
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Pharmacol. , 16 July 2020
Sec. Drugs Outcomes Research and Policies
Volume 11 - 2020 | https://doi.org/10.3389/fphar.2020.01028
Aim: To perform a systematic review on the application of artificial intelligence (AI) based knowledge discovery techniques in pharmacoepidemiology.
Study Eligibility Criteria: Clinical trials, meta-analyses, narrative/systematic review, and observational studies using (or mentioning articles using) artificial intelligence techniques were eligible. Articles without a full text available in the English language were excluded.
Data Sources: Articles recorded from 1950/01/01 to 2019/05/06 in Ovid MEDLINE were screened.
Participants: Studies including humans (real or simulated) exposed to a drug.
Results: In total, 72 original articles and 5 reviews were identified via Ovid MEDLINE. Twenty different knowledge discovery methods were identified, mainly from the area of machine learning (66/72; 91.7%). Classification/regression (44/72; 61.1%), classification/regression + model optimization (13/72; 18.0%), and classification/regression + features selection (12/72; 16.7%) were the three most frequent tasks in reviewed literature that machine learning methods has been applied to solve. The top three used techniques were artificial neural networks, random forest, and support vector machines models.
Conclusions: The use of knowledge discovery techniques of artificial intelligence techniques has increased exponentially over the years covering numerous sub-topics of pharmacoepidemiology.
Systematic Review Registration: Systematic review registration number in PROSPERO: CRD42019136552.
By definition, artificial intelligence is “the theory and development of computer systems able to perform tasks normally requiring human intelligence” (Oxford, 2019). The British logician Alan Turing reports the earliest work in the field in the second quarter of the 20th century. In 1935, Alan Turing proposed the basic concept of an intelligent machine commonly known as universal Turing Machine. He further elaborated his vision in 1947 by describing computer intelligence as “a machine that can learn from experience” (Turing, 1937). As human intelligence is a combination of diverse abilities (i.e., learning, reasoning, problem solving, perception, and using language), artificial (or machine) intelligence is also a composite of methods and techniques from different disciplines of science and engineering to assimilate them in machines (Figure 1). It is worthy to note that artificial intelligence is commonly confused with machine learning. Learning (Machine/Deep Learning) is a subfield in artificial intelligence that deals with methods and techniques to assimilate learning abilities in machines. One reason of machine (or deep) learning emerging as a dominant sub-field of artificial intelligence is the considerable advancement in computer technologies and impressive achievements in learning algorithms. By definition, machine learning is a multidisciplinary field, which involves methods and techniques from mathematics, statistics, and computer science to learn from experiences (historical data) with respect to some tasks (i.e., the nature of the problem), and measure the performance (performance matrix) and improve it (re-enforcement) (Michie et al., 1994). Today, machine learning algorithms based on the principal of reinforcement learning not only enhances the learning abilities of the machine but also complement the other aspects of intelligence such as appropriate reasoning, efficient problem solving, and factual perception. Traditionally, experimental design, observational data analysis (statistical data analysis), and computer science have always been integral constituents of research in biomedical sciences. However, in the past decade the sprightly ascent of machine learning based knowledge discovery methods in artificial intelligence sparked this trend conspicuously. For numerous medical fields, the contribution of knowledge discovery techniques in artificial intelligence have been described extensively. However, their level of infusion to pharmacoepidemiology is unknown. Acording to the international society of pharmacoepidemiology, this discipline may be defined as “the study of the utilization and effects of drugs in large numbers of people.” Considering this gap in knowledge, the objective of this systematic review is to provide an overview of the use of knowledge discovery techniques of artificial intelligence in pharmacoepidemiology.
An independent author (MS) registered the protocol of the systematic review in the PROSPERO International Prospective Register of Systematic Reviews database (identifier CRD42019136552).
We evaluated observational studies, meta-analyses, and clinical trials using artificial intelligence techniques and for which the exposure or the outcome of the study was a drug. Drugs include any substance approved on the pharmaceutical market having an anatomical therapeutic chemical classification code as proposed by the World Health Organization (WHO). Only studies for which the full text was available in the English language were considered as eligible. Abstracts sent to international or national conferences, letters to the editor, and case reports/series were considered ineligible along with articles evaluating natural language processing techniques. Reviews describing the use of natural language processing techniques are available elsewhere (Dreisbach et al., 2019). The reference list of narrative and systematic reviews included with our MEDLINE query were further screened for undetected records.
The main outcome was the frequency of studies published per year from January 1950 to May 2019, a narrative overview of their findings, and a lay description of knowledge discovery methods of artificial intelligence that were used. Secondary outcomes included the evaluation of 1) the medical field in which the aforementioned techniques were used and 2) the number and the type of artificial intelligence techniques that were used. Additionally, we assessed the frequency distribution of articles by 3) the study design; 4) type of data sources (e.g. primary/secondary or simulated); 5) the specific data source; 6) the purpose for using artificial intelligence based knowledge discovery techniques, and 7) the level of evidence provided by the study.
The purpose of using artificial intelligence based knowledge discovery techniques (outcome no. 6) was categorized as follows: 1) To predict clinical response following a pharmacological treatment; 2) To predict the needed dosage given the patient’s characteristics; 3) To predict the occurrence/severity of adverse drug reactions; 4) To predict diagnosis leading to a drug prescription; 5) To predict drug consumption, 6) To predict the propensity score; 7) To predict drug-induced lengths of stay in hospital; 8) To predict adherence to pharmacological treatments; 9) To optimize treatment regimen; 10) To identify subpopulation more at risk of drug inefficacy, and 11) To predict drug-drug interactions.
Ovid MEDLINE (from January 1950 to May 2019) was searched along with the references listed in the reviews identified with our research query (Supplementary Table 1). Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist is provided in Supplementary Table 2.
In the first screening procedure, titles and abstracts of retrieved record were screened by two independent researchers (MS and DL) for obvious exclusions. All articles that were considered eligible at the first screening procedure underwent a full-text evaluation. If disagreements arose during the two steps evaluation process, it was resolved by consensus.
A data extraction form was developed for this systematic review and it is shown in Supplementary Table 3. The scale proposed by Merlin et al. (2009) was used to establish the level of evidence of each study.
In total, 6,470 and 240 records were identified in Ovid MEDLINE and in the reference list of reviews retrieved with the search query, respectively. After title/abstract screening, 6,633 records were eliminated because of ineligibility and 77 articles (72 original articles and 5 reviews) underwent a full-text evaluation. The 77 articles were considered eligible to be included in this systematic review. The PRISMA flowchart of the selection process is shown in Figure 2 and the PRISMA checklist has been provided in Supplementary Table 2.
We observed increased use of artificial intelligence based knowledge discovery techniques in pharmacoepidemiology over the years as seen in Figure 3. In all, 17 medical fields were identified. The top four most prevalent medical fields were pure pharmacoepidemiology (16/72; 22.2%), oncology (15/72; 20.8%), infective medicine (8/72; 11.1%), and neurology (6/72; 8.3%) (Supplementary Table 4).
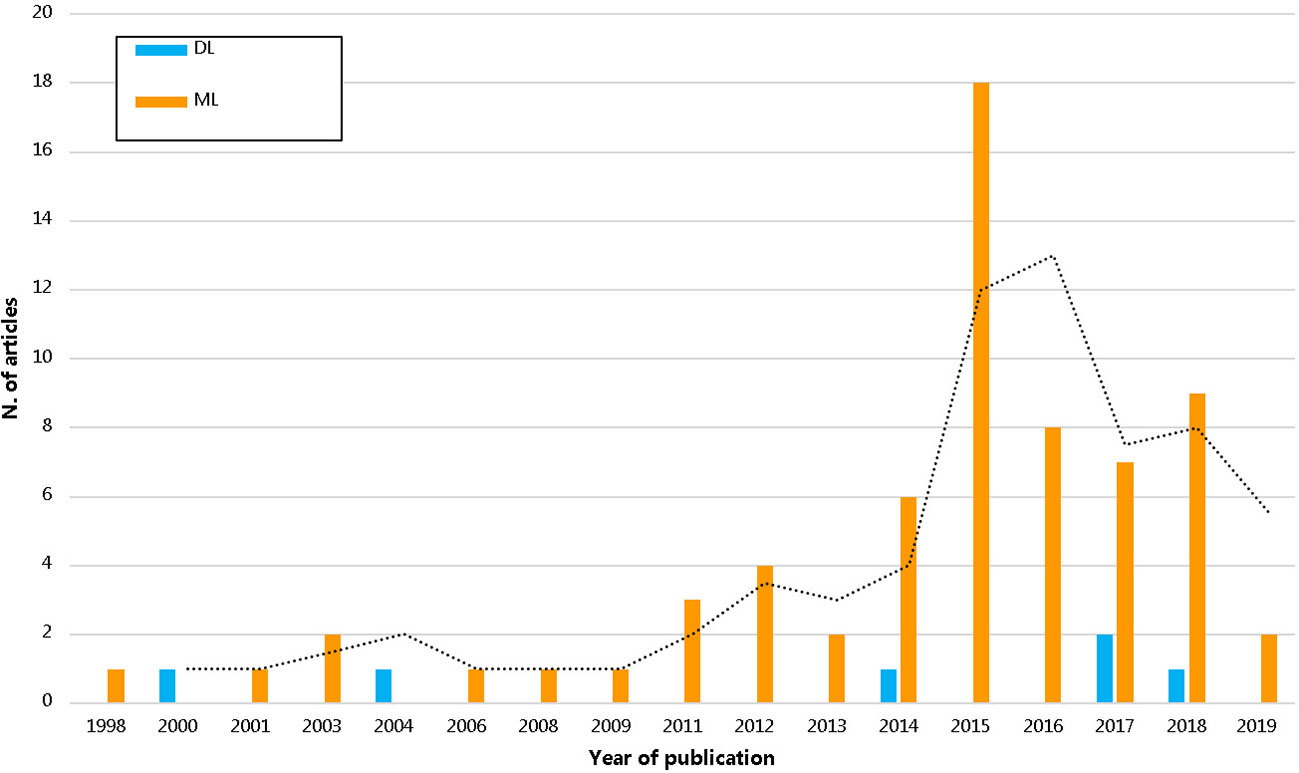
Figure 3 The trend of pharmacoepidemiological studies using artificial intelligence by years. DL, deep learning; ML, machine learning.
Fifty-five out of 72 articles (76.4%) used artificial intelligence techniques in the setting of a cohort study (Supplementary Figure 1). Most of the studies provided a medium-low level of evidence of III-3 (4/72; 5.6%), III-2 (49/72; 68.1%), and III-1 (16/72; 22.2%) while, a few articles provided a level of evidence of II (3/72; 4.1%).
In the 72 selected articles, the data sources included electronic health records (36.1%), ad-hoc databases from clinical studies (31.9%), administrative databases (29.2%), survey (1.4%), and simulated data (1.4%). The data sources were mainly secondary (59.8%) and primary sources (31.8%). Only in two articles (2.8%), researchers used both secondary sources and simulated data. Analogously, only in two articles (2.8%), researchers used simulated data (2.8%). The specific data sources used in selected articles are provided in Supplementary Table 5.
A narrative overview of the articles is provided in Table 1. The lay description of the knowledge discovery techniques that were used in retrieved articles is provided in Lay Description of the Knowledge Discovery Techniques of Artificial Intelligence Used in Pharmacoepidemiology.
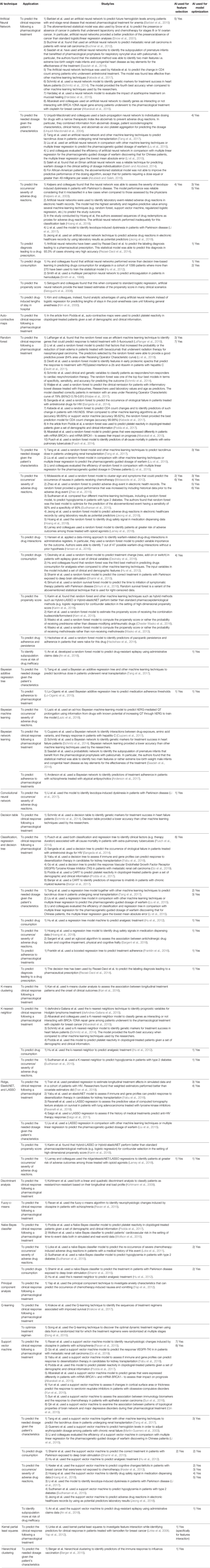
Table 1 Main applications of knowledge discovery methods of artificial intelligence (AI) in pharmacoepidemiology.
The main applications of artificial intelligence based knowledge discovery techniques in pharmacoepidemiology were classification/regression (44/72; 61.1%), classification/regression + model optimization (13/72; 18.0%), classification/regression + features selection (12/72; 16.7%), classification/regression + features interaction (1/72; 1.4%), and classification/regression + features selection + model optimization (2/72; 2.8%).
Classification and regression are two different types of predictive modeling where in the former the prediction is a label (class) whilst in the latter it is a quantity. For example, in classification, a patient can be classified as belonging to one of two classes: “having the disease” and “not having the disease” given a set of information from his/her medical history. In regression, instead, the researcher may try to predict the cholesterol level of a patient based on patient’s weight. Feature (variable) selection is a type of modeling in which the researcher constructs and trains statistical models by selecting relevant features to reduce overfitting and training time, and to improve accuracy. The main reason for feature selection is to improve the model performance that may be negatively impacted with the inclusion of partially relevant or irrelevant features as this leads to overfitting. Conversely, incorrectly excluding variables may lead to a bias in the model prediction (Heinze et al., 2018). Feature interaction, instead, is said to be relevant when the impact of any feature changes based on the levels of the other features hence rendering an additive model unsatisfactory. For a model with the lowest order interaction, the prediction is calculated based on a constant, a value for the first feature, a value for the second feature, and finally, the value for the interaction of the two features (Molnar, 2018).
In the retrieved articles, twenty different knowledge discovery techniques were used. Multiple techniques were used in the same article leading for a total of 122 applications. Random forest (30/122; 24.6%), artificial neural networks (22/122; 18.0%), and support vector machine (19/122; 15.6%) models were the three most used techniques (Table 1, Supplementary Figure 2). The top six purposes of using artificial intelligence techniques were to predict: 1) the clinical response following a pharmacological treatment (42.7%); 2) the occurrence/severity of adverse drug reactions (19.4%); 3) the needed dosage given the patient’s characteristics (14.5%); 4) drug consumption (9.7%), and 5) propensity score (4.8%) (Table 1).
An artificial neural network is a machine learning technique that tries to mimic neurons’ mechanisms of processing signals and is applicable to solve complex knowledge extraction tasks. In artificial neural networks, the input signals are characterized by the features variables (e.g., covariates) where each gets a different weight according to its importance in the knowledge extraction task (e.g., having or not having an adverse event). In its simplest form, as in the case of single-layer network, features represent the input nodes of the artificial neural networks, and all the input nodes are then arranged in one layer (e.g., skip-layer units) while the outcome represents the output node (Zhang, 2016a). Artificial neural networks can be split into two broad categories based on network topology, Feedforward and Feedback Artificial Neural Networks. The choice and applicability of the different network topology depend on the nature of problem. Convolutional Neural Network based on the principal of feedforward is well suited for the problems related to image analysis whereas problems such as speech recognition are better suited for the recurrent neural networks based on the feedback network topology. For this reason, the model has been used widely for computer vision task such as the automatic identification of patterns in medical images (Yamashita et al., 2018). Among studies selected in this systematic review, the artificial neural network was primarily used for Auto Contractive Maps (ACM). The ACM differs from the other artificial neural networks because it is able to learn from data without randomizing weight for each variable. In this technique, the weight of each variable is calculated based on their convergence criterion when all the output nodes become null. In particular, the model uses a data-driven mechanism to set-up weights based on the Euclidean space given the topological properties of each variable.
BART is a technique that combines several Bayesian regression trees and starts by building an individual regression tree for each variable that are subsequently summed. By definition, the BART model is flexible and able to evaluate non-linear effects and multi-way interactions automatically. For each node of the regression tree, the levels of the variable are separated into two sub-groups based on their predictive power for the outcome. By definition, Bayesian additive regression trees are able to capture additive effects among variables (Hernandez et al., 2018).
A Bayesian network is a special machine learning technique used in causal inference. Causal inference determines the probability of an outcome using evidence from prior observations. The model use prior knowledge from a causal diagram (direct acyclic graph) which describes the underlying joint probability distribution among variables with conditional dependencies (Sesen et al., 2013). The model incorporates prior knowledge about the topic and then learns from the data how the variables interact with each other in the network.
In the case of high dimensional datasets where the number of variables is bigger than the number of observations, least squares method (linear model) cannot be used. In such a scenario, the commonly used approach is to reduce dimensionality through regularization. In such a case, penalized regression can be the preferred choice to perform feature selection. In this case the coefficients are obtained through the minimization of the penalized residual sum of squares where the penalty is imposed on the regression coefficients and used as a tuning parameter. If the penalty is imposed on the sum of the squared coefficents, penalized regression is called the Ridge regression. If the penalty is imposed on the sum of the absolute values of the coeeficients, we have the Least Absolute Shrinkage and Selection Operator (LASSO) regression. The Elastic Net imposes the penalty on the combination of the both sum of the squared and absolute values of the coefficients. LASSO forces (shrinks) the coefficients of all the variables with a poor contribution to the prediction to be zero and, therefore, these variables are excluded from the final model. ElasticNET, instead, shrinks some of the coefficient towards zero but also preserve some of the variables with medium-low predictive power providing a less aggressive feature selection strategy (Kyung et al., 2010).
The naïve Bayes classifier is an artificial intelligence technique used for classification that relies on the Bayesian classification (Zhang, 2016c) based on the following principles: given the hypothesis h, a set of data D and a probability measure P, we can define P(h) as the probability that h is true. P(h) represents the prior knowledge on h; P(D) is the probability that the data in D will be observed; P(D|h) is the probability of observing the set D given that h is true; and P(h|D) is the probability that h true for a given data D, i.e., posterior probability of h. The theorem can be formalized as following: P(D|h) = P(D|h) P(h)/P(D). The theorem allows for calculating the posterior probability of h given D starting from the knowledge of the prior probabilities of D, and the conditional probability of D given h. Consequently, it is possible to calculate the maximum posterior hypothesis (MAP), or rather the most probable hypothesis of h given D. The naïve Bayes algorithm classifies the new data by assigning the most probable target value, or rather the MAP value, given the sequence of attributes (a1, a2,…, an) that describe the new data.
A discriminant analysis is used to group observations based on the similarities of their features. Suppose we have g groups D1, D2,…, Dg from which the observations are coming from. The objective of the discriminant analysis is to categorize an individual in one of these groups given a set of observations, x1, x2, … … … … ,xp (where p is the number of variables). For example, we want to discriminate between patients with or without diabetes mellitus type 2 (g = 2) based on observations of glycaemia, body weight, and age (p = 3) (in this case x1 = blood glucose concentration, x2 = body weight, and x3 = age). For the specific characteristics of the individuals of a group Di, we can compute a probability that describes the likelihood of belonging to the group i, given the observed variables. Linear discriminant analysis is a classification technique that uses linear combinations of features to categorize observations in groups. The model requires that the data are normally distributed, homoscedastic or have an identical covariate matrix among classes. Quadratic discriminant analysis, instead, relaxes the last assumption or rather does not require that classes have the same covariate matrix.
The principal component analysis is a technique that reduces the dimensionality of quantitative variables in the dataset through linear combinations of these variables, also known as the principal components. The principal components are selected so that the first principal component (first linear combination) has the highest variance, the second principal component has the second highest variance but also uncorrelated with the first principal component and so on. When the original variables are highly correlated, only a few principal components are retained as they would still explain a large portion of the variation in the data.
Q-learning is a reinforcement-learning algorithm used to optimize the solution of discrete time stochastic processes. The technique is “model-free” and “goal-oriented.” It provides at each stage of the process the optimal set of decisions to maximize a long-term reward. The algorithm is used in pharmacoepidemiology considering that many therapeutic processes are a set of actions that change over time and may be associated with a clinical outcome (i.e., a set of drugs administrated over time and the occurrence of an adverse drug reaction) (Song et al., 2015; Krakow et al., 2017).
Support vector machine (SVM) is a method used for classification. The SVM algorithm has three core components: i) A line; or a hyperplane as the “boundary” that separates data points, ii) A margin; i.e., the distance between the groups of data that are close to each other, and iii) Support vectors; i.e., the vectors to separate data points located within the margin of a hyperplane. In the presence of linearly separable data points, the algorithm finds among all straight lines or hyperplanes that separate the different groups those that maximize the margin value. In fact, a straight line or a hyperplane with maximum margin value allows minimizing the classification error. In non-linear classification, it is necessary to operate in two separate phases. In the first phase, data points are mapped on a large dimensional space to make them separable in a linear manner. Subsequently, the algorithm searches for a line or a hyperplane that maximizes the size of the margin, given that the instances are linearly separable. The support vector machine usually uses data transformations to transform a non-linear into a linear relationship of variables to simplify the delineation of boundaries. These data transformations usually use the kernel function (Noble, 2006). Sequential minimal optimization, instead, is an algorithm used to train the support vector machine (Platt, 1998).
A classification and regression tree (CART) is a model constructed by recursively partitioning variables based on their predictive power for the study outcome. The model starts by identifying the variable with the strongest predictive power. This variable is included in the model as the root node or rather the parent node from which all other splitting procedures will be performed. In the regression tree, each node represents a variable. The decision tree split each node into two levels to make them have the best separation for maximizing their predictive power of the variable. With this model, the user does not need to make any assumptions about the statistical distribution of the data (e.g., normality assumption). The model can handle both categorical and numerical data (Kingsford and Salzberg, 2008). The boosted regression tree incorporates the important advantages of tree‐based method described above. However, it overcomes the inclusion of a single tree by including boosting (a combination of simple models to improve the overall predicting performance) (Elith et al., 2008).
A decision table is a hierarchical (rule) table used for classification in which attributes of variables are paired. A decision table is composed of columns with the inputs and outputs of a decision and rows denoting rules. This technique allows for the detection of the interrelationship among variables and their attributes (Becker, 1998). Decision tables use the wrapper method that finds the best subset of features or rather it removes features with a poor contribution to the model. In this way, the algorithm reduces the probability of overfitting.
The k-means clustering algorithm uses unlabeled data to generate a fixed number (k) of clusters of data with similarities in attributes. The center of the clusters (k) is called centroids and are calculated by averaging data allocated to the cluster. The algorithm is composed of two steps: 1) Initialization, where the user sets the number of clusters, k, 2) the application of an algorithm (e.g. Lloyd’s algorithm) for which each data point is assigned to its closest cluster (Bock, 2007). The process iterates until the variation of data points in the cluster is minimized.
K-nearest neighbors is a machine learning technique used for both regression and classification. The k-Nearest Neighbor algorithm uses a training dataset with labeled data to classify new data points without labels. In the training dataset, the number of clusters (k) is identified based on their labels (e.g., having or not having a disease). The algorithm classifies a new data point by calculating its distance to each cluster of the training set until the closest cluster is identified. The technique does not make any assumption about the distribution of data (Zhang, 2016b).
The fuzzy c-means is an artificial intelligence technique for clustering based on the similarities in the features. The term fuzzy stands for indistinct, confused, and blurred. It is based on the assumption that the world around us is not dichotomous (e.g., black and white) but contains in itself all the infinite nuances that exist between these two extremes. This concept is expressed mathematically by a real number between zero and one that represents the degree of membership (membership function) of the object in question to one or the other group (e.g., how much a gray is white, or how much a gray is black).
Random forest is a machine leaning method based on the principle of ensemble learning. The key aspiration behind the random forest is to improve the performance of the indvidual tree learners with the help of bootstrap aggregating (or bagging). The technique builds each tree by bootstrapping a random sample from the data. To select the variables that need to be split in the decision tree, the random forest randomly selects features and uses scores (e.g., the decrease in Gini impurity score) as the splitting criterion. Gini impurity is a metric used in decision trees to determine which variable and at what threshold the data should be split into smaller groups. Gini Impurity measures misclassification of random records from the data set used to train the model. To understand the importance of each variable for classification/regression, the random forest classifies variables based on their importance for classification/regression in a parameter called “variable importance measure,” which has however been noted to be biased. Alternative measures are available to overcome this limitation, such as partial dependent plots. These plots provide an overview of how each variable influences the prediction of the study outcome when related to other variables selected by the random forest. Crucial parameters for the random forest are the number of trees generated in the random forest, the number of variables randomly selected for splitting in each decision tree, and the minimum size of each terminal node (Couronne et al., 2018).
Kernel partial least squares is a nonlinear partial least squares (PLS) method. PLS is a dimensionality reduction technique that models independent variables using latent variables (also known as components as in PCA). The aim is to find a few linear combinations of the original variables that are most correlated with the output. This technique is able to minimize multicollinearity among variables and it is useful in the set of high-dimensional datasets (Rosipal and Trejo, 2001).
Hierarchical clustering is a technique that performs a hierarchal decomposition of the data based on group similarities. The model builds up a distance matrix that computes the distance among data points. In particular, given a set of N observations to be grouped, and a distance (or similarity) matrix N × N, which defines the distance of the data points to each other, the basic process of hierarchical grouping is as follows:
1. The algorithm starts associating a cluster to each entity so it will have initially N clusters, each of which contains only one data point and then computes the distance (similarity) among the clusters.
2. Subsequently, it will look for the pair of clusters that are “close” to each other (more similar) and it will combine them in a single cluster. In this way, the number of clusters will be reduced by one unit.
3. It will calculate again the distance (similarity) between the new cluster and each of the old clusters.
4. It will repeat steps 2 and 3 until the entities are grouped in the desired cluster number (Johnson, 1967).
In the last decade, there has been increased use knowledge discovery techniques of artificial intelligence in pharmacoepidemiology. This result is in line with those of Koohy (2017) who showed an increased popularity of machine learning methods for biomedical research from 1990 to 2017. We strongly believe that one of the major consequences for the increased interest in applying machine learning techniques over the years is the dramatic growth in size and complexity of clinical and biological data that have led to the necessity of combining mathematics, statistics, and computer science to extract actionable insight. By using advanced algorithms that are capable of self-learning from the data, machine-learning techniques provide support for decision making to the final user (e.g. a researcher) without a pre-specific hypothesis (i.e., “hypothesis-free algorithms”). In this systematic review, we found that random forest, artificial neural network, and support vector machine were the most used techniques in the selected articles. The extensive use of artificial neural networks may be related to its first appearance in the scientific literature. In fact, this technique has existed for over 60 years (Jones et al., 2018). Random Forest instead, since its introduction in 2001 (Breiman, 2001), has rapidly gained popularity becoming a common “standard tool” to predict clinical outcomes with the advantage of being easily usable by scientists without any strong knowledge in statistics or machine learning (Couronne et al., 2018). Similarly, the support vector machine is considered to be one of the most powerful techniques for the recognition of subtle patterns in complex datasets (Huang et al., 2018). Interestingly, we observed that in the majority of the articles, researchers used more than one knowledge discover technique, which is a common approach in large data analytics. In fact, it is usually not possible to know beforehand the best algorithm for a specific classification/regression progress, and data scientist should rely on “past experience from other scientists” or benchmark multiple algorithms in order to determine the one that maximizes the accuracy of the model, an approach also known as “use trial and error” (Brownlee, 2014).
It should be highlighted that we found that secondary data were mostly used among selected articles. This is not surprising considering that electronic healthcare databases and administrative databases have revolutionized pharmacoepidemiology research in the last three decades. These data sources can be used by pharmacoepidemiologists to address clinical questions on drug use, drug effectiveness, and treatment optimization (Hennessy, 2006) carrying the advantage of being easier and less costly to reuse than primary data that, on the contrary, required to be collected anew (Schneeweiss and Avorn, 2005).
As expected, the majority of selected articles provided a medium-low level of evidence according to the Merlin scale (Merlin et al., 2009), a phenomenon that is a natural consequence of the level of evidence that is attributed to observational studies (Murad et al., 2016). In fact, among selected articles, the majority used a cohort or a case-control design, therefore, independently of the technique that was used to predict the study outcome the level of evidence was classified as medium-low.
In the selected articles, we identified 17 medical fields, of which the most prevalent were pure pharmacoepidemiology (mostly methodological studies in pharmacoepidemiology), oncology, infective medicine, and neurology. Clearly, the high frequency of articles investigating pure pharmacoepidemiology is related to the research query used for selecting the articles. Regarding the other medical fields, our findings are in accordance with the current scientific literature (Jiang et al., 2017). In fact, a recent article showed increased use of artificial intelligence in areas with a high prevalence of the disease of which an early diagnosis may guarantee a better prognosis or a reduced disease progression like oncology, neurology, and cardiology.
Finally, it is not surprising that the main purpose of using artificial intelligence techniques in this systematic review was related to the prediction of a clinical response to a treatment (i.e., supervised learning problems). Artificial intelligence and machine learning techniques have entailed some important methodological advancements in the analysis of “big data.” The utility of these techniques lies behind their potential for analysing large and complex data for making predictions that can improve and personalize the management and treatment of a disease, and improve the total well-being of an individual (Collins and Moons, 2019). As secondary purpose of using artificial intelligence techniques there was the prediction of occurrence/severity of adverse drug reactions. In this case, it can be related to the great impact of adverse drug reactions as iatrogenic disease that requires often a treatment and represents a cost to the health-care system.
The use of knowledge discovery techniques from artificial intelligence has increased exponentially over the years covering numerous sub-topics of pharmacoepidemiology. Random forest, artificial neural networks, and support vector machine models were the three most used techniques applied mainly on secondary data. The aforementioned techniques have been used mostly to predict the clinical response following a pharmacological treatment, the occurrence/severity of adverse drug reactions and the needed dosage is given the patient’s characteristics.
In the second part of this systematic review, we will summarize the evidence on the performance of artificial intelligence versus traditional pharmacoepidemiological techniques.
All authors drafted the paper, revised it for important intellectual content, and approved the final version of the manuscript to be published. MS and MA developed the concept and designed the study. MS, DL, MA, MK, and AK analyzed or interpreted the data. MS, DL, MA, MK, and AK wrote the paper.
Maurizio Sessa, David Liang, and Morten Andersen belong to the Pharmacovigilance Research Center, Department of Drug Design and Pharmacology, University of Copenhagen, supported by a grant from the Novo Nordisk Foundation (NNF15SA0018404).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.01028/full#supplementary-material
Albarakati, N., Abdel-Fatah, T. M. A., Doherty, R., Russell, R., Agarwal, D., Moseley, P., et al. (2015). Targeting BRCA1-BER deficient breast cancer by ATM or DNA-PKcs blockade either alone or in combination with cisplatin for personalized therapy. Mol. Oncol. 9, 204–217. doi: 10.1016/j.molonc.2014.08.001
Alzubiedi, S., Saleh, M., II (2016). Pharmacogenetic-guided Warfarin Dosing Algorithm in African-Americans. J. Cardiovasc. Pharmacol. 67, 86–92. doi: 10.1097/FJC.0000000000000317
An, S., Malhotra, K., Dilley, C., Han-Burgess, E., Valdez, J. N., Robertson, J., et al. (2018). Predicting drug-resistant epilepsy - A machine learning approach based on administrative claims data. Epilep. Behav. 89, 118–125. doi: 10.1016/j.yebeh.2018.10.013
Anderson, J. P., Icten, Z., Alas, V., Benson, C., Joshi, K. (2017). Comparison and predictors of treatment adherence and remission among patients with schizophrenia treated with paliperidone palmitate or atypical oral antipsychotics in community behavioral health organizations. BMC Psychiatry 17, 346. doi: 10.1186/s12888-017-1507-8
Banjar, H., Ranasinghe, D., Brown, F., Adelson, D., Kroger, T., Leclercq, T., et al. (2017). Modelling Predictors of Molecular Response to Frontline Imatinib for Patients with Chronic Myeloid Leukaemia. PloS One 12, e0168947. doi: 10.1371/journal.pone.0168947
Barbieri, C., Mari, F., Stopper, A., Gatti, E., Escandell-Montero, P., Martinez-Martinez, J. M., et al. (2015). A new machine learning approach for predicting the response to anemia treatment in a large cohort of End Stage Renal Disease patients undergoing dialysis. Comput. Biol. Med. 61, 56–61. doi: 10.1016/j.compbiomed.2015.03.019
Becker, B. G. (1998). “Visualizing decision table classifiers,” in Proceedings IEEE Symposium on Information Visualization (Cat. No. 98TB100258). (Research Triangle, CA, USA: IEEE), pp. 102–105. doi: 10.1109/INFVIS.1998.729565
Berger, C. T., Greiff, V., Mehling, M., Fritz, S., Meier, M. A., Hoenger, G., et al. (2015). Influenza vaccine response profiles are affected by vaccine preparation and preexisting immunity, but not HIV infection. Hum. Vaccin. Immunother. 11, 391–396. doi: 10.1080/21645515.2015.1008930
Bock, H. H. (2007). “Clustering Methods: A History of k-Means Algorithms,” in Selected Contributions in Data Analysis and Classification. Studies in Classification, Data Analysis, and Knowledge Organization. Eds. P. Brito, G. Cucumel, P. Bertrand, and F. de Carvalho (Berlin, Heidelberg: Springer), 161–172.
Brownlee, J. (2014). Machine learning mastery, Available at: http://machinelearningmastery.com/discover-feature-engineering-howtoengineer-features-and-how-to-getgood-at-it.
Buchner, A., Kendlbacher, M., Nuhn, P., Tullmann, C., Haseke, N., Stief, C. G., et al. (2012). Outcome assessment of patients with metastatic renal cell carcinoma under systemic therapy using artificial neural networks. Clin. Genitourin. Cancer 10, 37–42. doi: 10.1016/j.clgc.2011.10.001
Chester Wasko, M., Dasgupta, A., Ilse Sears, G., Fries, J. F., Ward, M. M. (2016). Prednisone Use and Risk of Mortality in Patients With Rheumatoid Arthritis: Moderation by Use of Disease-Modifying Antirheumatic Drugs. Arthritis Care Res. (Hoboken). 68, 706–710. doi: 10.1002/acr.22722
Collins, G. S., Moons, K. G. M. (2019). Reporting of artificial intelligence prediction models. Lancet (Lond. Engl.) 393, 1577–1579. doi: 10.1016/S0140-6736(19)30037-6
Couronne, R., Probst, P., Boulesteix, A.-L. (2018). Random forest versus logistic regression: a large-scale benchmark experiment. BMC Bioinf. 19, 270. doi: 10.1186/s12859-018-2264-5
Cuypers, L., Libin, P., Schrooten, Y., Theys, K., Di Maio, V. C., Cento, V., et al. (2017). Exploring resistance pathways for first-generation NS3/4A protease inhibitors boceprevir and telaprevir using Bayesian network learning. Infect. Genet. Evol. 53, 15–23. doi: 10.1016/j.meegid.2017.05.007
deAndres-Galiana, E. J., Fernandez-Martinez, J. L., Luaces, O., Del Coz, J. J., Fernandez, R., Solano, J., et al. (2015). On the prediction of Hodgkin lymphoma treatment response. Clin. Transl. Oncol. 17, 612–619. doi: 10.1007/s12094-015-1285-z
Devinsky, O., Dilley, C., Ozery-Flato, M., Aharonov, R., Goldschmidt, Y., Rosen-Zvi, M., et al. (2016). Changing the approach to treatment choice in epilepsy using big data. Epilep. Behav. 56, 32–37. doi: 10.1016/j.yebeh.2015.12.039
Devitt, E. J., Power, K. A., Lawless, M. W., Browne, J. A., Gaora, P. O., Gallagher, W. M., et al. (2011). Early proteomic analysis may allow noninvasive identification of hepatitis C response to treatment with pegylated interferon alpha-2b and ribavirin. Eur. J. Gastroenterol. Hepatol. 23, 177–183. doi: 10.1097/MEG.0b013e3283424e3e
Dreisbach, C., Koleck, T. A., Bourne, P. E., Bakken, S. (2019). A systematic review of natural language processing and text mining of symptoms from electronic patient-authored text data. Int. J. Med. Inform. 125, 37–46. doi: 10.1016/j.ijmedinf.2019.02.008
Elith, J., Leathwick, J. R., Hastie, T. (2008). A working guide to boosted regression trees. J. Anim. Ecol. 77, 802–813. doi: 10.1111/j.1365-2656.2008.01390.x
Franklin, J. M., Shrank, W. H., Lii, J., Krumme, A. K., Matlin, O. S., Brennan, T. A., et al. (2016). Observing versus Predicting: Initial Patterns of Filling Predict Long-Term Adherence More Accurately Than High-Dimensional Modeling Techniques. Health Serv. Res. 51, 220–239. doi: 10.1111/1475-6773.12310
Go, H., Kang, M. J., Kim, P.-J., Lee, J.-L., Park, J. Y., Park, J.-M., et al. (2019). Development of Response Classifier for Vascular Endothelial Growth Factor Receptor (VEGFR)-Tyrosine Kinase Inhibitor (TKI) in Metastatic Renal Cell Carcinoma. Pathol. Oncol. Res. 25, 51–58. doi: 10.1007/s12253-017-0323-2
Hackshaw, M. D., Nagar, S. P., Parks, D. C., Miller, L.-A. N. (2014). Persistence and compliance with pazopanib in patients with advanced renal cell carcinoma within a U.S. administrative claims database. J. Manage. Care Spec. Pharm. 20, 603–610. doi: 10.18553/jmcp.2014.20.6.603
Hansen, P. W., Clemmensen, L., Sehested, T. S. G., Fosbol, E. L., Torp-Pedersen, C., Kober, L., et al. (2016). Identifying Drug-Drug Interactions by Data Mining: A Pilot Study of Warfarin-Associated Drug Interactions. Circ. Cardiovasc. Qual. Outcomes 9, 621–628. doi: 10.1161/CIRCOUTCOMES.116.003055
Hardalac, F., Basaranoglu, M., Yuksel, M., Kutbay, U., Kaplan, M., Ozderin Ozin, Y., et al. (2015). The rate of mucosal healing by azathioprine therapy and prediction by artificial systems. Turk. J. Gastroenterol. 26, 315–321. doi: 10.5152/tjg.2015.0199
Heinze, G., Wallisch, C., Dunkler, D. (2018). Variable selection - A review and recommendations for the practicing statistician. Biom. J. 60, 431–449. doi: 10.1002/bimj.201700067
Hennessy, S. (2006). Use of health care databases in pharmacoepidemiology. Basic Clin. Pharmacol. Toxicol. 98, 311–313. doi: 10.1111/j.1742-7843.2006.pto_368.x
Hernandez, B., Raftery, A. E., Pennington, S. R., Parnell, A. C. (2018). Bayesian Additive Regression Trees using Bayesian Model Averaging. Stat. Comput. 28, 869–890. doi: 10.1007/s11222-017-9767-1
Hoang, T., Liu, J., Roughead, E., Pratt, N., Li, J. (2018). Supervised signal detection for adverse drug reactions in medication dispensing data. Comput. Methods Prog. Biomed. 161, 25–38. doi: 10.1016/j.cmpb.2018.03.021
Hu, Y.-J., Ku, T.-H., Jan, R.-H., Wang, K., Tseng, Y.-C., Yang, S.-F. (2012). Decision tree-based learning to predict patient controlled analgesia consumption and readjustment. BMC Med. Inform. Decis. Mak. 12, 131. doi: 10.1186/1472-6947-12-131
Huang, S., Cai, N., Pacheco, P. P., Narrandes, S., Wang, Y., Xu, W. (2018). Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics 15, 41–51. doi: 10.21873/cgp.20063
Jeong, E., Park, N., Choi, Y., Park, R. W., Yoon, D. (2018). Machine learning model combining features from algorithms with different analytical methodologies to detect laboratory-event-related adverse drug reaction signals. PloS One 13, e0207749. doi: 10.1371/journal.pone.0207749
Jiang, F., Jiang, Y., Zhi, H., Dong, Y., Li, H., Ma, S., et al. (2017). Artificial intelligence in healthcare: past, present and future. Stroke Vasc. Neurol. 2, 230–243. doi: 10.1136/svn-2017-000101
Johnson, S. C. (1967). Hierarchical clustering schemes. Psychometrika 32, 241–254. doi: 10.1007/BF02289588
Jones, L. D., Golan, D., Hanna, S. A., Ramachandran, M. (2018). Artificial intelligence, machine learning and the evolution of healthcare: A bright future or cause for concern? Bone Joint Res. 7, 223–225. doi: 10.1302/2046-3758.73.BJR-2017-0147.R1
Kan, H., Nagar, S., Patel, J., Wallace, D. J., Molta, C., Chang, D. J. (2016). Longitudinal Treatment Patterns and Associated Outcomes in Patients With Newly Diagnosed Systemic Lupus Erythematosus. Clin. Ther. 38, 610–624. doi: 10.1016/j.clinthera.2016.01.016
Karim, M. E., Pang, M., Platt, R. W. (2018). Can We Train Machine Learning Methods to Outperform the High-dimensional Propensity Score Algorithm? Epidemiology 29, 191–198. doi: 10.1097/EDE.0000000000000787
Kebede, M., Zegeye, D. T., Zeleke, B. M. (2017). Predicting CD4 count changes among patients on antiretroviral treatment: Application of data mining techniques. Comput. Methods Prog. Biomed. 152, 149–157. doi: 10.1016/j.cmpb.2017.09.017
Keijsers, N. L. W., Horstink, M. W., II, Gielen, S. C. A. M. (2003). Automatic assessment of levodopa-induced dyskinesias in daily life by neural networks. Mov. Disord. 18, 70–80. doi: 10.1002/mds.10310
Kern, D. M., Davis, J., Williams, S. A., Tunceli, O., Wu, B., Hollis, S., et al. (2015). Comparative effectiveness of budesonide/formoterol combination and fluticasone/salmeterol combination among chronic obstructive pulmonary disease patients new to controller treatment: a US administrative claims database study. Respir. Res. 16, 52. doi: 10.1186/s12931-015-0210-x
Kesler, S. R., Wefel, J. S., Hosseini, S. M. H., Cheung, M., Watson, C. L., Hoeft, F. (2013). Default mode network connectivity distinguishes chemotherapy-treated breast cancer survivors from controls. Proc. Natl. Acad. Sci. U. S. A. 110, 11600–11605. doi: 10.1073/pnas.1214551110
Kim, W. O., Kil, H. K., Kang, J. W., Park, H. R. (2000). Prediction on lengths of stay in the postanesthesia care unit following general anesthesia: preliminary study of the neural network and logistic regression modelling. J. Kor. Med. Sci. 15, 25–30. doi: 10.3346/jkms.2000.15.1.25
Kingsford, C., Salzberg, S. L. (2008). What are decision trees? Nat. Biotechnol. 26, 1011–1013. doi: 10.1038/nbt0908-1011
Kohlmann, M., Held, L., Grunert, V. P. (2009). Classification of therapy resistance based on longitudinal biomarker profiles. Biom. J. 51, 610–626. doi: 10.1002/bimj.200800157
Koohy, H. (2017). The rise and fall of machine learning methods in biomedical research. F1000Research 6, 2012. doi: 10.12688/f1000research.13016.2
Krakow, E. F., Hemmer, M., Wang, T., Logan, B., Arora, M., Spellman, S., et al. (2017). Tools for the Precision Medicine Era: How to Develop Highly Personalized Treatment Recommendations From Cohort and Registry Data Using Q-Learning. Am. J. Epidemiol. 186, 160–172. doi: 10.1093/aje/kwx027
Kyung, M., Gill, J., Ghosh, M., Casella, G. (2010). Penalized regression, standard errors, and Bayesian lassos. Bayesian Anal. 5, 369–411. doi: 10.1214/10-BA607
LaRanger, R., Karimpour-Fard, A., Costa, C., Mathes, D., Wright, W. E., Chong, T. (2019). Analysis of Keloid Response to 5-Fluorouracil Treatment and Long-Term Prevention of Keloid Recurrence. Plast. Reconstr. Surg. 143, 490–494. doi: 10.1097/PRS.0000000000005257
Larney, S., Hickman, M., Fiellin, D. A., Dobbins, T., Nielsen, S., Jones, N. R., et al. (2018). Using routinely collected data to understand and predict adverse outcomes in opioid agonist treatment: Protocol for the Opioid Agonist Treatment Safety (OATS) Study. BMJ Open 8, e025204. doi: 10.1136/bmjopen-2018-025204
Lazic, S. E., Edmunds, N., Pollard, C. E. (2018). Predicting Drug Safety and Communicating Risk: Benefits of a Bayesian Approach. Toxicol. Sci. 162, 89–98. doi: 10.1093/toxsci/kfx236
Li, X., Liu, R., Luo, Z.-Y., Yan, H., Huang, W.-H., Yin, J.-Y., et al. (2015). Comparison of the predictive abilities of pharmacogenetics-based warfarin dosing algorithms using seven mathematical models in Chinese patients. Pharmacogenomics 16, 583–590. doi: 10.2217/pgs.15.26
Li, M. H., Mestre, T. A., Fox, S. H., Taati, B. (2017). “Automated vision-based analysis of levodopa-induced dyskinesia with deep learning,” in Conf. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Conf, (Seogwipo, South Korea: IEEE) vol. 2017, 3377–3380. doi: 10.1109/EMBC.2017.8037580
Li, Y., Huang, X., Jiang, J., Hu, W., Hu, J., Cai, J., et al. (2018). Clinical Variables for Prediction of the Therapeutic Effects of Bevacizumab Monotherapy in Nasopharyngeal Carcinoma Patients With Radiation-Induced Brain Necrosis. Int. J. Radiat. Oncol. Biol. Phys. 100, 621–629. doi: 10.1016/j.ijrobp.2017.11.023
Linke, S. P., Bremer, T. M., Herold, C. D., Sauter, G., Diamond, C. (2006). A multimarker model to predict outcome in tamoxifen-treated breast cancer patients. Clin. Cancer Res. 12, 1175–1183. doi: 10.1158/1078-0432.CCR-05-1562
Liu, R., Li, X., Zhang, W., Zhou, H.-H. (2015). Comparison of Nine Statistical Model Based Warfarin Pharmacogenetic Dosing Algorithms Using the Racially Diverse International Warfarin Pharmacogenetic Consortium Cohort Database. PloS One 10, e0135784. doi: 10.1371/journal.pone.0135784
Lo-Ciganic, W.-H., Donohue, J. M., Thorpe, J. M., Perera, S., Thorpe, C. T., Marcum, Z. A., et al. (2015). Using machine learning to examine medication adherence thresholds and risk of hospitalization. Med. Care 53, 720–728. doi: 10.1097/MLR.0000000000000394
Loke, P. Y., Chew, L., Yap, C. W. (2011). Pilot study on developing a decision support tool for guiding re-administration of chemotherapeutic agent after a serious adverse drug reaction. BMC Cancer 11, 319. doi: 10.1186/1471-2407-11-319
Martin-Guerrero, J. D., Camps-Valls, G., Soria-Olivas, E., Serrano-Lopez, A. J., Perez-Ruixo, J. J., Jimenez-Torres, N. V. (2003). Dosage individualization of erythropoietin using a profile-dependent support vector regression. IEEE Trans. Biomed. Eng. 50, 1136–1142. doi: 10.1109/TBME.2003.816084
Merlin, T., Weston, A., Tooher, R. (2009). Extending an evidence hierarchy to include topics other than treatment: revising the Australian “levels of evidence”. BMC Med. Res. Methodol. 9, 34. doi: 10.1186/1471-2288-9-34
Michie, D., Spiegelhalter, D. J., Taylor, C. C., Campbell, J. (Eds.) (1994). Machine Learning, Neural and Statistical Classification. USA: Ellis Horwood. doi: 10.1080/00401706.1995
Molassiotis, A., Farrell, C., Bourne, K., Brearley, S. G., Pilling, M. (2012). An exploratory study to clarify the cluster of symptoms predictive of chemotherapy-related nausea using random forest modeling. J. Pain Symptom Manage. 44, 692–703. doi: 10.1016/j.jpainsymman.2011.11.003
Murad, M. H., Asi, N., Alsawas, M., Alahdab, F. (2016). New evidence pyramid. Evid. Based. Med. 21, 125–127. doi: 10.1136/ebmed-2016-110401
Noble, W. S. (2006). What is a support vector machine? Nat. Biotechnol. 24, 1565–1567. doi: 10.1038/nbt1206-1565
Oxford (2019). Artificial Intelligence. Oxford Dict. Available at: https://www.lexico.com/en/definition/artificial_intelligence [Accessed June 28, 2019].
Platt, J. (1998). Sequential minimal optimization: A fast algorithm for training support vector machines.
Podda, G. M., Grossi, E., Palmerini, T., Buscema, M., Femia, E. A., Della Riva, D., et al. (2017). Prediction of high on-treatment platelet reactivity in clopidogrel-treated patients with acute coronary syndromes. Int. J. Cardiol. 240, 60–65. doi: 10.1016/j.ijcard.2017.03.074
Pusch, T., Pasipanodya, J. G., Hall, R.G., Gumbo, T. (2014). Therapy duration and long-term outcomes in extra-pulmonary tuberculosis. BMC Infect. Dis. 14, 115. doi: 10.1186/1471-2334-14-115
Qin, J., Wei, M., Liu, H., Chen, J., Yan, R., Yao, Z., et al. (2015). Altered anatomical patterns of depression in relation to antidepressant treatment: Evidence from a pattern recognition analysis on the topological organization of brain networks. J. Affect. Disord. 180, 129–137. doi: 10.1016/j.jad.2015.03.059
Ravan, M., Hasey, G., Reilly, J. P., MacCrimmon, D., Khodayari-Rostamabad, A. (2015). A machine learning approach using auditory odd-ball responses to investigate the effect of Clozapine therapy. Clin. Neurophysiol. 126, 721–730. doi: 10.1016/j.clinph.2014.07.017
Ravanelli, M., Agazzi, G. M., Ganeshan, B., Roca, E., Tononcelli, E., Bettoni, V., et al. (2018). CT texture analysis as predictive factor in metastatic lung adenocarcinoma treated with tyrosine kinase inhibitors (TKIs). Eur. J. Radiol. 109, 130–135. doi: 10.1016/j.ejrad.2018.10.016
Rezaei-Darzi, E., Farzadfar, F., Hashemi-Meshkini, A., Navidi, I., Mahmoudi, M., Varmaghani, M., et al. (2014). Comparison of two data mining techniques in labeling diagnosis to Iranian pharmacy claim dataset: artificial neural network (ANN) versus decision tree model. Arch. Iran. Med. 17, 837–843.
Rosipal, R., Trejo, L. J. (2001). Kernel partial least squares regression in reproducing kernel hilbert space. J. Mach. Learn. Res. 2, 97–123. doi: 10.5555/944790.944806
Saadah, L. M., Chedid, F. D., Sohail, M. R., Nazzal, Y. M., Al Kaabi, M. R., Rahmani, A. Y. (2014). Palivizumab prophylaxis during nosocomial outbreaks of respiratory syncytial virus in a neonatal intensive care unit: predicting effectiveness with an artificial neural network model. Pharmacotherapy 34, 251–259. doi: 10.1002/phar.1333
Saigo, H., Altmann, A., Bogojeska, J., Muller, F., Nowozin, S., Lengauer, T. (2011). Learning from past treatments and their outcome improves prediction of in vivo response to anti-HIV therapy. Stat. Appl. Genet. Mol. Biol. 10. doi: 10.2202/1544-6115.1604
Saleh, M., II, Alzubiedi, S. (2014). Dosage individualization of warfarin using artificial neural networks. Mol. Diagn. Ther. 18, 371–379. doi: 10.1007/s40291-014-0090-7
Sangeda, R. Z., Mosha, F., Prosperi, M., Aboud, S., Vercauteren, J., Camacho, R. J., et al. (2014). Pharmacy refill adherence outperforms self-reported methods in predicting HIV therapy outcome in resource-limited settings. BMC Public Health 14, 1035. doi: 10.1186/1471-2458-14-1035
Sargent, L., Nalls, M., Amella, E. J., Mueller, M., Lageman, S. K., Bandinelli, S., et al. (2018). Anticholinergic Drug Induced Cognitive and Physical Impairment: Results from the InCHIANTI Study. J. Gerontol. A. Biol. Sci. Med. Sci. 75 (5), 995–1002. doi: 10.1093/gerona/gly289
Schmitz, B., De Maria, R., Gatsios, D., Chrysanthakopoulou, T., Landolina, M., Gasparini, M., et al. (2014). Identification of genetic markers for treatment success in heart failure patients: insight from cardiac resynchronization therapy. Circ. Cardiovasc. Genet. 7, 760–770. doi: 10.1161/CIRCGENETICS.113.000384
Schneeweiss, S., Avorn, J. (2005). A review of uses of health care utilization databases for epidemiologic research on therapeutics. J. Clin. Epidemiol. 58, 323–337. doi: 10.1016/j.jclinepi.2004.10.012
Sesen, M. B., Nicholson, A. E., Banares-Alcantara, R., Kadir, T., Brady, M. (2013). Bayesian networks for clinical decision support in lung cancer care. PloS One 8, e82349. doi: 10.1371/journal.pone.0082349
Setoguchi, S., Schneeweiss, S., Brookhart, M. A., Glynn, R. J., Cook, E. F. (2008). Evaluating uses of data mining techniques in propensity score estimation: a simulation study. Pharmacoepidemiol. Drug Saf. 17, 546–555. doi: 10.1002/pds.1555
Shamir, R. R., Dolber, T., Noecker, A. M., Walter, B. L., McIntyre, C. C. (2015). Machine Learning Approach to Optimizing Combined Stimulation and Medication Therapies for Parkinson’s Disease. Brain Stimul. 8, 1025–1032. doi: 10.1016/j.brs.2015.06.003
Simuni, T., Long, J. D., Caspell-Garcia, C., Coffey, C. S., Lasch, S., Tanner, C. M., et al. (2016). Predictors of time to initiation of symptomatic therapy in early Parkinson’s disease. Ann. Clin. Transl. Neurol. 3, 482–494. doi: 10.1002/acn3.317
Smith, B. P., Ward, R. A., Brier, M. E. (1998). Prediction of anticoagulation during hemodialysis by population kinetics and an artificial neural network. Artif. Organs 22, 731–739. doi: 10.1046/j.1525-1594.1998.06101.x
Snow, P. B., Brandt, J. M., Williams, R. L. (2001). Neural network analysis of the prediction of cancer recurrence following debulking laparotomy and chemotherapy in stages III and IV ovarian cancer. Mol. Urol. 5, 171–174. doi: 10.1089/10915360152745858
Song, R., Wang, W., Zeng, D., Kosorok, M. R. (2015). Penalized Q-Learning for Dynamic Treatment Regimens. Stat. Sin. 25, 901–920. doi: 10.5705/ss.2012.364
Sudharsan, B., Peeples, M., Shomali, M. (2015). Hypoglycemia prediction using machine learning models for patients with type 2 diabetes. J. Diabetes Sci. Technol. 9, 86–90. doi: 10.1177/1932296814554260
Sun, C.-Y., Su, T.-F., Li, N., Zhou, B., Guo, E.-S., Yang, Z.-Y., et al. (2016). A chemotherapy response classifier based on support vector machines for high-grade serous ovarian carcinoma. Oncotarget 7, 3245–3254. doi: 10.18632/oncotarget.6569
Tang, J., Liu, R., Zhang, Y.-L., Liu, M.-Z., Hu, Y.-F., Shao, M.-J., et al. (2017). Application of Machine-Learning Models to Predict Tacrolimus Stable Dose in Renal Transplant Recipients. Sci. Rep. 7, 42192. doi: 10.1038/srep42192
Tran, L., Yiannoutsos, C., Wools-Kaloustian, K., Siika, A., van der Laan, M., Petersen, M. (2019). Double Robust Efficient Estimators of Longitudinal Treatment Effects: Comparative Performance in Simulations and a Case Study. Int. J. Biostat. 15 (2). doi: 10.1515/ijb-2017-0054
Turing, A. M. (1937). On computable numbers, with an application to the Entscheidungsproblem. Proc. Lond. Math. Soc 2, 230–265. doi: 10.1112/plms/s2-42.1.230
Urquidi-Macdonald, M., Mager, D. E., Mascelli, M. A., Frederick, B., Freedman, J., Fitzgerald, D. J., et al. (2004). Abciximab pharmacodynamic model with neural networks used to integrate sources of patient variability. Clin. Pharmacol. Ther. 75, 60–69. doi: 10.1016/j.clpt.2003.09.008
Waljee, A. K., Sauder, K., Patel, A., Segar, S., Liu, B., Zhang, Y., et al. (2017). Machine Learning Algorithms for Objective Remission and Clinical Outcomes with Thiopurines. J. Crohns. Colitis 11, 801–810. doi: 10.1093/ecco-jcc/jjx014
Wasko, M. C. M., Dasgupta, A., Hubert, H., Fries, J. F., Ward, M. M. (2013). Propensity-adjusted association of methotrexate with overall survival in rheumatoid arthritis. Arthritis Rheumatol. 65, 334–342. doi: 10.1002/art.37723
Wolfson, J., Bandyopadhyay, S., Elidrisi, M., Vazquez-Benitez, G., Vock, D. M., Musgrove, D., et al. (2015). A Naive Bayes machine learning approach to risk prediction using censored, time-to-event data. Stat. Med. 34, 2941–2957. doi: 10.1002/sim.6526
Yabu, J. M., Siebert, J. C., Maecker, H. T. (2016). Immune Profiles to Predict Response to Desensitization Therapy in Highly HLA-Sensitized Kidney Transplant Candidates. PloS One 11, e0153355. doi: 10.1371/journal.pone.0153355
Yamashita, R., Nishio, M., Do, R. K. G., Togashi, K. (2018). Convolutional neural networks: an overview and application in radiology. Insights Imaging 9, 611–629. doi: 10.1007/s13244-018-0639-9
Yap, K. Y.-L., Low, X. H., Chui, W. K., Chan, A. (2012). Computational prediction of state anxiety in Asian patients with cancer susceptible to chemotherapy-induced nausea and vomiting. J. Clin. Psychopharmacol. 32, 207–217. doi: 10.1097/JCP.0b013e31824888a1
Yun, J.-Y., Jang, J. H., Kim, S. N., Jung, W. H., Kwon, J. S. (2015). Neural Correlates of Response to Pharmacotherapy in Obsessive-Compulsive Disorder: Individualized Cortical Morphology-Based Structural Covariance. Prog. Neuropsychopharmacol. Biol. Psychiatry 63, 126–133. doi: 10.1016/j.pnpbp.2015.06.009
Zhang, Z. (2016a). A gentle introduction to artificial neural networks. Ann. Transl. Med. 4, 370. doi: 10.21037/atm.2016.06.20
Zhang, Z. (2016b). Introduction to machine learning: k-nearest neighbors. Ann. Transl. Med. 4, 218. doi: 10.21037/atm.2016.03.37
Zhang, Z. (2016c). Naïve Bayes classification in R. Ann. Transl. Med. 4, 12. doi: 10.21037/atm.2016.03.38
Keywords: systematic review, pharmacoepidemiology, artificial intelligence, machine learning, deep learning
Citation: Sessa M, Khan AR, Liang D, Andersen M and Kulahci M (2020) Artificial Intelligence in Pharmacoepidemiology: A Systematic Review. Part 1—Overview of Knowledge Discovery Techniques in Artificial Intelligence. Front. Pharmacol. 11:1028. doi: 10.3389/fphar.2020.01028
Received: 23 October 2019; Accepted: 24 June 2020;
Published: 16 July 2020.
Edited by:
Irene Lenoir-Wijnkoop, Utrecht University, NetherlandsReviewed by:
Robert L. Lins, Independent researcher, Antwerp, BelgiumCopyright © 2020 Sessa, Khan, Liang, Andersen and Kulahci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Sessa, bWF1cml6aW8uc2Vzc2FAc3VuZC5rdS5kaw==
†ORCID: Maurizio Sessa, orcid.org/0000-0003-0874-4744
‡These authors share first authorship
§These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.