshly prepared Fenton’s reagent (30 mM H2O2, 50 mM Ascorbic acid, and 80 mM FeCl3) and pBR322 plasmid DNA, followed by incubation for 30 min at 37 °C. The samples were resolved onto 1% (w/v) agarose gel using electrophoresis unit at 60 V for 1 h. For this assay, rutin (100 µg/mL) was used as a standard. Immediately, DNA bands were viewed with Gel Doc XR (Bio-Rad, USA), and the band density was determined by using LabImage Bioimaging Platform (Version: 4.1.1) software.
The antimutagenic potential of Obeth from O. bracteata was studied using Ames test, with two Salmonella typhimurium strains as per the method given by Maron and Ames (1983) with few amendments recommended by Kaur et al. (2000). The strains of S. typhimurium were procured from Microbial Type Culture Collection (MTCC), Chandigarh. Under sterile conditions, direct-acting mutagens, i.e., 4- Nitro-o-phenylenediamine and sodium azide were used for TA98 and TA100, respectively. The antimutagenic activity of Obeth, at different concentration levels (25–250 μg/0.1 mL/plate), was studied using two experimental approaches viz. co-incubation and pre-incubation modes to distinguish between the bio-antimutagenicity and desmutagenicity, respectively. Negative control contains only bacterial culture, Obeth and top agar which showed that Obeth did not exhibit mutagenic characteristics. In co-incubation mode, mutagen with Obeth was blended into 2mL of top agar containing bacterial culture whereas, in pre-incubation, the mixture of Obeth and mutagen was incubated at 37 °C for 30 min before adding to the top agar with bacterial culture. The positive control contains bacterial culture and mutagens which help to assess the dosage rate of toxicity/mutagenicity. Obeth antimutagenicity was also tested using S9-mix in the top agar as per the procedure suggested by Garner et al. (1972). The protocol includes a similar method as previously mentioned, except with the inclusion of S9-mix among other components of top agar for the activation of indirect acting mutagens. The reverting colonies were counted using a colony counter.
Antimutagenic activity is defined by the percentage decrease in the number of colonies as shown below:
where,
a= number of mutagenic-induced colonies (positive control)
b= number of colonies induced with mutagen and different concentration of Obeth
c= number of colonies in (negative control).
This experiment was performed in compliance with the guideline of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSE), Ministry of Environment and Forestry, Government of India, and authorized by Institutional Animal Ethics Committee (IAEC), GNDU (approval number 226/CPCSEA/2014/06). As indicated by the IAEC, all rats were entitled to humane care.
Male Wistar rats, weighing between 150–240 g were purchased from the National Institute of Pharmaceutical Education and Research (NIPER), S.A.S. Nagar, Mohali, Punjab (India). The acclimatization of rats was conducted for one week in polypropylene cages containing husk in the aerated Central Animal House of Guru Nanak Dev University, Amritsar. Commercial rodent feed was provided for the rats, and water was given ad libitum with an environmentally regulated condition (55 ± 5 percent humidity, 23 ± 2° C) with a 12 h light/dark cycle photoperiod.
After acclimatization, rats were divided into 7 groups (n=6). Group, I served as control; Group II intoxicated with 1 mL kg-1 CCl4 (mixed in same volume of olive oil via intraperitoneal (i.p.) doses, 3 days in a week) which induced liver mutilation and is mostly used for analyzing the hepatoprotective potential of various drugs (Ritesh et al., 2015; Zamzami et al., 2019); Group III was provided pretreatment of 50 mg kg−1 body weight (b.wt.) of silymarin (natural chemopreventive compound) for 21 days via oral route accompanied by CCl4 (i.p.) doses, 3 days in week without fasting of rats; Group IV was administered with an oral dose of only Obeth (100 mg kg−1 b.wt.) of O. bracteata as negative control; Group V was provided with the Obeth (50 mg kg−1 b.wt.) accompanied by CCl4 (i.p.) doses; Group VI was provided Obeth (100 mg kg−1 b.wt.) intoxicated with CCl4; Group VII was given Obeth (200 mg kg− 1 b.wt.) accompanied with CCl4 (i.p) treatment. The experimental concentrations of Obeth have been decided based on the literature report on O. bracteata in in vivo studies (Asif et al., 2019).
The rats were kept on fasting overnight before termination of experiment, the blood was extracted from retro-orbital venous puncture into heparinized tubes under moderate diethyl-ether anesthesia. The blood samples were centrifuged at 3,000 rpm for 20 min for the separation of serum. The extracted serum was used for examining the different hepatic markers enzymes (Serum Glutamic Pyruvic Transaminase (SGPT), Serum Glutamic-Oxaloacetic Transaminase (SGOT), Alkaline Phosphatase (ALP), total bilirubin, total protein, Albumin, urea, creatinine, and cholesterol) using Erba diagnostics kits with Bene Sphera autoanalyzer.
The liver was dissected after euthanizing by cervical dislocation. The collected liver was washed with ice-cold saline solution. After this, the liver was rinsed with the mixture of (150 mM KCl + 10 mM Tris-HCl) buffer. Finally, 10 percent (w/v) of dried liver homogenate was mixed with chilled Tris-KCl buffer and centrifugation at 2,500 rpm for 10 min. Supernatant was separated and used for further analysis.
The standard protocol given by Bradford, 1976 was used for the quantification of protein concentration in the liver. In this experiment, 900 µL of Bradford reagent was mixed with 100 µL of homogenate. The reaction solution was then kept for 30 min at room temperature and the absorbance was calculated on ELISA plate reader at 595/450 nm.
To check lipid peroxidation in liver homogenate, TBARS (thiobarbituric reactive species) procedure recommended by Devasagayam et al. (1983) was used. 2 mL of TBA solution was mixed to 500 μL of the sample. The mixture was fully stirred and incubated for 30 min at 80°C followed by centrifugation for 15 min at 2,000 rpm. Lastly, supernatant reading was taken at 532 nm and 600 nm. MDA was used as a standard for measuring the amount of TBARS represented as MDA value (MDA equal μmol/g of tissue).
Lipid hydroperoxide generation in liver homogenate was assessed as a method proposed by Jiang et al. (1992). 900 µL of Fox reagent (0.1 M Xylenol orange, 0.025M of H2SO4, 0.25 M of (NH4)2 Fe(SO4)2.6H2O, and 0.004 M of C15H24O) was mixed to 100 µL of liver homogenate. This reaction mix was kept for 40 min at 37°C and final reading was measured at 560 nm. The lipid hydroperoxide generation is shown as nM H2O2 equivalent/g of tissue.
The reduced glutathione content in liver homogenate was detected as per the protocol given by Anderson (1985) with little changes. Potassium phosphate buffer (200 µM, pH 8) and DTNB solution (600 µM) were mixed to 100 µL homogenate, preceded by incubation for 10 min and the absorbance was recorded at 412 nm. Glutathione was used as a reference for making a standard curve and total GSH content was expressed as µmol of GSH content/g of tissue.
Phase I enzymes were determined as per the method performed by Choi et al. (2003). In this protocol, the liver homogenate was added into two 96-well plates. In one plate 15 μL of sodium hydrosulphite (500 mM) was added in each well preceded with 30–35 bubbles of carbon monoxide. Another plate was saturated with only 30–35 bubbles of carbon monoxide without adding sodium hydrosulphite. The absorbance was observed at 420 nm, 450 nm, and 490 nm.
The cytochrome b5 (Cyt b5) quantities were analyzed as the method suggested by Omura and Sato (1964). In this experiment, 500 µL of liver homogenate was added to 4.3 mL of Tris-HCl buffer. After that, 50 μL of this mixture was poured onto two separate 96-well plates. Now, 50 μL of NADPH (500 mM) was added in each well of one plate and 50 μL of Tris-HCl in each well in the other plate followed by incubation for 20 min. The absorbance was measured at 409 nm and 424 nm. The enzyme activity was determined by using the extinction coefficient (6.22 mM1 cm1).
The NADPH Cyt b5 was determined using procedure recommended by Mihara and Sato (1972) with little modification. For this, 200 µL of NADPH (100 µM) and 200 µL of Potassium ferricyanide (200 µM) were mixed with 500 µL of potassium phosphate buffer (300 mM). The process was initiated with the addition of 100 µL of homogenate. The NADPH Phase II Enzymes
Phase II metabolic isozyme glutathione-S-transferase (GST) was quantified as per the method suggested by Habig et al. (1974). In this experiment, 2.7 mL of sodium phosphate buffer (100 mM) was mixed with 100 µL each of 1-Chloro-2,4-dinitrobenzene (CDNB) (0.03 M) and 100 µL reduced glutathione (0.03 M) followed by incubation at 37°C for 5 min. After incubation, 100 µL of homogenate was added to initiate the reaction and enzyme activity was recorded at 340 nm at 30 s interval for 3 min.
Catalase was analyzed in the liver homogenate as the method described by Aebi (1984) with little modifications. 300 µL of H2O2 (150 mM) and 2.6 mL of 50 mM potassium phosphate buffer (50 mM) were mixed thoroughly. After this, homogenate was added to initiate the reaction. Eventually, the absorbance was recorded for the catalase activity at an interval of 15 s, for 2 min at 240 nm.
A slight modification was done to assess the GR level in the samples using the protocol given by Carlberg and Mannervik (1975). In this assay, the reaction mixture was prepared by adding 1.2 mL of sodium phosphate buffer (200 mM), 200 μL oxidized glutathione (10 mM), 200 μL EDTA (20 mM), and 200 μL of homogenate. Finally, 200 μL NADPH (1000 µM) was mixed in a reaction solution and at an interval of 15 s, absorbance was measured at 340 nm for 3 min.
To determine the lactate dehydrogenase (LDH) activity in liver homogenate, the protocol recommended by Kaur S. et al. (2019) with slight modifications was adopted. In this experiment, reaction mixture comprising of 2.3 mL of Tris HCl buffer (100 mM, pH 7.1), 300 μL of sodium pyruvate (100 mM), 100 μL Triton X (7.5%), and 300 μL of NADH (3 mM) was incubated at 30°C for 15 min. In this reaction mixer, 100 μL of homogenate was added and absorbance was measured at an interval of 10 s at 340 nm for 1 min.
Guaiacol peroxidase activity in liver homogenate was determined as per the method suggested by Gee et al. (1970) with slight modifications. The reaction mixture was prepared by adding 75 μL (20 mM) of guaiacol solution to 25 μL 100 mM phosphate buffer with 100 µL of liver homogenate and then 25 μL of H2O2 was added. The absorbance was taken at 436 nm.
To check the hepatocytes’ structural integrity, the isolated livers from the respective groups were fixed in a 10% buffered formalin solution, dipped in ethanol for dehydration and implanted in paraffin wax. The microtome was used to make sections of the livers (0.4 mm thick) using paraffin blocks and stained with H&E (hematoxylin and eosin) by the standard procedure. The slides were observed with a Nikon Eclipse E200 Compound microscope.
To evaluate the pattern of expression of p53 and cyclin D proteins (after treatment with CCl4 and Obeth) in hepatocytes, IHC analysis was performed. In this method, the liver sections (3-6 μm) were deparaffinized on glass slides. The sections were kept for 30 min in H2O2 (0.3% of methanol). The section slides were blocked with 5% skimmed milk for 30 min. Finally, slides were incubated overnight with anti-p53 and anti-cyclin D antibodies at 4°C. Slides were then washed twice with 1 x PBS followed by the addition of anti-rabbit IgG HRP- conjugated secondary antibody and incubated for 30 min and washed thrice with 1x PBS. The expression of proteins was developed by using 3-diaminobenzidine (DAB) for 2–3 min and washed with 1 x PBS. The slides were observed under a Nikon Eclipse E200, Japan microscope.
The comet assay (single-cell gel electrophoresis) was performed according to the protocol given by Collins and Dušinská (2002) with slight modifications. In this assay, 500 mg of rat liver was minced in trypsin solution and kept for 15–20 min at room temperature. The homogenate was passed through a muslin cloth to remove tissue debris followed by centrifugation at 2,500 rpm for 10 min to obtain a cell pellet. After that, 50 µL of cell suspension was added to a low melting point agar (LMPA) solution and transferred to normal melting point agar (NMPA) coated glass slides. The slides were kept in the fridge for 10–15 min. The slides were immersed in lysine buffer (100 mM EDTA, 10 mM Tris-HCl, 1% Triton X-100, 250 mM NaCl, 10 percent DMSO; pH 10) at 4°C for 3- 4 hr. Now slides were dipped in electrophoresis buffer for 10-15 min before starting electrophoresis (voltage = 25 V, current = 300 mA, time = 20 min). After electrophoresis, the slides were dried and kept in neutralization buffer for 15 min (thrice). After 24 h slides were stained with 100 μL (20 μg/mL) ethidium bromide and observed in the fluorescent microscope (Nikon, Tokyo, Japan). Per slide thirty cells were selected randomly to assess DNA impairment, tail length, tail DNA (%) and tail moment using Comet Assay Software Project (CASP) Lab version CASP1.2.3 beta 1.
The liver was dissected and rapidly stored according to respective groups at -80°C. The samples were homogenized in RIPA lysis buffer followed by centrifugation at 12,000 rpm for 20 min and the upper transparent solution was then poured into new tubes. The quantity of proteins was analyzed using Bradford method. Equal concentration of protein (40 μg) was resolved by SDS-PAGE and transferred to PVDF membrane through western bloting. After that, the membrane was blocked using BSA (5% in TBST, 0.1% Tween-20) for 2 h at 25°C and kept overnight with antibodies against p53 (1:1,000), NFκB (1:2,000) and COX-2 (1:500). After that, the membrane was washed thrice with TBST and HRP-conjugated secondary antibody (1:1,500) was added followed by incubation for 2 hr at room temperature. ECL reagent was used to develop blots and imaging was done using Image-Quant LAS 4000, GE Healthcare. Band densities were quantified with Alphaease FC Software (version 4.0). β-actin (1:500) as endogenous control was used for normalizing the expression of the protein of interest.
The filtered Obeth was analyzed for the presence of polyphenols using UHPLC Nexera system (Shimadzu, MA, USA), column (C18). The solvent used in mobile phase consisted of solvent A (0.2% acetic acid) and solvent B (methanol). The flow rate of 1.0 mL/min was sustained for 21 min of running time. The gradient flow used was: 20% B (0–10 min), 55% B (10–12 min), 70% B (12–14 min), 50% B (14–15 min), 40% B (15–17 min), and 30% B (17 –21 min). For testing, the injection volume of the sample was 10 μL with column temperature maintained at 25°C. The spectra were observed at 280 nm with a photodiode array (PDA) analyzer. The peaks of the compound were identified by comparing retention time and distribution of UV-VIS spectra of samples with those of standard compounds.
All results are represented as Mean ± SE of minimum three independent replicates. To compare difference among the means, one-way and two-way analysis of variance (ANOVA) with Tukey’s test was used at 5% confidence level (p-value ≤ 0.05).
Superoxide radical scavenging and lipid peroxidation activity of Obeth is shown in Figure 1, Table S1. SRS assay there is dose-dependent increase in free radical scavenging activity via NBT inhibition with EC50 (115.14 μg/mL) whereas EC50 of Rutin was observed to be 46.18 µg/mL. The lipid peroxidation activity of Rutin and Obeth was EC50 (115.68 μg/mL) and EC50 (199.33 μg/mL) respectively (Table S2) exhibiting Obeth with better antioxidant activity as compared to standard rutin.
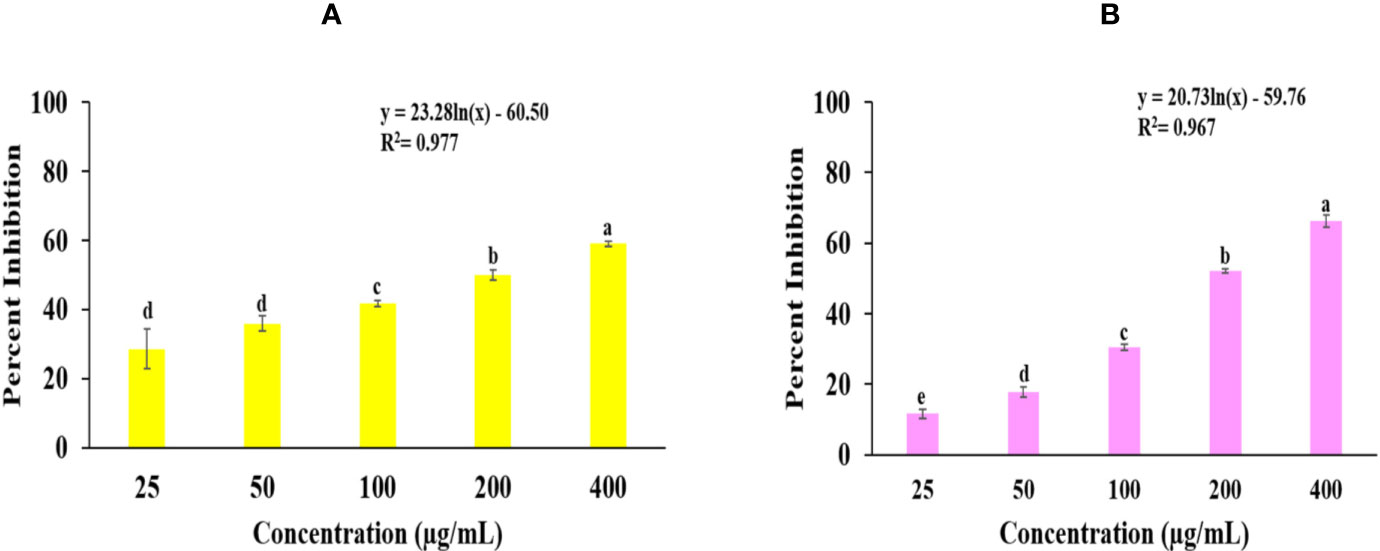
Figure 1 Graphs shows the in vitro antioxidant potential of Obeth from O. bracteata. (A) Superoxide radical scavenging assay. EC50 = 115.14 µg/mL; F-ratio = 187.19; HSD = 8.77. (B) Lipid peroxidation inhibition assay. EC50 = 199.33 µg/mL; F-ratio = 331.68; HSD = 5.89. Values are expressed as Mean ± SE at level of significance p ≤ 0.05. Data labels with different letters represent significant difference among them.
DNA protective effect of Obeth at various concentrations viz. 25, 50, 100 µg/mL was demonstrated in Figure 2. It was observed that the Obeth exhibits quite strong radical scavenging properties with the highest tested concentration (100 μg/mL) on pBR322 plasmid DNA. The experiment exhibited the formation of nicked DNA because of hydroxyl (•OH) radicals produced by Fenton’s reagent. The various concentrations of Obeth showed a dose-dependent minimization of supercoiled DNA to open circular.
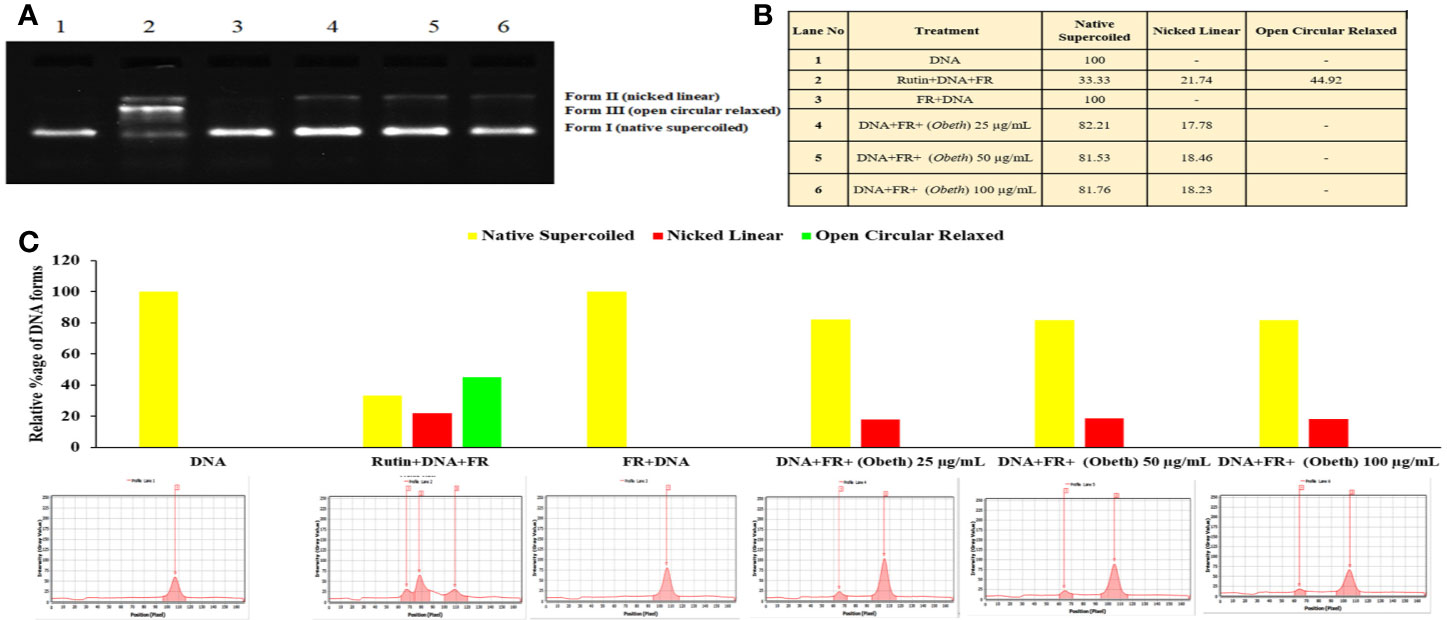
Figure 2 Effect of Obeth from O. bracteata on DNA protection against oxidative damage caused by Fenton’s reagent (FR) in DNA nicking assay. (A) Relative mobility of different forms of plasmid DNA exposed to Obeth extract in electrophoresis. Lane 1: Negative control (pBR322 DNA + distilled water); Lane 2: Positive control (pBR322 DNA + FR); Lane 3: pBR322 DNA + FR + 100 µg/mL rutin; Lanes 4, 5, 6: pBR322 DNA + FR + 25, 50, 100 µg/mL Obeth fraction respectively. (B) Densitometric analysis showing relative percentage of different DNA forms in plasmid after treatment with various concentrations of Obeth fraction in the form of table and (C) graph respectively (software used = Lab Image). CN = Control, FR=Fenton reagent.
The antimutagenic potential of Obeth was tested against sodium azide and 2-aminofluorene mutagenicity in S. typhimurium strains as shown in Table S3. The Obeth in TA100 strain (-S9 mix) displayed a high inhibition rate (82.30% at 250 μg/0.1 mL/plate) showing strong modulation of genotoxicity of base-pair substitution mutagen sodium azide as compared to NPD (frame-shift mutagen) in TA98 tester strain Table 1. The Obeth exhibited high antimutagenicity potential for preincubation mode than in co-incubation approach without -S9 in both TA100 and TA98 as shown in Figure 3A and with +S9 mix in Figure 3B. Minimum EC50 values were depicted by the extract against 2AF mutagenicity in pre-incubation mode in TA100 strain as shown in Table S3. In case of TA98 strain also, Obeth showed good antimutagenic potential in pre-incubation experiments as compared to co-incubation approach (Figure 3, Table 1). The calculated EC50 value showed the high suppression of mutagenicity with a minimum EC50 value (29.51 µg/0.1 mL/plate) with TA98 (+S9) in the pre-incubation approach of treatment as shown in Table S3. The evaluation between all treatments, with and without S9, is shown in Figure 3, Table S3 respectively. The assessment permits noticeable potential of Obeth +S9, even at minimum concentrations, as compared to -S9 treatments. Therefore, the complete investigation showed the conspicuous activity of Obeth, with S9 mix in pre-incubation approach. The findings were statistically evaluated by two-way ANOVA for statistical difference and the interactions as seen in Table 1, S3. The study revealed a statistically significant difference between co-incubation and pre-incubation methods of investigation, in addition to among different treatments used. So, the Obeth of O. bracteata exhibited marked antimutagenicity potential.
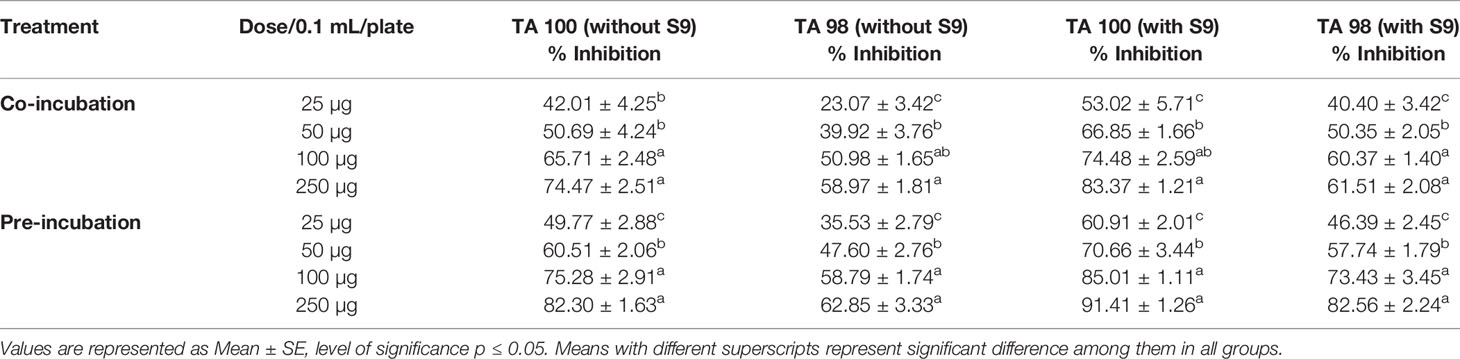
Table 1 Percentage inhibition of mutagenicity by Obeth extract of O. bracteata in TA98 and TA100 tester strains of Salmonella typhimurium with and without S9.
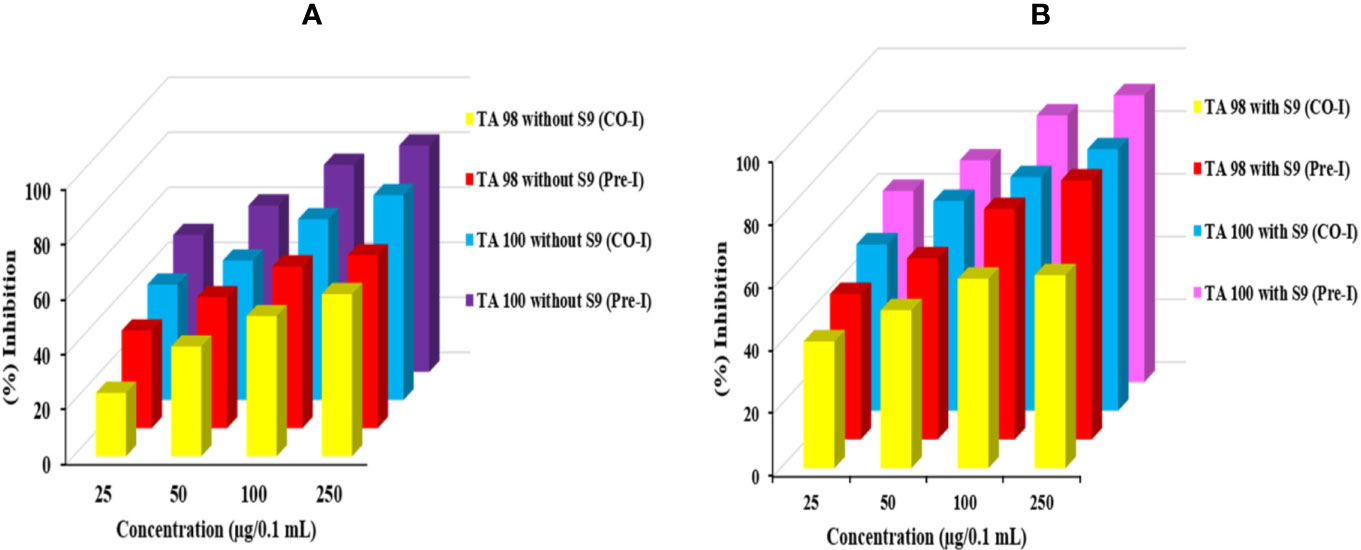
Figure 3 Graph represents the antimutagenic potential of Obeth using S. typhimurium in Ames assay. (A) Represents the TA100 and TA98 without S9 (Pre and Co-incubation). (B) Represents TA100 and TA98 with S9 (Pre and Co-incubation). (A) Represent the percent inhibition between co- and pre-incubation in TA98 and TA100 strain without S9. (B) Represent the percent inhibition between co- and pre-incubation in TA98 and TA100 strain with S9.
In the present study CCl4 treated group, the percentage fold decrease in b. wt. of rats was recorded after 3 weeks, and a statistically significant difference was observed in the b.wt. of CCl4 rats treated group (0.88-fold) relative to control group I (one-fold). Additionally, dose-dependent variability in the b.wt. of Obeth + CCl4 intoxicated groups was maintained b.wt. as the control group (Table S4).
Group II (CCl4) significantly increased the amount of SGOT, SGPT, and ALP to 351.45 ± 21.01, 133.86 ± 29.88, and 529.16 ± 56.74 IU/L respectively in comparison with Group I (control) at 5% level of significance (p ≤ 0.05) (Table S5). Increased concentration of these enzymes in serum, highlighted the poor functioning of the liver and liver injury because of their seepage into the blood. However, the results showed that the treatment of Obeth minimized the amount of these enzymes relative to the CCl4 Group II. III and IV exhibited the amounts of SGOT, SGPT, and ALP in serum in the acceptable limits, signifying reduction in liver injury (Figure 4A).
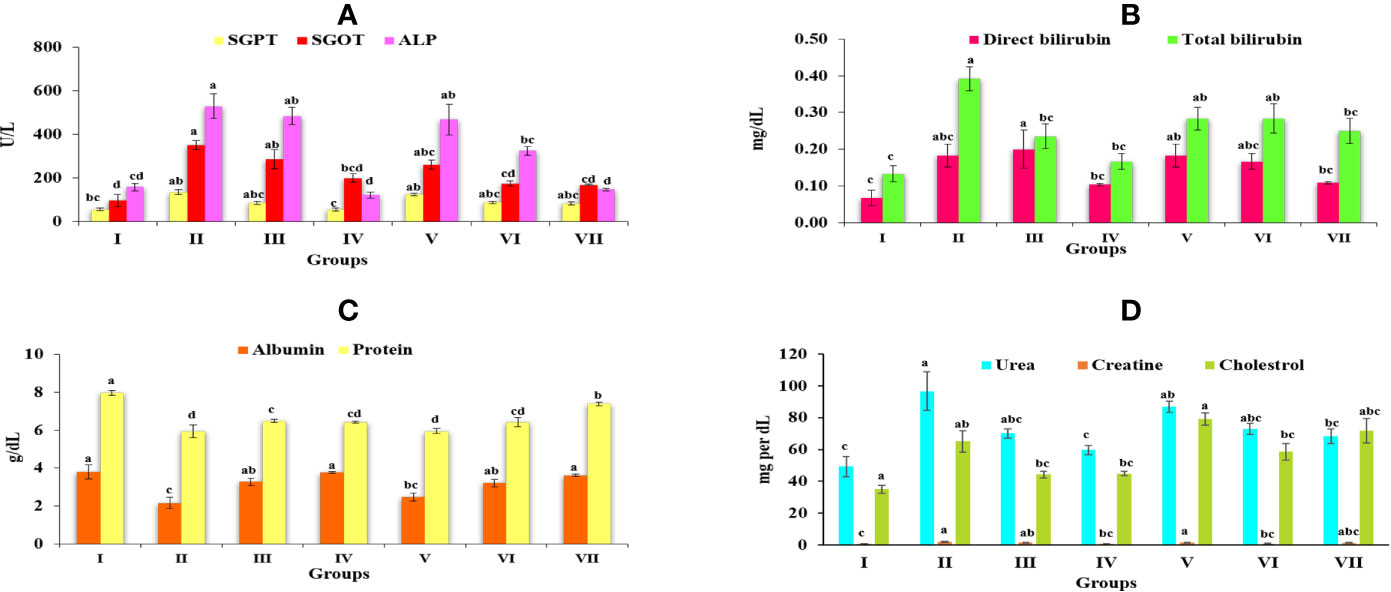
Figure 4 Graph represents the effect of Obeth of O. bracteata on level of serum enzymes level in CCl4-treated male Wistar rats. (A) Represent the levels of SGOT, SGPT, and ALP. (B) Represent the levels of direct bilirubin and total bilirubin. (C) Represent the levels of albumin and total protein. (D) Represent the levels of urea, creatine and cholesterol. Values are expressed as Mean ± SE at level of significance p ≤ 0.05. Data labels with different letters represent significant difference among them.
Obeth pretreatment followed by CCl4 doses in Groups V, VI, and VII was effective in maintaining natural level of direct bilirubin and total bilirubin disturbed in Group II (Figure 4B). In Group III and Obeth treated Group VII indicated an optimum capacity for regeneration by stopping a rise in these enzymes (Table S6).
CCl4 stimulated changes in the hepatic enzymes resulting in decreased albumin (2.15 ± 0.3 g/dL) and protein (5.93 ± 0.34 g/dL) content that lead to hepatic injury (Table S7). Pretreatment of Obeth followed by CCl4 intoxication exhibited substantial protective capacity retaining the normal levels of these enzymes as identified in Groups V, VI, and VII (Figure 4C).
The levels of urea, creatinine and cholesterol in the serum of various groups is given in Table S8. There was a significant increase in the level of urea, creatinine and cholesterol in CCl4 treated Group II. As depicted from the results of Group IV pretreated with Obeth followed by CCl4 showed a restored level of urea, creatinine and cholesterol, hence showing the recovered hepatic damage in a dose-dependent manner (Figure 4D).
The protein content in the CCl4-treated group (24.89 ± 4.16 mg/g tissue) was observed to decrease significantly as compared to the control group (52.74 ± 11.37 mg/g tissue) Table S9. Group IV, however, showed a high protein content of 53.38 ± 4.21 mg/g. On the other side, pretreated groups (V, VI, and VII) were shown to greatly recover the normal protein amount as seen in Figure 5A.
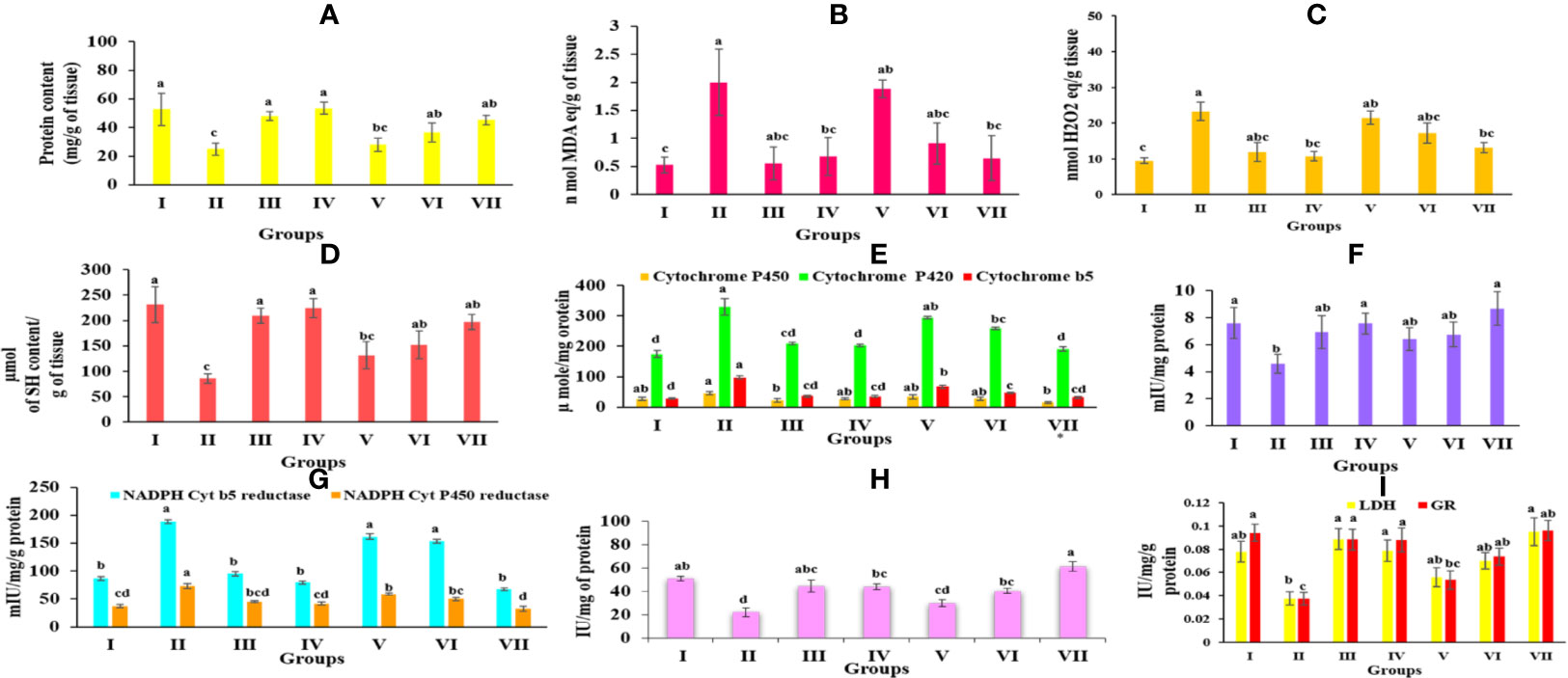
Figure 5 Graph represents the effect of Obeth of O. bracteata on level of enzymes in CCl4-treated male Wistar rats. (A) Represent the levels of protein in liver homogenates. (B) Represent the levels of malondialdehyde (MDA) content in liver homogenate. (C) Represent the levels of lipid hyperoxide in liver homogenates. (D) Represent the levels of reduced glutathione (GSH) in liver homogenate. (E) Represent the levels of NADH Cyt b5 and NADH Cyt P450 in liver homogenate of Wistar rats. (F) Represent the levels of GST in liver homogenate of Wistar rats. (G) Represent the levels of NADH Cyt b5 and NADH Cyt P450 in liver homogenate of Wistar rats. (H) Represent the levels of Catalase in liver homogenate. (I) Represent the levels of lactate dehydrogenase and glutathione reductase in liver homogenate. Values are expressed as Mean ± SE at level of significance p ≤ 0.05. Data labels with different letters represent significant difference among them.
The level of MDA content exhibited the level of lipid peroxidation. Group II revealed a significant increase (p ≤ 0.05) in the MDA content relative to the Group I. Group IV Obeth treated exhibited less concentration of MDA content (0.67 ± 0.33 nM MDA equivalent/g of tissue). However, pretreatment of Obeth followed by CCl4 resulted in a dose-dependent decrease in raised MDA content as seen in Figure 5B and Table S9.
It was observed that the group treated with CCl4 displayed a substantial increase in the lipid hydroperoxide content (23.24 ± 2.54 nM equivalent H2O2/g of tissue) relative to the control group (9.43 ± 0.79 nM equivalent H2O2/g of tissue). Pretreatment by Obeth maximum dose showed to decrease CCl4 toxic impact by exhibiting a maximal decrease in lipid peroxide content (Figure 5C and Table S9).
The group administered with silymarin displayed substantial improvement in GSH content (209.75 ± 14.71 μmol of SH content/g of tissue) compared with CCl4-treated group with GSH content of 85.99 ± 9.57 μmol of SH content/g of tissue (Table S9). Obeth recovered the reduced amount of GSH effectively as close to that of the control group (Figure 5D).
As shown in Table S10, Obeth exhibited a dose-dependent effect on cytochrome P450, b5 and P420 levels in the liver homogenate. CCl4-treated group exhibited reduction in the level of cytochrome P450 (45.90 ± 5.07 μmol) and P420 (329.18 ± 26.63 μmol) as compared to the control group. Pre-treatment of Obeth followed by CCl4, showed increasing cytochrome levels P450 and P420 showing an important defensive effect against hepatic toxicity (Figure 5E). A rise in the production of cytochrome b5 (98.32 ± 5.74 μmol) in CCl4 treated group compared to the control group (29.34 ± 2.27 μmol) was observed. NADH cytochrome P450 reductase activity showed an increase in the CCl4-intoxicated group relative to the control group. Silymarin (45.27 ± 1.91 mIU) and Obeth Group IV (41.82 ± 3.23 mIU) showed same activity as compared to the control group of rats (37.23 ± 2.96 mIU) whereas group VII showed decrease in level of enzyme (32.47 ± 4.99 mIU) as shown in Table S11. Moreover, the CCl4-treated group exhibited an increase in the specific action of NADH cytochrome b5 reductase to 188.43 ± 3.86 mIU in comparison to the control group (86.40 ± 3.17 mIU) whereas group VII exhibited minimization in level of enzyme (67.20 ± 2.36 mIU) as shown in (Figure 5G, Table S11).
CCl4 treated group showed decreased GST behavior (4.59 ± 0.684 mIU/mg protein) compared to the control group (7.6 ± 1.14 mIU) (Table S12) (Figure 5F). However, it was observed that there was a significant increase in GST activity in group III, IV, and VII which showed enhancement of hepatic antioxidant enzymes to combat the increased level of reactive oxygen species.
As shown in Table S13, the catalase activity decreased in the CCl4-intoxicated group (21.92 ± 3.76 IU/mg protein) as compared to control group (50.76 ± 1.95 IU/mg protein). The higher pretreated dose of Obeth (200 mg/kg b.wt.) followed by CCl4 showed an increase in catalase function over the untreated group, thereby offering protective activity against CCl4 intoxication (Figure 5H).
The enzymatic expression of GR in the CCl4-administered group showed 0.03 IU of GR level as compared to control group (0.09 IU). The increased GR specific activity was detected in groups VI and VII which suggested Obeth has a protective role against hepatic damage done by CCl4 (Table S13, Figure 5I).
The level of lactate dehydrogenase is correlated with the integrity of the cell membrane. The increased LDH level in the CCl4-intoxicated group (0.03 IU) mediated cell injury occurred due to the lethal metabolites created by Phase I and II enzyme stimulation. Pre-treated groups with different doses of Obeth were shown to significantly reduce the level of LDH equivalent to the control group (Table S13) and (Figure 5I).
The histopathological analysis showed supportive results for the biochemical and serum enzyme markers. The liver of the CCl4-treated group showed necrosis, inflammation and loss of hepatocytes with vacuolization when observed between central vein and portal traid area with hematoxylin and eosin staining (Figure 6B). In Figure 6, compared to other groups, CCl4-intoxicated group showed a maximum region of fibrotic markers and collagen localization. Obeth and Silymarin treated groups maintained normal liver hepatocyte architecture relative to control group around the central vein area (Figures 6C–G).
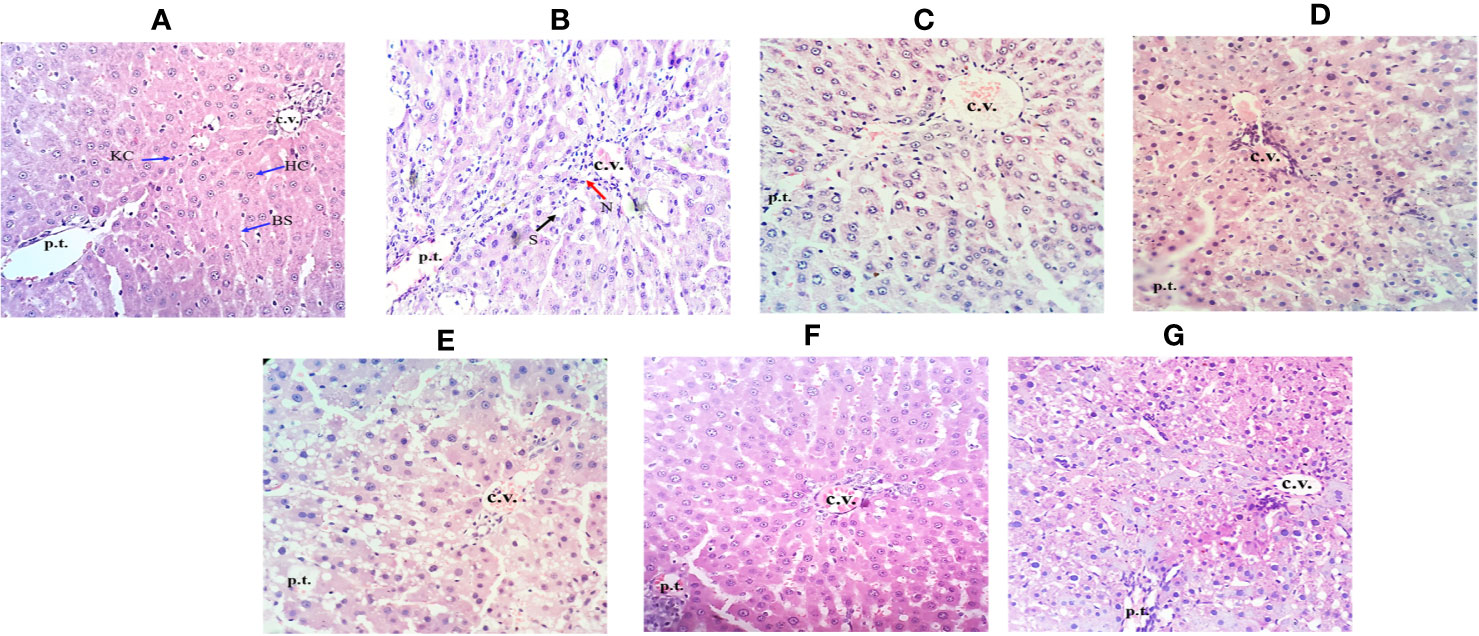
Figure 6 Effect of Obeth from O. bracteata on histology of liver tissues as assessed by H & E staining. (A) Control group (tap water ad libitum) (GI) showing normal lobular architecture with clear portal triad. (B) CCl4: Olive oil (1:1; 1mL kg-1b.wt.) induced necro-inflammatory activity in GII was noted. Treatment of Obeth extract was associated with diminution of portal hypertension with increasing dose. (C) Silymarin (100 mg kg-1b.wt.) + CCl4, (D) low dose (50 mg kg-1b.wt.) + CCl4, (E) medium dose (100 mg kg-1b.wt.) + CCl4, (F) High dose (200 mg kg-1b.wt.) + CCl4, (G) Negative (only extract 100 mg kg-1b.wt. Arrows indicate Kupffer cell (KC), central vein (CV), hepatic cells (HC), blood sinusoids (BS), steatosis (S) (black arrow), and necrosis (N) (red arrow).
Within hepatocytes, over-expression of p53 protein and cyclin-D contribute to hepatocellular carcinoma development. In this experiment, CCl4-treated group exhibited an upregulated expression level of cyclin D (Figure 7A) and p53 markers (Figure 7B) which finally show fibrosis and lobular inflammation whereas, treatment of Obeth decreased the CCl4 induced p53 and cyclin D protein expression dose-dependently in liver hepatocytes as compared to the toxic group (Figures 7A, c–g and 7B, c–g).
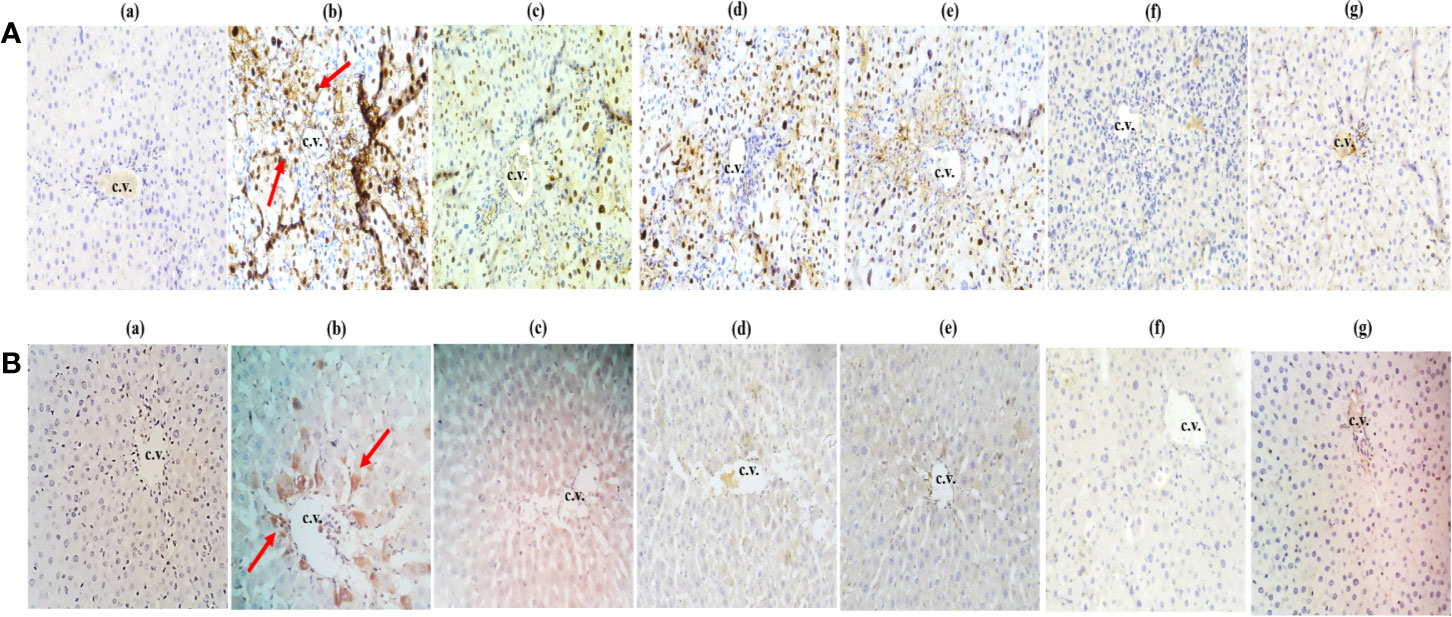
Figure 7 Effect of Obeth from O. bracteata on expression levels of proteins using immunohistochemical analysis of liver tissues. (A) shows cyclin (d) and (B) shows p53 antibodies. Arrows showed the increase in expression of cyclin D and p53 proteins in CCl4 treated liver samples (Original magnification = 40 x). Expression of these proteins was downregulated when observed in liver samples co-treated with CCl4 and Obeth (200 mg/kg b.wt.). Red arrow indicates the increase in the expression of protein in the CCl4 treated group. (b) Control group (tap water ad libitum) (GI) showing normal lobular architecture with clear portal triad. (b) CCl4: Olive oil (1:1; 1mL kg-1 b.wt.) induced elevation in the expression level of cyclin D was noted. Treatment of Obeth extract was associated with diminution of portal hypertension with increasing dose. (c) Silymarin (100 mg kg-1b.wt.) + CCl4, (d) low dose (50 mg kg-1b.wt.) + CCl4, (e) medium dose (100 mg kg-1b.wt.) + CCl4, (f) High dose (200 mg kg-1b.wt.) + CCl4, (g) Negative (only extract 100 mg kg-1 b.wt.).
As shown in Figure 8, larger Comet tail length in CCl4 treated groups indicated the higher necrosis, apoptosis and DNA fragmentation in hepatocytes. Whereas, reduced Comet tail length in Obeth + CCl4 treated groups (100 mg/kg b.wt. and 200 mg/kg b.wt.) showed inhibition of necrosis, apoptosis and DNA fragmentation.
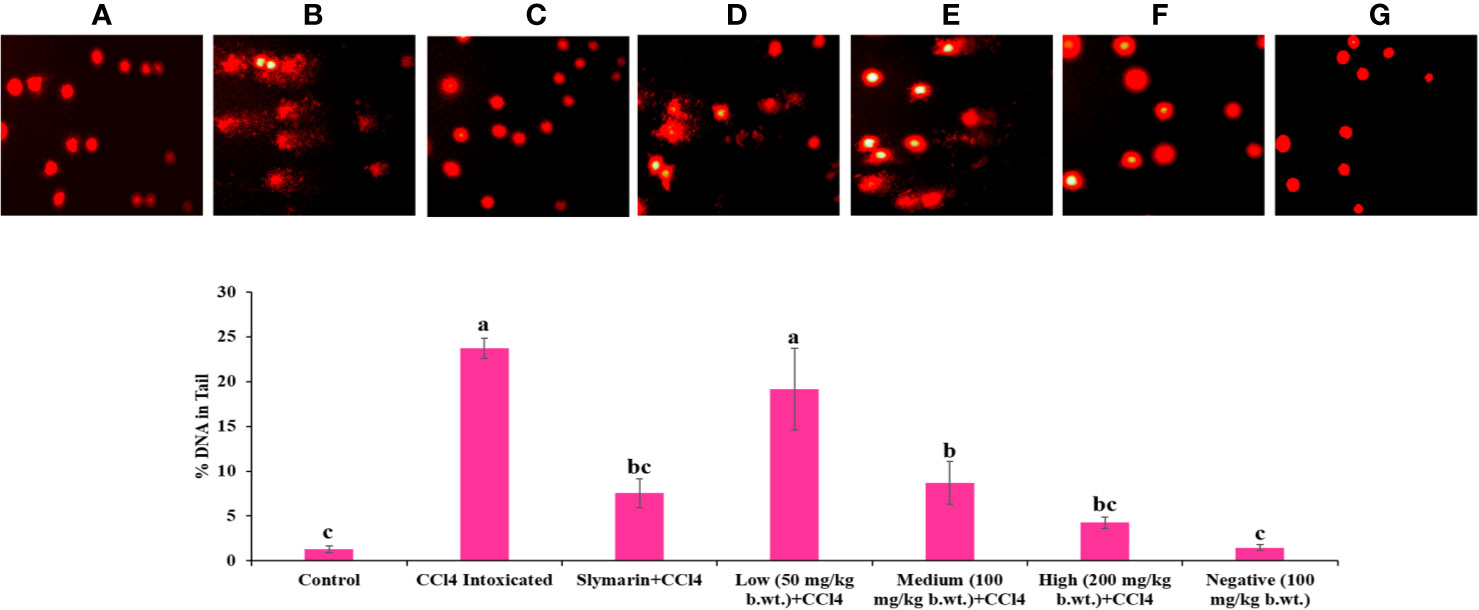
Figure 8 Single cell gel electrophoresis assay was used for determined DNA damage/protect in liver of rats. Results of the comet assay confirm that pretreatment with Obeth decreases CCl4-induced hepatotoxicity in rats. (A) Control group (tap water ad libitum) (GI), (B) CCl4: Olive oil (1:1; 1mL kg-1b.wt.) in GII, (C) Silymarin (100 mg kg-1b.wt.) + CCl4, (D) low dose (50 mg kg-1b.wt.) + CCl4, (E) medium dose (100 mg kg-1b.wt.) + CCl4, (F) High dose (200 mg kg-1b.wt.) + CCl4, (G) Negative (only extract 100 mg kg-1b.wt.). Histograms showing densitometric analysis of % of DNA in tail in comet assay. Values are expressed as Mean ± SE at level of significance p ≤ 0.05. Data labels with different letters represent significant difference among them.
To explore the relationship between p53, COX-2, and NfκB and the hepatoprotective effects of Obeth on toxicity induced by CCl4, we studied protein expression using western blotting. As shown in Figure 9. there was a significant increased (p < 0.05) in p53, Cox-2, and NfκB protein expression in CCl4 treated group as compared to the untreated control group. However, there was a dose-dependent decrease in the expression of proteins on administration of Obeth with CCl4 treated groups as compared to only CCl4 intoxicated group.
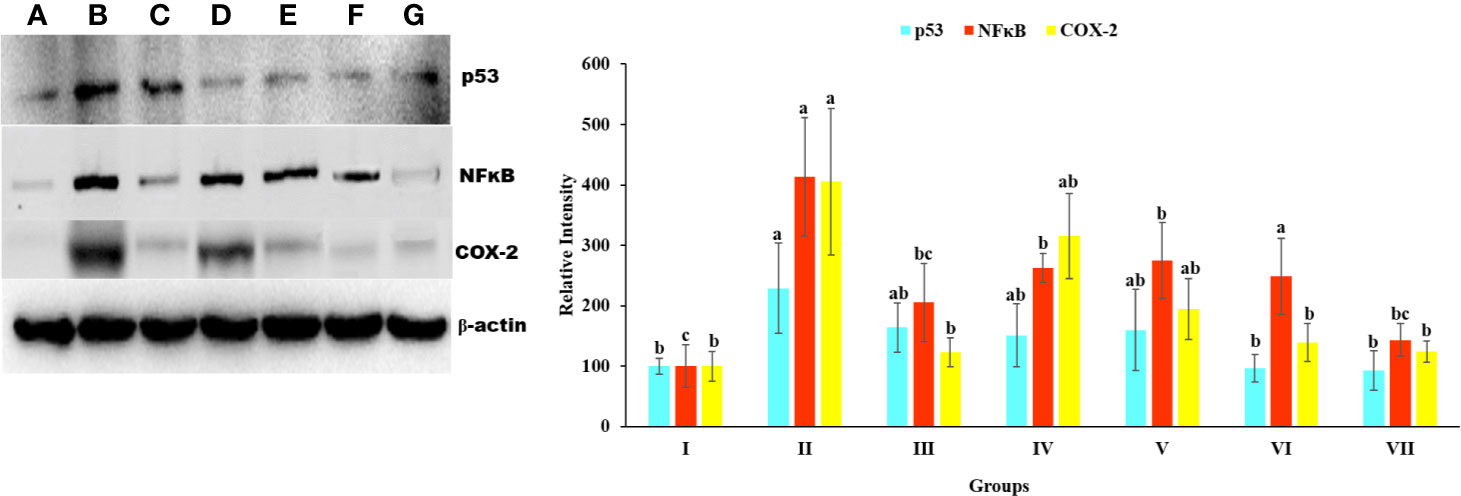
Figure 9 Expression level p53, NFκB, and COX-2 protein in liver homogenate of Wistar rats detected using Western blotting. (A) Control group (tap water ad libitum) (GI), (B) CCl4: Olive oil (1:1; 1mL kg-1b.wt.) induced elevation in the expression level of p53 was noted. (C) Silymarin (100 mg kg-1b.wt.) + CCl4, (D) low dose (50 mg kg-1b.wt.) + CCl4, (E) medium dose (100 mg kg-1b.wt.) + CCl4, (F) High dose (200 mg kg-1b.wt.) + CCl4, (G) Negative (only extract 100 mg kg-1 b/wt.). Histograms showing densitometric analysis of p53, NFκB, and COX-2 protein bands using western blotting. Band density was measured and normalized to that of β-actin. Values are expressed as mean ± SE at level of significance p ≤ 0.05. Data labels with different letters represent significant difference among them.
The phytochemical analysis by HPLC exhibited the occurrence of catechin, epicatechin, Onosmin A, rutin and kaempferol as major phytoconstituents in Obeth (Figure 10, Table 2).
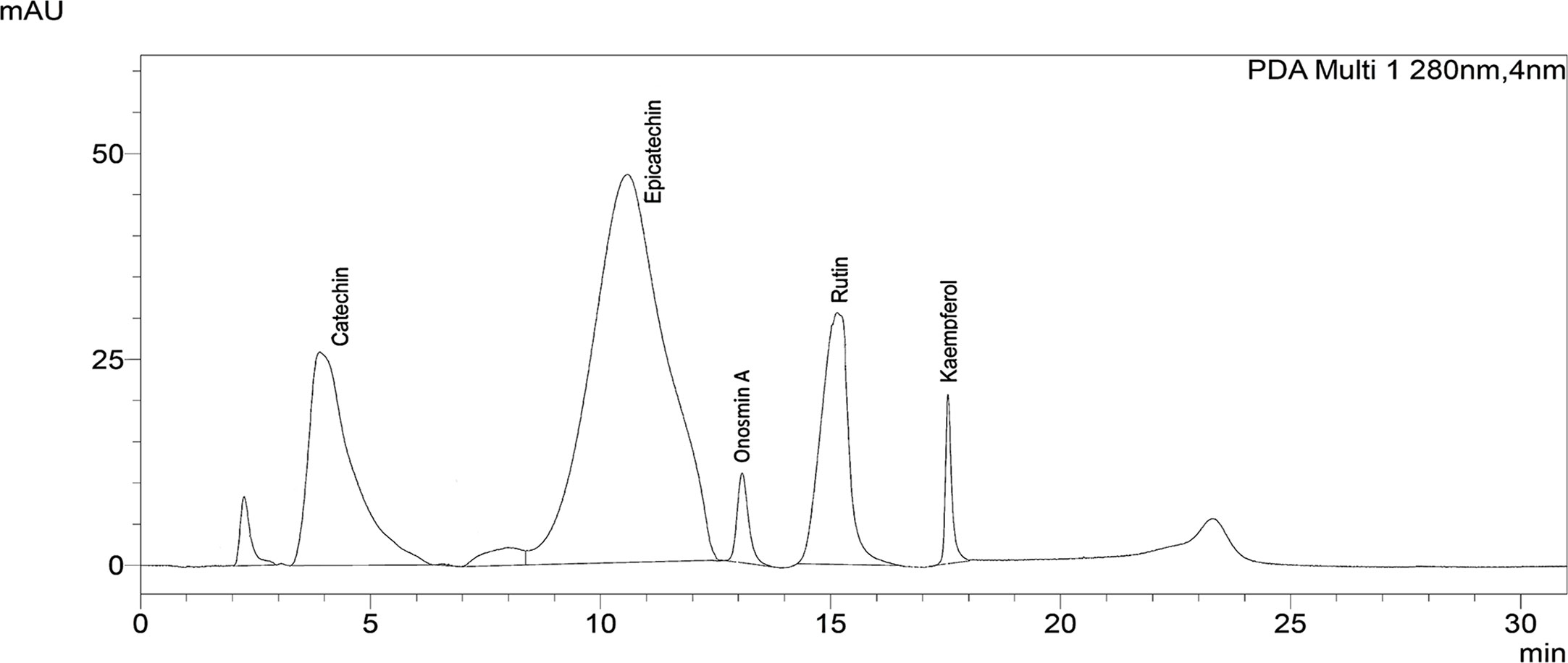
Figure 10 Chromatogram of Obeth of O. bracteata showing presence of polyphenols as identified using HPLC analysis by reference compound.

Table 2 Amount (mg/L) of different phytochemical quantified by HPLC in different Obeth of O. bracteata.
The active polyphenols present in the plants act as scavengers of ROS, DNA protective agents and help to maintain the expression of genes/proteins that break down toxins and keep cellular redox homeostasis (Liu et al., 2018). These polyphenols have the antioxidant, antimutagenic and chemopreventive properties that might help to protect and conserve the constancy of genome (Mehta et al., 2010; Rodríguez-García et al., 2019). Previous studies have reported the potential of Onosma genus as antioxidant compounds and their use in averting numerous pharmacological situations (Naz et al., 2006; Ebrahimzadeh et al., 2010; Ahmad et al., 2013; Imran et al., 2018; Farooq et al., 2019).
In the current study, we investigated the antioxidant, antimutagenic, genoprotective, and hepatoprotective properties of Obeth from Onosma bracteata along with evaluation of the expression of proteins involved in the inflammation and apoptotic induction in in vivo studies using male Wistar rats. In antioxidant assays, Obeth had shown potential to scavenge free radicals, namely in SRS and LPO assays. Obeth showed strong radical scavenging activity with EC50 (115.14 µg/mL) whereas LPO inhibition activity with EC50 (199.33 µg/mL) (Tables S1 and S2). Radical scavenging potential of Obeth might be correlated to the nature of phenolics, contributing to their electron transfer/hydrogen donating ability.
Plants contain natural antioxidants, primarily flavonoids, which are being explored to inhibit various diseases viz. chronic inflammation and cancer (Surh, 2003). Onosma genus plants are enriched with bioactive components viz. flavonoids, alkannin, and shikonin responsible for wound healing, analgesic and its antibacterial actions (Kumar et al., 2013). The HPLC analysis of Obeth revealed the existence of different phytoconstituents viz., kaempferol, Onosmin A, catechin and epicatechin. Numerous reports have shown that Onosma genus and its isolated compounds are responsible for excellent antioxidant activities (Riedl et al., 2004; Kandemir and Türkmen, 2008; Menghani et al., 2011). The ethyl acetate, methanol and water extracts from Onosma heterophylla Griseb. have an appreciable amount of kaempferol, caffeic acid, o-Coumaric acid, rutin and rosmarinic acid which are responsible for antioxidant activity (Ozer et al., 2018). Saravanakumar et al., 2019 also reported that the methanol extract of Onosma isaurica Boiss. & Heldr. and Onosma bracteosa Hausskn. & Bornm. showed effective free radical scavenging ability due to the presence of the phytoconstituents. This plant has been widely used because of its medicinal properties.
In Ames assay, the antimutagenic potential of Obeth against sodium azide, 4-nitro-o-phenylenediamine and 2- aminofluorene mutagens was evaluated against two Salmonella typhimurium strains, i.e., TA100 and TA98. The study showed that Obeth effectively modulated the mutagenicity of direct-acting base-pair mutagen, sodium azide with 82.30% inhibition at 250 μg/0.1 mL/plate (Table S3) in TA 100 while it showed moderate antimutagenic effect against 4NPD with 62.85% inhibition at the same concentration in TA98 tester strain. Obeth exhibited antimutagenicity against 2AF (S9 dependent) in both the tester strains. Kaur et al., 2001 reported that phenolic fractions isolated from Terminalia arjuna showed mutagen specificity via decreasing the frameshift mutagen NPD whereas it failed in the prevention of sodium azide (base pair substitution)-induced his+ revertants. Kaur et al., 2016 described that ethyl acetate fraction of Cassia fistula (CaFE) showed strong antimutagenic and antioxidant properties due to the presence of polyphenolic compounds (umbelliferone, catechin and epicatechin). Obeth also demonstrated the potential to protect plasmid DNA against free radicals of Fenton’s reagent in plasmid DNA nicking assay. Obeth at 100 µg/mL concentration efficiently retained DNA’s supercoiled form.
In in vivo studies, serum analysis showed that the oxidative stress serum markers (SGOT, SGPT, ALP, creatinine and direct bilirubin) were significantly increased, following the dosage of CCl4 to rats (Group II) (Tables S5, S6, and S8). The potential to produce albumin proteins (which function as transport molecules) vanished in damaged hepatocytes, and the capacity to make inflammatory proteins increased. CCl4 generated free radicals (trichloromethyl and peroxytrichloromethyl) that induced damage in hepatocytes resulting in leakage of cytoplasmic SGPT and SGOT into the bloodstream (Shah et al., 2015). Obeth ameliorated the toxic effects of CCl4 by enhancing the activity of antioxidant molecules in the liver. Obeth significantly decreased the damage caused by CCl4 (Group II) in cellular TBARS. A decrease in GSH enzymes and protein levels was observed in the CCl4 treated group (Group II), while an increase in these markers was observed following Obeth treatment (Table S9). Sabir et al. (2017) reported in their study that aqueous extracts of Zanthoxylum armatum DC. (syn. Zanthoxylum alatum Roxb.) restored the level of glutathione and showed hepatoprotective property against paracetamol-induced hepatic damage in albino mice which decreased the hepatic glutathione level. The activation of lipid hydroperoxides, Cytochrome P450, P420, and b5 were observed in CCl4 treated group whereas Obeth treatment declined the level of these markers (Table S10). The values of the above parameters in the Obeth-treated group were the same as of the control (GroupI) which reflects a high safety potential of Obeth. CCl4 treated group showed decline GST levels as compared to control but in a dose-dependent manner whereas Obeth treated groups exhibited restored level of GST. Allocati et al., 2018 reported that GSTs can protect DNA against oxidative stress and eliminate toxic molecules that can led to mutations or carcinogenesis. Laouar et al., 2017 reported that treatment of CCl4 lead to significantly reduced levels of hepatic GSH, GPx, and GST activities than control group in Wistar rats, whereas aqueous extract of Juniperus phoenicea L. berries treatment significantly restored the levels of these enzymes. The Group II showed a significant increase in the amount of NADPH cytochrome b5 and NADH cytochrome 450, whereas, Obeth treated groups exhibited a dose-dependent decline in the level of these enzymes (Table S11). The CCl4 treatment generates CCl3• radicals which not only damaged DNA but also caused necrosis and apoptosis as evidenced from increase in Comet tail length in CCl4 treated group whereas in case of Obeth -treated groups, the reduced Comet tail length showed the DNA protective activity of Obeth in a dose-dependent manner (Figure 8). Damage of cells by CCl3• free radical (toxic) formed by the reduction of CCl4, in hepatocytes has also been reported Brai et al. (2014) and Josan et al. (2015). The previous report from Younis et al., 2018 showed that CCl4 treated rats showed a high degree of DNA damage and remarkable changes in (comet, head, tail) length and %DNA in the tail, while methanolic extract of Fraxinus xanthoxyloides (G.Don) Wall. ex A.DC. ameliorates the effect of CCl4 similar to that of control group.
Histopathological studies showed that Obeth was itself non-toxic and showed close histological similarity to the control group and displayed the same histological arrangement with no sign of inflammation (Figure 6G). In contrast, CCl4 treated group displayed steatosis, necrosis, inflammation in blood sinusoids and vacuolization (Figure 6B). However, pretreatment of Obeth followed by CCl4 treatment effectively restored the regular hepatocytes’ architecture and also reduces the vacuoles formation between the central vein and portal triad indicating hepatoprotective potential of Obeth, as shown in Figures 6D–F. Earlier, Josan et al. (2015) reported that CCl4 treated Sprague Dawley rats showed necrosis of hepatocytes nearby central vein and increased the level of alanine transaminase (ALT) whereas control group showed normal architecture of hepatocytes. Ganaie et al., 2015 also reported that dose-dependent treatment of ethanolic extract of Rumex vesicarius L. showed a restored pattern of histological architecture, while silymarin treatment followed by CCl4-intoxication in Wistar rats showed less disarrangement of hepatocytes. The results of the Immunohistochemistry (IHC) studies showed that p53 protein and Cyclin D expressions in the liver tissues were maximum in the CCl4 intoxicated group. Upregulated expression of pro-apoptotic p53 in CCl4 treated group might be formed in response to hepatocyte inflammation and the induction of necrosis in liver cells (Figures 7A, B). Similar results were observed in western blotting studies showing p53 upregulation in the CCl4 group as compared with the control group. The tumor suppressor gene p53 is upregulated in response to DNA damage, and its upregulated expression is only found in cells that show mutant forms (Aly et al., 2019). The upregulated expression of Cyclin D protein in the CCl4 intoxicated group also highlighted the mutation in hepatocytes while Obeth treatment showed the downregulation of Cyclin D exhibiting its protective effects against CCl4 intoxication. Cyclin D has been described to have the main role in cell progression through the G1/S phase of the cell cycle, and its deregulation can lead to tumorigenesis (Ying et al., 2008; Chibazakura et al., 2011; Gao et al., 2016).
The expression of p53, COX-2, and NF-κB was upregulated due to cellular stress, inflammation and mutation in cellular DNA (Banerjee et al., 2002; Cha and DeMatteo, 2005). Hepatic cancer can be controlled by the suppression of COX-2, NF-κB and p53 which might offer an effective control approach. The investigation of the effects of Obeth towards stimulated forms of p53, cyclin-D, COX-2 and NF- κB proteins was performed. The expression levels of COX-2 and NF-κB proteins were upregulated after CCl4 administration (Figure 9) highlighting the production of mediator cytokines which facilitate tumor growth via inhibiting apoptosis and promoting cell proliferation (Brücher et al., 2019; Lerdwanangkun et al., 2019). Eltahir et al. (2020) reported that CCl4 treated Wistar rats showed significantly high expression levels of NF- κB and COX-2 while Boswellia serrata Roxb. (BS) gum resin treatment followed by CCl4 significantly restored the level of NF- κB and COX-2 expression due to the presences of a natural compound that exhibited anti-inflammatory effects by targeting multiple pathways. The stressed cells promoted the overactivation of signaling proteins or inflammatory cytokines including, IL-6, TNFα, and IL-1β that stimulate the overproduction of IL-6 in Kupffer cells leading to the enhanced proliferation of hepatocytes, some of which harbor oncogenic mutations (Sun and Karin, 2008). Obeth has efficient antioxidant properties that protect the hepatocytes against any damage by inducing antioxidative mechanisms. Previous reports have proved the connotation among natural compounds isolated from plants and cancer inhibition via antioxidant properties (Seufi et al., 2009). In the present study, it was found that the Obeth contains catechin, epicatechin, rutin, kaempferol, and Onosmin A (Figure 10, Table 2). All these compounds have antioxidant potential are known to ascertain their remedial effects by scavenging the free radical species due to the presence of H donating hydroxyl groups on the different polyphenolic rings of these molecules stabilize the negative charge on the different reactive species and hence, nullify their damaging consequences (Kaur P. et al., 2019). One such study showed catechin to regulate the expression of NF-κB and Cox-2 by scavenging the ROS and hence delivering the anti-inflammatory and antioxidant effects (Chu et al., 2017). Similarly, rutin has also proven as an effective hepatoprotective agent against CCl4 induced Wistar rat liver damage by altering the expression of IL-6/STAT3 pathway genes expression due to its antioxidative, anti-inflammatory and anti-apoptotic properties (Hafez et al., 2015). On the other hand, epicatechin has the potential to inhibit Sprague-Dawley rat liver inflammation via NFκB signaling pathway initiated by HSP60 (Huang et al., 2019). Zang et al., 2018 reported that soybean leaves and its isolated compound kaempferol galactoside and kaempferol have the ability to reduce serum parameters (serum aspartate aminotransferase and serum alanine aminotransferase) increased by CCl4 intoxication. In addition to this, all these compounds were also found to ameliorate the lipid peroxidation and regulate the levels of different liver enzymes (Sinha et al., 2012; Khan et al., 2012; Liu et al., 2015; Wang et al., 2015). The polyphenolics such as catechin, caffeine, epicatechin (EC), epigallocatechin gallate (EGCG), and epicatechin gallate (ECG) showed preventive effects of Liubao Tea against CCl4 -induced liver injury in Kunming mice, comparable to silymarin (Pan et al., 2018). Ahmad et al., 2005, reported that Onosmin A isolated from Onosma hispida Wall. ex G.Don has potential to inhibit lipogenase enzyme in a concentration-dependent manner. It can be stated that the chemical association of phytochemicals, presumable flavonoids in Obeth from O. bracteata have protective effects on liver damage via suppression of CCl4-induced oxidative stress (Figure 11). This study reported that antioxidant, antimutagenic, DNA protective, and anti-inflammatory properties of Obeth are correlated with the phytoconstituents present in and have also been shown to have modulatory impact on pro-carcinogenic proteins p53, NF-κB, and COX-2 for the first time.
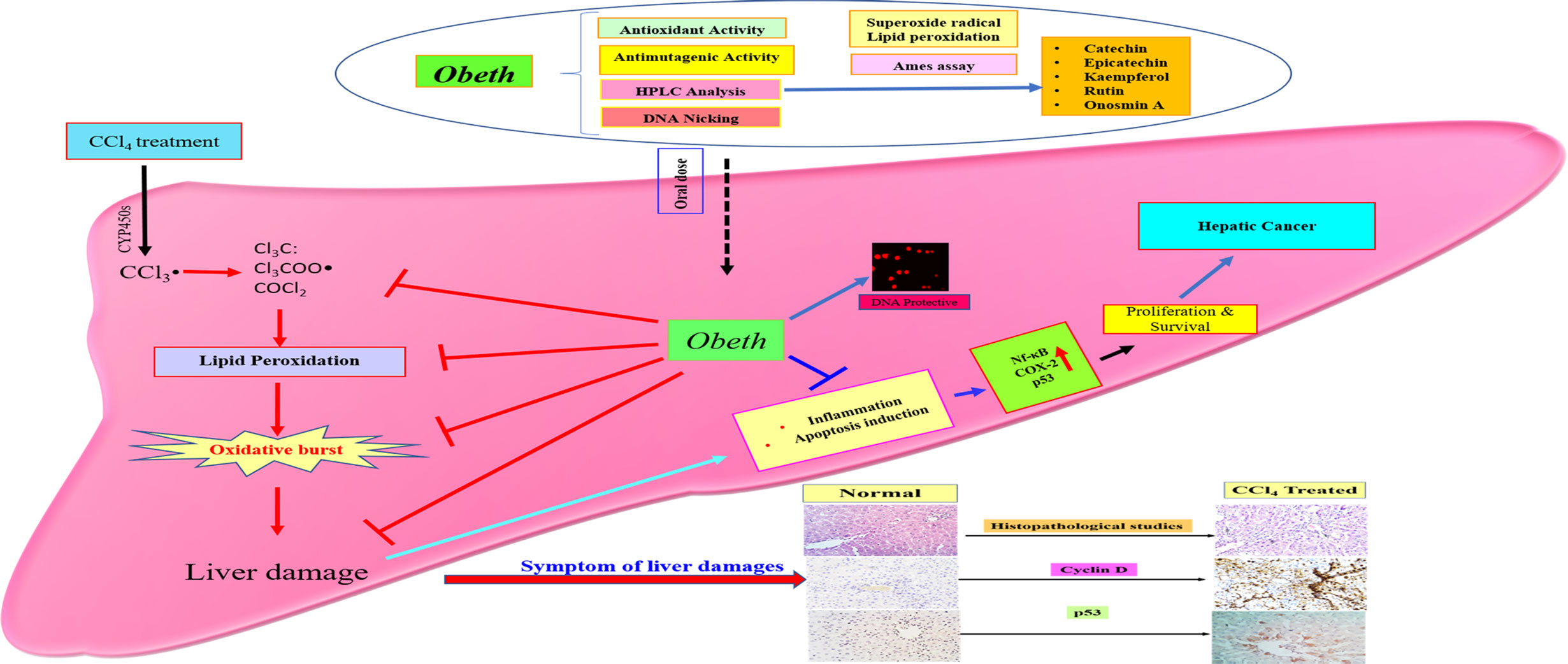
Figure 11 Schematic proposed mechanisms for hepatoprotective property of Obeth from O. bracteata. Obeth phytoconstituents ameliorated hepatic oxidative stress and suppressed inflammation, apoptosis, and DNA damage in CCl4-injured hepatocytes.
In the current study, Obeth of O. bracteata has shown significant antioxidant, genoprotective, antimutagenic and hepatoprotective properties. As revealed by HPLC investigations, Obeth harbors catechin, epicatechin, rutin, Onosmin A and kaempferol as major phytoconstituents. Obeth exerted its protective potential via its radical scavenging and DNA protective abilities, minimizing oxidative stress through induction of antioxidative enzymes and ameliorating the hepatic changes triggered by CCl4. The modulation of crucial proteins such as p53, cyclin D, Cox-2, and NF-κB demonstrated potential of Obeth as an effective chemotherapeutic agent against hepatic cancer.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
The animal study was reviewed and approved by Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSE), Ministry of Environment and Forestry, Government of India, and authorized by Institutional Animal Ethical Committee (IAEC), Guru Nanak Dev University, Amritsar, India (approval number 226/CPCSEA/2014/06).
AK: formal analysis, investigation, methodology, data curation, and writing—original draft. HT and KS: helped to revise the manuscript, reviewing, and editing. VK: intellectual contribution and reviewing manuscript. KP: formal analysis. SJ: conceptualization and project administration. SK: conceptualization, supervision, project administration, reviewing, editing, and resources. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are thankful to the University Grants Commission (UGC)- Basic Scientific Research (BSR), DST-PURSE, and DST-FIST (Grant No. SR/FST/LSI-691/2016(C) programme for providing financial assistance. We also like to acknowledge UGC, New Delhi for the instrumentation facility provided under UGC-DRS V (Grant No. F4-13/2015/DRS-II (SAP-II), CPEPA, RUSA 2.0 scheme, UPE program, and Centre of Emerging Life Sciences, Guru Nanak Dev University, Amritsar (India) for providing the required support and facilities.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.01301/full#supplementary-material
2AA, 2-aminoanthracene; 2-AF, 2-aminofluorene; AA, Ascorbic acid; BC, bacterial culture; b.wt., body weight; CCl4, carbon tetrachloride; COX-2, cyclooxygenase-2; CPCSEA, committee for the Purpose of Control and Supervision of Experiments on Animals; ANOVA, analysis of variance. CYP, cytochrome P450; df, Degree of freedom; EDTA, ethylenediamine tetraacetic acid; ECL, Enhanced chemiluminescence; FeCl3, Ferric chloride; GSH, reduced glutathione; GSSG, oxidized glutathione; HPLC, high-performance liquid chromatography; i.p, intraperitoneal; MDA, malondialdehyde; NBT, nitro blue tetrazolium; NF-κB, nuclear factor-kappaB; NPD, 4- Nitro-o-phenylenediamine; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate buffer saline; PMS, Phenazine methosulfate; PVDF, polyvinylidene fluoride; ROS, reactive oxygen species; rpm, revolutions per minute; SA, sodium azide; SDS, sodium dodecyl sulphate; TA, top agar; TCA, trichloroacetic acid; WB, western blotting.
Aebi, H. (1984). Catalase in vitro. In Methods Enzymology. (USA: Academic Press), 105, 121–126. doi: 10.1016/S0076-6879(84)05016-3
Ahmad, I., Nawaz, S. A., Afza, N., Malik, A., Fatima, I., Khan, S. B., et al. (2005). Isolation of onosmins A and B, lipoxygenase inhibitors from Onosma hispida. Chem. Pharm. Bull. 53 (8), 907–910. doi: 10.1248/cpb.53.907
Ahmad, S., Khan, M. A., Ayaz, S., Ahamd, I. (2013). Antibacterial and antifungal studies of the crude extract and solvent fractions of Onosma khyberianum. Pharmacologica 4 (9), 525–528. doi: 10.5567/pharmacologia.2013.525.528
Albaqami, J., Myles, L. E., Tiriveedhi, V. (2018). The Effect of Onosma bracteatum in cancer cells. MOJ Bioequiv. Availab. 5 (6), 321–325. doi: 10.15406/mojbb.2018.05.00122
Allocati, N., Masulli, M., Di Ilio, C., Federici, L. (2018). Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 7 (1), 1–15. doi: 10.1038/s41389-017-0025-3
Alqahtani, A., Khan, Z., Alloghbi, A., S Said Ahmed, T., Ashraf, M., Hammouda, M. (2019). Hepatocellular carcinoma: Molecular mechanisms and targeted therapies. Medicina 55 (9):526. doi: 10.3390/medicina55090526
Aly, S. M., Fetaih, H. A., Hassanin, A. A., Abomughaid, M. M., Ismail, A. A. (2019). Protective Effects of Garlic and Cinnamon Oils on Hepatocellular Carcinoma in Albino Rats. Anal. Cell. Pathol. 2019, 1–15. doi: 10.1155/2019/9895485
Anderson, M. E. (1985). Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymology 113, 548–555. doi: 10.1016/S0076-6879(85)13073-9.
Aravalli, R. N., Steer, C. J., Cressman, E. N. (2008). Molecular mechanisms of hepatocellular carcinoma. Hepatology 48 (6), 2047–2063. doi: 10.1002/hep.22580
Asif, H. M., Hayee, A., Aslam, M. R., Ahmad, K., Hashmi, A. S. (2019). Dose-dependent, antidepressant, and anxiolytic effects of a traditional medicinal plant for the management of behavioral dysfunctions in animal models. Dose-Response 17 (4), 1–6. doi: 10.1177/1559325819891262
Banerjee, T., Valacchi, G., Ziboh, V. A., Van Der Vliet, A. (2002). Inhibition of TNFα-induced cyclooxygenase-2 expression by amentoflavone through suppression of NF-κB activation in A549 cells. Mol. Cell. Biochem. 238 (1–2), 105–110. doi: 10.1023/a:1019963222510
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 (1–2), 248–254. doi: 10.1016/0003-2697(76)90527-3
Brai, B., II, Adisa, R. A., Odetola, A. A. (2014). Hepatoprotective properties of aqueous leaf extract of Persea americana, mill (lauraceae)’avocado’against CCl 4-induced damage in rats. Afr. J. Tradit. Complement. Altern. Med. 11 (2), 237–244. doi: 10.4314/ajtcam.v11i2.2
Brücher, B. L., Lang, F., Jamall, I. S. (2019). NF-κB signaling and crosstalk during carcinogenesis. 4open 2, 13. doi: 10.1051/fopen/2019010
Canli, Ö., Nicolas, A. M., Gupta, J., Finkelmeier, F., Goncharova, O., Pesic, M., et al. (2017). Myeloid cell-derived reactive oxygen species induce epithelial mutagenesis. Cancer Cell 32 (6), 869–883. doi: 10.1016/j.ccell.2017.11.004
Carlberg, I. N. C. E. R., Mannervik, B. E. N. G. T. (1975). Purification and characterization of the flavoenzyme glutathione reductase from rat liver. J. Biol. Chem. 250 (14), 5475–5480.
Cha, C., DeMatteo, R. P. (2005). Molecular mechanisms in hepatocellular carcinoma development. Best Pract. Res. Clin. Gastroenterol. 19 (1), 25–37. doi: 10.1016/j.bpg.2004.11.005
Chao, W. W., Chen, S. J., Peng, H. C., Liao, J. W., Chou, S. T. (2019). Antioxidant Activity of Graptopetalum paraguayense E. Walther Leaf Extract Counteracts Oxidative Stress Induced by Ethanol and Carbon Tetrachloride Co-Induced Hepatotoxicity in Rats. Antioxidants 8 (8), 251. doi: 10.3390/antiox8080251
Che, C. T., Zhang, H. (2019). Plant natural products for human health. Int. J. Mol. Sci. 20, 1–4. doi: 10.3390/ijms20040830
Chibazakura, T., Kamachi, K., Ohara, M., Tane, S., Yoshikawa, H., Roberts, J. M. (2011). Cyclin A promotes S-phase entry via interaction with the replication licensing factor Mcm7. Mol. Cell. Biol. 31 (2), 248–255. doi: 10.1128/MCB.00630-10
Choi, S. J., Kim, M. R., Kim, S., II, Jeon, J. K. (2003). Microplate assay measurement of cytochrome P450-carbon monoxide complexes. BMB Rep. 36 (3), 332–335. doi: 10.5483/BMBRep.2003.36.3.332
Chu, C., Deng, J., Man, Y., Qu, Y. (2017). Green tea extracts epigallocatechin-3-gallate for different treatments. BioMed Res. Int. 2017, 1–9. doi: 10.1155/2017/5615647
Collins, A. R., Dušinská, M. (2002). “Oxidation of cellular DNA measured with the comet assay”, In: Oxidative stress biomarkers and antioxidant protocols. 186, 147–159. doi: 10.1385/1-59259-173-6:147
Devasagayam, T. P., Pushpendran, C. K., Eapen, J. (1983). Differences in lipid peroxidation of rat liver rough and smooth microsomes. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 750 (1), 91–97. doi: 10.1016/0005-2760(83)90207-2
Ebrahimzadeh, M. A., Nabavi, S. F., Nabavi, S. M., Eslami, B., Asgarirad, H. (2010). In vitro antioxidant and free radical scavenging activity of Leonurus cardiaca subsp. Persicus, Grammosciadium platycarpum and Onosma demawendicum. Afr. J. Biotechnol. 9 (51), 8865–8871. doi: 10.5897/AJB10.782
Eltahir, H. M., Fawzy, M. A., Mohamed, E. M., Alrehany, M. A., Shehata, A. M., Abouzied, M. M. (2020). Antioxidant, anti-inflammatory and anti-fibrotic effects of Boswellia serrate gum resin in CCl4-induced hepatotoxicity. Exp. Ther. Med. 19 (2), 1313–1321. doi: 10.3892/etm.2019.8353
Fareed, S., Siddiqui, H. H., Haque, S. E., Khalid, M., Akhtar, J. (2012). Psychoimmunomodulatory Effects of Onosma bracteatum Wall. (Gaozaban) on Stress Model in Sprague Dawley Rats. J. Clin. Diagn. Res. 6 (7), 1356–1360.
Farooq, U., Pan, Y., Disasa, D., Qi, J. (2019). Novel Anti-Aging Benzoquinone Derivatives from Onosma bracteatum Wall. Molecules 24 (7), 1428. doi: 10.3390/molecules24071428
Ganaie, M. A., Khan, T. H., Siddiqui, N. A., Ansari, M. N. (2015). Ameliorative effect of methanol extract of Rumex vesicarius on CCl4-induced liver damage in Wistar albino rats. Pharm. Biol. 53 (8), 1163–1167. doi: 10.3109/13880209.2014.967782
Gao, X., Fan, L., Li, H., Li, J., Liu, X., Sun, R., et al. (2016). Hepatic injury is associated with cell cycle arrest and apoptosis with alteration of cyclin A and D1 in ammonium chloride-induced hyperammonemic rats. Exp. Ther. Med. 11 (2), 427–434. doi: 10.3892/etm.2015.2931
Garner, R. C., Miller, E. C., Miller, J. A. (1972). Liver microsomal metabolism of aflatoxin B1 to a reactive derivative toxic to Salmonella typhimurium TA 1530. Cancer Res. 32 (10), 2058–2066.
Gee, J. B. L., Vassallo, C. L., Bell, P., Kaskin, J., Basford, R. E., Field, J. B. (1970). Catalase-dependent peroxidative metabolism in the alveolar macrophage during phagocytosis. J. Clin. Invest. 49 (6), 1280–1287. doi: 10.1172/JCI106340
Green Solvents & Bio Solvents - Global Market Outlook (2017-2026) (2018). Available at: https://www.researchandmarkets.com/reports/4655796/green-solvents-and-biosolvents-global-market (Accessed May 05,2020).
Gu, X., Manautou, J. E. (2012). Molecular mechanisms underlying chemical liver injury. Expert Rev. Mol. Med. 14, 1–21. doi: 10.1017/S1462399411002110
Habig, W. H., Pabst, M. J., Jakoby, W. B. (1974). Glutathione S-transferase—the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249 (22), 7130–7139.
Hafez, M. M., Al-Harbi, N. O., Al-Hoshani, A. R., Al-Hosaini, K. A., Al Shrari, S. D., Al Rejaie, S. S., et al. (2015). Hepato-protective effect of rutin via IL-6/STAT3 pathway in CCl4-induced hepatotoxicity in rats. Biol. Res. 48 (1), 1–10. doi: 10.1186/s40659-015-0022-y
Halliwell, B., Gutteridge, J. M. C. (1989). Free radicals in biology and medicine. 2nd edn (Oxford, United Kingdom: Clarendon Press).
Huang, Z., Jing, X., Sheng, Y., Zhang, J., Hao, Z., Wang, Z., et al. (2019). (-)-Epicatechin attenuates hepatic sinusoidal obstruction syndrome by inhibiting liver oxidative and inflammatory injury. Redox Biol. 22, 101117. doi: 10.1016/j.redox.2019.101117
Imran, H., ur Rahman, A., Sohail, T., Taqvi, S., II, Yaqeen, Z. (2018). Onosma bracteatum wall: A Potent analgesic agent. Bangladesh J. Med. Sci. 17 (1), 36–41. doi: 10.3329/bjms.v17i1.35276
Jiang, Z. Y., Hunt, J. V., Wolff, S. P. (1992). Ferrous ion oxidation in the presence of xylenol orange for detection of lipid hydroperoxide in low density lipoprotein. Anal. Biochem. 202 (2), 384–389. doi: 10.1016/0003-2697(92)90122-N
Josan, S., Billingsley, K., Orduna, J., Park, J. M., Luong, R., Yu, L., et al. (2015). Assessing inflammatory liver injury in an acute CCl4 model using dynamic 3D metabolic imaging of hyperpolarized [1-13C] pyruvate. NMR Biomed. 28 (12), 1671–1677. doi: 10.1002/nbm.3431
Kandemir, A., Türkmen, Z. (2008). The Flora of Üzümlü-Sakaltutan (Erzincan-Gümüşhane). Turkish J. Bot. 32 (4), 265–304.
Karakus, E., Karadeniz, A., Simsek, N., Can, I., Kara, A., Yildirim, S., et al. (2011). Protective effect of Panax ginseng against serum biochemical changes and apoptosis in liver of rats treated with carbon tetrachloride (CCl4). J. Hazard. Mater. 195, 208–213. doi: 10.1016/j.jhazmat.2011.08.027
Kaur, S. J., Grover, I. S., Kumar, S. (2000). Modulatory effects of a tannin fraction isolated from Terminalia arjuna on the genotoxicity of mutagens in Salmonella typhimurium. Food Chem. Toxicol. 38 (12), 1113–1119. doi: 10.1016/s0278-6915(00)00104-6
Kaur, S., Grover, I. S., Kumar, S. (2001). Antimutagenic potential of extracts isolated from Terminalia arjuna. J. Environ. Pathol. Toxicol. Oncol. 20 (1), 1–6. doi: 10.1615/JEnvironPatholToxicolOncol.v20.i1.20
Kaur, S., Kumar, M., Kaur, P., Kaur, V., Kaur, S. (2016). Modulatory effects of Cassia fistula fruits against free radicals and genotoxicity of mutagens. Food Chem. Toxicol. 98, 220–231. doi: 10.1016/j.fct.2016.10.027
Kaur, V., Kumar, M., Kaur, P., Kaur, S., Singh, A. P., Kaur, S. (2017). Hepatoprotective activity of Butea monosperma bark against thioacetamide-induced liver injury in rats. Biomed. Pharmacother. 89, 332–341. doi: 10.1016/j.biopha.2017.01.165
Kaur, S., Sharma, D., Singh, A. P., Kaur, S. (2019). Amelioration of hepatic function, oxidative stress, and histopathologic damages by Cassia fistula L. fraction in thioacetamide-induced liver toxicity. Environ. Sci. Pollut. Res. 26 (29), 29930–29945. doi: 10.1007/s11356-019-06158-y
Kaur, P., Mehta, R. G., Singh, B., Arora, S. (2019). Development of aqueous-based multi-herbal combination using principal component analysis and its functional significance in HepG2 cells. BMC Complement. Altern. Med. 19 (1), 18. doi: 10.1186/s12906-019-2432-9
Khan, R. A., Khan, M. R., Sahreen, S. (2012). CCl 4-induced hepatotoxicity: protective effect of rutin on p53, CYP2E1 and the antioxidative status in rat. BMC Complement. Altern. Med. 12 (1), 178. doi: 10.1186/1472-6882-12-178
Kumar, N., Kumar, R., Kishore, K. (2013). Onosma L.: A review of phytochemistry and ethnopharmacology. Pharmacognosy Rev. 7 (14), 140. doi: 10.4103/0973-7847.120513
Laouar, A., Klibet, F., Bourogaa, E., Benamara, A., Boumendjel, A., Chefrour, A., et al. (2017). Potential antioxidant properties and hepatoprotective effects of Juniperus phoenicea berries against CCl4 induced hepatic damage in rats. Asian Pac. J. Trop. Med. 10 (3), 263–269. doi: 10.1016/j.apjtm.2017.03.005
Lee, J. C., Kim, H. R., Kim, J., Jang, Y. S. (2002). Antioxidant property of an ethanol extract of the stem of Opuntia ficus-indica var. saboten. J. Agric. Food Chem. 50 (22), 6490–6496. doi: 10.1021/jf020388c
Lerdwanangkun, P., Wonganan, P., Storer, R. J., Limpanasithikul, W. (2019). Combined effects of celecoxib and cepharanthine on human colorectal cancer cells in vitro. J. Appl. Pharm. Sci. 9 (04), 117–125. doi: 10.7324/JAPS.2019.90415
Liu, J., Lu, J. F., Wen, X. Y., Kan, J., Jin, C. H. (2015). Antioxidant and protective effect of inulin and catechin grafted inulin against CCl4-induced liver injury. Int. J. Biol. Macromol. 72, 1479–1484. doi: 10.1016/j.ijbiomac.2014.09.066
Liu, Z., Ren, Z., Zhang, J., Chuang, C. C., Kandaswamy, E., Zhou, T., et al. (2018). Role of ROS and nutritional antioxidants in human diseases. Front. Physiol. 9, 477. doi: 10.3389/fphys.2018.00477
Maron, D. M., Ames, B. N. (1983). Revised methods for the Salmonella mutagenicity test. Mutat. Research/Environmental Mutagenesis Relat. Subj. 113 (3-4), 173–215. doi: 10.1016/0165-1161(83)90010-9
Mehta, R. G., Murillo, G., Naithani, R., Peng, X. (2010). Cancer chemoprevention by natural products: how far have we come? Pharm. Res. 27 (6), 950–961. doi: 10.1007/s11095-010-0085-y
Menghani, E., Sudhanshu, R. N., Mittal, S. (2011). Free radical scavenging capacity and antioxidant activity of Onosma bracteatum. Int. J. Pharm. Res. Dev. 4, 16–20.
Mihara, K., Sato, R. (1972). Partial purification of NADH-cytochrome b5 reductase from rabbit liver microsomes with detergents and its properties. J. Biochem. 71 (4), 725–735. doi: 10.1093/oxfordjournals.jbchem.a129819
Misra, H. P., Fridovich, I. (1972). The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J. Biol. Chem. 247 (10), 3170–3175.
Naz, S., Ahmad, S., Rasool, S. A., Sayeed, S. A., Siddiqi, R. (2006). Antibacterial activity directed isolation of compounds from Onosma hispidum. Microbiol. Res. 161 (1), 43–48. doi: 10.1016/j.micres.2005.05.001
Nishikimi, M., Rao, N. A., Yagi, K. (1972). The occurrence of superoxide anion in the reaction of reduced phenazine methosulfate and molecular oxygen. Biochem. Biophys. Res. Commun. 46 (2), 849–854. doi: 10.1016/S0006-291X(72)80218-3
Omura, T., Sato, R. (1964). The carbon monoxide-binding pigment of liver microsomes I. Evidence for its hemoprotein nature. J. Biol. Chem. 239 (7), 2370–2378.
Ozer, M. S., Kirkan, B., Sarikurkcu, C., Cengiz, M., Ceylan, O., Atılgan, N., et al. (2018). Onosma heterophyllum: Phenolic composition, enzyme inhibitory and antioxidant activities. Ind. Crops Prod. 111, 179–184. doi: 10.1016/j.indcrop.2017.10.026
Pan, Y., Long, X., Yi, R., Zhao, X. (2018). Polyphenols in Liubao tea can prevent CCl4-induced hepatic damage in mice through its antioxidant capacities. Nutrients 10 (9), 1280. doi: 10.3390/nu10091280
Pizzino, G., Irrera, N., Cucinotta, M., Pallio, G., Mannino, F., Arcoraci, V., et al. (2017). Oxidative stress: harms and benefits for human health. Oxid. Med. Cell. Longevity 2017, 1–13. doi: 10.1155/2017/8416763
Rahal, A., Kumar, A., Singh, V., Yadav, B., Tiwari, R., Chakraborty, S., et al. (2014). Oxidative stress, prooxidants, and antioxidants: the interplay. BioMed. Res. Int. 1–19. doi: 10.1155/2014/761264
Riedl, H., Binzet, R., Orcan, N. (2004). A new species of Onosma (Boraginaceae–Lithospermeae) from southern Turkey. Edinburgh J. Bot. 61 (2-3), 127–130. doi: 10.1017/S0960428605000211
Ritesh, K. R., Suganya, A., Dileepkumar, H. V., Rajashekar, Y., Shivanandappa, T. (2015). A single acute hepatotoxic dose of CCl4 causes oxidative stress in the rat brain. Toxicol. Rep. 2, 891–895. doi: 10.1016/j.toxrep.2015.05.012
Robin, Arora, S., Vig, A. P. (2015). Inhibition of DNA oxidative damage and antimutagenic activity by dichloromethane extract of Brassica rapa var. rapa L. seeds. Ind. Crops Prod. 74, 585–591. doi: 10.1016/j.indcrop.2015.05.038
Rodríguez-García, C., Sánchez-Quesada, C., Gaforio, J. J. (2019). Dietary flavonoids as cancer chemopreventive agents: an updated review of human studies. Antioxidants 8 (5), 137. doi: 10.3390/antiox8050137
Sabir, S. M., Rocha, J. B. T., Boligon, A. A., Athayde, M. L. (2017). Hepatoprotective activity and phenolic profile of Zanthoxylum alatum Roxb. fruit extract. Pakistan J. Pharm. Sci. 30 (5), 1551–1556.
Saravanakumar, K., Sarikurkcu, C., Sarikurkcu, R. T., Wang, M. H. (2019). A comparative study on the phenolic composition, antioxidant and enzyme inhibition activities of two endemic Onosma species. Ind. Crops Prod. 142, 111878. doi: 10.1016/j.indcrop.2019.111878
Seufi, A. M., Ibrahim, S. S., Elmaghraby, T. K., Hafez, E. E. (2009). Preventive effect of the flavonoid, quercetin, on hepatic cancer in rats via oxidant/antioxidant activity: molecular and histological evidences. J. Exp. Clin. Cancer Res. 28 (1), 80. doi: 10.1186/1756-9966-28-80
Shah, M. D., Gnanaraj, C., Haque, A. E., Iqbal, M. (2015). Antioxidative and chemopreventive effects of Nephrolepis biserrata against carbon tetrachloride (CCl4)-induced oxidative stress and hepatic dysfunction in rats. Pharm. Biol. 53 (1), 31–39. doi: 10.3109/13880209.2014.909502
Sinha, M., Das, D. K., Manna, K., Datta, S., Ray, T., Sil, A. K., et al. (2012). Epicatechin ameliorates ionising radiation-induced oxidative stress in mouse liver. Free Radical Res. 46 (7), 842–849. doi: 10.3109/10715762.2012.684245
Snezhkina, A. V., Kudryavtseva, A. V., Kardymon, O. L., Savvateeva, M. V., Melnikova, N. V., Krasnov, G. S., et al. (2019). ROS Generation and antioxidant defense systems in normal and malignant cells. Oxid. Med. Cell. Longev. 2019, 1–17. doi: 10.1155/2019/6175804
Sun, B., Karin, M. (2008). NF-κB signaling, liver disease and hepatoprotective agents. Oncogene 27 (48), 6228–6244. doi: 10.1038/onc.2008.300
Surh, Y. J. (2003). Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 3 (10), 768–780. doi: 10.1038/nrc1189
Ved, D. K., Sureshchandra, S. T., Barve, V., Srinivas, V., Sangeetha, S., Ravikumar, K., et al. (2016). (envis. frlht. org/frlhtenvis. nic. in). (Bengaluru: FRLHT’s ENVIS Centre on Medicinal Plants), 1475–1484.
Wang, Y., Tang, C., Zhang, H. (2015). Hepatoprotective effects of kaempferol 3-O-rutinoside and kaempferol 3-O-glucoside from Carthamus tinctorius L. @ on CCl4-induced oxidative liver injury in mice. J. Food Drug Anal. 23 (2), 310–317. doi: 10.1016/j.jfda.2014.10.002
Wu, J. C., Lai, C. S., Tsai, M. L., Ho, C. T., Wang, Y. J., Pan, M. H. (2017). Chemopreventive effect of natural dietary compounds on xenobiotic-induced toxicity. J. Food Drug Anal. 25 (1), 176–186. doi: 10.1016/j.jfda.2016.10.019
Ying, J., Li, H., Yu, J., Ng, K. M., Poon, F. F., Wong, S. C. C., et al. (2008). WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/β-catenin signaling, and is frequently methylated in colorectal cancer. Clin. Cancer Res. 14 (1), 55–61. doi: 10.1158/1078-0432.CCR-07-1644
Younis, T., Rasul, A., Jabeen, F., Hussain, G., Altaf, J., Jafri, L., et al. (2018). Ameliorating role of methanolic leaves extract of Fraxinus xanthoxyloides against CCl4-challanged nephrotoxicity in rats. Pakistan J. Pharm. Sci. 31, 1475–1484.
Zamzami, M. A., Baothman, O. A., Samy, F., Abo-Golayel, M. K. (2019). Amelioration of CCl4-Induced Hepatotoxicity in Rabbits by Lepidium sativum Seeds. Evidence-Based Complement. Altern. Med. 2019, 1–17. doi: 10.1155/2019/5947234
Zang, Y., Hashimoto, S., Yu, C., Igarashi, K. (2018). Protective effects of dietary kaempferol glycoside components from unripe soybean (Edamame, Glycine max L. Merrill.’Jindai’) leaves and their serous metabolite on carbon tetrachloride-induced liver injury mice. J. Food Sci. Technol. 55 (11), 4515–4521. doi: 10.1007/s13197-018-3385-6
Keywords: antimutagenicity, apoptosis, carcinogenesis, hepatoprotective effect, Onosma bracteata, phytoconstituents
Citation: Kumar A, Kaur V, Pandit K, Tuli HS, Sak K, Jain SK and Kaur S (2020) Antioxidant Phytoconstituents From Onosma bracteata Wall. (Boraginaceae) Ameliorate the CCl4 Induced Hepatic Damage: In Vivo Study in Male Wistar Rats. Front. Pharmacol. 11:1301. doi: 10.3389/fphar.2020.01301
Received: 02 June 2020; Accepted: 05 August 2020;
Published: 21 August 2020.
Edited by:
Yan Xu, Cleveland State University, United StatesReviewed by:
Wentzel Christoffel Gelderblom, Cape Peninsula University of Technology, South AfricaCopyright © 2020 Kumar, Kaur, Pandit, Tuli, Sak, Jain and Kaur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satwinderjeet Kaur, c2F0d2luZGVyamVldC5ib3RlbnZAZ25kdS5hYy5pbg==; c2prYXVyMjAxMUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.